- Department of Medical Sciences, Molecular Biotechnology Center, University of Torino, Torino, Italy
Extracellular vesicles (EVs) are considered to be a novel complex mechanism of cell communication within the tumor microenvironment. EVs may act as vehicles for transcription factors and nucleic acids inducing epigenetic changes in recipient cells. Since tumor EVs may be present in patient biological fluids, it is important to investigate their function and molecular mechanisms of action. It has been shown that tumor cells release EVs, which are capable of regulating cell apoptosis, proliferation, invasion, and epithelial–mesenchymal transition, as well as to suppress activity of immune cells, to enhance angiogenesis, and to prepare a favorable microenvironment for metastasis. On the other hand, EVs derived from stromal cells, such as mesenchymal stem cells (MSCs), may influence the phenotype of tumor cells through reciprocal cross talk greatly influenced by the transcription factors and nucleic acids they carry. In particular, non-coding RNAs (ncRNAs), including microRNAs and long ncRNAs, have recently been identified as the main candidates for the phenotypic changes induced in the recipient cells by EVs. ncRNAs, which are important regulators of mRNA and protein expression, can function either as tumor suppressors or as oncogenes, depending on their targets. Herein, we have attempted to revise actual evidence reported in the literature on the role of EVs in tumor biology with particular regard to the cross talk of ncRNAs between cancer cells and MSCs.
Introduction
Extracellular vesicles (EVs) have recently been identified to be instrumental in intercellular communication through the exchange of biologically active molecules, in particular, non-coding RNAs (ncRNA) that can not only modulate gene expression locally but also systemically (1–3). EV molecular composition (nucleic acids and protein) is regulated by cell growth conditions, signal molecules, growth factors, etc. (4–6). Apart from healthy cells, tumor cells also release excessive amounts of tumoral extracellular vesicles (T-EVs) found to be rich in specific sets of ncRNAs different from normal cells, which circulate in different biological fluids in the body (7). The role of ncRNAs in the regulation of gene expression has been extensively studied, and various classes of ncRNAs, with different targets and functions, have been identified (8, 9).
Non-coding RNAs are usually divided into two major groups according to their length. These include small ncRNAs (below 200 nt) defined as microRNAs (miRNAs), and the long ncRNAs (lncRNAs; above 200 nt). miRNAs mostly act on RNA by silencing or post-transcriptionally regulating gene expression (10, 11), whereas lncRNA participate in imprinting and gene dosage regulation, using diverse molecular mechanisms. These include complement annealing with genome DNA, scaffolding histone-modifying complexes by acting as either a “sponge” for proteins and miRNAs, and/or as molecular guides within ribonucleoprotein complexes (12). Other important roles played by both ncRNAs include stem cell pluripotency, embryonic development, cell differentiation, and tumorigenesis.
Recent evidence demonstrates dramatic changes that happen in the level and pattern of RNA carried within circulating EVs during tumor development. Depending on their biological properties and content, EVs have been involved in cancer initiation, progression, and pre-metastatic niche formation (13). It has also become evident that EVs may transfer not only functional ncRNA but also DNA, thus modifying gene expression in recipient cells (14–16).
Tumors contain a heterogeneous population of cells, including mesenchymal stem cells (MSCs), endothelial cells, cancer-associated fibroblasts, immune inflammatory cells, and also cancer stem cells (CSCs). Communication between these cells and cells present in the normal surrounding tissue helps tumor-initiating cells to survive, proliferate, invade, and establish metastasis (17–19). This communication is performed not only by cytokines, hormones, and proteins, but also by transcription factors, ncRNAs, and DNA carried by EVs.
In this review, we mainly focus on the role of ncRNAs carried by T-EVs and by MSC-derived EVs (MSC-EVs) in modification of tumor microenvironment with particular regard to the interaction between cancer cells and MSCs.
Selected ncRNAs Detected Frequently in EVs During Cancer Development
The discovery of defined pattern of ncRNA expression in cancer patients has exposed the potential to exploit them as novel diagnostic markers as well as possible therapeutic targets. For instance, Lawrie and colleagues in 2007 reported the presence of miRNAs in the blood of cancer patients and demonstrated their potential as cancer biomarkers (20). Since then, the list of tumor-associated circulating miRNAs has grown evidently and the molecular function of miRNAs in the context of cancer widely elucidated. Interestingly, many of these ncRNAs were also detected within EVs derived from MSCs, therefore possibly indicating a bidirectional function in the communication between tumor cells and stem cells (see Figure 1). However, the functions of different miRNA are not univocal and depending on the context they may exert pro- or anti-tumorigenic activity. Here, we discuss the functions of some miRNAs, miRNA families (i.e., miRNAs sharing similar seed sequence), or clusters (i.e., miRNAs processed from one transcript), which have been reported to be present in both MSC-EVs and T-EVs.
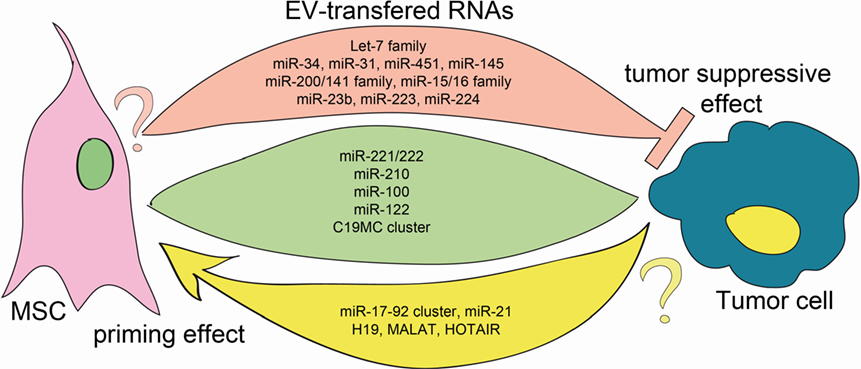
Figure 1. EV transfer of ncRNA between MSCs and tumor cells. Green, rose, and yellow flows represent the EV-mediated exchange of information between cells. All mentioned RNAs have been detected in both MSC-EVs and T-EVs. Green flow: RNAs that transferred function in both MSC and tumor cells. Rose flow: it depicts bidirectional transfer of tumor-suppressor miRNAs. Whereas the anti-tumoral action of these EV-carried miRNAs is well established, no data are available on their function on MSC (question mark) (5, 21–28). Yellow flow: it indicates traffic of oncomiRs and oncogenes, that were shown to prime MSCs toward a protumoral phenotype. The action of EVs carrying these RNAs has not been investigated in tumor cells (question mark) (6, 29–32).
Let-7 Family
Let-7 family consists of 13 different members, including let-7a-1, 7a-2, 7a-3, 7b, 7c, 7d, 7e, 7f-1, 7f-2, 7g, 7i, mir-98, and mir-202. Furthermore, the presence of these miRNAs has been detected in the circulation of patients with breast, prostate, colon, gastric, oral, and ovarian cancers (21). This cluster of miRNAs are considered to have tumor-suppressor properties as they not only repress genes responsible for self-renewal and “stem” characteristics in cells but also promote differentiation during normal development. Many oncogenes, such as RAS, MYC, HMGA2, and LIN28, are known to be direct targets of let-7 (33). Furthermore, low levels of let-7 have been identified to correlate with more aggressive tumors as well as CSCs (29, 34, 35). Whether let-7 deregulation is the cause of cancer initiation and development or vice versa is still unknown.
Extracellular release of let-7 in plasma of cancer patients or in conditioned medium of tumor cells and MSCs has been reported in several studies (36–38). There are two hypotheses about the function of extracellular let-7 in tumorigenesis: the first hypothesis proposes that the release of let-7 through EVs results in the depletion of cells of this miRNA consequently leading to the maintenance of oncogenesis and invasiveness of donor cell. The second theory considers extracellular let-7 as pro-oncogenic based on evidence that it targets caspase-3 (39) and BAX mRNAs (40). In the latter theory, EVs containing let-7 may cause resistance to apoptosis, thus favoring the acquisition of a malignant phenotype by recipient cells.
miR-21
miR-21 is one of the most frequently upregulated miRNA identified in various different cancer types, including lung, ovarian, breast, colon, gastric, and pancreatic cancers (30, 41, 42). miR-21 is highly enriched in EVs and is considered to be an oncogene, as it promotes tumor cell proliferation, migration, and invasiveness by targeting a number of tumor-suppressor genes, such as various components of the p53 network (43), PTEN (44), and antagonists of the RAS pathway PDCD4, BTG2, SPRY2, and others (45–47). Furthermore, miR-21 also exhibits angiogenic properties as EV-mediated transfer of miR-21 from tumor cells to recipient cells alters their phenotype and promote angiogenesis by enhancing the expression of VEGF (48–50).
Due to its upregulation in cancers and oncogenic properties, miR-21 levels in the plasma has been described as a marker for several types of tumors, such as breast, colorectal, prostate, gastric, ovarian cancer, B-cell lymphoma, glioblastoma, pancreatic cancer, and non-small cell lung cancer (51). Interestingly, this miRNA has also been detected in EVs derived from several stem cells, including MSCs (31). Since MSCs are recruited within the tumor, they may contribute to tumor angiogenesis by releasing miR-21containing EVs.
miR-17-92 Cluster
The cluster consists of seven different miRNAs: miR-17 (miR-17-5p and miR-17-3p), miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a. Overexpression of this cluster of miRNAs has been observed in several types of cancers (52–56). Numerous studies have shown the presence of these miRNAs in EVs present in plasma or in conditioned medium from cancer cell cultures (42, 51, 57, 58). The molecular mechanisms of pro-tumorigenic action of the miR-17-92 cluster include the targeting of E2F transcription factor family – a critical regulator of cell cycle and apoptosis (59); cyclin-dependent kinase inhibitor CDKN1A (p21) – a potent negative regulator of the G1-S checkpoint (60); and BCL2L11/BIM pro-apoptotic gene (61). Interestingly, it was reported recently that EVs from non-tumor astrocytes were rich in miR-19a and could promote metastatic transformation of recipient tumor cells by targeting PTEN (62). Furthermore, this cluster of miRNAs has also been identified to play a role in tumor angiogenesis by directly targeting anti-angiogenic factors, such as thrombospondin-1 and connective tissue growth factor (63), as well as several other pro-angiogenic proteins, including the integrin subunit alpha5 (64).
miR-92a is an example of miRNA with a dual role in angiogenesis depending on the cell of origin. For instance, when carried by T-EVs, it has been shown to be pro-angiogenic (58), whereas when carried by MSC-EVs it exhibits anti-angiogenic properties (57). This observation suggests that the function of a single miRNA should be considered within a more complex context depending on the interaction with multiple factors.
miR-15-16 Family
This family includes miR-15a/16-1 cluster (on chromosome 13q14), the miR-15b/16-2 cluster (on chromosome 3q25), and the miR-195/497 cluster (on chromosome 17p13).
The role of miR-15a/16-1 in cancer was suggested by the observation that these genes were down-regulated in B-cell chronic lymphocytic leukemia (65). These two miRNAs function as tumor suppressors, targeting Bcl2, MCL1, and Cyclin D1 genes (66). 15b/16-2 miRNAs are highly similar with miR-15a/16-1 cluster (miR-16-1 and miR-16-2 are identical), but their biological function is controversial, as this cluster has been reported to behave as either a tumor suppressor (67, 68) or oncogenic (69–71).
Expression of miR-15-16 cluster within plasma EVs has been shown by several research groups (51, 72). EVs containing miR-15a and miR-16 are released by different types of vascular cells (such as endothelial progenitor cells, vascular smooth muscle cells, and pericytes) as well as by MSCs. These miRNAs display an anti-angiogenic activity by targeting VEGF-A and AKT3 (73, 74).
miRNA-200 Family
The miRNA-200 family consists of five highly homologous members: miR-200a, miR-200b, miR-200c, miR-429, and miR-141. They are described as tumor-suppressor miRNAs, which are dysregulated in several malignancies (75, 76). For instance, miR-200c and miR-141 are strong epithelial differentiation inducers of undifferentiated cancer cells (77). They also tend to increase E-cadherin expression and, therefore, cell adhesion, as well as reduce cancer cell migration and invasion (78). Targets of this family of miRNAs include E2F3, E-cadherin suppressor targets, such as Zeb1, Zeb2, and SNAI2, which are important inducers of epithelial–mesenchymal transition and of subsequent tumor cell invasion (79). Additionally, these miRNAs repress Suz12 and Bmi1 stem cell markers whose expression leads to the formation of CSCs (80). Interestingly, other studies suggest a pro-tumorigenic activity of this family of miRNAs, since miR-141 is upregulated in the plasma during prostate (81) or ovarian cancer development (82) and miR-200b has been shown to stimulate epithelial–mesenchymal transition in human tongue cancer cells (70).
miR-122
miR-122 acts as a tumor suppressor by targeting genes involved in cell proliferation, migration, differentiation, apoptosis, and angiogenesis particularly in hepatocellular carcinoma (83). For instance, it directly down-regulates cyclin G1 that negatively regulates p53 protein stability (84). Other pro-tumorigenic targets of miR-122 include disintegrin, metalloprotease 10 (ADAM10), serum response factor (SRF), and insulin-like growth factor-1 receptor (IGF-1R) (85, 86).
On the other hand, high levels of circulating miR-122 has been shown in hepatocellular carcinoma (87) and, therefore, it is considered as a diagnostic marker for prediction of metastatic progression in other malignancies, including breast cancer (88). Furthermore, T-EVs carrying miR-122 downregulate the consumption of glucose in recipient cells by targeting pyruvate kinase PKM2 and Glucose transporter 1, leading to increased glucose availability for cancer cells. In addition, the same authors showed that T-EV secreted miR-122 also stimulated metastasis (89). It can, therefore, be suggested that miR-122 potentially plays a role during the early stages of tumor growth prior to angiogenesis, when the availability of nutrients is limited, and also when disseminated tumor cells reach distant tissue where they compete with surrounding normal cells for nutrients (89). Therefore, miR-122 has potentially a dual role in cancer development acting as inhibitor and or as an inducer of metastasis depending on the stage of tumor development.
miR-221/222
miR-221/222 have been characterized as both oncogenic and/or as a tumor suppressor, according to the type of cancer (90). These miRNAs target the oncogene KIT, thus inhibiting tumor growth (91). However, they also target important tumor suppressors, such as PTEN, p27, p57, and TIMP3 (92), and overexpression of miR-221 has been observed in CSCs and also during EMT (93). Furthermore, secretion of miR-221/222 has been shown in the plasma of patients with oral, colon, lung, and other cancers (51). Interestingly, EVs released by MSCs also contain miR-221 and have been implicated to exhibit anti-angiogenic effects (94).
C19MC Cluster
The miRNAs of C19MC cluster (“chromosome 19 microRNA cluster”) constitutes the largest human miRNA cluster known as of yet (95) as it encodes more than 50 mature miRNAs sharing common seed sequences (96). C19MC is exclusively expressed in the placenta and in undifferentiated cells, being the predominant miRNA species detected in placenta-derived EVs (97). Although the placenta is a normal tissue, it shares several common features with tumors, such as high cell proliferation, lack of cell-contact inhibition, and migratory and invasive properties. Both cancer and placental cells create a microenvironment supportive of immunologic privilege and angiogenesis (98, 99). Some C19MC-miRNAs were classified as oncomiRs as they were associated with invasion and metastasis (100, 101). One such example is miR-520 that was described to play a supportive role in proliferation and invasion due to its ability to suppress CD44 (102) and activate Ras/Raf/MEK/Erk signaling pathways (103). By contrast, this miRNA has also been reported to be involved in suppressing metastasis as it is also attributed toward the direct downregulation of TGFBR2 (104). Nevertheless, amplification of C19MC cluster is considered as a marker characteristic of pediatric brain tumors (100) and the emission of the related oncomiRs has been recently described (105). In addition, it has been shown that C19MC-carring EVs can also exert a possible antiviral effect (106, 107). The pattern of expression of C19MC-miRNAs in embryonic and tumor tissues suggests that exosomes carrying C19MC-miRNAs may play an important role in immunomodulation, cell reprograming, invasion, and angiogenesis.
Long-Non-Coding RNA
Recently, lncRNAs have been identified to be involved in various human cancers. Many of them have been detected within EVs present in biological fluids. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and HOX transcript antisense RNA (HOTAIR) are the most studied lncRNAs, which are deregulated in majority of cancers (108). They promote tumor growth by regulating cell cycle, invasiveness, migration, epithelial–mesenchymal transition, and tumor angiogenesis.
Metastasis-associated lung adenocarcinoma transcript 1 regulates alternative splicing of pre-mRNAs (109), and influences the expression of different metastasis-associated transcripts (110). HOTAIR downregulates various genes, including HOXA5 – differentiating factor involved in lung development (111); p21 – a mediator of p53-induced growth arrest and apoptosis (112); Wnt inhibitory factor 1 (WIF-1) – an inhibitor of the Wnt/β-catenin pathway that mediates EMT (113); as well as tumor suppressor PTEN – an inhibitor of EMT (114).
In some cases, lncRNAs carried by EVs could enhance its own expression by an autoregulatory loop, as it has been observed with highly upregulated in liver cancer (HULC) lncRNA (115). HULC inhibits expression of tumor suppressor gene p18 and promotes hepatoma cell proliferation. In turn, the inhibition of tumor suppressor gene p18 activity enhances expression of HULC (116).
The biological activity of lncRNAs carried by EVs has been described in CSCs studies. lncRNA H19 carried by T-EVs released from liver CSCs has been shown to promote angiogenesis and endothelial cell pro-adhesive functions (117). H19, as well as other lncRNAs, can not only function as a miRNA sponge for let-7 (118) but also as a precursor for miRNAs, or as an epigenetic modulator (119). Furthermore, H19 has been shown to play a role in tumor angiogenesis, upregulating VEGF and also stimulating heterotypic adhesion between endothelial cells and CSCs (117). Interestingly, the H19 lncRNA has been recently identified in EVs released from MSCs (6).
DNA Carried by EVs
Extracellular DNA is predominantly found within apoptotic bodies; however, it is also present within microvesicles and exosomes (120, 121) particularly more common in EVs released by tumor cells. Similar to other genetic cargo, EV DNA may also be incorporated and reused by recipient cells (122). Holmgren et al., for instance, reported that DNA secretion within EVs by tumor apoptotic cells occurred independently from nuclear condensation and apoptosis, and that only specific fractions of DNA fragments were present in apoptotic bodies. These fragments were enriched in oncogenes and had the ability to induce malignancy (123). Furthermore, DNA enriched in T-EVs was identified to reflect the genetic status of the tumor, for example, the amplification status of the oncogene c-Myc, as well as retrotransposons such as LINE-1 and Alu (124). This, therefore, means that T-EVs could transfer DNA between cells and insert it into recipient genome using a retrotransposon mechanism (124). It has also been shown that on transfecting non-tumorigenic immortalized rat intestinal epithelial cells (IEC-18) with human Ras oncogene increased the production of EVs rich in double-stranded DNA fragments spanning the entire host genome, including full-length human Ras and other rat oncogenes. In addition, exposure of non-transformed RAT-1 cells to EVs containing mutant H-ras DNA led to enhanced cell proliferation (125), therefore, implying that cancer cells could use EVs for the transfer of DNA oncogenes. Recently, mitochondrial DNA has also been reported to be present in plasma EVs that can be imported into the mitochondria of recipient cells with physiological or pathological consequences (126). Transferred DNA can, therefore, act as a template for both DNA and RNA synthesis (127).
Cancer Stem Cells Regulate Tumor Environment through EVs: Establishment of Tumor Niche, Metastasis, and Immunomodulation
T-EVs have a number of specific characteristics relevant to tumor development. The acidic microenvironment present around tumors stimulates the release of T-EVs with higher cell fusion capacity and elevated content of the tumor marker Caveolin-1 (128). A number of studies have shown that T-EVs facilitate the escape of tumor cells from the immune system (129). For instance, it has been show that T-EVs carry ligands for death receptors FasL and TRAIL that may induce apoptosis in lymphocytes (130, 131). Furthermore, these EVs could regulate monocyte differentiation, promoting the generation of myeloid immunosuppressive cells (132). Biological function of the circulated T-EVs and their horizontal transfer of nucleic acids between distant cells have been reported in various studies. For example, Cossetti et al. demonstrated that human melanoma cells [transfected with enhanced green fluorescent protein (EGFP) plasmid] when injected into mice, released exosomes carrying EGFP RNA, that was found not only in the circulation but also in spermatozoa (133). Furthermore, EVs have also been reported to have the ability to shuttle viruses, such as HIV and EBV (124, 133–135). These data, therefore, suggest that EVs may potentially participate in the dynamic regulation of whole body functions. Since T-EVs exhibit specific molecular patterns and are present in the circulation, saliva, urine, and other biological fluids, they are considered as a new diagnostic non-invasive tool, whose reliability has been demonstrated by diverse clinical studies reported in the literature (136, 137).
Cancer stem cells, also known as “tumor-initiating cells,” are a subpopulation of cancer cells that have the ability to self-renew and give rise to new tumors and metastasis. At present, CSCs were identified in many solid tumors, such as breast, renal, ovary, brain, pancreatic, prostate, colon, melanomas, and hepatocellular cancers (138, 139). Several studies indicate that CSCs release EVs that may contribute to tumor initiation and progression by stimulation of cell proliferation, invasion, angiogenesis, and metastasis formation, as well as by the promotion of tumor immune escape (29, 140–143). In our laboratory, we have observed that EVs derived from renal CSCs impaired dendritic cell maturation as well as T cell immune response through HLA-G, a known suppressor of immune cell function and an effector of cancer immune escape (140). Moreover, these EVs promoted angiogenesis and lung metastases. Possible effectors of the above observed effects by EVs can be attributed to miRNAs carried by EVs, which are implicated in angiogenesis, tumor progression and metastases, such as miR-200c, miR-146, miR-92, miR-301, miR-7g, and miR-130b (29).
Extracellular vesicles derived from breast CXCR4-positive CSCs were shown to promote proliferation, motility, and metastasis, generating an enhanced tumorigenic phenotype in tumor cells (144). These EVs were highly enriched in mRNA of genes related with stem cell differentiation and development, such as NANOG, NEUROD1, HTR7, KISS1R, and HOXC6. Enrichment of these mRNA was detected also within EVs from plasma of breast cancer patients with poor prognosis, suggesting that CSCs-derived EVs could enhance cancer development by transporting RNA (144). Alessandro et al. (117) observed that CD90-positive CSCs from hepatocellular carcinoma released EVs, enriched with lncRNA H19, which upregulated VEGF in endothelial cells, stimulated angiogenesis, and promoted the adhesion of cancer cells to the endothelial cell monolayer. Furthermore, EVs from different cancer cell types were shown to contain different miRNA patterns. For instance, Sanchez et al. (145) demonstrated that EVs from bulk prostate cancer cell line and from prostate CSCs contained 19 differentially expressed miRNAs and showed collaborative biological effect in tumorigenic niche formation. Moreover, breast cancer cell lines released EVs were identified to be rich in mir-130a, which contributes to tumorigenesis of cancer by regulating TGB-β/Smad signaling (146), mir-106b that promotes breast cancer invasion and metastasis by targeting BRMS1 and RB (147), miR-210 that promotes angiogenesis and metastasis in vivo (148), and several others miRNAs (149).
As single miRNA can regulate multiple targets in different cells or tissues, single miRNA could act as a tumor suppressor in one context and as an oncogene in another. Let-7, miR-15b, miR-122, and miR-100 are examples of miRNAs that were detected in plasma of cancer patients and exhibited dual role in cancer development.
Mesenchymal Stem Cells Could Regulate Tumor Growth via EVs
Tumor initiation and metastasis require formation of a favorable niche, which is a specific microenvironment that promotes tumor cell viability, proliferation, invasion, and involves several other types of cells, including MSCs. MSCs may enhance or suppress tumor progression and metastasis depending on doses and time of administration as well as on tumor model and stage (150). Their effects are considered to be dependent on paracrine mechanisms (150, 151) and EVs have been implicated as mediators of MSC actions (152).
Similar to MSCs, EVs derived from them also exhibit a controversial influence on the development of tumors. Several studies have shown that MSC-EVs promote tumor growth through different mechanisms, including miRNAs transfer. For example, Zhu et al. (153) showed that MSC-EVs enhanced VEGF expression in tumor cells by activating extracellular signal-regulated kinase1/2 (ERK1/2) pathway. Other research groups showed that MSC-EVs significantly increased survival of tumor cells and support tumor growth in vivo in part by mechanism involving miR-21 and miR-34a (6).
On the other hand, several studies have also demonstrated the inhibitory role of MSC-EVs on tumor growth. For instance, it has been described, that MSC-EVs markedly downregulated the expression of VEGF in tumor cells, inhibiting tumor angiogenesis, by a mechanism involving miR-16, which was abundant in MSC-EVs and predicted to target VEGF (74). In our laboratory, it was found that EVs derived from human bone marrow MSCs inhibited both in vitro and in vivo growth of HepG2, Kaposi, and Skov-3 cells (19). MSC-EVs induced a block in cell cycle progression in G0–G1 phase in all cell lines. Moreover, they induced apoptosis in HepG2 and Kaposi cells and necrosis in Skov-3 cells. The biological effect was explained by a differential regulation of genes involved in the control of cell cycle inducing arrest of proliferation, therefore causing cell death by apoptosis or necrosis. Similar inhibition of cell cycle progression with arrest in the G1 phase was reported for MSCs as well (154–156). This anti-proliferative effect of MSCs was related to soluble factors since the cell-to-cell contact was not required (157).
EVs derived from human liver stem cells (HLSCs) also exhibited an anti-tumor effect due to the transfer of anti-tumor miRNAs (miR-223, miR-24, miR-31, miR-125b, and miR-451). EVs obtained from Dicer knock-down HLSCs showed a significant reduction of anti-tumor activity both in vitro and in vivo. Furthermore, the inhibition of miR-451 and miR-31 reduced the observed pro-apoptotic activity of HLSC-EVs. When injecting HLSC-EVs or miR-31 or miR-451 mimics in vivo (intra-tumor), a regression in the tumor was observed (22). In another study, it has been shown that MSC-EVs carried miR-23b, which decreases proliferation and invasion of breast cancer cells through the inhibition of MARCKS promoter of cell motility and cycling (158). Notably, this miRNA was detected in T-EVs as well together with other anti-tumor miRNAs, such as miR-145, miR-143, miR-223, and miR-224. It has, therefore, been suggested that tumor cells possibly dismiss these miRNAs through EVs to achieve more metastatic properties (159).
This controversial data on the effect of MSC-EVs suggest that MSCs could release EVs that could have functions that are complex, diverse, and even opposite depending on the environment they are in. Even the same molecular pathways may respond in a different manner when stimulated by EVs. For instance, VEGF expression in tumor cells and consequent angiogenesis were shown either to be upregulated (153) or downregulated (74) after MSC-EV stimulation. The main difference between these studies showing anti-tumor or pro-tumor effect of MSC-EVs was the method of EV collection. When EVs were obtained from serum deprived MSCs they promoted tumor growth. In contrast, when EVs were collected from MSCs cultured with serum, they exhibited tumor-suppressive action. Indeed, Vallabhaneni et al. (6) showed that serum-deprivation may stress MSCs resulting in upregulation of miRNAs (miR-21 and miR-34) and lncRNA (lnc-Y1 and lnc-7SK) that are involved in cell survival and inhibition of apoptosis. These results suggest that cell stress stimulate production and secretion of molecules that may support tumor growth.
Another condition that changes the pro- or anti-tumoral characteristics of MSC-EVs is MSC priming by tumor cells. MSCs co-cultured with tumor cells or stimulated with T-EVs were shown to release EVs with tumorigenic properties. For example, in our laboratory, it was found that MSC stimulation with EVs from renal carcinoma CSCs leads to production of MSC-EVs that enhanced tumor cell migration and exhibited angiogenic properties (160). Another research group showed that MSC-EVs of patients with multiple myeloma-induced tumor growth in vivo and promoted dissemination of tumor cells to the bone morrow, whereas normal MSC-EVs exhibited an anti-tumor effect. Possible mechanism of this finding was the decreased content of tumor-suppressor miR-15a in EVs derived from multiple myeloma MSCs with respect to normal MSC-EVs (161).
As for the cells, the timing of EV administration may be critical (150). In fact, in the early phase of tumor growth, MSCs as well their EVs may facilitate the angiogenic shift of tumor favoring tumor initiation (162). By contrast, in established tumors, MSCs and MSC-EVs may promote apoptosis of endothelial cells and tumor regression (163). Therefore, MSC-EV influence on tumor growth may depend not only on type and stage of tumor but also on MSC culture conditions that may modify the cell secretome.
Conclusion
Through various studies reported in the current literature, it is quite evident that EVs derived from both MSCs and tumor cells display common ncRNAs; however, the functions they exhibit may be diverse depending on the cellular environment. Since ncRNAs interact with numerous molecular partners, their function is complex and depends on the cellular context. The response to EVs not only depends on their ncRNA/protein content but also on the metabolic pathways activated in recipient cells as well as their specific function. The same EVs, for instance, may trigger opposite actions in normal and tumor cells (22). Pandolfi and co-workers (164) have suggested that there is a RNA language that uses miRNAs response elements in different transcripts as “letters.” These diverse sets of miRNAs may form diverse “words,” thus performing different molecular and cellular functions. This hypothesis may, therefore, explain the different functions carried out by the same miRNAs in different contexts on tumor development. Taking into account that EV transport is bidirectional and that tumor cells can change the content of MSC-EVs and vice versa (160, 161), it is possible that the individual ncRNA display different and even opposite functions in vivo.
Author Contributions
All authors conceived and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia (2006) 20(9):1487–95. doi:10.1038/sj.leu.2404296
2. Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int (2010) 78(9):838–48. doi:10.1038/ki.2010.278
3. Quesenberry PJ, Aliotta J, Camussi G, Abdel-Mageed AB, Wen S, Goldberg L, et al. Potential functional applications of extracellular vesicles: a report by the NIH Common Fund Extracellular RNA Communication Consortium. J Extracell Vesicles (2015) 4:27575. doi:10.3402/jev.v4.27575
4. Lopatina T, Bruno S, Tetta C, Kalinina N, Porta M, Camussi G. Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhances their angiogenic potential. Cell Commun Signal (2014) 12:26. doi:10.1186/1478-811X-12-26
5. Lopatina T. The angiogenic potential of adipose mesenchymal stem cell-derived extracellular vesicles is modulated by basic fibroblast growth factor. J Stem Cell Res Ther (2014) 4(10):7. doi:10.4172/2157-7633.1000245
6. Vallabhaneni KC, Penfornis P, Dhule S, Guillonneau F, Adams KV, Mo YY, et al. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget (2015) 6(7):4953–67. doi:10.18632/oncotarget.3211
7. Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci (2010) 123(Pt 10):1603–11. doi:10.1242/jcs.064386
8. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet (2006) 15(Spec No 1):R17–29. doi:10.1093/hmg/ddl046
9. Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res (2011) 90(3):430–40. doi:10.1093/cvr/cvr097
10. Ambros V. The functions of animal microRNAs. Nature (2004) 431(7006):350–5. doi:10.1038/nature02871
11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi:10.1016/S0092-8674(04)00045-5
12. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev (2009) 23(13):1494–504. doi:10.1101/gad.1800909
13. Castellana D, Zobairi F, Martinez MC, Panaro MA, Mitolo V, Freyssinet JM, et al. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1-CX3CR1 axis. Cancer Res (2009) 69(3):785–93. doi:10.1158/0008-5472.CAN-08-1946
14. Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood (2007) 110(7):2440–8. doi:10.1182/blood-2007-03-078709
15. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol (2007) 9(6):654–9. doi:10.1038/ncb1596
16. Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem (2014) 289(7):3869–75. doi:10.1074/jbc.C113.532267
17. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature (2001) 414(6859):105–11. doi:10.1038/35102167
18. Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem (2012) 287(2):1397–405. doi:10.1074/jbc.M111.288662
19. Bruno S, Collino F, Deregibus MC, Grange C, Tetta C, Camussi G. Microvesicles derived from human bone marrow mesenchymal stem cells inhibit tumor growth. Stem Cells Dev (2013) 22(5):758–71. doi:10.1089/scd.2012.0304
20. Lawrie CH. MicroRNA expression in lymphoma. Expert Opin Biol Ther (2007) 7(9):1363–74. doi:10.1517/14712598.7.9.1363
21. Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer (2010) 17(1):F19–36. doi:10.1677/ERC-09-0184
22. Fonsato V, Collino F, Herrera MB, Cavallari C, Deregibus MC, Cisterna B, et al. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells (2012) 30(9):1985–98. doi:10.1002/stem.1161
23. Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer (2013) 108(2):361–9. doi:10.1038/bjc.2012.546
24. Kan CW, Hahn MA, Gard GB, Maidens J, Huh JY, Marsh DJ, et al. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer (2012) 12:627. doi:10.1186/1471-2407-12-627
25. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol (2008) 141(5):672–5. doi:10.1111/j.1365-2141.2008.07077.x
26. Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer (2012) 106(4):768–74. doi:10.1038/bjc.2011.595
27. Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One (2011) 6(3):e17745. doi:10.1371/journal.pone.0017745
28. Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One (2010) 5(7):e11803. doi:10.1371/journal.pone.0011803
29. Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res (2011) 71(15):5346–56. doi:10.1158/0008-5472.CAN-11-0241
30. Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med (2009) 13(1):39–53. doi:10.1111/j.1582-4934.2008.00556.x
31. Wang Y, Zhang L, Li Y, Chen L, Wang X, Guo W, et al. Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium. Int J Cardiol (2015) 192:61–9. doi:10.1016/j.ijcard.2015.05.020
32. Dal Bo M, Bomben R, Hernández L, Gattei V. The MYC/miR-17-92 axis in lymphoproliferative disorders: a common pathway with therapeutic potential. Oncotarget (2015) 6(23):19381–92. doi:10.18632/oncotarget.4574
33. Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med (2008) 14(9):400–9. doi:10.1016/j.molmed.2008.07.001
34. Cai WY, Wei TZ, Luo QC, Wu QW, Liu QF, Yang M, et al. The Wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci (2013) 126(Pt 13):2877–89. doi:10.1242/jcs.123810
35. Kong D, Heath E, Chen W, Cher ML, Powell I, Heilbrun L, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One (2012) 7(3):e33729. doi:10.1371/journal.pone.0033729
36. Ji H, Chen M, Greening DW, He W, Rai A, Zhang W, et al. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS One (2014) 9(10):e110314. doi:10.1371/journal.pone.0110314
37. Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One (2010) 5(10):e13247. doi:10.1371/journal.pone.0013247
38. Kobayashi M, Salomon C, Tapia J, Illanes SE, Mitchell MD, Rice GE. Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200. J Transl Med (2014) 12:4. doi:10.1186/1479-5876-12-4
39. Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis (2008) 13(10):1215–22. doi:10.1007/s10495-008-0256-z
40. Sun X, Fan C, Du N, Ren H. Possible carcinogenesis of tumor suppressor let-7. Med Hypotheses (2013) 81(3):410–3. doi:10.1016/j.mehy.2013.05.033
41. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene (2007) 26(19):2799–803. doi:10.1038/sj.onc.1210083
42. Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol (2013) 11:219. doi:10.1186/1477-7819-11-219
43. Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res (2008) 68(19):8164–72. doi:10.1158/0008-5472.CAN-08-1305
44. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology (2007) 133(2):647–58. doi:10.1053/j.gastro.2007.05.022
45. Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun (2009) 388(3):539–42. doi:10.1016/j.bbrc.2009.08.044
46. Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res (2009) 19(7):828–37. doi:10.1038/cr.2009.72
47. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell (2010) 18(3):282–93. doi:10.1016/j.ccr.2010.08.013
48. Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett (2016) 370(1):125–35. doi:10.1016/j.canlet.2015.10.011
49. Xu Y, Luo F, Liu Y, Shi L, Lu X, Xu W, et al. Exosomal miR-21 derived from arsenite-transformed human bronchial epithelial cells promotes cell proliferation associated with arsenite carcinogenesis. Arch Toxicol (2015) 89(7):1071–82. doi:10.1007/s00204-014-1291-x
50. Cappellesso R, Tinazzi A, Giurici T, Simonato F, Guzzardo V, Ventura L, et al. Programmed cell death 4 and microRNA 21 inverse expression is maintained in cells and exosomes from ovarian serous carcinoma effusions. Cancer Cytopathol (2014) 122(9):685–93. doi:10.1002/cncy.21442
51. Fernandez-Mercado M, Manterola L, Larrea E, Goicoechea I, Arestin M, Armesto M, et al. The circulating transcriptome as a source of non-invasive cancer biomarkers: concepts and controversies of non-coding and coding RNA in body fluids. J Cell Mol Med (2015) 19(10):2307–23. doi:10.1111/jcmm.12625
52. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature (2005) 435(7043):828–33. doi:10.1038/nature03552
53. Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res (2005) 65(21):9628–32. doi:10.1158/0008-5472.CAN-05-2352
54. Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, Rogounovitch TI, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci (2008) 99(6):1147–54. doi:10.1111/j.1349-7006.2008.00800.x
55. Tsuchida A, Ohno S, Wu W, Borjigin N, Fujita K, Aoki T, et al. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci (2011) 102(12):2264–71. doi:10.1111/j.1349-7006.2011.02081.x
56. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A (2006) 103(7):2257–61. doi:10.1073/pnas.0510565103
57. Kalinina N, Klink G, Glukhanyuk E, Lopatina T, Efimenko A, Akopyan Z, et al. miR-92a regulates angiogenic activity of adipose-derived mesenchymal stromal cells. Exp Cell Res (2015) 339(1):61–6. doi:10.1016/j.yexcr.2015.10.007
58. Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene (2013) 32(22):2747–55. doi:10.1038/onc.2012.295
59. O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature (2005) 435(7043):839–43. doi:10.1038/nature03677
60. Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol (2008) 28(7):2167–74. doi:10.1128/MCB.01977-07
61. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell (2008) 13(3):272–86. doi:10.1016/j.ccr.2008.02.013
62. Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature (2015) 527(7576):100–4. doi:10.1038/nature15376
63. Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet (2006) 38(9):1060–5. doi:10.1038/ng1855
64. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science (2009) 324(5935):1710–3. doi:10.1126/science.1174381
65. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A (2002) 99(24):15524–9. doi:10.1073/pnas.242606799
66. Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ (2015) 22(1):6–11. doi:10.1038/cdd.2014.87
67. Careccia S, Mainardi S, Pelosi A, Gurtner A, Diverio D, Riccioni R, et al. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene (2009) 28(45):4034–40. doi:10.1038/onc.2009.255
68. Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res (2012) 72(7):1865–77. doi:10.1158/0008-5472.CAN-11-2663
69. Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer (2010) 126(11):2553–62. doi:10.1002/ijc.24960
70. Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M, et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene (2012) 31(4):432–45. doi:10.1038/onc.2011.263
71. Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Sacca M, Memeo L, et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene (2011) 30(41):4231–42. doi:10.1038/onc.2011.140
72. Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Muller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology (2011) 77(5):.e9–16. doi:10.1016/j.urology.2011.01.020
73. Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res (2013) 112(2):335–46. doi:10.1161/CIRCRESAHA.111.300418
74. Lee JK, Park SR, Jung BK, Jeon YK, Lee YS, Kim MK, et al. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS One (2013) 8(12):e84256. doi:10.1371/journal.pone.0084256
75. Li XY, Li H, Bu J, Xiong L, Guo HB, Liu LH, et al. Prognostic role of MicroRNA-200c-141 cluster in various human solid malignant neoplasms. Dis Markers (2015) 2015:935626. doi:10.1155/2015/935626
76. Feng B, Wang R, Chen LB. Review of miR-200b and cancer chemosensitivity. Biomed Pharmacother (2012) 66(6):397–402. doi:10.1016/j.biopha.2012.06.002
77. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem (2008) 283(22):14910–4. doi:10.1074/jbc.C800074200
78. Butz H, Liko I, Czirjak S, Igaz P, Khan MM, Zivkovic V, et al. Down-regulation of Wee1 kinase by a specific subset of microRNA in human sporadic pituitary adenomas. J Clin Endocrinol Metab (2010) 95(10):E181–91. doi:10.1210/jc.2010-0581
79. Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor beta (TGF-beta) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem (2013) 288(15):10241–53. doi:10.1074/jbc.M112.443655
80. Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell (2010) 39(5):761–72. doi:10.1016/j.molcel.2010.08.013
81. Li Z, Ma YY, Wang J, Zeng XF, Li R, Kang W, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther (2016) 9:139–48. doi:10.2147/OTT.S95565
82. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol (2008) 110(1):13–21. doi:10.1016/j.ygyno.2008.04.033
83. Hu J, Xu Y, Hao J, Wang S, Li C, Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell (2012) 3(5):364–71. doi:10.1007/s13238-012-2036-3
84. Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res (2009) 69(14):5761–7. doi:10.1158/0008-5472.CAN-08-4797
85. Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology (2009) 49(5):1571–82. doi:10.1002/hep.22806
86. Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem (2009) 284(46):32015–27. doi:10.1074/jbc.M109.016774
87. Cho HJ, Kim JK, Nam JS, Wang HJ, Lee JH, Kim BW, et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem (2015) 48(16–17):1073–8. doi:10.1016/j.clinbiochem.2015.06.019
88. Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med (2012) 10:42. doi:10.1186/1479-5876-10-42
89. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol (2015) 17(2):183–94. doi:10.1038/ncb3094
90. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet (2009) 10(10):704–14. doi:10.1038/nrg2634
91. Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A (2005) 102(50):18081–6. doi:10.1073/pnas.0506216102
92. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res (2005) 65(16):7065–70. doi:10.1158/0008-5472.CAN-05-1783
93. Ke J, Zhao Z, Hong SH, Bai S, He Z, Malik F, et al. Role of microRNA221 in regulating normal mammary epithelial hierarchy and breast cancer stem-like cells. Oncotarget (2015) 6(6):3709–21. doi:10.18632/oncotarget.2888
94. Yang J, Gao F, Zhang Y, Liu Y, Zhang D. Buyang Huanwu Decoction (BYHWD) enhances angiogenic effect of mesenchymal stem cell by upregulating VEGF expression after focal cerebral ischemia. J Mol Neurosci (2015) 56(4):898–906. doi:10.1007/s12031-015-0539-0
95. Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet (2010) 19(18):3566–82. doi:10.1093/hmg/ddq272
96. Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells (2008) 26(6):1506–16. doi:10.1634/stemcells.2007-1081
97. Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod (2012) 18(8):417–24. doi:10.1093/molehr/gas013
98. Strickland S, Richards WG. Invasion of the trophoblasts. Cell (1992) 71(3):355–7. doi:10.1016/0092-8674(92)90503-5
99. Mullen CA. Review: analogies between trophoblastic and malignant cells. Am J Reprod Immunol (1998) 39(1):41–9. doi:10.1111/j.1600-0897.1998.tb00332.x
100. Kleinman CL, Gerges N, Papillon-Cavanagh S, Sin-Chan P, Pramatarova A, Quang DA, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet (2014) 46(1):39–44. doi:10.1038/ng.2849
101. Vaira V, Elli F, Forno I, Guarnieri V, Verdelli C, Ferrero S, et al. The microRNA cluster C19MC is deregulated in parathyroid tumours. J Mol Endocrinol (2012) 49(2):115–24. doi:10.1530/JME-11-0189
102. Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol (2008) 10(2):202–10. doi:10.1038/ncb1681
103. Liu P, Wilson MJ. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-kappaB factor in human fibrosarcoma cells. J Cell Physiol (2012) 227(2):867–76. doi:10.1002/jcp.22993
104. Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A, et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene (2012) 31(37):4150–63. doi:10.1038/onc.2011.571
105. Miura K, Higashijima A, Murakami Y, Tsukamoto O, Hasegawa Y, Abe S, et al. Circulating chromosome 19 miRNA cluster microRNAs in pregnant women with severe pre-eclampsia. J Obstet Gynaecol Res (2015) 41(10):1526–32. doi:10.1111/jog.12749
106. Delorme-Axford E, Bayer A, Sadovsky Y, Coyne CB. Autophagy as a mechanism of antiviral defense at the maternal-fetal interface. Autophagy (2013) 9(12):2173–4. doi:10.4161/auto.26558
107. Mouillet JF, Ouyang Y, Bayer A, Coyne CB, Sadovsky Y. The role of trophoblastic microRNAs in placental viral infection. Int J Dev Biol (2014) 58(2–4):281–9. doi:10.1387/ijdb.130349ys
108. Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) (2013) 91(7):791–801. doi:10.1007/s00109-013-1028-y
109. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell (2010) 39(6):925–38. doi:10.1016/j.molcel.2010.08.011
110. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res (2013) 73(3):1180–9. doi:10.1158/0008-5472.CAN-12-2850
111. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer (2013) 13:464. doi:10.1186/1471-2407-13-464
112. Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One (2013) 8(10):e77293. doi:10.1371/journal.pone.0077293
113. Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci (2013) 104(12):1675–82. doi:10.1111/cas.12296
114. Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, et al. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol (2013) 182(1):64–70. doi:10.1016/j.ajpath.2012.08.042
115. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res (2010) 38(16):5366–83. doi:10.1093/nar/gkq285
116. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem (2012) 287(31):26302–11. doi:10.1074/jbc.M112.342113
117. Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer (2015) 14:155. doi:10.1186/s12943-015-0426-x
118. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell (2013) 52(1):101–12. doi:10.1016/j.molcel.2013.08.027
119. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev (2014) 28(5):491–501. doi:10.1101/gad.234419.113
120. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res (2014) 24(6):766–9. doi:10.1038/cr.2014.44
121. Lazaro-Ibanez E, Sanz-Garcia A, Visakorpi T, Escobedo-Lucea C, Siljander P, Ayuso-Sacido A, et al. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate (2014) 74(14):1379–90. doi:10.1002/pros.22853
122. Holmgren L, Szeles A, Rajnavolgyi E, Folkman J, Klein G, Ernberg I, et al. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood (1999) 93(11):3956–63.
123. Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci U S A (2001) 98(11):6407–11. doi:10.1073/pnas.101129998
124. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun (2011) 2:180. doi:10.1038/ncomms1180
125. Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J. Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem Biophys Res Commun (2014) 451(2):295–301. doi:10.1016/j.bbrc.2014.07.109
126. Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science (2004) 303(5660):1007–10. doi:10.1126/science.1093133
127. Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum Mol Genet (2006) 15(1):143–54. doi:10.1093/hmg/ddi435
128. Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem (2009) 284(49):34211–22. doi:10.1074/jbc.M109.041152
129. Rivoltini L, Carrabba M, Huber V, Castelli C, Novellino L, Dalerba P, et al. Immunity to cancer: attack and escape in T lymphocyte-tumor cell interaction. Immunol Rev (2002) 188:97–113. doi:10.1034/j.1600-065X.2002.18809.x
130. Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med (2002) 195(10):1303–16. doi:10.1084/jem.20011624
131. Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology (2005) 128(7):1796–804. doi:10.1053/j.gastro.2005.03.045
132. Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, et al. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res (2006) 66(18):9290–8. doi:10.1158/0008-5472.CAN-06-1819
133. Cossetti C, Lugini L, Astrologo L, Saggio I, Fais S, Spadafora C. Soma-to-germline transmission of RNA in mice xenografted with human tumour cells: possible transport by exosomes. PLoS One (2014) 9(7):e101629. doi:10.1371/journal.pone.0101629
134. Canitano A, Venturi G, Borghi M, Ammendolia MG, Fais S. Exosomes released in vitro from Epstein-Barr virus (EBV)-infected cells contain EBV-encoded latent phase mRNAs. Cancer Lett (2013) 337(2):193–9. doi:10.1016/j.canlet.2013.05.012
135. Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem (2003) 278(52):52347–54. doi:10.1074/jbc.M309009200
136. Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One (2009) 4(4):e5219. doi:10.1371/journal.pone.0005219
137. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature (2015) 523(7559):177–82. doi:10.1038/nature14581
138. Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J (2013) 27(1):13–24. doi:10.1096/fj.12-218222
139. Bussolati B, Dekel B, Azzarone B, Camussi G. Human renal cancer stem cells. Cancer Lett (2013) 338(1):141–6. doi:10.1016/j.canlet.2012.05.007
140. Grange C, Tapparo M, Tritta S, Deregibus MC, Battaglia A, Gontero P, et al. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer (2015) 15(1):1009. doi:10.1186/s12885-015-2025-z
141. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature (2015) 527(7578):329–35. doi:10.1038/nature15756
142. Ye J, Wu D, Wu P, Chen Z, Huang J. The cancer stem cell niche: cross talk between cancer stem cells and their microenvironment. Tumour Biol (2014) 35(5):3945–51. doi:10.1007/s13277-013-1561-x
143. Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells (2014) 32(4):983–97. doi:10.1002/stem.1619
144. Rodriguez M, Silva J, Herrera A, Herrera M, Pena C, Martin P, et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget (2015) 6(38):40575–87. doi:10.18632/oncotarget.5818
145. Sanchez CA, Andahur EI, Valenzuela R, Castellon EA, Fulla JA, Ramos CG, et al. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget (2015) 7(4):3993–4008. doi:10.18632/oncotarget.6540
146. Liu L, Nie J, Chen L, Dong G, Du X, Wu X, et al. The oncogenic role of microRNA-130a/301a/454 in human colorectal cancer via targeting Smad4 expression. PLoS One (2013) 8(2):e55532. doi:10.1371/journal.pone.0055532
147. Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene (2012) 31(50):5162–71. doi:10.1038/onc.2012.11
148. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem (2013) 288(15):10849–59. doi:10.1074/jbc.M112.446831
149. Kruger S, Abd Elmageed ZY, Hawke DH, Worner PM, Jansen DA, Abdel-Mageed AB, et al. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer (2014) 14:44. doi:10.1186/1471-2407-14-44
150. Klopp AH, Gupta A, Spaeth E, Andreeff M, Marini F III. Concise review: dissecting a discrepancy in the literature: do mesenchymal stem cells support or suppress tumor growth? Stem Cells (2011) 29(1):11–9. doi:10.1002/stem.559
151. Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature (2005) 438(7069):820–7. doi:10.1038/nature04186
152. Biancone L, Bruno S, Deregibus MC, Tetta C, Camussi G. Therapeutic potential of mesenchymal stem cell-derived microvesicles. Nephrol Dial Transplant (2012) 27(8):3037–42. doi:10.1093/ndt/gfs168
153. Zhu W, Huang L, Li Y, Zhang X, Gu J, Yan Y, et al. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett (2012) 315(1):28–37. doi:10.1016/j.canlet.2011.10.002
154. Ramasamy R, Lam EW, Soeiro I, Tisato V, Bonnet D, Dazzi F. Mesenchymal stem cells inhibit proliferation and apoptosis of tumor cells: impact on in vivo tumor growth. Leukemia (2007) 21(2):304–10. doi:10.1038/sj.leu.2404489
155. Tian LL, Yue W, Zhu F, Li S, Li W. Human mesenchymal stem cells play a dual role on tumor cell growth in vitro and in vivo. J Cell Physiol (2011) 226(7):1860–7. doi:10.1002/jcp.22511
156. Cousin B, Ravet E, Poglio S, De Toni F, Bertuzzi M, Lulka H, et al. Adult stromal cells derived from human adipose tissue provoke pancreatic cancer cell death both in vitro and in vivo. PLoS One (2009) 4(7):e6278. doi:10.1371/journal.pone.0006278
157. Li L, Tian H, Chen Z, Yue W, Li S, Li W. Inhibition of lung cancer cell proliferation mediated by human mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai) (2011) 43(2):143–8. doi:10.1093/abbs/gmq118
158. Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi RU, et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal (2014) 7(332):ra63. doi:10.1126/scisignal.2005231
159. Ostenfeld MS, Jeppesen DK, Laurberg JR, Boysen AT, Bramsen JB, Primdal-Bengtson B, et al. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res (2014) 74(20):5758–71. doi:10.1158/0008-5472.CAN-13-3512
160. Lindoso RS, Collino F, Camussi G. Extracellular vesicles derived from renal cancer stem cells induce a pro-tumorigenic phenotype in mesenchymal stromal cells. Oncotarget (2015) 6(10):7959–69. doi:10.18632/oncotarget.3503
161. Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest (2013) 123(4):1542–55. doi:10.1172/JCI66517
162. Beckermann BM, Kallifatidis G, Groth A, Frommhold D, Apel A, Mattern J, et al. VEGF expression by mesenchymal stem cells contributes to angiogenesis in pancreatic carcinoma. Br J Cancer (2008) 99(4):622–31. doi:10.1038/sj.bjc.6604508
163. Otsu K, Das S, Houser SD, Quadri SK, Bhattacharya S, Bhattacharya J. Concentration-dependent inhibition of angiogenesis by mesenchymal stem cells. Blood (2009) 113(18):4197–205. doi:10.1182/blood-2008-09-176198
Keywords: exosomes, extracellular vesicles, non-coding RNA, microRNA, long non-coding RNA, tumor stem cells, mesenchymal stem cells
Citation: Lopatina T, Gai C, Deregibus MC, Kholia S and Camussi G (2016) Cross Talk between Cancer and Mesenchymal Stem Cells through Extracellular Vesicles Carrying Nucleic Acids. Front. Oncol. 6:125. doi: 10.3389/fonc.2016.00125
Received: 26 February 2016; Accepted: 09 May 2016;
Published: 23 May 2016
Edited by:
Renaud Seigneuric, UB/INSERM U866, FranceReviewed by:
Janusz Rak, McGill University, CanadaStefano Fais, Istituto Superiore di Sanità, Italy
Copyright: © 2016 Lopatina, Gai, Deregibus, Kholia and Camussi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanni Camussi, giovanni.camussi@unito.it
 Tatiana Lopatina
Tatiana Lopatina Chiara Gai
Chiara Gai Giovanni Camussi
Giovanni Camussi