- 1Department of Chemical Engineering, University of Michigan, Ann Arbor, MI, USA
- 2Department of Internal Medicine, University of Michigan, Ann Arbor, MI, USA
- 3Veterans Administration Ann Arbor Healthcare System, Ann Arbor, MI, USA
- 4Translational Oncology Program, University of Michigan, Ann Arbor, MI, USA
Circulating tumor cells (CTCs) have garnered a lot of attention in the past few decades. Isolation of these rare cells from the billions of blood cells has been a challenge until recent times. With the advent of new sensitive technologies that permit live cell isolation and downstream genomic analysis, the existing paradigm of CTC research has evolved to explore clinical utility of these cells. CTCs have been identified as prognostic and pharmacodynamic biomarkers in many solid tumors, including lung cancer. As a means of liquid biopsy, CTCs could play a major role in the development of personalized medicine and targeted therapies. This review discusses the state of various isolation strategies, cell separation techniques and key studies that illustrate the application of liquid biopsy to lung cancer.
Introduction
Lung cancer is the leading cause of cancer worldwide, accounting for 160,000 deaths in the United States in 2014 (1) with a 5-year survival rate of 20% (2). Approximately 224,000 new cases of lung cancer were reported in 2014 (2). Smoking is the leading risk factor (3). Non-small-cell lung cancer (NSCLC) constitutes 80% of all new lung cancer cases (4). Over 50% of patients are diagnosed initially with locally advanced or metastatic disease with worse outcomes. Survival is improved through screening, early diagnosis, and treatment (5). However, currently approved screening strategies involving low-dose CT scans have a low sensitivity and high false positive rates of >90% (5). There is an unmet need for additional biomarkers that can improve the sensitivity of low-dose CT screening, particularly in patients with indeterminate pulmonary nodules. Following a diagnosis of lung cancer and stage-specific therapy, currently, the only available techniques to monitor disease progression other than clinical symptoms are periodic CT scans done every 3–6 months. Earlier detection of recurrence in cases of earlier stages of lung cancer (stages I and II) is needed to direct certain subsets of patients for treatment of oligometastatic disease; this may include surgery and/or radiation therapy. In cases of patients with more advanced lung cancers, surveillance CT scans may demonstrate progression of disease; repeat biopsies of these recurrent/progressive lesions allow us to determine the underlying resistance mechanisms, in cases of lung cancers associated with specific molecular targets (6). This type of surveillance may miss early recurrence/resistance and identification of treatable oligometastatic disease. In addition, repeat biopsies are invasive and not without risk. There is an unmet need for earlier detection of resistance in this subgroup of patients using a minimally invasive approach, which could potentially serve as decision aid for subsequent alternative therapies targeting secondary mutations or alternative pathway activation (7, 8). Furthermore, a better non-invasive approach for serial monitoring is necessary to address other clinical and research unmet needs, not only for early response assessment for targeted therapies but also for novel immunotherapies where radiologic response may lag behind or be erroneous (e.g., pseudo-progression).
Emerging research in blood-based biomarkers provide new opportunities to diagnose lung cancer earlier and also to assist with detection of earlier recurrence. These biomarkers include circulating tumor cells (CTCs) (9) and circulating cell-free nucleic acids (CfNA) (10–12). CfNA are released from apoptotic or necrotic tumor cells (13). Besides plasma, tumor nucleic acids have also been detected in other fractions of blood, such as platelets (14), extracellular vesicle exosomes (15), and buffy coat (leukocyte-enriched) (16). New technologies are being developed to increase the sensitivity and specificity of cfNA detection in the blood (17). CTCs, on the other hand, have been extensively studied as prognostic and pharmacodynamic biomarkers in many cancers (18–22). Both cfNA and CTCs were demonstrated to correlate with tumor burden and revealed genetic signatures of primary and metastatic tumors (23, 24). CTC processing technologies, however, have unique advantages over cfNA, by permitting a vast array of molecular and functional studies, including cell culture, xenograft implantation, and ex vivo drug testing (Figure 1) (25). CTCs represent a subset of tumor cells that have acquired the ability to disseminate from the primary tumor and intravasate to the circulatory system (26). Sampling CTCs may be a viable non-invasive alternative to tissue biopsies for diagnosis of lung cancers. In many patients, however, CTCs are quite low in number, and need to be isolated from an overwhelming majority of blood cells (1 CTC: 1 billion blood cells). We have reported capability of detecting and characterizing CTCs from early stages of lung cancer (27). CTCs have demonstrated utility in surveillance of patients and their changing numbers predict progression-free survival (PFS) and overall survival (OS) in several cancers (19, 28). Additionally, CTCs have been proposed as surrogate biomarkers in a multitude of research areas, including the selection of neoadjuvant and adjuvant therapy, detection of recurrent disease, and as pharmacodynamic biomarkers of novel therapeutics (22, 26, 29–34). In this review, we summarize current technological and scientific advancements in CTC research specifically pertaining to lung cancer and discuss possible future directions (Figure 2).
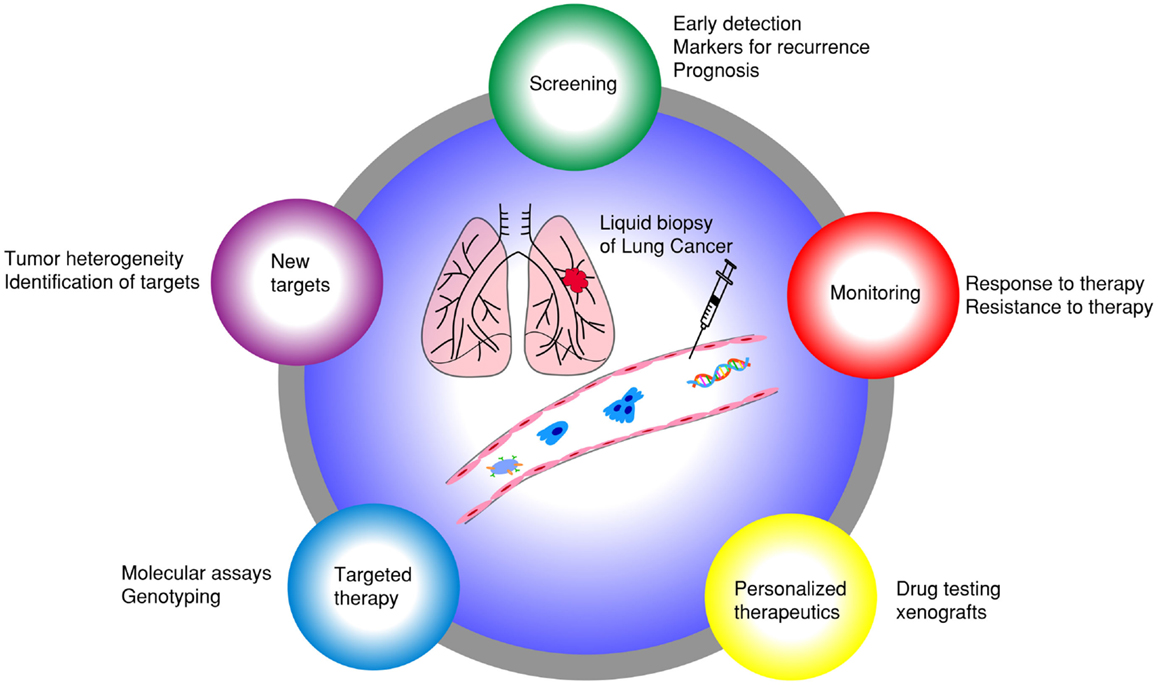
Figure 1. Liquid biopsy of lung cancer: different applications of using CTCs as surrogate biomarkers in lung cancer.
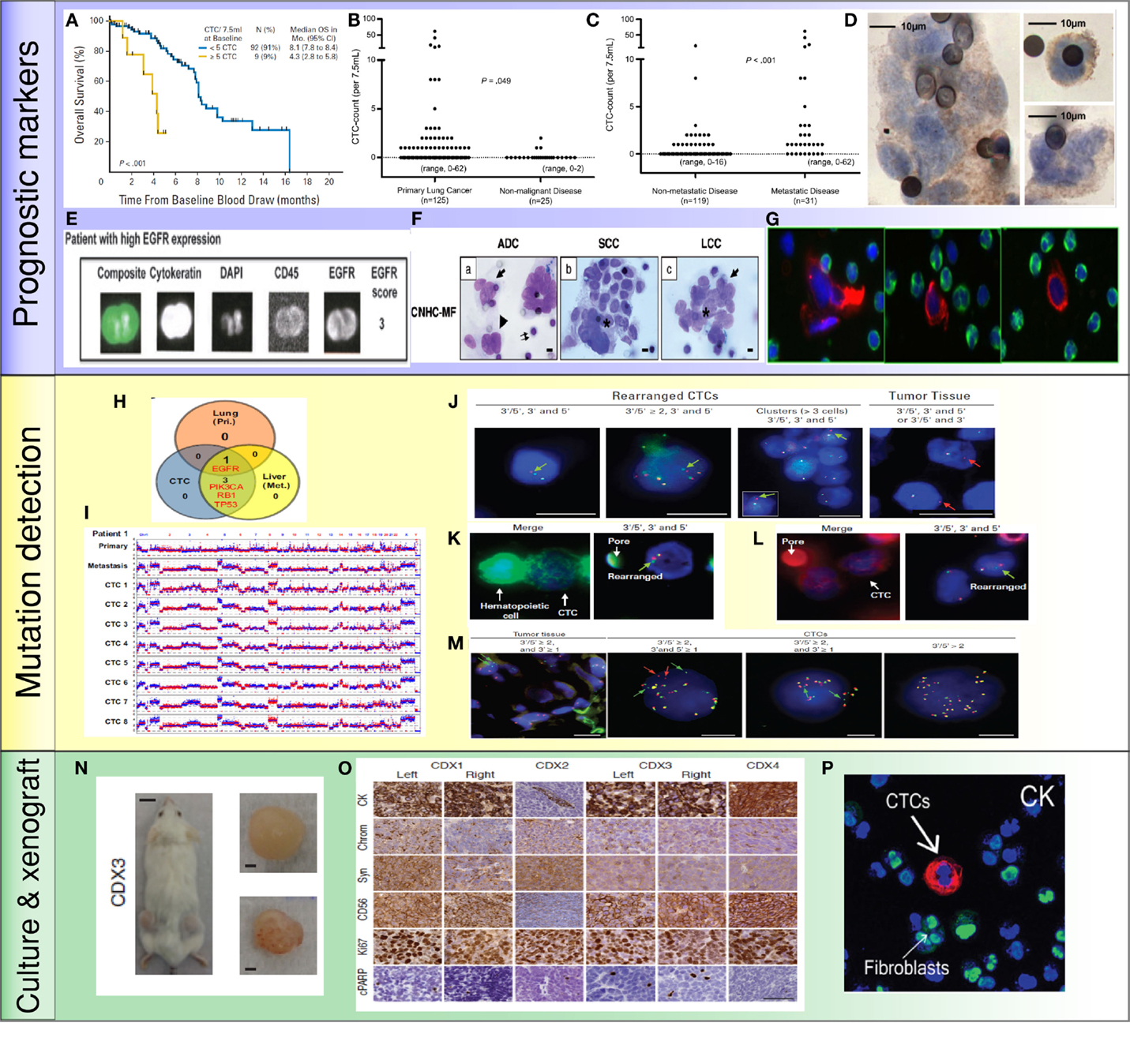
Figure 2. Application of CTCs in lung cancer: (A) less than 5 CTCs/7.5 ml of blood predicted improved survival by CellSearch system (19). (B,C) Higher numbers of CTCs were detected in metastatic lung cancer than cancer without distant metastasis (35). (D) NSCLC CTCs were detected by ISET technology and stained positive for EGFR (36). (E) NSCLC CTCs were isolated by CellSearch system and stained positive for EGFR and CK (33). (F) Morphologic features of CTCs from different histologies of NSCLC (37). (G) CTCs were detected by HD-CTC assay and stained positive for CK (red) and negative to CD45 (green) (38). (H,I) Mutations were detected in CTCs, primary tumors, and metastatic sites. Copy number variation patterns among single CTCs, primary tumor, and metastatic sites (39). (J) ALK rearrangement patterns in CTCs and primary tumor (40). (K,L) ALK rearranged CTCs stained positive for vimentin (K) and N-Cadherin (L) (40). (M) ROS1-rearranged CTCs were compared to primary tumor (41). (N) CTCs isolated from SCLC patients generated tumor in a mouse (42). (O) CTC-derived xenografts were stained for different protein markers (42). (P) NSCLC CTCs were isolated and expanded by a microfluidic co-culture model and stained positive for CK (27).
CTC Isolation Technologies
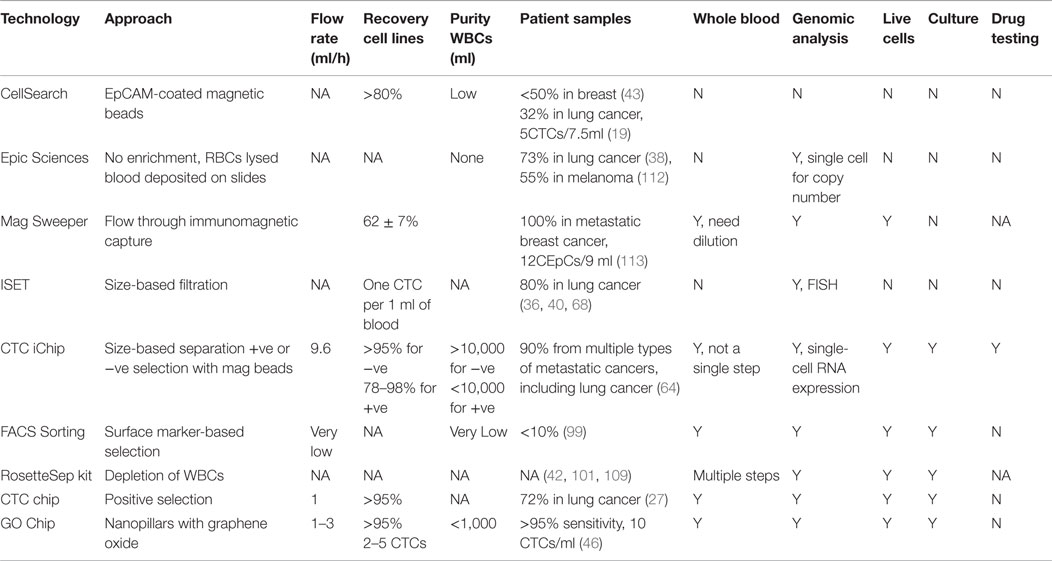
Circulating tumor cells have now been proposed as surrogate biomarkers in over 270 clinical trials (10). However, to date, CTCs have not been incorporated into routine clinical practice for management of patients with cancer. The efforts to identify biological relevance and clinical utility of CTCs parallel the development of CTC isolation technologies. There are several key parameters worthy of consideration when designing a method to isolate CTCs: (a) specificity, (b) sensitivity, (c) purity, (d) viability, and (e) throughput. All the downstream assays, such as molecular and genomic analysis and culturing for ex vivo drug testing, depend on these factors. We will discuss about pros and cons associated with current isolation technologies in general and specifically as they pertain to lung cancer (Table 1).
Collectively, there are two major approaches; one is anti-epithelial cell adhesion molecule (EpCAM) dependent while the other is EpCAM independent. The FDA approved CellSearch technology utilizes EpCAM-coated magnetic beads to isolate CTCs in a multitude of cancers in spite of limited detection efficiency (32% in lung cancer) (19, 43–45). Microfluidic-based technologies have changed the existing paradigm for recovery of CTCs. Microfluidic chips coated with EpCAM and microfluidic systems utilizing immunomagnetic principles have been shown to capture CTCs from lung cancer samples with 100% efficiency (46–49). These antibody-based microfluidic devices have the advantage of high sensitivity, low numbers of white blood cells contamination (can be as low as 1,500 WBCs), as well as preserving the viability of CTCs due to minimal handling of whole blood. The drawback is that they suffer from limited throughput due to low flow rates (1–3 ml/h) and a requirement for antibody–antigen interaction. Another problem with EpCAM-dependent methods is that they can only capture a subset of CTCs and miss cells undergoing epithelial–mesenchymal transition (EMT) (10). Wit et al. recovered lung CTCs by filtration from the waste of CellSearch system (50). The percentage of patients having more than 5 cells per 7.5 ml of blood increased from 15% (EpCAM positive) to 41% (EpCAM positive and negative). This suggested that including the EpCAM negative population increased CTC recovery.
By contrast, the label-free approaches to isolate CTCs do not rely on the expression of specific cell surface markers but instead on inherent CTC properties such as size, deformability, or dielectric susceptibility, and/or negative selection of WBCs (51–60). While improvements in size-based and other physical separation techniques have allowed higher throughput over the years, they suffer from limitations related to heterogeneity of tumor cells, contamination with blood cells and result in lower yield and specificity compared with the antigen-based systems (61). For example, CTCs within a patient may have a wide range of sizes (>4–30 μm) and many of them may overlap in size with blood cells (62). More recently, several new integrated platforms have emerged for CTC isolation. Liu et al. introduced an integrated device that separates blood cells and CTCs by deterministic lateral displacement, followed by an affinity-based enrichment (9.6 ml/h) (63). The CTC-iChip by Ozkumur et al. combines magnetic labeling and high throughput sorting of cells (8 ml/h) (64), which is based on the principle of conjugating capturing antibodies on magnetic particles and enriching rare cells by applying external magnetic forces (65). While EpCAM-independent systems allow high throughput and an unbiased surface marker-independent approach that can capture cells undergoing EMT (66), the need for multiplexing and pre-processing of blood samples make it cumbersome and time consuming. Chang et al. employed similar principles by labeling CTCs with antibody cocktail conjugated with magnetic beads followed by size-based filtration to trap CTCs on chip for immunofluorescence staining (67). This system also operates at high flow rates (2 ml/min) but requires RBC lysis and the average WBCs contamination was around 4000.
Other label-free technologies that are not microfluidic based are also employed in clinical evaluation of lung CTCs. The isolation by size of epithelial tumor cells (ISET) technology, isolating CTCs based on their larger size, is among the earliest developed EpCAM-independent approaches which filter CTCs from blood cells as they pass through a membrane filter (68). CTCs were detected in 80% of samples from stages IIIA–IV NSCLC patients using ISET compared with 23% using CellSearch (36). Using the same approach, CTCs were present in 65% of NSCLC patients in a more recent report (69). In another study, an automatic microscope scanning and analysis technology called high-definition CTC (HD-CTC) assay was utilized to examine CTCs from stages I–IV NSCLC patients (38, 70). This technology permits high-resolution imaging of CTCs and is not biased toward size or surface markers. Recently, DNA aptamers were utilized to isolate CTCs from NSCLC patients (71). CTCs were identified in 86% of the samples that were positive for aptamers and pan-CK. The ISET, HD-CTC assay, and aptamer approach require RBC lysis and have limited purity of isolated CTCs, therefore posing constraints on molecular and functional studies of the cells.
In summary, CTC technologies have evolved rapidly in the last decade, yet there is none that has FDA approval other than CellSearch. However, to incorporate CTCs into basic as well as small cohort clinical research, there are more tools than ever before, with microfluidic devices leading the way with higher sensitivity. Any ideal CTC technology should offer high throughput, minimal handling (whole blood) that can separate live CTCs with high sensitivity and specificity. Presently, there is no single technology that is optimal for every downstream analysis; the choice of technology is driven more by the end user application and ease of accessibility to the technology. Immunoaffinity-based technologies offer both sensitivity and specificity albeit with dependence on the known biomarker. A high throughput system that requires minimal pre-manipulation of whole blood and that can operate with either positive selection or negative depletion approach seems to be most promising for lung cancer CTC isolation. Furthermore, the efficiency of positive selection depends on the discovery of lung cancer-specific surface markers such that a cocktail of capturing antibodies can be applied to target a broader range of lung CTCs.
CTCs as Prognostic and Predictive Markers in Lung Cancer
Previously, the oncology community believed that there was little merit in diagnosing recurrence or progression earlier in patients who had surgery for earlier stages of lung cancer or following initial therapy for locally advanced/metastatic NSCLC. This was related to poor therapy choices at recurrence/progression that often does not alter clinically significant outcomes such as PFS and OS. There has been a rethink of this approach in a small, but significant minority of patients. This relates to the emerging field of therapy directed at oligometastatic disease such as local radiation or use of immunotherapy or newer biologics that may render patient disease free for a significant amount of time, even if OS is not affected. Many of these therapies are also better tolerated with broader therapeutic windows. We will, in this section, outline various studies relating CTCs to prognosis in lung cancer as well as studies that predict therapy response.
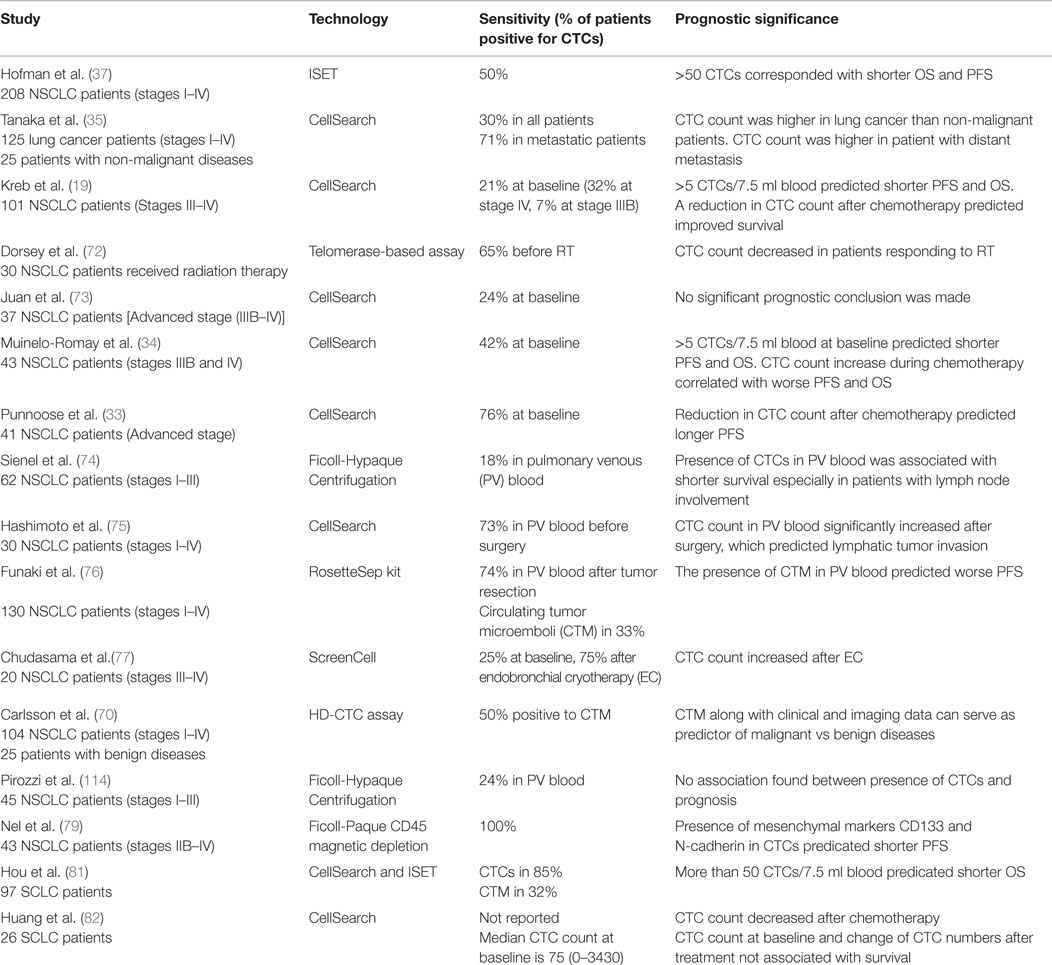
Hofman et al. used ISET technology to isolate CTCs from 208 NSCLC patients with stages I–IV cancer. Fifty percent of these patients had CTCs by morphological examination (37). A cut-off value of >50 corresponded to shorter PFS and OS. There was, however, no direct correlation between numbers of CTCs and disease stage, or other clinicopathologic parameters. Therefore, CTCs and tumor staging appeared to be independent prognostic factors. In another study using the CellSearch system, there were greater numbers of CTCs in metastatic lung cancer patients (P < 0.001) compared to patients without distant metastases (35). Similarly, another study using the CellSearch system found that in 101 patients with stage III/IV NSCLC, numbers of CTCs were higher in stage IV compared to stage III patients (19). With a threshold of 5 CTCs in 7.5 ml blood, patients were categorized into favorable and unfavorable groups. Both the PFS (6.8 vs. 2.4 months) and OS (8.1 vs. 4.3 months) were higher in the favorable group than the unfavorable (P < 0.001). Additionally, CTC numbers decreased with one cycle of chemotherapy. Reduction in numbers of CTCs with therapy correlated with improved PFS (6.9 vs. 2.4 months; P = 0.005) and OS (8.8 vs. 3.9 months; P < 0.001). This study highlighted that CTC numbers were not only prognostic, but also that a change in CTC number with therapy predicted disease progression dynamically.
Dorsey et al. investigated the change of CTC number in patients with localized NSCLC undergoing radiation treatment. Using a telomerase-based detection assay, 65% of the patients were positive for CTCs prior to treatment. CTC numbers significantly reduced after radiation (9.1 vs. 0.6 CTCs/ml; P < 0.001). This study suggested that analyzing CTC can serve as “real-time liquid biopsies” accompanying treatment to monitor tumor progression (72). Several studies examining CTCs in advanced NSCLC patients receiving chemotherapy have shown that >2 CTCs/7.5 ml or any increase in CTC numbers after therapy predicted lower OS and PFS (P = 0.05) (33, 34, 73). To improve detection sensitivity, CTCs from pulmonary vein blood were examined in patients undergoing surgery (74–76). Compared to peripheral blood CTCs (2 out of 30 positives), pulmonary CTCs were present in 22 out of 30 samples before surgery (0–1122 cells/2.5 ml, median, 4 cells/2.5 ml) (75). Surprisingly, the number of pulmonary CTCs increased significantly after surgical manipulation (0–1855 cells/2.5 ml, median, 60 cells/2.5 ml); this increase also correlated with pathological evidence of microscopic lymphatic invasion (P = 0.043). Chudasama et al. investigated the effect of endobronchial cryotherapy (EC) on shedding of CTCs before and after the procedure in peripheral blood (77). CTC count increased following cryotherapy in 15 out of 20 advanced stage patients (P = 0.0086) which predicted poor prognosis during follow-up. In summary, these studies suggest that monitoring change of CTC numbers during therapy is prognostic for NSCLC. An increase of CTC counts may entail additional follow-up examinations.
More recently, several groups have reported on the prognostic value of CTC clusters called circulating tumor microemboli (CTM) (36, 70). Krebs et al. observed the prevalence of CTM by ISET technology in 43% of patients with stage IIIB/IV NSCLC (36). In another study using HD-CTC assay, 50% of NSCLC patients with stages I–IV disease had CTM (70). It was shown that CTM can be used to diagnose lung cancer when combined with clinical and imaging data. The existence of CTMs was also observed in pulmonary venous (PV) blood of patients with NSCLC (76). Among 130 patients tested, 74% of them were positive for CTCs. CTMs were detected in 33% of samples which predicted tumor recurrence and worse disease-free survival rate (P < 0.01).
Other studies correlated prognosis to the presence/absence of protein expression of CTCs in NSCLC. As demonstrated by Wu et al., CTCs in multiple types of cancer, including lung cancer, harbored a mixed population of epithelial and mesenchymal phenotypes (78). Nel et al. stained CTCs for both epithelial markers such as EpCAM and pan-cytokeratin (CK) as well as mesenchymal markers such as N-cadherin and CD133 (79). Different subsets of CTC populations were identified with heterogeneous combinations of epithelial and mesenchymal characteristics. CD133 expression correlated positively with N-cadherin. The presence of these mesenchymal markers predicted shorter PFS (2 vs. 8 months, P = 0.003) likely due to emergence of chemoresistant populations.
Small-cell lung cancer (SCLC) accounts for 13% of newly diagnosed lung cancer and is considered aggressive with early dissemination and poor prognosis (80). Hou et al. demonstrated that CTCs were present in 85% of SCLC compared to 21% in NSCLC patients (19, 81). Higher CTC numbers were noted in SCLC than NSCLC; >50 CTCs/7.5 ml of blood predicted shorter PFS (4.6 vs. 8.8 months; 95% CI) and OS (5.4 vs. 11.5 months; 95% CI). A reduction in CTC number after chemotherapy was associated with longer PFS (9.6 vs. 4.1 months; 95% CI) and OS (10.4 vs. 4.1 months; 95% CI). Huang et al. evaluated prognostic significance of CTCs in SCLC. CTCs were enumerated before and after chemotherapy (82). A reduction of CTCs was observed in 16/26 patients after treatment. However, CTC count at baseline and the percentage change of CTCs were not statistically significantly associated with survival. A summary of the studies investigating the prognostic value of CTCs in lung cancer is shown in Table 2.
Taken together, several studies have demonstrated the prognostic utility of CTCs in lung cancer. CTC count and change of CTC number after surgery, radiation, and chemotherapy may serve as predictors of recurrence. At the current time, however, CTCs are not routinely used as prognostic or predictive markers in clinics. There are several reasons for this. Most of the previous studies used CellSearch or traditional approaches without pre-enrichment, which limited sensitivity of the tests in detecting CTCs. Many of the studies had small sample sizes (<100) limiting statistical significance. This resulted in contradictory or inconclusive findings. Given newer and extremely sensitive technologies that allow isolation and accurate characterization of CTCs, large numbers of patients within specific stages of lung cancer need to be enrolled. The stringent biomarker studies need to use training and test sets that will allow independent validation and reproducibility.
Applications of CTCs in the Era of Targeted Therapies in Lung Cancer
The past two decades have seen a large discovery effort such that lung cancer is not considered one homogeneous cancer. Over 64% of all lung cancers have an underlying driver mutation that is responsible for proliferation of the cancer and many of these mutations are mutually exclusive (83). Nearly 30% cases of these driver mutant lung cancers have an approved therapy (targeted therapy). The most common ones are adenocarcinomas (AC) that are associated with mutations in the EGFR gene or rearrangements in the ALK and ROS-1 gene (84). Additional genomic aberrations include those in BRAF, AKT1, ERRB2, PIK3CA, and fusions in RET (85). Detection of mutation by biopsy may not fully reflect intratumoral heterogeneity (86). In this regard, sampling CTCs as “liquid biopsy” may complement solid biopsy to inform effective targeted therapies. Liquid biopsy is also non-invasive allowing dynamic monitoring of disease progression (13).
One of the earliest investigations was identifying EGFR mutations in CTCs from metastatic NSCLC known to harbor these mutations. In 11 out of 12 patients, expected mutations were validated, including the appearance of the resistance mutation T790M. In this study, CTC numbers paralleled radiographic response and offered first insights into genomic profiling of CTCs as a way to monitor genotypic changes during therapy (24). Two recent studies examined EGFR mutation in advanced NSCLC patients. Marchetti et al. demonstrated that EGFR mutation was detected in CTCs of 84% of the patients carrying EGFR-mutant primary tumors (87). In 94% of the cases, mutations found in CTCs matched the mutations in tumor tissues. The unmatched mutations in CTCs and primary tumors were likely due to tumor heterogeneity between primary lesions and metastatic sites. Breitenbuecher et al. utilized a RT-PCR assay to detect in-frame deletions in the EGFR exon 19 (88). All eight EGFR-mutant patients demonstrated identical mutations in the CTCs. EGFR mutations were also detected from circulating DNA of advanced lung AC patients with 73% sensitivity (89). Both CTCs and cfDNA can be used in future research to determine the “best in class” EGFR tyrosine kinase inhibitors (TKI) for individual patients.
Other studies focused on investigating ALK rearrangement in CTCs (40, 90). By performing filter-adapted fluorescent in situ hybridization (FA-FISH), researchers identified unique ALK-rearranged pattern in CTCs with a mesenchymal phenotype. This unique population of CTCs may be highly invasive, behaving as metastasis initiation cells (91). Adapting the similar approach, ROS1 rearrangement was investigated in NSCLC CTCs and compared to tumor biopsy specimens (41). Among four patients tested, CTCs harbored similar split patterns as tumors but exhibited an increase in ROS1 copy number. The number of ROS1-rearranged CTCs increased in one patient who did not respond to crizotinib treatment. In another study, whole-genome amplification of single CTCs from lung cancer patients was performed followed by analyzing copy number variation (CNV) in addition to somatic mutations (39). It was demonstrated that CTCs obtained from the same patient exhibited similar CNV pattern but was distinguishable from CTCs obtained from a different histology of lung cancer. These studies suggest that profiling CTC genome can predict cancer progression as well as emergence of secondary resistant mechanisms to be further targeted by therapy.
CTCs as Biomarkers for Early Diagnosis of Lung Cancer
Sensitive detection of CTCs provides opportunities for early diagnosis of lung cancer. CTCs can be shed by primary tumor even at early stages of tumor development (92, 93). It was demonstrated that the presence of CTCs in 5 out of 168 chronic obstructive pulmonary disease patients predicted occurrence of lung nodules 1–4 years after initial detection of CTCs (94). In one study, CTCs were isolated from 84% of lung cancer patients of various stages, including early stage of lung cancer (57.1%) (95). CTCs were identified with CD45-FISH method that was reported to increase detection sensitivity by including cells deficient in epithelial markers like CK. Two studies utilized tumor-specific ligand folate and an oligonucleotide followed by qPCR and immunofluorescence staining to identify NSCLC CTCs (96, 97). CTCs were observed in more than 70% of all stages with 67.2% in stage I cancer. It was further demonstrated that CTCs can be more sensitive for early diagnosis of lung cancer than blood serum markers such as cyfra21-1 or CEA. More recently, one study evaluated CTCs from potential lung cancer patients to predict malignancy of the lung lesions as a way to circumvent sampling bias by solid biopsy (98). CTCs isolated shared similar morphological features and histology (72%) with biopsy specimens. In stage I patients tested (42%), the numbers of CTCs correlated with tumor size (P = 0.001). Our group also demonstrated that CTCs are detectable in early stages of lung cancer (68%) (27). Early diagnosis of cancer aided by liquid biopsy is challenging due to low abundance of CTCs present; therefore, it is necessary to develop more sensitive and specific technologies that allow more inclusive characterization methodologies that will aid early detection of lung cancer.
Current Applications: Culture of CTCs and Xenografts
While the exploration of innovative technologies for enhanced CTC isolation is always in the forefront of research, ex vivo culturing and in vivo xenograft models has gradually gained momentum in the field. Zhang et al. sorted a subset of breast CTCs to form cell lines and tumors in mice (99). This model helped to identify a novel gene signature associated with development of brain metastasis. Yu et al. successfully cultured breast CTCs followed by in vivo implantation and drug testing (100). Cayrefourcq et al. established and characterized a cell line derived from colon CTCs which served as a model for studying metastasis and testing treatment agents (101). Higher numbers of CTCs in SCLC allow ex vivo culture of these cells and formation of CTC-derived xenografts for drug screening and mutation detection. In the study by Hodgkinson et al., CTCs from SCLC patients were injected directly to mice (42). Samples with CTC count >400/7.5 ml successfully gave rise to tumors in mice in 2–4 months. This study demonstrated that CTCs from SCLC were tumorigenic and that CTC-derived xenografts termed CDXs mirrored the corresponding tumor biopsy specimens. CDXs can potentially serve as in vivo drug testing models with responses similar to those seen in patients. Next-generation sequencing (NGS) revealed similar genomic aberrations in CDXs as seen in SCLCs. Our group recently demonstrated capability of using a co-culture model to isolate and culture CTCs from patients with stages I–III NSCLC (27). A microfluidic co-culture model utilizing hydrogel and cancer-associated fibroblasts was developed to facilitate CTC culturing. This permitted histological characterizations as well as genomic comparison between CTCs and matched primary tumors. Culturing of CTCs can overcome a critical limitation related to the rarity of these cells. This will allow further in vivo and functional studies.
Future Directions
Over the past decade, the advancement of technological innovation to isolate CTCs has allowed investigation of their clinical utility (102, 103). We now understand that CTCs contain heterogeneous populations of both epithelial and mesenchymal phenotypes (104). They harbor genetic alterations that correspond to primary tumors and metastatic sites. The discordant or unique mutations carried by CTCs that are absent in primary tumors reflect heterogeneity in primary tumor or small amount of subclonal populations that are missed by conventional sequencing methods (105). Together with cfDNA, CTCs have been shown as promising surrogates of tumor burden and activating mutations for targeted therapies. Specifically, CTCs offer opportunities to perform biological studies such as phenotypic and histological characterization, invasion and migration assays, in vitro expansion, drug testing, and use as xenografts in animal models. Despite current advances in the field, CTC markers still fall short, when it comes to factoring inter-tumor and intra-tumor heterogeneity (106). We have seen this manifests as discordance in markers between primary/metastatic cancers and CTC genotypes or phenotypes. Additionally, there is still the problem of contaminating white blood cells, despite the emergence of several technologies that enable positive selection and negative depletion of leukocytes. These factors along with a low yield in earlier stages of lung cancer handicap functional studies related to CTCs [e.g., ability to detect metastasis initiation properties of lung CTCs as demonstrated in other cancers using EPISPOT assay (101, 107) or invasion capabilities (27, 108)]. In vivo studies of CTCs through generating CTC-derived xenografts generally require larger numbers of cells (42, 109) which are hard to obtain from early stage cancer patients; this can be to some extent overcome by increasing blood throughput and sensitivity of isolation methods. Biology of metastasis as related to a cascade of events has vast implications in drug development. The study of CTCs opens up a window for understanding this process. One study found that the WNT2 gene was enriched in breast CTCs and another study showed that genes involved in ECM were highly expressed in pancreatic CTCs (110, 111). These findings suggest that these genes and pathways can be targeted therapeutically to halt metastasis and likely improve survival. However, this type of study has not been done with lung cancer. Understanding metastasis initiating capabilities of CTCs from primary lung cancer will have a huge impact in providing specific adjuvant therapies targeting at these CTCs to reduce metastasis and improve survival. The next few years will allow us to further study these biological processes in depth and allow meaningful translation into the clinic.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Marisa L. Conte, Research and Data Informationist at the Taubman Health Sciences Library, for her help with the search for the relevant literature.
References
2. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin (2014) 64(1):9–29. doi:10.3322/caac.21208
3. Services USDoHaH. The Health Consequences of Smoking: A Report of the Surgeon General. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health (2004).
4. Namba Y, Kijima T, Yokota S, Niinaka M, Kawamura S, Iwasaki T, et al. Gefitinib in patients with brain metastases from non-small-cell lung cancer: review of 15 clinical cases. Clin Lung Cancer (2004) 6(2):123–8. doi:10.3816/CLC.2004.n.026
5. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med (2011) 365(5):395–409. doi:10.1056/NEJMoa1102873
6. Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer. J Natl Compr Canc Netw (2012) 10(10):1236–71.
7. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med (2005) 352(8):786–92. doi:10.1056/NEJMoa044238
8. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science (2007) 316(5827):1039–43. doi:10.1126/science.1141478
9. Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem (2013) 59(1):110–8. doi:10.1373/clinchem.2012.194258
10. Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer (2014) 14(9):623–31. doi:10.1038/nrc3820
11. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med (2014) 6(224):224ra24. doi:10.1126/scitranslmed.3007094
12. Shen J, Liao J, Guarnera MA, Fang H, Cai L, Stass SA, et al. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J Thorac Oncol (2014) 9(1):33–40. doi:10.1097/JTO.0000000000000025
13. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol (2014) 32(6):579–86. doi:10.1200/JCO.2012.45.2011
14. Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood (2011) 118(13):3680–3. doi:10.1182/blood-2011-03-344408
15. Gevensleben H, Garcia-Murillas I, Graeser MK, Schiavon G, Osin P, Parton M, et al. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin Cancer Res (2013) 19(12):3276–84. doi:10.1158/1078-0432.CCR-12-3768
16. Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med (2014) 20(4):430–5. doi:10.1038/nm.3511
17. Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem (2013) 59(12):1722–31. doi:10.1373/clinchem.2013.206359
18. Tjensvoll K, Nordgard O, Smaaland R. Circulating tumor cells in pancreatic cancer patients: methods of detection and clinical implications. Int J Cancer (2014) 134(1):1–8. doi:10.1002/ijc.28134
19. Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol (2011) 29(12):1556–63. doi:10.1200/JCO.2010.28.7045
20. Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol (2005) 23(7):1420–30. doi:10.1200/JCO.2005.08.140
21. Botteri E, Sandri MT, Bagnardi V, Munzone E, Zorzino L, Rotmensz N, et al. Modeling the relationship between circulating tumour cells number and prognosis of metastatic breast cancer. Breast Cancer Res Treat (2010) 122(1):211–7. doi:10.1007/s10549-009-0668-7
22. Kurihara T, Itoi T, Sofuni A, Itokawa F, Tsuchiya T, Tsuji S, et al. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J Hepatobiliary Pancreat Surg (2008) 15(2):189–95. doi:10.1007/s00534-007-1250-5
23. Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature (2009) 461(7265):809–U67. doi:10.1038/nature08489
24. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med (2008) 359(4):366–77. doi:10.1056/NEJMoa0800668
25. Pantel K, Alix-Panabieres C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res (2013) 73(21):6384–8. doi:10.1158/0008-5472.CAN-13-2030
26. Cen P, Ni X, Yang J, Graham DY, Li M. Circulating tumor cells in the diagnosis and management of pancreatic cancer. Biochim Biophys Acta (2012) 1826(2):350–6. doi:10.1016/j.bbcan.2012.05.007
27. Zhang Z, Shiratsuchi H, Lin J, Chen GA, Reddy RM, Azizi E, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget (2014) 5(23):12383–97.
28. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med (2004) 351(8):781–91. doi:10.1056/NEJMoa040766
29. Soeth E, Grigoleit U, Moellmann B, Roder C, Schniewind B, Kremer B, et al. Detection of tumor cell dissemination in pancreatic ductal carcinoma patients by CK 20 RT-PCR indicates poor survival. J Cancer Res Clin Oncol (2005) 131(10):669–76. doi:10.1007/s00432-005-0008-1
30. Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br J Cancer (2012) 106(3):508–16. doi:10.1038/bjc.2011.545
31. Hoffmann K, Kerner C, Wilfert W, Mueller M, Thiery J, Hauss J, et al. Detection of disseminated pancreatic cells by amplification of cytokeratin-19 with quantitative RT-PCR in blood, bone marrow and peritoneal lavage of pancreatic carcinoma patients. World J Gastroenterol (2007) 13(2):257–63. doi:10.3748/wjg.v13.i2.257
32. de Albuquerque A, Kubisch I, Breier G, Stamminger G, Fersis N, Eichler A, et al. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. Oncology (2012) 82(1):3–10. doi:10.1159/000335479
33. Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res (2012) 18(8):2391–401. doi:10.1158/1078-0432.CCR-11-3148
34. Muinelo-Romay L, Vieito M, Abalo A, Nocelo MA, Baron F, Anido U, et al. Evaluation of circulating tumor cells and related events as prognostic factors and surrogate biomarkers in advanced NSCLC patients receiving first-line systemic treatment. Cancers (Basel) (2014) 6(1):153–65. doi:10.3390/cancers6010153
35. Tanaka F, Yoneda K, Kondo N, Hashimoto M, Takuwa T, Matsumoto S, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res (2009) 15(22):6980–6. doi:10.1158/1078-0432.CCR-09-1095
36. Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol (2012) 7(2):306–15. doi:10.1097/JTO.0b013e31823c5c16
37. Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Flejou JF, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res (2011) 17(4):827–35. doi:10.1158/1078-0432.CCR-10-0445
38. Wendel M, Bazhenova L, Boshuizen R, Kolatkar A, Honnatti M, Cho EH, et al. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys Biol (2012) 9(1):016005. doi:10.1088/1478-3967/9/1/016005
39. Ni XH, Zhuo ML, Su Z, Duan JC, Gao Y, Wang ZJ, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. P Natl Acad Sci U S A (2013) 110(52):21083–8. doi:10.1073/pnas.1320659110
40. Pailler E, Adam J, Barthelemy A, Oulhen M, Auger N, Valent A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol (2013) 31(18):2273–81. doi:10.1200/JCO.2012.44.5932
41. Pailler E, Auger N, Lindsay CR, Vielh P, Islas-Morris-Hernandez A, Borget I, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol (2015) 26(7):1408–15. doi:10.1093/annonc/mdv165
42. Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med (2014) 20(8):897–903. doi:10.1038/nm.3600
43. Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res (2007) 13(3):920–8. doi:10.1158/1078-0432.CCR-06-1695
44. Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol (2010) 2010:617421. doi:10.1155/2010/617421
45. Chen X, Wang X, He H, Liu Z, Hu JF, Li W. Combination of circulating tumor cells with serum carcinoembryonic antigen enhances clinical prediction of non-small cell lung cancer. PLoS One (2015) 10(5):e0126276. doi:10.1371/journal.pone.0126276
46. Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol (2013) 8(10):735–41. doi:10.1038/nnano.2013.194
47. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature (2007) 450(7173):1235–9. doi:10.1038/nature06385
48. Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. P Natl Acad Sci U S A (2010) 107(43):18392–7. doi:10.1073/pnas.1012539107
49. Wang C, Ye M, Cheng L, Li R, Zhu W, Shi Z, et al. Simultaneous isolation and detection of circulating tumor cells with a microfluidic silicon-nanowire-array integrated with magnetic upconversion nanoprobes. Biomaterials (2015) 54:55–62. doi:10.1016/j.biomaterials.2015.03.004
50. Wit S, Dalum G, Lenferink AT, Tibbe AG, Hiltermann TJ, Groen HJ, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep (2015) 5:12270. doi:10.1038/srep12270
51. Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices (2011) 13(1):203–13. doi:10.1007/s10544-010-9485-3
52. De Giorgi V, Pinzani P, Salvianti F, Panelos J, Paglierani M, Janowska A, et al. Application of a filtration- and isolation-by-size technique for the detection of circulating tumor cells in cutaneous melanoma. J Invest Dermatol (2010) 130(10):2440–7. doi:10.1038/jid.2010.141
53. Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y, et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res (2011) 31(2):427–41.
54. Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park JM, et al. SSA-MOA: a novel CTC isolation platform using selective size amplification (SSA) and a multi-obstacle architecture (MOA) filter. Lab Chip (2012) 12(16):2874–80. doi:10.1039/c2lc40065k
56. Di Carlo D, Edd JF, Irimia D, Tompkins RG, Toner M. Equilibrium separation and filtration of particles using differential inertial focusing. Anal Chem (2008) 80(6):2204–11. doi:10.1021/ac702283m
57. Huang T, Jia CP, Jun Y, Sun WJ, Wang WT, Zhang HL, et al. Highly sensitive enumeration of circulating tumor cells in lung cancer patients using a size-based filtration microfluidic chip. Biosens Bioelectron (2014) 51:213–8. doi:10.1016/j.bios.2013.07.044
58. Hosokawa M, Yoshikawa T, Negishi R, Yoshino T, Koh Y, Kenmotsu H, et al. Microcavity array system for size-based enrichment of circulating tumor cells from the blood of patients with small-cell lung cancer. Anal Chem (2013) 85(12):5692–8. doi:10.1021/ac400167x
59. Hosokawa M, Kenmotsu H, Koh Y, Yoshino T, Yoshikawa T, Naito T, et al. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One (2013) 8(6):e67466. doi:10.1371/journal.pone.0067466
60. Fan X, Jia C, Yang J, Li G, Mao H, Jin Q, et al. A microfluidic chip integrated with a high-density PDMS-based microfiltration membrane for rapid isolation and detection of circulating tumor cells. Biosens Bioelectron (2015) 71:380–6. doi:10.1016/j.bios.2015.04.080
61. Sun YF, Yang XR, Zhou J, Qiu SJ, Fan J, Xu Y. Circulating tumor cells: advances in detection methods, biological issues, and clinical relevance. J Cancer Res Clin Oncol (2011) 137(8):1151–73. doi:10.1007/s00432-011-0988-y
62. Dong Y, Skelley AM, Merdek KD, Sprott KM, Jiang C, Pierceall WE, et al. Microfluidics and circulating tumor cells. J Mol Diagn (2013) 15(2):149–57. doi:10.1016/j.jmoldx.2012.09.004
63. Liu ZB, Zhang W, Huang F, Feng HT, Shu WL, Xu XP, et al. High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosens Bioelectron (2013) 47:113–9. doi:10.1016/j.bios.2013.03.017
64. Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med (2013) 5(179):179ra47. doi:10.1126/scitranslmed.3005616
65. Pappas D, Wang K. Cellular separations: a review of new challenges in analytical chemistry. Anal Chim Acta (2007) 601(1):26–35. doi:10.1016/j.aca.2007.08.033
66. Satelli A, Brownlee Z, Mitra A, Meng QH, Li S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin Chem (2015) 61(1):259–66. doi:10.1373/clinchem.2014.228122
67. Chang CL, Huang WF, Jalal SI, Chan BD, Mahmood A, Shahda S, et al. Circulating tumor cell detection using a parallel flow micro-aperture chip system. Lab Chip (2015) 15(7):1677–88. doi:10.1039/c5lc00100e
68. Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol (2000) 156(1):57–63. doi:10.1016/S0002-9440(10)64706-2
69. Mascalchi M, Falchini M, Maddau C, Salvianti F, Nistri M, Bertelli E, et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J Cancer Res Clin Oncol (2015). doi:10.1007/s00432-015-2021-3
70. Carlsson A, Nair VS, Luttgen MS, Keu KV, Horng G, Vasanawala M, et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J Thorac Oncol (2014) 9(8):1111–9. doi:10.1097/JTO.0000000000000235
71. Zamay GS, Kolovskaya OS, Zamay TN, Glazyrin YE, Krat AV, Zubkova O, et al. Aptamers selected to postoperative lung adenocarcinoma detect circulating tumor cells in human blood. Mol Ther (2015) 23(9):1486–96. doi:10.1038/mt.2015.108
72. Dorsey JF, Kao GD, MacArthur KM, Ju M, Steinmetz D, Wileyto EP, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non-small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: pilot study results. Cancer (2015) 121(1):139–49. doi:10.1002/cncr.28975
73. Juan O, Vidal J, Gisbert R, Munoz J, Macia S, Gomez-Codina J. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin Transl Oncol (2014) 16(7):637–43. doi:10.1007/s12094-013-1128-8
74. Sienel W, Seen-Hibler R, Mutschler W, Pantel K, Passlick B. Tumour cells in the tumour draining vein of patients with non-small cell lung cancer: detection rate and clinical significance. Eur J Cardio-Thorac (2003) 23(4):451–6. doi:10.1016/S1010-7940(02)00865-5
75. Hashimoto M, Tanaka F, Yoneda K, Takuwa T, Matsumoto S, Okumura Y, et al. Significant increase in circulating tumour cells in pulmonary venous blood during surgical manipulation in patients with primary lung cancer. Interact Cardiovasc Thorac Surg (2014) 18(6):775–83. doi:10.1093/icvts/ivu048
76. Funaki S, Sawabata N, Abulaiti A, Nakagiri T, Shintani Y, Inoue M, et al. Significance of tumour vessel invasion in determining the morphology of isolated tumour cells in the pulmonary vein in non-small-cell lung cancer. Eur J Cardiothorac Surg (2013) 43(6):1126–30. doi:10.1093/ejcts/ezs553
77. Chudasama D, Rice A, Soppa G, Anikin V. Circulating tumour cells in patients with lung cancer undergoing endobronchial cryotherapy. Cryobiology (2015) 71(1):161–3. doi:10.1016/j.cryobiol.2015.06.001
78. Wu S, Liu S, Liu Z, Huang J, Pu X, Li J, et al. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One (2015) 10(4):e0123976. doi:10.1371/journal.pone.0123976
79. Nel I, Jehn U, Gauler T, Hoffmann AC. Individual profiling of circulating tumor cell composition in patients with non-small cell lung cancer receiving platinum based treatment. Transl Lung Cancer Res (2014) 3(2):100–6. doi:10.3978/j.issn.2218-6751.2014.03.05
80. van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet (2011) 378(9804):1741–55. doi:10.1016/S0140-6736(11)60165-7
81. Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol (2012) 30(5):525–32. doi:10.1200/JCO.2010.33.3716
82. Huang CH, Wick JA, Sittampalam GS, Nirmalanandhan VS, Ganti AK, Neupane PC, et al. A multicenter pilot study examining the role of circulating tumor cells as a blood-based tumor marker in patients with extensive small-cell lung cancer. Front Oncol (2014) 4:271. doi:10.3389/fonc.2014.00271
83. Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA (2014) 311(19):1998–2006. doi:10.1001/jama.2014.3741
84. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature (2014) 511(7511):543–50. doi:10.1038/nature13385
85. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature (2012) 489(7417):519–25. doi:10.1038/nature11404
86. Abe H, Kawahara A, Azuma K, Taira T, Takase Y, Fukumitsu C, et al. Heterogeneity of anaplastic lymphoma kinase gene rearrangement in non-small-cell lung carcinomas: a comparative study between small biopsy and excision samples. J Thorac Oncol (2015) 10(5):800–5. doi:10.1097/JTO.0000000000000507
87. Marchetti A, Del Grammastro M, Felicioni L, Malatesta S, Filice G, Centi I, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One (2014) 9(8):e103883. doi:10.1371/journal.pone.0103883
88. Breitenbuecher F, Hoffarth S, Worm K, Cortes-Incio D, Gauler TC, Kohler J, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One (2014) 9(1):e85350. doi:10.1371/journal.pone.0085350
89. Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res (2011) 17(24):7808–15. doi:10.1158/1078-0432.CCR-11-1712
90. Faugeroux V, Pailler E, Auger N, Taylor M, Farace F. Clinical utility of circulating tumor cells in ALK-positive non-small-cell lung cancer. Front Oncol (2014) 4:281. doi:10.3389/fonc.2014.00281
91. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell (2009) 139(5):871–90. doi:10.1016/j.cell.2009.11.007
92. Weinberg RA. Mechanisms of malignant progression. Carcinogenesis (2008) 29(6):1092–5. doi:10.1093/carcin/bgn104
93. Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell (2012) 148(1–2):349–61. doi:10.1016/j.cell.2011.11.025
94. Ilie M, Hofman V, Long-Mira E, Selva E, Vignaud JM, Padovani B, et al. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One (2014) 9(10):e111597. doi:10.1371/journal.pone.0111597
95. Chen YY, Xu GB. Effect of circulating tumor cells combined with negative enrichment and CD45-FISH identification in diagnosis, therapy monitoring and prognosis of primary lung cancer. Med Oncol (2014) 31(12):240. doi:10.1007/s12032-014-0240-0
96. Yu Y, Chen Z, Dong J, Wei P, Hu R, Zhou C, et al. Folate receptor-positive circulating tumor cells as a novel diagnostic biomarker in non-small cell lung cancer. Transl Oncol (2013) 6(6):697–702. doi:10.1593/tlo.13535
97. Lou J, Ben S, Yang G, Liang X, Wang X, Ni S, et al. Quantification of rare circulating tumor cells in non-small cell lung cancer by ligand-targeted PCR. PLoS One (2013) 8(12):e80458. doi:10.1371/journal.pone.0080458
98. Fiorelli A, Accardo M, Carelli E, Angioletti D, Santini M, Di Domenico M. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg (2015) 99(6):1899–905. doi:10.1016/j.athoracsur.2014.11.049
99. Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med (2013) 5(180):180ra48. doi:10.1126/scitranslmed.3005109
100. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science (2014) 345(6193):216–20. doi:10.1126/science.1253533
101. Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res (2015) 75(5):892–901. doi:10.1158/0008-5472.CAN-14-2613
103. Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol (2011) 192(3):373–82. doi:10.1083/jcb.201010021
104. Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science (2013) 339(6119):580–4. doi:10.1126/science.1228522
105. Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res (2013) 73(10):2965–75. doi:10.1158/0008-5472.CAN-12-4140
106. Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med (2010) 16(9):398–406. doi:10.1016/j.molmed.2010.07.001
107. Deneve E, Riethdorf S, Ramos J, Nocca D, Coffy A, Daures JP, et al. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem (2013) 59(9):1384–92. doi:10.1373/clinchem.2013.202846
108. Fan T, Zhao Q, Chen JJ, Chen WT, Pearl ML. Clinical significance of circulating tumor cells detected by an invasion assay in peripheral blood of patients with ovarian cancer. Gynecol Oncol (2009) 112(1):185–91. doi:10.1016/j.ygyno.2008.09.021
109. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol (2013) 31(6):539–U143. doi:10.1038/nbt.2576
110. Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature (2012) 487(7408):510–3. doi:10.1038/nature11217
111. Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep (2014) 8(6):1905–18. doi:10.1016/j.celrep.2014.08.029
112. Ruiz C, Li JL, Luttgen MS, Kolatkar A, Kendall JT, Flores E, et al. Limited genomic heterogeneity of circulating melanoma cells in advanced stage patients. Phys Biol (2015) 12(1):016008. doi:10.1088/1478-3975/12/1/016008
113. Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. P Natl Acad Sci U S A (2009) 106(10):3970–5. doi:10.1073/pnas.0813188106
Keywords: circulating tumor cells, liquid biopsy, review of literature, prognostic biomarkers, non-small-cell lung cancer, small-cell lung cancer, lung cancer
Citation: Zhang Z, Ramnath N and Nagrath S (2015) Current status of CTCs as liquid biopsy in lung cancer and future directions. Front. Oncol. 5:209. doi: 10.3389/fonc.2015.00209
Received: 28 June 2015; Accepted: 08 September 2015;
Published: 30 September 2015
Edited by:
Taofeek K. Owonikoko, Emory University, USAReviewed by:
Yuhchyau Chen, University of Rochester Medical Center, USAJanaki Deepak, University of Maryland School of Medicine, USA
Nathan A. Pennell, Cleveland Clinic, USA
Copyright: © 2015 Zhang, Ramnath and Nagrath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nithya Ramnath, Department of Internal Medicine, University of Michigan, 1500 E Medical Center Dr SPC 5912, Ann Arbor, MI 48109, USA, nithyar@med.umich.edu;
Sunitha Nagrath, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA, snagrath@umich.edu
 Zhuo Zhang
Zhuo Zhang Nithya Ramnath
Nithya Ramnath Sunitha Nagrath
Sunitha Nagrath