- Department of Virology and Department of Pathogens & Inflammation, UPRES EA4266, SFR FED 4234, CHRU Besançon, University of Franche-Comté, Besançon, France
Breast cancer is the leading causes of cancer-related death among women. The vast majority of breast cancers are carcinomas that originate from cells lining the milk-forming ducts of the mammary gland. Numerous articles indicate that breast tumors exhibit diverse phenotypes depending on their distinct physiopathological signatures, clinical courses, and therapeutic possibilities. The human cytomegalovirus (HCMV) is a multifaceted highly host specific betaherpesvirus that is regarded as asymptomatic or mildly pathogenic virus in immunocompetent host. HCMV may cause serious in utero infections as well as acute and chronic complications in immunocompromised individual. The involvement of HCMV in late inflammatory complications underscores its possible role in inflammatory diseases and cancer. HCMV targets a variety of cell types in vivo, including macrophages, epithelial cells, endothelial cells, fibroblasts, stromal cells, neuronal cells, smooth muscle cells, and hepatocytes. HCMV can be detected in the milk after delivery and thereby HCMV could spread to adjacent mammary epithelial cells. HCMV also infects macrophages and induces an atypical M1/M2 phenotype, close to the tumor-associated macrophage phenotype, which is associated with the release of cytokines involved in cancer initiation or promotion and breast cancer of poor prognosis. HCMV antigens and DNA have been detected in tissue biopsies of breast cancers and elevation in serum HCMV IgG antibody levels has been reported to precede the development of breast cancer in some women. In this review, we will discuss the potential role of HCMV in the initiation and progression of breast cancer.
Introduction
Breast cancer is the most frequent cause of cancer-related death among women. According to world health organization more than 5 million deaths have been attributed to breast cancer in 2011 (Global Health Estimates, WHO 2013). Breast cancer represents a heterogeneous disease. Based on the transcriptomic profile, breast tumors have been categorized into at least five intrinsic subtypes (1, 2). These subtypes include basal like, ERBB2+, normal breast-like, luminal subtype A, and B (1). Information regarding the intrinsic subtype of breast cancer in patients has prognostic significance and may help in designing personalized therapy in future (3). Familial history of breast cancer, hormonal replacement therapies, alcohol consumption, and radiation exposure are few risk factors associated with breast cancer (4). Biological entities especially viruses are also known to trigger various cancers in human. There are at least one dozen of viruses with established role in human malignancies [comprehensively reviewed in Ref. (5)]. Few viruses are suspected to play a role in the initiation or promotion of breast cancer (6). One of such viruses is human cytomegalovirus (HCMV).
Human cytomegalovirus (also called human herpesvirus 5) is a member of herpesviridae family (subfamily betaherpesvirinae). HCMV infects nearly 50–90% of the population worldwide. HCMV infection is either asymptomatic or causes mild discomfort. In certain situations where immunity is either immature or immunocompromised or suppressed, HCMV infection causes significant morbidity and mortality (7, 8). In the last decade, presence of HCMV genome and antigens has been reported in several kinds of human cancers. These human malignancies include breast cancer, brain cancer, prostate cancer, colon cancer, and salivary gland cancer (9, 10). In addition, we and others have shown the oncogenic transforming potential of HCMV in vitro (11–13). In this review, we will focus reader’s attention on the potential link between HCMV infection and the initiation and/or development of breast cancer.
HCMV and Breast Cancer: A Volatile Relationship
Several attempts have been made to search for a virus responsible for breast cancer. In 1971, Moore and colleagues examined the milk samples from women with or without the history of breast cancer using electron microscopy. They found higher prevalence of virus like particles designated as “particle B” in the milk of women with familial history of breast cancer. Exact nature of “particle B” is still an enigma (14). With the rapid advancement in DNA and protein technology, presence of several viruses has been detected in breast cancer and normal tissue. The presence of human papilloma virus (HPV) (15, 16), Epstein–Barr virus (EBV) (17), human endogenous retroviruses (18), and more recently JC and BK human polyomaviruses (19) has been reported in patients derived breast cancer specimens (Table 1). However, contrasting findings are also available, which make the relation of viruses and breast cancer highly volatile.
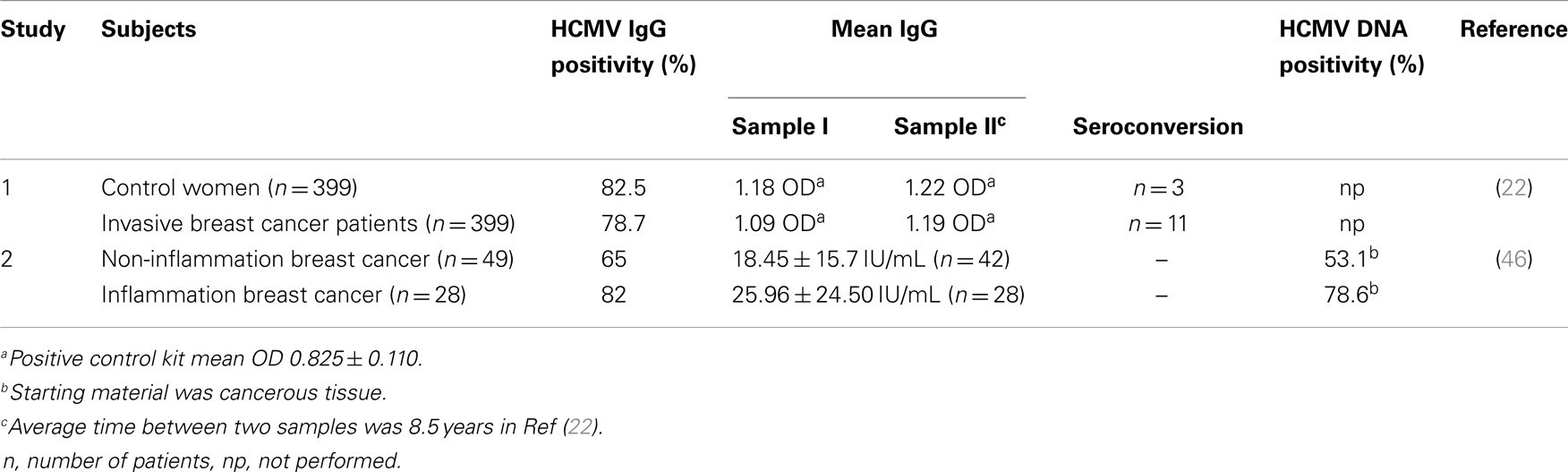
Human cytomegalovirus has large protein repertoire that can initiate or promote neoplastic changes in a cell. Richardson hypothesized that incidence of breast cancer could be raised by late exposure to HCMV (31). This hypothesis was based on the incidence of breast cancer and its correlation with the seroprevalence of HCMV. Cox and colleagues investigated the correlation between levels of HCMV IgG with the development of breast cancer. They enrolled 399 women with invasive breast cancer and 399 controls. Results of their study suggest a statistically significant correlation with the elevation of HCMV IgG levels and development of breast cancer in some women (22) (Table 2).
Breast milk is the predominant source of HCMV transmission in human. Presence of HCMV has been detected in more than 90% of milk samples derived from women seropositive for HCMV (32, 33). Using polymerase chain reaction and Southern analysis, presence of HCMV DNA was reported in normal breast tissue (34). Data from Cobb’s laboratory provide a more direct evidence of breast epithelium as an important reservoir for HCMV in humans. They detected the presence of HCMV antigens (specifically in glandular epithelium) in surgical biopsies of normal breast, breast with tumor, and normal breast tissue from the breast cancer patients using immunohistochemistry. Prevalence of HCMV antigens was relatively higher in neoplastic epithelium of patients with breast cancer (23).
A recent report by Soderberg-Naucler laboratory demonstrated the presence of HCMV proteins and DNA in breast cancer and sentinel lymph node metastases tissue using immunohistochemistry and PCR. In their study, presence of HCMV antigens was restricted to metastatic tumor cells that are not consistent with previous reports (23, 35).
There are several reports where no correlation has been found between HCMV and breast cancer. For example, Antonsson and coworkers screened for the presence of several viruses (including HCMV and EBV) in 54 fresh breast tissue samples using real time PCR. They did not detect the presence of HCMV in any sample (20). In another instance, Utrera-Barillas and colleagues investigated the association of presence of HCMV DNA with breast cancer progression in primary breast cancer biopsies using real time PCR (36). They detected HCMV DNA in only 2 cases out of 27 breast cancer specimens. They found no significant correlation between HCMV presence and breast cancer progression. Variation in tissue handling, sample size, PCR primer designing, and sites of tissue sampling could be responsible for the discrepancy in results among different laboratories (37).
Collectively, data from in vitro and in vivo suggest that HCMV may be involved in the initiation or progression of breast cancer. However, in order to obtained conclusive results, clinical findings need to be analyzed on large cohorts and in vitro findings need further validation in animal models.
HCMV Cell Tropism and Breast Cancer
Human cytomegalovirus is known to infect virtually most organs of the human body including blood, brain, breast, colon, eye, kidney, liver, and lung. Therefore, HCMV exhibits broader tropism. Several reports indicate the replication of HCMV in various cells ranging from monocytes to neural stem cells (23, 38–40). Monocytes and macrophages are widely recognized as important HCMV reservoirs responsible for the dissemination of the virus throughout of the body (41, 42). Furthermore, infection of monocytes by HCMV has potential to reprogram monocytes, resulting in polarization toward inflammatory macrophages (M1) that also exhibits properties of immunosuppressive macrophage (M2) (43) (Figure 1). This polarization is mediated by induction of NF-κB and PI3K activities in monocytes upon HCMV infection (43). M1 macrophages secrete inflammatory factors including TNFalpha, IL-6, and nitric oxide synthase 2 (Figure 1). Prolonged secretion of these cytokines is often linked with the development of cancer [reviewed in Ref. (44)]. We have also observed the positive correlation among the seroprevalence of HCMV IgG, elevated IL-6 levels, and incidence of liver cancer in a patient oriented study (45). In another instance, El-Shinawi and co workers investigated the prevalence of HCMV infection in patients with inflammatory breast cancer (IBC) and non-IBC invasive ductal carcinoma (IDC) patients. They observed the higher prevalence of HCMV IgG in patients with IBC than IDC patients (Table 2). Furthermore, they detected higher levels of DNA and activation of NF-κB in cancerous tissue isolated from IBC as compared to IDC patients. Increased activation of NF-κB can be a result of HCMV infection of breast cells or indirectly by cytokine production in the tumor microenvironment (46).
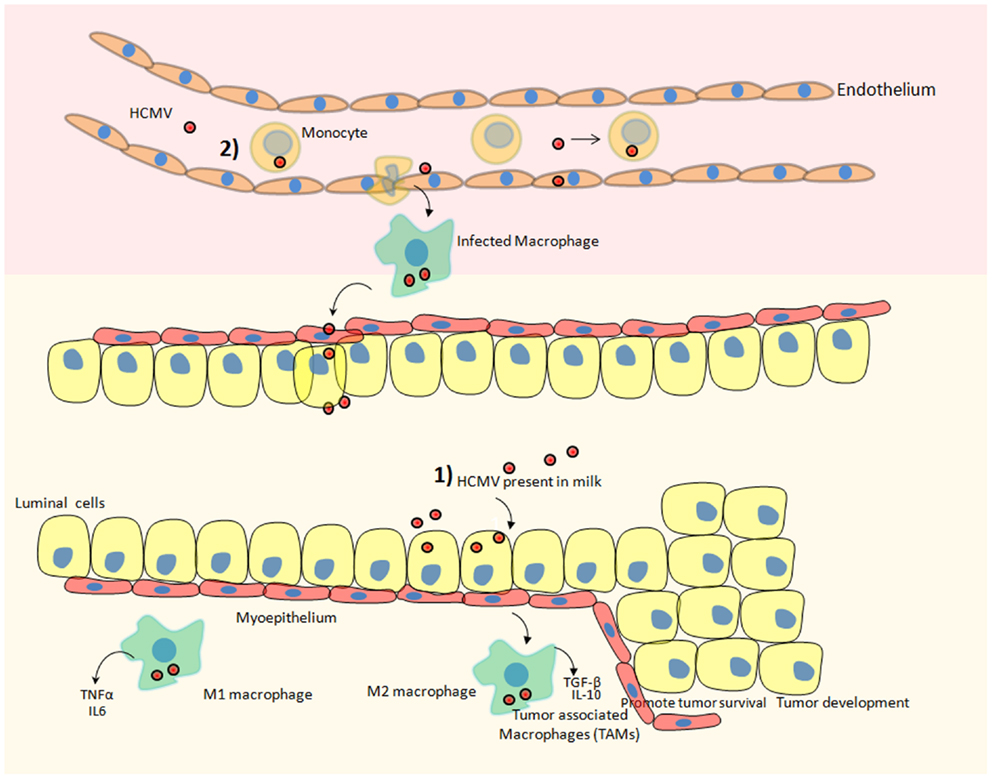
Figure 1. Hypothetical scheme of HCMV dispersal and its possible involvement in the development of breast cancer. Two potential scenarios could ultimately result in breast tumor after HCMV infection. (1) HCMV that is present in the milk could directly infect the mammary epithelial cells lining the duct responsible for converting most precursors into milk constituents and transporting them to the mammary lumen. Subsequently, macrophages present in breast tissue could be also infected by HCMV favoring a protumoral microenvironment. (2) HCMV present in blood (viremia) could infect circulating monocytes. Upon migration of infected monocytes into breast tissue, HCMV-infected macrophages could transmit the virus to mammary epithelial cells. Additionally, monocytes/macrophages are regarded as a prominent reservoir of HCMV infection. HCMV infection of monocytes/macrophages can reprogram them to acquire M1/M2 characteristics. M1 macrophages secrete pro-inflammatory cytokines and M2 macrophages secrete immuno-suppressive factors that can promote the progression of breast cancer. Tumor-associated macrophages (TAM) that are of poor prognosis during breast cancer and fuel the progression of the disease could be preferentially activated by HCMV.
Macrophages associated with the tumorous environment are known as tumor-associated macrophages (TAM). The predominant fractions of TAMs are M2 polarized macrophages. Upon infiltrating tumor surroundings of breast cancer cells, macrophages may acquire M2 state. These M2 macrophages secrete high levels of immunosuppressive cytokines, e.g., IL-10, TGF-beta, and little amount of pro-inflammatory cytokines [reviewed in Ref. (47)] (Figure 1). These M2 macrophages being immunosuppressive in nature indirectly favors the development of breast cancer. Since HCMV can infect normal breast and malignant breast tissue therefore the role of HCMV in favoring TAMs phenotype (by inducing M2 polarization) and breast cancer progression is highly speculated. For instance, the clinical isolate HCMV-DB displays preferential macrophage tropism, triggers M2 activation state, and stimulates the upregulation of the proto-oncogene Bcl-3 (38).
Indeed, TAMs are considered as an important therapeutic target in breast cancer. Luo and colleagues have identified legumain a stressed molecule overexpressed by TAMs. In addition, the application of DNA vaccine against legumain has been shown to dramatically reduce tumor angiogenesis in animal model (48).
Taken together, these primarily data suggest that the potential route of viral dispersal to breast tissue and involvement of HCMV in breast cancer progression.
Potential Oncogenes in HCMV Genome
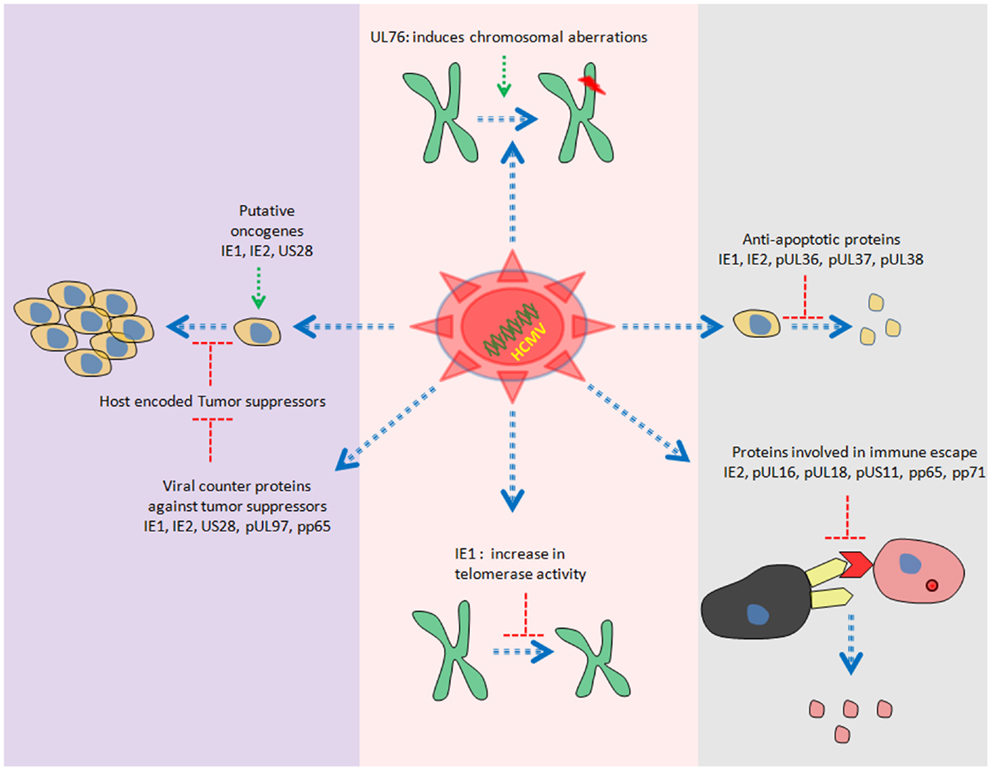
Human cytomegalovirus is a double stranded DNA virus with ~240 kb of genomic information. Recent study reveals the presence of more than 700 translated ORFs (49) in HCMV genome, which is more than double of previous predictions (50, 51). HCMV gene products have been reported to be involved in cell cycle dysfunction, genome instability, cell immortalization, inhibition of important cellular players involved in apoptosis and immune invasion (52–55) (Figure 2).
Several members of HCMV proteome have oncogenic properties. For instance, stable expression of US28 in NIH3T3 cells has been shown to induce transformed phenotypes. In addition, injection of NIH3T3 cells stably expressing US28 in mice leads to tumor formation (56). One of the responsible mechanisms for induction of tumor growth could be the activation of IL6–JAK1–signal transducer and activator of transcription 3 (STAT3) axis by US28 in vitro and in vivo (57). Furthermore, we have also observed similar findings in primary human hepatocytes and HepG2 cells upon HCMV infection (13).
Other gene products with oncogenic potential are immediate early (IE) 1 and IE2. Shen and colleagues have observed the transformation of primary baby rat kidney (BRK) cells upon transient expression of IE1, IE2, and adenovirus E1A proteins. Interestingly, they were not able to detect the presence of IE1/IE2 DNA in clonal cell lines derived from transformed BRK foci. The unconventional “hit and run” mechanism has been proposed to explain the transformation by IE1/IE2 (58). In addition, ectopic expression of IE1 induces telomerase mRNA and enhance telomerase activation in normal human diploid fibroblasts in vitro (59) (Figure 2).
Purified HCMV virions have reported to induce chromosomal breaks in primary human foreskin fibroblasts (HFF) (60). Data from Spector laboratory further revealed that only viral entry but not viral gene expression was prerequisite to induce chromosomal breaks in HFF (60). UL76 (one of the virion associated proteins) stable expression in human glioblastoma cells has been shown to induce chromosomal breaks (61) (Figure 2).
Taken together, data suggest that the presence of several HCMV proteins that have ability to induce transformation. Of note, transient or stable expression of a particular viral gene may not truly represent the natural HCMV infection scenario. Rigorous experimentations are needed to elucidate the exact function of these viral proteins in tumor tissue.
Antiviral Drugs Against HCMV as Novel Breast Cancer Therapies
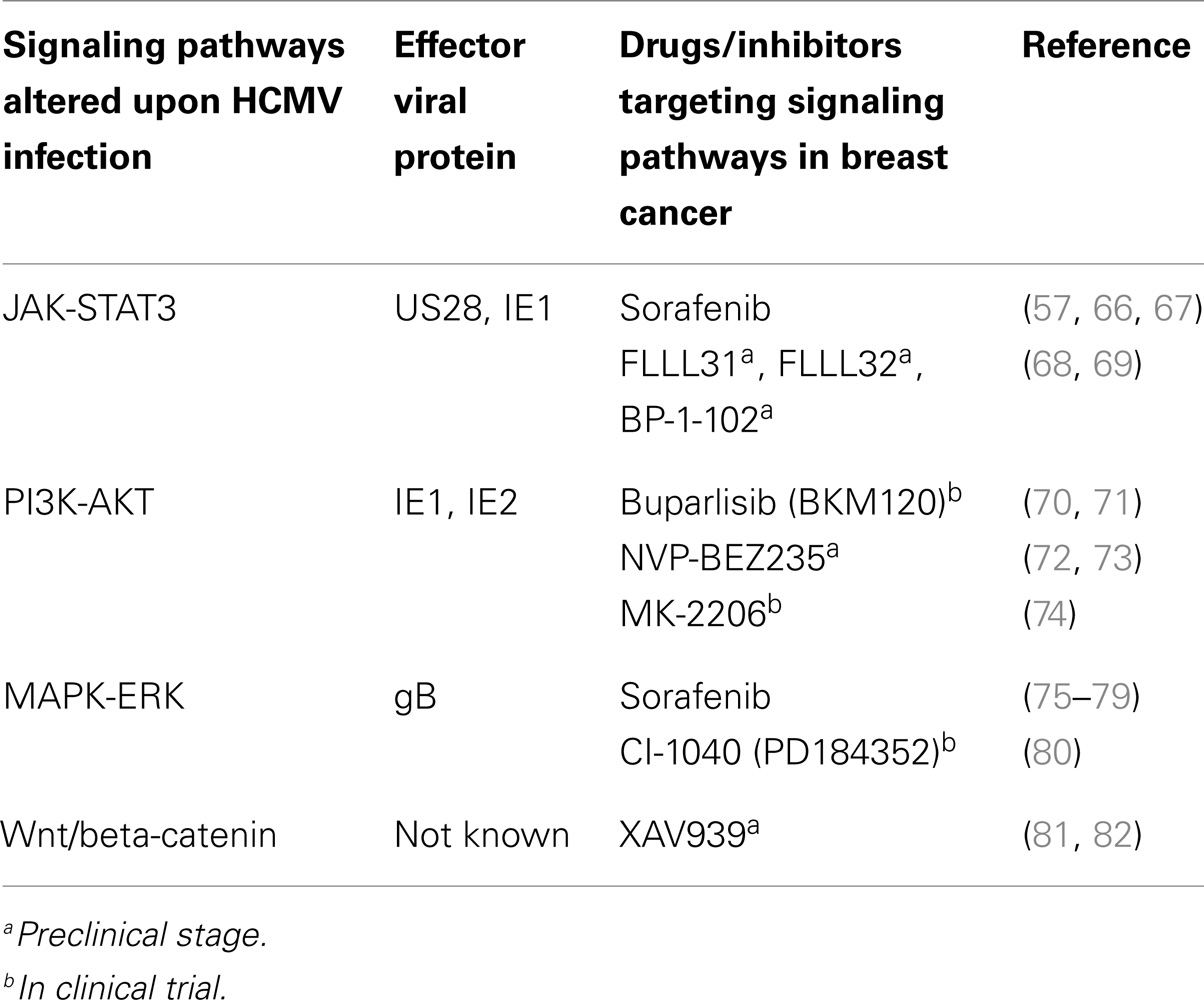
Presence of HCMV in various cancerous tissues raises the possibility of using anti-HCMV drugs in targeting cancer cells. In breast cancer and HCMV infection, there are several common pathways that are activated. For example, aberrant activity of STAT3, PI3K, NF-κB, MAPK, and Wnt driven cascade is observed in both HCMV infection and breast cancer (Table 3). Therefore, drugs that target these pathways should have significant impact on both HCMV infection and tumor progression. There are several compounds at preclinical or clinical trial stage that show significant impact on cancer development (Table 3). One of such FDA approved drug is sorafenib, a multi kinase inhibitor that affects several signaling cascades. Sorafenib is known to inhibit the replication of HCMV in several cell types in vitro (62). In addition, sorafenib inhibits cell death and induce apoptosis in several breast cancer cell lines including MCF-7 and MDA-MB-231 (63). In a phase II clinical trial involving patients with HER2 negative breast cancer, combination of sorafenib and cepacitabine has been reported to improved progression free survival in patients (64). Furthermore, phase III clinical trial (Clinicaltrials.gov, NCT01234337) with reduced dose of sorafenib has been also started (65).
Signal transducer and activator of transcription 3 is an important transcription factor that governs genes responsible for cell cycle progression and apoptosis. Upregulation of pSTAT3 has been reported in several kinds of malignancies including breast cancer (83). In addition, activation of IL6–JAK–STAT3 axis has been observed upon HCMV infection in several cell types (13, 57). Involvement of viral proteins IE1 and US28 in modulating this signaling axis has been suggested (57, 66) (Table 3). Moreover, recent work demonstrates the activation of STAT3 in a triple negative breast cancer cell line (MDA-MB-231) upon treatment of cmvIL-10 (84). cmvIL-10 is encoded by ORF UL11a and is homolog to human IL-10 (85). Furthermore, exposure of cmvIL-10 to these breast cancer cell lines resulted in increase in cell proliferation and decrease in apoptosis.
Lin and colleagues designed two inhibitors designated as “FLLL31” and “FLLL32” derived from curcumin. These inhibitors bind specifically to Janus kinase 2 and STAT3 Src homology-2 domain that are indispensable for STAT3 dimerization and downstream signal transduction (68). In vitro application of these inhibitors has shown to inhibit STAT3 activation, cell invasion, and colony formation in breast cancer cell lines (68). Even results were promising in animal models. Same group has recently developed another STAT3 inhibitor termed BP-1-102 that represses the tumor growth of breast cancer cells in xenografts (69).
Another important signaling axis pertaining to homeostasis is PI3K–Akt axis. PI3K driven signaling cascades play an important role in cell growth, differentiation, glucose metabolism, and chemotaxis (86). In breast cancer, aberrant expression of PI3K and its downstream signaling partners is frequently observed. Involvement of IE1 and IE2 in activation of PI3K pathway has been also suggested (70). Several PI3K inhibitors are being tested against various cancers including breast cancer in clinical trials (Table 3). However, impact of these inhibitors in HCMV replication is not assessed.
In addition to above mentioned signaling pathways, impact of HCMV infection on Wnt pathway has been investigated (81, 87). HCMV-infected fibroblasts and human placental extravillous trophoblasts (81) exhibited decreased levels of Wnt 5a/b, Wnt driven beta-catenin, and total as well as phosphorylated form of lipoprotein receptor related protein 6 (87). In addition, treatment of cells with Wnt modulators (monensin, nigericin, and salinomycin) resulted in inhibition of HCMV replication in fibroblasts, suggesting important role of Wnt signaling partners in HCMV replication. Wnt plays important role in cell growth, cell fate determination, and tumorigenesis. Upregulation of Wnt pathway genes have been observed in breast cancer cell lines and patient samples (88). Collectively, this makes Wnt pathways an attractive tool in targeting HCMV and breast cancerous cells simultaneously. For example, XAV939 a small molecule tankyrase inhibitor has been shown to attenuate Wnt pathways in several breast cancer cell lines (82). However, effect of XAV939 in HCMV replication is not known yet and needs further investigations. Development of resistance against a particular drug during cancer progression is frequent. Therefore, there is need to formulate feasible combinatorial therapies against breast cancer.
Conclusion
Several clinical and experimental investigations suggest the involvement of HCMV in various malignancies including breast cancer. In vitro and animal models suggest the presence of potent oncogenes genes in HCMV genome. Significance of higher prevalence of HCMV in breast cancer tissue is poorly understood. Whether it is just an “epiphenomenon” or crucial player in the progression of cancer needs further investigations.
Author Contributions
Georges Herbein and Amit Kumar wrote the manuscript. Both authors read and approved this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the University of Franche-Comté (UFC), the Région Franche-Comté (RECH-FON12-000013), and the FEDER to Georges Herbein. Amit Kumar is a recipient of a postdoctoral fellowship of the Région Franche-Comté.
References
1. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A (2001) 98:10869–74. doi: 10.1073/pnas.191367098
2. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature (2012) 490:61–70. doi:10.1038/nature11412
3. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol (2009) 27:1160–7. doi:10.1200/JCO.2008.18.1370
4. Ban KA, Godellas CV. Epidemiology of breast cancer. Surg Oncol Clin N Am (2014) 23:409–22. doi:10.1016/j.soc.2014.03.011
5. Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology (2009) 392:1–10. doi:10.1016/j.virol.2009.06.001
6. Alibek K, Kakpenova A, Mussabekova A, Sypabekova M, Karatayeva N. Role of viruses in the development of breast cancer. Infect Agent Cancer (2013) 8:32. doi:10.1186/1750-9378-8-32
7. Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin Infect Dis (2004) 39:155–61. doi:10.1086/421496
8. Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol (2008) 325:417–70. doi:10.1007/978-3-540-77349-8_23
9. Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia (2009) 11:1–9.
10. Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res (2011) 157:193–203. doi:10.1016/j.virusres.2010.10.026
11. Geder KM, Lausch R, O’Neill F, Rapp F. Oncogenic transformation of human embryo lung cells by human cytomegalovirus. Science (1976) 192:1134–7. doi:10.1126/science.179143
12. Clanton DJ, Jariwalla RJ, Kress C, Rosenthal LJ. Neoplastic transformation by a cloned human cytomegalovirus DNA fragment uniquely homologous to one of the transforming regions of herpes simplex virus type 2. Proc Natl Acad Sci U S A (1983) 80:3826–30. doi:10.1073/pnas.80.12.3826
13. Lepiller Q, Abbas W, Kumar A, Tripathy MK, Herbein G. HCMV activates the IL-6-JAK-STAT3 axis in HepG2 cells and primary human hepatocytes. PLoS One (2013) 8:e59591. doi:10.1371/journal.pone.0059591
14. Moore DH, Charney J, Kramarsky B, Lasfargues EY, Sarkar NH, Brennan MJ, et al. Search for a human breast cancer virus. Nature (1971) 229:611–4. doi:10.1038/229611a0
15. Gumus M, Yumuk PF, Salepci T, Aliustaoglu M, Dane F, Ekenel M, et al. HPV DNA frequency and subset analysis in human breast cancer patients’ normal and tumoral tissue samples. J Exp Clin Cancer Res (2006) 25:515–21.
16. Wang T, Zeng X, Li W, Zhu H, Wang G, Liu X, et al. Detection and analysis of human papillomavirus (HPV) DNA in breast cancer patients by an effective method of HPV capture. PLoS One (2014) 9:e90343. doi:10.1371/journal.pone.0090343
17. Grinstein S, Preciado MV, Gattuso P, Chabay PA, Warren WH, De Matteo E, et al. Demonstration of Epstein-Barr virus in carcinomas of various sites. Cancer Res (2002) 62:4876–8.
18. Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, et al. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin Cancer Res (2001) 7:1553–60.
19. Hachana M, Amara K, Ziadi S, Gacem RB, Korbi S, Trimeche M. Investigation of human JC and BK polyomaviruses in breast carcinomas. Breast Cancer Res Treat (2012) 133:969–77. doi:10.1007/s10549-011-1876-5
20. Antonsson A, Bialasiewicz S, Rockett RJ, Jacob K, Bennett IC, Sloots TP. Exploring the prevalence of ten polyomaviruses and two herpes viruses in breast cancer. PLoS One (2012) 7:e39842. doi:10.1371/journal.pone.0039842
21. Mazouni C, Fina F, Romain S, Ouafik L, Bonnier P, Brandone JM, et al. Epstein-Barr virus as a marker of biological aggressiveness in breast cancer. Br J Cancer (2011) 104:332–7. doi:10.1038/sj.bjc.6606048
22. Cox B, Richardson A, Graham P, Gislefoss RE, Jellum E, Rollag H. Breast cancer, cytomegalovirus and Epstein-Barr virus: a nested case-control study. Br J Cancer (2010) 102:1665–9. doi:10.1038/sj.bjc.6605675
23. Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, Wang W, et al. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae (2010) 1:8. doi:10.1186/2042-4280-1-8
24. Heng B, Glenn WK, Ye Y, Tran B, Delprado W, Lutze-Mann L, et al. Human papilloma virus is associated with breast cancer. Br J Cancer (2009) 101:1345–50. doi:10.1038/sj.bjc.6605282
25. Frega A, Lorenzon L, Bononi M, De Cesare A, Ciardi A, Lombardi D, et al. Evaluation of E6 and E7 mRNA expression in HPV DNA positive breast cancer. Eur J Gynaecol Oncol (2012) 33:164–7.
26. Ohba K, Ichiyama K, Yajima M, Gemma N, Nikaido M, Wu Q, et al. In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLoS One (2014) 9:e97787. doi:10.1371/journal.pone.0097787
27. Hachana M, Trimeche M, Ziadi S, Amara K, Korbi S. Evidence for a role of the simian virus 40 in human breast carcinomas. Breast Cancer Res Treat (2009) 113:43–58. doi:10.1007/s10549-008-9901-z
28. Wang Y, Melana SM, Baker B, Bleiweiss I, Fernandez-Cobo M, Mandeli JF, et al. High prevalence of MMTV-like env gene sequences in gestational breast cancer. Med Oncol (2003) 20:233–6. doi:10.1385/MO:20:3:233
29. Melana SM, Nepomnaschy I, Hasa J, Djougarian A, Djougarian A, Holland JF, et al. Detection of human mammary tumor virus proteins in human breast cancer cells. J Virol Methods (2010) 163:157–61. doi:10.1016/j.jviromet.2009.09.015
30. Golan M, Hizi A, Resau JH, Yaal-Hahoshen N, Reichman H, Keydar I, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia (2008) 10:521–33.
31. Richardson A. Is breast cancer caused by late exposure to a common virus? Med Hypotheses (1997) 48:491–7. doi:10.1016/S0306-9877(97)90118-3
32. Asanuma H, Numazaki K, Nagata N, Hotsubo T, Horino K, Chiba S. Role of milk whey in the transmission of human cytomegalovirus infection by breast milk. Microbiol Immunol (1996) 40:201–4. doi:10.1111/j.1348-0421.1996.tb03335.x
33. Hamprecht K, Maschmann J, Vochem M, Dietz K, Speer CP, Jahn G. Epidemiology of transmission of cytomegalovirus from mother to preterm infant by breastfeeding. Lancet (2001) 357:513–8. doi:10.1016/S0140-6736(00)04043-5
34. Tsai JH, Tsai CH, Cheng MH, Lin SJ, Xu FL, Yang CC. Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma, and thyroid tumor tissues. J Med Virol (2005) 75:276–81. doi:10.1002/jmv.20267
35. Taher C, de Boniface J, Mohammad AA, Religa P, Hartman J, Yaiw KC, et al. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One (2013) 8:e56795. doi:10.1371/journal.pone.0056795
36. Utrera-Barillas D, Valdez-Salazar HA, Gómez-Rangel D, Alvarado-Cabrero I, Aguilera P, Gómez-Delgado A, et al. Is human cytomegalovirus associated with breast cancer progression? Infect Agent Cancer (2013) 8:12. doi:10.1186/1750-9378-8-12
37. Lazzeroni M, Serrano D. Potential use of vaccines in the primary prevention of breast cancer in high-risk patients. Breast Care (Basel) (2012) 7:281–7. doi:10.1159/000342167
38. Khan KA, Coaquette A, Davrinche C, Herbein G. Bcl-3-regulated transcription from major immediate-early promoter of human cytomegalovirus in monocyte-derived macrophages. J Immunol (2009) 182:7784–94. doi:10.4049/jimmunol.0803800
39. Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A (2005) 102(50):18153–8. doi:10.1073/pnas.0509201102
40. Belzile JP, Stark TJ, Yeo GW, Spector DH. Human cytomegalovirus infection of human embryonic stem cell-derived primitive neural stem cells is restricted at several steps but leads to the persistence of viral DNA. J Virol (2014) 88:4021–39. doi:10.1128/JVI.03492-13
41. Chan G, Nogalski MT, Yurochko AD. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci U S A (2009) 106:22369–74. doi:10.1073/pnas.0908787106
42. Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci U S A (2010) 107(46):20039–44. doi:10.1073/pnas.1014509107
43. Chan G, Bivins-Smith ER, Smith MS, Yurochko AD. NF-kappaB and phosphatidylinositol 3-kinase activity mediates the HCMV-induced atypical M1/M2 polarization of monocytes. Virus Res (2009) 144:329–33. doi:10.1016/j.virusres.2009.04.026
44. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell (2010) 140:883–99. doi:10.1016/j.cell.2010.01.025
45. Lepiller Q, Tripathy MK, Di Martino V, Kantelip B, Herbein G. Increased HCMV seroprevalence in patients with hepatocellular carcinoma. Virol J (2011) 8:485. doi:10.1186/1743-422X-8-485
46. El-Shinawi M, Mohamed HT, El-Ghonaimy EA, Tantawy M, Younis A, Schneider RJ, et al. Human cytomegalovirus infection enhances NF-κB/p65 signaling in inflammatory breast cancer patients. PLoS One (2013) 8:e55755. doi:10.1371/journal.pone.0055755
47. Tang X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett (2013) 332:3–10. doi:10.1016/j.canlet.2013.01.024
48. Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest (2006) 116:2132–41. doi:10.1172/JCI27648
49. Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, et al. Decoding human cytomegalovirus. Science (2012) 338:1088–93. doi:10.1126/science.1227919
50. Murphy E, Rigoutsos I, Shibuya T, Shenk TE. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A (2003) 100:13585–90. doi:10.1073/pnas.1735466100
51. Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, et al. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol (2003) 84:17–28. doi:10.1099/vir.0.18606-0
52. Kalejta RF, Shenk T. Manipulation of the cell cycle by human cytomegalovirus. Front Biosci (2002) 7:d295–306. doi:10.2741/kalejta
53. Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol (1995) 69:7960–70.
54. Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, et al. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science (1994) 265:391–4. doi:10.1126/science.8023160
55. Bego MG, St Jeor S. Human cytomegalovirus infection of cells of hematopoietic origin: HCMV-induced immunosuppression, immune evasion, and latency. Exp Hematol (2006) 34:555–70. doi:10.1016/j.exphem.2005.11.012
56. Maussang D, Verzijl D, van Walsum M, Leurs R, Holl J, Pleskoff O, et al. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc Natl Acad Sci U S A (2006) 103:13068–73. doi:10.1073/pnas.0604433103
57. Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal (2010) 3:ra58. doi:10.1126/scisignal.2001180
58. Shen Y, Zhu H, Shenk T. Human cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci U S A (1997) 94:3341–5. doi:10.1073/pnas.94.7.3341
59. Strååt K, Liu C, Rahbar A, Zhu Q, Liu L, Wolmer-Solberg N, et al. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst (2009) 101:488–97. doi:10.1093/jnci/djp031
60. Fortunato EA, Dell’Aquila ML, Spector DH. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc Natl Acad Sci U S A (2000) 97:853–8. doi:10.1073/pnas.97.2.853
61. Siew VK, Duh CY, Wang SK. Human cytomegalovirus UL76 induces chromosome aberrations. J Biomed Sci (2009) 16:107. doi:10.1186/1423-0127-16-107
62. Michaelis M, Paulus C, Löschmann N, Dauth S, Stange E, Doerr HW, et al. The multi-targeted kinase inhibitor sorafenib inhibits human cytomegalovirus replication. Cell Mol Life Sci (2011) 68:1079–90. doi:10.1007/s00018-010-0510-8
63. Fumarola C, Caffarra C, La Monica S, Galetti M, Alfieri RR, Cavazzoni A, et al. Effects of sorafenib on energy metabolism in breast cancer cells: role of AMPK-mTORC1 signaling. Breast Cancer Res Treat (2013) 141:67–78. doi:10.1007/s10549-013-2668-x
64. Baselga J, Segalla JG, Roché H, Del Giglio A, Pinczowski H, Ciruelos EM, et al. Sorafenib in combination with capecitabine: an oral regimen for patients with HER2-negative locally advanced or metastatic breast cancer. J Clin Oncol (2012) 30:1484–91. doi:10.1200/JCO.2011.36.7771
65. Baselga J, Costa F, Gomez H, Hudis CA, Rapoport B, Roche H, et al. A phase 3 trial comparing capecitabine in combination with sorafenib or placebo for treatment of locally advanced or metastatic HER2-negative breast cancer (the RESILIENCE study): study protocol for a randomized controlled trial. Trials (2013) 14:228. doi:10.1186/1745-6215-14-228
66. Reitsma JM, Sato H, Nevels M, Terhune SS, Paulus C. Human cytomegalovirus IE1 protein disrupts interleukin-6 signaling by sequestering STAT3 in the nucleus. J Virol (2013) 87:10763–76. doi:10.1128/JVI.01197-13
67. Liu CY, Tseng LM, Su JC, Chang KC, Chu PY, Tai WT, et al. Novel sorafenib analogues induce apoptosis through SHP-1 dependent STAT3 inactivation in human breast cancer cells. Breast Cancer Res (2013) 15:R63. doi:10.1186/bcr3457
68. Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E, et al. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res (2010) 70:2445–54. doi:10.1158/0008-5472.CAN-09-2468
69. Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci U S A (2012) 109:9623–8. doi:10.1073/pnas.1121606109
70. Yu Y, Alwine JC. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3’-OH kinase pathway and the cellular kinase Akt. J Virol (2002) 76:3731–8. doi:10.1128/JVI.76.8.3731-3738.2002
71. Saura C, Bendell J, Jerusalem G, Su S, Ru Q, De Buck S, et al. Phase Ib study of buparlisib plus trastuzumab in patients with HER2-positive advanced or metastatic breast cancer that has progressed on trastuzumab-based therapy. Clin Cancer Res (2014) 20:1935–45. doi:10.1158/1078-0432.CCR-13-1070
72. Smith MS, Bentz GL, Smith PM, Bivins ER, Yurochko AD. HCMV activates PI(3)K in monocytes and promotes monocyte motility and transendothelial migration in a PI(3)K-dependent manner. J Leukoc Biol (2004) 76:65–76. doi:10.1189/jlb.1203621
73. Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res (2008) 68:8022–30. doi:10.1158/0008-5472
74. Sangai T, Akcakanat A, Chen H, Tarco E, Wu Y, Do KA, et al. Biomarkers of response to Akt inhibitor MK-2206 in breast cancer. Clin Cancer Res (2012) 18:5816–28. doi:10.1158/1078-0432.CCR-12-1141
75. Boyle KA, Pietropaolo RL, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol (1999) 19(5):3607–13.
76. Johnson RA, Huong SM, Huang ES. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection through two distinct pathways: a novel mechanism for activation of p38. J Virol (2000) 74:1158–67. doi:10.1128/JVI.74.3.1158-1167.2000
77. Rodems SM, Spector DH. Extracellular signal-regulated kinase activity is sustained early during human cytomegalovirus infection. J Virol (1998) 72:9173–80.
78. Reeves MB, Breidenstein A, Compton T. Human cytomegalovirus activation of ERK and myeloid cell leukemia-1 protein correlates with survival of latently infected cells. Proc Natl Acad Sci U S A (2012) 109:588–93. doi:10.1073/pnas.1114966108
79. Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res (2004) 64:7099–109. doi:10.1158/0008-5472.CAN-04-1443
80. Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol (2004) 22:4456–62. doi:10.1200/JCO.2004.01.185
81. Angelova M, Zwezdaryk K, Ferris M, Shan B, Morris CA, Sullivan DE. Human cytomegalovirus infection dysregulates the canonical Wnt/β-catenin signaling pathway. PLoS Pathog (2012) 8:e1002959. doi:10.1371/journal.ppat.1002959
82. Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Inhibition of tankyrases induces axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS One (2012) 7:e48670. doi:10.1371/journal.pone.0048670
83. Diaz N, Minton S, Cox C, Bowman T, Gritsko T, Garcia R, et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res (2006) 12:20–8. doi:10.1158/1078-0432.CCR-04-1749
84. Valle Oseguera CA, Spencer JV. cmvIL-10 stimulates the invasive potential of MDA-MB-231 breast cancer cells. PLoS One (2014) 9:e88708. doi:10.1371/journal.pone.0088708
85. Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci U S A (2000) 97:1695–700. doi:10.1073/pnas.97.4.1695
86. Lauring J, Park BH, Wolff AC. The phosphoinositide-3-kinase-Akt-mTOR pathway as a therapeutic target in breast cancer. J Natl Compr Canc Netw (2013) 11:670–8.
87. Kapoor A, He R, Venkatadri R, Forman M, Arav-Boger R. Wnt modulating agents inhibit human cytomegalovirus replication. Antimicrob Agents Chemother (2013) 57:2761–7. doi:10.1128/AAC.00029-13
Keywords: cytomegalovirus, breast cancer, macrophage, HCMV, inflammation
Citation: Herbein G and Kumar A (2014) The oncogenic potential of human cytomegalovirus and breast cancer. Front. Oncol. 4:230. doi: 10.3389/fonc.2014.00230
Received: 24 June 2014; Accepted: 08 August 2014;
Published online: 25 August 2014.
Edited by:
Charles Stringfellow Cobbs, Swedish Neuroscience Institute, USAReviewed by:
Mauro G. Tognon, University of Ferrara, ItalyPier Giorgio Petronini, University of Parma, Italy
Copyright: © 2014 Herbein and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georges Herbein, Department of Virology, Hôpital Saint-Jacques, University of Franche-Comté, 2 Place Saint-Jacques, Besançon Cedex F-25030, France e-mail:Z2Vvcmdlcy5oZXJiZWluQHVuaXYtZmNvbXRlLmZy
 Georges Herbein
Georges Herbein Amit Kumar
Amit Kumar