
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol. , 16 December 2013
Sec. Molecular and Cellular Oncology
Volume 3 - 2013 | https://doi.org/10.3389/fonc.2013.00308
This article is part of the Research Topic Chromosomal Instability, Aneuploidy, and Cancer View all 9 articles
Faithful action of the mitotic spindle segregates duplicated chromosomes into daughter cells. Perturbations of this process result in chromosome mis-segregation, leading to chromosomal instability and cancer development. Chromosomes are not simply passengers segregated by spindle microtubules but rather play a major active role in spindle assembly. The GTP bound form of the Ran GTPase (RanGTP), produced around chromosomes, locally activates spindle assembly factors. Recent studies have uncovered that chromosomes organize mitosis beyond spindle formation. They distinctly regulate other mitotic events, such as spindle maintenance in anaphase, which is essential for chromosome segregation. Furthermore, the direct function of chromosomes is not only to produce RanGTP but, in addition, to release key mitotic regulators from chromatin. Chromatin-remodeling factors and nuclear pore complex proteins, which have established functions on chromatin in interphase, dissociate from mitotic chromatin and function in spindle assembly or maintenance. Thus, chromosomes actively organize their own segregation using chromatin-releasing mitotic regulators as well as RanGTP.
In all organisms, chromosomes have to be segregated faithfully to daughter cells to stably transmit genetic information. Perturbation of this process can result in chromosome mis-segregation, which has a clear link to aneuploidy and cancer development in humans. For a long time, chromosomes were thought to be passengers segregated by spindle microtubules. But it is now evident that chromosomes play a major role in their own segregation during mitosis. The active role of chromatin was initially shown by an experiment, in which injection of bacteriophage lambda DNA into Xenopus eggs, arrested in metaphase, turned out to be sufficient to trigger MT formation (1). The “chromatin effect” was reproduced with plasmid DNA-coated beads incubated in Xenopus egg extracts. DNA-beads were converted into functional chromatin in extracts and induced spindle assembly (2). The chromatin-beads contained neither centrosomes nor kinetochores, showing that chromatin is sufficient to drive spindle assembly.
The Ran GTPase was identified as a factor essential for chromatin-driven spindle assembly (3–7). The guanine nucleotide exchange factor for Ran (RCC1) localizes to chromatin while the GTPase activating protein for Ran (RanGAP) resides in the mitotic cytoplasm. The specific localizations of RCC1 and RanGAP result in high concentrations of the GTP bound form of Ran (RanGTP) locally around chromosomes. RanGTP binds to the heterodimeric nuclear transport receptor importin α/β and dissociates nuclear localization signal (NLS)-containing proteins from the importins (8–10). Liberated NLS proteins function in spindle assembly around chromosomes (Figure 1).
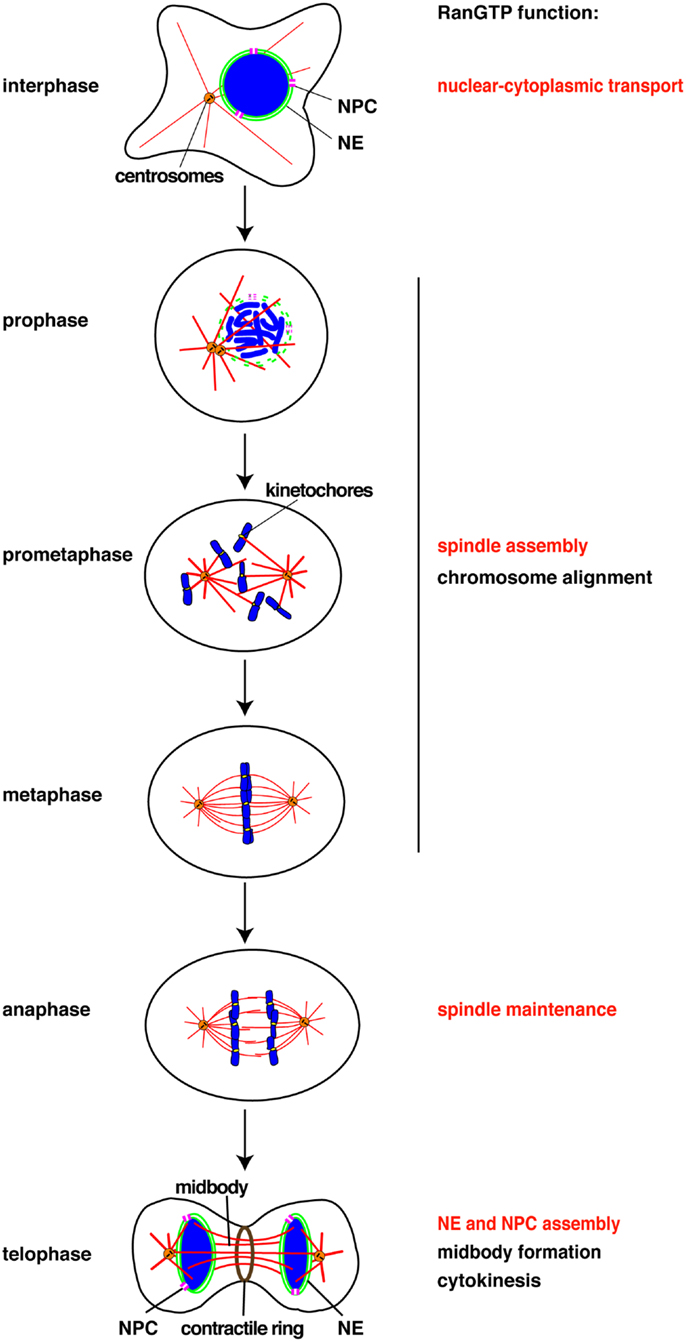
Figure 1. Chomatin and RanGTP function at each cell cycle stage. RanGTP directly stimulates the mitotic events displayed in red. Other events (black) may also be RanGTP-regulated, but have not yet been studied. NE, nuclear envelope, NPC, nuclear pore complex.
Several NLS proteins have since been identified as RanGTP-dependent spindle assembly factors (11). Among them, proteins such as NuMA do not have any reported functions in interphase, indicating that their nuclear localization separates them from microtubules in the cytoplasm. When the nuclear envelope breaks down upon mitotic entry, NuMA stimulates microtubule nucleation in a RanGTP-dependent manner (9, 10). It has been recently discovered, however, that some of the NLS proteins with established interphase functions play distinct roles in mitosis, including proteins dissociating from mitotic chromatin. Here we summarize this new class of NLS proteins as “chromatin-releasing mitotic regulators.”
Chromatin structure dramatically changes at the onset of mitosis, with the formation of highly ordered and condensed chromosomes. Some chromatin-binding proteins like condensin complex proteins specifically bind to mitotic chromatin while others, like cohesin components, dissociate (12). RCC1 binds to chromatin throughout the cell cycle, but more strongly in mitosis due to several mechanisms including its mitosis-specific phosphorylation by Cdk1 (13). Increased binding of RCC1 to chromatin is essential for producing a high RanGTP concentration around chromosomes and for spindle assembly. In parallel, it has become clear that some of the chromatin-binding proteins that dissociate during mitosis play important, mitosis-specific roles (Table 1).
The chromatin-remodeling ATPase ISWI binds chromatin during interphase, although its specific role in the nucleus remains unclear (12, 14). The majority of ISWI dissociates from mitotic chromatin and re-localizes to the spindle (15). ISWI directly binds microtubules in a RanGTP-dependent manner. The region that contains chromatin-binding domains and an NLS also mediates microtubule-binding of ISWI. ISWI is, however, not required for spindle assembly, but is essential for maintaining spindle microtubules during anaphase and in turn for chromosome segregation (15) (Figure 1). This microtubule stabilizing function of ISWI is independent of chromatin-remodeling. The release from chromatin is a prerequisite for ISWI to function as a microtubule-associated protein (MAP), but its microtubule-binding is further regulated by RanGTP.
Chromatin-remodeling ATPases share a SWI/SNF-type ATPase domain, but are classified into four different families (SWI/SNF, ISWI, INO80, and CHD) according to unique flanking domains (16). The INO80-family ATPase (Swr1 and Ino80)-containing complexes incorporate and remove the histone variant H2A.Z into the first nucleosome of genes, respectively, regulating gene expression (17–19). However, Ino80 associates with spindle microtubules during mitosis, and its depletion from human cells leads to defective spindle assembly and abnormal chromosome segregation (20). Ino80 directly binds to microtubules in vitro (21). It has not been determined if the Ino80 function in microtubule regulation is regulated by the RanGTP system. Pontin/RuvBL1, a component of the Ino80-containing chromatin-remodeling complex, was shown to localize at spindle poles and microtubules, to interact with γ-tubulin, and to be essential for spindle assembly (22).
The SWI/SNF-family ATPases, BRM, and BRG-1, are phosphorylated during mitosis and released from condensed chromosomes (23). It has been suggested that their dissociation could be the mechanism leading to transcriptional arrest during mitosis. Other research demonstrated that BAF180, a component of BRG-1-containing chromatin-remodeling complex, localizes at spindle poles and kinetochores (24), suggesting that the complex could participate in spindle regulation and cell division.
Taken together, these data indicate that there are general roles for chromatin-remodeling ATPases and complexes (16) to regulate microtubules in mitosis. Chromatin-remodeling alters nucleosome positions, evicts histones, or incorporates histone variants during DNA damage repair and DNA replication, maintaining genomic integrity (25). These efforts would be irrelevant if chromosomes were not properly segregated during mitosis. Chromatin-remodeling factors regulate spindle microtubules independent of chromatin-remodeling, but this regulation is for chromosome segregation and still serves to maintain genomic integrity.
The nuclear pore complex (NPC), composed of ~30 different proteins (nucleoporins or Nups), is critical to transport protein and RNA across the nuclear envelope in interphase (Figure 1). The envelope breaks down during mitosis in metazoan cells and the NPC disassembles into nucleoporin subcomplexes and dissociates from chromatin. It has been shown that some nucleoporins play important roles in mitosis (Table 1).
The nucleoporin Rae1/Gle2 directly binds microtubules in vitro, localizes to the mitotic spindle, and is required for spindle assembly (26). A Rae1-containing ribonucleoprotein complex nucleates or stabilizes microtubules in a RanGTP/importin β-regulated manner (26). Rae1 is known to form a nucleoporin subcomplex with Nup98 (27). Nup98 was also shown to directly bind microtubules as well as MCAK, a microtubule depolymerizing kinesin motor protein (28). Although localizing mostly in the mitotic cytoplasm, Nup98 is required for spindle assembly by antagonizing MCAK (28). It remains unclear if Rae1 and Nup98 function together or independently in spindle assembly.
The Nup107-160 complex, the largest subcomplex of the NPC, re-localizes to kinetochores and spindle microtubules during mitosis (29, 30). The components of the Nup107-160 complex are maintained in interphase and mitosis (30). Using Xenopus egg extract, it was shown that the Nup107-160 complex is required for spindle assembly independent of its interphase function at nuclear pores (30). In extracts depleted of the Nup107-160 complex, microtubules are nucleated from sperm centrosomes normally but eventually disassembled, leaving largely unattached sperm chromosomes (30). The molecular function of the Nup107-160 complex in spindle assembly remains unclear. When microtubule-kinetochore attachment is disrupted (e.g., by nocodazole treatment), the Nup107-160 complex recruits the γ-tubulin ring complex (γTuRC) to kinetochores and promotes microtubule nucleation there (31). Whether the complex recruits γTuRC to kinetochores and microtubules under physiological conditions is an open question.
RanBP2/Nup358 also re-localizes from NPCs to kinetochores and spindle microtubules and plays an essential role in microtubule-kinetochore interaction (32, 33). RanBP2 depletion in human cells by RNAi causes loss of stable kinetochore microtubules and accumulation of mitotic cells with multipolar spindles and unaligned chromosomes (33). The kinetochore localization of RanBP2 is dependent on RanGTP and the nuclear export factor Crm1 (34). However, the molecular function of RanBP2 at kinetochores remains to be determined.
The above findings demonstrate that chromatin-remodeling factors and nucleoporins play important, distinct roles during mitosis (Table 1). The mechanism by which these proteins dissociate from mitotic chromatin, however, remains largely unknown. Chromatin structures dramatically change during mitosis, including chromatin condensation, and these modifications may cause the release of certain chromatin-binding proteins (12). Another possibility is that chromatin proteins themselves may be regulated during mitosis by posttranslational modifications or/and interacting proteins. The phosphorylation of BRM and BRG-1, for instance, correlates with their dissociation from mitotic chromatin (23). Phosphorylation of Nup98 is crucial for NPC disassembly (35) and may lead to the dissociation of multiple nucleoporins from chromatin. Thus, chromatin either actively releases chromatin-binding proteins or makes use of proteins leaving chromatin for mitotic organization.
Dissociation from chromatin seems to be sufficient to activate chromatin proteins. In the mitotic cytoplasm, however, importins bind to NLS proteins and inhibit some of the proteins, while not affecting others (36). RanGTP locally activates the inhibited proteins around chromosomes. Thus, mitotic regulators that dissociate from chromatin function either locally around chromosomes or throughout the mitotic cytoplasm. Accumulating evidence indicates that this spatial regulation of individual proteins is critical for proper mitotic progression.
While metazoan cells segregate chromosomes in the mitotic cytoplasm after disassembling nuclear envelope and NPC (open mitosis), protozoa such as yeasts segregate inside the nucleus without disassembling envelope and NPC (closed mitosis) (37). In yeasts, RanGTP is indirectly required for spindle assembly by mediating nuclear import of the microtubule regulator Alp7/TACC (38). Therefore, the mitotic regulators that dissociate from chromatin and NPCs seem to have evolved in metazoa with the disassembly of the nuclear envelope and NPCs.
Chromosomes drive spindle assembly in early mitosis and later, through the function of ISWI, maintain spindle microtubules in anaphase (15) (Figure 1). ISWI depletion from Xenopus egg extracts or Drosophila cultured cells does not affect spindle assembly, but upon anaphase entry spindle microtubules depolymerize and chromosomes do not segregate (15). This finding suggests that chromosomes may regulate multiple mitotic events essential for their own segregation. Indeed, perturbation of the Ran pathway in Drosophila embryos by injecting recombinant proteins induced defects in mitotic spindle assembly, in chromosome alignment to the metaphase plate, in chromosome segregation, and in the assembly of the microtubule midbody (39). RNAi of components of the Ran pathway in C. elegans also resulted in multiple mitotic defects (40). Although these experiments did not discriminate between direct and indirect effects of the Ran pathway, they suggest possible new roles of chromosomes in their own alignment and in midbody formation (Figure 1).
Currently, all the known RanGTP targets are MAPs or their regulators (11). It has been shown in starfish oocytes that chromatin drives local actin polymerization to mediate chromosome congression (41). The MAP NabKin has been identified as a nuclear protein that binds fibrous actin in a RanGTP-dependent manner (42). NabKin localizes to the mitotic spindle as well as to cortical fibrous actin in Xenopus oocytes, and is essential for cytokinesis (42), although the molecular function of NabKin remains to be determined. Chromosomes and RanGTP thus also seem able to regulate the actin cytoskeleton in chromosome congression/alignment and cytokinesis (Figure 1).
Finally, RanGTP drives the assembly of a lamin B “matrix” around chromosomes in mitosis (43). The matrix is essential for proper spindle assembly in human cells and Xenopus egg extracts, in particular for the organization of assembled microtubules into bipolar spindles (43). The mechanism of lamin B matrix assembly is not understood but is certainly independent of microtubule assembly. It will be important to identify further RanGTP targets involved in the assembly of microtubules, actin filaments, and the lamin matrix.
Chromosomes induce the production of RanGTP in their vicinity, which is essential for mitotic progression. Here we propose the release of chromatin-bound mitotic regulators as the second key role of chromosomes. Released proteins function independently of chromatin during mitosis with or without spatial control by RanGTP. The list of the chromatin-releasing mitotic regulators seems certainly to increase in the future. Clarification of the role of each protein will significantly contribute to our understanding of how chromosomes regulate their own segregation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Iain Mattaj for critically reading the manuscript.
1. Karsenti E, Newport J, Kirschner M. Respective roles of centrosomes and chromatin in the conversion of microtubule arrays from interphase to metaphase. J Cell Biol (1984) 99:47s–54. doi: 10.1083/jcb.99.1.47s
2. Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, et al. Self organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature (1996) 382:420–5. doi:10.1038/382420a0
3. Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature (1999) 400:178–81. doi:10.1038/22133
4. Zhang C, Hughes M, Clarke PR. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci (1999) 112:2453–61.
5. Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol (1999) 9:481–4. doi:10.1016/S0960-9822(99)80213-9
6. Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science (1999) 284:1356–8. doi:10.1126/science.284.5418.1356
7. Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science (1999) 284:1359–62. doi:10.1126/science.284.5418.1359
8. Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin on TPX2 activity. Cell (2001) 104:83–93. doi:10.1016/S0092-8674(01)00193-3
9. Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell (2001) 104:95–106. doi:10.1016/S0092-8674(01)00194-5
10. Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science (2001) 291:653–6. doi:10.1126/science.1057661
11. Meunier S, Vernos I. Microtubule assembly during mitosis – from distinct origins to distinct functions? J Cell Sci (2012) 125:2805–14. doi:10.1242/jcs.092429
12. MacCallum DE, Losada A, Kobayashi R, Hirano T. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell (2002) 13:25–39. doi:10.1091/mbc.01-09-0441
13. Li HY, Zheng Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev (2004) 18:512–27. doi:10.1101/gad.1177304
14. Demeret C, Bocquet S, Lemaitre JM, Francon P, Mechali M. Expression of ISWI and its binding to chromatin during the cell cycle and early development. J Struct Biol (2002) 140:57–66. doi:10.1016/S1047-8477(02)00575-0
15. Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj IW, Karsenti E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J Cell Biol (2009) 187:813–29. doi:10.1083/jcb.200906020
16. Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem (2009) 78:273–304. doi:10.1146/annurev.biochem.77.062706.153223
17. Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science (2004) 303:343–8. doi:10.1126/science.1090701
18. Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell (2011) 144:200–13. doi:10.1016/j.cell.2010.12.021
19. Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell (2013) 154:1246–56. doi:10.1016/j.cell.2013.08.043
20. Hur SK, Park EJ, Han JE, Kim YA, Kim JD, Kang D, et al. Roles of human INO80 chromatin remodeling enzyme in DNA replication and chromosome segregation suppress genome instability. Cell Mol Life Sci (2010) 67:2283–96. doi:10.1007/s00018-010-0337-3
21. Park EJ, Hur SK, Lee HS, Lee SA, Kwon J. The human Ino80 binds to microtubule via the E-hook of tubulin: implications for the role in spindle assembly. Biochem Biophys Res Commun (2011) 416:416–20. doi:10.1016/j.bbrc.2011.11.069
22. Ducat D, Kawaguchi S, Liu H, Yates JR III, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol Biol Cell (2008) 19:3097–110. doi:10.1091/mbc.E07-11-1202
23. Muchardt C, Reyes JC, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J (1996) 15:3394–402.
24. Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A (2000) 97:13015–20. doi:10.1073/pnas.240208597
25. Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet (2013) 14:62–75. doi:10.1038/nrg3345
26. Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell (2005) 121:223–34. doi:10.1016/j.cell.2005.02.016
27. Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol (1999) 145:237–54. doi:10.1083/jcb.145.2.237
28. Cross MK, Powers MA. Nup98 regulates bipolar spindle assembly through association with microtubules and opposition of MCAK. Mol Biol Cell (2011) 22:661–72. doi:10.1091/mbc.E10-06-0478
29. Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J (2007) 26:1853–64. doi:10.1038/sj.emboj.7601642
30. Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, et al. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell (2006) 17:3806–18. doi:10.1091/mbc.E05-11-1061
31. Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol (2010) 12:164–9. doi:10.1038/ncb2016
32. Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol (2003) 162:991–1001. doi:10.1083/jcb.200304080
33. Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol (2004) 14:611–7. doi:10.1016/j.cub.2004.03.031
34. Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, et al. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol (2005) 7:626–32. doi:10.1038/ncb1263
35. Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell (2011) 144:539–50. doi:10.1016/j.cell.2011.01.012
36. Maresca TJ, Niederstrasser H, Weis K, Heald R. Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr Biol (2005) 15:1755–61. doi:10.1016/j.cub.2005.08.049
37. Boettcher B, Barral Y. The cell biology of open and closed mitosis. Nucleus (2013) 4:160–5. doi:10.4161/nucl.24676
38. Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature (2007) 447:334–7. doi:10.1038/nature05773
39. Silverman-Gavrila RV, Wilde A. Ran is required before metaphase for spindle assembly and chromosome alignment and after metaphase for chromosome segregation and spindle midbody organization. Mol Biol Cell (2006) 17:2069–80. doi:10.1091/mbc.E05-10-0991
40. Askjaer P, Galy V, Hannak E, Mattaj IW. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell (2002) 13:4355–70. doi:10.1091/mbc.E02-06-0346
41. Mori M, Monnier N, Daigle N, Bathe M, Ellenberg J, Lenart P. Intracellular transport by an anchored homogeneously contracting F-actin meshwork. Curr Biol (2011) 21:606–11. doi:10.1016/j.cub.2011.03.002
42. Samwer M, Dehne HJ, Spira F, Kollmar M, Gerlich DW, Urlaub H, et al. The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. EMBO J (2013) 32:1886–902. doi:10.1038/emboj.2013.108
43. Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science (2006) 311:1887–93. doi:10.1126/science.1122771
44. Corona DF, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, et al. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell (1999) 3:239–45. doi:10.1016/S1097-2765(00)80314-7
45. Kashiwaba S, Kitahashi K, Watanabe T, Onoda F, Ohtsu M, Murakami Y. The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem Biophys Res Commun (2010) 402:619–25. doi:10.1016/j.bbrc.2010.10.066
46. van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell (2004) 119:777–88. doi:10.1016/j.cell.2004.11.033
47. Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol (2008) 15:338–45. doi:10.1038/nsmb.1413
48. Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature (2000) 406:541–4. doi:10.1038/35020123
49. Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, et al. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskeleton (2003) 56:79–93. doi:10.1002/cm.10136
50. Grigoletto A, Lestienne P, Rosenbaum J. The multifaceted proteins Reptin and Pontin as major players in cancer. Biochim Biophys Acta (2010) 1815:147–57. doi:10.1016/j.bbcan.2010.11.002
51. Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell (2003) 14:600–10. doi:10.1091/mbc.E02-09-0582
52. Hulsmann BB, Labokha AA, Gorlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell (2012) 150:738–51. doi:10.1016/j.cell.2012.07.019
53. Xu S, Powers MA. Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis. Mol Biol Cell (2010) 21:1585–96. doi:10.1091/mbc.E09-07-0561
54. Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell (2003) 113:195–206. doi:10.1016/S0092-8674(03)00235-6
55. Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell (2003) 11:853–64. doi:10.1016/S1097-2765(03)00116-3
56. Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem (1995) 270:14209–13. doi:10.1074/jbc.270.23.14209
57. Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, et al. A giant nucleopore protein that binds Ran/TC4. Nature (1995) 376:184–8. doi:10.1038/376184a0
58. Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Walde S, et al. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol (2011) 194:597–612. doi:10.1083/jcb.201102018
59. Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol (2002) 156:595–602. doi:10.1083/jcb.200110109
Keywords: RanGTP, chromatin remodeler, spindle, nuclear pore complex proteins, mitosis, microtubules, microtubule-associated proteins
Citation: Yokoyama H and Gruss OJ (2013) New mitotic regulators released from chromatin. Front. Oncol. 3:308. doi: 10.3389/fonc.2013.00308
Received: 17 September 2013; Accepted: 04 December 2013;
Published online: 16 December 2013.
Edited by:
Samuel F. Bakhoum, Geisel School of Medicine at Dartmouth, USAReviewed by:
Enrico Vittorio Avvedimento, University Naples Federico II, ItalyCopyright: © 2013 Yokoyama and Gruss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hideki Yokoyama, Zentrum für Molekulare Biologie der Universität Heidelberg, DKFZ-ZMBH Alliance, Im Neuenheimer Feld 282, 69120 Heidelberg, Germany e-mail:aC55b2tveWFtYUB6bWJoLnVuaS1oZWlkZWxiZXJnLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.