
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Synaptic Neurosci., 30 June 2022
Volume 14 - 2022 | https://doi.org/10.3389/fnsyn.2022.910820
This article is part of the Research TopicBrain Serotonergic SystemView all 5 articles
 Gary C. Mouradian Jr.1,2*
Gary C. Mouradian Jr.1,2* Pengyuan Liu1
Pengyuan Liu1 Pablo Nakagawa1,3
Pablo Nakagawa1,3 Erin Duffy1
Erin Duffy1 Javier Gomez Vargas1,3
Javier Gomez Vargas1,3 Kirthikaa Balapattabi1,3
Kirthikaa Balapattabi1,3 Justin L. Grobe1,2,3,4,5
Justin L. Grobe1,2,3,4,5 Curt D. Sigmund1,2,3
Curt D. Sigmund1,2,3 Matthew R. Hodges1,2
Matthew R. Hodges1,2Acute regulation of CO2 and pH homeostasis requires sensory feedback from peripheral (carotid body) and central (central) CO2/pH sensitive cells – so called respiratory chemoreceptors. Subsets of brainstem serotonin (5-HT) neurons in the medullary raphe are CO2 sensitive or insensitive based on differences in embryonic origin, suggesting these functionally distinct subpopulations may have unique transcriptional profiles. Here, we used Patch-to-Seq to determine if the CO2 responses in brainstem 5-HT neurons could be correlated to unique transcriptional profiles and/or unique molecular markers and pathways. First, firing rate changes with hypercapnic acidosis were measured in fluorescently labeled 5-HT neurons in acute brainstem slices from transgenic, Dahl SS (SSMcwi) rats expressing T2/ePet-eGFP transgene in Pet-1 expressing (serotonin) neurons (SSePet1–eGFP rats). Subsequently, the transcriptomic and pathway profiles of CO2 sensitive and insensitive 5-HT neurons were determined and compared by single cell RNA (scRNAseq) and bioinformatic analyses. Low baseline firing rates were a distinguishing feature of CO2 sensitive 5-HT neurons. scRNAseq of these recorded neurons revealed 166 differentially expressed genes among CO2 sensitive and insensitive 5-HT neurons. Pathway analyses yielded novel predicted upstream regulators, including the transcription factor Egr2 and Leptin. Additional bioinformatic analyses identified 6 candidate gene markers of CO2 sensitive 5-HT neurons, and 2 selected candidate genes (CD46 and Iba57) were both expressed in 5-HT neurons determined via in situ mRNA hybridization. Together, these data provide novel insights into the transcriptional control of cellular chemoreception and provide unbiased candidate gene markers of CO2 sensitive 5-HT neurons. Methodologically, these data highlight the utility of the patch-to-seq technique in enabling the linkage of gene expression to specific functions, like CO2 chemoreception, in a single cell to identify potential mechanisms underlying functional differences in otherwise similar cell types.
The neural network coordinating respiratory muscle activity dynamically maintains blood gas homeostasis via modulation from hypoxic (low O2) and hypercapnic (high CO2) ventilatory chemoreflexes. Central respiratory chemoreceptor neurons in the hindbrain may encode local CO2/pH levels and adjust ventilatory drive accordingly. These are central respiratory chemoreceptor cells (neurons and/or glia) and include distinct subpopulations of phox2b+ glutamatergic neurons in the retrotrapezoid nucleus (RTN) and Pet-1+ serotonergic (5-HT) neurons. While still a topic of debate (Corcoran et al., 2009), CO2/pH responses in these hindbrain neurons are considered unique and intrinsic properties, where acidification increases firing rates independent of synaptic inputs to encode CO2/pH status and modulate breathing. Data continue to emerge regarding the mechanisms underlying cellular sensing of CO2/pH and associated changes to firing rate in RTN neurons, which appear to require the expression of a pH-sensitive potassium ion channel (TASK-2), G protein-coupled receptor 4 (Gpr4) (Kumar et al., 2015) and 5-HT receptor activation (Wu et al., 2019). Local RTN astrocytes may instead rely on pH-sensitive inwardly rectifying potassium ion channels (Kir4.1 and/or Kir5.1) to sense changes in local milieu CO2/pH levels (Gourine et al., 2010; Wenker et al., 2010). Less progress has been made in identifying molecular mechanisms of CO2 sensitivity in 5-HT neurons, but it appears unlikely that they rely on the same mechanisms as RTN neurons (Mulkey et al., 2007; Teran et al., 2014).
Whereas midbrain 5-HT neurons critically modulate higher brain functions such as sleep-wake cycles (Buchanan et al., 2008; Cui et al., 2016), hindbrain 5-HT neurons are critical for vital physiological functions including respiratory control (Feldman et al., 2003; Hodges and Richerson, 2008; Ptak et al., 2009). In addition to their pH/CO2 chemosensitive properties, these anatomically caudal groups of 5-HT neurons modulate ventilatory output via tonic release of excitatory 5-HT and co-released neuropeptides, substance P and thyrotropin-releasing hormone (Hodges and Richerson, 2008; Corcoran et al., 2009). Dysfunction of the brainstem 5-HT system is linked to the unexpected respiratory failure in Sudden Infant Death Syndrome (SIDS) and it may be linked to Sudden Unexpected Death in Epilepsy (SUDEP) (Buchanan, 2019). Thus, characterization of the functional contributions of distinct subsets of hindbrain 5-HT neurons (modulatory vs. CO2 sensitive) may lead to a better understanding of the pathogenesis of these devastating outcomes.
Subpopulations of brainstem 5-HT neurons increase their firing rates in response to hypercapnic acidosis, and experimentally induced dysfunction of 5-HT neurons reduces hypercapnic ventilatory responses in vivo (Hodges et al., 2007, 2008; Wu et al., 2008; Corcoran et al., 2009; Ray et al., 2011; Kaplan et al., 2016; Cerpa et al., 2017). Intersectional fate mapping in mice demonstrates that embryonic origins of pH-sensitive 5-HT neurons are rhombomere (R)5 (Pet1- and Egr2-expressing cell lineage)-specific (Brust et al., 2014), whereas other subpopulations of brainstem 5-HT neurons (R6 or Tac1/Pet1-expressing) are not CO2 sensitive but likely modulate respiratory motor outputs. All but rhombomere 4 5-HT neurons express Pet1 during development (Hendricks et al., 1999). These data indicate that subpopulations of hindbrain 5-HT neurons programmed during embryonic development for distinct post-natal functional contributions to ventilatory control. They also suggest CO2 sensitive 5-HT neurons express distinct sets of genes which may be determinants and/or biomarkers of their unique CO2 sensing properties. However, it remains unknown if postnatal phenotypically defined 5-HT neurons have similar transcriptional differences as 5-HT neurons defined by embryonic origin.
Here, the patch-to-seq technique was employed (Figure 1; Cadwell et al., 2016; Chen et al., 2016; Fuzik et al., 2016; van den Hurk et al., 2018) to determine if postnatal cellular CO2 responses in brainstem 5-HT neurons could be attributed to unique transcriptional profiles. In acute brainstem slice preparations from transgenic rats expressing GFP in all serotoninergic neurons, 5-HT neuronal firing rate was measured during control and hypercapnic acidosis using cell-attached patch clamp electrophysiology. The rat pups were derived from the Dahl SS (SSMcwi) strain which have a robust CO2 response by 18 days of age (Davis et al., 2006) and expressed the T2/ePet-eGFP transgene to direct enhanced GFP expression in Pet-1-expressing (serotonin) neurons (SSePet1: eGFP; RGD strain ID: 8661234). Subsequently, intracellular contents from the recorded neurons were collected for single cell RNA sequencing (scRNA-Seq; not to be confused with 10x genomics) and subsequent transcriptomic analyses. Combining these techniques provides a powerful approach for correlating electrophysiological responses to CO2/pH changes with transcriptional data on a cell-to-cell basis.
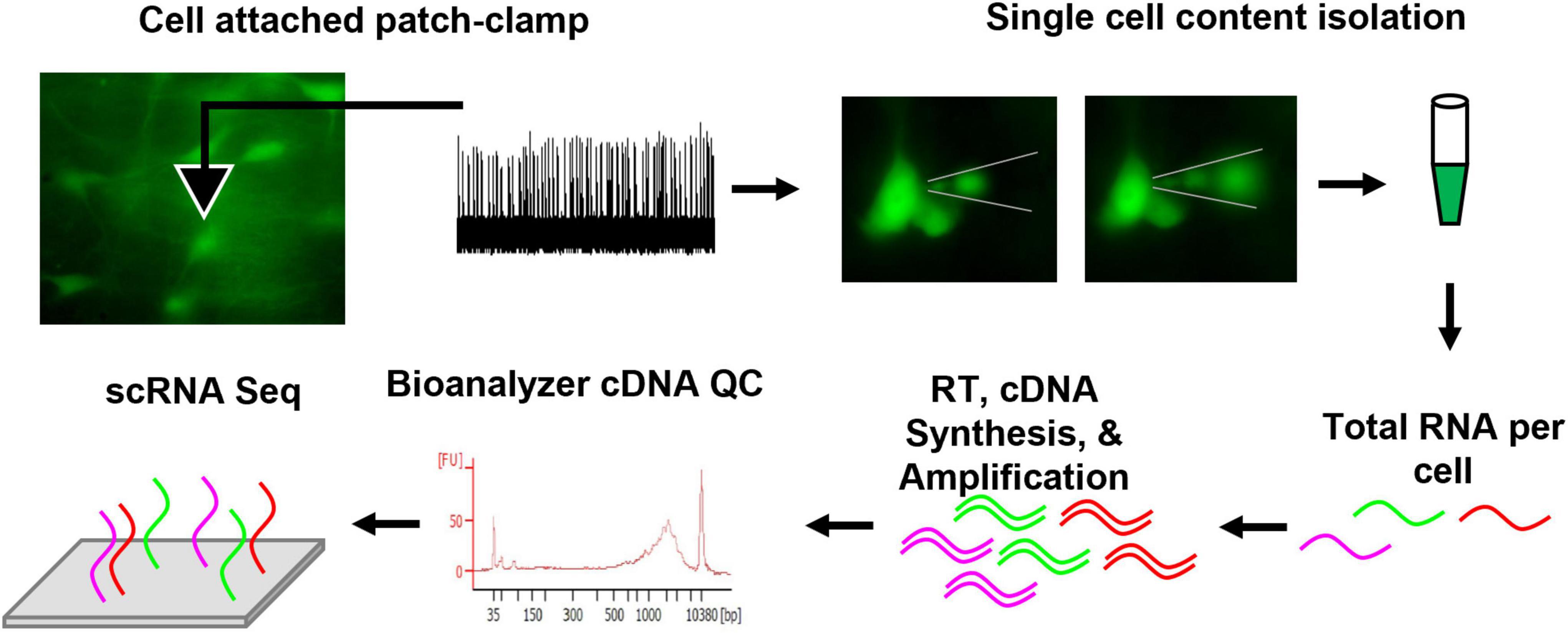
Figure 1. Methodological schematic outlining the patch-seq technique. In summary, eGFP 5-HT neurons were fluorescently identified from an acute brainstem slice, loose cell attached patch-clamp yielded firing rate measurements during control, CO2 challenge, and washout periods. Thereafter, the recorded neuron’s cell contents were isolated under fluorescence to visualize removal of intracellular content. The content was collected in sterile PBS and frozen. Total RNA per cell was reverse transcribed (RT), cDNA was synthesized, and amplified. Quality control (QC) was completed using an Agilent Bioanalyzer. Samples passing QC were submitted for sequencing. Further methodological details can be found in the section “Materials and Methods”.
SSePet1: eGFP rats (n = 72) that have restricted expression of eGFP to all CNS 5-HT neurons as previously described (Katter et al., 2013) were used between 18 and 23 days of age. Male and female rats were housed in the Biomedical Resource Center when not undergoing study with a standard 12:12 h light/dark cycle. Rats had ad libitum access to food (Dyets Inc., Bethlehem, PA, United States, 0.4% salt: #113755) and hyperchlorinated water. All experiments were approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee prior to experimental use.
Rat pups (P18-23) were anesthetized with 2.5% isoflurane and transcardially perfused with 5 mL of NMDG solution (in mM: 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 0.5 CaCl2-2H20, 7 MgSO4-7H20, 92 NMDG, 20 HEPES, 25 glucose, 2 thiourea, 5 Na-ascorbate, and 3 Na-pyruvate at 7.5 adjusted pH). The brain was quickly isolated and placed into NMDG solution. The first quarter of the frontal brain was grossly cut and then the brain was embedded in 4 mL of 0.04 g Agarose (Sigma-Aldrich, Burlington, MA, United States; A0576) dissolved in 0.9% NaCl solution. Coronal sections 200 μm thick were made caudal to rostral using a vibratome (Leica; VT 1200S) in room temperature bubbled NMDG solution. Brainstem slices were then incubated for 12 min in heated (32°C) NMDG solution and then for 10 min in heated (32°C) normal aCSF solution (in mM: 3.0 KCl, 2.0 CaCl2-2H2O, 1.0 MgCl2-6H20, 1.25 NaH2PO4, 123 NaCl, 25 NaHCO3, 10 glucose) before incubating at room temperature normal aCSF solution for 1 h prior to commencing electrophysiology recordings. All solutions were bubbled with 95% O2 and 5% CO2).
All working surfaces including the microscope, objectives, micromanipulators, pipette puller, beakers, and computer workstation were cleaned with DNA-OFF (Takara, San Jose, CA, United States; #9036) and RNase Zap (Life Technologies, San Jose, CA, United States; #AM9780). Then, brainstem slices inclusive of raphe magnus (approximately −12 to −10 mm from bregma) were placed in the recording chamber on a fixed-stage fluorescent microscope (Nikon; Eclipse FN1) equipped with DIC optics (Photometrics; CoolSNAP ES2). 5-HT neurons targeted for recording and single cell content isolation were identified by positive eGFP fluorescence.
Neuron recordings (n = 91) were performed using loose-cell attached patch-clamp recordings at a bath temperature of 37°C using pCLAMP recording software connected to a MultiClamp amplifier and a Digidata 1440A analog-to-digital converter (each from Molecular Devices). Patch clamp electrodes (2.5–6 MΩ) were backfilled with normal aCSF. Normal aCSF spiked with synaptic blockers was used in the perfused bath for recordings [in μM: 10 CNQX (Abcam; ab120017), 50 D-AP-5 (Abcam; ab120003), and 10 Gabazine (Abcam; ab120042)]. Firing rate was measured during control conditions (95% O2/5% CO2; pH: 7.363) for 5 min then during a hypercapnic challenge (85% O2/15% CO2; pH 7.092) for 10 min, followed by at least 10 min upon returning to control conditions. Bath pH was sampled by syringe and measured on a blood gas analyzer (Radiometer; ABL800 FLEX). The chemosensitivity index (C.I.) was calculated as the average for each of the two condition changes per experiment (from baseline to CO2 challenge and from CO2 challenge to recovery) using the following formula adapted from a prior report (Wang et al., 1998).
Where FR15 is the average firing rate at 15% CO2 challenge, FR5 is the average firing rate at 5% CO2 conditions (control and recovery), pH5 is the average pH at 5% CO2, and pH15 is the average pH at 15% CO2. The logarithms are in base 10. The 0.2 constant represents the 0.2 pH unit changes used to initially develop this formula as detailed previously (Wang et al., 1998). The C.I. was used to determine which 5-HT neurons were chemosensitive. Following cell recordings, a second micropipette was backfilled with <1 μl of modified normal aCSF [in mM unless stated otherwise: 123 K-gluconate, 12 KCl, 10 HEPES, 0.20 EGTA, 4 MgATP, 0.30 NaGTP, 10 Na-phosphocreatine, and 1U/μl RNAse inhibitor (40U/μl; Takara, San Jose, CA, United States; #2313A)] based on published Patch-Seq methods (Cadwell et al., 2016). Note, we omitted glycogen addition as it interfered with cDNA synthesis. The backfilled micropipette was advanced to the recorded neuron using slight positive pressure. Positive pressure was released upon nearing the recorded neuron (verified by location of recording pipette). Then, negative pressure was applied to isolate intracellular content monitored under live fluorescence imaging to visualize movement of eGFP+ content into the isolation micropipette (and shrinking of neuron). Fluorescent images before and after the isolation were obtained for documentation (Figure 1). The isolation pipette was quickly removed from the tissue and the tip broken inside a sterile RNAse/DNAse free 0.5 mL microcentrifuge tube and intracellular content was gently aspirated into 4 μl of sterile PBS (No Ca2+ no Mg2+). The sample was then flash frozen on dry ice and stored at −80°C until cDNA creation. Less than 3 min elapsed from isolation to flash freezing to minimize RNA degradation.
We converted total RNA to complementary DNA (cDNA) using the SMART-Seq v4 Ultra Low Input RNA Kit and associated protocol (Takara Bio USA, Inc., San Jose, CA, United States, #634890). Briefly, 4 μl of isolated sample was thawed on ice, spun down, and transferred into 0.2 mL RNAse/DNAse free PCR tubes. To begin first strand cDNA synthesis, volume was brought to 9.5 μl with nuclease-free water. 1 μl of 10x reaction buffer was added to each tube before adding 1 μl of 3′ SMART-Seq CDS Primer IIA. Samples were mixed well and spun down prior to incubating the tubes at 72°C in a preheated, hot-lid thermal cycler for 3 min and then immediately placed on ice for 2 min. Then, 7.5 μl of master mix, containing 5X Ultra Low First-Strand Buffer, SMART-Seq v4 Oligonucleotide, RNase Inhibitor, and SMARTScribe Reverse Transcriptase, was added to each sample for completion of first strand cDNA synthesis using the outlined thermal cycler program indicated in the protocol. Next, cDNA was amplified using 17 PCR cycles after the addition of PCR Master Mix (30 μl) containing 2X SeqAmp PCR Buffer, PCR Primer II A, and SeqAmp DNA Polymerase, was added to each sample. The number of PCR cycles was determined based on protocol recommendation and from earlier pilot experiments. The amplified cDNA was then purified using immobilization based clean up with Agencourt AMPure XP beads (Beckman Coulter Inc., Brea, CA, United States, #A63880) and fresh 80% ethanol. cDNA was then eluted (17 μl) with the Elution Buffer and transferred into new PCR tubes. An Agilent 2100 Bioanalyzer and an Agilent High Sensitivity DNA Kit (Agilent, Santa Clara, CA, United States; #5067-4626) was used to validate success of cDNA synthesis and amplification. All sequenced samples yielded a peak at ∼2,500 bp that spanned over 400 and 10,000 bp with over 3.5 ng of cDNA. Negative control samples yielded no peaks. cDNA library preparation was completed on validated cDNA samples using Illumina Library preparation protocols outlined in the NextEra XT DNA Library Preparation Kit (Illumina, Madison, WI, United States; #FC-131-1024). As suggested by the Takara SMARTER-Seq v4 protocol, 100–150 pg of amplified cDNA was used as the input volume for the NextEra XT DNA Library Preparation Kit. Libraries were fragmented and tagmented with adapters for sequencing according to the NextEra XT Index Kit (Illumina, Madison, WI, United States; #FC-131-2001). cDNA library sizes (250–1,500 base pairs) and concentration (>2 nM) were verified prior to sequencing using the Agilent High Sensitivity DNA Kit.
Paired end sequencing was completed on an Illumina HiSeq platform in-house. We performed sequence data analyses using established in-house pipelines for read mapping, alignment, transcript quantification, and statistical differential expression (Mouradian et al., 2016) and DESeq2 (Love et al., 2014) for batch-effect normalization of data obtained from two separate flow-cells. Briefly, the Tuxedo Suite was used for RNA Sequencing analysis. The Bowtie and TopHat v2 software components were used for sequence alignment, and Cufflinks for assembling reads into transcripts and differential gene expression using the Cuffdiff algorithm.
Hierarchical clustering was performed using the heatmap.2 function of the gplots R package as an alternative visualization method to multidimensional scaling by calculating Euclidean distances from logFPKM of the top 100 most variable genes. The heatmap.2 function produced a false colored image based on the top 100 most variable genes and attached a dendrogram based on the default parameters of the heatmap.2 function1.
Multidimensional scaling was performed and plotted using the R package limma using the plotMDS function to produce a principal coordinate plot which was used to visualize the distances between each sample and their gene expression profiles represented by the columns of x (Ritchie et al., 2015; Law et al., 2016). Samples are plotted on a two-dimensional scatterplot so that the distances on the plot approximate the typical log2 fold change between samples. The first dimension represents the leading-fold change that best separates samples and explains the greatest proportion of variation in the dataset. The subsequent plotted dimensions have a smaller effect and are orthogonal to the ones plotted prior.
Traditional support vector machine (SVM) learning with R packages such as “e1071” can be too noisy because there are too many genes, and the method is computationally intensive. Our study utilized the R package pathClass to narrow down the number of genes relevant to the outcome, which makes SVM cleaner and less computationally intensive. This method is known as SVM recursive feature elimination (SVM-RFE), which builds a model based on the current number of genes and removes a gene of interest to then check the accuracy of the model. Additionally, “e1071” was used to implement SVM to build a model to see what genes are important in prediction chemosensitivity. First, SVM-RFE was implemented using pathClass. PathClass reduced the number of genes used as predictors for the outcome (CO2 sensitive vs. insensitive) to reduce intensive computation and eliminate overfitting of data, a consequence too many predictors. After predictors were identified by pathClass, “e1071” was used to implement SVM to build a model using the predictor genes to visualize what genes are important in predicting chemosensitivity and to what accuracy. A radial basis function was used for the kernel (a kernel is simply a shape in high-dimensional space used to segregate the data [e.g., points (cells)] that fall inside or outside a circular shape in the case of a radial basis function). Four tests were performed to predict accuracy and for each test one predictor gene was used and seven random samples were left out of the analysis to function as a test group while the remaining samples (Kaplan et al., 2016) were processed as training data. The accuracy of the proposed model was tested to ensure that cells were classified accurately. The model used was: svm.model <- svm(Labels ∼ data = trainset, cost = 100, gamma = 1). Next, the model was looped 10,000 times to see how accurate the radial kernel SVM (predicting only based on one gene at a time due to large list of predictors) is in the long run. After the 10,000 trials, the radial kernel SVM was used to predict the percentage of accuracy. The next tests performed evaluated how altering the value of C (cost) affected the accuracy of the classifier. C was changed to 1,000 (high cost) and 10 (low cost). Again, after 10,000 trails looking at low and high cost, the radial kernel SVM was used to predict the percentage of accuracy. Finally, a polynomial kernel was used with 100,000 trials using the same model. SVM-RFE iteratively eliminated features that are less significant in determining the classification model (i.e., the gene or list of genes most able to predict if a 5-HT neuron is CO2 sensitive or not (Huang et al., 2014).
Brains were collected after transcardial perfusion with Dulbecco’s phosphate-buffered saline (DPBS, Gibco) containing 0.1% heparin followed by 4% paraformaldehyde (PFA, 4% in PBS, Biotium). The brains were dissected and post fixed in 4% PFA for 24 h and dehydrated with serial, 24-h sucrose solutions (10, 20, then 30% sucrose). The tissue was then place into a cryomold with Tissue-Tek optimal cutting temperature compound (O.C.T., Sakura) and snap frozen in dry ice and 4-methylbutane. Tissue was cryosectioned at 14 μM and stored at −80°C until use. Endogenous mRNA was detected using the in situ hybridization technique RNAscope® 2.5 High Definition – Fluorescent Multiplex Assay (Wang et al., 2012). Briefly, tissues were placed in 10% neutral buffered formalin (10% NBF, Sigma-Aldrich, Burlington, MA, United States) for 15 min at 4°C, dehydrated in a serial ethanol dilution up to 100% ethanol before blocking endogenous peroxidase activity with H2O2 and treatment with protease plus (ACD Bio). Slides were then incubated for 2 h at 40°C with probes identifying tryptophan hydroxylase 2 (Tph2; Rn-Tph2 – 316411, ACD Bio), CD46 (CD46; Rn-CD46-C2 – 847661-C2, ACD Bio), Iron-Sulfur Cluster Assembly Factor IBA57 (Iba57; Rn-Iba57-C2 – 847651-C2, ACD Bio) or positive control probe peptidylprolyl isomerase B (Ppib; Mm-Ppib – 313911, ACD Bio), RNA polymerase II subunit A (Mm-Polr2a-C2 – 312471-C2, ACD Bio), and negative control probe 4-hydroxy-tetrahydrodipicolinate reductase (dapB; 310043, ACD Bio). RNAscope® fluorescent multiplex assay protocol was followed for all other amplifications and development steps. Slides were stained with DAPI and cover slipped with ProLong Gold (P10144, Thermo Fisher Scientific, Coraopolis, PA, United States).
Max projection images were acquired by a blinded investigator from 5 fluorescent z-sections (each 2 μm apart) from tissue at 4 and 20× magnification via a Nikon A1 confocal microscope using NIS elements software. Imaging settings (laser power, gain, offset, and photomultiplier tube) were consistent throughout the experiment. Images were loaded in ImageJ (FIJI) for analysis (Schindelin et al., 2012). Each Tph2+ cell was manually outlined. Tph2 images were autothresholded while all CD46 and Iba57 images were thresholded at 9 and 10 on a 255 white-black scale, respectively. Then, the Tph2+ cell outlines were overlaid onto the thresholded CD46 or Iba57 images to count the number of Tph2+ cells expressing CD46 or Iba57 and the total number of Tph2+ cells. A similar quantification was repeated within the raphe magnus outlined guided by the rat brain atlas (Paxinos and Watson, 1998). Samples were then unmasked and identified. Data are reported as CD46 or Iba57 as a percentage of Tph2 + cells, either throughout the section or within the raphe magnus.
Statistical analyses of data were performed using Prism (GraphPad Prism 9.0, GraphPad Software, LLC, San Diego, CO, United States). Data are reported as mean ± SEM unless otherwise specified. Differences between groups in electrophysiological parameters were detected using an unpaired Student’s t-test or across conditions with one- or two-way Repeated Measures ANOVA with Sidak’s post hoc tests using log normalized values to achieve parametric assumptions. Un-normalized electrophysiology data are reported in the section “Results.” Simple linear regressions were used to determine correlations between gene expression and a given electrophysiologic metric. Samples were determined outliers if its value was greater than two standard deviations from the mean. Alpha (p-value) < 0.05 is statistically significant.
All eGFP-expressing 5-HT neurons showed spontaneous action potentials under control conditions (5% CO2, pH = 7.363), which ensured cell viability and membrane integrity in the cell-attached configuration (Figure 2A). Switching superfusate to CO2-enriched aCSF (15% CO2, pH = 7.092) reversibly increased firing rates (FRs) in some eGFP + 5-HT neurons. Using a calculated chemosensitivity index (C.I.; see section “Materials and Methods”), each of the 91 5-HT neurons recorded were classified as CO2 sensitive (C.I. >120; n = 48) or CO2 insensitive (C.I. <120; n = 43; Figure 2B). The average C.I. for CO2 sensitive 5-HT neurons was 169.8 ± 8.3 (vs. 100.1 ± 1.4 for insensitive; P < 0.0001; Figure 2C). FRs in individual CO2 sensitive 5-HT neurons significantly increased during the challenge and then decreased upon washout (recovery; Figure 2D) but remained slightly higher than control (P < 0.0001 for baseline FR vs. CO2 challenge; P = 0.0004 for baseline vs. recovery). In contrast, FRs in CO2 insensitive 5-HT neurons changed little during the CO2 challenge and in recovery (Figure 2E; P > 0.05 for baseline FR vs. CO2 challenge; P = 0.0092 for CO2 challenge vs. recovery). CO2 sensitive and insensitive 5-HT neurons differed in their responses to hypercapnic acidosis and had distinct basal FRs which were lower in CO2 sensitive 5-HT neurons vs. insensitive 5-HT neurons at baseline (P = 0.0012) and recovery conditions (Figure 2F; P = 0.0010). There were no FR differences between CO2 sensitive and CO2 insensitive 5-HT neurons during the CO2 challenge (P = 0.3475). See Supplementary Figures 1A–F for raw firing rate values per condition per neuron. Such distinct basal FRs are highly predictive of 5-HT neuron chemosensitivity (Supplementary Figures 1G–J). Among the 91 neurons recorded, 21 neurons (n = 11 CO2 sensitive; n = 10 CO2 insensitive) yielded sufficient RNA of high quality for manual, not 10x Genomics, scRNAseq. This subset of 5-HT neurons from each group reflected electrophysiologic properties consistent with data from the total populations recorded (Figures 2G–J) where CO2 sensitive neurons had a higher mean C.I. than insensitive neurons (169.1 ± 14.2 vs. 94.8 ± 2.4, respectively; P < 0.0001; Figure 2G), consistently showed reversible increases in FR (Figure 2H) or no change (Figure 2I), had lower FRs under basal and recovery conditions (Figure 2J) where basal FRs were predictive of 5-HT neuron chemosensitivity (Supplementary Figures 1D,E).
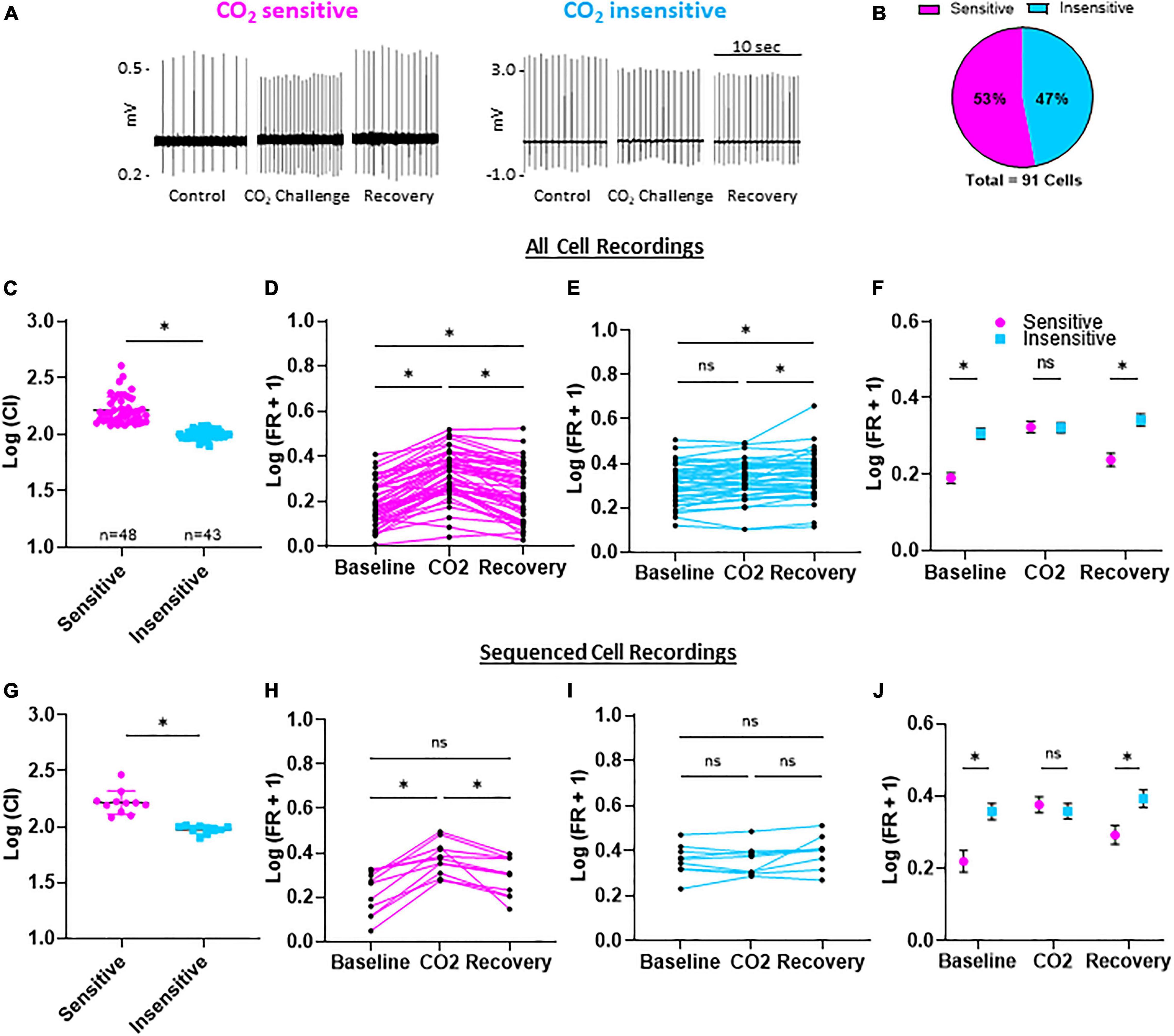
Figure 2. Firing rates and chemosensitivity index (C.I.) of CO2 sensitive and insensitive 5-HT neurons. Representative, condensed recording traces during control (95% O2/5% CO2; 7.363 pH), CO2 challenge (85% O2/15% CO2; 7.092 pH), and recovery (95% O2/5% CO2; 7.363 pH) (A). Distribution of CO2 sensitive or insensitive 5-HT neurons among 91 neurons (B). Log transformed chemosensitivity index (C.I.) of CO2 sensitive and insensitive 5-HT neurons (C). Log transformed firing rates during baseline, CO2 challenge, and recovery compared among all CO2 sensitive (D), CO2 insensitive (E), and between CO2 sensitive and insensitive (F) 5-HT neurons. The sub-population of neurons used for scRNA-Seq had representative log transformed C.I. values, and firing rate differences during baseline, CO2 challenges, and recovery (G–J). Notably, baseline and recovery firing rates were lower in CO2 sensitive vs. insensitive 5-HT neurons while firing rate during the CO2 challenge was similar between CO2 sensitive and insensitive neurons. Unpaired, two-tailed student’s t-test (C,G). One-way repeated measures ANOVA with Sidak’s multiple comparisons test (D,E,H,I). ANOVA results: Condition p < 0.0001 and Individual Cell p < 0.001 (D); Condition and Individual Cell p < 0.0001 (E); Condition p = 0.0002 and Individual Cell p = 0.0025 (H); Condition p = 0.0539 and Individual Cell p < 0.0001 (I). Two-way repeated measures ANOVA with Sidak’s multiple comparisons test (F,J). Condition p < 0.0001, Cell Phenotype p = 0.0005, and Interaction p < 0.0001 (F); Condition p = 0.0002, Cell Phenotype p = 0.0279, and Interaction p < 0.0001 (J). *p < 0.05 and ns, not significant.
Sequenced cDNA libraries from the recorded 5-HT neurons (n = 21) yielded a high-quality read summary (Table 1). Among the CO2 sensitive neurons, on average there were 5.690 million bases, over 46 million reads, over 82% of all bases had a Phred-like quality score of 30 or greater (higher indicates less probability of incorrectly identified base, e.g., Q30, 1 in 1,000 incorrect), an average quality score of 32.7, over 36 million of reads were mapped (∼75% of all reads), and 8,583 genes detected (Table 1A). Among the CO2 insensitive neurons, on average there were 6.090 million bases, over 50 million reads, over 82% of all bases had a Phred-like quality score of 30 or greater, an average quality score of 32.6, over 36 million of reads were mapped (∼75% of all reads), and 7,230 genes detected (Table 1B). We found higher normalized expression of canonical serotonergic genes compared to pan-neuronal and glial genes (15.26 ± 0.25, 10.34 ± 0.35, and 1.60 ± 0.25, respectively; P < 0.0001 for each comparison; Figure 3A inset). Therefore, the sequencing data is of high quality and contain high expression of 5-HT and neuronal markers with little evidence of contamination by local glia in 5-HT neurons.

Figure 3. 5-HT neuron RNA sequencing data are highly representative of 5-HT neurons. Expression [Log2(FPKM + 1)] of 5-HT, neuronal, and glial cell markers are indicated for each of the 21 neuron cDNA libraries that were sequenced (A) and 5-HT neuronal purity is statistically significant [(A), inset]. Number and characteristics of differentially expressed genes (DEGs) among sequenced cells (B). Subcellular location and molecular types represented across the 166 DEGs (C). One-way ANOVA (P < 0.05) with Sidak’s post hoc tests (A). *P < 0.0001.
There were 166 differentially expressed genes (DEGs) identified based on a p-value corrected for multiple comparisons (q-value) < 0.05 (Figure 3B), 127 of which had increased, and 39 had decreased expression levels in CO2 sensitive 5-HT neurons relative to insensitive 5-HT neurons. 91 genes were uniquely expressed in CO2 sensitive 5-HT neurons and 24 genes uniquely expressed in CO2 insensitive 5-HT neurons. 51 genes were expressed in at least 1 neuron from both phenotypes. Ingenuity Pathway Analysis identified “cytoplasm” as the most represented cellular location for the gene products of identified DEGs across the groups, where ∼20% of all DEGs were categorized to encode an “enzyme” and ∼40% of DEGS were classified in the “other” category (Figure 3C).
We next gauged the capacity for multi-dimensional scaling (MDS) and unbiased clustering analyses to segregate CO2 sensitive from insensitive 5-HT neurons based on transcriptomic data. Using the 166 DEGs led to visually distinct hierarchical clustering (Figure 4A) and MDS clusters (Figure 4B). However, this was not the case when we included all of the genes (DEGs and non-DEGs) detected from each 5-HT neuron. Omission of two cells (one with the highest dim 2 value and one with the lowest dim 1 value from Figure 4B) did not improve clustering patterns using either analysis (not shown). Rather, hierarchical clustering and MDS using all genes revealed two distinct groups of 5-HT neurons unrelated to previous chemosensitivity grouping criteria. Repeating the DEG analysis between these new groupings, Group 1 and 2, yielded over 3,000 DEGs (Figures 4C,D). Thus, a priori identification of the CO2 sensitivity phenotype using the patch-to-seq technique provided the necessary input to differentiate the 5-HT neurons and identify DEGs between CO2 sensitive and insensitive 5-HT neurons. These results allowed subsequent bioinformatic analyses to gauge predictive capacity of these DEGs in determining the CO2 sensitivity status of a 5-HT neuron.
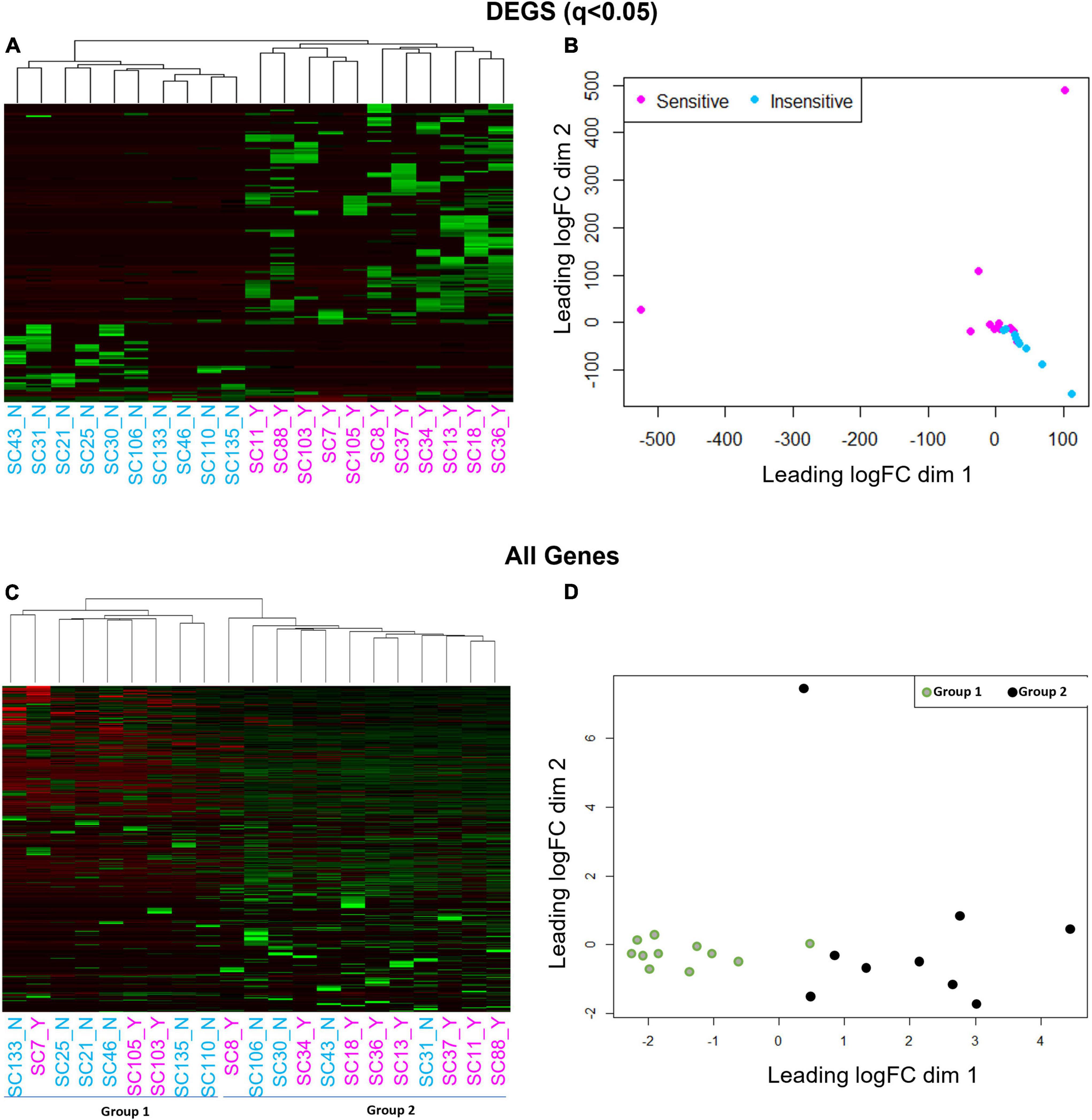
Figure 4. Hierarchical clustering (A,C) and multi-dimensional scaling [MDS; (B,D)] using all genes reveals a cell grouping unrelated to chemosensitivity versus grouping based on chemosensitivity with 166 DEGs. Hierarchical clustering and MDS using 166 DEGs result in distinct CO2 sensitive vs. insensitive 5-HT neuron grouping by hierarchical clustering and MDS (A,B). Hierarchical clustering and MDS using all genes measured by scRNA-Seq results in two distinct cell groups un-related to CO2 chemosensitivity by hierarchical clustering and MDS (C,D).
Ingenuity Pathway Analyses can be a powerful approach for determining how multiple genes are linked within known biological processes and signaling pathways. To enhance the number of identified biological functions, canonical pathways, and predicted upstream regulators unique to CO2 sensitive neurons, the analyses from the previously identified 166 DEGs were expanded to include additional DEGs with more relaxed significance criteria (p < 0.01; Supplementary Figure 2). This generated significantly [−log (p-value) > 1.3] downregulated and upregulated canonical pathways (always CO2 sensitive relative to insensitive) as indicated by the predicted z-scores (Supplementary Figure 2B). Many of the identified pathways were related to immune system function (ICOS Signaling in T Helper Cells, NF-κB Activation by Viruses, Th1 Pathway, Induction of Apoptosis by HIV1, and CD40 signaling) with unknown biological relevance to 5-HT neuron function. However, many of the same DEGs contributed to significant z-scores greater than 2.0 across the identified canonical pathways (Figure 5A).
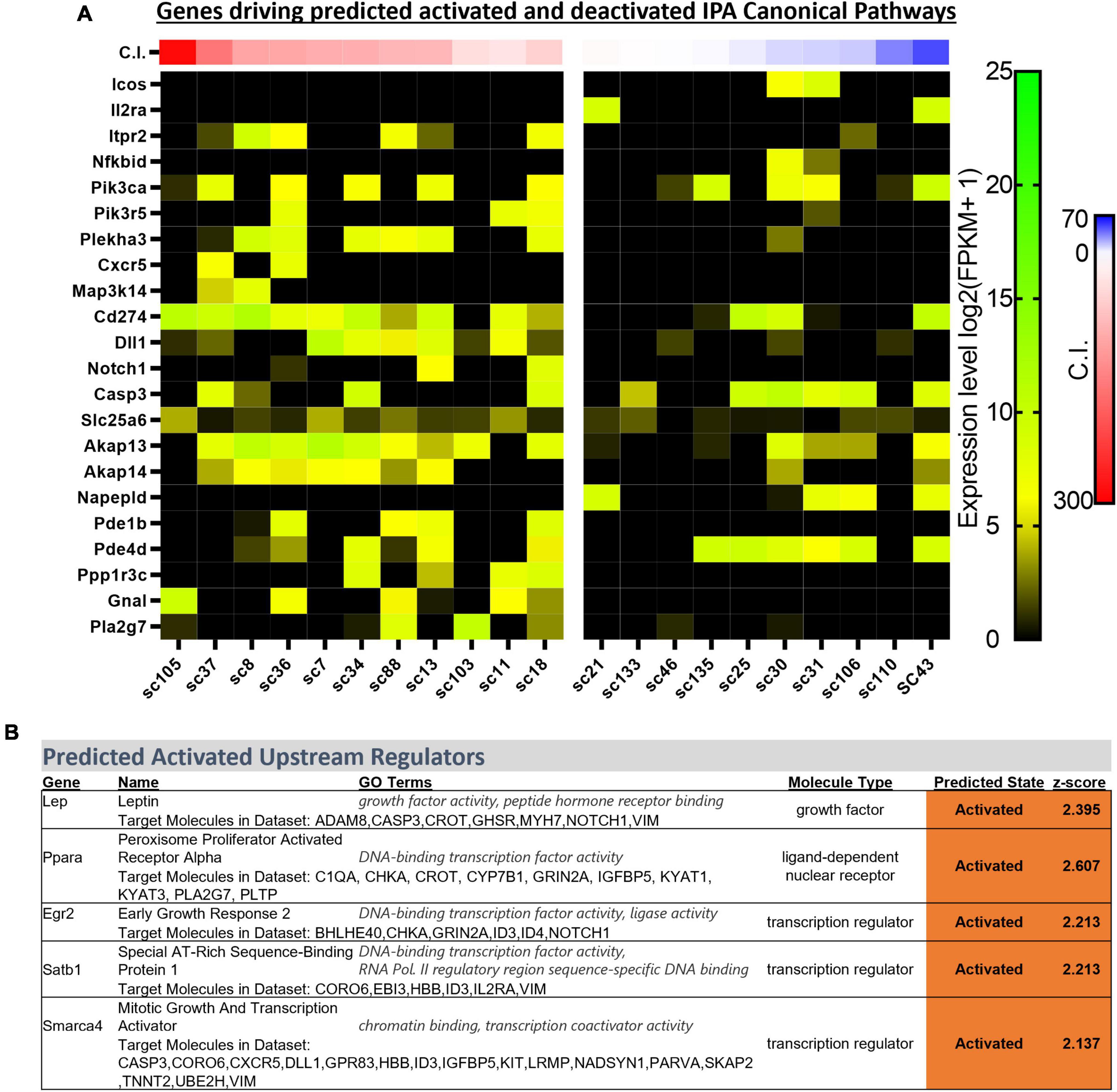
Figure 5. Heatmap of gene expression for each DEG contributing to activated z-scores across identified ingenuity pathway analysis (IPA) canonical pathways (indicated in the text) across all sequenced cells categorized by chemosensitivity index (C.I.) (A). IPA predicted activated upstream regulators of 166 DEGs indicate Egr2 and Lep, among others, as activated in CO2 sensitive 5-HT neurons (B).
Upstream Regulator analyses identified 5 gene regulators predicted to be activated in CO2 sensitive neurons. Among these were 2 protein coding genes involved in metabolic regulation were predicted to be activated in CO2 sensitive neurons, Lep (Leptin; z-score: 2.395) and Ppara (Ppar alpha; z-score: 2.607; Figure 5B). These data suggest that CO2 sensitive 5-HT neurons exhibit increased activation of genes responsive to leptin, which to our understanding has not previously been linked to CO2 sensitive 5-HT raphe neurons. Furthermore, Egr2 (z-score: 2.213), which is highly relevant given prior reports that mature CO2 sensitive 5-HT neurons are derived from 5-HT neurons that expressed Egr2 (and Pet-1 which is also known as Fev) during embryonic development (Brust et al., 2014). Prior fate mapping experiments in mice determined 5-HT neurons arise from unique cell lineages distinguished by the embryological expression of unique transcription factors. Specifically, rhombomere 5 (r5) and rhombomere r6p (posterior) give rise to the 5-HT neurons in the raphe magnus and obscurus, respectively. Electrophysiologic recordings in postnatal (P25-32) mice revealed the majority of r5 5-HT neurons were CO2 sensitive while most r6p 5-HT neurons were CO2 insensitive and that r5 and r6p neurons express distinct gene expression markers (Brust et al., 2014; Okaty et al., 2015). Whether r5 and r6p enriched genes had utility to distinguish the current rat neurons, and thus gauge if they might be embryologically determined, was assessed next. Expression of general 5-HT specific marker genes were consistent between the two data sets suggesting general similarities between mouse and rat 5-HT neurons (Figure 6). However, expression levels of the rat genes associated with R6P, R5, or r5/R6P enriched mouse genes did not cause any noticeable phenotypic separations (Figure 6). Rather, all the rat 5-HT neurons had similar gene expression levels as R6P-enriched genes. These data suggest that embryological gene markers may represent more anatomical separations of CO2 sensitive and insensitive 5-HT neurons and therefore most CO2 sensitive neurons are found in the r5 derived lineage, but not all. Thus, the current data represent a more specific transcriptional comparison between CO2 sensitive and insensitive 5-HT neurons because they were postnatally separated based on chemosensitivity and not based on embryological gene expression patterns. Therefore, a series of manual gene analyses and application of machine learning algorithms were employed to identify the most useful gene markers of CO2 sensitive and insensitive 5-HT rat neurons.
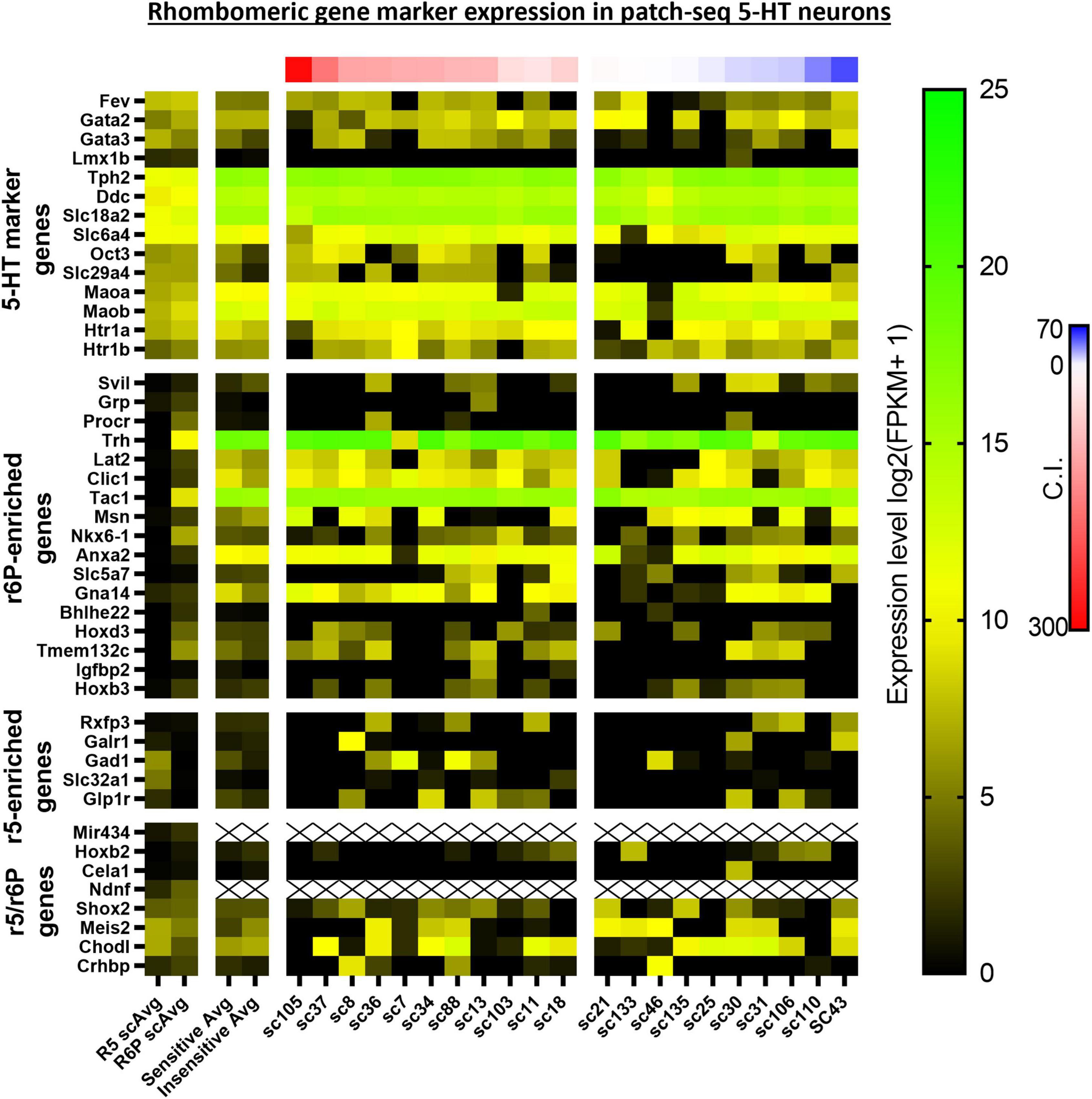
Figure 6. Heatmap of averaged single cell RNA Seq gene expression data derived from rhombomeric 5 (r5; i.e., CO2 sensitive 5-HT mouse neurons) and 6 (r6; i.e., CO2 insensitive 5-HT mouse neurons) in mice from a prior study by Okaty et al. (2015) compared to both averaged and single cell data of our 5-HT CO2 sensitive and insensitive rat neurons indicate differences in transcriptomic regulation of chemosensitivity between species. 5-HT marker genes (top horizontal panel) are enriched in both data sets. R6p-enriched genes (top middle horizontal panel) are highly expressed across all rat 5-HT neurons regardless of chemosensitivity while r5-enriched genes (lower middle horizontal panel) are undifferentiated between rat CO2 sensitive and insensitive 5-HT neurons. r5/r6p genes (bottom horizontal panel) are similarly expressed between CO2 sensitive and insensitive 5-HT rat neurons, consistent with the expression differences between R5 and R6p averages.
Support vector machine-recursive feature elimination (see section “Materials and Methods” for details) bioinformatic analyses were used to refine and validate the predictive capability of select DEGs with greatest probabilities of differentiating between 5-HT CO2 sensitive and insensitive 5-HT neurons. The SVM-RFE performed on the 166 DEG list resulted in 4 candidate genes (CD46, Iba57, Maz, and Chd5), that each had high predictive accuracies (68.7, 71.2, 72.0, and 74.0%, respectively) in determining whether a 5-HT neuron is CO2 sensitive or insensitive (Figure 7). CD46, Iba57, and Maz, were expressed only in CO2 sensitive neurons while Chd5 had expression in both types of neurons. CD46 and Iba57 were the only two genes from the SVM-RFE analysis that were identified in the top 10 CO2-enriched genes among the 166 DEGs (no SVM-RFE genes were observed in the top 10 DEGs or top 10 CO2-insensitive genes; Supplementary Figure 3). Two selected genes, Cd46 and Iba57, had gene expression observed throughout entire brainstem tissue sections inclusive of the raphe magnus (RMg) using RNAScope. Whether Cd46 and Iba57 are strictly co-localized to neurons was not assessed, but they were indeed co-expressed in serotonergic (Tph2+) neurons, consistent with RNA-Seq derived data (Supplementary Figures 4, 5). Performing SVM-RFE using all identified genes resulted in 2 candidate genes, Hnrnpk and Diablo, which have predictive accuracies of 81.0 and 87.9%, respectively.
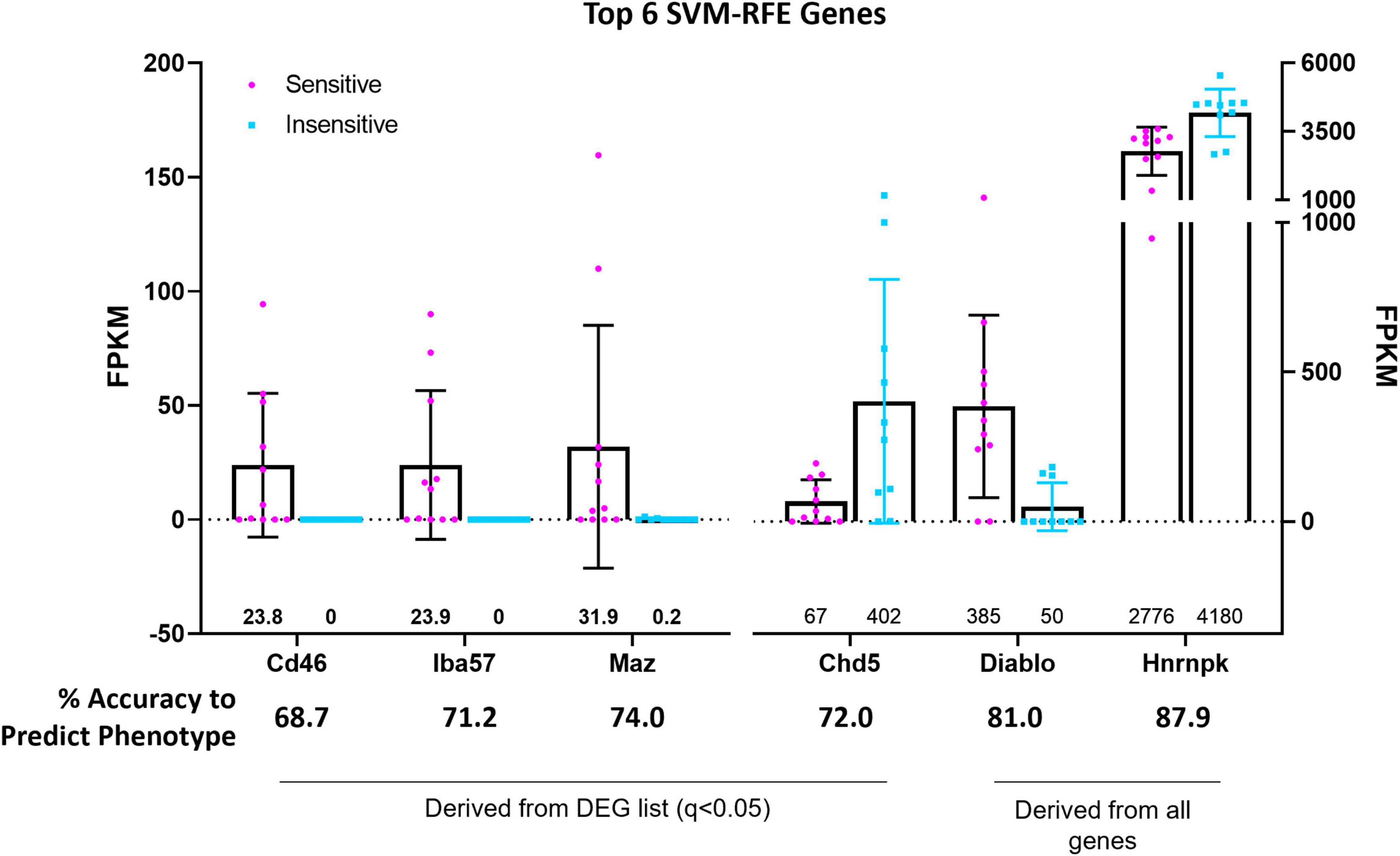
Figure 7. Support vector machine, recursive feature elimination (SVM-RFE) analyses identified the genes CD46, Iba57, Chd5, and Maz amongst 166 DEGs and the genes Hnrnpk and Diablo among all identified and sequenced genes as most accurate predictors of 5-HT neuron CO2 sensitivity (sensitive vs. insensitive).
Diagnostic ability of the six candidate genes derived from SVM-RFE was assessed using logistic regression and a receiver operator characteristic (ROC) curve. These methods measured the associated degrees of specificity (true negative rate) and sensitivity (true positive rate) in assessing whether a 5-HT neuron is CO2 sensitive or insensitive. The area under the curve (AUC) for Chd5 (0.77 ± 0.11; p = 0.0346), Maz (0.78 ± 0.11; p = 0.0290), Hnrnpk (0.87 ± 0.88; p = 0.0039) and Diablo (0.88 ± 0.084; p = 0.0031) were comparable to the predictive accuracies measured using SVM-RFE (Figure 8A). FPKM values were plotted against ROC derived predictive probabilities to determine if greater gene expression improved or detracted from the predictive accuracies for each gene (Figure 8B). Whereas increasing Chd5 and Hnrnpk gene expression reduces predictive probability, increasing Maz and Diablo gene expression increases predictive probabilities. Thus, Chd5 and Hnrnpk are better suited to identify CO2 insensitive 5-HT neurons because they are expressed at higher levels in CO2 insensitive vs. sensitive 5-HT neurons. Maz and Diablo gene expressions are better suited to identify CO2 sensitive 5-HT neurons because they are expressed at higher levels in CO2 sensitive than insensitive 5-HT neurons Logistic regression, ROC curves, and the AUC could not be calculated for CD46 and Iba57 because of perfect separation of gene expression (i.e., only CO2 sensitive 5-HT neurons expressed CD46 and Iba57). Such perfect separation indicates that CD46 and Iba57 have high predictive ability to determine if 5-HT neurons are CO2 sensitive.
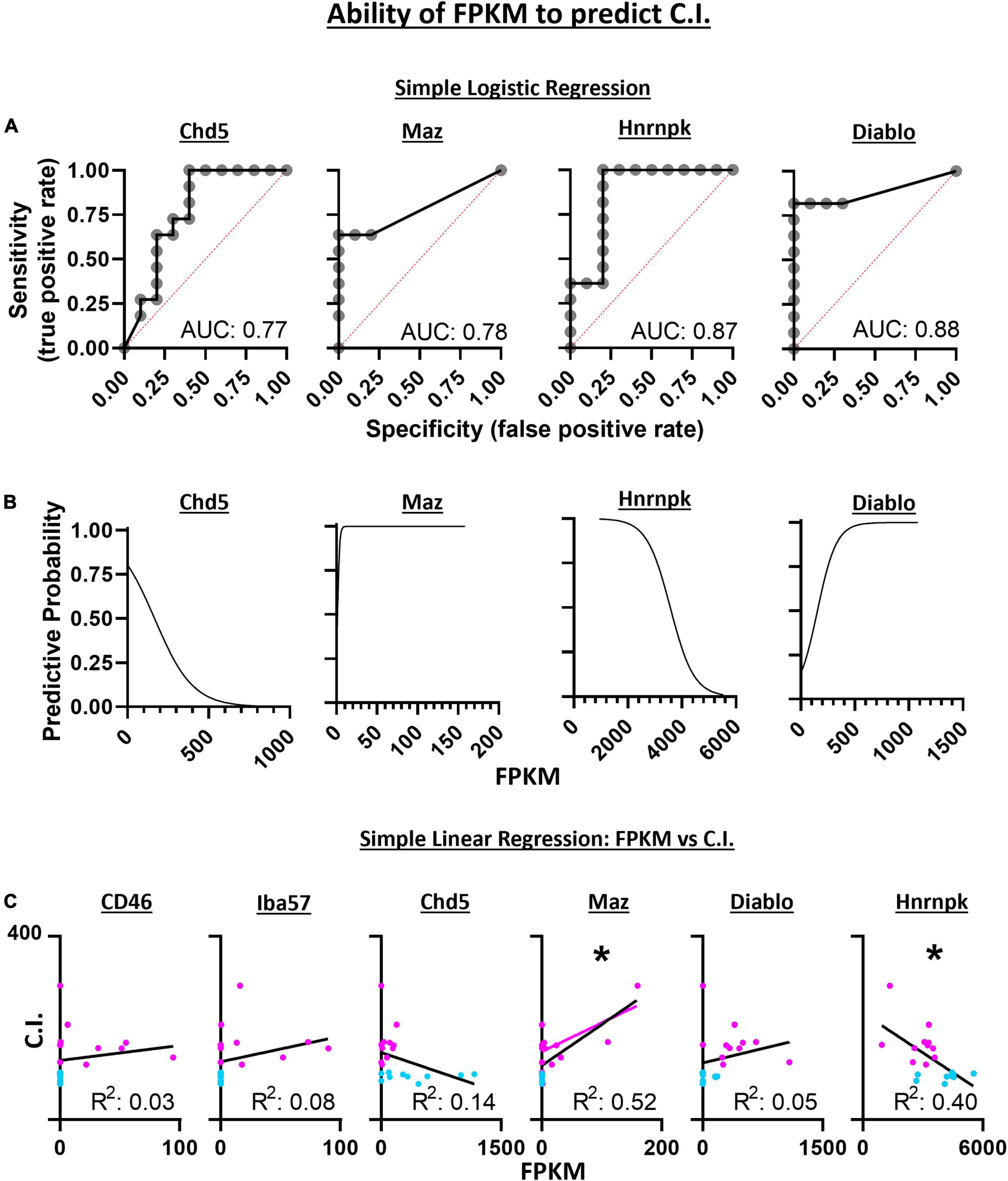
Figure 8. Simple logistic regression indicates the expression level (FPKM) of Chd5, Maz, Hnrnpk, and Diablo have high sensitivity and specificity with predictive accuracies (area under the curve) similar to the SVM-RFE analyses (A). With increasing gene expression (FPKM), the predictive accuracy for assessing the chemosensitivity each neuron decreases for Chd5 and Hnrnpk but the accuracy increases within increasing Maz and Diablo gene expression (B). The expression levels of Maz and Hnrnpk but not CD46, Iba57, Chd5, or Diablo predict the degree of chemosensitivity of each 5-HT neuron assess by simple linear regression (C). Maz and Hnrnpk have significant R2 values indicating their FPKM values account for a significant degree of the C.I. variability with a significant positive and negative Pearson r correlation, respectively. *P < 0.002; Pearson r correlation.
Linear regression was used to assess the ability for gene expression levels to predict the degree of chemosensitivity. Maz was found to have a significant best fit line (R2 = 0.52; P = 0.0002) as was Hnrnpk (R2 = 0.40; P = 0.0021) indicating Maz and Hnrnpk gene expression can account for a large degree of C.I. variability. A two-tailed Pearson r correlation analysis found Maz to have a significant positive correlation (r = 0.72; P = 0.0002) and Hnrnpk to have a significant negative correlation (r = −0.63; P = 0.0021) between gene expression and C.I. (Figure 8C).
In this study, we used the patch-to-seq technique and a combination of supervised, unsupervised, and software-based bioinformatic analysis techniques to identify functional and transcriptomic predictors of CO2 sensitive 5-HT neurons in the brainstem. We determined favorable diagnostic probabilities of using baseline firing rate as a predictor of 5-HT neuron chemosensitivity, suggesting low baseline firing rates may be a functional marker of CO2 sensitive 5-HT neurons. We identified 166 DEGs among CO2 sensitive and insensitive 5-HT neurons validating the original hypothesis that these subpopulations of 5-HT neurons are transcriptionally distinct. Bioinformatic pathway analyses revealed functional pathways not previously associated with neuronal chemosensitivity including a possible role of leptin and early growth response factor 2 (Egr2) in mediating gene expression in CO2 sensitive but not CO2 insensitive 5-HT neurons. Embryologic expression of Egr2 gives rise to 5-HT CO2 sensitive neurons (Brust et al., 2014) and facilitate the adult hypercapnic ventilatory response (dependent on cellular CO2 chemoreception) (Ray et al., 2013). Additional bioinformatic analyses identified 6 novel candidate gene markers of CO2 sensitive 5-HT neurons that have a high accuracy (68.7–87.9%) in predicting if a 5-HT neuron is CO2 sensitive or insensitive, where two candidate gene markers, CD46 and Iba57, were confirmed expressed in 5-HT neurons by RNA scope.
The rationale that functionally distinct subpopulations are reflected by distinct transcriptomes stems from prior studies where intersectional fate mapping and single cell RNA sequencing clearly delineate rhombomere 5 (R5)-derived 5-HT neurons from the posterior rhombomere 6 (R6P) subgroups (Okaty et al., 2015). The r5 lineage gives rise to the 5-HT neurons that populate raphe magnus, which contained the overwhelming majority of CO2 sensitive 5-HT neurons compared to the raphe obscurus (derived from the r6 lineage). Indeed, the identification of 166 DEGs among CO2 sensitive and insensitive 5-HT neurons support the hypothesis that these functionally distinct 5-HT neuron subpopulations are transcriptionally distinct. Additionally, transcriptional analyses using unsupervised MDS and hierarchical clustering accurately grouped neurons by CO2 sensitivity. However, using more relaxed significance criteria and removing outlier cells to enhance pathway analyses led to more than 3000 DEGs that when used with the same unsupervised MDS and hierarchical clustering analyses, failed to group cells based on CO2 sensitivity. Instead, two other distinct groups of cells were observed indicating that a phenotype other than CO2 sensitivity is more distinguishable at the transcriptional level in these 5-HT neurons. Thus, the differences in cell grouping resulting from these two sets of analyses highlight the utility of the patch-seq technique in coupling cellular phenotypes to group cells to identify transcriptional differences that may underly functionally distinct cells.
We also reasoned that gene expression differences measured between CO2 sensitive and insensitive 5-HT neurons here might represent specific rhombomeric lineages. Previous studies show that rhombomeric origin was predictive of CO2 sensitivity in 5-HT neurons. Here we asked if functional distinctions among brainstem 5-HT neurons in rats aligned with gene markers of the R5 and R6P lineages in mice. While the R5 and R6P gene markers did not provide utility in distinguishing CO2 sensitive from insensitive 5-HT rat neurons, there were similar expression patterns of canonical 5-HT markers between datasets. Note, canonical 5-HT markers are similarly expressed across all 5-HT neurons despite lineage and/or phenotype. Despite confirming similar expression patterns of 5-HT neuron gene markers amongst both data sets, rhombomere-specific gene markers do not distinguish CO2 sensitive from insensitive 5-HT rat neurons. Thus, rhombomeric gene markers, while providing predicative power in whether a 5-HT neuron will develop into a CO2 sensitive or insensitive 5-HT neuron is clear from prior studies, the same genes do not appear to have a role in driving functional differences in postnatal 5-HT neurons. While this conclusion assumes rhombomere gene markers are similar in mice and rats, there is support that this may be the case given the very similar expression levels and profiles of the 5-HT marker genes across rat and mouse 5-HT neurons.
Our prior study identified specific pH sensitive potassium ion channels as putative mechanisms of cellular CO2 sensitivity in 5-HT neurons (Puissant et al., 2017). However, the previous technique suffered from a lack of functional data and neuronal specificity due to incomplete cellular dissociation. Our current approach yielded highly specific 5-HT neuron contents and functional data, again highlighting a strength in the patch-seq approach. However, the data generated by the patch-to-seq data failed to identify differential expression of previously identified pH sensitive K channel genes among CO2 sensitive and insensitive 5-HT neurons. In addition, the DEGs identified herein do not obviously functionally contribute to this cellular property based on what is known about their protein function. Pathway analyses did yield novel information about gene sets that strongly associate with CO2 chemosensitivity, including immune-related signaling pathways as previously found using an alternative approach (Mouradian et al., 2016). Predicted upstream regulators of these immune-related pathways showed 5 genes that were all predicted to be activated in CO2 sensitive neurons whereas there were no upstream regulators (activated or deactivated) in CO2 insensitive neurons. These data further suggest CO2 sensitivity in 5-HT neurons is transcriptionally regulated by upstream regulators whose function involves immune related functions, but if or how these specific upstream regulators contribute to the cellular properties of chemosensitivity to CO2/pH remains unclear.
The transcription factor Egr2 has key roles in fate determination of 5-HT neurons and 5-HT neurons with a history of Egr2 expression have cellular CO2 responses (Brust et al., 2014; Okaty et al., 2015). Pharmacogenetic inhibition of neurons with a history of Egr2 expression in adult mice inhibits the hypercapnic ventilatory response, further indicating that Egr2 is a marker of central respiratory chemoreceptor neurons (Ray et al., 2013). Thus, Egr2 could be an alternative marker to specifically distinguish CO2 sensitive from insensitive 5-HT neurons, but Egr2 in these neurons may only be expressed during embryologic development given that Egr2 gene expression was not detected in the sequencing data. Also identified as an upstream regulator with high activation in CO2 sensitive 5-HT neurons was leptin, a key regulator of energy homeostasis. This suggests that CO2 sensitive 5-HT neurons are differentially responsive to leptin signaling compared to CO2 insensitive 5-HT neurons. Given that leptin is a key metabolic regulator, it’s potential to influence CO2 sensitive 5-HT neuronal gene expression may provide a link between metabolic and respiratory control. Indeed, CO2 sensitivity at the whole-animal level, measured by the hypercapnic ventilatory response is impaired long-term (measured from ∼30 to 230 days of age) in leptin-deficient (ob/ob) mice which is improved 3 days after leptin administration directly to the CNS (via 4th ventricle) but not subcutaneously (Tankersley et al., 1996; Bassi et al., 2012, 2015). Also, direct delivery of leptin into the nucleus of the solitary tract in the brainstem, a key site for integration of peripheral chemosensory afferents, causes an increase in CO2 chemosensitivity in normal rats (Inyushkin et al., 2009). Together, these data highlight an important stimulatory effect of CNS but not peripheral leptin on CO2 chemoreception.; While long-term obesity from genetic leptin deficiency causes long-term reductions in CO2 hypercapnic ventilatory responses, whether CO2 sensitivity at the neuronal level, and specifically in 5-HT neurons, is affected due to sustained elevated leptin levels as occurs in obesity remains unknown. However, leptin does have roles in a variety of biological functions, including reproduction, angiogenesis, blood pressure, and immune functions, the latter of which are predicted to be differentially regulated in CO2 sensitive 5-HT neurons (Zhang et al., 2005). Together, the 166 DEGs are putative mechanisms contributing to intrinsic chemosensitivity in 5-HT neurons and/or are markers of CO2 sensitive and insensitive 5-HT neurons.
We reasoned that the most practical molecular and histological markers of CO2 sensitive 5-HT neurons are those most strongly expressed in one cell type and not the other. Results indicate markers of CO2 sensitive 5-HT neurons would be better suited than markers of CO2 insensitive 5-HT neurons. Thus, using a combination of manual and machine learning analyses, 2 genes were identified that follow this expression pattern from the DEG list, CD46 and Iba57. Gene expression of CD46 and Iba57 are limited to CO2 sensitive 5-HT neurons. Selection of these genes as histological markers within 5-HT neurons is supported by their high accuracy to predict the CO2 sensitivity phenotype calculated by SVM-RFE. Though simple linear regression is unable to correlate the expression level with C.I. for either CD46 or Iba57 independently, the majority (8 of 11) of CO2 sensitive 5-HT neurons express one or both genes while neither of the genes are expressed in any CO2 insensitive 5-HT neuron, making this pair of gene markers a viable histologic screening tool for identifying CO2 sensitive 5-HT neurons. Indeed, RNAScope verified expression of CD46 and Iba57 in 5-HT neurons (it did not test for co-expression among other cell types), though RNAScope did not validate the binary expression pattern of CD46+ and Iba57+ as suggested by the high percentage of 5-HT neurons expressing CD46 and Iba57. This discrepancy between the sequencing results and RNA Scope may reflect the inability for total cell isolation using the patch-seq technique while RNA Scope can probe the contents of entire cells.
Our electrophysiologic data show that CO2 sensitive neurons have an average baseline firing rate (0.70 Hz) lower than CO2 insensitive neurons (1.306 Hz). These results are consistent with prior results derived from the perfused, in situ brainstem preparation [0.82 Hz vs. 1.82 Hz, respectively (Iceman et al., 2013)], acute brainstem slices (∼0.50 Hz vs. ∼1.5 Hz) (Brust et al., 2014) and primary cultures (Wang et al., 2001). While lower firing rates have been shown to be a feature of CO2 sensitive 5-HT neurons (Brust et al., 2014), the diagnostic criteria for baseline firing rate to predict if a 5-HT neuron is chemosensitive was previously unknown. A logistic regression test demonstrated significant sensitivity (true positive rate) for determining if a 5-HT neuron is CO2 sensitive with high confidence (79% sensitivity) when the baseline firing rate criterion was <0.4481 Hz indicating a practical means to estimate if a 5-HT neuron is CO2 sensitive.
In summary, via the patch-seq technique, functionally distinct brainstem 5-HT neurons were identified to be transcriptionally distinct aiding subsequent identification of novel genes associated with cellular CO2/pH sensitive neurons. These differentially expressed genes may contribute to cellular CO2 chemoreception, serve as distinct cellular markers of CO2 sensitive 5-HT neurons, and may provide important links to the 5-HT system dysfunction associated with human pathologies such as SIDS and SUDEP.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA820863.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin.
GM and MH contributed to the conception and design of the study, and wrote the first draft of the manuscript. GM, PL, ED, and JLG contributed to statistical analyses. JG wrote RNA-Scope methods sections of the manuscript. All authors contributed to manuscript revision.
Parker B. Francis Foundation, American Heart Association Career Development Award 20CDA35310121, Advancing Healthier Wisconsin Endowment (GM); NIH K01HL153101 and Advancing a Healthier Wisconsin Endowment (PN); American Physiological Society and American Heart Association (898067) postdoctoral fellowships (KB); NIH HL134850 and American Heart Association 18EIA33890055 (JG); NIH HL084207 and HL144807 (CS); NIH HL122358 (MH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnsyn.2022.910820/full#supplementary-material
Supplementary Figure 1 | Firing rates of all CO2 sensitive (A) and insensitive (B) 5-HT neurons. Average firing rates during baseline, CO2 challenge, and recovery compared among all CO2 sensitive and CO2 insensitive 5-HT neurons (C). The sub-population of neurons used for scRNA-Seq had representative firing rate differences during baseline, CO2 challenges, and recovery albeit one CO2 insensitive cell had oddly very high firing rates (D–F). Notably, baseline and recovery firing rates were lower in CO2 sensitive vs. insensitive 5-HT neurons while firing rate during the CO2 challenge was similar between CO2 sensitive and insensitive neurons. One-way repeated measures ANOVA with Sidak’s multiple comparisons test (A,B,D,E). ANOVA results: Condition p < 0.0001 and Individual Cell p < 0.001 (A); Condition and Individual Cell p < 0.0001 (B); Condition p < 0.0001 and Individual Cell p = 0.001 (D); Condition p = 0.0510 and Individual Cell p < 0.0001 (E). Two-way repeated measures ANOVA with Sidak’s multiple comparisons test (C,F). Condition p < 0.0001, Cell Phenotype p = 0.0040, and Interaction p < 0.0001 (C); Condition p < 0.0001, Cell Phenotype p = 0.0379, and Interaction p < 0.0001 (F). *p < 0.05 and ns, not significant Simple logistic regression using all cell recordings (G) or recordings from only cells that were sequenced (I) indicates that baseline firing rate of 5-HT medullary raphe neurons has high sensitivity and specificity with predictive accuracies (area under the curve) in determining if a 5-HT neuron is CO2 sensitive. With lower baseline firing rates, the predictive accuracy for assessing the chemosensitivity phenotype of 5-HT neurons increases (H,J). **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Figure 2 | Ingenuity pathway analysis (IPA) canonical pathways significantly represented, but not predicted to be activated, by the 166 differentially expressed genes between CO2 sensitive and insensitive 5-HT neurons determined by q < 0.05 (A). IPA canonical pathways significantly predicted to be down regulated (blue) or upregulated (orange) in CO2 sensitive neurons versus CO2 insensitive 5-HT neurons (B).
Supplementary Figure 3 | Top 10 differentially expressed genes (DEGs, top panel), CO2 sensitive enriched genes (middle panel), and CO2 insensitive enriched genes (lower panel). Data indicate most regularity in gene expression for CO2 sensitive enriched genes compared to Top 10 DEGs and CO2 insensitive enriched genes (i.e., CO2 sensitive enriched genes are only expressed in CO2 sensitive neurons).
Supplementary Figure 4 | RNAScope immunofluorescence validates co-expression of CD46 in Tph2+ 5-HT neurons within the rat brainstem including the raphe magnus (RMg). 4x objective overlay z-stack image outlining an area of the RMg that includes Tph2-expressing (green) cells (brightness enhanced for visualization of tissue outline) (A). Zoom in of RMg region outlined in (A) captured with a 20x objective showing Tph2+ (green; left), CD46+ (red; middle; contrast enhanced 40%), and an overlay (right; contrast enhanced by 40%) (B). Inset in overlay is enhanced zoom of a single Tph2+ neuron identified by the white arrow. Percentage of RMg and all Tph2+ cell also expressing at least 1 CD46 pixel (C). (A) Scale bar = 100 μm, (B) scale bar = 50 μm.
Supplementary Figure 5 | RNAScope immunofluorescence validates co-expression of Iba57 in Tph2 + 5-HT neurons within the rat brainstem including the raphe magnus (RMg). 4x objective overlay z-stack image outlining an area of the RMg that includes Tph2-expressing (green) cells (brightness enhanced for visualization of tissue outline) (A). Zoom in of RMg region outlined in (A) captured with a 20x objective showing Tph2+ (green; left), Iba57+ (red; middle; contrast enhanced by 40%), and an overlay (right; contrast enhanced by 40%). Inset in overlay is enhanced zoom of a single Tph2+ neuron identified by the white arrow (B). Percentage of RMg and all Tph2+ cell also expressing at least 1 Iba57 pixel (C). (A) Scale bar = 100 μm, (B) scale bar = 50 μm.
Bassi, M., Furuya, W. I., Zoccal, D. B., Menani, J. V., Colombari, E., Hall, J. E., et al. (2015). Control of respiratory and cardiovascular functions by leptin. Life Sci. 125, 25–31. doi: 10.1016/j.lfs.2015.01.019
Bassi, M., Giusti, H., Leite, C. M., Anselmo-Franci, J. A., do Carmo, J. M., da Silva, A. A., et al. (2012). Central leptin replacement enhances chemorespiratory responses in leptin-deficient mice independent of changes in body weight. Pflügers Arch. 464, 145–153. doi: 10.1007/s00424-012-1111-1
Brust, R. D., Corcoran, A. E., Richerson, G. B., Nattie, E., and Dymecki, S. M. (2014). Functional and developmental identification of a molecular subtype of brain serotonergic neuron specialized to regulate breathing dynamics. Cell Rep. 9, 2152–2165. doi: 10.1016/j.celrep.2014.11.027
Buchanan, G. F. (2019). Impaired CO2-Induced Arousal in SIDS and SUDEP. Trends Neurosci. 42, 242–250.
Buchanan, G. F., Hodges, M. R., Richerson, G. B., Monti, J. M., Prandi-Perumal, S. R., Jacobs, B. L., et al. (2008). “Contributions of chemosensitive serotonergic neurons to interactions between the sleep-wake cycle and respiratory control”. Serotonin and Sleep: Molecular, Functional and Clinical Aspects. eds B. L. Jacobs, D. J. Nutt, J. M. Monti, and S. R. Pandi-Perumal. Switzerland: Birkhauser Verlag, 529–554. doi: 10.1016/s0034-5687(01)00289-4
Cadwell, C. R., Palasantza, A., Jiang, X., Berens, P., Deng, Q., Yilmaz, M., et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol. 34, 199–203. doi: 10.1038/nbt.3445
Cerpa, V. J., Wu, Y., Bravo, E., Teran, F. A., Flynn, R. S., and Richerson, G. B. (2017). Medullary 5-HT neurons: Switch from tonic respiratory drive to chemoreception during postnatal development. Neuroscience. 344, 1–14. doi: 10.1016/j.neuroscience.2016.09.002
Chen, X., Zhang, K., Zhou, L., Gao, X., Wang, J., Yao, Y., et al. (2016). Coupled electrophysiological recording and single cell transcriptome analyses revealed molecular mechanisms underlying neuronal maturation. Protein Cell. 7, 175–186. doi: 10.1007/s13238-016-0247-8
Corcoran, A. E., Hodges, M. R., Wu, Y., Wang, W., Wylie, C. J., Deneris, E. S., et al. (2009). Medullary serotonin neurons and central CO2 chemoreception. Respir. Physiol. Neurobiol. 168, 49–58. doi: 10.1016/j.resp.2009.04.014
Cui, S.-Y., Li, S.-J., Cui, X.-Y., Zhang, X.-Q., Yu, B., Huang, Y.-L., et al. (2016). Ca2 + in the dorsal raphe nucleus promotes wakefulness via endogenous sleep-wake regulating pathway in the rats. Mol. Brain 9:71. doi: 10.1186/s13041-016-0252-0
Davis, S. E., Solhied, G., Castillo, M., Dwinell, M., Brozoski, D., and Forster, H. V. (2006). Postnatal developmental changes in CO2 sensitivity in rats. J. Appl. Physiol. 101, 1097–1103. doi: 10.1152/japplphysiol.00378.2006
Feldman, J. L., Mitchell, G. S., and Nattie, E. E. (2003). Breathing: rhythmicity, plasticity, chemosensitivity. Annu. Rev. Neurosci. 26, 239–266. doi: 10.1146/annurev.neuro.26.041002.131103
Fuzik, J., Zeisel, A., Mate, Z., Calvigioni, D., Yanagawa, Y., Szabo, G., et al. (2016). Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol. 34, 175–183. doi: 10.1038/nbt.3443
Gourine, A. V., Kasymov, V., Marina, N., Tang, F., Figueiredo, M. F., Lane, S., et al. (2010). Astrocytes control breathing through pH-dependent release of ATP. Science 329, 571–575. doi: 10.1126/science.1190721
Hendricks, T., Francis, N., Fyodorov, D., and Deneris, E. S. (1999). The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J. Neurosci. 19, 10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999
Hodges, M. R., and Richerson, G. B. (2008). Contributions of 5-HT neurons to respiratory control: Neuromodulatory and trophic effects. Respir. Physiol. Neurobiol. 164, 222–232. doi: 10.1016/j.resp.2008.05.014
Hodges, M. R., Tattersall, G. J., Harris, M. B., McEvoy, S. D., Richerson, D. N., Deneris, E. S., et al. (2008). Defects in breathing and thermoregulation in mice with near-complete absence of central serotonin neurons. J. Neurosci. 28, 2495–2505. doi: 10.1523/JNEUROSCI.4729-07.2008
Hodges, M. R., Wang, W., Chen, Z. F., Deneris, E. S., Johnson, R. L., and Richerson, G. B. (2007). Adult mice with genetic deletion of 5-HT neurons exhibit a severe loss of central chemoreception. FASEB J. 27, 14128–14138
Huang, M. L., Hung, Y. H., Lee, W. M., Li, R. K., and Jiang, B. R. S. V. M.-R. F. E. (2014). based feature selection and Taguchi parameters optimization for multiclass SVM classifier. Sci. World J. 2014:795624. doi: 10.1155/2014/795624
Iceman, K. E., Richerson, G. B., and Harris, M. B. (2013). Medullary serotonin neurons are CO2 sensitive in situ. J. Neurophysiol. 110, 2536–2544. doi: 10.1152/jn.00288.2013
Inyushkin, A. N., Inyushkina, E. M., and Merkulova, N. A. (2009). Respiratory responses to microinjections of leptin into the solitary tract nucleus. Neurosci. Behav. Physiol. 39, 231–240.
Kaplan, K., Echert, A. E., Massat, B., Puissant, M. M., Palygin, O., Geurts, A. M., et al. (2016). Chronic central serotonin depletion attenuates ventilation and body temperature in young but not adult Tph2 knockout rats. J. Appl. Physiol. 120, 1070–1081. doi: 10.1152/japplphysiol.01015.2015
Katter, K., Geurts, A. M., Hoffmann, O., Mates, L., Landa, V., Hiripi, L., et al. (2013). Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. FASEB J. 27, 930–941. doi: 10.1096/fj.12-205526
Kumar, N. N., Velic, A., Soliz, J., Shi, Y., Li, K., Wang, S., et al. (2015). PHYSIOLOGY. Regulation of breathing by CO(2) requires the proton-activated receptor GPR4 in retrotrapezoid nucleus neurons. Science 348, 1255–1260. doi: 10.1126/science.aaa0922
Law, C. W., Alhamdoosh, M., Su, S., Dong, X., Tian, L., Smyth, G. K., et al. (2016). RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res 5, ISCBCommJ–1408. doi: 10.12688/f1000research.9005.3
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mouradian, G. C. Jr., Liu, P., and Hodges, M. R. (2016). Raphe gene expression changes implicate immune-related functions in ventilatory plasticity following carotid body denervation in rats. Exp. Neurol. 287, 102–112 doi: 10.1016/j.expneurol.2016.04.017
Mulkey, D. K., Talley, E. M., Stornetta, R. L., Siegel, A. R., West, G. H., Chen, X., et al. (2007). TASK channels determine pH sensitivity in select respiratory neurons but do not contribute to central respiratory chemosensitivity. J. Neurosci. 27, 14049–14058. doi: 10.1523/JNEUROSCI.4254-07.2007
Okaty, B. W., Freret, M. E., Rood, B. D., Brust, R. D., Hennessy, M. L., deBairos, D., et al. (2015). Multi-Scale Molecular Deconstruction of the Serotonin Neuron System. Neuron 88, 774–791. doi: 10.1016/j.neuron.2015.10.007
Paxinos, G., and Watson, C. (1998). The Rat Brain in Stereotaxic Coordinates, 4th Edn. San Diego: Academic Press.
Ptak, K., Yamanishi, T., Aungst, J., Milescu, L. S., Zhang, R., Richerson, G. B., et al. (2009). Raphe neurons stimulate respiratory circuit activity by multiple mechanisms via endogenously released serotonin and substance P. J. Neurosci. 29, 3720–37 doi: 10.1523/JNEUROSCI.5271-08.2009
Puissant, M. M., Mouradian, G. C. Jr., Liu, P., and Hodges, M. R. (2017). Identifying Candidate Genes that Underlie Cellular pH Sensitivity in Serotonin Neurons Using Transcriptomics: A Potential Role for Kir5.1 Channels. Front. Cell Neurosci. 11:34. doi: 10.3389/fncel.2017.00034
Ray, R. S., Corcoran, A. E., Brust, R. D., Kim, J. C., Richerson, G. B., Nattie, E., et al. (2011). Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science 333, 637–642.
Ray, R. S., Corcoran, A. E., Brust, R. D., Soriano, L. P., Nattie, E. E., and Dymecki, S. M. (2013). Egr2-neurons control the adult respiratory response to hypercapnia. Brain Res. 1511, 115–125. doi: 10.1016/j.brainres.2012.12.017
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Tankersley, C., Kleeberger, S., Russ, B., Schwartz, A., and Smith, P. (1996). Modified control of breathing in genetically obese (ob/ob) mice. J. Appl. Physiol. 81, 716–723. doi: 10.1152/jappl.1996.81.2.716
Teran, F. A., Massey, C. A., and Richerson, G. B. (2014). Serotonin neurons and central respiratory chemoreception: where are we now? Prog. Brain Res. 209, 207–233. doi: 10.1016/B978-0-444-63274-6.00011-4
van den Hurk, M., Erwin, J. A., Yeo, G. W., Gage, F. H., and Bardy, C. (2018). Patch-Seq Protocol to Analyze the Electrophysiology, Morphology and Transcriptome of Whole Single Neurons Derived From Human Pluripotent Stem Cells. Front. Mol. Neurosci. 11:261. doi: 10.3389/fnmol.2019.00150
Wang, F., Flanagan, J., Su, N., Wang, L. C., Bui, S., Nielson, A., et al. (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29. doi: 10.1016/j.jmoldx.2011.08.002
Wang, W., Pizzonia, J. H., and Richerson, G. B. (1998). Chemosensitivity of rat medullary raphe neurones in primary tissue culture. J. Physiol. 511(Pt 2), 433–450.
Wang, W., Tiwari, J. K., Bradley, S. R., Zaykin, R. V., and Richerson, G. B. (2001). Acidosis-stimulated neurons of the medullary raphe are serotonergic. J. Neurophysiol. 85, 2224–2235. doi: 10.1152/jn.2001.85.5.2224
Wenker, I. C., Kreneisz, O., Nishiyama, A., and Mulkey, D. K. (2010). Astrocytes in the retrotrapezoid nucleus sense H+ by inhibition of a Kir4.1-Kir5.1-like current and may contribute to chemoreception by a purinergic mechanism. J. Neurophysiol. 104, 3042–3052. doi: 10.1152/jn.00544.2010
Wu, Y., Hodges, M. R., and Richerson, G. B. (2008). Stimulation by hypercapnic acidosis in mouse 5-HT neurons in vitro is enhanced by age and increased temperature. Soc. Neurosci. Abstr. 34, 383.9.
Wu, Y., Proch, K. L., Teran, F. A., Lechtenberg, R. J., Kothari, H., and Richerson, G. B. (2019). Chemosensitivity of Phox2b-expressing retrotrapezoid neurons is mediated in part by input from 5-HT neurons. J. Physiol. 597, 2741–2766. doi: 10.1113/JP277052
Keywords: CO2 chemoreception, 5-HT neurons, patch-seq, PH sensitivity, brainstem 5-HT, raphe magnus, transcriptomic analysis, medullary raphe
Citation: Mouradian GC Jr, Liu P, Nakagawa P, Duffy E, Gomez Vargas J, Balapattabi K, Grobe JL, Sigmund CD and Hodges MR (2022) Patch-to-Seq and Transcriptomic Analyses Yield Molecular Markers of Functionally Distinct Brainstem Serotonin Neurons. Front. Synaptic Neurosci. 14:910820. doi: 10.3389/fnsyn.2022.910820
Received: 01 April 2022; Accepted: 10 May 2022;
Published: 30 June 2022.
Edited by:
Fu-Ming Zhou, University of Tennessee Health Science Center (UTHSC), United StatesReviewed by:
Patrice G. Guyenet, University of Virginia, United StatesCopyright © 2022 Mouradian, Liu, Nakagawa, Duffy, Gomez Vargas, Balapattabi, Grobe, Sigmund and Hodges. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gary C. Mouradian Jr., Z21vdXJhZGlAbWN3LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.