- 1Department of Psychology, University of Minnesota, Minneapolis, MN, United States
- 2Department of Medicine, University of Minnesota, Minneapolis, MN, United States
- 3Hennepin Healthcare Research Institute, Minneapolis, MN, United States
- 4Department of Psychology, Arizona State University, Tempe, AZ, United States
Substance use disorder (SUD) represents a large and growing global health problem. Despite the strong addictive potency of drugs of abuse, only a minority of those exposed develop SUDs. While certain life experiences (e.g., childhood trauma) may increase subsequent vulnerability to SUDs, mechanisms underlying these effects are not yet well understood. Given the chronic and relapsing nature of SUDs, and the length of time that can elapse between prior life events and subsequent drug exposure, changes in SUD vulnerability almost certainly involve long-term epigenetic dysregulation. To validate this idea, functional effects of specific epigenetic modifications in brain regions mediating reinforcement learning (e.g., nucleus accumbens, prefrontal cortex) have been investigated in a variety of animal models of SUDs. In addition, the effects of epigenetic modifications produced by prior life experiences on subsequent SUD vulnerability have been studied, but mostly in a correlational manner. Here, we review how epigenetic mechanisms impact SUD-related behavior in animal models and summarize our understanding of the relationships among life experiences, epigenetic regulation, and future vulnerability to SUDs. Despite variations in study design, epigenetic modifications that most consistently affect SUD-related behavior are those that produce predominantly unidirectional effects on gene regulation, such as DNA methylation and histone phosphorylation. Evidence explicitly linking environmentally induced epigenetic modifications to subsequent SUD-related behavior is surprisingly sparse. We conclude by offering several directions for future research to begin to address this critical research gap.
1 Introduction
Substance use disorders (SUDs) impose severe burdens on individuals and society (Sharma et al., 2019; Strang et al., 2020; Ignaszewski, 2021; Peterson et al., 2021). Despite the addictive potency of drugs of abuse, a large majority of the population that experiments with drugs does not develop SUDs (Vowles et al., 2015; Reynolds et al., 2021). The trajectory that leads some into a SUD and others to avoid it is assumed to be a product of heritable variations in DNA sequence (e.g., single nucleotide and structural variants) and environmental factors, including an individual’s current circumstances and the cumulative effects of their past life experiences. With regard to the latter, epidemiological research has documented the persistent and cumulative effects of multiple adverse circumstances that increase susceptibility to SUDs and other psychopathologies. Most notable among them is the enduring impact of trauma, especially when experienced during critical developmental periods (Alati et al., 2006; Radliff et al., 2012; Zarse et al., 2019).
The long-term nature of such effects implicates epigenetic adaptation - or molecular regulation of gene transcription - within the brain as an underlying mechanism (Mews et al., 2018; Hamilton and Nestler, 2019; Werner et al., 2021). Some studies have documented epigenetic changes resulting from prior life experiences, such as early-life drug exposure or stress, while others have implicated epigenetic modifications in the addictive effects of drugs of abuse. Surprisingly few studies, however, have demonstrated a causal relationship between the presumed epigenetic “scars” left by major life events and future vulnerability to SUDs (Figure 1).
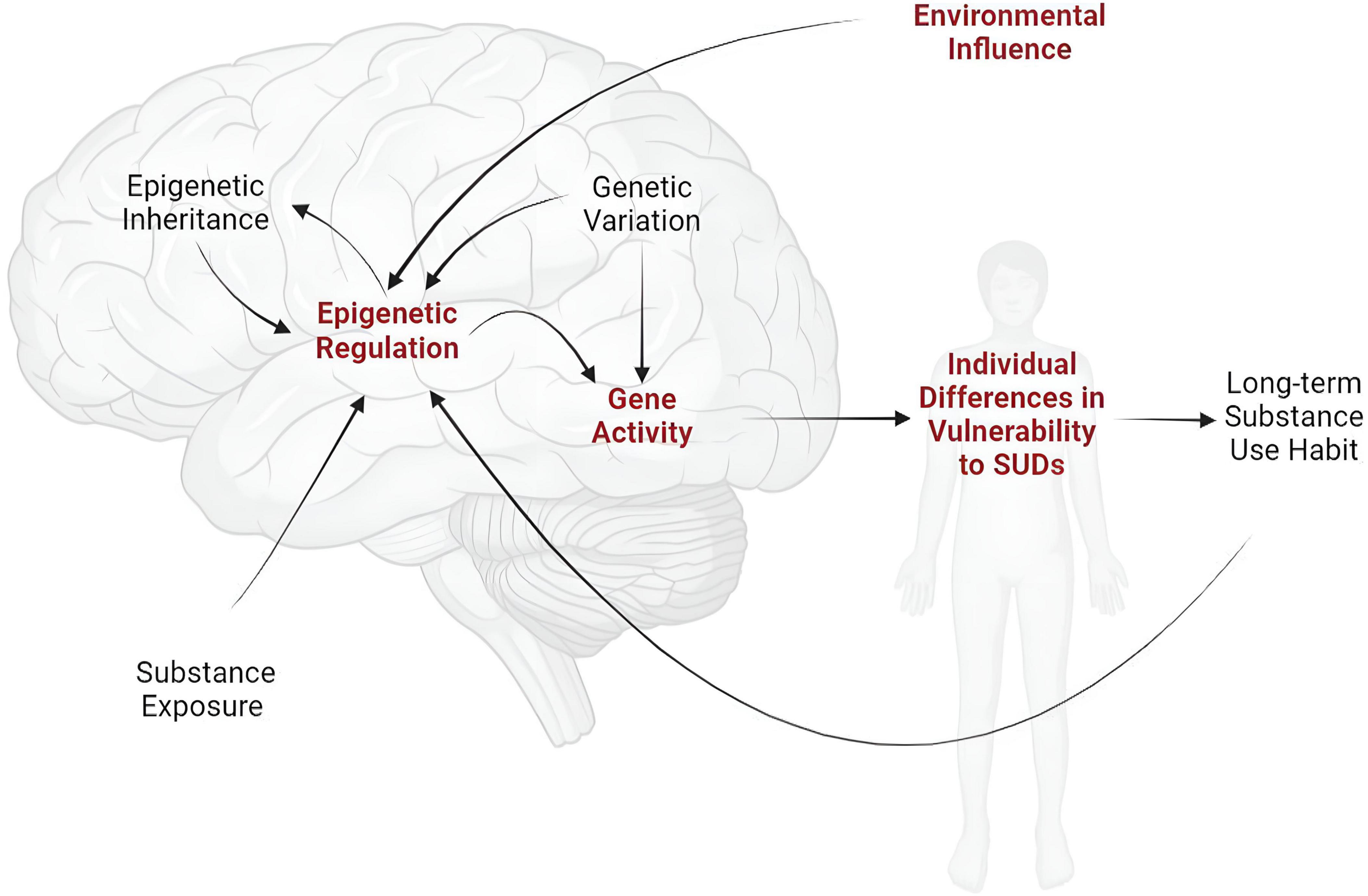
Figure 1. Individual vulnerability to SUDs is shaped by inherited genetic variation and by current and past environmental experiences, including substance exposure. Given the enduring nature of SUDs, these effects are almost certainly produced and sustained mechanistically through molecular pathways that regulate gene expression (i.e., epigenetics). While research has focused intensively and extensively on the effects of genetic variation and substance exposure and use on gene expression, relatively little is known about the epigenetic mechanisms through which long-term environmental factors affect susceptibility to SUDs (indicated in red).
While human studies have been critical in implicating epigenetic mechanisms in SUD vulnerability, variations in factors such as stress exposure, parenting styles, the severity of drug use, or developmental period (e.g., prenatal, adolescent), often make it difficult to characterize the finer details of relationships between specific forms of experience and SUD-related outcomes. Furthermore, epigenetic variance is tissue-specific, which can only be assayed in human brain tissue post-mortem. The harvesting of animal brain tissue, clearly, is less subject to this constraint. Furthermore, animal studies are designed to be fully experimental, randomized, and controlled, allowing greater precision and specificity in evaluating the effects of environmental factors or of different levels of drug exposure on epigenetic changes and SUD vulnerability.
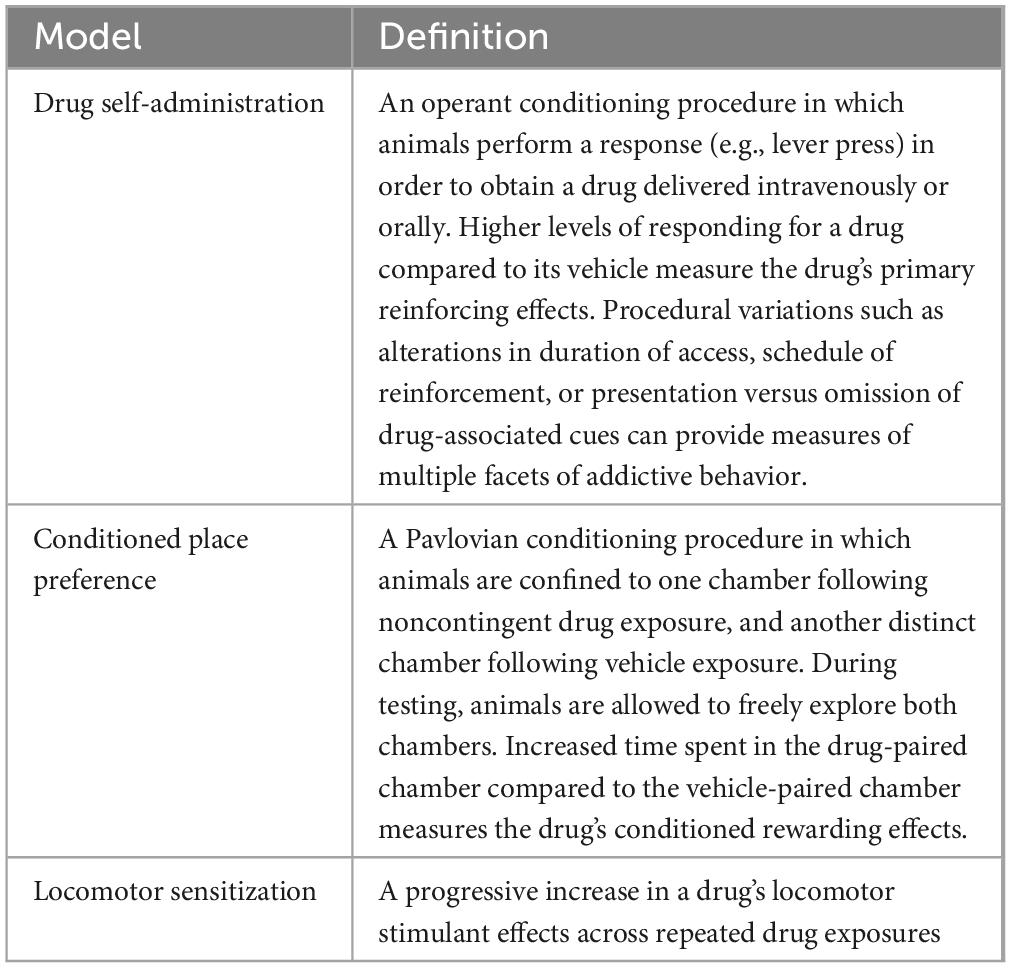
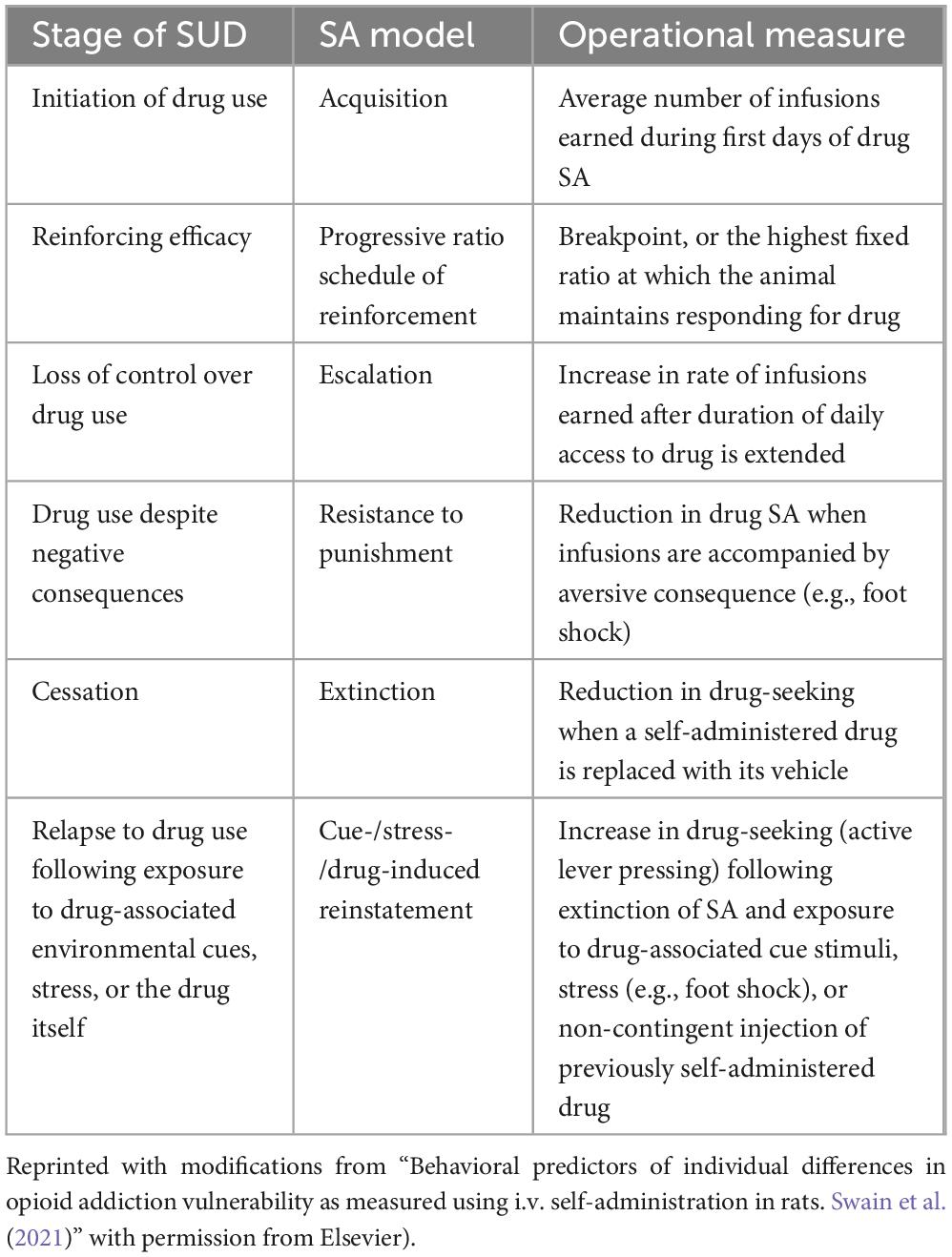
A variety of preclinical behavioral models of SUDs have been developed (Table 1). Among these, drug self-administration (SA) is often considered the “gold standard” for measuring SUD-like behavior because it involves volitional drug taking as occurs in humans and can be used to model various phases of the SUD trajectory (e.g., acquisition, relapse, etc., see Table 2). However, data from other models (e.g., conditioned place preference, CPP) can also provide important insights into the mechanisms underlying SUD. In this review, we will outline the forms of epigenetic modification that regulate SUD-related behavior in animals, summarize our understanding of their roles in mediating environmentally driven vulnerability to SUDs, and highlight several promising future research directions.
2 Epigenetic modifications mediating addictive effects of substances
Epigenetic regulation affects both transcriptional and post-transcriptional processes, with the latter including alternative splicing and RNA silencing (Wong et al., 2011). Such regulation occurs through a complex set of molecular processes, including DNA and histone modification, chromatin remodeling, and the actions of non-coding RNAs (ncRNAs). While preclinical studies have enumerated ways in which chromosomal modifications are up- or down-regulated by exposure to addictive substances, few have examined the causal effects of specific forms of epigenetic modification on SUD-related behavior. As described below, and summarized in Supplementary Table 1, such studies have produced complex and often inconsistent results.
2.1 DNA modification
2.1.1 Molecular mechanisms
Covalent modifications of the nucleotide bases of DNA regulate gene transcription without altering the genetic code itself. The most commonly studied form is 5-methylcytosine (5mC), which occurs through DNA methyltransferase (DNMT) enzymes. 70% of 5mC moieties in the adult brain are found on cytosine-guanine (CpG) sites (Lister et al., 2013). CpG-rich regions of around 1,000 base-pairs (bp), CpG islands (CGIs), contain 70% of gene promoters and are typically hypomethylated (6–8% methylation) (Illingworth et al., 2008). Methylation at CGIs commonly reduces binding of transcription factors (TFs), thereby suppressing gene expression (Deaton and Bird, 2011). Conversely, 5mCs located at the gene body are associated with gene activation and alternative splicing (Jones, 2012).
A second DNA modification is 5-hydroxymethylcytosine (5hmC), formed through the oxidation of 5mC by ten-eleven translocation methylcytosine dioxygenase (TET) enzymes (Goll and Bestor, 2005; Ito et al., 2011). 5hmC is highly enriched in the CNS (∼17% of the methylated genome) relative to the periphery (Lister et al., 2013). While the functions of hydroxymethylation are complex, promotion of gene expression is the most prominent (Ehrlich and Ehrlich, 2014). 5hmC is sequentially converted to 5-formyl and 5-carboxyl cytosine, before its eventual dissociation from cytosine residues (Shi et al., 2017).
2.1.2 Behavioral effects
The effects of systemic or brain region-specific demethylation through DNMT inhibitors (DMNTis) on behaviors associated with addictive drugs have been fairly consistent. Most studies report that reduced DNA methylation decreased animals’ locomotor sensitivity, drug SA (e.g., cue- and drug-induced reinstatement; see Table 2), or conditioned place preference (CPP; see Supplementary Table 1) for cocaine, opioids or alcohol (Anier et al., 2010; Han et al., 2010; Warnault et al., 2013; Barbier et al., 2015; Massart et al., 2015; Hong et al., 2021) (but see Laplant et al., 2010). In contrast, increasing methylation through methyl donors has yielded mixed results, depending on the donor administered and the substance studied. While systemic injection of the methyl donor methionine (MET) decreased rodents’ locomotor sensitivity, SA (drug-induced reinstatement) or CPP for cocaine (Laplant et al., 2010; Tian et al., 2012; Wright et al., 2015), S-adenosylmethionine (SAM) has opposite effects (Anier et al., 2013; Massart et al., 2015). Similarly, systemic administration of MET increased opioid SA (drug-induced reinstatement) in one study but had no significant effect on opioid CPP in another (Tian et al., 2012; Hong et al., 2021). In addition, altering the balance between methylation and hydroxymethylation through knockdown of the TET1 enzyme in the nucleus accumbens (NAc), increased CPP for cocaine in mice, consistent with the changes induced by repeated cocaine administration (Feng et al., 2015).
2.2 Histone modification
The basic structural and functional unit of chromatin is the nucleosome, composed of ∼147-bp DNA wrapped around an octamer containing 2 copies each of the histones H2A, H2B, H3, and H4 (Andrews and Luger, 2011). Post-translational histone monomers can be covalently modified to incorporate various functional groups at their N-terminals. At least 18 different forms of histone modification have been identified, most commonly at lysine (K) residues on histones H3 and H4 (Zhao and Garcia, 2015). Among these, acetylation, methylation, and phosphorylation have been most frequently studied in behavioral models of SUD.
2.2.1 Histone acetylation
2.2.1.1 Molecular mechanisms
Acetylation of lysine residues of the N-terminal histone tail is closely associated with transcriptional activation. While thought to encourage recruitment of chromatin remodeling proteins, the causal roles of acetylation in gene transcription are still not well established. For example it has still to be determined whether the strongest indicator of transcriptional activation, H3K27ac, is a transcriptional effector itself or simply provides a close readout of transcriptional activation (Shvedunova and Akhtar, 2022). Histone acetyltransferases (HATs) and histone deacetylases (HDACs) mediate histone acetylation and deacetylation, respectively (Shahbazian and Grunstein, 2007).
2.2.1.2 Behavioral effects
Stimulants tend to increase acetylation of histones (Renthal and Nestler, 2009; McCowan et al., 2015). Upregulation of histone acetylation by other means has been shown to enhance stimulant-associated behaviors in most (Levine et al., 2005; Kalda et al., 2007; Renthal et al., 2007; Shen et al., 2008; Sun et al., 2008; Wang et al., 2010; Malvaez et al., 2011; Rogge et al., 2013; Campbell et al., 2021), but not all studies (Romieu et al., 2008; Ferguson et al., 2013; Kennedy et al., 2013; Taniguchi et al., 2017; Campbell et al., 2021). Notably, Campbell et al. (2021) demonstrated that the same manipulation could produce opposite effects depending on the specific behavioral paradigm: reducing HDAC3 activity in NAc dopamine receptor D1 (DRD1)-containing medium spiny neurons decreased cocaine seeking during extinction of SA (see Table 2), but enhancing cocaine-CPP. In contrast, increased histone acetylation inhibits alcohol- and nicotine-associated behaviors, including voluntary alcohol intake (Warnault et al., 2013; Sakharkar et al., 2014; Bohnsack et al., 2022), alcohol withdrawal-induced anxiety-like behavior (Sakharkar et al., 2014; Bohnsack et al., 2022), and nicotine CPP (Pastor et al., 2011). The effects of altering histone acetylation on opioid addiction have been the least conclusive, with behavioral effects in both directions (Ferguson et al., 2013; Chen et al., 2016; Saberian et al., 2021; Anderson et al., 2023).
2.2.2 Histone phosphorylation
2.2.2.1 Molecular mechanisms
Like histone acetylation, phosphorylation reduces the positive charge on histone molecules thereby promoting gene expression. Mediated by protein kinases, histone phosphorylation most frequently occurs at serine (S) residues on the histone N-terminal tails, but also at threonine (T) and tyrosine (Y) residues (Rossetto et al., 2012). Histone phosphorylation at certain sites is closely associated with nearby acetylation, a process referred to as phosphoacetylation (Clements et al., 2003; Rossetto et al., 2012). For example, phosphorylation at H3S10, H3T11 and H3S28 facilitates acetylation at H3K14 by forming a HAT/CoA/histone complex, leading to increased HAT activity (Lo et al., 2000).
2.2.2.2 Behavioral effects
Few studies have specifically investigated the effects of histone phosphorylation or phosphoacetylation on SUD-associated behavior, although an increase in histone phosphorylation or phosphoacetylation most commonly enhances SUD-related behavior (Kumar et al., 2005; Stipanovich et al., 2008; Besnard et al., 2011; Sheng et al., 2011). Conversely, a reduction of histone H3 phosphorylation and phosphoacetylation decreased cocaine-induced locomotor sensitization but enhanced cocaine-CPP (Brami-Cherrier et al., 2005). In addition, some research has demonstrated that pharmacologically inhibiting extracellular signal-regulated kinases 1 and 2 (ERK1/2), which phosphorylate a wide range of proteins including histones and their epigenetic modifiers, generally reduce behavioral responses to psychostimulants and marijuana (Pascoli et al., 2014; Sun et al., 2016).
2.2.3 Histone methylation
2.2.3.1 Molecular mechanisms
Histone methylation most frequently occurs at lysine and arginine (R) sites. Mediated by histone N-methyltransferases (HMTs), lysine residues can be mono-, di-, or trimethylated, and arginine residues can be mono- or di-methylated (Miller and Grant, 2013). Methyl groups can be removed by lysine-specific histone demethylases (KDMs) and arginine demethylases (Bannister and Kouzarides, 2003; Greer and Shi, 2012). Histone methylation is more stable than either acetylation or phosphorylation, suggesting longer-lasting effects on gene regulation (Zee et al., 2010; Mews et al., 2014). In contrast to the permissive effects of histone acetylation and phosphorylation, histone methylation can either permit or repress gene transcription, depending on the target residue, its degree of methylation and, in some cases, the presence of other marks in its vicinity (Kouzarides, 2002). For example, H3K4me3 (permissive) and H3K27me3 (repressive) marks interact bivalently, presenting a potential target for studying dynamic gene regulation in response to changes in environmental conditions (Blanco et al., 2020).
2.2.3.2 Behavioral effects
Given their complex nature, it is not surprising that the effects of histone methylation on SUD-related behavior are not uniform. For example, approximately half of the studies on psychostimulants have found that an increase in a permissive histone methylation mark (e.g., H3K4me3, H4R3me2a) or decrease in a repressive mark (e.g., H3K9me2) heightens responses to stimulants (Maze et al., 2010; Kennedy et al., 2013; Aguilar-Valles et al., 2014; Heller et al., 2014; Li et al., 2015; Zhang et al., 2018), while the remainder have reported the reverse (e.g., permissive: H3K4me3; repressive: H3K9me2, H3K36me3, H3R2me2a) (Aguilar-Valles et al., 2014; Damez-Werno et al., 2016; Anderson et al., 2018b,2018a; Xu et al., 2021). One study suggested that decreased enrichment of a repressive methylation mark (H3K9me1) led to increased alcohol consumption (Barbier et al., 2016). Moreover, HDACi increased both histone acetylation and repressive methylation, leading to decreased locomotor sensitization to cocaine (Kennedy et al., 2013). This example shows the complicated interaction between chromatin marks with different functions.
2.3 Chromatin remodeling
2.3.1 Molecular mechanisms
Repositioning of nucleosomes causes surrounding chromatin to assume more open or closed configurations that promote or depress gene expression, respectively. Changes in DNA and histone modification can induce alterations in chromatin structure by recruiting ATPase-containing chromatin remodeling enzymes (SWI/SNF, CHD, ISWI and INO80 families, Längst and Manelyte, 2015).
2.3.2 Behavioral effects
Promotion of open-chromatin heightens locomotor sensitization and drug preference for cocaine (Wang et al., 2016; Salery et al., 2017; Werner et al., 2019). These changes in chromatin conformation have been achieved by altering the abundance and functionality of the chromatin remodelers SWI/SNF and INO80, and of activity-regulated cytoskeleton-associated protein, which interacts with the SWI/SNF complex (Shan et al., 2020; Hargreaves, 2021). Results of this approach have not yet been reported for other addictive substances.
2.4 ncRNAs
ncRNAs comprise microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), circular noncoding RNAs (circRNAs) and long noncoding RNAs (lncRNAs). Unlike mRNAs that serve as templates for translation, these molecules regulate gene transcription (Chekulaeva and Rajewsky, 2019).
2.4.1 miRNA: molecular mechanisms and behavioral effects
Among ncRNAs, the role of miRNAs in the various effects of addictive drugs are the best understood. miRNAs are ∼22-nucleotide (nt)-long molecules that can form an RNA-induced silencing complex (RISC) to bind to complementary target mRNAs (Shang et al., 2023). This process induces cleavage of the mRNA transcript and inhibition of protein translation, affecting SUD-like behavior, possibly through alterations in synaptic signaling (Bartel, 2004; Chandrasekar and Dreyer, 2009; Jonkman and Kenny, 2012; Kenny, 2022). This mechanism has been explored extensively in animal models of cocaine use. Conditional knockout of RISC protein AGO2 in Drd2 neurons abolished the acquisition of cocaine SA (acquisition) and CPP in mice (Schaefer et al., 2010). In addition, overexpression of each of the miRNAs miR-124, -212, -495, and let-7d reduced behavioral responses to cocaine (Hollander et al., 2010; Chandrasekar and Dreyer, 2011; Bastle et al., 2017), while overexpression of miR-181a enhanced cocaine-CPP (Chandrasekar and Dreyer, 2011). Interestingly, the effects of miRNA may be at least in part cell-type-specific, since overexpression of miR-1 in Drd1 neurons increased cocaine SA (cue-induced reinstatement), while overexpression in Drd2 neurons reduced cocaine SA (breakpoint) (Forget et al., 2021). In studies of alcohol use disorder, elevated levels or activity of miR-30a-5p, miR-124a, miR-137, miR-206 and miR-411 increased consumption, whereas overexpression of let-7d decreased consumption (Bahi and Dreyer, 2013, 2020; Darcq et al., 2014; Tapocik et al., 2014; Kyzar et al., 2019; Most et al., 2019).
2.4.2 snoRNA: molecular mechanisms and behavioral effects
snoRNAs range from 60 to 300 nt in length and carry out diverse cellular functions (Fafard-Couture et al., 2021). While their major function is to guide chemical modifications of the target precursor RNA (pre-RNA) molecules, snoRNAs can also affect alternative splicing of pre-RNAs through complementary binding (Kiss, 2002). Likely by promoting incorporation of exon Vb into the serotonin receptor 2C (5HT2CR) transcript, overexpression of one such molecule, MBII-52, attenuated cocaine-induced CPP and locomotor sensitization (Chen et al., 2014).
2.4.3 circRNA: molecular mechanisms and behavioral effects
circRNAs form a covalent loop structure through a non-canonical “back splicing” process (Rybak-Wolf et al., 2015). They are abundantly expressed, especially in the brain. circRNAs are transcribed from the same parent gene as their relevant mRNAs; however, their circular structure makes them resistant to exonuclease degradation. Additionally, circRNAs can be recognized by miRNAs that also target their respective mRNAs. As such, circRNAs may affect gene activity by protecting mRNAs from cleavage (Jeck et al., 2013). Overexpression of the circRNA circTmeff-1 in the NAc core enhanced morphine-CPP in mice while circTmeff-1 knockdown produced the opposite effect (Yu et al., 2021). Similarly, knockdown of circTmeff-1 in mouse NAc core reduced cocaine-CPP, potentially through regulation of miR-206 (Shen et al., 2022).
2.4.4 lncRNA: molecular mechanisms and behavioral effects
Finally, untranslated linear RNA transcripts greater than 200 nt in length are defined as lncRNAs. Widely distributed in various tissue types, lncRNAs regulate either proximal (cis) genes or distal (trans) genes. lncRNAs can affect chromatin structure by binding with nucleosomes, neutralizing histone charges and recruiting chromatin modifiers (Statello et al., 2020). In two addiction-related studies of lncRNA function to date, overexpression of lncRNA Gas5 in the NAc decreased cocaine-CPP and SA (e.g., breakpoint, extinction) in mice (Xu et al., 2020), and knockdown of the lncRNA Lrap increased alcohol intake in rats (Saba et al., 2021).
2.5 Conclusion
The above studies indicate that epigenetic modifications can regulate behaviors induced by addictive drugs, but the direction of many effects has differed across studies (Table 2). Given the inherent complexity of the epigenetic regulation and the variation among study designs (e.g., behavioral paradigm, dosing regimen, manipulation method, brain region), such inconsistencies are not surprising. A further examination of the variables involved in these studies points out several that could contribute to such inconsistency. First, research on different substances produces behavioral results with different levels of consistency. For example, opioids have produced more mixed results compared to cocaine, alcohol, and nicotine. Secondly, the choice of epigenetic manipulation method can affect the behavioral outcome. For example, although reducing DNA methylation by administering DNMTi produced fairly consistent results, attempts to increase DNA methylation by injecting methyl donors (e.g., MET, SAM) failed to do so. Thirdly, the specificity of the sites where these manipulations occur (e.g., brain regions, cell types) can sometimes produce contrasting effects (Forget et al., 2021). Lastly, even when other variables are held constant, the choices of behavioral paradigm can still affect behavioral outcomes (Campbell et al., 2021). However, it is noteworthy that epigenetic modifications that have largely unidirectional effects on gene transcription, such as DNA methylation and histone phosphorylation, generally produce more consistent effects on SUD-related behaviors.
3 Environment and vulnerability to SUDs: role of epigenetics
Epidemiological analyses have revealed strong associations between significant early-life events, such as substance exposure and childhood trauma, on the incidence of SUDs (Enoch, 2011; McCabe et al., 2022). In the absence of a viable alternative, the most plausible mechanism is that environmental stimulation stably alters the epigenetic regulation of gene expression to affect physiological and psychological responses upon subsequent exposure to addictive substances. These changes could further lead to development and maintenance of SUDs or increase the risk of relapse after abstinence. In this section, we review studies employing animal models that have sought to establish the mechanistic relationships among environment, epigenome, and future SUD-related behavior from several perspectives.
3.1 Substance exposure across developmental stages
3.1.1 Parental substance exposure and epigenetic inheritance
An adverse preconception environment may produce long-lasting effects on offspring (intergenerational) and future generations (transgenerational). In clinical studies, smoking and drinking in fathers are associated with adverse physical and psychological outcomes in the offspring, including asthma, sleep problems, anxiety, depression, and behavioral dysfunction (Svanes et al., 2017; Luan et al., 2022). More surprisingly, grandparental tobacco and alcohol use has been linked to neuropsychological problems in grandchildren with sex-specific effects, suggesting transgenerational inheritance (Golding et al., 2017; Kendler et al., 2018). Implicating a specific epigenetic mechanism, fathers’ preconception smoking, particularly in puberty, is associated with differential DNA methylation in the offspring of genes associated with asthma, inflammation, bipolar disorder, and binge eating (McElroy et al., 2018; Kitaba et al., 2023). This points to the possibility of environmentally induced epigenetic modifications affecting SUD vulnerability in one or more familial generations, consistent with recent evidence of the persistence of transgenerational epigenetic marks despite epigenome-wide reprogramming during gametogenesis and fertilization (Takahashi et al., 2023).
In animal studies, this has been tested primarily in sires to eliminate in utero effects on fetal development caused by maternal preconception substance exposure (Lassi et al., 2014). Prolonged cocaine SA in male rats during spermatogenesis reduced cocaine consumption exclusively in male offspring, with an increase in H3 acetylation at the Bdnf promoter observed in both the sperm of the sires and the mPFC of the offspring (Vassoler et al., 2012). Consistent with this, male offspring of cocaine-exposed fathers exhibited increased expression of Bdnf, which was found to suppress cocaine seeking behavior (Berglind et al., 2007).
Similarly, alcohol consumption was decreased in male offspring after paternal alcohol exposure (Finegersh and Homanics, 2014; Rompala et al., 2017). Persistent hypomethylation at the Bdnf promoter was detected in both the sperm of alcohol-exposed sires and the ventral tegmental area (VTA) of the offspring, accompanied by an increase in VTA Bdnf expression, which was implicated in alcohol preference and sensitivity (Moonat et al., 2013; Ting-A-Kee et al., 2013; Raivio et al., 2014).
These studies, while limited in number, suggest that heritable epigenetic modification induced by environmental exposure affects cellular and behavioral responses to addictive substances in the offspring, However, it is noteworthy that none of these epigenetic studies examined substance exposure prior to spermatogenesis or effects beyond the F1 generation. Therefore, the extent to which paternal substance exposure induces epigenetic changes that are stably maintained in the germline requires further inquiry.
3.1.2 Gestational substance exposure
Most addictive substances and/or their metabolites can pass through the placenta (Rosen and Johnson, 1988; Schenker et al., 1993), interfering with brain development including the reward signaling system. Consequently, infants born to substance-dependent mothers are at risk for developmental and neurobehavioral deficits (Lesser-Katz, 1982). Furthermore neonates with gestational substance exposure can display withdrawal syndromes due to the abrupt cessation of substance exposure upon birth (Nichols, 1967; Vagnarelli et al., 2006; Patrick et al., 2020). Moreover, gestational substance exposure can lead to neuroinflammation and elevated permeability of the blood-brain barrier, increasing vulnerability to future toxin exposure (Kousik et al., 2012). These various outcomes could contribute to increased vulnerability to substance use later in life.
Two rodent models support the hypothesis that gestational substance exposure increases adult SUD vulnerability through epigenetic regulation, although causality has not been fully demonstrated. Δ-9-tetrahydrocannabinol (THC) exposure during pregnancy increased morphine-CPP in adult offspring, accompanied by a decrease of DRD2 receptor density in the NAc. Strikingly, this was also observed in aborted human fetal brain tissue from THC-consuming mothers. Further examination revealed an increase in the repressive H3K9me2 mark, a reduction in the permissive H3K4me3 mark, and a reduction in RNA polymerase binding at the Drd2 gene. All three mechanisms may contribute in combination to downregulation of the DRD2 receptor (Dinieri et al., 2011; Sadeghzadeh et al., 2017). A change in the balance between permissive and repressive epigenetic marks was also associated with increased cocaine vulnerability in a mouse model of gestational methamphetamine (METH) exposure (Itzhak et al., 2014). Gestational METH exposure altered DNA methylation in the hippocampi of adult offspring, and increased cocaine-CPP and locomotor sensitization. Interestingly, these effects on DNA methylation opposed histone modifications at the same site. For example, DNA hypomethylation (permissive) was observed at the same sites modified by H3K27me3 (repressive), and DNA hypermethylation (repressive) was found at H3K4me3 (permissive) modified sites. The co-existence of these counteracting regulatory mechanisms highlights a complex balance of epigenetic modifications that underlie behavior abnormalities.
3.1.3 Adolescent substance use
Adolescence is a second critical period when the brain undergoes rapid development and changes in plasticity, resulting in further maturation of higher-order executive and cognitive abilities (Aoki et al., 2017; Larsen and Luna, 2018). External perturbations during this period, including the use of addictive substances, increase vulnerability to psychiatric disorders, such as depression, anxiety, and SUDs (Reynolds et al., 2019; Volkow and Wargo, 2022). Adults with a history of adolescent substance use are more likely to acquire long-term, severe substance use problems (Breslau et al., 1993; Odgers et al., 2008; McCabe et al., 2022). These observations suggest that addictive substances produce distinct effects on brain development during adolescence, which may contribute to vulnerability to SUDs in adulthood.
Adolescent alcohol consumption increased alcohol-preference in adulthood in rodent models through epigenetic regulation. Kyzar et al. (2019) demonstrated that adolescent alcohol exposure increased miR-137 levels in the adult rat amygdala, which downregulated its target, lysine-specific demethylase 1 (LSD1). This resulted in enrichment of H3K9me2 (repressive) at the Bdnf4 promoter, thereby decreasing Bdnf4 transcription. Demonstrating its functional significance, intra-amygdala infusion of miR-137 antagomir rescued behavioral, transcriptional, and epigenetic changes induced by adolescent alcohol exposure, whereas intra-amygdala knockdown of LSD1 diminished the rescuing effects of miR-137 antagomir. In contrast, adolescent alcohol exposure in mice increased histone H4 acetylation (a permissive mark) at the Bdnf promoter and Bdnf transcription in the mPFC (Montesinos et al., 2016). These transcriptional and epigenetic changes were reversed by blocking the toll-like receptor 4 (TLR4) pathway, normalizing alcohol-preference in adults. Adolescent alcohol exposure also elevated drinking behavior in adult rats through altered enrichment of H3K27ac in the amygdala, since targeted change of this chromatin mark ameliorated excessive drinking in adulthood (Bohnsack et al., 2022). Taken together, these studies suggest that adolescent alcohol exposure induces changes in the epigenome, gene expression, and increased vulnerability to SUD in adulthood.
3.2 Stress
Severe acute or chronic stress induced by unpredictable or uncontrollable adverse events causes molecular, cellular, cognitive and behavioral abnormalities (Mineka and Kihlstrom, 1978; Taylor, 2010). Children who have experienced early-life adverse events such as bullying are more likely to use addictive substances in adulthood (Ttofi et al., 2016). More specifically, exposure to adverse childhood experiences is proportionally associated with increased odds of lifetime substance abuse, early initiation, and low cessation rates (Zarse et al., 2019). In adults, post-traumatic stress disorder is associated with increased chances of developing short-term or lifetime SUDs (Goldstein et al., 2016), while perceived work-related stress exhibits a positive correlation with alcohol and tobacco dependence (Peretti-Watel et al., 2009).
3.2.1 Early-life stress (ELS)
Early-life stress has been modeled in rodents through maternal separation or low-quality maternal care (e.g., fostering, early weaning, limited bedding/nesting material) (Murthy and Gould, 2018). Consistent with the substantial body of evidence in clinical studies that ELS alters DNA methylation (Kinnally et al., 2011; Szyf and Bick, 2013; Catale et al., 2020), research with animal models also implicates DNA methylation in conferring vulnerability to adult SUDs after ELS. Maternal separation enhanced behavioral sensitivity to cocaine in adult rat offspring, along with hypermethylation of the Pp1c gene promoter in the NAc (Anier et al., 2014). These molecular and behavioral effects were mediated at least in part by increased expression of DNMT and were reversed by DNMT inhibition (Anier et al., 2010). In contrast, recipients of high-quality maternal care (Bilbo et al., 2007) exhibited lower morphine-CPP in adulthood compared to those receiving standard maternal care. The same group found decreased DNA methylation at the promoter of the anti-inflammatory cytokine IL-10 coding gene (Il10), which increased IL-10 expression in the NAc, and protected against morphine-induced neuroinflammation in glial cells (Schwarz et al., 2011). Thus, these findings implicate DNA methylation as a potential mediator between ELS and susceptibility to SUDs.
3.2.2 Chronic stress in adulthood
Drug-naïve adult animals exposed to chronic stress subsequently display increased vulnerability to SUD-like behavior, accompanied by stress-induced epigenetic changes. Chronic stress significantly enhanced morphine-CPP 24 h after repeated daily foot shock in adult male rats. Striatal enrichment of the permissive mark H3K4me2 at the Fosb promoter was elevated in the shocked group prior to morphine exposure, which potentiated the expression of Fosb during the morphine-CPP sessions. These effects may be mediated by the glucocorticoid receptor (GR) signaling pathway, since co-administration of the GR antagonist mifepristone diminished or normalized behavioral, transcriptional, and epigenetic alterations produced by the chronic foot shock (Chen et al., 2019). This suggests that chronic stress exposure sensitizes responses to opioids through alterations in GR-mediated epigenetic regulation.
3.3 Physical activity
Physical exercise mitigates many psychological disorders and generally improves the quality of life (Fossati et al., 2021). Physical exercise in SUD treatment programs has been shown to reduce drug craving, ease symptoms of withdrawal, reduce depression and anxiety, and increase rates of abstinence (Wang et al., 2014; Dai et al., 2020). Preclinical studies reveal that physical activity is intrinsically rewarding, potentially serving as an alternative reinforcer to addictive substances (Cosgrove et al., 2002; Greenwood et al., 2011; Robertson et al., 2016; Robison et al., 2018).
In animal models, physical exercise regulates key genes involved in SUD-like behavior. Peterson et al. (2014) reported that physical activity dose-dependently reduced rats’ cocaine-seeking behavior and Bdnf4 expression in the PFC, potentially through a reduction of histone acetylation. Since cocaine induces Bdnf expression (McCarthy et al., 2012), which in turn enhances cocaine-induced locomotor activity and cocaine SA (Schoenbaum et al., 2007), physical exercise may epigenetically counteract the overexpression of Bdnf4 in the PFC and therefore mitigate the addictive potency of cocaine.
3.4 Conclusion
An individual’s interactions with external factors such as previous substance use, stress, and physical activity, can affect their vulnerability to future SUDs. These changes in SUD vulnerability have been proposed to involve the kinds of the long-term changes in regulation of the genome that occur in human and animals after prior experiences (Cadet, 2016; Ajonijebu et al., 2017; Hamilton and Nestler, 2019). And yet, as reviewed here, direct evidence to support this contention is surprisingly limited. This likely reflects the practical challenges in empirically demonstrating the mediating effects of epigenetic mechanisms on the relationship between prior environmental events and subsequent SUD-related behavior. To do so effectively, it is critical to design models that capture environmental factors and SUDs, ands to select appropriate transcriptional and epigenetic targets for validation. Given the likely role of prior environmental exposure in the development of SUDs, further elaboration of the manner in which this occurs would be invaluable. For example, most studies reviewed in this section could benefit from targeted epigenetic editing (further discussed in the next section) at genes of interest to further strengthen the case for a causal link between prior life events and later vulnerability to SUD-like behaviors. Nevertheless, even from our limited dataset, some intriguing findings have emerged. Most notably, and contrary to expectations, paternal preconception drug exposure epigenetically reduced male offspring’s drug taking in several studies (Vassoler et al., 2012; Finegersh and Homanics, 2014; Rompala et al., 2017).
4 Discussion
Our review has underscored both the sparsity and inconsistency of findings on the role of epigenetic regulation on SUD-related behavior and on precisely how it mediates environmentally predisposed SUD vulnerability. We conclude by offering several directions for future research that may aid in further elaborating the epigenetic landscape that lies between prior experience and development of SUDs.
4.1 Need for systematic and coordinated preclinical research on epigenetic mechanisms of SUD
As reviewed in section 2 “Epigenetic modifications mediating addictive effects of substances,” one prominent issue with preclinical research on epigenetic mechanisms underlying SUD is the inconsistency of findings regarding the functions of each epigenetic modification. This is not surprising given the molecular complexity of epigenetic regulation and the experimental designs that vary across studies. However, without a more detailed understanding of how the various forms of modification contribute to SUD vulnerability, our characterization of environmental influences and our search for new therapeutic tools to mitigate them will remain elusive. To these ends, it would be beneficial to adopt a more constrained set of parameters, and thereby increase comparability, across studies. For example, among the studies reviewed in section 2 “Epigenetic modifications mediating addictive effects of substances,” animals’ SUD-related behaviors were modeled by 4 measures in the CPP paradigm and 12 measures in the SA paradigm. While the choices of SUD-related measures in individual studies are no doubt grounded in sound rationales, findings would be more easily integrated if researchers were to prioritize the use of behaviors that show relatively robust differential responses to epigenetic manipulations, such as the acquisition phase in CPP and the drug-induced reinstatement phase in the SA paradigm. Furthermore, most of the current mechanistic studies focus on a single level of epigenetic modification. Therefore, little is known about how different mechanisms interact in producing a particular outcome.
4.2 Need for further preclinical studies on environment-induced epigenetic regulation in SUDs
Given the necessarily correlative nature of most human studies, animal studies offer the best opportunity to tease apart causal relationships between adversity, the epigenome, and later SUD-like behavior. That said, it is surprising how few studies have taken full advantage of animal models to establish causal relationships between environmentally induced epigenetic changes and their long-term effects on SUD vulnerability. First off, only a small number of animal models that capture environmentally induced epigenetic regulation have been employed in SUD studies. For example, no studies have evaluated effects of acute-stress induced epigenetic changes on subsequent SUD vulnerability in animal models. However, both clinical and preclinical studies have reported that acute stress alters the epigenome and may also regulate long-term SUD vulnerability (Vaisvaser et al., 2016; Carter et al., 2017; Rusconi and Battaglioli, 2018). In addition, early-life nutritional status mediates psychiatric outcomes later in life, including schizophrenia, major affective disorder and personality disorders (Brown et al., 2000; Xu et al., 2009; Hock et al., 2018). For example, prenatal folic acid (a methyl donor) supplementation shows promise in alleviating fetal alcohol spectrum disorder (Gupta et al., 2016). Although injections of the methyl donors MET and SAM altered animals’ addiction-like responses to cocaine (see section 2.1.2 “Behavioral effects”), dietary methyl donor supplementation (e.g., choline) that has more clinical relevance has not been tested. Moreover, a limited number of addictive substances have been examined in each paradigm. Stimulants, especially cocaine, have been the most studied class of substances, followed by alcohol. In contrast, findings regarding opioids and nicotine are relatively sparse. This is unfortunate, given the former being the most lethal and the latter the most prevalent of abused substances (Reynolds et al., 2021; United Nations Office on Drugs and Crime, 2023). Given the compelling evidence that environmental factors and life experiences induce substantial epigenetic modifications, which may in turn alter molecular, cellular, and behavioral processes, it is past time to expand the range of SUDs investigated in these epigenetic/behavioral paradigms.
4.3 Need for advanced functional and mechanistic studies
Many studies have documented the impact of environmental exposure on gene regulation and expression in reward signaling pathways in human SUDs or animal models (Szutorisz and Hurd, 2018; Walters and Kosten, 2019; Wanner et al., 2019; Smith et al., 2020; Swenson et al., 2020; Siomek-Gorecka et al., 2021). Nevertheless, their contribution to our understanding of underlying mechanisms has been limited. Many animal studies on the relations between epigenetic mechanisms and SUD vulnerability have focused on the epigenetic effects of substance exposure per se, rather than the effects of these epigenetic marks on future SUD vulnerability. Similarly, most studies that have associated prior environmental exposure with subsequent SUD by comparing their shared transcriptomic and epigenomic effects have not established a causal relationship. Evidence of correlations also does not help in elucidating whether substance-induced epigenetic changes are adaptive, or further, protective responses against adverse exposures or dysfunctional alterations that elevate SUD risks in the future (Rogers and Leslie, 2024). Therefore, the field would benefit from more studies explicitly designed to identify causal epigenetic relationships between environmental factors and SUD-related behavioral outcomes. For example, epigenetic changes induced by early-life events could be manipulated prior to SUD-like behavioral tests in animal models to confirm their functions in mediating SUD vulnerability. More detailed epigenetic regulatory mechanisms that mediate these processes could be investigated using cell-type specific gene knockdown or overexpression (Colby et al., 2003; Yamaguchi et al., 2018). Modifying DNA methylation and histone phosphorylation have emerged thus far as most consistently affecting relevant behaviors and could be prioritized in such investigations.
Newly developed techniques for targeted epigenome editing promise to accelerate the discovery of the details of epigenetic regulation underling SUD vulnerability. As with targeted editing of the genome, the epigenome can be precisely modified by introducing a fusion protein that contains an epigenetic catalyzing domain and a DNA-recognizing and -binding complex. The most commonly used platform is the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system. Fusing epigenetic modifiers (e.g., DNMTs, TETs, HATs, HMTs) to nuclease-deactivated Cas9 (dCas9) protein allows the epigenetic modifiers to be directed to the target sites by single-guide RNA (sgRNA) where they alter local epigenomic marks and regulate gene activity (Gjaltema and Rots, 2020). Compared to broad-based modification of epigenetic marks by means of viral vectors or perturbations of enzymatic activity, epigenome editing offers the prospect of more subtle interrogation of molecular targets and regulatory pathways implicated in correlational studies.
4.4 Comprehensive epigenetic profiling
Studies reviewed in section 3 “Environment and vulnerability to SUDs: role of epigenetics” primarily examined epigenetic marks and their gene targets that have been implicated in earlier addiction research, such as Fosb, Bdnf, and Drd2 (Larson et al., 2010; Gorwood et al., 2012; Li and Wolf, 2015). While examining molecules that are known to be involved in SUDs could further validate earlier findings, this does not offer new insights into the role of the vast number of genes and gene networks that have not been investigated in SUD research. On the other hand, epigenetic sequencing techniques that have matured in recent years (e.g., CUT&RUN, CUT&Tag, Third-generation sequencing) allow for comprehensive assessments of genomic sites that are regulated by epigenetic marks of interest. In addition, the studies reviewed above investigated changes in epigenetic modifications and their targeted genes in “bulk” brain tissue (i.e., undifferentiated according to cell type), therefore preventing dissection of regulatory processes specific to each cell type. To this end, single-cell transcriptomic and epigenomic sequencing offers the opportunity to profile high-resolution, cell type-specific epigenetic changes (Mazan-Mamczarz et al., 2022). Comprehensive epigenetic profiling provides opportunities to discover new gene targets and gene networks that are regulated by environmentally induced epigenetic changes and may influence subsequent SUD vulnerability.
4.5 Employing behavioral predictors of SUD vulnerability
Similar to human populations, lab animals exhibit different levels of vulnerability to addictive substances due to differential genetic composition and environmental exposures (Solberg Woods and Palmer, 2019; Carrette et al., 2021). However, vulnerability in these studies is typically evaluated retrospectively by degree of drug taking, sometimes indexed across multiple measures (Navandar et al., 2021; Kaplan et al., 2022). This leaves open the possibility that epigenetic differences identified between vulnerable and resilient animals are a consequence of differential drug consumption and not a cause of it. Therefore, it would be beneficial to further explore prospective measures of SUD vulnerability that do not themselves involve drug exposure or that equate drug exposure across subjects (Swain et al., 2021). One exciting recent development is UCSD’s “RATTACA” project, which can provide researchers with genetically diverse Heterogenous Stock rats that have been selected for a behavioral trait, such as vulnerability to a given form of SUD, based on their global genotypic profile (Johnson et al., 2023).
4.6 Integration of preclinical and clinical studies
The rationale for conducting animal studies of SUD and other pathological disorders is to further our understanding of the human conditions such studies seek to model. But this depends on both the construct validity of a given behavioral paradigm and the extent to which the brain’s anatomical, physiological and molecular milieux are conserved across species, neither of which can be taken for granted (Everitt et al., 2018; Rydell-Törmänen and Johnson, 2019; Uliana et al., 2022). Opportunities to test the clinical relevance of findings from the animal lab are limited, which means that considerable resources may be invested in studying mechanisms that may be ultimately of little clinical significance. This fundamental problem can be mitigated by selecting epigenetic targets that are deemed more likely to apply across species on a priori grounds. Such filtering can be facilitated, at least to a degree, through statistical integration of the results of large-scale genomic and epigenomic studies that have been conducted in parallel across humans and other species. Thus, overlapping lists of genes differentially expressed in humans and other species exposed to opioids have been compared through Rank-rank Hypergeometric Overlap analysis (Liu et al., 2021; Browne et al., 2023), a threshold-free algorithm to calculate concordance between two complete gene expression profiles (Plaisier et al., 2010). More recently, a comprehensive platform, Mergeomics, has been developed that can utilize a full spectrum of multiomic datasets across species, as well as across tissue and cell types, epigenetic marks and genotypes, to reveal commonalities in gene networks and molecular pathways underlying pathophysiology in humans and other species (Shu et al., 2016; Ding et al., 2021).
4.7 Conclusion
Given the associations between environmental adversity and future SUD vulnerability, it is critical to understand the molecular mechanisms that mediate them. Epigenetic modifications are primary candidates since they are sensitive to environmental stimuli as well as genetic variation, can be long-lasting and, as reviewed here, can impact SUD-related behaviors in preclinical models. While current theories are still largely inferential, the adoption of novel and more systematic approaches in preclinical models will allow more precise and comprehensive characterization of the role of interacting epigenetic mechanisms in vulnerability to SUD, from regulation of individual genes to reshaping gene expression networks and brain circuitry. This, in turn, will yield dividends in the development of novel interventions to mitigate the devastating impact of addictive drugs on individuals and society.
Author contributions
SL: Writing – original draft, Writing – review and editing. AH: Writing – review and editing. JG: Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funding for this study was provided by the NIH/NIDA grant U01 DA051993 (Gewirtz JC and Harris AC, co-PIs), the Hennepin Healthcare Research Institute Career Development Award (Harris AC), and University of Minnesota Doctoral Dissertation Fellowship (Liu SX).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2024.1462769/full#supplementary-material
References
Aguilar-Valles, A., Vaissière, T., Griggs, E. M., Mikaelsson, M. A., Takács, I. F., Young, E. J., et al. (2014). Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biol. Psychiatry 76, 57–65. doi: 10.1016/J.BIOPSYCH.2013.09.014
Ajonijebu, D. C., Abboussi, O., Russell, V. A., Mabandla, M. V., and Daniels, W. M. U. (2017). Epigenetics: A link between addiction and social environment. Cell Mol. Life Sci. 74, 2735–2747. doi: 10.1007/S00018-017-2493-1
Alati, R., Al Mamun, A., Williams, G. M., O’Callaghan, M., Najman, J. M., and Bor, W. (2006). In utero alcohol exposure and prediction of alcohol disorders in early adulthood: A birth cohort study. Arch. Gen. Psychiatry 63, 1009–1016. doi: 10.1001/ARCHPSYC.63.9.1009
Anderson, E. M., Sun, H., Guzman, D., Taniguchi, M., Cowan, C. W., Maze, I., et al. (2018b). Knockdown of the histone di-methyltransferase G9a in nucleus accumbens shell decreases cocaine self-administration, stress-induced reinstatement, and anxiety. Neuropsychopharmacology 44, 1370–1376. doi: 10.1038/s41386-018-0305-4
Anderson, E. M., Larson, E. B., Guzman, D., Wissman, A. M., Neve, R. L., Nestler, E. J., et al. (2018a). Overexpression of the histone dimethyltransferase g9a in nucleus accumbens shell increases cocaine self-administration, stress-induced reinstatement, and anxiety. J. Neurosci. 38, 803–813. doi: 10.1523/JNEUROSCI.1657-17.2017
Anderson, E. M., Tsvetkov, E., Galante, A., DeVries, D., McCue, L. M., Wood, D., et al. (2023). Epigenetic function during heroin self-administration controls future relapse-associated behavior in a cell type-specific manner. Proc. Natl. Acad. Sci. U.S.A. 120:e2210953120. doi: 10.1073/PNAS.2210953120
Andrews, A. J., and Luger, K. (2011). Nucleosome structure(s) and stability: Variations on a theme. Annu. Rev. Biophys. 40, 99–117. doi: 10.1146/ANNUREV-BIOPHYS-042910-155329
Anier, K., Malinovskaja, K., Aonurm-Helm, A., Zharkovsky, A., and Kalda, A. (2010). DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology 35, 2450–2461. doi: 10.1038/npp.2010.128
Anier, K., Malinovskaja, K., Pruus, K., Aonurm-Helm, A., Zharkovsky, A., and Kalda, A. (2014). Maternal separation is associated with DNA methylation and behavioural changes in adult rats. Eur. Neuropsychopharmacol. 24, 459–468. doi: 10.1016/J.EURONEURO.2013.07.012
Anier, K., Zharkovsky, A., and Kalda, A. (2013). S-adenosylmethionine modifies cocaine-induced DNA methylation and increases locomotor sensitization in mice. Int. J. Neuropsychopharmacol. 16, 2053–2066. doi: 10.1017/S1461145713000394
Aoki, C., Romeo, R. D., and Smith, S. S. (2017). Adolescence as a critical period for developmental plasticity. Brain Res. 1654, 85–86. doi: 10.1016/J.BRAINRES.2016.11.026
Bahi, A., and Dreyer, J. L. (2013). Striatal modulation of BDNF expression using microRNA124a-expressing lentiviral vectors impairs ethanol-induced conditioned-place preference and voluntary alcohol consumption. Eur. J. Neurosci. 38, 2328–2337. doi: 10.1111/EJN.12228
Bahi, A., and Dreyer, J. L. (2020). Lentiviral-mediated up-regulation of let-7d microRNA decreases alcohol intake through down-regulating the dopamine D3 receptor. Eur. Neuropsychopharmacol. 37, 70–81. doi: 10.1016/J.EURONEURO.2020.06.011
Bannister, A. J., and Kouzarides, T. (2003). Histone methylation: Recognizing the methyl mark. Methods Enzymol. 376, 269–288. doi: 10.1016/S0076-6879(03)76018-2
Barbier, E., Johnstone, A. L., Khomtchouk, B. B., Tapocik, J. D., Pitcairn, C., Rehman, F., et al. (2016). Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol. Psychiatry 22, 1746–1758. doi: 10.1038/mp.2016.131
Barbier, E., Tapocik, J. D., Juergens, N., Pitcairn, C., Borich, A., Schank, J. R., et al. (2015). DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J. Neurosci. 35, 6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bastle, R. M., Oliver, R. J., Gardiner, A. S., Pentkowski, N. S., Bolognani, F., Allan, A. M., et al. (2017). In silico identification and in vivo validation of miR-495 as a novel regulator of motivation for cocaine that targets multiple addiction-related networks in the nucleus accumbens. Mol. Psychiatry 23, 434–443. doi: 10.1038/mp.2016.238
Berglind, W. J., See, R. E., Fuchs, R. A., Ghee, S. M., Whitfield, T. W., Miller, S. W., et al. (2007). A BDNF infusion into the medial prefrontal cortex suppresses cocaine seeking in rats. Eur. J. Neurosci. 26, 757–766. doi: 10.1111/J.1460-9568.2007.05692.X
Besnard, A., Bouveyron, N., Kappes, V., Pascoli, V., Pages, C., Heck, N., et al. (2011). Alterations of molecular and behavioral responses to cocaine by selective inhibition of Elk-1 phosphorylation. J. Neurosci. 31, 14296–14307. doi: 10.1523/JNEUROSCI.2890-11.2011
Bilbo, S. D., Newsum, N. J., Sprunger, D. B., Watkins, L. R., Rudy, J. W., and Maier, S. F. (2007). Differential effects of neonatal handling on early life infection-induced alterations in cognition in adulthood. Brain Behav. Immun. 21, 332–342. doi: 10.1016/J.BBI.2006.10.005
Blanco, E., González-Ramírez, M., Alcaine-Colet, A., Aranda, S., and Di Croce, L. (2020). The bivalent genome: Characterization, structure, and regulation. Trends Genet. 36, 118–131. doi: 10.1016/J.TIG.2019.11.004
Bohnsack, J. P. W., Zhang, H., Wandling, G. M., He, D., Kyzar, E. J., Lasek, A. W., et al. (2022). Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure. Sci. Adv. 8:2748. doi: 10.1126/SCIADV.ABN2748/SUPPL_FILE/SCIADV.ABN2748_SM.PDF
Brami-Cherrier, K., Valjent, E., Hervé, D., Darragh, J., Corvol, J. C., Pages, C., et al. (2005). Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J. Neurosci. 25, 11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005
Breslau, N., Fenn, N., and Peterson, E. L. (1993). Early smoking initiation and nicotine dependence in a cohort of young adults. Drug Alcohol Depend. 33, 129–137. doi: 10.1016/0376-8716(93)90054-T
Brown, A. S., Van Os, J., Driessens, C., Hoek, H. W., and Susser, E. S. (2000). Further evidence of relation between prenatal famine and major affective disorder. Am. J. Psychiatry 157, 190–195. doi: 10.1176/APPI.AJP.157.2.190/ASSET/IMAGES/LARGE/AV7T1.JPEG
Browne, C. J., Futamura, R., Minier-Toribio, A., Hicks, E. M., Ramakrishnan, A., Martínez-Rivera, F. J., et al. (2023). Transcriptional signatures of heroin intake and relapse throughout the brain reward circuitry in male mice. Sci. Adv. 9:eadg8558. doi: 10.1126/SCIADV.ADG8558
Cadet, J. L. (2016). Epigenetics of stress, addiction, and resilience: Therapeutic implications. Mol. Neurobiol. 53:545. doi: 10.1007/S12035-014-9040-Y
Campbell, R. R., Kramár, E. A., Pham, L., Beardwood, J. H., Augustynski, A. S., López, A. J., et al. (2021). HDAC3 activity within the nucleus accumbens regulates cocaine-induced plasticity and behavior in a cell-type-specific manner. J. Neurosci. 41, 2814–2827. doi: 10.1523/JNEUROSCI.2829-20.2021
Carrette, L. L. G., de Guglielmo, G., Kallupi, M., Maturin, L., Brennan, M., Boomhower, B., et al. (2021). The cocaine and oxycodone biobanks, two repositories from genetically diverse and behaviorally characterized rats for the study of addiction. eNeuro 8, doi: 10.1523/ENEURO.0033-21.2021
Carter, S. D., Mifsud, K. R., and Reul, J. M. H. M. (2017). Acute stress enhances epigenetic modifications but does not affect the constitutive binding of pCREB to immediate-early gene promoters in the rat hippocampus. Front. Mol. Neurosci. 10:416. doi: 10.3389/FNMOL.2017.00416
Catale, C., Bussone, S., Lo Iacono, L., Viscomi, M. T., Palacios, D., Troisi, A., et al. (2020). Exposure to different early-life stress experiences results in differentially altered DNA methylation in the brain and immune system. Neurobiol. Stress 13:100249. doi: 10.1016/J.YNSTR.2020.100249
Chandrasekar, V., and Dreyer, J. L. (2009). microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 42, 350–362. doi: 10.1016/J.MCN.2009.08.009
Chandrasekar, V., and Dreyer, J. L. (2011). Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36:1149. doi: 10.1038/NPP.2010.250
Chekulaeva, M., and Rajewsky, N. (2019). Roles of long noncoding RNAs and circular RNAs in translation. Cold Spring Harb. Perspect. Biol. 11:2680. doi: 10.1101/CSHPERSPECT.A032680
Chen, H., Qiang, H., Fan, K., Wang, S., and Zheng, Z. (2014). The snoRNA MBII-52 regulates cocaine-induced conditioned place preference and locomotion in mice. PLoS One 9:e99986. doi: 10.1371/JOURNAL.PONE.0099986
Chen, M., Zhang, X., and Hao, W. (2019). H3K4 dimethylation at FosB promoter in the striatum of chronic stressed rats promotes morphine-induced conditioned place preference. PLoS One 14:e0221506. doi: 10.1371/JOURNAL.PONE.0221506
Chen, W. S., Xu, W. J., Zhu, H. Q., Gao, L., Lai, M. J., Zhang, F. Q., et al. (2016). Effects of histone deacetylase inhibitor sodium butyrate on heroin seeking behavior in the nucleus accumbens in rats. Brain Res. 1652, 151–157. doi: 10.1016/J.BRAINRES.2016.10.007
Clements, A., Poux, A. N., Lo, W. S., Pillus, L., Berger, S. L., and Marmorstein, R. (2003). Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12, 461–473. doi: 10.1016/S1097-2765(03)00288-0
Colby, C. R., Whisler, K., Steffen, C., Nestler, E. J., and Self, D. W. (2003). Striatal cell type-specific overexpression of ΔFosB enhances incentive for cocaine. J. Neurosci. 23, 2488. doi: 10.1523/JNEUROSCI.23-06-02488.2003
Cosgrove, K. P., Hunter, R. G., and Carroll, M. E. (2002). Wheel-running attenuates intravenous cocaine self-administration in rats: Sex differences. Pharmacol. Biochem. Behav. 73, 663–671. doi: 10.1016/S0091-3057(02)00853-5
Dai, C.-L., Chen, C.-C., Richardson, G. B., and Gordon, H. R. D. (2020). Managing substance use disorder through a walking/running training program. Subst. Abuse 14:1178221820936681. doi: 10.1177/1178221820936681
Damez-Werno, D. M., Sun, H. S., Scobie, K. N., Shao, N., Rabkin, J., Dias, C., et al. (2016). Histone arginine methylation in cocaine action in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 113, 9623–9628. doi: 10.1073/PNAS.1605045113
Darcq, E., Warnault, V., Phamluong, K., Besserer, G. M., Liu, F., and Ron, D. (2014). MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol. Psychiatry 20, 1240–1250. doi: 10.1038/mp.2014.120
Deaton, A. M., and Bird, A. (2011). CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022. doi: 10.1101/GAD.2037511
Ding, J., Blencowe, M., Nghiem, T., Ha, S. M., Chen, Y. W., Li, G., et al. (2021). Mergeomics 2.0: A web server for multi-omics data integration to elucidate disease networks and predict therapeutics. Nucleic Acids Res. 49, W375–W387. doi: 10.1093/NAR/GKAB405
Dinieri, J. A., Wang, X., Szutorisz, H., Spano, S. M., Kaur, J., Casaccia, P., et al. (2011). Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol. Psychiatry 70, 763–769. doi: 10.1016/J.BIOPSYCH.2011.06.027
Ehrlich, M., and Ehrlich, K. C. (2014). DNA cytosine methylation and hydroxymethylation at the borders. Epigenomics 6:563. doi: 10.2217/EPI.14.48
Enoch, M. A. (2011). The role of early life stress as a predictor for alcohol and drug dependence. Psychopharmacology 214:17. doi: 10.1007/S00213-010-1916-6
Everitt, B. J., Giuliano, C., and Belin, D. (2018). Addictive behaviour in experimental animals: Prospects for translation. Philos. Trans. R. Soc. B Biol. Sci. 373:27. doi: 10.1098/RSTB.2017.0027
Fafard-Couture, É, Bergeron, D., Couture, S., Abou-Elela, S., and Scott, M. S. (2021). Annotation of snoRNA abundance across human tissues reveals complex snoRNA-host gene relationships. Genome Biol. 22, 1–24. doi: 10.1186/s13059-021-02391-2
Feng, J., Shao, N., Szulwach, K. E., Vialou, V., Huynh, J., Zhong, C., et al. (2015). Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 18, 536–544. doi: 10.1038/nn.3976
Ferguson, D., Koo, J. W., Feng, J., Heller, E., Rabkin, J., Heshmati, M., et al. (2013). Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J. Neurosci. 33, 16088–16098. doi: 10.1523/JNEUROSCI.1284-13.2013
Finegersh, A., and Homanics, G. E. (2014). Paternal alcohol exposure reduces alcohol drinking and increases behavioral sensitivity to alcohol selectively in male offspring. PLoS One 9:e99078. doi: 10.1371/JOURNAL.PONE.0099078
Forget, B., Garcia, E. M., Godino, A., Rodriguez, L. D., Kappes, V., Poirier, P., et al. (2021). Cell-type- and region-specific modulation of cocaine seeking by micro-RNA-1 in striatal projection neurons. Mol. Psychiatry 27, 918–928. doi: 10.1038/s41380-021-01328-2
Fossati, C., Torre, G., Vasta, S., Giombini, A., Quaranta, F., Papalia, R., et al. (2021). physical exercise and mental health: The routes of a reciprocal relation. Int. J. Environ. Res. Public Health 18:2364. doi: 10.3390/IJERPH182312364
Gjaltema, R. A. F., and Rots, M. G. (2020). Advances of epigenetic editing. Curr. Opin. Chem. Biol. 57, 75–81. doi: 10.1016/J.CBPA.2020.04.020
Golding, J., Ellis, G., Gregory, S., Birmingham, K., Iles-Caven, Y., Rai, D., et al. (2017). Grand-maternal smoking in pregnancy and grandchild’s autistic traits and diagnosed autism. Sci. Rep. 7:46179. doi: 10.1038/SREP46179
Goldstein, R. B., Smith, S. M., Chou, S. P., Saha, T. D., Jung, J., Zhang, H., et al. (2016). The epidemiology of DSM-5 posttraumatic stress disorder in the United States: Results from the national epidemiologic survey on alcohol and related conditions-III. Soc. Psychiatry Psychiatr. Epidemiol. 51, 1137–1148. doi: 10.1007/S00127-016-1208-5/TABLES/5
Goll, M. G., and Bestor, T. H. (2005). Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514. doi: 10.1146/ANNUREV.BIOCHEM.74.010904.153721
Gorwood, P., Le Strat, Y., Ramoz, N., Dubertret, C., Moalic, J. M., and Simonneau, M. (2012). Genetics of dopamine receptors and drug addiction. Hum. Genet. 131, 803–822. doi: 10.1007/S00439-012-1145-7
Greenwood, B. N., Foley, T. E., Le, T. V., Strong, P. V., Loughridge, A. B., Day, H. E. W., et al. (2011). Long-term voluntary wheel running is rewarding and produces plasticity in the mesolimbic reward pathway. Behav. Brain Res. 217, 354–362. doi: 10.1016/J.BBR.2010.11.005
Greer, E. L., and Shi, Y. (2012). Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357. doi: 10.1038/nrg3173
Gupta, K. K., Gupta, V. K., and Shirasaka, T. (2016). An update on fetal alcohol syndrome-pathogenesis, risks, and treatment. Alcohol Clin. Exp. Res. 40, 1594–1602. doi: 10.1111/ACER.13135
Hamilton, P. J., and Nestler, E. J. (2019). Epigenetics and addiction. Curr. Opin. Neurobiol. 59, 128–136. doi: 10.1016/J.CONB.2019.05.005
Han, J., Li, Y., Wang, D., Wei, C., Yang, X., and Sui, N. (2010). Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur. J. Pharmacol. 642, 93–98. doi: 10.1016/J.EJPHAR.2010.05.050
Hargreaves, D. C. (2021). Chromatin openness requires continuous SWI/SNF activity. Nat. Genet. 53, 263–264. doi: 10.1038/S41588-021-00781-7
Heller, E. A., Cates, H. M., Peña, C. J., Sun, H., Shao, N., Feng, J., et al. (2014). Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci. 17, 1720–1727. doi: 10.1038/nn.3871
Hock, R. S., Bryce, C. P., Fischer, L., First, M. B., Fitzmaurice, G. M., Costa, P. T., et al. (2018). Childhood malnutrition and maltreatment are linked with personality disorder symptoms in adulthood: Results from a Barbados lifespan cohort. Psychiatry Res. 269:301. doi: 10.1016/J.PSYCHRES.2018.05.085
Hollander, J. A., Im, H. I., Amelio, A. L., Kocerha, J., Bali, P., Lu, Q., et al. (2010). Striatal microRNA controls cocaine intake through CREB signalling. Nature 466, 197–202. doi: 10.1038/NATURE09202
Hong, Q., Xu, W., Lin, Z., Liu, J., Chen, W., Zhu, H., et al. (2021). Role of GABRD gene methylation in the nucleus accumbens in heroin-seeking behavior in rats. Front. Pharmacol. 11:2210. doi: 10.3389/FPHAR.2020.612200/BIBTEX
Ignaszewski, M. J. (2021). The epidemiology of drug abuse. J. Clin. Pharmacol. 61, S10–S17. doi: 10.1002/jcph.1937
Illingworth, R., Kerr, A., DeSousa, D., Jørgensen, H., Ellis, P., Stalker, J., et al. (2008). A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 6:e22. doi: 10.1371/JOURNAL.PBIO.0060022
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi: 10.1126/SCIENCE.1210597/SUPPL_FILE/ITO.SOM.PDF
Itzhak, Y., Ergui, I., and Young, J. I. (2014). Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring. Mol. Psychiatry 20, 232–239. doi: 10.1038/mp.2014.7
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/RNA.035667.112
Johnson, B. B., Sanches, T. M., Okamoto, M. H., Nguyen, K.-M., Ortez, C. A., Polesskaya, O., et al. (2023). RATTACA: Genetic predictions in heterogeneous stock rats offer a new tool for genetic correlation and experimental design. bioRxiv [Preprint] doi: 10.1101/2023.09.18.558279 bioRxiv: 2023.09.18.558279.
Jones, P. A. (2012). Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi: 10.1038/NRG3230
Jonkman, S., and Kenny, P. J. (2012). Molecular, cellular, and structural mechanisms of cocaine addiction: A key role for MicroRNAs. Neuropsychopharmacology 2013, 198–211. doi: 10.1038/npp.2012.120
Kalda, A., Heidmets, L. T., Shen, H. Y., Zharkovsky, A., and Chen, J. F. (2007). Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav. Brain Res. 181, 76–84. doi: 10.1016/J.BBR.2007.03.027
Kaplan, G., Xu, H., Abreu, K., and Feng, J. (2022). DNA epigenetics in addiction susceptibility. Front. Genet. 13:806685. doi: 10.3389/FGENE.2022.806685/BIBTEX
Kendler, K. S., Ohlsson, H., Sundquist, J., and Sundquist, K. (2018). Transmission of alcohol use disorder across three generations: A Swedish national study. Psychol. Med. 48, 33–42. doi: 10.1017/S0033291717000794
Kennedy, P. J., Feng, J., Robison, A. J., Maze, I., Badimon, A., Mouzon, E., et al. (2013). Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat. Neurosci. 16, 434–440. doi: 10.1038/nn.3354
Kenny, P. J. (2022). Epigenetics, microRNA, and addiction. Dialogues Clin. Neurosci. 16, 335–344. doi: 10.31887/DCNS.2014.16.3/PKENNY
Kinnally, E. L., Feinberg, C., Kim, D., Ferguson, K., Leibel, R., Coplan, J. D., et al. (2011). DNA methylation as a risk factor in the effects of early life stress. Brain Behav. Immun. 25, 1548–1553. doi: 10.1016/J.BBI.2011.05.001
Kiss, T. (2002). Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 246–259. doi: 10.1016/S0092-8674(02)00718-3
Kitaba, N. T., Knudsen, G. T. M., Johannessen, A., Rezwan, F. I., Malinovschi, A., Oudin, A., et al. (2023). Fathers’ preconception smoking and offspring DNA methylation. Clin. Epigenet. 15, 1–16. doi: 10.1186/S13148-023-01540-7
Kousik, S. M., Napier, T. C., and Carvey, P. M. (2012). The effects of psychostimulant drugs on blood brain barrier function and neuroinflammation. Front. Pharmacol. 3:121. doi: 10.3389/FPHAR.2012.00121/BIBTEX
Kouzarides, T. (2002). Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12, 198–209. doi: 10.1016/S0959-437X(02)00287-3
Kumar, A., Choi, K. H., Renthal, W., Tsankova, N. M., Theobald, D. E. H., Truong, H. T., et al. (2005). Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314. doi: 10.1016/J.NEURON.2005.09.023
Kyzar, E. J., Bohnsack, J. P., Zhang, H., and Pandey, S. C. (2019). MicroRNA-137 drives epigenetic reprogramming in the adult amygdala and behavioral changes after adolescent alcohol exposure. eNeuro 6, doi: 10.1523/ENEURO.0401-19.2019
Längst, G., and Manelyte, L. (2015). Chromatin remodelers: From function to dysfunction. Genes 6:299. doi: 10.3390/GENES6020299
Laplant, Q., Vialou, V., Covington, H. E., Dumitriu, D., Feng, J., Warren, B. L., et al. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137–1143. doi: 10.1038/NN.2619
Larsen, B., and Luna, B. (2018). Adolescence as a neurobiological critical period for the development of higher-order cognition. Neurosci. Biobehav. Rev. 94:179. doi: 10.1016/J.NEUBIOREV.2018.09.005
Larson, E. B., Akkentli, F., Edwards, S., Graham, D. L., Simmons, D. L., Alibhai, I. N., et al. (2010). Striatal regulation of ΔFosB, FosB, and cFos during cocaine self-administration and withdrawal. J. Neurochem. 115, 112–122. doi: 10.1111/J.1471-4159.2010.06907.X
Lassi, Z. S., Imam, A. M., Dean, S. V., and Bhutta, Z. A. (2014). Preconception care: Caffeine, smoking, alcohol, drugs and other environmental chemical/radiation exposure. Reprod. Health 11, 1–12. doi: 10.1186/1742-4755-11-S3-S6
Lesser-Katz, M. (1982). Some effects of maternal drug addiction on the neonate. Int. J. Addict. 17, 887–896. doi: 10.3109/10826088209056335
Levine, A. A., Guan, Z., Barco, A., Xu, S., Kandel, E. R., and Schwartz, J. H. (2005). CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc. Natl. Acad. Sci. U.S.A. 102, 19186–19191. doi: 10.1073/PNAS.0509735102
Li, X., and Wolf, M. E. (2015). Multiple faces of BDNF in cocaine addiction. Behav. Brain Res. 10:240. doi: 10.1016/J.BBR.2014.11.018
Li, Y., Zhu, R., Wang, W., Fu, D., Hou, J., Ji, S., et al. (2015). Arginine methyltransferase 1 in the nucleus accumbens regulates behavioral effects of cocaine. J. Neurosci. 35, 12890–12902. doi: 10.1523/JNEUROSCI.0246-15.2015
Lister, R., Mukamel, E. A., Nery, J. R., Urich, M., Puddifoot, C. A., Johnson, N. D., et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905. doi: 10.1126/SCIENCE.1237905
Liu, S. X., Gades, M. S., Swain, Y., Ramakrishnan, A., Harris, A. C., Tran, P. V., et al. (2021). Repeated morphine exposure activates synaptogenesis and other neuroplasticity-related gene networks in the dorsomedial prefrontal cortex of male and female rats. Drug Alcohol Depend. 221:108598. doi: 10.1016/j.drugalcdep.2021.108598
Lo, W. S., Trievel, R. C., Rojas, J. R., Duggan, L., Hsu, J. Y., Allis, C. D., et al. (2000). Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5, 917–926. doi: 10.1016/S1097-2765(00)80257-9
Luan, M., Zhang, X., Fang, G., Liang, H., Yang, F., Song, X., et al. (2022). Preconceptional paternal alcohol consumption and the risk of child behavioral problems: A prospective cohort study. Sci. Rep. 12, 1–11. doi: 10.1038/s41598-022-05611-2
Malvaez, M., Mhillaj, E., Matheos, D. P., Palmery, M., and Wood, M. A. (2011). CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J. Neurosci. 31, 16941–16948. doi: 10.1523/JNEUROSCI.2747-11.2011
Massart, R., Barnea, R., Dikshtein, Y., Suderman, M., Meir, O., Hallett, M., et al. (2015). Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J. Neurosci. 35:8042. doi: 10.1523/JNEUROSCI.3053-14.2015
Mazan-Mamczarz, K., Ha, J., De, S., and Sen, P. (2022). Single-cell analysis of the transcriptome and epigenome. Methods Mol. Biol. 2399:21. doi: 10.1007/978-1-0716-1831-8_3
Maze, I., Covingtoni, H. E., Dietz, D. M., Laplant, Q., Renthal, W., Russo, S. J., et al. (2010). Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216. doi: 10.1126/SCIENCE.1179438/SUPPL_FILE/MAZE.SOM.PDF
McCabe, S. E., Schulenberg, J. E., Schepis, T. S., McCabe, V. V., and Veliz, P. T. (2022). longitudinal analysis of substance use disorder symptom severity at age 18 years and substance use disorder in adulthood. JAMA Netw. Open 5:e225324. doi: 10.1001/JAMANETWORKOPEN.2022.5324
McCarthy, D. M., Brown, A. N., and Bhide, P. G. (2012). Regulation of BDNF expression by cocaine. Yale J. Biol. Med. 85:437.
McCowan, T. J., Dhasarathy, A., and Carvelli, L. (2015). The epigenetic mechanisms of amphetamine. J. Addict. Prev. 2015:1. doi: 10.13188/2330-2178.S100001
McElroy, S. L., Winham, S. J., Cuellar-Barboza, A. B., Colby, C. L., Ho, A. M. C., Sicotte, H., et al. (2018). Bipolar disorder with binge eating behavior: A genome-wide association study implicates PRR5-ARHGAP8. Transl. Psychiatry 8:40. doi: 10.1038/S41398-017-0085-3
Mews, P., Walker, D. M., and Nestler, E. J. (2018). Epigenetic priming in drug addiction. Cold Spring Harb. Symp. Quant. Biol. 83:131. doi: 10.1101/SQB.2018.83.037663
Mews, P., Zee, B. M., Liu, S., Donahue, G., Garcia, B. A., and Berger, S. L. (2014). Histone methylation has dynamics distinct from those of histone acetylation in cell cycle reentry from quiescence. Mol. Cell Biol. 34, 3968–3980. doi: 10.1128/MCB.00763-14
Miller, J. L., and Grant, P. A. (2013). The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 61:289. doi: 10.1007/978-94-007-4525-4_13
Mineka, S., and Kihlstrom, J. F. (1978). Unpredictable and uncontrollable events: A new perspective on experimental neurosis. J. Abnorm. Psychol. 87, 256–271. doi: 10.1037/0021-843X.87.2.256
Montesinos, J., Pascual, M., Rodríguez-Arias, M., Miñarro, J., and Guerri, C. (2016). Involvement of TLR4 in the long-term epigenetic changes, rewarding and anxiety effects induced by intermittent ethanol treatment in adolescence. Brain Behav. Immun. 53, 159–171. doi: 10.1016/J.BBI.2015.12.006
Moonat, S., Sakharkar, A. J., Zhang, H., Tang, L., and Pandey, S. C. (2013). Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol. Psychiatry 73, 763–773. doi: 10.1016/J.BIOPSYCH.2013.01.012
Most, D., Salem, N. A., Tiwari, G. R., Blednov, Y. A., Mayfield, R. D., and Harris, R. A. (2019). Silencing synaptic MicroRNA-411 reduces voluntary alcohol consumption in mice. Addict. Biol. 24, 604–616. doi: 10.1111/ADB.12625
Murthy, S., and Gould, E. (2018). Early life stress in rodents: Animal models of illness or resilience? Front. Behav. Neurosci. 12:157. doi: 10.3389/FNBEH.2018.00157/BIBTEX
Navandar, M., Martín-García, E., Maldonado, R., Lutz, B., Gerber, S., and Ruiz, et al. (2021). Transcriptional signatures in prefrontal cortex confer vulnerability versus resilience to food and cocaine addiction-like behavior. Sci. Rep. 11, 1–11. doi: 10.1038/s41598-021-88363-9
Nichols, M. M. (1967). Acute alcohol withdrawal syndrome in a newborn. Am. J. Dis. Child. 113, 714–715. doi: 10.1001/archpedi.1967.02090210128015
Odgers, C. L., Caspi, A., Nagin, D. S., Piquero, A. R., Slutske, W. S., Milne, B. J., et al. (2008). Is it important to prevent early exposure to drugs and alcohol among adolescents? Psychol. Sci. 19, 1037–1044. doi: 10.1111/j.1467-9280.2008.02196.x
Pascoli, V., Cahill, E., Bellivier, F., Caboche, J., and Vanhoutte, P. (2014). Extracellular signal-regulated protein kinases 1 and 2 activation by addictive drugs: A signal toward pathological adaptation. Biol. Psychiatry 76, 917–926. doi: 10.1016/J.BIOPSYCH.2014.04.005
Pastor, V., Host, L., Zwiller, J., and Bernabeu, R. (2011). Histone deacetylase inhibition decreases preference without affecting aversion for nicotine. J. Neurochem. 116, 636–645. doi: 10.1111/J.1471-4159.2010.07149.X
Patrick, S. W., Barfield, W. D., and Poindexter, B. B. (2020). Neonatal opioid withdrawal syndrome. Pediatrics 146:a039669. doi: 10.1542/PEDS.2020-029074/75310
Peretti-Watel, P., Constance, J., Seror, V., and Beck, F. (2009). Working conditions, job dissatisfaction and smoking behaviours among French clerks and manual workers. J. Occup. Environ. Med. 51, 343–350. doi: 10.1097/JOM.0B013E31819464FE
Peterson, A. B., Abel, J. M., and Lynch, W. J. (2014). Dose-dependent effects of wheel running on cocaine-seeking and prefrontal cortex Bdnf exon IV expression in rats. Psychopharmacology 231, 1305–1314. doi: 10.1007/S00213-013-3321-4
Peterson, C., Li, M., Xu, L., Mikosz, C. A., and Luo, F. (2021). Assessment of annual cost of substance use disorder in us hospitals. JAMA Netw. Open 4, 1–8. doi: 10.1001/jamanetworkopen.2021.0242
Plaisier, S. B., Taschereau, R., Wong, J. A., and Graeber, T. G. (2010). Rank–rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38:e169. doi: 10.1093/NAR/GKQ636
Radliff, K. M., Wheaton, J. E., Robinson, K., and Morris, J. (2012). Illuminating the relationship between bullying and substance use among middle and high school youth. Addict. Behav. 37, 569–572. doi: 10.1016/J.ADDBEH.2012.01.001
Raivio, N., Miettinen, P., and Kiianmaa, K. (2014). Innate BDNF expression is associated with ethanol intake in alcohol-preferring AA and alcohol-avoiding ANA rats. Brain Res. 1579, 74–83. doi: 10.1016/J.BRAINRES.2014.07.006
Renthal, W., and Nestler, E. J. (2009). Histone acetylation in drug addiction. Semin. Cell Dev. Biol. 20, 387–394. doi: 10.1016/J.SEMCDB.2009.01.005
Renthal, W., Maze, I., Krishnan, V., Covington, H. E., Xiao, G., Kumar, A., et al. (2007). Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56, 517–529. doi: 10.1016/J.NEURON.2007.09.032
Reynolds, A. J., Eales, L., Suh-Ruu, Ou, M., Mondi, C. F., Giovanelli, A., et al. (2021). Prevalence of substance use disorders by time since first substance use among young people in the US. JAMA Pediatr. 175, 640–643. doi: 10.1001/JAMAPEDIATRICS.2020.6981
Reynolds, L. M., Yetnikoff, L., Pokinko, M., Wodzinski, M., Epelbaum, J. G., Lambert, L. C., et al. (2019). Early adolescence is a critical period for the maturation of inhibitory behavior. Cereb. Cortex 29:3676. doi: 10.1093/CERCOR/BHY247
Robertson, C. L., Ishibashi, K., Chudzynski, J., Mooney, L. J., Rawson, R. A., Dolezal, B. A., et al. (2016). Effect of exercise training on striatal dopamine D2/D3 receptors in methamphetamine users during behavioral treatment. Neuropsychopharmacology 41, 1629–1636. doi: 10.1038/NPP.2015.331
Robison, L. S., Swenson, S., Hamilton, J., and Thanos, P. K. (2018). Exercise reduces dopamine D1R and increases D2R in rats: Implications for addiction. Med. Sci. Sports Exerc. 50, 1596–1602. doi: 10.1249/MSS.0000000000001627
Rogers, A., and Leslie, F. (2024). Addiction neurobiologists should study resilience. Addict. Neurosci. 11:100152. doi: 10.1016/J.ADDICN.2024.100152
Rogge, G. A., Singh, H., Dang, R., and Wood, M. A. (2013). HDAC3 is a negative regulator of cocaine-context-associated memory formation. J. Neurosci. 33, 6623–6632. doi: 10.1523/JNEUROSCI.4472-12.2013
Romieu, P., Host, L., Gobaille, S., Sandner, G., Aunis, D., and Zwiller, J. (2008). Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats. J. Neurosci. 28, 9342–9348. doi: 10.1523/JNEUROSCI.0379-08.2008
Rompala, G. R., Finegersh, A., Slater, M., and Homanics, G. E. (2017). Paternal preconception alcohol exposure imparts intergenerational alcohol-related behaviors to male offspring on a pure C57BL/6J background HHS public access. Alcohol 60, 169–177. doi: 10.1016/j.alcohol.2016.11.001
Rosen, T. S., and Johnson, H. L. (1988). Drug-addicted mothers, their infants, and SIDS. Ann. N. Y. Acad. Sci. 533, 89–95.
Rossetto, D., Avvakumov, N., and Côté, J. (2012). Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 7:1098. doi: 10.4161/EPI.21975
Rusconi, F., and Battaglioli, E. (2018). Acute stress-induced epigenetic modulations and their potential protective role toward depression. Front. Mol. Neurosci. 11:361214. doi: 10.3389/FNMOL.2018.00184/BIBTEX
Rybak-Wolf, A., Stottmeister, C., Glažar, P., Jens, M., Pino, N., Giusti, S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885. doi: 10.1016/J.MOLCEL.2015.03.027
Rydell-Törmänen, K., and Johnson, J. R. (2019). The applicability of mouse models to the study of human disease. Methods Mol. Biol. 1940, 3–22. doi: 10.1007/978-1-4939-9086-3_1
Saba, L. M., Hoffman, P. L., Homanics, G. E., Mahaffey, S., Daulatabad, S. V., Janga, S. C., et al. (2021). A long non-coding RNA (Lrap) modulates brain gene expression and levels of alcohol consumption in rats. Genes Brain Behav. 20, e12698. doi: 10.1111/GBB.12698
Saberian, H., Asgari Taei, A., Torkaman-Boutorabi, A., Riahi, E., Aminyavari, S., Naghizadeh, A., et al. (2021). Effect of histone acetylation on maintenance and reinstatement of morphine-induced conditioned place preference and ΔFosB expression in the nucleus accumbens and prefrontal cortex of male rats. Behav. Brain Res. 414:113477. doi: 10.1016/j.bbr.2021.113477
Sadeghzadeh, F., Babapour, V., and Haghparast, A. (2017). Food deprivation facilitates reinstatement of morphine-induced conditioned place preference: Role of intra-accumbal dopamine D2-like receptors in associating reinstatement of morphine CPP with stress. Synapse 71:51. doi: 10.1002/SYN.21951
Sakharkar, A. J., Zhang, H., Tang, L., Baxstrom, K., Shi, G., Moonat, S., et al. (2014). Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: A role in anxiety-like and alcohol-drinking behaviours. Int. J. Neuropsychopharmacol. 17, 1207–1220. doi: 10.1017/S1461145714000054
Salery, M., Dos Santos, M., Saint-Jour, E., Moumné, L., Pagès, C., Kappès, V., et al. (2017). Activity-regulated cytoskeleton-associated protein accumulates in the nucleus in response to cocaine and acts as a brake on chromatin remodeling and long-term behavioral alterations. Biol. Psychiatry 81, 573–584. doi: 10.1016/J.BIOPSYCH.2016.05.025
Schaefer, A., Im, H. I., Venø, M. T., Fowler, C. D., Min, A., Intrator, A., et al. (2010). Argonaute 2 in dopamine 2 receptor–expressing neurons regulates cocaine addiction. J. Exp. Med. 207:1843. doi: 10.1084/JEM.20100451
Schenker, S., Yang, Y., Johnson, R. F., Downing, J. W., Schenken, R. S., Henderson, G. I., et al. (1993). The transfer of cocaine and its metabolites across the term human placenta. Clin. Pharmacol. Ther. 53, 329–339. doi: 10.1038/CLPT.1993.29
Schoenbaum, G., Stalnaker, T. A., and Shaham, Y. (2007). A role for BDNF in cocaine reward and relapse. Nat. Neurosci. 10:935. doi: 10.1038/NN0807-935
Schwarz, J. M., Hutchinson, M. R., and Bilbo, S. D. (2011). Early-life experience decreases drug-induced reinstatement of morphine CPP in adulthood via microglial-specific epigenetic programming of anti-inflammatory IL-10 expression. J. Neurosci. 31, 17835–17847. doi: 10.1523/JNEUROSCI.3297-11.2011
Shahbazian, M. D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Shan, C. M., Bao, K., Diedrich, J., Chen, X., Lu, C., Yates, J. R., et al. (2020). The INO80 complex regulates epigenetic inheritance of heterochromatin. Cell Rep. 33:8561. doi: 10.1016/J.CELREP.2020.108561
Shang, R., Lee, S., Senavirathne, G., and Lai, E. C. (2023). microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 24, 816–833. doi: 10.1038/s41576-023-00611-y
Sharma, A., Sharma, A., Gupta, S., and Thapar, S. (2019). Study of family burden in substance dependence: A tertiary care hospital-based study. Indian J. Psychiatry 61:131. doi: 10.4103/PSYCHIATRY.INDIANJPSYCHIATRY_123_15
Shen, H. Y., Kalda, A., Yu, L., Ferrara, J., Zhu, J., and Chen, J. F. (2008). Additive effects of histone deacetylase inhibitors and amphetamine on histone H4 acetylation, cAMP responsive element binding protein phosphorylation and ΔFosB expression in the striatum and locomotor sensitization in mice. Neuroscience 157, 644–655. doi: 10.1016/J.NEUROSCIENCE.2008.09.019
Shen, Q., Xie, B., Galaj, E., Yu, H., Li, X., Lu, Y., et al. (2022). CircTmeff-1 in the nucleus accumbens regulates the reconsolidation of cocaine-associated memory. Brain Res. Bull. 185, 64–73. doi: 10.1016/J.BRAINRESBULL.2022.04.010
Sheng, J., Lv, Z. G., Wang, L., Zhou, Y., and Hui, B. (2011). Histone H3 phosphoacetylation is critical for heroin-induced place preference. Neuroreport 22, 575–580. doi: 10.1097/WNR.0B013E328348E6AA
Shi, D. Q., Ali, I., Tang, J., and Yang, W. C. (2017). New insights into 5hmC DNA modification: Generation, distribution and function. Front. Genet. 8:100. doi: 10.3389/FGENE.2017.00100/BIBTEX
Shu, L., Zhao, Y., Kurt, Z., Byars, S. G., Tukiainen, T., Kettunen, J., et al. (2016). Mergeomics: Multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genom. 17:1–16. doi: 10.1186/S12864-016-3198-9
Shvedunova, M., and Akhtar, A. (2022). Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349. doi: 10.1038/s41580-021-00441-y
Siomek-Gorecka, A., Dlugosz, A., and Czarnecki, D. (2021). The molecular basis of alcohol use disorder (AUD). Genetics, epigenetics, and nutrition in AUD: An amazing triangle. Int. J. Mol. Sci. 22:4262. doi: 10.3390/IJMS22084262
Smith, A., Kaufman, F., Sandy, M. S., and Cardenas, A. (2020). Cannabis exposure during critical windows of development: Epigenetic and molecular pathways implicated in neuropsychiatric disease. Curr. Environ. Health Rep. 7, 325–342. doi: 10.1007/S40572-020-00275-4
Solberg Woods, L. C., and Palmer, A. A. (2019). Using heterogeneous stocks for fine-mapping genetically complex traits. Methods Mol. Biol. 2018:233. doi: 10.1007/978-1-4939-9581-3_11
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2020). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi: 10.1038/s41580-020-00315-9
Stipanovich, A., Valjent, E., Matamales, M., Nishi, A., Ahn, J. H., Maroteaux, M., et al. (2008). A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature 453, 879–884. doi: 10.1038/NATURE06994
Strang, J., Volkow, N. D., Degenhardt, L., Hickman, M., Johnson, K., Koob, G. F., et al. (2020). Opioid use disorder. Nat. Rev. Dis. Prim. 6:2589. doi: 10.1038/S41572-019-0137-5
Sun, J., Wang, L., Jiang, B., Hui, B., Lv, Z., and Ma, L. (2008). The effects of sodium butyrate, an inhibitor of histone deacetylase, on the cocaine- and sucrose-maintained self-administration in rats. Neurosci. Lett. 441, 72–76. doi: 10.1016/J.NEULET.2008.05.010
Sun, W. L., Quizon, P. M., and Zhu, J. (2016). Molecular mechanism: ERK signaling, drug addiction, and behavioral effects. Prog. Mol. Biol. Transl. Sci. 137, 1–40. doi: 10.1016/BS.PMBTS.2015.10.017
Svanes, C., Koplin, J., Skulstad, S. M., Johannessen, A., Bertelsen, R. J., Benediktsdottir, B., et al. (2017). Father’s environment before conception and asthma risk in his children: A multi-generation analysis of the respiratory health in northern europe study. Int. J. Epidemiol. 46, 235–245. doi: 10.1093/IJE/DYW151
Swain, Y., Gewirtz, J. C., and Harris, A. C. (2021). Behavioral predictors of individual differences in opioid addiction vulnerability as measured using i.v. self-administration in rats. Drug Alcohol Depend. 221:8561. doi: 10.1016/J.DRUGALCDEP.2021.108561
Swenson, S., Blum, K., McLaughlin, T., Gold, M. S., and Thanos, P. K. (2020). The therapeutic potential of exercise for neuropsychiatric diseases: A review. J. Neurol. Sci. 412:6763. doi: 10.1016/J.JNS.2020.116763
Szutorisz, H., and Hurd, Y. L. (2018). High times for cannabis: Epigenetic imprint and its legacy on brain and behavior. Neurosci. Biobehav. Rev. 85:93. doi: 10.1016/J.NEUBIOREV.2017.05.011
Szyf, M., and Bick, J. (2013). DNA methylation: A mechanism for embedding early life experiences in the genome. Child Dev. 84, 49–57. doi: 10.1111/J.1467-8624.2012.01793.X
Takahashi, Y., Morales Valencia, M., Yu, Y., Ouchi, Y., Takahashi, K., Shokhirev, M. N., et al. (2023). Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell 186:715–731.e19. doi: 10.1016/J.CELL.2022.12.047
Taniguchi, M., Carreira, M. B., Cooper, Y. A., Bobadilla, A. C., Heinsbroek, J. A., Koike, N., et al. (2017). HDAC5 and its target gene, Npas4, function in the nucleus accumbens to regulate cocaine-conditioned behaviors. Neuron 96:130–144.e6. doi: 10.1016/J.NEURON.2017.09.015
Tapocik, J. D., Barbier, E., Flanigan, M., Solomon, M., Pincus, A., Pilling, A., et al. (2014). microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J. Neurosci. 34, 4581–4588. doi: 10.1523/JNEUROSCI.0445-14.2014
Taylor, S. E. (2010). Mechanisms linking early life stress to adult health outcomes. Proc. Natl. Acad. Sci. U.S.A. 107, 8507–8512. doi: 10.1073/PNAS.1003890107
Tian, W., Zhao, M., Li, M., Song, T., Zhang, M., Quan, L., et al. (2012). Reversal of cocaine-conditioned place preference through methyl supplementation in mice: Altering global DNA methylation in the prefrontal cortex. PLoS One 7:e33435. doi: 10.1371/JOURNAL.PONE.0033435
Ting-A-Kee, R., Vargas-Perez, H., Bufalino, M. R., Bahi, A., Dreyer, J. L., Tyndale, R. F., et al. (2013). Infusion of brain-derived neurotrophic factor into the ventral tegmental area switches the substrates mediating ethanol motivation. Eur. J. Neurosci. 37, 996–1003. doi: 10.1111/EJN.12105
Ttofi, M. M., Farrington, D. P., Lösel, F., Crago, R. V., and Theodorakis, N. (2016). School bullying and drug use later in life: A meta-analytic investigation. Sch. Psychol. Q. 31, 8–27. doi: 10.1037/SPQ0000120
Uliana, D. L., Zhu, X., Gomes, F. V., and Grace, A. A. (2022). Using animal models for the studies of schizophrenia and depression: The value of translational models for treatment and prevention. Front. Behav. Neurosci. 16:935320. doi: 10.3389/FNBEH.2022.935320
United Nations Office on Drugs and Crime (2023). Executive summary, world drug report. Vienna: United Nations Office on Drugs and Crime.
Vagnarelli, F., Amarri, S., Scaravelli, G., Pellegrini, M., Garcia-Algar, O., and Pichini, S. (2006). TDM grand rounds: Neonatal nicotine withdrawal syndrome in an infant prenatally and postnatally exposed to heavy cigarette smoke. Ther. Drug Monit. 28, 585–588. doi: 10.1097/01.FTD.0000245391.56176.AD
Vaisvaser, S., Modai, S., Farberov, L., Lin, T., Sharon, H., Gilam, A., et al. (2016). Neuro-epigenetic indications of acute stress response in humans: The case of MicroRNA-29c. PLoS One 11:e0146236. doi: 10.1371/JOURNAL.PONE.0146236
Vassoler, F. M., White, S. L., Schmidt, H. D., Sadri-Vakili, G., and Christopher Pierce, R. (2012). Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci. 16, 42–47. doi: 10.1038/nn.3280
Volkow, N. D., and Wargo, E. M. (2022). Association of severity of adolescent substance use disorders and long-term outcomes. JAMA Netw. Open 5:e225656. doi: 10.1001/JAMANETWORKOPEN.2022.5656
Vowles, K. E., McEntee, M. L., Julnes, P. S., Frohe, T., Ney, J. P., and Van Der Goes, D. N. (2015). Rates of opioid misuse, abuse, and addiction in chronic pain: A systematic review and data synthesis. Pain 156, 569–576. doi: 10.1097/01.J.PAIN.0000460357.01998.F1
Walters, H., and Kosten, T. A. (2019). Early life stress and the propensity to develop addictive behaviors. Int. J. Dev. Neurosci. 78, 156–169. doi: 10.1016/J.IJDEVNEU.2019.06.004
Wang, D., Wang, Y., Wang, Y., Li, R., and Zhou, C. (2014). Impact of physical exercise on substance use disorders: A meta-analysis. PLoS One 9:e110728. doi: 10.1371/JOURNAL.PONE.0110728
Wang, L., Lv, Z., Hu, Z., Sheng, J., Hui, B., Sun, J., et al. (2010). Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology 35, 913–928. doi: 10.1038/NPP.2009.193
Wang, Z. J., Martin, J. A., Mueller, L. E., Caccamise, A., Werner, C. T., Neve, R. L., et al. (2016). BRG1 in the nucleus accumbens regulates cocaine-seeking behavior. Biol. Psychiatry 80, 652–660. doi: 10.1016/J.BIOPSYCH.2016.04.020
Wanner, N. M., Colwell, M. L., and Faulk, C. (2019). The epigenetic legacy of illicit drugs: Developmental exposures and late-life phenotypes. Environ. Epigenet. 5:dvz022. doi: 10.1093/EEP/DVZ022
Warnault, V., Darcq, E., Levine, A., Barak, S., and Ron, D. (2013). Chromatin remodeling – a novel strategy to control excessive alcohol drinking. Transl. Psychiatry 3:e231. doi: 10.1038/tp.2013.4
Werner, C. T., Altshuler, R. D., Shaham, Y., and Li, X. (2021). Epigenetic mechanisms in drug relapse. Biol. Psychiatry 89:331. doi: 10.1016/J.BIOPSYCH.2020.08.005
Werner, C. T., Mitra, S., Martin, J. A., Stewart, A. F., Lepack, A. E., Ramakrishnan, A., et al. (2019). Ubiquitin-proteasomal regulation of chromatin remodeler INO80 in the nucleus accumbens mediates persistent cocaine craving. Sci. Adv. 5:eaay0351. doi: 10.1126/sciadv.aay0351
Wong, C. C. Y., Mill, J., and Fernandes, C. (2011). Drugs and addiction: An introduction to epigenetics. Addiction 106, 480–489. doi: 10.1111/J.1360-0443.2010.03321.X
Wright, K. N., Hollis, F., Duclot, F., Dossat, A. M., Strong, C. E., Chase Francis, T., et al. (2015). Methyl supplementation attenuates cocaine-seeking behaviors and cocaine-induced c-Fos activation in a DNA methylation-dependent manner. J. Neurosci. 35, 8948–8958. doi: 10.1523/JNEUROSCI.5227-14.2015
Xu, H., Brown, A. N., Waddell, N. J., Liu, X., Kaplan, G. J., Chitaman, J. M., et al. (2020). Role of long noncoding RNA Gas5 in cocaine action. Biol. Psychiatry 88, 758–766. doi: 10.1016/J.BIOPSYCH.2020.05.004
Xu, M. Q., Sun, W. S., Liu, B. X., Feng, G. Y., Yu, L., Yang, L., et al. (2009). Prenatal malnutrition and adult schizophrenia: Further evidence from the 1959-1961 Chinese famine. Schizophr. Bull. 35, 568–576. doi: 10.1093/SCHBUL/SBN168
Xu, S. J., Lombroso, S. I., Fischer, D. K., Carpenter, M. D., Marchione, D. M., Hamilton, P. J., et al. (2021). Chromatin-mediated alternative splicing regulates cocaine-reward behavior. Neuron 109:2943–2966.e8. doi: 10.1016/J.NEURON.2021.08.008
Yamaguchi, H., Hopf, F. W., Li, S., Bin, and de Lecea, L. (2018). In vivo cell type-specific CRISPR knockdown of dopamine beta hydroxylase reduces locus coeruleus evoked wakefulness. Nat. Commun. 9:5211. doi: 10.1038/S41467-018-07566-3
Yu, H., Xie, B., Zhang, J., Luo, Y., Galaj, E., Zhang, X., et al. (2021). The role of circTmeff-1 in incubation of context-induced morphine craving. Pharmacol. Res. 170:105722. doi: 10.1016/J.PHRS.2021.105722
Zarse, E. M., Neff, M. R., Yoder, R., Hulvershorn, L., Chambers, J. E., and Chambers, R. A. (2019). The adverse childhood experiences questionnaire: Two decades of research on childhood trauma as a primary cause of adult mental illness, addiction, and medical diseases. Cogent. Med. 6:1581447. doi: 10.1080/2331205X.2019.1581447
Zee, B. M., Levin, R. S., Xu, B., LeRoy, G., Wingreen, N. S., and Garcia, B. A. (2010). In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285, 3341–3350. doi: 10.1074/JBC.M109.063784
Zhang, Y. X., Akumuo, R. C., España, R. A., Yan, C. X., Gao, W. J., and Li, Y. C. (2018). The histone demethylase KDM6B in the medial prefrontal cortex epigenetically regulates cocaine reward memory. Neuropharmacology 141, 113–125. doi: 10.1016/J.NEUROPHARM.2018.08.030
Keywords: substance use disorder, epigenetics, environment, vulnerability, animal models
Citation: Liu SX, Harris AC and Gewirtz JC (2024) How life events may confer vulnerability to addiction: the role of epigenetics. Front. Mol. Neurosci. 17:1462769. doi: 10.3389/fnmol.2024.1462769
Received: 10 July 2024; Accepted: 02 September 2024;
Published: 18 September 2024.
Edited by:
Mayur Doke, University of Miami Health System, United StatesReviewed by:
Venkataraghavan Ramamoorthy, Baptist Health South Florida, United StatesSushesh Srivatsa Palakurthi, Texas A&M University, United States
Copyright © 2024 Liu, Harris and Gewirtz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan C. Gewirtz, Sm9uYXRoYW4uR2V3aXJ0ekBhc3UuZWR1
 Shirelle X. Liu
Shirelle X. Liu Andrew C. Harris
Andrew C. Harris Jonathan C. Gewirtz1,4*
Jonathan C. Gewirtz1,4*