- 1Department for Neuroscience, Croatian Institute for Brain Research, University of Zagreb Medical School, Zagreb, Croatia
- 2Reta Lila Weston Institute, Department of Clinical and Movement Neuroscience, UCL Queen Square Institute of Neurology, London, United Kingdom
- 3Division of Molecular Medicine, Ruđer Bošković Institute, Zagreb, Croatia
- 4Department of Biology and Human Genetics, School of Medicine, University of Split, Split, Croatia
- 5Department of Molecular Diagnostics and Genetics, Dubrava University Hospital, Zagreb, Croatia
- 6Laboratory for Neurobiochemistry, Department of Laboratory Diagnostics, University Hospital Centre Zagreb, Zagreb, Croatia
- 7Department of Neurology, University Hospital Centre Zagreb, Kišpatićeva, Zagreb, Croatia
- 8School of Medicine, University of Zagreb, Zagreb, Croatia
- 9Department for Functional Genomics, Centre for Translational and Clinical Research, University of Zagreb Medical School, Zagreb, Croatia
- 10Department of Nuclear Medicine, University Hospital Split, Split, Croatia
Introduction: Genetic studies have shown that variants in the microtubule-associated protein tau (MAPT) gene, which encodes tau protein, can increase the risk for Alzheimer’s disease (AD). Additionally, two haplotypes of the MAPT gene (H1 and H2) are associated with various neurodegenerative disorders, including AD. This study aimed to test the association of MAPT haplotypes (H1 and H2) and MAPT haplotype-tagging polymorphisms (rs1467967, rs242557, rs3785883, rs2471738, del–In9, rs7521) with AD.
Methods: The study included 964 individuals: 113 with AD, 53 with mild cognitive impairment (MCI), 54 with other dementias, and 744 healthy controls.
Results: The results showed that individuals carrying the A allele in the MAPT rs1467967 polymorphism, the GG genotype in the MAPT rs7521 polymorphism, and the G allele in the MAPT rs242557 polymorphism had worse performance on various neuropsychological tests. Carriers of the C allele in MAPT rs2471738 polymorphism and CC homozygotes also showed worse performance on neuropsychological tests and pathological levels of several cerebrospinal fluid (CSF) biomarkers. However, T allele carriers in the MAPT rs2471738 polymorphism were more represented among patients with dementia and apolipoprotein E (APOE) ɛ4 carriers. Carriers of the H2 MAPT haplotype had worse performance on various neuropsychological tests, consistent with our previous study, which associated the H2 MAPT haplotype with pathological levels of CSF AD biomarkers. Regarding the MAPT rs3785883 polymorphism, further research is needed since both the AA and GG genotypes were associated with pathological levels of CSF and plasma AD biomarkers.
Discussion: In conclusion, further genetic studies are needed to elucidate the role of MAPT haplotypes and MAPT haplotype-tagging polymorphisms in the development of AD.
1 Introduction
Alzheimer’s disease (AD) is a complex condition influenced by both genetic and environmental factors. Analysis of tau-positron emission tomography (tau-PET) scans from 1,612 individuals identified four different AD subtypes based on the spread of tau pathology: (1) limbic (apolipoprotein E ɛ4+ [APOE ɛ4+], less tau, amnestic, older onset, 32.7% of cases), (2) lateral temporal (more tau, rapidly progressive, multi-domain impairment, asymmetric, 18.1% of cases), (3) medial temporal lobe-sparing (APOE ɛ4-, younger onset, dysexecutive, 30.2% of cases), and (4) posterior (older onset, slowly-progressing, visuospatial impairment, 19% of cases, also called posterior cortical atrophy or atypical AD) (Vogel et al., 2021). These findings indicate that additional studies are necessary to elucidate AD’s complex interplay between genetic and environmental factors. According to AlzGene (Bertram et al., 2007), 1,395 studies identified 695 genes associated with AD. In the sporadic form of AD, which accounts for about 99% of disease cases, the main genetic factor that increases the relative risk of developing AD is the ɛ4 variant of the APOE gene. One ɛ4 allele increases the risk of developing AD by about three times, while both ɛ4 alleles increase the relative risk by eight to twelve times (Alzheimer's Association, 2020). Numerous other genes associated with sporadic AD have been discovered, but their influence on the risk of the disease is much smaller than that of the APOE gene. These genes include BIN1, CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, MS4A4E, CD2AP (Bertram et al., 2007), TREM2 (Jonsson et al., 2013), and PLD3 (Cruchaga et al., 2014), among others. These genes were mostly identified in genome-wide association studies (GWAS) that compared many single nucleotide polymorphisms (SNPs) between AD patients and healthy controls (HC).
The microtubule-associated protein tau (MAPT) gene, located on chromosome 17q21.3, encodes the tau protein, which, in addition to amyloid β (Aβ), plays a major role in AD. Although AD is considered a secondary tauopathy (i.e., it is not caused by the mutations in the MAPT gene, as is the case in primary tauopathies), certain MAPT variants have been shown to increase the risk for AD (Myers et al., 2005; Laws et al., 2007; Chang et al., 2014; Yuan et al., 2017; Zhou and Wang, 2017). Moreover, two MAPT haplotypes (H1 and H2), which can be defined by six haplotype-tagging SNPs (rs1467967, rs242557, rs3785883, rs2471738, del–In9, rs7521) (Pittman et al., 2005), have been associated with various neurodegenerative disorders, including AD (Myers et al., 2005; Laws et al., 2007; Kauwe et al., 2008; Di Maria et al., 2010; Pastor et al., 2016; Sánchez-Juan et al., 2019). The MAPT H1 haplotype has been shown to increase risk, while MAPT H2 haplotype reduces the risk for several neurodegenerative disorders (Strickland et al., 2020). In our previous preliminary study, we observed that the levels of cerebrospinal fluid (CSF) biomarkers differed between patients with different genotypes in MAPT SNPs (specifically, rs1467967 and rs2471738) and different MAPT haplotypes (Babić Leko et al., 2018). This study aimed to further validate the association of MAPT haplotype-tagging polymorphisms with AD. We tested whether there is a difference in the results of neuropsychological tests, levels of CSF and plasma biomarkers, and the frequency of APOE genotypes among patients with different genotypes in MAPT haplotype-tagging polymorphisms. We also examined whether there is a difference in the distribution of MAPT haplotype-tagging polymorphisms and MAPT haplotypes between patients with AD and HC. Moreover, we analyzed additional AD biomarkers, such as chitinase 3-like protein 1 (YKL-40), S100 calcium-binding protein B (S100B), neurofilament light chain (NfL), and p-tau181/Aβ1-42 ratio.
2 Materials and methods
2.1 Subjects
This study included 964 subjects, of whom 220 were recruited at the University Hospital Centre Zagreb. These 220 participants suffered from various types of primary dementia causes: AD, mild cognitive impairment (MCI), frontotemporal dementia (FTD), vascular cognitive impairment/vascular dementia (VaD), mixed dementia (AD+VaD), non-specific dementia (ND), corticobasal syndrome (CBS), and Parkinson’s disease (PD). AD was diagnosed using the criteria of the National Institutes on Aging - Alzheimer’s Association (NIA-AA) (McKhann et al., 2011). The following criteria were used for the diagnosis of other dementia types: for MCI, the criteria of Petersen et al. (1999) and Albert et al. (2011); for FTD, the criteria of Neary et al. (1998); and for VaD, the Hachinski Ischemic Score (Hachinski et al., 1975) and the criteria of the National Institute for Neurological Disorders and Stroke – Association Internationale pour la Recherche et l’Enseignement en Neurosciences (NINCDS-AIREN) (Román et al., 1993).
All patients included in this study provided informed consent for lumbar puncture and participation in the study. Patients underwent neurological examinations with complete blood tests, including levels of thyroid hormones, vitamin B12 and B9, and serology for syphilis and Lyme disease. The Ethical Committee of the Clinical Hospital Centre Zagreb (case no. 02/21 AG, class 8.1–18/82–2 from April 24, 2018) and the Central Ethical Committee of the University of Zagreb Medical School (case no. 380–59–10,106-18-111/126, class 641–01/18–02/01 from June 20, 2018) approved all procedures, which were conducted in accordance with the Helsinki Declaration (World Medical Association, 2013).
A larger cohort of 744 HC older than 65 years of age included participants from the “10,001 Dalmatians project,” part of the Croatian Biobank program (Rudan et al., 2009). Participants provided informed consent for participation in the study, and the Ethical Board of the University of Split, School of Medicine (case no. 2181-198-03-04/10-11-0008 and 2181-198-03-04-19-0022) approved all procedures.
Information on demographic data, determined biomarkers, and the number of APOE and MAPT genotypes and MAPT haplotypes in all included cohorts is presented in Table 1.
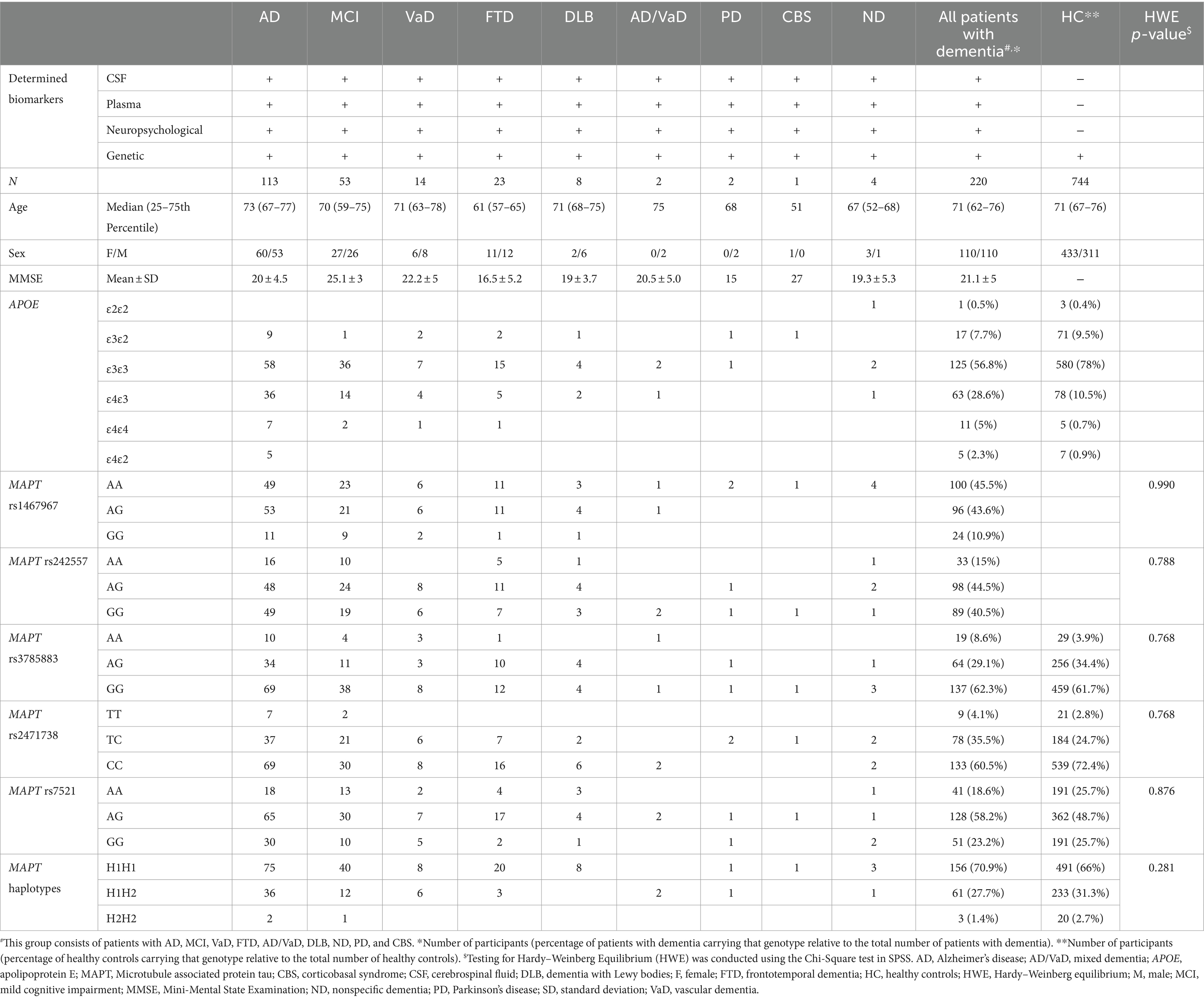
Table 1. Information on demographic data, determined biomarkers, and the number of APOE and MAPT genotypes in different cohorts.
2.2 Analysis of cerebrospinal fluid and plasma biomarkers
This part of the analysis was done in 220 participants recruited at the University Hospital Centre Zagreb. All biomarkers were analyzed in CSF, while some were also analyzed in plasma. CSF was collected by lumbar puncture between intervertebral spaces L3/L4 or L4/L5, centrifuged at 2,000 g for 10 min, and stored at −80°C in polypropylene tubes. Thrombocyte-free plasma was extracted from venous blood through a series of centrifugations (first at 1,100 g for 3 min, followed by centrifugation at 5,087 g for 15 min) and stored at −20°C.
The following enzyme-linked immunosorbent assays (ELISA) were used for the determination of biomarkers: tau phosphorylated at threonine 181 (p-tau181) (Innotest Phospho-Tau [181P], Fujirebio), Aβ1-42 (Innotest β-amyloid1–42, Fujirebio, Gent, Belgium), NfL (Human NF-L/NEFL ELISA Kit, LifeSpan BioSciences, Seattle, WA, USA), S100B (Human S100B, R&D Systems, Minneapolis, MN, USA), and YKL-40 (Chitinase 3-like 1 Quantikine ELISA Kit, R&D Systems). All biomarkers were measured only in CSF, except for S100B and NfL, which were measured in both CSF and plasma.
2.3 Determination of polymorphisms
The salting-out method by Miller et al. (1988) was used for the isolation of DNA from venous blood. In the 220 participants recruited at the University Hospital Centre Zagreb, MAPT rs1467967, rs242557, rs3785883, rs2471738, del–In9, rs7521, and APOE rs7412 and rs429358 SNPs were determined using TaqMan SNP Genotyping Assays (Applied Biosystems) on an ABI Prism 7,300 Real-Time PCR System apparatus (Applied Biosystems, Foster City, CA). The MAPT H1/H2 haplotype is defined by the del-In9 deletion in MAPT intron 9.
In 744 participants recruited as part of the “10,001 Dalmatians project,” MAPT rs3785883, rs2471738, rs7521, rs1800547, and APOE rs7412 and rs429358 were determined using Illumina genotyping platforms (CNV370v1, CNV370-Quadv3, and OmniExpressExome-8v1-2_A, Illumina, San Diego, CA). In this cohort, MAPT haplotypes were defined by the MAPT rs1800547 SNP. MAPT rs1467967 and rs242557 polymorphisms were not determined in this cohort of healthy controls. APOE ε2, ε3, and ε4 variants were determined by the following combination of APOE SNPs: variant ε2 (rs429358 T allele and rs7412 T allele), variant ε3 (rs429358 T allele and rs7412 C allele), and variant ε4 (rs429358 C allele and rs7412 C allele).
2.4 Neuropsychological testing
Participants recruited at the University Hospital Centre Zagreb were tested with various neuropsychological assessments, including the Mini-Mental State Examination (MMSE), the modified MMSE test, the Alzheimer’s Disease Assessment Scale-Cognitive subscale (ADAS-Cog), the Picture Pairs Learning and Recall (PPLR) test, the Word Pairs Learning and Recall (WPLR) test, the Visual Association Test (VAT), the Rey–Osterrieth Complex Figure Test (ROCFT), the Clock Drawing Test (CDT), the California Verbal Learning Test (CVLT), the NeuroPsychiatric Inventory (NPI), the Reverse Naming test, the Stroop test, the Visual Reaction Time (VRT) test, and the Boston Naming Test (BNT).
2.5 Statistical analysis
Statistical analysis was performed using SPSS 19.0.1 (SPSS, Chicago, IL, USA) and R statistical software (R Core Team, Vienna, Austria). The level of statistical significance was set at α = 0.05.
The frequencies of APOE genotypes and different diagnoses among subjects with different MAPT genotypes and haplotypes were analyzed using a χ2-test (Chi-squared test), with Bonferroni correction applied for pairwise comparisons.
Levels of all fluid biomarkers and the majority of variables collected by neuropsychological testing deviated from a normal distribution (p < 0.05), except for ADAS-Cog (p = 0.07), BNT (p = 0.153), PPLR corr (p = 0.193), PPLR incorr (p = 0.126), PPLR t corr (p = 0.470), and PPLR t incorr (p = 0.288). Thus, for comparisons of CSF and plasma biomarkers and scores on neuropsychological tests between groups, we used non-parametric tests. First, all variables were compared between groups of patients with different MAPT genotypes using the Kruskal-Wallis test (denoted by the letter “H”), followed by post-hoc Dunn’s test, which corrects p-values for multiple comparisons. In addition to statistical analyses between all genotypes (for example, AA vs. AG vs. GG), MAPT genotypes were also grouped, and additional analyses were conducted (for example, AA vs. G allele carriers and GG vs. A allele carriers). CSF and plasma biomarkers and scores on neuropsychological tests were compared between two groups (for example, AA vs. G allele carriers) using the Mann–Whitney U test. All statistical analyses were conducted in the group of all patients with dementia (1), AD and MCI patients (2), and AD patients (3).
Variables were analyzed using linear regression when the influence of possible covariates had to be considered. Since the distribution of the majority of variables (CSF and plasma biomarkers and scores on neuropsychological tests) deviated from a normal distribution, variables were logarithmically transformed before linear regression. CSF and plasma biomarkers and scores on neuropsychological tests were considered as dependent variables, while MAPT genotypes and haplotypes (and covariates) were considered as independent variables.
3 Results
Before analyses, we tested if the variables were associated with sex, age, and APOE genotype (ɛ4+ vs. ɛ4-). Regarding neuropsychological tests, none of the variables were associated with sex, age, or APOE genotype. Thus, there was no need to consider sex, age, or APOE genotype as covariates, and analyses were not controlled for the influence of these factors. On the other hand, fluid biomarkers showed an association with sex, age, and APOE genotype. More precisely, CSF YKL-40 (rS = 0.339, p < 0.001), CSF NfL (rS = 0.199, p = 0.019), and plasma NfL (rS = 0.413, p < 0.001) were positively correlated with the age of participants. Moreover, NfL plasma levels were significantly increased in males compared to females (U = 2076, Z = −2.392, p = 0.017), while the p-tau181/Aβ1-42 ratio was significantly increased in carriers of the ɛ4 APOE genotype (U = 3,875, Z = −4.056, p < 0.001).
Additionally, we tested if the interaction between each MAPT polymorphism and APOE genotype was significantly associated with the tested variables. It was shown that the interaction between rs242557 and APOE significantly predicted scores on ADAS-Cog (β = 0.163, SE = 0.077, p = 0.037). The interaction between rs1467967 and APOE predicted scores on ROCFTD (β = −0.315, SE = 0.14, p = 0.028). The interaction between rs1467967 and APOE (β = 0.201, SE = 0.081, p = 0.015) and rs242557 and APOE (β = 0.211, SE = 0.081, p = 0.010) significantly predicted VAT time. Additionally, the interaction between rs2471738 and APOE predicted CSF NfL levels (β = 0.168, SE = 0.070, p = 0.017). The interaction between MAPT haplotype and APOE predicted p-tau181/Aβ1-42 ratio (β = −0.419, SE = 0.115, p < 0.001). Thus, for these variables, the aforementioned interactions were considered as covariates.
Scores on neuropsychological tests and levels of CSF and plasma biomarkers in patients with different MAPT genotypes and haplotypes are presented in Supplementary Tables S1, S2, respectively.
3.1 MAPT rs1467967 genotype
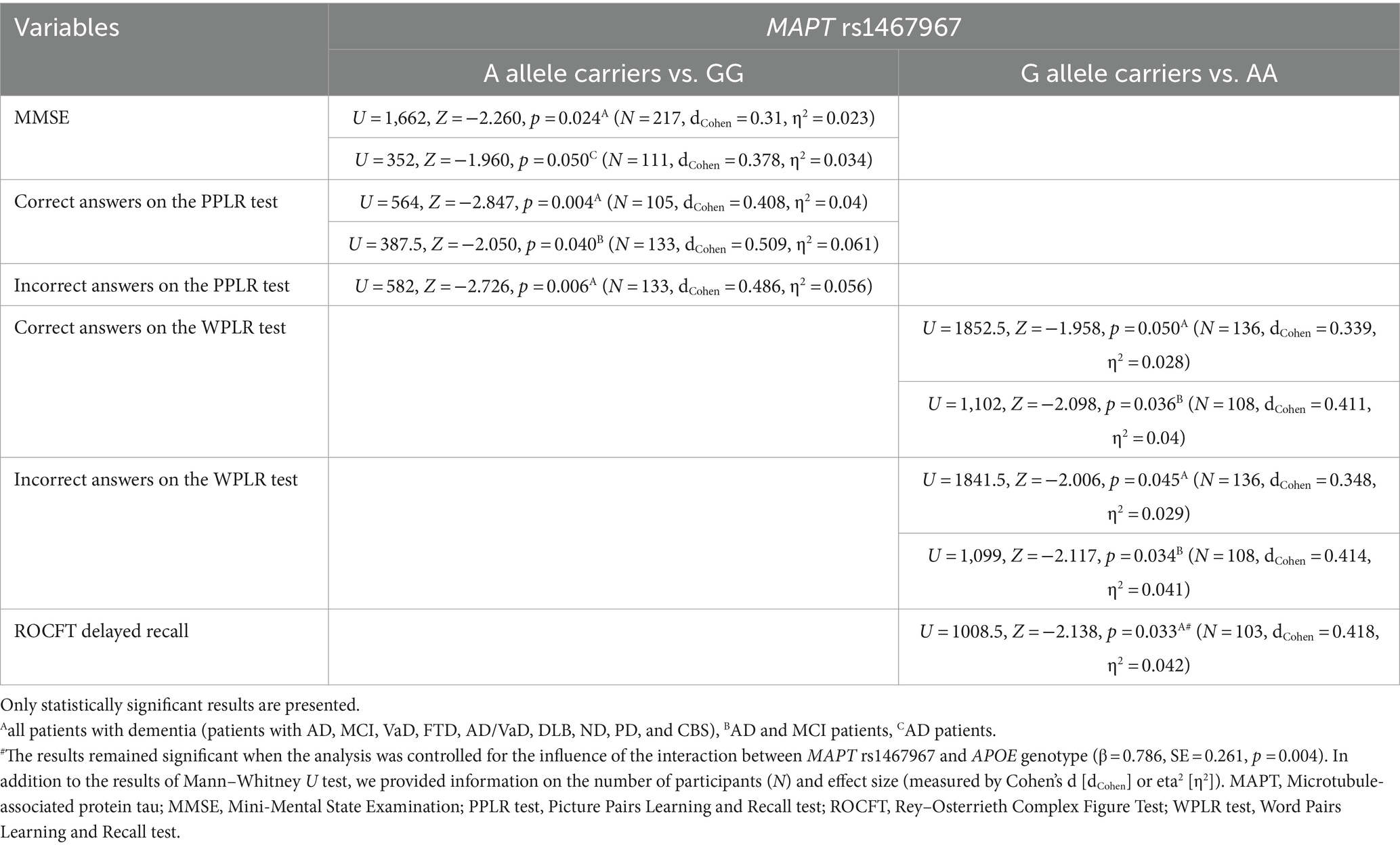
Carriers of the A allele in the MAPT rs1467967 polymorphism had significantly lower MMSE scores compared to GG homozygotes. Carriers of the A allele had a significantly lower number of correct answers on the PPLR test, and a higher number of incorrect answers compared to GG homozygotes. Moreover, AA homozygotes had a significantly lower number of correct answers on the WPLR test and a higher number of incorrect answers compared to G allele carriers. AA homozygotes also had poorer performance on ROCFT, with lower scores on ROCFT delayed recall compared to G allele carriers (Figure 1; Table 2).
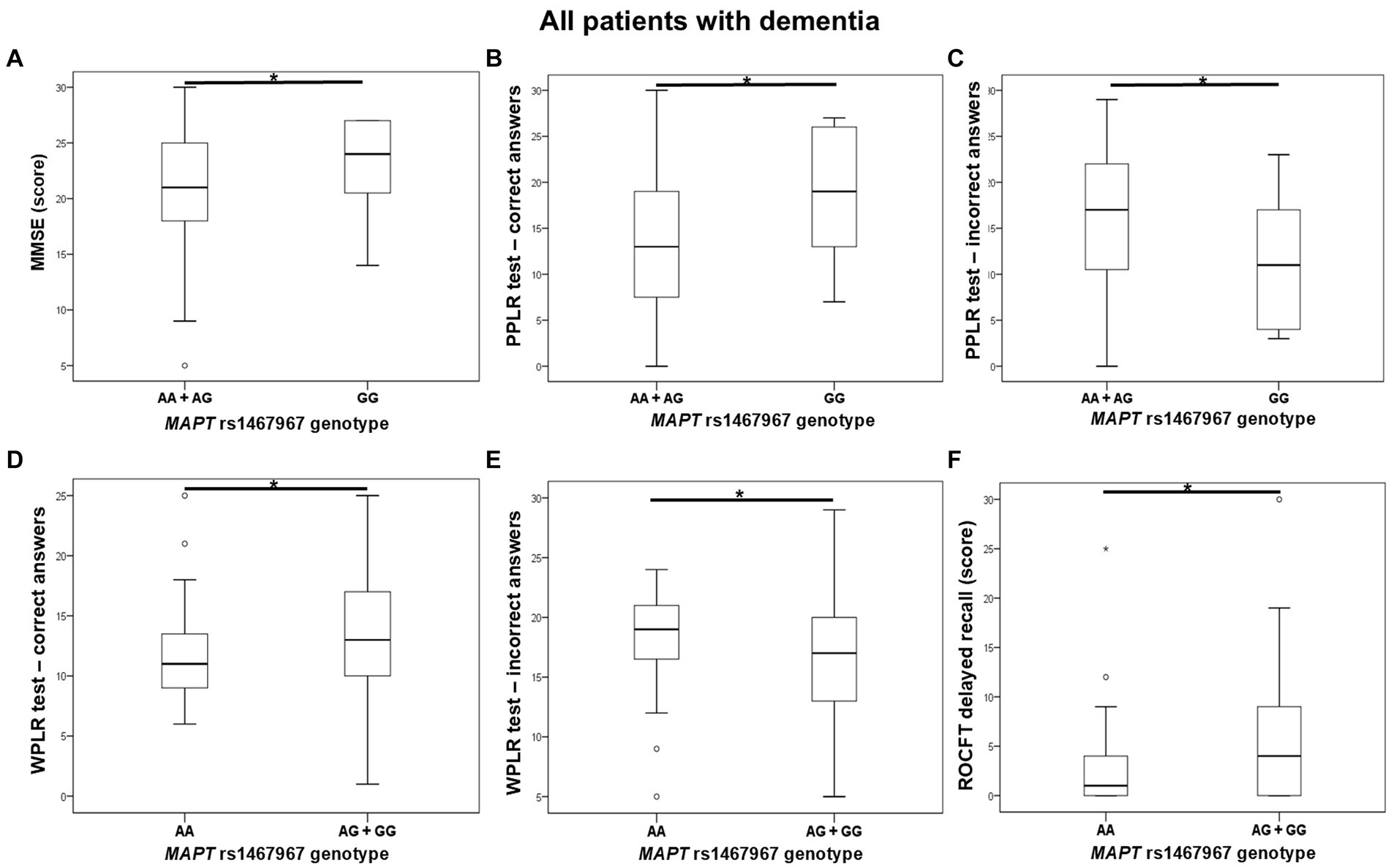
Figure 1. AA homozygotes and A allele carriers in the MAPT rs1467967 polymorphism showed poorer performance on (A) MMSE, (B,C) PPLR test, (D,E) WPLR test, and (F) ROCFT. The group “all patients with dementia” includes patients with AD, MCI, VaD, FTD, AD/VaD, DLB, ND, PD, and CBS. The box represents the interquartile range (between 25th and 75th percentiles), while the whiskers represent the range between the minimum and maximum values.
Plasma and CSF levels of S100B and NfL, as well as CSF p-tau181/Aβ1-42 ratio and YKL-40 levels, did not differ significantly between patients with different MAPT rs1467967 genotypes.
3.2 MAPT rs242557 genotype
G allele carriers in the MAPT rs242557 polymorphism had worse performance on the CVLT test compared to AA homozygotes. Specifically, levels of CVLT d and CVLT 1–5 were significantly decreased in G allele carriers compared to AA homozygotes. According to the NPI assessment, GG homozygotes were more depressed than A allele carriers. Moreover, worse performance on VAT (higher levels of VAT, VAT time, and VAT t average) was shown for G allele carriers compared to AA homozygotes (Figure 2; Table 3). PPLR t incorr levels were also higher in carriers of the G allele compared to AA homozygotes.
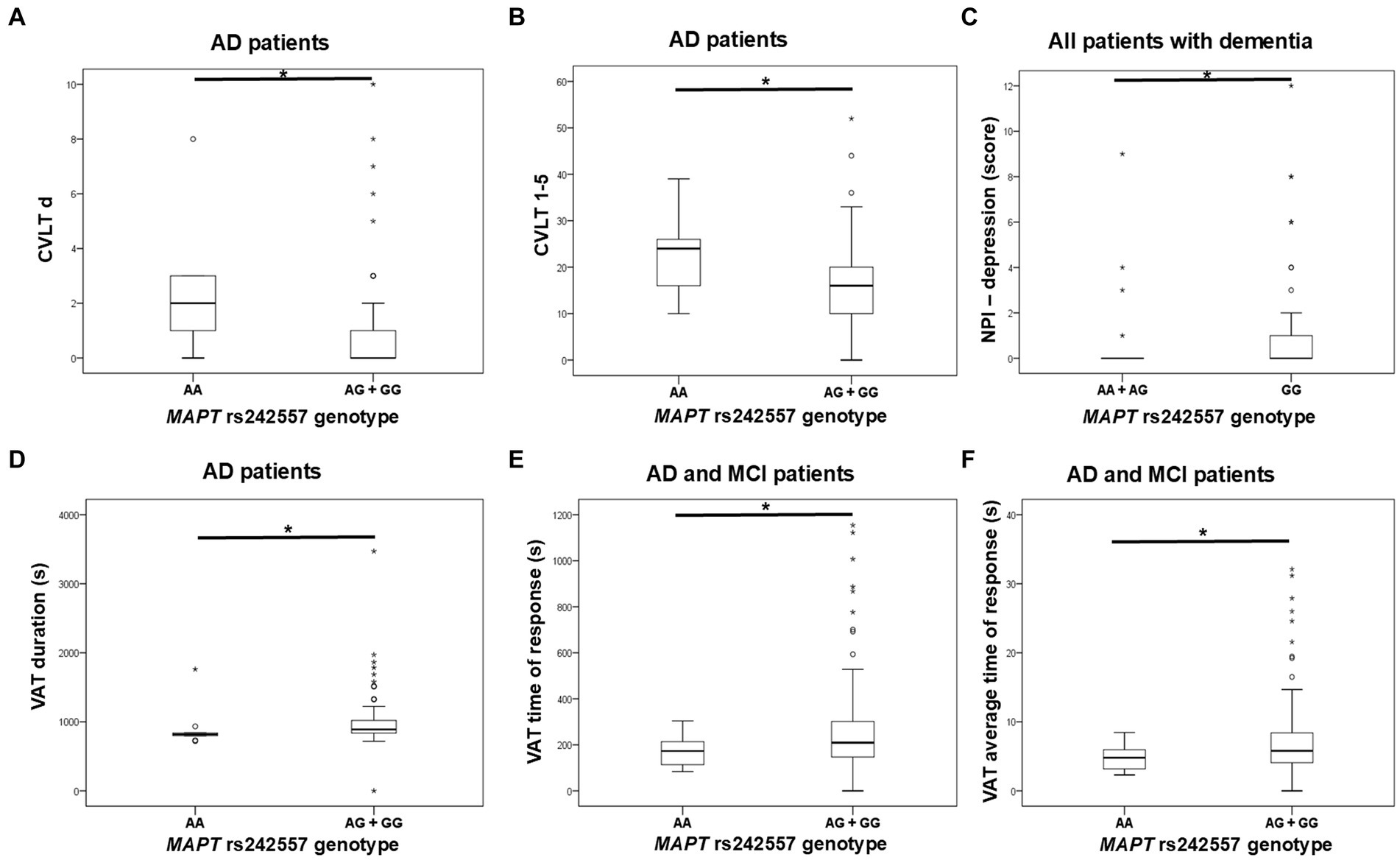
Figure 2. GG homozygotes and G allele carriers in the MAPT rs242557 polymorphism showed poorer performance on (A,B) CVLT, (C) NPI assessment, and (D–F) VAT test. The group “all patients with dementia” includes patients with AD, MCI, VaD, FTD, AD/VaD, DLB, ND, PD, and CBS. The box represents the interquartile range (between 25th and 75th percentiles), while the whiskers represent the range between the minimum and maximum values.
No significant difference in plasma and CSF levels of S100B and NfL, as well as CSF p-tau181/Aβ1-42 ratio and YKL-40 levels, was observed between patients with different MAPT rs242557 genotypes.
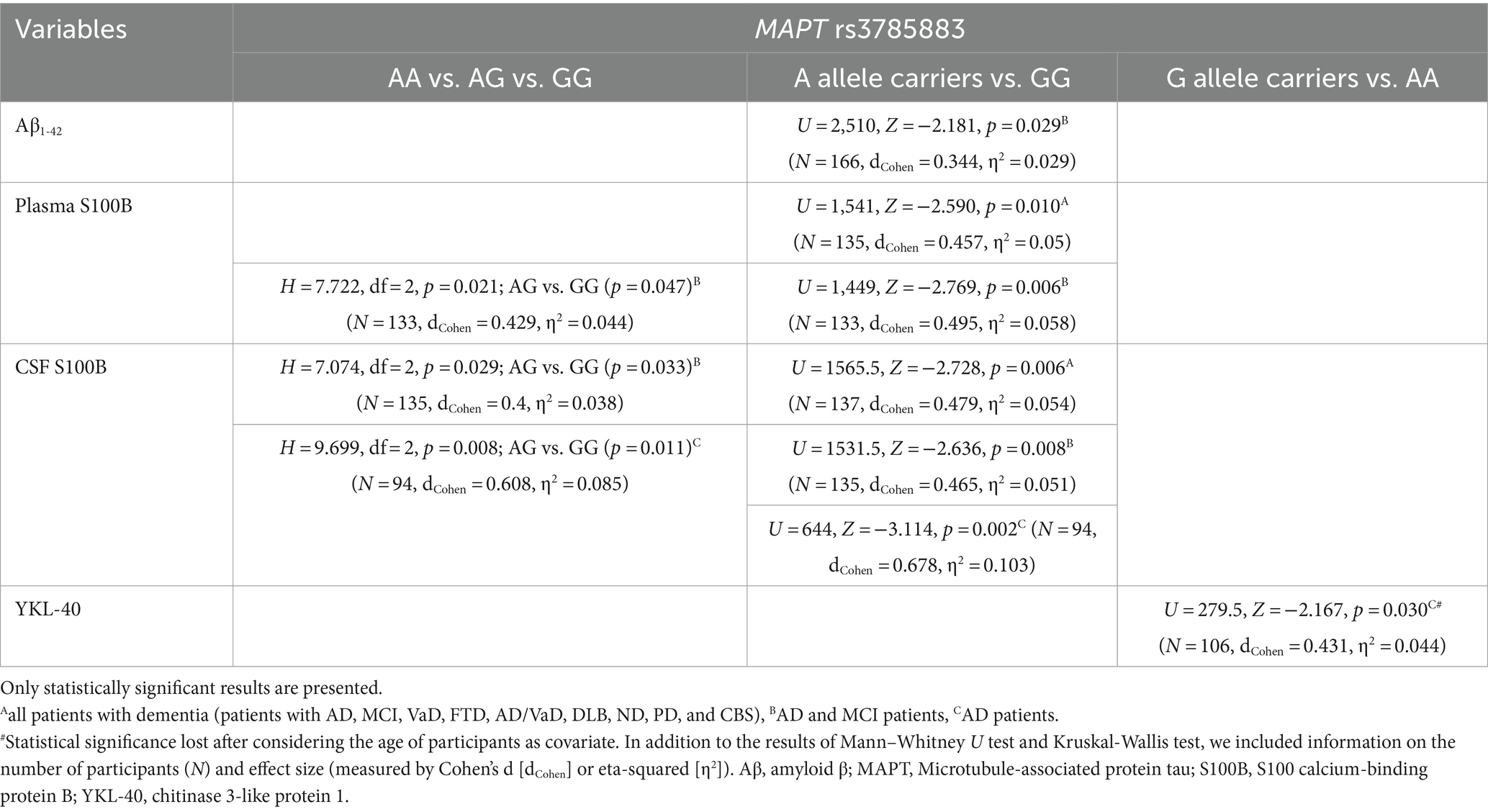
3.3 MAPT rs3785883 genotype
Aβ1-42 levels were significantly decreased in A allele carriers compared to GG MAPT rs3785883 homozygotes (Figure 3; Table 4).
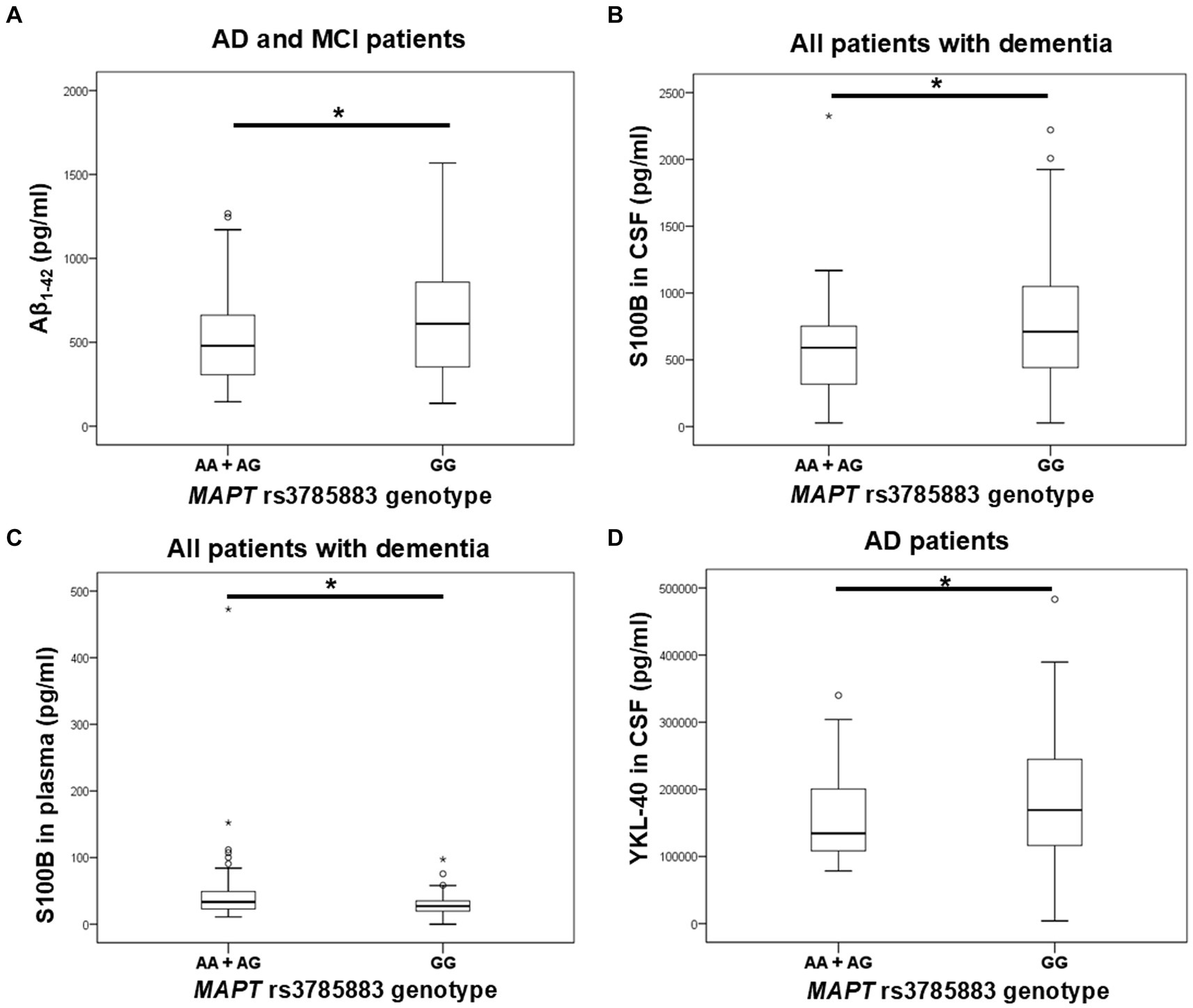
Figure 3. A allele carriers in the MAPT rs3785883 polymorphism had significantly decreased Aβ1-42 levels (A), and had significantly increased plasma S100B levels (C). Conversely, GG homozygotes in the MAPT rs3785883 polymorphism had significantly increased CSF S100B levels (B), and CSF YKL-40 (D) levels. The group “all patients with dementia” includes patients with AD, MCI, VaD, FTD, AD/VaD, DLB, ND, PD, and CBS. The box represents the interquartile range (between 25th and 75th percentiles), while the whiskers represent the range between the minimum and maximum values.
There was a significant difference in the distribution of MAPT rs3785883 genotypes between HC and patients with dementia (χ2 = 9.2, df = 1, p = 0.004). Specifically, a higher frequency of AA homozygotes was observed among patients with dementia (p = 0.018), while G allele carriers were more represented among HC (p = 0.018).
Additionally, A allele carriers had significantly increased S100B plasma levels compared to GG homozygotes (Figure 3; Table 4). Moreover, S100B in plasma was increased in carriers of the AG genotype compared to GG homozygotes (Table 4).
S100B in CSF was significantly increased in carriers of the GG genotype compared to A allele carriers (Figure 3; Table 4). In fact, S100B in CSF was increased in carriers of GG homozygotes compared to carriers of the AG genotype (Table 4). In AD patients, an increase in CSF YKL-40 levels was observed in G allele carriers compared to AA homozygotes (Figure 3; Table 4).
No significant difference in scores on neuropsychological tests was observed between patients with different MAPT rs3785883 genotypes.
3.4 MAPT rs2471738 genotype
TC heterozygotes in the MAPT rs2471738 genotype had a lower rate of disinhibition compared to TT homozygotes. C allele carriers had worse performance on the CVLT test (lower CVLT d and CVLT d cue scores). Finally, CC homozygotes made a higher number of errors on the Stroop test compared to T allele carriers (Figure 4; Table 5).
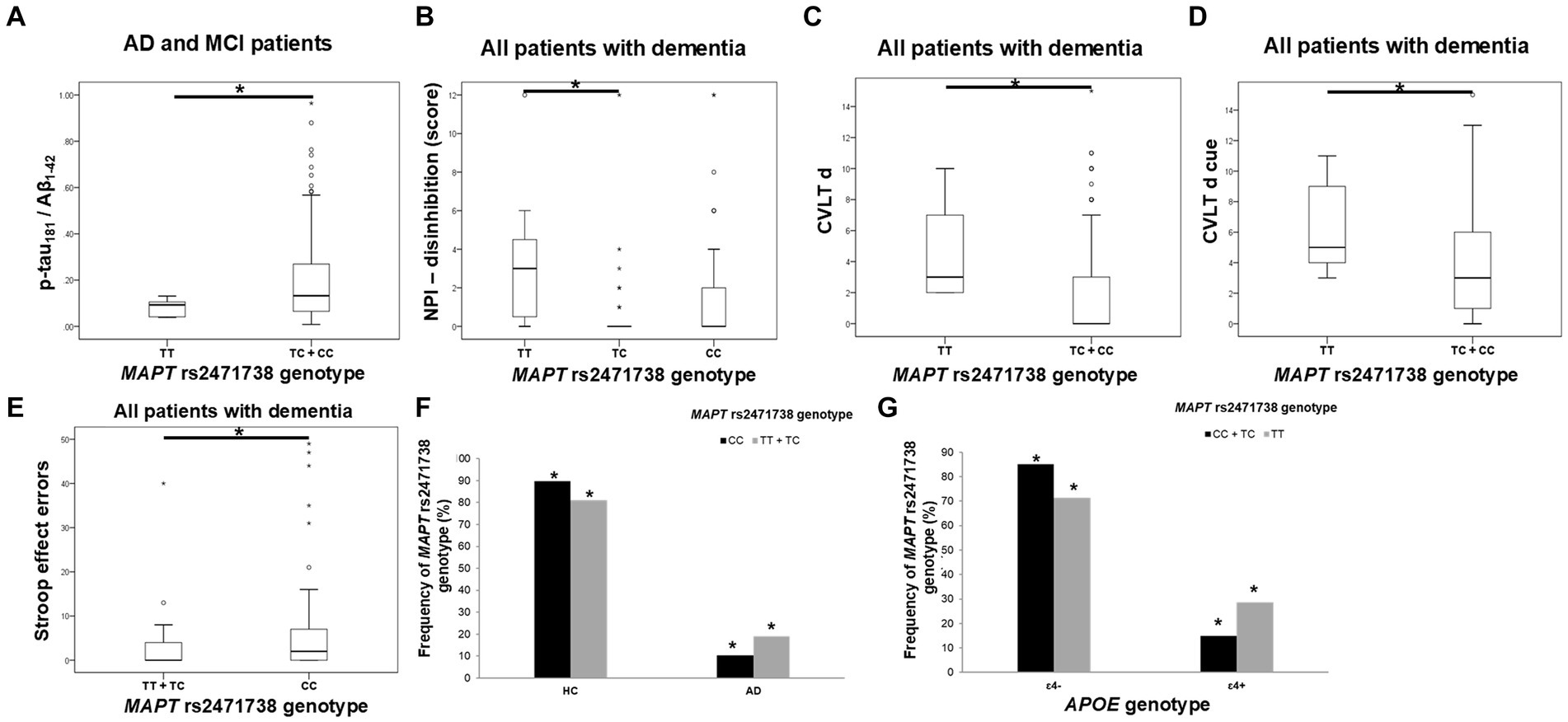
Figure 4. C allele carriers in the MAPT rs2471738 genotype had higher levels of p-tau181/Aβ1-42 ratio (A). TC heterozygotes had a lower rate of disinhibition (B) compared to TT homozygotes. C allele carriers and CC homozygotes had worse performance on the CVLT test (C,D) and the Stroop test (E). T allele carriers were more frequent among patients with AD (F), and a higher number of APOE ɛ4 carriers was observed among TT homozygotes (G). The group “all patients with dementia” includes patients with AD, MCI, VaD, FTD, AD/VaD, DLB, ND, PD, and CBS. The box represents the interquartile range (between 25th and 75th percentiles), while the whiskers represent the range between the minimum and maximum values.
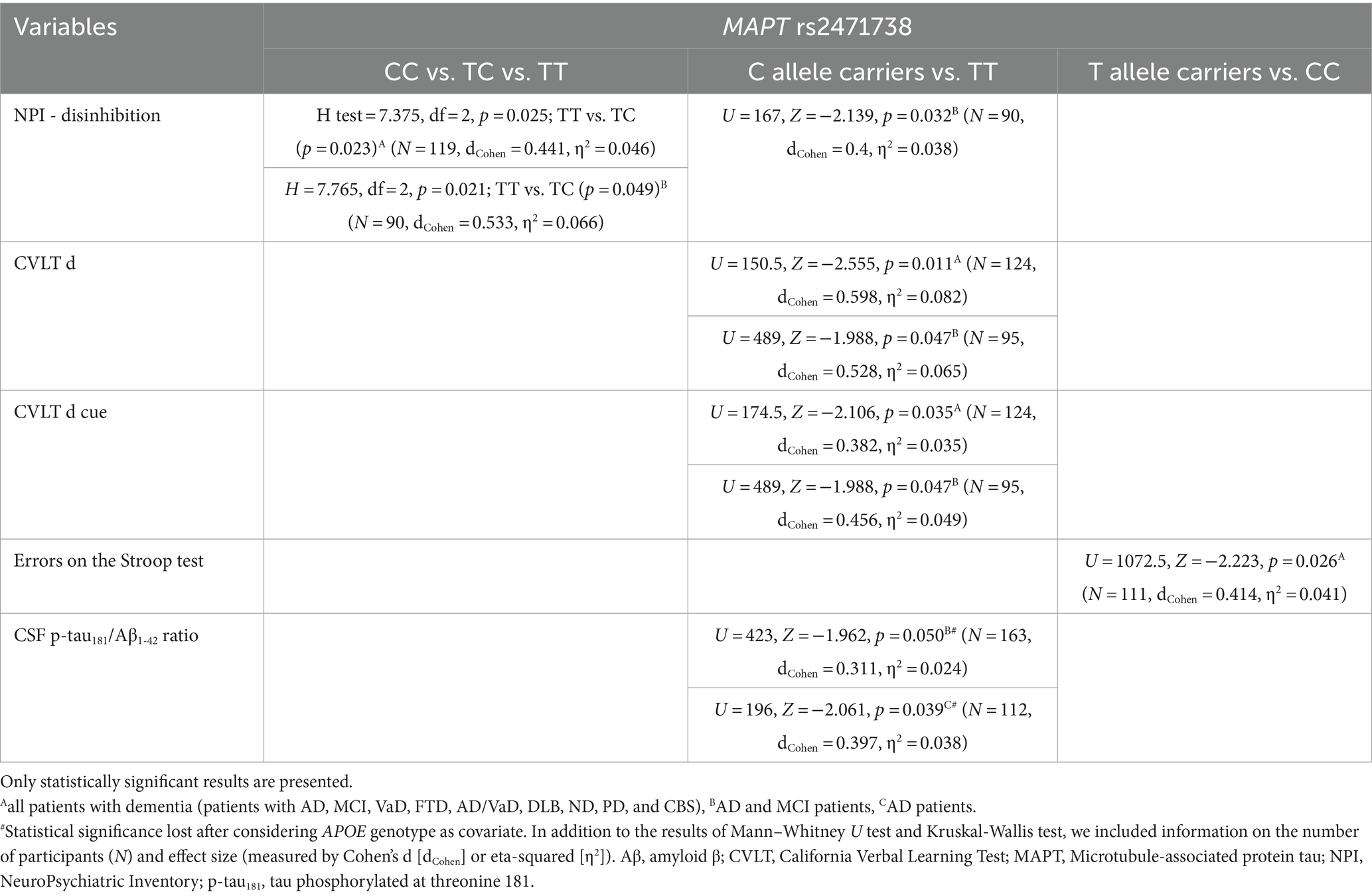
Table 5. Association of MAPT rs2471738 polymorphism with the results on neuropsychological tests and the levels of fluid biomarkers.
The CSF p-tau181/Aβ1-42 ratio was significantly higher in C allele carriers compared to TT homozygotes (Figure 4; Table 5).
There was a significant difference in the distribution of MAPT rs2471738 genotypes between HC and patients with AD (χ2 = 11.889, df = 1, p = 0.001), AD and MCI (χ2 = 16.631, df = 1, p < 0.001), and patients with dementia (χ2 = 16.226, df = 1, p < 0.001). Specifically, a higher frequency of TT homozygotes and T allele carriers was observed among patients with AD (p < 0.001, p = 0.006, respectively), AD and MCI (p < 0.001, p = 0.001, respectively), and patients with dementia (p < 0.001, p = 0.002, respectively). Moreover, among TT homozygotes (χ2 = 6.391, df = 1, p = 0.024), there was a higher number of APOE ɛ4 carriers (p = 0.046) (when considering only individuals older than 65 years of age, and after excluding ɛ4ɛ2 genotype from analysis) (Figure 4).
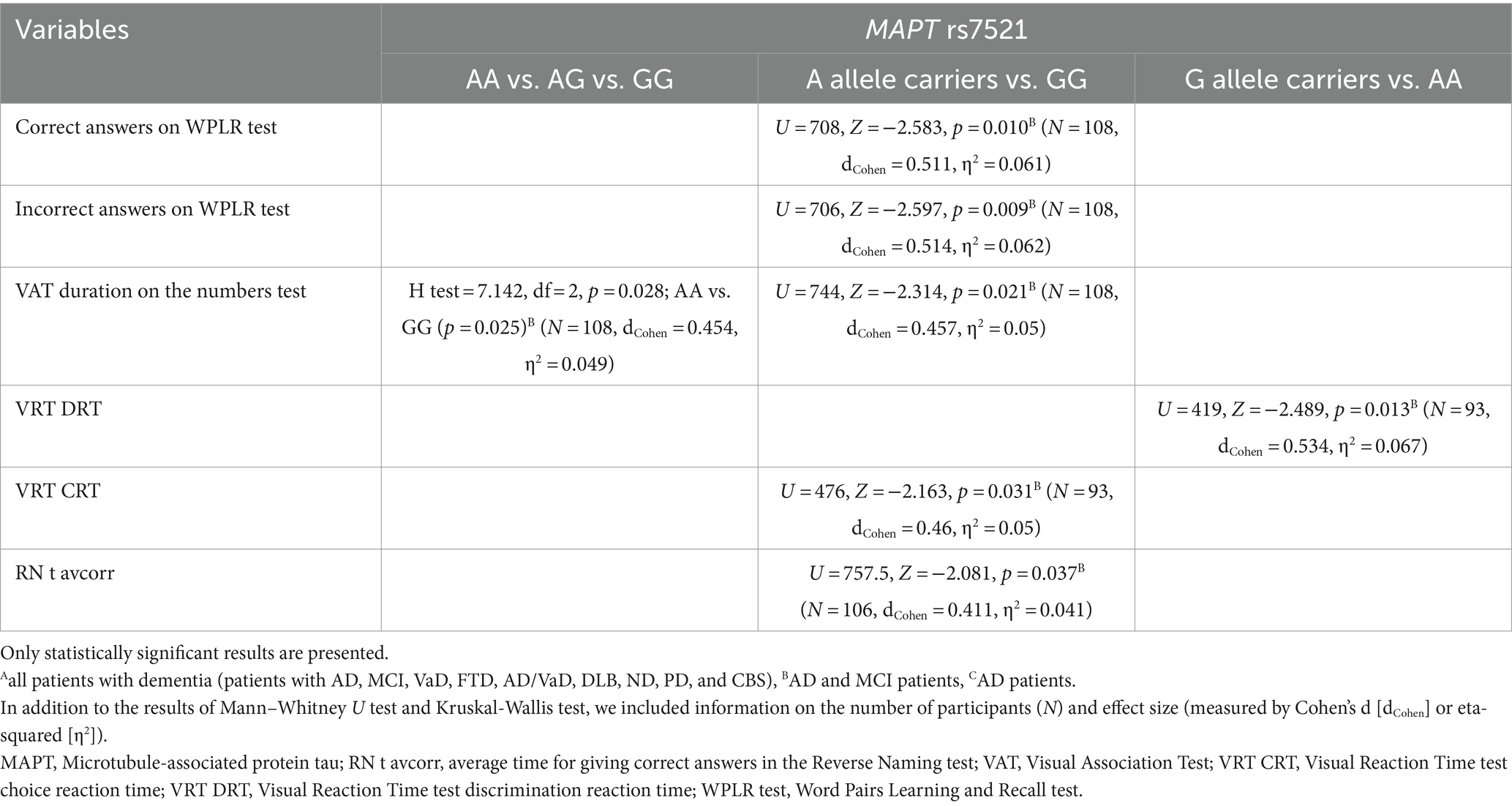
3.5 MAPT rs7521 genotype
GG homozygotes in the MAPT rs7521 genotype exhibited worse performance on the WPLR test. They had a significantly lower number of correct answers on the WPLR test and a higher number of incorrect answers compared to A allele carriers. Furthermore, GG homozygotes had a longer VAT duration on the numbers test compared to AA homozygotes. G allele carriers and GG homozygotes showed poorer performance on the VRT test. Specifically, VRT discrimination reaction time (DRT) was longer in G allele carriers compared to AA homozygotes. The VRT choice reaction time (CRT) was longer in GG homozygotes compared to A allele carriers. Finally, GG homozygotes had a longer average time for giving correct answers in the Reverse Naming test compared to A allele carriers (RN t avcorr) (Figure 5; Table 6).
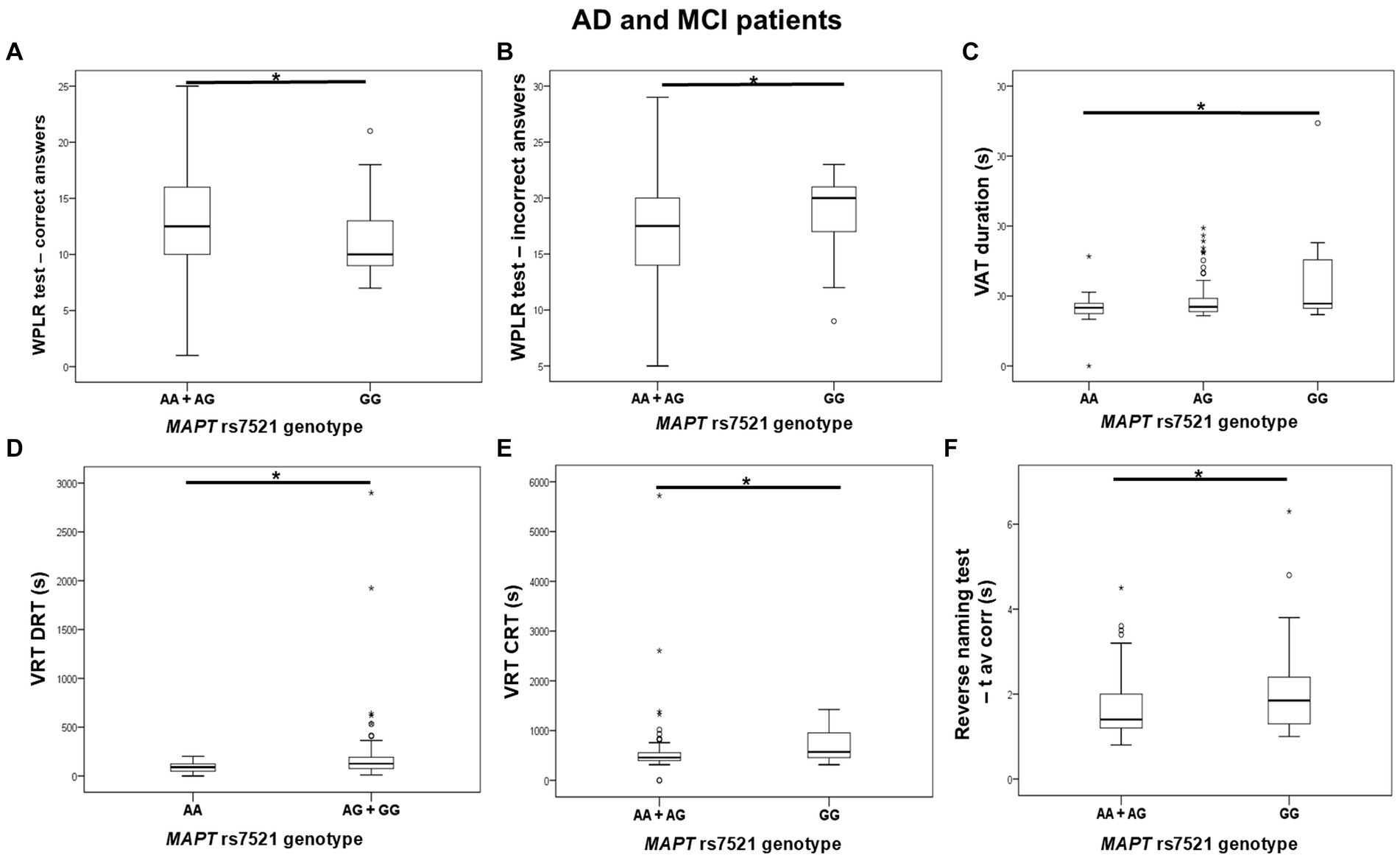
Figure 5. GG homozygotes in the MAPT rs7521 genotype had worse performance on the WPLR test (A,B), VAT test (C), VRT test (D,E), and Reverse Naming test (F) compared to A allele carriers. The box represents the interquartile range (between 25th and 75th percentiles), while the whiskers represent the range between the minimum and maximum values.
There was no significant difference in the distribution of MAPT rs7521 genotypes between AD patients (and also all patients with dementia) and HC. Plasma and CSF levels of S100B and NfL, as well as CSF p-tau181/Aβ1-42 ratio and YKL-40 levels, did not differ significantly between patients with different MAPT rs7521 genotypes.
3.6 MAPT haplotypes
Carriers of the H1H1 MAPT haplotype exhibited better performance, specifically a higher number of correct answers on the BNT, compared to carriers of the H2 haplotype (H1H2 + H2H2). H1H1 carriers also had fewer incorrect answers on the PPLR test compared to H2 carriers. Finally, H1H1 carriers performed better on the VRT test, with shorter VRT CRT compared to H2 carriers (Figure 6; Table 7). However, H1H1 carriers had higher plasma NfL levels compared to H2 carriers (Figure 6; Table 7).
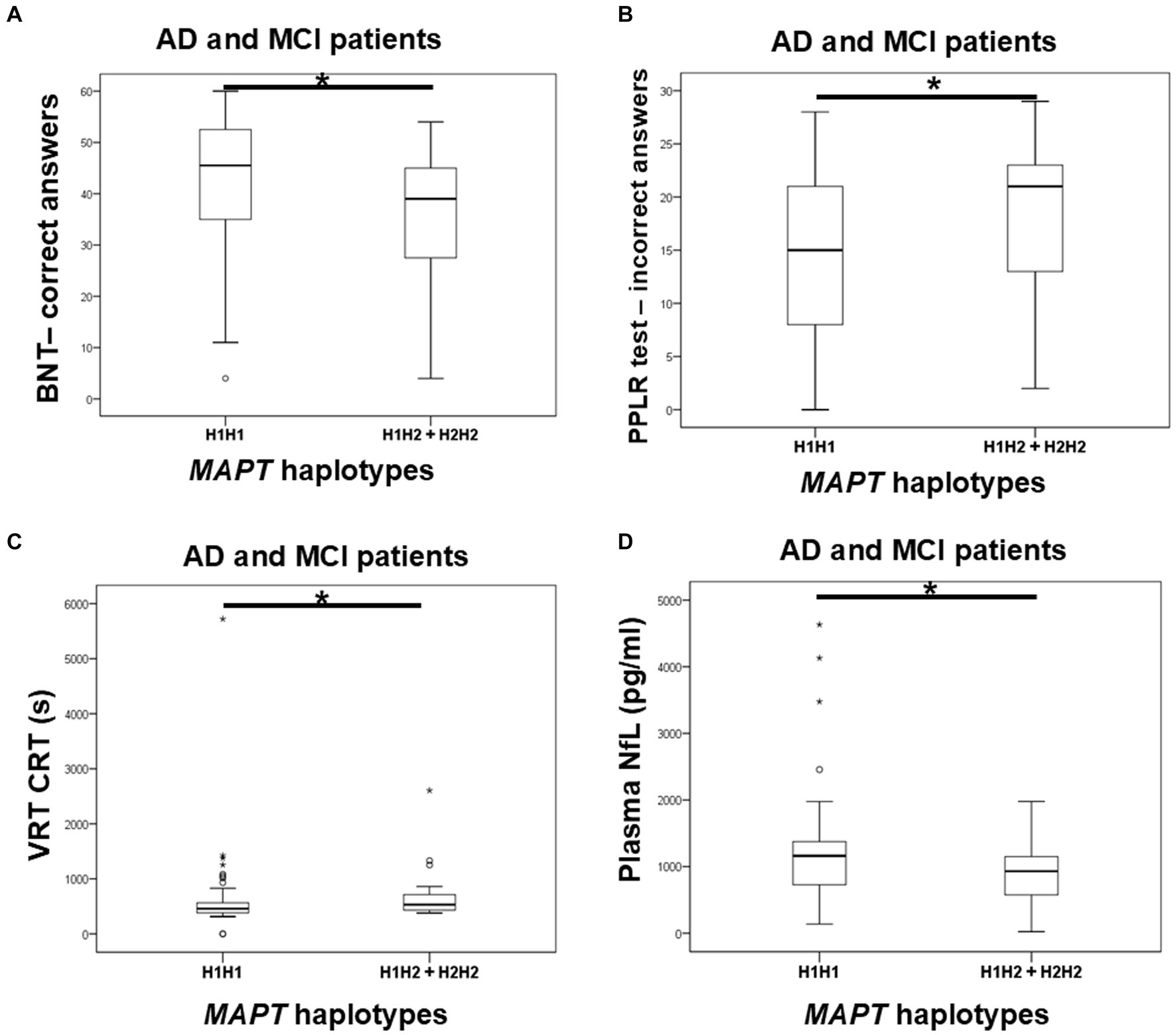
Figure 6. Carriers of the H1H1 MAPT haplotype had better performance on the BNT (A), PPLR test (B), and VRT test (C). However, carriers of the H1H1 MAPT haplotype had higher plasma NfL levels (D) compared to carriers of the H2 MAPT haplotype. The box represents the interquartile range (between 25th and 75th percentiles), while the whiskers represent the range between the minimum and maximum values.
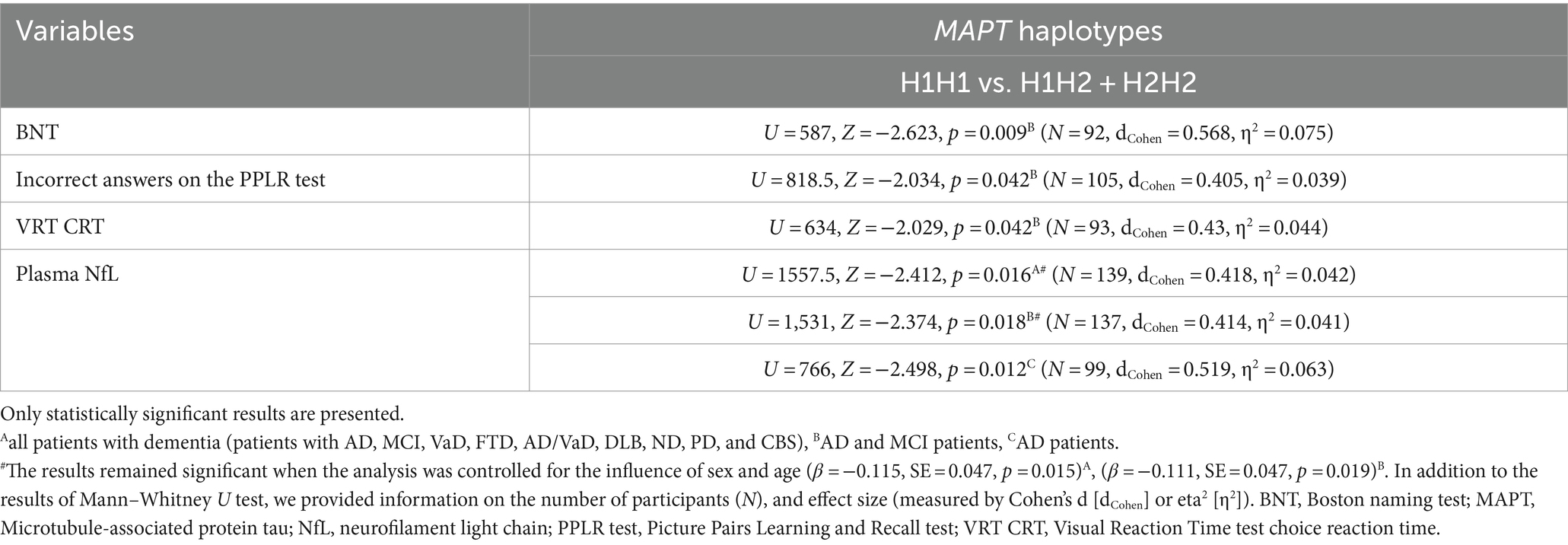
Table 7. Association of MAPT haplotypes with the results on neuropsychological tests and the levels of fluid biomarkers.
There was no significant difference in the distribution of MAPT haplotypes between AD patients (and also all patients with dementia) and HC. Plasma and CSF levels of S100B, as well as CSF NfL, p-tau181/Aβ1-42 ratio, and YKL-40 levels, did not differ significantly between patients with different MAPT haplotypes.
4 Discussion
In the current study, we tested the association of MAPT haplotype-tagging polymorphisms and MAPT haplotypes with AD. We investigated whether there is a difference in the levels of CSF biomarkers, the results of neuropsychological tests, and the frequency of APOE genotypes among patients with different MAPT genotypes and haplotypes. Multiple GWA studies were conducted using various CSF AD biomarkers as quantitative traits.1 Additionally, various plasma AD biomarkers were analyzed as quantitative traits in GWAS, including plasma p-tau181, NfL, Aβ1-42, Aβ1-40, and t-tau (Stevenson-Hoare et al., 2023). Scores on neuropsychological tests were also used as quantitative traits in AD-associated GWAS (Homann et al., 2022). Due to genetic variability between populations, it is important to replicate studies in different populations. Since GWAS using the aforementioned quantitative traits had not previously been conducted in the Croatian population, we aimed to perform preliminary screening in this study to determine if some of the CSF and plasma biomarkers, as well as scores on neuropsychological tests, are associated with MAPT haplotype-tagging polymorphisms and MAPT haplotypes.
In this study, we observed that carriers of the A allele and AA genotype in the MAPT rs1467967 polymorphism had worse performance on MMSE test, PPLR test, WPLR test, and ROCFT. These results align with our previous study, which showed significantly increased levels of CSF t-tau in AA homozygotes and increased levels of CSF t-tau and p-tau181 in carriers of the AG MAPT rs1467967 genotype (Babić Leko et al., 2018). Several studies (Mukherjee et al., 2007; Abraham et al., 2009) and two meta-analyses (Yuan et al., 2017; Zhou and Wang, 2017) failed to show an association of the MAPT rs1467967 polymorphism with AD. Similarly, the association of the MAPT rs1467967 polymorphism with CSF AD biomarkers was not confirmed in studies of Elias-Sonnenschein et al. (2013) and Kauwe et al. (2008). However, Laws et al. (2007) did show an association between the MAPT rs1467967 A allele and AD. Ning et al. also noted that the distribution of rs1467967 genotypes differed between VaD patients and HC, suggesting that the MAPT rs1467967 polymorphism could serve as a genetic biomarker for VaD (Ning et al., 2011).
GG homozygotes in the MAPT rs242557 polymorphism had worse performance on the CVLT test and exhibited more depression than carriers of the A allele. Additionally, G allele carriers had worse performance on the VAT and PPLR tests. In contrast to our observed results, most previous studies associated the A allele of the MAPT rs242557 polymorphism with AD. Compta et al. observed increased CSF t-tau and p-tau181 levels in PD patients carrying the A allele in MAPT rs242557 polymorphism (Compta et al., 2011), while Laws et al. observed increased CSF t-tau levels in AD patients carrying AA MAPT rs242557 genotype (Laws et al., 2007). The A allele in the MAPT rs242557 polymorphism has been associated with a higher risk for AD (Myers et al., 2005; Laws et al., 2007; Chang et al., 2014; Yuan et al., 2017; Zhou and Wang, 2017), although some studies failed to observe this association (Mukherjee et al., 2007; Abraham et al., 2009; Feulner et al., 2010; Setó-Salvia et al., 2011; Allen et al., 2014). A recent study involving young healthy adults showed that A allele MAPT rs242557 carriers had a thinner cortex in brain regions vulnerable to early tau pathology (Huang et al., 2023). Another study conducted in cognitively healthy elders showed that A allele MAPT rs242557 carriers had higher retention of the tau-PET tracer 18F-AV1451 in the hippocampus, making them more likely to develop tau pathology (Shen et al., 2019). However, a study by Tang et al. conducted in 3072 individuals from China showed that plasma tau levels were increased in participants carrying the G allele in the MAPT rs242557 SNP, and GG homozygotes were 1.47 times more likely to develop cognitive impairment compared to AG heterozygotes (Tang et al., 2021).
Carriers of the A allele in the MAPT rs3785883 polymorphism had significantly decreased CSF Aβ1-42 levels and increased plasma S100B levels. Additionally, a higher frequency of AA homozygotes was observed among patients with dementia. Conversely, GG homozygotes showed a significant increase in CSF S100B and YKL-40 levels. The MAPT rs3785883 A allele was associated with an increased risk for AD (Allen et al., 2014), higher levels of CSF p-tau (Kauwe et al., 2008; Cruchaga et al., 2010), earlier age of disease onset (Kauwe et al., 2008), and disease progression (Cruchaga et al., 2010; Peterson et al., 2014). However, some studies reported the association of the MAPT rs3785883 G allele with AD (Myers et al., 2005, 2007) or failed to observe any association between MAPT rs3785883 and AD (Mukherjee et al., 2007; Abraham et al., 2009; Setó-Salvia et al., 2011). In this study, we confirmed the association of the MAPT rs3785883 A allele with CSF Aβ1-42 levels, as observed by Kauwe et al. (2008). However, given the significant increase in CSF S100B and YKL-40 levels in GG homozygotes, further validation of this SNP’s association with AD is necessary using a larger cohort of patients. Despite this, compared to other biomarkers, CSF Aβ1-42 is considered a much more sensitive biomarker of AD. In our opinion, the association of the A allele with this biomarker is more closely related to the pathological changes that occur in AD.
TC heterozygotes in the MAPT rs2471738 genotype showed less disinhibition than TT homozygotes. C allele carriers performed worse on the CVLT test and had higher CSF p-tau181/Aβ1-42 ratios, while CC homozygotes made more Stroop test errors. More TT homozygotes and T allele carriers were found in patients with AD, AD and MCI, and dementia, with APOE ɛ4 carriers being more common among TT homozygotes. Our current study also found more TT homozygotes and T allele carriers among dementia patients and APOE ɛ4 carriers, but C allele carriers and CC homozygotes still showed poorer neuropsychological test performance and pathological CSF biomarker levels.
Although MAPT rs7521 polymorphism was not previously linked to AD (Mukherjee et al., 2007; Allen et al., 2014; Chang et al., 2014; Yuan et al., 2017; Zhang et al., 2017), we found that GG homozygotes performed worse on the WPLR test, VAT, VRT, and Reverse naming test.
H2 MAPT carriers performed worse on the BNT, PPLR, and VRT tests, supporting our previous study results, which showed significantly increased levels of CSF t-tau and p-tau231 among carriers of the H2 haplotype (Babić Leko et al., 2018). However, since carriers of H1H1 MAPT haplotype showed higher plasma NfL levels, further validation on larger cohorts is needed. Most studies link the H1 MAPT haplotype to increased AD risk (Myers et al., 2005; Laws et al., 2007; Kauwe et al., 2008; Di Maria et al., 2010; Pastor et al., 2016; Sánchez-Juan et al., 2019) or suggest the H2 haplotype is protective against AD (Allen et al., 2014; Zhang et al., 2017), though some found no association (Baker et al., 2000; Russ et al., 2001; Mukherjee et al., 2007; Abraham et al., 2009). Interestingly, a recent study involving 338 brain samples of Pick’s disease, a relatively rare and predominantly sporadic form of FTD classified as a primary tauopathy, linked the H2 MAPT haplotype to an increased risk of Pick’s disease (Valentino et al., 2023). Geographical differences in MAPT haplotype frequency may also help explain the discrepancies in results. In Europe, the H2 MAPT haplotype frequency is the highest in Mediterranean countries (Donnelly et al., 2010). In Croatia, which is part of the Mediterranean Basin, the H2 haplotype frequency is over 30% (around 33%). Thus, although we did not observe a significant difference in the distribution of MAPT haplotypes between HC and patients with dementia, carriers of the H2 MAPT haplotype showed worse performance on neuropsychological tests and pathological levels of various CSF biomarkers. This suggests that some effects of the H2 MAPT haplotype on disease progression might not be observed in other populations due to the smaller frequency of H2 MAPT haplotype carriers, which is not the case in the Croatian population.
The main strength of our study is the large number of participants and the inclusion of various biomarkers (such as CSF and plasma biomarkers, neuropsychological test, and APOE genotype). The main limitation is the cross-sectional design of the study, which does not allow for longitudinal tracking of participants. Additionally, CSF and plasma biomarkers and neuropsychological tests were determined in only 220 participants out of the total number of 964 participants. CSF biomarkers and neuropsychological tests were also determined only in patients with dementia since our “healthy control” group was recruited within a different project, and they have only genetic data.
In conclusion, our study showed that participants with the A allele in the MAPT rs1467967 polymorphism, the G allele in the MAPT rs242557 polymorphism, and the GG genotype in the MAPT rs7521 polymorphism performed worse on various neuropsychological tests. Although T allele carriers in MAPT rs2471738 polymorphism were more represented among patients with dementia and APOE ɛ4 carriers, C allele carriers and CC homozygotes still showed worse performance on neuropsychological tests and pathological levels of various CSF biomarkers. Carriers of the H2 MAPT haplotype also performed worse on various neuropsychological tests, supporting our previous findings that associated the H2 MAPT haplotype with pathological CSF AD biomarkers (Babić Leko et al., 2018). Regarding the MAPT rs3785883 polymorphism, further research is needed as both AA genotype and GG genotype showed associations with CSF and plasma AD biomarkers, although AA homozygotes were more represented among patients with dementia. Our study further confirmed the association of MAPT haplotype-tagging polymorphisms and MAPT haplotypes with AD. Some of our results align with mainstream findings, such as the association of the A allele in MAPT rs1467967 and rs3785883 polymorphisms with worse performance on neuropsychological tests and pathological CSF Aβ1-42 levels, respectively. However, some results differ from most studies, such as the association of the G allele in MAPT rs242557 polymorphism and the H2 MAPT haplotype with worse performance on neuropsychological tests. Therefore, further research on the association of MAPT haplotype-tagging polymorphisms and MAPT haplotypes with AD is necessary.
Data availability statement
The data presented in this study are available on request from the corresponding authors.
Ethics statement
The Ethical Committee of the Clinical Hospital Center Zagreb (case no. 02/21 AG, class 8.1-18/82-2, dated April 24, 2018) and the Central Ethical Committee of the University of Zagreb Medical School (case no. 380-59-10,106-18-111/126, class 641-01/18-02/01, dated June 20, 2018) approved all procedures conducted in this research. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
MBL: Formal analysis, Investigation, Methodology, Project administration, Writing – review & editing, Conceptualization, Data curation, Funding acquisition, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. EŠ: Investigation, Methodology, Writing – review & editing, Formal analysis, Project administration. NW: Investigation, Methodology, Writing – review & editing. MN: Investigation, Methodology, Writing – review & editing. NP: Investigation, Methodology, Software, Writing – review & editing. KZ: Investigation, Methodology, Writing – review & editing. LL: Investigation, Methodology, Writing – review & editing. ŽV: Data curation, Investigation, Methodology, Writing – review & editing. MB: Data curation, Investigation, Methodology, Writing – review & editing. FB: Investigation, Writing – review & editing. TZ: Investigation, Writing – review & editing. RS: Data curation, Investigation, Methodology, Supervision, Validation, Writing – review & editing. GŠ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Recovery and Resilience Plan G.A. NPOO.C3.2.R3-I1.04.0257 to GŠ (“Detection and validation of molecular markers of inflammation in Alzheimer’s and Parkinson’s disease, multiple sclerosis, and schizophrenia”), University of Zagreb grant no. 10106–24-1526 to GŠ (“Inflammasome, tau protein, and nucleocytoplasmic transport in Alzheimer’s disease”), Croatian Science Foundation grant no. IP-2019-04-3584 to GŠ (“Role of the blood–brain barrier, innate immunity, and tau protein oligomerization in the pathogenesis of Alzheimer’s disease”), and Croatian Science Foundation grant no. IP-2019-04-2593 to TZ (“Regulation of thyroid and parathyroid function and blood calcium homeostasis”).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2024.1456670/full#supplementary-material
Footnotes
References
Abraham, R., Sims, R., Carroll, L., Hollingworth, P., O’Donovan, M. C., Williams, J., et al. (2009). An association study of common variation at the MAPT locus with late-onset Alzheimer’s disease. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 150B, 1152–1155. doi: 10.1002/ajmg.b.30951
Albert, M. S., DeKosky, S. T., Dickson, D., Dubois, B., Feldman, H. H., Fox, N. C., et al. (2011). The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 270–279. doi: 10.1016/j.jalz.2011.03.008
Allen, M., Kachadoorian, M., Quicksall, Z., Zou, F., Chai, H., Younkin, C., et al. (2014). Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimers Res. Ther. 6:39. doi: 10.1186/alzrt268
Alzheimer's Association (2020). 2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 16, 391–460. doi: 10.1002/ALZ.12068
Babić Leko, M., Willumsen, N., Nikolac Perković, M., Klepac, N., Borovečki, F., Hof, P. R., et al. (2018). Association of MAPT haplotype-tagging polymorphisms with cerebrospinal fluid biomarkers of Alzheimer’s disease: a preliminary study in a Croatian cohort. Brain Behav. 8:e01128. doi: 10.1002/brb3.1128
Baker, M., Graff-Radford, D., Wavrant Devrièze, F., Graff-Radford, N., Petersen, R. C., Kokmen, E., et al. (2000). No association between TAU haplotype and Alzheimer’s disease in population or clinic based series or in familial disease. Neurosci. Lett. 285, 147–149. doi: 10.1016/S0304-3940(00)01057-0
Bertram, L., McQueen, M. B., Mullin, K., Blacker, D., and Tanzi, R. E. (2007). Systematic meta-analyses of Alzheimer disease genetic association studies: the Alz gene database. Nat. Genet. 39, 17–23. doi: 10.1038/NG1934
Chang, C. W., Hsu, W. C., Pittman, A., Wu, Y. R., Hardy, J., and Fung, H. C. (2014). Structural study of the microtubule-associated protein tau locus of Alzheimer’s disease in Taiwan. Biom. J. 37, 127–132. doi: 10.4103/2319-4170.117891
Compta, Y., Ezquerra, M., Muñoz, E., Tolosa, E., Valldeoriola, F., Rios, J., et al. (2011). High cerebrospinal tau levels are associated with the rs 242557 tau gene variant and low cerebrospinal β-amyloid in Parkinson's disease. Neurosci. Lett. 487, 169–173. doi: 10.1016/j.neulet.2010.10.015
Cruchaga, C., Karch, C. M., Jin, S. C., Benitez, B. A., Cai, Y., Guerreiro, R., et al. (2014). Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature 505, 550–554. doi: 10.1038/nature12825
Cruchaga, C., Kauwe, J. S. K., Mayo, K., Spiegel, N., Bertelsen, S., Nowotny, P., et al. (2010). SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer’s disease. PLoS Genet. 6:e1001101. doi: 10.1371/journal.pgen.1001101
Di Maria, E., Cammarata, S., Parodi, M. I., Borghi, R., Benussi, L., Galli, M., et al. (2010). The H1 haplotype of the tau gene (MAPT) is associated with mild cognitive impairment. J. Alzheimers Dis. 19, 909–914. doi: 10.3233/JAD-2010-1285
Donnelly, M. P., Paschou, P., Grigorenko, E., Gurwitz, D., Mehdi, S. Q., Kajuna, S. L. B., et al. (2010). The distribution and most recent common ancestor of the 17q21 inversion in humans. Am. J. Hum. Genet. 86, 161–171. doi: 10.1016/J.AJHG.2010.01.007
Elias-Sonnenschein, L. S., Helisalmi, S., Natunen, T., Hall, A., Paajanen, T., Herukka, S.-K., et al. (2013). Genetic loci associated with Alzheimer’s disease and cerebrospinal fluid biomarkers in a Finnish case-control cohort. PLoS One 8:e59676. doi: 10.1371/journal.pone.0059676
Feulner, T. M., Laws, S. M., Friedrich, P., Wagenpfeil, S., Wurst, S. H. R., Riehle, C., et al. (2010). Examination of the current top candidate genes for Alzheimer's disease in a genome-wide association study. Mol. Psychiatry 15, 756–766. doi: 10.1038/MP.2008.141
Hachinski, V. C., Iliff, L. D., Zilhka, E., Du Boulay, G. H., McAllister, V. L., Marshall, J., et al. (1975). Cerebral blood flow in dementia. Arch. Neurol. 32, 632–637. doi: 10.1001/archneur.1975.00490510088009
Homann, J., Osburg, T., Ohlei, O., Dobricic, V., Deecke, L., Bos, I., et al. (2022). Genome-wide association study of Alzheimer’s disease brain imaging biomarkers and neuropsychological phenotypes in the European medical information framework for Alzheimer’s disease multimodal biomarker discovery dataset. Front. Aging Neurosci. 14:37. doi: 10.3389/FNAGI.2022.840651/FULL
Huang, W., Zeng, J., Jia, L., Zhu, D., O’Brien, J., Ritchie, C., et al. (2023). Genetic risks of Alzheimer’s by APOE and MAPT on cortical morphology in young healthy adults. Brain Commun. 5:234. doi: 10.1093/BRAINCOMMS/FCAD234
Jonsson, T., Stefansson, H., Steinberg, S., Jonsdottir, I., Jonsson, P. V., Snaedal, J., et al. (2013). Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116. doi: 10.1056/NEJMoa1211103
Kauwe, J. S. K., Cruchaga, C., Mayo, K., Fenoglio, C., Bertelsen, S., Nowotny, P., et al. (2008). Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc. Natl. Acad. Sci. 105, 8050–8054. doi: 10.1073/pnas.0801227105
Laws, S. M., Friedrich, P., Diehl-Schmid, J., Müller, J., Eisele, T., Bäuml, J., et al. (2007). Fine mapping of the MAPT locus using quantitative trait analysis identifies possible causal variants in Alzheimer’s disease. Mol. Psychiatry 12, 510–517. doi: 10.1038/sj.mp.4001935
McKhann, G. M., Knopman, D. S., Chertkow, H., Hyman, B. T., Jack, C. R., Kawas, C. H., et al. (2011). The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 263–269. doi: 10.1016/j.jalz.2011.03.005
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. doi: 10.1093/nar/16.3.1215
Mukherjee, O., Kauwe, J. S., Mayo, K., Morris, J. C., and Goate, A. M. (2007). Haplotype-based association analysis of the MAPT locus in late onset Alzheimer’s disease. BMC Genet. 8:3. doi: 10.1186/1471-2156-8-3
Myers, A. J., Kaleem, M., Marlowe, L., Pittman, A. M., Lees, A. J., Fung, H. C., et al. (2005). The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum. Mol. Genet. 14, 2399–2404. doi: 10.1093/hmg/ddi241
Myers, A. J., Pittman, A. M., Zhao, A. S., Rohrer, K., Kaleem, M., Marlowe, L., et al. (2007). The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol. Dis. 25, 561–570. doi: 10.1016/J.NBD.2006.10.018
Neary, D., Snowden, J. S., Gustafson, L., Passant, U., Stuss, D., Black, S., et al. (1998). Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 51, 1546–1554. doi: 10.1212/wnl.51.6.1546
Ning, M., Zhang, Z., Chen, Z., Zhao, T., Zhang, D., Zhou, D., et al. (2011). Genetic evidence that vascular dementia is related to Alzheimer’s disease: genetic association between tau polymorphism and vascular dementia in the Chinese population. Age Ageing 40, 125–128. doi: 10.1093/ageing/afq131
Pastor, P., Moreno, F., Clarimón, J., Ruiz, A., Combarros, O., Calero, M., et al. (2016). MAPT H1 haplotype is associated with late-onset Alzheimer’s disease risk in APOEɛ4 noncarriers: results from the dementia genetics Spanish consortium. J. Alzheimers Dis. 49, 343–352. doi: 10.3233/JAD-150555
Petersen, R. C., Smith, G. E., Waring, S. C., Ivnik, R. J., Tangalos, E. G., and Kokmen, E. (1999). Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 56, 303–308. doi: 10.1001/archneur.56.3.303
Peterson, D., Munger, C., Crowley, J., Corcoran, C., Cruchaga, C., Goate, A. M., et al. (2014). Variants in PPP3R1 and MAPT are associated with more rapid functional decline in Alzheimer’s disease: the Cache County dementia progression study. Alzheimers Dement. 10, 366–371. doi: 10.1016/J.JALZ.2013.02.010
Pittman, A. M., Myers, A. J., Abou-Sleiman, P., Fung, H. C., Kaleem, M., Marlowe, L., et al. (2005). Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J. Med. Genet. 42, 837–846. doi: 10.1136/jmg.2005.031377
Román, G. C., Tatemichi, T. K., Erkinjuntti, T., Cummings, J. L., Masdeu, J. C., Garcia, J. H., et al. (1993). Vascular dementia: diagnostic criteria for research studies. Report of the NINDS-AIREN international workshop. Neurology 43, 250–260. doi: 10.1212/wnl.43.2.250
Rudan, I., Marušić, A., Janković, S., Rotim, K., Boban, M., Lauc, G., et al. (2009). “10 001 Dalmatians:” Croatia launches its national biobank. Croat. Med. J. 50, 4–6. doi: 10.3325/cmj.2009.50.4
Russ, C., Powell, J. F., Zhao, J., Baker, M., Hutton, M., Crawford, F., et al. (2001). The microtubule associated protein tau gene and Alzheimer’s disease - an association study and meta-analysis. Neurosci. Lett. 314, 92–96. doi: 10.1016/s0304-3940(01)02289-3
Sánchez-Juan, P., Moreno, S., de Rojas, I., Hernández, I., Valero, S., Alegret, M., et al. (2019). The MAPT H1 haplotype is a risk factor for Alzheimer’s disease in APOE ε4 non-carriers. Front. Aging Neurosci. 11:327. doi: 10.3389/FNAGI.2019.00327
Setó-Salvia, N., Clarimón, J., Pagonabarraga, J., Pascual-Sedano, B., Campolongo, A., Combarros, O., et al. (2011). Dementia risk in Parkinson's disease: disentangling the role of MAPT haplotypes. Arch. Neurol. 68, 359–364. doi: 10.1001/ARCHNEUROL.2011.17
Shen, X. N., Miao, D., Li, J. Q., Tan, C. C., Cao, X. P., Tan, L., et al. (2019). MAPT rs 242557 variant is associated with hippocampus tau uptake on 18F-AV-1451 PET in non-demented elders. Aging (Albany NY) 11, 874–884. doi: 10.18632/AGING.101783
Stevenson-Hoare, J., Heslegrave, A., Leonenko, G., Fathalla, D., Bellou, E., Luckcuck, L., et al. (2023). Plasma biomarkers and genetics in the diagnosis and prediction of Alzheimer’s disease. Brain 146, 690–699. doi: 10.1093/BRAIN/AWAC128
Strickland, S. L., Reddy, J. S., Allen, M., N’songo, A., Burgess, J. D., Corda, M. M., et al. (2020). MAPT haplotype-stratified GWAS reveals differential association for AD risk variants. Alzheimers Dement. 16, 983–1002. doi: 10.1002/ALZ.12099
Tang, X., Liu, S., Cai, J., Chen, Q., Xu, X., Mo, C. B., et al. (2021). Effects of gene and plasma tau on cognitive impairment in rural Chinese population. Curr. Alzheimer Res. 18, 56–66. doi: 10.2174/1567205018666210324122840
Valentino, R. R., Scotton, W. J., Roemer, S. F., Lashley, T., Heckman, M. G., Shoai, M., et al. (2023). Creating the Pick’s disease international consortium: association study of MAPT H2 haplotype with risk of Pick’s disease. med Rxiv. doi: 10.1101/2023.04.17.23288471
Vogel, J. W., Young, A. L., Oxtoby, N. P., Smith, R., Ossenkoppele, R., Strandberg, O. T., et al. (2021). Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat. Med. 27, 871–881. doi: 10.1038/s41591-021-01309-6
World Medical Association (2013). World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310, 2191–2194. doi: 10.1001/jama.2013.281053
Yuan, H., Du, L., Ge, P., Wang, X., and Xia, Q. (2017). Association of microtubule-associated protein tau gene polymorphisms with the risk of sporadic Alzheimer’s disease: a meta-analysis. Int. J. Neurosci. 128, 1–22. doi: 10.1080/00207454.2017.1400972
Zhang, C.-C., Zhu, J.-X., Wan, Y., Tan, L., Wang, H.-F., Yu, J.-T., et al. (2017). Meta-analysis of the association between variants in MAPT and neurodegenerative diseases. Oncotarget 8, 44994–45007. doi: 10.18632/oncotarget.16690
Keywords: Alzheimer’s disease, APOE gene, biomarkers, cerebrospinal fluid, dementia, MAPT gene, MAPT haplotypes, MAPT polymorphisms
Citation: Babić Leko M, Španić Popovački E, Willumsen N, Nikolac Perković M, Pleić N, Zubčić K, Langer Horvat L, Vogrinc Ž, Boban M, Borovečki F, Zemunik T, de Silva R and Šimić G (2024) Further validation of the association between MAPT haplotype-tagging polymorphisms and Alzheimer’s disease: neuropsychological tests, cerebrospinal fluid biomarkers, and APOE genotype. Front. Mol. Neurosci. 17:1456670. doi: 10.3389/fnmol.2024.1456670
Edited by:
Marija Heffer, Josip Juraj Strossmayer University of Osijek, CroatiaReviewed by:
Natalie A. Prowse, Carleton University, CanadaGamze Guven, Istanbul University, Türkiye
Copyright © 2024 Babić Leko, Španić Popovački, Willumsen, Nikolac Perković, Pleić, Zubčić, Langer Horvat, Vogrinc, Boban, Borovečki, Zemunik, de Silva and Šimić. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mirjana Babić Leko, bWJhYmljbGVrb0BnbWFpbC5jb20=; Goran Šimić, Z3NpbWljQGhpaW0uaHI=
 Mirjana Babić Leko
Mirjana Babić Leko Ena Španić Popovački1
Ena Španić Popovački1 Matea Nikolac Perković
Matea Nikolac Perković Klara Zubčić
Klara Zubčić Lea Langer Horvat
Lea Langer Horvat Željka Vogrinc
Željka Vogrinc Marina Boban
Marina Boban Fran Borovečki
Fran Borovečki Tatijana Zemunik
Tatijana Zemunik Rohan de Silva
Rohan de Silva Goran Šimić
Goran Šimić