
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci., 26 October 2023
Sec. Molecular Signalling and Pathways
Volume 16 - 2023 | https://doi.org/10.3389/fnmol.2023.1279232
This article is part of the Research TopicGlobal Excellence in Ion Channel Pharmacology and Channelopathies: A Latin American PerspectiveView all 5 articles
Introduction: Oligodendrocyte progenitor cells (OPCs) are vital for neuronal myelination and remyelination in the central nervous system. While the molecular mechanisms involved in OPCs’ differentiation and maturation are not completely understood, GABA is known to positively influence these processes through the activation of GABAA receptors (GABAARs). The molecular identity of GABAARs expressed in human OPCs remains unknown, which restricts their specific pharmacological modulation to directly assess their role in oligodendrocytes’ maturation and remyelination.
Methods: In this study, we conducted a transcriptomic analysis to investigate the molecular stoichiometry of GABAARs in OPCs from the human brain. Using eight available transcriptomic datasets from the human brain cortex of control individuals, we analyzed the mRNA expression of all 19 known GABAARs subunit genes in OPCs, with variations observed across different ages.
Results: Our analysis indicated that the most expressed subunits in OPCs are α1–3, β1–3, γ1–3, and ε. Moreover, we determined that the combination of any α with β2 and γ2 is likely to form heteropentameric GABAARs in OPCs. Importantly, we also found a strong correlation between GABAAR subunits and transcripts for postsynaptic scaffold proteins, suggesting the potential postsynaptic clustering of GABAARs in OPCs.
Discussion: This study presents the first transcriptional-level identification of GABAAR subunits expressed in human OPCs, providing potential receptor combinations. Understanding the molecular composition of GABAARs in OPCs not only enhances our knowledge of the underlying mechanisms in oligodendrocyte maturation but also opens avenues for targeted pharmacological interventions aimed at modulating these receptors to promote remyelination in neurological disorders.
Oligodendrocyte progenitor cells (OPCs) are among the most abundant glial cells distributed throughout the brain. These cells are known for maintaining the myelination of the central nervous system throughout an individual’s lifespan (Dawson et al., 2003; Trotter et al., 2010; Richardson et al., 2011; Gallo and Deneen, 2014; Fernandez-Castaneda and Gaultier, 2016). Under normal circumstances, acute myelin loss is compensated through a sequence of events involving the activation, proliferation, migration, and differentiation of OPCs into the damaged zone where myelin was compromised (van Tilborg et al., 2018; Baydyuk et al., 2020). It is suggested that this sequence begins with the involvement of GABAA receptors (GABAARs) expressed in OPCs. When these receptors are activated by GABA—released at direct synaptic contacts with axons (Lin and Bergles, 2004; Jabs et al., 2005; Gallo et al., 2008)– it leads to membrane depolarization and an increase in cytosolic Ca2+ (Kirchhoff and Kettenmann, 1992; Arellano et al., 2016), thereby setting off the distinct stages of OPC maturation and remyelination (Cheli et al., 2016; Santiago González et al., 2017; Marisca et al., 2020). However, in some life-threatening conditions, including genetic disorders (leukodystrophies), traumatic brain injuries, and autoimmune diseases, there appears to be a decrease of response of OPCs to demyelination, which correlates with the clinical presentation of these disorders (Raabe et al., 2018; Butt et al., 2019; Gruchot et al., 2019; Huntemer-Silveira et al., 2020).
We propose that the control of GABAARs activity and its downstream signaling in OPCs may be critical for the development of drugs aimed at treating demyelinating diseases (Reyes-Haro et al., 2021). However, the stoichiometry of GABAARs in human OPCs remains unknown. GABAAR pentamers are commonly assembled from subunits of three different families, encoded by a pool of 19 genes (α1–6, β1–3, γ1–3, δ, ε, π, θ, ρ1–3), and they are usually comprised of two α, two β, and one γ or δ subunits (McKernan and Whiting, 1996; Petri et al., 2002). Through transcriptional and functional studies, we recently observed that most GABAARs in OPCs from neonate (P0-P14) rats and mice are most likely made up of α3β2γ1 subunits (Ordaz et al., 2021), and in one human dataset, we found that the most expressed subunits were α3, β1 and 3, γ1, γ2, and ε (Serrano-Regal et al., 2020), suggesting potential differences between humans and rodents. To better determine the potential diversity of GABAARs subunit expression in the human brain, we used multiple publicly available human RNA sequencing datasets, and we explored the likely stoichiometry of GABAARs in OPCs, their enriched molecular pathways and protein networks, their correlation with accessory proteins, and their potential differences in expression as a function of age.
We downloaded three single-cell (sc) and five single-nucleus (sn) RNA-sequencing datasets from the Gene Expression Omnibus (GEO) repository1 and the Allen Brain Map2 (Supplementary Table 1). We chose datasets derived from healthy cortical tissue (frontal and temporal cortex) from fetal (n = 8 subjects), pediatric (n = 9), adolescent (n = 1), and adult (n = 42) human populations. To identify OPCs among all other cell types, we selected cells expressing the platelet-derived growth factor α type receptor (PDGFRA), a marker enriched in OPCs (Pringle et al., 1992; Pringle and Richardson, 1993; Miller et al., 1999). In this selection, we found a total of 12,822 OPCs, with 11% (1,419 OPCs) expressing at least one of the 19 GABAAR subunits. More specifically, in adults, 20% (1,007/4,796) of PDGFRA+ cells expressed at least one GABAAR subunit, compared to 0.6% (20/3,086 OPCs) in adolescent, 6% (268/4,502 OPCs) in pediatric, and 28% (124/438 OPCs) in fetal populations (Supplementary Table 2).
Given that GABAARs are typically present in OPCs, the low percentage of GABAAR subunit expression was unexpected. We hypothesized that this might be related to different selection criteria among research groups during quality control. Consequently, we also performed an analysis in CellRanger using FASTQ files from four out of the eight datasets we obtained from the GEO repository and the Allen Brain Map (specifically PRJNA5776618, PRJNA673712, PRJNA589018, PRJNA674571). The FASTQ files for Jäkel et al. (2019) (GSE118257) and Hodge et al. (2019)3 were not publicly accessible. Although Darmanis et al. (2015) (PRJNA281204) and Lake et al. (2018) (PRJNA383372) had FASTQ files available, they were incompatible with CellRanger due to the unique UMI/Cell Barcode/Sequencing index used in their Drop-seq platform. From our analysis of the four compatible FASTQ datasets, we categorized OPCs as PDGFRA+ cells and examined their GABAAR subunit gene expression in the adult healthy control cortex. Interestingly, out of 635 OPCs (PDGFRA+ cells) we identified, 325 (51%) expressed at least one of the 19 GABAAR subunits (Supplementary Table 2). This contrasts with results from the same four datasets directly sourced from the GEO repository, where 4,002 OPCs were identified, but only 447 (11%) exhibited expression of any of the 19 GABAAR subunits (Supplementary Table 2). This data implies that selection criteria for quality control can significantly influence the percentage of recognized OPCs. However, it remains unclear whether this discrepancy affects the contribution of GABAAR subunit genes to OPCs.
To integrate the GABAAR subunits from all seven GEO datasets and one from the Allen Brain Map, we transformed the raw data expressed as unique molecular identifiers (UMIs) or FPKMs, into fractional contribution (FC), which is the percentage of the sum of the expression levels of each subunit gene in each cell over the sum of all 19 subunit genes. This metric reflects the contribution of each subunit to the total mRNA available to form GABAARs (Sequeira et al., 2019). The FC from each of the eight datasets indicated that the family of β subunits has the largest contribution, whereas ρ and π subunits have minimal contribution. The subunits α1/3 and γ2/3, showed more contribution compared to other subunits from their respective families (Figure 1A).
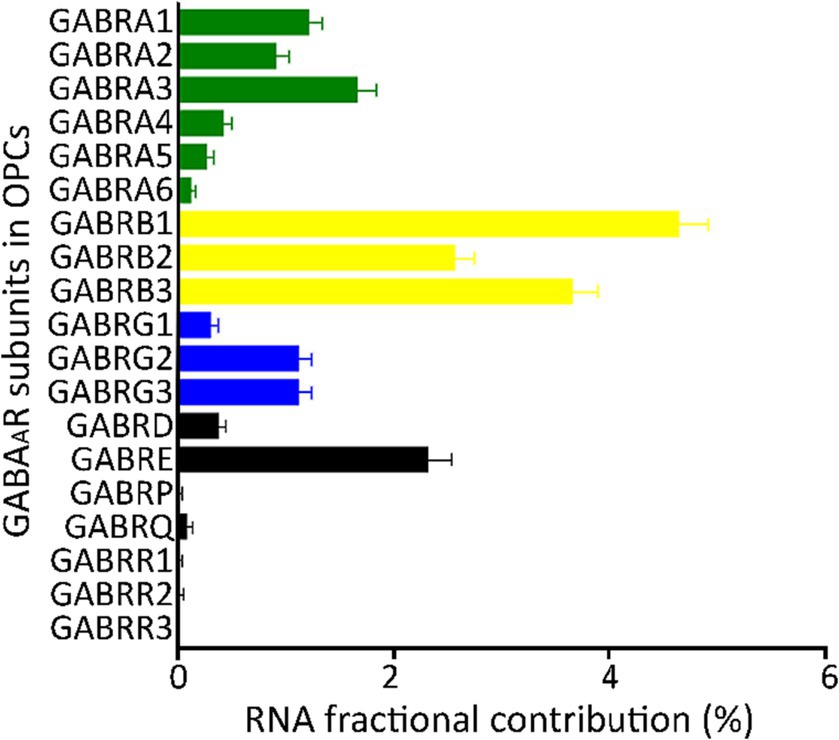
Figure 1. Fractional contribution of GABAAR subunit genes. Average fractional contribution (FC) of each GABAAR subunit gene, derived from 1,007 PDGFRA+ cells (OPCs) across 8 datasets from the human cortex (GSE138852, GSE155488, GSE67835, GSE118257, GSE140231, GSE160813, GSE97930, https://celltypes.brain-map.org/rnaseq). Bars depict the mean FC of GABAAR subunit gene across the eight datasets, and standard error (SE) is indicated for each bar.
In our analysis of four individual GEO datasets, we observed variations in subunit contributions between them (Supplementary Figures 1A,C,E,G). This diverse contribution of different subunits across datasets suggests the existence of multiple GABAAR subunit stoichiometries in human OPCs. When using the FC analysis on these datasets with available FASTQ files, there were also noticeable differences in subunit contributions. However, in many instances, these contributions remained consistent (Supplementary Figures 1B,D,F,H). For the OPCs in these four FASTQ datasets, the most dominant GABAAR subunits were α1–3, β1–3, γ2, and δ (Supplementary Figure 2A). Similarly, when examining the same four datasets from the GEO repository, the primary contributors were α1–3, β1–3, γ2/3, and ε (Supplementary Figure 2B).
Importantly, single-cell data tends to be highly variable. The coverage level within a single cell is approximately 5%–40% of what is observed in bulk mRNA (Lun et al., 2016; Van den Berge et al., 2018; Choi et al., 2020; Ding et al., 2020). This disparity can largely be attributed to a significant number of cells that show little to no expression of certain subunits. Given these considerations, we incorporated two alternative methods to compute the FC (refer to “Materials and methods: Determination of FCs” for details): FC2 was derived from the expression level of each subunit gene relative to the sum of all 19 gene subunits per dataset, and FC3 was based on the percentage of cells expressing each subunit gene relative to the total number of cells (Supplementary Table 3). By employing these analytical approaches, we identified contributions of the majority of the 19 GABAAR subunit genes in human OPCs. Notably, subunits α1–3, β1–3, γ2/3, and ε showed the highest contributions (Figure 1; Supplementary Figures 3A,B). As a result, our validation indicates that for most subunits, any of the three FC methods can be reliably employed to normalize the expression levels across datasets.
The expression pattern of GABAAR subunits in cortical neurons has been well established (Sieghart and Sperk, 2002; Olsen and Sieghart, 2009). To understand if these patterns are consistent in different cell types, we compared OPCs and neurons (identified by their unique barcodes) from five GEO datasets (GSE14023, GSE67835, GSE138852, GSE118257, GSE97930). Our analysis revealed that both cell types have high contributions of subunits α1–4, β1–3, γ2/3, and δ. However, within the OPCs, α3, β1, and γ3 dominated their respective subunit families (Figure 2A). In contrast, neurons predominantly had α1, β2/3, and γ2/3 subunit contributions (Figure 2B), consistent with previous studies (Serrano-Regal et al., 2020; Ordaz et al., 2021). The contribution of other subunits was reduced in both cell types. To determine if each cell type possesses a distinct molecular signature of GABAAR subunits, we conducted a two-way hierarchical clustering analysis comparing OPCs and neurons. This was based on data from the five GEO datasets (GSE14023, GSE67835, GSE138852, GSE118257, GSE97930) which contains OPC and neurons, with cells identifiable by their unique barcodes. The analysis revealed two distinct clusters: one for OPCs and another for neurons across all datasets (Figure 2C). This underscores that each cell type has its unique molecular profile for GABAARs. We observed that the α, β, and γ subunits were particularly abundant, and these subunits consistently clustered together. While all subunits were present in OPCs, the α1–3, β1–3, and γ1–3 subunits contributed the most RNA and formed pronounced clusters. This suggests that the GABAAR stoichiometry in OPCs predominantly involves these subunit families.
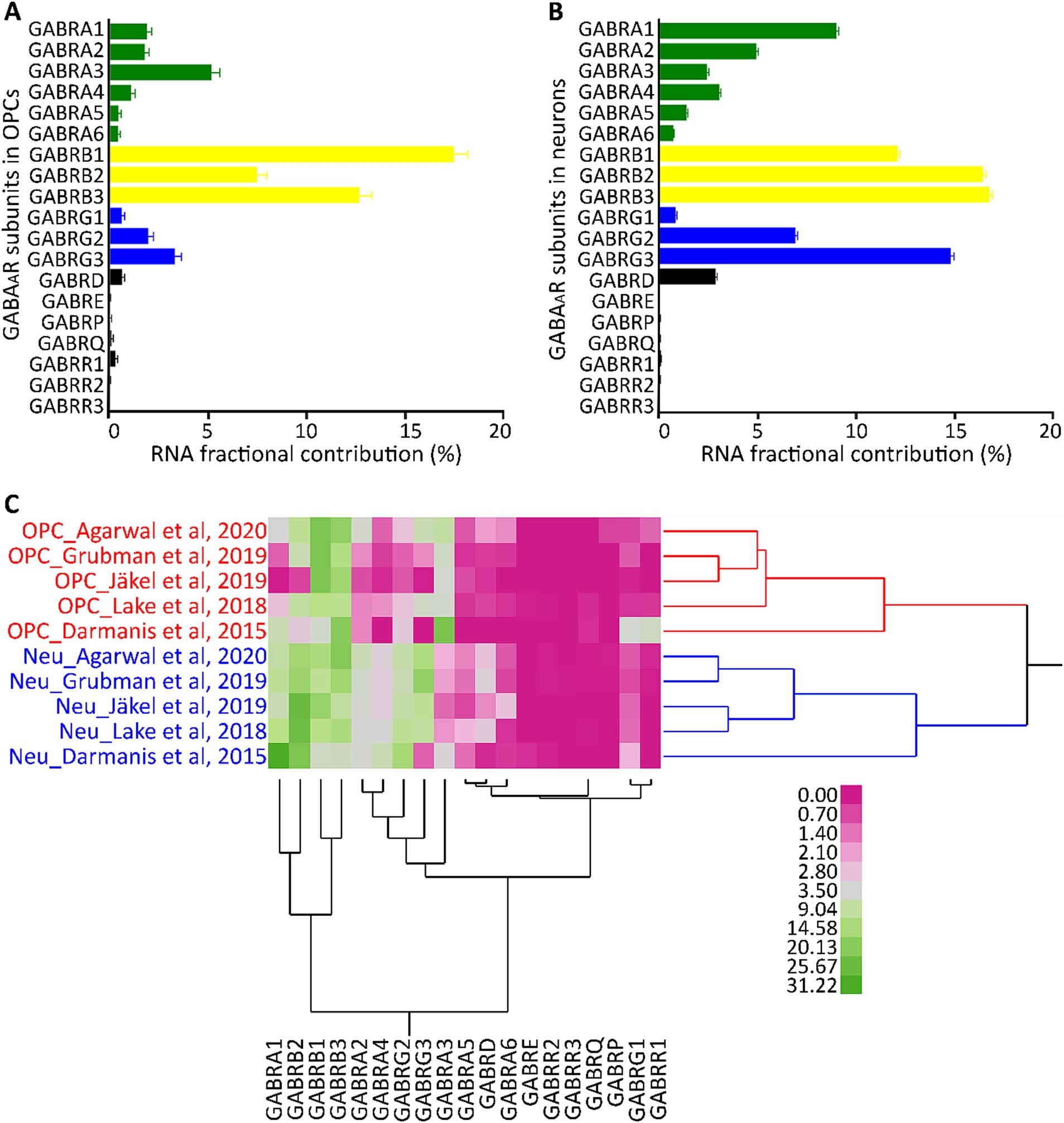
Figure 2. Segregation analysis of GABAAR subunit genes in OPCs and neurons. Average FC for each GABAAR subunit gene based on 2,431 barcoded OPCs (A) and 26,155 barcoded neurons (B) across five datasets from the human cortex (GSE14023, GSE67835, GSE138852, GSE118257, GSE97930). Bars indicate the mean FC of GABAAR subunit genes from the five datasets, and SE is shown for each bar. (C) Cell type segregation analysis, using the mean FC of each GABAAR subunit gene from the five datasets with barcoded OPCs and neurons. This is illustrated by a heatmap with a dendrogram using robust standardization, produced by JMP software (JMP, RRID:SCR_014242). GABAAR subunit gene names are found in the columns, while datasets (cell type_author) are listed in rows. Pink and green represent the abundance levels of GABAAR subunit genes.
In neurons, it is well-known that GABAARs are heteropentameric complexes composed of two α, two β, and one γ subunit (McKernan and Whiting, 1996; Petri et al., 2002). Using this knowledge as a foundation, we examined the correlations formed by these subunits in the GABAARs of human OPCs.
We conducted a multivariate analysis using the mean of the FC from each GEO and Allen Brain Map dataset for PDGFRA+ cells derived from healthy adult human cortical tissue. We found strong correlations among several subunits (Figure 3). As reported in previous literature, we observed correlations between α4 and α6 with the δ subunit which suggest the presence of receptors similar to the extrasynaptic receptors found in neurons (Mody, 2001). Further, we observed correlations between all α subunits with γ2, and between α4 and α6 with γ3. Complexes of α5 with γ2 have been observed in the hippocampal GABAAR in mice (Ghafari et al., 2017). However, correlations between α4 and α6 with γ subunits have not been previously described (Mody, 2001). Based on our observations and existing literature, potential GABAAR subunit combinations in OPCs may include α1β2γ1/2, α2β2γ1/2, α3β1γ1/2, and α5β2γ2.
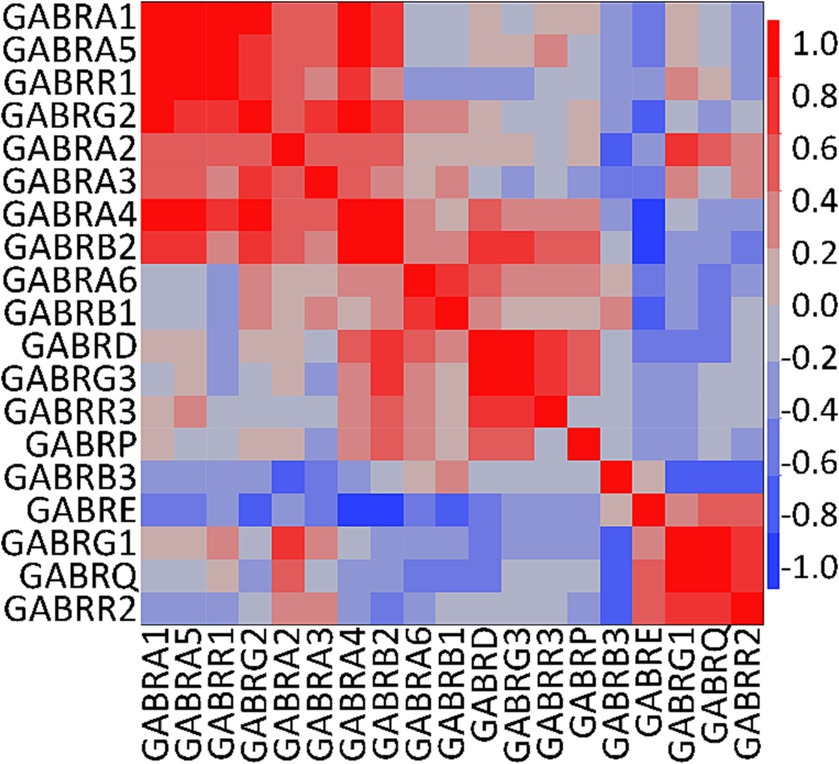
Figure 3. Correlation analysis of human GABAAR subunit genes in OPCs. Multivariate analysis using the mean of the fractional contribution of human GABAAR subunit genes from 1,007 PDGFRA+ cells (OPCs) across eight datasets of human cortex (GSE138852, GSE155488, GSE67835, GSE118257, GSE140231, GSE160813, GSE97930, https://celltypes.brain-map.org/rnaseq). The analysis was executed using JMP software (JMP, RRID:SCR_014242). Rows and columns are clustered based on correlation distance and average linkage. The correlation level of GABAAR subunit genes is represented by blue and red.
To further understand the cellular and biological processes that the expression of GABAAR subunits in OPCs might influence, we conducted a Gene Ontology (GO) analysis using all GEO and Allen Brain Map datasets. We identified genes with the strongest correlations to the GABAAR subunit genes and carried out the GO analysis on the first 1,000 genes that demonstrated a significant correlation (p < 4.2e-322; Supplementary Table 4).
The analysis revealed that this set of genes are involved in neural pathways. Importantly, these genes contribute to the signal recognition particle (SRP)-dependent cotranslational protein targeting to the membrane, are implicated in Huntington’s disease, and are involved in focal adhesion and postsynapse pathways (Figure 4; Supplementary Tables 4, 5). Specifically, the gene encoding radixin (RDX), was found among the first thousand genes showing the strongest correlation with GABAAR subunit genes (p = 3.54293E-139; Supplementary Table 4). Additionally, robust correlations were found for other genes encoding scaffold proteins such as neuroligin 2 (NLGN2; p = 4.19956e-322), collybistin (ARHGEF9; p = 3.54293E-139), and GABA type A receptor associated protein like 1 (GABARAPL1; p = 3.69304E-139; highlighted in Supplementary Table 4). As these genes encode for scaffold proteins in postsynaptic membranes, their strong correlations with GABAAR subunit genes suggest “postsynaptic” clustering of GABAARs in OPCs. Surprisingly, despite its conventional association with GABAergic synapses, the correlation with gephyrin (GPHN) was not statistically significant (p = 0.5367362811).

Figure 4. GO analysis of human GABAAR subunit genes in OPCs. A twenty most significantly (p ≤ 0.05) enriched GO terms in biological process, cellular components, and molecular functions impacted by GABAAR subunit genes using the Metascape GO enrichment platform. These terms were derived from the top 1,000 genes showing the strongest correlation with GABAAR subunit genes across eight datasets (GSE138852, GSE155488, GSE67835, GSE118257, GSE140231, GSE160813, GSE97930, https://celltypes.brain-map.org/rnaseq). All statistically significant values of the terms were transformed using a base 10 logarithm and are displayed as negative values. The terms are color-coded according to their respective p-values.
To assess age-related differences in the expression of GABAAR subunits, we used two GEO datasets (GSE155488 and GSE160813) spanning three age groups. We compared the expression levels of the GABAAR subunits in PDGFRA+ cells (OPCs) from fetal tissue derived from the telencephalon, pediatric tissue from the central area, and adult tissue from both the supratentorial region and the temporal cortex (Supplementary Table 1).
From our analysis of the FC for each subunit, we observed that adults predominantly exhibited higher expression levels of the ε subunit. Furthermore, besides ε, the notable subunits in adults were α2, β3, and γ3. In contrast, the fetal and pediatric populations mainly expressed α3 and β3 (Figure 5).
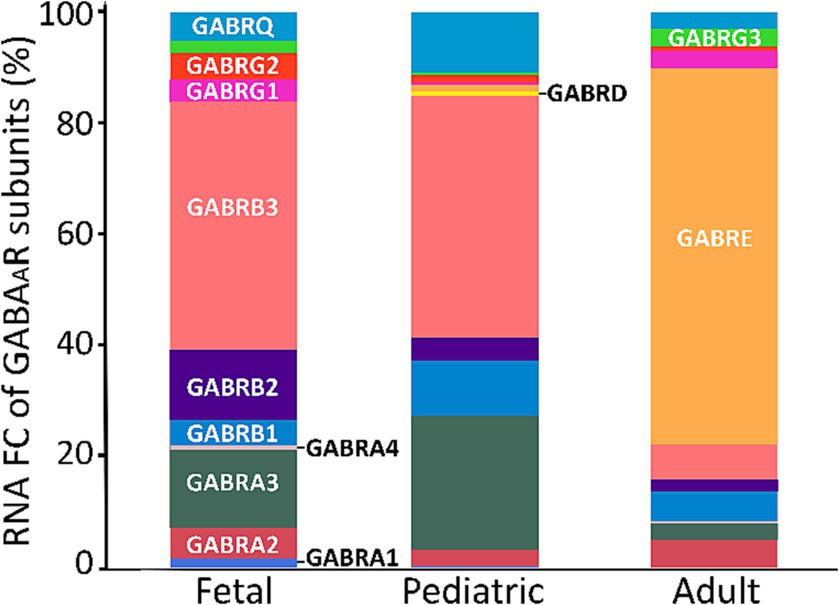
Figure 5. FC of GABAAR subunits across age span. A cross-sectional analysis of GABAAR subunits FC was conducted using 125 fetal, 268 pediatric, and 1,007 adult PDGFRA+ cells (OPCs) derived from two datasets of the human cortex (GSE155488 and GSE160813). The bars represent the mean percentage of RNA FC from both datasets for each GABAAR subunit.
To the best of our knowledge, this is the first report that aims to identify the most probable subunit combinations that constitute the GABAAR in human OPCs based on transcriptomic analysis. We found that while OPCs show varied expression of GABAAR subunits, the α, β, and γ subunits are predominantly expressed. However, among these expressed subunits, α1–3, β1–3, γ1–3, ε, and δ subunits demonstrate the highest FCs. Given the significant FC of various subunits within the same group (e.g., α1–3), it suggests the existence of multiple stoichiometries for the receptor. Notably, certain α, β, and γ subunits exhibited strong correlations in our computational analysis. Although these correlations do not directly indicate physiological interactions, they do highlight the need for further functional characterization. Building on prior experimental findings in rats, we expect that at least some of these receptor stoichiometries will be confirmed in vivo.
GABAARs in oligodendrocyte (OL) lineage cells respond similarly to the same pharmacological agents as neurons, indicating shared receptor subunits. However, as evidenced in rat OL electrophysiological studies (Arellano et al., 2016; Cisneros-Mejorado et al., 2020) they occasionally exhibit differential responses to specific drugs. For example, benzodiazepines such as diazepam potentiate GABAARs in both OL lineage cells and neurons (Möhler, 2002). Yet some β-carbolines like β-CCB, which inhibit the GABA-response of neurons, enhance GABAARs in OLs. Additionally, Zn2+ has no effect on neuronal GABAARs containing the γ2 subunit, but inhibits OL lineage cells, implicating the γ1 subunit (Ordaz et al., 2021). These differential responses confirm that each cell type has its unique GABAAR stoichiometry. Previous studies have proposed stoichiometries for GABAARs in rat OPCs based on their pharmacological profiles, highlighting the incorporation of α3, β2, and γ1 (Arellano et al., 2016; Ordaz et al., 2021). In contrast, our human transcriptomic analysis revealed strong correlations between all α subunits with both β2 and γ2, and a reduced correlation of α subunits with β1 and γ1. Two of our datasets showed high ε subunit expression levels (Allen Brain Map and GSE155488; Hodge et al., 2019; Perlman et al., 2020). The ε subunit can form a GABAAR pentamer subtype with two α and two β subunits in the brain and liver (Erlitzki et al., 2000). Intriguingly, this subtype is insensitive to the effects of intravenous anesthetic agents (Davies et al., 1997) and is significantly increased in the cerebellum of patients with schizophrenia, bipolar disorder, and major depression (Fatemi et al., 2013). However, our analysis did not find any correlations of ε with other subunits. Nevertheless, we found correlations of δ with α4/6 and β2, a combination consistent with the expression of extrasynaptic GABAARs. These extrasynaptic GABAARs (Jones et al., 1997; Peng et al., 2002), documented in electrophysiological studies on murine OLs cocultured with neurons, are of particular interest due to their strong sensitivity to neurosteroids (Belelli et al., 2002; Belelli and Lambert, 2005). These have been suggested to have a positive effect on myelination in various animal models (Ghoumari et al., 2003; Shaw et al., 2019).
As anticipated, the GO analysis showed GABAAR subunits’ involvement in neural pathways. Expression of GABAARs in OPCs has been well-documented (Bergles et al., 2010). Specifically, the GABAARs γ2 subunit has been suggested to be postsynaptic in OPCs from murines (Passlick et al., 2013; Balia et al., 2015; Orduz et al., 2015; McKenzie et al., 2018). We also found a strong correlation between GABAARs and ER translocation (R-HSA-1799339: SRP-dependent cotranslational protein targeting to membrane). Our findings confirm a previous transcriptomic analysis using induced pluripotent stem cell-derived O4+ OL lineage cells from an individual with the Parkinson’s Disease genetic variant SNCA, which demonstrated a correlation with SRP-dependent co-translational protein targeting to the membrane (Azevedo et al., 2022). This suggests that misfolding of GABAAR subunits could potentially disrupt the OPC maturation signaling cascade in various neurodegenerative diseases, including Parkinson’s Disease. This is also supported by evidence that alterations in human αSynuclein, the pathogenic hallmark of all synucleinopathies, impact OPC maturation (Ettle et al., 2014; Azevedo et al., 2022). Additionally, our GO analysis highlighted active participation in postsynaptic pathways, a finding we confirmed by detecting transcripts for postsynaptic proteins including radixin, neuroligin 2, collybistin, and GABA type A receptor-associated protein like 1. These findings align with a recent study by Patt et al. (2023). Importantly, our analysis revealed that gephyrin did not exhibit a significant correlation with GABAAR subunits in OPCs, setting it apart from the other identified postsynaptic proteins.
By analyzing the FASTQ file datasets using the CellRanger and Seurat pipelines, coupled with our quality control measures, we noted an increase in the number of PDGFRA+ cells compared to those in the GEO datasets. However, these differences did not impact the FC. The first three α, β, and γ subunits consistently showed the highest FCs. Hence, the quality control measures employed by the authors for the GEO datasets did not significantly affect the overall findings.
In our analysis of GABAAR subunits development, we relied on just two datasets (GSE155488 and GSE160813; Perlman et al., 2020; Fernandes et al., 2021) with RNA-seq data covering different ages. Yet, our findings showed the dynamic expression of GABAAR subunits in OPCs throughout various developmental stages. This suggests a persistently evolving role for these cells and likely functional significance during brain maturation. The ε subunit demonstrated a notably higher expression in adults compared to fetal and pediatric groups. This variation implies a potential shift in GABAAR function within OPCs as the brain matures. The increased representation of the ε subunit in adults might relate to distinct physiological roles or response dynamics unique to mature OPCs However, it is worth noting that of the original eight GEO adult datasets, only the one from Fernandes et al. (2021) (GSE160813) indicated substantial expression and contribution of the ε subunit. This warrants further analysis to determine the role, if any, of this subunit in GABAARs in adult human OPCs. Conversely, the dominant expression of α3 and β3 subunits in fetal and pediatric stages could emphasize their critical role during early brain development. In addition, we noted a distinctive pattern concerning the γ2 subunit, which was predominantly present in fetal populations, followed by pediatric populations, and was least prevalent in adults. This suggests a likelihood of decreasing γ2 subunits in adulthood. This finding aligns with a recognized characteristic of OPCs: as these cells age, they become less responsive to specific drugs like diazepam and zolpidem, indicating a reduction in the γ2 subunit of GABAARs (Vélez-Fort et al., 2010; Balia et al., 2015; Patt et al., 2023). Similarly, we observed a parallel trend with the δ subunit, where its presence decreases in adulthood. In contrast, the α3 subunit remained consistent across all age groups. Notably, previous research indicated that the α5 subunit becomes more prominent in the adult hippocampus, reflecting alterations in receptor subunits during development (Patt et al., 2023). However, in our datasets, α5 was not as prominent as α1–3 subunits. Despite this, our correlation analysis demonstrated that α5 is correlated with other subunits, suggesting the possibility of GABAARs containing α5 combined with other subunits in humans, with no discernible age-related changes. These observed shifts in receptor subunits indicate a change of GABAARs in OPCs across developmental stages.
Our study demonstrates that GABAARs in human OPCs likely have multiple stoichiometries due to the considerable variation in the expression of different subunits. The significant correlation we observed between subunits suggests the presence of several distinct receptor forms, each potentially contributing to the pharmacological profile of GABAARs in OPCs. These findings enrich our understanding of GABAAR diversity, emphasizing the need for further functional characterization of these receptors in OPCs from human. Understanding the specific compositions of GABAARs in OPCs could shed light on the physiological role of these cells in neurodevelopment and disease. For example, in the datasets we examined, the one provided by Jäkel et al. (2019) (GSE118257) contained RNA-sequencing data on patients with multiple sclerosis (MS), a severe demyelinating disorder. It would have been intriguing to investigate potential differences in GABAARs under such condition. However, this dataset only included 14 PDGFRA+ cells from active lesions of MS out of 295 total cells. This limited sample size lacked the statistical power necessary to discern alterations in GABAAR stoichiometry within OPCs during pathology. The scarcity of RNA-sequencing datasets from human tissues affected by demyelinating diseases currently restricts our ability to conduct a comprehensive analysis in this regard. Additionally, exploring a different scenario could be equally enlightening. For instance, studying musicians who have higher intracortical myelination (Kim and Knösche, 2016) might reveal if there are variations in GABAAR stoichiometry that contribute to enhanced myelination. This investigation could give us insights into the potential role of OPCs in neural plasticity and activation. Nevertheless, this avenue of research remains unexplored, leaving a promising area for future studies to unravel the complexities of the function of GABAARs in OPCs in diverse physiological and pathological contexts. Furthermore, it may lead the way for developing targeted therapies for demyelinating diseases, neurological disorders, and other conditions linked to oligodendrocyte dysfunction.
We retrieved three scRNA-sequencing and five snRNA-sequencing libraries from the Gene Expression Omnibus and Allen Brain Map repositories. The specimens used for sequencing were sourced either fresh from surgical resections or frozen from brain tissue banks, covering the frontal cortex, temporal cortex, visual cortex, and entorhinal cortex (refer to Supplementary Table 1). The sequencing data used in our study came from control fetal, pediatric, adolescent and adult subjects, with no known neurological or psychiatric illnesses. We extended the age range from fetuses to adults to explore the differences in subunit expression during development. For surgical specimens, we only included sequencing data derived from tissue devoid of pathological features. The datasets contained barcodes, genes, and matrix files, which we merged using R software. We sorted OPCs in each dataset by selecting PDGFRA+ cells, followed by selecting the GABAAR subunit genes and examining their expression levels. We used JMP software for these processes (JMP, RRID:SCR_014242).
We normalized the different units (unique molecular identifier counts, fragments per kilobase million reads, and raw read values) found in the raw data by calculating the FC of individual subunits. The FC of each GABAAR subunit per cell type is the percentage of expression level of each subunit gene to the total pool of subunit genes within each cell type. For this, the sum of unique molecular identifier counts, fragments per kilobase million reads, and raw read values mapped reads in RNA-Seq data, of all 19 genes per cell type was 100% (Sequeira et al., 2019). Additionally, we explored the use of other two FC methods: For FC2, we obtained the percentage of expression level of each subunit gene over the sum of all 19 gene subunits per dataset. And for FC3, we obtained the percentage of number of cells expressing each subunit gene over the total number of cells. Thus, FC1 represents the mean ± SD of the FC for each dataset (Fcd), where Fcd is the corresponding mean value of the FC in each cell (Fcc), FC2 represent the single subunit FC value for all the dataset and FC3 represents the probability of the subunit to be found in the dataset. The same methods were applied to neurons as a validating control group.
We performed a two-way hierarchical clustering with the FCs of five datasets (GSE14023, GSE67835, GSE138852, GSE118257, GSE97930) featuring barcoded OPCs and neurons. JMP software facilitated this analysis, with data being robustly standardized.
We conducted a multiple Pearson’s correlation analysis of the subunits using the mean of the FCs across all datasets. After generating co-clustering heatmaps for each combination of subunit gene expression values, we reorganized the subunits into representative clusters using JMP software.
All datasets were included in this analysis. We defined OPCs as those cells positive for PDGFRA. We calculated the sum of all GABAAR subunits per cell across all the included datasets. Then, using JMP software, we performed a response screening test to compare the GABAAR subunit genes with the rest of the available genes, thereby predicting the best gene combinations. We selected the top 1,000 genes with the lowest value of p (cutoff p < 4.2e-322) from the resulting analysis and investigated them using the Metascape GO enrichment platform (Zhou et al., 2019). We designated H. sapiens for both species input and analysis. Enrichment analysis, encompassing pathways and processes, was undertaken using these ontology sources: GO Biological Processes, GO Cellular Components, KEGG Pathway, Reactome Gene Sets, Canonical Pathways, CORUM, WikiPathways, and PANTHER Pathway.
We downloaded FASTQ data from four datasets available in this format (PRJNA5776618, PRJNA673712, PRJNA589018, PRJNA674571) using the SRAtoolkits library. The quality of the downloaded files was verified using FastQC software. The data were then uploaded and analyzed in CellRanger to align reads, generate feature-barcode matrices, and perform clustering and gene expression analysis. We employed the Seurat pipeline for quality control, using the following parameters: min.cells = 3, min.features = 200, subset = nFeature_RNA < 3,000 and percent.mt < 5. These excluded cells expressing more than 5,000 genes and those with at least 500. It also filtered out cells where mitochondrial transcripts accounted for more than 5% of the total transcripts. Subsequently, we normalized the data using the command NormalizeData (seurat, normalization.method = “LogNormalize”) in Seurat. Following this quality control process, we identified OPCs as PDGFRA+ cells and examined the expression of their GABAAR subunit genes.
We compared the means of all groups for continuous variables. For nonparametric comparisons, we applied the Wilcoxon test for multiple pairwise comparisons and the Wilcoxon/Kruskal-Wallis test for simple comparisons. We set statistical significance at p < 0.05.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
BAG: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. JG-C: Investigation, Software, Writing – review & editing, Methodology. ROA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. AL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. BAG was supported as a Becario de la Dirección General de Calidad y Educación en Salud, Secretaría de Salud, México, in partial fulfillment of her social service for an MD degree. ROA and JG-C were supported by CONAHCYT through the grant number 319740 and PAPIIT-UNAM IN205822. AL was supported by RO1 grant AG070255.
The authors would like to express our gratitude to Luis Concha from the Instituto de Neurobiología, Campus UNAM Juriquilla, for his assistance with accessing a remote computer to manage large dataset files and for his guidance on using R software. We extend our gratitude to Jerome Verleyen for providing us with access to the HPC infrastructure at the Unidad Universitaria de Secuenciación Masiva y Bioinformática, Instituto de Biotecnología (UNAM), a vital resource that is part of the Laboratorio Nacional de Apoyo Tecnológico a las Ciencias Genómicas (CONAHCyT). Additionally, we would like to acknowledge the Instituto de Biotecnología-UNAM for generously granting us access to its computer cluster, a resource that significantly contributed to the successful completion of our research. We are also deeply thankful to Edith Garay from the Instituto de Neurobiología, Campus UNAM Juriquilla, for her expert technical support, which significantly enriched the quality of our research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1279232/full#supplementary-material
Arellano, R. O., Sánchez-Gómez, M. V., Alberdi, E., Canedo-Antelo, M., Chara, J. C., Palomino, A., et al. (2016). Axon-to-glia interaction regulates GABAA receptor expression in oligodendrocytes. Mol. Pharmacol. 89, 63–74. doi: 10.1124/mol.115.100594
Azevedo, C., Teku, G., Pomeshchik, Y., Reyes, J. F., Chumarina, M., Russ, K., et al. (2022). Parkinson’s disease and multiple system atrophy patient iPSC-derived oligodendrocytes exhibit alpha-synuclein-induced changes in maturation and immune reactive properties. Proc. Natl. Acad. Sci. U. S. A. 119:e2111405119. doi: 10.1073/pnas.2111405119
Balia, M., Vélez-Fort, M., Passlick, S., Schäfer, C., Audinat, E., Steinhäuser, C., et al. (2015). Postnatal down-regulation of the GABAA receptor γ2 subunit in neocortical NG2 cells accompanies synaptic-to-extrasynaptic switch in the GABAergic transmission mode. Cereb. Cortex 25, 1114–1123. doi: 10.1093/cercor/bht309
Baydyuk, M., Morrison, V. E., Gross, P. S., and Huang, J. K. (2020). Extrinsic factors driving oligodendrocyte lineage cell progression in CNS development and injury. Neurochem. Res. 45, 630–642. doi: 10.1007/s11064-020-02967-7
Belelli, D., Casula, A., Ling, A., and Lambert, J. J. (2002). The influence of subunit composition on the interaction of neurosteroids with GABA(a) receptors. Neuropharmacology 43, 651–661. doi: 10.1016/s0028-3908(02)00172-7
Belelli, D., and Lambert, J. J. (2005). Neurosteroids: endogenous regulators of the GABA(a) receptor. Nat. Rev. Neurosci. 6, 565–575. doi: 10.1038/nrn1703
Bergles, D. E., Jabs, R., and Steinhäuser, C. (2010). Neuron-glia synapses in the brain. Brain Res. Rev. 63, 130–137. doi: 10.1016/j.brainresrev.2009.12.003
Butt, A. M., De La Rocha, I. C., and Rivera, A. (2019). Oligodendroglial cells in Alzheimer’s disease. Adv. Exp. Med. Biol. 1175, 325–333. doi: 10.1007/978-981-13-9913-8_12
Cheli, V. T., Santiago González, D. A., Namgyal Lama, T., Spreuer, V., Handley, V., Murphy, G. G., et al. (2016). Conditional deletion of the L-type calcium channel Cav1.2 in oligodendrocyte progenitor cells affects postnatal myelination in mice. J. Neurosci. 36, 10853–10869. doi: 10.1523/JNEUROSCI.1770-16.2016
Choi, K., Chen, Y., Skelly, D. A., and Churchill, G. A. (2020). Bayesian model selection reveals biological origins of zero inflation in single-cell transcriptomics. Genome Biol. 21:183. doi: 10.1186/s13059-020-02103-2
Cisneros-Mejorado, A. J., Garay, E., Ortiz-Retana, J., Concha, L., Moctezuma, J. P., Romero, S., et al. (2020). Demyelination-remyelination of the rat caudal cerebellar peduncle evaluated with magnetic resonance imaging. Neuroscience 439, 255–267. doi: 10.1016/j.neuroscience.2019.06.042
Darmanis, S., Sloan, S. A., Zhang, Y., Enge, M., Caneda, C., Shuer, L. M., et al. (2015). A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U S A 112, 7285–90. doi: 10.1073/pnas.1507125112
Davies, P. A., Hanna, M. C., Hales, T. G., and Kirkness, E. F. (1997). Insensitivity to anaesthetic agents conferred by a class of GABA(a) receptor subunit. Nature 385, 820–823. doi: 10.1038/385820a0
Dawson, M. R. L., Polito, A., Levine, J. M., and Reynolds, R. (2003). NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci. 24, 476–488. doi: 10.1016/s1044-7431(03)00210-0
Ding, J., Adiconis, X., Simmons, S. K., Kowalczyk, M. S., Hession, C. C., Marjanovic, N. D., et al. (2020). Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746. doi: 10.1038/s41587-020-0465-8
Erlitzki, R., Gong, Y., Zhang, M., and Minuk, G. (2000). Identification of gamma-aminobutyric acid receptor subunit types in human and rat liver. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G733–G739. doi: 10.1152/ajpgi.2000.279.4.G733
Ettle, B., Reiprich, S., Deusser, J., Schlachetzki, J. C. M., Xiang, W., Prots, I., et al. (2014). Intracellular alpha-synuclein affects early maturation of primary oligodendrocyte progenitor cells. Mol. Cell. Neurosci. 62, 68–78. doi: 10.1016/j.mcn.2014.06.012
Fatemi, S. H., Folsom, T. D., Rooney, R. J., and Thuras, P. D. (2013). Expression of GABAA α2-, β1-and ε-receptors are altered significantly in the lateral cerebellum of subjects with schizophrenia, major depression and bipolar disorder. Transl. Psychiatry 3:e303. doi: 10.1038/tp.2013.64
Fernandes, M. G. F., Luo, J. X. X., Cui, Q. L., Perlman, K., Pernin, F., Yaqubi, M., et al. (2021). Age-related injury responses of human oligodendrocytes to metabolic insults: link to BCL-2 and autophagy pathways. Commun Biol 4:20. doi: 10.1038/s42003-020-01557-1
Fernandez-Castaneda, A., and Gaultier, A. (2016). Adult oligodendrocyte progenitor cells—multifaceted regulators of the CNS in health and disease. Brain Behav. Immun. 57, 1–7. doi: 10.1016/j.bbi.2016.01.005
Gallo, V., and Deneen, B. (2014). Glial development: the crossroads of regeneration and repair in the CNS. Neuron 83, 283–308. doi: 10.1016/j.neuron.2014.06.010
Gallo, V., Mangin, J. M., Kukley, M., and Dietrich, D. (2008). Synapses on NG2-expressing progenitors in the brain: multiple functions? J. Physiol. 586, 3767–3781. doi: 10.1113/JPHYSIOL.2008.158436
Ghafari, M., Falsafi, S. K., Szodorai, E., Kim, E. J., Li, L., Höger, H., et al. (2017). Formation of GABAA receptor complexes containing α1 and α5 subunits is paralleling a multiple T-maze learning task in mice. Brain Struct. Funct. 222, 549–561. doi: 10.1007/S00429-016-1233-X
Ghoumari, A. M., Ibanez, C., El-Etr, M., Leclerc, P., Eychenne, B., O’Malley, B. W., et al. (2003). Progesterone and its metabolites increase myelin basic protein expression in organotypic slice cultures of rat cerebellum. J. Neurochem. 86, 848–859. doi: 10.1046/j.1471-4159.2003.01881.x
Gruchot, J., Weyers, V., Göttle, P., Förster, M., Hartung, H.-P., Küry, P., et al. (2019). The molecular basis for remyelination failure in multiple sclerosis. Cells 8:825. doi: 10.3390/cells8080825
Hodge, R. D., Bakken, T. E., Miller, J. A., Smith, K. A., Barkan, E. R., Graybuck, L. T., et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. doi: 10.1038/s41586-019-1506-7
Huntemer-Silveira, A., Patil, N., Brickner, M. A., and Parr, A. M. (2020). Strategies for oligodendrocyte and myelin repair in traumatic CNS injury. Front. Cell. Neurosci. 14:619707. doi: 10.3389/fncel.2020.619707
Jabs, R., Pivneva, T., Hüttmann, K., Wyczynski, A., Nolte, C., Kettenmann, H., et al. (2005). Synaptic transmission onto hippocampal glial cells with hGFAP promoter activity. J. Cell Sci. 118, 3791–3803. doi: 10.1242/JCS.02515
Jäkel, S., Agirre, E., Mendanha Falcão, A., van Bruggen, D., Lee, K. W., Knuesel, I., et al. (2019). Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547. doi: 10.1038/s41586-019-0903-2
Jones, A., Korpi, E. R., McKernan, R. M., Pelz, R., Nusser, Z., Mäkelä, R., et al. (1997). Ligand-gated ion channel subunit partnerships: GABAA receptor alpha6 subunit gene inactivation inhibits delta subunit expression. J. Neurosci. 17, 1350–1362. doi: 10.1523/JNEUROSCI.17-04-01350.1997
Kim, S. G., and Knösche, T. R. (2016). Intracortical myelination in musicians with absolute pitch: quantitative morphometry using 7-T MRI. Hum. Brain Mapp. 37, 3486–3501. doi: 10.1002/HBM.23254
Kirchhoff, F., and Kettenmann, H. (1992). GABA triggers a [Ca2+]i increase in murine precursor cells of the oligodendrocyte lineage. Eur. J. Neurosci. 4, 1049–1058. doi: 10.1111/j.1460-9568.1992.tb00131.x
Lake, B. B., Chen, S., Sos, B. C., Fan, J., Kaeser, G. E., Yung, Y. C., et al. (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80. doi: 10.1038/nbt.4038
Lin, S., and Bergles, D. E. (2004). Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. Nat. Neurosci. 7, 24–32. doi: 10.1038/nn1162
Lun, A. T. L., Bach, K., and Marioni, J. C. (2016). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17:75. doi: 10.1186/s13059-016-0947-7
Marisca, R., Hoche, T., Agirre, E., Hoodless, L. J., Barkey, W., Auer, F., et al. (2020). Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 23, 363–374. doi: 10.1038/s41593-019-0581-2
McKenzie, A. T., Wang, M., Hauberg, M. E., Fullard, J. F., Kozlenkov, A., Keenan, A., et al. (2018). Brain cell type specific gene expression and co-expression network architectures. Sci. Rep. 8:8868. doi: 10.1038/s41598-018-27293-5
McKernan, R. M., and Whiting, P. J. (1996). Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci. 19, 139–143. doi: 10.1016/s0166-2236(96)80023-3
Miller, R. H., Hayes, J. E., Dyer, K. L., and Sussman, C. R. (1999). Mechanisms of oligodendrocyte commitment in the vertebrate CNS. Int. J. Dev. Neurosci. 17, 753–763. doi: 10.1016/S0736-5748(99)00068-4
Mody, I. (2001). Distinguishing between GABA(a) receptors responsible for tonic and phasic conductances. Neurochem. Res. 26, 907–913. doi: 10.1023/A:1012376215967
Möhler, H. (2002). Pathophysiological aspects of diversity in neuronal inhibition: a new benzodiazepine pharmacology. Dialogues Clin. Neurosci. 4, 261–269. doi: 10.31887/DCNS.2002.4.3/hmoehler
Olsen, R. W., and Sieghart, W. (2009). GABA a receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology 56, 141–148. doi: 10.1016/j.neuropharm.2008.07.045
Ordaz, R. P., Garay, E., Limon, A., Pérez-Samartín, A., Sánchez-Gómez, M. V., Robles-Martínez, L., et al. (2021). GABA a receptors expressed in oligodendrocytes cultured from the neonatal rat contain α 3 and γ 1 subunits and present differential functional and pharmacological properties. Mol. Pharmacol. 99, 133–146. doi: 10.1124/molpharm.120.000091
Orduz, D., Maldonado, P. P., Balia, M., Vélez-Fort, M., de Sars, V., Yanagawa, Y., et al. (2015). Interneurons and oligodendrocyte progenitors form a structured synaptic network in the developing neocortex. Elife 4:6953. doi: 10.7554/eLife.06953
Passlick, S., Grauer, M., Schafer, C., Jabs, R., Seifert, G., and Steinhauser, C. (2013). Expression of the γ2-subunit distinguishes synaptic and extrasynaptic GABAA receptors in NG2 cells of the hippocampus. J. Neurosci. 33, 12030–12040. doi: 10.1523/JNEUROSCI.5562-12.2013
Patt, L., Tascio, D., Domingos, C., Timmermann, A., Jabs, R., Henneberger, C., et al. (2023). Impact of developmental changes of GABAA receptors on interneuron-NG2 glia transmission in the Hippocampus. Int. J. Mol. Sci. 24:13490. doi: 10.3390/IJMS241713490
Peng, Z., Hauer, B., Mihalek, R. M., Homanics, G. E., Sieghart, W., Olsen, R. W., et al. (2002). GABA(a) receptor changes in delta subunit-deficient mice: altered expression of alpha4 and gamma2 subunits in the forebrain. J. Comp. Neurol. 446, 179–197. doi: 10.1002/cne.10210
Perlman, K., Couturier, C. P., Yaqubi, M., Tanti, A., Cui, Q.-L. Q., Pernin, F., et al. (2020). Developmental trajectory of oligodendrocyte progenitor cells in the human brain revealed by single cell RNA sequencing. Glia 68, 1291–1303. doi: 10.1002/glia.23777
Petri, S., Krampfl, K., Dengler, R., Bufler, J., Weindl, A., and Arzberger, T. (2002). Human GABA a receptors on dopaminergic neurons in the pars compacta of the substantia nigra. J. Comp. Neurol. 452, 360–366. doi: 10.1002/cne.10379
Pringle, N. P., Mudhar, H. S., Collarini, E. J., and Richardson, W. D. (1992). PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development 115, 535–551. doi: 10.1242/DEV.115.2.535
Pringle, N. P., and Richardson, W. D. (1993). A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development 117, 525–533. doi: 10.1242/DEV.117.2.525
Raabe, F. J., Galinski, S., Papiol, S., Falkai, P. G., Schmitt, A., and Rossner, M. J. (2018). Studying and modulating schizophrenia-associated dysfunctions of oligodendrocytes with patient-specific cell systems. NPJ Schizophr. 4:23. doi: 10.1038/s41537-018-0066-4
Reyes-Haro, D., Cisneros-Mejorado, A., and Arellano, R. O. (2021). Therapeutic potential of GABAergic signaling in myelin plasticity and repair. Front. Cell Dev. Biol. 9:662191. doi: 10.3389/fcell.2021.662191
Richardson, W. D., Young, K. M., Tripathi, R. B., and McKenzie, I. (2011). NG2-glia as multipotent neural stem cells: fact or fantasy? Neuron 70, 661–673. doi: 10.1016/j.neuron.2011.05.013
Santiago González, D. A., Cheli, V. T., Zamora, N. N., Lama, T. N., Spreuer, V., Murphy, G. G., et al. (2017). Conditional deletion of the L-type calcium channel Cav1.2 in NG2-positive cells impairs remyelination in mice. J. Neurosci. 37, 10038–10051. doi: 10.1523/JNEUROSCI.1787-17.2017
Sequeira, A., Shen, K., Gottlieb, A., and Limon, A. (2019). Human brain transcriptome analysis finds region-and subject-specific expression signatures of GABAAR subunits. Commun Biol 2:153. doi: 10.1038/s42003-019-0413-7
Serrano-Regal, M. P., Bayón-Cordero, L., Ordaz, R. P., Garay, E., Limon, A., Arellano, R. O., et al. (2020). Expression and function of GABA receptors in myelinating cells. Front. Cell. Neurosci. 14:256. doi: 10.3389/fncel.2020.00256
Shaw, J. C., Dyson, R. M., Palliser, H. K., Gray, C., Berry, M. J., and Hirst, J. J. (2019). Neurosteroid replacement therapy using the allopregnanolone-analogue ganaxolone following preterm birth in male guinea pigs. Pediatr. Res. 85, 86–96. doi: 10.1038/s41390-018-0185-7
Sieghart, W., and Sperk, G. (2002). Subunit composition, distribution and function of GABA(a) receptor subtypes. Curr. Top. Med. Chem. 2, 795–816. doi: 10.2174/1568026023393507
Trotter, J., Karram, K., and Nishiyama, A. (2010). NG2 cells: properties, progeny and origin. Brain Res. Rev. 63, 72–82. doi: 10.1016/j.brainresrev.2009.12.006
Van den Berge, K., Perraudeau, F., Soneson, C., Love, M. I., Risso, D., Vert, J.-P., et al. (2018). Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biol. 19:24. doi: 10.1186/s13059-018-1406-4
van Tilborg, E., de Theije, C. G. M., van Hal, M., Wagenaar, N., de Vries, L. S., Benders, M. J., et al. (2018). Origin and dynamics of oligodendrocytes in the developing brain: implications for perinatal white matter injury. Glia 66, 221–238. doi: 10.1002/glia.23256
Vélez-Fort, M., Maldonado, P. P., Butt, A. M., Audinat, E., and Angulo, M. C. (2010). Postnatal switch from synaptic to extrasynaptic transmission between interneurons and NG2 cells. J. Neurosci. 30, 6921–6929. doi: 10.1523/JNEUROSCI.0238-10.2010
Keywords: PDGFRA, fractional contribution, single cell, single nucleus, RNA-sequencing, oligodendrocyte, OPC, GABA receptor
Citation: Gutierrez BA, González-Coronel JM, Arellano RO and Limon A (2023) Transcriptional and bioinformatic analysis of GABAA receptors expressed in oligodendrocyte progenitor cells from the human brain. Front. Mol. Neurosci. 16:1279232. doi: 10.3389/fnmol.2023.1279232
Received: 18 August 2023; Accepted: 12 October 2023;
Published: 26 October 2023.
Edited by:
Carlos Fernández-Peña, St. Jude Children's Research Hospital, United StatesReviewed by:
Li-Jin Chew, Brown University, United StatesCopyright © 2023 Gutierrez, González-Coronel, Arellano and Limon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rogelio O. Arellano, YXJlbGxhbm8ub3N0b2FAY29tdW5pZGFkLnVuYW0ubXg=; Agenor Limon, YWdsaW1vbnJAdXRtYi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.