
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 27 April 2023
Sec. Neuroplasticity and Development
Volume 16 - 2023 | https://doi.org/10.3389/fnmol.2023.1170708
This article is part of the Research Topic The Commonality in Converged Pathways and Mechanisms Underpinning Neurodevelopmental and Psychiatric Disorders View all 8 articles
Introduction: Schizophrenia is a severe and chronic psychiatric disorder with hereditary risk up to 80% as previous studies indicated. Several researches have demonstrated a significant association between schizophrenia and microduplications that overlap the vasoactive intestinal peptide receptor 2 gene (VIPR2).
Methods: To further investigate potential causal VIPR2 gene variants, all exons and un-translated portions of the VIPR2 gene were sequenced using amplicon targeted resequencing in 1804 Chinese Han patients with schizophrenia and 996 healthy counterparts in the present study.
Results: Nineteen rare non-synonymous mutations and 1 frameshift deletion was identified for schizophrenia, among which 5 variants have never been reported so far. Frequencies of rare non-synonymous mutations were significantly different between the two groups. Specifically, the non-synonymous mutation rs78564798 (Pallele = 0.006) as well as two rare variations in the VIPR2 gene’s introns (rs372544903, Pallele = 0.026 and a novel mutation, chr7:159034078, GRCh38, Pallele = 0.048) were significantly associated with schizophrenia.
Discussion: Our findings add new evidence that the functional and probable causative variants of VIPR2 gene may play an important role in susceptibility to schizophrenia. Further studies on validations of VIPR2’s function in the etiology of schizophrenia are warranted.
Schizophrenia (SCZ) is a severe psychiatric disorder that typically manifests in the late teens and early twenties and affects almost 1% of the world population (Marder and Cannon, 2019). Hallucinations, delusions, disorganized talk, diminished motivation and expression, and cognitive deficits are the core symptoms of SCZ (Joyce and Roiser, 2007; Owen et al., 2016). More than 50% of patients with SCZ experience long-term intermittent psychiatric problems, and about 20% experience chronic symptoms or even disability (Barbato, 1998). SCZ is currently one of the top 10 causes of disability worldwide (Fleischhacker et al., 2014), and those who have it typically live 10–20 years less than the general population (Chesney et al., 2014). Although the etiology and pathogenesis of SCZ have not been completely understood, it is generally acknowledged that heritable and environmental factors affect. Previous twin and family studies suggest heritability accounting for 60% ~ 80% of the risk of SCZ (Sullivan et al., 2003).
Multiple researches on the genetics of SCZ have shown numerous single-nucleotide polymorphisms (SNPs; Ripke et al., 2014; Li et al., 2017; Trubetskoy et al., 2022), rare coding variations (Purcell et al., 2014), and rare copy number variations (CNVs; Li et al., 2016; Marshall et al., 2017) that are linked to the risk of developing SCZ. It was first reported in 2011 that microduplications that overlapped the VIPR2 gene or its upstream were significantly associated with SCZ (Levinson et al., 2011; Vacic et al., 2011). Subsequent researches conducted on the Han Chinese population further confirmed the significant association between SCZ and the VIPR2 CNV (Yuan et al., 2014; Li et al., 2016). The VIPR2 gene was also found to be differently methylated between SCZ patients and controls by genome-wide DNA methylation study on post-mortem human brain tissue (Wockner et al., 2014). Additionally, several studies showed that the VIPR2 gene is associated with other psychiatric disorders, such as higher frequencies of CNVs in autism spectrum disorder (Firouzabadi et al., 2017), hypomethylation at CpG sites in attention deficit and hyperactivity disorder (ADHD; Wilmot et al., 2016) and different frequencies of SNP (rs885861) between patients with mood disorders (MD) and controls (Soria et al., 2010).
The VIPR2 gene is located on chromosome 7q36.3 and harbors 14 exons, which encodes a class B 7-transmembrane G-protein-coupled receptor (GPCR) called VPAC2. Three VPAC2 isoforms are depicted in Figure 1, including NP 003373 (438aa), NP 001295188 (422aa), and NP 001291451 (358aa). In humans, the VPAC2 receptor is widely present in multiple organs and brain. It is highly expressed in neurons of the cerebral cortex as well as the thalamus and hypothalamus, particularly in the suprachiasmatic nuclei (Sheward et al., 1995; An et al., 2012; Ago et al., 2021). Vasoactive intestinal peptide (VIP) and pituitary adenylate cyclase-activating polypeptide (PACAP) are two natural ligands that interact with the VPAC2 receptor which plays a crucial role in the physiology of the central nervous system (CNS) (Laburthe et al., 2007). As a GPCR, the VPAC2 receptor mainly triggers the activation of adenylate cyclase (AC), through coupling with Gs-type trimeric G-proteins, which subsequently produces cAMP and activates protein kinase A (PKA). Additionally, it has been discovered that the VPAC2 receptor also activates phospholipase C (PLC) through G proteins of Gq and Gi/Go families (MacKenzie et al., 2001; Laburthe et al., 2002, 2007).
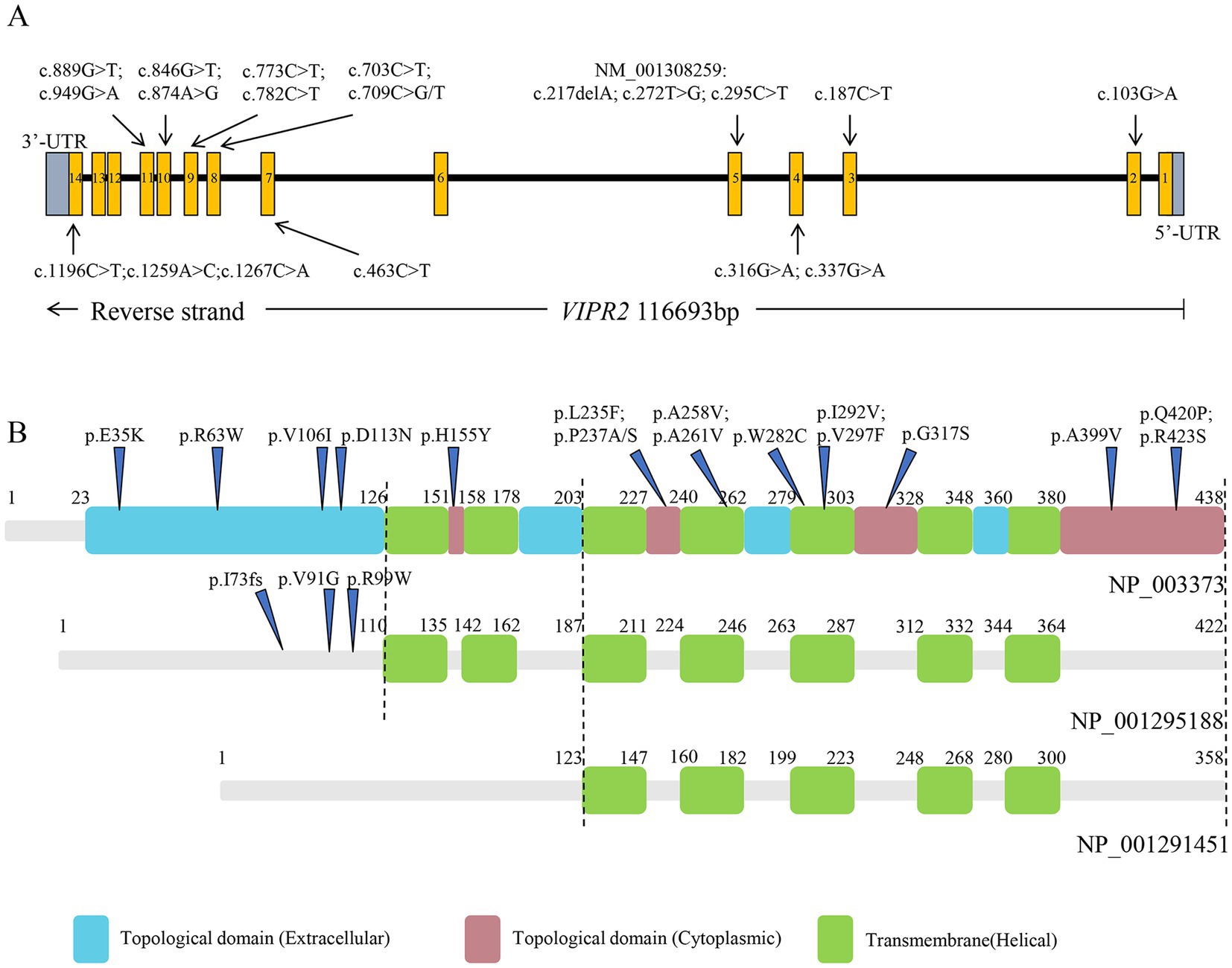
Figure 1. Structure of VIPR2 gene and VPAC2 isoforms. (A) VIPR2 gene structure, boxes 1 ~ 14 indicate the protein-coding exons. (B) VPAC2 isoforms structure based on NP_003373, NP_001295188, and NP_001291451.
Studies with VPAC2-overexpressing and VPAC2-deficient mice models have revealed that VPAC2 receptors are crucial for regulating circadian rhythms (Shen et al., 2000; Harmar et al., 2002) and have an effect on fear cognition (Ago et al., 2017). According to studies, VPAC2-deficient mice exhibited altered synaptic structure in their prefrontal cortex and selective deficits in fear extinction, which is a primary symptom of post-traumatic stress disorder (Milad et al., 2009; Ago et al., 2017). According to research on the spatiotemporal expression of VPAC2 in postpartum mice, the maturation of circuits that are involved in cognition may be disrupted by excessive, incorrectly timed or ectopic activation of VPAC2 receptors (Waschek et al., 1996; Ago et al., 2021). Recently, it is reported that the conditional human VIPR2 CNV BAC transgenic mouse model of VIPR2 CNV, known as hVIPR2-BAC tg mice, demonstrated cognitive, social behavior, and sensorimotor gating deficits as well as a decline in the complexity of the projection neurons (Tian et al., 2019; Ago et al., 2021). In cell culture models, as a response to the VPAC2 receptor and VIP agonists BAY 55–9,837, accumulation of VIPR2 mRNA and cAMP was observed, highlighting the functional importance of the microduplications (Vacic et al., 2011).
Aforementioned evidence consistently indicates that the VIPR2 gene is a risk gene for SCZ, and study results in cell culture and mice models suggest that VPAC2 receptor may impair cognitive function and sensory information processing by influencing neurological development, synaptic plasticity, and neuronal maturation, which may help explaining the potential role of VIPR2 gene in the pathogenic mechanisms of SCZ. To further discover potential pathogenic variants of VIPR2 gene for SCZ, amplicon targeted resequencing was performed for all exons and un-translated region (UTR) of VIPR2 gene in 1804 SCZ patients and 996 healthy controls in the present study, and associations between SNPs, single nucleotide variants (SNVs), short insertions and deletions (InDels) of the VIPR2 gene and SCZ were investigated.
A total of 1804 SCZ patients (693 women and 1,111 men; mean age = 44.51 years, SD = 12.13) were enrolled for this study and 996 healthy controls (559 women and 437 men; mean age = 43.41 years, SD = 20.17) without a history of mental health disorders or neurological diseases were recruited from local communities by advertisement (Table 1).
All participants were independent Chinese Han from the Jiangsu Province of China and provided signed informed consent prior to participating in the study. All SCZ patients were recruited by the Wuxi Mental Health Center from 2015 to 2017 and diagnosed by at least two independent psychiatrists according to the DSM-IV criteria. Patients were excluded if they have suffered hypertension; diabetes; neurological illness; mood disorder; mental retardation; history of substance use and psychotic disorder due to general medical condition. The study was approved by the local Ethics Committees, and conducted according to the principles of the Declaration of Helsinki 1975.
Peripheral blood samples were collected from all participants using K2EDTA tubes, and DNA was extracted through LifeFeng Genomic DNA Purification Kit (Lifefeng Biotech Co., Ltd., Shanghai, China). DNA quality control was performed utilizing a NanoDrop 1,000 Spectrophotometer (Thermo Scientific, United States). Thirty-two pairs of primers divided into two pools were designed to cover the VIPR2 gene. The primers’ sequences and their targeted regions are shown in Supplementary Table S1. Sequence libraries preparation was performed in a two-staged PCR process (Shanghai DynastyGene Co. Ltd). The size distribution of DNA library fragments was determined using the High Sensitivity DNA kit (Agilent Technologies, United States) on the Agilent 2,100 Bioanalyzer. Hundred and fifty bp paired-end reads were run on an Illumina HiSeq X ten platform (Illumina, United States).
Each data set was independently put through the pipeline of the Genome Analysis Toolkit (GATK) Best Practices1 for detecting germline short variants. Raw reads were aligned to the human reference genome (GRCh38) using the Burrows-Wheeler Aligner (BWA; Li and Durbin, 2010), SNPs and InDels were called using the GATK haplotypercaller, and the variations were annotated using Annovar (Wang et al., 2010). Alignments involving multiple species were carried out using the online Clustal Omega program.2 Rare non-synonymous mutations were confirmed by Sanger sequencing.
The SHEsisPlus online software platform3 (Yong and He, 2005; Li et al., 2009; Shen et al., 2016) was used to perform all single locus tests, including Hardy–Weinberg equilibrium (HWE) and association tests. Associations of allelic and genotypic polymorphisms with SCZ were analyzed using the chi-square test or Fisher’s exact test. All tests were two-tailed, and the p-values were calibrated using the false discovery rate (FDR-BH), which was regarded as statistical significance as being less than 0.05. Pairwise linkage disequilibrium (LD) analysis was performed for the common variants by Haploview software version 4.24 and the algorithm of four gamete rule was used to define haplotype blocks. The QC and analysis pipeline are shown in Figure 2.
We performed Sanger sequencing validation for 57 cases containing non-synonymous mutations or frameshift deletion detected by next-generation sequencing. Twenty variants harbored by 33 cases among them that all have next-generation sequencing depths deeper than 40, were confirmed to be true positives. The remaining 24 samples were all found to be false positives, 22 of which had depths lower than 40. Therefore, variants considered in the subsequent analysis were only those with a depth greater than 40.
A total of 173 high-quality variations, including 61 3’-UTR variants, 45 coding exon variants, 65 intron variants, one splicing area variant, and one downstream variant, were identified in the SCZ and control groups (Supplementary Table S2). In all, 147 loci with minor allele frequency (MAF) below 0.01 in across all databases, including the 1,000 Genomes Project, Exome Aggregation Consortium, and NHLBI Exome Sequencing Project, were chosen as rare variants. Of these, 70 loci were reported for the first time, and details of the other 24 common SNPs are provided in Supplementary Table S3.
Thirty-nine distinct variant loci, comprising two frameshift deletions, 13 synonymous mutations, and 25 non-synonymous variations that include two polymorphisms at the rs78564798 locus, were identified in the coding exons. Among whom, 5 synonymous mutations, 6 non-synonymous variants, and 2 frameshift deletions were initially reported. Figure 1 depicts the 19 non-synonymous variants and one frameshift deletion observed in cases. Sanger sequencing was utilized to confirm them and it was determined that all variants were heterozygous (Figure 3; Supplementary Figure S1).
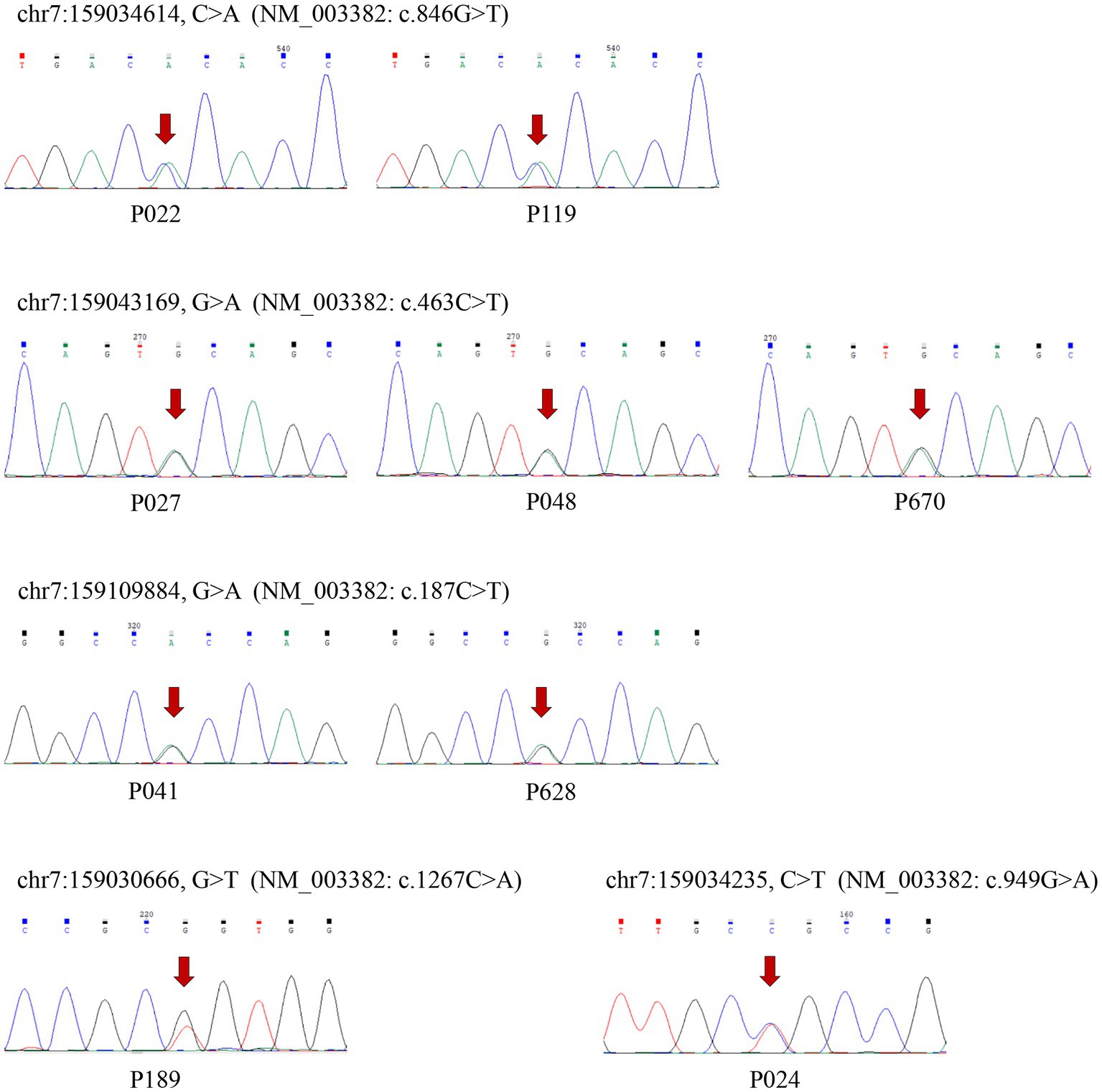
Figure 3. The results of Sanger sequence verifying the rare non-synonymous variants. The six variants were newly reported and arrows indicate the mutation sites.
The pathogenicity of the non-synonymous variants was predicted using SIFT, PolyPhen-2,5 MutationTaster,6 and LRT (Chun and Fay, 2009) in Annovar (Table 2). Only five non-synonymous variants (NM_003382: c.846G > T, c.463C > T, c.1267C > A, c.949G > A, c.187C > T) were predicted to be “D” (Deleterious for SIFT and LRT, Probably damaging for Polyphen-2, disease causing for MutationTaster) by at least three sorts of software. All of them were only detected in cases except rs534492909 (NM_003382: c.463C > T), and two of the five non-synonymous variants were firstly reported. Six variants which were only identified in control group were anticipated to be “D” by two or fewer pathogenicity prediction tools. The CADD (Combined Annotation Dependent Depletion) score was available at https://cadd.gs.washington.edu/snv for all variants. Scores of the five detrimental non-synonymous variants are all greater than 20, ranking them in the top 1% of deleterious variations in the human genome.
The amino acid variants for the five detrimental non-synonymous variants are VIPR2 (NP_003373): p.(Trp282Cys), p.(His155Tyr), p.(Arg423Ser), p.(Gly317Ser), and p.(Arg63Trp), respectively. Multiple alignments of VPAC2 receptor sequences from human, mouse, pig, cattle, and chicken showed that all the five loci are mostly conserved across evolution (Figure 4).

Figure 4. Multiple alignments of VPAC2 protein sequences of various species. The sites of non-synonymous variants were box out.
Hardy–Weinberg equilibrium analysis was performed for all the loci, and seven loci, including two rare and five common variants, were eliminated from further study because their HWE p-values in control group were less than 0.05. The results of pairwise linkage disequilibrium analysis showed that there was no haplotype block identified (Figure 5).
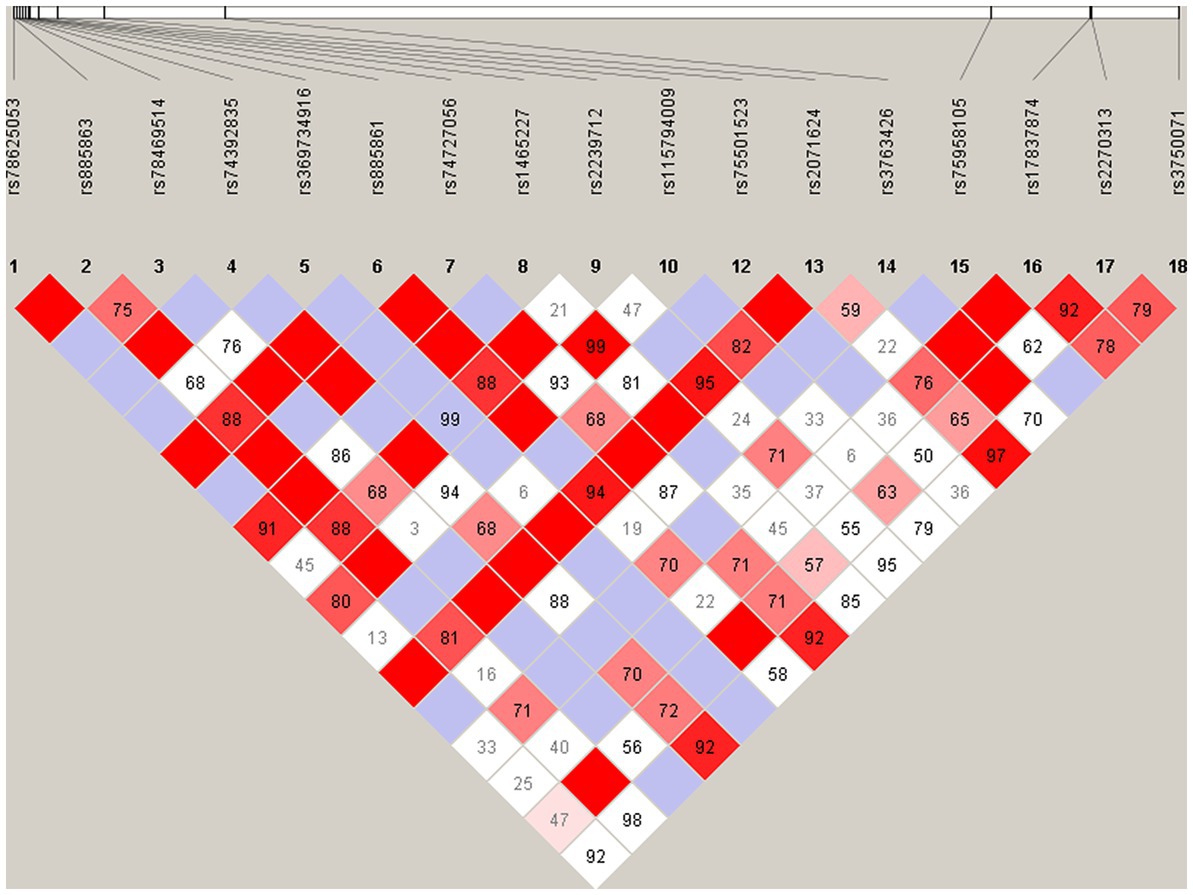
Figure 5. Pairwise linkage disequilibrium plot for the common variants in VIPR2 gene. The pairwise D′ values are presented in the matrices, Deep red implicates relatively strong linkage disequilibrium, and vice versa.
After FDR-BH correction, three rare mutations, including one non-synonymous variant and two intron variants, achieved significant association with SCZ (Table 3). Two alter alleles of the non-synonymous variant, rs78564798 [NM_003382: c.709C > G/T, p.(Pro237Ala)/p.(Pro237Ser)], were identified in two different cases while the alter allele “C” was carried by ten control samples (Pallele = 0.006, Pgenotype = 0.006 after FDR-BH correction). One of the significant intron variants (chr7:159034078, GRCh38) was novel reported and the variant only detected in 14 cases suggesting that the alter allele “C” in this locus may be a risk factor of SCZ (Pallele = 0.048, Pgenotype = 0.048 after FDR-BH correction). The other significant intron variant, rs372544903 (Pallele = 0.026, Pgenotype = 0.028 after FDR-BH correction), was only found in the control group indicating that the alter allele “T” may be a protective factor of SCZ. Additionally, there was a nominally significant association between SCZ and the rare non-synonymous variant rs199630455 (Pallele = 0.009, Pgenotype = 0.009, OR for “C” [95% CI] = 0.206 [0.054 ~ 0.778]), and the alter allele “C” may be a protective factor of SCZ, while the significance disappeared after correction.
We also carried out gene-based association studies for frameshift deletions and non-synonymous variations. In total, non-synonymous variants were detected in 54 patients with SCZ and 49 control samples. The cumulative numbers of non-synonymous mutation carriers in the case group and control group were substantially different according to the chi-square test (chi2 = 6.72, p = 0.009, Supplementary Table S4).
In this study, a total of 24 non-synonymous mutations and two frameshift deletions on the VIPR2 gene were identified by amplicon targeted resequencing. Among them, six non-synonymous mutations and one frameshift deletion were detected only in the control group, as well as 11 non-synonymous mutations and one frameshift deletion were detected only in patients with SCZ. Only variants detected in the control group were predicted to be benign (tolerated or neutral) by at least two pathogenicity prediction tools suggesting that some variants on the VIPR2 gene may have little effect on the expression, structure, and function of the VPAC2 receptor. Additionally, a gene-based association study found that the control group carried significantly more non-synonymous mutations and frameshift deletions than the case group, indicating that the presence of non-synonymous mutations on the VIPR2 gene, the majority of which are predicted to be benign, may prevent the VIPR2 gene’s overexpression or overactivation of its downstream pathways, offering protection against SCZ.
Four detrimental non-synonymous variants that predicted by at least three tools, namely, VIPR2 (NP_003373): p.(Trp282Cys), p.(Arg423Ser), p.(Gly317Ser), and p.(Arg63Trp), were only detected in patients with SCZ. The fact that these variations are highly conserved across species raises the possibility that the amino acid changes at these locations may have a significant effect on the structure and functionality of the VPAC2 receptor and subsequently contribute to the molecular pathogenesis of SCZ. One of them, p.(Arg63Trp), is found in the extracellular domain of the protein, and this mutation may have an impact on how the receptor binds to its ligand. Mutations p.(Arg423Ser) and p.(Gly317Ser) in the protein’s intracellular domain may impact the activation of the downstream pathway, while p.(Trp282Cys) in the helical transmembrane domain may affect the protein’s location on the cell membrane. Additionally, the SCHEMA browser database7 also reported the variants p.(Val297Phe), p.(Trp282Cys), p.(Asp113Asn), and p.(Val106Ile), which were discovered in individuals with SCZ in our investigation. We conducted meta-analysis for these loci, but no significance was found. Recently, five missense SNVs in the VIPR2 gene were identified in 516 patients with SCZ (Chen et al., 2022). Only p.(Thr189Lys) was found in our sample’s control group, and none of the loci found in our cases were reported in their research. Our research adds to the body of information on exon sequencing of the VIPR2 gene in the Chinese Han population and presents a comprehensive mutation spectrum of the VIPR2 gene for SCZ.
Our study identified several variants in the VIPR2 gene that may affect the expression and function of VPAC2, while the role of VPAC2 in postnatal maturation of the nervous system has been demonstrated, which is vital for the normal development of circuits crucial to cognition. VPAC2 and PAC1 are the receptors for PACAP and VIP. It has been confirmed in vitro using cultured neurons that VPAC2 activation by PACAP and VIP inhibits axons and dendrites growth while PAC1 receptor activation by PACAP stimulates growth (Ago et al., 2021). It has been reported that PACAP as a ligand for VPAC2, is significantly associated with SCZ and risperidone can reverse the aberrant behaviors shown in PACAP−/− mice, which are thought to represent schizophrenia-like phenotypes in rodents (Hashimoto et al., 2007). Additionally, mice who were continuously treated with PCP and utilized as prospective animal models for SCZ had considerably less PACAP mRNA expressed in the frontal cortex (Hashimoto et al., 2007). On the other hand, VIPR2 is involved in cAMP signaling and the activation of PKA, which in turn affects the hippocampus’s N-methyl-D-aspartate (NMDA) receptor-mediated synaptic transmission (Yang et al., 2009; Tam et al., 2015). VIP is another ligand for VPAC2, and Vip-deficient mice display a decline in hippocampus-dependent associative memory (Chaudhury et al., 2008), a cognitive impairment frequently observed in patients with SCZ (Libby et al., 2013; Tam et al., 2015).
Our results provided additional evidence that VIPR2 was a susceptibility gene of SCZ. However, there are two limitations of our study. The first is the small sample size, and the second is the absence of functional validation. To further understand the etiology associated with the VIPR2 gene in SCZ, targeted sequencing for the VIPR2 gene in more patients with the disorder and more functional validations are thought to be necessary.
In conclusions, the comprehensive mutation spectrum of the VIPR2 gene in SCZ was revealed by our research. In VIPR2, we discovered a significant difference between the overall number of rare non-synonymous mutation carriers in SCZ cases and healthy controls. Significant associations between SCZ and the non-synonymous mutation rs78564798 as well as two newly discovered rare variations in the VIPR2 gene’s introns have been found. Six novel rare non-synonymous variants and two novel frameshift deletions were reported in our study. The amino acid sequences of the five loci that were found in the case group were highly conserved, and according to functional prediction, their mutations may influence the structure and function of VPAC2 and thus be involved in the pathogenesis of SCZ. All the results further supported the hypothesis that the VIPR2 gene is a susceptibility gene for SCZ, but further functional validations are required to understand the role of VIPR2 in the etiology of SCZ.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Nanjing Medical University. The reference number is [2019]166. The patients/participants provided their written informed consent to participate in this study.
JMY and ZZ designed and supervised the whole research process. JJY and JZ carried out all experiments and managed the literature searches. FF and SY conducted DNA extraction and Sanger sequencing. JJY and JW undertook the statistical analysis. JMY and JJY conducted the patient recruitment and diagnosis as well as sample collection. JZ wrote the first draft of the manuscript. JJY, JMY, and ZZ proof read the article. All authors contributed to and approved the final manuscript.
This study was supported by Wuxi Taihu Talent Project (Nos. WXTTP2020008 and WXTTP2021), Wuxi Municipal Health Commission (Q202131), and Science and Technology Development Foundation of Nanjing Medical University (NMUB2020292), Medical Research Project of Jiangsu Province Health Commission (M2020078).
We thank all of patients and healthy controls who were involved in this study. And we are grateful for the help from psychiatrists during recruitment and diagnosis of SCZ patients.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1170708/full#supplementary-material
VIPR2, vasoactive intestinal peptide receptor 2 gene; SCZ, schizophrenia; ADHD, attention deficit and hyperactivity disorder; MD, mood disorders; SNPs, single-nucleotide polymorphisms; CNVs, copy number variations; GPCR, G-protein-coupled receptor; VIP, vasoactive intestinal peptide; PACAP, pituitary adenylate cyclase-activating polypeptide; CNS, the central nervous system; AC, adenylate cyclase; PKA, protein kinase A; PLC, phospholipase C; SNVs, single nucleotide variants; UTR, un-translated region; GATK, Genome Analysis Toolkit; BWA, Burrows-Wheeler Aligner; InDels, short insertions and deletions; HWE, Hardy–Weinberg equilibrium; FDR, false discovery rate; LD, linkage disequilibrium; MAF, minor allele frequency; CADD, Combined Annotation Dependent Depletion; NMDA, N-methyl-D-aspartate.
1. ^https://software.broadinstitute.org/gatk
2. ^http://www.ebi.ac.uk/Tools/msa/clustalo/
3. ^http://shesisplus.bio-x.cn/SHEsis.html
4. ^https://www.broadinstitute.org/haploview
5. ^http://genetics.bwh.harvard.edu/pph2
Ago, Y., Asano, S., Hashimoto, H., and Waschek, J. A. (2021). Probing the VIPR2 microduplication linkage to schizophrenia in animal and cellular models. Front. Neurosci. 15:717490. doi: 10.3389/fnins.2021.717490
Ago, Y., Hayata-Takano, A., Kawanai, T., Yamauchi, R., Takeuchi, S., Cushman, J. D., et al. (2017). Impaired extinction of cued fear memory and abnormal dendritic morphology in the prelimbic and infralimbic cortices in VPAC2 receptor (VIPR2)-deficient mice. Neurobiol. Learn. Mem. 145, 222–231. doi: 10.1016/j.nlm.2017.10.010
An, S., Tsai, C., Ronecker, J., Bayly, A., and Herzog, E. D. (2012). Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J. Comp. Neurol. 520, 2730–2741. doi: 10.1002/cne.23078
Barbato, A. (1998). Psychiatry in transition: outcomes of mental health policy shift in Italy. Aust. N. Z. J. Psychiatry 32, 673–679. doi: 10.3109/00048679809113122
Chaudhury, D., Loh, D. H., Dragich, J. M., Hagopian, A., and Colwell, C. S. (2008). Select cognitive deficits in vasoactive intestinal peptide deficient mice. BMC Neurosci. 9:63. doi: 10.1186/1471-2202-9-63
Chen, C.-H., Cheng, M.-C., Hu, T.-M., Ping, L.-Y., Kushima, I., and Aleksic, B. (2022). Identification of rare mutations of the vasoactive intestinal peptide receptor 2 gene in schizophrenia. Psychiatr. Genet. 32:3. doi: 10.1097/YPG.0000000000000313
Chesney, E., Goodwin, G. M., and Fazel, S. (2014). Risks of all-cause and suicide mortality in mental disorders: a meta-review. World Psychiatry 13, 153–160. doi: 10.1002/wps.20128
Chun, S., and Fay, J. C. (2009). Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561. doi: 10.1101/gr.092619.109
Firouzabadi, S. G., Kariminejad, R., Vameghi, R., Darvish, H., Ghaedi, H., Banihashemi, S., et al. (2017). Copy number variants in patients with autism and additional clinical features: report of VIPR2 duplication and a novel microduplication syndrome. Mol. Neurobiol. 54, 7019–7027. doi: 10.1007/s12035-016-0202-y
Fleischhacker, W. W., Arango, C., Arteel, P., Barnes, T. R. E., Carpenter, W., Duckworth, K., et al. (2014). Schizophrenia—time to commit to policy change. Schizophr. Bull. 40, S165–S194. doi: 10.1093/schbul/sbu006
Harmar, A. J., Marston, H. M., Shen, S., Spratt, C., West, K. M., Sheward, W. J., et al. (2002). The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cells 109, 497–508. doi: 10.1016/S0092-8674(02)00736-5
Hashimoto, R., Hashimoto, H., Shintani, N., Chiba, S., Hattori, S., Okada, T., et al. (2007). Pituitary adenylate cyclase-activating polypeptide is associated with schizophrenia. Mol. Psychiatry 12, 1026–1032. doi: 10.1038/sj.mp.4001982
Joyce, E. M., and Roiser, J. P. (2007). Cognitive heterogeneity in schizophrenia. Curr. Opin. Psychiatry 20:3. doi: 10.1097/YCO.0b013e3280ba4975
Laburthe, M., Couvineau, A., and Marie, J. C. (2002). VPAC receptors for VIP and PACAP. Recept. Channels 8, 137–153. doi: 10.1080/10606820213680
Laburthe, M., Couvineau, A., and Tan, V. (2007). Class II G protein-coupled receptors for VIP and PACAP: structure, models of activation and pharmacology. Peptides 28, 1631–1639. doi: 10.1016/j.peptides.2007.04.026
Levinson, D. F., Duan, J., Oh, S., Wang, K., Sanders, A. R., Shi, J., et al. (2011). Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am. J. Psychiatry 168, 302–316. doi: 10.1176/appi.ajp.2010.10060876
Li, Z., Chen, J., Xu, Y., Yi, Q., Ji, W., Wang, P., et al. (2016). Genome-wide analysis of the role of copy number variation in schizophrenia risk in Chinese. Biol. Psychiatry 80, 331–337. doi: 10.1016/j.biopsych.2015.11.012
Li, Z., Chen, J., Yu, H., He, L., Xu, Y., Zhang, D., et al. (2017). Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49, 1576–1583. doi: 10.1038/ng.3973
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., et al. (2009). A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 19:519–523. doi: 10.1038/cr.2009.33
Libby, L. A., Yonelinas, A. P., Ranganath, C., and Ragland, J. D. (2013). Recollection and familiarity in schizophrenia: a quantitative review. Biol. Psychiatry 73, 944–950. doi: 10.1016/j.biopsych.2012.10.027
MacKenzie, C. J., Lutz, E. M., Johnson, M. S., Robertson, D. N., Holland, P. J., and Mitchell, R. (2001). Mechanisms of phospholipase C activation by the vasoactive intestinal polypeptide/pituitary adenylate cyclase-activating polypeptide type 2 receptor. Endocrinology 142, 1209–1217. doi: 10.1210/endo.142.3.8013
Marder, S. R., and Cannon, T. D. (2019). Schizophrenia. N. Engl. J. Med. 381, 1753–1761. doi: 10.1056/NEJMra1808803
Marshall, C. R., Howrigan, D. P., Merico, D., Thiruvahindrapuram, B., Wu, W., Greer, D. S., et al. (2017). Contribution of copy number variants to schizophrenia from a genome-wide study of 41, 321 subjects. Nat. Genet. 49, 27–35. doi: 10.1038/ng.3725
Milad, M. R., Pitman, R. K., Ellis, C. B., Gold, A. L., Shin, L. M., Lasko, N. B., et al. (2009). Neurobiological basis of failure to recall extinction memory in posttraumatic stress disorder. Biol. Psychiatry 66, 1075–1082. doi: 10.1016/j.biopsych.2009.06.026
Owen, M. J., Sawa, A., and Mortensen, P. B. (2016). Schizophrenia. Lancet 388, 86–97. doi: 10.1016/S0140-6736(15)01121-6
Purcell, S. M., Moran, J. L., Fromer, M., Ruderfer, D., Solovieff, N., Roussos, P., et al. (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. doi: 10.1038/nature12975
Ripke, S., Neale, B. M., Corvin, A., Walters, J. T. R., Farh, K.-H., Holmans, P. A., et al. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427.
Shen, J., Li, Z., Chen, J., Song, Z., Zhou, Z., and Shi, Y. (2016). SHEsisPlus, a toolset for genetic studies on polyploid species. Sci. Rep. 6:24095. doi: 10.1038/srep24095
Shen, S., Spratt, C., Sheward, W. J., Kallo, I., West, K., Morrison, C. F., et al. (2000). Overexpression of the human VPAC2 receptor in the suprachiasmatic nucleus alters the circadian phenotype of mice. Proc. Natl. Acad. Sci. 97, 11575–11580. doi: 10.1073/pnas.97.21.11575
Sheward, W. J., Lutz, E. M., and Harmar, A. J. (1995). The distribution of vasoactive intestinal peptide2 receptor messenger RNA in the rat brain and pituitary gland as assessed by in situ hybridization. Neuroscience 67, 409–418. doi: 10.1016/0306-4522(95)00048-N
Soria, V., Martínez-Amorós, È., Escaramís, G., Valero, J., Pérez-Egea, R., García, C., et al. (2010). Differential Association of Circadian Genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 35, 1279–1289. doi: 10.1038/npp.2009.230
Sullivan, P. F., Kendler, K. S., and Neale, M. C. (2003). Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192. doi: 10.1001/archpsyc.60.12.1187
Tam, S. K. E., Pritchett, D., Brown, L. A., Foster, R. G., Bannerman, D. M., and Peirson, S. N. (2015). Chapter fifteen-sleep and circadian rhythm disruption and recognition memory in schizophrenia. Methods Enzymol. 552, 325–349. doi: 10.1016/bs.mie.2014.10.008
Tian, X., Richard, A., El-Saadi, M. W., Bhandari, A., Latimer, B., Van Savage, I., et al. (2019). Dosage sensitivity intolerance of VIPR2 microduplication is disease causative to manifest schizophrenia-like phenotypes in a novel BAC transgenic mouse model. Mol. Psychiatry 24, 1884–1901. doi: 10.1038/s41380-019-0492-3
Trubetskoy, V., Pardiñas, A. F., Qi, T., Panagiotaropoulou, G., Awasthi, S., Bigdeli, T. B., et al. (2022). Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508. doi: 10.1038/s41586-022-04434-5
Vacic, V., McCarthy, S., Malhotra, D., Murray, F., Chou, H.-H., Peoples, A., et al. (2011). Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 471, 499–503. doi: 10.1038/nature09884
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi: 10.1093/nar/gkq603
Waschek, J. A., Ellison, J., Bravo, D. T., and Handley, V. (1996). Embryonic expression of vasoactive intestinal peptide (VIP) and VIP receptor genes. J. Neurochem. 66, 1762–1765.
Wilmot, B., Fry, R., Smeester, L., Musser, E. D., Mill, J., and Nigg, J. T. (2016). Methylomic analysis of salivary DNA in childhood ADHD identifies altered DNA methylation in VIPR2. J. Child Psychol. Psychiatry 57, 152–160. doi: 10.1111/jcpp.12457
Wockner, L. F., Noble, E. P., Lawford, B. R., Young, R. M., Morris, C. P., Whitehall, V. L., et al. (2014). Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Translational Psychiatry. 4:e339. doi: 10.1038/tp.2013.111
Yang, K., Trepanier, C. H., Li, H., Beazely, M. A., Lerner, E. A., Jackson, M. F., et al. (2009). Vasoactive intestinal peptide acts via multiple signal pathways to regulate hippocampal NMDA receptors and synaptic transmission. Hippocampus 19, 779–789. doi: 10.1002/hipo.20559
Yong, Y., and He, L. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–98. doi: 10.1038/sj.cr.7290272
Keywords: VIPR2 gene, schizophrenia, amplicon targeted resequencing, association study, causative variants
Citation: Yin J, Zhou J, Fang F, Yu S, Wang J, Yuan J and Zhou Z (2023) Identification of VIPR2 rare and common variants in the Chinese Han population with schizophrenia. Front. Mol. Neurosci. 16:1170708. doi: 10.3389/fnmol.2023.1170708
Received: 21 February 2023; Accepted: 07 April 2023;
Published: 27 April 2023.
Edited by:
Bing Lang, Central South University, ChinaReviewed by:
Xinyu Fang, Nanjing Brain Hospital Affiliated to Nanjing Medical University, ChinaCopyright © 2023 Yin, Zhou, Fang, Yu, Wang, Yuan and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmin Yuan, eWptcGFwZXJAMTYzLmNvbQ==; Zhenhe Zhou, emhvdXpoQG5qbXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.