- 1Department of Pharmacology and Toxicology, Virginia Commonwealth University, Richmond, VA, United States
- 2VCU Alcohol Research Center, Virginia Commonwealth University, Richmond, VA, United States
Adolescence is marked in part by the ongoing development of the prefrontal cortex (PFC). Binge ethanol use during this critical stage in neurodevelopment induces significant structural changes to the PFC, as well as cognitive and behavioral deficits that can last into adulthood. Previous studies showed that adolescent binge ethanol causes lasting deficits in working memory, decreases in the expression of chromatin remodeling genes responsible for the methylation of histone 3 lysine 36 (H3K36), and global decreases in H3K36 in the PFC. H3K36me3 is present within the coding region of actively-transcribed genes, and safeguards against aberrant, cryptic transcription by RNA Polymerase II. We hypothesize that altered methylation of H3K36 could play a role in adolescent binge ethanol-induced memory deficits. To investigate this at the molecular level, ethanol (4 g/kg, i.g.) or water was administered intermittently to adolescent mice. RNA-and ChIP-sequencing were then performed within the same tissue to determine gene expression changes and identify genes and loci where H3K36me3 was disrupted by ethanol. We further assessed ethanol-induced changes at the transcription level with differential exon-use and cryptic transcription analysis – a hallmark of decreased H3K36me3. Here, we found ethanol-induced changes to the gene expression and H3K36me3-regulation of synaptic-related genes in all our analyses. Notably, H3K36me3 was differentially trimethylated between ethanol and control conditions at synaptic-related genes, and Snap25 and Cplx1 showed evidence of cryptic transcription in males and females treated with ethanol during adolescence. Our results provide preliminary evidence that ethanol-induced changes to H3K36me3 during adolescent neurodevelopment may be linked to synaptic dysregulation at the transcriptional level, which may explain the reported ethanol-induced changes to PFC synaptic function.
1. Introduction
Adolescent development is characterized by a number of physiological, behavioral, emotional, and cognitive changes (Spear, 2000). These changes give rise to heightened reward sensitivity, sensation seeking, and impulsivity, and allow for increased participation in risky behaviors, such as the initiation and escalation of alcohol use (Spear, 2000; Casey et al., 2008; Romer et al., 2017; Lees et al., 2020). In a 2020 report, 16.1% of Americans between the age of 12–20 reported participation in alcohol consumption, with an estimated 9.2% participating in binge drinking (SAMHSA, 2020). Binge alcohol consumption, common during adolescence, is defined as having five or more drinks on one occasion (SAMHSA, 2020). Adolescents respond differently to alcohol than adults, showing increased sensitivity to alcohol’s rewarding aspects, and decreased sensitivity to alcohol’s aversive aspects (Spear, 2014). This altered sensitivity enables adolescents to consume more alcohol, giving rise to the binge drinking commonly seen among this age group, while subsequently increasing the risk for developing an alcohol use disorder (AUD) later in life (Grant and Dawson, 1997; Spear, 2000). Adolescent binge drinking is particularly harmful as the brain is still undergoing development throughout this period (Spear, 2000). Alcohol use can disrupt ongoing developmental processes, resulting in structural, cognitive, and behavioral effects (De Bellis et al., 2005; Medina et al., 2008; Bava et al., 2013; Squeglia et al., 2014; Lees et al., 2020; El Marroun et al., 2021), some of which last into adulthood (Pascual et al., 2007; Coleman et al., 2011, 2014; Vargas et al., 2014; Crews et al., 2016; Marco et al., 2017; Wolstenholme et al., 2017).
The prefrontal cortex (PFC) integrates information from several brain regions to regulate attention, executive function, and working memory (Spear, 2000). The PFC is one of the last brain regions to mature, and undergoes significant changes in adolescent neurodevelopment – evidenced by increased synaptic pruning and myelination (Gogtay et al., 2004; Barnea-Goraly et al., 2005; Drzewiecki et al., 2017). Human imaging studies show that adolescent alcohol use is associated with alterations in PFC structure and myelination (De Bellis et al., 2005; Medina et al., 2008; Pfefferbaum et al., 2018; El Marroun et al., 2021), and similar effects have been shown in rodents (Vargas et al., 2014; Montesinos et al., 2015; Vetreno et al., 2016; Wolstenholme et al., 2017; Tavares et al., 2019). This ethanol-induced damage to the PFC may play a role in the behavioral effects seen in response to adolescent binge ethanol, specifically in terms of memory function (Bekinschtein and Weisstaub, 2014; Vargas et al., 2014; Montesinos et al., 2015; Warburton and Brown, 2015; Marco et al., 2017; Wolstenholme et al., 2017; Macht et al., 2020; Bent et al., 2022; van Hees et al., 2022).
Binge ethanol use during adolescence can result in diminished cognitive function, including deficits in conditioned discrimination, reversal learning, and memory (Pascual et al., 2007, 2021; Coleman et al., 2011, 2014; Hanson et al., 2011; Mota et al., 2013; Vargas et al., 2014; Montesinos et al., 2015; Vetreno and Crews, 2015; Vetreno et al., 2016, 2020; Carbia et al., 2017; Marco et al., 2017; Wolstenholme et al., 2017; Contreras et al., 2019; Drissi et al., 2020; Macht et al., 2020; Peñasco et al., 2020; Bent et al., 2022; van Hees et al., 2022). In rodents, adolescent binge ethanol leads to long-term deficits in working memory (Vargas et al., 2014; Marco et al., 2017; Wolstenholme et al., 2017; Bent et al., 2022) and cognitive flexibility (Coleman et al., 2011; Vetreno and Crews, 2012; Acheson et al., 2013; Coleman et al., 2014; Gass et al., 2014). Moreover, it has been shown in rodent (Markwiese et al., 1998; White et al., 2000; Younis et al., 2019; Bent et al., 2022) and human (Acheson et al., 1998) studies that ethanol-induced behavioral effects are more substantial when ethanol exposure takes place during adolescence than in adulthood, suggesting that adolescence is a critical time for this damage to occur. Memory deficits in a novel object recognition (NOR) task occurred in adolescent mice exposed to binge ethanol (Wolstenholme et al., 2017; Bent et al., 2022), but did not occur when ethanol exposure occurred in adulthood (Bent et al., 2022). While the behavioral and cognitive effects of adolescent binge ethanol have been widely studied, the mechanisms behind how these behavioral effects occur is largely unknown.
Recent findings have implicated histone methylation as playing a critical role in regulating psychiatric disorders, including AUD (Tsankova et al., 2007; Robison and Nestler, 2012; Krishnan et al., 2014; Barbier et al., 2017; Berkel and Pandey, 2017; Pandey et al., 2017), and ethanol dysregulates histone methylation in both human (Ponomarev et al., 2012) and rodent models (Finegersh and Homanics, 2014; Montesinos et al., 2016; Barbier et al., 2017; Kyzar et al., 2017; Wolstenholme et al., 2017). The stable, yet highly dynamic nature of these histone methylation marks also makes them particularly suitable for involvement in learning and memory processes (Parkel et al., 2013). Given these data, ethanol exposure during the critical adolescent period could alter histone methylation in specific brain regions to affect behavioral outcomes into adulthood. While a recent study has utilized epigenomic editing to modulate adolescent ethanol-induced behavioral changes in rats (Bohnsack et al., 2022), the connection between ethanol-induced epigenetic changes in the brain and behavioral deficits requires additional investigation.
Previously, it was shown that adolescent binge ethanol leads to persistent memory deficits in a NOR task (Montesinos et al., 2015; Vetreno and Crews, 2015; Vetreno et al., 2016; Marco et al., 2017; Wolstenholme et al., 2017; Drissi et al., 2020; Macht et al., 2020; Peñasco et al., 2020; Pascual et al., 2021; Bent et al., 2022; van Hees et al., 2022), alterations in adult spatial memory (Contreras et al., 2019; Bent et al., 2022), dysregulation of histone methyltransferases (HMTs) specific for H3K36(Wolstenholme et al., 2017), and global decreases of H3K36 levels in the PFC (Wolstenholme et al., 2017). Dysregulation of H3K36 may underlie the memory deficits associated with adolescent binge ethanol, due to the role H3K36 methylation plays in supporting transcriptional fidelity and memory consolidation (Butler and Dent, 2012; Gräff et al., 2012). H3K36me3 is enriched within transcribed regions of genes, and acts as a safeguard to protect against aberrant transcription (Smolle et al., 2013). After RNA Polymerase II (RNA pol II) transcribes a sequence, chromatin is “reset” to maintain proper transcriptional fidelity (Smolle et al., 2013). H3K36me3 plays a key role in this, recruiting other factors to de-acetylate histones H3 and H4, and maintaining ordered chromatin over open reading frames to repress unwanted transcription from occurring (Carrozza et al., 2005; Smolle and Workman, 2013). When H3K36me3 is not present, chromatin remains open and becomes subject to what is considered “cryptic” or aberrant transcription – RNA pol II can begin transcription from the middle of the coding region of the gene, instead of beginning at the correct transcription start site, ultimately producing an incorrect, truncated transcript (Wei et al., 2019). These effects can be seen with knockdown of Setd2, the specific HMT for H3K36me3. In mouse embryonic stem cells, Setd2 knockdown resulted in increased number of cryptic transcripts, hallmarks of which include transcription of infrequently transcribed genes, dysregulation of intragenic transcriptional initiation, and alternative splicing patterns (Zhang et al., 2014). A reduction in H3K36me3 in a mouse model also gave rise to impaired splicing, altered gene expression, and ultimately, cognitive deficits (Sessa et al., 2019). Cryptic transcripts have the potential to interfere with the full-length gene product, leading to gene expression discrepancies (Butler and Dent, 2012). H3K36 dysregulation has been associated with a number of neurological diseases, such as Autism Spectrum Disorder (Sessa et al., 2019), and H3K36me3-mediated alternative splicing was found to play a role in cocaine reward behavior (Xu et al., 2021). Given this data, we hypothesized that adolescent binge ethanol negatively affects trimethylation of H3K36 within the PFC, leading to transcriptional changes in genes impacting memory-related processes.
Using RNA-seq and chromatin immunoprecipitation coupled to sequencing (ChIP-seq), we determined which H3K36me3-regulated loci were differentially affected by adolescent binge ethanol, and how binge ethanol affects gene expression. Here, we found that adolescent binge ethanol impacted the regulation and expression of genes important for synaptic function and memory processes. Ultimately, this work identified novel genes that may contribute to the persistent memory deficits associated with binge ethanol.
2. Results
We conducted molecular analyses on chromatin and gene expression responses to adolescent binge ethanol exposure in male and female DBA/2J mice at multiple levels of resolution, including whole transcript, exon-utilization, and cryptic transcription. These analyses were done within the same tissue to determine within-sample changes at both the chromatin regulation and gene expression levels (Figure 1). We found divergent responses between male and female animals with consistent, but fewer common responses that occurred across the sexes. We therefore describe our findings below to explicitly highlight these sex-specific responses. While both sexes showed commonality between gene ontology terms, the genes contained within these categories were sometimes different, suggesting the possibility that ethanol may act on common pathways, but through different genes or signaling cascades.
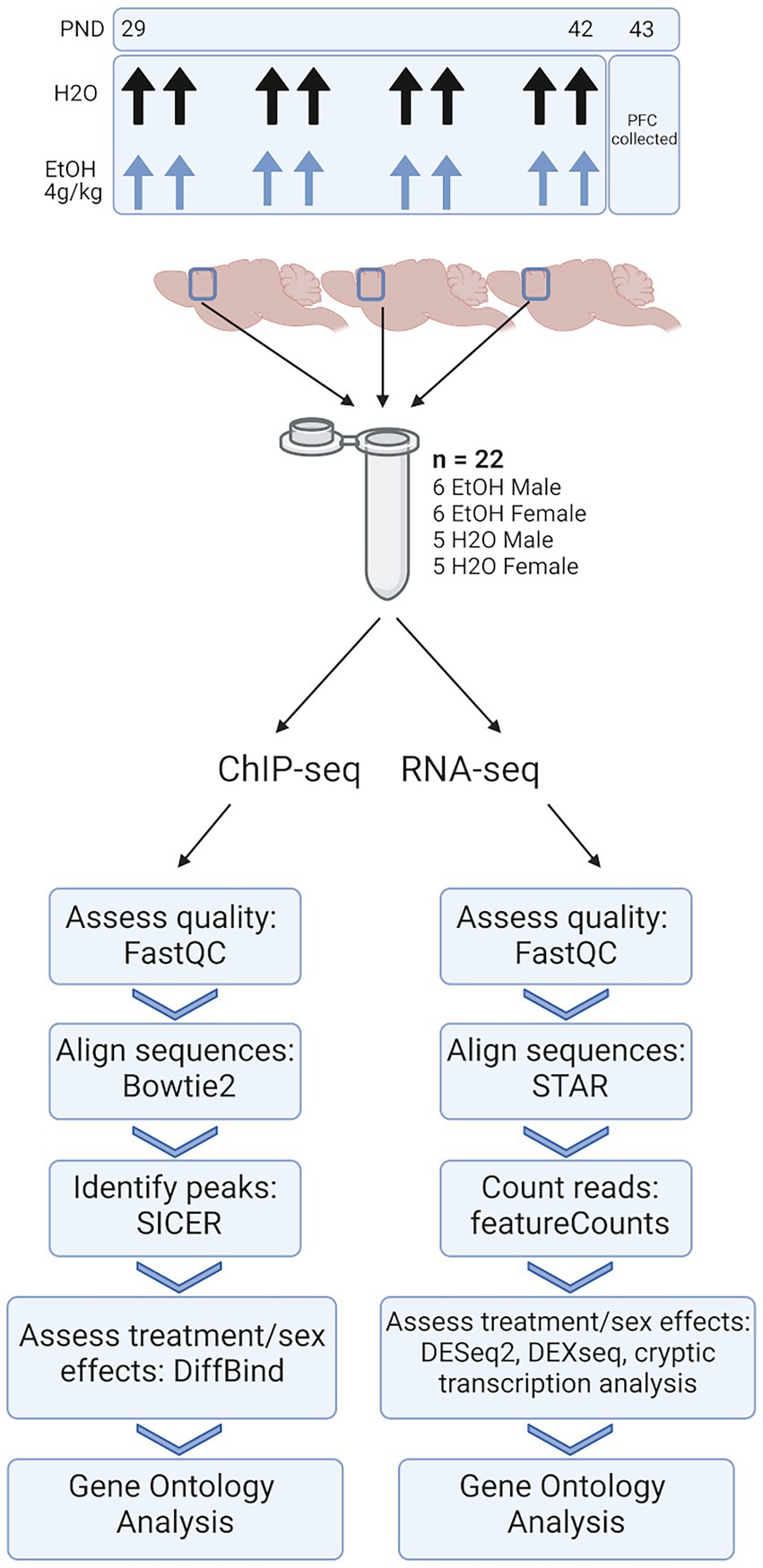
Figure 1. Methods. Adolescent DBA/2J mice were dosed with water or ethanol (4 g/kg) from PND 29–42. PFC was collected 24 h after the last dose of ethanol. Three PFCs from each treatment group were homogenized together, divided, and subjected to ChIP-seq and RNA-seq analysis.
2.1. Adolescent binge ethanol induces changes to extracellular matrix and axon guidance-related genes in males
To identify genes that were differentially regulated by repeated binge ethanol in the PFC of DBA/2J male mice, we used DESeq2 with a value of p < 0.01 without additional filtering for fold-change to generate a gene list of sufficient length for gene discovery using downstream gene ontology analyses. Adolescent binge ethanol differentially regulated 342 total genes in males, with 175 being up-regulated and 167 down-regulated (Supplementary Table 1). Two hundred and ninety-seven of these genes were unique to males (Figure 2A) and served as the input for male-specific gene ontology over-representation analysis using ToppFun. Results of the gene ontology analysis are shown in Figure 2B. The first 15 molecular function and biological process categories were selected to represent our results. The full gene ontology results can be found in Supplementary Table 2. The top significant categories for molecular function included semaphorin binding, extracellular matrix structural constituent, tubulin binding, and calmodulin-dependent kinase activity. For biological process, top significant categories included negative regulation of cell growth, male sex differentiation, and axon guidance.
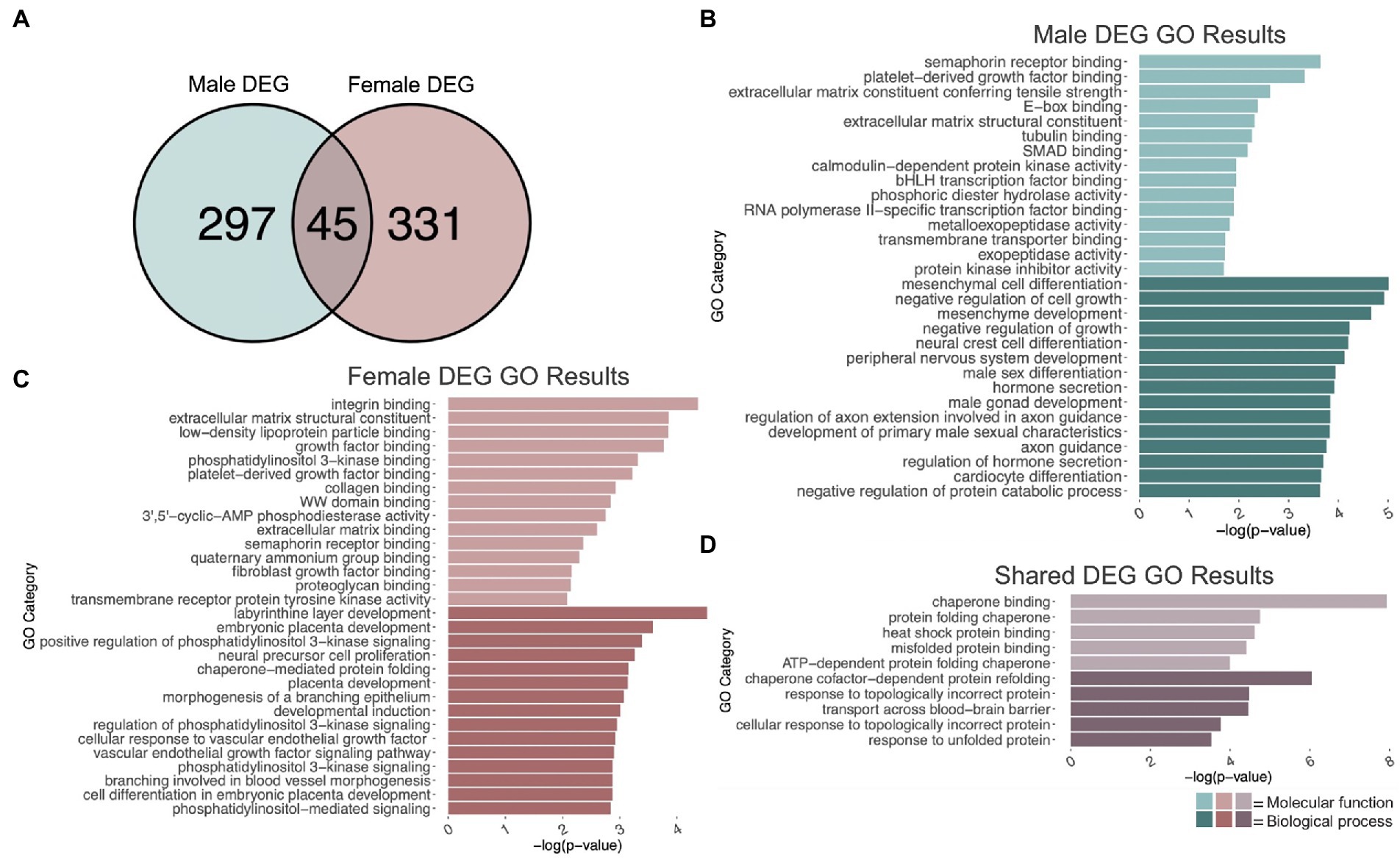
Figure 2. GO analysis of DEGs. (A) Number of genes differentially expressed due to adolescent binge ethanol in males and females at p < 0.01. Fisher’s exact test determined male and female gene overlap to be significant, p = 5.7e-34. (B) GO analysis of DEGs unique to males. (C) GO analysis of DEGs unique to females. (D) GO analysis of DEGs that are shared between males and females.
2.2. Adolescent binge ethanol induces changes to growth factor binding and extracellular matrix organization in females
Differential gene expression and gene ontology analysis were carried out for females as described above. At a value of p < 0.01, 376 genes were differentially expressed due to binge ethanol in females, with 215 up-regulated and 161 down-regulated (Supplementary Table 1). Three hundred and thirty-one genes were uniquely differentially regulated in female PFC (Figure 2A) and served as input for female-specific gene ontology analysis. Results of the female gene ontology analysis are shown in Figure 2C. Molecular function gene categories that were significantly altered by ethanol in females include extracellular matrix structural constituent, growth factor binding, and semaphorin receptor binding, similar categories to our male differential gene expression analysis. The top significant categories for biological process included positive regulation of phosphatidylinositol 3 kinase activity, neural precursor cell differentiation, and chaperone mediated protein folding. Although not included within the top 15 biological process categories, females also showed gene expression changes in genes involved in oligodendrocyte differentiation (p = 0.01, Supplementary Table 3). Reduced oligodendrocyte differentiation is thought to be an underlying cause of ethanol-induced myelin loss (Bichenkov and Ellingson, 2009; Kim et al., 2015; Darbinian et al., 2021; Guo et al., 2021), and has shown to be epigenetically regulated (Emery and Lu, 2015; Liu et al., 2015; Guo et al., 2021). Our lab has previously seen reduced myelin-related gene expression and reduced levels of H3K9me3, which was shown to regulate differentiation of oligodendrocyte precursor cells into mature oligodendrocytes (Liu et al., 2015; Wolstenholme et al., 2017).
While the extracellular matrix structural constituent category (GO:0005201) was found in both the male and female gene ontology analysis, the genes contained in each of these categories differed by sex. In males, Ltbp4, Col4a1, Col4a2, Col9a3, Abi3bp, Tnc, Col6a6, and Agrn in this category were differentially expressed, while in females, Col24a1, Fbn1, Nid1, Ntn1, Col5a1, Hspg2, Creld2, Chadl, Thbs1, Igfbp7, and Eln in this category were differentially expressed. Similarly, both male and female gene ontology analysis returned the semaphorin receptor binding category (GO:0030215) but with different genes represented. In males, Sema3a, Sema5b, Sema4a, and Sema4b were differentially expressed due to binge ethanol. In females, Sh3bp1, Sema6a, and Sema3d were differentially expressed. These results imply that the extracellular matrix and semaphorin binding is altered by adolescent binge ethanol in both males and females, but may be altered through a different mechanism in each sex.
2.3. Adolescent binge ethanol induces changes to unfolded protein response-related genes in males and females
Three hundred forty-two genes were differentially expressed in males, and 376 genes were differentially expressed in females due to adolescent binge ethanol at a value of p < 0.01. Of those, only 45 genes were differentially expressed in both males and females (p = 5.7e-34, Figure 2A). Due to the smaller number of genes for our shared gene ontology analysis, we selected the first five molecular function and biological process categories to represent our results (Figure 2D). Gene ontology analysis showed that these overlapping genes were related to chaperone binding (molecular function), misfolded protein binding (molecular function), chaperone-cofactor-dependent protein refolding (biological process), and transport across blood–brain barrier (biological process). The full gene ontology results can be found in Supplementary Table 4.
2.4. Adolescent binge ethanol induces differential exon use of dendritic spine-related genes in males and females
Given H3K36me3’s proposed role in alternative splicing (Zhou et al., 2014; Xu et al., 2021), we next performed an exon-level analysis of binge ethanol gene expression in males and females using DEXSeq. At a value of p < 0.001, males showed 595 unique differentially used exons within 501 genes due to binge ethanol treatment (Supplementary Table 5). Of those, 460 genes were unique to males, and were used as input for male-specific gene ontology analysis (Figure 3A). Significantly over-represented gene ontology categories for molecular function included tubulin binding, histone binding, and extracellular matrix structural constituent (Figure 3B). Top biological process categories included dendrite development, dendritic spine development, and positive regulation of GTPase activity (Figure 3B).
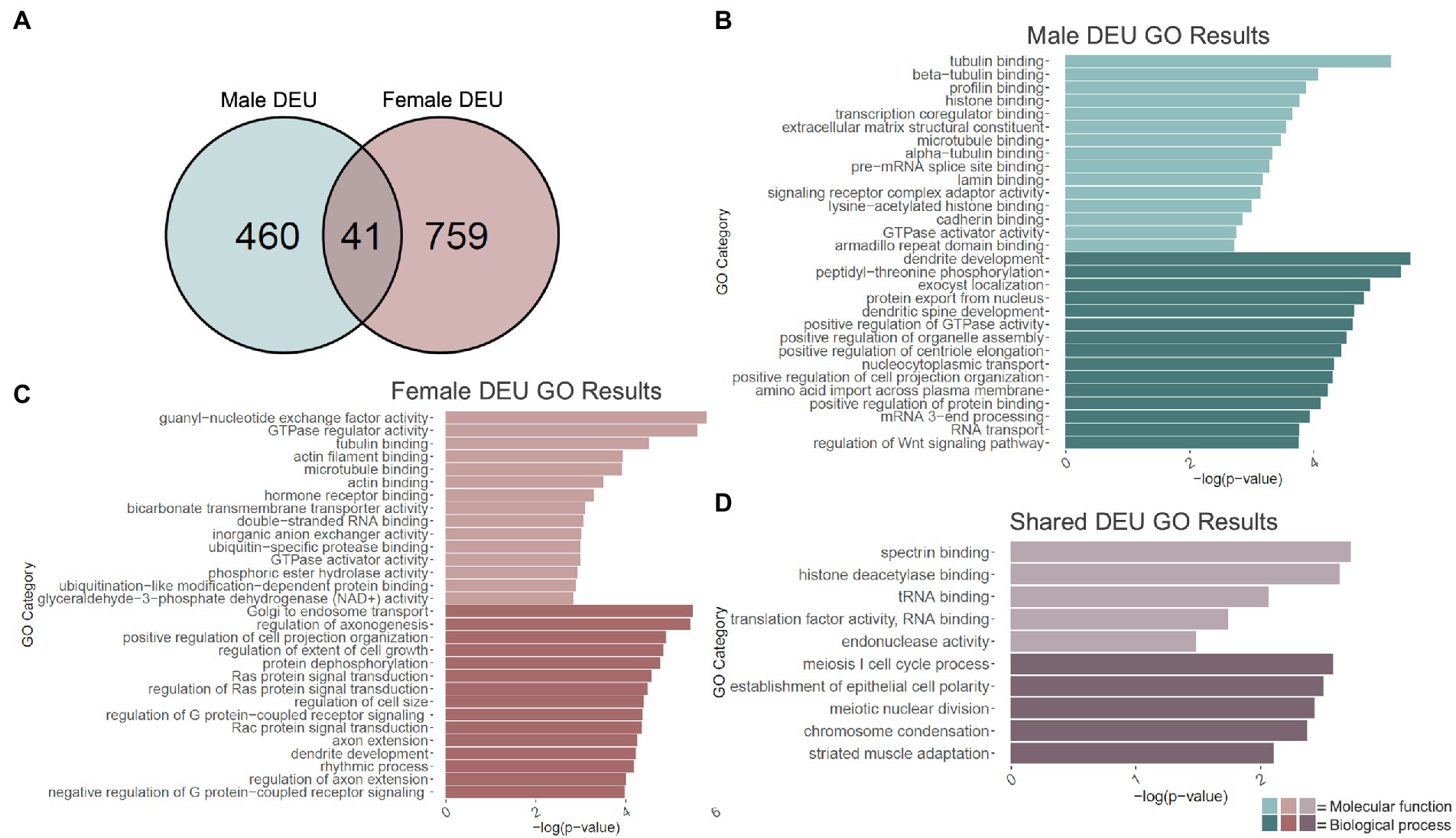
Figure 3. GO analysis of differential exon use (DEU) genes. (A) Number of genes showing differential exon use due to adolescent binge ethanol in males and females at p < 0.001. Fisher’s exact test determined male and female gene overlap to be significant, p = 1.7e-11. (B) GO analysis of genes with differentially used exons unique to males. (C) GO analysis of genes with differentially used exons unique to females. (D) GO analysis of genes with differentially used exons that are shared between males and females.
Exon-level analysis was carried out identically in females. At a value of p < 0.001, females showed 980 differentially used exons in 800 genes, almost double the number in males (Supplementary Table 5). Of those 759 genes were unique to females and were used as input into the female-specific gene ontology analysis (Figure 3A). Molecular function categories that were significantly over-represented included guanyl-nucleotide exchange factor activity, tubulin binding, actin filament binding, and microtubule binding (Figure 3C). Biological process categories that were over-represented included positive regulation of cell projection organization, axon extension, and dendrite development (Figure 3C).
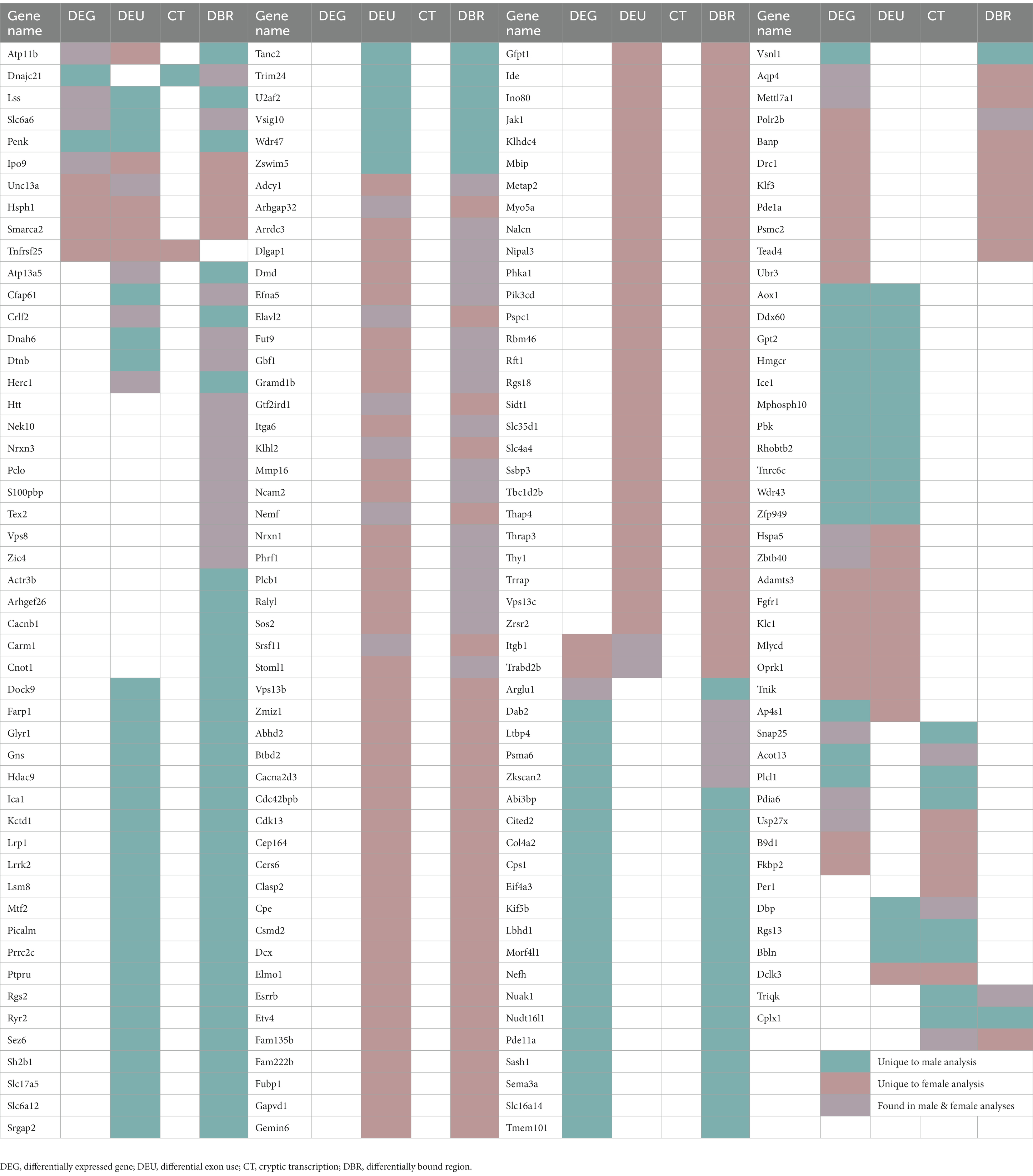
DEXSeq analysis showed that after adolescent ethanol treatment, 501 genes had differentially used exons in males, and 800 genes showed differentially used exons in females at a value of p < 0.001. Of those, 41 genes showed differentially used exons in both males and females (p = 1.7e-11, Figure 3A). Significantly over-represented gene categories included histone deacetylase binding (molecular function), translation factor activity, RNA binding (molecular function), and chromatin condensation (biological process; Figure 3D). Tnfrsf25 was the only gene in females that was differentially expressed due to binge ethanol, showed differential exon usage, and also showed evidence of cryptic transcription (Table 1). Full gene ontology results for males, females, and shared genes that showed differential exon usage can be found in Supplementary Tables 6–8.
2.5. Adolescent binge ethanol induces cryptic transcription of Snap25 and Cplx1 in males and females
Adolescent binge ethanol led to the cryptic transcription of 30 genes in males and 26 in females (Figure 4A; Supplementary Table 9). Of these, 4 genes were cryptically transcribed in both sexes (p = 8.2e-9): Ccdc124, Per1, Snap25, and Cplx1. Interestingly, Snap25 and Cplx1 have been implicated in memory function (Hou et al., 2004; Söderqvist et al., 2010; Gao et al., 2015; Ramos-Miguel et al., 2017; Ying et al., 2017; Irfan et al., 2019; Wang et al., 2019; Gopaul et al., 2020), and play a role in brain development (Bark et al., 1995; Johansson et al., 2008; Salimi et al., 2008). In both males and females, Snap25 also showed a significant decrease in gene expression due to binge ethanol treatment (Supplementary Table 1). Per1 has also been previously shown to influence ethanol consumption (Gamsby et al., 2013) and was shown in a GWAS study to predict problematic alcohol use (Baranger et al., 2016).
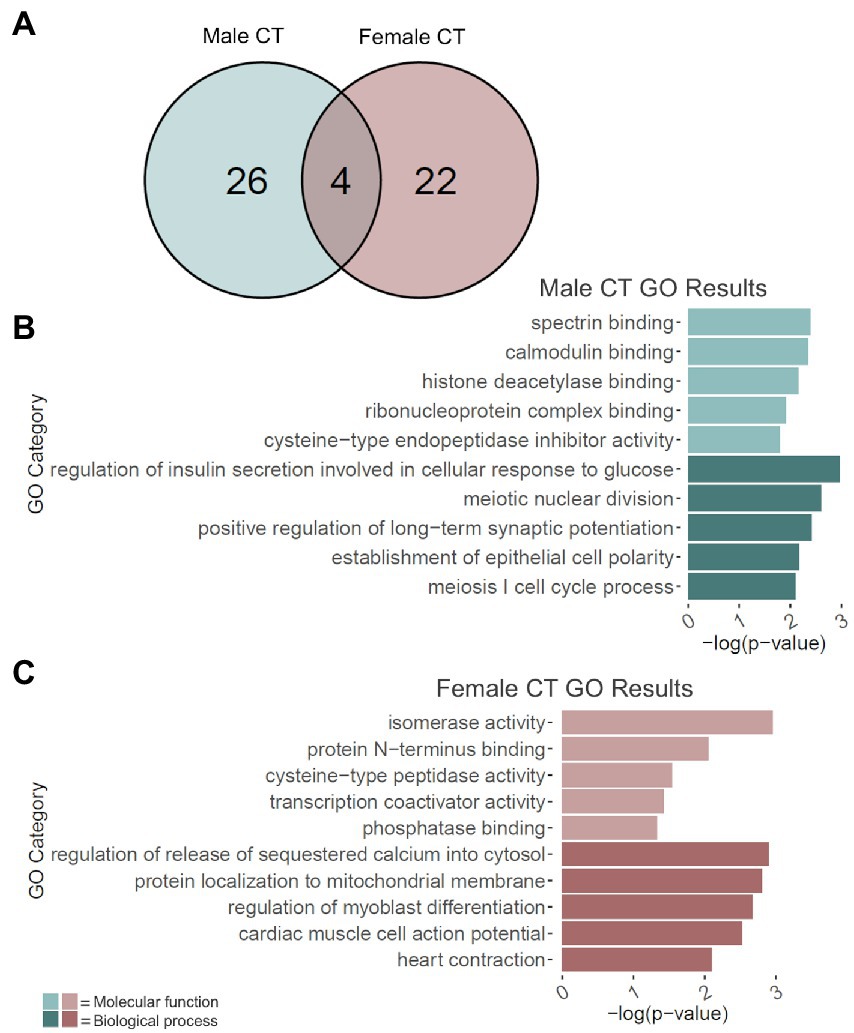
Figure 4. GO analysis of cryptically transcribed (CT) genes. (A) Number of genes cryptically transcribed due to adolescent binge ethanol in males and females at p < 0.05. Fisher’s exact test determined male and female gene overlap to be significant, p = 8.2e-09. (B) GO analysis of cryptically transcribed genes with unique to males. (C) GO analysis of cryptically transcribed genes unique to females. Snap25, Cplx1, Ccdc124, and Per1 were cryptically transcribed in both males and females and were excluded from the male and female GO analyses to identify categories unique to each sex.
The 26 genes that were uniquely cryptically transcribed in males, and the 22 genes that were uniquely cryptically transcribed in females were used as input for male-and female-specific gene ontology analysis. In males, gene categories that were over-represented due to binge ethanol treatment included calmodulin binding (molecular function), histone deacetylase binding (molecular function), and positive regulation of long-term potentiation (biological process; Figure 4B). Gene categories over-represented in females included transcription coactivator activity (molecular function) and regulation of release of sequestered calcium into cytosol (biological process; Figure 4C). Full gene ontology results for males and females can be found in Supplementary Table 10.
2.6. Adolescent binge ethanol induces differential H3K36 trimethylation of genes related to calcium channel activity and action potential in males
Changes in H3K36me3 due to adolescent binge ethanol were assessed with DiffBind. Using a cutoff of value of p < 0.05 to generate a gene list with a length sufficient for downstream analysis, 1,522 total genes were differentially bound in male PFC (Supplementary Table 11). 1,231 of these genes were unique to males (Figure 5A) and served as the input for male-specific gene ontology analysis. As with our RNA-seq dataset, the first 15 molecular function and biological process categories were selected to represent our ChIP-seq gene ontology results (Figure 5B). The full gene ontology results can be found in Supplementary Table 12. Molecular function categories of genes that were over-represented due to adolescent binge ethanol in males included calcium channel activity, long non-coding RNA binding, and nuclear receptor coactivator activity. Top biological process categories included action potential, regulation of neurotransmitter levels, and signal release from synapse. Although a Fisher’s Exact test showed the gene lists significantly overlapped, we were surprised to see only 27 genes were differentially expressed in our RNA-seq analysis and also showed altered H3K36me3 regulation (p = 5e-03, Table 1). Forty-nine genes that showed differential exon usage also showed altered H3K36me3 regulation (p = 7.5e-07, Table 1). Three genes that showed altered H3K36me3 regulation were cryptically transcribed in males (Triqk, Dnajc21, Dclk3, p = 0.16, Table 1). In males, Penk, Lss, and Slc6a6 were the only genes showed differential trimethylation of H3K36, differential gene expression, and differential exon use (Table 1). No genes were shared across all four analyses.
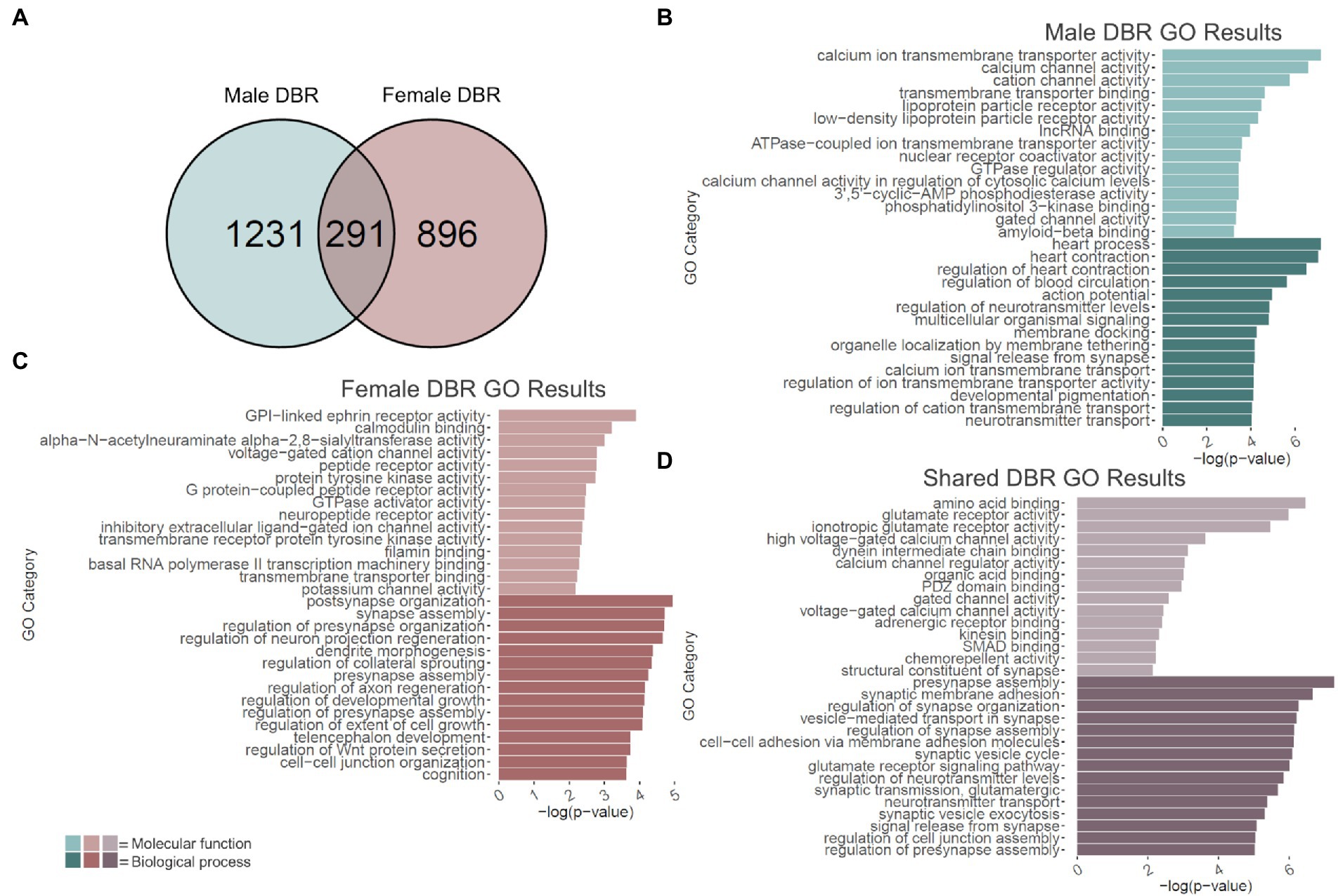
Figure 5. GO analysis of genes showing differentially bound H3K36me3 regions (DBR). (A) Number of genes showing differential binding of H3K36me3 due to adolescent binge ethanol in males and females at p < 0.05. Fisher’s exact test determined male and female gene overlap to be significant, p = 2.9e-131. (B) GO analysis of genes showing differential binding of H3K36me3 unique to males. (C) GO analysis of genes showing differential binding of H3K36me3 unique to females. (D) GO analysis of genes showing differential binding of H3K36me3 that are shared between males and females.
2.7. Adolescent binge ethanol induces differential H3K36 trimethylation at calmodulin binding, synapse assembly, and cognition related genes in females
In females, DiffBind analysis showed that adolescent binge ethanol caused 1,187 total genes to be differentially associated with H3K36me3 in female PFC (Supplementary Table 11). Eight hundred ninety-six unique genes showed altered H3K36me3 associations in females, and these were used as input for female-specific gene ontology analysis (Figure 5A). Molecular function categories that were over-represented due to binge ethanol treatment included calmodulin binding, GPI-linked ephrin receptor activity, and inhibitory extracellular ligand-gated ion channel activity (Figure 5C). Over-represented biological process categories included synapse assembly, dendrite morphogenesis, and cognition (Figure 5C). Notably, adolescent binge ethanol impacted 20 genes associated with calmodulin binding and 30 genes associated with cognition (Figure 5C and Supplementary Table 13). Calmodulin plays a critical role in long-term potentiation and memory function (Ataei et al., 2015), and thus, disruption to the regulation of these genes could contribute to ethanol’s impact on memory. Similar to our male comparison, in females only 17 genes that were differentially expressed in our RNA-seq analysis showed altered H3K36me3 (p = 0.2, Table 1). Seventy-five genes that showed differential exon usage showed altered H3K36me3 (p = 5.5e-14, Table 1), and only Cplx1 was differentially trimethylated at H3K36 and cryptically transcribed (p = 0.61, Table 1). In female ethanol-treated animals, four genes were found significant in our differential H3K36me3 analysis, differential gene expression analysis, and differential exon use analysis: Smarca2, Hsph1, Unc13a, and Ipo9 (Table 1). No genes were shared across all four analyses.
2.8. Adolescent binge ethanol induces dysregulation of H3K36me3 at synaptic-related genes in both males and females
Of the 1,522 genes that showed altered H3K36me3 in males and 1,187 genes that showed altered H3K36me3 in females, 291 of those genes were shared between both sexes (p = 2.9e-131, Figure 5A). Gene ontology analysis of these 291 genes indicated that they overwhelmingly played a role in synaptic transmission and membrane excitability (Figure 5D). Molecular function categories included glutamate receptor activity, voltage-gated calcium channel activity, and structural constituent of synapse. Biological process categories included presynapse assembly, synaptic membrane adhesion, glutamate receptor signaling pathway, and signal release from synapse. While not within the top 15 biological function categories, adolescent binge ethanol also induced changes to H3K36 trimethylation in males and females at 16 genes related to learning or memory (GO:0007611): Phb2, Adcy1, Ptgs2, Adcy8, Nrxn3, Nrxn1, Plcb1, Mme, Cacna1c, Amfr, Dcdc2, Gria1, Grm7, Brsk1, Ep300, Htt and eight genes related to calmodulin binding (GO:0005516): Adcy1, Adcy8, Plcb1, Cacna1a, Cacna1c, Ubr4, Grm7, Invs (Supplementary Table 14). Interestingly, both categories were also significantly over-represented in the separate male and female analyses, although each sex saw differential H3K36me3 antibody binding at different genes (Supplementary Tables 12, 13). This gene-level functional analysis shows that adolescent binge ethanol produces a striking response of H3K36me3 dysregulation at genes critical for synaptic function. Given the role of H3K36me3 in memory-related processes described above (Butler and Dent, 2012; Sessa et al., 2019), and the ethanol-induced changes in H3K36me3 regulation we see in genes relating to membrane excitability, synaptic activity, and memory, these results provide evidence that link adolescent binge ethanol to H3K36me3 changes, and may link H3K36me3 changes to adolescent binge ethanol-induced memory deficits.
3. Discussion
Epigenetic regulation of gene expression provides an attractive mechanism for driving long-term behavioral and cognitive changes following binge ethanol. Epigenetic modifications can positively or negatively impact gene transcription, and are dynamic marks that are affected by environmental stimuli, such as stress or alcohol and illicit drug use (Jaenisch and Bird, 2003; Liu et al., 2008; Karlić et al., 2010; Hitchcock and Lattal, 2014). By altering transcription, epigenetic alterations provide a mechanism for regulating downstream gene expression, which in turn affects behavioral outcomes (Lester et al., 2011). To our knowledge, the studies here are the first to assess concurrent changes to gene expression and epigenetic regulation in the PFC of binge ethanol exposed adolescents. We assessed differential gene expression and ethanol-induced changes to H3K36me3, an epigenetic mark associated with memory function, alternative splicing, and cryptic transcription. These analyses were all carried out within the same tissue samples, to get a within-animal comparison of ethanol effects at both the chromatin and mRNA regulation levels. Our work shows that adolescent binge ethanol altered the regulation and expression of genes important for synaptic function.
Males and females both showed ethanol-induced changes to H3K36me3 at genes related to synaptic transmission and membrane excitability. Dysregulation of these genes could impede cell signaling pathways or synaptic pruning processes required for ongoing PFC development throughout adolescence. Previous studies have shown accelerated gray matter loss in the frontal cortex after adolescent alcohol exposure, indicating that synaptic pruning may be disrupted by binge levels of ethanol (Luciana et al., 2013; Squeglia et al., 2014; Pfefferbaum et al., 2018; El Marroun et al., 2021). A prior report showed that during a cognitive task, adolescents who binge drank showed differential brain region utilization despite similar performance, which may reflect the need to recruit more neural activity for successful performance (Squeglia et al., 2011). Males and females showed different responses, with male binge drinkers showing greater BOLD responses than controls, and female binge drinkers showing decreased BOLD responses compared to controls within the bilateral frontal, anterior cingulate, temporal, and cerebellar cortices (Squeglia et al., 2011). Altered regulation of genes involved in synaptic transmission and membrane excitability could potentially explain these ethanol-induced neuroimaging and behavioral changes. Notably, females showed much smaller fold changes in the genes where H3K36me3 was altered by ethanol compared to males, supporting our previous ELISA data where only ethanol-treated males showed global H3K36 decreases in the PFC (Wolstenholme et al., 2017).
Additionally, both sexes also showed decreased gene expression and cryptic transcription of Snap25 with binge ethanol exposure. Snap25 is part of the SNARE complex, and aids in calcium-induced release of neurotransmitters into the synaptic cleft (Najera et al., 2019). It has been associated with psychiatric outcomes such as ADHD and schizophrenia, both characterized in part by deficits in working memory (Najera et al., 2019; Kofler et al., 2020). In a post-mortem study, AUD patients showed lower protein levels of the Snap25b isoform. Snap25 switches in expression from the Snap-25a isoform in early life to the Snap-25b isoform in adulthood (Bark et al., 1995), and this switch is influenced by sex (Irfan et al., 2019). Compared to males, females showed increased protein levels of the SNAP-25a isoform remaining at 4 weeks of age in the hippocampus (Irfan et al., 2019). Snap25 deficiency studies have shown that the transition to Snap-25b is crucial for proper neurodevelopment – elimination of Snap-25b expression leads to impaired short-term plasticity (Johansson et al., 2008), and reduced magnitude of LTP (Irfan et al., 2019; Gopaul et al., 2020). The ethanol-induced altered expression of Snap25 could directly induce the long-term working memory deficits seen after adolescent binge ethanol exposure. Not only could a change in the shift of Snap25 isoforms induce changes in brain development and structure, but a polymorphism in Snap25 has also been directly implicated in reduced gray matter of the cingulate cortex (Söderqvist et al., 2010) and reduced and working memory function in children and adolescents (Söderqvist et al., 2010; Gao et al., 2015).
Cplx1 also showed evidence of cryptic transcription in males and females after exposure to adolescent binge ethanol. Complexins are SNARE-binding proteins and modulate calcium-induced vesicle release (McMahon et al., 1995; Reim et al., 2001; Chen et al., 2002). Cplx1 is crucial for neurodevelopment, and similar to Snap25, also displays an age-dependent shift in expression. A study in the dorsolateral PFC showed that Cplx1 increased in expression from 0 to 25 years – associated with an increase in synaptic maturation (Salimi et al., 2008). Interestingly, gene expression of complexins were found to be decreased in the PFC after prenatal (Barr et al., 2005) and perinatal (Zink et al., 2009) ethanol exposure. Given its role in both neurotransmitter release and PFC development, ethanol-induced changes to Cplx1 could also contribute to ethanol’s impacts on memory.
H3K36me3 plays a role in memory consolidation within the PFC (Gräff et al., 2012). H3K36me3 is subjected to modulation by experience, as its level increased in the PFC after testing mice for recent (24 h) and remote (7 day) memory with a NOR task (Gräff et al., 2012). When histone modifications were pharmacologically blocked, memory consolidation was impaired, and this impairment was associated with a decrease in H3K36me3 levels (Gräff et al., 2012). Additionally, increases in global H3K36me3 levels were seen in transgenic animals with increased memory function, and when training time prior to NOR was increased, suggesting the level of H3K36me3 may determine how well a memory is consolidated (Gräff et al., 2012). Furthermore, a separate study showed that when H3K36me3 levels were reduced in a mouse model, deficits in NOR and decreased expression of memory-related genes occurred (Sessa et al., 2019). Ultimately, these data presented here suggest that adolescent binge ethanol perturbs H3K36me3’s regulation of genes important for synaptic function and memory processes and/or induces cryptic transcription of memory-related genes Snap25 and Cplx1. These molecular changes could potentially be the mechanism by which ethanol induces persistent memory deficits.
Notably, a number of synaptic and memory-related gene ontology categories were repeated in the male, female, and the shared differential H3K36me3 analyses – although not all categories were within the top 15 for each analysis, calmodulin binding (GO:0005516), dendrite morphogenesis (GO:0048813), glutamatergic synaptic transmission (GO:0035249), negative regulation of synaptic transmission (GO:0050805), axon guidance (GO:0007411), learning or memory (GO:0007611) and cognition (GO:0050890) were seen significantly over-represented in each analysis (Supplementary Tables 12–14). This pattern was also seen in our RNA-seq differential genes expression analyses. For example, males and females showed different genes that were differentially expressed, but were all contained within the extracellular matrix structural constituent GO category (GO:0005201, Supplementary Tables 2–3). However, four of the same genes (Kdr, Eng, Adamts6, and P4ha1) in the extracellular matrix category (GO:0030198) were differentially expressed in both males and females (Supplementary Table 4). This may suggest that ethanol impacts the same pathways in males and females, but may do so through different mechanisms, or could suggest that ethanol influences a number of genes related to a pathway in both sexes, but these genes influence the expression of other downstream genes in a sex-specific manner. One study suggests that transcription factors have sex-biased regulatory targets (Lopes-Ramos et al., 2020) – because ethanol can alter transcription factor activity (Miles, 1995), it is possible that ethanol-induced changes to transcription factors may influence different genes in males and females. Another study saw sexual dimorphism in glutamatergic signaling-related genes within the nucleus accumbens after binge ethanol exposure, indicating the transcriptional response to ethanol is influenced by sex (Finn et al., 2018). Alternatively, our results may reflect that the same altered processes in males and females are altered, but due to stochastic variation in gene expression and possible low sample numbers, different genes, but the same biological pathways or molecular functions are identified between the sexes. While many RNA-sequencing studies have supported our results in finding sex-specific effects and/or little overlap between male and female gene lists (Labonté et al., 2017; Finn et al., 2018; Hitzemann et al., 2020), many other studies only include male subjects (Mulligan et al., 2017; Erickson et al., 2019; Brenner et al., 2020; Farris et al., 2020; Krishnan et al., 2022), and more studies including females will be required to parse out ethanol’s sex-dependent influence on gene expression.
Both males and females showed ethanol-induced changes to genes involved in the extracellular matrix. The extracellular matrix regulates cellular migration and axonal growth, and stabilizes myelinated tracts (Zimmermann and Dours-Zimmermann, 2008) – thus underlying its role in adolescent PFC development characterized by synaptic pruning and increased myelination (Spear, 2000). During early and adolescent brain development, the extracellular matrix supports neurogenesis, cell migration, axonal growth, and synaptogenesis, but in adulthood inhibits these reorganization processes (Gundelfinger et al., 2010). This restriction in adult plasticity is induced by the development of perineural nets, and corresponds with the end of the critical period where neuronal circuits are shaped by experience (Fawcett, 2009). Given its role in brain development and plasticity, alterations in the structure and function of the extracellular matrix could lead to lasting changes in the brain and have been suggested to underlie mental health, neuropsychiatric, or neurodegenerative disease (Lubbers et al., 2014). Previous studies have shown ethanol’s negative impact on the extracellular matrix in both adolescents (Coleman et al., 2014; Nato et al., 2014) and adults (Hitzemann et al., 2020), with extracellular matrix-related genes being identified in a number of AUD or substance use disorder GWAS studies (Adkins et al., 2017; Peng et al., 2021). Our study helps to support ethanol’s impact to the extracellular matrix.
Males and females both showed differential expression of a number of genes relating to the unfolded protein response (Figure 2D). Previous studies have shown that decreased levels of H3K36me3 allow for the production of cryptic transcripts (Carrozza et al., 2005; Butler and Dent, 2012; Carvalho et al., 2013), which associate with ribosomes (Wei et al., 2019), and likely become misfolded proteins due to their truncated nature. Therefore, this ethanol-induced response may be related to increased levels of cryptic transcription occurring.
We chose to do a differential exon use analysis using DEX-seq, due to H3K36me3’s proposed role in alternative splicing (Zhou et al., 2014; Xu et al., 2021). Here, we saw that genes showing evidence of differential exon use were involved in gene ontology categories related to tubulin binding and dendrite development in both males and females. When comparing the gene lists across analyses, we saw the most overlap in genes that showed differential exon use due to binge ethanol and genes where H3K36me3 was impacted by ethanol. In males, 49 genes showed ethanol-induced changes to H3K36me3 and differential exon use (p = 7.5e-07), and in females, 75 genes showed this overlap (p = 5.5e-14). This overlap could potentially be due to H3K36me3’s proposed role in alternative splicing (Zhou et al., 2014; Xu et al., 2021). We do, however, caution that given our sequencing protocol (poly-A selected, 150 bp reads), there may be some level of bias in alignments given our relatively short 150 bp read length. The impact of adolescent binge drinking on alternative splicing, and whether this is mediated through a H3K36me3 mechanism will need to be more thoroughly investigated in the future, perhaps with specific genes that showed both ethanol-induced differential exon use and altered H3K36me3 regulation or with long read RNA sequencing.
Given the critical role H3K36me3 plays in regulating cryptic transcription, we initially hypothesized that genes showing evidence of cryptic transcription would largely overlap with the same genes where we saw ethanol-induced differential binding of H3K36me3. However, there was little overlap of gene loci where the H3K36me3 antibody was differentially bound and genes that were cryptically transcribed – in males, only 3 genes that showed ethanol-induced changes to H3K36me3 were cryptically transcribed (p = 0.16), and in females, only one gene was found in both analyses (p = 0.61, Table 1). Similarly, there were less than 30 genes that were both differentially expressed and differentially bound by H3K36me3 in males (p = 5e-03) or females (p = 0.2, Table 1). H3K36me3 is deposited co-transcriptionally, and unlike most other histone marks, it does not strictly activate or repress gene transcription due to its presence at the gene promotor (DiFiore et al., 2020). The influence of H3K36me3 on gene expression levels has yet to be fully elucidated – gene-body marking by H3K36me3 does not always correlate with gene expression levels (Pu et al., 2015), and in one study, most DEGs did not exhibit obvious changes in H3K36me3 levels (Zhang et al., 2021). Some have suggested that decreases in H3K36me3 could leave the chromatin primed for greater flexibility in chromatin accessibility (Simon et al., 2014; Pu et al., 2015; Zhang et al., 2021). In two studies, high gene-body levels of H3K36me3 have been associated with transcriptional stability and few changes in gene expression, whereas low gene-body levels of H3K36me3 were associated with more gene expression changes (Pu et al., 2015; Zhang et al., 2021). It is possible that ethanol did not disrupt H3K36me3 gene-body levels to the point of eliciting strong impacts to gene expression levels, and thus we did not see a large overlap of genes where levels of H3K36me3 were altered and genes that were differentially expressed due to binge ethanol. However, suggested by the studies above, decreases in H3K36me3 gene-body levels could still lead to altered chromatin regulation.
A study using a similar approach to generate ChIP-seq and RNA-seq data from within the same tissue also suggested that effects at the chromatin regulation level do not always accurately predict levels of gene expression (Reshetnikov et al., 2021). This could potentially be explained by the differences in H3K36me3 half-life versus the half-life of mRNA. Zheng et al. (2014) estimated the half-life of H3K36me3 in HeLa cells at about 57 h. The half-life of mRNA varies based on a variety of factors, but is estimated to last up until 10 h in mammalian cells (Yang et al., 2003; Chen et al., 2008; Sharova et al., 2009; Tani et al., 2012). This could suggest H3K36me3-regulated genes might not accurately represent the full mRNA profile, i.e., H3K36me3 may remain on the chromatin long after the associated mRNA has been degraded. Thus, H3K36me3 may provide a “molecular memory” of previous transcriptional events, similar to those seen with H3K4 methylation in Ng et al. (2003). Another study that assessed chromatin and gene expression changes within the same PFC tissue saw that H3K4me3 showed low correlation with gene expression, but that correlation improved when looking at specific cell types (Reshetnikov et al., 2020, 2021). Thus, an alternate explanation for the small overlap between genes where H3K36me3 levels were altered and differentially expressed genes could be that the data was obscured by looking at all cells comprising the PFC instead of cell-specific populations.
4. Materials and methods
4.1. Animals
Male and female DBA/2J mice (n = 66) from Jackson Laboratory arrived in the Virginia Commonwealth University vivarium at postnatal day 19–21 (Bar Harbor, ME, United States). DBA/2J mice were used as they show a more robust acute response to binge ethanol than the more commonly used C57BL/6J strain – DBA/2J mice show increased sensitivity to ethanol in a loss of righting reflex test (Linsenbardt et al., 2009), as well as increased locomotor activity compared to C57/B6 mice (Phillips et al., 1994; Kerns et al., 2005). Importantly, this strain also shows a more prominent decrease in myelin gene expression (Kerns et al., 2005; Goudriaan et al., 2020) – important to our model as myelin decreases have been widely shown in human studies of adolescent binge drinking (De Bellis et al., 2005; Medina et al., 2008; Pfefferbaum et al., 2018; El Marroun et al., 2021). Mice were housed 4/cage in same sex cages in an AALAC-accredited facility under 12-h light/dark cycles with food and water available ad libitum for the entire experiment. After a week acclimation to the animal facility, mice were habituated to the gavage procedure with 0.1% saccharin on PND 27 and 28 and then divided into two treatment groups: ethanol treated and control. Mice were dosed with 4 g/kg ethanol (25% w/v in water by gavage) or water intermittently (2 days on/2 days off) on PND 29, 30, 33, 34, 37, 38, 41, and 42. PFC tissue was collected 24 h after the last dose of ethanol (3–6 h into the animals’ light cycle). A triangle-shaped wedge of tissue above the corpus callosum was collected anterior to Bregma 0.5 mm to represent the PFC, consistent with typical PFC dissection regions for the Wolstenholme lab (Wolstenholme et al., 2011, 2017). This wedge contains prelimbic, infralimbic and medial PFC as well as anterior cingulate. Tissue was collected 24 h after the last dose based on a previous study in our lab, where we saw significant decreases in global H3K36 and H3K36-related gene expression at this timepoint (Wolstenholme et al., 2017). Collecting tissue at this timepoint allowed us to investigate the immediate impacts of adolescent binge ethanol use to see whether molecular changes during a critical developmental period could inform the long-term cognitive deficits seen in adulthood. At this dose, blood ethanol concentrations reached 313 mg/dL 1 h after gavage (Wolstenholme et al., 2017). All animal housing and care was conducted with the approval of the Virginia Commonwealth University IACUC Committee and in accordance with the NIH Guide for the Care and Use of Laboratory Animals (National Research Council, 2011).
For our ChIP-seq and RNA-seq studies, 66 total animals were used. To ensure enough DNA input for high quality sequencing data, we combined three PFCs from each treatment group and sex into a single sample. Each combined tissue sample was homogenized and divided in half between ChIP-seq and RNA-seq sample preps so that we were able to link chromatin-level changes with gene expression-level changes from within the same tissue. Five biological replicates (of three pooled PFCs) were obtained for water-treated male and female groups, and six biological replicates (of three pooled PFCs) were obtained for ethanol-treated males and females, resulting in n = 22 pooled samples for each of our sequencing studies (Figure 1).
Sex-specific responses to ethanol have been well-noted (Squeglia et al., 2011; Labonté et al., 2017; Wolstenholme et al., 2017; Finn et al., 2018; Tavares et al., 2019; Flores-Bonilla and Richardson, 2020; Hitzemann et al., 2020; Radke et al., 2021; Bent et al., 2022), and because of this, we had an a priori hypothesis that ethanol induces different molecular changes in males and females. Thus, our male and female analyses were run separately. These sex-specific responses are exemplified with previous ELISA data from our lab showing that global H3K36 methylation levels were significantly decreased after adolescent binge ethanol treatment in males only, and that males had more robust decreases in myelin-related gene expression (Wolstenholme et al., 2017). These sex-specific responses have been seen specifically in other RNA-seq studies, where males and females show little overlap in differentially expressed genes (Labonté et al., 2017; Finn et al., 2018). Another recent ethanol-associated RNA-seq study also chose to analyze the sexes separately, given the sex-specific effects in alcohol use (Hitzemann et al., 2020).
4.2. RNA isolation
Homogenized and divided tissue was immediately pelleted and flash frozen on liquid nitrogen. Total RNA was isolated from PFC tissue using STAT60 Reagent (Tel-Test, Friendswood, TX) and miRNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNA was quantified by measuring absorbance at 260 nm using a NanoDrop, and RNA integrity was assessed by electrophoresis using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). All samples obtained an RNA Integrity Number (RIN) between 8.4 and 9.3.
4.3. RNA-seq library preparation and sequencing
RNA-seq data have been deposited with the Gene Expression Omnibus resource (Accession #GSE220746). Isolated RNA of pooled samples was sequenced by Novogene. To avoid non-biological experimental variation that could arise from sample batches, similar group representation was ensured prior to each processing stage (RNA isolation, library preparation, and lane assignment). Preparation of polyA-enriched cDNA libraries was conducted following standard protocols using NEBNext Ultra™ II Directional RNA Library Prep Kit (Illumina). Libraries were sequenced on an Illumina NovaSeq 6000 (2 × 150 bp paired-end reads). A summary of RNA-seq metrics can be found in Supplementary Table 15.
4.4. RNA-seq alignment and differential gene expression (DEG) analysis
Quality of FASTQ formatted samples files were assessed with FastQC (Andrews, 2010), and aligned using STAR v2.7.8a (Dobin et al., 2013) with GRCm38/mm10 reference genome. Annotations were obtained from ENSEMBL. Only reads that mapped to a single location were used in subsequent analysis. Raw read counts were compiled using featureCounts within the Bioconductor package Subread v2.0.2 (Liao et al., 2014). A pre-filtering step was applied to eliminate very low expressing genes prior to differential gene expression analysis, where at least two samples were required to have >10 counts. Normalization and differential expression analysis were carried out with DESeq2 v1.32.0 (Love et al., 2014). DESeq2 internally normalizes read counts using a median of ratios method (Love et al., 2014). Genes with a value of p < 0.01 were considered significantly altered and used in downstream bioinformatic analysis.
4.5. Differential exon usage
GFF annotation file containing collapsed exon counting bins was prepared from the ENSEMBL GRCm38/mm10 GTF file using the DEXSeq v1.38.0 (Anders et al., 2012) Python script dexseq_prepare_annotation.py with gene aggregation disabled. The number of reads overlapping each exon bin was then counted using the DEXSeq Python script dexseq_count.py, the GFF file, and each sample’s SAM file. Differential exon usage (DEU) analysis was then carried to compare ethanol-treated animals to water-treated animals using the DEXSeq R package standard analysis workflow. Genes with transcripts possessing at least one differentially utilized exon bin with a value of p < 0.001 were considered to be significantly altered and were used in downstream bioinformatic analysis.
4.6. Cryptic transcription analysis
To identify genes that displayed evidence of cryptic transcription in males and females, we followed the protocol outlined in Carvalho et al. (2013). Exon read counts generated with DEXSeq (Anders et al., 2012) were used to compare ethanol-treated and control animals for each sex. Briefly, we first identified all transcripts with significantly higher read counts for at least one exon in the EtOH treated group. Second, transcripts with significantly higher read counts for the first annotated and expressed exon (likely corresponding to upregulated full-length mRNAs) were discarded. Third, the new cryptic TSS was defined as the first significantly higher exon, and all transcripts that showed significantly higher read counts for <60% of the downstream exons were filtered out. In addition to Carvalho’s parameters, we required the first differentially used exon to be among the first half of all exons. The resulting gene lists of cryptically transcribed genes were used for downstream bioinformatics analyses.
4.7. Chromatin immunoprecipitation
Prior to ChIP, the H3K36me3 antibody (Active Motif #61101, Carlsbad, CA) was validated using qPCR to verify H3K36me3 presence or absence at specific genomic locations against IgG (Supplementary Figure 1A). The same antibody lot number was used for all ChIP reactions. H3K36me3-ChIP was performed on PFC tissue using the MAGNA ChIP Hi-Sens kit (Millipore, Burlington, MA), following the manufacturer’s instructions. Briefly, tissue from 3 combined PFCs was dounce homogenized on ice, divided in half, and pelleted. One half of each sample was flash frozen and reserved for RNA-seq. The other half of each sample was subjected to fixation, where DNA was cross-linked to histones by incubating cells in a 1% formaldehyde solution at 37°C for 10 min. The formaldehyde reaction was quenched using glycine, and tissue was washed 3x with PBS containing protease inhibitors (Millipore, Burlington, MA). Cell lysis buffer was added to isolate nuclei. Prepared chromatin was sonicated using a Bioruptor Pico (Diagenode, Denville, NJ) for 12 cycles (30 s on/30 s off). Fragment size was verified on 2% agarose gel to be between 200 and 1,000 bp. After sonication, samples were stored at −20°C to await immunoprecipitation, and a portion of sonicated chromatin from each sample was reserved as an input reference. For immunoprecipitation, sonicated chromatin was incubated overnight with 2 μg of H3K36me3 antibody (Active Motif, Carlsbad, CA) bound to Protein A/G magnetic beads with end-over-end rotation. After incubation, DNA was eluted from beads in elution buffer with proteinase K. DNA was extracted using phenol:chloroform:isoamyl alcohol (Thermo Fisher, Waltham, MA) and purified with ethanol precipitation. Pellets were resuspended in 20 μL TE buffer and stored at −20°C. To obtain sufficient DNA for sequencing, 8 technical replicates from each sonicated sample were subjected to immunoprecipitation. DNA from all 8 technical replicates were combined and concentrated using a speed-vacuum prior to library preparation. Independent control samples were run alongside each set of ChIP reactions to ensure successful chromatin immunoprecipitation.
4.8. ChIP-seq library preparation and sequencing
ChIP-seq data have been deposited with the Gene Expression Omnibus resource (Accession #GSE220746). To avoid non-biological experimental variation that could arise from sample batches, similar group representation was ensured prior to each processing stage (sonication, ChIP, library preparation, and lane assignment). Input and H3K36me3-enriched samples were sequenced by the VCU Genomics Core Facility for library preparation and sequencing. Preparation of libraries were conducted following standard protocols using the Accel-NGS 2S PLUS DNA Library kit (Swift). Samples were sequenced on a NextSeq 2000 (Illumina) using 150 base pair single-end reads at greater than 50 million reads. A summary of ChIP-seq metrics can be found in Supplementary Table 15.
4.9. ChIP-seq alignment and differential binding analysis
Quality of FASTQ formatted samples files were assessed with FastQC. Adapter sequences and reads with quality scores <20 were removed using CutAdapt (Martin, 2011) and aligned using Bowtie2v2.4.1 (Langmead and Salzberg, 2012) with GRCm38/mm10 reference genome. To prevent inaccurate representation of repetitive genomic regions, reads that aligned to blacklisted regions were discarded, as defined by ENCODE to be a set of regions in the genome that have anomalous, unstructured, or high signal in next-generation sequencing experiments independent of cell line or experiment (Amemiya et al., 2019). Only reads that mapped to a single location were used in subsequent analysis. Peaks were identified using SICERv1.1 (W200, G600) (Xu et al., 2014). As with our RNA-seq analysis, male and female analyses were run separately. Normalization and differential peak binding analysis were carried out using the Bioconductor package DiffBind v3.4.11, and annotations were obtained using the Bioconductor package ChIPSeekerv1.28.3 (Yu et al., 2015). Genes with a value of p < 0.05 were considered significantly altered and used in downstream bioinformatic analysis to expand our gene list for hypothesis-generating analyses. Representative samples of H3K36me3 read coverage are shown in Supplementary Figure 1B.
4.10. Bioinformatics analysis
Downstream bioinformatics analysis was carried out similarly for gene lists derived from differential expression, differential exon use, cryptic transcription, and differential binding analyses. Gene ontology over-representation analysis was determined using ToppFun (Chen et al., 2009). Gene sets were filtered based on the number of genes within each category (min = 4, max = 500) and value of p of <0.05. Lists were further filtered by gene list hits. For analyses with ≥100 genes, categories containing ≥3 hits were included and the top 15 molecular function and biological process categories are represented in our figures. For analyses with ≤100 genes, categories containing ≥2 hits were included, and the top 5 molecular function and biological process categories are represented. Categories that had identical query gene lists and similar category names were removed to reduce repetitiveness. Full gene ontology tables can be found in Supplementary Tables 2–4, 6–8, 10, 12–14. Within each analysis, male and female gene lists were compared using a Fisher’s exact test to determine whether there was significant gene overlap of the two lists in comparison to the total number of genes in the mm10 genome. This same test was used to determine if there was significant overlap between each analysis within each sex. The Bioconductor package GeneOverlap v1.32.0 was used for these overlap calculations.
5. Conclusion
Overall, this study unveiled potential genes that may be contributing to adolescent binge ethanol-related memory deficits. As H3K36me3 plays an important role in memory, and was previously found to be decreased in the PFC after adolescent binge ethanol (Wolstenholme et al., 2017), we assessed ethanol-induced changes to this histone mark in the developing adolescent PFC. We further assessed ethanol-induced changes at the transcription level with differential gene expression and differential exon-use analyses, as well as cryptic transcription analysis – a marker of decreased H3K36me3. Importantly, all analyses were completed within the same tissue to view changes at both the epigenetic and gene expression levels.
Through this set of experiments, we found changes to synaptic-related genes in all our analyses. Notably, H3K36me3 was dysregulated at synaptic-related genes. Dysregulation of synaptic-related gene expression could impede cell signaling pathways required for ongoing PFC development throughout adolescence and provide an avenue for cognitive deficits to occur in adulthood (Figure 6). We also found that Snap25 and Cplx1 showed evidence of cryptic transcription and that the former showed decreased gene expression in both males and females. These genes play a strong role in memory function and brain development, and similarly, their specific dysregulation could underlie adolescent ethanol-induced cognitive phenotypes (Johansson et al., 2008; Salimi et al., 2008; Söderqvist et al., 2010; Gao et al., 2015; Irfan et al., 2019; Gopaul et al., 2020). Further experiments will be required to assess the exact role of these genes in ethanol-induced memory deficits, and whether they are implicated in other ethanol-related behaviors.
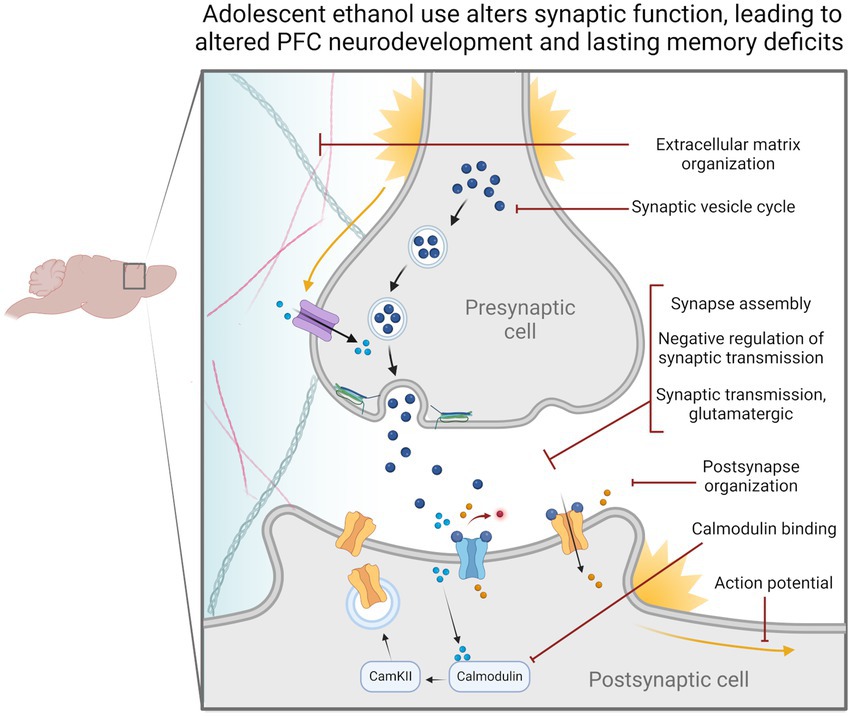
Figure 6. Adolescent binge ethanol alters the regulation and expression of genes relating to synaptic function. Our differential gene expression, differential exon usage, cryptic transcription and differential H3K36me3 bound loci analyses identified a number of gene ontology categories related to synaptic structure and function. These changes are likely reflected in presynaptic and postsynaptic cells as well as within the extracellular matrix and may be why persistent cognitive deficits occur after adolescent binge ethanol exposure. Categories shown were significantly over-represented in at least one analysis in both males and females, p < 0.05. Extracellular matrix organization – GO:0030198, Synaptic vesicle cycle – GO:0099504, Synapse assembly – GO:0007416, Negative regulation of synaptic transmission – GO:0050805, Synaptic transmission, glutamatergic – GO:0035249, Postsynapse organization – GO:0099173, Calmodulin binding – GO:0005516, Action potential – GO:0001508.
Data availability statement
The datasets presented in this study have been deposited with the Gene Expression Omnibus resource (Accession #GSE220746). Full gene lists can be found in Supplementary material.
Ethics statement
The animal study was reviewed and approved by the Virginia Commonwealth University IACUC Committee and in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Author contributions
EB and JW conceived and designed the study. EB executed, analyzed, and wrote the study with revision and editing assistance from JW. JW provided resources, experimental interpretation, and critical review of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Institutes of Health, National Institute on Alcohol Abuse and Alcoholism, USA 1F31AA029259-01A1, R01AA026347, and P50AA022537.
Acknowledgments
The ChIP-seq data included in this study was generated at the Genomics Core facility at Virginia Commonwealth University. We thank Mikhail G. Dozmorov for his help with our cryptic transcription analysis, and Rachel Stevenson for her help in developing our ChIP protocol. Figures 1, 6 were made with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1082104/full#supplementary-material
SUPPLEMENTARY TABLE 1 | Differential gene expression results for male and female PFC.
SUPPLEMENTARY TABLE 2 | Gene ontology analysis for the 342 genes differentially expressed in the PFC after adolescent binge ethanol in males only, p < 0.05.
SUPPLEMENTARY TABLE 3 | Gene ontology analysis for the 376 genes differentially expressed in the PFC after adolescent binge ethanol in females only, p < 0.05.
SUPPLEMENTARY TABLE 4 | Gene ontology analysis for the 45 genes differentially expressed in the PFC after adolescent binge ethanol in both males and females, p < 0.05.
SUPPLEMENTARY TABLE 5 | Differential exon use results in male and female PFC.
SUPPLEMENTARY TABLE 6 | Gene ontology analysis for the 461 genes showing differential exon usage in the PFC after adolescent binge ethanol in males only, p < 0.05.
SUPPLEMENTARY TABLE 7 | Gene ontology analysis for the 759 genes showing differential exon usage in the PFC after adolescent binge ethanol in females only, p < 0.05.
SUPPLEMENTARY TABLE 8 | Gene ontology analysis for the 41 genes showing differential exon usage in the PFC after adolescent binge ethanol in both males and females, p < 0.05.
SUPPLEMENTARY TABLE 9 | Cryptically transcribed genes in male and female PFC after adolescent binge ethanol, p < 0.05.
SUPPLEMENTARY TABLE 10 | Gene ontology analysis for the 26 cryptically transcribed genes in the PFC after adolescent binge ethanol in males and the 23 cryptically transcribed genes in the PFC after adolescent binge ethanol in females, p < 0.05.
SUPPLEMENTARY TABLE 11 | Differential trimethylation of H3K36 results in male and female PFC. In males, 1522 total genes showed H3K36me3 changes after adolescent binge ethanol treatment (p < 0.05); 182 of these genes showed significant changes in H3K36me3 at multiple sites. In females, 1187 total genes showed H3K36me3 changes (p < 0.05), 113 of these genes showed significant changes in H3K36me3 at multiple sites.
SUPPLEMENTARY TABLE 12 | Gene ontology analysis for the 1231 unique genes that showed differential trimethylation of H3K36 in the PFC after adolescent binge ethanol in males only, p < 0.05.
SUPPLEMENTARY TABLE 13 | Gene ontology analysis for the 896 unique genes that showed differential trimethylation of H3K36 in the PFC after adolescent binge ethanol in females only, p < 0.05.
SUPPLEMENTARY TABLE 14 | Gene ontology analysis for the 291 genes that showed differential trimethylation of H3K36 in the PFC after adolescent binge ethanol in both males and females, p < 0.05.
SUPPLEMENTARY TABLE 15 | Summary of sample and alignment information for RNA-seq and ChIP-seq data.
SUPPLEMENTARY FIGURE 1 | H3K36me3 ChIP antibody validation and sequencing coverage. A. H3K36me3 antibody (Active Motif) was validated using positive (Aldoa) and negative (Chr12dsrt) primers to visualize regions of expected high and low binding, respectively, and was compared to IgG (Millipore) to identify enrichment over non-specific binding. Graph shows representative sample. Aldo1a F: AGGTGGATGCACCATCACATT, Aldo1a R: TGCATTAGAGATGCGGAGCA, Chr12dst F: CACTGACACCTGACCTCGAT, Chr12dsrt R: ATGCCACAGCCCAAGTATGG. B. Representative ChIP read coverage of three genes that showed ethanol-induced changes to H3K36me3: Cacna1b, Lrrk2, and Ryr2.
References
Acheson, S. K., Bearison, C., Risher, M. L., Abdelwahab, S. H., Wilson, W. A., and Swartzwelder, H. S. (2013). Effects of acute or chronic ethanol exposure during adolescence on behavioral inhibition and efficiency in a modified water maze task. PLoS One 8, –e77768. doi: 10.1371/journal.pone.0077768
Acheson, S. K., Stein, R. M., and Swartzwelder, H. S. (1998). Impairment of semantic and figural memory by acute ethanol: age-dependent effects. Alcohol. Clin. Exp. Res. 22, 1437–1442. doi: 10.1111/j.1530-0277.1998.tb03932.x
Adkins, A. E., Hack, L. M., Bigdeli, T. B., Williamson, V. S., McMichael, G. O., Mamdani, M., et al. (2017). Genomewide association study of alcohol dependence identifies risk loci altering ethanol-response behaviors in model organisms. Alcohol. Clin. Exp. Res. 41, 911–928. doi: 10.1111/acer.13362
Amemiya, H. M., Kundaje, A., and Boyle, A. P. (2019). The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9:9354. doi: 10.1038/s41598-019-45839-z
Anders, S., Reyes, A., and Huber, W. (2012). Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017. doi: 10.1101/gr.133744.111
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Ataei, N., Sabzghabaee, A. M., and Movahedian, A. (2015). Calcium/calmodulin-dependent protein kinase II is a ubiquitous molecule in human long-term memory synaptic plasticity: a systematic review. Int. J. Prev. Med. 6:88. doi: 10.4103/2008-7802.164831
Baranger, D. A. A., Ifrah, C., Prather, A. A., Carey, C. E., Corral-Frías, N. S., Drabant Conley, E., et al. (2016). PER1 rs3027172 genotype interacts with early life stress to predict problematic alcohol use, but not reward-related ventral striatum activity. Front. Psychol. 7:464. doi: 10.3389/fpsyg.2016.00464
Barbier, E., Johnstone, A. L., Khomtchouk, B. B., Tapocik, J. D., Pitcairn, C., Rehman, F., et al. (2017). Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol. Psychiatry 22, 1746–1758. doi: 10.1038/mp.2016.131
Bark, I. C., Hahn, K. M., Ryabinin, A. E., and Wilson, M. C. (1995). Differential expression of SNAP-25 protein isoforms during divergent vesicle fusion events of neural development. Proc. Natl. Acad. Sci. U. S. A. 92, 1510–1514. doi: 10.1073/pnas.92.5.1510
Barnea-Goraly, N., Menon, V., Eckert, M., Tamm, L., Bammer, R., Karchemskiy, A., et al. (2005). White matter development during childhood and adolescence: a cross-sectional diffusion tensor imaging study. Cereb. Cortex 15, 1848–1854. doi: 10.1093/cercor/bhi062
Barr, A. M., Hofmann, C. E., Phillips, A. G., Weinberg, J., and Honer, W. G. (2005). Prenatal ethanol exposure in rats decreases levels of complexin proteins in the frontal cortex. Alcohol. Clin. Exp. Res. 29, 1915–1920. doi: 10.1097/01.alc.0000187806.68957.0a
Bava, S., Jacobus, J., Thayer, R. E., and Tapert, S. F. (2013). Longitudinal changes in white matter integrity among adolescent substance users. Alcohol. Clin. Exp. Res. 37, E181–E189. doi: 10.1111/j.1530-0277.2012.01920.x
Bekinschtein, P., and Weisstaub, N. (2014). Role of PFC during retrieval of recognition memory in rodents. J. Physiol. 108, 252–255. doi: 10.1016/j.jphysparis.2014.03.001
Bent, M. A. M., Pais, A. C., and Wolstenholme, J. T. (2022). Comparing behavior following binge ethanol in adolescent and adult DBA/2 J mice. Behav. Brain Res. 419:113703. doi: 10.1016/j.bbr.2021.113703
Berkel, T. D., and Pandey, S. C. (2017). Emerging role of epigenetic mechanisms in alcohol addiction. Alcohol. Clin. Exp. Res. 41, 666–680. doi: 10.1111/acer.13338
Bichenkov, E., and Ellingson, J. S. (2009). Ethanol alters the expressions of c-Fos and myelin basic protein in differentiating oligodendrocytes. Alcohol 43, 627–634. doi: 10.1016/j.alcohol.2009.09.026
Bohnsack, J. P., Zhang, H., Wandling, G. M., He, D., Kyzar, E. J., Lasek, A. W., et al. (2022). Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure. Sci. Adv. 8:eabn2748. doi: 10.1126/sciadv.abn2748
Brenner, E., Tiwari, G. R., Kapoor, M., Liu, Y., Brock, A., and Mayfield, R. D. (2020). Single cell transcriptome profiling of the human alcohol-dependent brain. Hum. Mol. Genet. 29, 1144–1153. doi: 10.1093/hmg/ddaa038
Butler, J. S., and Dent, S. Y. R. (2012). Taking stock of transcriptional elongation and histones: increased security through decreased exchange. Nat. Struct. Mol. Biol. 19, 863–864. doi: 10.1038/nsmb.2370
Carbia, C., Cadaveira, F., Caamaño-Isorna, F., Rodríguez-Holguín, S., and Corral, M. (2017). Binge drinking during adolescence and young adulthood is associated with deficits in verbal episodic memory. PLoS One 12:e0171393. doi: 10.1371/journal.pone.0171393
Carrozza, M. J., Li, B., Florens, L., Suganuma, T., Swanson, S. K., Lee, K. K., et al. (2005). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cells 123, 581–592. doi: 10.1016/j.cell.2005.10.023
Carvalho, S., Raposo, A. C., Martins, F. B., Grosso, A. R., Sridhara, S. C., Rino, J., et al. (2013). Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41, 2881–2893. doi: 10.1093/nar/gks1472
Casey, B. J., Getz, S., and Galvan, A. (2008). The adolescent brain. Dev. Rev. 28, 62–77. doi: 10.1016/j.dr.2007.08.003
Chen, J., Bardes, E. E., Aronow, B. J., and Jegga, A. G. (2009). ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311. doi: 10.1093/nar/gkp427
Chen, C. A., Ezzeddine, N., and Shyu, A.-B. (2008). Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 448, 335–357. doi: 10.1016/S0076-6879(08)02617-7
Chen, X., Tomchick, D. R., Kovrigin, E., Araç, D., Machius, M., Südhof, T. C., et al. (2002). Three-dimensional structure of the complexin/SNARE complex. Neuron 33, 397–409. doi: 10.1016/S0896-6273(02)00583-4
Coleman, L. G. Jr., He, J., Lee, J., Styner, M., and Crews, F. T. (2011). Adolescent binge drinking alters adult brain neurotransmitter gene expression, behavior, brain regional volumes, and neurochemistry in mice. Alcohol. Clin. Exp. Res. 35, 671–688. doi: 10.1111/j.1530-0277.2010.01385.x
Coleman, L. G., Liu, W., Oguz, I., Styner, M., and Crews, F. T. (2014). Adolescent binge ethanol treatment alters adult brain regional volumes, cortical extracellular matrix protein and behavioral flexibility. Pharmacol. Biochem. Behav. 116, 142–151. doi: 10.1016/j.pbb.2013.11.021
Contreras, A., Polín, E., Miguéns, M., Pérez-García, C., Pérez, V., Ruiz-Gayo, M., et al. (2019). Intermittent-excessive and chronic-moderate ethanol intake during adolescence impair spatial learning, memory and cognitive flexibility in the adulthood. Neuroscience 418, 205–217. doi: 10.1016/j.neuroscience.2019.08.051
Crews, F. T., Vetreno, R. P., Broadwater, M. A., and Robinson, D. L. (2016). Adolescent alcohol exposure persistently impacts adult neurobiology and behavior. Pharmacol. Rev. 68, 1074–1109. doi: 10.1124/pr.115.012138
Darbinian, N., Darbinyan, A., Merabova, N., Bajwa, A., Tatevosian, G., Martirosyan, D., et al. (2021). Ethanol-mediated alterations in oligodendrocyte differentiation in the developing brain. Neurobiol. Dis. 148:105181. doi: 10.1016/j.nbd.2020.105181
De Bellis, M. D., Narasimhan, A., Thatcher, D. L., Keshavan, M. S., Soloff, P., and Clark, D. B. (2005). Prefrontal cortex, thalamus, and cerebellar volumes in adolescents and young adults with adolescent-onset alcohol use disorders and comorbid mental disorders. Alcohol. Clin. Exp. Res. 29, 1590–1600. doi: 10.1097/01.alc.0000179368.87886.76
DiFiore, J. V., Ptacek, T. S., Wang, Y., Li, B., Simon, J. M., and Strahl, B. D. (2020). Unique and shared roles for histone H3K36 methylation states in transcription regulation functions. Cell Rep. 31:107751. doi: 10.1016/j.celrep.2020.107751
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Drissi, I., Deschamps, C., Fouquet, G., Alary, R., Peineau, S., Gosset, P., et al. (2020). Memory and plasticity impairment after binge drinking in adolescent rat hippocampus:GluN2A/GluN2B NMDAreceptor subunits imbalance throughHDAC2. Addict. Biol. 25, 1–15. doi: 10.1111/adb.12760
Drzewiecki, C. M., Willing, J., and Juraska, J. M. (2017). Synaptic number changes in the medial prefrontal cortex across adolescence in male and female rats: a role for pubertal onset. Physiol. Behav. 176, 139–148. doi: 10.1002/syn.21909
El Marroun, H., Klapwijk, E. T., Koevoets, M., Brouwer, R. M., Peters, S., van't Ent, D., et al. (2021). Alcohol use and brain morphology in adolescence: a longitudinal study in three different cohorts. Eur. J. Neurosci. 54, 6012–6026. doi: 10.1111/ejn.15411
Emery, B., and Lu, Q. R. (2015). Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb. Perspect. Biol. 7:a020461. doi: 10.1101/cshperspect.a020461
Erickson, E. K., Blednov, Y. A., Harris, R. A., and Mayfield, R. D. (2019). Glial gene networks associated with alcohol dependence. Sci. Rep. 9:10949. doi: 10.1038/s41598-019-47454-4
Farris, S. P., Tiwari, G. R., Ponomareva, O., Lopez, M. F., Mayfield, R. D., and Becker, H. C. (2020). Transcriptome analysis of alcohol drinking in non-dependent and dependent mice following repeated cycles of forced swim stress exposure. Brain Sci. 10:275. doi: 10.3390/brainsci10050275
Fawcett, J. (2009). Molecular control of brain plasticity and repair. Prog Brain Res. 175, 501–509. doi: 10.1016/S0079-6123(09)17534-9
Finegersh, A., and Homanics, G. (2014). Acute ethanol alters multiple histone modifications at model gene promoters in the cerebral cortex. Alcohol. Clin. Exp. Res. 38, 1865–1873. doi: 10.1111/acer.12465
Finn, D. A., Hashimoto, J. G., Cozzoli, D. K., Helms, M. L., Nipper, M. A., Kaufman, M. N., et al. (2018). Binge ethanol drinking produces sexually divergent and distinct changes in nucleus accumbens signaling cascades and pathways in adult C57BL/6J mice. Front. Genet. 9:325. doi: 10.3389/fgene.2018.00325
Flores-Bonilla, A., and Richardson, H. N. (2020). Sex differences in the neurobiology of alcohol use disorder. Alcohol Res. 40:04. doi: 10.35946/arcr.v40.2.04
Gamsby, J. J., Templeton, E. L., Bonvini, L. A., Wang, W., Loros, J. J., Dunlap, J. C., et al. (2013). The circadian Per1 and Per2 genes influence alcohol intake, reinforcement, and blood alcohol levels. Behav. Brain Res. 249, 15–21. doi: 10.1016/j.bbr.2013.04.016
Gao, Q., Liu, L., Chen, Y., Li, H., Yang, L., Wang, Y., et al. (2015). Synaptosome-related (SNARE) genes and their interactions contribute to the susceptibility and working memory of attention-deficit/hyperactivity disorder in males. Prog. Neuro Psychopharmacol. Biol. Psychiatry 57, 132–139. doi: 10.1016/j.pnpbp.2014.11.001
Gass, J. T., Glen, W. B., McGonigal, J. T., Trantham-Davidson, H., Lopez, M. F., Randall, P. K., et al. (2014). Adolescent alcohol exposure reduces behavioral flexibility, promotes disinhibition, and increases resistance to extinction of ethanol self-administration in adulthood. Neuropsychopharmacology 39, 2570–2583. doi: 10.1038/npp.2014.109
Gogtay, N., Giedd, J. N., Lusk, L., Hayashi, K. M., Greenstein, D., Vaituzis, A. C., et al. (2004). Dynamic mapping of human cortical development during childhood through early adulthood. Proc. Natl. Acad. Sci. U. S. A. 101, 8174–8179. doi: 10.1073/pnas.0402680101
Gopaul, K. R., Irfan, M., Miry, O., Vose, L. R., Moghadam, A., Subah, G., et al. (2020). Developmental time course of SNAP-25 isoforms regulate hippocampal long-term synaptic plasticity and hippocampus-dependent learning. Int. J. Mol. Sci. 21:1448. doi: 10.3390/ijms21041448
Goudriaan, A., Loos, M., Spijker, S., Smit, A. B., and Verheijen, M. H. G. (2020). Genetic variation in CNS myelination and functional brain connectivity in recombinant inbred mice. Cells 9:2119. doi: 10.3390/cells9092119
Gräff, J., Woldemichael, B. T., Berchtold, D., Dewarrat, G., and Mansuy, I. M. (2012). Dynamic histone marks in the hippocampus and cortex facilitate memory consolidation. Nat. Commun. 3:991. doi: 10.1038/ncomms1997
Grant, B. F., and Dawson, D. A. (1997). Age at onset of alcohol use and its association with DSM-IV alcohol abuse and dependence: results from the National Longitudinal Alcohol Epidemiologic Survey. J. Subst. Abus. 9, 103–110. doi: 10.1016/S0899-3289(97)90009-2
Gundelfinger, E. D., Frischknecht, R., Choquet, D., and Heine, M. (2010). Converting juvenile into adult plasticity: a role for the brain’s extracellular matrix. Eur. J. Neurosci. 31, 2156–2165. doi: 10.1111/j.1460-9568.2010.07253.x
Guo, F., Zhang, Y.-F., Liu, K., Huang, X., Li, R. X., Wang, S. Y., et al. (2021). Chronic exposure to alcohol inhibits new myelin generation in adult mouse brain. Front. Cell. Neurosci. 15:732602. doi: 10.3389/fncel.2021.732602
Hanson, K. L., Cummins, K., Tapert, S. F., and Brown, S. A. (2011). Changes in neuropsychological functioning over 10 years following adolescent substance abuse treatment. Psychol. Addict. Behav. 25, 127–142. doi: 10.1037/a0022350
Hanson, K. L., Medina, K. L., Padula, C. B., Tapert, S. F., and Brown, S. A. (2011). Impact of adolescent alcohol and drug use on neuropsychological functioning in young adulthood: 10-year outcomes. J. Child Adolesc. Subst. Abuse 20, 135–154. doi: 10.1080/1067828X.2011.555272
Hitchcock, L. N., and Lattal, K. M. (2014). Histone-mediated epigenetics in addiction. Prog. Mol. Biol. Transl. Sci. 128, 51–87. doi: 10.1016/B978-0-12-800977-2.00003-6
Hitzemann, R., Phillips, T. J., Lockwood, D. R., Darakjian, P., and Searles, R. P. (2020). Phenotypic and gene expression features associated with variation in chronic ethanol consumption in heterogeneous stock collaborative cross mice. Genomics 112, 4516–4524. doi: 10.1016/j.ygeno.2020.08.004
Hou, Q., Gao, X., Zhang, X., Kong, L., Wang, X., Bian, W., et al. (2004). SNAP-25 in hippocampal CA1 region is involved in memory consolidation. Eur. J. Neurosci. 20, 1593–1603. doi: 10.1111/j.1460-9568.2004.03600.x
Irfan, M., Gopaul, K. R., Miry, O., Hökfelt, T., Stanton, P. K., and Bark, C. (2019). SNAP-25 isoforms differentially regulate synaptic transmission and long-term synaptic plasticity at central synapses. Sci. Rep. 9:6403. doi: 10.1038/s41598-019-42833-3
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. doi: 10.1038/ng1089
Johansson, J. U., Ericsson, J., Janson, J., Beraki, S., Stanić, D., Mandic, S. A., et al. (2008). An ancient duplication of exon 5 in the Snap25 gene is required for complex neuronal development/function. PLoS Genet. 4:e1000278. doi: 10.1371/journal.pgen.1000278
Karlić, R., Chung, H. R., Lasserre, J., Vlahoviček, K., and Vingron, M. (2010). Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U. S. A. 107, 2926–2931. doi: 10.1073/pnas.0909344107
Kerns, R. T., Ravindranathan, A., Hassan, S., Cage, M. P., York, T., Sikela, J. M., et al. (2005). Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J. Neurosci. 25, 2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005
Kim, A., Zamora-Martinez, E. R., Edwards, S., and Mandyam, C. D. (2015). Structural reorganization of pyramidal neurons in the medial prefrontal cortex of alcohol dependent rats is associated with altered glial plasticity. Brain Struct. Funct. 220, 1705–1720. doi: 10.1007/s00429-014-0755-3
Kofler, M. J., Singh, L. J., Soto, E. F., Chan, E. S. M., Miller, C. E., Harmon, S. L., et al. (2020). Working memory and short-term memory deficits in ADHD: a bifactor modeling approach. Neuropsychology 34, 686–698. doi: 10.1037/neu0000641
Krishnan, H. R., Sakharkar, A. J., Teppen, T. L., Berkel, T. D. M., and Pandey, S. C. (2014). The epigenetic landscape of alcoholism. Epigenetics 115, 75–116. doi: 10.1016/B978-0-12-801311-3.00003-2
Krishnan, H. R., Zhang, H., Chen, Y., Bohnsack, J. P., Shieh, A. W., Kusumo, H., et al. (2022). Unraveling the epigenomic and transcriptomic interplay during alcohol-induced anxiolysis. Mol. Psychiatry 27, 4624–4632. doi: 10.1038/s41380-022-01732-2
Kyzar, E. J., Zhang, H., Sakharkar, A. J., and Pandey, S. C. (2017). Adolescent alcohol exposure alters lysine demethylase 1 (LSD1) expression and histone methylation in the amygdala during adulthood. Addict. Biol. 22, 1191–1204. doi: 10.1111/adb.12404
Labonté, B., Engmann, O., Purushothaman, I., Menard, C., Wang, J., Tan, C., et al. (2017). Sex-specific transcriptional signatures in human depression. Nat. Med. 23, 1102–1111. doi: 10.1038/nm.4386
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lees, B., Meredith, L. R., Kirkland, A. E., Bryant, B. E., and Squeglia, L. M. (2020). Effect of alcohol use on the adolescent brain and behavior. Pharmacol. Biochem. Behav. 192:172906:172906. doi: 10.1016/j.pbb.2020.172906
Lester, B. M., Tronick, E., Nestler, E., Abel, T., Kosofsky, B., Kuzawa, C. W., et al. (2011). Behavioral epigenetics. Ann. N. Y. Acad. Sci. 1226, 14–33. doi: 10.1111/j.1749-6632.2011.06037.x
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Linsenbardt, D. N., Moore, E. M., Gross, C. D., Goldfarb, K. J., Blackman, L. C., and Boehm, S. L. II. (2009). Sensitivity and tolerance to the hypnotic and ataxic effects of ethanol in adolescent and adult C57BL/6J and DBA/2J mice. Alcohol. Clin. Exp. Res. 33, 464–476. doi: 10.1111/j.1530-0277.2008.00857.x
Liu, L., Li, Y., and Tollefsbol, T. O. (2008). Gene-environment interactions and epigenetic basis of human diseases. Curr. Issues Mol. Biol. 10, 25–36.
Liu, X. J., Magri, L., Zhang, F., Marsh, N. O., Albrecht, S., Huynh, J. L., et al. (2015). Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. J. Neurosci. 35, 352–365. doi: 10.1523/JNEUROSCI.2606-14.2015
Lopes-Ramos, C. M., Chen, C.-Y., Kuijjer, M. L., Paulson, J. N., Sonawane, A. R., Fagny, M., et al. (2020). Sex differences in gene expression and regulatory networks across 29 human tissues. Cell Rep. 31:107795. doi: 10.1016/j.celrep.2020.107795
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lubbers, B. R., Smit, A. B., Spijker, S., and van den Oever, M. C. (2014). Neural ECM in addiction, schizophrenia, and mood disorder. Prog Brain Res. 214, 263–284. doi: 10.1016/B978-0-444-63486-3.00012-8
Luciana, M., Collins, P. F., Muetzel, R. L., and Lim, K. O. (2013). Effects of alcohol use initiation on brain structure in typically developing adolescents. Am. J. Drug Alcohol Abuse 39, 345–355. doi: 10.3109/00952990.2013.837057
Macht, V., Elchert, N., and Crews, F. (2020). Adolescent alcohol exposure produces protracted cognitive-behavioral impairments in adult male and female rats. Brain Sci. 10:785. doi: 10.3390/brainsci10110785
Marco, E. M., Peñasco, S., Hernández, M. D., Gil, A., Borcel, E., Moya, M., et al. (2017). Long-term effects of intermittent adolescent alcohol exposure in male and female rats. Front. Behav. Neurosci. 11, 1–13. doi: 10.3389/fnbeh.2017.00233
Markwiese, B. J., Acheson, S. K., Levin, E. D., Wilson, W. A., and Swartzwelder, H. S. (1998). Differential effects of ethanol on memory in adolescent and adult rats. Alcohol. Clin. Exp. Res. 22, 416–421. doi: 10.1111/j.1530-0277.1998.tb03668.x
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17:10. doi: 10.14806/ej.17.1.200
McMahon, H. T., Missler, M., Li, C., and Südhof, T. C. (1995). Complexins: cytosolic proteins that regulate SNAP receptor function. Cells 83, 111–119. doi: 10.1016/0092-8674(95)90239-2
Medina, K. L., McQueeny, T., Nagel, B. J., Hanson, K. L., Schweinsburg, A. D., and Tapert, S. F. (2008). Prefrontal cortex volumes in adolescents with alcohol use disorders: unique gender effects. Alcohol. Clin. Exp. Res. 32, 386–394. doi: 10.1111/j.1530-0277.2007.00602.x
Montesinos, J., Pascual, M., Pla, A., Maldonado, C., Rodríguez-Arias, M., Miñarro, J., et al. (2015). TLR4 elimination prevents synaptic and myelin alterations and long-term cognitive dysfunctions in adolescent mice with intermittent ethanol treatment. Brain Behav. Immun. 45, 233–244. doi: 10.1016/j.bbi.2014.11.015
Montesinos, J., Pascual, M., Rodríguez-Arias, M., Miñarro, J., and Guerri, C. (2016). Involvement of TLR4 in the long-term epigenetic changes, rewarding and anxiety effects induced by intermittent ethanol treatment in adolescence. Brain Behav. Immun. 53, 159–171. doi: 10.1016/j.bbi.2015.12.006
Mota, N., Parada, M., Crego, A., Doallo, S., Caamaño-Isorna, F., Rodríguez Holguín, S., et al. (2013). Binge drinking trajectory and neuropsychological functioning among university students: a longitudinal study. Drug Alcohol Depend. 133, 108–114. doi: 10.1016/j.drugalcdep.2013.05.024
Mulligan, M. K., Mozhui, K., Pandey, A. K., Smith, M. L., Gong, S., Ingels, J., et al. (2017). Genetic divergence in the transcriptional engram of chronic alcohol abuse: a laser-capture RNA-seq study of the mouse mesocorticolimbic system. Alcohol 58, 61–72. doi: 10.1016/j.alcohol.2016.09.001
Najera, K., Fagan, B. M., and Thompson, P. M. (2019). SNAP-25 in major psychiatric disorders: a review. Neuroscience 420, 79–85. doi: 10.1016/j.neuroscience.2019.02.008
National Research Council (2011). Guide for the Care and Use of Laboratory Animals, 8th Edn. Washington, DC: National Academies Press.
Nato, A. Q., Ul Mustafa, H. A., Patel, R. K., Sexton, H. G., Moore, S. D., Denvir, J., et al. (2014). RNA-Seq data analysis of a rodent model of adolescent binge drinking reveals pathways and candidate genes involved in neuronal remodeling and neuroimmune activation. bioRxiv. doi: 10.1101/2020.11.02.365841
Ng, H. H., Robert, F., Young, R. A., and Struhl, K. (2003). Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719. doi: 10.1016/S1097-2765(03)00092-3
Pandey, S. C., Kyzar, E. J., and Zhang, H. (2017). Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology 122, 74–84. doi: 10.1016/j.neuropharm.2017.02.002
Parkel, S., Lopez-Atalaya, J. P., and Barco, A. (2013). Histone H3 lysine methylation in cognition and intellectual disability disorders. Learn. Mem. 20, 570–579. doi: 10.1101/lm.029363.112
Pascual, M., Blanco, A. M., Cauli, O., Miñarro, J., and Guerri, C. (2007). Intermittent ethanol exposure induces inflammatory brain damage and causes long-term behavioural alterations in adolescent rats. Eur. J. Neurosci. 25, 541–550. doi: 10.1111/j.1460-9568.2006.05298.x
Pascual, M., López-Hidalgo, R., Montagud-Romero, S., Ureña-Peralta, J. R., Rodríguez-Arias, M., and Guerri, C. (2021). Role of mTOR-regulated autophagy in spine pruning defects and memory impairments induced by binge-like ethanol treatment in adolescent mice. Brain Pathol. 31, 174–188. doi: 10.1111/bpa.12896
Peñasco, S., Rico-Barrio, I., Puente, N., Fontaine, C. J., Ramos, A., Reguero, L., et al. (2020). Intermittent ethanol exposure during adolescence impairs cannabinoid type 1 receptor-dependent long-term depression and recognition memory in adult mice. Neuropsychopharmacology 45, 309–318. doi: 10.1038/s41386-019-0530-5
Peng, Q., Wilhelmsen, K. C., and Ehlers, C. L. (2021). Common genetic substrates of alcohol and substance use disorder severity revealed by pleiotropy detection against GWAS catalog in two populations. Addict. Biol. 26:e12877. doi: 10.1111/adb.12877
Pfefferbaum, A., Kwon, D., Brumback, T., Thompson, W. K., Cummins, K., Tapert, S. F., et al. (2018). Altered brain developmental trajectories in adolescents after initiating drinking. Am. J. Psychiatry 175, 370–380. doi: 10.1176/appi.ajp.2017.17040469
Phillips, T. J., Dickinson, S., and Burkhart-Kasch, S. (1994). Behavioral sensitization to drug stimulant effects in C57BL/6J and DBA/2J inbred mice. Behav. Neurosci. 108, 789–803. doi: 10.1037/0735-7044.108.4.789
Ponomarev, I., Wang, S., Zhang, L., Harris, R. A., and Mayfield, R. D. (2012). Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. Neurobiol. Dis. 32, 1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012
Pu, M., Ni, Z., Wang, M., Wang, X., Wood, J. G., Helfand, S. L., et al. (2015). Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 29, 718–731. doi: 10.1101/gad.254144.114
Radke, A. K., Sneddon, E. A., and Monroe, S. C. (2021). Studying sex differences in rodent models of addictive behavior. Curr Protoc 1:e119. doi: 10.1002/cpz1.119
Ramos-Miguel, A., Sawada, K., Jones, A. A., Thornton, A. E., Barr, A. M., Leurgans, S. E., et al. (2017). Presynaptic proteins complexin-I and complexin-II differentially influence cognitive function in early and late stages of Alzheimer’s disease. Acta Neuropathol. 133, 395–407. doi: 10.1007/s00401-016-1647-9
Reim, K., Mansour, M., Varoqueaux, F., McMahon, H. T., Südhof, T. C., Brose, N., et al. (2001). Complexins regulate a late step in Ca2+−dependent neurotransmitter release. Cells 104, 71–81. doi: 10.1016/S0092-8674(01)00192-1
Reshetnikov, V. V., Kisaretova, P. E., Ershov, N. I., Merkulova, T. I., and Bondar, N. P. (2020). Data of correlation analysis between the density of H3K4me3 in promoters of genes and gene expression: data from RNA-seq and ChIP-seq analyses of the murine prefrontal cortex. Data Brief 33:106365:106365. doi: 10.1016/j.dib.2020.106365
Reshetnikov, V. V., Kisaretova, P. E., Ershov, N. I., Merkulova, T. I., and Bondar, N. P. (2021). Social defeat stress in adult mice causes alterations in gene expression, alternative splicing, and the epigenetic landscape of H3K4me3 in the prefrontal cortex: an impact of early-life stress. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 106:110068. doi: 10.1016/j.pnpbp.2020.110068
Robison, A. J., and Nestler, E. J. (2012). Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637. doi: 10.1038/nrn3111
Romer, D., Reyna, V. F., and Satterthwaite, T. D. (2017). Beyond stereotypes of adolescent risk taking: placing the adolescent brain in developmental context. Dev. Cogn. Neurosci. 27, 19–34. doi: 10.1016/j.dcn.2017.07.007
Salimi, K., Glantz, L. A., Hamer, R. M., German, T. T., Gilmore, J. H., and Jarskog, L. F. (2008). Regulation of complexin 1 and complexin 2 in the developing human prefrontal cortex. Synapse 62, 273–282. doi: 10.1002/syn.20492
SAMHSA (2020). Key substance use and mental health indicators in the United States: Results from the 2020 National Survey on drug use and health. HHS Publication No PEP21-07-01-003, NSDUH Series H-56.
Sessa, A., Fagnocchi, L., Mastrototaro, G., Massimino, L., Zaghi, M., Indrigo, M., et al. (2019). SETD5 regulates chromatin methylation state and preserves global transcriptional Fidelity during brain development and neuronal wiring. Neuron 104, 271–289.e13. doi: 10.1016/j.neuron.2019.07.013
Sharova, L. V., Sharov, A. A., Nedorezov, T., Piao, Y., Shaik, N., and Ko, M. S. H. (2009). Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16, 45–58. doi: 10.1093/dnares/dsn030
Simon, J. M., Hacker, K. E., Singh, D., Brannon, A. R., Parker, J. S., Weiser, M., et al. (2014). Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 24, 241–250. doi: 10.1101/gr.158253.113
Smolle, M., and Workman, J. L. (2013). Transcription-associated histone modifications & cryptic transcription. Biochim. Biophys. Acta 1829, 84–97. doi: 10.1016/j.bbagrm.2012.08.008
Smolle, M., Workman, J. L., and Venkatesh, S. (2013). reSETting chromatin during transcription elongation. Epigenetics. doi: 10.4161/epi.23333, [Epub-ahead of print]
Söderqvist, S., McNab, F., Peyrard-Janvid, M., Matsson, H., Humphreys, K., Kere, J., et al. (2010). The SNAP25 gene is linked to working memory capacity and maturation of the posterior cingulate cortex during childhood. Biol. Psychiatry 68, 1120–1125. doi: 10.1016/j.biopsych.2010.07.036
Spear, L. (2000). The adolescent brain and age-related behavioral manifestations. Neurosci. Biobehav. Rev. 24, 417–463. doi: 10.1016/S0149-7634(00)00014-2
Spear, L. (2014). Adolescents and alcohol: acute sensitivities, enhanced intake, and later consequences. Neurotoxicol. Teratol. 41, 51–59. doi: 10.1016/j.ntt.2013.11.006
Squeglia, L. M., Jacobus, J., and Tapert, S. F. (2014). The effect of alcohol use on human adolescent brain structures and systems. Handb. Clin. Neurol. 125, 501–510. doi: 10.1016/B978-0-444-62619-6.00028-8
Squeglia, L. M., Schweinsburg, A. D., Pulido, C., and Tapert, S. F. (2011). Adolescent binge drinking linked to abnormal spatial working memory brain activation: differential gender effects. Alcohol. Clin. Exp. Res. 35, 1831–1841. doi: 10.1111/j.1530-0277.2011.01527.x
Tani, H., Mizutani, R., Salam, K. A., Tano, K., Ijiri, K., Wakamatsu, A., et al. (2012). Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956. doi: 10.1101/gr.130559.111
Tavares, E. R., Silva-Gotay,, Riad, W. V., Bengston, L., and Richardson, H. N. (2019). Sex differences in the effect of alcohol drinking on myelinated axons in the anterior cingulate cortex of adolescent rats. Brain Sci. 9, 1–15. doi: 10.3390/brainsci9070167
Tsankova, N., Renthal, W., Kumar, A., and Nestler, E. J. (2007). Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 8, 355–367. doi: 10.1038/nrn2132
van Hees, L., Didone, V., Charlet-Briart, M., van Ingelgom, T., Alexandre, A., Quertemont, E., et al. (2022). Voluntary alcohol binge-drinking in adolescent C57Bl6 mice induces delayed appearance of behavioural defects in both males and females. Addict. Biol. 27:e13102. doi: 10.1111/adb.13102
Vargas, W. M., Bengston, L., Gilpin, X. N. W., Whitcomb, B. W., and Richardson, H. N. (2014). Alcohol binge drinking during adolescence or dependence during adulthood reduces prefrontal myelin in male rats. J. Neurosci. 34, 14777–14782. doi: 10.1523/JNEUROSCI.3189-13.2014
Vetreno, R. P., Bohnsack, J. P., Kusumo, H., Liu, W., Pandey, S. C., and Crews, F. T. (2020). Neuroimmune and epigenetic involvement in adolescent binge ethanol-induced loss of basal forebrain cholinergic neurons: restoration with voluntary exercise. Addict. Biol. 25, 1–14. doi: 10.1111/adb.12731
Vetreno, R. P., and Crews, F. T. (2012). Adolescent binge drinking increases expression of the danger signal receptor agonist HMGB1 and toll-like receptors in the adult prefrontal cortex. Neuroscience 226, 475–488. doi: 10.1016/j.neuroscience.2012.08.046
Vetreno, R. P., and Crews, F. T. (2015). Binge ethanol exposure during adolescence leads to a persistent loss of neurogenesis in the dorsal and ventral hippocampus that is associated with impaired adult cognitive functioning. Front. Neurosci. 9:35. doi: 10.3389/fnins.2015.00035
Vetreno, R. P., Yaxley, R., Paniagua, B., and Crews, F. T. (2016). Diffusion tensor imaging reveals adolescent binge ethanol-induced brain structural integrity alterations in adult rats that correlate with behavioral dysfunction. Addict. Biol. 21, 939–953. doi: 10.1111/adb.12232
Wang, X., Zhang, J., Zhou, L., Xu, B., Ren, X., He, K., et al. (2019). Long-term iron exposure causes widespread molecular alterations associated with memory impairment in mice. Food Chem. Toxicol. 130, 242–252. doi: 10.1016/j.fct.2019.05.038
Warburton, E. C., and Brown, M. W. (2015). Neural circuitry for rat recognition memory. Behav. Brain Res. 285, 131–139. doi: 10.1016/j.bbr.2014.09.050
Wei, W., Hennig, B. P., Wang, J., Zhang, Y., Piazza, I., Pareja Sanchez, Y., et al. (2019). Chromatin-sensitive cryptic promoters putatively drive expression of alternative protein isoforms in yeast. Genome Res. 29, 1974–1984. doi: 10.1101/gr.243378.118
White, A. M., Ghia, A. J., Levin, E. D., and Swartzwelder, H. S. (2000). Binge pattern ethanol exposure in adolescent and adult rats: differential impact on subsequent responsiveness to ethanol. Alcohol. Clin. Exp. Res. 24, 1251–1256. doi: 10.1111/j.1530-0277.2000.tb02091.x
Wolstenholme, J. T., Mahmood, T., Harris, G. M., Abbas, S., and Miles, M. F. (2017). Intermittent ethanol during adolescence leads to lasting behavioral changes in adulthood and alters gene expression and histone methylation in the PFC. Front. Mol. Neurosci. 10:307. doi: 10.3389/fnmol.2017.00307
Wolstenholme, J. T., Warner, J. A., Capparuccini, M. I., Archer, K. J., Shelton, K. L., and Miles, M. F. (2011). Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57Bl/6 mice. PLoS One 6:e21100. doi: 10.1371/journal.pone.0021100
Xu, S., Grullon, S., Ge, K., and Peng, W. (2014). Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 1150, 97–111. doi: 10.1007/978-1-4939-0512-6_5
Xu, S.-J., Lombroso, S. I., Fischer, D. K., Carpenter, M. D., Marchione, D. M., Hamilton, P. J., et al. (2021). Chromatin-mediated alternative splicing regulates cocaine-reward behavior. Neuron 109, 2943–2966.e8. doi: 10.1016/j.neuron.2021.08.008
Yang, E., van Nimwegen, E., Zavolan, M., Rajewsky, N., Schroeder, M., Magnasco, M., et al. (2003). Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 13, 1863–1872. doi: 10.1101/gr.1272403
Ying, M., Sui, X., Zhang, Y., Sun, Q., Qu, Z., Luo, X., et al. (2017). Identification of novel key molecules involved in spatial memory impairment in triple transgenic mice of Alzheimer’s disease. Mol. Neurobiol. 54, 3843–3858. doi: 10.1007/s12035-016-9959-2
Younis, R. M., Wolstenholme, J. T., Bagdas, D., Bettinger, J. C., Miles, M. F., and Damaj, M. I. (2019). Adolescent but not adult ethanol binge drinking modulates ethanol behavioral effects in mice later in life. Pharmacol. Biochem. Behav. 184:172740. doi: 10.1016/j.pbb.2019.172740
Yu, G., Wang, L.-G., and He, Q.-Y. (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. doi: 10.1093/bioinformatics/btv145
Zhang, L.-Q., Liu, J.-J., Liu, L., Fan, G. L., Li, Y. N., and Li, Q. Z. (2021). The impact of gene-body H3K36me3 patterns on gene expression level changes in chronic myelogenous leukemia. Gene 802:145862:145862. doi: 10.1016/j.gene.2021.145862
Zhang, Y., Xie, S., Zhou, Y., Xie, Y., Liu, P., Sun, M., et al. (2014). H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep. 8, 1989–2002. doi: 10.1016/j.celrep.2014.08.031
Zheng, Y., Tipton, J. D., Thomas, P. M., Kelleher, N. L., and Sweet, S. M. M. (2014). Site-specific human histone H3 methylation stability: fast K4me3 turnover. Proteomics 14, 2190–2199. doi: 10.1002/pmic.201400060
Zhou, H.-L., Luo, G., Wise, J. A., and Lou, H. (2014). Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 42, 701–713. doi: 10.1093/nar/gkt875
Zimmermann, D. R., and Dours-Zimmermann, M. T. (2008). Extracellular matrix of the central nervous system: from neglect to challenge. Histochem. Cell Biol. 130, 635–653. doi: 10.1007/s00418-008-0485-9
Keywords: adolescent ethanol, alcohol, PFC, epigenetics, H3K36me3, RNA-seq, ChIP-seq, cryptic transcription
Citation: Brocato ER and Wolstenholme JT (2023) Adolescent binge ethanol impacts H3K36me3 regulation of synaptic genes. Front. Mol. Neurosci. 16:1082104. doi: 10.3389/fnmol.2023.1082104
Edited by:
Kimberly Frances Raab-Graham, Wake Forest University, United StatesReviewed by:
R. Dayne Mayfield, The University of Texas at Austin, United StatesLaura B. Ferguson, The University of Texas at Austin, United States
Copyright © 2023 Brocato and Wolstenholme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer T. Wolstenholme, amVubmlmZXIud29sc3RlbmhvbG1lQHZjdWhlYWx0aC5vcmc=
 Emily R. Brocato
Emily R. Brocato Jennifer T. Wolstenholme
Jennifer T. Wolstenholme