
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 12 September 2022
Sec. Molecular Signalling and Pathways
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.988803
 Hui Zhen1
Hui Zhen1 Mingyue Zheng2
Mingyue Zheng2 Huazhi Geng3
Huazhi Geng3 Qian Song1
Qian Song1 Lili Gao1
Lili Gao1 Zuoqing Yuan1
Zuoqing Yuan1 Hongkuan Deng1
Hongkuan Deng1 Qiuxiang Pang1
Qiuxiang Pang1 Bosheng Zhao1*
Bosheng Zhao1*Disturbances in the excitatory/inhibitory balance of brain neural circuits are the main source of encephalopathy during neurodevelopment. Changes in the function of neural circuits can lead to depolarization or repeat rhythmic firing of neurons in a manner similar to epilepsy. GABAergic neurons are inhibitory neurons found in all the main domains of the CNS. Previous studies suggested that DjCamkII and DjCaln play a crucial role in the regulation of GABAergic neurons during planarian regeneration. However, the mechanisms behind the regeneration of GABAergic neurons have not been fully explained. Herein, we demonstrated that DjCamkII and DjCaln were mutual negative regulation during planarian head regeneration. DjNFAT exerted feedback positive regulation on both DjCaln and DjCamkII. Whole-mount in situ hybridization (WISH) and fluorescence in situ hybridization (FISH) revealed that DjNFAT was predominantly expressed in the pharynx and parenchymal cells in intact planarian. Interestingly, during planarian head regeneration, DjNFAT was predominantly located in the newborn brain. Down-regulation of DjNFAT led to regeneration defects in the brain including regenerative brain became small and the lateral nerves cannot be regenerated completely, and a decreasein the number of GABAergic neurons during planarian head regeneration. These findings suggest that the feedback loop between DjCaln, DjCamkII, and DjNFAT is crucial for the formation of GABAergic neurons during planarian head regeneration.
Disturbances in the excitatory/inhibitory balance of brain neural circuits are the main source of encephalopathy during neurodevelopment. Once the function of neural circuits changes, the neurons in the corresponding circuits will depolarize or repeatedly fire rhythmically, which is a pattern closely related to the occurrence of epilepsy. γ-Aminobutyric acid (GABA) is the major inhibitory neurotransmitter in the central nervous system (CNS) and thereby plays a crucial role in the balance between inhibitory and excitatory neural circuits (Watanabe et al., 2002). GABAergic neurons are produced in all the main domains of the CNS, where they develop from discrete regions of the neuroepithelium. Therefore, it is expected that dysfunctions in the GABAergic system lead to neurodegenerative diseases, such as Parkinson’s disease (PD) and Huntington’s disease (HD) (Kleppner and Tobin, 2001; Galvan and Wichmann, 2007; Gajcy et al., 2010). Therefore, a certain degree of GABAergic neuron regeneration is essential to maintain the excitatory/inhibitory balance. In addition, it is widely accepted that continuous neurogenesis takes place in the adult hippocampus throughout the life of mammals, including humans (Eriksson et al., 1998; Zhao et al., 2008). The newborn neurons mostly differentiate into excitatory granular cells and functionally integrate into the preexisting hippocampal neural circuitry (Cameron and McKay, 2001; van Praag et al., 2002; Toni et al., 2007). However, little is known about the number of newborn cells that can regenerate and differentiate into inhibitory GABAergic interneurons in the damaged brain.
Calcineurin (Caln), also called protein phosphatase B (PP2B), is a Ca2+/calmodulin-dependent serine/threonine phosphatase that was first described in a bovine brain 40 years ago (Ho et al., 1976; Wang and Desai, 1976). The protein has a 60 kDa catalytic subunit (Caln A) and a 19 kDa regulatory subunit (Caln B) (Rusnak and Mertz, 2000; Li et al., 2011). Caln is located in the brain abundantly (Goto et al., 1986; Kuno et al., 1992). In neurons, Caln is localized in multiple organelles, such as cytoplasm, endoplasmic reticulum, and Golgi apparatus (Bram et al., 1993; Mehta and Zhang, 2014; Cardenas and Marengo, 2016). Studies indicate that GABA is regulated by Caln in synaptogenesis, for the reorganization and development of an adult CNS (Hayama et al., 2013; Toyoda et al., 2015; Wang et al., 2015).
Nuclear factor of activated T-cells (NFAT) can be dephosphorylated by Caln in order to translocate into the nucleus and become activated (Shaw et al., 1988; Schulz and Yutzey, 2004; Boothby, 2010). In the peripheral nervous system, Caln/NFAT signaling plays a critical role in the survival, proliferation, and differentiation of neural and glial precursor cells, especially during tissue regeneration (Serrano-Perez et al., 2015). Moreover, the neurotrophin nerve growth factor (NGF) regulates the up-regulation of the plasminogen activation system and synaptic protein plasminogen activator inhibitor 1 (PAI-1) in primary mouse hippocampal neurons through Caln/NFAT (Stefos et al., 2013). In mice, GABAA receptors promote anxiety behavior and hippocampal neurogenesis through Caln/NFAT4, indicating that this signaling pathway may serve as a potential drug target for the treatment of mood disorders (Quadrato et al., 2014). Meanwhile, the transcription factor NFAT5 is expressed in the anterior blastema of tail fragments after 3 days into regeneration in planarian Schmidtea mediterranea (Suzuki-Horiuchi et al., 2021).
In human T Cells, activation of Ca2+/calmodulin-dependent protein kinase II (CamKII) can reduce the efficiency of NFAT by nearly 35% (Hama et al., 1995). In cardiomyocytes, the activation of cytoplasmic CamKII inhibits calmodulin and NFAT through phosphorylation (MacDonnell et al., 2009). In budding yeast, a lack of the CamKII homologue CMK2 increased the level of calcium/calcineurin signaling, as well as the expression levels of PMR1 and PMC1. The expression of target genes including CMK2, RCN1, PMR1, and PMC1 is dependent on the activation of Crz1 (Mehta et al., 2009; Xu et al., 2019). These results indicate that CamKII is a negative feedback controller of the Caln/NFAT signaling pathway. However, whether there is a mutual regulation relationship between CamKII, Caln and NFAT or not in the neurons regeneration process has not been previously reported.
Planarians are often used as model organisms in the study of CNS regeneration due to their powerful regeneration ability (Newmark and Sanchez Alvarado, 2002; Elliott and Sanchez Alvarado, 2013). The planarian brain comprises of neurons similar to those found in humans, including dopaminergic, cholinergic, GABAergic, and serotonergic neurons (Nishimura et al., 2007a,b, 2008, 2010). Ca2+ signaling plays an important role in neuromuscular signaling and anterior-posterior patterning in planarians regeneration (Nogi et al., 2009). The knockdown of certain voltage-operated Ca2+ channels results in different regenerative polarities (Zhang et al., 2011). And knockdown of a specific voltage-operated Ca2+ channel (Cav1B) that impairs muscle function creates an environment permissive for anteriorization in planarian regeneration (Chan et al., 2017). As previously stated, we found that DjCamKII and DjCaln were both expressed in the brain. Furthermore, DjCamKII (RNAi) or DjCaln (RNAi) regenerated brains became slim and could not regenerate their lateral nerve. As such, down-regulation of DjCamKII or DjCaln led to a significant decrease in GAD and affected the number of GABAergic neurons during the planarian head regeneration (Zhen et al., 2020). However, the mechanism behind how CamKII and Caln regulate the formation of GABAergic neurons during the planarian head regeneration is not clear. The present study aims to examine how Caln, CamkII, and NFAT affect the number of GABAergic neurons during head regeneration in the planarian Dugesia japonica.
A clonal strain of the planarian D. japonica, originally obtained from Boshan, China, was established in our laboratory and used in all experiments (Zhen et al., 2020). The D. japonica planarians were cultured in Lushan spring water at 20°C. Before use in experiments, all planarians were starved for at least 7 days. For the regeneration experiments, the planarian heads were removed and the fragments were used for experiments at 1, 3, 5, 7, and 10 days after amputation.
Whole-mount in situ hybridization, digoxigenin (DIG)-labeled antisense RNA probes, and fluorescence in situ hybridization (FISH), Fluorescein-labeled antisense RNA probes, were synthesized using an in vitro labeling kit (Roche, Basel, Switzerland). The RNA probes-DjCaln, DjCamKII, DjNFAT, and DjGAD primers were designed by Primer Premier 5.0 (Supplementary Table 1). Hybridizations were carried out by incubation with the antisense RNA probe (1 ng/μl) at 56°C for 16 h. After hybridization, 10% horse serum was blocked at room temperature for 2 h after maleic acid buffer washing. Then, antibodies were diluted in maleic acid buffer containing Tween 20 (MABT) with 10% of horse serum for WISH (anti-DIG-AP, 1:2,000, Roche, Basel, Switzerland) and FISH (anti-DIG-POD, 1:500, and anti-FITC-POD, 1:500, Roche, Basel, Switzerland). Then, a mixture of 5 bromo, 4 chloro, 3-indolyl phosphate, and nitroblue tetrazolium (Roche, Basel, Switzerland) was utilized for color development in WISH, while Cy3-Tyramide and FITC Tyramide (Roche, Basel, Switzerland) [0.001% H2O2 in PBST (0.01% Tween 20)] was utilized in FISH. The numbers of gad+ and TH+ neurons in the brain region were counted manually under a confocal microscope (Laser confocal, Leica TCS SP8 MP, Germany) (Hill and Petersen, 2015; Brown et al., 2018). Particles of H3p+ and TPH+ cells were counted using ImageJ, version 6.0 (National Institutes of Health, United States), and Chat+ cells were counted from a zseries of images using three-dimensional segmentation software in Imaris (Hill and Petersen, 2015). Cell counts were statistically analyzed using GraphPad Prism 8 software.
The template of double-stranded RNA was synthesized by PCR. The dsRNA-DjCaln, DjCamKII, and DjNFAT primers were designed by Primer Premier 5.0 (Supplementary Table 1). dsRNA was synthesized using MEGAscriptTM RNAi Kit (Invitrogen, United States), and then planarians were soaked in 50 ng/μl dsRNA for 5 h and then transferred to the culture medium (Orii et al., 2003). Control animals were treated with dsRNA of the green fluorescent protein (GFP) (Fraguas et al., 2021).
Immunostaining with the mouse monoclonal antibody anti-SYNAPSIN (1:300, 3C11, AB Company, United States) and anti-phosphohistone-H3 (S10) (1:200, Cell Signaling Technology, Danvers, United States) were performed as previously described (Cowles et al., 2012; Lu et al., 2017). First, the planarians were euthanized with 2% of HCl and fixed in paraformaldehyde with phosphate-buffered saline (PBS) at 37°C for 1 h. Then, planarians were dehydrated with 100% of methanol and incubated with the primary antibodies anti-SYNAPSIN and anti-phosphohistone-H3. Incubation with secondary antibodies (1:200, rabbit anti-mouse, and goat anti-rabbit, marked with HRP, Bioss, United States) was performed overnight at 4°C. The samples were then observed using a laser confocal microscope (Leica TCS SP8 MP).
Protein expressions of DjCaln and DjCamkII were quantified with Western blot using the primary antibodies anti-DjCaln and anti-DjCamkII antibodies at 1:500 dilutions. Western blotting was performed as previously described (Gao et al., 2017). Briefly, planarian protein was extracted into PBS buffer, boiled for 20 min, and subjected to SDS-PAGE analysis. After the protein transfer to polyvinylidene fluoride membranes, membranes were blocked with 5% skim milk for 2 h at room temperature (RT). Then, membranes were incubated with primary antibodies (anti-DjCaln and anti-DjCamkII) followed by secondary antibodies (goat anti-rabbit horseradish peroxidase (HRP)-conjugated antibody, 1:5,000, Bioss, United States) overnight at 4°C. Protein signals were observed after treatment with enhanced chemiluminescence solution (Thermo, United States). The quantitation was performed by measuring the optical density of bands with ImageJ, version 6.0 (National Institutes of Health).
Expressions of DjCamKII, DjCaln, and DjNFAT mRNA were quantified using qPCR as previously described (Lu et al., 2017). DjCamKII, DjCaln, and DjNFAT primers were designed by Primer Premier, version 5.0 (Supplementary Table 1). The β-actin gene was used as the internal reference. Briefly, total RNA was extracted with TRIol reagent (Invitrogen, United States). The reaction was carried out under the following conditions: initial denaturation at 95°C for 10 s followed by 35 cycles of denaturation at 95°C for 30 s, annealing for 30 s at 55°C, and extension at 72°C for 45 s. The qPCR amplification cycles were performed on the ABI 7,500 real-time PCR system (Applied Biosystems, United States) using the quick-start Universal SYBR Green Master (Roche, Switzerland). Relative expressions compared to β-actin were calculated using the 2–ΔΔct method.
Results are presented as mean ± SD. Comparisons between groups were carried out using multiple t-tests. For all analyses, *P < 0.05 and **P < 0.01 were considered statistically significant. Statistical analyses were carried out using SPSS statistical software for Windows, version 16.0 (SPSS).
To study the regulation between DjCaln and DjCamkII during planarian regeneration, the mRNA and protein levels of DjCaln and DjCamkII were quantified by qPCR and Western blot. Down-regulation of DjCaln led to significant up-regulation of the expression of DjCamkII mRNA at 5, 7, and 10 days during planarian head regeneration (Figure 1A). Down-regulation of DjCamkII also led to significant up-regulation of the expression of DjCaln mRNA at 5, 7, and 10 days of during planarian head regeneration (Figure 1B). Western blot results showed that when the expression of DjCaln protein decreased, the expression of DjCamkII protein significantly increased at 5, 7, and 10 days of during planarian head regeneration (Figures 2A,B). When the expression of DjCamkII protein decreased, the expression of DjCaln protein significantly increased at 5, 7, and 10 days of during planarian head regeneration (Figures 2C,D). These results revealed that DjCamkII and DjCaln were mutual negative regulation during planarian head regeneration.
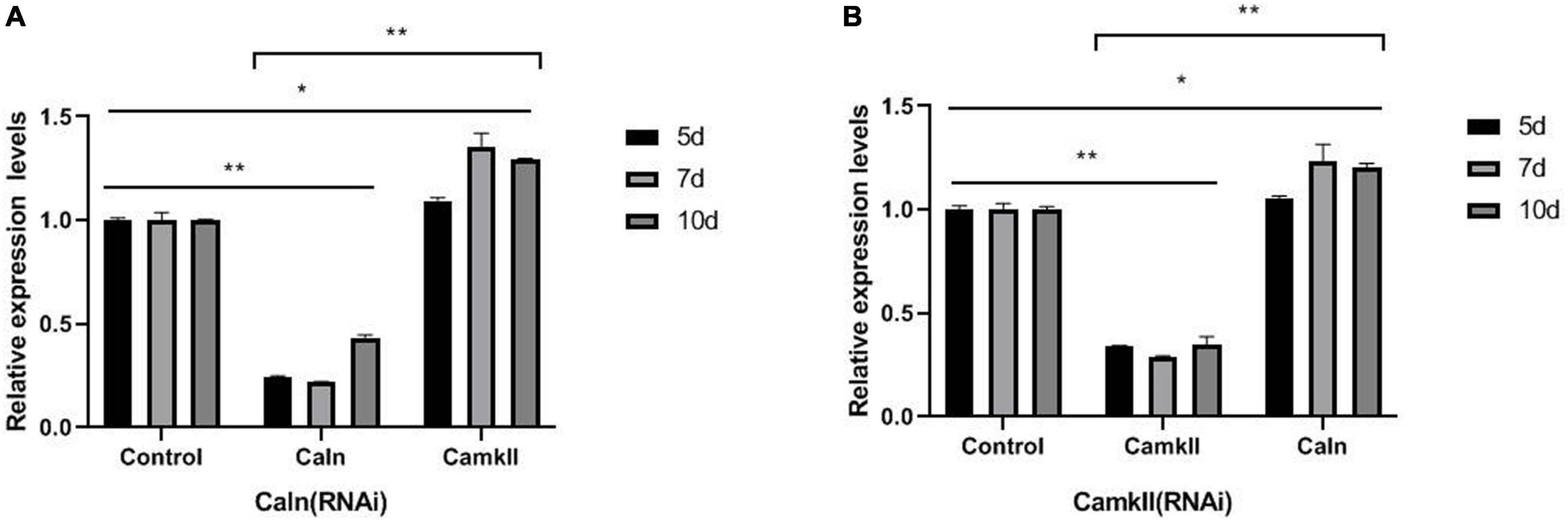
Figure 1. Regulatory relationship between DjCaln and DjCamkII mRNA during planarian head regeneration. (A) DjCamkII mRNA levels in DjCaln (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 4. (B) DjCaln mRNA levels in DjCamkII (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 4, *p < 0.05, **P < 0.01.
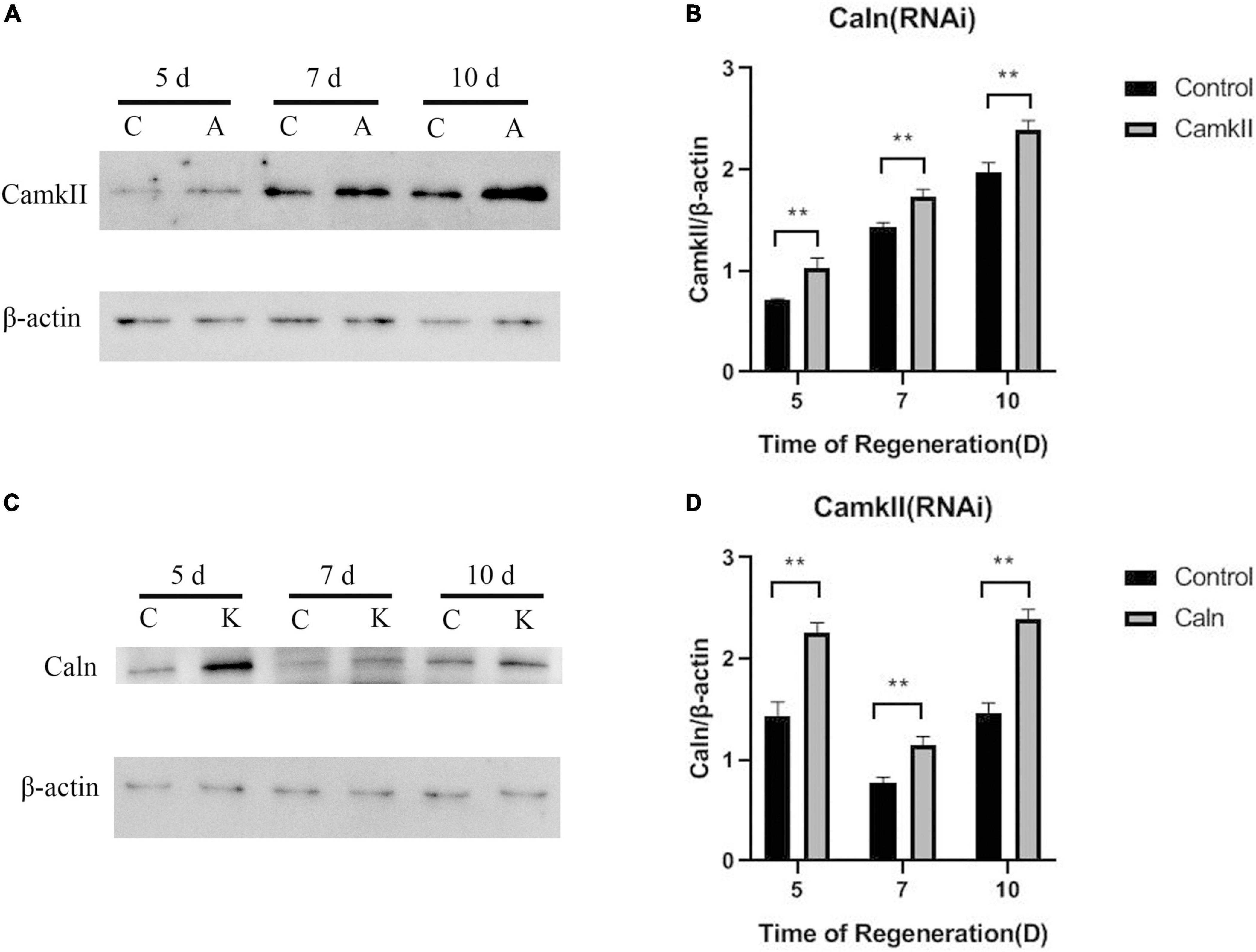
Figure 2. Regulatory relationship between DjCaln and DjCamkII protein during planarian head regeneration. (A,B) DjCamkII protein levels in DjCaln (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 5. (C,D) DjCaln protein levels in DjCamkII (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 5, **P < 0.01. C, control; A, DjCaln (RNAi); K, DjCamkII (RNAi).
Caln activated NFATs by dephosphorylating multiple N-terminal phosphoserine residues in the regulatory domain (Muller et al., 2009). To explore the relationship between DjCaln, DjCamkII, and DjNFAT, qPCR was performed in DjCaln (RNAi), DjCamkII (RNAi), or DjNFAT (RNAi) regenerative planarians. The results demonstrated that the expression of DjNFAT was significantly decreased in DjCaln (RNAi) or DjCamkII (RNAi) regenerative planarians (Figures 3A,B). Down-regulation of DjNFAT led to a significant decrease in the expression of DjCaln and DjCamkII mRNA (Figure 4A), as well as DjCaln and DjCamkII protein levels (Figures 4B–E) at 5, 7, and 10 days during planarian head regeneration. These results illustrated that DjNFAT promoted the expression of DjCaln and DjCamkII by positive feedback during planarian head regeneration.
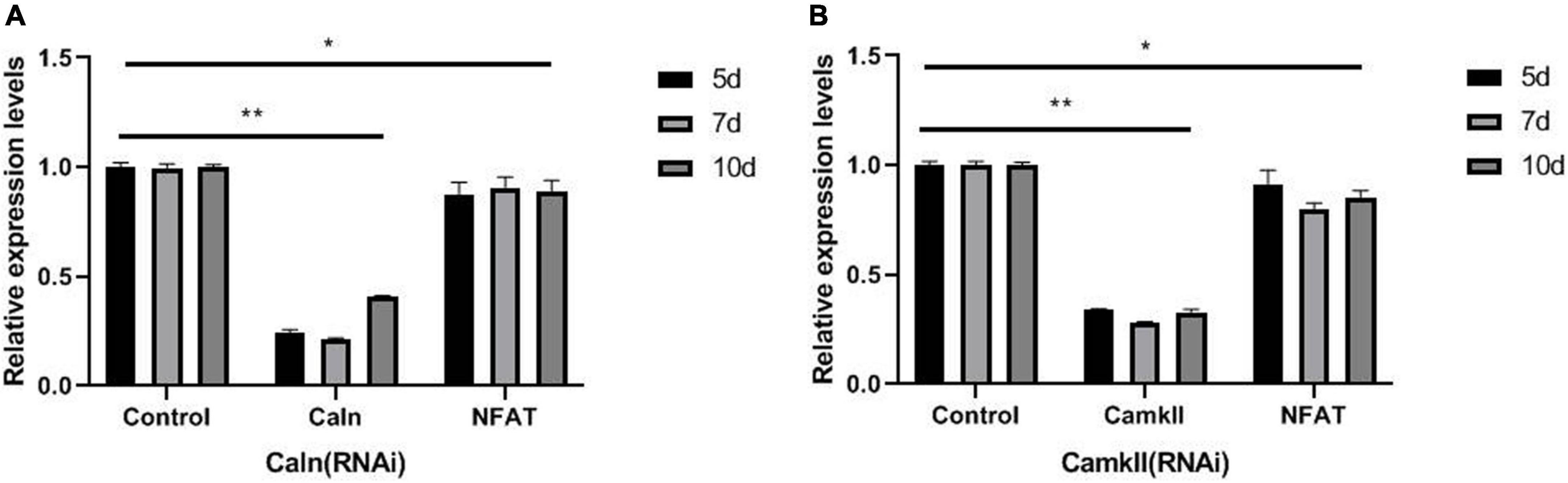
Figure 3. DjNFAT mRNA levels in DjCaln (RNAi) and DjCamkII (RNAi) during planarian head regeneration. (A) DjNFAT mRNA levels in DjCaln (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 4. (B) DjNFAT mRNA levels in DjCamkII (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 4. *p < 0.05, **p < 0.01.

Figure 4. DjNFAT regulates DjCaln and DjCamkII by feedback positive regulation during planarian head regeneration. (A) DjCaln and DjCamkII mRNA level in DjNFAT (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 4. (B,C) Relative DjCaln protein level in DjNFAT (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 5. (D,E) Relative DjCamkII protein level in DjNFAT (RNAi) planarians at 5, 7, and 10 days during planarian head regeneration, n = 5, *P < 0.05, **P < 0.01. C, control; A, DjNFAT (RNAi).
During the regeneration, DjCamKII and DjCaln were abundant in the regenerated brain (Zhen et al., 2020). To determine whether DjNFAT was involved in brain regeneration, the expression of DjNFAT was examined using WISH in decapitated animals as they formed a new brain. The results showed that, in intact planarians, DjNFAT was mainly expressed in the pharynx and parenchymal cells (Figure 5A). In head regeneration, at 1 and 3 days into regeneration, the positive signal of DjNFAT was predominantly detected at the wound (Figures 5C,D). Then the primordium of the brain continued to change as the regenerated brain began to undergo a pattern formation, in which a positive signal of DjNFAT was detected at 5 days of regeneration (Figure 5E). The regeneration of the brain was basically completed at 7 days of regeneration, and the DjNFAT positive signal was further strengthened (Figure 5F). At 10 days of regeneration, the DjNFAT positive signal of the regenerated brain reached the strongest level, and the regeneration of the lateral nerves was complete (Figure 5G). Using qPCR, changes in the expression of DjNFAT mRNA during the head regenerative process were detected, and the results showed that over time, the expression of DjNFAT gradually increased, peaking at 10 days of regeneration, which was consistent with the results of WISH (P < 0.01) (Figure 5B). In general, the above results indicated that DjNFAT was widely expressed in intact and regenerative planarians.

Figure 5. Expression patterns of DjNFAT in the intact and regenerating planarians after decapitation. (A) Dorsal view of an intact planarian shows an abundant expression in the pharynx and parenchymal cell (arrows). (B) qPCR analysis of DjNFAT expressions in the regenerating fragments at 0, 1, 3, 5, 7, and 10 days during planarian head regeneration, n = 4, *P < 0.05, **P < 0.01. d, day. (C–G) WISH showing the expressions of DjNFAT in regenerating planarians at 1, 3, 5, 7, and 10 days during planarian head regeneration (arrows), n = 6. Scale bars: 0.5 mm.
Trunk fragments of control (RNAi) and DjNFAT (RNAi) planarians were stained with anti-phosphohistone H3 (H3P) on days 1, 3, and 5 of regeneration (Brown et al., 2018). The number of dividing cells during regeneration was determined and no significant differences were found between control (RNAi) and DjNFAT (RNAi), suggesting that a gross proliferation defect was not present (Supplementary Figure 2). DjNFAT (RNAi) regenerating trunk fragments showed a normal regionalized expression of ston2 (Supplementary Figure 1; Zeng et al., 2018), suggesting that DjNFAT did not affect neoblast proliferation and brain patterning.
The knockdown of DjCamKII and DjCaln led to defects in regenerated brains including incompact phenotypes at the posterior of the new brain, and lateral branches that could not regenerate (Zhen et al., 2020). To study the function of the DjNFAT gene in brain regeneration, RNAi was carried out by socking technique commonly used in planarians research. Immunostaining with an anti-SYNORF1 antibody against synapsin (Cebria, 2008) revealed that DjNFAT-RNAi treatment caused apparent brain defects at 7 and 10 days of regeneration (Figure 6). Compared to control animals (Figures 6A–C), DjNFAT-RNAi regenerated animals were found that the regenerative brain became small and the lateral nerves cannot be regenerated completely (Figures 6D–F). Hence, these results implied that DjNFAT was required for the formation of a functional brain during planarian head regeneration.
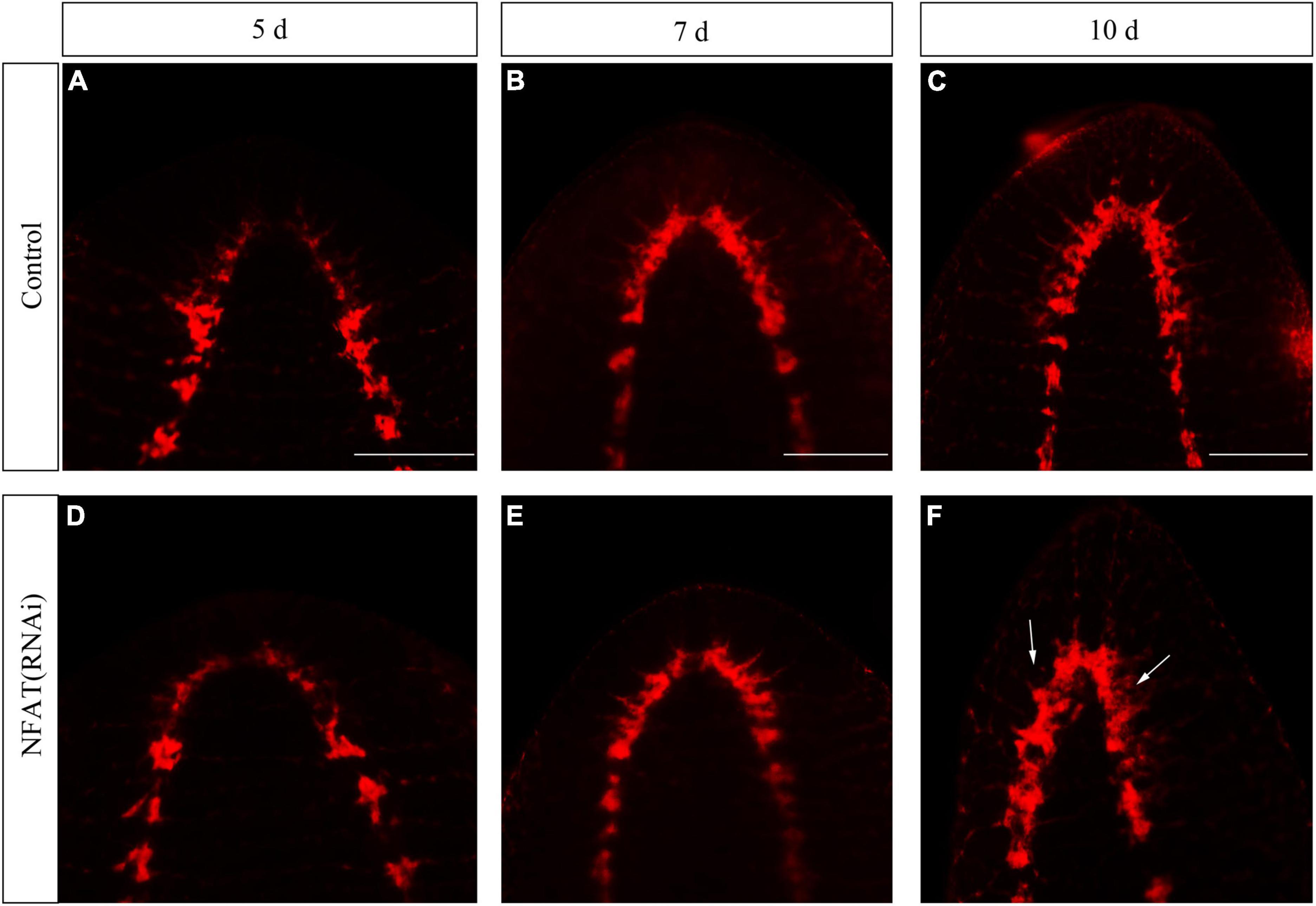
Figure 6. Immunostaining with anti-SYNORF1 in DjNFAT(RNAi) treated planarians during planarian head regeneration. (A–C) Normal regenerated brains (control), n = 5. (D–F) The phenotypes of the regenerated brain in the DjNFAT(RNAi) treated planarians at 5, 7, and 10 days after amputation, n = 6. Anterior is at the front. Scale bars: 0.5 mm.
To confirm whether or not DjNFAT might affect the number of certain types of neurons at 5, 7, and 10 days during neurogenesis, the neurotransmitter synthesizing enzymes were used as the neuron markers in FISH to measure the numbers of neurons in decapitated planarians after DjNFAT-RNAi treatment. The results revealed that the numbers of choline acetyltransferase (DjChat, cholinergic neurons), tyrosine hydroxylase (DjTH, dopaminergic neurons), tryptophan hydroxylase (DjTPH, serotonergic neurons) positive-neurons were similar to the controls in the regenerated planarians, suggesting that DjNFAT-RNAi could not influence the regeneration of planarian cholinergic, dopaminergic or serotonergic neurons (Supplementary Figure 3).
As previously presented, knockdown of DjCamKII or DjCaln led to GABAergic neurons decreased during planarian head regeneration (Zhen et al., 2020). To confirm whether DjNFAT also affected the number of GABAergic neurons at 5, 7, and 10 days during neurogenesis, WISH was used to measure the number of GABAergic neurons after DjNFAT (RNAi), with the help of neurotransmitter synthase GAD as GABAergic neuron marker. Compared with the control group, the number of DjGAD-positive neurons in planarian regenerated significantly decreased (Figures 7A,B). FISH analysis showed that the number of DjGAD-expressing cells in the newborn brain area was significantly reduced at 5, 7, and 10 days after DjNFAT RNAi (Figure 7B). The expression level of DjGAD protein in DjNFAT (RNAi) planarians was detected by Western blot. Compared with control animals, the results showed that DjGAD protein decreased in the regeneration period (Figures 7C,D), which indicated that DjNFAT may affect the number of GABAergic neurons during planarian head regeneration through transcriptional regulation of GAD.
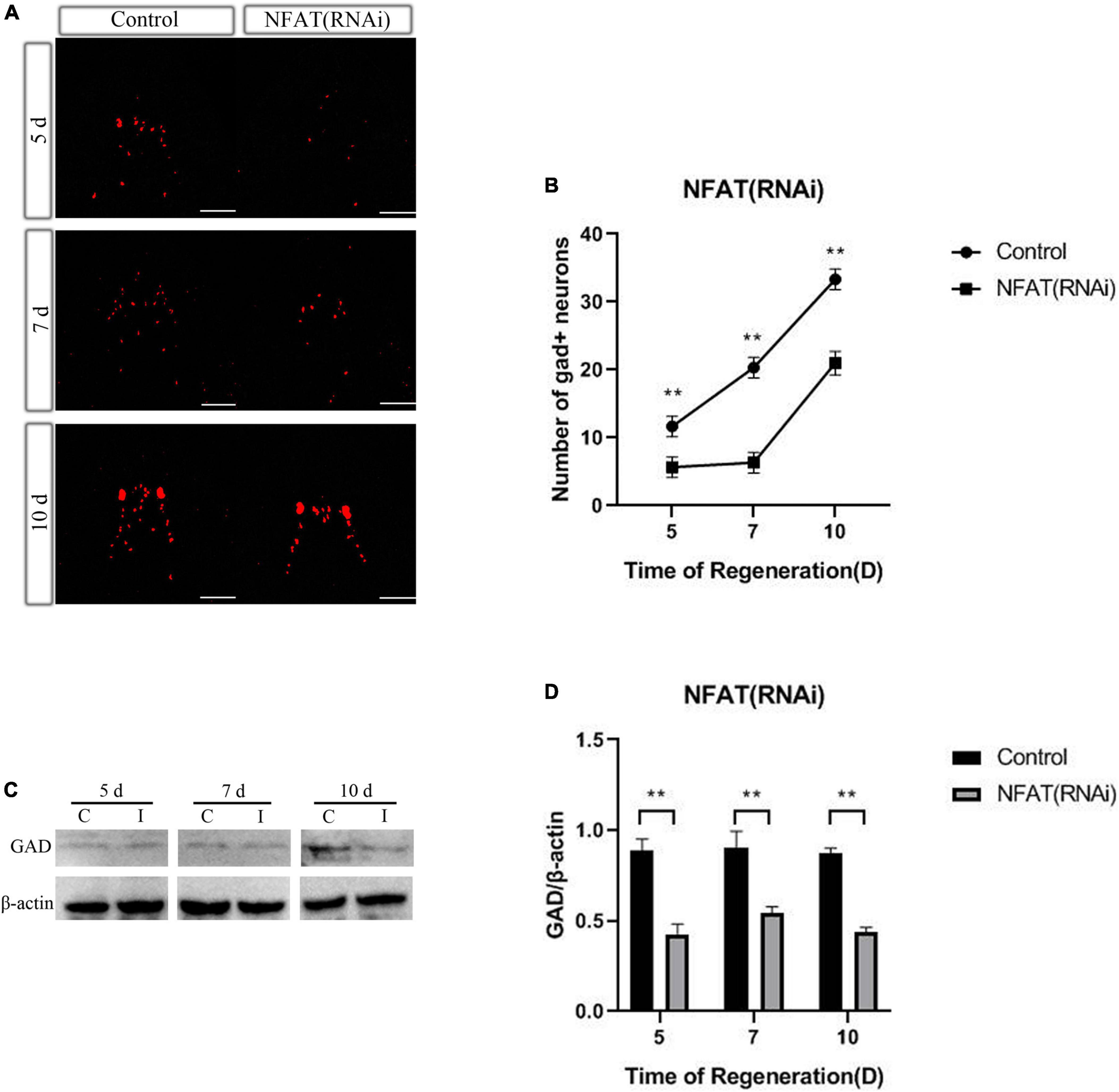
Figure 7. DjNFAT inhibition influences the number of GABAergic neurons during planarian head regeneration. (A) FISH showing the expression of DjGAD in regenerating planarians at 5, 7, and 10 days after amputation, n = 5. Scale bars: 0.2 mm. (B) The number of DjGAD-expressing neurons. Error bars represent the SD (**P < 0.01). Anterior is at the front. d, day. (C) Western blotting results of DjGAD protein expression in the regenerated head of control and DjNFAT-RNAi planarians, n = 4. (D) The expression levels of GAD protein in regenerated planarians. C, control; I, DjNFAT (RNAi).
The most remarkable characteristic of the planarian resides in its strong regeneration ability. The planarian brain comprises of different types of neurons, including the same type of neurons found in humans, such as dopaminergic, cholinergic, GABAergic, and serotonergic neurons (Nishimura et al., 2007a,b, 2008, 2010). As such, understanding the neural regeneration mechanism of planarians can shed light on how humans make and reintegrate new neurons. As previously explained, down-regulation of DjCamKII or DjCaln led to the number of GABAergic neurons decreased during planarian head regeneration (Zhen et al., 2020). In this study, the mechanism by which Wnt/Ca2+ signaling pathway regulates the formation of GABAergic neurons during planarian neurogenesis was examined. The relationship between DjCamKII and DjCaln suggests a mutual negative regulation during planarian head regeneration. Additionally, knockdown of DjCaln or DjCamKII reduced the expression of DjNFAT, while knockdown of DjNFAT could reduce both the mRNA and protein expressions of DjCaln and DjCamKII. DjNFAT exerted feedback positive regulation on both DjCaln and DjCamkII. DjNFAT was predominantly expressed in the pharynx and parenchymal cells of the intact planarian, but located in the newborn brain during planarian head regeneration. Down-regulation of DjNFAT affected the number of GABAergic neurons and the expression level of DjGAD protein during planarian head regeneration. These findings suggest that the feedback loop between DjCaln, DjCamkII, and DjNFAT is essential for the number of GABAergic neurons during planarian head regeneration.
Previous studies demonstrated that CamKII can mediate phosphorylation of Caln (Hashimoto et al., 1988; Hashimoto and Soderling, 1989; Martensen et al., 1989) and CaMKIIδc can directly phosphorylate Caln to negatively regulate Calcineurin/NFAT signaling in cardiac myocytes (MacDonnell et al., 2009). In budding yeast, the lack of CMK2 up-regulated the level of calcium/calcineurin signaling and augmented the expression levels of Crz1-dependent PMR1 and PMC1 (Xu et al., 2019). In Human T Cells, CaMKII down-regulated both Calcineurin and Protein Kinase C mediated pathways for cytokine gene transcription (Hama et al., 1995). Consistent with previous reports, down-regulation of DjCaln led to increase in the expressions of DjCamkII mRNA and protein at 5, 7, and 10 days during planarian head regeneration. Down-regulation of DjCamkII also led to the increase in the expression of DjCaln mRNA and protein decreased at 5, 7, and 10 days during planarian head regeneration (Figures 1, 2). These results indicated that DjCamKII and DjCaln suggest a mutual negative regulation during planarian head regeneration. Knockdown of DjCaln or DjCamKII reduced the expression of DjNFAT, while the expression of DjCamkII and DjCaln decreased after down-regulation of DjNFAT (Figures 3, 4), which was a feedback positive regulation of DjCamkII and DjCaln by DjNFAT. These results indicated that DjNFAT exerted feedback positive regulation on both DjCaln and DjCamkII during planarian head regeneration (Figure 8).
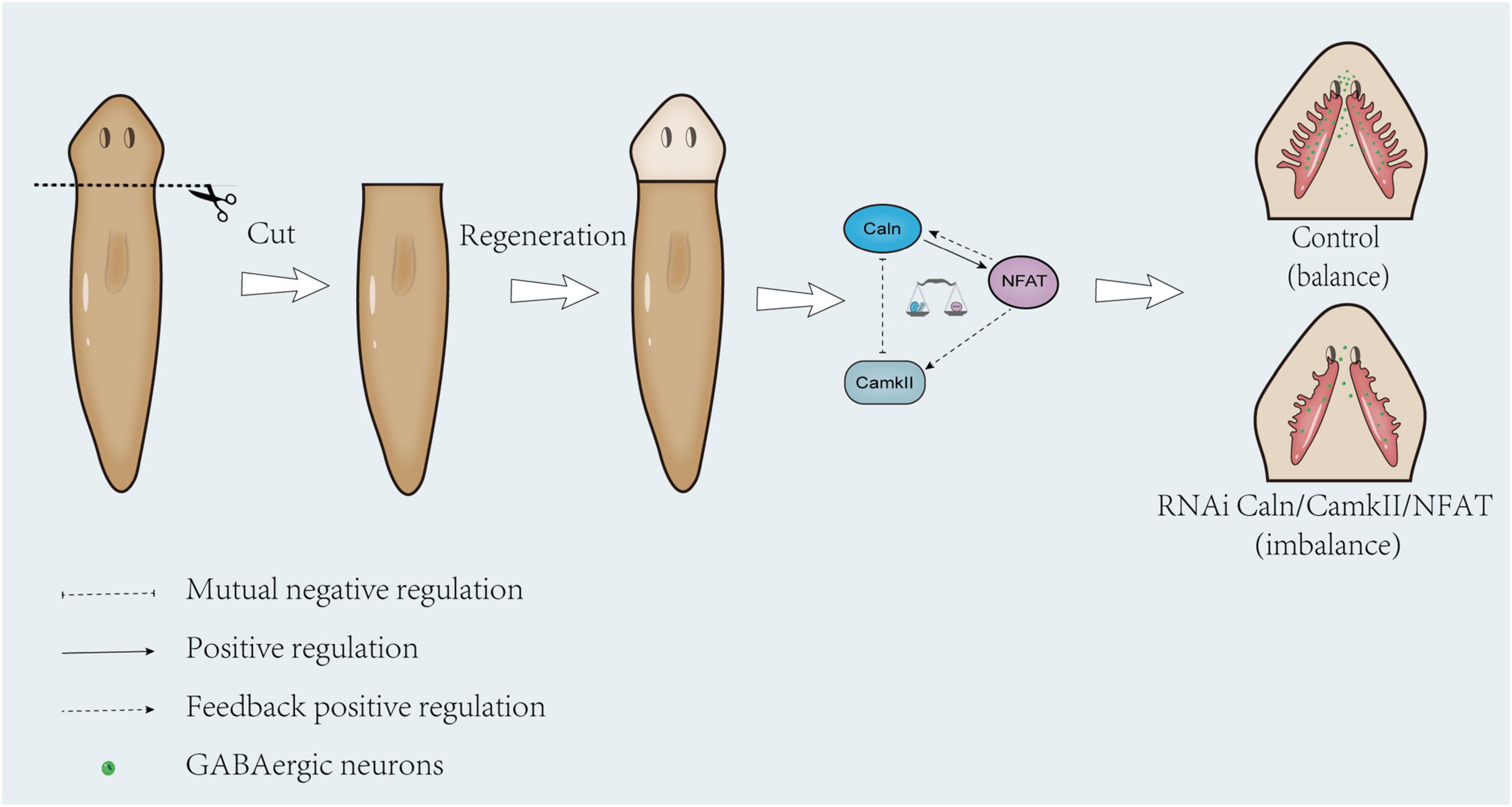
Figure 8. Proposed model illustrating feedback loop between DjCaln, DjCamkII, and DjNFAT that regulates the number of GABAergic neurons during planarian head regeneration. In the control group, the mutual regulation of DjCaln, DjCamkII, and DjNFAT was in balance, and the GABAergic neurons could be normal formed during the planarian head regeneration. In the absence of adequate regulation of DjCaln or DjCamkII after DjNFAT RNAi, the number of GABAergic neurons would be reduced during planarian head regeneration.
The NFAT protein is part of the family of transcription factors, which usually exists in a hyperphosphorylated state in the cytoplasm (Shaw et al., 1988; Schulz and Yutzey, 2004; Boothby, 2010). The NFATs play a role in many vertebrate developmental systems, including the nervous system (Crabtree and Olson, 2002; Schulz and Yutzey, 2004; Macian, 2005). NFATs (NFATc1–c4) are expressed in neurons (Canellada et al., 2008). For example, NFATc3 and NFATc4 are highly expressed in primary hippocampal neurons (Vihma et al., 2016). The transcription factor Nfat5 was expressed in the anterior blastema of the tail fragments after 3 days into regeneration in planarian S. mediterranea (Suzuki-Horiuchi et al., 2021). In contrast to previous studies, in this paper, the results showed that DjNFAT was mainly expressed in the pharynx and parenchymal cells in intact planarians (Figure 5A). During planarian head regeneration, DjNFAT was located in the blastema at 3 days and perceptible in the brain at 10 days during head regeneration in planarian D. japonica (Figures 5C–G). As previously mentioned, DjNFAT was difficult to detect in the brain under homeostatic conditions. However, its expression became readily apparent soon after amputation, indicating that DjNFAT might participate in newborn brain regeneration.
Studies found that calcineurin and NFAT are essential for neuregulin and ErbB signaling, neural crest diversification, and differentiation of Schwann cells (Kao et al., 2009). Caln/NFAT4 signaling pathway is a key mechanism for the disruption of synaptic remodeling and homeostasis in the hippocampus after acute injury (Furman et al., 2016). Caln/NFAT is also essential in driving glutamate dysregulation and neuronal hyperactivity during AD (Sompol et al., 2017). Moreover, Caln/NFAT signaling plays an important role in the shaping of the synaptic connectivity of thalamocortical and nucleus reticularis thalami GABAergic neurons mediated by slow-wave sleep (Pigeat et al., 2015). As previously described, knockdown of DjCaln or DjCamkII led to regenerated brains defects including partial deletions and lateral branches not regenerated. And the number of GABAergic neurons also decreased during planarian head regeneration (Zhen et al., 2020). In this study, down-regulation of DjNFAT caused defects in regenerated brains including newborn brain small and lateral nerve cannot regenerate (Figure 6). The number of GABAergic neurons in DjNFAT (RNAi) planarians was significantly reduced (Figures 7A,B), which is consistent with DjCaln-RNAi or DjCamkII-RNAi animals (Zhen et al., 2020). In addition, the expression level of DjGAD protein in DjNFAT knockdown planarians was down-regulated (Figures 7C,D), which means that DjNFAT could regulate the GABAergic neurons during planarian head regeneration. Thus, we conclude that the feedback loop between DjCaln, DjCamkII, and DjNFAT regulates the number of GABAergic neurons during planarian head regeneration (Figure 8).
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
BZ designed this work. BZ and HZ wrote the manuscript. HZ performed the experiments. HZ, MZ, HG, QS, LG, ZY, HD, QP, and BZ analyzed the data. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant number: 31970430).
We thank Wang for its valuable linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.988803/full#supplementary-material
Supplementary Figure 1 | The expression of DjSton2 in control and DjNFAT(RNAi) regenerated planarians visualized by FISH.
Supplementary Figure 2 | Immunostaining of phosphohistone-H3 on days 1, 3, and 5 of regeneration in control and DjNFAT(RNAi) regenerated planarians.
Supplementary Figure 3 | The different neuronal types in control and DjNFAT(RNAi) regenerated planarians visualized by FISH. (A) FISH of neuronal populations that regenerate properly after DjNFAT(RNAi). Cholinergic neurons (Chat). (B) FISH of neuronal populations that regenerate properly after DjNFAT(RNAi). Dopaminergic neurons (TH). (C) FISH of neuronal populations that regenerate properly after DjNFAT(RNAi). Serotonergic neurons (TPH). (n = 10). Scale bars = 0.2 mm. Anterior is at the front. ns, no significant.
Boothby, M. (2010). CRACking the code without Rosetta: Molecular regulation of calcium-stimulated gene transcription after T cell activation. J. Immunol. 185, 4969–4971. doi: 10.4049/jimmunol.1090097
Bram, R. J., Hung, D. T., Martin, P. K., Schreiber, S. L., and Crabtree, G. R. (1993). Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: Roles of calcineurin binding and cellular location. Mol. Cell Biol. 13, 4760–4769. doi: 10.1128/mcb.13.8.4760-4769.1993
Brown, D. D. R., Molinaro, A. M., and Pearson, B. J. (2018). The planarian TCF/LEF factor Smed-tcf1 is required for the regeneration of dorsal-lateral neuronal subtypes. Dev Biol 433, 374–383. doi: 10.1016/j.ydbio.2017.08.024
Cameron, H. A., and McKay, R. D. (2001). Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J. Comp. Neurol. 435, 406–417. doi: 10.1002/cne.1040
Canellada, A., Ramirez, B. G., Minami, T., Redondo, J. M., and Cano, E. (2008). Calcium/calcineurin signaling in primary cortical astrocyte cultures: Rcan1-4 and cyclooxygenase-2 as NFAT target genes. Glia 56, 709–722. doi: 10.1002/glia.20647
Cardenas, A. M., and Marengo, F. D. (2016). How the stimulus defines the dynamics of vesicle pool recruitment, fusion mode, and vesicle recycling in neuroendocrine cells. J Neurochem 137, 867–879. doi: 10.1111/jnc.13565
Cebria, F. (2008). Organization of the nervous system in the model planarian Schmidtea mediterranea: An immunocytochemical study. Neurosci Res 61, 375–384. doi: 10.1016/j.neures.2008.04.005
Chan, J. D., Zhang, D., Liu, X., Zarowiecki, M., Berriman, M., and Marchant, J. S. (2017). Utilizing the planarian voltage-gated ion channel transcriptome to resolve a role for a Ca(2+) channel in neuromuscular function and regeneration. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1036–1045. doi: 10.1016/j.bbamcr.2016.10.010
Cowles, M. W., Hubert, A., Zayas, R. M., and Lissencephaly-, A. (2012). 1 homologue is essential for mitotic progression in the planarian Schmidtea mediterranea. Dev. Dyn. 241, 901–910. doi: 10.1002/dvdy.23775
Crabtree, G. R., and Olson, E. N. (2002). NFAT signaling: Choreographing the social lives of cells. Cell 109(Suppl), S67–S79. doi: 10.1016/s0092-8674(02)00699-2
Elliott, S. A., and Sanchez Alvarado, A. (2013). The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2, 301–326. doi: 10.1002/wdev.82
Eriksson, P. S., Perfilieva, E., Bjork-Eriksson, T., Alborn, A. M., Nordborg, C., Peterson, D. A., et al. (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317.
Fraguas, S., Carcel, S., Vivancos, C., Molina, M. D., Gines, J., Mazariegos, J., et al. (2021). CREB-binding protein (CBP) gene family regulates planarian survival and stem cell differentiation. Dev. Biol. 476, 53–67.
Furman, J. L., Sompol, P., Kraner, S. D., Pleiss, M. M., Putman, E. J., Dunkerson, J., et al. (2016). Blockade of astrocytic calcineurin/NFAT signaling helps to normalize hippocampal synaptic function and plasticity in a rat model of traumatic brain injury. J. Neurosci. 36, 1502–1515. doi: 10.1523/JNEUROSCI.1930-15.2016
Gajcy, K., Lochynski, S., and Librowski, T. (2010). A role of GABA analogues in the treatment of neurological diseases. Curr. Med. Chem. 17, 2338–2347.
Galvan, A., and Wichmann, T. (2007). GABAergic circuits in the basal ganglia and movement disorders. Prog. Brain Res. 160, 287–312.
Gao, L., Han, Y., Deng, H., Hu, W., Zhen, H., Li, N., et al. (2017). The role of a novel C-type lectin-like protein from planarian in innate immunity and regeneration. Dev. Comp. Immunol. 67, 413–426. doi: 10.1016/j.dci.2016.08.010
Goto, S., Matsukado, Y., Mihara, Y., Inoue, N., and Miyamoto, E. (1986). The distribution of calcineurin in rat brain by light and electron microscopic immunohistochemistry and enzyme-immunoassay. Brain Res. 397, 161–172. doi: 10.1016/0006-8993(86)91381-8
Hama, N., Paliogianni, F., Fessler, B. J., and Boumpas, D. T. (1995). Calcium/calmodulin-dependent protein kinase II downregulates both calcineurin and protein kinase C-mediated pathways for cytokine gene transcription in human T cells. J. Exp. Med. 181, 1217–1222. doi: 10.1084/jem.181.3.1217
Hashimoto, Y., and Soderling, T. R. (1989). Regulation of calcineurin by phosphorylation. Identification of the regulatory site phosphorylated by Ca2+/calmodulin-dependent protein kinase II and protein kinase C. J. Biol. Chem. 264, 16524–16529.
Hashimoto, Y., King, M. M., and Soderling, T. R. (1988). Regulatory interactions of calmodulin-binding proteins: Phosphorylation of calcineurin by autophosphorylated Ca2+/calmodulin-dependent protein kinase II. Proc. Natl. Acad. Sci. U.S.A. 85, 7001–7005.
Hayama, T., Noguchi, J., Watanabe, S., Takahashi, N., Hayashi-Takagi, A., Ellis-Davies, G. C., et al. (2013). GABA promotes the competitive selection of dendritic spines by controlling local Ca2+ signaling. Nat. Neurosci. 16, 1409–1416. doi: 10.1038/nn.3496
Hill, E. M., and Petersen, C. P. (2015). Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142, 4217–4229. doi: 10.1242/dev.123612
Ho, C., Teo, T. S., Desai, R., and Wang, J. H. (1976). Catalytic and regulatory properties of two forms of bovine heart cyclic nucleotide phosphodiesterase. Biochim. Biophys. Acta 429, 461–473. doi: 10.1016/0005-2744(76)90294-1
Kao, S. C., Wu, H., Xie, J., Chang, C. P., Ranish, J. A., Graef, I. A., et al. (2009). Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science 323, 651–654. doi: 10.1126/science.1166562
Kleppner, S. R., and Tobin, A. J. (2001). GABA signalling: Therapeutic targets for epilepsy, Parkinson’s disease and Huntington’s disease. Expert Opin. Ther. Targets 5, 219–239. doi: 10.1517/14728222.5.2.219
Kuno, T., Mukai, H., Ito, A., Chang, C. D., Kishima, K., Saito, N., et al. (1992). Distinct cellular expression of calcineurin A alpha and A beta in rat brain. J. Neurochem. 58, 1643–1651. doi: 10.1111/j.1471-4159.1992.tb10036.x
Li, H., Rao, A., and Hogan, P. G. (2011). Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103.
Lu, Q., Wu, S., Zhen, H., Deng, H., Song, Q., Ma, K., et al. (2017). 14-3-3 alpha and 14-3-3 zeta contribute to immune responses in planarian Dugesia japonica. Gene 615, 25–34. doi: 10.1016/j.gene.2017.03.017
MacDonnell, S. M., Weisser-Thomas, J., Kubo, H., Hanscome, M., Liu, Q., Jaleel, N., et al. (2009). CaMKII negatively regulates calcineurin-NFAT signaling in cardiac myocytes. Circ. Res. 105, 316–325. doi: 10.1161/CIRCRESAHA.109.194035
Macian, F. (2005). NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484. doi: 10.1038/nri1632
Martensen, T. M., Martin, B. M., and Kincaid, R. L. (1989). Identification of the site on calcineurin phosphorylated by Ca2+/CaM-dependent kinase II: Modification of the CaM-binding domain. Biochemistry 28, 9243–9247. doi: 10.1021/bi00450a002
Mehta, S., and Zhang, J. (2014). Using a genetically encoded FRET-based reporter to visualize calcineurin phosphatase activity in living cells. Methods Mol. Biol. 1071, 139–149. doi: 10.1007/978-1-62703-622-1_11
Mehta, S., Li, H., Hogan, P. G., and Cunningham, K. W. (2009). Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol. Cell Biol. 29, 2777–2793. doi: 10.1128/MCB.01197-08
Muller, M. R., Sasaki, Y., Stevanovic, I., Lamperti, E. D., Ghosh, S., Sharma, S., et al. (2009). Requirement for balanced Ca/NFAT signaling in hematopoietic and embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 7034–7039. doi: 10.1073/pnas.0813296106
Newmark, P. A., and Sanchez Alvarado, A. (2002). Not your father’s planarian: A classic model enters the era of functional genomics. Nat. Rev. Genet. 3, 210–219. doi: 10.1038/nrg759
Nishimura, K., Kitamura, Y., Inoue, T., Umesono, Y., Sano, S., Yoshimoto, K., et al. (2007a). Reconstruction of dopaminergic neural network and locomotion function in planarian regenerates. Dev. Neurobiol. 67, 1059–1078. doi: 10.1002/dneu.20377
Nishimura, K., Kitamura, Y., Inoue, T., Umesono, Y., Yoshimoto, K., Takeuchi, K., et al. (2007b). Identification and distribution of tryptophan hydroxylase (TPH)-positive neurons in the planarian Dugesia japonica. Neurosci. Res. 59, 101–106. doi: 10.1016/j.neures.2007.05.014
Nishimura, K., Kitamura, Y., Taniguchi, T., and Agata, K. (2010). Analysis of motor function modulated by cholinergic neurons in planarian Dugesia japonica. Neuroscience 168, 18–30. doi: 10.1016/j.neuroscience.2010.03.038
Nishimura, K., Kitamura, Y., Umesono, Y., Takeuchi, K., Takata, K., Taniguchi, T., et al. (2008). Identification of glutamic acid decarboxylase gene and distribution of GABAergic nervous system in the planarian Dugesia japonica. Neuroscience 153, 1103–1114. doi: 10.1016/j.neuroscience.2008.03.026
Nogi, T., Zhang, D., Chan, J. D., and Marchant, J. S. (2009). A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel beta subunits: Subversion of flatworm regenerative polarity. PLoS Negl. Trop Dis. 3:e464. doi: 10.1371/journal.pntd.0000464
Orii, H., Mochii, M., and Watanabe, K. (2003). A simple “soaking method” for RNA interference in the planarian Dugesia japonica. Dev. Genes Evol. 213, 138–141.
Pigeat, R., Chausson, P., Dreyfus, F. M., Leresche, N., and Lambert, R. C. (2015). Sleep slow wave-related homo and heterosynaptic LTD of intrathalamic GABAAergic synapses: Involvement of T-type Ca2+ channels and metabotropic glutamate receptors. J. Neurosci. 35, 64–73. doi: 10.1523/JNEUROSCI.2748-14.2015
Quadrato, G., Elnaggar, M. Y., Duman, C., Sabino, A., Forsberg, K., and Di Giovanni, S. (2014). Modulation of GABAA receptor signaling increases neurogenesis and suppresses anxiety through NFATc4. J. Neurosci. 34, 8630–8645. doi: 10.1523/JNEUROSCI.0047-14.2014
Schulz, R. A., and Yutzey, K. E. (2004). Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev. Biol. 266, 1–16.
Serrano-Perez, M. C., Fernandez, M., Neria, F., Berjon-Otero, M., Doncel-Perez, E., Cano, E., et al. (2015). NFAT transcription factors regulate survival, proliferation, migration, and differentiation of neural precursor cells. Glia 63, 987–1004. doi: 10.1002/glia.22797
Shaw, J. P., Utz, P. J., Durand, D. B., Toole, J. J., Emmel, E. A., and Crabtree, G. R. (1988). Identification of a putative regulator of early T cell activation genes. Science 241, 202–205.
Sompol, P., Furman, J. L., Pleiss, M. M., Kraner, S. D., Artiushin, I. A., Batten, S. R., et al. (2017). Calcineurin/NFAT signaling in activated astrocytes drives network hyperexcitability in abeta-bearing mice. J. Neurosci. 37, 6132–6148. doi: 10.1523/JNEUROSCI.0877-17.2017
Stefos, G. C., Soppa, U., Dierssen, M., and Becker, W. (2013). NGF upregulates the plasminogen activation inhibitor-1 in neurons via the calcineurin/NFAT pathway and the Down syndrome-related proteins DYRK1A and RCAN1 attenuate this effect. PLoS One 8:e67470. doi: 10.1371/journal.pone.0067470
Suzuki-Horiuchi, Y., Schmitz, H., Barlassina, C., Eccles, D., Sinn, M., Ortmeier, C., et al. (2021). Transcription factors active in the anterior blastema of Schmidtea mediterranea. Biomolecules 11:1782. doi: 10.3390/biom11121782
Toni, N., Teng, E. M., Bushong, E. A., Aimone, J. B., Zhao, C., Consiglio, A., et al. (2007). Synapse formation on neurons born in the adult hippocampus. Nat. Neurosci. 10, 727–734.
Toyoda, H., Saito, M., Sato, H., Tanaka, T., Ogawa, T., Yatani, H., et al. (2015). Enhanced desensitization followed by unusual resensitization in GABAA receptors in phospholipase C-related catalytically inactive protein-1/2 double-knockout mice. Pflugers Arch. 467, 267–284. doi: 10.1007/s00424-014-1511-5
van Praag, H., Schinder, A. F., Christie, B. R., Toni, N., Palmer, T. D., and Gage, F. H. (2002). Functional neurogenesis in the adult hippocampus. Nature 415, 1030–1034.
Vihma, H., Luhakooder, M., Pruunsild, P., and Timmusk, T. (2016). Regulation of different human NFAT isoforms by neuronal activity. J Neurochem 137, 394–408.
Wang, C. Y., Lin, H. C., Song, Y. P., Hsu, Y. T., Lin, S. Y., Hsu, P. C., et al. (2015). Protein kinase C-dependent growth-associated protein 43 phosphorylation regulates gephyrin aggregation at developing GABAergic synapses. Mol. Cell Biol. 35, 1712–1726. doi: 10.1128/MCB.01332-14
Wang, J. H., and Desai, R. (1976). A brain protein and its effect on the Ca2+-and protein modulator-activated cyclic nucleotide phosphodiesterase. Biochem. Biophys. Res. Commun. 72, 926–932. doi: 10.1016/s0006-291x(76)80220-3
Watanabe, M., Maemura, K., Kanbara, K., Tamayama, T., and Hayasaki, H. (2002). GABA and GABA receptors in the central nervous system and other organs. Int. Rev. Cytol. 213, 1–47.
Xu, H., Fang, T., Yan, H., and Jiang, L. (2019). The protein kinase Cmk2 negatively regulates the calcium/calcineurin signalling pathway and expression of calcium pump genes PMR1 and PMC1 in budding yeast. Cell Commun. Signal. 17:7. doi: 10.1186/s12964-019-0320-z
Zeng, A., Li, H., Guo, L., Gao, X., McKinney, S., Wang, Y., et al. (2018). Prospectively isolated Tetraspanin(+) neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593–1608.e20. doi: 10.1016/j.cell.2018.05.006
Zhang, D., Chan, J. D., Nogi, T., and Marchant, J. S. (2011). Opposing roles of voltage-gated Ca2+ channels in neuronal control of regenerative patterning. J. Neurosci. 31, 15983–15995. doi: 10.1523/JNEUROSCI.3029-11.2011
Zhao, C., Deng, W., and Gage, F. H. (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660.
Keywords: Caln, CamkII, NFAT, GABAergic neurons, planarian, regeneration
Citation: Zhen H, Zheng M, Geng H, Song Q, Gao L, Yuan Z, Deng H, Pang Q and Zhao B (2022) The feedback loop between calcineurin, calmodulin-dependent protein kinase II, and nuclear factor of activated T-cells regulates the number of GABAergic neurons during planarian head regeneration. Front. Mol. Neurosci. 15:988803. doi: 10.3389/fnmol.2022.988803
Received: 07 July 2022; Accepted: 24 August 2022;
Published: 12 September 2022.
Edited by:
Artem Mikhailovich Kosenkov, Institute of Cell Biophysics (RAS), RussiaCopyright © 2022 Zhen, Zheng, Geng, Song, Gao, Yuan, Deng, Pang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bosheng Zhao, emhhb2Jvc2hlbmdAc2R1dC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.