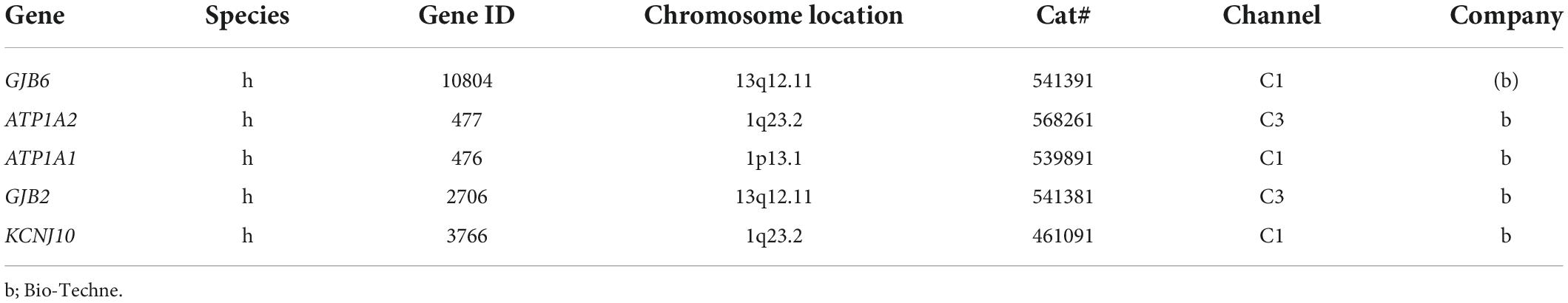
- Department of Surgical Sciences, Head and Neck Surgery, Section of Otolaryngology, Uppsala University Hospital, Uppsala, Sweden
Background: Gap junction (GJ) proteins, connexin26 and 30, are highly prevalent in the human cochlea (HC), where they are involved in transcellular signaling, metabolic supply, and fluid homeostasis. Their genes, GJB2 and GJB6, are both located at the DFNB1 locus on chromosome 13q12. Mutations in GJB2 may cause mild to profound non-syndromic deafness. Here, we analyzed for the first time the various expressions of GJB2 and GJB6 gene transcripts in the different cell networks in the HC using the RNAscope technique.
Materials and methods: Archival paraformaldehyde-fixed sections of surgically obtained HC were used to label single mRNA oligonucleotides using the sensitive multiplex RNAscope® technique with fluorescent-tagged probes. Positive and negative controls also included the localization of ATP1A1, ATP1A2, and KCNJ10 gene transcripts in order to validate the specificity of labeling.
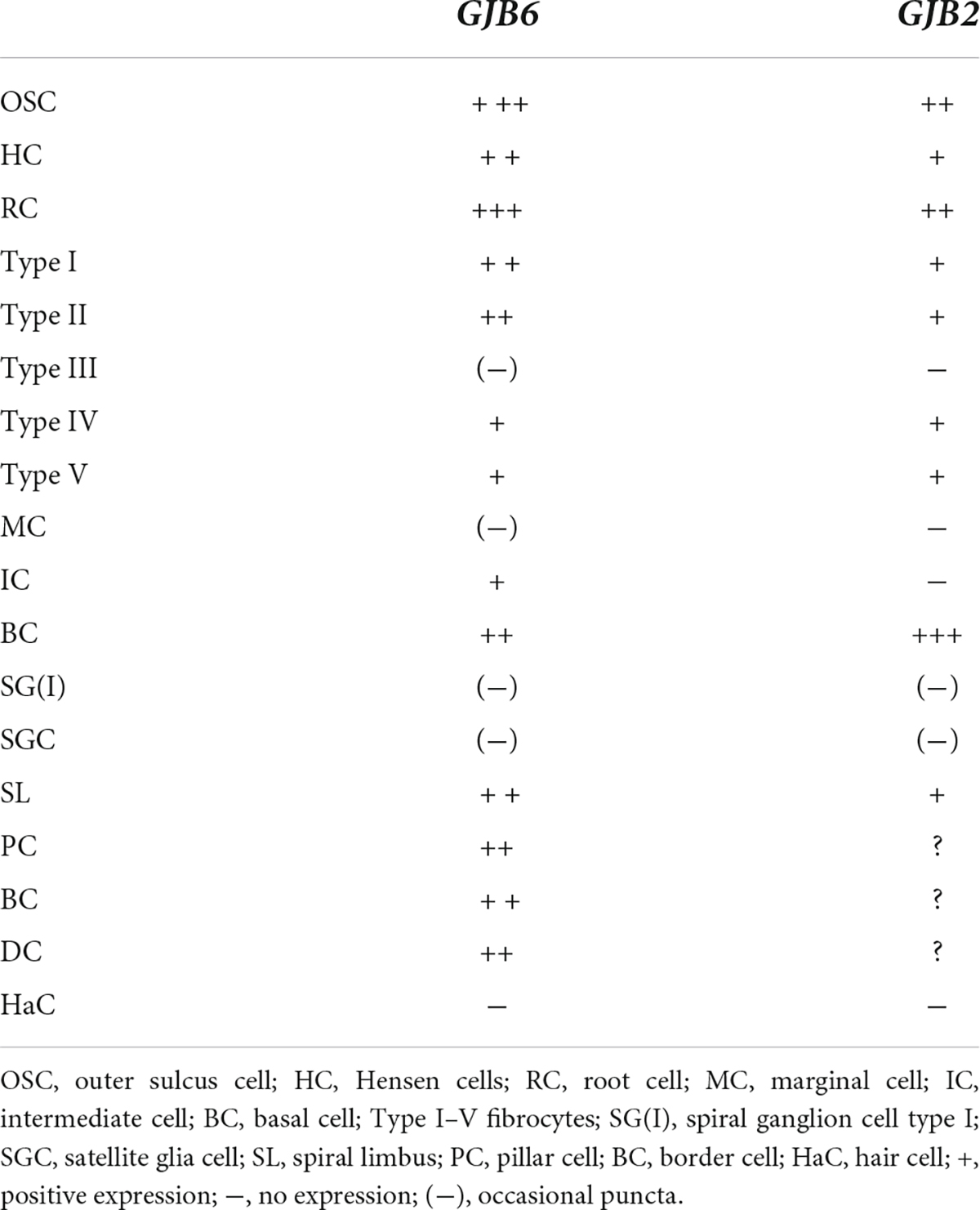
Results: Confocal and super-resolution structured illumination microscopy (SR-SIM) detected single gene transcripts as brightly stained puncta. The GJB2 and GJB6 gene transcripts were distributed in the epithelial and connective tissue systems in all three cochlear turns. The largest number of GJB2 and GJB6 gene transcripts was in the outer sulcus, spiral ligament, and stria vascularis (SV). Oligonucleotides were present in the supporting cells of the organ of Corti (OC), spiral limbus fibrocytes, and the floor of the scala vestibuli. Multiplex gene data suggest that cells in the cochlear lateral wall contain either GJB2 or GJB6 gene transcripts or both. The GJB6, but not GJB2, gene transcripts were found in the intermediate cells but none were found in the marginal cells. There were no GJB2 or GJB6 gene transcripts found in the hair cells and only a few in the spiral ganglion cells.
Conclusion: Both GJB2 and GJB6 mRNA gene transcripts were localized in cells in the adult HC using RNAscope® in situ hybridization (ISH) and high resolution microscopy. Generally, GJB6 dominated over GJB2, except in the basal cells. Results suggest that cells may contain either GJB2 or GJB6 gene transcripts or both. This may be consistent with specialized GJ plaques having separate channel permeability and gating properties. A reduction in the number of GJB2 gene transcripts was found in the basal turn. Such information may be useful for future gene therapy.
Introduction
Gap junctions (GJs) consist of paired protein hexamers forming direct bridges between cells. Such joined hemichannels or connexons may allow the transfer of electric impulses, small molecules, second messengers, nutrients, and microRNAs (miRNAs) between cells (Leybaert et al., 2003; Beltramello et al., 2004; Chang et al., 2008; Mammano, 2019). The gap junction networks are extensively developed in the epithelial and connective tissue systems in the mammalian cochlea (Kikuchi et al., 1995). Cx26 and Cx30 proteins are the dominating connexins and play essential roles in the hearing process in adults as well as during organ morphogenesis (Kikuchi et al., 1995; Lautermann et al., 1998; Xia et al., 1998; Jagger and Forge, 2015; Mammano and Bortolozzi, 2018; Mammano, 2019).
Mutations in the GJB2 and GJB6 genes that encode the Cx26 and Cx30 proteins are the most frequent causes of prelingual hereditary deafness of varying severity, and give rise to nearly half of the cases of autosomal recessive non-syndromic deafness (Kelsell et al., 1997; Cohn and Kelley, 1999; Grifa et al., 1999; Petit et al., 2001). Most of these mutations are present in the GJB2 gene located in the DFNB1 locus (Lerer et al., 2001; del Castillo et al., 2002; Pallares-Ruiz et al., 2002; Del Castillo et al., 2005; Boulay et al., 2013). The GJB6 gene is positioned near the GJB2 on chromosome 13. Moreover, immunostaining and co-immunoprecipitation indicate that Cx26 and Cx30 proteins commonly co-assemble, presumably into heteromeric or heterotypic molecular arrangements (Lautermann et al., 1998; Zhao and Santos-Sacchi, 2000; Ahmad et al., 2003; Forge et al., 2003; Sun et al., 2005; Zhao and Yu, 2006). Such molecular configurations could explain the human ear’s unique vulnerability to various genetic disruptions.
There is an active search for therapeutic options to restore auditory function that is disrupted by genetic connexin-related hearing loss. A major hurdle is the short and variable time frames for potential gene therapy due to the proteins’ essential role in early cochlear morphogenesis, their co-regulation, and inter-dependence. More knowledge is necessary regarding Cx26 and Cx30 gene expression, transcription, and protein composition within the composite cellular networks along the cochlear spiral. Assessment of the GJB2 and GJB6 gene expressions may improve our understanding of the function of different cellular networks in the human cochlea (HC) and how cell-specific changes and mutations may interact and cause Cx26/Cx30-based deafness. Recent human studies are partly contrasted with previous findings suggesting a separate expression of the two proteins in different domains, as revealed by super resolution structured illumination microscopy (SR-SIM) (Liu et al., 2016, 2017a).
Here, we strained to analyze GJB2 and GJB6 mRNA transcripts in the adult HC using RNAscope® in situ hybridization (ISH) of archival paraformaldehyde-fixed sections. It was combined with immunohistochemical analyses of Cx26 and Cx30 proteins. In an earlier investigation, we analyzed Na/K-ATPase (NKA) gene transcripts—ATP1A1, ATP1A3, and ATP1B1—encoding the Na/K-ATPase α1, α3, and β1 isoforms together with GJB6 and fractalkine gene transcripts (Liu et al., 2021a; Liu and Rask-Andersen, 2022). In this study, we extended these analyses to include GJB2, KCNJ10, and ATP1A2. KCNJ10, ATP1A1 and ATP1A2 were used as additional markers to accurately locate connexin gene transcripts in the stria vascularis cell components. A RNAscope® employing the multiplex technique was used to examine whether GJB2 and GJB6 gene transcripts occur separately or in conjunction with each other in the cellular networks. Single mRNA gene transcripts appeared as bright foci, and results showed that the largest number of Cx26/Cx30 gene transcripts was in the outer sulcus epithelium, basal cells of the SV, and types I and II fibrocytes. A reduced number of GJB2 gene transcripts were found in the basal turn. Multiplex gene data suggest that cells in the lateral wall may contain either GJB2 or GJB6 gene transcripts or both. These findings, together with protein localization, suggest that cells and GJ plaques may contain different channels with various levels of permeability and gating characteristics in the cochlear domains. The Cx26/Cx30 proteins may play a major role for K+ cycling along medial and lateral transcellular pathways. Further documentation of gene expression in the developing cochlea is warranted.
Materials and methods
RNAscope protocol
The technique used for the RNAscope® analyses of HC sections was recently described (Liu and Rask-Andersen, 2022). In short, fixed-frozen human tissue sections underwent pretreatment with H2O2 (10 min, RT) and protease III (30 min, 40°C). After protease III incubation, the sections were exposed to a RNA-scope hybridization assay. Bio-Techne produced the probes. Hybridization was initiated by adding the RNA probe(s) (a mixture of probes termed C1, C2, and C3 channels) to the sections. Incubation was performed in a HybEZ™ Oven (Bio-Techne) for 2 h at 40°C. After hybridization, the slides were washed using 1x RNA-scope® Wash Buffer. The sections were incubated with RNA-scope® Multiplex FL v2 Amp 1, 2, and 3 (for 30 min/30 min/15 min, respectively) sequentially at 40°C to amplify the signal. For the RNA-scope® Multiplex study, RNAscope® Multiplex FL v2, HRP-C1, HRP-C2, and HRP-C3 were sequentially added to the sections (time 15 min). For signal detection, TSA-diluted Opal 520, 570, and 690 fluorophores were added to the sections after HRP-C1, C2, and C3, incubating the sections for 30 min each at 40°C. When the three Opal fluorophores were assigned to each channel, in present representation, two channels—C1 and C2—were assigned to GJB2 and GJB6, KCNJ10 and GJB2, and Na/K-ATP1A1 and Na/K-ATP1A2 (Table 1). After each fluorophore incubation and rinse with 1x RNA-scope® Wash Buffer, the RNA-scope® Multiplex FL v2 HRP blocker was added and incubated in an oven for 15 min at 40°C. Then, sections were counterstained withxs 4’,6-diamidino-2-phenylindole (DAPI), and the slides were cover-slipped with ProLong® Glass Antifade Mountant (Thermo Fisher Scientific, Waltham, MA, United States). The RNA-scope ISH produces a puncta of signals that represent a single mRNA transcript (Grabinski et al., 2015). The RNAscope® Multiplex Fluorescent v2 assay allows simultaneous detection of up to four RNA targets (Wang et al., 2015). The standard RNAscope probes have 20 zz pairs with each consisting of 35-50 nucleotides and resulting in 1000bp coverage for any given transcript. Both the probes target different regions and they do not cross-detect each other. The GJB6 is targeting exon 51 and GJB2 is targeting exon 22.
Semi-thin sectioning of surgically obtained human cochlea
Four archival human specimens were sectioned semi-thin, stained, photographed, and re-analyzed. The specimens were fixed in 3% glutaraldehyde and decalcified in 10% Na-EDTA. The bone was post-fixed in 1% osmic acid (OsO4, 75632 Sigma-Aldrich), and dehydrated and embedded in Epon (Resolution Performance Products, Houston, TX, United States). The technique was described earlier (Tylstedt et al., 1997). Photo images of various turns were examined in parallel with the mRNAscope®. Sections included different turns of the cochlea and were compared with the RNAscope findings at different regions (Figure 1).
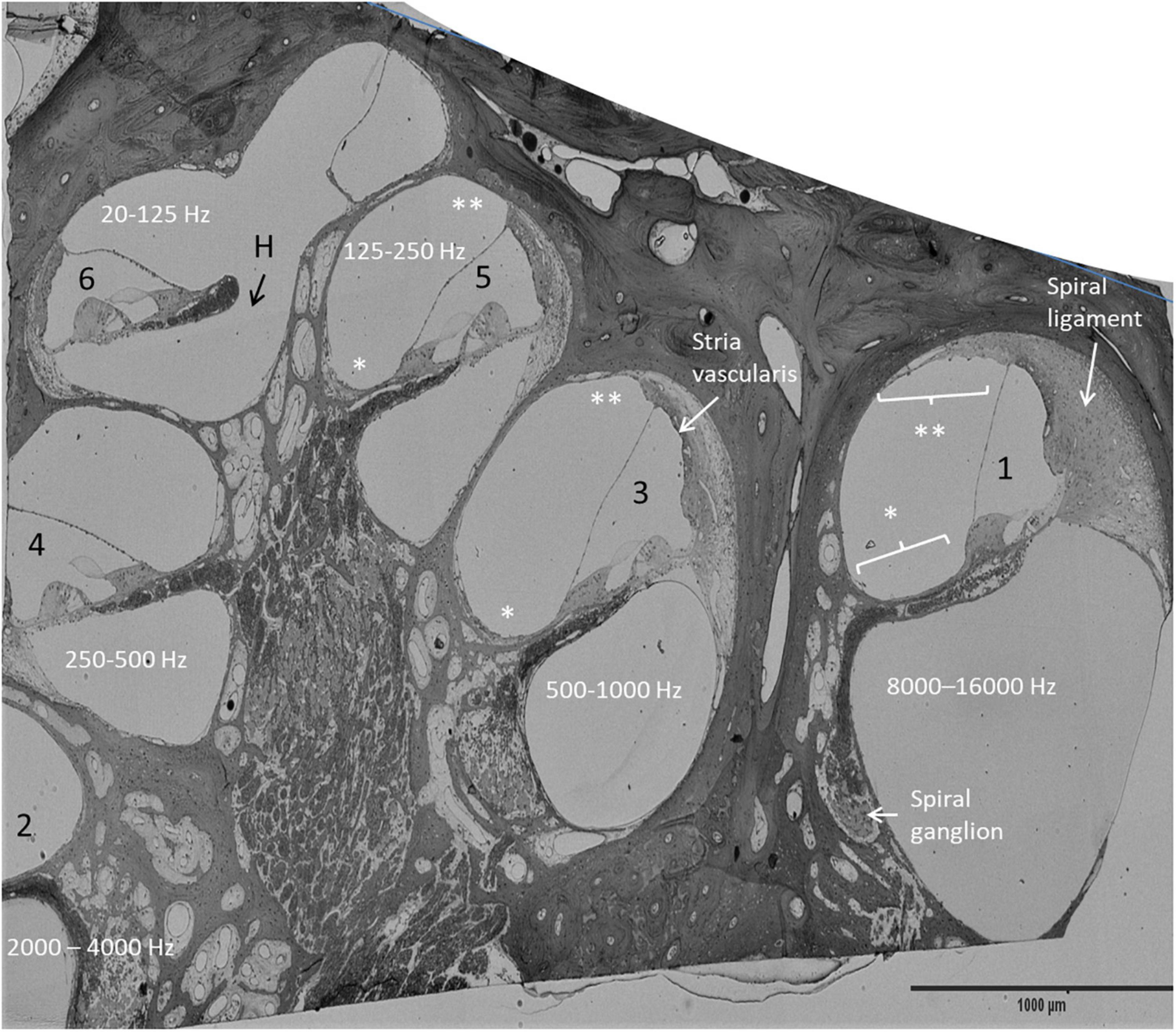
Figure 1. Mid-modiolar, semi-thin section of the right HC. Frequency distribution was estimated by assessing the number of cochlear turns. The anatomy of the OC and the lateral wall vary along the cochlear axis. The spiral ligament is more prominent in the high frequency region (1). *: mesothelium lining the floor of the scala vestibuli, **: suprastrial fibrocytes, H, helicotrema.
Immunohistochemistry
Techniques used for fixation, decalcification, embedment, and cryo-sectioning for immunohistochemistry were described in earlier publications (Liu et al., 2016, 2017a,b). Here, we used data obtained from studies on archival sections and previously obtained material. This included immune labeling of Na/K-ATPase (NKA) α1, α2, β1, NKCC, occludin, lamininβ2, Claudin-11, Cx30, and Cx26 (Table 1).
Imaging and photography
The RNA-scope sections were analyzed using an inverted fluorescence microscope (Nikon TE2000; Nikon, Tokyo, Japan) and a three-filter digital camera (emission spectra maxima at 358, 461, and 555 nm). NIS Element BR-3.2 (Nikon) imaging software was used. Laser confocal microscopy was performed with a three-channel laser emission system. The software used was Nikon EZ-C1 (ver. 3.80), SR-SIM [Elyra S.1 SIM system with a 63x/1.4 oil Plan-Apochromat objective (Zeiss), sCMOS camera (PCO Edge), and ZEN 2012 software]. Multichannel SR-SIM imaging was performed using a laser and filter setup: 405 nm laser of excitation coupled with BP 420–480 + LP 750 filter, 488 nm laser of excitation with BP 495–550 + LP750 filter, 561 nm laser of excitation with BP 570–620 + LP 750 filter, and 647 nm laser of excitation with LP 655 filter. The resolution was measured with sub-resolution fluorescent beads (40 nm, Zeiss) in the green channel (BP 495–550 + LP750). An average point spread function (PSF) value was obtained from multiple beads using the built-in experimental PSF algorithm of the ZEN software. The resolution of the SR-SIM gave a lateral precision of 80 nm (Gustafsson et al., 2008). The volume resolution of three-dimensional (3-D) SIM is almost eightfold that of conventional microscopy (Schermelleh et al., 2008). The 3-D reconstructions and rendering of Cx26 (red) and Cx30 (green) protein expression were performed in spot detection mode using Imaris 9.5.1 (Bitplane, Zurich, Switzerland). The grid size of the frame on the 3-D images was 5μm. The number of GJB2 gene puncta was counted using ImageJ particle analysis in the lateral wall at all three turns. Numbers were difficult to assess and had to be estimated in areas of transcription that burst in the cell nuclei. Both the total number of puncta and the number per cell (DAPI) were calculated and compared. Distortion of tissue may have influenced the results and focus remained primarily on obtaining comparative data of the three different turns.
Results
Immunohistochemistry and super resolution structured illumination microscopy
An archival, semi-thin section of a decalcified HC fixed in 3% glutaraldehyde is demonstrated in Figure 1. This image shows the different anatomy of the OC and the lateral cochlear wall in the three turns. The spiral limbus and tectorial membrane are smaller and the spiral ligament is thicker against the basal high frequency region. Frequency mapping was estimated based on the number of cochlear turns. Confocal microscopy of Cx26 and Cx30 antibody co-labeling of the middle turn in an adult cochlea is shown in Figures 2A–C. Cx26 and Cx30 are both separately and co-expressed in the spiral limbus and lateral cochlear wall. Cx30 expression dominates in the OC (Figure 2B, not co-stained). SR-SIM using spot detection mode shows the distribution of Cx26 and Cx30 in the lateral wall (Figure 2D). There is a separate expression, with a band of Cx26 corresponding to the basal cell layer and the junction at the type I fibrocytes of the SV. Higher magnification confirms a close association between the two channel proteins (Figure 2D’,D”). The molecular arrangements of connexons (homomeric/heteromeric/heterotypic) cannot be assessed with certainty since the microscope resolution of 80 nm is far above the diameter of a Cx26 channel (Maeda et al., 2009). Cx30 is expressed solely in the intermediate cells, suggesting a homomeric molecular design. No GJ proteins were expressed in the marginal cells.
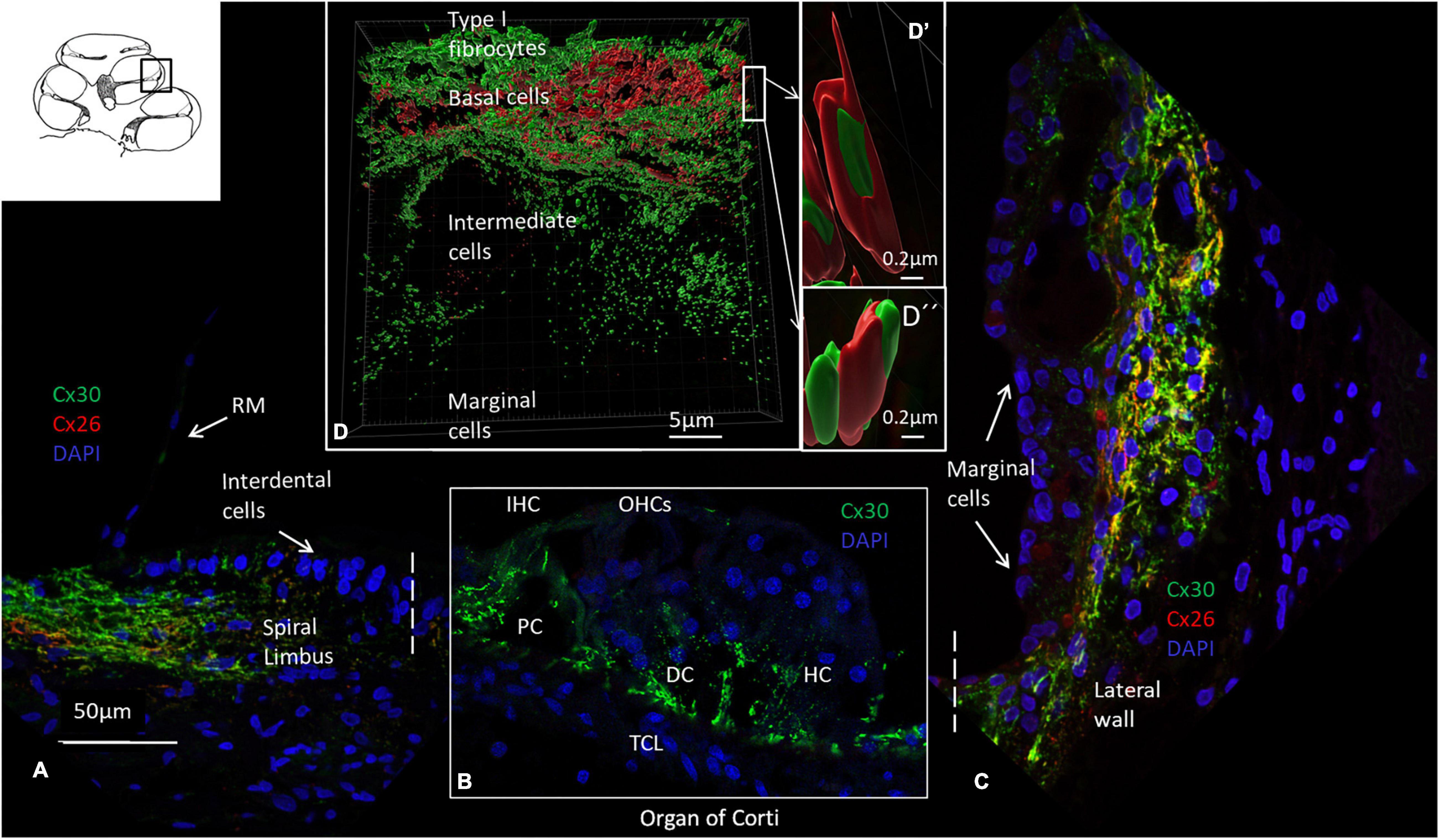
Figure 2. Confocal microscopy, SR-SIM, and Cx26 and Cx30 antibody labeling of the organ of Corti and lateral wall. (A) Cx26 and Cx30 are expressed in the spiral limbus fibrocytes. There is no expression in Reissner’s membrane (RM). (B) The OC expresses both Cx26 and Cx30, but in Figure B, only Cx30 is shown. IHC: inner hair cell. OHCs: outer hair cells. PC: pillar cell. HC, Hensen cell; DC, Deiters cell. TCL: tympanic covering layer. (C) Cx26 and Cx30 are both jointly and separately expressed in the lateral wall. Cx30 dominates except at the basal cell area of the stria vascularis. There is no expression in the marginal cells or in cells laterally margining the bone (type III fibrocytes). (D) SR-SIM using spot detection mode (Imaris 9.5.1) demonstrates separate and closely associated protein labeling with geometric arrangements in the lateral cochlear wall (D’ and D”). Cx30, but not Cx26, is expressed in the intermediate cells. The cochlear turn is not given.
RNAscope – confocal and super resolution structured illumination microscopy
The RNAscope® and ISH identified the single GJB2 and GJB6 mRNA oligonucleotide transcripts in the formaldehyde-fixed sections of the HC. Decalcification maintained the cell integrity, and gene transcripts could be localized in the cell nuclei and cytoplasm. Na/K-ATPase and Kir4.1 gene transcripts (ATPA1, ATP1A2, and KCNJ10) were used as additional controls. ATPA1 was only expressed in the marginal cells of the SV, while KCNJ10 was expressed in the intermediate cells and ATP1A2 in the basal cells (Figure 3).
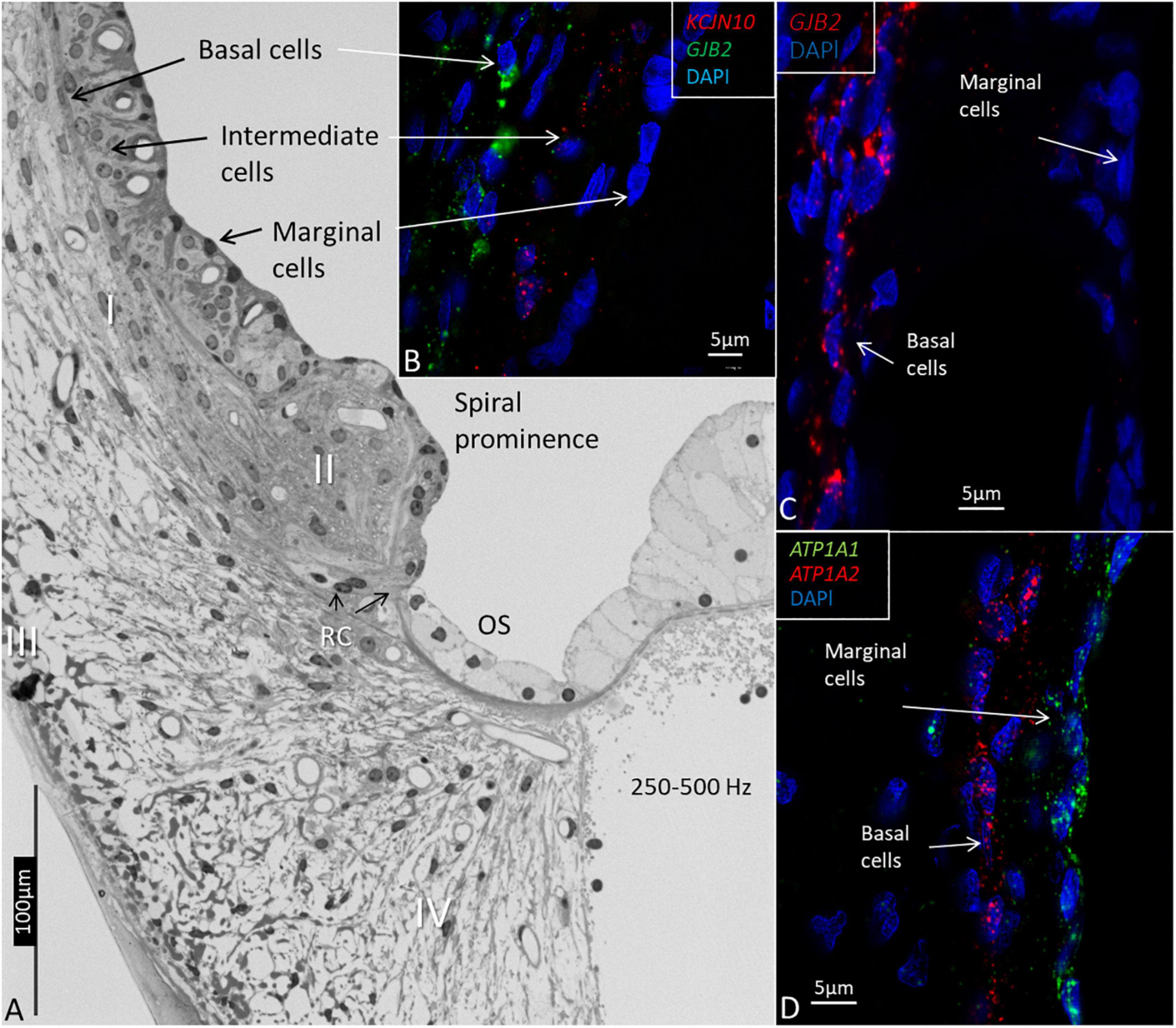
Figure 3. Light microscopy and confocal microscopy of the lateral wall of the upper-middle cochlear turn (frequency location 250–500Hz) showing marginal, intermediate, and basal cells of the stria vascularis. (A) The marginal cells are believed to be true epithelial cells, but there is no continuous basal lamina. Type I fibrocytes (I) of the spiral ligament lie close to the basal cells. The spiral prominence epithelium is flatter and is related to the type II fibrocytes. The outer sulcus (OS) epithelium, and Claudius and Hensen cells have a light cytoplasm and are fairly organelle-free. Root cells (RCs) project into the subepithelial space. Type III fibrocytes are located near the bone, and type IV faces the scala tympani. (B) RNAscope shows the distribution of KCNJ10 and GJB2 gene transcripts in intermediate and basal cells. (C) GJB2 transcripts are located in the basal cells, but not in the marginal cells. (D) ATP1A1 gene transcripts are seen in the marginal cells, while the basal cells contain ATP1A2.
The GJB2 and GJB6 gene transcripts were demonstrated in the cells of the spiral limbus, OC, and lateral wall in all three turns (Figures 4–8 and Table 2). The largest numbers of GJB2 and GJB6 puncta were detected in the outer sulcus epithelium, root cells, basal cells, and types II and I fibrocytes. In the SV, many GJB2 and some GJB6 gene transcript puncta were noted in the basal cells. Intermediate cells contained GJB6, but did not contain GJB2 puncta. At the suprastrial region and the junction between the Reissner’s membrane and the SV, type V fibrocytes displayed both GJB2 and GJB6 transcripts. Type I fibrocytes contained a larger number of GJB6 as well as GJB2 gene transcripts, especially the cells located near the basal cells. While the upper and middle parts of the spiral prominence epithelium contained few GJB2/GJB6 transcripts, the lower slope contained a large number of both GJB2 and GJB6 gene transcripts. The GJB2-containing basal cells merged with the spiral prominence epithelium and marginal cells in the apical turn (Figure 4D). A similar situation existed at the lateral insertion of the Reissner’s membrane (Figure 4C). A corresponding, but slightly different, anatomy was present in the middle and lower turns of the HC (Figures 5, 6).
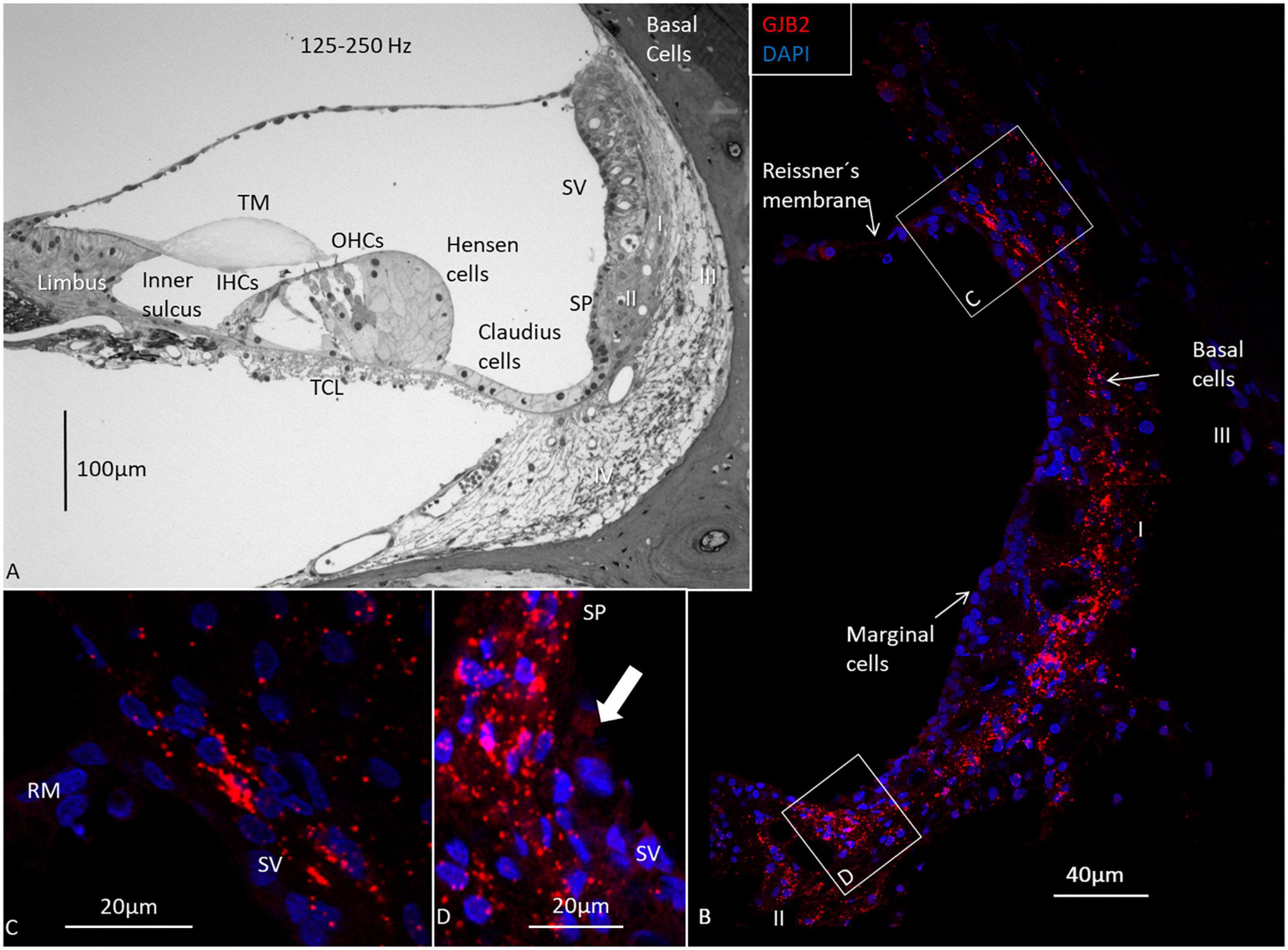
Figure 4. Light microscopy and confocal microscopy of the apical turn of the human cochlea at the frequency region 125–250 Hz. (A) A semi-thin section shows the stria vascularis (SV) and the relatively thin spiral ligament containing four different types of fibrocytes (I-IV). There is no suprastrial connective tissue layer containing type V fibrocytes. A thin layer of basal cells can be seen next to the type I fibrocytes. Type II fibrocytes lie beneath the spiral prominence (SP) epithelium. (B) Distribution of GJB2 gene transcripts in the lateral wall. GJB2 gene transcripts are concentrated at the basal cell (BC) region, while the marginal cells show no gene transcripts. A few red puncta can be observed at the intermediate cells. Gene transcripts are located in type I fibrocytes. Framed areas are shown in higher magnification in (C,D). (C) Expression of GJB2 at the angle between the Reissner’s membrane (RM) and SV. There are no transcripts in RM. (D) Enlargement of the angle between the SV with transcript-containing basal cells reaching the SP epithelium (arrow). IHCs, inner hair cells; OHCs, outer hair cells; TM, tectorial membrane; Limbus, spiral limbus.
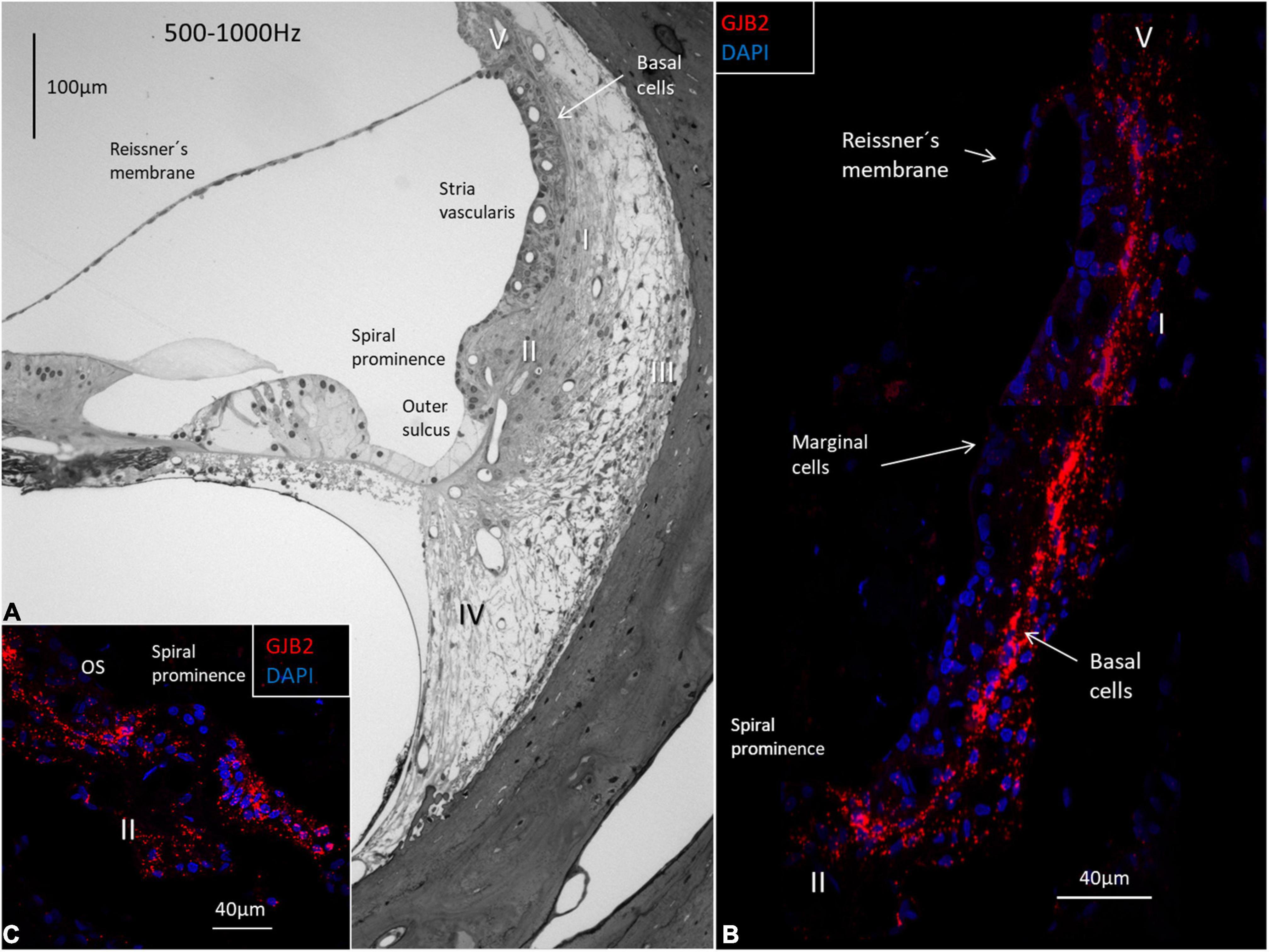
Figure 5. Light microscopy of the human OC and lateral wall at the frequency region 500-1000 Hz and confocal microscopy of GJB2. (A) The semi-thin section shows the stria vascularis and the spiral ligament with five different types of fibrocytes (I-V). Basal cells face the type I fibrocytes. The spiral prominence epithelium is thin and changes into a light epithelium in the outer sulcus (OS). (B) A large number of GJB2 gene transcripts are concentrated in the basal cell region. There are no transcripts in the marginal cells. (C) There are few transcripts in the spiral prominence epithelium. A large number of transcripts are seen in the outer sulcus.
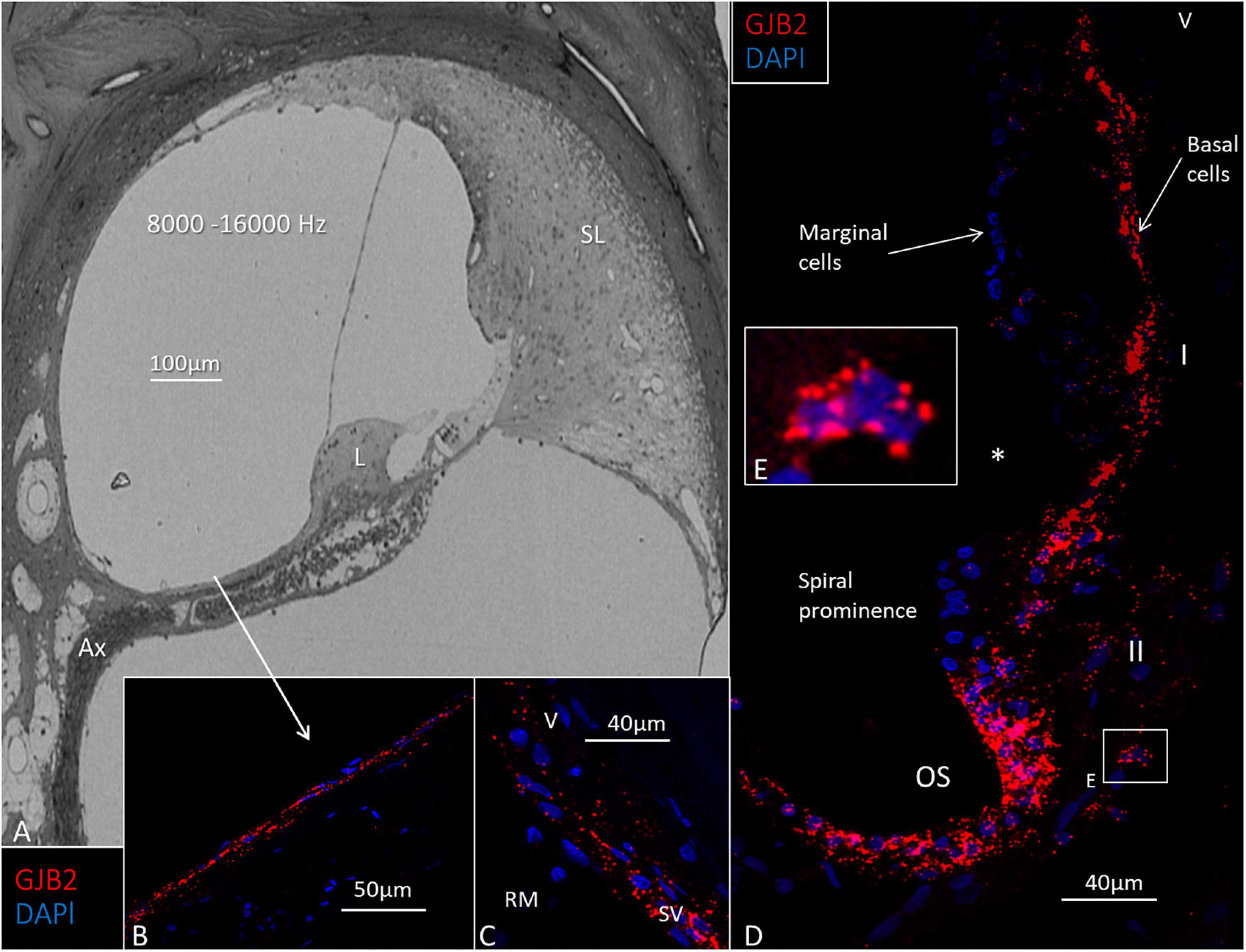
Figure 6. Light microscopy and GJB2 in the basal turn of the cochlea at the high frequency region 8000–16000 Hz). (A) The spiral limbus (L) and tectorial membrane near the round window are smaller at this location, and the spiral ligament (SL) is more voluminous. Ax: peripheral axons. (B) GJB2 gene transcripts in mesothelial cells lining the floor of the scala vestibuli. (C) GJB2 gene transcripts at the lateral insertion of the Reissner’s membrane (RM). SV: stria vascularis. V: type V fibrocytes. (D) A large number of GJB2 gene transcripts in the basal cells extend from the RM to the spiral prominence. *: disrupted marginal cells. The outer sulcus (OS) contains many GJB2 gene transcripts. (E) Magnification of the framed area in (D) shows a type II fibrocyte rich in GJB2 transcripts. I, II: fibrocytes types I and II.
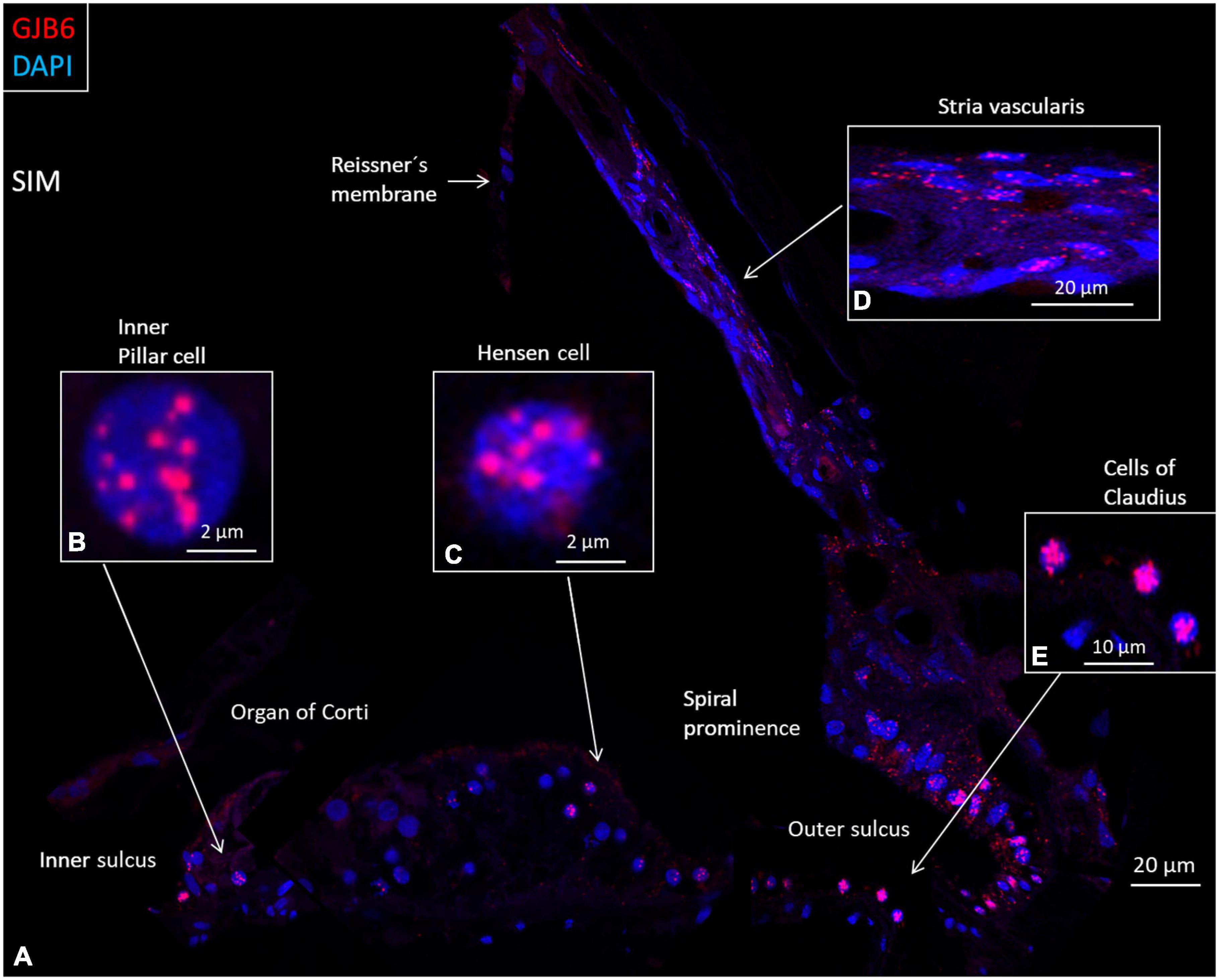
Figure 7. SR-SIM composite micrograph of GJB6 gene transcripts in the basal turn of the human cochlea. (A) Many GJB6 transcripts are seen in the outer sulcus and type I fibrocytes. Some Claudius cell nuclei contain large numbers of GJB6 gene transcripts. A few supporting cells in the OC contain gene puncta. (B) Higher magnification of an inner pillar cell nucleus contains GJB6 transcripts. (C) Higher magnification of a Hensen cell nucleus contains GJB6 transcripts. (D) Higher magnification shows GJB6 transcripts, mainly in the basal cells. (E) Claudius cells containing many gene transcripts. Cells in the tympanic covering layer contain no gene puncta.
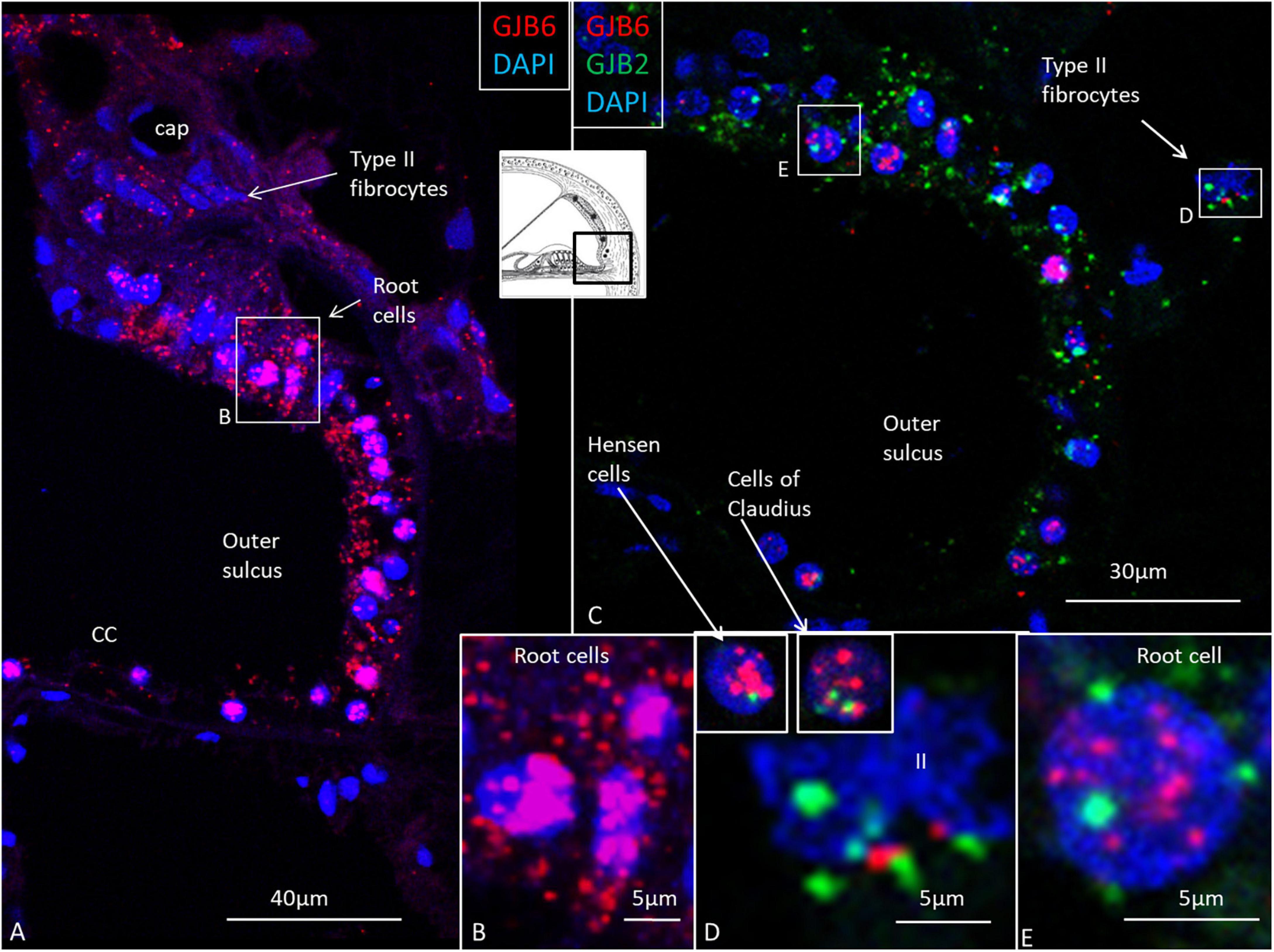
Figure 8. GJB2 and GJB6 gene transcripts in the outer sulcus in the basal turn of the cochlea. (A) Outer sulcus epithelium and root cells contain a large number of GJB6 transcripts. Type II fibrocytes contain a moderate number of GJB6 transcripts. The framed area is magnified in (B). (B) Root cell nuclei are crowded with GJB6 gene transcripts. (C) Multiplex RNAscope shows both GJB2 and GJB6 transcripts co-express in the outer sulcus epithelium. Framed areas are magnified in (D,E), and show both gene transcripts present in a type II fibrocyte (II) and root cell nuclei. Insets in (D) show Hensen and Claudius’s cell nuclei expressing both transcripts, but with GJB6 dominant. The diameter of the cell nuclei is around 5 microns.
Relatively few gene transcripts were found in type IV fibrocytes, and none were detected in the type III fibrocytes. Type I and II fibrocytes, Hensen, Claudius, and outer sulcus cells contained many GJs on transmission electron microscopy (TEM). The spiral limbus contained many GJB2 and GJB6 gene transcripts, and many were contained in the cells lining the scala vestibuli floor (Figures 6A–C). A composite micrograph of the OC and lateral wall is shown in Figure 7. Claudius cell nuclei were crowded with GJB6 gene transcripts, but there were only a few in the cytoplasm (Figure 7A). None were found in Reissner’s membrane or hair cells. In Deiters’, Claudius, and outer sulcus epithelial cells, the number of transcripts were also concentrated to the cell nuclei.
GJB6 dominates over GJB2 except in the stria vascularis’s basal cells
Multiplex labeling of GJB2 and GJB6 transcripts in the HC shows that both transcripts are generally present in the epithelial and connective tissue cells (Figures 8, 9), except in the intermediate cells that only contained GJB6 transcripts. An exceptional large number of GJB2 and GJB2 gene transcripts were noted in the outer sulcus and root cells. Both GJB2 and GJB6 were present in these cell nuclei (Figures 8B,D,E). Generally, the GJB6 dominated over GJB2 except in the basal cells where GJB2 exceeded GJB6 (Figure 9). We tried to assess whether cells may contain either GJB2 or GJB6 transcripts separately or if both are transcribed in the same cell by analyzing serial stacks of images. These results showed that some cell nuclei seemed to contain only one of the two transcripts while others contained both transcripts (Figures 9B–D).

Figure 9. (A) SR-SIM and multiplex labeling of GJB2 and GJB6 transcripts in the human SV. (B–D) Intermediate cells solely contain GJB6, while basal cells contain either GJB2 and GJB6 transcripts separately or together in the same cell. This may suggest that GJ plaques consist of GJ channels with different properties and molecular arrangements. V: vessels. Arrow in B shows a perinuclear GJB2 transcript.
GJB2 gene transcripts in various cochlear turns
Results of ImageJ particle analysis of the number of GJB2 gene puncta in the lateral wall in the three turns are shown in Table 3. Results showed that there were fewer gene transcripts in the basal turn. This did not seem to result from a reduction of transcripts in each cell, but from a lower number of cells (DAPI) in the SV and spiral ligament at this level.
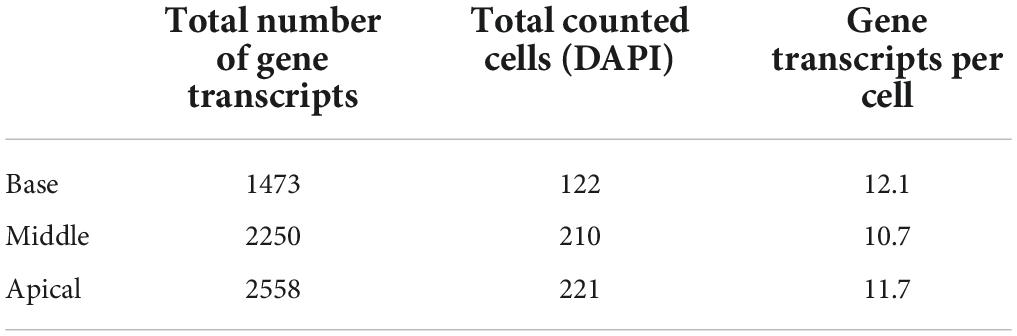
Table 3. Quantification of GJB2 gene transcripts in the lateral wall of the three turns in a human cochlear section.
To better understand the relationship between the GJB2-containing basal cells and the spiral prominence epithelium, we analyzed the co-expression of the tight junction (TJ) protein Claudin-11 and Cx26 (Figure 10A). It showed that basal cells jointly expressed TJ and GJ proteins and that TJ-expressing cells formed a thin layer that separated the basal and marginal cells from each other (Figures 10B,C).
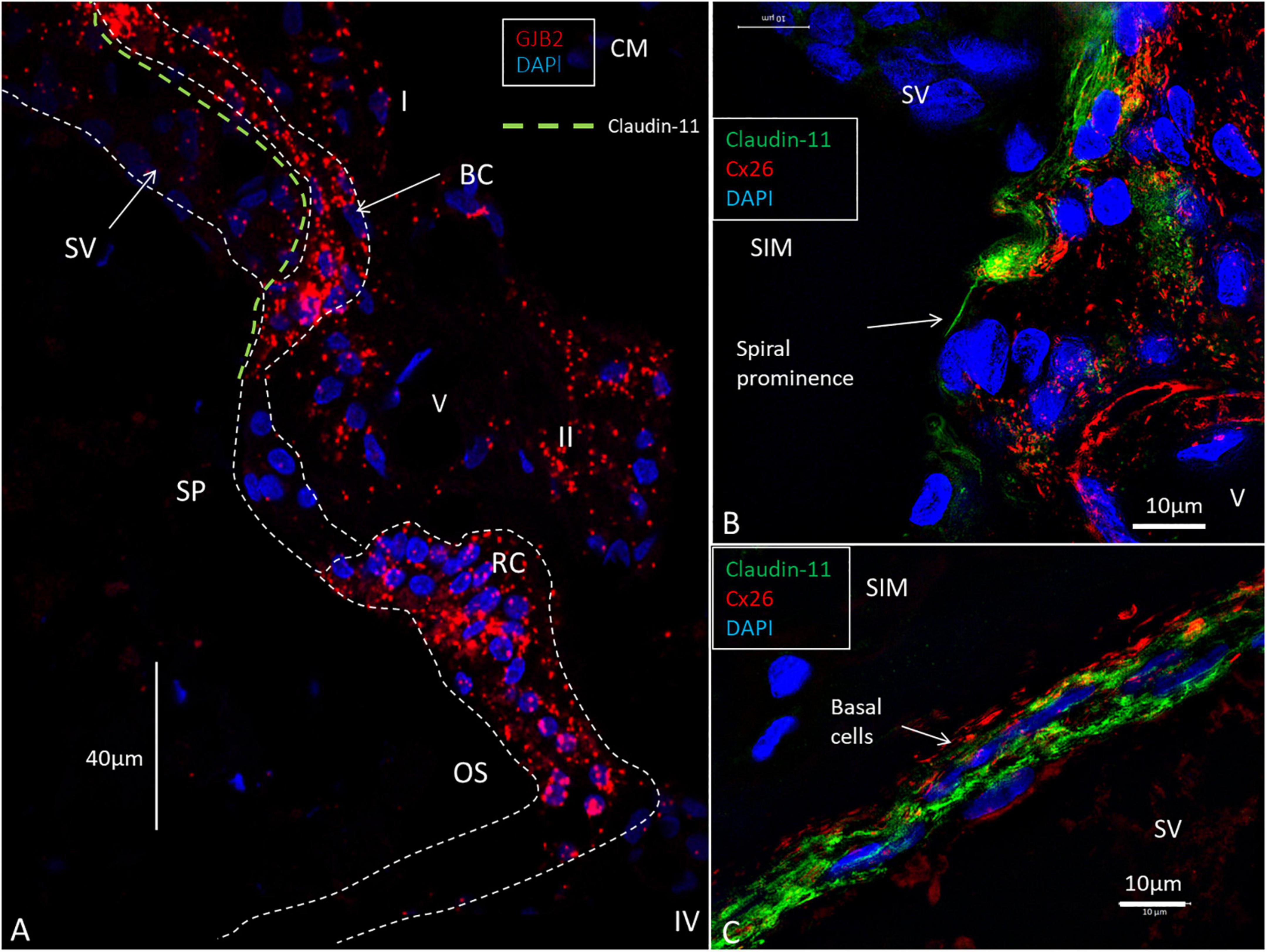
Figure 10. RNAscope of GJB2 transcripts in the spiral prominence and outer sulcus. (A) The outer sulcus (OS), root cells (RC), and basal cells (BC) are rich in GJB2 gene transcripts, while the spiral prominence epithelium virtually lacks gene puncta. The type I and II fibrocytes contain moderate numbers of gene transcripts. The green broken line shows the location of the Claudin-11 TJ barrier. (B) SR-SIM of Cx26 and Claudin-11 expression at the transitional zone between the stria vascularis (SV) and spiral prominence. A layer of the tight junction protein can extend to the spiral prominence (arrow). (C) SR-SIM of Cx26 and Claudin-11 protein expression in the basal cells of the SV. V: vessel. I: type I fibrocytes. II: type II fibrocytes. CM: Confocal microscopy. SIM: structured illumination microscopy.
We found a few GJB2 and GJB6 transcript puncta present in the type I spiral ganglion cells as well as in the satellite glia cells (SGCs) (Figure 11). Larger stained areas were also seen in the peripheral cytoplasm of type I cells recognized as conceivably unspecific.
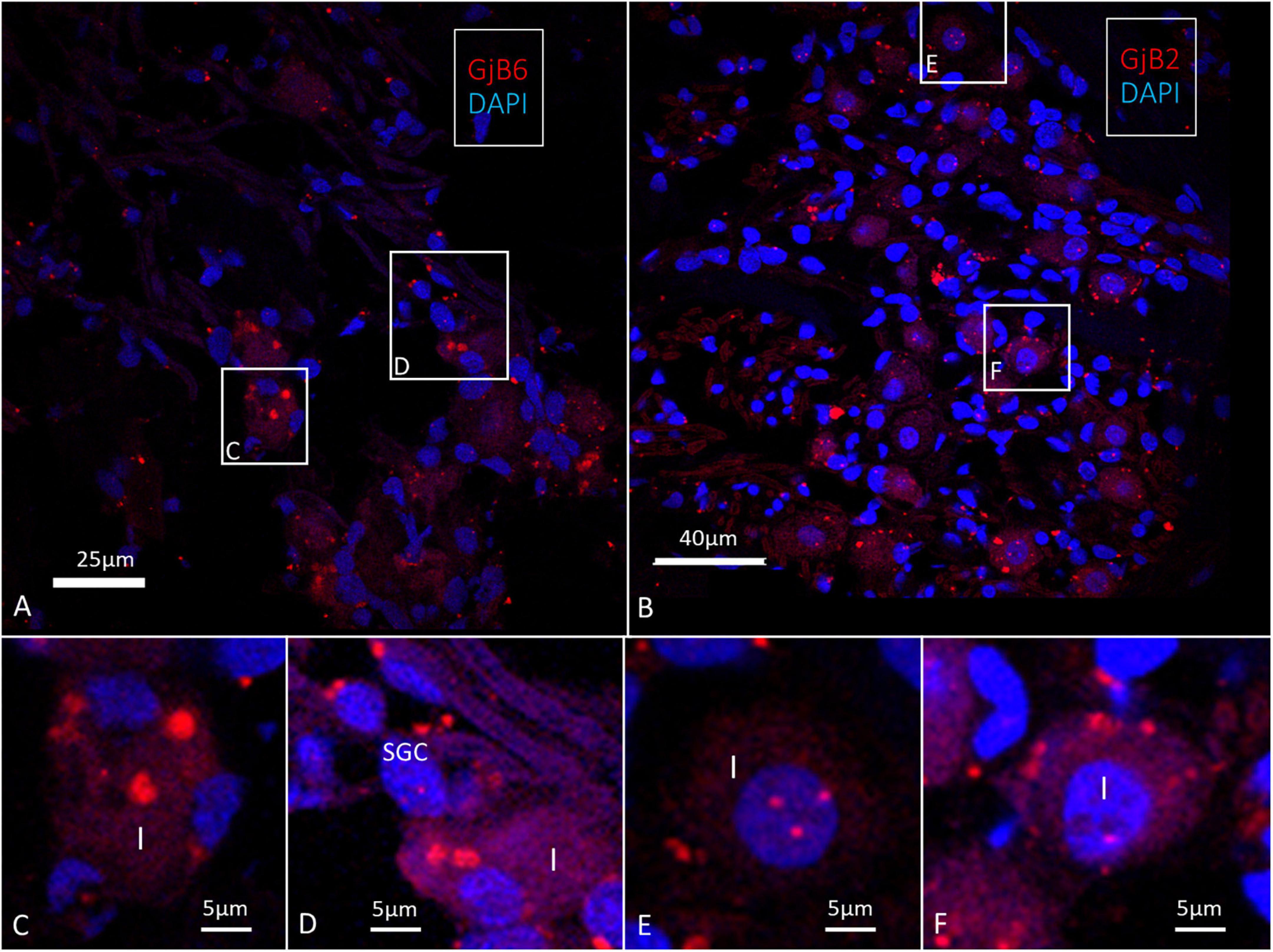
Figure 11. Confocal microscopy of GJB2 and GJB6 transcripts in the human spiral ganglion. (A) Occasional GJB6 transcript puncta are seen in the type I ganglion cells and a few puncta can be seen in the surrounding cells, including the satellite glia cells (SGC). Framed areas are magnified in (C,D). (B) A few GJB2 gene transcript puncta are seen in the type I ganglion cell soma. Framed areas are shown in higher magnification in (E,F). (E) Type I cell nucleus contains GJB6 transcripts (F). A type I cell contains GJB6 puncta. Larger red-stained areas (non-puncta) are considered to be unspecific.
Discussion
The RNAscope® ISH of archival paraformaldehyde-fixed sections identified single GJB2 and GJB6 gene transcripts in the epithelial and connective tissue networks in surgically obtained HC using confocal microscopy and SR-SIM. A study limitation was the restricted amount of human tissue for more systematic gene localization. The results seem to be consistent with the distribution and expression of Cx26 and Cx30 isoforms described in recent investigations (Liu et al., 2017a,b).
The total number of GJB2 gene transcripts was less in the base in accordance with the proteins in the guinea pig (Zhao and Yu, 2006). The specificity of the labeling was validated by using alternate gene biomarkers characterizing various cell domains in the SV. The KCNJ10 encodes the inward rectifying potassium channel 4.1 (Kir4.1) that was specifically located in the intermediate cells (Wangemann et al., 1995; Hibino et al., 1997) while ATP1A1 encoding Na/K-ATPase α1 was restricted to the marginal cells (Liu and Rask-Andersen, 2022). The basal cells expressed ATP1A2 encoding Na/K-ATPase α2 and could be used to validate the various gene transcripts’ locations.
The GJs are composed of different connexins that post-translationally oligomerize into hexameric hemichannels before being inserted into the plasma membrane. They form direct intercellular passageways for transporting ions, ATP, glucose, miRNA, and second messengers (Dermietzel et al., 1990; Goodenough et al., 2003; Evans and Martin, 2009; Zhu et al., 2015; Laird and Lampe, 2018). Several mutations affecting the GJB2 gene are known to give rise to hearing loss of varying severities, and there is an active search for therapeutic options. Deafness-causing mutations in the GJB2 gene may give rise to both non-syndromic and syndromic deafness, but are mostly autosomal recessive point mutations (Wang et al., 2003). Large deletions in the DFNB1 locus may also affect GJB2 transcription and regulatory sequences as well as GJB6, and result in profound hearing loss (Del Castillo and Del Castillo, 2017). The molecular and structural arrangements of GJs may affect phenotypic expression and the consequences of different mutations.
Our knowledge of the structure of cochlear GJs is mostly based on animal models, which have suggested that Cx26 and Cx30 generally co-assemble, particularly in mesenchymal fibrocytes where they are believed to form unique heteromeric or heterotypic assemblies (Lautermann et al., 1998; Zhao and Santos-Sacchi, 2000; Forge et al., 2003; Sun et al., 2005). The GJ channels containing multiple connexins could have separate gating properties that contribute to functional diversity in intercellular communication (Sosinsky and Caspar, 1995; White and Bruzzone, 1996). Co-immunostaining and co-immunoprecipitation have indicated that proteins could even co-assemble in the same plaque (Ahmad et al., 2003). Such arrangements could enhance intercellular Ca2+ signaling that is essential for inner ear homeostasis and explains the ear’s vulnerability to connexin derangements.
Hybrid plaques
There is uncertainty of whether human GJ channels in the OC and lateral wall of the cochlea are organized as heteromeric or homomeric connexons. Recent studies submit that Cx26 and Cx30 can be separately expressed, forming closely associated GJ plaques with Cx26 subdomains that occasionally appear as “hybrid plaques” (Liu et al., 2016, 2017a). Different GJ channels could populate the same GJ plaque or closely interact with different plaques. This was determined by freeze-fracture immunogold labeling (Fujimoto, 1995) and astrocytes expressing Cx26, Cx30, and Cx43 in rats (Rash et al., 2001). The present study also supports this arrangement. A high degree of complexity of the transcriptional co-regulation mechanism may be present in these sophisticated human cellular networks (Kim et al., 2012). SR-SIM showed that both GJB2 and GJB6 gene transcripts were separately localized in different cells, but could also appear in the same cell. Cx26 and GJB2 gene transcripts were enriched in the basal cell layer, which forms a critical stratum for generating the endo-cochlear potential (EP) by passive movement of potassium ions across its apical membranes (Tasaki and Spyropoulos, 1959; Salt et al., 1987). Cx26 and Cx30 co-staining showed a distinct layer of Cx26-positive cells with less Cx30 (Figure 2). The marginal cells contained neither GJB2 nor GJB6 gene transcripts. The basal cells may comprise preferentially homomeric or homotypic Cx26 and Cx30 hemichannels, while a heterotypic molecular arrangement seems less likely considering the IHC and GJB2 and GJB6 transcript distribution. It may favor the presence of molecularly separate GJs and possibly hybrid junctions with plaques consisting of both Cx26 and Cx30 channels (Liu et al., 2017a,b). They may have different properties and act synergistically, including the generation of the EP, a prerequisite for human hearing. The rich content of the Cx26 protein is reflected by the large number of GJB2 gene transcripts and could clarify the major effects caused by GJB2 gene disruption on human hearing and EP that is not readily compensated for by Cx30. Cochlear cell degeneration of cCx26 null mice was more extensive and rapid than in Cx30 null mice (Sun et al., 2009). One would expect that genetic deletion of one of the genes would result in a similar pathology if it was heterotypically arranged. The dominating effect of Cx26 was also shown by the fact that hearing can be normal without Cx30 in cases where Cx26 levels are preserved (Ahmad et al., 2007; Boulay et al., 2013; Jagger and Forge, 2015).
Hemichannels
Notably, intermediate cells only contained GJB6 gene transcripts and Cx30 protein. Transcripts encircled the blood vessels, which is consistent with murine studies showing that Cx30, but not Cx26, is present in the SV’s intermediate cells (Cohen-Salmon et al., 2002). They found that, in homozygous Cx30 mutations, a down-regulation of betaine homocysteine S-methyltransferase in the stria capillaries resulted in endothelial dysfunction and hearing loss. It is not known whether these channels represent GJs or hemichannels that form routes between the extracellular space and cytosol. Such channels have been implicated in paracrine signaling through plasma membrane release of various messengers, such as ATP and glutamate, nicotinamide adenine dinucleotide, and prostaglandins (Wang et al., 2013). They may also be involved in regulating inflammatory responses (Baroja-Mazo et al., 2013; Willebrords et al., 2016).
Epithelial GJB2 and GJB6 and K+ recirculation
Supporting cells in the OC, including Hensen and Claudius cells, contain a moderate number of GJB2 and GJB6 gene transcripts, mostly located in the same cell nuclei. The precise number was difficult to assess since fluorescent puncta coalesced into larger assemblies, possibly representing transcriptional bursts. No transcripts were found in the hair cells or in the cells of the tympanic covering layer (TCL) located beneath the basilar membrane. The large number of transcripts in the outer sulcus epithelium, root cells, type I and II fibrocytes, and proteins may support the view that there is a lateral flow of K+ ions from the hair cell region to the lateral wall and back to the SV. Many cells contained a large number of transcripts, both in the cell nuclei and cytoplasm, while others contained a lower number. This suggests that cells show a variable transcription activity. SR-SIM and Cx30 immune staining indicated that open hemichannels are also present in the supporting cells in the human OC (Liu et al., 2017a). Inner and outer pillars and Deiters’ cells face cortilymph, and hemichannels could mediate autocrine/paracrine signaling by releasing ATP (Leybaert et al., 2003). They could also play an important role in maintaining low extracellular glutamate levels in order to avoid neuronal death from glutamate excitotoxicity and thereby maintain synaptic transmission (Ye et al., 2003). Cx30 greatly dominates Cx26 in the adult human OC.
GJB2 and GJB6 in the scala vestibuli
GJB2 and GJB6 gene transcripts were also detected in the mesothelial lining of the floor of the scala vestibule and supra-limbus cells. These cells expressed Cx26 and Cx30 (Liu et al., 2016), suggesting that both proteins have the capacity to trap the sequestered K+ ions and bring them to the suprastrial fibrocytes in the lateral wall. This could be an additional route for recirculating K+ from the inner hair cell region, inner sulcus, and spiral limbus contributing to endolymph production from perilymph (Sterkers et al., 1982; Marcus et al., 2002; Konishi et al., 1978). The GJs were shown in both the supralimbal and mesenchymal cells lining the scala vestibuli in the rat (Kikuchi et al., 1995). More studies are needed in order to verify whether GJs are present between these cells in the HC.
GJB2 and GJB6 gene transcripts and the spiral prominence
Researchers resembled the basal cell layer with the placenta barrier, where both GJs and TJs regulate the exchange of fluids and small metabolites between blood supplies (Metz et al., 1978; Kikuchi et al., 1995). A breach of this barrier could be detrimental, and a reciprocal regulation between GJ and TJ proteins was suggested (Giepmans and Moolenaar, 1998; Kojima et al., 2002). Gap junctions were shown to influence TJ permeability, such as Claudin and Occludin which also co-immuno-precipitate (Kojima et al., 2001; Dbouk et al., 2009). A similar interaction between GJ and TJ may occur in the cellular networks of the SV. Claudin-11 was shown to be indispensable for hearing and for maintaining the EP, but not of a high K+ concentration in the endolymph (Kitajiri et al., 2004a,b). Human studies show that Cx26 partly co-assembles with the TJ Claudin-11 along the basal cell layer. Unexpectedly, GJB2-containing basal cells reached the upper slope of the spiral prominence epithelium and seemed to face endolymph. Staining showed that Claudin-11 insulates basal cells from spiral prominence and endolymph (Liu et al., 2017b) (Figures 10A–C). The TJ protein forms laminar insulations between the GJB2-positive basal cells and spiral prominence epithelium (Figure 10). A similar arrangement exists at the insertion of Reissner’s membrane. This compartmentalization could be pivotal to separating the spiral prominence from the SV in order to polarize K+ flow into it. The spiral prominence may be critical as a circuit for ions entering and leaving this region.
Few GJB2 and GJB6 transcripts in the human spiral ganglion
There were few GJB2 and GJB6 gene transcripts puncta that were detectable in the spiral ganglion (Figure 11). This is surprising since IHC showed positive staining of both proteins (Liu et al., 2009, 2019, 2021b). Cx30 was localized in discrete deposits in type I spiral ganglion cell bodies while Cx26 was more diffusely stained. Differences may depend on the antibodies that were used. In an earlier study, larger areas were found with assumed GJB6 expression (Liu et al., 2021b). A similar pattern was noted in the present study, but with few transcript puncta present in type I spiral ganglion cells and satellite cells (Figure 11). Further studies using additional molecular techniques are necessary in order to establish the degree to which GJB2 and GJB6 transcripts are present in the human spiral ganglion cells. There was no positive Cx30 staining after double-labeling with TUJ1 or in parvalbumin-positive nerve endings beneath the hair cells. This suggests that Cx30 may not be involved in nerve excitation in the human OC. Cx36 is the principal neuronal connexin in the mammalian CNS, but the Cx36 gene transcript, GJD1, was not detected in an earlier investigation (Liu et al., 2021b).
GJB2 and GJB6 transcription rates
A noteworthy number of GJB2 and GJB6 gene transcripts in outer sulcus and spiral ligament cells may suggest a high transcription rate. The GJ connexins have been shown to have a remarkably short half-life of just a few hours (Laird, 2006). Connexin proteins are continually synthesized and degraded depending on physiological demands, and genes may be upregulated with increased protein levels and altered localization (Risek et al., 1990). Gap junctions can remodel, such as in cardiac disease where Cx43 serves to electrically convey impulses between myocytes (Dhein and Salameh, 2021). Connexin channels may also change between an active and inactive state, regulated by phosphorylation processes (Bukauskas et al., 2000). A high turnover rate indicates that there is a rapid synthesis and replenishment essential to upholding function. Cell-cell coupling was previously considered stationary, but it is now realized that gene expression and transcription can be highly irregular, with spontaneous variability occurring in some cells due to environmental conditions. Such “burst” transcription has been described in several mammalian tissues (Bahar Halpern et al., 2015). Insults may remodel GJ coupling with rapid degradation in the plasma membrane, removal, and turnover, which are critical for function. Noise- and age-dependent hearing loss have been associated with Cx26 dysregulation (Wang et al., 2014; Wu et al., 2014; Zhou et al., 2016; Fetoni et al., 2018). We found a reduction in the number of expressed genes in the basal turn (Table 3), supporting an earlier finding of diminished Cx26 and Cx30 activity at the high-frequency level (Zhao and Yu, 2006). Further elucidation is also need on whether this relates to age-related hearing loss. Inflammatory changes in spiral ligament cells could also interrupt GJ K+ recycling and cause sensorineural hearing loss. Methods to pharmacologically target and modify GJ trafficking have been suggested as a way to restore altered coupling and transcellular electrical activity in other organs. Whether such intriguing strategies could also be applied to the inner ear to treat hearing impairment remains to be determined.
Conclusion
The present study shows for the first time the combined distribution of GJB2 and GJB6 gene transcripts in various turns of the HC using RNAscope technique (Figure 12). The transcripts are often co-localized, but were also separately expressed with a heterogeneous localization in the stria vascularis. The GJB6 transcripts, but not the GJB2, were found in the intermediate cells while the GJB2 transcripts dominated over GJB6 in the basal cells. The largest amount of gene transcripts were found in the outer sulcus epithelium, type I and II fibrocytes, and basal cells. A reduction in the number of GJB2 gene transcripts were noted in the basal turn. There were no gene transcripts in the sensory cells or Reissner’s membrane. Few GJB2 and GJB6 gene transcripts were distinguished in the spiral ganglion. The findings suggest that cells and GJ plaques may be specialized and contain different channels with various permeability and gating characteristics in the cochlear domains.
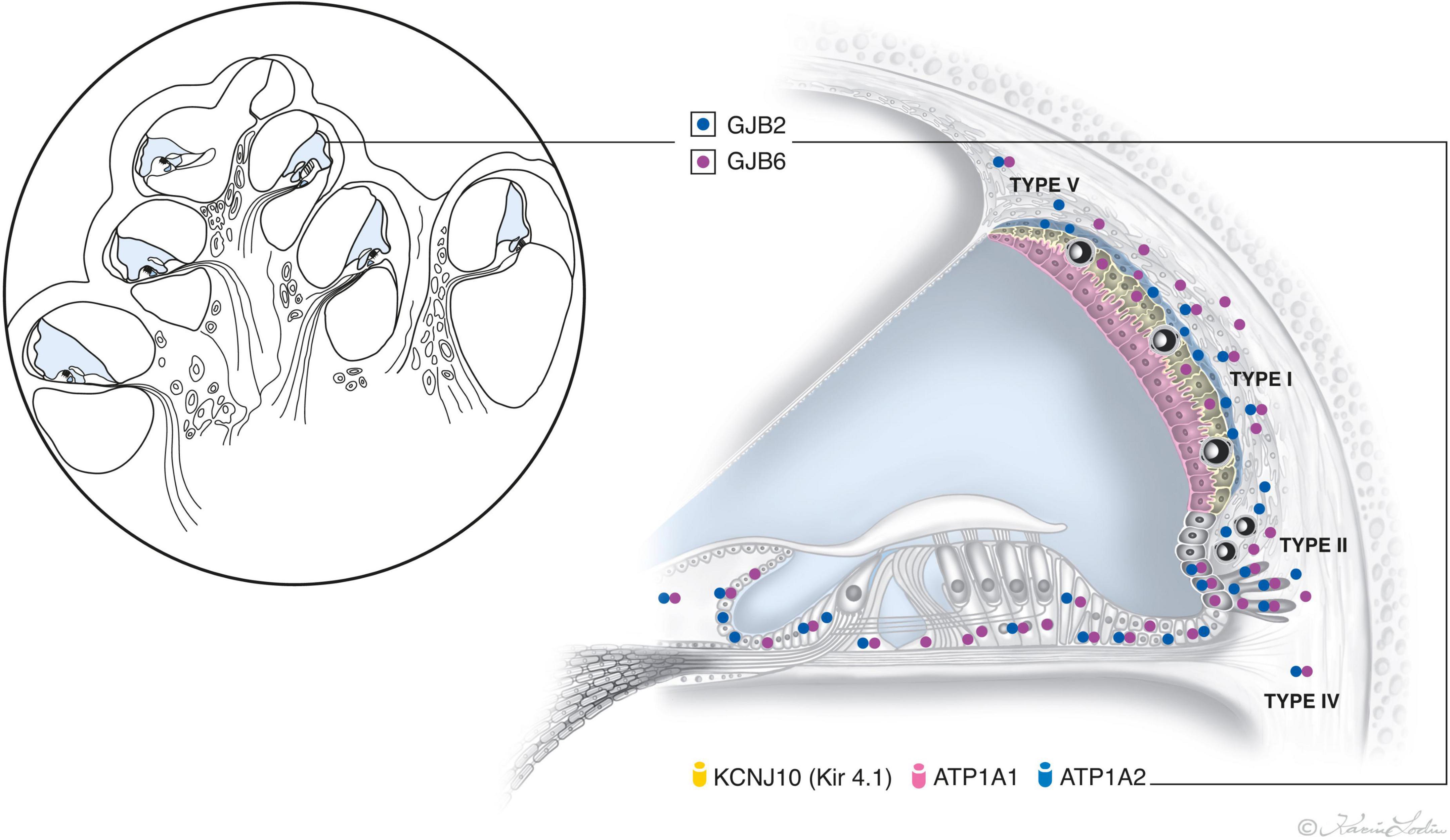
Figure 12. Illustration showing the relative distribution of GJB2 and GJB6 gene transcript puncta in the epithelial and connective tissue networks in the human cochlea. The location and number of transcripts vary among cells in the lateral wall, with the largest number appearing in the outer sulcus cells, root cells, and types I and II fibrocytes and basal cells. GJB2 dominates in the basal cell layer. The intermediate cells only express GJB6. No GJB2 and GJB6 gene transcripts are expressed in the marginal and sensory hair cells. Two GJ systems exist in the HC – Epithelial and connective tissue. The epithelial system consists of interdental cells of the spiral limbus, supporting cells of the organ of Corti (OC), bordering cells, phalangeal cells, Hensen cells, Deiters’ cells, inner and outer sulcus cells with root cells projecting into the spiral ligament, and marginal cells of the stria vascularis. The connective tissue system consists of fibrocytes of the spiral limbus, lateral wall including the basal and intermediate cells of the stria vascularis (SV), suprastrial fibrocytes, and mesenchymal cells lining the scala vestibuli and supralimbal cells (Kikuchi et al., 1995).
Data availability statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Human cochlea tissue used in prior studies was reused, semi-thin sectioned, and photographed (Tylstedt et al., 1997). The study was approved by the Local Ethics Committee (no. 99398, 22/9 1999, cont., 2003, no. C254/4; no. C45/7 2007, Dnr. 2013/190). The study adhered to the rules of the Declaration of Helsinki. Specimens were earlier obtained from patients suffering from life-threatening petroclival meningioma undergoing transcochlear surgery. The patients/participants provided written informed consent to participate in this study.
Author contributions
WL performed IHC and RNAscope. HR-A and WL documented the results, interpreted data, and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
ALF grants from Uppsala University Hospital and Uppsala University, the Tysta Skolan Foundation, and the Swedish Deafness Foundation (hrf) supported this study. MED-EL GmbH Innsbruck, Austria supported this project under an agreement and contract with Uppsala University. We also acknowledge the kind donations of private funds made by Arne Sundström, Sweden. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We are grateful to SciLife Laboratories and the Bio-Vis Platform at Uppsala University for providing the SR-SIM microscope equipment and personal support throughout the duration of the study. For semi-and thin sectioning of human cochlea we are indebted to Karin Staxäng at the BioVis Platform EM, Uppsala University, Uppsala, Sweden. We gratefully thank MED-EL GmbH Innsbruck, Austria. We especially thank Susanne Braun and Carolyn Garnham of MED-EL Austria.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/nuccore/NM_001110219.2
- ^ https://www.ncbi.nlm.nih.gov/nuccore/NM_004004.5
References
Ahmad, S., Chen, S., Sun, J., and Lin, X. (2003). Connexins 26 and 30 are co-assembled to form gap junctions in the cochlea of mice. Biochem. Biophys. Res. Commun. 307, 362–368. doi: 10.1016/S0006-291X(03)01166-5
Ahmad, S., Tang, W., Chang, Q., Qu, Y., Hibshman, J., Li, Y., et al. (2007). Restoration of connexin26 protein level in the cochlea completely rescues hearing in a mouse model of human connexin30-linked deafness. Proc. Natl. Acad. Sci. U.S.A. 104, 1337–1341. doi: 10.1073/pnas.0606855104
Bahar Halpern, K., Tanami, S., Landen, S., Chapal, M., Szlak, L., Hutzler, A., et al. (2015). Bursty gene expression in the intact mammalian liver. Mol. Cell 58, 147–156. doi: 10.1016/J.MOLCEL.2015.01.027/ATTACHMENT/B4FA4D3D-BFF7-4540-83ED-478CFC3CBDB8/MMC3.XLSX
Baroja-Mazo, A., Barberà-Cremades, M., and Pelegrín, P. (2013). The participation of plasma membrane hemichannels to purinergic signaling. Biochim. Biophys. Acta Biomembr. 1828, 79–93. doi: 10.1016/J.BBAMEM.2012.01.002
Beltramello, M., Piazza, V., Bukauskas, F. F., Pozzan, T., and Mammano, F. (2004). Impaired permeability to Ins(1,4,5)P3 in a mutant connexin underlies recessive hereditary deafness. Nat. Cell Biol. 71, 63–69. doi: 10.1038/ncb1205
Boulay, A. C., del Castillo, F. J., Giraudet, F., Hamard, G., Giaume, C., Petit, C., et al. (2013). Hearing is normal without connexin30. J. Neurosci. 33, 430–434. doi: 10.1523/JNEUROSCI.4240-12.2013
Bukauskas, F. F., Jordan, K., Bukauskiene, A., Bennett, M. V. L., Lampe, P. D., Laird, D. W., et al. (2000). Clustering of connexin 43-enhanced green fluorescent protein gap junction channels and functional coupling in living cells. Proc. Natl. Acad. Sci. U. S. A. 97, 2556–2561. doi: 10.1073/PNAS.050588497/ASSET/29B990C6-359E-4B19-B8BA-2802D5A5C300/ASSETS/GRAPHIC/PQ0505884004.JPEG
Chang, Q., Tang, W., Ahmad, S., Zhou, B., and Lin, X. (2008). Gap Junction Mediated Intercellular Metabolite Transfer in the Cochlea Is Compromised in Connexin30 Null Mice. PLoS One 3:e4088. doi: 10.1371/JOURNAL.PONE.0004088
Cohen-Salmon, M., Ott, T., Michel, V., Hardelin, J. P., Perfettini, I., Eybalin, M., et al. (2002). Targeted ablation of connexin26 in the inner ear epithelial gap junction network causes hearing impairment and cell death. Curr. Biol. 12, 1106–1111. doi: 10.1016/S0960-9822(02)00904-1
Cohn, E. S., and Kelley, P. M. (1999). Clinical phenotype and mutations in connexin 26 (DFNB1/GJB2), the most common cause of childhood hearing loss. Am. J. Med. Genet. 89, 130–136. doi: 10.1002/(SICI)1096-8628(19990924)89:3<130::AID-AJMG3<3.0.CO;2-M
Dbouk, H. A., Mroue, R. M., El-Sabban, M. E., and Talhouk, R. S. (2009). Connexins: A myriad of functions extending beyond assembly of gap junction channels. Cell Commun. Signal 7:4. doi: 10.1186/1478-811X-7-4
Del Castillo, F. J., and Del Castillo, I. (2017). DFNB1 non-syndromic hearing impairment: Diversity of mutations and associated phenotypes. Front. Mol. Neurosci. 10:428. doi: 10.3389/fnmol.2017.00428
Del Castillo, F. J., Rodríguez-Ballesteros, M., Álvarez, A., Hutchin, T., Leonardi, E., De Oliveira, C. A., et al. (2005). A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 42, 588–594. doi: 10.1136/jmg.2004.028324
del Castillo, I., Villamar, M., Moreno-Pelayo, M. A., del Castillo, F. J., Alvarez, A., Telleria, D., et al. (2002). A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N. Engl. J. Med. 346, 243–249. doi: 10.1056/NEJMoa012052
Dermietzel, R., Hwang, T. K., and Spray, D. S. (1990). The gap junction family: Structure, function and chemistry. Anat. Embryol. 182, 517–528. doi: 10.1007/BF00186458
Dhein, S., and Salameh, A. (2021). Remodeling of Cardiac Gap Junctional Cell-Cell Coupling. Cells 10:2422. doi: 10.3390/CELLS10092422
Evans, W. H., and Martin, P. E. M. (2009). Gap junctions: Structure and function (Review). Mol. Membr. Biol. 19, 121–136. doi: 10.1080/09687680210139839
Fetoni, A. R., Zorzi, V., Paciello, F., Ziraldo, G., Peres, C., Raspa, M., et al. (2018). Cx26 partial loss causes accelerated presbycusis by redox imbalance and dysregulation of Nfr2 pathway. Redox Biol. 19, 301–317. doi: 10.1016/j.redox.2018.08.002
Forge, A., Marziano, N. K., Casalotti, S. O., Becker, D. L., and Jagger, D. (2003). The inner ear contains heteromeric channels composed of Cx26 and Cx30 and deafness-related mutations in Cx26 have a dominant negative effect on Cx30. Cell Commun. Adhes. 10, 341–346. doi: 10.1080/cac.10.4-6.341.346
Fujimoto, K. (1995). Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 108, 3443–3449. doi: 10.1242/JCS.108.11.3443
Giepmans, B. N. G., and Moolenaar, W. H. (1998). The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr. Biol. 8, 931–934. doi: 10.1016/S0960-9822(07)00375-2
Goodenough, D. A., Goliger, J. A., and Paul, D. L. (2003). CONNEXINS, CONNEXONS, AND INTERCELLULAR COMMUNICATION. Annu. Rev. Biochem. 65, 475–502. doi: 10.1146/ANNUREV.BI.65.070196.002355
Grabinski, T. M., Kneynsberg, A., Manfredsson, F. P., and Kanaan, N. M. (2015). A method for combining rnascope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS One 10:e0120120. doi: 10.1371/journal.pone.0120120
Grifa, A., Wagner, C., D’Ambrosio, L., Melchionda, S., Bernardi, F., Lopez-Bigas, N., et al. (1999). Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nat. Genet. 23, 16–18. doi: 10.1038/12612
Gustafsson, M. G. L., Shao, L., Carlton, P. M., Wang, C. J. R., Golubovskaya, I. N., Cande, W. Z., et al. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970. doi: 10.1529/biophysj.107.120345
Hibino, H., Horio, Y., Inanobe, A., Doi, K., Ito, M., Yamada, M., et al. (1997). An ATP-Dependent Inwardly Rectifying Potassium Channel, KAB-2 (Kir4.1), in Cochlear Stria Vascularis of Inner Ear: Its Specific Subcellular Localization and Correlation with the Formation of Endocochlear Potential. J. Neurosci. 17:4711. doi: 10.1523/JNEUROSCI.17-12-04711.1997
Jagger, D. J., and Forge, A. (2015). Connexins and gap junctions in the inner ear it’s not just about K+ recycling. Cell Tissue Res. 360, 633–644. doi: 10.1007/s00441-014-2029-z
Kelsell, D. P., Dunlop, J., Stevens, H. P., Lench, N. J., Liang, J. N., Parry, G., et al. (1997). Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387, 80–83. doi: 10.1038/387080a0
Kikuchi, T., Kimura, R. S., Paul, D. L., and Adams, J. C. (1995). Gap junctions in the rat cochlea: Immunohistochemical and ultrastructural analysis. Anat. Embryol. 191, 101–118. doi: 10.1007/BF00186783
Kim, J., Choi, M., Kim, J. R., Jin, H., Kim, V. N., and Cho, K. H. (2012). The co-regulation mechanism of transcription factors in the human gene regulatory network. Nucleic Acids Res. 40, 8849–8861. doi: 10.1093/NAR/GKS664
Kitajiri, S. I., Furuse, M., Morita, K., Saishin-Kiuchi, Y., Kido, H., Ito, J., et al. (2004a). Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear. Res. 187, 25–34. doi: 10.1016/S0378-5955(03)00338-1
Kitajiri, S. I., Miyamoto, T., Mineharu, A., Sonoda, N., Furuse, K., Hata, M., et al. (2004b). Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J. Cell Sci. 117, 5087–5096. doi: 10.1242/jcs.01393
Kojima, T., Kokai, Y., Chiba, H., Yamamoto, M., Mochizuki, Y., and Sawada, N. (2001). Cx32 but not Cx26 is associated with tight junctions in primary cultures of rat hepatocytes. Exp. Cell Res. 263, 193–201. doi: 10.1006/EXCR.2000.5103
Kojima, T., Spray, D. C., Kokai, Y., Chiba, H., Mochizuki, Y., and Sawada, N. (2002). Cx32 formation and/or Cx32-mediated intercellular communication induces expression and function of tight junctions in hepatocytic cell line. Exp. Cell Res. 276, 40–51. doi: 10.1006/EXCR.2002.5511
Konishi, T., Hamrick, P. E., and Walsh, P. J. (1978). Ion transportin guinea pig cochlea. I. Potassium and sodium transport. Acta Otolaryngol. (Stockh) 86, 22–34.
Laird, D. W. (2006). Life cycle of connexins in health and disease. Biochem. J. 394, 527–543. doi: 10.1042/BJ20051922
Laird, D. W., and Lampe, P. D. (2018). Therapeutic strategies targeting connexins. Nat. Rev. Drug Discov. 1712, 905–921. doi: 10.1038/nrd.2018.138
Lautermann, J., Ten Cate, W. J. F., Altenhoff, P., Jahnke, K., Grümmer, R., Winterhager, E., et al. (1998). Expression of the gap-junction connexins 26 and 30 in the rat cochlea. Cell Tissue Res. 294, 415–420. doi: 10.1007/S004410051192
Lerer, I., Sagi, M., Ben-Neriah, Z., Wang, T., Levi, H., and Abeliovich, D. (2001). A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: A novel founder mutation in Ashkenazi Jews. Hum. Mutat. 18:460. doi: 10.1002/HUMU.1222
Leybaert, L., Braet, K., Vandamme, W., Cabooter, L., Martin, P. E. M., and Evans, W. H. (2003). Connexin channels, connexin mimetic peptides and ATP release. Cell Commun. Adhes. 10, 251–257. doi: 10.1080/CAC.10.4-6.251.257
Liu, W., Boström, M., Kinnefors, A., and Rask-Andersen, H. (2009). Unique expression of connexins in the human cochlea. Hear. Res. 250, 55–62. doi: 10.1016/j.heares.2009.01.010
Liu, W., Danckwardt-Lillieström, N., Schrott-Fischer, A., Glueckert, R., and Rask-Andersen, H. (2021a). Distribution of Immune Cells Including Macrophages in the Human Cochlea. Front. Neurol. 12:2092. doi: 10.3389/fneur.2021.781702
Liu, W., Luque, M., Li, H., Schrott-Fischer, A., Glueckert, R., Tylstedt, S., et al. (2021b). Spike Generators and Cell Signaling in the Human Auditory Nerve: An Ultrastructural, Super-Resolution, and Gene Hybridization Study. Front. Cell. Neurosci. 15:642211. doi: 10.3389/fncel.2021.642211
Liu, W., Edin, F., Blom, H., Magnusson, P., Schrott-Fischer, A., Glueckert, R., et al. (2016). Super-resolution structured illumination fluorescence microscopy of the lateral wall of the cochlea: The Connexin26/30 proteins are separately expressed in man. Cell Tissue Res. 365, 13–27. doi: 10.1007/s00441-016-2359-0
Liu, W., Li, H., Edin, F., Brännström, J., Glueckert, R., Schrott-Fischer, A., et al. (2017a). Molecular composition and distribution of gap junctions in the sensory epithelium of the human cochlea—a super-resolution structured illumination microscopy (SR-SIM) study. Ups. J. Med. Sci. 122, 160–170. doi: 10.1080/03009734.2017.1322645
Liu, W., Schrott-Fischer, A., Glueckert, R., Benav, H., and Rask-Andersen, H. (2017b). The Human “Cochlear Battery” – Claudin-11 Barrier and Ion Transport Proteins in the Lateral Wall of the Cochlea. Front. Mol. Neurosci. 10:239 doi: 10.3389/fnmol.2017.00239
Liu, W., Luque, M., Glueckert, R., Danckwardt-Lillieström, N., Nordström, C. K., Schrott-Fischer, A., et al. (2019). Expression of Na/K-ATPase subunits in the human cochlea: A confocal and super-resolution microscopy study with special reference to auditory nerve excitation and cochlear implantation. Ups. J. Med. Sci. 124, 168–179. doi: 10.1080/03009734.2019.1653408
Liu, W., and Rask-Andersen, H. (2022). Na/K-ATPase Gene Expression in the Human Cochlea: A Study Using mRNA in situ Hybridization and Super-Resolution Structured Illumination Microscopy. Front. Mol. Neurosci. 15:108. doi: 10.3389/fnmol.2022.857216
Maeda, S., Nakagawa, S., Suga, M., Yamashita, E., Oshima, A., Fujiyoshi, Y., et al. (2009). Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458, 597–602. doi: 10.1038/NATURE07869
Mammano, F. (2019). Inner Ear Connexin Channels: Roles in Development and Maintenance of Cochlear Function. Cold Spring Harb. Perspect. Med. 9:a033233. doi: 10.1101/CSHPERSPECT.A033233
Mammano, F., and Bortolozzi, M. (2018). Ca2+ signaling, apoptosis and autophagy in the developing cochlea: Milestones to hearing acquisition. Cell Calcium 70, 117–126. doi: 10.1016/J.CECA.2017.05.006
Marcus, D. C., Wu, T., Wangemann, P., and Kofuji, P. (2002). KCNJ10 (Kir4.1) potassium channel knockout abolishes endocochlear potential. Am. J. Physiol. Cell Physiol. 282, C403–C407. doi: 10.1152/ajpcell.00312.2001
Metz, J., Aoki, A., and Forssmann, W. G. (1978). Studies on the ultrastructure and permeability of the hemotrichorial placenta. Cell Tissue Res. 1923, 391–407. doi: 10.1007/BF00212321
Pallares-Ruiz, N., Blanchet, P., Mondain, M., Claustres, M., and Roux, A. F. (2002). A large deletion including most of GJB6 in recessive non syndromic deafness: A digenic effect? Eur. J. Hum. Genet. 10, 72–76. doi: 10.1038/SJ.EJHG.5200762
Petit, C., Levilliers, J., and Hardelin, J. P. (2001). Molecular genetics of hearing loss. Annu. Rev. Genet. 35, 589–646. doi: 10.1146/annurev.genet.35.102401.091224
Rash, J. E., Yasumura, T., Davidson, K. G. V., Furman, C. S., Dudek, F. E., and Nagy, J. I (2001). Identification of Cells Expressing Cx43, Cx30, Cx26, Cx32 and Cx36 in Gap Junctions of Rat Brain and Spinal Cord. Cell Commun. Adhes. 8, 315–320. doi: 10.3109/15419060109080745
Risek, B., Guthrie, S., Kumar, N., and Gilula, N. B. (1990). Modulation of gap junction transcript and protein expression during pregnancy in the rat. J. Cell Biol. 110, 269–282. doi: 10.1083/JCB.110.2.269
Salt, A. N., Melichar, I., and Thalmann, R. (1987). Mechanisms of endocochlear potential generation by stria vascularis. Laryngoscope 97, 984–991. doi: 10.1288/00005537-198708000-00020
Schermelleh, L., Carlton, P. M., Haase, S., Shao, L., Winoto, L., Kner, P., et al. (2008). Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336. doi: 10.1126/science.1156947
Sosinsky, G., and Caspar, D. L. D. (1995). Mixing of connexins in gap junction membrane channels. Proc. Natl. Acad. Sci. U. S. A. 92:9210. doi: 10.1073/PNAS.92.20.9210
Sterkers, O., Saumon, G., Tran Ba Huy, P., and Amiel, C. (1982). K, Cl, and H2O entry in endolymph, perilymph, and cerebrospinal fluid of the rat. Am. J. Physiol. Renal. Fluid Electrolyte Physiol. 243, F173–F180.
Sun, J., Ahmad, S., Chen, S., Tang, W., Zhang, Y., Chen, P., et al. (2005). Cochlear gap junctions coassembled from Cx26 and 30 show faster intercellular Ca 2+ signaling than homomeric counterparts. Am. J. Physiol. Physiol. 288, C613–C623. doi: 10.1152/ajpcell.00341.2004
Sun, Y., Tang, W., Chang, Q., Wang, Y., Kong, W., and Lin, X. (2009). Connexin30 null and conditional connexin26 null mice display distinct pattern and time course of cellular degeneration in the cochlea. J. Comp. Neurol. 516, 569–579. doi: 10.1002/cne.22117
Tasaki, I., and Spyropoulos, C. S. (1959). Stria vascularis as source of endocochlear potential. J. Neurophysiol. 22, 149–155. doi: 10.1152/jn.1959.22.2.149
Tylstedt, S., Kinnefors, A., and Rask-Andersen, H. (1997). Neural interaction in the human spiral ganglion: A TEM study. Acta Otolaryngol. 117, 505–512. doi: 10.3109/00016489709113429
Wang, H., Su, N., Wang, L. C., Wu, X., Bui, S., Chang, K. J., et al. (2015). “Multiplex fluorescent RNA in situ hybridization via RNAscope,” in Situ Hybridization Methods, ed G. Hauptmann (New York: Springer), 405–414. doi: 10.1007/978-1-4939-2303-8_21
Wang, H. L., Chang, W. T., Li, A. H., Yeh, T. H., Wu, C. Y., Chen, M. S., et al. (2003). Functional analysis of connexin-26 mutants associated with hereditary recessive deafness. J. Neurochem. 84, 735–742. doi: 10.1046/j.1471-4159.2003.01555.x
Wang, N., De Bock, M., Decrock, E., Bol, M., Gadicherla, A., Vinken, M., et al. (2013). Paracrine signaling through plasma membrane hemichannels. Biochim. Biophys. Acta Biomembr. 1828, 35–50. doi: 10.1016/j.bbamem.2012.07.002
Wang, S. L., Yu, L. G., Liu, R. P., Zhu, W. Z., Gao, W. M., Xue, L. P., et al. (2014). Gene-Gene Interaction of GJB2, SOD2, and CAT on Occupational Noise-induced Hearing Loss in Chinese Han Population. Biomed. Environ. Sci. 27, 965–968. doi: 10.3967/BES2014.131
Wangemann, P., Liu, J., and Marcus, D. C. (1995). Ion transport mechanisms responsible for K+ secretion and the transepithelial voltage across marginal cells of stria vascularis in vitro. Hear. Res. 84, 19–29. doi: 10.1016/0378-5955(95)00009-S
White, T. W., and Bruzzone, R. (1996). Multiple connexin proteins in single intercellular channels: Connexin compatibility and functional consequences. J. Bioenerg. Biomembr. 28, 339–350. doi: 10.1007/BF02110110
Willebrords, J., Crespo Yanguas, S., Maes, M., Decrock, E., Wang, N., Leybaert, L., et al. (2016). Connexins and their channels in inflammation. Crit. Rev. Biochem. Mol. Biol. 51, 413–439. doi: 10.1080/10409238.2016.1204980
Wu, X., Wang, Y., Sun, Y., Chen, S., Zhang, S., Shen, L., et al. (2014). Reduced expression of Connexin26 and its DNA promoter hypermethylation in the inner ear of mimetic aging rats induced by d-galactose. Biochem. Biophys. Res. Commun. 452, 340–346. doi: 10.1016/J.BBRC.2014.08.063
Xia, J. H., Liu, C. Y., Tang, B. S., Pan, Q., Huang, L., Dai, H. P., et al. (1998). Mutations in the gene encoding gap junction protein β-3 associated with autosomal dominant hearing impairment. Nat. Genet. 204, 370–373. doi: 10.1038/3845
Ye, Z. C., Wyeth, M. S., Baltan-Tekkok, S., and Ransom, B. R. (2003). Functional hemichannels in astrocytes: A novel mechanism of glutamate release. J. Neurosci. 23, 3588–3596. doi: 10.1523/JNEUROSCI.23-09-03588.2003
Yu, Q., Wang, Y., Chang, Q., Wang, J., Gong, S., Li, H., et al. (2014). Virally-expressed connexin26 restores gap junction function in the cochlea of conditional GJB2 knockout mice. Gene Ther. 21, 71–80. doi: 10.1038/gt.2013.59
Zhao, H. B., and Santos-Sacchi, J. (2000). Voltage gating of gap junctions in cochlear supporting cells: Evidence for nonhomotypic channels. J. Membr. Biol. 175, 17–24. doi: 10.1007/s002320001051
Zhao, H. B., and Yu, N. (2006). Distinct and gradient distributions of connexin26 and connexin30 in the cochlear sensory epithelium of guinea pigs. J. Comp. Neurol. 499, 506–518. doi: 10.1002/cne.21113
Zhou, X. X., Chen, S., Xie, L., Ji, Y. Z., Wu, X., Wang, W. W., et al. (2016). Reduced Connexin26 in the Mature Cochlea Increases Susceptibility to Noise-Induced Hearing Loss in Mice. Int. J. Mol. Sci. 17:301. doi: 10.3390/IJMS17030301
Keywords: human, cochlea, GJB2, GJB6, RNAscope, in situ hybridization
Citation: Liu W and Rask-Andersen H (2022) GJB2 and GJB6 gene transcripts in the human cochlea: A study using RNAscope, confocal, and super-resolution structured illumination microscopy. Front. Mol. Neurosci. 15:973646. doi: 10.3389/fnmol.2022.973646
Received: 20 June 2022; Accepted: 01 August 2022;
Published: 20 September 2022.
Edited by:
Laura Astolfi, University of Padua, ItalyReviewed by:
Francisco Javier del Castillo, Ramón y Cajal University Hospital, SpainGerald Seifert, University Hospital Bonn, Germany
Delfo D’Alessandro, University of Pisa, Italy
Copyright © 2022 Liu and Rask-Andersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Liu, bHdvbzI0QGdtYWlsLmNvbQ==
 Wei Liu
Wei Liu Helge Rask-Andersen
Helge Rask-Andersen