- 1Institute of Experimental Neurology, IRCCS San Raffaele Hospital and Vita-Salute San Raffaele University, Milan, Italy
- 2Department of Drug Sciences, Pharmacology Section, University of Pavia, Pavia, Italy
- 3Department of Pharmacy (DiFar), School of Medical and Pharmaceutical Sciences, University of Genoa, Genoa, Italy
- 4Center of Excellence for Biomedical Research, 3Rs Center, University of Genoa, Genoa, Italy
- 5Centro 3R (Inter-University Center for the Promotion of the 3Rs Principles in Teaching and Research), Italy
The circadian molecular machinery is a fine timekeeper with the capacity to harmonize physiological and behavioral processes with the external environment. This tight-knit regulation is coordinated by multiple cellular clocks across the body. In this review, we focus our attention on the molecular mechanisms regulated by the clock in different brain areas and within different cells of the central nervous system. Further, we discuss evidence regarding the role of circadian rhythms in the regulation of neuronal activity and neurotransmitter systems. Not only neurons, but also astrocytes and microglia actively participate in the maintenance of timekeeping within the brain, and the diffusion of circadian information among these cells is fine-tuned by neurotransmitters (e.g., dopamine, serotonin, and γ-aminobutyric acid), thus impacting on the core clock machinery. The bidirectional interplay between neurotransmitters and the circadian clockwork is fundamental in maintaining accuracy and precision in daily timekeeping throughout different brain areas. Deepening the knowledge of these correlations allows us to define the basis of drug interventions to restore circadian rhythms, as well as to predict the onset of drug treatment/side effects that might promote daily desynchronization. Furthermore, it may lead to a deeper understanding of the potential impacts of modulations in rhythmic activities on the pace of aging and provide an insight in to the pathogenesis of psychiatric diseases and neurodegenerative disorders.
Introduction
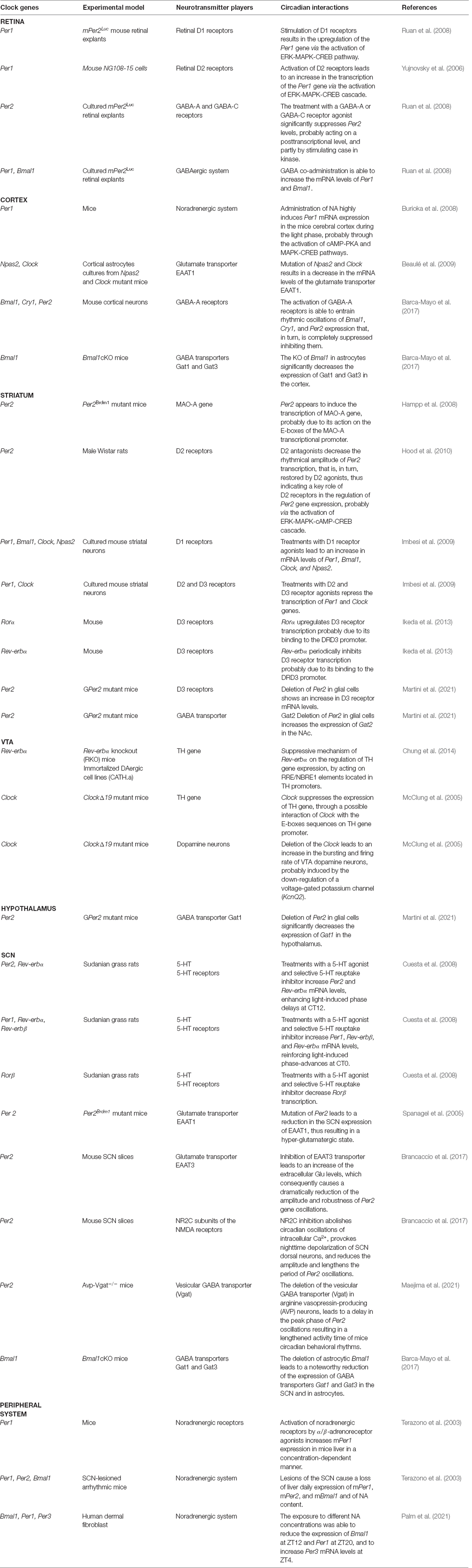
Circadian rhythms are the basis of daily life, enabling the adaptation and adjustment of physiological and behavioral processes—from the brain to the peripheral organs—with the external environment. The entrainment and synchronization of the endogenous circadian oscillator rely on external timing cues, also called Zeitgebers, which allow the internal molecular clockwork to align with the environment. Light represents the main external cue that, through the retinohypothalamic tract (RHT; Gooley et al., 2001; Chen et al., 2011), is transmitted to the hypothalamic suprachiasmatic nucleus (SCN), the master circadian pacemaker (Stephan and Zucker, 1972; Ralph et al., 1990; Hastings et al., 2003). The SCN contains approximately 10,000 neurons, each of them displaying a cell-autonomous circadian clock that is able to coordinate all the other “slave” and peripheral oscillators located in nearly every mammalian organ. Noteworthy, the synchronization of peripheral clocks is not only hierarchically orchestrated by the hypothalamic circadian pacemaker through direct neuronal innervations but also through multiple soluble molecular signals, i.e., daily rhythms of glucocorticoids (Dibner et al., 2010). At the molecular level, the internal cellular clockworks are known as autoregulatory transcriptional–translational feedback loops (TTFLs; as schematized in Figure 1). The circadian locomotor output cycles kaput (Clock) and the brain and muscle Arnt-like protein 1 (Bmal1) are central players in the regulation of this loop. As positive regulators of the circadian cycle, they interact together to form the Clock:Bmal1 heterodimer in the cytoplasm. The latter translocates into the nucleus where it binds the Enhancer Box (E-box) response elements on the promoters of Period (Per1, Per2, Per3) and Cryptochrome (Cry1, Cry2), the core clock components of the negative arm of the TTFL (Hastings et al., 2018; Cox and Takahashi, 2019). Upon translation and nuclear accumulation, Per and Cry proteins inhibit the transcriptional activity of Clock:Bmal1 heterodimer. During night-time hours, Per and Cry proteins are progressively phosphorylated by the casein kinase 1ε/δ (CK1ε/δ) and adenosine 3’,5’-monophosphate (AMP) kinase (AMPK) respectively, ubiquitinated by their specific E3 ubiquitin ligase complexes, and finally degraded by the proteasome (Shirogane et al., 2005), thus turning off their inhibitory effect on Clock:Bmal1 and allowing for the onset of a new oscillatory cycle (Koike et al., 2012). In addition to the core loop, further regulators are represented by the nuclear receptors Rev-erbα (also known as Nr1d1), Rev-erbβ (also known as Nr1d2), and the retinoic acid orphan receptors Rorα, Rorβ, and Rorγ (Rorα/β/γ), which together form a second TTFL that steadies the robustness of the main TTFL, by regulating the rhythmic oscillation of Bmal1 expression throughout the day. In particular, Rorα/β/γ positively regulate the transcription of Bmal1 gene, whereas Rev-erbα/β act as transcriptional repressors of its expression, by both competing for binding to Retinoic acid receptor-related Orphan Receptor Element (RORE) sites present on Bmal1 gene promoter (Preitner et al., 2002; Koike et al., 2012).
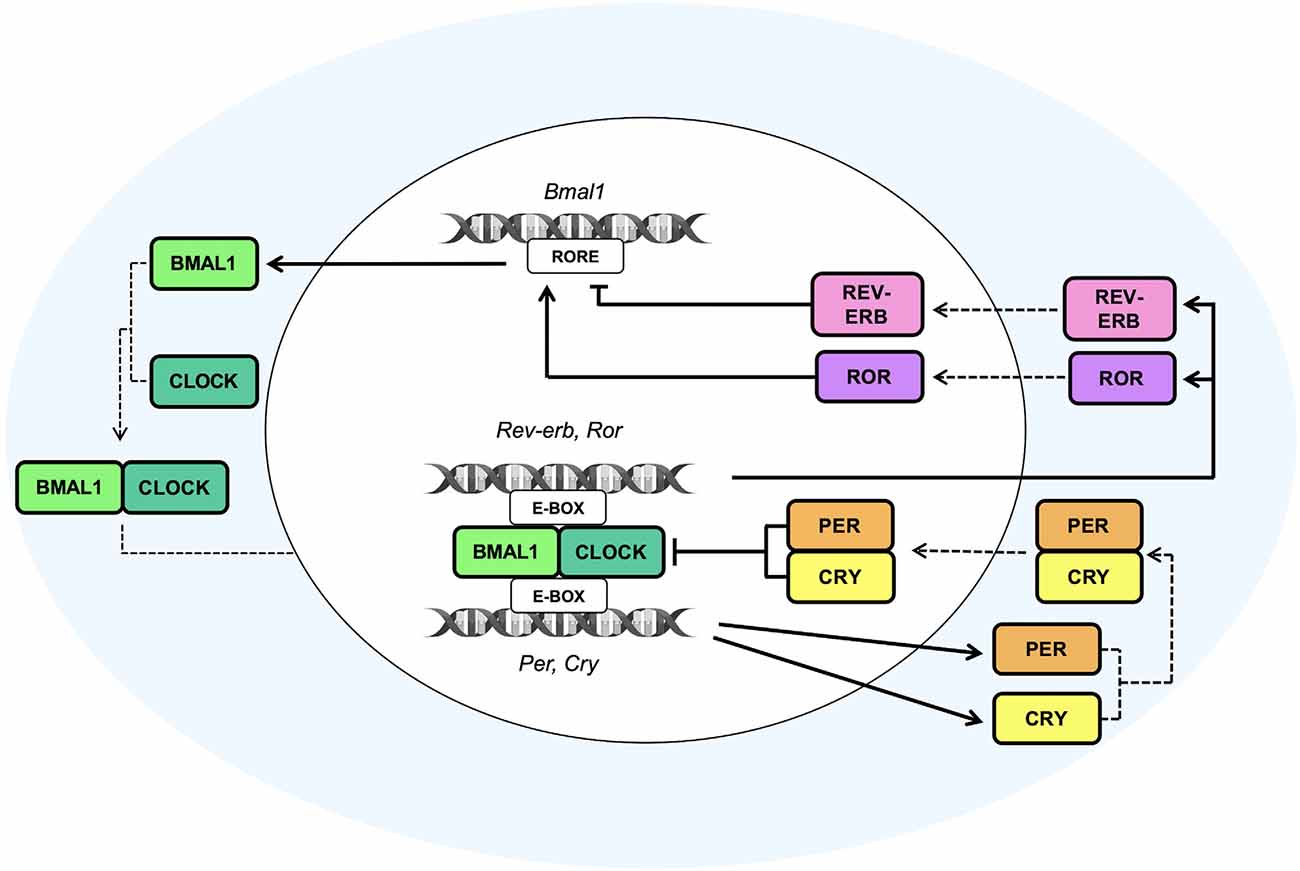
Figure 1. The molecular clock machinery. Schematic view of the molecular transcriptional–translational feedback loops (TTFLs) that coordinate the multiple cellular clocks across the mammalian body. The heterodimeric complexes given by the brain and muscle Arnt-like protein 1 (Bmal1) and circadian locomotor output cycles kaput (Clock) bind the Enhancer Box response element (E-box) on Period (Per) and Cryptochrome (Cry) gene promoters to induce the daytime expression of Per and Cry proteins, which, in turn, inhibit the transcriptional activity of Clock:Bmal1 heterodimer. The subsequent nocturnal degradation of Per and Cry proteins suppresses their inhibitory effect on Clock:Bmal1 heterodimer and allows to start a new oscillatory cycle. An additional feedback loop, consisting of the nuclear receptors Rev-erb and the retinoic acid orphan receptors Ror, is involved in the establishment of the rhythmic expression of Bmal1 throughout the day. Ror and Rev-erb are both transcribed by the binding of the heterodimer Clock and Bmal1 to the E-box sequences on their gene promoters. Particularly, Ror positively regulates the transcription of the Bmal1 gene, whereas Rev-erb acts as a transcriptional repressor of its expression, by both competing for binding Retinoic acid receptor-related Orphan Receptor Element (RORE) sites present on the Bmal1 gene promoter. The cooperation and synchronization of all of these loops, that are the basis of the molecular clock machinery, contribute to the generation of daily rhythm.
At the same time, also the intracellular calcium influx, membrane depolarization, and cAMP signaling, which all follow a daily rhythmical pattern, play a key role in the regulation of the biological transcriptional clock and in the establishment of the neuronal firing rhythms (Lundkvist et al., 2005; O’Neill et al., 2008). Indeed, through a cascade pathway these additional players are able to induce the phosphorylation of the calcium/cAMP response element binding protein (CREB) which, in turn, binds calcium/cAMP regulatory elements (CREs) sequences present on Per1 and Per2 gene promoters (Travnickova-Bendova et al., 2002; O’Neill et al., 2008), thus regulating their transcription and creating additional positive feedback loops involved in the generation of daily rhythm.
The pathways described above, however, are not the only mechanisms involved in the generation of rhythm. The TTFL, in fact, regulates the oscillations of approximately 20% of all the genome (Panda et al., 2002). Many other levels of regulation, such as post-transcriptional, translational, and post-translational modifications, play a critical role in coordinating rhythmicity. These modifications impact cellular localization, protein stability, and protein-protein interactions, and are essential for the modulation of multiple biological responses and for the establishment of a fine-tuning rhythmicity (Brenna and Albrecht, 2020). Besides, alterations or mutations of the kinases or on the target sequences responsible for these post-translation modifications can result in several diseases strictly linked to the circadian system, such as sleep disorders, neurodegenerative, and psychiatric diseases, and also cancer (Schwab et al., 2000; Xu et al., 2005; Fang et al., 2015; Brenna and Albrecht, 2020).
Since the circadian molecular machinery acts as a temporal variable, orchestrating a number of physiological functions and processes in the central nervous system (CNS), it is fundamental to acquire an understanding of the molecular mechanisms regulated by the clock in the different brain areas and within the different CNS cells. Therefore, in this review, we aim to critically discuss evidence regarding the role of circadian rhythms in the regulation of neuronal activity and neurotransmitter systems.
The Circadian Electrical Activity
In the SCN, circadian timekeeping is a dynamic process characterized by reciprocal interactions between the molecular oscillations of the clock machinery and the electrical activity of neurons. TTFLs are able to orchestrate, in a circadian manner, the variations of Na+ and K+ currents (Flourakis et al., 2015), as well as the intracellular concentration of Ca2+ ([Ca2+]i; Brancaccio et al., 2013), responsible, in turn, for the regulation of neuronal activity. Remarkably, the [Ca2+]i, moving into cells through voltage-operated calcium channels activated by electrical firing (VOOC), NMDA-type glutamate receptors (NMDARs), or released by intracellular stores (Harvey et al., 2020), plays a pivotal role in the modulation of the biological clockwork (Brancaccio et al., 2013). Indeed, the electrical activation due to the peak of [Ca2+]i triggered by a light signal has been found to induce the expression of Per1 and Per2 genes, thereby affecting the expression of the positive TTFL components (Brancaccio et al., 2013). Notably, at circadian time 6 (CT6) light signals induce Ca2+ influx and activate a phosphorylation cascade pathway involving the protein kinase A and G (PKA and PKG), the Ca2+/calmodulin-dependent protein kinase (CaMK), the mitogen-activated protein kinases (MAPK) and the extracellular-signal-regulated kinases (ERK). This pathway leads to the phosphorylation of CREB, which, after being activated by the cAMP-regulated transcriptional co-activator 1 (CRTC1), heterodimerizes with the histone acetyltransferase CREB binding protein (CBP) and then, at CT9, binds the CRE sequences present on Per gene promoters (Gau et al., 2002; Parra-Damas et al., 2017), leading to the consequent transcription of Per1 (at CT10) and Per2 (at CT12; Brancaccio et al., 2013). Moreover, Brenna et al. recently demonstrated that Per2 supports the stimulus-dependent induction of the Per1 gene by regulating the CREB/CRTC1/CBP complex. Accordingly, upon light or forskolin stimuli, Per2 acts as a co-factor of CREB promoting the formation of a transactivation complex on the CRE element of the Per1 gene. In particular, Per2 has been shown to facilitate the interaction between CREB and its co-regulator CRTC1, thereby inducing complex formation (Brenna et al., 2021).
Furthermore, since the release of [Ca2+]i follows a circadian pattern orchestrated by the TTFLs, the bidirectional interlink between the clockwork and the neural signaling creates a closed loop that drives the rest-activity cycle of the neuronal activity (Brancaccio et al., 2013; Figure 2). These findings demonstrate that TTFL is strongly sensitive to the electrical state, and that the coupling between the clock machinery and Ca2+ activity is fundamental for a proper generation of rhythm and for a robust timekeeping within the brain. Hence, the electrical activity seems to play a key role in re-setting the intracellular clock, contributing to the suppression of neuronal activity during the night, and to the induction of a subsequent electrical firing peak with the coming of the next circadian day.
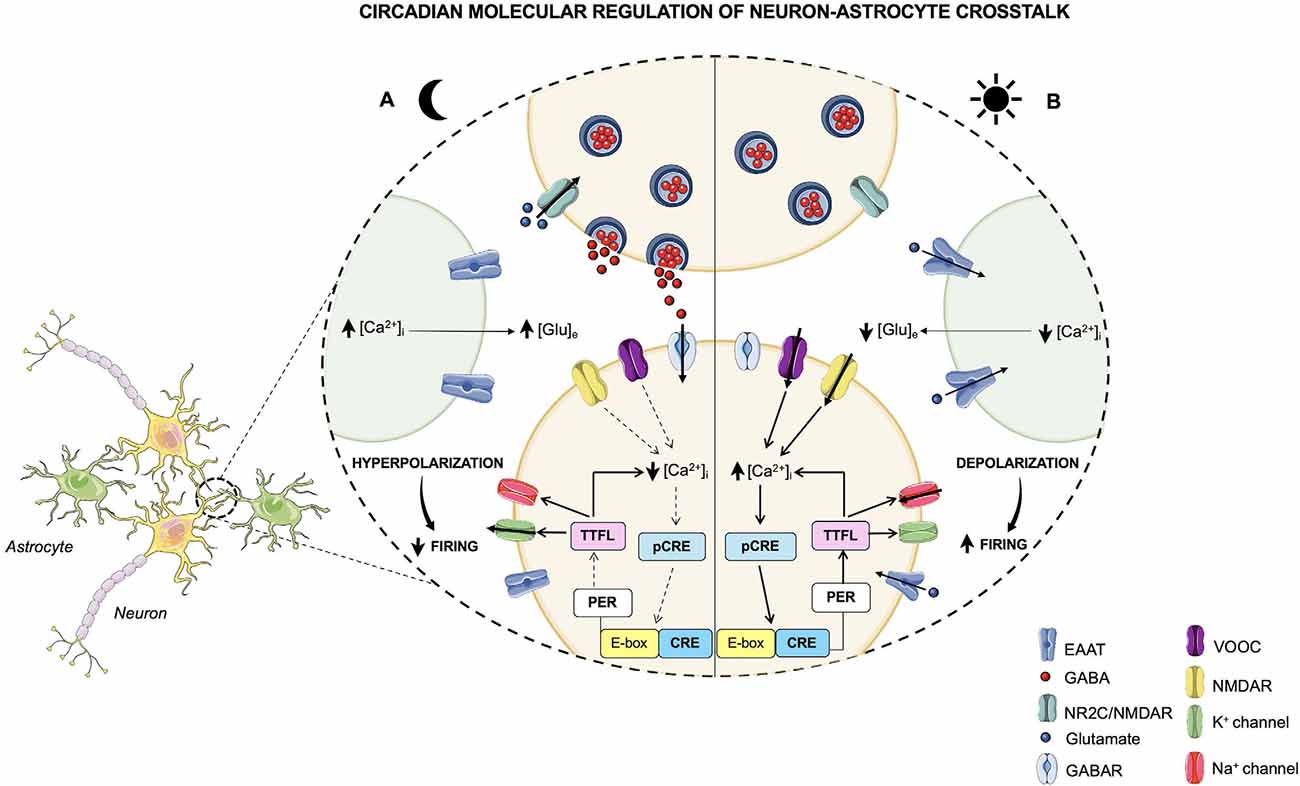
Figure 2. Circadian molecular regulation of neuron-astrocyte crosstalk. The circadian timekeeper in the SCN is dictated by reciprocal interactions between the molecular oscillations of the clock machinery and the electrical activity of neurons, which lead to the establishment of a daily rhythm. (A) During the night, astrocytes are active and their [Ca2+]i is high, leading to an increased release of extracellular Glu, which, in turn, through the NR2C/NMDA receptors, is able to induce the release of GABA from presynaptic neurons. GABA acts on the postsynaptic GABA receptors and exerts its inhibitory effect on neurons that during the night are less active. In fact, the neuronal [Ca2+]i is low, and, as a result, the loop that impacts the TTFLs is blunted. However, the TTFL activates the K+conductances, thus leading to a hyperpolarization of the cell membrane and to a dampened firing. (B) During the daytime, when astrocytes are quiescent with a low [Ca2+]i, Glu is re-uptaken by astrocytes and neurons and, consequently, its extracellular levels are decreased. On the other hand, neurons are active and neuronal [Ca2+]i, whose oscillations are controlled by the TTFL, is high. The neuronal electrical activity stimulates the activation of the Ca2+/cAMP-responsive elements (CREs) on Per genes, thus promoting the transcription of Per1 and Per2 genes. Subsequently, the transcription of these TTFL components leads to a further rise in the [Ca2+]i and to the activation of Na+ currents that results in cell membrane depolarization and a heightened firing.
The Clock in Neurotransmission
Data concerning the synthesis and the uptake of neurotransmitters and their correlation with the expression/functions of circadian pacemakers support the role of certain transmitters (e.g., dopamine -DA-, serotonin -5-HT-, and γ-aminobutyric acid -GABA-) as fine-tuners of daily rhythmicity (Terazono et al., 2003; Imbesi et al., 2009; Nakamaru-Ogiso et al., 2012; Barca-Mayo et al., 2017; Brancaccio et al., 2017; Maejima et al., 2021; Palm et al., 2021). Deepening the knowledge of these correlations may lay the foundation of drug interventions aimed at restoring circadian rhythms, but also at predicting the onset of side effects related to drug treatment/abuse that might cause daily desynchronization. Besides the events discussed so far, other specific mechanisms dictating the strength and the specificity of chemical transmission would deserve attention due to their potential role in circadian rhythmicity. In relation to the latter, we will briefly discuss how the mode of synaptic communication, the co-localization of different neurotransmitters, and the intensity of the releasing stimulus (Albers et al., 2017) may be relevant to the rhythmicity of the neurotransmitter-clock protein crosstalk.
Regarding the synaptic communication, catecholaminergic and indolaminergic projections originate from a limited number of neurons in defined central areas, i.e., the locus coeruleus (LC) for noradrenaline (NA), the substantia nigra (SN), and the ventral tegmental area (VTA) for DA or the raphe nucleus for 5-HT (Descarries and Mechawar, 2000; Vizi et al., 2004). Once released, the neurotransmitter diffuses through the biophase to reach receptors located postsynaptically, a process named “volume transmission” to assure the communication among sites within the same brain area or even in adjacent brain regions (Fuxe et al., 2012; Olivero et al., 2020). In this regard, DA, which controls the daily rhythmicity in mammals (Imbesi et al., 2009; Hood et al., 2010), is actively released from dopaminergic projections and reaches the postsynaptic structures by “volume transmission” (Descarries and Mechawar, 2000), which are located also in vicinal regions (i.e., the striatum, the SCN). Here, the activation of dopaminergic receptors (i.e., the D1 and D2 receptors) elicits functional responses, including the tuning of clock proteins, i.e., the Per1 and 2 proteins (Imbesi et al., 2009; Hood et al., 2010). Therefore, DA diffusion allows the propagation of the dopaminergic input during the diurnal period, favoring a widespread circadian regulation of biological processes. However, the efficiency of the DA “volume transmission” strictly depends on the continuous replenishment of the presynaptic vesicle, that is ensured by the storage of the newly-synthetized transmitter or by the re-uptake of the released amine, which are both efficiently controlled by clock proteins (see Section “The Dopaminergic System”). Besides the SN and the VTA, dopaminergic cells also represent the largest percentage of the resident cells in the retina. Here, the highest concentration of DA is reached during the day due to multiple excitatory signals, including light, and it is controlled by melatonin. DA diffuses by “volume transmission” also in the brain regions to enable the communication between the amacrine cells (that are dopaminergic in nature) and the horizontal and the bipolar cells, as well as the photoreceptor-containing processes (Witkovsky, 2004). The mechanism of “volume transmission” in the CNS dictates a virtuous “clock protein-transmitter loop” that favors the diurnal control of some circadian regulators. An interesting feature of the retinal amacrine cells (Hirasawa et al., 2009) and in general of the DA-positive neuronal processes in the SCN and striatum (Romei et al., 2016), is that these cells are also specialized for GABA. Dopaminergic cells cannot synthesize GABA, but can efficiently recapture this amino acid, mainly at nerve endings (Romei et al., 2016), where it is co-stored with DA. The uptake of GABA in dopaminergic terminals, which is an event that would occur preferentially when the [GABA]out exceeds the physiological level, triggers a concomitant release of the catecholamine. Since GABA acts preferentially as a nocturnal controller of circadian rhythms and has an opposite outcome with respect to DA in controlling the expression of the clock genes [i.e., Per2 (Ruan et al., 2008)], it is possible that the GABA-induced carrier-mediated release of DA may have a role in permitting the transition from the nocturnal to the diurnal phase of the daily circadian rhythmicity.
The SCN is composed of GABAergic neuronal subtypes and glial cells. As well as many neurons, a large part of the GABAergic SNC neurons also contains neuropeptides. Therefore, depending on the stimulus, these neurons preferentially release both GABA and neuropeptides. When the concomitant release of neuropeptides and GABA occurs, this may affect the GABA outcomes, causing an indirect impact on the daily timekeeping due to the modifications of the hierarchical organization of the neuronal control of the clock oscillation. Interestingly, the co-release of neuropeptides and GABA in the SCN and in some other hypothalamic regions has been recently related to the dyssynchronization of the clock circadian molecules, particularly Clock and Period, that occurs in stress conditions (Wong and Schumann, 2012).
In the mammalian brain, serotonin availability in the biophase varies following a circadian pattern from day to night. The serotonergic projections from the medial raphe nucleus converge in the SCN and in the retino-hypothalamic tract, where the indolaminergic signal is mainly mediated by presynaptic 5-HT1B receptors (Selim et al., 1993; Pickard et al., 1999; Smith et al., 2001). 5-HT1B receptors are also largely expressed in GABA terminals, particularly in the SCN neurons and in the RHT, where their activation inhibits the exocytosis of the inhibitory amino acid. 5HT modulates the effect of light on the clock at both early and late night (Morin, 1999) time points, with variable effects when applied at night. Furthermore, a recent study using a rat in vivo microdialysis revealed that diurnal changes in 5-hydroxyindole-acetic acid (5-HIAA) levels, the principal 5-HT metabolite, could be observed also in other brain regions that receive the serotonergic projections, in particular, in the prefrontal cortex (PFC), but not in the hippocampus; this metabolic activation might be related to enhanced neuronal activity of 5-HT (Daszuta and Barrit, 1982; Nakayama, 2002). The contribution of 5-HT to the expression and function of clock proteins might depend on the phase of the day and on brain areas but again largely involves the control of the GABAergic innervation.
Notably, based on evidence from the literature suggesting a functional crosstalk between neurotransmitter systems and the circadian clockwork, we will summarize evidence showing such bidirectional interplay (as schematized in Table 1). Accordingly, robustness and precision in daily timekeeping have been reported to rely on the punctual intercellular communication, mediated by neurotransmitters, among CNS cells (Herzog et al., 2004; Hastings et al., 2018).
The Dopaminergic System
Dopamine (DA) is produced in midbrain neurons of the substantia nigra (SN) and ventral tegmental area (VTA) and it is involved in the regulation of locomotion, sleep-wake cycle, learning, motivation, and reward. DA levels have been shown to display a circadian oscillation in several brain areas, such as the retina, midbrain, striatum, olfactory bulb, and hypothalamus (Korshunov et al., 2017).
In particular, the retina is the first region of CNS to deal with light information and it is responsible for its transmission to SCN through the retino-hypothalamic pathway (Gooley et al., 2001; Hattar et al., 2006; Prigge et al., 2016). Studies on mammalian retinas show that DA has the highest peak of synthesis during the daytime, whereas it is reduced during the nighttime. Doyle et al. (2002) found that in rats with normal and dystrophic retinas, DA content and turnover, as observed by measuring its two major metabolites 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA), follow a daily rhythmicity with the peak during the light phase. Besides, they also reported that cones and rodes are not required for generating DA circadian rhythmicity, since the latter was maintained also in rats with dystrophic retinas (Doyle et al., 2002). Moreover, dopamine has been reported to stimulate the expression of the core clock component Per1 in retinal neurons, especially through the activation of D1 and D2 receptors, whose specific role is currently a matter of debate. In intact mPer2Luc retinal explants, stimulation of D1 receptors resulted in Per1 upregulation by activating the extracellular signal-regulated kinase (ERK)-mitogen-activated protein kinase (MAPK) pathway. Enhanced phosphorylation of the cAMP-responsive element-binding protein (CREB) has been found to promote the cAMP-response elements (CRE)-mediated transcription of Per1 gene (Ruan et al., 2008). In line with such evidence, Yujnovsky et al. (2006) reported an increase in Per1 transcription in mouse NG108–15 cells, induced by the activation of dopamine D2 receptor (D2R) by D2 agonists, such as quinpirole. Indeed, the stimulation of D2 receptors led to the activation of the MAPK transduction cascade, to an increase in CREB phosphorylation and the recruitment of the Clock:Bmal1 heterodimer, thus resulting in an enhanced Per1 transcription. A marked reduction of the Clock:Bmal1 activation and, consequently, of Per1 levels was also observed in the retina of D2R-null mice, thus indicating the importance of D2 receptors in mediating retinal circadian expression (Yujnovsky et al., 2006).
Besides the retina, the circadian activity of the dopaminergic signaling system has also been reported in the midbrain and striatum, possibly transmitted through the neuronal projections from the SCN to these areas, such as via the orexinergic system (ORX) in the lateral hypothalamus (LH), the lateral habenula (LHb), the paraventricular thalamic nucleus (PVT), and the medial preoptic nucleus (MLPO; Abrahamson and Moore, 2001; Mendoza and Challet, 2014). These innervations from the SCN are able to regulate DA firing, expression, and metabolism. Notably, the LHb acts as an inhibitory pathway on the VTA. Accordingly, its activation has been found to decrease the firing of dopaminergic neurons (Bourdy and Barrot, 2012; Lecca et al., 2012). Furthermore, activation of the ORX system in the VTA has been shown to be implicated in the regulation of the plasticity of the dopaminergic system, as well as in locomotion, reward, motivation, and addiction (Borgland et al., 2006; Narita et al., 2006). Consequently, the rhythmical dopaminergic activity in the striatum relies on this SCN-VTA pathway. All these findings suggest that the intricate circuit of interconnections between SCN and the different brain areas plays a pivotal role in establishing the proper circadian rhythmicity of the dopamine system in the VTA and in the striatum.
DA signaling, synthesis, transport, and degradation are all under circadian regulation. Promoters of genes encoding for tyrosine hydroxylase (TH, the rate-limiting enzyme of DA synthesis), dopamine transporter (DAT), and monoamine oxidase A (MAO-A, the key enzyme that regulates DA degradation) have been found to contain E-Box sequences and to display circadian rhythmicity in their expression (Yoon and Chikaraishi, 1994; Sleipness et al., 2007; Hampp et al., 2008). In this regard, Sleipness and coworkers (Sleipness et al., 2007) demonstrated that electrolytic lesions of the SCN in rats alter the day-night variations in the expression of the DA transporter and synthetase in the midbrain. Remarkably, many studies revealed that the firing of dopaminergic neurons and the expression of TH protein in the VTA change during the day, with a peak during the daytime (Webb et al., 2009). In particular, Clock and Rev-Erbα have been found to be implicated in the reduction of TH expression in the VTA (McClung et al., 2005; Chung et al., 2014). Notably, Chung et al. (2014) demonstrated, in in vitro and in vivo studies on wild-type and Rev-erbα knockout (RKO) mice, an increase in TH mRNA expression and DA extracellular content in RKO compared to WT mice. These findings indicate a loss of rhythmicity and a parallel hyper-activation of the dopaminergic system in RKO mice, suggesting that Rev-erbα may negatively regulate TH gene expression. In particular, Rev-erbα has been shown to control TH expression by acting on RRE/NBRE1 (NGFI-B response element) elements located in TH promoters through the recruitment of the histone deacetylase 3 (HDAC3) complexes. The latter negatively regulates permissive histone acetylation, resulting in the inhibition of the transcriptional activation of the TH gene promoter (Chung et al., 2014). Furthermore, McClung and coworkers revealed that TH expression in mice was downregulated by Clock. Indeed, TH mRNA expression and protein levels were strongly increased in the VTA of ClockΔ19 mutant mice compared with WT (McClung et al., 2005). Although the mechanism by which Clock modulates TH expression is unknown, Clock has been suggested to bind E-box sequences on the TH gene promoter. Moreover, ClockΔ19 mutant mice have shown an increase in bursting and firing rates of VTA dopamine neurons. Such an effect is probably mediated by the downregulation of a voltage-gated potassium channel (KcnQ2), induced by the loss of Clock in mutants, thus indicating the key implication of Clock in affecting the excitability of DA neurons (McClung et al., 2005).
In the striatum, the rhythmical DA and MAO-A content has been found to be regulated by Per2. Per2Brdm1 mutant mice have been observed to display a reduction in MAO-A expression in the striatum and a consequent increase in extracellular DA levels, thus suggesting that Per2 may promote MAO-A transcription (Hampp et al., 2008). As a feedback loop, DA has been found to regulate the rhythm of Per2 expression via activation of D2 receptors. Indeed, rats treated with selective D2 antagonists (i.e., raclopide) showed a significant decrease in the rhythmical amplitude of Per2 transcription in the striatum, which, conversely, was not observed upon treatment with D1 antagonists. In turn, the reduction of Per2 levels, induced by the administration of 6-hydroxydopamine (6-OHDA), was restored by using D2 agonists, such as quinpirole, whereas the use of D1 agonists had no effect (Hood et al., 2010).
Taken together, these results suggest that fluctuation in extracellular DA levels may be involved in the maintenance of the rhythmical amplitude of Per2 transcription in the dorsal striatum via D2 receptors. Such modulation has been thought to involve the ERK-MAPK intracellular pathway, which is regulated by DA signaling, and leads to the activation of the cAMP-CREB pathway that results in the augmented transcription of Per2 gene (Hood et al., 2010). Moreover, Imbesi and coworkers, investigating the implication of DA receptors in the regulation of neuronal clock genes expression in cultured mouse striatal neurons, found that treatment with D1 receptor agonists led to an increase in Per1, Bmal1, Clock, and Npas2 mRNA levels. On the other hand, treatments with D2 and D3 receptor agonists, such as quinpirole, repressed the transcription of Per1 and Clock genes, whereas Npas2 and Bmal1 mRNA expression was not affected (Imbesi et al., 2009). Additionally, in mouse ventral striatum, a 24-h variation in the expression of dopamine D3 receptor has been reported. Interestingly, Rora and Rev-erbα have been found to modulate these oscillations, by possibly binding the D3 receptor promoter. In particular, Rora has been found to upregulate D3 receptor transcription, whereas Rev-erbα has been reported to periodically inhibit its expression (Ikeda et al., 2013). Moreover, in the nucleus accumbens (NAc), D3 receptors have been found to be regulated also by glial Per2 (Martini et al., 2021). Indeed, mice with a deletion of Per2 in glial cells showed an increase in D3 receptor mRNA levels compared to control animals (Martini et al., 2021). In particular, Per2 has been suggested to interact with several nuclear receptors, such as Rev-erbα and, to a lesser extent, with Rora (Schmutz et al., 2010), both interacting with the D3 receptor’s promoter and consequently modulating D3 receptor expression (Ikeda et al., 2013). Together, these studies highlight the putative links between the dopaminergic system and the striatal clockwork.
The Serotonergic System
Serotonin (5-HT) is an important neurotransmitter involved in the coordination and synchronization of the circadian system and melatonin synthesis. The serotonergic system and the master pacemaker are interconnected with each other through several projections, which are probably at the root of their reciprocal regulation. SCN projects indirectly to the median raphe nucleus (MRN) and the dorsal raphe nucleus (DRN) via the dorsomedial hypothalamic nucleus (DMH). In turn, the raphe nuclei project directly back to the SCN from the MRN, whereas indirectly via the intergeniculate leaflet (IGL) from the DRN (Azmitia and Segal, 1978; Hay-Schmidt et al., 2003; Deurveilher and Semba, 2005).
Clock genes have been found to regulate the expression of genes implicated in the serotonergic pathway in a circadian manner. The Tph2 gene, encoding for the tryptophan hydroxylase, has been found to exhibit a rhythmical expression in the midbrain of rats, with the highest peak arising 2 h before the light-dark transition, thus resulting in a circadian secretion of 5-HT (Malek et al., 2005). However, Tph2 mRNA rhythm seems to be indirectly mediated by the SCN through daily glucocorticoid variation. Indeed, Malek et al. (2007) demonstrated that, in rodent MRN and DRN, the loss of corticosterone daily fluctuations, induced by adrenalectomy, abolished the rhythmical expression of Thp2 mRNA completely. In contrast, the Thp2 daily expression pattern was fully restored after the readjustment of plasma corticosterone levels. Nevertheless, the precise mechanisms involved in the circadian regulation of Thp2 transcription by glucocorticoids are unknown (Malek et al., 2007). Furthermore, both serotonin transporter SERT expression and activity in mouse raphe nuclei have been reported to significantly change in a time-dependent manner, displaying higher levels during the dark phase (Ushijima et al., 2012). Likewise, studies on hamsters showed that 5-hydroxyindole-acetic acid (5-HIAA), the principal 5-HT metabolite, is mainly released during the dark phase (Glass et al., 1992), thus suggesting that the release of 5-HT is greater during the active phase and demonstrating that both its synthesis and release follow a circadian rhythmical pattern.
At the same time, the capability of 5-HT, not only to be controlled by the biological clock but in turn to influence circadian functions has been proven by Nakamaru-Ogiso et al. (2012) demonstrating that 5-HT deficiency in rats, induced by an acute tryptophan depletion, led to the disruption of circadian sleep-wake rhythm and disintegrated fragmental patterns. 5-HT plays an important role also in the regulation of photic entrainment of the SCN and it is able to modulate light-induced phase shifts in locomotor activity (Mistlberger et al., 2000; Cuesta et al., 2008). Indeed, 5-HT works as an inhibitor of the retinal output to the central clock, performing both a presynaptic action through 5-HT1B receptors on retinal afferent terminals, as well as a postsynaptic action through 5-HT7 receptors on SCN neurons (Smith et al., 2001). The activation of 5-HT1B receptors has also been associated with the suppression of the inhibitory effect of light on pineal melatonin production (Rea and Pickard, 2000) and with significant light-induced phase shifts, which led to a variation of SCN gene expression. The serotonergic stimulation, obtained through the treatment of the rodent model Arvicanthis ansorgei [a diurnal model used for the study of circadian rhythms (Hubbard et al., 2015)] with a 5-HT agonist and selective 5-HT reuptake inhibitors (SSRIs, such as fluoxetine), resulted in behavioral phase-advances and in an increase of light-induced phase-delays and advances (Cuesta et al., 2008). At the molecular level, such light-induced phase shifts, anticipated by serotoninergic activation, have been found to be correlated with changes in clock gene expression in the SCN. Notably, at CT12, Per2, and Rev-erbα mRNA levels were higher compared to those obtained after a light pulse alone, thus suggesting that they may play a role in the enhancement of light-induced phase-delays. Whereas, due to increased expression of Per1, Rev-erbα, and Rev-erbβ at CT0, these clock genes seemed to be implicated in the reinforcement of light-induced phase advances. On the contrary, Rorβ transcription was found to be reduced at CT0 (Cuesta et al., 2008).
Further evidence of the strong interconnection between circadian and serotonergic networks is related to the influence that many serotonergic drugs have on the circadian system. For example, as described before, SSRIs deeply impact the response of the circadian system to environmental light and have been seen to phase advance the firing rate of SCN neurons in rats’ brain slice cultures (Sprouse et al., 2006).
The Noradrenergic System
Noradrenaline (NA) is a neurotransmitter mainly synthesized in the locus coeruleus (LC), and it is implicated in the modulation of alertness, arousal, cognition, executive functions, and mammalian circadian rhythmicity (Palm et al., 2021). The release of NA from the LC is a very dynamic process fundamental for the correct alternation between the rapid eye movement (REM) phase and the non-rapid eye movement (NREM) phase of sleep. Recent studies have revealed that the activity of LC in releasing NA is higher during wakefulness and NREM sleep, whereas it is lower during REM sleep, thus suggesting the key role of LC in sleep architecture and functions (Osorio-Forero et al., 2022). Moreover, in the LC of rats, the expression of MAO-A and MAO-B is under circadian control due to their rhythmical oscillations that peaks at nighttime, probably thanks to the presence of E-box sequences on their gene promoters (Chevillard et al., 1981).
Animal studies demonstrated that alterations of clock genes expression in the SCN, which also impact clock mRNA levels in the peripheral tissues, are associated with noradrenaline pathways. Notably, NA is able to modulate the physiological expression of Per1, Per2, and Bmal1 in mouse livers (Terazono et al., 2003). Administration of adrenaline, both in vitro and in vivo, increased mPer1 expression in mouse liver in a concentration-dependent manner. In SCN-lesioned arrhythmic mice, a loss in the liver daily expression of mPer1, mPer2, and mBmal1 and of NA content was further observed, and restored by adrenaline injections. Moreover, in human dermal fibroblasts, the exposure to NA reduced the expression of Bmal1 at ZT12 and Per1 at ZT20 after incubation with NA compared to controls. Meanwhile, the expression of Per3 at CT4 was increased after the incubation with NA compared to cultures without NA treatments (Palm et al., 2021). In addition, in mice, administration of NA for 6 days highly induced Per1 mRNA transcription in the cerebral cortex during the light phase (Burioka et al., 2008). Together, these findings demonstrate the adrenergic role in regulating clock gene expression and in conveying central clock information to peripheral organs by NA release, probably through the involvement of cAMP-PKA and MAPK-CREB pathways (Terazono et al., 2003; Burioka et al., 2008; Palm et al., 2021).
The Glutamatergic System
Glutamate (Glu) is the primary excitatory neurotransmitter in the mammalian nervous system and it is essential for the maintenance of higher brain functions such as cognition, learning, and memory. Furthermore, it is implicated in brain cellular metabolism and in the regulation of sleep-wake states (Danbolt, 2001; Jones, 2017). Interestingly, astrocytes are primarily responsible for the rhythmic oscillation of extracellular Glu levels, which, in turn, are necessary for molecular timekeeping in the SCN. In order to regulate the balance between Glu uptake and release, astrocytes are responsible for the uptake of the majority of extracellular Glu through glutamate transporters on the surface of the astrocytic peri-synaptic processes, such as the excitatory amino acid transporters 1 and 2 (EAAT1 and EAAT2; Lehre and Danbolt, 1998). After being re-uptaken, Glu in astrocytes can be metabolized to glutamine by glutamine synthetase and used by neurons as a precursor for the synthesis of Glu or GABA (Waniewski and Martin, 1986). Otherwise, Glu can be oxidatively metabolized to α-ketoglutarate, which is involved in ATP production (Farinelli and Nicklas, 1992). On the other hand, Glu release from astrocytes is mediated by metabotropic glutamate receptors (mGluRs), which, once activated, induce an increase in the astrocytic [Ca2+]i, thus triggering Glu release from astrocytes (Hamilton and Attwell, 2010). The astroglial-released Glu activates extra-synaptic N-methyl-D-aspartate (NMDA) receptors on adjacent excitatory neurons, thus synchronizing excitatory neuronal firing (Fellin et al., 2004; Figure 2). Accordingly, the glutamatergic connections to the SCN are mediated by pre-synaptic NMDA/NR2C receptors expressed in the dorsal SCN, which, in turn, once activated by extracellular Glu, increase the GABAergic tone. Indeed, Brancaccio et al. demonstrated that inhibition of NR2C subunits of the NMDA receptors, obtained by treating mouse SCN slices with an NR2C-selective antagonist (i.e., DPQ-1105), led to an alteration of molecular and electrical circadian oscillations in the SCN. Notably, NR2C inhibition abolished circadian oscillations of intracellular Ca2+, induced nighttime depolarization of SCN dorsal neurons, reduced the amplitude, and lengthened the period of Per2 oscillations (Brancaccio et al., 2017). Such evidence highlights that the astrocytic-Glu/NMDA-NR2C/SCN axis may represent a novel pathway required for the synchronization of SCN neurons. Moreover, in ex vivo studies, astrocytes of mouse SCN were found to be active and release higher Glu levels during the circadian nighttime (Brancaccio et al., 2017). Accordingly, another study demonstrated that glutamate concentration is under circadian control, and in rat brain it peaks during the dark phase of the day, mainly in the striatum and nucleus accumbens (Castañeda et al., 2004).
A circadian variation has also been seen in the expression of some receptors and transporters involved in the glutamate pathway. For example, Elmenhorst et al. (2016) in an in vivo PET experiment on rat brains, found approximately a 10% increase in the availability of metabotropic glutamate receptor mGluR5 during the light phase in different brain regions, such as the cortex, amygdala, caudate putamen, and nucleus accumbens. Meanwhile, in rodents, the glutamate transporter EAAT3, involved in Glu uptake, has been found to follow a circadian expression within the SCN, showing a peak around the dark-light transition (Cagampang et al., 1996). Brancaccio et al. (2017) demonstrated that EAAT3 inhibition, obtained by treating mouse SCN slices with a selective antagonist, led to an increase in the extracellular Glu levels, and a reduction in amplitude and robustness of Per2 gene oscillations, resulting in an alteration of the neuronal timekeeping. Similarly, excitatory amino acid transporter 1 (EAAT1) mRNA and protein levels in the SCN, and consequently the glutamate uptake, have been found to follow a diurnal rhythm and to be strongly modulated by clock genes expression. Spanagel and coworkers found that Per2Brdm1 mutant mice showed a reduction in the SCN expression of EAAT1 compared to wild-type mice, thus resulting in a reduced glutamate uptake by astrocytes and in a hyper-glutamatergic state (Spanagel et al., 2005). Likewise, studies on cortical astrocyte cultures from Npas2 and Clock mutant mice revealed a significant reduction in glutamate uptake compared to wild-type mice due to a decrease in EAAT1 mRNA levels caused by mutations (Beaulé et al., 2009). These results suggest that both Glu release and uptake are orchestrated in a circadian manner, however, the molecular mechanisms by which the clock genes modulate the total Glu uptake are unknown.
The GABAergic System
γ-aminobutyric acid (GABA) is synthesized from Glu and it is the principal inhibitory neurotransmitter in the brain. The timing of GABA synthesis, release, and uptake seems to have a crucial role in the orchestration of SCN rhythm. Notably, the glutamic acid decarboxylase (GAD, the enzyme responsible for GABA synthesis), especially the isoform GAD65, has been reported to show a circadian oscillation. In rat SCN slices, autoradiography revealed higher levels of GAD65 mRNA in the light period rather than in the dark period, with a peak of expression during the transition from dark to light (Huhman et al., 1996). Furthermore, Maejima and coworkers demonstrated the importance of the GABAergic system for the performance of circadian behavior at the correct time of the day through the regulation of SCN neuronal activity. They found that in the SCN of Avp-Vgat−/− mice (mutation resulting in a decrease of GABA release from the vesicular GABA transporter (Vgat) in arginine vasopressin-producing (AVP) neurons), the frequency of GABA-A receptor-mediated postsynaptic currents (mGPSCs) was strongly reduced during daytime compared to control mice. Besides, in the SCN of Avp-Vgat−/− mice, a delay in the peak phase of Per2:Luc oscillations has been observed, thus leading to a lengthened activity time of circadian behavioral rhythms. Thus, these results suggest the key role of the GABAergic transmission from AVP neurons in directing the correct timekeeping of SCN outputs from molecular clocks to behavior (Maejima et al., 2021).
Further evidence for GABA involvement in the induction of a rhythmic expression of the core clock genes in neurons, through the activation of GABA-A receptors, comes from Barca-Mayo et al. (2017). In in vitro cortical neurons of GABA-treated mice, they found that GABA was able to entrain rhythmic oscillations of Bmal1, Cry1, and Per2 expression, and that the inhibition of GABA-A receptor signaling suppressed such rhythms. In Bmal1cKO mice (inducible knockout mouse model of Bmal1 in astrocytes), extracellular GABA levels have been found to be altered, resulting in an over-inhibition of the signaling pathways involved in cognitive functions and in a delay of the active phase. The impairment of GABA levels has been related to the deletion of astrocytic Bmal1 that decreased the expression of GABA transporters Gat1 and Gat3 in the SCN and in astrocytes, in turn reducing extracellular GABA clearance (Barca-Mayo et al., 2017). Furthermore, also the glial deletion of Per2 gene in mice (GPer2 mouse model) has been found to alter Gats mRNA levels (Martini et al., 2021). Notably, the expression of Gat1 significantly decreased in the hypothalamus of GPer2 mice, whereas that of Gat2 has been reported to increase in the NAc of this mouse model compared to controls (Martini et al., 2021). Together, these findings suggest that astrocytic Bmal1 and glial Per2 may regulate the extracellular GABA concentration through the modulation of Gats mRNA levels.
Moreover, GABA is also important in the regulation of retinal clock genes fluctuations, since, in cultured mPer2Luc retinal explants, it decreased and suppressed the rhythmic amplitude of Per2 oscillations in a dose-dependent manner, possibly through the activation of GABA-A and GABA-C receptors (Ruan et al., 2008). Only after treating mPer2Luc retinal explants with a GABA-A or GABA-C receptor agonist, the amplitude of Per2:Luc signals significantly decreased, whereas no effects have been observed with GABA-B receptor agonists (Ruan et al., 2008). Moreover, GABA was able to increase Per1 and Bmal1, but not Per2 and Clock mRNA levels, thus indicating its ability to suppress Per2 levels acting at the posttranscriptional level, by stimulating casein kinase, which is able to phosphorylate Per2 for its ubiquitin-mediated proteasomal degradation (Ruan et al., 2008).
The Role of Glia in The Generation of Circadian Rhythmicity
The existence of an endogenous neuronal circadian timekeeper with the capacity to orchestrate and coordinate physiological and behavioral processes with the external environment is now well established. However, studies on circadian timekeeping are moving beyond the neuronal cell population, since the capability of encoding rhythmical information is not only restricted to neurons but also extends to glial cells. Indeed, as discussed above, the devolution of the control of the glutamatergic and, consequently, of the GABAergic tone to astrocytes, since they are the primarily responsible for the rhythmic oscillation of extracellular Glu levels, suggests that such glial cells synergistically communicate with neurons to establish the 24 h daily rhythm (Figure 2). Hence, since astrocytes and microglia are integral parts of the timepiece, the correct functioning of the glial clock system is fundamental for the maintenance of a healthy brain. Recent studies indicate that misalignments in the expression of glial clock genes contribute to the development of different pathological outcomes. Particularly, astrocytic Clock, Per1, and Per2 have been found to be involved in the modulation of ATP release (Marpegan et al., 2011) and, subsequently, in the regulation of energy metabolism and glial activity. Therefore, mutations of these clock genes may result in metabolic diseases. Moreover, microglial Bmal1 has been proven to be an important molecular player in the inflammatory response and in different pathological processes, such as neurological diseases and sleep disorders. Notably, mouse microglial Bmal1 deficiency has been found to decrease the expression of pro-inflammatory cytokines (i.e., IL-1β, IL-6, and TNFα) and increase that of anti-inflammatory cytokines (i.e., IL-10; Wang et al., 2020), probably owing to the presence of E-boxes sequences on cytokine gene promoters. Likewise, the deletion of mouse astrocytic Bmal1 leads to the impairment of cognitive functions, majorly affecting the declarative memory (Barca-Mayo et al., 2017), as well as to an imbalance of the GABAergic system (Barca-Mayo et al., 2017), resulting in the development of neurological and sleep disorders. In this regard, another core glial clock component was revealed to play an important role in the regulation of mood-related behaviors, such as anxiety and depression (Martini et al., 2021). GPer2 mice, in fact, showed significantly less immobility in the forced swim test, thus displaying reduced despair, one of the main manifestations of depression (Martini et al., 2021). Besides, they spent more time in the open field (an anxiety-provoking space) of the elevated O-maze, indicating a reduced anxiety-like behavior in mice lacking Per2 in glial cells (Martini et al., 2021). Nevertheless, glial Per2 KO had no influence on circadian parameters since GPer2 mice did not display any differences in the wheel-running activity and in body temperature fluctuations compared to control animals, thus indicating that the deletion of Per2 in glial cells did not affect free-running rhythms in mice (Martini et al., 2021). An interesting observation is that, contrary to glial Per2 KO animals, total Per2 KO mice exhibited a shortening of the free running period (Martini et al., 2021). Therefore, these data suggest the importance of glial Per2 in the regulation of mood behaviors through a process which, given the normal maintenance of circadian parameters, is not associated with the circadian clock machinery, but is probably related to the action of glial Per2 on the glutamatergic, GABAergic, and dopaminergic signaling pathways (Martini et al., 2021).
Collectively, these findings suggest that an abnormal circadian system in glial cells may degenerate into metabolic diseases, immune system dysfunctions, and psychiatric illnesses. Therefore, deepening the knowledge of the role of the glial population in the generation of rhythm may be important for the discovery of new pathways and interactors involved in the development of these pathological disorders. In the following section, we will summarize recent findings reporting the existence of an autonomous rhythmical pattern in glia cells, demonstrating that astrocytes and microglia are not passive participants in the orchestration of rhythm, but play a key role in the regulation of circadian rhythms and in the maintenance of timekeeping within the brain (Hayashi et al., 2013; Fonken et al., 2015; Barca-Mayo et al., 2017; Brancaccio et al., 2017, 2019; Nakazato et al., 2017; Sominsky et al., 2021).
Astrocytes-Neurons Interaction in the Circadian Rhythmicity
Considerable evidence supports the primary involvement of astrocytes in the generation of rhythm, affirming their capacity to influence the molecular clock machinery of the central circadian pacemaker (Barca-Mayo et al., 2017; Brancaccio et al., 2017, 2019). Astrocytes express clock genes and can act as autonomous circadian oscillators (Ewer et al., 1992; Prolo et al., 2005). Ewer et al. (1992) first demonstrated the existence of a circadian clock in glia, by reporting the presence of the core clock component period in the astrocytes in Drosophila melanogaster. Also, primary cultures of cortical astrocytes derived from Per2::luciferase knock-in mice and Per1::luciferase transgenic rats were found to express circadian rhythms in the activity of these two clock genes, with a longer average period in the activity of Per1::luc than Per2::luc (Prolo et al., 2005). Furthermore, beyond Glu, also some gliotransmitters, such as adenosine triphosphate (ATP), a neuronal transmitter involved in synaptic communication between astrocytes and neurons, are released following a circadian pattern (Womac et al., 2009; Marpegan et al., 2011). In particular, similarly to that observed in SCN cell cultures, extracellular ATP release and accumulation followed a daily rhythm in primary cultures of rat cortical astrocytes, thus suggesting that ATP might represent an important output of the mammalian molecular clock (Womac et al., 2009). Indeed, extracellular ATP levels have been found to strictly depend on clock genes expression. In this context, mouse astrocytes carrying mutations on Clock, Per1, and Per2 genes showed a constitutively blunted ATP fluctuation during the day compared to wild-type cultures (Marpegan et al., 2011). Moreover, astrocytes have been suggested to be active during the circadian night, in contrast to neurons, whose peak of activity has been registered during the circadian day. These findings are based on data reporting that, in mouse SCN astrocytes, Glu release peaked during the nighttime in phase with astrocyte [Ca2+]i levels, that were anti-phasic to those of neuronal [Ca2+]i (Brancaccio et al., 2017). Overall, these results underline the ability of the astrocyte clock to impose its own rhythm autonomously, thus indicating that astrocytes are actively involved in the maintenance and control of molecular timekeeping within the brain.
However, such rhythmicity of astroglia is partly entrained and sustained by the SCN master clock (Prolo et al., 2005). In co-cultures of mouse cortical astrocytes with adult SCN explants, the amplitude of Per1 rhythmicity has been observed to be significantly greater than that of primary astrocytes alone (Prolo et al., 2005), thus suggesting the existence of a crosstalk between the master neuronal clock in the SCN and the glial clock. In particular, in addition to SCN neurons, also astrocytes are able to encode circadian information and have been observed to strongly influence the central circadian pathway and the master clock machinery (Brancaccio et al., 2017, 2019; Tso et al., 2017). Notably, the increased release of Glu by astrocytes during the night has been found to be accomplished by a decrease in Per2 expression rhythms in the SCN, thus denoting the active role played by astrocytes in promoting correct molecular timekeeping in the SCN via the glutamatergic signaling (Brancaccio et al., 2017). Furthermore, Brancaccio et al. (2019) using SCN slices of Cry1/2-null mice, demonstrated that the expression of Cry1 alone in astrocytes was sufficient to initiate self-sustained circadian rhythms of Per2 in the SCN, thus restoring the central molecular clock that was previously repressed, though with a longer time extent than neuronal Cry1. The crosstalk between SCN and astrocytes has also been suggested by data on astrocytes affecting the circadian pattern of behavior in mice. Although astrocytes are not directly connected to motor centers, the rhythmicity of locomotor activity, completely lost in Cry1/2-null mice, was re-established after the Cry1 transfection of SCN astrocytes (Brancaccio et al., 2019), thus speculating that the astrocytic TTFL indirectly drives new circadian behaviors by hiring and restoring the neuronal TTFL in the SCN, probably by recruiting the E-box-based TTFL of neurons through the driving of neuronal [Ca2+]i oscillations (Brancaccio et al., 2019).
In addition, Bmal1 has also been reported to follow a rhythmical pattern and to strictly impact the circadian communications between astrocytes and neurons (Barca-Mayo et al., 2017; Tso et al., 2017). Bmal1 deletion in astrocytes has been found to lead to a lengthening of Per2 and locomotor circadian periodicity (Tso et al., 2017). Moreover, when maintained in constant darkness, a delay and reduction in the active period have been observed in Bmal1 KO mice, as well as an impairment in cognitive functions mainly affecting the declarative memory (Barca-Mayo et al., 2017). Indeed, Bmal1 KO mice were subjected to the novel object recognition (NOR) test, where they were exposed to two identical objects for 10 min, and they were subsequently tested by changing one of the familiar objects with a new one after 1 h and 24 h, with the aim to assess short-term and long-term memory respectively (Barca-Mayo et al., 2017). As a result, Bmal1 KO animals displayed a significant reduction in the discrimination index after both 1 and 24 h, thus demonstrating both short-term and long-term memory being compromised (Barca-Mayo et al., 2017). Notably, oscillations of Cry1, Bmal1, and Per2 expression were significantly impaired in the cortex and hippocampus of astrocyte Bmal1 KO mice, thus demonstrating the existence of a strong intercellular communication and interplay between astrocytes and neurons that influence the circadian time-keeping within the brain and is fundamental for the maintenance of a healthy brain (Barca-Mayo et al., 2017).
Further demonstration of the ability of astrocytes to encode circadian information, and thus to influence the neuronal clock machinery, was provided by the finding that, in co-cultures of mouse synchronous astrocytes with asynchronous primary cortical neurons, the former was able to re-establish the rhythmical expression of Bmal1, Cry1, and Per2 in the arrhythmic cortical neurons (Barca-Mayo et al., 2017). In contrast, astrocytes with Bmal1 deletion failed to induce rhythmicity in the clock genes expression of asynchronous cortical neurons, thus demonstrating the pivotal role of an active astrocyte clock for adequately affecting the neuronal clockwork (Barca-Mayo et al., 2017). Accordingly, the deletion of astrocytic Bmal1 has been found to impact the neuronal clock through GABA signaling, which acts as a mediator between neuron and astrocyte communication. In co-cultures of astrocytes with asynchronous cortical neurons, the inhibition of the GABA-A receptors prevented the astrocytic induction of a sustained rhythm in neurons, suggesting that the GABAergic pathway is required for the transmission of rhythm from astroglia to neurons (Barca-Mayo et al., 2017). Indeed, in astrocytes with Bmal1 KO, the expression of GABA transporters Gat1 and Gat3 significantly decreased in the cortex and in the SCN compared to WT astrocytes, thus impairing the extracellular levels of GABA, providing the explanation for the reason behind the failure of the arrhythmic astrocytes to synchronize the clock of asynchronous cortical neurons (Barca-Mayo et al., 2017). All these data suggest that the loss of daily rhythm in astrocytes due to the deletion of astrocytic Bmal1 is sufficient to lengthen and impair circadian rhythms in neurons and in behavior, both in vitro and in vivo, demonstrating that astrocytes may actively contribute to the generation of SCN neuronal output rhythm and to internal clock synchronization.
Microglia: The Rhythmicity Around Immune and Inflammatory Response
Circadian clock genes are rhythmically expressed in microglia in several brain areas, such as the hippocampus and the cortex (Hayashi et al., 2013; Fonken et al., 2015; Nakazato et al., 2017). In hippocampal microglia isolated from adult rats, Per1 and Per2 expression was found to reach its peak in the middle of the light phase, Rev-erb at the end of the light phase, whereas the higher level of Bmal1 expression was observed at the onset of the light period and in the middle of the dark phase (Fonken et al., 2015). Furthermore, also murine cortical microglia showed a fluctuant expression of Per1, Per2, Rev-erbα, and Npas2, thus supporting the idea of the existence of an autonomous clock machinery in microglia (Nakazato et al., 2011; Hayashi et al., 2013; Nakazato et al., 2017). The presence of the core clock proteins in microglia may also be responsible for the rhythmicity of a variety of microglial functions, such as immune and inflammatory response, by tuning the rhythmic fluctuations in basal inflammatory genes expression, such as cytokines and chemokines (Saijo and Glass, 2011). In rat hippocampal microglia, the expression of interleukin-1β (IL-1β), interleukin-6 (IL6), interleukin 1R1 (IL1R1), and tumor necrosis factor α (TNFα) was strictly affected by the time of the day, and, similarly to clock genes, they were strongly expressed during the light phase (Fonken et al., 2015). In addition, Bmal1 has been found to be a critical player in the immunological activity of microglial cells, possibly due to the presence of E-box sequences in gene promoters of several cytokines (Nakazato et al., 2017; Wang et al., 2020). In particular, microglial Bmal1 was found to be a key mediator for the regulation of the expression of IL6 (Nakazato et al., 2017). Accordingly, Bmal1 deficiency has been found to decrease the expression of pro-inflammatory cytokines (i.e., IL-1β, IL-6, and TNFα) and increase that of anti-inflammatory cytokines (i.e., IL-10) in mouse microglia (Wang et al., 2020). These data support the involvement of the microglial clock system in the regulation of inflammatory responses. Another microglia-associated function with diurnal rhythmicity is the regulation of synaptic strength via the circadian secretion of cathepsin S (CatS). CatS is a microglia-specific lysosomal cysteine protease rhythmically secreted during the dark phase, fundamental for the maintenance of neuronal circuitry (Hayashi et al., 2013). Hayashi and coworkers found that CatS expression followed a circadian pattern driven by the intrinsic microglial molecular clock (Hayashi et al., 2013). Accordingly, in WT mouse cortical microglia, CatS transcripts showed a peak at CT14, whereas, in the cortical microglia of ClockΔ19 mutant mice, no diurnal fluctuation of CatS expression was observed, thus suggesting that its transcription was activated and mediated by the binding of Clock:Bmal1 heterodimer to the E-boxes sequences on the CatS gene promoter (Hayashi et al., 2013). All these data demonstrate the existence of a microglial clock involved in the regulation of microglia-mediated functions, including the neuroimmune roles and the control of the synaptic strength.
Beyond increasing knowledge regarding the presence of an intrinsic clock in microglial cells, data from literature report that, in turn, the microglial clock system may also influence the neuronal clock machinery and rhythms among the CNS. Indeed, ablation of microglia resulted in the disruption of the central circadian system both at molecular and physiological levels (Sominsky et al., 2021). Notably, using Cx3cr1-Dtr rats, a rat model in which microglia is depleted, aberrations in clock gene expression were observed (Sominsky et al., 2021). In the SCN, Per1 mRNA levels were increased and a reduction in Bmal1 protein was revealed, whereas, in the hippocampus, the microglial depletion led to a suppression of Bmal1 expression and to an increase of Per1 and Per2 mRNA levels (Sominsky et al., 2021). Moreover, ablation of microglial cells from the CNS led to a disruption of body temperature rhythms, energy metabolism, and diurnal activity profiles. To such effect, Cx3cr1-Dtr rats showed a significant reduction in total energy expenditure during their active phase and also in their diurnal rhythms compared to WT (Sominsky et al., 2021). Accordingly, Sominsky et al. (2021) also observed a total loss of diurnal activity rhythms and a changing in temperature rhythms, with a pattern completely inverse to that of energy expenditure and activity changes. Considered together, these findings suggest that microglia may play a key role in diurnal rhythm generation since its ablation leads to the concomitant disruption of diurnal profiles and to the dysregulation of the expression of certain clock genes in different brain domains. In contrast, Barahona and collaborators showed that, in the cortex, ablation of microglia did not affect the rhythmical pattern of clock genes expression, since in Cx3cr1-Dtr rats clock genes all maintained their diurnal rhythmical expression (Barahona et al., 2022). Due to these conflicting data, further studies on this glia population are required to elucidate its role in the generation of rhythms throughout the brain.
A Matter of Time: Why Understanding The Functioning of The Biological Clock in The CNS Is Crucial?
Circadian rhythms are dictated by highly specialized cells of specific brain areas that orchestrate a complex network of coupled self-sustained clocks both in the brain and in peripheral organs. Remarkably, neurons are not the only CNS cell population to orchestrate the rhythm, but also astrocytes and microglia have been reported to harbor an intrinsic autonomous clock and to actively participate in the maintenance of timekeeping within the brain. Moreover, since neurotransmitters play a key role in the diffusion of circadian information among CNS cells and impact the core clock machinery, they have emerged as fine-tuners of daily rhythmicity. Indeed, the bidirectional interplay between neurotransmitters and the circadian clockwork is fundamental to maintain the accuracy and precision in daily timekeeping among different brain areas. In this regard, the network reported in Figure 3 shows the complex interactions taking place between clock proteins and several neurotransmitter players, as depicted by the literature discussed above, with the aim of illustrating this intricate bidirectional crosstalk (Figure 3). However, much is still unknown about the deeper molecular interconnections involved in the orchestration of the temporal variable among CNS cells. Therefore, expanding our knowledge about this complex network appears fundamental to better understand the functioning of the biological clockwork, which represents an extremely dynamic process across the lifespan. Indeed, rhythmic activities have been observed to significantly change during the pace of aging, also contributing to the acceleration of the process. Furthermore, due to the sexually dimorphic nature of some neurotransmitters systems [e.g., the NA system (Joshi and Chandler, 2020), the DA system (Rial et al., 2018), and the 5HT system (Näslund et al., 2013)], the connection between chemical transmission and expression and functions of clock genes in the CNS might account for sex-dependent variability in the circadian timing system. In this regard, seasonality, as well as most of the sex and gender-related-mechanisms, is known to affect the efficiency of learning, memory, and mood control processes (Bailey and Silver, 2014; Urban et al., 2021). Although so far poorly investigated, the interplay linking sex-neurotransmitters-clock genes deserves of course attention, since if disrupted or malfunctioning, might subserve the development of central disorders including those that so far preferentially attracted the interest of the researchers as discussed below.
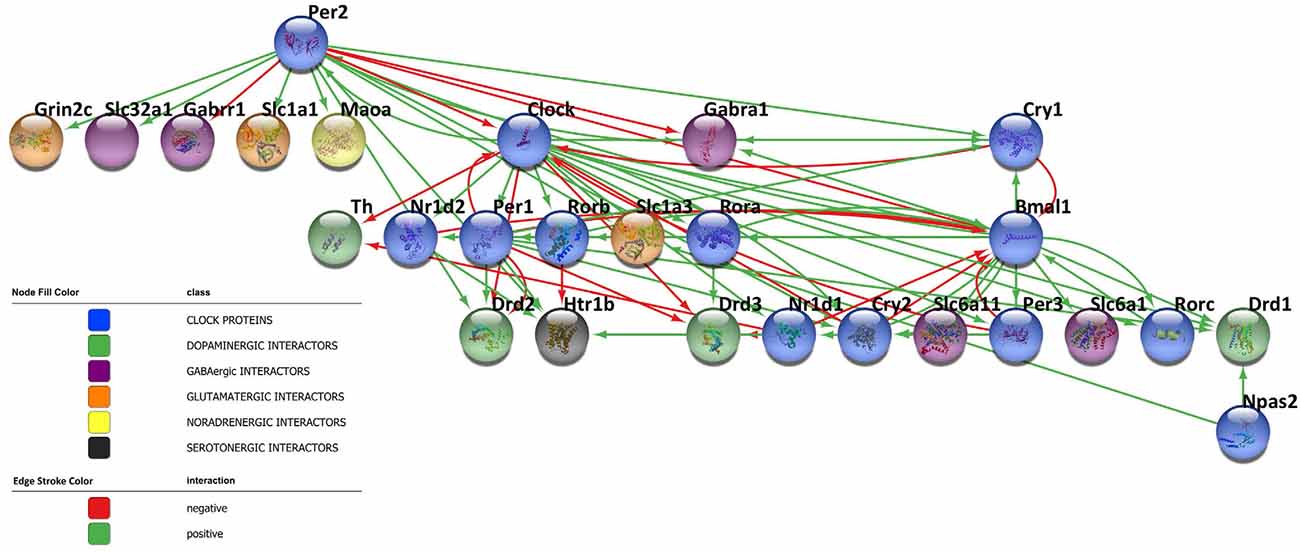
Figure 3. Bidirectional crosstalk between clock proteins and neurotransmitter players. The network contains 27 nodes and the edges represent the crosstalk between the clock proteins and neurotransmitter players depicted by the literature (positive regulation: green line, negative regulation: red line). The different biological processes investigated are shown in the network according to the color legend. The network has been visualized using Cytoscape 3.9.1 (Shannon et al., 2003).
Noteworthy, although future longitudinal studies are necessary to corroborate such preliminary evidence, the circadian system seems to play a key role in the pathogenesis of psychiatric diseases and neurodegenerative disorders. In particular, circadian rhythms disruption has been described not only as an early warning sign of neurodegenerative diseases but also as a potential risk factor. Evidence from literature suggests the existence of a strict correlation between several neurodegenerative diseases, specifically Alzheimer’s disease (AD) and Parkinson’s disease (PD), and circadian rhythms destruction (CRD; Videnovic et al., 2014). Accordingly, AD and PD share common features and symptoms ascribable to an abnormal and disrupted circadian pattern (Videnovic et al., 2014). For example, patients with neurodegenerative diseases show frequent disorders in the sleep/wake cycle, with sleep fragmentation and an increase in activity levels in the nighttime, and a prevalence of sleepiness during daytime (Merlino et al., 2010; Breen et al., 2014). Notably, it has been confirmed that an alteration of the molecular clockwork leads to an increase in the oxidative injury of the brain, thus promoting neuronal damage, cell death, and neurodegeneration. Interestingly, several studies on mice found that both global and brain-specific deletion of Bmal1 induced higher neuronal oxidative damage, synaptic degeneration, severe astrogliosis, impaired neuronal network functional connectivity, and impaired learning and memory, compared to age-matched wild type controls, resulting in premature aging, reduced lifespan, and pathogenesis of neurodegeneration (Musiek et al., 2013). Furthermore, the majority of neurodegenerative disorders are associated with the accumulation of intra- and extra-cellular aggregates (i.e., amyloid-β, phosphorylated tau protein, α-sinuclein), whose clearance is partly controlled by the glymphatic flow. Recent studies on mouse models have found that the glymphatic flow follows a rhythmical circadian pattern with an important upregulation during sleep, thus facilitating the removal of metabolic products accumulated during wakefulness (Hablitz et al., 2020). Remarkably, in mouse models of neurodegenerative disorders (such as APP/PS1 mice, a model of AD), where circadian rhythms are disturbed, a reduced brain clearance has been reported, due to the presence of disrupted flow patterns of the glymphatic system, thus resulting in a continuous and augmented accumulation of aggregates that, in turn, may promote cell death and degeneration of brain structures (Peng et al., 2016; Rasmussen et al., 2018). In conclusion, expanding our knowledge about the molecular regulation of circadian rhythmicity in CNS cells within different brain areas represents a crucial challenge, with a potential impact on the biological comprehension of the onset of neurodegenerative processes.
Author Contributions
FF and CL: conceived the idea. FF, EB, AP, and CL: wrote the manuscript. EP: network analysis. FF, EB, AP, EP, CS, SG, and CL: critical discussion. All authors contributed to the article and approved the submitted version.
Funding
Research has been supported by the University of Pavia (grant from PRIN 2020SCBBN2_005 to CL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahamson, E. E., and Moore, R. Y. (2001). Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191. doi: 10.1016/s0006-8993(01)02890-6
Albers, H. E., Walton, J. C., Gamble, K. L., McNeill, J. K., 4th, and Hummer, D. L. (2017). The dynamics of GABA signaling: revelations from the circadian pacemaker in the suprachiasmatic nucleus. Front. Neuroendocrinol. 44, 35–82. doi: 10.1016/j.yfrne.2016.11.003
Azmitia, E. C., and Segal, M. (1978). An autoradiographic analysis of the differential ascending projections of the dorsal and median raphe nuclei in the rat. J. Comp. Neurol. 179, 641–667. doi: 10.1002/cne.901790311
Bailey, M., and Silver, R. (2014). Sex differences in circadian timing systems: implications for disease. Front. Neuroendocrinol. 35, 111–139. doi: 10.1016/j.yfrne.2013.11.003
Barahona, R. A., Morabito, S., Swarup, V., and Green, K. N. (2022). Cortical diurnal rhythms remain intact with microglial depletion. Sci. Rep. 12:114. doi: 10.1038/s41598-021-04079-w
Barca-Mayo, O., Pons-Espinal, M., Follert, P., Armirotti, A., Berdondini, L., and De Pietri Tonelli, D. (2017). Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 8:14336. doi: 10.1038/ncomms14336
Beaulé, C., Swanstrom, A., Leone, M. J., and Herzog, E. D. (2009). Circadian modulation of gene expression, but not glutamate uptake, in mouse and rat cortical astrocytes. PLoS One 4:e7476. doi: 10.1371/journal.pone.0007476
Borgland, S. L., Taha, S. A., Sarti, F., Fields, H. L., and Bonci, A. (2006). Orexin A in the VTA is critical for the induction of synaptic plasticity and behavioral sensitization to cocaine. Neuron 49, 589–601. doi: 10.1016/j.neuron.2006.01.016
Bourdy, R., and Barrot, M. (2012). A new control center for dopaminergic systems: pulling the VTA by the tail. Trends Neurosci. 35, 681–690. doi: 10.1016/j.tins.2012.06.007
Brancaccio, M., Edwards, M. D., Patton, A. P., Smyllie, N. J., Chesham, J. E., Maywood, E. S., et al. (2019). Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192. doi: 10.1126/science.aat4104
Brancaccio, M., Maywood, E. S., Chesham, J. E., Loudon, A. S. I., and Hastings, M. H. (2013). A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78, 714–728. doi: 10.1016/j.neuron.2013.03.011
Brancaccio, M., Patton, A. P., Chesham, J. E., Maywood, E. S., and Hastings, M. H. (2017). Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 93, 1420–1435.e5. doi: 10.1016/j.neuron.2017.02.030
Breen, D. P., Vuono, R., Nawarathna, U., Fisher, K., Shneerson, J. M., Reddy, A. B., et al. (2014). Sleep and circadian rhythm regulation in early Parkinson disease. JAMA Neurol. 71, 589–595. doi: 10.1001/jamaneurol.2014.65
Brenna, A., and Albrecht, U. (2020). Phosphorylation and circadian molecular timing. Front. Physiol. 11:612510. doi: 10.3389/fphys.2020.612510
Brenna, A., Ripperger, J. A., Saro, G., Glauser, D. A., Yang, Z., and Albrecht, U. (2021). PER2 mediates CREB-dependent light induction of the clock gene Per1. Sci. Rep. 11:21766. doi: 10.1038/s41598-021-01178-6
Burioka, N., Koyanagi, S., Endo, M., Takata, M., Fukuoka, Y., Miyata, M., et al. (2008). Clock gene dysfunction in patients with obstructive sleep apnoea syndrome. Eur. Respir. J. 32, 105–112. doi: 10.1183/09031936.00138207
Cagampang, F. R., Rattray, M., Powell, J. F., Chong, N. W., Campbell, I. C., and Coen, C. W. (1996). Circadian variation of EAAC1 glutamate transporter messenger RNA in the rat suprachiasmatic nuclei. Brain Res. Mol. Brain Res. 35, 190–196. doi: 10.1016/0169-328x(95)00203-5
Castañeda, T. R., de Prado, B. M., Prieto, D., and Mora, F. (2004). Circadian rhythms of dopamine, glutamate and GABA in the striatum and nucleus accumbens of the awake rat: modulation by light. J. Pineal Res. 36, 177–185. doi: 10.1046/j.1600-079x.2003.00114.x
Chen, S.-K., Badea, T. C., and Hattar, S. (2011). Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476, 92–95. doi: 10.1038/nature10206
Chevillard, C., Barden, N., and Saavedra, J. M. (1981). Twenty-four hour rhythm in monoamine oxidase activity in specific areas of the rat brain stem. Brain Res. 223, 205–209. doi: 10.1016/0006-8993(81)90825-8
Chung, S., Lee, E. J., Yun, S., Choe, H. K., Park, S.-B., Son, H. J., et al. (2014). Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell 157, 858–868. doi: 10.1016/j.cell.2014.03.039
Cox, K. H., and Takahashi, J. S. (2019). Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 63, R93–R102. doi: 10.1530/JME-19-0153
Cuesta, M., Mendoza, J., Clesse, D., Pévet, P., and Challet, E. (2008). Serotonergic activation potentiates light resetting of the main circadian clock and alters clock gene expression in a diurnal rodent. Exp. Neurol. 210, 501–513. doi: 10.1016/j.expneurol.2007.11.026
Danbolt, N. C. (2001). Glutamate uptake. Prog. Neurobiol. 65, 1–105. doi: 10.1016/s0301-0082(00)00067-8
Daszuta, A., and Barrit, M. C. (1982). Endogenous serotonin (5-HT) and 5-hydroxyindole acetic acid (5-HIAA) levels in large regions and in discrete brain areas of C57BL and BALBc mice at three times of the day. Brain Res. Bull. 8, 477–482. doi: 10.1016/0361-9230(82)90004-1
Descarries, L., and Mechawar, N. (2000). Ultrastructural evidence for diffuse transmission by monoamine and acetylcholine neurons of the central nervous system. Prog. Brain Res. 125, 27–47. doi: 10.1016/S0079-6123(00)25005-X
Deurveilher, S., and Semba, K. (2005). Indirect projections from the suprachiasmatic nucleus to major arousal-promoting cell groups in rat: implications for the circadian control of behavioural state. Neuroscience 130, 165–183. doi: 10.1016/j.neuroscience.2004.08.030
Dibner, C., Schibler, U., and Albrecht, U. (2010). The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549. doi: 10.1146/annurev-physiol-021909-135821
Doyle, S. E., McIvor, W. E., and Menaker, M. (2002). Circadian rhythmicity in dopamine content of mammalian retina: role of the photoreceptors. J. Neurochem. 83, 211–219. doi: 10.1046/j.1471-4159.2002.01149.x
Elmenhorst, D., Mertens, K., Kroll, T., Oskamp, A., Ermert, J., Elmenhorst, E.-M., et al. (2016). Circadian variation of metabotropic glutamate receptor 5 availability in the rat brain. J. Sleep Res. 25, 754–761. doi: 10.1111/jsr.12432
Ewer, J., Frisch, B., Hamblen-Coyle, M. J., Rosbash, M., and Hall, J. C. (1992). Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J. Neurosci. 12, 3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992
Fang, L., Yang, Z., Zhou, J., Tung, J.-Y., Hsiao, C.-D., Wang, L., et al. (2015). Circadian clock gene CRY2 degradation is involved in chemoresistance of colorectal cancer. Mol. Cancer Ther. 14, 1476–1487. doi: 10.1158/1535-7163.MCT-15-0030
Farinelli, S. E., and Nicklas, W. J. (1992). Glutamate metabolism in rat cortical astrocyte cultures. J. Neurochem. 58, 1905–1915. doi: 10.1111/j.1471-4159.1992.tb10068.x
Fellin, T., Pascual, O., Gobbo, S., Pozzan, T., Haydon, P. G., and Carmignoto, G. (2004). Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron 43, 729–743. doi: 10.1016/j.neuron.2004.08.011
Flourakis, M., Kula-Eversole, E., Hutchison, A. L., Han, T. H., Aranda, K., Moose, D. L., et al. (2015). A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848. doi: 10.1016/j.cell.2015.07.036
Fonken, L. K., Frank, M. G., Kitt, M. M., Barrientos, R. M., Watkins, L. R., and Maier, S. F. (2015). Microglia inflammatory responses are controlled by an intrinsic circadian clock. Brain. Behav. Immun. 45, 171–179. doi: 10.1016/j.bbi.2014.11.009
Fuxe, K., Borroto-Escuela, D. O., Romero-Fernandez, W., Ciruela, F., Manger, P., Leo, G., et al. (2012). On the role of volume transmission and receptor-receptor interactions in social behaviour: focus on central catecholamine and oxytocin neurons. Brain Res. 1476, 119–131. doi: 10.1016/j.brainres.2012.01.062
Gau, D., Lemberger, T., von Gall, C., Kretz, O., Le Minh, N., Gass, P., et al. (2002). Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 34, 245–253. doi: 10.1016/s0896-6273(02)00656-6
Glass, J. D., Randolph, W. W., Ferreira, S. A., Rea, M. A., Hauser, U. E., Blank, J. L., et al. (1992). Diurnal variation in 5-hydroxyindole-acetic acid output in the suprachiasmatic region of the Siberian hamster assessed by in vivo microdialysis: evidence for nocturnal activation of serotonin release. Neuroendocrinology 56, 582–590. doi: 10.1159/000126277
Gooley, J. J., Lu, J., Chou, T. C., Scammell, T. E., and Saper, C. B. (2001). Melanopsin in cells of origin of the retinohypothalamic tract. Nat. Neurosci. 4:1165. doi: 10.1038/nn768
Hablitz, L. M., Plá, V., Giannetto, M., Vinitsky, H. S., Stæger, F. F., Metcalfe, T., et al. (2020). Circadian control of brain glymphatic and lymphatic fluid flow. Nat. Commun. 11:4411. doi: 10.1038/s41467-020-18115-2
Hamilton, N. B., and Attwell, D. (2010). Do astrocytes really exocytose neurotransmitters? Nat. Rev. Neurosci. 11, 227–238. doi: 10.1038/nrn2803
Hampp, G., Ripperger, J. A., Houben, T., Schmutz, I., Blex, C., Perreau-Lenz, S., et al. (2008). Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr. Biol. 18, 678–683. doi: 10.1016/j.cub.2008.04.012
Harvey, J. R. M., Plante, A. E., and Meredith, A. L. (2020). Ion channels controlling circadian rhythms in suprachiasmatic nucleus excitability. Physiol. Rev. 100, 1415–1454. doi: 10.1152/physrev.00027.2019
Hastings, M. H., Maywood, E. S., and Brancaccio, M. (2018). Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469. doi: 10.1038/s41583-018-0026-z
Hastings, M. H., Reddy, A. B., and Maywood, E. S. (2003). A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661. doi: 10.1038/nrn1177
Hattar, S., Kumar, M., Park, A., Tong, P., Tung, J., Yau, K.-W., et al. (2006). Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J. Comp. Neurol. 497, 326–349. doi: 10.1002/cne.20970
Hayashi, Y., Koyanagi, S., Kusunose, N., Okada, R., Wu, Z., Tozaki-Saitoh, H., et al. (2013). The intrinsic microglial molecular clock controls synaptic strength via the circadian expression of cathepsin S. Sci. Rep. 3:2744. doi: 10.1038/srep02744
Hay-Schmidt, A., Vrang, N., Larsen, P. J., and Mikkelsen, J. D. (2003). Projections from the raphe nuclei to the suprachiasmatic nucleus of the rat. J. Chem. Neuroanat. 25, 293–310. doi: 10.1016/s0891-0618(03)00042-5
Herzog, E. D., Aton, S. J., Numano, R., Sakaki, Y., and Tei, H. (2004). Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythms 19, 35–46. doi: 10.1177/0748730403260776
Hirasawa, H., Puopolo, M., and Raviola, E. (2009). Extrasynaptic release of GABA by retinal dopaminergic neurons. J. Neurophysiol. 102, 146–158. doi: 10.1152/jn.00130.2009
Hood, S., Cassidy, P., Cossette, M.-P., Weigl, Y., Verwey, M., Robinson, B., et al. (2010). Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J. Neurosci. 30, 14046–14058. doi: 10.1523/JNEUROSCI.2128-10.2010
Hubbard, J., Ruppert, E., Calvel, L., Robin-Choteau, L., Gropp, C.-M., Allemann, C., et al. (2015). Arvicanthis ansorgei, a novel model for the study of sleep and waking in diurnal rodents. Sleep 38, 979–988. doi: 10.5665/sleep.4754
Huhman, K. L., Hennessey, A. C., and Albers, H. E. (1996). Rhythms of glutamic acid decarboxylase mRNA in the suprachiasmatic nucleus. J. Biol. Rhythms 11, 311–316. doi: 10.1177/074873049601100404
Ikeda, E., Matsunaga, N., Kakimoto, K., Hamamura, K., Hayashi, A., Koyanagi, S., et al. (2013). Molecular mechanism regulating 24-hour rhythm of dopamine D3 receptor expression in mouse ventral striatum. Mol. Pharmacol. 83, 959–967. doi: 10.1124/mol.112.083535
Imbesi, M., Yildiz, S., Dirim Arslan, A., Sharma, R., Manev, H., and Uz, T. (2009). Dopamine receptor-mediated regulation of neuronal “clock” gene expression. Neuroscience 158, 537–544. doi: 10.1016/j.neuroscience.2008.10.044
Jones, B. E. (2017). Principal cell types of sleep-wake regulatory circuits. Curr. Opin. Neurobiol. 44, 101–109. doi: 10.1016/j.conb.2017.03.018
Joshi, N., and Chandler, D. (2020). Sex and the noradrenergic system. Handb. Clin. Neurol. 175, 167–176. doi: 10.1016/B978-0-444-64123-6.00012-6
Koike, N., Yoo, S.-H., Huang, H.-C., Kumar, V., Lee, C., Kim, T.-K., et al. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. doi: 10.1126/science.1226339
Korshunov, K. S., Blakemore, L. J., and Trombley, P. Q. (2017). Dopamine: a modulator of circadian rhythms in the central nervous system. Front. Cell. Neurosci. 11:91. doi: 10.3389/fncel.2017.00091
Lecca, S., Melis, M., Luchicchi, A., Muntoni, A. L., and Pistis, M. (2012). Inhibitory inputs from rostromedial tegmental neurons regulate spontaneous activity of midbrain dopamine cells and their responses to drugs of abuse. Neuropsychopharmacology 37, 1164–1176. doi: 10.1038/npp.2011.302
Lehre, K. P., and Danbolt, N. C. (1998). The number of glutamate transporter subtype molecules at glutamatergic synapses: chemical and stereological quantification in young adult rat brain. J. Neurosci. 18, 8751–8757. doi: 10.1523/JNEUROSCI.18-21-08751.1998
Lundkvist, G. B., Kwak, Y., Davis, E. K., Tei, H., and Block, G. D. (2005). A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J. Neurosci. 25, 7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005
Maejima, T., Tsuno, Y., Miyazaki, S., Tsuneoka, Y., Hasegawa, E., Islam, M. T., et al. (2021). GABA from vasopressin neurons regulates the time at which suprachiasmatic nucleus molecular clocks enable circadian behavior. Proc. Natl. Acad. Sci. U S A 118:e2010168118. doi: 10.1073/pnas.2010168118
Malek, Z. S., Dardente, H., Pevet, P., and Raison, S. (2005). Tissue-specific expression of tryptophan hydroxylase mRNAs in the rat midbrain: anatomical evidence and daily profiles. Eur. J. Neurosci. 22, 895–901. doi: 10.1111/j.1460-9568.2005.04264.x
Malek, Z. S., Sage, D., Pévet, P., and Raison, S. (2007). Daily rhythm of tryptophan hydroxylase-2 messenger ribonucleic acid within raphe neurons is induced by corticoid daily surge and modulated by enhanced locomotor activity. Endocrinology 148, 5165–5172. doi: 10.1210/en.2007-0526
Marpegan, L., Swanstrom, A. E., Chung, K., Simon, T., Haydon, P. G., Khan, S. K., et al. (2011). Circadian regulation of ATP release in astrocytes. J. Neurosci. 31, 8342–8350. doi: 10.1523/JNEUROSCI.6537-10.2011
Martini, T., Ripperger, J. A., Stalin, J., Kores, A., Stumpe, M., and Albrecht, U. (2021). Deletion of the clock gene Period2 (Per2) in glial cells alters mood-related behavior in mice. Sci. Rep. 11:12242. doi: 10.1038/s41598-021-91770-7
McClung, C. A., Sidiropoulou, K., Vitaterna, M., Takahashi, J. S., White, F. J., Cooper, D. C., et al. (2005). Regulation of dopaminergic transmission and cocaine reward by the clock gene. Proc. Natl. Acad. Sci. U S A 102, 9377–9381. doi: 10.1073/pnas.0503584102
Mendoza, J., and Challet, E. (2014). Circadian insights into dopamine mechanisms. Neuroscience 282, 230–242. doi: 10.1016/j.neuroscience.2014.07.081
Merlino, G., Piani, A., Gigli, G. L., Cancelli, I., Rinaldi, A., Baroselli, A., et al. (2010). Daytime sleepiness is associated with dementia and cognitive decline in older Italian adults: a population-based study. Sleep Med. 11, 372–377. doi: 10.1016/j.sleep.2009.07.018
Mistlberger, R. E., Antle, M. C., Glass, J. D., and Miller, J. D. (2000). Behavioral and serotonergic regulation of circadian rhythms. Biol. Rhythm Res. 31, 240–283. doi: 10.1076/0929-1016(200007)31:3;1-K;FT240
Morin, L. P. (1999). Serotonin and the regulation of mammalian circadian rhythmicity. Ann. Med. 31, 12–33. doi: 10.3109/07853899909019259
Musiek, E. S., Lim, M. M., Yang, G., Bauer, A. Q., Qi, L., Lee, Y., et al. (2013). Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Invest. 123, 5389–5400. doi: 10.1172/JCI70317
Nakamaru-Ogiso, E., Miyamoto, H., Hamada, K., Tsukada, K., and Takai, K. (2012). Novel biochemical manipulation of brain serotonin reveals a role of serotonin in the circadian rhythm of sleep-wake cycles. Eur. J. Neurosci. 35, 1762–1770. doi: 10.1111/j.1460-9568.2012.08077.x
Nakayama, K. (2002). Diurnal rhythm in extracellular levels of 5-hydroxyindoleacetic acid in the medial prefrontal cortex of freely moving rats: an in vivo microdialysis study. Prog. Neuropsychopharmacol. Biol. Psychiatry 26, 1383–1388. doi: 10.1016/s0278-5846(02)00304-4
Nakazato, R., Hotta, S., Yamada, D., Kou, M., Nakamura, S., Takahata, Y., et al. (2017). The intrinsic microglial clock system regulates interleukin-6 expression. Glia 65, 198–208. doi: 10.1002/glia.23087
Nakazato, R., Takarada, T., Yamamoto, T., Hotta, S., Hinoi, E., Yoneda, Y., et al. (2011). Selective upregulation of Per1 mRNA expression by ATP through activation of P2X7 purinergic receptors expressed in microglial cells. J. Pharmacol. Sci. 116, 350–361. doi: 10.1254/jphs.11069fp
Narita, M., Nagumo, Y., Hashimoto, S., Narita, M., Khotib, J., Miyatake, M., et al. (2006). Direct involvement of orexinergic systems in the activation of the mesolimbic dopamine pathway and related behaviors induced by morphine. J. Neurosci. 26, 398–405. doi: 10.1523/JNEUROSCI.2761-05.2006
Näslund, J., Studer, E., Nilsson, K., Westberg, L., and Eriksson, E. (2013). Serotonin depletion counteracts sex differences in anxiety-related behaviour in rat. Psychopharmacology (Berl). 230, 29–35. doi: 10.1007/s00213-013-3133-6
O’Neill, J. S., Maywood, E. S., Chesham, J. E., Takahashi, J. S., and Hastings, M. H. (2008). cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–953. doi: 10.1126/science.1152506
Olivero, G., Vergassola, M., Cisani, F., Roggeri, A., and Pittaluga, A. (2020). Presynaptic release-regulating metabotropic glutamate receptors: an update. Curr. Neuropharmacol. 18, 655–672. doi: 10.2174/1570159X17666191127112339
Osorio-Forero, A., Cherrad, N., Banterle, L., Fernandez, L. M. J., and Lüthi, A. (2022). When the locus coeruleus speaks up in sleep: recent insights, emerging perspectives. Int. J. Mol. Sci. 23:5028. doi: 10.3390/ijms23095028
Palm, D., Uzoni, A., Simon, F., Tucha, O., Thome, J., Faltraco, F., et al. (2021). Norepinephrine influences the circadian clock in human dermal fibroblasts from study participants with a diagnosis of attention-deficit hyperactivity disorder. J. Neural Transm. 128, 1147–1157. doi: 10.1007/s00702-021-02376-2
Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., et al. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320. doi: 10.1016/s0092-8674(02)00722-5
Parra-Damas, A., Rubió-Ferrarons, L., Shen, J., and Saura, C. A. (2017). CRTC1 mediates preferential transcription at neuronal activity-regulated CRE/TATA promoters. Sci. Rep. 7:18004. doi: 10.1038/s41598-017-18215-y
Peng, W., Achariyar, T. M., Li, B., Liao, Y., Mestre, H., Hitomi, E., et al. (2016). Suppression of glymphatic fluid transport in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 93, 215–225. doi: 10.1016/j.nbd.2016.05.015
Pickard, G. E., Smith, B. N., Belenky, M., Rea, M. A., Dudek, F. E., Sollars, P. J., et al. (1999). 5-HT1B receptor-mediated presynaptic inhibition of retinal input to the suprachiasmatic nucleus. J. Neurosci. 19, 4034–4045. doi: 10.1523/JNEUROSCI.19-10-04034.1999
Preitner, N., Damiola, F., Lopez-Molina, L., Zakany, J., Duboule, D., Albrecht, U., et al. (2002). The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. doi: 10.1016/s0092-8674(02)00825-5
Prigge, C. L., Yeh, P.-T., Liou, N.-F., Lee, C.-C., You, S.-F., Liu, L.-L., et al. (2016). M1 ipRGCs influence visual function through retrograde signaling in the retina. J. Neurosci. 36, 7184–7197. doi: 10.1523/JNEUROSCI.3500-15.2016
Prolo, L. M., Takahashi, J. S., and Herzog, E. D. (2005). Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 25, 404–408. doi: 10.1523/JNEUROSCI.4133-04.2005
Ralph, M. R., Foster, R. G., Davis, F. C., and Menaker, M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978. doi: 10.1126/science.2305266
Rasmussen, M. K., Mestre, H., and Nedergaard, M. (2018). The glymphatic pathway in neurological disorders. Lancet Neurol. 17, 1016–1024. doi: 10.1016/S1474-4422(18)30318-1
Rea, M. A., and Pickard, G. E. (2000). A 5-HT(1B) receptor agonist inhibits light-induced suppression of pineal melatonin production. Brain Res. 858, 424–428. doi: 10.1016/s0006-8993(99)02462-2
Rial, R. V., Canellas, F., Gamundí, A., Akaârir, M., and Nicolau, M. C. (2018). Pleasure: the missing link in the regulation of sleep. Neurosci. Biobehav. Rev. 88, 141–154. doi: 10.1016/j.neubiorev.2018.03.012
Romei, C., Bonifacino, T., Milanese, M., Usai, C., and Raiteri, L. (2016). Colocalization of neurotransmitter transporters on the plasma membrane of the same nerve terminal may reflect cotransmission. Brain Res. Bull. 127, 100–110. doi: 10.1016/j.brainresbull.2016.08.013
Ruan, G.-X., Allen, G. C., Yamazaki, S., and McMahon, D. G. (2008). An autonomous circadian clock in the inner mouse retina regulated by dopamine and GABA. PLoS Biol. 6:e249. doi: 10.1371/journal.pbio.0060249
Saijo, K., and Glass, C. K. (2011). Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol. 11, 775–787. doi: 10.1038/nri3086
Schmutz, I., Ripperger, J. A., Baeriswyl-Aebischer, S., and Albrecht, U. (2010). The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24, 345–357. doi: 10.1101/gad.564110
Schwab, C., DeMaggio, A. J., Ghoshal, N., Binder, L. I., Kuret, J., McGeer, P. L., et al. (2000). Casein kinase 1 delta is associated with pathological accumulation of tau in several neurodegenerative diseases. Neurobiol. Aging 21, 503–510. doi: 10.1016/s0197-4580(00)00110-x
Selim, M., Glass, J. D., Hauser, U. E., and Rea, M. A. (1993). Serotonergic inhibition of light-induced fos protein expression and extracellular glutamate in the suprachiasmatic nuclei. Brain Res. 621, 181–188. doi: 10.1016/0006-8993(93)90105-v
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shirogane, T., Jin, J., Ang, X. L., and Harper, J. W. (2005). SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863–26872. doi: 10.1074/jbc.M502862200
Sleipness, E. P., Sorg, B. A., and Jansen, H. T. (2007). Diurnal differences in dopamine transporter and tyrosine hydroxylase levels in rat brain: dependence on the suprachiasmatic nucleus. Brain Res. 1129, 34–42. doi: 10.1016/j.brainres.2006.10.063
Smith, B. N., Sollars, P. J., Dudek, F. E., and Pickard, G. E. (2001). Serotonergic modulation of retinal input to the mouse suprachiasmatic nucleus mediated by 5-HT1B and 5-HT7 receptors. J. Biol. Rhythms 16, 25–38. doi: 10.1177/074873040101600104
Sominsky, L., Dangel, T., Malik, S., De Luca, S. N., Singewald, N., Spencer, S. J., et al. (2021). Microglial ablation in rats disrupts the circadian system. FASEB J. 35:e21195. doi: 10.1096/fj.202001555RR
Spanagel, R., Pendyala, G., Abarca, C., Zghoul, T., Sanchis-Segura, C., Magnone, M. C., et al. (2005). The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat. Med. 11, 35–42. doi: 10.1038/nm1163
Sprouse, J., Braselton, J., and Reynolds, L. (2006). Fluoxetine modulates the circadian biological clock via phase advances of suprachiasmatic nucleus neuronal firing. Biol. Psychiatry 60, 896–899. doi: 10.1016/j.biopsych.2006.03.003
Stephan, F. K., and Zucker, I. (1972). Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U S A 69, 1583–1586. doi: 10.1073/pnas.69.6.1583
Terazono, H., Mutoh, T., Yamaguchi, S., Kobayashi, M., Akiyama, M., Udo, R., et al. (2003). Adrenergic regulation of clock gene expression in mouse liver. Proc. Natl. Acad. Sci. U S A 100, 6795–6800. doi: 10.1073/pnas.0936797100
Travnickova-Bendova, Z., Cermakian, N., Reppert, S. M., and Sassone-Corsi, P. (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. U S A 99, 7728–7733. doi: 10.1073/pnas.102075599
Tso, C. F., Simon, T., Greenlaw, A. C., Puri, T., Mieda, M., Herzog, E. D., et al. (2017). Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr. Biol. 27, 1055–1061. doi: 10.1016/j.cub.2017.02.037
Urban, M. W., Lo, C., Bodinayake, K. K., Brunswick, C. A., Murakami, S., Heimann, A. C., et al. (2021). The circadian clock gene Per1 modulates context fear memory formation within the retrosplenial cortex in a sex-specific manner. Neurobiol. Learn. Mem. 185:107535. doi: 10.1016/j.nlm.2021.107535
Ushijima, K., Koyanagi, S., Sato, Y., Ogata, T., Matsunaga, N., Fujimura, A., et al. (2012). Role of activating transcription factor-4 in 24-hour rhythm of serotonin transporter expression in the mouse midbrain. Mol. Pharmacol. 82, 264–270. doi: 10.1124/mol.112.079079
Videnovic, A., Lazar, A. S., Barker, R. A., and Overeem, S. (2014). “The clocks that time us”–circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 10, 683–693. doi: 10.1038/nrneurol.2014.206
Vizi, E. S., Kiss, J. P., and Lendvai, B. (2004). Nonsynaptic communication in the central nervous system. Neurochem. Int. 45, 443–451. doi: 10.1016/j.neuint.2003.11.016
Wang, X.-L., Wolff, S. E. C., Korpel, N., Milanova, I., Sandu, C., Rensen, P. C. N., et al. (2020). Deficiency of the circadian clock gene bmal1 reduces microglial immunometabolism. Front. Immunol. 11:586399. doi: 10.3389/fimmu.2020.586399
Waniewski, R. A., and Martin, D. L. (1986). Exogenous glutamate is metabolized to glutamine and exported by rat primary astrocyte cultures. J. Neurochem. 47, 304–313. doi: 10.1111/j.1471-4159.1986.tb02863.x
Webb, I. C., Baltazar, R. M., Wang, X., Pitchers, K. K., Coolen, L. M., Lehman, M. N., et al. (2009). Diurnal variations in natural and drug reward, mesolimbic tyrosine hydroxylase and clock gene expression in the male rat. J. Biol. Rhythms 24, 465–476. doi: 10.1177/0748730409346657
Witkovsky, P. (2004). Dopamine and retinal function. Doc. Ophthalmol. 108, 17–40. doi: 10.1023/b:doop.0000019487.88486.0a
Womac, A. D., Burkeen, J. F., Neuendorff, N., Earnest, D. J., and Zoran, M. J. (2009). Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur. J. Neurosci. 30, 869–876. doi: 10.1111/j.1460-9568.2009.06874.x
Wong, C. C. P., and Schumann, G. (2012). Integration of the circadian and stress systems: influence of neuropeptides and implications for alcohol consumption. J. Neural Transm. 119, 1111–1120. doi: 10.1007/s00702-012-0829-4
Xu, Y., Padiath, Q. S., Shapiro, R. E., Jones, C. R., Wu, S. C., Saigoh, N., et al. (2005). Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644. doi: 10.1038/nature03453
Yoon, S. O., and Chikaraishi, D. M. (1994). Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J. Biol. Chem. 269, 18453–18462.
Keywords: circadian rhythms, microglia, astrocyte, neurotransmitters, brain areas, neuron
Citation: Fagiani F, Baronchelli E, Pittaluga A, Pedrini E, Scacchi C, Govoni S and Lanni C (2022) The Circadian Molecular Machinery in CNS Cells: A Fine Tuner of Neuronal and Glial Activity With Space/Time Resolution. Front. Mol. Neurosci. 15:937174. doi: 10.3389/fnmol.2022.937174
Received: 05 May 2022; Accepted: 07 June 2022;
Published: 01 July 2022.
Edited by:
Urs Albrecht, Université de Fribourg, SwitzerlandReviewed by:
Shimon Amir, Concordia University, CanadaTomaz Martini, Swiss Federal Institute of Technology Lausanne, Switzerland
Copyright © 2022 Fagiani, Baronchelli, Pittaluga, Pedrini, Scacchi, Govoni and Lanni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Lanni, Y3Jpc3RpbmEubGFubmlAdW5pcHYuaXQ=
† These authors have contributed equally to this work
 Francesca Fagiani
Francesca Fagiani Eva Baronchelli
Eva Baronchelli Anna Pittaluga3,4
Anna Pittaluga3,4 Chiara Scacchi
Chiara Scacchi Stefano Govoni
Stefano Govoni Cristina Lanni
Cristina Lanni