- Child and Adolescent Psychiatry, Center of Mental Health, University Hospital of Würzburg, Würzburg, Germany
Members of the family of metabotropic glutamate receptors are involved in the pathomechanism of several disorders of the nervous system. Besides the well-investigated function of dysfunctional glutamate receptor signaling in neurodegenerative diseases, neurodevelopmental disorders (NDD), like autism spectrum disorders (ASD) and attention-deficit and hyperactivity disorder (ADHD) might also be partly caused by disturbed glutamate signaling during development. However, the underlying mechanism of the type III metabotropic glutamate receptor 8 (mGluR8 or GRM8) involvement in neurodevelopment and disease mechanism is largely unknown. Here we show that the expression pattern of the two orthologs of human GRM8, grm8a and grm8b, have evolved partially distinct expression patterns in the brain of zebrafish (Danio rerio), especially at adult stages, suggesting sub-functionalization of these two genes during evolution. Using double in situ hybridization staining in the developing brain we demonstrate that grm8a is expressed in a subset of gad1a-positive cells, pointing towards glutamatergic modulation of GABAergic signaling. Building on this result we generated loss-of-function models of both genes using CRISPR/Cas9. Both mutant lines are viable and display no obvious gross morphological phenotypes making them suitable for further analysis. Initial behavioral characterization revealed distinct phenotypes in larvae. Whereas grm8a mutant animals display reduced swimming velocity, grm8b mutant animals show increased thigmotaxis behavior, suggesting an anxiety-like phenotype. We anticipate that our two novel metabotropic glutamate receptor 8 zebrafish models may contribute to a deeper understanding of its function in normal development and its role in the pathomechanism of disorders of the central nervous system.
Introduction
The main excitatory neurotransmitter in the vertebrate nervous system is L-glutamate, which mediates its function through binding to ionotropic (AMPA, NMDA, and kainate receptors), and metabotropic receptors. Whereas ionotropic receptors are responsible for the fast action of glutamate, metabotropic receptors have slower and longer-lasting effects on neurotransmission. The latter group of eight subtypes of metabotropic glutamate receptors (mGlu receptors or GRMs) identified in mammalian species so far can be subdivided into three distinct classes due to pharmacological properties, sequence homology, cellular localization, and G protein coupling (Niswender and Conn, 2010). The family type I receptors (mGluR1 and mGluR5) are mainly found at postsynaptic sites where they couple to Gαq and Gα11 in order to activate downstream phospholipase Cβ1 and release intracellular Ca2+ and activate protein kinase C (PKC). In contrast, type II (mGluR2 and mGluR3) and type III (mGluR4, mGluR6, mGluR7 and mGluR8) receptors are coupled to Gαi/o and act mainly on the adenylyl cyclase to control cAMP levels. While mGluR type II and mGluR type III receptors are associated with similar downstream signaling pathways, they differ substantially in their synaptic localization. Type III mGluRs (except for mGluR6) are predominantly expressed at the presynapse, thereby regulating excitatory (glutamatergic), inhibitory (GABAergic) and neuromodulatory transmitter release. In contrast, type II mGluRs are located outside of the release zone of the pre- and postsynapse (Niswender and Conn, 2010; Bodzęta et al., 2021).
All mGluRs contain a large extracellular domain that comprises the agonist-binding Venus fly trap (VFT) domain and a cysteine-rich domain that connects to the highly conserved seven-pass trans-membrane domain (Pin and Bettler, 2016). mGluRs are constitutive dimers which are linked via disulfide bonds in the proximity of the VFT (Niswender and Conn, 2010). The complexity of mGluRs function and mode of action in the nervous system is enhanced by alternative splicing to generate multiple isoforms of single mGluRs, homo- and heterodimerization of different mGluRs to change its signaling properties, cell type-specific protein interactions and varying downstream signaling cascades (Niswender and Conn, 2010; Pin and Bettler, 2016; Bodzęta et al., 2021; McCullock and Kammermeier, 2021).
Over the past years, evidence indicates that mGluRs are involved in several disorders of the nervous system. In neurodegenerative disorders like Parkinson’s disease, Alzheimer’s disease and Huntington’s disease involvement of mGluRs, including the type III receptor GRM8, were documented frequently (Ribeiro et al., 2017). In neurodevelopmental disorders, an association of copy number variation of the GRM8 locus with ADHD was found (Elia et al., 2012; Akutagava-Martins et al., 2014; Hawi et al., 2015; Liu et al., 2021). A 1.9 Mb microdeletion on chromosome 7 q including the GRM8 locus was associated with intellectual disability and signs of autism (Sangu et al., 2017). Using GWAS, an association of GRM8 genotype and trait depression was detected (Terracciano et al., 2010). In a Han Chinese population, a link with major depression and schizophrenia was demonstrated for SNPs located in the GRM8 gene locus (Li et al., 2016). A similar result was found in an Iranian population for GRM8 and schizophrenia (Tavakkoly-Bazzaz et al., 2018). The association of the GRM8 locus and major depression was later confirmed in a massive meta-analysis of several GWAS studies (Howard et al., 2019). In substance use conditions, associations with GRM8 were shown as well (Long et al., 2015; Bauer and Covault, 2020).
Several animal models have been developed to elucidate mGluR8 function. The very first study on mGluR8 knockout mice revealed an anxiolytic-like phenotype in a conditioned fear model (Gerlai et al., 2002). Further studies have either demonstrated an anxiogenic-like behavior (Linden et al., 2002, 2003; Robbins et al., 2007; Duvoisin et al., 2010a,b) or no increase in anxiety-like behavior (Fendt et al., 2010; Davis et al., 2013), probably being an age, developmental stage or background strain dependent effect in the different models generated. For instance, in older mGluR8 knockout mice (6 month of age) it was shown that increased measures of anxiety were present (Linden et al., 2002; Duvoisin et al., 2005), whereas in younger mice (2–4 months of age) no differences were found in the same test (Raber and Duvoisin, 2015). Another behavioral dimension often characterized to investigate psychiatric conditions like ADHD are activity-related phenotypes. The mGluR8 knock-out mice are either being hypoactive (Duvoisin et al., 2005), hyperactive (Gerlai et al., 2002) or have not shown any alteration of motor behavior (Linden et al., 2002; Robbins et al., 2007; Raber and Duvoisin, 2015). Besides genetic models, pharmacological models have been developed to investigate mGluR8 function, demonstrating alterations in terms of alcohol dependence and anxiety-like phenotype (Bahi, 2017). Therefore, the currently available animal models do not draw a consistent picture of mGluR8 function in psychiatric relevant behavioral modulation.
Zebrafish (Danio rerio), an increasingly accepted vertebrate model organism, is widely used in for instance toxicology, pharmacology, neuroscience and developmental research. The conserved vertebrate brain organization and neurochemistry, the rich behavioral repertoire, the high number of offspring, the comparably fast development, the short generation times and the genetic homology to human makes zebrafish an attractive model organism (Stewart et al., 2014, 2015). Especially, screening for new compounds for pharmacological interventions on larger scales is a huge benefit over other vertebrate model organisms (Rennekamp and Peterson, 2015).
The aim of this study is to explore the consequences of mGluR8 deficiency to understand its role in neurodevelopment and potential contribution to behaviors relevant for neurodevelopmental psychiatric disorders using zebrafish larvae. In the zebrafish genome there are two mGluR8 paralogs encoded by grm8a and grm8b. We characterize the expression pattern of grm8a and grm8b at early and late developmental stages until adulthood with a particular focus on the nervous system to understand spatial and temporal differences. This information will be valuable to evaluate behavioral differences and gain insight into neuronal dysfunction of both proteins. Next, we generate loss-of-function mutants of the two paralogs using CRISPR/Cas9. The mutants are viable without any gross morphological abnormalities which makes them suitable for further behavioral characterizations. An initial basic behavioral assessment at larval stage concludes our analysis where we found first hints that mGluR8-deficiency can cause psychiatric relevant phenotypes in zebrafish.
Experimental Procedures
Animal Handling
All experiments were performed in the zebrafish (Danio rerio) AB/AB wildtype strain background (zfin id.: ZDB-GENO-960809-7). Monoaminergic cells expressing grm8a were identified in the enhancer trap line Tg(Etvmat2:GFP) (Wen et al., 2008). Larvae were raised at 28°C with a light/dark cycle of 14/10 h in Danieau’s solution with or without methylene blue (Cold Spring Harb. Protoc, 2011)1. To suppress pigmentation for whole-mount RNA in situ hybridization (ISH) and immunohistochemistry, Danieau’s solution containing 0.2 mM 1-phenyl-2-thiourea was used. Embryo staging was performed according to Kimmel et al. (1995). Embryos were manually dechorionated and fixed in 4% paraformaldehyde (PFA) in 1x phosphate-buffered saline (PBS). For adult brain preparations, fish were euthanized with an overdose of MS-222 and decapitated. The heads were fixed over night at 4°C in 4% PFA before the brains were dissected and post-fixed for another 4 h at room temperature (RT). All animal handling was performed in accordance with the regulations for animal welfare of the District Government of Lower Franconia, Germany.
Whole-Mount RNA in situ Hybridization
A grm8a target sequence was reverse transcription-PCR amplified with RevertAid reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, United States) and an oligodT primer (for gene-specific primer sequences see Supplementary Table 1), cloned into pCRII-TOPO vector (Thermo Fisher Scientific, Waltham, MA, United States) and verified by Sanger sequencing. Cloned full-length grm8b cDNA was a kind gift from Marion Haug and Stephan Neuhauss (Haug et al., 2013). The resulting plasmids were linearized (NotI, Thermo Fisher Scientific, Waltham, MA, United States) and purified (GenElute PCR Clean-Up Kit, Merck KGaA, Darmstadt, Germany) before in vitro transcribed with SP6 RNA polymerase and digoxiginin (DIG) RNA Labeling Mix (Merck KGaA, Darmstadt, Germany). The final RNA probes were purified by LiCl and ethanol precipitation. Before usage, the grm8b probe was hydrolyzed for 20 min at 60°C and 30 min in –20°C and precipitated by LiCl and ethanol again.
Whole-mount RNA ISH was performed according to the methods published previously (Thisse and Thisse, 2008; Lechermeier et al., 2019, 2020; Lüffe et al., 2021). Briefly, specimens were fixed in 4% PFA, washed in PBS containing 0.1% Tween-20 (PBST) and dehydrated in methanol (MeOH) and finally stored in 100% MeOH at –20°C. For ISH, the samples were rehydrated and then permeabilized by Proteinase K treatment (10 μg/ml). The specimens were then post-fixed in 4% PFA, washed in PBST and were incubated in hybridization buffer containing 5 mg/ml torula yeast RNA type VI (Merck KGaA, Darmstadt, Germany) for 1 h at 65°C. Subsequently, the samples were incubated in hybridization buffer containing the RNA ISH probe (dilution 1:100) overnight at 65°C. On the next day, the samples were passed through stringency washes at 65°C in hybridization buffer with saline-sodium citrate (SSC) buffer with a last step in 0.05 × SSC (in PBST) for 1 h at 65°C. For subsequent immuno detection of DIG, the samples were incubated in ISH blocking buffer [PBST with 2% normal sheep serum (NSS) and 2 mg/ml bovine serum albumin (BSA)] and then for 2 h at RT in sheep anti-DIG Fab fragments conjugated with Alkaline Phosphatase (AP; anti-DIG-AP, Merck KGaA, Darmstadt, Germany) diluted 1:5,000. After extensive washes in PBST, the samples were rinsed in alkaline tris-buffer (pH 9.5) for 30 min at RT. The AP activity was visualized with nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) (Merck KGaA, Darmstadt, Germany). Finally, the enzymatic reaction was stopped, and samples were transferred into 80% glycerol in PBST for long-term storage in darkness.
Dissected adult brains were processed as described above. Before incubating in ISH blocking buffer, the brains were embedded in 3% agarose in PBS and cut into transverse sections of 80 μm using a vibratome (Vibratome Series 1,000 Sectioning System). Subsequently, the brain slices were processed as described for whole-mount RNA ISH above.
For two-color RNA ISH, a gad1a (previously gad67a) containing plasmid (Martin et al., 1998) was linearized (EcoRI, Thermo Fisher Scientific, Waltham, MA, United States), purified (GenElute PCR Clean-Up Kit, Merck KGaA, Darmstadt, Germany) and in vitro transcribed using T3 RNA polymerase and fluorescein (FLUO) RNA Labeling Mix (Merck KGaA, Darmstadt, Germany). DIG- and FLUO-labeled probes were diluted together (1:100 each) in hybridization buffer. After hybridization, stringency washes and blocking, the samples were incubated in sheep anti-FLUO-AP Fab fragments diluted 1:2,000 in ISH blocking buffer. Subsequent to several washes in tris-buffer (pH 8.2) the AP activity was visualized with SIGMAFAST fast red TR/naphthol AS-MX phosphate tablets (4-chloro-2-methylbenzenediazonium/3-hydroxy-2-naphthoic acid 2,4-dimethylanilide phosphate) in 0.1 M Trizma buffer (Merck KGaA, Darmstadt, Germany). The reaction was stopped in PBST, and the anti-FLUO-AP Fab fragments were detached in PBST at 68°C for 2 h. Afterward, blocking, anti-DIG-AP immunolabeling and visualization of AP activity was performed as described before.
Cryosections
Samples were washed and cryoprotected in PBS containing 15% sucrose overnight at 4°C. Embedding was done in 7.5% gelatine dissolved in 15% sucrose in PBS. Blocks were cut and snap frozen in 2-methylbutane pre-cooled with liquid nitrogen. On a cryostat (Microm HM 500 OM), cryoblocks were cut transversally with a thickness of 20 μm and sections were mounted on slides and cover slipped with 80% glycerol. The samples were stored in darkness at 4°C until image acquisition.
Generation and Genotyping of Grm8a and Grm8b Mutant Lines
The selection of the CRISPR/Cas9 target sites were done with CHOPCHOP (Labun et al., 2019). Oligonucleotides for the synthesis of the single guide RNA (sgRNA) (Supplementary Table 1) were annealed and ligated with the linearized (Eco31I/BsaI, Thermo Fisher Scientific, Waltham, MA, United States) pDR274 vector (Addgene #42250). Sequences were verified with Sanger sequencing (Eurofins Genomics, Ebersberg, Germany). The plasmid containing the sgRNA sequence was linearized (DraI, Thermo Fisher Scientific, Waltham, MA, United States) and in vitro transcribed with a custom-made T7 RNA polymerase (a kind gift from Thomas Ziegenhals and Utz Fischer). The sgRNA was purified by Roti-phenol/chloroform/isoamylalcohol (Carl Roth GmbH & Co., KG, Karlsruhe, Germany) and injected (100 ng/μl) together with the Cas9-NLS protein (300 ng/μl, S. pyogenes, New England Biolabs, Ipswich, MA, United States) into the animal pole of fertilized one-cell stage zebrafish eggs. Successful targeting events were confirmed by PCR on extracted gDNA (primer sequences in Supplementary Table 1) and subsequent Sanger sequencing (Eurofins Genomics, Ebersberg, Germany). Germline transmission of indel mutations were verified by outcrossing with AB/AB wildtypes and subsequent genotyping of F1 larvae. Positive F0 fish were used to obtain the F1 generation by additional outcrossing. Individual grm8a and grm8b F1 adult mutants were used to analyze the induced mutations and establish the lines. Finally, the lines containing the deletion mutations were selected. For final mutation verification, gDNA of grm8a–/– and grm8b–/– F3 mutants were PCR amplified with primers flanking the sgRNA target site and Sanger sequenced. For the experiments described below, F3 embryos and larvae were used.
Immunohistochemistry
Whole-mount immunohistochemical stainings were performed on embryos at 24 hpf with the yolk removed. The genotype of immuno-stained grm8a and grm8b mutants was determined using the tail of each embryo for gDNA extraction and subsequent genotyping as described above.
Anti-cleaved Caspase 3 (cCasp3)
After fixation in 4% PFA for 3 h at RT embryos were dehydrated through a MeOH series [in PBS with 0.8% Triton X-100 (0.8% PBST)] and finally stored in 100% MeOH. Following permeabilization in 100% acetone for 7 min at –20°C, the samples were rehydrated in 50% MeOH in 0.8% PBT for 1 h at –20°C and subsequently washed in ddH2O and 0.8% PBST at RT. Blocking was done for 1 h at RT in blocking buffer (0.8% PBST with 1% DMSO, 10% NSS and 2 mg/ml BSA), before the samples were labeled for 3 days at 4°C with a polyclonal rabbit anti-cleaved Caspase-3 (Asp175) primary antibody (#9661, Cell Signaling Technology, Danvers, MA, United States; RRID:AB_2341188) diluted 1:500 in blocking buffer. After several washing steps in 0.8% PBT, the samples were exposed to secondary goat anti-rabbit IgG (H + L) conjugated to Alexa Fluor 488 (A-11034, Thermo Fisher Scientific, Waltham, MA, United States; RRID:AB_2576217) diluted 1:1,000 in blocking buffer for 2 days at 4°C. Finally, after extensive washing in 0.8% PBT the samples were stored in 80% glycerol (in PBST) in darkness at 4°C.
Quantitative Real-Time PCR
Quantitative real-time RT-PCR (qPCR) was performed on 5 dpf old –/–, +/– and +/+ siblings of grm8a and grm8b mutant lines. Genomic DNA extraction was done on tail cuts for subsequent genotyping of individual embryos. The whole body including the head was used for total RNA extraction and subsequent cDNA synthesis using standard methods. RNA from 10 pooled embryos were isolated for each genotype. For the qPCR analysis each target gene is represented by three technical replicates of each biological sample (n = 3) for each genotype. Additionally, a no RT control (NRT) and no template control (NTC) served as negative controls. Quantification and calculation were done with the comparative Ct (2–ΔΔCt) method using actb1 and gapdh as housekeeping genes (Supplementary Table 1). Group differences were determined by applying a one-way ANOVA with a significance level of 0.05.
Image Acquisition and Processing
A Zeiss Axiophot light microscope equipped with a Zeiss AxioCam MRc digital camera and the AxioVision Rel.4.8 Ink. software was used for image acquisition of RNA ISH labeling on embryonic and larval whole-mounts as well as on adult brain sections. For adult brain sections, single images were merged using a grid/collection ImageJ plugin (Preibisch et al., 2009). Size measurements were done on images taken at a Leica M205 FA fluorescence microscope equipped with a Leica DFC 420C digital camera and the Leica Application Suite V3.8 Ink. software. Images were recorded through a Leica Planapo 1.0x objective (Leica Camera AG, Wetzlar, Germany). Size measurements were done on fixed embryos (24 hpf) using the ImageJ image processing package Fiji 2.0.0 (Schindelin et al., 2012). Head area was traced using the midbrain-hindbrain boundary as posterior boundary. Yolk diameter was defined as the distance between most ventral center of the eye and the edge between circular and elongated yolk sac. Length measurements were calculated between the dorsal part of the midbrain-hindbrain boundary and the most posterior end of the tail not considering the fin. Size parameters are all given in squared pixels (pixel2). Image acquisition of anti-cCasp3 stainings was performed at a Zeiss LSM 780 confocal microscope equipped with a Lasos Argon 488 nm laser and the ZEN 2012 SP1 software. Images were taken through a Plan APO 20x/0.8 objective. Brightness adjustments of confocal images were done in Fiji 2.0.0. Images were arranged into final figures by using the vector graphics software Inkscape 1.0.1.
Locomotor Tracking
Locomotion tracking was performed on 5 dpf old larvae using the semi-automatic system ZebraBox and the corresponding commercial software ZebraLab (ViewPoint, Lyon, France). For the grm8a mutant line larvae from 6 clutches were analyzed on 6 different days. For the grm8b mutant line larvae from 10 clutches were used on 10 different days. Individual larvae were placed in 12-well plates containing 1 ml of Danieau’s solution. Temperature was kept constant at 28°C and swimming tracks were recorded in the dark (infrared illumination of 850 nm) with an infrared camera with 30 fps (frames per second). Three activity levels were defined with the following thresholds: inactive, < 0.2 cm/s; low activity, 0.2 cm/s < and < 1 cm/s; high activity, > 1 cm/s. Larvae were tracked for a total duration of 10 min, separated into a 5 min habituation and a 5 min test phase. Only data collected during the test phase was considered for final group comparisons. Locomotor activity was determined based on four different parameters: Total distance swum, mean velocity during low/high activity or both (total mean velocity), duration of inactivity, low or high activity and the number of events during inactive, low, and high activity phase.
To analyze thigmotaxis behavior, data obtained during locomotor assays were replayed using the commercial tracking software. Therefore, each region of interest/well (ROI) was virtually divided into two different zones: An outer zone with a width of 4 mm and an inner zone with a radius of 7.35 mm. An increased percentage of time spent in the outer zone of the ROI was defined as increased thigmotaxis behavior.
Data Analysis and Statistics
RStudio (RStudio 1.3.959, at https://rstudio.com/) was used for data analysis. Normal distribution was checked with the Shapiro-Wilk’s test. According to distribution, group differences were calculated by one-way ANOVA or Kruskal-Wallis rank sum test followed by Tukey’s HSD post-hoc test for parametric and Dunn’s post hoc test for non-parametric data. The Benjamini-Hochberg adjustment was used to control the false discovery rate (FDR). The general significance level was set to 0.05. Z-score transformation was used to compare data obtained with two different versions of the tracking software ZebraLab. Effect sizes were calculated by the Cliff’s delta (Supplementary Table 4) using the effsize package in RStudio (Torchiano, 2016). Appropriate sample sizes were determined by using the software G*Power 3.1.9.4 (Faul et al., 2007, 2009) with α and β set to 0.05.
Results
Two Orthologs of the Human GRM8 Gene Are Present in the Zebrafish Genome
Due to a probable genome duplication in the teleost lineage during evolution (Meyer and Schartl, 1999; Glasauer and Neuhauss, 2014) two orthologs of the human GRM8 gene (ENSG00000179603) are present in the zebrafish genome (Haug et al., 2013). The human gene is located on chromosome 7 and consists of 11 exons, encoding a protein of 908 aa and a molecular mass of 101.7 kDa. The two zebrafish orthologs are located on chromosome 4 (grm8a, ENSDARG00000077654) and chromosome 25 (grm8b, ENSDARG00000076508), respectively. grm8a is a 10 exon gene producing a protein of 909 aa with a molecular mass of 101.7 kDa. grm8b consists of 11 exons and encodes a protein of 907 aa with a molecular mass of 100.9 kDa. Both zebrafish protein sequences are matching the human sequence to 81.08% for Grm8a and 80.26% for Grm8b. The overall protein structure of zebrafish Grm8a and Grm8b are similar and share 86.55% sequence identity, whereas especially in the N-terminal part of the two proteins the biggest sequence variation can be detected (data not shown). Interestingly, using protein sequences from multiple species we detected a loss of an evolutionary conserved tyrosine residue in the C-terminal domain of Grm8b, despite an overall similar structural conservation of both zebrafish proteins across species (data not shown).
Early Developmental Expression of Grm8a and Grm8b
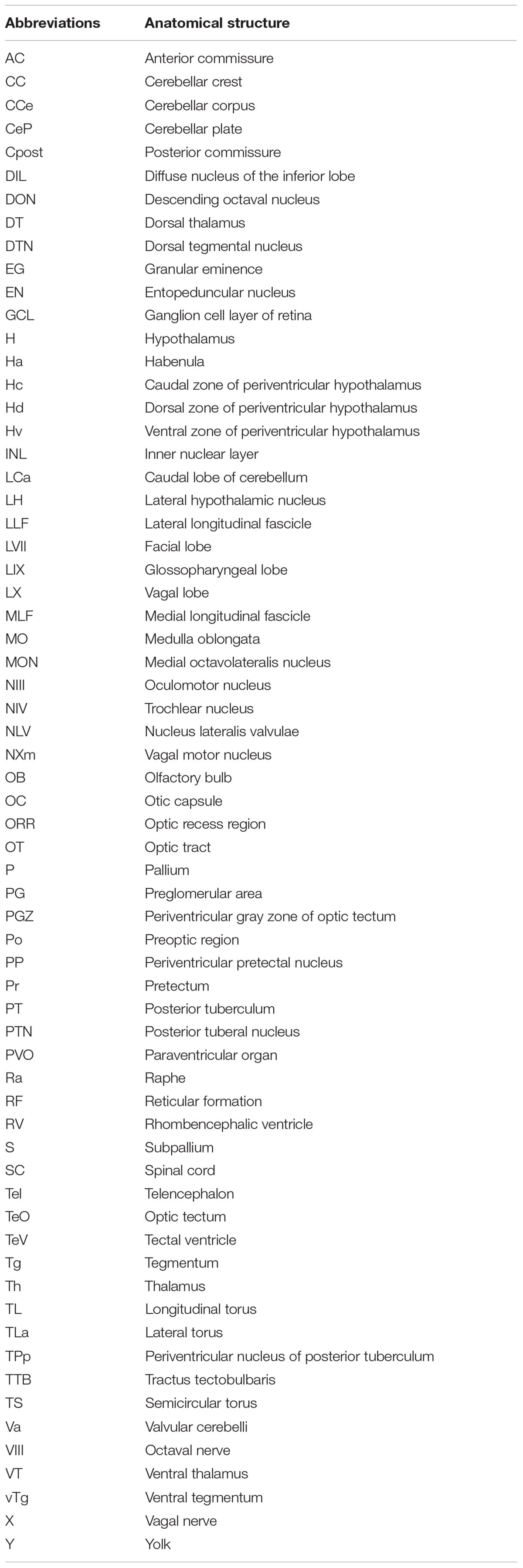
Expression of grm8a and grm8b were shown in the CNS of 3 dpf and 5 dpf old zebrafish before (Haug et al., 2013). However, a detailed description of the developmental trajectory including adult stages is missing. This information is crucial to understand the potential impact of null mutations of both genes on neurodevelopment and behavior. Therefore, we used RNA in situ hybridization (ISH) to characterize the expression patterns of both genes. We focused our analysis first on early developmental stages and found restricted expression at early and broad expression at advanced developmental stages for both paralogs, although the temporal differences are more pronounced for grm8a compared to grm8b. Transcripts for both paralogs were detected first at 24 hpf in the telencephalon and ventral tegmentum, and for grm8b in the hypothalamus and the medulla oblongata (Figures 1A,B and Supplementary Figures 1A–S, 2A–S). Similar or partly overlapping expression patterns for grm8a and grm8b were observed in the telencephalon (30–72 hpf), the ventral tegmentum (24–72 hpf), the optic tectum (72 hpf), and the ganglion and inner nuclear layer of the retina (72 hpf) whereas expression in the preoptic region (grm8a: 36–72 hpf; grm8b: 30–72 hpf) and the hypothalamus (grm8a: 30–72 hpf; grm8b: 24–72 hpf) shows temporal discrepancies potentially attributable to staining quality.
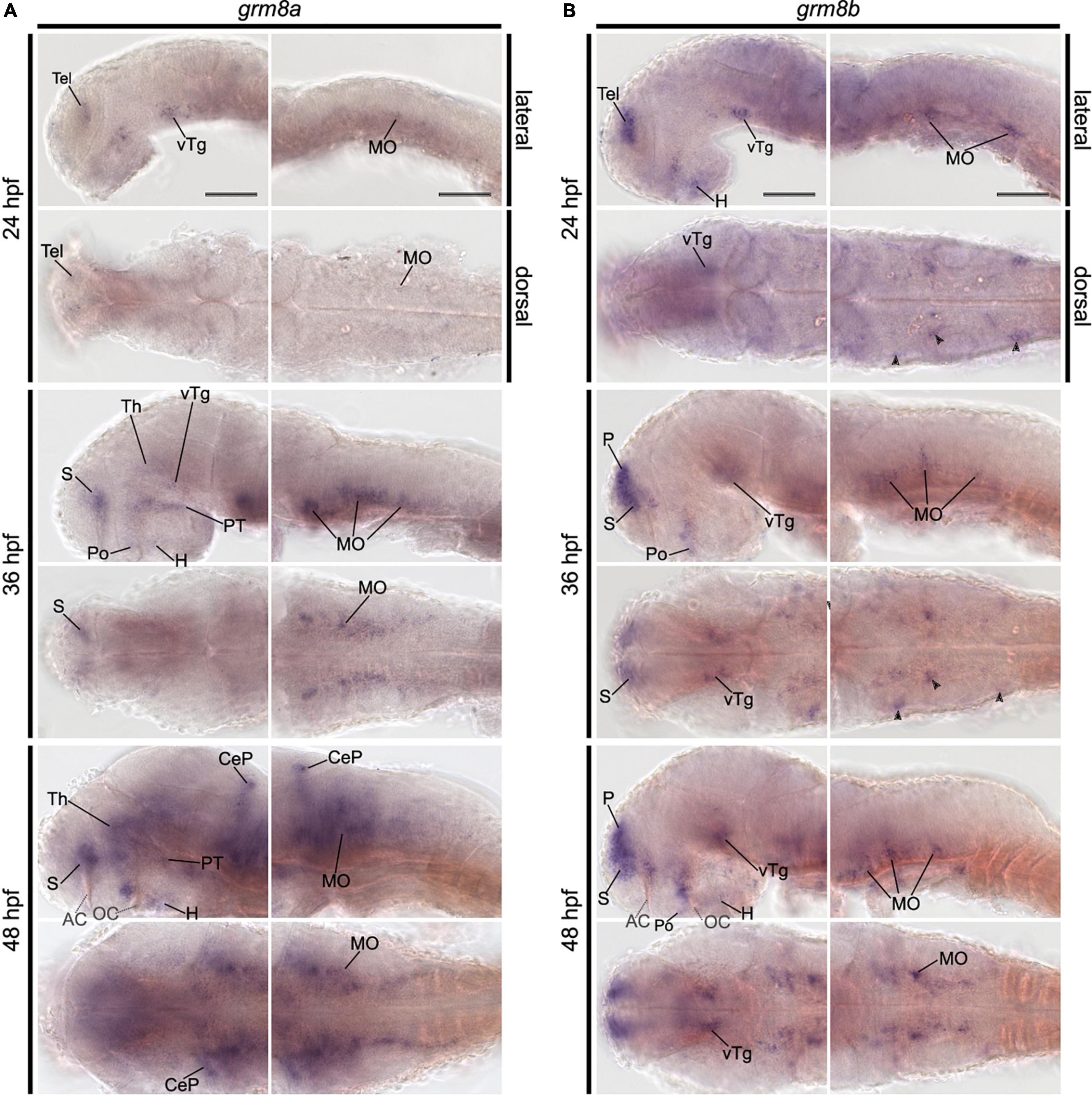
Figure 1. Pattern of grm8a and grm8b expression in the developing zebrafish revealed by whole-mount RNA in situ hybridization. (A) Grm8a expression at 24, 36, and 48 hpf in the developing zebrafish. Lateral (top) and dorsal (bottom) overviews indicate expression as early as 24 hpf in the central nervous system. (B) Grm8b expression at 24, 36, and 48 hpf in the developing zebrafish. Lateral (top) and dorsal (bottom) overviews indicate prominent expression as early as 24 hpf in the central nervous system. Details on the individual expression patterns are described in the main text. Arrowheads indicate transcript labeling in individual cell cluster of the medulla oblongata (MO). All images are oriented with anterior to the left. Abbreviations are listed in Table 1. Scale bar is 100 μm.
Further brain regions with prominent temporal discrepancies between grm8a and grm8b expression comprise the thalamus and the olfactory bulb. grm8a expression in thalamic regions is detected at 36 hpf (Figure 1A) in contrast to first grm8b transcript labeling at 72 hpf (Supplementary Figures 2I,O). Further, until 72 hpf distinct grm8b transcript labeling is detected in the most anterior telencephalon (comprising the developing olfactory bulbs) (Supplementary Figures 2B,D,F,H,J), whereas grm8a expression is absent or only detected faintly for earlier stages than 72 hpf (Supplementary Figures 1B,D,F,H,J).
Differences in both temporal and spatial distribution were observed for expression in the medulla oblongata. While grm8a transcripts were labeled in a bilateral stripe-like pattern from 30 hpf (Figure 1A and Supplementary Figures 1C,D,K–N), grm8b transcript labeling is restricted to scattered cell clusters at 24, 30, and 36 hpf (Figure 1B and Supplementary Figures 2A–F,K–N). At 48 hpf, grm8b expression also adapts a stripe-like pattern (Figure 1B and Supplementary Figures 2G,H). Differences in the spatial distribution comprises grm8a expression in the medial anterior medulla oblongata at 72 hpf (Supplementary Figure 1R), a subregion suggested to be implicated in the integration of sensory and modulatory input and in providing regulatory output onto (pre-) motor areas (Naumann et al., 2016).
Although many brain regions show similar expression patterns or only minor temporal or spatial differences in the localization of the two transcripts, distinct expression of grm8a or grm8b is also detected for some areas in the central nervous system. For instance, the posterior tuberculum is exclusively labeled for grm8a with expression starting as early as 30 hpf (Supplementary Figure 1K), the cerebellum with first detection at 48 hpf (Figure 1A), and the pretectum with transcripts labeled at 72 hpf (Supplementary Figures 1O,Q). Notably, like previously mentioned for the medial anterior MO, all identified regions with distinct grm8a expression were demonstrated to be involved in sensorimotor integration and/or motor control in the past (Ahrens et al., 2012; Naumann et al., 2016; Jha and Thirumalai, 2020). In the pallium distinct grm8b expression was observed (Figure 1B and Supplementary Figure 2K). Interestingly, this area is suggested to comprise homologous structures to the tetrapod cortex, the amygdala, and the hippocampus (Mueller et al., 2011; Ganz et al., 2014).
Taken together, the presented findings confirm grm8a and grm8b expression in the CNS of developing zebrafish (Supplementary Table 2). Besides, temporal, and spatial discrepancies in the expression patterns suggest that grm8a and grm8b may take over common and distinct functional roles during (neuro-) development.
Adult Expression of Grm8a and Grm8b in the Zebrafish Brain
The previously distinct expression pattern for grm8a or grm8b become increasingly similar with further development into adulthood. This can be best illustrated in the pallium, the thalamus, and the posterior tuberculum. Distinct expression domains, observed during early embryonic development (Figures 1A,B and Supplementary Figures 1A–H, 2A–H) align partially by 72 hpf (Supplementary Figures 1I,J, 2I,J) and fully when reaching adulthood (Figures 2, 3). At adult stage, grm8a and grm8b transcripts are both detected along the ventricular side of the pallium (P, Figures 2A–E, 3A–D), in the ventral (VT, Figures 2E, 3D,E) and dorsal thalamus (DT, Figures 2F,G, 3F,G) and the periventricular nucleus of the posterior tuberculum (TPp, Figures 2F,G, 3F,G) and the area comprising the posterior tubercular nucleus or the paraventricular organ (PTN/PVO, Figures 2F–H, 3F–H).
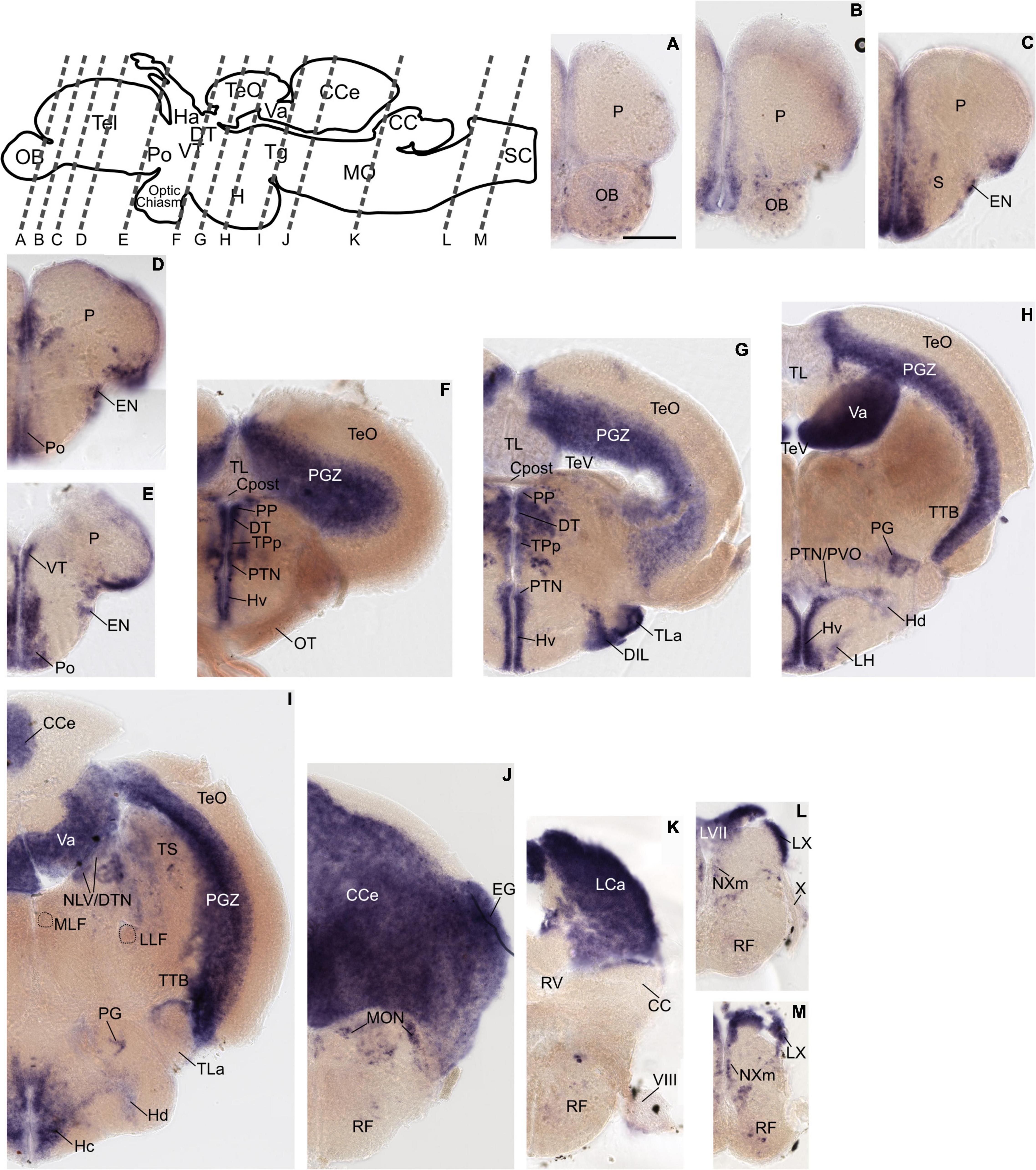
Figure 2. Grm8a expression pattern in adult zebrafish brain labeled by RNA in situ hybridization. (A–M) Grm8a transcript labeling by RNA in situ hybridization on cross-sections of an adult zebrafish brain. Images are arranged from anterior to posterior as indicated by the scheme at the top. Details on grm8a expression pattern are described in the main text. Anatomical abbreviations are listed in Table 1. Scale bar is 200 μm.
Figure 3. Grm8b expression pattern in adult zebrafish brain labeled by RNA in situ hybridization. (A–L) Grm8b transcript labeling by RNA in situ hybridization on cross-sections of an adult zebrafish brain. Images are arranged from anterior to posterior as indicated by the scheme at the top. Details on grm8b expression pattern are described in the main text. Anatomical abbreviations are listed in Table 1. Scale bar is 200 μm.
Expression domains with similar expression of grm8a and grm8b in the early developmental phase maintain the expression similarity of both paralogs till adulthood. This applies for instance to expression in the subpallium (S) revealed in the central, ventral, dorsal, and entopeduncular nuclei (EN, Figures 2C–E, 3B–D), the preoptic region (Po, Figures 2D,E, 3D), the hypothalamus with expression in the diffuse nucleus of the inferior lobe (DIL, Figures 2G, 3G), the lateral hypothalamic nucleus (LH, Figures 2H, 3H) and the ventral, dorsal and caudal zone of the periventricular hypothalamus (Hv, Hd, Hc, Figures 2F–I, 3E–J), the tegmentum [with expression in the area encompassing the lateral valvulae and dorsal tegmental nuclei (NLV/DTN, Figures 2I, 3I)] and the optic tectum [with expression in the periventricular gray zone (PGZ, Figures 2F–I, 3F–J)].
Not only expression similarities are maintained into adulthood, discrepancies are too. For instance, the less pronounced grm8b expression detected in the developing cerebellum is still observed in the mature brain [with expression in the corpus cerebelli (CCe, Figures 2I–K, 3H–K), the valvular cerebelli (Va, Figures 2H,I, 3G–I), the medial octavolateralis nucleus (MON, Figures 2J, 3K) and the eminentia granularis (EG, Figures 2J, 3K)]. Similarly, the weak grm8b expression is detectable in the developing and mature pretectum [periventricular pretectal nucleus (PP, Figures 2F,G, 3F,G)]. Notably, other regions in the mature brain with exclusive expression of grm8a such as the reticular formation (RF), the vagal lobe (LX), and the torus semicularis (TS) (Figures 2I–M) were demonstrated to be connected to the pretectum and/or cerebellum (Kaslin and Brand, 2016; Kramer et al., 2019). The restricted distribution of grm8b transcripts observed in the developing MO is maintained into adulthood as well. While the reticular formation (RF, Figures 2J–M), the vagal motor nucleus (NXm, Figures 2L,M) and the vagal (LX, Figures 2L,M) and facial lobe (LVII, Figure 2L) all express grm8a, expression of grm8b was only detected in the facial (LVII, Figure 2L) and the glossopharyngeal lobe (LIX, Figure 3L).
Other brain structures with comparably weak expression of grm8b include nuclei of the posterior tuberculum such as the torus lateralis (TLa, Figures 2G, 3G) and the preglomerular nuclei (PG, Figures 2H,I, 3G). While labeling of grm8a transcripts appear generally stronger and broader at 72 hpf (Supplementary Figures 1I,J,O–S) and in the adult brain (Figure 2), the opposite is true for transcript expression in the olfactory bulb throughout development (OB, Figure 3A).
Grm8a Is Partly Expressed by a Subset of Gad1a-Positive Cells
Grm8, like other type III metabotropic glutamate receptors, is involved in the regulation of neurotransmitter release of various neuronal cell types. Therefore, it is crucial to determine the transmitter identity of Grm8-expressing cells in order to assess the effect of Grm8 loss-of-function on different transmitter systems.
Comparing to our previous findings we noted that the developmental distribution of grm8a resembles the one of gad1a and gad1b at 36 hpf (Lüffe et al., 2021), especially in the medulla oblongata (MO, Figures 4A,B). Hence, we hypothesized that grm8a expression might correlate with the distribution of GABAergic markers as seen for other species and brain regions before (Ferraguti et al., 2005). Therefore, we decided to investigate the distribution of gad1a, a well-known GABAergic marker in relation to grm8a expression. To confirm colocalization in single cells we used double RNA ISH for gad1a (red) and grm8a (blue) at 36 hpf on whole-mounts and cryosections. Due to the largely similar pattern of gad1a and gad1b expression (Lüffe et al., 2021), the technical benefits and the broader expression of grm8a compared to grm8b (Figures 1A,B, 2, 3), we decided to assess the colocalization of grm8a and gad1a expression.
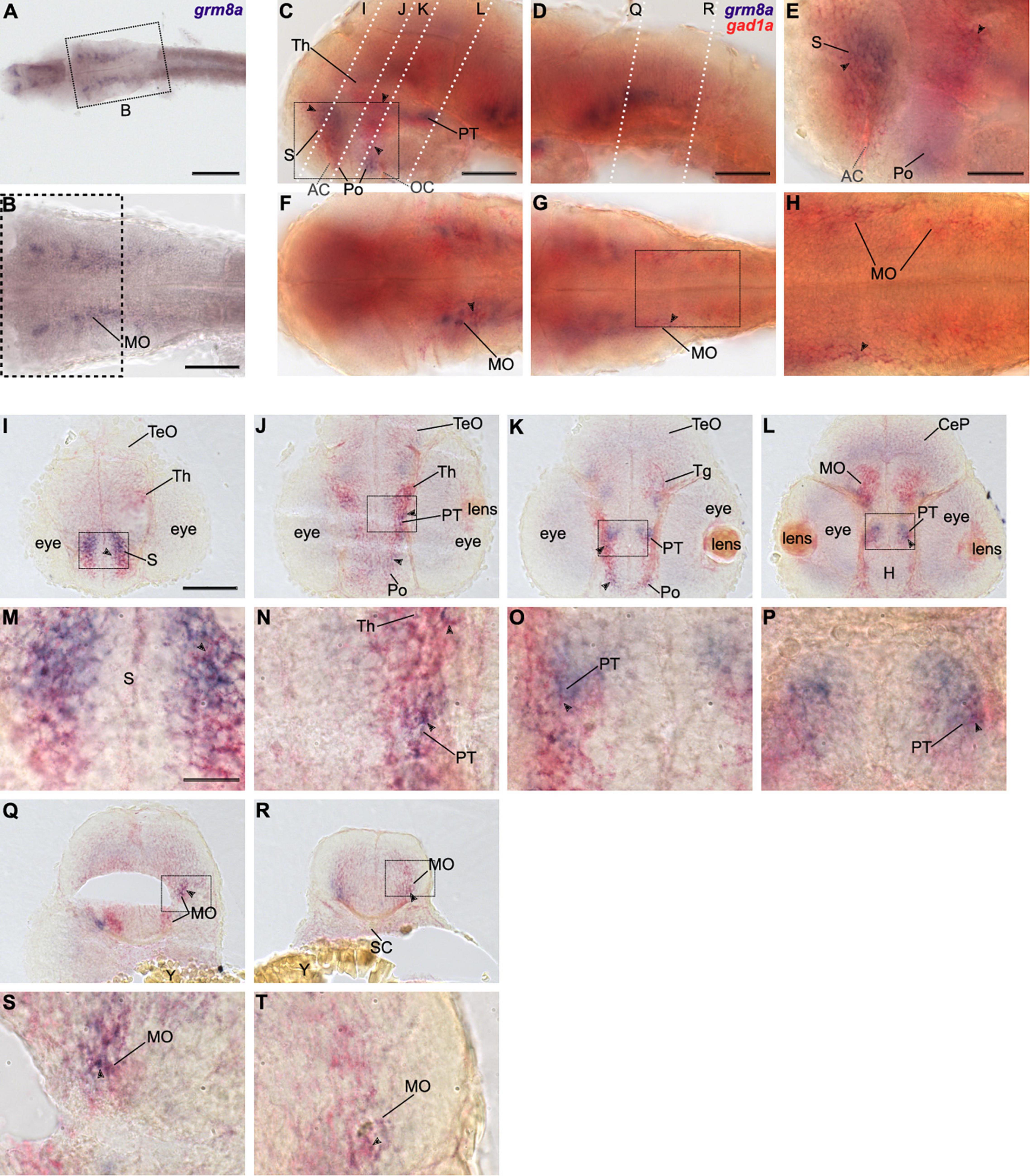
Figure 4. Grm8a is partly expressed in gad1a positive neurons in the developing zebrafish brain. (A) Whole-mount RNA in situ hybridization of grm8a expression at 36 hpf. (B) Magnified picture of the region boxed in (A). Scale bar, 200 μm (overview) and 100 μm (magnification). (C–T) Double labeling of grm8a (blue) and gad1a (red) expression in 36 hpf old embryos using two-color RNA in situ hybridization. Lateral (C,D) and dorsal (F,G) overview of embryonic CNS with anterior to the left. (E,H) Magnifications of boxed areas in (C,G). Dashed lines in (C,D) indicate cutting sites for cross sections shown in (I–R). (M–P,S,T) Magnifications of boxed areas in (I–L,Q,R), respectively. Arrowheads indicate sites of co-localization. Abbreviations are listed in Table 1. Scale bars: 100 μm (overview), 50 μm (magnified images).
Colocalization between grm8a and gad1a expression was detected throughout development in the subpallium (S, Figures 4C,E,I,M), the thalamus (Th, Figures 4C,I,J,N) and the medulla oblongata (MO, Figures 4F–H,L,Q–T). Further, the posterior tuberculum (PT, Figures 4C,J–L,N–P) and the preoptic region (Po, Figures 4C,E,J,K) showed an overlap of both expression patterns at 36 and 48 hpf. To further compare grm8a expression with other neuronal markers we used the Tg(Etvmat2:GFP) reporter line, which labels monoaminergic neurons (Wen et al., 2008). Interestingly, while grm8a and gad1a expression overlap extensively across several brain regions, the colocalization of grm8a-positive punctae and vmat2-expressing cells was restricted to only a few cells of the rostral/intermediate and caudal hypothalamus and the raphe nuclei (Supplementary Figure 3). Taken together, prominent colocalization of grm8a with gad1a was observed with high incidence in previously mentioned brain regions associated with motor functions, including the subpallium, the posterior tuberculum, the thalamus, the cerebellum, and the medulla oblongata.
Generation of Grm8a and Grm8b Knock-Out Lines Using CRISPR/Cas9
Based on the expression of grm8a and grm8b transcripts in the brain of developing and mature zebrafish we hypothesized that both paralogs may have functional roles in the CNS of zebrafish throughout development. Until now, insights into Grm8 function exclusively originate from a few studies in mammals (Linden et al., 2002, 2003; Duvoisin et al., 2005; Schmid and Fendt, 2006; Woo et al., 2021) whereas functional data on Grm8 in zebrafish is missing. We therefore decided to generate both grm8a and grm8b knockout lines using CRISPR/Cas9.
Several CRISPR/Cas9 mutant lines were generated and one for each paralog was screened for morphological and behavioral phenotypes. The line for grm8a, which was chosen to be characterized further, contains a 17 bp deletion in grm8a exon 4 (Figure 5A and Supplementary Figure 4A). For grm8b, a line with a 13 bp deletion in exon 2 was selected (Figure 5C and Supplementary Figure 4D). In both cases, the CRISPR/Cas9-induced deletion mutation induces a frameshift and premature stop codon, leading to a termination of translation upstream (grm8b) or at the beginning of (grm8a) the ligand-binding domain in the very N-terminal region of the respective proteins potentially generating protein fragments of 217 and 71 aa, respectively (Figures 5B,D). The seven transmembrane and other functional domains are located more downstream of the target sites and are therefore missing in the truncated gene products (Figures 5B,D) making it unlikely that residual Grm8 activity is generated from the mutant alleles. We therefore consider these lines as potential null alleles.
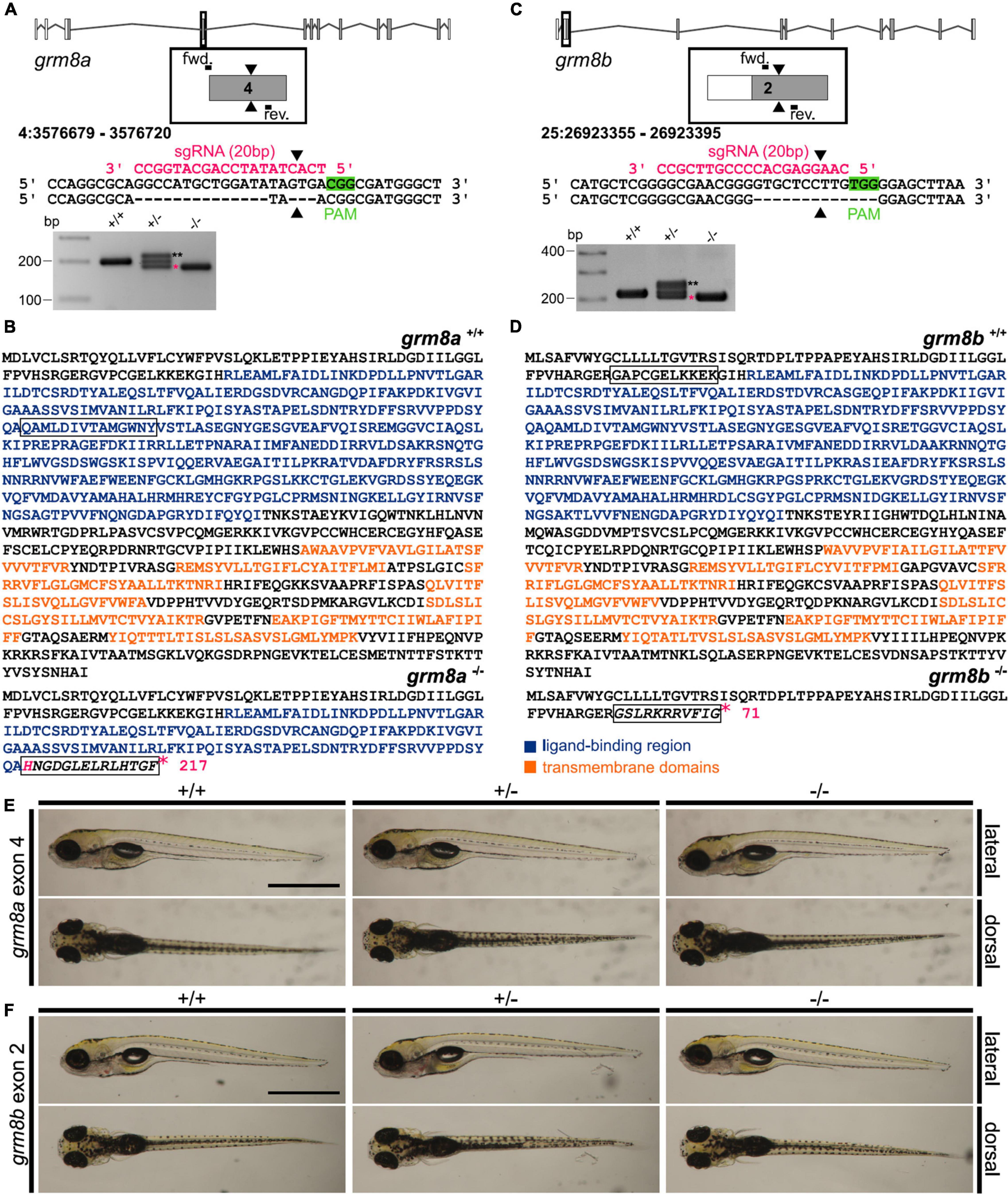
Figure 5. Generation of grm8a and grm8b knock-out lines using CRISPR/Cas9. (A) Grm8a exon-intron structure (top) with coding (gray) and non-coding exons (white). The sgRNA (pink) targeting grm8a exon 4 and the primer binding sites for genotyping PCR are displayed. The CRISPR/Cas9-induced strand break (black arrowheads) caused a 17 bp deletion represented by a smaller PCR product (pink asterisk) in grm8a± and grm8a–/– animals (bottom). The third band of ∼220 bp corresponds to a heterodimer of wildtype and mutated PCR product (black asterisks). (B) The predicted amino acid sequence of the wildtype (top) and mutated allele (bottom) show that the frameshifted sequence (black box) is interrupted by a premature stop codon (pink asterisk) in the ligand binding domain. (C) Grm8b exon-intron structure (top) with coding (gray) and non-coding exons (white). The sgRNA (pink) targeting grm8b exon 2 and the primer binding sites for genotyping are indicated. The CRISPR/Cas9-induced strand break (black arrowheads) caused a 13 bp deletion represented by a smaller PCR product (pink asterisk) in grm8b± and grm8b–/– animals (bottom). The third band of ∼250 bp corresponds to a heterodimer of wildtype and mutated PCR product (black asterisks). (D) The predicted amino acid sequence of the wildtype (top) and mutated allele (bottom) shows that the frameshifted sequence (black box) is interrupted by a premature stop codon (pink asterisk) in the N-terminal domain. (E) Dorsal (top) and ventral (bottom) views of 5 dpf old grm8a+/+, grm8a± and grm8a–/– zebrafish. Anterior is to the left. (F) Dorsal (top) and ventral (bottom) views of 5 dpf old grm8b+/+, grm8b± and grm8b–/– zebrafish. Scale bar, 1 mm.
We found that the gross morphology of grm8a (Figure 5E) and grm8b (Figure 5F) mutants is almost normal. Size measurements detect no significant alterations in head size, yolk diameter and body length of mutant embryos (Supplementary Figures 4B,E) except for a slight but not significant reduction in body length of grm8b mutants (Supplementary Figure 4E). Likewise, anti-cleaved Caspase 3 staining reveal no evidence for differences in cell apoptosis between mutants and wildtype siblings (Supplementary Figures 4C,F). In addition, there are no indications for early lethality in either of the mutant lines.
Next, we quantified the expression of both grm8 transcripts in mutant animals. Whereas in grm8a mutants an allele-specific loss of grm8a transcript level was detected, indicating a probable non-sense-mediated decay, the grm8b transcript levels were unchanged, except for a slight, but not significant increase in grm8a–/– animals using the second primer pair located more downstream of the transcript in comparison to the first primer pair (Supplementary Figure 5A). However, in grm8b–/– animals a significant increase of grm8b expression using the second primer pair was detected, while grm8a transcript levels were unchanged (Supplementary Figure 5B). Due to the partial colocalization of grm8a with gad1a transcripts detected before (Figure 4), we decided to quantify expression levels of gad1a, gad1b and gad2 in both mutant lines. In grm8a–/– animals a significant reduction of gad1a transcript level was detected, whereas gad1b and gad2 were unchanged (Supplementary Figure 5A). In grm8b mutant animals no differences in gad1a, gad1b and gad2 expression levels were found (Supplementary Figure 5B).
In conclusion, the overall wildtype-like appearance of grm8a and grm8b CRISPR/Cas9 mutants suggests that each of the Grm8 paralogs are not essential for cell survival, anatomical development, and survival in general.
Grm8a and Grm8b Mutants Show Differential Locomotor and Thigmotaxis Phenotypes
In order to investigate the behavioral consequences of grm8a and grm8b loss-of-function we performed locomotor tracking at 5 dpf as described previously (Lüffe et al., 2021). Briefly, we placed larvae in a 12-well dish and recorded locomotor activity in a commercial setup for 5 min after 5 min of habituation. Activity levels were analyzed for the following parameters: mean velocity (low, high and total), total distance traveled, duration (inactive, low and high activity) and number of events counted for inactive, low and high activity for +/+, +/– and –/– animals (Supplementary Table 3). For grm8a, significant differences in mean velocity were detected in mutant animals (Figure 6A), they exhibit a hypoactive phenotype characterized by a reduced “high” velocity and a general reduction in total mean velocity. A similar trend was observed for total distance swum and the duration and number of “high active” swimming events albeit without reaching statistical significance (Supplementary Table 3). In grm8b mutant animals, no differences in locomotor activity were detected (Figure 6B). Analysis of thigmotaxis behavior which is the tendency of animals to stay close to the walls of the wells and is usually considered to be associated with anxiety-like behavior were not altered for grm8a mutants compared to wildtype animals (Figure 6C). However, the grm8b mutants spend significantly more time in the outer ring compared to wildtypes suggesting an anxiety-like phenotype (Figures 6B,C). This initial characterization of grm8a and grm8b mutant behavior in zebrafish larvae points toward hypo-locomotion in grm8a mutants and an increase in fear-response in grm8b mutants, demonstrating that these mutant lines can be used to further characterize Grm8 function and elucidate its potential contribution to behavioral phenotypes with relevance for psychiatric disorders.
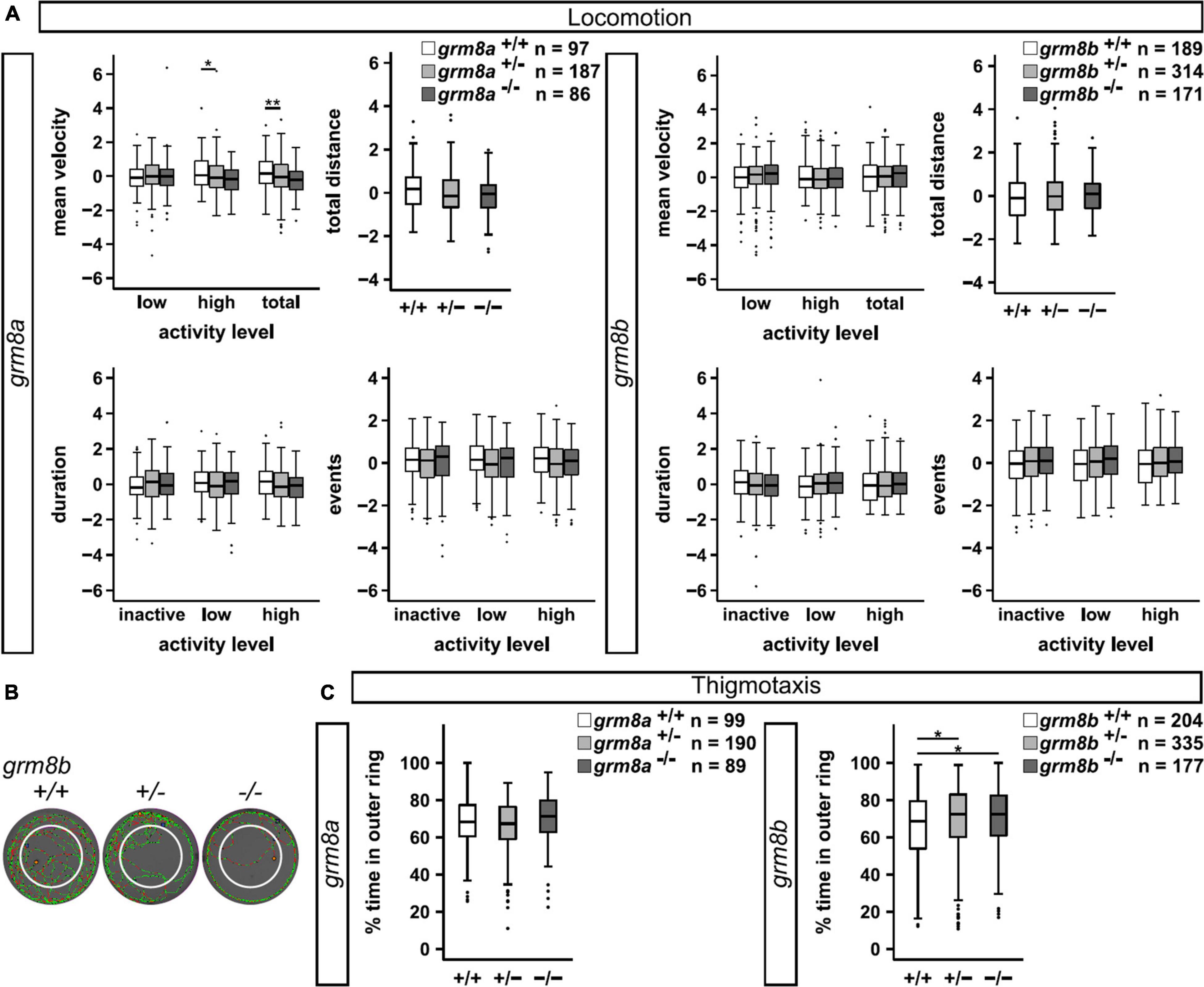
Figure 6. Behavioral characterization of grm8a and grm8b larvae. (A) Locomotor activity of grm8a (left panel) and grm8b mutant animals (right panel) were analyzed for mean velocity (top left) in low or high activity, or combined (total), total distance swum (top right), duration (bottom left) and events (bottom right) of inactivity, low and high activity. Raw data was standardized using z-score transformation. *P < 0.05, **P < 0.01. (B) Example traces of grm8b+/+, grm8b+/–, and grm8b–/– animals analyzed for thigmotaxis behavior. Note the reduced movement of mutant animals in the inner zone. Green represents slow, red fast movement. (C) Analysis of thigmotaxis behavior in grm8a (left panel) and grm8b (right panel) mutant animals. Time spent in the outer ring was analyzed for all genotypes and raw data standardized using z-score transformation. *P < 0.05.
Discussion
The current knowledge about Grm8 function in psychiatric disorders is scarce because the field lacks validated models in different species to address open questions. The present work provides such a model, describing the stepwise generation, validation and initial behavioral characterization of two paralog-specific mutant lines, on which future studies can now build. Further, with the presented colocalization of grm8a and gad1a expression, in particular in motor-related brain regions, the data highlights the GABAergic system as an interesting target for future investigations on the molecular link between grm8a/grm8b disruption and behavioral phenotypes implicated in neurodevelopmental and psychiatric conditions involving altered motor functions.
To study the role of Grm8 in neuronal circuits and its contribution to behavioral alterations in zebrafish, it is imperative to understand the spatial and temporal expression dynamics of both paralogs. We used whole-mount RNA ISH to identify spatial differences and similarities in the expression pattern of both genes. Since we are particularly interested in the potential role during neural development, we used early and late embryonic stages as well as adult brain tissue to monitor temporal changes in expression. Our data is in line with and complements the earlier study of Haug et al. (2013) in which embryonic expression was already demonstrated. Here, we provide a more detailed description to cover additional developmental stages and also include the expression pattern in the adult brain, which was not described before. Further, our research reveals a mutual expression in developing and adult brain regions involved in motor functions such as the subpallium, the posterior tuberculum, the thalamus, the cerebellum, and the medulla oblongata (with reticular formation, vagal and facial lobe). These results complement previous studies in zebrafish and other vertebrate species (Duvoisin et al., 1995; Saugstad et al., 1997; Shigemoto et al., 1997; Messenger et al., 2002; Haug et al., 2013; Ribeiro et al., 2017). Interestingly, we detected expression differences between grm8a and grm8b especially in the adult brain suggesting partial sub-functionalization of these two genes during evolution (Postlethwait et al., 2004). In contrast to grm8b, grm8a is strongly expressed in the reticular formation, the vagal nucleus, vagal and facial lobe. On the other hand, grm8b transcript is restricted to the facial and glossopharyngeal lobe, while a consistently stronger labeling of grm8b over grm8a can be exclusively found in the olfactory bulb. Thus, both grm8a and grm8b expression patterns will help to interpret phenotypic differences in grm8a and grm8b mutants in the future.
As the expression patterns for grm8a and grm8b partly resemble the distribution of GABAergic cells detected in previous studies (Lechermeier et al., 2019, 2020; Lüffe et al., 2021), we decided to use gad1a as a marker to prove the GABAergic identity of grm8a positive cells. We detected a partial colocalization of both transcripts at larval stages in brain regions associated with motor functions, including the subpallium, the posterior tuberculum, the thalamus, the cerebellum, and the medulla oblongata. There is evidence from the literature that indeed some GABAergic cells may express Grm8 protein. For instance, in mice and rat hippocampus it was shown that a subset of mGluR8a positive cells were of GABAergic origin and that presynaptic expression of mGluR8 in GABAergic boutons may modulate its inhibition depending on glutamatergic activity in this network (Ferraguti et al., 2005; Katona et al., 2020). Therefore, further functional experiments are needed to demonstrate that Grm8 is involved in the regulation of GABAergic activity also in zebrafish. In addition, our study has investigated the localization of grm8a and gad1a transcripts only. Subsequent research will have to include the paralogs as well.
In order to generate loss-of-function models we have chosen the guide RNA target sites to be located very early in the coding sequences of both genes. Even if translation would occur from the mutant transcripts, these fragments lack for instance the transmembrane regions so that both proteins would not reach their proper destination in the cell, would not get post translational modifications as needed and would not locate on the cell surface to act as a glutamate receptor. We therefore consider both lines to be functional null alleles. Due to the lack of proper antibodies recognizing zebrafish Grm8, we are not able to demonstrate the definite loss of protein expression in the mutant animals. Considering the sequencing results and expression differences detected with qPCR and the behavioral phenotypes we are confident that the mutants are indeed loss-of-function models.
The fact that both mutant lines are viable and do not display any gross morphological abnormalities or increased cell apoptosis indicates that, due to the similar expression pattern, the remaining grm8-gene might compensate for the loss of the other and therefore may prevent harsher phenotypes. This was partly reflected in our qPCR experiments where we observed a small, but not significant upregulation of grm8b transcript in grm8a–/– animals. Further, upregulation of grm8b transcripts was detected in grm8b–/– animals too, indicating a potential compensation for the loss of functional Grm8b protein in this mutant. Similar experiments with double mutant animals (grm8a–/–; grm8b–/–) in comparison to single mutants are needed to investigate this phenomenon further. In addition, other type III-mGluRs like mGluR4 or mGluR7 should be considered as potential compensators in future expression analyses due to the substantial similarities in their amino acid sequence and overlapping expression domains (Haug et al., 2013). The significant reduction of gad1a transcripts in grm8a–/– animals suggests that the GABAergic system might be affected by the Grm8a loss. The mechanistic background of this observation is currently unknown and needs to be addressed in future experiments.
To characterize the behavioral consequences of grm8a or grm8b loss, we performed an initial basic locomotor assay in the mutant lines. We detected a significant reduction in velocity in grm8a mutant larvae, indicating a hypoactivity phenotype. Hypoactivity was seen before in one mGluR8-knockout mouse model (Duvoisin et al., 2005), whereas other mouse models display no change in activity or hyperactivity phenotypes (Gerlai et al., 2002; Linden et al., 2002; Robbins et al., 2007; Raber and Duvoisin, 2015). The grm8a model could be useful in investigating motor related phenotypes further and resolving this conflicting data. Further unraveling the underlying mechanism(s) of this phenotype is of particular interest with respect to the observed grm8a and gad1a expression colocalization in brain regions involved in motor functions. The grm8b mutant animals did not show signs of motor activity alterations. Instead, they display a mild thigmotaxis phenotype. Thigmotaxis is considered to be an indicator for anxiety-like behavior, a phenotype which is not visible in grm8a mutants. Anxiety-like phenotypes were demonstrated for instance in a pharmacological model, in which mice were treated with the mGluR8-specific agonist (S)-3,4-dicarboxyphenylglycine. This induced decreased alcohol intake and produced an anxiolytic-like effect in the light-dark-box and open field assays. These effects were reversed with the type III-mGluR-specific antagonist (S)-2-amino-2-methyl-4- phosphonobutyric (Bahi, 2017), indicating that modulating mGluR8 activity with pharmacotherapy might be relevant in alcohol dependent disorders and anxiety. Also, the combination of mGluR8 with mGluR4 loss-of-function models could deliver stronger phenotypes and give deeper insights into disease mechanisms associated with type III-mGluRs (Raber and Duvoisin, 2015). In addition, it supports the proposed investigation of grm4 transcript levels in grm8a and grm8b mutant lines within future research efforts. Altered fear responses are a frequent observation in mGluR8 knockout mice (Linden et al., 2002; Duvoisin et al., 2005; Robbins et al., 2007) and are correlated with the Grm8 deficiency in the amygdala (Schmid and Fendt, 2006; Fendt et al., 2013). Interestingly, grm8a and predominantly grm8b expression was detected in the zebrafish ventral/lateral pallium, which is assumed to comprise the homologous structure to the mammalian amygdala (Mueller et al., 2011; Ganz et al., 2014), opening the possibility that fear-related behavior in zebrafish larvae might be associated with grm8a/grm8b expression in the pallium. Therefore, our zebrafish grm8b model may be useful in elucidating the mechanism behind anxiety-like behavior associated with reduced Grm8 activity.
Conclusion
Taken together, the two here newly developed animal models of Grm8 loss-of-function may be useful for the investigation of the mechanistic backgrounds of behavioral phenotypes with relevance to psychiatric disorders. Further research on double mutants and the characterization of adult animals are now the next logic steps. Several psychiatric disorders, including neurodevelopmental disorders, are linked to cellular processes that are thought to disturb the equilibrium of excitation and inhibition in the nervous system of patients, known as E/I imbalance (Canitano and Pallagrosi, 2017; Lopatina et al., 2019; Mamiya et al., 2021). This phenomenon might also be experimentally accessible in our loss-of-function lines, especially since we revealed expression of grm8a in GABAergic cell types. We anticipate that our two novel metabotropic glutamate receptor 8 zebrafish models may contribute to a deeper understanding of its function in development and disease.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Regierung von Unterfranken (RUF 55.2.2 2532 2 622 10).
Author Contributions
TL performed the experiments. ZG, MB, DÖ, CD, and CL contributed to experimental support. MR, CL, and CD contributed to conception and design of the study and supervised the project. TL and CD wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was funded by the Interdisciplinary Center for Clinical Research (IZKF) of the University of Würzburg, Germany (project number N-320, and Z-4-149), the Verein zur Durchführung Neurowissenschaftlicher Tagungen e.V and the Faculty of Biology, University of Würzburg. This publication was supported by the Open Access Publication Fund of the University of Würzburg.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Manfred Schartl, Eva Klopocki and Thomas Haaf for generously providing laboratory and fish facility space. We would also like to thank Zuzana Fouskova for excellent technical assistance and Christine Krempl and the CoreUnit for Confocal Microscopy at the Institute for Virology and Immune Biology for access to and support with confocal imaging. TL was a member of the Graduate School of Life Sciences (GSLS) at the University of Würzburg. This publication uses data collected within the framework of the Ph.D. thesis “Behavioral and pharmacological validation of genetic zebrafish models for ADHD” of Teresa Lüffe published in 2021 at the Graduate School of Life Sciences, Julius-Maximilians Universität Würzburg, Section: Neuroscience (Lüffe, 2021).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.901309/full#supplementary-material
Footnotes
References
Ahrens, M. B., Li, J. M., Orger, M. B., Robson, D. N., Schier, A. F., Engert, F., et al. (2012). Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485, 471–477. doi: 10.1038/nature11057
Akutagava-Martins, G. C., Salatino-Oliveira, A., Genro, J. P., Contini, V., Polanczyk, G., Zeni, C., et al. (2014). Glutamatergic copy number variants and their role in attention-deficit/hyperactivity disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 165, 502–509. doi: 10.1002/ajmg.b.32253
Bahi, A. (2017). Decreased anxiety, voluntary ethanol intake and ethanol-induced CPP acquisition following activation of the metabotropic glutamate receptor 8 “mGluR8”. Pharmacol. Biochem. Behav. 155, 32–42. doi: 10.1016/j.pbb.2017.03.004
Bauer, L. O., and Covault, J. M. (2020). GRM8 genotype is associated with externalizing disorders and greater inter-trial variability in brain activation during a response inhibition task. Clin. Neurophysiol. 131, 1180–1186. doi: 10.1016/j.clinph.2020.02.031
Bodzęta, A., Scheefhals, N., and MacGillavry, H. D. (2021). Membrane trafficking and positioning of mGluRs at presynaptic and postsynaptic sites of excitatory synapses. Neuropharmacology 200:108799. doi: 10.1016/j.neuropharm.2021.108799
Canitano, R., and Pallagrosi, M. (2017). Autism spectrum disorders and schizophrenia spectrum disorders: excitation/inhibition imbalance and developmental trajectories. Front. Psychiatry 8:69. doi: 10.3389/fpsyt.2017.00069
Davis, M. J., Duvoisin, R. M., and Raber, J. (2013). Related functions of mGlu4 and mGlu8. Pharmacol. Biochem. Behav. 111, 11–16. doi: 10.1016/j.pbb.2013.07.022
Duvoisin, R. M., Pfankuch, T., Wilson, J. M., Grabell, J., Chhajlani, V., Brown, D. G., et al. (2010a). Acute pharmacological modulation of mGluR8 reduces measures of anxiety. Behav. Brain Res. 212, 168–173. doi: 10.1016/j.bbr.2010.04.006
Duvoisin, R. M., Villasana, L., Pfankuch, T., and Raber, J. (2010b). Sex-dependent cognitive phenotype of mice lacking mGluR8. Behav. Brain Res. 209, 21–26. doi: 10.1016/j.bbr.2010.01.006
Duvoisin, R. M., Zhang, C., and Ramonell, K. (1995). A novel metabotropic glutamate receptor expressed in the retina and olfactory bulb. J. Neurosci. 15, 3075–3083. doi: 10.1523/jneurosci.15-04-03075.1995
Duvoisin, R. M., Zhang, C., Pfankuch, T. F., O’Connor, H., Gayet-Primo, J., Quraishi, S., et al. (2005). Increased measures of anxiety and weight gain in mice lacking the group III metabotropic glutamate receptor mGluR8. Eur. J. Neurosci. 22, 425–436. doi: 10.1111/j.1460-9568.2005.04210.x
Elia, J., Glessner, J. T., Wang, K., Takahashi, N., Shtir, C. J., Hadley, D., et al. (2012). Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat. Genet. 44, 78–84. doi: 10.1038/ng.1013
Faul, F., Erdfelder, E., Buchner, A., and Lang, A. G. (2009). Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160. doi: 10.3758/brm.41.4.1149
Faul, F., Erdfelder, E., Lang, A. G., and Buchner, A. (2007). G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191. doi: 10.3758/bf03193146
Fendt, M., Bürki, H., Imobersteg, S., van der Putten, H., McAllister, K., Leslie, J. C., et al. (2010). The effect of mGlu8 deficiency in animal models of psychiatric diseases. Genes Brain Behav. 9, 33–44. doi: 10.1111/j.1601-183X.2009.00532.x
Fendt, M., Imobersteg, S., Peterlik, D., Chaperon, F., Mattes, C., Wittmann, C., et al. (2013). Differential roles of mGlu(7) and mGlu(8) in amygdala-dependent behavior and physiology. Neuropharmacology 72, 215–223. doi: 10.1016/j.neuropharm.2013.04.052
Ferraguti, F., Klausberger, T., Cobden, P., Baude, A., Roberts, J. D., Szucs, P., et al. (2005). Metabotropic glutamate receptor 8-expressing nerve terminals target subsets of GABAergic neurons in the hippocampus. J. Neurosci. 25, 10520–10536. doi: 10.1523/jneurosci.2547-05.2005
Ganz, J., Kroehne, V., Freudenreich, D., Machate, A., Geffarth, M., Braasch, I., et al. (2014). Subdivisions of the adult zebrafish pallium based on molecular marker analysis. F1000Res 3:308. doi: 10.12688/f1000research.5595.2
Gerlai, R., Adams, B., Fitch, T., Chaney, S., and Baez, M. (2002). Performance deficits of mGluR8 knockout mice in learning tasks: the effects of null mutation and the background genotype. Neuropharmacology 43, 235–249. doi: 10.1016/s0028-3908(02)00078-3
Glasauer, S. M., and Neuhauss, S. C. (2014). Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289, 1045–1060. doi: 10.1007/s00438-014-0889-2
Haug, M. F., Gesemann, M., Mueller, T., and Neuhauss, S. C. (2013). Phylogeny and expression divergence of metabotropic glutamate receptor genes in the brain of zebrafish (Danio rerio). J. Comp. Neurol. 521, 1533–1560. doi: 10.1002/cne.23240
Hawi, Z., Cummins, T. D., Tong, J., Johnson, B., Lau, R., Samarrai, W., et al. (2015). The molecular genetic architecture of attention deficit hyperactivity disorder. Mol. Psychiatry 20, 289–297. doi: 10.1038/mp.2014.183
Howard, D. M., Adams, M. J., Clarke, T.-K., Hafferty, J. D., Gibson, J., Shirali, M., et al. (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352. doi: 10.1038/s41593-018-0326-7
Jha, U., and Thirumalai, V. (2020). Neuromodulatory selection of motor neuron recruitment patterns in a visuomotor behavior increases speed. Curr. Biol. 30, 788–801. doi: 10.1016/j.cub.2019.12.064
Katona, L., Hartwich, K., Tomioka, R., Somogyi, J., Roberts, J. D. B., Wagner, K., et al. (2020). Synaptic organisation and behaviour-dependent activity of mGluR8a-innervated GABAergic trilaminar cells projecting from the hippocampus to the subiculum. Brain Struct. Funct. 225, 705–734. doi: 10.1007/s00429-020-02029-2
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kramer, A., Wu, Y., Baier, H., and Kubo, F. (2019). Neuronal architecture of a visual center that processes optic flow. Neuron 103, 118–132. doi: 10.1016/j.neuron.2019.04.018
Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174. doi: 10.1093/nar/gkz365
Lechermeier, C. G., D’Orazio, A., Romanos, M., Lillesaar, C., and Drepper, C. (2020). Distribution of transcripts of the GFOD gene family members gfod1 and gfod2 in the zebrafish central nervous system. Gene. Expr. Patt. 2020:119111. doi: 10.1016/j.gep.2020.119111
Lechermeier, C. G., Zimmer, F., Luffe, T. M., Lesch, K. P., Romanos, M., Lillesaar, C., et al. (2019). Transcript Analysis of Zebrafish GLUT3 Genes, slc2a3a and slc2a3b, define overlapping as well as distinct expression domains in the zebrafish (danio rerio) central nervous system. Front. Mol. Neurosci. 12:199. doi: 10.3389/fnmol.2019.00199
Li, W., Ju, K., Li, Z., He, K., Chen, J., Wang, Q., et al. (2016). Significant association of GRM7 and GRM8 genes with schizophrenia and major depressive disorder in the Han Chinese population. Eur. Neuropsychopharmacol. 26, 136–146. doi: 10.1016/j.euroneuro.2015.05.004
Linden, A. M., Baez, M., Bergeron, M., and Schoepp, D. D. (2003). Increased c-Fos expression in the centromedial nucleus of the thalamus in metabotropic glutamate 8 receptor knockout mice following the elevated plus maze test. Neuroscience 121, 167–178. doi: 10.1016/s0306-4522(03)00393-2
Linden, A. M., Johnson, B. G., Peters, S. C., Shannon, H. E., Tian, M., Wang, Y., et al. (2002). Increased anxiety-related behavior in mice deficient for metabotropic glutamate 8 (mGlu8) receptor. Neuropharmacology 43, 251–259. doi: 10.1016/s0028-3908(02)00079-5
Liu, Y., Qu, H. Q., Chang, X., Nguyen, K., Qu, J., Tian, L., et al. (2021). Deep learning prediction of attention-deficit hyperactivity disorder in African Americans by copy number variation. Exp. Biol. Med. 246, 2317–2323. doi: 10.1177/15353702211018970
Long, E. C., Aliev, F., Wang, J. C., Edenberg, H. J., Nurnberger, J. Jr., Hesselbrock, V., et al. (2015). Further analyses of genetic association between GRM8 and alcohol dependence symptoms among young adults. J. Stud. Alcohol Drugs 76, 414–418. doi: 10.15288/jsad.2015.76.414
Lopatina, O. L., Malinovskaya, N. A., Komleva, Y. K., Gorina, Y. V., Shuvaev, A. N., Olovyannikova, R. Y., et al. (2019). Excitation/inhibition imbalance and impaired neurogenesis in neurodevelopmental and neurodegenerative disorders. Rev. Neurosci. 30, 807–820. doi: 10.1515/revneuro-2019-0014
Lüffe, T. M. (2021). Behavioral and pharmacological validation of genetic zebrafish models for ADHD. Würzburg: Julius-Maximilians Universität Würzburg.
Lüffe, T. M., D’Orazio, A., Bauer, M., Gioga, Z., Schoeffler, V., Lesch, K. P., et al. (2021). Increased locomotor activity via regulation of GABAergic signalling in foxp2 mutant zebrafish-implications for neurodevelopmental disorders. Transl. Psychiatry 11:529. doi: 10.1038/s41398-021-01651-w
Mamiya, P. C., Arnett, A. B., and Stein, M. A. (2021). Precision Medicine Care in ADHD: the case for neural excitation and inhibition. Brain Sci. 11:91. doi: 10.3390/brainsci11010091
Martin, S. C., Heinrich, G., and Sandell, J. H. (1998). Sequence and expression of glutamic acid decarboxylase isoforms in the developing zebrafish. J. Comp. Neurol. 396, 253–266.
McCullock, T. W., and Kammermeier, P. J. (2021). The evidence for and consequences of metabotropic glutamate receptor heterodimerization. Neuropharmacology 199:108801. doi: 10.1016/j.neuropharm.2021.108801
Messenger, M. J., Dawson, L. G., and Duty, S. (2002). Changes in metabotropic glutamate receptor 1-8 gene expression in the rodent basal ganglia motor loop following lesion of the nigrostriatal tract. Neuropharmacology 43, 261–271. doi: 10.1016/s0028-3908(02)00090-4
Meyer, A., and Schartl, M. (1999). Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11, 699–704. doi: 10.1016/s0955-0674(99)00039-3
Mueller, T., Dong, Z., Berberoglu, M. A., and Guo, S. (2011). The dorsal pallium in zebrafish, Danio rerio (Cyprinidae, Teleostei). Brain Res. 1381, 95–105. doi: 10.1016/j.brainres.2010.12.089
Naumann, E. A., Fitzgerald, J. E., Dunn, T. W., Rihel, J., Sompolinsky, H., and Engert, F. (2016). From whole-brain data to functional circuit models: the zebrafish optomotor response. Cell 167, 947–960. doi: 10.1016/j.cell.2016.10.019
Niswender, C. M., and Conn, P. J. (2010). Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322. doi: 10.1146/annurev.pharmtox.011008.145533
Pin, J.-P., and Bettler, B. (2016). Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68. doi: 10.1038/nature20566
Postlethwait, J., Amores, A., Cresko, W., Singer, A., and Yan, Y. L. (2004). Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20, 481–490. doi: 10.1016/j.tig.2004.08.001
Preibisch, S., Saalfeld, S., and Tomancak, P. (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465. doi: 10.1093/bioinformatics/btp184
Raber, J., and Duvoisin, R. M. (2015). Novel metabotropic glutamate receptor 4 and glutamate receptor 8 therapeutics for the treatment of anxiety. Expert. Opin. Invest. Drugs 24, 519–528. doi: 10.1517/13543784.2014.986264
Rennekamp, A. J., and Peterson, R. T. (2015). 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 24, 58–70. doi: 10.1016/j.cbpa.2014.10.025
Ribeiro, F. M., Vieira, L. B., Pires, R. G., Olmo, R. P., and Ferguson, S. S. (2017). Metabotropic glutamate receptors and neurodegenerative diseases. Pharmacol. Res. 115, 179–191. doi: 10.1016/j.phrs.2016.11.013
Robbins, M. J., Starr, K. R., Honey, A., Soffin, E. M., Rourke, C., Jones, G. A., et al. (2007). Evaluation of the mGlu8 receptor as a putative therapeutic target in schizophrenia. Brain Res. 1152, 215–227. doi: 10.1016/j.brainres.2007.03.028
Sangu, N., Shimojima, K., Takahashi, Y., Ohashi, T., Tohyama, J., and Yamamoto, T. (2017). A 7q31.33q32.1 microdeletion including LRRC4 and GRM8 is associated with severe intellectual disability and characteristics of autism. Hum. Genome Var. 4:17001. doi: 10.1038/hgv.2017.1
Saugstad, J. A., Kinzie, J. M., Shinohara, M. M., Segerson, T. P., and Westbrook, G. L. (1997). Cloning and expression of rat metabotropic glutamate receptor 8 reveals a distinct pharmacological profile. Mol. Pharmacol. 51, 119–125. doi: 10.1124/mol.51.1.119
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schmid, S., and Fendt, M. (2006). Effects of the mGluR8 agonist (S)-3,4-DCPG in the lateral amygdala on acquisition/expression of fear-potentiated startle, synaptic transmission, and plasticity. Neuropharmacology 50, 154–164. doi: 10.1016/j.neuropharm.2005.08.002
Shigemoto, R., Kinoshita, A., Wada, E., Nomura, S., Ohishi, H., Takada, M., et al. (1997). Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J. Neurosci. 17, 7503–7522. doi: 10.1523/jneurosci.17-19-07503.1997
Stewart, A. M., Braubach, O., Spitsbergen, J., Gerlai, R., and Kalueff, A. V. (2014). Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 37, 264–278. doi: 10.1016/j.tins.2014.02.011
Stewart, A. M., Ullmann, J. F., Norton, W. H., Parker, M. O., Brennan, C. H., Gerlai, R., et al. (2015). Molecular psychiatry of zebrafish. Mol. Psychiatry 20, 2–17. doi: 10.1038/mp.2014.128
Tavakkoly-Bazzaz, J., Azarnezhad, A., Mousavi, N., Salehipour, P., Shahsavand Ananloo, E., and Alizadeh, F. (2018). TCF4 and GRM8 gene polymorphisms and risk of schizophrenia in an Iranian population: a case-control study. Mol. Biol. Rep. 45, 2403–2409. doi: 10.1007/s11033-018-4406-2
Terracciano, A., Tanaka, T., Sutin, A. R., Sanna, S., Deiana, B., Lai, S., et al. (2010). Genome-wide association scan of trait depression. Biol. Psychiatry 68, 811–817. doi: 10.1016/j.biopsych.2010.06.030
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Torchiano, M. (2016). Effsize - a package for efficient effect size computation. Genève: Zenodo. doi: 10.5281/ZENODO.1480624
Wen, L., Wei, W., Gu, W., Huang, P., Ren, X., Zhang, Z., et al. (2008). Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev. Biol. 314, 84–92. doi: 10.1016/j.ydbio.2007.11.012
Keywords: nervous system, brain disorders, psychiatric disorders, brain development, excitatory/inhibitory imbalance, metabotropic glutamate (mGlu) receptor
Citation: Lüffe TM, Bauer M, Gioga Z, Özbay D, Romanos M, Lillesaar C and Drepper C (2022) Loss-of-Function Models of the Metabotropic Glutamate Receptor Genes Grm8a and Grm8b Display Distinct Behavioral Phenotypes in Zebrafish Larvae (Danio rerio). Front. Mol. Neurosci. 15:901309. doi: 10.3389/fnmol.2022.901309
Received: 21 March 2022; Accepted: 10 May 2022;
Published: 13 June 2022.
Edited by:
Wietske van der Ent, University of Oslo, NorwayReviewed by:
Ruey-Kuang Cheng, Institute of Molecular and Cell Biology (A*STAR), SingaporeNicolas Gervasi, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright © 2022 Lüffe, Bauer, Gioga, Özbay, Romanos, Lillesaar and Drepper. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carsten Drepper, ZHJlcHBlcl9jQHVrdy5kZQ==
 Teresa M. Lüffe
Teresa M. Lüffe Moritz Bauer
Moritz Bauer Zoi Gioga
Zoi Gioga Duru Özbay
Duru Özbay Marcel Romanos
Marcel Romanos Christina Lillesaar
Christina Lillesaar Carsten Drepper
Carsten Drepper