
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci. , 22 June 2022
Sec. Molecular Signalling and Pathways
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.898717
This article is part of the Research Topic Targeting Ion Channels for Drug Discovery: Emerging Challenges for High Throughput Screening Technologies View all 11 articles
 Kadla R. Rosholm1*
Kadla R. Rosholm1* Beatrice Badone1
Beatrice Badone1 Stefania Karatsiompani1
Stefania Karatsiompani1 David Nagy2
David Nagy2 Fitzwilliam Seibertz3,4
Fitzwilliam Seibertz3,4 Niels Voigt3,4,5
Niels Voigt3,4,5 Damian C. Bell1
Damian C. Bell1In the Hollywood blockbuster “The Curious Case of Benjamin Button” a fantastical fable unfolds of a man’s life that travels through time reversing the aging process; as the tale progresses, the frail old man becomes a vigorous, vivacious young man, then man becomes boy and boy becomes baby. The reality of cellular time travel, however, is far more wondrous: we now have the ability to both reverse and then forward time on mature cells. Four proteins were found to rewind the molecular clock of adult cells back to their embryonic, “blank canvas” pluripotent stem cell state, allowing these pluripotent stem cells to then be differentiated to fast forward their molecular clocks to the desired adult specialist cell types. These four proteins – the “Yamanaka factors” – form critical elements of this cellular time travel, which deservedly won Shinya Yamanaka the Nobel Prize for his lab’s work discovering them. Human induced pluripotent stem cells (hiPSCs) hold much promise in our understanding of physiology and medicine. They encapsulate the signaling pathways of the desired cell types, such as cardiomyocytes or neurons, and thus act as model cells for defining the critical ion channel activity in healthy and disease states. Since hiPSCs can be derived from any patient, highly specific, personalized (or stratified) physiology, and/or pathophysiology can be defined, leading to exciting developments in personalized medicines and interventions. As such, hiPSC married with high throughput automated patch clamp (APC) ion channel recording platforms provide a foundation for significant physiological, medical and drug discovery advances. This review aims to summarize the current state of affairs of hiPSC and APC: the background and recent advances made; and the pros, cons and challenges of these technologies. Whilst the authors have yet to finalize a fully functional time traveling machine, they will endeavor to provide plausible future projections on where hiPSC and APC are likely to carry us. One future projection the authors are confident in making is the increasing necessity and adoption of these technologies in the discovery of the next blockbuster, this time a life-enhancing ion channel drug, not a fantastical movie.
When Takahashi and Yamanaka discovered four genes (c-Myc, Oct3/4, Sox2, and Klf4), whose protein products were shown to reprogram (induce) somatic cells into pluripotent stem cells (Takahashi and Yamanaka, 2006), these Yamanaka factors were heralded as a breakthrough in biomedical research (Press release, 2012). For the first time, pluripotency, the ability to form a range of specialized cell types from a pluripotent cell, was possible from mature, adult somatic cells, no longer requiring embryonic stem cells (Shi et al., 2017; Yoshida and Yamanaka, 2017).
The limited availability of embryonic tissue to derive embryonic stem cells coupled with legal and ethical regulations, always stymied stem cell research development and progress (Lo and Parham, 2009; Jha, 2011; Moradi et al., 2019). With the discovery of the Yamanaka factors, somatic cells could be taken from adult tissue and reprogrammed, turning back the clock on mature cells to an embryonic-like, pluripotent state. The processes where pluripotent stem cells (PSCs) are differentiated and matured, allows fast-forwarding of cells into specialized cell lines providing model cells for in vitro research (Rowe and Daley, 2019).
The use of human induced pluripotent stem cells (hiPSCs) has a number of advantages: it reduces the use of animal tissue; compared to cells overexpressing recombinant protein [e.g., Chinese hamster ovary (CHO)] human cells are more physiologically relevant and translatable models; in comparison with primary cells, such as tissue dissociated cardiomyocytes and neurons, they are easier to access (typically from a blood or skin sample) and to maintain in culture; hiPSCs can be derived from both healthy and patient populations, providing both human “wild type” control and diseased models; and, since hiPSCs can be derived from any of us, the options to study a personal genotype and phenotype are broad, allowing for truly personalized (also termed precision or stratified) medicines and interventions (Matsa et al., 2016; Hamazaki et al., 2017; Doss and Sachinidis, 2019). This technology, however, has limitations worth considering, such as: the fetal nature of hiPSCs; the lifespan in culture; the long-term maturation process (for details on the challenges and limitations see the following section “Maturity and Subtype Specificity of hiPSC-Derived Cell Lines”).
Whilst these seminal developments in generating and differentiating hiPSCs were taking place, similar strides were being made in ion channel recording techniques. The dazzlingly brilliant but highly time consuming and technically challenging manual patch clamp (MPC) technique (Hamill et al., 1981) was simplified, made faster and more efficient, through a combination of planar patch clamp and automation (Bell and Dallas, 2018). By switching from manual micro-manipulation of a single recording electrode down onto an adherent cell in MPC, to the application of cell suspensions by robot pipettors to arrays of electrodes in automated patch clamp (APC), the technical hurdles were lowered whilst vastly increasing throughput of ion channel recordings.
Thus, with hiPSCs and APC, researchers now have the capabilities to characterize ion channel populations in hiPSCs and their differentiated cell lines, for instance exploring the effect of different reprogramming, differentiation and maturation conditions and introducing quantifiable metrics for quality assessment. Furthermore, as the hiPSC technology develops, APC will be applicable for safety pharmacology and drug screens in highly translatable hiPSC model cell lines from healthy and disease patient populations.
In this review, the authors aim to describe advances in the generation of mature hiPSC-derived cell lines; developments in cell suspension preparation for APC measurements; APC techniques and technologies that have made these once challenging ion channel recordings more successful, physiologically relevant, and amenable to high throughput screening; and the key challenges solved thus far and those to be faced. Finally, we provide examples of novel applications of hiPSCs and APC. These techniques offer huge potential to advance biomedical research and drug discovery, and the future they project looks to be bright and filled with many novel and exciting findings.
During the last decade, substantial effort has gone into developing protocols for generating specialized hiPSC-derived cells and tissues for applications within a range of areas, such as blood, smooth muscle, liver, heart, and brain (Bellin et al., 2012; Shi et al., 2017; Rowe and Daley, 2019).
In general, the formation of different specialized hiPSC derived cells follow the same sequence of events: (1) the reprogramming (induction) of human unipotent somatic cells into hiPSCs by directed expression of certain transcriptional factors; and, (2) the differentiation and maturation of hiPSCs into specialized cells, typically by using signaling pathway agonists and antagonists in specific concentrations to induce stepwise progression through the embryonic development stages (Bellin et al., 2012; Figure 1).
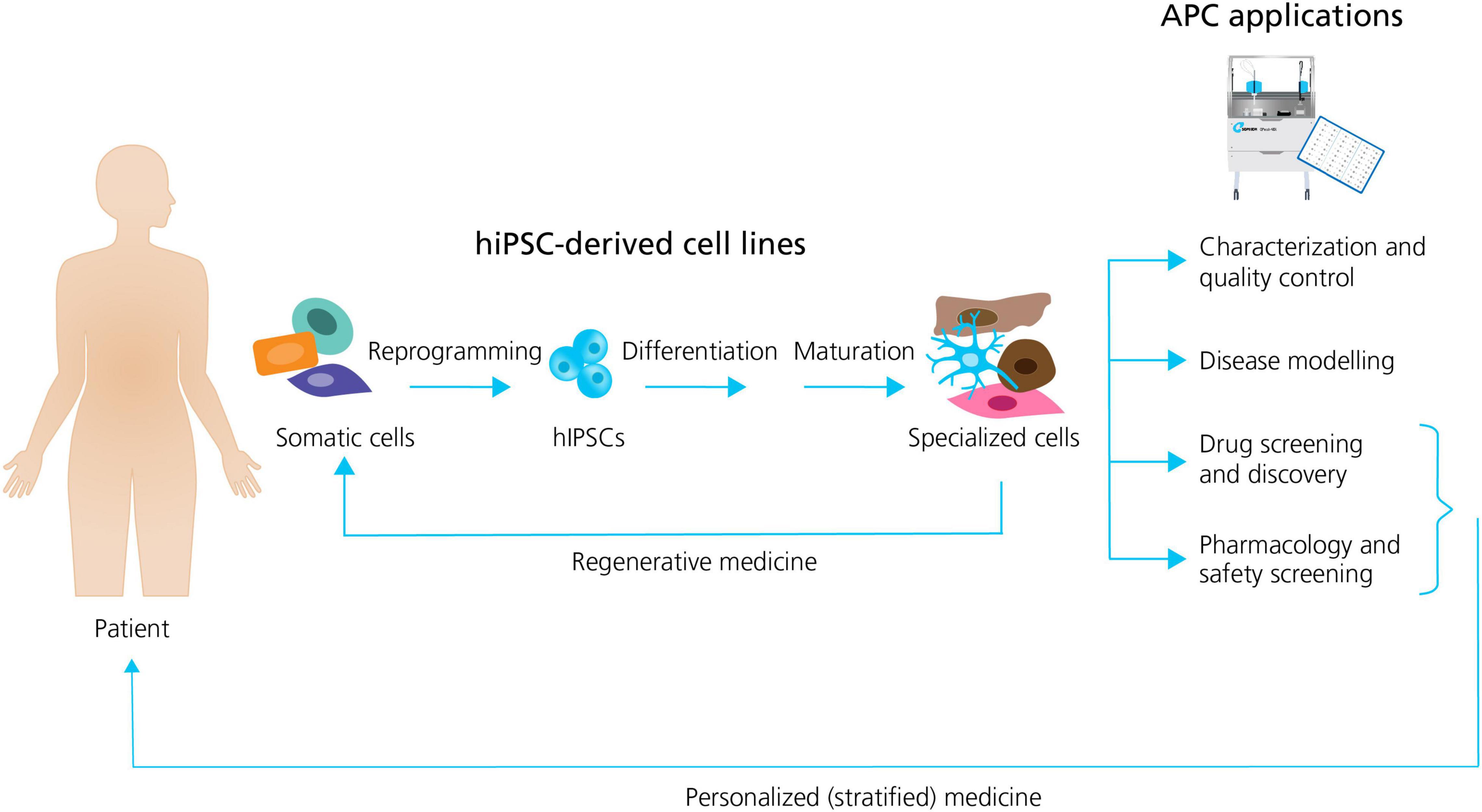
Figure 1. Generation of specialized hiPSC-derived cells and APC applications. Somatic patient cells can be reprogrammed into hiPSCs, followed by the differentiation and maturation into specialized cells. The specialized cells can be used directly in regenerative medicine, or they can be employed together with APC for a range of applications including: characterization and quality control; disease modeling; drug screening and discovery; and pharmacology and safety screening. Patient hiPSCs can be used as personalized drug screening and pharmacology models in trials. Adapted from Bellin et al. (2012), with permission from publisher Springer Nature.
In this section we aim to summarize the latest methodological advances of hiPSC generation and differentiation and discuss the key challenges in the field.
A challenge with hiPSC-derived cell lines is the genetic variability existing within and between hiPSC populations, and an ongoing task is to control and maintain genome integrity during reprogramming and differentiation (Assou et al., 2018; Popp et al., 2018). An important development in this regard was the introduction of non-integrating methods, such as Sendai virus (Fusaki et al., 2009), for introducing reprogramming factors, which aim to reduce the risk of variability or spurious gene activation/inactivation in hiPSC-derived cell lines (Shi et al., 2017; Berry et al., 2019).
Another focus is the inherent biological variability between donors stemming from their individual genetic and epigenetic background (Berry et al., 2019; Rowe and Daley, 2019), which has been estimated to be responsible for up to 46% of phenotype variability and also influences differentiation efficiency (Kilpinen et al., 2017). A strategy for circumventing inter-donor variability is to apply gene-editing technologies, such as CRISPR/Cas9 (Jehuda et al., 2018) to generate isogenic controls, which allow the investigation of disease-causing mutations with control cells having the same genetic background (Yoshida and Yamanaka, 2017).
A further key challenge is to improve the maturity and subtype specificity of hiPSC-derived cell lines. Despite improved protocols for producing specific somatic cell types and subtypes, the vast majority of hiPSC-directed differentiations produce populations that most closely resemble fetal or neonatal cells (Berry et al., 2019), including the extensively studied hiPSC-derived cardiomyocytes (hiPSC-CMs; Kolanowski et al., 2017; Di Baldassarre et al., 2018; Ahmed et al., 2020), and hiPSC-derived neurons (hiPSC-neurons; Casalia et al., 2021; de Leeuw et al., 2021; Giacomelli et al., 2022). In both cases there is a further complexity in creating pure subtype populations, for example atrial or ventricular hiPSC-CMs (Cyganek et al., 2018; Feyen et al., 2020) or defining the desired hiPSC-neuron subtype (e.g., sensory, motor, or dopaminergic neurons; Davis-Dusenbery et al., 2014; Hartfield et al., 2014; Young et al., 2014).
Strategies to improve cell maturation during differentiation are many (see Figure 2) and include: the addition of specific chemical cues during differentiation (Burridge et al., 2012; Yoshida and Yamanaka, 2017; Ahmed et al., 2020); replacing the energy source, for instance the addition of fatty acids as a substitute for glucose in the culturing media, has been shown to improve hiPSC-CM maturation by activating the required metabolic pathways (Sharma et al., 2015; Yoshida and Yamanaka, 2017; Yang et al., 2019); prolonged time in culture which does, however, also increase the risk of contamination and mutation accumulation (Blokzijl et al., 2016; Odawara et al., 2016; Breckwoldt et al., 2017; Yoshida and Yamanaka, 2017; Biendarra-Tiegs et al., 2019; Ahmed et al., 2020); electrical and mechanical stimulation (pacing) have been reported as valid methods to improve the maturity and function of hiPSC-CMs and -neurons (Ahmed et al., 2020; Giacomelli et al., 2022); and finally modulating the chemical and spatial microenvironment of the differentiating cell culture, for example changing the adherence matrix (Feaster et al., 2015; Depalma et al., 2021), cell culture geometry, e.g., using 3D culture (Lemoine et al., 2017; Tiburcy et al., 2017; Di Baldassarre et al., 2018; de Leeuw et al., 2021) or preparing co-cultures with supportive cells (Odawara et al., 2016; Andersen et al., 2020; Zafeiriou et al., 2020).
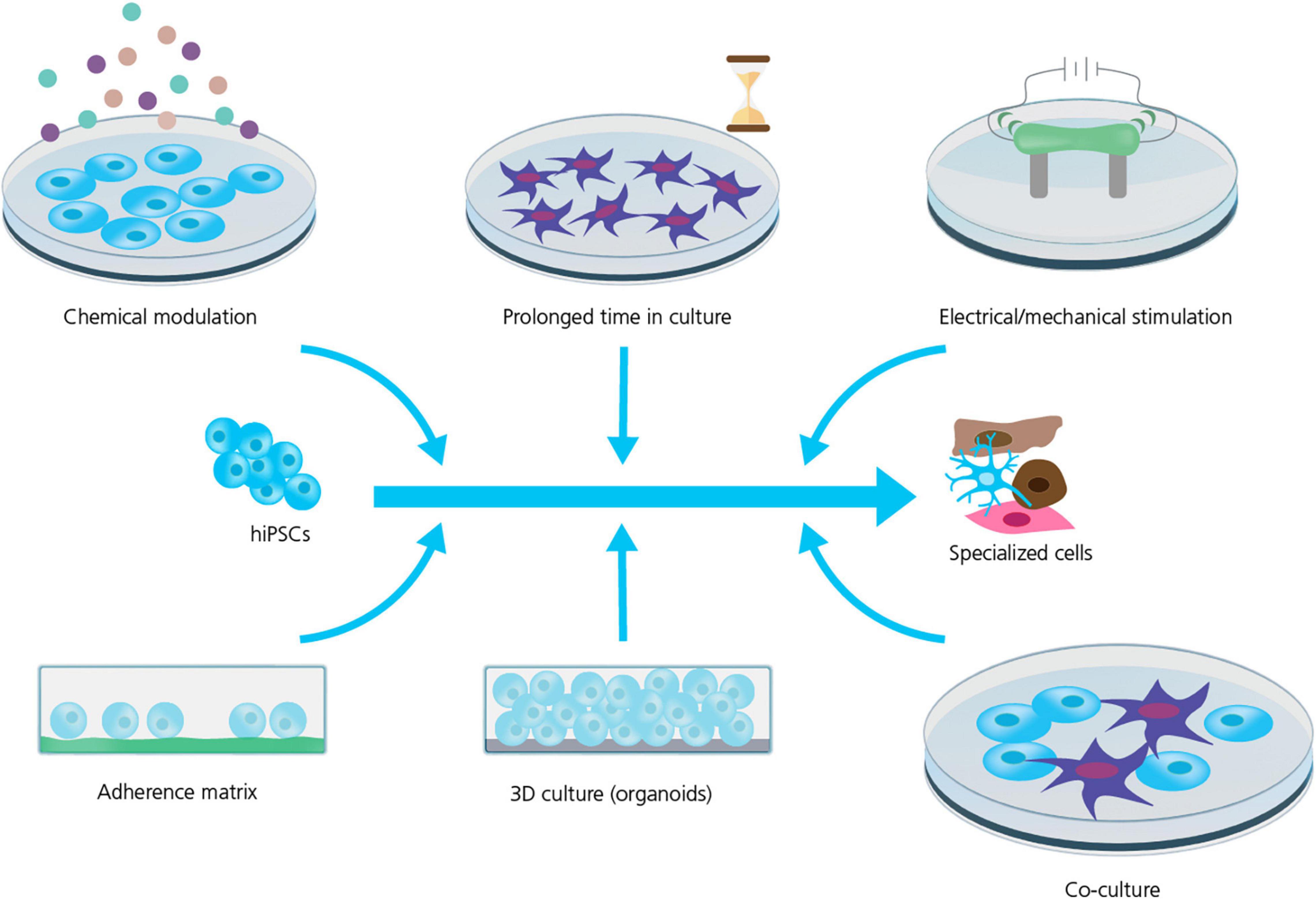
Figure 2. Strategies for improving hiPSC maturity. Several strategies are used to improve the maturity and phenotype specificity of hiPSC-derived cell lines. However, the mechanism and efficiency of these strategies are still being explored and requires rigorous characterization and quality control. Adapted from Ahmed et al. (2020), with permission under Creative Commons Attribution License (CC BY).
Cell reprogramming, differentiation and maturation are manipulations that lead to many intracellular changes that can affect the lifespan of cells. Indeed, a somatic cell obtained from a patient which is “rejuvenated” into a stem-cell like phenotype, shows intracellular signatures such as an active telomerase activity and immature mitochondria, as well as a decrease in senescence markers [e.g., senescence marker p21, (Strässler et al., 2018)]. After differentiation, although the cellular aging process may restart, certain improved characteristics remain compared to the parent cell (e.g., longer telomeres).
Due to these intracellular changes, additional challenges arise when referring to how the age of patients could influence the functionality and maturity of hiPSC-derived models and whether it is feasible to apply this technology in age-related diseases. There is also an increasing need to perform safety pharmacology in “aged” senescent cells to functionally model and translate the complex physiology, pathophysiology and pharmacology of the aging, senior population, particularly in light of the growth of this demographic (Fermini and Bell, 2022).
In addressing these challenges, there is a need to define quality standards during hiPSC reprogramming and differentiation, as well as rigorous and statistically sound characterization of the final product (Engle et al., 2018). The specific composition of ion channels is an important marker of cell maturity and subtype specificity, for example in hiPSC-and -neurons (Jonsson et al., 2012; Bardy et al., 2016; Goversen et al., 2018b), rendering APC important as a high-throughput (HT) characterization method providing ion channel maturation metrics. Thus, we envision molecular and genetic approaches in combination with APC electrophysiological characterization will generate authentic, translatable and quantifiable hiPSC models for disease investigation and drug-screening.
Ion channels represent attractive targets in hiPSC-derived cell lines, making electrophysiological studies of hiPSC-derived cell lines important not only for characterization (as mentioned above), but also for advances in biomedical research including drug discovery and development. The HT nature of APC that enables recordings of large datasets within a few days or even hours, makes this method ideal for applications such as: quantification of inter-patient or inter-batch variability; identification of cell sub-populations within the same batch, based on their ion channel content; and HT drug screening.
In this section we summarize the latest advances and challenges toward making such APC measurements possible.
Electrophysiological methods, such as MPC, have been extensively employed for characterization of hiPSC-derived cell lines over the years (Chen et al., 2014; Jones et al., 2014; Lemoine et al., 2017; Giannetti et al., 2021). However, the inherently labor intensive and low throughput nature of MPC, lead electrophysiologists to consider more time and resource efficient alternatives. Consequently, increasing efforts have gone into transferring MPC experimentation and measurements to APC systems (Haythornthwaite et al., 2012; Franz et al., 2017; Li et al., 2019; Jung et al., 2022). A key element that allows this transfer from MPC to APC is the provision of unicellular cell suspensions (see Figure 3). The quality (homogenous, unicellular populations) and health (minimal enzymatic and mechanical membrane damage and cytotoxicity) of cells in suspension are critical to achieving high success recording rates on APC. Using dissociated cell suspensions literally turns the ion channel current recording format on its head: in MPC the recording electrode is moved to an adherent cell, contrasting with APC where dissociated, single cells in suspension are moved by robotic pippetors to arrays of wells containing fixed, planar recording electrodes.
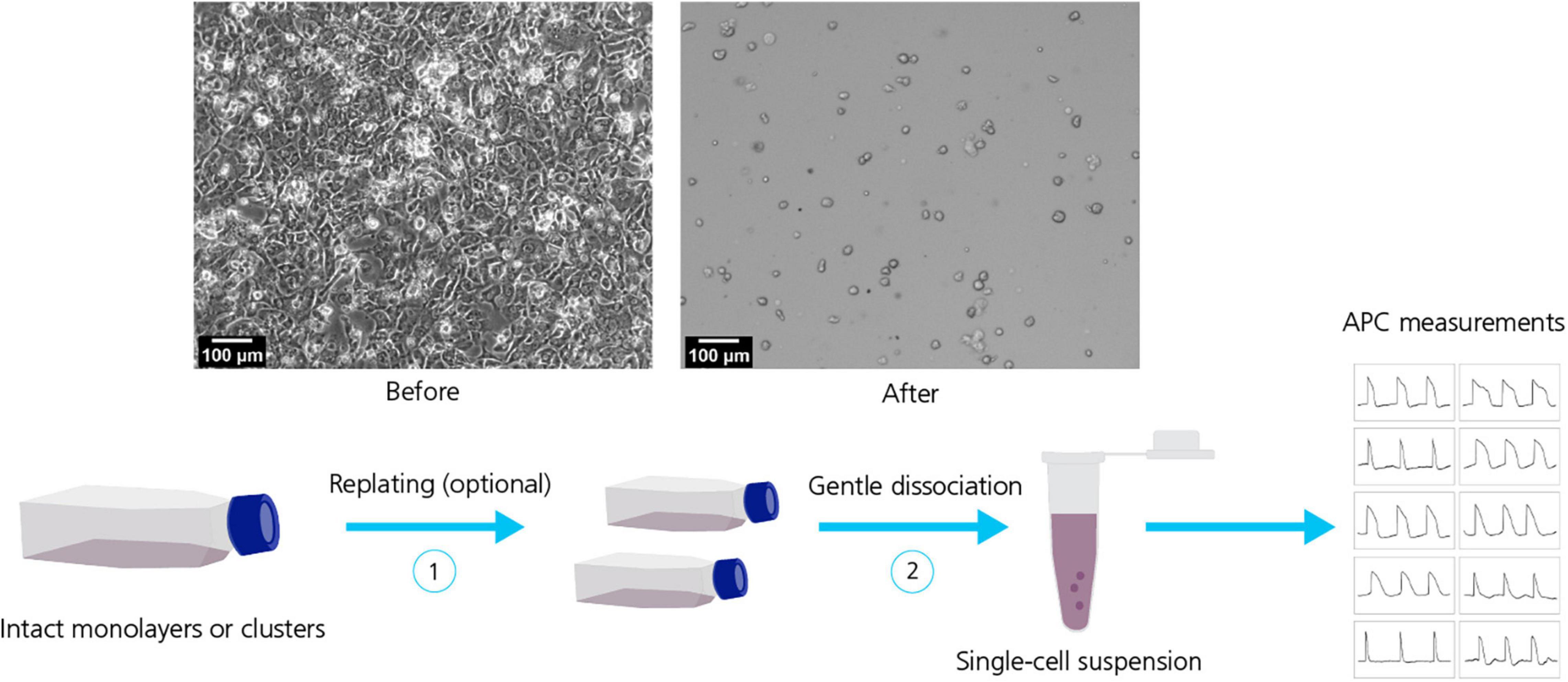
Figure 3. Dissociation of hiPSC-derived cell lines. Top row: Micrographs of hiPSC-CMs before (left) and after (right) dissociation. Bottom row: Dissociation of hiPSC-derived cell lines typically include a sequence of steps for example replating (1) followed by gentle dissociation methods (2) to obtain a homogeneous, healthy single-cell suspension suitable for APC measurements. Adapted from Rajamohan et al. (2016), with permission under Creative Commons Attributions License (CC BY).
Initial with hiPSC-derived cell lines focused on APC assay development, including cell-handling on the machine; intra- and extra-cellular recording solutions; and, the suction protocols employed to position the cell and rupture the membrane, the so-called whole-cell conformation, to allow electrical access to the interior of the cell. However, progress has been slow, with only minor improvements in experiment success rate and reproducibility. It has become increasingly evident that working with hiPSC-derived cell lines demands a greater understanding of the cell population than working with clonal, homogenous immortalized cell lines. This includes the process of generating hiPSC-derived cell lines (as discussed in the section above), characterization and quality control of the final product and specialized dissociation methods for generating purer, more homogenous cell suspensions.
Whilst measurements on hiPSC-derived cell lines have progressed slowly, the technological development of APC features have advanced at a faster pace. This includes: the exchange of external and internal solutions; fast compound addition and wash out; perforated patch methods (Rosholm et al., 2021a); current clamp and dynamic clamp recordings, e.g., enabling measurements of action potentials (Becker et al., 2020; Rosholm, 2021); and temperature control for physiologically relevant temperature experimentation; all in a high throughput fashion that provides the statistical power necessary for drawing accurate and correct conclusions (Bell and Dallas, 2018; Bell and Fermini, 2021).
One major difference of APC compared to other methods typically used for hiPSC measurements, such as MPC, multielectrode-array (Odawara et al., 2016) or fluorescent plate readers (Sube and Ertel, 2017; Kilfoil et al., 2021), is that APC measures single cells that have been dissociated from 2D or 3D tissue culture. Whilst 2D/3D multi-cell or tissue recordings provide information on tissue/population behavior (including, e.g., morphology changes and network signaling), single cell APC recordings have increased temporal resolution providing crucial information regarding inter-cell variability in, e.g., compound sensitivity, different ion channel expression levels and biological activity. It is imperative to be aware of and understand which questions can be answered using dissociated cells, because several important properties such as cell morphology and ion channel expression might change due to cell handling and dissociation from culture flasks (Raman and Bean, 1997). To fill this potential information gap that can arise in cell dissociated suspensions used in APC, a complementary technology would be single cell dye-based imaging of adherent cell populations (e.g., see Vala Sciences), though this, too, has its limitations: the dye(s) used can introduce new, non-biological variables to the imaging measurements and is an indirect, proxy measure of the ion channel activity.
The preferred harvest protocol will depend on the cell type, quality and culturing days and will likely evolve continuously (also, see Figure 3). Dissociating hiPSC-derived cell lines into single-cell suspensions is often challenging (especially CMs and neurons), due to the formation of tight cellular “tissue”-like structures and networks that must be disrupted. In general, the longer the cells have been cultured, the tighter the connections become, increasing the possibility of damage during cell harvest. For APC it is of utmost importance to work with a healthy, homogenous cell suspension that is free of debris. Improvements involve: using gentle dissociating enzymes (e.g., TrypLE™, Gibco, United States; Accutase™, Stemcell Technologies, United States; or Papain, Sigma-Aldrich, United States); cold-harvest and/or incubation; addition of protective chemicals to ensure membrane integrity (e.g., ROCK inhibitor, blebbistatin) and prevent stickiness (DNase); replating for 24 h – 1 week; filtering to remove aggregates and possibly positive selection based on surface markers (Rajamohan et al., 2016; Obergrussberger et al., 2017; Li et al., 2021b). Finally, the use of fluoride containing seal enhancing solutions on many APC platforms can be problematic for some measurements, e.g., cardiac action potentials, that rely on physiologically accurate calcium kinetics. Consequently, where possible, these measurements should be performed on systems using physiological recording solutions (Mann et al., 2019; Rosholm, 2021).
Even with the aforementioned challenges, the field is now rapidly advancing and with the existing cell quality and suspension preparation methods researchers have made significant progress in exploring and developing the possibilities of measuring hiPSC-derived cell lines using APC (Rajamohan et al., 2016; Mann et al., 2019; Potet et al., 2020; Toh et al., 2020; Li et al., 2021a).
Originally hiPSC-derived cell lines were mainly considered for applications in regenerative medicine (Laustriat et al., 2010; Doss and Sachinidis, 2019) but their extensive range of advantages have also made them interesting for the development of cell-based assays in the fields of drug-discovery, pharmaceutical research and personalized medicine (Yu et al., 2007; Di Baldassarre et al., 2018; Berry et al., 2019). Due to the improved quality and cell handling methods, ongoing development of APC features, and initiatives such as the Comprehensive in vitro Proarrhythmia Assay for cardiac ion channel safety pharmacology (Colatsky et al., 2016), the field is now making good progress developing new APC applications for the characterization and pharmacology of hiPSC-derived cell lines.
In this section we provide examples of some of these novel APC applications for studies of hiPSC-derived cell lines.
A challenge of hiPSC-derived cells is that they typically display heterogeneous current expression. This might be due to different maturation states, different cell types, genetic, and/or biological variability. In APC, currents are recorded in parallel from many cells. This makes APC well suited to characterize cell populations and identify subpopulations (Haythornthwaite et al., 2012; Franz et al., 2017). A representative example is shown in Figure 4 in which a subpopulation of hiPSC-derived cortical neurons is selected that expresses both Nav and a mixture of delayed rectifier and A-type Kv channels. Properties of hiPSC-neurons, such as ion channel expression, AP firing characteristics and the resting membrane potential (RMP), have shown to approach that of mature adult neuronal cells by increased time in culture (Prè et al., 2014; Berry et al., 2015; Odawara et al., 2016). APC recordings can assist the evaluation of neuron population purity and maturity by comparing ion channel expression at different culturing time points or across different batches (see Figure 4).
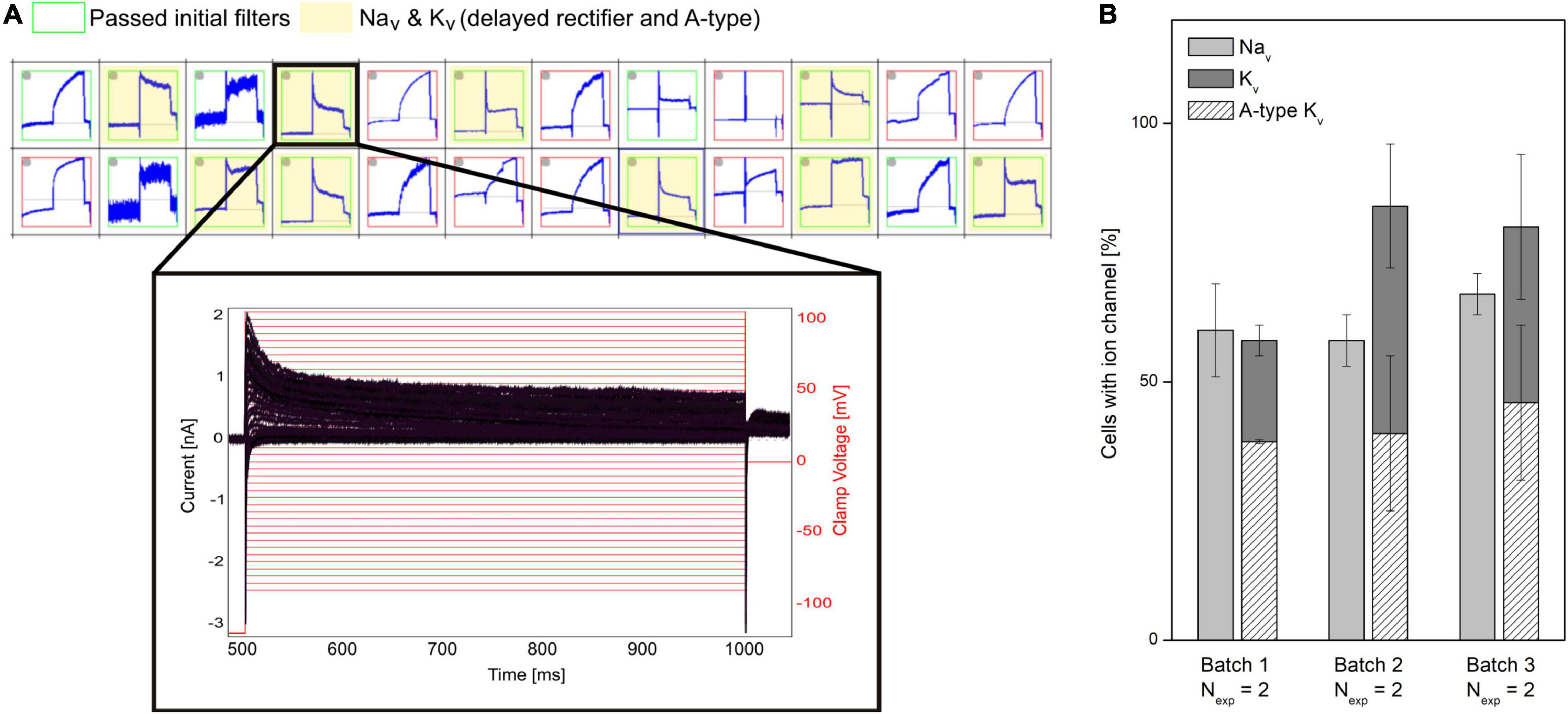
Figure 4. Characterizing mixed ion channel populations in hiPSC-derived cortical neurons. (A) Quality control filters allow selection of cells expressing the ion channels of interest. Top: Cut-out of the measurement plate view showing 24 of 384 experiments. Red and green borders correspond to experiments that failed and passed, respectively, the initial filter criteria (Rmem > 500 MΩ, Ccell > 1.5 pF). The experiments highlighted in yellow passed filter criteria for Nav and Kv currents. Bottom: Example of a population of current traces (black) elicited by + 5 mV voltage-steps (red) recorded from a neuron, expressing Nav channels and a mixture of delayed rectifier and A-type Kv channels. (B) The percentage (average ± SEM) of investigated cells containing Nav and Kv channels were displayed for three different differentiation batches of hiPSC cortical neurons (Nexp = 2 experiment days, with > 48 measurement sites per experiment). Of the cells expressing Kv channels, the percentage of Kv expressing cells also having A-type Kv currents are overlayed (diagonal pattern). No significant differences were observed between the three batches (Rosholm et al., 2021b). Adapted from Rosholm et al. (2021b), with permission from Sophion Bioscience A/S.
Measurements of cardiac APs are of special interest for disease modeling, drug discovery and cardiac safety (Braam et al., 2009; Mummery, 2018; Rowe and Daley, 2019; Bell and Fermini, 2021). However, the immature phenotype and heterogeneous ion channel expression of hiPSC-derived CMs, as compared to acutely isolated, primary CMs has limited the physiological relevance of their electrophysiological characterization, in general, and high throughput measurements, in particular (Casini et al., 2017; Mann et al., 2019; Vaidyanathan et al., 2021). This includes the presence of the pacemaker current If (routinely expressed in immature CMs and lost in adult CMs) and reduced densities of hyperpolarizing current IK1 (Jonsson et al., 2012; Goversen et al., 2018b). Highlighting these differences, Figure 5 provides a comparison between hiPSC-derived CMs and acutely isolated, primary adult CMs showing the relative size and contribution of ion channel currents that form the AP in each.
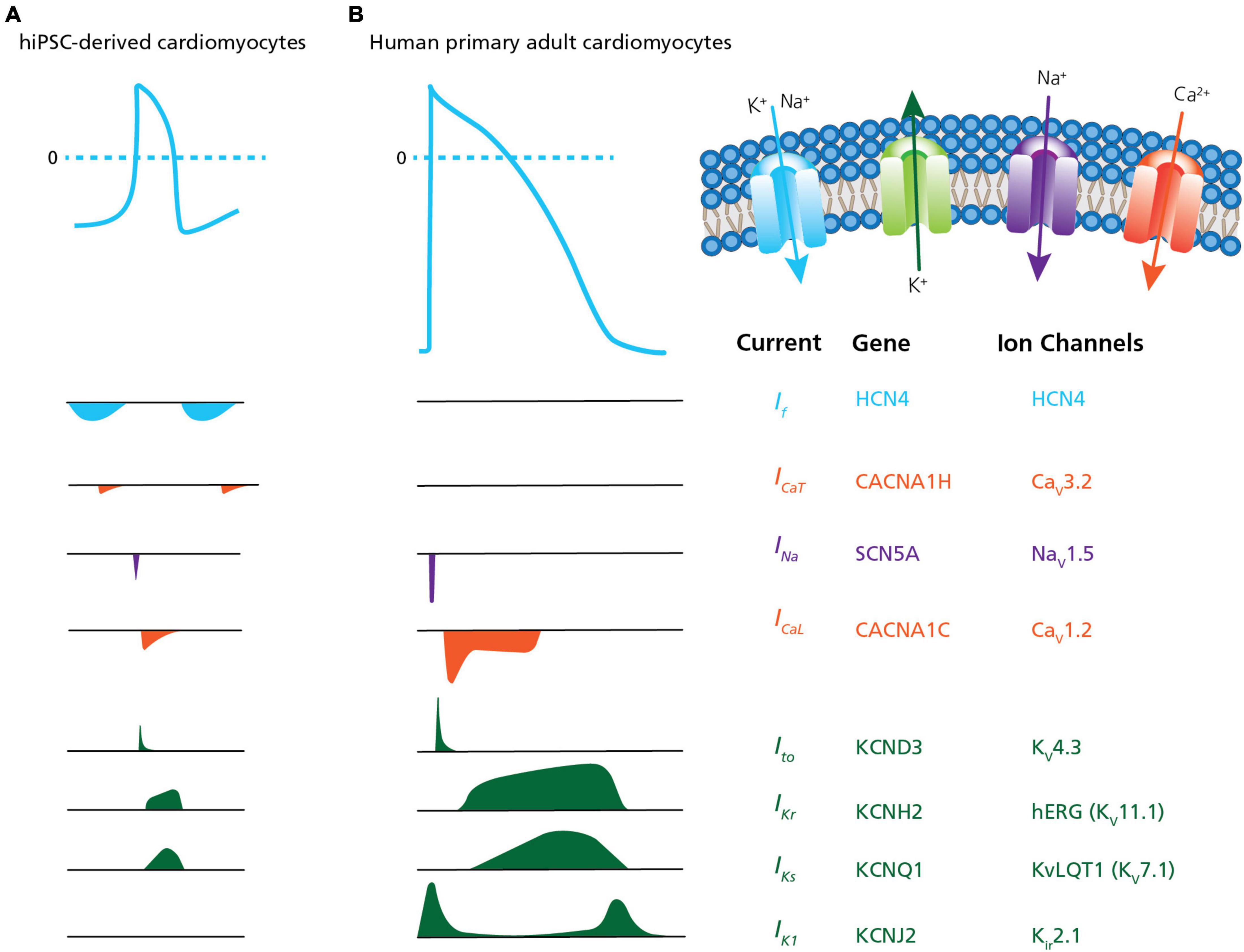
Figure 5. Ion channel content in hiPSC-CMs versus human primary CMs. A comparison of action potential and ion channel currents recorded in (A) hiPSC-CMs and (B) human primary adult CMs. The associated genes and ion channels are listed on the right. From the top the currents are: If, pacemaker or funny current; ICaT, T-type calcium channel current; INa, sodium current; ICaL, L-type calcium channel current; Ito, transient outward potassium current; IKr, rapid delayed- rectifier potassium current; IKs, slow delayed-rectifier potassium current; and IK1, inward-rectifier potassium current. The traces highlight differences traditionally observed in hiPSC-CMs, such as the presence of the pacemaker current If (routinely expressed in immature CMs and lost in adult CMs) and reduced densities of hyperpolarizing current IK1. Adapted from Karbassi et al. (2020), with permission from publisher Springer Nature.
Different methods have been employed to compensate for missing currents. For example, a method that is increasingly being used is enhancing missing currents using viral transfection (Jones et al., 2014; Vaidyanathan et al., 2021). Another strategy that has been employed in APC is dynamic clamp, which is a method that models and electronically introduces ion currents into cells, to compensate for the lack of expression (Becker et al., 2013; Verkerk et al., 2017). For instance, this electronic compensation can be used to add the missing hyperpolarizing current IK1, bringing the depolarized RMP often observed in dissociated hiPSC-CMs closer to the physiological, hyperpolarized potential, which results in AP durations and waveforms comparable to mature CMs (Li et al., 2019; Becker et al., 2020). Drawbacks of dynamic clamp are that the compensating current is only as accurate as the assumption-based model of the mature CM, and ideally each cell’s current phenotype and derived model would need to be measured and performed individually with real-time compensation during experimentation, a potential computer processing bottleneck for the ms temporal resolution of 100 s of simultaneous recordings in APC.
Finally, the maturation of hiPSC-CMs has improved recently, and the paradigm of impaired IK1 expression in hiPSC-CMs has been challenged (Horváth et al., 2018). In Figure 6 we display APC measurements of “mature” IK1 currents in hiPSC-CMs.
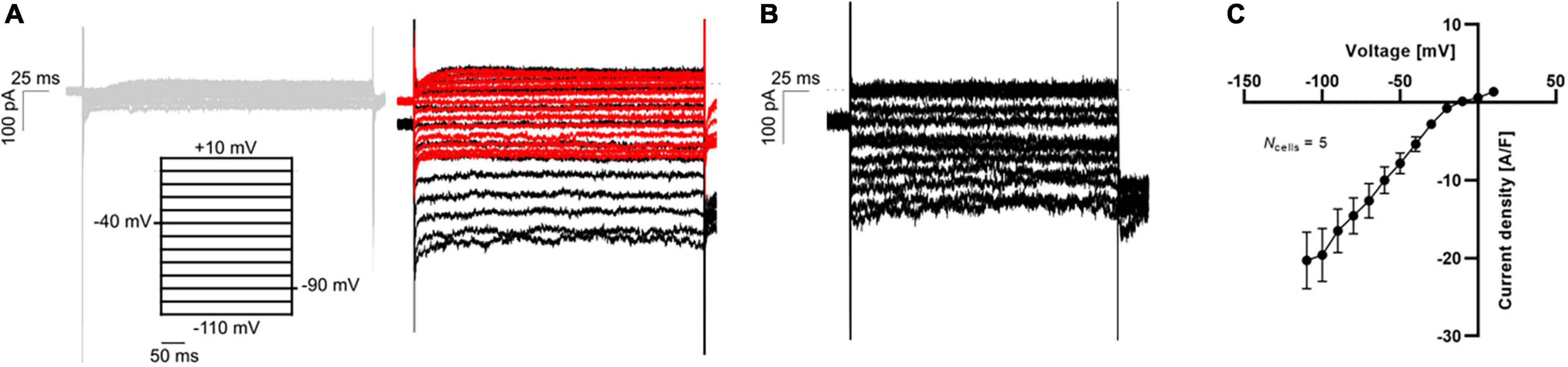
Figure 6. APC measurements of IK1 current-voltage relationship in hiPSC-CMs using physiological solutions. (A) Representative K+ current traces in response to a voltage step protocol from –110 mV to +10 mV with 10 mV increment step size (black). The current was measured in 5 mM extracellular K+ (gray, left) and 75 mM extracellular K+ before (black) and after (red) addition of 1 mM BaCl2 (right). (B) The BaCl2 sensitive IK1 was calculated by subtracting the insensitive current (red trace in A) from the total IK (black trace in A). (C) The IK1 current-voltage relationship was plotted by extracting the average steady-state sensitive current (shown in B) and plotting as a function of step voltage. Adapted from Rosholm et al. (2022), with permission from Sophion Bioscience A/S.
As observed with hiPSC-CMs, hiPSC-neurons (including several subtypes, such as motor neurons and sensory neurons) do not completely replicate the adult neuron phenotype in terms of morphology, electrical function, and critically, in resembling neurodegenerative disease features (Casalia et al., 2021; Giacomelli et al., 2022). Although iPSC-neurons fail to entirely capture all the characteristics of fully mature brain cells, they express functional ion channels and are capable of displaying primary neuronal functions such as firing action potentials (Franz et al., 2017). Recent developments of more sophisticated and reproducible differentiation protocols combined with advanced genome engineering, longer maturation in culture and improved dissociation procedures have improved iPSC-neurons which now constitute a promising cell model for APC recordings despite their relatively immature phenotype (Shi et al., 2017; Giacomelli et al., 2022).
The high throughput of APC allows sufficient data generation to accurately define heterogeneous ion channel populations in hiPSC-CMs, correlating channel levels to the resulting AP firing properties (Rosholm, 2021). Presently, optimized hiPSC-CM maturation and harvest protocols have made it possible to measure APs and capture compound effects in physiological solutions using APC (see Figure 7). The data were very similar to data obtained with a complementary optical assay (Seibertz et al., 2020). These advances hold great promise that robust and consistent AP assays for cardiac safety and compound screens will soon be feasible and routine.
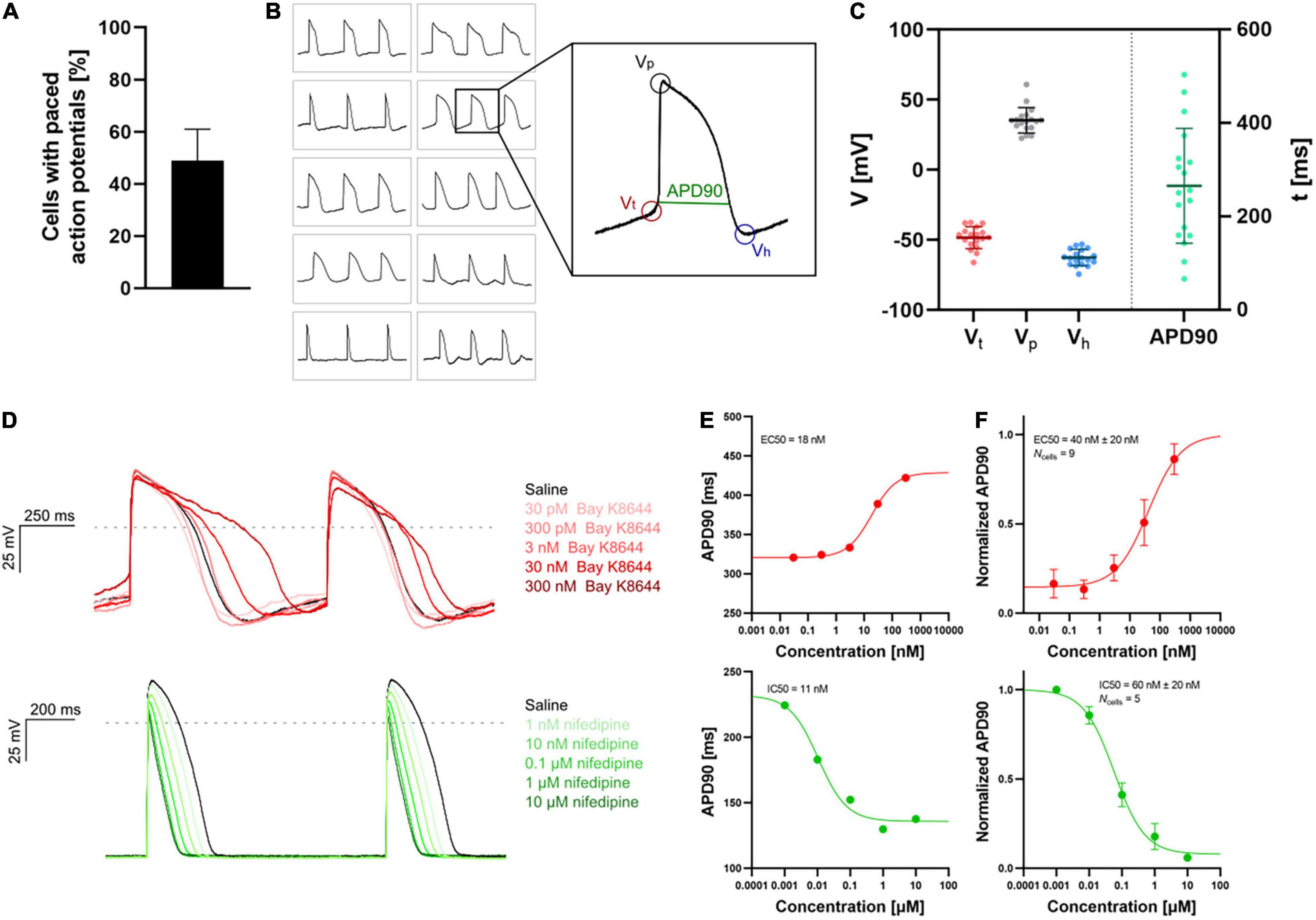
Figure 7. APC current-clamp measurements of paced action potentials and APD90 concentration-response plots. (A) The percentage of successful experiments displaying paced action potentials [Vp > 0 mV, resting membrane potential (RMP) < –40 mV]. Data is avg ± SD of three measurement plates (each of 48 recording sites). Assay success rate was 10%–30% out of 48 sites. (B) Paced action potentials from 10 individual hiPSC-CMs within a single measurement plate (left) and an expanded action potential displaying the extracted parameters: threshold potential (Vt), peak potential (Vp), hyperpolarization potential (Vh), and action potential duration at 90% repolarization (APD90). (C) Plot of extracted parameters, Vt (red), Vp (gray), Vh (blue), and APD90 (green, ms) for 18 individual hiPSC-CMs with the avg ± SD (solid lines). (D) Paced action potentials for saline controls (black traces) and in response to increasing concentrations of Bay K8644 (top, traces in shades of red) and nifedipine (bottom, traces in shades of green). (E) Plots of APD90 versus compound concentration with Boltzmann fits including EC50 or IC50 calculated values, recorded in a single cell. (F) Cumulative concentration-response (average, APD90 normalized to pre compound concentration) relationships with Boltzmann fits including EC50 or IC50 calculated values. Data points are avg ± SEM of Ncells. Adapted from Rosholm (2021) and Rosholm et al. (2022), with permission from Sophion Bioscience A/S.
As the quality of hiPSC-derived cell lines increases, an obvious next step will be to utilize the HT capability of APC for studying disease models, drug-screening and personalized (or stratified) medicine (Chen et al., 2016). hiPSC disease models, characterization and drug-screening assays employing these cells have been developed for a range of diseases including neurodegenerative diseases (Kim, 2015; Lee and Huang, 2017) such as Alzheimer’s Disease (Medda et al., 2016; Brownjohn et al., 2017), and cardiovascular diseases (Braam et al., 2009; Sube and Ertel, 2017; Mummery, 2018; Wu et al., 2020) such as long-QT syndrome (Friedrichs et al., 2015; Jouni et al., 2015; Chen et al., 2016).
Although examples of APC applications within personalized biomedicine are still scarce, their need and value mean they are in active development (Friedrichs et al., 2015; Goversen et al., 2018a), with expectations high that we will see more in the near future. For an example of such an application see Rosholm (2019) in which an hiPSC motor neuron disease model, derived from a Spinal Muscular Atrophy patient (Faravelli et al., 2015) was characterized together with control cells from healthy subjects. The results showed the expected overactivity of Nav channels, which could be rescued by a Nav antagonistic compound addition (SMN-C3), see Figure 8.
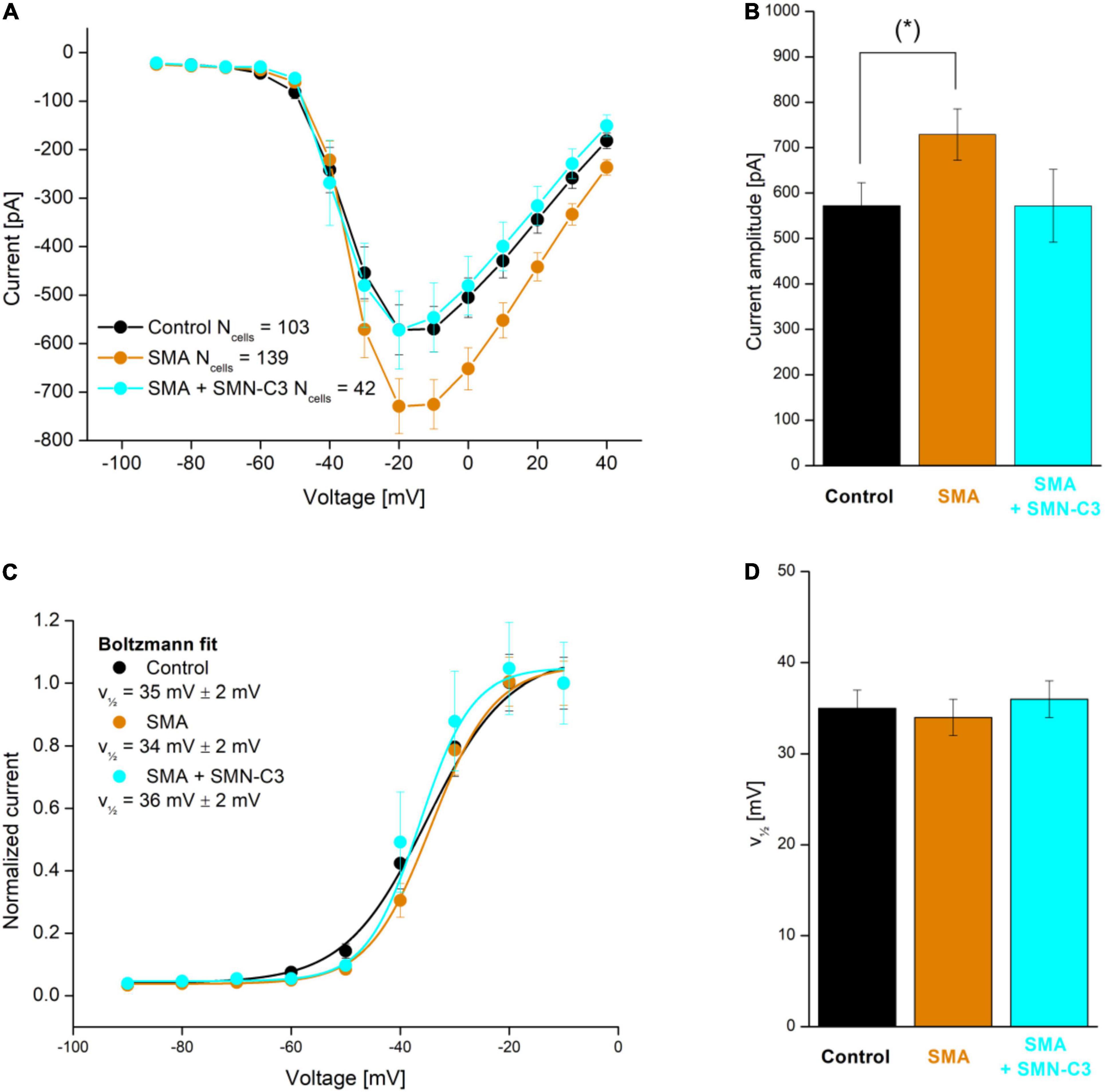
Figure 8. APC evaluation of compound effects on Nav channel properties in SMA hiPSC-neurons. Parallel recordings of control neurons (black), SMA neurons (orange), and SMA neurons treated with SMN-C3 during culturing (blue). (A) Average Nav current versus step voltage. (B) Quantification of the current amplitude at –20 mV. The current amplitude is significantly larger in SMA cells than in control cells [Student’s t-test, p < 0.05 (*), 95% confidence interval]. (C) Average Nav current normalized to the current recorded at V = –20 mV. Fitting of a Boltzmann function (solid lines) yielded the half-activation voltage (, mV). (D) Half-activation voltage () values. Error bars are SEM. Adapted from Rosholm (2019), with permission from Sophion Bioscience A/S.
As highlighted in this review, the advances made in our labs and amongst APC and stem cell researchers across the world in the last few years have catapulted this niche, often challenging, area of research into the limelight, and is becoming widely adopted amongst the stem cell and drug discovery communities (Rajamohan et al., 2016; Franz et al., 2017). With experimental success rates climbing, improved protocols and assays developed using physiological recording conditions, the large, objective data sets generated are furthering our understanding of ion channel physiology and pathophysiology. We believe these advances in APC in vitro electrophysiology, married with cross-correlated combinations of animal and human iPSC models, in time will reduce and circumvent the expensive use (and sometimes limited translatability) of primary animal organ, tissue and cell models (Ebert et al., 2012; Deng, 2017; Pessôa et al., 2020). The advances made to date are providing tools and assays for significant progress in the accuracy and efficiency of safety pharmacology, drug discovery and development, and regenerative and personalized medicines.
Jumping into the fabled time traveling DeLorean car and heading back to the future: one area that is likely to feature heavily in the future of stem cell and ion channel research are the possibilities afforded by patient-derived hiPSCs. Measuring and defining an individual patient’s hiPSCs (such as their ion channel composition, pharmacology and biophysics) will allow the design of personalized, targeted medical interventions (Matsa et al., 2016; Hamazaki et al., 2017; Doss and Sachinidis, 2019), thus improving the efficacy of medicinal regimes and limiting side-effects.
Taking this concept of measuring and defining patient hiPSCs a step further: by identifying patient genotypes (by gene or genome sequencing) involved in a panel of potential health issues specific to the patient, these can be correlated to ion channel phenotypes determined using APC recordings of patient-derived hiPSCs, across a range of cell types. Such patient genotype/phenotype correlation (“phenotypic screening”) will allow existing health issues to be identified and addressed (Muñoz-Martín and Matsa, 2021). Combining this ion channel phenotypic screening with other typical phenotype screens (such as defining patient metabolic pathways or “metabolome”) will allow potential future health issues to be flagged, enabling pre-emptive, prophylactic medical interventions or lifestyle changes to be implemented. In addition, high throughput APC would allow for the application of artificial intelligence (or machine learning) tools to study multidimensional high-volume data for particular phenotypes, which may allow for identification of novel cellular biomarkers as a basis for robust drug discovery research. With time and concomitant “big data” analyses, the correlation between genotype and phenotype is likely to become sufficiently predictive allowing gene sequencing alone to be used in forecasting the ion channel targeting medicinal regimes that best suit a designated genotype patient cohort or sub-group within the general population.
In conclusion, the marriage of hiPSC and APC offers much potential for ion channel biomedical research and drug discovery. The possibilities of defining ion channel activity, biophysics and pharmacology in specialized cells (like CMs and neuronal cells), including cells derived from healthy or diseased patients, provide model cells to efficiently perform APC driven in vitro studies and high throughput screens that increasingly translate to human clinical studies (Rowe and Daley, 2019; Muñoz-Martín and Matsa, 2021). In writing this review, the authors aim to convey the recent adventures and advances in cellular time travel and the vast potential for these techniques and technologies. After reading, we hope the reader, like us, is now flexing their muscles in the starting blocks for the bright biomedical future of the brave new world we are soon to be sprinting toward.
KR structured the manuscript and coordinated the writing process, and produced the experimental data included in the manuscript. KR, BB, and DB prepared the first version of the manuscript. SK, DN, NV, and FS reviewed the manuscript. All authors contributed to the article and approved the submitted version.
The work of NV is supported by the Deutsche Forschungsgemeinschaft (DFG; VO 1568/3-1, VO 1568/4-1, IRTG1816 project 12, SFB1002 project A13, and under Germany’s Excellence Strategy – EXC 2067/1- 390729940) and the DZHK (German Center for Cardiovascular Research, 81X300189, 81X4300102, 81X4300115, and 81X4300112).
KR, BB, SK, and DB are employed by Sophion Bioscience A/S, Denmark. DN was employed by Sophion Bioscience Inc., United States.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge Jane Lucas for assistance with figure preparation. hiPSC-neurons were provided by Boston Children’s Hospital and BrainXell. HiPSC-CMs were provided by NV, Göttingen.
APs, action potentials; APC, automated patch clamp; CMs, cardiomyocytes; hiPSC, human induced pluripotent stem cell; HT, high throughput; MPC, manual patch clamp; PSCs, pluripotent stem cells.
Ahmed, R. E., Anzai, T., Chanthra, N., and Uosaki, H. (2020). A brief review of current maturation methods for human induced pluripotent stem cells-derived cardiomyocytes. Front. Cell Dev. Biol. 8:178. doi: 10.3389/fcell.2020.00178
Andersen, J., Revah, O., Miura, Y., Thom, N., Amin, N. D., Kelley, K. W., et al. (2020). Generation of functional human 3D cortico-motor assembloids. Cell 183, 1913–1929.e26. doi: 10.1016/j.cell.2020.11.017
Assou, S., Bouckenheimer, J., and De Vos, J. (2018). Concise review: Assessing the genome integrity of human induced pluripotent stem cells: What quality control metrics? Stem Cells 36, 814–821. doi: 10.1002/stem.2797
Bardy, C., Van Den Hurk, M., Kakaradov, B., Erwin, J. A., Jaeger, B. N., Hernandez, R. V., et al. (2016). Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Mol. Psychiatry 21, 1573–1588. doi: 10.1038/mp.2016.158
Becker, N., Horváth, A., De Boer, T., Fabbri, A., Grad, C., Fertig, N., et al. (2020). Automated dynamic clamp for simulation of IK1 in human induced pluripotent stem cell–derived cardiomyocytes in real time using patchliner dynamite8. Curr. Protoc. Pharmacol. 88, 1–23. doi: 10.1002/cpph.70
Becker, N., Stoelzle, S., Göpel, S., Guinot, D., Mumm, P., Haarmann, C., et al. (2013). Minimized cell usage for stem cell-derived and primary cells on an automated patch clamp system. J. Pharmacol. Toxicol. Methods 68, 82–87. doi: 10.1016/j.vascn.2013.03.009
Bell, D. C., and Dallas, M. L. (2018). Using automated patch clamp electrophysiology platforms in pain-related ion channel research: insights from industry and academia. Br. J. Pharmacol. 175, 2312–2321. doi: 10.1111/bph.13916
Bell, D. C., and Fermini, B. (2021). Use of automated patch clamp in cardiac safety assessment: past, present & future perspectives. J. Pharmacol. Toxicol. Methods 111:107114. doi: 10.1016/j.vascn.2021.107114
Bellin, M., Marchetto, M. C., Gage, F. H., and Mummery, C. L. (2012). Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 13, 713–726. doi: 10.1038/nrm3448
Berry, B. J., Akanda, N., Smith, A. S. T., Long, C. J., Schnepper, M. T., Guo, X., et al. (2015). Morphological and functional characterization of human induced pluripotent stem cell-derived neurons (iCell Neurons) in defined culture systems. Biotechnol. Prog. 31, 1613–1622. doi: 10.1002/btpr.2160
Berry, B. J., Smith, A. S. T., Young, J. E., and Mack, D. L. (2019). Advances and current challenges associated with the use of human induced pluripotent stem cells in modeling neurodegenerative disease. Cells Tissues Organs 205, 331–349. doi: 10.1159/000493018
Biendarra-Tiegs, S. M., Li, X., Ye, D., Brandt, E. B., Ackerman, M. J., and Nelson, T. J. (2019). Single-Cell RNA-Sequencing and Optical Electrophysiology of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Reveal Discordance between Cardiac Subtype-Associated Gene Expression Patterns and Electrophysiological Phenotypes. Stem Cells Dev. 28, 659–673. doi: 10.1089/scd.2019.0030
Blokzijl, F., De Ligt, J., Jager, M., Sasselli, V., Roerink, S., Sasaki, N., et al. (2016). Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264. doi: 10.1038/nature19768
Braam, S. R., Passier, R., and Mummery, C. L. (2009). Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery. Trends Pharmacol. Sci. 30, 536–545. doi: 10.1016/j.tips.2009.07.001
Breckwoldt, K., Letuffe-Brenière, D., Mannhardt, I., Schulze, T., Ulmer, B., Werner, T., et al. (2017). Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 12, 1177–1197. doi: 10.1038/nprot.2017.033
Brownjohn, P. W., Smith, J., Portelius, E., Serneels, L., Kvartsberg, H., De Strooper, B., et al. (2017). Phenotypic screening identifies modulators of amyloid precursor protein processing in human stem cell models of Alzheimer’s Disease. Stem Cell Rep. 8, 870–882. doi: 10.1016/j.stemcr.2017.02.006
Burridge, P. W., Keller, G., Gold, J. D., and Wu, J. C. (2012). Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10, 16–28. doi: 10.1016/j.stem.2011.12.013
Casalia, M. L., Casabona, J. C., García, C., Cavaliere Candedo, V., Quintá, H. R., Farías, M. I., et al. (2021). A familiar study on self-limited childhood epilepsy patients using hIPSC-derived neurons shows a bias towards immaturity at the morphological, electrophysiological and gene expression levels. Stem Cell Res. Ther. 12:590. doi: 10.1186/s13287-021-02658-2
Casini, S., Verkerk, A. O., and Remme, C. A. (2017). Human iPSC-derived cardiomyocytes for investigation of disease mechanisms and therapeutic strategies in inherited arrhythmia syndromes: strengths and limitations. Cardiovasc. Drugs Ther. 31, 325–344. doi: 10.1007/s10557-017-6735-0
Chen, H., Qian, K., Du, Z., Cao, J., Petersen, A., Liu, H., et al. (2014). Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 14, 796–809. doi: 10.1016/j.stem.2014.02.004
Chen, I. Y., Matsa, E., and Wu, J. C. (2016). Induced pluripotent stem cells: at the heart of cardiovascular precision medicine. Nat. Rev. Cardiol. 13, 333–349. doi: 10.1038/nrcardio.2016.36
Colatsky, T., Fermini, B., Gintant, G., Pierson, J. B., Sager, P., Sekino, Y., et al. (2016). The comprehensive in vitro proarrhythmia assay (CiPA) initiative — Update on progress. J. Pharmacol. Toxicol. Methods 81, 15–20. doi: 10.1016/j.vascn.2016.06.002
Cyganek, L., Tiburcy, M., Sekeres, K., Gerstenberg, K., Bohnenberger, H., Lenz, C., et al. (2018). Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight 3:e99941. doi: 10.1172/jci.insight.99941
Davis-Dusenbery, B. N., Williams, L. A., Klim, J. R., and Eggan, K. (2014). How to make spinal motor neurons. Development 141, 491–501. doi: 10.1242/dev.097410
de Leeuw, S. M., Davaz, S., Wanner, D., Milleret, V., Ehrbar, M., Gietl, A., et al. (2021). Increased maturation of iPSC-derived neurons in a hydrogel-based 3D culture. J. Neurosci. Methods 360:109254. doi: 10.1016/j.jneumeth.2021.109254
Deng, B. (2017). Mouse models and induced pluripotent stem cells in researching psychiatric disorders. Stem Cell Invest. 4, 4–6. doi: 10.21037/sci.2017.06.10
Depalma, S. J., Davidson, C. D., Stis, A. E., Helms, A. S., and Baker, B. M. (2021). Microenvironmental determinants of organized iPSC-cardiomyocyte tissues on synthetic fibrous matrices. Biomater. Sci. 9, 93–107. doi: 10.1039/d0bm01247e
Di Baldassarre, A., Cimetta, E., Bollini, S., Gaggi, G., and Ghinassi, B. (2018). Human-induced pluripotent stem cell technology and cardiomyocyte generation: Progress and clinical applications. Cells 7:48. doi: 10.3390/cells7060048
Doss, M. X., and Sachinidis, A. (2019). Current Challenges of iPSC-Based Disease Modeling and Therapeutic Implications. Cells 8:403. doi: 10.3390/cells8050403
Ebert, A. D., Liang, P., and Wu, J. C. (2012). Induced pluripotent stem cells as a disease modeling and drug screening platform. J. Cardiovasc. Pharmacol. 60, 408–416. doi: 10.1097/FJC.0b013e318247f642
Engle, S. J., Blaha, L., and Kleiman, R. J. (2018). Best practices for translational disease modeling using human iPSC-derived neurons. Neuron 100, 783–797. doi: 10.1016/j.neuron.2018.10.033
Faravelli, I., Nizzardo, M., Comi, G. P., and Corti, S. (2015). Spinal muscular atrophy-recent therapeutic advances for an old challenge. Nat. Rev. Neurol. 11, 351–359. doi: 10.1038/nrneurol.2015.77
Feaster, T. K., Cadar, A. G., Wang, L., Williams, C. H., Chun, Y. W., Hempel, J. E., et al. (2015). A method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 117, 995–1000. doi: 10.1161/CIRCRESAHA.115.307580
Fermini, B., and Bell, D. C. (2022). On the perspective of an aging population and its potential impact on drug attrition and pre-clinical cardiovascular safety assessment. J. Pharmacol. Toxicol. Methods. doi: 10.1016/j.vascn.2022.107184
Feyen, D. A. M., McKeithan, W. L., Bruyneel, A. A. N., Spiering, S., Hörmann, L., Ulmer, B., et al. (2020). Metabolic maturation media improve physiological function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 32:107925. doi: 10.1016/j.celrep.2020.107925
Franz, D., Olsen, H. L., Klink, O., and Gimsa, J. (2017). Automated and manual patch clamp data of human induced pluripotent stem cell-derived dopaminergic neurons. Nat. Sci. Data 4:170056. doi: 10.1038/sdata.2017.56
Friedrichs, S., Malan, D., Voss, Y., and Sasse, P. (2015). Scalable Electrophysiological Investigation of iPS Cell-Derived Cardiomyocytes Obtained by a Lentiviral Purification Strategy. J. Clin. Med. 4, 102–123. doi: 10.3390/jcm4010102
Fusaki, N., Ban, H., Nishiyama, A., Saeki, K., and Hasegawa, M. (2009). Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 348–362. doi: 10.2183/pjab.85.348
Giacomelli, E., Vahsen, B. F., Calder, E. L., Xu, Y., Scaber, J., Gray, E., et al. (2022). Human stem cell models of neurodegeneration: from basic science of amyotrophic lateral sclerosis to clinical translation. Cell Stem Cell 29, 11–35. doi: 10.1016/j.stem.2021.12.008
Giannetti, F., Benzoni, P., Campostrini, G., Milanesi, R., Bucchi, A., Baruscotti, M., et al. (2021). A detailed characterization of the hyperpolarization-activated “funny” current (I f) in human-induced pluripotent stem cell (iPSC)–derived cardiomyocytes with pacemaker activity. Pflugers Arch. 473, 1009–1021. doi: 10.1007/s00424-021-02571-w
Goversen, B., van der Heyden, M. A. G., van Veen, T. A. B., and de Boer, T. P. (2018b). The immature electrophysiological phenotype of iPSC-CMs still hampers in vitro drug screening: special focus on I K1. Pharmacol. Ther. 183, 127–136. doi: 10.1016/j.pharmthera.2017.10.001
Goversen, B., Becker, N., Stoelzle-Feix, S., Obergrussberger, A., Vos, M. A., van Veen, T. A. B., et al. (2018a). A hybrid model for safety pharmacology on an automated patch clamp platform: Using dynamic clamp to join iPSC-derived cardiomyocytes and simulations of Ik1 ion channels in real-time. Front. Physiol. 8:1094. doi: 10.3389/fphys.2017.01094
Hamazaki, T., El Rouby, N., Fredette, N. C., Santostefano, K. E., and Terada, N. (2017). Concise review: induced pluripotent stem cell research in the era of precision medicine. Stem Cells 35, 545–550. doi: 10.1002/STEM.2570
Hamill, O. P., Marty, A., Neher, E., Sakmann, B., and Sigworth, F. J. (1981). Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 391, 85–100. doi: 10.1007/BF00656997
Hartfield, E. M., Yamasaki-Mann, M., Ribeiro Fernandes, H. J., Vowles, J., James, W. S., Cowley, S. A., et al. (2014). Physiological characterisation of human iPS-derived dopaminergic neurons. PLoS One 9:e87388. doi: 10.1371/journal.pone.0087388
Haythornthwaite, A., Stoelzle, S., Hasler, A., Kiss, A., Mosbacher, J., George, M., et al. (2012). Characterizing human ion channels in induced pluripotent stem cell – derived neurons. J. Biomol. Screen. 17, 1264–1272. doi: 10.1177/1087057112457821
Horváth, A., Lemoine, M. D., Löser, A., Mannhardt, I., Flenner, F., Uzun, A. U., et al. (2018). Low resting membrane potential and low inward rectifier potassium currents are not inherent features of hiPSC-Derived Cardiomyocytes. Stem Cell Rep. 10, 822–833. doi: 10.1016/j.stemcr.2018.01.012
Jehuda, B. R., Shemer, Y., and Binah, O. (2018). Genome editing in induced pluripotent stem cells using CRISPR/Cas9. Stem Cell Rev. Rep. 14, 323–336. doi: 10.1007/s12015-018-9811-3
Jha, A. (2011). Look, No Embryos! The Future of Ethical Stem Cells. Guardian. Available online at: https://www.theguardian.com/science/2011/mar/13/ips-reprogrammed-stem-cells (accessed May 23, 2022).
Jones, D. K., Liu, F., Vaidyanathan, R., Eckhardt, L. L., Trudeau, M. C., and Robertson, G. A. (2014). hERG 1b is critical for human cardiac repolarization. Proc. Natl. Acad. Sci. U.S.A. 111, 18073–18077. doi: 10.1073/pnas.1414945111
Jonsson, M. K. B., Vos, M. A., Mirams, G. R., Duker, G., Sartipy, P., De Boer, T. P., et al. (2012). Application of human stem cell-derived cardiomyocytes in safety pharmacology requires caution beyond hERG. J. Mol. Cell. Cardiol. 52, 998–1008. doi: 10.1016/j.yjmcc.2012.02.002
Jouni, M., Si-Tayeb, K., Es-Salah-Lamoureux, Z., Latypova, X., Champon, B., Caillaud, A., et al. (2015). Toward personalized medicine: using cardiomyocytes differentiated from urine-derived pluripotent stem cells to recapitulate electrophysiological characteristics of type 2 long QT syndrome. J. Am. Heart Assoc. 4, 1–13. doi: 10.1161/JAHA.115.002159
Jung, P., Seibertz, F., Ignatyeva, N., Sampathkumar, S., Ritter, M., Li, H., et al. (2022). Increased cytosolic calcium buffering contributes to a cellular arrhythmogenic substrate in iPSC-cardiomyocytes from patients with dilated cardiomyopathy. Basic Res. Cardiol. 117:5.
Karbassi, E., Fenix, A., Marchiano, S., Muraoka, N., Nakamura, K., Yang, X., et al. (2020). Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 17, 341–359. doi: 10.1038/s41569-019-0331-x
Kilfoil, P., Feng, S. L., Bassyouni, A., Lee, T., Leishman, D., Li, D., et al. (2021). Characterization of a high throughput human stem cell cardiomyocyte assay to predict drug-induced changes in clinical electrocardiogram parameters. Eur. J. Pharmacol. 912:174584. doi: 10.1016/j.ejphar.2021.174584
Kilpinen, H., Goncalves, A., Leha, A., Afzal, V., Alasoo, K., Ashford, S., et al. (2017). Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. doi: 10.1038/nature22403
Kim, C. (2015). iPSC technology-Powerful hand for disease modeling and therapeutic screen. BMB Rep. 48, 256–265. doi: 10.5483/BMBRep.2015.48.5.100
Kolanowski, T. J., Antos, C. L., and Guan, K. (2017). Making human cardiomyocytes up to date: derivation, maturation state and perspectives. Int. J. Cardiol. 241, 379–386. doi: 10.1016/j.ijcard.2017.03.099
Laustriat, D., Gide, J., and Peschanski, M. (2010). Human pluripotent stem cells in drug discovery and predictive toxicology. Biochem. Soc. Trans. 38, 1051–1057. doi: 10.1042/BST0381051
Lee, S., and Huang, E. J. (2017). Modeling ALS and FTD with iPSC-derived neurons. Brain Res. 1656, 88–97. doi: 10.1016/j.brainres.2015.10.003
Lemoine, M. D., Mannhardt, I., Breckwoldt, K., Prondzynski, M., Flenner, F., Ulmer, B., et al. (2017). Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci. Rep. 7:5464. doi: 10.1038/s41598-017-05600-w
Li, W., Luo, X., Ulbricht, Y., and Guan, K. (2021b). Blebbistatin protects iPSC-CMs from hypercontraction and facilitates automated patch-clamp based electrophysiological study. Stem Cell Res. 56:102565. doi: 10.1016/j.scr.2021.102565
Li, W., Luo, X., Ulbricht, Y., and Guan, K. (2021a). Applying automated patch-clamp to disease modeling: recapitulate phenotypes of Brugada syndrome using iPSC-CMs. bioRvix [Preprint]. doi: 10.1101/2021.06.07.447362
Li, W., Luo, X., Ulbricht, Y., Wagner, M., Piorkowski, C., El-Armouche, A., et al. (2019). Establishment of an automated patch-clamp platform for electrophysiological and pharmacological evaluation of hiPSC-CMs. Stem Cell Res. 41:101662. doi: 10.1016/j.scr.2019.101662
Lo, B., and Parham, L. (2009). Ethical issues in stem cell research. Endocr. Rev. 30, 204–213. doi: 10.1210/ER.2008-0031
Mann, S. A., Heide, J., Knott, T., Airini, R., Epureanu, F. B., Deftu, A. F., et al. (2019). Recording of multiple ion current components and action potentials in human induced pluripotent stem cell-derived cardiomyocytes via automated patch-clamp. J. Pharmacol. Toxicol. Methods 100:106599. doi: 10.1016/j.vascn.2019.106599
Matsa, E., Ahrens, J. H., and Wu, J. C. (2016). Human induced pluripotent stem cells as a platform for personalized and precision cardiovascular medicine. Physiol. Rev. 96, 1093–1126. doi: 10.1152/physrev.00036.2015
Medda, X., Mertens, L., Versweyveld, S., Diels, A., Barnham, L., Bretteville, A., et al. (2016). Development of a scalable, high-throughput-compatible assay to detect tau aggregates using iPSC-derived cortical neurons maintained in a three-dimensional culture format. J. Biomol. Screen. 21, 804–815. doi: 10.1177/1087057116638029
Moradi, S., Mahdizadeh, H., Šarić, T., Kim, J., Harati, J., Shahsavarani, H., et al. (2019). Research and therapy with induced pluripotent stem cells (iPSCs): social, legal, and ethical considerations. Stem Cell Res. Ther. 10:341. doi: 10.1186/s13287-019-1455-y
Mummery, C. L. (2018). Perspectives on the use of human induced pluripotent stem cell-derived cardiomyocytes in biomedical research. Stem Cell Rep. 11, 1306–1311. doi: 10.1016/j.stemcr.2018.11.011
Muñoz-Martín, N., and Matsa, E. (2021). Benefits & key considerations of using human iPSC-derived disease models in drug discovery. Int. Biopharm. Ind. 4, 26–29.
Obergrussberger, A., Haarmann, C., Stölzle-feix, S., Becker, N., Ohtsuki, A., Brüggemann, A., et al. (2017). “Automated patch clamp recordings of human stem cell- derived cardiomyocytes,” in Stem Cell-Derived Models in Toxicology, eds M. Clements and L. Roquemore (New York, NY: Humana Press), 57–82. doi: 10.1007/978-1-4939-6661-5
Odawara, A., Katoh, H., Matsuda, N., and Suzuki, I. (2016). Physiological maturation and drug responses of human induced pluripotent stem cell-derived cortical neuronal networks in long-term culture. Sci. Rep. 6:26181. doi: 10.1038/srep26181
Pessôa, L. V., de, F., Pieri, N. C. G., Recchia, K., and Bressan, F. F. (2020). “Induced pluripotent stem cells from animal models: applications on translational research,” in Novel Perspectives of Stem Cell Manufacturing and Therapies, eds D. Kitala and A. C. Maurício (Rijeka: IntechOpen). doi: 10.5772/INTECHOPEN.94199
Popp, B., Krumbiegel, M., Grosch, J., Sommer, A., Uebe, S., Kohl, Z., et al. (2018). Need for high-resolution Genetic Analysis in iPSC: results and lessons from the forips consortium. Sci. Rep. 8:17201. doi: 10.1038/s41598-018-35506-0
Potet, F., Egecioglu, D. E., Burridge, P. W., and George, A. L. (2020). GS-967 and eleclazine block sodium channels in human induced pluripotent stem cell-derived cardiomyocytes. Mol. Pharmacol. 98, 540–547. doi: 10.1124/MOLPHARM.120.000048
Prè, D., Nestor, M. W., Sproul, A. A., Jacob, S., Koppensteiner, P., Chinchalongporn, V., et al. (2014). A time course analysis of the electrophysiological properties of neurons differentiated from human induced Pluripotent Stem Cells (iPSCs). PLoS One 9:e103418. doi: 10.1371/journal.pone.0103418
Rajamohan, D., Kalra, S., Hoang, M. D., George, V., Staniforth, A., Russell, H., et al. (2016). Automated electrophysiological and pharmacological evaluation of human pluripotent stem cell-derived cardiomyocytes. Stem Cells Dev. 25, 439–452. doi: 10.1089/scd.2015.0253
Raman, I. M., and Bean, B. P. (1997). Resurgent sodium current and action potential formation in dissociated cerebellar Purkinje neurons. J. Neurosci. 17, 4517–4526. doi: 10.1523/jneurosci.17-12-04517.1997
Rosholm, K. R. (2019). Electrophysiological Characterization of Human iPSC-Derived Motor Neurons Using Qube 384 and QPatch. Sophion Application Report. Available online at: https://sophion.com/wp-content/uploads/2019/09/Electrophys-characterization-of-hiSPC-derived-motor-neurons-Qube-QPatch_AR_Public28033-5.pdf (accessed May 23, 2022).
Rosholm, K. R. (2021). hiPSC-Derived Cardiomyocyte Recordings Using Physiological Solutions on QPatch II. Sophion Application Report. Available online at: https://sophion.com/wp-content/uploads/2021/12/hiPSC-derived-cardiomyocyte-recordings-using-physiological-solutions-on-QPatch-II_AR_PUBLIC32856-3.pdf (accessed May 23, 2022).
Rosholm, K. R., Boddum, K., and Lindquist, A. (2021a). Perforated whole-cell recordings in automated patch clamp electrophysiology. Methods Mol. Biol. 2188, 93–108. doi: 10.1007/978-1-0716-0818-0_5
Rosholm, K. R., Chen, I. P., Sauter, D. R., Lindquist, A., Forman, T. E., Dwyer, S. A., et al. (2021b). Electrophysiological Characterization of iPSC-derived Cortical Neurons Using Automated Patch Clamp. Available online at: https://sophion.com/wp-content/uploads/2021/02/Electrophysiological-characterization-of-iPSC-derived-cortical-neurons-using-APC.pdf (accessed May 23, 2022).
Rosholm, K. R., Nagy, D., Liutkute, A., Seibertz, F., Voigt, N., and Lindquist, A. (2022). Electrophysiological Characterization of hiPSC-Derived Cardiomyocytes, Including Voltage-Gated Ion Channels and Action Potential Measurements, Using Automated Patch Clamp. Available online at: https://sophion.com/wp-content/uploads/2022/02/Electrophysiological-characterization-of-hiPSC-derived-CMs_BPS2022.pdf (accessed May 23, 2022).
Rowe, R. G., and Daley, G. Q. (2019). Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 20, 377–388. doi: 10.1038/s41576-019-0100-z
Seibertz, F., Reynolds, M., and Voigt, N. (2020). Single-cell optical action potential measurement in human induced pluripotent stem cell-derived cardiomyocytes. J. Vis. Exp. 166:e61890. doi: 10.3791/61890
Sharma, A., Li, G., Rajarajan, K., Hamaguchi, R., Burridge, P. W., and Wu, S. M. (2015). Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J. Vis. Exp. 97:52628. doi: 10.3791/52628
Shi, Y., Inoue, H., Wu, J. C., and Yamanaka, S. (2017). Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 16, 115–130. doi: 10.1038/nrd.2016.245
Strässler, E. T., Aalto-Setälä, K., Kiamehr, M., Landmesser, U., and Kränkel, N. (2018). Age is relative—Impact of donor age on induced pluripotent stem cell-derived cell functionality. Front. Cardiovasc. Med. 5:4. doi: 10.3389/fcvm.2018.00004
Sube, R., and Ertel, E. A. (2017). Cardiomyocytes derived from human induced pluripotent stem cells: an in-vitro model to predict cardiac effects of drugs. J. Biomed. Sci. Eng. 10, 527–549. doi: 10.4236/jbise.2017.1011040
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Press release (2012). The 2012 Nobel Prize in Physiology or Medicine – Press release – NobelPrize.Org. Available online at: https://www.nobelprize.org/prizes/medicine/2012/press-release/ (accessed January 14, 2022).
Tiburcy, M., Hudson, J. E., Balfanz, P., Schlick, S., Meyer, T., Liao, M. L. C., et al. (2017). Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation 135, 1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145
Toh, M. F., Brooks, J. M., Strassmaier, T., Haedo, R. J., Puryear, C. B., Roth, B. L., et al. (2020). Application of High-Throughput Automated Patch-Clamp Electrophysiology to Study Voltage-Gated Ion Channel Function in Primary Cortical Cultures. SLAS Discov. 25, 447–457. doi: 10.1177/2472555220902388
Vaidyanathan, R., Markandeya, Y. S., Kamp, T. J., Makielski, J. C., January, C. T., and Eckhardt, L. L. (2021). I K1 -enhanced human-induced pluripotent stem cell-derived cardiomyocytes: an improved cardiomyocyte model to investigate inherited arrhythmia syndromes. Am. J. Physiol. Heart Circ. Physiol. 310, 1611–1621. doi: 10.1152/ajpheart.00481.2015
Verkerk, A. O., Veerman, C. C., Zegers, J. G., Mengarelli, I., Bezzina, C. R., and Wilders, R. (2017). Patch-clamp recording from human induced pluripotent stemcell-derived cardiomyocytes: improving action potential characteristics through dynamic clamp. Int. J. Mol. Sci. 18:1873. doi: 10.3390/ijms18091873
Wu, J. C., Garg, P., Yoshida, Y., Yamanaka, S., Gepstein, L., Hulot, J. S., et al. (2020). Towards precision medicine with human iPSCs for cardiac Channelopathies. Circ. Res. 125, 653–658. doi: 10.1161/CIRCRESAHA.119.315209
Yang, X., Rodriguez, M. L., Leonard, A., Sun, L., Fischer, K. A., Wang, Y., et al. (2019). Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports 13, 657–668. doi: 10.1016/j.stemcr.2019.08.013
Yoshida, Y., and Yamanaka, S. (2017). Induced pluripotent stem cells 10 years later. Circ. Res. 120, 1958–1968. doi: 10.1161/CIRCRESAHA.117.311080
Young, G. T., Gutteridge, A., Fox, H. D. E., Wilbrey, A. L., Cao, L., Cho, L. T., et al. (2014). Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol. Ther. 22, 1530–1543. doi: 10.1038/mt.2014.86
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. doi: 10.1126/science.1151526
Keywords: hiPSC, stem cells, automated patch clamp, cardiomyocytes, neurons, ion channels
Citation: Rosholm KR, Badone B, Karatsiompani S, Nagy D, Seibertz F, Voigt N and Bell DC (2022) Adventures and Advances in Time Travel With Induced Pluripotent Stem Cells and Automated Patch Clamp. Front. Mol. Neurosci. 15:898717. doi: 10.3389/fnmol.2022.898717
Received: 17 March 2022; Accepted: 13 May 2022;
Published: 22 June 2022.
Edited by:
Marc Rogers, Metrion Biosciences Limited, United KingdomReviewed by:
Edward Humphries, AstraZeneca, United KingdomCopyright © 2022 Rosholm, Badone, Karatsiompani, Nagy, Seibertz, Voigt and Bell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kadla R. Rosholm, a3JyQHNvcGhpb24uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.