- 1Department of Geriatrics, The Second Xiangya Hospital of Central South University, Changsha, China
- 2Institute of Aging and Age-Related Disease Research, Central South University, Changsha, China
Aging-related neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS), are gradually becoming the primary burden of society and cause significant health-care concerns. Aging is a critical independent risk factor for neurodegenerative diseases. The pathological alterations of neurodegenerative diseases are tightly associated with mitochondrial dysfunction, inflammation, and oxidative stress, which in turn stimulates the further progression of neurodegenerative diseases. Given the potential research value, lncRNAs have attracted considerable attention. LncRNAs play complex and dynamic roles in multiple signal transduction axis of neurodegeneration. Emerging evidence indicates that lncRNAs exert crucial regulatory effects in the initiation and development of aging-related neurodegenerative diseases. This review compiles the underlying pathological mechanisms of aging and related neurodegenerative diseases. Besides, we discuss the roles of lncRNAs in aging. In addition, the crosstalk and network of lncRNAs in neurodegenerative diseases are also explored.
Introduction
Neurodegenerative diseases are a range of disorders characterized by irreversible neuron or myelin sheath loss and gliosis, which deteriorate over time and result in dysfunction. Aging significantly enhances the risk of developing neurodegenerative diseases (Hou et al., 2019). The world population over the age of 60 is expected to double to 22% by 2050. An increase in morbidity and mortality has been noted among the aging individuals (Divo et al., 2018). With aging populations, aging-related neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS), are prevalent worldwide. Therefore, it is important to gain a deeper insight into biological mechanisms underlying these diseases.
Although the function of protein aggregation in neurodegenerative diseases have received considerable attention, increasing evidence also focus on RNAs as contributing factors in these diseases. Because proteins are vital functional expression of genetic code, messenger RNAs (mRNAs) have been explored more intensively than non-coding RNAs (ncRNAs). Nevertheless, extensive research into the function of ncRNAs has broadened our understanding of diverse biological and pathological processes over the past few decades. Among the subtypes of ncRNAs, long non-coding RNAs (lncRNAs) account for a large proportion, which are a momentous source of molecular regulatory factors in eukaryotic nuclei and are involved in modulating gene expression, including chromatin structure, transcription, and translation (Xiang et al., 2014). LncRNAs are increasingly regarded as indispensable molecules in diverse cellular processes such as differentiation, proliferation, apoptosis and senescence (Mercer and Mattick, 2013; McHugh et al., 2015). Moreover, it has been shown that lncRNAs are associated with various pathological processes of aging-associated diseases, such as neurodegeneration, metabolic imbalances, and cancer (Greco et al., 2015). This review summarizes the pathological mechanisms of aging-related neurodegenerative diseases, including AD, PD, HD, and ALS. Then, we mainly concentrate on the functions of lncRNAs in the progression of these neurodegenerative diseases.
The Pathological Mechanisms of Aging-Related Neurodegenerative Diseases
With aging, the central nervous system (CNS) gradually undergoes degeneration, which is featured by a chronic and temporary loss of the function and structure of neuronal substances, eventually leading to mental and functional impairment (Campbell et al., 1999). Increasing evidence demonstrates that mitochondrial dysfunction, oxidative stress, and inflammation are primarily pathophysiological mechanisms of aging-related neurodegenerative diseases (Figure 1).
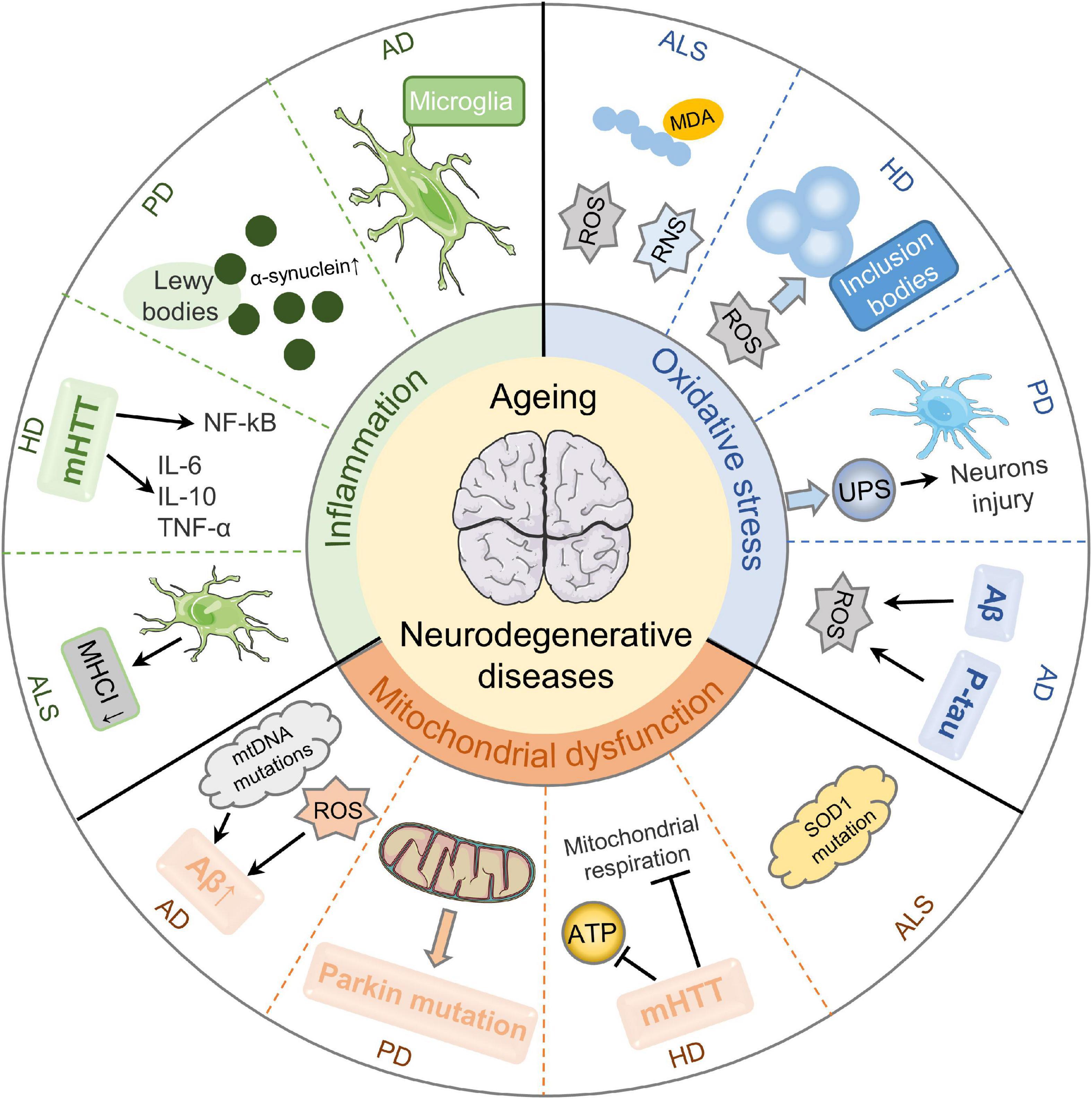
Figure 1. Pathological mechanisms of aging-related neurodegenerative diseases. Mitochondrial dysfunction, oxidative stress, and inflammation are essential cellular and molecular events in the pathogenesis of aging-related neurodegenerative diseases. Mitochondrial dysfunction results in Aβ deposition by stimulating mtDNA mutations and ROS production in AD. It can also aggravate the functions and mutations of Parkin, causing PD development. Besides, mHTT impairs mitochondrial ATP production and mitochondrial respiration in HD. SOD1 dysfunction is a cause of sporadic ALS. Secondly, oxidative stress is a key player in aging-related neurodegenerative diseases. Aβ-induced oxidative imbalance and P-tau protein are critical in the neurodegeneration of AD with oxidative stress. Enhanced oxidative stress causes UPS dysfunction and further aggravates the injury of dopaminergic neurons in PD substantia nigra. ROS plays a role in blocking neurotransmitter transmission in HD through stimulating protein misfolding and forming inclusion bodies. Besides, ROS/RNS overproduction is evident in ALS patients. Thirdly, neuroinflammation is implicated in the pathologic processes of aging-related neurodegenerative diseases. The inflammation of AD is mainly regulated by microglia in the innate immune response of the central nervous system. The accumulation and aggregation of α-synuclein in Lewy bodies are involved in the development of PD. The release of inflammatory cytokines induced by mHTT and the activation of NF-kB signaling pathway are the main inflammatory mechanisms of HD. The increased microgliosis and astrocytosis result in decreased MHCI level and ultimately contribute to neurotoxicity in ALS.
Mitochondrial Dysfunction
Mitochondria are pivotal modulators of cellular lifecycle, exerting an important role in aging-related neurodegenerative disorders. Emerging evidence suggests that mitochondrial dysfunction shows causal effects in the pathogenesis of these diseases.
Mitochondrial Dysfunction and Aging
Mitochondrial dysfunction accelerates aging primarily through two mechanisms, including stimulation of mitochondrial DNA (mtDNA) mutations and generation of reactive oxygen species (ROS). mtDNA mutations, such as point mutations or massive deletions, accumulate with age (Corral-Debrinski et al., 1992). The polymerase chain reaction (PCR) strategy revealed an average of two point mutations per 10 kb in the two protein-coding regions of mtDNA in the elderly subjects, compared with one point mutation per 10 kb in the young brains (Lin et al., 2002). The aggregation of mtDNA mutations with aging is relevant to the mitochondrial dysfunction. In addition, net production of ROS is another momentous mechanism and is considered to be important for mitochondria aging (Munro and Pamenter, 2019).
Mitochondrial Dysfunction and Alzheimer’s Disease
Studies have found that the typical histopathological changes of AD are amyloid deposition and neurofibrillary tangles (NFTs). There are many theories trying to explain this change, including the amyloid-β peptide (Aβ) waterfall theory, hyperphosphorylated tau (P-tau) protein theory, neurovascular hypothesis and so on (Ballard et al., 2011). Emerging evidence suggests that mitochondrial dysfunction is associated with AD pathogenesis (Lin and Beal, 2006). In transgenic amyloid precursor protein (APP) mice, oxidative damage preceded Aβ deposition and was associated with upregulation of apoptosis and mitochondrial metabolism related genes (Reddy et al., 2004).
Mitochondrial Dysfunction and Parkinson’s Disease
The level of dopaminergic (DA) neurons in the substantia nigra striatum decreases with age. Lewy bodies are unique cytoplasmic inclusion, immunostaining for α-synuclein and ubiquitin. Several causative genes, including DJ-1, PTEN-induced kinase 1 (PINK1), and Parkin (PARK2), strongly support mitochondria dysfunction as a crucial pathogenesis in PD (Lin and Beal, 2006). For example, Chung et al. (2004) found that the functions and mutations of Parkin could be exacerbated by mitochondrial dysfunction.
Mitochondrial Dysfunction and Huntington’s Disease
Huntington’s disease is caused by the amplification of CAG triplet repeat in the first exon of huntingtin (HTT) gene, resulting in abnormal forms of the protein to clump together and form aggregates in brain cells. Mitochondrial dysfunction is involved in HD process. Mitochondrial respiration and adenosine triphosphate (ATP) production of striatal cells were obviously impaired in mutant HTT (mHTT) embryos (Milakovic and Johnson, 2005). mHTT can directly act on mitochondria or indirectly affect mitochondrial function via altering transcription (Panov et al., 2002; Sadri-Vakili and Cha, 2006).
Mitochondrial Dysfunction and Amyotrophic Lateral Sclerosis
There are 2 types of ALS, among them about 90% are sporadic ALS (SALS) and 10% are familial ALS (FALS). Mutations in superoxide dismutase 1 (SOD1) gene cause 15% of FALS, while SOD1 dysfunction may also be a causal factor of sporadic ALS (Abati et al., 2020). Pathological biopsy of nerves and muscles revealed abnormal mitochondrial structure and localization in ALS. SOD1 mutations cause toxic aggregates formation in mitochondria, thereby disrupting several cellular processes and causing diverse detrimental effects (Hayashi et al., 2016). Animal studies showed that SOD1 mutation overexpression resulted in impaired mitochondrial energy metabolism (Mattiazzi et al., 2002).
Oxidative Stress
Oxidative stress is an imbalance between biological oxidative and antioxidative systems, which is caused by excessive ROS production (Newsholme et al., 2016). It plays deleterious effects in modulating the function of biomolecules that are sensitive to ROS/reactive nitrogen species (RNS), thus involving neuronal deterioration (Islam, 2017). In addition, the function of heavy metals as antioxidants in oxidative stress and their harmful effects on CNS are indisputable. Oxidative stress is associated with the occurrence and development of aging-related neurodegenerative diseases.
Oxidative Stress and Aging
Emerging evidence demonstrates that the free radical theory of aging has gradually become a major mechanism. Oxidative stress cause telomere dysfunction and shortening, which ultimately leads to cell senescence. ROS generation and response to oxidative stress are crucial factors in determining longevity (Finkel and Holbrook, 2000). Antioxidants, such as mental iron, copper, and zinc, are bioaccumulated by various activities. Decreased antioxidants and elevated ROS levels contribute to cellular senescence and thereby lead to various aging-related neurodegenerative diseases.
Oxidative Stress and Alzheimer’s Disease
The significance of oxidative stress and ROS in AD through harmful effects on biomolecules has been indicated. It has been reported that accumulation of Aβ aggregates exerts a crucial role in oxidative stress, causing energy failure and mitochondrial dysfunction (Tönnies and Trushina, 2017). Moreover, Aβ-induced oxidative imbalance is associated with increased levels of DNA/RNA oxidation, lipid peroxidation, and protein oxidation, suggesting a central role in AD (Wang X. et al., 2014; Butterfield and Boyd-Kimball, 2018). In addition, emerging evidence proves that metals accumulate in brain with aging and are involved in the pathogenesis of AD. Moreover, P-tau protein, the main constituent of NFTs, is involved in the cognitive decline and neurodegeneration with oxidative stress in AD (Goedert and Spillantini, 2006).
Oxidative Stress and Parkinson’s Disease
Motor dysfunction in PD is caused by dopamine depletion in the substantia nigra striatum pathway and dopaminergic neurons loss in the substantia nigra pars compacta (SNpc) (Ramalingam et al., 2019). Oxidative stress exhibits an inevitable effect in progressive neurodegenerative PD (Trist et al., 2019). Dopaminergic neurons are sensitive to mitochondrial ROS (Chen et al., 2019). ROS production is regulated by dopamine metabolism and glutathione in the SNpc (Smeyne and Smeyne, 2013). Patients with PD have enhanced levels of oxidized molecules and reduced glutathione (Homma and Fujii, 2015). The ubiquitin-proteasome system (UPS) reduces oxidative free radicals production (Collier et al., 2011). Enhanced oxidative stress lead to UPS dysfunction and further aggravate the injury and damage of dopaminergic neurons in PD substantia nigra (Betarbet et al., 2005).
Oxidative Stress and Huntington’s Disease
Although the primary cause of HD has been proved to be the toxicity of mHTT, diverse other processes have also been shown to be associated with HD (e.g., oxidative stress). Singh et al. (2019) revealed that oxidative stress exerts a vital role in neuronal degeneration cascade in HD. ROS causes the formation of inclusion bodies by inducing protein misfolding, which clump together in neurons and block neurotransmitter delivery (Rubinsztein and Carmichael, 2003). Repair of damaged DNA may result in instability and amplification of CAG nucleotide repeats in mHTT (Kolli et al., 2017).
Oxidative Stress and Amyotrophic Lateral Sclerosis
Studies have shown that oxidative stress regulated a range of cellular biological and pathological processes, including lipid peroxidation, protein injury, as well as DNA and RNA oxidation in ALS (Morimoto et al., 2020). SOD1 mutations and ROS/RNS overproduction are evident in ALS (Baltazar et al., 2014). Besides, the oxidative stress biomarkers can bind to biomolecules, such as malondialdehyde (MDA) modified protein and lipid peroxidation product in ALS (Mendez and Sattler, 2015).
Inflammation
Neurodegeneration, featured by the loss and progressive dysfunction of axons and neurons, is the main pathological feature of neurodegenerative disorders (Amor et al., 2010). Recent studies have shown the function of neuroinflammation in neurodegeneration, especially in the elderly. There is a growing awareness that inflammation is involved in the pathological process of aging-related neurodegenerative diseases. Although triggering events are varied, chronic immune activation is a common feature. Immune activation in the CNS exerts a pivotal role in the modulation of brain homeostasis during development and aging. When responding to tissue damage and pathogen invasion, nerve cells continuously survey the microenvironment and promote the inflammatory response, thereby further engaging self-limiting reaction through the immune system and starting tissue repair (Wyss-Coray and Mucke, 2002).
Inflammation and Aging
Inflammation plays a double-edged sword in the process of aging. Advantageously, it is closely associated with immunity by resisting pathogen invasion. Detrimentally, an excessive inflammatory response can disrupt the balance of the organism, which may eventually lead to disease. “Neuroinflammatory aging” is related with an obvious reduction of neuron numbers, neuronal dendrites, cortex and spine volume. The brain barrier is a structure that maintain the normal functional activities of neurons in the CNS, which is composed of cerebrospinal fluid-brain barrier (CBB), blood–brain barrier (BBB), and blood-cerebrospinal fluid barrier (BCB) (Abbott et al., 2010). The neuroinflammation sensitivity of different barriers and brain regions is significantly different (Stephenson et al., 2018). Changes in cells activation during aging explain the incremental susceptibility of the elderly to neuroinflammation and neurodegenerative diseases (Benz and Liebner, 2020).
Inflammation and Alzheimer’s Disease
Considerable evidence suggests that systemic inflammation is closely related to the pathogenesis of AD (Holmes, 2013). Systemic inflammation in AD could increase the expression of central proinflammatory cytokines, the death of neuronal cells, and the generation of ROS (Perry et al., 2010), thereby exacerbating the clinical symptoms. Moreover, the permeability of BBB gradually increases the progression of AD (Farrall and Wardlaw, 2009). The harmful effects of peripheral inflammation are associated with central inflammatory response because of the crosstalk between the periphery and the CNS. Therefore, it suggests that peripheral inflammation is a potential risk factor for AD progression. Inflammation in AD is primarily regulated by microglia in the innate immune responses of the CNS.
Inflammation and Parkinson’s Disease
Since PD-like symptoms were first observed in individuals infected with influenza virus, the function of inflammation in PD has been widely studied (Lutters et al., 2018). Inflammation contributes to the occurrence and development of PD both directly or indirectly. Studies have found that some viral proteins could promote the accumulation and aggregation of α-synuclein in Lewy bodies (Maries et al., 2003), further suggesting brain inflammation in the pathogenesis of PD. In addition, neurotropic pathogens could reach the basal ganglia via diverse pathways, and ultimately cause a series of neuroinflammation and neurodegenerative disorders in the nigrostriatal tract (Hawkes et al., 2007).
Inflammation and Huntington’s Disease
There is a growing interest of inflammation in HD progression. It has been demonstrated the higher expression of mHTT in monocytes and microglia (Moscovitch-Lopatin et al., 2010). Increasing evidence proves that mHTT influences the function of monocytes and microglia by mediating inflammation in HD (Valadão et al., 2020). On the one hand, mHTT directly induces the release of inflammatory cytokines (Gupta et al., 2021). On the other hand, mHTT promotes the release of inflammatory chemokines or cytokines by modulating nuclear factor-κB (NF-κB) signaling pathway (Khoshnan et al., 2004). Neuronal damage itself can cause a vicious cycle of inflammatory response and neurodegenerative events, which in turn leads to more neuronal death in HD (Zuccato et al., 2010). Moreover, a pioneer study observed the high levels of interleukin-6 (IL-6), IL-8, IL-10 and tumor necrosis factor-α (TNF-α) in HD patients, all of which were associated with disease progression (Silvestroni et al., 2009).
Inflammation and Amyotrophic Lateral Sclerosis
Neuroinflammation is a key modulator of ALS progression, and is featured by astroglia, microglia, infiltrating lymphocytes and peripheral monocytes in CNS (Lyon et al., 2019). Emerging evidence shows that inflammation of the innate immune system is linked to ALS (Lu et al., 2016). Major histocompatibility complex class I (MHCI) expression is increased in neuromuscular junction (NMJ) and peripheral motor axons and NMJs in SOD1 mutation mice (Nardo et al., 2016). Increased MHCI level exhibits a neuroprotective effect in peripheral nerves, depending on the removal of motor axon debris by immune cells (Thonhoff et al., 2018). MHCI level is decreased because of the increased microgliosis and astrocytosis in SOD1 mutation mice and ALS patients, ultimately promoting neurotoxicity (Song et al., 2016).
LncRNAs and Aging
Aging is a natural phenomenon featured by accumulation of degenerative alterations and damage. It is a primary risk factor in the etiology and development of various diseases, such as metabolic disruptions, neurodegenerative disorders, cardiovascular malfunctions, and cancers. Progressive loss of the functions of multiple cells, tissues, or organs is positively associated with aging. Though diverse mechanisms of aging have been studied, such as telomere shortening, free radicals, accumulated mutations, defective DNA repair, and increased DNA damage, the aging process remains widely unknown (Sinha et al., 2014). In fact, the current knowledge underscores the significance of multiple theories of aging. Therefore, it is best defined as a multifactorial process that involves complex interacting molecular and cellular mechanisms. Accordingly, there is currently no single measure that can be qualified as a specific biomarker or hallmarks of aging. A growing number of studies are exploring multiple biomarkers of aging at different levels, providing potential prospects for clinical diagnosis and therapy.
Cellular senescence is a causative process of aging and is responsible for aging-related diseases. It is a permanent state of cell cycle arrest, accompanied by an enhanced secretory phenotype and resistance to cell death. Cellular senescence can be induced by telomere attrition, DNA damage, mitochondrial dysfunction, chromosome destabilization, and oncogene activation (Wei and Ji, 2018). There are no specific or universal hallmarks for senescent cells. Several senescence biomarkers can be used to assess cellular senescence, such as higher activities of senescence-associated β-galactosidase (SA-β-gal), changes in senescence-related proteins (p16, p21, p27, and p53), alterations in cellular senescence-related morphology, and production of senescence-associated secretory phenotype (SASP) (Calcinotto et al., 2019; Ni et al., 2020). There is growing interest in discovering novel markers of senescence.
LncRNAs, a class of ncRNAs ≥ 200 nt in length, play an important role in many biological processes, such as transcription, post-transcriptional processing, and chromatin modification. According to their genomic location and orientation, they can be divided into sense, antisense, bidirectional, intergenic, intron, enhancer, and promoter lncRNAs (He et al., 2018; Cui et al., 2021). Mammalian transcription generates various lncRNAs involved in organ development (Rayner and Liu, 2016), differentiation (Chen and Zhang, 2016), synaptic formation (Maag et al., 2015), learning and memory (Gudenas and Wang, 2015). Increasing evidence has shown that a large number of lncRNAs are involved in cellular senescence at diverse stages of the cell cycle (Puvvula, 2019). With the accumulated evidence, we hereby discuss the association and correlation between lncRNAs and aging.
Firstly, lncRNAs play a role in cellular senescence and organismal aging by regulating cell cycle. For example, studies have demonstrated that depletion of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) could induce G1 or G1/S arrest, thereby enhancing senescence phenotype and inhibiting cell growth (Tripathi et al., 2013). LncRNA H19 is necessary in cell proliferation, growth, and senescence. Ratajczak (2012) proved that imprinting deletion of insulin-like growth factor-2 (Igf2)-H19 locus was involved in cellular senescence. Besides, p21-associated ncRNA DNA damage activated (PANDA) has been reported to trigger DNA damage via p53, leading to G1 cell cycle arrest (Wang Y. et al., 2017). In addition, several other lncRNAs are associated with aging process by affecting cell cycle, including Gadd7, 7SL, FAL1, etc. (Liu et al., 2012; Kour and Rath, 2016).
Secondly, lncRNAs participate in the progression of SASP and promote the secretion of inflammatory factor. LncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) is regarded as a novel inflammatory modulator. It can influence the formation of paraspeckles and subsequently regulate cellular senescence (McCluggage and Fox, 2021). Moreover, lncRNA LET stimulates the accumulation of nuclear factor 90 (NF90), which inhibits the translation of several SASP factors (Tominaga-Yamanaka et al., 2012; Yang et al., 2013). Cui et al. (2014) revealed that lnc-IL7R could alleviate the inflammatory response induced by lipopolysaccharide, suggesting an involvement in SASP production. Additionally, lincRNA-Cox2, Lethe, and THRIL have been shown to contribute to the generation of SASP (Panda et al., 2017).
Thirdly, lncRNAs is involved in telomere dynamics and attrition. TElomere Repeat-containing RNA (TERRA) is a lncRNA transcribed by telomeric DNA sequences. Many studies have been conducted on how TERRA transcription modulates telomere structure (Aguado et al., 2020). Yehezkel et al. (2013) confirmed an association between TERRA and aging process. They found that TERRA expression was upregulated, telomere shortening was accelerated, and replicative senescence enters prematurely in immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome type I. Moreover, telomeric RNA component (TERC), a 451 nt lncRNA, is the core component of telomerase with classical function to provide template for the extension of telomerase. Introduction of TERC rescued premature aging phenotype of telomerase deficient mice, suggesting a role of TERC in senescence and aging (Samper et al., 2001).
Lastly, lncRNAs contribute to recruitment of chromatin remodeling complexes during senescence and aging. HOX antisense intergenic RNA (HOTAIR) plays a role in the recruitment and binding of chromatin remodeling complexes to the HOX sites, leading to retargeting of polycomb repressive complex 2 (PRC2) (Rinn et al., 2007). Besides, KCNQ-overlapping transcript 1 (Kcnq1ot1) has been reported to affect Kcnq1 locus by interacting with PRC2 complexes (Pandey et al., 2008). Additionally, recent studies have demonstrated that several other lncRNAs, such as Air, H19, and TERRA are also involved in chromatin remodeling (Zhu et al., 2013; He et al., 2018).
Roles of lncRNAs in Aging-Related Neurodegenerative Diseases
Aging-related neurodegenerative diseases are a range of progressive atrophy and loss of neurological function of neurons and neural tissues, eventually leading to cognitive or motor impairments. Although common mechanisms may result in neuron loss in a variety of disorders, different pathological characteristics are caused by specific toxic aggregation of particular proteins and/or genetic mutations. For example, Aβ aggregation and P-tau in AD, α-synuclein Lewy bodies in PD, mHTT aggregates in HD and SOD1 in ALS (Chung et al., 2011). Increasing evidence indicates that diverse lncRNAs are involved in neural function, and their related RNA networks may influence neurodegeneration (Wu and Kuo, 2020). Since the pathology of neurodegenerative diseases are related to accumulation of certain proteins, studies have suggested that lncRNAs are associated with different protein aggregation events and disease pathogenesis. Abnormal regulation of lncRNAs is related to the aging process and aging-related diseases. In the following sections, we summarize the roles of lncRNAs in aging-related AD, PD, HD, and ALS.
Roles of lncRNAs in Alzheimer’s Disease
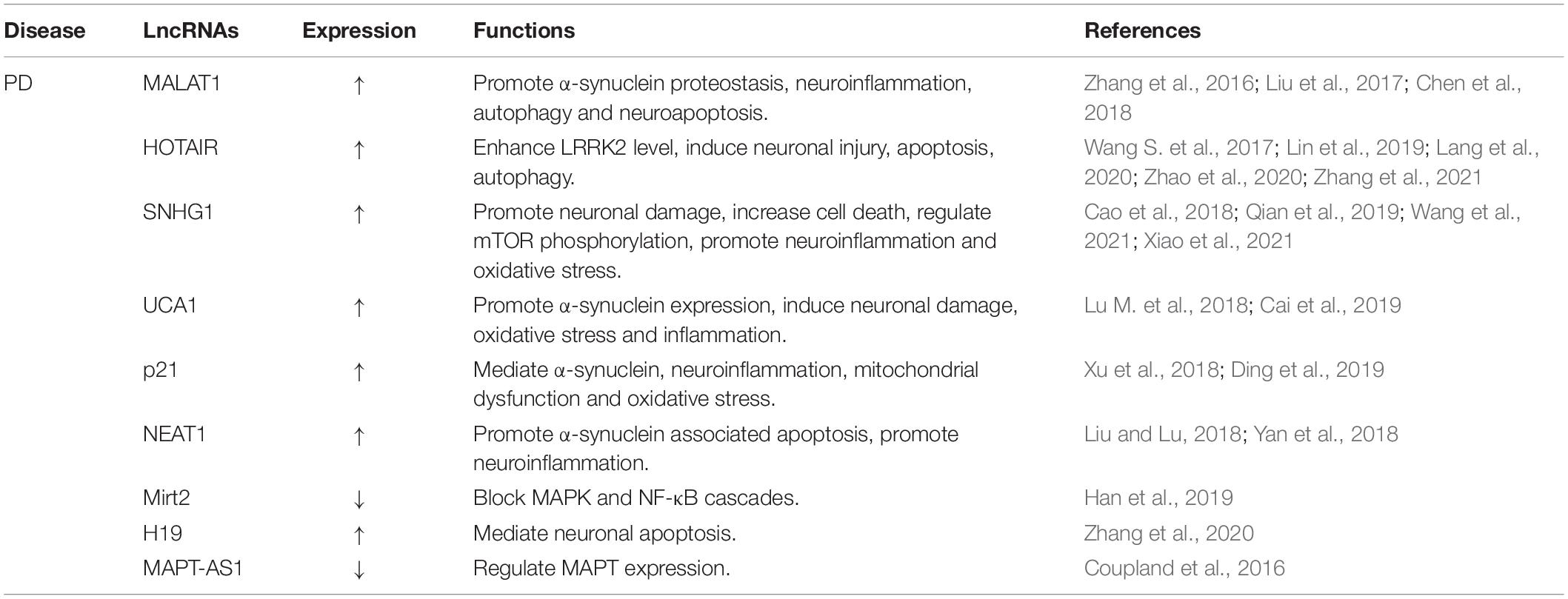
Alzheimer’s disease, the major etiology of dementia, is characterized by aggregation of Aβ peptides and P-tau in neurofibrillary tangles. APP is a transmembrane protein that produces Aβ by sequential division of β-site APP cleaving enzyme-1 (BACE1) and γ-secretase (Karran et al., 2011). The balanced ratio of Aβ42/Aβ40 is disrupted and cause the amyloid plaques in AD brain (Ballard et al., 2011). It has been demonstrated that multiple lncRNAs are involved in AD pathology (Figure 2). In this section, we compile the roles of lncRNAs in aging-related AD (Table 1).
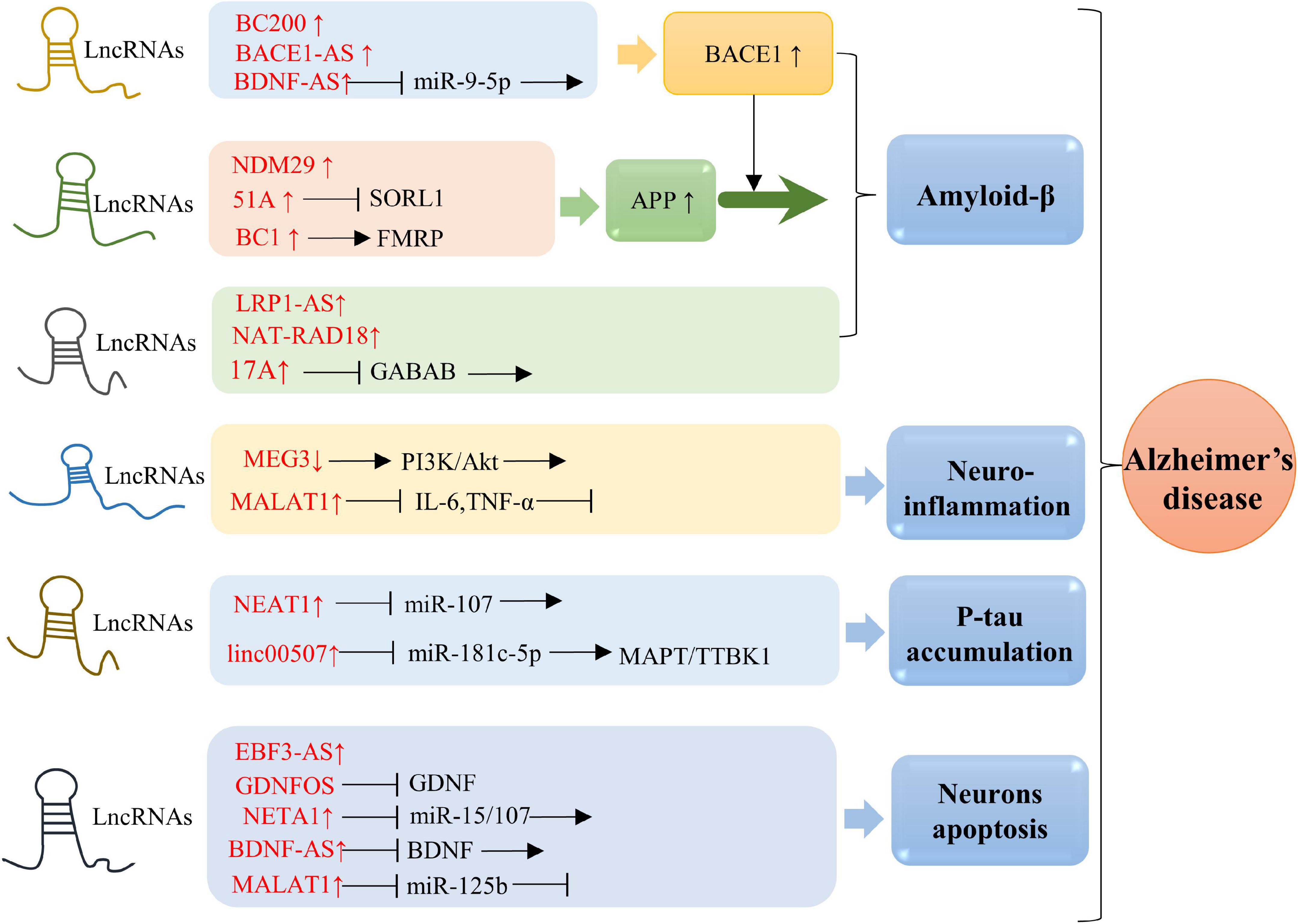
Figure 2. Molecular mechanisms of lncRNAs in AD. Overexpression of BACE1-AS, BC200, and BDNF-AS is associated with BACE1 activity. Elevated level of 51A in AD patient altered splicing mode of SORL1, causing damage to APP processing and leading to promotion of the Aβ deposition. Upregulation of BC1 induces APP mRNA translation by binding to FMRP. Enhanced expression of NDM29 induces APP synthesis. Overexpression of LRP1-AS is involved in regulating Aβ accumulation. NAT-RAD18 level is upregulated in response to Aβ40. Overexpression of 17A is tightly associated with Aβ secretion and Aβ42 production. Down-expression of MEG3 promotes neuroinflammation via PI3/Akt pathway. Significantly upregulated expression of MALAT1 inhibits neuroinflammation through reducing IL-6 and TNF-α. NEAT1 and linc00507 have been characterized to be involved in Tau. Upregulation of NEAT1 promoted Tau protein phosphorylation by sponging miR-107, while elevated level of linc00507 enhances hyperphosphorylation of Tau protein through regulating miR-181c-5p/MAPT/TTBK1 pathway. Upregulation of NETA1 and MALAT1 promoted cell apoptosis by targeting miR-15/107 and miR-125b, respectively. In addition, EBF3-AS exhibits a role in regulating neurons apoptosis.
BACE1 has been extensively recognized as the potential to develop Aβ-lowering drug therapy for AD. BACE1 antisense transcript (BACE1-AS) is a conserved lncRNA transcribed from the antisense protein-coding BACE1 gene (Fotuhi et al., 2019). Faghihi et al. (2008) reported that BACE1-AS realizes its function via raising BACE1 mRNA stability and then producing additional Aβ42, suggesting the driving function of BACE1-AS in AD pathologic process. They also demonstrated that BACE1-AS could prevent binding to miR-485-5p. HuD, one of the main neuronal RNA-binding proteins (RBPs), is involved in mediating neuronal maintenance, differentiation and plasticity, thus regulating memory and learning. HuD could interact with BACE1 mRNA, APP mRNA, and BACE1-AS to prolong the half-lives of the mRNA and enhance the expression of BACE1 (Deschênes-Furry et al., 2006). Kang et al. (2014) found elevated expressions of HuD and BACE1-AS in AD brains.
Brain cytoplasmic 200 (BC200) is a major cytoplasmic lncRNA, which is mainly expressed in neurons and can be transported to dendrites. It is a translational modulator that selectively targets the somatic dendrite domain of neurons. BC200 is believed to act as a regulator of local protein synthesis in the postsynaptic dendrite microdomain and is involved in maintaining long-term synaptic plasticity (Mus et al., 2007). BC200 has been reported to exhibit important effects in human lung, esophagus, and cervix tumors (Hu and Lu, 2015). Mus et al. (2007) proved that BC200 RNA expression was markedly specific increased in AD brains. They also indicated that elevated BC200 levels was paralleled with the severity of AD. In addition, studies have shown that BC200 upregulation directly promote BACE1 level and impair cell viability subsequently, thereby increasing Aβ42 expression (Feng et al., 2018; Li et al., 2018).
Brain cytoplasmic 1 (BC1) contains a 5′ stem-loop domain, followed by a single-stranded central homopolymer A-rich region and a 3′-stem-loop domain (Lin et al., 2008). BC1 acts as a translational repressor, which is modulated by the adjacent A-rich region and 3′ stem-loop through interactions with poly(A)-binding protein (PABP), eukaryotic initiation factor 4A (eIF4A), and eIF4B (Kondrashov et al., 2005; Lee et al., 2020). It is abundant in the synapse and inhibits translation at initiation. BC1 is a cytoplasmic lncRNA in neurons that promotes APP mRNA translation through the interaction with the fragile X syndrome protein (FMRP) (Aleshkina et al., 2021). Zhang et al. (2018) demonstrated that inhibition of BC1 blocked the accumulation and aggregation of Aβ in AD mice brains and protected them from memory and learning deficits. In contrast, exogenous overexpression of BC1 caused Aβ peptides aggregation and induced learning and memory disorders.
NEAT1 consists of two subtype transcripts, NEAT1v1 and NEAT1v2, which are essential components of para-nuclear plaque formation (Koyama et al., 2020). LncRNA NEAT1 has been reported to be widely expressed in many mammalian cells. Increasing evidence demonstrates that NEAT1 plays crucial roles in multiple pathophysiological processes including neurodegenerative diseases (An et al., 2018), cancers (Yu et al., 2017), immune disorders (Taheri et al., 2020), and etc. Previous studies have shown that NEAT1 is involved in the occurrence of AD. Ke et al. (2019) found that NEAT1 exacerbated P-tau expression, Aβ level, and neuron damage via sponging miR-107, thus boosting AD progression. Besides, it has been reported that NEAT1 is involved in AD through modulating the miR-124/BACE1 signaling pathway (Zhao et al., 2019). Additionally, Spreafico et al. (2018) indicated that NEAT1 facilitated neuronal cell death via regulating the expression of microRNA-15/107 family in postmortem AD tissues. MALAT1, also known as NEAT2, has recently been implicated in neurodegenerative diseases due to anti-inflammatory property (Masoumi et al., 2019). Ma et al. (2019) indicated that MALAT1 interacted with miR-125b to promote neurite growth while inhibit neuronal apoptosis and suppress inflammatory cytokines in AD.
LncRNA 17A is synthesized by RNA polymerase III and located at intron 3 of g protein-coupled receptor 51 gene (Luo and Chen, 2016). LncRNA 17A could damage gamma-aminobutyric acid type B (GABAB) signal transduction by generating non-functional receptor isoforms, enhance the Aβ42/Aβ40 peptide ratio, and promote neurodegeneration (Massone et al., 2011). Moreover, lncRNA 17A expression has been found to be upregulated in AD patients. Besides, inflammation of brain tissue triggers 17A expression and complicates the AD process (Wang X. et al., 2019).
LncRNA 51A locates in an antisense configuration on intron 1 of the neuronal sortilin-related receptor (SORL1) gene. SORL1 has been reported to interact with APP, affect transport and proteolysis, and participate in AD pathogenesis (Ciarlo et al., 2013). Decreased SORL1 level shifts APP from the reverse transcriptional cycle to the β-secretase cleavage pathway, thus increasing APP secretion and subsequent Aβ formation. It has been proposed that overexpression of 51A promotes Aβ formation by decreasing SORL1 variant A, thereby increasing susceptibility to AD (Ma et al., 2009).
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophic factor family, is a widely studied growth factor which includes neurotrophin-3 (NT3), neurotrophin-4 (NT4), and nerve growth factor (NGF) (Björkholm and Monteggia, 2016). BDNF has a profound impact on brain morphology, development, and function because it is widely expressed in the CNS. BDNF-antisense (BDNF-AS) is a lncRNA with dozens of alternate splicing variants transcribed from 11p14 cytogenetic band. LncRNA BDNF-AS continuously reduces endogenous BDNF protein levels and functions by altering the chromatin structure of the BDNF region, thereby inhibiting the expression of BDNF sense transcripts (Modarresi et al., 2012). It is involved in a variety of processes in the CNS, including synaptic plasticity, synapse formation, and neuronal maturation. Abnormal regulation of BDNF-AS is implicated in AD process. Guo et al. (2018) found an increased level of BDNF-AS and a reduced expression of BDNF, accompanied by enhanced apoptosis induction and decreased cell viability in an AD cell model established by Aβ25–35 exposure to PC12 cells. They also demonstrated that BDNF-AS silencing had crucial beneficial effects on enhancing cell viability, and suppressing oxidative stress and apoptosis through negative modulation of BDNF. Ding et al. (2021) uncovered an elevated expression of BDNF-AS in the peripheral blood of AD patients. Further mechanism studies showed that BDNF-AS acted as a ceRNA to competitively bind miR-9-5p to induce BACE1 expression, ultimately promoting neurotoxicity.
The human SOX2 gene encodes a protein of 317 amino acids (Stevanovic et al., 1994). The structural core of SOX2 is its high-mobility-group (HMG) domain, which contains a nuclear export signal and a nuclear localization in addition to binding to specific DNA consensus sequences (Novak et al., 2020). The transcription factor SOX2 is a single exon protein, which exerts an essential and pleiotropic role in development and homeostasis. LncRNA SOX2 overlapping transcript (SOX2OT) is located at 3q26.3 on human chromosome and is constitutive of more than two transcription start sites and ten exons (Wang et al., 2020). The intronic region of SOX2OT contains a SOX2 structure and shares the same transcriptional direction (Fantes et al., 2003). Arisi et al. (2011) revealed that SOX2OT reduces neurogenesis by mediating SOX2 gene expression in AD. It has been proved that abnormal expression or activation of 10–11 translocation-2 (TET2) is closely associated with AD (Carrillo-Jimenez et al., 2019). Li L. et al. (2021) demonstrated that TET2 was involved in neuron formation via modulating several lncRNAs (SOX2OT, MALAT1, etc.).
Glial cell line-derived neurotrophic factor (GDNF) is a 134 amino acid protein in the GDNF family ligands (GFLs) including artemin, persephin, and neurturin (Cintrón-Colón et al., 2020). It is an effective nutrient factor for central norepinephrine neurons, midbrain dopaminergic neurons, spinal motoneurons and peripheral neurons. GDNF opposite strand (GDNFOS), as its name suggests, is transcribed from the opposite strand of GDNF gene in the primate genome. GDNFOS isoforms have been found to be differentially expressed in tissue expression patterns and could regulate the level of endogenous GDNF in AD brains (Airavaara et al., 2011).
Early B cell factor 3 (EBF3) belongs to the Collier/Olf/EBF (COE) transcription factor family. It is an evolutionarily conserved atypical transcription factor thought to affect the stratification of cerebral cortex. Gu et al. (2018) revealed that lncRNA EBF3 antisense (EBF3-AS) expression was enhanced in hippocampus of AD mice. Moreover, they also found that EBF3-AS was involved in the regulation of neurons apoptosis in AD cell models induced by okadaic acid (OA) and Aβ25–35.
Neuroblastoma differentiation marker 29 (NDM29) is a lncRNA transcribed by RNA polymerase III. NDM29 markedly enhances APP level, which in turn increases generation of the two major Aβ isoforms (Li D. et al., 2021). LncRNA NDM29 was reported to induce APP synthesis and accelerate the cleavage through γ-secretase and BACE1, which can be restrained by anti-inflammatory agents and accelerated by inflammatory stimulation (Massone et al., 2012).
Low-density lipoprotein receptor (LDLR)-related protein (LRP1) is a large endophagocytic and signal transduction receptor in the LDLR gene family. It is widely expressed in the brain. Apolipoprotein E (ApoE), the ligand of LRP1, is involved in senile plaques in AD brains (Rebeck et al., 1993), implicating a role for LRP1 in the accumulation of Aβ. LRP1-antisense (LRP1-AS) negatively mediates LRP1 expression at both protein and RNA levels. It was reported that LRP1-AS expression was significantly increased in the AD brain (Yamanaka et al., 2015).
In addition, a variety of other lncRNAs exhibit an indispensable role AD progression. For instance, Yan et al. (2020) demonstrated that elevated linc00507 level in AD models promoted P-tau accumulation through miR-181c-5p/microtubule-associated protein tau (MAPT)/tau-tubulin kinase-1 (TTBK1) network. LncRNA MEG3 is an imprinted gene mapped in human chromosome 14 and mouse chromosome 12. Yi et al. (2019) found a downregulated level of MEG3 in AD mice. They proved that upregulation of MEG3 could decrease Aβ expression, reduce inflammation injury, and protect neurons by phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway. RAD18 is a member of the chromosome family responsible for repairing DNA damage (Hedglin and Benkovic, 2015). NAT-RAD18 is a lncRNA of the natural antisense transcript against RAD18. It was found that RAD18 expression was down-regulated, while NAT-RAD18 level was up-regulated in response to Aβ40 (Parenti et al., 2007).
Roles of lncRNAs in Parkinson’s Disease
Parkinson’s disease is pathologically characterized by α-synuclein aggregation in Lewy bodies. Several site mutations are recognized to cause hereditary PD, including α-synuclein, Parkin, DJ-1, LRRK2, PINK1, and ATP13A2 (Majidinia et al., 2016). Neurotoxins such as 6-hydroxydopamine (6-OHDA) and MPTP/MPP+ have been often utilized to induce degeneration dopaminergic neuron degeneration and reproduce pathological features of PD (Nagel et al., 2009). Accumulating evidence proposes that lncRNAs are closely related to the development of PD (Figure 3). In this section, we discuss the roles of lncRNAs in aging-related PD (Table 2).
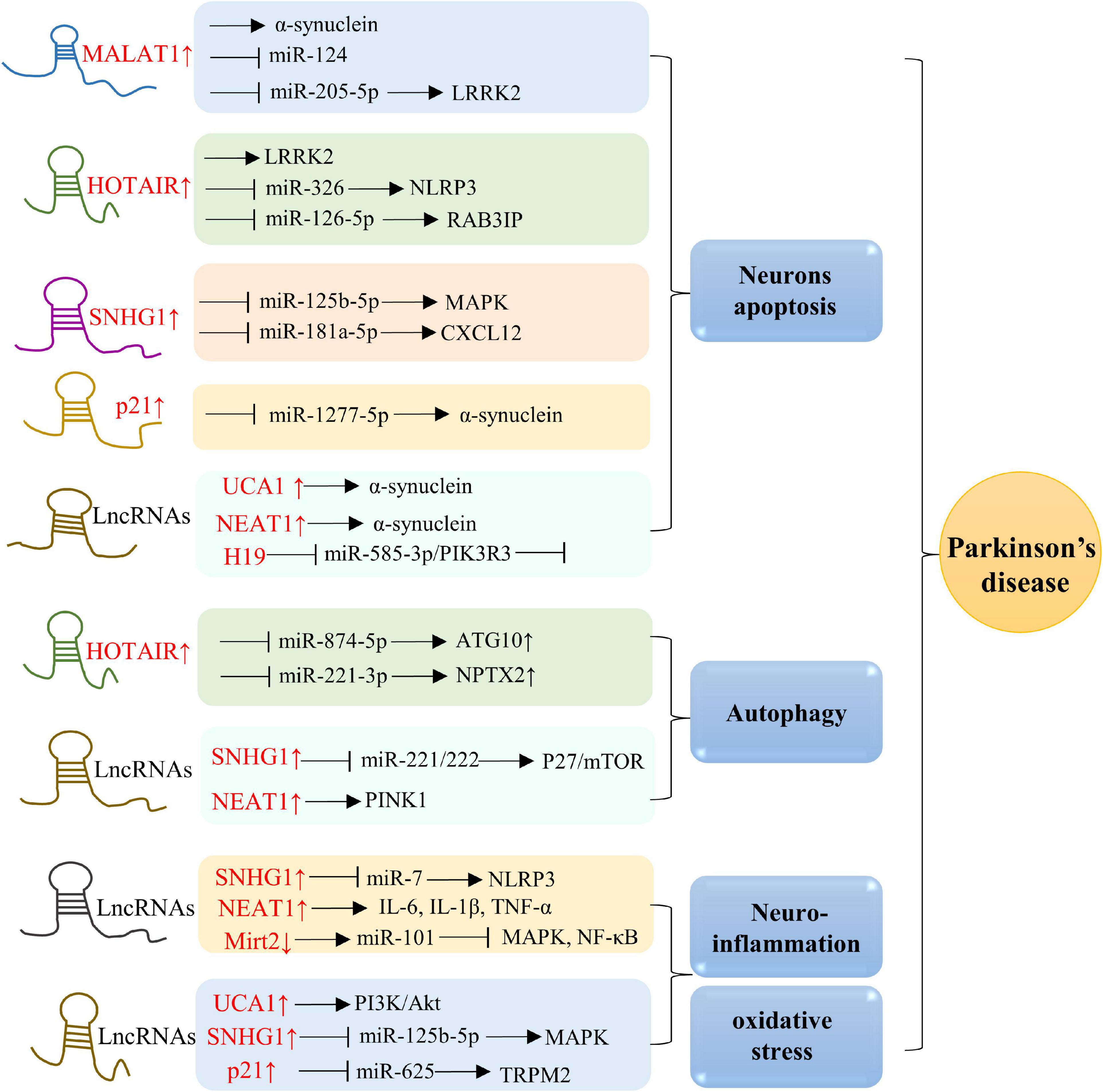
Figure 3. Molecular mechanisms of lncRNAs in PD. Overexpression of MALAT1, HOTAIR, SNHG1, p21, UCA1, NEAT1, and H19 are in associations with neurons apoptosis. Besides, lncRNAs, including HOTAIR, SNHG1, and NEAT1, also play a role in modulating autophagy. Additionally, elevated levels of SNHG1, NEAT1, Mirt2, UCA1, and p21 in AD are positively related to oxidative stress and neuroinflammation.
The original sequence of MALAT1 gene is over 8,000 bp, which is highly conserved in 33 species of mammals (Ji et al., 2003). It is also named as NEAT2. Given that MALAT1 is expressed in a variety of nerve cells in the brain, it is not surprising that MALAT1 exerts diverse roles in normal brain physiology. Increasing evidence observes dysregulated MALAT1 in PD progression. MALAT1 is upregulated in MPP+ induced PD cells and MTPT-induced PD mice (Kraus et al., 2017). Studies have found that MALAT1 participates in PD pathology through regulating various mechanisms, including α-synuclein proteostasis, neuroinflammation, autophagy, and neuroapoptosis. LRRK2 mutation is one of the causes for inherited and sporadic PD (Jankovic and Tan, 2020). Liu et al. (2017) found a reversed expression between MALAT1 and miR-124 in MPTP-induced PD mice. Further mechanistic studies showed that MALAT1 promoted apoptosis by interacting with miR-124 and negatively modulating its expression. Zhang et al. (2016) proved that MALAT1 could increase the expression of a-synuclein protein by binding with it to enhance the stability. Besides, Chen et al. (2018) revealed elevated levels of MALAT1 and LRRK2, and decreased expression of miR-205-5p in MPTP-induced PD mice. They proposed that the MALAT1/miR-205-5p axis modulates cellular apoptosis by targeting LRRK2.
LncRNA HOTAIR is transcribed from the antisense chain of homologous frame C locus on chromosome 12. HOTAIR has been extensively explored in human cancer (Rajagopal et al., 2020). Molecular mechanisms of HOTAIR in cancer progression include recruitment of lysine specific demethylase 1 (LSD1) complexes and PRC2, histone 3 lysine 4 (H3K4) demethylation and histone 3 lysine 27 (H3K27) methylation (Qu et al., 2019). Recent studies have found the functions of HOTAIR in PD progression. HOTAIR has been reported to accelerate MPP+-induced neuron damage by mediating the miR-874-5p/autophagy-related 10 (ATG10) axis in PD (Zhao et al., 2020). Besides, Wang S. et al. (2017) demonstrated that HOTAIR could enhance the expression and stability of LRRK2, thus involving in PD process. Rab3a interacting protein (RAB3IP), is a Rab-specific guanine nucleotide exchange factor (GEF) (Tong et al., 2021). Lin et al. (2019) indicated the function of the cellular HOTAIR/miR-126-5p/RAB3IP signaling pathway in PD. NOD-like receptor family pyrin domain containing 3 (NLRP3) is a prominent inflammasome in immune system and contributes to several disease. HOTAIR targeted miR-326 to promote the neuronal injury through facilitating NLRP3 mediated apoptosis in PD (Zhang et al., 2021). Additionally, Lang et al. (2020) explored that HOTAIR enhanced neuronal pentraxin II (NPTX2) through targeting miR-221-3p, thereby driving autophagy in PD mouse models.
LncRNA small nucleolar RNA host gene 1 (SNHG1), also known as linc00057, is approximately 3,927 bases in length (Tycowski et al., 1994). Studies has focused on its roles as a competing endogenous RNA (ceRNA) to modulate tumorigenesis (Lu Q. et al., 2018). SNHG1 attenuates p53 expression, thus promoting cell migration, proliferation, and invasion (Shen et al., 2017). Notably, SNHG1 has been recognized as a key modulator of PD neurotoxicity. The C-X-C motif chemokine ligand 12 (CXCL12) binds to its receptors to induce downstream signaling pathways (Shi et al., 2020). Wang et al. (2021) demonstrated that SNHG1 stimulated MPP+ induced neuronal damage through mediating miR-181a-5p/CXCL12 axis. Besides, Qian et al. (2019) indicated another mechanistic pathway by which SNHG1 targets miR-221/222 and subsequently modulates p27/mammalian target of rapamycin (mTOR) expression in MPP+-induced PD models. Moreover, SNHG1 regulates PD by participating in miR-7/NLRP3 pathway to promote neuroinflammation (Cao et al., 2018). Moreover, Xiao et al. (2021) proved that SNHG1 could also promote oxidative stress, apoptosis, and inflammation by modulating the miR-125b-5p/mitogen-activated protein kinase 1 (MAPK1) axis in models of PD.
LncRNA urothelial carcinoma-associated 1 (UCA1) was first identified in human bladder transitional cell line (Wang et al., 2006). Increasing evidence demonstrates that UCA1 is involved not only in many tumor diseases, but also in regulating neurodegenerative disorders. LncRNA UCA1 has been widely studied in the pathogenesis and development of PD. Studies have demonstrated that UCA1 participates in regulating α-synuclein aggregation, dopaminergic neuroapoptosis, and neuroinflammation. Lu M. et al. (2018) explored that UCA1 promoted the expression of a-synuclein in PD development. Besides, Cai et al. (2019) showed that UCA1 could aggravate dopaminergic neuronal damage, inflammation, and oxidative stress by promoting the PI3K/Akt axis, while reduction of UCA1 exerted opposite effects in a PD rat model.
LncRNA-p21 has been studied to regulate PD via mediating α-synuclein, mitochondrial dysfunction, oxidative stress and neuroinflammation. Xu et al. (2018) indicated that lncRNA-p21 stimulated cellular apoptosis and suppressed cell viability by sponging miR-1277-5p and indirectly enhancing α-synuclein level in PD. The transient receptor melastatin 2 (TRPM2) is a non-selective Ca2+ osmotic channel elevated in PD brains (Belrose and Jackson, 2018). It has been found that lncRNA-p21-miR-625-TRPM2 axis has pivotal effects in oxidative stress and neuroinflammation in PD models (Ding et al., 2019).
LncRNA NEAT1 is also referred to PD progression. Overexpression of NEAT1 was positively correlated with the concentration of MPTP and promoted the stability of PINK1 protein (Yan et al., 2018). Mechanistically, NEAT1 positively modulated PINK1 level through stabilizing PINK1 protein and suppressing PINK1 protein degradation. The levels of TNF-α, IL-6, and IL-1β, were up-regulated in MPP+-induced PD models, implying that NEAT1 is closely correlated with neuroinflammation (Yan et al., 2018). In addition, Liu and Lu (2018) showed that NEAT1 could promote α-synuclein-related apoptosis in PD.
In addition, many other lncRNAs play important roles PD process. For example, myocardial infraction associated transcript 2 (Mirt2) exerts anti-inflammatory effects in many cell types, while PD is often associated with excessive inflammation. Therefore, Han et al. (2019) proved that miR-101 inhibited by Mirt2 led to the blocking of MAPK and NF-κB cascades, which might be important in the treatment of PD. Besides, Zhang et al. (2020) demonstrated that lncRNA H19 mitigated neuronal apoptosis by targeting miR-585-3p/phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3) in MPP+ treated neuroblastoma cells and MPTP-induced PD mice. LncRNA microtubule-associated protein tau antisense RNA 1 (MAPT-AS1) locates on the antisense chain of the promoter region of MAPT, which is believed to associated with disease state of PD (Wang D. et al., 2019). It has been found that MAPT-AS1 is an underlying epigenetic modulator of MAPT expression in PD (Coupland et al., 2016).
Roles of lncRNAs in Huntington’s Disease
Huntington’s disease is a neurodegeneration caused by a CAG repeat in the gene encoding HTT protein (Jimenez-Sanchez et al., 2017). Brain-derived neurotrophic factor (BDNF), a neurotransmitter regulator, is involved in neuroplasticity and is critical for its survival and growth (Sharma et al., 2021). It has been found that BDNF expression reduces in HD (Humbert and Saudou, 2005). Repressor element 1-silencing transcription factor (REST) has the capacity to modulate the epigenome and transcriptome (Buckley et al., 2010). HTT blocks REST-mediated transcriptional inhibition through the cytoplasmic complex of HTT-associated protein 1 (HAP1). Therefore, REST can assemble the repressor complex in the nucleus in mHTT (Hwang and Zukin, 2018). PRC2 catalyzes mono-, di-, and trimethylation of H3K27, acting as a chromatin associated methyltransferase (Laugesen et al., 2019). Notably, major targets of these genes are p53 and REST, while some of which interact with the PRC2 complex. Emerging evidence indicates that some lncRNAs are closely associated with the etiology of HD.
LncRNA human accelerated region 1 (HAR1), a direct target of REST, is a region where the sequence has been obviously changed. It is a constituent of two overlapping lncRNA loci, including HAR1A and HAR1B (Waters et al., 2021). Johnson et al. (2010) found HAR1 expression was significantly lower in HD patients. Further mechanistic study revealed that HAR1 was repressed by REST by specific DNA regulatory motifs, suggesting that the roles of HAR1 in HD progression.
MEG3 is also known as gene trap locus 2 (Gtl2). LncRNA MEG3 regulates a range of aging-related neurodegenerative diseases. Inactivation of Meg3 results in a marked increase in microvascular formation and angiogenesis-promoting gene expressions in the brain (Zhou et al., 2012). MEG3 is another target of REST and is associated with PRC2 complex (Johnson, 2012; Chang et al., 2017). Chanda et al. (2018) found that after MEG3 knockout in HD cell models, mHTT aggregates were markedly reduced and endogenous Tp53 levels were down-regulated.
Taurine-upregulated gene 1 (TUG1) is actively involved in numerous physiological processes, including modulating genes at epigenetics, transcription, post-transcription, translation, and post-translation (Guo et al., 2020). Studies have demonstrated that lncRNA TUG1 is closely associated with PRC2 among HD-related lncRNAs. Since the interaction of epigenetic regulatory complex of PRC2, the altered expression of TUG1 is related to diverse molecular pathways in HD brain. Khalil et al. (2009) indicated that TUG1 regulated the cytotoxicity of mHTT through p53.
LncRNA DiGeorge syndrome critical region gene 5 (DGCR5), also known as linc00037, is a REST-regulated lncRNA in neurodegeneration (Dong et al., 2018). The downregulation of DGCR5 in HD brain implies that DGCR5 is closely associated with transcriptional regulation in the progression of HD (Johnson et al., 2009).
NEAT1 is essential for structural integrity of the nuclear paraspeckle. A reanalysis of microarray data revealed an improvement in HD patients compared to the control group (Hodges et al., 2006). Cheng et al. (2018) confirmed that NEAT1_2 detected a threefold increase in the brains of HD patients. Besides, it has been proved that NEAT1 is overexpressed of in HD model brain of transgenic mice (Chanda et al., 2018). Moreover, the roles of NEAT1 were also demonstrated in cell models of HD (Sunwoo et al., 2017).
Roles of lncRNAs in Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis primarily targets motor neurons, leading to serious disability and ultimately death from respiratory failure (Hardiman et al., 2017). RBPs mainly include fused in sarcoma/translated in liposarcoma (FUS/TLS) and TAR DNA-binding domain protein 43 (TDP43), which is involved in regulating RNA metabolism (Tischbein et al., 2019). It has been revealed that the abnormal aggregation of TDP43 and FUS/TLS directly causes the misfolding of wild-type SOD1 (wtSOD1) in ALS (Pokrishevsky et al., 2012). In FALS and SALS, the most common candidate mutations are SOD1, FUS, and C9orf72 gene both (DeJesus-Hernandez et al., 2011; Ajroud-Driss and Siddique, 2015). There is mounting evidence indicates that lncRNAs exerts a vital role in ALS pathogenesis.
LncRNA NEAT1_2 has been shown to be associated with the early course of ALS (Nishimoto et al., 2013). They also explored that the interaction between NEAT1_2 and ALS-associated RBPs, and found that FUS/TLS and TDP43 were enriched in paraspeckles. Besides, Hutchinson et al. (2007) investigated markedly raised frequency of paraplaque formation in the early stage of ALS pathology, implying that NEAT1_2 could serve as the scaffold of RBPs in the ALS motor nucleus.
Postmortem ALS tissues were examined by iCHIP, and NEAT1 was found to interact with FUS/TLS and TDP43, MALAT1 with TDP43, and MEG3 with FUS (Tollervey et al., 2011; Lagier-Tourenne et al., 2012). Moreover, Biscarini et al. (2018) found that three of the identified conservative lncRNAs, including lncMN-1, lncMN-2, and Lhx1os, were affected by FUS/TLS in ALS mouse models.
Conclusion and Perspectives
LncRNAs knowledge has changed dramatically over the past decade and is likely to continue to in the future. It has been demonstrated that lncRNAs own diverse molecular functions, such as translation, post-translation, and epigenetic modification. Massive evidence proves that lncRNAs are involved in aging-related neurodegenerative diseases. As the aging population continues to increase, aging-related neurodegenerative diseases will become a heavy burden of society. Therefore, it is essential to further understanding of mechanisms and roles of lncRNAs in these diseases. However, there exists some challenges regarding its clinical application. In addition, the functions of lncRNAs at molecular or cellular levels remain elusive. In-depth understanding of the mechanisms and networks of lncRNAs from different perspectives will provide new insights into the prevention, diagnosis and treatment of aging-related neurodegenerative diseases.
Author Contributions
Y-QN wrote the manuscript. HX drew the figures. Y-SL conceived the idea and supervised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 82071593 and 81770833) and the Fundamental Research Funds for the Central Universities of Central South University (No. 2019zzts354).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abati, E., Bresolin, N., Comi, G., and Corti, S. (2020). Silence superoxide dismutase 1 (SOD1): a promising therapeutic target for amyotrophic lateral sclerosis (ALS). Expert Opin. Ther. Targets 24, 295–310. doi: 10.1080/14728222.2020.1738390
Abbott, N. J., Patabendige, A. A., Dolman, D. E., Yusof, S. R., and Begley, D. J. (2010). Structure and function of the blood-brain barrier. Neurobiol. Dis. 37, 13–25. doi: 10.1016/j.nbd.2009.07.030
Aguado, J., d’Adda di Fagagna, F., and Wolvetang, E. (2020). Telomere transcription in ageing. Ageing Res. Rev. 62:101115. doi: 10.1016/j.arr.2020.101115
Airavaara, M., Pletnikova, O., Doyle, M. E., Zhang, Y. E., Troncoso, J. C., and Liu, Q. R. (2011). Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J. Biol. Chem. 286, 45093–45102. doi: 10.1074/jbc.M111.310250
Ajroud-Driss, S., and Siddique, T. (2015). Sporadic and hereditary amyotrophic lateral sclerosis (ALS). Biochim. Biophys. Acta 1852, 679–684. doi: 10.1016/j.bbadis.2014.08.010
Aleshkina, D., Iyyappan, R., Lin, C. J., Masek, T., Pospisek, M., and Susor, A. (2021). ncRNA BC1 influences translation in the oocyte. RNA Biol. 18, 1893–1904. doi: 10.1080/15476286.2021.1880181
Amor, S., Puentes, F., Baker, D., and van der Valk, P. (2010). Inflammation in neurodegenerative diseases. Immunology 129, 154–169. doi: 10.1111/j.1365-2567.2009.03225.x
An, H., Williams, N. G., and Shelkovnikova, T. A. (2018). NEAT1 and paraspeckles in neurodegenerative diseases: a missing lnc found? Noncoding RNA Res. 3, 243–252. doi: 10.1016/j.ncrna.2018.11.003
Arisi, I., D’Onofrio, M., Brandi, R., Felsani, A., Capsoni, S., Drovandi, G., et al. (2011). Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J. Alzheimers Dis. 24, 721–738. doi: 10.3233/jad-2011-101881
Ballard, C., Gauthier, S., Corbett, A., Brayne, C., Aarsland, D., and Jones, E. (2011). Alzheimer’s disease. Lancet 377, 1019–1031. doi: 10.1016/s0140-6736(10)61349-9
Baltazar, M. T., Dinis-Oliveira, R. J., de Lourdes Bastos, M., Tsatsakis, A. M., Duarte, J. A., and Carvalho, F. (2014). Pesticides exposure as etiological factors of Parkinson’s disease and other neurodegenerative diseases–a mechanistic approach. Toxicol. Lett. 230, 85–103. doi: 10.1016/j.toxlet.2014.01.039
Belrose, J., and Jackson, M. (2018). TRPM2: a candidate therapeutic target for treating neurological diseases. Acta Pharmacol. Sin. 39, 722–732. doi: 10.1038/aps.2018.31
Benz, F., and Liebner, S. (2020). Structure and function of the blood-brain barrier (BBB). Handb. Exp. Pharmacol. 37, 13–25. doi: 10.1007/164_2020_404
Betarbet, R., Sherer, T. B., and Greenamyre, J. T. (2005). Ubiquitin-proteasome system and Parkinson’s diseases. Exp. Neurol. 191, (Suppl. 1), S17–S27. doi: 10.1016/j.expneurol.2004.08.021
Biscarini, S., Capauto, D., Peruzzi, G., Lu, L., Colantoni, A., Santini, T., et al. (2018). Characterization of the lncRNA transcriptome in mESC-derived motor neurons: implications for FUS-ALS. Stem Cell Res. 27, 172–179. doi: 10.1016/j.scr.2018.01.037
Björkholm, C., and Monteggia, L. M. (2016). BDNF - a key transducer of antidepressant effects. Neuropharmacology 102, 72–79. doi: 10.1016/j.neuropharm.2015.10.034
Buckley, N. J., Johnson, R., Zuccato, C., Bithell, A., and Cattaneo, E. (2010). The role of REST in transcriptional and epigenetic dysregulation in Huntington’s disease. Neurobiol. Dis. 39, 28–39. doi: 10.1016/j.nbd.2010.02.003
Butterfield, D., and Boyd-Kimball, D. (2018). Oxidative stress, amyloid-β peptide, and altered key molecular pathways in the pathogenesis and progression of Alzheimer’s disease. J. Alzheimers Dis. 62, 1345–1367. doi: 10.3233/jad-170543
Cai, L., Tu, L., Li, T., Yang, X., Ren, Y., Gu, R., et al. (2019). Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int. Immunopharmacol. 75:105734. doi: 10.1016/j.intimp.2019.105734
Calcinotto, A., Kohli, J., Zagato, E., Pellegrini, L., Demaria, M., and Alimonti, A. (2019). Cellular senescence: aging. Cancer Inj. Physiol. Rev. 99, 1047–1078. doi: 10.1152/physrev.00020.2018
Campbell, I., Krucker, T., Steffensen, S., Akwa, Y., Powell, H., Lane, T., et al. (1999). Structural and functional neuropathology in transgenic mice with CNS expression of IFN-alpha. Brain Res. 835, 46–61. doi: 10.1016/s0006-8993(99)01328-1
Cao, B., Wang, T., Qu, Q., Kang, T., and Yang, Q. (2018). Long noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s disease via regulating miR-7/NLRP3 pathway. Neuroscience 388, 118–127. doi: 10.1016/j.neuroscience.2018.07.019
Carrillo-Jimenez, A., Deniz, Ö, Niklison-Chirou, M. V., Ruiz, R., Bezerra-Salomão, K., Stratoulias, V., et al. (2019). TET2 regulates the neuroinflammatory response in microglia. Cell Rep. 29, 697–713. doi: 10.1016/j.celrep.2019.09.013
Chanda, K., Das, S., Chakraborty, J., Bucha, S., Maitra, A., Chatterjee, R., et al. (2018). Altered levels of long NcRNAs Meg3 and Neat1 in cell and animal models of Huntington’s disease. RNA Biol. 15, 1348–1363. doi: 10.1080/15476286.2018.1534524
Chang, K. H., Wu, Y. R., and Chen, C. M. (2017). Down-regulation of miR-9* in the peripheral leukocytes of Huntington’s disease patients. Orphanet. J. Rare Dis. 12:185. doi: 10.1186/s13023-017-0742-x
Chen, D., Zhang, X. Y., Sun, J., Cong, Q. J., Chen, W. X., Ahsan, H. M., et al. (2019). Asiatic acid protects dopaminergic neurons from neuroinflammation by suppressing mitochondrial ros production. Biomol. Ther. 27, 442–449. doi: 10.4062/biomolther.2018.188
Chen, L., and Zhang, S. (2016). Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 366, 509–521. doi: 10.1007/s00441-016-2451-5
Chen, Q., Huang, X., and Li, R. (2018). lncRNA MALAT1/miR-205-5p axis regulates MPP(+)-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am. J. Transl. Res. 10, 563–572.
Cheng, C., Spengler, R. M., Keiser, M. S., Monteys, A. M., Rieders, J. M., Ramachandran, S., et al. (2018). The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum. Mol. Genet. 27, 4303–4314. doi: 10.1093/hmg/ddy331
Chung, D., Rudnicki, D., Yu, L., and Margolis, R. (2011). A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477. doi: 10.1093/hmg/ddr263
Chung, K., Thomas, B., Li, X., Pletnikova, O., Troncoso, J., Marsh, L., et al. (2004). S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science 304, 1328–1331. doi: 10.1126/science.1093891
Ciarlo, E., Massone, S., Penna, I., Nizzari, M., Gigoni, A., Dieci, G., et al. (2013). An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 6, 424–433. doi: 10.1242/dmm.009761
Cintrón-Colón, A. F., Almeida-Alves, G., Boynton, A. M., and Spitsbergen, J. M. (2020). GDNF synthesis, signaling, and retrograde transport in motor neurons. Cell Tissue Res. 382, 47–56. doi: 10.1007/s00441-020-03287-6
Collier, T. J., Kanaan, N. M., and Kordower, J. H. (2011). Ageing as a primary risk factor for Parkinson’s disease: evidence from studies of non-human primates. Nat. Rev. Neurosci. 12, 359–366. doi: 10.1038/nrn3039
Corral-Debrinski, M., Horton, T., Lott, M., Shoffner, J., Beal, M., and Wallace, D. (1992). Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 2, 324–329. doi: 10.1038/ng1292-324
Coupland, K. G., Kim, W. S., Halliday, G. M., Hallupp, M., Dobson-Stone, C., and Kwok, J. B. (2016). Role of the long non-coding RNA MAPT-AS1 in regulation of microtubule associated protein tau (MAPT) expression in Parkinson’s disease. PLoS One 11:e0157924. doi: 10.1371/journal.pone.0157924
Cui, H., Xie, N., Tan, Z., Banerjee, S., Thannickal, V. J., Abraham, E., et al. (2014). The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 44, 2085–2095. doi: 10.1002/eji.201344126
Cui, X. Y., Zhan, J. K., and Liu, Y. S. (2021). Roles and functions of antisense lncRNA in vascular aging. Ageing Res. Rev. 72:101480. doi: 10.1016/j.arr.2021.101480
DeJesus-Hernandez, M., Mackenzie, I. R., Boeve, B. F., Boxer, A. L., Baker, M., Rutherford, N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. doi: 10.1016/j.neuron.2011.09.011
Deschênes-Furry, J., Perrone-Bizzozero, N., and Jasmin, B. J. (2006). The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays 28, 822–833. doi: 10.1002/bies.20449
Ding, X. M., Zhao, L. J., Qiao, H. Y., Wu, S. L., and Wang, X. H. (2019). Long non-coding RNA-p21 regulates MPP(+)-induced neuronal injury by targeting miR-625 and derepressing TRPM2 in SH-SY5Y cells. Chem. Biol. Interact. 307, 73–81. doi: 10.1016/j.cbi.2019.04.017
Ding, Y., Luan, W., Shen, X., Wang, Z., and Cao, Y. (2021). LncRNA BDNF-AS as ceRNA regulates the miR-9-5p/BACE1 pathway affecting neurotoxicity in Alzheimer’s disease. Arch. Gerontol. Geriatr. 99:104614. doi: 10.1016/j.archger.2021.104614
Divo, M., Celli, B., Poblador-Plou, B., Calderón-Larrañaga, A., de-Torres, J., Gimeno-Feliu, L., et al. (2018). Chronic obstructive pulmonary disease (COPD) as a disease of early aging: evidence from the epichron cohort. PLoS One 13:e0193143. doi: 10.1371/journal.pone.0193143
Dong, H. X., Wang, R., Jin, X. Y., Zeng, J., and Pan, J. (2018). LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J. Cell Physiol. 233, 4126–4136. doi: 10.1002/jcp.26215
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Fantes, J., Ragge, N. K., Lynch, S. A., McGill, N. I., Collin, J. R., Howard-Peebles, P. N., et al. (2003). Mutations in SOX2 cause anophthalmia. Nat. Genet. 33, 461–463. doi: 10.1038/ng1120
Farrall, A. J., and Wardlaw, J. M. (2009). Blood-brain barrier: ageing and microvascular disease–systematic review and meta-analysis. Neurobiol. Aging 30, 337–352. doi: 10.1016/j.neurobiolaging.2007.07.015
Feng, L., Liao, Y., He, J., Xie, C., Chen, S., Fan, H., et al. (2018). Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol. 18:4. doi: 10.1186/s12883-017-1008-x
Finkel, T., and Holbrook, N. J. (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247. doi: 10.1038/35041687
Fotuhi, S. N., Khalaj-Kondori, M., Hoseinpour Feizi, M. A., and Talebi, M. (2019). Long non-coding RNA BACE1-AS may serve as an Alzheimer’s disease blood-based biomarker. J. Mol. Neurosci. 69, 351–359. doi: 10.1007/s12031-019-01364-2
Goedert, M., and Spillantini, M. G. (2006). A century of Alzheimer’s disease. Science 314, 777–781. doi: 10.1126/science.1132814
Greco, S., Gorospe, M., and Martelli, F. (2015). Noncoding RNA in age-related cardiovascular diseases. J. Mol. Cell. Cardiol. 83, 142–155. doi: 10.1016/j.yjmcc.2015.01.011
Gu, C., Chen, C., Wu, R., Dong, T., Hu, X., Yao, Y., et al. (2018). Long noncoding RNA EBF3-AS promotes neuron apoptosis in Alzheimer’s disease. DNA Cell Biol. 37, 220–226. doi: 10.1089/dna.2017.4012
Gudenas, B., and Wang, L. (2015). Gene coexpression networks in human brain developmental transcriptomes implicate the association of long noncoding RNAs with intellectual disability. Bioinform. Biol. Insights 9, 21–27. doi: 10.4137/bbi.s29435
Guo, C., Qi, Y., Qu, J., Gai, L., Shi, Y., and Yuan, C. (2020). Pathophysiological functions of the lncRNA TUG1. Curr. Pharm. Des. 26, 688–700. doi: 10.2174/1381612826666191227154009
Guo, C. C., Jiao, C. H., and Gao, Z. M. (2018). Silencing of LncRNA BDNF-AS attenuates Aβ(25-35)-induced neurotoxicity in PC12 cells by suppressing cell apoptosis and oxidative stress. Neurol. Res. 40, 795–804. doi: 10.1080/01616412.2018.1480921
Gupta, S., Khan, A., Vishwas, S., Gulati, M., Gurjeet Singh, T., Dua, K., et al. (2021). Demethyleneberberine: A possible treatment for Huntington’s disease. Med. Hypotheses. 153:110639. doi: 10.1016/j.mehy.2021.110639
Han, Y., Kang, C., Kang, M., Quan, W., Gao, H., and Zhong, Z. (2019). Long non-coding RNA Mirt2 prevents TNF-α-triggered inflammation via the repression of microRNA-101. Int. Immunopharmacol. 76:105878. doi: 10.1016/j.intimp.2019.105878
Hardiman, O., Al-Chalabi, A., Chio, A., Corr, E. M., Logroscino, G., Robberecht, W., et al. (2017). Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3:17071. doi: 10.1038/nrdp.2017.71
Hawkes, C. H., Del Tredici, K., and Braak, H. (2007). Parkinson’s disease: a dual-hit hypothesis. Neuropathol. Appl. Neurobiol. 33, 599–614. doi: 10.1111/j.1365-2990.2007.00874.x
Hayashi, Y., Homma, K., and Ichijo, H. (2016). SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS. Adv Biol. Regul. 60, 95–104. doi: 10.1016/j.jbior.2015.10.006
He, J., Tu, C., and Liu, Y. (2018). Role of lncRNA in aging and age-related diseases. Aging Med. 1, 158–175. doi: 10.1002/agm2.12030
Hedglin, M., and Benkovic, S. J. (2015). Regulation of Rad6/Rad18 activity during DNA damage tolerance. Annu. Rev. Biophys. 44, 207–228. doi: 10.1146/annurev-biophys-060414-033841
Hodges, A., Strand, A. D., Aragaki, A. K., Kuhn, A., Sengstag, T., Hughes, G., et al. (2006). Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 15, 965–977. doi: 10.1093/hmg/ddl013
Holmes, C. (2013). Review: systemic inflammation and Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 39, 51–68. doi: 10.1111/j.1365-2990.2012.01307.x
Homma, T., and Fujii, J. (2015). Application of glutathione as anti-oxidative and anti-aging drugs. Curr. Drug Metab. 16, 560–571. doi: 10.2174/1389200216666151015114515
Hou, Y., Dan, X., Babbar, M., Wei, Y., Hasselbalch, S., Croteau, D., et al. (2019). Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581. doi: 10.1038/s41582-019-0244-7
Hu, T., and Lu, Y. R. (2015). BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 15:36. doi: 10.1186/s12935-015-0183-3
Humbert, S., and Saudou, F. (2005). [Huntington’s disease: intracellular signaling pathways and neuronal death]. J. Soc. Biol. 199, 247–251. doi: 10.1051/jbio:2005026
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B., and Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 8:39. doi: 10.1186/1471-2164-8-39
Hwang, J., and Zukin, R. (2018). REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 48, 193–200. doi: 10.1016/j.conb.2017.12.008
Islam, M. T. (2017). Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol. Res. 39, 73–82. doi: 10.1080/01616412.2016.1251711
Jankovic, J., and Tan, E. K. (2020). Parkinson’s disease: etiopathogenesis and treatment. J. Neurol. Neurosurg. Psychiatry 91, 795–808. doi: 10.1136/jnnp-2019-322338
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041. doi: 10.1038/sj.onc.1206928
Jimenez-Sanchez, M., Licitra, F., Underwood, B. R., and Rubinsztein, D. C. (2017). Huntington’s disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb. Perspect. Med. 7:a024240. doi: 10.1101/cshperspect.a024240
Johnson, R. (2012). Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 46, 245–254. doi: 10.1016/j.nbd.2011.12.006
Johnson, R., Richter, N., Jauch, R., Gaughwin, P. M., Zuccato, C., Cattaneo, E., et al. (2010). Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol. Genomics 41, 269–274. doi: 10.1152/physiolgenomics.00019.2010
Johnson, R., Teh, C., Jia, H., Vanisri, R., Pandey, T., Lu, Z., et al. (2009). Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15, 85–96. doi: 10.1261/rna.1127009
Kang, M. J., Abdelmohsen, K., Hutchison, E. R., Mitchell, S. J., Grammatikakis, I., Guo, R., et al. (2014). HuD regulates coding and noncoding RNA to induce APP→Aβ processing. Cell Rep. 7, 1401–1409. doi: 10.1016/j.celrep.2014.04.050
Karran, E., Mercken, M., and De Strooper, B. (2011). The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 10, 698–712. doi: 10.1038/nrd3505
Ke, S., Yang, Z., Yang, F., Wang, X., Tan, J., and Liao, B. (2019). Long noncoding RNA NEAT1 aggravates Aβ-induced neuronal damage by targeting miR-107 in Alzheimer’s disease. Yonsei Med. J. 60, 640–650. doi: 10.3349/ymj.2019.60.7.640
Khalil, A., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Khoshnan, A., Ko, J., Watkin, E. E., Paige, L. A., Reinhart, P. H., and Patterson, P. H. (2004). Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J. Neurosci. 24, 7999–8008. doi: 10.1523/jneurosci.2675-04.2004
Kolli, N., Lu, M., Maiti, P., Rossignol, J., and Dunbar, G. L. (2017). CRISPR-Cas9 mediated gene-silencing of the mutant huntingtin gene in an in vitro model of Huntington’s disease. Int. J. Mol. Sci. 18:754. doi: 10.3390/ijms18040754
Kondrashov, A. V., Kiefmann, M., Ebnet, K., Khanam, T., Muddashetty, R. S., and Brosius, J. (2005). Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353, 88–103. doi: 10.1016/j.jmb.2005.07.049
Kour, S., and Rath, P. C. (2016). Long noncoding RNAs in aging and age-related diseases. Ageing Res. Rev. 26, 1–21. doi: 10.1016/j.arr.2015.12.001
Koyama, S., Tsuchiya, H., Amisaki, M., Sakaguchi, H., Honjo, S., Fujiwara, Y., et al. (2020). NEAT1 is required for the expression of the liver cancer stem cell marker CD44. Int. J. Mol. Sci. 21:1927. doi: 10.3390/ijms21061927
Kraus, T. F. J., Haider, M., Spanner, J., Steinmaurer, M., Dietinger, V., and Kretzschmar, H. A. (2017). Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report. Mol. Neurobiol. 54, 2869–2877. doi: 10.1007/s12035-016-9854-x
Lagier-Tourenne, C., Polymenidou, M., Hutt, K., Vu, A., Baughn, M., Huelga, S., et al. (2012). Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15, 1488–1497. doi: 10.1038/nn.3230
Lang, Y., Li, Y., Yu, H., Lin, L., Chen, X., Wang, S., et al. (2020). HOTAIR drives autophagy in midbrain dopaminergic neurons in the substantia nigra compacta in a mouse model of Parkinson’s disease by elevating NPTX2 via miR-221-3p binding. Aging 12, 7660–7678. doi: 10.18632/aging.103028
Laugesen, A., Højfeldt, J. W., and Helin, K. (2019). Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell. 74, 8–18. doi: 10.1016/j.molcel.2019.03.011
Lee, Y., Lee, H. S., Kim, M., and Shin, H. (2020). Brain cytoplasmic RNAs in neurons: from biosynthesis to function. Biomolecules 10:313. doi: 10.3390/biom10020313
Li, D., Zhang, J., Li, X., Chen, Y., Yu, F., and Liu, Q. (2021). Insights into lncRNAs in Alzheimer’s disease mechanisms. RNA Biol. 18, 1037–1047. doi: 10.1080/15476286.2020.1788848
Li, L., Miao, M., Chen, J., Liu, Z., Li, W., Qiu, Y., et al. (2021). Role of Ten eleven translocation-2 (Tet2) in modulating neuronal morphology and cognition in a mouse model of Alzheimer’s disease. J. Neurochem. 157, 993–1012. doi: 10.1111/jnc.15234
Li, H., Zheng, L., Jiang, A., Mo, Y., and Gong, Q. (2018). Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport 29, 1061–1067. doi: 10.1097/wnr.0000000000001057
Lin, D., Pestova, T. V., Hellen, C. U., and Tiedge, H. (2008). Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell Biol. 28, 3008–3019. doi: 10.1128/mcb.01800-07
Lin, M., Simon, D., Ahn, C., Kim, L., and Beal, M. (2002). High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum. Mol. Genet. 11, 133–145. doi: 10.1093/hmg/11.2.133
Lin, M. T., and Beal, M. F. (2006). Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795. doi: 10.1038/nature05292
Lin, Q., Hou, S., Dai, Y., Jiang, N., and Lin, Y. (2019). LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol. Chem. 400, 1217–1228. doi: 10.1515/hsz-2018-0431
Liu, W., Zhang, Q., Zhang, J., Pan, W., Zhao, J., and Xu, Y. (2017). Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 7:19. doi: 10.1186/s13578-017-0147-5
Liu, X., Li, D., Zhang, W., Guo, M., and Zhan, Q. (2012). Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 31, 4415–4427. doi: 10.1038/emboj.2012.292
Liu, Y., and Lu, Z. (2018). Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clin. Exp. Pharmacol. Physiol. 45, 841–848. doi: 10.1111/1440-1681.12932
Lu, C. H., Allen, K., Oei, F., Leoni, E., Kuhle, J., Tree, T., et al. (2016). Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 3:e244. doi: 10.1212/nxi.0000000000000244
Lu, M., Sun, W. L., Shen, J., Wei, M., Chen, B., Qi, Y. J., et al. (2018). LncRNA-UCA1 promotes PD development by upregulating SNCA. Eur Rev. Med. Pharmacol. Sci. 22, 7908–7915. doi: 10.26355/eurrev_201811_16417
Lu, Q., Shan, S., Li, Y., Zhu, D., Jin, W., and Ren, T. (2018). Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 32, 3957–3967. doi: 10.1096/fj.201701237RR
Luo, Q., and Chen, Y. (2016). Long noncoding RNAs and Alzheimer’s disease. Clin. Interv. Aging 11, 867–872. doi: 10.2147/cia.S107037
Lutters, B., Foley, P., and Koehler, P. J. (2018). The centennial lesson of encephalitis lethargica. Neurology 90, 563–567. doi: 10.1212/wnl.0000000000005176
Lyon, M. S., Wosiski-Kuhn, M., Gillespie, R., Caress, J., and Milligan, C. (2019). Inflammation, Immunity, and amyotrophic lateral sclerosis: I. Etiology and pathology. Muscle Nerve 59, 10–22. doi: 10.1002/mus.26289
Ma, P., Li, Y., Zhang, W., Fang, F., Sun, J., Liu, M., et al. (2019). Long non-coding RNA MALAT1 inhibits neuron apoptosis and neuroinflammation while stimulates neurite outgrowth and its correlation with MiR-125b mediates PTGS2, CDK5 and FOXQ1 in Alzheimer’s disease. Curr. Alzheimer Res. 16, 596–612. doi: 10.2174/1567205016666190725130134
Ma, Q. L., Galasko, D. R., Ringman, J. M., Vinters, H. V., Edland, S. D., Pomakian, J., et al. (2009). Reduction of SorLA/LR11, a sorting protein limiting beta-amyloid production, in Alzheimer disease cerebrospinal fluid. Arch. Neurol. 66, 448–457. doi: 10.1001/archneurol.2009.22
Maag, J., Panja, D., Sporild, I., Patil, S., Kaczorowski, D., Bramham, C., et al. (2015). Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front. Neurosci. 9:351. doi: 10.3389/fnins.2015.00351
Majidinia, M., Mihanfar, A., Rahbarghazi, R., Nourazarian, A., Bagca, B., and Avci, ÇB. (2016). The roles of non-coding RNAs in Parkinson’s disease. Mol. Biol. Rep. 43, 1193–1204. doi: 10.1007/s11033-016-4054-3
Maries, E., Dass, B., Collier, T. J., Kordower, J. H., and Steece-Collier, K. (2003). The role of alpha-synuclein in Parkinson’s disease: insights from animal models. Nat. Rev. Neurosci. 4, 727–738. doi: 10.1038/nrn1199
Masoumi, F., Ghorbani, S., Talebi, F., Branton, W. G., Rajaei, S., Power, C., et al. (2019). Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 328, 50–59. doi: 10.1016/j.jneuroim.2018.11.013
Massone, S., Ciarlo, E., Vella, S., Nizzari, M., Florio, T., Russo, C., et al. (2012). NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochim. Biophys. Acta 1823, 1170–1177. doi: 10.1016/j.bbamcr.2012.05.001
Massone, S., Vassallo, I., Fiorino, G., Castelnuovo, M., Barbieri, F., Borghi, R., et al. (2011). 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317. doi: 10.1016/j.nbd.2010.09.019
Mattiazzi, M., D’Aurelio, M., Gajewski, C., Martushova, K., Kiaei, M., Beal, M., et al. (2002). Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 277, 29626–29633. doi: 10.1074/jbc.M203065200
McCluggage, F., and Fox, A. H. (2021). Paraspeckle nuclear condensates: global sensors of cell stress? Bioessays 43:e2000245. doi: 10.1002/bies.202000245
McHugh, C., Chen, C., Chow, A., Surka, C., Tran, C., McDonel, P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. doi: 10.1038/nature14443
Mendez, E., and Sattler, R. (2015). Biomarker development for C9orf72 repeat expansion in ALS. Brain Res. 1607, 26–35. doi: 10.1016/j.brainres.2014.09.041
Mercer, T., and Mattick, J. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Milakovic, T., and Johnson, G. (2005). Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J. Biol. Chem. 280, 30773–30782. doi: 10.1074/jbc.M504749200
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. doi: 10.1038/nbt.2158
Morimoto, S., Ishikawa, M., Watanabe, H., Isoda, M., Takao, M., Nakamura, S., et al. (2020). Brain transcriptome analysis links deficiencies of stress-responsive proteins to the pathomechanism of Kii ALS/PDC. Antioxidants 9:423. doi: 10.3390/antiox9050423
Moscovitch-Lopatin, M., Weiss, A., Rosas, H. D., Ritch, J., Doros, G., Kegel, K. B., et al. (2010). Optimization of an HTRF assay for the detection of soluble mutant huntingtin in human buffy coats: a potential biomarker in blood for huntington disease. PLoS Curr. 2:Rrn1205. doi: 10.1371/currents.RRN1205
Munro, D., and Pamenter, M. (2019). Comparative studies of mitochondrial reactive oxygen species in animal longevity: technical pitfalls and possibilities. Aging Cell 18:e13009. doi: 10.1111/acel.13009
Mus, E., Hof, P. R., and Tiedge, H. (2007). Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 104, 10679–10684. doi: 10.1073/pnas.0701532104
Nagel, F., Bähr, M., and Dietz, G. P. (2009). Tyrosine hydroxylase-positive amacrine interneurons in the mouse retina are resistant against the application of various parkinsonian toxins. Brain Res. Bull. 79, 303–309. doi: 10.1016/j.brainresbull.2009.04.010
Nardo, G., Trolese, M. C., and Bendotti, C. (2016). Major histocompatibility complex I expression by motor neurons and its implication in amyotrophic lateral sclerosis. Front. Neurol. 7:89. doi: 10.3389/fneur.2016.00089
Newsholme, P., Cruzat, V. F., Keane, K. N., Carlessi, R., and de Bittencourt, P. I. Jr. (2016). Molecular mechanisms of ROS production and oxidative stress in diabetes. Biochem. J. 473, 4527–4550. doi: 10.1042/bcj20160503c
Ni, Y. Q., Lin, X., Zhan, J. K., and Liu, Y. S. (2020). Roles and functions of exosomal non-coding RNAs in vascular aging. Aging Dis. 11, 164–178. doi: 10.14336/ad.2019.0402
Nishimoto, Y., Nakagawa, S., Hirose, T., Okano, H. J., Takao, M., Shibata, S., et al. (2013). The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 6:31. doi: 10.1186/1756-6606-6-31
Novak, D., Hüser, L., Elton, J. J., Umansky, V., Altevogt, P., and Utikal, J. (2020). SOX2 in development and cancer biology. Semin. Cancer Biol. 67(Pt 1), 74–82. doi: 10.1016/j.semcancer.2019.08.007
Panda, A. C., Abdelmohsen, K., and Gorospe, M. (2017). SASP regulation by noncoding RNA. Mech. Ageing Dev. 168, 37–43. doi: 10.1016/j.mad.2017.05.004
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246. doi: 10.1016/j.molcel.2008.08.022
Panov, A., Gutekunst, C., Leavitt, B., Hayden, M., Burke, J., Strittmatter, W., et al. (2002). Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 5, 731–736. doi: 10.1038/nn884
Parenti, R., Paratore, S., Torrisi, A., and Cavallaro, S. (2007). A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur. J. Neurosci. 26, 2444–2457. doi: 10.1111/j.1460-9568.2007.05864.x
Perry, V. H., Nicoll, J. A., and Holmes, C. (2010). Microglia in neurodegenerative disease. Nat. Rev. Neurol. 6, 193–201. doi: 10.1038/nrneurol.2010.17
Pokrishevsky, E., Grad, L. I., Yousefi, M., Wang, J., Mackenzie, I. R., and Cashman, N. R. (2012). Aberrant localization of FUS and TDP43 is associated with misfolding of SOD1 in amyotrophic lateral sclerosis. PLoS One 7:e35050. doi: 10.1371/journal.pone.0035050
Puvvula, P. K. (2019). LncRNAs regulatory networks in cellular senescence. Int. J. Mol. Sci. 20:2615. doi: 10.3390/ijms20112615
Qian, C., Ye, Y., Mao, H., Yao, L., Sun, X., Wang, B., et al. (2019). Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222/p27/mTOR pathway in Parkinson’s disease. Exp. Cell Res. 384:111614. doi: 10.1016/j.yexcr.2019.111614
Qu, X., Alsager, S., Zhuo, Y., and Shan, B. (2019). HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 454, 90–97. doi: 10.1016/j.canlet.2019.04.016
Rajagopal, T., Talluri, S., Akshaya, R. L., and Dunna, N. R. (2020). HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin. Chim. Acta 503, 1–18. doi: 10.1016/j.cca.2019.12.028
Ramalingam, M., Huh, Y. J., and Lee, Y. I. (2019). The impairments of α-synuclein and mechanistic target of rapamycin in rotenone-induced SH-SY5Y cells and mice model of Parkinson’s disease. Front. Neurosci. 13:1028. doi: 10.3389/fnins.2019.01028
Ratajczak, M. Z. (2012). Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem. Cytobiol. 50, 171–179. doi: 10.5603/fhc.2012.0026
Rayner, K., and Liu, P. (2016). Long noncoding RNAS in the heart: the regulatory roadmap of cardiovascular development and disease. Circ. Cardiovasc. Genet. 9, 101–103. doi: 10.1161/circgenetics.116.001413
Rebeck, G. W., Reiter, J. S., Strickland, D. K., and Hyman, B. T. (1993). Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron 11, 575–580. doi: 10.1016/0896-6273(93)90070-8
Reddy, P. H., McWeeney, S., Park, B. S., Manczak, M., Gutala, R. V., Partovi, D., et al. (2004). Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer’s disease. Hum. Mol. Genet. 13, 1225–1240. doi: 10.1093/hmg/ddh140
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Rubinsztein, D. C., and Carmichael, J. (2003). Huntington’s disease: molecular basis of neurodegeneration. Expert Rev. Mol. Med. 5, 1–21. doi: 10.1017/s1462399403006549
Sadri-Vakili, G., and Cha, J. H. (2006). Mechanisms of disease: histone modifications in Huntington’s disease. Nat. Clin. Pract. Neurol. 2, 330–338. doi: 10.1038/ncpneuro0199
Samper, E., Flores, J. M., and Blasco, M. A. (2001). Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc-/- mice with short telomeres. EMBO Rep. 2, 800–807. doi: 10.1093/embo-reports/kve174
Sharma, E., Behl, T., Mehta, V., Kumar, A., Setia, D., Uddin, M. S., et al. (2021). Exploring the various aspects of brain-derived neurotropic factor (BDNF) in diabetes mellitus. CNS Neurol. Disord. Drug Targets 20, 22–33. doi: 10.2174/1871527319666201014125642
Shen, Y., Liu, S., Fan, J., Jin, Y., Tian, B., Zheng, X., et al. (2017). Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep. 18, 536–548. doi: 10.15252/embr.201643139
Shi, Y., Riese, D. J. II, and Shen, J. (2020). The role of the CXCL12/CXCR4/CXCR7 chemokine axis in cancer. Front. Pharmacol. 11:574667. doi: 10.3389/fphar.2020.574667
Silvestroni, A., Faull, R. L., Strand, A. D., and Möller, T. (2009). Distinct neuroinflammatory profile in post-mortem human Huntington’s disease. Neuroreport 20, 1098–1103. doi: 10.1097/WNR.0b013e32832e34ee
Singh, A., Kukreti, R., Saso, L., and Kukreti, S. (2019). Oxidative stress: a key modulator in neurodegenerative diseases. Molecules 24:1583. doi: 10.3390/molecules24081583
Sinha, J., Ghosh, S., Swain, U., Giridharan, N., and Raghunath, M. (2014). Increased macromolecular damage due to oxidative stress in the neocortex and hippocampus of WNIN/Ob, a novel rat model of premature aging. Neuroscience 269, 256–264. doi: 10.1016/j.neuroscience.2014.03.040
Smeyne, M., and Smeyne, R. J. (2013). Glutathione metabolism and Parkinson’s disease. Free Radic. Biol. Med. 62, 13–25. doi: 10.1016/j.freeradbiomed.2013.05.001
Song, S., Miranda, C. J., Braun, L., Meyer, K., Frakes, A. E., Ferraiuolo, L., et al. (2016). Major histocompatibility complex class I molecules protect motor neurons from astrocyte-induced toxicity in amyotrophic lateral sclerosis. Nat. Med. 22, 397–403. doi: 10.1038/nm.4052
Spreafico, M., Grillo, B., Rusconi, F., Battaglioli, E., and Venturin, M. (2018). Multiple layers of CDK5R1 regulation in Alzheimer’s disease implicate long non-coding RNAs. Int. J. Mol. Sci. 19:2022. doi: 10.3390/ijms19072022
Stephenson, J., Nutma, E., van der Valk, P., and Amor, S. (2018). Inflammation in CNS neurodegenerative diseases. Immunology 154, 204–219. doi: 10.1111/imm.12922
Stevanovic, M., Zuffardi, O., Collignon, J., Lovell-Badge, R., and Goodfellow, P. (1994). The cDNA sequence and chromosomal location of the human SOX2 gene. Mamm. Genome 5, 640–642. doi: 10.1007/bf00411460
Sunwoo, J. S., Lee, S. T., Im, W., Lee, M., Byun, J. I., Jung, K. H., et al. (2017). Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol. Neurobiol. 54, 1577–1586. doi: 10.1007/s12035-016-9928-9
Taheri, M., Eghtedarian, R., Dinger, M. E., and Ghafouri-Fard, S. (2020). Exploring the role of non-coding RNAs in the pathophysiology of systemic lupus erythematosus. Biomolecules 10:937. doi: 10.3390/biom10060937
Thonhoff, J. R., Simpson, E. P., and Appel, S. H. (2018). Neuroinflammatory mechanisms in amyotrophic lateral sclerosis pathogenesis. Curr. Opin. Neurol. 31, 635–639. doi: 10.1097/wco.0000000000000599
Tischbein, M., Baron, D. M., Lin, Y. C., Gall, K. V., Landers, J. E., Fallini, C., et al. (2019). The RNA-binding protein FUS/TLS undergoes calcium-mediated nuclear egress during excitotoxic stress and is required for GRIA2 mRNA processing. J. Biol. Chem. 294, 10194–10210. doi: 10.1074/jbc.RA118.005933
Tollervey, J., Curk, T., Rogelj, B., Briese, M., Cereda, M., Kayikci, M., et al. (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458. doi: 10.1038/nn.2778
Tominaga-Yamanaka, K., Abdelmohsen, K., Martindale, J. L., Yang, X., Taub, D. D., and Gorospe, M. (2012). NF90 coordinately represses the senescence-associated secretory phenotype. Aging 4, 695–708. doi: 10.18632/aging.100497
Tong, S. J., Wall, A. A., Hung, Y., Luo, L., and Stow, J. L. (2021). Guanine nucleotide exchange factors activate Rab8a for Toll-like receptor signalling. Small GTPases 12, 27–43. doi: 10.1080/21541248.2019.1587278
Tönnies, E., and Trushina, E. (2017). Oxidative stress, synaptic dysfunction, and Alzheimer’s disease. J. Alzheimers Dis. 57, 1105–1121. doi: 10.3233/jad-161088
Tripathi, V., Shen, Z., Chakraborty, A., Giri, S., Freier, S. M., Wu, X., et al. (2013). Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 9:e1003368. doi: 10.1371/journal.pgen.1003368
Trist, B. G., Hare, D. J., and Double, K. L. (2019). Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell 18:e13031. doi: 10.1111/acel.13031
Tycowski, K. T., Shu, M. D., and Steitz, J. A. (1994). Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 266, 1558–1561. doi: 10.1126/science.7985025
Valadão, P. A. C., Santos, K. B. S., Ferreira, E. V. T. H., Macedo, E. C. T., Teixeira, A. L., Guatimosim, C., et al. (2020). Inflammation in Huntington’s disease: a few new twists on an old tale. J. Neuroimmunol. 348:577380. doi: 10.1016/j.jneuroim.2020.577380
Wang, D., Li, J., Cai, F., Xu, Z., Li, L., Zhu, H., et al. (2019). Overexpression of MAPT-AS1 is associated with better patient survival in breast cancer. Biochem. Cell Biol. 97, 158–164. doi: 10.1139/bcb-2018-0039
Wang, X., Zhang, M., and Liu, H. (2019). LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer’s disease. Biosci. Biotechnol. Biochem. 83, 609–621. doi: 10.1080/09168451.2018.1562874
Wang, H., Wang, X., Zhang, Y., and Zhao, J. (2021). LncRNA SNHG1 promotes neuronal injury in Parkinson’s disease cell model by miR-181a-5p/CXCL12 axis. J. Mol. Histol. 52, 153–163. doi: 10.1007/s10735-020-09931-3
Wang, S., Zhang, X., Guo, Y., Rong, H., and Liu, T. (2017). The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget 8, 24449–24456. doi: 10.18632/oncotarget.15511
Wang, Y., Zhang, M., Xu, H., Wang, Y., Li, Z., Chang, Y., et al. (2017). Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget 8, 72182–72196. doi: 10.18632/oncotarget.20053
Wang, X., Wang, W., Li, L., Perry, G., Lee, H., and Zhu, X. (2014). Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim. Biophys. Acta 1842, 1240–1247. doi: 10.1016/j.bbadis.2013.10.015
Wang, X. S., Zhang, Z., Wang, H. C., Cai, J. L., Xu, Q. W., Li, M. Q., et al. (2006). Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 12, 4851–4858. doi: 10.1158/1078-0432.Ccr-06-0134
Wang, Y., Wu, N., Luo, X., Zhang, X., Liao, Q., and Wang, J. (2020). SOX2OT, a novel tumor-related long non-coding RNA. Biomed. Pharmacother. 123:109725. doi: 10.1016/j.biopha.2019.109725
Waters, E., Pucci, P., Hirst, M., Chapman, S., Wang, Y., Crea, F., et al. (2021). HAR1: an insight into lncRNA genetic evolution. Epigenomics 13, 1831–1843. doi: 10.2217/epi-2021-0069
Wei, W., and Ji, S. (2018). Cellular senescence: molecular mechanisms and pathogenicity. J. Cell Physiol. 233, 9121–9135. doi: 10.1002/jcp.26956
Wu, Y. Y., and Kuo, H. C. (2020). Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 27:49. doi: 10.1186/s12929-020-00636-z
Wyss-Coray, T., and Mucke, L. (2002). Inflammation in neurodegenerative disease–a double-edged sword. Neuron 35, 419–432. doi: 10.1016/s0896-6273(02)00794-8
Xiang, J., Yin, Q., Chen, T., Zhang, Y., Zhang, X., Wu, Z., et al. (2014). Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531. doi: 10.1038/cr.2014.35
Xiao, X., Tan, Z., Jia, M., Zhou, X., Wu, K., Ding, Y., et al. (2021). Long noncoding RNA SNHG1 knockdown ameliorates apoptosis, oxidative stress and inflammation in models of Parkinson’s disease by inhibiting the miR-125b-5p/MAPK1 axis. Neuropsychiatr. Dis. Treat. 17, 1153–1163. doi: 10.2147/ndt.S286778
Xu, X., Zhuang, C., Wu, Z., Qiu, H., Feng, H., and Wu, J. (2018). LincRNA-p21 inhibits cell viability and promotes cell apoptosis in Parkinson’s disease through activating α-synuclein expression. Biomed. Res. Int. 2018:8181374. doi: 10.1155/2018/8181374
Yamanaka, Y., Faghihi, M. A., Magistri, M., Alvarez-Garcia, O., Lotz, M., and Wahlestedt, C. (2015). Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell. Rep. 11, 967–976. doi: 10.1016/j.celrep.2015.04.011
Yan, W., Chen, Z. Y., Chen, J. Q., and Chen, H. M. (2018). LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 496, 1019–1024. doi: 10.1016/j.bbrc.2017.12.149
Yan, Y., Yan, H., Teng, Y., Wang, Q., Yang, P., Zhang, L., et al. (2020). Long non-coding RNA 00507/miRNA-181c-5p/TTBK1/MAPT axis regulates tau hyperphosphorylation in Alzheimer’s disease. J. Gene Med. 22:e3268. doi: 10.1002/jgm.3268
Yang, F., Huo, X. S., Yuan, S. X., Zhang, L., Zhou, W. P., Wang, F., et al. (2013). Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 49, 1083–1096. doi: 10.1016/j.molcel.2013.01.010
Yehezkel, S., Shaked, R., Sagie, S., Berkovitz, R., Shachar-Bener, H., Segev, Y., et al. (2013). Characterization and rescue of telomeric abnormalities in ICF syndrome type I fibroblasts. Front. Oncol. 3:35. doi: 10.3389/fonc.2013.00035
Yi, J., Chen, B., Yao, X., Lei, Y., Ou, F., and Huang, F. (2019). Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer’s disease through inactivating the PI3K/Akt signaling pathway. J. Cell. Biochem. 120, 18053–18065. doi: 10.1002/jcb.29108
Yu, X., Li, Z., Zheng, H., Chan, M. T., and Wu, W. K. (2017). NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 50:e12329. doi: 10.1111/cpr.12329
Zhang, H. N., Xu, Q. Q., Thakur, A., Alfred, M. O., Chakraborty, M., Ghosh, A., et al. (2018). Endothelial dysfunction in diabetes and hypertension: role of microRNAs and long non-coding RNAs. Life Sci. 213, 258–268. doi: 10.1016/j.lfs.2018.10.028
Zhang, Q., Huang, X. M., Liao, J. X., Dong, Y. K., Zhu, J. L., He, C. C., et al. (2021). LncRNA HOTAIR promotes neuronal damage through facilitating NLRP3 mediated-pyroptosis activation in Parkinson’s disease via regulation of miR-326/ELAVL1 axis. Cell Mol. Neurobiol. 41, 1773–1786. doi: 10.1007/s10571-020-00946-8
Zhang, Q. S., Wang, Z. H., Zhang, J. L., Duan, Y. L., Li, G. F., and Zheng, D. L. (2016). Beta-asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting α-synuclein protein expression. Biomed. Pharmacother. 83, 153–159. doi: 10.1016/j.biopha.2016.06.017
Zhang, Y., Xia, Q., and Lin, J. (2020). LncRNA H19 attenuates apoptosis in MPTP-induced Parkinson’s disease through regulating miR-585-3p/PIK3R3. Neurochem. Res. 45, 1700–1710. doi: 10.1007/s11064-020-03035-w
Zhao, J., Li, H., and Chang, N. (2020). LncRNA HOTAIR promotes MPP+-induced neuronal injury in Parkinson’s disease by regulating the miR-874-5p/ATG10 axis. Excli J. 19, 1141–1153. doi: 10.17179/excli2020-2286
Zhao, M. Y., Wang, G. Q., Wang, N. N., Yu, Q. Y., Liu, R. L., and Shi, W. Q. (2019). The long-non-coding RNA NEAT1 is a novel target for Alzheimer’s disease progression via miR-124/BACE1 axis. Neurol. Res. 41, 489–497. doi: 10.1080/01616412.2018.1548747
Zhou, Y., Zhang, X., and Klibanski, A. (2012). MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 48, R45–R53. doi: 10.1530/jme-12-0008
Zhu, J., Fu, H., Wu, Y., and Zheng, X. (2013). Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 56, 876–885. doi: 10.1007/s11427-013-4553-6
Keywords: long non-coding RNAs, ageing, neurodegenerative diseases, Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis
Citation: Ni Y-Q, Xu H and Liu Y-S (2022) Roles of Long Non-coding RNAs in the Development of Aging-Related Neurodegenerative Diseases. Front. Mol. Neurosci. 15:844193. doi: 10.3389/fnmol.2022.844193
Received: 27 December 2021; Accepted: 09 February 2022;
Published: 14 March 2022.
Edited by:
Erquan Zhang, National Institute of Biological Sciences (NIBS), ChinaReviewed by:
Shuaipeng Ma, University of Texas Southwestern Medical Center, United StatesMarco Venturin, University of Milan, Italy
Copyright © 2022 Ni, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: You-Shuo Liu, bGl1eW91c2h1b0Bjc3UuZWR1LmNu
 Yu-Qing Ni1,2
Yu-Qing Ni1,2 Hui Xu
Hui Xu You-Shuo Liu
You-Shuo Liu