
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 02 August 2022
Sec. Methods and Model Organisms
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.840966
This article is part of the Research Topic Molecular and Cellular Mechanisms of Sensory Functions in Insect Models View all 6 articles
The ATP-dependent chromatin remodeling factor CHD1 (chromodomain-helicase-DNA binding protein 1) is involved in both the de novo assembly and the remodeling of chromatin. Recently, we discovered a crucial role of CHD1 in the incorporation of the histone variant H3.3 in the fly brain illustrated by widespread transcriptional upregulation and shortened lifespan in Chd1-mutant animals. Because many genes linked to sensory perception were dysregulated in Chd1-mutant heads, we studied the role of CHD1 in these processes. Here we show that Chd1-mutant flies have severe defects in their response behavior to olfactory and gustatory but not visual stimuli. Further analyses suggested that poor performance in gustatory response assays was caused by reduced motivation for foraging and feeding rather than defects in taste perception. Moreover, we show that shortened lifespan of Chd1-mutant flies is accompanied by indications of premature functional aging as suggested by defects in negative geotaxis and exploratory walking assays. The latter phenotype was rescued by neuronal re-expression of Chd1, while the olfactory defects were not. Interestingly, we found evidence for indirect regulation of the non-neuronal expression of odorant binding proteins (Obp) by neuronal expression of Chd1. Together, these results emphasize the crucial role of CHD1 activity controlling diverse neuronal processes thereby affecting healthy lifespan.
The SWI/SNF family member CHD1 (chromodomain-helicase-DNA binding protein 1) is an ATP-dependent chromatin remodeling and assembly factor (Marfella and Imbalzano, 2007; Clapier and Cairns, 2009). Since a major function of chromatin remodeling factors is to effect changes of histone-DNA interactions within or between nucleosomes, they are critical components of all processes that require access to the DNA substrate, most prominently transcription, replication, and DNA damage repair. Together with histone chaperones, such as NAP1, CHD1 can facilitate in vitro the assembly of nucleosomes into regularly spaced arrays (Lusser et al., 2005). In vivo, it has diverse roles that include functions in transcription regulation, transcription-independent chromatin assembly as well as in DNA damage repair (Simic et al., 2003; Konev et al., 2007; Simsiii et al., 2007; Petesch and Lis, 2008; Srinivasan et al., 2008; Gaspar-Maia et al., 2009; Skene et al., 2009; Morettini et al., 2011; Guzman-Ayala et al., 2015; Siggens et al., 2015; de Dieuleveult et al., 2016; Kari et al., 2016; Lee et al., 2017; Rüthemann et al., 2017; Shenoy et al., 2017).
A transcription- and replication-independent role of CHD1 is the assembly of histone H3.3 into paternal pronuclear chromatin in Drosophila, which is achieved in concert with the histone chaperone HIRA (Loppin et al., 2005; Konev et al., 2007). The H3.3 variant is considered a “replacement”-type histone, because it is deposited in a replication-independent manner, for instance during nucleosome re-assembly in the wake of transcribing polymerases (Talbert and Henikoff, 2010). Recently, we have also implicated CHD1 in H3.3 assembly in the fly brain. Its deletion resulted in reduced chromatin-associated H3.3 levels and widespread upregulation of transcription. Moreover, sugar, fatty acid, and amino acid metabolism was severely impaired, and Chd1-mutant flies exhibited reduced food intake and a strong shortening of lifespan. Consistent with this, we found that many genes linked to the regulation of hunger and satiety were dysregulated in the heads of mutant flies. Re-expression of CHD1 only in neurons rescued all these phenotypes (Schoberleitner et al., 2021) emphasizing the critical role of this chromatin assembly factor for brain function.
Food intake requires the recognition of appropriate food, which is to a large part conferred by the ability to taste and smell and also requires vision to evaluate and discriminate sumptuous food sources from dangerous substances in the environment. Because we found genes linked to sensory perception to be enriched among the genes dysregulated in Chd1-mutant flies, we set out to investigate the contribution of CHD1 to sensory perception and locomotory behavior. The results revealed that CHD1 is not required for the response to light, but that its absence causes impaired gustatory and olfactory response as well as compromised locomotory behavior. Furthermore, we found that the lack of response to the repelling odorant benzaldehyde was neither dependent on Chd1 expression in neurons, nor was it caused by the dysregulation of odorant binding proteins (Obp) observed in Chd1-mutant heads suggesting that functions in other cell types might be responsible for the olfactory defect.
Fly stocks were maintained at 25°C and 60% humidity in a 12/12 h light/dark cycle in batches of 20 flies on sugar-cornmeal media as described previously (Sebald et al., 2016). All mutations and transgenes were studied in a w1118 background. Chd1 deficient (Chd1–/–) flies were obtained by crossing Df(2L)Chd11/CyO with Df(2L)Exel7014/CyO. Chd1WT/WT flies were obtained from crosses of Df(2L)Chd11,P{Chd1WT}/CyO and Df(2L)Exel7014,P{Chd1WT}/CyO. For pan neural induction of Chd1 in Chd1 mutant flies (Chd1–/–; elav-Gal4>UAS-Chd1WT; termed Chd1elav), the lines Df(2L)Chd11,P{UASt-Chd1WT}/CyO and Df(2L)Exel7014/CyO; elav-Gal4/TM3 were combined. Elav-Gal4 (Stock ID 8760) line was obtained from the Bloomington Stock Center. For detailed description of strain genotypes see Schoberleitner et al. (2021).
For all assays described below, batches of female virgin flies in 3 or 5 technical replicates (20 flies each) at different ages were tested at the same time of day (10 a.m.) in a uniformly illuminated area at constant temperature unless otherwise stated.
To test the response to visual light we followed a procedure by Vang et al. (2012). Briefly, flies were starved in vials containing 1.5% agar for 18 h. Then they were transferred to an empty vial by tapping, and the vial was connected to a 25 cm long test tube in a dark room 30 min prior to the test to adapt to darkness. The horizontal test tube was then gently pounded down to position the flies at one end, away from the light source. A perpendicular light source (15 cm distance) was placed at the other end of the test tube (Figure 2A). To start the test, the cold light source (Schott KL 1500 LCD, 15V, 150W, 3300K, position 5) was switched on thereby establishing a light gradient of about 90 lux at the nearest point to about 3 lux at the furthest point. Every minute in the 10 min test duration, the number of flies in each third of the test tube was scored. Data were expressed as attraction response index (RI%) corresponding to the percentage of flies in the segment closest to the light source relative to the total number of flies placed in the test tube. The test was repeated three times and data were plotted as mean ± SEM.
Response to volatile repellents was assayed as described in Vang et al. (2012). Briefly, flies were starved in vials containing 1.5% agar for 18 h before transferring them to an empty test vial 30 min prior to the test. To start the test, the vial was connected to a test tube containing 100 mM or 10 mM benzaldehyde in 1.5% agar at one end and the flies were gently tapped toward the repellent (Figure 2C). The number of flies in each third of the test tube was scored every minute for 10 min. Data were expressed as repulsion response index (RI%), corresponding to the percentage of flies in the segment furthest from the attractant relative to the total number of flies placed in the test tube. The test was repeated three times and data were plotted as mean ± SEM.
Response to non-volatile chemicals was tested as described by Vang et al. (2012). Briefly, flies were starved in vials containing 1.5% agar for 18 h before transferring them by tapping to an empty test vial 30 min prior to the test. The test was started by connecting the vial with a 25 cm long test tube containing 100 mM sucrose in 1.5% agar at one end and gently tapping the flies to the end of the empty vial (Figure 2E). Every minute for 10 min the number of flies in each third of the test tube was scored. Data were expressed as attraction response index (RI%), corresponding to the percentage of flies in the segment closest to the attractant relative to the total number of flies placed in the test tube. The test was repeated three times and data were plotted as mean ± SEM.
To evaluate the flies’ ability to display reflex-like response to attractive substances in the food and motivation to feed, we assayed the proboscis extension reflex. Thirty starved (24 h) female virgin flies of each genotype were presented with glass capillaries containing a solution of sucrose to the proboscis by making contact, and the number of proboscis extension responses was determined as described in Shiraiwa and Carlson (2007) and Qi et al. (2015). Presentation of stimulus was repeated in 10 trials per fly (n = 30/genotype). The experiment was performed once, and data were plotted as mean ± SD.
The startle-induced negative geotaxis assay was performed as described in Ismail et al. (2015) (Figure 3A). Flies of the indicated ages were transferred to climbing vials (two empty vials connected face to face) without anesthesia and left to acclimatize for 1 h. Testing was performed by tapping down the flies and counting the number of flies that climb past a mark on the vials (8 cm from the bottom) within 60 sec using a custom-made apparatus. In each experiment, the test was repeated 10 times (trials) with 1 min breaks in between. The results were analyzed as% of climbing flies at each time point (n = 5 batches of 10–12 flies/genotype/experiment). The experiment was repeated three times and data were plotted as mean ± SEM.
To assess walking behavior the exploratory walking assay was performed as previously described by Ismail et al. (2015) (Figure 3C). Briefly, female virgin flies were placed individually into the center of a 14.5 cm petri dish with a 1 cm square grid. The number of grid-line crossings during 1 min was scored and graphically presented (n > 40/genotype/experiment). The experiment was repeated three times and data were plotted as mean ± SEM.
RNA-seq data from virgin female Drosophila heads (Schoberleitner et al., 2021); GEO accession number (GSE146392) were subjected to gene ontology analysis using the GOrilla tool (Eden et al., 2009) by comparing the unranked target list (differentially expressed genes with log2 fold change ≥1, adjusted p-value ≤ 0.05 and base mean ≥20) with a background list containing all transcripts identified in the sequencing analysis. Heatmaps of significantly enriched gene categories were generated using the R package pheatmap (Kolde, 2019).
Graphing and statistics were performed using GraphPad Prism software v.8.2.1. The level for statistical significance was set at p ≤ 0.05 for all statistical tests and significant differences were marked (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns, not significant).
Data from photo- and chemotaxis experiments were quantified and interpreted as response toward the non-volatile and volatile chemicals as well as light. To calculate the statistical significance of the experimental clustering (interaction trial × age × genotype effect), 3-way ANOVA analysis based on distance matrices (distances within all replicates versus distances between all replicates) was performed. Data from proboscis extension reflex (PER) experiments were presented as response frequency, i.e., percentage of responding flies per stimulation trial (total of 10) (Figure 2H). The experiment was performed once (n = 30/genotype) for each age (4 and 14 days). To estimate the statistical significance of the response frequency clustering (stimulation trial × genotype effect), 2-way ANOVA analysis based on distance matrices (distances within all replicates versus distances between all replicates) for each age timepoint (4 and 14 days) was performed.
Results obtained from the negative geotaxis assay were illustrated as climbing capability, and statistical analysis of differences between the two fly lines at the respective age and each given sequence cycle was performed by 3-way ANOVA based on distance matrices (distances within all replicates versus distances between all replicates). Exploratory walking behavior was scored as number of grid-line crossings per minute, and all data are graphically presented as individual values (n > 40/genotype/experiment, three experiments). Statistical analysis of explorative walking differences between the two fly lines at the respective age was performed by unpaired Student’s t-test.
To examine if and how CHD1 might affect sensory perception in Drosophila, we turned to our previously generated RNA-seq data from fly heads (Schoberleitner et al., 2021). The data set contains gene expression profiles from Chd1-deletion mutant flies (termed Chd1–/–), from Chd1-deletion mutant flies rescued by transgenic expression of Chd1 under the control of its native promoter (termed Chd1WT/WT) and from mutant flies that were rescued by neuron-specific expression of Chd1 under the control of the elav promoter (termed Chd1elav). We intersected the expression profiles of these lines and performed gene ontology (GO) term enrichment analysis with the 1,878 genes that were upregulated in Chd1–/– compared to Chd1WT/WT and Chd1elav fly heads. Among the highly enriched GO categories were genes associated with sensory perception including sensory perception of light, mechanical and chemical stimuli (Figure 1A). For instance, rhodopsin genes, genes encoding Obp and defective proboscis extension response (dpr) genes as well as genes encoding N-methyl-D-aspartate (NMDA) receptors were significantly upregulated in Chd1–/– flies compared to Chd1WT/WT or Chd1elav (Figures 1B–D). Furthermore, many G-protein coupled signaling-linked genes had increased transcription in Chd1–/– compared to Chd1WT/WT or Chd1elav heads (Figure 1E; Schoberleitner et al., 2021). These included genes related to serotonin-, dopamine-, GABA-, or octopaminergic signaling, along with allostatic, acetylcholine, rhodopsin, and tachykinin associated signaling (Figure 1E). The dysregulation of these genes suggested that sensory abilities of Chd1-deficient flies might be compromised.
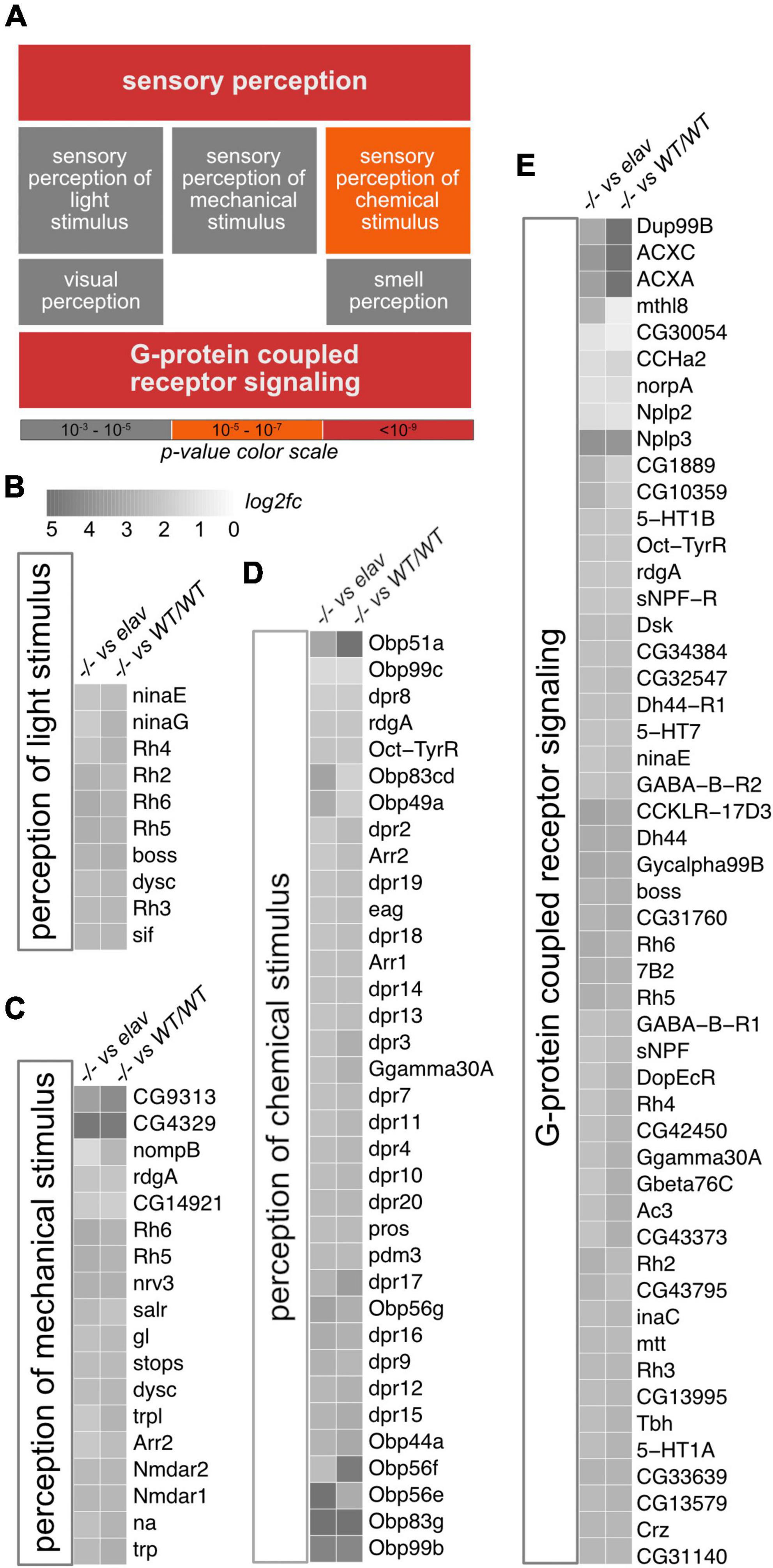
Figure 1. Gene ontology (GO) enrichment analysis of upregulated genes in Chd1–/– heads. (A) Significantly enriched GO terms from the analysis of 1897 genes that were upregulated in Chd1–/– versus Chd1WT/WT as well as Chd1–/– versus Chd1elav (log2 fold change ≥1, adjusted p ≤ 0.05, base mean ≥20). Color code signifies significance of enrichment as indicated. (B–E) Heatmaps of differentially regulated genes in enriched categories. Color scale in panel B, indicating log2 fold change (log2fc), applies to all subpanels.
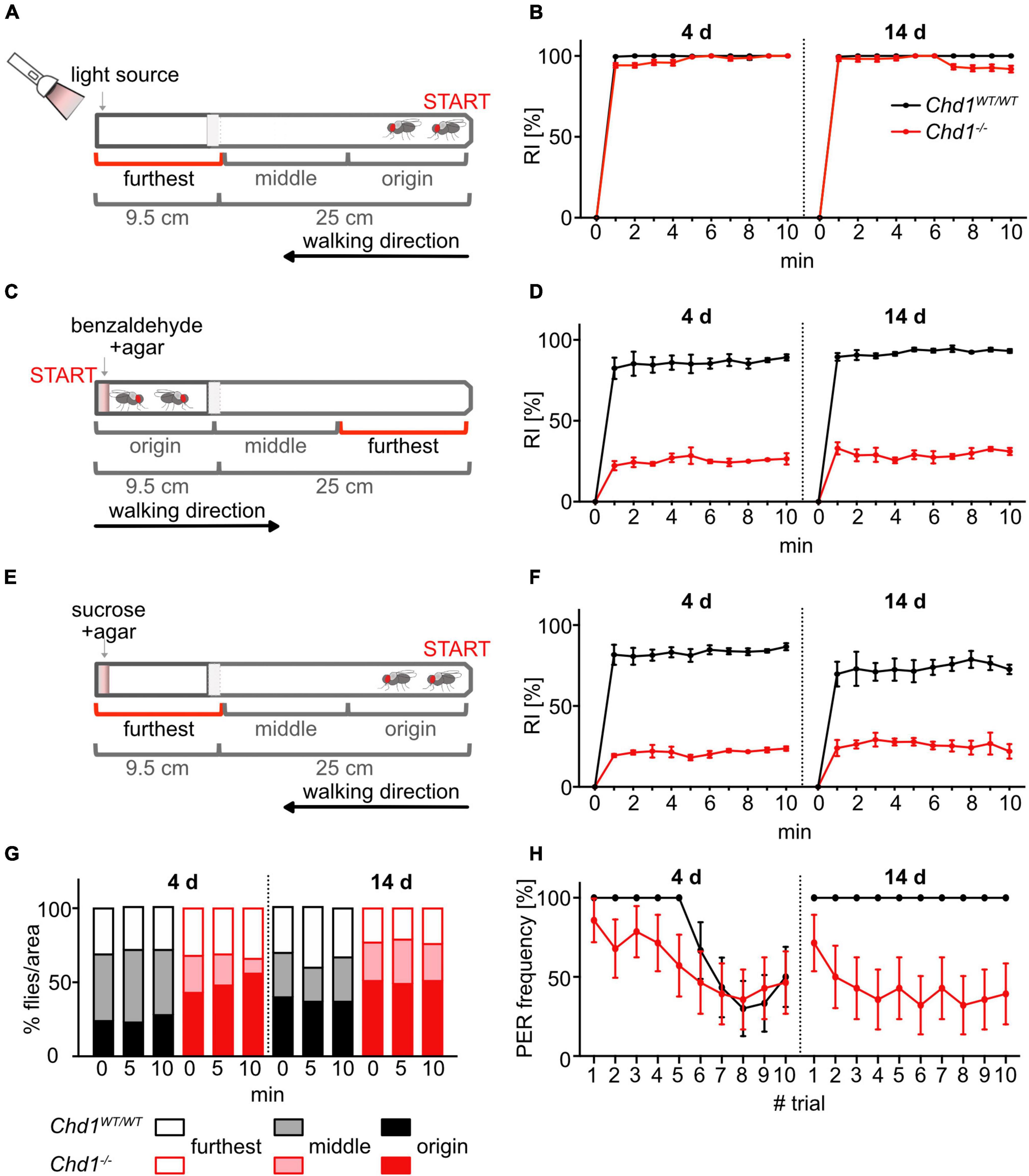
Figure 2. Absence of CHD1 impairs olfactory and gustatory perception. (A) Experimental set-up of the assay used to assess positive phototaxis. (B) Positive phototaxis response indices (RI) of 4 and 14 days old Chd1WT/WT and Chd1–/– flies. Mean ± SEM of six replicates (20 flies per group) are shown. (C,E) Experimental set-up to test olfactory behavior in response to the repellent odorant benzaldehyde (C) and gustatory response behavior to sucrose (E). (D,F) Repulsion (D) and attraction (F) response indices of 4 and 14 days old Chd1WT/WT and Chd1–/– flies. Mean ± SEM of six technical replicates (20 flies per group) and three independent experiments are shown. 3-way ANOVA of (B,D,F): genotype main effect: (B) F(1,8) = 124.66, p < 0.0001; (D) F(1,88) = 3809, p < 0.0001; (F) F(1,88) = 1680, p < 0.0001; age main effect: (B) F(1,8) = 3.267, ns; (D) F(1,88) = 28.41, p < 0.0001; (F) F(1,88) = 3.500, p = 0.0647; trial main effect: (B) F(10,80) = 5872, p < 0.0001; (D) F(10,88) = 135.8, p < 0.0001, (F) F(10,88) = 58.59, p < 0.0001; interaction (age × genotype) effect: (B) F(1,8) = 3.474, p = 0.0993; (D) F(1,88) = 1.289, p = 0.2594; (F) F(1,88) = 28.10, p < 0.0001; interaction (trial × age × genotype) effect: (B) F(10,80) = 4.458, p < 0.0001; (D) F(10,88) = 0.3576, p = 0.9613; (F) F(10,88) = 0.34789, p = 0.8996, (G) Behavior of flies in the test set-up shown in panels C,E in the absence of any stimulus. Percentage of flies in the different sectors at 0, 5, and 10 min of the test. Mean ± SEM of 3 technical replicates (20 flies per group) and 3 independent experiments are shown. 2-way ANOVA of interaction (genotype × time lapse) effect: F(22,684) = 887713; genotype main effect: F(11,684) = 517.0; time lapse effect: F(2,684) = 2934590; age main effect: F(9,27) = 3.535, p = 0.0052; genotype main effect: F(3,27) = 28.22. (H) Proboscis extension response (PER) frequency in 10 sequential tastant offerings with ingestion permitted (n = 30 per genotype and age). 2-way ANOVA of age main effect: F(9,27) = 3.535; genotype main effect: F(3,27) = 28.22.
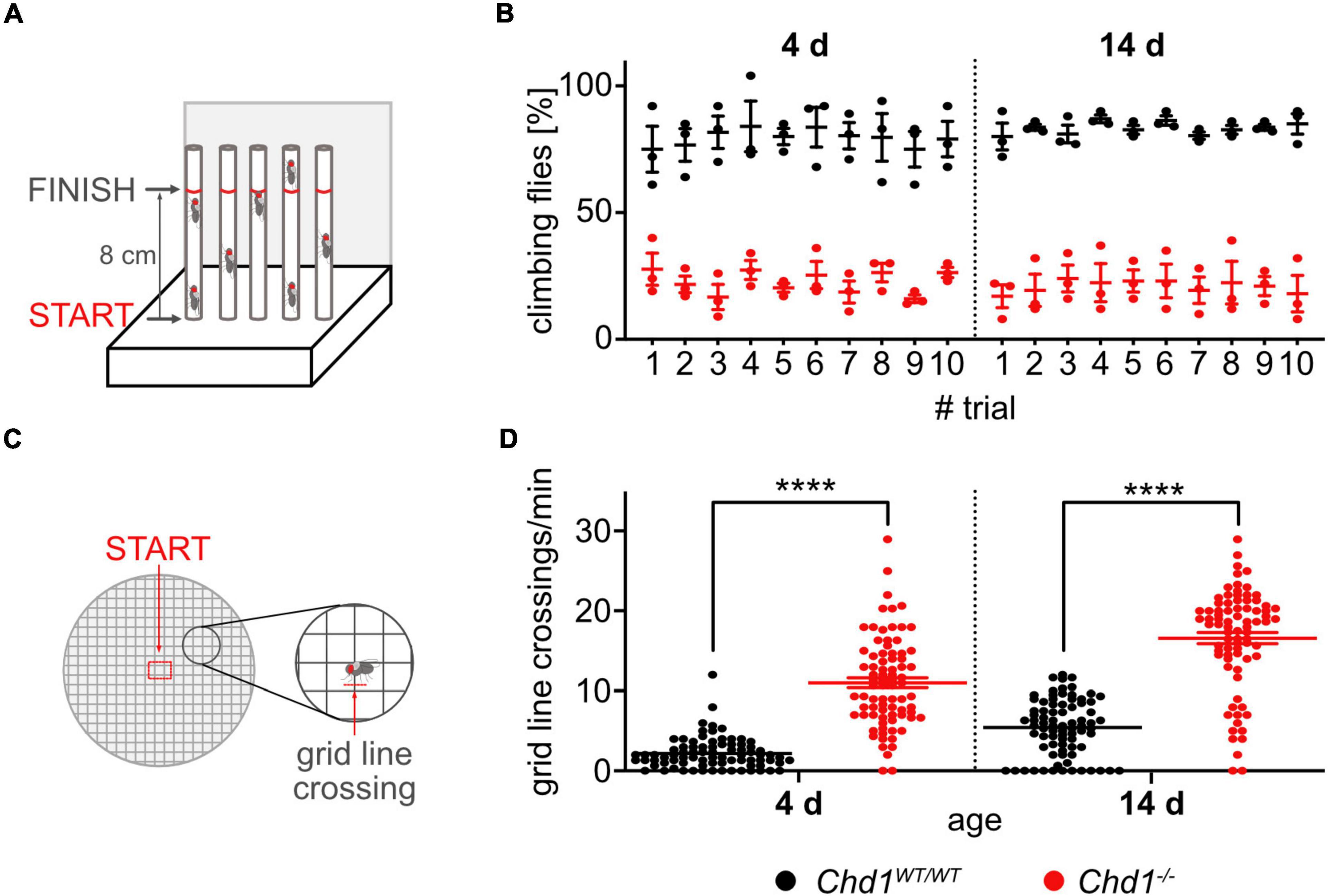
Figure 3. Compromised climbing and exploratory walking behavior of Chd1-deficient flies. (A) Experimental set-up of the negative geotaxis assay. (B) Percentage of 4 and 14 days old flies climbing to 8 cm height within 60 s in 10 trials. Mean ± SEM of 5 cohorts of 10–12 flies per genotype from 3 independent experiments is shown. 3-way ANOVA results for genotype main effect: F(1,8) = 197.2, p < 0.0001; age main effect: F(1,8) = 0.05742, p = 0.8167. (C) Experimental set-up for exploratory walking behavior. (D) Gridline crossings of individual 4 and 14 days old flies during 1 min were scored. Three independent experiments of n > 40 per genotype/experiment were performed. Statistical significance was determined by unpaired Student’s t-test. ****p < 0.0001.
To examine this idea, we performed various experiments to assess the behavioral response of the flies to sensory cues. We first tested response to light using a simple phototaxis assay (Vang et al., 2012). Mated female flies (4 or 14 days old) were placed in a 34 cm long tube, gently tapped to the bottom of the vial and scored for their tendency to move toward a light source that was placed at the opposite end of the tube (Figure 2A). The number of flies present in each area of the test tube, i.e., at the origin, in the middle and the furthest part (Figure 2A), was counted every minute. Interestingly, the experiment revealed no difference between Chd1–/– and Chd1WT/WT (Figure 2B). Young w1118 flies (wildtype background control) also responded to the light by immediately walking toward it, but then exhibited a tendency to move backward again accounting for the slightly lower RI values detected with this line (Supplementary Figure 1A). Thus, Chd1 deletion did not affect light reception and positive phototaxis behavior, although we cannot rule out that other aspects of vision are compromised.
Next, we tested olfactory abilities by observing the behavior of Chd1-deficient and Chd1-wildtype flies in a similar experimental set-up, except that the strongly repellent odor benzaldehyde was added to an agar disc that was placed at one end of the apparatus (Figure 2C). As expected, wildtype flies (w1118, Canton S) and Chd1WT/WT flies at both ages showed strong repulsion by benzaldehyde by walking away from the repellent after exposure (Figure 2D and Supplementary Figure 1B). By contrast, about 70% of 4- and 14-day-old Chd1-deficient flies remained close to the benzaldehyde source, where they had been placed at the start of the assay (Figure 2D). Because Chd1–/– flies showed similar walking behavior in the phototaxis assay as the wildtype flies, it is not likely that a potential impairment of locomotory abilities acted as a confounding parameter in this assay. Thus, the results suggest that absence of CHD1 strongly impacts on the sense of smell.
To assess the gustatory abilities of the flies, sucrose was offered as an attractant in the assay set-up shown in Figure 2E. Sucrose must be tasted because it is not volatile. Starved Chd1WT/WT flies of both ages departed from the non-sucrose end, and the majority accumulated in the compartment nearest to the sucrose similar to w1118 flies (Figure 2F and Supplementary Figure 1C). By contrast, the behavior of Chd1–/– flies was comparable to the smell test with only about 20.2 ± 0.3% of the 4-day-old and 27.9 ± 0.5% of the 14-day-old mutant flies moving close to the sucrose source (Figure 2F). To ascertain that the observed aberrant behavior of Chd1-deficient flies was caused by defective gustatory perception and not by reduced motivation for foraging, we subjected the flies to the same assay set-up yet omitting the taste stimulus. The 4 days old starved Chd1WT/WT flies showed roughly equal distribution across all sections of the tube at each time point with a slight preference for the middle part, whereas older wildtype flies slightly preferred the origin and furthest sections. Chd1–/– flies were similar to the older wildtype flies except for a slight preference of the origin section at both ages (Figure 2G) suggesting that reduced motivation to forage might indeed contribute to the poor gustatory response observed in the tube assay. To examine taste abilities of the flies by another method, we employed the PER assay (Shiraiwa and Carlson, 2007). When the taste neuron-innervated sensilla of the fly’s labellum make contact to an attractive substance, such as sucrose solution, the proboscis is extended to consume the food. Monitoring the frequency of PER across 10 experimental trials, we observed that in young flies, the PER frequency progressively decreased in both Chd1WT/WT and Chd1–/– flies most likely reflecting progressive satiety (Wang et al., 2004; Shiraiwa and Carlson, 2007; Slone et al., 2007; Chen and Amrein, 2014). At 14 days of age, Chd1-wildtype flies showed 100% response frequency throughout the experiment, whereas Chd1-mutant flies started with about 75% response which gradually decreased to about 50% response (Figure 2H). Given that young Chd1-deficient flies exhibited almost normal PER, it is likely that their gustatory abilities are principally intact, at least at this young age. At 14 days of age, most of the flies initially responded to the stimulus but very quickly turned to satiety-like behavior. Together, the results from the gustatory tube assay and the PER assay suggest that absence of CHD1 does not cause a (complete) loss of taste perception. Rather, it appears that the flies can still taste but might have diminished motivation to feed. This is consistent with our previous observations that Chd1-deficient flies show strongly reduced food intake correlating with dysregulation of many neuropeptide genes responsible for feeding and foraging behavior (Schoberleitner et al., 2021).
Although walking abilities of Chd1–/– flies appeared similar to wildtype flies in the phototaxis assay described above, we performed two additional locomotory assays. The negative geotaxis assay measures the startle-induced reflexive tendency of flies to move against gravity (Figure 3A), while in the exploratory walking assay (Figure 3C), more complex behavior involving the regulation of walking parameters, such as speed and direction, as well as orientation is assessed. Both, reflexive as well as decision-based locomotion, however, are substantially affected by age-related decline and are therefore also used to test for functional senescence (Grotewiel et al., 2005; Ismail et al., 2015).
We found that both 4 and 14 days old Chd1–/– flies performed significantly worse in the negative geotaxis assay compared to wildtype (Figure 3B and Supplementary Figure 1D). Chd1-deficient flies were slow climbing upward and showed numerous direction changes compared to Chd1-wildtype and w1118 flies that were moving only upward.
For the exploratory walking assay, flies were placed individually into the middle of a graduated petri dish, and their movement was monitored by counting the number of grid-line crossings (Figure 3D). Typically, the physical borders of an open field (borders of the Petri dish) are of main interest for the fly, and it will normally strive to head there (Soibam et al., 2012). Accordingly, Chd1-wildtype flies after placement into the center of the dish either remained rooted to the spot observing the environment for a short time or walked by the shortest path toward the border of the Petri dish and remained there. A similar, albeit not as pronounced behavior was displayed by w1118 flies (Supplementary Figure 1E). By contrast, Chd1-mutant flies immediately started to walk in circles (many grid-line crossings) but did not head for the border (Figure 3D). When 14 days old flies were tested, we observed that the tendency to go to the border of the dish by the shortest way decreased also for Chd1-wildtype flies, but the difference between the genotypes remained (Figure 3D). The reduced performance of the flies in these test paradigms in the absence of CHD1 suggest an earlier entry into a state of age-related decline compared to wildtype flies.
The RNA-seq data had shown that most dysregulated genes in Chd1–/– heads were rescued by both neuron-specific expression of Chd1 or expression under the control of its native promoter (Schoberleitner et al., 2021). To examine, if neuronal expression of Chd1 in the Chd1-deficient background would affect the sensory and locomotory abilities of the fly, we performed negative geotaxis, exploratory walking, gustatory, and olfactory assays also with Chd1elav flies.
The results show that negative geotaxis, exploratory walking abilities, and gustatory response were completely rescued by expression of Chd1 with the elav driver (Figures 4A–C) indicating that activity of CHD1 in neurons is responsible for the respective defects in Chd1–/– flies. Intriguingly, however, neuronal expression of Chd1 was not sufficient to rescue the olfactory deficits of Chd1–/– flies (Figure 4D). Hence, for proper olfactory response, CHD1 activity in other cell types in addition to neurons is required.
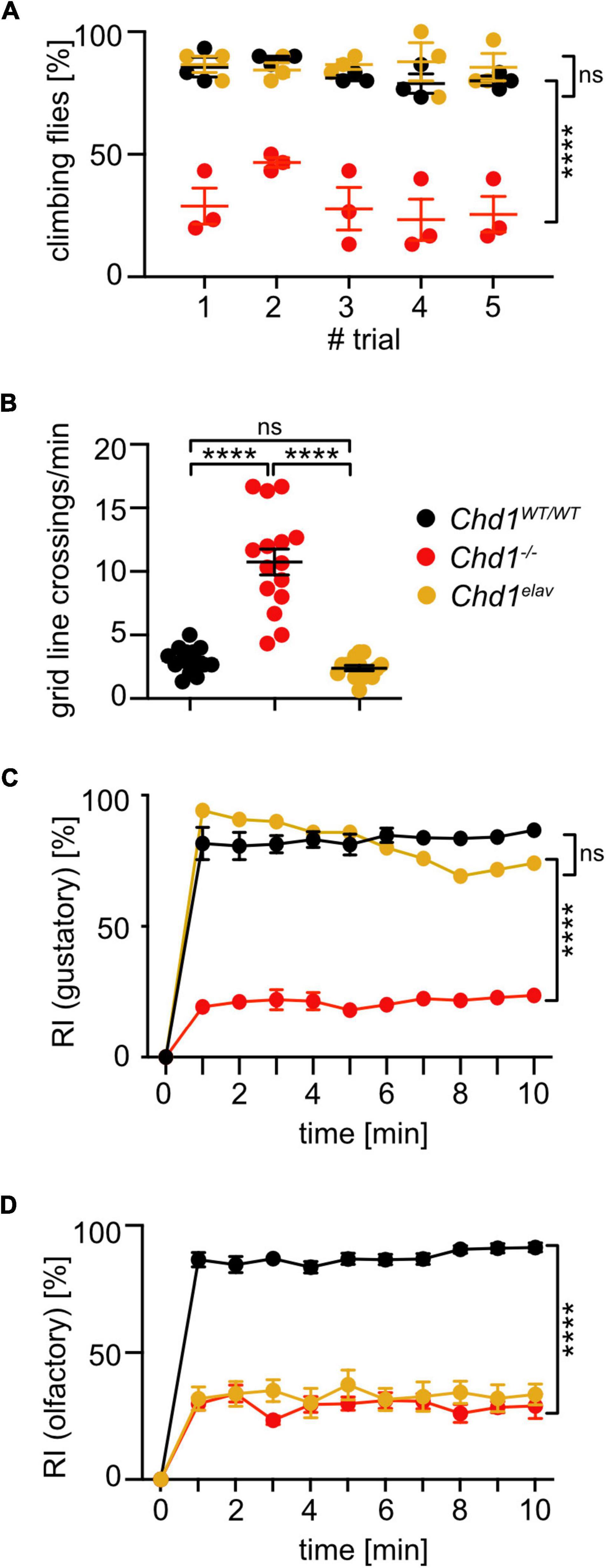
Figure 4. Pan-neuronal expression of Chd1 rescues locomotory and gustatory impairments but has no effect on olfactory response behavior. (A) Negative geotaxis assay to assess startle-induced climbing behavior. Percentage of 7 days old flies climbing to 8 cm height within 60 s was determined in 5 trials. Mean ± SEM of 3 cohorts of 10 flies each per genotype/experiment from 3 independent experiments is shown. Significant differences between fly lines was determined by unpaired Student’s t-test. ****p < 0.0001, ns, not significant. (B) Gridline crossings in exploratory walking assay of individual 7 days old flies during 1 min were scored. Three independent experiments of n > 15 per genotype/experiment were performed. Statistical significance was determined by unpaired Student’s t-test. ****p < 0.0001. (C) Attraction response index (RI) to sucrose of 7 days old flies was tested as described in Figure 2. Mean ± SEM of three technical replicates (20 flies per group) and three independent experiments are shown, except for Chd1elav flies which were tested in 6 technical replicates (20 flies each) and one experiment. (D) Benzaldehyde of 7 days old flies was tested as described in Figure 2. Mean ± SEM of three technical replicates (20 flies per group) and three independent experiments are shown. Statistical significance was determined by 2-way ANOVA: Genotype main effect: (C) F(2,44) = 44.46, ****p < 0.0001, and (D) F(7,704) = 457.6, ****p < 0.0001; trial main effect: (C) F(10,44) = 254.4, ****p < 0.0001, and (D) F(10,704) = 110.4, ****p < 0.0001; trial × genotype interaction effect: (C) F(20,44) = 3910, ****p < 0.0001, and (D) F(70,704) = 5.453, ****p < 0.0001.
We have previously shown that CHD1 expression in the brain is required for normal regulation of several metabolic parameters, such as sugar, fatty acid, and amino acid homeostasis. Lack of CHD1 results in reduced food intake, increased global inflammation and premature death. On the molecular level, these phenotypes are linked to reduced chromatin-associated levels of the histone variant H3.3, which is incorporated into chromatin mostly during transcription-related processes that lead to the loss of histones. Consequently, Chd1–/– flies exhibit increased global transcription presumably due to a general relief of chromatin-mediated repression (Schoberleitner et al., 2021). Here we expand on these findings and show that absence of CHD1 results in olfaction deficits, reduced gustatory response, which is most likely caused by reduced motivation for feeding, as well as age-related locomotory impairments. GO enrichment analyses revealed that these phenotypic deficits are in agreement with the dysregulation of genes known to be involved in these processes. Re-expression of Chd1 in neurons rescued the expression of most of the dysregulated genes (Figure 1) as well as the climbing, exploratory walking and gustatory abilities of the flies. Given that the two locomotion assays are often used to assess functional aging (Grotewiel et al., 2005; Ismail et al., 2015), these results, in combination with our previous findings revealing that neuronal expression of Chd1 rescues shortened lifespan and chronic inflammation, support the notion that CHD1 plays an important role in the prevention of premature aging.
The reasons for the complete failure to restore olfactory perception by neuronal Chd1 expression, however, remain unknown. Deletion of Chd1 caused the upregulation of 16 out of 34 Obp genes that were detected in our RNA-seq analysis suggesting an apparently obvious mechanism to explain the olfactory response phenotype. However, even though transcription of almost all dysregulated Obp genes was restored in Chd1elav flies, olfactory response was still impaired. Diverse functions have been ascribed to OBPs in insects, ranging from roles in the transport of odorants to odorant receptors, the sequestration of odorants from the sensillum lymph, the function as co-ligands at neuronal receptors or the protection of odorants from degradation (Scheuermann and Smith, 2019). Even though most studies have examined the functional effects of OBP ablation rather than of overexpression as we observe here, reduction of certain OBPs sometimes resulted in increased response to certain odorants whereas response to others was lost indicating that the mode of action of OBPs is complex and may be different for different OBPs, different odorants and different cell types (Swarup et al., 2011; He et al., 2019; Scheuermann and Smith, 2019). Interestingly, a recent study reported that deletion of the ten most abundant Obp genes in antennal basiconic sensilla did not affect olfactory abilities of the flies (Xiao et al., 2019) leading the authors to conclude that olfaction is not dependent on the abundant OBPs or that minor OBPs can compensate (Xiao et al., 2019). Our results would support this notion in the sense that the dysregulation/rescue of Obp expression had no effect on the flies’ olfactory response, at least the response to benzaldehyde. It is possible, however, that the reactions are different for other odorant types. Regardless of the potential role of OBPs in the olfaction process, our results also suggest that functions of CHD1 outside of neurons must be critical for the failure of Chd1elav flies to respond to benzaldehyde.
Another intriguing observation from our study is that neuronal expression of Chd1 is sufficient to restore expression of Obp genes. OBPs are synthesized by non-neuronal chemosensory support cells in antennal and other sensilla and secreted into the lymph of the sensillum. Thus, the rescue of Chd1 expression in Chd1elav flies, which is supposed to occur only in the sensory neurons, actually should not affect the regulation of Obp genes in the support cells. A hint toward a potential mechanism to explain this apparent discrepancy comes from a recent study on the function of Obp69a in translating social interaction into sex-specific behavior involving the male-specific pheromone 11-cis-Vaccenyl acetate (cVA). The authors found that expression of Obp69a in the auxiliary cells was regulated by active neurotransmission of the cVA sensing neuron to the second order olfactory neuron (Bentzur et al., 2018). In analogy to this, we speculate that the activity of sensory neurons in Chd1–/– flies might be altered, which is also illustrated by the dysregulation of multiple genes linked to G-protein-coupled receptor signaling and neurotransmission (Figure 1E; Schoberleitner et al., 2021). This may affect the regulation of Obp genes in the support cells. Upon neuronal re-expression of Chd1 (Chd1elav), neuronal function is restored (Figure 1E), which may cause the downregulation of the Obp genes in the support cells.
In summary, we have characterized CHD1 as an important factor contributing to neuronal function and regulating sensory and locomotory behavior. As a chromatin regulator, the molecular mechanism behind these functions involves regulation of transcription including, but not limited to, histone variant H3.3 incorporation thereby maintaining chromatin integrity (Schoberleitner et al., 2021). Considering that CHD1 as well as its significance for brain function are conserved between insects and mammals (Piatti et al., 2015; Schoberleitner et al., 2019; Cardoso et al., 2021), these findings may also be relevant for the study of age-related decline processes in humans.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
IS and AL conceived the project, designed the experiments, and wrote the manuscript. IS and BM performed experiments and analyzed the data. IB analyzed the RNA-seq data. All authors read and approved the final manuscript.
Research on this project was funded by the Austrian Science Fund (FWF) [P31377-B30] to AL.
We thank Gabriele Salcher-Scheran for technical help and Georg Dechant (Medical University of Innsbruck) for critical reading and suggestions on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.840966/full#supplementary-material
Bentzur, A., Shmueli, A., Omesi, L., Ryvkin, J., Knapp, J. M., Parnas, M., et al. (2018). Odorant binding protein 69a connects social interaction to modulation of social responsiveness in Drosophila. PLoS Genet. 14:e1007328. doi: 10.1371/journal.pgen.1007328
Cardoso, A. R., Lopes-Marques, M., Oliveira, M., Amorim, A., Prata, M. J., and Azevedo, L. (2021). Genetic variability of the functional domains of chromodomains helicase DNA-Binding (CHD) proteins. Genes (Basel) 12:1827. doi: 10.3390/genes12111827
Chen, Y., and Amrein, H. (2014). Enhancing perception of contaminated food through acid-mediated modulation of taste neuron responses. Curr. Biol. 24, 1969–1977. doi: 10.1016/j.cub.2014.07.069
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
de Dieuleveult, M., Yen, K., Hmitou, I., Depaux, A., Boussouar, F., Bou Dargham, D., et al. (2016). Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530, 113–116. doi: 10.1038/nature16505
Eden, E., Navon, R., Steinfeld, I., Lipson, D., and Yakhini, Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. doi: 10.1186/1471-2105-10-48
Gaspar-Maia, A., Alajem, A., Polesso, F., Sridharan, R., Mason, M. J., Heidersbach, A., et al. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868. doi: 10.1038/nature08212
Grotewiel, M. S., Martin, I., Bhandari, P., and Cook-Wiens, E. (2005). Functional senescence in Drosophila melanogaster. Ageing Res. Rev. 4, 372–397. doi: 10.1016/j.arr.2005.04.001
Guzman-Ayala, M., Sachs, M., Koh, F. M., Onodera, C., Bulut-Karslioglu, A., Lin, C.-J., et al. (2015). Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development (Cambridge, England) 142, 118–127. doi: 10.1242/dev.114843
He, Z., Luo, Y., Shang, X., Sun, J. S., and Carlson, J. R. (2019). Chemosensory sensilla of the Drosophila wing express a candidate ionotropic pheromone receptor. PLoS Biol. 17:e2006619. doi: 10.1371/journal.pbio.2006619
Ismail, M. Z. B. H., Hodges, M. D., Boylan, M., Achall, R., Shirras, A., and Broughton, S. J. (2015). The Drosophila insulin receptor independently modulates lifespan and locomotor senescence. PLoS One 10:e0125312. doi: 10.1371/journal.pone.0125312
Kari, V., Mansour, W. Y., Raul, S. K., Baumgart, S. J., Mund, A., Grade, M., et al. (2016). Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 17, 1609–1623. doi: 10.15252/embr.201642352
Kolde, R. (2019). “pheatmap: Pretty Heatmaps”(RRID:SCR_016418)
Konev, A. Y., Tribus, M., Park, S. Y., Podhraski, V., Lim, C. Y., Emelyanov, A. V., et al. (2007). CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science (New York, N.Y.) 317, 1087–1090. doi: 10.1126/science.1145339
Lee, Y., Park, D., and Iyer, V. R. (2017). The ATP-dependent chromatin remodeler Chd1 is recruited by transcription elongation factors and maintains H3K4me3/H3K36me3 domains at actively transcribed and spliced genes. Nucleic Acids Res. 45, 7180–7190. doi: 10.1093/nar/gkx321
Loppin, B., Bonnefoy, E., Anselme, C., Laurençon, A., Karr, T. L., and Couble, P. (2005). The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390. doi: 10.1038/nature04059
Lusser, A., Urwin, D. L., and Kadonaga, J. T. (2005). Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12, 160–166. doi: 10.1038/nsmb884
Marfella, C. G. A., and Imbalzano, A. N. (2007). The Chd family of chromatin remodelers. Mut. Res. 618, 30–40. doi: 10.1016/j.mrfmmm.2006.07.012
Morettini, S., Tribus, M., Zeilner, A., Sebald, J., Campo-Fernandez, B., Scheran, G., et al. (2011). The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res. 39, 3103–3115. doi: 10.1093/nar/gkq1298
Petesch, S. J., and Lis, J. T. (2008). Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84. doi: 10.1016/j.cell.2008.05.029
Piatti, P., Lim, C. Y., Nat, R., Villunger, A., Geley, S., Shue, Y. T., et al. (2015). Embryonic stem cell differentiation requires full length Chd1. Sci. Rep. 5:8007. doi: 10.1038/srep08007
Qi, W., Yang, Z., Lin, Z., Park, J. Y., Suh, G. S., and Wang, L. (2015). A quantitative feeding assay in adult Drosophila reveals rapid modulation of food ingestion by its nutritional value. Mol. Brain 8:87. doi: 10.1186/s13041-015-0179-x
Rüthemann, P., Balbo Pogliano, C., Codilupi, T., Garajovà, Z., and Naegeli, H. (2017). Chromatin remodeler CHD1 promotes XPC-to-TFIIH handover of nucleosomal UV lesions in nucleotide excision repair. EMBO J. 36, 3372–3386. doi: 10.15252/embj.201695742
Scheuermann, E. A., and Smith, D. P. (2019). Odor-specific deactivation defects in a Drosophila odorant-binding protein mutant. Genetics 213, 897–909. doi: 10.1534/genetics.119.302629
Schoberleitner, I., Bauer, I., Huang, A., Andreyeva, E. N., Sebald, J., Pascher, K., et al. (2021). CHD1 controls H3.3 incorporation in adult brain chromatin to maintain metabolic homeostasis and normal lifespan. Cell Rep. 37:109769. doi: 10.1016/j.celrep.2021.109769
Schoberleitner, I., Mutti, A., Sah, A., Wille, A., Gimeno-Valiente, F., Piatti, P., et al. (2019). Role for chromatin remodeling factor Chd1 in learning and memory. Front. Mol. Neurosci. 12:3. doi: 10.3389/fnmol.2019.00003
Sebald, J., Willi, M., Schoberleitner, I., Krogsdam, A., Orth-Höller, D., Trajanoski, Z., et al. (2016). Impact of the chromatin remodeling factor CHD1 on gut microbiome composition of Drosophila melanogaster. PLoS One 11:e0153476. doi: 10.1371/journal.pone.0153476
Shenoy, T. R., Boysen, G., Wang, M. Y., Xu, Q. Z., Guo, W., Koh, F. M., et al. (2017). CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann. Oncol. 28, 1495–1507. doi: 10.1093/annonc/mdx165
Shiraiwa, T., and Carlson, J. R. (2007). Proboscis extension response (PER) assay in Drosophila. J. Vis. Exp. 193. doi: 10.3791/193 [Epub ahead of print].
Siggens, L., Cordeddu, L., Rönnerblad, M., Lennartsson, A., and Ekwall, K. (2015). Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions. Epigenetics Chromatin 8:4. doi: 10.1186/1756-8935-8-4
Simic, R., Lindstrom, D. L., Tran, H. G., Roinick, K. L., Costa, P. J., Johnson, A. D., et al. (2003). Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22, 1846–1856. doi: 10.1093/emboj/cdg179
Simsiii, R., Millhouse, S., Chen, C., Lewis, B., Erdjumentbromage, H., Tempst, P., et al. (2007). Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and Pre-mRNA splicing. Mol. Cell 28, 665–676. doi: 10.1016/j.molcel.2007.11.010
Skene, P. J., Hernandez, A. E., Groudine, M., and Henikoff, S. (2009). The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife 3:e02042. doi: 10.7554/eLife.02042
Slone, J., Daniels, J., and Amrein, H. (2007). Sugar receptors in Drosophila. Curr. Biol. 17, 1809–1816. doi: 10.1016/j.cub.2007.09.027
Soibam, B., Mann, M., Liu, L., Tran, J., Lobaina, M., Kang, Y. Y., et al. (2012). Open-field arena boundary is a primary object of exploration for Drosophila. Brain Behav. 2, 97–108. doi: 10.1002/brb3.36
Srinivasan, S., Dorighi, K. M., and Tamkun, J. W. (2008). Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 4:e1000217. doi: 10.1371/journal.pgen.1000217
Swarup, S., Williams, T. I., and Anholt, R. R. (2011). Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 10, 648–657. doi: 10.1111/j.1601-183X.2011.00704.x
Talbert, P. B., and Henikoff, S. (2010). Histone variants — ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275. doi: 10.1038/nrm2861
Vang, L. L., Medvedev, A. V., and Adler, J. (2012). Simple ways to measure behavioral responses of Drosophila to stimuli and use of these methods to characterize a novel mutant. PLoS One 7:e37495. doi: 10.1371/journal.pone.0037495
Wang, Z., Singhvi, A., Kong, P., and Scott, K. (2004). Taste representations in the Drosophila brain. Cell 117, 981–991. doi: 10.1016/j.cell.2004.06.011
Keywords: chromatin remodeling factor, neuron, perception, olfaction, locomotion, transcriptional regulation, histone variant
Citation: Schoberleitner I, Mertens B, Bauer I and Lusser A (2022) Regulation of sensory perception and motor abilities by brain-specific action of chromatin remodeling factor CHD1. Front. Mol. Neurosci. 15:840966. doi: 10.3389/fnmol.2022.840966
Received: 21 December 2021; Accepted: 07 July 2022;
Published: 02 August 2022.
Edited by:
Youngseok Lee, Kookmin University, South KoreaReviewed by:
Jae Young Kwon, Sungkyunkwan University, South KoreaCopyright © 2022 Schoberleitner, Mertens, Bauer and Lusser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra Lusser, QWxleGFuZHJhLmx1c3NlckBpLW1lZC5hYy5hdA==
†Present Address: Ines Schoberleitner, Medical University of Innsbruck, Innsbruck, Austria
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.