
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 22 February 2023
Sec. Pain Mechanisms and Modulators
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.1027063
This article is part of the Research Topic Molecular Mechanism of Neuroimmune Modulation and Synaptic Plasticity in Acute and Chronic Pain View all 19 articles
Introduction: Long non-coding RNAs (lncRNAs) act as key regulators in multiple human diseases. In particular, the dysfunction of lncRNAs in dorsal root ganglion (DRG) contributes to the pathogenesis of neuropathic pain (NP). Nevertheless, the role and mechanism of most lncRNAs in NP remain unclear.
Methods: Two classic chronic NP models, including L4 spinal nerve ligation (SNL) model and chronic constriction injury (CCI) of the sciatic nerve, were performed. Mechanical allodynia and heat hyperalgesia were used to evaluate neuropathic pain. DRG microinjection was used to deliver agents into DRG. qRT-PCR, immunofluorescence, immunoprecipitation, western blotting, siRNA transfection, AAV transduction were performed to investigate the phenotypes and molecular basis.
Results: Here, we discovered that Rmst as a lncRNA was specifically expressed in Atf3+ injured DRG neurons and significantly upregulated following peripheral nerve damage. Rmst overexpression by direct DRG injection of AAV5-Rmst causes neuropathic symptoms in the absence of nerve damage. Conversely, blocking Rmst expression in injured DRGs alleviated nerve injury-induced pain hypersensitivities and downregulated Dnmt3a expression. Furthermore, we found peripheral nerve damage induced Rmst increase could interact with RNA-binding protein HuR to stabilize the Dnmt3a mRNA.
Conclusion: Our findings reveal a crucial role of Rmst in damaged DRG neurons under NP condition and provide a novel target for drug development against NP.
Chronic pain has been a perplexing problem for decades. Globally, approximately one-fifth of people suffer from chronic pain (Tompkins et al., 2017), and about thirty percent of these patients have gone through the symptoms of neuropathic pain (NP) (van Hecke et al., 2014). Patients with NP have detrimental effects on their quality of life and ability. In addition to personal suffering, chronic pain is a substantial financial burden on society, and lies billions of dollars every year (van Velzen et al., 2020). However, there are limited treatments available for NP. Dorsal root ganglia (DRG) neuron is responsible for conveying the nociception in peripheral nerve injury. Peripheral nerve damage-induced maladaptive alterations at transcriptional and translational levels of pain-associated genes in the primary afferents of DRG neurons contribute to these abnormal spontaneous activities and subclinical behavioral patterns (Li et al., 2020; Pan et al., 2021). Hence, identifying new targets and mechanisms in damaged DRGs could open a novel avenue against NP.
As potent and multifaceted roles of long non-coding RNAs (lncRNAs) in gene regulation, lncRNAs are paid much attention in many human illnesses, including NP (Wu et al., 2016, 2019). However, the function of most lncRNAs in NP is not thoroughly understood. A lncRNA called rhabdomyosarcoma 2-associated transcript (Rmst) was essential for neuronal differentiation (Ng et al., 2012) and neurogenesis (Ng et al., 2013). Although previous studies showed that Rmst was involved in neurological disorders (Hou and Cheng, 2018; Ma et al., 2021; Zhao et al., 2021; Li et al., 2022), little is known about its role in NP.
DNA methyltransferase 3 alpha gene encodes the Dnmt3a protein and is responsible for catalyzing 5-methylcytosine methylation. The aberrant regulation of Dnmt3a is implicated in multiple nervous diseases (Feng et al., 2005; Clemens and Gabel, 2020), especially in NP (Guo et al., 2019). Increased Dnmt3a have been found in injured DRG (Shao et al., 2017; Zhao et al., 2017). What’s more, the increased-Dnmt3a was able to mediate the epigenetic inaction of the voltage-dependent potassium channel subunit (Kcna2) via DNA hypermethylation of Kcna2 promoter region (Zhao et al., 2017). Depletion of Dnmt3a in the injured DRGs effectively attenuated NP by restoring Kcna2 expression (Zhao et al., 2017, p. 3). These studies suggested that Dnmt3a plays a pivotal role in regulating DNA methylation of nerve damage related gene alterations. Nevertheless, it is unclear how peripheral nerve damage triggers the activation of Dnmt3a in injured DRG.
Here, we sought to characterize a lncRNA rhabdomyosarcoma 2-associated transcript (Rmst) in injured DRG neurons was substantially increased in NP. The nerve damage triggered Rmst expression in injured DRGs contributes to regulating Dnmt3a through interaction with RNA binding protein HuR in NP. Thus, the role and mechanism of Rmst may provide novel and insightful directions for NP management in clinic.
RNA sequencing dataset for mouse DRG after peripheral nerve injury was obtained from previous research (Wu et al., 2016). The mouse DRG was harvested 7 days after L4 spinal nerve ligation (SNL) model and sequencing was performed on the Illumina HiSeq2500 platform with 2 × 100-bp paired-end reads.
For scRNA-seq dataset for mouse DRG after peripheral nerve injury, we obtained the cell count matrix and metadata from Gene Expression Omnibus (GEO) with the series record GSE155622 (Wang K. et al., 2021). The mouse DRG was harvested for Smart-seq2 as smart-seq2 was better to captured low abundance transcripts as well as more lncRNAs (Wang X. et al., 2021). All metadata was also obtained from GSE155622.
C57BL/6J adult mice were from SPF Biotechnology Co., Ltd (Beijing, China). A 12-h light-dark cycle environment with unlimited access to food and water was used to house mice. All processes were endorsed by the Animal Care and Use Committee at Guangzhou Medical University.
Two classic chronic NP models, including L4 SNL model and chronic constriction injury (CCI) of the sciatic nerve, were performed as described previously (Li et al., 2020). SNL model was established by tightly ligating L4 spinal nerve distal to DRG with 7-0 silk suture. L4 spinal nerve was exposed in the matched sham mice, however, the L4 spinal nerve was neither ligated nor transected. CCI model was established by loosely ligated the sciatic nerve at three spots with 1 mm-intervals by 7-0 silk suture. Sham animals did not receive the ligation of the sciatic nerve.
As previous described, mechanical allodynia was quantified by measuring paw withdrawal frequency by low (0.07 g) and median (0.4 g) von Frey filaments (Stoelting Co., Wood Dale, IL, USA) (Li et al., 2020). We use two calibrated plastic filaments to stimuli the central of plantar surface of hind paws. A positive response is quick withdrawal of the paw. Mice were totally received 10 applications. The paw withdrawal frequency describes the positive withdrawal responses within 10 applications.
Heat hyperalgesia was quantified by measuring paw withdrawal latencies after heat stimulation as described (Li et al., 2020). All mice before behavior test were left in a glass surface in individual plexiglas cages. A beam of light was applied to the central of hind paw. The performance of a positive response is a swift raise of the hind paw. The Model 336 Analgesia Meter (IITC Inc., Life Science Instruments. Woodland Hills, CA, USA) was automatically records the withdrawal latency from heating source. 4–5 trials on each side were performed at intervals of 5 min.
As described previously, DRG microinjection was carried out (Li et al., 2020). After 3-cm-long skin incision, we firstly exposed the corresponding spinal nerve (L4 and/or L3). After that, we used rongeur to remove the unilateral articular processes for DRG microinjection. The glass micropipette was carefully inserted into the exposed ipsilateral L4 and/or L3 DRGs and 1 μl of either siRNA solution or viral solution was injected into L4 and/or L3 DRG. The injection was performed at a rate of 10 nl/s. After the injection was completed, the pipette was left in place for 10 min before removal to allow the fluid to distribute and the pressure within the DRG to equalize. The skin was sutured with 6-0 silk and mice were kept on a heating pad. All reagent and surgical instruments are sterilized in advance.
We performed DRG neuron cultures as described (Li et al., 2020). We first prepared complete neurobasal medium (CNM) including 10% fetal bovine serum, and 1x antibiotics, 2% B-27 supplement, and 1% GluMax supplement. 3–4 weeks mice were used for collecting DRGs. The collected DRGs were incubated with collagenase solution including dispase, collagenase type I in HBSS. All reagents are from Thermofisher Scientific Company (Waltham, MA, USA).
The TRIzol Reagent (Cat. No:15596026, Invitrogen Corporation, Carlsbad, CA, USA) was used for extracting RNA from DRGs, followed by reverse transcription using PrimeScript RT Master Mix (Cat. No: RR036A, Takara Bio Inc, Shiga, Japan). Quantitative PCR were performed using TB Green Premix Ex Taq II (Tli RNaseH Plus) (Cat. No: RR820A, Takara Bio Inc, Shiga, Japan) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Finally, relative fold changes of each gene were calculated by ΔCt method (2–ΔΔCt). Supplementary Table 1 included all primer information.
Primary neurons grew in 6-well plate and were treated with actinomycin D (Cat. No:A1410, Sigma-Aldrich, Burlington, MA, USA) for testing the mRNA stability. After that, we collected neurons at 0, 2, 4, 8, and 12 h after actinomycin D treatment for RNA extraction.
Cytoplasmic and Nuclear RNA purification Kit were purchased from Norgen Biteck Corp. (Cat. No: NGB-21000, Thorold, ON, Canada). After Nuclear and cytoplasmic RNA fraction isolation, various gene expression levels in both nuclear and cytoplasmic fractions of all samples were calculated by RT-PCR as protocol above.
Nuclear and cytoplasmic protein were separated using the NE-PER nuclear and cytoplasmic extraction reagents (Cat. No: 78833, Thermofisher Scientific Company, Waltham, MA, USA) following the manufacturer’s instructions. The collected protein was aliquoted and stored at −80°C.
The pAAV-CMV-mRmst-:WPRE vector (vector ID: VB220510-1032tdc) and AAV5-Rmst virus packaging was designed and constructed to overexpress the Rmst expression by VectorBuilder Company (Chicago, IL, USA). Briefly, the full-length cDNA of Rmst (NR_028262.1) was amplified by RT-PCR. After that, double enzyme-digested PCR products were ligated into the mammalian ncRNA expression AAV vector as a plasmid. AAV5-Gfp (vector ID: VB150925-10026) is used as negative control. SiRNA for Rmst and Dnmt3a were designed and produced by Tsingke Biotechnology Co., Ltd. (Beijing, China). All siRNA sequencing used in this work were listed at Supplementary Table 1.
Mice were perfused with 4% PFA after deep isoflurane before being analyzed by immunohistochemistry. The DRGs were collected and post-fixed in 4% PFA overnight, followed by dehydrating in 30% sucrose for two nights at 4°C. Finally, the DRGs were sectioned at 15–20 μm and kept them in −80°C refrigerator.
Before primary antibody incubation, the section was blocked in 1X PBS with 10% donkey serum and 0.3% Triton X-100. The sections were then incubated with anti-DNMT3a (Santa Cruz, Dallas, TX, USA) overnight at 4°C followed by incubating secondary antibody conjugated to Cy3 (1:500, Jackson ImmunoResearch, West Grove, PA, USA) for 2 h. Finally, the sections were mounted using Fluoroshield™ with DAPI (Cat. No: F6057, Sigma-Aldrich, Burlington, MA, USA).
The Magna RIP Kit were purchased from EMD Millipore (Burlington, MA, USA) company for RIP assay. A anti-HuR antibody (Santa Cruz, Dallas, TX, USA) was used in RIP assay. After purification of RNA, RT-PCR was performed following the previous protocol.
The collected protein was firstly separated using SDS-PAGE electrophoresis on the basis of size, followed by moving to PVDF membranes with appropriate size. The blot was then immediately placed in 5% fresh non-fat milk powder for blocking for 1 h. Next, the appropriate primary and secondary antibodies were used to incubate the transferred membrane according to the recommended dilution and time in datasheet. The rabbit anti-DNMT3b (1:500), rabbit anti-DNMT3a (1:500), and rabbit anti-histone H3 (1:1,000) were purchased from Cell Signaling Technology (Danvers, MA, USA). The mouse anti-HuR (1:500) and rabbit anti-GAPDH (1:1,000) were purchased from Santa Cruz company (Dallas, TX, USA). Membranes were visualized by the Clarity Western ECL Substrate (Cat. No: 170-5060, Bio-Rad Laboratories, Inc., Hercules, CA, USA), exposed by ChemiDoc Touch (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and analyzed by Image J.
Sample size estimation was based on certain assumptions, including significance level, expected placebo mean, expected treatment mean, standard deviation, power, expected dropout. In our case, expected placebo mean, expected treatment mean, standard deviation, and expected dropout were depended on our pilot studies and previous report in the field (Li et al., 2020; Pan et al., 2021). After that, sample sizes were calculated using nQUERY software, assuming a significance level of 0.05, 90% power and homogeneous variances for the 2 samples to be compared, with the means and SEM for different parameters predicted from pilot study. The data is presented as mean + SEM and analyzed by GraphPad Prism 8. The Shapiro–Wilk test was used for normal distribution test. If the data passed the normality test, the t-test or ANOVA was used in this study by GraphPad Prism 8 software according to experimental design and the detailed statistical method was listed in figure legend. The interaction of factors after ANOVA is provided in result. A P-value of less than 0.05 was considered statistical significant.
As the critical role of lncRNAs during the formation of NP, we extracted the 10 most up-and downregulated lncRNAs in L4 DRGs 7 days after SNL model (Figure 1A) from the previous RNA sequencing dataset (Wu et al., 2016). Among them, we found Rmst, a previously reported brain specific lncRNA (Ng et al., 2012), was also expressed in damaged DRGs (Figure 1A). As previous research has been found that Rmst was specific to neuron (Briese et al., 2016), we are wondering the distribution of Rmst in the neuron subtype after nerve injury. Thus, we analyzed the scRNA sequencing dataset from DRGs with peripheral nerve damage (Wang K. et al., 2021). As the smart-seq2 technology was better to captured low abundance transcripts as well as more lncRNAs (Wang X. et al., 2021), we focused on the smart-seq2 analysis to explore DRG neuron subtype. In t-distributed stochastic neighbor embedding (tSNE), 6 traditional classification neuronal subtype was found (Figure 1B), including S100b+ neurofilament (NF), Mgpra3+ non-peptidergic neurons (NP), Th+ tyrosine hydroxylase (TH), Tac1+ and Sst+ peptidergic neurons (PEP), and Atf3+ injured neuron (Figures 1B,C). Interestingly, we found Rmst was specifically expressed in Atf3+ injured neuron (Figure 1C), suggesting that Rmst may be involved in NP.
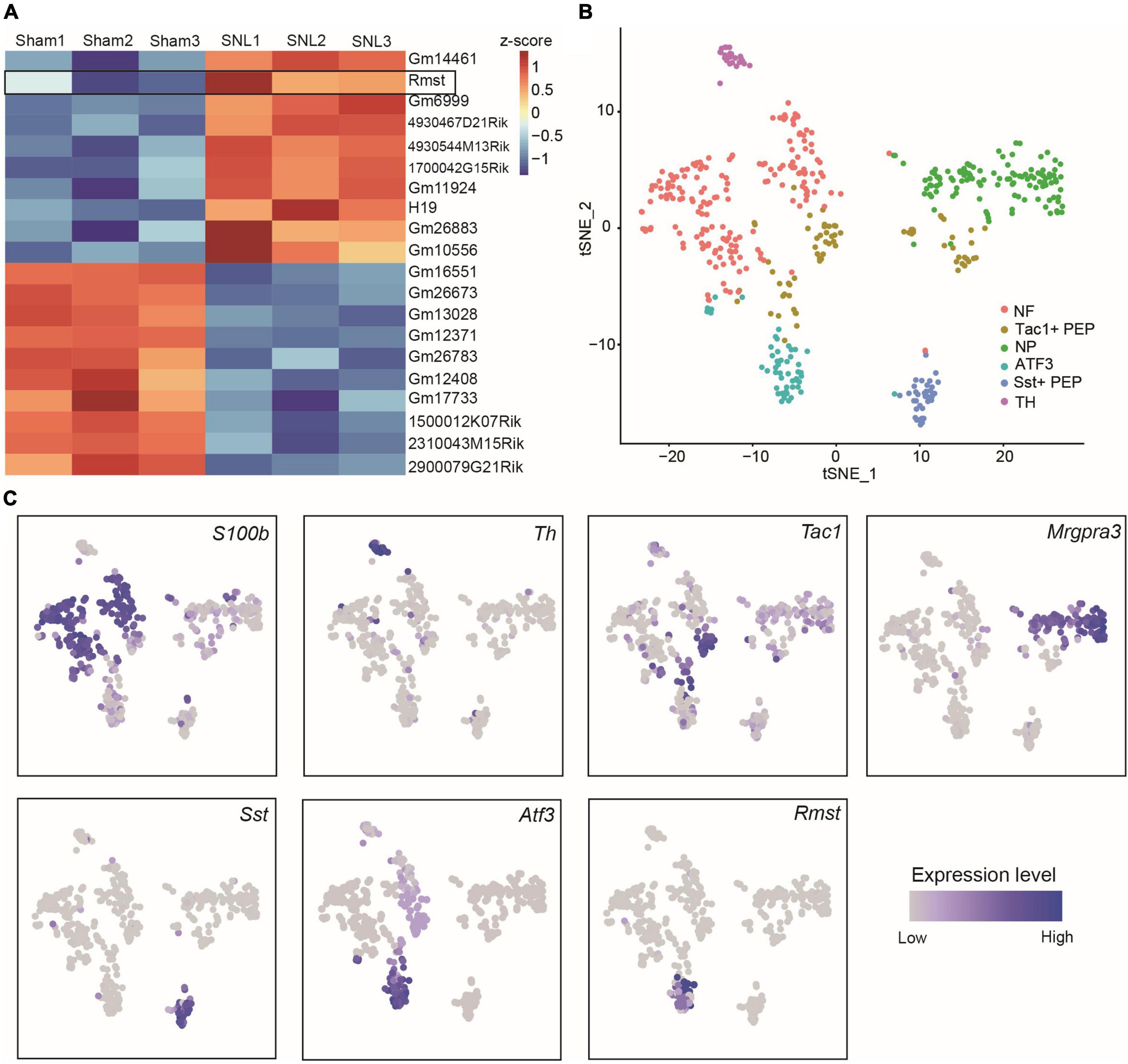
Figure 1. Rhabdomyosarcoma 2-associated transcript (Rmst) was specifically expressed in Atf3+ injured neurons. (A) The 10 most up- and downregulated lncRNAs in L4 DRGs after SNL model. The heatmap was created using Z-score values obtained from RNA-seq dataset. (B) TSNE plot of all cells from DRGs after nerve injury. Each dot was color-coded and annotated by neural subtypes. NF: neurofilament, NP: non-peptidergic neurons, TH: tyrosine hydroxylase, PEP: peptidergic neurons. (C) TSNE plots showing scRNA-seq data, colored by gene expression level, showing S100b, Th, Tac1, Mrgpra3, Sst, Atf3, and Rmst expression.
To provide the adequate evidence, we performed animal neuropathic pain model to validate the result. We found lncRNA Rmst expression was increased early and persistently at least 28 days in damaged DRGs after SNL surgery but not sham surgery (Figure 2A) [F(5,36) = 5.942]. To be specific, Rmst expression was elevated 1.58-fold on day 3, 2.07-fold on day 7, 2.01-fold on day 14, 1.99-fold on day 21, 1.88-fold on day 28 after SNL but not in the contralateral L4 DRGs and L3 DRGs (Figures 2A–D) [Figure 2B: F(5,36) = 1.896; Figure 2C: F(5,36) = 0.7694; Figure 2D: F(5,36) = 2.980]. A similar phenomenon was observed in another NP mouse model called CCI model (Figures 2E,F) [Figure 2E: F(5,36) = 3.530]; [Figure 2F: F(5,36) = 0.8966]. The Rmst expression in damaged DRGs were elevated 1.97-fold on day 3, 2.23-fold on day 7, 2.23-fold on day 14, 2.46-fold on day 21, 2.3-fold on day 28 after CCI model (Figure 2E). Taken together, our data showed peripheral nerve damage could trigger the elevated Rmst expression in the damaged DRGs that maintained at least until 1-month post-SNL, suggesting that Rmst may take part in the nerve damage-induced NP.
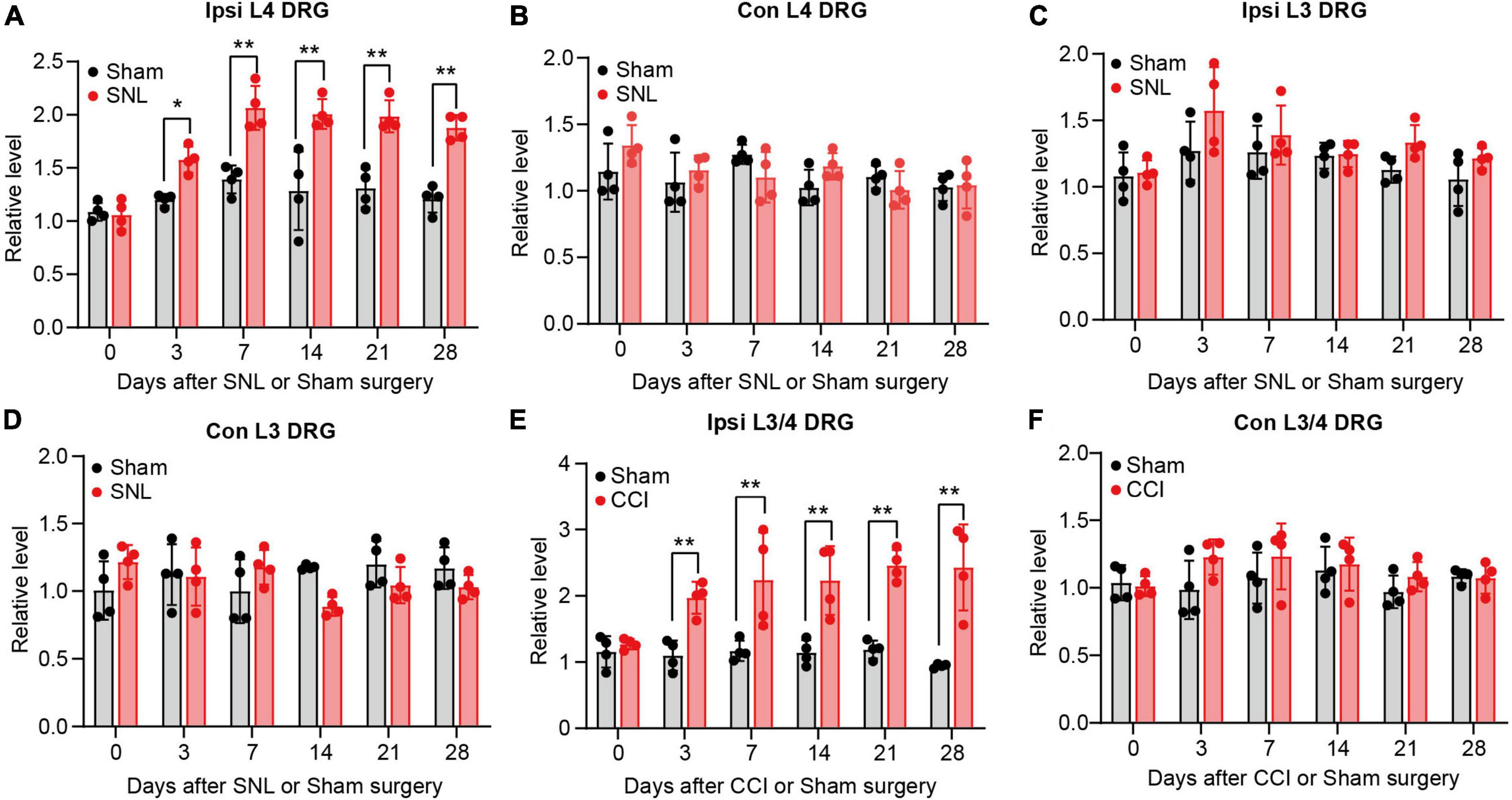
Figure 2. Rhabdomyosarcoma 2-associated transcript (Rmst) upregulation was found in damaged DRG of mice after SNL or CCI model. (A–D) Level of Rmst in ipsilateral L4 DRGs (A), contralateral L4 DRGs (B), ipsilateral L3 DRGs (C), and contralateral L3 DRGs (D) after SNL or sham surgery n = 16. (E,F) Levels of Rmst in the ipsilateral (E) and contralateral (F) side L3/4 DRGs after CCI or sham surgery n = 8. Two-way ANOVA followed by post-hoc Tuckey test. *P < 0.05 and **P < 0.01 compared with sham group at each time point.
As the apparent change of Rmst in damaged DRGs, we further ask whether blocking Rmst expression in injured DRG could alleviate pain hypersensitivity. We first confirmed that DRG microinjection of siRmst, but not Scr, 3 days before SNL could block SNL-induced increased Rmst expression (Figure 3A) [F(3,8) = 65.29]. Microinjection of siRmst also slightly reduced basal expression of Rmst expression (Figure 3A). More importantly, we found pre-microinjection of siRmst could ameliorate SNL-induced nociceptive hypersensitivities, including mechanical allodynia (Figures 3B,C) [Figure 3B: F(12,80) = 9.633; Figure 3C: F(12,80) = 10.15] and heat hyperalgesia (Figure 3D) [Figure 3D: F(12,80) = 17.99]. Neither Scr nor siRmst changed basal paw response to mechanical or heat stimuli while mice received sham surgery (Figures 3B–D). We then ask the role of Rmst during the maintenance period after SNL model. We first confirmed that DRG microinjection of siRmst in maintenance period after SNL (Figure 3E) [F(3,8) = 30.85]. As expected, siRmst delivery through DRG microinjection on day 7 post-SNL rescued pain hypersensitivity (Figures 3F–H) [Figure 3F: F(18,112) = 3.614; Figure 3G: F(18,112) = 3.312; Figure 3H: F(18,112) = 20.60]. Our data strongly support that nerve damage triggered nociceptive hypersensitivity may be attributed to elevated Rmst expression in the damaged DRG.
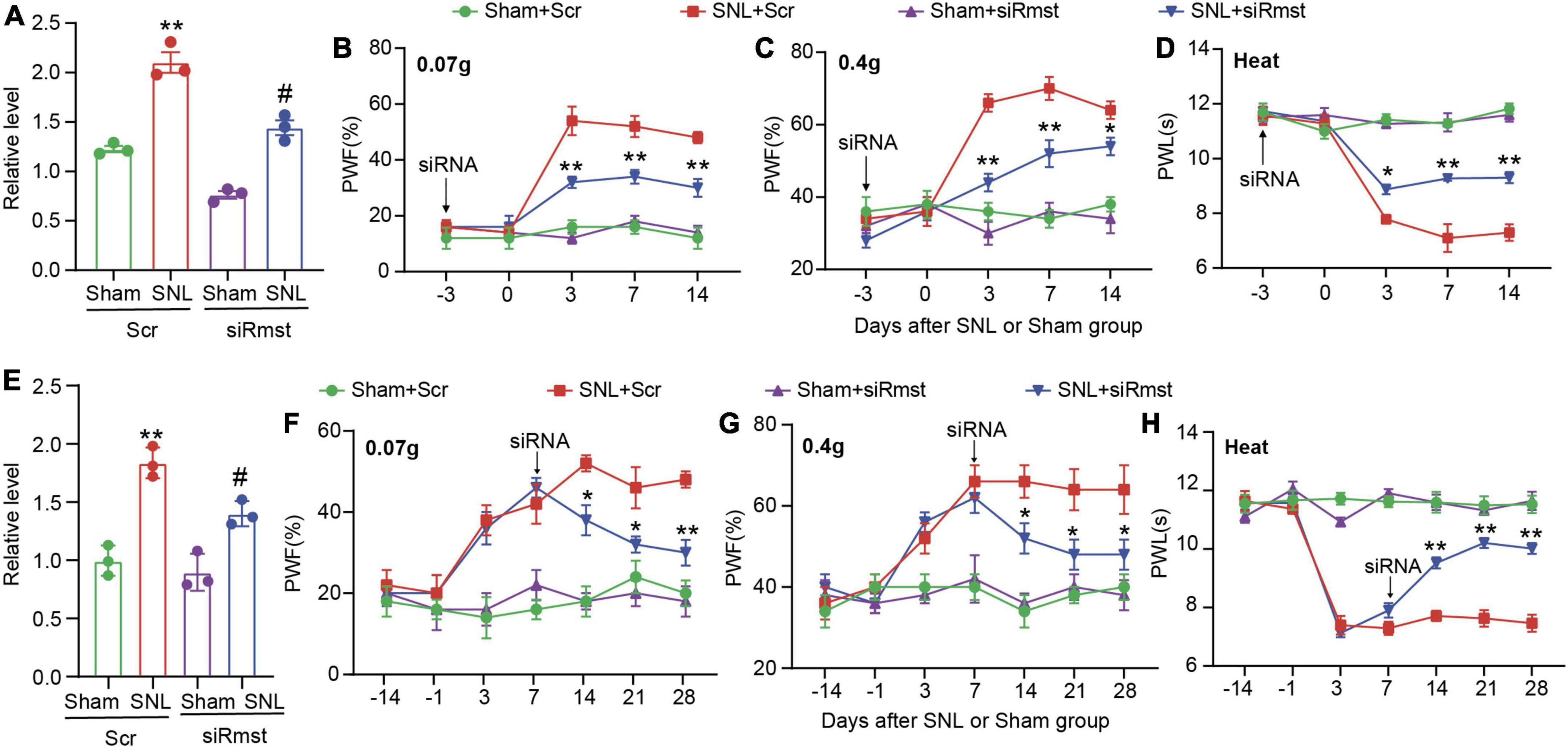
Figure 3. Blocking nerve damage triggered rhabdomyosarcoma 2-associated transcript (Rmst) expression in DRG mitigated the nerve damaged-induced nociceptive hypersensitivity. (A) Level of Rmst in the L4 DRGs on day 14 after nerve injury model in presence of siRmst or Scr. n = 12 mice, One-way ANOVA followed by post-hoc Tukey test, **P < 0.01 compared with Sham + Scr group and #P < 0.05 compared with SNL + Scr group. (B–D) The PWF to von Frey filament (B,C) and the PWL to thermal stimuli (D) on the development period of NP. n = 5 mice, two-way ANOVA followed by post-hoc Tukey test, *P < 0.05 and **P < 0.01 compared with the SNL + Scr group. (E) Level of Rmst in the L4 DRGs on day 28 after nerve injury model in presence of siRmst or Scr. n = 12 mice, One-way ANOVA followed by post-hoc Tukey test, **P < 0.01 compared with Sham + Scr group and #P < 0.05 compared with SNL + Scr group. (F–H) The PWF to von Frey filament (F,G) and the PWL to thermal stimuli (H) on the maintenance period of NP. n = 5 mice, two-way ANOVA followed by post-hoc Tukey test, *P < 0.05 and **P < 0.01 compared with SNL + Scr group. Paw withdrawal frequency: PWF; Paw withdrawal latency: PWL.
Next, we asked if DRG Rmst overexpression in neuron is sufficient for NP production. As the previous paper has reported Rmst was specifically expressed in the neuron (Briese et al., 2016), we utilized Adeno-associated Virus 5 (AAV5) following the previous papers (Li et al., 2020) to package Rmst full-length vector (AAV5-Rmst) and microinjected to the ipsilateral DRG to overexpress the Rmst expression. As a proof of concept, we found DRG microinjection of AAV5-Rmst in the ipsilateral side could particularly increase Rmst expression in ipsilateral side but neither contralateral side nor AAV5-Gfp (Figure 4A) [F(3,8) = 84.17]. More importantly, pain symptoms (Figures 4B–D) [Figure 4B: F(15,96) = 5.679; Figure 4C: F(15,96) = 3.432; Figure 4D: F(15,96) = 14.98] were induced third weeks after Rmst overexpression in DRG of naïve mice. It means that, even without nerve damage, DRG Rmst overexpression in neuron could result in NP-like symptoms.
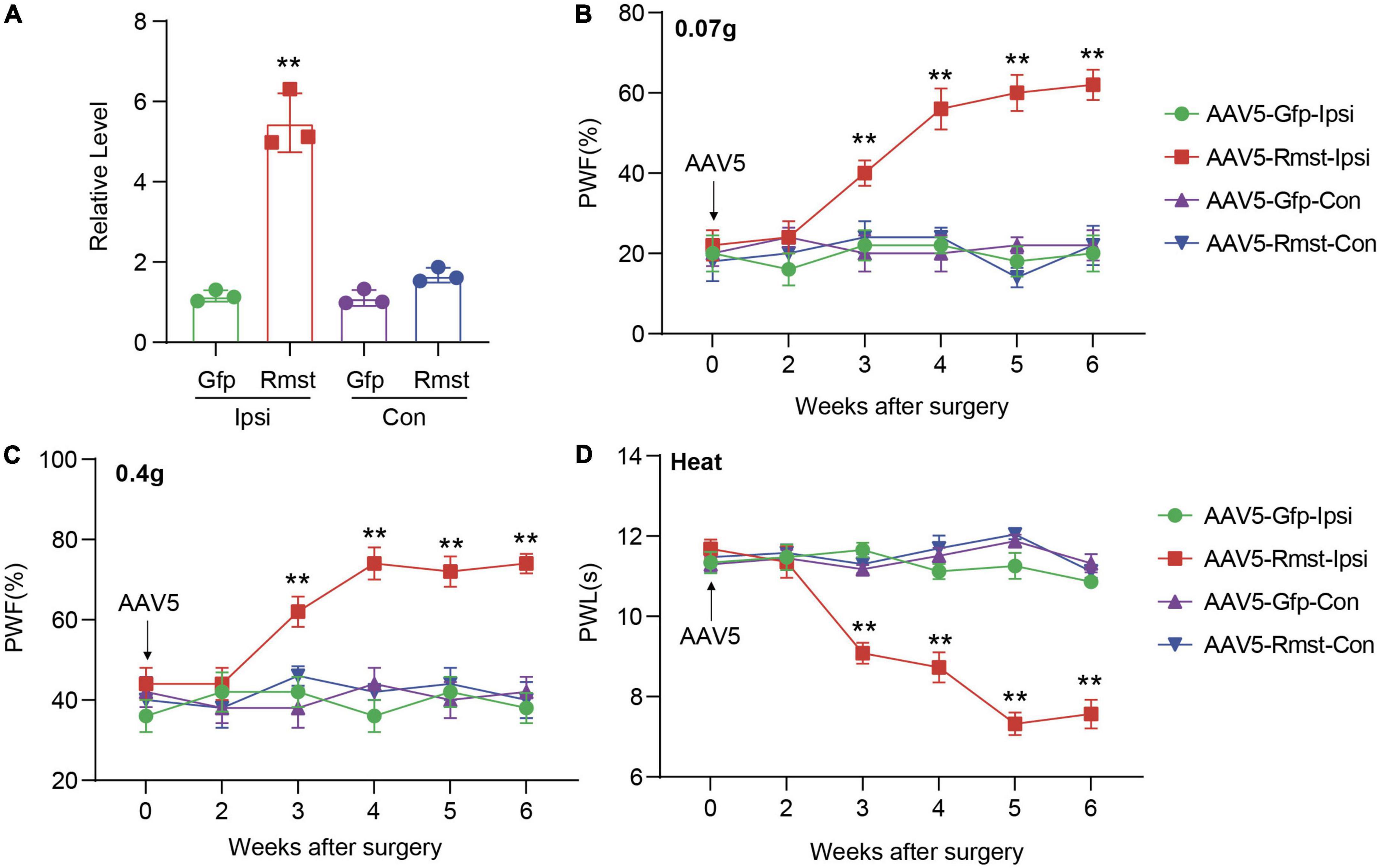
Figure 4. Overexpression of DRG rhabdomyosarcoma 2-associated transcript (Rmst) produced nociceptive hypersensitivity in naïve mice. (A) Rmst expression in L3/4 DRGs 6 weeks after receiving AAV5-Rmst or AAV5-Gfp. n = 6 mice, two-tailed unpaired Student’s t-test, **P < 0.01 compared with AAV5-Gfp group. (B–D) The PWF to von Frey filament (B,C) and the PWL to thermal stimuli (D) on mice with DRG microinjection of AAV5-Rmst or AAV5-Gfp. n = 6, two-way ANOVA followed by post-hoc Tukey test, **P < 0.01 compared with AAV5-Gfp-Ipsi group.
Next, the detailed mechanism of Rmst involved in NP was investigated. As the subcellular location of lncRNA can provide significant information on its function, we utilized cytoplasmic and nucleus RNA extraction protocol to purify populations of subcellar RNA fractions. Consistent with previously report in vitro (Zhao et al., 2021), Rmst in naïve mouse DRG was distributed predominantly in the cytoplasm (Figure 5A). Cytoplasmic lncRNAs can function in the posttranscriptional gene expression through mRNA stability and translation (Rashid et al., 2016). It has been reported that Rmst could upregulate DNA methyltransferase 3 (Dnmt3) by increasing the stability for its mRNA (Peng et al., 2020) in MCF7 cells, a breast cancer related epithelial cell line. We then examined whether overexpression of Rmst could also increase the Dnmt3a and Dnmt3b mRNA in primary DRG neuron. Surprisingly, only Dnmt3a mRNA as well as protein level increased but not Dnmt3b was observed in cultured neurons co-transduced with AAV5-Rmst (Figures 5B–D). In fact, Dnmt3a in primary afferent neurons was reported to participate in NP by repressing the potassium voltage-gated channel Kv1.2 encoded by Kcna2 (Zhao et al., 2017). To elucidate whether Rmst participated in regulating Dnmt3a mRNA stability, actinomycin D was used to inhibit the RNA synthesis. We found that Rmst could stabilize Dnmt3a mRNA transcripts (Figure 5E). The half-life of Dnmt3a mRNA was around 8.5 h for Rmst overexpressed neurons, as compared to 6.5 h for Gfp control (Figure 5E) [F(4,20) = 2.384]. Next, we found microinjection of siRmst, but not Scr, could abolish the SNL- induced Dnmt3a increases (Figure 5F) [F(3,8) = 17.11]. Decreased Dnmt3a protein in damaged DRG with the siRmst microinjection after SNL was observed in the nucleus (Figures 5G,H) [F(3,8) = 45.02].
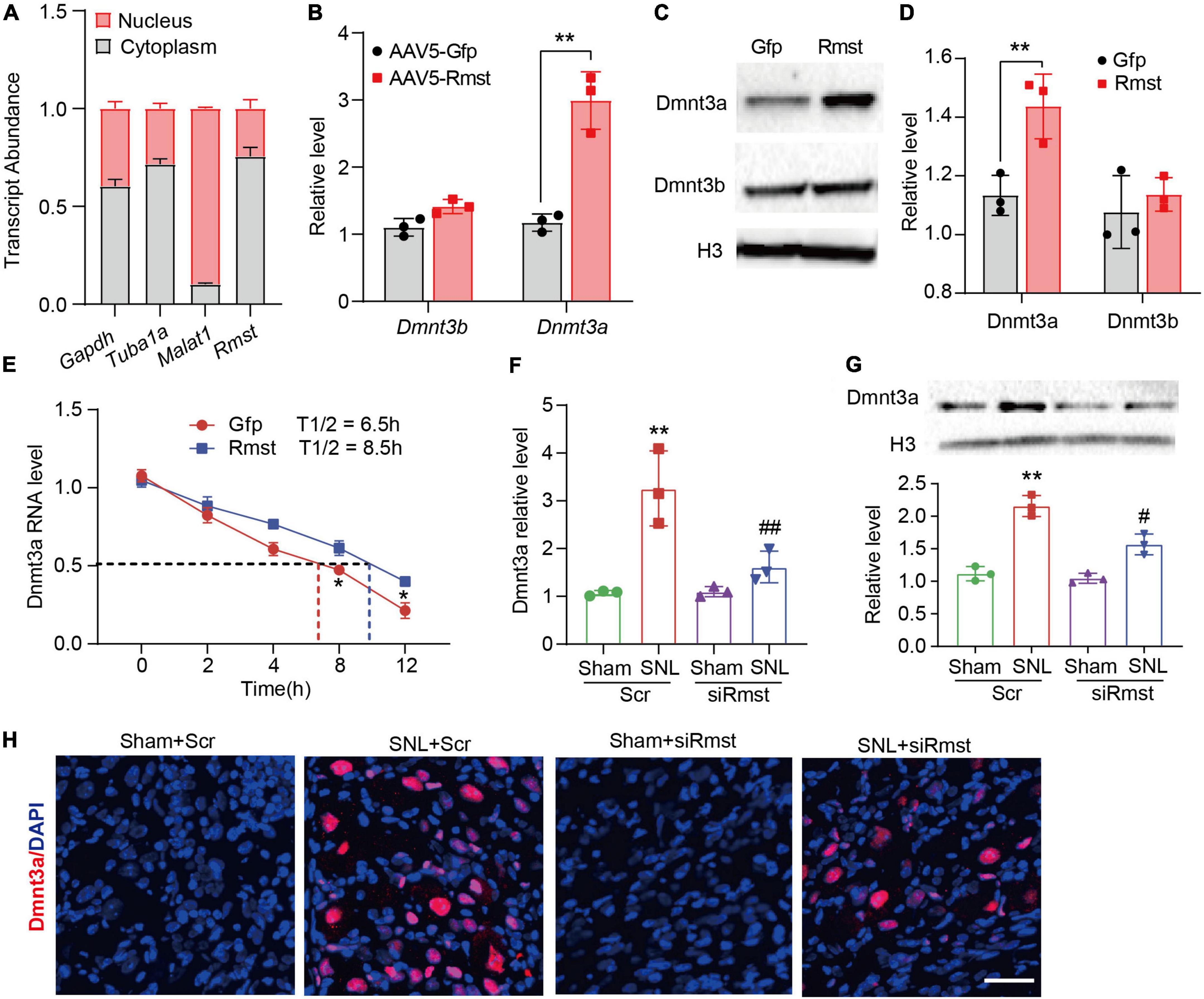
Figure 5. Dorsal root ganglion (DRG) rhabdomyosarcoma 2-associated transcript (Rmst) participated in the nerve damage induced Dnmt3a expression by stabilizing Dnmt3a mRNA. (A) Transcript abundance of cytoplasmic and nuclear RNA fractions for Gapdh mRNA, Tuba1a mRNA, Malat1 mRNA, and Rmst mRNA from mouse DRGs. n = 3 mice. (B) Level of Dnmt3a and Dnmt3b mRNA expression in DRG neuron with AAV5-Rmst. T-test was used for statistic analysis. **P < 0.01 versus AAV5-Gfp group. (C,D) Dnmt3a and Dnmt3b protein expression in DRG neuron with AAV5-Rmst. T-test was used for statistic analysis. **P < 0.01 versus AAV5-Gfp group. (E) Primary DRG neurons with the treatment of ActD (5 μg/mL) at multiple time point as shown were treated with AAV5-Rmst or AAV5-Gfp. n = 3 biological repeats. *P < 0.05 versus AAV5-Rmst group. (F,G) Levels of Dnmt3a mRNA (F) and protein expression (G) in ipsilateral L4 DRGs of SNL mice pre-received with siRmst or Scr. n = 12, one-way ANOVA followed by post-hoc Tukey test, **P < 0.01 versus Sham + Scr group and #P < 0.05, ##P < 0.01 versus SNL + Scr group. (H) Immunostaining for the ipsilateral L4 DRGs showed Dnmt3a protein expression in mice post-SNL and pre-delivered with siRmst or Scr. Scale bar = 40 μm.
To further confirm whether Rmst is responsible for stabilizing Dnmt3a mRNA in the damaged DRG, we inhibited Dnmt3a expression through delivering Dnmt3a siRNA into the DRG after AAV5-Rmst microinjection. We found blocking Rmst overexpression-induced Dnmt3a increase could not lower Rmst expression (Figure 6A) [F(2,6) = 19.28] but lead to a fall of Dnmt3a mRNA and protein (Figures 6B,C) [Figure 6B: F(2,6) = 62.27; Figure 6C: F(2,6) = 31.06]. More importantly, blocking Dnmt3a expression attenuated the Rmst induced-nociceptive hypersensitivity (Figures 6D–F) [Figure 6D: F(8,60) = 13.12; Figure 6E: F(8,60) = 3.500; Figure 6F: F(8,59) = 10.35]. Collectively, nerve injury induced Rmst upregulation participates in NP by stabilizing the Dnmt3a mRNA expression.
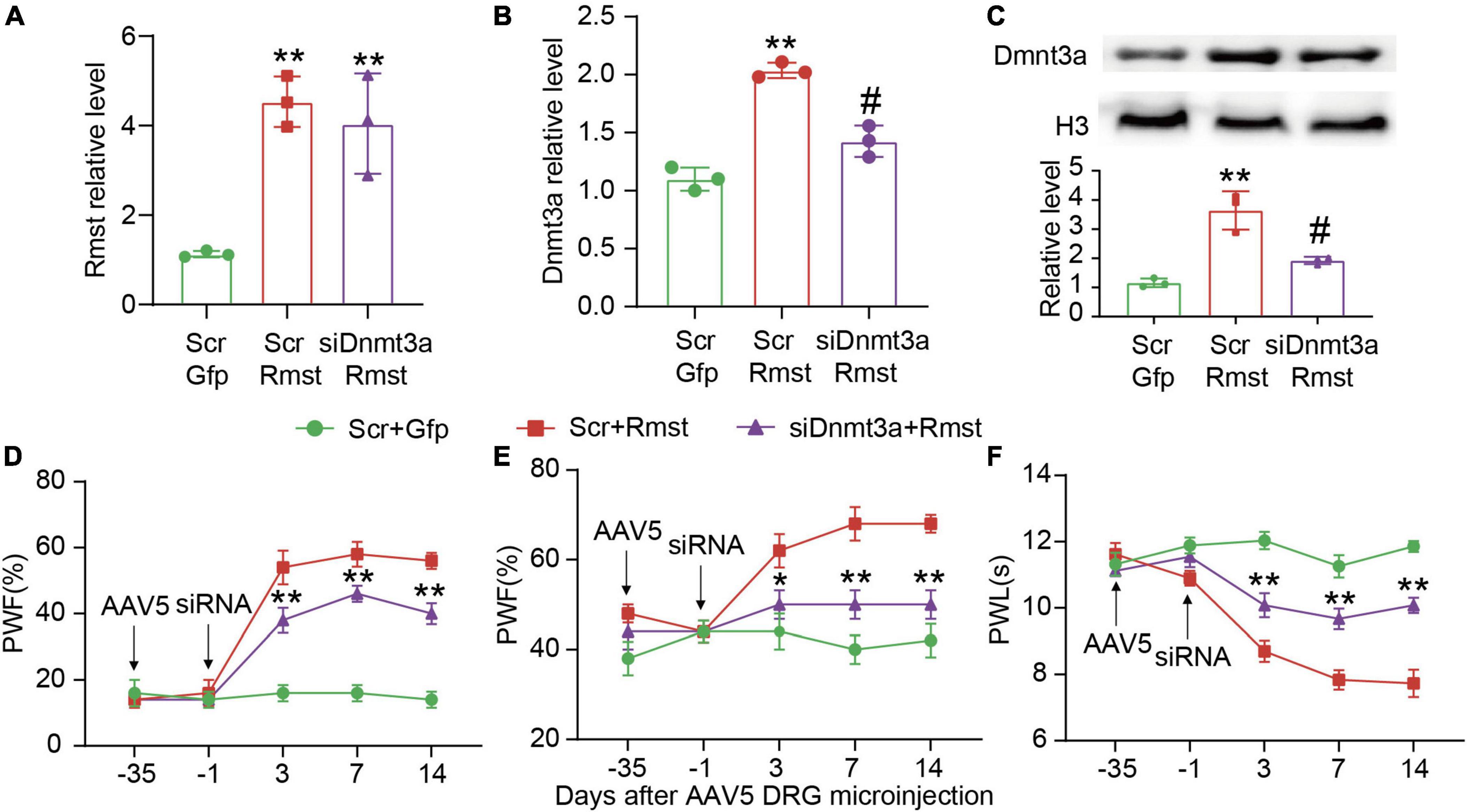
Figure 6. Blocking DNA methyltransferase 3 alpha (Dnmt3a) expression mitigated the rhabdomyosarcoma 2-associated transcript (Rmst) triggered-mechanical allodynia and heat hyperalgesia. (A–C) Rmst mRNA (A), Dnmt3a mRNA (B), and Dnmt3a protein (C) in mice after microinjection with siDnmt3a (Scr as control) and AAV5-Rmst (AAV5-Gfp as control). **P < 0.01 versus AAV5-Gfp + Scr group and #P < 0.05 versus AAV5-Rmst + Scr group. (D–F) The ipsilateral PWF to von Frey filament (D,E) and the PWL to thermal stimuli (F) after microinjection of siDnmt3a or Scr in mice pre-received with AAV5-Rmst or AAV5-Gfp. **P < 0.01 versus AAV5-Rmst + Scr group.
Finally, we asked the potential mechanism of Rmst induced Dnmt3a upregulation in injured DRG neuron. Given that RNA-binding proteins (RBPs) affect the targeted mRNA stability, we focused on one of well-characterized RBPs, HuR. Notably, HuR was reported as a contributor to nociceptive pain (Kunder et al., 2022) and an anti-HuR could alleviate nerve-injury induced NP (Borgonetti and Galeotti, 2021). In particular, it was HuR reported to stabilize the Dnmt3 mRNA (Peng et al., 2020). Therefore, under NP condition, we asked whether HuR contributed to Rmst-mediated increased Dnmt3a.
We first determined the alteration of HuR in DRGs after SNL or Rmst overexpression. Unexpectedly, we found neither SNL nor Rmst could regulate the HuR expression in injured DRG (Figure 7A). In fact, in many cancerous settings, HuR was increased subcellular localization within the cytoplasm to stabilize various prosurvival mRNA (Schultz et al., 2020). Therefore, we examine whether SNL induced cytoplasmic accumulation of HuR. We found SNL caused a steep increase of HuR protein in the cytoplasm of the damaged DRG while Rmst overexpression in DRG was found similar phenomenon (Figure 7B). This may suggest that under NP condition, HuR is transported from the nucleus to cytoplasm. Next, RNA immunoprecipitation (RIP) assay revealed that HuR was capable to enrich Rmst and Dnmt3a in SNL group, approximately 20-fold, and 60-fold, respectively, compared to sham group (Figure 7C). Finally, we found overexpression of Rmst in primary neuron promoted the binding between Dnmt3a mRNA and HuR (Figure 7D). Thus, Rmst appeared to be the essential regulator that promoted Dnmt3a expression through interacting with HuR in DRG neuron.
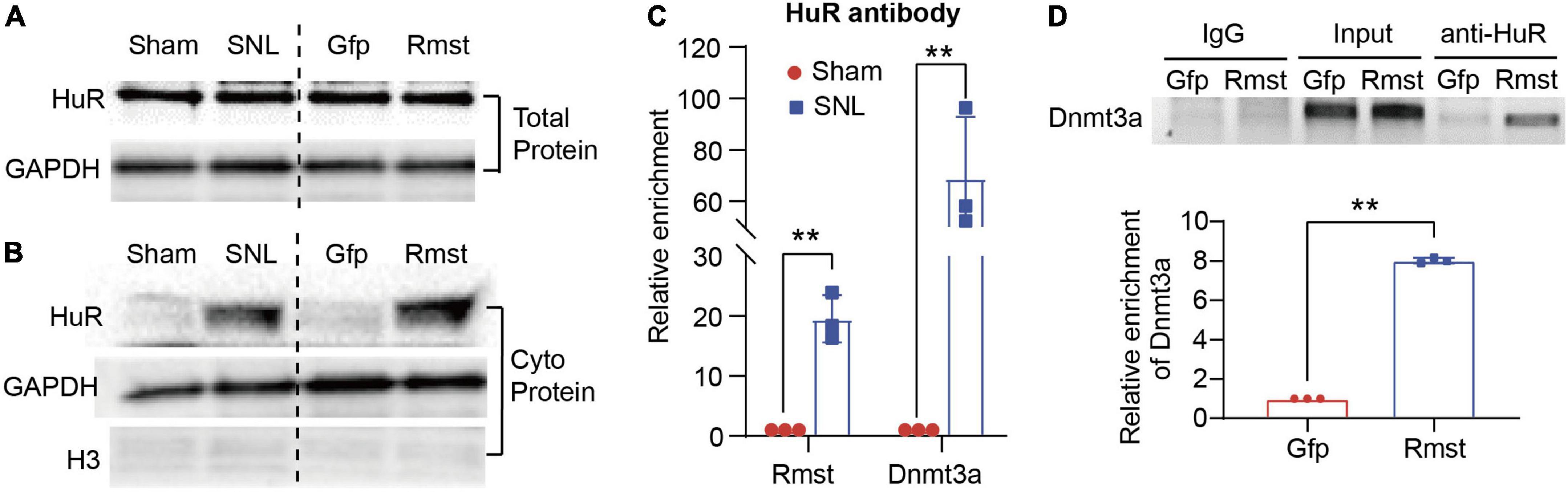
Figure 7. Rhabdomyosarcoma 2-associated transcript (Rmst) was interaction with HuR to regulate the Dnmt3a mRNA Stability Under NP Condition. (A,B) Level of HuR expression in total protein (A) and cytoplasm protein (B) in DRG with SNL model and AAV5-Rmst (AAV5-Gfp as control) DRG microinjection. (C) Level of Rmst or Dnmt3a mRNA immunoprecipitated by anti-HuR on day 7 post-SNL surgery n = 3 biological repeats. **P < 0.01 versus sham group. (D) Level of Dnmt3a immunoprecipitated by HuR in DRG neuron with AAV5-Rmst. n = 3 biological repeats. **P < 0.01 compared with AAV5-Gfp group.
This is the first study to examine the molecular and cellular function of Rmst, a lncRNA in injured DRG neuron that modulates NP. Specifically, the increased Rmst was positively regulated Dnmt3a by promoting its mRNA stability and interacting with HuR, which leads to NP. Blocking the elevation of Rmst could reverse nerve injury-induced Dnmt3a upregulation and alleviate pain hypersensitivities (Figure 8). The present study suggests that Rmst in DRGs is likely a key regulator in NP.
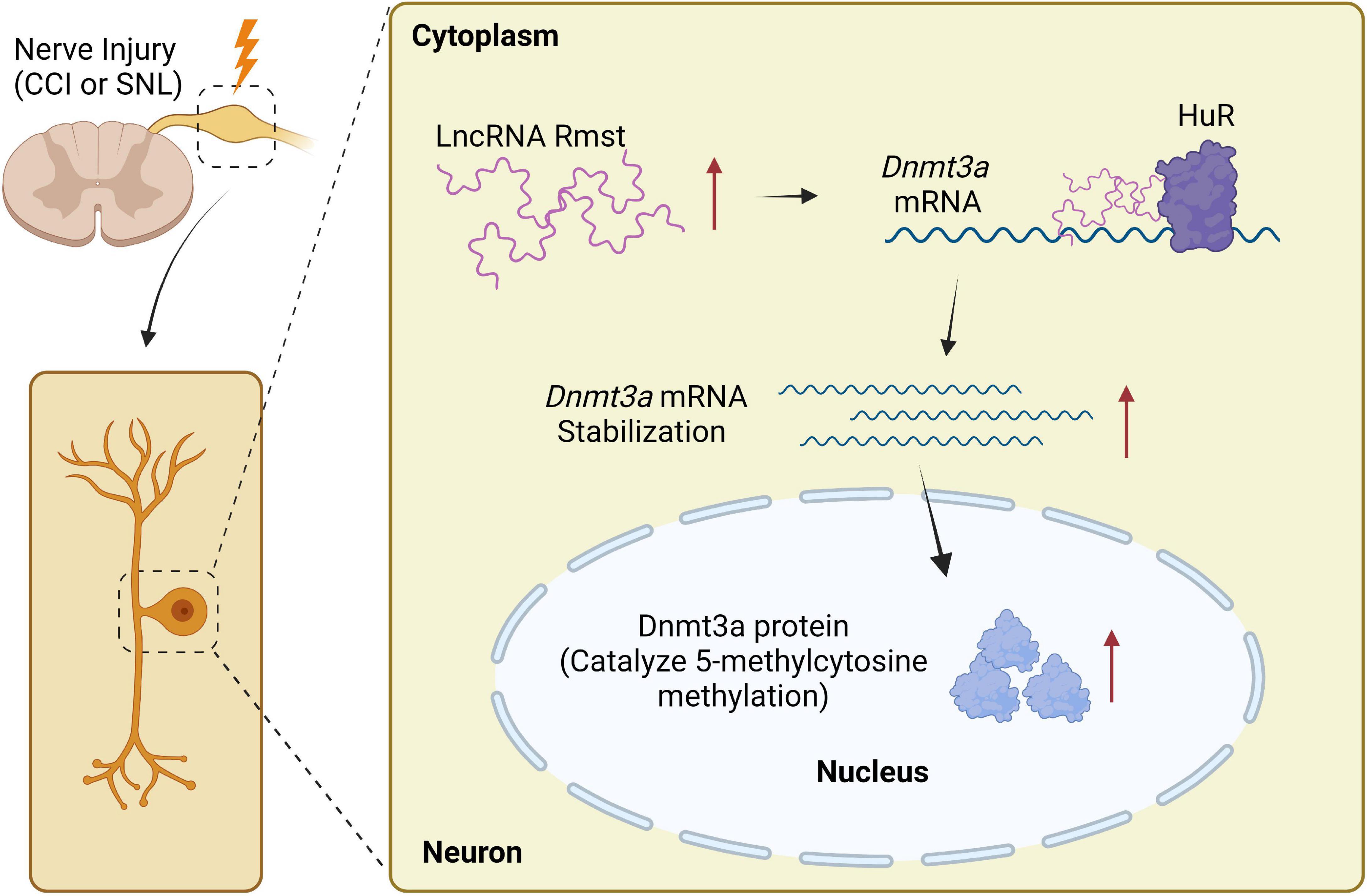
Figure 8. Proposed mechanism on rhabdomyosarcoma 2-associated transcript (Rmst) in NP. Nerve injury-triggered Rmst upregulation interacts with HuR in injured DRGs, resulting in stabilizing Dnmt3a mRNA and protein. The latter one has been reported as a critical NP regulator in DRG neurons. In contrast, in normal DRG neuron, Dnmt3a mRNA would be degraded without Rmst.
In 2013, the first lncRNA involved in NP was reported in detail (Zhao et al., 2013, p. 2). After that, the regulation of lncRNA on NP are popping up over the past few years (Wu et al., 2019). And we are the first to report lncRNA Rmst is involved in the NP. In fact, Rmst was first identified as non-coding RNA in 2012, and it was essential for neuronal specification in human embryonic stem cells (Ng et al., 2012). Under normal condition, RMST in both human and mouse occurs primarily in CNS (Uhde et al., 2010; Ng et al., 2012) and, more importantly, is abundant in neuron (Julian et al., 2013, p. 2; Briese et al., 2016). RMST physically interacts with SOX2 and regulates neural fate by regulating neurogenesis related genes (Ng et al., 2013, p. 2). RMST deficiency in neural stem cells resulted in glia differentiation (Ng et al., 2013), indicating that RMST is important for neural differentiation in particularly during brain development period. However, increased Rmst expression was reported in CNS diseases, including stroke (Zhao et al., 2021; Li et al., 2022) and Parkinson’s disease (Ma et al., 2021). Blocking Rmst expression could protect from neuronal apoptosis and improve neurological function (Ma et al., 2021; Zhao et al., 2021; Li et al., 2022), which suggests that excessive Rmst expression may cause CNS disease aggravations. Our present study found Rmst was also increased expressed in DRG neuron under NP condition. More importantly, blocking Rmst expression in injured DRGs could mitigate nerve injury-induced nociceptive hypersensitivity. However, why Rmst siRNA in DRGs did not alter response to mechanical and heat stimuli is unclear, which may be due to low Rmst expression in physiological condition. Together, the strong evidence indicates that the dysregulation of Rmst in neuron may contribute to NP development.
Evidence has been emerged lncRNAs mediates DNA methylation in various pathological condition (Huang et al., 2022), including schizophrenia (Ni et al., 2021, p. 006), diabetic retinopathy (He et al., 2021, p. 3), colon cancer (Merry et al., 2015) and so on. In particular, RMST was characterized as a positive regulator for DNMT3 but not DNMT1 by increasing DNMT3 mRNA stability in cancer (Peng et al., 2020). In our study, Rmst upregulation may contribute to the nerve damage-triggered Dnmt3a increase by stabilizing its mRNA in NP. Notably, when Rmst was overexpressed in DRG neuron, only DNMT3a but not DNMT3b appear to generate, and blocking DRG Rmst expression abolished SNL-induced DNMT3a upregulation, which was consistent with previous study (Peng et al., 2020, p. 3). Furthermore, Rmst did not directly modulate nociceptive hypersensitivity as in absence of Dnmt3a expression in DRG neuron the overexpression of Rmst failed to completely mimic nerve damage-induced nociceptive hypersensitivity. In fact, it has been reported DNMT3a in DRG neurons is involved in the NP (Guo et al., 2019). Knockout DNMT3a in DRG significantly attenuated nociceptive hypersensitivity (Zhao et al., 2017). What’s more, in NP, transcriptional factors, such as CREB (Yang et al., 2021) and Oct1 (Zhao et al., 2017), could bind to the promoter region of Dnmt3a to boost Dnmt3a expression. Our study demonstrated that Rmst is required for the stability of Dnmt3a mRNA, enhancing DNMT3a expression. It should also be noted that Rmst also regulates other NP related genes including sex-determining region Y-box2 (Sox2) (Ng et al., 2013, p. 2; Zhang et al., 2019, p. 2) and heterogeneous nuclear ribonucleoprotein D (hnRNPD) (Liu et al., 2020; Feng et al., 2021). Whether these genes are also regulated by Rmst in NP remains to be determined.
Mechanistically, nerve injury-induced elevated Rmst could recruit HuR protein, thereby stabilizing the Dnmt3a mRNA and reducing the Dnmt3a mRNA degradation. HuR, as an RNA-binding protein, is capable to stabilize AU-rich elements (AREs)-containing reporter mRNA in the cytoplasm through binding AREs sequences (Gallouzi et al., 2000). The new finding has pointed out anti-HuR delivery has been proven effective to relieve pain hypersensitivity by inhibiting spinal neuroinflammation (Borgonetti and Galeotti, 2021). Therefore, the effectiveness of analgesics of anti-HuR may also be due to the degradation of Dnmt3a mRNA. However, further experiments are needed.
In conclusion, our study indicated that blocking Rmst expression in injured DRGs mitigated NP at least in part through enhancing degradation of Dnmt3a mRNA. Thus, Rmst may become a promising target and provide insightful directions for NP treatment.
The RNA sequencing dataset for mouse DRG after peripheral nerve injury was obtained from previous research (https://doi.org/10.1177/1744806916629048). The ScRNA-seq dataset for mouse DRG after peripheral nerve injury was from Gene Expression Omnibus (GEO) with the series record GSE155622.
This animal study was reviewed and approved by Guangzhou Medical University.
XG and XS conceptualized and designed the study. XG and WC contributed to write the manuscript. GZ and FH performed animal model and behavior test. XG and JQ performed molecular experiments. All authors read and approved the final manuscript.
This work of XS was supported by the National Natural Science Foundation of China (No. 81870823) and Guangzhou Institute of Pediatric/Guangzhou Women and Children’s Medical Center Funds (No. GCP-2018-001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer B-CJ declared a past co-authorship with one of the author XG to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.1027063/full#supplementary-material
Borgonetti, V., and Galeotti, N. (2021). Intranasal delivery of an antisense oligonucleotide to the RNA-binding protein HuR relieves nerve injury-induced neuropathic pain. Pain 162, 1500–1510. doi: 10.1097/j.pain.0000000000002154
Briese, M., Saal, L., Appenzeller, S., Moradi, M., Baluapuri, A., Sendtner, M., et al. (2016). Whole transcriptome profiling reveals the RNA content of motor axons. Nucleic Acids Res. 44:e33. doi: 10.1093/nar/gkv1027
Clemens, A. W., and Gabel, H. W. (2020). Emerging insights into the distinctive neuronal methylome. Trends Genet. 36, 816–832. doi: 10.1016/j.tig.2020.07.009
Feng, J., Chang, H., Li, E., and Fan, G. (2005). Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79, 734–746. doi: 10.1002/jnr.20404
Feng, X., Niu, L. J., Long, M. T., Luo, K., Huang, X., Chen, M., et al. (2021). Transcranial ultrasound stimulation of the anterior cingulate cortex reduces neuropathic pain in mice. Evid. Based Complement. Alternat. Med. 2021:651038. doi: 10.1155/2021/6510383
Gallouzi, I.-E., Brennan, C. M., Stenberg, M. G., Swanson, M. S., Eversole, A., Maizels, N., et al. (2000). HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. U.S.A. 97, 3073–3078. doi: 10.1073/pnas.97.7.3073
Guo, X., Yao, Y., and Tao, Y.-X. (2019). “Role of DNA methylation in chronic pain,” in Epigenetics of chronic pain, eds G. Bai and K. Ren (Amsterdam: Elsevier), 99–110. doi: 10.1016/B978-0-12-814070-3.00005-3
He, Y., Dan, Y., Gao, X., Huang, L., Lv, H., Chen, J., et al. (2021). DNMT1-mediated lncRNA MEG3 methylation accelerates endothelial-mesenchymal transition in diabetic retinopathy through the PI3K/Akt/mTOR signaling pathway. Am. J. Physiol. Endocrinol. Metab. 320, E598–E608. doi: 10.1152/ajpendo.00089.2020
Hou, X.-X., and Cheng, H. (2018). Long non-coding RNA RMST silencing protects against middle cerebral artery occlusion (MCAO)-induced ischemic stroke. Biochem. Biophys. Res. Commun. 495, 2602–2608. doi: 10.1016/j.bbrc.2017.12.087
Huang, W., Li, H., Yu, Q., Xiao, W., and Wang, D. O. (2022). LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 41:100. doi: 10.1186/s13046-022-02319-z
Julian, L. M., Vandenbosch, R., Pakenham, C. A., Andrusiak, M. G., Nguyen, A. P., McClellan, K. A., et al. (2013). Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell 12, 440–452. doi: 10.1016/j.stem.2013.02.001
Kunder, N., de la Pena, J. B., Lou, T. F., Chase, R., Stanowick, A., Black, B. J., et al. (2022). A conserved RNA-binding protein contributes to nociceptive pain. J. Pain 23:14. doi: 10.1016/j.jpain.2022.03.056
Li, J., Wang, N., Nie, H., Wang, S., Jiang, T., Ma, X., et al. (2022). Long non-coding RNA RMST worsens ischemic stroke via microRNA-221-3p/PIK3R1/TGF-β signaling pathway. Mol. Neurobiol. 59, 2808–2821. doi: 10.1007/s12035-021-02632-2
Li, Y., Guo, X., Sun, L., Xiao, J., Su, S., Du, S., et al. (2020). N6-methyladenosine demethylase FTO contributes to neuropathic pain by stabilizing G9a expression in primary sensory neurons. Adv. Sci. 7:1902402. doi: 10.1002/advs.201902402
Liu, C., Peng, Z., Li, P., Fu, H., Feng, J., Zhang, Y., et al. (2020). lncRNA RMST suppressed GBM cell mitophagy through enhancing FUS SUMOylation. Mol. Ther. Nucleic Acids 19, 1198–1208. doi: 10.1016/j.omtn.2020.01.008
Ma, X., Wang, Y., Yin, H., Hua, L., Zhang, X., Xiao, J., et al. (2021). Down-regulated long non-coding RNA RMST ameliorates dopaminergic neuron damage in Parkinson’s disease rats via regulation of TLR/NF-κB signaling pathway. Brain Res. Bull. 174, 22–30. doi: 10.1016/j.brainresbull.2021.04.026
Merry, C. R., Forrest, M. E., Sabers, J. N., Beard, L., Gao, X. H., Hatzoglou, M., et al. (2015). DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 24, 6240–6253. doi: 10.1093/hmg/ddv343
Ng, S.-Y., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi: 10.1016/j.molcel.2013.07.017
Ng, S.-Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors: LncRNAs involved in neuronal differentiation. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Ni, C., Jiang, W., Wang, Z., Wang, Z., Zhang, J., Zheng, X., et al. (2021). LncRNA-AC006129.1 reactivates a SOCS3-mediated anti-inflammatory response through DNA methylation-mediated CIC downregulation in schizophrenia. Mol. Psychiatry 26, 4511–4528. doi: 10.1038/s41380-020-0662-3
Pan, Z., Du, S., Wang, K., Guo, X., Mao, Q., Feng, X., et al. (2021). Downregulation of a dorsal root ganglion-specifically enriched long noncoding RNA is required for neuropathic pain by negatively regulating RALY-triggered Ehmt2 expression. Adv. Sci. 8:e2004515. doi: 10.1002/advs.202004515
Peng, W.-X., Koirala, P., Zhang, W., Ni, C., Wang, Z., Yang, L., et al. (2020). lncRNA RMST enhances DNMT3 expression through interaction with HuR. Mol. Ther. 28, 9–18. doi: 10.1016/j.ymthe.2019.09.024
Rashid, F., Shah, A., and Shan, G. (2016). Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics 14, 73–80. doi: 10.1016/j.gpb.2016.03.005
Schultz, C. W., Preet, R., Dhir, T., Dixon, D. A., and Brody, J. R. (2020). Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip. Rev. RNA 11:e1581. doi: 10.1002/wrna.1581
Shao, C., Gao, Y., Jin, D., Xu, X., Tan, S., Yu, H., et al. (2017). DNMT3a methylation in neuropathic pain. J. Pain Res. 10, 2253–2262. doi: 10.2147/JPR.S130654
Tompkins, D. A., Hobelmann, J. G., and Compton, P. (2017). Providing chronic pain management in the “Fifth Vital Sign” era: Historical and treatment perspectives on a modern-day medical dilemma. Drug Alcohol Depend. 173, S11–S21. doi: 10.1016/j.drugalcdep.2016.12.002
Uhde, C. W., Vives, J., Jaeger, I., and Li, M. (2010). Rmst is a novel marker for the mouse ventral mesencephalic floor plate and the anterior dorsal midline cells. PLoS One 5:e8641. doi: 10.1371/journal.pone.0008641
van Hecke, O., Austin, S. K., Khan, R. A., Smith, B. H., and Torrance, N. (2014). Neuropathic pain in the general population: A systematic review of epidemiological studies. Pain 155, 654–662. doi: 10.1016/j.pain.2013.11.013
van Velzen, M., Dahan, A., and Niesters, M. (2020). Neuropathic pain: Challenges and opportunities. Front. Pain Res. 1:1. doi: 10.3389/fpain.2020.00001
Wang, K., Wang, S., Chen, Y., Wu, D., Hu, X., Lu, Y., et al. (2021). Single-cell transcriptomic analysis of somatosensory neurons uncovers temporal development of neuropathic pain. Cell Res. 31, 904–918. doi: 10.1038/s41422-021-00479-9
Wang, X., He, Y., Zhang, Q., Ren, X., and Zhang, Z. (2021). Direct comparative analyses of 10X genomics chromium and smart-seq2. Genomics Proteomics Bioinformatics 19, 253–266. doi: 10.1016/j.gpb.2020.02.005
Wu, S., Bono, J., and Tao, Y.-X. (2019). Long noncoding RNA (lncRNA): A target in neuropathic pain. Expert Opin. Ther. Targets 23, 15–20. doi: 10.1080/14728222.2019.1550075
Wu, S., Marie Lutz, B., Miao, X., Liang, L., Mo, K., Chang, Y. J., et al. (2016). Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Mol. Pain 12:174480691662904. doi: 10.1177/1744806916629048
Yang, Y., Wen, J., Zheng, B., Wu, S., Mao, Q., Liang, L., et al. (2021). CREB participates in paclitaxel-induced neuropathic pain genesis through transcriptional activation of Dnmt3a in primary sensory neurons. Neurotherapeutics 18, 586–600. doi: 10.1007/s13311-020-00931-5
Zhang, L., Xie, R., Yang, J., Zhao, Y., Qi, C., Bian, G., et al. (2019). Chronic pain induces nociceptive neurogenesis in dorsal root ganglia from Sox2-positive satellite cells. Glia 67, 1062–1075. doi: 10.1002/glia.23588
Zhao, J.-Y., Liang, L., Gu, X., Li, Z., Wu, S., Sun, L., et al. (2017). DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat. Commun. 8:14712. doi: 10.1038/ncomms14712
Zhao, L., Zhang, M., Yan, F., and Cong, Y. (2021). Knockdown of RMST impedes neuronal apoptosis and oxidative stress in OGD/R-induced ischemic stroke via depending on the miR-377/SEMA3A signal network. Neurochem. Res. 46, 584–594. doi: 10.1007/s11064-020-03194-w
Keywords: neuropathic pain, dorsal root ganglia, Rmst lncRNA, HuR, DNA methyltransferase 3 alpha (DNMT3A), mRNA stability
Citation: Guo X, Zhang G, Cai W, Huang F, Qin J and Song X (2023) Long non-coding RNA rhabdomyosarcoma 2-associated transcript contributes to neuropathic pain by recruiting HuR to stabilize DNA methyltransferase 3 alpha mRNA expression in dorsal root ganglion neuron. Front. Mol. Neurosci. 15:1027063. doi: 10.3389/fnmol.2022.1027063
Received: 24 August 2022; Accepted: 16 November 2022;
Published: 22 February 2023.
Edited by:
Yize Li, Tianjin Medical University General Hospital, ChinaReviewed by:
Jingjing Yuan, First Affiliated Hospital of Zhengzhou University, ChinaCopyright © 2023 Guo, Zhang, Cai, Huang, Qin and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingrong Song, c3hqZXNzQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.