- Department of Biochemistry and Molecular Biology, Sidney Kimmel Medical College, Thomas Jefferson University, Philadelphia, PA, United States
Spinal and bulbar muscular atrophy (SBMA) is a neurodegenerative and neuromuscular genetic disease caused by the expansion of a polyglutamine-encoding CAG tract in the androgen receptor (AR) gene. The AR is an important transcriptional regulator of the nuclear hormone receptor superfamily; its levels are regulated in many ways including by ubiquitin-dependent degradation. Ubiquitination is a post-translational modification (PTM) which plays a key role in both AR transcriptional activity and its degradation. Moreover, the ubiquitin-proteasome system (UPS) is a fundamental component of cellular functioning and has been implicated in diseases of protein misfolding and aggregation, including polyglutamine (polyQ) repeat expansion diseases such as Huntington’s disease and SBMA. In this review, we discuss the details of the UPS system, its functions and regulation, and the role of AR ubiquitination and UPS components in SBMA. We also discuss aspects of the UPS that may be manipulated for therapeutic effect in SBMA.
Introduction
The ubiquitin-proteasome system (UPS) is of central importance to overall protein quality control in eukaryotic cells. It encompasses a vast, complex network of proteins that carry out a multitude of functions to preserve proteostasis. These functions include regulating intracellular protein levels by mediating their destruction, trafficking of proteins within and across subcellular compartments, regulating gene transcription, facilitating DNA damage repair, and mediating immune responses. Therefore, it is not surprising that dysfunction of UPS components has been implicated in diseases of neurodegeneration, viral infection, cancer, and other disorders (Rao et al., 2015; Budroni and Versteeg, 2021; Hwang et al., 2022; Park et al., 2022). The UPS also connects to the cell’s other protein degradation and quality control pathways, the unfolded protein response (UPR), endoplasmic reticulum-associated degradation (ERAD), and the autophagy-lysosome pathway (ALP), processes that also have been implicated in various neurodegenerative diseases (Chen et al., 2019; Lopata et al., 2020; Qu et al., 2021). The androgen receptor (AR), an important nuclear hormone receptor that plays a causative role in the neuromuscular disease spinal and bulbar muscular atrophy (SBMA), is regulated by the UPS for both its transcriptional functions and its degradation. This role of the UPS in AR function and metabolism, as well as its role in additional aspects of neuromuscular function in SBMA, is the focus of this review.
Spinal and bulbar muscular atrophy
Spinal and bulbar muscular atrophy (SBMA; also known as Kennedy’s disease) is an adult-onset, slowly progressive, neuromuscular disease affecting 1 in 40,000 males (Fischbeck, 1997; Dejager et al., 2002; Pradat et al., 2020). It is one of nine polyglutamine (polyQ) repeat expansion neurodegenerative disorders that include Huntington’s disease (HD), dentatorubral-pallidoluysian atrophy, and the spinocerebellar ataxias 1, 2, 3, 6, 7, and 17 (Lieberman et al., 2019). An X-linked disease, SBMA is caused by expansion of a polyQ-encoding, polymorphic, CAG repeat within exon 1 of the AR gene to a number greater than 37 (La Spada et al., 1991).
SBMA primarily affects men (Müller et al., 2021); studies of cell and mouse models of SBMA reveal the requirement for circulating testosterone for the emergence and progression of disease symptoms (Katsuno et al., 2002, 2003; Takeyama et al., 2002; Chevalier-Larsen et al., 2004; Sopher et al., 2004; Yu et al., 2006). Patients experience facial, tongue, and proximal muscle weakness that is accompanied by cramps, intention tremor, and fasciculations (Dejager et al., 2002; Atsuta et al., 2006; Rhodes et al., 2009; Guber et al., 2018). Speech, swallowing, and mobility are progressively impaired (Guber et al., 2018), and many patients become wheelchair-bound (Atsuta et al., 2006). Aspiration pneumonia is a common cause of death for SBMA patients (Atsuta et al., 2006). In addition to neuromuscular symptoms, patients also experience signs of mild androgen insensitivity, as well as metabolic and sensory disturbances (Arnold and Merry, 2019).
Although SBMA patients frequently exhibit signs of partial androgen insensitivity, including gynecomastia, testicular atrophy, and infertility, such noted partial loss of AR function does not explain the mechanism for SBMA neuromuscular symptoms (Pinsky et al., 1992); rather, these symptoms in SBMA result primarily from mutant AR proteotoxicity (Merry et al., 1998; Katsuno et al., 2002). Nonetheless, it is possible that reduced AR transcriptional function in the neuromuscular system upon polyQ expansion may exacerbate the proteotoxic effects of mutant, misfolded, AR, since knockout of endogenous AR exacerbated symptoms in a mouse model of SBMA (Thomas et al., 2006). Moreover, AR is known to play a trophic role in motor neurons and an important anabolic role in muscle; both are the direct result of the transcriptional function of androgen-bound AR (Kurz et al., 1986; Marron et al., 2005; Fargo et al., 2008a; MacLean et al., 2008; Little et al., 2009). These positive AR functions in spinal motor neurons and muscle are reduced in SBMA mouse models, as evidenced by modified AR transcriptional functions in both motor neurons and muscle (Lieberman, 2018). The production in SBMA patients of only mutant AR protein, produced from this single mutant allele, results in AR protein with characteristics of both proteotoxicity and partial loss of function.
SBMA pathology is characterized by the loss of lower motor neurons (LMN) from the brainstem and anterior horn of the spinal cord in postmortem tissues (Sobue et al., 1989) and by muscle fiber atrophy, fiber-type grouping, and centralized nuclei (Sobue et al., 1989; Jokela et al., 2016), evidence of both neurogenic and myogenic causes. Moreover, the significant elevation of serum creatine kinase (CK) level (Chahin and Sorenson, 2009; Querin et al., 2016), implicates a primary involvement of muscle atrophy in disease pathogenesis (reviewed in: Arnold and Merry, 2019). In support of this idea, muscle tissue-specific deletion of polyQ-expanded AR alleviated signs of disease in animal models of SBMA, illustrating an important myogenic contribution to disease (Soraru et al., 2008; Cortes et al., 2014; Lieberman et al., 2014). Finally, muscle biomarkers in SBMA patients correlate more strongly with disease severity than do neuronal biomarkers (Lombardi et al., 2019).
In addition to these characteristic pathologies, affected SBMA tissues exhibit misfolding and predominantly, but not exclusively, nuclear aggregation of the mutant AR (Li et al., 1998a,b; Adachi et al., 2005; Suzuki et al., 2007). These neuronal intranuclear inclusions (NII) in patient tissue contain truncated N-terminal fragments of polyQ-expanded AR and ubiquitin (Li et al., 1998a,b, 2007; Heine et al., 2015). Additionally, chaperone proteins Hsp40, Hsp70, and Hsp90, and UPS components, including 20S proteasome, NEDD8, PA700, and REGγ, are detected in inclusions in cell and mouse models (Stenoien et al., 1999; Abel et al., 2001; Bailey et al., 2002; Yersak et al., 2017). P62 also accumulates in NII, as detected in patient tissue and a mouse model of SBMA (Doi et al., 2013).
Despite the fundamental knowledge of the genetic cause of this disorder, the growing understanding of the diverse symptom range and underlying molecular mechanisms, a number of well-characterized in vitro and in vivo disease models, and several potential interventions in clinical trials, there is still no effective treatment for SBMA (Arnold and Merry, 2019).
The ubiquitin-proteasome system
The proteasome
The UPS is responsible for the majority of intracellular protein degradation (Rock et al., 1994; Collins and Goldberg, 2017). The proteasome is a large (2.4 MDa), highly conserved, multisubunit protein complex with ATP-dependent proteolytic activity. It is found in both the nuclear and cytosolic compartments of eukaryotic cells. It consists of a cylindrical 20S core particle that can be found either unbound as a free 20S proteasome, or in complex with a regulatory particle flanking it on either or both sides. The constitutive 26S proteasome is comprised of a 20S core particle bound to a single 19S regulatory particle (19S–20S), while the 30S proteasome contains two 19S regulatory particles, one on either side (19S-20S-19S) (Coux et al., 1996; Baumeister et al., 1998; Demartino and Gillette, 2007). Although not bound by any regulatory particle, the free 20S proteasome is also capable of degrading polypeptides, particularly during periods of cellular stress (Raynes et al., 2016).
The 20S core particle of the proteasome is composed of 4 stacked rings, two outer α-rings and two catalytic inner β-rings, each comprised of 7 subunits. Located in the inner β-rings are the three active sites, each with distinct amino acid cleavage specificity and each bearing an active site threonine amino acid. The β1 caspase-like subunit cleaves after acidic sites, the β2 trypsin-like subunit cleaves after basic residues, and the β5 chymotrypsin-like subunit cleaves after large hydrophobic sites (Wilk and Orlowski, 1983; Voges et al., 1999).
The regulatory particle of the proteasome recognizes and binds polyubiquitin-tagged substrate proteins, cleaves the ubiquitin chain, unfolds the protein in an ATP-dependent manner, and feeds the polypeptide into the center of the 20S core. Within the 20S core particle, the movement-coupled proteolysis occurs in an ATP-dependent manner. The regulatory particles in an assembled proteasome complex determine its structure and function, resulting in distinct proteasome types (Collins and Goldberg, 2017).
Ubiquitination
Ubiquitination is a post-translational modification (PTM) wherein a single ubiquitin protein (monoubiquitination) or a chain of multiple, linked ubiquitin proteins (polyubiquitination) is covalently attached to a substrate protein and alters its cellular fate (Hershko, 1988). Ubiquitin is a highly conserved and very small (8.6 kDa) protein, found “ubiquitously” in all cell types. Mammals have four distinct genes (UBB, UBC, UBA52, and RPS27A) encoding ubiquitin precursor proteins. The ubiquitin precursors are polymeric proteins which are cleaved by specific deubiquitinases to produce free monoubiquitin (Finley et al., 1989; Grou et al., 2015). UBB and UBC code for polyubiquitin (Lund et al., 1985; Wiborg et al., 1985) while UBA52 and RPS27A code for fusion proteins consisting of a single ubiquitin and a ribosomal protein, L40 and S27a, respectively (Redman and Rechsteiner, 1989; Baker and Board, 1991).
Three classes of enzymes work in concert to mediate protein ubiquitination in a multi-step process. First, an ATP-dependent E1 activating enzyme forms a thioester bond with the C-terminus of a free ubiquitin. Next, the activated ubiquitin is transferred to the active cysteine site of an E2 conjugating enzyme. In the third and final step, an E3 ubiquitin ligase catalyzes the ubiquitin transfer to a substrate protein, forming an isopeptide bond (Hershko, 1988; Ciechanover, 1998). This process can occur once for monoubiquitin, or multiple times sequentially to result in a polyubiquitin chain. A ubiquitin chain is typically attached to lysine residues on substrate proteins; however, non-lysine ubiquitination on cysteine, serine, and threonine residues, and at the N-terminus of substrate proteins has also been documented (McDowell and Philpott, 2016; McClellan et al., 2019).
E3 ubiquitin ligases
There are two known human E1 activating enzymes: Ube1 and Uba6 (Schulman and Wade Harper, 2009; reviewed in: Wang and Zhao, 2019), approximately 40 known human E2 conjugating enzymes (Michelle et al., 2009; Wenzel et al., 2011b), and over 600 known human E3 ubiquitin ligases (Li et al., 2008; Medvar et al., 2016). Additionally, E3/E4 elongation ligases are capable of adding polyubiquitin chains en bloc (Koegl et al., 1999). E3 ubiquitin ligases represent an important layer of substrate specificity for ubiquitination. They are classified into four main structurally and mechanistically distinct families (Buetow and Huang, 2016; Morreale and Walden, 2016; Zheng and Shabek, 2017; Yang et al., 2021).
Members of the HECT (Homologous to the E6-AP Carboxyl Terminus) family of ubiquitin E3 ligases possess a HECT domain in the C-terminus and ubiquitinate substrate proteins in a two-step process. The N-terminus of the HECT E3 ligase binds both the substrate protein and the ubiquitin-bearing E2-conjugating enzyme. In the first step of ubiquitination, a ubiquitin is transferred from the E2 enzyme to a catalytic cysteine residue in the conserved C-terminal HECT domain. In the next step, ubiquitin is transferred from the cysteine site to the substrate protein (Huibregtse et al., 1995). Based on N-terminal structural differences that impart specificity, the HECT family can be further divided into the Nedd4, HERC, and “other HECT” E3 ligase sub-families (Rotin and Kumar, 2009; Weber et al., 2019).
The RING (Really Interesting New Gene) E3 ubiquitin ligases, the largest family of E3 ligases, contain an N-terminal RING domain that binds the E2 conjugating enzyme. Ubiquitination takes place in a single-step reaction, with ubiquitin being transferred directly from the E2 enzyme to the substrate protein. The RING E3 ligase family can be further divided based on functional structure, i.e., monomeric, dimeric (for example, RNF4), multisubunit complex. The multiprotein RING E3 ligases include the cullin-RING ligase (CRLs) protein complexes. These are composed of a RING-protein, an adaptor protein, and a substrate receptor protein. Some multisubunit complex RING E3 ligases can be very large, such as the APC/C (Anaphase Promoting Complex/Cyclosome) which can be comprised of 19 proteins, and the SCF (Skp1, Cullin1, and F-box) E3 ligase complexes (Chang and Barford, 2014; Metzger et al., 2014; Buetow and Huang, 2016; Yang et al., 2021).
The U-box and RBR E3 ubiquitin ligases are the two smaller families of E3 ligases. U-box E3 ligases contain a conserved C-terminal U-box domain that is structurally similar to a RING domain (Aravind and Koonin, 2000; Hatakeyama et al., 2001). In addition to E3 ligase activity, they are capable of E4 elongation activity; that is, they work in conjunction with another E3 ligase to add polyubiquitin chains to an existing ubiquitin chain. U-box E3 ligases include E4B (also known as UBE4A) (Koegl et al., 1999) and CHIP (Imai et al., 2002).
The RBR (RING-Between-RING) E3 ligases contain two RING domains separated by an IBR (in-between-RING) domain (Aguilera et al., 2000). RBR E3 ligases bind to the activated E2 enzyme via the RING1 domain. Ubiquitination is a two-step reaction. In the initial step, ubiquitin is transferred to a catalytic RING2 domain; in the next and final step, ubiquitin is transferred from the RING2 domain to the substrate protein. RBR E3 ubiquitin ligases include Parkin, the E3 ligase implicated in Parkinson’s disease (Wenzel et al., 2011a).
Deubiquitinases
Deubiquitinases (DUBs) are isopeptidases that hydrolyze the amide bond between a ubiquitinated protein and its ubiquitin molecule or chain, and thus catalyze the removal of ubiquitin from substrate proteins. In the cell, DUBs can be found either associated with or independent of the proteasome. Proteasome-independent DUBs modify the intracellular fate of ubiquitinated substrate proteins and provide an additional layer of specificity and regulation of ubiquitination (Komander and Rape, 2012).
Six structurally distinct families of deubiquitinating enzymes have been identified; these include four cysteine protease DUBs, the zinc metalloprotease DUBs, and the motif-interacting-with-ubiquitin (MIU)–containing DUB family (MINDYs). The four cysteine protease DUB families include the ubiquitin-specific proteases (USP), the ubiquitin C-terminal hydrolases (UCH), the ovarian tumor proteases (OTU), and the Machado-Joseph disease protein domain proteases (MJD or Josephin). The largest and best studied of the DUB protein families is the USP family (Amerik and Hochstrasser, 2004; Mevissen and Komander, 2017).
In addition to having substrate protein specificity, some DUBs exhibit preferences for cleaving certain linkage types (Mevissen and Komander, 2017). Moreover, DUBs may work in tandem with an E3 ligase to exert opposing effects on a substrate protein to regulate its abundance or subcellular localization (Kim and Sixma, 2017; Mevissen and Komander, 2017). For example, the intricate mutual associations of the DUB USP7, its substrate protein p53, and the E3 ligase MDM2 have been well-studied. Tumor suppressor p53 is a transcription factor involved in cell cycle arrest and apoptosis, among other cellular functions (Vogelstein et al., 2000). MDM2 targets it for proteasomal degradation, maintaining a relatively short half-life of p53 (Maki et al., 1996). The DUB USP7 is capable of binding and deubiquitinating p53 or MDM2, thereby protecting them from proteasomal degradation. USP7 has greater binding affinity for MDM2 than for p53 (Hu et al., 2006; Sheng et al., 2006), and at cellular homeostasis, it deubiquitinates and stabilizes MDM2 (Brooks et al., 2007). However, during periods of cellular stress, USP7 preferentially binds p53, protecting it from degradation. Additionally, USP7-MDM2-p53 form a multiprotein complex (Brooks et al., 2007). Other examples of DUB-E3 ligase interactions and tandem function on the same substrate protein include USP39/TRIM26 (Li et al., 2021) and USP18/Skp2 (Tan et al., 2018).
Functions of the ubiquitin-proteasome system
The intracellular functions of the UPS can be broadly categorized as proteolytic or non-proteolytic. To a great extent, these functions depend on the ubiquitin code—a large diversity of ubiquitin chain and linkage types associated with different cellular fates. For many years, the role of polyubiquitin modification in regulating proteasomal degradation was thought to be solely due to K48-linkages, with all other chain types serving a non-proteolytic role. However, the ubiquitin code is revealing itself to be far more versatile and complex than was initially believed. A thorough review of the cellular processes associated with distinct ubiquitin linkages was recently published (Blount et al., 2020); only a short overview is thus provided here.
Polyubiquitin chains consist of ubiquitin proteins linked via one of seven lysines (K6, K11, K27, K29, K33, K48, K63) (Peng et al., 2003) or its N-terminal methionine residue (M1). Polyubiquitin chains can be homotypic, i.e., made up of only one linkage type, or heterotypic/mixed (Mevissen and Komander, 2017). Proteins may also be modified by monoubiquitin; this modification is associated with protein trafficking, including endocytosis (Marmor and Yarden, 2004) and nuclear export (Li et al., 2003), histone modification and regulation of transcriptional activity (Zhang, 2003), and more recently, with proteasomal degradation (Braten et al., 2016; Livneh et al., 2017).
The K48-linked tetraubiquitin chain topology is the most abundant and is associated with degradation mediated by the 26S proteasome (Chau et al., 1989; Gregori et al., 1990; Peng et al., 2003). However, a notable exception is found in the K48-ubiquitination of Met4, which regulates its transcriptional activity (Flick et al., 2004) in the absence of degradation. The ubiquitin code is further complicated in that, along with monoubiquitin and K48 linkages, K11, K29, and K63-linked polyubiquitin chains can also serve as degradation signals (Komander and Rape, 2012). It is also important to note that the proteasome can mediate protein degradation in a ubiquitin-independent manner (Driscoll and Goldberg, 1989; Buneeva and Medvedev, 2018).
Non-proteolytic roles for polyubiquitination include, for K6 linkages, mitochondrial homeostasis (Durcan et al., 2014) and the DNA damage response (Wu-Baer et al., 2003; Elia et al., 2015; Blount et al., 2020), and for K27 linkages, cellular proliferation (Shearer et al., 2022) and the innate immune response (Yin et al., 2019). K29 polyubiquitin linkages play a role in the cellular stress response and in regulating the cell cycle (Yu et al., 2021) while K33-linked chains are involved in T cell activation and signaling (Huang et al., 2010; Yang et al., 2015). K63-linked polyubiquitin chains play important roles in intracellular trafficking (Lauwers et al., 2009; Erpapazoglou et al., 2014), cell signaling (Deng et al., 2000; Wang et al., 2001), and DNA damage repair (Liu et al., 2018). They are also associated with autophagic targeting, and particularly with the selective autophagy of aggregates (aggrephagy) (Olzmann et al., 2007; Olzmann and Chin, 2008; Tan et al., 2008). Finally, linear polyubiquitin chains are unique in that, instead of linking via a lysine residue of ubiquitin, they consist of a ubiquitin protein linked to the C-terminal glycine of another ubiquitin at its N-terminal methionine (M1) (Kirisako et al., 2006). They are formed by the E3 ligase linear ubiquitin chain assembly complex (LUBAC) and play a role in NF-kappaB activation and immune signaling (Shimizu et al., 2015).
Regulation of the ubiquitin-proteasome system
Multiple layers of regulation of both degradative and non-degradative ubiquitination have been discovered. Many of these mechanisms are substrate-specific. E3 ubiquitin ligases and DUBs contain specific substrate recognition domains. Additionally, substrate proteins may contain general recognition signals for components of the UPS. Degrons or degradative signals are short amino acid sequences on substrate proteins associated with ubiquitination and proteasomal degradation. Both N-terminal and C-terminal degrons are well-studied (Varshavsky, 2008, 2019). Some proteins that harbor a PEST sequence—an amino acid sequence enriched in proline (P), glutamic acid (E), serine (S), and threonine (T)—have been found to undergo rapid degradation (Rogers et al., 1986). Intrinsically disordered regions of proteins represent targeting domains for proteasomal degradation (Guharoy et al., 2016). Additionally, misfolded proteins are specifically ubiquitinated and then degraded by the UPS or ALP (Amm et al., 2014).
PTMs play a critical role in UPS regulation by affecting the ubiquitination status of substrate proteins. E3 ligases and DUBs are also subject to regulation by PTMs. Finally, PTMs on ubiquitin itself can impact polyubiquitin chain formation; for example, ubiquitin acetylation inhibits chain elongation (Ohtake et al., 2015; Song and Luo, 2019). Importantly, many E3 ligases are capable of autoubiquitination, and thus negatively regulate their own intracellular levels (De Bie and Ciechanover, 2011).
Subcellular compartmentalization of substrate proteins and of the different UPS components also contributes to the regulation of the UPS. Some DUBs are predominantly nuclear or cytosolic, while others localize in both compartments. This impacts their ability to deubiquitinate particular substrate proteins based on their localization (Urbé et al., 2012). Similarly, E3 ligases are affected by localization as well (Checquolo et al., 2010; Breckel and Hochstrasser, 2021). Additionally, some E3 ligases localize primarily at the endoplasmic reticulum (ER) and are involved in ER-associated degradation (ERAD), while others are associated with Golgi bodies, the cell membrane, or mitochondria and peroxisomes, as reviewed by Rusilowicz-Jones et al. (2021).
Finally, ubiquitin-like proteins include SUMO (small ubiquitin-related modifier) and NEDD8 (neural precursor cell expressed, developmentally downregulated 8). In addition to modifying substrate proteins, they can also modify existing polyubiquitin chains (Herrmann et al., 2007). Other factors impacting protein ubiquitination, like the maintenance of the free ubiquitin pool and transcriptional regulation of ubiquitin genes are detailed by Kimura and Tanaka (2010).
Androgen receptor lifecycle and ubiquitination of androgen receptor
Androgen receptor
In order to understand the role of ubiquitination in SBMA, a discussion of wild-type AR (WT AR) is warranted. The AR gene (AR; gene: NR3C4) that encodes the 920-amino acid, 110 kDa protein, is located on the X chromosome at Xq11.2-Xq12 (Brown et al., 1989). Its primary physiological ligands are testosterone and its metabolite 5α-dihydrotestosterone (DHT) (Georget et al., 1997). AR is widely expressed in mammalian tissues, including in the central and peripheral nervous systems, muscle, and male reproductive organs (Rana et al., 2014). A transcription factor, it controls the expression of a multitude of target genes (Jia et al., 2008; Jiang et al., 2009; Massie et al., 2011; Jin et al., 2013), including genes responsible for embryonic sex differentiation and development of male secondary sex characteristics (Quigley et al., 1995; Hiort, 2013). AR and androgens play important roles in neurons, including during development and in response to injury (Pérez and Kelley, 1996; Fargo et al., 2008b; Davey et al., 2017; Mhaouty-Kodja, 2018). In skeletal muscle, AR and androgens play an important anabolic role, including effects on muscle mass and strength (Sheffield-Moore et al., 1999; Chen et al., 2005; Kadi, 2008; Leblanc et al., 2011; Davey et al., 2017).
The AR is composed of four primary domains: the intrinsically disordered N-terminal domain (NTD) (Lavery and McEwan, 2008; Sheikhhassani et al., 2022), a DNA-binding domain containing two zinc fingers (Shaffer et al., 2004), a hinge region, and a C-terminal ligand-binding domain (LBD) (Brinkmann et al., 1989). The AR contains three transactivation domains, of which the most potent is the AF1 in the NTD (Christiaens et al., 2002). The other transactivation domains, AF2 and BF3, are located in the LBD and are only exposed upon ligand-binding (He et al., 2004b; Estébanez-Perpiñá et al., 2007; Buzon et al., 2012). AR translocation to the nucleus is facilitated by a bipartite nuclear localization signal (NLS) within the hinge region (Zhou et al., 1994). Nuclear export may be regulated by a nuclear export signal (NESAR) in the carboxyl-terminal region (amino acids 744-818) (Saporita et al., 2003; Gong et al., 2012) although a specific NES has not been defined. The polymorphic glutamine repeat is located in the NTD (La Spada et al., 1991).
Lifecycle of wild-type AR
A steroid hormone nuclear receptor, AR exists in the cytoplasm in an inactive and monomeric form in a multiprotein aporeceptor complex along with p23, immunophilins, and chaperone proteins such as Hsp70, Hsp40, and Hsp90. Ligand-binding induces a conformational change and reorganization of the aporeceptor complex (Ni et al., 2013) and induces the interaction between the N-terminal FQNLF motif and AF2 domain in an intramolecular N/C interaction (He et al., 2004b). Ligand-binding induces dissociation of importin 7 (Ni et al., 2013) and chaperone proteins Hsp70 and Hsp40 (Eftekharzadeh et al., 2019). The intramolecular N/C interaction exposes the hinge region NLS, allowing α-importin to bind AR and facilitate nuclear translocation (Zhou et al., 1994; Cutress et al., 2008; van Royen et al., 2012).
Within the nucleus, AR forms homodimers on DNA (Langley et al., 1995; El Kharraz et al., 2021). While both monomeric and dimeric AR are found in the nucleus (van Royen et al., 2012), it is the dimeric AR that predominantly interacts with DNA at androgen response elements (AREs) in promoter or enhancer regions of target genes and recruits coregulatory proteins (Wong et al., 1993; Shaffer et al., 2004). There, N/C interactions and LBD interactions facilitate AR dimerization in a unique head-to-head and tail-to-tail conformation, revealed by cryo-EM (Yu et al., 2020). AR assembles a transcriptional complex containing co-regulators, RNA polymerase II, and histone-modifying enzymes (Chang and McDonnell, 2005; Yu et al., 2020). Over 270 co-activators and co-repressors of AR have been identified (DePriest et al., 2016), including steroid receptor coactivator (SRC) family proteins (Bevan et al., 1999), CREB-binding protein (CBP) (Aarnisalo et al., 1998), p300 (Zhong et al., 2014), and UBC9 (Poukka et al., 1999).
Details of AR nuclear export are lacking; however, it is known that the wild-type AR is primarily degraded in the cytoplasm by the 26S proteasome (Sheflin et al., 2000; Gong et al., 2012).
E3 ligases mediating ubiquitination of wild-type AR
A number of proteins are involved in regulating the ubiquitination status of AR. The functions of these proteins are not solely degradative; both proteolytic and non-proteolytic ubiquitinating activities have been identified. These activities regulate AR stability, intracellular localization, and transactivation (Jaworski, 2006). An overview of the E3 ubiquitin ligases and DUBs known to act on AR and alter its ubiquitination state is shown in Figure 1.
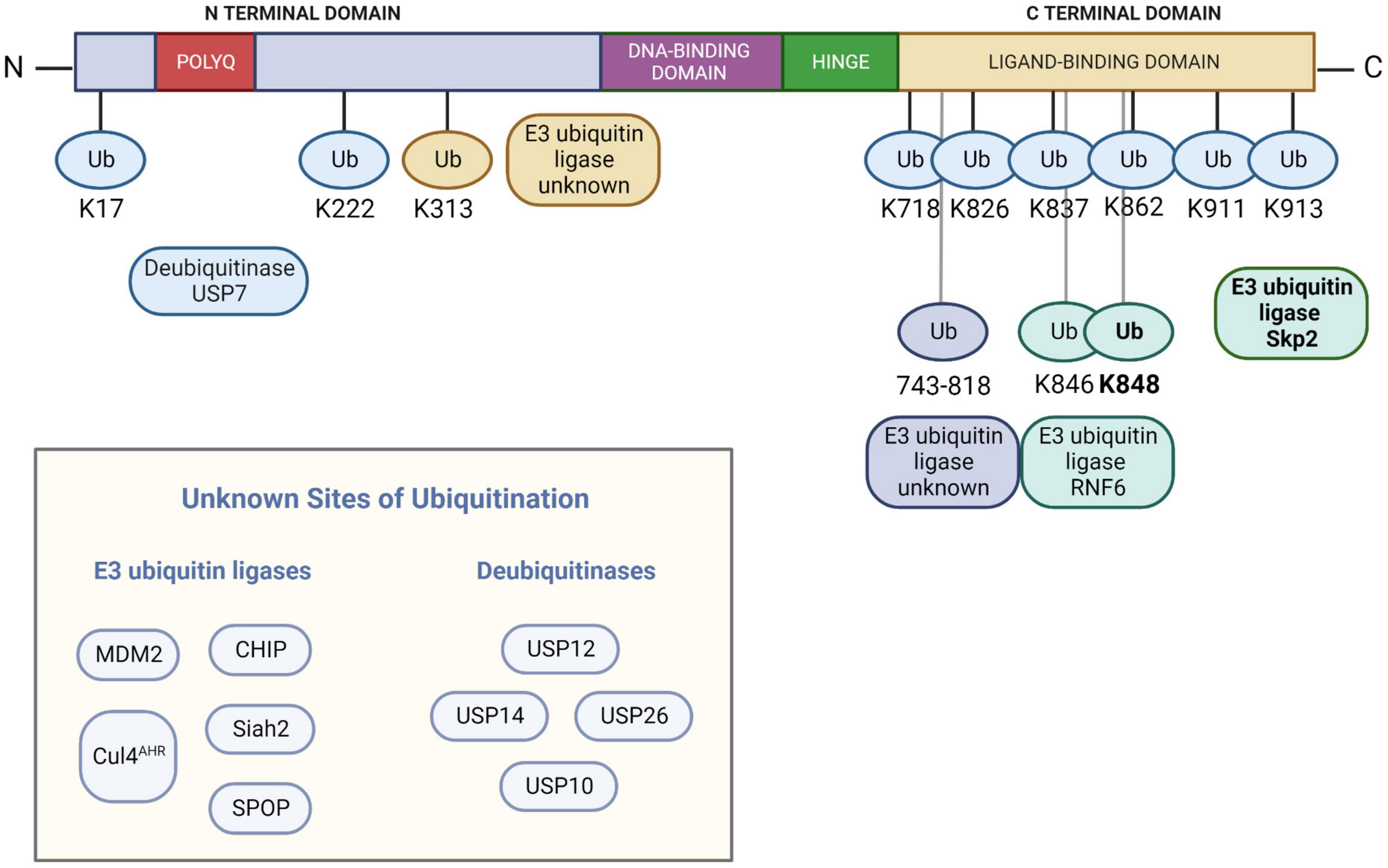
Figure 1. The known E3 ubiquitin ligases and deubiquitinases that impact the ubiquitination status of the androgen receptor, along with sites of ubiquitin modification (if known). Note that Skp2 modifies K848 with K48 polyubiquitin chains while RNF2 modifies both K846 and K848 with K6, K27, and K63 polyubiquitin chains. AR numbering is based on NCBI reference sequence NM_000044.6 and UniProtKB/Swiss-Prot: P10275.3. Figure created with BioRender.com.
Several E3 ubiquitin ligases have been described that modify WT AR. The best-studied E3 ubiquitin ligase acting on AR is the U-Box protein COOH terminus of the Hsp70-interacting protein (CHIP). CHIP polyubiquitinates AR and targets it for proteasomal degradation (He et al., 2004a; Sarkar et al., 2017). CHIP-mediated ubiquitination requires the presence of chaperone proteins such as Hsp90 or Hsp70 and Hsp40 (Murata et al., 2001; Wang et al., 2010; Chymkowitch et al., 2011). Since CHIP predominantly interacts with cytoplasmic AR (He et al., 2004a), it is likely to be at least partly responsible for the reduced stabilization of AR in the absence of hormone (Kemppainen et al., 1992).
RING-finger E3 ubiquitin ligase MDM2 polyubiquitinates AR and mediates its degradation (Lin et al., 2002; Xu et al., 2009; Vummidi Giridhar et al., 2019). Its actions on AR are dependent on Akt-regulated phosphorylation (Lin et al., 2002). MDM2 acts on AR while it is bound to DNA (Lv et al., 2021). In addition to polyubiquitinating AR and targeting it for proteasomal degradation, MDM2 may monoubiquitinate DNA-bound AR and enhance its transcriptional activity (Gaughan et al., 2005).
Other proteins that polyubiquitinate AR and target it for proteasomal degradation have been identified. The cullin 3 (CUL3)-based E3 ligase SPOP polyubiquitinates AR in a manner that is regulated by S648 and T649 in the hinge region (An et al., 2014). In addition to targeting AR for proteasomal degradation (Khan et al., 2006; Rees et al., 2006; Chen et al., 2022), SPOP also polyubiquitinates and mediates the degradation of AR co-regulator SRC-3 (Li et al., 2011; Geng et al., 2014), thus impacting AR transcriptional activity in two ways. Siah2 adds K48-linked polyubiquitin chains to regulate the level of transcriptionally inactive AR (NCORC1-bound AR) (Qi et al., 2013; Jing et al., 2018). Skp2 polyubiquitinates AR at K848 (Li et al., 2014) and targets it for proteasomal degradation (Li et al., 2014; Chen et al., 2022). AHR acts on AR as part of a CUL4 complex (Ohtake et al., 2007, 2009). Exported AR is polyubiquitinated at the NESAR and targeted for proteasomal degradation (Gong et al., 2012), although the E3 ligase involved in this ubiquitination is unknown.
Other E3 ligases acting on AR directly impact AR transactivation. E3 ligase RNF6 polyubiquitinates AR at K846 and K848. Unlike the role of Skp2, however, RNF6 promotes AR transcriptional activity on specific target genes via K6, K27, and K63 linkages and appears to have no impact on AR degradation (Xu et al., 2009; Sarkar et al., 2014; Lim et al., 2017). Ubiquitination at K313 also facilitates AR transactivation (McClurg et al., 2017).
Deubiquitinases mediating deubiquitination of wild-type AR
Several cysteine protease DUBs have been reported to cleave ubiquitin from AR. USP10 deubiquitinates DNA-bound AR and promotes its transcriptional activity (Faus et al., 2005). USP12 (Burska et al., 2013; McClurg et al., 2014) and the proteasome-associated DUB USP14 (Liao et al., 2017, 2018; Xia et al., 2019) deubiquitinate and stabilize AR. USP7 deubiquitinates AR on AREs on AR target genes, stabilizing it and promoting its transcriptional activity (Chen et al., 2015). USP26 deubiquitinates nuclear AR, abrogating the ubiquitinating effects of MDM2 (Dirac and Bernards, 2010; Wang et al., 2020). Notably, both USP26 (Lahav-Baratz et al., 2017) and USP7 (Marine et al., 2006) also exert their deubiquitinating activity on MDM2.
Regulation of androgen receptor ubiquitination
PTMs such as phosphorylation and acetylation are involved in the regulation of ubiquitination of AR for proteasomal degradation. Phosphorylation, in particular, has a pivotal role in the selection of E3 ligases. Phosphorylation driven by the kinase Akt at S215, S516, and S792 (Lin et al., 2001, 2002; Palazzolo et al., 2007, 2009; Chymkowitch et al., 2011) plays a role in MDM2-mediated ubiquitination and subsequent AR degradation. Phosphorylation by PAK6 at S579 also facilitates MDM2-mediated ubiquitination and proteasomal degradation (Liu et al., 2013).
CHIP interacts with and ubiquitinates AR in both a phosphorylation-dependent (Rees et al., 2006) and phosphorylation-independent manner (He et al., 2004a; Chymkowitch et al., 2011). Another report suggests that the activity of MDM2 or CHIP on AR is dependent on AR phosphorylation status (Chymkowitch et al., 2011). Additionally, the chaperone protein Hsp70 and its co-chaperone Hsp40 are important for CHIP-mediated ubiquitination of Hsp90 client proteins like AR (Wang et al., 2010; Eftekharzadeh et al., 2019). On the other hand, p300-mediated acetylation of AR prevents its ubiquitination and degradation, and promotes its transcriptional activity (Zhong et al., 2014). Other UPS components regulate AR activity and/or levels, although the mechanistic details of their roles have not been elucidated.
Polyglutamine-expanded androgen receptor and spinal and bulbar muscular atrophy
The discovery of the critical role for AR hormone binding in SBMA led to an understanding of the role of specific hormone-dependent aspects of AR metabolism in disease. Nuclear localization of the mutant AR is required for its toxicity (Montie et al., 2009; Nedelsky et al., 2010); retention of the AR in the cytoplasm promotes its autophagic degradation and reduces its toxicity (Montie and Merry, 2009; Montie et al., 2009). On the other hand, promoting the nuclear export of polyQ-expanded AR led to enhanced degradation via the proteasome (Arnold et al., 2019). In addition, the N/C interaction is required for disease, both in vitro (Orr et al., 2010) and in vivo (Zboray et al., 2015). Inhibiting the N/C interaction promoted phosphorylation at S16, a PTM required for this neuroprotection (Zboray et al., 2015). Moreover, both AR arginine methylation (Scaramuzzino et al., 2015) and AR acetylation (Montie et al., 2011) represent PTMs of the mutant AR that are required for toxicity; in addition, roles for NLK and CK2 in modulating both the phosphorylation and toxicity of mutant AR indicate a role for AR phosphorylation in SBMA (Todd et al., 2015; Polanco et al., 2016). PolyQ-expanded AR exhibits increased DNA-binding (Belikov et al., 2015) and both increased and decreased transcriptional activity compared to WT AR (Mhatre et al., 1993; Kazemi-Esfarjani et al., 1995; Choong et al., 1996; Nakajima et al., 1996; Irvine, 2000; Lieberman et al., 2002; Sheppard et al., 2011; Belikov et al., 2015; Scaramuzzino et al., 2015; Badders et al., 2018). PolyQ-expanded AR also exhibits differential protein interactions vs. WT AR (Pluciennik et al., 2021), including its interactions with UPS components USP26 and USP7.
The role of polyglutamine-expanded androgen receptor ubiquitination and ubiquitin-proteasome system components in spinal and bulbar muscular atrophy
Not only is the wild-type AR primarily degraded by the proteasome (Sheflin et al., 2000), proteasome activity is required for the clearance of polyQ-expanded AR as well (Bailey et al., 2002; Nath et al., 2018; Arnold et al., 2019). In addition, a role for the proteasome was observed in the nuclear inclusion-associated proteolysis of polyQ-expanded AR; this role appeared late in the maturation of nuclear inclusions, suggesting that AR proteolysis within inclusions likely does not contribute to toxicity (Heine et al., 2015). Nonetheless, this observation of nuclear mutant AR proteolysis indicates that proteasomes can act on polyQ-expanded AR, albeit inefficiently, within the nuclear compartment. Although proteasomes are found in both the nuclear and cytoplasmic subcellular compartments, the relative localization may vary by cell type and cell cycle stage (Wojcik and DeMartino, 2003; Kito et al., 2020; Guo, 2022). Moreover, not only are catalytically active proteasomes primarily found in the cytoplasm (Dang et al., 2016), but many nuclear proteins require export to the cytoplasm for their proteasomal degradation (Chen and Madura, 2014), demonstrating the importance of cytoplasmic proteasomes in cellular protein degradation.
Despite the demonstrated role of cytoplasmic proteasomes in the degradation of many nuclear proteins, including polyQ-expanded AR, a role for nuclear proteasomes in the degradation of misfolded proteins has been demonstrated (Prasad et al., 2010; Park et al., 2013). Moreover, recent evidence indicates that nuclear AR can be efficiently degraded by nuclear proteasomes in the absence of androgens in models of prostate cancer (Lv et al., 2021). Despite this finding of efficient nuclear proteasomal degradation of AR in prostate cancer models, the observation that the inefficient proteasomal degradation of polyQ-AR contributes to its aggregation (Heine et al., 2015), and that enhancing nuclear export of polyQ-AR substantially enhances its proteasomal degradation (Arnold et al., 2019), suggests that polyQ-expanded AR is more efficiently degraded by proteasomes within the cytoplasmic compartment. The role of hormone binding and release and the mechanisms that impact the differences in efficiency between nuclear and cytoplasmic proteasomal degradation of polyQ-expanded AR remain to be determined.
A role for AR ubiquitination in the pathogenesis of SBMA derives from the finding that the DUB USP7 is required for disease manifestations in vitro and in vivo (Pluciennik et al., 2021). USP7 was found to preferentially interact with polyQ-expanded AR and deubiquitinate it at 8 specific lysine residues, K17, K222, K718, K826, K837, K862, K911, and K913. USP7 depletion decreased mutant AR aggregation and toxicity in cell models and in both transgenic Drosophila and mouse SBMA models (Pluciennik et al., 2021). Additionally, USP7 preferentially interacted with soluble, monomeric, and aggregated toxicity-associated species of polyQ-expanded AR (Berger et al., 2015; Heine et al., 2015; Pluciennik et al., 2021). USP7 knockdown reduced the level of this aggregated species as well as of a population of monomeric AR. Mutation of one of the eight USP7-regulated ubiquitination sites (K17) substantially increased polyQ-expanded AR aggregation, suggesting the importance of this site for mutant AR homeostasis. USP7 also interacted with polyQ-expanded mHtt and polyQ-expanded Ataxin-3 and was shown to attenuate pathology induced by polyQ-expanded Ataxin-3 (Pluciennik et al., 2021).
The U-Box E3 ligase CHIP polyubiquitinates both WT and polyQ-expanded AR; however, its overexpression preferentially ubiquitinated and cleared polyQ-expanded AR, leading to phenotypic improvements in cultured neurons and transgenic polyQ-expanded AR mouse models of SBMA (Adachi et al., 2007). Like the DUB USP7 (Pluciennik et al., 2021), CHIP also interacts with mHtt and polyQ-expanded Ataxin-3 (Jana et al., 2005), and its overexpression increased the ubiquitination and degradation of other polyQ-expanded proteins such as mHtt, mutant Ataxin-1, and mutant Ataxin-3 (Jana et al., 2005; Miller, 2005; Al-Ramahi et al., 2006; Williams et al., 2009). CHIP-mediated ubiquitination is dependent on the Hsp90/Hsp70 chaperone machinery (Wang et al., 2010). Indeed, both the ubiquitinating U-box domain and the chaperone protein-interacting TPR domain facilitate CHIP’s ubiquitinating ability.
The regulation of AR ubiquitination by PTMs leads to potentially tractable therapeutic approaches. For example, the Akt pathway promotes phosphorylation of AR at S215 and S792. Overexpression of insulin-like growth factor (IGF-1) activated Akt in skeletal muscle of AR97Q transgenic mice, resulting in increased polyQ-AR phosphorylation at S215 and its increased proteasomal degradation. PolyQ-expanded AR aggregation was reduced, motor defects were attenuated, onset of disease was delayed, and mouse survival was extended (Palazzolo et al., 2007, 2009; Rinaldi et al., 2012). In contrast, phosphorylation at S96 increased polyQ-expanded AR aggregation and toxicity; its inhibition lowered levels of polyQ-expanded AR in a proteasome-dependent manner, and ameliorated SBMA phenotypes in cell and knock-in mouse models (Polanco et al., 2016).
The role of additional ubiquitin-proteasome system components in spinal and bulbar muscular atrophy
Nrf1 represents an important transcriptional regulator of proteasome subunit expression during UPS impairment or proteasome stress, thereby regulating proteasome homeostasis (Radhakrishnan et al., 2010; Sha and Goldberg, 2014; Lehrbach et al., 2019). Nrf1 is itself regulated by its proteolytic cleavage at the ER by DDI2, thus enabling its nuclear translocation for transactivation (Koizumi et al., 2016; Lehrbach and Ruvkun, 2016; Northrop et al., 2020). Both Nrf1 and DDI2 levels were reduced in skeletal muscle of a knock-in mouse model of SBMA (Nath et al., 2018). Importantly, activation of Nrf1 not only increased the levels of many proteasome subunits, including 20S CP, 19S RP, and 11S RP components, it also increased the ubiquitination of polyQ-expanded AR and mediated its proteasomal degradation in SBMA patient fibroblasts and in Drosophila and mouse models of SBMA (Bott et al., 2016), indicating a therapeutic approach to enhance mutant AR degradation.
The predominantly nuclear 11S proteasome activator REGγ was observed to impact polyQ-expanded AR in a cell-specific manner. In a PC12 model of SBMA, REGγ enhanced polyQ-expanded AR aggregation and cell toxicity in manner that was independent of its ability to cap the 20S proteasome and correlated with its inhibition of MDM2-mediated polyubiquitination of polyQ-expanded AR. However, REGγ had a distinct effect in cultured SBMA mouse motor neurons, rescuing cellular toxicity in a proteasome binding-dependent manner (Yersak et al., 2017).
Components of the ubiquitination cascade are altered in muscles of SBMA mouse models. For example, the E2 ubiquitin conjugating enzyme UBE2Q1 was upregulated (Rusmini et al., 2015) and the E2 enzyme Ube2T was suppressed in disease-relevant skeletal muscle of SBMA knock-in mice (Nath et al., 2018). Although several muscle-specific E3 ligases are known to be induced during skeletal muscle atrophy (Bodine et al., 2001; Sartori et al., 2013; Bodine and Baehr, 2014; Milan et al., 2015; Baehr et al., 2022), the alterations in SBMA mouse muscle indicate the activation of a UPS pathway that is distinct from that observed upon general skeletal muscle atrophy (Rusmini et al., 2015; Nath et al., 2020; Forouhan et al., 2022). In addition to these changes, several UPS-related genes were reduced in diseased tissues of SBMA mouse models, including both core and cap proteasomal subunits and E2 ubiquitin conjugating enzymes (Tokui et al., 2009; Nath et al., 2018). In contrast, elevated proteasome activity was observed in quadriceps muscle of the same knock-in mouse model of SBMA (Rocchi et al., 2016), perhaps indicating a compensatory mechanism. The importance of these changes in disease manifestation is suggested by the observation that either surgical castration or skeletal muscle-specific AR ASO treatment rescued the expression of many of these UPS genes, concomitant with a restoration of motor function (Nath et al., 2018).
Despite alterations in the expression of some UPS components, the evidence from the above studies does not indicate the presence of a global, organism-wide impairment of the UPS in SBMA. Rather, the changes in proteasomal expression and activity in SBMA appear in a tissue-specific manner, with greater changes in skeletal muscle than in neuronal tissues. Additionally, the pathways impacting muscular atrophy appear to be distinct from the known skeletal muscle atrophy profile. Regardless, the above studies demonstrate that altering various components of the UPS can result in attenuation of the disease phenotype in SBMA model systems and suggest that the modulating UPS components could be a viable approach to developing disease treatment.
Research gaps and points of consideration in the development of ubiquitin-proteasome system-dependent therapeutics for spinal and bulbar muscular atrophy
Several studies have evaluated the role of E3 ligases, DUBs, or other UPS components in disease pathogenesis. Regulation of protein degradation by the UPS can be manipulated to promote clearance of misfolded and pathogenic proteins and rescue the phenotypes in disease models. Such an approach represents a potentially powerful one for the treatment of SBMA and other polyQ disorders. New selective ubiquitination-based technologies such as proteolysis-targeting chimeras (PROTACs) and selective AR degraders (SARDs) are being developed for treating AR-related disorders by the targeted degradation of AR (Auvin et al., 2019; Beretta and Zaffaroni, 2019; Hwang et al., 2019; Ponnusamy et al., 2019; Alabi and Crews, 2021; Zeng et al., 2021; Hu and Crews, 2022).
Due to the keen interest in this therapeutic area, some important points may be considered during the development of therapeutics that rely on the UPS. First, target selection is of primary importance. Some components of the UPS facilitate AR transcriptional activity or impact its intracellular levels. Due to the vast number of AR target genes and their functions, it may be desirable to preserve AR function during treatment to ensure overall improved quality of life for patients. Similarly, while some UPS components clearly contribute to SBMA pathogenesis, these proteins may also serve important functions independently of the AR and their depletion could have unintended consequences on health. Depletion of E3 ligase RACK1 rescued aggregation and/or cell toxicity in Drosophila models of polyQ disease (Xie et al., 2021), but overexpression of E3 ligases WWP1 and TRAF6 increased mHtt aggregation (Zucchelli et al., 2011; Lin et al., 2016). Indeed, one E3 ligase acts on Htt in two very different ways. K48-polyubiquitination by Ube3a mediated degradation of Htt; however, K63-polyubiquitination by the same E3 ligase promoted Htt aggregation (Bhat et al., 2014).
Additionally, the specific levels of UPS components such as E3 ligases and DUBs may differentially impact mono- and polyubiquitination or have distinct effects on substrate proteins. For example, the E3 ligase MDM2 regulates its substrate protein, p53, in two ways. Low levels of MDM2 promote p53 monoubiquitination and nuclear export, while higher levels promote p53 polyubiquitination and proteasomal degradation in the nucleus (Li et al., 2003). A partial knockdown of the DUB USP7 in a cellular model of SBMA rescued AR-associated aggregation and cell death; yet, a near-complete knockdown failed to do so (Pluciennik et al., 2021), likely due to confounding effects on other proteins that impact AR levels.
The functional redundancy of E3 ligases is another aspect to consider. Different E3 ligases are capable of acting on and ubiquitinating the same substrate protein. Thus, another E3 ligase may compensate for a depleted one. This phenomenon has been observed in plants (Feke et al., 2019), yeast (Breckel and Hochstrasser, 2021), and also specifically in the context of ubiquitinating polyQ-expanded disease proteins (Morishima et al., 2008). This is a key aspect of the role of the E3 ligase in preserving cellular proteostasis. However, it adds another challenge for suppressing the activity of E3 ligases and must inform the research and development of therapeutics targeting proteins related to ubiquitination and the UPS (Garcia-Barcena et al., 2020).
Author contributions
MS, AP, and DM developed the structure of the manuscript. MS wrote the initial drafts of the manuscript. AP and DM edited and contributed to the writing of the final version of the manuscript. All authors approved the submitted version of the manuscript.
Funding
This work was funded by R01NS090335 to DM from the National Institute of Neurological Disorders and Stroke/National Institutes of Health.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aarnisalo, P., Palvimo, J. J., and Jänne, O. A. (1998). CREB-binding protein in androgen receptor-mediated signaling. Proc. Natl. Acad. Sci. U.S.A. 95, 2122–2127.
Abel, A., Walcott, J., Woods, J., Duda, J., and Merry, D. E. (2001). Expression of expanded repeat androgen receptor produces neurologic disease in transgenic mice. Hum. Mol. Genet. 10, 107–116.
Adachi, H., Katsuno, M., Minamiyama, M., Waza, M., Sang, C., Nakagomi, Y., et al. (2005). Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain 128(Pt 3), 659–670. doi: 10.1093/brain/awh381
Adachi, H., Waza, M., Tokui, K., Katsuno, M., Minamiyama, M., Tanaka, F., et al. (2007). CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J. Neurosci. 27, 5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007
Aguilera, M., Oliveros, M., Martínez-Padrón, M., Barbas, J. A., and Ferrús, A. (2000). Ariadne-1: A Vital Drosophila gene is required in development and defines a new conserved family of RING-finger proteins. Genetics 155, 1231–1244. doi: 10.1093/genetics/155.3.1231
Alabi, S. B., and Crews, C. M. (2021). Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J. Biol. Chem. 296:100647. doi: 10.1016/j.jbc.2021.100647
Al-Ramahi, I., Lam, Y. C., Chen, H. K., De Gouyon, B., Zhang, M., Pérez, A. M., et al. (2006). CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J. Biol. Chem. 281, 26714–26724. doi: 10.1074/jbc.M601603200
Amerik, A. Y., and Hochstrasser, M. (2004). Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207. doi: 10.1016/j.bbamcr.2004.10.003
Amm, I., Sommer, T., and Wolf, D. H. (2014). Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 182–196. doi: 10.1016/j.bbamcr.2013.06.031
An, J., Wang, C., Deng, Y., Yu, L., and Huang, H. (2014). Destruction of Full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 6, 657–669. doi: 10.1016/j.celrep.2014.01.013
Aravind, L., and Koonin, E. V. (2000). The U box is a modified RING finger–a common domain in ubiquitination. Curr. Biol. 10, R132–R134. doi: 10.1016/s0960-9822(00)00398-5
Arnold, F. J., and Merry, D. E. (2019). Molecular mechanisms and therapeutics for SBMA/Kennedy’s disease. Neurotherapeutics 16, 928–947. doi: 10.1007/s13311-019-00790-9
Arnold, F. J., Pluciennik, A., and Merry, D. E. (2019). Impaired nuclear export of polyglutamine-expanded androgen receptor in spinal and bulbar muscular atrophy. Sci. Rep. 9:119. doi: 10.1038/s41598-018-36784-4
Atsuta, N., Watanabe, H., Ito, M., Banno, H., Suzuki, K., Katsuno, M., et al. (2006). Natural history of spinal and bulbar muscular atrophy (SBMA): A study of 223 Japanese patients. Brain 129, 1446–1455. doi: 10.1093/brain/awl096
Auvin, S., Öztürk, H., Abaci, Y. T., Mautino, G., Meyer-Losic, F., Jollivet, F., et al. (2019). A molecule inducing androgen receptor degradation and selectively targeting prostate cancer cells. Life Sci. Alliance 2:e201800213. doi: 10.26508/lsa.201800213
Badders, N. M., Korff, A., Miranda, H. C., Vuppala, P. K., Smith, R. B., Winborn, B. J., et al. (2018). Selective modulation of the androgen receptor AF2 domain rescues degeneration in spinal bulbar muscular atrophy. Nat. Med. 24, 427–437. doi: 10.1038/nm.4500
Baehr, L. M., Hughes, D. C., Waddell, D. S., and Bodine, S. C. (2022). SnapShot: Skeletal muscle atrophy. Cell 185, 1618–1618.e1. doi: 10.1016/j.cell.2022.03.028
Bailey, C. K., Andriola, I. F., Kampinga, H. H., and Merry, D. E. (2002). Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 11, 515–523. doi: 10.1093/hmg/11.5.515
Baker, R. T., and Board, P. G. (1991). The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res. 19, 1035–1040. doi: 10.1093/nar/19.5.1035
Baumeister, W., Walz, J., Zühl, F., and Seemüller, E. (1998). The proteasome: Paradigm of a self-compartmentalizing protease. Cell 92, 367–380. doi: 10.1016/s0092-8674(00)80929-0
Belikov, S., Bott, L. C., Fischbeck, K. H., and Wrange, Ö (2015). The polyglutamine-expanded androgen receptor has increased DNA binding and reduced transcriptional activity. Biochem. Biophys. Rep. 3, 134–139. doi: 10.1016/j.bbrep.2015.07.014
Beretta, G. L., and Zaffaroni, N. (2019). Androgen receptor-directed molecular conjugates for targeting prostate cancer. Front. Chem. 7:369. doi: 10.3389/fchem.2019.00369
Berger, T. R., Montie, H. L., Jain, P., Legleiter, J., and Merry, D. E. (2015). Identification of novel polyglutamine-expanded aggregation species in spinal and bulbar muscular atrophy. Brain Res. 1628, 254–264. doi: 10.1016/j.brainres.2015.09.033
Bevan, C. L., Hoare, S., Claessens, F., Heery, D., and Parker, M. (1999). The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 19, 8383–8392. doi: 10.1128/MCB.19.12.8383
Bhat, K. P., Yan, S., Wang, C. E., Li, S., and Li, X. J. (2014). Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. U.S.A. 111, 5706–5711. doi: 10.1073/pnas.1402215111
Blount, J. R., Johnson, S. L., and Todi, S. V. (2020). Unanchored ubiquitin chains, revisited. Front. Cell Dev. Biol. 8:582361. doi: 10.3389/fcell.2020.582361
Bodine, S. C., and Baehr, L. M. (2014). Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 307, E469–E484. doi: 10.1152/ajpendo.00204.2014
Bodine, S. C., Stitt, T. N., Gonzalez, M., Kline, W. O., Stover, G. L., Bauerlein, R., et al. (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019. doi: 10.1038/ncb1101-1014
Bott, L. C., Badders, N. M., Chen, K.-L., Harmison, G. G., Bautista, E., Shih, C. C. Y., et al. (2016). A small-molecule Nrf1 and Nrf2 activator mitigates polyglutamine toxicity in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 25, 1979–1989. doi: 10.1093/hmg/ddw073
Braten, O., Livneh, I., Ziv, T., Admon, A., Kehat, I., Caspi, L. H., et al. (2016). Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 113, E4639–E4647. doi: 10.1073/pnas.1608644113
Breckel, C. A., and Hochstrasser, M. (2021). Ubiquitin ligase redundancy and nuclear-cytoplasmic localization in yeast protein quality control. Biomolecules 11:1821. doi: 10.3390/biom11121821
Brinkmann, A. O., Faber, P. W., van Rooij, H. C., Kuiper, G. G., Ris, C., Klaassen, P., et al. (1989). The human androgen receptor: Domain structure, genomic organization and regulation of expression. J. Steroid Biochem. 34, 307–310. doi: 10.1016/0022-4731(89)90098-8
Brooks, C. L., Li, M., Hu, M., Shi, Y., and Gu, W. (2007). The p53–Mdm2–HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 26, 7262–7266. doi: 10.1038/sj.onc.1210531
Brown, C. J., Goss, S. J., Lubahn, D. B., Joseph, D. R., Wilson, E. M., French, F. S., et al. (1989). Androgen receptor locus on the human X chromosome: Regional localization to Xq11-12 and description of a DNA polymorphism. Am. J. Hum. Genet. 44, 264–269.
Budroni, V., and Versteeg, G. A. (2021). Negative Regulation of the innate immune response through proteasomal degradation and deubiquitination. Viruses 13:584. doi: 10.3390/v13040584
Buetow, L., and Huang, D. T. (2016). Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642. doi: 10.1038/nrm.2016.91
Buneeva, O. A., and Medvedev, A. E. (2018). Ubiquitin-independent degradation of proteins in proteasomes. Biochem. (Moscow) Suppl. Ser. B Biomed. Chem. 12, 203–219. doi: 10.1134/s1990750818030022
Burska, U. L., Harle, V. J., Coffey, K., Darby, S., Ramsey, H., O’Neill, D., et al. (2013). Deubiquitinating enzyme Usp12 Is a Novel Co-activator of the androgen receptor. J. Biol. Chem. 288, 32641–32650. doi: 10.1074/jbc.m113.485912
Buzon, V., Carbo, L. R., Estruch, S. B., Fletterick, R. J., and Estebanez-Perpina, E. (2012). A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol. Cell. Endocrinol. 348, 394–402. doi: 10.1016/j.mce.2011.08.012
Chahin, N., and Sorenson, E. J. (2009). Serum creatine kinase levels in spinobulbar muscular atrophy and amyotrophic lateral sclerosis. Muscle Nerve 40, 126–129. doi: 10.1002/mus.21310
Chang, C.-Y., and McDonnell, D. P. (2005). Androgen receptor–cofactor interactions as targets for new drug discovery. Trends Pharmacol. Sci. 26, 225–228. doi: 10.1016/j.tips.2005.03.002
Chang, L., and Barford, D. (2014). Insights into the anaphase-promoting complex: A molecular machine that regulates mitosis. Curr. Opin. Struct. Biol. 29, 1–9. doi: 10.1016/j.sbi.2014.08.003
Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K., et al. (1989). A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583. doi: 10.1126/science.2538923
Checquolo, S., Palermo, R., Cialfi, S., Ferrara, G., Oliviero, C., Talora, C., et al. (2010). Differential subcellular localization regulates c-Cbl E3 ligase activity upon Notch3 protein in T-cell leukemia. Oncogene 29, 1463–1474. doi: 10.1038/onc.2009.446
Chen, L., and Madura, K. (2014). Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics 197, 193–197. doi: 10.1534/genetics.114.163824
Chen, M., Lingadahalli, S., Narwade, N., Lei, K. M. K., Liu, S., Zhao, Z., et al. (2022). TRIM33 drives prostate tumor growth by stabilizing androgen receptor from Skp2-mediated degradation. EMBO Rep. 23:e53468. doi: 10.15252/embr.202153468
Chen, R.-H., Chen, Y.-H., and Huang, T.-Y. (2019). Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 26:80. doi: 10.1186/s12929-019-0569-y
Chen, S.-T., Okada, M., Nakato, R., Izumi, K., Bando, M., and Shirahige, K. (2015). The deubiquitinating enzyme USP7 Regulates androgen receptor activity by modulating its binding to chromatin. J. Biol. Chem. 290, 21713–21713. doi: 10.1074/JBC.M114.628255
Chen, Y., Zajac, J. D., and Maclean, H. E. (2005). Androgen regulation of satellite cell function. J. Endocrinol. 186, 21–31. doi: 10.1677/joe.1.05976
Chevalier-Larsen, E. S., O’Brien, C. J., Wang, H., Jenkins, S. C., Holder, L., Lieberman, A. P., et al. (2004). Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci. 24, 4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004
Choong, C. S., Kemppainen, J. A., Zhou, Z. X., and Wilson, E. M. (1996). Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol. Endocrinol. 10, 1527–1535. doi: 10.1210/mend.10.12.8961263
Christiaens, V., Bevan, C. L., Callewaert, L., Haelens, A., Verrijdt, G., Rombauts, W., et al. (2002). Characterization of the two coactivator-interacting surfaces of the androgen receptor and their relative role in transcriptional control*. J. Biol. Chem. 277, 49230–49237. doi: 10.1074/jbc.m209322200
Chymkowitch, P., Le May, N., Charneau, P., Compe, E., and Egly, J.-M. (2011). The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 30, 468–479. doi: 10.1038/emboj.2010.337
Ciechanover, A. (1998). The ubiquitin–proteasome pathway: On protein death and cell life. EMBO J. 17, 7151–7160.
Collins, G. A., and Goldberg, A. L. (2017). The logic of the 26S proteasome. Cell 169, 792–806. doi: 10.1016/j.cell.2017.04.023
Cortes, C. J., Ling, S. C., Guo, L. T., Hung, G., Tsunemi, T., Ly, L., et al. (2014). Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron 82, 295–307. doi: 10.1016/j.neuron.2014.03.001
Coux, O., Tanaka, K., and Goldberg, A. L. (1996). Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847.
Cutress, M. L., Whitaker, H. C., Mills, I. G., Stewart, M., and Neal, D. E. (2008). Structural basis for the nuclear import of the human androgen receptor. J. Cell Sci. 121, 957–968. doi: 10.1242/jcs.022103
Dang, F. W., Chen, L., and Madura, K. (2016). Catalytically active proteasomes function predominantly in the cytosol. J. Biol. Chem. 291, 18765–18777. doi: 10.1074/jbc.M115.712406
Davey, R. A., Clarke, M. V., Russell, P. K., Rana, K., Seto, J., Roeszler, K. N., et al. (2017). Androgen action via the androgen receptor in neurons within the brain positively regulates muscle mass in male mice. Endocrinology 158, 3684–3695. doi: 10.1210/en.2017-00470
De Bie, P., and Ciechanover, A. (2011). Ubiquitination of E3 ligases: Self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18, 1393–1402. doi: 10.1038/cdd.2011.16
Dejager, S., Bry-Gauillard, H., Bruckert, E., Eymard, B., Salachas, F., LeGuern, E., et al. (2002). A comprehensive endocrine description of Kennedy’s Disease revealing androgen insensitivity linked to CAG repeat length. J. Clin. Endocrinol. Metab. 87, 3893–3901. doi: 10.1210/jcem.87.8.8780
Demartino, G. N., and Gillette, T. G. (2007). Proteasomes: Machines for all reasons. Cell 129, 659–662. doi: 10.1016/j.cell.2007.05.007
Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., et al. (2000). Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361. doi: 10.1016/s0092-8674(00)00126-4
DePriest, A. D., Fiandalo, M. V., Schlanger, S., Heemers, F., Mohler, J. L., Liu, S., et al. (2016). Regulators of androgen action resource: A one-stop shop for the comprehensive study of androgen receptor action. Database (Oxford) 2016:bav125. doi: 10.1093/database/bav125
Dirac, A. M. G., and Bernards, R. (2010). The deubiquitinating enzyme USP26 Is a Regulator of androgen receptor signaling. Mol. Cancer Res. 8, 844–854. doi: 10.1158/1541-7786.mcr-09-0424
Doi, H., Adachi, H., Katsuno, M., Minamiyama, M., Matsumoto, S., Kondo, N., et al. (2013). p62/SQSTM1 differentially removes the toxic mutant androgen receptor via autophagy and inclusion formation in a spinal and bulbar muscular atrophy mouse model. J. Neurosci. 33, 7710–7727. doi: 10.1523/jneurosci.3021-12.2013
Driscoll, J., and Goldberg, A. L. (1989). Skeletal muscle proteasome can degrade proteins in an ATP-dependent process that does not require ubiquitin. Proc. Natl. Acad. Sci. U.S.A. 86, 787–791. doi: 10.1073/pnas.86.3.787
Durcan, T. M., Tang, M. Y., Pérusse, J. R., Dashti, E. A., Aguileta, M. A., McLelland, G. L., et al. (2014). USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 33, 2473–2491. doi: 10.15252/embj.201489729
Eftekharzadeh, B., Banduseela, V. C., Chiesa, G., Martínez-Cristóbal, P., Rauch, J. N., Nath, S. R., et al. (2019). Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. Nat. Commun. 10, 1–14. doi: 10.1038/s41467-019-11594-y
El Kharraz, S., Dubois, V., van Royen, M. E., Houtsmuller, A. B., Pavlova, E., Atanassova, N., et al. (2021). The androgen receptor depends on ligand-binding domain dimerization for transcriptional activation. EMBO Rep. 22:e52764. doi: 10.15252/embr.202152764
Elia, A. E., Boardman, A. P., Wang, D. C., Huttlin, E. L., Everley, R. A., Dephoure, N., et al. (2015). Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell 59, 867–881. doi: 10.1016/j.molcel.2015.05.006
Erpapazoglou, Z., Walker, O., and Haguenauer-Tsapis, R. (2014). Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 3, 1027–1088. doi: 10.3390/cells3041027
Estébanez-Perpiñá, E., Arnold, L. A., Nguyen, P., Rodrigues, E. D., Mar, E., Bateman, R., et al. (2007). A surface on the androgen receptor that allosterically regulates coactivator binding. Proc. Natl. Acad. Sci. U.S.A. 104, 16074–16079.
Fargo, K. N., Alexander, T. D., Tanzer, L., Poletti, A., and Jones, K. J. (2008a). Androgen regulates neuritin mRNA levels in an in vivo model of steroid-enhanced peripheral nerve regeneration. J. Neurotrauma 25, 561–566. doi: 10.1089/neu.2007.0466
Fargo, K. N., Galbiati, M., Foecking, E. M., Poletti, A., and Jones, K. J. (2008b). Androgen regulation of axon growth and neurite extension in motoneurons. Horm. Behav. 53, 716–728. doi: 10.1016/j.yhbeh.2008.01.014
Faus, H., Meyer, H. A., Huber, M., Bahr, I., and Haendler, B. (2005). The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol. Cell Endocrinol. 245, 138–146. doi: 10.1016/j.mce.2005.11.011
Feke, A., Liu, W., Hong, J., Li, M.-W., Lee, C.-M., Zhou, E. K., et al. (2019). Decoys provide a scalable platform for the identification of plant E3 ubiquitin ligases that regulate circadian function. eLife 8:e44558. doi: 10.7554/elife.44558
Finley, D., Bartel, B., and Varshavsky, A. (1989). The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401. doi: 10.1038/338394a0
Fischbeck, K. H. (1997). Kennedy disease. J. Inherit. Metab. Dis. 20, 152–158. doi: 10.1023/a:1005344403603
Flick, K., Ouni, I., Wohlschlegel, J. A., Capati, C., McDonald, W. H., Yates, J. R., et al. (2004). Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 6, 634–641. doi: 10.1038/ncb1143
Forouhan, M., Lim, W. F., Zanetti-Domingues, L. C., Tynan, C. J., Roberts, T. C., Malik, B., et al. (2022). AR cooperates with SMAD4 to maintain skeletal muscle homeostasis. Acta Neuropathol. 143, 713–731. doi: 10.1007/s00401-022-02428-1
Garcia-Barcena, C., Osinalde, N., Ramirez, J., and Mayor, U. (2020). How to inactivate human ubiquitin E3 ligases by mutation. Front. Cell Dev. Biol. 8:39. doi: 10.3389/fcell.2020.00039
Gaughan, L., Logan, I. R., Neal, D. E., and Robson, C. N. (2005). Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 33, 13–26. doi: 10.1093/nar/gki141
Geng, C., Rajapakshe, K., Shah, S. S., Shou, J., Eedunuri, V. K., Foley, C., et al. (2014). Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 74, 5631–5643. doi: 10.1158/0008-5472.can-14-0476
Georget, V., Lobaccaro, J. M., Terouanne, B., Mangeat, P., Nicolas, J. C., and Sultan, C. (1997). Trafficking of the androgen receptor in living cells with fused green fluorescent protein-androgen receptor. Mol. Cell. Endocrinol. 129, 17–26. doi: 10.1016/s0303-7207(97)04034-3
Gong, Y., Wang, D., Dar, J. A., Singh, P., Graham, L., Liu, W., et al. (2012). Nuclear export signal of androgen receptor (NESAR) regulation of androgen receptor level in human prostate cell lines via ubiquitination and proteasome-dependent degradation. Endocrinology 153, 5716–5725. doi: 10.1210/en.2012-1841
Gregori, L., Poosch, M. S., Cousins, G., and Chau, V. (1990). A uniform isopeptide-linked multiubiquitin chain is sufficient to target substrate for degradation in ubiquitin-mediated proteolysis. J. Biol. Chem. 265, 8354–8357. doi: 10.1016/s0021-9258(19)38890-8
Grou, C. P., Pinto, M. P., Mendes, A. V., Domingues, P., and Azevedo, J. E. (2015). The de novo synthesis of ubiquitin: Identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 5:12836. doi: 10.1038/srep12836
Guber, R. D., Kokkinis, A. D., Schindler, A. B., Bendixen, R. M., Heatwole, C. R., Fischbeck, K. H., et al. (2018). Patient-identified impact of symptoms in spinal and bulbar muscular atrophy. Muscle Nerve 57, 40–44. doi: 10.1002/mus.25957
Guharoy, M., Bhowmick, P., and Tompa, P. (2016). Design principles involving protein disorder facilitate specific substrate selection and degradation by the ubiquitin-proteasome system. J. Biol. Chem. 291, 6723–6731. doi: 10.1074/jbc.r115.692665
Guo, X. (2022). Localized proteasomal degradation: From the nucleus to cell periphery. Biomolecules 12:229. doi: 10.3390/biom12020229
Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K.-I. (2001). U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111–33120. doi: 10.1074/jbc.m102755200
He, B., Bai, S., Hnat, A. T., Kalman, R. I., Minges, J. T., Patterson, C., et al. (2004a). An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J. Biol. Chem. 279, 30643–30653. doi: 10.1074/jbc.m403117200
He, B., Gampe, R. T. Jr., Kole, A. J., Hnat, A. T., Stanley, T. B., An, G., et al. (2004b). Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell 16, 425–438. doi: 10.1016/j.molcel.2004.09.036
Heine, E. M., Berger, T. R., Pluciennik, A., Orr, C. R., Zboray, L., and Merry, D. E. (2015). Proteasome-mediated proteolysis of the polyglutamine-expanded androgen receptor is a late event in spinal and bulbar muscular atrophy (SBMA) pathogenesis. J. Biol. Chem. 290, 12572–12584. doi: 10.1074/jbc.M114.617894
Herrmann, J., Lerman, L. O., and Lerman, A. (2007). Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 100, 1276–1291. doi: 10.1161/01.res.0000264500.11888.f0
Hershko, A. (1988). Ubiquitin-mediated protein degradation. J. Biol. Chem. 263, 15237–15240. doi: 10.1016/s0021-9258(19)37575-1
Hiort, O. (2013). The differential role of androgens in early human sex development. BMC Med. 11:152. doi: 10.1186/1741-7015-11-152
Hu, M., Gu, L., Li, M., Jeffrey, P. D., Gu, W., and Shi, Y. (2006). Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: Implications for the regulation of the p53–MDM2 pathway. PLoS Biol. 4:e27. doi: 10.1371/journal.pbio.0040027
Hu, Z., and Crews, C. M. (2022). Recent developments in PROTAC-Mediated protein degradation: From bench to clinic. Chembiochem 23:e202100270. doi: 10.1002/cbic.202100270
Huang, H., Jeon, M.-S., Liao, L., Yang, C., Elly, C., Yates, J. R., et al. (2010). K33-Linked polyubiquitination of T cell Receptor-ζ regulates proteolysis-independent T cell signaling. Immunity 33, 60–70. doi: 10.1016/j.immuni.2010.07.002
Huibregtse, J. M., Scheffner, M., Beaudenon, S., and Howley, P. M. (1995). A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563–2567. doi: 10.1073/pnas.92.7.2563
Hwang, D. J., He, Y., Ponnusamy, S., Mohler, M. L., Thiyagarajan, T., McEwan, I. J., et al. (2019). New generation of selective androgen receptor degraders: Our initial design. synthesis, and biological evaluation of new compounds with enzalutamide-resistant prostate cancer activity. J. Med. Chem. 62, 491–511. doi: 10.1021/acs.jmedchem.8b00973
Hwang, J.-T., Lee, A., and Kho, C. (2022). Ubiquitin and ubiquitin-like proteins in cancer, neurodegenerative disorders, and heart diseases. Int. J. Mol. Sci. 23:5053. doi: 10.3390/ijms23095053
Imai, Y., Soda, M., Hatakeyama, S., Akagi, T., Hashikawa, T., Nakayama, K.-I., et al. (2002). CHIP is associated with parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol. Cell 10, 55–67. doi: 10.1016/s1097-2765(02)00583-x
Irvine, R. A. (2000). Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum. Mol. Genet. 9, 267–274. doi: 10.1093/hmg/9.2.267
Jana, N. R., Dikshit, P., Goswami, A., Kotliarova, S., Murata, S., Tanaka, K., et al. (2005). Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 280, 11635–11640. doi: 10.1074/jbc.m412042200
Jaworski, T. (2006). Degradation and beyond: Control of androgen receptor activity by the proteasome system. Cell. Mol. Biol. Lett. 11, 109–131. doi: 10.2478/s11658-006-0011-9
Jia, L., Berman, B. P., Jariwala, U., Yan, X., Cogan, J. P., Walters, A., et al. (2008). Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS One 3:e3645. doi: 10.1371/journal.pone.0003645
Jiang, M., Ma, Y., Chen, C., Fu, X., Yang, S., Li, X., et al. (2009). Androgen-Responsive gene database: Integrated knowledge on androgen-responsive genes. Mol. Endocrinol. 23, 1927–1933. doi: 10.1210/me.2009-0103
Jin, H.-J., Kim, J., and Yu, J. (2013). Androgen receptor genomic regulation. Transl. Androl. Urol. 2, 158–177. doi: 10.3978/j.issn.2223-4683.2013.09.01
Jing, Y., Nguyen, M. M., Wang, D., Pascal, L. E., Guo, W., Xu, Y., et al. (2018). DHX15 promotes prostate cancer progression by stimulating Siah2-mediated ubiquitination of androgen receptor. Oncogene 37, 638–650. doi: 10.1038/onc.2017.371
Jokela, M., Huovinen, S., Raheem, O., Lindfors, M., Palmio, J., Penttilä, S., et al. (2016). Distinct muscle biopsy findings in genetically defined adult-onset motor neuron disorders. PLoS One 11:e0151376. doi: 10.1371/journal.pone.0151376
Kadi, F. (2008). Cellular and molecular mechanisms responsible for the action of testosterone on human skeletal muscle. A basis for illegal performance enhancement. Br. J. Pharmacol. 154, 522–528. doi: 10.1038/bjp.2008.118
Katsuno, M., Adachi, H., Doyu, M., Minamiyama, M., Sang, C., Kobayashi, Y., et al. (2003). Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy. Nat. Med. 9, 768–773. doi: 10.1038/nm878
Katsuno, M., Adachi, H., Kume, A., Li, M., Nakagomi, Y., Niwa, H., et al. (2002). Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron 35, 843–854. doi: 10.1016/s0896-6273(02)00834-6
Kazemi-Esfarjani, P., Trifiro, M. A., and Pinsky, L. (1995). Evidence for a repressive function of the long polyglutamine tract in the human androgen receptor: Possible pathogenetic relevance for the (CAG)n-expanded neuronopathies. Hum. Mol. Genet. 4, 523–527. doi: 10.1093/hmg/4.4.523
Kemppainen, J. A., Lane, M. V., Sar, M., and Wilson, E. M. (1992). Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. J. Biol. Chem. 267, 968–974. doi: 10.1016/s0021-9258(18)48380-9
Khan, O. Y., Fu, G., Ismail, A., Srinivasan, S., Cao, X., Tu, Y., et al. (2006). Multifunction steroid receptor coactivator, E6-associated protein, is involved in development of the prostate gland. Mol. Endocrinol. 20, 544–559. doi: 10.1210/me.2005-0110
Kim, R. Q., and Sixma, T. K. (2017). Regulation of USP7: A high incidence of E3 complexes. J. Mol. Biol. 429, 3395–3408. doi: 10.1016/j.jmb.2017.05.028
Kimura, Y., and Tanaka, K. (2010). Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147, 793–798. doi: 10.1093/jb/mvq044
Kirisako, T., Kamei, K., Murata, S., Kato, M., Fukumoto, H., Kanie, M., et al. (2006). A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887. doi: 10.1038/sj.emboj.7601360
Kito, Y., Matsumoto, M., Hatano, A., Takami, T., Oshikawa, K., Matsumoto, A., et al. (2020). Cell cycle-dependent localization of the proteasome to chromatin. Sci. Rep. 10:5801. doi: 10.1038/s41598-020-62697-2
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999). A novel ubiquitination factor. E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644.
Koizumi, S., Irie, T., Hirayama, S., Sakurai, Y., Yashiroda, H., Naguro, I., et al. (2016). The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife 5:e18357. doi: 10.7554/elife.18357
Komander, D., and Rape, M. (2012). The ubiquitin code. Ann. Rev. Biochem. 81, 203–229. doi: 10.1146/annurev-biochem-060310-170328
Kurz, E. M., Sengelaub, D. R., and Arnold, A. P. (1986). Androgens regulate the dendritic length of mammalian motoneruons in adulthood. Science 232, 395–398.
La Spada, A. R., Wilson, E. M., Lubahn, D. B., Harding, A. E., and Fischbeck, K. H. (1991). Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79. doi: 10.1038/352077a0
Lahav-Baratz, S., Kravtsova-Ivantsiv, Y., Golan, S., and Ciechanover, A. (2017). The testis-specific USP26 is a deubiquitinating enzyme of the ubiquitin ligase Mdm2. Biochem. Biophys. Res. Commun. 482, 106–111. doi: 10.1016/j.bbrc.2016.10.135
Langley, E., Zhou, Z. X., and Wilson, E. M. (1995). Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270, 29983–29990. doi: 10.1074/jbc.270.50.29983
Lauwers, E., Jacob, C., and André, B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502. doi: 10.1083/jcb.200810114
Lavery, D. N., and McEwan, I. J. (2008). Structural characterization of the native NH2-Terminal transactivation domain of the human androgen receptor: A collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry 47, 3360–3369. doi: 10.1021/bi702221e
Leblanc, E. S., Wang, P. Y., Lee, C. G., Barrett-Connor, E., Cauley, J. A., Hoffman, A. R., et al. (2011). Higher testosterone levels are associated with less loss of lean body mass in older men. J. Clin. Endocrinol. Metab. 96, 3855–3863. doi: 10.1210/jc.2011-0312
Lehrbach, N. J., and Ruvkun, G. (2016). Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLife 5:e17721. doi: 10.7554/elife.17721
Lehrbach, N. J., Breen, P. C., and Ruvkun, G. (2019). Protein sequence editing of SKN-1A/Nrf1 by peptide:N-Glycanase controls proteasome gene expression. Cell 177, 737:750.e15. doi: 10.1016/j.cell.2019.03.035
Li, B., Lu, W., Yang, Q., Yu, X., Matusik, R. J., and Chen, Z. (2014). Skp2 regulates androgen receptor through ubiquitin-mediated degradation independent of Akt/mTOR pathways in prostate cancer. Prostate 74, 421–432. doi: 10.1002/pros.22763
Li, C., Ao, J., Fu, J., Lee, D. F., Xu, J., Lonard, D., et al. (2011). Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 30, 4350–4364. doi: 10.1038/onc.2011.151
Li, M., Brooks, C. L., Wu-Baer, F., Chen, D., Baer, R., and Gu, W. (2003). Mono–Versus polyubiquitination: Differential control of p53 fate by Mdm2. Science 302, 1972–1975. doi: 10.1126/science.1091362
Li, M., Chevalier-Larsen, E. S., Merry, D. E., and Diamond, M. I. (2007). soluble androgen receptor oligomers underlie pathology in a mouse model of spinobulbar muscular atrophy. J. Biol. Chem. 282, 3157–3164. doi: 10.1074/jbc.m609972200
Li, M., Miwa, S., Kobayashi, Y., Merry, D. E., Yamamoto, M., Tanaka, F., et al. (1998a). Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann. Neurol. 44, 249–254. doi: 10.1002/ana.410440216
Li, M., Nakagomi, Y., Kobayashi, Y., Merry, D. E., Tanaka, F., Doyu, M., et al. (1998b). Nonneural nuclear inclusions of androgen receptor protein in spinal and bulbar muscular atrophy. Am. J. Pathol. 153, 695–701. doi: 10.1016/s0002-9440(10)65612-x
Li, W., Bengtson, M. H., Ulbrich, A., Matsuda, A., Reddy, V. A., Orth, A., et al. (2008). Genome-Wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial e3 that regulates the organelle’s dynamics and signaling. PLoS One 3:e1487. doi: 10.1371/journal.pone.0001487
Li, X., Yuan, J., Song, C., Lei, Y., Xu, J., Zhang, G., et al. (2021). Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 2021, 1–18. doi: 10.1038/s41418-021-00754-7
Liao, Y., Liu, N., Hua, X., Cai, J., Xia, X., Wang, X., et al. (2017). Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis. 8:e2585. doi: 10.1038/cddis.2016.477
Liao, Y., Xia, X., Liu, N., Cai, J., Guo, Z., Li, Y., et al. (2018). Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene 37, 1896–1910. doi: 10.1038/s41388-017-0069-z
Lieberman, A. P. (2018). “Spinal and bulbar muscular atrophy,” in. Handb. Clin. Neurol. 148, 625–632.
Lieberman, A. P., Harmison, G., Strand, A. D., Olson, J. M., and Fischbeck, K. H. (2002). Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 11, 1967–1976.
Lieberman, A. P., Shakkottai, V. G., and Albin, R. L. (2019). Polyglutamine repeats in neurodegenerative diseases. Annu. Rev. Pathol. 14, 1–27. doi: 10.1146/annurev-pathmechdis-012418-012857
Lieberman, A. P., Yu, Z., Murray, S., Peralta, R., Low, A., Guo, S., et al. (2014). Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 7, 774–774. doi: 10.1016/j.Celrep.2014.02.008
Lim, J. J., Han, C. Y., Lee, D. R., and Tsang, B. K. (2017). Ring finger protein 6 mediates androgen-induced granulosa cell proliferation and follicle growth via modulation of androgen receptor signaling. Endocrinology 158, 993–1004. doi: 10.1210/en.2016-1866
Lin, H.-K., Wang, L., Hu, Y.-C., Altuwaijri, S., and Chang, C. (2002). Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21, 4037–4048. doi: 10.1093/emboj/cdf406
Lin, H.-K., Yeh, S., Kang, H.-Y., and Chang, C. (2001). Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. 98, 7200–7205. doi: 10.1073/pnas.121173298
Lin, L., Jin, Z., Tan, H., Xu, Q., Peng, T., and Li, H. (2016). Atypical ubiquitination by E3 ligase WWP1 inhibits the proteasome-mediated degradation of mutant huntingtin. Brain Res. 1643, 103–112. doi: 10.1016/j.Brainres.2016.03.027
Little, C. M., Coons, K. D., and Sengelaub, D. R. (2009). Neuroprotective effects of testosterone on the morphology and function of somatic motor neurons following the death of neighboring motoneurons. J. Comp. Neurol. 512, 359–372.
Liu, P., Gan, W., Su, S., Hauenstein, A. V., Fu, T. M., Brasher, B., et al. (2018). K63-linked polyubiquitin chains bind to DNA to facilitate DNA damage repair. Sci. Signal. 11:eaar8133. doi: 10.1126/scisignal.Aar8133
Liu, T., Li, Y., Gu, H., Zhu, G., Li, J., Cao, L., et al. (2013). p21-activated kinase 6 (PAK6) inhibits prostate cancer growth via phosphorylation of androgen receptor and tumorigenic E3 ligase murine double minute-2 (Mdm2). J. Biol. Chem. 288, 3359–3369. doi: 10.1074/jbc.m112.384289
Livneh, I., Kravtsova-Ivantsiv, Y., Braten, O., Kwon, Y. T., and Ciechanover, A. (2017). Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. BioEssays 39:1700027. doi: 10.1002/bies.201700027
Lombardi, V., Querin, G., Ziff, O. J., Zampedri, L., Martinelli, I., Heller, C., et al. (2019). Muscle and not neuronal biomarkers correlate with severity in spinal and bulbar muscular atrophy. Neurology 92, e1205–e1211. doi: 10.1212/WNL.0000000000007097
Lopata, A., Kniss, A., Löhr, F., Rogov, V. V., and Dötsch, V. (2020). Ubiquitination in the ERAD process. Int. J. Mol. Sci. 21:5369. doi: 10.3390/ijms21155369
Lund, P. K., Moats-Staats, B. M., Simmons, J. G., Hoyt, E., D’Ercole, A. J., Martin, F., et al. (1985). Nucleotide sequence analysis of a cDNA encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J. Biol. Chem. 260, 7609–7613. doi: 10.1016/s0021-9258(17)39652-7
Lv, S., Song, Q., Chen, G., Cheng, E., Chen, W., Cole, R., et al. (2021). Regulation and targeting of androgen receptor nuclear localization in castration-resistant prostate cancer. J. Clin. Invest. 131:e141335. doi: 10.1172/JCI141335
MacLean, H. E., Chiu, W. S. M., Notini, A. J., Axell, A.-M., Davey, R. A., McManus, J. F., et al. (2008). Impaired skeletal muscle development and function in male, but not female, genomic androgen receptor knockout mice. FASEB J. 22, 2676–2689.
Maki, C. G., Huibregtse, J. M., and Howley, P. M. (1996). In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 56, 2649–2654.
Marine, J. C., Francoz, S., Maetens, M., Wahl, G., Toledo, F., and Lozano, G. (2006). Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 13, 927–934. doi: 10.1038/sj.cdd.4401912
Marmor, M. D., and Yarden, Y. (2004). Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23, 2057–2070. doi: 10.1038/sj.onc.1207390
Marron, T. U., Guerini, V., Rusmini, P., Sau, D., Brevini, T. A., Martini, L., et al. (2005). Androgen-induced neurite outgrowth is mediated by neuritin in motor neurones. J. Neurochem. 92, 10–20. doi: 10.1111/j.1471-4159.2004.02836.x
Massie, C. E., Lynch, A., Ramos-Montoya, A., Boren, J., Stark, R., Fazli, L., et al. (2011). The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 30, 2719–2733. doi: 10.1038/emboj.2011.158
McClellan, A. J., Laugesen, S. H., and Ellgaard, L. (2019). Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol. 9:190147. doi: 10.1098/rsob.190147
McClurg, U. L., Cork, D. M. W., Darby, S., Ryan-Munden, C. A., Nakjang, S., Mendes Cortes, L., et al. (2017). Identification of a novel K311 ubiquitination site critical for androgen receptor transcriptional activity. Nucleic Acids Res. 45, 1793–1804. doi: 10.1093/nar/gkw1162
McClurg, U. L., Summerscales, E. E., Harle, V. J., Gaughan, L., and Robson, C. N. (2014). Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget 5, 7081–7092. doi: 10.18632/oncotarget.2162
McDowell, G. S., and Philpott, A. (2016). New insights into the role of ubiquitylation of proteins. Int. Rev. Cell Mol. Biol. Elsevier 325, 35–88.
Medvar, B., Raghuram, V., Pisitkun, T., Sarkar, A., and Knepper, M. A. (2016). Comprehensive database of human E3 ubiquitin ligases: Application to aquaporin-2 regulation. Physiol. Genomics 48, 502–512. doi: 10.1152/physiolgenomics.00031.2016
Merry, D. E., Kobayashi, Y., Bailey, C. K., Taye, A. A., and Fischbeck, K. H. (1998). Cleavage, aggregation, and toxicity of the expanded androgen receptor in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 7, 693–701.
Metzger, M. B., Pruneda, J. N., Klevit, R. E., and Weissman, A. M. (2014). RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60. doi: 10.1016/j.bbamcr.2013.05.026
Mevissen, T. E. T., and Komander, D. (2017). Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192. doi: 10.1146/annurev-biochem-061516-044916
Mhaouty-Kodja, S. (2018). Role of the androgen receptor in the central nervous system. Mol. Cell. Endocrinol. 465, 103–112. doi: 10.1016/j.mce.2017.08.001
Mhatre, A. N., Trifiro, M. A., Kaufman, M., Kazemi-Esfarjani, P., Figlewicz, D., Rouleau, G., et al. (1993). Reduced transcriptional regulatory competence of the androgen receptor in X–linked spinal and bulbar muscular atrophy. Nat. Genet. 5, 184–188. doi: 10.1038/ng1093-184
Michelle, C., Vourc’H, P., Mignon, L., and Andres, C. R. (2009). What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 68, 616–628. doi: 10.1007/s00239-009-9225-6
Milan, G., Romanello, V., Pescatore, F., Armani, A., Paik, J.-H., Frasson, L., et al. (2015). Regulation of autophagy and the ubiquitin–proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 6:6670. doi: 10.1038/ncomms7670
Miller, V. M. (2005). CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 25, 9152–9161. doi: 10.1523/jneurosci.3001-05.2005
Montie, H. L., and Merry, D. E. (2009). Autophagy and access: Understanding the role of androgen receptor subcellular localization in SBMA. Autophagy 5, 1194–1197. doi: 10.4161/auto.5.8.9726
Montie, H. L., Cho, M. S., Holder, L., Liu, Y., Tsvetkov, A. S., Finkbeiner, S., et al. (2009). Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 18, 1937–1950. doi: 10.1093/hmg/ddp115
Montie, H. L., Pestell, R. G., and Merry, D. E. (2011). SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J. Neurosci. 31, 17425–17436. doi: 10.1523/JNEUROSCI.3958-11.2011
Morishima, Y., Wang, A. M., Yu, C., Pratt, W. B., Osawa, Y., and Lieberman, A. P. (2008). CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum. Mol. Genet. 17, 3942–3952. doi: 10.1093/hmg/ddn296
Morreale, F. E., and Walden, H. (2016). Types of ubiquitin ligases. Cell 165, 248.e–248.e. doi: 10.1016/j.cell.2016.03.003
Müller, K. I, Nilssen, Ø, Nebuchenykh, M., Løseth, S., Jonsrud, C., Hoem, G., et al. (2021). Kennedy disease in two sisters with biallelic CAG expansions of the androgen receptor gene. Neuromuscul. Disord. 32, 75–79. doi: 10.1016/j.nmd.2021.11.007
Murata, S., Minami, Y., Minami, M., Chiba, T., and Tanaka, K. (2001). CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2, 1133–1138. doi: 10.1093/embo-reports/kve246
Nakajima, H., Kimura, F., Nakagawa, T., Furutama, D., and Ohsawa, N. (1996). Transcriptional activation by the androgen receptor in X-linked spinal and bulbar muscular atrophy. J. Neurol. Sci. 142, 12–16.
Nath, S. R., Lieberman, M. L., Yu, Z., Marchioretti, C., Jones, S. T., Danby, E. C. E., et al. (2020). MEF2 impairment underlies skeletal muscle atrophy in polyglutamine disease. Acta Neuropathol. 140, 63–80. doi: 10.1007/s00401-020-02156-4
Nath, S. R., Yu, Z., Gipson, T. A., Marsh, G. B., Yoshidome, E., Robins, D. M., et al. (2018). Androgen receptor polyglutamine expansion drives age-dependent quality control defects and muscle dysfunction. J. Clin. Invest. 128, 3630–3641. doi: 10.1172/JCI99042
Nedelsky, N. B., Pennuto, M., Smith, R. B., Palazzolo, I., Moore, J., Nie, Z., et al. (2010). Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron 67, 936–952. doi: 10.1016/j.neuron.2010.08.034
Ni, L., Llewellyn, R., Kesler, C. T., Kelley, J. B., Spencer, A., Snow, C. J., et al. (2013). Androgen induces a switch from cytoplasmic retention to nuclear import of the androgen receptor. Mol. Cell. Biol. 33, 4766–4778. doi: 10.1128/mcb.00647-13
Northrop, A., Vangala, J. R., Feygin, A., and Radhakrishnan, S. K. (2020). Disabling the protease DDI2 attenuates the transcriptional activity of NRF1 and potentiates proteasome inhibitor cytotoxicity. Int. J. Mol. Sci. 21:327. doi: 10.3390/ijms21010327
Ohtake, F., Baba, A., Takada, I., Okada, M., Iwasaki, K., Miki, H., et al. (2007). Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446, 562–566. doi: 10.1038/nature05683
Ohtake, F., Fujii-Kuriyama, Y., and Kato, S. (2009). AhR acts as an E3 ubiquitin ligase to modulate steroid receptor functions. Biochem. Pharmacol. 77, 474–484. doi: 10.1016/j.bcp.2008.08.034
Ohtake, F., Saeki, Y., Sakamoto, K., Ohtake, K., Nishikawa, H., Tsuchiya, H., et al. (2015). Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 16, 192–201. doi: 10.15252/embr.201439152
Olzmann, J. A., and Chin, L.-S. (2008). Parkin-mediated K63-linked polyubiquitination: A signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy 4, 85–87. doi: 10.4161/AUTO.5172
Olzmann, J. A., Li, L., Chudaev, M. V., Chen, J., Perez, F. A., Palmiter, R. D., et al. (2007). Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178, 1025–1038. doi: 10.1083/jcb.200611128
Orr, C. R., Montie, H. L., Liu, Y., Bolzoni, E., Jenkins, S. C., Wilson, E. M., et al. (2010). An interdomain interaction of the androgen receptor is required for its aggregation and toxicity in spinal and bulbar muscular atrophy. J. Biol. Chem. 285, 35567–35577. doi: 10.1074/jbc.M110.146845
Palazzolo, I., Burnett, B. G., Young, J. E., Brenne, P. L., La Spada, A. R., Fischbeck, K. H., et al. (2007). Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum. Mol. Genet. 16, 1593–1603. doi: 10.1093/hmg/ddm109
Palazzolo, I., Stack, C., Kong, L., Musaro, A., Adachi, H., Katsuno, M., et al. (2009). Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron 63, 316–328. doi: 10.1016/j.neuron.2009.07.019
Park, E.-S., Dezhbord, M., Lee, A. R., and Kim, K.-H. (2022). The roles of ubiquitination in pathogenesis of influenza virus infection. Int. J. Mol. Sci. 23:4593. doi: 10.3390/ijms23094593
Park, S. H., Kukushkin, Y., Gupta, R., Chen, T., Konagai, A., Hipp, M. S., et al. (2013). PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145. doi: 10.1016/j.cell.2013.06.003
Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., et al. (2003). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926. doi: 10.1038/nbt849
Pérez, J., and Kelley, D. B. (1996). Trophic effects of androgen: Receptor expression and the survival of laryngeal motor neurons after axotomy. J. Neurosci. 16, 6625–6633. doi: 10.1523/jneurosci.16-21-06625.1996
Pinsky, L., Trifiro, M., Kaufman, M., Beitel, L. K., Mhatre, A., Kazemi-Esfarjani, P., et al. (1992). Androgen resistance due to mutation of the androgen receptor. Clin. Invest. Med. 15, 456–472.
Pluciennik, A., Liu, Y., Molotsky, E., Marsh, G. B., Ranxhi, B., Arnold, F. J., et al. (2021). Deubiquitinase USP7 contributes to the pathogenicity of spinal and bulbar muscular atrophy. J. Clin. Invest. 131:e134565. doi: 10.1172/JCI134565
Polanco, M. J., Parodi, S., Piol, D., Stack, C., Chivet, M., Constestabile, A., et al. (2016). Adenylyl cyclase activating polypeptide reduces phosphorylation and toxicity of the polyglutamine-expanded androgen receptor in spinobulbar muscular atrophy. Sci. Transl. Med. 8:370ra181. doi: 10.1126/scitranslmed.aaf9526
Ponnusamy, S., He, Y., Hwang, D. J., Thiyagarajan, T., Houtman, R., Bocharova, V., et al. (2019). Orally bioavailable androgen receptor degrader, potential next-generation therapeutic for enzalutamide-resistant prostate cancer. Clin. Cancer Res. 25, 6764–6780. doi: 10.1158/1078-0432.Ccr-19-1458
Poukka, H., Aarnisalo, P., Karvonen, U., Palvimo, J. J., and Janne, O. A. (1999). Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 274, 19441–19446. doi: 10.1074/jbc.274.27.19441
Pradat, P. F., Bernard, E., Corcia, P., Couratier, P., Jublanc, C., Querin, G., et al. (2020). The french national protocol for Kennedy’s disease (SBMA): Consensus diagnostic and management recommendations. Orphanet J. Rare Dis. 15:90. doi: 10.1186/s13023-020-01366-z
Prasad, R., Kawaguchi, S., and Ng, D. T. (2010). A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell 21, 2117–2127. doi: 10.1091/mbc.E10-02-0111
Qi, J., Tripathi, M., Mishra, R., Sahgal, N., Fazil, L., Ettinger, S., et al. (2013). The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell 23, 332–346. doi: 10.1016/j.ccr.2013.02.016
Qu, J., Zou, T., and Lin, Z. (2021). The roles of the ubiquitin–proteasome system in the endoplasmic reticulum stress pathway. Int. J. Mol. Sci. 22:1526. doi: 10.3390/ijms22041526
Querin, G., Bertolin, C., Da Re, E., Volpe, M., Zara, G., Pegoraro, E., et al. (2016). Non-neural phenotype of spinal and bulbar muscular atrophy: Results from a large cohort of Italian patients. J. Neurol. Neurosurg. Psychiatry 87, 810–816. doi: 10.1136/jnnp-2015-311305
Quigley, C. A., Bellis, A. D., Marschke, K. B., El-Awady, M. K., Wilson, E. M., and French, F. S. (1995). Androgen receptor defects: Historical. Clin. Mol. Perspect. Endocr. Rev. 16, 271–321. doi: 10.1210/edrv-16-3-271
Radhakrishnan, S. K., Lee, C. S., Young, P., Beskow, A., Chan, J. Y., and Deshaies, R. J. (2010). Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28. doi: 10.1016/j.molcel.2010.02.029
Rana, K., Davey, R. A., and Zajac, J. D. (2014). Human androgen deficiency: Insights gained from androgen receptor knockout mouse models. Asian .J Androl. 16, 169–177. doi: 10.4103/1008-682x.122590
Rao, G., Croft, B., Teng, C., and Awasthi, V. (2015). Ubiquitin-proteasome system in neurodegenerative disorders. J. Drug Metab. Toxicol. 6:187. doi: 10.4172/2157-7609.1000187
Raynes, R., Pomatto, L. C. D., and Davies, K. J. A. (2016). Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Aspects Med. 50, 41–55. doi: 10.1016/j.mam.2016.05.001
Redman, K. L., and Rechsteiner, M. (1989). Identification of the long ubiquitin extension as ribosomal protein S27a. Nat. 338, 438–440. doi: 10.1038/338438a0
Rees, I., Lee, S., Kim, H., and Tsai, F. T. (2006). The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim. Biophys. Acta 1764, 1073–1079. doi: 10.1016/j.bbapap.2006.03.013
Rhodes, L. E., Freeman, B. K., Auh, S., Kokkinis, A. D., La Pean, A., Chen, C., et al. (2009). Clinical features of spinal and bulbar muscular atrophy. Brain 132, 3242–3251. doi: 10.1093/brain/awp258
Rinaldi, C., Bott, L. C., Chen, K.-L., Harmison, G. G., Katsuno, M., Sobue, G., et al. (2012). Insulinlike growth factor (IGF)-1 administration ameliorates disease manifestations in a mouse model of spinal and bulbar muscular atrophy. Mol. Medicine 18, 1261–1268. doi: 10.2119/molmed.2012.00271
Rocchi, A., Milioto, C., Parodi, S., Armirotti, A., Borgia, D., Pellegrini, M., et al. (2016). Glycolytic-to-oxidative fiber-type switch and mTOR signaling activation are early-onset features of SBMA muscle modified by high-fat diet. Acta Neuropathol. 132, 127–144. doi: 10.1007/s00401-016-1550-4
Rock, K. L., Gramm, C., Rothstein, L., Clark, K., Stein, R., Dick, L., et al. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771. doi: 10.1016/s0092-8674(94)90462-6
Rogers, S., Wells, R., and Rechsteiner, M. (1986). Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 234, 364–368. doi: 10.1126/science.2876518
Rotin, D., and Kumar, S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409. doi: 10.1038/nrm2690
Rusilowicz-Jones, E. V., Brazel, A. J., Frigenti, F., Urbe, S., and Clague, M. J. (2021). Membrane compartmentalisation of the ubiquitin system. Semin. Cell Dev. Biol. S1084-9521, 292–295. doi: 10.1016/j.semcdb.2021.11.016
Rusmini, P., Polanco, M. J., Cristofani, R., Cicardi, M. E., Meroni, M., Galbiati, M., et al. (2015). Aberrant autophagic response in the muscle of a knock-in mouse model of spinal and bulbar muscular atrophy. Sci. Rep. 5:15174. doi: 10.1038/srep15174
Saporita, A. J., Zhang, Q., Navai, N., Dincer, Z., Hahn, J., Cai, X., et al. (2003). Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J. Biol. Chem. 278, 41998–42005. doi: 10.1074/jbc.M302460200
Sarkar, S., Brautigan, D. L., and Larner, J. M. (2017). Aurora kinase A promotes AR degradation via the E3 ligase CHIP. Mol. Cancer Res. 15, 1063–1072. doi: 10.1158/1541-7786.mcr-17-0062
Sarkar, S., Brautigan, D. L., Parsons, S. J., and Larner, J. M. (2014). Androgen receptor degradation by the E3 ligase CHIP modulates mitotic arrest in prostate cancer cells. Oncogene 33, 26–33. doi: 10.1038/onc.2012.561
Sartori, R., Schirwis, E., Blaauw, B., Bortolanza, S., Zhao, J., Enzo, E., et al. (2013). BMP signaling controls muscle mass. Nat. Genet. 45, 1309–1318. doi: 10.1038/ng.2772
Scaramuzzino, C., Casci, I., Parodi, S., Lievens, P. M., Polanco, M. J., Milioto, C., et al. (2015). Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron 85, 88–100. doi: 10.1016/j.neuron.2014.12.031
Schulman, B. A., and Wade Harper, J. (2009). Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331. doi: 10.1038/nrm2673
Sha, Z., and Goldberg, A. L. (2014). Proteasome-Mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 24, 1573–1583. doi: 10.1016/j.cub.2014.06.004
Shaffer, P. L., Jivan, A., Dollins, D. E., Claessens, F., and Gewirth, D. T. (2004). Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. U.S.A. 101, 4758–4763. doi: 10.1073/pnas.0401123101
Shearer, R. F., Typas, D., Coscia, F., Schovsbo, S., Kruse, T., Mund, A., et al. (2022). K27-linked ubiquitylation promotes p97 substrate processing and is essential for cell proliferation. EMBO J. 41:e110145. doi: 10.15252/embj.2021110145
Sheffield-Moore, M., Urban, R. J., Wolf, S. E., Jiang, J., Catlin, D. H., Herndon, D. N., et al. (1999). Short-Term oxandrolone administration stimulates net muscle protein synthesis in young men1. J. Clin. Endocrinol. Metab. 84, 2705–2711. doi: 10.1210/jcem.84.8.5923
Sheflin, L., Keegan, B., Zhang, W., and Spaulding, S. W. (2000). Inhibiting proteasomes in human HepG2 and LNCaP cells increases endogenous androgen receptor levels. Biochem. Biophys. Res. Commun. 276, 144–150. doi: 10.1006/bbrc.2000.3424
Sheikhhassani, V., Scalvini, B., Ng, J., Heling, L. W. H. J., Ayache, Y., Evers, T. M. J., et al. (2022). Topological dynamics of an intrinsically disordered N-terminal domain of the human androgen receptor. Protein Sci. 31:e4334. doi: 10.1002/pro.4334
Sheng, Y., Saridakis, V., Sarkari, F., Duan, S., Wu, T., Arrowsmith, C. H., et al. (2006). Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 13, 285–291. doi: 10.1038/nsmb1067
Sheppard, R. L., Spangenburg, E. E., Chin, E. R., and Roth, S. M. (2011). Androgen receptor polyglutamine repeat length affects receptor activity and C2C12 cell development. Physiol. Genomics 43, 1135–1143. doi: 10.1152/physiolgenomics.00049.2011
Shimizu, Y., Taraborrelli, L., and Walczak, H. (2015). Linear ubiquitination in immunity. Immunol. Rev. 266, 190–207. doi: 10.1111/imr.12309
Sobue, G., Hashizume, Y., Mukai, E., Hirayama, M., Mitsuma, T., and Takahashi, A. (1989). X-Linked recessive bulbospinal neuropathy: A clinopathological study. Brain 112, 209–232. doi: 10.1093/brain/112.1.209
Song, L., and Luo, Z.-Q. (2019). Post-translational regulation of ubiquitin signaling. J. Cell Biol. 218, 1776–1786. doi: 10.1083/jcb.201902074
Sopher, B. L., Thomas, P. S. Jr., LaFevre-Bernt, M. A., Holm, I. E., Wilke, S. A., Ware, C. B., et al. (2004). Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron 41, 687–699. doi: 10.1016/s0896-6273(04)00082-0
Soraru, G., D’Ascenzo, C., Polo, A., Palmieri, A., Baggio, L., Vergani, L., et al. (2008). Spinal and bulbar muscular atrophy: Skeletal muscle pathology in male patients and heterozygous females. J. Neurol. Sci. 264, 100–105. doi: 10.1016/j.jns.2007.08.012
Stenoien, D. L., Cummings, C. J., Adams, H. P., Mancini, M. G., Patel, K., Demartino, G. N., et al. (1999). Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum. Mol. Genet. 8, 731–741. doi: 10.1093/hmg/8.5.731
Suzuki, K., Katsuno, M., Banno, H., Takeuchi, Y., Atsuta, N., Ito, M., et al. (2007). CAG repeat size correlates to electrophysiological motor and sensory phenotypes in SBMA. Brain 131, 229–239. doi: 10.1093/brain/awm289
Takeyama, K., Ito, S., Yamamoto, A., Tanimoto, H., Furutani, T., Kanuka, H., et al. (2002). Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron 35, 855–864. doi: 10.1016/s0896-6273(02)00875-9
Tan, J. M. M., Wong, E. S. P., Kirkpatrick, D. S., Pletnikova, O., Ko, H. S., Tay, S.-P., et al. (2008). Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 17, 431–439. doi: 10.1093/hmg/ddm320
Tan, Y., Zhou, G., Wang, X., Chen, W., and Gao, H. (2018). USP18 promotes breast cancer growth by upregulating EGFR and activating the AKT/Skp2 pathway. Int. J. Oncol. 53, 371–383. doi: 10.3892/ijo.2018.4387
Thomas, P. S. Jr., Fraley, G. S., Damian, V., Woodke, L. B., Zapata, F., Sopher, B. L., et al. (2006). Loss of endogenous androgen receptor protein accelerates motor neuron degeneration and accentuates androgen insensitivity in a mouse model of X-linked spinal and bulbar muscular atrophy. Hum. Mol. Genet. 14, 2225–2238. doi: 10.1093/hmg/ddl148
Todd, T. W., Kokubu, H., Miranda, H. C., Cortes, C. J., La Spada, A. R., and Lim, J. (2015). Nemo-like kinase is a novel regulator of spinal and bulbar muscular atrophy. Elife 4:e08493. doi: 10.7554/eLife.08493
Tokui, K., Adachi, H., Waza, M., Katsuno, M., Minamiyama, M., Doi, H., et al. (2009). 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum. Mol. Genet. 18, 898–910. doi: 10.1093/hmg/ddn419
Urbé, S., Liu, H., Hayes, S. D., Heride, C., Rigden, D. J., and Clague, M. J. (2012). Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol. Biol. Cell 23, 1095–1103. doi: 10.1091/mbc.e11-08-0668
van Royen, M. E., van Cappellen, W. A., de Vos, C., Houtsmuller, A. B., and Trapman, J. (2012). Stepwise androgen receptor dimerization. J. Cell Sci. 125, 1970–1979. doi: 10.1242/jcs.096792
Varshavsky, A. (2008). Discovery of cellular regulation by protein degradation. J. Biol. Chem. 283, 34469–34489. doi: 10.1074/jbc.x800009200
Varshavsky, A. (2019). N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366. doi: 10.1073/pnas.1816596116
Vogelstein, B., Lane, D., and Levine, A. J. (2000). Surfing the p53 network. Nature 408, 307–310. doi: 10.1038/35042675
Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068. doi: 10.1146/annurev.biochem.68.1.1015
Vummidi Giridhar, P., Williams, K., Vonhandorf, A. P., Deford, P. L., and Kasper, S. (2019). Constant degradation of the androgen receptor by MDM2 conserves prostate cancer stem cell integrity. Cancer Res. 79, 1124–1137. doi: 10.1158/0008-5472.can-18-1753
Wang, A. M., Morishima, Y., Clapp, K. M., Peng, H.-M., Pratt, W. B., Gestwicki, J. E., et al. (2010). Inhibition of Hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J. Biol. Chem. 285, 15714–15723. doi: 10.1074/jbc.m109.098806
Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J.-i., and Chen, Z. J. (2001). TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351. doi: 10.1038/35085597
Wang, F., and Zhao, B. (2019). UBA6 and its bispecific pathways for ubiquitin and FAT10. Int. J. Mol. Sci. 20:2250. doi: 10.3390/ijms20092250
Wang, J., Zhao, X., and Hong, R. (2020). USP26 deubiquitinates androgen receptor (AR) in the maintenance of sperm maturation and spermatogenesis through the androgen receptor signaling pathway. Adv. Clin. Exp. Med. 29, 1153–1160. doi: 10.17219/acem/123355
Weber, J., Polo, S., and Maspero, E. (2019). HECT E3 ligases: A tale with multiple facets. Front. Physiol. 10:370. doi: 10.3389/fphys.2019.00370
Wenzel, D. M., Lissounov, A., Brzovic, P. S., and Klevit, R. E. (2011a). UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108. doi: 10.1038/nature09966
Wenzel, D. M., Stoll, K. E., and Klevit, R. E. (2011b). E2s: Structurally economical and functionally replete. Biochem. J. 433, 31–42. doi: 10.1042/bj20100985
Wiborg, O., Pedersen, M. S., Wind, A., Berglund, L. E., Marcker, K. A., and Vuust, J. (1985). The human ubiquitin multigene family: Some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 4, 755–759. doi: 10.1002/j.1460-2075.1985.tb03693.x
Wilk, S., and Orlowski, M. (1983). Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. J. Neurochem. 40, 842–849. doi: 10.1111/j.1471-4159.1983.tb08056.x
Williams, A. J., Knutson, T. M., Colomer Gould, V. F., and Paulson, H. L. (2009). In vivo suppression of polyglutamine neurotoxicity by C-terminus of Hsp70-interacting protein (CHIP) supports an aggregation model of pathogenesis. Neurobiol. Dis. 33, 342–353. doi: 10.1016/j.nbd.2008.10.016
Wojcik, C., and DeMartino, G. N. (2003). Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579–589. doi: 10.1016/s1357-2725(02)00380-1
Wong, C. I., Zhou, Z. X., Sar, M., and Wilson, E. M. (1993). Steroid requirement for androgen receptor dimerization and DNA binding. Modulation by intramolecular interactions between the NH2-terminal and steroid-binding domains. J. Biol. Chem. 268, 19004–19012. doi: 10.1016/s0021-9258(17)46727-5
Wu-Baer, F., Lagrazon, K., Yuan, W., and Baer, R. (2003). The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743–34746. doi: 10.1074/jbc.c300249200
Xia, X., Huang, C., Liao, Y., Liu, Y., He, J., Guo, Z., et al. (2019). Inhibition of USP14 enhances the sensitivity of breast cancer to enzalutamide. J. Exp. Clin. Cancer Res. 38:220. doi: 10.1186/s13046-019-1227-7
Xie, J., Han, Y., and Wang, T. (2021). RACK1 modulates polyglutamine-induced neurodegeneration by promoting ERK degradation in Drosophila. PLoS Genet. 17:e1009558. doi: 10.1371/journal.pgen.1009558
Xu, K., Shimelis, H., Linn, D. E., Jiang, R., Yang, X., Sun, F., et al. (2009). Regulation of androgen receptor transcriptional activity and specificity by RNF6-Induced ubiquitination. Cancer Cell 15, 270–282. doi: 10.1016/j.Ccr.2009.02.021
Yang, M., Chen, T., Li, X., Yu, Z., Tang, S., Wang, C., et al. (2015). K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8+ T cell activation. Nat. Immunol. 16, 1253–1262. doi: 10.1038/ni.3258
Yang, Q., Zhao, J., Chen, D., and Wang, Y. (2021). E3 ubiquitin ligases: Styles, structures and functions. Mol. Biomed. 2:23. doi: 10.1186/s43556-021-00043-2
Yersak, J. M., Montie, H. L., Chevalier-Larsen, E. S., Liu, Y., Huang, L., Rechsteiner, M., et al. (2017). The 11S proteasomal activator REGγ impacts polyglutamine-expanded androgen receptor aggregation and motor neuron viability through distinct mechanisms. Front. Mol. Neurosci. 10:159. doi: 10.3389/fnmol.2017.00159
Yin, Q., Han, T., Fang, B., Zhang, G., Zhang, C., Roberts, E. R., et al. (2019). K27-linked ubiquitination of BRAF by ITCH engages cytokine response to maintain MEK-ERK signaling. Nat. Commun. 10:1870. doi: 10.1038/s41467-019-09844-0
Yu, X., Yi, P., Hamilton, R. A., Shen, H., Chen, M., Foulds, C. E., et al. (2020). Structural insights of transcriptionally active, full-length androgen receptor coactivator complexes. Mol. Cell 79, 812.e–823.e. doi: 10.1016/j.molcel.2020.06.031
Yu, Y., Zheng, Q., Erramilli, S. K., Pan, M., Park, S., Xie, Y., et al. (2021). K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle. Nat. Chem. Biol. 17, 896–905. doi: 10.1038/s41589-021-00823-5
Yu, Z., Dadgar, N., Albertelli, M., Gruis, K., Jordan, C., Robins, D. M., et al. (2006). Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J. Clin. Invest. 116, 2663–2672. doi: 10.1172/JCI28773
Zboray, L., Pluciennik, A., Curtis, D., Liu, Y., Berman-Booty, L. D., Orr, C., et al. (2015). Preventing the androgen receptor N/C interaction delays disease onset in a mouse model of SBMA. Cell Rep. 13, 2312–2323. doi: 10.1016/j.celrep.2015.11.019
Zeng, S., Huang, W., Zheng, X., Liyan, C., Zhang, Z., Wang, J., et al. (2021). Proteolysis targeting chimera (PROTAC) in drug discovery paradigm: Recent progress and future challenges. Eur. J. Med. Chem. 210:112981. doi: 10.1016/j.ejmech.2020.112981
Zhang, Y. (2003). Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 17, 2733–2740. doi: 10.1101/gad.1156403
Zheng, N., and Shabek, N. (2017). Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157. doi: 10.1146/annurev-biochem-060815-014922
Zhong, J., Ding, L., Bohrer, L. R., Pan, Y., Liu, P., Zhang, J., et al. (2014). p300 Acetyltransferase regulates androgen receptor degradation and PTEN-Deficient prostate tumorigenesis. Cancer Res. 74, 1870–1880. doi: 10.1158/0008-5472.can-13-2485
Zhou, Z. X., Sar, M., Simental, J. A., Lane, M. V., and Wilson, E. M. (1994). A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J. Biol. Chem. 269, 13115–13123. doi: 10.1016/s0021-9258(17)36806-0
Zucchelli, S., Marcuzzi, F., Codrich, M., Agostoni, E., Vilotti, S., Biagioli, M., et al. (2011). Tumor necrosis factor receptor-associated factor 6 (TRAF6) associates with huntingtin protein and promotes its atypical ubiquitination to enhance aggregate formation. J. Biol. Chem. 286, 25108–25117. doi: 10.1074/jbc.m110.187591
Keywords: spinal and bulbar muscular atrophy, Kennedy’s disease, polyglutamine, androgen receptor, ubiquitin, proteasome, neurodegenerative
Citation: Sengupta M, Pluciennik A and Merry DE (2022) The role of ubiquitination in spinal and bulbar muscular atrophy. Front. Mol. Neurosci. 15:1020143. doi: 10.3389/fnmol.2022.1020143
Received: 15 August 2022; Accepted: 20 September 2022;
Published: 06 October 2022.
Edited by:
Matt Scaglione, Duke University, United StatesReviewed by:
Constanza J. Cortes, University of Alabama at Birmingham, United StatesMaria Pennuto, University of Padua, Italy
Copyright © 2022 Sengupta, Pluciennik and Merry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diane E. Merry, ZGlhbmUubWVycnlAamVmZmVyc29uLmVkdQ==
 Medha Sengupta
Medha Sengupta Anna Pluciennik
Anna Pluciennik Diane E. Merry
Diane E. Merry