
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 13 December 2021
Sec. Pain Mechanisms and Modulators
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.779024
This article is part of the Research Topic Transcriptional Changes in the Dorsal Root Ganglia View all 4 articles
Peripheral nerve injury induces genome-wide transcriptional reprogramming of first-order neurons and auxiliary cells of dorsal root ganglia (DRG). Accumulating experimental evidence suggests that onset and mechanistic principles of post-nerve injury processes are sexually dimorphic. We examined largely understudied aspects of early transcriptional events in DRG within 24 h after sciatic nerve axotomy in mice of both sexes. Using high-depth RNA sequencing (>50 million reads/sample) to pinpoint sexually dimorphic changes related to regeneration, immune response, bioenergy, and sensory functions, we identified a higher number of transcriptional changes in male relative to female DRG. In males, the decline in ion channel transcripts was accompanied by the induction of innate immune cascades via TLR, chemokine, and Csf1-receptor axis and robust regenerative programs driven by Sox, Twist1/2, and Pax5/9 transcription factors. Females demonstrated nerve injury-specific transcriptional co-activation of the actinin 2 network. The predicted upstream regulators and interactive networks highlighted the role of novel epigenetic factors and genetic linkage to sex chromosomes as hallmarks of gene regulation post-axotomy. We implicated epigenetic X chromosome inactivation in the regulation of immune response activity uniquely in females. Sexually dimorphic regulation of MMP/ADAMTS metalloproteinases and their intrinsic X-linked regulator Timp1 contributes to extracellular matrix remodeling integrated with pro-regenerative and immune functions. Lexis1 non-coding RNA involved in LXR-mediated lipid metabolism was identified as a novel nerve injury marker. Together, our data identified unique early response triggers of sex-specific peripheral nerve injury regulation to gain mechanistic insights into the origin of female- and male-prevalent sensory neuropathies.
Sensory deficits arising after peripheral nerve injury (PNI) or disease can impact touch, vibration, position, temperature, and pain senses. Regeneration of the damaged sensory neurons is a complex process orchestrated in part by transcriptional events at the level of the neuron cell bodies localized in dorsal root ganglia (DRG; Gordon et al., 2003; McDonald et al., 2006; Zochodne, 2012). Each DRG contains thousands of cell bodies of phenotypically distinct primary afferent neurons, each enveloped by a layer of satellite glia and surrounded by resident and infiltrating cells of immune, stromal, endothelial, and other origins (Devor, 1999; Lawson, 2002; Takeda et al., 2009). The diversity of cell populations ensures dynamic DRG transcriptomes in humans (Flegel et al., 2015; LaPaglia et al., 2018; Ray et al., 2018; North et al., 2019). Rodent models of PNI have provided a valuable tool to capture extensive transcriptional reprogramming gradually unfolding at the site of nerve injury and the corresponding DRG and spinal cord tissues (Kubo et al., 2002; Xiao et al., 2002; Bosse et al., 2006; Costigan et al., 2009; Michaelevski et al., 2010; LaCroix-Fralish et al., 2011; Kim et al., 2012; Li et al., 2013; Yao et al., 2013; Jiang et al., 2014; Perkins et al., 2014; Chernov et al., 2015, 2020; Chandran et al., 2016; He and Jin, 2016; Mahar and Cavalli, 2018; Renthal et al., 2020). In addition, the transcriptional makeup of DRG is enriched by axonally trafficked (Devor, 1999; Takeda et al., 2009) and exosome-delivered (Simeoli et al., 2017) coding and non-coding (nc) RNAs in a dynamic manner in the course of PNI response.
Sex is increasingly recognized as a critical biological variable in the functional repair of damaged sensory neurons. Inflammatory, neuropathic, and idiopathic hypersensitivity and persistent pain conditions often have greater prevalence and symptom severity in women as compared to men (Unruh, 1996; Greenspan et al., 2007; Fillingim et al., 2009; Mogil, 2012; Sorge and Totsch, 2017; Boerner et al., 2018). Transcriptome RNA-sequencing (RNA-seq)-based analyses of human DRG at baseline (Ray et al., 2019) and in patients with radicular/neuropathic pain (North et al., 2019) highlighted DRG’s central role in sex-specific gene regulation. Likewise, emerging research has revealed sexual dimorphism in rodent DRG transcriptomes in response to peripheral nerve injury (Stephens et al., 2019; Ahlström et al., 2021), hyperalgesic priming by interleukin 6 (Il-6; Paige et al., 2020), sciatic nerve injection of myelin basic protein (MBP) derived peptides (Chernov et al., 2020), and other stimuli. Transcriptome differences in DRG have been implicated in female-prevalent hypersensitivity partly due to the activity of prostaglandin signaling and neuroendocrine mechanisms involving prolactin receptors (North et al., 2019; Ray et al., 2019; Chernov et al., 2020; Mecklenburg et al., 2020; Paige et al., 2020; Tavares-Ferreira et al., 2020).
Rapid initiation of regeneration is essential to the successful functional repair of sensory neurons in PNI (Gordon et al., 2003; McDonald et al., 2006; Zochodne, 2012). Within 24 h after sciatic nerve transection in rodents, DRG transcriptome induces neuron-specific regeneration associated genes (RAGs), including activating transcription factor 3 (Atf3), and growth-associated protein (Gap)43 (Bolin et al., 1995; Tsujino et al., 2000; Bosse et al., 2001; Snider et al., 2002; Sun and He, 2010; Van Kesteren et al., 2011; Ma and Willis, 2015; Chandran et al., 2016; Rao and Pearse, 2016; Renthal et al., 2020; Shiers et al., 2020; Suzuki and Ochi, 2020; Lee and Cho, 2021). Shortly after peripheral nerve injury, the expression of genes encoding cytokines, chemokines, Toll-like receptors (TLRs), matrix metalloproteinases (MMPs), and other mechanistically related proteins increased (Kim et al., 2012; Chernov et al., 2015). Both resident and infiltrating cell types in DRG can promote sex-specific responses to nerve injury (Mapplebeck et al., 2016, 2017). The injured sensory neurons engage molecular crosstalk with DRG macrophages via colony-stimulating factor 1 (Csf1) to stimulate male-specific macrophage expansion at day four post-PNI (spared nerve injury) (Yu et al., 2020). As a result, these processes contribute to the initiation and maintenance of hypersensitivity in a sex-specific manner.
It remains unknown which early-response DRG transcriptome changes display sexual dimorphism in response to PNI. Using high-depth RNA-seq, the present study has identified significant sex-specific early-response genome-wide transcriptional changes in coding and ncRNA in male and female DRG at 24 h after sciatic nerve axotomy in mice. Applying predictive bioinformatics, we determined signaling pathways, particularly those related to regenerative, immune, metabolic, sex hormone, and X/Y chromosome-linked systems.
To identify sexually dimorphic early-response PNI-specific molecular signatures, we conducted sciatic nerve complete axotomy (Anderson, 1902; Ramon y Cajal, 1928) in female and male mice according to our established procedure (Chattopadhyay and Shubayev, 2009; Chernov et al., 2015). At 24 h after sham or axotomy surgery (n = 6/group; pooled n = 2/sample), ipsilateral lumbar (L)4 and L5 (pooled) DRG were processed using RNA-seq. Genome-wide gene regulation and gene ontology analyses were performed as specified below.
A total of 140,789 individual transcripts were identified in DRG across four groups, corresponding to 54,309 annotated genes. Following exclusion of genes with low abundance measurements [count per million (CPM) <10 across four groups], 26,518 genes were analyzed for differential regulation between axotomy and sham groups in female and male DRG.
DESeq2 workflow used the Wald test (Love et al., 2014) to quantify significance parameters and predict differentially expressed genes (DEGs; Figure 1A). In addition, an adaptive t-prior shrinking (apeglm) allowed to rank DEGs by the “effect size” and reduce noise associated with higher fold change (FC) from low abundance genes (Zhu et al., 2019). Over 10,600 significant DEGs with adjusted p-values (padj) below a false discovery rate (FDR) cutoff (padj < 0.1) were further analyzed (Supplementary Table 1). We identified 2,418 upregulated and 2,303 downregulated DEGs in females, and 4,314 upregulated and 4,251 downregulated DEGs in males (Figure 1B). A total of 3,664 DEGs exhibited monomorphic regulation in both sexes, including 1,776 and 1,819 up- and downregulated genes, respectively. Of all DEGs, 642 and 2,538 genes were uniquely upregulated in females or males, respectively; 484 and 2,432 genes were uniquely downregulated in females or males, respectively. Over 42% of DEGs exhibited FC above 1.5 or below −1.5.
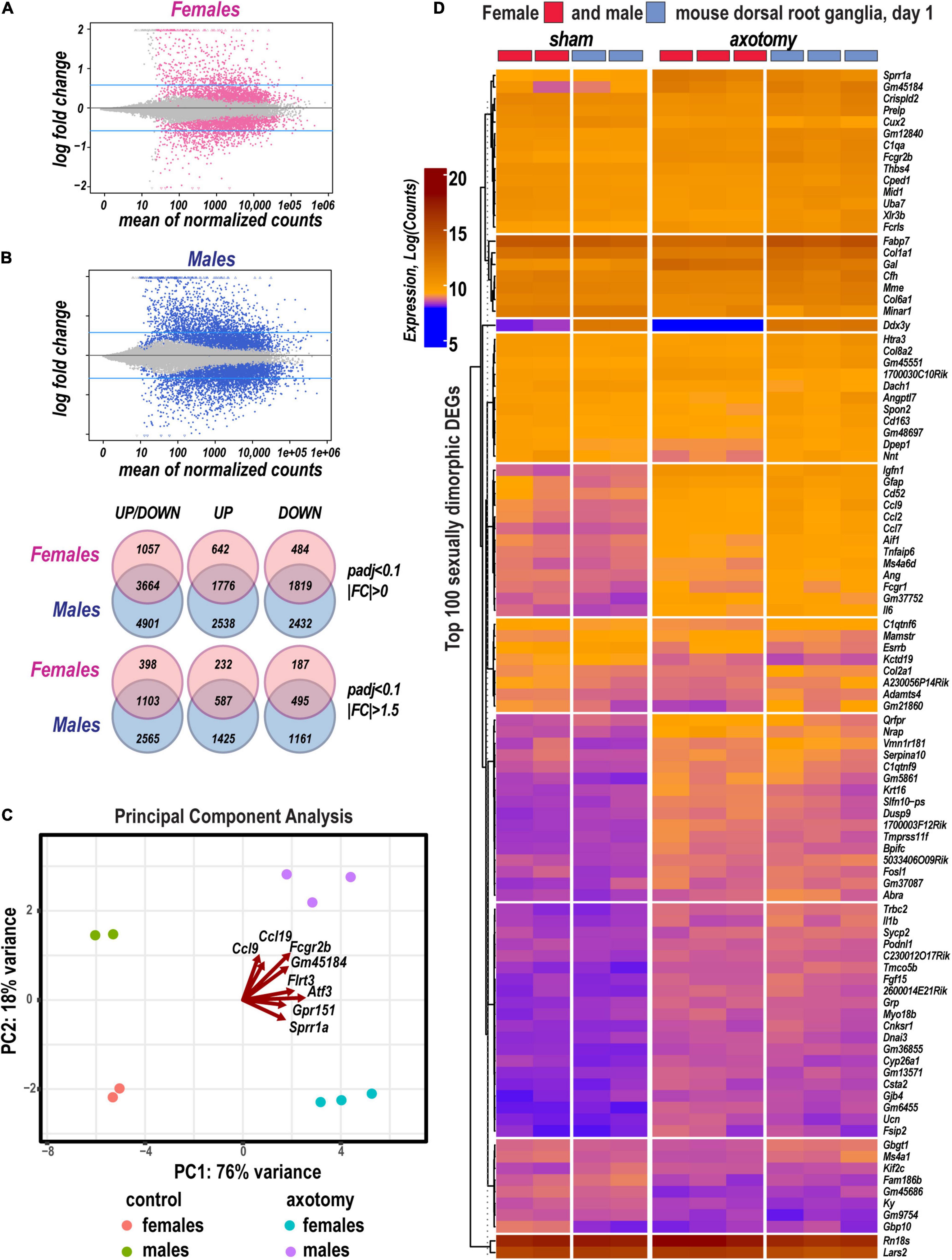
Figure 1. Sexually dimorphic transcriptional changes in murine female and male DRG 24 h after axotomy. (A) Bland-Altman plots of shrunken log2FC and log2 of Mean Average counts (MAplot function in the DESeq2 suit). Adjusted log2FC were calculated using the adaptive t-prior apeglm method (lfcShrink function in DESeq2 suit). Female and male DEGs with padj < 0.1 are marked as red or blue points, respectively. Points are colored gray if padj > 0.1. Points that fall out of the displayed window are plotted as open triangles pointing either up or down. Blue horizontal lines indicate Log2FC > 0.58 and Log2FC < −0.58 thresholds. (B) Venn diagrams (left to right) demonstrate a number of total identified DEGs (padj < 0.1, log2FC≠0), upregulated DEGs [padj < 0.1, Log2FC > 0 (top graph) or Log2FC > 0.58 (bottom graphs)], or downregulated DEGs [padj < 0.1, Log2FC < 0 (top graph) or Log2FC < −0.58 (bottom graphs)] in female (red circles) and male (blue circles) mice. The intersections of circles correspond to unidirectionally changed DEGs. (C) Principal component (PC) analysis of DEGs in three replicate groups of female and male DRG and the respective sham-operated controls. Blue and red indicate males and females, respectively. Red arrows indicate genes with the most significant weight. (D) Hierarchical clusters of the most significant up- and downregulated DEGs (padj < 0.1). Heatmap colors correspond to gene abundance (Log counts) following variance stabilizing transformation (DESeq2).
Based on the principal component analysis (PCA) of 1,000 top-regulated DEGs (Figure 1C), we attributed 76% (PC1) and 18% (PC2) variance to PNI and biological sex, respectively. In PNI, the variance due to sexually dimorphic regulation was higher than in sham. Hierarchical clustering (Figure 1D) highlighted genes related to nerve regeneration, ion channels, transporters, extracellular matrix (ECM), immune, metabolic, and transcriptional regulators. The statistically significant protein-coding and ncRNA DEGs prevailing in each sex are visualized on volcano plots (Figure 2). We conducted in-depth analyses of DEGs within specific gene ontology (GO) categories, gene families with mechanistic relevance to PNI response (Figure 3 and Supplementary Table 1).
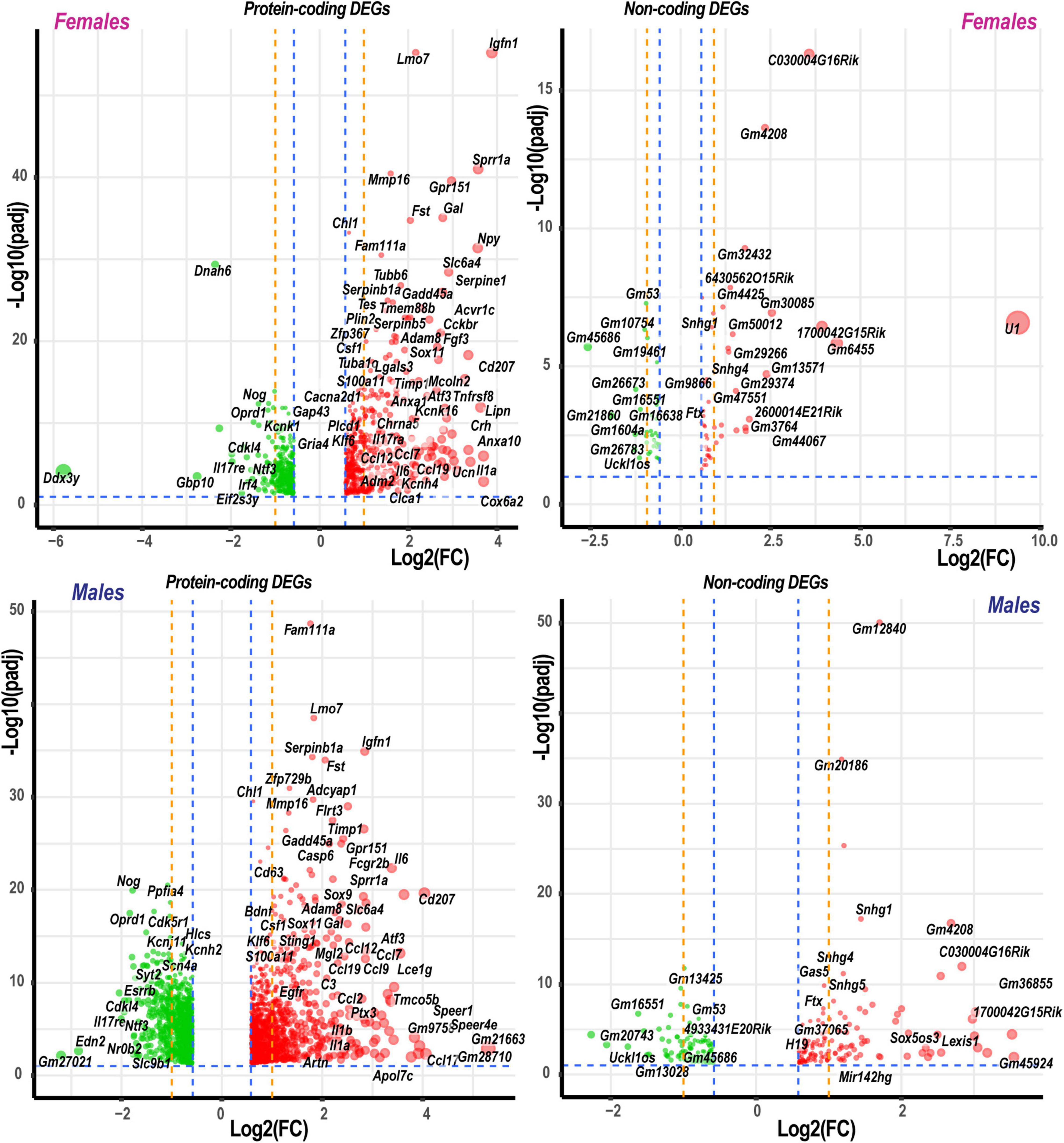
Figure 2. Significant DEGs in females and males. Significance [–log10(padj)] versus log2 fold change (FC) volcano plots of protein-coding and ncRNA DEGs in females and males. Red and green colors indicated up- and downregulated DEGs, respectively. The diameter of each circle is proportional to the respective log2FC. Only data above thresholds (padj < 0.1, | Log2FC| > 0.58, indicated by blue dashed lines) are displayed. Dotted orange lines indicate stringent | Log2FC| > 1 (| FC| > 2) thresholds. Select DEGs are labeled.
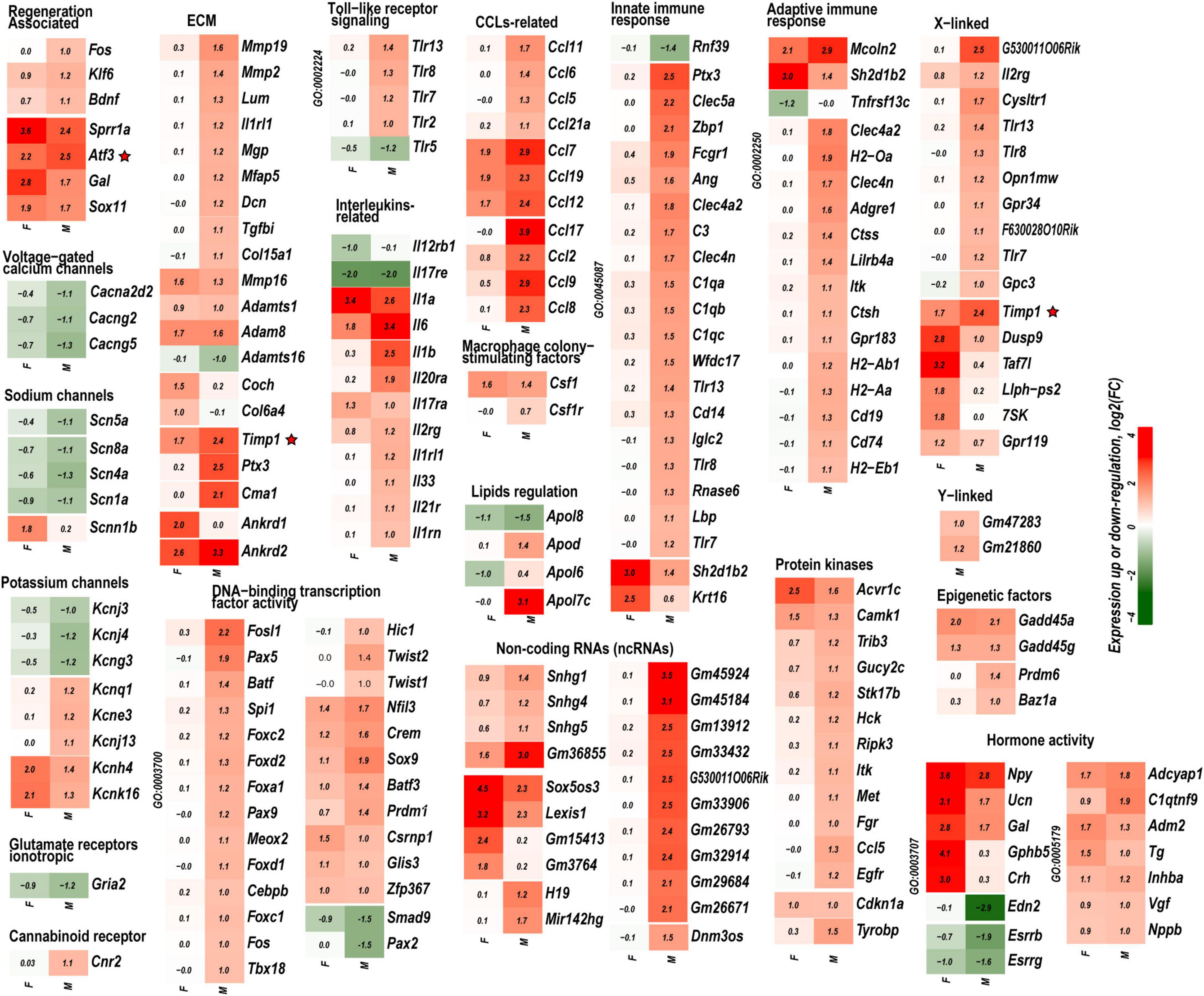
Figure 3. Mono- and dimorphic regulation of select gene families grouped by GO terms, functional similarity, structure, genomic co-localization, and published study datasets. Only DEGs with significance scores (padj < 0.1) and | Log2FC| > 1 (| FC| > 2) are shown. Up- and downregulated DEGs are colored according to the respective color scales of log2FC. GO term identification numbers are shown then applicable on the left side of each heatmap. Red stars indicate genes investigated by protein immunoblotting. A complete list of DEGs is included in Supplementary Table 1.
Regeneration associated genes are neuron-intrinsic factors that stimulate axonal outgrowth after PNI (Bolin et al., 1995; Tsujino et al., 2000; Bosse et al., 2001; Snider et al., 2002; Abe and Cavalli, 2008; Sun and He, 2010; Van Kesteren et al., 2011; Ma and Willis, 2015; Chandran et al., 2016; Rao and Pearse, 2016; Renthal et al., 2020; Shiers et al., 2020; Suzuki and Ochi, 2020; Lee and Cho, 2021). RAG genes encoding the activating protein 1 (AP-1) complex and the Sox (sex-determining region Y box) family were upregulated in both sexes. Atf3 gene was upregulated 2.2-fold and 2.5-fold in both male and female DRG, respectively. This expression pattern consistently correlated with the increase of Atf3 and Atf3b protein isoforms (Figure 4A) encoded by the Atf3 gene. Sox11-regulated Sprr1a gene encoding a small proline-rich protein (Jing et al., 2012) demonstrated more significant upregulation in females. In contrast, brain-derived neurotrophic factor (Bdnf), a stimulator of sciatic nerve axon growth (Zhou and Shine, 2003), was expressed higher in males than in females. Other male-prevalent RAGs included the AP-1 component c-Fos (Fos). RAGs were consistently upregulated at later stages of PNI, including sciatic nerve chronic constriction injury (CCI; Stephens et al., 2019; Ahlström et al., 2021). We suggest that their early activation and sustained long-term expression are indispensable for a robust response to injury.
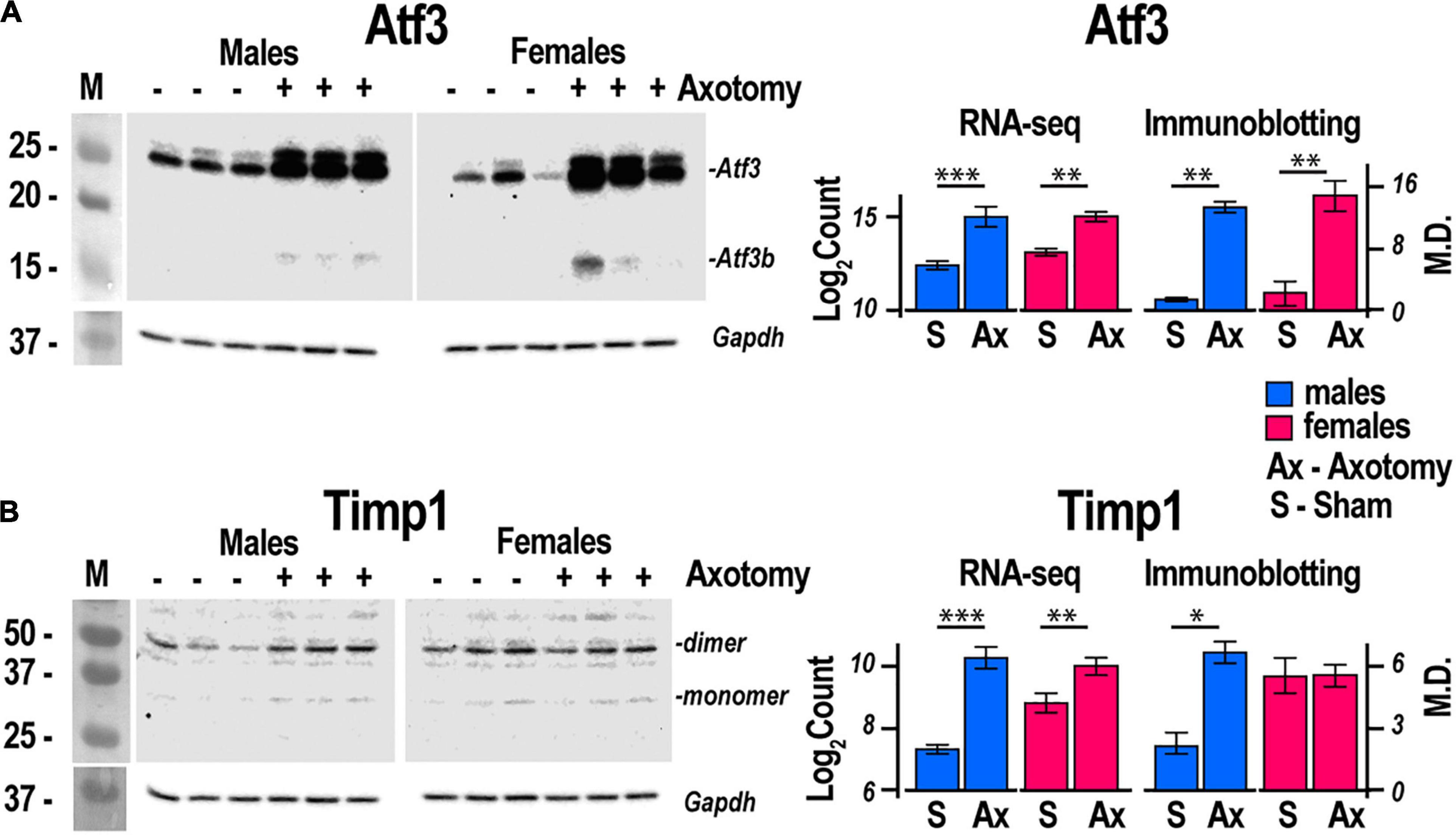
Figure 4. Atf3 and Timp1 protein expression correlated with transcriptional activity of respective genes. Axotomy or sham surgeries were conducted in male and female mice (n = 3/group). After 24 h, DRGs ipsilateral to the surgery site were dissected and total proteins were analyzed by immunoblotting. Primary antibodies to mouse (A) Atf3 and (B) Timp1 were used. Gapdh served as loading control. Right panels, comparison of absolute expression Log2Count of (A) Atf3, and (B) Timp1 with densitometry analysis of protein bands by immunoblotting. Digitized immunoblotting data from three individual replicates were used to calculate means and standard deviation. ANOVA with post hoc Tukey test was applied to determine statistical significance: * p < 0.01; ** p < 0.001; *** p < 0.0001. M, molecular weight marker; Log2Count, Log2 normalized transcript counts; M.D., mean density; S, Sham, Ax, Axotomy.
The reduced expression of voltage-gated ion channels mRNA abundance persisted in both male and female DRG; however, the magnitude of the decrease was more significant in males. Subunits of voltage-dependent calcium channel subunits Cacna2d2, Cacng2, and Cacng5 were downregulated. mRNA levels of voltage-gated sodium channels Scn1a (Nav1.1), Scn4a (Nav1.4), and Scn5a (Nav1.5), including DRG-specific channels Scn8a (Nav1.6), have decreased, except female-specific upregulation of epithelial sodium channel (Scnn1b). Potassium channel genes Kcnj3, Kcnj4, and Kcng3 were downregulated. Notably, Kcnk16 and Kcnh4 were upregulated in both sexes, whereas Kcnj13, Kcnq1, and Kcne3 were upregulated only in males. Ionotropic glutamate receptors AMPA (Gria2) was downregulated higher in males. We observed a male-specific increase of the immune cells-associated cannabinoid receptor (CBR) Cnr2 mRNA.
Males exhibited a more significant number of the upregulated ECM genes relative to females. Mmp2, Mmp19, Adamts4, Adam10, and Adam17 were upregulated in males but did not change in females. Mmp16, Adam8, and Adamts1 increased in both sexes. Adamts16 decreased in males. The tissue inhibitor of metalloproteinases Timp1 gene, a stoichiometric activity regulator, was induced in both sexes, albeit the increase was higher in males. The Timp1 expression pattern correlated with the increase of Timp1 protein in male DRG. Importantly, a higher absolute level of Timp1 mRNA and protein expression was detected in female sham DRG as compared to males (Figure 4B). In addition, we observed sexually dimorphic regulation of genes encoding the ECM structural molecules, including collagen and fibronectin species. TNF-inducible pentraxin-related protein Ptx3 gene was induced in males. Ankyrin repeat protein Ankrd2 upregulated in females and Ankrd1 upregulated in both sexes. We uncovered an interactive protein network of serine proteases, including the extracellular chymotryptic serine protease Cma1 (Figure 5A). According to the Ingenuity Pathway Analysis (IPA), the Cma1 network can coordinate male-specific signaling receptor binding (P = 3.17e-5), integrins binding (P = 1.2e-5), cytokine activity (P = 5.56e-5), and microglial cell/macrophage activation (P = 1.6e-3).
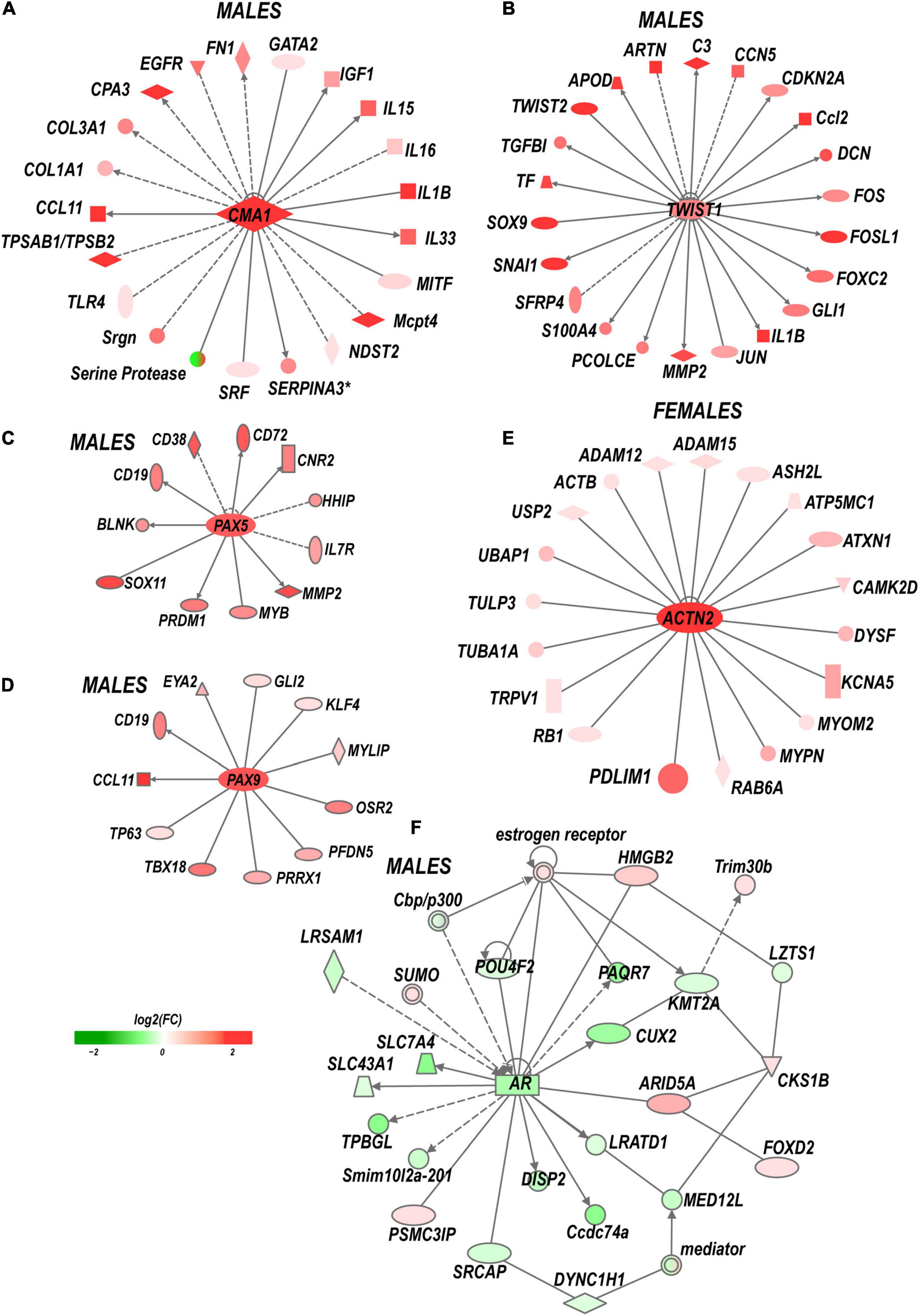
Figure 5. Sex-specific co-activation networks predicted by IPA. (A) Chimase (Cma1) network in males. (B) Twist1 network in males. (C) Pax5 and (D) Pax9 networks in males. (E) Actinin 2 (Actn2) network in females. (F) Co-downregulation of androgen receptor network in males. Gray arrow lines indicate the directionality of regulatory signaling. The intensity of red and green colors indicates up- and downregulation corresponding to Log2FC. Solid and dashed lines indicate direct and indirect interactions, respectively.
Crucial genes encoding pattern-recognition receptors (PRPs) and TLRs exhibited male-specific upregulation. Cell surface Tlr2 and all X-linked endosomal TLRs (Tlr7, Tlr8, and Tlr13) were upregulated in males. Tlr5 was downregulated in males. mRNA levels encoding proteins that constitute the TLR Signaling axis: TIR-domain containing Myd88 and Trif (Ticam1), Irak3/4 kinases, interferon regulatory factors (Irf)-3/4/8/9, and NFkB family members Nfkb1 and Nfkb2 increased moderately in males.
The upregulation of cytokine and chemokine genes was generally more significant in males, affecting interleukin ligands (Il1b, Il6, and Il33) and interleukin receptors (Il1rl1, Il1rn, Il2rg, 1l17rb, and others). Remarkably, only interleukins Il6, Il1a/b, and Il17ra receptor genes were upregulated in females, consistent with the female-specific activity of Il17a activity in macrophage-sensory neuronal crosstalk (Luo et al., 2021). Ccl2 [chemokine (C–C motif) ligand 2] exhibited robust upregulation in males. Ccl2 change in females was lower, suggesting that Ccl2 is a male-specific transcript activated by PNI events. Similarly, other chemokines, including Ccl5, Ccl6, Ccl8, Ccl9, Ccl11, Ccl17, and Ccl21a, were predominantly upregulated in males. Ccl7, Ccl12, and Ccl19 were upregulated in both sexes. The macrophage colony-stimulating factor 1 (Csf1) gene was upregulated in both sexes, whereas the Csf1 receptor (Csf1r) gene expression moderately increased only in male DRG. Notably, multiple genes related to the adaptive and innate immune responses according to GO terms GO:0045087 and GO:0002250, respectively, exhibited sexually dimorphic regulation.
Lipid carrier apolipoproteins Apol6, Apo7c, and apolipoprotein B receptor (Apobr) decreased in female DRG. ApoL6, Apo7c, Apobr, and Apolipoprotein D (Apod) genes were upregulated in male DRG. Ch25h gene, encoding the cholesterol-25-hydroxylase enzyme, responsible for the biosynthesis of the LXR ligand oxysterol (25-hydroxycholesterol), increased only in females.
Both female and males DRG exhibited increased expression of multiple components of the complexes I, II, IV, and V (ATP synthases). Females specifically upregulated cytochrome c oxidase of complex IV (Cox6a2).
Upstream regulators. Upstream regulators affect the expression of multiple downstream genes, thus representing sex-specific signaling hubs in early PNI response. Upstream regulators were predicted using IPA, as follows:
(i) Transcription factors (TFs). Multiple sexually dimorphic TFs were identified in our RNA-seq dataset. Sox7 displayed upregulation in female DRG. Male DRG exhibited moderate upregulation of Sox8 and Sox10, fork-head box (Fox a1/c1/c2/d1/d2/s1), and interferon activated (Ifi209/207) TFs. In males, co-regulatory networks centers around basic helix-loop-helix TFs Twist1/2 (Figure 5B). According to our IPA predictions, the activated Twist1/2 network can promote macrophages-mediated inflammatory response (P = 1.7e-7). Two upregulated networks involving members of the paired box (Pax5 and Pax9) TFs were predicted (Figures 5C,D). The top canonical pathways regulated by these networks include B Cell Development (P = 2.1e-4), IL-7 (P = 7.0e-4) and IL-17A (P = 3.4e-2) Signaling pathways, PI3K Signaling in B Lymphocytes (P = 2.3e-3), Role of JAK1/JAK3 in Cytokines signaling (P = 5.5e-4), Primary Immunodeficiency (P = 2.6e-6), and Sonic Hedgehog signaling (P = 1.5e-2). Smad9 and Pax2 were downregulated in males.
(ii) ncRNAs. We identified over 2500 significantly regulated ncRNA in both sexes, including 260 ncRNAs that changed expression >1.5-fold. Notably, the imprinted trans-regulator H19 and the precursor gene for hematopoietic cell-specific Mir124 exhibited male-specific regulation. Snhg1, Snhg4, and Snhg5 host ncRNAs, each comprising one or more small nucleolar RNAs (snoRNAs), increased predominantly in males. In females, Sox5os3 (an antisense transcript of Sox5 gene) and Lexis1 (a lipid-responsive LXR-induced inhibitor of cholesterol synthesis) ncRNAs were upregulated to a higher degree. It is worth noting that many ncRNAs are poorly characterized to date; therefore, their engagement in PNI responses will require future reassessment.
(iii) Kinases. We observed male-specific upregulation of protein tyrosine kinases (PTK) encoded by Fgr, Hck, and Itk genes. The Syk kinase-interacting TYRO (protein tyrosine kinase)-binding protein encoded by the Tyrobp gene (also known as Dap12) was upregulated in males. Most serine/threonine kinases, including mitogen-activated protein kinases (MAPK), protein kinases A (PKA), protein kinases B (AKT), protein kinases C (PKC), cyclin-dependent kinases (CDK), and Ca2+/calmodulin-dependent protein kinases (CAMK), did not significantly change or downregulated in both sexes. CAMK encoded by Camk1, and cyclin-dependent kinase inhibitor p21Cip1 (Cdkn1a) increased in both sexes.
(iv) X/Y chromosome-linked genes. We identified a higher upregulation of X-linked genes in males than females. Over 330 PNI-specific DEGs were linked to the sex chromosomes. Thirteen X-linked genes upregulated in females (Taf7l, Dusp9, Llph-ps2, Timp1, Gpr119, Il2rg, and others). At least 44 genes, including Timp1, were upregulated in males. Y-linked Gm21860 lncRNA and the protein-coding Gm47283 genes exhibited an increase in males.
(v) Epigenetic factors. Stress-sensing Gadd45 genes (Gadd45a and Gadd45a) upregulated in both sexes. Epigenetic writers and readers of histone code were differentially regulated as well. In males, the putative histone-lysine N-methyltransferase Prdm6 and a bromodomain protein Baz1a genes were upregulated.
(vi) Sex hormone receptors. In females, the level of estrogen receptor (ESR) Esr2 moderately increased. Activation of nociceptive signaling via ESR signaling has been implicated in female-specific pain hypersensitivity (Khomula et al., 2017; Chernov et al., 2020). In addition, because of the ESR’s direct interaction with the Nrip1 nuclear receptor (Cavaillès et al., 1995), the axonal outgrowth in females is likely mediated by the estrogen/ESR axis via co-regulated Nrip1, Pdlim1/7 (PDZ and LIM domain-containing proteins), alpha-actinin 2 (Actn2) genes, and other interacting molecules (Figure 5E). In male DRG, the level of the ESRs did not significantly increase, whereas the androgen receptor (Ar) gene was downregulated. IPA-predicted co-downregulated Ar-interacting factors included Pou4F2, progestin and adipoQ receptor Paqr7, neuron-specific TF Cux2, sterol-sensing domains containing Disp2, Coiled-coil domain-containing Ccsd74a, E3 ubiquitin-protein ligase Lrsam1, and others (Figure 5F).
The prediction of relevant canonical signaling pathways induced in DRG at 24 h post-PNI (Figure 6) was conducted in IPA using a standard null hypothesis of the regulation randomness, a right-tailed Fisher’s exact test, and stringent statistical thresholds (padj < 0.1, | FC| > 2, activation | z-score| > 2).
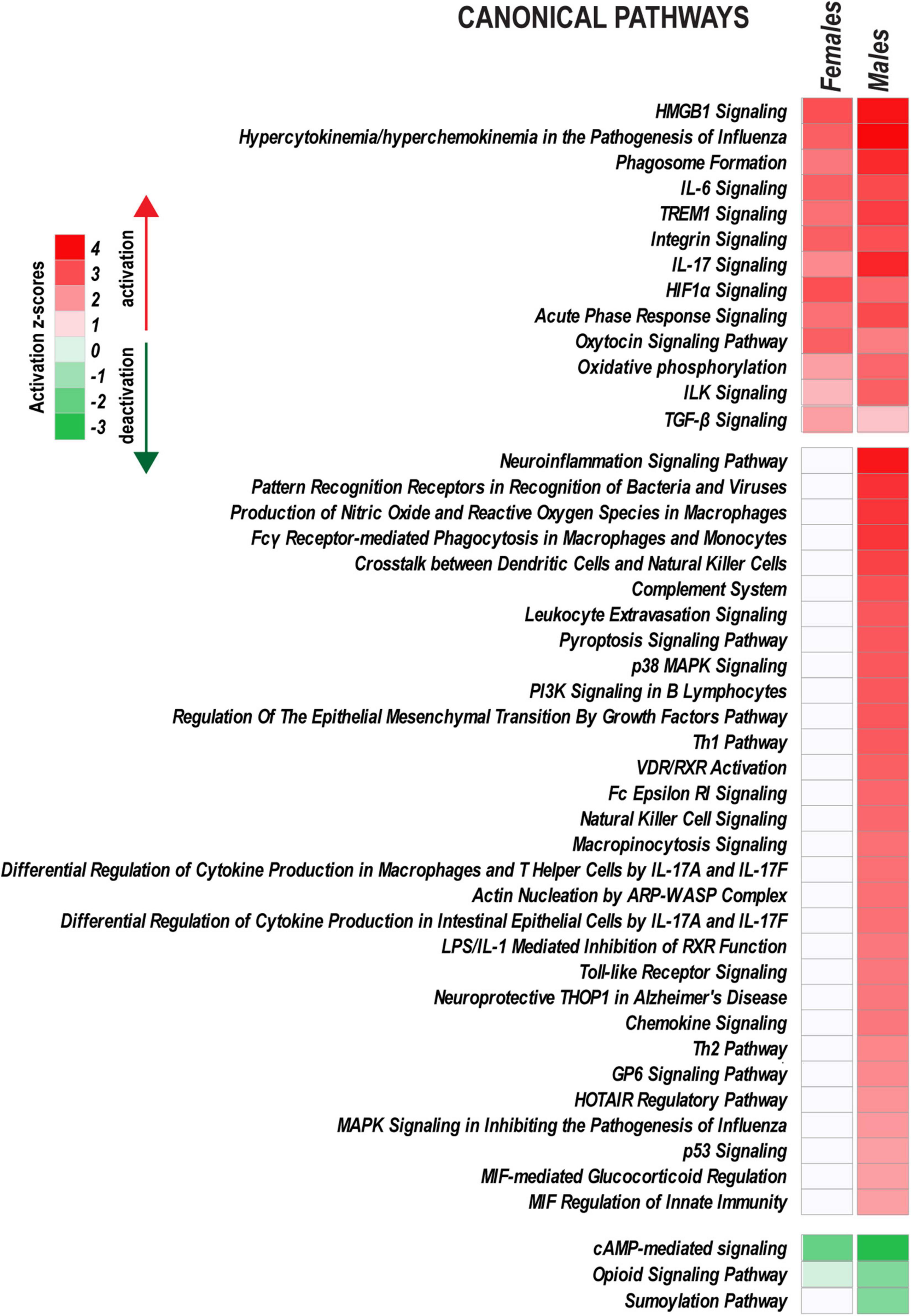
Figure 6. Ingenuity Pathway Analysis highlights sex-specific versus monomorphic early response mechanisms leveraged by PNI. Canonical pathways with |z-score| > 2.0 (identified by Fisher’s exact test) are shown on heatmaps. Positive and negative z-scores indicate upregulation (z > 2.0) or downregulation (z < –2.0) of respective pathways. Color intensity corresponds to higher (red) or lower (green) z-scores.
Peripheral nerve injury activated a characteristic set of signaling pathways in both sexes, including HMGB1 Signaling, TREM1 signaling, IL-6 Signaling, IL-17 Signaling, Integrin signaling, HIF1α Signaling, Oxytocin Signaling, and ILK Signaling. Immunity responses (Toll-like Receptor signaling, Chemokine signaling, Leukocyte extravasation signaling, Fcγ Receptor-mediated Phagocytosis in Macrophages and others), PI3K/AKT Signaling, HOTAIR regulatory pathway, and VDR/RXR Signaling also increased in males. cAMP-mediated signaling, Opioid Signaling, and Sumoylation pathways were downregulated.
Recent transcriptomics data in animals identified sex-specific PNI responses in DRG (North et al., 2019; Ray et al., 2019; Stephens et al., 2019; Chernov et al., 2020; Mecklenburg et al., 2020; Paige et al., 2020; Tavares-Ferreira et al., 2020; Yu et al., 2020). However, sex differences in the immediate post-injury early-response transcriptional events remain not well understood. Herein, taking advantage of the sensitivity provided by high-depth RNA-seq analysis, we strived to untangle sexually dimorphic regulatory transcriptional mechanisms in murine DRG that develop at a 24 h time point after sciatic nerve axotomy. Other biological variables and experimental conditions, including species, strains, age, injury models, and timepoint, beyond the scope of this study, are expected to influence DRG transcriptomics.
Nerve regenerative program in damaged PNS afferent axons is orchestrated by neuron-specific RAG genes expressed in DRG (Bolin et al., 1995; Tsujino et al., 2000; Bosse et al., 2001; Snider et al., 2002; Abe and Cavalli, 2008; Sun and He, 2010; Van Kesteren et al., 2011; Ma and Willis, 2015; Chandran et al., 2016; Rao and Pearse, 2016; Renthal et al., 2020; Shiers et al., 2020; Suzuki and Ochi, 2020; Lee and Cho, 2021). In agreement, we observed upregulation of critical RAGs, including Atf3, Sox11, Klf6, Sprr1a, and Gal. Most transcriptional changes of RAGs were sexually monomorphic, except Sprr1a and galanin (Gal) that exhibited a higher increase in females as compared to males. Promoted by Sox11, Sprr1a is known to accelerate peripheral nerve regeneration (Jing et al., 2012). Cross-analysis with single-cell RNA-seq data in a spinal nerve injury model in mice (Renthal et al., 2020) indicated similar upregulation of Atf3, Sox11, Sprr1a, and other RAGs specific to injured neuron cells. At least two Sox-family genes exhibited upregulation. Sox9 and Sox11. Sox proteins are evolutionarily conserved transcription factors sharing a similar HMG-box domain with the Y chromosome-linked Sry protein responsible for male sex determination. Sox proteins are architectural components of chromatin (Pevny and Lovell-Badge, 1997), which govern neuronal and glial lineage specification during development. In the regenerating adult PNS, Sox members are pioneer TFs that poise downstream TFs for transcriptional regulation (Stevanovic et al., 2021).
Neuroinflammation signaling pathway was induced within 24 h post-PNI in DRG of both sexes. However, males exhibited more robust upregulation of chemokines, stimulation of Neuroinflammation Signaling, and Toll-like Receptor Signaling. Males, but not females, exhibited robust DRG expression of at least 11 chemokine genes, including Ccl2, a known CCI-responding gene (Fu et al., 2010; LaCroix-Fralish et al., 2011) implicated in neuroinflammatory processes. Strong upregulation of Tlr genes in males included the X-linked endosomal Tlr7, Tlr8, and Tlr13, as well as moderate upregulation of Irf(s), Myd88, Irak3/4, and Nfkb genes. Remarkably, Tlr5 decreased in males. Tlrs has been shown to mediate pain preferentially in males by activating cytokine and membrane lipid raft remodeling (Stokes et al., 2013; Ramachandran et al., 2019; Miller et al., 2020). The early-response activation of the TLR Signaling in male DRG can promote pro-inflammatory responses and drive chemokines and cytokines. Il6, expressed by neuronal, glial, and immune cells in male (Dubový et al., 2010) and female DRG (Ko et al., 2016), is induced at 24 h post-PNI. It is worth noting that neuronal Il6 is also known as an axon growth-promoting RAG (Zhong et al., 1999; Cao et al., 2006; Ma and Willis, 2015). An important recent study demonstrated that IL17a directly activates nociceptor in mediating mechanical hypersensitivity selectively in female mice (Luo et al., 2021), consistent with the present finding of immediate and female-biased IL17ra induction in DRG.
Our data demonstrated monomorphic upregulation of injury-specific colony stimulation factor (Csf1) that is involved in macrophage proliferation and pain (Guan et al., 2016). However, the Csf1-receptor (Csf1r) and its downstream transmembrane adapter Tyrobp (Dap12) were upregulated in males. These findings may provide mechanistic insights into the mechanism of Csf1-induced macrophage expansion and pain regulation in male DRG reported on day 4 after spared nerve injury (Yu et al., 2020).
Sensory function is transcriptionally regulated by gene expression and axonal transport of chief neurotransmitter receptors, transporter, and ion channel genes. Their remarkable decline observed in both sexes is likely attributed to anterograde axonal transport to the regenerating nerve stump (Cavalli et al., 2005). The upregulated ion channels in both sexes, including Kcnk16, Kcnh4, and epithelial ENaC (Scnn1b), undergo axonal transport from DRG to peripheral terminals to participate in the transduction of painful sensation (García-Añoveros et al., 2001).
Under acute pathological conditions, estrogens can regulate a large spectrum of neuronal (Hucho et al., 2006) and immune functions (Straub, 2007) relevant to PNI. Estrogens (e.g., 17β-estradiol) activate nuclear Esr1 and Esr2 as well as membrane-bound GPCRs. While both female and male DRG display relatively high baseline Esr1 and Esr2 levels, the downstream signaling is likely influenced by the naturally higher 17β-estradiol levels in females. In the nervous system, rapid attenuation of estrogens in DRG can be complemented by neuron-specific biosynthesis (Evrard and Balthazart, 2004) in addition to estrogens of gonadal origin.
Extracellular matrix regulation in DRG affects markers of neurite growth, including ankyrin repeat protein Ankrd2 (Tsukamoto et al., 2008) in females and Ankrd1 (Stam et al., 2007) in both sexes. Male-specific upregulation of chimase (Cma1), extracellular Mmp19, Adamts4, and Mmp2, is coordinated with the robust expression of their stoichiometric regulator Timp1. Male-specific upregulation of the Timp1 gene and higher baseline mRNA and protein levels in females relative to males strongly suggest distinct sexually dimorphic regulation of Timp1 expression. We previously implicated epigenetic mechanisms in Mmp/Timp gene regulation (Chernov et al., 2009, 2010; Chernov and Strongin, 2011), resulting in rapid Timp1 induction at the site of PNI and DRG (Kim et al., 2012; Chernov et al., 2015; Liu et al., 2015). Accordingly, we proposed that epigenetically controlled MMPs, ADAMTSs, and TIMPs remodel the ECM to promote axonal growth, cell differentiation, migration and survival, and neurovascular permeability to immune cells in a sex-specific manner (Shubayev and Strongin, 2018).
Epigenetic cues of sex-specific PNI response are robust in DRG. Previously considered as “junk” (Richard Boland, 2017), ncRNAs perform as potent epigenetic regulators of post-PNI remodeling (Yao and Yu, 2019; Li P. et al., 2021). A portion of ncRNA in DRG is trafficked by axonal transport (Cavalli et al., 2005) to the injury site to participate in regenerative processes. We identified over 740 regulated ncRNA in DRG transcriptomes in both sexes. The imprinted trans-regulator H19 that modulates PI3K Signaling (Matouk et al., 2014) exhibited male-specific upregulation. Male-specific Mir142hg, the precursor gene for hematopoietic cell-specific Mir124, orchestrates innate immunity (Sharma, 2017; Shrestha et al., 2017) and neuropathic pain (Zhang et al., 2018; Li X. et al., 2021). Snhg1, Snhg4, and Snhg5 host ncRNAs, each encoding one or more small nucleolar RNAs (snoRNAs), increased predominantly in males. In females, Sox5os3 (an antisense transcript of the Sox5 gene) was distinctly upregulated. We identified a novel marker of early PNI in both sexes, the Lexis1 ncRNA, that may play a role in cholesterol metabolism and LXR pathway regulation (Sallam et al., 2016).
Upregulated Gadd45a/Gadd45g can guide epigenetic mechanisms responsible for Atf3, c-Jun, and Bdnf expression (Sultan and Sweatt, 2013). In addition, due to Gadd(s)/TET-dependent DNA demethylation, a broader gene activation may persist. Further studies of upstream regulators and DNA/histone methylation profiling are needed to establish sexually dimorphic characteristics of these epigenetic mechanisms in PNI.
Genomic localization on X/Y chromosomes likely contributes to sex-specific gene regulation in PNI. In contrast to males, in females, the RNA dosage is controlled by chromosome-wide epigenetic inactivation of one of the two X chromosomes (Xi), thus shutting off most Xi-linked transcripts. Because the X chromosome encodes the largest number of immunity-related genes (Bianchi et al., 2012), female immune cells rapidly leverage the expression of immune genes by partial reactivation of Xi (Syrett et al., 2017, 2019). We identified over 330 PNI-specific DEGs linked to the sex chromosomes, including female-specific upregulation of the X-linked TATA-box binding protein Taf7l (CT40) gene. The encoded Taf7l TF interacts with BORIS (a CTCF-family protein) (Rivero-Hinojosa et al., 2017) to promote long-range chromatin looping and regulation of large gene clusters (Zhou et al., 2014). These data imply a role of dynamic chromatin architecture in sexually dimorphic epigenetic regulation post-PNI. In males, transcripts on the Y chromosome, except for a short segment called the pseudoautosomal region (PAR), are not matched in the female genome. Two, albeit not well established, Y-linked genes were induced post-PNI in male DRG.
Additional considerations were given to the timing of sexually dimorphic changes, which directly relate to the temporal coordination of biological and behavioral processes. Generally, male DRG displayed a significantly higher number of differentially expressed genes, both up- and downregulated, in response to PNI within 24 h, than on day 7 (Ahlström et al., 2021) and day 14 PNI timepoints (Stephens et al., 2019). By comparing the most prominent sexually dimorphic transcripts at the early and later stages of PNI response, we observed consistent regulation of genes related to immunological response, neuronal transmission, regeneration, and plasticity obtained on day 7 (Ahlström et al., 2021) and day 14 PNI timepoints in rats (Stephens et al., 2019). However, as outlined in the present study, many sexually dimorphic and monomorphic transcripts were specific for the early stages of PNI response.
In conclusion, the present data provide insights into the regulation of the early-response transcriptional landscapes in DRG in female and male DRG after PNI. Epigenetic mechanisms emerged as essential hallmarks of sexually dimorphic regulation in early PNI response. The identity of the contributing cells in which these changes occur requires future studies to elucidate the functional activity of the respective genes and regulatory pathways in the context of specific cell types.
Reagents and resources are listed in the resource table (Supplementary Table 2).
Female and male C57BL6/J mice (6–8 weeks old, Jackson Labs) were randomly assigned to axotomy (n = 6/group) and sham (n = 4/group). The mice were housed in a temperature-controlled room (∼22°C), on a 12-h light/dark cycle, and free access to food and water. All procedures were conducted between 8 and 12 daytime. Under isoflurane anesthesia, the left sciatic nerve was exposed at the mid-thigh level. The entire width of the nerve was axotomized using sterile microsurgery scissors. In a sham surgery animal cohort, nerves were exposed using surgical scissors without transection. The muscle was then sutured, and the skin stapled. At 24 h after surgery, lumbar L4 and L5 DRG were collected for RNA isolation. All animal procedures were performed according to the Policy on Humane Care and Use of Laboratory Animals and the protocol approved by the Institutional Animal Care and Use Committee at the VA San Diego Healthcare System.
All surgical and tissue harvesting instruments were sterilized and repeatedly treated with RNAse Away reagent followed by a rinse in RNAse-free water. Tissues were immediately submerged in 500 μl RNAlater Stabilization Solution to preserve RNA integrity, placed at 4°C overnight, and then transferred for storage at −20°C. All sample groups were processed in parallel to minimize batch effects.
Dorsal root ganglia tissues from two animals were pooled to obtain at least 500 ng of total RNAs. Tissues were transferred in Trizol solution and disrupted by mechanical homogenization. Total RNAs were purified using RNeasy RNA purification reagents. RNA concentrations and quality were determined using Nanodrop absorbance ratios at 260/280 nm and 260/230 nm. RNA integrity was determined using the Agilent Bioanalyzer Nano RNA chip; 500 ng of total RNA samples (three replicates/group) with RIN ≥ 7.0 were used for RNA-seq.
mRNA libraries were generated following the TruSeq Stranded mRNA library preparation protocol (Illumina). In brief, the poly-A enriched mRNAs were purified using poly-T oligo coupled magnetic beads, followed by mRNA fragmentation, first and second strand synthesis, cleaning on AMPure XP beads, and 3′-adenylation. Ligation of TruSeq dual-index adapters was used for barcoding. The quality of RNA-seq libraries was validated using qPCR. Libraries were sized on Agilent Bioanalyzer DNA high sensitivity chip and normalized. RNA-seq was performed using the paired-end 100 cycle program on the NovaSeq 6000 system at the Genomics High Throughput Facility (University of California Irvine). Base calls were recorded and converted to FASTQ files containing sequencing reads and the corresponding quality scores using Illumina software. Sequencing was conducted until we acquired at least 50 million paired-end reads per sample.
FASTQ files were uploaded to the Amazon S3 server and processed on Elastic Compute Cloud instances [EC2; Amazon Web Services (AWS)] running Ubuntu Server 20.04 LTS (64-bit ARM). Data analysis steps are summarized in Supplementary Figure 1 and Supplementary Table 2. FASTQ files were filtered to remove low-quality bases, TruSeq dual-index adapter sequences, and unpaired reads using Trimmomatic (Bolger et al., 2014). Transcript-level quantification was performed using Salmon (Patro et al., 2017) in quasi-mapping mode using version M27 of the Gencode mouse genome. To correct systematic biases commonly present in RNA-seq data, both -seqBias and -gcBias features were used. Transcript- to gene-level conversion was done using Tximeta (Love et al., 2020). RNA-seq quantification data quality was assessed using MultiQC (Ewels et al., 2016).
Gene count matrices were imported into the DESeq2 package (Love et al., 2014). Outliers were identified by Cook’s distance method and excluded from further analysis. Dataset’s normalization was conducted using trimmed M-values (TMM) included in the DESeq2 package. Log2FC values were calculated using the Wald test. The adjusted (shrunken) log2FC values were calculated using the adaptive t-prior apeglm method (Zhu et al., 2019). Significant DEGs were identified by padj values below a false discovery rate (FDR) cutoff (padj < 0.1) (Supplementary Table 1). Padj < 0.1 were used in downstream analyses unless otherwise noted. Batch effects were controlled using removeBatchEffect (Ritchie et al., 2015) and RUVseq (Risso et al., 2014) functions. DEGs were visualized using ggVennDiagramm, PCAtools, ComplexHeatmap, and EnhancedVolcano R packages.
Biological interpretation of the regulated signaling pathways in female and male animals was conducted by the Ingenuity Pathway Analysis (IPA) based on causal network approaches (Krämer et al., 2014). Cut-off filtering was applied (padj < 0.1, |Log2FC| > 0.58) for IPA. Signaling pathway regulation directionality was estimated based on statistical z-scores. Gene ontology terms were retrieved from Geneontology1.
DRG tissues were ground on ice in the lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 0.1% SDS, and 5 mM EDTA) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific). Insoluble debris was removed by centrifugation (12,000 × g; 10 min). Protein concentration was measured using BCA protein assay. Lysates (15 μg of total protein) were separated by SDS-PAGE in 15% Tris-glycine gels (Bio-Rad). Proteins were transferred onto a PVDF membrane. The membrane was blocked in 5% non-fat milk (Bio-Rad) and incubated for 18 h at 4°C with monoclonal rabbit anti-Atf3 (1:1000 dilutions) or polyclonal goat anti-mouse Timp1 (1:1000 dilutions) followed by incubation for 1 h at ambient temperature with anti-rabbit IgG (1:2000 dilutions) or anti-goat (1:1000 dilutions) horseradish peroxidase-conjugated secondary antibody. For loading control, the membranes were re-probed using anti-GAPDH antibody (1:2000 dilutions). The blots were developed using SuperSignal West Dura Extended Duration Substrate kit (Thermo Fisher Scientific). Bands were digitized and quantitated using Chemidoc System (Bio-Rad).
Digitized immunoblotting data were used to calculate means and standard deviation. Analysis of variance (ANOVA) with post hoc Tukey test was conducted in R (version 4.1.1)2 to determine statistical significance.
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182318.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at the VA San Diego Healthcare System.
AC: conceptualization, methodology, software, formal analysis, investigation, data curation, writing – original draft, writing – review and editing, and visualization. VS: conceptualization, resources, project administration, funding acquisition, writing – original draft, and review and editing. Both authors contributed to the article and approved the submitted version.
This research was supported by the National Institutes of Health (NIH) RO1 DE022757 (to VS) and the Department of Veterans Affairs Merit Review Award 5I01BX000638 (to VS).
The content is solely the authors’ responsibility and does not necessarily represent the official views of the funding agencies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Jennifer Dolkas, Megh Jariwala, and Mila Angert for their expert technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.779024/full#supplementary-material
Supplementary Figure 1 | RNA-seq analysis workflow.
Supplementary Table 1 | Post-axotomy (24 h) DEGs in murine female and male DRG.
Supplementary Table 2 | Key resources table.
CNS, central nervous system; DEG, differentially expressed gene; DRG, dorsal root ganglia; FC, fold change; FDR, false discovery rate; IPA, Ingenuity Pathway Analysis; ncRNA, non-coding RNAs; padj, adjusted p-values; PNI, peripheral nerve injury; PNS, peripheral nervous system; RAG, regeneration-associated gene; RNA-seq, RNA sequencing; TF, transcription factor; TLR, Toll-like receptors.
Abe, N., and Cavalli, V. (2008). Nerve injury signaling. Curr. Opin. Neurobiol. 18, 276–283. doi: 10.1016/j.conb.2008.06.005
Ahlström, F. H. G., Mätlik, K., Viisanen, H., Blomqvist, K. J., Liu, X., Lilius, T. O., et al. (2021). Spared nerve injury causes sexually dimorphic mechanical allodynia and differential gene expression in spinal cords and dorsal root ganglia in rats. Mol. Neurobiol. 58, 5396–5419. doi: 10.1007/s12035-021-02447-1
Anderson, H. K. (1902). The nature of the lesions which hinder the development of nerve-cells and their processes. J. Physiol. 28, 499–513. doi: 10.1113/jphysiol.1902.sp000931
Bianchi, I., Lleo, A., Gershwin, M. E., and Invernizzi, P. (2012). The X chromosome and immune associated genes. J. Autoimmun. 38, J187–J192. doi: 10.1016/j.jaut.2011.11.012
Boerner, K. E., Chambers, C. T., Gahagan, J., Keogh, E., Fillingim, R. B., and Mogil, J. S. (2018). Conceptual complexity of gender and its relevance to pain. Pain 159, 2137–2141. doi: 10.1097/j.pain.0000000000001275
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolin, L. M., Verity, A. N., Silver, J. E., Shooter, E. M., and Abrams, J. S. (1995). Interleukin-6 production by Schwann cells and induction in sciatic nerve injury. J. Neurochem. 64, 850–858. doi: 10.1046/j.1471-4159.1995.64020850.x
Bosse, F., Hasenpusch-Theil, K., Kury, P., and Muller, H. W. (2006). Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J. Neurochem. 96, 1441–1457. doi: 10.1111/j.1471-4159.2005.03635.x
Bosse, F., Küry, P., and Müller, H. W. (2001). Gene expression profiling and molecular aspects in peripheral nerve regeneration. Restor. Neurol. Neurosci. 19, 5–18.
Cao, Z., Gao, Y., Bryson, J. B., Hou, J., Chaudhry, N., Siddiq, M., et al. (2006). The cytokine interleukin-6 is sufficient but not necessary to mimic the peripheral conditioning lesion effect on axonal growth. J. Neurosci. 26, 5565–5573. doi: 10.1523/jneurosci.0815-06.2006
Cavaillès, V., Dauvois, S., L’Horset, F., Lopez, G., Hoare, S., Kushner, P. J., et al. (1995). Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J 14, 3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x
Cavalli, V., Kujala, P., Klumperman, J., and Goldstein, L. S. (2005). Sunday driver links axonal transport to damage signaling. J. Cell Biol. 168, 775–787. doi: 10.1083/jcb.200410136
Chandran, V., Coppola, G., Nawabi, H., Omura, T., Versano, R., Huebner, E. A., et al. (2016). A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89, 956–970. doi: 10.1016/j.neuron.2016.01.034
Chattopadhyay, S., and Shubayev, V. I. (2009). MMP-9 controls Schwann cell proliferation and phenotypic remodeling via IGF-1 and ErbB receptor-mediated activation of MEK/ERK pathway. Glia 57, 1316–1325. doi: 10.1002/glia.20851
Chernov, A. V., Baranovskaya, S., Golubkov, V. S., Wakeman, D. R., Snyder, E. Y., Williams, R., et al. (2010). Microarray-based transcriptional and epigenetic profiling of matrix metalloproteinases, collagens, and related genes in cancer. J. Biol. Chem. 285, 19647–19659. doi: 10.1074/jbc.M109.088153
Chernov, A. V., Dolkas, J., Hoang, K., Angert, M., Srikrishna, G., Vogl, T., et al. (2015). The calcium-binding proteins S100A8 and S100A9 initiate the early inflammatory program in injured peripheral nerves. J. Biol. Chem. 290, 11771–11784. doi: 10.1074/jbc.M114.622316
Chernov, A. V., Hullugundi, S. K., Eddinger, K. A., Dolkas, J., Remacle, A. G., Angert, M., et al. (2020). A myelin basic protein fragment induces sexually dimorphic transcriptome signatures of neuropathic pain in mice. J. Biol. Chem. 295, 10807–10821. doi: 10.1074/jbc.RA120.013696
Chernov, A. V., Sounni, N. E., Remacle, A. G., and Strongin, A. Y. (2009). Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J. Biol. Chem. 284, 12727–12734. doi: 10.1074/jbc.M900273200
Chernov, A. V., and Strongin, A. Y. (2011). Epigenetic regulation of matrix metalloproteinases and their collagen substrates in cancer. Biomol. Concepts 2, 135–147. doi: 10.1515/BMC.2011.017
Costigan, M., Moss, A., Latremoliere, A., Johnston, C., Verma-Gandhu, M., Herbert, T. A., et al. (2009). T-cell infiltration and signaling in the adult dorsal spinal cord is a major contributor to neuropathic pain-like hypersensitivity. J. Neurosci. 29:14415. doi: 10.1523/JNEUROSCI.4569-09.2009
Devor, M. (1999). Unexplained peculiarities of the dorsal root ganglion. Pain 82, S27–S35. doi: 10.1016/s0304-3959(99)00135-9
Dubový, P., Klusáková, I., Svízenská, I., and Brázda, V. (2010). Satellite glial cells express IL-6 and corresponding signal-transducing receptors in the dorsal root ganglia of rat neuropathic pain model. Neuron Glia Biol. 6, 73–83. doi: 10.1017/s1740925x10000074
Evrard, H. C., and Balthazart, J. (2004). Rapid regulation of pain by estrogens synthesized in spinal dorsal horn neurons. J. Neurosci. 24, 7225–7229. doi: 10.1523/jneurosci.1638-04.2004
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fillingim, R. B., King, C. D., Ribeiro-Dasilva, M. C., Rahim-Williams, B., and Riley, J. L. III (2009). Sex, gender, and pain: a review of recent clinical and experimental findings. J. Pain 10, 447–485. doi: 10.1016/j.jpain.2008.12.001
Flegel, C., Schöbel, N., Altmüller, J., Becker, C., Tannapfel, A., Hatt, H., et al. (2015). RNA-Seq analysis of human trigeminal and dorsal root ganglia with a focus on chemoreceptors. PLoS One 10:e0128951. doi: 10.1371/journal.pone.0128951
Fu, E. S., Zhang, Y. P., Sagen, J., Candiotti, K. A., Morton, P. D., Liebl, D. J., et al. (2010). Transgenic inhibition of glial NF-kappa B reduces pain behavior and inflammation after peripheral nerve injury. Pain 148, 509–518. doi: 10.1016/j.pain.2010.01.001
García-Añoveros, J., Samad, T. A., Zuvela-Jelaska, L., Woolf, C. J., and Corey, D. P. (2001). Transport and localization of the DEG/ENaC ion channel BNaC1alpha to peripheral mechanosensory terminals of dorsal root ganglia neurons. J. Neurosci. 21, 2678–2686. doi: 10.1523/jneurosci.21-08-02678.2001
Gordon, T., Sulaiman, O., and Boyd, J. G. (2003). Experimental strategies to promote functional recovery after peripheral nerve injuries. J. Peripher. Nerv. Syst. 8, 236–250.
Greenspan, J. D., Craft, R. M., LeResche, L., Arendt-Nielsen, L., Berkley, K. J., Fillingim, R. B., et al. (2007). Studying sex and gender differences in pain and analgesia: a consensus report. Pain 132 Suppl 1, S26–S45. doi: 10.1016/j.pain.2007.10.014
Guan, Z., Kuhn, J. A., Wang, X., Colquitt, B., Solorzano, C., Vaman, S., et al. (2016). Injured sensory neuron-derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat. Neurosci. 19, 94–101. doi: 10.1038/nn.4189
He, Z., and Jin, Y. (2016). Intrinsic control of axon regeneration. Neuron 90, 437–451. doi: 10.1016/j.neuron.2016.04.022
Hucho, T. B., Dina, O. A., Kuhn, J., and Levine, J. D. (2006). Estrogen controls PKCepsilon-dependent mechanical hyperalgesia through direct action on nociceptive neurons. Eur. J. Neurosci. 24, 527–534. doi: 10.1111/j.1460-9568.2006.04913.x
Jiang, N., Li, H., Sun, Y., Yin, D., Zhao, Q., Cui, S., et al. (2014). Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural. Regen. Res. 9, 1186–1194. doi: 10.4103/1673-5374.135325
Jing, X., Wang, T., Huang, S., Glorioso, J. C., and Albers, K. M. (2012). The transcription factor Sox11 promotes nerve regeneration through activation of the regeneration-associated gene Sprr1a. Exp. Neurol. 233, 221–232. doi: 10.1016/j.expneurol.2011.10.005
Khomula, E. V., Ferrari, L. F., Araldi, D., and Levine, J. D. (2017). Sexual dimorphism in a reciprocal interaction of ryanodine and IP3 receptors in the induction of hyperalgesic priming. J. Neurosci. 37, 2032–2044. doi: 10.1523/JNEUROSCI.2911-16.2017
Kim, Y., Remacle, A. G., Chernov, A. V., Liu, H., Shubayev, I., Lai, C., et al. (2012). The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS One 7:e33664. doi: 10.1371/journal.pone.0033664
Ko, J. S., Eddinger, K. A., Angert, M., Chernov, A. V., Dolkas, J., Strongin, A. Y., et al. (2016). Spinal activity of interleukin 6 mediates myelin basic protein-induced allodynia. Brain Behav. Immun. 56, 378–389. doi: 10.1016/j.bbi.2016.03.003
Krämer, A., Green, J., Pollard, J. Jr., and Tugendreich, S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530. doi: 10.1093/bioinformatics/btt703
Kubo, T., Yamashita, T., Yamaguchi, A., Hosokawa, K., and Tohyama, M. (2002). Analysis of genes induced in peripheral nerve after axotomy using cDNA microarrays. J. Neurochem. 82, 1129–1136.
LaCroix-Fralish, M. L., Austin, J. S., Zheng, F. Y., Levitin, D. J., and Mogil, J. S. (2011). Patterns of pain: meta-analysis of microarray studies of pain. Pain 152, 1888–1898. doi: 10.1016/j.pain.2011.04.014
LaPaglia, D. M., Sapio, M. R., Burbelo, P. D., Thierry-Mieg, J., Thierry-Mieg, D., Raithel, S. J., et al. (2018). RNA-Seq investigations of human post-mortem trigeminal ganglia. Cephalalgia 38, 912–932. doi: 10.1177/0333102417720216
Lawson, S. N. (2002). Phenotype and function of somatic primary afferent nociceptive neurones with C-, Aδ- or Aα/β-Fibres. Exp. Physiol. 87, 239–244. doi: 10.1113/eph8702350
Lee, J., and Cho, Y. (2021). Potential roles of stem cell marker genes in axon regeneration. Exp. Mol. Med. 53, 1–7. doi: 10.1038/s12276-020-00553-z
Li, P., Jia, Y., Tang, W., Cui, Q., Liu, M., and Jiang, J. (2021). Roles of non-coding RNAs in central nervous system axon regeneration. Front. Neurosci. 15:630633. doi: 10.3389/fnins.2021.630633
Li, S., Liu, Q., Wang, Y., Gu, Y., Liu, D., Wang, C., et al. (2013). Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One 8:e57000. doi: 10.1371/journal.pone.0057000
Li, X., Wang, S., Yang, X., and Chu, H. (2021). miR-142-3p targets AC9 to regulate sciatic nerve injury-induced neuropathic pain by regulating the cAMP/AMPK signalling pathway. Int. J. Mol. Med. 47, 561–572. doi: 10.3892/ijmm.2020.4824
Liu, H., Angert, M., Nishihara, T., Shubayev, I., Dolkas, J., and Shubayev, V. I. (2015). Spinal glia division contributes to conditioning lesion–induced axon regeneration into the injured spinal cord. J. Neuropathol. Exp. Neurol. 74, 500–511. doi: 10.1097/nen.0000000000000192
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Love, M. I., Soneson, C., Hickey, P. F., Johnson, L. K., Pierce, N. T., Shepherd, L., et al. (2020). Tximeta: reference sequence checksums for provenance identification in RNA-seq. PLoS Comput. Biol. 16:e1007664. doi: 10.1371/journal.pcbi.1007664
Luo, X., Chen, O., Wang, Z., Bang, S., Ji, J., Lee, S. H., et al. (2021). IL-23/IL-17A/TRPV1 axis produces mechanical pain via macrophage-sensory neuron crosstalk in female mice. Neuron 109, 2691–2706.e5. doi: 10.1016/j.neuron.2021.06.015
Ma, T. C., and Willis, D. E. (2015). What makes a RAG regeneration associated? Front. Mol. Neurosci. 8:43. doi: 10.3389/fnmol.2015.00043
Mahar, M., and Cavalli, V. (2018). Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323–337. doi: 10.1038/s41583-018-0001-8
Mapplebeck, J. C., Beggs, S., and Salter, M. W. (2017). Molecules in pain and sex: a developing story. Mol. Brain 10:9. doi: 10.1186/s13041-017-0289-8
Mapplebeck, J. C. S., Beggs, S., and Salter, M. W. (2016). Sex differences in pain: a tale of two immune cells. Pain 157 Suppl 1, S2–S6. doi: 10.1097/j.pain.0000000000000389
Matouk, I. J., Raveh, E., Abu-lail, R., Mezan, S., Gilon, M., Gershtain, E., et al. (2014). Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta 1843, 1414–1426. doi: 10.1016/j.bbamcr.2014.03.023
McDonald, D., Cheng, C., Chen, Y., and Zochodne, D. (2006). Early events of peripheral nerve regeneration. Neuron Glia Biol. 2, 139–147.
Mecklenburg, J., Zou, Y., Wangzhou, A., Garcia, D., Lai, Z., Tumanov, A. V., et al. (2020). Transcriptomic sex differences in sensory neuronal populations of mice. Sci. Rep. 10:15278. doi: 10.1038/s41598-020-72285-z
Michaelevski, I., Segal-Ruder, Y., Rozenbaum, M., Medzihradszky, K. F., Shalem, O., Coppola, G., et al. (2010). Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 3:ra53. doi: 10.1126/scisignal.2000952
Miller, Y. I., Navia-Pelaez, J. M., Corr, M., and Yaksh, T. L. (2020). Lipid rafts in glial cells: role in neuroinflammation and pain processing. J. Lipid Res. 61, 655–666. doi: 10.1194/jlr.TR119000468
Mogil, J. S. (2012). Sex differences in pain and pain inhibition: multiple explanations of a controversial phenomenon. Nat. Rev. Neurosci. 13, 859–866. doi: 10.1038/nrn3360
North, R. Y., Li, Y., Ray, P., Rhines, L. D., Tatsui, C. E., Rao, G., et al. (2019). Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 142, 1215–1226. doi: 10.1093/brain/awz063
Paige, C., Barba-Escobedo, P. A., Mecklenburg, J., Patil, M., Goffin, V., Grattan, D. R., et al. (2020). Neuroendocrine mechanisms governing sex differences in hyperalgesic priming involve prolactin receptor sensory neuron signaling. J. Neurosci. 40, 7080–7090. doi: 10.1523/jneurosci.1499-20.2020
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Perkins, J. R., Antunes-Martins, A., Calvo, M., Grist, J., Rust, W., Schmid, R., et al. (2014). A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat. Mol. Pain 10:7. doi: 10.1186/1744-8069-10-7
Pevny, L. H., and Lovell-Badge, R. (1997). Sox genes find their feet. Curr. Opin. Genet. Dev. 7, 338–344. doi: 10.1016/s0959-437x(97)80147-5
Ramachandran, R., Wang, Z., Saavedra, C., DiNardo, A., Corr, M., Powell, S. B., et al. (2019). Role of Toll-like receptor 4 signaling in mast cell-mediated migraine pain pathway. Mol. Pain 15:1744806919867842. doi: 10.1177/1744806919867842
Ramon y Cajal, S. (1928). Degeneration and Regeneration of the Nervous System. Oxford: Clarendon Press.
Rao, S. N., and Pearse, D. D. (2016). Regulating axonal responses to injury: the intersection between signaling pathways involved in axon myelination and the inhibition of axon regeneration. Front. Mol. Neurosci. 9:33. doi: 10.3389/fnmol.2016.00033
Ray, P., Torck, A., Quigley, L., Wangzhou, A., Neiman, M., Rao, C., et al. (2018). Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 159, 1325–1345. doi: 10.1097/j.pain.0000000000001217
Ray, P. R., Khan, J., Wangzhou, A., Tavares-Ferreira, D., Akopian, A. N., Dussor, G., et al. (2019). Transcriptome analysis of the human tibial nerve identifies sexually dimorphic expression of genes involved in pain, inflammation, and neuro-immunity. Front. Mol. Neurosci. 12:37. doi: 10.3389/fnmol.2019.00037
Renthal, W., Tochitsky, I., Yang, L., Cheng, Y. C., Li, E., Kawaguchi, R., et al. (2020). Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron 108, 128–144.e9. doi: 10.1016/j.neuron.2020.07.026
Richard Boland, C. (2017). Non-coding RNA: It’s Not Junk. Dig. Dis. Sci. 62, 1107–1109. doi: 10.1007/s10620-017-4506-1
Risso, D., Ngai, J., Speed, T. P., and Dudoit, S. (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902. doi: 10.1038/nbt.2931
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Rivero-Hinojosa, S., Kang, S., Lobanenkov, V. V., and Zentner, G. E. (2017). Testis-specific transcriptional regulators selectively occupy BORIS-bound CTCF target regions in mouse male germ cells. Sci. Rep. 7:41279. doi: 10.1038/srep41279
Sallam, T., Jones, M. C., Gilliland, T., Zhang, L., Wu, X., Eskin, A., et al. (2016). Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128. doi: 10.1038/nature17674
Sharma, S. (2017). Immunomodulation: a definitive role of microRNA-142. Dev. Comp. Immunol. 77, 150–156. doi: 10.1016/j.dci.2017.08.001
Shiers, S., Klein, R. M., and Price, T. J. (2020). Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain 161, 2410–2424. doi: 10.1097/j.pain.0000000000001973
Shrestha, A., Mukhametshina, R. T., Taghizadeh, S., Vásquez-Pacheco, E., Cabrera-Fuentes, H., Rizvanov, A., et al. (2017). MicroRNA-142 is a multifaceted regulator in organogenesis, homeostasis, and disease. Dev. Dyn. 246, 285–290. doi: 10.1002/dvdy.24477
Shubayev, V. I., and Strongin, A. Y. (2018). Tissue inhibitors of metalloproteases strike a nerve. Neural Regen. Res. 13, 1890–1892. doi: 10.4103/1673-5374.239437
Simeoli, R., Montague, K., Jones, H. R., Castaldi, L., Chambers, D., Kelleher, J. H., et al. (2017). Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat. Commun. 8:1778. doi: 10.1038/s41467-017-01841-5
Snider, W. D., Zhou, F. Q., Zhong, J., and Markus, A. (2002). Signaling the pathway to regeneration. Neuron 35, 13–16. doi: 10.1016/s0896-6273(02)00762-6
Sorge, R. E., and Totsch, S. K. (2017). Sex differences in pain. J. Neurosci. Res. 95, 1271–1281. doi: 10.1002/jnr.23841
Stam, F. J., MacGillavry, H. D., Armstrong, N. J., de Gunst, M. C., Zhang, Y., van Kesteren, R. E., et al. (2007). Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur. J. Neurosci. 25, 3629–3637. doi: 10.1111/j.1460-9568.2007.05597.x
Stephens, K. E., Zhou, W., Ji, Z., Chen, Z., He, S., Ji, H., et al. (2019). Sex differences in gene regulation in the dorsal root ganglion after nerve injury. BMC Genomics 20:147. doi: 10.1186/s12864-019-5512-9
Stevanovic, M., Drakulic, D., Lazic, A., Ninkovic, D. S., Schwirtlich, M., and Mojsin, M. (2021). SOX transcription factors as important regulators of neuronal and glial differentiation during nervous system development and adult neurogenesis. Front. Mol. Neurosci. 14:654031. doi: 10.3389/fnmol.2021.654031
Stokes, J. A., Cheung, J., Eddinger, K., Corr, M., and Yaksh, T. L. (2013). Toll-like receptor signaling adapter proteins govern spread of neuropathic pain and recovery following nerve injury in male mice. J. Neuroinflammation 10:148. doi: 10.1186/1742-2094-10-148
Straub, R. H. (2007). The complex role of estrogens in inflammation. Endocr. Rev. 28, 521–574. doi: 10.1210/er.2007-0001
Sultan, F. A., and Sweatt, J. D. (2013). The role of the Gadd45 family in the nervous system: a focus on neurodevelopment, neuronal injury, and cognitive neuroepigenetics. Adv. Exp. Med. Biol. 793, 81–119. doi: 10.1007/978-1-4614-8289-5_6
Sun, F., and He, Z. (2010). Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr. Opin. Neurobiol. 20, 510–518. doi: 10.1016/j.conb.2010.03.013
Suzuki, N., and Ochi, H. (2020). Regeneration enhancers: a clue to reactivation of developmental genes. Dev. Growth Differ. 62, 343–354. doi: 10.1111/dgd.12654
Syrett, C. M., Paneru, B., Sandoval-Heglund, D., Wang, J., Banerjee, S., Sindhava, V., et al. (2019). Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 4:e126751. doi: 10.1172/jci.insight.126751
Syrett, C. M., Sindhava, V., Hodawadekar, S., Myles, A., Liang, G., Zhang, Y., et al. (2017). Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 13:e1007050. doi: 10.1371/journal.pgen.1007050
Takeda, M., Takahashi, M., and Matsumoto, S. (2009). Contribution of the activation of satellite glia in sensory ganglia to pathological pain. Neurosci. Biobehav. Rev. 33, 784–792. doi: 10.1016/j.neubiorev.2008.12.005
Tavares-Ferreira, D., Ray, P. R., Sankaranarayanan, I., Mejia, G. L., Wangzhou, A., Shiers, S., et al. (2020). Sex differences in nociceptor translatomes contribute to divergent prostaglandin signaling in male and female mice. bioRxiv [Preprint]. doi: 10.1101/2020.07.31.231753
Tsujino, H., Kondo, E., Fukuoka, T., Dai, Y., Tokunaga, A., Miki, K., et al. (2000). Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol. Cell. Neurosci. 15, 170–182. doi: 10.1006/mcne.1999.0814
Tsukamoto, Y., Hijiya, N., Yano, S., Yokoyama, S., Nakada, C., Uchida, T., et al. (2008). Arpp/Ankrd2, a member of the muscle ankyrin repeat proteins (MARPs), translocates from the I-band to the nucleus after muscle injury. Histochem. Cell Biol. 129, 55–64. doi: 10.1007/s00418-007-0348-9
Van Kesteren, R., Mason, M., MacGillavry, H., Smit, A., and Verhaagen, J. (2011). A gene network perspective on axonal regeneration. Front. Mol. Neurosci. 4:46. doi: 10.3389/fnmol.2011.00046
Xiao, H. S., Huang, Q. H., Zhang, F. X., Bao, L., Lu, Y. J., Guo, C., et al. (2002). Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc. Natl. Acad. Sci. U. S. A. 99, 8360–8365. doi: 10.1073/pnas.122231899
Yao, C., and Yu, B. (2019). Role of long noncoding RNAs and circular RNAs in nerve regeneration. Front. Mol. Neurosci. 12:165. doi: 10.3389/fnmol.2019.00165
Yao, D., Li, M., Shen, D., Ding, F., Lu, S., Zhao, Q., et al. (2013). Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci. Bull. 29, 321–332. doi: 10.1007/s12264-013-1340-0
Yu, X., Liu, H., Hamel, K. A., Morvan, M. G., Yu, S., Leff, J., et al. (2020). Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat. Commun. 11:264. doi: 10.1038/s41467-019-13839-2
Zhang, Y., Mou, J., Cao, L., Zhen, S., Huang, H., and Bao, H. (2018). MicroRNA-142-3p relieves neuropathic pain by targeting high mobility group box 1. Int. J. Mol. Med. 41, 501–510. doi: 10.3892/ijmm.2017.3222
Zhong, J., Dietzel, I. D., Wahle, P., Kopf, M., and Heumann, R. (1999). Sensory impairments and delayed regeneration of sensory axons in interleukin-6-deficient mice. J. Neurosci. 19, 4305–4313. doi: 10.1523/jneurosci.19-11-04305.1999
Zhou, H., Wan, B., Grubisic, I., Kaplan, T., and Tjian, R. (2014). TAF7L modulates brown adipose tissue formation. eLife 3:e02811. doi: 10.7554/eLife.02811
Zhou, L., and Shine, H. D. (2003). Neurotrophic factors expressed in both cortex and spinal cord induce axonal plasticity after spinal cord injury. J. Neurosci. Res. 74, 221–226. doi: 10.1002/jnr.10718
Zhu, A., Ibrahim, J. G., and Love, M. I. (2019). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092. doi: 10.1093/bioinformatics/bty895
Keywords: Mus musculus, sexual dimorphism, peripheral nerve injury, axotomy, RNA-seq, dorsal root ganglia
Citation: Chernov AV and Shubayev VI (2021) Sexual Dimorphism of Early Transcriptional Reprogramming in Dorsal Root Ganglia After Peripheral Nerve Injury. Front. Mol. Neurosci. 14:779024. doi: 10.3389/fnmol.2021.779024
Received: 17 September 2021; Accepted: 19 November 2021;
Published: 13 December 2021.
Edited by:
Wolfgang Liedtke, Regeneron Pharmaceuticals, Inc., United StatesReviewed by:
Temugin Berta, University of Cincinnati, United StatesCopyright © 2021 Chernov and Shubayev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrei V. Chernov, YWNoZXJub3ZAaGVhbHRoLnVjc2QuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.