
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mol. Neurosci. , 12 November 2021
Sec. Brain Disease Mechanisms
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.756613
This article is part of the Research Topic The Role of Epigenetics in Neuropsychiatric Disorders View all 9 articles
The lack of early diagnostic biomarkers for schizophrenia greatly limits treatment options that deliver therapeutic agents to affected cells at a timely manner. While previous schizophrenia biomarker research has identified various biological signals that are correlated with certain diseases, their reliability and practicality as an early diagnostic tool remains unclear. In this article, we discuss the use of atypical epigenetic and/or consequent transcriptional alterations (ETAs) as biomarkers of early-stage schizophrenia. Furthermore, we review the viability of discovering and applying these biomarkers through the use of cutting-edge technologies such as human induced pluripotent stem cell (iPSC)-derived neurons, brain models, and single-cell level analyses.
Neuropsychiatric diseases, such as autism, depression, and schizophrenia are neurological and psychiatric disorders that severely compromise diverse brain functions associated with normal activities. Despite decades of research, well-validated objective biological markers (measurable signals indicative of disease, infection, or injury) for neuropsychiatric diseases have remained stubbornly out of reach. This lack of objective biomarkers has relegated clinicians to diagnose neuropsychiatric diseases on the basis of phenomenological criteria (symptoms, signs, and course of illness), which, though useful in describing the disease in a rough and ready way, continues to produce heated controversies regarding the efficacy of properly diagnosing various, if not all, neuropsychiatric diseases (Bhati, 2013; Malaspina et al., 2013; Wakefield, 2016; Parker, 2019). In addition, the absence of biomarkers indicative of early-stage neuropsychiatric diseases undermines timely delivery of therapeutic agents to diseased neurons before neural impairment becomes so severe the damage cannot be reversed. Thus, perhaps unsurprisingly, temporally specific disease type-targeted treatment options for neuropsychiatric diseases remain largely unavailable.
The main cause for the lack of objective neuropsychiatric biomarkers is arguably in the absence of proper disease models (which can be used for studying/identifying early signals indicative of neuropsychiatric diseases) that accurately reflect the development and symptoms of early-stage neuropsychiatric diseases. Traditionally, modeling early-stage neuropsychiatric diseases has been exceptionally difficult as the model system must (1) reliably retain both genetic and environmental risk factors associated with neuropsychiatric diseases, (2) faithfully reproduce disease-specific early-stage neural aberrations, and (3) accommodate practical and ethical considerations. As such, these challenges have prevented the rigorous biological investigation of early-stage biomarkers for neuropsychiatric disorders.
Induced pluripotent stem cell (iPSC) and organoid technology are novel and emerging methods that largely bypass the difficulties laid out above. Hence, in this article, we highlight studies that model schizophrenia, a neuropsychiatric disorder characterized by episodes of psychosis, using patient-derived neural organoids. In addition, we propose utilizing single-cell level analyses in combination with organoid technology to discover disease-associated epigenetic and/or consequent transcriptional alterations (ETAs)-based biomarkers, a novel class of biomarkers previously proposed to be applicable in diagnosing neurodegenerative diseases (Lee et al., 2020). Finally, we discuss various obstacles in diagnosing asymptomatic patients using newly discovered ETA-based biomarkers and suggest solutions for these challenges.
Schizophrenia is a devastating psychiatric illness that affects approximately 1% of the population worldwide (Saha et al., 2008; Stilo and Murray, 2010). The disease consists of varying degrees of positive (hallucinations, delusions, disorganized speech and behavior, psychomotor agitation, and hypersensitivity to psychoactive drugs such as ketamine, phencyclidine, amphetamine, and cocaine), negative (social withdrawal, anhedonia, flattened affect, and alogia), and cognitive (deficits in attention, working memory, executive function, and reversal learning) symptoms (Perez and Lodge, 2014; Sigurdsson, 2016). The majority of these symptoms, aside from certain positive symptoms (Carbon and Correll, 2014; Leucht et al., 2020), currently have few or no effective pharmacological treatments.
Although the etiology of schizophrenia remains largely elusive, post-mortem studies of schizophrenic individuals have suggested defects in neurodevelopmental processes may be a cause. This is largely due to the discovery of macroscopic and histologic alterations in the brains of schizophrenic individuals. These alterations include: diminished brain volume, increased ventricle size, and abnormal cortical and hippocampal neural structures (Brown et al., 1986; Bogerts et al., 1993a,b), without clear pathological inclusion bodies, dystrophic neurites, or neurodegeneration (Wong and Van Tol, 2003). Hence, it remains possible that the detected structural abnormalities may not be directly considered a cause for schizophrenia, but merely a consequence of the disease.
Accumulating evidence suggests abnormal neural development caused by neurotransmitter dysfunction in multiple brain regions is likely to underpin schizophrenia (Lewis and Levitt, 2002; Fatemi and Folsom, 2009; Sigurdsson and Duvarci, 2015; Zamanpoor, 2020). The dopamine hypothesis, which remains the central dogma of schizophrenia for decades, posits that excessive subcortical dopamine signaling in the mesolimbic and nigrostriatal pathways is responsible for positive disease symptoms (Meltzer and Stahl, 1976; Weinberger, 1987; Davis et al., 1991; McCutcheon et al., 2019). However, the failure of dopamine blockage to relieve some patient symptoms suggests other neurotransmitters may play a role. Studies found that impaired glutamate signaling may also contribute to schizophrenia by causing functional deficits in inhibitory interneurons, resulting in disruption of the excitation-inhibition balance in the cortical and hippocampal regions (Benes and Berretta, 2001; Beasley et al., 2002; Hashimoto et al., 2003; Goto and Grace, 2005; Lodge and Grace, 2007; Lodge et al., 2009; Boley et al., 2014). This imbalance consequently leads to hyperactivity in the downstream dopamine system as well as abnormal neural circuit functions in other brain regions, which may culminate in the development of schizophrenia (Perez and Lodge, 2014; Sigurdsson, 2016). Lastly, some studies also implicate the serotonin pathway in schizophrenia development (Abdolmaleky et al., 2004; Alex and Pehek, 2007; Stepnicki et al., 2018). It is important to note, however, that the dopamine, glutamate, or serotonin hypotheses are not mutually exclusive and can be connected at the neural circuit level. Furthermore, studies suggesting neurotransmitter dysfunction contributes to schizophrenia also implicate that certain cell-types associated with the production and/or acquisition of neurotransmitters are central in the pathogenesis of schizophrenia.
While the underlying causes for abnormal neural development are currently unknown, genetic studies, including gene mapping, linkage analysis, genome-wide association studies, and next-generation sequencing, have uncovered schizophrenia susceptibility-associated single-nucleotide polymorphisms and schizophrenia-risk genes over the years (Zamanpoor, 2020). With a heritability rate of 79–81%, one may assume that schizophrenia can largely be explained through aberrations in genetic makeup and can be diagnosed through screening of key disease-associated genetic loci. Although genetically engineered animal models have reported evidence establishing the potential association between genetic risk factors and the disease (Winship et al., 2019), it is generally accepted that schizophrenia results from the interaction between increased susceptibility caused by genetic risk factors and environmental events that trigger the psychotic episodes (Ayhan et al., 2016). Thus, the diagnosis of schizophrenia through the screening of disease-associated genetic risk factors alone has proven to be extremely challenging. To diagnose schizophrenia properly, it is imperative that we find other ways to separate the healthy from the vulnerable.
Because the treatment of schizophrenia and its symptoms are currently unavailable, scientists have alternatively attempted to alleviate the disorder by intervening the disease development in its early-stage, thereby ultimately preventing the accumulation of disease-associated neural impairments. This approach has garnered considerable attention as studies have consistently shown early identification and treatment of psychotic symptoms markedly improves clinical outcomes (Kulhara et al., 2008; Cheng and Schepp, 2016). For example, previous studies have shown a statistical correlation between prolonged Durations of Untreated Psychosis (DUPs) and poor clinical outcomes (McGlashan, 1999; Compton, 2004; Qin et al., 2014). Given the importance of early treatment for pre-symptomatic schizophrenic patients, a great deal of research has been conducted to find objective, well-validated, biomarkers for schizophrenia.
Recent efforts in search for schizophrenia biomarkers have yielded various potential signs that signal both disease presence and severity (Table 1). For example, neuroimaging-based studies have identified strong candidate biomarkers based on dopamine hyperactivity, hippocampal hyperactivity, immune dysregulation, N-methyl-D-aspartate receptor hypofunction, and cortical gray matter volume loss (Bogerts et al., 1990; Suddath et al., 1990; Shenton et al., 1992; Dierks et al., 1999; Hirayasu et al., 2000; Kasai et al., 2003; Koo et al., 2006; Ongur et al., 2006; Talati et al., 2014; Tregellas et al., 2014; Torres et al., 2016; Kraguljac et al., 2021). Additionally, research suggests some physiological complications observed in schizophrenic patients may also be used as potential biomarkers. For example, early auditory information processing (EAIP), has been proposed as a physiological biomarker of schizophrenia, as auditory discrimination-based testing is a highly quantitative and reliable measure of brain activity (Javitt et al., 1997; Perez et al., 2017). Finally, bio-fluids, such as blood (Lai et al., 2016) and cerebrospinal fluid (Huang et al., 2006), have been used as patient-derived sources for biomarkers to diagnose schizophrenia. Research indicates that the analysis of patient-derived blood anolytes (small molecules and proteins) distinguishes healthy controls from schizophrenic patients with up to 83% sensitivity and specificity (Schwarz et al., 2010). However, it is important to note that the biomarkers described above need significant improvements before being used generally. In the following sections, we will discuss what improvements must be made as well as potential ways in which these improvements can be achieved.
The primary concern with the biomarkers described previously is their questionable utility as a schizophrenia biomarker in the early stages of disease development. While the latest renditions of schizophrenia biomarkers are becoming increasingly practical and accurate, most, if not all, current biomarkers are inapplicable to pre-symptomatic patients. Schizophrenia, as well as many other neurological diseases, is easily identifiable in structured interviews in its post-onset stages. Therefore, biomarkers that are used in post-onset patient-derived samples, run the risk of being redundant by nature, unless the biomarker can provide additional information on top of the rough and ready interview-based diagnosis. For instance, biomarkers high in specificity can distinguish schizophrenia by subtype and help decide the administration of patient-specific treatments. Instead, to fully exploit the utility of biomarkers, they should be used to detect disease-specifying, ideally disease subtype-specifying, signals before the phenotypic onset occurs to administer disease-specific therapeutics as early as possible. For this, it is not only essential that the selected biomarkers themselves be observable in early-stage schizophrenia, but also necessary that such biomarkers be found in specific cell-types. We have named these biomarker containing cells first-responding disease-associated cell types (FDCs).
To achieve this, we suggest the use of disease-associated ETAs be used as early-stage schizophrenia biomarkers, as they have high potential to be exploited as an early signal for schizophrenia. Additionally, we propose that (1) these biomarkers and FDCs should be discovered in patient-derived neural organoids (such as synthesized miniature brains) and (2) found biomarkers should be applied in discovered FDCs generated through patient-derived iPSCs. By using these cutting-edge methods, we believe reliably identifying pre-symptomatic schizophrenic patients is possible. We further delineate our proposal in the following sections.
Before the first onset of a frank schizophrenic episode, patients experience clinical features such as mild physical anomalies, poor motor coordination, mild cognitive impairments, social deficits, anxiety, sadness, lability, irritability, sleep disturbances, cognitive impairment in attention, concentration, mild illusions, suspiciousness, magical thinking, substance use, social withdrawal, and preoccupations (McGlashan, 1996; McGlashan and Johannessen, 1996; Lieberman et al., 2001). Since these premorbid and prodromal pathological symptoms are generally mild in nature, it is no surprise that these symptoms cannot be considered as biomarkers for diagnostics (Erlenmeyer-Kimling and Cornblatt, 1987). This becomes particularly clear when considering these phenomena largely overlap with range of mental and behavioral experiences of young adults who do not subsequently develop schizophrenia.
Research, however, indicates that pre-symptomatic patients undergo a series of neurological perturbations that render the brain at “high-risk.” The pathophysiological processes during the premorbid and prodromal stages are largely categorized as neurodevelopmental (inductive, patterning, and synaptogenetic anomalies) and neuroplastic (post-pubertal hormonal effects, myelination, and synaptic regression) dysfunctions. Both pathological dysfunctions and their phenotypic manifestations are highly diverse, suggesting that the biological mechanism underpinning such broad aberrations may be associated with system-wide atypical cellular changes in certain specific disease-associated cell-types, such as FDCs. Here, ETAs are highlighted as alterations in ETAs are likely responsible for these system-wide changes.
It must be pointed out that research on the early stages of schizophrenia is mostly incomplete. Perhaps the most obvious reason underlying this shortcoming is in the lack of disease models that retain genetic and environmental risk factors associated with schizophrenia while validly reproducing early-stage biological insults. Animal models of schizophrenia, while can be helpful in investigating some contributions of schizophrenia-associated genetic aberrations and/or environmental factors, are largely inadequate in recapitulating the complex, neuron-specific, interactions between genetic, epigenetic, and transcriptional factors observed in schizophrenic patients.
These complications have led researchers to model schizophrenia using patient-derived iPSC cellular models. Schizophrenia iPSC lines were first established from Disrupted in schizophrenia 1 (DISC1) mutation-carrying chronic paranoid schizophrenia patient in 2011 (Chiang et al., 2011). Since then, iPSC and its derivative technologies, such as patient-derived organoids, have steadily enabled a wide range of studies investigating schizophrenia. We have summarized key studies that modeled schizophrenia using iPSC or its derivative technologies in Table 2.
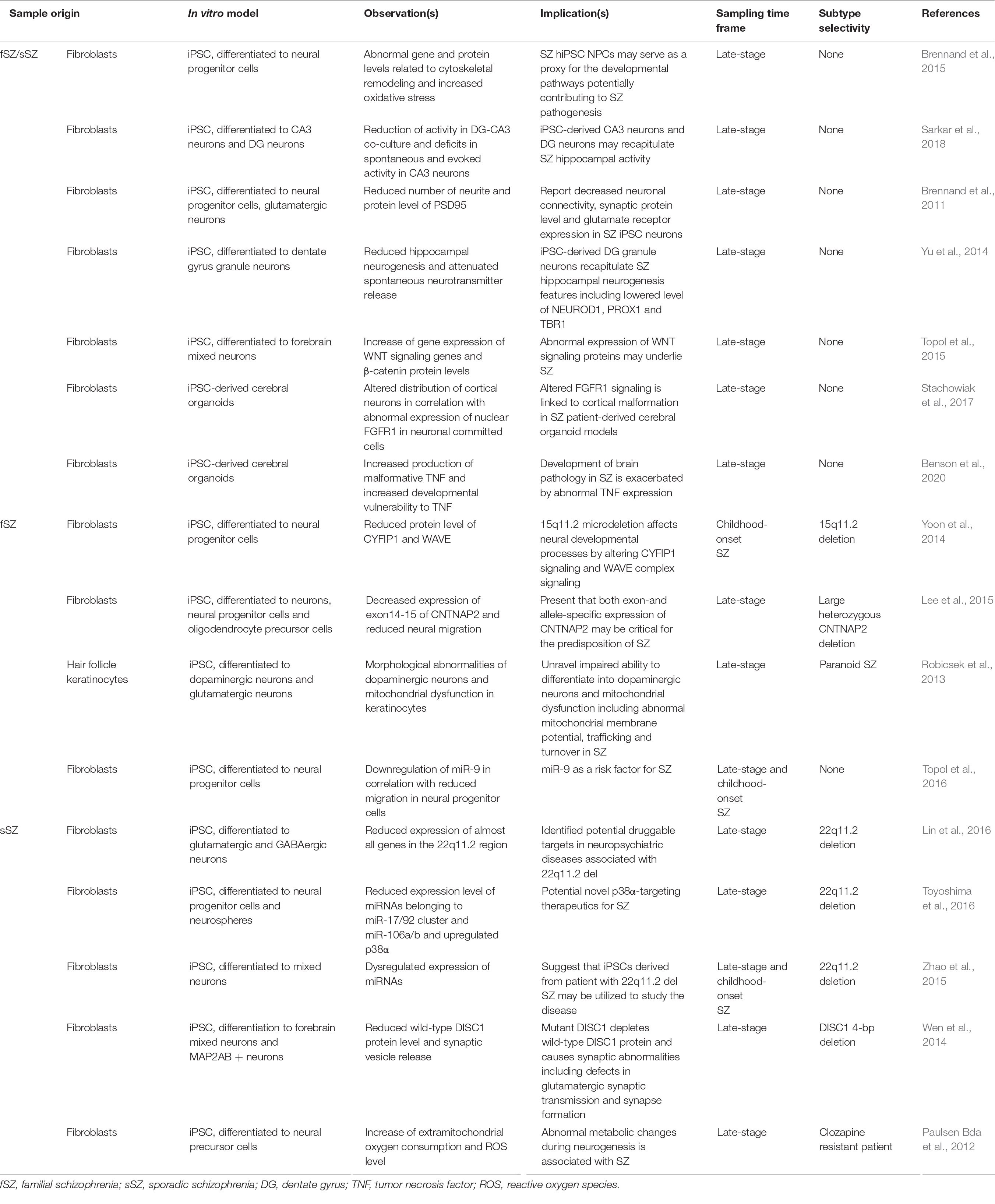
Table 2. Selected reports utilizing iPSC models and organoid technologies for the characterization of schizophrenia.
No matter how advanced iPSC modeling of schizophrenia becomes, the utility of iPSC or its derivative technologies as a method to sample ETA-based biomarker sources relies on whether patient-derived samples have measurable differences in epigenetic, or other transcriptional profiles, as compared to healthy controls. Increasingly, more and more studies continue to provide strong evidence for this notion. For example, an early study by Brennand et al. (2011) showed that differentiated iPSC neurons from a schizophrenic patient had impaired neuronal connectivity, decreased neurite number, expression of synaptic proteins such as PSD-95, and, importantly, altered transcript expression. Similarly, multiple studies have shown reduced levels of neuronal differentiation-associated genes in neural progenitor cells from schizophrenic patient-derived iPSCs (Yu et al., 2014), suggesting a link between altered microRNA profiling (Zhao et al., 2015; Topol et al., 2016; Akkouh et al., 2021), chromatin openness in which disease variants affecting neurodevelopment reside (Forrest et al., 2017; Zhang et al., 2020), and protein expression patterns (Tiihonen et al., 2019). These results demonstrate (1) iPSC-derived neurons, but not other cell-types, from schizophrenia patients faithfully recapitulate many pathological disease features and (2) iPSC-derived neurons may be used to detect uniquely altered transcriptional, epigenetic, and proteome signals that could be utilized as potential biomarkers.
The fact that ETAs are observed in iPSC lines differentiated into neurons, but not other non-disease-associated cell- types strongly suggests that schizophrenia primarily targets neural cells, which, as previously mentioned above, is already widely accepted (Iritani, 2013). This becomes particularly important in the search for cell-types that exhibits early manifestation of measurable ETA-based signals because it suggests that among various cell-types intercommunicating within the brain, neurons are particularly sensitive to schizophrenia-associated perturbations. Here, single-cell level analyses are highlighted, as these technologies, combined with patient-derived organoids, could allow investigations into various specific cell-subtypes in a wide range of brain cells early in its disease development. In fact, single-cell level analyses have already attracted attention as methods that allow improved characterization of schizophrenia (Skene et al., 2018; Sawada et al., 2020). Bulk-level tissue sampling provides limited resolution for identifying cell-type-specific abnormalities. However, with single-cell analysis, cells are characterized by their RNA and/or transcriptome and grouped by their cell state and type. Exemplifying the capability of single-cell RNA sequencing analysis, Sawada et al. (2020), showed developmental excitation-inhibition imbalance in neural organoids derived from monozygotic twins discordant for psychosis. The authors further demonstrated that GABAergic neurons, but not other cell-types, showed alterations in their RNA profiles. Interestingly, these results suggest iPSC-derived GABAergic neurons may be FDCs that could be used to identify ETA-based biomarkers, such as those observed by Sawada et al. (2020). Collectively, we suggest that the discovery of both ETA-based biomarkers and FDCs is possible through the combined use of organoid technology and single cell-level analyses.
For our strategy to be used as a method to diagnose schizophrenia to the general public, it is important to consider the cost of such approach. Unfortunately, iPSC generation and single-cell sequencing are among the most difficult and expensive techniques in modern biomedicine. To put it in numbers, iPSC reprogramming of 20,000 cells costs an estimated $3,000 ∼ $9,000 per sample ($0.1.5 ∼ $0.45 per cell). On top of that, if the generation of iPSC-derived neurons produces not only pure FDCs but also heterogeneous cell-types, each sample will cost an additional estimated $2,000 ∼ $8,000 for single-cell analysis. Predicting the cost of our strategy is further complicated by the fact that the expenditure of iPSC generation and single-cell sequencing is determined by various relevant factors. For example, the cost of iPSC generation is determined by the source of peripheral cell (fibroblast, blood cell), reprogramming methods (viral, non-viral) as well as the target differentiation cell-type (cerebral organoid, mid-brain, dopaminergic neuron, GABAergic neuron). The cost of single-cell sequencing is determined by protocols involving the preparation of cells (methods such as single-nucleus RNA sequencing, drop-sequencing), the number of cells analyzed per sample, as well as the number of sequencing reads per cells. If the price of our method is too high, the clinical application loses practical merit regardless of the prospects of diagnosing schizophrenia in its early phase.
Despite these obstacles, we are still optimistic in predicting that the technologies will decrease in price for the following three reasons. First, much like many other technological advances, the price for newly developed technologies will likely decrease after the quality of product reaches a certain threshold. Between now and when iPSC generation technology and single-cell sequencing were first introduced (2006 and 2009, respectively; Takahashi and Yamanaka, 2006; Tang et al., 2009), the technological advancement has been focused in increasing both quality and purity of product, instead of decreasing the cost of the technologies. Therefore, to project the cost of single-cell sequencing and iPSC generation out into the future with past data points in cost may be mis-leading. Second, compiled meta-analysis and data sharing technologies may help bypass additional sampling required for the discovery of new ETA-based biomarkers. Third, assuming that technological advancement reaches to a point where reliable generation of pure FDCs are produced at a high quantity, diagnosing asymptomatic schizophrenic patients would only require targeted generation of FDCs using cell type-specific iPSC without the use of expensive single-cell analysis technologies (Figure 1). To sum, we believe that the cost concern surrounding iPSC generation and single-cell level analysis is quite manageable given the rapid technological advancement we have seen in the past decade.
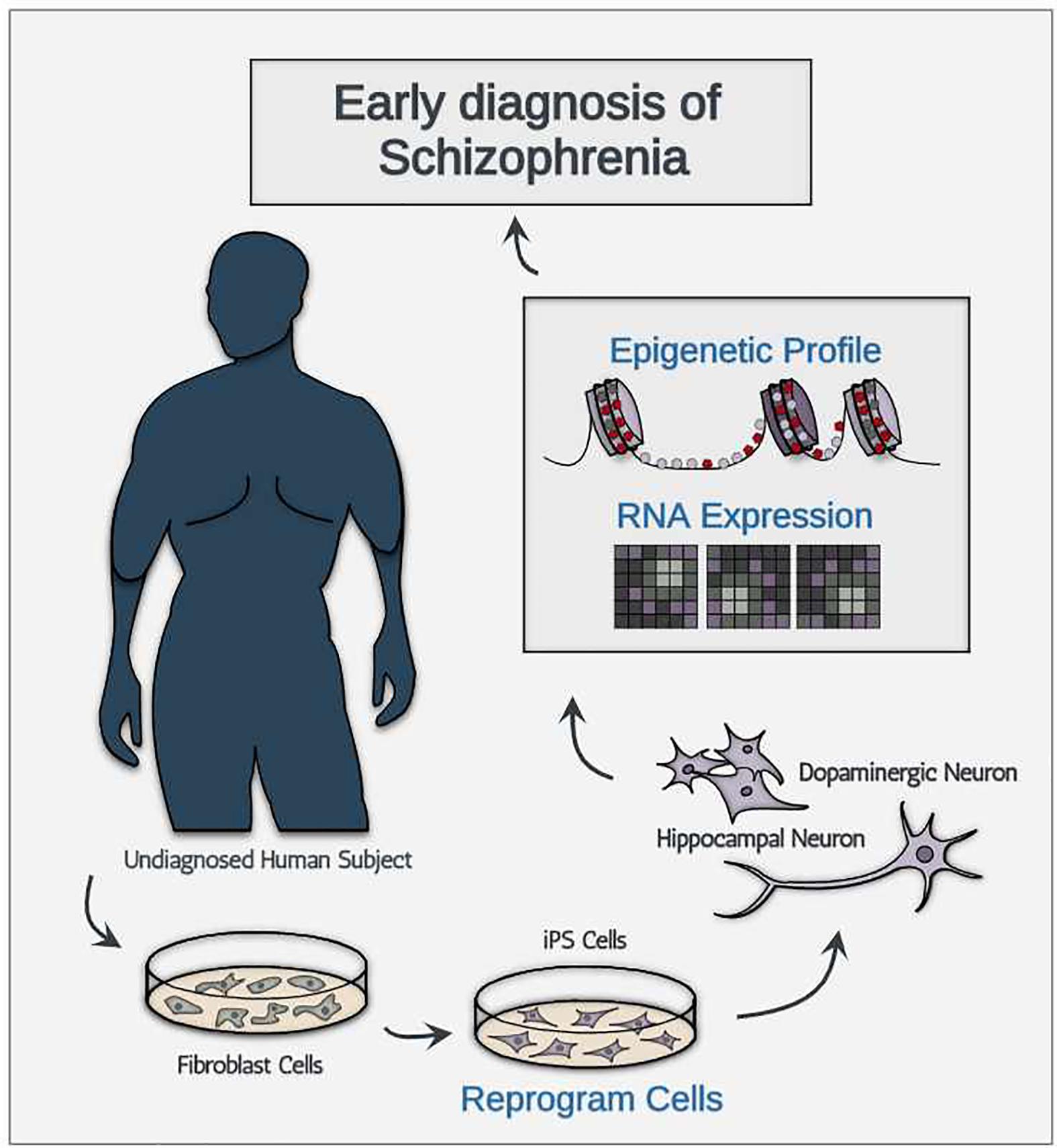
Figure 1. A schematic illustration showing the conceptual diagnostic workflow consisting of iPSC-derived neurons for early diagnosis of schizophrenia.
Despite the clear advantages of biomarkers, psychiatrists and clinicians have yet to adopt any of the previously devised neuroimaging-, physiological-, or blood sample-based biomarkers for diagnosing schizophrenia. As noted by the developers of a blood sample-based biomarker for schizophrenia (VeriPsych), this reluctance may be due to the fact that “most psychiatrists believe that they are very good at diagnosing schizophrenia patients using a basic clinical interview” (Bahn et al., 2011). Unfortunately, this shows that (1) research has failed to generate a practically applicable early-stage biomarker that is more reliable in diagnosing schizophrenia than structured interviews and (2) clinicians are largely not benefiting from diagnosing schizophrenia based on raw biological data (which can specify schizophrenia by subtype and help decide the administration of patient-specific treatments). It is for these reasons we developed our strategy to practically meet the demands of practitioners who treat schizophrenia. Although our proposed strategy may help early diagnosis and, thus, intervention of schizophrenia, there are potential concerns surrounding our strategy. Below, we describe and address these concerns.
Given that our proposed strategy requires ETA-based biomarkers originating from disease-associated genetic aberrations, our model could be considered only applicable to inherited forms of schizophrenia. From this consideration, two concerns emerge: (1) the redundancy of ETA-based biomarkers and (2) the limited generalizability of our strategy. Concerns about the redundancy of ETA-based biomarkers stem from a pragmatic point of view where reliable early schizophrenia diagnosis is likely best achieved through a concerted effort combining multiple biomarkers including: genomics, physiology, and ETAs. However, if ETA profile alterations are observed only in inherited schizophrenia cases and abnormal ETA profiles develop from genetic differences between healthy and schizophrenic patients, identifying high-risk schizophrenic patients could be achieved by simply screening genetic loci associated with schizophrenia without using ETA-based biomarkers. While the concern surrounding the redundancy of our proposal is, to an extent, legitimate, careful examination of ETA-based biomarkers may provide significant advantages over traditional genomics-based schizophrenia diagnosis.
For instance, biomarkers that exploit altered disease-specific ETA profiles may help identify high-risk schizophrenic patients with hidden or otherwise unknown genetic risk factors. While genome-wide association studies have identified more than 100 schizophrenia susceptibility genes and predicted more than 1,000 (Shaw et al., 1998; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Li et al., 2017; Pardinas et al., 2018), the rate to which familial genetic aberrations are linked to phenotypic psychosis is strikingly low despite the near 80% inheritance rate when either parent is diagnosed with schizophrenia (Cardno and Gottesman, 2000). This implies that not only is screening for genetic loci associated with schizophrenia not practical but that some cases of inherited schizophrenia are caused by currently unknown genetic changes. ETA-based biomarkers may be effective in identifying high-risk schizophrenic patients that were assigned low-risk using genetic screening alone, assuming these unknown genetic changes lead to disease-associated ETAs.
In fact, the advantages of ETA-based biomarkers over traditional genomics-based biomarker are also closely connected to the concerns about the possible limited generalizability of our strategy. Schizophrenia affects approximately 1% of the population worldwide. About 40% of the disease is thought to be familial cases, with the other 60% occurring sporadically in individuals with no family history of the disease. Interestingly, research indicates that approximately 10% of these sporadic cases are linked to de novo germline mutations (Xu et al., 2008). This suggests that, if these various de novo germline mutations can produce stereotypical, easily measurable, alterations in ETA profiles, ETA-based biomarkers could be used to identify sporadic schizophrenia cases. It is worth pointing out, however, that since it is currently unknown how much, and how distinct, disease-specific genetic abnormalities translate directly into alterations in ETA profiles, further studies investigating the unique ETA profiles of schizophrenic patients are needed.
In this review, we have presented our perspectives on the discovery and the practical application of ETA-based biomarkers for early diagnosis of schizophrenia based on recent advances in patient-derived iPSC and organoids. Although potential issues such as initial high cost may prevent widespread use of ETA-based biomarkers, we are firmly optimistic that our proposed strategy will be generally useful, especially considering the rapid pace at which these technologies continue to advance.
DL, JS, HL, and SL conceptualized the theme of the review and wrote the manuscript together. HJ contributed to Tables 1, 2. All authors contributed to the article and approved the submitted version.
This work was supported by Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science and Information and Communications Technology (ICT) (2021R1A2C1003817 and 2019R1A4A1024278); the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) and Korea Dementia Research Center (KDRC), funded by the Ministry of Health & Welfare and Ministry of Science and ICT, Republic of Korea (HU21C0027) (to SL); the KBRI basic research program through Korea Brain Research Institute funded by Ministry of Science and ICT (21-BR-03-02 to SL; 21-BR-03-03 to HL); the DGIST R&D Program of the Ministry of Science and ICT (21-IJRP-01) (to HL); and the National Research Foundation of Korea (NRF-2017M3C7A1047640) (to JS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdolmaleky, H. M., Faraone, S. V., Glatt, S. J., and Tsuang, M. T. (2004). Meta-analysis of association between the T102C polymorphism of the 5HT2a receptor gene and schizophrenia. Schizophr. Res. 67, 53–62.
Abi-Dargham, A., Rodenhiser, J., Printz, D., Zea-Ponce, Y., Gil, R., Kegeles, L. S., et al. (2000). Increased baseline occupancy of D2 receptors by dopamine in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 97, 8104–8109. doi: 10.1073/pnas.97.14.8104
Akkouh, I. A., Hughes, T., Steen, V. M., Glover, J. C., Andreassen, O. A., Djurovic, S., et al. (2021). Transcriptome analysis reveals disparate expression of inflammation-related miRNAs and their gene targets in iPSC-astrocytes from people with schizophrenia. Brain Behav. Immun. 94, 235–244. doi: 10.1016/j.bbi.2021.01.037
Alex, K. D., and Pehek, E. A. (2007). Pharmacologic mechanisms of serotonergic regulation of dopamine neurotransmission. Pharmacol. Ther. 113, 296–320. doi: 10.1016/j.pharmthera.2006.08.004
Ayhan, Y., McFarland, R., and Pletnikov, M. V. (2016). Animal models of gene-environment interaction in schizophrenia: a dimensional perspective. Prog. Neurobiol. 136, 1–27. doi: 10.1016/j.pneurobio.2015.10.002
Bahn, S., Noll, R., Barnes, A., Schwarz, E., and Guest, P. C. (2011). Challenges of introducing new biomarker products for neuropsychiatric disorders into the market. Int. Rev. Neurobiol. 101, 299–327. doi: 10.1016/B978-0-12-387718-5.00012-2
Beasley, C. L., Zhang, Z. J., Patten, I., and Reynolds, G. P. (2002). Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biol. Psychiatry 52, 708–715. doi: 10.1016/s0006-3223(02)01360-4
Benes, F. M., and Berretta, S. (2001). GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology 25, 1–27. doi: 10.1016/S0893-133X(01)00225-1
Benson, C. A., Powell, H. R., Liput, M., Dinham, S., Freedman, D. A., Ignatowski, T. A., et al. (2020). Immune factor, TNFalpha, disrupts human brain organoid development similar to schizophrenia-schizophrenia increases developmental vulnerability to TNFalpha. Front. Cell Neurosci. 14:233. doi: 10.3389/fncel.2020.00233
Bhati, M. T. (2013). Defining psychosis: the evolution of DSM-5 schizophrenia spectrum disorders. Curr. Psychiatry Rep. 15:409. doi: 10.1007/s11920-013-0409-9
Bloemen, O. J., de Koning, M. B., Gleich, T., Meijer, J., de Haan, L., Linszen, D. H., et al. (2013). Striatal dopamine D2/3 receptor binding following dopamine depletion in subjects at Ultra High Risk for psychosis. Eur. Neuropsychopharmacol. 23, 126–132. doi: 10.1016/j.euroneuro.2012.04.015
Bogerts, B., Ashtari, M., Degreef, G., Alvir, J. M., Bilder, R. M., and Lieberman, J. A. (1990). Reduced temporal limbic structure volumes on magnetic resonance images in first episode schizophrenia. Psychiatry Res. 35, 1–13. doi: 10.1016/0925-4927(90)90004-p
Bogerts, B., Falkai, P., Greve, B., Schneider, T., and Pfeiffer, U. (1993a). The neuropathology of schizophrenia: past and present. J. Hirnforsch. 34, 193–205.
Bogerts, B., Lieberman, J. A., Ashtari, M., Bilder, R. M., Degreef, G., Lerner, G., et al. (1993b). Hippocampus-amygdala volumes and psychopathology in chronic schizophrenia. Biol. Psychiatry 33, 236–246. doi: 10.1016/0006-3223(93)90289-p
Boley, A. M., Perez, S. M., and Lodge, D. J. (2014). A fundamental role for hippocampal parvalbumin in the dopamine hyperfunction associated with schizophrenia. Schizophr. Res. 157, 238–243. doi: 10.1016/j.schres.2014.05.005
Brennand, K., Savas, J. N., Kim, Y., Tran, N., Simone, A., Hashimoto-Torii, K., et al. (2015). Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry 20, 361–368. doi: 10.1038/mp.2014.22
Brennand, K. J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., et al. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225. doi: 10.1038/nature09915
Brown, R., Colter, N., Corsellis, J. A., Crow, T. J., Frith, C. D., Jagoe, R., et al. (1986). Postmortem evidence of structural brain changes in schizophrenia. Differences in brain weight, temporal horn area, and parahippocampal gyrus compared with affective disorder. Arch. Gen. Psychiatry 43, 36–42. doi: 10.1001/archpsyc.1986.01800010038005
Carbon, M., and Correll, C. U. (2014). Thinking and acting beyond the positive: the role of the cognitive and negative symptoms in schizophrenia. CNS Spectr. 19(Suppl. 1), 38–52. doi: 10.1017/S1092852914000601
Cardno, A. G., and Gottesman, I. I. (2000). Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am. J. Med. Genet. 97, 12–17.
Cazzullo, C. L., Scarone, S., Grassi, B., Vismara, C., Trabattoni, D., Clerici, M., et al. (1998). Cytokines production in chronic schizophrenia patients with or without paranoid behaviour. Prog. Neuropsychopharmacol. Biol. Psychiatry 22, 947–957. doi: 10.1016/s0278-5846(98)00059-1
Cheng, S. C., and Schepp, K. G. (2016). Early intervention in schizophrenia: a literature review. Arch. Psychiatr. Nurs. 30, 774–781. doi: 10.1016/j.apnu.2016.02.009
Chiang, C. H., Su, Y., Wen, Z., Yoritomo, N., Ross, C. A., Margolis, R. L., et al. (2011). Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol. Psychiatry 16, 358–360. doi: 10.1038/mp.2011.13
Compton, M. T. (2004). Duration of untreated psychosis significantly associated with positive symptoms one year after treatment. Evid. Based. Ment. Health 7:101. doi: 10.1136/ebmh.7.4.101
Davis, K. L., Kahn, R. S., Ko, G., and Davidson, M. (1991). Dopamine in schizophrenia: a review and reconceptualization. Am. J. Psychiatry 148, 1474–1486. doi: 10.1176/ajp.148.11.1474
Dierks, T., Linden, D. E., Jandl, M., Formisano, E., Goebel, R., Lanfermann, H., et al. (1999). Activation of Heschl’s gyrus during auditory hallucinations. Neuron 22, 615–621. doi: 10.1016/s0896-6273(00)80715-1
Dracheva, S., Marras, S. A., Elhakem, S. L., Kramer, F. R., Davis, K. L., and Haroutunian, V. (2001). N-methyl-D-aspartic acid receptor expression in the dorsolateral prefrontal cortex of elderly patients with schizophrenia. Am. J. Psychiatry 158, 1400–1410. doi: 10.1176/appi.ajp.158.9.1400
Erlenmeyer-Kimling, L., and Cornblatt, B. (1987). High-risk research in schizophrenia: a summary of what has been learned. J. Psychiatr. Res. 21, 401–411. doi: 10.1016/0022-3956(87)90087-2
Fatemi, S. H., and Folsom, T. D. (2009). The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr. Bull. 35, 528–548. doi: 10.1093/schbul/sbn187
Forrest, M. P., Zhang, H., Moy, W., McGowan, H., Leites, C., Dionisio, L. E., et al. (2017). Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell 21, 305.e8–318.e8. doi: 10.1016/j.stem.2017.07.008
Garey, L. J., Ong, W. Y., Patel, T. S., Kanani, M., Davis, A., Mortimer, A. M., et al. (1998). Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J. Neurol. Neurosurg. Psychiatry 65, 446–453. doi: 10.1136/jnnp.65.4.446
Goto, Y., and Grace, A. A. (2005). Dopaminergic modulation of limbic and cortical drive of nucleus accumbens in goal-directed behavior. Nat. Neurosci. 8, 805–812. doi: 10.1038/nn1471
Hashimoto, T., Volk, D. W., Eggan, S. M., Mirnics, K., Pierri, J. N., Sun, Z., et al. (2003). Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J. Neurosci. 23, 6315–6326.
Hirayasu, Y., Potts, G. F., O’Donnell, B. F., Kwon, J. S., Arakaki, H., Akdag, S. J., et al. (1998). Auditory mismatch negativity in schizophrenia: topographic evaluation with a high-density recording montage. Am. J. Psychiatry 155, 1281–1284. doi: 10.1176/ajp.155.9.1281
Hirayasu, Y., Shenton, M. E., Salisbury, D. F., and McCarley, R. W. (2000). Hippocampal and superior temporal gyrus volume in first-episode schizophrenia. Arch. Gen. Psychiatry 57, 618–619. doi: 10.1001/archpsyc.57.6.618
Hope, S., Ueland, T., Steen, N. E., Dieset, I., Lorentzen, S., Berg, A. O., et al. (2013). Interleukin 1 receptor antagonist and soluble tumor necrosis factor receptor 1 are associated with general severity and psychotic symptoms in schizophrenia and bipolar disorder. Schizophr. Res. 145, 36–42. doi: 10.1016/j.schres.2012.12.023
Howes, O. D., Montgomery, A. J., Asselin, M. C., Murray, R. M., Valli, I., Tabraham, P., et al. (2009). Elevated striatal dopamine function linked to prodromal signs of schizophrenia. Arch. Gen. Psychiatry 66, 13–20. doi: 10.1001/archgenpsychiatry.2008.514
Howes, O. D., Williams, M., Ibrahim, K., Leung, G., Egerton, A., McGuire, P. K., et al. (2013). Midbrain dopamine function in schizophrenia and depression: a post-mortem and positron emission tomographic imaging study. Brain 136, 3242–3251. doi: 10.1093/brain/awt264
Huang, J. T., Leweke, F. M., Oxley, D., Wang, L., Harris, N., Koethe, D., et al. (2006). Disease biomarkers in cerebrospinal fluid of patients with first-onset psychosis. PLoS Med. 3:e0030428. doi: 10.1371/journal.pmed.0030428
Iritani, S. (2013). What happens in the brain of schizophrenia patients: an investigation from the viewpoint of neuropathology. Nagoya J. Med. Sci. 75, 11–28.
Javitt, D. C., Strous, R. D., Grochowski, S., Ritter, W., and Cowan, N. (1997). Impaired precision, but normal retention, of auditory sensory (”echoic”) memory information in schizophrenia. J. Abnorm. Psychol. 106, 315–324. doi: 10.1037//0021-843x.106.2.315
Kaalund, S. S., Newburn, E. N., Ye, T., Tao, R., Li, C., Deep-Soboslay, A., et al. (2014). Contrasting changes in DRD1 and DRD2 splice variant expression in schizophrenia and affective disorders, and associations with SNPs in postmortem brain. Mol. Psychiatry 19, 1258–1266. doi: 10.1038/mp.2013.165
Kasai, K., Shenton, M. E., Salisbury, D. F., Hirayasu, Y., Onitsuka, T., Spencer, M. H., et al. (2003). Progressive decrease of left Heschl gyrus and planum temporale gray matter volume in first-episode schizophrenia: a longitudinal magnetic resonance imaging study. Arch. Gen. Psychiatry 60, 766–775. doi: 10.1001/archpsyc.60.8.766
Kim, Y. K., Kim, L., and Lee, M. S. (2000). Relationships between interleukins, neurotransmitters and psychopathology in drug-free male schizophrenics. Schizophr. Res. 44, 165–175. doi: 10.1016/s0920-9964(99)00171-1
Koo, M. S., Dickey, C. C., Park, H. J., Kubicki, M., Ji, N. Y., Bouix, S., et al. (2006). Smaller neocortical gray matter and larger sulcal cerebrospinal fluid volumes in neuroleptic-naive women with schizotypal personality disorder. Arch. Gen. Psychiatry 63, 1090–1100. doi: 10.1001/archpsyc.63.10.1090
Kraguljac, N. V., McDonald, W. M., Widge, A. S., Rodriguez, C. I., Tohen, M., and Nemeroff, C. B. (2021). Neuroimaging biomarkers in schizophrenia. Am. J. Psychiatry 178, 509–521. doi: 10.1176/appi.ajp.2020.20030340
Kraguljac, N. V., White, D. M., Reid, M. A., and Lahti, A. C. (2013). Increased hippocampal glutamate and volumetric deficits in unmedicated patients with schizophrenia. JAMA Psychiatry 70, 1294–1302. doi: 10.1001/jamapsychiatry.2013.2437
Kristiansen, L. V., Beneyto, M., Haroutunian, V., and Meador-Woodruff, J. H. (2006). Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol. Psychiatry 11, 737–750. doi: 10.1038/sj.mp.4001844
Kulhara, P., Banerjee, A., and Dutt, A. (2008). Early intervention in schizophrenia. Indian J. Psychiatry 50, 128–134. doi: 10.4103/0019-5545.42402
Kung, L., and Roberts, R. C. (1999). Mitochondrial pathology in human schizophrenic striatum: a postmortem ultrastructural study. Synapse 31, 67–75.
Lai, C. Y., Scarr, E., Udawela, M., Everall, I., Chen, W. J., and Dean, B. (2016). Biomarkers in schizophrenia: a focus on blood based diagnostics and theranostics. World J. Psychiatry 6, 102–117. doi: 10.5498/wjp.v6.i1.102
Laruelle, M., Abi-Dargham, A., Gil, R., Kegeles, L., and Innis, R. (1999). Increased dopamine transmission in schizophrenia: relationship to illness phases. Biol. Psychiatry 46, 56–72. doi: 10.1016/s0006-3223(99)00067-0
Lee, D., Choi, Y. H., Seo, J., Kim, J. K., and Lee, S. B. (2020). Discovery of new epigenomics-based biomarkers and the early diagnosis of neurodegenerative diseases. Ageing Res. Rev. 61:101069. doi: 10.1016/j.arr.2020.101069
Lee, I. S., Carvalho, C. M., Douvaras, P., Ho, S. M., Hartley, B. J., Zuccherato, L. W., et al. (2015). Characterization of molecular and cellular phenotypes associated with a heterozygous CNTNAP2 deletion using patient-derived hiPSC neural cells. NPJ Schizophr. 1:15019. doi: 10.1038/npjschz.2015.19
Leucht, S., Crippa, A., Siafis, S., Patel, M. X., Orsini, N., and Davis, J. M. (2020). Dose-response meta-analysis of antipsychotic drugs for acute schizophrenia. Am. J. Psychiatry 177, 342–353. doi: 10.1176/appi.ajp.2019.19010034
Lewis, D. A., and Levitt, P. (2002). Schizophrenia as a disorder of neurodevelopment. Annu. Rev. Neurosci. 25, 409–432. doi: 10.1146/annurev.neuro.25.112701.142754
Li, Y., Tong, S., Liu, D., Gai, Y., Wang, X., Wang, J., et al. (2008). Abnormal EEG complexity in patients with schizophrenia and depression. Clin. Neurophysiol. 119, 1232–1241. doi: 10.1016/j.clinph.2008.01.104
Li, Z., Chen, J., Yu, H., He, L., Xu, Y., Zhang, D., et al. (2017). Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49, 1576–1583. doi: 10.1038/ng.3973
Liddle, P. F., Friston, K. J., Frith, C. D., Hirsch, S. R., Jones, T., and Frackowiak, R. S. (1992). Patterns of cerebral blood flow in schizophrenia. Br. J. Psychiatry 160, 179–186. doi: 10.1192/bjp.160.2.179
Lieberman, J. A., Perkins, D., Belger, A., Chakos, M., Jarskog, F., Boteva, K., et al. (2001). The early stages of schizophrenia: speculations on pathogenesis, pathophysiology, and therapeutic approaches. Biol. Psychiatry 50, 884–897. doi: 10.1016/s0006-3223(01)01303-8
Lin, M., Pedrosa, E., Hrabovsky, A., Chen, J., Puliafito, B. R., Gilbert, S. R., et al. (2016). Integrative transcriptome network analysis of iPSC-derived neurons from schizophrenia and schizoaffective disorder patients with 22q11.2 deletion. BMC Syst. Biol. 10:105. doi: 10.1186/s12918-016-0366-0
Lodge, D. J., Behrens, M. M., and Grace, A. A. (2009). A loss of parvalbumin-containing interneurons is associated with diminished oscillatory activity in an animal model of schizophrenia. J. Neurosci. 29, 2344–2354. doi: 10.1523/JNEUROSCI.5419-08.2009
Lodge, D. J., and Grace, A. A. (2007). Aberrant hippocampal activity underlies the dopamine dysregulation in an animal model of schizophrenia. J. Neurosci. 27, 11424–11430. doi: 10.1523/JNEUROSCI.2847-07.2007
Malaspina, D., Owen, M. J., Heckers, S., Tandon, R., Bustillo, J., Schultz, S., et al. (2013). Schizoaffective disorder in the DSM-5. Schizophr. Res. 150, 21–25. doi: 10.1016/j.schres.2013.04.026
McCutcheon, R. A., Abi-Dargham, A., and Howes, O. D. (2019). Schizophrenia, dopamine and the striatum: from biology to symptoms. Trends Neurosci. 42, 205–220. doi: 10.1016/j.tins.2018.12.004
McGlashan, T. H. (1996). Early detection and intervention in schizophrenia: research. Schizophr. Bull. 22, 327–345. doi: 10.1093/schbul/22.2.327
McGlashan, T. H. (1999). Duration of untreated psychosis in first-episode schizophrenia: marker or determinant of course? Biol. Psychiatry 46, 899–907. doi: 10.1016/s0006-3223(99)00084-0
McGlashan, T. H., and Johannessen, J. O. (1996). Early detection and intervention with schizophrenia: rationale. Schizophr. Bull. 22, 201–222. doi: 10.1093/schbul/22.2.201
Meltzer, H. Y., and Stahl, S. M. (1976). The dopamine hypothesis of schizophrenia: a review. Schizophr. Bull. 2, 19–76. doi: 10.1093/schbul/2.1.19
Mittleman, B. B., Castellanos, F. X., Jacobsen, L. K., Rapoport, J. L., Swedo, S. E., and Shearer, G. M. (1997). Cerebrospinal fluid cytokines in pediatric neuropsychiatric disease. J. Immunol. 159, 2994–2999.
Muller, N., Dobmeier, P., Empl, M., Riedel, M., Schwarz, M., and Ackenheil, M. (1997). Soluble IL-6 receptors in the serum and cerebrospinal fluid of paranoid schizophrenic patients. Eur. Psychiatry 12, 294–299. doi: 10.1016/S0924-9338(97)84789-X
Ohnuma, T., Kato, H., Arai, H., Faull, R. L., McKenna, P. J., and Emson, P. C. (2000). Gene expression of PSD95 in prefrontal cortex and hippocampus in schizophrenia. Neuroreport 11, 3133–3137. doi: 10.1097/00001756-200009280-00019
Ong, W. Y., and Garey, L. J. (1993). Ultrastructural features of biopsied temporopolar cortex (area 38) in a case of schizophrenia. Schizophr. Res. 10, 15–27. doi: 10.1016/0920-9964(93)90072-q
Ongur, D., Cullen, T. J., Wolf, D. H., Rohan, M., Barreira, P., Zalesak, M., et al. (2006). The neural basis of relational memory deficits in schizophrenia. Arch. Gen. Psychiatry 63, 356–365. doi: 10.1001/archpsyc.63.4.356
Owen, F., Cross, A. J., Crow, T. J., Longden, A., Poulter, M., and Riley, G. J. (1978). Increased dopamine-receptor sensitivity in schizophrenia. Lancet 2, 223–226. doi: 10.1016/s0140-6736(78)91740-3
Pandey, G. N., Ren, X., Rizavi, H. S., and Zhang, H. (2015). Proinflammatory cytokines and their membrane-bound receptors are altered in the lymphocytes of schizophrenia patients. Schizophr. Res. 164, 193–198. doi: 10.1016/j.schres.2015.02.004
Pardinas, A. F., Holmans, P., Pocklington, A. J., Escott-Price, V., Ripke, S., Carrera, N., et al. (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389. doi: 10.1038/s41588-018-0059-2
Parker, G. (2019). How well does the DSM-5 capture schizoaffective disorder? Can. J. Psychiatry 64, 607–610. doi: 10.1177/0706743719856845
Paulsen Bda, S., de Moraes Maciel, R., Galina, A., Souza da Silveira, M., dos Santos Souza, C., Drummond, H., et al. (2012). Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant 21, 1547–1559. doi: 10.3727/096368911X600957
Perez, S. M., and Lodge, D. J. (2014). New approaches to the management of schizophrenia: focus on aberrant hippocampal drive of dopamine pathways. Drug Des. Dev. Ther. 8, 887–896. doi: 10.2147/DDDT.S42708
Perez, V. B., Tarasenko, M., Miyakoshi, M., Pianka, S. T., Makeig, S. D., Braff, D. L., et al. (2017). Mismatch negativity is a sensitive and predictive biomarker of perceptual learning during auditory cognitive training in schizophrenia. Neuropsychopharmacology 42, 2206–2213. doi: 10.1038/npp.2017.25
Pilowsky, L. S., Bressan, R. A., Stone, J. M., Erlandsson, K., Mulligan, R. S., Krystal, J. H., et al. (2006). First in vivo evidence of an NMDA receptor deficit in medication-free schizophrenic patients. Mol. Psychiatry 11, 118–119. doi: 10.1038/sj.mp.4001751
Qin, H., Zhang, J., Wang, Z., Min, H., Yan, C., Chen, F., et al. (2014). Duration of untreated psychosis and clinical outcomes of first-episode schizophrenia: a 4-year follow-up study. Shanghai Arch. Psychiatry 26, 42–48. doi: 10.3969/j.issn.1002-0829.2014.01.006
Roberts, R. C., Conley, R., Kung, L., Peretti, F. J., and Chute, D. J. (1996). Reduced striatal spine size in schizophrenia: a postmortem ultrastructural study. Neuroreport 7, 1214–1218. doi: 10.1097/00001756-199604260-00024
Robicsek, O., Karry, R., Petit, I., Salman-Kesner, N., Muller, F. J., Klein, E., et al. (2013). Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol. Psychiatry 18, 1067–1076. doi: 10.1038/mp.2013.67
Rothermundt, M., Arolt, V., Weitzsch, C., Eckhoff, D., and Kirchner, H. (1996). Production of cytokines in acute schizophrenic psychosis. Biol. Psychiatry 40, 1294–1297. doi: 10.1016/S0006-3223(96)00360-5
Saha, S., Chant, D., and McGrath, J. (2008). Meta-analyses of the incidence and prevalence of schizophrenia: conceptual and methodological issues. Int. J. Methods Psychiatr. Res. 17, 55–61. doi: 10.1002/mpr.240
Sarkar, A., Mei, A., Paquola, A. C. M., Stern, S., Bardy, C., Klug, J. R., et al. (2018). Efficient generation of CA3 neurons from human pluripotent stem cells enables modeling of hippocampal connectivity In vitro. Cell Stem Cell 22, 684–697. doi: 10.1016/j.stem.2018.04.009
Sawada, T., Chater, T. E., Sasagawa, Y., Yoshimura, M., Fujimori-Tonou, N., Tanaka, K., et al. (2020). Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol. Psychiatry 25, 2695–2711. doi: 10.1038/s41380-020-0844-z
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. doi: 10.1038/nature13595
Schwarz, E., Izmailov, R., Spain, M., Barnes, A., Mapes, J. P., Guest, P. C., et al. (2010). Validation of a blood-based laboratory test to aid in the confirmation of a diagnosis of schizophrenia. Biomark Insights 5, 39–47. doi: 10.4137/bmi.s4877
Seeman, P., Guan, H. C., and Van Tol, H. H. (1993). Dopamine D4 receptors elevated in schizophrenia. Nature 365, 441–445. doi: 10.1038/365441a0
Shaw, S. H., Kelly, M., Smith, A. B., Shields, G., Hopkins, P. J., Loftus, J., et al. (1998). A genome-wide search for schizophrenia susceptibility genes. Am. J. Med. Genet. 81, 364–376.
Shenton, M. E., Kikinis, R., Jolesz, F. A., Pollak, S. D., LeMay, M., Wible, C. G., et al. (1992). Abnormalities of the left temporal lobe and thought disorder in schizophrenia. A quantitative magnetic resonance imaging study. N. Engl. J. Med. 327, 604–612. doi: 10.1056/NEJM199208273270905
Sigurdsson, T. (2016). Neural circuit dysfunction in schizophrenia: insights from animal models. Neuroscience 321, 42–65. doi: 10.1016/j.neuroscience.2015.06.059
Sigurdsson, T., and Duvarci, S. (2015). Hippocampal-prefrontal interactions in cognition, behavior and psychiatric disease. Front. Syst. Neurosci. 9:190. doi: 10.3389/fnsys.2015.00190
Silbersweig, D. A., Stern, E., Frith, C., Cahill, C., Holmes, A., Grootoonk, S., et al. (1995). A functional neuroanatomy of hallucinations in schizophrenia. Nature 378, 176–179. doi: 10.1038/378176a0
Skene, N. G., Bryois, J., Bakken, T. E., Breen, G., Crowley, J. J., Gaspar, H. A., et al. (2018). Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833. doi: 10.1038/s41588-018-0129-5
Stachowiak, E. K., Benson, C. A., Narla, S. T., Dimitri, A., Chuye, L. E. B., Dhiman, S., et al. (2017). Cerebral organoids reveal early cortical maldevelopment in schizophrenia-computational anatomy and genomics, role of FGFR1. Transl. Psychiatry 7:6. doi: 10.1038/s41398-017-0054-x
Stepnicki, P., Kondej, M., and Kaczor, A. A. (2018). Current concepts and treatments of schizophrenia. Molecules 23:2087. doi: 10.3390/molecules23082087
Stilo, S. A., and Murray, R. M. (2010). The epidemiology of schizophrenia: replacing dogma with knowledge. Dialog. Clin. Neurosci. 12, 305–315.
Suddath, R. L., Christison, G. W., Torrey, E. F., Casanova, M. F., and Weinberger, D. R. (1990). Anatomical abnormalities in the brains of monozygotic twins discordant for schizophrenia. N. Engl. J. Med. 322, 789–794. doi: 10.1056/NEJM199003223221201
Sweet, R. A., Pierri, J. N., Auh, S., Sampson, A. R., and Lewis, D. A. (2003). Reduced pyramidal cell somal volume in auditory association cortex of subjects with schizophrenia. Neuropsychopharmacology 28, 599–609. doi: 10.1038/sj.npp.1300120
Tada, M., Nagai, T., Kirihara, K., Koike, S., Suga, M., Araki, T., et al. (2016). Differential alterations of auditory gamma oscillatory responses between pre-onset high-risk individuals and first-episode schizophrenia. Cereb. Cortex 26, 1027–1035. doi: 10.1093/cercor/bhu278
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Talati, P., Rane, S., Kose, S., Blackford, J. U., Gore, J., Donahue, M. J., et al. (2014). Increased hippocampal CA1 cerebral blood volume in schizophrenia. Neuroimage Clin. 5, 359–364. doi: 10.1016/j.nicl.2014.07.004
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382. doi: 10.1038/nmeth.1315
Tiihonen, J., Koskuvi, M., Storvik, M., Hyotylainen, I., Gao, Y., Puttonen, K. A., et al. (2019). Sex-specific transcriptional and proteomic signatures in schizophrenia. Nat. Commun. 10:3933. doi: 10.1038/s41467-019-11797-3
Topol, A., Zhu, S., Hartley, B. J., English, J., Hauberg, M. E., Tran, N., et al. (2016). Dysregulation of miRNA-9 in a subset of schizophrenia patient-derived neural progenitor cells. Cell Rep. 15, 1024–1036. doi: 10.1016/j.celrep.2016.03.090
Topol, A., Zhu, S., Tran, N., Simone, A., Fang, G., and Brennand, K. J. (2015). Altered WNT signaling in human induced pluripotent stem cell neural progenitor cells derived from four schizophrenia patients. Biol. Psychiatry 78, e29–e34. doi: 10.1016/j.biopsych.2014.12.028
Torres, U. S., Duran, F. L., Schaufelberger, M. S., Crippa, J. A., Louza, M. R., Sallet, P. C., et al. (2016). Patterns of regional gray matter loss at different stages of schizophrenia: a multisite, cross-sectional VBM study in first-episode and chronic illness. Neuroimage Clin. 12, 1–15. doi: 10.1016/j.nicl.2016.06.002
Toyoshima, M., Akamatsu, W., Okada, Y., Ohnishi, T., Balan, S., Hisano, Y., et al. (2016). Analysis of induced pluripotent stem cells carrying 22q11.2 deletion. Transl. Psychiatry 6:e934. doi: 10.1038/tp.2016.206
Tregellas, J. R., Smucny, J., Harris, J. G., Olincy, A., Maharajh, K., Kronberg, E., et al. (2014). Intrinsic hippocampal activity as a biomarker for cognition and symptoms in schizophrenia. Am. J. Psychiatry 171, 549–556. doi: 10.1176/appi.ajp.2013.13070981
Uranova, N., Orlovskaya, D., Vikhreva, O., Zimina, I., Kolomeets, N., Vostrikov, V., et al. (2001). Electron microscopy of oligodendroglia in severe mental illness. Brain Res. Bull. 55, 597–610. doi: 10.1016/s0361-9230(01)00528-7
Wakefield, J. C. (2016). Diagnostic issues and controversies in DSM-5: return of the false positives problem. Annu. Rev. Clin. Psychol. 12, 105–132. doi: 10.1146/annurev-clinpsy-032814-112800
Weinberger, D. R. (1987). Implications of normal brain development for the pathogenesis of schizophrenia. Arch. Gen. Psychiatry 44, 660–669. doi: 10.1001/archpsyc.1987.01800190080012
Wen, Z., Nguyen, H. N., Guo, Z., Lalli, M. A., Wang, X., Su, Y., et al. (2014). Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418. doi: 10.1038/nature13716
Winship, I. R., Dursun, S. M., Baker, G. B., Balista, P. A., Kandratavicius, L., Maia-de-Oliveira, J. P., et al. (2019). An overview of animal models related to schizophrenia. Can. J. Psychiatry 64, 5–17. doi: 10.1177/0706743718773728
Wong, A. H., and Van Tol, H. H. (2003). Schizophrenia: from phenomenology to neurobiology. Neurosci. Biobehav. Rev. 27, 269–306. doi: 10.1016/s0149-7634(03)00035-6
Xu, B., Roos, J. L., Levy, S., van Rensburg, E. J., Gogos, J. A., and Karayiorgou, M. (2008). Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 40, 880–885. doi: 10.1038/ng.162
Yoon, K. J., Nguyen, H. N., Ursini, G., Zhang, F., Kim, N. S., Wen, Z., et al. (2014). Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91. doi: 10.1016/j.stem.2014.05.003
Yu, D. X., Di Giorgio, F. P., Yao, J., Marchetto, M. C., Brennand, K., Wright, R., et al. (2014). Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Rep. 2, 295–310. doi: 10.1016/j.stemcr.2014.01.009
Zamanpoor, M. (2020). Schizophrenia in a genomic era: a review from the pathogenesis, genetic and environmental etiology to diagnosis and treatment insights. Psychiatr. Genet. 30, 1–9. doi: 10.1097/YPG.0000000000000245
Zhang, S., Zhang, H., Zhou, Y., Qiao, M., Zhao, S., Kozlova, A., et al. (2020). Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science 369, 561–565. doi: 10.1126/science.aay3983
Zhao, D., Lin, M., Chen, J., Pedrosa, E., Hrabovsky, A., Fourcade, H. M., et al. (2015). MicroRNA profiling of neurons generated using induced pluripotent stem cells derived from patients with schizophrenia and schizoaffective disorder, and 22q11.2 Del. PLoS One 10:e0132387. doi: 10.1371/journal.pone.0132387
Keywords: schizophrenia, iPSC, organoid, epigenetic alteration, transcriptional alteration
Citation: Lee D, Seo J, Jeong HC, Lee H and Lee SB (2021) The Perspectives of Early Diagnosis of Schizophrenia Through the Detection of Epigenomics-Based Biomarkers in iPSC-Derived Neurons. Front. Mol. Neurosci. 14:756613. doi: 10.3389/fnmol.2021.756613
Received: 10 August 2021; Accepted: 20 October 2021;
Published: 12 November 2021.
Edited by:
Margret Shirinian, American University of Beirut, LebanonCopyright © 2021 Lee, Seo, Jeong, Lee and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyosang Lee, aHlvc2FuZzIyQGRnaXN0LmFjLmty; Sung Bae Lee, c2JsZWVAZGdpc3QuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.