
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 09 September 2021
Sec. Methods and Model Organisms
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.718241
 Ian A. Taukulis1
Ian A. Taukulis1 Rafal T. Olszewski1
Rafal T. Olszewski1 Soumya Korrapati1
Soumya Korrapati1 Katharine A. Fernandez2
Katharine A. Fernandez2 Erich T. Boger3
Erich T. Boger3 Tracy S. Fitzgerald4
Tracy S. Fitzgerald4 Robert J. Morell3
Robert J. Morell3 Lisa L. Cunningham2
Lisa L. Cunningham2 Michael Hoa1*
Michael Hoa1*The endocochlear potential (EP) generated by the stria vascularis (SV) is necessary for hair cell mechanotransduction in the mammalian cochlea. We sought to create a model of EP dysfunction for the purposes of transcriptional analysis and treatment testing. By administering a single dose of cisplatin, a commonly prescribed cancer treatment drug with ototoxic side effects, to the adult mouse, we acutely disrupt EP generation. By combining these data with single cell RNA-sequencing findings, we identify transcriptional changes induced by cisplatin exposure, and by extension transcriptional changes accompanying EP reduction, in the major cell types of the SV. We use these data to identify gene regulatory networks unique to cisplatin treated SV, as well as the differentially expressed and druggable gene targets within those networks. Our results reconstruct transcriptional responses that occur in gene expression on the cellular level while identifying possible targets for interventions not only in cisplatin ototoxicity but also in EP dysfunction.
The stria vascularis (SV) is a stratified epithelial tissue located in the lateral wall of the mammalian cochlea that generates and maintains the endocochlear potential (EP) and regulates the secretion of endolymph (Wangemann, 2002; Chang et al., 2015). Endolymph possesses a high potassium concentration that creates a positive potential of 80–120 mV in the endolymph-containing scala media relative to the perilymph-containing compartments of the cochlea. The EP is the driving force for hair cell mechanoelectrical transduction and hearing. SV cell types, which express several critical ion channels, work together to generate the EP. Importantly, mutations in SV ion channel genes related to EP generation result in hearing loss in humans (Wangemann, 2002).
More broadly, mutations in genes expressed by SV cell types result in hearing loss in mammals. In mouse, mutations in genes expressed by marginal cells of the SV, including Slc12a2, Atp1b2, Kcnq1, and Kcne1, result in a loss or reduction of EP and deafness (Delpire et al., 1999; Dixon et al., 1999; Wangemann, 2002; Chang et al., 2015; Faridi et al., 2019; McNeill et al., 2020; Mutai et al., 2020). Slc12a2 encodes Na+/K+/Cl– symporter 1 (NKCC1), which is expressed at the basolateral membrane of SV marginal cells and is responsible for transporting potassium from the intrastrial space into marginal cells (Wangemann, 2002). The loss of SLC12A2 results in deafness in mouse models (Delpire et al., 1999; Dixon et al., 1999; Wangemann, 2002) and has recently been implicated in human hearing loss (McNeill et al., 2020; Mutai et al., 2020). Atp1b2 encodes the β2 subunit of Na+, K+-ATPase (NKA) protein, which has been shown to be expressed in the basolateral membranes of mouse SV marginal cells (Konishi et al., 1978; Schulte and Steel, 1994; Wangemann et al., 1995; Wangemann, 2002). Similar to NKCC1, NKA pumps out cations from the SV to maintain a high potassium concentration and positive EP in the endolymph in the scala media (Nishiyama et al., 1994; Furukawa et al., 1996; Lang et al., 2007; Nin et al., 2008). ATP1A1 and ATP1B2, encoded by Atp1b2, form the Na+/K+-ATPase that is essential for maintaining the low potassium concentration in the intrastrial space and is important for the generation of the +80 mV endocochlear potential in the scala media (Wangemann, 2002). Decline in EP is associated with the loss of Na+/K+-ATPase activity in the stria vascularis (Gratton et al., 1996) and direct inhibition of the Na+/K+-ATPase with ouabain has been shown to reduce the EP (Nin et al., 2008). These data suggest that Atp1b2 is essential for EP generation.
Kcnq1 and Kcne1 encode the voltage-gated potassium channel Kv7.1, which is expressed on the apical surface of the marginal cell and plays a crucial role in secreting potassium into the endolymph and maintaining the EP. Conditional Kcnq1 null mice exhibit a collapse of Reissner’s membrane, loss of EP, and are deaf (Chang et al., 2015). Mutations in KCNQ1/KCNE1 result in human deafness and have been reviewed previously (Faridi et al., 2019). Similarly, genes expressed by intermediate cells including Kcnj10 and Met cause hearing loss in humans (Scholl et al., 2009; Reichold et al., 2010; Freudenthal et al., 2011; Mujtaba et al., 2015; Alabdullatif et al., 2017). Notably, Kcnj10 encodes Kir4.1, which is an inwardly rectifying potassium channel expressed on the apical (marginal cell-facing) membranes of intermediate cells, that has been shown to be critical for EP generation with its loss resulting in hearing loss in both mice and humans (Marcus et al., 2002; Wangemann, 2002; Wangemann et al., 2004; Scholl et al., 2009; Reichold et al., 2010; Freudenthal et al., 2011; Chen and Zhao, 2014). Furthermore, loss of Ednrb has been shown to result in the reduction of EP (da-Eto et al., 2011). While loss of Met expression has not been demonstrated to specifically result in loss or reduction of EP, dysfunctional HGF/MET signaling resulting from a knock-in mouse of a human mutation of HGF has been implicated in the reduction of EP through a reduction in intermediate cells (Morell et al., 2020). Finally, mutations in genes expressed by SV basal cells, including Cldn11, Gjb2 and Gjb6 have been implicated in the reduction of EP and hearing loss. Loss of Cldn11, which encodes the CLDN11 protein that is expressed in tight junctions of SV basal cells, has been shown to result in deafness and reduced EP in Cldn11 null mice (Gow et al., 2004; Kitajiri et al., 2004a). Gjb2 and Gjb6 encode gap junctions expressed by SV basal cells and, to a lesser extent, intermediate cells (Kikuchi et al., 1995; Xia et al., 1999, 2001). Mutations of GJB2 and GJB6 are associated with deafness in humans (Kelsell et al., 1997; Grifa et al., 1999) and have been shown to result in hearing loss and a reduction in EP in mouse (Teubner et al., 2003; Wingard and Zhao, 2015; Mei et al., 2017; Lin X. et al., 2019; Mammano, 2019; Wu et al., 2019). Thus, existing literature emphasizes the importance of genes and proteins expressed by SV cell types in both hearing and EP generation.
Previously, our group has utilized single-cell RNA-sequencing to characterize the gene regulatory networks of the major SV cell types, including marginal, intermediate, and basal cells, that underlie SV homeostatic functions, including EP generation in the adult mouse (Korrapati et al., 2019). Gene regulatory networks (GRNs) define and maintain cell type-specific transcriptional states, which in turn underlie cellular morphology and function (Aibar et al., 2017; Fiers et al., 2018). From this perspective, cells and their respective states are defined by a combination of transcription factors (TFs) that interact with a set of target genes to produce a specific gene expression profile. Perturbation of cell state through genetic, epigenetic, or pharmacologic means may provide insight into the contribution of these GRNs to cell function (Liberali et al., 2015; Li et al., 2018; Musa et al., 2018; Caldera et al., 2019). Diseases or pharmacologic treatments represent perturbations of GRNs and investigation of these states at the single cell level may shed light on critical GRNs that underlie human disease or responses to treatment by potentially identifying key regulators in these GRNs (del Sol et al., 2010; Fiers et al., 2018; Caldera et al., 2019).
Cisplatin is commonly used to treat patients with a variety of cancers and results in permanent hearing loss in 35–100% of treated adults due to its ototoxicity as reviewed by Paken et al. (2019). Recently, Breglio et al. (2017) demonstrated long term retention of cisplatin in the SV of both mice and humans, as measured by platinum concentration over time. These authors also demonstrated accompanying declines in EP in a mouse model of cisplatin ototoxicity that mimics the treatment and time course seen in humans undergoing cisplatin treatment for cancer (Breglio et al., 2017). In this model, cisplatin treatment results in temporary decline in the EP after the 1st cycle that gradually recovers but eventually becomes permanent after three cycles of cisplatin and accompanies hearing loss (Breglio et al., 2017; Fernandez et al., 2019). SV retention of cisplatin combined with the decline in EP and its known ototoxic effects in humans prompted us to investigate the underlying transcriptional changes in SV cell types associated with cisplatin exposure to identify GRNs involved in the loss of EP-generating function in the SV.
To investigate the acute transcriptional response of the SV to cisplatin instead of the cumulative effects of cisplatin exposure, a mouse model for acute cisplatin-induced ototoxicity consisting of the dose equivalent (14 mg/kg, IP) to one cycle of cisplatin was utilized based on the cisplatin-induced ototoxicity mouse model by Fernandez et al. (2019). Using this mouse model for acute cisplatin-induced ototoxicity to better understand the molecular mechanisms underlying EP generation and maintenance, we demonstrate that the decline in EP associated with cisplatin treatment is associated with changes in known SV cell type-specific EP-related genes at the protein and RNA levels by fluorescence immunohistochemistry and digital droplet qPCR (ddPCR), respectively.
To further investigate transcriptional changes associated with a decline in EP in response to cisplatin treatment, we performed single-cell RNA-sequencing (scRNA-Seq) of cisplatin-treated adult SV and compared these transcriptional profiles to our previously published unperturbed adult SV scRNA-Seq dataset (Korrapati et al., 2019). In addition to corroborating our observations with known EP-related genes, we identify cell type-specific gene regulatory networks associated with differentially expressed EP-related genes. Among the major SV cell types, we demonstrate that marginal and intermediate cells are differentially affected by cisplatin treatment and we define differentially expressed genes as they relate to select EP-related GRNs affected by cisplatin. Finally, we utilize Pharos1 to identify druggable gene targets in these GRNs that are preferentially affected by cisplatin. We pinpoint potentially repurposable FDA-approved drugs that may counteract transcriptional changes induced by cisplatin. More generally, analyses of these data serve as an example of a bioinformatic pipeline designed to identify druggable targets and potentially repurposable drugs to reverse not only cisplatin-induced hearing loss but also hearing loss related to stria vascularis dysfunction.
Cisplatin-treated mice have a reduced EP compared to control mice (Control mean EP = 86.96 mV, Cisplatin-treated mean EP = 58.43 mV, p = 0.0012) (Figure 1A). EP measurements were obtained 24 h after treatment in control (n = 12) and cisplatin-treated (n = 10) mice that were given a single IP injection of saline or 14 mg/kg cisplatin, respectively. Given the constraints of performing auditory brainstem response (ABR) and EP recordings at the same time point, ABR thresholds (Figure 1B) and hair cell counts (Supplementary Figures S1A,B) were analyzed in a parallel cohort of control (n = 6) and cisplatin-treated (n = 6) mice. ABR thresholds measured at the 8, 16, 32, and 40 kHz frequencies were unchanged between control and cisplatin-treated mice (two-way ANOVA, p > 0.9999) (Figure 1B). Inner (two-way ANOVA, p = 0.6434) and outer (two-way ANOVA, p = 0.6423) hair cell counts between cisplatin-treated and control cochlea in the apical and basal turns were comparable and did not demonstrate a significant difference between conditions (Supplementary Figures S1A,B). DPOAE amplitudes between control and cisplatin-treated mice were unchanged in both left and right ears (Supplementary Figures S1C,D). Additionally, SV cross-sectional area in the apical, medial, and basal turns of the cochlea was unchanged between control and cisplatin-treated mice (two-way ANOVA, p = 0.7680) (Figure 1C). Similarly, SV thickness in the apical, medial, and basal turns of the cochlea was unchanged between control and cisplatin-treated mice (two-way ANOVA, p = 0.0554) (Figure 1D). Total nucleus counts in the SV in the apical, medial, and basal turns of the cochlea were unchanged between control and cisplatin-treated mice (two-way ANOVA, p = 0.3974) (Figure 1E). Furthermore, the number of TUNEL-positive cell nuclei, calculated as a percentage of the total nuclei, was unchanged between control and cisplatin-treated mice across apical, medial, and basal cochlear turns (two-way ANOVA, p = 0.8282) (Figure 1F). Representative TUNEL staining in SV cross-section co-labeled with DAPI and phalloidin are shown (Figure 1G). Together these data indicate that a single injection of 14 mg/kg cisplatin resulted in reduced EP and suggest that hearing loss and hair cell death at this time point is not significant.
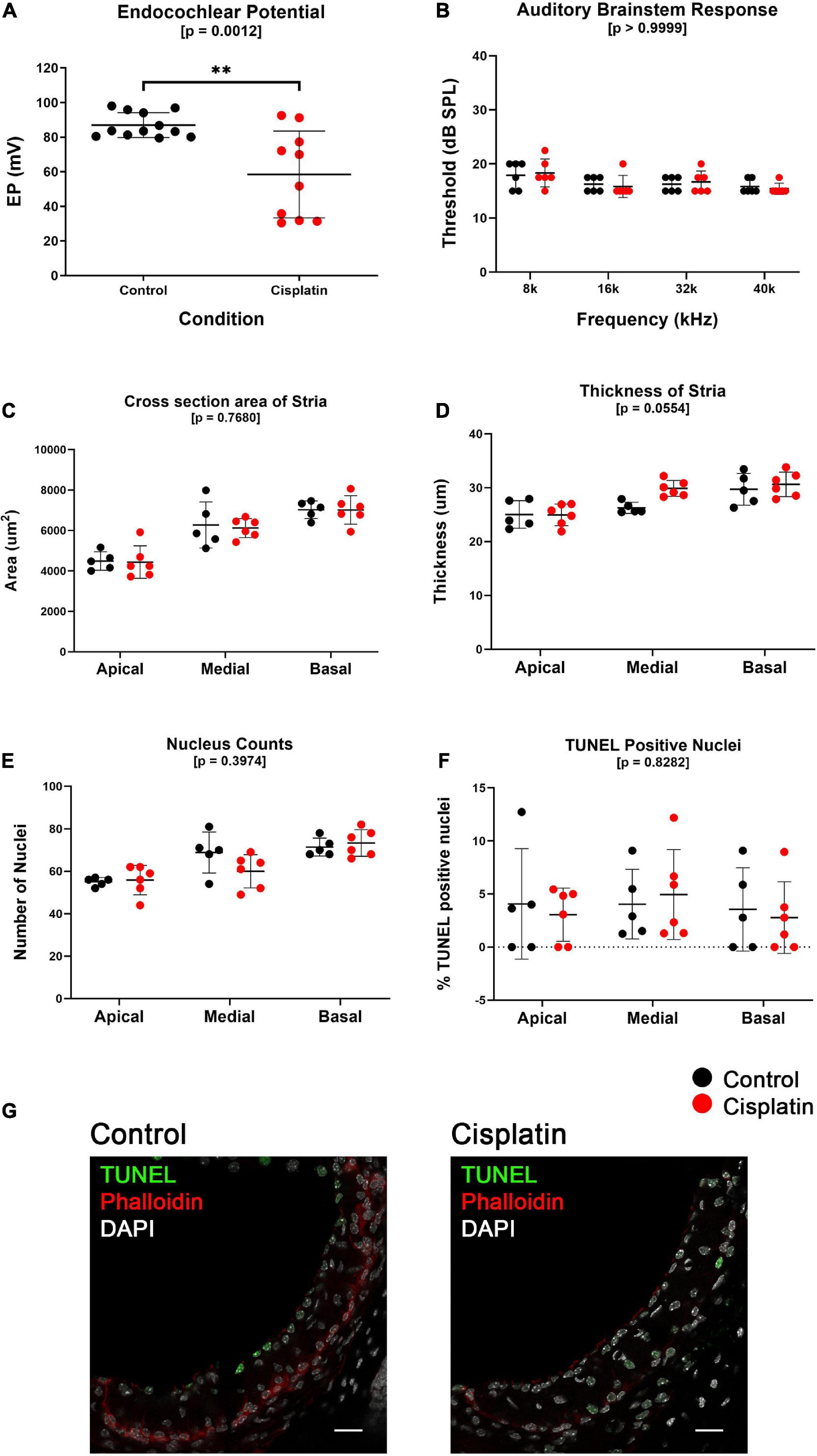
Figure 1. A single dose of cisplatin (14 mg/kg, IP) results in a reduction in EP after 24 h with no reduction in ABR threshold, minimal structural change, and no evidence of increased apoptosis. (A), EP measurements obtained 24 h after treatment in control (n = 12) and cisplatin-treated (n = 10) demonstrate that cisplatin-treated mice have a reduced EP compared to control mice (p = 0.0012). (B) In a separate cohort of mice, ABR thresholds measured at the 8, 16, 32, and 40 kHz frequencies were unchanged between control (n = 6) and cisplatin-treated (n = 6) mice (two-way ANOVA, p > 0.9999). (C) SV cross-sectional area was unchanged between conditions in all cochlear turns (two-way ANOVA, p = 0.7680). (D) SV thickness was unchanged between conditions in all cochlear turns (two-way ANOVA, p = 0.0554). (E) Total nucleus counts in the SV were unchanged between conditions in all cochlear turns (two-way ANOVA, p = 0.3974). (F) TUNEL-positive cell nuclei, calculated as a percentage of the total nuclei, was unchanged between conditions in all cochlear turns (two-way ANOVA, p = 0.8282). (G) Representative TUNEL staining in SV cross-sections. For (B–G), one cohort of mice were used, control (n = 6) and cisplatin-treated (n = 6). Scale bars: 20 μm. **p < 0.01.
SLC12A2 is expressed at the basolateral membrane of SV marginal cells, transports potassium from the intrastrial space into marginal cells and is critical for EP generation (Wangemann, 2002). We observed that at 24 h after cisplatin injection, SLC12A2 immunofluorescence intensity was significantly reduced in the apical and basal turns of the cochlea (two-way ANOVA, p < 0.0001; Sidak’s multiple comparisons test, q = 0.0068 and q = 0.0022, respectively) (Figure 2A), with representative cross sections of control (Figure 2B) and cisplatin-treated mice (Figure 2C). Immunofluorescence staining intensity of KCNJ10, which is known to be expressed on the apical (marginal cell-facing) membranes of intermediate cells (Marcus et al., 2002; Wangemann, 2002; Chen and Zhao, 2014), was unchanged between control and cisplatin-treated mice (two-way ANOVA, p = 0.1784) (Figure 2D), with representative cross sections of control (Figure 2E) and cisplatin-treated mice (Figure 2F). Finally, CLDN11 (claudin 11) immunofluorescence staining intensity, a tight junction protein expressed by SV basal cells, was reduced in the basal turn of cisplatin-treated compared to control mice (two-way ANOVA, p = 0.0002; Sidak’s multiple comparisons test q = 0.0418) (Figure 2G), with cross sections of control (Figure 2H) and cisplatin-treated mice (Figure 2I).
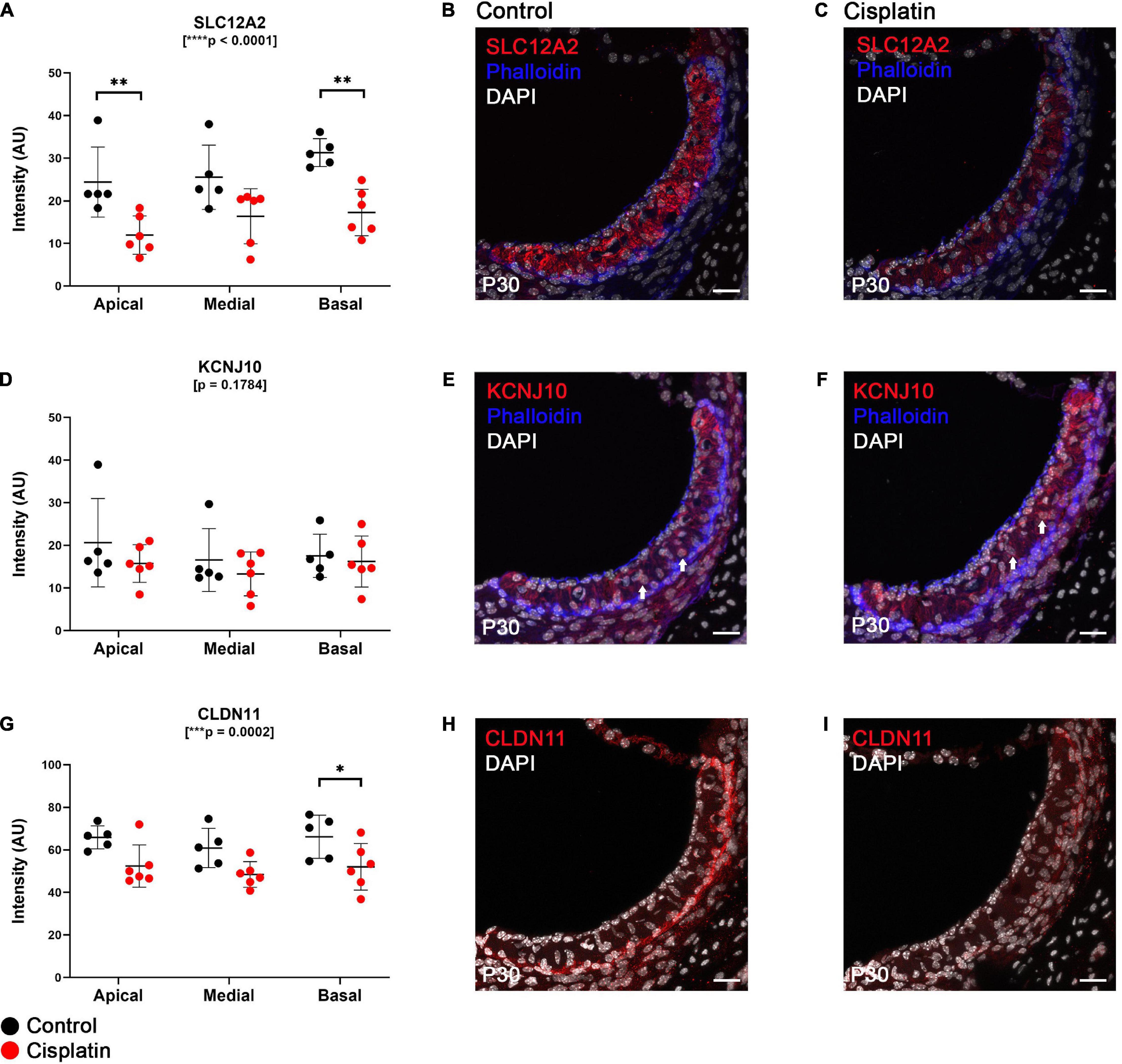
Figure 2. Expression of key EP-related cell-type specific proteins is reduced in the SV after cisplatin treatment. (A) Twenty four hours after cisplatin injection, SLC12A2 immunofluorescence intensity was significantly reduced in the apical and basal turns of the cochlea (two-way ANOVA, p < 0.0001; Sidak’s multiple comparisons test, q = 0.0068 and q = 0.0022, respectively). (B,C) Representative SLC12A2 immunostaining of SV cross sections of control and cisplatin-treated mice. (D) Immunofluorescence staining intensity of KCNJ10 was unchanged between control and cisplatin-treated mice (two-way ANOVA, p = 0.1784). (E,F) Representative KCNJ10 immunostaining of SV cross sections of control and cisplatin-treated mice. White arrows point to representative intermediate cell nuclei. (G) Immunofluorescence staining intensity of CLDN11 was reduced in the basal turn of cisplatin-treated compared to control mice (two-way ANOVA, p = 0.0002; Sidak’s multiple comparisons test q = 0.0418). (H,I) Representative CLDN11 immunostaining of SV cross sections of control and cisplatin-treated mice. Scale bars: 20 μm. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
As we have described, a number of EP-related genes have been identified including Kcne1, Kcnq1, Atp1b2, Slc12a2, Kcnj10, Met, Cldn11, Gjb2, and Gjb6 (Wangemann, 2002; Gow et al., 2004; Kitajiri et al., 2004b; Morell et al., 2020). We utilized ddPCR of whole isolated SV to quantify the effect of cisplatin-treatment on the expression of these EP-related genes in the SV to gain a better sense of the transcriptional response with respect to EP-generating mechanisms. In whole isolated SV, we observed a significant reduction in selected EP-related genes specific to marginal cells: Kcne1 (q = 0.0020), Kcnq1 (q = 0.0161), Atp1b2 (q = 0.0020), and Slc12a2 (q = 0.0020) (unpaired multiple t-test, Benjamini–Hochberg correction) (Figure 3). A significant reduction in an EP-related gene specific to intermediate cells: Met (uncorrected p = 0.0064) was seen between control and cisplatin-treated mice (unpaired multiple t-test, Benjamini–Hochberg correction) (Figure 3). Expression in whole dissected SV of other known EP-related intermediate cell-specific genes including Kcnj10 (p = 0.1746) and Ednrb (p = 0.5309) were not reduced in cisplatin-treated compared to control. Ednrb expression has previously been identified in the stria vascularis (Matsushima et al., 2002; Stanchina et al., 2006; Xu et al., 2010; da-Eto et al., 2011) and is identified as an intermediate cell-specific gene in the dataset published by Korrapati et al. (2019) as shown in the gene Expression Analysis Resource (gEAR)2. Finally, expression of EP-related genes in basal cells, including Gjb2 (q = 0.0207) was significantly reduced, while Gjb6 (q = 0.1086), and Cldn11 (q = 0.4012), were not decreased in cisplatin-treated compared to control mice (unpaired multiple t-test, Benjamini–Hochberg correction) (Figure 3). Both GJB2 and GJB6 protein have been previously shown to be expressed by basal cells and intermediate cells (Lang et al., 2007; Wangemann et al., 2007; Nickel and Forge, 2008; Liu W. et al., 2009; Locher et al., 2015; Liu W. et al., 2016). CLDN11 is a well-established specific marker for basal cells in the stria vascularis (Gow et al., 2004; Kitajiri et al., 2004a; Liu W. et al., 2017; Korrapati et al., 2019). The limited sensitivity of ddPCR to changes at the cell type level when assaying whole SV tissue may account for the inability to detect changes in cell type-specific genes between control and cisplatin-treated mice. Nonetheless, ddPCR analysis of cisplatin-treated SV suggest that RNA transcription related to EP generation is adversely affected by cisplatin. Due to our limited ability to resolve cell type-specific responses in the SV in response to cisplatin and to gain a more comprehensive understanding of the transcriptional response to cisplatin at the cellular level in the SV, we then utilized scRNA-Seq to perform transcriptional profiling of the SV in cisplatin-treated adult mice.
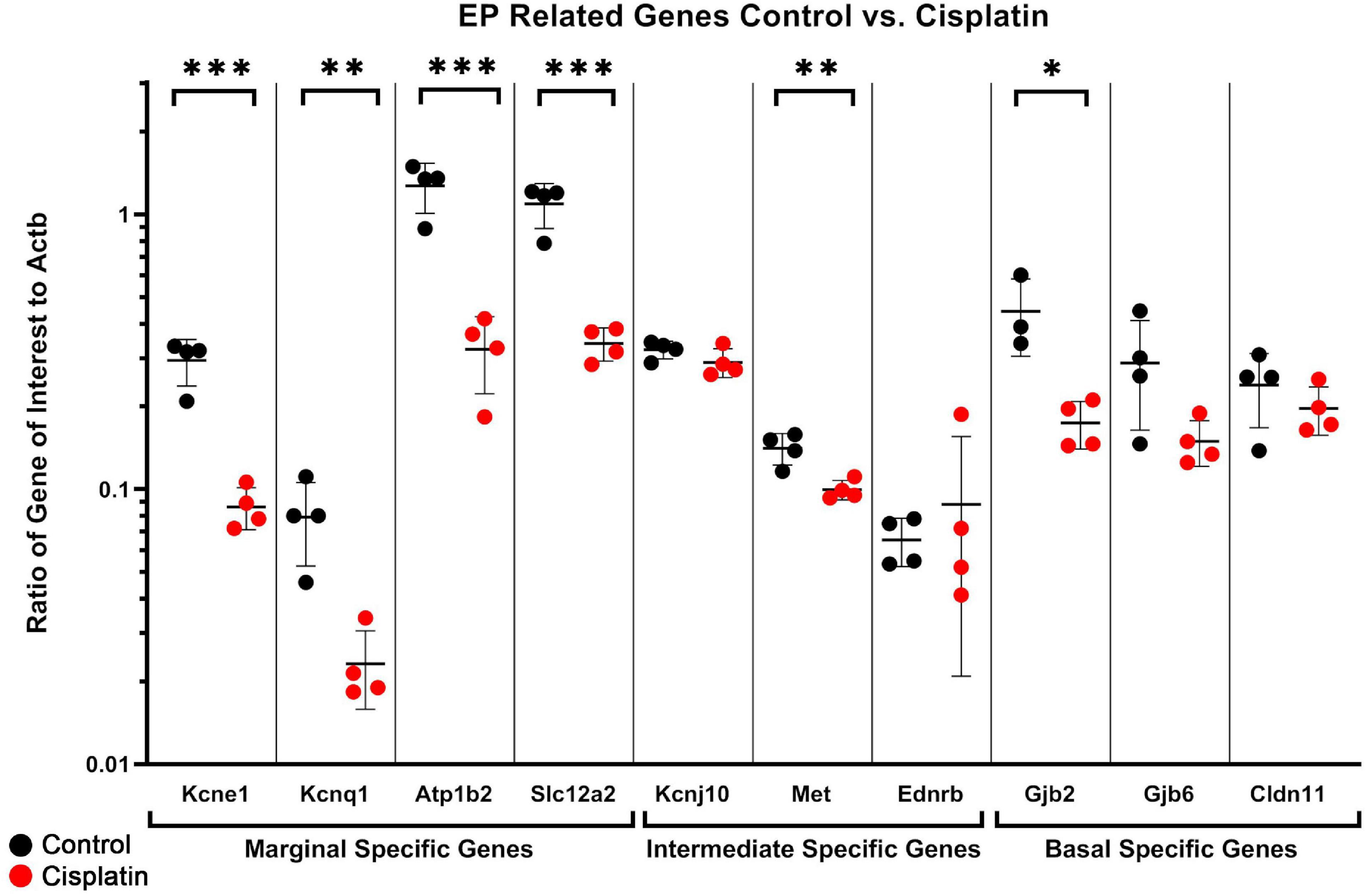
Figure 3. ddPCR of dissected SV demonstrates a reduction in several EP-related genes after cisplatin treatment. In whole dissected SV, we observed a significant reduction in selected EP-related genes specific to marginal cells: Kcne1 (q = 0.0020), Kcnq1 (q = 0.0161), Atp1b2 (q = 0.0020), and Slc12a2 (q = 0.0020) (unpaired multiple t-test, Benjamini–Hochberg correction). A significant reduction in two EP-related genes specific to intermediate cells: Met (uncorrected p = 0.0064) and Kcnj13 (uncorrected p = 0.0117) was seen between control and cisplatin-treated mice (unpaired multiple t-test, Benjamini–Hochberg correction). Expression of other known intermediate cell-specific genes including Kcnj10 (p = 0.1746) and Ednrb (p = 0.5309) were not reduced in cisplatin-treated compared to control. Expression of EP-related genes in basal cells, including Gjb2 (q = 0.0207) was significantly reduced, while Gjb6 (q = 0.1086), and Cldn11 (q = 0.4012), were not decreased in cisplatin-treated compared to control mice (unpaired multiple t-test, Benjamini–Hochberg correction). *p < 0.05; **p < 0.01; ***p < 0.001.
Major cell types in cisplatin-treated SV were identified using published markers (Korrapati et al., 2019), demonstrating the capture of marginal, intermediate, and basal cells in pink, green, and blue, respectively, using a uniform manifold and projection (UMAP) clustering (Figure 4). The genes Kcne1 and Kcnq1 were used as markers to identity marginal cells, Kcnj10 and Cd44 were used to identify intermediate cells, and Cldn11 and Tjp1 were used to identify basal cells. Additional clusters were identified using published markers and included cell types adjacent to and within the structure of the stria. These included the fibrocytes of the lateral wall, cells of Reissner’s membrane that separate the scala media from the scala vestibuli, the spindle cells at the edges of the SV, and various lymphoid, myeloid, and erythroid populations including neutrophils, B-cells, macrophages, and red blood cells (RBCs) (Figure 4). Bioinformatic clustering revealed similar SV cell type-specific clusters across both cisplatin-treated and untreated SV single cell RNA-Seq datasets (Korrapati et al., 2019). Cell type-specific gene expression in both the cisplatin-treated and control SV datasets is provided as a supplement (Supplementary Table S1).
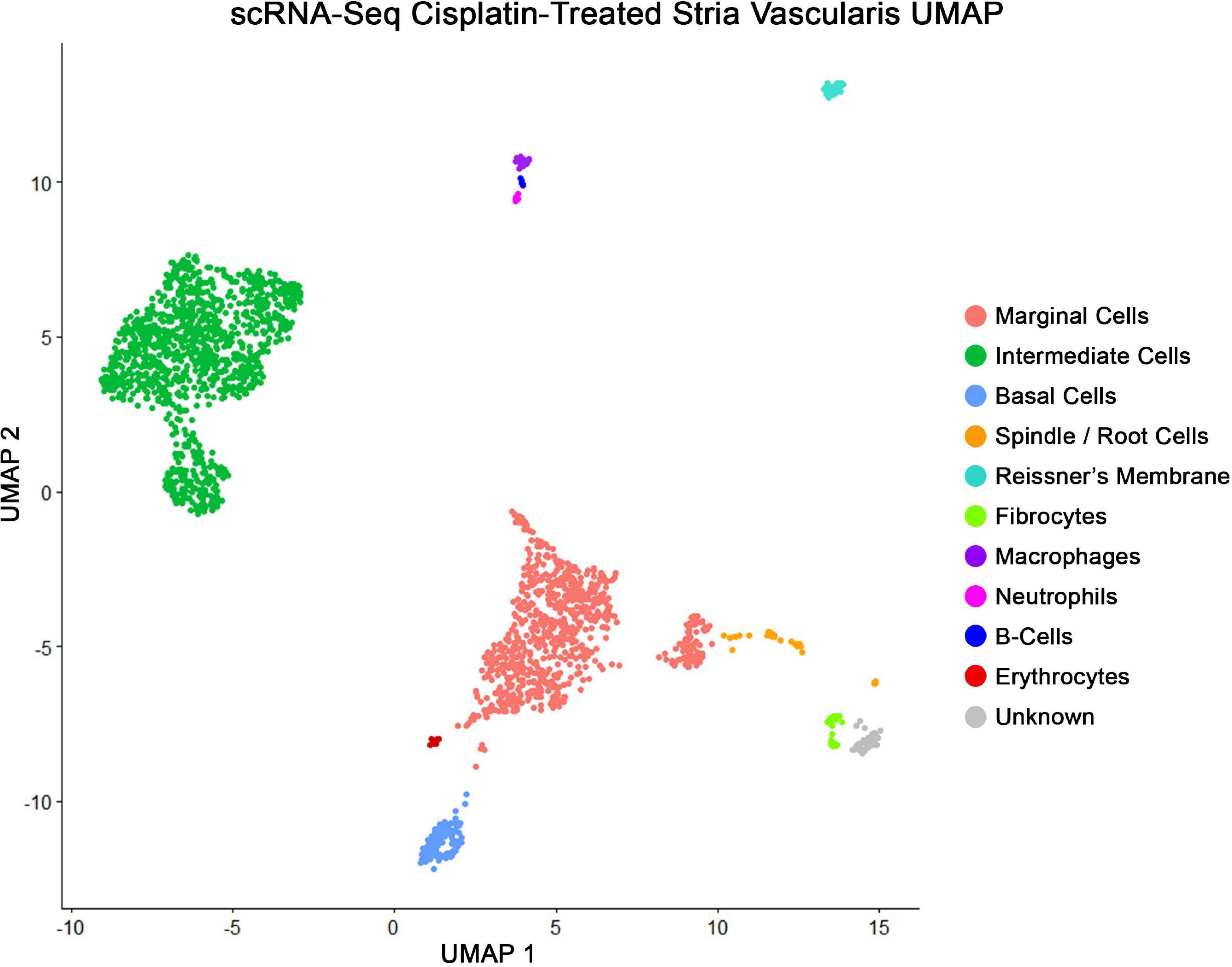
Figure 4. scRNA-Seq of cisplatin-treated adult SV demonstrates all the expected cell populations as untreated adult SV. Uniform manifold and projection (UMAP) clusters identified using published markers, showing marginal, intermediate, and basal cells in pink, green, and blue, respectively. Each dot represents a single whole cell. Additional cell types identified were spindle cells, fibrocytes, Reissner’s membrane cells, neutrophils, B-cells, macrophages, and red blood cells (RBCs).
To examine the transcriptional effects of cisplatin treatment on SV cell types, we performed three separate differential expression (DE) analyses on marginal, intermediate, and basal cells between cisplatin-treated and control SV cells (Figure 5). Volcano plots depict differential gene expression of the marginal (Figure 5A), intermediate (Figure 5B), and basal (Figure 5C) cell types. Volcano plots depict expression of genes, represented by filled circles, enriched in cisplatin-treated SV cells to the right and genes enriched in control SV cells to the left. Fold-change thresholds in log2 scale (fold-change = ± 1.25) correspond to the vertical dotted lines along the horizontal axis and the p-value threshold in log10 scale (p-value = 0.0001) corresponds to the horizontal dotted line along the vertical axis. The higher along the vertical axis, the more significant the p-value and the further along in either the negative or positive direction from the dotted line, respectively, the greater the fold-change expression in either the control or cisplatin treated SV cells. Circles in red depict genes that met set threshold criteria (fold-change threshold >1.25, p < 0.0001). DE analysis of major SV cell types suggests that cisplatin has a greater effect on SV marginal and intermediate cells as supported by the larger number of differentially expressed genes (Supplementary Table S3). On closer examination, EP-related genes specific for SV marginal cells (Slc12a2, Kcne1, Atp1b2, Kcnq1) are enriched in control cells suggesting downregulation of these genes in response to cisplatin in marginal cells (Figure 5A). Furthermore, control SV intermediate cells demonstrate an enrichment for many EP-related genes (Gjb2, Gjb6, Kcnj10, Met) suggesting a downregulation of these genes in response to cisplatin (Figure 5B). In contrast to SV marginal and intermediate cells, basal cells demonstrated few differentially expressed genes (Figure 5C). Gene ontology (GO) biological process analysis for genes upregulated in cisplatin-treated cells compared to control cells revealed an enrichment for genes involved in signal recognition particle (SRP)-dependent cotranslational protein targeting to membrane (GO:0006614), ATP synthesis coupled electron transport (GO:0042775), and negative regulation of viral entry into host cell (GO:0046597) in marginal, intermediate, and basal cells, respectively. GO biological process analysis for downregulated genes revealed an enrichment for genes involved in membrane repolarization (GO:0086009), peptidyl-serine phosphorylation (GO:0018105), and mitochondrial ATP synthesis coupled electron transport (GO:0042775) in marginal, intermediate, and basal cells, respectively. GO analysis for both upregulated and downregulated genes suggest that cisplatin has differential effects on cells in the stria vascularis and suggest future areas for potential investigation. Together, these data suggest that SV marginal and intermediate cells demonstrate preferential effects on their transcriptome in response to cisplatin. We provide a list of differentially expressed genes (DEGs) for SV marginal, intermediate, and basal cells between cisplatin-treated and control adult mice (Supplementary Table S3).
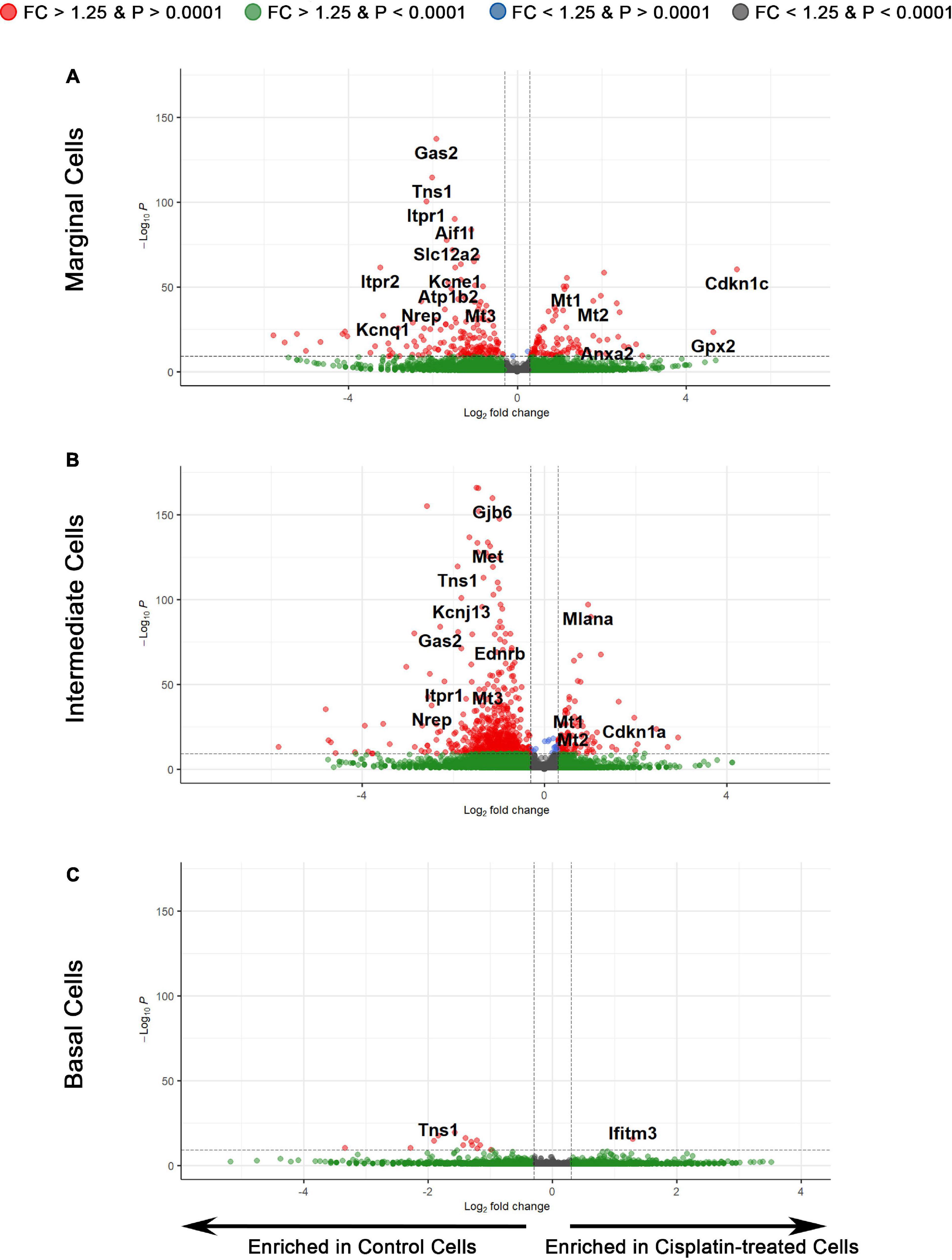
Figure 5. Differential expression analysis reveals the transcriptomic changes in individual cell types of the SV after cisplatin treatment. (A) Volcano plot depicting differential gene expression between the scRNA-Seq data of marginal cells of control and cisplatin-treated mice. Genes are represented by circles, with genes enriched in cisplatin-treated SV cells to the right and genes enriched in control SV cells to the left. Positive and negative fold-change thresholds (FC ± 1.25) in log2 scale are represented by the vertical dotted lines. The p-value threshold (p-value ± 0.0001) in –log10 scale is represented by the horizontal dotted line. Red circles are genes that exceed both fold change and p-value threshold criteria. Green circles are genes that exceed the fold change threshold but not the p-value threshold. Blue circles are genes that exceed the p-value threshold but not the fold change threshold. Gray circles are genes that do not meet either threshold criteria. (B) Differential gene expression between the scRNA-Seq data of intermediate cells of control and cisplatin-treated mice. (C) Differential gene expression between the scRNA-Seq data of basal cells of control and cisplatin-treated mice.
Utilizing a larger group of upregulated and downregulated genes, we utilized ddPCR of cisplatin-treated and control whole isolated SV to validate the dataset (Supplementary Figure S2). Upregulated genes that demonstrated consistency between ddPCR and scRNA-Seq included Cdkn1c, Kcnj13, Mt1, and Mt2 (Supplementary Figure S2A). Despite a trend toward increased expression, the apparent inconsistency for Anxa2, Cdkn1a, Gpx2, Ifitm, and Mlana between ddPCR and scRNA-Seq could be due to the insensitivity of a whole tissue assay to changes at the single cell level. Downregulated genes including Aes1, Aif1l, Gas2, Itpr1, Itpr2, Mt3, Nrep, and Tns1 demonstrated consistency between ddPCR and scRNA-Seq (Supplementary Figure S2B). Overall, these findings validate downregulation of genes associated with cisplatin treatment.
Regulons are defined as transcription factors and their motif-enriched downstream target genes. We have previously utilized SCENIC to identify homeostatic cell type-specific regulons in the adult SV (Korrapati et al., 2019). SCENIC analysis along with regulon specificity score (RSS) (Supplementary Figure S3 and Supplementary Table S4) analysis of cisplatin-treated adult SV single cell transcriptomes reveals regulons specific for marginal, intermediate, and basal cells. RSS is a measurement of a regulon’s degree of specificity to a cell type within a single cell data set. Fourteen, twelve, and nine cell-type specific regulons were identified corresponding to marginal, intermediate, and basal cells, respectively. Of these regulons, eight marginal cell regulons, four intermediate cell regulons, and five basal cell regulons were unique to the cisplatin-treated SV. Due to the apparent preferential effect on the transcriptional profiles of marginal and intermediate cells, we focused our attention on marginal and intermediate cell regulons in cisplatin-treated SV. We selected two regulons identified in the cisplatin-treated scRNA-Seq data as examples for our pipeline of cell-type specific regulatory intervention: Kruppel like factor 10 (Klf10) in marginal cells and T-Box transcription factor 2 (Tbx2) in intermediate cells. In both cases, their RSS scores are above the threshold for specificity for their respective cell types (Supplementary Figure S3A for the Klf10 regulon and Supplementary Figure S3B for the Tbx2 regulon), the transcription factor for each regulon is differentially expressed between control and cisplatin-treated cochlea, and there are multiple downstream targets with FDA approved drugs that either promote or inhibit expression. We utilized ddPCR to quantify the expression of Klf10 and select downstream targets, including Atp1b2, Tspan1, and Cldn3 (Supplementary Figure S4A), as well as the expression of Tbx2 and select downstream targets, including Kcnn4, Ndufb2, and Tubb5 (Supplementary Figure S4B) in cisplatin-treated SV to provide supportive evidence for the existence of these regulons. These analyses allowed us to identify cell type-specific regulons that are present in the cisplatin-treated adult SV.
Given that SV marginal and intermediate cells appeared to be preferentially affected by cisplatin treatment compared to SV basal cells, we focused on selected regulons for SV marginal and intermediate cells based on high RSS scores and presence of EP-related genes. These regulons and their downstream targets serve as examples of potential cell type-specific therapeutic targets. SCENIC analysis identifies the Kruppel like factor 10 (Klf10) regulon as a regulon that is present specifically in marginal cells of cisplatin-treated SV. SCENIC analysis did not show presence of the Klf10 regulon in the control RNA-Seq dataset. Klf10 has been broadly implicated in epithelial to mesenchymal transition (EMT) as well as TGFβ signaling and is thought to function as a tumor suppressor gene (Venkov et al., 2011; Memon and Lee, 2018). Differential expression (DE) analysis of scRNA-Seq data suggests that transcripts for Klf10, Claudin 3 (Cldn3), and Tetraspanin 1 (Tspan1) are enriched in cisplatin-treated marginal cells. Conversely, DE analysis suggests that transcripts for activated leukocyte cell adhesion molecule (Alcam), ATPase Na+/K+ transporting subunit beta 2 (Atp1b2), Carbonic anhydrase 12 (Ca12), Glutamyl aminopeptidase (Enpep), Protein-arginine deiminase type-2 (Padi2), and Secreted phosphoprotein 1 (Spp1) are downregulated in cisplatin-treated marginal cells (Supplementary Table S3).
In intermediate cells, SCENIC analysis identified that the T-Box transcription factor 2 (Tbx2) regulon is present in cisplatin-treated SV. Tbx2 has been broadly implicated in cell proliferation, induction of EMT, and senescence bypass (Wang et al., 2012; Decaesteker et al., 2018; Crawford et al., 2019). Based on DE analysis, regulon-specific transcripts that were enriched in cisplatin-treated intermediate cells include NADH dehydrogenase 1 beta subcomplex subunit 2 (Ndufb2), and Tubulin beta class I (Tubb5). Tbx2 regulon-specific transcripts that were downregulated in response to cisplatin include Tbx2, Tyrosine-protein kinase (Abl1), ATPase Na+/K+ transporting subunit beta 1 (Atp1b1), Endothelin receptor type B (Ednrb), Hepatocyte growth factor receptor (Met), and cAMP-specific 3’,5’-cyclic phosphodiesterase 4B (Pde4b) (Supplementary Table S3). Altogether, these analyses suggest that EMT and cell proliferation processes may play a role in the response to cisplatin in the SV marginal and intermediate cells and identify possible druggable gene targets to modulate the response to cisplatin.
This study utilized a mouse model of cisplatin ototoxicity in which a single dose of cisplatin was used to reduce the EP. Our data using this model reveals cell-type specific transcriptional responses in the EP-generating cells of the stria vascularis in response to cisplatin. We demonstrate that SV marginal and intermediate cells are preferentially affected by cisplatin exposure, which is consistent with the role of these cells in generating the EP. Finally, we also demonstrate a pipeline for identifying druggable targets and repurposable drugs using a model of SV dysfunction, which might identify candidates for intervening in cisplatin-related and strial-related hearing loss.
We suggest that genes involved in SV function that are downregulated in response to cisplatin may be reasonable targets for augmentation to mitigate the effects of cisplatin on the SV. Therapeutic gene targets that are downregulated in response to cisplatin include Alcam, Atp1b2, Spp1, and Car12. Reduced Alcam expression has been previously associated with disruption of the blood-brain barrier and its expression may be implicated in blood-labyrinth barrier function (Lécuyer et al., 2017). Atp1b2, Spp1, and Car12 are known to be expressed in different cell types in the stria vascularis and lateral wall, notably marginal cells (Lopez et al., 1995; Wangemann, 2002; Wu et al., 2013). Atp1b2 and Car12 have been previously implicated in EP generation (Ikeda et al., 1987; Wangemann, 2002). ddPCR validate downregulation of Atp1b2, a gene critical to EP generation, is downregulated in SV marginal cells in response to cisplatin (Figure 3). On the other hand, Spp1, or secreted phosphoprotein 1, has not been implicated previously in EP generation. Schmitt and colleagues examined cochlear effects of loss of Spp1 in response to cisplatin and found no change in Spp1 expression by immunohistochemistry and no difference in auditory brainstem responses between Spp1 wild-type and Spp1 null mice 72 h after cisplatin treatment, concluding that Spp1 did not appear to mitigate cisplatin ototoxicity (Schmitt and Rubel, 2013). Our scRNA-Seq data demonstrates decreased expression of Spp1 in marginal cells in response to cisplatin (Supplementary Table S3). Reduced expression of the SPP1 protein after cisplatin treatment was validated using immunofluorescence intensity analysis (Supplementary Figure S5). Properties of Spp1 include cell death inhibition, which presents a rationale for examining whether augmentation of Spp1 levels in marginal cells could mitigate cisplatin-induced ototoxicity (Schmitt and Rubel, 2013). Thus, augmenting expression of genes downregulated in response to cisplatin in cell types responsible for EP generation may be an avenue for pursuing otoprotection.
Conversely, we suggest that certain genes that are upregulated in the presence of cisplatin may be reasonable targets for inhibition to mitigate the effects of cisplatin on the SV. Novel therapeutic gene targets that are upregulated in response to cisplatin include Klf10, Cldn3 encoding claudin 3, and Tspan1 encoding Tetraspanin 1, which are part of the Klf10 regulon. While not previously characterized in the inner ear, Klf10 overexpression has been implicated in renal podocyte dysfunction in diabetic nephropathy with suppression of Klf10 shown to reduce diabetes-induced proteinuria and kidney injury in mice (Lin C. et al., 2019). Despite similarities between the kidney and the stria vascularis (Spector and Carr, 1979; Walker et al., 1990; Beitz et al., 1999; Yamamoto et al., 2009), the known interacting partners of Klf10 in the kidney (Lin C. et al., 2019), including Kdm6a, nephrin (Npsh1), and methyltransferase Dnmt1, are not differentially expressed in the stria vascularis (see Supplementary Table S3). Our Klf10 regulon analysis identifies Tspan1 as a potential downstream target that is differentially regulated in marginal cells in response to cisplatin (Supplementary Figure S6A). Tetraspanins are thought to act as molecular scaffolds for adhesion, signaling, and adaptor proteins (Termini and Gillette, 2017). While the role of Tspan1 as well as the larger family of tetraspanins remains largely undefined in the inner ear, increased expression of Tspan1 has been observed in a wide variety of cancers including gastric, colon, pancreatic, liver and esophageal cancer and has been implicated in cancer cell migration and invasion (Zhang et al., 2019). Whether these upregulated genes act as compensatory versus detrimental adaptations in response to cisplatin remains to be determined. If detrimental, these gene targets may be candidates for inhibition to mitigate ototoxicity.
In order to enhance the robustness of our drug repurposing analyses, we utilized two druggable genome databases, Pharos (Nguyen et al., 2017) and DrugBank (Wishart et al., 2018). Utilizing these databases, druggability of differentially expressed genes in the Tbx2 regulon revealed potential therapeutic gene targets in intermediate cells in the form of target genes that were druggable by FDA approved drugs (Supplementary Figure S6B). Pharos has a specific focus on drugs that target G-protein coupled receptors, protein kinases, and ion channels (Nguyen et al., 2017), and has been utilized previously by our group to identify potential druggable gene targets (Korrapati et al., 2019). Within Pharos, target genes that are druggable by at least one FDA approved drug are designated as Tclin target genes. Gene targets with FDA approved drugs in DrugBank are searchable and are shown along with known and unknown interactions of FDA-approved drugs. As a result of our druggable targets analysis of differentially expressed genes in the intermediate cell-specific Tbx2 regulon, we identify metformin as a potential repurposable drug candidate targeting Ndufb2 (Chen et al., 2019; Muri et al., 2019) to test for potential to mitigate cisplatin-induced ototoxicity as an example of this repurposing drug candidate approach. Ndufb2 is part of the mitochondrial respiratory chain complex I assembly and has been implicated previously in bacterial sepsis (Li et al., 2017) and Alzheimer’s disease (Manners et al., 2018), suggesting that it may play a role in inflammation. Metformin, a well-known medication used to treat type 2 diabetes, has both anti-inflammatory (Martin-Montalvo et al., 2013; Kelly et al., 2015) and antitumor (Coyle et al., 2016; Hosoya et al., 2017; Zi et al., 2018; Feng et al., 2020) properties with several clinical trials currently underway to study its potential benefit in the setting of cancer (Chae et al., 2016). The antitumor properties of metformin are thought to be potentially mediated through the MTOR signaling pathway (Zi et al., 2018). Ndufb2 encodes the B2 subunit of NADH:ubiquinone oxidoreductase. Anti-inflammatory properties of metformin may in part be mediated by its ability to limit the production of reactive oxygen species by NADH:Ubiquinone Oxidoreductase, which may limit induction of pro-inflammatory interleukin-1β and boost anti-inflammatory interleukin-10 (Kelly et al., 2015). Recently, metformin has been found to have some beneficial properties as it relates to hearing protection in preclinical and clinical studies against a variety of insults to the inner ear including radiation, noise, and cisplatin, as well as in the recovery of hearing in the setting of sudden sensorineural hearing loss (Chang et al., 2014; Hosoya et al., 2017; Kesici et al., 2018; Chen et al., 2019; Muri et al., 2019; Feng et al., 2020). These observations suggest the possibility that metformin may have broader therapeutic applications as it relates to hearing.
In the model utilized in this study, we examined the acute changes that occur in the cochlea in the presence of a reduced EP. Outer hair cells are more susceptible to cisplatin-induced death than inner hair cells (Cheng et al., 2005; Rybak et al., 2007; Li et al., 2011; Breglio et al., 2017; Fernandez et al., 2019), as is the case in other models in which reduced EP precedes hair cell death. In genetic models of EP dysfunction (Liu H. et al., 2016; Huebner et al., 2019; Morell et al., 2020) without a known ototoxin, outer hair cell loss occurs in a delayed fashion after the observed reduction in EP despite the appearance of normally developed hair cells. As the SV generates the EP and establishes the environment for proper hair cell functioning, we speculate that hearing loss prior to outer hair cell degeneration in the setting of EP dysfunction may be due to the sustained effect of a reduced EP on active cochlear amplification by outer hair cells and that the loss of EP may be involved indirectly in the degeneration of outer hair cells (Anniko and Wróblewski, 1986; Jacob et al., 2011; Lukashkina et al., 2017; Zhao, 2017; Morell et al., 2020). Alternatively, work by Sewell demonstrates that reductions in EP resulting from furosemide administration are accompanied by elevations in in auditory nerve thresholds and suggest the loss of EP may have an effect at the auditory nerve (Sewell, 1984). While acknowledging this as speculative, work by our group and others suggests that alterations in the local extracellular ionic composition of the endolymph surrounding the outer hair cells may have an impact on their function and status (Anniko and Wróblewski, 1986; Coffin et al., 2009; Jacob et al., 2011). Alternatively, alterations in EP may initially impact cochlear function prior to effects on the hair cells by impacting auditory nerve fiber activity. Regardless, understanding how disruption of critical SV functions, including EP generation, are associated with hair cell loss, albeit delayed, may be important in identifying ways to prevent hair cell loss and, ultimately, hearing loss.
We utilized a single dose of cisplatin as a model of SV disease and EP generation dysfunction prior to the onset of deleterious effects of the drug on hearing and hair cells and then examined the transcriptional changes within specific SV cell types. Druggable target databases were then used to predict drugs that might mitigate or reverse these transcriptional changes. Our data at the RNA and protein levels indicate that cisplatin impairs stria vascularis function. We have identified cell-type specific regulatory pathways involved in the response to cisplatin, and these pathways have been used to suggest existing drugs that may mitigate cisplatin ototoxicity. In this way, we identify transcriptional changes that occur apparently prior to the onset of hearing loss that may be therapeutically targeted to prevent cisplatin-related ototoxicity.
Inbred CBA/J male and female mice were purchased from JAX (Stock No. 000656). P30 mice obtained from breeding pairs were used for all experiments including EP measurements, immunohistochemistry, digital droplet qPCR (ddPCR) and single-cell RNA-sequencing (scRNA-Seq).
Experimental mice were given an intraperitoneal (IP) injection of 14 mg/kg cisplatin 24 h prior to EP measurement or sacrifice. Control mice were given an equal volume IP injection of saline 24 h prior to EP measurement or sacrifice. All mice were age P30 for these experiments.
Methods have been previously described (Korrapati et al., 2019; Morell et al., 2020). Briefly, inner ears from P30 CBA/J mice were dissected and placed in 4% paraformaldehyde (PFA) overnight. Fixed adult mouse inner ears were then decalcified in 0.25 M EDTA for 4–5 days, transferred to 30% sucrose in PBS at 4°C overnight, followed by immersion in half 30% sucrose in PBS and super cryoembedding medium (SCEM) (C-EM001, Section-Lab Co, Ltd.; Hiroshima, Japan), followed by embedding in 100% SCEM for 2 h at room temperature, frozen in a cryomold biopsy square.
For cryosectioning, adhesive film (C-FUF303, Section-Lab Co, Ltd.) was fastened to the cut surface of the sample in order to support the section and sectioned slowly using a CM3050S cryostat microtome (Leica, Vienna, Austria) to obtain 10 μm thickness sections. The adhesive film with sections attached was submerged for 60 s in 100% ethanol, then transferred to distilled water. The adhesive film prevents specimen shrinkage and detachment. This methodology allows for high quality anatomic preservation of the specimen and sectioning at a thickness of 0.5 μm. Sections were mounted with SCMM mounting medium (Section-Lab, Hiroshima, Japan) and imaged using a 1.4 N.A. objective.
Fluorescence immunohistochemistry for known SV cell-type markers was performed as follows. Mid-modiolar sections were washed in PBS then permeabilized and blocked for 2 h at room temperature in PBS with 0.2% Triton X-100 (PBS-T) with 10% fetal bovine serum (A3840001, ThermoFisher Scientific, Waltham, MA, United States). Samples were then incubated in the appropriate primary antibodies in PBS-T with 10% fetal bovine serum, followed by three rinses in PBS-T and labeling with AlexaFluor-conjugated secondary antibodies (1:250, Life Technologies) in PBS-T for 1 h at room temperature. Where indicated, 4,6-diamidino-2-phenylindole (DAPI, 1:10,000, Life Technologies) was included with the secondary antibodies to detect nuclei. Organs were washed in PBS three times and mounted in SlowFade Gold (S36937, Invitrogen, ThermoFisher). Specimens were imaged using a Zeiss LSM710 confocal microscope. Sections were mounted with SCEM tissue embedding medium (C-EM001, Section-Lab Co, Ltd.). Primary antibodies used included rabbit anti-KCNJ10 (RRID:AB_2040120, Alomone Labs, APC-035, polyclonal, dilution 1:200), rabbit anti-CLDN11 (RRID:AB_2533259, Life Technologies, 364500, polyclonal, dilution 1:200), goat anti-SLC12A2 (RRID:AB_2188633, Santa Cruz Biotech, sc-21545, polyclonal, dilution 1:200), and Phalloidin AlexaFluor 647 (RRID: AB_2620155, Invitrogen, A22287, dilution 1:250).
Methods for endocochlear potential (EP) measurement have been described (Wangemann et al., 2004, 2007) and have been previously performed by our group (Imtiaz et al., 2018; Sharlin et al., 2018; Morell et al., 2020). Here, adult CBA/J mice were anesthetized with 2,2,2-tribromoethanol (T4842, Sigma-Aldrich, St. Louis, MO, United States) at a dose of 0.35 mg/g body weight. EP measurements were made using glass microelectrodes inserted into the round window and through the basilar membrane of the first turn of the cochlea. Induction of anoxia, allowing measurement of anoxic-state EP, was accomplished by intramuscular injection of succinylcholine chloride (0.1 μg/g, NDC-0409-6629-02, Pfizer, New York, NY, United States) after establishment of deep anesthesia followed by additional injection of 2,2,2-Tribromoethanol (T4842, Sigma-Aldrich, St. Louis, MO, United States). Anoxic-state EP provides an indicator of the lowest EP and sensory hair cell function. In the presence of functional hair cells, the anoxic-state EP is negative, whereas the EP is zero if the hair cells are not functional. Data were recorded digitally (Digidata 1440A and AxoScope 10; Axon Instruments) and analyzed using Clampfit10 (RRID:SCR_011323, Molecular Devices, San Jose, CA, United States). For EP measurements, control (n = 12 mice) and cisplatin-treated (n = 10 mice) mice were utilized with each mouse serving as a biological replicate.
Methods have been previously described (Morell et al., 2020). Briefly, auditory brainstem responses (ABRs) were measured in both ears for P30 control (n = 6) and cisplatin-treated (n = 6) CBA/J mice. Mice were anesthetized with an intraperitoneal injection of ketamine (56 mg/kg) and dexdomitor (0.375 mg/kg) and placed on a heating pad connected to a temperature controller (TC-2000, World Precision Instruments) inside a sound-attenuated booth (Acoustic Systems). The heating pad and thermometer placed under the mouse body were used to maintain body temperature near 37°C. Recordings were obtained using Tucker-Davis Technologies hardware (RZ6 Processor) and software (BioSigRZ, version 5.7.5). For ABR testing, subdermal needle electrodes (Rhythmlink) were placed at the vertex, under the test ear, and under the contralateral ear (ground). Blackman-gated tone burst stimuli (3 ms, 29.9/s, alternating polarity) were presented to the test ear at 8, 16, 32, and 40 kHz via a closed-field Tucker-Davis Technologies MF-1 speaker. Responses were amplified (20×), filtered (0.3–3 kHz), and digitized (25 kHz) with 512–1024 artifact-free responses per waveform. For each frequency, testing began at 80 dB SPL and decreased in 10 dB steps until the ABR waveform was no longer discernable. Once the response was lost, testing continued in 5 dB steps with a minimum of two waveforms per stimulus level to verify repeatability of ABR waves. ABR thresholds were determined by visual inspection of stacked waveforms for the lowest stimulus level that yielded repeatable waves.
Distortion-product otoacoustic emissions (DPOAEs) were measured in both ears for P30 control (n = 6) and cisplatin-treated (n = 6) adult CBA/J mice using Tucker-Davis Technologies hardware (RZ6 Multi I/O processor, MF-1 speakers) and software (BioSigRz, version 5.7.5) in conjunction with an Etymotic ER-10B + microphone. Two tones were presented simultaneously at levels of f1 = 65 dB SPL and f2 = 55 dB SPL with the higher frequency tone (f2) set between 4-44.8 kHz (5 points per octave) and f2/f1 = 1.25. Mean noise floors were calculated from levels at six frequencies surrounding the 2f1-f2 DPOAE frequency.
ImageJ was utilized to calculate the cross-sectional area and thickness of the SV in mid-modiolar sections of both control (n = 5 mice) and cisplatin-treated (n = 6 mice) CBA/J mice at postnatal day 30 (P30) as previously described (Sharlin et al., 2018; Morell et al., 2020). Briefly, midmodiolar cross-sections of adult mouse cochleae were imaged using a Zeiss LSM710 confocal microscope at 40X magnification at each turn of the cochlea. Using phalloidin staining as an outline for the SV, the images for each apical, medial, and basal turn were measured for changes in total SV cross-sectional area (μm2), changes in SV thickness (μm) measured at the thickest point, and changes in the number of nuclei between control and experimental mice.
TUNEL assay (Invitrogen, C10617) was performed per manufacturer provided protocol. Briefly, after washing with PBS, samples were incubated with a 1X Proteinase K in PBS solution for 15 min at room temperature and then washed again in PBS. The samples were then covered in terminal deoxynucleotidyl transferase (TdT) reaction buffer and incubated for 10 min at 37°C. The TdT reaction buffer was removed and replaced with a solution of TdT reaction buffer, EdUTP, and TdT enzyme and incubated for 60 min at 37°C. The samples were rinsed in DI water and washed with a solution of 3% BSA and 0.1% Triton X-100 in PBS for 5 min at room temperature. Samples were then incubated with a solution of Click-iTTM Plus TUNEL Supermix and 10X Click-iTTM Plus TUNEL Reaction buffer additive and incubated for 30 min at 37°C, away from light. The samples were then washed in 3% BSA in PBS for 5 min at room temperature and rinsed in 1X PBS. Samples were then washed with PBST three times and incubated for 1 h at room temperature with Phalloidin AlexaFluor 647 (Invitrogen, A22287, dilution 1:250) and DAPI diluted with 10% Fetal Bovine Serum (FBS) in PBST. Samples were then washed with PBST three times and mounted.
TUNEL-positive nuclei were used as an indicator of apoptosis in cells. The apoptotic rate was calculated as the quotient of TUNEL-positive nuclei over the total number of nuclei labeled with DAPI, multiplied by 100, to calculate a percentage of total apoptotic cells within each turn.
Control (n = 5) and experimental (n = 6) adult CBA/J mice were given saline and 14 mg/kg cisplatin IP injections 24 h prior to sacrifice and dissection. To prepare samples for freezing, inner ears were dissected and placed in 4% paraformaldehyde (PFA) overnight. Ears were then placed in 0.25 M ethylenediaminetetraacetic acid (EDTA) in phosphate-buffered saline (PBS) at 4°C overnight. Next, they were placed in 30% sucrose in PBS at 4°C overnight, followed by immersion in half 30% sucrose in PBS and super cryoembedding medium (SCEM, SECTION-LAB Co. Ltd.) for 2 h. Finally, they were placed in full SCEM for 2 h, frozen in a cryomold biopsy square, and sectioned onto slides using a cryostat.
For immunofluorescence staining, dried slides were placed on slide warmer for 15 min and then washed with phosphate-buffered saline with tween 20 (PBST) for 10 min. Samples were blocked with 10% FBS in PBST for 2 h at room temperature. Primary antibodies were diluted in 10% FBS in PBST and slides incubated overnight at 4°C. Samples were washed with PBST three times and incubated for 1 h at room temperature with AlexaFluor-conjugated secondary antibodies and DAPI diluted with 10% Fetal Bovine Serum (FBS) in PBST. Samples were then washed with PBST three times and mounted. Primary antibodies used included goat anti-SLC12A2 (Santa Cruz Biotech, sc-21545, polyclonal, dilution 1:100), rabbit anti-KCNJ10 (Alomone Labs, APC-035, polyclonal, dilution 1:100), rabbit anti-CLDN11 (Life Technologies, 364500, polyclonal, dilution 1:100), and Phalloidin AlexaFluor 647 (Invitrogen, A22287, dilution 1:250).
Immunofluorescence intensity analysis was performed as previously described (Sharlin et al., 2018; Morell et al., 2020). Briefly, imaging was performed using a Zeiss LSM710 confocal microscope at 40X magnification at each turn of the cochlea. After converting images to grayscale, fluorescence intensity quantification was performed in ImageJ by calculating the fluorescence intensity of the outlined region of the SV as previously described (Hartig, 2013). Fluorescence intensity was normalized by comparing the SV fluorescence intensity to that of a corresponding region in the scala media. Measurements for the upper (apical), middle, and lower (basal) turns of the cochlea were obtained. These measurements were obtained for known SV cell types including marginal cells (SLC12A2), intermediate cells (KCNJ10), and basal cells (CLDN11). KCNJ10 fluorescence intensity in the spiral ganglion neurons served as a control for immunofluorescence signal intensity measurements. Spiral ganglion fluorescence intensity was unchanged between cisplatin-treated and untreated mice with no statistically significant difference between fluorescence intensity measurements (data not shown). A two-way ANOVA comparison was used to measure statistical significance between conditions for each antibody.
Cochlear whole mounts from P30 control (n = 6) and cisplatin-treated (n = 6) adult CBA/J mice were immuno-stained with anti-MYO7A antibody (Axxora, LLC) for hair cells and DAPI (Life Technologies) for cell nuclei. Organ of Corti sections were microdissected and mounted on slides. Inner and outer hair cell counts were performed from two 210 μm length regions from two cochlear turns (apical and basal) and the mean hair cell count was determined.
Control (n = 4) and experimental (n = 4) adult (P30) CBA/J mice were given a single IP injection of saline or 14 mg/kg cisplatin, respectively, and sacrificed after 24 h. To prepare samples for ddPCR, inner ears were dissected in PBST over ice and SV tissue carefully removed to avoid including unwanted cell types. RNA was retrieved using the Arcturus PicoPure RNA isolation kit and converted to cDNA. TaqMan probes of 17 genes of interest were designed and ordered from Applied Biosystems. Three 96-well plates were prepared to include samples from four control and four experimental mice in triplicate. PCR was done in a 20 μL total volume of 10 μL Bio-Rad ddPCR Supermix for Probes (No dUTP), 1 μL probe of gene of interest, 1 μL reference gene Actb, 1 μL diluted cDNA, and 7 μL molecular grade water. The ddPCR Droplets were generated using the QX200 AutoDG Droplet Digital. PCR was performed as described in the QX200 ddPCR EvaGreen Supermix instructions. Droplets were read with a QX200 Droplet Reader (BioRad) and analyzed with QuantaSoft software (BioRad). Primer sequences utilized for ddPCR are provided in the supplement (Supplementary Table S2).
A group of P30 CBA/J mice (n = 6, three female, three male) were given a single IP injection of 14 mg/kg cisplatin and sacrificed after 24 h. SV from these mice were micro-dissected as previously described (Korrapati et al., 2019) and utilized to generate single cell transcriptomes. To prepare samples for scRNA-Seq, inner ears were dissected in Dulbecco’s Modified Eagle Medium (DMEM) over ice and SV tissue carefully removed to avoid including unwanted cell types. SV tissue was then placed in a tube containing 200 μL DMEM. 10 μL trypsin was added to the tube and incubated for 7 min at 37°C. After incubation 150 μL liquid was removed and replaced with 150 μL 10% FBS in DMEM. The tissue was triturated for 2 min to dissociate single cells and then passed through a 20 μm filter. Two μL of supernatant was removed, leaving a 100 μL final volume. The concentration was then measured using a cell counter and the final concentration used to determine the volume to be placed in the well of a 10 × 8-channel microfluidics chip. Within this chip droplets were created that each contained a single cell and single uniquely barcoded bead. The cells were then lysed, and RT performed to convert mRNA to cDNA. Single cell libraries were prepared, and the results sequenced on an Illumina sequencer. The details regarding scRNA-Seq workflow are as described in Zheng et al. (2017). After scRNA-Seq protocol and library prep, the resultant sparse matrix was imported into R and clustered using the Seurat package.
Following Illumina sequencing of the scRNA-Seq results, three files were created: a list of individually barcoded cells, a list of gene symbols with their Ensembl IDs, and a sparse matrix of the read counts of each gene in each cell. These files were then imported into R and converted to a dgTMatrix; a form suitable for analysis using the Seurat package in R. A total of 2,681 cells were captured in the experiment, with 2,236 cells used in the final analysis following filters to minimize doublets and debris. The median number of genes identified per cell was 1,312, while the median number of reads per cell was 3,669. The clusters created by the Seurat package were identified and labeled using published markers of cell types within the SV and lateral wall of the murine cochlea. The methods behind Seurat clustering and differential expression have been previously described (Satija et al., 2015).
Cisplatin-treated adult SV single-cell transcriptomes were compared to a previously published dataset consisting of unperturbed adult SV single-cell transcriptomes (Korrapati et al., 2019) (GEO Accession ID: GSE136196; see footnote 2). The two datasets were combined using the MergeSeurat function and normalized after the merge. Cellular identities were carried over from the individual Seurat clustering as discussed in the previous section. Differential expression analysis was performed using the FindAllMarkers function in Seurat (Satija et al., 2015) and the DEsingle package (Miao et al., 2018). DESingle is a differential expression package that utilizes raw reads from single cell experiments and performs its own normalization and batch correction. The results of the differential expression analyses were visualized using the EnhancedVolcano package (Blighe et al., 2018) in R.
Single-cell regulatory network inference (SCENIC) and clustering analysis was performed on the post-Seurat analysis object using the methods described initially by Aibar et al. (2017) and applied to the analysis of single cell transcriptomes of the adult SV (Korrapati et al., 2019). Cisplatin-treated SV cells analyzed in the SCENIC pipeline were given identities based on clustering analysis from Seurat to allow for post hoc identification of cell types. Briefly, cell type-specific regulons were identified in cisplatin-treated adult SV, regulon activity was quantified, and a regulon activity matrix was constructed to identify cisplatin-treated cell states. Expression of the genes within these regulons was then compared against the genes demonstrating differential expression between cisplatin-treated and control SV single cell transcriptomes obtained by using DESingle (Miao et al., 2018).
To identify druggable targets within this list of differentially expressed cell type-specific regulon genes, Pharos (see footnote 1) was utilized as previously described (Korrapati et al., 2019). Briefly, Pharos is a web interface for the Target Central Resource Database (TCRD) created by the “Illuminating the Druggable Genome” program, which collates known drug-protein interactions, including both FDA-approved drugs and small molecules (Nguyen et al., 2017). Pharos categorizes each protein with a “target developmental level” according to how much is known on its “druggability” (Owens, 2007). Targets are classified according to available knowledge: (1) Tclin, where there is known interaction with FDA approved drugs, (2) Tchem, where genes have known and unknown interactions with ligands, (3) Tbio, where genes have a known function but no known interactions, and (4) Tdark, where there is very little known either about function or interaction. To focus on the most clinically relevant targets, we identified Tclin and Tchem genes that were differentially expressed between control and cisplatin-treated scRNA-Seq datasets that were also present in the cell-type specific regulons identified by the SCENIC program. The goal was to identify possible drug interventions in genes and regulatory networks known to be involved in the maintenance of the EP that were also either up or down regulated in the presence of cisplatin.
For pairwise comparisons between control and cisplatin-treated CBA/J mice, an unpaired two-tailed Student’s t-test was used. Comparisons of multiple assays across the same tissue were corrected for multiple comparisons using a Benjamini–Hochberg correction with a false discovery rate (FDR or q value) of 5%. Two-way ANOVA and Sidak’s multiple comparisons test were used for comparisons between control and cisplatin-treated mice involving cochlear turns. All statistical analyses were performed using GraphPad Prism version 6.0 (RRID:SCR_002798 GraphPad) for PC. For measurements of strial thickness, cross-sectional area and fluorescent intensity, all values are means ± standard deviations (SD). Both males and females were tested, and there was no evidence of a significant effect of sex on any measures. Therefore, the data displayed in all graphs are from males and females combined.
All data generated in these studies have been deposited in the Gene Expression Omnibus (GEO) database (GSE165662) and can be found on GEO at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165662. Single-cell data have been uploaded into the gene Expression Analysis Resource (gEAR), a website for visualization and comparative analysis of multiomic data, with an emphasis on hearing research (https://umgear.org/p?l=d22c4230). All other data associated with this study are available in the main text or the Supplementary Material.
The animal study was reviewed and approved by Animal Care and Use Committee of the National Institute of Neurological Diseases and Stroke and the National Institute on Deafness and Other Communication Disorders, National Institutes of Health.
IT, RO, SK, and KF contributed to isolation of single cells for single cell RNA-Seq (scRNA-Seq). EB and RJM were responsible for sequencing and alignment of scRNA-Seq and snRNA-Seq datasets. IT and MH contributed to scRNA-Seq bioinformatic data analysis. IT and RO were responsible for smFISH and immunohistochemistry. IT, KF, EB, RJM, LLC, and MH contributed to writing and revising the manuscript. All the authors read and approved final manuscript.
This research was supported (in part) by the Intramural Research Program of the NIH, NIDCD to MH (ZIA DC000088), LLC (ZIA DC000079), TF (ZIC DC000080), and RJM (ZIC DC000086).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge Hui Cheng for her guidance on statistical analyses and error testing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.718241/full#supplementary-material
Supplementary Figure 1 | A single large dose of cisplatin shows no loss of inner or outer hair cells or elevated DPOAE threshold in either ear after 24 h. (A) Inner hair cell counts of control (n = 6) and cisplatin-treated (n = 6) mice were unchanged in either apical or basal halves of the cochlea between conditions (two-way ANOVA, p = 0.6434). (B) Outer hair cell counts were unchanged in either apical or basal halves of the cochlea between conditions (two-way ANOVA, p = 0.6423). (C) Left ear DPOAE levels were unchanged between conditions (paired t-test, p = 0.1823). (D) Right ear DPOAE levels were unchanged between conditions (paired t-test, p = 0.0911).
Supplementary Figure 2 | DDPCR validation of upregulated and downregulated genes in control and cisplatin-treated whole stria vascularis as identified by DESingle. (A) Upregulated genes: Anxa2 (unpaired t-test, p = 0.1392), Cdkn1a (unpaired t-test, p = 0.0490), Cdkn1c (unpaired t-test, p = 0.0001), Gpx2 (unpaired t-test, p = 0.3055), Ifitm3 (unpaired t-test, p = 0.0961), Mlana (unpaired t-test, p = 0.9097), Mt1 (unpaired t-test, p = 0.0002), Mt2 (unpaired t-test, p = 0.0002). (B) Downregulated genes: Aes1 (unpaired t-test, p = 0.0101), Aif1l (unpaired t-test, p = 0.0017), Gas2 (unpaired t-test, p = 0.0005), Itpr1 (unpaired t-test, p = 0.0004), Itpr2 (unpaired t-test, p = 0.0048), Mt3 (unpaired t-test, p = 0.0123), Nrep (unpaired t-test, p = 0.0087), Tns1 (unpaired t-test, p = 0.0002).
Supplementary Figure 3 | Regulon specificity scores ranked according to their specificity to major cell types of the SV. Heavy dotted line is the mean RSS score and the lighter dotted line is one standard deviation from the mean. Regulons marked in blue meet the standard deviation threshold for specificity. (A) Marginal cell regulons. (B) Intermediate cell regulons and (C) Basal cell regulons.
Supplementary Figure 4 | DDPCR measurement of the presence of (A), Klf10 and (B), Tbx2 regulons. Transcription factors and downstream targets were measured against housekeeping gene Actb.
Supplementary Figure 5 | Protein expression of SPP1 is significantly reduced in cisplatin-treated SV as measured by immunofluorescence intensity analysis.
Supplementary Figure 6 | Pharos druggable targets analysis combined with DE analysis identifies potential therapeutic gene targets and repurposable drugs for cisplatin-induced ototoxicity. (A) Illustration depicting the differentially expressed Tclin and Tchem target genes of the marginal cell specific regulon Klf10. Genes are colored according to their Pharos gene family identification and interacting drugs and ligands are colored according to their drug-protein interaction. (B) The differentially expressed Tclin and Tchem target genes of the intermediate cell specific regulon Tbx2.
Supplementary Table 1 | Marker genes of UMAP clusters of the cisplatin-treated and control scRNA-Seq datasets as identified by the Seurat FindMarkers function.
Supplementary Table 2 | Forward, reverse, and probe primer sequences for ddPCR probes.
Supplementary Table 3 | DEsingle results for marginal, intermediate, and basal cells between cisplatin-treated and control scRNA-Seq datasets.
Supplementary Table 4 | Gene regulatory networks (regulons) identified in the cisplatin-treated scRNA-Seq dataset with their regulon specificity scores.
Aibar, S., González-Blas, C. B., Moerman, T., Huynh-Thu, V. A., Imrichova, H., Hulselmans, G., et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. doi: 10.1038/nmeth.4463
Alabdullatif, M. A., Al Dhaibani, M. A., Khassawneh, M. Y., and El-Hattab, A. W. (2017). Chromosomal microarray in a highly consanguineous population: diagnostic yield, utility of regions of homozygosity, and novel mutations. Clin. Genet. 91, 616–622. doi: 10.1111/cge.12872
Anniko, M., and Wróblewski, R. (1986). Ionic environment of cochlear hair cells. Hear. Res. 22, 279–293. doi: 10.1016/0378-5955(86)90104-8
Beitz, E., Kumagami, H., Krippeit-Drews, P., Ruppersberg, J. P., and Schultz, J. E. (1999). Expression pattern of aquaporin water channels in the inner ear of the rat. The molecular basis for a water regulation system in the endolymphatic sac. Hear. Res. 132, 76–84. doi: 10.1016/s0378-5955(99)00036-2
Blighe, K., Rana, S., and Lewis, M. (2018). EnhancedVolcano: Publication-Ready Volcano Plots With Enhanced Colouring and Labeling. Available online at: https://github.com/kevinblighe/EnhancedVolcano (accessed June 6, 2019).
Breglio, A. M., Rusheen, A. E., Shide, E. D., Fernandez, K. A., Spielbauer, K. K., McLachlin, K. M., et al. (2017). Cisplatin is retained in the cochlea indefinitely following chemotherapy. Nat. Commun. 8:1654.
Caldera, M., Müller, F., Kaltenbrunner, I., Licciardello, M. P., Lardeau, C. H., Kubicek, S., et al. (2019). Mapping the perturbome network of cellular perturbations. Nat. Commun. 10:5140.
Chae, Y. K., Arya, A., Malecek, M. K., Shin, D. S., Carneiro, B., Chandra, S., et al. (2016). Repurposing metformin for cancer treatment: current clinical studies. Oncotarget 7, 40767–40780. doi: 10.18632/oncotarget.8194
Chang, J., Jung, H. H., Yang, J. Y., Lee, S., Choi, J., Im, G. J., et al. (2014). Protective effect of metformin against cisplatin-induced ototoxicity in an auditory cell line. JARO - J. Assoc. Res. Otolaryngol. 15, 149–158. doi: 10.1007/s10162-013-0431-y
Chang, Q., Wang, J., Li, Q., Kim, Y., Zhou, B., Wang, Y., et al. (2015). Virally mediated Kcnq1 gene replacement therapy in the immature scala media restores hearing in a mouse model of human Jervell and Lange-Nielsen deafness syndrome. EMBO Mol. Med. 7, 1077–1086. doi: 10.15252/emmm.201404929
Chen, H. C., Chung, C. H., Lu, C. H., and Chien, W. C. (2019). Metformin decreases the risk of sudden sensorineural hearing loss in patients with diabetes mellitus: a 14-year follow-up study. Diabetes Vasc. Dis. Res. 16, 324–327. doi: 10.1177/1479164119826292
Chen, J., and Zhao, H. B. (2014). The role of an inwardly rectifying K+ channel (Kir4.1) in the inner ear and hearing loss. Neuroscience 265, 137–146. doi: 10.1016/j.neuroscience.2014.01.036
Cheng, A. G., Cunningham, L. L., and Rubel, E. W. (2005). Mechanisms of hair cell death and protection. Curr. Opin. Otolaryngol. Head Neck Surg. 13, 343–348. doi: 10.1097/01.moo.0000186799.45377.63
Coffin, A. B., Reinhart, K. E., Owens, K. N., Raible, D. W., and Rubel, E. W. (2009). Extracellular divalent cations modulate aminoglycoside-induced hair cell death in the zebrafish lateral line. Hear. Res. 253, 42–51. doi: 10.1016/j.heares.2009.03.004
Coyle, C., Cafferty, F. H., Vale, C., and Langley, R. E. (2016). Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 27, 2184–2195. doi: 10.1093/annonc/mdw410
Crawford, N. T., McIntyre, A. J., McCormick, A., D’Costa, Z. C., Buckley, N. E., and Mullan, P. B. (2019). TBX2 interacts with heterochromatin protein 1 to recruit a novel repression complex to EGR1-targeted promoters to drive the proliferation of breast cancer cells. Oncogene. 38, 5971–5986. doi: 10.1038/s41388-019-0853-z
da-Eto, M. I., Ohgami, N., Iida, M., Yajima, I., Kumasaka, M. Y., Takaiwa, K., et al. (2011). Partial requirement of endothelin receptor B in spiral ganglion neurons for postnatal development of hearing. J. Biol. Chem. 286:34.
Decaesteker, B., Denecker, G., Van Neste, C., Dolman, E. M., Van Loocke, W., Gartlgruber, M., et al. (2018). TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat. Commun. 9:4866.
del Sol, A., Balling, R., Hood, L., and Galas, D. (2010). Diseases as network perturbations. Curr. Opin. Biotechnol. 21, 566–571. doi: 10.1016/j.copbio.2010.07.010
Delpire, E., Lu, J., England, R., Dull, C., and Thorne, T. (1999). Deafness and imbalance associated with inactivation of the secretory Na- K-2Cl co-transporter. Nat. Genet. 22, 192–195. doi: 10.1038/9713
Dixon, M. J., Gazzard, J., Chaudhry, S. S., Sampson, N., Schulte, B. A., and Steel, K. P. (1999). Mutation of the Na-K-Cl co-transporter gene Slc12a2 results in deafness in mice. Hum. Mol. Genet. 8, 1579–1584. doi: 10.1093/hmg/8.8.1579
Faridi, R., Tona, R., Brofferio, A., Hoa, M., Olszewski, R., Schrauwen, I., et al. (2019). Mutational and phenotypic spectra of KCNE1 deficiency in Jervell and Lange-Nielsen Syndrome and Romano-Ward Syndrome. Hum. Mutat. 40, 162–176.
Feng, A. Y., Enriquez-Marulanda, A., Kouhi, A., Ali, N. E. S., Moore, J. M., and Vaisbuch, Y. (2020). Metformin potential impact on the growth of vestibular schwannomas. Otol. Neurotol. 38, 237–239.
Fernandez, K., Wafa, T., Fitzgerald, T. S., and Cunningham, L. L. (2019). An optimized, clinically relevant mouse model of cisplatin-induced ototoxicity. Hear. Res. 375, 66–74. doi: 10.1016/j.heares.2019.02.006
Fiers, M. W. E. J., Minnoye, L., Aibar, S., González-Blas, C. B., Atak, Z. K., and Aerts, S. (2018). Mapping gene regulatory networks from single-cell omics data. Brief. Funct. Genomics. 17, 246–254. doi: 10.1093/bfgp/elx046
Freudenthal, B., Kulaveerasingam, D., Lingappa, L., Shah, M. A., Brueton, L., Wassmer, E., et al. (2011). KCNJ10 mutations disrupt function in patients with east syndrome. Nephron - Physiol. 119, 40–48.
Furukawa, M., Ikeda, K., Takeuchi, S., Oshima, T., Kikuchi, T., and Takasaka, T. (1996). Na+,K+-ATPase activity in the cochlear lateral wall of the gerbil. Neurosci. Lett. 213, 165–168. doi: 10.1016/0304-3940(96)12862-7
Gow, A., Davies, C., Southwood, C. M., Frolenkov, G., Chrustowski, M., Ng, L., et al. (2004). Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J. Neurosci. 24, 7051–7062. doi: 10.1523/jneurosci.1640-04.2004
Gratton, M. A., Schmiedt, R. A., and Schulte, B. A. (1996). Age-related decreases in endocochlear potential are associated with vascular abnormalities in the stria vascularis. Hear. Res. 94, 116–124. doi: 10.1016/0378-5955(96)00011-1
Grifa, A., Wagner, C. A., D’Ambrosio, L., Melchionda, S., Bernardi, F., Lopez-Bigas, N., et al. (1999). Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nat. Genet. 23, 16–18. doi: 10.1038/12612
Hartig, S. M. (2013). Basic image analysis and manipulation in imageJ. Curr. Protoc. Mol. Biol. 102, 14.15.1–14.15.12.
Hosoya, M., Fujioka, M., Sone, T., Okamoto, S., Akamatsu, W., Ukai, H., et al. (2017). Cochlear cell modeling using disease-specific ipscs unveils a degenerative phenotype and suggests treatments for congenital progressive hearing loss. Cell Rep. 18, 68–81. doi: 10.1016/j.celrep.2016.12.020
Huebner, A. K., Maier, H., Maul, A., Nietzsche, S., Herrmann, T., Praetorius, J., et al. (2019). Early hearing loss upon disruption of Slc4a10 in C57BL/6 mice. JARO - J. Assoc. Res. Otolaryngol. 20, 233–245. doi: 10.1007/s10162-019-00719-1
Ikeda, K., Kusakari, J., Takasaka, T., and Saito, Y. (1987). Early effects of acetazolamide on anionic activities of the guinea pig endolymph: evidence for active function of carbonic anhydrase in the cochlea. Hear. Res. 31, 211–216. doi: 10.1016/0378-5955(87)90189-4
Imtiaz, A., Belyantseva, I. A., Beirl, A. J., Fenollar-Ferrer, C., Bashir, R., Bukhari, I., et al. (2018). CDC14A phosphatase is essential for hearing and male fertility in mouse and human. Hum. Mol. Genet. 27, 780–798. doi: 10.1093/hmg/ddx440
Jacob, S., Pienkowski, M., and Fridberger, A. (2011). The endocochlear potential alters cochlear micromechanics. Biophys. J. 100, 2586–2594. doi: 10.1016/j.bpj.2011.05.002
Kelly, B., Tannahill, G. M., Murphy, M. P., and O’Neill, L. A. J. (2015). Metformin inhibits the production of reactive oxygen species from NADH: ubiquinone oxidoreductase to limit induction of interleukin-1β (IL-1β) and boosts interleukin-10 (IL-10) in lipopolysaccharide (LPS)-activated macrophages. J. Biol. Chem. 290, 20348–20359. doi: 10.1074/jbc.m115.662114
Kelsell, D. P., Dunlop, J., Stevens, H. P., Lench, N. J., Liang, J. N., Parry, G., et al. (1997). Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387, 80–83. doi: 10.1038/387080a0
Kesici, G. G., Öcal, F. C. A., Gürgen, S. G., Erdem, S. R., Öğüş, E., Erbek, H. S., et al. (2018). The protective effect of metformin against the noise-induced hearing loss. Eur. Arch. Oto-Rhino-Laryngology. 275, 2957–2966. doi: 10.1007/s00405-018-5161-7
Kikuchi, T., Kimura, R. S., Paul, D. L., and Adams, J. C. (1995). Gap junctions in the rat cochlea: immunohistochemical and ultrastructural analysis. Anat. Embryol. (Berl). 191, 101–118.
Kitajiri, S. I., Furuse, M., Morita, K., Saishin-Kiuchi, Y., Kido, H., Ito, J., et al. (2004a). Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear. Res. 187, 25–34. doi: 10.1016/s0378-5955(03)00338-1
Kitajiri, S. I., Miyamoto, T., Mineharu, A., Sonoda, N., Furuse, K., Hata, M., et al. (2004b). Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J. Cell Sci. 117, 5087–5096. doi: 10.1242/jcs.01393
Konishi, T., Hamrick, P. E., and Walsh, P. J. (1978). Ion transport in Guinea pig cochlea: I. Potassium and sodium transport. Acta Otolaryngol. 86, 22–34. doi: 10.3109/00016487809124717
Korrapati, S., Taukulis, I., Olszewski, R., Pyle, M., Gu, S., Singh, R., et al. (2019). Single cell and single nucleus RNA-Seq reveal cellular heterogeneity and homeostatic regulatory networks in adult mouse stria vascularis. Front. Mol. Neurosci. 12:316. doi: 10.3389/fnmol.2019.00316
Lang, F., Vallon, V., Knipper, M., and Wangemann, P. (2007). Functional significance of channels and transporters expressed in the inner ear and kidney. Am. J. Physiol. - Cell Physiol. 293, 1187–1208.
Lécuyer, M. A., Saint-Laurent, O., Bourbonnière, L., Larouche, S., Larochelle, C., Michel, L., et al. (2017). Dual role of ALCAM in neuroinflammation and blood-brain barrier homeostasis. Proc. Natl. Acad. Sci. U.S.A. 114, 524–533.
Li, Y., Ding, D., Jiang, H., Fu, Y., and Salvi, R. (2011). Co-administration of cisplatin and furosemide causes rapid and massive loss of cochlear hair cells in mice. Neurotox. Res. 20, 307–319. doi: 10.1007/s12640-011-9244-0
Li, Y., McGrail, D. J., Xu, J., Mills, G. B., Sahni, N., and Yi, S. (2018). Gene regulatory network perturbation by genetic and epigenetic variation. Trends Biochem. Sci. 43, 576–592. doi: 10.1016/j.tibs.2018.05.002
Li, Z., Zhang, Y., Liu, Y., Liu, Y., and Li, Y. (2017). Identification of key genes in Gram-positive and Gram-negative sepsis using stochastic perturbation. Mol. Med. Rep. 16, 3133–3146. doi: 10.3892/mmr.2017.7013
Liberali, P., Snijder, B., and Pelkmans, L. (2015). Single-cell and multivariate approaches in genetic perturbation screens. Nat. Rev. Genet. 16, 18–32. doi: 10.1038/nrg3768
Lin, C., Hsu, Y., Huang, Y., Shih, Y., Wang, C., Chiang, W., et al. (2019). A KDM6A–KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction. EMBO Mol. Med. 11:e9828.
Lin, X., Li, G., Zhang, Y., Zhao, J., Lu, J., Gao, Y., et al. (2019). Hearing consequences in Gjb2 knock-in mice: implications for human p.V37I mutation. Aging (Albany. NY). 11, 7416–7441. doi: 10.18632/aging.102246
Liu, H., Li, Y., Chen, L., Zhang, Q., Pan, N., Nichols, D. H., et al. (2016). Organ of corti and stria vascularis: is there an interdependence for survival? PLoS One 11:e0168953. doi: 10.1371/journal.pone.0168953
Liu, W., Boström, M., Kinnefors, A., and Rask-Andersen, H. (2009). Unique expression of connexins in the human cochlea. Hear. Res. 250, 55–62. doi: 10.1016/j.heares.2009.01.010
Liu, W., Edin, F., Blom, H., Magnusson, P., Schrott-Fischer, A., Glueckert, R., et al. (2016). Super-resolution structured illumination fluorescence microscopy of the lateral wall of the cochlea: the Connexin26/30 proteins are separately expressed in man. Cell Tissue Res. 365, 13–27. doi: 10.1007/s00441-016-2359-0
Liu, W., Schrott-Fischer, A., Glueckert, R., Benav, H., and Rask-Andersen, H. (2017). The human “cochlear battery” – claudin-11 barrier and ion transport proteins in the lateral wall of the cochlea. Front. Mol. Neurosci. 10:239. doi: 10.3389/fnmol.2017.00239
Locher, H., de Groot, J. C. M. J., van Iperen, L., Huisman, M. A., Frijns, J. H. M., and Chuva de Sousa Lopes, S. M. (2015). Development of the stria vascularis and potassium regulation in the human fetal cochlea: insights into hereditary sensorineural hearing loss. Dev. Neurobiol. 75, 1219–1240. doi: 10.1002/dneu.22279
Lopez, C. A., Olson, E. S., Adams, J. C., Mou, K., Denhardt, D. T., and Davis, R. L. (1995). Osteopontin expression detected in adult cochleæ and inner ear fluids. Hear. Res. 85, 210–222. doi: 10.1016/0378-5955(95)00046-7
Lukashkina, V. A., Levic, S., Lukashkin, A. N., Strenzke, N., and Russell, I. J. (2017). A connexin30 mutation rescues hearing and reveals roles for gap junctions in cochlear amplification and micromechanics. Nat. Commun. 8:14530.
Mammano, F. (2019). Inner ear connexin channels: roles in development and maintenance of cochlear function. Cold Spring Harb. Perspect. Med. 9:a033233. doi: 10.1101/cshperspect.a033233
Manners, H. N., Roy, S., and Kalita, J. K. (2018). Intrinsic-overlapping co-expression module detection with application to Alzheimer’s Disease. Comput. Biol. Chem. 77, 373–389. doi: 10.1016/j.compbiolchem.2018.10.014
Marcus, D. C., Wu, T., Wangemann, P., and Kofuji, P. (2002). KCNJ10 (kir4.1) potassium channel knockout abolishes endocochlear potential. Am. J. Physiol. - Cell Physiol. 282, 403–407.
Martin-Montalvo, A., Mercken, E. M., Mitchell, S. J., Palacios, H. H., Mote, P. L., Scheibye-Knudsen, M., et al. (2013). Metformin improves healthspan and lifespan in mice. Nat. Commun. 4:2192.
Matsushima, Y., Shinkai, Y., Kobayashi, Y., Sakamoto, M., Kunieda, T., and Tachibana, M. (2002). A mouse model of Waardenburg syndrome type 4 with a new spontaneous mutation of the endothelin-B receptor gene. Mamm. Genome 13, 30–35. doi: 10.1007/s00335-001-3038-2
McNeill, A., Iovino, E., Mansard, L., Vache, C., Baux, D., Bedoukian, E., et al. (2020). SLC12A2 variants cause a neurodevelopmental disorder or cochleovestibular defect. Brain 143, 2380–2387. doi: 10.1093/brain/awaa176
Mei, L., Chen, J., Zong, L., Zhu, Y., Liang, C., Jones, R. O., et al. (2017). mechanism of digenic Cx26 (GJB2) and Cx30 (GJB6) mutations: reduction of endocochlear potential by impairment of heterogeneous gap junctional function in the cochlear lateral wall. Neurobiol. Dis. 108, 195–203. doi: 10.1016/j.nbd.2017.08.002
Memon, A., and Lee, W. K. (2018). KLF10 as a tumor suppressor gene and its TGF-β signaling. Cancers (Basel) 10:161. doi: 10.3390/cancers10060161
Miao, Z., Deng, K., Wang, X., and Zhang, X. (2018). DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics 34, 3223–3224. doi: 10.1093/bioinformatics/bty332
Morell, R. J., Olszewski, R., Tona, R., Leitess, S., Wafa, T. T., Taukulis, I., et al. (2020). Noncoding microdeletion in mouse HGF disrupts neural crest migration into the stria vascularis, reduces the endocochlear potential, and suggests the neuropathology for human nonsyndromic deafness DFNB39. J. Neurosci. 40, 2976–2992. doi: 10.1523/jneurosci.2278-19.2020
Mujtaba, G., Schultz, J. M., Imtiaz, A., Morell, R. J., Friedman, T. B., and Naz, S. (2015). A mutation of MET, encoding hepatocyte growth factor receptor, is associated with human DFNB97 hearing loss. J. Med. Genet. 52, 548–552. doi: 10.1136/jmedgenet-2015-103023
Muri, L., Le, N. D., Zemp, J., Grandgirard, D., and Leib, S. L. (2019). Metformin mediates neuroprotection and attenuates hearing loss in experimental pneumococcal meningitis. J. Neuroinflammation 16:156.
Musa, A., Ghoraie, L. S., Zhang, S. D., Glazko, G., Yli-Harja, O., Dehmer, M., et al. (2018). A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 19, 506–523.
Mutai, H., Wasano, K., Momozawa, Y., Kamatani, Y., Miya, F., Masuda, S., et al. (2020). Variants encoding a restricted carboxy-terminal domain of SLC12A2 cause hereditary hearing loss in humans. PLoS Genet. 16:e1008643. doi: 10.1371/journal.pgen.1008643
Nguyen, D. T., Mathias, S., Bologa, C., Brunak, S., Fernandez, N., Gaulton, A., et al. (2017). Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 45, 995–1002.
Nickel, R., and Forge, A. (2008). Gap junctions and connexins in the inner ear: their roles in homeostasis and deafness. Curr. Opin. Otolaryngol. Head Neck Surg. 16, 452–457. doi: 10.1097/moo.0b013e32830e20b0
Nin, F., Hibino, H., Doi, K., Suzuki, T., Hisa, Y., and Kurachi, Y. (2008). The endocochlear potential depends on two K+ diffusion potentials and an electrical barrier in the stria vascularis of the inner ear. Proc. Natl. Acad. Sci. U.S.A. 105, 1751–1756. doi: 10.1073/pnas.0711463105
Nishiyama, S., Okada, T., Kobayashi, T., Garcia Del Saz, E., and Seguchi, H. (1994). Na-K-ATPase activity in the guinea pig stria vascularis in experimentally-induced endolymphatic hydrops. Histol. Histopathol. 9, 205–209.
Paken, J., Govender, C. D., Pillay, M., and Sewram, V. (2019). A review of cisplatin-associated ototoxicity. Semin. Hear. 40, 108–121. doi: 10.1055/s-0039-1684041
Reichold, M., Zdebik, A. A., Lieberer, E., Rapedius, M., Schmidt, K., Bandulik, S., et al. (2010). KCNJ10 gene mutations causing EAST syndrome (epilepsy, ataxia, sensorineural deafness, and tubulopathy) disrupt channel function. Proc. Natl. Acad. Sci. U.S.A. 107, 14490–14495. doi: 10.1073/pnas.1003072107
Rybak, L. P., Whitworth, C. A., Mukherjea, D., and Ramkumar, V. (2007). Mechanisms of cisplatin-induced ototoxicity and prevention. Hear. Res. 226, 157–167. doi: 10.1016/j.heares.2006.09.015
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F., and Regev, A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. doi: 10.1038/nbt.3192
Schmitt, N. C., and Rubel, E. W. (2013). Osteopontin does not mitigate cisplatin ototoxicity or nephrotoxicity in adult mice. Otolaryngol. - Head Neck Surg. 149, 614–620. doi: 10.1177/0194599813498218
Scholl, U. I., Choi, M., Liu, T., Ramaekers, V. T., Häusler, M. G., Grimmer, J., et al. (2009). Seizures, sensorineural deafness, ataxia, mental retardation, and electrolyte imbalance (SeSAME syndrome) caused by mutations in KCNJ10. Proc. Natl. Acad. Sci. U.S.A. 106, 5842–5847. doi: 10.1073/pnas.0901749106
Schulte, B. A., and Steel, K. P. (1994). Expression of α and β subunit isoforms of Na, K-ATPase in the mouse inner ear and changes with mutations at the Wv or Sld loci. Hear. Res. 78, 65–76. doi: 10.1016/0378-5955(94)90045-0
Sewell, W. F. (1984). The effects of furosemide on the endocochlear potential and auditory-nerve fiber tuning curves in cats. Hear. Res. 14, 305–314. doi: 10.1016/0378-5955(84)90057-1
Sharlin, D. S., Ng, L., Verrey, F., Visser, T. J., Liu, Y., Olszewski, R. T., et al. (2018). Deafness and loss of cochlear hair cells in the absence of thyroid hormone transporters Slc16a2 (Mct8) and Slc16a10 (Mct10). Sci. Rep. 8:4403.
Spector, G. J., and Carr, C. (1979). The ultrastructural cytochemistry of peroxisomes in the guinea pig cochlea: a metabolic hypothesis for the stria vascularis. Laryngoscope 89, 1–38. doi: 10.1288/00005537-197906001-00001
Stanchina, L., Baral, V., Robert, F., Pingault, V., Lemort, N., Pachnis, V., et al. (2006). Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev. Biol. 295, 232–249. doi: 10.1016/j.ydbio.2006.03.031
Termini, C. M., and Gillette, J. M. (2017). Tetraspanins function as regulators of cellular signaling. Front. Cell Dev. Biol. 5:34. doi: 10.3389/fcell.2017.00034
Teubner, B., Michel, V., Pesch, J., Lautermann, J., Cohen-Salmon, M., Söhl, G., et al. (2003). Connexin30 (Gjb6)-deficiency causes severe hearing impairment and lack of endocochlear potential. Hum. Mol. Genet. 12, 13–21. doi: 10.1093/hmg/ddg001
Venkov, C., Plieth, D., Ni, T., Karmaker, A., Bian, A., George, A. L., et al. (2011). Transcriptional networks in epithelial-mesenchymal transition. PLoS One 6:e25354. doi: 10.1371/journal.pone.0025354
Walker, E. M., Fazekas-May, M. A., and Bowen, W. R. (1990). Nephrotoxic and ototoxic agents. Clin. Lab. Med. 10, 323–354. doi: 10.1016/s0272-2712(18)30572-9
Wang, B., Lindley, L. E., Fernandez-Vega, V., Rieger, M. E., Sims, A. H., and Briegel, K. J. (2012). The T box transcription factor TBX2 promotes epithelial-mesenchymal transition and invasion of normal and malignant breast epithelial cells. PLoS One 7:e41355. doi: 10.1371/journal.pone.0041355
Wangemann, P. (2002). K+ cycling and the endocochlear potential. Hear. Res. 165, 1–9. doi: 10.1016/s0378-5955(02)00279-4
Wangemann, P., Itza, E. M., Albrecht, B., Wu, T., Jabba, S. V., Maganti, R. J., et al. (2004). Loss of KCNJ10 protein expression abolishes endocochlear potential and causes deafness in Pendred syndrome mouse model. BMC Med. 2:30. doi: 10.1186/1741-7015-2-30
Wangemann, P., Liu, J., and Marcus, D. C. (1995). Ion transport mechanisms responsible for K+ secretion and the transepithelial voltage across marginal cells of stria vascularis in vitro. Hear. Res. 84, 19–29. doi: 10.1016/0378-5955(95)00009-s
Wangemann, P., Nakaya, K., Wu, T., Maganti, R. J., Itza, E. M., Sanneman, J. D., et al. (2007). Loss of cochlear HCO3- secretion causes deafness via endolymphatic acidification and inhibition of Ca2+ reabsorption in a Pendred syndrome mouse model. Am. J. Physiol. - Ren. Physiol. 292, 1345–1353.
Wingard, J. C., and Zhao, H. B. (2015). Cellular and deafness mechanisms underlying connexin mutation-induced hearing loss – A common hereditary deafness. Front. Cell. Neurosci. 9:202. doi: 10.3389/fncel.2015.00202
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, 1074–1082.
Wu, L., Sagong, B., Choi, J. Y., Kim, U. K., and Bok, J. (2013). A systematic survey of carbonic anhydrase mRNA expression during mammalian inner ear development. Dev. Dyn. 242, 269–280. doi: 10.1002/dvdy.23917
Wu, X., Zhang, W., Li, Y., and Lin, X. (2019). Structure and function of cochlear gap junctions and implications for the translation of cochlear gene therapies. Front. Cell. Neurosci. 13:529. doi: 10.3389/fncel.2019.00529
Xia, A. P., Katori, Y., Oshima, T., Watanabe, K., Kikuchi, T., and Ikeda, K. (2001). Expression of connexin 30 in the developing mouse cochlea. Brain Res. 898, 364–367. doi: 10.1016/s0006-8993(01)02216-8
Xia, A. P., Kikuchi, T., Hozawa, K., Katori, Y., and Takasaka, T. (1999). Expression of connexin 26 and Na, K-ATPase in the developing mouse cochlear lateral wall: functional implications. Brain Res. 846, 106–111. doi: 10.1016/s0006-8993(99)01996-4
Xu, D. Y., Zhang, Q. X., Ma, Y. Q., Zheng, X. L., and Liu, S. X. (2010). Immunohistochemical localisation of endothelin receptor subtypes in the cochlear lateral wall. J. Laryngol. Otol. 124, 1073–1077. doi: 10.1017/s0022215110001428
Yamamoto, H., Shi, X., and Nuttall, A. L. (2009). The influence of loud sound stress on expression of osmotic stress protein 94 in the murine inner ear. Neuroscience 158, 1691–1698. doi: 10.1016/j.neuroscience.2008.10.063
Zhang, X., Shi, G., Gao, F., Liu, P., Wang, H., and Tan, X. (2019). TSPAN1 upregulates MMP2 to promote pancreatic cancer cell migration and invasion via PLCγ. Oncol. Rep. 41, 2117–2125.
Zhao, H. B. (2017). Hypothesis of K+-recycling defect is not a primary deafness mechanism for Cx26 (GJB2) deficiency. Front. Mol. Neurosci. 10:162. doi: 10.3389/fnmol.2017.00162
Zheng, G. X. Y., Terry, J. M., Belgrader, P., Ryvkin, P., Bent, Z. W., Wilson, R., et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049.
Keywords: stria vascularis, endocochlear potential, inner ear, scRNA-Seq, cisplatin, differential expression, gene regulatory networks
Citation: Taukulis IA, Olszewski RT, Korrapati S, Fernandez KA, Boger ET, Fitzgerald TS, Morell RJ, Cunningham LL and Hoa M (2021) Single-Cell RNA-Seq of Cisplatin-Treated Adult Stria Vascularis Identifies Cell Type-Specific Regulatory Networks and Novel Therapeutic Gene Targets. Front. Mol. Neurosci. 14:718241. doi: 10.3389/fnmol.2021.718241
Received: 31 May 2021; Accepted: 17 August 2021;
Published: 09 September 2021.
Edited by:
Fabio Mammano, University of Padua, ItalyReviewed by:
Andy Groves, Baylor College of Medicine, United StatesCopyright © 2021 Taukulis, Olszewski, Korrapati, Fernandez, Boger, Fitzgerald, Morell, Cunningham and Hoa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Hoa, bWljaGFlbC5ob2FAbmloLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.