- 1VIB-KU Leuven Center For Brain & Disease Research, Leuven, Belgium
- 2Laboratory for the Research of Neurodegenerative Diseases, Department of Neurosciences, Leuven Brain Institute (LBI), KU Leuven, Leuven, Belgium
- 3Neuroscience Discovery, Janssen Research & Development, Janssen Pharmaceutica N.V., Beerse, Belgium
- 4UK Dementia Research Institute, University College London, London, United Kingdom
Recent advances in RNA sequencing technologies helped to uncover the existence of tens of thousands of long non-coding RNAs (lncRNAs) that arise from the dark matter of the genome. These lncRNAs were originally thought to be transcriptional noise but an increasing number of studies demonstrate that these transcripts can modulate protein-coding gene expression by a wide variety of transcriptional and post-transcriptional mechanisms. The spatiotemporal regulation of lncRNA expression is particularly evident in the central nervous system, suggesting that they may directly contribute to specific brain processes, including neurogenesis and cellular homeostasis. Not surprisingly, lncRNAs are therefore gaining attention as putative novel therapeutic targets for disorders of the brain. In this review, we summarize the recent insights into the functions of lncRNAs in the brain, their role in neuronal maintenance, and their potential contribution to disease. We conclude this review by postulating how these RNA molecules can be targeted for the treatment of yet incurable neurological disorders.
Introduction
The sequencing of the ∼3.1 billion base pairs of the human genome marked the completion of the Human Genome Project in April 2003 after a little over 10 years of research. One of the main findings of this large scale project was that only a fraction of the genome encoded in total ∼22,300 protein-coding genes, whereas the remaining fraction was considered “junk DNA” (Human Genome Project, 2003). Since then, our perception of the complexity of the human genome changed dramatically (Frankish et al., 2021). In the past 20 years, advances in RNA sequencing technology pointed out that approximately 80% of our genome is actually transcribed, whereas only around 2% is subsequently translated into proteins (Carninci et al., 2005; Dunham et al., 2012; Hon et al., 2017; Zhao et al., 2021). Besides protein-coding messenger RNAs (mRNAs), transfer RNA (tRNAs) and ribosomal RNAs (rRNAs), other essential RNA species include long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), and small non-coding RNAs (sncRNA) such as microRNAs (miRNAs) and small-interfering RNAs (siRNAs) (Mattick and Rinn, 2015; Quan et al., 2017; Lorenzi et al., 2021).
Remarkably, the majority of our DNA generates a large number of lncRNAs (Iyer et al., 2015; Bonetti and Carninci, 2017; Yao et al., 2019; Zhao et al., 2021). By definition, lncRNAs contain more than 200 nucleotides in length and lack protein-coding potential. Based on their genomic location and orientation relative to neighboring protein-coding genes, lncRNAs are broadly categorized into intergenic lncRNAs, intronic lncRNAs, bidirectional lncRNAs, sense lncRNAs, antisense lncRNAs and enhancer RNAs (Figure 1) (Ma et al., 2013; Youse et al., 2020).
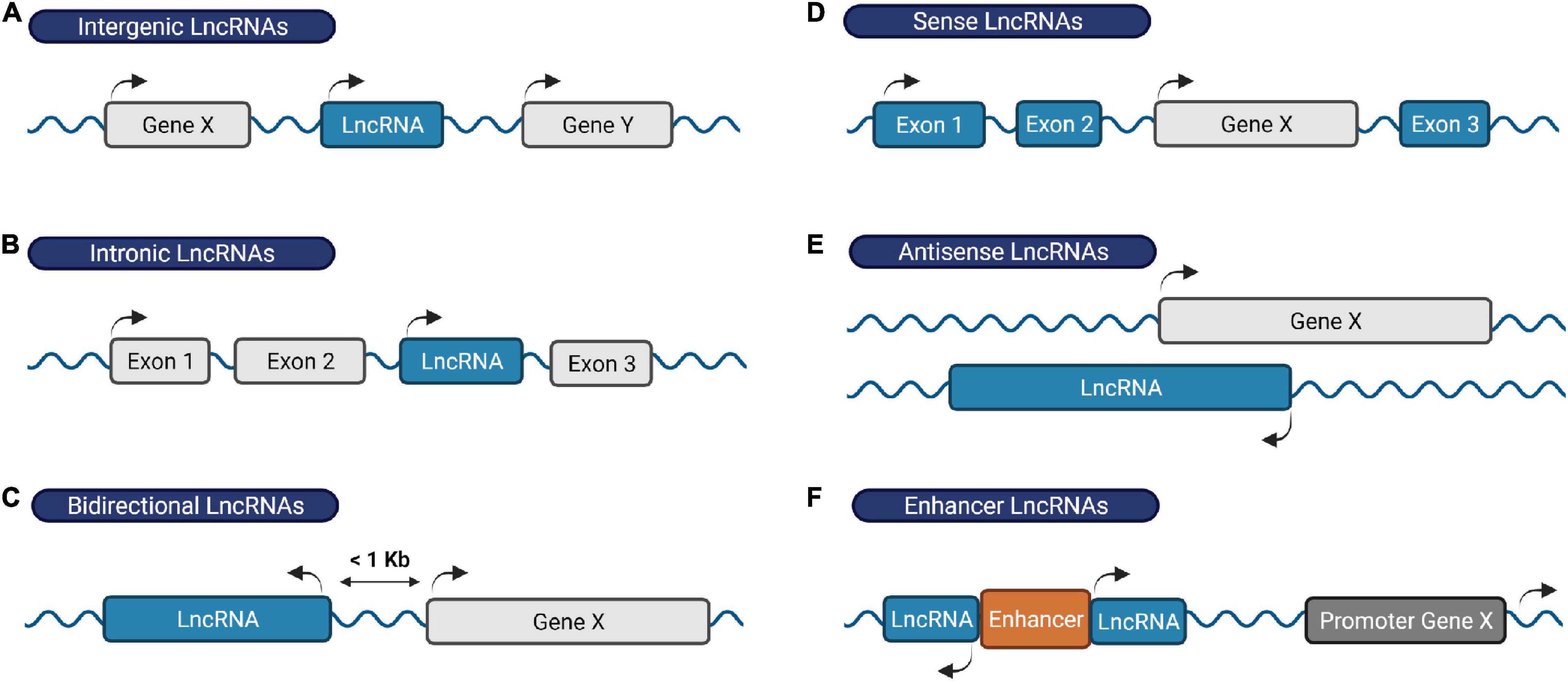
Figure 1. Classification of lncRNAs based on their genomic location. (A) Intergenic RNAs are located between two protein-coding genes, (B) intronic lncRNAs are transcribed within an intronic region of a protein-coding gene, (C) bidirectional lncRNAs are located on the opposite strand of a protein-coding gene whose transcription initiates less than 1,000 base pairs away, (D) sense lncRNAs are transcribed from and overlap with the sense strand of a protein-coding gene, (E) antisense lncRNAs originate from the antisense strand of a protein coding-gene, and (F) enhancer RNAs derive from enhancer regions and play a role in gene transcription activation. Protein-coding genes/exons shown in gray; LncRNA genes/exons shown in blue.
LncRNA promoters are structurally similar to those from protein-coding genes, exhibiting characteristic profiles of transcriptional activity markers (e.g., histone acetylation, methylation, ubiquitination, etc.) around their transcription start sites (Derrien et al., 2012; Djebali et al., 2012; Roberts et al., 2014). Consistent with their promoter structure, lncRNAs are generally transcribed by RNA polymerase II (Hon et al., 2017). Additionally, many lncRNAs share some features of messenger RNAs (mRNAs) at the transcript level including the presence of a 5′-cap, 3′-polyadenylation, and the occurrence of alternative splicing events that give rise to alternative transcript isoforms (Derrien et al., 2012; Djebali et al., 2012; Roberts et al., 2014). The mechanisms that regulate these post-transcriptional events in lncRNAs are not well documented; the limited amount of evidence available suggests that the same mechanisms are used as for protein-coding genes albeit with different efficiency (Gil and Ulitsky, 2018; Statello et al., 2021). There are also marked differences between lncRNAs and protein-coding genes. LncRNAs are generally expressed at lower levels although there are examples of highly expressed lncRNAs (e.g., TUG1 and MALAT1) (Derrien et al., 2012; Washietl et al., 2014; Jiang et al., 2016). LncRNAs also exhibit a more highly specific spatiotemporal expression pattern, and display poorer sequence conservation across species compared to protein-coding genes (Derrien et al., 2012; Djebali et al., 2012; Jiang et al., 2016). The high level of orchestrated regulation at the transcriptional and post-transcriptional level suggests that lncRNAs are at least as functionally relevant as protein-coding genes despite low conservation and expression levels.
The lncRNA Xist (X-inactive-specific transcript) was one of the first lncRNAs identified when it was isolated in the early 1990s from a female placental complementary DNA library screening (Brown et al., 1991). This lncRNA remains the most studied nuclear lncRNA to date. Xist is exclusively expressed from one of the two X chromosomes in females during early embryonic development. The transcript “coats” the X chromosome in cis and triggers a series of events that results in chromosome-wide transcriptional silencing and heterochromatinization acting in concert with many other lncRNAs, RNA binding proteins and DNA (Rocha and Heard, 2017). As such, Xist ensures X chromosome inactivation and dosage compensation between females and males in mammals (Brown et al., 1991).
Xist is an archetypical example on the functional relevance of lncRNAs. Evolutionary biologists uncovered that the amount of transcribed DNA (as lncRNAs) correlates with the organisms’ complexity and genome size (Rubin et al., 2000; Mattick, 2001; Taft et al., 2007; Liu et al., 2013; Kapusta and Feschotte, 2014). These observations further suggest that despite low sequence conservation across species, lncRNAs may contribute to the development of complex organisms and organs including the central nervous system (CNS) (Aprea and Calegari, 2015). In this review, we will summarize the current knowledge on the role of lncRNAs in brain physiology and disease; and discuss the future perspectives and challenges of using lncRNAs as RNA-based therapeutic targets in neurological disorders.
LncRNAs in the CNS
The observation that most of the mammalian genome is actively transcribed, that much of this pervasive transcription is likely functional, and that lncRNA loci are linked to key biological functions has boosted the interest in lncRNAs (Dunham et al., 2012; Kopp and Mendell, 2018; Lorenzi et al., 2021; Statello et al., 2021; Zhao et al., 2021). However, the first challenge in understanding the physiological relevance of lncRNAs is the difficulty to annotate these transcripts (Uszczynska-Ratajczak et al., 2018). The development of recent sequencing technologies has been key for the emergence of different annotation methods and databases that continue to improve our knowledge of these genes. A few examples are the GENCODE geneset, which combines manual with automated annotation from two pipelines (Ensembl-HAVANA and Ensembl-Genebuild) for protein-coding genes annotation, but uses mostly manual annotation for lncRNA genes (Frankish et al., 2021); NONCODE, an integrative database exclusively dedicated to the annotation of non-coding RNAs, in particular lncRNAs in animals (Zhao et al., 2021); or FANTOM CAT, a meta-assembly mostly based on CAGE (Cap Analysis of Gene Expression) data and annotations from diverse sources (Figure 2) (Hon et al., 2017). Thus, while the number of functional lncRNAs is still a matter of debate, between 15,000 and 100,000 lncRNAs have recently been reported to exist in the human genome. Similar numbers have been annotated in the mouse genome (Uszczynska-Ratajczak et al., 2018). In comparison, approximately 20,000 protein-coding genes have been mapped (Salzberg, 2018). Thus, given their relative quantities, it is hypothesized that lncRNAs contribute to the organism’s complexity.
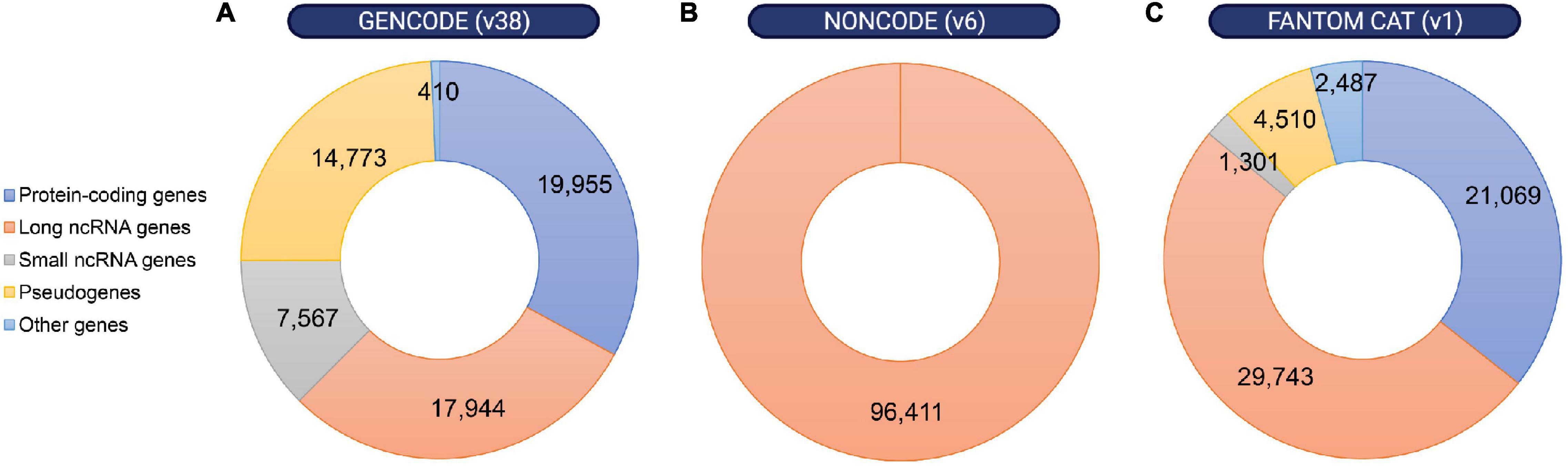
Figure 2. Distribution of annotated loci in the human genome according to different gene map databases. Despite the discrepancies between different annotation methods, non-coding RNA loci are remarkably abundant within the human genome. Number of protein-coding genes, long or small non-coding RNA genes, pseudogenes and other genes were obtained from (A) GENCODE release 38 (https://www.gencodegenes.org/human/stats.html), (B) NON-CODE v6.0 (http://www.noncode.org/analysis.php), and (C) FANTOM CAT v1 (https://fantom.gsc.riken.jp/cat/v1/#/genes).
The development and maintenance of the nervous system in particular is a complex process that must rely on highly coordinated spatiotemporal gene expression programs to finetune the balance between cell proliferation and differentiation and ensure these cell populations give rise to a functional network. A remarkable amount of 40% of all annotated tissue-specific lncRNA genes are specifically enriched in particular brain regions or cell types and participate in many aspects of brain function and development (Derrien et al., 2012; Francescatto et al., 2014; Briggs et al., 2015; Zimmer-Bensch, 2019). In this section, we will discuss some features of lncRNAs that support a widespread functional role for this RNA subclass in mediating CNS development and function.
Brain LncRNAs Are Evolutionary Conserved
Accumulating data suggest that conserved lncRNAs are more likely to be functionally relevant. While lncRNAs exhibit higher sequence conservation in their promoter regions, splice-junction motifs and small functional domains, lncRNA gene bodies exhibit an overall lower evolutionary conservation relative to protein-coding genes (Chodroff et al., 2010; Ulitsky et al., 2011; Derrien et al., 2012; Necsulea and Kaessmann, 2014; Nitsche et al., 2015). A possible explanation is the fact that many lncRNAs have a recent evolutionary origin, and approximately one third of human lncRNAs has been reported to be restricted to the primate lineage (Derrien et al., 2012; Necsulea and Kaessmann, 2014; Washietl et al., 2014). Accordingly, these lncRNAs seem to have less exonic sequence constraints compared to ancestral and conserved lncRNAs (Necsulea and Kaessmann, 2014; Washietl et al., 2014). Moreover, several lncRNAs have been shown to be under positive evolutionary selection in human compared to other mammalian species (Tay et al., 2009; Lindblad-Toh et al., 2011; Grossman et al., 2013; Francescatto et al., 2014; Washietl et al., 2014; Sarropoulos et al., 2019).
Nonetheless, brain-specific lncRNAs display the strongest evolutionary conservation compared to those expressed in other tissues (Ponjavic et al., 2009; He et al., 2014), and their levels of sequence conservation correlate very well with the levels of brain complexity (Johnson et al., 2015). Intergenic lncRNAs that show strong evolutionary conservation (when evaluating mouse and human genomes) are particularly enriched in the brain (Ponjavic et al., 2009). Moreover, these enriched lncRNAs are located adjacent to protein-coding genes that are co-expressed in the brain and involved in transcriptional regulation or CNS development (Ponjavic et al., 2009).
Other studies indicate that some lncRNAs with relevant functional roles in the brain have been subjected to higher sequence constraints throughout evolution. For instance, the lncRNAs linc-Brn1b, Dali, and Pnky are 3 conserved lncRNAs located in close proximity of and co-expressed with Brn1 and Brn2 (Sauvageau et al., 2013; Chalei et al., 2014; Ramos et al., 2015). These two genes encode the POU-homeodomain transcription factors POU3F3 and POU3F2, respectively, that play crucial roles in neocortex development (Sugitani et al., 2002). Linc-Brn1b and Dali are located downstream of Brn1 and loss-of-function studies demonstrated that these lncRNAs are required for correct brain development (Sauvageau et al., 2013; Chalei et al., 2014). Pnky is a lncRNA transcribed in the opposite strand of Brn2 with two regions conserved in vertebrates, and its expression is upregulated in neural stem cells in the developing mouse and human cortex. Pnky controls neuronal differentiation in dividing neural stem cells by interacting with PTBP1, a splicing factor that represses the inclusion of neural exons in non-neural cells (Ramos et al., 2015). Additional examples of well-conserved lncRNA genes with a role in neurogenesis include the lncRNA Rmst and Tuna; knockdown of these lncRNAs suppresses neuronal differentiation in mouse embryonic stem cells (Ng et al., 2013; Lin et al., 2014).
Structural features rather than the primary sequence can contribute to functional relevance of lncRNAs (Torarinsson et al., 2006; Seemann et al., 2012; Smith et al., 2013; Diederichs, 2014; Johnsson et al., 2014; Kapusta and Feschotte, 2014). For example, the lncRNA MALAT1 contains a large number of helices that are highly conserved across vertebrates and even more in mammalian MALAT1 homologs, indicating the presence of an evolutionary conserved core with predicted functional roles in mammals and vertebrates (McCown et al., 2019).
LncRNA Genes Exhibit Highly Specific Expression Patterns in the CNS
The CNS is one of the most complex organs in mammals, exhibiting a tight spatial and temporal organization and gene expression profile during development and homeostasis. Several studies showed that lncRNA expression is spatially more restricted than mRNAs in multiple brain regions suggesting that lncRNAs may play crucial roles in the coordination of the complex gene expression in distinct CNS regions (Belgard et al., 2011; Luo et al., 2013; Ramos et al., 2013; Kadakkuzha et al., 2015).
A systematic in situ hybridization analysis demonstrated that the expression of the majority of lncRNAs in the mouse brain is restricted to distinct neuroanatomical loci (Mercer et al., 2008). The lncRNA MIAT/Gomafu was identified as a nuclear retained lncRNA that is specifically expressed in a distinct set of postmitotic neurons within the mouse nervous system (Sone et al., 2007). Gomafu-deficient mice develop normally but demonstrate mild hyperactivity associated with an increase in dopamine levels specifically in the nucleus accumbens (Ip et al., 2016). Recently, Abhd11os (ABHD11-AS1 in human) has been identified as a striatal-specific lncRNA and proposed to play a neuroprotective role against mutant Huntingtin (HTT), a protein implicated in Huntington’s Disease (Francelle et al., 2015). Studies combining bulk tissue RNA sequencing with single-cell RNA sequencing approaches confirmed cell-type and region specificity (Liu et al., 2016; Fan et al., 2020).
Taken together, expression analyses and functional studies have demonstrated that the spatial expression of a large number of lncRNAs is tightly regulated in various brain regions. However, the mechanisms that control this restricted expression pattern are not yet fully understood.
LncRNAs Are Dynamically Regulated in the CNS
The expression of many lncRNAs is temporally regulated during CNS development and several of them are linked to the regulation of protein-coding genes that play crucial roles in neurodevelopment (Briggs et al., 2015). Expression profiling of both protein-coding and non-coding transcripts during differentiation of mouse embryonic forebrain-derived neural stem cells indicated that 5% of approximately 3,600 analyzed lncRNAs are differentially expressed during neuronal-glial fate specification and oligodendrocyte lineage maturation (Mercer et al., 2010). Many of these lncRNAs exhibit coordinated expression with protein-coding genes involved in neuronal and glial lineage differentiation at distinct developmental stages, suggesting that these ncRNAs might regulate the expression of their associated protein-coding genes (Mercer et al., 2010).
Several reports also demonstrated that lncRNA expression is thoroughly regulated in processes such as synaptogenesis and in response to neuronal activity and plasticity (Zalfa et al., 2003; Bernard et al., 2010; Barry et al., 2014). For instance, the lncRNA ADEPTR is upregulated in an activity-dependent manner and consequently transported to synapses where it modulates the structural plasticity of dendritic spines in mouse hippocampal neurons (Grinman et al., 2021). Keihani et al. (2019) also identified NeuroLNC as a nuclear and neuron-specific lncRNA involved in the regulation of genes with crucial roles in neuronal physiology including neurotransmitter release, synapse organization and neuronal migration in rat models.
Is the expression of lncRNAs also temporally regulated in the human CNS? Lipovich et al. (2014) profiled the expression of thousands of lncRNAs in the human neocortex using microarray technology and identified 8 lncRNA genes with specific developmental expression patterns. The majority of these lncRNAs are located antisense and/or close to known protein-coding genes. Moreover, these loci exhibit primate-specific gene structure features. A transcriptomic analysis of human neurons derived from induced pluripotent stem cells revealed the presence of more than 1,500 lncRNAs whose expression is regulated during their transition to early differentiating neurons (Lin et al., 2011). Many of these lncRNAs are associated with chromatin-remodeling complexes such as RE1-Silencing Transcription factor (REST), REST corepressor 1 (CoREST) and Polycomb Repressive Complex 2 (PRC2) (Lin et al., 2011).
Widespread changes in the transcriptome, including in the expression of lncRNAs, also occur during aging (Wood et al., 2013; Barry et al., 2015; Kour and Rath, 2015; Chen et al., 2017). Several studies demonstrated that lncRNAs can be either up- or downregulated in senescent cells. These lncRNAs are typically associated to protein-coding genes that play a role in cell cycle arrest, cellular growth/tumor suppression, telomere organization and p53 signaling (Grammatikakis et al., 2014; Quinodoz and Guttman, 2014; Angrand et al., 2015; Xu C. L. et al., 2020). These molecular functions all have a clear link to aging. Finally, genome-wide association studies identified single-nucleotide polymorphisms associated with neurological diseases including Schizophrenia, Bipolar Disorder and Autism Spectrum Disorder, and that map to lncRNA genes that are dynamically expressed in the CNS (Lin et al., 2011). Together, the expression of lncRNAs in the brain are tightly regulated during neuronal development and aging. Changes in their expression levels during aging and in neurodegenerative diseases further imply that lncRNAs can play crucial roles in pathological events in the brain and underscore their functional importance in brain development and homeostasis.
Mechanisms of lncRNAs in Neurological Disorders
Perhaps the most intriguing feature of lncRNAs is their association with disease. Emerging evidence emphasizes the importance of lncRNAs in CNS development and (dys)function. Genome-wide association studies and comparative transcriptomic analyses link lncRNA deregulation and dysfunction to multiple human diseases, including a wide range of neurodevelopmental, neuropsychiatric, and neurodegenerative conditions (Salta and De Strooper, 2017; Salvatori et al., 2020). The identification of multiple natural antisense transcripts (NATs), a class of lncRNAs, at distinct human loci associated with hereditary neurodegenerative disorders including Alzheimer’s Disease, Frontotemporal Dementia, Parkinson’s Disease, Amyotrophic Lateral Sclerosis and Huntington’s Disease further illustrates the potential role of lncRNAs in the expression regulation of neurodegeneration-related genes (Zucchelli et al., 2019).
However, the association of altered expression of specific lncRNAs in brain disorders is not hard proof of biological relevance, and functional studies investigating the role of these lncRNAs are still needed. Since lncRNA function is intrinsically influenced by their subcellular localization (Chen, 2016; Bridges et al., 2021), we will separately discuss lncRNAs that exert their function at the epigenetic or transcriptional level in the nucleus, and those responsible for post-transcriptional regulatory mechanisms in the cytoplasm (Table 1 and Figure 3). Nevertheless, some lncRNAs might be present in both compartments and exert different functions depending on their location (Bridges et al., 2021).
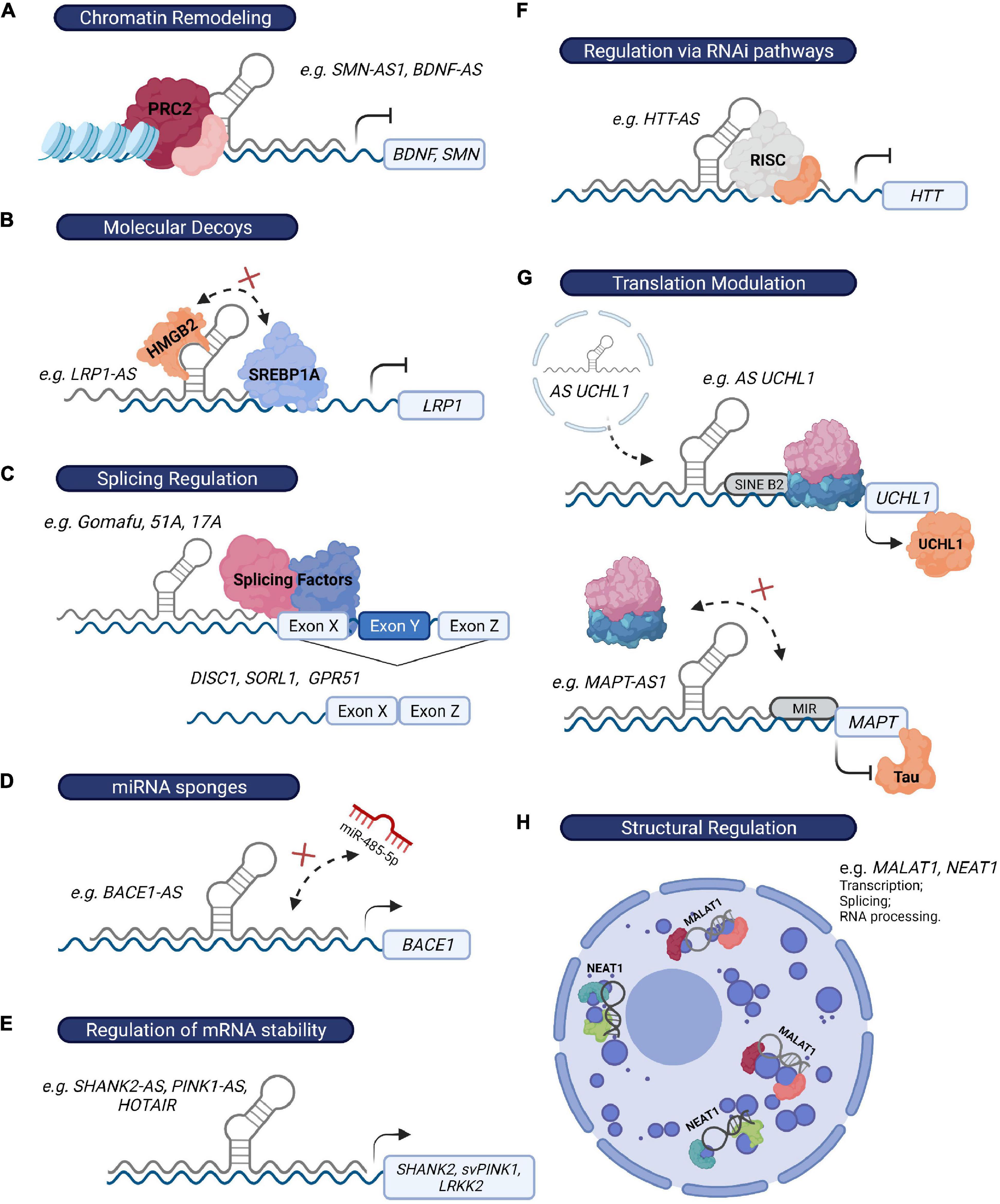
Figure 3. LncRNAs regulate gene expression through multiple mechanisms. (A) LncRNAs can recruit chromatin modifiers (e.g., PRC2 complex) to their target-gene promoters and epigenetically influence their expression (e.g., BDNF-AS and SMN-AS1). (B) Alternatively, they can modulate transcription by acting as molecular decoys and sequester specific chromatin and non-chromatin modulators, transcription factors, or other regulatory proteins from the promoters of their target genes (e.g., LRP1-AS). (C) LncRNAs can also influence transcription by affecting the alternative splicing of their target genes (e.g., Gomafu, 51A, 17A). (D) Some lncRNAs regulate mRNA turnover by acting as miRNA “sponges” (e.g., BACE1-AS), as they prevent miRNAs from binding their target genes. (E) Additionally, they can directly bind to their target mRNAs, leading to the formation of an RNA-RNA duplex and therefore influencing mRNA stability (SHANK2-AS, PINK1-AS, and HOTAIR). (F) LncRNAs can also exert their function via recruitment of the RISC complex to their target mRNA (e.g., HTT-AS). (G) Some lncRNAs directly modulate translation efficiency of their target genes through the presence of specific sequences that either promote (e.g., SINE B2 element in AS UCHL1) or prevent (e.g., MIR sequence in MAPT-AS1) the association of ribosome machinery with their target mRNAs. (H) Finally, NEAT1 and MALAT1 exert a crucial structural function in the formation of paraspeckles and speckles, respectively, two nuclear structures that regulate multiple mechanisms, including RNA transcription, splicing and processing.
Epigenetic Regulation
Numerous nuclear lncRNAs with potential roles in neurological disorders associate with chromatin where they interact with a wide variety of proteins to enhance or repress their binding and activity at specific DNA regions (Guttman et al., 2011; Roberts et al., 2014; Yao et al., 2019). Specifically, lncRNAs can recruit chromatin modifiers to their target-gene promoters and activate or inhibit their transcription in cis (close to their transcription sites) or in trans (regulation is exerted at distant loci). Alternatively, they can act as molecular decoys, sequestering specific chromatin modulators from the promoters of target genes. Finally, they can directly interact with DNA and generate DNA-RNA hybrid structures, such as R-loops, which ultimately influence chromatin accessibility and remodeling (Yao et al., 2019; Statello et al., 2021).
LncRNAs Interact With the PRC2 Complex
Many lncRNAs interact with the PRC2 complex, constituted by a group of polycomb proteins that mediate epigenetic silencing in diverse biological processes (Davidovich and Cech, 2015; Balas et al., 2021). The NAT BDNF-AS is transcribed antisense to the BDNF gene, which encodes a neurotrophic factor essential for neuronal development and maintenance (Modarresi et al., 2012; Miranda et al., 2019). Moreover, BDNF protein levels are reduced in various neurodegenerative disorders including Alzheimer’s Disease (Laske et al., 2011), Huntington’s Disease (Ferrer et al., 2000; Zuccato and Cattaneo, 2007), and Major Depressive Disorder (Molendijk et al., 2011; Emon et al., 2020). BDNF-AS knockdown leads to an increase in BDNF mRNA and protein levels both in vitro and in vivo by reducing the recruitment of the histone lysine methyltransferase EZH2, a component of PRC2, and consequently decreasing the levels of the repressive chromatin mark in the BDNF promoter region. Even though it remains unclear how BDNF-AS recruits the PRC2 components to the BDNF locus, this interaction could be of relevance in disorders where an upregulation of BDNF protein levels in the brain is needed (Modarresi et al., 2012).
Two independent studies identified SMN-AS1 as an antisense lncRNA that arises from the SMN locus and recruits the PRC2 complex to transcriptionally repress SMN expression and to have relevant implications for Spinal Muscular Atrophy (SMA) (d’Ydewalle et al., 2017; Woo et al., 2017). SMA is a genetic disorder caused by an autosomal recessive mutation of deletion of the SMN1 gene which results in atrophy of skeletal muscles due to a progressive motor neuron loss. Increasing the expression of the homologous SMN2 gene has been shown to functionally compensate for the loss of SMN1 and ameliorate disease severity by upregulating SMN protein levels (d’Ydewalle and Sumner, 2015). Knockdown of SMN-AS1 with antisense oligonucleotides (ASOs) can dissociate PRC2 from the SMN promoter and increase SMN expression in both in vitro and in vivo SMA models (d’Ydewalle et al., 2017; Woo et al., 2017). Combining SMN2 splice-switching oligonucleotides and ASOs targeting SMN-AS1 further boosted the levels of SMN protein and improved survival of SMA mice compared to either therapy by itself. These findings suggest that targeting this lncRNA combined with the FDA approved SMN2 splice-switching oligonucleotides (Spinraza) could represent a valuable therapeutic approach for severe SMA cases characterized by very low SMN levels (U.S. Food & Drug Administration, 2016; d’Ydewalle et al., 2017).
LncRNAs Interact With Other Chromatin Modifiers
Chromatin-remodeling complexes other than PRC2 might be involved in neurological disorders through their interaction with lncRNAs. Huntington’s Disease (HD) is a polyglutamine (PolyQ)-related disorder caused by an expanded CAG repeat (>36) in the first exon of the HTT gene. The repeat expansion results in the production of a mutant neurotoxic form of the HTT protein that triggers a progressive degeneration of cortical and striatal neurons. Although the mechanisms behind neuronal loss in HD are not yet completely understood, aberrant chromatin remodeling and transcriptional dysregulation seem to represent key features in the pathogenesis (Benn et al., 2008). For instance, during HD pathology, the transcriptional repressor REST is abnormally relocated to the nucleus by a mutant HTT-dependent mechanism, resulting in the repression of many REST target genes (Zuccato et al., 2007). The HAR1 non-coding locus is directly targeted by REST, which might explain the reduction in HAR1F and HAR1R transcript levels observed in the striatum of HD patients (Johnson et al., 2010). The expression of other lncRNAs with potential epigenetic regulatory functions is dysregulated in the brains of HD patients (Johnson, 2012). These include the putative REST targets DGCR5, NEAT1, and MEG3 (Myers et al., 2011), and TUG1 (Johnson, 2012). Interestingly, TUG1 and MEG3 can interact with PRC2 (Khalil et al., 2009). Therefore, it is likely that altered levels of some of these lncRNAs might influence gene expression patterns during disease. Similarly, DGCR5 has recently also been proposed to regulate the expression of genes associated with Schizophrenia (Meng et al., 2018).
Transcriptional Regulation
Multiple lncRNAs regulate their target genes at the transcriptional level. Importantly, two mechanisms may play a role in this regulation: the lncRNA transcript can influence the transcription of neighboring loci, and/or the act of transcription of the lncRNA itself can drive chromatin remodeling and affect the expression of other genes (Statello et al., 2021).
Is It the Transcript or Transcription?
UBE3A-ATS is a nuclear lncRNA that has been implicated in Angelman syndrome (AS), a severe neurodevelopmental disorder caused by a maternal deficiency of the imprinted gene UBE3A (Meng et al., 2012). Although patients carry at least one functional copy of the paternal UBE3A, in neurons this allele is silenced by the antisense lncRNA. Whether silencing of UBE3A by UBE3A-ATS occurs due to the presence of the antisense lncRNA or due to transcription of this locus is unclear (Meng et al., 2012). Nonetheless, reducing UBE3A-ATS levels to restore UBE3A protein levels has been proposed as a potential therapeutic intervention for AS (Meng et al., 2013, 2015; Wolter et al., 2020).
LncRNAs Act as Molecular Decoys
LncRNAs can also act as molecular decoys for other RNA-binding proteins (RBPs) including transcription and splicing factors. For example, Yamanaka et al. (2015) identified a NAT associated with the LRP1 gene, designated as LRP1-AS (Lrp1-AS in mouse), which has been implicated in Alzheimer’s Disease (AD) pathology. AD is the main cause of dementia worldwide and two hallmarks of this neurodegenerative disorder are the accumulation of extracellular amyloid-β (Aβ)-containing plaques and intracellular neurofibrillary tangles composed of hyperphosphorylated and aggregated Tau protein within the patients’ brains (Scheltens et al., 2021). LRP1 protein plays a major role in different aspects of AD (Tachibana et al., 2019; Rauch et al., 2020). Modulation of Lrp1-AS levels in a mouse cell line revealed that this lncRNA negatively regulates Lrp1 expression both at the mRNA and protein levels. Lrp1-AS directly binds to HMGB2, a non-histone chromatin modifier, inhibiting its ability to promote SREBP1A-dependent transcription of Lrp1. Accordingly, increased levels of LRP1-AS and a reduction in LRP1 expression in human AD brain samples compared to age-matched controls were observed (Yamanaka et al., 2015). However, further studies are needed to specifically investigate the role of LRP1-AS in the transcriptional regulation of LRP1 in the human brain and its functional effects in AD pathogenesis.
Development of Fragile X Syndrome (FXS) or the related disease Fragile X-associated Tremor/Ataxia Syndrome (FXTAS) is caused by the presence of a triple CGG repeat motif in the 5′ untranslated region (5′-UTR) of the FMR1 gene (Khalil et al., 2008; Peschansky et al., 2015; Huang et al., 2019). Unaffected individuals typically carry 5–54 repeats while 55–200 repeats are considered a premutation leading to the development of FXTAS. A full mutation occurs when > 200 repeats are present, and this is necessary to develop FXS. In most FXS patients, FMR1 is hypermethylated and transcriptionally silenced resulting in decreased FMR1 mRNA and FMRP protein levels, impacting neurogenesis (Sunamura et al., 2018). Intriguingly, the FMR1 locus is particularly complex and encodes several lncRNAs. The expression of these lncRNAs are differentially affected by the repeat expansion mutations suggesting that they could potentially explain the distinct clinical features of FXS and FXTAS (Ladd et al., 2007; Khalil et al., 2008; Pastori et al., 2014; Vittal et al., 2018; Huang et al., 2019). One of these lncRNAs, FMR4, overlaps with the repeat region and is silenced in FXS patients but upregulated in premutation carriers (Khalil et al., 2008; Peschansky et al., 2015). Previously, Khalil et al. (2008) have shown that even though FMR4 does not regulate FMR1 expression, it exhibits anti-apoptotic functions in human cell lines. Later on, and consistent with a role at the transcriptional level, FMR4 was found to be primarily associated with chromatin, and a discordant expression between the lncRNA and MBD4, a transcriptional repressor involved in DNA repair mechanisms and regulation of apoptosis (Bellacosa, 2001), was observed during human neuronal precursor cells development. This suggests a trans-acting regulatory role for FMR4 independent of FMR1 which might be impaired in Fragile X repeat expansion-associated diseases (Peschansky et al., 2015).
The presence of hexanucleotide GGGGCC (G4C2) repeat expansions in the C9orf72 gene represents the most common genetic cause of Frontotemporal Dementia (FTD) and Amyotrophic Lateral Sclerosis (ALS), two devastating neurodegenerative disorders commonly designated as c9FTD/ALS (Barker et al., 2017). While the mechanisms by which these repeat expansions lead to c9FTD/ALS are not fully understood, both sense and antisense RNA foci comprising C9orf72 RNA are widely distributed across the CNS of these patients (Gendron et al., 2013; Zu et al., 2013; Cooper-Knock et al., 2015). A potential mechanism by which RNA foci are thought to cause neurotoxicity is by directly sequestering specific RBPs and disrupting their function (Barker et al., 2017). Ultimately, this leads to a wide range of RNA misprocessing events, including aberrant alternative splicing. In support of this, Lee et al. (2013) transfected human neuroblastoma cells with different G4C2 repeat lengths and revealed that longer repeat lengths originate RNA foci that co-localize with a subset of RBPs involved in alternative splicing, which include SF2, SC35 and hnRNP-H. Furthermore, hnRNP-H directly binds to G4C2 repeat sequences and associate with RNA foci in both transfected cells and in the brains of c9FTD/ALS patients (Lee et al., 2013).
LncRNAs Influence Alternative Splicing
Another mechanism by which lncRNAs can influence transcription is by affecting the alternative splicing of their target genes (Yao et al., 2019). Schizophrenia is a complex mental disorder that affects about 1% of the population, and it is likely to result from different genetic, epigenetic and environmental factors that culminate in neurodevelopmental abnormalities and brain dysfunction (Morikawa and Manabe, 2010; Barry et al., 2014). Many genes associated with Schizophrenia are aberrantly spliced, including DISC1 (Nakata et al., 2009) and ERBB4 (Law et al., 2007). In line with this, Gomafu can act as a splicing factor scaffold, directly binding to the splicing factors QKI and SRSF1 to regulate alternative splicing of DISC1 and ERBB4 in human neurons derived from induced pluripotent stem cells (Barry et al., 2014). Notably, abnormal alternative splicing patterns upon ASO-mediated knockdown of Gomafu in vitro match those observed for these two genes in post-mortem brains from individuals affected by Schizophrenia. Finally, Gomafu levels are reduced in cortical samples from patients compared to controls further suggesting a role for Gomafu in Schizophrenia pathology (Barry et al., 2014).
In AD, the lncRNAs 51A/SORL1-AS and 17A have both been suggested to interfere with the alternative splicing of their neighboring genes - SORL1 encoding Sortilin-1 and GPR51 encoding GABA receptor B2, respectively - and their expression is upregulated in the brain tissue from AD patients (Massone et al., 2011; Ciarlo et al., 2013).
Post-transcriptional Regulation
LncRNAs can be exported to the cytoplasm, where they regulate gene expression at the post-transcriptional level by influencing mRNA turnover, modulating translation or by interfering with post-translational modifications (Yao et al., 2019; Salvatori et al., 2020).
LncRNAs “Sponge” miRNAs
Some lncRNAs regulate mRNA turnover of their target genes by competing with miRNA binding sites by acting as miRNA “sponges” (Su et al., 2018; Yao et al., 2019). For instance, BACE1-AS has been identified as an antisense lncRNA associated with the BACE1 gene (Faghihi et al., 2008). BACE1-AS regulates the levels of BACE1 mRNA by masking BACE1 mRNA binding site for miR-485-5p (Faghihi et al., 2010). BACE1 encodes a secretase involved in the biosynthesis of Aβ and plays a central role in the amyloid cascade in AD pathophysiology (Hampel et al., 2020). In the presence of cell stressors including Aβ1–42, BACE1-AS levels increase thereby raising the levels of both BACE1 mRNA and protein. This in turn stimulates the production of Aβ1–42, at least in vitro, which may lead to a detrimental accumulation of toxic Aβ aggregates (Faghihi et al., 2008). Moreover, different brain regions from AD patients exhibit increased levels of this lncRNA compared to control subjects indicating a direct role for BACE1-AS in driving AD pathology (Faghihi et al., 2008).
LncRNAs Regulate mRNA Stability
Another possibility is that lncRNA transcripts directly bind to their target mRNA, originating an RNA duplex and affecting mRNA stability by either recruiting proteins that promote mRNA degradation or, on the contrary, by acting as molecular decoys for RBPs involved in mRNA decay (Yao et al., 2019). Autism Spectrum Disorder (ASD) comprises a range of heterogeneous neurodevelopmental disorders characterized by cognitive, social and sensory impairments (Lord et al., 2020). Many are co-expressed with ASD risk genes in the developing brain (Cogill et al., 2018). For instance, the expression SHANK2-AS, a lncRNA transcribed antisense to the SHANK2 gene, is upregulated and its levels negatively correlate with SHANK2 mRNA and protein levels in ASD patients compared to control individuals (Wang et al., 2015; Luo et al., 2018). Mutations in the SHANK2 gene, which encodes a post-synaptic density scaffold protein, have been identified as risk factors for ASD (Sato et al., 2012; Zaslavsky et al., 2019) and Schizophrenia (Peykov et al., 2015). SHANK2-AS can directly bind to SHANK2 mRNA, consequently decreasing its expression in vitro (Luo et al., 2018). Furthermore, overexpression of SHANK2-AS or downregulation of SHANK2 reduces neurite length and number in a human neuronal cell line suggesting that abnormal expression of SHANK2-AS may affect neuronal structure and growth by downregulating SHANK2 expression, and thus directly contribute to ASD pathology (Luo et al., 2018).
Parkinson’s Disease (PD) is a neurodegenerative disorder generally characterized by the accumulation of α-synuclein (encoded by the SNCA gene) aggregates within Lewy bodies or Lewy neurites and degeneration of dopaminergic neurons, particularly in the substantia nigra. While most PD cases are sporadic, familial forms of the disease can occur and result from mutations in a group of genes that include LRRK2, PARK2, PARK7, PINK1 or the SNCA gene itself (Bandres-Ciga et al., 2020). The antisense lncRNA PINK1-AS stabilizes the expression of a PINK1 splice variant (svPINK1) in vivo through the formation of an RNA duplex (Scheele et al., 2007). This interaction might have important implications in multiple disorders besides PD (Wilhelmus et al., 2011), as PINK1 is thought to have a neuroprotective role against stress-induced mitochondrial dysfunction, oxidative stress and apoptosis (Deas et al., 2009). In addition, the lncRNA HOTAIR has also been implicated in PD pathology by increasing LRKK2 mRNA stability and inducing dopaminergic neuronal apoptosis in a human neuroblastoma cell line (Wang S. et al., 2017). There are also several lncRNAs identified that can arise from the antisense strand in the SNCA locus either overlapping with the SNCA gene in the 5′ or 3′end (Zucchelli et al., 2019). Although their role in regulating SNCA expression is still enigmatic, expression analysis confirmed that some of these lncRNAs are co-expressed with SNCA, including in the substantia nigra (Zucchelli et al., 2019).
LncRNAs Act via the RNAi Pathway
The lncRNA HTT-AS originates from the HD repeat locus containing the repeat region and represses mutant HTT expression via an RNA-induced silencing complex (RISC)-dependent mechanism (Chung et al., 2011). In human cells, overexpression of the HTT-AS transcript reduces HTT mRNA levels, while knocking down the lncRNA upregulates HTT transcripts. Interestingly, reduced levels of HTT-AS were observed in the frontal cortex from HD patients compared to control individuals suggesting a potential protective role in HD in which the presence of expanded repeats reduces HTT-AS expression, removing its inhibitory effect on HTT expression (Chung et al., 2011).
LncRNAs Modulate Translation
Finally, some lncRNAs directly modulate translation efficiency of genes associated with neurological disorders (Muddashetty et al., 2002; Salta and De Strooper, 2017). Carrieri et al. (2012) have identified AS Uchl1 as an antisense lncRNA associated with the Uchl1 gene. UCHL1 is a highly abundant neuronal protein involved in neuronal development (Reinicke et al., 2019), dopaminergic neuron differentiation (Carrieri et al., 2015) and regulation of the ubiquitin-proteasome pathway (Yichin et al., 2002). UCHL1 gene variants are associated with susceptibility to PD (Ying et al., 2015), and oxidative modifications and downregulation of UCHL1 protein are observed in sporadic cases of both AD and PD patients (Choi et al., 2004). AS Uchl1 shuttles from the nucleus to the cytoplasm upon rapamycin induced mTOR activity in a dopaminergic cellular model, where it targets Uchl1 mRNA to active polysomes to facilitate its translation and upregulate Uchl1 protein levels (Carrieri et al., 2012). Owing to the presence of an embedded repetitive sequence SINE B2 in the inverted orientation at the non-overlapping part of the transcript, AS Uchl1 has been considered the representative member of a recently identified functional NAT class designated as SINEUPs. These molecules require a SINE B2 sequence to UP-regulate translation of their target mRNA. Importantly, due to their ability to increase the translation of virtually any gene of interest, the development of synthetic SINEUPs that target the antisense sequence of the target mRNA might constitute an interesting therapeutic approach to enhance protein synthesis (Zucchelli et al., 2015).
The very recent identification of NATs with embedded mammalian-wide interspersed repeat (MIR) sequences – referred to as MIR-NATs – associated with protein-coding genes tied to neurodegenerative disorders has revealed a new class of lncRNAs that mediate gene regulation at the translational level through a MIR-dependent mechanism (Simone et al., 2021). One of these MIR-NATs, MAPT-AS1, is associated with the MAPT gene – encoding the Tau protein - and has been suggested to repress Tau translation by competing for ribosomal RNA pairing with the MAPT mRNA internal ribosome entry site (Simone et al., 2021).
The Nucleus Takes It All
NEAT1 and MALAT1 (also known as NEAT2) are two human lncRNAs conserved within the mammalian lineage which regulate key nuclear functions (Hutchinson et al., 2007; Zhang et al., 2017; An et al., 2018). Despite displaying an adjacent genomic location, these two RNAs exert their function in related but distinct nuclear subdomains (West et al., 2014). MALAT1 is localized in nuclear speckles which are enriched in serine- and arginine-rich (SR) splicing factors, while NEAT1 is a structural component of nuclear bodies called paraspeckles, also composed by multiple proteins with reported roles in transcription and RNA processing (West et al., 2014). Therefore, both lncRNAs play important regulatory roles in many cellular pathways that are commonly affected in neurological diseases (Zhang et al., 2017; An et al., 2018). The expression of both MALAT1 and NEAT1 is markedly upregulated in the brains from Frontotemporal Lobar Degeneration associated with TDP-43 (FTLD-TDP) patients (Tollervey et al., 2011). Moreover, this differential expression can explain an increased binding of TDP-43 protein to these lncRNAs in FTLD-TDP patient samples, supporting a role for lncRNAs and TDP-43 in the regulation of splicing in the brain with direct implications for neurodegenerative diseases (Tollervey et al., 2011).
Although a large proportion of the genetic risk for familial ALS (fALS) is still elusive, mutations in SOD1, TARDBP (TDP-43), FUS and C9orf72 genes are linked to the onset of fALS (Mejzini et al., 2019). Both TDP-43 and FUS/TLS are enriched in paraspeckles in cultured cells where they directly bind NEAT1_2 transcript. Analysis of human spinal motor neurons revealed upregulated levels of NEAT1_2, and enhanced paraspeckle formation was confirmed in two independent cohorts of ALS cases compared to controls (Nishimoto et al., 2013; Shelkovnikova et al., 2018). The formation of paraspeckles has been linked to the loss of TDP-43 function in cells, suggesting a protective role for these structures (Shelkovnikova et al., 2018).
A microarray analysis demonstrated upregulated levels of NEAT1 in human HD postmortem brain patients and in a mouse model of HD (Sunwoo et al., 2017). Transfection of NEAT1 in a mouse cell line increases cell viability under oxidative stress, an observation that aligns with a previous report describing the involvement of NEAT1 in cell survival pathways under stress conditions (Choudhry et al., 2015). Thus, these studies support the idea of a protective role for this lncRNA in non-physiological settings (Sunwoo et al., 2017). Conversely, expression of NEAT1 is reduced in cortical brain regions from Schizophrenia patients. Furthermore, RNA-seq analysis performed in the frontal cortex of NEAT1 deficient mice indicated that pathways related to oligodendrocytes differentiation and RNA post-translational modifications are significantly impacted (Katsel et al., 2019). These results are in line with previously discussed data indicating that NEAT1 expression is dramatically changed during mouse oligodendrocyte-lineage specification (Mercer et al., 2010). Additionally, NEAT1–/– mice displayed a significant reduction in the numbers of oligodendrocyte-lineage cells and impaired expression of genes related to myelination, supporting a role for NEAT1 in oligodendrocyte function and related abnormalities in Schizophrenia pathology (Haroutunian et al., 2014; Katsel et al., 2019).
Finally, MALAT1 was found to bind to α-synuclein protein and increase its stability in a human neuroblastoma cell line (Zhang et al., 2016). Accordingly, inhibition of MALAT1 using resveratrol was found to increase miR-129 levels, consequently downregulating SNCA expression and improving disease-related phenotypes in a PD mouse model (Xia et al., 2019).
Targeting lncRNAs in Neurological Disorders: Trash or Treasure?
Around 30 years ago, gene transcription was perceived as a process mostly regulated by protein transcription factors and RNA was largely seen as an intermediary between DNA and protein; RNA had merely a supportive role in the translation of genetic information into diverse functional programs within the cells. However, the recent identification of multiple endogenous RNA classes with unexpected functions in gene regulation, including lncRNAs, has raised the interest to exploit RNA-based therapies (Wahlestedt, 2013; Roovers et al., 2018). To date, the biological functions of many annotated lncRNAs remain largely unexplored. The high unmet need for efficacious disease-modifying therapies for many of the neurodegenerative diseases mentioned in this review underscores the necessity to take bold steps and invest more in lncRNA research in order to open up innovative therapeutic opportunities for disorders of the brain.
Targeting LncRNAs at the DNA Level
Recent progress in genome-editing techniques, such as CRISPR-based methods including CRISPR-interference (CRISPRi) and CRISPR-activation (CRISPRa), has brought the exciting possibility of transcriptionally silence or activate lncRNA expressing loci, and clearly demonstrate that these tools will be crucial for a better understanding of lncRNA biology (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013; Qi et al., 2013; Gilbert et al., 2014; Thakore et al., 2015; Zhu et al., 2016; Abudayyeh et al., 2017; Liu S. J. et al., 2017; Chen et al., 2019; Phelan and Staudt, 2020; Wolter et al., 2020; Xu D. et al., 2020; Zhang et al., 2021). One way to achieve such transcriptional modulation is to use a dead-Cas9 approach. In this approach, a mutant form of Cas9 without endonuclease activity is fused to transcriptional repressors or activators to achieve transcriptional silencing or activation, respectively, of a specific gene promoter (Liu S. J. et al., 2017; Arun et al., 2018) (Figure 4A). Furthermore, Cas9-based gene therapy was shown to successfully reduce the expression of UBE3A-ATS and activate the paternal UBE3A in a mouse model of AS. Early treatment with an adeno-associated viral (AAV) delivery system designed to activate the expression of paternal UBE3A for at least 17 months ameliorated disease phenotype in AS mice and provided proof of concept that this approach is therapeutically relevant (Wolter et al., 2020). Onasemnogene abeparvovec, an AAV-based therapy carrying a functional copy of the SMN gene, was approved in May 2019 as the first gene therapy for SMA in the United States and illustrates that AAV approaches can lead to clinical successes for devastating neurological diseases (Hoy, 2019). While recent advances in CRISPR-based techniques demonstrate great potential for the discovery of disease mechanisms and identification of new therapeutic targets, there are still some challenges and risks that need to be considered prior to their use as therapy for neurological disorders. These include finding an effective CNS delivery method, the irreversibility of DNA editing, and safety concerns related to undesired on-target and off-target effects (Sun and Roy, 2021). For instance, a genome-wide association analysis found that almost two-thirds of lncRNA loci are at risk of inadvertently influencing the expression of neighboring genes upon CRISPR-mediated targeting (Goyal et al., 2017). Thus, future research will be crucial to explore the full potential of these modalities as therapeutics.
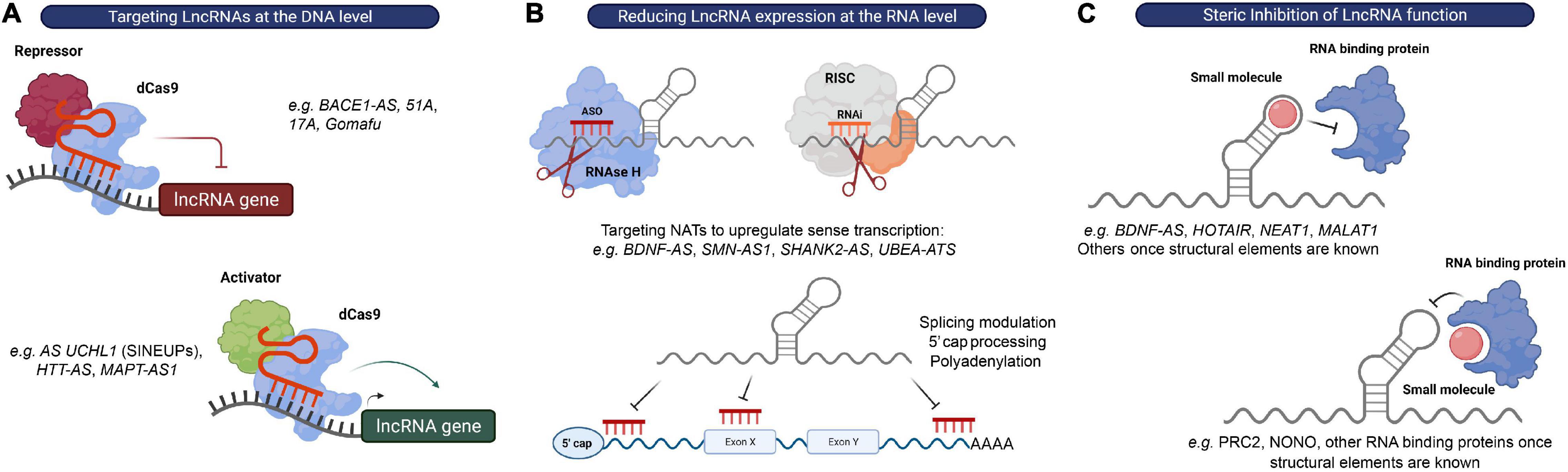
Figure 4. Strategies to target lncRNAs. (A) DNA editing: CRISPRi and CRISPRa tools can be used to transcriptionally silence or activate lncRNA expressing loci, respectively. (B) Modulation of RNA levels: ASOs and siRNAs can be used to reduce lncRNA levels in order to alter the expression of their associated protein-coding genes. recruits RBPs that mediate RNA processing events, such as 5′ capping, splicing or polyadenylation, to modulate expression or RNAse H to promote degradation of the lncRNA transcript, respectively, while RNAi induces RISC-mediated cleavage of the lncRNA transcript. (C) Steric inhibition: small molecules target secondary and tertiary structures of lncRNAs and/or their binding partners to block their interaction.
Reducing LncRNA Expression at the RNA Level
The possibility of modulating RNA expression with oligonucleotides provides a relatively straightforward strategy to impact virtually any RNA of interest and to target previously “undruggable” portions of our genome (Arun et al., 2018). At the moment, there are two major strategies employing oligonucleotide-based therapeutics: ASOs and RNA-mediated interference (RNAi), which share the fundamental principle of exerting their catalytic activity by binding their target RNA through Watson-Crick base pairing (Watts and Corey, 2012). ASOs are single stranded nucleotide sequences that bind RNA primary sequences to either affect RNA processing events, such as 5′-cap formation, splicing and polyadenylation, induce RNA degradation via the recruitment of Ribonuclease H (RNase H), or repress translation (in the case of protein-coding genes) (DeVos and Miller, 2013). Alternatively, a complementary hybrid RNA strand will engage with the target RNA in the RISC complex to initiate its degradation in the RNAi mechanism (Hannon and Rossi, 2004) (Figure 4B).
Remarkably, both ASO- and siRNA-based strategies have been proven successful in reducing the expression of several lncRNAs with potential roles in neurological disorders (Scheele et al., 2007; Chung et al., 2011; Carrieri et al., 2012; Modarresi et al., 2012; Barry et al., 2014; Meng et al., 2015; Peschansky et al., 2015; Yamanaka et al., 2015; d’Ydewalle et al., 2017; Wang X. et al., 2017; Woo et al., 2017; Luo et al., 2018). However, their efficiency is at least partially dependent on the subcellular compartment in which the target lncRNA localizes (Lennox and Behlke, 2016). Specifically, while lncRNAs primarily located within the nucleus were shown to be easier to target using ASOs, the expression of cytoplasmic lncRNAs was more efficiently reduced using RNAi methods. Additionally, both strategies were able to suppress the levels of lncRNAs residing in both compartments, although ASOs performed overall better in this case (Lennox and Behlke, 2016). In line with these observations, RNAse H1-dependent ASOs robustly exert their function in both the nucleus and the cytoplasm (Liang et al., 2017). These studies highlight the importance of better understanding how and where lncRNAs play their functional roles to select the most appropriate therapeutic approach to target these molecules.
A particular attractive class of candidate targets for these approaches is the previously mentioned NATs. Most of these NATs affect the transcription and/or translation of their neighboring genes. Furthermore, many of these antisense lncRNAs have been shown to act as repressors of sense coding genes in a locus-specific manner (e.g., SMN-AS1, BDNF-AS, SHANK2-AS, and UBE3A-ATS). Since there is still a considerable lack of therapeutic approaches to upregulate gene expression, inhibition of these transcripts with siRNAs or ASOs (called “antagoNATs”) might be of particular interest to increase the expression of genes found downregulated in CNS disorders (Wahlestedt, 2013) (Figure 4B).
The recent approval of several ASOs and siRNAs for clinical intervention clearly demonstrates the huge therapeutic potential of RNA-based therapies for a wide range of human diseases, including HD, ALS, AD and FTD (Roberts et al., 2020). However, two main limitations need to be considered. First, systemic delivery to the CNS remains a limitation since oligonucleotides are unable to cross the blood-brain barrier (Roberts et al., 2020). Additionally, due to the presence of complex secondary and tertiary structures, their incorporation into large protein complexes, or their specific intracellular location, some lncRNAs might be hard to target using these tools (Gutschner et al., 2011; Wilusz et al., 2012; Brown et al., 2014; Pegueroles and Gabaldón, 2016).
Steric Inhibition of LncRNA Function
Interfering with lncRNA function(s) rather than modulating its expression levels may represent an alternative therapeutic approach. An increasing body of evidence suggests that the function of many lncRNAs is largely mediated by their interaction with RBPs or protein complexes. Thus, a promising therapeutic option for targeting lncRNAs is the use of steric hindering antisense oligonucleotides or small molecules that affect these interactions by either (1) blocking the RNA-binding domain of the protein, or (2) directly binding to and disrupting secondary and tertiary structures in the lncRNA molecule (Meyer et al., 2020) (Figure 4C).
The variety of lncRNA binding partners and their unique structural features offer exceptional opportunities to target these RNAs using small molecules. Importantly, the development of new RNA sequencing techniques and structure determination assays such as SHAPE (Wilkinson et al., 2006), SHAPE-MaP (Smola et al., 2015), PARIS (Lu et al., 2016), or CROSSalign (Ponti et al., 2018) have made it possible to map the secondary and tertiary structures of lncRNAs. In addition, a database tool called LNCmap has been recently developed to explore correlations among diseases, small molecules and lncRNA signatures (Yang et al., 2017). Indeed, structural domains of several lncRNAs that interact with proteins or protein complexes, continue to be revealed (Smith et al., 2013; Mondal et al., 2015; Somarowthu et al., 2015; Liu F. et al., 2017; McCown et al., 2019; Balas et al., 2021).
MALAT1 has been proposed to contain a bipartite triple helix at the 3′ end and single-point mutations that destabilize this structure have been shown to reduce MALAT1 levels in cells, implying a crucial role for this structure in enabling MALAT1 expression (Brown et al., 2014, 2016). The secondary structure of the inverted SINE B2 element embedded in the mouse AS Uchl1 has also been recently revealed by Podbevšek et al. (2018). The authors found that removal of a structural motif containing a short hairpin abolishes the ability of AS Uchl1 to upregulate UCHL1 protein levels, highlighting the importance of specific structural determinants of the SINE B2 sequence in the functionality of AS Uchl1 (Podbevšek et al., 2018). Furthermore, the RBP NONO is an important component of paraspeckles and was shown to be recruited to these structures by specifically binding to highly abundant and conserved G-quadruplex motifs in the lncRNA NEAT1 (Simko et al., 2020). These motifs represent the main structural element recognized by the catalytic subunit of PRC2 (Wang X. et al., 2017), which we already discussed as an important binding partner for many lncRNAs, including HOTAIR (Rinn et al., 2007), XIST (Bousard et al., 2019), SMN-AS1 (d’Ydewalle et al., 2017), BDNF-AS (Modarresi et al., 2012), and others. Moreover, the observation that stable G-quadruplexes are present at the G-rich region of C9orf72 repeat RNA suggests a link between these motifs and neurodegeneration (Cammas and Millevoi, 2017). Several groups are currently exploring the potential of small molecules to modulate the function of lncRNAs, including some lncRNAs previously implicated in neurological disorders such as BDNF-AS, HOTAIR, NEAT1 or MALAT1 (Pedram Fatemi et al., 2015; Donlic et al., 2018; Abulwerdi et al., 2019; Ren et al., 2019; Simko et al., 2020; Khaled et al., 2021). While the field of RNA-targeting small molecules is still in its infancy, efforts toward understanding the fundamental dynamics between small molecule and lncRNAs recognition together with methodology development will likely contribute to unlock the full potential of using these compounds to treat complex brain disorders by targeting lncRNAs.
Although still early, it has been shown that small molecules can target several classes of RNA besides lncRNAs, including miRNAs, repeat expansion regions, mRNAs encoding for intrinsically disordered proteins, splicing modifiers and motifs located at the 5′- and 3′- untranslated regions (5′- and 3′- UTRs, respectively) (Meyer et al., 2020; Yu et al., 2020). Recently, Risdiplam received FDA approval for the treatment of SMA (U.S. Food & Drug Administration, 2020), and Branaplam is undergoing clinical trials as a therapy for the same disease as well as a for HD (ClinicalTrials. gov, 2021). Both Risdiplam and Branaplam increase SMN protein levels by acting as SMN2 splicing modulators (Meyer et al., 2020). Other small molecules have been identified as potential therapeutic agents for neurological disorders but are still in early discovery stages. These include Synucleozid for PD (Zhang et al., 2020), Mitoxantrone for primary tauopathies (Zheng et al., 2009), and at least one compound with the ability to target expanded repeat regions of FMR1 mRNA and C9orf72 mRNA, involved in FXTAS and c9FTD/ALS, respectively (Colak et al., 2014; Su et al., 2014; Wang et al., 2019).
Conclusion and Future Perspectives
Supported by their characteristics including high cell- and specificity and dynamic regulation of specific cellular pathways at the transcriptional and post-transcriptional level, lncRNA-based therapies could bring important clinical advantages. These include their superior potential as new targets, reduced toxic effects derived from off-target mechanisms, and the possibility to have a biologically meaningful effect with lower compound doses due to their lack of translation, fast turnover, and general low expression levels (Barry, 2014; Bonetti and Carninci, 2017). The fact that most disease-linked single-nucleotide polymorphisms have been shown to map to the non-coding genome further emphasizes the point that exploring non-coding loci can be relevant to identify new potential therapeutic targets (Tak and Farnham, 2015).
However, there are still some challenges to overcome in the field of lncRNAs research. First and foremost, more basic research is needed to address the exact roles of lncRNAs in the brain and identify their related disease-relevant signaling pathways. Second, and related to the first point, further investigation is needed to develop therapeutic strategies that efficiently alter lncRNA transcript levels or repress their function(s) using ASOs and siRNAs provided that the delivery of these molecules to the CNS is tackled. Though impressive progress occurred in the past years regarding the use of oligonucleotide-based therapies, a deeper appreciation of lncRNA structural features and their interactions with DNA, RNA and proteins will open the exciting possibility of targeting these RNAs using small molecules with unprecedented specificity as agonists or antagonists. On the long term, development of orally available, brain-penetrant lncRNA-targeting small molecules could represent a new therapeutic modality for genomic imprinting diseases, neurodevelopmental and neurodegenerative diseases. As new RNA-targeted therapeutic strategies continue to emerge, we believe that unraveling the functional roles of lncRNAs will pave the way to transform lncRNAs originally perceived as “junk” DNA to a therapeutic treasure for patients affected by CNS diseases.
Author Contributions
RP, AS, and Cd’Y were responsible for the conception of the article. RP did the scientific literature review, created the figures, and wrote the first draft of the review. Cd’Y contributed to the scientific literature review. AS and Cd’Y critically revised the entire review. All authors approved the final version.
Funding
RP holds a doctoral student fellowship from VLAIO, the Flemish Agency for Innovation and Entrepeneurship (Vlaams Agentschap Innoveren en Ondernemen). This work was supported by VLAIO (R&D grants HBC.2018.2290 and HBC.2020.3236) and VIB-KU Leuven.
Conflict of Interest
Cd’Y was employed by company Janssen Pharmaceutica N.V.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Illustrations were created with BioRender.com.
References
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. doi: 10.1038/nature24049
Abulwerdi, F. A., Xu, W., Ageeli, A. A., Yonkunas, M. J., Arun, G., Nam, H., et al. (2019). Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 14, 223–235. doi: 10.1021/acschembio.8b00807
An, H., Williams, N. G., and Shelkovnikova, T. A. (2018). NEAT1 and paraspeckles in neurodegenerative diseases: a missing lnc found? Non-coding. RNA Res. 3, 243–252. doi: 10.1016/j.ncrna.2018.11.003
Angrand, P.-O., Vennin, C., Le Bourhis, X., and Adriaenssens, E. (2015). The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 6:165. doi: 10.3389/fgene.2015.00165
Aprea, J., and Calegari, F. (2015). Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. EMBO J. 34, 2865–2884. doi: 10.15252/embj.201592655
Arun, G., Diermeier, S. D., and Spector, D. L. (2018). Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 24, 257–277. doi: 10.1016/j.molmed.2018.01.001
Balas, M. M., Hartwick, E. W., Barrington, C., Roberts, J. T., Wu, S. K., Bettcher, R., et al. (2021). Establishing RNA-RNA interactions remodels lncRNA structure and promotes PRC2 activity. Sci. Adv. 7:eabc9191. doi: 10.1126/sciadv.abc9191
Bandres-Ciga, S., Diez-Fairen, M., Kim, J. J., and Singleton, A. B. (2020). Genetics of Parkinson’s disease: an introspection of its journey towards precision medicine. Neurobiol. Dis. 137:104782. doi: 10.1016/j.nbd.2020.104782
Barker, H. V., Niblock, M., Lee, Y., and Shaw, C. E. (2017). RNA misprocessing in C9orf72 - linked neurodegeneration. Front. Cell. Neurosci. 11:195. doi: 10.3389/fncel.2017.00195
Barry, G. (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry 19, 410–416. doi: 10.1038/mp.2013.196
Barry, G., Briggs, J. A., Vanichkina, D. P., Poth, E. M., Beveridge, N. J., Ratnu, V. S., et al. (2014). The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494. doi: 10.1038/mp.2013.45
Barry, G., Guennewig, B., Fung, S., Kaczorowski, D., and Weickert, C. S. (2015). Long non-coding RNA expression during aging in the human subependymal zone. Front. Neurol. 6:45. doi: 10.3389/fneur.2015.00045
Belgard, T. G., Marques, A. C., Oliver, P. L., Abaan, H. O., Sirey, T. M., Hoerder-Suabedissen, A., et al. (2011). A transcriptomic atlas of mouse neocortical layers. Neuron 71, 605–616. doi: 10.1016/j.neuron.2011.06.039
Bellacosa, A. (2001). Role of MED1 (MBD4) Gene in DNA repair and human cancer. J. Cell. Physiol. 187, 137–144. doi: 10.1002/jcp.1064
Benn, C. L., Sun, T., Sadri-Vakili, G., McFarland, K. N., DiRocco, D. P., Yohrling, G. J., et al. (2008). Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J. Neurosci. 28, 10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Bonetti, A., and Carninci, P. (2017). From bench to bedside: the long journey of long. Curr. Opin. Syst. Biol. 3, 119–124. doi: 10.1016/j.coisb.2017.04.016
Bousard, A., Raposo, A. C., Zylicz, J. J., Picard, C., Pires, V. B., Qi, Y., et al. (2019). The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 20, 1–18. doi: 10.15252/embr.201948019
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell Biol. 220, 1–17. doi: 10.1083/JCB.202009045
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L., and Barry, G. (2015). Mechanisms of Long Non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 88, 861–877. doi: 10.1016/j.neuron.2015.09.045
Brown, C. J., Ballabio, A., Rupert, J. L., Lafreniere, R. G., Grompe, M., Tonlorenzi, R., et al. (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. doi: 10.1038/349038a0
Brown, J. A., Bulkley, D., Wang, J., Valenstein, M. L., Yario, T. A., Steitz, T. A., et al. (2014). Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 21, 633–640. doi: 10.1038/nsmb.2844
Brown, J. A., Kinzig, C. G., Degregorio, S. J., and Steitz, J. A. (2016). Hoogsteen-position pyrimidines promote the stability and function of the MALAT1 RNA triple helix. RNA 22, 743–749. doi: 10.1261/rna.055707.115.1
Cammas, A., and Millevoi, S. (2017). RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acid Ther. 45, 1584–1595. doi: 10.1093/nar/gkw1280
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi: 10.1126/science.1112014
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. doi: 10.1038/nature11508
Carrieri, C., Forrest, A. R. R., Santoro, C., Persichetti, F., Carninci, P., Zucchelli, S., et al. (2015). Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front. Cell. Neurosci. 9:114. doi: 10.3389/fncel.2015.00114
Chalei, V., Sansom, S. N., Kong, L., Lee, S., Montiel, J. F., Vance, K. W., et al. (2014). The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife 3, 1–24. doi: 10.7554/eLife.04530
Chen, B. J., Ueberham, U., Mills, J. D., Kirazov, L., Kirazov, E., Knobloch, M., et al. (2017). RNA sequencing reveals pronounced changes in the noncoding transcriptome of aging synaptosomes. Neurobiol. Aging 56, 67–77. doi: 10.1016/j.neurobiolaging.2017.04.005
Chen, L.-L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41, 761–772. doi: 10.1016/j.tibs.2016.07.003
Chen, W., Zhang, G., Li, J., Zhang, X., Huang, S., Xiang, S., et al. (2019). CRISPRlnc: a manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 47, D63–D68. doi: 10.1093/nar/gky904
Chodroff, R. A., Goodstadt, L., Sirey, T. M., Oliver, P. L., Davies, K. E., Green, E. D., et al. (2010). Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 11:R72. doi: 10.1186/gb-2010-11-7-r72
Choi, J., Levey, A. I., Weintraub, S. T., Rees, H. D., Gearing, M., Chin, L.-S., et al. (2004). Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J. Biol. Chem. 279, 13256–13264. doi: 10.1074/jbc.M314124200
Choudhry, H., Albukhari, A., Morotti, M., Haider, S., Moralli, D., Smythies, J., et al. (2015). Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34, 4482–4490. doi: 10.1038/onc.2014.378
Chung, D. W., Rudnicki, D. D., Yu, L., and Margolis, R. L. (2011). A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477. doi: 10.1093/hmg/ddr263
Ciarlo, E., Massone, S., Penna, I., Nizzari, M., Gigoni, A., Dieci, G., et al. (2013). An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. DMM Dis. Model. Mech. 6, 424–433. doi: 10.1242/dmm.009761
ClinicalTrials. gov (2021). Available online at: https://clinicaltrials.gov/ct2/show/results/NCT02268552 (accessed May 20, 2021).
Cogill, S. B., Srivastava, A. K., Yang, M. Q., and Wang, L. (2018). Co-expression of long non-coding RNAs and autism risk genes in the developing human brain. BMC Syst. Biol. 12:91. doi: 10.1186/s12918-018-0639-x
Colak, D., Zaninovic, N., Cohen, M. S., Rosenwaks, Z., Yang, W.-Y., Gerhardt, J., et al. (2014). Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343, 1002–1005. doi: 10.1126/science.1245831
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cooper-Knock, J., Higginbottom, A., Stopford, M. J., Highley, J. R., Ince, P. G., Wharton, S. B., et al. (2015). Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. 130, 63–75. doi: 10.1007/s00401-015-1429-9
Davidovich, C., and Cech, T. R. (2015). The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–2022. doi: 10.1261/rna.053918.115.Kuzmichev
Deas, E., Plun-Favreau, H., and Wood, N. W. (2009). PINK1 function in health and disease. EMBO Mol. Med. 1, 152–165. doi: 10.1002/emmm.200900024
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
DeVos, S. L., and Miller, T. M. (2013). Antisense oligonucleotides: treating neurodegeneration at the level of RNA. Neurotherapeutics 10, 486–497. doi: 10.1007/s13311-013-0194-5
Diederichs, S. (2014). The four dimensions of noncoding RNA conservation. Trends Genet. 30, 121–123. doi: 10.1016/j.tig.2014.01.004
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Donlic, A., Morgan, B. S., Xu, J. L., Liu, A., Roble, C., and Hargrove, A. E. (2018). Discovery of small molecule ligands for MALAT1 by tuning an RNA-binding scaffold. Angew. Chemie Int. Ed. 57, 13242–13247. doi: 10.1002/anie.201808823
Dunham, I., Kundaje, A., Aldred, S. F., Collins, P. J., Davis, C. A., Doyle, F., et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
d’Ydewalle, C., Ramos, D. M., Pyles, N. J., Ng, S. Y., Gorz, M., Pilato, C. M., et al. (2017). The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron 93, 66–79. doi: 10.1016/j.neuron.2016.11.033
d’Ydewalle, C., and Sumner, C. J. (2015). Spinal muscular atrophy therapeutics: where do we stand? Neurotherapeutics 12, 303–316. doi: 10.1007/s13311-015-0337-y
Emon, P. Z., Das, R., Nishuty, N. L., Qusar, M. M. A. S., Bhuiyan, M. A., and Islam, R. (2020). Reduced serum BDNF levels are associated with the increased risk for developing MDD: a case-control study with or without antidepressant therapy. BMC Res. Notes 13:9. doi: 10.1186/s13104-020-04952-3
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. doi: 10.038/nm1784
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van der Brug, M. P., Nalls, M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, 1–13. doi: 10.1186/gb-2010-11-5-r56
Fan, X., Fu, Y., Zhou, X., Sun, L., Yang, M., Wang, M., et al. (2020). Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci. Adv. 6, 1–16. doi: 10.1126/sciadv.aaz2978
Ferrer, I., Goutan, E., Marín, C., Rey, M. J., and Ribalta, T. (2000). Brain-derived neurotrophic factor in Huntington disease. Brain Res. 866, 257–261. doi: 10.1016/s0006-8993(00)02237-x
Francelle, L., Galvan, L., Gaillard, M., Petit, F., Bernay, B., Guillermier, M., et al. (2015). Neurobiology of Aging The striatal long noncoding RNA Abhd11os is neuroprotective against an N-terminal fragment of mutant huntingtin in vivo. Neurobiol. Aging 36, e7–e16. doi: 10.1016/j.neurobiolaging.2014.11.014
Francescatto, M., Vitezic, M., Heutink, P., and Saxena, A. (2014). Brain-specific noncoding RNAs are likely to originate in repeats and may play a role in up-regulating genes in cis. Int. J. Biochem. Cell Biol. 54, 331–337. doi: 10.1016/j.biocel.2014.06.014
Frankish, A., Diekhans, M., Jungreis, I., Lagarde, J., Loveland, J. E., Mudge, J. M., et al. (2021). Gencode 2021. Nucleic Acids Res. 49, D916–D923. doi: 10.1093/nar/gkaa1087
Gendron, T. F., Bieniek, K. F., Zhang, Y.-J., Jansen-West, K., Ash, P. E. A., Caulfield, T., et al. (2013). Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844. doi: 10.1007/s00401-013-1192-8
Gil, N., and Ulitsky, I. (2018). Production of Spliced Long Noncoding RNAs specifies regions with increased enhancer activity. Cell Syst. 7, 537.e3–547.e3. doi: 10.1016/j.cels.2018.10.009
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. doi: 10.1016/j.cell.2014.09.029
Goyal, A., Myacheva, K., Groß, M., Klingenberg, M., Duran Arqué, B., and Diederichs, S. (2017). Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 45:e12. doi: 10.1093/nar/gkw883
Grammatikakis, I., Panda, A. C., Abdelmohsen, K., and Gorospe, M. (2014). Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging 6, 992–1009. doi: 10.18632/aging.100710
Grinman, E., Nakahata, Y., Avchalumov, Y., Espadas, I., Swarnkar, S., Yasuda, R., et al. (2021). Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 7:eabf0605. doi: 10.1126/sciadv.abf0605
Grossman, S. R., Andersen, K. G., Shlyakhter, I., Tabrizi, S., Winnicki, S., Yen, A., et al. (2013). Identifying recent adaptations in large-scale genomic data. Cell 152, 703–713. doi: 10.1016/j.cell.2013.01.035
Gutschner, T., Baas, M., and Diederichs, S. (2011). Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 21, 1944–1954. doi: 10.1101/gr.122358.111
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., et al. (2011). LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. doi: 10.1038/nature10398
Hampel, H., Vassar, R., De Strooper, B., Hardy, J., Willem, M., Singh, N., et al. (2020). The β-Secretase BACE1 in Alzheimer’s disease. Biol. Psychiatry 89, 745–756. doi: 10.1016/j.biopsych.2020.02.001
Hannon, G. J., and Rossi, J. J. (2004). Unlocking the potential of the human genome with RNA interference. Nature 431, 371–378. doi: 10.1038/nature02870
Haroutunian, V., Katsel, P., Roussos, P., Davis, K. L., Altshuler, L. L., and Bartzokis, G. (2014). Myelination, oligodendrocytes, and serious mental illness. Glia 62, 1856–1877. doi: 10.1002/glia.22716
He, Z., Bammann, H., Han, D., and Xie, G. (2014). Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. RNA 20, 1103–1111. doi: 10.1261/rna.043075.113
Hon, C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J. L., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5’ ends. Nat. Publ. Gr. 543, 199–204. doi: 10.1038/nature21374
Hoy, S. M. (2019). Onasemnogene abeparvovec: first global approval. Drugs 79, 1255–1262. doi: 10.1007/s40265-019-01162-5
Huang, G., Zhu, H., Wu, S., Cui, M., and Xu, T. (2019). Long noncoding RNA can be a probable mechanism and a novel target for diagnosis and therapy in fragile X syndrome. Front. Genet. 10:446. doi: 10.3389/fgene.2019.00446
Human Genome Project (2003). Available online at: https://www.genome.gov/human-genome-project (accessed April 26, 2021).
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B., and Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. doi: 10.1186/1471-2164-8-39
Ip, J. Y., Sone, M., Nashiki, C., Pan, Q., Kitaichi, K., Yanaka, K., et al. (2016). Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 6:27204. doi: 10.1038/srep27204
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. doi: 10.1038/ng.3192
Jiang, C., Li, Y., Zhao, Z., Lu, J., Chen, H., Ding, N., et al. (2016). Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 7, 7120–7133. doi: 10.18632/oncotarget.6859
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Johnson, M. B., Wang, P. P., Atabay, K. D., Murphy, E. A., Doan, R. N., Hecht, J. L., et al. (2015). Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 18, 637–646. doi: 10.1038/nn.3980
Johnson, R. (2012). Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 46, 245–254. doi: 10.1016/j.nbd.2011.12.006
Johnson, R., Richter, N., Jauch, R., Gaughwin, P. M., Zuccato, C., Cattaneo, E., et al. (2010). Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol. Genomics 41, 269–274. doi: 10.1152/physiolgenomics.00019.2010
Johnsson, P., Lipovich, L., Grandér, D., and Morris, K. V. (2014). Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 1840, 1063–1071. doi: 10.1016/j.bbagen.2013.10.035
Kadakkuzha, B. M., Liu, X. A., McCrate, J., Shankar, G., Rizzo, V., Afinogenova, A., et al. (2015). Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell. Neurosci. 9:63. doi: 10.3389/fncel.2015.00063
Kapusta, A., and Feschotte, C. (2014). Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 30, 439–452. doi: 10.1016/j.tig.2014.08.004
Katsel, P., Roussos, P., Fam, P., Khan, S., Tan, W., Hirose, T., et al. (2019). The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophr. 5:3. doi: 10.1038/s41537-019-0071-2
Keihani, S., Kluever, V., Mandad, S., Bansal, V., Rahman, R., Fritsch, E., et al. (2019). The long noncoding RNA neuroLNC regulates presynaptic activity by interacting with the neurodegeneration-associated protein TDP-43. Sci. Adv. 5:eaay2670. doi: 10.1126/sciadv.aay2670
Khaled, H. G., Feng, H., Hu, X., Sun, X., Zheng, W., Li, P. P., et al. (2021). A high-throughput screening to identify small molecules that suppress huntingtin promoter activity or activate huntingtin-antisense promoter activity. Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-85279-2
Khalil, A. M., Faghihi, M. A., Modarresi, F., Brothers, S. P., and Wahlestedt, C. (2008). A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One 3:e0001486. doi: 10.1371/journal.pone.0001486
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. PNAS 106, 11667–11672. doi: 10.1073/pnas.0904715106
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Kour, S., and Rath, P. C. (2015). Age-dependent differential expression profile of a novel intergenic long noncoding RNA in rat brain. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 46, 55–66. doi: 10.1016/j.ijdevneu.2015.07.011
Ladd, P. D., Smith, L. E., Rabaia, N. A., Moore, J. M., Georges, S. A., Hansen, R. S., et al. (2007). An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16, 3174–3187. doi: 10.1093/hmg/ddm293
Laske, C., Stellos, K., Hoffmann, N., Stransky, E., Straten, G., Eschweiler, G. W., et al. (2011). Higher BDNF serum levels predict slower cognitive decline in Alzheimer’s disease patients. Int. J. Neuropsychopharmacol. 14, 399–404. doi: 10.1017/S1461145710001008
Law, A. J., Kleinman, J. E., Weinberger, D. R., and Weickert, C. S. (2007). Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum. Mol. Genet. 16, 129–141. doi: 10.1093/hmg/ddl449
Lee, Y.-B., Chen, H.-J., Joao, N., Peres, Gomez-Deza, J., Attig, J., et al. (2013). Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5, 1178–1186. doi: 10.1016/j.celrep.2013.10.049
Lennox, K. A., and Behlke, M. A. (2016). Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 44, 863–877. doi: 10.1093/nar/gkv1206
Liang, X. H., Sun, H., Nichols, J. G., and Crooke, S. T. (2017). RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol. Ther. 25, 2075–2092. doi: 10.1016/j.ymthe.2017.06.002
Lin, M., Pedrosa, E., Shah, A., Hrabovsky, A., Maqbool, S., Zheng, D., et al. (2011). RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 6:e0023356. doi: 10.1371/journal.pone.0023356
Lin, N., Chang, K., Li, Z., Gates, K., Rana, Z. A., Dang, J., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019. doi: 10.1016/j.molcel.2014.01.021
Lindblad-Toh, K., Garber, M., Zuk, O., Lin, M. F., Parker, B. J., Washietl, S., et al. (2011). A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482. doi: 10.1038/nature10530
Lipovich, L., Tarca, A. L., Cai, J., Jia, H., Chugani, H. T., Sterner, K. N., et al. (2014). Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb. Cortex 24, 1451–1459. doi: 10.1093/cercor/bhs414
Liu, F., Somarowthu, S., and Pyle, A. M. (2017). Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 13, 282–289. doi: 10.1038/nchembio.2272
Liu, G., Mattick, J. S., and Taft, R. J. (2013). A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 12, 2061–2072. doi: 10.4161/cc.25134
Liu, S. J., Horlbeck, M. A., Cho, S. W., Birk, H. S., Malatesta, M., He, D., et al. (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355:aah7111. doi: 10.1126/science.aah7111
Liu, S. J., Nowakowski, T. J., Pollen, A. A., Lui, J. H., Horlbeck, M. A., Attenello, F. J., et al. (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17:67. doi: 10.1186/s13059-016-0932-1
Lord, C., Brugha, T. S., Charman, T., Cusack, J., Dumas, G., Frazier, T., et al. (2020). Autism spectrum disorder. Nat. Rev. Dis. Prim. 6:5. doi: 10.1038/s41572-019-0138-4
Lorenzi, L., Chiu, H., Cobos, F. A., Gross, S., Volders, P., Cannoodt, R., et al. (2021). The RNA Atlas expands the catalog of human non-coding RNAs. Nat. Biotechnol. [Epub ahead of print]. doi: 10.1038/s41587-021-00936-1
Lu, Z., Zhang, Q. C., Lee, B., Flynn, R. A., Smith, M. A., Robinson, J. T., et al. (2016). RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279. doi: 10.1016/j.cell.2016.04.028
Luo, H., Sun, S., Li, P., Bu, D., Cao, H., and Zhao, Y. (2013). Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS One 8:e0070835. doi: 10.1371/journal.pone.0070835
Luo, T., Liu, P., Wang, X.-Y., Li, L., Li-Ping, Zao, Huang, J., et al. (2018). Effect of the autism-associated lncRNA Shank2-AS on architecture and growth of neurons. J. Cell. Biochem. [Epub ahead of print]. doi: 10.1002/jcb.27471
Ma, L., Bajic, V. B., Zhang, Z., Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. doi: 10.4161/rna.24604
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Massone, S., Vassallo, I., Fiorino, G., Castelnuovo, M., Barbieri, F., Borghi, R., et al. (2011). 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317. doi: 10.1016/j.nbd.2010.09.019
Mattick, J. S. (2001). Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2, 986–991. doi: 10.1093/embo-reports/kve230
Mattick, J. S., and Rinn, J. L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7. doi: 10.1038/nsmb.2942
McCown, P. J., Wang, M. C., and Jaeger, L. (2019). Secondary structural model of human MALAT1 reveals multiple structure - function relationships. Int. J. Mol. Sci. 20, 1–19.
Mejzini, R., Flynn, L. L., Pitout, I. L., Fletcher, S., Wilton, S. D., and Akkari, P. A. (2019). ALS genetics, mechanisms, and therapeutics: where are we now? Front. Neurosci. 13:1310. doi: 10.3389/fnins.2019.01310
Meng, L., Person, R. E., and Beaudet, A. L. (2012). Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum. Mol. Genet. 21, 3001–3012. doi: 10.1093/hmg/dds130
Meng, L., Person, R. E., Huang, W., Zhu, P. J., Costa-Mattioli, M., and Beaudet, A. L. (2013). Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the angelman syndrome mouse model. PLoS Genet. 9:e1004039. doi: 10.1371/journal.pgen.1004039
Meng, L., Ward, A. J., Chun, S., Bennett, C. F., Beaudet, A. L., and Rigo, F. (2015). Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518, 409–412. doi: 10.1038/nature13975
Meng, Q., Wang, K., Brunetti, T., Xia, Y., Jiao, C., Dai, R., et al. (2018). The DGCR5 long noncoding RNA may regulate expression of several schizophrenia-related genes. Sci. Transl. Med. 10, 1–11. doi: 10.1126/scitranslmed.aat6912
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. PNAS 105, 716–721. doi: 10.1073/pnas.0706729105
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G., Mattick, J. S., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Meyer, S. M., Williams, C. C., Akahori, Y., Tanaka, T., Aikawa, H., Tong, Y., et al. (2020). Small molecule recognition of disease-relevant RNA structures. Chem. Soc. Rev. 49, 7167–7199. doi: 10.1039/d0cs00560f
Miranda, M., Morici, J. F., Zanoni, M. B., and Bekinschtein, P. (2019). Brain-derived neurotrophic factor: a key molecule for memory in the healthy and the pathological brain. Front. Cell. Neurosci. 13:363. doi: 10.3389/fncel.2019.00363
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. doi: 10.1038/nbt.2158
Molendijk, M. L., Bus, B. A. A., Spinhoven, P., Penninx, B., Kenis, G., and Prickaerts, J. (2011). Serum levels of brain-derived neurotrophic factor in major depressive disorder: state-trait issues, clinical features and pharmacological treatment. Mol. Psychiatry 16, 1088–1095. doi: 10.1038/mp.2010.98
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Mitra, S., et al. (2015). MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6:7743. doi: 10.1038/ncomms8743
Morikawa, T., and Manabe, T. (2010). Aberrant regulation of alternative pre-mRNA splicing in schizophrenia. Neurochem. Int. 57, 691–704. doi: 10.1016/j.neuint.2010.08.012
Muddashetty, R., Khanam, T., Kondrashov, A., Bundman, M., Iacoangeli, A., Kremerskothen, J., et al. (2002). Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321, 433–445. doi: 10.1016/s0022-2836(02)00655-1
Myers, R. M., Stamatoyannopoulos, J., Snyder, M., Dunham, I., Hardison, R. C., Bernstein, B. E., et al. (2011). A user’s guide to the Encyclopedia of DNA elements (ENCODE). PLoS Biol. 9:e1001046. doi: 10.1371/journal.pbio.1001046
Nakata, K., Lipska, B. K., Hyde, T. M., Ye, T., Newburn, E. N., Morita, Y., et al. (2009). DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. PNAS 106, 15873–15878. doi: 10.1073/pnas.0903413106
Necsulea, A., and Kaessmann, H. (2014). Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 15, 734–748. doi: 10.1038/nrg3802
Ng, S., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi: 10.1016/j.molcel.2013.07.017
Nishimoto, Y., Nakagawa, S., Hirose, T., Okano, H. J., Takao, M., Shibata, S., et al. (2013). The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 6:31. doi: 10.1186/1756-6606-6-31
Nitsche, A., Rose, D., Fasold, M., Reiche, K., and Stadler, P. F. (2015). Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 21, 801–812. doi: 10.1261/rna.046342.114
Pastori, C., Peschansky, V. J., Barbouth, D., Mehta, A., Silva, J. P., and Wahlestedt, C. (2014). Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum. Genet. 133, 59–67. doi: 10.1007/s00439-013-1356-6
Pedram Fatemi, R., Salah-Uddin, S., Modarresi, F., Khoury, N., Wahlestedt, C., and Faghihi, M. A. (2015). Screening for small-molecule modulators of long noncoding RNA-protein interactions using alphascreen. J. Biomol. Screen. 20, 1132–1141. doi: 10.1177/1087057115594187
Pegueroles, C., and Gabaldón, T. (2016). Secondary structure impacts patterns of selection in human lncRNAs. BMC Biol. 14:13. doi: 10.1186/s12915-016-0283-0
Peschansky, V. J., Pastori, C., Zeier, Z., Motti, D., Katya, W., Velmeshev, D., et al. (2015). Changes in expression of the long non-coding RNA FMR4 associate with altered gene expression during differentiation of human neural precursor cells. Front. Genet. 6:263. doi: 10.3389/fgene.2015.00263
Peykov, S., Berkel, S., Schoen, M., Weiss, K., Degenhardt, F., Strohmaier, J., et al. (2015). Identification and functional characterization of rare SHANK2 variants in schizophrenia. Mol. Psychiatry 20, 1489–1498. doi: 10.1038/mp.2014.172
Phelan, J. D., and Staudt, L. M. (2020). CRISPR-based technology to silence the expression of IncRNAs. PNAS 117, 8225–8227. doi: 10.1073/pnas.2003702117
Podbevšek, P., Fasolo, F., Bon, C., Cimatti, L., Reißer, S., Carninci, P., et al. (2018). Structural determinants of the SINE B2 element embedded in the long non-coding RNA activator of translation AS Uchl1. Sci. Rep. 8:3189. doi: 10.1038/s41598-017-14908-6
Ponjavic, J., Oliver, P. L., Lunter, G., and Ponting, C. P. (2009). Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5:e1000617. doi: 10.1371/journal.pgen.1000617
Ponti, R. D., Armaos, A., Marti, S., and Tartaglia, G. G. (2018). A method for RNA structure prediction shows evidence for structure in lncRNAs. Front. Mol. Biosci. 5:111. doi: 10.3389/fmolb.2018.00111
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Quan, Z., Zheng, D., and Qing, H. (2017). Regulatory roles of long non-coding rnas in the central nervous system and associated neurodegenerative diseases. Front. Cell. Neurosci. 11:175. doi: 10.3389/fncel.2017.00175
Quinodoz, S., and Guttman, M. (2014). Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 24, 651–663. doi: 10.1016/j.tcb.2014.08.009
Ramos, A. D., Andersen, R. E., Kriegstein, A. R., Lim, D. A., Ramos, A. D., Andersen, R. E., et al. (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Stem Cell 16, 439–447. doi: 10.1016/j.stem.2015.02.007
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K. Y., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi: 10.1016/j.stem.2013.03.003
Rauch, J. N., Luna, G., Guzman, E., Audouard, M., Challis, C., Sibih, Y. E., et al. (2020). LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385. doi: 10.1038/s41586-020-2156-5
Reinicke, A. T., Laban, K., Sachs, M., Kraus, V., Walden, M., Damme, M., et al. (2019). Ubiquitin C-terminal hydrolase L1 (UCH-L1) loss causes neurodegeneration by altering protein turnover in the first postnatal weeks. PNAS 116, 7963–7972. doi: 10.1073/pnas.1812413116
Ren, Y., Wang, Y. F., Zhang, J., Wang, Q. X., Han, L., Mei, M., et al. (2019). Targeted design and identification of AC1NOD4Q to block activity of HOTAIR by abrogating the scaffold interaction with EZH2. Clin. Epigenetics 11, 1–16. doi: 10.1186/s13148-019-0624-2
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Roberts, T. C., Langer, R., and Wood, M. J. A. (2020). Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694. doi: 10.1038/s41573-020-0075-7
Roberts, T. C., Morris, K. V., and Wood, M. J. A. (2014). The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. B Biol. Sci. 369:20130507. doi: 10.1098/rstb.2013.0507
Rocha, S. T., and Heard, E. (2017). Novel players in X inactivation: insights into Xist -mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 24, 197–204. doi: 10.1038/nsmb.3370
Roovers, J., De Jonghe, P., and Weckhuysen, S. (2018). Expert opinion on therapeutic targets the therapeutic potential of RNA regulation in neurological disorders. Expert Opin. Ther. Targets 22, 1017–1028. doi: 10.1080/14728222.2018.1542429
Rubin, G. M., Yandell, M. D., Wortman, J. R., Gabor, G. L., Miklos, Nelson, C. R., et al. (2000). Comparative genomics of the eukaryotes. Science 287, 2204–2215. doi: 10.1126/science.287.5461.2204
Salta, E., and De Strooper, B. (2017). Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 18, 627–640. doi: 10.1038/nrn.2017.90
Salvatori, B., Biscarini, S., and Morlando, M. (2020). Non-coding RNAs in nervous system development and disease. Front. Cell Dev. Biol. 8:273. doi: 10.3389/fcell.2020.00273
Salzberg, S. L. (2018). Open questions: how many genes do we have? BMC Biol. 16:12. doi: 10.1186/s12915-018-0564-x
Sarropoulos, I., Marin, R., Cardoso-Moreira, M., and Kaessmann, H. (2019). Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571, 510–514. doi: 10.1038/s41586-019-1341-x
Sato, D., Lionel, A. C., Leblond, C. S., Prasad, A., Pinto, D., Walker, S., et al. (2012). SHANK1 deletions in males with autism spectrum disorder. Am. J. Hum. Genet. 90, 879–887. doi: 10.1016/j.ajhg.2012.03.017
Sauvageau, M., Goff, L. A., Lodato, S., Bonev, B., Groff, A. F., Gerhardinger, C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2:e01749. doi: 10.7554/eLife.01749
Scheele, C., Petrovic, N., Faghihi, M. A., Lassmann, T., Fredriksson, K., Rooyackers, O., et al. (2007). The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 8:74. doi: 10.1186/1471-2164-8-74
Scheltens, P., De Strooper, B., Kivipelto, M., Holstege, H., Chételat, G., Teunissen, C. E., et al. (2021). Alzheimer’s disease. Lancet 397, 1577–1590. doi: 10.1016/S0140-6736(20)32205-4
Seemann, S. E., Sunkin, S. M., Hawrylycz, M. J., Ruzzo, W. L., and Gorodkin, J. (2012). Transcripts with in silico predicted RNA structure are enriched everywhere in the mouse brain. BMC Genomics 13:214. doi: 10.1186/1471-2164-13-214
Shelkovnikova, T. A., Kukharsky, M. S., An, H., Dimasi, P., Alexeeva, S., Shabir, O., et al. (2018). Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 13:30. doi: 10.1186/s13024-018-0263-7
Simko, E. A. J., Liu, H., Zhang, T., Velasquez, A., Teli, S., Haeusler, A. R., et al. (2020). G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 48, 7421–7438. doi: 10.1093/nar/gkaa475
Simone, R., Javad, F., Emmett, W., Wilkins, O. G., Almeida, F. L., Barahona-Torres, N., et al. (2021). MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration. Nature 594, 117–123. doi: 10.1038/s41586-021-03556-6
Smith, M. A., Gesell, T., Stadler, P. F., and Mattick, J. S. (2013). Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 41, 8220–8236. doi: 10.1093/nar/gkt596
Smola, M. J., Rice, G. M., Busan, S., Siegfried, N. A., and Weeks, K. M. (2015). Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669. doi: 10.1038/nprot.2015.103
Somarowthu, S., Legiewicz, M., Chillón, I., Marcia, M., Liu, F., and Pyle, A. M. (2015). HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361. doi: 10.1016/j.molcel.2015.03.006
Sone, M., Hayashi, T., Tarui, H., Agata, K., Takeichi, M., and Nakagawa, S. (2007). The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 120, 2498–2506. doi: 10.1242/jcs.009357
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi: 10.1038/s41580-020-00315-9
Su, L., Wang, C., Zheng, C., Wei, H., and Song, X. (2018). A meta-analysis of public microarray data identifies biological regulatory networks in Parkinson’s disease. BMC Med. Genomics 11:40. doi: 10.1186/s12920-018-0357-7
Su, Z., Zhang, Y., Gendron, T. F., Bauer, P. O., Chew, J., Yang, W.-Y., et al. (2014). Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 84:239. doi: 10.1016/j.neuron.2014.09.019
Sugitani, Y., Nakai, S., Minowa, O., Nishi, M., Jishage, K.-I., Kawano, H., et al. (2002). Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16, 1760–1765. doi: 10.1101/gad.978002
Sun, J., and Roy, S. (2021). Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 24, 297–311. doi: 10.1038/s41593-020-00778-1
Sunamura, N., Iwashita, S., Enomoto, K., Kadoshima, T., and Isono, F. (2018). Loss of the fragile X mental retardation protein causes aberrant differentiation in human neural progenitor cells. Sci. Rep. 8:11585. doi: 10.1038/s41598-018-30025-4
Sunwoo, J.-S., Lee, S.-T., Im, W., Lee, M., Byun, J.-I., Jung, K.-H., et al. (2017). Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol. Neurobiol. 54, 1577–1586. doi: 10.1007/s12035-016-9928-9
Tachibana, M., Holm, M., Liu, C., Shinohara, M., Aikawa, T., Oue, H., et al. (2019). APOE4-mediated amyloid-β pathology depends on its neuronal receptor LRP1. J. Clin. Invest. 129, 1272–1277. doi: 10.1172/jci124853
Taft, R. J., Pheasant, M., and Mattick, J. S. (2007). The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29, 288–299. doi: 10.1002/bies.20544
Tak, Y. G., and Farnham, P. J. (2015). Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8, 1–18. doi: 10.1186/s13072-015-0050-4
Tay, S.-K., Blytheb, J., and Lipovicha, L. (2009). Global discovery of primate-specific genes in the human genome. PNAS 106:14180. doi: 10.1073/pnas.0908685106
Thakore, P. I., D’Ippolito, A. M., Song, L., Safi, A., Shivakumar, N. K., Kabadi, A. M., et al. (2015). Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149. doi: 10.1038/nmeth.3630
Tollervey, J. R., Curk, T., Rogelj, B., Briese, M., Cereda, M., Kayikci, M., et al. (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458. doi: 10.1038/nn.2778
Torarinsson, E., Sawera, M., Havgaard, J. H., Fredholm, M., and Gorodkin, J. (2006). Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 16, 885–889. doi: 10.1101/gr.5226606
U.S. Food & Drug Administration (2016). Available online at: https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-spinal-muscular-atrophy (accessed May 11, 2021).
U.S. Food & Drug Administration (2020). Available online at: https://www.fda.gov/news-events/press-announcements/fda-approves-oral-treatment-spinal-muscular-atrophy (accessed May 11, 2021).
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H., and Bartel, D. P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550. doi: 10.1016/j.cell.2011.11.055
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R., and Johnson, R. (2018). Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 19, 535–548. doi: 10.1038/s41576-018-0017-y
Vittal, P., Pandya, S., Sharp, K., Berry-kravis, E., Zhou, L., Ouyang, B., et al. (2018). ASFMR1 splice variant. A predictor of fragile X-associated tremor/ataxia syndrome. Neurol. Genet. 4:e246. doi: 10.1212/NXG.0000000000000246
Wahlestedt, C. (2013). Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 12, 433–446. doi: 10.1038/nrd4018
Wang, S., Zhang, X., Guo, Y., Rong, H., and Liu, T. (2017). The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget 8, 24449–24456. doi: 10.18632/oncotarget.15511
Wang, X., Goodrich, K. J., Gooding, A. R., Youmans, D. T., Cech, T. R., Wang, X., et al. (2017). Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines article targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell 65, 1056.e5–1067.e5. doi: 10.1016/j.molcel.2017.02.003
Wang, Y., Zhao, X., Ju, W., Flory, M., Zhong, J., Jiang, S., et al. (2015). Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl. Psychiatry 5:e660. doi: 10.1038/tp.2015.144
Wang, Z.-F., Ursu, A., Childs-Disney, J. L., Guertler, R., Yang, W.-Y., Bernat, V., et al. (2019). The Hairpin Form of r(G(4)C(2))(exp) in c9ALS/FTD Is Repeat-Associated Non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol. 26, 179.e12–190.e12. doi: 10.1016/j.chembiol.2018.10.018
Washietl, S., Kellis, M., and Garber, M. (2014). Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 24, 616–628. doi: 10.1101/gr.165035.113
Watts, J. K., and Corey, D. R. (2012). Silencing disease genes in the laboratory and the clinic. J. Pathol. 226, 365–379. doi: 10.1002/path.2993
West, J. A., Davis, C. P., Sunwoo, H., Simon, M. D., Sadreyev, R. I., Wang, P. I., et al. (2014). The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802. doi: 10.1016/j.molcel.2014.07.012
Wilhelmus, M. M. M., Van Der Pol, S. M. A., Jansen, Q., Witte, M. E., Van Der Valk, P., Rozemuller, A. J. M., et al. (2011). Association of Parkinson disease-related protein PINK1 with Alzheimer disease and multiple sclerosis brain lesions. Free Radic. Biol. Med. 50, 469–476. doi: 10.1016/j.freeradbiomed.2010.11.033
Wilkinson, K. A., Merino, E. J., and Weeks, K. M. (2006). Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616. doi: 10.1038/nprot.2006.249
Wilusz, J. E., JnBaptiste, C. K., Lu, L. Y., Kuhn, C. D., Joshua-Tor, L., and Sharp, P. A. (2012). A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 26, 2392–2407. doi: 10.1101/gad.204438.112
Wolter, J. M., Mao, H., Fragola, G., Simon, J. M., Krantz, J. L., Bazick, H. O., et al. (2020). Cas9 gene therapy for Angelman syndrome traps Ube3a-ATS long non-coding RNA. Nature 587, 281–284. doi: 10.1038/s41586-020-2835-2
Woo, C. J., Maier, V. K., Davey, R., Brennan, J., Li, G., Brothers, J., et al. (2017). Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. PNAS 114, E1509–E1518. doi: 10.1073/pnas.1616521114
Wood, S. H., Craig, T., Li, Y., Merry, B., and de Magalhães, J. P. (2013). Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age 35, 763–776. doi: 10.1007/s11357-012-9410-1
Xia, D., Sui, R., and Zhang, Z. (2019). Administration of resveratrol improved Parkinson’s disease-like phenotype by suppressing apoptosis of neurons via modulating the MALAT1/miR-129/SNCA signaling pathway. J. Cell. Biochem. 120, 4942–4951. doi: 10.1002/jcb.27769
Xu, C. L., Sang, B., Liu, G. Z., Li, J. M., Zhang, X. D., Liu, L. X., et al. (2020). SENEBLOC, a long non-coding RNA suppresses senescence via p53-dependent and independent mechanisms. Nucleic Acids Res. 48, 3089–3102. doi: 10.1093/nar/gkaa063
Xu, D., Cai, Y., Tang, L., Han, X., Gao, F., Cao, H., et al. (2020). A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 10:1794. doi: 10.1038/s41598-020-58104-5
Yamanaka, Y., Faghihi, M. A., Magistri, M., Alvarez-Garcia, O., Lotz, M., and Wahlestedt, C. (2015). Antisense RNA Controls LRP1 sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 344, 1173–1178. doi: 10.1126/science.1249098
Yang, H., Shang, D., Xu, Y., Zhang, C., Feng, L., Sun, Z., et al. (2017). The LncRNA connectivity map: using LncRNA signatures to connect small molecules, LncRNAs, and diseases. Sci. Rep. 7, 1–6. doi: 10.1038/s41598-017-06897-3
Yao, R., Wang, Y., and Chen, L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551.
Yichin, L., Fallon, L., Lashuel, H. A., Liu, Z., and Lansbury, P. T. J. (2002). The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell 111, 209–218. doi: 10.1016/s0092-8674(02)01012-7
Ying, L., Chen, Y.-Y., Liu, H., Yao, C.-J., Zhu, X.-X., Chen, D.-J., et al. (2015). Association between ubiquitin carboxy-terminal hydrolase-L1 S18Y variant and risk of Parkinson’s disease: the impact of ethnicity and onset age. Neurol. Sci. 36, 179–188. doi: 10.1007/s10072-014-1987-y
Youse, H., Fatemeh, M., Ladan, M., Amir, M., and Aref, R. (2020). Long noncoding RNAs and exosomal lncRNAs: classi fi cation, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 39, 953–974. doi: 10.1038/s41388-019-1040-y
Yu, A. M., Choi, Y. H., and Tu, M. J. (2020). RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol. Rev. 72, 862–898. doi: 10.1124/pr.120.019554
Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., et al. (2003). The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327. doi: 10.1016/s0092-8674(03)00079-5
Zaslavsky, K., Zhang, W.-B., McCready, F. P., Rodrigues, D. C., Deneault, E., Loo, C., et al. (2019). SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nat. Neurosci. 22, 556–564. doi: 10.1038/s41593-019-0365-8
Zhang, P., Park, H.-J., Zhang, J., Junn, E., Andrews, R. J., Velagapudi, S. P., et al. (2020). Translation of the intrinsically disordered protein α-synuclein is inhibited by a small molecule targeting its structured mRNA. PNAS 117, 1457–1467. doi: 10.1073/pnas.1905057117
Zhang, Q.-S., Wang, Z.-H., Zhang, J.-L., Duan, Y.-L., Li, G.-F., and Zheng, D.-L. (2016). Beta-asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting α-synuclein protein expression. Biomed. Pharmacother. 83, 153–159. doi: 10.1016/j.biopha.2016.06.017
Zhang, X., Hamblin, M. H., and Yin, K. J. (2017). The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 14, 1705–1714. doi: 10.1080/15476286.2017.1358347
Zhang, Z., Chen, J., Zhu, Z., Zhu, Z., Liao, X., Wu, J., et al. (2021). CRISPR-Cas13-Mediated Knockdown of lncRNA-GACAT3 Inhibited Cell Proliferation and Motility, and Induced Apoptosis by Increasing p21, Bax, and E-cadherin expression in bladder cancer. Front. Mol. Biosci. 7:433. doi: 10.3389/fmolb.2020.627774
Zhao, L., Wang, J., Li, Y., Song, T., Wu, Y., Fang, S., et al. (2021). NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171. doi: 10.1093/nar/gkaa1046
Zheng, S., Chen, Y., Donahue, C. P., Wolfe, M. S., and Varani, G. (2009). Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone). Chem. Biol. 16, 557–566. doi: 10.1016/j.chembiol.2009.03.009
Zhu, S., Li, W., Liu, J., Chen, C. H., Liao, Q., Xu, P., et al. (2016). Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 34, 1279–1286. doi: 10.1038/nbt.3715
Zimmer-Bensch, G. (2019). Emerging roles of long non-coding RNAs as drivers of brain evolution. Cells 8:1399. doi: 10.3390/cells8111399
Zu, T., Liu, Y., Bañez-Coronel, M., Reid, T., Pletnikova, O., Lewis, J., et al. (2013). RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. PNAS 110, E4968–E4977. doi: 10.1073/pnas.1315438110
Zuccato, C., Belyaev, N., Conforti, P., Ooi, L., Tartari, M., Papadimou, E., et al. (2007). Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J. Neurosci. 27, 6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007
Zuccato, C., and Cattaneo, E. (2007). Role of brain-derived neurotrophic factor in Huntington’s disease. Prog. Neurobiol. 81, 294–330. doi: 10.1016/j.pneurobio.2007.01.003
Zucchelli, S., Cotella, D., Takahashi, H., Carrieri, C., Cimatti, L., Fasolo, F., et al. (2015). SINEUPs: a new class of natural and synthetic antisense long non-coding RNAs that activate translation. RNA Biol. 12, 771–779. doi: 10.1080/15476286.2015.1060395
Keywords: long non-coding RNAs, central nervous system, neuronal development, neurological disorders, gene regulation
Citation: Policarpo R, Sierksma A, De Strooper B and d’Ydewalle C (2021) From Junk to Function: LncRNAs in CNS Health and Disease. Front. Mol. Neurosci. 14:714768. doi: 10.3389/fnmol.2021.714768
Received: 25 May 2021; Accepted: 25 June 2021;
Published: 19 July 2021.
Edited by:
Inmaculada Segura, Ludwig Maximilian University of Munich, GermanyReviewed by:
Massimiliano Caiazzo, University of Naples Federico II, ItalyMasato Yano, Niigata University, Japan
Copyright © 2021 Policarpo, Sierksma, De Strooper and d’Ydewalle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Constantin d’Ydewalle, Y3Zhbm91dHJASVRTLkpOSi5jb20=
 Rafaela Policarpo
Rafaela Policarpo Annerieke Sierksma1,2
Annerieke Sierksma1,2 Constantin d’Ydewalle
Constantin d’Ydewalle