- 1Institute of Neuroscience, Lobachevsky University of Nizhny Novgorod, Nizhny Novgorod, Russia
- 2Charité Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Institute for Cell- and Neurobiology, Berlin, Germany
Transcriptional regulation is essential for the correct functioning of cells during development and in postnatal life. The basic Helix-loop-Helix (bHLH) superfamily of transcription factors is well conserved throughout evolution and plays critical roles in tissue development and tissue maintenance. A subgroup of this family, called neural lineage bHLH factors, is critical in the development and function of the central nervous system. In this review, we will focus on the function of one subgroup of neural lineage bHLH factors, the Neurod family. The Neurod family has four members: Neurod1, Neurod2, Neurod4, and Neurod6. Available evidence shows that these four factors are key during the development of the cerebral cortex but also in other regions of the central nervous system, such as the cerebellum, the brainstem, and the spinal cord. We will also discuss recent reports that link the dysfunction of these transcription factors to neurological disorders in humans.
Introduction
The interest to understand the molecular mechanisms that generate our central nervous system has never been greater, as the intensive work of clinicians, neurologists, and developmental biologists demonstrate that several naturally occurring neurological disorders originate from deficits impairing brain development in humans (Ross and Walsh, 2001; Subramanian et al., 2019). This can be particularly seen in disorders affecting the development of the cerebral cortex, which are frequently associated with seizures both in childhood and in adult life (Subramanian et al., 2019). Furthermore, cognitive disorders ranging from mild to severe intellectual disability and autism are also concomitant features of cortical neurodevelopmental disorders (Guerrini and Dobyns, 2014). The advent of novel and powerful human genetics is greatly contributing to the identification of rare and common disease-causing variants disrupting the normal development of the nervous system (Ku et al., 2010; McCarthy and MacArthur, 2017; Niemi et al., 2018; Momozawa and Mizukami, 2021). Many of these underlie the elementary mechanisms acting on neurogenesis, neuronal differentiation, fate acquisition, dendritogenesis, axonal navigation, and synapse formation (Cardoso et al., 2019; Wang et al., 2019; Parenti et al., 2020).
Development of the central nervous system in humans, as in many other species, is an elaborated process that begins during an early fetal stage, for instance in the third gestational week in humans or by embryonic day 11 in mice (Bayer and Altman, 2007). It initiates with the formation of the neural tube and the differentiation and specification of neural progenitor cells that, subsequently, lead to the genesis of differentiated neurons in a process called neurogenesis that culminates in the early postnatal life in humans, but can span throughout the adult life in other species, such as rodents (Altman and Das, 1965; Johnson, 2001; Bayer and Altman, 2007; Stiles and Jernigan, 2010; Silbereis et al., 2016; Sorrells et al., 2018; Buffalo et al., 2019; Petrik and Encinas, 2019). The specification of neural progenitor cells and their activation to self-renew and/or to differentiate in more committed progenitors and neurons is mediated by extrinsic and intrinsic molecular mechanisms (Götz and Sommer, 2005; Urbán and Guillemot, 2014; Götz et al., 2016; Oproescu et al., 2021). The intrinsic mechanisms that direct neural progenitor cell progression and differentiation rely on the coordinated function of multiple transcription factors that determine their identity and, simultaneously, the suppression of their progenitor cell programs (Schuurmans et al., 2004; Britz et al., 2006; Hevner et al., 2006; Davidson, 2010; Hodge and Hevner, 2011; Busskamp et al., 2014; Ware et al., 2016; Mall et al., 2017; Lee et al., 2019).
In the developing nervous system, proneural basic Helix-loop-Helix (bHLH) transcription factors are master regulators of cell proliferation and key in neuronal differentiation and specification (Dokucu et al., 1996; Sommer et al., 1996; Bertrand et al., 2002). Among these factors, the Neurod family stands as a critical regulator of neuronal progenitor cell differentiation and neuronal specification in the cerebral cortex, as well as in other regions of the nervous system such as the cerebellum, the brainstem, and the spinal cord. The Neurod family is composed of four members, Neurod1, Neurod2, Neurod4, and Neurod6.
In this review, we will discuss on the function of bHLH factors in neuronal development and particularly focus on the Neurod family in the development of cerebral cortex (including neuronal fate specification, dendritogenesis, and axonal navigation), as well as on the function of these factors in the development of other areas of the central nervous system. In addition, we will discuss Neurod disease-causing variants found in patients presenting with neurological disorders, such as Alzheimer’s disease.
bHLH Transcription Factors and The Neurod Family
The bHLH superfamily of transcription factors contains numerous genes crucial for the regulation of gene expression in most eukaryotic organisms. These factors are classified according to the similarities in their protein structure and the characteristic presence of a basic domain that directly binds to chromatin as well as a Helix-loop-Helix (HLH) domain that comprises of a non-conserved loop region connecting to alpha-helices (Chien et al., 1996; Bertrand et al., 2002). The bHLH domain was first identified by Murre and colleagues using early, but sophisticated, oligonucleotide screening procedures on lgt11 expression clones (Murre et al., 1989). The protein sequence characteristic of the bHLH domain consists of about 60–100 amino acids. bHLH factors are known to heterodimerize with other bHLH factors, using their non-conserved loop region, to form a functional DNA binding unit. Upon forming heterodimers, bHLH transcription factors are capable to bind to E-box motifs on chromatin, which display the consensus sequence CANNTG (Longo et al., 2008). The central “NN” and flanking nucleotides are believed to confer the DNA-binding specificity shown by bHLH proteins (Ellenberger et al., 1994; Bertrand et al., 2002).
The bHLH superfamily of transcription factors is well conserved throughout evolution and was first identified in animals, although recent investigations have started to reveal their presence and function in other organisms that range from yeast to plants (Murre et al., 1994; Zhang T. et al., 2018). Phylogenetic analysis of the bHLH superfamily using seven different species (human, mouse, rat, worm, fly, yeast, and plant Arabidopsis) has revealed over 600 members belonging to this family (Stevens et al., 2008). Unsurprisingly, the number of bHLH genes increases with the complexity of the organism, for instance, the smaller number of bHLH genes, 38, is found in Caenorhabditis elegans, around 58 in Drosophila melanogaster, 117 in the Mus musculus, and approximately 130 in Homo sapiens (Ledent et al., 2002; Skinner et al., 2010).
In 1989, Murrey and others first classified bHLH transcription factors according to their expression pattern into two classes: a class A (with ubiquitous expression) and a class B (with tissue-specific expression; Murre et al., 1989). This classification has been further expanded using large–scale phylogenetic analyses comparing the bHLH domains (Sun and Baltimore, 1991; Atchley and Fitch, 1997; Meredith and Johnson, 2000; Dennis et al., 2019). A more recent phylogenetic classification done by Skinner and colleagues has related bHLH factors into five distinct clades, in which clade A contains neural lineage genes such as Neurod, Neurog1, Ascl1, and Atoh1; or clade C that contains muscle-specific genes such as Myod1 or Myf5 (Skinner et al., 2010).
Neural lineage bHLH transcription factors participate in the regulation of cell survival, differentiation, migration, and fate specification during neural development and in postnatal life. These factors oftentimes have overlapping functions but can be further subdivided into: (i) proneural or determination factors (usually expressed in progenitor cells) and (ii) differentiation factors [predominantly expressed in postmitotic neurons (Bertrand et al., 2002)].
(i) Proneural factors. They represent a small subset of the neural lineage bHLH factors which are preferentially expressed in multipotent precursor cells. They control and direct progenitor cell decisions as well as the cellular fate choices to undergo glial or neuronal differentiation. An interesting trait of these transcription factors is their pioneering function to remodel chromatin and their capacity to reprogram non-neuronal differentiated cells into neurons (Wapinski et al., 2013, 2017; Pataskar et al., 2016; Guillemot and Hassan, 2017). Members of this group include the genes Neurod1, Neussrod4, Ascl1, Neurog1, and Neurog2.
(ii) Differentiation factors. These genes encompass most of the neural lineage bHLH transcription factors and are predominantly expressed by differentiated neurons, in which they regulate fate specification and neuronal identity maintenance. Members of this group include Neurod1, Neurod2, Neurod6, Bhlhe22.
The Neurod family contains four closely related proteins: Neurod1, Neurod2, Neurod4, and Neurod6. The expression pattern of these genes is highly overlapping but not identical in the developing cerebral cortex (Figure 1). Expression of these genes is abundantly present in the neuroepithelium of the dorsal telencephalon in early development and is sustained in the adult neocortex, hippocampus, and cerebellum (Schwab et al., 1998). The expression of Neurod1 can be first detected around embryonic day 12 in the dorsal ventricular zone (VZ) of mice (Bormuth et al., 2013). In the developing cerebral cortex, Neurod1 is also expressed by mitotic and early-postmitotic neuronal cells that reside in the subventricular zone (SVZ), which contains transit-amplifying progenitors that contribute to the generation of most of the excitatory neurons that form the mature cortex. In the postnatal life, Neurod1 expression is retained in the cerebral cortex, particularly in most excitatory pyramidal neurons that form the upper-most layers of the cortex (Lee et al., 1995; Bormuth et al., 2013; D’Amico et al., 2013). Neurod4 expression has been reported to be confined to the ventricular zone of the dorsal telencephalon during development and can be detected around embryonic day 12/13 in mice (Mattar et al., 2008). The other two members of the Neurod family, Neurod2 and Neurod6, display a highly overlapping expression pattern that appears in the mouse cerebral cortex around embryonic day 12 (Bormuth et al., 2013). Interestingly, both transcription factors are abundantly expressed by postmitotic pyramidal neurons during embryonic development, albeit their expression levels decline in the postnatal life. Neurod6 seems to be selectively expressed in a subset of pyramidal neurons, specifically those residing in the deeper layers of the adult mouse cortex; whereas Neurod2 is expressed by all cortical pyramidal neurons irrespectively of their laminar position (Bormuth et al., 2013).
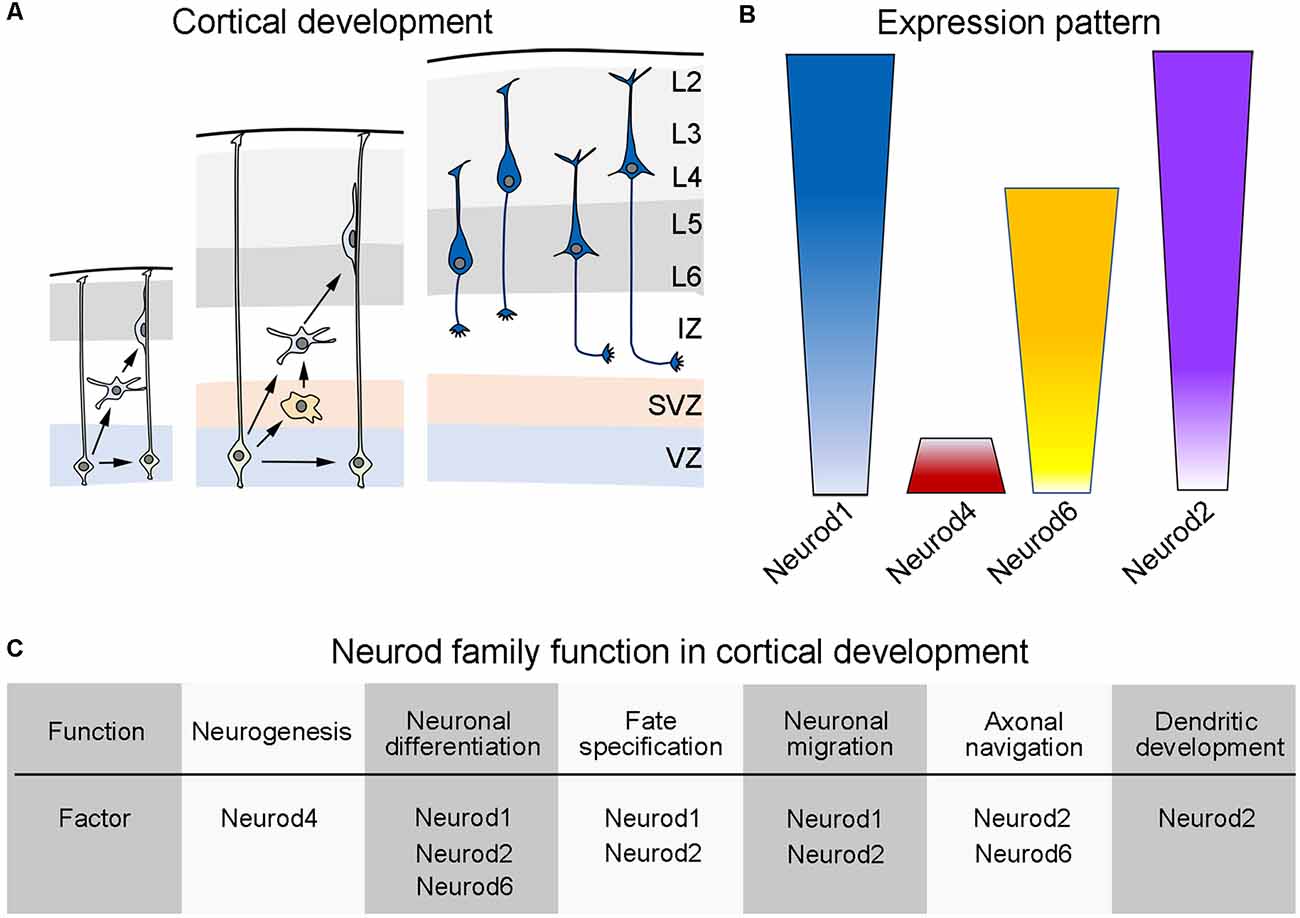
Figure 1. Developmental expression of the Neurod family during cortical development of mice. (A) Schematic representation of the cortical development in mice. (Left) During early cortical development, neural progenitor cells (light green, also called radial glial cells) located in the dorsal ventricular zone (VZ) make early decisions as to self-renew or differentiate into early born neurons (light blue). (Middle) As cortical development progress, neuronal progenitor cells increase their choices and can then self-renew or differentiate into more committed progenitors (light orange) that populate an emerging subventricular zone (SVZ) or into differentiated neurons (light blue). Upon differentiation, neurons radially migrate throughout an intermediate zone (IZ) in order to populate the developing cortical plate (gray areas), using radial glial fibers as a scaffold. (Right) According to the time of birth, cortical pyramidal neurons (dark blue) settle into their appropriate cortical layer and start the elaboration of dendritic trees and the elongation or their axonal processes (see text). (B) Schematic display of the expression pattern exhibited by the different members of the Neurod family during cortical development (see text). (C) Table summarizing the known functions of the different members of the Neurod family during cortical development (see text). Panel (A) is inspired from Guo et al. (2015).
Neurod Family in Cortical Development and Cortical Function
Pyramidal neurons (also known as cortical projection cells) are generated from progenitor cells located in the ventricular and subventricular zone of the dorsal telencephalon (see Figure 1; reviewed in Götz and Sommer, 2005; Elston and Fujita, 2014; Agirman et al., 2017). Soon after leaving the cell cycle, pyramidal neurons initiate a radial migration using the fibers of neighboring radial glial cells as a scaffold and cross an intermediate zone (IZ) on their way to reach their final destination within the developing cortex (reviewed in Rakic, 1995; Kriegstein and Noctor, 2004; He et al., 2015). After completing radial migration, pyramidal neurons settle in the cortical plate (CP) and undergo terminal differentiation (Gutierrez et al., 2004; Bianchi et al., 2013; Elston and Fujita, 2014). An interesting trait in cortical development is that the distinct projection neurons that form the mature cerebral cortex do not develop simultaneously but rather they are generated and migrate in a temporal order to populate the cortical plate in an inside-first and outside-last manner (reviewed in Angevine and Sidman, 1961; Rakic, 1974; Noctor et al., 2001; Buchsbaum and Cappello, 2019). This means that neurons residing in the deepest layers of the mature cerebral cortex are generated first during development and settle in the deepest part of the developing cortex. After pyramidal neurons form the deep layers of the cortex, superficially located projection neurons are generated and radially migrate to populate their corresponding upper layers (Rakic, 1974, 1995; Noctor et al., 2001; Kriegstein and Noctor, 2004; He et al., 2015). Once in the cortical plate, pyramidal neurons initiate dendritic arborizations and the projection of their axonal processes in a stereotyped manner, that is, they start with their axon outgrowth, fasciculation, pathfinding, and targeting of their appropriated neuronal partners (Figure 1).
bHLH genes cooperate to control transcriptional programs that select different aspects of neural progenitor cell biology and effectively determine neuron subtype identity. Proneural bHLH genes in the telencephalon, such as Neurog1 and Neurog2, activate transcriptional cascades of gene expression in progenitor cells that eventually lead to their terminal neuronal differentiation (Ge et al., 2006). Neurog1 and Neurog2 also contribute with the dorsalization of the telencephalon by suppressing the ventralization factor Ascl1 (Fode et al., 2000). The expression of Neurog 1 and Neurog2 is primarily restricted to the dorsal ventricular zone, although a few Neurog2 expressing cells can be observed outside this area (Ge et al., 2006). An important instructive function of Neurog2 is to promote a cortical neuron identity in differentiating cells of the dorsal telencephalon. Neurod4 is a known target of Neurog2 and dimerizes with it, forming Neurog2/Neurod4 heterodimers (Mattar et al., 2008). Neurog2/Neurod4 heterodimers accelerate the expression of particular transcriptional programs in the cortex that regulate neurogenesis. Mattar et al. (2008) have also shown that NeuroD4 and Neurog2 can independently act to regulate gene expression, albeit with a temporal delay. Similarly, Neurod4 can also form heterodimers with Neurog1 which are required for habenular neurogenesis. In the habenula, Neurod4 and Neurog1 depend on Pax6 expression downstream of Sonic hedgehog (Halluin et al., 2016).
Interestingly, the phosphorylation of Neurod4 limits its ability to drive neuronal differentiation during neurogenesis, which implies that post-transcriptional modifications finely tune the activity of bHLH transcription factors, such as Neurod4 (Hardwick and Philpott, 2015; Hardwick et al., 2019). In this context, a phospho-mutant Neurod4 increases its protein stability and enhances its chromatin binding when compared to wild-type Neurod4, which results in a transcriptional up-regulation of a wide range of target genes (Hardwick and Philpott, 2015). Lastly, Neurod4 has also been reported to be capable to reprogram human and mouse astrocytes, and when it is co-expressed with Insm1 it is capable of driving glutamatergic neuron maturation (Masserdotti et al., 2015).
In the developing neocortex, Neurod1 has been shown to promote terminal neuronal differentiation in progenitor cells, although there exists a hierarchy in the sequential activation of transcription factors that regulate the transition from precursor cells to differentiated pyramidal neurons (Hevner et al., 2001, 2006; Muzio et al., 2002a, b; Hodge et al., 2008; Hodge and Hevner, 2011). Indeed, the sequential expression of the transcription factors Pax6 → Neurog2 → Tbr2 → Neurod1 → Tbr1 correlates with the transition from primary progenitor cells into intermediate progenitors and ultimately into the generation of newborn glutamatergic pyramidal neurons. Outside the cerebral cortex, Neurod1 has also been shown to induce terminal neuronal differentiation. For instance, Boutin et al. (2010), using the olfactory periglomerular neuron lineage in vitro, showed that expression of Neurod1 alone suffices to induce terminal differentiation in olfactory periglomerular progenitor cells (Boutin et al., 2010). In vivo, Neurod1 overexpression in the periventricular region leads to the rapid appearance of postmitotic cells with morphological and molecular characteristics of mature neurons both in the subventricular zone and rostral migratory stream (Boutin et al., 2010). The function of Neurod1 in promoting terminal neuronal differentiation seems to be conserved in evolution and has been reported even in lower organisms like the worm C. teleta (Sur et al., 2017). Environmental enrichment also seems to induce Neurod1 expression in the forebrain and to enhance neuronal activity. For example, studies in the juvenile Atlantic salmon (Salmo salar) showed that environmental enrichment upregulates Neurod1 expression in their forebrain, which greatly improves their learning abilities (Salvanes et al., 2013). In mice, environmental enrichment leads to an increase in hippocampal volume and enhances dorsal-ventral differences in DNA methylation, including binding sites recognize by Neurod1, which seem to greatly promote adult neurogenesis (Zhang T.-Y. et al., 2018). In adult humans, NEUROD1 expression increases in the cerebral cortex after a traumatic injury, which might indicate a protective mechanism play by Neurod1 in the postnatal cerebral cortex (Zheng et al., 2013).
Neurog1 and Neurog2 play an important function in suppressing RhoA expression just as cortical progenitor cells are about to leave the cell cycle, and this suppression is maintained in postmitotic neurons by the direct action of Neurod1 (Ge et al., 2006). The suppression of RhoA is critical for the migration of pyramidal neurons into the cortical plate (Ge et al., 2006). On the way to the cortical plate, pyramidal neurons also require Reelin, a protein secreted by the Cajal-Retzius cells that locate in the superficial marginal zone of the developing cortex. Neurod2 has been shown to control pyramidal neuron migration and Reelin signaling by direct regulation of Cdk5r1, Lrp8 and the transcription factor Cux1, which in turn controls the differentiation of the upper layer (2/3 and 4) neurons (Bayam et al., 2015).
Neurod2 and Neurod6 are essential regulators of axonal navigation and axonal fasciculation in the mouse neocortex. For instance, the anterior commissure and the corpus callosum fiber tracts, which communicate the two cerebral hemispheres, are completely absent in Neurod2 and Neurod6 double mutant mice (Bormuth et al., 2013). Detailed inspections in Neurod2 and Neurod6 double mutant mice showed that callosal axons defasciculate in the subventricular zone during development and follow random trajectories into the ipsilateral cortex rather than growing toward the midline to contralaterally decussate (Bormuth et al., 2013). These axonal defects have been correlated with the dysregulation of the cell adhesion protein Cntn2 and the axonal receptor Robo1 (Bormuth et al., 2013). Furthermore, Neurod2 mutant mice also exhibit deficits in their thalamocortical projections to different cortical areas, such as the somatosensory barrel cortex (Ince-Dunn et al., 2006). Furthermore, the ablation of Neurod2 and Neurod6 also results in reduced numbers of functional glutamatergic synapses and, consequently, in a diminished excitatory cortical network (Bormuth et al., 2013).
The development of dendrites and synapses is a fundamental process in the establishment of neuronal polarity and connectivity. In this regard, Neurod2 has been shown to regulate the structural and functional maturation of the hippocampal mossy fiber synapses via the regulation of the synaptic scaffolding protein PSD95 (Wilke et al., 2012). Neurod1 and Neurod2 can abrogate GABAergic differentiation directed by Ascl1, a well-known bHLH transcription factor critical for GABAergic neuron development. Furthermore, the forced expression of Neurod2 in progenitor cells of the ventral telencephalon is sufficient to prevent their normal differentiation into GABAergic neurons (Roybon et al., 2010). In addition, Neurod2 regulates calcium signaling and homeostasis of mature neurons by controlling the expression of the Stim1 gene that encodes for an ER calcium sensor (Guner et al., 2017). Abnormal dendritic spine remodeling and turnover from postnatal day 30, and onwards, was reported in Neurod2 mutant mice, particularly in apical tuft dendrites of pyramidal layer 5 projection neurons of the somatosensory cortex (Runge et al., 2020a). Thus, Neurod2 is a nexus in the gene network that controls spine turnover in the postnatal cortex (Runge et al., 2020b).
Neural progenitor cells in the dorsal ventricular zone of the telencephalon also express Neurod6, some of which move into the subventricular zone and undergo multiple rounds of symmetrical and/or asymmetrical cell divisions to produce the set of neurons that reside in the upper cortical layers. Neurod6 positive progenitor cells in the subventricular zone are committed to generate glutamatergic neurons and might have evolved to expand the number of pyramidal neurons in the mammalian forebrain (Wu et al., 2005). Neurod6 has been shown to be central in the mitochondrial biogenesis during the early stages of neuronal differentiation. At these stages, Neurod6 appears to stimulate a maximal mitochondria mass accumulation which correlates with the onset of differentiation and lamellipodia formation in the axonal growth cone, as well as at the regions of axonal branching. This seems to be achieved by the transcriptional regulation of Neurod6 on genes encoding for cytoskeletal proteins, mitochondrial trafficking, regulators of membrane potential, and mitochondria chaperones (Uittenbogaard and Chiaramello, 2002, 2004, 2014; Kathleen Baxter et al., 2009; Uittenbogaard et al., 2010a, b; Baxter et al., 2012).
Neurod6 might also confer cellular tolerance to mitochondrial stressors and oxidative stress, which is critical to prevent neurodevelopmental disorders and neurodegenerative diseases, such as the autism spectrum disorder or Parkinson’s disease. The long–term consequences of early life stress on adult pathological states are associated with significant changes in DNA methylation and deregulation of miRNAs. miR-30a-5p regulates hundreds of downstream targets, including Neurod6, which may represent an important biological signature associated with the risk to develop psychiatric disorders as a consequence of exposure to early life adversities (Cattaneo et al., 2020). Neurod1 has also been shown to be critical for neuronal plasticity and increased levels of Neurod1 expression are triggered in the murine hippocampus after chronic or mild stress (Boulle et al., 2014). During neuronal differentiation, DNA demethylation-reprograming events are also associated with Neurod2 genome-wide binding (Hahn et al., 2019). In particular, it has been shown that highly methylated genomic regions in neuronal progenitor cells become demethylated after the onset of Neurod2 expression, and this coincides with the transition from proliferative progenitor state to differentiated neurons (Hahn et al., 2019). Furthermore, it has also been recently reported that maternal hyperglycemia increases H3K14 acetylation levels at Neurog1 and Neurod2 binding sites. Enhanced and premature expression of Neurog1 and Neurod2 eventually leads to an earlier differentiation of progenitor cells, which accelerates the genesis of newborn neurons in the cerebral cortex (Ji et al., 2019). Therefore, Neurod factors appear to display a pioneer function in remodeling chromatin. In keeping with this, Pataskar et al. (2016) recently demonstrated the pioneer function of Neurod1 in chromatin remodeling (Pataskar et al., 2016). The pioneer function of Neurod1 seems to be responsible for the potentiation of mineralocorticoid receptor-mediated transcription in the hippocampus, which has been suggested to act as a neuronal protective mechanism against the development of psychopathologies and, in particular, mood disorders (van Weert et al., 2019). In addition, Neurod1 has also been shown to reprogram striatal non-reactive astrocytes into neurons, albeit the reprogramming function of Neurod1 seems to be less efficient in cortical non-reactive astrocytes (Agirman et al., 2017; Liu et al., 2020).
Over two decades ago, Schwab et al. demonstrated that Neurod1 and Neurod6 are required for terminal neuronal differentiation in the hippocampus (Schwab et al., 1998, 2000). In Neurod1 and Neurod6 double mutant mice, the granule cells that are destined to populate the hippocampal dentate gyrus can be generated, but they fail to properly mature and display several phenotypes that include the lack of normal sodium currents, small dendritic arborization, and alterations of the entorhinal and commissural axonal projections (Schwab et al., 2000). Neurod1 has also been reported to play key functions in the survival and differentiation of newborn neurons in the subgranular and subventricular zones of the adult hippocampus (Gao et al., 2009). In keeping with this data, Roybon and others have also reported a key regulatory role for the Neurog2 and Neurod1 heterodimer complexes in controlling neuronal commitment and hippocampal neuroblast formation both in embryonic and in postnatal neurogenesis (Roybon et al., 2009). Specifically, Neurog2 and Neurod1 heterodimers control progenitor cell production and the amplification of granule neuron progenitors, but they are not required for the acquisition of their granule cell identity (Roybon et al., 2009). In the hippocampus, Neurod1 seems to induce the cell cycle exit of progenitor cells and to promote a rapid neuronal maturation of their progeny, maturation that seems to be reinforced by the expression of Neurod2 in differentiated hippocampal neurons (Roybon et al., 2009).
Neurod Family in The Development of The Posterior Neural Tube
The Neurod family also plays critical functions in the development of the posterior nervous system, that is the cerebellum, brainstem, and the spinal cord. After the onset of neural induction, the brainstem and spinal cord adopt their posterior identity by responding to instructive patterning signals generated from specialized cell centers located within the neural tube as wells as in surrounding tissues. These patterning centers produce fibroblast growth factors, bone morphogenetic proteins, retinoids, and Wnt proteins, which are capable to diffuse over long distances to carry out their instructive function (Doniach, 1995; Lumsden and Krumlauf, 1996; Stern, 2005). The most salient outcome of this early patterning is the generation of distinct anterior-posterior segments characterized by and overlapping as well as differential expression of transcription factors, predominantly members of the Hox family (Philippidou and Dasen, 2013). In the brainstem, for example, seven units called rhombomeres (rhombomere 1–7) develop, whereas four units (cervical, thoracic, lumbar, and sacral) are specified in the spinal cord of humans and mice (Trainor and Krumlauf, 2000). Each rhombomeric and spinal cord unit is further patterned along their dorsoventral axis by the action of diffusible morphogens emanating from a dorsal and a ventral group of specialized cells that act as organizers, the roof, and floor plate. These two organizers antagonistically act and exert their function via the secretion of Sonic hedgehog (by the floor plate) and bone morphogenic proteins and Wnt proteins (by the roof plate; Roelink et al., 1995; Liem et al., 1997; Ulloa and Marti, 2010). These signals diffuse from the roof and floor plate forming concentration gradients that differentially act upon progenitor cells located at different distances from the signal source and along the dorsal-ventral axis of the neural tube. It is thus the spatial position of progenitor cells within the neural tube that determines their response to morphogenic signals. Progenitor cells then respond to these signals, in a dose-dependent manner, and differentially express particular sets of transcription factors, among these the Neurod family and several other bHLH factors.
This is the case of the cerebellum that develops from the dorsal part of rhombomere 1 (known as the cerebellar anlage), which directly receives instructive signals from the roof plate (Millet et al., 1996; Broccoli et al., 1999; Chizhikov et al., 2006; Butts et al., 2014a, b). The cerebellar anlage contains two distinct germinal zones, the ventricular zone and the rhombic lip, which produce all GABAergic and glutamatergic cerebellar neurons, respectively (Hallonet et al., 1990; Alder et al., 1996; Wingate and Hatten, 1999; Hoshino et al., 2005; Millen et al., 2014; Yamada et al., 2014). Rhombic lip progenitor cells generate three distinct neuronal populations in a stereotyped temporal order. The first generation of glutamatergic neurons occurs between embryonic days 10.5 and 13.5 (in mice), and during this period deep cerebellar neurons are generated. A subsequent generation of glutamatergic neurons occurs between embryonic day 13.5 and birth, throughout this time external granular cell layer cells become specified, these cells are the precursors of the granule cells that develop in the early postnatal life. Lastly, unipolar brush cells become specified from the rhombic lip between embryonic day 15.5 and the first day of postnatal life (Ben-Arie et al., 1997; Gazit et al., 2004; Machold and Fishell, 2005; Englund et al., 2006; Fink et al., 2006; Machold et al., 2011; Yamada et al., 2014).
The generation of deep cerebellar neurons seems to largely depend on the action of the bHLH factor Olig3 (Lowenstein et al., 2021), whereas the production of external granular layer cells and unipolar brush cells is regulated by the bHLH factor Atoh1 (Ben-Arie et al., 1997; Gazit et al., 2004; Machold and Fishell, 2005; Wang et al., 2005). Neurod1 has long been known to play a critical role in the differentiation of granule cells, mainly in postnatal life (Gao et al., 2009). Deletion of Neurod1 greatly disrupts differentiation of these cells by prolonging the proliferation of their external granular layer cell progenitors and, in parallel, by inducing apoptosis in the developing cerebellum (Miyata et al., 1999; Pan et al., 2009). The extended proliferation and lethality of Neurod1-deficient external granular layer cell progenitors might result from the loss of the pioneer and proneural function that Neurod1 exerts in these progenitor cells by mediating, among other molecular cascades, the expression of different elements of the Notch signaling pathway (Pataskar et al., 2016). Indeed, expression of Neurod1 is known to drive terminal neuronal differentiation in external granular layer cell progenitor cells both in development and in the postnatal life of mammals and other vertebrates, such as in Xenopus (Cho and Tsai, 2004; Boutin et al., 2010; D’Amico et al., 2013; Butts et al., 2014a; Hanzel et al., 2019). Furthermore, a recent report shows that elevated levels of Neurod1 expression are sufficient to drive medulloblastoma cells into granule cell differentiation, which demonstrates that Neurod1 overrides oncogenic mutations present in medulloblastoma cells (Cheng et al., 2020). In spite of the substantial knowledge gained from the study of Neurod1 function in cerebellar development, less is known about the function of other members of the Neurod family in cerebellar development and cerebellar function. However, an early study using Neurod2 mutant mice showed that these mutants correctly develop until about the second week of postnatal life, after which they began exhibiting ataxia and the failure to thrive, which seems to be indicative of a cerebellar dysfunction (Olson et al., 2001). More recently, Pieper and colleagues (2019) reported on a critical function of Neurod2 in promoting survival of both granule cells and inhibitory neurons (particularly basket and stellate cells) that originate from the ventricular zone (Pieper et al., 2019). The analysis of Neurod2 mutant mice seems to indicate that Neurod2 might have an important function in cerebellar inhibitory neuron function, as well as in the axonogenesis and synaptic formation of inhibitory cerebellar neurons onto Purkinje cells (Pieper et al., 2019).
Work over the last two decades has also shown the great influence of the Neurod family in other regions of the posterior nervous system, which include the midbrain, the hindbrain, and the spinal cord. Progenitor cells in the ventral midbrain express high levels of Neurod1, and the combinatorial expression of Neurod1 with other bHLH factors sub-specifies different neuronal populations emanating from this area, some of which retain Neurod1 expression (Park et al., 2006; Arimura et al., 2019; Ásgrímsdóttir and Arenas, 2020; Poulin et al., 2020). Furthermore, Neurod1 and Neurod6 have been recently reported to have a critical function in the development of particular dopaminergic midbrain neurons (Khan et al., 2017). Specifically, Khan and colleagues analyzed Neurod6 and Neurod1 double mutant mice, and found that these genes are required for the survival of dopaminergic midbrain neurons located in the ventral tegmental area, particularly those that project to the intermediate and dorsal regions of the septum (Khan et al., 2017).
Unlike the neurogenic function that the Neurod family has in the developing cortex, cerebellum, and the midbrain; in the hindbrain and spinal cord Neurod1, Neurod2 and Neurod6 seem to mainly regulate the correct specification of discrete subpopulations of inhibitory interneurons (Hernandez-Miranda et al., 2017). In the hindbrain and spinal cord, Ptf1a-expressing progenitor cells generate all inhibitory neurons, which are known to co-express the homeodomain proteins Lbx1, Pax2, and Lhx1/5. Interestingly, these inhibitory interneurons do not uniformly maintain the expression of these transcription factors during their maturation and greatly vary in their expression, indicating that differential expression of such factors might reflect distinct subpopulations of inhibitory neurons (Pillai et al., 2007). Indeed, available evidence illustrates that Neurod1, Neurod2, and Neurod6 secure the specification of dynorphin+ and galanin+ inhibitory interneurons, whereas Lhx1/5 instruct a NPY+ inhibitory fate (Bröhl et al., 2008).
Neurod Genes in Human Neurological Disorders
The first report of human patients with a mutation on a gene of the Neurod family came in 2010 by Rubio-Cabezas and others (Rubio-Cabezas et al., 2010). In this study, the authors reported on two distinct homozygous mutations in NEUROD1 that were found in two unrelated probands diagnosed with permanent neonatal diabetes and neurological abnormalities. The identified mutations correspond to frameshift mutations that predictably generate truncated proteins, without affecting their DNA-binding domain. Neurologically, both patients presented with learning difficulties, cerebellar hypoplasia, profound sensorineural deafness, and visual impairment due to severe myopia and retinal dystrophy. Thus, the deficits observed in NEUROD1-deficient patients resemble those seen in Neurod1 mutant mice, which include pronounced cortical, cerebellar, brainstem, and spinal cord impairments (see above). More recently, Sega and colleagues found de novo mutations in NEUROD2 in two unrelated children diagnosed with early infantile epileptic encephalopathy and developmental delay (Sega et al., 2019). In keeping with this, an early onset of epilepsy has also been described in Neurod2 deficient mice (Chen et al., 2016). In this context, Chen et al. suggested that Neurod2 tightly controls the inhibition/excitation balance of neuronal transmission in the mature cortex. Furthermore, deficiencies of Neurod2 function in the mouse brain cause a decrease in the cell-intrinsic excitability of excitatory pyramidal neurons (Chen et al., 2016). There are two transcriptional targets of Neurod2 that may contribute to these processes: gastrin-releasing peptide (GRP) and the small conductance, calcium-activated potassium channel, Sk2 (Kcnn2). The expression of both genes is greatly decreased in Neurod2 mutant mice (Chen et al., 2016). Very recently, Runge et al. (2020b) described seven families with pathogenic NEUROD2 mutations causing a variety of neurological disorders, such as autism spectrum disorders, intellectual disability, and speech delay. The authors of this study also suggest that behavioral deficits in social behavior, which are reminiscent of autistic disorders, can be found in Neurod2 mutant mice. In addition, Spellman et al. identified a direct association of NEUROD2 gene polymorphisms with changes in cognitive functions present in schizophrenic patients treated with antipsychotic drugs (Spellmann et al., 2017). It has also been shown that the lateral and basolateral amygdala nuclei fail to form in Neurod2 mutant mice, and that these mice display deficits in emotional learning (Lin et al., 2005). In particular, Lin et al., found that Neurod2 is required for amygdala development and the regulation of the AMPA receptor, the γ subunit of the GABAA receptor, and the gene Ulip1, which are all involved in emotional learning (Lin et al., 2005).
The most salient phenotype arising from the loss of NEUROD1 in humans is epilepsy. In mice, the ablation of Neurod1 produces an epileptogenic phenotype associated with a malformation of the hippocampal dentate granule cell layer, which seems to result from an excessive cell death of the neurons forming this layer (Liu et al., 2020). Impaired neurogenesis and decreased expression of NEUROD1/Neurod1 have also been demonstrated in the hippocampus of the Huntington’s disease R6/2 mouse model and in differentiated neural cultures derived from Huntington’s disease patients (Fedele et al., 2011). NEUROD6 has been recently identified as a possible biomarker for the diagnosis of Alzheimer’s disease. Indeed, low expression levels of NEUROD6/Neurod6 have been detected in postmortem Alzheimer’s patients and in Alzheimer’s mouse models using RNA sequencing datasets, microarray datasets, and meta-analysis (Hokama et al., 2014; Satoh et al., 2014; Li et al., 2015).
Despite the fact that NEUROD1 expression levels have been reported not to be significantly changed in Alzheimer’s patients (Satoh et al., 2014), its overexpression into hippocampal progenitor cells increases dendritic spine density of hippocampal newborn neurons and results in a great improvement in spatial memory in the Alzheimer’s disease mouse model APPxPS1 (Richetin et al., 2015). Further studies by Richetin and others have also shown that Neurod1 promotes spinogenesis and mitochondrial availability at the vicinity of mature spines, and that this improves the integration and survival of adult-generated hippocampal neurons, which are severely impaired in the APPxPS1 mouse model (Richetin et al., 2017). These results provide a potential therapeutic approach to patients affected with Alzheimer’s disease. The half-life of Neurod1 can be increased by blocking its ubiquitin-dependent proteasomal degradation, which enhances the transcriptional programs mediated by Neurod1 during neuronal differentiation, but also those involved in neuronal maturation and synaptic transmission (de Wilde et al., 2016; Lee et al., 2020; Pomeshchik et al., 2020). Neurod1 has also been used to successfully reprogram reactive glial cells functional cortical neurons in stab-injured or Alzheimer’s disease mouse models and in adult non-human primates after ischemic stroke, which again offers the possibility to develop new therapeutical approaches for patients affected with Alzheimer’s disease (Guo et al., 2014; Ge et al., 2020).
Conclusion
The bHLH superfamily of transcription factors is well conserved throughout evolution and plays critical roles in tissue development and tissue maintenance. Whereas many bHLH transcription factors display ubiquitous expression, a small fraction of them has a tissue-specific expression. The question of how different members of this superfamily were selected to carry out common and divergent cellular functions remains to be elucidated. In the developing nervous system, the subfamily of neural lineage bHLH transcription factors regulates a variety of biological functions that range from progenitor cell proliferation and survival to neuronal differentiation, neuronal migration, fate specification, axonal navigation, dendritic elongation, and synaptic formation. Some members of this subfamily (called proneural, including Ascl1, Neurog1, Neurog2, Neurod1, and Neurod4) have been shown to be able to remodel chromatin and induce neuronal differentiation in progenitor cells, but they are also capable of reprogramming differentiated non-neuronal cells into neurons (Castro et al., 2011; Wapinski et al., 2013, 2017; Chanda et al., 2014; Treutlein et al., 2016; Rao et al., 2021). While Ascl1, Neurog1, Neurog2, and Neurod4 are predominantly expressed in progenitor cells, other factors like Neurod1, Neurod2, and Neurod6 are expressed both in progenitors and retained in postmitotic neurons. This raises the question of whether the function of these factors differs in progenitor cells and in differentiated neurons. Future research may elucidate whether post-transcriptional regulations, such as phosphorylation, on Neurod1, Neurod2, and Neurod6 account for their functional restriction at different points in the life of a neuron. Recent work on Ascl1 (in neurons) or Myod1 (in muscle cells) has shown that these transcription factors have an oscillatory expression which accounts for the proliferation of progenitors cells, whereas the sustained expression of these factors drives cell differentiation (Imayoshi et al., 2013; Vasconcelos and Castro, 2014; Lahmann et al., 2019; Sueda et al., 2019; Zhang et al., 2021), whether this oscillatory behavior is common for all bHLH transcription factors is unknown. However, the Neurod family offers the possibility to deepen into the expression dynamics of bHLH factors as they are expressed both in progenitors and in their progeny. An early diagnosis of neurological diseases is central in their management. Recent discoveries suggest that the expression of distinct members of the NEUROD family could serve as biomarkers at the onset of various neurological diseases, such as Alzheimer’s disease, and also serve in the development of patient-oriented gene therapies.
Author Contributions
ST, VT, and LH-M wrote the original draft and reviewed the literature. ST, VT, and LH-M revised the original draft. LH-M edited the final draft. All authors contributed to the article and approved the submitted final version.
Funding
This research work was funded by Russian Science Foundation, grant no. 19-14-00345 to VT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agirman, G., Broix, L., and Nguyen, L. (2017). Cerebral cortex development: an outside-in perspective. FEBS Lett. 591, 3978–3992. doi: 10.1002/1873-3468.12924
Alder, J., Cho, N. K., and Hatten, M. E. (1996). Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron 17, 389–399. doi: 10.1016/s0896-6273(00)80172-5
Altman, J., and Das, G. D. (1965). Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335. doi: 10.1002/cne.901240303
Angevine, J. B. Jr., and Sidman, R. L. (1961). Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766–768. doi: 10.1038/192766b0
Arimura, N., Dewa, K. I., Okada, M., Yanagawa, Y., Taya, S. I., Hoshino, M., et al. (2019). Comprehensive and cell-type-based characterization of the dorsal midbrain during development. Genes Cells 24, 41–59. doi: 10.1111/gtc.12656
Ásgrímsdóttir, E. S., and Arenas, E. (2020). Midbrain dopaminergic neuron development at the single cell level: in vivo and in stem cells. Front. Cell. Dev. Biol. 8:463. doi: 10.3389/fcell.2020.00463
Atchley, W. R., and Fitch, W. M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U S A 94, 5172–5176. doi: 10.1073/pnas.94.10.5172
Baxter, K. K., Uittenbogaard, M., and Chiaramello, A. (2012). The neurogenic basic helix-loop-helix transcription factor NeuroD6 enhances mitochondrial biogenesis and bioenergetics to confer tolerance of neuronal PC12-NeuroD6 cells to the mitochondrial stressor rotenone. Exp. Cell Res. 318, 2200–2214. doi: 10.1016/j.yexcr.2012.07.004
Bayam, E., Sahin, G. S., Guzelsoy, G., Guner, G., Kabakcioglu, A., Ince-Dunn, G., et al. (2015). Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation. BMC Genomics 16:681. doi: 10.1186/s12864-015-1882-9
Bayer, S. A., and Altman, J. (2007). The Human Brain During the Early First Trimester, 1st Edn. Boca Raton, FL: CRC Press.
Ben-Arie, N., Bellen, H. J., Armstrong, D. L., McCall, A. E., Gordadze, P. R., Guo, Q., et al. (1997). Math1 is essential for genesis of cerebellar granule neurons. Nature 390, 169–172. doi: 10.1038/36579
Bertrand, N., Castro, D. S., and Guillemot, F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. doi: 10.1038/nrn874
Bianchi, S., Stimpson, C. D., Duka, T., Larsen, M. D., Janssen, W. G., Collins, Z., et al. (2013). Synaptogenesis and development of pyramidal neuron dendritic morphology in the chimpanzee neocortex resembles humans. Proc. Natl. Acad. Sci. U S A 110, 10395–10401. doi: 10.1073/pnas.1301224110
Bormuth, I., Yan, K., Yonemasu, T., Gummert, M., Zhang, M., Wichert, S., et al. (2013). Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J. Neurosci. 33, 641–651. doi: 10.1523/JNEUROSCI.0899-12.2013
Boulle, F., Massart, R., Stragier, E., Païzanis, E., Zaidan, L., Marday, S., et al. (2014). Hippocampal and behavioral dysfunctions in a mouse model of environmental stress: normalization by agomelatine. Transl. Psychiatry 4:e485. doi: 10.1038/tp.2014.125
Boutin, C., Hardt, O., de Chevigny, A., Coré, N., Goebbels, S., Seidenfaden, R., et al. (2010). NeuroD1 induces terminal neuronal differentiation in olfactory neurogenesis. Proc. Natl. Acad. Sci. U S A 107, 1201–1206. doi: 10.1073/pnas.0909015107
Britz, O., Mattar, P., Nguyen, L., Langevin, L. M., Zimmer, C., Alam, S., et al. (2006). A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex 16, i138–i151. doi: 10.1093/cercor/bhj168
Broccoli, V., Boncinelli, E., and Wurst, W. (1999). The caudal limit of Otx2 expression positions the isthmic organizer. Nature 401, 164–168. doi: 10.1038/43670
Bröhl, D., Strehle, M., Wende, H., Hori, K., Bormuth, I., Nave, K. A., et al. (2008). A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev. Biol. 322, 381–393. doi: 10.1016/j.ydbio.2008.08.002
Buchsbaum, I. Y., and Cappello, S. (2019). Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development 146:dev163766. doi: 10.1242/dev.163766
Buffalo, E. A., Movshon, J. A., and Wurtz, R. H. (2019). From basic brain research to treating human brain disorders. Proc. Natl. Acad. Sci. U S A 116, 26167–26172. doi: 10.1073/pnas.1919895116
Busskamp, V., Lewis, N. E., Guye, P., Ng, A. H., Shipman, S. L., Byrne, S. M., et al. (2014). Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 10:760. doi: 10.15252/msb.20145508
Butts, T., Green, M. J., and Wingate, R. J. (2014a). Development of the cerebellum: simple steps to make a ‘little brain’. Development 141, 4031–4041. doi: 10.1242/dev.106559
Butts, T., Hanzel, M., and Wingate, R. J. (2014b). Transit amplification in the amniote cerebellum evolved via a heterochronic shift in NeuroD1 expression. Development 141, 2791–2795. doi: 10.1242/dev.101758
Cardoso, A. R., Lopes-Marques, M., Silva, R. M., Serrano, C., Amorim, A., Prata, M. J., et al. (2019). Essential genetic findings in neurodevelopmental disorders. Hum. Genomics 13:31. doi: 10.1186/s40246-019-0216-4
Castro, D. S., Martynoga, B., Parras, C., Ramesh, V., Pacary, E., Johnston, C., et al. (2011). A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 25, 930–945. doi: 10.1101/gad.627811
Cattaneo, A., Suderman, M., Cattane, N., Mazzelli, M., Begni, V., Maj, C., et al. (2020). Long-term effects of stress early in life on microRNA-30a and its network: preventive effects of lurasidone and potential implications for depression vulnerability. Neurobiol. Stress 13:100271. doi: 10.1016/j.ynstr.2020.100271
Chanda, S., Ang, C. E., Davila, J., Pak, C., Mall, M., Lee, Q. Y., et al. (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3, 282–296. doi: 10.1016/j.stemcr.2014.05.020
Chen, F., Moran, J. T., Zhang, Y., Ates, K. M., Yu, D., Schrader, L. A., et al. (2016). The transcription factor NeuroD2 coordinates synaptic innervation and cell intrinsic properties to control excitability of cortical pyramidal neurons. J. Physiol. 594, 3729–3744. doi: 10.1113/JP271953
Cheng, Y., Liao, S., Xu, G., Hu, J., Guo, D., Du, F., et al. (2020). NeuroD1 dictates tumor cell differentiation in medulloblastoma. Cell Rep. 31:107782. doi: 10.1016/j.celrep.2020.107782
Chien, C. T., Hsiao, C. D., Jan, L. Y., and Jan, Y. N. (1996). Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl. Acad. Sci. U S A 93, 13239–13244. doi: 10.1073/pnas.93.23.13239
Chizhikov, V. V., Lindgren, A. G., Currle, D. S., Rose, M. F., Monuki, E. S., Millen, K. J., et al. (2006). The roof plate regulates cerebellar cell-type specification and proliferation. Development 133, 2793–2804. doi: 10.1242/dev.02441
Cho, J.-H., and Tsai, M.-J. (2004). The role of BETA2/NeuroD1 in the development of the nervous system. Mol. Neurobiol. 30, 35–47. doi: 10.1385/MN:30:1:035
D’Amico, L. A., Boujard, D., and Coumailleau, P. (2013). The neurogenic factor NeuroD1 is expressed in post-mitotic cells during juvenile and adult Xenopus neurogenesis and not in progenitor or radial glial cells. PLoS One 8:e66487. doi: 10.1371/journal.pone.0066487
Davidson, E. H. (2010). Emerging properties of animal gene regulatory networks. Nature 468, 911–920. doi: 10.1038/nature09645
de Wilde, M. C., Overk, C. R., Sijben, J. W., and Masliah, E. (2016). Meta-analysis of synaptic pathology in Alzheimer’s disease reveals selective molecular vesicular machinery vulnerability. Alzheimers Dement. 12, 633–644. doi: 10.1016/j.jalz.2015.12.005
Dennis, D. J., Han, S., and Schuurmans, C. (2019). bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 1705, 48–65. doi: 10.1016/j.brainres.2018.03.013
Dokucu, M. E., Zipursky, S. L., and Cagan, R. L. (1996). Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 122, 4139–4147. doi: 10.1242/dev.122.12.4139
Doniach, T. (1995). Basic FGF as an inducer of anteroposterior neural pattern. Cell 83, 1067–1070. doi: 10.1016/0092-8674(95)90133-7
Ellenberger, T., Fass, D., Arnaud, M., and Harrison, S. C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980. doi: 10.1101/gad.8.8.970
Elston, G. N., and Fujita, I. (2014). Pyramidal cell development: postnatal spinogenesis, dendritic growth, axon growth, and electrophysiology. Front. Neuroanat. 8:78. doi: 10.3389/fnana.2014.00078
Englund, C., Kowalczyk, T., Daza, R. A., Dagan, A., Lau, C., Rose, M. F., et al. (2006). Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J. Neurosci. 26, 9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006
Fedele, V., Roybon, L., Nordstrom, U., Li, J. Y., and Brundin, P. (2011). Neurogenesis in the R6/2 mouse model of Huntington’s disease is impaired at the level of NeuroD1. Neuroscience 173, 76–81. doi: 10.1016/j.neuroscience.2010.08.022
Fink, A. J., Englund, C., Daza, R. A., Pham, D., Lau, C., Nivison, M., et al. (2006). Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J. Neurosci. 26, 3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006
Fode, C., Ma, Q., Casarosa, S., Ang, S. L., Anderson, D. J., Guillemot, F., et al. (2000). A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14, 67–80. doi: 10.1101/gad.14.1.67
Gao, Z., Ure, K., Ables, J. L., Lagace, D. C., Nave, K. A., Goebbels, S., et al. (2009). Neurod1 is essential for the survival and maturation of adult-born neurons. Nat. Neurosci. 12, 1090–1092. doi: 10.1038/nn.2385
Gazit, R., Krizhanovsky, V., and Ben-Arie, N. (2004). Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway. Development 131, 903–913. doi: 10.1242/dev.00982
Ge, W., He, F., Kim, K. J., Blanchi, B., Coskun, V., Nguyen, L., et al. (2006). Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U S A 103, 1319–1324. doi: 10.1073/pnas.0510419103
Ge, L. J., Yang, F. H., Li, W., Wang, T., Lin, Y., Feng, J., et al. (2020). in vivo neuroregeneration to treat ischemic stroke through NeuroD1 AAV-based gene therapy in adult non-human primates. Front. Cell. Dev. Biol. 8:590008. doi: 10.3389/fcell.2020.590008
Götz, M., Nakafuku, M., and Petrik, D. (2016). Neurogenesis in the developing and adult brain-similarities and key differences. Cold Spring Harb. Perspect. Biol. 8:a018853. doi: 10.1101/cshperspect.a018853
Götz, M., and Sommer, L. (2005). Cortical development: the art of generating cell diversity. Development 132, 3327–3332. doi: 10.1242/dev.01931
Guerrini, R., and Dobyns, W. B. (2014). Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 13, 710–726. doi: 10.1016/S1474-4422(14)70040-7
Guillemot, F., and Hassan, B. A. (2017). Beyond proneural: emerging functions and regulations of proneural proteins. Curr. Opin. Neurobiol. 42, 93–101. doi: 10.1016/j.conb.2016.11.011
Guner, G., Guzelsoy, G., Isleyen, F. S., Sahin, G. S., Akkaya, C., Bayam, E., et al. (2017). NEUROD2 regulates Stim1 expression and store-operated calcium entry in cortical neurons. eNeuro 4:ENEURO.0255-16.2017. doi: 10.1523/ENEURO.0255-16.2017
Guo, J., Higginbotham, H., Li, J., Nichols, J., Hirt, J., Ghukasyan, V., et al. (2015). Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 6:7857. doi: 10.1038/ncomms8857
Guo, Z., Zhang, L., Wu, Z., Chen, Y., Wang, F., Chen, G., et al. (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14, 188–202. doi: 10.1016/j.stem.2013.12.001
Gutierrez, H., Dolcet, X., Tolcos, M., and Davies, A. (2004). HGF regulates the development of cortical pyramidal dendrites. Development 131, 3717–3726. doi: 10.1242/dev.01209
Hahn, M. A., Jin, S.-G., Li, A. X., Liu, J., Huang, Z., Wu, X., et al. (2019). Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation. Sci. Adv. 5:eaax0080. doi: 10.1126/sciadv.aax0080
Hallonet, M. E., Teillet, M. A., and Le Douarin, N. M. (1990). A new approach to the development of the cerebellum provided by the quail-chick marker system. Development 108, 19–31. doi: 10.1242/dev.108.1.19
Halluin, C., Madelaine, R., Naye, F., Peers, B., Roussigne, M., Blader, P., et al. (2016). Habenular neurogenesis in zebrafish is regulated by a Hedgehog, Pax6 proneural gene cascade. PLoS One 11:e0158210. doi: 10.1371/journal.pone.0158210
Hanzel, M., Rook, V., and Wingate, R. J. T. (2019). Mitotic granule cell precursors undergo highly dynamic morphological transitions throughout the external germinal layer of the chick cerebellum. Sci. Rep. 9:15218. doi: 10.1038/s41598-019-51532-y
Hardwick, L. J. A., Davies, J. D., and Philpott, A. (2019). Multi-site phosphorylation controls the neurogenic and myogenic activity of E47. Biochem. Biophys. Res. Commun. 511, 111–116. doi: 10.1016/j.bbrc.2019.02.045
Hardwick, L. J., and Philpott, A. (2015). Multi-site phosphorylation regulates NeuroD4 activity during primary neurogenesis: a conserved mechanism amongst proneural proteins. Neural. Dev. 10:15. doi: 10.1186/s13064-015-0044-8
He, S., Li, Z., Ge, S., Yu, Y. C., and Shi, S. H. (2015). Inside-out radial migration facilitates lineage-dependent neocortical microcircuit assembly. Neuron 86, 1159–1166. doi: 10.1016/j.neuron.2015.05.002
Hernandez-Miranda, L. R., Müller, T., and Birchmeier, C. (2017). The dorsal spinal cord and hindbrain: from developmental mechanisms to functional circuits. Dev. Biol. 432, 34–42. doi: 10.1016/j.ydbio.2016.10.008
Hevner, R. F., Hodge, R. D., Daza, R. A., and Englund, C. (2006). Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci. Res. 55, 223–233. doi: 10.1016/j.neures.2006.03.004
Hevner, R. F., Shi, L., Justice, N., Hsueh, Y., Sheng, M., Smiga, S., et al. (2001). Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366. doi: 10.1016/s0896-6273(01)00211-2
Hodge, R. D., and Hevner, R. F. (2011). Expression and actions of transcription factors in adult hippocampal neurogenesis. Dev. Neurobiol. 71, 680–689. doi: 10.1002/dneu.20882
Hodge, R. D., Kowalczyk, T. D., Wolf, S. A., Encinas, J. M., Rippey, C., Enikolopov, G., et al. (2008). Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J. Neurosci. 28, 3707–3717. doi: 10.1523/JNEUROSCI.4280-07.2008
Hokama, M., Oka, S., Leon, J., Ninomiya, T., Honda, H., Sasaki, K., et al. (2014). Altered expression of diabetes-related genes in Alzheimer’s disease brains: the Hisayama study. Cereb. Cortex 24, 2476–2488. doi: 10.1093/cercor/bht101
Hoshino, M., Nakamura, S., Mori, K., Kawauchi, T., Terao, M., Nishimura, Y. V., et al. (2005). Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47, 201–213. doi: 10.1016/j.neuron.2005.06.007
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208. doi: 10.1126/science.1242366
Ince-Dunn, G., Hall, B. J., Hu, S.-C., Ripley, B., Huganir, R. L., Olson, J. M., et al. (2006). Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron 49, 683–695. doi: 10.1016/j.neuron.2006.01.031
Ji, S., Zhou, W., Li, X., Liu, S., Wang, F., Li, X., et al. (2019). Maternal hyperglycemia disturbs neocortical neurogenesis via epigenetic regulation in C57BL/6J mice. Cell Death Dis. 10:211. doi: 10.1038/s41419-019-1438-z
Johnson, M. H. (2001). Functional brain development in humans. Nat. Rev. Neurosci. 2, 475–483. doi: 10.1038/35081509
Kathleen Baxter, K., Uittenbogaard, M., Yoon, J., and Chiaramello, A. (2009). The neurogenic basic helix-loop-helix transcription factor NeuroD6 concomitantly increases mitochondrial mass and regulates cytoskeletal organization in the early stages of neuronal differentiation. ASN Neuro 1:e00016. doi: 10.1042/AN20090036
Khan, S., Stott, S. R., Chabrat, A., Truckenbrodt, A. M., Spencer-Dene, B., Nave, K. A., et al. (2017). Survival of a novel subset of midbrain dopaminergic neurons projecting to the lateral septum is dependent on NeuroD proteins. J. Neurosci. 37, 2305–2316. doi: 10.1523/JNEUROSCI.2414-16.2016
Kriegstein, A. R., and Noctor, S. C. (2004). Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 27, 392–399. doi: 10.1016/j.tins.2004.05.001
Ku, C. S., Loy, E. Y., Salim, A., Pawitan, Y., and Chia, K. S. (2010). The discovery of human genetic variations and their use as disease markers: past, present and future. J. Hum. Genet. 55, 403–415. doi: 10.1038/jhg.2010.55
Lahmann, I., Bröhl, D., Zyrianova, T., Isomura, A., Czajkowski, M. T., Kapoor, V., et al. (2019). Oscillations of MyoD and Hes1 proteins regulate the maintenance of activated muscle stem cells. Genes Dev. 33, 524–535. doi: 10.1101/gad.322818.118
Ledent, V., Paquet, O., and Vervoort, M. (2002). Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 3:RESEARCH0030. doi: 10.1186/gb-2002-3-6-research0030
Lee, T.-Y., Cho, I.-S., Bashyal, N., Naya, F. J., Tsai, M. J., Yoon, J. S., et al. (2020). ERK regulates NeuroD1-mediated neurite outgrowth via proteasomal degradation. Exp. Neurobiol. 29, 189–206. doi: 10.5607/en20021
Lee, J. E., Hollenberg, S. M., Snider, L., Turner, D. L., Lipnick, N., Weintraub, H., et al. (1995). Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268, 836–844. doi: 10.1126/science.7754368
Lee, J., Taylor, C. A., Barnes, K. M., Shen, A., Stewart, E. V., Chen, A., et al. (2019). A Myt1 family transcription factor defines neuronal fate by repressing non-neuronal genes. eLife 8:e46703. doi: 10.7554/eLife.46703
Li, X., Long, J., He, T., Belshaw, R., and Scott, J. (2015). Integrated genomic approaches identify major pathways and upstream regulators in late onset Alzheimer’s disease. Sci. Rep. 5:12393. doi: 10.1038/srep12393
Liem, K. F. Jr., and Tremml, G., and Jessell, T. M. (1997). A role for the roof plate and its resident TGFbeta-related proteins in neuronal patterning in the dorsal spinal cord. Cell 91, 127–138. doi: 10.1016/s0092-8674(01)80015-5
Lin, C.-H., Hansen, S., Wang, Z., Storm, D. R., Tapscott, S. J., Olson, J. M., et al. (2005). The dosage of the neuroD2 transcription factor regulates amygdala development and emotional learning. Proc. Natl. Acad. Sci. U S A 102, 14877–14882. doi: 10.1073/pnas.0506785102
Liu, M.-H., Li, W., Zheng, J.-J., Xu, Y.-G., He, Q., Chen, G., et al. (2020). Differential neuronal reprogramming induced by NeuroD1 from astrocytes in grey matter versus white matter. Neural Regen. Res. 15, 342–351. doi: 10.4103/1673-5374.265185
Longo, A., Guanga, G. P., and Rose, R. B. (2008). Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry 47, 218–229. doi: 10.1021/bi701527r
Lowenstein, E. D., Rusanova, A., Stelzer, J., Hernaiz-Llorens, M., Schroer, A. E., Epifanova, E., et al. (2021). Olig3 regulates early cerebellar development. eLife 10:e64684. doi: 10.7554/eLife.64684
Lumsden, A., and Krumlauf, R. (1996). Patterning the vertebrate neuraxis. Science 274, 1109–1115. doi: 10.1126/science.274.5290.1109
Machold, R., and Fishell, G. (2005). Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48, 17–24. doi: 10.1016/j.neuron.2005.08.028
Machold, R., Klein, C., and Fishell, G. (2011). Genes expressed in Atoh1 neuronal lineages arising from the r1/isthmus rhombic lip. Gene Expr. Patterns 11, 349–359. doi: 10.1016/j.gep.2011.03.007
Mall, M., Kareta, M. S., Chanda, S., Ahlenius, H., Perotti, N., Zhou, B., et al. (2017). Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544, 245–249. doi: 10.1038/nature21722
Masserdotti, G., Gillotin, S., Sutor, B., Drechsel, D., Irmler, M., Jorgensen, H. F., et al. (2015). Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17, 74–88. doi: 10.1016/j.stem.2015.05.014
Mattar, P., Langevin, L. M., Markham, K., Klenin, N., Shivji, S., Zinyk, D., et al. (2008). Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol. Cell. Biol. 28, 1456–1469. doi: 10.1128/MCB.01510-07
McCarthy, M. I., and MacArthur, D. G. (2017). Human disease genomics: from variants to biology. Genome Biol. 18:20. doi: 10.1186/s13059-017-1160-z
Meredith, A., and Johnson, J. E. (2000). Negative autoregulation of Mash1 expression in CNS development. Dev. Biol. 222, 336–346. doi: 10.1006/dbio.2000.9697
Millen, K. J., Steshina, E. Y., Iskusnykh, I. Y., and Chizhikov, V. V. (2014). Transformation of the cerebellum into more ventral brainstem fates causes cerebellar agenesis in the absence of Ptf1a function. Proc. Natl. Acad. Sci. U S A 111, E1777–E1786. doi: 10.1073/pnas.1315024111
Millet, S., Bloch-Gallego, E., Simeone, A., and Alvarado-Mallart, R. M. (1996). The caudal limit of Otx2 gene expression as a marker of the midbrain/hindbrain boundary: a study using in situ hybridisation and chick/quail homotopic grafts. Development 122, 3785–3797. doi: 10.1242/dev.122.12.3785
Miyata, T., Maeda, T., and Lee, J. E. (1999). NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13, 1647–1652. doi: 10.1101/gad.13.13.1647
Momozawa, Y., and Mizukami, K. (2021). Unique roles of rare variants in the genetics of complex diseases in humans. J. Hum. Genet. 66, 11–23. doi: 10.1038/s10038-020-00845-2
Murre, C., Bain, G., van Dijk, M. A., Engel, I., Furnari, B. A., Massari, M. E., et al. (1994). Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta 1218, 129–135. doi: 10.1016/0167-4781(94)90001-9
Murre, C., McCaw, P. S., and Baltimore, D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783. doi: 10.1016/0092-8674(89)90682-x
Muzio, L., Di Benedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., Mallamaci, A., et al. (2002a). Conversion of cerebral cortex into basal ganglia in Emx2−/− Pax6Sey/Sey double-mutant mice. Nat. Neurosci. 5, 737–745. doi: 10.1038/nn892
Muzio, L., Di Benedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., Mallamaci, A., et al. (2002b). Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb. Cortex 12, 129–139. doi: 10.1093/cercor/12.2.129
Niemi, M. E. K., Martin, H. C., Rice, D. L., Gallone, G., Gordon, S., Kelemen, M., et al. (2018). Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature 562, 268–271. doi: 10.1038/s41586-018-0566-4
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S., and Kriegstein, A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720. doi: 10.1038/35055553
Olson, J. M., Asakura, A., Snider, L., Hawkes, R., Strand, A., Stoeck, J., et al. (2001). NeuroD2 is necessary for development and survival of central nervous system neurons. Dev. Biol. 234, 174–187. doi: 10.1006/dbio.2001.0245
Oproescu, A. M., Han, S., and Schuurmans, C. (2021). New insights into the intricacies of proneural gene regulation in the embryonic and adult cerebral cortex. Front. Mol. Neurosci. 14:642016. doi: 10.3389/fnmol.2021.642016
Pan, N., Jahan, I., Lee, J. E., and Fritzsch, B. (2009). Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 337, 407–428. doi: 10.1007/s00441-009-0826-6
Parenti, I., Rabaneda, L. G., Schoen, H., and Novarino, G. (2020). Neurodevelopmental disorders: from genetics to functional pathways. Trends Neurosci. 43, 608–621. doi: 10.1016/j.tins.2020.05.004
Park, C.-H., Kang, J. S., Kim, J.-S., Chung, S., Koh, J.-Y., Yoon, E.-H., et al. (2006). Differential actions of the proneural genes encoding Mash1 and neurogenins in Nurr1-induced dopamine neuron differentiation. J. Cell Sci. 119, 2310–2320. doi: 10.1242/jcs.02955
Pataskar, A., Jung, J., Smialowski, P., Noack, F., Calegari, F., Straub, T., et al. (2016). NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 35, 24–45. doi: 10.15252/embj.201591206
Petrik, D., and Encinas, J. M. (2019). Perspective: of mice and men—how widespread is adult neurogenesis? Front. Neurosci. 13:923. doi: 10.3389/fnins.2019.00923
Philippidou, P., and Dasen, J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80, 12–34. doi: 10.1016/j.neuron.2013.09.020
Pieper, A., Rudolph, S., Wieser, G. L., Gotze, T., Miessner, H., Yonemasu, T., et al. (2019). NeuroD2 controls inhibitory circuit formation in the molecular layer of the cerebellum. Sci. Rep. 9:1448. doi: 10.1038/s41598-018-37850-7
Pillai, A., Mansouri, A., Behringer, R., Westphal, H., and Goulding, M. (2007). Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development 134, 357–366. doi: 10.1242/dev.02717
Pomeshchik, Y., Klementieva, O., Gil, J., Martinsson, I., Hansen, M. G., de Vries, T., et al. (2020). Human iPSC-derived hippocampal spheroids: an innovative tool for stratifying Alzheimer disease patient-specific cellular phenotypes and developing therapies. Stem Cell Rep. 15, 256–273. doi: 10.1016/j.stemcr.2020.06.001
Poulin, J.-F., Gaertner, Z., Moreno-Ramos, O. A., and Awatramani, R. (2020). Classification of midbrain dopamine neurons using single-cell gene expression profiling approaches. Trends Neurosci. 43, 155–169. doi: 10.1016/j.tins.2020.01.004
Rakic, P. (1974). Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425–427. doi: 10.1126/science.183.4123.425
Rakic, P. (1995). Radial versus tangential migration of neuronal clones in the developing cerebral cortex. Proc. Natl. Acad. Sci. U S A 92, 11323–11327. doi: 10.1073/pnas.92.25.11323
Rao, Z., Wang, R., Li, S., Shi, Y., Mo, L., Han, S., et al. (2021). Molecular mechanisms underlying ascl1-mediated astrocyte-to-neuron conversion. Stem Cell Rep. 16, 534–547. doi: 10.1016/j.stemcr.2021.01.006
Richetin, K., Leclerc, C., Toni, N., Gallopin, T., Pech, S., Roybon, L., et al. (2015). Genetic manipulation of adult-born hippocampal neurons rescues memory in a mouse model of Alzheimer’s disease. Brain 138, 440–455. doi: 10.1093/brain/awu354
Richetin, K., Moulis, M., Millet, A., Arrazola, M. S., Andraini, T., Hua, J., et al. (2017). Amplifying mitochondrial function rescues adult neurogenesis in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 102, 113–124. doi: 10.1016/j.nbd.2017.03.002
Roelink, H., Porter, J. A., Chiang, C., Tanabe, Y., Chang, D. T., Beachy, P. A., et al. (1995). Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445–455. doi: 10.1016/0092-8674(95)90397-6
Ross, M. E., and Walsh, C. A. (2001). Human brain malformations and their lessons for neuronal migration. Annu. Rev. Neurosci. 24, 1041–1070. doi: 10.1146/annurev.neuro.24.1.1041
Roybon, L., Hjalt, T., Stott, S., Guillemot, F., Li, J.-Y., Brundin, P., et al. (2009). Neurogenin2 directs granule neuroblast production and amplification while NeuroD1 specifies neuronal fate during hippocampal neurogenesis. PLoS One 4:e4779. doi: 10.1371/journal.pone.0004779
Roybon, L., Mastracci, T. L., Ribeiro, D., Sussel, L., Brundin, P., Li, J. Y., et al. (2010). GABAergic differentiation induced by Mash1 is compromised by the bHLH proteins Neurogenin2, NeuroD1, and NeuroD2. Cereb. Cortex 20, 1234–1244. doi: 10.1093/cercor/bhp187
Rubio-Cabezas, O., Minton, J. A., Kantor, I., Williams, D., Ellard, S., Hattersley, A. T., et al. (2010). Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes 59, 2326–2331. doi: 10.2337/db10-0011
Runge, K., Cardoso, C., and de Chevigny, A. (2020a). Dendritic spine plasticity: function and mechanisms. Front. Synaptic Neurosci. 12:36. doi: 10.3389/fnsyn.2020.00036
Runge, K., Mathieu, R., Bugeon, S., Lafi, S., Beurrier, C., Sahu, S., et al. (2020b). Disruption of the transcription factor NEUROD2 causes an autism syndrome via cell-autonomous defects in cortical projection neurons. bioRxiv [Preprint]. doi: 10.1101/296889
Salvanes, A. G., Moberg, O., Ebbesson, L. O., Nilsen, T. O., Jensen, K. H., Braithwaite, V. A., et al. (2013). Environmental enrichment promotes neural plasticity and cognitive ability in fish. Proc. Biol. Sci. 280:20131331. doi: 10.1098/rspb.2013.1331
Satoh, J.-I., Yamamoto, Y., Asahina, N., Kitano, S., and Kino, Y. (2014). RNA-Seq data mining: downregulation of NeuroD6 serves as a possible biomarker for Alzheimer’s disease brains. Dis. Markers 2014:123165. doi: 10.1155/2014/123165
Schuurmans, C., Armant, O., Nieto, M., Stenman, J. M., Britz, O., Klenin, N., et al. (2004). Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 23, 2892–2902. doi: 10.1038/sj.emboj.7600278
Schwab, M. H., Bartholomae, A., Heimrich, B., Feldmeyer, D., Druffel-Augustin, S., Goebbels, S., et al. (2000). Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J. Neurosci. 20, 3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000
Schwab, M. H., Druffel-Augustin, S., Gass, P., Jung, M., Klugmann, M., Bartholomae, A., et al. (1998). Neuronal basic helix-loop-helix proteins (NEX, neuroD, NDRF): spatiotemporal expression and targeted disruption of the NEX gene in transgenic mice. J. Neurosci. 18, 1408–1418. doi: 10.1523/JNEUROSCI.18-04-01408.1998
Sega, A. G., Mis, E. K., Lindstrom, K., Mercimek-Andrews, S., Ji, W., Cho, M. T., et al. (2019). De novo pathogenic variants in neuronal differentiation factor 2 (NEUROD2) cause a form of early infantile epileptic encephalopathy. J. Med. Genet. 56, 113–122. doi: 10.1136/jmedgenet-2018-105322
Silbereis, J. C., Pochareddy, S., Zhu, Y., Li, M., and Sestan, N. (2016). The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268. doi: 10.1016/j.neuron.2015.12.008
Skinner, M. K., Rawls, A., Wilson-Rawls, J., and Roalson, E. H. (2010). Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 80, 1–8. doi: 10.1016/j.diff.2010.02.003
Sommer, L., Ma, Q., and Anderson, D. J. (1996). neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8, 221–241. doi: 10.1006/mcne.1996.0060
Sorrells, S. F., Paredes, M. F., Cebrian-Silla, A., Sandoval, K., Qi, D., Kelley, K. W., et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381. doi: 10.1038/nature25975
Spellmann, I., Riedel, M., Stadtler, J., Zill, P., Obermeier, M., Cerovecki, A., et al. (2017). Associations of NEUROD2 polymorphisms and change of cognitive dysfunctions in schizophrenia and schizoaffective disorder after eight weeks of antipsychotic treatment. Cogn. Neuropsychiatry 22, 280–297. doi: 10.1080/13546805.2017.1322502
Stern, C. D. (2005). Neural induction: old problem, new findings, yet more questions. Development 132, 2007–2021. doi: 10.1242/dev.01794
Stevens, J. D., Roalson, E. H., and Skinner, M. K. (2008). Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation 76, 1006–1022. doi: 10.1111/j.1432-0436.2008.00285.x
Stiles, J., and Jernigan, T. L. (2010). The basics of brain development. Neuropsychol. Rev. 20, 327–348. doi: 10.1007/s11065-010-9148-4
Subramanian, L., Calcagnotto, M. E., and Paredes, M. F. (2019). Cortical malformations: lessons in human brain development. Front. Cell. Neurosci. 13:576. doi: 10.3389/fncel.2019.00576
Sueda, R., Imayoshi, I., Harima, Y., and Kageyama, R. (2019). High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 33, 511–523. doi: 10.1101/gad.323196.118
Sun, X. H., and Baltimore, D. (1991). An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell 64, 459–470. doi: 10.1016/0092-8674(91)90653-g
Sur, A., Magie, C. R., Seaver, E. C., and Meyer, N. P. (2017). Spatiotemporal regulation of nervous system development in the annelid Capitella teleta. Evodevo 8:13. doi: 10.1186/s13227-017-0076-8
Trainor, P., and Krumlauf, R. (2000). Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat. Cell Biol. 2, 96–102. doi: 10.1038/35000051
Treutlein, B., Lee, Q. Y., Camp, J. G., Mall, M., Koh, W., Shariati, S. A., et al. (2016). Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395. doi: 10.1038/nature18323
Uittenbogaard, M., Baxter, K. K., and Chiaramello, A. (2010a). NeuroD6 genomic signature bridging neuronal differentiation to survival via the molecular chaperone network. J. Neurosci. Res. 88, 33–54. doi: 10.1002/jnr.22182
Uittenbogaard, M., Baxter, K. K., and Chiaramello, A. (2010b). The neurogenic basic helix-loop-helix transcription factor NeuroD6 confers tolerance to oxidative stress by triggering an antioxidant response and sustaining the mitochondrial biomass. ASN Neuro 2:e00034. doi: 10.1042/AN20100005
Uittenbogaard, M., and Chiaramello, A. (2002). Constitutive overexpression of the basic helix-loop-helix Nex1/MATH-2 transcription factor promotes neuronal differentiation of PC12 cells and neurite regeneration. J. Neurosci. Res. 67, 235–245. doi: 10.1002/jnr.10119
Uittenbogaard, M., and Chiaramello, A. (2004). Expression profiling upon Nex1/MATH-2-mediated neuritogenesis in PC12 cells and its implication in regeneration. J. Neurochem. 91, 1332–1343. doi: 10.1111/j.1471-4159.2004.02814.x
Uittenbogaard, M., and Chiaramello, A. (2014). Mitochondrial biogenesis: a therapeutic target for neurodevelopmental disorders and neurodegenerative diseases. Curr. Pharm. Des. 20, 5574–5593. doi: 10.2174/1381612820666140305224906
Ulloa, F., and Marti, E. (2010). Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev. Dyn. 239, 69–76. doi: 10.1002/dvdy.22058
Urbán, N., and Guillemot, F. (2014). Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 8:396. doi: 10.3389/fncel.2014.00396
van Weert, L., Buurstede, J. C., Sips, H. C. M., Mol, I. M., Puri, T., Damsteegt, R., et al. (2019). Mechanistic insights in NeuroD potentiation of mineralocorticoid receptor signaling. Int. J. Mol. Sci. 20:1575. doi: 10.3390/ijms20071575
Vasconcelos, F. F., and Castro, D. S. (2014). Transcriptional control of vertebrate neurogenesis by the proneural factor Ascl1. Front. Cell. Neurosci. 8:412. doi: 10.3389/fncel.2014.00412
Wang, W., Corominas, R., and Lin, G. N. (2019). De novo mutations from whole exome sequencing in neurodevelopmental and psychiatric disorders: from discovery to application. Front. Genet. 10:258. doi: 10.3389/fgene.2019.00258
Wang, V. Y., Rose, M. F., and Zoghbi, H. Y. (2005). Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron 48, 31–43. doi: 10.1016/j.neuron.2005.08.024
Wapinski, O. L., Lee, Q. Y., Chen, A. C., Li, R., Corces, M. R., Ang, C. E., et al. (2017). Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20, 3236–3247. doi: 10.1016/j.celrep.2017.09.011
Wapinski, O. L., Vierbuchen, T., Qu, K., Lee, Q. Y., Chanda, S., Fuentes, D. R., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. doi: 10.1016/j.cell.2013.09.028
Ware, M., Hamdi-Roze, H., Le Friec, J., David, V., and Dupe, V. (2016). Regulation of downstream neuronal genes by proneural transcription factors during initial neurogenesis in the vertebrate brain. Neural. Dev. 11:22. doi: 10.1186/s13064-016-0077-7
Wilke, S. A., Hall, B. J., Antonios, J. K., Denardo, L. A., Otto, S., Yuan, B., et al. (2012). NeuroD2 regulates the development of hippocampal mossy fiber synapses. Neural. Dev. 7:9. doi: 10.1186/1749-8104-7-9
Wingate, R. J., and Hatten, M. E. (1999). The role of the rhombic lip in avian cerebellum development. Development 126, 4395–4404. doi: 10.1242/dev.126.20.4395
Wu, S. X., Goebbels, S., Nakamura, K., Nakamura, K., Kometani, K., Minato, N., et al. (2005). Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc. Natl. Acad. Sci. U S A 102, 17172–17177. doi: 10.1073/pnas.0508560102
Yamada, M., Seto, Y., Taya, S., Owa, T., Inoue, Y. U., Inoue, T., et al. (2014). Specification of spatial identities of cerebellar neuron progenitors by ptf1a and atoh1 for proper production of GABAergic and glutamatergic neurons. J. Neurosci. 34, 4786–4800. doi: 10.1523/JNEUROSCI.2722-13.2014
Zhang, T.-Y., Keown, C. L., Wen, X., Li, J., Vousden, D. A., Anacker, C., et al. (2018). Environmental enrichment increases transcriptional and epigenetic differentiation between mouse dorsal and ventral dentate gyrus. Nat. Commun. 9:298. doi: 10.1038/s41467-017-02748-x
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018). Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18:235. doi: 10.1186/s12870-018-1441-z
Zhang, Y., Lahmann, I., Baum, K., Shimojo, H., Mourikis, P., Wolf, J., et al. (2021). Oscillations of Delta-like1 regulate the balance between differentiation and maintenance of muscle stem cells. Nat. Commun. 12:1318. doi: 10.1038/s41467-021-21631-4
Keywords: bHLH factor, neurod family, brain development, neurological diseases, transcription factors
Citation: Tutukova S, Tarabykin V and Hernandez-Miranda LR (2021) The Role of Neurod Genes in Brain Development, Function, and Disease. Front. Mol. Neurosci. 14:662774. doi: 10.3389/fnmol.2021.662774
Received: 01 February 2021; Accepted: 11 May 2021;
Published: 09 June 2021.
Edited by:
Veronica Martinez Cerdeño, University of California, Davis, United StatesCopyright © 2021 Tutukova, Tarabykin and Hernandez-Miranda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis R. Hernandez-Miranda, bHVpcy5oZXJuYW5kZXotbWlyYW5kYUBjaGFyaXRlLmRl; Victor Tarabykin, dmljdG9yLnRhcmFieWtpbkBjaGFyaXRlLmRl
 Svetlana Tutukova
Svetlana Tutukova Victor Tarabykin
Victor Tarabykin Luis R. Hernandez-Miranda
Luis R. Hernandez-Miranda