
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci. , 15 February 2021
Sec. Neuroplasticity and Development
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.642016
This article is part of the Research Topic Transcription Regulation - Brain Development and Homeostasis - A Finely Tuned and Orchestrated Scenario in Physiology and Pathology View all 12 articles
Historically, the mammalian brain was thought to lack stem cells as no new neurons were found to be made in adulthood. That dogma changed ∼25 years ago with the identification of neural stem cells (NSCs) in the adult rodent forebrain. However, unlike rapidly self-renewing mature tissues (e.g., blood, intestinal crypts, skin), the majority of adult NSCs are quiescent, and those that become ‘activated’ are restricted to a few neurogenic zones that repopulate specific brain regions. Conversely, embryonic NSCs are actively proliferating and neurogenic. Investigations into the molecular control of the quiescence-to-proliferation-to-differentiation continuum in the embryonic and adult brain have identified proneural genes encoding basic-helix-loop-helix (bHLH) transcription factors (TFs) as critical regulators. These bHLH TFs initiate genetic programs that remove NSCs from quiescence and drive daughter neural progenitor cells (NPCs) to differentiate into specific neural cell subtypes, thereby contributing to the enormous cellular diversity of the adult brain. However, new insights have revealed that proneural gene activities are context-dependent and tightly regulated. Here we review how proneural bHLH TFs are regulated, with a focus on the murine cerebral cortex, drawing parallels where appropriate to other organisms and neural tissues. We discuss upstream regulatory events, post-translational modifications (phosphorylation, ubiquitinylation), protein–protein interactions, epigenetic and metabolic mechanisms that govern bHLH TF expression, stability, localization, and consequent transactivation of downstream target genes. These tight regulatory controls help to explain paradoxical findings of changes to bHLH activity in different cellular contexts.
One-hundred years ago, Santiago Ramon y Cajal likened the pyramidal cells of the cerebral cortex to “a garden filled with innumerable trees…which can multiply their branches thanks to intelligent cultivation, send their roots deeper, and produce more exquisite flowers and fruits every day” (Jones, 1994). In his 1942 book Man on His Nature, neurophysiologist Sir Charles Sherrington compared the electrical activity of the cortex to “an enchanted loom” (Sherrington, 2009). The field of neuroscience is ripe with such metaphors that remark upon the exquisite architecture and cellular diversity of the cerebral cortex. It follows, then, that one of the most dominant inquiries in developmental neuroscience has been how this enormous cellular diversity is established and choreographed during brain development. Detangling this great mystery has important implications for our understanding of neurological disorders and diseases, as well as for the future design of therapeutic strategies to replace lost/dysfunctional neural cells.
This review centers on the regulatory events that govern proneural gene function in the developing and adult cerebral cortex. To provide a contextual framework, we first provide a high-level overview, not meant to be comprehensive, of the cellular context in which these genes function. The cerebral cortex, which is the seat of higher order cognitive functioning and sensory processing, is comprised of a six-layered neocortex and several three- or four-layered allocortical territories, including the hippocampal formation and paleocortex. Cortical territories are found in all mammals, but display enormous structural diversity across species, transitioning during evolution between smooth (lissencephalic) structures in smaller mammals such as rodents, to highly folded (gyrencephalic) structures in most extant primates and larger mammals (Lewitus et al., 2014). These gross structural differences arise due to species-specific differences in the regulatory events that control self-renewal, proliferation, mode of division (symmetric, asymmetric) and differentiation properties of neural stem cells (NSCs) and their daughter neural progenitor cells (NPCs). Distinguishing features of NSCs include maintenance into adulthood, the capacity to self-renew, and multipotency, which refers to their tri-lineage potential, or the capacity to give rise to neurons, astrocytes and oligodendrocytes. Conversely, NPCs do not self-renew, are more restricted in their proliferative potential, and may have reduced developmental potential as they acquire lineage biases.
To understand how the cerebral cortex acquires species-specific forms, it is essential to elucidate how NSC/NPC (hereafter NPC for simplicity) fate decisions are controlled. Proneural genes, which encode basic-helix-loop-helix (bHLH) transcription factors (TFs), are critical pieces to the puzzle as they control NPC decisions to divide or differentiate while also specifying neural subtype identities (Bertrand et al., 2002; Wilkinson et al., 2013; Guillemot and Hassan, 2017; Dennis et al., 2019). At face value, the functions of proneural genes appear simplistic, but their activities are tightly regulated by both cell intrinsic and extrinsic influences. Here we review the regulatory mechanisms that govern proneural gene function in embryonic and adult cortical domains, drawing parallels to other bHLH genes, brain regions, non-neural tissues, and non-mammalian species when comparison is informative. Of note, unless otherwise specified, the animal work cited was conducted using murine transgenic models, tissues or cells.
Proneural genes encode type II, tissue-specific bHLH TFs that are expressed in the nervous system and have evolutionarily conserved roles in promoting neural cell fate specification and differentiation (Bertrand et al., 2002; Wilkinson et al., 2013; Guillemot and Hassan, 2017; Dennis et al., 2019). Proneural genes were first identified and characterized in Drosophila melanogaster where they belong to two main families that each specify distinct neural cell fates: achaete-scute complex (AS-C) and atonal-related genes (Bertrand et al., 2002; Huang et al., 2014). In the fly, bHLH genes that are defined as proneural are expressed in uncommitted ectodermal precursors and have the ability to: (1) select single ectodermal precursors within a proneural cluster to become neural by activating Notch/Delta-mediated lateral inhibition, and (2) specify neural precursor identity by activating generic and subtype-specific neuronal differentiation genes. While vertebrate and invertebrate proneural genes share several features, a major difference is that in vertebrates, proneural gene expression initiates in NPCs that are already specified as neural. With this difference in mind, vertebrate bHLH genes are defined as proneural if they: (1) are expressed in dividing NPCs, usually those at the apex of lineage hierarchies, (2) drive NPCs to differentiate into neuronal or glial cells, (3) specify neural subtype identities, and (4) activate Notch signaling in neighboring NPCs by inducing the expression of Notch ligands, such as Dll1 and Dll3 (Bertrand et al., 2002; Wilkinson et al., 2013; Guillemot and Hassan, 2017; Dennis et al., 2019). Based on these criteria, four proneural genes are expressed in the developing and/or adult cerebral cortex: Neurogenin (Neurog) 1, Neurog2, Neurod4 (aka Math3), and Achaete-scute family bHLH transcription factor 1 (Ascl1; aka Mash1) (Bertrand et al., 2002; Wilkinson et al., 2013; Guillemot and Hassan, 2017; Dennis et al., 2019). All other commonly studied neural bHLH genes, such as Neurod1, Neurod2, Neurod6 and others are instead properly termed ‘neuronal differentiation’ genes because of their later expression/function in neural lineages, either in later-stage progenitors with a restricted proliferative and differentiation potential [e.g., Neurod1 (Pleasure et al., 2000)], and/or in postmitotic neurons [e.g., Neurod2, Neurod6 (Bormuth et al., 2013; Guzelsoy et al., 2019)]. In this review we mainly focus on the cortical functions of Neurog1, Neurog2 and Ascl1, which have been most extensively studied.
To bind DNA, proneural bHLH TFs must dimerize, either with other proneural TFs or with type I bHLH factors, also known as E-proteins (Murre et al., 1989). E-proteins, which have more ubiquitous expression patterns than class II bHLH TFs, are encoded by three genes: Tcf4 (aka E2-2), Tcf12 (aka HEB), and Tcf3 (aka E2A), the latter encoding E12 and E47 splice variants (Bertrand et al., 2002; Wang and Baker, 2015). Proneural TFs can also dimerize with HLH proteins of the Id (inhibitor of DNA-binding) family, which lack the basic DNA-binding domain and thus form non-functional heterodimers (Wang and Baker, 2015). To activate transcription, bHLH dimers bind to Ephrussi-box (E-box) sequences (CANNTG) in regulatory regions of the genome (Murre et al., 1989; Wang and Baker, 2015). ChIP-seq analyses have revealed that different proneural TF binding sites have differential enrichment of the central two E-box residues; Neurog2 favors CAKMTG motifs (K: G/T nucleotides, M: A/C nucleotides), with the CAGATG motif predominant, while Ascl1 preferentially binds sites with CAGSTG motifs (S: G/C nucleotides), with the CAGCTG motif predominant (Wapinski et al., 2013; Raposo et al., 2015; Aydin et al., 2019). Despite these known biases, the binding of proneural TF hetero- or homo-dimers to their cognate sites is highly context-specific and tightly regulated, which is the subject of this review.
To set the stage for the embryonic context in which proneural TFs function, we briefly outline critical developmental transitions. The neurons and macroglial cells (oligodendrocytes, astrocytes) that make up the adult cerebral cortex are derived from multipotent NPCs located in the dorsal telencephalon (cortex), with some additional contributions from ventral telencephalic (subcortical) NPCs. Telencephalic NPCs are parcellated into apical and basal compartments (Taverna et al., 2014). Apical NPCs reside in the ventricular zone (VZ), a single cell-layered neurepithelium that appears pseudostratified due to interkinetic nuclear migration, with G2/M-phase nuclei moving to the apical surface whereas S-phase nuclei move basally (Taverna et al., 2014). Apical NPCs are termed neuroepithelial cells (NECs) prior to neurogenesis and initially divide symmetrically to expand the NPC pool (Gotz and Huttner, 2005). When neurogenesis begins, at approximately embryonic day (E) 11 in mouse, NECs transform into apical radial glia (aRG), which remain in the VZ, but switch to self-renewing asymmetric neurogenic divisions to give rise to one aRG and either one new neuron (direct neurogenesis) or one basal progenitor (indirect neurogenesis) (Gotz and Huttner, 2005; Bultje et al., 2009). aRG and NECs differ at the transcriptomic level, with aRGs initiating the expression of several glial markers (Taverna et al., 2014). In rodents, basal progenitors, which form a subventricular zone (SVZ), predominantly include neuronal-committed intermediate progenitor cells (INPs) that have a limited proliferative capacity (1–2 divisions) and undergo terminal symmetric neurogenic divisions (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2004; Englund et al., 2005; Kowalczyk et al., 2009; Taverna et al., 2014). Further stratifications of these apical and basal NPC pools have been made based on morphological and gene expression criteria and are reviewed elsewhere (Taverna et al., 2014).
Cortical NPCs give rise in a sequential fashion to excitatory glutamatergic neurons that form the six layers of the cortical plate between E11-E17 in mouse (Caviness, 1982; Caviness et al., 1995; Takahashi et al., 1999), followed by astrocytes, beginning at E16 (Bayraktar et al., 2014), and then oligodendrocytes, beginning early postnatally (Kessaris et al., 2006). The earliest-born cortical neurons form a preplate that is later split into an overlying marginal zone (layer I) and an underlying subplate (layer VII), the latter a transient neuronal layer that nevertheless plays important roles in thalamocortical axonal pathfinding and in guiding neuronal migration (Ohtaka-Maruyama, 2020). Layer VI corticothalamic neurons are born next, followed by the sequential differentiation of layer V subcerebral and callosal neurons, layer IV internal granular layer neurons, and finally, layer II/III corticocortical neurons, two layers that are fused in mouse (Caviness, 1982; Kast and Levitt, 2019). GABAergic interneurons and oligodendrocytes also populate cortical domains, but they are born in the ventral telencephalic (subcortical) VZ/SVZ and enter the cortex via tangential migration (Peyre et al., 2015).
The progressive nature of laminar fate determination raises the question of how cortical NPCs change over time (Pearson and Doe, 2004). Seminal studies involving heterochronic transplantation experiments in ferrets revealed that early-stage cortical NPCs are multipotent, responding to new environmental signals to generate alternative laminar identities post-transplant, but only when in S-phase of the cell cycle, whereas later stage cortical NPCs lose their ability to respond to early environmental signals (McConnell and Kaznowski, 1991; Frantz and McConnell, 1996; Bohner et al., 1997; Desai and McConnell, 2000). These findings were corroborated by retroviral lineage tracing experiments, which confirmed that early cortical NPCs are multipotent and give rise to neuronal clones that span cortical layers, whereas late NPCs are fate restricted and only generate upper layer neurons (Luskin et al., 1988; Price and Thurlow, 1988; Walsh and Cepko, 1988). More recently, genetic lineage tracing experiments using various Cre drivers (Franco et al., 2012; Guo et al., 2013; Eckler et al., 2015) and Mosaic Analysis with Double Markers (MADM) (Gao et al., 2014) have confirmed that cortical NPCs are multipotent at the population and clonal level, although some fate-restricted NPCs may also exist (Franco et al., 2012; Gil-Sanz et al., 2015). How NPCs give rise to such diverse neural cell types in a stereotypically defined manner has been the subject of study for several decades now (Pearson and Doe, 2004). The importance of intrinsic factors was demonstrated by plating cortical NPCs at clonal density, which generated stereotyped lineage trees that matched those seen in vivo (Qian et al., 2000). Since then, revolutionary new technologies such as FlashTag and single cell (sc) RNA-seq have identified sequential transcriptional waves that successively define apical and basal NPCs and daughter neurons (Telley et al., 2016). Further studies with these techniques identified two axes of NPC transcriptional organization throughout the neurogenic period: a “birthdate axis” in which the transcriptional state varies depending on embryonic age, and a “differentiation axis,” which drives NPCs to differentiate in a conserved sequence regardless of neuronal birthdate (Telley et al., 2019). Interestingly, this work showed that late-stage apical NPCs (E14/E15) have predominantly environment-sensing transcriptional properties, with activation of genetic programs related to ion transport and cell-cell or cell-matrix interaction-related processes, as opposed to the cell-intrinsic transcriptional programs in earlier apical NPCs (Telley et al., 2019).
From these pioneering studies of cortical NPCs, Neurog2 was highlighted as a critical ‘neurogenic’ (actually, proneural) gene as it is expressed at high levels in apical and basal NPCs and at low levels in newborn neurons (Telley et al., 2016), consistent with earlier immunostaining studies (Hand et al., 2005). Functional assays demonstrating that Neurog1 and Neurog2 are true proneural genes pre-dated these studies by a decade or more and involved classical loss- and gain-of-function assays (Fode et al., 2000; Parras et al., 2002; Schuurmans et al., 2004; Mattar et al., 2008; Dixit et al., 2011, 2014; Kovach et al., 2013; Han et al., 2018). From these studies, Neurog2 and Neurog1 were shown to be necessary and sufficient to specify the excitatory, glutamatergic neuronal identity of early-born (layer V, VI) cortical neurons, as well as Cajal-Retzius neurons, which populate layer I (Dixit et al., 2014). In contrast, Ascl1, which is expressed at the highest levels in subcortical NPCs, is necessary and sufficient to specify a GABAergic neuronal or oligodendrocyte fate in the ventral telencephalon (Casarosa et al., 1999; Horton et al., 1999; Schuurmans et al., 2004; Parras et al., 2007). Interestingly, Ascl1 is also expressed at lower levels in cortical NPCs (Britz et al., 2006), where it also biases NPCs toward an oligodendrocyte fate (Han et al., 2020). In addition, Ascl1 is also required for the generation of a subset of glutamatergic Cajal-Retzius neurons, as opposed to the GABAergic fates specified by this TF in ventral telencephalic domains, highlighting the importance of cell context in dictating how these proneural genes function (Dixit et al., 2011). Finally, the transient expression of Neurog2 and Ascl1 in newborn neurons also has functional consequences, as these genes play a role in guiding neuronal migration by regulating expression of the Rho GTPases, Rnd2 and Rnd3, respectively (Heng et al., 2008; Pacary et al., 2011, 2013).
Studies of non-human primates (NHP) and human cortices have revealed that the apical NPC pool has expanded to include both aRG and outer or basal RG (bRG), the latter forming a large outer SVZ (oSVZ) not present in rodents (Lukaszewicz et al., 2005; Zecevic et al., 2005; Bayatti et al., 2008; Martinez-Cerdeno et al., 2012; Dehay et al., 2015). Like aRG, bRG are self-renewing and generate neurons by giving rise to transit-amplifying INPs (Fietz et al., 2010; Hansen et al., 2010; Reillo et al., 2011). However, INPs divide several more times in gyrencephalic species than in rodents to generate more later-born, upper-layer or supragranular neurons that make primate cortices larger with many folds (Molnar et al., 2011; Falk and Hofman, 2012; Stahl et al., 2013; Pollen et al., 2015) (Figure 1A). Several genes that promote basal NPC expansion can induce cortical folding in lissencephalic mammals, such as mice, or alter folding in gyrencephalic species, such as ferrets (Hansen et al., 2010; Fietz et al., 2012; Stahl et al., 2013; de Juan Romero et al., 2015; Florio et al., 2015, 2018; Ju et al., 2016; Wang et al., 2016; Fiddes et al., 2018; Suzuki et al., 2018; Chizhikov et al., 2019). Conversion from a lissencephalic to gyrencephalic cortex is also associated with alterations of the simple radial trajectories of migrating neurons in lissencephalic species, to more circuitous, tangential routes in gyrencephalic species (Del Toro et al., 2017; Llinares-Benadero and Borrell, 2019). Recent studies have revealed an unexpected role for Neurog2 and Ascl1 co-expression in sustaining a lissencephalic form in the rodent cortex due to the essential role that double+ NPCs play in patterning Notch signaling, which impacts the symmetry of radial glial trajectories (Han et al., 2020).
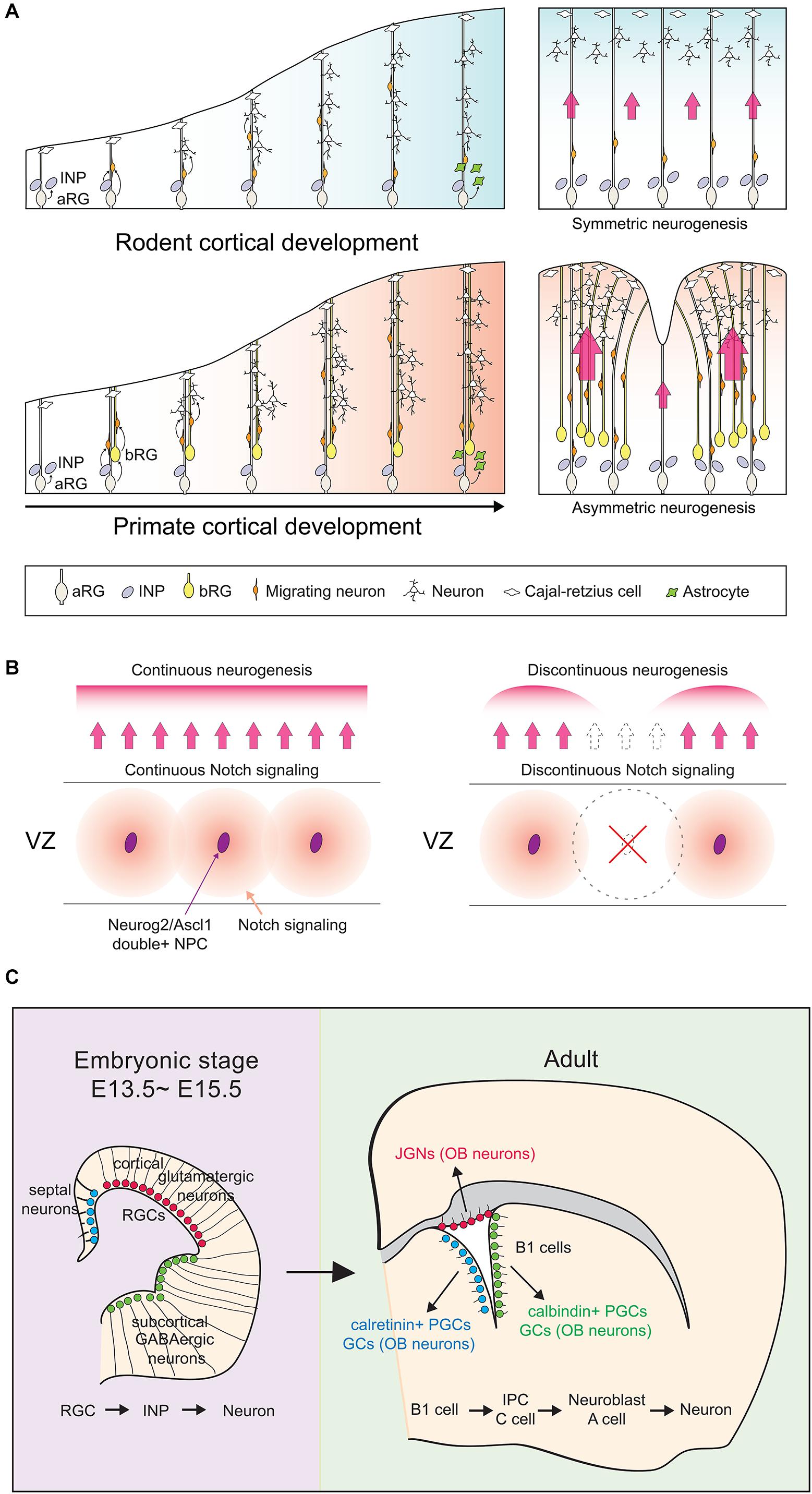
Figure 1. Cortical development between species and developmental stages. (A) In both lissencephalic and gyrencephalic species, apical radial glial (aRG) cells can divide asymmetrically to give rise to another aRG and either a nascent neuron (direct neurogenesis) or a neuronal-committed intermediate neuronal progenitor (INP; indirect neurogenesis). Gyrencephalic species like primates have an additional population of radial glial cells, the basal RG (bRG), which contribute more dividing INP cells to in turn generate more upper layer neurons in primate cortices. In the panels on the right, red arrows signify migratory routes of nascent neurons traveling along aRG processes. Larger arrows signify cortical regions with increased neurogenesis. (B) Neurog2/Ascl1 double-positive NPCs (purple ovals) act as ‘niche’ cells in the ventricular zone (VZ) of the rodent cortex, preventing the formation of cortical folds by maintaining continuous Notch signaling patterns. (C) Unlike the majority of actively dividing apical radial glia in E13.5–15.5 cortices, a subset remains quiescent and is set aside to become adult neural stem cells, the ‘B1’ cells. B1 cells reside in the adult cortical VZ and retain their regional identities. For adult B1 cells to exit quiescence and become neurogenic, upregulation of a cortical proneural gene like Neurog2 or Ascl1 must occur. aRG, apical radial glia; INP, intermediate neuronal progenitor; bRG, basal radial glia; JGN, juxtaglomerular neuron; OB, olfactory bulb; PGC, periglomerular cell; GC, granule cell.
While NSCs persist into adulthood, they differ from embryonic NSCs in several ways. Firstly, the adult NSC transcriptional profile is more closely related to astrocytes than to embryonic NSCs (Beckervordersandforth et al., 2010). Secondly, most embryonic NSCs are actively dividing and neurogenic, whereas adult NSCs are mainly quiescent and gliogenic (Gotz et al., 2016). Indeed, up to 90% of adult NSCs are quiescent in the adult brain at any given time, with cell cycle times ranging from 1 day to 3 months (Ponti et al., 2013; Reeve et al., 2017). Adult NSCs that become ‘activated’ are also restricted to a few neurogenic zones and repopulate only specific brain regions. For instance, the ventricular-subventricular zone (V-SVZ) repopulates the murine olfactory bulb and human striatum, while the subgranular zone (SGZ) repopulates the mouse/human dentate gyrus (Spalding et al., 2013; Ernst et al., 2014; Urban and Guillemot, 2014; Gotz et al., 2016; Boldrini et al., 2018; Ruddy and Morshead, 2018; Sorrells et al., 2018). Outside of these niches, the adult NSC response is limited.
Conversely, most embryonic NSCs divide rapidly in vivo, with cell cycle times of 8–18 h (Takahashi et al., 1995). However, a small but important pool of embryonic NSCs is slow-dividing; these are the embryonic precursors of adult NSCs, the origins of which had remained elusive until recently (Furutachi et al., 2013, 2015; Fuentealba et al., 2015). Using barcoding, a genetic lineage tracing method that can identify clonal relationships between widely distributed cells, it was revealed that a subset of E13.5-E15.5 aRG, termed ‘pre-B1 cells,’ are set aside as slow-dividing NPCs that will later become adult B1 cells (Fuentealba et al., 2015) (Figure 1B). B1 cells are adult NSCs, which when activated give rise to transit amplifying intermediate precursor cells (IPCs, C cells) that generate neuroblasts (A cells) that migrate through the rostral migratory stream (RMS) to the olfactory bulb (Li and Clevers, 2010). B1 cells retain regional identities; dorsal NSCs give rise to glutamatergic juxtaglomerular neurons (JGNs), ventral NSCs to calbindin+ periglomerular cells (PGCs) and granule cells (GCs), and septal NSCs to calretinin+ PGCs and GCs (Brill et al., 2009; Fuentealba et al., 2015). With respect to the focus of this review, for adult NSCs to become activated and neurogenic, neural determinants such as Neurog2 and Ascl1, which are expressed at high levels in embryonic NSCs and low levels in adult NSCs, must be upregulated (Gotz et al., 2016; Guillemot and Hassan, 2017). We discuss the associated regulatory mechanisms herein.
Neurog2 and Ascl1 are classical proneural genes, rapidly inducing NPC cell cycle exit and differentiation when misexpressed in the embryonic cortex (Britz et al., 2006; Mattar et al., 2008; Kovach et al., 2013; Wilkinson et al., 2013). Yet curiously, Neurog2 (Hagey and Muhr, 2014) and Ascl1 (Castro et al., 2011; Li et al., 2014) can also induce proliferation when expressed in some cellular contexts. Moreover, during normal development, Neurog1, Neurog2 and Ascl1 are mainly expressed in dividing NPCs (Britz et al., 2006). These findings raise the question of how proneural gene expression is compatible with both pro-proliferative and pro-differentiative NPC phenotypes. This conundrum was partially resolved in ground-breaking studies that demonstrated that Neurog2 and Ascl1 are expressed in 2–3 hr oscillatory cycles in dividing NPCs versus at sustained levels in NPCs that differentiate (Shimojo et al., 2008; Imayoshi et al., 2013; Ochi et al., 2020).
Notch signaling is the driving force behind oscillatory proneural gene expression (Kageyama et al., 2008, 2020) (Figure 2). In a process known as ‘lateral inhibition,’ NPCs that express high levels of the proneural TFs transactivate the expression of cell-membrane tethered Notch ligands such as Dll1 and Dll3 (Castro et al., 2006; Henke et al., 2009), which bind Notch receptors on neighboring NPCs. Upon ligand binding, Notch is proteolytically cleaved to form a Notch intracellular domain (NICD) that translocates to the nucleus where it binds to Rbpj, a DNA binding protein. NICD-Rbpj complexes transcribe downstream genes, including hairy and enhancer of split (Hes) 1 and Hes5, which encode bHLH transcriptional repressors that recruit Groucho/TLE co-repressors and bind to N-boxes (CACNAG), directly repressing proneural gene transcription to form a lateral inhibitory loop (Kageyama et al., 2007, 2008, 2020; Kovach et al., 2012; Huang et al., 2014). Hes1 is also expressed in 2–3 h oscillatory cycles, and Hes1 protein drives its dynamic expression through direct repression of its own transcription, as well as indirectly driving oscillatory expression of the proneural genes through transcriptional repression (Shimojo et al., 2008, 2011). Consequently, Hes and proneural genes are expressed out-of-phase with one another in ‘salt-and-pepper’ expression profiles, referring to their scattered expression when captured at individual time points (Kageyama et al., 2008). Notably, while these oscillatory cycles are transcriptionally driven, proneural proteins also oscillate as they have short intracellular half-lives (< 30 min) and are rapidly degraded with each transcriptional cycle (Nguyen et al., 2006; Kovach et al., 2013; Urban et al., 2016).
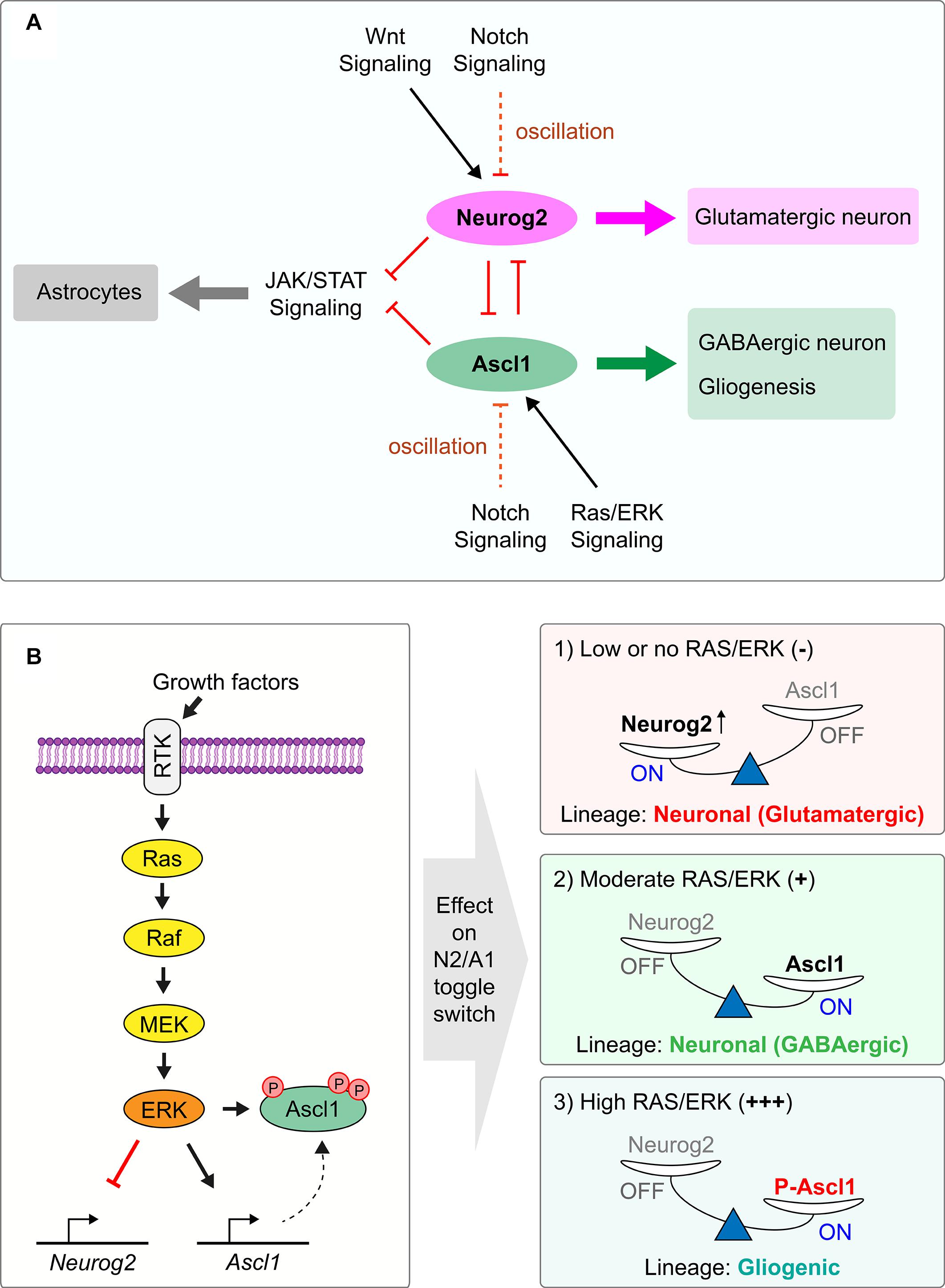
Figure 2. Regulation of the Neurog2/Ascl1 proneural gene toggle switch by extracellular signaling pathways. (A) Proneural TFs Neurog2 and Ascl1 are competing lineage determinants in the cortex, specifying glutamatergic pyramidal neurons and GABAergic interneurons, respectively. Neurog2 and Ascl1 have cross-repressive interactions with each other and form a bistable toggle switch, preventing lineage commitment in double-positive NPCs. Environmental signals regulate the expression of each gene to turn on the expression of one proneural TF and turn off the other. Notch signaling controls expression of both Neurog2 and Ascl1 through lateral inhibition, with Hes1 protein driving the 2–3 h transcriptional oscillatory cycles of the proneural genes. Extracellular Wnt promotes Neurog2 and suppresses Ascl1 expression, acting in early neurogenesis. Conversely, Ras/ERK signaling favors Ascl1 over Neurog2 expression. (B) Ras/ERK signaling cascade activation is achieved by ligand binding to RTK, culminating in the phosphorylation of Ascl1 protein and activation of Ascl1 expression, tipping the Neurog2/Ascl1 toggle switch in favor of Ascl1. At moderate Ras/ERK activation, this leads to GABAergic neuronal specification by Ascl1, while at higher levels of Ras/ERK activation, this leads to gliogenic specification by phosphorylated Ascl1.
While sustained proneural TF expression biases NPCs toward differentiation, it can also maintain the NPC pool by allowing neighboring NPCs with activated Notch signaling to continue to proliferate. Hes1/5-mediated repression of proneural genes is essential to maintain the NPC pool, with co-deletion of Hes1/5 or Rbpj, their upstream regulator (Son et al., 2020), leading to precocious neurogenesis and NPC pool depletion (Ohtsuka et al., 1999; Hatakeyama et al., 2004). Strikingly, proneural genes also regulate the patterning of Notch signaling, with NPCs that co-express Neurog2 and Ascl1 acting as Notch-ligand expressing niche cells, the deletion of which disrupts the continuity of Notch signaling, resulting in cortical folding (Han et al., 2020) (Figure 1B).
Notably, there is also evidence for a Notch-independent mode of Rbpj function in regulation of bHLH TFs. While Rbpj suppresses Neurog1 transcription in NPCs, it positively regulates Neurog1 expression in migrating postmitotic neurons independent of Notch pathway activation (Son et al., 2020). Thus, as shown for Neurog2 (Hand et al., 2005), Neurog1 is expressed in dividing NPCs and newborn neurons, but distinct regulatory mechanisms drive its expression in the two cell types (Son et al., 2020). Interestingly, Rbpj also binds a conserved binding motif in the Ascl1 promoter in the locus coeruleus (Shi et al., 2012), and Rbpj directly represses Atoh7, another bHLH proneural gene, in a Notch-independent fashion in the retina (Miesfeld et al., 2018). Further studies are required to elucidate the extent to which Rbpj regulates Neurog1, Neurog2, and Ascl1 expression through Notch-dependent and -independent modes in the embryonic cortex.
During embryogenesis, cortical NPCs differentiate into glutamatergic neurons and later astrocytes, but retain the potential to divert to embryonic subcortical fates (GABAergic neurons, oligodendrocytes), as revealed by the mutation of several cortical transcription factors (Theil et al., 1999; Stoykova et al., 2000; Tole et al., 2000; Muzio et al., 2002a, b; Schuurmans et al., 2004; Kroll and O’Leary, 2005), or when Ras/ERK signaling is ectopically activated (Chandran et al., 2003; Gabay et al., 2003; Hack et al., 2004; Kessaris et al., 2006; Li et al., 2014) (Figure 2). These events all induce a Neurog2 to Ascl1 transition and drive a dorsal-to-ventral re-specification of NPCs, indicating a lineage bifurcation point regulated by Neurog2 and Ascl1. While Neurog2 and Ascl1 both function as transcriptional activators themselves (Castro and Guillemot, 2011; Kovach et al., 2013), they are mutually transcriptionally cross-repressive; in Neurog2 null mutants, Ascl1 is upregulated and subcortical phenotypes are generated in the cortex (Fode et al., 2000; Schuurmans et al., 2004), while conversely, Ascl1 can repress Neurog2 expression when misexpressed in cortical NPCs (Han et al., 2020). Neurog2 is also required to repress Ascl1 expression in multipotent retinal progenitor cells (Hufnagel et al., 2010). Given that Neurog2 functions as a transcriptional activator, the mechanism for its repression of Ascl1 transcript and protein expression is indirect, and remains to be fully elucidated. Partial features include that Neurog2 acts through a yet unknown transcriptional regulator to repress Etv1 expression, which indirectly regulates Ascl1 expression through repression of Hes5, a known transcriptional repressor of Ascl1 (Kovach et al., 2013). Notably, proneural gene cross-repression in the cortex may be limited to competing lineage determinants such as Neurog2 and Ascl1 (Han et al., 2020), as Neurog2 is instead required to positively regulate the transcription of the functionally related proneural gene, Neurog1 (Fode et al., 2000). However, mutant analyses in the retina revealed that in the absence of Neurog2, Ascl1 or Neurod4, the other two bHLH genes are upregulated (Akagi et al., 2004), indicative of cross-repressive interactions that further support context-specific functions of these genes.
Strikingly, Neurog2 and Ascl1 are also cross-repressive at the functional level; Neurog2 inhibits the ability of Ascl1 to promote a glioblast fate, while Ascl1 inhibits the ability of Neurog2 to specify a glutamatergic neuronal identity in cortical NPCs (Han et al., 2020) (Figure 2). This cross-repression at the protein level may be mediated by the formation of less transcriptionally active Neurog-Ascl1 heterodimers, reviewed in greater detail below. Taken together, these findings invoke comparisons to other stem cell systems in which pairs of TFs that specify different cell fates are in some instances co-expressed in the same progenitor cell, and their mutual cross-antagonism prevents fate specification and differentiation to maintain cellular bi- or multi-potency (Chickarmane et al., 2009; Dillon, 2012; Okawa et al., 2016; Brand and Morrissey, 2020). The co-expression of distinct lineage determinants has the added purpose of ‘priming’ progenitor cells for subsequent lineage selection, as downstream genes in either lineage can be readily transcribed. In the lingo of computational biologists, antagonistic TF pairs form a gene regulatory network motif known as a toggle switch (Huang et al., 2007; Chickarmane et al., 2009; Enver et al., 2009; Zandi et al., 2010; Strasser et al., 2012). Based on these operational criteria, Neurog2 and Ascl1 form a toggle switch to prevent lineage commitment in the embryonic cortex (Han et al., 2020).
Ras/ERK signaling is a critical regulator of the Neurog2-Ascl1 toggle switch, and therefore it is important to understand how this signal transduction pathway is regulated in the embryonic cortex (Li et al., 2014). Ras/ERK signaling is activated by both pro-proliferative growth factors, such as epidermal growth factor (Egf) and fibroblast growth factor (Fgf) (Ghosh and Greenberg, 1995; Vaccarino et al., 1999; Raballo et al., 2000; Lukaszewicz et al., 2002; Imamura et al., 2008; Wang et al., 2009), and by pro-differentiative factors, including platelet-derived growth factor (PDGF) (Menard et al., 2002), nerve growth factor (Ngf) (Greene and Tischler, 1976; Vaudry et al., 2002), neurotrophin 3 (Ntf3) (Lukaszewicz et al., 2002; Ohtsuka et al., 2009), and brain derived neurotrophic factor (Bdnf) (Barnabe-Heider and Miller, 2003; Ito et al., 2003; Medina et al., 2004; Fukumitsu et al., 2006; Bartkowska et al., 2007). Each of these signals bind receptor tyrosine kinase (RTK) receptors. The kinetics of RTK/ERK signaling is critical to its function, in that the apparently divergent effects of RTK/ERK signaling on proliferation versus differentiation are explained by the ability of Ngf/Ntrk1 to activate ERK in a sustained manner, whereas Fgf induces strong, transient ERK activation (Marshall, 1995; York et al., 1998). Mechanistic insights have also been gained into how Fgf activation biases NPCs to acquire an oligodendrocyte fate, both in the telencephalon and spinal cord (Gabay et al., 2003; Furusho et al., 2011; Li et al., 2014; Farreny et al., 2018), where Fgf acts in combination with Shh in an evolutionarily conserved manner (Esain et al., 2010). Mechanistically, downstream activation of ERK directly phosphorylates Ascl1, and higher levels of RAS/ERK activation biases this proneural TF to preferentially transactivate glioblast genes instead of promoting a GABAergic neuronal identity (Li et al., 2014).
During cortical development, activation of Ras/ERK signaling is spatially and temporally regulated, as revealed by the dynamic expression of phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), which is initially detected in the antihem adjacent to the lateral pallium where neurogenesis is first initiated in the cortex (Miyama et al., 1997), before spreading across the VZ by E14.5 (Li et al., 2014). Notably, the expression of pErk1/2 matches the pattern of expression of fibroblast growth factor receptor 3 (FGFR3) and a set of ets-domain transcription factors activated downstream of RTK signaling, including Etv1, Etv4 and Etv5 (Hasegawa et al., 2004; Li et al., 2014). The Etv transcription factors act as downstream effectors of FGF signaling and participate in regulating the Neurog2-Ascl1 toggle switch; Neurog2 indirectly represses Etv1, which in turn indirectly represses Ascl1 as described above (Kovach et al., 2013). Taken together, these studies highlight the multiple points of intersection between the RAS/ERK signal transduction pathways and proneural genes.
Consistent with a role for canonical Wingless/INT (Wnt) signaling in specifying a cortical identity, two transgenic reporters for this pathway, BAT-gal (Maretto et al., 2003) and TCF-lacZ (Liu et al., 2006), are both expressed at higher levels in the dorsal versus ventral telencephalon (Backman et al., 2005; Machon et al., 2005; Li et al., 2012). Upon Wnt binding to LRP/Frizzled receptor complexes, β-catenin (encoded by Ctnnb1) is stabilized and translocates to the nucleus where it forms active transcriptional complexes with Tcf1. Conditional knock-out (cKO) of Ctnnb1 in early cortical NPCs, prior to neurogenesis, downregulates Neurog2 and upregulates Ascl1 expression (Backman et al., 2005). Conversely, the addition of exogenous Wnts allows dissociated dorsal telencephalic chick cells or murine cortical neurospheres, which normally ventralize rapidly (Gabay et al., 2003), to maintain their dorsal identity in vitro (Gunhaga et al., 2003; Machon et al., 2005; Watanabe et al., 2005). Similarly, misexpression of Ctnnb1 in subcortical NPCs induces ectopic Neurog1/2 expression and suppresses Ascl1 (Hirabayashi et al., 2004; Backman et al., 2005). Thus, the Wnt pathway also controls the Neurog2-Ascl1 toggle switch, biasing NPCs toward Neurog2 expression and a cortical cell fate (Figure 2).
Wnt reporter activity drops off dramatically in cortical NPCs in mid-neurogenesis (E15.5-E16.5), correlating with the time when Neurog2 function is attenuated (Backman et al., 2005; Machon et al., 2005; Li et al., 2012). In the absence of Wnts, glycogen synthase kinase (GSK) is activated, forming a destruction complex with axin, APC and other molecules that phosphorylates and targets β-catenin for degradation. GSK3 also directly phosphorylates Neurog2 during mid-late corticogenesis through phosphorylation (Li et al., 2012), which promotes the formation of Neurog2-E47 heterodimers at the expense of more transcriptionally active Neurog2-Neurog2 homodimers (Li et al., 2012). Notably, Neurog2-E47 heterodimers have longer half-lives than Neurog2-Neurog2 homodimers, so their reduced transcriptional activity is not due to enhanced degradation (Li et al., 2012), but rather due to DNA binding preferences, as discussed further below. Therefore, Wnt signaling intersects Neurog2 function at a few levels, not only promoting Neurog2 expression, but also regulating its activity.
Several signaling pathways induce cortical NPCs to differentiate into astrocytes (Stipursky et al., 2012), including: (1) cytokines, such as cardiotrophin-1 (CT-1), leukemia inhibitory factor (LIF), and ciliary neurotrophic factor (CNTF), all of which activate JAK/STAT signaling (Bonni et al., 1997; Koblar et al., 1998; Barnabe-Heider et al., 2005; He et al., 2005); (2) bone morphogenetic proteins (BMPs) (Bonni et al., 1997) and transforming growth factor beta (Tgfb), which function through downstream Smad effector proteins to promote astrocyte maturation (Gross et al., 1996; Bonaguidi et al., 2005); and (3) Notch-Delta signaling, as described above (Gaiano et al., 2000; Ge et al., 2002; Grandbarbe et al., 2003; Kamakura et al., 2004; Wu et al., 2017).
Interestingly, cortical NPCs expressing Neurog1/2 and/or Ascl1 are biased against an astrocytic fate (Han et al., 2020) (Figure 2), with several mechanisms of action identified. Firstly, Neurog1, which declines in expression when astrocyte differentiation begins at E15.5 (He et al., 2005; Han et al., 2018), sequesters transcriptional co-activators (CBP/p300) away from Stat1/3 and Smad1 TFs, preventing the transactivation of downstream astrocytic genes such as GFAP by cytokine and BMP/Tgfb signaling (Sun et al., 2001). Secondly, Neurog1 induces the transcription of miR-9, which downregulates the expression of genes in the JAK/STAT pathway (Zhao et al., 2015). Conversely, signaling pathways promoting astrocytic fate impair the ability of proneural TFs to induce neuronal differentiation. BMP7, which is secreted from the dorsal telencephalic midline (Furuta et al., 1997) induces Id1 or Id2 expression in spinal cord and cortical NPCs (Vinals et al., 2004; Le Dreau et al., 2018). Id proteins inhibit proneural gene function by sequestering E proteins to prevent their heterodimerization with bHLH TFs (Le Dreau et al., 2018). Furthermore, Id1 induced by BMP4 promotes Ascl1 protein degradation to prevent this TF from promoting neuronal differentiation (Vinals et al., 2004).
Another important aspect of astrocyte differentiation is the timing of when NPCs switch from neurogenesis to gliogenesis. Cytokines are critical regulators of this switch (Barnabe-Heider et al., 2005), but the proneural genes are also involved, as gliogenesis occurs precociously in Neurog2–/–;Ascl1–/– cortices (Nieto et al., 2001). Notably, a similar precocious differentiation of glial cells is seen in Neurod4–/–;Ascl1–/– cortices in the tectum, hindbrain and spinal cord (Tomita et al., 2000), suggesting similar processes may be at play in other brain regions. One interpretation of these data is that in the absence of two proneural genes, neurogenesis cannot take place and instead, gliogenesis ensues. Another interpretation is that Neurog2 and Ascl1 regulate temporal identity transitions through the co-dependent activation of a unique set of downstream genes. Consistent with the latter interpretation, Neurog2 and Ascl1 are also together required to regulate the timing of cortical neurogenesis, as evidenced by the precocious differentiation of supragranular neurons (Dennis et al., 2017) in Neurog2–/–;Ascl1–/– cortices (Dennis et al., 2017). Mechanistically, Neurog2 and Ascl1 regulate the timing of cortical neurogenesis as both proteins are required to transactivate Fezf2, a critical component of the de-repression circuit that specifies laminar identities. How these genes regulate the timing of cortical gliogenesis is less clear. A simple competitive model may explain these findings, as highlighted above, with the loss of Neurog2 and Ascl1 preventing the sequestration of transcriptional co-activators away from Stat1/3 and Smad1 TFs. However, the recent identification of a slew of transcriptional targets that are co-bound and co-regulated by Neurog2 and Ascl1 in cortical NPCs may shed new light into this process (Han et al., 2020).
Several studies suggest that Neurog2 and Ascl1 fate specification activities are temporally regulated. For example, Neurog2 is only necessary and sufficient to specify a glutamatergic neuronal identity in cortical NPCs before E14.5 (Schuurmans et al., 2004; Li et al., 2012), whereas it promotes NPC progenitor transitions from aRG to INP (Britz et al., 2006) and neuronal migration (Heng et al., 2008) after E14.5. Ascl1 is also normally expressed in embryonic cortical NPCs (Britz et al., 2006; Han et al., 2020), albeit at lower levels than in subcortical domains, but it does not induce the differentiation of these cells into GABAergic neurons or oligodendrocytes, although it may transactivate oligodendrocyte genes postnatally (Han et al., 2020). Temporally constrained, Ascl1 is upregulated in Neurog2–/– cortical NPCs throughout the neurogenic period but can only respecify these cells to a GABAergic fate before E14.5 (Britz et al., 2006). Temporal changes in Neurog2 cortical function are not surprising, considering that several differences in early and late cortical NPCs have previously been documented. For example, only early, pre-neurogenic cortices respond to the proliferative activity of Wnts (Viti et al., 2003b) and the ventralizing activity of Shh (Kohtz et al., 1998), while conversely, only late-stage NPCs respond to the gliogenic activity of CNTF (Molne et al., 2000; Takizawa et al., 2001; Viti et al., 2003a; Song and Ghosh, 2004). Several non-mutually exclusive molecular regulatory mechanisms controlling Neurog2 and Ascl1 functions in the cortex help explain these confounding findings.
There is a tendency to consider the presence of gene transcripts as an indication that a gene is ‘active’ in a particular cell type, but there are many downstream regulatory events that must also be considered. The first consideration is whether transcripts are translated into proteins. Early studies revealed that Neurog1, Neurog2 and Ascl1 transcripts are present in many more telencephalic cells than the proteins, but the mechanisms of translational control were not elucidated until recently. A ground-breaking study found that a large host of transcribed neuronal differentiation genes are not translated in the developing cortex (Yang et al., 2014). This study showed that eukaryotic initiation factor 4E1 (eIF4E1) and the eIF4E-Binding Protein, 4E-T, components of the eukaryotic translational machinery, form P-body-like complexes that bind proneural bHLH mRNAs to inhibit their translation, a mechanism of translational control critical for controlling the timing of cortical neurogenesis (Yang et al., 2014) (Figure 3A). Since then, many additional proteins have been identified that control the translation of proneural and neural differentiation genes, including other components of the translational machinery and critical RNA binding proteins (Amadei et al., 2015; Zahr et al., 2018, 2019). Future work will be required to identify specific RNA binding proteins that control the stability and translation of proneural gene transcripts.
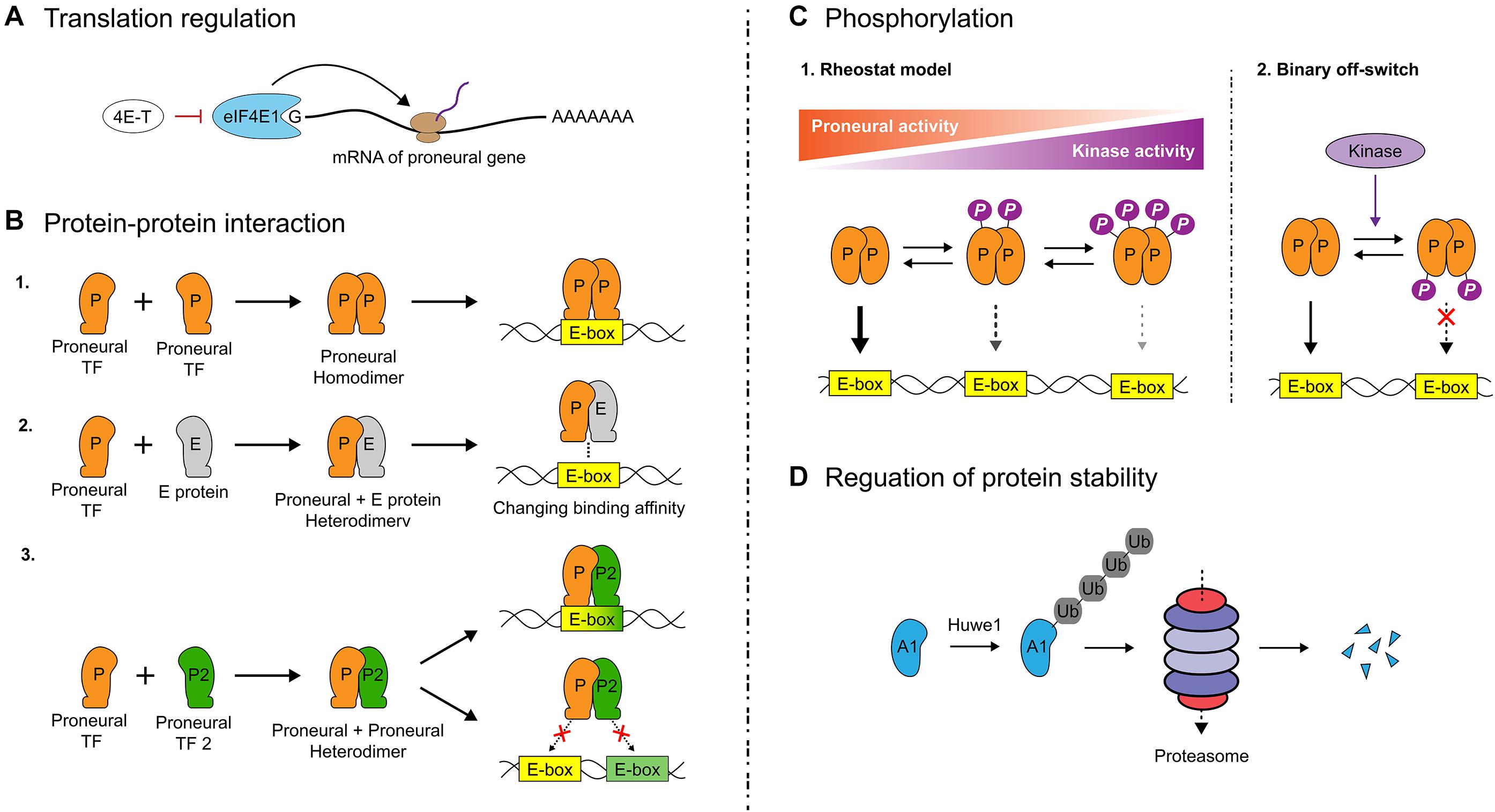
Figure 3. Translational and post-translational regulation of proneural gene function. (A) The timing of cortical neurogenesis is controlled by eukaryotic initiation factor 4E1 (eIF4E1) and eIF4E-Binding Protein 4E-T, components of the eukaryotic translational machinery. These factors bind proneural gene transcripts, inhibiting proneural genes translation. (B) To bind DNA, bHLH TFs must dimerize. Proneural TF homodimers bind to regions with their cognate E-box sequences. Heterodimers with E proteins may enhance proneural TF binding to DNA if the proneural TF and E protein share a preferred E-box motif; otherwise, DNA binding is impaired. Heterodimerization between proneural TFs similarly may enhance or impede DNA binding depending on the E-box content of a target gene. (C) Beyond regulating proneural TF-mediated cell fate choices and neuronal migration, phosphorylation of proneural TFs generally decreases their transcriptional activity. The classical rheostat model holds that progressive phosphorylation of serines or threonines by proline directed serine-threonine kinases (e.g., Cdk, Erk, and Gsk3) at serine-proline (SP) or threonine-proline (TP) sites in proneural TFs decreases their activity depending on the number of sites phosphorylated. More recently, a single conserved residue has been discovered in proneural TFs at the loop/helix-2 junction, which when phosphorylated acts as binary off switch for proneural TF activity, with this mechanism overriding the rheostat mechanism. (D) Proneural TFs have short intracellular half-lives and are degraded through the ubiquitin-proteasome pathway. Recently, the E3 ligase Huwe1 was identified as a critical regulator of Ascl1 stability in the adult V-SVZ, with loss of Huwe1 leading to NPC depletion due to sustained Ascl1 expression inducing continuous neuronal differentiation.
The requirement for dimerization presents numerous opportunities for combinatorial control of bHLH transcriptional activity (Figure 3B). A given bHLH dimer will have its own E-box specificity, and while a comprehensive picture of the differential binding patterns of homo- and heterodimers is not yet known, preferred binding motifs for certain proneural TFs have been discovered in mouse and fish (Seo et al., 2007; Lin et al., 2010; Wapinski et al., 2013; Raposo et al., 2015; Pfurr et al., 2017; Aydin et al., 2019) and certain bHLH dimers are less transcriptionally active than others. As mentioned earlier in this review, Ascl1 preferentially binds to CAGCTG motifs in genomic regulatory regions (Wapinski et al., 2013; Raposo et al., 2015; Aydin et al., 2019), while Neurog1 and Neurog2 preferentially bind CADATG motifs (where D = A/G/T) (Seo et al., 2007; Madelaine and Blader, 2011; Aydin et al., 2019). E47 has been shown to preferentially bind CAGSTG motifs (where S = C/G) (Lin et al., 2010; Pfurr et al., 2017). Recently it has been shown that E proteins alter the neurogenic strength of proneural TFs through physical interactions in a context-specific, E-box-dependent manner, by either synergizing with Ascl1 on CAGSTG motifs or impeding Neurog2’s binding to CADATG motifs (Le Dreau et al., 2018). For example, misexpression of E47 and Ascl1 in spinal cord NPCs increases differentiation relative to Ascl1 alone (Le Dreau et al., 2018), but the opposite effect is observed for co-electroporation of E47 with Neurog2, either in spinal (Le Dreau et al., 2018) or cortical (Li et al., 2012) NPCs. However, there are other regulatory considerations, including that E47 heterodimerization enhances Neurog2 (Li et al., 2012) and Ascl1 (Vinals et al., 2004) protein stability, which can influence their transactivation of some target sites. The finding of E-box-dependent cooperativity of E proteins with proneural TFs leads us to the important consideration that proneural TF activity at downstream targets is modulated by the availability of appropriate dimerization partners. Indeed, in the chick spinal cord, when E protein availability is limited due to its sequestration by Id proteins, Ascl1 proneural strength is negatively impacted due to the reduction in Ascl1∼E47 heterodimers, which effectively transactivate CAGSTG E-box-containing downstream targets (Le Dreau et al., 2018). Conversely, Neurog2 transactivation of CADATG E-box-containing downstream targets is stabilized by a reduction in E47 availability due to Id sequestration (Le Dreau et al., 2018).
Heterodimers can also form between two proneural bHLH TFs. Examples include Neurog1∼Neurog2 heterodimers, which form in E12.5 cortical NPCs (Han et al., 2018) and are likely functionally important, as evidenced by the reduced Hes5 expression (i.e., Notch signaling) and precocious neurogenesis that occurs in E12.5 Neurog1–/– cortices (Schuurmans et al., 2004; Han et al., 2018). Mechanistically, Neurog1∼Neurog2 heterodimers have a reduced capacity to induce neurogenesis compared to Neurog2∼Neurog2 homodimers, leading to the conclusion that Neurog1 slows the pace of cortical neurogenesis at early stages (∼E12.5) when there are higher levels of Neurog1/Neurog2 co-expression (Han et al., 2018). Interestingly, co-expression of Neurog2 with Neurod4 accelerates cortical neurogenesis (Mattar et al., 2008), and while protein–protein interactions were not assessed, it remains possible that either Neurog2∼Neurod4 heterodimers have an enhanced capacity to transactivate target genes, or these bHLH TFs form other dimers that bind to distinct E-boxes located in the same regulatory regions of a target gene. Notably, there are other examples of bHLH proneural genes and differentiation genes having co-operative functions, including in the developing retina (Akagi et al., 2004) and hippocampus (Schwab et al., 2000).
Neurog2∼Ascl1 heterodimers have also been identified (Gradwohl et al., 1996; Han et al., 2020), with evidence suggesting that they are non-functional, as predicted by the differential enrichment of bound E-box motifs for each TF identified in ChIP-seq experiments (Aydin et al., 2019). Indeed, co-expression of Neurog2 and Ascl1 blocks the transactivation of promoters specific to each TF in vitro in transcriptional reporter assays, as well as blocking the ability of Ascl1 to induce in vivo proliferation and Sox9 expression (a glioblast marker in cortical NPCs) as well as the ability of Neurog2 to induce in vivo glutamatergic neuron formation (Han et al., 2020). Despite the inhibitory interactions between Neurog2 and Ascl1, and while most of the genes activated by Neurog2 and Ascl1 do not overlap, there are also some commonly regulated genes (Masserdotti et al., 2015; Aydin et al., 2019). Moreover, the gene regulatory network (GRN) that is associated with Neurog2/Ascl1 double+ cortical NPCs is distinct from the GRNs associated with single+ NPCs. Neurog2 and Ascl1 could regulate a distinct repertoire of genes when in combination through two potential modes of action. Firstly, they could act on gene regulatory elements that contain both Neurog2- and Ascl1-specific binding sites, as exemplified by Dll1, which has two distinct enhancers that are specifically activated by Neurog2 (DeltaN) or Ascl1 (DeltaM) (Castro et al., 2006). Alternatively, enhancers may contain hybrid E-boxes that are bound equally well by Neurog2 and Ascl1, including when they form heterodimers, as exemplified by the Dll3 promoter (Henke et al., 2009). Further studies on commonly regulated targets of Neurog2 and Ascl1 will aid our understanding of how cortical NPC fate decisions are regulated.
Finally, proneural TFs also form physical interactions with other non-bHLH TFs that play critical roles in cortical development. For example, Neurog2 synergizes with the T-box TF Tbr2, expressed in INPs, to control the radial migration of cortical neurons (Sessa et al., 2017). Neurog2 and Tbr2 control migration by synergistically transactivating Rnd2, a critical regulator of cortical neuron migration (Sessa et al., 2017). Other TFs have also been shown to associate with proneural TFs to regulate their functions. For instance, Ascl1 and the POU domain TFs Brn1 and Brn2 cooperatively bind the Dll1 promoter (Castro et al., 2006). Similarly, in the fly, senseless cooperates with atonal to regulate proneural activity (Nolo et al., 2000), and Myt1 is required for optimal Neurog2 proneural activity in Xenopus (Quan et al., 2004). The future identification of additional proneural TF binding partners in cortical NSCs/NPCs will aid in our understanding of the complex GRNs that underlie development of this brain region.
Intracellular kinases are key intermediaries between the environment and the cell nucleus, so understanding their impact on proneural TFs can reveal how environmental cues regulate cortical neurogenesis. Interestingly, DNA-binding proteins and TFs are often natively unfolded and intrinsically disordered (Ward et al., 2004), with disordered regions targeted by twice as many kinases as structured domains (Gsponer et al., 2008). Neurog2 is an example of an intrinsically disordered TF that is targeted by various kinases that modulate its activity in a context-dependent manner (McDowell et al., 2014). In general, N- and C-terminal phosphorylation outside of the bHLH domain has inhibitory effects on bHLH proneural activity, but other processes can be promoted by phosphorylation, as highlighted below.
The rheostat model holds that progressive phosphorylation of TFs results in a graded, finely tuned reduction in DNA binding and hence, transcriptional activity (Pufall et al., 2005) (Figure 3C). This model has garnered support with regards to proneural TFs from experimental work in mouse and xenopus (Ali et al., 2011; Hindley et al., 2012; McDowell et al., 2014; Hardwick and Philpott, 2015). Proneural TFs are phosphorylated by a host of proline-directed serine/threonine (S/T) kinases, including cyclin-dependent kinases (Cdk – on Neurog2 and Neurod4) (Ali et al., 2011; Hindley et al., 2012; McDowell et al., 2014; Hardwick and Philpott, 2015), GSK3 (on Neurog2) (Li et al., 2012) and ERK (on Ascl1) (Li et al., 2014). These S/T kinases can progressively phosphorylate nine serine-proline (SP) sites in Neurog2, six SPs in Ascl1 and a combined seven threonine-proline (TP) and SP sites in Neurod4. In xenopus, the progressive phosphorylation of Neurog2 SP phosphoacceptor sites by Cdks limits its ability to drive neurogenesis, with the number of serine-proline sites phosphorylated more important than their location (McDowell et al., 2014). Based on the “cell cycle length hypothesis,” NPCs that differentiate have a longer G1 phase, and the prediction is that Cdk activity would be reduced in these cells so that Neurog2 would be underphosphorylated, thereby in a permissive state to initiate transcription of neurogenesis-associated target genes (Calegari and Huttner, 2003). Conversely, Cdk levels would rise in dividing NPCs, increasing proneural TF phosphorylation, and inhibiting transactivation of downstream gene (Ali et al., 2011). Notably, Cdk inhibits Neurog2-mediated transactivation of Neurod1, a neuronal differentiation gene, more robustly than Dll1, which induces neighboring NPCs to proliferate (Hindley et al., 2012), suggesting Cdk plays a critical role in regulating neural development. Accordingly, in the developing cortex, the proneural competence of Neurog2 also declines during late neurogenesis due to increasing levels of GSK3-mediated phosphorylation (Li et al., 2012).
Phosphorylation by SP kinases not only controls the decision to proliferate or differentiate, but also influences cell fate choices that are important in normal development but can also impact tumor formation. In the spinal cord, phosphorylation of Neurog2 S231 and S234 (SP sites) promotes the formation of TF complexes between Neurog2 and the adaptor protein Ldb1, which recruits LIM-homeodomain TFs Isl1 and Lhx3 to form a complex that transactivates motor neuron specific genes (Ma et al., 2008). Similarly, in the embryonic telencephalon, intermediate vs high RAS/ERK activation levels dictate whether Ascl1 selects GABA vs OPC transcriptional targets, respectively (Li et al., 2014) (Figure 2B). Notably, there is also a correlation between higher pERK levels and more glial cells in pilocytic astrocytomas, compared to lower levels of pERK and fewer glial cells in ganglioglioma, despite these two tumor types sharing the same bRAFv600e mutation (Li et al., 2014). It is interesting to speculate that ERK-mediated phosphorylation of ASCL1 controls, at least in part, the different cellular features of these genetically similar tumors. Similarly, the tumorigenicity of bHLH TF Olig2 is driven by its phosphorylation status, with phosphomimetic mutations rendering it more tumorigenic, and phospho-dead mutations non-tumorigenic (Sun et al., 2011). In line with this, phosphorylation of a conserved triple serine motif in Olig2 promotes its unorthodox ‘antineural’ pro-proliferative functions, instead of the ‘proneural-like’ activity of inducing an oligodendrocyte fate (Sun et al., 2011). These data highlight the importance of phosphorylation events of bHLH TFs not only for normal development, but also in tumorigenesis.
A single conserved S/T residue at the Loop/Helix 2 (L-H2) junction acts as an evolutionarily conserved, binary ‘off’ switch for both vertebrate and invertebrate proneural TFs (Quan et al., 2016) (Figure 3C). 3D modeling revealed that the conserved S/T residue faces the DNA backbone such that addition of a negatively charged phosphate group would generate electrostatic repulsion between the TF and DNA, effectively rendering the TF a null mutant. At the L-H2 junction, Drosophila ato and vertebrate Atoh1 are phosphorylated on S292 by protein kinase A (PKA), while Neurog2 is phosphorylated on T149 by MARK1 and PLK1. A phosphomimetic mutation (T149D) destabilized Neurog2 binding to DNA and abolished its ability to induce neurogenesis in cortical NPCs in vivo (Quan et al., 2016). Strikingly, this binary off-switch essentially ‘trumps’ the rheostat model of control, as introduction of a single phosphomimetic mutation in the conserved L-H2 region of Ascl1 and Neurog2 prevents their proneural activities, even when ‘activating’ phospho-null mutations are introduced in SP and TP sites throughout the proteins (Hardwick and Philpott, 2018a, b). The speculation that these different regulatory modes may come into play at different developmental time points depending on an NPC’s ‘kinase environment’ is interesting due to the possibility of rapidly halting proneural activity to ensure that correct neuronal numbers are generated (Hardwick and Philpott, 2018a).
Neurog2 is also phosphorylated on Tyr241, a residue outside the bHLH domain that is dispensable for proneural activity, but required to specify a polarized neuronal phenotype and establish appropriate radial migration patterns (Hand et al., 2005). Mutation of Y241 leads to defects in neuronal migration and neuronal morphogenesis defects in the neocortex (Hand et al., 2005), in part by preventing the association between Neurog2 and CBP, a transcriptional co-activator protein that is required for Neurog2 to transactivate genes that control neuronal migration and dendritic polarity, such as Dcx (Ge et al., 2006). Notably, the ability of Neurog2 to sequester CBP is also proposed to be important for the indirect repression of RhoA, which must be downregulated for cortical neurons to migrate appropriately (Ge et al., 2006).
It is now well established that both fly (Kiparaki et al., 2015) and vertebrate (Nguyen et al., 2006; Ali et al., 2011; Li et al., 2012; Kovach et al., 2013) proneural TFs have very short intracellular half-lives (∼20–40 min). In the fly, two destabilizing motifs were found in the proneural TF encoded by scute (Sc); the transactivation domain (TAD) and an SPTSS motif, including a phosphoacceptor site for proline-directed S/T kinases (Kiparaki et al., 2015). Notably, S-A mutations in the SPTSS motif dramatically stabilized fly Sc (Kiparaki et al., 2015), and similarly, there was an ∼2-fold increase in Neurog2 stability when all 9 SP sites were mutated to SA in Xenopus (Ali et al., 2011). Removal of the C-terminal TAD domain also dramatically stabilizes murine Neurog2 (Li et al., 2012) and fly Sc (Kiparaki et al., 2015) proteins. However, while the forced tethering of mouse Neurog2 to E47 (Li et al., 2012), or E12 to human ASCL1 (Sriuranpong et al., 2002) stabilizes these proneural TFs, Sc heterodimerization with fly Daughterless (Da), the E-protein homolog, promotes further degradation (Kiparaki et al., 2015). Thus, there are critical differences in how proneural protein stability is regulated, but nevertheless, in all species, proneural TFs have short intracellular half-lives.
There is growing evidence that proneural protein degradation is mediated by the ubiquitin-proteasome degradation system (UPS) (Figure 3D). Ubiquitin moieties form isopeptide bonds with lysine residues in substrate proteins that are targeted for degradation through the actions of three enzymes; ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes (Komander and Rape, 2012). Polyubiquitylated substrates undergo degradation through the UPS (Johnson et al., 1995). Proneural TFs regulated by UPS include murine (Vinals et al., 2004) and human (Sriuranpong et al., 2002) Ascl1, xenopus Neurog2 (Vosper et al., 2007, 2009) and the fly protein Sc (Kiparaki et al., 2015). Recently, the E3-ligase Huwe1 (HECT, UBA and WWE domain containing 1) was identified as a critical destabilizer of Ascl1 in the postnatal hippocampus (Urban et al., 2016) (Figure 3D). Huwe1 maintains adult NSCs in quiescence by targeting Ascl1 for degradation, with excess NSCs entering the cell cycle upon conditional Huwe1 deletion, resulting in a depletion of the NSC pool (Urban et al., 2016). Spatial resolution is also emerging in the picture of Ascl1 regulation by Huwe1 (Gillotin et al., 2018). Cytoplasmic Ascl1 is predominantly attached to longer polyubiquitin chains on lysines within the bHLH region and is rapidly targeted for degradation by the UPS, while chromatin-bound Ascl1 is ubiquitylated with shorter chains on N-terminal and bHLH lysines but is not targeted for degradation (Gillotin et al., 2018).
While comparable E3-ligases have yet to be identified for Neurog1 and Neurog2 in the cortex, Fbxo9 is an E3-ligase that destabilizes Neurog2 in the developing dorsal root ganglia (Liu et al., 2020). Mechanistically, in Xenopus, Neurog2 is stabilized by Cdk inhibitor p27Xic1 (Nguyen et al., 2006), and p27Xic1 promotes neurogenesis partially due to its stabilizing effect on Neurog2 (Vernon et al., 2003), but whether there is a direct involvement with UPS is not known.
Transcription factors that act as cell fate determinants generally transactivate lineage-specific target genes only in certain cellular contexts (Gascon et al., 2017). For instance, the glutamatergic neuronal fate-specifying properties of Neurog2 are temporally restricted; in the embryonic cortex, Neurog2 only efficiently induces neurogenesis before E14.5 (Li et al., 2012). Regional restrictions also occur, with Neurog2 efficiently able to induce neurogenesis in the dorsal and not ventral telencephalon (Mattar et al., 2008). Proneural genes have also emerged as critical architects of neuronal reprogramming (Wilkinson et al., 2013). However, in keeping with their tight contextual regulation, they are not active in all cell types; Ascl1 is a potent neuronal reprogramming factor in fibroblasts (Vierbuchen et al., 2010; Caiazzo et al., 2011; Kim et al., 2011; Pang et al., 2011; Pfisterer et al., 2011; Son et al., 2011), hepatocytes (Marro et al., 2011), cardiomyocytes (Chuang et al., 2017), astrocytes (Rivetti di Val Cervo et al., 2017) and pluripotent stem cells (Yang et al., 2017), and not in the adult neocortex (Grande et al., 2013), hippocampus or spinal cord (Ohori et al., 2006; Jessberger et al., 2008). Conversely, Neurog2 has a more limited ability to convert astrocytes to neurons (Grande et al., 2013; Gascon et al., 2016, 2017; Russo et al., 2020; Stricker and Gotz, 2020) and is used less often for neuronal reprogramming as it must be combined with other signals to become a potent lineage converter (Gascon et al., 2016; Russo et al., 2020).
Understanding how the lineage determination activities of Neurog2 and Ascl1 are restricted requires an understanding of how they interact with factors that remodel chromatin. In the field of cellular reprogramming, it is widely held that epigenetic regulators act as ‘gatekeepers’ to prevent cells from transiting from one cell fate to another other organisms, controlling genome accessibility to lineage-specifying TFs (Tursun et al., 2011; Cheloufi et al., 2015; Gascon et al., 2017). Notably, chromatin structure, and hence the accessibility of promoters/enhancers, changes during cortical development (Kishi et al., 2012). Moreover, even within the cortical NPC pool at a single age, there are distinct NPC populations defined as Neurog2/Ascl1 negative, single+ or double+ NPCs that each have distinct chromatin landscapes (Han et al., 2020). Below we review how proneural bHLH TFs intersect with chromatin modifiers to influence the genome architecture, ultimately affecting their ability to bind and transactivate target genes.
Ascl1- and Neurog2 form homo- or hetero-dimers with other bHLH proteins to bind specific E-box motifs in the genome. Neurog2 and Ascl1 are termed ‘pioneer factors’ based on their ability to bind ‘closed’ (nucleosome-bound) chromatin and facilitate the opening of these sites for TF binding (Wapinski et al., 2013; Aydin et al., 2019) (Figure 4A). leading to the opening of distinct chromatin regions which render downstream differentiation genes accessible (Aydin et al., 2019). Several studies have begun to unravel how Neurog2 and Ascl1 influence the chromatin landscape through interactions with different epigenetic modifiers and pathways, as summarized below:
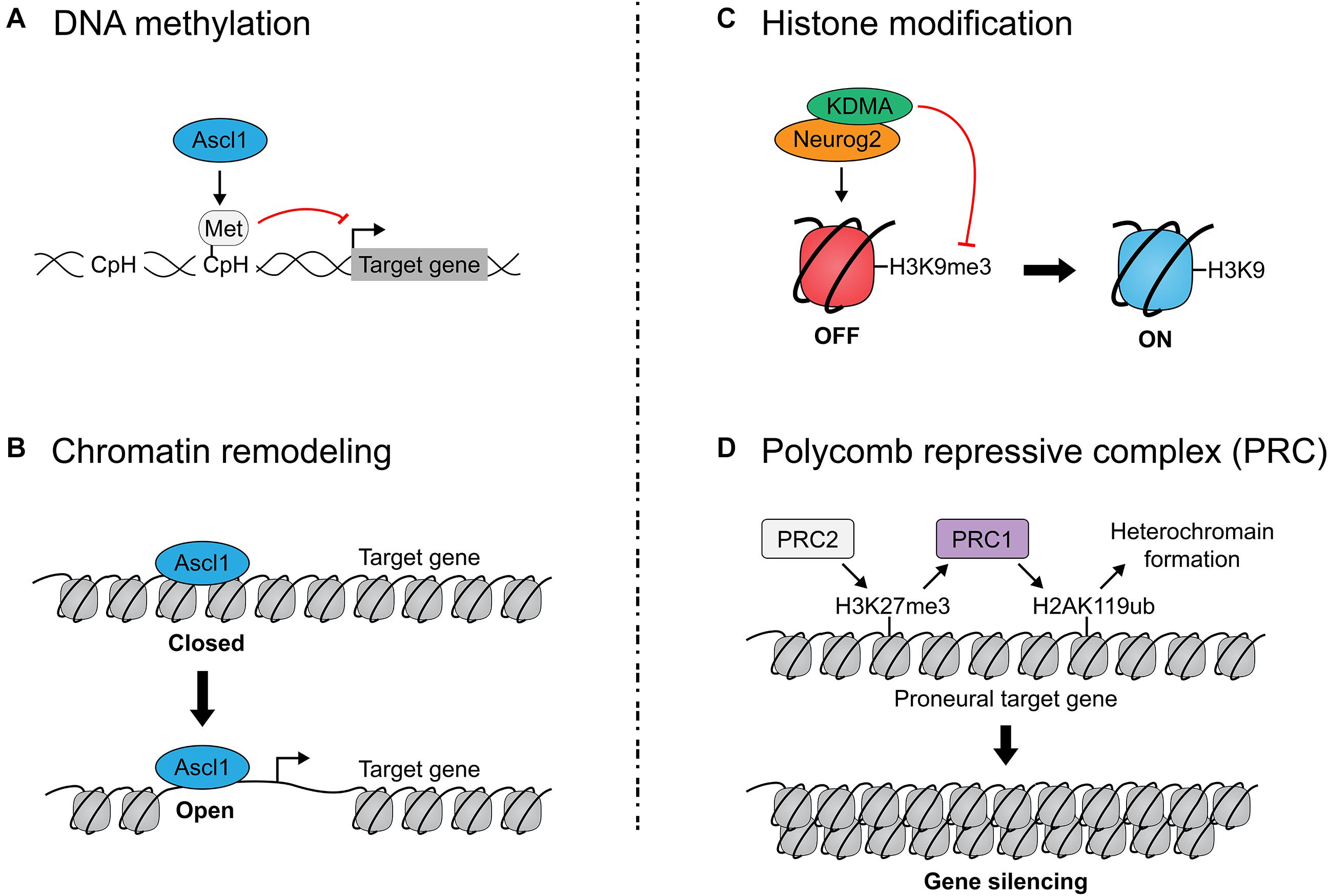
Figure 4. Epigenetic regulation of proneural transcription factor activity. (A) Ascl1 induces non-CpG (CpH) methylation of fibroblast-specific genes during reprogramming of mouse embryonic fibroblasts to neurons. DNA methylation represses gene expression by compacting the genome and rendering it inaccessible. (B) Ascl1 is a pioneer factor which can bind nucleosome-bound DNA (in a ‘closed’ state) and facilitate the de-compacting of these sites to permit gene expression. Neurog2 has also been identified as a pioneer factor. (C) Neurog2 interacts with the H3K9 demethylase to open chromatin and promote neurogenesis. (D) Polycomb repressive complexes (PRC) close chromatin to render target genes inaccessible to proneural TFs. PRC1 ubiquitylates K119 in Histone H2A, while PRC2 methylates Lys27 in Histone H3, altogether leading to heterochromatin formation. Mutation of PRC1/2 genes prolongs proneural TF expression, implicating PRC1/2 in the temporal regulation of proneural TF activity.
DNA hypermethylation of cytosine residues in CpG sequences, which are typically found in promoters/enhancers, represses gene expression by compacting the genome and rendering it inaccessible to TF binding. By changing DNA methylation patterns, chromatin accessibility is altered, as are downstream gene expression patterns. Notably, DNA can also be methylated on non-CpG sequences (CpH) that are prevalent throughout the genome, including in non-regulatory regions, but these modifications similarly repress gene expression (Guo et al., 2014). Strikingly, in a direct neuronal reprogramming study in which Ascl1 was overexpressed in mouse embryonic fibroblasts (MEFs), Ascl1 induced CpH methylation of fibroblast-specific genes, but cooperation with other TFs (Brn2, Myt1l) (Wapinski et al., 2013), which increase reprogramming efficiency, was required for methylation patterns to faithfully recapitulate those seen in cortical neurons (Luo et al., 2019) (Figure 4B).
Ascl1-mediated trans-differentiation of MEFs to neurons showed that Ascl1 induces widespread chromatin remodeling (Wapinski et al., 2017). During this switch, there is substantial Ascl1-driven genome-wide remodeling of chromatin architecture near Ascl1 binding sites, leading to a stabilized nucleosome configuration at day 5 which facilitates the stable expression of mature neuronal genes. Notably, the presence of a swift and concerted chromatin switch in this trans-differentiation protocol contrasts with the classical ‘step-wise’ view of in vivo neuronal development and iPSC reprogramming, emphasizing the limitations of using direct somatic cell reprogramming to model development. In addition, Neurog1 interacts with Brg1, a SWI/SNF chromatin remodeler to aid neurogenesis (Seo et al., 2005).
bHLH TFs act as lineage determinants in multiple tissues, and include Myod1, a master regulator of a skeletal muscle fate (Lee et al., 2020). Strikingly, when misexpressed in MEFs, Ascl1 and Myod1 bind very similar target genes, but to a different degree, and can only induce chromatin opening of sites involved in neuronal and muscle lineage reprogramming, respectively (Lee et al., 2020). However, when Myod1 is overexpressed with Myt1l, which inhibits the acquisition of a muscle identity, Myod1 can induce neuronal differentiation (Lee et al., 2020).
A common property of TFs that act as transactivators is their association with co-activator proteins that function as histone acetyl transferases (HATs), as exemplified by p300/CBP (Sheikh, 2014). p300/CBP preferentially acetylates lysine residues (K) on histone H3/H4 tails (e.g., H3K27ac), and ‘opens’ chromatin by electrostatic repulsion between negatively charged acetyl groups and DNA. Conversely, histone deacetylases (HDACs) remove these acetyl groups and compact the chromatin. Several papers have documented associations between the proneural bHLH genes and HAT co-activators (Koyano-Nakagawa et al., 1999; Sun et al., 2001; Vojtek et al., 2003; Seo et al., 2005, 2007; Ge et al., 2006). The proneural genes are also thought to indirectly repress gliogenesis by sequestering HAT proteins away from gliogenic genes (Sun et al., 2001; Ge et al., 2006). Interestingly, the bHLH protein Hes1 switches from binding a TLE-HDAC co-repressor complex to HAT binding as neurogenesis proceeds (Ju et al., 2004), highlighting the importance of dynamic interactions between these factors in regulating the timing of neurogenesis.
Opening of the chromatin to provide access to TF binding is also associated with methylation of H3 histone tails, but here the specific lysine (K) residues are critical. For instance, while RNAPol II and trimethylation (me3) of histone H3K4 cluster at transcription start sites of actively transcribed genes, and histone H3K36me3 is in the body of actively transcribed genes, other chromatin marks are found in silenced regions of the genome (H3K9me3, H3K27me3) (Ringrose et al., 2004; Bernstein et al., 2006; Pavri et al., 2006; Sims and Reinberg, 2006; Muller and Verrijzer, 2009; Tavares et al., 2012). Notably, Neurog2 forms a complex with KDMA, an H3K9 demethylase, to open chromatin and promote neurogenesis (Lin et al., 2017) (Figure 4C). Additionally, Tbr2 physically interacts with JMJD3 histone demethylase, upregulating neuronal-specific genes when co-expressed, potentially by directing JMJD3 to remove repressive H3K27me3 marks (Sessa et al., 2017). Interestingly, Neurog2 physically associates with and shares a majority of its bound genomic target sequences with Tbr2 and acts synergistically on shared target genes in equimolar quantities, so perhaps Neurog2 interacts with and directs JMJD3 activity as well (Sessa et al., 2017).
Only a handful of epigenetic gatekeepers are known; most influence chromatin structure, some via interactions with Polycomb group (PcG) proteins, which close chromatin (Tursun et al., 2011; Cheloufi et al., 2015; Gascon et al., 2017). PcG proteins modify chromatin to confer transcriptional repression and exist in two repressive complexes: PRC1 (Ring1a, Ring1b, etc.) and PRC2 (Eed, Suz12, Ezh1/2, etc.). PRC1 catalyzes the ubiquitylation of K119 in Histone H2A (H2AK119ub), while PRC2 catalyzes the methylation of Lys27 in Histone H3 (H3K27me3), altogether leading to downregulation of nearby genes (Figure 4D).
PRC1 and PRC2 control temporal NPC fate competence by regulating proneural gene expression and proneural TF target gene availability (Figure 5). PRC1 controls the temporal window of Neurog1, but not Neurog2, expression, with mutation of Ring1b extending Neurog1 expression into late-stage neurogenesis (e.g., E17) (Hirabayashi et al., 2009). Extending Neurog1 gene expression and derepressing the expression of neuronal lineage genes, Ring1B and Ezh2 deletions also delayed the onset of gliogenesis, indicating that PRC1 and PRC2 control the neurogenic-to-gliogenic fate switch in cortical NPCs (Hirabayashi et al., 2009). Recently, Ring1b was also shown to control the spatial expression pattern of Neurog1, with Ring1b deletion expanding Neurog1 expression further ventrally to overlap with Ascl1 expression in the murine E10 telencephalon (Eto et al., 2020). Surprisingly, while Neurog1/2 have been shown to repress Ascl1 expression in the cortex, establishing mutually exclusive expression patterns, Ring1b deletion markedly increased Neurog1/Ascl1 double+ NPCs, suggesting that PcG proteins may also regulate mutual exclusivity of proneural TF expression (Eto et al., 2020). Ring1b has also been shown to temporally limit the production of subcerebral neurons by NPCs through repression of Fezf2 expression (Morimoto-Suzki et al., 2014), which is a known transcriptional target of Neurog2. Furthermore, PRC2 components Eed and Suz12 and PRC1 components Ring1a and Ring1b suppress the ability of Neurog2 to induce a corticothalamic differentiation program by repressing downstream target, Foxp2 (Oishi et al., 2020). In this way, PRC1 and PRC2 progressively regulate targets of proneural TF-induced differentiation programs to control NPC temporal fate competence.
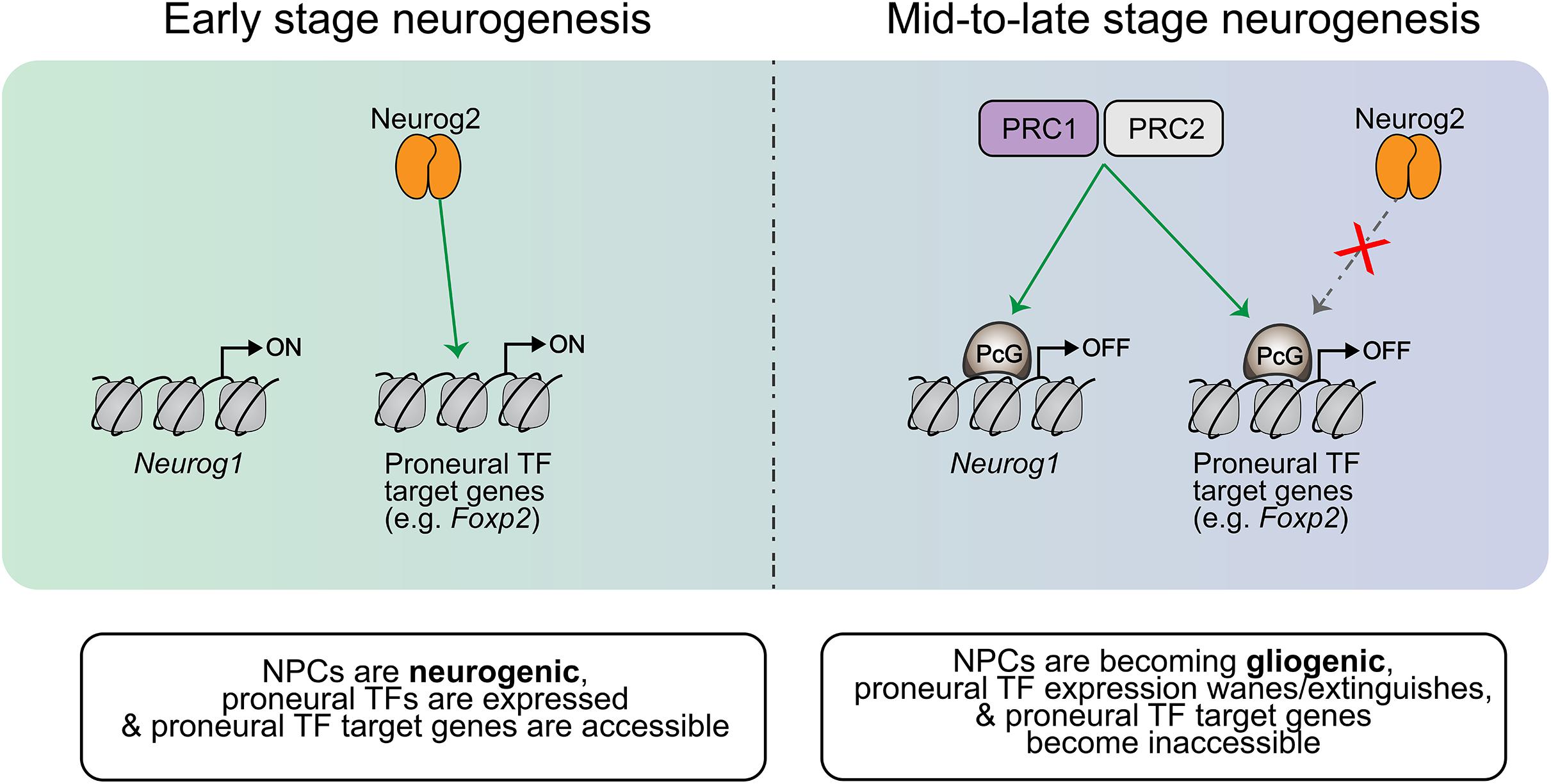
Figure 5. Temporal regulation of proneural gene expression and target gene availability. In early NPCs, Neurog1 expression is turned on and proneural TF target genes are available (not repressed). As the cortical neurogenic period progresses, PRC1/2 catalyze the addition of PcG proteins to repress certain genes which are part of the neuronal differentiation cascade. For example, PRC1/2 repress Foxp2 in mid-neurogenesis, making it inaccessible to Neurog2 and thereby terminating the Neurog2-mediated specification of corticothalamic neurons past this period. This PRC1/2 activity thus controls the temporal competence of NPCs, dictating the differentiation programs that may be initiated at different stages of development. PRC1/2 also extinguish the expression of proneural TFs like Neurog1 later on in the neurogenic period, and this contributes to the induction of gliogenic differentiation programs within NPCs as Neurog1 has been shown to prevent the induction of such programs.
Recent RNA-Seq evidence from FlashTag-isolated apical NPCs further substantiates the claim that PRC2 temporally regulates NPC competence, with mutation of Eed (inactivation of PRC2) leading to precocious neurogenesis (Telley et al., 2019). Eed mutants further displayed precocious generation of typically late-born neurons and increased cell cycle exit in early neurogenesis, leading to a terminal decrease in cortical thickness (which indicates a shorter neurogenic period) (Telley et al., 2019). Similarly, the PRC2 catalytic component Ezh2 was shown to control developmental rate, with loss of Ezh2 shifting NPCs toward differentiation over proliferation as well as an earlier induction of gliogenesis (Pereira et al., 2010).
Overall, these findings implicate PcG proteins as crucial temporal regulators of proneural transcriptional programs, operating to limit the timeframe of subtype specification by proneural TFs by occluding proneural TF target genes. These critical epigenetic regulators do not act alone, though; temporal NPC fate specification is simultaneously and robustly regulated by other mechanisms such as declining proneural transcriptional activity.
The global metabolic changes associated with neuronal differentiation have only recently begun to be elucidated (Agostini et al., 2016), and new work is emerging investigating the metabolic regulators of Neurog2- or Ascl1- driven direct neuronal conversion strategies in both human and mouse cells. As we have expounded in this review, proneural TF function is context dependent, and thus the efficiency at which proneural TFs induce neuronal conversion must also be contingent on the bio-energetic differences of starting cell types in conversion protocols. Indeed, it has been shown that oxidative stress (Gascon et al., 2016) and fatty acid B-oxidation (Russo et al., 2020) impose major hurdles in the reprogramming of astrocytes, which rely on glycolytic metabolism (McKay et al., 1983; Tsacopoulos and Magistretti, 1996), to neurons, which rely on oxidative metabolism (Herrero-Mendez et al., 2009). Thus, genetic or pharmacological manipulations to reduce oxidative stress, such as Bcl-2 overexpression or the addition of forskolin, vitamin E, and calcitriol, increases the efficiency (both the speed and number of converted cells) of proneural TF-driven neuronal conversion (Gascon et al., 2016). Bcl-2 functions independently of its canonical anti-apoptotic role to reduce reactive oxygen species (ROS) and facilitate more efficient proneural TF-driven fate transitions; hence, co-transduction with Ascl1 or Neurog2 significantly improves astrocyte-to-neuron conversion efficiencies and neuronal maturation in vitro, as well as in vivo in injured mouse cortex (Gascon et al., 2016). Interestingly, microarray analysis of Ascl1-transduced MEFs revealed that forskolin treatment enriched BMP and Wnt signaling pathway genes (Gascon et al., 2016), both of which influence proneural TF expression, though it is unknown if the reprogramming enhancement seen in this study is caused by direct effects on proneural TF function or by creating a more permissive cellular environment for reprogramming to occur. Further underscoring the importance of bioenergetics in the regulation of proneural TF function, one-fifth of the mitochondrial proteome differs between astrocytes and cortical neurons, and CRISPRa-mediated induction of neuronal, but not astrocytic, mitochondrial proteins enhances the efficiency of Ascl1-driven reprogramming (Russo et al., 2020). The most robust enhancement of Ascl1-driven reprogramming occurs with co-transduction of neuronal-specific antioxidant protein Sod1, leading to increases in recruitment of cells for reprogramming, speed of conversion into neurons, and lifespan of converted neurons (Russo et al., 2020). These new findings add an important layer of complexity to the web of proneural TF regulation, and when considered together with the other regulatory mechanisms detailed in this review, will allow for the design of more efficient proneural TF-driven neuronal reprogramming protocols in the future.
Proneural TFs are critical regulators of neural cell differentiation and subtype specification, contributing to the enormous cellular diversity observed in the cortex. It therefore holds that proneural TFs are themselves tightly regulated, and these interplaying mechanisms of regulation are being elucidated by the scientific community. A coherent picture of robust regulation is emerging, one with overlapping mechanisms that limit proneural TF actions to certain temporal windows in development. Altogether, the current literature suggests a temporal sequence of regulation as follows. Initially, proneural gene transcript expression is induced by early morphogenetic signals and modulated in undifferentiated NPCs by antagonistic Notch signaling and by other cross-repressive proneural TFs. In NPCs where proneural gene transcript expression is permitted, translation may still be prevented by repressive eIF4E-4E-T complexes. Once proneural gene expression becomes sustained in Notch(-) NPCs, proneural TFs may then be translated into TFs which are regulated in their stability and transcriptional activity by different dimerization partners and by phosphorylation of different residues within their bHLH, N- and C-terminal domains. When the actions of a proneural TF are no longer needed, the protein may be degraded proteolytically by polyubiquitylation (for which exact enzymatic candidates are emerging), or by broader inhibition at the epigenetic level (with mechanisms like repressive PcG proteins limiting proneural TF access to downstream targets).
In the future, the ultimate application of the multifaceted picture of proneural gene regulation that we have painted in this review would aid in the refinement and improvement of neuronal reprogramming strategies incorporating these proneural genes for regenerative medicine purposes.
A-MO and CS: conceptualization. A-MO, SH, and CS: writing—review and editing. A-MO and SH: artwork. CS: funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by operating grants to CS from the Canadian Institutes of Health Research (CIHR PJT – 162108) and a Canada-Israel Health Research Initiative, jointly funded by the CIHR, the Israel Science Foundation, the International Development Research Centre, Canada, and the Azrieli Foundation (IDRC 108875).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
A-MO was supported by a Canada Graduate Scholarship – Master’s NSERC Studentship. SH was supported by Scholarships from the University of Calgary Cumming School of Medicine, Ontario Graduate Scholarship, University of Toronto Vision Science Research Program (VSRP), Peterborough K.M. Hunter Charitable Foundation, and a Margaret and Howard GAMBLE Research Grant. CS holds the Dixon Family Chair in Ophthalmology Research.
Agostini, M., Romeo, F., Inoue, S., Niklison-Chirou, M. V., Elia, A. J., Dinsdale, D., et al. (2016). Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 23, 1502–1514. doi: 10.1038/cdd.2016.36
Akagi, T., Inoue, T., Miyoshi, G., Bessho, Y., Takahashi, M., Lee, J. E., et al. (2004). Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem. 279, 28492–28498. doi: 10.1074/jbc.m400871200
Ali, F., Hindley, C., McDowell, G., Deibler, R., Jones, A., Kirschner, M., et al. (2011). Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138, 4267–4277. doi: 10.1242/dev.067900
Amadei, G., Zander, M. A., Yang, G., Dumelie, J. G., Vessey, J. P., Lipshitz, H. D., et al. (2015). A Smaug2-based translational repression complex determines the balance between precursor maintenance versus differentiation during mammalian neurogenesis. J. Neurosci. 35, 15666–15681. doi: 10.1523/jneurosci.2172-15.2015
Aydin, B., Kakumanu, A., Rossillo, M., Moreno-Estelles, M., Garipler, G., Ringstad, N., et al. (2019). Proneural factors Ascl1 and Neurog2 contribute to neuronal subtype identities by establishing distinct chromatin landscapes. Nat. Neurosci. 22, 897–908. doi: 10.1038/s41593-019-0399-y
Backman, M., Machon, O., Mygland, L., van den Bout, C. J., Zhong, W., Taketo, M. M., et al. (2005). Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev. Biol. 279, 155–168. doi: 10.1016/j.ydbio.2004.12.010
Barnabe-Heider, F., and Miller, F. D. (2003). Endogenously produced neurotrophins regulate survival and differentiation of cortical progenitors via distinct signaling pathways. J. Neurosci. 23, 5149–5160. doi: 10.1523/jneurosci.23-12-05149.2003
Barnabe-Heider, F., Wasylnka, J. A., Fernandes, K. J., Porsche, C., Sendtner, M., Kaplan, D. R., et al. (2005). Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron 48, 253–265. doi: 10.1016/j.neuron.2005.08.037
Bartkowska, K., Paquin, A., Gauthier, A. S., Kaplan, D. R., and Miller, F. D. (2007). Trk signaling regulates neural precursor cell proliferation and differentiation during cortical development. Development 134, 4369–4380. doi: 10.1242/dev.008227
Bayatti, N., Moss, J. A., Sun, L., Ambrose, P., Ward, J. F., Lindsay, S., et al. (2008). A molecular neuroanatomical study of the developing human neocortex from 8 to 17 postconceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cereb. Cortex 18, 1536–1548. doi: 10.1093/cercor/bhm184
Bayraktar, O. A., Fuentealba, L. C., Alvarez-Buylla, A., and Rowitch, D. H. (2014). Astrocyte development and heterogeneity. Cold Spring Harb. Perspect. Biol. 7:a020362.
Beckervordersandforth, R., Tripathi, P., Ninkovic, J., Bayam, E., Lepier, A., Stempfhuber, B., et al. (2010). In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7, 744–758. doi: 10.1016/j.stem.2010.11.017
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Bertrand, N., Castro, D. S., and Guillemot, F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. doi: 10.1038/nrn874
Bohner, A. P., Akers, R. M., and McConnell, S. K. (1997). Induction of deep layer cortical neurons in vitro. Development 124, 915–923.
Boldrini, M., Fulmore, C. A., Tartt, A. N., Simeon, L. R., Pavlova, I., Poposka, V., et al. (2018). Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 22, 589–599.e5.
Bonaguidi, M. A., McGuire, T., Hu, M., Kan, L., Samanta, J., and Kessler, J. A. (2005). LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development 132, 5503–5514. doi: 10.1242/dev.02166
Bonni, A., Sun, Y., Nadal-Vicens, M., Bhatt, A., Frank, D. A., Rozovsky, I., et al. (1997). Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278, 477–483. doi: 10.1126/science.278.5337.477
Bormuth, I., Yan, K., Yonemasu, T., Gummert, M., Zhang, M., Wichert, S., et al. (2013). Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J. Neurosci. 33, 641–651. doi: 10.1523/jneurosci.0899-12.2013
Brand, M., and Morrissey, E. (2020). Single-cell fate decisions of bipotential hematopoietic progenitors. Curr. Opin. Hematol. 27, 232–240. doi: 10.1097/moh.0000000000000592
Brill, M. S., Ninkovic, J., Winpenny, E., Hodge, R. D., Ozen, I., Yang, R., et al. (2009). Adult generation of glutamatergic olfactory bulb interneurons. Nat. Neurosci. 12, 1524–1533. doi: 10.1038/nn.2416
Britz, O., Mattar, P., Nguyen, L., Langevin, L. M., Zimmer, C., Alam, S., et al. (2006). A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex 16(Suppl. 1), i138–i151.
Bultje, R. S., Castaneda-Castellanos, D. R., Jan, L. Y., Jan, Y. N., Kriegstein, A. R., and Shi, S. H. (2009). Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron 63, 189–202. doi: 10.1016/j.neuron.2009.07.004
Caiazzo, M., Dell’Anno, M. T., Dvoretskova, E., Lazarevic, D., Taverna, S., Leo, D., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227. doi: 10.1038/nature10284
Calegari, F., and Huttner, W. B. (2003). An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J. Cell Sci. 116, 4947–4955. doi: 10.1242/jcs.00825
Casarosa, S., Fode, C., and Guillemot, F. (1999). Mash1 regulates neurogenesis in the ventral telencephalon. Development 126, 525–534.
Castro, D. S., and Guillemot, F. (2011). Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle 10, 4026–4031. doi: 10.4161/cc.10.23.18578
Castro, D. S., Martynoga, B., Parras, C., Ramesh, V., Pacary, E., Johnston, C., et al. (2011). A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 25, 930–945. doi: 10.1101/gad.627811
Castro, D. S., Skowronska-Krawczyk, D., Armant, O., Donaldson, I. J., Parras, C., Hunt, C., et al. (2006). Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell 11, 831–844. doi: 10.1016/j.devcel.2006.10.006
Caviness, V. S. Jr. (1982). Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Brain Res. 256, 293–302. doi: 10.1016/0165-3806(82)90141-9
Caviness, V. S. Jr., Takahashi, T., and Nowakowski, R. S. (1995). Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 18, 379–383. doi: 10.1016/0166-2236(95)93933-o
Chandran, S., Kato, H., Gerreli, D., Compston, A., Svendsen, C. N., and Allen, N. D. (2003). FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development 130, 6599–6609. doi: 10.1242/dev.00871
Cheloufi, S., Elling, U., Hopfgartner, B., Jung, Y. L., Murn, J., Ninova, M., et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224. doi: 10.1038/nature15749
Chickarmane, V., Enver, T., and Peterson, C. (2009). Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput. Biol. 5:e1000268. doi: 10.1371/journal.pcbi.1000268
Chizhikov, V. V., Iskusnykh, I. Y., Steshina, E. Y., Fattakhov, N., Lindgren, A. G., Shetty, A. S., et al. (2019). Early dorsomedial tissue interactions regulate gyrification of distal neocortex. Nat. Commun. 10:5192.
Chuang, W., Sharma, A., Shukla, P., Li, G., Mall, M., Rajarajan, K., et al. (2017). Partial reprogramming of pluripotent stem cell-derived cardiomyocytes into neurons. Sci. Rep. 7:44840.
de Juan Romero, C., Bruder, C., Tomasello, U., Sanz-Anquela, J. M., and Borrell, V. (2015). Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J. 34, 1859–1874. doi: 10.15252/embj.201591176
Dehay, C., Kennedy, H., and Kosik, K. S. (2015). The outer subventricular zone and primate-specific cortical complexification. Neuron 85, 683–694. doi: 10.1016/j.neuron.2014.12.060
Del Toro, D., Ruff, T., Cederfjall, E., Villalba, A., Seyit-Bremer, G., Borrell, V., et al. (2017). Regulation of cerebral cortex folding by controlling neuronal migration via FLRT adhesion molecules. Cell 169, 621–635.e16.
Dennis, D. J., Han, S., and Schuurmans, C. (2019). bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 1705, 48–65. doi: 10.1016/j.brainres.2018.03.013
Dennis, D. J., Wilkinson, G., Li, S., Dixit, R., Adnani, L., Balakrishnan, A., et al. (2017). Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc. Natl. Acad. Sci. U.S.A. 114, E4934–E4943.
Desai, A. R., and McConnell, S. K. (2000). Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development 127, 2863–2872.
Dillon, N. (2012). Factor mediated gene priming in pluripotent stem cells sets the stage for lineage specification. Bioessays 34, 194–204. doi: 10.1002/bies.201100137
Dixit, R., Wilkinson, G., Cancino, G. I., Shaker, T., Adnani, L., Li, S., et al. (2014). Neurog1 and Neurog2 control two waves of neuronal differentiation in the piriform cortex. J. Neurosci. 34, 539–553. doi: 10.1523/jneurosci.0614-13.2014
Dixit, R., Zimmer, C., Waclaw, R. R., Mattar, P., Shaker, T., Kovach, C., et al. (2011). Ascl1 participates in Cajal-Retzius cell development in the neocortex. Cereb. Cortex 21, 2599–2611. doi: 10.1093/cercor/bhr046
Eckler, M. J., Nguyen, T. D., McKenna, W. L., Fastow, B. L., Guo, C., Rubenstein, J. L. R., et al. (2015). Cux2-positive radial glial cells generate diverse subtypes of neocortical projection neurons and macroglia. Neuron 86, 1100–1108. doi: 10.1016/j.neuron.2015.04.020
Englund, C., Fink, A., Lau, C., Pham, D., Daza, R. A., Bulfone, A., et al. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247–251. doi: 10.1523/jneurosci.2899-04.2005
Enver, T., Pera, M., Peterson, C., and Andrews, P. W. (2009). Stem cell states, fates, and the rules of attraction. Cell Stem Cell 4, 387–397. doi: 10.1016/j.stem.2009.04.011
Ernst, A., Alkass, K., Bernard, S., Salehpour, M., Perl, S., Tisdale, J., et al. (2014). Neurogenesis in the striatum of the adult human brain. Cell 156, 1072–1083. doi: 10.1016/j.cell.2014.01.044
Esain, V., Postlethwait, J. H., Charnay, P., and Ghislain, J. (2010). FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9. Development 137, 33–42. doi: 10.1242/dev.038026
Eto, H., Kishi, Y., Yakushiji-Kaminatsui, N., Sugishita, H., Utsunomiya, S., Koseki, H., et al. (2020). The Polycomb group protein Ring1 regulates dorsoventral patterning of the mouse telencephalon. Nat. Commun. 11:5709.
Falk, D., and Hofman, M. A. (2012). Evolution of the primate brain. Preface. Prog. Brain Res. 195, vii–ix.
Farreny, M. A., Agius, E., Bel-Vialar, S., Escalas, N., Khouri-Farah, N., Soukkarieh, C., et al. (2018). FGF signaling controls Shh-dependent oligodendroglial fate specification in the ventral spinal cord. Neural Dev. 13:3.
Fiddes, I. T., Lodewijk, G. A., Mooring, M., Bosworth, C. M., Ewing, A. D., Mantalas, G. L., et al. (2018). Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22.
Fietz, S. A., Kelava, I., Vogt, J., Wilsch-Brauninger, M., Stenzel, D., Fish, J. L., et al. (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699. doi: 10.1038/nn.2553
Fietz, S. A., Lachmann, R., Brandl, H., Kircher, M., Samusik, N., Schroder, R., et al. (2012). Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. U.S.A. 109, 11836–11841. doi: 10.1073/pnas.1209647109
Florio, M., Albert, M., Taverna, E., Namba, T., Brandl, H., Lewitus, E., et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470. doi: 10.1126/science.aaa1975
Florio, M., Heide, M., Pinson, A., Brandl, H., Albert, M., Winkler, S., et al. (2018). Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife 7:e32332.
Fode, C., Ma, Q., Casarosa, S., Ang, S. L., Anderson, D. J., and Guillemot, F. (2000). A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14, 67–80.
Franco, S. J., Gil-Sanz, C., Martinez-Garay, I., Espinosa, A., Harkins-Perry, S. R., Ramos, C., et al. (2012). Fate-restricted neural progenitors in the mammalian cerebral cortex. Science 337, 746–749. doi: 10.1126/science.1223616
Frantz, G. D., and McConnell, S. K. (1996). Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron 17, 55–61. doi: 10.1016/s0896-6273(00)80280-9
Fuentealba, L. C., Rompani, S. B., Parraguez, J. I., Obernier, K., Romero, R., Cepko, C. L., et al. (2015). Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655. doi: 10.1016/j.cell.2015.05.041
Fukumitsu, H., Ohtsuka, M., Murai, R., Nakamura, H., Itoh, K., and Furukawa, S. (2006). Brain-derived neurotrophic factor participates in determination of neuronal laminar fate in the developing mouse cerebral cortex. J. Neurosci. 26, 13218–13230. doi: 10.1523/jneurosci.4251-06.2006
Furusho, M., Kaga, Y., Ishii, A., Hebert, J. M., and Bansal, R. (2011). Fibroblast growth factor signaling is required for the generation of oligodendrocyte progenitors from the embryonic forebrain. J. Neurosci. 31, 5055–5066. doi: 10.1523/jneurosci.4800-10.2011
Furuta, Y., Piston, D. W., and Hogan, B. L. (1997). Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124, 2203–2212.
Furutachi, S., Matsumoto, A., Nakayama, K. I., and Gotoh, Y. (2013). p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 32, 970–981. doi: 10.1038/emboj.2013.50
Furutachi, S., Miya, H., Watanabe, T., Kawai, H., Yamasaki, N., Harada, Y., et al. (2015). Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18, 657–665. doi: 10.1038/nn.3989
Gabay, L., Lowell, S., Rubin, L. L., and Anderson, D. J. (2003). Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron 40, 485–499. doi: 10.1016/s0896-6273(03)00637-8
Gaiano, N., Nye, J. S., and Fishell, G. (2000). Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26, 395–404. doi: 10.1016/s0896-6273(00)81172-1
Gao, P., Postiglione, M. P., Krieger, T. G., Hernandez, L., Wang, C., Han, Z., et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788. doi: 10.1016/j.cell.2014.10.027
Gascon, S., Masserdotti, G., Russo, G. L., and Gotz, M. (2017). Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell 21, 18–34. doi: 10.1016/j.stem.2017.06.011
Gascon, S., Murenu, E., Masserdotti, G., Ortega, F., Russo, G. L., Petrik, D., et al. (2016). Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell 18, 396–409. doi: 10.1016/j.stem.2015.12.003
Ge, W., He, F., Kim, K. J., Blanchi, B., Coskun, V., Nguyen, L., et al. (2006). Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U.S.A. 103, 1319–1324. doi: 10.1073/pnas.0510419103
Ge, W., Martinowich, K., Wu, X., He, F., Miyamoto, A., Fan, G., et al. (2002). Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci. Res. 69, 848–860. doi: 10.1002/jnr.10364
Ghosh, A., and Greenberg, M. E. (1995). Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron 15, 89–103. doi: 10.1016/0896-6273(95)90067-5
Gillotin, S., Davies, J. D., and Philpott, A. (2018). Subcellular localisation modulates ubiquitylation and degradation of Ascl1. Sci. Rep. 8:4625.
Gil-Sanz, C., Espinosa, A., Fregoso, S. P., Bluske, K. K., Cunningham, C. L., Martinez-Garay, I., et al. (2015). Lineage tracing using Cux2-Cre and Cux2-CreERT2 mice. Neuron 86, 1091–1099. doi: 10.1016/j.neuron.2015.04.019
Gotz, M., and Huttner, W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788.
Gotz, M., Nakafuku, M., and Petrik, D. (2016). Neurogenesis in the developing and adult brain-similarities and key differences. Cold Spring Harb. Perspect. Biol. 8:a018853. doi: 10.1101/cshperspect.a018853
Gradwohl, G., Fode, C., and Guillemot, F. (1996). Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 180, 227–241. doi: 10.1006/dbio.1996.0297
Grandbarbe, L., Bouissac, J., Rand, M., Hrabe de Angelis, M., Artavanis-Tsakonas, S., and Mohier, E. (2003). Delta-Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development 130, 1391–1402. doi: 10.1242/dev.00374
Grande, A., Sumiyoshi, K., Lopez-Juarez, A., Howard, J., Sakthivel, B., Aronow, B., et al. (2013). Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat. Commun. 4:2373.
Greene, L. A., and Tischler, A. S. (1976). Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U.S.A. 73, 2424–2428. doi: 10.1073/pnas.73.7.2424
Gross, R. E., Mehler, M. F., Mabie, P. C., Zang, Z., Santschi, L., and Kessler, J. A. (1996). Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 17, 595–606. doi: 10.1016/s0896-6273(00)80193-2
Gsponer, J., Futschik, M. E., Teichmann, S. A., and Babu, M. M. (2008). Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322, 1365–1368. doi: 10.1126/science.1163581
Guillemot, F., and Hassan, B. A. (2017). Beyond proneural: emerging functions and regulations of proneural proteins. Curr. Opin. Neurobiol. 42, 93–101. doi: 10.1016/j.conb.2016.11.011
Gunhaga, L., Marklund, M., Sjodal, M., Hsieh, J. C., Jessell, T. M., and Edlund, T. (2003). Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat. Neurosci. 6, 701–707. doi: 10.1038/nn1068
Guo, C., Eckler, M. J., McKenna, W. L., McKinsey, G. L., Rubenstein, J. L., and Chen, B. (2013). Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron 80, 1167–1174. doi: 10.1016/j.neuron.2013.09.037
Guo, J. U., Su, Y., Shin, J. H., Shin, J., Li, H., Xie, B., et al. (2014). Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222. doi: 10.1038/nn.3607
Guzelsoy, G., Akkaya, C., Atak, D., Dunn, C. D., Kabakcioglu, A., Ozlu, N., et al. (2019). Terminal neuron localization to the upper cortical plate is controlled by the transcription factor NEUROD2. Sci. Rep. 9:19697.
Hack, M. A., Sugimori, M., Lundberg, C., Nakafuku, M., and Gotz, M. (2004). Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol. Cell. Neurosci. 25, 664–678. doi: 10.1016/j.mcn.2003.12.012
Hagey, D. W., and Muhr, J. (2014). Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 9, 1908–1920. doi: 10.1016/j.celrep.2014.11.013
Han, S., Dennis, D. J., Balakrishnan, A., Dixit, R., Britz, O., Zinyk, D., et al. (2018). A non-canonical role for the proneural gene Neurog1 as a negative regulator of neocortical neurogenesis. Development 145:dev157719. doi: 10.1242/dev.157719
Han, S., Wilkinson, G. A., Okawa, S., Adnani, L., Dixit, R., Faisal, I., et al. (2020). Proneural genes define ground state rules to regulate neurogenic patterning and cortical folding. bioRxiv [Preprint]. doi: 10.1101/2020.09.22.307058
Hand, R., Bortone, D., Mattar, P., Nguyen, L., Heng, J. I., Guerrier, S., et al. (2005). Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48, 45–62. doi: 10.1016/j.neuron.2005.08.032
Hansen, D. V., Lui, J. H., Parker, P. R., and Kriegstein, A. R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. doi: 10.1038/nature08845
Hardwick, L. J., and Philpott, A. (2015). Multi-site phosphorylation regulates NeuroD4 activity during primary neurogenesis: a conserved mechanism amongst proneural proteins. Neural Dev. 10:15.
Hardwick, L. J. A., and Philpott, A. (2018a). Interaction between opposing modes of phospho-regulation of the proneural proteins Ascl1 and Ngn2. Wellcome Open Res. 3:129. doi: 10.12688/wellcomeopenres.14848.1
Hardwick, L. J. A., and Philpott, A. (2018b). The N terminus of Ascl1 underlies differing proneural activity of mouse and Xenopus Ascl1 proteins. Wellcome Open Res. 3:125. doi: 10.12688/wellcomeopenres.14842.1
Hasegawa, H., Ashigaki, S., Takamatsu, M., Suzuki-Migishima, R., Ohbayashi, N., Itoh, N., et al. (2004). Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J. Neurosci. 24, 8711–8719. doi: 10.1523/jneurosci.3070-04.2004
Hatakeyama, J., Bessho, Y., Katoh, K., Ookawara, S., Fujioka, M., Guillemot, F., et al. (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539–5550. doi: 10.1242/dev.01436
Haubensak, W., Attardo, A., Denk, W., and Huttner, W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 3196–3201. doi: 10.1073/pnas.0308600100
He, F., Ge, W., Martinowich, K., Becker-Catania, S., Coskun, V., Zhu, W., et al. (2005). A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 8, 616–625. doi: 10.1038/nn1440
Heng, J. I., Nguyen, L., Castro, D. S., Zimmer, C., Wildner, H., Armant, O., et al. (2008). Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118. doi: 10.1038/nature07198
Henke, R. M., Meredith, D. M., Borromeo, M. D., Savage, T. K., and Johnson, J. E. (2009). Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev. Biol. 328, 529–540. doi: 10.1016/j.ydbio.2009.01.007
Herrero-Mendez, A., Almeida, A., Fernandez, E., Maestre, C., Moncada, S., and Bolanos, J. P. (2009). The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat. Cell Biol. 11, 747–752. doi: 10.1038/ncb1881
Hindley, C., Ali, F., McDowell, G., Cheng, K., Jones, A., Guillemot, F., et al. (2012). Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development 139, 1718–1723. doi: 10.1242/dev.077552
Hirabayashi, Y., Itoh, Y., Tabata, H., Nakajima, K., Akiyama, T., Masuyama, N., et al. (2004). The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131, 2791–2801. doi: 10.1242/dev.01165
Hirabayashi, Y., Suzki, N., Tsuboi, M., Endo, T. A., Toyoda, T., Shinga, J., et al. (2009). Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613. doi: 10.1016/j.neuron.2009.08.021
Horton, S., Meredith, A., Richardson, J. A., and Johnson, J. E. (1999). Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol. Cell. Neurosci. 14, 355–369. doi: 10.1006/mcne.1999.0791
Huang, C., Chan, J. A., and Schuurmans, C. (2014). Proneural bHLH genes in development and disease. Curr. Top. Dev. Biol. 110, 75–127. doi: 10.1016/b978-0-12-405943-6.00002-6
Huang, S., Guo, Y. P., May, G., and Enver, T. (2007). Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 305, 695–713. doi: 10.1016/j.ydbio.2007.02.036
Hufnagel, R. B., Le, T. T., Riesenberg, A. L., and Brown, N. L. (2010). Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev. Biol. 340, 490–503. doi: 10.1016/j.ydbio.2010.02.002
Imamura, O., Satoh, Y., Endo, S., and Takishima, K. (2008). Analysis of extracellular signal-regulated kinase 2 function in neural stem/progenitor cells via nervous system-specific gene disruption. Stem Cells 26, 3247–3256. doi: 10.1634/stemcells.2008-0578
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208. doi: 10.1126/science.1242366
Ito, H., Nakajima, A., Nomoto, H., and Furukawa, S. (2003). Neurotrophins facilitate neuronal differentiation of cultured neural stem cells via induction of mRNA expression of basic helix-loop-helix transcription factors Mash1 and Math1. J. Neurosci. Res. 71, 648–658. doi: 10.1002/jnr.10532
Jessberger, S., Toni, N., Clemenson, G. D. Jr., Ray, J., and Gage, F. H. (2008). Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci. 11, 888–893. doi: 10.1038/nn.2148
Johnson, E. S., Ma, P. C., Ota, I. M., and Varshavsky, A. (1995). A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456. doi: 10.1074/jbc.270.29.17442
Jones, E. G. (1994). Santiago Ramon y Cajal and the Croonian Lecture, March 1894. Trends Neurosci. 17, 190–192. doi: 10.1016/0166-2236(94)90100-7
Ju, B. G., Solum, D., Song, E. J., Lee, K. J., Rose, D. W., Glass, C. K., et al. (2004). Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell 119, 815–829. doi: 10.1016/j.cell.2004.11.017
Ju, X. C., Hou, Q. Q., Sheng, A. L., Wu, K. Y., Zhou, Y., Jin, Y., et al. (2016). The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. eLife 5:e18197.
Kageyama, R., Ochi, S., Sueda, R., and Shimojo, H. (2020). The significance of gene expression dynamics in neural stem cell regulation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 96, 351–363. doi: 10.2183/pjab.96.026
Kageyama, R., Ohtsuka, T., and Kobayashi, T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251. doi: 10.1242/dev.000786
Kageyama, R., Ohtsuka, T., Shimojo, H., and Imayoshi, I. (2008). Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247–1251. doi: 10.1038/nn.2208
Kamakura, S., Oishi, K., Yoshimatsu, T., Nakafuku, M., Masuyama, N., and Gotoh, Y. (2004). Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 6, 547–554. doi: 10.1038/ncb1138
Kast, R. J., and Levitt, P. (2019). Precision in the development of neocortical architecture: From progenitors to cortical networks. Prog. Neurobiol. 175, 77–95. doi: 10.1016/j.pneurobio.2019.01.003
Kessaris, N., Fogarty, M., Iannarelli, P., Grist, M., Wegner, M., and Richardson, W. D. (2006). Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 9, 173–179. doi: 10.1038/nn1620
Kim, J., Su, S. C., Wang, H., Cheng, A. W., Cassady, J. P., Lodato, M. A., et al. (2011). Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell 9, 413–419. doi: 10.1016/j.stem.2011.09.011
Kiparaki, M., Zarifi, I., and Delidakis, C. (2015). bHLH proteins involved in Drosophila neurogenesis are mutually regulated at the level of stability. Nucleic Acids Res. 43, 2543–2559. doi: 10.1093/nar/gkv083
Kishi, Y., Fujii, Y., Hirabayashi, Y., and Gotoh, Y. (2012). HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat. Neurosci. 15, 1127–1133. doi: 10.1038/nn.3165
Koblar, S. A., Turnley, A. M., Classon, B. J., Reid, K. L., Ware, C. B., Cheema, S. S., et al. (1998). Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc. Natl. Acad. Sci. U.S.A. 95, 3178–3181. doi: 10.1073/pnas.95.6.3178
Kohtz, J. D., Baker, D. P., Corte, G., and Fishell, G. (1998). Regionalization within the mammalian telencephalon is mediated by changes in responsiveness to Sonic Hedgehog. Development 125, 5079–5089.
Kovach, C., Dixit, R., Li, S., Mattar, P., Wilkinson, G., Elsen, G. E., et al. (2012). Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb. Cortex 23, 1884–1900. doi: 10.1093/cercor/bhs176
Kovach, C., Dixit, R., Li, S., Mattar, P., Wilkinson, G., Elsen, G. E., et al. (2013). Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb. Cortex 23, 1884–1900. doi: 10.1093/cercor/bhs176
Kowalczyk, T., Pontious, A., Englund, C., Daza, R. A., Bedogni, F., Hodge, R., et al. (2009). Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb. Cortex 19, 2439–2450. doi: 10.1093/cercor/bhn260
Koyano-Nakagawa, N., Wettstein, D., and Kintner, C. (1999). Activation of Xenopus genes required for lateral inhibition and neuronal differentiation during primary neurogenesis. Mol. Cell. Neurosci. 14, 327–339. doi: 10.1006/mcne.1999.0783
Kroll, T. T., and O’Leary, D. D. (2005). Ventralized dorsal telencephalic progenitors in Pax6 mutant mice generate GABA interneurons of a lateral ganglionic eminence fate. Proc. Natl. Acad. Sci. U.S.A. 102, 7374–7379. doi: 10.1073/pnas.0500819102
Le Dreau, G., Escalona, R., Fueyo, R., Herrera, A., Martinez, J. D., Usieto, S., et al. (2018). E proteins sharpen neurogenesis by modulating proneural bHLH transcription factors’ activity in an E-box-dependent manner. eLife 7:e37267.
Lee, Q. Y., Mall, M., Chanda, S., Zhou, B., Sharma, K. S., Schaukowitch, K., et al. (2020). Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes. Nat. Cell Biol. 22, 401–411. doi: 10.1038/s41556-020-0490-3
Lewitus, E., Kelava, I., Kalinka, A. T., Tomancak, P., and Huttner, W. B. (2014). An adaptive threshold in mammalian neocortical evolution. PLoS Biol. 12:e1002000. doi: 10.1371/journal.pbio.1002000
Li, L., and Clevers, H. (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545. doi: 10.1126/science.1180794
Li, S., Mattar, P., Dixit, R., Lawn, S. O., Wilkinson, G., Kinch, C., et al. (2014). RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J. Neurosci. 34, 2169–2190. doi: 10.1523/jneurosci.4077-13.2014
Li, S., Mattar, P., Zinyk, D., Singh, K., Chaturvedi, C. P., Kovach, C., et al. (2012). GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 32, 7791–7805. doi: 10.1523/jneurosci.1309-12.2012
Lin, H., Zhu, X., Chen, G., Song, L., Gao, L., Khand, A. A., et al. (2017). KDM3A-mediated demethylation of histone H3 lysine 9 facilitates the chromatin binding of Neurog2 during neurogenesis. Development 144, 3674–3685. doi: 10.1242/dev.144113
Lin, Y. C., Jhunjhunwala, S., Benner, C., Heinz, S., Welinder, E., Mansson, R., et al. (2010). A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643. doi: 10.1038/ni.1891
Liu, H., Thurig, S., Mohamed, O., Dufort, D., and Wallace, V. A. (2006). Mapping canonical Wnt signaling in the developing and adult retina. Invest. Ophthalmol. Vis. Sci. 47, 5088–5097. doi: 10.1167/iovs.06-0403
Liu, J. A., Tai, A., Hong, J., Cheung, M. P. L., Sham, M. H., Cheah, K. S. E., et al. (2020). Fbxo9 functions downstream of Sox10 to determine neuron-glial fate choice in the dorsal root ganglia through Neurog2 destabilization. Proc. Natl. Acad. Sci. U.S.A. 117, 4199–4210. doi: 10.1073/pnas.1916164117
Llinares-Benadero, C., and Borrell, V. (2019). Deconstructing cortical folding: genetic, cellular and mechanical determinants. Nat. Rev. Neurosci. 20, 161–176. doi: 10.1038/s41583-018-0112-2
Lukaszewicz, A., Savatier, P., Cortay, V., Giroud, P., Huissoud, C., Berland, M., et al. (2005). G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47, 353–364. doi: 10.1016/j.neuron.2005.06.032
Lukaszewicz, A., Savatier, P., Cortay, V., Kennedy, H., and Dehay, C. (2002). Contrasting effects of basic fibroblast growth factor and neurotrophin 3 on cell cycle kinetics of mouse cortical stem cells. J. Neurosci. 22, 6610–6622. doi: 10.1523/jneurosci.22-15-06610.2002
Luo, C., Lee, Q. Y., Wapinski, O., Castanon, R., Nery, J. R., Mall, M., et al. (2019). Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. eLife 8:e40197.
Luskin, M. B., Pearlman, A. L., and Sanes, J. R. (1988). Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron 1, 635–647. doi: 10.1016/0896-6273(88)90163-8
Ma, Y. C., Song, M. R., Park, J. P., Henry Ho, H. Y., Hu, L., Kurtev, M. V., et al. (2008). Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron 58, 65–77. doi: 10.1016/j.neuron.2008.01.037
Machon, O., Backman, M., Krauss, S., and Kozmik, Z. (2005). The cellular fate of cortical progenitors is not maintained in neurosphere cultures. Mol. Cell. Neurosci. 30, 388–397. doi: 10.1016/j.mcn.2005.08.003
Madelaine, R., and Blader, P. (2011). A cluster of non-redundant Ngn1 binding sites is required for regulation of deltaA expression in zebrafish. Dev. Biol. 350, 198–207. doi: 10.1016/j.ydbio.2010.10.025
Maretto, S., Cordenonsi, M., Dupont, S., Braghetta, P., Broccoli, V., Hassan, A. B., et al. (2003). Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 3299–3304. doi: 10.1073/pnas.0434590100
Marro, S., Pang, Z. P., Yang, N., Tsai, M. C., Qu, K., Chang, H. Y., et al. (2011). Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9, 374–382. doi: 10.1016/j.stem.2011.09.002
Marshall, C. J. (1995). Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185. doi: 10.1016/0092-8674(95)90401-8
Martinez-Cerdeno, V., Cunningham, C. L., Camacho, J., Antczak, J. L., Prakash, A. N., Cziep, M. E., et al. (2012). Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS One 7:e30178. doi: 10.1371/journal.pone.0030178
Masserdotti, G., Gillotin, S., Sutor, B., Drechsel, D., Irmler, M., Jorgensen, H. F., et al. (2015). Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17, 74–88. doi: 10.1016/j.stem.2015.05.014
Mattar, P., Langevin, L. M., Markham, K., Klenin, N., Shivji, S., Zinyk, D., et al. (2008). Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol. Cell. Biol. 28, 1456–1469. doi: 10.1128/mcb.01510-07
McConnell, S. K., and Kaznowski, C. E. (1991). Cell cycle dependence of laminar determination in developing neocortex. Science 254, 282–285. doi: 10.1126/science.1925583
McDowell, G. S., Hindley, C. J., Lippens, G., Landrieu, I., and Philpott, A. (2014). Phosphorylation in intrinsically disordered regions regulates the activity of Neurogenin2. BMC Biochem. 15:24. doi: 10.1186/s12858-014-0024-3
McKay, N. D., Robinson, B., Brodie, R., and Rooke-Allen, N. (1983). Glucose transport and metabolism in cultured human skin fibroblasts. Biochim. Biophys. Acta 762, 198–204. doi: 10.1016/0167-4889(83)90071-x
Medina, D. L., Sciarretta, C., Calella, A. M., Von Bohlen Und Halbach, O., Unsicker, K., and Minichiello, L. (2004). TrkB regulates neocortex formation through the Shc/PLCgamma-mediated control of neuronal migration. EMBO J. 23, 3803–3814. doi: 10.1038/sj.emboj.7600399
Menard, C., Hein, P., Paquin, A., Savelson, A., Yang, X. M., Lederfein, D., et al. (2002). An essential role for a MEK-C/EBP pathway during growth factor-regulated cortical neurogenesis. Neuron 36, 597–610. doi: 10.1016/s0896-6273(02)01026-7
Miesfeld, J. B., Moon, M. S., Riesenberg, A. N., Contreras, A. N., Kovall, R. A., and Brown, N. L. (2018). Rbpj direct regulation of Atoh7 transcription in the embryonic mouse retina. Sci. Rep. 8:10195.
Miyama, S., Takahashi, T., Nowakowski, R. S., and Caviness, V. S. Jr. (1997). A gradient in the duration of the G1 phase in the murine neocortical proliferative epithelium. Cereb. Cortex 7, 678–689. doi: 10.1093/cercor/7.7.678
Miyata, T., Kawaguchi, A., Saito, K., Kawano, M., Muto, T., and Ogawa, M. (2004). Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131, 3133–3145. doi: 10.1242/dev.01173
Molnar, Z., Vasistha, N. A., and Garcia-Moreno, F. (2011). Hanging by the tail: progenitor populations proliferate. Nat. Neurosci. 14, 538–540. doi: 10.1038/nn.2817
Molne, M., Studer, L., Tabar, V., Ting, Y. T., Eiden, M. V., and McKay, R. D. (2000). Early cortical precursors do not undergo LIF-mediated astrocytic differentiation. J. Neurosci. Res. 59, 301–311. doi: 10.1002/(sici)1097-4547(20000201)59:3<301::aid-jnr3>3.0.co;2-h
Morimoto-Suzki, N., Hirabayashi, Y., Tyssowski, K., Shinga, J., Vidal, M., Koseki, H., et al. (2014). The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development. Development 141, 4343–4353. doi: 10.1242/dev.112276
Muller, J., and Verrijzer, P. (2009). Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150–158. doi: 10.1016/j.gde.2009.03.001
Murre, C., McCaw, P. S., Vaessin, H., Caudy, M., Jan, L. Y., Jan, Y. N., et al. (1989). Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58, 537–544. doi: 10.1016/0092-8674(89)90434-0
Muzio, L., DiBenedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., and Mallamaci, A. (2002a). Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb. Cortex 12, 129–139. doi: 10.1093/cercor/12.2.129
Muzio, L., DiBenedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., and Mallamaci, A. (2002b). Conversion of cerebral cortex into basal ganglia in Emx2(-/-) Pax6(Sey/Sey) double-mutant mice. Nat. Neurosci. 5, 737–745. doi: 10.1038/nn892
Nguyen, L., Besson, A., Heng, J. I., Schuurmans, C., Teboul, L., Parras, C., et al. (2006). p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 20, 1511–1524. doi: 10.1101/gad.377106
Nieto, M., Schuurmans, C., Britz, O., and Guillemot, F. (2001). Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29, 401–413. doi: 10.1016/s0896-6273(01)00214-8
Noctor, S. C., Martinez-Cerdeno, V., Ivic, L., and Kriegstein, A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144. doi: 10.1038/nn1172
Nolo, R., Abbott, L. A., and Bellen, H. J. (2000). Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102, 349–362. doi: 10.1016/s0092-8674(00)00040-4
Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H., and Kageyama, R. (2020). Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 147:dev182204. doi: 10.1242/dev.182204
Ohori, Y., Yamamoto, S., Nagao, M., Sugimori, M., Yamamoto, N., Nakamura, K., et al. (2006). Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord. J. Neurosci. 26, 11948–11960. doi: 10.1523/jneurosci.3127-06.2006
Ohtaka-Maruyama, C. (2020). Subplate neurons as an organizer of mammalian neocortical development. Front. Neuroanat. 14:8. doi: 10.3389/fnana.2020.00008
Ohtsuka, M., Fukumitsu, H., and Furukawa, S. (2009). Neurotrophin-3 stimulates neurogenetic proliferation via the extracellular signal-regulated kinase pathway. J. Neurosci. Res. 87, 301–306. doi: 10.1002/jnr.21855
Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F., and Kageyama, R. (1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207. doi: 10.1093/emboj/18.8.2196
Oishi, K., van den Berg, D. L. C., and Guillemot, F. (2020). Temporal control of cortico-thalamic neuron specification by regulation of Neurogenin activity and Polycomb repressive complexes. bioRxiv [Preprint]. doi: 10.1101/431684
Okawa, S., Nicklas, S., Zickenrott, S., Schwamborn, J. C., and Del Sol, A. (2016). A generalized gene-regulatory network model of stem cell differentiation for predicting lineage specifiers. Stem Cell Rep. 7, 307–315. doi: 10.1016/j.stemcr.2016.07.014
Pacary, E., Azzarelli, R., and Guillemot, F. (2013). Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nat. Commun. 4:1635.
Pacary, E., Heng, J., Azzarelli, R., Riou, P., Castro, D., Lebel-Potter, M., et al. (2011). Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron 69, 1069–1084. doi: 10.1016/j.neuron.2011.02.018
Pang, Z. P., Yang, N., Vierbuchen, T., Ostermeier, A., Fuentes, D. R., Yang, T. Q., et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223. doi: 10.1038/nature10202
Parras, C. M., Hunt, C., Sugimori, M., Nakafuku, M., Rowitch, D., and Guillemot, F. (2007). The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J. Neurosci. 27, 4233–4242. doi: 10.1523/jneurosci.0126-07.2007
Parras, C. M., Schuurmans, C., Scardigli, R., Kim, J., Anderson, D. J., and Guillemot, F. (2002). Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16, 324–338. doi: 10.1101/gad.940902
Pavri, R., Zhu, B., Li, G., Trojer, P., Mandal, S., Shilatifard, A., et al. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717. doi: 10.1016/j.cell.2006.04.029
Pearson, B. J., and Doe, C. Q. (2004). Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 20, 619–647. doi: 10.1146/annurev.cellbio.19.111301.115142
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M. W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962. doi: 10.1073/pnas.1002530107
Peyre, E., Silva, C. G., and Nguyen, L. (2015). Crosstalk between intracellular and extracellular signals regulating interneuron production, migration and integration into the cortex. Front. Cell. Neurosci. 9:129. doi: 10.3389/fncel.2015.00129
Pfisterer, U., Kirkeby, A., Torper, O., Wood, J., Nelander, J., Dufour, A., et al. (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 108, 10343–10348.
Pfurr, S., Chu, Y. H., Bohrer, C., Greulich, F., Beattie, R., Mammadzada, K., et al. (2017). The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development. Development 144, 3917–3931. doi: 10.1242/dev.145698
Pleasure, S. J., Collins, A. E., and Lowenstein, D. H. (2000). Unique expression patterns of cell fate molecules delineate sequential stages of dentate gyrus development. J. Neurosci. 20, 6095–6105. doi: 10.1523/jneurosci.20-16-06095.2000
Pollen, A. A., Nowakowski, T. J., Chen, J., Retallack, H., Sandoval-Espinosa, C., Nicholas, C. R., et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. doi: 10.1016/j.cell.2015.09.004
Ponti, G., Obernier, K., Guinto, C., Jose, L., Bonfanti, L., and Alvarez-Buylla, A. (2013). Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc. Natl. Acad. Sci. U.S.A. 110, E1045–E1054.
Price, J., and Thurlow, L. (1988). Cell lineage in the rat cerebral cortex: a study using retroviral-mediated gene transfer. Development 104, 473–482.
Pufall, M. A., Lee, G. M., Nelson, M. L., Kang, H. S., Velyvis, A., Kay, L. E., et al. (2005). Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science 309, 142–145. doi: 10.1126/science.1111915
Qian, X., Shen, Q., Goderie, S. K., He, W., Capela, A., Davis, A. A., et al. (2000). Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28, 69–80.
Quan, X. J., Denayer, T., Yan, J., Jafar-Nejad, H., Philippi, A., Lichtarge, O., et al. (2004). Evolution of neural precursor selection: functional divergence of proneural proteins. Development 131, 1679–1689. doi: 10.1242/dev.01055
Quan, X. J., Yuan, L., Tiberi, L., Claeys, A., De Geest, N., Yan, J., et al. (2016). Post-translational control of the temporal dynamics of transcription factor activity regulates neurogenesis. Cell 164, 460–475. doi: 10.1016/j.cell.2015.12.048
Raballo, R., Rhee, J., Lyn-Cook, R., Leckman, J. F., Schwartz, M. L., and Vaccarino, F. M. (2000). Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 20, 5012–5023. doi: 10.1523/jneurosci.20-13-05012.2000
Raposo, A. A., Vasconcelos, F. F., Drechsel, D., Marie, C., Johnston, C., Dolle, D., et al. (2015). Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 10, 1544–1556. doi: 10.1016/j.celrep.2015.02.025
Reeve, R. L., Yammine, S. Z., Morshead, C. M., and van der Kooy, D. (2017). Quiescent Oct4+ neural stem cells (NSCs) repopulate ablated glial fibrillary acidic protein+ NSCs in the adult mouse brain. Stem Cells 35, 2071–2082. doi: 10.1002/stem.2662
Reillo, I., de Juan Romero, C., Garcia-Cabezas, M. A., and Borrell, V. (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–1694. doi: 10.1093/cercor/bhq238
Ringrose, L., Ehret, H., and Paro, R. (2004). Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16, 641–653. doi: 10.1016/j.molcel.2004.10.015
Rivetti di Val Cervo, P., Romanov, R. A., Spigolon, G., Masini, D., Martin-Montanez, E., Toledo, E. M., et al. (2017). Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 35, 444–452. doi: 10.1038/nbt.3835
Ruddy, R. M., and Morshead, C. M. (2018). Home sweet home: the neural stem cell niche throughout development and after injury. Cell Tissue Res. 371, 125–141. doi: 10.1007/s00441-017-2658-0
Russo, G. L., Sonsalla, G., Natarajan, P., Breunig, C. T., Bulli, G., Merl-Pham, J., et al. (2020). CRISPR-mediated induction of neuron-enriched mitochondrial proteins boosts direct glia-to-neuron conversion. Cell Stem Cell. doi: 10.1016/j.stem.2020.10.015 [Epub ahead of print].
Schuurmans, C., Armant, O., Nieto, M., Stenman, J. M., Britz, O., Klenin, N., et al. (2004). Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 23, 2892–2902. doi: 10.1038/sj.emboj.7600278
Schwab, M. H., Bartholomae, A., Heimrich, B., Feldmeyer, D., Druffel-Augustin, S., Goebbels, S., et al. (2000). Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J. Neurosci. 20, 3714–3724. doi: 10.1523/jneurosci.20-10-03714.2000
Seo, S., Lim, J. W., Yellajoshyula, D., Chang, L. W., and Kroll, K. L. (2007). Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 26, 5093–5108. doi: 10.1038/sj.emboj.7601923
Seo, S., Richardson, G. A., and Kroll, K. L. (2005). The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 132, 105–115. doi: 10.1242/dev.01548
Sessa, A., Ciabatti, E., Drechsel, D., Massimino, L., Colasante, G., Giannelli, S., et al. (2017). The Tbr2 molecular network controls cortical neuronal differentiation through complementary genetic and epigenetic pathways. Cereb. Cortex 27, 3378–3396.
Sheikh, B. N. (2014). Crafting the brain - role of histone acetyltransferases in neural development and disease. Cell Tissue Res. 356, 553–573. doi: 10.1007/s00441-014-1835-7
Shi, M., Hu, Z. L., Zheng, M. H., Song, N. N., Huang, Y., Zhao, G., et al. (2012). Notch-Rbpj signaling is required for the development of noradrenergic neurons in the mouse locus coeruleus. J. Cell Sci. 125, 4320–4332. doi: 10.1242/jcs.102152
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. doi: 10.1016/j.neuron.2008.02.014
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2011). Dynamic expression of notch signaling genes in neural stem/progenitor cells. Front. Neurosci. 5:78. doi: 10.3389/fnins.2011.00078
Sims, R. J. III, and Reinberg, D. (2006). Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 20, 2779–2786. doi: 10.1101/gad.1468206
Son, A. I., Mohammad, S., Sasaki, T., Ishii, S., Yamashita, S., Hashimoto-Torii, K., et al. (2020). Dual role of Rbpj in the maintenance of neural progenitor cells and neuronal migration in cortical development. Cereb. Cortex 30, 6444–6457. doi: 10.1093/cercor/bhaa206
Son, E. Y., Ichida, J. K., Wainger, B. J., Toma, J. S., Rafuse, V. F., Woolf, C. J., et al. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218. doi: 10.1016/j.stem.2011.07.014
Song, M. R., and Ghosh, A. (2004). FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 7, 229–235. doi: 10.1038/nn1192
Sorrells, S. F., Paredes, M. F., Cebrian-Silla, A., Sandoval, K., Qi, D., Kelley, K. W., et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381. doi: 10.1038/nature25975
Spalding, K. L., Bergmann, O., Alkass, K., Bernard, S., Salehpour, M., Huttner, H. B., et al. (2013). Dynamics of hippocampal neurogenesis in adult humans. Cell 153, 1219–1227.
Sriuranpong, V., Borges, M. W., Strock, C. L., Nakakura, E. K., Watkins, D. N., Blaumueller, C. M., et al. (2002). Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol. Cell. Biol. 22, 3129–3139. doi: 10.1128/mcb.22.9.3129-3139.2002
Stahl, R., Walcher, T., De Juan Romero, C., Pilz, G. A., Cappello, S., Irmler, M., et al. (2013). Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell 153, 535–549. doi: 10.1016/j.cell.2013.03.027
Stipursky, J., Spohr, T. C., Sousa, V. O., and Gomes, F. C. (2012). Neuron-astroglial interactions in cell-fate commitment and maturation in the central nervous system. Neurochem. Res. 37, 2402–2418. doi: 10.1007/s11064-012-0798-x
Stoykova, A., Treichel, D., Hallonet, M., and Gruss, P. (2000). Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 20, 8042–8050. doi: 10.1523/jneurosci.20-21-08042.2000
Strasser, M., Theis, F. J., and Marr, C. (2012). Stability and multiattractor dynamics of a toggle switch based on a two-stage model of stochastic gene expression. Biophys. J. 102, 19–29. doi: 10.1016/j.bpj.2011.11.4000
Stricker, S. H., and Gotz, M. (2020). Epigenetic regulation of neural lineage elaboration: Implications for therapeutic reprogramming. Neurobiol. Dis. 148, 105174. doi: 10.1016/j.nbd.2020.105174
Sun, Y., Meijer, D. H., Alberta, J. A., Mehta, S., Kane, M. F., Tien, A. C., et al. (2011). Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron 69, 906–917. doi: 10.1016/j.neuron.2011.02.005
Sun, Y., Nadal-Vicens, M., Misono, S., Lin, M. Z., Zubiaga, A., Hua, X., et al. (2001). Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376. doi: 10.1016/s0092-8674(01)00224-0
Suzuki, I. K., Gacquer, D., Van Heurck, R., Kumar, D., Wojno, M., Bilheu, A., et al. (2018). Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384.e16.
Takahashi, T., Goto, T., Miyama, S., Nowakowski, R. S., and Caviness, V. S. Jr. (1999). Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J. Neurosci. 19, 10357–10371. doi: 10.1523/jneurosci.19-23-10357.1999
Takahashi, T., Nowakowski, R. S., and Caviness, V. S. Jr. (1995). The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci. 15, 6046–6057. doi: 10.1523/jneurosci.15-09-06046.1995
Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yanagisawa, M., et al. (2001). DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1, 749–758. doi: 10.1016/s1534-5807(01)00101-0
Tavares, L., Dimitrova, E., Oxley, D., Webster, J., Poot, R., Demmers, J., et al. (2012). RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678. doi: 10.1016/j.cell.2011.12.029
Taverna, E., Gotz, M., and Huttner, W. B. (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502. doi: 10.1146/annurev-cellbio-101011-155801
Telley, L., Agirman, G., Prados, J., Amberg, N., Fievre, S., Oberst, P., et al. (2019). Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364:eaav2522. doi: 10.1126/science.aav2522
Telley, L., Govindan, S., Prados, J., Stevant, I., Nef, S., Dermitzakis, E., et al. (2016). Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446. doi: 10.1126/science.aad8361
Theil, T., Alvarez-Bolado, G., Walter, A., and Ruther, U. (1999). Gli3 is required for Emx gene expression during dorsal telencephalon development. Development 126, 3561–3571.
Tole, S., Ragsdale, C. W., and Grove, E. A. (2000). Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J). Dev. Biol. 217, 254–265. doi: 10.1006/dbio.1999.9509
Tomita, K., Moriyoshi, K., Nakanishi, S., Guillemot, F., and Kageyama, R. (2000). Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 19, 5460–5472. doi: 10.1093/emboj/19.20.5460
Tsacopoulos, M., and Magistretti, P. J. (1996). Metabolic coupling between glia and neurons. J. Neurosci. 16, 877–885. doi: 10.1523/jneurosci.16-03-00877.1996
Tursun, B., Patel, T., Kratsios, P., and Hobert, O. (2011). Direct conversion of C. elegans germ cells into specific neuron types. Science 331, 304–308. doi: 10.1126/science.1199082
Urban, N., and Guillemot, F. (2014). Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 8:396. doi: 10.3389/fncel.2014.00396
Urban, N., van den Berg, D. L., Forget, A., Andersen, J., Demmers, J. A., Hunt, C., et al. (2016). Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353, 292–295. doi: 10.1126/science.aaf4802
Vaccarino, F. M., Schwartz, M. L., Raballo, R., Rhee, J., and Lyn-Cook, R. (1999). Fibroblast growth factor signaling regulates growth and morphogenesis at multiple steps during brain development. Curr. Top. Dev. Biol. 46, 179–200. doi: 10.1016/s0070-2153(08)60329-4
Vaudry, D., Stork, P. J., Lazarovici, P., and Eiden, L. E. (2002). Signaling pathways for PC12 cell differentiation: making the right connections. Science 296, 1648–1649. doi: 10.1126/science.1071552
Vernon, A. E., Devine, C., and Philpott, A. (2003). The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development 130, 85–92. doi: 10.1242/dev.00193
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Sudhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. doi: 10.1038/nature08797
Vinals, F., Reiriz, J., Ambrosio, S., Bartrons, R., Rosa, J. L., and Ventura, F. (2004). BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 23, 3527–3537. doi: 10.1038/sj.emboj.7600360
Viti, J., Feathers, A., Phillips, J., and Lillien, L. (2003a). Epidermal growth factor receptors control competence to interpret leukemia inhibitory factor as an astrocyte inducer in developing cortex. J. Neurosci. 23, 3385–3393. doi: 10.1523/jneurosci.23-08-03385.2003
Viti, J., Gulacsi, A., and Lillien, L. (2003b). Wnt regulation of progenitor maturation in the cortex depends on Shh or fibroblast growth factor 2. J. Neurosci. 23, 5919–5927. doi: 10.1523/jneurosci.23-13-05919.2003
Vojtek, A. B., Taylor, J., DeRuiter, S. L., Yu, J. Y., Figueroa, C., Kwok, R. P., et al. (2003). Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation. Mol. Cell. Biol. 23, 4417–4427. doi: 10.1128/mcb.23.13.4417-4427.2003
Vosper, J. M., Fiore-Heriche, C. S., Horan, I., Wilson, K., Wise, H., and Philpott, A. (2007). Regulation of neurogenin stability by ubiquitin-mediated proteolysis. Biochem. J. 407, 277–284. doi: 10.1042/bj20070064
Vosper, J. M., McDowell, G. S., Hindley, C. J., Fiore-Heriche, C. S., Kucerova, R., Horan, I., et al. (2009). Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 284, 15458–15468. doi: 10.1074/jbc.m809366200
Walsh, C., and Cepko, C. L. (1988). Clonally related cortical cells show several migration patterns. Science 241, 1342–1345. doi: 10.1126/science.3137660
Wang, B., Waclaw, R. R., Allen, Z. J. II, Guillemot, F., and Campbell, K. (2009). Ascl1 is a required downstream effector of Gsx gene function in the embryonic mouse telencephalon. Neural Dev. 4:5.
Wang, L., Hou, S., and Han, Y. G. (2016). Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nat. Neurosci. 19, 888–896. doi: 10.1038/nn.4307
Wang, L. H., and Baker, N. E. (2015). E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev. Cell 35, 269–280. doi: 10.1016/j.devcel.2015.10.019
Wapinski, O. L., Lee, Q. Y., Chen, A. C., Li, R., Corces, M. R., Ang, C. E., et al. (2017). Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20, 3236–3247. doi: 10.1016/j.celrep.2017.09.011
Wapinski, O. L., Vierbuchen, T., Qu, K., Lee, Q. Y., Chanda, S., Fuentes, D. R., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. doi: 10.1016/j.cell.2013.09.028
Ward, J. J., Sodhi, J. S., McGuffin, L. J., Buxton, B. F., and Jones, D. T. (2004). Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645. doi: 10.1016/j.jmb.2004.02.002
Watanabe, K., Kamiya, D., Nishiyama, A., Katayama, T., Nozaki, S., Kawasaki, H., et al. (2005). Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 8, 288–296.
Wilkinson, G., Dennis, D., and Schuurmans, C. (2013). Proneural genes in neocortical development. Neuroscience 253, 256–273. doi: 10.1016/j.neuroscience.2013.08.029
Wu, Z. Q., Li, D., Huang, Y., Chen, X. P., Huang, W., Liu, C. F., et al. (2017). Caspr controls the temporal specification of neural progenitor cells through notch signaling in the developing mouse cerebral cortex. Cereb. Cortex 27, 1369–1385.
Yang, G., Smibert, C. A., Kaplan, D. R., and Miller, F. D. (2014). An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron 84, 723–739. doi: 10.1016/j.neuron.2014.10.022
Yang, N., Chanda, S., Marro, S., Ng, Y. H., Janas, J. A., Haag, D., et al. (2017). Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 14, 621–628. doi: 10.1038/nmeth.4291
York, R. D., Yao, H., Dillon, T., Ellig, C. L., Eckert, S. P., McCleskey, E. W., et al. (1998). Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature 392, 622–626. doi: 10.1038/33451
Zahr, S. K., Kaplan, D. R., and Miller, F. D. (2019). Translating neural stem cells to neurons in the mammalian brain. Cell Death Differ. 26, 2495–2512. doi: 10.1038/s41418-019-0411-9
Zahr, S. K., Yang, G., Kazan, H., Borrett, M. J., Yuzwa, S. A., Voronova, A., et al. (2018). A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97, 520-537.e6.
Zandi, S., Bryder, D., and Sigvardsson, M. (2010). Load and lock: the molecular mechanisms of B-lymphocyte commitment. Immunol. Rev. 238, 47–62. doi: 10.1111/j.1600-065x.2010.00950.x
Zecevic, N., Chen, Y., and Filipovic, R. (2005). Contributions of cortical subventricular zone to the development of the human cerebral cortex. J. Comp. Neurol. 491, 109–122. doi: 10.1002/cne.20714
Keywords: Neurog1, Neurog2, Ascl1, phosphorylation, protein–protein interactions, protein stability, epigenetic control, translational control
Citation: Oproescu A-M, Han S and Schuurmans C (2021) New Insights Into the Intricacies of Proneural Gene Regulation in the Embryonic and Adult Cerebral Cortex. Front. Mol. Neurosci. 14:642016. doi: 10.3389/fnmol.2021.642016
Received: 15 December 2020; Accepted: 26 January 2021;
Published: 15 February 2021.
Edited by:
Estela Maris Muñoz, CONICET Dr. Mario H. Burgos Institute of Histology and Embryology (IHEM), ArgentinaReviewed by:
Alessandro Sessa, San Raffaele Hospital (IRCCS), ItalyCopyright © 2021 Oproescu, Han and Schuurmans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carol Schuurmans, Y2Fyb2wuc2NodXVybWFuc0BzdW5ueWJyb29rLmNh; Y3NjaHV1cm1Ac3JpLnV0b3JvbnRvLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.