- 1Department of Neurology, Jinshan Hospital, Fudan University, Shanghai, China
- 2Department of Neurology, Shanghai Medical College, Fudan University, Shanghai, China
Epilepsy is a common neurologic disorder. The underlying pathological processes include synaptic strength, inflammation, ion channels, and apoptosis. Acting as epigenetic factors, non-coding RNAs (ncRNAs) participate in the regulation of pathophysiologic processes of epilepsy and are dysregulated during epileptogenesis. Aberrant expression of ncRNAs are observed in epilepsy patients and animal models of epilepsy. Furthermore, ncRNAs might also be used as biomarkers for diagnosis and the prognosis of treatment response in epilepsy. In this review, we will summarize the role of ncRNAs in the pathophysiology of epilepsy and the putative utilization of ncRNAs as diagnostic biomarkers and therapeutic targets.
Epilepsy is a chronic neurologic disorder which is characterized by recurring unprovoked seizures and affects more than 50 million people worldwide. In China, the prevalence rate is around 2.89%, and 40–50% of these patients do not receive the necessary treatment. China has more than nine million existing cases and about 45,000 new patients suffering from epilepsy each year (Gu et al., 2013). Epilepsy entails recurrent seizures, an increased mortality rate, a huge economic burden, and decreased quality of life. At present, the main treatment is antiepileptic drugs (AEDs), but ~30% of epilepsy patients do not respond to AEDs, a condition known as refractory epilepsy. None of the AEDs currently in use focus on the pathogenesis of epilepsy. Therefore, it is necessary to search for new ways to treat epilepsy. Currently, the pathogenesis of epilepsy is not completely clear. A growing number of studies have shown that ncRNAs participate in pathological and physiological processes in the nervous system, for instance the development of the nervous system, synaptic plasticity, learning and memory, oxidative stress and so on (Aksoy-Aksel et al., 2014; Nwaobi et al., 2014; Karnati et al., 2015; Loke et al., 2015). These characteristics suggest that ncRNAs may play an important role in the pathogenesis of epilepsy. NcRNAs refer to small endogenous RNAs that do not code for proteins, or function without being translated into a protein. Only 1.5% of the human genome appears to code for proteins; about 80% of the rest of the genome transcribes into ncRNAs (Huang et al., 2016). A number of studies have shown that ncRNAs play important roles in epigenetic modifications (Molina, 2017; Redis and Calin, 2017). NcRNAs are divided into many subtypes such as rRNA (ribosomal RNA), tRNA (transfer RNA), snRNAs (small nuclear RNA), snoRNA (small nucleolar RNA), miRNA (microRNA), siRNA (small interfering RNA), piRNA (piwi-interacting RNA), eRNA (enhancer associated RNA), lncRNA (long non-coding RNA), and circRNA (circular RNA) (Redis and Calin, 2017). In this review, we will highlight recent studies examining the latest advances in ncRNAs and their relations with epilepsy.
MicroRNA and Epilepsy
miRNA is a class of small non-coding RNA (ncRNA) which participates in post-transcriptional regulation. The primary transcript is first produced in the nucleus and then processed into hairpin RNA (pre-miRNA) by the Drosha microprocessor complex. Exportin-5 transfers pre-miRNA out of the nucleus for its final processing into mature miRNA by the RNAase III enzyme Dicer (O'Carroll and Schaefer, 2013). Mature miRNA forms RNA-induced silencing complex (RISC) with Argonaute (Ago) proteins, which can form sequence-specific base-pairings with the target mRNA, usually with the 3′ untranslated region (UTR) (Chandradoss et al., 2015; Lin and Gregory, 2015). This formation can lead to mRNA degradation or translational inhibition and then regulates the expression of target genes. Recent studies have demonstrated that miRNA can also regulate promoters with CpG island methylation at the transcriptional level (Benhamed et al., 2012; Tan et al., 2013).
Aberrant Expression of miRNAs in Epilepsy
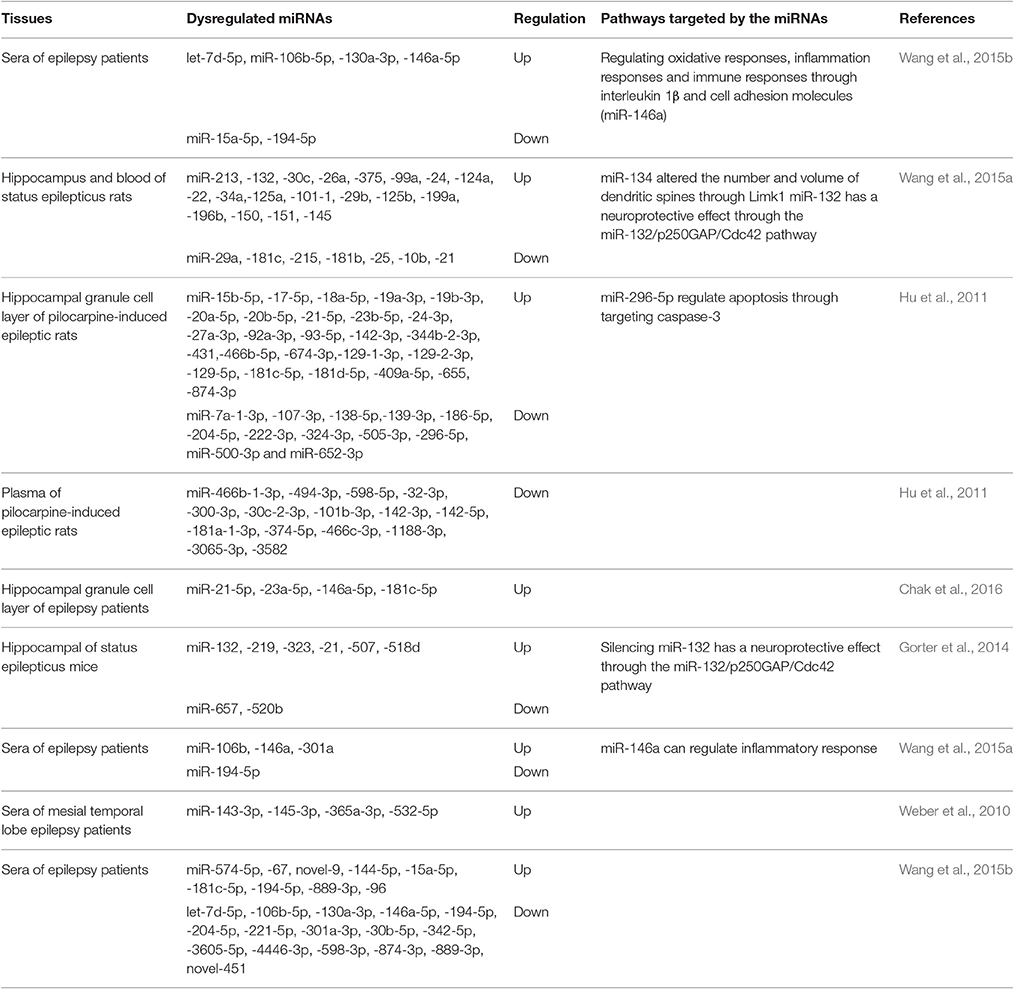
Important roles for miRNA in brain development have been observed, as well as roles in tissue-specific expression in the brain. The aberrant expression of miRNAs was observed in the blood and brain tissues of animal models of epilepsy and epileptic patients (Table 1; Wang et al., 2015b; Zhu et al., 2015). Studies have demonstrated that these aberrant expressions of miRNAs link to the mechanisms of epileptogenesis through regulating ion channels, neuronal morphology, synaptic plasticity, inflammatory response, and neuronal apoptosis. Through comparing the sera miRNA expression of 117 epilepsy patients and 112 healthy controls, it has been found that let-7d-5p, miR-106b-5p, miR-130a-3p, and miR-146a-5p were up-regulated in the sera of epilepsy patients, whereas miR-15a-5p and miR-194-5p were down-regulated (Wang et al., 2015a,b). Another study found that the expression of miR-34a, miR-22, miR-125a, and miR-21 in blood and the hippocampus were changed 24 h after the onset of status epilepticus (SE) (Hu et al., 2011). Roncon et al. (2015) detected miRNA expression in the hippocampal granule cell layer and in plasma at a different phase of the development of pilocarpine-induced epileptic rats. Sixty-three differentially expressed miRNAs were found in the granular cell layer (GCL). When validating miRNAs which were up-regulated in the chronic phase of epileptic rats in brain tissues from epileptic patients, they found that miR-21-5p, miR-23a-5p, miR-146a-5p, and miR-181c-5p were also up-regulated in human brain tissues. They also found that 27 miRNAs were differentially expressed in the plasma samples of epileptic patients. Chak et al. (2016) reported that miR-21 is up-regulated in epilepsy and that pre-miR-21 may attenuate miR-21-mediated suppression following SE and could potentially lead to prolonged TGF-β receptor expression, which can impact epileptogenesis. Gorter et al. (2014) detected miRNA expression in the CA1, dentate gyrus, and parahippocampal cortex regions after electrically-induced SE at 1 day, 1 week, and 3–4 months in a rat model for temporal lobe epilepsy. The expression of miRNAs exhibited dynamic changes after SE and changed differently in different regions. In CA1, 18 miRNAs were up at 1 day, 16 at 1 week, and 7 in the chronic phase. In dentate gyrus, 20 miRNAs were up at 1 day, 15 at 1 week, and 37 at 3 months after SE. In parahippocampal cortex, 31 miRNAs were up at 1 day, 37 at 1 week, and 22 in the chronic stage.
Araujo et al. (2016) performed a miRNA microarray of the hippocampi of Wistar rats 24 h after intra-hippocampal pilocarpine-induced SE. A total of 73 miRNAs were found to be significantly dysregulated, of which 36 were up-regulated and 37 were down-regulated. Among these miRNAs, the expression level of miR-352 and 196b-5p were over-expressed and miR-128a-3p were under-expressed. They also evaluated the three miRNAs at three time points: 0 h, 24 h and chronic phase after SE. They found that the expression of miR-128a-3p was significantly down-regulated at the three time points compared to the control group, miR-352 was significantly up-regulated at 24 h post-SE and in chronic phase, while miR-196b-5p was significantly upregulated only at 24 h post-SE. They found that the expression levels of miRNAs show similar trends to the rat models when compared with the hippocampi of epileptic patients with control group.
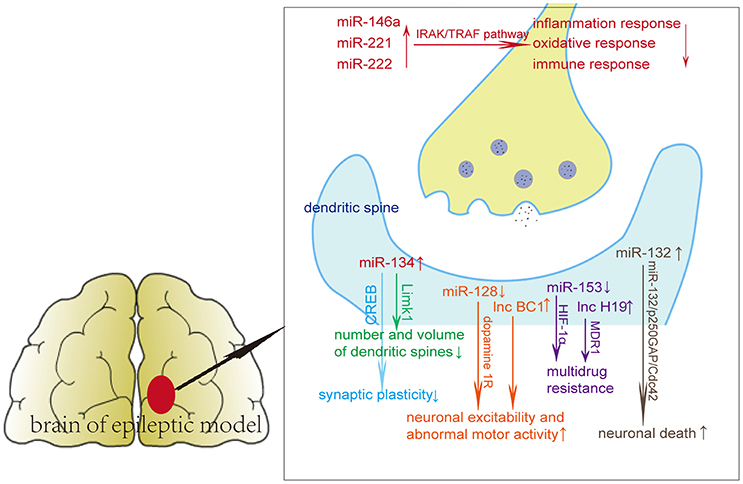
miR-134 is a brain-specific miRNA, and the high expression of miR-134 was observed after the occurrence of epilepsy and SE. Silencing of miR-134 expression could reduce the density of the CA3 pyramidal neuron dendrite spine in the hippocampus and suppress the injury caused by seizures and prolonged seizures. Morphometric analysis of dendritic spines revealed that miR-134 could increase neuron volume and decrease spine volume (Jimenez-Mateos et al., 2012; Wang et al., 2014). It has been demonstrated that LIM kinase-1 (Limk1) plays important roles in dendritic spine morphogenesis through phosphorylating and inactivating cofilin (Meng et al., 2002). Loss of Limk1 results in abnormal spine morphology. Studies found that miR-134 inhibits LIM kinase-1 (Limk1) mRNA, thus preventing protein translation of Limk1. Pretreatment of mice with miR-134 antagomirs before pilocarpine administration reduced the number of mice that developed SE, attenuated the degree of epileptic seizures and increased survival rate (Jimenez-Mateos et al., 2015). In addition, other studies observed expression of miR-134 decreased after the occurrence of epilepsy. miR-134 may prevent synaptic plasticity by inhibiting CREB and p-CREB expressions, thereby exerting neuroprotective qualities (Zhu et al., 2015).
miR-128 is abundant in the brains of humans and adult mice, and the expression of miR-128 increases gradually during the process of growth, peaking in adulthood. miR-128 is encoded by miR-128-1 and miR-128-2. miR-128-2 plays a major role. miR-128-2−/− mice progress rapidly to fatal epilepsy and death. The high expression of miR-128 in mice can reduce seizures through suppressing neuronal excitability and abnormal motor activity. Studies suggest that miR-128 may function through suppressing the dopamine 1 receptor, which can increase neuronal excitability and abnormal motor activity (Tan et al., 2013). Yuan et al. (2016a) reported that miR-128 was down-regulated in glioblastoma, and knockout of the miR-128a could induce epilepsy in a mouse model. Furthermore, dysregulation of miR-128 expression may be associated with glioma-associated epilepsy in low-grade glioma.
miR-146a has been widely studied in the inflammatory response. High expression of miR-146a was observed in both epileptic animal models and epilepsy patients (Matos et al., 2014; Roncon et al., 2015). Studies found that miR-146a expression was at its lowest level at acute seizure phase and at its highest level at the latent phase. Another study revealed that expression of miR-146a was increased in the chronic phase of epileptic rats and remained at high levels 1 week after SE (Gorter et al., 2014; He et al., 2016). Functional polymorphisms of the gene miR-146a may also play a role in the regulation of epilepsy. Rs2910164 and rs57095329 are two SNPs in the gene miR-146a, which can modify the expression level of mature miR-146a. Studies found that rs57095329 may be associated with drug resistance and frequency of seizures (Cui et al., 2015).
Li et al. (2016) found that miR-153 was down expressed in plasma and temporal cortexes resected from surgical mTLE patients compared with control patients. miR-153 might down-regulate HIF-1α expression through binding with two sites in the 3′UTR region of HIF-1α transcript. miR-153 may serve as a putative regulator of hypoxia-inducible factor-1α (HIF-1α), which participates in multidrug resistance in refractory epilepsy. Functional experiments showed that miR-153 mimics can inhibit HIF-1α expression in a pharmacoresistant astrocyte model. miR-153 may regulate the expression of HIF-1α in mTLE and serve as a putative biomarker and treatment target for epilepsy.
Yuan et al. (2016) induced epileptic neurons by exposing the neurons to magnesium-free medium for 3 h, which were considered as a useful in vitro model of refractory SE. They found that silencing miR-132 has a neuroprotective effect on cultured epileptic neurons, including inhibiting the electrical excitability level of cultured epileptic neurons. Using a lithium-pilocarpine-induced epileptic mouse model, they found that silencing miR-132 can inhibit the aberrant formation of dendritic spines and chronic spontaneous seizures. Another study found that pretreatment with miR-132 antagomirs can reduce hippocampus injuries after SE. Upregulation of miR-132 may be associated with neuronal death (Jimenez-Mateos et al., 2011).
miRNAs and Neurogenesis in Epilepsy
Neurogenesis impairment participate in the pathophysiology of epilepsy in humans and also observed in animal models (Mendonca et al., 2017). Aberrant hippocampal neurogenesis can cause epilepsy and seizures can also effect hippocampal neurogenesis. Activation, migration, integration of neural stem cells, and expression of brain plasticity-associated proteins in hippocampus may be required for the maintenance of the kindling criterion (Retchkiman et al., 1996; Schmoll et al., 2003; Buga et al., 2012; Uemori et al., 2017). More and more evidence suggests that miRNAs participate in neurogenesis, which play an important role in the pathophysiological mechanism of epilepsy. miR-124 has been reported to regulate Neuron Restrictive Silencer Factor (NRSF). NRSF was widely expressed in neural stem cells, and regulates the differentiation, diversity and plasticity of neural stem cells. NRSF can also repress the genes HCN1 and KCC2, which can regulate neural activity through ion channels (Brennan et al., 2016). Franzoni et al. (2015) found that miR-128 participates in neuronal migration and intrinsic excitability. miR-146a was reported to participate in the epileptogenesis and progression of seizures through tGCLhe regulation of inflammation and immune responses (He et al., 2016).
Pathways Targeted by miRNAs
A series of functional studies found that miRNAs affect seizures via affecting neuroinflammation or apoptosis. An individual miRNA can have different targets, regulating single genes in several pathways or several genes in a single pathway (Ebert and Sharp, 2012). The dysregulation of miRNAs probably affects various molecular and cellular pathways in epilepsy, including inflammation, oxidative stress, immune responses, axon guidance, cell differentiation, migration, and proliferation (Figure 1, Table 1). miR-134 regulates the number and volume of dendritic spines, presumably through its target Limk1, which plays important roles in dendritic spine morphogenesis through phosphorylating and inactivating cofilin (Jimenez-Mateos et al., 2015). miR-146a, miR-221, and miR-222 can participate in epilepsy via regulating oxidative responses, inflammation responses and immune responses through targets such as interleukin 1β and cell adhesion molecules (Aronica et al., 2010; Kan et al., 2012).
Yan et al. (2016) found that the potential target genes of miR-3613-5p, miR-4668-5p, miR-8071, miR-197-5p, miR-4322, and miR-6781-5p, predicted by bioinformatics analysis, were involved in biological processes, molecular functions, and cellular components through affecting the calcium signaling pathway, the MAPK signaling pathway and the PI3K-Akt signaling pathway. These miRNAs may regulate seizure development in mesial temporal lobe epilepsy with hippocampal sclerosis (mTLE-HS). Yuan et al. (2016) found that silencing miR-132 has a neuroprotective effect on cultured epileptic neurons and lithium-pilocarpine-induced epileptic mouse models through the miR-132/p250GAP/Cdc42 pathway by regulating the morphology and electrophysiology of dendritic spines. Over-expression of miR-184 was observed in mesial temporal lobe epilepsy patients with hippocampal sclerosis compared with mesial temporal lobe epilepsy patients without hippocampal sclerosis. miR-184 may regulate inflammatory responses through regulating inflammatory signal transduction and apoptosis (Danis et al., 2016).
miRNAs as Putative Biomarkers of Epilepsy
A series of studies have reported that changes of miRNAs observed in certain biological fluids correlate with various pathological conditions suggesting that circulating miRNAs might be useful and informative biomarkers to reflect the pathological status of the body (Valentino et al., 2017; Wang, 2017; Zendjabil et al., 2017; Zhao et al., 2017). Evidence has emerged that miRNAs in blood or cerebrospinal fluid may serve as potential biomarkers of brain injury (Weber et al., 2010; Kulstein et al., 2016; Martinez and Peplow, 2016; Sirker et al., 2016; Wang, 2017). These miRNAs in biological fluids may come through the damaged blood–brain barrier after epilepsy onset or originate from controlled release in exosomes (Choi et al., 2017; Gourlay et al., 2017). These miRNAs circulate in blood or cerebrospinal fluid after release due to encapsulation in extracellular vesicles or complexing with proteins (Turchinovich et al., 2012). Animal studies in epileptic rat models suggest that specific miRNA in blood plasma and sera reflected different types of brain injury, including dynamic change after seizure onset and a pattern unique to prolonged seizures (Gorter et al., 2014; Yan et al., 2016).
Wang et al. (2015b) found that miR-106b-5p was up-regulated in sera from epilepsy patients compared with healthy controls, and miR-106b-5p had 80.3% sensitivity and 81.2% specificity in diagnosis of epilepsy. Roncon et al. (2015) reported that the alterations of miR-9a-3p, miR-466b-1-3p, miR-494-3p, and miR-598-5p occurred earlier than the onset of the first spontaneous seizure in a rat model. These miRNAs may be proposed as putative biomarkers of epileptogenesis. Gorter et al. (2014) compared the expression levels of miR-21-5p, miR-146-5p, and 142-5p in plasma with brain tissue in as SE rat model. They found that the expression pattern of miR-21-5p at different time points was similar to that observed in brain tissue. However, the pattern of expression of miR-146a-5p and 142-5p was different from the pattern observed in brain areas. miR-21-5p may serve as a potential biomarker in plasma to reflect dynamic changes in brain tissue. Sun et al. (2016) found that miR-30a was upregulated in the sera of epileptic patients and the expression level of miR-30a was positively associated with seizure frequency at the onset of seizures. Of a total of 50 microRNAs, 2 were increased and 48 were decreased and found to be differentially expressed in mTLE-HS compared with healthy controls. Among these miRNAs, miR-8071 had 83.33% sensitivity and 96.67% specificity in mTLE-HS diagnoses and was associated with seizure severity (Yan et al., 2016). Surges et al. (2016) found that more than 200 miRNAs were differentially expressed within 30 min of seizure onset in the sera of patients. Among these miRNAs, miR-143, miR-145, miR-532, and miR-365a were significantly deregulated. They also found 10 miRNAs to be differentially expressed 20–28 h after seizures in patients with seizures occurring during sleep. miR-663b was significantly deregulated among these 10 miRNAs. Detectable transient miRNA alterations in blood sera were detected after single seizures in the early postictal phase. Li et al. (2016) found that miR-153 was down-regulated in temporal cortexes resected from surgical mTLE patients compared with control patients. Down-regulation of miR-153 in the plasma of mTLE patients was also observed in an independent validation cohort. These studies suggest that the expression level of miRNAs in blood or cerebrospinal fluid is deregulated after epilepsy onset, and some may relate to the severity and frequency of epilepsy. Furthermore, the expression of these miRNAs circulate in biofluids are stable. Due to these characteristics miRNAs may serve as potential biomarkers of epilepsy in the future.
Prospects for miRNAs Therapeutics
At present, diagnosis of epilepsy is mainly based on the clinical symptoms, neuroimaging and electroencephalograms. Early diagnosis is closely related to the clinical prognosis of epilepsy patients. miRNAs may be used as a biomarker in the early diagnosis of epilepsy due to its tissue-specific and stable expression. However, the multi-targeting and multi-pathway actions of miRNAs make prediction and therapeutic function difficult. Several studies have reported the potential therapeutic targets of miRNAs; these studies provide a novel prospect of epilepsy treatment and have improved our understanding of the targets of these miRNAs. A series of functional studies have reported the potential therapeutic targets of epilepsy on miRNA levels through administration of mimics or antagomirs (Jimenez-Mateos et al., 2012; Wang et al., 2014; Yuan et al., 2016). Pre-administration of miR-134 antagomirs to pilocarpine mice reduced the number of mice that triggered SE, attenuated the seizure degree of epileptic mice and increased survival rate (Jimenez-Mateos et al., 2015). Yuan et al. (2016a) reported that dysregulation of miR-128 expression may participate in glioma-associated epilepsy in low-grade glioma, and that knockout of the miR-128a could induce epilepsy in mouse models. Yuan et al. (2016) found that silencing miR-132 decreased the electrical excitability level in epileptic neurons, inhibited the aberrant formation of dendritic spines and attenuated chronic spontaneous seizures in a lithium-pilocarpine-induced epileptic mouse models. Upregulation of miR-132 may be associated with neuronal death and pretreatment with miR-132 antagomirs can attenuate hippocampus injuries after SE (Jimenez-Mateos et al., 2011). These results suggest that silencing miR-132 has a neuroprotective effect and that miR-132 may serve as a putative target for developing antiepileptic treatment.
Further studies focusing on the effect of miRNAs in animal models or epileptic patients are needed in the future.
Long Non-coding RNA
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that have little or no protein-coding capacity, and have drawn intense research efforts recently (Schaukowitch and Kim, 2014). While lncRNAs were at first considered transcriptional noises, recent studies have demonstrated that they play essential roles in biological pathways, including X inactivation, imprinting, development, and differentiation (Danis et al., 2016; Valentino et al., 2017; Zendjabil et al., 2017). lncRNAs can regulate many processes in mammalian and gene expression through a diversity of mechanisms and at different levels, but the available research tools are still insufficient. Therefore, their functions in epilepsy remain unclear. It is known that lncRNAs can serve as molecular scaffolds for combinations of chromatins and proteins, such as homeobox A1 (HOXA1). lncRNAs can also act as enhancers, for instance as enhancer RNAs (eRNAs). They can also repress transcription through inhibiting RNA polymerase II or mediating chromatin remodeling and histone modification (Hewson et al., 2016).
Lee et al. (2015) performed a microarray analysis to compare lncRNAs expression in pilocarpine and kainate models with control mice to study epileptic mechanisms. They found 384 aberrant lncRNAs in the pilocarpine model and 279 aberrant lncRNAs in the kainate model. These dysregulated lncRNAs may participate in epileptic mechanisms. lncRNAs may regulate the occurrence and development of epilepsy through a diversity of mechanisms, such as neurogenesis, regulation of neurotransmitter, ion channels, and synaptic plasticity (Ng et al., 2013). Xiao et al. (2017) reported that aberrantly methylated lncRNA and pathway targets might be involved in TLE development and progression. Abnormal development of the nervous system may cause epilepsy. lncRNAs participate in embryonic morphogenesis and neuronal differentiation in the embryonic period (Mercer et al., 2010). For example, Dlx1as can modulate the expression of neighboring homeobox genes, thereby regulating neuronal differentiation (Ramos et al., 2013). Evf2 plays a role in the regulation of homeodomain transcription factors and the formation of GABA-dependent neuronal circuitry in the developing mouse forebrain (Bond et al., 2009). lncRNAs also participate in synaptic plasticity. lncRNA Malat1 can increase the density of dendritic spines and thereby modulate synaptic plasticity and neuronal regeneration (Wu et al., 2013). The lncRNAs BC1 and BC200 modulate protein synthesis in postsynaptic dendritic microdomains and are thought to play roles in signal transduction and synaptic plasticity. Studies have found that deficiency of BC1 increases neuronal excitability and facilitates the progression of epilepsy (Gitaí et al., 2011). Studies on cancer have found that lncRNA H19 increases multidrug resistance 1 (MDR1) expression and MDR1-associated drug resistance in liver cancer cells through regulating MDR1 promoter methylation. It is considered that MDR1 also plays important roles in the drug resistance of refractory epilepsy. It may be supposed, therefore, that lncRNAs may also participate in refractory epilepsy.
Circular RNA
CircRNAs are a novel type of noncoding RNA differing from linear RNAs. A covalent bond linking the 3′ and 5′ ends of circRNA form closed loop structures (Jeck et al., 2013; Chen et al., 2015). Nuclease hydrolyzes target the tails of linear RNAs, while circular RNAs form closed loop structures; therefore, circRNAs are not susceptible to degradation by RNA exonuclease or RNase (Chen et al., 2016). Due to the stability of circRNAs, expression of circRNAs is more abundant than the corresponding linear mRNAs in plasma (Li Y. et al., 2015; Qin et al., 2016; Shao and Chen, 2016). CircRNAs are highly enriched in eukaryotic organisms and display elevated sequence conservation with specific expression in various tissues during different developmental stages (Memczak et al., 2013; Tan et al., 2017). CircRNAs can act as competitive endogenous RNAs (ceRNAs); the temporal and spatial specificity of circRNA expression supports this possibility (Dong et al., 2016; Ebbesen et al., 2016; Gruner et al., 2016; Szabo et al., 2016). Some circRNAs contain miRNA response elements (MREs) and can interact with miRNAs as miRNA sponges (Hansen et al., 2013; Li F. et al., 2015). Recently gathered evidence indicates that circRNAs participate in the micro-regulation of miRNA expression levels (Zhao et al., 2016; Xue et al., 2017). CircRNAs can perturb miRNA inhibitory functions on target mRNAs by competitive binding with miRNAs, and they can then regulate the expression of target genes (Peng et al., 2016; Zhao et al., 2016; Tang et al., 2017). For instance, sex-determining region Y (SRY) can act as a natural miRNA sponge to suppress miR-138 activity (Yeh et al., 2013; Granados-Riveron and Aquino-Jarquin, 2014; Zhao and Shen, 2015). CircRNAs can modulate the expression of other RNAs through partial base pairing with target RNAs. For example, CDR1as can increase CDR1 mRNA stability through complementary base pairing with CDR1 mRNA. CircRNA can recruit the components of multiprotein complexes or directly combine with proteins and regulate their activity. For example, CDR1as can bind with the protein Argonaute (AGO). Recent studies have found that circRNAs can act as translation templates to encode proteins (Xu et al., 2015; Geng et al., 2016).
CircRNAs may participate in diseases through competitive binding with target miRNAs based on their characteristics (Yu et al., 2016; Xu et al., 2017). Many studies have demonstrated that aberrant circRNAs are associated with the initiation and development of various diseases, but much of the effort in these studies into circRNAs has initially been devoted to cancer research (Zhao and Shen, 2015; Nair et al., 2016; Li et al., 2017). The relationship with epilepsy has not been reported, but the regulatory role of circRNAs is considered widespread. Studies have found that circRNAs are abundant in brain tissues and have specific distribution (Dong et al., 2016; Kumar et al., 2016). For instance, circRNAs are highly expressed in neuropils, especially in dendrites, and participate in regulating synaptic function (You et al., 2015). This suggests that circRNAs may participate in the regulation of synaptic function and neuroplasticity, which play an important role in the development of epilepsy. The antisense to the cerebellar degeneration-related protein1 transcript (CDR1as) acts as a natural miRNA sponge, and can negatively regulate miR-7. High CDR1as expression can decrease miR-7 activity by binding to it and thus increasing target gene expression (Geng et al., 2016). miR-7 is an important regulatory factor in signaling pathways, and it can regulate many other regulatory factors such as epidermal growth factor receptor (EGFR), insulin receptor substrate-1 (IRS-1), AKT3, IRS-2,UBE2A, CTGF, p21-activated kinase- 1 (Pak1), and Raf1(Reddy et al., 2008; Peng et al., 2016; Zhao et al., 2016; Tang et al., 2017). Both CDR1as and miR-7 are abundant in brain tissues, and these regulatory factors play important roles in epilepsy. Thus, we speculate that circRNAs may participate in the epileptogenesis and development of epilepsy. Studies have reported that some circRNAs seem to have virus miRNA binding sites and can affect immune responses. Wang et al. (2015) reported that circular RNA100783 may be involved in chronic CD28-associated CD8 (+) T cell aging and global immunosenescence. miR-138 has a protective effect on brain injury and can regulate T helper 1 (Th1) and T helper 2 (Th2) expressions via inhibiting the function of runt-related transcription factor 3 (RUNX3) and regulate IL-1β through targeting Forkhead Box C1 (Fu et al., 2015; Tang et al., 2016; Yuan et al., 2016b). Circular RNA SRY can act as natural miRNA sponges to suppress miR-138 activity (Granados-Riveron and Aquino-Jarquin, 2014). The above studies suggest that circRNA may participate in inflammatory reactions that induce neuropathy, therefore participating in the epileptogenesis and development of epilepsy.
Conclusions
There are still significant gaps in our current understanding of ncRNAs compared with coding RNAs, and many functions remain unclear. Due to the abundance and stability of ncRNAs in circulating fluids, they may be considered as clinical diagnosis biomarkers in the future. Further study of ncRNAs will improve our understanding regarding epileptogenesis and pathogenesis of epilepsy and lead to new methods of diagnosis and treatment.
Author Contributions
YS conceived the content and wrote the critical review. YC provided the ideas and supervised the work. Both authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported through grants from the National Natural Science Foundation of China (81571261) and the Shanghai Committee of Science and Technology (14411972500).
References
Aksoy-Aksel, A., Zampa, F., and Schratt, G. (2014). microRNAs and synaptic plasticity–a mutual relationship. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130515. doi: 10.1098/rstb.2013.0515
Araujo, M. A., Marques, T. E., Octacilio-Silva, S., Arroxelas-Silva, C. L., Pereira, M. G., Peixoto-Santos, J. E., et al. (2016). Identification of microRNAs with dysregulated expression in status epilepticus induced epileptogenesis. PLoS ONE 11:e0163855. doi: 10.1371/journal.pone.0163855
Aronica, E., Fluiter, K., Iyer, A., Zurolo, E., Vreijling, J., van Vliet, E. A., et al. (2010). Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 31, 1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x
Benhamed, M., Herbig, U., Ye, T., Dejean, A., and Bischof, O. (2012). Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 14, 266–275. doi: 10.1038/ncb2443
Bond, A. M., Vangompel, M. J. W., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. doi: 10.1038/nn.2371
Brennan, G. P., Dey, D., Chen, Y., Patterson, K. P., Magnetta, E. J., Hall, A. M., et al. (2016). Dual and opposing roles of microRNA-124 in epilepsy are mediated through inflammatory and NRSF-dependent gene networks. Cell Rep. 14, 2402–2412. doi: 10.1016/j.celrep.2016.02.042
Buga, A. M., Vintilescu, R., Balseanu, A. T., Pop, O. T., Streba, C., Toescu, E., et al. (2012). Repeated PTZ treatment at 25-day intervals leads to a highly efficient accumulation of doublecortin in the dorsal hippocampus of rats. PLoS ONE 7:e39302. doi: 10.1371/journal.pone.0039302
Chak, K., Roy-Chaudhuri, B., Kim, H. K., Kemp, K. C., Porter, B. E., and Kay, M. A. (2016). Increased precursor microRNA-21 following status epilepticus can compete with mature microRNA-21 to alter translation. Exp. Neurol. 286, 137–146. doi: 10.1016/j.expneurol.2016.10.003
Chandradoss, S. D., Schirle, N. T., Szczepaniak, M., MacRae, I. J., and Joo, C. (2015). A dynamic search process underlies microRNA targeting. Cell 162, 96–107. doi: 10.1016/j.cell.2015.06.032
Chen, I., Chen, C. Y., and Chuang, T. J. (2015). Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 6, 563–579. doi: 10.1002/wrna.1294
Chen, Y., Li, C., Tan, C., and Liu, X. (2016). Circular RNAs: a new frontier in the study of human diseases. J. Med. Genet. 53, 359–365. doi: 10.1136/jmedgenet-2016-103758
Choi, J. W., Kim, S. C., Hong, S. H., and Lee, H. J. (2017). Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dent. Res. 96, 458–466. doi: 10.1177/0022034516685071
Cui, L., Tao, H., Wang, Y., Liu, Z., Xu, Z., Zhou, H., et al. (2015). A functional polymorphism of the microRNA-146a gene is associated with susceptibility to drug-resistant epilepsy and seizures frequency. Seizure Eur. J. Epilepsy 27, 60–65. doi: 10.1016/j.seizure.2015.02.032
Danis, B., van Rikxoort, M., Kretschmann, A., Zhang, J., Godard, P., Andonovic, L., et al. (2016). Differential expression of miR-184 in temporal lobe epilepsy patients with and without hippocampal sclerosis - influence on microglial function. Sci. Rep. 6:33943. doi: 10.1038/srep33943
Dong, R., Ma, X. K., Chen, L. L., and Yang, L. (2016). Increased complexity of circRNA expression during species evolution. RNA Biol. doi: 10.1080/15476286.2016.1269999. [Epub ahead of print].
Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2016). Insights into circular RNA biology. RNA Biol. doi: 10.1080/15476286.2016.1271524. [Epub ahead of print].
Ebert, M. S., and Sharp, P. A. (2012). Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524. doi: 10.1016/j.cell.2012.04.005
Franzoni, E., Booker, S. A., Parthasarathy, S., Rehfeld, F., Grosser, S., Srivatsa, S., et al. (2015). miR-128 regulates neuronal migration, outgrowth and intrinsic excitability via the intellectual disability gene Phf6. Elife 4:e04263. doi: 10.7554/eLife.04263
Fu, D., Yu, W., Li, M., Wang, H., Liu, D., Song, X., et al. (2015). MicroRNA-138 regulates the balance of Th1/Th2 via targeting RUNX3 in psoriasis. Immunol. Lett. 166, 55–62. doi: 10.1016/j.imlet.2015.05.014
Geng, H. H., Li, R., Su, Y. M., Xiao, J., Pan, M., Cai, X. X., et al. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE 11:e0151753. doi: 10.1371/journal.pone.0151753
Gitaí, D. L. G., Fachin, A. L., Mello, S. S., Elias, C. F., Bittencourt, J. C., Leite, J. P., et al. (2011). non-coding RNA BC1 is down-regulated in the hippocampus of Wistar Audiogenic Rat (WAR) strain after audiogenic kindling. Brain Res. 1367, 114–21. doi: 10.1016/j.brainres.2010.10.069
Gorter, J. A., Iyer, A., White, I., Colzi, A., van Vliet, E. A., Sisodiya, S., et al. (2014). Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol. Dis. 62, 508–520. doi: 10.1016/j.nbd.2013.10.026
Gourlay, J., Morokoff, A. P., Luwor, R. B., Zhu, H. J., Kaye, A. H., and Stylli, S. S. (2017). The emergent role of exosomes in glioma. J. Clin. Neurosci. 35, 13–23. doi: 10.1016/j.jocn.2016.09.021
Granados-Riveron, J. T., and Aquino-Jarquin, G. (2014). Does the linear Sry transcript function as a ceRNA for miR-138? The sense of antisense. F1000Res. 3, 90. doi: 10.12688/f1000research.3872.1
Gruner, H., Cortes-Lopez, M., Cooper, D. A., Bauer, M., and Miura, P. (2016). CircRNA accumulation in the aging mouse brain. Sci. Rep. 6:38907. doi: 10.1038/srep38907
Gu, L., Liang, B., Chen, Q., Long, J., Xie, J., Wu, G., et al. (2013). Prevalence of epilepsy in the people's republic of China: a systematic review. Epilepsy Res. 105, 195–205. doi: 10.1016/j.eplepsyres.2013.02.002
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
He, F., Liu, B., Meng, Q., Sun, Y., Wang, W., and Wang, C. (2016). Modulation of miR-146a/complement factor H-mediated inflammatory responses in a rat model of temporal lobe epilepsy. Biosci. Rep. 36, e00433. doi: 10.1042/BSR20160290
Hewson, C., Capraro, D., Burdach, J., Whitaker, N., and Morris, K. V. (2016). Extracellular vesicle associated long non-coding RNAs functionally enhance cell viability. Noncoding RNA Res. 1, 3–11. doi: 10.1016/j.ncrna.2016.06.001
Hu, K., Zhang, C., Long, L., Long, X., Feng, L., Li, Y., et al. (2011). Expression profile of microRNAs in rat hippocampus following lithium–pilocarpine-induced status epilepticus. Neurosci. Lett. 488, 252–257. doi: 10.1016/j.neulet.2010.11.040
Huang, J., Eilbeck, K., Smith, B., Blake, J. A., Dou, D., Huang, W., et al. (2016). The development of non-coding RNA ontology. Int. J. Data Min. Bioinform. 15, 214–232. doi: 10.1504/IJDMB.2016.077072
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Jimenez-Mateos, E. M., Bray, I., Sanz-Rodriguez, A., Engel, T., McKiernan, R. C., Mouri, G., et al. (2011). miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 179, 2519–2532. doi: 10.1016/j.ajpath.2011.07.036
Jimenez-Mateos, E. M., Engel, T., Merino-Serrais, P., Fernaud-Espinosa, I., Rodriguez-Alvarez, N., Reynolds, J., et al. (2015). Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain Struct. Funct. 220, 2387–2399. doi: 10.1007/s00429-014-0798-5
Jimenez-Mateos, E. M., Engel, T., Merino-Serrais, P., McKiernan, R. C., Tanaka, K., Mouri, G., et al. (2012). Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 18, 1087–1094. doi: 10.1038/nm.2834
Kan, A. A., van Erp, S., Derijck, A. A., de Wit, M., Hessel, E. V., O'Duibhir, E., et al. (2012). Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell. Mol. Life Sci. 69, 3127–3145. doi: 10.1007/s00018-012-0992-7
Karnati, H. K., Panigrahi, M. K., Gutti, R. K., Greig, N. H., and Tamargo, I. A. (2015). miRNAs: key players in neurodegenerative disorders and epilepsy. J. Alzheimers Dis. 48, 563–580. doi: 10.3233/JAD-150395
Kulstein, G., Marienfeld, R., Miltner, E., and Wiegand, P. (2016). Automation of DNA and miRNA co-extraction for miRNA-based identification of human body fluids and tissues. Electrophoresis 37, 2742–2750. doi: 10.1002/elps.201600365
Kumar, L., Shamsuzzama, Haque, R., Baghel, T., and Nazir, A. (2016). Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol. Neurobiol. 1–11 doi: 10.1007/s12035-016-0213-8. [Epub ahead of print].
Lee, D. Y., Moon, J., Lee, S. T., Jung, K. H., Park, D. K., Yoo, J. S., et al. (2015). Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem. Biophys. Res. Commun. 462, 433–440. doi: 10.1016/j.bbrc.2015.04.149
Li, P., Chen, H., Chen, S., Mo, X., Li, T., Xiao, B., et al. (2017). Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer 116, 626–633. doi: 10.1038/bjc.2016.451
Li, Y., Huang, C., Feng, P., Jiang, Y., Wang, W., Zhou, D., et al. (2016). Aberrant expression of miR-153 is associated with overexpression of hypoxia-inducible factor-1alpha in refractory epilepsy. Sci. Rep. 6:32091. doi: 10.1038/srep32091
Li, F., Zhang, L., Li, W., Deng, J., Zheng, J., An, M., et al. (2015). Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 6, 6001–6013. doi: 10.18632/oncotarget.3469
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984. doi: 10.1038/cr.2015.82
Lin, S., and Gregory, R. I. (2015). MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333. doi: 10.1038/nrc3932
Loke, Y. J., Hannan, A. J., and Craig, J. M. (2015). The role of epigenetic change in autism spectrum disorders. Front. Neurol. 6:107. doi: 10.3389/fneur.2015.00107
Martinez, B., and Peplow, P. V. (2016). Blood microRNAs as potential diagnostic and prognostic markers in cerebral ischemic injury. Neural Regen. Res. 11, 1375–1378. doi: 10.4103/1673-5374.191196
Matos, G., Scorza, F. A., Mazzotti, D. R., Guindalini, C., Cavalheiro, E. A., Tufik, S., et al. (2014). The effects of sleep deprivation on microRNA expression in rats submitted to pilocarpine-induced status epilepticus. Prog. Neuropsychopharmacol. Biol. Psychiatry 51, 159–165. doi: 10.1016/j.pnpbp.2014.02.001
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Mendonca, F. N., Santos, L. E., Rodrigues, A. M., Gomes, D. S. S., Arida, R. M., Da, S. G., et al. (2017). Physical exercise restores the generation of newborn neurons in an animal model of chronic epilepsy. Front. Neurosci. 11:98. doi: 10.3389/fnins.2017.00098
Meng, Y., Zhang, Y., Tregoubov, V., Janus, C., Cruz, L., Jackson, M., et al. (2002). Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 35, 121–133. doi: 10.1016/S0896-6273(02)00758-4
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G., Mattick, J. S., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Molina, F. E. (2017). Role of ncRNAs in development, diagnosis and treatment of human cancer. Recent Pat. Anticancer Drug Discov. 12, 128–135. doi: 10.2174/1574892812666170105113415
Nair, A. A., Niu, N., Tang, X., Thompson, K. J., Wang, L., Kocher, J. P., et al. (2016). Circular RNAs and their associations with breast cancer subtypes. Oncotarget 7, 80967–80979. doi: 10.18632/oncotarget.13134
Ng, S., Lin, L., Soh, B. S., and Stanton, L. W. (2013). Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 29, 461–468. doi: 10.1016/j.tig.2013.03.002
Nwaobi, S. E., Lin, E., Peramsetty, S. R., and Olsen, M. L. (2014). DNA methylation functions as a critical regulator of Kir4.1 expression during CNS development. Glia 62, 411–427. doi: 10.1002/glia.22613
O'Carroll, D., and Schaefer, A. (2013). General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacol 38, 39–54. doi: 10.1038/npp.2012.87
Peng, L., Chen, G., Zhu, Z., Shen, Z., Du, C., Zang, R., et al. (2016). Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget 8, 808–818. doi: 10.18632/oncotarget.13656
Qin, M., Liu, G., Huo, X., Tao, X., Sun, X., Ge, Z., et al. (2016). Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 16, 161–169. doi: 10.3233/CBM-150552
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi: 10.1016/j.stem.2013.03.003
Reddy, S. D., Ohshiro, K., Rayala, S. K., and Kumar, R. (2008). MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 68, 8195–8200. doi: 10.1158/0008-5472.CAN-08-2103
Redis, R. S., and Calin, G. A. (2017). Snapshot: non-coding RNAs and metabolism. Cell Metab. 25, 220–220.e1. doi: 10.1016/j.cmet.2016.12.012
Retchkiman, I., Fischer, B., Platt, D., and Wagner, A. P. (1996). Seizure induced C-Fos mRNA in the rat brain: comparison between young and aging animals. Neurobiol. Aging 17, 41–44. doi: 10.1016/0197-4580(95)02022-5
Roncon, P., Soukupovà, M., Binaschi, A., Falcicchia, C., Zucchini, S., Ferracin, M., et al. (2015). MicroRNA profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy–comparison with human epileptic samples. Sci. Rep. 5:14143. doi: 10.1038/srep14143
Schaukowitch, K., and Kim, T. K. (2014). Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience 264, 25–38. doi: 10.1016/j.neuroscience.2013.12.009
Schmoll, H., Badan, I., Grecksch, G., Walker, L., Kessler, C., and Popa-Wagner, A. (2003). Kindling status in sprague-dawley rats induced by pentylenetetrazole: involvement of a critical development period. Am. J. Pathol. 162, 1027–1034. doi: 10.1016/S0002-9440(10)63897-7
Shao, Y., and Chen, Y. (2016). Roles of circular RNAs in neurologic disease. Front. Mol. Neurosci. 9:25. doi: 10.3389/fnmol.2016.00025
Sirker, M., Fimmers, R., Schneider, P. M., and Gomes, I. (2016). Evaluating the forensic application of 19 target microRNAs as biomarkers in body fluid and tissue identification. Forensic Sci. Int. Genet. 27, 41–49. doi: 10.1016/j.fsigen.2016.11.012
Sun, J., Cheng, W., Liu, L., Tao, S., Xia, Z., Qi, L., et al. (2016). Identification of serum miRNAs differentially expressed in human epilepsy at seizure onset and post-seizure. Mol. Med. Rep. 14, 5318–5324. doi: 10.3892/mmr.2016.5906
Surges, R., Kretschmann, A., Abnaof, K., van Rikxoort, M., Ridder, K., Frohlich, H., et al. (2016). Changes in serum miRNAs following generalized convulsive seizures in human mesial temporal lobe epilepsy. Biochem. Biophys. Res. Commun. 481, 13–18. doi: 10.1016/j.bbrc.2016.11.029
Szabo, L., Morey, R., Palpant, N. J., Wang, P. L., Afari, N., Jiang, C., et al. (2016). Erratum to: statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 17, 263. doi: 10.1186/s13059-016-1123-9
Tan, C. L., Plotkin, J. L., Veno, M. T., von Schimmelmann, M., Feinberg, P., Mann, S., et al. (2013). MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258. doi: 10.1126/science.1244193
Tan, W. L., Lim, B. T., Anene-Nzelu, C. G., Ackers-Johnson, M., Dashi, A., See, K., et al. (2017). A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 113, 298–309. doi: 10.1093/cvr/cvw250
Tang, C. M., Zhang, M., Huang, L., Hu, Z. Q., Zhu, J. N., Xiao, Z., et al. (2017). CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 7:40342. doi: 10.1038/srep40342
Tang, X. J., Yang, M. H., Cao, G., Lu, J. T., Luo, J., Dai, L. J., et al. (2016). Protective effect of microRNA-138 against cerebral ischemia/reperfusion injury in rats. Exp. Ther. Med. 11, 1045–1050. doi: 10.3892/etm.2016.3021
Turchinovich, A., Weiz, L., and Burwinkel, B. (2012). Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 37, 460–465. doi: 10.1016/j.tibs.2012.08.003
Uemori, T., Toda, K., and Seki, T. (2017). Seizure severity-dependent selective vulnerability of the granule cell layer and aberrant neurogenesis in the rat hippocampus. Hippocampus. doi: 10.1002/hipo.22752. [Epub ahead of print].
Valentino, A., Reclusa, P., Sirera, R., Giallombardo, M., Camps, C., Pauwels, P., et al. (2017). Exosomal microRNAs in liquid biopsies: future biomarkers for prostate cancer. Clin. Transl. Oncol. 19, 651–657. doi: 10.1007/s12094-016-1599-5
Wang, J., Tan, L., Tan, L., Tian, Y., Ma, J., Tan, C. C., et al. (2015a). Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci. Rep. 5:10201. doi: 10.1038/srep10201
Wang, J., Yu, J., Tan, L., Tian, Y., Ma, J., Tan, C., et al. (2015b). Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci. Rep. 5:9522. doi: 10.1038/srep09522
Wang, Y. H., Yu, X. H., Luo, S. S., and Han, H. (2015). Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun. Ageing 12, 17. doi: 10.1186/s12979-015-0042-z
Wang, K. (2017). The ubiquitous existence of microRNA in body fluids. Clin. Chem. 63, 784–785. doi: 10.1373/clinchem.2016.267625
Wang, X., Jia, R., Wei, D., Cui, W., and Jiang, W. (2014). miR-134 blockade prevents status epilepticus like-activity and is neuroprotective in cultured hippocampal neurons. Neurosci. Lett. 572, 20–25. doi: 10.1016/j.neulet.2014.04.049
Weber, J. A., Baxter, D. H., Zhang, S., Huang, D. Y., Huang, K. H., Lee, M. J., et al. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741. doi: 10.1373/clinchem.2010.147405
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 97, 69–80. doi: 10.1016/j.brainresbull.2013.06.001
Xiao, W., Cao, Y., Long, H., Luo, Z., Li, S., Deng, N., et al. (2017). Genome-Wide DNA methylation patterns analysis of noncoding RNAs in temporal lobe epilepsy patients. Mol. Neurobiol. 1–11. doi: 10.1007/s12035-016-0353-x. [Epub ahead of print].
Xu, H., Guo, S., Li, W., and Yu, P. (2015). The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 5:12453. doi: 10.1038/srep12453
Xu, L., Zhang, M., Zheng, X., Yi, P., Lan, C., and Xu, M. (2017). The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 143, 17–27. doi: 10.1007/s00432-016-2256-7
Xue, J., Liu, Y., Luo, F., Lu, X., Xu, H., Liu, X., et al. (2017). Circ100284, via miR-217 regulation of EZH2, is involved in the arsenite-accelerated cell cycle of human keratinocytes in carcinogenesis. Biochim. Biophys. Acta 1863, 753–763. doi: 10.1016/j.bbadis.2016.12.018
Yan, S., Zhang, H., Xie, W., Meng, F., Zhang, K., Jiang, Y., et al. (2016). Altered microRNA profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget 8, 4136–4146. doi: 10.18632/oncotarget.13744
Yeh, Y. M., Chuang, C. M., Chao, K. C., and Wang, L. H. (2013). MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF- α. Int. J. Cancer 133, 867–878. doi: 10.1002/ijc.28086
You, X., Vlatkovic, I., Babic, A., Will, T., Epstein, I., Tushev, G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610. doi: 10.1038/nn.3975
Yu, L., Gong, X., Sun, L., Zhou, Q., Lu, B., and Zhu, L. (2016). The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS ONE 11:e0158347. doi: 10.1371/journal.pone.0158347
Yuan, J., Huang, H., Zhou, X., Liu, X., Ou, S., Xu, T., et al. (2016). MicroRNA-132 Interact with p250GAP/Cdc42 pathway in the hippocampal neuronal culture model of acquired epilepsy and associated with epileptogenesis process. Neural Plast. 2016:5108489. doi: 10.1155/2016/5108489
Yuan, Y., Xiang, W., Yanhui, L., Ruofei, L., Yunhe, M., Jiewen, L., et al. (2016a). Dysregulation of microRNA-128 expression in WHO grades 2 glioma is associated with glioma-associated epilepsy: down-regulation of miR-128 induces glioma-associated seizure. Epilepsy Res. 127, 6–11. doi: 10.1016/j.eplepsyres.2016.08.005
Yuan, Y., Zhang, G. Q., Chai, W., Ni, M., Xu, C., and Chen, J. Y. (2016b). Silencing of microRNA-138-5p promotes IL-1beta-induced cartilage degradation in human chondrocytes by targeting FOXC1: miR-138 promotes cartilage degradation. Bone Joint Res. 5, 523–530. doi: 10.1302/2046-3758.510.BJR-2016-0074.R2
Zendjabil, M., Favard, S., Tse, C., Abbou, O., and Hainque, B. (2017). [The microRNAs as biomarkers: what prospects?]. C. R. Biol. 340, 114–131. doi: 10.1016/j.crvi.2016.12.001
Zhao, Y., Alexandrov, P. N., Jaber, V., and Lukiw, W. J. (2016). Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's Disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes Basel 7, 116. doi: 10.3390/genes7120116
Zhao, Y., Song, Y., Yao, L., Song, G., and Teng, C. (2017). Circulating microRNAs: promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. 36, 77–94. doi: 10.1089/dna.2016.3426
Zhao, Z. J., and Shen, J. (2015). Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 14, 514–521. doi: 10.1080/15476286.2015.1122162
Keywords: non-coding RNA, epilepsy, microRNA, long non-coding RNA, circular RNA
Citation: Shao Y and Chen Y (2017) Pathophysiology and Clinical Utility of Non-coding RNAs in Epilepsy. Front. Mol. Neurosci. 10:249. doi: 10.3389/fnmol.2017.00249
Received: 21 April 2017; Accepted: 25 July 2017;
Published: 10 August 2017.
Edited by:
Andrei Surguchov, Kansas University of Medical Center Research Institute, United StatesReviewed by:
Aurel Popa-Wagner, University of Rostock, GermanyTobias Engel, Royal College of Surgeons in Ireland, Ireland
Federica Calore, The Ohio State University Columbus, United States
Copyright © 2017 Shao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinghui Chen, eWluZ2h1aWNoZW5AZnVkYW4uZWR1LmNu
 Yiye Shao
Yiye Shao Yinghui Chen
Yinghui Chen