
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci. , 07 June 2017
Sec. Molecular Signalling and Pathways
Volume 10 - 2017 | https://doi.org/10.3389/fnmol.2017.00168
This article is part of the Research Topic The Molecular Mechanism Behind Synaptic Transmission View all 22 articles
Membrane fusion is one of the most fundamental physiological processes in eukaryotes for triggering the fusion of lipid and content, as well as the neurotransmission. However, the architecture features of neurotransmitter release machinery and interdependent mechanism of synaptic membrane fusion have not been extensively studied. This review article expounds the neuronal membrane fusion processes, discusses the fundamental steps in all fusion reactions (membrane aggregation, membrane association, lipid rearrangement and lipid and content mixing) and the probable mechanism coupling to the delivery of neurotransmitters. Subsequently, this work summarizes the research on the fusion process in synaptic transmission, using electron microscopy (EM) and molecular simulation approaches. Finally, we propose the future outlook for more exciting applications of membrane fusion involved in synaptic transmission, with the aid of stochastic optical reconstruction microscopy (STORM), cryo-EM (cryo-EM), and molecular simulations.
Neurotransmission is composed of the delivery of neurotransmitters from presynaptic neuron to another neuron, and the feedback of postsynaptic neuron (Jahn and Scheller, 2006; Burnstock, 2007). It is a chemical event which is involved in the transmission of the impulse (Xu et al., 2017), and relies on: the availability of the neurotransmitter; the neurotransmitter release (exocytosis); the binding of the neurotransmitter to the postsynaptic receptor, the excitatory-inhibitory interaction in the postsynaptic cell (Bonifacino and Glick, 2004); and the subsequent removing or deactivating of the neurotransmitter (Iversen, 1971; Heuser and Reese, 1973). Hence, this process requires the controlled release of neurotransmitter from synaptic vesicles by membrane fusion with the presynaptic plasma membrane (Martens and Mcmahon, 2008). Soluble N-ethylmaleimidesensitive factor attachment protein receptors (SNAREs) are the core constituents of the protein machinery which is responsible for synaptic membrane fusion (Jahn and Scheller, 2006). In general, the SNAREs-mediated fusion event is thought to involve a hemifusion diaphragm between the fusion talk and the fusion pore (hemifusion intermediate, Figure 1), where the outer lipid bilayers have been fused, whereas not the inner ones (Zimmerberg, 1987; Brunger et al., 2015). Beyond that, there exists a direct pathway where pre-fusion contact translates into fusion pore without the hemifusion state (Gerst, 1999; Xu et al., 2005; Brunger et al., 2015).
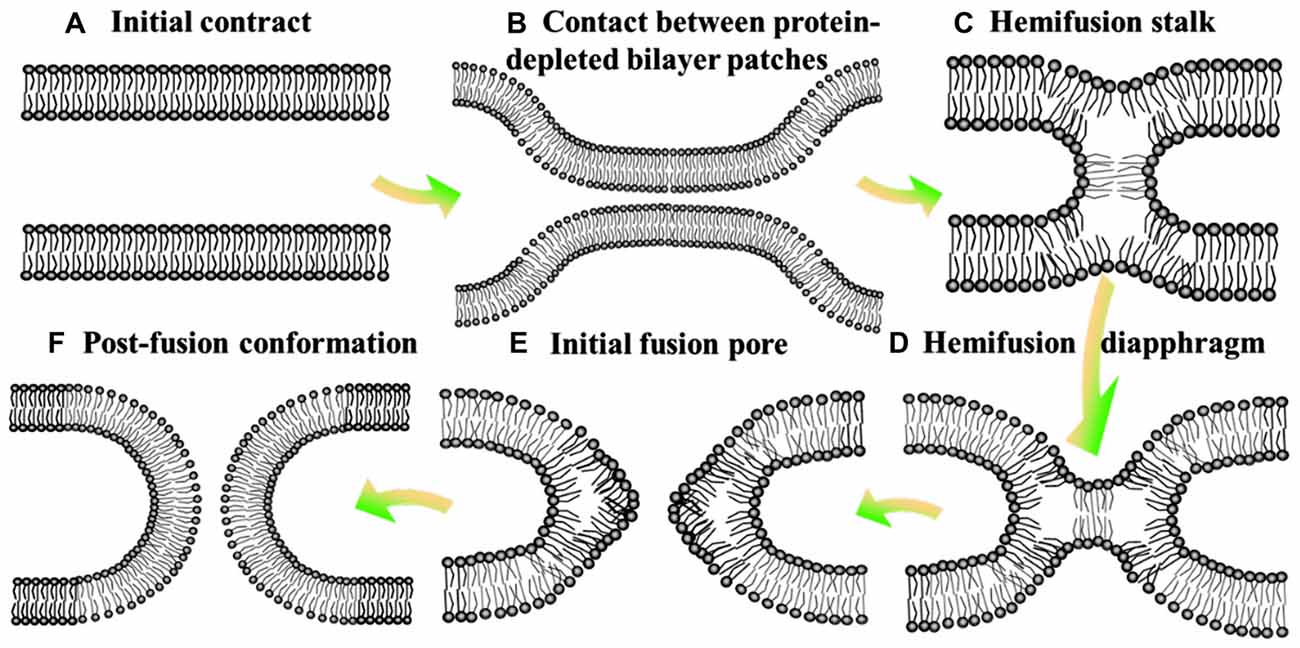
Figure 1. Hemifusion intermediate during the process of membrane fusion. At the state of initial contact (A), lipid bilayers move apart to allow local close contact between two membrane bilayers which proteins mediate membrane binding and fusion (B) and a merger of their contacting leaflets into a stalk like hemifusion connection (C) that expands into a small hemifusion diaphragm (D). An initial fusion pore opens in a HD (E). This pore gives rise to an hourglass fusion pore (F), expansion of which completes the fusion reaction. There show the bilayer surfaces formed by lipid polar heads.
An integral part in the SNAREs-mediated fusion is the assembly of synaptic vesicle transmembrane protein (synaptobrevin) with the target plasma membrane proteins synaptosome-associated protein with relative molecular mass 25 K (SnAP25) and syntaxin (Bommert et al., 1993), which is thought to provide the driving force for the fusion (Haucke et al., 2011). During this process, the extended α-helices of these proteins trend to assemble together, with the formation of four-helix bundles (Mehta et al., 1996). The formed trans-SNARE complex then facilitates the close proximity of vesicular and plasma membranes (~3–4 nm), and induces the membrane fusion (Martens and Mcmahon, 2008).
The spatial regulation of membrane shape, curvature and fluidity are strongly influenced by the lipid composition and topology, during the processes of membrane fusion and fission. Effective neurotransmission requires the precise spatial regulation of lipid-protein interactions for synaptic vesicle targeting, docking, priming and fusion at the active zone (Rohrbough and Broadie, 2005). For an in-depth understanding of the neuronal membrane fusion, various points and states will be summarized in this article, including the process and mechanism of membrane fusion, electron microscope (EM) approaches and molecular simulation results.
Membrane fusion is identified as a process where two separate phospholipid bilayers merge into an interconnected structure. It is a fundamental physiological and pathological process at the level of cell, organelle and vesicle, resulting in the mixing of the two bilayers of lipids and proteins, as well as the mixing of the contents (Jahn et al., 2003).
Despite derived by diverse proteins, all fusion reactions processes four fundamental steps (Jahn and Südhof, 1999): membrane aggregation (approaching each other), membrane association (coming into a very close apposition), lipid rearrangement (highly-localized lipid rearrangements of adjacent two bilayers) and lipid and content mixing (complete fusion; Figure 1; Wilschut and Hoekstra, 1986; Plattner et al., 1992; Blijleven et al., 2016).
Hemifusion of lipid bilayers is an important intermediate state in the membrane fusion, which might be boosted by negative spontaneous curvature of monolayer (monolayer trends to bulge toward the hydrophobic tails) and deformation of monolayer induced by the distortion of lipid monolayer (inclusion of amphiphilic peptides; Figure 1; Chernomordik et al., 2006). In addition, bilayers hemifuse when brought to distances of the polar heads of lipids much smaller than the one in the equilibrium state, by adding polyethylene glycol to draw water from the contact zone or by a direct dehydration of multileveled lipid sample. To complete the fusion process, the hemifusion state should proceed to a full fusion pore (Geisow and Fisher, 1986). The pore might open directly from a fusion stalk (stalk-pore pathway) or from a hemifusion state with discernible hemifusion intermediates (hemifusion-fusion pathway; Chernomordik et al., 2006). The formation and closure of the fusion pore are usually regulated by the conformational change with a high activation energy and phase separated lipids, respectively (Oberhauser et al., 1992). The hemifusion diaphragm is a possible intermediate between the stalk and the final fusion pore (Chernomordik et al., 2006). However, Diao et al. (2012) observed more fast fusion (on the ms scale) upon Ca2+ addition starting from a hemifusion-free state. This discovery revealed that the neurotransmitter release (especially the fastest event) is more dependent on the immediate pathway, and then stimulated a substantially revised membrane fusion paradigm for the membrane fusion (Wickner and Rizo, 2017).
The major constituent of the most bilayer lipid, especially the membranes of mammalian cells, is phosphatidylcholine (PC) which has no spontaneous fusion for hours or days (Wang, 2010). Only the tension (or dehydration contact zones) of these bilayers could fuse by the interposition of polyethylene glycol (Sharma and Lindau, 2016). A monolayer protruding into the layer of polar heads seems to consist of molecules with a reasonable inverted cone (Epand, 1998) and positive spontaneous curvature (Chernomordik et al., 2006). A lipid monolayer that orients toward the hydrocarbon tails usually possesses a negative curvature, composed by cone shaped lipid molecules (Chizmadzhev, 2004). A possible explain is that the action of proteins in fusion process do not directly facilitate the formation of fusion intermediates, also generate fusogenic lipids (sphingomyelinase and phospholipase; Epand, 2000; Chernomordik and Kozlov, 2008). It is promoted by the defects created of the bilayers referring membrane perturbation, including the vicinity of lipid phase transition, the separation of lateral phase or the generation of domain, the high local curvature of membrane, osmotic or electric stress in or on the membrane; the present of amphipaths or macromolecules within the membrane, etc. (Cevc and Richardsen, 1999). High concentrations of lysolipids, which increase the intrinsic curvature of the monolayer, inhibit several biological membrane fusion processes (Shangguan et al., 1996; Söllner, 2004).
The context of membrane fusion in vivo is more complicated since biological fusion is always mediated by protein. However, the specific mechanisms of these processes still remain elusive, especially on proteins which promote the development of hemifusion and fusion pore. Viral membrane fusion and synaptic membrane fusion are widely studied among current protein-mediated membrane fusions. The former promotes the combine between the viral membrane and the host cell membrane, then inducing the release of viral genome into the cytoplasm, as well as the replication cycle of virus. There are two mechanisms of activating viral fusion proteins: exposure to low pH and pH-independent (Earp et al., 2005).
The latter carries the neurotransmitter across the synapses (neurotransmitter release) and plays an important role in the signals traveling in the central nervous system. Neurotransmitter release during the synaptic membrane fusion process requires a protein family that have termed SNAREs which can be divided into four categories (Diao et al., 2012; Hughson, 2013): (1) vesicle-anchored (v) and target-membrane–anchored (t) SNAREs; (2) N-ethylmaleimide–sensitive factor (NSF) and NSF attachment proteins (SNAPs); (3) Rab GTPases and multicomponent vesicle tethering complexes; and (4) Sec1/Munc18 (SM) proteins. So far, the mechanism of this family still has a few basic doubts, such as function, conformational changes, etc. The SNARE-mediated membrane fusion was conducted the zippering mechanism which pulls two membranes together (Diao et al., 2013b). In general, synaptic membrane fusion requires a consecutive two-step pathway. First, the N-terminal domain of the vesicle (v-) SNARE, synaptobrevin-2, docks to the target membrane (t-) SNARE, thereby results in a conformational rearrangement of a half-zippered SNARE complex. Then, the assembled SNARE complex locks the C-terminal portion of the t-SNARE into the same way as the four-helix bundle, which is formed with syntaxin and SNAP-25. Besides, this fusion is greatly accelerated by synaptotagmin-Ca2+ (Lai et al., 2013). Reconstitutions of synaptic vesicle fusion indicated that the interactions between the Ca2+-binding loops of the synaptotagmin-1 and phospholipids are critical to release of neurotransmission, when little content mixing occurs in the absence of Ca2+ (Diao et al., 2013b; Wickner and Rizo, 2017).
In general, neuronal communication is mediated by neurotransmitters release induced by Ca2+-induced synaptic vesicle exocytosis. It is a long-sought goal that understanding the mechanism of synaptic vesicle fusion, associated with the development of in vivo synthetic system. With the advent of modern electron microscopic techniques, we could particularly investigate the neurotransmission and the consequent membrane alterations.
During neurotransmitter release, several SNARE protein complexes involve with synaptobrevin (vesicle (v-) SNAREs) and syntaxin and SNAP-25 (target membrane (t-) SNAREs) to mediate the fusion of two membranes. Meanwhile, this vesicle membrane fusion is acutely triggered in a Ca2+-dependent manner (Figure 2). Munc18 (also called neuronal Sec1) forms a tight complex with syntaxin, with a closed conformation that is unable to bind other SNAREs. Regarding as Munc13, a synaptic vesicle “priming” protein, catalyzes the transition from syntaxin-Munc18 complex to fully assembled v/t-SNARE complex (syntaxin—SNAP-25—synaptobrevin), via bridging the vesicle and plasma membranes and controlling vesicle tethering (Hughson, 2013; Ma et al., 2013; Wickner and Rizo, 2017). Chen et al. (2002) explored the atomic structure of the complexin/SNARE complex, using the X-ray and TROSY-based NMR methods. The results revealed that complexin presents an antiparallel helical conformation, stabilizes the interface between two helices of synaptobrevin and syntaxin, thus enables the Ca2+-evoked neurotransmitter release with the exquisitely high speed.
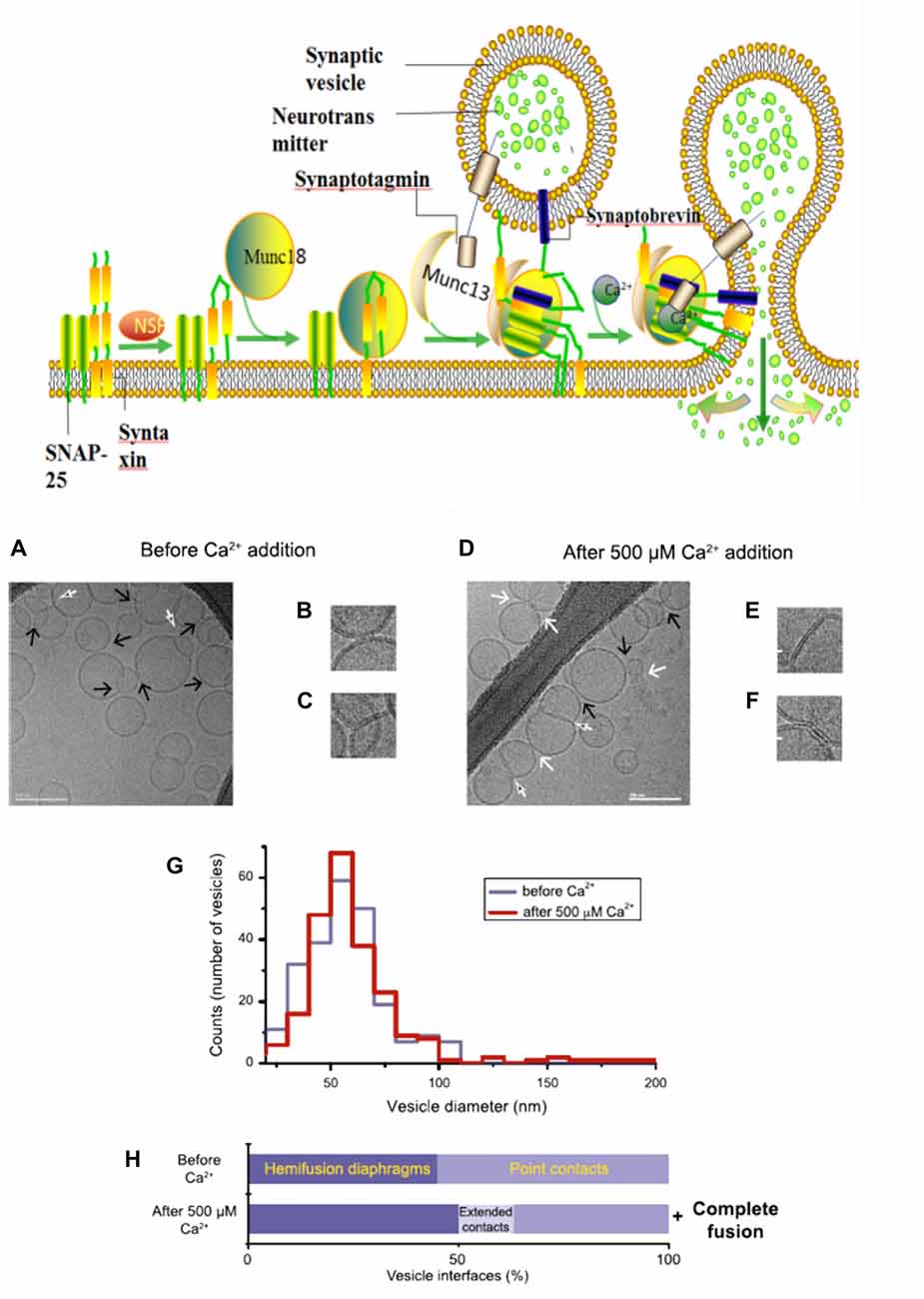
Figure 2. A model with Muncs (Upside) the Soluble N-ethylmaleimidesensitive factor attachment protein receptor (SNARE) complex, fused by v-liposomes (containing synaptobrevin) and t-liposomes (containing SNAP-25 and syntaxin), was disassembly derived under the N-ethylmaleimide–sensitive factor (NSF) with the consuming of adenosine triphosphate (ATP). The entry of Munc18-bound syntaxin into SNARE complexes was catalyzed by Munc13, with the assembly of ternary v/t-SNARE complexes. Then Synaptotagmin-Ca2+ induce the release of neurotransmitter, thereby drive membrane fusion. Imaging of donor/acceptor interface morphologies by cryo-electron microscopy (EM) before and after Ca2+ addition (Downside; Diao et al., 2012).
Over the past 10 years, cryo-electron microscopy (cryo-EM) has been developing rapidly, which combines the potential of three-dimensional (3D) imaging at molecular resolution with a close-to-life preservation of biological samples. Rapid freezing followed by the investigation of the frozen-hydrated samples avoids the artifacts caused by chemical fixation and dehydration procedures. Furthermore, the biological material is observed directly, without heavy metal staining, avoiding artifacts caused by the unpredictable accumulation of staining material (Lucić et al., 2005). The vesicle clusters induced by Ca2+-bound C2 domains of synaptptagmin-1 was visualized by the Cryo-EM method, and the tomographic 3D reconstruction of a vesicle cluster revealed that this process might be induced by Ca2+-dependent phospholipid binding of the C2AB fragment, where the C2B domain cooperates with the SNAREs bring the membranes together, as well as the multiple interactions between the C2B domain and phospholipids (diacyglycerol and phosphatidylinositol 4,5-bisphosphate (PIP2); Araç et al., 2006).
In fact, there is still an active debate regarding whether SNAREs are linked to pre-fusion contact to a fusion pore or participate later in the fusion process by facilitating hemifusion, through the formation of tight SNARE complexes and gathering of the vesicle and plasma membranes. With the aid of single-vesicle fluorescence fusion assay and EPR, the direct observation of two-faceted functions of complexin revealed the formation of a complex substrate (SNARE complexes, complexins and phospholipids) for Ca2+ and Ca2+-sensing fusion effectors in the release process of neurotransmitter (Yoon et al., 2008). Neuron firing gives rising of the intracellular Ca2+ concentration, with the triggering of synaptic vesicles fusion to carry neurotransmitter molecules. Diao et al. (2012) used recently developed Cyro-EM method to monitor the temporal sequence of both content and lipid exchange upon Ca2+-triggering between single pairs of donor and acceptor vesicles on a 100-ms time scale (Diao et al., 2012). Their system performed a quantitative analysis of all observed cryo-EM images (before and after Ca2+-injection) and achieved a Ca2+ sensitivity in the 250–500 μM range (Figure 2). During their experiments, hemifusion diaphragms were observed, as well as points where liposomes contacted each other without the shape change of membrane. Extended tight contacts of membrane were not observed, without the present of Ca2+. With the addition of Ca2+, there merely exists the process from point-contacts to fast fusion (Diao et al., 2012). It was found that alone neuronal SNAREs cannot efficiently induce the complete fusion. The combination of SNAREs with selected components (small-head group lipids, Munc18-1, Munc 13 and synaptotagmin-Ca2+) could lower the activation barriers during the fusion process, because of enhancing the kinetic control by complexin (Kyoung et al., 2011; Wickner and Rizo, 2017). Bharat et al. (2014) performed reconstitutes synaptic fusion and applied large-scale, automated cryo-electron tomography to observe this in vitro system. Afterwards docking and priming of vesicles with the fast Ca2+-triggered fusion, a local protrusion in the plasma membrane will be induced by the SNARE proteins, with the direction towards the primed vesicle and allowing synchronous and instantaneous fusion upon the complexin clamp release (Zhang et al., 2015b).
Many efforts have been devoted to modeling the membrane fusion process involved in synaptic transmission via molecular simulations, such as Coarse-grained (CG) molecular dynamics (MD) simulations. The results of these studies revealed the mechanics of membrane fusion involved in synaptic transmission and some key physical properties of lipid monolayers and related proteins.
The SNARE complex between opposing membranes promotes membrane fusion of synaptic transmission (Mayer, 1999; Pfeffer, 1999). In vivo, the formation of complex connects the opposing membranes and pulls two membranes together using their α-helical transmembrane domains (TMD; Ossig et al., 2000). In addition, SNARE complexes are also deemed to overcome the fusion barriers and to accelerate the fusion process (Chen and Scheller, 2001; Hong, 2005; Risselada and Grubmüeller, 2012).
The SNARE complex is represented by a twisted bundle of four α-helices which generally consists of SNAP-25, syntaxin-1, and synaptobrevin-2, confirmed by coarse-grain MD simulations (Durrieu et al., 2009; Tekpinar and Zheng, 2014) There is mechanistically link between the conformational flexibility of SNARE TMD helices and their ability to induce lipid mixing (Nagy et al., 2005; Stelzer et al., 2008). The basic residues (positive charged) at the C terminal of SNAP-25 is benefit for the tight zippering of SNARE complex and the binding with negatively charged lipid head groups, improving the high frequency and clipping neurotransmitter release (Fang et al., 2015). Besides, the transmembrane domain of synaptobrevin II (sybII TMD) influences both the natural helicity and flexibility of SNARE (Gao et al., 2012; Zheng, 2014; Han et al., 2016). The assemblage of SNARE complex is regulated by complexin, a cytoplasmic neuronal protein and the MD results suggest that the α-accessory helix of complexin (Cpx AH) make partially unzipped state of the SNARE bundle being stabilized by its functions in relate in the clamping of synaptic vesicle fusion (Ghahremanpour et al., 2010; Bykhovskaia et al., 2013; Lai et al., 2014, 2016; Gong et al., 2016).
The composition of several parts of the SNARE completes its function of modulate membrane fusion (Figure 3). MD simulations have also been used in the study of other functions in relation with membrane fusion. During the synaptic transmission, vesicles filled with neurotransmitter molecules are required to be docked to the membrane (Knecht and Grubmüller, 2003; Bock et al., 2010; Lai et al., 2015). Therefore, the SNARE complex use attractive forces to counterbalance the long-range repulsion between the vesicle and membrane (Diao et al., 2013a; Fortoul et al., 2015). More recently, the formation of a transient pore by using the MD method was first reported. The close contact of two membranes gives rise to a high local transmembrane voltage. The decrease of the distance of the opposed bilayers brings out the increase of the transmembrane voltage. When the distance is under a critical value, the local transmembrane voltage is enough high to induce the transient of membrane pores (Figure 4; Ribrault et al., 2011; Bu et al., 2016). Finally, some findings have offered new structural and dynamic details of SNARE disassembly mechanism based on CG modeling (Zheng, 2016).
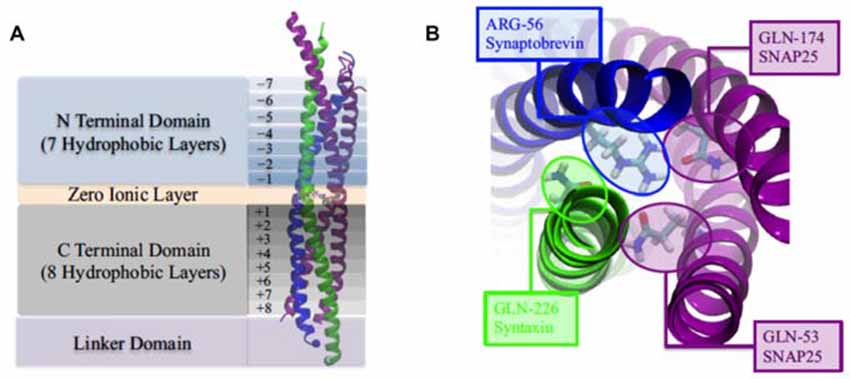
Figure 3. (A) The four domains of neuronal SNARE protein: N Terminal Domian, Zero Ionic Layer, C Terminal Domain, Linker Domain; and involved four helices: snaptobrevin (blue), syntaxin (green), synaptosome-associated protein with relative molecular mass 25 K (SNAP25; magenta). (B) Top view of zero ionic layer. Reproduced with permission from Tekpinar and Zheng (2014).
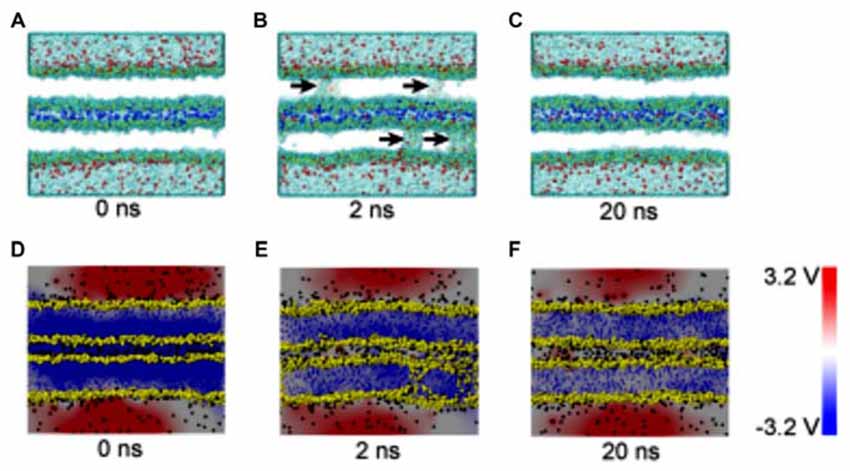
Figure 4. Snapshots and electrical potential distributions during the process of fusion pore (membrane contact-pore formation-membrane healing) through the molecular dynamics (MD) simulations (A–C): the process of fusion pore formation. (D–F): electric potential alteration during the process (Bu et al., 2016).
The controlled release of neurotransmitter by membrane fusion, from synaptic vesicles to presynaptic cell, is an important step in the synaptic transmission (Trimbuch and Rosenmund, 2016). This universal fusion can be accelerated by synaptotagmin-Ca2+, with the assembly of specialized proteins (such as SNAREs) within the opposing membrane bilayers. Electron density map, 3-D topography and simulation studies of the SNARE ring complex, advance that membrane-associated SNAREs overcome repulsive forces to process the two membranes being close to each other (just 2.8 Å apart; Chen and Scheller, 2001). However, people are actually working on an assumption that all proteins are at the right position for inducing membrane fusion. Since all proteins are unlabeled, one is not able to tell the real story on the protein side, which is also an important part for the study of membrane fusion in synaptic transmission. Thus, in the future, it requires the high spatial resolution techniques in order to monitor these interactions simultaneously during the fusion processes, such as stochastic optical reconstruction microscopy (STORM), is required (Diao et al., 2011). In STORM, single biomolecules containing photo-switchable fluorophores are turned on and off repeatedly, to find their positions precisely with ~20 nm resolution by determining the center position of the point-spread function from reconstructed images for each biomolecule (Rust et al., 2006).
Cryo-electron microscopy, abbreviated as “cryo-EM”, is a form of transmission EM (TEM) technique which observes the sample (generally biological sample) at cryogenic temperatures in order to void the ultrastructural changes (Doerr, 2016). It has been rapidly developed in the decade, with increasing popularity in structural biology (Callaway, 2015; Nogales et al., 2016). As cryo-EM became matured, it has been adopted by an ever-increasing range of disciplines to offer tools for providing a cell-like yet simplified environment for investigating the membrane fusion, especially the dynamic structural change of important proteins and the dynamic mechanism of fusion process. In particular, optimized negative-staining (OpNS) EM images have revealed several important physical attributes of CETP and substantial molecular basis for the CETP-mediated lipid exchange (Zhang et al., 2012, 2015a).
Active zones of synaptic plasma membranes are known to concentrate the components that drive membrane fusion, such as the SNAREs, Munc 18-1, Munc 13-1 and small-head group lipids (e.g., diacyglycerol and PIP2), while the participation and characteristic of these macromolecular complexes are still not fully understood (Rizo and Xu, 2015; Ryu et al., 2016; Wickner and Rizo, 2017). Strikingly, current experimental techniques do not achieve a resolution better than ms/μs in time, and neuronal membrane fusion normally occurs at ms timescale (Diao et al., 2012). CG MD simulation is one of effective solutions to overcome the time-scale gap between computational and experimental methods (Bhushan, 2016). With the aid of residue-based and shape-based CG approaches, the regulatory mechanisms of SNARE proteins have been briefly outlined, focused on the intricate molecular mechanisms between proteins and membranes (Bu et al., 2016; Zhang et al., 2016; Han et al., 2017). However, the application of CG models sacrifice degrees of freedom and accurate molecular interactions to get the requirement of less resources (Bhushan, 2016). Though the prohibitive computational cost usually limits the simulation times and system sizes of all-atom models less than 1000 ns and 10 nm, it will provide the description of SNARE-mediated membrane fusions with all-atom detail, such as the specific lipid properties for stimulating fusion, the tethering/SM protein complex, the lipid-protein (such as Munc18-1, Munc13-1, and complexin) interactions, and membrane architecture. Nevertheless, molecular simulation selectivity leads to the factitious results of synaptic membrane fusion, therefore, the computational methods and initial models should be amending continuously by the sufficient basic parameters derived from the non-invasive experimental observations (such as STORM and cryo-EM; Diao et al., 2013b; Wickner and Rizo, 2017).
As STORM, cryo-EM, and all-atoms molecular simulations continue to develop through advances in technological innovation, the combination of the three techniques will be a powerful tool for the in-depth investigation on the regulatory mechanisms of synaptic membrane fusion at atomistic resolution, uncovering the recruitment process of Sec1-Munc 18 family proteins to catalyze SNARE assembly, specific lipid properties which be crucial for fusion, and the intricate balance of protein-lipid interactions. We expect to see more exciting applications of synaptic membrane fusion with continued advances in these methods.
The manuscript was initially drafted by ZY, SZ and LZ and then further edited after discussion with LG, SC and NL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RS and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
We thank the anonymous reviewers for the helpful suggestions. This research was supported by the National Natural Science Foundation of China (No. 11504287, 11374237), Fundamental Research Funds for the Central Universities, China Postdoctoral Science Foundation (2017M613147) and Shaanxi Province Postdoctoral Science Foundation (2015). LZ acknowledges Young Talent Support Plan of Xi’an Jiaotong University.
Araç, D., Chen, X., Khant, H. A., Ubach, J., Ludtke, S. J., Kikkawa, M., et al. (2006). Close membrane-membrane proximity induced by Ca2+-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat. Struct. Mol. Biol. 13, 209–217. doi: 10.1038/nsmb1056
Bharat, T. A., Malsam, J., Hagen, W. J., Scheutzow, A., Söllner, T. H., and Briggs, J. A. (2014). SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium-triggered fusion. EMBO Rep. 15, 308–314. doi: 10.1002/embr.201337807
Bhushan, B. (Eds). (2016). “Coarse-grained molecular dynamics,” in Encyclopedia of Nanotechnology (Dordrecht: Springer), 604–604.
Blijleven, J. S., Boonstra, S., Onck, P. R., van der Giessen, E., and van Oijen, A. M. (2016). Mechanisms of influenza viral membrane fusion. Semin. Cell Dev. Biol. 60, 78–88. doi: 10.1016/j.semcdb.2016.07.007
Bock, L. V., Hutchings, B., Grubmüller, H., and Woodbury, D. J. (2010). Chemomechanical regulation of SNARE proteins studied with molecular dynamics simulations. Biophys. J. 99, 1221–1230. doi: 10.1016/j.bpj.2010.06.019
Bommert, K., Charlton, M. P., Debello, W. M., Chin, G. J., Betz, H., and Augustine, G. J. (1993). Inhibition of neurotransmitter release by C2-domain peptides implicates synaptotagmin in exocytosis. Nature 363, 163–165. doi: 10.1038/363163a0
Bonifacino, J. S., and Glick, B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166. doi: 10.1016/s0092-8674(03)01079-1
Brunger, A. T., Cipriano, D. J., and Diao, J. (2015). Towards reconstitution of membrane fusion mediated by SNAREs and other synaptic proteins. Crit. Rev. Biochem. Mol. Biol. 50, 231–241. doi: 10.3109/10409238.2015.1023252
Bu, B., Tian, Z., Li, D., and Ji, B. (2016). High transmembrane voltage raised by close contact initiates fusion pore. Front. Mol. Neurosci. 9:136. doi: 10.3389/fnmol.2016.00136
Burnstock, G. (2007). Physiology and pathophysiology of purinergic neurotransmission. Physiol. Rev. 87, 659–797. doi: 10.1152/physrev.00043.2006
Bykhovskaia, M., Jagota, A., Gonzalez, A., Vasin, A., and Littleton, J. T. (2013). Interaction of the complexin accessory helix with the C-terminus of the SNARE complex: molecular-dynamics model of the fusion clamp. Biophys. J. 105, 679–690. doi: 10.1016/j.bpj.2013.06.018
Callaway, E. (2015). The revolution will not be crystallized: a new method sweeps through structural biology. Nature 525, 172–174. doi: 10.1038/525172a
Cevc, G., and Richardsen, H. (1999). Lipid vesicles and membrane fusion. Adv. Drug Deliv. Rev. 38, 207–232. doi: 10.1016/S0169-409X(99)00030-7
Chen, X., Tomchick, D. R., Kovrigin, E., Araç, D., Machius, M., Südhof, T. C., et al. (2002). Three-dimensional structure of the complexin/SNARE complex. Neuron 33, 397–409. doi: 10.1016/S0896-6273(02)00583-4
Chen, Y. A., and Scheller, R. H. (2001). SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2, 98–106. doi: 10.1038/35052017
Chernomordik, L. V., and Kozlov, M. M. (2008). Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15, 675–683. doi: 10.1038/nsmb.1455
Chernomordik, L. V., Zimmerberg, J., and Kozlov, M. M. (2006). Membranes of the world unite! J. Cell Biol. 175, 201–207. doi: 10.1083/jcb.200607083
Chizmadzhev, Y. A. (2004). The mechanisms of lipid-protein rearrangements during viral infection. Bioelectrochemistry 63, 129–136. doi: 10.1016/j.bioelechem.2003.10.016
Diao, J., Cipriano, D. J., Zhao, M., Zhang, Y., Shah, S., Padolina, M. S., et al. (2013a). Complexin-1 enhances the on-rate of vesicle docking via simultaneous SNARE and membrane interactions. J. Am. Chem. Soc. 135, 15274–15277. doi: 10.1021/ja407392n
Diao, J., Zhao, M., Zhang, Y., Kyoung, M., and Brunger, A. T. (2013b). Studying protein-reconstituted proteoliposome fusion with content indicators in vitro. Bioessays 35, 658–665. doi: 10.1002/bies.201300010
Diao, J., Grob, P., Cipriano, D. J., Kyoung, M., Zhang, Y., Shah, S., et al. (2012). Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. Elife 1:e00109. doi: 10.7554/eLife.00109
Diao, J., Ishitsuka, Y., and Bae, W. R. (2011). Single-molecule FRET study of SNARE-mediated membrane fusion. Biosci. Rep. 31, 457–463. doi: 10.1042/BSR20110011
Doerr, A. (2016). Single-particle cryo-electron microscopy. Nat. Methods 13:23. doi: 10.1038/nmeth.3700
Durrieu, M.-P., Bond, P. J., Sansom, M. S. R., Lavery, R., and Baaden, M. (2009). Coarse-grain simulations of the R-SNARE fusion protein in its membrane environment detect long-lived conformational sub-states. Chemphyschem 10, 1548–1552. doi: 10.1002/cphc.200900216
Earp, L. J., Delos, S. E., Park, H. E., and White, J. M. (2005). The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285, 25–66. doi: 10.1007/3-540-26764-6_2
Epand, R. M. (1998). Lipid polymorphism and protein-lipid interactions. Biochim. Biophys. Acta 1376, 353–368. doi: 10.1016/S0304-4157(98)00015-X
Fang, Q., Zhao, Y., Herbst, A. D., Kim, B. N., and Lindau, M. (2015). Positively charged amino acids at the SNAP-25 C terminus determine fusion rates, fusion pore properties and energetics of tight SNARE complex zippering. J. Neurosci. 35, 3230–3239. doi: 10.1523/JNEUROSCI.2905-14.2015
Fortoul, N., Singh, P., Hui, C.-Y., Bykhovskaia, M., and Jagota, A. (2015). Coarse-grained model of SNARE-mediated docking. Biophys. J. 108, 2258–2269. doi: 10.1016/j.bpj.2015.03.053
Gao, Y., Zorman, S., Gundersen, G., Xi, Z. Q., Ma, L., Sirinakis, G., et al. (2012). Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337, 1340–1343. doi: 10.1126/science.1224492
Geisow, M. J., and Fisher, D. (1986). Molecular mechanisms of membrane-fusion. Biochem. Soc. Trans. 14, 241–242.
Gerst, J. E. (1999). SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell. Mol. Life Sci. 55, 707–734. doi: 10.1007/s000180050328
Ghahremanpour, M. M., Mehrnejad, F., and Moghaddam, M. E. (2010). Structural studies of SNARE complex and its interaction with complexin by molecular dynamics simulation. Biopolymers 93, 560–570. doi: 10.1002/bip.21397
Gong, J. H., Lai, Y., Li, X. H., Wang, M. X., Leitz, J., Hu, Y. C., et al. (2016). C-terminal domain of mammalian complexin-1 localizes to highly curved membranes. Proc. Natl. Acad. Sci. U S A 113, E7590–E7599. doi: 10.1073/pnas.1609917113
Han, J., Pluhackova, K., and Böckmann, R. A. (2017). The multifaceted role of SNARE proteins in membrane fusion. Front. Physiol. 8:5. doi: 10.3389/fphys.2017.00005
Han, J., Pluhackova, K., Bruns, D., and Böeckmann, R. A. (2016). Synaptobrevin transmembrane domain determines the structure and dynamics of the SNARE motif and the linker region. Biochim. Biophys. Acta 1858, 855–865. doi: 10.1016/j.bbamem.2016.01.030
Haucke, V., Neher, E., and Sigrist, S. J. (2011). Protein scaffolds in the coupling of synaptic exocytosis and endocytosis. Nat. Rev. Neurosci. 12, 127–138. doi: 10.1038/nrn2948
Heuser, J. E., and Reese, T. S. (1973). Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. J. Cell Biol. 57, 315–344. doi: 10.1083/jcb.57.2.315
Hong, W. (2005). SNAREs and traffic. Biochim. Biophys. Acta 1744, 120–144. doi: 10.1016/j.bbamcr.2005.03.014
Hughson, F. M. (2013). Neuroscience. Chaperones that SNARE neurotransmitter release. Science 339, 406–407. doi: 10.1126/science.1233801
Iversen, L. L. (1971). Role of transmitter uptake mechanisms in synaptic neurotransmission. Br. J. Pharmacol. 41, 571–591. doi: 10.1111/j.1476-5381.1971.tb07066.x
Jahn, R., Lang, T., and Südhof, T. C. (2003). Membrane fusion. Cell 112, 519–533. doi: 10.1016/S0092-8674(03)00112-0
Jahn, R., and Scheller, R. H. (2006). SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643. doi: 10.1038/nrm2002
Jahn, R., and Südhof, T. C. (1999). Membrane fusion and exocytosis. Annu. Rev. Biochem. 68, 863–911. doi: 10.1146/annurev.biochem.68.1.863
Knecht, V., and Grubmüller, H. (2003). Mechanical coupling via the membrane fusion SNARE protein syntaxin 1A: a molecular dynamics study. Biophys. J. 84, 1527–1547. doi: 10.1016/S0006-3495(03)74965-0
Kyoung, M., Srivastava, A., Zhang, Y., Diao, J., Vrljic, M., Grob, P., et al. (2011). in vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc. Natl. Acad. Sci. U S A 108, E304–313. doi: 10.1073/pnas.1107900108
Lai, Y., Choi, U. B., Zhang, Y., Zhao, M., Pfuetzner, R. A., Wang, A. L., et al. (2016). N-terminal domain of complexin independently activates calcium-triggered fusion. Proc. Natl. Acad. Sci. U S A 113, E4698–E4707. doi: 10.1073/pnas.1604348113
Lai, Y., Diao, J., Liu, Y., Ishitsuka, Y., Su, Z., Schulten, K., et al. (2013). Fusion pore formation and expansion induced by Ca2+ and synaptotagmin 1. Proc. Natl. Acad. Sci. U S A 110, 1333–1338. doi: 10.1073/pnas.1218818110
Lai, Y., Diao, J. J., Cipriano, D. J., Zhang, Y. X., Pfuetzner, R. A., Padolina, M. S., et al. (2014). Complexin inhibits spontaneous release and synchronizes Ca2+-triggered synaptic vesicle fusion by distinct mechanisms. Elife 3:e03756. doi: 10.3410/f.718529800.793499007
Lai, Y., Lou, X., Diao, J., and Shin, Y.-K. (2015). Molecular origins of synaptotagmin 1 activities on vesicle docking and fusion pore opening. Sci. Rep. 5:9267. doi: 10.1038/srep09267
Lucić, V., Förster, F., and Baumeister, W. (2005). Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74, 833–865. doi: 10.1146/annurev.biochem.73.011303.074112
Ma, C., Su, L. J., Seven, A. B., Xu, Y. B., and Rizo, J. (2013). Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425. doi: 10.1126/science.1230473
Martens, S., and Mcmahon, H. T. (2008). Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9, 543–556. doi: 10.1038/nrm2417
Mayer, A. (1999). Intracellular membrane fusion: SNAREs only? Curr. Opin. Cell Biol. 11, 447–452. doi: 10.1016/S0955-0674(99)80064-7
Mehta, P. P., Battenberg, E., and Wilson, M. C. (1996). SNAP-25 and synaptotagmin involvement in the final Ca2+-dependent triggering of neurotransmitter exocytosis. Proc. Natl. Acad. Sci. U S A 93, 10471–10476. doi: 10.1073/pnas.93.19.10471
Nagy, G., Milosevic, I., Fasshauer, D., Müller, E. M., De Groot, B. L., Lang, T., et al. (2005). Alternative splicing of SNAP-25 regulates secretion through nonconservative substitutions in the SNARE domain. Mol. Biol. Cell 16, 5675–5685. doi: 10.1091/mbc.e05-07-0595
Nogales, E., Louder, R. K., and He, Y. (2016). Cryo-EM in the study of challenging systems: the human transcription pre-initiation complex. Curr. Opin. Struct. Biol. 40, 120–127. doi: 10.1016/j.sbi.2016.09.009
Oberhauser, A. F., Monck, J. R., and Fernandez, J. M. (1992). Events leading to the opening and closing of the exocytotic fusion pore have markedly different temperature dependencies. kinetic analysis of single fusion events in patch-clamped mouse mast cells. Biophys. J. 61, 800–809. doi: 10.1016/S0006-3495(92)81884-2
Ossig, R., Schmitt, H. D., De Groot, B., Riedel, D., Keränen, S., Ronne, H., et al. (2000). Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J. 19, 6000–6010. doi: 10.1093/emboj/19.22.6000
Pfeffer, S. R. (1999). Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1, E17–E22. doi: 10.1038/8967
Plattner, H., Knoll, G., and Erxleben, C. (1992). The mechanics of biological membrane fusion. merger of aspects from electron microscopy and patch-clamp analysis. J. Cell Sci. 103, 613–618.
Ribrault, C., Reingruber, J., Petkovic, M., Galli, T., Ziv, N. E., Holcman, D., et al. (2011). Syntaxin1A lateral diffusion reveals transient and local SNARE interactions. J. Neurosci. 31, 17590–17602. doi: 10.1523/JNEUROSCI.4065-11.2011
Risselada, H. J., and Grubmüeller, H. (2012). How SNARE molecules mediate membrane fusion: recent insights from molecular simulations. Curr. Opin. Struct. Biol. 22, 187–196. doi: 10.1016/j.sbi.2012.01.007
Rizo, J., and Xu, J. (2015). The synaptic vesicle release machinery. Annu. Rev. Biophys. 44, 339–367. doi: 10.1146/annurev-biophys-060414-034057
Rohrbough, J., and Broadie, K. (2005). Lipid regulation of the synaptic vesicle cycle. Nat. Rev. Neurosci. 6, 139–150. doi: 10.1038/nrn1608
Rust, M. J., Bates, M., and Zhuang, X. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795. doi: 10.1038/nmeth929
Ryu, J. K., Jahn, R., and Yoon, T. Y. (2016). Review: progresses in understanding N-ethylmaleimide sensitive factor (NSF) mediated disassembly of SNARE complexes. Biopolymers 105, 518–531. doi: 10.1002/bip.22854
Shangguan, T., Alford, D., and Bentz, J. (1996). Influenza-virus-liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry 35, 4956–4965. doi: 10.1021/bi9526903
Sharma, S., and Lindau, M. (2016). The mystery of the fusion pore. Nat. Struct. Mol. Biol. 23, 5–6. doi: 10.1038/nsmb.3157
Söllner, T. H. (2004). Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 16, 429–435. doi: 10.1016/j.ceb.2004.06.015
Stelzer, W., Poschner, B. C., Stalz, H., Heck, A. J., and Langosch, D. (2008). Sequence-specific conformational flexibility of SNARE transmembrane helices probed by hydrogen/deuterium exchange. Biophys. J. 95, 1326–1335. doi: 10.1529/biophysj.108.132928
Tekpinar, M., and Zheng, W. (2014). Unzipping of neuronal snare protein with steered molecular dynamics occurs in three steps. J. Mol. Model. 20:2381. doi: 10.1007/s00894-014-2381-7
Trimbuch, T., and Rosenmund, C. (2016). Should I stop or should I go? the role of complexin in neurotransmitter release. Nat. Rev. Neurosci. 17, 118–125. doi: 10.1038/nrn.2015.16
Wang, X. J. (2010). The molecular mechanism of herpesvirus membrane fusion. Prog. Biochem. Biophys. 37, 583–588. doi: 10.3724/sp.j.1206.2010.00037
Wickner, W., and Rizo, J. (2017). A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell. 28, 707–711. doi: 10.1091/mbc.E16-07-0517
Wilschut, J., and Hoekstra, D. (1986). Membrane fusion: lipid vesicles as a model system. Chem. Phys. Lipids 40, 145–166. doi: 10.1016/0009-3084(86)90068-x
Xu, J., Camacho, M., Xu, Y., Esser, V., Liu, X., Trimbuch, T., et al. (2017). Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. Elife 6:e22567. doi: 10.7554/eLife.22567
Xu, Y., Zhang, F., Su, Z., Mcnew, J. A., and Shin, Y. K. (2005). Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 12, 417–422. doi: 10.1038/nsmb921
Yoon, T. Y., Lu, X., Diao, J., Lee, S. M., Ha, T., and Shin, Y. K. (2008). Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 15, 707–713. doi: 10.1038/nsmb.1446
Zhang, L., Yan, F., Zhang, S. L., Lei, D. S., Charles, M. A., Cavigiolio, G., et al. (2012). Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat. Chem. Biol. 8, 342–349. doi: 10.1038/nchembio.796
Zhang, M., Charles, R., Tong, H., Zhang, L., Patel, M., Wang, F., et al. (2015a). HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation. Sci. Rep. 5:8741. doi: 10.1038/srep08741
Zhang, X., Rebane, A. A., Ma, L., Li, F., Jiao, J., Qu, H., et al. (2016). Stability, folding dynamics and long-range conformational transition of the synaptic t-SNARE complex. Proc. Natl. Acad. Sci. U S A 113, E8031–E8040. doi: 10.1073/pnas.1605748113
Zhang, Y. X., Diao, J. J., Colbert, K. N., Lai, Y., Pfuetzner, R. A., Padolina, M. S., et al. (2015b). Munc18a does not alter fusion rates mediated by neuronal SNAREs, synaptotagmin and complexin. J. Biol. Chem. 290, 10518–10534. doi: 10.1074/jbc.M114.630772
Zheng, W. (2014). All-atom and coarse-grained simulations of the forced unfolding pathways of the SNARE complex. Proteins 82, 1376–1386. doi: 10.1002/prot.24505
Zheng, W. (2016). Probing the structural dynamics of the SNARE recycling machine based on coarse-grained modeling. Proteins Struct. Funct. Bioinform. 84, 1055–1066. doi: 10.1002/prot.25052
Keywords: membrane fusion, neurotransmission, neurotransmitter release machinery, electron microscope, molecular simulation
Citation: Yang Z, Gou L, Chen S, Li N, Zhang S and Zhang L (2017) Membrane Fusion Involved in Neurotransmission: Glimpse from Electron Microscope and Molecular Simulation. Front. Mol. Neurosci. 10:168. doi: 10.3389/fnmol.2017.00168
Received: 21 March 2017; Accepted: 15 May 2017;
Published: 07 June 2017.
Edited by:
Jiajie Diao, University of Cincinnati, United StatesReviewed by:
Dechang Li, Beijing Institute of Technology, ChinaCopyright © 2017 Yang, Gou, Chen, Li, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengli Zhang, emhhbmdzbEBtYWlsLnhqdHUuZWR1LmNu
Lei Zhang, emhhbmdsZWlvQG1haWwueGp0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.