- 1Centre National de la Recherche Scientifique (CNRS), UMR-5203, Institut de Génomique Fonctionnelle, Montpellier, France
- 2Institut National de la Santé et de la Recherche Médicale (INSERM), U1191, Montpellier, France
- 3Université de Montpellier, UMR-5203, Montpellier, France
- 4Department of Neuroscience, Memory and Brain Research Center, Baylor College of Medicine, Houston, TX, USA
Repeated psychostimulant exposure induces persistent gene expression modifications that contribute to enduring changes in striatal GABAergic spiny projecting neurons (SPNs). However, it remains unclear whether changes in the control of mRNA translation are required for the establishment of these durable modifications. Here we report that repeated exposure to D-amphetamine decreases global striatal mRNA translation. This effect is paralleled by an enhanced phosphorylation of the translation factors, eIF2α and eEF2, and by the concomitant increased translation of a subset of mRNAs, among which the mRNA encoding for the activity regulated cytoskeleton-associated protein, also known as activity regulated gene 3.1 (Arc/Arg3.1). The enrichment of Arc/Arg3.1 mRNA in the polysomal fraction is accompanied by a robust increase of Arc/Arg3.1 protein levels within the striatum. Immunofluorescence analysis revealed that this increase occurred preferentially in D1R-expressing SPNs localized in striosome compartments. Our results suggest that the decreased global protein synthesis following repeated exposure to D-amphetamine favors the translation of a specific subset of mRNAs in the striatum.
Introduction
Repetitive behaviors observed following repeated exposure to psychostimulant drugs result in part from imbalanced activity of striatal neural circuits (Canales and Graybiel, 2000). Convergent evidence suggests that long-lasting molecular changes leading to persistent alterations of synaptic properties and spines morphology of striatal projection neurons (SPNs) contribute to the distortion of these circuits (Russo et al., 2010; Luscher and Malenka, 2011). While transcriptional and epigenetic modifications are clearly necessary for the establishment of these persistent changes (Robison and Nestler, 2011), less is known about the ability of repeated psychostimulant exposure to impact on the regulation of protein synthesis through the control of the translational machinery.
Initiation is the rate-limiting step of translation and is tightly controlled by two main mechanisms. The first involves the formation of the eukaryotic initiation factor 4 F (eIF4F) complex, which is regulated by the mechanistic target of rapamycin complex 1 (mTORC1). The second mechanism controls the availability of the ternary complex through the phosphorylation of the α subunit of the eukaryotic initiation factor 2 (eIF2α). Phosphorylated eIF2α decreases general mRNA translation and upregulates the translation of a subset of selective mRNAs containing upstream open reading frames (uORFs) in their 5′ untranslated region (UTR; Buffington et al., 2014). Besides initiation, elongation represents another step of translational control. Phosphorylation of the eukaryotic elongation factor 2 (eEF2) at T56 impairs its binding to the ribosome, thereby decreasing the rate of protein elongation (Nairn et al., 1985; Ryazanov et al., 1988; Price et al., 1991) but also promotes the translation of a subset of mRNAs involved in the control of synaptic plasticity (Scheetz et al., 2000; Park et al., 2008; Verpelli et al., 2010).
By using ribopuromycylation assay, polysome profiling combined with qRT-PCR, and western blot analysis to study the phosphorylation state of translation factors, we investigated the consequence of repeated D-amphetamine exposure on the control of mRNA translation within the striatum.
Materials and Methods
Animals
Eight week-old male C57BL/6 were purchased from Charles River Laboratories (France). Male and female Drd2-eGFP heterozygous mice (C57BL/6) were generated as described previously (Gong et al., 2003). Animals were housed under standardized conditions with a 12 h light/dark cycle, stable temperature (22 ± 1°C), controlled humidity (55 ± 10%), and food and water ad libitum. All experiments were in accordance with the guidelines of the French Agriculture and Forestry Ministry for handling animals and were approved by the local Ethic Committee (D34-172-13).
Drugs and Treatments
(+)-α-Methylphenethylamine [D-amphetamine (D-amph)] sulfate salt (10 mg/kg) from Sigma–Aldrich (St.-Quentin-Fallavier, France) was dissolved in 0.9% (w/v) NaCl (saline) and injected intraperitoneally (i.p) in a volume of 10 ml/kg, one injection per day during 5 days. Mice were habituated to handling and saline injection for 3 consecutive days before the day of the experiment.
Western Blot
After pharmacological manipulation, the striatum (including the nucleus accumbens and dorsal striatum) of one hemisphere was extracted as previously described (Puighermanal et al., 2016a), sonicated in 300 μl of 10% sodium dodecyl sulfate (SDS), and boiled at 100°C for 10 min. Protein quantification and western blots were performed as described (Biever et al., 2015). Primary antibodies against p-eIF2α (Ser51) (1:1000; Cell Signaling, #3398), eIF2α (1:1000; Cell Signaling, #5324), p-eEF2 (Thr56) (1:1000; Cell Signaling, #2331), p-p70S6K (Thr389) (1:1000; Cell Signaling, #9234), p-4EBP1 (Thr37/46) (1:500; Cell Signaling, #2855), 4EBP1 (1:500; Cell Signaling, #9644), OPHN1 (1:1000; Cell Signaling, #11939), ATF4 (1:1000; NeuroMab, #75-345), MAP2 (1:2000; Sigma, #M4403) from Sigma, CaMKIIa (1:1000; Millipore, #05-532), puromycin [1:1000; (David et al., 2012)], and β-actin (1:40000; Abcam, #AB6276) were used. The optical density of the relevant immunoreactive bands (or for all the bands for puromycin staining) was quantified after acquisition on a ChemiDoc XRS System (Bio-Rad) controlled by Image Lab software version 3.0 (Bio-Rad).
Immunofluorescence
After pharmacological manipulation, tissue preparation and immunofluorescence were performed as described (Bertran-Gonzalez et al., 2008). Primary antibodies against GFP (1:1000; Life Technologies, #A10262), Calbindin-D28k (1:1000; Swant, #CB38), and Arc/Arg3.1 (1:500; Santa Cruz Biotechnology, #sc17839) were used.
Puromycin Incorporation in Whole Striatal Lysates
Puromycin incorporation was performed as described previously (Biever et al., 2015). Briefly, striata were rapidly dissected on an ice-cooled dish and homogenized using 20 up-and-down strokes of a prechilled glass homogenizer with 800 μl of polysomal buffer containing 50 mM Tris pH 7.8, 240 mM KCl, 10 mM MgCl2, 250 mM D-sucrose, 2% Triton X-100, 20 μl/ml emetine, 5 mM DTT, 100 U/ml RNasin (Promega), and protease inhibitor cocktail (Roche). Samples were centrifuged for 5 min at 16,000 × g at 4°C and supernatant was incubated with 100 μg/ml of puromycin for 10 min at 4°C and then boiled for 10 min at 100°C. Protein concentrations were determined using BCA protein assay (Pierce, Rockford, IL, USA) and samples were stored at -20°C for further western blot analyses.
Polysome Profiling
The polysome profiling approach was performed as described previously (Biever et al., 2015). RNA from fractions <2 ribosomes (referred to as non-polysomal ‘NP’) and fractions with ≥2 ribosomes (referred to as polysomal ‘P’) was extracted using the TRIZOL (Thermo Fischer) protocol according to the manufacturer’s instructions. The carrier glycoblue was added before RNA precipitation step during the TRIZOL protocol. To remove potential DNA contamination, fractions were treated with DNAse (Ambion) according to the manufacturer’s instruction. RNA integrity was tested using Fragment Analyzer (Advanced Analytical).
cDNA Synthesis and Quantitative Real-Time PCR
RNA from non-polysomal and polysomal fractions was reverse transcribed to first strand cDNA using the SuperScript® VILOTM cDNA synthesis kit (Invitrogen). Resulting cDNA was used for quantitative real-time PCR (qRT-PCR), using 2X SYBR Green Mix and LC480 Real-Time PCR System (Roche) as described (Puighermanal et al., 2016b). Analysis was performed using LightCycler® 480 Software (Roche). Results are presented as linearized Cp-values normalized to the stably-expressed genes β-actin or gapdh and the ΔCP method was used to give the fold change. The primer sequences used in this study are detailed in Table 1.
Statistical Analysis
All statistical analyses were performed using one-way analysis of variance (ANOVA) for multiple comparisons, followed by Bonferroni post hoc test. Student t-test with equal variances was used for groups of two, when relevant. Statistical significance was determined as p < 0.05. Prism 5.0 software was used to perform statistical analyses.
Results
Repeated Exposure to D-Amphetamine Reduces Protein Synthesis in the Striatum
To investigate whether repeated exposure to D-amphetamine (10 mg/kg, once daily for 5 days) could alter global mRNA translation in the striatum, we performed polysome profile analysis on striatal lysates at 60 min following the last injection of D-amphetamine. We observed an increase in the amplitude of the ‘vacant’ 80S monosome peak along with a reduction in the polysome population in mice treated with D-amphetamine compared to saline-treated mice (Figure 1A). Consequently, the polysome to monosome ratio was significantly decreased in D-amphetamine-treated mice (Figure 1A, inset).
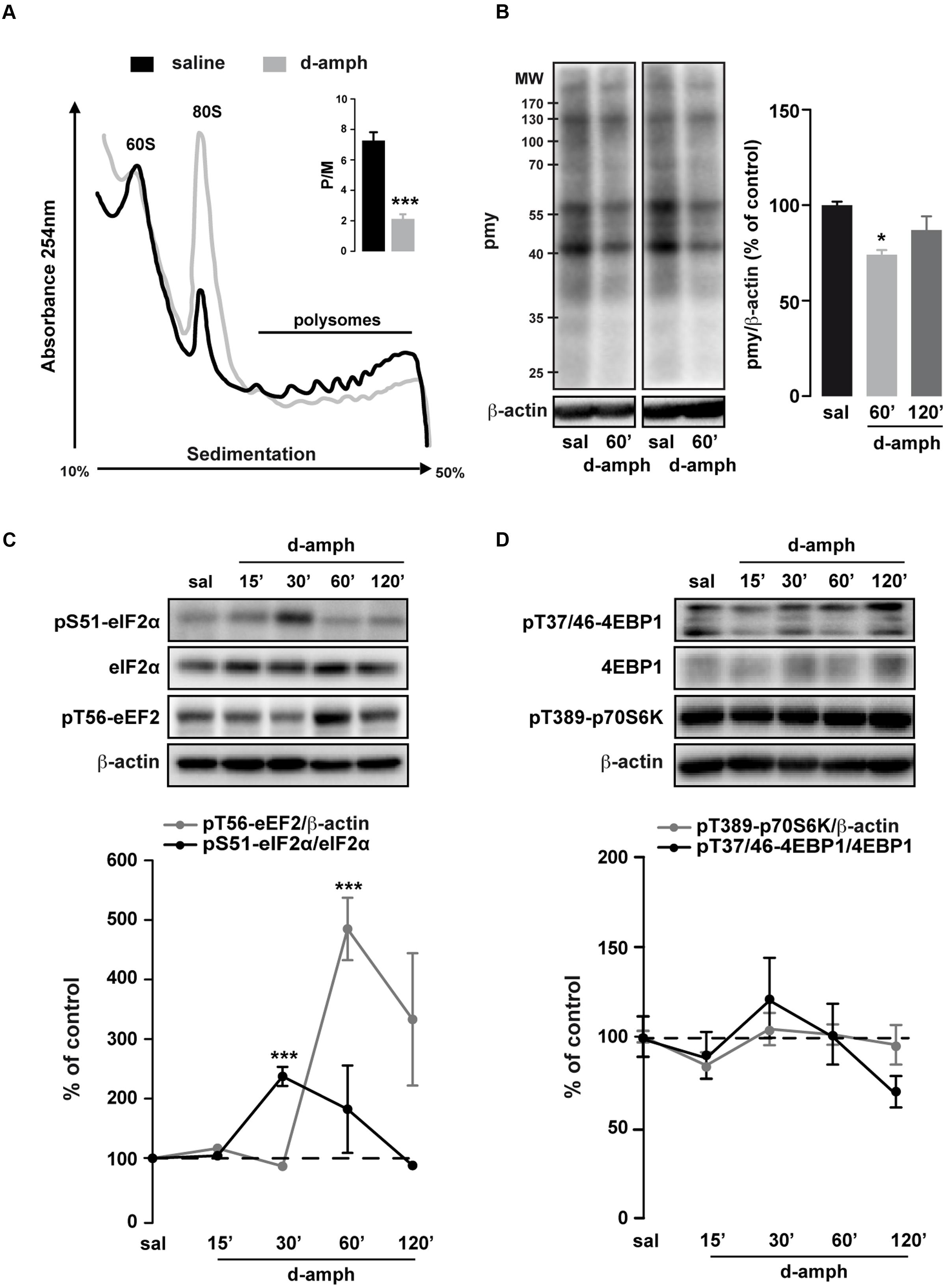
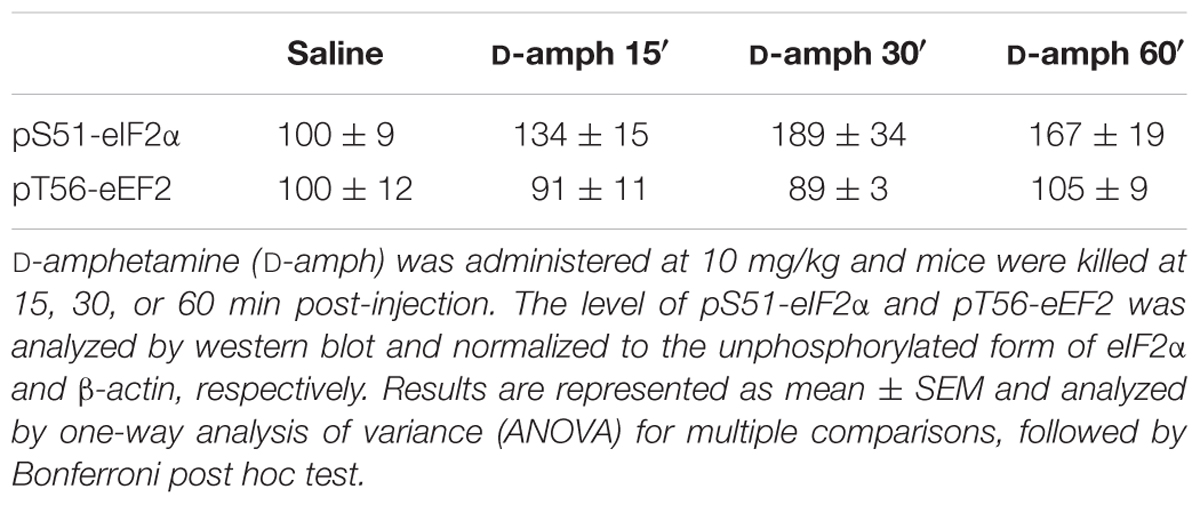
FIGURE 1. Repeated D-amphetamine administration decreases global protein synthesis and induces the phosphorylation of the translation factors eIF2α and eEF2. (A) Polysome profiles of whole striatal lysates from mice repeatedly treated with saline or D-amphetamine (10 mg/kg, once daily for 5 days) and killed 60 min after the last injection. Inset: Polysomes (P) to monosome ratio (M) from mice chronically treated with saline or D-amphetamine (mean ± SEM, ∗∗∗p < 0.001 by unpaired Student t-test, n = 3 mice/group; right). (B) Representative and quantified western blot analysis of puromycin (pmy) staining (normalized to β-actin) of whole striatal lysates incubated 10 min with puromycin from mice killed 60 min after the last injection of saline (sal) or D-amphetamine (D-amph; n = 5 mice/group). Results are represented as mean ± SEM. ∗p < 0.05, by one-way ANOVA followed by Bonferroni post hoc test. (C) Representative and quantified western blot analysis of phospho-eIF2α (Ser51; normalized to eIF2α) and phospho-eEF2 (Thr56; normalized to β-actin) in striatal lysates of mice treated chronically with saline or D-amphetamine (10 mg/kg, once daily for 5 days) and killed at different time points after injection. (D) Phospho-4EBP1 (Thr37/46; normalized to 4EBP1) and phospho-p70S6K (Thr389; normalized to β-actin) similar to (C). Data are expressed as a percentage of saline control (n = 5–9 mice/group). Results are represented as mean ± SEM. ∗∗∗p < 0.001 by one-way ANOVA followed by Bonferroni post hoc test.
Polysome profiling is a representation of the steady-state ribosomes engaged in translation. To determine whether repeated exposure to D-amphetamine could modulate de novo protein synthesis, we performed an assay adapted from the ribopuromycylation method (David et al., 2012; Biever et al., 2015). We found a transient decrease in puromycin incorporation in striatal lysates of mice treated with D-amphetamine (Figure 1B). Altogether, these results indicate that, in the striatum, global mRNA translation was decreased in mice repeatedly exposed to D-amphetamine.
Repeated Exposure to D-Amphetamine Enhances eIF2α and eEF2 Phosphorylation in the Striatum
The regulation of mRNA translation is tightly controlled by the phosphorylation of translation initiation and elongation factors (Buffington et al., 2014). We therefore analyzed the phosphorylation state of the initiation factor eIF2α and the elongation factor eEF2 in the striatum of mice repeatedly exposed to D-amphetamine. Western blot analysis of whole striatal lysates at different time points (15, 30, 60, and 120 min) following the last D-amphetamine administration revealed a robust enhancement of pS51-eIF2α and pT56-eEF2 at 30 and 60 min post-injection, respectively (Figure 1C). The increase in pS51-eIF2α and pT56-eEF2, which was not observed following a single D-amphetamine exposure (Table 2), does not result from an accumulation of phosphorylation over the 5 days of the injection protocol as no change was found at earlier time points (e.g., 15 min post-injection for pS51-eIF2α; 15 and 30 min post-injection for pT56-eEF2; Figure 1C). The mTORC1 pathway, known to regulate the initiation of translation (Ma and Blenis, 2009), was not activated following repeated D-amphetamine exposure (Figure 1D). Together, these results revealed that the decrease in global mRNA translation observed in mice repeatedly administered with D-amphetamine is paralleled by an enhanced eIF2α and eEF2 phosphorylation.
Repeated Exposure to D-Amphetamine Does Not Induce Endoplasmic Reticulum (ER) Stress or Neurotoxicity in the Striatum
Enhanced eIF2α phosphorylation is observed in response to ER stress (Walter and Ron, 2011). To test whether repeated D-amphetamine exposure promoted ER stress in the striatum, the levels of ER stress markers including PERK (PKR-like ER kinase), PDI (protein disulfide isomerase), IRE1α (inositol requiring enzyme 1 α), and BiP (Binding immunoglobulin protein) were analyzed 30, 60, or 120 min following the last D-amphetamine administration. While striatal levels of PERK, PDI, and IRE1α were not altered by D-amphetamine (Table 3), BiP levels were transiently decreased only 60 min after the last D-amphetamine administration (Table 3).
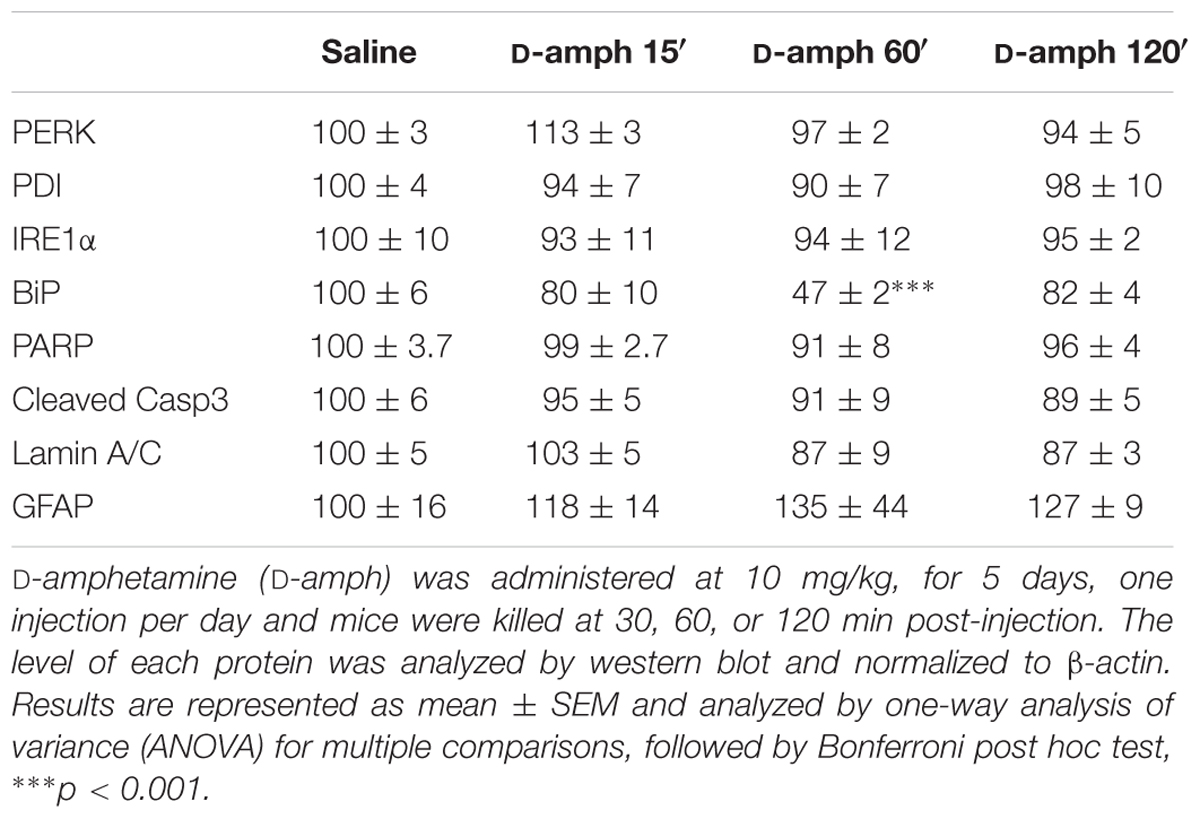
TABLE 3. Effect of repeated D-amphetamine administration on endoplasmic reticulum stress and neurotoxicity in the striatum.
To determine whether repeated D-amphetamine exposure produced apoptosis or gliosis, we analyzed the levels of PARP [Poly (ADP-ribose) polymerase], cleaved caspase 3, cleaved lamin A/C, and GFAP (glial fibrillary acid protein). As summarized in Table 3, none of these markers were changed in mice repeatedly treated with D-amphetamine. Moreover, no persistent damage of dopamine terminals was observed as suggested by the stable levels of tyrosine hydroxylase between saline- (100 ± 4) and D-amphetamine-treated (30 min, 95 ± 2; 60 min, 102 ± 16; 120 min, 93 ± 4) mice. Altogether, these results indicate that the increased eIF2α phosphorylation observed in the striatum following repeated D-amphetamine exposure is not accompanied by ER stress or neurotoxicity.
Enhanced Translation of Selective uORF-Bearing mRNAs by D-Amphetamine
Enhanced eIF2α phosphorylation represses global translation but coincidently promotes the translation of mRNAs containing uORFs in their 5′UTRs (Dever, 2002; Buffington et al., 2014). To determine whether repeated exposure to D-amphetamine could increase the translation of specific uORF-bearing mRNAs, we performed polysome profiling combined with qRT-PCR to analyze mRNA levels in the non-polysomal fraction (poorly or not translated mRNAs) and in the polysomal fraction (actively translated mRNAs) in saline and D-amphetamine-treated mice. To correct for the change in the mRNA abundance in the monosomal fraction, mRNA levels in polysomal fractions were systemically normalized to levels in non-polysomal fractions (P/NP ratio). We first analyzed the level of Atf4 and Ophn1, two mRNAs encoding for the activating transcription factor 4 (ATF4) and oligophrenin 1 (OPHN1), respectively, in polysomal fractions from whole striatal lysates. As shown in Figures 2A,B, Atf4 and Ophn1 mRNAs were not enriched in polysomal fractions suggesting that repeated D-amphetamine exposure did not enhance the translation of Atf4 and Ophn1 within the striatum. In line with these results, ATF4 and OPHN1 protein levels, analyzed 60 and 120 min following the last administration of D-amphetamine, remained unchanged (Figures 2C,D). The analysis of Ppp1r15a and Ddit3 mRNAs, two other uORF-bearing mRNAs encoding for the protein phosphatase regulatory subunit 15A and the DNA damage-inducible transcript 3, respectively, revealed an enrichment in polysomal fractions of D-amphetamine-treated mice indicating that these two mRNAs were translationally upregulated (Figures 2E,G). However, while Ppp1r15a mRNA was also enriched in non-polysomal fractions suggesting that Ppp1r15a was both transcriptionally and translationally enhanced (Figure 2F), Ddit3 mRNA was only enriched in polysomal fractions (Figure 2H). Altogether, these results indicate that repeated D-amphetamine exposure represses global protein synthesis but concomitantly upregulates the translational efficiency of selective uORF-bearing mRNAs.
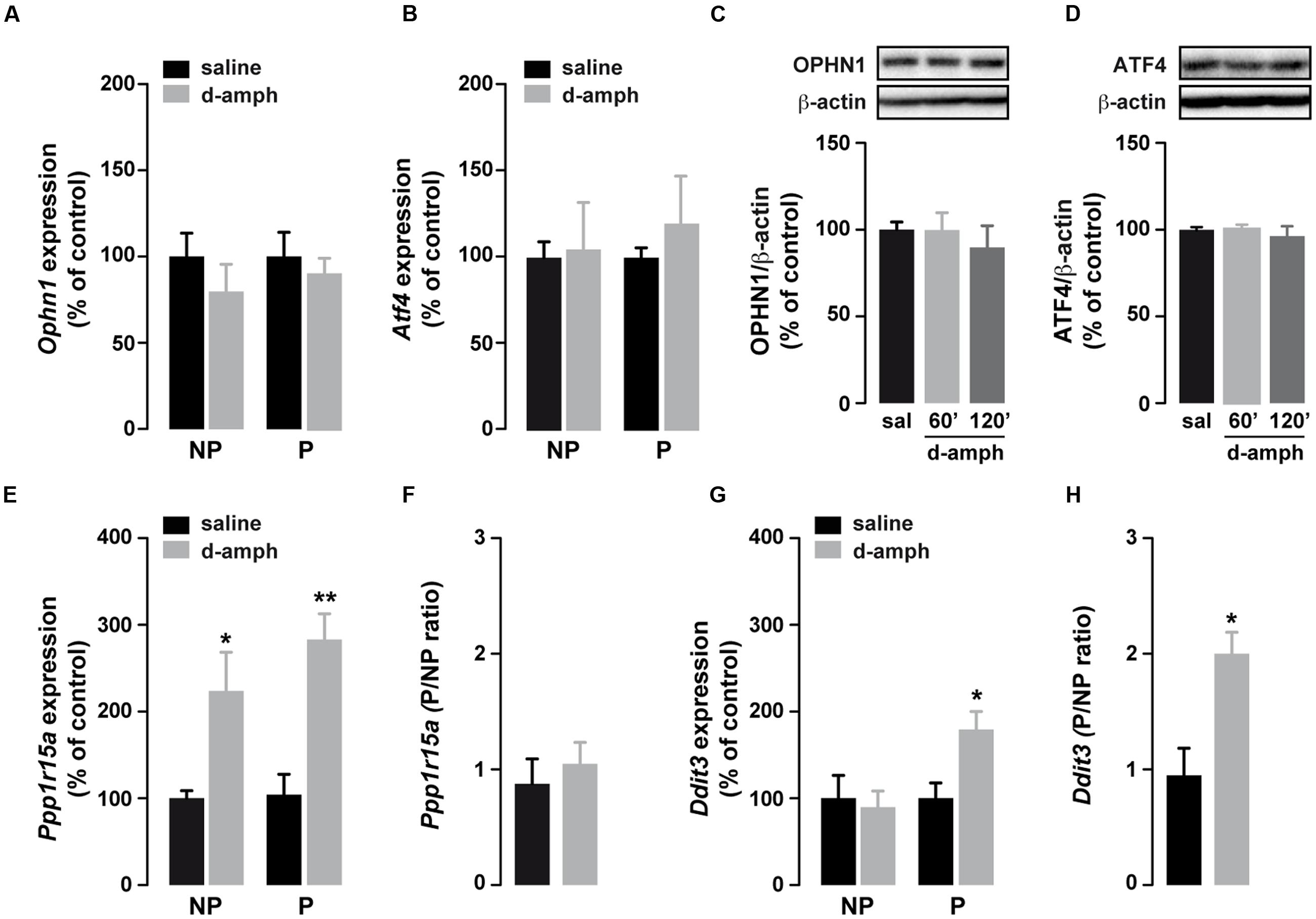
FIGURE 2. Repeated d-amphetamine administration increases the translation of a subset of uORF-containing mRNAs. (A,B,E,G) Relative mRNA expression levels of Ophn1 (A), Atf4 (B), Ppp1r15a (E), and Ddit3 (G) in non-polysomal (NP) and polysomal (P) fractions analyzed by qRT-PCR in the striatum of mice chronically treated with D-amphetamine (10 mg/kg, once daily for 5 days) or saline (n = 5 mice/group). All candidate mRNAs were normalized to β-actin or Gapdh mRNA and expressed as a percentage of saline control. (C,D) Representative western blot (top) and quantification (bottom) of OPHN1 (C) and ATF4 (D) (normalized to β-actin) expression levels in the striatum 60 or 120 min after the last injection of saline (sal) or D-amphetamine (D-amph). Data are expressed as a percentage of saline control (n = 5 mice/group). (F,H) Ratio of non-polysomal (NP) and polysomal (P) fractions of Ppp1r15a (F) and Ddit3 (H) mRNAs from the results represented in (E,G), respectively. Results are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01 by unpaired Student t-test (saline versus D-amphetamine).
Enhanced Striatal Translation of Arc/Arg3.1 mRNA by D-Amphetamine
Besides the slowing of elongation, eEF2 phosphorylation at T56 increases the translation of dendrite-localized mRNAs such as Camk2a and Arc/Arg3.1 (Scheetz et al., 2000; Park et al., 2008; Kenney et al., 2016). We therefore tested whether the enhanced eEF2 phosphorylation induced by D-amphetamine was accompanied by increased binding of Camk2a, Map2, and Arc/Arg3.1 mRNAs to polysomes. No changes were observed in the abundance of Map2 mRNA in non-polysomal and polysomal fractions between saline and D-amphetamine-treated mice (Figure 3A). In addition, striatal MAP2 protein levels remained unchanged at 60 and 120 min following the last administration of D-amphetamine (Figure 3B). The analysis of Camk2a mRNAs revealed an enrichment in both non-polysomal and polysomal fractions from mice repeatedly treated with D-amphetamine (Figure 3C). However, no significant changes were found in the Camk2a P/NP ratio (saline = 1.8 ± 0.58, D-amphetamine = 1.6 ± 0.07; p = 0.14) as well as at the protein level (Figure 3D). On the other hand, although Arc/Arg3.1 mRNAs were also enriched in both non-polysomal and polysomal fractions (Figure 3E), we observed a threefold increase in P/NP ratio of Arc/Arg3.1 mRNA levels (Figure 3F). To determine whether the enrichment of Arc/Arg3.1 mRNAs in the polysomal fractions was accompanied by an increase at the protein level, we monitored Arc/Arg3.1-positive cells 60 min after the last administration of saline or D-amphetamine. Immunofluorescence analysis revealed a robust increase in Arc/Arg3.1 immunoreactivity in the dorsal striatum and the nucleus accumbens corresponding to both Arc/Arg3.1-positive neurons and neuropil (most presumably dendritic processes; Figures 3G,H). This increase occurred preferentially in striosomes/patches (identified as calbindin-D28k-poor zones) and only sparsely in the matrix compartment (Figure 3I). Finally, double immunofluorescence analysis of Arc/Arg3.1 and GFP in Drd2-eGFP mice showed that D-amphetamine evoked Arc/Arg3.1 expression predominantly in GFP-negative neurons, presumably accounting for D1R-expressing neurons (Figures 3J,K). Together, our results suggest that repeated D-amphetamine administration enhances Arc/Arg3.1 mRNA translation preferentially in a subset of striosomal D1R-expressing neurons.
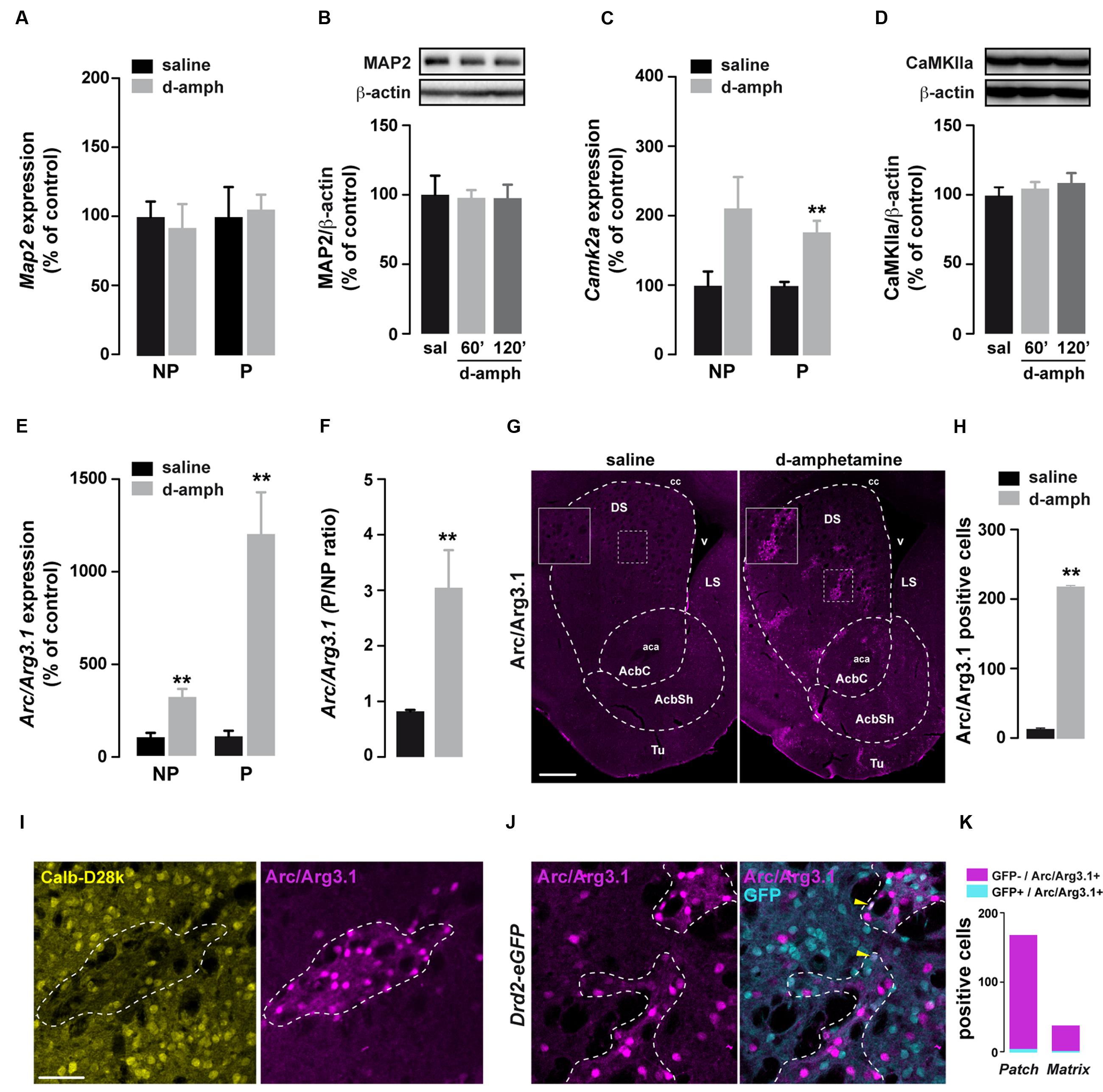
FIGURE 3. Repeated D-amphetamine treatment enhances the translation of Arc/Arg3.1. (A,C,E) Relative striatal mRNA expression levels of Map2 (A), Camk2a (C), and Arc/Arg3.1 (E) in non-polysomal (NP) and polysomal (P) fractions analyzed by qRT-PCR 60 min after the last injection of saline or D-amphetamine (10 mg/kg, once daily for 5 days). All candidate mRNAs were normalized to β-actin or Gapdh mRNA and expressed as a percentage of saline control (n = 5 mice/group). (B,D) Quantification and representative western blot of MAP2 (B) and CaMKIIa (D) (normalized to β-actin) of mice killed 60 or 120 min after the last injection of saline (sal) or D-amphetamine (D-amph; n = 5 mice/group). Data are expressed as a percentage of saline control. (F) Ratio of non-polysomal (NP) and polysomal (P) fractions of Arc/Arg3.1 mRNA. (G) Single immunostaining for Arc/Arg3.1 in coronal sections of the striatum from mice chronically treated with D-amphetamine (10 mg/kg, once daily for 5 days) or saline and killed 60 min after the last injection. Scale bar: 400 μm. (H) Quantification of Arc/Arg3.1-positive cells in the striatum 60 min after the last injection of saline or D-amphetamine (D-amph; n = 3 mice/group) (I) Double immunostaining for Calbindin-D28k (Calb-D28k; yellow) and Arc/Arg3.1 (magenta) in coronal striatal sections of mice chronically treated with D-amphetamine (10 mg/kg, once daily for 5 days) and killed 60 min after the last injection. Scale bar: 60 μm. (J) Double immunostaining for GFP (cyan) and Arc/Arg3.1 (magenta) in coronal striatal sections of Drd2-eGFP mice chronically treated with D-amphetamine (10 mg/kg, once daily for 5 days) and killed 60 min after the last injection. Note the presence of Arc/Arg3.1 staining in few GFP-positive cells (yellow arrowhead). (K) Quantification of Arc/Arg3.1-positive/GFP-positive and Arc/Arg3.1-positive/GFP-negative cells within patch or matrix compartments in the striatum of Drd2-eGFP killed at 60 min after the last injection of saline or D-amphetamine (D-amph; n = 3 mice/group).
Discussion
The present study reveals that, in addition to transcriptional and epigenetic modifications, repeated exposure to D-amphetamine can also alter the regulation of translation within the striatum. While global and TOP mRNA translations were not affected following a single D-amphetamine administration (Biever et al., 2015), the use of polysome profiling and ribopuromycylation-based assay revealed that repeated D-amphetamine administration triggers a transient decrease of steady-state ribosomes engaged in translation as well as de novo protein synthesis within the striatum. This reduced efficiency of global mRNA translation was accompanied by an enhanced phosphorylation of the translation initiation factor eIF2α and the elongation factor eEF2, known to slow the initiation and the elongation, respectively (Buffington et al., 2014; Kenney et al., 2014). Interestingly, convergent observations indicate that a reduction of initiation most likely account for the decrease in overall protein synthesis observed in mice repeatedly exposed to D-amphetamine. Indeed, unlike the slowing of elongation, which has been associated with increased polysome formation, the attenuation of the initiation is correlated with a reduction of the polysomal population (Hershey et al., 2012). Moreover, while the increase in phosphorylation of eIF2α occurred rapidly (30 min) after D-amphetamine administration, eEF2 phosphorylation was only detectable at 60 min suggesting that this last event may rather contribute to coordinate the rate of elongation with reduced initiation. Further experiments are required to firmly establish that enhanced phosphorylation of eIF2α and/or eEF2 is causally linked to the reduced global protein synthesis observed in the striatum of mice repeatedly administered with D-amphetamine.
Phosphorylation of eIF2α delays the delivery of initiator tRNAs to initiating ribosomes, thereby reducing global protein synthesis (Pavitt et al., 1998; Dever, 1999; Sonenberg and Hinnebusch, 2009). Paradoxically, this phenomenon favors the translation of mRNAs bearing uORFs located at the 5′ leader of the coding sequence (Lu et al., 2004; Vattem and Wek, 2004). Although the mechanisms of enhanced translation remain to be established, we identified two uORF-containing mRNAs whose translation was increased. The first one, Ddit3, encodes the transcription factor CHOP (CCAAT/enhancer-binding protein homologous protein; Ubeda et al., 1996). The enrichment of Ddit3 mRNA only in the polysomal fraction suggests that Ddit3 mRNAs did not accumulate with the repeated exposure to D-amphetamine. The second uORF-bearing mRNA that was translationally enhanced encodes for the protein phosphatase 1 (PP-1) regulatory subunit 15A also known as GADD34 (growth arrest and DNA damage-inducible protein; Lee et al., 2009). Interestingly, the selective translation of Ddit3 and Ppp1r15a has been previously shown to rely on enhanced eIF2α phosphorylation, allowing scanning ribosomes to bypass inhibitory uORFs in order to translate the main ORFs (Palam et al., 2011; Young et al., 2015, 2016). Thus, D-amphetamine-induced eIF2α phosphorylation may facilitate bypassing the inhibitory uORF within the Ddit3 and Ppp1r15a mRNAs, thereby explaining the increased binding of these mRNAs to polysomes following drug treatment. In contrast to Ddit3, Ppp1r15a mRNA was also enriched in the non-polysomal fraction suggesting that Ppp1r15a mRNA was also regulated at the transcriptional level by repeated D-amphetamine. As Ppp1r15a is a transcriptional target of CHOP (Marciniak et al., 2004), increased CHOP levels could promote a transcriptional upregulation of Ppp1r15a by D-amphetamine. By interacting with the catalytic subunit of PP-1, GADD34 forms a phosphatase complex involved in eIF2α dephosphorylation (Connor et al., 2001; Novoa et al., 2001). Thus, the increased transcription and translation of GADD34 could contribute to the establishment of a negative feedback regulatory mechanism aiming to reduce the exacerbated eIF2α phosphorylation induced by D-amphetamine. Strikingly, we did not find changes in the translational efficiency of Atf4 and Ophn1, two uORF-bearing mRNAs, which are translated in an eIF2α-dependent fashion in the hippocampus and in the ventral tegmental area (Costa-Mattioli et al., 2005, 2007; Di Prisco et al., 2014; Huang et al., 2016). Several hypotheses could account for these differences. First, an enhanced initiation at these two mRNAs through phosphorylated eIF2α shortly followed by an inhibition of translation elongation by eEF2 phosphorylation could explain why the synthesis of ATF4 and OPHN1 proteins remains unchanged following D-amphetamine treatment. Alternatively, the expression of uORF-lacking splice variants of these genes in the striatum could account for our findings. Indeed, numerous proteins are encoded by mRNAs with diverse 5′UTRs that are generated by alternative splicing and impose different modes of translational regulation (Martineau et al., 2004; Baranick et al., 2008; Al-Fageeh and Smales, 2009; Riley et al., 2010). Finally, depending on the stimuli triggering eIF2α phosphorylation, the brain areas and the cell types where it occurs, different uORF-bearing mRNAs could be translated. Such possibilities remain to be investigated.
Increased eIF2α phosphorylation is observed when unfolded protein response (UPR) is engaged in response to the accumulation of unfolded or misfolded proteins in the lumen of the ER (Walter and Ron, 2011). The expression of GADD34 and CHOP is increased during this process. While GADD34 comprises a negative feedback loop to reverse the translational attenuation mediated by the enhanced eIF2α phosphorylation, the transcription factor CHOP promotes the expression of genes involved in apoptosis (Wang et al., 1998; Zinszner et al., 1998; Novoa et al., 2001). Previous experiments performed in rats indicate that repeated exposure to psychostimulants, including cocaine, D-amphetamine, or methamphetamine, activates the ER stress response, which through the engagement of apoptotic pathways leads to striatal neurotoxicity (Jayanthi et al., 2004, 2009; Krasnova et al., 2005; Beauvais et al., 2011; Go et al., 2016). Our results clearly highlight that the effects induced by repeated exposure to D-amphetamine are radically different in mice. Thus, despite the increase of translation efficiency of Ddit3 and Ppp1r15a mRNAs observed, several evidence suggest that in our condition the ER stress response is not activated. First, the striatal levels of ER stress markers including PERK, PDI, and IRE1α remained unchanged in mice repeatedly administered with D-amphetamine. Second, the translation of the transcription activator of the integrated stress response ATF4 was not enhanced (Harding et al., 2000). Finally, none of the apoptosis or gliosis markers tested were altered suggesting that the regulation of translation by D-amphetamine most likely contributes to persistent modifications altering striatal plasticity rather than representing a protective mechanism to cope with an insult.
Previous studies indicate that eEF2 phosphorylation may promote the translation of a subset of mRNAs among which the dendritic-localized including Camk2a, Map2, and Arc/Arg3.1 (Scheetz et al., 2000; Belelovsky et al., 2005; Park et al., 2008; Kenney et al., 2016). While the translation of Camk2a and Map2 within the striatum was unchanged, we found that D-amphetamine enhances the transcription and translation of Arc/Arg3.1. Indeed, mice repeatedly treated with D-amphetamine displayed a strong enrichment of Arc/Arg3.1 mRNA in both non-polysomal and polysomal fractions, which was accompanied by an increase of Arc/Arg3.1 protein levels. Although pharmacological or genetic manipulations of eEF2 phosphorylation are required to causally link this event to the D-amphetamine-induced Arc/Arg3.1 mRNA translation, alternative molecular mechanisms could contribute to the regulation of Arc/Arg3.1 translation. Indeed, enhanced Arc/Arg3.1 synthesis can occur through ERK/Mnk1 signaling and independently of eEF2 phosphorylation in the hippocampus (Panja et al., 2009, 2014). Increased Arc/Arg3.1 levels were preferentially found in the D1R-expressing SPNs located in striosome compartments. These results extend earlier studies showing that acute psychostimulant administration upregulates Arc/Arg3.1 transcripts and protein levels in the striatum (Fosnaugh et al., 1995; Tan et al., 2000; Klebaur et al., 2002; Fumagalli et al., 2006; Salery et al., 2016). While Arc/Arg3.1 expression is often used as a marker of neuronal activity, convergent evidence suggests that the induction of this plasticity-associated gene would sustain homeostatic responses. Thus, by interacting with endophilin 2/3 and dynamin, Arc/Arg3.1 promotes AMPA receptor endocytosis, thereby contributing to synaptic scaling in the hippocampus (Chowdhury et al., 2006; Park et al., 2008). Future studies will be required to determine whether this mechanism could provide a molecular basis for the decreased AMPAR/NMDAR ratio observed in the striatum following repeated exposure to psychostimulants (Thomas et al., 2001; Kourrich et al., 2007; Kasanetz et al., 2010) and could account for the development of increased motor responses induced by D-amphetamine.
Conclusion
Our study provides evidence that repeated administration of D-amphetamine modulates striatal gene expression not only through the regulation of transcription but also by controlling the translational machinery. Moreover, our results further support the hypothesis that a transient decrease of global mRNA translation efficiently favors the translation of a selective subset of mRNAs (Di Prisco et al., 2014; Huang et al., 2016).
Author Contributions
AB and EP conducted all the experiments. IG-R and LC helped with biochemical studies. JB-V performed qRT-PCR experiments. AB, EP, and EV designed research. AB and EP analyzed the results. AB, EP, and EV wrote the manuscript with the help of MC-M.
Funding
This work was supported by Inserm, Fondation pour la Recherche Médicale (EV), and a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (EP). AB is supported by the Fonds National de la Recherche, Luxembourg (3977033). EP is a recipient of Marie Curie Intra-European Fellowship IEF327648.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Alexandre David (Inserm, U1191) for provinding the anti-puromycin antibody and the gradient fractionation system for polysome profile analysis. The authors thank Sanjeev Khatiwada for his comments.
References
Al-Fageeh, M. B., and Smales, C. M. (2009). Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA 15, 1164–1176. doi: 10.1261/rna.1179109
Baranick, B. T., Lemp, N. A., Nagashima, J., Hiraoka, K., Kasahara, N., and Logg, C. R. (2008). Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl. Acad. Sci. U.S.A. 105, 4733–4738. doi: 10.1073/pnas.0710650105
Beauvais, G., Atwell, K., Jayanthi, S., Ladenheim, B., and Cadet, J. L. (2011). Involvement of dopamine receptors in binge methamphetamine-induced activation of endoplasmic reticulum and mitochondrial stress pathways. PLoS ONE 6:e28946. doi: 10.1371/journal.pone.0028946
Belelovsky, K., Elkobi, A., Kaphzan, H., Nairn, A. C., and Rosenblum, K. (2005). A molecular switch for translational control in taste memory consolidation. Eur. J. Neurosci. 22, 2560–2568. doi: 10.1111/j.1460-9568.2005.04428.x
Bertran-Gonzalez, J., Bosch, C., Maroteaux, M., Matamales, M., Herve, D., Valjent, E., et al. (2008). Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. J. Neurosci. 28, 5671–5685. doi: 10.1523/JNEUROSCI.1039-08.2008
Biever, A., Puighermanal, E., Nishi, A., David, A., Panciatici, C., Longueville, S., et al. (2015). PKA-dependent phosphorylation of ribosomal protein S6 does not correlate with translation efficiency in striatonigral and striatopallidal medium-sized spiny neurons. J. Neurosci. 35, 4113–4130. doi: 10.1523/JNEUROSCI.3288-14.2015
Buffington, S. A., Huang, W., and Costa-Mattioli, M. (2014). Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 37, 17–38. doi: 10.1146/annurev-neuro-071013-014100
Canales, J. J., and Graybiel, A. M. (2000). A measure of striatal function predicts motor stereotypy. Nat. Neurosci. 3, 377–383. doi: 10.1038/73949
Chowdhury, S., Shepherd, J. D., Okuno, H., Lyford, G., Petralia, R. S., Plath, N., et al. (2006). Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459. doi: 10.1016/j.neuron.2006.08.033
Connor, J. H., Weiser, D. C., Li, S., Hallenbeck, J. M., and Shenolikar, S. (2001). Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 21, 6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001
Costa-Mattioli, M., Gobert, D., Harding, H., Herdy, B., Azzi, M., Bruno, M., et al. (2005). Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436, 1166–1173. doi: 10.1038/nature03897
Costa-Mattioli, M., Gobert, D., Stern, E., Gamache, K., Colina, R., Cuello, C., et al. (2007). eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206. doi: 10.1016/j.cell.2007.01.050
David, A., Dolan, B. P., Hickman, H. D., Knowlton, J. J., Clavarino, G., Pierre, P., et al. (2012). Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 197, 45–57. doi: 10.1083/jcb.201112145
Dever, T. E. (1999). Translation initiation: adept at adapting. Trends Biochem. Sci. 24, 398–403. doi: 10.1016/S0968-0004(99)01457-7
Dever, T. E. (2002). Gene-specific regulation by general translation factors. Cell 108, 545–556. doi: 10.1016/S0092-8674(02)00642-6
Di Prisco, G. V., Huang, W., Buffington, S. A., Hsu, C. C., Bonnen, P. E., Placzek, A. N., et al. (2014). Translational control of mGluR-dependent long-term depression and object-place learning by eIF2alpha. Nat. Neurosci. 17, 1073–1082. doi: 10.1038/nn.3754
Fosnaugh, J. S., Bhat, R. V., Yamagata, K., Worley, P. F., and Baraban, J. M. (1995). Activation of arc, a putative “effector” immediate early gene, by cocaine in rat brain. J. Neurochem. 64, 2377–2380. doi: 10.1046/j.1471-4159.1995.64052377.x
Fumagalli, F., Bedogni, F., Frasca, A., Di Pasquale, L., Racagni, G., and Riva, M. A. (2006). Corticostriatal up-regulation of activity-regulated cytoskeletal-associated protein expression after repeated exposure to cocaine. Mol. Pharmacol. 70, 1726–1734. doi: 10.1124/mol.106.026302
Go, B. S., Kim, J., Yang, J. H., and Choe, E. S. (2016). Psychostimulant-induced endoplasmic reticulum stress and neurodegeneration. Mol. Neurobiol. doi: 10.1007/s12035-016-9969-0 [Epub ahead of print].
Gong, S., Zheng, C., Doughty, M. L., Losos, K., Didkovsky, N., Schambra, U. B., et al. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. doi: 10.1038/nature02033
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., et al. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi: 10.1016/S1097-2765(00)00108-8
Hershey, J. W., Sonenberg, N., and Mathews, M. B. (2012). Principles of translaional control: an overview. Cold Spring Harb. Perspect. Biol. 4:a011528. doi: 10.1101/cshperspect.a011528
Huang, W., Placzek, A. N., Viana Di Prisco, G., Khatiwada, S., Sidrauski, C., Krnjevic, K., et al. (2016). Translational control by eIF2alpha phosphorylation regulates vulnerability to the synaptic and behavioral effects of cocaine. Elife 5:e12052. doi: 10.7554/eLife.12052
Jayanthi, S., Deng, X., Noailles, P. A., Ladenheim, B., and Cadet, J. L. (2004). Methamphetamine induces neuronal apoptosis via cross-talks between endoplasmic reticulum and mitochondria-dependent death cascades. FASEB J. 18, 238–251. doi: 10.1096/fj.03-0295com
Jayanthi, S., McCoy, M. T., Beauvais, G., Ladenheim, B., Gilmore, K., Wood, W. III, et al. (2009). Methamphetamine induces dopamine D1 receptor-dependent endoplasmic reticulum stress-related molecular events in the rat striatum. PLoS ONE 4:e6092. doi: 10.1371/journal.pone.0006092
Kasanetz, F., Deroche-Gamonet, V., Berson, N., Balado, E., Lafourcade, M., Manzoni, O., et al. (2010). Transition to addiction is associated with a persistent impairment in synaptic plasticity. Science 328, 1709–1712. doi: 10.1126/science.1187801
Kenney, J. W., Genheden, M., Moon, K. M., Wang, X., Foster, L. J., and Proud, C. G. (2016). Eukaryotic elongation factor 2 kinase regulates the synthesis of microtubule-related proteins in neurons. J. Neurochem. 136, 276–284. doi: 10.1111/jnc.13407 15as16
Kenney, J. W., Moore, C. E., Wang, X., and Proud, C. G. (2014). Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv. Biol. Regul. 55, 15–27. doi: 10.1016/j.jbior.2014.04.003
Klebaur, J. E., Ostrander, M. M., Norton, C. S., Watson, S. J., Akil, H., and Robinson, T. E. (2002). The ability of amphetamine to evoke arc (Arg 3.1) mRNA expression in the caudate, nucleus accumbens and neocortex is modulated by environmental context. Brain Res. 930, 30–36. doi: 10.1016/S0006-8993(01)03400-X
Kourrich, S., Rothwell, P. E., Klug, J. R., and Thomas, M. J. (2007). Cocaine experience controls bidirectional synaptic plasticity in the nucleus accumbens. J. Neurosci. 27, 7921–7928. doi: 10.1523/JNEUROSCI.1859-07.2007
Krasnova, I. N., Ladenheim, B., and Cadet, J. L. (2005). Amphetamine induces apoptosis of medium spiny striatal projection neurons via the mitochondria-dependent pathway. FASEB J. 19, 851–853.
Lee, Y. Y., Cevallos, R. C., and Jan, E. (2009). An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J. Biol. Chem. 284, 6661–6673. doi: 10.1074/jbc.M806735200
Lu, P. D., Harding, H. P., and Ron, D. (2004). Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33. doi: 10.1083/jcb.200408003
Luscher, C., and Malenka, R. C. (2011). Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron 69, 650–663. doi: 10.1016/j.neuron.2011.01.017
Ma, X. M., and Blenis, J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318. doi: 10.1038/nrm2672
Marciniak, S. J., Yun, C. Y., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., et al. (2004). CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077. doi: 10.1101/gad.1250704
Martineau, Y., Le Bec, C., Monbrun, L., Allo, V., Chiu, I. M., Danos, O., et al. (2004). Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol. Cell. Biol. 24, 7622–7635. doi: 10.1128/MCB.24.17.7622-7635.2004
Nairn, A. C., Bhagat, B., and Palfrey, H. C. (1985). Identification of calmodulin-dependent protein kinase III and its major Mr 100,000 substrate in mammalian tissues. Proc. Natl. Acad. Sci. U.S.A. 82, 7939–7943. doi: 10.1073/pnas.82.23.7939
Novoa, I., Zeng, H., Harding, H. P., and Ron, D. (2001). Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153, 1011–1022. doi: 10.1083/jcb.153.5.1011
Palam, L. R., Baird, T. D., and Wek, R. C. (2011). Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949. doi: 10.1074/jbc.M110.216093
Panja, D., Dagyte, G., Bidinosti, M., Wibrand, K., Kristiansen, A. M., Sonenberg, N., et al. (2009). Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J. Biol. Chem. 284, 31498–31511. doi: 10.1074/jbc.M109.056077
Panja, D., Kenney, J. W., D’Andrea, L., Zalfa, F., Vedeler, A., Wibrand, K., et al. (2014). Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK. Cell Rep. 9, 1430–1445. doi: 10.1016/j.celrep.2014.10.016
Park, S., Park, J. M., Kim, S., Kim, J. A., Shepherd, J. D., Smith-Hicks, C. L., et al. (2008). Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 59, 70–83. doi: 10.1016/j.neuron.2008.05.023
Pavitt, G. D., Ramaiah, K. V., Kimball, S. R., and Hinnebusch, A. G. (1998). eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 12, 514–526. doi: 10.1101/gad.12.4.514
Price, N. T., Redpath, N. T., Severinov, K. V., Campbell, D. G., Russell, J. M., and Proud, C. G. (1991). Identification of the phosphorylation sites in elongation factor-2 from rabbit reticulocytes. FEBS Lett. 282, 253–258. doi: 10.1016/0014-5793(91)80489-P
Puighermanal, E., Biever, A., and Valjent, E. (2016a). Synaptoneurosome preparation from C57BL/6 striata. Bio Protoc. 6:e1735. doi: 10.21769/BioProtoc.1735
Puighermanal, E., Cutando, L., Boubaker-Vitre, J., Honore, E., Longueville, S., Herve, D., et al. (2016b). Anatomical and molecular characterization of dopamine D1 receptor-expressing neurons of the mouse CA1 dorsal hippocampus. Brain Struct. Funct. doi: 10.1007/s00429-016-1314-x [Epub ahead of print].
Riley, A., Jordan, L. E., and Holcik, M. (2010). Distinct 5’UTRs regulate XIAP expression under normal growth conditions and durinig cellular stress. Nucleic Acids Res. 38, 4665–4674. doi: 10.1093/nar/gkq241
Robison, A. J., and Nestler, E. J. (2011). Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637. doi: 10.1038/nrn3111
Russo, S. J., Dietz, D. M., Dumitriu, D., Morrison, J. H., Malenka, R. C., and Nestler, E. J. (2010). The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 33, 267–276. doi: 10.1016/j.tins.2010.02.002
Ryazanov, A. G., Natapov, P. G., Shestakova, E. A., Severin, F. F., and Spirin, A. S. (1988). Phosphorylation of the elongation factor 2: the fifth Ca2+/calmodulin-dependent system of protein phosphorylation. Biochimie 70, 619–626. doi: 10.1016/0300-9084(88)90245-3
Salery, M., Dos Santos, M., Saint-Jour, E., Moumne, L., Pages, C., Kappes, V., et al. (2016). Activity-regulated cytoskeleton-associated protein accumulates in the nucleus in response to cocaine and acts as a brake on chromatin remodeling and long-term behavioral alterations. Biol. Psychiatry doi: 10.1016/j.biopsych.2016.05.025 [Epub ahead of print].
Scheetz, A. J., Nairn, A. C., and Constantine-Paton, M. (2000). NMDA receptor-mediated control of protein synthesis at developing synapses. Nat. Neurosci. 3, 211–216. doi: 10.1038/72915
Sonenberg, N., and Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. doi: 10.1016/j.cell.2009.01.042
Tan, A., Moratalla, R., Lyford, G. L., Worley, P., and Graybiel, A. M. (2000). The activity-regulated cytoskeletal-associated protein arc is expressed in different striosome-matrix patterns following exposure to amphetamine and cocaine. J. Neurochem. 74, 2074–2078. doi: 10.1046/j.1471-4159.2000.0742074.x
Thomas, M. J., Beurrier, C., Bonci, A., and Malenka, R. C. (2001). Long-term depression in the nucleus accumbens: a neural correlate of behavioral sensitization to cocaine. Nat. Neurosci. 4, 1217–1223. doi: 10.1038/nn757
Ubeda, M., Wang, X. Z., Zinszner, H., Wu, I., Habener, J. F., and Ron, D. (1996). Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell. Biol. 16, 1479–1489. doi: 10.1128/MCB.16.4.1479
Vattem, K. M., and Wek, R. C. (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274. doi: 10.1073/pnas.0400541101
Verpelli, C., Piccoli, G., Zibetti, C., Zanchi, A., Gardoni, F., Huang, K., et al. (2010). Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J. Neurosci. 30, 5830–5842. doi: 10.1523/JNEUROSCI.0119-10.2010
Walter, P., and Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. doi: 10.1126/science.1209038
Wang, X. Z., Kuroda, M., Sok, J., Batchvarova, N., Kimmel, R., Chung, P., et al. (1998). Identification of novel stress-induced genes downstream of chop. EMBO J. 17, 3619–3630. doi: 10.1093/emboj/17.13.3619
Young, S. K., Palam, L. R., Wu, C., Sachs, M. S., and Wek, R. C. (2016). Ribosome elongation stall directs gene-specific translation in the integrated stress response. J. Biol. Chem. 291, 6546–6558. doi: 10.1074/jbc.M115.705640
Young, S. K., Willy, J. A., Wu, C., Sachs, M. S., and Wek, R. C. (2015). Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J. Biol. Chem. 290, 28257–28271. doi: 10.1074/jbc.M115.693184
Keywords: D-amphetamine, protein synthesis, striatum, translation factors, eIF2α, Arc/Arg3.1
Citation: Biever A, Boubaker-Vitre J, Cutando L, Gracia-Rubio I, Costa-Mattioli M, Puighermanal E and Valjent E (2017) Repeated Exposure to D-Amphetamine Decreases Global Protein Synthesis and Regulates the Translation of a Subset of mRNAs in the Striatum. Front. Mol. Neurosci. 9:165. doi: 10.3389/fnmol.2016.00165
Received: 25 October 2016; Accepted: 20 December 2016;
Published: 10 January 2017.
Edited by:
Jason D. Shepherd, University of Utah, USAReviewed by:
Clive R. Bramham, University of Bergen, NorwayDavid M. Lovinger, National Institute on Alcohol Abuse and Alcoholism, USA
Copyright © 2017 Biever, Boubaker-Vitre, Cutando, Gracia-Rubio, Costa-Mattioli, Puighermanal and Valjent. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emmanuel Valjent, ZW1tYW51ZWwudmFsamVudEBpZ2YuY25ycy5mcg==; ZW1tYW51ZWwudmFsamVudEBnbWFpbC5jb20=
†These authors have contributed equally to this work.
 Anne Biever
Anne Biever Jihane Boubaker-Vitre
Jihane Boubaker-Vitre Laura Cutando
Laura Cutando Irene Gracia-Rubio
Irene Gracia-Rubio Mauro Costa-Mattioli
Mauro Costa-Mattioli Emma Puighermanal
Emma Puighermanal Emmanuel Valjent
Emmanuel Valjent