- Center for Epigenetics and Metabolism, School of Medicine, University of California at Irvine, Irvine, CA, USA
Epigenetic changes, such as DNA methylation or histone modification, can remodel the chromatin and regulate gene expression. Remodeling of chromatin provides an efficient mechanism of transducing signals, such as light or nutrient availability, to regulate gene expression. CLOCK:BMAL1 mediated activation of clock-controlled genes (CCGs) is coupled to circadian changes in histone modification at their promoters. Several chromatin modifiers, such as the deacetylases SIRT1 and HDAC3 or methyltransferase MLL1, have been shown to be recruited to the promoters of the CCGs in a circadian manner. Interestingly, the central element of the core clock machinery, the transcription factor CLOCK, also possesses histone acetyltransferase activity. Rhythmic expression of the CCGs is abolished in the absence of these chromatin modifiers. Recent research has demonstrated that chromatin remodeling is at the cross-roads of circadian rhythms and regulation of metabolism and aging. It would be of interest to identify if similar pathways exist in the epigenetic regulation of memory formation.
Epigenetic Regulation of Memory Formation: Is the Circadian Clock Involved?
“Epigenetics” literally means “above genetics.” It is defined as the study of heritable changes in gene expression that does not involve any change to the DNA sequence. Such changes in gene expression can be brought about by a variety of mechanism that involves a combination of post-translational modifications of histones, remodeling of chromatin, incorporation of histone variants, or methylation of DNA on CpG islands. Histone acetylation is a mark for activation of transcription, which is achieved by remodeling the chromatin to make it more accessible to the transcription machinery (Jenuwein and Allis, 2001). Histone methylation, on the other hand, acts as a signal for recruitment of chromatin remodeling factors which can either activate or repress transcription. DNA methylation leads to compaction of the chromatin and causes gene silencing. Many of these epigenetic events are crucial in brain function (Zocchi and Sassone-Corsi, 2010).
Transcription of genes is critical in synaptic plasticity and establishment of long-term memory. The role of histone modifications and DNA methylation has been elucidated in memory formation (Barrett and Wood, 2008; Day and Sweatt, 2011). CREB-binding protein (CBP) is a well characterized histone acetyltransferase (HAT) that is recruited by CREB and plays a role in learning and memory. Several mouse models harboring mutations in CBP have been generated and all of them have demonstrated varied defects in learning and memory (Barrett and Wood, 2008). Demonstrating the importance of DNA methylation, impairment in memory was also observed in mice deficient in DNA methyltransferase (DNMT) 1 and DNMT 3a in the forebrain excitatory neurons (Feng et al., 2010).
A time-of-day effect is observed in memory formation, thereby linking the circadian clock to this biological process (Gerstner and Yin, 2010). Long-term potentiation (LTP) in the hippocampus has been demonstrated to undergo circadian changes (Barnes et al., 1977; Chaudhury et al.,2005). Arrhythmic per0 mutant Drosophila melanogaster have defects in long-term memory formation (Sakai et al., 2004). Interestingly, phosphorylation of mitogen-activated protein kinase (MAPK) displays rhythmicity in the hippocampus and inhibition of this oscillation leads to impairment in the persistence of long-term memory (Eckel-Mahan et al., 2008).
Circadian clocks are present in almost all the tissues in mammals. The master or “central” clock is located in the hypothalamic suprachiasmatic nucleus (SCN) containing 10–15,000 neurons. “Peripheral clocks” are present in almost all other mammalian tissues, such as liver, heart, lung, and kidney where they maintain circadian rhythms and regulate tissue-specific gene expression. There is evidence that a functional clock exists in many parts in the brain, including the hippocampus. Suggesting the presence of an autonomous clock, Per2 expression was found to be rhythmic in isolated hippocampus (Wang et al., 2009). What remains to be determined is whether the genes involved in memory formation are regulated by the circadian clock.
The Clock Machinery
The molecular machinery that regulates circadian rhythms comprises of a set of genes, known as “clock” genes, the products of which interact to generate and maintain the rhythms. A conserved feature among many organisms is the regulation of the circadian clock by a negative feedback loop (Sahar and Sassone-Corsi, 2009). Positive regulators induce the transcription of clock-controlled genes (CCGs), some of which encode proteins that feedback on their own expression by repressing the activity of the positive regulators. CLOCK and BMAL1 are the positive regulators of the mammalian clock machinery which regulate the expression of the negative regulators: cryptochrome (CRY1 and CRY2) and period (PER1, PER2, and PER3) families. CLOCK and BMAL1 are transcription factors that heterodimerize through the PAS domain and induce the expression of CCGs by binding to their promoters at E-boxes (CACGTG). Once a critical concentration of the Per and Cry proteins is accumulated, these proteins translocate into the nucleus and form a complex to inhibit CLOCK–BMAL1 mediated transcription, thereby closing the negative feedback loop. In order to start a new transcriptional cycle, the CLOCK–BMAL1 complex needs to be de-repressed through the proteolytic degradation of Per and Cry. Some CCGs are transcription factors, such as albumin D-box binding protein (DBP), RORα, and REV-ERBα, which can then regulate cyclic expression of other genes. DBP binds to D-boxes [TTA(T/C)GTAA], whereas RORα and REV-ERBα bind to the Rev-Erb/ROR-binding element, or RRE [(A/T)A(A/T)NT(A/G)GGTCA]. Approximately 10% of the transcriptome displays robust circadian rhythmicity (Akhtar et al., 2002; Panda et al., 2002). Interestingly, most transcripts that oscillate in one tissue (e.g., SCN) do not oscillate in another (e.g., liver; Akhtar et al., 2002; Panda et al., 2002; Miller et al., 2007; Masri and Sassone-Corsi, 2010). Further research is required to identify the genes that display rhythmicity in its expression in other areas of the brain and are involved in memory formation.
Epigenetics and Clock Genes
Genes encoding circadian clock proteins are regulated by epigenetic mechanisms, such as histone phosphorylation, acetylation, and methylation, which have been shown to follow circadian rhythm (Crosio et al., 2000; Etchegaray et al., 2003; Ripperger and Schibler, 2006; Masri and Sassone-Corsi, 2010). Exposure to light causes rapid phosphorylation of Histone 3 on Serine 10 (H3-S10) in the SCN. This phosphorylation parallels induction of immediate early genes such as c-fos and Per1, thereby indicating that light-mediated signaling can regulate circadian gene expression by remodeling the chromatin (Crosio et al., 2000).
CLOCK:BMAL1 mediated activation of CCGs has been shown to be coupled to circadian changes in histone acetylation at their promoters (Etchegaray et al., 2003). The central element of the core clock machinery, the transcription factor CLOCK, also possesses intrinsic HAT activity (Doi et al., 2006). Since CLOCK binds to E-box regions of DNA, the HAT activity of CLOCK can selectively remodel chromatin at the promoters of CCGs. The HAT function of CLOCK is essential for circadian gene expression. The enzymatic activity of CLOCK also allows it to acetylate non-histone substrates such as its own binding partner, BMAL1 (Hirayama et al., 2007). CLOCK specifically acetylates BMAL1 which facilitates CRY-dependent repression. Histone methylation is also important for circadian gene expression. MLL1, a methyltransferase that methylates histone H3 at lysine 4 (H3-K4) associates with CLOCK, and get recruited to promoters of CCGs in a circadian manner (Katada and Sassone-Corsi, 2010). H3-K4 methylation at these promoters also displayed rhythmicity (Katada and Sassone-Corsi, 2010). These findings underscore the importance of epigenetic mechanisms in circadian regulation and reveal the molecular pathways by which such essential control is achieved (Figure 1).
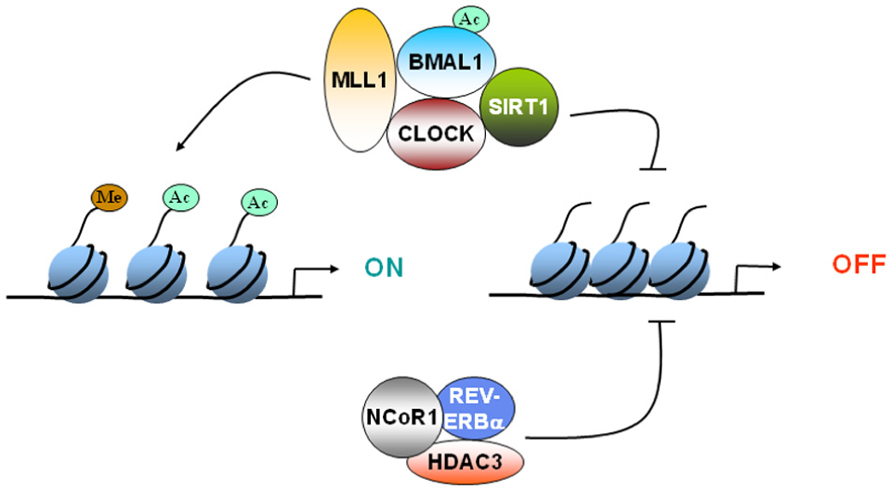
FIGURE 1. Epigenetic regulation of gene expression by circadian clock. CLOCK can acetylate histones to induce gene expression. CLOCK interacts with MLL1 (a histone methyltransferase) and SIRT1 (a deacetylase).These epigenetic regulators can modify the chromatin according the environmental stimuli, such as nutrient availability. Furthermore, REV-ERBα, a clock-controlled gene, can cause recruitment of HDAC3 and deacetylate histones. Circadian regulation of either the expression or the activity of these epigenetic regulators determines whether the gene gets turned “ON” or “OFF.”
The regulatory function of REV-ERBα is controlled by the nuclear receptor corepressor 1 (NCoR1), a corepressor that recruits HDAC3, a histone deacetylase, to mediate transcriptional repression of target genes, such as Bmal1. When the NCoR1– HDAC3 association is genetically disrupted in mice, circadian and metabolic defects develop (Alenghat et al., 2008). These mice demonstrate a shorter period, increased energy expenditure and are resistant to diet-induced obesity (Alenghat et al., 2008). HDAC3 recruitment to the genome was recently shown to be rhythmic in liver (high during the day and low at night; Feng et al., 2011). At these HDAC3 binding sites, REV-ERBα and NCoR1 recruitment were in phase with HDAC3 recruitment, whereas histone acetylation and RNA Polymerase II recruitment were anti-phasic. Although the circadian recruitment of HDAC3 has not been demonstrated in brain, it is interesting to note that inhibition of HDAC3 leads to enhancement of long-term memory formation (McQuown et al., 2011). These results raise the possibility that HDAC3 might be a key player in the integration of histone modification, circadian clock, and memory formation (Figure 1).
SIRT1: A Possible Mediator of Circadian Rhythms and Memory Formation
Role of SIRT1 in Regulation of Circadian Rhythms
The finding of a circadian HAT opened the search for a counterbalancing histone deacetylase (HDAC). Recently, SIRT1 was identified to be a critical modulator of the circadian clock machinery (Asher et al., 2008; Nakahata et al., 2008). SIRT1 belongs to the family of sirtuins, which constitutes the so-called class III of HDACs. These are the only HDACs whose enzymatic activity is NAD+-dependent, and that has been directly linked to the control of metabolism and aging (Bishop and Guarente, 2007). Histone H3 Lys9 and Lys14 at circadian promoters, BMAL1 and PER2 are specific targets of SIRT1. The CLOCK–BMAL1 complex interacts with SIRT1 and recruits it to the circadian promoters. Importantly, circadian gene expression and BMAL1 acetylation are compromised in liver-specific SIRT1 mutant mice (Nakahata et al., 2008). These findings led to the concept that SIRT1 operates as a rheostat of the circadian machinery, modulating the amplitude and “tightness” of CLOCK-mediated acetylation and consequent transcription cycles (Nakahata et al., 2008; Figure 1).
Circadian oscillation of SIRT1 activity suggested that cellular NAD+ levels may also oscillate. Circadian clock controls the expression of nicotinamide phosphoribosyltransferase (NAMPT), a key rate-limiting enzyme in the salvage pathway of NAD+ biosynthesis (Nakahata et al., 2009; Ramsey et al., 2009). The rhythmicity in the expression of this enzyme drives the oscillation in NAD+ levels (Nakahata et al., 2009; Ramsey et al., 2009). CLOCK, BMAL1, and SIRT1 are recruited to the NAMPT promoter in a circadian time-dependent manner. The oscillatory expression of NAMPT is abolished in clock/clock mice, which results in drastically reduced levels of NAD+ in MEFs derived from these mice (Nakahata et al., 2009). These results make a compelling case for the existence of an enzymatic/transcriptional feedback loop, wherein SIRT1 regulates the levels of its own cofactor. Interestingly, mice deficient of NAD+ hydrolase CD38 displayed altered rhythmicity of NAD+. Very high levels of NAD+ in tissues such as the brain and liver have been reported in the CD38-null mice (Aksoy et al., 2006). The high, chronic levels of NAD+ results in several anomalies in circadian behavior and metabolism (Sahar et al., 2011). CD38-null mice display a shortened period length of locomotor activity and alteration in the rest-activity rhythm (Sahar et al., 2011). The question that still remains is whether NAD+ levels oscillate in the SCN or in other parts of the brain, such as hippocampus.
Protection from Neurodegeneration by SIRT1
The finding that SIRT1 acts as a rheostat of circadian acetylation is of interest as it may be linked to other, recently described functions of this regulator in aging and neurodegeneration (Gan and Mucke, 2008). SIRT1 was shown to deacetylate and coactivate retinoic acid receptor β (RARβ), which leads to induction of the expression of Adam10, a gene that encodes α-secretase (Donmez et al., 2010). Cleavage of the amyloid precursor protein by α-secretase prevents the production of the toxic amyloid β peptides that cause Alzheimer’s disease (AD). Thus, SIRT1 appears to have a neuroprotective role (Donmez et al., 2010). SIRT1 also deacetylates tau and prevents tauopathy that is evident in several neurodegenerative diseases (Min et al.,2010). Moreover, treatment with SIRT1 activator resveratrol or ectopic expression of SIRT1 was shown to prevent neuronal cell death (Kim et al., 2007).
SIRT1 and Memory Formation
Recent reports suggest that SIRT1 also plays a major role in synaptic plasticity and memory formation (Gao et al., 2010; Michan et al., 2010). Brain-specific SIRT1 mutant mice (Gao et al., 2010) or the whole body SIRT1 knockout mice (Michan et al., 2010) displayed defects in learning and memory. Brain-specific SIRT1 mutant mice display lower levels of CREB protein expression in the hippocampus (Gao et al., 2010). CREB expression was found to be downregulated by a microRNA, miR-134. SIRT1 negatively regulates the expression of miR-134, and thus, in turn, regulates CREB expression (Gao et al., 2010). miR-134 overexpression in the hippocampus mimics the loss of SIRT1, whereas knocking down miR-134 in hippocampus ameliorates memory defects in the SIRT1 mutant mice (Gao et al., 2010). If SIRT1 activity is rhythmic in the hippocampus (as it is in the liver), it will be interesting to determine whether the circadian clock can modulate CREB expression through SIRT1.
Conclusion
The importance of epigenetic events is becoming clear in the regulation of circadian rhythms and memory. Current data suggests that many epigenetic regulators themselves are regulated in a circadian manner, at least in some tissues. The challenge ahead is to understand whether these epigenetic events follow a rhythmic pattern in tissues that are involved in the process of learning and memory, such as hippocampus, cortex, and amygdala.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to apologize to all colleagues whose work, because of lack of space, could not be cited. We thank all the members of the Paolo Sassone-Corsi laboratory for helpful discussions. Work in our laboratory is supported by the National Institute of Health and the Institut National de la Sante et de la Recherche Medicale (France) and Sirtris Pharmaceuticals, Inc.
References
Akhtar, R. A., Reddy, A. B., Maywood, E. S., Clayton, J. D., King, V. M., Smith, A. G., Gant, T. W., Hastings, M. H., and Kyriacou, C. P. (2002). Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550.
Aksoy, P., White, T. A., Thompson, M., and Chini, E. N. (2006). Regulation of intracellular levels of NAD: a novel role for CD38. Biochem. Biophys. Res. Commun. 345, 1386–1392.
Alenghat, T., Meyers, K., Mullican, S. E., Leitner, K., Adeniji-Adele, A., Avila, J., Bućan, M., Ahima, R. S., Kaestner, K. H., and Lazar, M. A. (2008). Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000.
Asher, G., Gatfield, D., Stratmann, M., Reinke, H., Dibner, C., Kreppel, F., Mostoslavsky, R., Alt, F. W., and Schibler, U. (2008). SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317– 328.
Barnes, C. A., McNaughton, B. L., Goddard, G. V., Douglas, R. M., and Adamec, R. (1977). Circadian rhythm of synaptic excitability in rat and monkey central nervous system. Science 197, 91–92.
Barrett, R. M., and Wood, M. A. (2008). Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn. Mem. 15, 460–467.
Bishop, N. A., and Guarente, L. (2007). Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 8, 835–844.
Chaudhury, D., Wang, L. M., and Colwell, C. S. (2005). Circadian regulation of hippocampal long-term potentiation. J. Biol. Rhythms 20, 225–236.
Crosio, C., Cermakian, N., Allis, C. D., and Sassone-Corsi, P. (2000). Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 3, 1241–1247.
Day, J. J., and Sweatt, J. D. (2011). Cognitive neuroepigenetics: a role for epigenetic mechanisms in learning and memory. Neurobiol. Learn. Mem. 96, 2–12.
Doi, M., Hirayama, J., and Sassone-Corsi, P. (2006). Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508.
Donmez, G., Wang, D., Cohen, D. E., and Guarente, L. (2010). SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142, 320–332.
Eckel-Mahan, K. L., Phan, T., Han, S., Wang, H., Chan, G. C. K., Scheiner, Z. S., and Storm, D. R. (2008). Circadian oscillation of hippocampal MAPK activity and cAmp: implications for memory persistence. Nat. Neurosci. 11, 1074–1082.
Etchegaray, J. P., Lee, C., Wade, P. A., and Reppert, S. M. (2003). Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182.
Feng, D., Liu, T., Sun, Z., Bugge, A., Mullican, S. E., Alenghat, T., Liu, X. S., and Lazar, M. A. (2011). A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319.
Feng, J., Zhou, Y., Campbell, S. L., Le, T., Li, E., Sweatt, J. D., Silva, A. J., and Fan, G. (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430.
Gan, L., and Mucke, L. (2008). Paths of convergence: sirtuins in aging and neurodegeneration. Neuron 58, 10–14.
Gao, J., Wang, W. Y., Mao, Y. W., Gräff, J., Guan, J., Pan, L., Mak, G., Kim, D., Su, S., and Tsai, L. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Gerstner, J. R., and Yin, J. C. (2010). Circadian rhythms and memory formation. Nat. Rev. Neurosci. 11, 577–588.
Hirayama, J., Sahar, S., Grimaldi, B., Tamaru, T., Takamatsu, K., Nakahata, Y., and Sassone-Corsi, P. (2007). CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090.
Katada, S., and Sassone-Corsi, P. (2010). The histone methyltransferase MLL1permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 17, 1414–1421.
Kim, D., Nguyen, M. D., Dobbin, M. M., Fischer, A., Sananbenesi, F., Rodgers, J. T., Delalle, I., Baur, J. A., Sui, G., Armour, S. M., Puigserver, P., Sinclair, D. A., and Tsai, L. H. (2007). SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 26, 3169–3179.
Masri, S., and Sassone-Corsi, P. (2010). Plasticity and specificity of the circadian epigenome. Nat. Neurosci. 13, 1324–1329.
McQuown, S. C., Barrett, R. M., Matheos, D. P., Post, R. J., Rogge, G. A., Alenghat, T., Mullican, S. E., Jones, S., Rusche, J. R., Lazar, M. A., and Wood, M. A. (2011). HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 31, 764–774.
Michan, S., Li, Y., Chou, M. M.-H., Parrella, E., Ge, H., Long, J. M., Allard, J. S., Lewis, K., Miller, M., Xu, W., Mervis, R. F., Chen, J., Guerin, K. I., Smith, L. E. H., McBurney, M. W., Sinclair, D. A., Baudry, M., de Cabo, R., and Longo, V. D. (2010). SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 30, 9695–9707.
Miller, B. H., McDearmon, E. L., Panda, S., Hayes, K. R., Zhang, J., Andrews, J. L., Antoch, M. P., Walker, J. R., Esser, K. A., Hogenesch, J. B., and Takahashi, J. S. (2007). Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 104, 3342–3347.
Min, S. W., Cho, S. H., Zhou, Y., Schroeder, S., Haroutunian, V., Seeley, W. W., Huang, E. J., Shen, Y., Masliah, E., Mukherjee, C., Meyers, D., Cole, P. A., Ott, M., and Gan, L. (2010). Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966.
Nakahata, Y., Kaluzova, M., Grimaldi, B., Sahar, S., Hirayama, J., Chen, D., Guarente, L. P., and Sassone-Corsi, P. (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340.
Nakahata, Y., Sahar, S., Astarita, G., Kaluzova, M., and Sassone-Corsi, P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657.
Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., Schultz, P. G., Kay, S. A., Takahashi, J. S., and Hogenesch, J. B. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320.
Ramsey, K. M., Yoshino, J., Brace, C. S., Abrassart, D., Kobayashi, Y., Marcheva, B., Hong, H.-K., Chong, J. L., Buhr, E. D., Lee, C., Takahashi, J. S., Imai, S.-I., and Bass, J. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654.
Ripperger, J. A., and Schibler, U. (2006). Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374.
Sahar, S., Nin, V., Barbosa, M. T., Chini, E. N., and Sassone-Corsi, P. (2011). Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging 3, 794–802.
Sahar, S., and Sassone-Corsi, P. (2009). Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer 9, 886–896.
Sakai, T., Tamura, T., Kitamoto, T., and Kidokoro, Y. (2004). A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 101, 16058–16063.
Wang, L. M.-C., Dragich, J. M., Kudo, T., Odom, I. H., Welsh, D. K., O’Dell, T. J., and Colwell, C. S. (2009). Expression of the circadian clock gene Period2 in the hippocampus: possible implications for synaptic plasticity and learned behaviour. ASN Neuro 1, e00012. doi: 10.1042/AN20090020
Keywords: memory, epigenetics, circadian rhythms, clock
Citation: Sahar S and Sassone-Corsi P (2012) Circadian rhythms and memory formation: regulation by chromatin remodeling. Front. Mol. Neurosci. 5:37. doi: 10.3389/fnmol.2012.00037
Received: 01 October 2011; Paper pending published: 17 November 2011;
Accepted: 12 March 2012; Published online: 26 March 2012.
Edited by:
Kristin Eckel-Mahan, University of California at Irvine, USACopyright: © 2012 Sahar and Sassone-Corsi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Saurabh Sahar, Center for Epigenetics and Metabolism, School of Medicine, University of California at Irvine, Irvine, CA 92697-4625, USA. e-mail:c3NhaGFyQHVjaS5lZHU=

