
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci., 01 July 2011
Sec. Molecular Signalling and Pathways
Volume 4 - 2011 | https://doi.org/10.3389/fnmol.2011.00007
This article is part of the Research TopicThe tumor and the embryo: oxygen and oxygen dependent pathways in developmentView all 6 articles
Mammalian gene expression patterns change profoundly in response to low oxygen levels. These changes in gene expression programs are strongly influenced by post-transcriptional mechanisms mediated by mRNA-binding factors: RNA-binding proteins (RBPs) and microRNAs (miRNAs). Here, we review the RBPs and miRNAs that modulate mRNA turnover and translation in response to hypoxic challenge. RBPs such as HuR (human antigen R), PTB (polypyrimidine tract-binding protein), heterogeneous nuclear ribonucleoproteins (hnRNPs), tristetraprolin, nucleolin, iron-response element-binding proteins (IRPs), and cytoplasmic polyadenylation-element-binding proteins (CPEBs), selectively bind to numerous hypoxia-regulated transcripts and play a major role in establishing hypoxic gene expression patterns. MiRNAs including miR-210, miR-373, and miR-21 associate with hypoxia-regulated transcripts and further modulate the levels of the encoded proteins to implement the hypoxic gene expression profile. We discuss the potent regulation of hypoxic gene expression by RBPs and miRNAs and their integrated actions in the cellular hypoxic response.
In response to hypoxic challenge, mammalian cells mount a well-orchestrated response that includes specific changes in the collective of expressed proteins. The altered gene expression program allows the cell to respond to hypoxic conditions, triggering adaptive processes that include changes in cell division, survival, motility, or differentiation.
Changes in transcription directly affect the abundance of some hypoxia-regulated proteins. The central mediator of hypoxic transcriptional gene control is the hypoxia-inducible factor (HIF; Rankin and Giaccia, 2008). HIF is a heterodimer transcription factor composed of a hypoxia-inducible α subunit and a constitutively expressed β subunit, each consisting of three isoforms (Bracken et al., 2003; Semenza, 2008). Among the HIF α isoforms, HIF-1α, and HIF-2α regulate similar sets of target genes, but only HIF-1α is expressed constitutively (Semenza, 2003; Schofield and Ratcliffe, 2004). In low oxygen (typically <2% O2), HIF-1 transcriptionally activates many genes encoding proteins implicated in angiogenesis, glucose metabolism, and cell proliferation and survival; many of these genes have hypoxia-response elements (HREs) in their regulatory regions (Wenger et al., 2005). HIF-1 can also regulate gene transcription in an HRE-independent manner, via its interaction histone acetyltransferases p300, CBP, and SRC-1 (Carrero et al., 2000). Other transcription factors that participate in the transcriptional response to hypoxia include the nuclear factor (NF)-κB complex, the activator protein 1 (AP1) complex, and the activating transcription factor ATF-4 (reviewed in Shih and Claffey, 1998; Gardner and Corn, 2008).
On a global scale, hypoxia markedly decreases total de novo transcription and total RNA levels, accompanied by changes in histone acetylation and methylation consistent with the notion that chromatin modifications helps to suppress overall gene transcription (Johnson and Barton, 2007; Johnson et al., 2008). Hypoxia also triggers the ubiquitination and other post-translational modifications of RNA polymerase II which alter its activity (Ignacak et al., 2009).
Since transcription is overall reduced with hypoxia, as it occurs in response to many damaging stimuli, the post-transcriptional regulation of pre-existing mRNAs is particularly important. The main post-transcriptional mechanisms affecting the levels of expressed proteins during hypoxia are mRNA turnover and translational control. These processes are modulated efficiently by both RNA-binding proteins (RBPs) and an emerging group of non-coding RNAs (ncRNAs) among which the best-known members are microRNAs. MicroRNAs are ∼22-nt long ncRNAs which associate with cellular mRNAs and typically repress gene expression by reducing their half-life and/or inhibiting their translation (Fabian et al., 2010). In this review, we will focus on the regulation of hypoxic gene expression by RBPs (Table 1) and miRNAs (Table 2).
Different stimuli can trigger wide changes in the rates of mRNA turnover in the cell. Rapid increases or decreases in mRNA stability can alter dramatically the levels of mRNA present in the cell, often in the absence of transcriptional changes (Mitchell and Tollervey, 2001; Wilusz et al., 2001). In response to hypoxia, several RBPs and miRNAs have been found to affect the half-lives of many specific mRNAs, as reviewed in detail below.
Accelerated mRNA decay is observed when the relative half-life of an mRNA is reduced in response to a given stimulus. Although the mechanisms of mRNA degradation are not fully understood, they involve enzymatic structures such as the exosome, processing (P)-bodies, and the RNA-induced silencing complex (RISC). The exosome is a large multi-protein complex that carries out 3′ → 5′ degradation of labile mRNAs following the recruitment by decay-promoting RBPs such as AUF1 [also named hnRNP D (heterogeneous nuclear ribonucleoprotein)], TTP (tristetraprolin), BRF1 (butyrate response factor-1), and KSRP (KH domain-containing RBP; Chen et al., 2001a; Butler, 2002; Mukherjee et al., 2002; Gherzi et al., 2004). P-bodies (PBs) are cytoplasmic structures that contain proteins involved in mRNA decapping and 5′ → 3′ degradation (Cougot et al., 2004; Kedersha et al., 2005; Parker and Sheth, 2007). RISC-loaded miRNAs target specific mRNAs through sequence similarity; the endoribonucleolytic activity of Argonaute 2 and other enzymes initiates the degradation of target mRNAs (Fabian et al., 2010). RBPs and miRNAs can recruit mRNAs to these degradation machineries (reviewed by Butler, 2002; Kedersha and Anderson, 2007; Fabian et al., 2010), but their specific involvement in the hypoxia-triggered changes in mRNA turnover in mammalian systems awaits further study. MicroRNAs and several RBPs such as TTP can modulate mRNA stability in response to hypoxia.
Increased mRNA stability, the converse process, consists in the accumulation of an mRNA through the reduction of its rate of decay. A stabilizing RBP typically associates with an mRNA and enhances its half-life by competing with, and hence preventing the binding of, degradation-promoting RBPs and possibly also degradation-promoting miRNAs (e.g., Zhao et al., 2000, reviewed by Abdelmohsen et al., 2008a). RBPs that promote mRNA stabilization include Hu proteins [the ubiquitous HuR (HuA) and the primarily neuronal HuB (Hel-N1), HuC, and HuD], NF90, the α-complex protein 1 (αCP1, also known as Poly(C)-binding protein or PCBP1), nucleolin, heterogenous ribonucleoprotein (hnRNP) C1/C2, PTB, CUG-binding protein 2(CUG-BP2), the poly(A)-binding protein-interacting protein 2 (PAIP2), and erythropoietin mRNA-binding protein (ERBP; Kiledjian et al., 1995; Rondon et al., 1995; McGary et al., 1997; Onesto et al., 2004; Shi et al., 2007; Abdelmohsen et al., 2008a; Hinman and Lou, 2008; Sawicka et al., 2008). The hypoxia-induced binding of RBPs HuR, polypyrimidine tract-binding protein (PTB), and ERBP to target mRNAs, and the hypoxia-triggered dissociation of hnRNP A2 and hnRBP L from target mRNAs, all have been implicated in mRNA stabilization, as discussed below.
The relative utilization of a given mRNA for translation can also be regulated by increasing or decreasing the loading of ribosomes onto the mRNA (the translation initiation step) or the rates of polypeptide synthesis and dissociation from the ribosome (the elongation and termination steps, respectively). The metabolic consequence of energy depletion in hypoxia is a fast and general inhibition of protein synthesis. Early global inhibition of mRNA translation is attributed to eIF2α (eukaryotic translation initiation factor 2α) phosphorylation, while under sustained hypoxic conditions the mammalian target of rapamycin (mTOR) has a pivotal role for the control of global mRNA translation (Koritzinsky et al., 2006). For an excellent review of the hypoxia-regulated pathways that alter global translation, see Wouters et al. (2005).
Translational inhibition occurs when, despite a lack of change in steady-state mRNA abundance, translation of an mRNA declines. RBPs that lower the translation of specific target mRNAs include the T-cell intracellular antigen-1 (TIA-1), the TIA-1-related protein (TIAR), NF90, HuR, CUG-BP1, hnRNP A2, hnRNP L, and the iron-response proteins (IRP1, IRP2; Hamilton et al., 1999; López de Silanes et al., 2005; Kim et al., 2007; Leipuviene and Theil, 2007; Abdelmohsen et al., 2008a). Many mammalian miRNAs studied to date also repress translation, whether or not they also affect mRNA stability (Fabian et al., 2010). RBPs and miRNAs can block translation by mobilizing mRNAs to subcytoplasmic domains where translation is inhibited; such sites include PBs, neuronal RNA granules, and stress granules (SGs), cytoplasmic foci that form transiently in response to cell damage including hypoxia (Gottschald et al., 2010) and harbor mRNAs that are typically stable and not translated. In general, these RNA granules contain RBPs and miRNAs that can direct specific mRNAs to translation and decay machineries (Anderson and Kedersha, 2006; Kedersha and Anderson, 2007). The action of hypoxia upon these RBPs and miRNAs is described below.
Translational activation occurs when an mRNA is more efficiently translated in the absence of changes in mRNA abundance. For instance, internal ribosome entry sites (IRES) have been proposed to promote mRNA translation during hypoxia. IRESs are cis-acting elements present in some 5′untranslated regions (UTRs) that mediate binding of ribosomes directly in front of the AUG start codon, independent of the 5′ cap recognition (Le Quesne et al., 2010); although IRESs are often highly structured, sometimes they lack secondary structure (Baird et al., 2007; Gilbert et al., 2007; Xia and Holcik, 2009). RBPs acting as IRES trans-acting factors (ITAFs) have also been proposed to play a role during hypoxia by supporting translational initiation. However, the functional relevance of IRESs during hypoxia is the subject of ongoing debate and alternative regulatory mechanisms have been discussed (Fähling, 2009a).
RBPs that promote translation include HuR, PTB, the cytoplasmic polyadenylation-element-binding protein (CPEB), nucleolin, and hnRNP A18 (Yang et al., 2006; Abdelmohsen et al., 2008a; Sawicka et al., 2008; Hägele et al., 2009); in specific instances, miRNAs can also promote translation (Vasudevan et al., 2007), but this does not appear to occur in response to hypoxia. RBPs can promote translation by competing with (or otherwise preventing the action of) RBPs or miRNAs that repress translation. Hypoxic conditions increase the levels or activity of numerous RBPs (e.g., HuR, PTB, nucleolin, hnRNP A18, CPEB), thereby ensuring the selective upregulation of proteins necessary for the hypoxic response, even though global translation is potently repressed by the shutting off of the general translational machinery (Wouters et al., 2005). At the same time, hypoxia can relieve the inhibitory action of some repressive RBPs, such as IRPs, hnRNP A2, and hnRNP L, a process named “post-transcriptional derepression,” and thus enhance translation of specific mRNAs.
The product of the gene ELAVL1 (embryonic lethal, abnormal vision, Drosophila-like 1), HuR is a 36-kDa RBP with three RNA-recognition motifs (RRMs), through which it binds to numerous mRNAs bearing U- or AU-rich sequences, typically present in their 3′-UTRs (López de Silanes et al., 2004; Hinman and Lou, 2008). Although HuR is predominantly nuclear, its influence on the post-transcriptional fate of target mRNAs is linked to its translocation to the cytoplasm, a process that is modulated by numerous transport factors (reviewed in Abdelmohsen et al., 2008a; Hinman and Lou, 2008). In the cytoplasm, HuR stabilizes many target mRNAs and can modulate the translation of several target mRNAs, enhancing the translation of many target mRNAs and repressing the translation of other target mRNAs (reviewed by Hinman and Lou, 2008; Srikantan and Gorospe, 2011). Through its influence on the patterns of expressed proteins, HuR can affect key cellular processes such as proliferation, differentiation, carcinogenesis, senescence, apoptosis, and the response to immune and environmental stresses (Wang et al., 2000, 2001; Gorospe, 2003; López de Silanes et al., 2003; Lal et al., 2005; Abdelmohsen et al., 2007a, b; Hinman and Lou, 2008; Srikantan and Gorospe, 2011).
In low oxygen, HuR potently elevates the expression of two major hypoxia-inducible proteins, vascular endothelial growth factor (VEGF) and HIF-1α, and likely regulates the levels and/or translation of other target mRNAs encoding hypoxia-inducible proteins (below). How hypoxia upregulates HuR function is unclear. Treatment with the hypoxia mimetic CoCl2 did not increase HuR cytoplasmic presence (Galbán et al., 2008), even though many other stress agents did trigger this elevation (Wang et al., 2000). Instead, HuR actions on hypoxia-regulated targets might be influenced by other post-translational modifications of HuR, including phosphorylation by kinases Chk2, PKC, or p38, which modulate HuR’s ability to bind target mRNAs (Abdelmohsen et al., 2007b; Doller et al., 2007, 2008; Lafarga et al., 2009); hypoxia activates Chk2 and PKC (Sahai et al., 1997; Xu et al., 2004; Gibson et al., 2005), but it remains to be studied if these kinases modulate HuR activity during hypoxia.
The association of HuR with the HIF-1α 5′UTR promoted HIF-1α translation in human cervical carcinoma cells (Galbán et al., 2008). In normoxia, HuR overexpression increased the levels of HIF-1α mRNA in actively translating polysomes and the de novo HIF-1α translation. In cells treated with CoCl2, HuR binding to the HIF-1α 5′UTR increased dramatically and HuR was necessary for the induction of HIF-1α by CoCl2 treatment. Although the HIF-1α 5′UTR has an IRES that enhances the constitutive translation of HIF-1α, this IRES did not appear to enhance HIF-1α translation after hypoxia (Galbán et al., 2008), nor did it seem to be affected by the presence of HuR or PTB (below). HuR binding to HIF-1α 3′UTR elevated HIF-1α mRNA levels in response to androgens (Sheflin et al., 2004). Although HIF-1α abundance is strongly increased by translational upregulation, the stabilization of HIF-1α protein is the best-understood mechanism to raise HIF-1α levels. In normoxia, HIF-1α is highly labile (with a half-life of <10 min) through the hydroxylation of conserved prolyl and asparagyl residues that are targeted for decay by the von Hippel–Lindau protein (pVHL), the recognition factor of an E3 ubiquitin ligase complex (Kim and Kaelin, 2004; Schofield and Ratcliffe, 2004). During hypoxia, these hydroxylases are quickly inhibited, resulting in the rapid and robust stabilization of HIF-1α (Schofield and Ratcliffe, 2004). Interestingly, pVHL levels are also regulated by HuR, which binds to the VHL mRNA, stabilizes it, and increases pVHL protein levels (Abdelmohsen et al., 2008b, 2009; Danilin et al., 2009). As hypoxia increases pVHL levels (Turcotte et al., 2004), it is plausible that HuR contributes to elevating pVHL under such conditions.
A key factor in neoangiogenesis, VEGF induces the proliferation of vascular endothelial cells and critically enhances tumor growth (Yancopoulos et al., 2000). The VEGF mRNA is highly unstable in normoxia, but its half-life increases rapidly in response to cytokines and stress conditions such as hypoxia (Levy et al., 1998; Pagès et al., 2000). In hypoxia, VEGF mRNA is transcriptionally upregulated by HIF-1 and related factors (Ema et al., 1997), but it is also potently upregulated at the post-transcriptional level, via the interaction of HuR with the VEGF 3′UTR that stabilizes the VEGF mRNA (Levy et al., 1998).
Numerous other hypoxia-inducible proteins are also encoded by HuR target mRNAs, although HuR’s role in their hypoxia-triggered increase has not been reported as-yet. The abundance of GLUT1 (glucose transporter 1, a protein critical for glucose uptake) increases by hypoglycemia and hypoxia, thereby ensuring the availability of intracellular glucose at times of low energy levels (Ouiddir et al., 1999; Qi and Pekala, 1999). While HIF-1 increases the expression of GLUT1 transcriptionally (Chen et al., 2001b), HuR binding to the GLUT1 mRNA enhances its translation (Gantt et al., 2006), although it is not yet known if HuR promotes GLUT1 expression during hypoxia. Similarly, hypoxia also increases the levels of HIF-2α, tissue growth factor (TGF)-β, c-Myc, and p53; although HuR interacts with these mRNAs (Mazan-Mamczarz et al., 2003; Abdelmohsen et al., 2008a; Srikantan and Gorospe, 2011), its role in enhancing the levels of the encoded proteins during hypoxia has not been investigated. Collectively, these proteins influence processes such as cell survival, cell proliferation, carcinogenesis, and differentiation.
Through its influence on target mRNAs, HuR could be envisioned to control hypoxic gene expression patterns in two stages. Immediately after hypoxia, HuR helps to elevate expression of HIF-1α and HIF-2α, which in turn increase the transcription of hypoxia-inducible genes (e.g., VEGF, TGF-β, p53, c-myc, GLUT1). During this time, HuR could further assist in upregulating these genes by binding to the corresponding mRNAs and enhancing their stability and/or translation. At later times, however, HuR likely helps to “shut off” hypoxic gene expression by enhancing the levels of pVHL, which targets HIF-1α for degradation, thereby helping to restore the levels of hypoxia-inducible proteins to basal conditions.
A 57-kDa protein also known as hnRNP I, PTB contains four RRMs through which it binds pyrimidine-rich sequences, preferentially those contained within longer pyrimidine tracts with cytosines (Auweter et al., 2007). PTB can bind to distant sites on the same RNA molecule and thereby it can introduce large RNA loops; such conformational changes are believed to be critical for PTB to recruit ribosomes onto IRESs to initiate translation and for modulating the interaction of factors required for splicing (Sawicka et al., 2008). PTB can also affect polyadenylation, cap-dependent translation, and mRNA stability (Sawicka et al., 2008). In response to hypoxia, PTB promotes HIF-1α mRNA translation and stabilizes the VEGF and insulin mRNAs, as described below.
PTB was shown to interact with the HIF-1α 5′UTR polypyrimidine tract and promoted HIF-1α translation in human embryonic kidney cells. Although PTB was proposed to function by promoting the formation of an IRES that facilitated HIF-1α translation during hypoxia (Schepens et al., 2005), the HIF-1α IRES did not mediate the hypoxia-triggered increase in HIF-1α translation (Galbán et al., 2008). In human cervical carcinoma cells, PTB also associated with the endogenous HIF-1α mRNA, primarily at the HIF-1α 3′UTR, and this interaction strongly increased HIF-1α translation after hypoxia or CoCl2 treatment (Galbán et al., 2008). The PTB-triggered increase in HIF-1α translation was strongly dependent on the presence of HuR, as HuR silencing severely reduced both the binding of PTB to HIF-1α mRNA after CoCl2 treatment and the PTB-stimulated translation of HIF-1α. Conversely, lowering PTB reduced both HuR binding to the HIF-1α 3′UTR and HIF-1α translation. This evidence supports a model whereby PTB and HuR, likely binding on multiple sites of the HIF-1α mRNA, together promote HIF-1α translation (Sheflin et al., 2004; Schepens et al., 2005; Galbán et al., 2008).
PTB formed a complex with the cold shock domain (CSD) protein and the PTB/CSD complex promoted the stability of numerous mRNAs, including VEGF mRNA in both normoxa and hypoxia (Coles et al., 2004). Since CSD also regulates VEGF transcription, the PTB/CSD complex was proposed to regulate in tandem the transcription and stabilization of target mRNAs (Coles et al., 2004). In addition, the VEGF 5′UTR contains two IRES (IRES A and IRES B) which contribute to enhancing VEGF translation after hypoxia; PTB associated with IRES B, but this interaction did not appear to impact upon VEGF translation (Huez et al., 1998).
The insulin mRNA is highly abundant in pancreatic β cells, where it constitutes ∼30% of the cellular mRNA (Tillmar et al., 2002). The insulin mRNA is highly stable, with a half-life of >24 h in resting conditions and an even longer half-life in the presence of high glucose levels (Welsh et al., 1985; Tillmar et al., 2002). The 3′UTR of insulin II mRNA was critical for the β-cell-specific glucose-regulated expression (Wicksteed et al., 2001). PTB mRNA levels increased markedly in pancreatic β cells cultured in high glucose; through binding to the rat insulin 3′UTR polypyrimidine-rich sequence (ins-PRS), PTB was proposed to increase insulin mRNA stability (Tillmar and Welsh, 2002; Tillmar et al., 2002). This interaction was inhibited by rapamycin, suggesting that signaling through mTOR (mammalian target of rapamycin) controlled this process (Tillmar and Welsh, 2004). Hypoxia also elevated insulin mRNA levels in a manner that was independent of rapamycin but dependent on the increased binding of PTB to the rat ins-PRS (Tillmar and Welsh, 2004; Fred and Welsh, 2005). Other effects of PTB, such as its ability to promote polyadenylation (Sawicka et al., 2008; Millevoi et al., 2009), may also contribute to the increased stability of the insulin mRNA.
The tristetraprolin family consists of three proteins with characteristic tandem zinc-finger domains, including TTP (also known as zinc-finger protein 36, ZFP36), ZFP36L1 or TIS11B (tetradecanoylphorbol acetate-inducible sequence 11B), and ZFP36L2 or TIS11D. TTP is inducible by different stimuli, including hypoxia, is predominantly cytoplasmic, and promotes the decay of target mRNAs in a variety of systems (Carballo et al., 1998; Kim et al., 2010). TTP was found to interact with the HIF-1α 3′UTR and promoted the degradation of HIF-1α mRNA (Kim et al., 2010). The generally reduced levels of TTP in many cancers was further proposed to contribute to the increased HIF-1α levels seen in many cancers (Brennan et al., 2009; Kim et al., 2010). In macrophages, the joint presence of hypoxia and lipopolysaccharide (LPS) led to decreased tumor necrosis factor (TNF)-α mRNA stability and TNF-α production, as well as to the reduced half-life and steady-state levels of mRNAs encoding other proinflammatory cytokines [macrophage inflammatory protein 2 (MIP2), interleukin (IL)-6, and granulocyte macrophage colony-stimulating factor (GM-CSF)]. This reduction was due, at least in part, to the decreased activity of p38, a kinase that phosphorylates and thereby inactivates TTP in hypoxic, LPS-treated cells (Werno et al., 2010). Hypoxia also increased the levels of MKP-3 [the mitogen-activated protein (MAP) kinase phosphatase-3], a dual-specificity phosphatase for MAP kinases ERK1/2. As TTP associated with the MKP-3 3′UTR and lowered the stability of an MKP-3 3′UTR reporter transcript, MKP-3 upregulation was attributed to the reduced decay-promoting influence of TTP upon MKP-3 mRNA during hypoxia (Bermudez et al., 2011).
The TTP-related protein TIS11B was found to interact with the 3′UTR of the VEGF mRNA and lowered VEGF mRNA stability (Ciais et al., 2004). Very recently, the levels of TIS11B were shown to be controlled by pVHL. In normoxic renal cell carcinoma (RCC) cells ectopically expressing pVHL, increased abundance of the microRNA hsa-miR-29b was responsible for decreasing TIS11B mRNA stability and TIS11B protein levels (Sinha et al., 2009). Hypoxia also increased TIS11B expression in pVHL-expressing RCC cells, although the specific mechanisms were not identified.
Iron-regulatory proteins (IRPs) interact with iron-responsive elements (IREs), specialized stem-loop structures present on mRNAs that are post-transcriptionally regulated in response to changes in iron concentration (Leipuviene and Theil, 2007). IREs present in the 3′UTR can alter mRNA stability, as shown for the transferrin receptor (TfR) and divalent metal transporter 1 (DMT1) mRNAs, while 5′UTR IREs block translation in a cap- and IRES-independent manner, as shown for ferritin (Ft) and mitochondrial aconitase (m-Aco) mRNAs (Schneider and Leibold, 2003; Leipuviene and Theil, 2007). IREs were identified in the 5′UTRs of HIF-2α and HIF-1α, and the association of IRP1 and IRP2 with the HIF-2α IRE repressed HIF-2α translation (Sanchez et al., 2007; Zimmer et al., 2008). Although exposure to hypoxia increased HIF-2α expression and function, in conditions of iron deficiency the interaction of IRP2 with the HIF-2α IRE led to strong repression of HIF-2α translation (Sanchez et al., 2007). Hypoxia increased HIF-2α expression by a process of translational derepression, elicited by relieving the interaction of IRP1 with IRE. Several drug compounds were identified which strengthened IRP/IRE interactions, thereby blocking the hypoxia-triggered increase in HIF-2α translation and HIF-2α levels (Zimmer et al., 2008). Whether IRP/IRE interactions through the HIF-1α 5′UTR (Leipuviene and Theil, 2007; Zimmer et al., 2008) are similarly implicated in the regulation of HIF-1α expression by hypoxia has not been reported.
The 100-kDa RBP nucleolin has four RRMs through which it modulates gene expression during cell proliferation, carcinogenesis, and viral infection. Exposure of human fibrosarcoma cells to an iron chelator (2,2-dipyridyl), a treatment that recapitulates some aspects of hypoxia, enhanced the translation of matrix metalloprotease-9 (MMP-9) without altering MMP-9 mRNA levels. This increase was attributed to nucleolin’s increased association with MMP-9 3′UTR (Fähling et al., 2005). In the same cell system, long-term hypoxia increased the association of a 64-kDa nucleolin cleavage product with the 5′UTR and 3′UTR of the collagen prolyl 4-hydroxylase-alpha(I) (C-P4H) mRNA (Fähling et al., 2006). Through these interactions, the 64-kDa nucleolin enhanced translation of the C-P4H mRNA and elevated C-P4H abundance during hypoxia. Under conditions of stress, nucleolin (together with the ribosomal protein RPL26) associated with the 5′UTR of the mRNA encoding the tumor suppressor p53 and promoted its translation (Takagi et al., 2005), although it has not been shown if hypoxia employs a similar mechanism of p53 upregulation.
The cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 affect translation by changing the length of the 3′ poly(A) tail of target mRNAs. CPEB1 and CPEB2 associated with the HIF-1α 3′UTR and repressed HIF-1α translation in untreated neuroblastoma cells (Hägele et al., 2009). In response to insulin treatment, however, both CPEBs enhanced the hypoxia increased HIF-1α levels by elevating HIF-1α translation via its 3′UTR, as shown using a heterologous reporter RNA. The increase in HIF-1α translation by insulin was accompanied by an elongation of the HIF-1α mRNA poly(A) tail (Hägele et al., 2009).
The levels of the RBP T-cell intracellular antigen-1 (TIA-1)-related protein (TIAR) were potently upregulated in a rat model of brain ischemia, associated with deprivation of oxygen, glucose, and serum (Jin et al., 2000); conversely, TIAR levels were reduced by anti-apoptotic drugs in a model of retinal ischemia (Osborne et al., 1998). These studies did not identify specific TIAR target mRNAs but they linked TIAR to the cell death caused by hypoxia and nutrient deprivation. TIA-1 and TIAR can aggregate within stress granules (SGs) under specific stress conditions, including hypoxia, and moreover HIF-1α expression was blocked in cells displaying TIAR/TIA-1 SGs (Gottschald et al., 2010). Under potent hypoxia, TIAR and TIA-1 inhibited HIF-1α expression, an effect that was linked to an AU-rich element in the HIF-1α 3′UTR that was the target of translational repression by TIAR. Other RBPs implicated in mRNA turnover and translation linked to SG metabolism (e.g., G3BP, HuR, CPEB, FMRP, FXR1, PMR1, TTP, BRF1, YB-1, ZBP, reviewed by Anderson and Kedersha, 2008) may also be involved in controlling gene expression in response to hypoxia.
Several hnRNPs have been implicated in the post-transcriptional regulation of gene expression during hypoxia. HnRNP A18 associated with the thioredoxin (TRX) mRNA 3′UTR and enhanced its translation (Yang et al., 2006); formation of this complex was induced via phosphorylation of hnRNP A18 by the hypoxia-activated kinase GSK3β (glycogen synthase kinase 3β). HnRNP A18 was proposed to enhance translation by interacting with eIF4G, a component of the basic translational machinery that interacts with the 5′ end of the mRNA, helping to form a loop between the 3′ and 5′ ends of the mRNA, and thereby translated thioredoxin more efficiently (Yang et al., 2006). It is unknown at present whether other hnRNP A18 target mRNAs, many of which encode stress-response proteins (Yang and Carrier, 2001), are similarly induced by hnRNP A18 during hypoxia.
Both hnRNP A2 and hnRNP L associated with the AU-rich elements in the GLUT1 3′UTR in numerous cell types and repressed GLUT1 mRNA stability and translation (Hamilton et al., 1999). Hypoxia enhanced GLUT1 expression by selectively decreasing the interaction of GLUT1 mRNA with hnRNP A2 and hnRNP L, thus relieving the repressive influence of these hnRNPs (Hamilton et al., 1999).
Recently, VEGF expression was found to be repressed by microRNAs miR-297 and miR-299 acting through a 3′UTR CA-rich element (CARE) in the VEGF mRNA. Hypoxia promoted the export of hnRNP L to the cytoplasm, where it interacted with the VEGF CARE, competing with miR-297/miR-299 and preventing microRNA-mediated repression (Jafarifar et al., 2011).
The levels of the erythropoietin (EPO) mRNA-binding protein (ERBP) increased in response to hypoxia and its binding activity was modulated by sulfhydryl groups (Rondon et al., 1995). ERBP stabilized the EPO mRNA in normoxic cells, but had a more potent stabilizing influence under low oxygen conditions (McGary et al., 1997).
Hypoxia increased the expression levels of two highly homologous cold stress proteins, RNA-binding motif protein 3 (RBM3), and cold-inducible RNA-binding protein (CIRP; Wellmann et al., 2004). These glycine-rich RBPs have been implicated in modulating global protein translation and function as RNA chaperones that facilitate translation upon the perception of cold stress (Fujita, 1999; Smart et al., 2007). RBM3 and CIRP also interacted with the 3′UTR of specific target mRNAs [e.g., those encoding the replication protein A (RPA2), TRX, and cyclooxygenase (COX)2] likely promoting their translation, although this effect was not studied in hypoxia (Lleonart, 2010).
The enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was shown to associate with AU-rich elements in lymphokine mRNAs (Nagy and Rigby, 1995). In conditions of hypoxia, GAPDH was found to interact with the 3′UTR of CCN2/connective tissue growth factor (CTGF) mRNA and promoted expression of CCN2, an inducer of tumor angiogenesis, in human chondrosarcoma-derived cells (Kondo et al., 2011).
Table 1 lists the collection of RBPs implicated in modulating the stability and translation of hypoxia-response genes.
MicroRNAs provide an important, newly recognized layer of complexity to the post-transcriptional regulation of mammalian gene expression. They exert their actions via pairing of a “seed” region to partially complementary sites generally within the 3′UTR of the target mRNA. The outcome of this interaction is most frequently post-transcriptional repression via mRNA degradation or/and decreased translational efficiency (Bartel, 2009; Fabian et al., 2010). The relative contribution of these two mechanisms varies for individual [microRNA-target mRNA] pairs.
Consistent with their demonstrated involvement in most physiological processes, microRNAs have been proposed to fine-tune the cell’s adaption to low oxygen tension. During the past 5 years, multiple groups have reported specific alterations in microRNA profiles upon hypoxic stress (Hua et al., 2006; Donker et al., 2007; Kulshreshtha et al., 2007a; Camps et al., 2008; Crosby et al., 2009a). Collectively, these studies have identified over 50 microRNAs that were either upregulated or downregulated in low oxygen; however most of these seem to be cell type-specific. Kulshreshtha et al. (2007a) identified the first group of hypoxia-inducible microRNAs, which included miR-210, miR-93, miR-30b, miR-181b, miR-181c, miR-192, miR-195, and miR-21. Many subsequent studies confirmed miR-210 as a microRNA widely implicated in the hypoxic response, and yet others identified miR-373, miR-498, mimR-181b, and miR-30b (Kulshreshtha et al., 2007b; Crosby et al., 2009a,b; Fasanaro et al., 2010). However, some microRNA expression patterns in response to hypoxia vary among systems. For example, some members of the let-7 family were induced in squamous cell carcinoma, but downregulated in nasopharyngeal carcinoma cells (Hua et al., 2006). The limited consistency between microRNA expression profiles is likely due to a combination of factors, including differences in detection technology and thresholds, cellular context, and depth and duration of hypoxia. Below we discuss the involvement of several prominent hypoxia-regulated microRNAs in post-transcriptional gene regulation under low oxygen (Table 2).
In recent years, a number of studies have identified miR-210 as a prototypical “hypoxia microRNA” (Hua et al., 2006; Kulshreshtha et al., 2007a; Camps et al., 2008; Fasanaro et al., 2008; Giannakakis et al., 2008; Pulkkinen et al., 2008; Zhang et al., 2009). miR-210 induction in hypoxia is widely accepted as a hallmark HIF-mediated response. The recruitment of HIF-1α to the promoter of the miR-210 gene has been extensively validated by multiple approaches, including ChIP (chromatin immunoprecipitation), HIF-1α knockdown and HIF-1α overexpression experiments (Kulshreshtha et al., 2007a; Camps et al., 2008; Crosby et al., 2009a). Together with bioinformatic analyses of clinical samples confirming strong associations between the expression of miR-210 and that of conventional HIF-1α targets (Camps et al., 2008), the data collectively support the notion that HIF-1α promotes miR-210 upregulation during hypoxia.
Searches for bona fide miR-210 targets have been undertaken using a variety of prediction programs [PicTar, TargetScan, DianaLab, Microcosm], together with integrative mRNA and microRNAs profiling approaches. Using a novel strategy based on the immunoprecipitation of RISC, in combination with microarrays and proteomics, Fasanaro et al. (2009) identified a diverse spectrum of potential miR-210 targets, including genes involved in proliferation, apoptosis, DNA repair, chromatin remodeling, angiogenesis, and metabolism.
A key feature of adaptation to low oxygen is the shift in energy metabolism from mitochondrial oxidative phosphorylation to glycolysis, a shift that is classically governed by HIF activation. Several reports independently demonstrated that miR-210 contributes to this metabolic response by reducing the levels of a key target protein, the evolutionarily conserved Iron–Sulfur Cluster Scaffold Homolog (ISCU; Chan et al., 2009; Fasanaro et al., 2009; Chen et al., 2010; Favaro et al., 2010). ISCU facilitates the assembly of Fe-S clusters that are incorporated into enzymes involved in energy production, including mitochondrial complex 1 and aconitase (Tong and Rouault, 2006). The miR-210-elicited downregulation of ISCU in response to hypoxia leads to decreased mitochondrial electron transport activity and increased reliance on glycolysis. Additional studies on miR-210 suggest that this microRNA orchestrates an “assault” on mitochondria during hypoxia, repressing aerobic respiration components such as NDUFA4 (NADH dehydrogenase ubiquinone 1 α subcomplex 4), SDHD (succinate dehydrogenase complex subunit D), and COX10 (Giannakakis et al., 2008; Chen et al., 2010; Puisségur et al., 2011). Expanding on these findings, two feed-forward loops between miR-210 and HIF-1α have been proposed. In the first, Puisségur et al. (2011) provided evidence that repression of SDHD in hypoxia by miR-210 contributes to the maintenance of HIF-1α activity in hypoxia. In the second, Kelly et al. (2011) showed that hypoxia-induced miR-210 targets the GPD1L mRNA, which encodes glycerol-3-phosphate dehydrogenase 1-like, a newly identified inhibitor of HIF-1α, in turn stabilizing HIF-1α protein.
The network of miR-210 effects was recently expanded to hypoxia-driven angiogenesis. Blockade of miR-210 in human endothelial cells inhibited the formation of capillary-like structures; the underlying mechanism proposed was the miR-210-mediated repression of ephrin A3 (EFNA3), a negative modulator of VEGF-dependent endothelial cell migration and tubulogenesis (Fasanaro et al., 2008; Pulkkinen et al., 2008). The influence of miR-210 was also expanded to include another feature of hypoxic cells, defective DNA repair, particularly downregulation of homology-dependent DNA repair (HDR) genes (Bindra et al., 2004). Inhibition of HDR during hypoxia likely involves miR-210-lowered levels of RAD52 (Crosby et al., 2009a; Fasanaro et al., 2009), a protein that assists with the loading of RAD51 onto DNA (Benson et al., 1998). However, it is unclear whether induction of miR-210 is sufficient to promote DNA damage or genetic instability in hypoxic conditions.
While a lack of miR-210 compromises cell survival in response to severe hypoxia (Kulshreshtha et al., 2007b; Fasanaro et al., 2008; Favaro et al., 2010), it is still unclear if this occurs primarily as a result of direct targeting of apoptotic pathways. The only apoptotic target gene identified to date is CASP8AP2, which is highly similar to FLASH, a mouse apoptotic protein that interacts with the death-effector domain of Caspase 8 to promote cell death. In mesenchymal stem cells following ischemic preconditioning, repression of this pathway by miR-210 appears to be critical for the maintenance of cell viability (Kim et al., 2009). Whether the protective role of miR-210 during hypoxia in cancer cells is mainly the consequence of the metabolic shift described above, the regulation of CASP8AP2 expression, or the impact of miR-210 on multiple pathways remains an open question.
Finally, miR-210 has also been proposed to act as regulator of the cell division cycle, but this effect appears to be context-dependent. In ovarian cancers, contrary to other solid tumors, miR-210 is often underexpressed as a result of genomic deletions, leading to increased expression of the transcription factor E2F3, which in turn accelerates cell cycle progression (Giannakakis et al., 2008). This paradigm seems to mirror the results with xenografts overexpressing miR-210, which grow slower than control xenografts (Huang et al., 2009). Another cell cycle effect was proposed by Zhang et al. (2009) who found that miR-210 overexpression in cancer cell lines bypasses hypoxia-induced cell cycle arrest by downregulating MNT, a c-MYC antagonist. The relative contribution of these mechanisms to cell cycle alterations in the hypoxic tumor is unknown at present.
The induction of miR-373 by hypoxia has been reported in HeLa, MCF-7, and squamous cell carcinoma cell lines (Hebert et al., 2007; Crosby et al., 2009a). Similarly to miR-210, the increase in miR-373 levels by hypoxia appears to be HIF-1α-dependent, with at least one functional HRE identified in its promoter. Consistently, HIF-1α knockdown by shRNA blocks miR-373 induction in hypoxic cells (Crosby et al., 2009a).
Given that miR-373 expression has been correlated with increased proliferation and tumorigenesis, a mechanistic understanding of its function and targets is highly relevant for cancer biology (Voorhoeve et al., 2006). MiR-373 was found to directly transactivate E-cadherin gene expression through pairing with complementary promoter sequences. This potentially novel mechanism of action may be directly relevant to invasion and metastasis, two processes that are known to be promoted by tumor hypoxia (Berx and Van Roy, 2001; Place et al., 2008). Recent work demonstrated that forced expression of miR-373 leads to a reduction in the RAD23B nucleotide excision repair protein, as well as in RAD52 (Crosby et al., 2009a). As with miR-210, it is not currently known whether induction of miR-373 is sufficient to promote genotoxic damage during hypoxia.
Hypoxia-mediated induction of miR-21 has been reported in several cancer cell lines, in pulmonary artery smooth muscle (PDAC), and in human vein endothelial cells (HUVEC; Kulshreshtha et al., 2007a; Pulkkinen et al., 2008; Sarkar et al., 2010). In contrast, miR-21 levels decreased upon exposure of cardiac myocytes to hypoxia and remained unchanged in breast cancer cells (Camps et al., 2008; Sayed et al., 2010).
MiR-21 is arguably the most frequently upregulated onco-microRNA (oncomir) in human tumors (Volinia et al., 2006; Iorio et al., 2007; Lui et al., 2007; Tetzlaff et al., 2007; Tran et al., 2007). MiR-21 is believed to play a pro-survival role in deeply hypoxic/anoxic cancer microenvironment and in ischemic disorders, such as acute myocardial infarction, where miR-21 prevents hypoxia-induced cell apoptosis by suppressing expression of the programmed cell death protein 4 (PDCD4). It may also be required for cell viability during ischemia/reperfusion (Dong et al., 2009; Cheng et al., 2010). MiR-21 may also inhibit proliferation under hypoxia, as suggested by its inhibition of the expression of Cdc25A, a key cell cycle progression protein (de Oliveira et al., 2009). In human pulmonary artery smooth muscle cells (PASMC), elevated miR-21 participates in hypoxia-triggered proliferation and cell migration by reducing the expression levels of targets such as PDCD4, Sprouty 2 (SPRY2), and peroxisome proliferator-activated receptor-α (PPARα; Sarkar et al., 2010).
A role for HIF in directly upregulating miR-21 seems unlikely, and whether miR-21 can be regarded as a bona fide hypoxia-regulated transcript remains unclear. Among the factors that may elevate miR-21 expression in tumors and/or hypoxic cells, the leading candidate is AP-1, itself a hypoxia-response transcription factor (Fujita et al., 2008; Nanduri et al., 2008).
MicroRNAs miR-20b, miR-199a, and miR-424 are also relevant to the hypoxic response by affecting HIF-1α expression. miR-20b directly suppressed HIF-1α expression in MCF-7 breast cancer cells (Cascio et al., 2010). In contrast, miR-199a is acutely downregulated in cardiac myocytes in an AKT-dependent manner under low oxygen conditions; in turn, this downregulation allows a rapid increase of its target, HIF-1α mRNA (Rane et al., 2009, 2010). An indirect feed-back mechanism was identified for miR-424 in endothelial cells, where PU.1-upregulated miR-424 leads to a reduction in the expression of Cullin 2 (CUL2), a critical component of the ubiquitin ligase system that degrades HIF-1α (Ghosh et al., 2010) and thereby elevates HIF-1α. Collectively, the recent work on hypoxia-related microRNAs has added a new dimension to the complexity of the hypoxic response, with implications in metabolism, cell cycle control, apoptosis, and angiogenesis. The integration of several RBPs and microRNAs in the hypoxic response elicited by HIF-1α is presented in Figure 1.
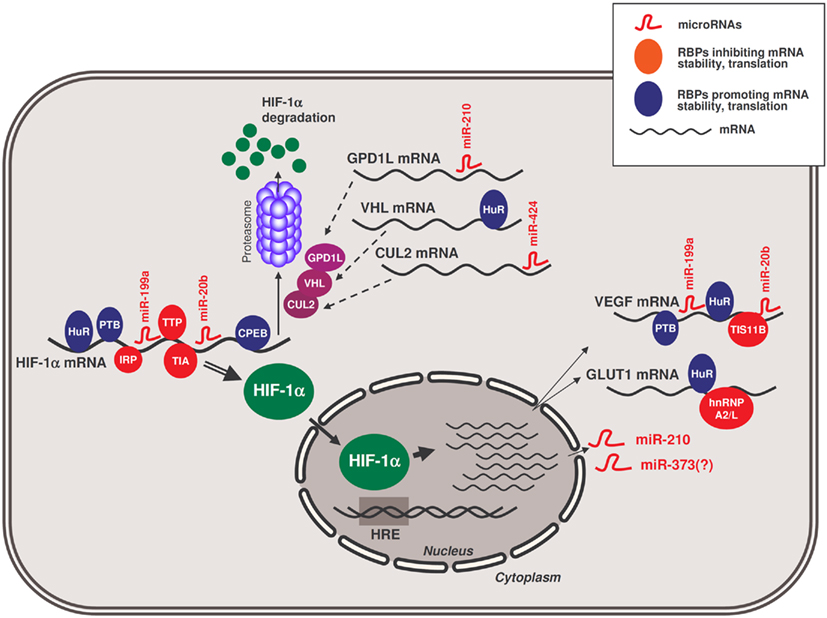
Figure 1. Schematic of RBPs and microRNAs influencing the hypoxic response by HIF-1α. RBPs and microRNAs that interact with HIF-1α mRNA and modulate the production of HIF-1α protein in hypoxia. In the nucleus, HIF-1α transcriptionally upregulates the expression of target genes bearing hypoxia-response elements (HREs) in their promoters, including those that encode GLUT1 and VEGF mRNAs and those that encode microRNAs like miR-210 and likely also miR-373. In turn, miR-210 can modulate the production of HIF-1α by repressing the production of GPD1L; other RBPs and other microRNAs assist in modulating the expression of additional HIF-1α stability regulators (e.g., VHL, CUL2), and other hypoxia-response proteins like GLUT1 and VEGF.
The complexity of the regulation of gene expression in response to low oxygen tension is striking. Many recent studies of gene regulation in hypoxia have uncovered diverse feed-back loops of repression and activation at the transcriptional and post-transcriptional levels. Transcriptional activation of hypoxia-inducible factors is critical for long-term adaptation, and its integration with post-transcriptional networks gives rise to a rich variety of modifications that are essential for responses by tissues with specific requirements.
The proteomic constellation of each cell is unique – it is determined by its state of differentiation and tissue environment. Cellular hypoxic adaptation and survival depends on the manner the cell senses oxygen levels and its ability to respond to lower ATP concentrations. The gene expression program and ensuing metabolic changes activated by oxygen deprivation are influenced by the assortment of pre-existing trans-acting factors, as well as by second messenger cascades activated by extracellular stimuli. Cells communicate by signaling molecules delivered to the cell in paracrine or endocrine fashion. Depending on the types of receptors expressed in the cell, different sets of signals can be activated and interfere with hypoxia-inducible second messenger cascades and transcriptional/post-transcriptional responses. The close relationship between oxygen and iron metabolism (Chepelev and Willmore, 2011) and the modulation of hypoxia responses by insulin or lipopolysaccharides are only some examples of the influence of stimuli upon the specific transcriptional and post-transcriptional responses to hypoxia. The data gathered so far suggest the existence of distinct cell type-specific usage of hypoxic gene regulatory sites located in the DNA (e.g., promoter, enhancer, silencer regions) and/or in the mRNA UTRs. In addition, RBPs and miRNAs could help to balance the expression pattern of hypoxia-inducible genes. For instance, the most extensively investigated hypoxia response, the HIF-activated gene expression system, is primarily triggered by changes in oxygen concentration. However, HIF activity is modulated by several other pathways and secondary cascades, including reactive oxygen species, AMPK, and the phosphatidylinositol 3-kinase (PI3K)/Akt pathway (Fähling, 2009b). The influence of these signaling systems upon the RBPs and microRNAs discussed in this review remains to be investigated in depth.
In closing, it should be noted that although several trans-binding factors have been identified that modulate the expression of hypoxia-response genes in cultured cells, particularly in cancer cells, their function under hypoxic conditions in tissues, organs, and organisms remains largely unknown. It is well known that tumors become hypoxic, that highly proliferating cells need more energy (and hence oxygen), and that inhibition of tumor vascularization is able to suppress tumor growth. However, compared to differentiated cells, cancer cells elicit markedly distinct adaptive responses to hypoxia, including different changes in gene expression, energy usage, proliferation, and apoptosis. In addition, hypoxia is also important during the adaptation to stress conditions and embryonic development (Dunwoodie, 2009). In light of the fact that hypoxia may elicit different responses depending on the species, sex, in vivo or in vitro conditions, oxygen tension, cellular origin and differentiation state, it is challenging to describe a universal pattern of adaptive gene expression in the hypoxic response. In summary, a great deal of knowledge has been gathered about the trans-acting RBPs and miRNAs that control hypoxic gene expression patterns. The stage is now ready to study how their actions are integrated within the context of specific hypoxic cell environments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Myriam Gorospe and Kumiko Tominaga were supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health. Mircea Ivan and Xue Wu are supported by an American Cancer Society Research Scholar Grant. Michael Fähling is supported by the Deutsche Forschungsgemeinschaft (DFG, grant FA845/2-2).
Abdelmohsen, K., Kuwano, Y., Kim, H. H., and Gorospe, M. (2008a). Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389, 243–255.
Abdelmohsen, K., Srikantan, S., Kuwano, Y., and Gorospe, M. (2008b). miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U.S.A. 105, 20297–20302.
Abdelmohsen, K., Lal, A., Kim, H. H., and Gorospe, M. (2007a). Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 6, 1288–1292.
Abdelmohsen, K., Pullmann, R. Jr., Lal, A., Kim, H. H., Galban, S., Yang, X., Blethrow, J., Walker, M., Shubert, J., Gillespie, D. A., Furneaux, H., and Gorospe, M. (2007b). Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25, 543–557.
Abdelmohsen, K., Srikantan, S., Yang, X., Lal, A., Kim, H. H., Kuwano, Y., Galban, S., Becker, K. G., Kamara, D., de Cabo, R., and Gorospe, M. (2009). Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28, 1271–1282.
Anderson, P., and Kedersha, N. (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008; 33, 141–150.
Auweter, S. D., Oberstrass, F. C., and Allain, F. H. T. (2007). Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J. Mol. Biol. 367, 174–186.
Baird, S. D., Lewis, S. M., Turcotte, M., and Holcik, M. (2007). A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 35, 4664–4677.
Benson, F. E., Baumann, P., and West, S. C. (1998). Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391, 401–404.
Bermudez, O., Jouandin, P., Rottier, J., Bourcier, C., Pagès, G., and Gimond, C. (2011). Post-transcriptional regulation of the DUSP6/MKP-3 phosphatase by MEK/ERK signaling and hypoxia. J. Cell Physiol. 226, 276–284.
Berx, G., and Van Roy, F. (2001). The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 3, 289–293.
Bindra, R. S., Schaffer, P. J., Meng, A., Woo, J., Måseide, K., Roth, M. E., Lizardi, P., Hedley, D. W., Bristow, R. G., and Glazer, P. M. (2004). Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell. Biol. 24, 8504–8518.
Bracken, C. P., Whitelaw, M. L., and Peet, D. J. (2003). The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell. Mol. Life Sci. 60, 1376–1393.
Brennan, S. E., Kuwano, Y., Alkharouf, N., Blackshear, P. J., Gorospe, M., and Wilson, G. M. (2009). The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 69, 5168–5176.
Camps, C., Buffa, F. M., Colella, S., Moore, J., Sotiriou, C., Sheldon, H., Harris, A. L., Gleadle, J. M., and Ragoussis, J. (2008). hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 14, 1340–1348.
Carballo, E., Lai, W. S., and Blackshear, P. J. (1998). Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281, 1001–1005.
Carrero, P., Okamoto, K., Coumailleau, P., O’Brien, S., Tanaka, H., and Poellinger, L. (2000). Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell. Biol. 20, 402–415.
Cascio, S., D’Andrea, A., Ferla, R., Surmacz, E., Gulotta, E., Amodeo, V., Bazan, V., Gebbia, N., and Russo, A. (2010). miR-20b modulates VEGF expression by targeting HIF-1α and STAT3 in MCF-7 breast cancer cells. J. Cell. Physiol. 224, 242–249.
Chan, S. Y., Zhang, Y. Y., Hemann, C., Mahoney, C. E., Zweier, J. L., and Loscalzo, J. (2009). MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 10, 273–284.
Chen, C. Y., Gherzi, R., Ong, S. E., Chan, E. L., Raijmakers, R., Pruijn, G. J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. (2001a). AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451–464.
Chen, C., Pore, N., Behrooz, A., Ismail-Beigi, F., and Maity, A. (2001b). Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J. Biol. Chem. 276, 9519–9525.
Chen, Z., Li, Y., Zhang, H., Huang, P., and Luthra, R. (2010). Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29, 4362–4368.
Cheng, Y., Zhu, P., Yang, J., Liu, X., Dong, S., Wang, X., Chun, B., Zhuang, J., and Zhang, C. (2010). Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc. Res. 87, 431–439.
Chepelev, N. L., and Willmore, W. G. (2011). Regulation of iron pathways in response to hypoxia. Free Radic. Biol. Med. 50, 645–666.
Ciais, D., Cherradi, N., Bailly, S., Grenier, E., Berra, E., Pouyssegur, J., Lamarre, J., and Feige, J. J. (2004). Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene 23, 8673–8680.
Coles, L. C., Bartley, M. A., Bert, A., Hunter, J., Polyak, S., Diamond, P., Vadas, M. A., and Goodall, G. J. (2004). A multi-protein complex containing cold shock domain (Y-box) and polypyrimidine tract binding proteins forms on the VEGF mRNA. Eur. J. Biochem. 271, 648–660.
Cougot, N., Babajko, S., and Seraphin, B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165, 31–40.
Crosby, M. E., Kulshreshtha, R., Ivan, M., and Glazer, P. M. (2009a). MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 69, 1221–1229.
Crosby, M. E., Glazer, P. M., and Ivan, M. (2009b). “Micro”-management of DNA repair genes by hypoxia. Cell Cycle 8, 4009–4010.
Danilin, S., Sourbier, C., Thomas, L., Rothhut, S., Lindner, V., Helwig, J. J., Jacqmin, D., Lang, H., and Massfelder, T. (2009). von Hippel-Lindau tumor suppressor gene-dependent mRNA stabilization of the survival factor parathyroid hormone-related protein in human renal cell carcinoma by the RNA-binding protein HuR. Carcinogenesis 30, 387–396.
de Oliveira, P. E., Zhang, L., Wang, Z., and Lazo, J. S. (2009). Hypoxia-mediated regulation of Cdc25A phosphatase by p21 and miR-21. Cell Cycle 8, 3157–3164.
Doller, A., Akool, el-S., Huwiler, A., Müller, R., Radeke, H. H., Pfeilschifter, J., and Eberhardt, W. (2008). Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell. Biol. 28, 2608–2625.
Doller, A., Huwiler, A., Müller, R., Radeke, H. H., Pfeilschifter, J., and Eberhardt, W. (2007). Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell 18, 2137–2148.
Dong, S., Cheng, Y., Yang, J., Li, J., Liu, X., Wang, X., Wang, D., Krall, T. J., Delphin, E. S., and Zhang, C. (2009). MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 284, 29514–29525.
Donker, R. B., Mouillet, J. F., Nelson, D. M., and Sadovsky, Y. (2007). The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol. Hum. Reprod. 13, 273–279.
Dunwoodie, S. L. (2009). The role of hypoxia in development of the Mammalian embryo. Dev. Cell 17, 755–773.
Ema, M., Taya, S., Yokotani, N., Sogawa, K., Matsuda, Y., and Fujii-Kuriyama, Y. (1997). A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1 regulates VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. U.S.A. 94, 4273–4278.
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379.
Fähling, M. (2009a). Surviving hypoxia by modulation of mRNA translation rate. J. Cell. Mol. Med. 13, 2770–2779.
Fähling, M. (2009b). Cellular oxygen sensing, signalling and how to survive translational arrest in hypoxia. Acta Physiol. (Oxf.) 195, 205–230.
Fähling, M., Mrowka, R., Steege, A., Nebrich, G., Perlewitz, A., Persson, P. B., and Thiele, B. J. (2006). Translational control of collagen prolyl 4-hydroxylase-(I) gene expression under hypoxia. J. Biol. Chem. 281, 26089–26101.
Fähling, M., Steege, A., Perlewitz, A., Nafz, B., Mrowka, R., Persson, P. B., and Thiele, B. J. (2005). Role of nucleolin in posttranscriptional control of MMP-9 expression. Biochim. Biophys. Acta 1731, 32–40.
Fasanaro, P., D’Alessandra, Y., Di Stefano, V., Melchionna, R., Romani, S., Pompilio, G., Capogrossi, M. C., and Martelli, F. (2008). MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 283, 15878–15883.
Fasanaro, P., Greco, S., Ivan, M., Capogrossi, M. C., and Martelli, F. (2010). microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol. Ther. 125, 92–104.
Fasanaro, P., Greco, S., Lorenzi, M., Pescatori, M., Brioschi, M., Kulshreshtha, R., Banfi, C., Stubbs, A., Calin, G. A., Ivan, M., Capogrossi, M. C., and Martelli, F. (2009). An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 2009. 284, 35134–35143.
Favaro, E., Ramachandran, A., McCormick, R., Gee, H., Blancher, C., Crosby, M., Devlin, C., Blick, C., Buffa, F., Li, J. L., Vojnovic, B., Pires das Neves, R., Glazer, P., Iborra, F., Ivan, M., Ragoussis, J., and Harris, A. L. (2010). MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE 5, e10345. doi: 10.1371/journal.pone.0010345
Fred, R. G., and Welsh, N. (2005). Increased expression of polypyrimidine tract binding protein results in higher insulin mRNA levels. Biochem. Biophys. Res. Commun. 328, 38–42.
Fujita, J. (1999). Cold shock response in mammalian cells. J. Mol. Microbiol. Biotechnol. 1, 243–255.
Fujita, S., Ito, T., Mizutani, T., Minoguchi, S., Yamamichi, N., Sakurai, K., and Iba, H. (2008). miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 378, 492–504.
Galbán, S., Kuwano, Y., Pullmann, R. Jr., Martindale, J. L., Kim, H. H., Lal, A., Abdelmohsen, K., Yang, X., Dang, Y., Liu, J. O., Lewis, S. M., Holcik, M., and Gorospe, M. (2008). RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol. Cell. Biol. 28, 93–107.
Gantt, K. R., Cherry, J., Richardson, M., Karschner, V., Atasoy, U., and Pekala, P. H. (2006). The regulation of glucose transporter (GLUT1) expression by the RNA binding protein HuR. J. Cell. Biochem. 99, 565–574.
Gardner, L. B., and Corn, P. G. (2008). Hypoxic regulation of mRNA expression. Cell Cycle 7, 1916–1924.
Gherzi, R., Lee, K. Y., Briata, P., Wegmüller, D., Moroni, C., Karin, M., and Chen, C. Y. (2004). A KH-domain RNA-binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14, 571–583.
Ghosh, G., Subramanian, I. V., Adhikari, N., Zhang, X., Joshi, H. P., Basi, D., Chandrashekhar, Y. S., Hall, J. L., Roy, S., Zeng, Y., and Ramakrishnan, S. (2010). Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J. Clin. Invest. 120, 4141–4154.
Giannakakis, A., Sandaltzopoulos, R., Greshock, J., Liang, S., Huang, J., Hasegawa, K., Li, C., O’Brien-Jenkins, A., Katsaros, D., Weber, B. L., Simon, C., Coukos, G., and Zhang, L. (2008). miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 7, 255–264.
Gibson, S. L., Bindra, R. S., and Glazer, P. M. (2005). Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res. 65, 10734–10741.
Gilbert, W. V., Zhou, K., Butler, T. K., and Doudna, J. A. (2007). Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317, 1224–1227.
Gorospe, M. (2003). HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle 2, 412–414.
Gottschald, O. R., Malec, V., Krasteva, G., Hasan, D., Kamlah, F., Herold, S., Rose, F., Seeger, W., and Hänze, J. (2010). TIAR and TIA-1 mRNA-binding proteins co-aggregate under conditions of rapid oxygen decline and extreme hypoxia and suppress the HIF-1α pathway. J. Mol. Cell Biol. 2, 345–356.
Hägele, S., Kühn, U., Böning, M., and Katschinski, D. M. (2009). Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1α mRNA 3′-UTR and modulate HIF-1α protein expression. Biochem. J. 417, 235–246.
Hamilton, B. J., Nichols, R. C., Tsukamoto, H., Boado, R. J., Pardridge, W. M., and Rigby, W. F. (1999). hnRNP A2 and hnRNP L bind the 3′UTR of glucose transporter 1 mRNA and exist as a complex in vivo. Biochem. Biophys. Res. Commun. 261, 646–651.
Hebert, C., Norris, K., Scheper, M. A., Nikitakis, N., and Sauk, J. J. (2007). High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer 6, 5.
Hinman, M. N., and Lou, H. (2008). Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65, 3168–3181.
Hua, Z., Lv, Q., Ye, W., Wong, C. K., Cai, G., Gu, D., Ji, Y., Zhao, C., Wang, J., Yang, B. B., and Zhang, Y. (2006). MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE 1, e116. doi: 10.1371/journal.pone.0000116
Huang, X., Ding, L., Bennewith, K. L., Tong, R. T., Welford, S. M., Ang, K. K., Story, M., Le, Q. T., and Giaccia, A. J. (2009). Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell 35, 856–867.
Huez, I., Créancier, L., Audigier, S., Gensac, M. C., Prats, A. C., and Prats, H. (1998). Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 18, 6178–6190.
Ignacak, M. L., Harbaugh, S. V., Dayyat, E., Row, B. W., Gozal, D., and Czyzyk-Krzeska, M. F. (2009). Intermittent hypoxia regulates RNA polymerase II in hippocampus and prefrontal cortex. Neuroscience 158, 1436–1445.
Iorio, M. V., Visone, R., Di Leva, G., Donati, V., Petrocca, F., Casalini, P., Taccioli, C., Volinia, S., Liu, C. G., Alder, H., Calin, G. A., Ménard, S., and Croce, C. M. (2007). MicroRNA signatures in human ovarian cancer. Cancer Res. 67, 8699–8707.
Jafarifar, F., Yao, P., Eswarappa, S. M., and Fox, P. L. (2011). Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 30, 1324–1334.
Jin, K., Li, W., Nagayama, T., He, X., Sinor, A. D., Chang, J., Mao, X., Graham, S. H., Simon, R. P., and Greenberg, D. A. (2000). Expression of the RNA-binding protein TIAR is increased in neurons after ischemic cerebral injury. J. Neurosci. Res. 59, 767–774.
Johnson, A. B., and Barton, M. C. (2007). Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat. Res. 618, 149–162.
Johnson, A. B., Denko, N., and Barton, M. C. (2008). Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res. 640, 174–179.
Kedersha, N., and Anderson, P. (2007). Mammalian stress granules and processing bodies. Meth. Enzymol. 431, 61–81.
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., Scheuner, D., Kaufman, R. J., Golan, D. E., and Anderson, P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884.
Kelly, T. J., Souza, A. L., Clish, C. B., and Puigserver, P. (2011). A hypoxia induced positive feedback loop promotes HIF-1α stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol. Cell. Biol. 31, 2696–2706.
Kiledjian, M., Wang, X., and Liebhaber, S. A. (1995). Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 14, 4357–4364.
Kim, H. S., Kuwano, Y., Zhan, M., Pullmann, R. Jr., Mazan-Mamczarz, K., Li, H., Kedersha, N., Anderson, P., Wilce, M. C., Gorospe, M., and Wilce, J. A. (2007). Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 27, 6806–6817.
Kim, H. W., Haider, H. K., Jiang, S., and Ashraf, M. (2009). Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J. Biol. Chem. 284, 33161–33168.
Kim, T. W., Yim, S., Choi, B. J., Jang, Y., Lee, J. J., Sohn, B. H., Yoo, H. S., Yeom, Y. I., and Park, K. C. (2010). Tristetraprolin regulates the stability of HIF-1α mRNA during prolonged hypoxia. Biochem. Biophys. Res. Commun. 391, 963–968.
Kim, W. Y., and Kaelin, W. G. (2004). Role of VHL gene mutation in human cancer. J. Clin. Oncol. 22, 4991–5004.
Kondo, S., Kubota, S., Mukudai, Y., Nishida, T., Yoshihama, Y., Shirota, T., Shintani, S., and Takigawa, M. (2011). Binding of glyceraldehyde-3-phosphate dehydrogenase to the cis-acting element of structure-anchored repression in ccn2 mRNA. Biochem. Biophys. Res. Commun. 405, 382–387.
Koritzinsky, M., Magagnin, M. G., van den Beucken, T., Seigneuric, R., Savelkouls, K., Dostie, J., Pyronnet, S., Kaufman, R. J., Weppler, S. A., Voncken, J. W., Lambin, P., Koumenis, C., Sonenberg, N., and Wouters, B. G. (2006). Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 25, 1114–1125.
Kulshreshtha, R., Ferracin, M., Wojcik, S. E., Garzon, R., Alder, H., Agosto-Perez, F. J., Davuluri, R., Liu, C. G., Croce, C. M., Negrini, M., Calin, G. A., and Ivan, M. (2007a). A microRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867.
Kulshreshtha, R., Ferracin, M., Negrini, M., Calin, G. A., Davuluri, R. V., and Ivan, M. (2007b). Regulation of microRNA expression: the hypoxic component. Cell Cycle 6, 1426–1431.
Lafarga, V., Cuadrado, A., Lopez de Silanes, I., Bengoechea, R., Fernandez-Capetillo, O., and Nebreda, A. R. (2009). p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol. Cell. Biol. 29, 4341–4351.
Lal, A., Kawai, T., Yang, X., Mazan-Mamczarz, K., and Gorospe, M. (2005). Anti-apoptotic function of RNA binding HuR effected through prothymosin-α. EMBO J. 24, 1852–1862.
Le Quesne, J. P., Spriggs, K. A., Bushell, M., and Willis, A. E. (2010). Dysregulation of protein synthesis and disease. J. Pathol. 220, 140–151.
Leipuviene, R., and Theil, E. C. (2007). The family of iron responsive RNA structures regulated by changes in cellular iron and oxygen. Cell Mol. Life Sci. 64, 2945–2955.
Levy, N. S., Chung, S., Furneaux, H., and Levy, A. P. (1998). Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273, 6417–6423.
Lleonart, M. E. (2010). A new generation of proto-oncogenes: cold-inducible RNA binding proteins. Biochim. Biophys. Acta 1805, 43–52.
López de Silanes, I., Fan, J., Yang, X., Yang, X., Zonderman, A. B., Potapova, O., Pizer, E. S., and Gorospe, M. (2003). Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22, 7146–7154.
López de Silanes, I., Galban, S., Martindale, J. L., Yang, X., Mazan-Mamczarz, K., Indig, F. E., Falco, G., Zhan, M., and Gorospe, M. (2005). Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 25, 9520–9531.
López de Silanes, I., Zhan, M., Lal, A., Yang, X., and Gorospe, M. (2004). Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992.
Lui, W. O., Pourmand, N., Patterson, B. K., and Fire, A. (2007). Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 67, 6031–6043.
Mazan-Mamczarz, K., Galbán, S., López de Silanes, I., Martindale, J. L., Atasoy, U., Keene, J. D., and Gorospe, M. (2003). RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. U.S.A. 100, 8354–8359.
McGary, E. C., Rondon, I. J., and Beckman, B. S. (1997). Post-transcriptional regulation of erythropoietin mRNA stability by erythropoietin mRNA-binding protein. J. Biol. Chem. 272, 8628–8634.
Millevoi, S., Decorsière, A., Loulergue, C., Iacovoni, J., Bernat, S., Antoniou, M., and Vagner, S. (2009). A physical and functional link between splicing factors promotes pre-mRNA 3′ end processing. Nucleic Acids Res. 37, 4672–4683.
Mukherjee, D., Gao, M., O’Connor, J. P., Raijmakers, R., Pruijn, G., Lutz, C. S., and Wilusz, J. (2002). The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21, 165–174.
Nagy, E., and Rigby, W. F. (1995). Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold). J. Biol. Chem. 270, 2755–2763.
Nanduri, J., Yuan, G., Kumar, G. K., Semenza, G. L., and Prabhakar, N. R. (2008). Transcriptional responses to intermittent hypoxia. Respir. Physiol. Neurobiol. 164, 277–281.
Onesto, C., Berra, E., Grépin, R., and Pagès, G. (2004). Poly(A)-binding protein-interacting protein 2, a strong regulator of vascular endothelial growth factor mRNA. J. Biol. Chem. 279, 34217–34226.
Osborne, N. N., Nash, M. S., and Wood, J. P. (1998). Melatonin counteracts ischemia-induced apoptosis in human retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 39, 2374–2383.
Ouiddir, A., Planès, C., Fernandes, I., VanHesse, A., and Clerici, C. (1999). Hypoxia upregulates activity and expression of the glucose transporter GLUT1 in alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 21, 710–718.
Pagès, G., Berra, E., Milanini, J., Levy, A. P., and Pouysségur, J. (2000). Stress-activated protein kinases (JNK and p38/HOG) are essential for vascular endothelial growth factor mRNA stability. J. Biol. Chem. 275, 26484–26491.
Parker, R., and Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646.
Place, R. F., Li, L. C., Pookot, D., Noonan, E. J., and Dahiya, R. (2008). MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U.S.A. 105, 1608–1613.
Puisségur, M. P., Mazure, N. M., Bertero, T., Pradelli, L., Grosso, S., Robbe-Sermesant, K., Maurin, T., Lebrigand, K., Cardinaud, B., Hofman, V., Fourre, S., Magnone, V., Ricci, J. E., Pouysségur, J., Gounon, P., Hofman, P., Barbry, P., and Mari, B. (2011). miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–478.
Pulkkinen, K., Malm, T., Turunen, M., Koistinaho, J., and Ylä-Herttuala, S. (2008). Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 582, 2397–2401.
Qi, C., and Pekala, P. H. (1999). The influence of mRNA stability on glucose transporter (GLUT1) gene expression. Biochem. Biophys. Res. Commun. 263, 265–269.
Rane, S., He, M., Sayed, D., Vashistha, H., Malhotra, A., Sadoshima, J., Vatner, D. E., Vatner, S. F., and Abdellatif, M. (2009). Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 104, 879–886.
Rane, S., He, M., Sayed, D., Yan, L., Vatner, D., and Abdellatif, M. (2010). An antagonism between the AKT and beta-adrenergic signaling pathways mediated through their reciprocal effects on miR-199a-5p. Cell. Signal. 22, 1054–1062.
Rankin, E. B., and Giaccia, A. J. (2008). The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 15, 678–685.
Rondon, I. J., Scandurro, A. B., Wilson, R. B., and Beckman, B. S. (1995). Changes in redox affect the activity of erythropoietin RNA binding protein. FEBS Lett. 359, 267–270.
Sahai, A., Mei, C., Pattison, T. A., and Tannen, R. L. (1997). Chronic hypoxia induces proliferation of cultured mesangial cells: role of calcium and protein kinase C. Am. J. Physiol. Renal Physiol. 273, 954–960.
Sanchez, M., Galy, B., Muckenthaler, M. U., and Hentze, M. W. (2007). Iron-regulatory proteins limit hypoxia-inducible factor-2α expression in iron deficiency. Nat. Struct. Mol. Biol. 14, 420–426.
Sarkar, J., Gou, D., Turaka, P., Viktorova, E., Ramchandran, R., and Raj, J. U. (2010). MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am. J. Physiol. Lung Cell Mol. Physiol. 299, L861–L871.
Sawicka, K., Bushell, M., Spriggs, K. A., and Willis, A. E. (2008). Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 36, 641–647.
Sayed, D., He, M., Hong, C., Gao, S., Rane, S., Yang, Z., and Abdellatif, M. (2010). MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J. Biol. Chem. 285, 20281–20290.
Schepens, B., Tinton, S. A., Bruynooghe, Y., Beyaert, R., and Cornelis, S. (2005). The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic Acids Res. 33, 6884–6894.
Schneider, B. D., and Leibold, E. A. (2003). Effects of iron regulatory protein regulation on iron homeostasis during hypoxia. Blood 102, 3404–3411.
Schofield, C. J., and Ratcliffe, P. J. (2004). Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354.
Sheflin, L. G., Zou, A. P., and Spaulding, S. W. (2004). Androgens regulate the binding of endogenous HuR to the AU-rich 3′UTRs of HIF-1α and EGF mRNA. Biochem. Biophys. Res. Commun. 322, 644–651.
Shi, L., Godfrey, W. R., Lin, J., Zhao, G., and Kao, P. N. (2007). NF90 regulates inducible IL-2 gene expression in T cells. J. Exp. Med. 204, 971–977.
Shih, S. C., and Claffey, K. P. (1998). Hypoxia-mediated regulation of gene expression in mammalian cells. Int. J. Exp. Pathol. 79, 347–357.
Sinha, S., Dutta, S., Datta, K., Ghosh, A. K., and Mukhopadhyay, D. (2009). Von Hippel-Lindau gene product modulates TIS11B expression in renal cell carcinoma: impact on vascular endothelial growth factor expression in hypoxia. J. Biol. Chem. 284, 32610–32618.
Smart, F., Aschrafi, A., Atkins, A., Owens, G. C., Pilotte, J., Cunningham, B. A., and Vanderklish, P. W. (2007). Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J. Neurochem. 101, 1367–1379.
Takagi, M., Absalon, M. J., McLure, K. G., and Kastan, M. B. (2005). Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63.
Tetzlaff, M. T., Liu, A., Xu, X., Master, S. R., Baldwin, D. A., Tobias, J. W., Livolsi, V. A., and Baloch, Z. W. (2007). Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr. Pathol. 18, 163–173.
Tillmar, L., Carlsson, C., and Welsh, N. (2002). Control of insulin mRNA stability in rat pancreatic islets: regulatory role of a 3′-UTR pyrimidine-rich sequence. J. Biol. Chem. 277, 1099–1106.
Tillmar, L., and Welsh, N. (2004). Glucose-induced binding of the polypyrimidine tract-binding protein (PTB) to the 3′-untranslated region of the insulin mRNA (ins-PRS) is inhibited by rapamycin. Mol. Cell Biochem. 260, 85–90.
Tillmar, L., and Welsh, N. (2002). Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol Med. 8, 263–272.
Tong, W. H., and Rouault, T. A. (2006). Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3, 199–210.
Tran, N., McLean, T., Zhang, X., Zhao, C. J., Thomson, J. M., O’Brien, C., and Rose, B. (2007). MicroRNA expression profiles in head and neck cancer cell lines. Biochem. Biophys. Res. Commun. 358, 12–17.
Turcotte, S., Desrosiers, R. R., and Béliveau, R. (2004). Hypoxia upregulates von Hippel-Lindau tumor-suppressor protein through RhoA-dependent activity in renal cell carcinoma. Am. J. Physiol. Renal Physiol. 286, F338–F348.
Vasudevan, S., Tong, Y., and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934.
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., Iorio, M., Roldo, C., Ferracin, M., Prueitt, R. L., Yanaihara, N., Lanza, G., Scarpa, A., Vecchione, A., Negrini, M., Harris, C. C., and Croce, C. M. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261.
Voorhoeve, P. M., le Sage, C., Schrier, M., Gillis, A. J., Stoop, H., Nagel, R., Liu, Y. P., van Duijse, J., Drost, J., Griekspoor, A., Zlotorynski, E., Yabuta, N., De Vita, G., Nojima, H., Looijenga, L. H., and Agami, R. (2006). A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181.
Wang, W., Furneaux, H., Cheng, H., Caldwell, M. C., Hutter, D., Liu, Y., Holbrook, N. J., and Gorospe, M. (2000). HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760–769.
Wang, W., Yang, X., Cristofalo, V. J., Holbrook, N. J., and Gorospe, M. (2001). Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol. Cell. Biol. 21, 5889–5898.
Wellmann, S., Bührer, C., Moderegger, E., Zelmer, A., Kirschner, R., Koehne, P., Fujita, J., and Seeger, K. (2004). Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J. Cell Sci. 117, 1785–1794.
Welsh, M., Nielsen, D. A., MacKrell, A. J., and Steiner, D. F. (1985). Control of insulin gene expression in pancreatic beta cells and in an insulin-producing cell line, RIN-5F cells. II. Regulation of insulin mRNA stability. J. Biol. Chem. 260, 13590–13594.
Wenger, R. H., Stiehl, D. P., and Camenisch, G. (2005). Integration of oxygen signaling at the consensus HRE. Sci. STKE 306, re12.
Werno, C., Schmid, T., Schnitzer, S. E., Peters, K., Milke, L., and Brüne, B. (2010). A combination of hypoxia and lipopolysaccharide activates tristetraprolin to destabilize proinflammatory mRNAs such as tumor necrosis factor-α. Am. J. Pathol. 177, 1104–1112.
Wicksteed, B., Herbert, T. P., Alarcon, C., Lingohr, M. K., Moss, L. G., and Rhodes, C. J. (2001). Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. J. Biol. Chem. 276, 22553–22558.
Wilusz, C. J., Wormington, M., and Peltz, S. W. (2001). The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237–246.
Wouters, B. G., van den Beucken, T., Magagnin, M. G., Koritzinsky, M., Fels, D., and Koumenis, C. (2005). Control of the hypoxic response through regulation of mRNA translation. Semin. Cell Dev. Biol. 16, 487–501.
Xia, X., and Holcik, M. (2009). Strong eukaryotic IRESs have weak secondary structure. PLoS ONE 4, e4136. doi: 10.1371/journal.pone.0004136
Xu, L., Pathak, P. S., and Fukumura, D. (2004). Hypoxia-induced activation of p38 mitogen-activated protein kinase and phosphatidylinositol 3′-kinase signaling pathways contributes to expression of interleukin 8 in human ovarian carcinoma cells. Clin. Cancer Res. 10, 701–707.
Yancopoulos, G. D., Davis, S., Gale, N. W., Rudge, J. S., Wiegand, S. J., and Holash, J. (2000). Vascular-specific growth factors and blood vessel formation. Vascular-specific growth factors and blood vessel formation. Nature 407, 242–248.
Yang, C., and Carrier, F. (2001). The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 276, 47277–47284.
Yang, R., Weber, D. J., and Carrier, F. (2006). F. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res. 34, 1224–1236.
Zhang, Z., Sun, H., Dai, H., Walsh, R. M., Imakura, M., Schelter, J., Burchard, J., Dai, X., Chang, A. N., Diaz, R. L., Marszalek, J. R., Bartz, S. R., Carleton, M., Cleary, M. A., Linsley, P. S., and Grandori, C. (2009). MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 8, 2756–2768.
Zhao, Z., Chang, F. C., and Furneaux, H. M. (2000). The identification of an endonuclease that cleaves within an HuR binding site in mRNA. Nucleic Acids Res. 28, 2695–2701.
Keywords: hypoxia, post-transcriptional gene regulation, microRNAs, RNA-binding proteins, mRNA turnover, translational control, ribonucleoprotein complex, untranslated regions
Citation: Gorospe M, Tominaga K, Wu X, Fähling M and Ivan M (2011) Post-transcriptional control of the hypoxic response by RNA-binding proteins and microRNAs. Front. Mol. Neurosci. 4:7. doi: 10.3389/fnmol.2011.00007
Received: 16 May 2011; Paper pending published: 03 June 2011;
Accepted: 17 June 2011; Published online: 01 July 2011.
Edited by:
Jochen C. Meier, Max Delbrück Center for Molecular Medicine, GermanyReviewed by:
Jeffrey Wilusz, Colorado State University, USACopyright: © 2011 Gorospe, Tominaga, Wu, Fähling and Ivan. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Myriam Gorospe, RNA Regulation Section, Laboratory of Molecular Biology and Immunology, National Institute on Aging-Intramural Research Program, National Institutes of Health, 251 Bayview Boulevard, Baltimore, MD 21224, USA. e-mail:bXlyaWFtLWdvcm9zcGVAbmloLmdvdg==; Michael Fähling, Institut für Vegetative Physiologie, Charité – Universitätsmedizin Berlin, Hessische Strasse 3-4, D-10115 Berlin, Germany. e-mail:bWljaGFlbC5mYWVobGluZ0BjaGFyaXRlLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.