- 1School of Computer Science and Technology, Harbin Institute of Technology, Harbin, China
- 2Beidahuang Industry Group General Hospital, Harbin, China
- 3Beidahuang Group Neuropsychiatric Hospital, Jiamusi, China
Introduction: Growing evidence highlights a potential genetic overlap between Alzheimer's disease (AD) and Parkinson's disease (PD); however, the role of the PD risk variant rs6430538 in AD remains unclear.
Methods: In Stage 1, we investigated the risk associated with the rs6430538C allele in seven large-scale AD genome-wide association study (GWAS) cohorts. In Stage 2, we performed expression quantitative trait loci (eQTL) analysis to calculate the cis-regulated effect of rs6430538 on TMEM163 in both AD and neuropathologically normal samples. Stage 3 involved evaluating the differential expression of TMEM163 in 4 brain tissues from AD cases and controls. Finally, in Stage 4, we conducted a transcriptome-wide association study (TWAS) to identify any association between TMEM163 expression and AD.
Results: The results showed that genetic variant rs6430538C allele might increase the risk of AD. eQTL analysis revealed that rs6430538 up-regulated TMEM163 expression in AD brain tissue, but down-regulated its expression in normal samples. Interestingly, TMEM163 showed differential expression in entorhinal cortex (EC) and temporal cortex (TCX). Furthermore, the TWAS analysis indicated strong associations between TMEM163 and AD in various tissues.
Discussion: In summary, our findings suggest that rs6430538 may influence AD by regulating TMEM163 expression. These discoveries may open up new opportunities for therapeutic strategies targeting AD.
1 Background
Potential genetic association may exist between common neurodegenerative diseases (Small and Petsko, 2015; Brainstorm et al., 2018; Guo et al., 2022). Alzheimer's disease (AD) and Parkinson's disease (PD) are both classic age-related neurodegenerative diseases (Scheltens et al., 2016; Poewe et al., 2017; Hu et al., 2021; Qiu et al., 2022a). Although the pathogenesis of AD and PD differs, they share certain similar pathways in the development of neurodegeneration (Xie et al., 2014). One of the most important pathological features of AD is the deposition of amyloid-β (Aβ), which is also present in the brains of PD patients (Compta et al., 2011; Yu et al., 2014). Similarly, the pathological features of PD are also observed in cases of AD (Lippa et al., 1997). Currently, the shared susceptibility loci of AD and PD, such as APOE and MAPT, have been identified, indicating a possible overlap in genetic background and pathological characteristics (Laws et al., 2007; Zhu et al., 2017). However, further exploration of additional genetic links is necessary.
Members of the transmembrane (TMEM) protein family have been identified as being associated with various neurodegenerative disorders (Chang et al., 2017; Nalls et al., 2019; Li et al., 2020). Among these genes, TMEM163 serves as an important susceptibility locus for PD. It plays a regulatory role in ATP-evoked behavior in neurons, as well as binding to Zn2+ and recruiting them to vesicular organelles (Chang et al., 2017; Salm et al., 2020; Kia et al., 2021; Mammadova-Bach and Braun, 2021). Notably, brain zinc has been found to be involved in the pathogenesis of AD by influencing amyloid metabolism (Sensi et al., 2018; Xu et al., 2019). It is plausible to hypothesize that rs6430538, through its impact on the expression level of zinc transporters, contributes to the aggravation of Aβ deposition and toxicity by regulating nearby genes. In an attempt to identify shared pathogenic genetic variants between PD and AD, Zhu et al. replicated the association of rs6430538 (ACMSD-TMEM163) with AD in a population study (Zhu et al., 2017). They investigated a sample comprising 992 sporadic AD patients and 1,358 controls from northern China and observed a protective effect of rs6430538 on AD (OR = 0.340, P = 0.015) (Zhu et al., 2017). Unfortunately, upon regression adjustment for confounding factors including age, gender, and APOE ε4 status, rs6430538 did not achieve statistical significance (P = 0.072) (Zhu et al., 2017). It is important to note, however, that the sample size of the independent case-control study was relatively small compared to genome-wide association studies (GWAS). Furthermore, previous investigations solely focused on exploring the potential risk association between rs6430538 and AD, neglecting the possibility that rs6430538 may influence AD through the regulation of gene expression.
In this study, we aimed to delve deeper into the association between the PD risk variant rs6430538 and AD. Specifically, we sought to investigate the impact of rs6430538 on AD by analyzing its influence on gene expression using comprehensive datasets, including large-scale GWAS, expression quantitative trait loci (eQTL), and RNA-seq datasets. By doing so, our findings would contribute to a better understanding of the functional role of non-coding region variants, such as rs6430538, and their implications for disease regulation.
2 Materials and methods
2.1 AD GWAS datasets
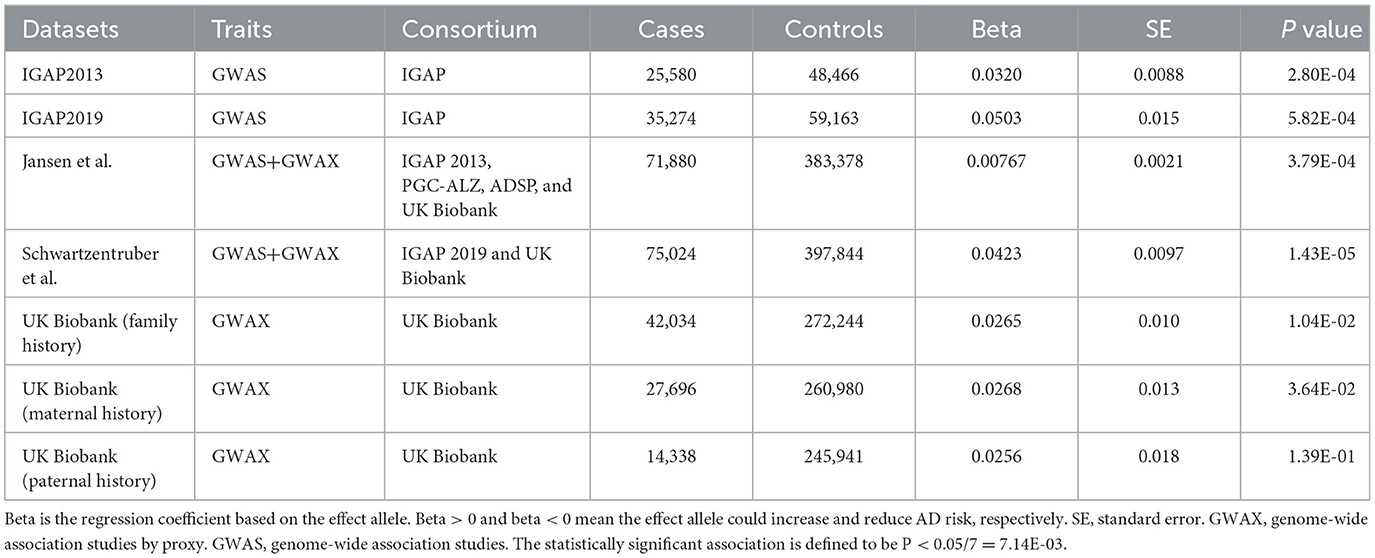
We investigated the risk of rs6430538 in seven large-scale GWAS for AD, including two GWAS meta-analyses by International Genomics of Alzheimer's Project (IGAP), three genome-wide association studies by proxy (GWAX) by UK Biobank, and two meta-analyses of GWAS and GWAX (Lambert et al., 2013; Marioni et al., 2018; Buniello et al., 2019; Jansen et al., 2019; Kunkle et al., 2019; Schwartzentruber et al., 2021). UK Biobank used self-reported family history of Alzheimer's disease as the standard of cases, while IGAP used Aβ and tau levels as the standard of cases (the NINCDS-ADRDA criteria or DSM-IV guidelines) (Lambert et al., 2013). Detailed GWAS information was shown in the original studies and Table 1.
2.2 eQTL datasets
We obtained eQTL datasets of normal brains in 13 normal brain tissues from Genotype-Tissue Expression (GTEx), including amygdala, anterior cingulate cortex, caudate, cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, spinal cord and substantia nigra. The donors were of multiple descents including European (85.3%), African (12.3%), Asian (1.4%), etc. and only about 1.2% of the donors died of neurological diseases (GTEx Consortium, 2017).
We obtained eQTL datasets of AD and non-AD samples from Mayo RNAseq Study (MAYO) and Religious Orders Study and Memory and Aging Project (ROSMAP) (Bennett et al., 2012a,b; Zou et al., 2012; GTEx Consortium, 2017; Ng et al., 2017). Mayo RNAseq Study collected the cerebellar (CER) (197 cases and 177 controls) and temporal cortex (TCX) (202 cases and 197 controls) tissues of AD individuals and non-AD individuals, respectively (Zou et al., 2012). Non-AD subjects contained several brain pathologies, such as frontotemporal dementia (FTD), multiple system atrophy and vascular dementia (Zou et al., 2012). ROSMAP collected samples from the dorsolateral prefrontal cortex (DLPFC) of 494 participants (Ng et al., 2017). 97% of the samples were diagnosed with pathological or clinical AD (Ng et al., 2017). The donors were all of European descent.
2.3 RNA expression datasets
RNA-seq datasets for AD and controls were generated from AlzData and gene expression omnibus (GEO) databases, including entorhinal cortex (EC) (GSE26927, GSE26972, GSE48350, and GSE5281), hippocampus (HIP) (GSE28146, GSE29378, GSE36980, GSE48350, and GSE5281), TCX (GSE29652, GSE36980, GSE37263, and GSE5281), and Frontal Cortex (FCTX) (GSE12685, GSE36980, GSE48350, GSE5281, GSE53890, and GSE66333) (Xu et al., 2018; Zhang et al., 2019). All differential expression results were adjusted for age and sex of samples.
2.4 Statistical analysis
2.4.1 Genetic association of rs6430538 with AD
We used seven GWAS datasets for AD to evaluate the genetic association between rs6430538C allele and AD. In addition, we carried out gender stratification analysis using GWAX datasets diagnosed by paternal history and maternal history from UK Biobank (Marioni et al., 2018). The statistically significant association is defined to be P < 5E-08 after adjusting for multiple testing.
2.4.2 eQTL analysis of rs6430538 on TMEM163
Disease states are known to alter the expression of specific genes (Gratuze et al., 2018; Liu et al., 2019; Hu et al., 2020; Ma and Qiu, 2022; Qiu et al., 2022b,c). Here, we calculated the regulated effect of rs6430538C allele on TMEM163 in neuropathologically normal and neuropathologically disease individuals, respectively, by applying linear regression based on an additive model (Hu et al., 2020; Qiu et al., 2022b). According to the additive model, each allele has an independent effect on the trait. We coded the possible genotypes of rs6430538 (CC = 2, CT = 1, TT = 0), where C is an effect allele and T is a non-effect allele. The statistically significant association is defined to be P < 0.05/18 = 2.78E-03.
2.4.3 Gene expression analysis of TMEM163
We evaluated the differential expression of TMEM163 in four brain tissues between AD cases and controls. All differential expression results were adjusted for sex with an FDR of 0.05.
2.4.4 Transcriptome-wide association study analysis
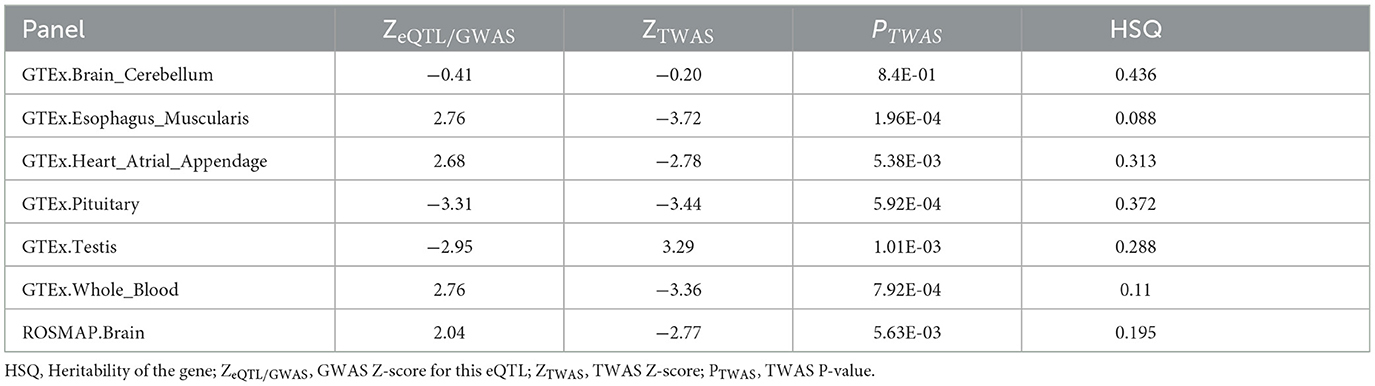
TWAS is a test for a significant association between cis components of gene expression and traits (Gusev et al., 2016). TWAS integrates GWAS and eQTL datasets, not only mining significant correlations between eQTLs and SNPs, but also capturing complete cis-SNP signals. We looked for evidence of the association between TMEM163 and AD in TWAS using FUSION software (Gusev et al., 2016). Herein, we integrated AD GWAS with gene expression dataset from GTEx and ROSMAP for TWAS analysis (Gusev et al., 2016; GTEx Consortium, 2017; Kunkle et al., 2019). The statistically significant association is defined to be P < 5E-08 after multiple testing.
3 Results
3.1 Rs6430538 regulated the expression of TMEM163 and increased AD risk
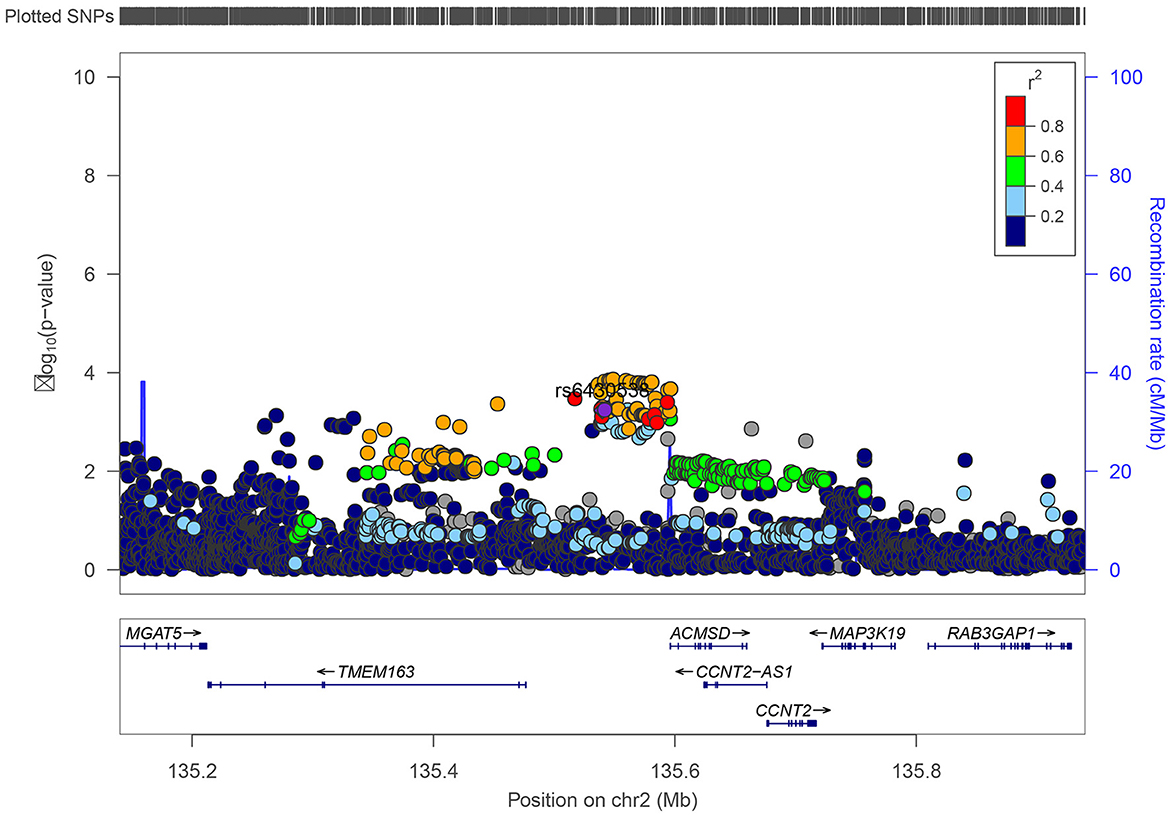
We inquired proxy SNPs of rs6430538 and the annotations information on noncoding genome using HaploReg4.1 (Ward and Kellis, 2016). Rs6430538 is located in the intergenic region on chromosome 2, with eight genetic variants highly linked to rs6430538 (r2 > 0.8) (Figure 1, Supplementary Table 1) (Pruim et al., 2010). In addition to the GWAX studies by UK Biobank, rs6430538C allele was suggestively associated with the risk of AD in four large-scale GWAS meta-analyses (Table 1). Moreover, rs6430538 showed no sex-specific differences in AD risk (Table 1).
Based on all brain eQTL studies, rs6430538 involved in regulating the expression of TMEM163, CCNT2, VDAC2P4, CCNT2-AS1. Herein, we focused on TMEM163, the shared regulated gene, in all the eQTL studies. The eQTL analysis showed that rs6430538 regulated TMEM163 overexpression in AD patients (Beta = 0.325, P = 1.31E-13), while rs6430538 inhibited TMEM163 expression in non-AD individuals (Table 2). The opposing effects of the same variant may be explained if there is dysregulation of TMEM163 in disease conditions.
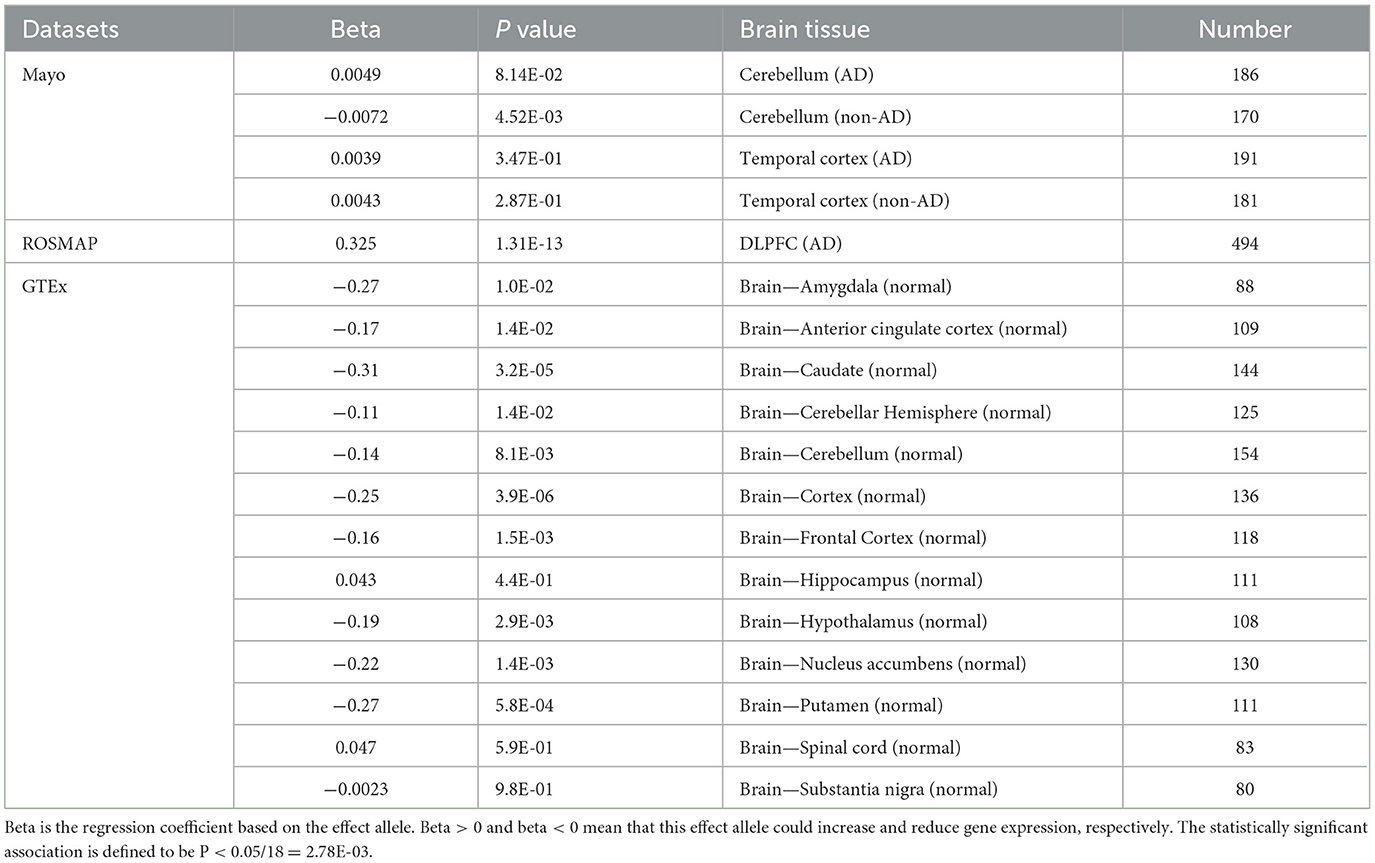
Table 2. The regulatory effects of rs6430538 variant C allele on TMEM163 in both AD and neuropathologically normal individuals.
3.2 TMEM163 was associated with AD and differentially expressed in AD and normal individuals
TMEM163 was differentially expressed in EC ( = −0.44, P = 0.01) and TCX ( = −0.52, P = 4.32E-05) of AD vs. controls (Figure 2, Supplementary Table 2). However, TMEM163 was not expressed in FCTX. Moreover, we used TWAS to prioritize potential susceptibility genes for AD, and TMEM163 was suggestively associated with AD in 5 GTEx tissues (whole blood, esophagus muscularis, heart atrial appendage, pituitary and testis) and brain tissues (Table 3). Importantly, TMEM163 was a potential casual gene of AD in brain (ROSMAP) (ZTWAS = −3.36, PTWAS = 5.63E-03) and whole blood (ZTWAS = −2.77, PTWAS = 7.92E-04) (Table 3).
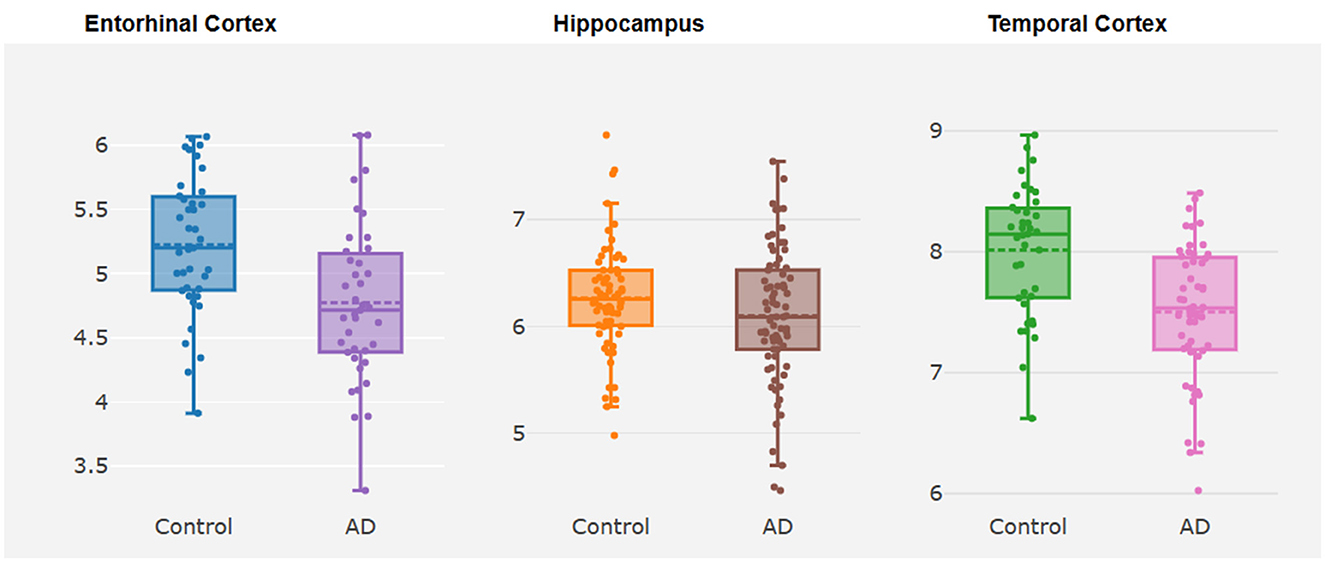
Figure 2. Box plots about differential expression analysis of TMEM163. The y-axis represents the expression level of TMEM163, while the x-axis represents different brain tissues.
4 Discussion
Neurodegenerative diseases, such as AD and PD, share certain genetic associations and pathological characteristics, despite differences in their pathogenesis (Majd et al., 2015; Zhu et al., 2017; Han et al., 2018). The identification of shared susceptibility loci, such as APOE and MAPT, suggests a potential overlap in genetic background and pathological mechanisms between these diseases. However, further exploration is required to uncover additional genetic links.
Based on the hypothesis that rs6430538, a risk variant associated with PD, may impact AD through the regulation of nearby genes, we aimed to investigate the influence of rs6430538 on AD by analyzing its effect on gene expression using large-scale datasets. To examine the genetic association of rs6430538 with AD, we analyzed seven GWAS datasets, including those from the IGAP and UK Biobank. Our findings indicated that the rs6430538C allele was associated with an increased risk of AD. Additionally, gender stratification analysis revealed no sex-specific differences in the association between rs6430538 and AD risk. Understanding gender-specific and nongender-specific effects could guide the development of sex-specific therapeutic strategies, taking into account potential differences in disease mechanisms and responses to treatment between males and females.
Although GWAS analysis indicated only suggestive association between rs6430538 and AD, stringent GWAS thresholds might obscure some genetic variants that affect AD through regulating gene expression. Therefore, we investigated the impact of rs6430538 on the expression of TMEM163 and other nearby genes using eQTL datasets. The results demonstrated that rs6430538 played a role in regulating the expression of TMEM163, CCNT2, VDAC2P4, and CCNT2-AS1. Specifically, rs6430538 exhibited a regulatory effect on TMEM163 expression, with overexpression observed in AD patients and inhibited expression in non-AD individuals. As revealed in previous studies, genetic variants linked to AD risk that modulate the expression of genes in human tissues, emphasizing the importance of non-coding regions in disease pathogenesis (Raj et al., 2014). To prioritize potential susceptibility genes for AD, we conducted a TWAS using GWAS and eQTL datasets. Our results indicated that TMEM163 was strongly associated with AD in multiple tissues, including brain tissues and whole blood.
However, our study had certain limitations. First of all, gender, age (or age of death), disease status (or cause of death) and ancestry, might lead to different gene expression levels in different eQTL datasets. We could not control all the variables. For example, 1.2% of the donors in GTEx died of neurological diseases, and these samples with abnormal neuropathology might have some effects. Similarly, due to the lack of sex stratified eQTLs and original genotype data, it was difficult to deeply explore the contribution of female genotype and gene expression in AD. Furthermore, although the PD susceptibility locus TMEM163 significantly affected AD in brain tissue and blood tissue, to our knowledge, none of the known methods claimed to find causal gene (Gusev et al., 2016; Wainberg et al., 2019; Zhu et al., 2021). Novel methods of exploring causal genes may be more powerful evidence for our research.
5 Conclusions
Genetic variants located in non-coding region may affect traits by regulating gene expression. Here, we comprehensively analyzed the effect of a PD risk variant rs6430538 on AD through regulating gene expression. We demonstrated that different neuropathological samples played different roles in the regulation of gene expression by genetic variants. These findings highlighted the possible genetic overlap between neurodegenerative diseases.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SQ: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. MS: Conceptualization, Writing – review & editing. YX: Formal analysis, Writing – review & editing. YH: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No: 62371161).
Acknowledgments
We thank the International Genomics of Alzheimer's Project (IGAP) and UK Biobank (http://www.ukbiobank.ac.uk/) for AD GWAS summary statistics. The investigators within IGAP provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. We thank the Mayo RNAseq Study (MAYO), Religious Orders Study and Memory and Aging Project (ROSMAP), Mount Sinai Brain Bank (MSBB), and Genotype-Tissue Expression (GTEx) for the eQTLs dataset resources. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The results published here are in part based on data obtained from Agora, a platform initially developed by the NIA-funded AMP-AD consortium that shares evidence in support of AD target discovery. Agora is available at: https://agora.adknowledgeportal.org/.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2024.1277187/full#supplementary-material
References
Bennett, D. A., Schneider, J. A., Arvanitakis, Z., and Wilson, R. S. (2012b). Overview and findings from the religious orders study. Curr. Alzheimer Res. 9, 628–645. doi: 10.2174/156720512801322573
Bennett, D. A., Schneider, J. A., Buchman, A. S., Barnes, L. L., Boyle, P. A., and Wilson, R. S. (2012a). Overview and findings from the rush Memory and Aging Project. Curr. Alzheimer Res. 9, 646–663. doi: 10.2174/156720512801322663
Brainstorm, C., Bulik-Sullivan, S., Finucane, H., Walters, R., Bras, J., Duncan, L., et al. (2018). Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757. doi: 10.1126/science.aap8757
Buniello, A., MacArthur, J. A. L., Cerezo, M., Harris, L. W., Hayhurst, J., Malangone, C., et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics (2019). Nucleic Acids Res. 47, D1005–D1012. doi: 10.1093/nar/gky1120
Chang, D., Nalls, M., Hallgrímsdóttir, I. B, Hunkapiller, J., van der Brug, M., Cai, F., et al. (2017). A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet. 49, 1511–1516. doi: 10.1038/ng.3955
Compta, Y., Parkkinen, L., O'Sullivan, S. S., Vandrovcova, J., Holton, J. L., Collins, C., et al. (2011). Lewy- and Alzheimer-type pathologies in Parkinson's disease dementia: which is more important? Brain 134, 1493–1505. doi: 10.1093/brain/awr031
Gratuze, M., Leyns, C. E. G., and Holtzman, D. M. (2018). New insights into the role of TREM2 in Alzheimer's disease. Mol. Neurodegener. 13, 66. doi: 10.1186/s13024-018-0298-9
GTEx Consortium Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI. (2017). Genetic effects on gene expression across human tissues. Nature 550, 204–213. doi: 10.1038/nature24277
Guo, P., Gong, W., Li, Y., Liu, L., Yan, R., Wang, Y., et al. (2022). Pinpointing novel risk loci for Lewy body dementia and the shared genetic etiology with Alzheimer's disease and Parkinson's disease: a large-scale multi-trait association analysis. BMC Med. 20, 214. doi: 10.1186/s12916-022-02404-2
Gusev, A., Ko, A., Shi, H., Bhatia, G., Chung, W., Penninx, B. W., et al. (2016). Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252. doi: 10.1038/ng.3506
Han, Z., Tian, R., Ren, P., Zhou, W., Wang, P., Luo, M., et al. (2018). Parkinson's disease and Alzheimer's disease: a Mendelian randomization study. BMC Med. Genet. 19, 215. doi: 10.1186/s12881-018-0721-7
Hu, Y., Sun, J.-Y., Zhang, Y., Zhang, H., Gao, S., Wang, T., et al. (2021). rs1990622 variant associates with Alzheimer's disease and regulates TMEM106B expression in human brain tissues. BMC Med. 19, 11. doi: 10.1186/s12916-020-01883-5
Hu, Y., Zhang, H., Liu, B., Gao, S., Wang, T., Han, Z., et al. (2020). rs34331204 regulates TSPAN13 expression and contributes to Alzheimer's disease with sex differences. Brain 143, e95. doi: 10.1093/brain/awaa302
Jansen, I. E., Savage, J. E., Watanabe, K., Bryois, J., Williams, D. M., Steinberg, S., et al. (2019). Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat. Genet. 51, 404–413. doi: 10.1038/s41588-018-0311-9
Kia, D. A., Zhang, D., Guelfi, S., Manzoni, C., Hubbard, L., Reynolds, R. H., et al. (2021). Identification of candidate parkinson disease genes by integrating genome-wide association study, expression, and epigenetic data Sets. JAMA Neurol. 78, 464–472. doi: 10.1001/jamaneurol.2020.5257
Kunkle, B. W., Grenier-Bole, B., Sims, R., Bis, J., Damotte, V., Naj, A., et al. (2019). Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 51, 414–430. doi: 10.1038/s41588-019-0358-2
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458. doi: 10.1038/ng.2802
Laws, S. M., Friedrich, P., Diehl-Schmid, J., Müller, J., Eisele, T., Bäuml, J., et al. (2007). Fine mapping of the MAPT locus using quantitative trait analysis identifies possible causal variants in Alzheimer's disease. Mol. Psychiatry 12, 510–517. doi: 10.1038/sj.mp.4001935
Li, C., Ou, R., Chen, Y., Gu, X., Wei, Q., Cao, B., et al. (2020). Mutation analysis of TMEM family members for early-onset Parkinson's disease in Chinese population. Neurobiol Aging, 101, 299.e1–299.e6. doi: 10.1016/j.neurobiolaging.2020.11.005
Lippa, C. F., Pulaski-Salo, D., Dickson, D. W., and Smith, T. W. (1997). Alzheimer's disease, Lewy body disease and aging: a comparative study of the perforant pathway. J. Neurol. Sci. 147, 161–166. doi: 10.1016/S0022-510X(96)05321-X
Liu, G., Hu, Y., and Jiang, Q. (2019). Population difference and disease status affect the association between genetic variants and gene expression. gastroenterology 157, 894–896. doi: 10.1053/j.gastro.2019.01.278
Ma, J., and Qiu, S. (2022). Genetic variant rs11136000 upregulates clusterin expression and reduces Alzheimer's disease risk. Front. Neurosci. 16, 926830. doi: 10.3389/fnins.2022.926830
Majd, S., Power, J. H., and Grantham, H. J. (2015). Neuronal response in Alzheimer's and Parkinson's disease: the effect of toxic proteins on intracellular pathways. BMC Neurosci. 16, 69. doi: 10.1186/s12868-015-0211-1
Mammadova-Bach, E., and Braun, A. (2021). Platelet life without TMEM163: no dense granules. Blood 137, 1708–1709. doi: 10.1182/blood.2021010691
Marioni, R. E., Harris, S. E., Zhang, Q., McRae, A. F., Hagenaars, S. P., Hill, W. D., et al. (2018). GWAS on family history of Alzheimer's disease. Transl. Psychiatry 8, 99. doi: 10.1038/s41398-018-0150-6
Nalls, M. A., Blauwendraat, C., Vallerga, C. L., Heilbron, K., Bandres-Ciga, S., Chang, D., et al. (2019). Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102. doi: 10.1016/S1474-4422(19)30320-5
Ng, B., White, C. C., Klein, H.-U., Sieberts, S. K., McCabe, C., Patrick, E., et al. (2017). An xQTL map integrates the genetic architecture of the human brain's transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426. doi: 10.1038/nn.4632
Poewe, W., Seppi, K., Tanner, C., Halliday, G., Brudin, P., Volkmann, J., et al. (2017). Parkinson disease. Nat. Rev. Dis. Primers 3, 17013. doi: 10.1038/nrdp.2017.13
Pruim, R. J., Welch, R. P., Sanna, S., Teslovich, T. M., Chines, P. S., Gliedt, T. P., et al. (2010). LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337. doi: 10.1093/bioinformatics/btq419
Qiu, S., Hu, Y., and Cheng, L. (2022c). BIN1 rs744373 located in enhancers of brain tissues upregulates BIN1 mRNA expression, thereby leading to Alzheimer's disease. Alzheimers. Dement. 18, 1587–1588. doi: 10.1002/alz.12548
Qiu, S., Hu, Y., and Cheng, L. A. (2022a). Genome-wide cross-trait analysis highlights the shared genetic structure between COVID-19 and Alzheimer's disease. J. Infect. 84, e1–e2. doi: 10.1016/j.jinf.2021.08.038
Qiu, S., Hu, Y., Zou, Q., and Liu, G. (2022b). Genetic variant rs9848497 up-regulates MST1R expression, thereby influencing leadership phenotypes. Proc. Natl. Acad. Sci. USA. 119, e2207847119. doi: 10.1073/pnas.2207847119
Raj, T., Ryan, K. J., Replogle, J. M., Chibnik, L. B., Rosenkrantz, L., Tang, A., et al. (2014). CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer's disease susceptibility. Hum. Mol. Genet. 23, 2729–2736. doi: 10.1093/hmg/ddt666
Salm, E. J., Dunn, P. J., Shan, L., Yamasaki, M., Malewicz, N. M., Miyazaki, T., et al. (2020). TMEM163 regulates ATP-gated P2X receptor and behavior. Cell Rep. 31, 107704. doi: 10.1016/j.celrep.2020.107704
Scheltens, P., Blennow, K., Breteler, M., de Strooper, B., Frisoni, G., Salloway, S., et al. (2016). Alzheimer's disease. Lancet 388, 505–517. doi: 10.1016/S0140-6736(15)01124-1
Schwartzentruber, J., Cooper, S., Liu, J. Z., Barrio-Hernandez, I., Bello, E., Kumasaka, N., et al. (2021). Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer's disease risk genes. Nature Genet. 53, 392. doi: 10.1038/s41588-020-00776-w
Sensi, S. L., Granzotto, A., Siotto, M., and Squitti, R. (2018). Copper and zinc dysregulation in Alzheimer's disease. Trends Pharmacol. Sci. 39, 1049–1063. doi: 10.1016/j.tips.2018.10.001
Small, S. A., and Petsko, G. A. (2015), Retromer in Alzheimer disease, Parkinson disease other neurological disorders. Nat. Rev. Neurosci. 16, 126–132. doi: 10.1038/nrn3896
Wainberg, M., Sinnott-Armstrong, N., Mancuso, N., Barbeira, A. N., Knowles, D. A., Golan, D., et al. (2019). Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599. doi: 10.1038/s41588-019-0385-z
Ward, L. D., and Kellis, M. (2016). HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881. doi: 10.1093/nar/gkv1340
Xie, A., Gao, J., Xu, L., and Meng, D. (2014). Shared mechanisms of neurodegeneration in Alzheimer's disease and Parkinson's disease. Biomed Res. Int. 2014, 648740. doi: 10.1155/2014/648740
Xu, M., Zhang, D.-F., Luo, R., Wu, Y., Zhou, H., Kong, L.-L., et al. (2018). A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer's disease. Alzheimers. Dement. 14, 215–229. doi: 10.1016/j.jalz.2017.08.012
Xu, Y., Xiao, G., Liu, L., and Lang, M. (2019). Zinc transporters in Alzheimer's disease. Mol. Brain 12, 106. doi: 10.1186/s13041-019-0528-2
Yu, J. T., Tan, L., and Hardy, J. (2014). Apolipoprotein E in Alzheimer's disease: an update. Annu. Rev. Neurosci. 37, 79–100. doi: 10.1146/annurev-neuro-071013-014300
Zhang, D.-F., Fan, Y., Xu, M., Wang, G., Wang, D., Li, J., et al. (2019). Complement C7 is a novel risk gene for Alzheimer's disease in Han Chinese. Natl Sci Rev 6, 257–274. doi: 10.1093/nsr/nwy127
Zhu, A., Matoba, N., Wilson, E. P., Tapia, A. L., Li, Y., Ibrahim, J. G., et al. (2021). MRLocus: Identifying causal genes mediating a trait through Bayesian estimation of allelic heterogeneity. PLoS Genet. 17, e1009455. doi: 10.1371/journal.pgen.1009455
Zhu, X.-C., Cao, L., Tan, M.-S., Jiang, T., Wang, H.-F., Lu, H., et al. (2017). Association of Parkinson's disease GWAS-linked loci with Alzheimer's disease in han Chinese. Mol. Neurobiol. 54, 308–318. doi: 10.1007/s12035-015-9649-5
Keywords: TMEM163, Alzheimer's disease, rs6430538, genome-wide association study, expression quantitative trait loci, multi-omics data
Citation: Qiu S, Sun M, Xu Y and Hu Y (2024) Integrating multi-omics data to reveal the effect of genetic variant rs6430538 on Alzheimer's disease risk. Front. Neurosci. 18:1277187. doi: 10.3389/fnins.2024.1277187
Received: 14 August 2023; Accepted: 26 February 2024;
Published: 18 March 2024.
Edited by:
Zhijie Han, Chongqing Medical University, ChinaReviewed by:
Min Xu, Chinese Academy of Sciences (CAS), ChinaIsmael Al-Ramahi, Baylor College of Medicine, United States
Copyright © 2024 Qiu, Sun, Xu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Hu, aHV5YW5nQGhpdC5lZHUuY24=
†These authors have contributed equally to this work
 Shizheng Qiu
Shizheng Qiu Meili Sun2†
Meili Sun2† Yang Hu
Yang Hu