
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 05 March 2024
Sec. Sleep and Circadian Rhythms
Volume 17 - 2023 | https://doi.org/10.3389/fnins.2023.1303727
This article is part of the Research Topic Women in Sleep and Circadian Rhythms View all 4 articles
Multiple studies have documented sex differences in sleep behaviour, however, the molecular determinants of such differences remain unknown. Furthermore, most studies addressing molecular mechanisms have been performed only in males, leaving the current state of knowledge biased towards the male sex. To address this, we studied the differences in the transcriptome of the cerebral cortex of male and female C57Bl/6 J mice after 6 h of sleep deprivation. We found that several genes, including the neurotrophin growth factor Bdnf, immediate early genes Fosb and Fosl2, and the adenylate cyclase Adcy7 are differentially upregulated in males compared to females. We identified the androgen-receptor activating transcription factor EZH2 as the upstream regulatory element specifying sex differences in the sleep deprivation transcriptome. We propose that the pathways downstream of these transcripts, which impact on cellular re-organisation, synaptic signalling, and learning may underpin the differential response to sleep deprivation in the two sexes.
Sleep is fundamental to many biological processes including metabolism, immunity, and memory formation. The mechanisms of sleep control are not fully understood, but it has been posited that sleep is controlled by a homeostatic process (Process S) and a circadian process (Process C), that interact to regulate sleep/wake cycles (Borbély, 1982; Borbély et al., 2016). Process S, or the homeostatic sleep drive, depends on the intensity and duration of the preceding period of wakefulness. Whereas Process C is controlled by the master circadian pacemaker/clock located in the suprachiasmatic nucleus (SCN), for which the key zeitgeber is light-information received from intrinsically photosensitive retinal ganglion cells (ipRGCs; Eban-Rothschild et al., 2018).
Circadian and sleep disruption can impact health, including cognitive function in mammals (Simon et al., 2022). The transcriptomic changes associated with the homeostatic response to sleep deprivation have been examined in rodents using microarrays and RNA-seq with results showing that immediate early genes (IEGs) Homer Scaffolding Protein 1 (Homer1), Activity Regulated Cytoskeleton Associated Protein (Arc), and brain-derived neurotrophic factor (Bdnf) are consistently upregulated (Maret et al., 2007; Hinard et al., 2012). Sleep deprivation has also been shown to regulate circadian clock genes with upregulation of Period 1/2 (Per1/2) and downregulation of D-Box Binding PAR BZIP Transcription Factor (Dbp) (Cirelli et al., 2004; Terao et al., 2006; Mackiewicz et al., 2007; Wisor et al., 2008; Thompson et al., 2010; Gerstner et al., 2016). However, one caveat of these studies is that they only used male rodents. Under-representation of females in neuroscience research is common due to the notion that inclusion of female rodents would lead to greater variability in comparison to male rodents. However, one meta-analysis looking at 9,932 traits has disputed this assumption, highlighting how there is no substantial difference between female and male variability (Prendergast et al., 2014). This bias has therefore limited our understanding of sex differences, across the field of neurobiology, and more specifically within sleep and circadian research.
In sleep and circadian research, there is growing evidence for sex differences in sleep and circadian physiology in both humans (Mong and Cusmano, 2016; Santhi et al., 2016; Carrier et al., 2017; Manuel et al., 2022) and murine models (Dib et al., 2021). In humans, these sex differences in sleep can be summarised as women reporting poorer sleep quality with more frequent insomnia symptoms than men (Li et al., 2002; Mong et al., 2011). However, quantitative studies have shown that women exhibit an overall longer sleep time than men, with EEG results showing women have higher EEG power in most frequencies during the two main sleep states [non-rapid eye movement (NREM) and rapid eye movement (REM)] suggesting higher quality NREM sleep and lower quality REM sleep (Dijk et al., 1989; Carrier et al., 2001; Long et al., 2021). These differences are dependent on other factors including age, and phase of estrous cycle, and species. In mice, studies indicate that males exhibit reduced wakefulness, but when in proestrus, females show increased wakefulness compared to males during the light and early dark period (Swift et al., 2020). Rodent sleep studies investigating sex differences have demonstrated that gonadectomy (GDX) eliminates sex differences in wakefulness by significantly decreasing the disparity in females, thus suggesting ovarian hormones contribute to time spend sleeping in rodents with female-specific brain organisation (Paul et al., 2006; Cusmano et al., 2014). EEG studies in mice provide conflicting results as to whether males spend more or less time in NREM sleep than females (Paul et al., 2006; Nichols et al., 2020; Choi et al., 2021), but the majority report higher NREMS sleep in males (Dib et al., 2021). Further studies focusing on the contribution of sex chromosomes demonstrate that males with either XX or XY chromosomes spend more time in NREM than females with XX or XY chromosomes, suggesting gonadal hormones regulate NREM sleep (Ehlen et al., 2013; Nichols et al., 2020).
Despite this evidence, our mechanistic understanding of these differences is poor. Current research suggests the involvement of both hormone-dependent and hormone independent elements (Paul et al., 2009). Previous rodent studies using acute sleep deprivation (6–8 h) found that females exhibit either a stronger (Paul et al., 2006) or weaker (Nichols et al., 2020; Choi et al., 2021) NREM rebound in the recovery sleep, under different experimental conditions. Again, these differences were reduced by GDX (Paul et al., 2006), providing more support for ovarian hormone contribution to sex differences in homeostatic sleep regulation. To explore the transcriptomic basis for these differences, we performed RNA-seq on the cerebral cortex of acutely sleep-deprived mice of both sexes.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, AJ (YWFydGkuamFnYW5uYXRoQG5kY24ub3guYWMudWs=).
This study did not generate new unique reagents.
All RNA-Seq data have been deposited on NCBI SRA and will be publicly available at https://www.ncbi.nlm.nih.gov/sra/PRJNA1071110 as of the date of publication. No original code was used in this study. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
All studies were conducted using male and female C57BL/6 J mice over 8 weeks of age and, unless otherwise indicated, animals were group housed with ad libitum access to food and water under a 12h/12h light/dark cycle (100 lux from white LED lamps). All animal procedures were conducted in accordance with the United Kingdom Home Office regulations [Guidance on the Operation of Animals (Scientific Procedures Act) 1986] and the University of Oxford’s Policy on the Use of Animals in Scientific research, following the principles of the 3Rs.
For the Sleep Deprivation (SD) experiments, animals were kept awake for 6 h between ZT0 (Zeitgeber time 0 or lights on within the 12 h/12 h light/dark cycle) and ZT6 (Zeitgeber time 6 or 6 h into the light period within the 12 h/12 h light/dark cycle) by providing novel objects to elicit exploratory behaviour, as previously described (Huber et al., 2000). The animals were then sacrificed (four males and four females per condition), and somatosensory cortical tissue punches collected. Control animals were allowed to sleep ad libitum between ZT0 and ZT6. Variation in sleep/wake and circadian behaviour through the oestrous cycle (Dib et al., 2021; Joye and Evans, 2022) is often a reason to exclude female mice during experiments and oestrous cycle monitoring can be cumbersome, which can lead many experimentalists to avoid using females. We did not monitor oestrous cycle, in order to capture variation as would be expected if oestrous stage is not controlled.
Total RNA from cortical punches was extracted using TRIzol and the RNeasy Mini Kit (Qiagen). Cortical tissue was mechanically disrupted in 700 μL of TRIzol and 140 μL of chloroform was added and the sample thoroughly mixed. Following a 3 min incubation at RT, the sample was then centrifuged for 15 min at 15,000 × g, 4°C. The clear top layer was then carefully collected, mixed with an equal volume of 70% ethanol and RNA extracted using the RNeasy Mini Kit, with on-column DNase digestion, following the manufacturer’s instructions. RNA was eluted in water and RNA concentration and quality were measured using a TapeStation system (Agilent) with the High Sensitivity RNA ScreenTape assay. mRNA purification and cDNA synthesis for the sequencing library were performed according to the Illumina Stranded mRNA Prep protocol (20040534) using the following index kit: IDT for Illumina RNA UD Indexes Set A, Ligation (20040553). Quality and concentration of the final libraries were checked with the KAPA Library Quantification Kit (Roche Diagnostics) in a StepOnePlus thermal cycler (Applied Biosystems) according to manufacturer’s instructions. All cDNA libraries were sequenced using a paired-end strategy (read length 150 bp) on an Illumina NovaSeq platform.
Raw RNA-Seq data processing (quality control, trimming, mapping to the genome, and read counting) was performed using tools embedded in Galaxy (v21.05) (Jalili et al., 2020). The fastqsanger files containing the raw sequencing data were uploaded to the public Galaxy server at usegalaxy.org. FastQC (v0.11.8; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for quality control of sequencing data. For quality and adapter trimming, Trim Galore! (v0.6.3; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was employed to remove low-quality bases, short reads, and Illumina adapters. High quality reads were then mapped to the Mus musculus (mm10) reference genome using HISAT2 (v2.1.0) (Kim et al., 2019), specifying the strand information as reverse. featureCounts (v2.0.1) (Liao et al., 2014) was run to quantify the number of reads mapped to each gene. The featureCounts built-in mm10 gene annotation file was selected and under paired-end reads options, the option to count fragments instead of reads was enabled. The generated counts files were converted to CSV and downloaded for downstream differential gene expression analysis in R. MultiQC (v1.9; https://multiqc.info; Ewels et al., 2016) was used to aggregate FastQC, HISAT2, and featureCounts results.
To identify differentially expressed genes in males versus females under baseline conditions and after sleep deprivation, the DESeq2 package (v1.32.0) (Love et al., 2014) was used in R (v4.2.0, https://www.r-project.org). DESeq2 corrects for multiple testing using the Benjamini-Hochberg (BH) method, and only genes with a BH adjusted p value <0.05 were considered statistically significant. To map differences in the response to sleep deprivation in the two sexes, we studied those genes that were differentially expressed in only one sex AND displayed a difference of greater that 0.5 log2-fold change after sleep deprivation. Heatmaps were drawn using the pheatmap function from the pheatmap package (v1.0.12). Volcano plots were generated using the ggplot2 package (v.3.3.5). STRING (database of known and predicted protein–protein interactions) interaction network mapping and functional annotation was conducted using string-db.org.
A comparison of the sleep deprivation regulated transcriptome (Supplementary Table 1) of the cortex in males vs. female mice showed that broadly, the top 100 differential transcripts were similarly regulated in both sexes. For example, Homer1, Iodothyronine deiodinase 2 (Dio2), Dual Specificity Phosphatase 4 (Dusp4), Cyclin Dependent Kinase Inhibitor 1a (Cdkn1a), Dbp, and Cold Inducible RNA Binding Protein (Cirbp), all shown previously to be regulated by sleep deprivation, changed consistently in both sexes (Figure 1A). 11 transcripts were identified as significantly different between males and females in a two-way comparison at baseline (Male Sham vs. Female Sham) and 12 after sleep deprivation (Figure 1B). Many of these genes are on sex chromosomes (e.g., Ddx3y, Eif2s3y, and Eif2s3x), but several were not—including the Acer2 (alkaline ceramiase 2 which catalyses the hydrolysis of ceramides into sphingoid bases), Lcat (extracellular cholesterol esterifying enzyme lecithin-cholesterol acyltransferase), and Scn7a (voltage gated sodium channel protein type 7 subunit alpha), which lie on chromosomes 4, 8, and 2, respectively. These results demonstrate sex-specific gene regulation that is not related to the sex chromosomes.
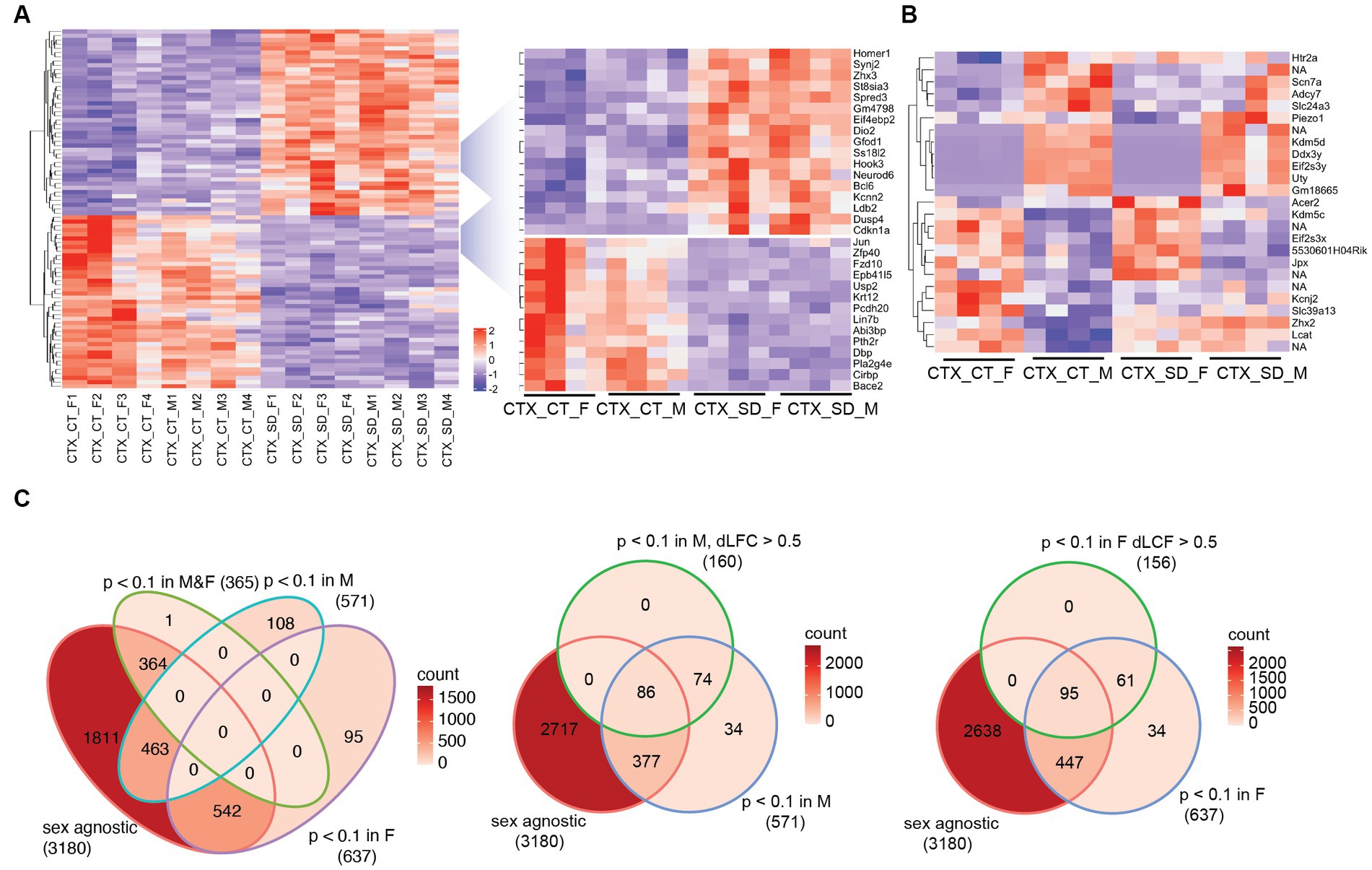
Figure 1. The transcriptional response in the cortex in response to sleep deprivation: (A) The top 100 differentially expressed transcripts in the cortex (CTX) under control conditions (CT) or 6 h sleep deprivation (SD) in four males (M1–4) and four females (F1–4). Key upregulated and downregulated clusters highlighted on the right. (B) Transcripts that are significantly different in a direct comparison across the two sexes, either after sleep deprivation (SD) or under control (CT) conditions. (C) Venn diagrams showing the transcript numbers that are significantly different after sleep deprivation in the different conditions shown [sex agnostic, padj < 0.1 in males only (M), females only (F), males and females (M&F)]. The central and right panel show those transcripts that are padj < 0.1 in males only (M) or females only (F), but also differ between males and females by a log fold change of >0.5 (dLFC > 0.5). NA is non-coding transcript with no associated genes.
We next assessed the relative response to sleep deprivation (SD vs. sham), between males and females. In total, 3,180 transcripts were seen to change in response to sleep deprivation (padj. < 0.1) when sex was not included as a factor in the analysis. Of these, 364 were significant in both sexes, with approximately 600 genes significantly different in only one sex and not the other (Figure 1C). However, it is important to consider that the power of the experiment is reduced when analysing the two sexes separately (n = 4 for each sex vs. n = 8 for both sexes). It is possible that the direction and general magnitude of change in both sexes is similar, but passes the statistical significance threshold for only one sex. To account for this, we identified those transcripts which were both significantly differential in only one sex and also different between the sexes by greater than 0.5 log2 fold change. In this manner, 316 transcripts were identified as changing in a sex-specific manner: 160 in males, and 156 in females (Figure 1C; Supplementary Table 2). Several of these transcripts are illustrated in Figure 2A.
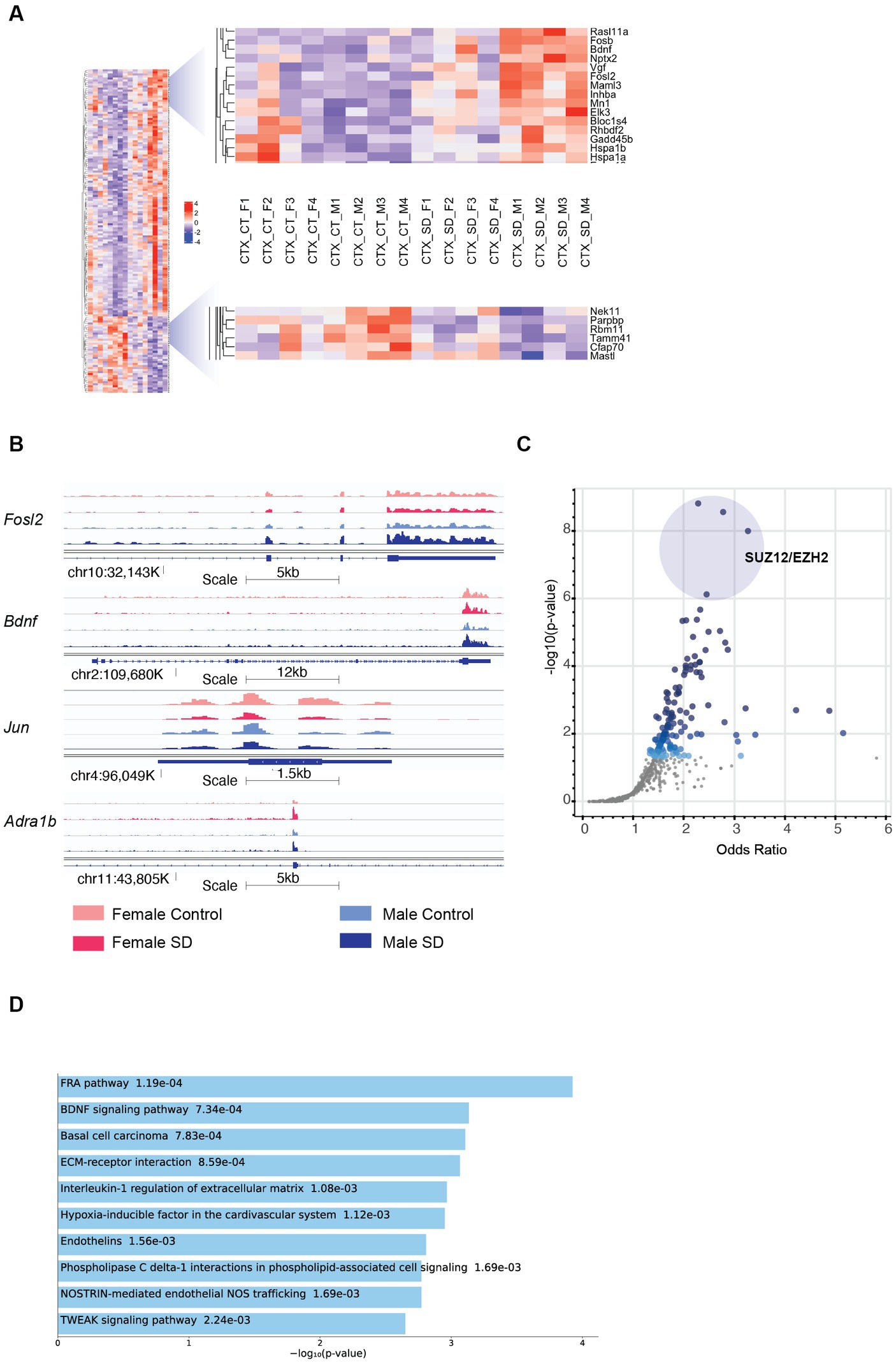
Figure 2. Sex-specific patterns of transcription in the cortex in response to sleep deprivation: (A) The transcripts with sex-specific regulation in response to sleep deprivation, i.e., padj < 0.1 in males only (M) OR females only (F), and also transcripts that differ between males and females by a log fold change of >0.5 (dLFC > 0.5). Legend for samples as in Figure 1. (B) RNA-Seq tracks for genes of interest as labelled, genomic position and scale indicated. (C) Transcription Factor prediction for the transcripts in (A) using CHEA Transcription Factor Targets dataset on Enrichr. Binding sites for EZH2 and SUZ12 are significantly enriched, p value indicated on Y-axis and X-axis the odds ratio (z-score assessing the rank of a term from a Fisher’s exact test against the expected rank of the term in the gene-set library). (D) Functional annotation of the transcripts in panel (A) from the Enrichr platform, Bioplanet 2019 results reported, the number alongside the term reports p value.
Comparison between males and females shows that Brain Derived Neurotrophic Factor (Bdnf; key to plasticity, learning, and memory) (Parkhurst et al., 2013; Hill and Martinowich, 2016) and its targets (immediate early genes Egr3, Fosb, and Fosl2) are upregulated more in males following sleep deprivation, as confirmed using pathway analysis (p = 9.3e−5; Figures 2A,B). Increased transcription around exon 4 (Bdnf variant 4) is observed in both sexes after SD, but transcription from the coding exons in increased more strongly in males (Supplementary Figure S2C). The same pattern of higher upregulation in males is seen with heat shock proteins (Hsp1a and Hsp1b; Figure 2A). In contrast, Jun, and the adrenergic receptor Adra1b show significantly stronger downregulation and upregulation, respectively, in females (Figure 2B). Transcription factor Jun is one of the primary effectors of hypoxia inducible factor 1a (HIF1a), for which interestingly pathway analysis shows that other HIF signalling transcripts such as Nostrin (upregulated) and endothelin1/3 (downregulated) are differentially expressed in females following sleep deprivation, but not males. Other genes of interest include the RNA binding protein Rbm11, which shows stronger downregulation in males (Figure 2A). Clock genes showed broadly similar responses to sleep deprivation as described previously, and in both sexes (Wisor et al., 2002; Mongrain et al., 2010).
To identify upstream regulatory elements that could explain these sex specific differences, transcription factor binding analysis using CHEA Transcription Factor Targets dataset was conducted. This analysis showed that binding sites for Enhancer of zeste homologue 2 (EZH2) and SUZ12 were significantly enriched in males (Figure 2C), and included transcripts such as Adcy7, Bdnf, and Fosl2 which are all upregulated only in males. EZH2 is a histone methyltransferase which binds to regulatory subunits, one of which is SUZ12, to form the polycomb repressor complex 2 (PRC2) which plays a role in gene silencing (Blackledge and Klose, 2021). EZH2 has multiple binding partners through which it binds to DNA, one of which includes E2F6. Inspection of the promoter regions using ENCODE Registry of candidate cis-Regulatory Elements (cCREs) and JASPAR Transcription Factor Binding Site Database from USCS browser identified E2F6 binding sites in both Bdnf and Adra1B promoters (Table 1; Figure 2). Similar analysis revealed both Androgen receptor (AR) and Estrogen receptor 1/2 (ESR1/2) binding sites in both Bdnf and Adra1B promoters (Table 1).
To understand the functional relevance of the genes that demonstrated sex-specific changes in their response to sleep deprivation, we conducted pathway analysis on the Enrichr platform (Chen et al., 2013), from which Bioplanet 2019 results are reported in Figure 2D. BDNF, HIF, and NOS signalling are all significantly different between the sexes. STRING analysis to generate Protein–Protein Interaction Networks and to assess functional enrichment within connected networks (Figure 3). We found that the network had a hub at JUN which linked to a cluster containing BDNF, EGR3, FOSB, FOSL2, and HSP1A, all of which are upregulated in males but not females (See Figures 1A,B). Other clusters included sex-chromosome specific genes (FDR = 0.006, Supplementary Figure S1A), vascular endothelin signalling genes (FDR = 0.01, Supplementary Figure S1B), and collagen-containing extracellular matrix genes (FDR = 0.0001, Supplementary Figure S1C) with Fn1 (Fibronectin 1) at the hub, a gene which is upregulated only in males.
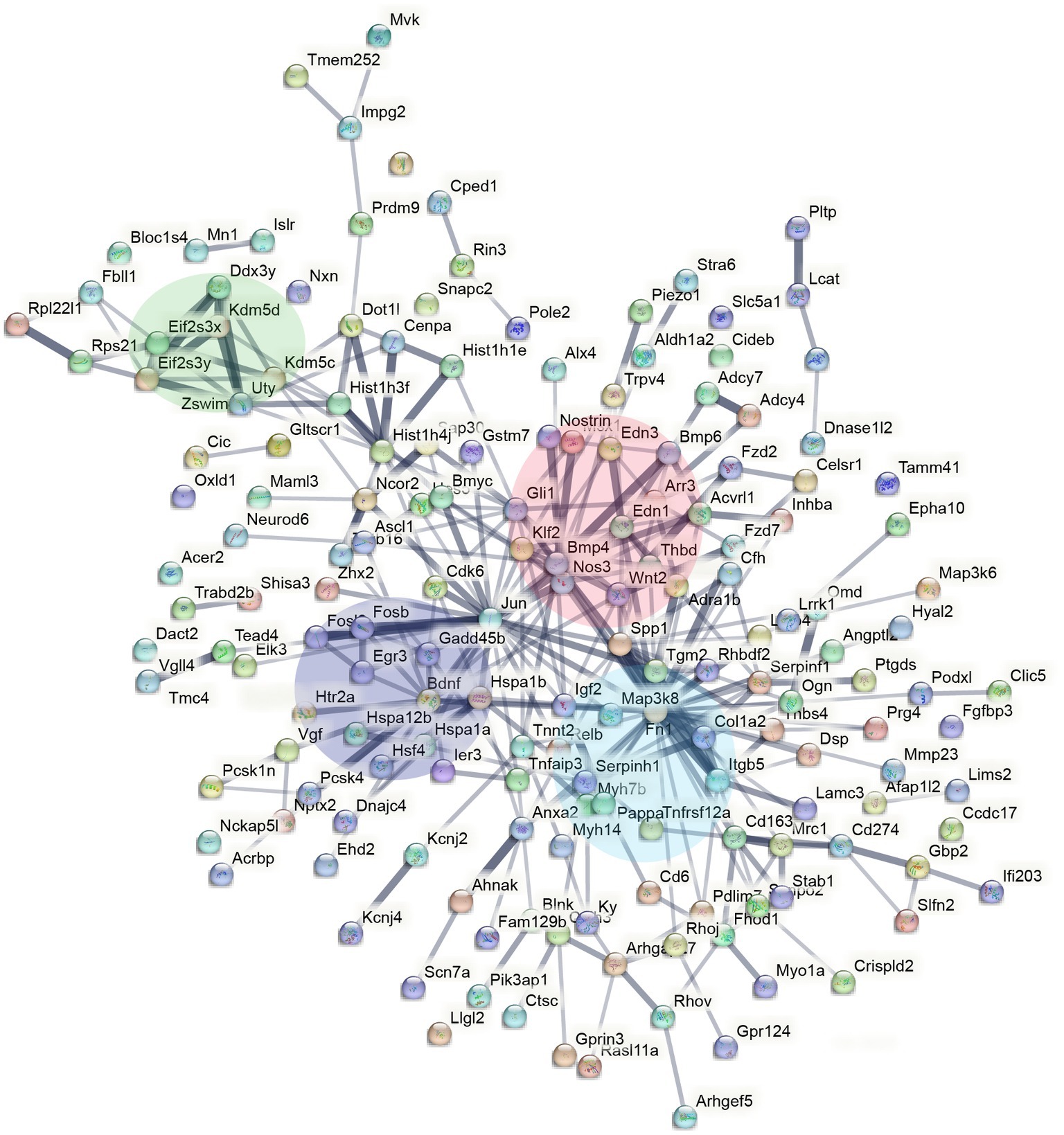
Figure 3. STRING interaction network mapping of the transcripts in Figure 2A showing clusters centering around BDNF signalling (purple), sex-chromosome specific genes (green), endothelin signalling (red), and FN1/extracellular matrix (blue).
The molecular mechanisms that underlie sex differences in sleep phenotypes remain largely unknown. To this end, our studies show that there are subtle, but important differences in male and female cortical transcription profiles following acute sleep deprivation in mice. Most of the genes long-established to be differentially regulated following sleep deprivation, such as DBP, demonstrate no sex difference (Figure 1A). However, a small number of genes show differential regulation in only one sex, such as FOSL2. These genes appear to cluster around BDNF, JUN, FN1, and BMP4 in the protein–protein interaction network predicted by STRING.
The first of the molecular pathways differentially regulated in both sexes following sleep deprivation involves BDNF signalling. BDNF has neurotrophic plasticity roles (Bramham and Messaoudi, 2005) related to neuronal survival, learning, memory, and sleep (Giese et al., 2014; Bathina and Das, 2015). Unsurprisingly, reduced levels are associated with neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease, but also depression and stress-related mental disorders for which sleep problems are a common symptom. This study shows that transcription of BDNF is increased significantly in males following sleep deprivation, suggesting the role of BDNF is reduced in females which would affect synapse plasticity. This differential signalling mechanism offers a rationale for the observed sex-difference in which female rodents experience a greater impact on memory and cognitive performance due to sleep deprivation compared to males (Fernandes-Santos et al., 2012; Hajali et al., 2012).
Bone morphogenetic protein 4 (Bmp4) serves as the hub for a cluster of genes related to endothelial signalling. Many elements of this pathway are differentially regulated in a sex-specific manner after sleep deprivation (Supplementary Figure S1B). The gene alkaline ceramidase 2 (Acer2) is upregulated in females only following sleep deprivation (Figure 1B; Supplementary Figure S2B), it encodes an enzyme that hydrolyses ceramides to generate the bioactive lipid sphindosine-1-phosphate (S1P; Li et al., 2018). S1P regulates HIF-1a activity (Kalhori et al., 2013) and is known to be a major regulator of endothelial function (Supplementary Figure S2A). These findings suggest that lipid signalling is different in males and females and given the growing evidence demonstrating a correlation between lipid profile and amount of sleep (Kaneita et al., 2008), the upregulation of Acer2 exclusively in females suggests an interesting sex-specific role of S1P signalling in regulating sleep.
A major hub of the network that is regulated by sleep in a sex-specific manner is Fibronectin 1 (Fn1), which regulates extracellular matrix assembly and function (Parisi et al., 2020). This is surprising as not much is known about its role in sleep regulation, although some evidence points to the dysfunction of extracellular matrix molecules as part of the mechanism underlying synaptic dysfunction in psychiatric disorders and memory consolidation during sleep (Gisabella et al., 2021). Overall, further investigation is required to establish the functional relevance of these findings.
Similarly, not much is known about the role of ADRA1B in sleep regulation. Using linkage analysis and whole-exome sequencing, a mutation in Adra1b gene has been identified in affected people with familial natural short sleep 2 (FNSS2) who have a lifelong reduction in sleep duration without suffering the cognitive consequences of sleep deprivation (Shi et al., 2019). Results of subsequent mice studies suggest a role of ADRA1B in activating neurons during REM sleep and wakefulness. Our study shows that expression of the normal gene is upregulated in females following sleep deprivation, it would be interesting to determine if the mutated form exhibits any sex-bias and whether this affects the phenotype of women with FNSS.
Through the promoter analysis, a network can start to be formed with EZH2 playing an important role at the top of the pathway. A study has shown that, separate to its PRC-and methylation-roles, EZH2 binds directly to the Androgen receptor (Ar) promoter and acts as a transcriptional activator of the Ar gene (Kim et al., 2018). Ar is a nuclear sex hormone receptor, which along with ESR1/2 (estrogen receptors 1/2), mediate downstream signalling events that control gene expression (Fuentes and Silveyra, 2019). JASPER promoter analysis identified multiple binding sites for these transcription factors in the promoter of Bdnf and Adra1b. Since EZH2 recruits Ar directly, enrichment of EZH2 binding sites following sleep deprivation correlates with male-only increase in expression of genes, such as BDNF, for which both Ar and EZH2 can act as a transcriptional activator. In support, we identified binding sites for Ar and EZH2 in key genes that were regulated in sex-specific manner (Table 1). Interestingly, a study investigating the sex differences in liver transcription programmes showed that EZH2 also plays a key transcriptional role leading to sex-bias in susceptibility to fibrosis and other liver diseases (Lau-Corona et al., 2020). Whilst this supports our identification of EZH2 as a potential mediator of sex-specific transcriptional programmes in response to sleep deprivation, the exact influence of this transcription factor requires further investigation.
This study is limited by the fact that we have not carried out any functional validation of the transcriptomic changes. Furthermore, transcriptomic changes are not necessarily reflected at the level of the protein, where translational and post-translational regulation can significantly alter the outcomes from transcriptional changes. Nevertheless, the goal of this study was to provide a description of transcriptional pathways that would inform future studies. It revealed some intriguing molecular pathways that are differentially regulated in both sexes following sleep deprivation that may provide important insight into the mechanisms that underlie sleep regulation and the response to loss of sleep. Additionally, this data describes, for the first time, how male and female mice react differently at the transcriptomic level to sleep deprivation. These small subtle differences could have big functional implications and shows that females process the signals from sleep deprivation differently, with the identified genes providing a groundwork for further investigation to uncover the molecular pathways that underlie sex differences in sleep regulation. To conclude, although traditionally neuroscience research is conducted exclusively in males, this study has shown that many of the key transcripts associated with sleep deprivation respond similarly in both sexes with differences not widespread and therefore females can be easily factored into analysis. This has strengthened the case for the inclusion of females in future research.
All sequencing data have been deposited at NCBI SRA at the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA1071110.
The animal study was approved by United Kingdom Home Office regulations [Guidance on the Operation of Animals (Scientific Procedures Act) 1986] and the University of Oxford’s Policy on the Use of Animals in Scientific research. The study was conducted in accordance with the local legislation and institutional requirements.
TS: Formal Analysis, Writing – original draft, Visualization. QD: Formal Analysis, Investigation, Writing – review & editing. LT: Investigation, Supervision, Writing – review & editing. AJ: Supervision, Conceptualization, Formal Analysis, Funding acquisition, Writing – original draft, Writing – review & editing. IS: Formal Analysis, Writing – review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the following sources of funding: BB/N01992X/1 David Phillips fellowship from the BBSRC to AJ, and Oxford-Elysium Cellular Health Fellowship to LT.
AJ is a founder Circadian Therapeutics Ltd. and hold shares in the company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2023.1303727/full#supplementary-material
Bathina, S., and Das, U. N. (2015). Brain-derived neurotrophic factor and its clinical implications. Arch. Med. Sci. 11, 1164–1178. doi: 10.5114/aoms.2015.56342
Blackledge, N. P., and Klose, R. J. (2021). The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833. doi: 10.1038/s41580-021-00398-y
Borbély, A. A., Daan, S., Wirz-Justice, A., and Deboer, T. (2016). The two-process model of sleep regulation: A reappraisal. J. Sleep Res. 25, 131–143. doi: 10.1111/jsr.12371
Bramham, C. R., and Messaoudi, E. (2005). BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog. Neurobiol. 76, 99–125. doi: 10.1016/j.pneurobio.2005.06.003
Carrier, J., Land, S., Buysse, D. J., Kupfer, D. J., and Monk, T. H. (2001). The effects of age and gender on sleep EEG power spectral density in the middle years of life (ages 20–60 years old). Psychophysiology 38, 232–242. doi: 10.1111/1469-8986.3820232
Carrier, J., Semba, K., Deurveilher, S., Drogos, L., Cyr-Cronier, J., Lord, C., et al. (2017). Sex differences in age-related changes in the sleep-wake cycle. Front. Neuroendocrinol. 47, 66–85. doi: 10.1016/j.yfrne.2017.07.004
Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G. V., et al. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14. doi: 10.1186/1471-2105-14-128
Choi, J., Kim, S. J., Fujiyama, T., Miyoshi, C., Park, M., Suzuki-Abe, H., et al. (2021). The role of reproductive hormones in sex differences in sleep homeostasis and arousal response in mice. Front. Neurosci. 15:739236. doi: 10.3389/fnins.2021.739236
Cirelli, C., Gutierrez, C. M., and Tononi, G. (2004). Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41, 35–43. doi: 10.1016/S0896-6273(03)00814-6
Cusmano, D. M., Hadjimarkou, M. M., and Mong, J. A. (2014). Gonadal steroid modulation of sleep and wakefulness in male and female rats is sexually differentiated and neonatally organized by steroid exposure. Endocrinology 155, 204–214. doi: 10.1210/en.2013-1624
Dib, R., Gervais, N. J., and Mongrain, V. (2021). A review of the current state of knowledge on sex differences in sleep and circadian phenotypes in rodents. Neurobiol. Sleep Circad. Rhyth. 11:100068. doi: 10.1016/j.nbscr.2021.100068
Dijk, J., Beersma, D. G. M., and Bloem, M. (1989). Sex differences in the sleep EEG of young adults: visual scoring and spectral analysis. Sleep 12, 500–507. doi: 10.1093/sleep/12.6.500
Eban-Rothschild, A., Appelbaum, L., and de Lecea, L. (2018). Neuronal mechanisms for sleep/wake regulation and modulatory drive. Neuropsychopharmacology 43, 937–952. doi: 10.1038/npp.2017.294
Ehlen, J. C., Hesse, S., Pinckney, L., and Paul, K. N. (2013). Sex chromosomes regulate nighttime sleep propensity during recovery from sleep loss in mice. PLoS One 8:e62205. doi: 10.1371/journal.pone.0062205
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/BIOINFORMATICS/BTW354
Fernandes-Santos, L., Patti, C. L., Zanin, K. A., Fernandes, H. A., Tufik, S., Andersen, M. L., et al. (2012). Sleep deprivation impairs emotional memory retrieval in mice: influence of sex. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 38, 216–222. doi: 10.1016/j.pnpbp.2012.03.014
Fuentes, N., and Silveyra, P. (2019). “Estrogen receptor signaling mechanisms” in Advances in Protein Chemistry and Structural Biology, vol. 116 (Academic Press Inc.), 135–170.
Gerstner, J. R., Koberstein, J. N., Watson, A. J., Zapero, N., Risso, D., Speed, T. P., et al. (2016). Removal of unwanted variation reveals novel patterns of gene expression linked to sleep homeostasis in murine cortex. BMC Genomics 17:727. doi: 10.1186/s12864-016-3065-8
Giese, M., Unternährer, E., Hüttig, H., Beck, J., Brand, S., Calabrese, P., et al. (2014). BDNF: An indicator of insomnia. Mol. Psychiatry 19, 151–152. doi: 10.1038/mp.2013.10
Gisabella, B., Babu, J., Valeri, J., Rexrode, L., and Pantazopoulos, H. (2021). Sleep and memory consolidation dysfunction in psychiatric disorders: evidence for the involvement of extracellular matrix molecules. Front. Neurosci. 15:569. doi: 10.3389/fnins.2021.646678
Hajali, V., Sheibani, V., Esmaeili-Mahani, S., and Shabani, M. (2012). Female rats are more susceptible to the deleterious effects of paradoxical sleep deprivation on cognitive performance. Behav. Brain Res. 228, 311–318. doi: 10.1016/j.bbr.2011.12.008
Hill, J. L., and Martinowich, K. (2016). Activity-dependent signaling: influence on plasticity in circuits controlling fear-related behavior. Curr. Opin. Neurobiol. 36, 59–65. doi: 10.1016/j.conb.2015.10.001
Hinard, V., Mikhail, C., Pradervand, S., Curie, T., Houtkooper, R. H., Auwerx, J., et al. (2012). Key electrophysiological, molecular, and metabolic signatures of sleep and wakefulness revealed in primary cortical cultures. J. Neurosci. 32, 12506–12517. doi: 10.1523/JNEUROSCI.2306-12.2012
Huber, R., Deboer, T., and Tobler, I. (2000). Effects of sleep deprivation on sleep and sleep EEG in three mouse strains: empirical data and simulations. Brain Res. 857, 8–19. doi: 10.1016/S0006-8993(99)02248-9
Jalili, V., Afgan, E., Gu, Q., Clements, D., Blankenberg, D., Goecks, J., et al. (2020). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 48, W395–W402. doi: 10.1093/NAR/GKAA434
Joye, D. A. M., and Evans, J. A. (2022). Sex differences in daily timekeeping and circadian clock circuits. Sem. Cell Dev. Biol. 126, 45–55. doi: 10.1016/j.semcdb.2021.04.026
Kalhori, V., Kemppainen, K., Asghar, M. Y., Bergelin, N., Jaakkola, P., and Törnquist, K. (2013). Sphingosine-1-phosphate as a regulator of hypoxia-induced factor-1α in thyroid follicular carcinoma cells. PLoS One 8:e66189. doi: 10.1371/journal.pone.0066189
Kaneita, Y., Uchiyama, M., Yoshiike, N., and Ohida, T. (2008). Associations of usual sleep duration with serum lipid and lipoprotein levels. Sleep 31, 645–652. doi: 10.1093/sleep/31.5.645
Kim, J., Lee, Y., Lu, X., Song, B., Fong, K. W., Cao, Q., et al. (2018). Polycomb-and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 25, 2808–2820.e4. doi: 10.1016/j.celrep.2018.11.035
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Lau-Corona, D., Bae, W. K., Hennighausen, L., and Waxman, D. J. (2020). Sex-biased genetic programs in liver metabolism and liver fibrosis are controlled by EZH1 and EZH2. PLoS Genet. 16:e1008796. doi: 10.1371/journal.pgen.1008796
Li, R. H. Y., Wing, Y. K., Ho, S. C., and Fong, S. Y. Y. (2002). Gender differences in insomnia-a study in the Hong Kong Chinese population. J. Psychosom. Res. 53, 601–609. doi: 10.1016/S0022-3999(02)00437-3
Li, F., Xu, R., Low, B. E., Lin, C. L., Garcia-Barros, M., Schrandt, J., et al. (2018). Alkaline ceramidase 2 is essential for the homeostasis of plasma sphingoid bases and their phosphates. FASEB J. 32, 3058–3069. doi: 10.1096/fj.201700445RR
Liao, Y., Smyth, G. K., and Shi, W. (2014). Feature counts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Long, S., Ding, R., Wang, J., Yu, Y., Lu, J., and Yao, D. (2021). Sleep quality and electroencephalogram delta power. Front. Neurosci. 15:803507. doi: 10.3389/fnins.2021.803507
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mackiewicz, M., Shockley, K. R., Romer, M. A., Galante, R. J., Zimmerman, J. E., Naidoo, N., et al. (2007). Macromolecule biosynthesis: a key function of sleep. Physiol. Genomics 31, 441–457. doi: 10.1152/physiolgenomics.00275.2006
Manuel, S., Nayantara, S., Amrita, A., Dorothee, F., Lilian, H., Natasha, A. K., et al. (2022). Sex differences and sex bias in human circadian and sleep physiology research. elife 11:e65419. doi: 10.7554/eLife.65419
Maret, S., Dorsaz, S., Gurcel, L., Pradervand, S., Petit, B., Pfister, C., et al. (2007). Homer 1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. U. S. A. 104, 20090–20095. doi: 10.1073/pnas.0710131104
Mong, J. A., Baker, F. C., Mahoney, M. M., Paul, K. N., Schwartz, M. D., Semba, K., et al. (2011). Sleep, rhythms, and the endocrine brain: influence of sex and gonadal hormones. J. Neurosci. 31, 16107–16116. doi: 10.1523/JNEUROSCI.4175-11.2011
Mong, J. A., and Cusmano, D. M. (2016). Sex differences in sleep: impact of biological sex and sex steroids. Philos. Trans. Roy. Soc. B. Biol. Sci. 371:20150110. doi: 10.1098/rstb.2015.0110
Mongrain, V., Hernandez, S. A., Pradervand, S., Dorsaz, S., Curie, T., Hagiwara, G., et al. (2010). Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep 33, 1147–1157. doi: 10.1093/sleep/33.9.1147
Nichols, I., Vincent, S., Hesse, S., Ehlen, J. C., Brager, A., and Paul, K. (2020). Sleep deprivation alters the influence of biological sex on active-phase sleep behavior. bioRxiv [Preprint]. doi: 10.1101/2020.02.21.958231
Parisi, L., Toffoli, A., Ghezzi, B., Mozzoni, B., Lumetti, S., and Macaluso, G. M. (2020). A glance on the role of fibronectin in controlling cell response at biomaterial interface. Jpn. Dental Sci. Rev. 56, 50–55. doi: 10.1016/j.jdsr.2019.11.002
Parkhurst, C. N., Yang, G., Ninan, I., Savas, J. N., Yates, J. R., Lafaille, J. J., et al. (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cells 155, 1596–1609. doi: 10.1016/j.cell.2013.11.030
Paul, K. N., Dugovic, C., Turek, F. W., and Laposky, A. D. (2006). Diurnal sex differences in the sleep-wake cycle of mice are dependent on gonadal function. Sleep 29, 1211–1223. doi: 10.1093/sleep/29.9.1211
Paul, K. N., Laposky, A. D., and Turek, F. W. (2009). Reproductive hormone replacement alters sleep in mice. Neurosci. Lett. 463, 239–243. doi: 10.1016/j.neulet.2009.07.081
Prendergast, B. J., Onishi, K. G., and Zucker, I. (2014). Female mice liberated for inclusion in neuroscience and biomedical research. Neurosci. Biobehav. Rev. 40, 1–5.
Santhi, N., Lazar, A. S., McCabe, P. J., Lo, J. C., Groeger, J. A., and Dijk, D. J. (2016). Sex differences in the circadian regulation of sleep and waking cognition in humans. Proc. Natl. Acad. Sci. U. S. A. 113, E2730–E2739. doi: 10.1073/pnas.1521637113
Shi, G., Xing, L., Wu, D., Bhattacharyya, B. J., Jones, C. R., McMahon, T., et al. (2019). A rare mutation of β1-adrenergic receptor affects sleep/wake behaviors. Neuron 103, 1044–1055.e7. doi: 10.1016/j.neuron.2019.07.026
Simon, K. C., Nadel, L., and Payne, J. D. (2022). The functions of sleep: A cognitive neuroscience perspective. Proc. Natl. Acad. Sci. 119:e2201795119. doi: 10.1073/pnas.2201795119
Swift, K. M., Keus, K., Echeverria, C. G., Cabrera, Y., Jimenez, J., Holloway, J., et al. (2020). Sex differences within sleep in gonadally intact rats. Sleep 43:zsz289. doi: 10.1093/sleep/zsz289
Terao, A., Wisor, J. P., Peyron, C., Apte-Deshpande, A., Wurts, S. W., Edgar, D. M., et al. (2006). Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix gene Chip study. Neuroscience 137, 593–605. doi: 10.1016/j.neuroscience.2005.08.059
Thompson, C. L., Wisor, J. P., Lee, C.-K., Pathak, S. D., Gerashchenko, D., Smith, K. A., et al. (2010). Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front. Neurosci. 4:165. doi: 10.3389/fnins.2010.00165
Wisor, J. P., O’Hara, B. F., Terao, A., Selby, C. P., Kilduff, T. S., Sancar, A., et al. (2002). A role of cryptochromes in sleep regulation. BMC Neurosci. 3:20. doi: 10.1186/1471-2202-3-20
Wisor, J. P., Pasumarthi, R. K., Gerashchenko, D., Thompson, C. L., Pathak, S., Sancar, A., et al. (2008). Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J. Neurosci. 28, 7193–7201. doi: 10.1523/JNEUROSCI.1150-08.2008
Keywords: sleep, cortex, transcriptomics, gene regulation, sex differences
Citation: Shi T, Shah I, Dang Q, Taylor L and Jagannath A (2024) Sex-specific regulation of the cortical transcriptome in response to sleep deprivation. Front. Neurosci. 17:1303727. doi: 10.3389/fnins.2023.1303727
Received: 28 September 2023; Accepted: 07 December 2023;
Published: 05 March 2024.
Edited by:
Victoria Revell, University of Surrey, United KingdomReviewed by:
Julie Seibt, University of Surrey, United KingdomCopyright © 2024 Shi, Shah, Dang, Taylor and Jagannath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aarti Jagannath, YWFydGkuamFnYW5uYXRoQG5kY24ub3guYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.