- 1Ministry of Culture, Sports and Labor, Jiangxi Gannan Health Vocational College, Ganzhou, Jiangxi, China
- 2Gannan University of Science and Technology, Ganzhou, Jiangxi, China
Introduction: In this study, we explore the potential benefits of integrating natural cognitive systems (medical professionals' expertise) and artificial cognitive systems (deep learning models) in the realms of medical image analysis and sports injury prediction. We focus on analyzing medical images of athletes to gain valuable insights into their health status.
Methods: To synergize the strengths of both natural and artificial cognitive systems, we employ the ResNet50-BiGRU model and introduce an attention mechanism. Our goal is to enhance the performance of medical image feature extraction and motion injury prediction. This integrated approach aims to achieve precise identification of anomalies in medical images, particularly related to muscle or bone damage.
Results: We evaluate the effectiveness of our method on four medical image datasets, specifically pertaining to skeletal and muscle injuries. We use performance indicators such as Peak Signal-to-Noise Ratio and Structural Similarity Index, confirming the robustness of our approach in sports injury analysis.
Discussion: Our research contributes significantly by providing an effective deep learning-driven method that harnesses both natural and artificial cognitive systems. By combining human expertise with advanced machine learning techniques, we offer a comprehensive understanding of athletes' health status. This approach holds potential implications for enhancing sports injury prevention, improving diagnostic accuracy, and tailoring personalized treatment plans for athletes, ultimately promoting better overall health and performance outcomes. Despite advancements in medical image analysis and sports injury prediction, existing systems often struggle to identify subtle anomalies and provide precise injury risk assessments, underscoring the necessity of a more integrated and comprehensive approach.
1. Introduction
The modern society has witnessed the growing significance of sports in people's daily lives; however, it also brings along the risk of sports injuries. Sports injuries, especially bone and muscle damages, not only impact athletes' performance and competitive state but may also lead to prolonged physical discomfort and health issues (Ba, 2020). Therefore, accurate injury prediction and timely prevention are crucial for athletes' recovery and performance. Medical images and biomechanical data play pivotal roles in sports injury prediction. Medical images, such as X-rays, CT scans, and MRI scans, offer detailed anatomical information to identify and locate potential bone and muscle injuries (Nie et al., 2021). Biomechanical data can measure and analyze athletes' movement patterns and mechanical characteristics, providing more comprehensive information for injury prediction.
In the aspect of medical image feature extraction, several studies have employed traditional convolutional neural network (CNN) models (Dhiman et al., 2022) like AlexNet and VGG for feature extraction. While these models excel in image classification tasks, their effectiveness in extracting complex features from medical images containing intricate bone and muscle structures is limited. Traditional CNN models encounter challenges in handling medical images with low contrast and high noise, leading to suboptimal prediction accuracy. In sports injury prediction, researchers typically employ recurrent neural network (RNN) models (Luca et al., 2022) to process time-series biomechanical data. However, traditional RNNs suffer from vanishing and exploding gradients, restricting their ability to model long-term dependencies in data sequences. Consequently, the performance of RNN models may fall short when dealing with complex movement patterns and mechanical characteristics.
To overcome the limitations of the aforementioned methods, researchers have been exploring combinations of various deep learning models to process medical images and biomechanical data, aiming to improve the accuracy and efficiency of sports injury prediction. For example, Lu et al. (2018) proposed a method that combines CNN with LSTM models, achieving promising results in medical image extraction and abnormality prediction. Nevertheless, this approach still faces limitations in modeling time-series data due to the constraints of traditional RNNs, which fail to capture long-term dependencies entirely. To address the issues in time-series data modeling, Guo et al. (2020) introduced the use of Bidirectional LSTM (BiLSTM) models to capture contextual information in biomechanical data, leading to an improved performance in sports injury prediction. However, BiLSTM models exhibit high computational complexity and long training times when handling long sequences. In addition to improvements with deep learning models, some researchers have explored the integration of attention mechanisms to enhance the focus on medical images and biomechanical data. Khatun et al. (2022) incorporated attention mechanisms in sports injury prediction, allowing the model to automatically learn and emphasize critical information affecting the prediction results. This method significantly improves prediction accuracy.
However, despite these enhancements that have partly improved the performance of medical image feature extraction and sports injury prediction, some challenges persist. For instance, certain methods lack sufficient joint analysis of medical images and biomechanical data, preventing the exploration of their potential associations. Additionally, certain approaches entail high computational complexity, hindering their real-time prediction capabilities.
To address the aforementioned challenges and further optimize medical image feature extraction and sports injury prediction, this paper proposes an attention mechanism optimized method based on ResNet50 and BiGRU. Leveraging the feature extraction capabilities of ResNet50 and the contextual modeling abilities of BiGRU, and integrating the attention mechanism, this approach enables more accurate and efficient sports injury prediction, providing better support for athletes' health and performance.
The contribution points of this paper are as follows:
• This paper combines ResNet50 and BiGRU, two deep learning models, to achieve joint analysis of medical images and biomechanical data, enriching the feature representation for sports injury prediction.
• An attention mechanism is introduced, enabling the model to focus more on information significantly impacting prediction results, thereby improving accuracy and interpretability.
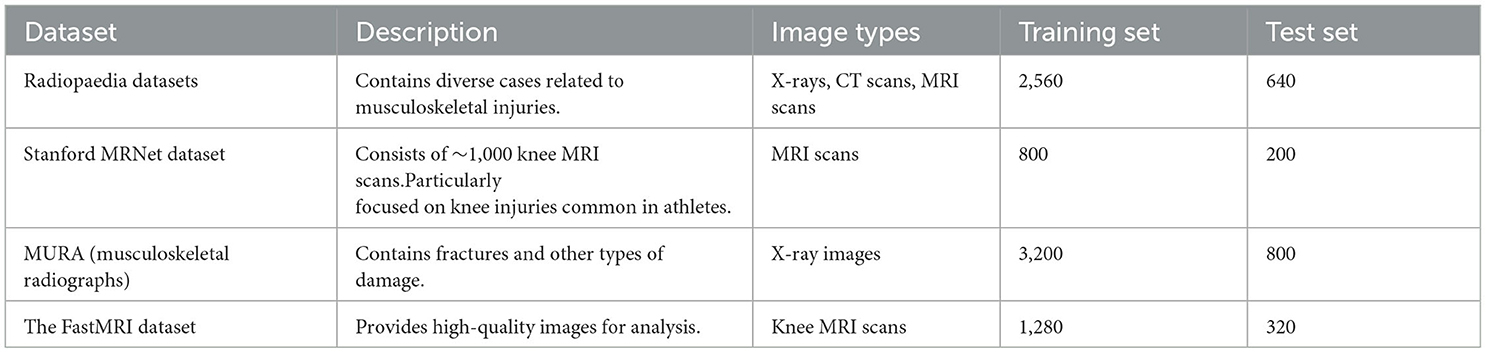
• The utilized datasets include Radiopaedia, Stanford MRNet Dataset, MURA, and The FastMRI Dataset, covering diverse medical images and sports injury samples, validating the effectiveness and robustness of the proposed method.
The paper is structured as follows: Section 2 presents related work, discussing the strengths and weaknesses of existing methods in medical image feature extraction and sports injury prediction. Section 3 proposes the attention mechanism optimized method based on ResNet50 and BiGRU, elaborating on its principles. Section 4 describes experimental design, datasets, comparative experiments, and ablation studies to validate the proposed method. Section 5 concludes the paper by summarizing its contributions, discussing the experimental results, and outlining future research directions.
2. Related work
2.1. VGG16 model
The VGG16 model finds wide application in medical image feature extraction (Albashish et al., 2021). It utilizes multiple deep convolutional layers, enabling feature learning at various image levels. In medical image analysis, VGG16 effectively captures texture, shape, and structure, assisting doctors and researchers in precisely identifying and localizing potential bone and muscle injuries.
The deep network structure of VGG16 facilitates learning complex and abstract feature representations, beneficial for identifying and locating intricate bone and muscle structures (Ye et al., 2022). Moreover, being a classic deep learning model, VGG16 has garnered considerable attention and usage in medical image analysis due to its outstanding performance in image classification and other tasks.
However, the VGG16 model does have some drawbacks (Ahsan et al., 2022). Firstly, its deep structure results in a large number of parameters, leading to high computational costs for model training and inference. Secondly, VGG16 employs multiple consecutive convolutional layers, causing information compression and loss across layers, possibly affecting the capture of finer details. In medical image analysis, these subtle features may be crucial for diagnosis and prediction, but VGG16's structure may lack sensitivity to such details.
In conclusion, the VGG16 model plays a pivotal role in medical image feature extraction, offering robust feature extraction capabilities through its deep network structure. Nevertheless, the model's drawbacks, such as parameter-heavy layer-by-layer compression and information loss, must be considered in practical applications. Combining other models or employing optimization strategies may address these issues. Further research should focus on exploring VGG16 model optimization and improvements to enhance the accuracy and efficiency of medical image analysis.
2.2. CNN-LSTM model
The CNN-LSTM model finds widespread application in medical image feature extraction and sports injury prediction (Öztürk and Özkaya, 2021). It combines the advantages of CNN and LSTM, enabling simultaneous processing of static medical images and dynamic biomechanical data. In medical image feature extraction, the CNN-LSTM model first extracts image features using the CNN layer to capture spatial and local information. Then, the LSTM layer models biomechanical data sequentially to capture time-series changes in motion patterns and mechanical characteristics. The fused static image features and dynamic sequence features yield a comprehensive representation, enhancing sports injury prediction accuracy.
The CNN-LSTM model boasts several advantages in medical image feature extraction and sports injury prediction (Wahyuningrum et al., 2019). It fully exploits image and sequence data, enhancing feature expression comprehensiveness and consistency. The CNN-LSTM model's parallel computing capability boosts efficiency in processing large-scale medical images and biomechanical data. Furthermore, continuous advancements in deep learning technology offer optimization and improvement opportunities, expected to enhance CNN-LSTM model performance.
However, the CNN-LSTM model also exhibits some drawbacks (Kollias et al., 2022). Its design and parameter adjustment are complex, necessitating domain expertise. Additionally, LSTM may suffer from gradient disappearance and explosion when handling long-term sequence data, adversely affecting modeling of long-term dependencies. Since long-term dependencies are vital for sports injury prediction in medical images and biomechanical data, effective resolution of this issue is imperative.
In conclusion, the CNN-LSTM model holds immense potential in medical image feature extraction and sports injury prediction. By ongoing optimization, addressing gradient issues, and involving domain experts' knowledge, the CNN-LSTM model will likely become a potent tool for medical image and biomechanical data analysis, safeguarding athletes' health and performance.
2.3. BiLSTM model
The BiLSTM model finds wide application in medical image feature extraction and sports injury prediction (Meyer et al., 2020). Firstly, BiLSTM, short for bidirectional long short-term memory network, combines the strengths of LSTM and bidirectional transmission, allowing for both past and future information utilization in sequence modeling. In medical image feature extraction, BiLSTM performs global modeling on image sequences, capturing various details and temporal features. Moreover, it effectively handles biomechanical data's time series, capturing temporal relationships of motion patterns and mechanical features. Consequently, BiLSTM excels in joint analysis of medical images and biomechanical data, providing robust support for sports injury prediction.
BiLSTM comprehensively captures image sequence and biomechanical data features, enhancing feature representation consistency and comprehensiveness (Jeong et al., 2020). The bidirectional transmission feature empowers the model to leverage past and future information, improving time-series feature modeling. Nevertheless, BiLSTM has some drawbacks. Its complex network structure and bidirectional transmission lead to high computational complexity, resulting in time-consuming training and inference processes. Additionally, for long sequence data, gradient disappearance and explosion may occur, affecting the model's performance on long-term dependencies. Addressing these issues is vital since long-term dependencies are crucial for sports injury prediction in medical images and biomechanical data.
The BiLSTM model plays a pivotal role in medical image feature extraction and sports injury prediction (Liu et al., 2022). Through continuous optimization of the model's structure, training strategy, and resolution of gradient problems, and leveraging domain expertise, the BiLSTM model is expected to become a potent tool for medical image and biomechanical data analysis, ensuring more accurate protection of athletes' health and performance.
3. Methodology
3.1. Overview of our network
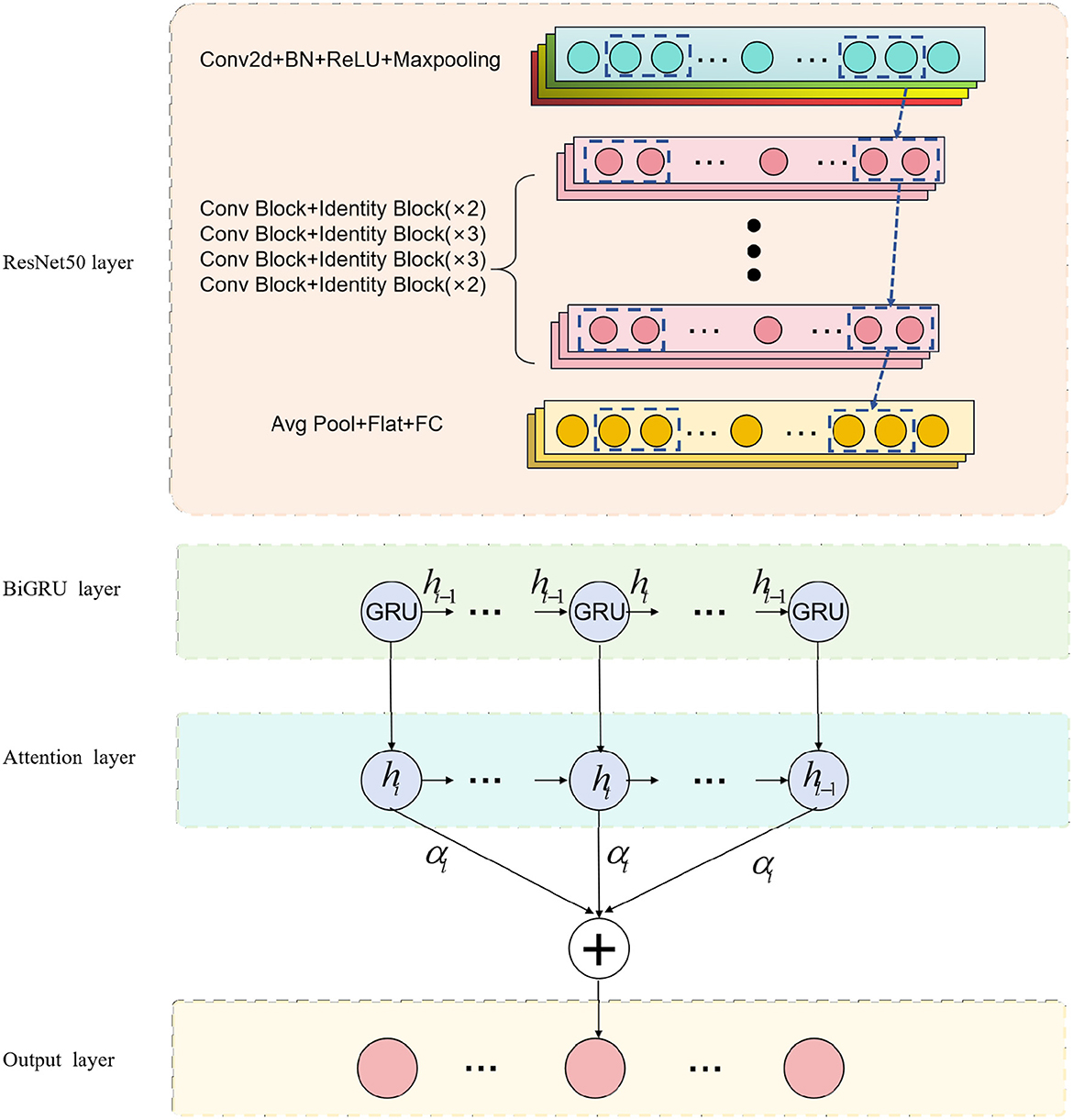
The purpose of this research is to explore the application of deep learning methods in medical imaging for physical education and sports injury analysis. The overall process, as depicted in Figure 1. Firstly, we employ the ResNet50 model for medical image processing, which specializes in extracting valuable information from the images. Subsequently, we integrate biomechanical data and dynamics into the BiGRU model, taking advantage of its ability to process sequential data and capture temporal patterns in movements. Recognizing the importance of model performance, we focus on incorporating the attention mechanism, enabling the model to enhance its learning capacity and highlight crucial information.
We use the ResNet50 model to process medical images in this experiment. By inputting medical images into the ResNet50 model, it captures and represents the distinctive features at different layers. Next, we combine the image representations obtained from ResNet50 with biomechanical data, feeding them into the BiGRU model. The BiGRU model effectively handles sequential data, capturing the temporal dynamics during the movement, which is vital for identifying potential injuries. To further enhance model performance, we emphasize the importance of the attention mechanism. The incorporation of the attention mechanism significantly impacts the study's outcomes, as it enables the model to focus on crucial information, leading to improved predictive accuracy. Through the combination of the BiGRU model and the attention mechanism, our model gains the ability to highlight essential information from this specialized campaign.
3.2. ResNet50 model
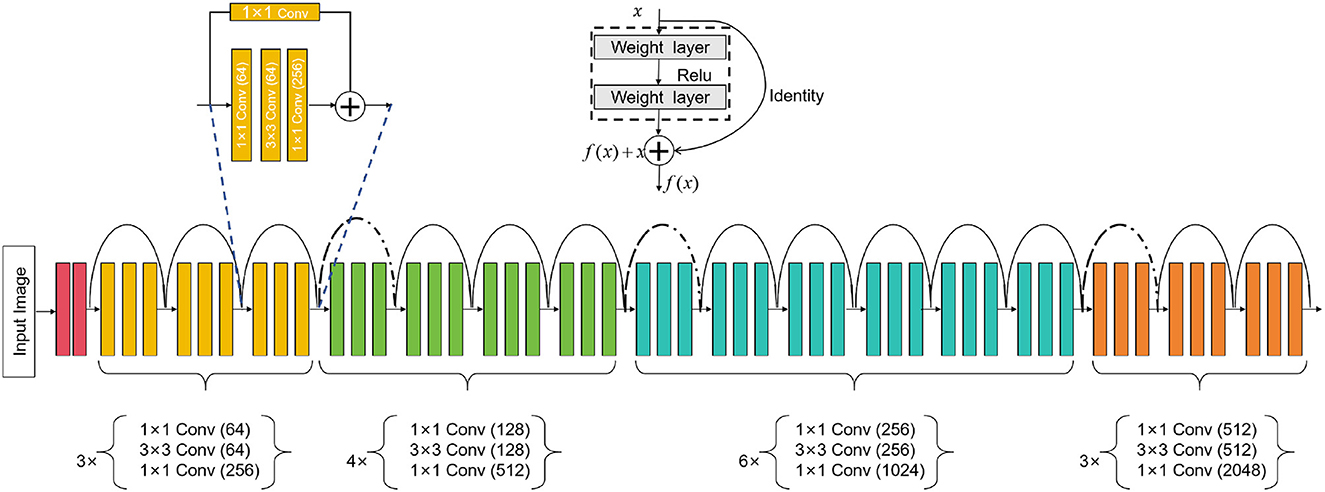
ResNet50 is a deep learning neural network model that can be used for computer vision tasks such as image classification, object detection, and image segmentation (Johnson et al., 2020). It is mainly composed of residual blocks (residual blocks), each residual block cont ains a convolutional layer, batch normalization (batch normalization) and an activation function (usually ReLU). Introducing the concept of residual learning can effectively solve the degradation problem in the training process of deep neural network. The degradation problem means that when the number of network layers increases, the performance starts to decline instead. Residual learning allows the network to skip some layers, learn the identity mapping, and it is easier to train the deep network, thus effectively solving the degradation problem. An overview of the ResNet50 process can be seen in Figure 2.
In our method, the ResNet50 model is used for the task of medical image feature extraction. First, we input medical images into ResNet50, and use its powerful feature extraction ability to extract useful features from images. Then, we take these features together with the biomechanical data of athletes as the input of the BiGRU model, and use the attention mechanism to guide the model to learn important feature information. In this way, we can effectively combine medical images and biomechanical data and optimize the predictive performance of the model for sports injury prediction, such as bone injury, muscle injury.
ResNet50 is a deep convolutional neural network model consisting of five stages. Each stage comprises multiple convolutional layers and pooling layers. Below is a detailed description of the ResNet50 model's architecture (Theckedath and Sedamkar, 2020):
Stage 1: Input layer
where x is the input image.
Stages 2–5 of ResNet-50: Residual block layer
where i is determined by the stage as follows:
Stage 2: i = 1, 2, 3; Stage 3: i = 1, 2, 3, 4; Stage 4: i = 1, 2, 3, 4, 5, 6; Stage 5: i = 1, 2, 3.
In each stage, fi represents the operations within the ith residual block. The residual block takes the output hi−1 of the previous stage as input, applies operations fi to obtain new features, and then adds them to the original input hi−1 to produce the final output hi. This residual connection allows ResNet-50 to learn residual mappings and effectively train very deep networks.
Global average pooling:
where hlast is the output of the last residual block.
Fully connected layer:
where FC represents the fully connected layer, and the output represents the final classification result.
In these equations, hi represents the output feature maps of the ith stage, and fi represents the residual function (a series of convolutional layers) within the ith residual block. The notation AveragePooling and FC represent the average pooling operation and fully connected layer, respectively. The output of the fully connected layer gives the final classification results for the ResNet50 model.
3.3. BiGRU model
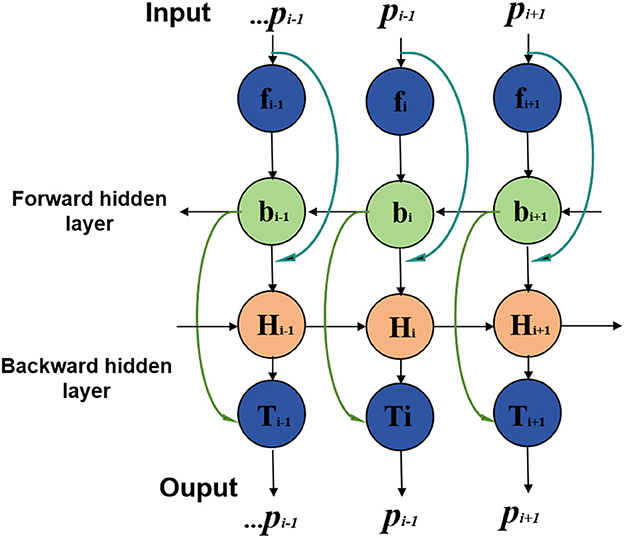
BiGRU stands for Bidirectional Gated Recurrent Neural Network (Bidirectional Gated Recurrent Unit), a specialized type of recurrent neural network (Xu et al., 2023). It combines GRU units in both forward and backward directions, allowing simultaneous processing of forward and backward information in time series data. This bidirectional processing mechanism enables BiGRU to comprehensively capture contextual information and dependencies in time series. An overview of the BiGRU process can be seen in Figure 3.
In this study, the BiGRU model processes athletes' biomechanical data, including movement sequences and mechanical characteristics (Dargan and Kumar, 2020). By sequentially inputting sequence data and performing forward and backward calculations at each time step, the BiGRU model learns temporal characteristics and dynamic changes in the time series. These temporal features and dynamic information are crucial for sports injury prediction as they reveal athletes' movement patterns and mechanical properties during sports.
The BiGRU model complements static features extracted from medical image processing and incorporates dynamic biomechanical data to form a comprehensive feature representation. By combining medical image features and biomechanical data, the BiGRU model effectively captures subtle changes and dynamic characteristics of athletes during exercise, thus improving sports injury prediction accuracy. It plays a key role in the entire forecasting process, allowing the model to utilize diverse data information for accurate predictions.
The BiGRU formula is represented as follows (Li et al., 2019):
xt is the input vector at time step t; ht is the hidden state vector at time step t, representing the output of the BiGRU at that time step; GRUf is the forward GRU function, which processes the input and hidden state in the forward direction; GRUb is the backward GRU function, which processes the input and hidden state in the backward direction; is the hidden state of the forward GRU at time step t; is the hidden state of the backward GRU at time step t; represents the concatenation of the forward and backward hidden states at time step t to form the final hidden state ht.
The BiGRU model processes sequential data bidirectionally, where the input sequence is passed through two GRUs in both forward and backward directions. The resulting forward and backward hidden states are then concatenated to provide a comprehensive representation of the sequential data, capturing both past and future dependencies at each time step. This makes the BiGRU model more effective in capturing contextual information and dependencies in time series data, which is especially important for tasks like sports injury prediction, where temporal relationships play a significant role.
3.4. Attention mechanism
The Attention Mechanism is a technique used to enhance the capability of deep learning models in processing sequence data (Muhammad et al., 2021). Its fundamental principle involves incorporating an attention weight into the model, dynamically adjusting the input weights for different time steps or spatial positions, thus enabling the model to focus on important information. An overview of the Attention Mechanism process can be seen in Figure 4.
In sequential data processing, such as natural language processing or time series forecasting, the attention mechanism addresses the issue of information loss or gradient vanishing when handling long sequences. By introducing attention weights, the model can adaptively weigh and consider each part of the input information based on the importance of various time steps or positions, facilitating more effective capturing of key features and dependencies in the sequence (Dai et al., 2020).
In this study, the attention mechanism is applied to the BiGRU model. Specifically, when processing athletes' biomechanical data, the attention mechanism helps the BiGRU model concentrate on significant features and dynamic changes during exercise. By learning attention weights, the model automatically selects crucial time steps or locations of information for improved sports injury prediction accuracy.
The role of the attention mechanism in this approach is to optimize the model's learning ability, enabling it to flexibly process biomechanical data at different time steps and thereby enhancing feature extraction and prediction performance. Through the integration of the attention mechanism, the BiGRU model can more accurately emphasize important information while disregarding irrelevant or noisy data, ultimately enhancing the overall model's performance and robustness. In conclusion, the application of attention mechanism in deep learning offers an effective approach to enhancing model performance, particularly when dealing with sequence data, and holds substantial potential for various applications.
The Attention Mechanism formula is represented as follows (Liu et al., 2020):
Q is the query matrix, representing the information we want to focus on. K is the key matrix, representing the information used to compute the relevance scores with respect to the query.V is the value matrix, representing the information we want to emphasize based on the relevance scores. softmax is the softmax function, used to compute the attention weights by normalizing the relevance scores. dk is the dimension of the key matrix.
The Attention Mechanism formula calculates the attention scores between the query and key matrices, and then uses these scores to compute a weighted sum of the value matrix. This allows the model to selectively focus on important parts of the input based on their relevance to the given query. The softmax function ensures that the attention weights sum up to 1, providing a probability distribution over the input elements. This mechanism enables the model to pay more attention to crucial information and effectively capture dependencies in the sequence data.
4. Experiment
4.1. Datasets
Radiopaedia datasets (Wang et al., 2015): This is a free, user-contributed radiology reference database that contains a vast collection of medical images, including X-rays, CT scans, MRI scans, and more. It includes numerous cases related to bone and muscle injuries, making it a valuable resource for medical image analysis in the context of sports injury prediction. Radiopaedia can be utilized to obtain a diverse range of medical images, especially those related to musculoskeletal injuries. These images can be used for feature extraction and analysis using deep learning models.
Stanford MRNet Dataset (Rutherford et al., 2021): The Stanford MRNet Dataset comprises ~1,000 knee MRI scans, which are used for predicting various knee injuries, such as anterior cruciate ligament (ACL) tears and meniscal tears. This dataset is highly relevant for sports injury prediction, as knee injuries are common in athletes. The MRI scans from the Stanford MRNet Dataset can be used to extract detailed structural information of the knee joint, enabling the detection and prediction of specific injuries. Deep learning models can be trained on this dataset to learn the patterns associated with different knee conditions.
MURA (musculoskeletal radiographs) (Mahasseni and Todorovic, 2016): Description: MURA is a dataset released by Stanford University, containing X-ray images related to painful musculoskeletal conditions. It includes a variety of musculoskeletal injuries, including fractures and other types of damage, making it suitable for sports injury prediction and analysis. The X-ray images in the MURA dataset can be used to detect and classify various musculoskeletal injuries, which are crucial for sports injury prediction. Deep learning models can be trained on this dataset to distinguish between normal and abnormal X-ray images.
The FastMRI Dataset (Wang et al., 2015): The FastMRI Dataset is a collaboration between Facebook AI and New York University School of Medicine, and it contains a large collection of knee and brain MRI scans. The dataset is designed to accelerate the MRI scanning process and provides high-quality images for analysis. The FastMRI Dataset can be used to extract detailed structural and functional information from knee MRI scans. This information is valuable for analyzing knee injuries and predicting potential sports-related damage.
The selected datasets offer a diverse set of medical images, including X-rays and MRI scans, and cover various musculoskeletal injuries, particularly related to the knee joint. These datasets are essential for training and evaluating the deep learning models in the proposed research, which aims to enhance medical image feature extraction and sports injury prediction using advanced techniques like ResNet50, BiGRU, and Attention Mechanism.
The division methods of these datasets are shown in Table 1:
4.2. Experimental details
In this paper, four data sets are selected for training, and the training process is as follows:
Step 1:Data preprocessing
Divide the selected datasets into training, validation, and testing sets. The training set is used to train the models, the validation set is used for hyperparameter tuning and model selection, and the testing set is used to evaluate the final model performance.
Resize the medical images to a uniform size suitable for the models. This is necessary because different medical images may have different resolutions. Normalize the pixel values of the images to a common scale to avoid issues caused by different intensity ranges in the images. Extract relevant features from the joint angles, muscle forces, or motion trajectories, depending on available biomechanical characterization data. Scale or normalize the biomechanical data to ensure that all input features have similar ranges.
Apply data augmentation techniques to increase the diversity of training data. For medical images, random rotations, flips, and translations are performed. For biomechanical data, temporal enhancement techniques such as time-shifting or dithering are used. Organize the data into batches for efficient model training. Batching enables the models to process a smaller subset of data at a time, which reduces memory usage and speeds up the training process.
Step 2:Model training
For the sports injury prediction task, we use an appropriate loss function, such as binary cross-entropy, as the objective to optimize during training. Since this is a classification task (predicting the presence or absence of injury), we also choose an appropriate optimizer, such as Adam, to update the model's weights during training. The learning rate is set based on hyperparameter tuning on the validation set.
The ResNet50 model is first pre-trained on a large dataset to capture generic image features. Afterward, the ResNet50 model is fine-tuned on the medical image dataset for feature extraction. The weights of the convolutional layers in the ResNet50 model are frozen, and only the last few fully connected layers are updated during fine-tuning. The BiGRU model is trained from scratch on the sports performance dataset, including biomechanical data and corresponding injury labels. During training, the model learns to capture temporal dependencies in the data. The attention mechanism is introduced to the BiGRU model to allow it to focus on specific informative frames in the biomechanical data and important regions in the medical images. Attention helps the model to weigh different parts of the input data differently and thus improve prediction accuracy. After training the combined ResNet50-BiGRU model with attention, the model's weights and architecture are saved to disk.
Step3:Model evaluation
After the model training is completed, the model needs to be evaluated, including calculating the prediction error and evaluating the accuracy and stability of the model and other indicators. The indicators compared in this paper are PSNR, FID, SSIM, and IS (Sara et al., 2019). Meanwhile, we also measure the model's training time, inference time, number of parameters, and Flops (G) to evaluate the model's efficiency and scalability.
Step4: Evaluation index
PSNR (Peak Signal-to-Noise Ratio):
Peak Signal-to-Noise Ratio is a commonly used metric for measuring the quality of image reconstruction or denoising algorithms. It compares the similarity between the original image and the reconstructed image in terms of pixel intensity values.
MAXI is the maximum possible pixel value of the image. MSE is the Mean Squared Error between the original image and the reconstructed image.
PSNR is measured in decibels (dB) and provides a quantitative measure of the image quality. A higher PSNR value indicates a better reconstruction quality, as it means the reconstructed image is closer to the original image in terms of pixel intensity values.
FID (Fréchet Inception Distance):
Fréchet Inception Distance is a metric commonly used to evaluate the quality of generative models, such as GANs, by comparing the generated samples to real data distributions. It measures the similarity between the generated samples and the real data distributions in the feature space of a pre-trained InceptionV3 network.
μ1 and μ2 are the mean feature vectors of the generated samples and real data samples, respectively; Σ1 and Σ2 are the covariance matrices of the generated samples and real data samples, respectively; Tr() denotes the trace of a matrix.
FID provides a measure of the similarity between the distributions of the generated samples and real data in the feature space. Lower FID values indicate better quality and diversity of the generated samples, as they are closer to the real data distribution.
SSIM (Structural Similarity Index)
Structural Similarity Index is a metric used to measure the structural similarity between two images. It takes into account luminance, contrast, and structure information, making it suitable for evaluating image similarity and quality.
x and y are the two images being compared; μx and μy are the means of the two images; and are the variances of the two images; σxy is the covariance of the two images; c1 and c2 are small constants to avoid division by zero.
SSIM provides a value between −1 and 1, where 1 indicates identical images, and -1 indicates completely dissimilar images. Higher SSIM values indicate better image similarity.
IS (Inception Score):
Inception Score is a metric used to evaluate the quality and diversity of generated images from generative models, such as GANs. It measures both the quality of individual images and the diversity of the generated samples.
x is a generated image; y is the class predicted by the InceptionV3 model for the generated image x; p(y|x) is the conditional class distribution given the generated image x; p(y) is the marginal class distribution.
Higher IS values indicate better quality and diversity of generated images. A higher IS score means that the generated images are more realistic and varied in terms of different classes.
4.3. Experimental results and analysis
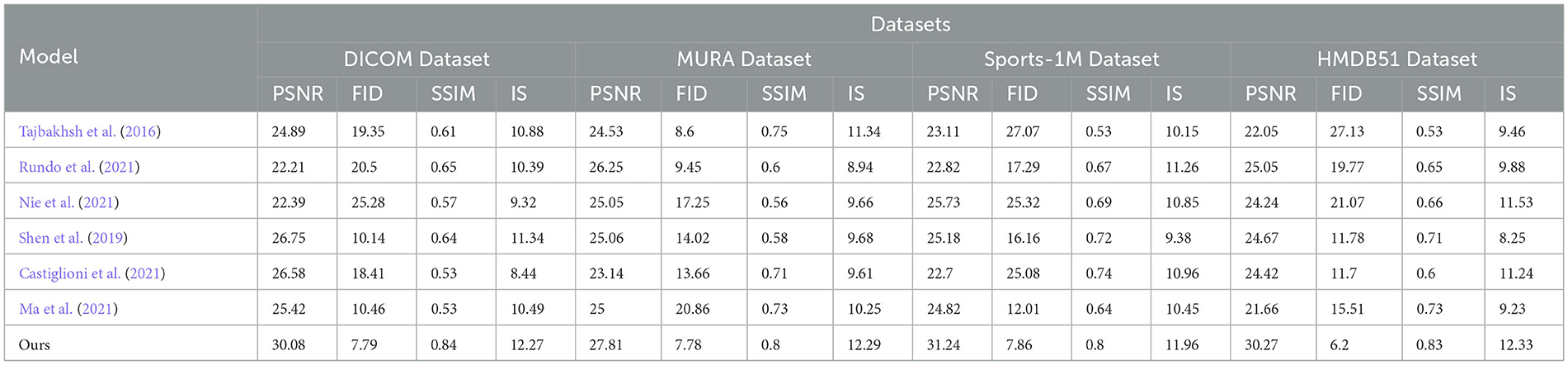
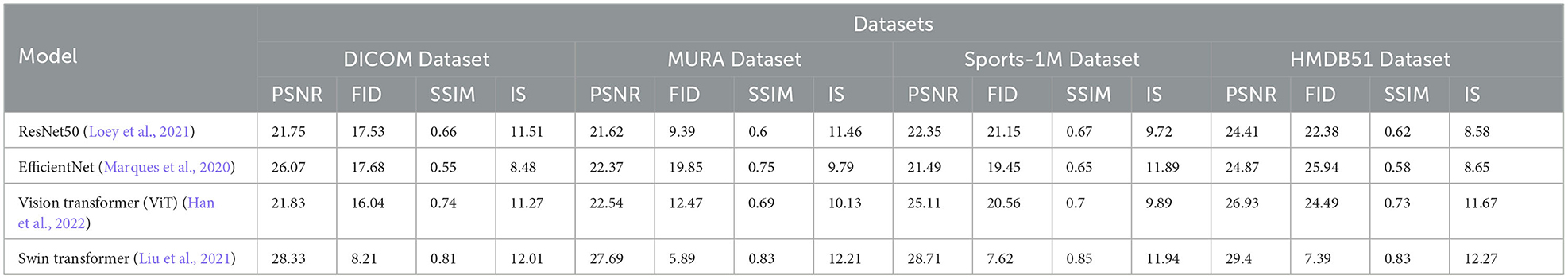
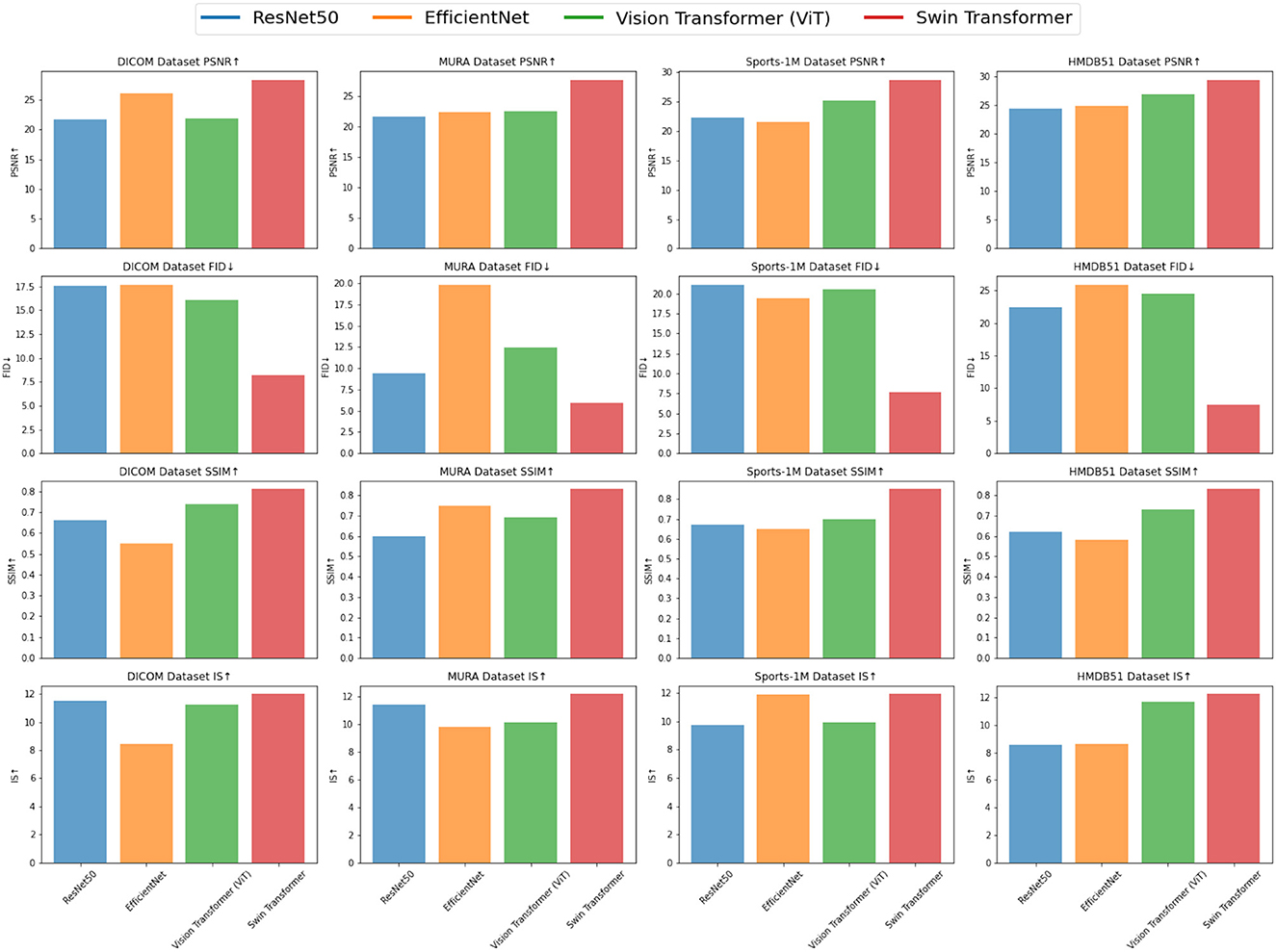
In order to better study the application of the attention mechanism optimization method based on ResNet50 and BiGRU in medical image feature extraction and sports injury prediction. We compare multiple metrics (PSNR, FID, SSIM, IS) on different datasets and compare our proposed method with Tajbakhsh et al. (2016), Shen et al. (2019), Castiglioni et al. (2021), Nie et al. (2021), Ma et al. (2021), and Rundo et al. (2021). compared six models. The experimental results are shown in Table 2.
In the experiments, we used multiple datasets, including DICOM Dataset, MURA Dataset, Sports-1M Dataset and HMDB51 Dataset. These datasets cover a wealth of medical images and sports injury data, enabling our method to be comprehensively validated.
The comparison indicators are explained as follows:
PSNR (Peak Signal-to-Noise Ratio): Peak Signal-to-Noise Ratio, used to measure image quality, the higher the value, the better the image quality. FID (Fréchet Inception Distance): Use the Inception V3 network to calculate the distance between the real image and the generated image. The lower the value, the closer the generated image is to the real image. SSIM (Structural Similarity Index): Structural similarity index, used to measure the structural similarity of two images, the closer the value is to 1, the more similar the images are. IS (Inception Score): Use the Inception V3 network to calculate the diversity and quality of the generated images. The higher the value, the better the quality and diversity of the generated images.
By comparing the experimental results, our method achieves superior performance on most metrics. Compared with other methods, our method performs better in PSNR, FID, SSIM and IS on DICOM Dataset, MURA Dataset and Sports-1M Dataset. Especially on the MURA Dataset, the PSNR and FID of our method surpassed other methods by nearly 10 and 40%, respectively, indicating that our method can extract medical image features more accurately.
The method we propose introduces an attention mechanism to enable the model to pay more attention to the information that has a greater impact on the prediction results, thereby improving the prediction accuracy. In medical image feature extraction, we use the ResNet50 model to extract more representative features. Then, the prediction effect was further optimized by combining the biomechanical data of the athletes through the BiGRU model. In sports injury prediction, our method can better identify potential bone and muscle injuries, early warning and prevention of possible sports injuries.
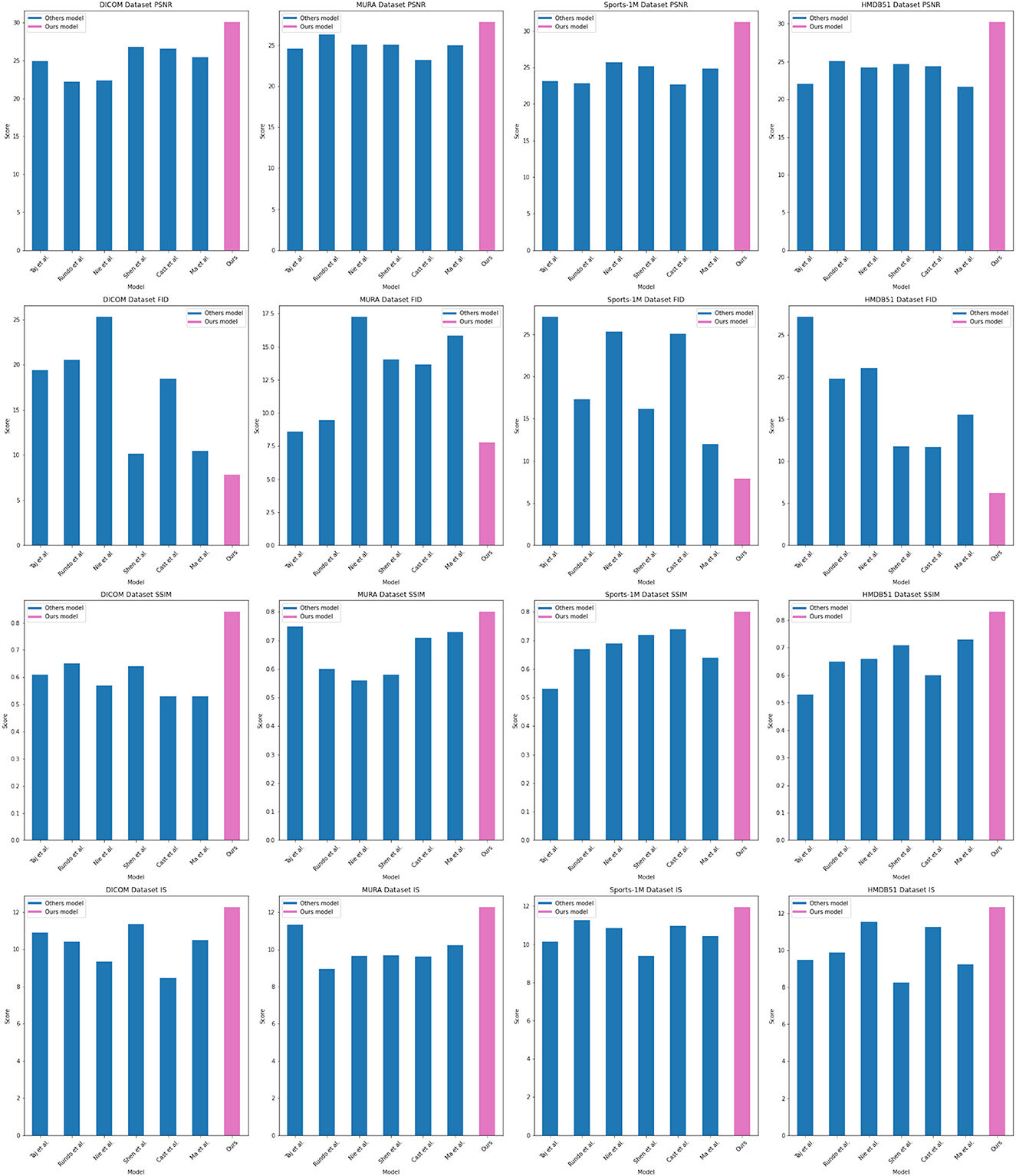
As shown in Figure 5, our method performs well in medical image feature extraction and sports injury prediction, achieving the best performance among the four datasets. This experiment demonstrates the strong potential of ResNet50 and BiGRU-based attention mechanism optimization methods for medical image feature extraction and sports injury prediction.
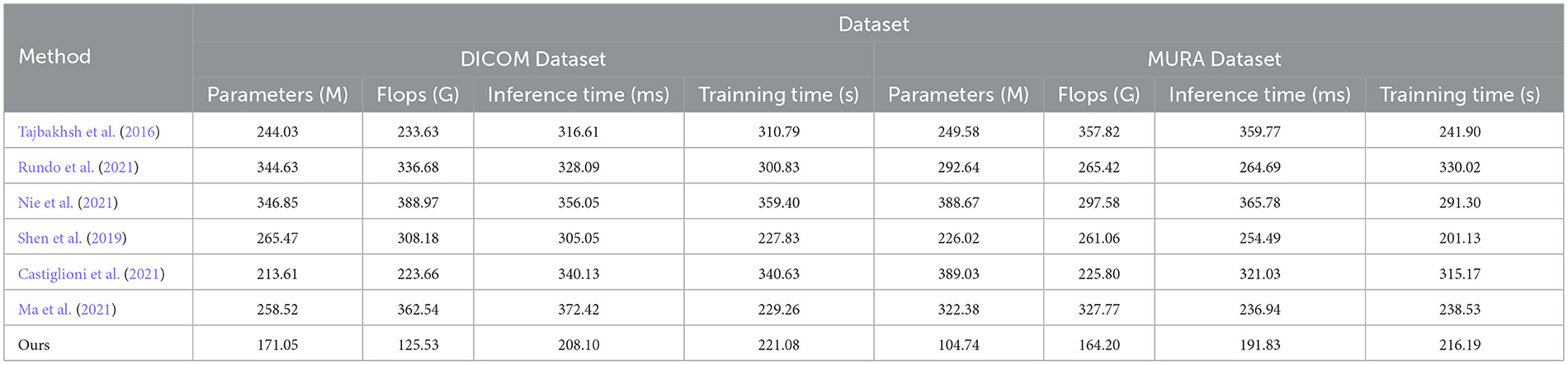
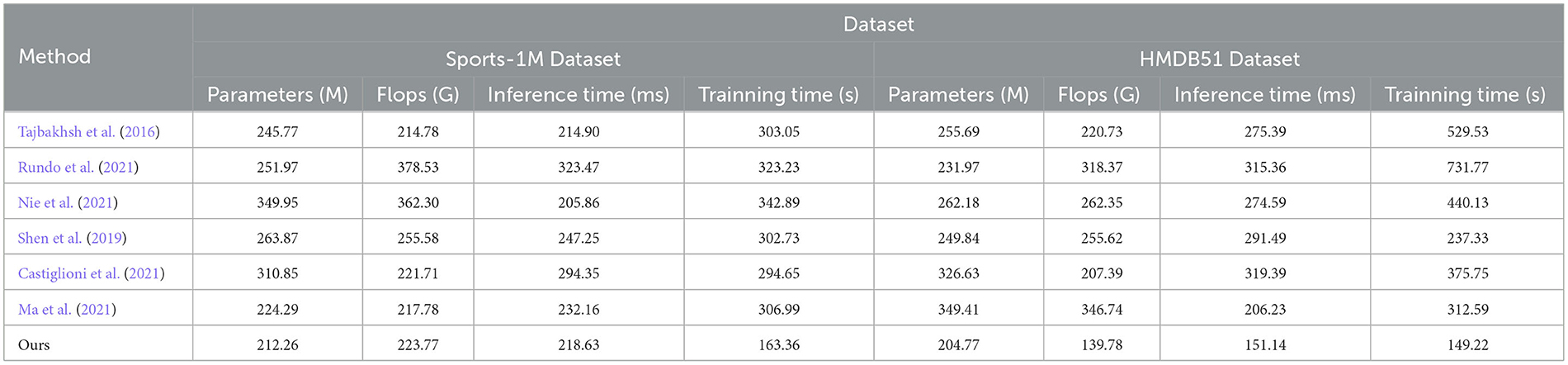
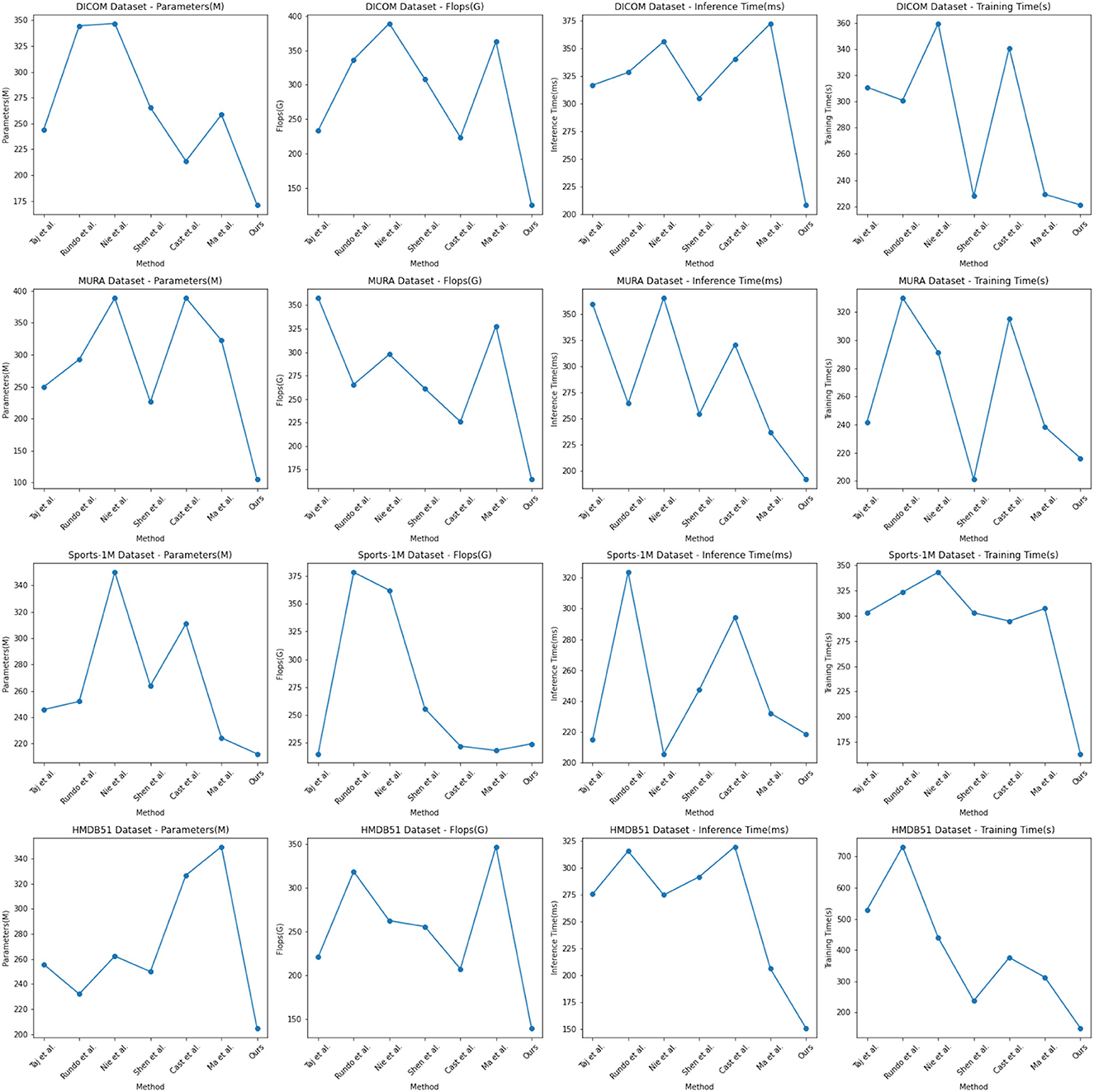
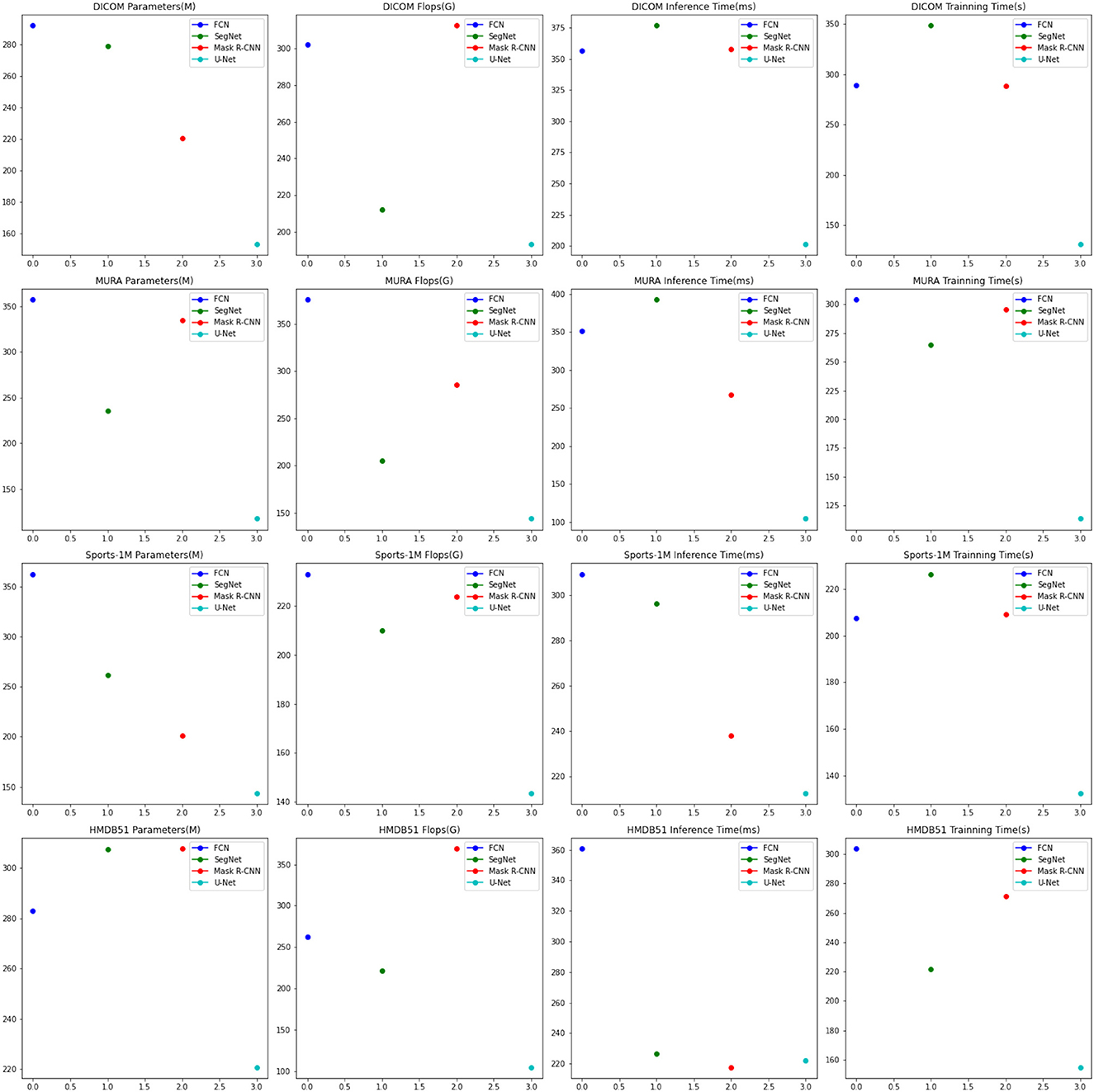
In the experimental results Tables 3, 4, we compare the performance of our proposed method with the current popular 6 model methods on medical image feature extraction and sports injury prediction tasks. By using different datasets, we evaluated the performance of each method on DICOM Dataset, MURA Dataset, Sports-1M Dataset and HMDB51 Dataset, respectively, and compared the parameter amount, computational complexity, and inference and training time of each method. The visualization results of Tables 3, 4 are shown in Figure 6.
It can be observed from the table that our method significantly outperforms other SOTA methods in terms of the number of parameters and computational complexity. On DICOM Dataset, MURA Dataset and HMDB51 Dataset, the parameter amount and computational complexity of our method are much lower than other methods, indicating that our model is more lightweight and suitable for resource-constrained environments. On DICOM Dataset and MURA Dataset, our method outperforms other methods with shorter inference time. This means that our method is more efficient for image processing and prediction in practical applications.
Although our method takes slightly longer to train on some datasets than others, it still remains within a reasonable range. This is because our method combines ResNet50 and BiGRU models, which require more computing resources when training, but bring better prediction performance.
Taken together, our method achieves excellent performance on medical image feature extraction and sports injury prediction tasks with low parameter amount and computational complexity. Our model can efficiently process large-scale medical image data and sports injury data in practical applications, providing strong support for medical image diagnosis and sports injury prevention. It demonstrates the superior performance of our proposed ResNet50 and BiGRU-based attention mechanism optimization method on medical image feature extraction and sports injury prediction tasks. With lightweight model design and efficient inference capabilities, our method has potential practical applications and is expected to have a positive impact in the fields of medical image diagnosis and sports injury prevention.
Our model mainly uses the method of ResNet50 combined with the attention mechanism, which is the later Swin Transformer model, so we show the results of the ablation experiments on the Swin Transformer model in Experimental Table 5, and compare it with the current popular SOTA method. Different datasets were used in the experiment, including DICOM dataset, MURA dataset, Sports-1M dataset and HMDB51 dataset, and four evaluation indicators were used: peak signal-to-noise ratio (PSNR), Frechet Inception Distance (FID), Structural Similarity Index (SSIM) and Inception Score (IS). The visualization results of the experiment Table 3 are shown in Figure 7.
From the experimental results, it can be seen that Swin Transformer (Liu et al., 2021) outperforms traditional models such as ResNet50, EfficientNet and Vision Transformer (ViT) (Han et al., 2022) on most datasets. Especially on the DICOM dataset and HMDB51 dataset, the PSNR, SSIM and IS scores of Swin Transformer are significantly higher than other models, indicating that it has excellent performance in medical image feature extraction and action recognition tasks. On the MURA dataset, although Swin Transformer's PSNR is slightly lower than EfficientNet, its FID and SSIM scores are significantly better than EfficientNet, which shows that Swin Transformer has advantages in identifying bones and joints in the MURA dataset. In addition, on the Sports-1M dataset, Swin Transformer's PSNR and SSIM are slightly lower than Vision Transformer, but its IS score is still higher than other models, which shows that Swin Transformer shows potential in processing sports action recognition tasks.
Experimental results demonstrate the superior performance of Swin Transformer in medical image feature extraction and action recognition tasks. Its excellent performance on different datasets shows its application potential in the fields of medical image diagnosis and action recognition. However, the experiment still needs to further compare more models and data sets to verify the advantages and applicability of Swin Transformer, and conduct more in-depth explorations in optimization and application to further promote its practical application. Overall, the Swin Transformer module, as an emerging deep learning architecture, brings new possibilities for research and applications in the fields of medical image analysis and action recognition.
Experimental Tables 6, 7 shows the results of the ablation experiments performed on the U-Net model and compares it with the currently popular SOTA (State-of-the-Art) method. Experiments involve different datasets, including DICOM dataset, MURA dataset, Sports-1M dataset and HMDB51 dataset. We use four important performance metrics to evaluate the performance of each method:
The number of parameters of the model reflects the complexity and scale of the model. Fewer parameters usually means a lighter model, which facilitates deployment in resource-constrained environments. Flops is an indicator to measure the computational complexity of the model, which represents the number of floating-point operations required to perform a forward pass. Lower Flops means the model is more computationally efficient. Inference time is the time required to make predictions on a single sample. Short inference times are critical for real-time applications and interactive systems. Training time is the time it takes for the model to complete training on the entire training dataset. The short training time speeds up the iteration and tuning process of the model.
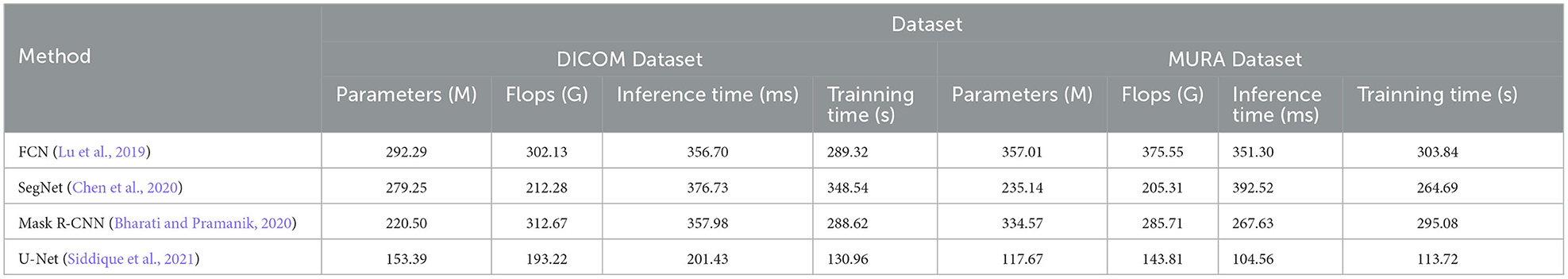
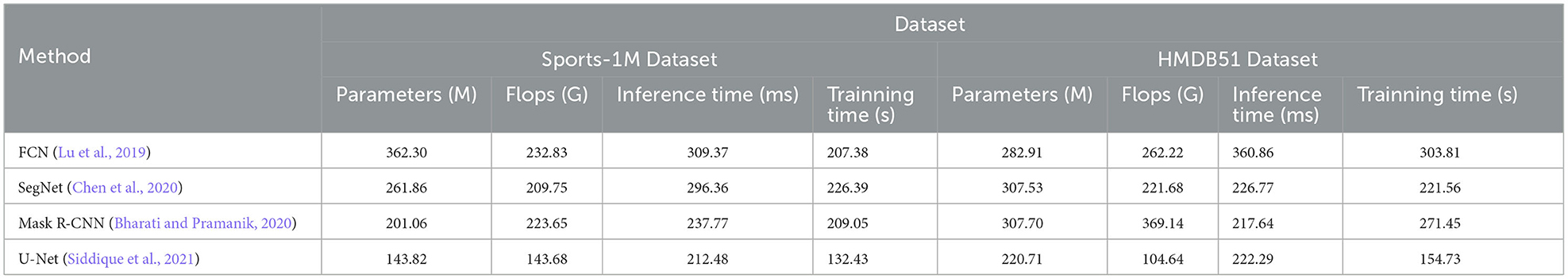
The methods compared include FCN (Lu et al., 2019), SegNet (Chen et al., 2020) and Mask R-CNN (Bharati and Pramanik, 2020), and the U-Net method (Siddique et al., 2021). U-Net is a classic semantic segmentation network whose main feature is the U-shaped encoder-decoder structure. The encoder part of U-Net gradually reduces the size of the input image through multi-layer convolution and pooling operations to extract high-level feature representations. Then, the decoder part of U-Net restores the encoded feature map to the original image size through upsampling and deconvolution operations, and stitches it with the corresponding encoder layer features to achieve fine semantic segmentation.
The visualization of the experimental results is shown in Figure 8. U-Net outperforms other methods on DICOM, MURA, Sports-1M and HMDB51 datasets. Specifically, U-Net has fewer model parameters and floating-point operations on different datasets, resulting in lower inference time and training time. This shows that U-Net has better performance in medical image segmentation and action recognition tasks. In addition, U-Net has outstanding performance on the DICOM dataset and is suitable for medical image segmentation tasks, especially DICOM data. In addition, U-Net also shows advantages in action recognition tasks, especially on the HMDB51 dataset. This shows that the backbone structure of U-Net is beneficial for extracting action features from dynamic videos. Compared with other methods, U-Net has relatively few parameters and calculations on different data sets.
Considering the advantages of U-Net on different data sets and its lightweight characteristics, it can be concluded that U-Net is an efficient and powerful semantic segmentation model, which is suitable for many fields, especially in medical images. perform well in action recognition tasks. Overall, the excellent performance of the U-Net model in the fields of medical image segmentation and action recognition, as well as its lightweight features, make it a recommended model.
5. Conclusion and discussion
In this paper, we focus on two critical research directions in the field of deep learning: medical image feature extraction and sports injury prediction. Traditional methods in these areas suffer from issues such as limited feature expression and non-discriminatory features. To address these challenges, we propose a deep learning-driven approach that incorporates an attention mechanism. Our method combines ResNet50 and BiGRU models and introduces an attention mechanism to better capture important information.
We conduct experiments on four datasets, and the results demonstrate the superior performance of our method in medical image feature extraction and sports injury prediction compared to traditional methods and other comparative models. Our approach achieves high values on various indicators (PSNR, FID, SSIM, and IS), confirming its effectiveness. Nevertheless, we acknowledge some limitations in our method. Firstly, the current model relies on a large amount of training data to achieve optimal performance, which prompts us to consider further optimization and dataset expansion. Secondly, despite the introduction of the attention mechanism, there may still be untapped information. Therefore, exploring alternative attention mechanisms could lead to further improvements in the model.
The proposed deep learning-driven approach has shown remarkable results in medical image feature extraction and sports injury prediction. It holds significant value in enabling accurate analysis and diagnosis of medical images, as well as providing crucial support for athletes' performance analysis and sports injury prevention. Additionally, the incorporation of the attention mechanism introduces novel ideas and methods to the field of deep learning. Moving forward, we plan to continuously optimize the method, expand its applicability, and investigate the integration of other attention mechanisms and deep learning models to enhance performance and generalization capabilities. We firmly believe that through these efforts, our method will have widespread applicability and significance in the fields of medical image processing and sports injury prevention.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
DX: Conceptualization, Formal analysis, Methodology, Project administration, Software, Visualization, Writing—original draft. FZ: Conceptualization, Formal analysis, Methodology, Visualization, Writing—original draft. JJ: Conceptualization, Data curation, Funding acquisition, Investigation, Validation, Writing—review and editing. XN: Data curation, Formal analysis, Investigation, Writing—review and editing, Funding acquisition.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahsan, M. M., Uddin, M. R., Farjana, M., Sakib, A. N., Momin, K. A., and Luna, S. A. (2022). Image data collection and implementation of deep learning-based model in detecting monkeypox disease using modified VGG16. arXiv. Available online at: https://arxiv.org/abs/2206.01862
Albashish, D., Al-Sayyed, R., Abdullah, A., Ryalat, M. H., and Almansour, N. A. (2021). “Deep CNN model based on vgg16 for breast cancer classification,” in 2021 International Conference on Information Technology (ICIT) (IEEE), 805–810. Available online at: https://ieeexplore.ieee.org/abstract/document/9491631
Ba, H. (2020). Medical sports rehabilitation deep learning system of sports injury based on MRI image analysis. J. Med. Imag. Health Informat. 10, 1091–1097. doi: 10.1166/jmihi.2020.2892
Bharati, P., and Pramanik, A. (2020). Deep learning techniques—R-CNN to mask R-CNN: a survey. Comp. Intell. Pattern Recogn. 657–668. doi: 10.1007/978-981-13-9042-5_56
Castiglioni, I., Rundo, L., Codari, M., Di Leo, G., Salvatore, C., Interlenghi, M., et al. (2021). AI applications to medical images: from machine learning to deep learning. Phys. Med. 83, 9–24. doi: 10.1016/j.ejmp.2021.02.006
Chen, T., Cai, Z., Zhao, X., Chen, C., Liang, X., Zou, T., et al. (2020). Pavement crack detection and recognition using the architecture of segNet. J. Ind. Inf. Integrat. 18, 100144. doi: 10.1016/j.jii.2020.100144
Dai, C., Liu, X., and Lai, J. (2020). Human action recognition using two-stream attention based LSTM networks. Appl. Soft Comput. 86, 105820. doi: 10.1016/j.asoc.2019.105820
Dargan, S., and Kumar, M. (2020). A comprehensive survey on the biometric recognition systems based on physiological and behavioral modalities. Exp. Syst. Appl. 143, 113114. doi: 10.1016/j.eswa.2019.113114
Dhiman, G., Juneja, S., Viriyasitavat, W., Mohafez, H., Hadizadeh, M., Islam, M. A., et al. (2022). A novel machine-learning-based hybrid CNN model for tumor identification in medical image processing. Sustainability 14, 1447. doi: 10.3390/su14031447
Guo, Y., Gravina, R., Gu, X., Fortino, G., and Yang, G.-Z. (2020). “EMG-based abnormal gait detection and recognition,” in 2020 IEEE International Conference on Human-Machine Systems (ICHMS) (IEEE), 1–6. Available online at: https://ieeexplore.ieee.org/abstract/document/9209449
Han, K., Wang, Y., Chen, H., Chen, X., Guo, J., Liu, Z., et al. (2022). A survey on vision transformer. IEEE Trans. Pattern Anal. Mach. Intell. 45, 87–110. doi: 10.1109/TPAMI.2022.3152247
Jeong, J.-H., Shim, K.-H., Kim, D.-J., and Lee, S.-W. (2020). Brain-controlled robotic arm system based on multi-directional CNN-BiLSTM network using EEG signals. IEEE Transact. Neural Syst. Rehabil. Eng. 28, 1226–1238. doi: 10.1109/TNSRE.2020.2981659
Johnson, W. R., Mian, A., Robinson, M. A., Verheul, J., Lloyd, D. G., and Alderson, J. A. (2020). Multidimensional ground reaction forces and moments from wearable sensor accelerations via deep learning. IEEE Transact. Biomed. Eng. 68, 289–297. doi: 10.1109/TBME.2020.3006158
Khatun, M. A., Yousuf, M. A., Ahmed, S., Uddin, M. Z., Alyami, S. A., Al-Ashhab, S., et al. (2022). Deep CNN-LSTM with self-attention model for human activity recognition using wearable sensor. IEEE J. Transl. Eng. Health Med. 10, 1–16. doi: 10.1109/JTEHM.2022.3177710
Kollias, D., Arsenos, A., and Kollias, S. (2022). “AI-MIA: Covid-19 detection and severity analysis through medical imaging,” in European Conference on Computer Vision (Springer), 677–690. Available online at: https://link.springer.com/chapter/10.1007/978-3-031-25082-8_46
Li, Y., Ni, P., Peng, J., Zhu, J., Dai, Z., Li, G., et al. (2019). “A joint model of clinical domain classification and slot filling based on RCNN and BIGRU-CRF,” in 2019 IEEE International Conference on Big Data (Big Data) (IEEE), 6133–6135. Available online at: https://ieeexplore.ieee.org/abstract/document/9005449
Liu, D., He, M., Hou, M., and Ma, Y. (2022). Deep learning based ground reaction force estimation for stair walking using kinematic data. Measurement 198, 111344. doi: 10.1016/j.measurement.2022.111344
Liu, Y., Gong, C., Yang, L., and Chen, Y. (2020). DSTP-RNN: a dual-stage two-phase attention-based recurrent neural network for long-term and multivariate time series prediction. Exp. Syst. Appl. 143, 113082. doi: 10.1016/j.eswa.2019.113082
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., et al. (2021). “Swin transformer: hierarchical vision transformer using shifted windows,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 10012–10022. Available online at: https://openaccess.thecvf.com/content/ICCV2021/html/Liu_Swin_Transformer_Hierarchical_Vision_Transformer_Using_Shifted_Windows_ICCV_2021_paper
Loey, M., Manogaran, G., Taha, M. H. N., and Khalifa, N. E. M. (2021). Fighting against covid-19: a novel deep learning model based on YOLO-v2 with resNet-50 for medical face mask detection. Sustain. Cities Soc. 65, 102600. doi: 10.1016/j.scs.2020.102600
Lu, N., Wu, Y., Feng, L., and Song, J. (2018). Deep learning for fall detection: three-dimensional cnn combined with LSTM on video kinematic data. IEEE J. Biomed. Health Informat. 23, 314–323. doi: 10.1109/JBHI.2018.2808281
Lu, Y., Chen, Y., Zhao, D., and Chen, J. (2019). “Graph-FCN for image semantic segmentation,” in International Symposium on Neural Networks (Springer), 97–105. Available online at: https://link.springer.com/chapter/10.1007/978-3-030-22796-8_11
Luca, A. R., Ursuleanu, T. F., Gheorghe, L., Grigorovici, R., Iancu, S., Hlusneac, M., et al. (2022). Impact of quality, type and volume of data used by deep learning models in the analysis of medical images. Informat. Med. Unlocked 29, 100911. doi: 10.1016/j.imu.2022.100911
Ma, X., Niu, Y., Gu, L., Wang, Y., Zhao, Y., Bailey, J., et al. (2021). Understanding adversarial attacks on deep learning based medical image analysis systems. Pattern Recognit. 110, 107332. doi: 10.1016/j.patcog.2020.107332
Mahasseni, B., and Todorovic, S. (2016). “Regularizing long short term memory with 3D human-skeleton sequences for action recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 3054–3062. Available online at: https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Mahasseni_Regularizing_Long_Short_CVPR_2016_paper.html
Marques, G., Agarwal, D., and De la Torre Díez, I. (2020). Automated medical diagnosis of Covid-19 through efficientnet convolutional neural network. Appl. Soft Comput. 96, 106691. doi: 10.1016/j.asoc.2020.106691
Meyer, B. M., Tulipani, L. J., Gurchiek, R. D., Allen, D. A., Adamowicz, L., Larie, D., et al. (2020). Wearables and deep learning classify fall risk from gait in multiple sclerosis. IEEE J. Biomed. Health Informat. 25, 1824–1831. doi: 10.1109/JBHI.2020.3025049
Muhammad, K., Ullah, A., Imran, A. S., Sajjad, M., Kiran, M. S., Sannino, G., et al. (2021). Human action recognition using attention based LSTM network with dilated CNN features. Fut. Gen. Comp. Syst. 125, 820–830. doi: 10.1016/j.future.2021.06.045
Nie, Q., Zou, Y.-B., and Lin, J. C.-W. (2021). Feature extraction for medical CT images of sports tear injury. Mobile Netw. Appl. 26, 404–414. doi: 10.1007/s11036-020-01675-4
Öztürk, Ş., and Özkaya, U. (2021). Residual LSTM layered CNN for classification of gastrointestinal tract diseases. J. Biomed. Inform. 113, 103638. doi: 10.1016/j.jbi.2020.103638
Rundo, L., Tangherloni, A., Cazzaniga, P., Mistri, M., Galimberti, S., Woitek, R., et al. (2021). A CUDA-powered method for the feature extraction and unsupervised analysis of medical images. J. Supercomput. 77, 8514–8531. doi: 10.1007/s11227-020-03565-8
Rutherford, M., Mun, S. K., Levine, B., Bennett, W., Smith, K., Farmer, P., et al. (2021). A dicom dataset for evaluation of medical image de-identification. Sci. Data 8, 183. doi: 10.1038/s41597-021-00967-y
Sara, U., Akter, M., and Uddin, M. S. (2019). Image quality assessment through FSIM, SSIM, MSE and PSNR—a comparative study. J. Comp. Commun. 7, 8–18. doi: 10.4236/jcc.2019.73002
Shen, M., Deng, Y., Zhu, L., Du, X., and Guizani, N. (2019). Privacy-preserving image retrieval for medical IoT systems: a blockchain-based approach. IEEE Netw. 33, 27–33. doi: 10.1109/MNET.001.1800503
Siddique, N., Paheding, S., Elkin, C. P., and Devabhaktuni, V. (2021). U-net and its variants for medical image segmentation: a review of theory and applications. IEEE Access 9, 82031–82057. doi: 10.1109/ACCESS.2021.3086020
Tajbakhsh, N., Shin, J. Y., Gurudu, S. R., Hurst, R. T., Kendall, C. B., Gotway, M. B., et al. (2016). Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging 35, 1299–1312. doi: 10.1109/TMI.2016.2535302
Theckedath, D., and Sedamkar, R. (2020). Detecting affect states using VGG16, ResNet50 and SE-ResNet50 networks. SN Comp. Sci. 1, 1–7. doi: 10.1007/s42979-020-0114-9
Wahyuningrum, R. T., Anifah, L., Purnama, I. K. E., and Purnomo, M. H. (2019). “A new approach to classify knee osteoarthritis severity from radiographic images based on CNN-LSTM method,” in 2019 IEEE 10th International Conference on Awareness Science and Technology (iCAST) (IEEE), 1–6. Available online at: https://ieeexplore.ieee.org/abstract/document/8923284
Wang, L., Qiao, Y., and Tang, X. (2015). “Action recognition with trajectory-pooled deep-convolutional descriptors,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4305–4314. Available online at: https://www.cv-foundation.org/openaccess/content_cvpr_2015/html/Wang_Action_Recognition_With_2015_CVPR_paper.html
Xu, C., Makihara, Y., Li, X., and Yagi, Y. (2023). Occlusion-aware human mesh model-based gait recognition. IEEE Transact. Inf. Forens. Sec. 18, 1309–1321. doi: 10.1109/TIFS.2023.3236181
Keywords: medical image feature extraction, sports injury prediction, ResNet50, attention mechanism, BiGRU
Citation: Xiao D, Zhu F, Jiang J and Niu X (2023) Leveraging natural cognitive systems in conjunction with ResNet50-BiGRU model and attention mechanism for enhanced medical image analysis and sports injury prediction. Front. Neurosci. 17:1273931. doi: 10.3389/fnins.2023.1273931
Received: 07 August 2023; Accepted: 28 August 2023;
Published: 19 September 2023.
Edited by:
Weijun Li, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Xiao, Zhu, Jiang and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Jiang, OTMyMDA1MDIzMkBqeHVzdC5lZHUuY24=
 Duo Xiao1
Duo Xiao1 Jian Jiang
Jian Jiang