- 1Division of Neuroscience, School of Biosciences, Cardiff University, Cardiff, United Kingdom
- 2European Cancer Stem Cell Research Institute, Cardiff University School of Biosciences, Cardiff, United Kingdom
Neural precursors generate neurons in the embryonic brain and in restricted niches of the adult brain in a process called neurogenesis. The precise control of cell proliferation and differentiation in time and space required for neurogenesis depends on sophisticated orchestration of gene transcription in neural precursor cells. Much progress has been made in understanding the transcriptional regulation of neurogenesis, which relies on dose- and context-dependent expression of specific transcription factors that regulate the maintenance and proliferation of neural progenitors, followed by their differentiation into lineage-specified cells. Here, we review some of the most widely studied neurogenic transcription factors in the embryonic cortex and neurogenic niches in the adult brain. We compare functions of these transcription factors in embryonic and adult neurogenesis, highlighting biochemical, developmental, and cell biological properties. Our goal is to present an overview of transcriptional regulation underlying neurogenesis in the developing cerebral cortex and in the adult brain.
Introduction
Neurogenesis happens primarily during embryonic stages, while the nervous system develops, although some regions in the adult brain retain the capacity to generate new neurons throughout life (Jurkowski et al., 2020). In both cases, neural progenitors need to balance their own proliferation with the production of differentiated cells to ensure that appropriate numbers of neurons and glia are made. During embryonic neurogenesis, progenitors first proliferate through symmetric divisions until about E11.5, when they change their division mode and start producing neurons through asymmetric divisions. Once all necessary neurons have been generated, they will begin generating glial cells in an irreversible switch that signifies the end of embryonic neurogenesis (Martynoga et al., 2012). Because transitions between phases cannot be reversed, accurate control of proliferation vs. differentiation is paramount to ensure the correct development of the nervous system. Postnatally, some radial glial cells become the specialized neural stem cells (NSCs) for postnatal and adult neurogenesis (Bond et al., 2020). In neurogenic regions [subventricular zone, hippocampus and hypothalamus, reviewed in Jurkowski et al., 2020], NSCs generate intermediate progenitor cells through asymmetric division. Intermediate progenitor cells expand rapidly and eventually differentiate into neuronal progenitor cells that migrate to their destination where they integrate into neuronal circuitry upon terminal differentiation. Contrastingly to embryonic neurogenesis, adult NSCs can simultaneously generate glial cells through a much less understood process.
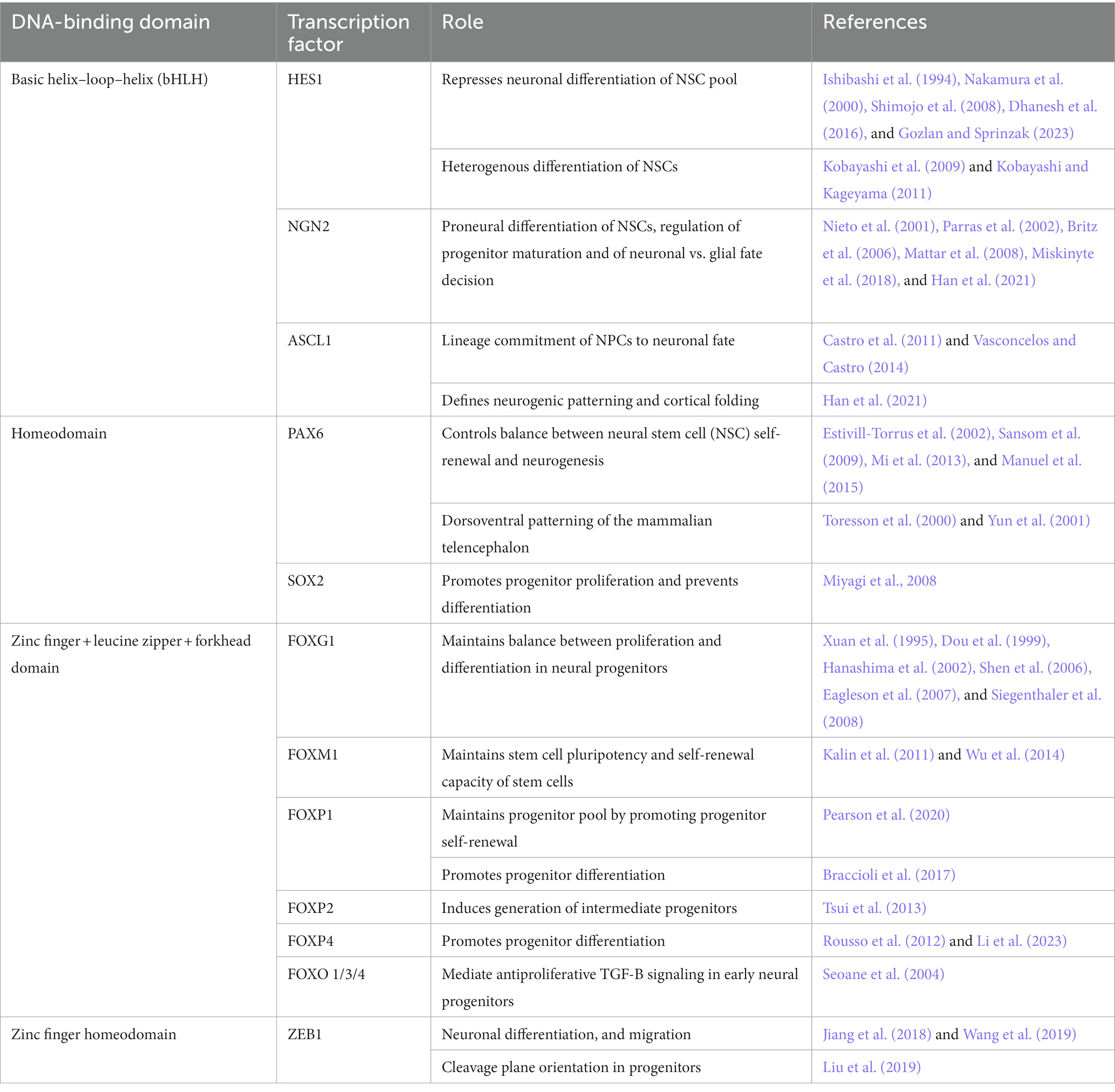
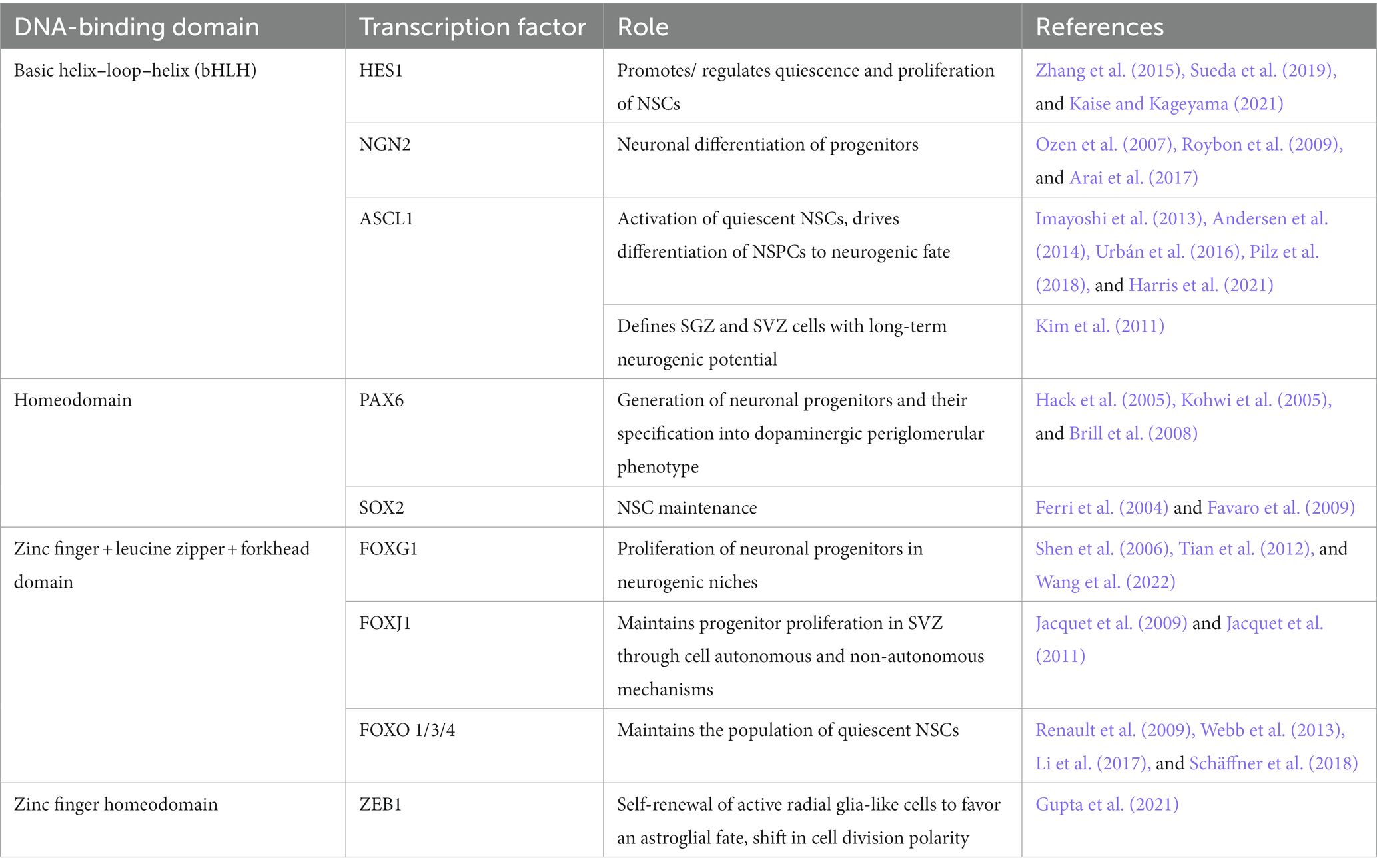
The balance between proliferation and differentiation of neural stem and progenitor cells requires exquisite control at the transcriptional level. Transcriptional control of embryonic and adult neurogenesis relies on shared transcription factors (TFs) that become spatially confined over time, are expressed at specific timepoints, or both. The cell type-specific transcriptional activity of such neurogenic TFs is mediated by epigenetic signatures, chromatin changes, and other protein partners. In this mini-review, we provide a brief overview focusing on the role of some of the best-characterized TFs that control neurogenesis in the embryonic dorsal telencephalon (Table 1) and in the adult subgranular zone (SGZ) of the hippocampus and the subventricular zone (SVZ) of the lateral ventricles (Table 2). For more comprehensive analyses of the role of specific TFs, we refer the reader to appropriate reviews.
bHLH transcription factors in neurogenesis
Transcription factors of the bHLH (basic helix loop helix) superfamily work as dimers and bind DNA through a basic domain at their amino terminal end (Jones, 2004). Several bHLH TFs play important and sometimes opposing roles during embryonic and adult neurogenesis.
HES (Hairy and Enhancer of Split homologs) family members of the bHLH TF family are effectors of the Notch signaling pathway (Ohtsuka et al., 1999). During corticogenesis, they regulate cell proliferation, differentiation, and fate specification by maintaining stemness of progenitors and controlling the timing of differentiation (Kageyama et al., 2007; Gozlan and Sprinzak, 2023) in neuroepithelial and radial glial cells. Of the different Hes genes, Hes1 is the most widely studied in the context of corticogenesis. HES1 levels in cortical neuronal progenitors experience cyclic oscillations, which are essential for the maintenance of neuronal progenitors (Shimojo et al., 2008). These oscillations result from the combination of Hes1 expression induction by Notch signaling, an autoinhibitory effect of HES1 on its own transcription and the great instability of the Hes1 mRNA and protein (Takebayashi et al., 1994; Hirata et al., 2002). In its capacity as an antineurogenic bHLH repressor (Nakamura et al., 2000), HES1 acts in two different ways. First, it represses expression of its target genes by directly binding to their promotors in a complex with co-repressors like Groucho/TLE-1 (Jiménez et al., 1997; Dhanesh et al., 2016). Second, HES1 interferes with the transcriptional activity of target TFs by binding to and sequestering E proteins such as E47, which are required by TFs like ASCL1 to function (Sasai et al., 1992; Dhanesh et al., 2016). Downstream targets of HES1 include cell-cycle regulators like the CDK inhibitor Cdkn1B (Murata et al., 2005), Gadd45g, cyclins D2 and E2, and the Notch ligand Dll1 (Shimojo et al., 2008). In addition, HES1 also inhibits expression of several proneural bHLH TFs, including Ascl1 and Neurog2 (Shimojo et al., 2008). HES1 fluctuations drive oscillatory expression of these TFs and help maintain the progenitor population, especially during early stages of corticogenesis (Shimojo et al., 2011). In turn, expression, or lack thereof of the proneural bHLH TFs Ascl1 and Neurog2 define four different progenitor states, with expression of both TFs representing the least lineage restricted progenitors and those expressing only Neurog2 committed to a neuronal lineage (Han et al., 2021). Furthermore, combined expression of Ascl1 and Neurog2 leads to cross-repression and to the production of Notch ligands that maintain proliferation in neighboring cells (Han et al., 2021).
During adult neurogenesis, sustained levels of HES1 are needed to keep aNSCs in the SVZ and SGZ in a quiescent state (Sueda et al., 2019), as constant, high HES1 indirectly leads to increased CDKN1A levels, inhibiting cell cycle progression (Maeda et al., 2023). This is accomplished through the interaction of HES1 with ID1, which represses HES1 autoinhibition (Bai et al., 2007). As NSCs activate, oscillating expression of HES1 drives a concomitant oscillatory expression of ASCL1, which is critical for NSC activation (Andersen et al., 2014). In fact, lower levels of ASCL1 are linked to higher numbers of resting NSCs (Urbán et al., 2016) and a proliferation vs. differentiation bias in progenitors (Imayoshi et al., 2013), while ASCL1 protein levels drop over time to ensure the maintenance of the aNSC pool (Harris et al., 2021).
Homeobox transcription factors in neurogenesis
There are 11 different classes of homeobox transcription factors, characterized by a helix-turn-helix homeodomain motif that mediates their binding to DNA (Holland et al., 2007). We discuss PAX6 and SOX2 here, but the roles of 21 homeobox TFs in vertebrate forebrain development have been comprehensively reviewed elsewhere (Leung et al., 2022).
PAX6 belongs to the paired-box homeodomain transcription factor family, harboring a second DNA binding domain, the paired box, in addition to the homeodomain (Dahl et al., 1997). PAX6 is one of the main regulators of cortical neurogenesis, controlling cell cycle length and exit in a dose and context dependent manner [reviewed in Manuel et al. (2015)]. As such, loss of Pax6 leads to shorter cell cycle length and a premature switch from proliferative to neurogenic divisions during early neurogenesis, with more pronounced effects in areas of higher Pax6 expression (Estivill-Torrus et al., 2002; Mi et al., 2013). However, at later stages, Pax6 loss leads to a longer cell cycle (Estivill-Torrus et al., 2002) and its overexpression decreases the number of proliferating progenitors in rostral and medial areas at E15.5 (Manuel et al., 2006). These results highlight the context dependent actions of this TF, which is needed both for progenitor proliferation and for neurogenesis. Interestingly, the effects of PAX6 during corticogenesis, except for its patterning role, are mediated by the paired-box, and not by the homeodomain (Haubst et al., 2004). PAX6 regulates progenitor cell proliferation in part by controlling expression of several genes involved in the G1/S transition, including cyclins and Cdks (Sansom et al., 2009; Mi et al., 2013). PAX6 has been shown to directly inhibit Cdk6 expression, thereby reducing Rb phosphorylation and slowing down G1 progression (Mi et al., 2013). Regarding neurogenesis, PAX6 directly induces expression of Tbr2, which confers intermediate progenitor identity (Quinn et al., 2007; Sansom et al., 2009). In addition, PAX6 also stimulates expression of Neurog2, and participates in a transcriptional network with NEUROG2, ASCL1 and HES1 to control the outcome of neural progenitor cell division (Sansom et al., 2009).
During adult neurogenesis, PAX6 seems to play a similar role controlling proliferation and neuronal differentiation of aNSCs (Hack et al., 2005; Maekawa et al., 2005). In the SGZ, PAX6 induces expression of Neurog2 and NeuroD1 (Xu et al., 2021), which are needed to maintain NSC progenitors and induce neuronal fate, respectively (Roybon et al., 2009). It also acts through FABP7 to maintain NSC and progenitor cell proliferation and prevent exhaustion of the stem cell pool (Osumi et al., 2008). In the SVZ and the rostral migratory stream (RMS), PAX6 is needed to regulate neuronal precursor proliferation and for periglomerular neuron fate (Hack et al., 2005).
SOX2 (SRY-box binding transcription factor 2) is a member of the Sox family of transcription factors, which consists of 9 subfamilies (SoxA, SoxB1, SoxB2, SoxC, SoxD, SoxE, SoxF, SoxG, SoxH). Sox2 is part of the SoxB1 subgroup (together with Sox1 and Sox3; Wegner, 2010) and is a master regulator of stemness in development and adult tissues (Sarkar and Hochedlinger, 2013). It is a pioneer factor that can initiate transcription in epigenetically silenced chromatin regions (Dodonova et al., 2020). SOX transcription factors bind the consensus sequence TTGT through their high-mobility-group (HMG) box (Wegner, 2010), with specificity of individual SOX factors conveyed by DNA regions flanking the consensus motif (Sarkar and Hochedlinger, 2013). SOX2 is expressed throughout embryonic and adult neurogenesis, as well as in pluripotent embryonic stem cells and Sox2 knockout (KO) is lethal during early embryogenesis (Avilion et al., 2003). In the developing brain SOX2 promotes progenitor proliferation and prevents cell differentiation, functions that overlap with SOX1 and SOX3 (Wegner and Stolt, 2005; Miyagi et al., 2008). Interestingly, Sox2 hypomorphism also affects differentiation into GABAergic interneurons in the cortex and olfactory bulb at E17.5 (Cavallaro et al., 2008).
In the adult CNS, SOX2 is expressed in all neurogenic niches, and conditional deletion of Sox2 results in impaired NSC proliferation, increased apoptosis, and reduced neurogenesis in the SVZ and SGZ (Ferri et al., 2004; Favaro et al., 2009). The wide-ranging functions of SOX2 in the brain are reviewed in more detail in Pevny and Nicolis (2010) and Mercurio et al. (2019).
Forkhead transcription factors in neurogenesis
Forkhead transcription factors are characterized by the presence of the so-called forkhead domain, which mediates their interaction with DNA. This domain consists of three α-helices and three β-sheets surrounded by two loops that form the “winged” region (Hannenhalli and Kaestner, 2009). Forkhead family members are classified into 19 subfamilies from FoxA to FoxS (Jackson et al., 2010). Members of the FoxG, FoxJ, FoxM, FoxO, and FoxP subfamilies have been implicated in embryonic and/or adult neurogenesis and play sometimes opposing roles in the regulation of neural stem cell behavior.
Foxg1 KO animals die at birth with severe brain hypoplasia (Xuan et al., 1995; Dou et al., 1999) and heterozygous animals display decreased cortical, hippocampal and striatal size, along with reduced numbers of TBR2+ intermediate progenitors (Shen et al., 2006; Eagleson et al., 2007; Siegenthaler et al., 2008). Those changes reflect the role of FOXG1 in maintaining the correct balance between proliferation and differentiation in neural progenitors, with lack of Foxg1 leading to lengthening of the cell cycle and premature cell cycle exit (Xuan et al., 1995; Hanashima et al., 2002). At the molecular level, FOXG1 antagonizes TGF-B signaling by repressing the expression of TGF-B family members BMP2, 4, 6, and 7, which are all ectopically upregulated in Foxg1 KOs. This repression requires the DNA binding domain of FOXG1 (Dou et al., 1999; Hanashima et al., 2002). FOXG1 also interferes with the ability of the TGF-B signaling effectors SMADs to promote expression of CDK inhibitors. The SMAD partner FAST-2 is needed for the transcriptional activation of Cdkn2b, but binding of FOXG1 to FAST-2 interferes with TGF-B signaling and antagonizes its growth inhibition effects (Dou et al., 2000). To activate Cdkn1a expression, SMAD proteins need to form a complex with members of the FOXO subfamily (Seoane et al., 2004). FOXG1 can reduce Cdkn1a expression levels by repressing expression of Foxo1 (Vezzali et al., 2016). Furthermore, FOXG1 interacts with FOXO at the protein level, forming a ternary complex with SMADs that can no longer activate Cdkn1a expression (Seoane et al., 2004). In addition, FOXG1 inhibition of Cdkn1a expression can also be mediated by its interaction with the polycomb protein BMI-1 (Fasano et al., 2009). FOXG1 could also potentially interfere with the expression of Cdkn1b, as its expression is stimulated by BMP treatment (Nakamura et al., 2003; Sharov et al., 2006) and by expression of Foxo1, 3 and 4 (Medema et al., 2000).
FOXG1 has also been indirectly linked to Notch signaling, as it interacts with TLE1, which enhances the repressive ability of FOXG1 (Yao et al., 2001). This interaction has been shown in vitro and in the E15.5 developing telencephalon. Moreover, TLE1 enables the interaction between FOXG1 and HES1, which increases HES1-mediated transcriptional repression (Yao et al., 2001), suggesting that FOXG1 might act to amplify the effect of Notch signaling in early neural progenitors, as all three proteins are expressed in cultures derived from E12.5 telencephalic progenitors.
Other forkhead family members are also involved in neurogenesis. FOXM1 stimulates expression of genes needed for G1/S transition and DNA replication, while simultaneously diminishing protein stability of CDK inhibitors (Kalin et al., 2011). These roles could explain why cortical progenitors derived from E14 ERT2Cre FoxM1fl/fl animals display a reduction in the number of neurospheres formed after tamoxifen addition (Wang et al., 2011). FOXM1 also regulates expression of Sox2 and Bmi1, which are necessary for neural progenitor self-renewal (Wang et al., 2011). However, conditional deletion of Foxm1 does not lead to major abnormalities in the brain (Schüller et al., 2007), suggesting the presence of compensatory mechanisms. From the FOXP family members, FOXP1 works to maintain the progenitor pool by promoting progenitor self-renewal, at least in part through the induction of vertical division angles and symmetric divisions (Pearson et al., 2020). However, FOXP1 has also been shown to inhibit Notch signaling in the developing cortex, thereby promoting progenitor differentiation (Braccioli et al., 2017). FOXP2 might regulate the generation of TBR2+ intermediate progenitors (Tsui et al., 2013) and FOXP4 promotes neuronal differentiation of neural progenitors by repressing N-Cadherin expression, therefore favoring detachment from the ventricular zone (Rousso et al., 2012; Li et al., 2023).
FOXG1 is strongly expressed in the SGZ of the dentate gyrus and the lateral ventricle SVZ (Shen et al., 2006; Schäffner et al., 2023). In aNSCs of the DG, FOXG1 plays a similar role of balancing proliferation and differentiation as it does in embryonic progenitors (Wang et al., 2022), with partial or total loss leading to defects in size and morphology of this anatomical structure. Progressive loss of progenitors, altered neuronal differentiation and reduced neuronal survival have been described in these mutant animals, as well as a failure to form the secondary radial glia scaffold (Shen et al., 2006; Tian et al., 2012). Remarkably, generation of olfactory interneurons in the SVZ does not seem to be affected by heterozygous lack of Foxg1 (Shen et al., 2006), suggesting a region-specific function of FOXG1 in adult neurogenesis.
FOXO1 and FOXO3 are also expressed in aNSCs of the SVZ and the SGZ (Paik et al., 2009; Renault et al., 2009). In a Foxo1/3/4 triple mutant, aNSCs get depleted over time due to decreased self-renewal and increased activation of progenitors early on (Paik et al., 2009; Schäffner et al., 2018). These effects are due to increased expression of cyclins and CDKs and decreased expression of CDK inhibitors, as well as derepression of the centrosomal gene Aspm, a known regulator of NSCs divisions (Paik et al., 2009). Very similar results are obtained in Foxo3 single KO animals (Renault et al., 2009). Transcriptional analysis has revealed that FOXO3 targets are enriched in cell quiescence-related genes, oxidative stress response and cell metabolism, further supporting the notion that FOXO proteins are necessary to maintain the population of quiescent NSCs over the lifespan of the animals by preventing excessive cell cycle reentry (Renault et al., 2009; Ro et al., 2013). This transcriptional control is mediated in part by the interaction of FOXO3 with the methylcytosine dioxygenase TET2 (Li et al., 2017). Moreover, FOX3 restricts the neurogenic effects of ASCL1 in adult NPCs by preventing ASCL1-dependent transcription (Webb et al., 2013) Additionally, lack of Foxo3 also impacts the outcome of aNSC progeny, with a bias toward astrocytes and reduced production of neurons and oligodendrocytes (Renault et al., 2009).
Finally, FOXJ1 has also been linked to adult neurogenesis in the SVZ of the lateral ventricle. This TF is required for ependymal cell specification during the transition to postnatal stages (Jacquet et al., 2009), but it also defines a subpopulation of progenitors that rely on FOXJ1 expression for its proliferative ability (Jacquet et al., 2011). FOXJ1 deficient progenitors produce fewer neurospheres and are biased toward a glial fate, with defective neurogenic potential. Interestingly, beyond the cell autonomous effect of FOXJ1 in the FOXJ1+ lineage, an additional non-autonomous effect on the remaining aNSCs in the SEZ has been described (Jacquet et al., 2011).
ZEB1 in neurogenesis
The transcription factor ZEB1 (zinc finger E-box binding homeobox 1) is emerging as a new regulator of self-renewal and fate choice in the CNS. The ZEB family of TFs consists of two members, Zeb1 and Zeb2, which are both core regulators of epithelial-mesenchymal transition (Vandewalle et al., 2009). Epithelial-mesenchymal transition is developmental program that has more recently garnered attention for its role in stemness and lineage regulation (Goossens et al., 2017). Zeb1-mutant mice show aberrant T cell development, underlining its involvement in lineage regulation (Higashi et al., 1997). Structurally, ZEB proteins comprise of two C2H2-type zinc finger domains that flank a central homeodomain. The zinc finger domains are necessary for DNA binding, with each zinc finger independently binding to separate E-box motifs in gene promoters with the consensus sequence 5′-CACCT(G)-3; Sekido et al., 1996, 1997; Remacle et al., 1999). The homeodomain mediates interaction with other proteins (e.g., CTBP, YAP) that are necessary for transcriptional regulation (Furusawa et al., 1999; Feldker et al., 2020). Depending on their interaction partners, ZEB TFs can activate or repress transcription, with E-cadherin repression and Vimentin activation being the best-known examples (Vandewalle et al., 2009). Phosphorylation of Thr-867 is necessary for nuclear import of ZEB1 (Llorens et al., 2016), otherwise the effects of post-transcriptional ZEB1 modifications are poorly understood.
In embryonal neurogenesis, ZEB1 is expressed in the subventricular zone and overlaps with proliferating progenitor cells between E14 and E18 (Yen et al., 2001). Constitutive deletion of Zeb1 causes defects in proliferation of embryonic neural progenitors in the ventricular zone of the lateral ventricles and the hypothalamus at E15.5 (Liu et al., 2008). ZEB1 blocks neuronal lineage progression as well as migration of cortical neuroblasts (Wang et al., 2019). Conditional loss of Zeb1 at E14.5 does not affect cell proliferation or radial glia cell maintenance but causes premature neuronal differentiation (Wang et al., 2019). Zeb1 overexpression at E14.5 results in reduced neurogenesis, migration defects and subcortical band heterotopia (Wang et al., 2019).
In the adult brain, ZEB1 is important for the self-renewal of adult hippocampal radial glia-like (RGL) cells. Zeb1 loss in RGL cells results in their precocious differentiation into the neuronal lineage (Gupta et al., 2021). This is accompanied by reduced differentiation into the astroglial lineage, but it remains to be resolved whether this is due to an as-yet unspecified role of ZEB1 in glial fate determination or a natural consequence of the increased neurogenesis. Hence, ZEB1 blocks neuronal lineage progression during embryonal and adult neurogenesis. In adult neural stem/progenitor cells, ZEB1 is associated with activation and proliferation, and loss of Zeb1 results in depletion of the stem cell pool. Contrastingly, ZEB1 functions in neural progenitors during embryonic neurogenesis appear to be separated in time, with Zeb1 loss affecting proliferation of progenitors in a constitutive knockout model, but not of later radial glia cells when deleted at E14.5 (Liu et al., 2008; Wang et al., 2019). In both adult and embryonic neurogenesis, Zeb1 loss is associated with a change in cell division type (symmetric vs. asymmetric) which promotes differentiation of the stem/progenitor cell pool. Interestingly, Zeb1 loss during embryogenesis promoted asymmetric divisions that prevented expansion of neural progenitors (and therefore caused premature differentiation), whereas in the adult hippocampus Zeb1 KO causes increased symmetric divisions of neural stem/progenitor cells which are necessary for self-renewal, thus promoting their differentiation (Liu et al., 2019; Gupta et al., 2021).
Conclusion
Although neurogenic transcription factors are expressed during embryonic and adult neurogenesis their functions often show differences during both processes. These differences include increased spatial confinement and spatial heterogeneity in adult neurogenic niches, different activities in neural progenitor cells at various developmental stages, and/or different effects on downstream progenitor cells. It is important to consider epigenetic modifications, post-translational modifications, and differential expression of interacting partners at different developmental stages to unravel the functions of each neurogenic transcription factor at specific points in time and space. For example, changes in ASCL1 post-translational degradation result in different behavior of adult neural stem cells in juvenile and adult hippocampal neurogenesis (Harris et al., 2021). Integrated analysis of neurogenic transcription factors across development and aging is needed to reveal the specific co-factors contributing to the differential functions in embryonic and adult neurogenesis.
Data availability statement
No new data was created during this study.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
NS was the recipient of a Biotechnology and Biological Sciences Research Council (BBSRC) PhD studentship (BB/T008741/1). FS was supported by MRC grant MR/S007709/1. IM-G was supported by funding from the BBSRC (BB/S002359/1). Open access publication fees were covered by the Cardiff University’s Open Access Institutional Fund.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andersen, J., Urbán, N., Achimastou, A., Ito, A., Simic, M., Ullom, K., et al. (2014). A Transcriptional Mechanism Integrating Inputs from Extracellular Signals to Activate Hippocampal Stem Cells. Neuron 83, 1085–1097. doi: 10.1016/j.neuron.2014.08.004
Arai, M. D., Zhan, B., Maruyama, A., Matsui-Harada, A., Horinouchi, K., and Komai, S. (2017). Enriched environment and Mash1 transfection affect neural stem cell differentiation after transplantation into the adult somatosensory cortex. J. Neurol. Sci. 373, 73–80. doi: 10.1016/j.jns.2016.12.013
Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N., and Lovell-Badge, R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140. doi: 10.1101/gad.224503
Bai, G., Sheng, N., Xie, Z., Bian, W., Yokota, Y., Benezra, R., et al. (2007). Id Sustains Hes1 Expression to Inhibit Precocious Neurogenesis by Releasing Negative Autoregulation of Hes1. Dev. Cell 13, 283–297. doi: 10.1016/j.devcel.2007.05.014
Bond, A. M., Ming, G., and Song, H. (2020). Ontogeny of adult neural stem cells in the mammalian brain. Curr. Top. Dev. Biol. 142, 67–98. doi: 10.1016/bs.ctdb.2020.11.002
Braccioli, L., Vervoort, S. J., Adolfs, Y., Heijnen, C. J., Basak, O., Pasterkamp, R. J., et al. (2017). FOXP1 Promotes Embryonic Neural Stem Cell Differentiation by Repressing Jagged1 Expression. Stem Cell Rep 9, 1530–1545. doi: 10.1016/j.stemcr.2017.10.012
Brill, M. S., Snapyan, M., Wohlfrom, H., Ninkovic, J., Jawerka, M., Mastick, G. S., et al. (2008). A Dlx2- and Pax6-Dependent Transcriptional Code for Periglomerular Neuron Specification in the Adult Olfactory Bulb. J. Neurosci. 28, 6439–6452. doi: 10.1523/JNEUROSCI.0700-08.2008
Britz, O., Mattar, P., Nguyen, L., Langevin, L.-M., Zimmer, C., Alam, S., et al. (2006). A Role for Proneural Genes in the Maturation of Cortical Progenitor Cells. Cereb. Cortex 16, i138–i151. doi: 10.1093/cercor/bhj168
Castro, D. S., Martynoga, B., Parras, C., Ramesh, V., Pacary, E., Johnston, C., et al. (2011). A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 25, 930–945. doi: 10.1101/gad.627811
Cavallaro, M., Mariani, J., Lancini, C., Latorre, E., Caccia, R., Gullo, F., et al. (2008). Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 135, 541–557. doi: 10.1242/dev.010801
Dahl, E., Koseki, H., and Balling, R. (1997). Pax genes and organogenesis. Bioessays 19, 755–765. doi: 10.1002/bies.950190905
Dhanesh, S. B., Subashini, C., and James, J. (2016). Hes1: the maestro in neurogenesis. Cell. Mol. Life Sci. 73, 4019–4042. doi: 10.1007/s00018-016-2277-z
Dodonova, S. O., Zhu, F., Dienemann, C., Taipale, J., and Cramer, P. (2020). Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 580, 669–672. doi: 10.1038/s41586-020-2195-y
Dou, C., Lee, J., Liu, B., Liu, F., Massague, J., Xuan, S., et al. (2000). BF-1 Interferes with Transforming Growth Factor β Signaling by Associating with Smad Partners. Mol. Cell. Biol. 20, 6201–6211. doi: 10.1128/MCB.20.17.6201-6211.2000
Dou, C.-L., Li, S., and Lai, E. (1999). Dual Role of Brain Factor-1 in Regulating Growth and Patterning of the Cerebral Hemispheres. Cereb. Cortex 9, 543–550. doi: 10.1093/cercor/9.6.543
Eagleson, K. L., McFadyen-Ketchum, L. J. S., Ahrens, E. T., Mills, P. H., Does, M. D., Nickols, J., et al. (2007). Disruption of Foxg1 expression by knock-in of Cre recombinase: Effects on the development of the mouse telencephalon. Neuroscience 148, 385–399. doi: 10.1016/j.neuroscience.2007.06.012
Estivill-Torrus, G., Pearson, H., Heyningen, V., Price, D. J., and Rashbass, P. (2002). Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development 129, 455–466. doi: 10.1242/dev.129.2.455
Fasano, C. A., Phoenix, T. N., Kokovay, E., Lowry, N., Elkabetz, Y., Dimos, J. T., et al. (2009). Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 23, 561–574. doi: 10.1101/gad.1743709
Favaro, R., Valotta, M., Ferri, A. L. M., Latorre, E., Mariani, J., Giachino, C., et al. (2009). Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 12, 1248–1256. doi: 10.1038/nn.2397
Feldker, N., Ferrazzi, F., Schuhwerk, H., Widholz, S. A., Guenther, K., Frisch, I., et al. (2020). Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. EMBO J. 39:e103209. doi: 10.15252/embj.2019103209
Ferri, A. L. M., Cavallaro, M., Braida, D., Cristofano, A. D., Canta, A., Vezzani, A., et al. (2004). Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–3819. doi: 10.1242/dev.01204
Furusawa, T., Moribe, H., Kondoh, H., and Higashi, Y. (1999). Identification of CtBP1 and CtBP2 as Corepressors of Zinc Finger-Homeodomain Factor δEF1. Mol. Cell. Biol. 19, 8581–8590. doi: 10.1128/MCB.19.12.8581
Goossens, S., Vandamme, N., Vlierberghe, P. V., and Berx, G. (2017). EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochimica Et Biophys Acta Bba Rev Cancer 1868, 584–591. doi: 10.1016/j.bbcan.2017.06.006
Gozlan, O., and Sprinzak, D. (2023). Notch signaling in development and homeostasis. Development 150:1138. doi: 10.1242/dev.201138
Gupta, B., Errington, A. C., Jimenez-Pascual, A., Eftychidis, V., Brabletz, S., Stemmler, M. P., et al. (2021). The transcription factor ZEB1 regulates stem cell self-renewal and cell fate in the adult hippocampus. Cell Rep. 36:109588. doi: 10.1016/j.celrep.2021.109588
Hack, M. A., Saghatelyan, A., Chevigny, A., Pfeifer, A., Ashery-Padan, R., Lledo, P.-M., et al. (2005). Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat. Neurosci. 8, 865–872. doi: 10.1038/nn1479
Han, S., Okawa, S., Wilkinson, G. A., Ghazale, H., Adnani, L., Dixit, R., et al. (2021). Proneural genes define ground-state rules to regulate neurogenic patterning and cortical folding. Neuron 109, 2847–2863.e11. doi: 10.1016/j.neuron.2021.07.007
Hanashima, C., Shen, L., Li, S. C., and Lai, E. (2002). Brain Factor-1 Controls the Proliferation and Differentiation of Neocortical Progenitor Cells through Independent Mechanisms. J. Neurosci. 22, 6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002
Hannenhalli, S., and Kaestner, K. H. (2009). The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233–240. doi: 10.1038/nrg2523
Harris, L., Rigo, P., Stiehl, T., Gaber, Z. B., Austin, S. H. L., Masdeu, M. D. M., et al. (2021). Coordinated changes in cellular behavior ensure the lifelong maintenance of the hippocampal stem cell population. Cell Stem Cell 28, 863–876.e6. doi: 10.1016/j.stem.2021.01.003
Haubst, N., Berger, J., Radjendirane, V., Graw, J., Favor, J., Saunders, G. F., et al. (2004). Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development 131, 6131–6140. doi: 10.1242/dev.01524
Higashi, Y., Moribe, H., Takagi, T., Sekido, R., Kawakami, K., Kikutani, H., et al. (1997). Impairment of T Cell Development in δEF1 Mutant Mice. J. Exp. Med. 185, 1467–1480. doi: 10.1084/jem.185.8.1467
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., et al. (2002). Oscillatory Expression of the bHLH Factor Hes1 Regulated by a Negative Feedback Loop. Science 298, 840–843. doi: 10.1126/science.1074560
Holland, P. W., Booth, H. A. F., and Bruford, E. A. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5:47. doi: 10.1186/1741-7007-5-47
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory Control of Factors Determining Multipotency and Fate in Mouse Neural Progenitors. Science 342, 1203–1208. doi: 10.1126/science.1242366
Ishibashi, M., Moriyoshi, K., Sasai, Y., Shiota, K., Nakanishi, S., and Kageyama, R. (1994). Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 13, 1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x
Jackson, B. C., Carpenter, C., Nebert, D. W., and Vasiliou, V. (2010). Update of human and mouse forkhead box (FOX) gene families. Hum. Genomics 4:345. doi: 10.1186/1479-7364-4-5-345
Jacquet, B. V., Muthusamy, N., Sommerville, L. J., Xiao, G., Liang, H., Zhang, Y., et al. (2011). Specification of a Foxj1-dependent lineage in the forebrain is required for embryonic-to-postnatal transition of neurogenesis in the olfactory bulb. J Neurosci Official J Soc Neurosci 31, 9368–9382. doi: 10.1523/jneurosci.0171-11.2011
Jacquet, B. V., Salinas-Mondragon, R., Liang, H., Therit, B., Buie, J. D., Dykstra, M., et al. (2009). FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136, 4021–4031. doi: 10.1242/dev.041129
Jiang, Y., Yan, L., Xia, L., Lu, X., Zhu, W., Ding, D., et al. (2018). Zinc finger E-box–binding homeobox 1 (ZEB1) is required for neural differentiation of human embryonic stem cells. J. Biol. Chem. 293, 19317–19329. doi: 10.1074/jbc.RA118.005498
Jiménez, G., Paroush, Z., and Ish-Horowicz, D. (1997). Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 11, 3072–3082. doi: 10.1101/gad.11.22.3072
Jones, S. (2004). An overview of the basic helix-loop-helix proteins. Genome Biol. 5:226. doi: 10.1186/gb-2004-5-6-226
Jurkowski, M. P., Bettio, L., Woo, E. K., Patten, A., Yau, S.-Y., and Gil-Mohapel, J. (2020). Beyond the Hippocampus and the SVZ: Adult Neurogenesis Throughout the Brain. Front. Cell. Neurosci. 14:576444. doi: 10.3389/fncel.2020.576444
Kageyama, R., Ohtsuka, T., and Kobayashi, T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251. doi: 10.1242/dev.000786
Kaise, T., and Kageyama, R. (2021). Hes1 oscillation frequency correlates with activation of neural stem cells. Gene Expr. Patterns 40:119170. doi: 10.1016/j.gep.2021.119170
Kalin, T. V., Ustiyan, V., and Kalinichenko, V. V. (2011). Multiple faces of FoxM1 transcription factor. Cell Cycle 10, 396–405. doi: 10.4161/cc.10.3.14709
Kim, E. J., Ables, J. L., Dickel, L. K., Eisch, A. J., and Johnson, J. E. (2011). Ascl1 (Mash1) Defines Cells with Long-Term Neurogenic Potential in Subgranular and Subventricular Zones in Adult Mouse Brain. PLoS One 6:e18472. doi: 10.1371/journal.pone.0018472
Kobayashi, T., and Kageyama, R. (2011). Hes1 Oscillations Contribute to Heterogeneous Differentiation Responses in Embryonic Stem Cells. Genes 2, 219–228. doi: 10.3390/genes2010219
Kobayashi, T., Mizuno, H., Imayoshi, I., Furusawa, C., Shirahige, K., and Kageyama, R. (2009). The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870–1875. doi: 10.1101/gad.1823109
Kohwi, M., Osumi, N., Rubenstein, J. L. R., and Alvarez-Buylla, A. (2005). Pax6 Is Required for Making Specific Subpopulations of Granule and Periglomerular Neurons in the Olfactory Bulb. J. Neurosci. 25, 6997–7003. doi: 10.1523/JNEUROSCI.1435-05.2005
Leung, R. F., George, A. M., Roussel, E. M., Faux, M. C., Wigle, J. T., and Eisenstat, D. D. (2022). Genetic Regulation of Vertebrate Forebrain Development by Homeobox Genes. Front. Neurosci. 16:843794. doi: 10.3389/fnins.2022.843794
Li, X., Yao, B., Chen, L., Kang, Y., Li, Y., Cheng, Y., et al. (2017). Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun. 8:15903. doi: 10.1038/ncomms15903
Li, X., Zou, S., Tu, X., Hao, S., Jiang, T., and Chen, J.-G. (2023). Inhibition of Foxp4 Disrupts Cadherin-based Adhesion of Radial Glial Cells, Leading to Abnormal Differentiation and Migration of Cortical Neurons in Mice. Neurosci. Bull. 1–15:7. doi: 10.1007/s12264-022-01004-7
Liu, Y., El-Naggar, S., Darling, D. S., Higashi, Y., and Dean, D. C. (2008). Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135, 579–588. doi: 10.1242/dev.007047
Liu, J., Liu, Y., Shao, J., Li, Y., Qin, L., Shen, H., et al. (2019). Zeb1 is important for proper cleavage plane orientation of dividing progenitors and neuronal migration in the mouse neocortex. Cell Death Differ. 26, 2479–2492. doi: 10.1038/s41418-019-0314-9
Llorens, M. C., Lorenzatti, G., Cavallo, N. L., Vaglienti, M. V., Perrone, A. P., Carenbauer, A. L., et al. (2016). Phosphorylation Regulates Functions of ZEB1 Transcription Factor. J. Cell. Physiol. 231, 2205–2217. doi: 10.1002/jcp.25338
Maeda, Y., Isomura, A., Masaki, T., and Kageyama, R. (2023). Differential cell-cycle control by oscillatory versus sustained Hes1 expression via p21. Cell Rep. 42:112520. doi: 10.1016/j.celrep.2023.112520
Maekawa, M., Takashima, N., Arai, Y., Nomura, T., Inokuchi, K., Yuasa, S., et al. (2005). Pax6 is required for production and maintenance of progenitor cells in postnatal hippocampal neurogenesis. Genes Cells 10, 1001–1014. doi: 10.1111/j.1365-2443.2005.00893.x
Manuel, M., Georgala, P. A., Carr, C. B., Chanas, S., Kleinjan, D. A., Martynoga, B., et al. (2006). Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development 134, 545–555. doi: 10.1242/dev.02764
Manuel, M. N., Mi, D., Mason, J. O., and Price, D. J. (2015). Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 9:70. doi: 10.3389/fncel.2015.00070
Martynoga, B., Drechsel, D., and Guillemot, F. (2012). Molecular Control of Neurogenesis: A View from the Mammalian Cerebral Cortex. Cold Spring Harb. Perspect. Biol. 4:a008359. doi: 10.1101/cshperspect.a008359
Mattar, P., Langevin, L. M., Markham, K., Klenin, N., Shivji, S., Zinyk, D., et al. (2008). Basic Helix-Loop-Helix Transcription Factors Cooperate To Specify a Cortical Projection Neuron Identity. Mol. Cell. Biol. 28, 1456–1469. doi: 10.1128/MCB.01510-07
Medema, R. H., Kops, G. J., Bos, J. L., and Burgering, B. M. (2000). AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404, 782–787. doi: 10.1038/35008115
Mercurio, S., Serra, L., and Nicolis, S. K. (2019). More than just Stem Cells: Functional Roles of the Transcription Factor Sox2 in Differentiated Glia and Neurons. Int. J. Mol. Sci. 20:4540. doi: 10.3390/ijms20184540
Mi, D., Carr, C. B., Georgala, P. A., Huang, Y.-T., Manuel, M. N., Jeanes, E., et al. (2013). Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb. Neuron 78, 269–284. doi: 10.1016/j.neuron.2013.02.012
Miskinyte, G., Hansen, M. G., Monni, E., Lam, M., Bengzon, J., Lindvall, O., et al. (2018). Transcription factor programming of human ES cells generates functional neurons expressing both upper and deep layer cortical markers. PLoS One 13:e0204688. doi: 10.1371/journal.pone.0204688
Miyagi, S., Masui, S., Niwa, H., Saito, T., Shimazaki, T., Okano, H., et al. (2008). Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 582, 2811–2815. doi: 10.1016/j.febslet.2008.07.011
Murata, K., Hattori, M., Hirai, N., Shinozuka, Y., Hirata, H., Kageyama, R., et al. (2005). Hes1 Directly Controls Cell Proliferation through the Transcriptional Repression of p27Kip1. Mol. Cell. Biol. 25, 4262–4271. doi: 10.1128/MCB.25.10.4262-4271.2005
Nakamura, Y., Ozaki, T., Koseki, H., Nakagawara, A., and Sakiyama, S. (2003). Accumulation of p27KIP1 is associated with BMP2-induced growth arrest and neuronal differentiation of human neuroblastoma-derived cell lines. Biochem Bioph Res 307, 206–213. doi: 10.1016/S0006-291X(03)01138-0
Nakamura, Y., Sakakibara, S., Miyata, T., Ogawa, M., Shimazaki, T., Weiss, S., et al. (2000). The bHLH Gene Hes1 as a Repressor of the Neuronal Commitment of CNS Stem Cells. J. Neurosci. 20, 283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000
Nieto, M., Schuurmans, C., Britz, O., and Guillemot, F. (2001). Neural bHLH Genes Control the Neuronal versus Glial Fate Decision in Cortical Progenitors. Neuron 29, 401–413. doi: 10.1016/S0896-6273(01)00214-8
Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F., and Kageyama, R. (1999). Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207. doi: 10.1093/emboj/18.8.2196
Osumi, N., Shinohara, H., Numayama-Tsuruta, K., and Maekawa, M. (2008). Concise Review: Pax6 Transcription Factor Contributes to both Embryonic and Adult Neurogenesis as a Multifunctional Regulator. Stem Cells 26, 1663–1672. doi: 10.1634/stemcells.2007-0884
Ozen, I., Galichet, C., Watts, C., Parras, C., Guillemot, F., and Raineteau, O. (2007). Proliferating neuronal progenitors in the postnatal hippocampus transiently express the proneural gene Ngn2. Eur. J. Neurosci. 25, 2591–2603. doi: 10.1111/j.1460-9568.2007.05541.x
Paik, J., Ding, Z., Narurkar, R., Ramkissoon, S., Muller, F., Kamoun, W. S., et al. (2009). FoxOs Cooperatively Regulate Diverse Pathways Governing Neural Stem Cell Homeostasis. Cell Stem Cell 5, 540–553. doi: 10.1016/j.stem.2009.09.013
Parras, C. M., Schuurmans, C., Scardigli, R., Kim, J., Anderson, D. J., and Guillemot, F. (2002). Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16, 324–338. doi: 10.1101/gad.940902
Pearson, C. A., Moore, D. M., Tucker, H. O., Dekker, J. D., Hu, H., Miquelajáuregui, A., et al. (2020). Foxp1 Regulates Neural Stem Cell Self-Renewal and Bias Toward Deep Layer Cortical Fates. Cell Rep. 30, 1964–1981.e3. doi: 10.1016/j.celrep.2020.01.034
Pevny, L. H., and Nicolis, S. K. (2010). Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 42, 421–424. doi: 10.1016/j.biocel.2009.08.018
Pilz, G. A., Bottes, S., Betizeau, M., Jörg, D. J., Carta, S., Simons, B. D., et al. (2018). Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662. doi: 10.1126/science.aao5056
Quinn, J. C., Molinek, M., Martynoga, B. S., Zaki, P. A., Faedo, A., Bulfone, A., et al. (2007). Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev. Biol. 302, 50–65. doi: 10.1016/j.ydbio.2006.08.035
Remacle, J. E., Kraft, H., Lerchner, W., Wuytens, G., Collart, C., Verschueren, K., et al. (1999). New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J. 18, 5073–5084. doi: 10.1093/emboj/18.18.5073
Renault, V. M., Rafalski, V. A., Morgan, A. A., Salih, D. A. M., Brett, J. O., Webb, A. E., et al. (2009). FoxO3 Regulates Neural Stem Cell Homeostasis. Cell Stem Cell 5, 527–539. doi: 10.1016/j.stem.2009.09.014
Ro, S.-H., Liu, D., Yeo, H., and Paik, J. (2013). FoxOs in neural stem cell fate decision. Arch. Biochem. Biophys. 534, 55–63. doi: 10.1016/j.abb.2012.07.017
Rousso, D. L., Pearson, C. A., Gaber, Z. B., Miquelajauregui, A., Li, S., Portera-Cailliau, C., et al. (2012). Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron 74, 314–330. doi: 10.1016/j.neuron.2012.02.024
Roybon, L., Hjalt, T., Stott, S., Guillemot, F., Li, J.-Y., and Brundin, P. (2009). Neurogenin2 Directs Granule Neuroblast Production and Amplification while NeuroD1 Specifies Neuronal Fate during Hippocampal Neurogenesis. PLoS One 4:e4779. doi: 10.1371/journal.pone.0004779
Sansom, S. N., Griffiths, D. S., Faedo, A., Kleinjan, D.-J., Ruan, Y., Smith, J., et al. (2009). The Level of the Transcription Factor Pax6 Is Essential for Controlling the Balance between Neural Stem Cell Self-Renewal and Neurogenesis. PLoS Genet. 5:e1000511. doi: 10.1371/journal.pgen.1000511
Sarkar, A., and Hochedlinger, K. (2013). The Sox Family of Transcription Factors: Versatile Regulators of Stem and Progenitor Cell Fate. Cell Stem Cell 12, 15–30. doi: 10.1016/j.stem.2012.12.007
Sasai, Y., Kageyama, R., Tagawa, Y., Shigemoto, R., and Nakanishi, S. (1992). Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 6, 2620–2634. doi: 10.1101/gad.6.12b.2620
Schäffner, I., Minakaki, G., Khan, M. A., Balta, E.-A., Schlötzer-Schrehardt, U., Schwarz, T. J., et al. (2018). FoxO Function Is Essential for Maintenance of Autophagic Flux and Neuronal Morphogenesis in Adult Neurogenesis. Neuron 99, 1188–1203.e6. doi: 10.1016/j.neuron.2018.08.017
Schäffner, I., Wittmann, M.-T., Vogel, T., and Lie, D. C. (2023). Differential vulnerability of adult neurogenic niches to dosage of the neurodevelopmental-disorder linked gene Foxg1. Mol. Psychiatry 28, 497–514. doi: 10.1038/s41380-022-01497-8
Schüller, U., Zhao, Q., Godinho, S. A., Heine, V. M., Medema, R. H., Pellman, D., et al. (2007). Forkhead Transcription Factor FoxM1 Regulates Mitotic Entry and Prevents Spindle Defects in Cerebellar Granule Neuron Precursors. Mol. Cell. Biol. 27, 8259–8270. doi: 10.1128/MCB.00707-07
Sekido, R., Murai, K., Kamachi, Y., and Kondoh, H. (1997). Two mechanisms in the action of repressor δEF1: binding site competition with an activator and active repression. Genes Cells 2, 771–783. doi: 10.1046/j.1365-2443.1997.1570355.x
Sekido, R., Takagi, T., Okanami, M., Moribe, H., Yamamura, M., Higashi, Y., et al. (1996). Organization of the gene encoding transcriptional repressor δEF1 and cross-species conservation of its domains. Gene 173, 227–232. doi: 10.1016/0378-1119(96)00185-0
Seoane, J., Le, H.-V., Shen, L., Anderson, S. A., and Massagué, J. (2004). Integration of Smad and Forkhead Pathways in the Control of Neuroepithelial and Glioblastoma Cell Proliferation. Cells 117, 211–223. doi: 10.1016/S0092-8674(04)00298-3
Sharov, A. A., Sharova, T. Y., Mardaryev, A. N., Vignano, A. T., Atoyan, R., Weiner, L., et al. (2006). Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc Natl Acad Sci 103, 18166–18171. doi: 10.1073/pnas.0608899103
Shen, L., Nam, H., Song, P., Moore, H., and Anderson, S. A. (2006). FoxG1 haploinsufficiency results in impaired neurogenesis in the postnatal hippocampus and contextual memory deficits. Hippocampus 16, 875–890. doi: 10.1002/hipo.20218
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. doi: 10.1016/j.neuron.2008.02.014
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2011). Dynamic Expression of Notch Signaling Genes in Neural Stem/Progenitor Cells. Front. Neurosci. 5:78. doi: 10.3389/fnins.2011.00078
Siegenthaler, J. A., Tremper-Wells, B. A., and Miller, M. W. (2008). Foxg1 Haploinsufficiency Reduces the Population of Cortical Intermediate Progenitor Cells: Effect of Increased p21 Expression. Cereb. Cortex 18, 1865–1875. doi: 10.1093/cercor/bhm209
Sueda, R., Imayoshi, I., Harima, Y., and Kageyama, R. (2019). High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 33, 511–523. doi: 10.1101/gad.323196.118
Takebayashi, K., Sasai, Y., Sakai, Y., Watanabe, T., Nakanishi, S., and Kageyama, R. (1994). Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J. Biol. Chem. 269, 5150–5156. doi: 10.1016/S0021-9258(17)37668-8
Tian, C., Gong, Y., Yang, Y., Shen, W., Wang, K., Liu, J., et al. (2012). Foxg1 has an essential role in postnatal development of the dentate gyrus. J Neurosci Official J Soc Neurosci 32, 2931–2949. doi: 10.1523/JNEUROSCI.5240-11.2012
Toresson, H., Potter, S. S., and Campbell, K. (2000). Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development 127, 4361–4371. doi: 10.1242/dev.127.20.4361
Tsui, D., Vessey, J. P., Tomita, H., Kaplan, D. R., and Miller, F. D. (2013). FoxP2 Regulates Neurogenesis during Embryonic Cortical Development. J. Neurosci. 33, 244–258. doi: 10.1523/JNEUROSCI.1665-12.2013
Urbán, N., Berg, D. L. C., Forget, A., Andersen, J., Demmers, J. A. A., Hunt, C., et al. (2016). Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353, 292–295. doi: 10.1126/science.aaf4802
Vandewalle, C., Roy, F. V., and Berx, G. (2009). The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 66, 773–787. doi: 10.1007/s00018-008-8465-8
Vasconcelos, F. F., and Castro, D. S. (2014). Transcriptional control of vertebrate neurogenesis by the proneural factor Ascl1. Front. Cell. Neurosci. 8:412. doi: 10.3389/fncel.2014.00412
Vezzali, R., Weise, S. C., Hellbach, N., Machado, V., Heidrich, S., and Vogel, T. (2016). The FOXG1/FOXO/SMAD network balances proliferation and differentiation of cortical progenitors and activates Kcnh3 expression in mature neurons. Oncotarget 7, 37436–37455. doi: 10.18632/oncotarget.9545
Wang, Z., Park, H. J., Carr, J. R., Chen, Y., Zheng, Y., Li, J., et al. (2011). FoxM1 in Tumorigenicity of the Neuroblastoma Cells and Renewal of the Neural Progenitors. Cancer Res. 71, 4292–4302. doi: 10.1158/0008-5472.CAN-10-4087
Wang, H., Xiao, Z., Zheng, J., Wu, J., Hu, X.-L., Yang, X., et al. (2019). ZEB1 Represses Neural Differentiation and Cooperates with CTBP2 to Dynamically Regulate Cell Migration during Neocortex Development. Cell Rep. 27, 2335–2353.e6. doi: 10.1016/j.celrep.2019.04.081
Wang, J., Zhai, H.-R., Ma, S.-F., Shi, H.-Z., Zhang, W.-J., Yun, Q., et al. (2022). FOXG1 Contributes Adult Hippocampal Neurogenesis in Mice. Int. J. Mol. Sci. 23:14979. doi: 10.3390/ijms232314979
Webb, A. E., Pollina, E. A., Vierbuchen, T., Urbán, N., Ucar, D., Leeman, D. S., et al. (2013). FOXO3 Shares Common Targets with ASCL1 Genome-wide and Inhibits ASCL1-Dependent Neurogenesis. Cell Rep. 4, 477–491. doi: 10.1016/j.celrep.2013.06.035
Wegner, M. (2010). All purpose Sox: The many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 42, 381–390. doi: 10.1016/j.biocel.2009.07.006
Wegner, M., and Stolt, C. C. (2005). From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 28, 583–588. doi: 10.1016/j.tins.2005.08.008
Wu, X., Gu, X., Han, X., Du, A., Jiang, Y., Zhang, X., et al. (2014). A Novel Function for Foxm1 in Interkinetic Nuclear Migration in the Developing Telencephalon and Anxiety-Related Behavior. J. Neurosci. 34, 1510–1522. doi: 10.1523/JNEUROSCI.2549-13.2014
Xu, C., Fan, W., Zhang, Y., Loh, H. H., and Law, P. (2021). Kappa opioid receptor controls neural stem cell differentiation via a miR-7a/Pax6 dependent pathway. Stem Cells 39, 600–616. doi: 10.1002/stem.3334
Xuan, S., Baptista, C. A., Balas, G., Tao, W., Soares, V. C., and Lai, E. (1995). Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14, 1141–1152. doi: 10.1016/0896-6273(95)90262-7
Yao, J., Lai, E., and Stifani, S. (2001). The Winged-Helix Protein Brain Factor 1 Interacts with Groucho and Hes Proteins To Repress Transcription. Mol. Cell. Biol. 21, 1962–1972. doi: 10.1128/MCB.21.6.1962-1972.2001
Yen, G., Croci, A., Dowling, A., Zhang, S., Zoeller, R. T., and Darling, D. S. (2001). Developmental and functional evidence of a role for Zfhep in neural cell development. Mol. Brain Res. 96, 59–67. doi: 10.1016/S0169-328X(01)00267-4
Yun, K., Potter, S., and Rubenstein, J. L. (2001). Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128, 193–205. doi: 10.1242/dev.128.2.193
Keywords: neurogenesis, transcription factor, embryonic, adult, bHLH, homeodomain, forkhead, Zeb1
Citation: Singh N, Siebzehnrubl FA and Martinez-Garay I (2023) Transcriptional control of embryonic and adult neural progenitor activity. Front. Neurosci. 17:1217596. doi: 10.3389/fnins.2023.1217596
Edited by:
Chitra D. Mandyam, United States Department of Veterans Affairs, United StatesReviewed by:
Igor Iskusnykh, University of Tennessee Health Science Center (UTHSC), United StatesCopyright © 2023 Singh, Siebzehnrubl and Martinez-Garay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabel Martinez-Garay, bWFydGluZXpnYXJheWlAY2FyZGlmZi5hYy51aw==
 Niharika Singh1,2
Niharika Singh1,2 Florian A. Siebzehnrubl
Florian A. Siebzehnrubl Isabel Martinez-Garay
Isabel Martinez-Garay