
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurosci., 03 May 2023
Sec. Auditory Cognitive Neuroscience
Volume 17 - 2023 | https://doi.org/10.3389/fnins.2023.1177791
This article is part of the Research TopicGene Therapy for Hearing Loss: From Mechanism to Clinic, volume IIView all 15 articles
 Jipeng Wang1†
Jipeng Wang1† Jianwei Zheng2†
Jianwei Zheng2† Haiyan Wang3†
Haiyan Wang3† Haoying He4
Haoying He4 Shuang Li3
Shuang Li3 Ya Zhang3
Ya Zhang3 You Wang5*
You Wang5* Xiaoxiang Xu3*
Xiaoxiang Xu3* Shuyi Wang1*
Shuyi Wang1*Sensorineural hearing loss is typically caused by damage to the cochlear hair cells (HCs) due to external stimuli or because of one’s genetic factors and the inability to convert sound mechanical energy into nerve impulses. Adult mammalian cochlear HCs cannot regenerate spontaneously; therefore, this type of deafness is usually considered irreversible. Studies on the developmental mechanisms of HC differentiation have revealed that nonsensory cells in the cochlea acquire the ability to differentiate into HCs after the overexpression of specific genes, such as Atoh1, which makes HC regeneration possible. Gene therapy, through in vitro selection and editing of target genes, transforms exogenous gene fragments into target cells and alters the expression of genes in target cells to activate the corresponding differentiation developmental program in target cells. This review summarizes the genes that have been associated with the growth and development of cochlear HCs in recent years and provides an overview of gene therapy approaches in the field of HC regeneration. It concludes with a discussion of the limitations of the current therapeutic approaches to facilitate the early implementation of this therapy in a clinical setting.
Deafness is the most common neurological disorder in humans, which has seriously affected the normal life of human beings. According to the World Hearing Report published by the World Health Organization, almost 1.5 billion people worldwide have varying degrees of hearing loss, and 430 million people are at a level of severe hearing loss that requires rehabilitation (Chadha et al., 2021). Deafness can be categorized as conductive, sensorineural, and mixed deafness (Cunningham and Tucci, 2017). The more common type of deafness is sensorineural deafness caused by death or functional loss of cochlear hair cells (HCs). HCs are the most critical cells for sound perception and transmission in the inner ear sensory cells, and their function is to convert the mechanical signals of sound coming in from the environment into electrical signals that the nervous system can perceive (Deans, 2021). HCs are the most critical cells in the mammalian inner ear sensory epithelium. Studies have shown that (Fujioka et al., 2015) compared with nonmammals (birds and reptiles), HCs cannot regenerate spontaneously in mammals; thus, HC damage often results in permanent hearing loss.
Gene therapy involves transferring an external normal or therapeutic gene, via a vector, to a target cell in the body, causing the target cell to express the relevant gene or to modify the pertinent gene as a therapeutic approach. It has now become a potential treatment for genetic deafness. In several animal models, gene therapy has been used to transfer several genes such as Syne4 (Taiber et al., 2021), Tmc1 (Marcovich et al., 2022), and Clarin-1 (Dulon et al., 2018) moved into the cochlea and has significantly improved the degree of hearing impairment in the study animals. During the developmental differentiation and maturation of inner ear HCs, there is also the regulation of multiple genes (Bermingham et al., 1999; Hertzano et al., 2004; Ikeda et al., 2015; Hou et al., 2019; Ding et al., 2020; Jen et al., 2022) and signaling pathways (Benito-Gonzalez and Doetzlhofer, 2014; Waqas et al., 2016; Ebeid and Huh, 2017; Bai et al., 2021). By interfering with these, the normal differentiation of HCs can be restored, and support cells (SCs) can be stimulated to re-differentiate and produce HCs (Menendez et al., 2020). The aim is to treat hearing loss associated with HC damage. In this review, we highlight how gene therapy can promote hair cell regeneration as a way to alleviate the hearing loss in patients and provide an outlook for future research in this area.
During inner ear development, many transcription factors, including Atoh1, are involved in the proliferation and differentiation of HCs (Figure 1). In a mouse model of inner ear development, Atoh1 was first expressed in the basal progenitor HCs at embryonic stage (E) 13.5 d, and gradually increased until the cochlear spiral matured at E17.5, and gradually decreased after postnatal (P) 0 d. After P7, Atoh1 expression could not be measured in the spiral (Lumpkin et al., 2003; Cotanche and Kaiser, 2010; Cai et al., 2013). In contrast, the change of Atoh1-related downstream targeting factor Gfi1 was consistent with the change of Atoh1, which started to be expressed at E12.5 and also gradually decreased in expression with the end of embryonic stage (Wallis et al., 2003). Conversely, Pou4f3 and Barhl1 were detected in cochlear basal HCs only at E13.5 and E14.5, respectively, and continued to be expressed after birth (Xiang et al., 1997; Hou et al., 2019; Figure 2).
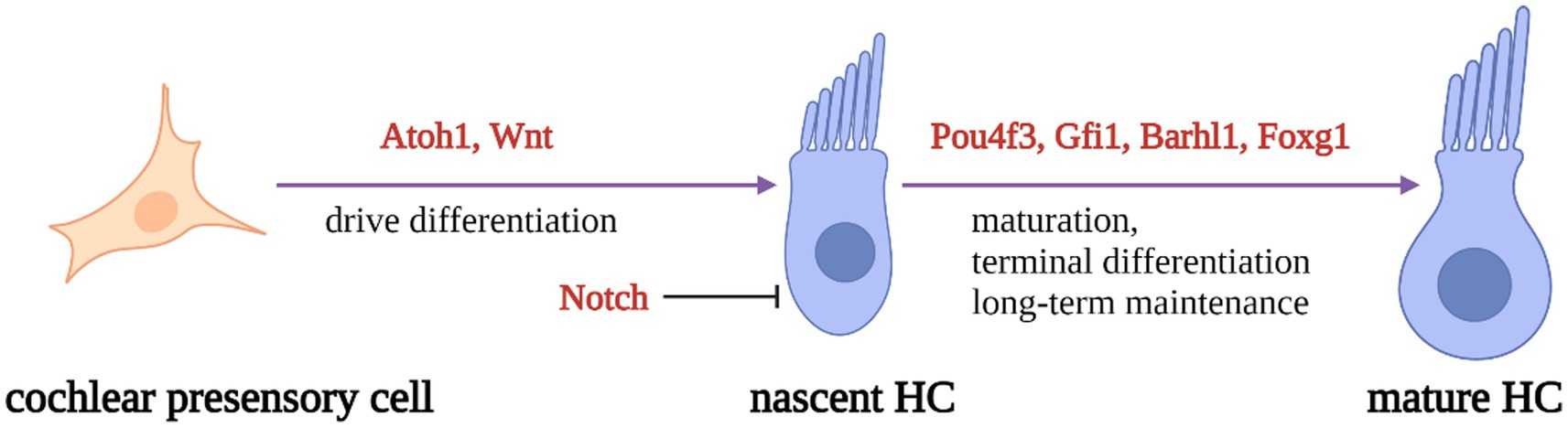
Figure 1. Schematic diagram of HC development process. Regulatory factors Atoh1 and Wnt signaling pathways are necessary for presensory cells to differentiate into initial HCs. Atoh1 downstream targeting factors (Pouf4, Gfi1, Barhl3) and Foxg1 play essential roles in nascent HCs maturation and long-term maintenance. At the same time, Notch signal pathway can inhibit the expression of Atoh1 in presensory cells and regulate the differentiation of HCs.
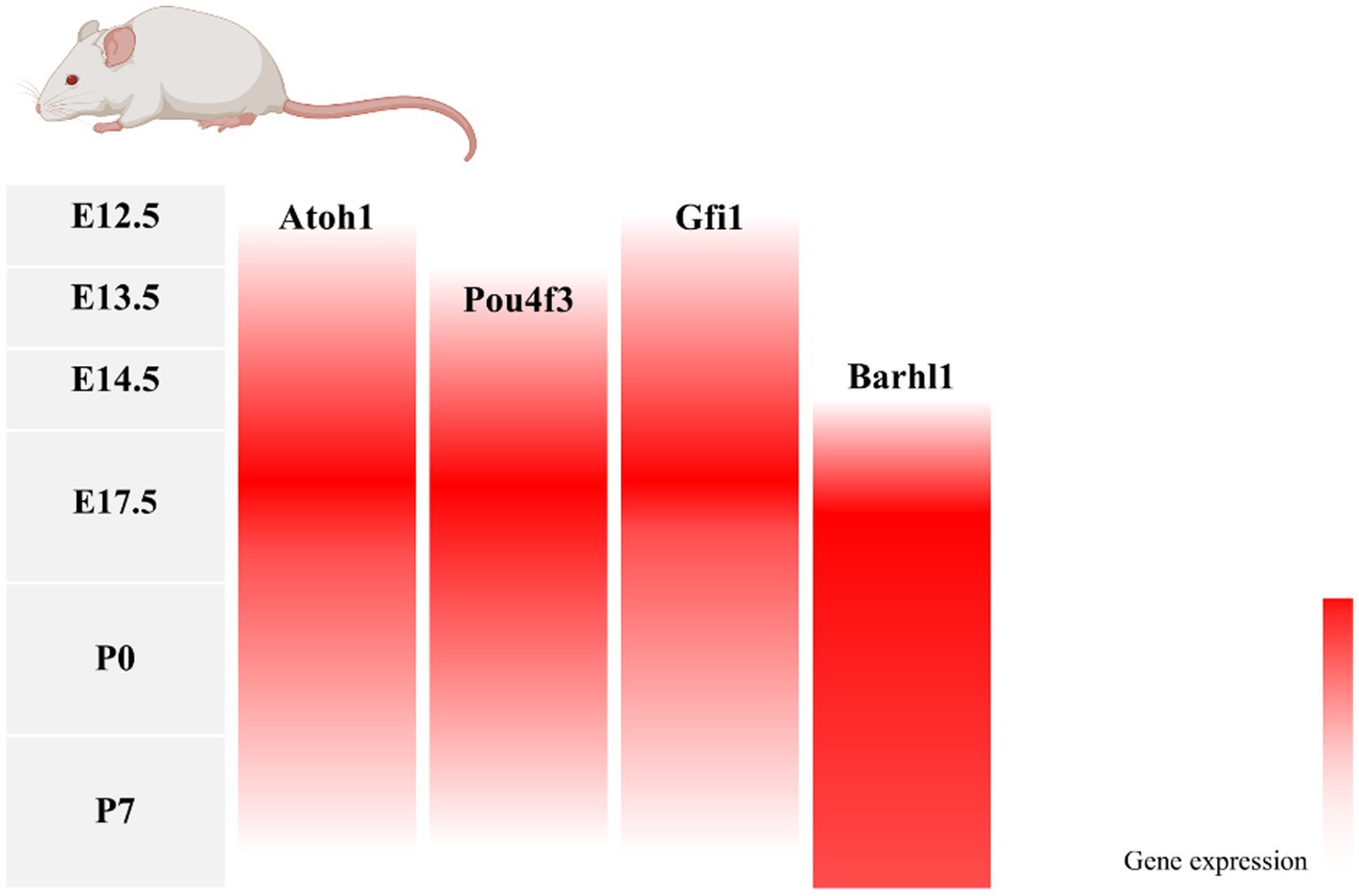
Figure 2. Changes in gene expression with age in a mouse model of inner ear development. During mouse embryonic cochlea development, Atoh1 and its downstream target transcription factors Pou4f3, Gfi1, and Barhl1 were successively expressed, with Atoh1, Pou4f3, and Gfi1 decreasing in expression after birth as the cochlea matured, and Barhl1 continuing to be expressed.
Atoh1, also known as Math1, is a helix–loop–helix (bHLH) family transcription factor with a coding sequence of 1.053 kb, encoding a protein of size 17.9 kDa. Atoh1 was the first transcription factor identified in differentiated HC progenitors and is essential for HCs growth and differentiation (Bermingham et al., 1999). In Atoh1 mutant mice, all inner ear sensory regions do not differentiate to produce HCs (Pan et al., 2011). Further studies revealed that the dependence of HCs on Atoh1 diminishes as sensory cells in the cochlea develop and mature (Chonko et al., 2013). However, it is not the case that the HCs are unaffected by Atoh1 after cochlear growth, as Atoh1 deficiency also disrupts the standard hair bundle structure of the auditory system and eventually leads to the delayed death of HCs (Cai et al., 2013; Cheng et al., 2016). In contrast, the enhanced expression of Atoh1 promotes the normal development of HCs and improves hearing (Izumikawa et al., 2005; Luo et al., 2022). Thus, the entire auditory system, from the developmental to mature stages, is inseparable from the regulation of Atoh1.
Due to the importance of Atoh1 in HCs, identifying the downstream targeting factors of Atoch1 is crucial to investigate developmental mechanisms. Atoh1 target groups were identified in mouse cerebellum and cochlea development was studied using genome-wide Atoh1 sequencing methods (Klisch et al., 2011; Cai et al., 2015). The direct Atoh1 target genes Pou4f3, Gfi1, and Barhl1 are associated with the normal differentiation and regeneration of HCs (Wallis et al., 2003; Zhong et al., 2018; Chen et al., 2021). The Atoh1 target group has been identified in the cochlea.
Pou4f3, a Pou family transcription factor, is the dominant nonsyndromic deafness 15 (DFNA15) deafness-causative gene (Vahava et al., 1998) and a downstream target of Atoh1 activation (Ikeda et al., 2015). During HC differentiation, there is a feed-forward synergy between Atoh1 and Pou4f3, with Atoh1 first stimulating Pou4f3 expression, which releases Atoh1-related elements in a closed state to activate a series of HC-specific enhancers (Yu et al., 2021). Gfi1 is a zinc-finger transcription factor. Studies have shown that Gfi1 expression is regulated by Pou4f3 (Hertzano et al., 2004). Gfi1 represses neuronal gene expression early in the development of HCs, and in the absence of Gfi1, cochlear maturation is stalled (Matern et al., 2020). Barhl1 is a BarH-like homologous domain transcription factor explicitly expressed in all HCs in the cochlear (Bulfone et al., 2000). Mice lacking Barhl1 developed severe age-related hearing loss. Further studies have found that HC death in Barhl1-null mice begins after 6 days of life and progresses slowly over several months (Li et al., 2002), suggesting that Barhl1 may be involved in the terminal differentiation and long-term maintenance of HCs.
In conclusion, Atoh1 is a crucial transcription factor in the formation of HCs, and Atoh1 mutants lose the ability to generate HC progenitors; Pou4f3 and Gfi1, the genes downstream of Atoh1, are required for the late developmental maturation of progenitors into HCs, and delayed degeneration of HCs occurs in Pou4f3 and Gfi1 mutants; Barhl1 is associated with the long-term maintenance of HCs. In Barhl1 mutants, HCs mature but eventually die within a certain period.
Foxg1, a member of the FOX family, is known to regulate ATP synthesis and metabolism in mitochondria (Pancrazi et al., 2015). Foxg1 is essential for proper development and formation of the inner ear. In Foxg1-null mice, severe inner ear malformations, including shortened cochleae with multiple rows of HCs and supporting cells and reduced or even absent cristae have been reported (Pauley et al., 2006; Hwang et al., 2009). Mechanistically, deletion of Foxg1 causes inhibition of Notch, Wnt, IGF, and EGF signaling pathways, production of HCs, and induction of their subsequent apoptosis (He et al., 2019). In addition, Foxg1 regulates auditory degeneration through the regulation of autophagy. In the Foxg1 downregulated group, the autophagic pathway was significantly inhibited, and reactive oxygen species levels were significantly increased, ultimately leading to the apoptosis of HCs (He et al., 2021). Similarly, Foxg1 downregulation also considerably increased the sensitivity of HCs to lipopolysaccharide-induced inflammation and accelerated the apoptosis of HCs under inflammatory conditions (He et al., 2020).
Due to the critical role that individual genes play in the differentiation and development of HCs, developmental failure during HC differentiation occurs after the deletion of relevant genes. Therefore, inducing re-differentiation to generate new HCs by reprogramming HC-related genes in the inner ear SCs is a potential way to improve HC-related hearing impairment (Costa et al., 2015; Ni et al., 2016; Shibata et al., 2020). HCs regenerate mainly through two pathways: one activates non-sensory cell activation to re-enter the cell cycle and further divide and differentiate into HCs; the other directly induces non-sensory cells to transdifferentiate into HCs without mitosis (Figure 3).
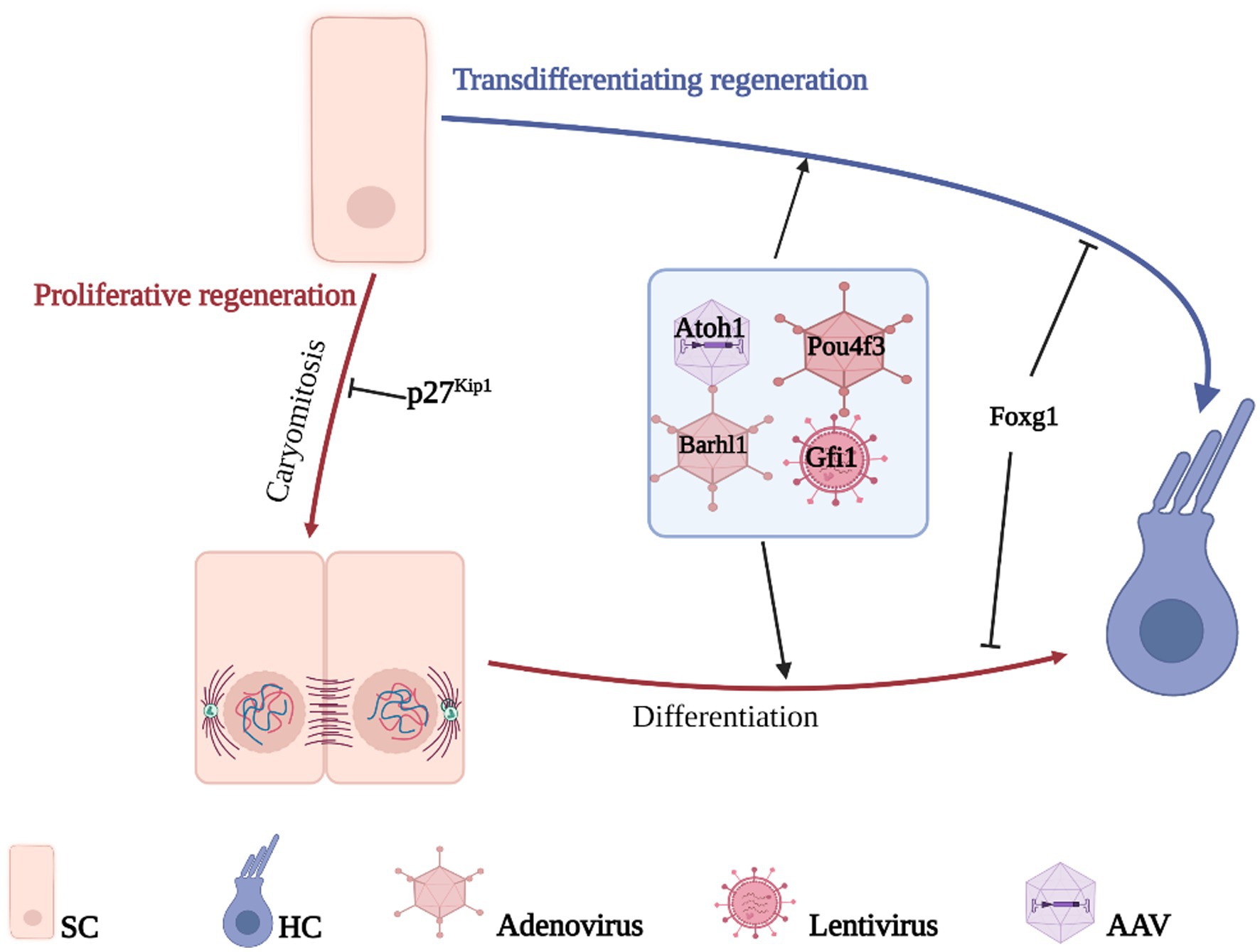
Figure 3. Gene therapy promotes HCs regeneration. HCs regenerate mainly through two pathways, direct transdifferentiation of SCs or proliferative differentiation of SCs, the difference between the two is whether mitosis is performed or not. The genes related to the growth, development, and maturation of HCs are transferred to the target cells by gene vector so that the non-sensory cells in the cochlea can differentiate into HCs and realize the regeneration of HCs.
Cell cycle inhibitors are critical for maintaining cells in a quiescent state after mitosis, and therefore activation of non-sensory cells to re-enter the cell cycle requires regulation of the corresponding inhibitors. P27Kip1 (p27), a member of the Cip/Kip family of cell cyclin-dependent protein kinase inhibitors, is significantly upregulated in dormant cells (Bencivenga et al., 2021) and has been shown to be a common cell cycle inhibitor for sensory and non-sensory cells in the inner ear (Chen and Segil, 1999; Löwenheim et al., 1999). Knocking down p27 in isolated mouse cochlear cells can effectively activate the proliferation of SCs in cochlea to re-enter the cell cycle, and mitotically generated SCs retain the ability to redifferentiate into HCs (Löwenheim et al., 1999; White et al., 2006; Ono et al., 2009; Maass et al., 2013). Further studies revealed that in p27 knockout mice, more than just SCs broke out of cell cycle quiescence, HCs also gained some proliferative capacity (Walters et al., 2014), and similar effects were achieved using Retinoic acid to inhibit p27 (Rubbini et al., 2015). Combined with p27 knockdown, the transdifferentiation of Atoh1 to produce HCs is not limited to the embryonic period and enables the regeneration of HCs in the mature mouse cochlea (Walters et al., 2017). Unfortunately, the production of HCs by mitotic re-differentiation of SCs does not function properly in mammals, but p27 remains a potential target for the regeneration of cochlear HCs.
Atoh1, the first HC development–related transcription factor to be identified, plays an irreplaceable role in HC regeneration. In ex vivo experiments in normal rats and guinea pigs, Atoh1 overexpression enabled nonsensory cells of the cochlea to acquire the ability to produce new HCs (Kawamoto et al., 2003; Shou et al., 2003). In a guinea pig model of deafness generated by ototoxic drug-induced HC death, Atoh1 was injected into the cochlea of deaf animals via an adenoviral vector to increase its expression in nonsensory cells, showing that new HCs were produced at the original site of cochlear trauma. Hearing was restored to some extent in deaf animals as measured based on the auditory brainstem response (ABR) thresholds (Izumikawa et al., 2005). The results showed that new HCs were produced at the original site of trauma in the cochlea and that deaf animals had some hearing recovery as measured using the ABR thresholds.
In contrast, in a model of aminoglycoside-induced profound deafness, although Atoh1 gene therapy induced the conversion of nonsensory cells in the cochlea into HCs, the resulting HCs failed to mature fully and did not improve hearing in the treated animals (Atkinson et al., 2014). This finding suggests that a combination of gene therapy modalities is required to maximize hearing function in patients. In the Mouse embryonic stem cells cultured in vitro, various transcription factors (Six1, Atoh1, Pou4f3, and Gfi1) reprogrammed mouse embryonic fibroblasts and expressed the corresponding HC markers. The resulting HCs that were induced were morphologically and physiologically similar to and susceptible to ototoxic drugs as in the case of primary HCs (Costa et al., 2015; Menendez et al., 2020). Similarly, the overexpression of Gfi1, Pou4f3, and Atoh1 in human fibroblasts resulted in cells expressing some markers of HCs (Duran Alonso et al., 2018). In drug-treated mouse cochlear sensory epithelial cells, the damage caused by HC death can be reversed by the cotransfection of Pax2 and Atoh1, with Pax2 promoting the proliferation of SCs and Atoh1 promoting the regeneration of HCs (Chen et al., 2013). In addition, HC-like cells were generated 4.1-fold more efficiently after cotransfection with Atoh1 and Gfi1 than with Atoh1 alone (Lee et al., 2020); Atoh1 and Ikzf2 overexpression induced the transformation of SCs into cochlear outer HCs in the adult mouse cochlea (Sun et al., 2021). The expression of Atoh1, Gfi1, and Pou4f3 increased the potency of HC transformation in aged animals (Iyer et al., 2022).
Wnt and Notch pathways play an essential role in cell proliferation and differentiation, including regulating HC differentiation in the cochlea (Ni et al., 2016; Waqas et al., 2016; Wu et al., 2016; Samarajeewa et al., 2019). Disruption of the Rbpsuh gene in neonatal mice or treatment of mouse inner ear cells with γ-secretase inhibitor resulted in inhibition of Notch/RBP-J pathway signaling, which in turn led to downregulation of Hes5 expression and upregulation of Atoh1 expression, ultimately producing ectopic HCs (Yamamoto et al., 2006; Mizutari et al., 2013; Ren et al., 2016; Luo et al., 2017). Using siRNA to downregulate Hes1/Hes5 can also achieve Atoh1 upregulation and increase the efficiency of conversion of HCs by SCs (Du et al., 2013; Jung et al., 2013). Adenovirus carrying human Myc and Cre recombinase genes was injected into the cochlea of adult mice, and an increase in HC numbers was observed along with the inhibition of the Notch pathway (Shu et al., 2019). The expression patterns of the hypermethylated 1 (HIC1) transcriptional repressor and Prox1 genes do not overlap with Atoh1 and related downstream genes, and studies confirm that both have a repressive effect on Atoh1 and are responsible for the decrease in Atoh1 expression in postnatal mice, whereas knockdown of HIC1 or Prox1 reverses the repression of Atoh1 expression and ultimately promotes the differentiation of HCs (Kirjavainen et al., 2008; Abdul-Aziz et al., 2021). Meanwhile, an increase in Atoch1 expression was induced by the Atoch1 enhancer or small activating RNA to regulate HC regeneration (Luo et al., 2022; Zhang et al., 2022).
Moreover, recent studies on Foxg1 have demonstrated its potential as a new target for the regeneration of HCs using gene therapy. In mice with Foxg1 was knocked out in the inner ear SCs, HC numbers were significantly increased compared to those in normal mice, and the survival time was greatly increased (Zhang Y. et al., 2020; Zhang S. et al., 2020).
In conclusion, with a clear understanding of the mechanism of developmental differentiation of HCs, the regeneration of HCs can be achieved by interference with the relevant genes and pathways, thus reversing hearing loss caused by HC damage.
As the third generation of gene editing technology after zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), the CRISPR/Cas system has the advantages of clear targeting, short RNA sequences, and simultaneous operation of multiple genetic loci, of which the type II CRISPR/Cas9 system is the most widely applied (Bhatia et al., 2023; van der Oost and Patinios, 2023; Wang and Doudna, 2023). Cas9 protein with nucleic acid endonuclease function and single guide RNA (sgRNA) shear the target genome to generate double-strand breaks (DSB), which in turn enables knockdown or knock-in of the target gene by Homology-directed repair (HDR) or Non-homologous end joining (NHEJ) to achieve knockdown or knock-in of target genes (Figure 4). Previous studies have used the CRISPR/Cas9 system to establish transgenic mouse models of deafness to investigate the importance of target genes for the development and maintenance of normal hearing in the inner ear HCs (Li et al., 2018, 2019; Zhu et al., 2018; Cui et al., 2020; Zhang L. et al., 2020; Tu et al., 2021; Xue et al., 2022). CRISPR-Cas9 technology is now also showing great potential in clinically blocking dominant and recessive mutations in deafness and improving hearing impairment (György et al., 2019; Farooq et al., 2020; Ding et al., 2021).
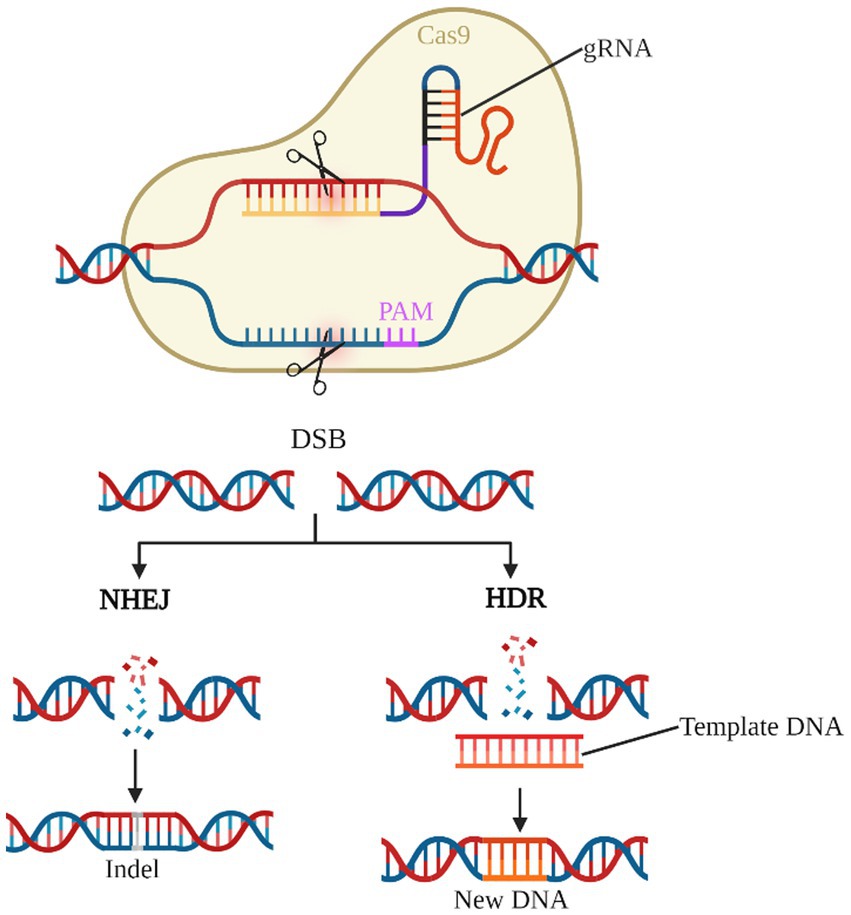
Figure 4. CRISPR/Cas9 gene editing system. The gRNA-Cas9 complex enters the cell and identifies the target gene corresponding to the PAM and shears it into a DSB. NHEJ causes the target gene shift mutation to achieve gene knock-out. HDR, on the other hand, repairs the target gene according to the exogenous template DNA and completes the gene knock-in.
Beethoven deaf mice are deafened by a point mutation (T into A) in the Tmc1 gene at locus 1,235, causing hearing impairment associated with reduced HCs in the inner ear and successfully targeting the Tmc1 gene by the lipid-mediated entry of the Cas9-gRNA complex into the mouse cochlea, resulting in a shift mutation and eventual loss of function due to a random insertion–deletion, which improves the survival of HCs while improved the hearing of mice (Gao et al., 2018). Efficient knockdown of the Htra2 gene associated with apoptosis by transfection of three gRNAs into in vitro cochlear explants and in vivo scala medium via the CRISPR/Cas9 system improved hearing loss caused by neomycin-induced hair cell apoptosis (Gu et al., 2021). In addition, CRISPR-Cas9 knockdown of Kcnq4 and myosin VI (MYO6) mutant genes have been shown to rescue inherited hearing impairment (Noh et al., 2022; Xue et al., 2022).
Although the CRISPR/Cas9 system can accurately and efficiently edit target genes, it also has limitations in hearing damage rescue studies. The presence of a short DNA sequence called the pro-spacer adjacent motif (PAM) near the complementary region of the gRNA and the target gene. The PAM sequences are mainly used to identify targets, and the presence or absence of PAM sequences in target nucleotides is a critical factor in the precise targeting of the CRISPR/Cas9 system (Manghwar et al., 2019).
The two main types of vectors for gene therapy are viral vectors and non-viral vectors, namely, viral vectors including adenovirus (AdV), adeno-associated virus (AAV), lentivirus, and retroviruses; and nonviral vectors including electroporation, liposomes, nanoparticles, and exosomes (Cring and Sheffield, 2022). The choice of gene therapy vector is significant, as it needs to deliver the exogenous gene safely and effectively to the cells of the inner ear without causing a robust immune response and to sustain its action.
AdVs were the first gene delivery vectors to be used; they are now used in various fields for HC regeneration (Syyam et al., 2022). In vivo or in vitro experiments involving the injection of AdVs carrying different target genes into target cell tissues can effectively transduce nonsensory cells into HCs, with SCs being the main ones transduced (Kawamoto et al., 2003; Shou et al., 2003; Izumikawa et al., 2005; Yamamoto et al., 2006; Chen et al., 2013; Atkinson et al., 2014; Shu et al., 2019; Lee et al., 2020). However, AdVs have a significant immunogenic effect, and their role in gene therapy is somewhat limited. In contrast, AAVs have a much lower immunoreactivity and have gradually become the vehicle of choice for gene therapy in different fields. AAVs have demonstrated their safety and efficacy in gene therapy for the regeneration of HCs. Injecting AAV8 in normal neonatal and adult mice did not cause damage to HCs in the inner ear or hearing loss (Kang et al., 2020). In addition, AAV-mediated gene delivery effectively ameliorates apoptosis and hearing loss of HCs in a drug-induced mouse model of deafness in the long term (Brigande et al., 2009; He et al., 2020; Gu et al., 2021; Xu et al., 2021; Xue et al., 2022). The results of this study are summarized below. Recent studies have shown that AAV-inner ear, a variant of AAV, can more safely and efficiently transduce Atoh1 into SCs and may be the best vehicle for future gene therapy to combat hearing loss (Tan et al., 2019; Tao et al., 2022). In addition, lentiviruses and retroviruses can also be used to deliver HC regeneration–related genes; however, their safety profile needs to be improved (Costa et al., 2015; Menendez et al., 2020).
Other nonviral gene delivery methods have also been used to regenerate HCs. Hes1 siRNA delivered by propylene-co-glycolate nanoparticles can reduce cochlear Hes1 mRNA while upregulating Atoh1 mRNA expression and, in doing so, promote the ability of SCs to acquire redifferentiated HCs (du et al., 2013). In addition, electroporation was influential in transducing plasmids encoding target genes such as Tub and Znf532 into the epithelial progenitor cells of the ear, activating the regeneration of HCs mediated by genes such as Atoh1 (Brigande et al., 2009; Xu et al., 2021).
HCs in the cochlea, as key members of the auditory conduction system, transform incoming mechanical signals into electrical signals for the body to perceive. They do not regenerate spontaneously in mammals, resulting in the associated hearing impairment being poorly treated. An exploration of the developmental maturation mechanisms of HCs reveals that the HC regulatory gene Atoh1 and its downstream targeting factors activate the ability of nonsensory SCs to differentiate into HCs. Emerging gene therapies can deliver external DNA or RNA into target cells via vectors to alter the gene expression of target cells and improve relevant functions. After addressing congenital hearing impairment, gene-based therapies can be used to treat other types of hearing impairment with the help of HC regeneration mechanisms (Table 1).
Many challenges remain in inducing regeneration of HCs in clinical settings based on gene therapy. First, the growth and development of HCs are regulated by multiple genes and pathways, and a single gene alone cannot bring about the differentiation of SCs into fully functional mature HCs. Second, the choice of vectors for gene delivery is also essential, as it is necessary to deliver the gene to the target cells efficiently and accurately without inducing a robust immune response in the body. Finally, enhancing the efficiency of HCs regeneration while ensuring high targeting requires innovation in multiple steps of gene therapy. Recent studies using the CRISPR/Cas9 system in combination with AAV vectors have shown great advantages (Kang et al., 2020; Zhao et al., 2020).
In conclusion, HC regeneration–based gene therapy shows immense potential in treating sensorineural hearing impairment. It is expected to be used in a clinical setting after further research on the mechanism of HC regeneration and optimizing targeted gene delivery methods.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported in part by grants from the National Natural Science Foundation of China (Grant No. 82271181) and Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund, Project CXPY2022092.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YZ declared a shared parent affiliation with the authors JW, HW, HH, SL, YZ, YW, XX, and SW at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdul-Aziz, D., Hathiramani, N., Phung, L., Sykopetrites, V., and Edge, A. (2021). HIC1 represses Atoh1 transcription and hair cell differentiation in the cochlea. Stem Cell Reports 16, 797–809. doi: 10.1016/j.stemcr.2021.02.022
Atkinson, P. J., Wise, A. K., Flynn, B. O., Nayagam, B. A., and Richardson, R. T. (2014). Hair cell regeneration after ATOH1 gene therapy in the cochlea of profoundly deaf adult guinea pigs. PLoS One 9:e102077. doi: 10.1371/journal.pone.0102077
Bai, H., Yang, S., Xi, C., Wang, X., Xu, J., Weng, M., et al. (2021). Signaling pathways (Notch, Wnt, Bmp and Fgf) have additive effects on hair cell regeneration in the chick basilar papilla after streptomycin injury in vitro: additive effects of signaling pathways on hair cell regeneration. Hear. Res. 401:108161. doi: 10.1016/j.heares.2020.108161
Bencivenga, D., Stampone, E., Roberti, D., Della Ragione, F., and Borriello, A. (2021). p27(Kip1), an intrinsically unstructured protein with scaffold properties. Cells 10:2254. doi: 10.3390/cells10092254
Benito-Gonzalez, A., and Doetzlhofer, A. (2014). Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of hedgehog signaling. J. Neurosci. 34, 12865–12876. doi: 10.1523/JNEUROSCI.1494-14.2014
Bermingham, N. A., Hassan, B. A., Price, S. D., Vollrath, M. A., Ben-Arie, N., Eatock, R. A., et al. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837–1841. doi: 10.1126/science.284.5421.1837
Bhatia, S., Pooja,, and Yadav, S. K. (2023). CRISPR-Cas for genome editing: classification, mechanism, designing and applications. Int. J. Biol. Macromol. 238:124054. doi: 10.1016/j.ijbiomac.2023.124054
Brigande, J. V., Gubbels, S. P., Woessner, D. W., Jungwirth, J. J., and Bresee, C. S. (2009). Electroporation-mediated gene transfer to the developing mouse inner ear. Methods Mol. Biol. 493, 125–139. doi: 10.1007/978-1-59745-523-7_8
Bulfone, A., Menguzzato, E., Broccoli, V., Marchitiello, A., Gattuso, C., Mariani, M., et al. (2000). Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum. Mol. Genet. 9, 1443–1452. doi: 10.1093/hmg/9.9.1443
Cai, T., Jen, H. I., Kang, H., Klisch, T. J., Zoghbi, H. Y., and Groves, A. K. (2015). Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 35, 5870–5883. doi: 10.1523/JNEUROSCI.5083-14.2015
Cai, T., Seymour, M. L., Zhang, H., Pereira, F. A., and Groves, A. K. (2013). Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J. Neurosci. 33, 10110–10122. doi: 10.1523/JNEUROSCI.5606-12.2013
Chadha, S., Kamenov, K., and Cieza, A. (2021). The world report on hearing, 2021. Bull. World Health Organ. 99, 242–242A. doi: 10.2471/BLT.21.285643
Chen, Y., Gu, Y., Li, Y., Li, G. L., Chai, R., Li, W., et al. (2021). Generation of mature and functional hair cells by co-expression of Gfi1, Pou4f3, and Atoh1 in the postnatal mouse cochlea. Cell Rep. 35:109016. doi: 10.1016/j.celrep.2021.109016
Chen, P., and Segil, N. (1999). p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581–1590.
Chen, Y., Yu, H., Zhang, Y., Li, W., Lu, N., Ni, W., et al. (2013). Cotransfection of Pax2 and Math1 promote in situ cochlear hair cell regeneration after neomycin insult. Sci. Rep. 3:2996. doi: 10.1038/srep02996
Cheng, Y. F., Tong, M., and Edge, A. S. (2016). Destabilization of Atoh1 by E3 ubiquitin ligase Huwe1 and casein kinase 1 is essential for Normal sensory hair cell development. J. Biol. Chem. 291, 21096–21109. doi: 10.1074/jbc.M116.722124
Chonko, K. T., Jahan, I., Stone, J., Wright, M. C., Fujiyama, T., Hoshino, M., et al. (2013). Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev. Biol. 381, 401–410. doi: 10.1016/j.ydbio.2013.06.022
Costa, A., Sanchez-Guardado, L., Juniat, S., Gale, J. E., Daudet, N., and Henrique, D. (2015). Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142, 1948–1959. doi: 10.1242/dev.119149
Cotanche, D. A., and Kaiser, C. L. (2010). Hair cell fate decisions in cochlear development and regeneration. Hear. Res. 266, 18–25. doi: 10.1016/j.heares.2010.04.012
Cring, M. R., and Sheffield, V. C. (2022). Gene therapy and gene correction: targets, progress, and challenges for treating human diseases. Gene Ther. 29, 3–12. doi: 10.1038/s41434-020-00197-8
Cui, L., Zheng, J., Zhao, Q., Chen, J. R., Liu, H., Peng, G., et al. (2020). Mutations of MAP1B encoding a microtubule-associated phosphoprotein cause sensorineural hearing loss. JCI Insight 5:e136046. doi: 10.1172/jci.insight.136046
Cunningham, L. L., and Tucci, D. L. (2017). Hearing loss in adults. N. Engl. J. Med. 377, 2465–2473. doi: 10.1056/NEJMra1616601
Deans, M. R. (2021). Conserved and divergent principles of planar polarity revealed by hair cell development and function. Front. Neurosci. 15:742391. doi: 10.3389/fnins.2021.742391
Ding, N., Lee, S., Lieber-Kotz, M., Yang, J., and Gao, X. (2021). Advances in genome editing for genetic hearing loss. Adv. Drug Deliv. Rev. 168, 118–133. doi: 10.1016/j.addr.2020.05.001
Ding, Y., Meng, W., Kong, W., He, Z., and Chai, R. (2020). The role of FoxG1 in the inner ear. Front. Cell Dev. Biol. 8:614954. doi: 10.3389/fcell.2020.614954
Du, X., Li, W., Gao, X., West, M. B., Saltzman, W. M., Cheng, C. J., et al. (2013). Regeneration of mammalian cochlear and vestibular hair cells through Hes1/Hes5 modulation with siRNA. Hear. Res. 304, 91–110. doi: 10.1016/j.heares.2013.06.011
Dulon, D., Papal, S., Patni, P., Cortese, M., Vincent, P. F. Y., Tertrais, M., et al. (2018). Clarin-1 gene transfer rescues auditory synaptopathy in model of usher syndrome. J. Clin. Invest. 128, 3382–3401. doi: 10.1172/JCI94351
Duran Alonso, M. B., Lopez Hernandez, I., de la Fuente, M. A., Garcia-Sancho, J., Giraldez, F., and Schimmang, T. (2018). Transcription factor induced conversion of human fibroblasts towards the hair cell lineage. PLoS One 13:e0200210. doi: 10.1371/journal.pone.0200210
Ebeid, M., and Huh, S. H. (2017). FGF signaling: diverse roles during cochlear development. BMB Rep. 50, 487–495. doi: 10.5483/BMBRep.2017.50.10.164
Farooq, R., Hussain, K., Tariq, M., Farooq, A., and Mustafa, M. (2020). CRISPR/Cas9: targeted genome editing for the treatment of hereditary hearing loss. J. Appl. Genet. 61, 51–65. doi: 10.1007/s13353-019-00535-6
Fujioka, M., Okano, H., and Edge, A. S. (2015). Manipulating cell fate in the cochlea: a feasible therapy for hearing loss. Trends Neurosci. 38, 139–144. doi: 10.1016/j.tins.2014.12.004
Gao, X., Tao, Y., Lamas, V., Huang, M., Yeh, W. H., Pan, B., et al. (2018). Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221. doi: 10.1038/nature25164
Gu, X., Wang, D., Xu, Z., Wang, J., Guo, L., Chai, R., et al. (2021). Prevention of acquired sensorineural hearing loss in mice by in vivo Htra2 gene editing. Genome Biol. 22:86. doi: 10.1186/s13059-021-02311-4
György, B., Nist-Lund, C., Pan, B., Asai, Y., Karavitaki, K. D., Kleinstiver, B. P., et al. (2019). Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat. Med. 25, 1123–1130. doi: 10.1038/s41591-019-0500-9
He, Z., Fang, Q., Li, H., Shao, B., Zhang, Y., Zhang, Y., et al. (2019). The role of FOXG1 in the postnatal development and survival of mouse cochlear hair cells. Neuropharmacology 144, 43–57. doi: 10.1016/j.neuropharm.2018.10.021
He, L., Guo, J. Y., Qu, T. F., Wei, W., Liu, K., Peng, Z., et al. (2020). Cellular origin and response of flat epithelium in the vestibular end organs of mice to Atoh1 overexpression. Hear. Res. 391:107953. doi: 10.1016/j.heares.2020.107953
He, Z. H., Li, M., Fang, Q. J., Liao, F. L., Zou, S. Y., Wu, X., et al. (2021). FOXG1 promotes aging inner ear hair cell survival through activation of the autophagy pathway. Autophagy 17, 4341–4362. doi: 10.1080/15548627.2021.1916194
He, Z. H., Zou, S. Y., Li, M., Liao, F. L., Wu, X., Sun, H. Y., et al. (2020). The nuclear transcription factor FoxG1 affects the sensitivity of mimetic aging hair cells to inflammation by regulating autophagy pathways. Redox Biol. 28:101364. doi: 10.1016/j.redox.2019.101364
Hertzano, R., Montcouquiol, M., Rashi-Elkeles, S., Elkon, R., Yücel, R., Frankel, W. N., et al. (2004). Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum. Mol. Genet. 13, 2143–2153. doi: 10.1093/hmg/ddh218
Hou, K., Jiang, H., Karim, M. R., Zhong, C., Xu, Z., Liu, L., et al. (2019). A critical E-box in Barhl1 3′ enhancer is essential for auditory hair cell differentiation. Cells 8:458. doi: 10.3390/cells8050458
Hwang, C. H., Simeone, A., Lai, E., and Wu, D. K. (2009). Foxg1 is required for proper separation and formation of sensory cristae during inner ear development. Dev. Dyn. 238, 2725–2734. doi: 10.1002/dvdy.22111
Ikeda, R., Pak, K., Chavez, E., and Ryan, A. F. (2015). Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol. Neurobiol. 51, 672–684. doi: 10.1007/s12035-014-8801-y
Iyer, A. A., Hosamani, I., Nguyen, J. D., Cai, T., Singh, S., McGovern, M. M., et al. (2022). Cellular reprogramming with ATOH1, GFI1, and POU4F3 implicate epigenetic changes and cell-cell signaling as obstacles to hair cell regeneration in mature mammals. Elife 11:e79712. doi: 10.7554/eLife.79712
Izumikawa, M., Minoda, R., Kawamoto, K., Abrashkin, K. A., Swiderski, D. L., Dolan, D. F., et al. (2005). Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 11, 271–276. doi: 10.1038/nm1193
Jen, H. I., Singh, S., Tao, L., Maunsell, H. R., Segil, N., and Groves, A. K. (2022). GFI1 regulates hair cell differentiation by acting as an off-DNA transcriptional co-activator of ATOH1, and a DNA-binding repressor. Sci. Rep. 12:7793. doi: 10.1038/s41598-022-11931-0
Jung, J. Y., Avenarius, M. R., Adamsky, S., Alpert, E., Feinstein, E., and Raphael, Y. (2013). siRNA targeting Hes5 augments hair cell regeneration in aminoglycoside-damaged mouse utricle. Mol. Ther. 21, 834–841. doi: 10.1038/mt.2013.18
Kang, W., Zhao, X., Sun, Z., Dong, T., Jin, C., Tong, L., et al. (2020). Adeno-associated virus vector enables safe and efficient Cas9 activation in neonatal and adult Cas9 knockin murine cochleae. Gene Ther. 27, 392–405. doi: 10.1038/s41434-020-0124-1
Kawamoto, K., Ishimoto, S., Minoda, R., Brough, D. E., and Raphael, Y. (2003). Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J. Neurosci. 23, 4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003
Kirjavainen, A., Sulg, M., Heyd, F., Alitalo, K., Ylä-Herttuala, S., Möröy, T., et al. (2008). Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev. Biol. 322, 33–45. doi: 10.1016/j.ydbio.2008.07.004
Klisch, T. J., Xi, Y., Flora, A., Wang, L., Li, W., and Zoghbi, H. Y. (2011). In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc. Natl. Acad. Sci. U. S. A. 108, 3288–3293. doi: 10.1073/pnas.1100230108
Lee, S., Song, J. J., Beyer, L. A., Swiderski, D. L., Prieskorn, D. M., Acar, M., et al. (2020). Combinatorial Atoh1 and Gfi1 induction enhances hair cell regeneration in the adult cochlea. Sci. Rep. 10:21397. doi: 10.1038/s41598-020-78167-8
Li, W., Feng, Y., Chen, A., Li, T., Huang, S., Liu, J., et al. (2019). Elmod3 knockout leads to progressive hearing loss and abnormalities in cochlear hair cell stereocilia. Hum. Mol. Genet. 28, 4103–4112. doi: 10.1093/hmg/ddz240
Li, S., Price, S. M., Cahill, H., Ryugo, D. K., Shen, M. M., and Xiang, M. (2002). Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development 129, 3523–3532. doi: 10.1242/dev.129.14.3523
Li, P., Wen, Z., Zhang, G., Zhang, A., Fu, X., and Gao, J. (2018). Knock-in mice with Myo3a Y137C mutation displayed progressive hearing loss and hair cell degeneration in the inner ear. Neural Plast. 2018:4372913. doi: 10.1155/2018/4372913
Löwenheim, H., Furness, D. N., Kil, J., Zinn, C., Gültig, K., Fero, M. L., et al. (1999). Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of corti. Proc. Natl. Acad. Sci. U. S. A. 96, 4084–4088. doi: 10.1073/pnas.96.7.4084
Lumpkin, E. A., Collisson, T., Parab, P., Omer-Abdalla, A., Haeberle, H., Chen, P., et al. (2003). Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–395. doi: 10.1016/S1567-133X(03)00089-9
Luo, Z., du, Y., Li, S., Zhang, H., Shu, M., Zhang, D., et al. (2022). Three distinct Atoh1 enhancers cooperate for sound receptor hair cell development. Proc. Natl. Acad. Sci. U. S. A. 119:e2119850119. doi: 10.1073/pnas.2119850119
Luo, W. W., Han, Z., Ren, D. D., Wang, X. W., Chi, F. L., and Yang, J. M. (2017). Notch pathway inhibitor DAPT enhances Atoh1 activity to generate new hair cells in situ in rat cochleae. Neural Regen. Res. 12, 2092–2099. doi: 10.4103/1673-5374.221169
Maass, J. C., Berndt, F. A., Cánovas, J., and Kukuljan, M. (2013). p27Kip1 knockdown induces proliferation in the organ of Corti in culture after efficient shRNA lentiviral transduction. J. Assoc. Res. Otolaryngol. 14, 495–508. doi: 10.1007/s10162-013-0383-2
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants.2019.09.006
Marcovich, I., Baer, N. K., Shubina-Oleinik, O., Eclov, R., Beard, C. W., and Holt, J. R. (2022). Optimized AAV vectors for TMC1 gene therapy in a humanized mouse model of DFNB7/11. Biomol. Ther. 12:914. doi: 10.3390/biom12070914
Matern, M. S., Milon, B., Lipford, E. L., McMurray, M., Ogawa, Y., Tkaczuk, A., et al. (2020). GFI1 functions to repress neuronal gene expression in the developing inner ear hair cells. Development 147:dev186015. doi: 10.1242/dev.186015
Menendez, L., Trecek, T., Gopalakrishnan, S., Tao, L., Markowitz, A. L., Yu, H. V., et al. (2020). Generation of inner ear hair cells by direct lineage conversion of primary somatic cells. Elife 9:9. doi: 10.7554/eLife.55249
Mizutari, K., Fujioka, M., Hosoya, M., Bramhall, N., Okano, H. J., Okano, H., et al. (2013). Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77, 58–69. doi: 10.1016/j.neuron.2012.10.032
Ni, W., Lin, C., Guo, L., Wu, J., Chen, Y., Chai, R., et al. (2016). Extensive supporting cell proliferation and mitotic hair cell generation by in vivo genetic reprogramming in the neonatal mouse cochlea. J. Neurosci. 36, 8734–8745. doi: 10.1523/JNEUROSCI.0060-16.2016
Noh, B., Rim, J. H., Gopalappa, R., Lin, H., Kim, K. M., Kang, M. J., et al. (2022). In vivo outer hair cell gene editing ameliorates progressive hearing loss in dominant-negative Kcnq4 murine model. Theranostics 12, 2465–2482. doi: 10.7150/thno.67781
Ono, K., Nakagawa, T., Kojima, K., Matsumoto, M., Kawauchi, T., Hoshino, M., et al. (2009). Silencing p27 reverses post-mitotic state of supporting cells in neonatal mouse cochleae. Mol. Cell. Neurosci. 42, 391–398. doi: 10.1016/j.mcn.2009.08.011
Pan, N., Jahan, I., Kersigo, J., Kopecky, B., Santi, P., Johnson, S., et al. (2011). Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear. Res. 275, 66–80. doi: 10.1016/j.heares.2010.12.002
Pancrazi, L., di Benedetto, G., Colombaioni, L., Della Sala, G., Testa, G., Olimpico, F., et al. (2015). Foxg1 localizes to mitochondria and coordinates cell differentiation and bioenergetics. Proc. Natl. Acad. Sci. U. S. A. 112, 13910–13915. doi: 10.1073/pnas.1515190112
Pauley, S., Lai, E., and Fritzsch, B. (2006). Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 235, 2470–2482. doi: 10.1002/dvdy.20839
Ren, H., Guo, W., Liu, W., Gao, W., Xie, D., Yin, T., et al. (2016). DAPT mediates atoh1 expression to induce hair cell-like cells. Am. J. Transl. Res. 8, 634–643.
Rubbini, D., Robert-Moreno, À., Hoijman, E., and Alsina, B. (2015). Retinoic acid signaling mediates hair cell regeneration by repressing p27kip and sox2 in supporting cells. J. Neurosci. 35, 15752–15766. doi: 10.1523/JNEUROSCI.1099-15.2015
Samarajeewa, A., Jacques, B. E., and Dabdoub, A. (2019). Therapeutic potential of Wnt and notch signaling and epigenetic regulation in mammalian sensory hair cell regeneration. Mol. Ther. 27, 904–911. doi: 10.1016/j.ymthe.2019.03.017
Shibata, S. B., West, M. B., Du, X., Iwasa, Y., Raphael, Y., and Kopke, R. D. (2020). Gene therapy for hair cell regeneration: review and new data. Hear. Res. 394:107981. doi: 10.1016/j.heares.2020.107981
Shou, J., Zheng, J. L., and Gao, W. Q. (2003). Robust generation of new hair cells in the mature mammalian inner ear by adenoviral expression of Hath1. Mol. Cell. Neurosci. 23, 169–179. doi: 10.1016/S1044-7431(03)00066-6
Shu, Y., Li, W., Huang, M., Quan, Y. Z., Scheffer, D., Tian, C., et al. (2019). Renewed proliferation in adult mouse cochlea and regeneration of hair cells. Nat. Commun. 10:5530. doi: 10.1038/s41467-019-13157-7
Sun, S., Li, S., Luo, Z., Ren, M., He, S., Wang, G., et al. (2021). Dual expression of Atoh1 and Ikzf2 promotes transformation of adult cochlear supporting cells into outer hair cells. Elife 10:e66547. doi: 10.7554/eLife.66547
Syyam, A., Nawaz, A., Ijaz, A., Sajjad, U., Fazil, A., Irfan, S., et al. (2022). Adenovirus vector system: construction, history and therapeutic applications. BioTechniques 73, 297–305. doi: 10.2144/btn-2022-0051
Taiber, S., Cohen, R., Yizhar-Barnea, O., Sprinzak, D., Holt, J. R., and Avraham, K. B. (2021). Neonatal AAV gene therapy rescues hearing in a mouse model of SYNE4 deafness. EMBO Mol. Med. 13:e13259. doi: 10.15252/emmm.202013259
Tan, F., Chu, C., Qi, J., Li, W., You, D., Li, K., et al. (2019). AAV-ie enables safe and efficient gene transfer to inner ear cells. Nat. Commun. 10:3733. doi: 10.1038/s41467-019-11687-8
Tao, Y., Liu, X., Yang, L., Chu, C., Tan, F., Yu, Z., et al. (2022). AAV-ie-K558R mediated cochlear gene therapy and hair cell regeneration. Signal Transduct. Target. Ther. 7:109. doi: 10.1038/s41392-022-00938-8
Tu, H., Zhang, A., Fu, X., Xu, S., Bai, X., Wang, H., et al. (2021). SMPX deficiency causes stereocilia degeneration and progressive hearing loss in CBA/CaJ mice. Front. Cell Dev. Biol. 9:750023. doi: 10.3389/fcell.2021.750023
Vahava, O., Morell, R., Lynch, E. D., Weiss, S., Kagan, M. E., Ahituv, N., et al. (1998). Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science 279, 1950–1954. doi: 10.1126/science.279.5358.1950
van der Oost, J., and Patinios, C. (2023). The genome editing revolution. Trends Biotechnol. 41, 396–409. doi: 10.1016/j.tibtech.2022.12.022
Wallis, D., Hamblen, M., Zhou, Y., Venken, K. J. T., Schumacher, A., Grimes, H. L., et al. (2003). The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221–232. doi: 10.1242/dev.00190
Walters, B. J., Coak, E., Dearman, J., Bailey, G., Yamashita, T., Kuo, B., et al. (2017). In vivo interplay between p27(Kip1), GATA3, ATOH1, and POU4F3 converts non-sensory cells to hair cells in adult mice. Cell Rep. 19, 307–320. doi: 10.1016/j.celrep.2017.03.044
Walters, B. J., Liu, Z., Crabtree, M., Coak, E., Cox, B. C., and Zuo, J. (2014). Auditory hair cell-specific deletion of p27Kip1 in postnatal mice promotes cell-autonomous generation of new hair cells and normal hearing. J. Neurosci. 34, 15751–15763. doi: 10.1523/JNEUROSCI.3200-14.2014
Wang, J. Y., and Doudna, J. A. (2023). CRISPR technology: a decade of genome editing is only the beginning. Science 379:eadd8643. doi: 10.1126/science.add8643
Waqas, M., Zhang, S., He, Z., Tang, M., and Chai, R. (2016). Role of Wnt and Notch signaling in regulating hair cell regeneration in the cochlea. Front. Med. 10, 237–249. doi: 10.1007/s11684-016-0464-9
White, P. M., Doetzlhofer, A., Lee, Y. S., Groves, A. K., and Segil, N. (2006). Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984–987. doi: 10.1038/nature04849
Wu, J., Li, W., Lin, C., Chen, Y., Cheng, C., Sun, S., et al. (2016). Co-regulation of the notch and Wnt signaling pathways promotes supporting cell proliferation and hair cell regeneration in mouse utricles. Sci. Rep. 6:29418. doi: 10.1038/srep29418
Xiang, M., Gan, L., Li, D., Chen, Z. Y., Zhou, L., O’Malley, B. W. Jr., et al. (1997). Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc. Natl. Acad. Sci. U. S. A. 94, 9445–9450. doi: 10.1073/pnas.94.17.9445
Xu, Z., Rai, V., and Zuo, J. (2021). TUB and ZNF532 promote the Atoh1-mediated hair cell regeneration in mouse cochleae. Front. Cell. Neurosci. 15:759223. doi: 10.3389/fncel.2021.759223
Xue, Y., Hu, X., Wang, D., Li, D., Li, Y., Wang, F., et al. (2022). Gene editing in a Myo6 semi-dominant mouse model rescues auditory function. Mol. Ther. 30, 105–118. doi: 10.1016/j.ymthe.2021.06.015
Yamamoto, N., Tanigaki, K., Tsuji, M., Yabe, D., Ito, J., and Honjo, T. (2006). Inhibition of notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J. Mol. Med. 84, 37–45. doi: 10.1007/s00109-005-0706-9
Yu, H. V., Tao, L., Llamas, J., Wang, X., Nguyen, J. D., Trecek, T., et al. (2021). POU4F3 pioneer activity enables ATOH1 to drive diverse mechanoreceptor differentiation through a feed-forward epigenetic mechanism. Proc. Natl. Acad. Sci. U. S. A. 118:e2105137118. doi: 10.1073/pnas.2105137118
Zhang, L., Gao, Y., Zhang, R., Sun, F., Cheng, C., Qian, F., et al. (2020). THOC1 deficiency leads to late-onset nonsyndromic hearing loss through p53-mediated hair cell apoptosis. PLoS Genet. 16:e1008953. doi: 10.1371/journal.pgen.1008953
Zhang, Y. L., Kang, M., Wu, J. C., Xie, M. Y., Xue, R. Y., Tang, Q., et al. (2022). Small activating RNA activation of ATOH1 promotes regeneration of human inner ear hair cells. Bioengineered 13, 6729–6739. doi: 10.1080/21655979.2022.2045835
Zhang, S., Zhang, Y., Dong, Y., Guo, L., Zhang, Z., Shao, B., et al. (2020). Knockdown of Foxg1 in supporting cells increases the trans-differentiation of supporting cells into hair cells in the neonatal mouse cochlea. Cell. Mol. Life Sci. 77, 1401–1419. doi: 10.1007/s00018-019-03291-2
Zhang, Y., Zhang, S., Zhang, Z., Dong, Y., Ma, X., Qiang, R., et al. (2020). Knockdown of Foxg1 in Sox9+ supporting cells increases the trans-differentiation of supporting cells into hair cells in the neonatal mouse utricle. Aging 12, 19834–19851. doi: 10.18632/aging.104009
Zhao, X., Jin, C., Dong, T., Sun, Z., Zheng, X., Feng, B., et al. (2020). Characterization of promoters for adeno-associated virus mediated efficient Cas9 activation in adult Cas9 knock-in murine cochleae. Hear. Res. 394:107999. doi: 10.1016/j.heares.2020.107999
Zhong, C., Chen, Z., Luo, X., Wang, C., Jiang, H., Shao, J., et al. (2018). Barhl1 is required for the differentiation of inner ear hair cell-like cells from mouse embryonic stem cells. Int. J. Biochem. Cell Biol. 96, 79–89. doi: 10.1016/j.biocel.2018.01.013
Keywords: hair cell regeneration, gene therapy, Atoh1, inner ear, sensorineural hearing loss
Citation: Wang J, Zheng J, Wang H, He H, Li S, Zhang Y, Wang Y, Xu X and Wang S (2023) Gene therapy: an emerging therapy for hair cells regeneration in the cochlea. Front. Neurosci. 17:1177791. doi: 10.3389/fnins.2023.1177791
Received: 02 March 2023; Accepted: 12 April 2023;
Published: 03 May 2023.
Edited by:
Xiaolong Fu, Southeast University, ChinaReviewed by:
Yuanyuan Zhang, Renmin Hospital of Wuhan University, ChinaCopyright © 2023 Wang, Zheng, Wang, He, Li, Zhang, Wang, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: You Wang, d2FuZ3lvdTAxMjBAMTI2LmNvbQ==; Xiaoxiang Xu, em4wMDM1MDRAd2h1LmVkdS5jbg==; Shuyi Wang, c2h1eWl3YW5nQHdodS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.