
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 18 April 2023
Sec. Translational Neuroscience
Volume 17 - 2023 | https://doi.org/10.3389/fnins.2023.1145307
This article is part of the Research Topic Brain Injury Associated Secondary Injury and Remote Organ Injury View all 7 articles
 Xintao Huang1*†
Xintao Huang1*† Xinjuan Xu2†
Xinjuan Xu2† Ce Wang1,3
Ce Wang1,3 Yi Wang1,3
Yi Wang1,3 Yajun Yang1,3
Yajun Yang1,3 Tianle Yao1
Tianle Yao1 Rui Bai1
Rui Bai1 Xile Pei1
Xile Pei1 Feirong Bai1,3
Feirong Bai1,3 Panpan Li1,3
Panpan Li1,3Introduction: Traumatic brain injury (TBI) is considered the most common traumatic neurological disease, is associated with high mortality and long-term complications, and is a global public health issue. However, there has been little progress in serum markers for TBI research. Therefore, there is an urgent need for biomarkers that can sufficiently function in TBI diagnosis and evaluation.
Methods: Exosomal microRNA (ExomiR), a stable circulating marker in the serum, has aroused widespread interest among researchers. To explore the level of serum ExomiR after TBI, we quantified ExomiR expression levels in serum exosomes extracted from patients with TBI using next-generation sequencing (NGS) and explored potential biomarkers using bioinformatics screening.
Results: Compared with the control group, there were 245 ExomiR (136 up-regulated and 109 down-regulated) in the serum of the TBI group that changed significantly. We observed serum ExomiRs expression profiles associated with neurovascular remodeling, the integrity of the blood-brain barrier, neuroinflammation, and a cascade of secondary injury, including eight up-regulated ExomiRs (ExomiR-124-3p, ExomiR-137-3p, ExomiR-9-3p, ExomiR-133a-5p, ExomiR-204-3p, ExomiR-519a-5p, ExomiR-4732-5p, and ExomiR-206) and 2 down-regulated ExomiR (ExomiR-21-3p and ExomiR-199a-5).
Discussion: The results revealed that serum ExomiRs might become a new research direction and breakthrough for the diagnosis and pathophysiological treatment of patients with TBI.
Traumatic brain injury (TBI) is a disease of the most complex organ. The fatality rate associated with TBI has been increasing annually and has become a critical public health issue worldwide, with nearly 90% of post-injury deaths occurring in low- and middle-income countries based on a survey by the World Health Organization (WHO) (Rubiano et al., 2015; Kamal et al., 2016; Dewan et al., 2019). Epidemiologically, the mortality rate of TBI is approximately 13 cases per 100,000 people annually in China (Jiang et al., 2019). From 2001 to 2016, approximately 120,000 patients suffered from acute TBI, according to 18 retrospective clinical studies in China (Jiang et al., 2007; Xu et al., 2016; Cheng et al., 2017; Li et al., 2017). Research has greatly clarified TBI’s mechanisms, involving a primary insult caused by direct biomechanical forces and a secondary insult, resulting in brain damage and even death (Werner and Engelhard, 2007). Following a primary mechanical injury in TBI, the literature suggests the presence of a delayed secondary injury involving various neuroinflammatory changes. Several signaling molecules and metabolic derangements disrupt the blood-brain barrier (BBB) from hours to days following a TBI, leading to the extravasation of immune cells and cerebral edema. Secondary brain injury, which is the focus of current research and clinical treatment, involves many molecular mechanisms, including the neuroinflammatory response, apoptosis, oxidative damage, BBB damage, and regulation of gene expression (Cornelius et al., 2013; Crupi et al., 2020).
Exosomes are disc-shaped vesicles of 30–150 nm diameter secreted by various cells and are widely distributed in the blood, urine, saliva, cerebrospinal fluid, and other extracellular fluids. Intracellular vesicles are formed by endocytosis, which fuses with the cell membrane and is secreted to form extracellular vesicles. They mediate cellular communication by transporting proteins, lipids, miRs, messenger RNA (mRNA), and DNA from the parent cell to the recipient cell (Zhang et al., 2015). Therefore, they may serve as diagnostic and prognostic serum biomarkers. After trauma, brain-cell-derived exosomes containing miR, mRNA, or DNA can cross the BBB to the peripheral blood. MiRs are endogenous non-coding RNAs with a length of about 22 nucleotides that regulate gene expression by mediating translation inhibition or mRNA degradation. Exosomal miRs are involved in the occurrence and development of a variety of central nervous system diseases, such as TBI (Ghai et al., 2020), spinal cord injury (Bao et al., 2018), stroke (Zhou et al., 2018), and neurodegenerative diseases (Nie et al., 2020). Therefore, exosomal miRs might be necessary to transfer information, regulate signal pathways, and provide a new diagnosis and treatment strategy for central nervous system (CNS) diseases.
In this study, exosomes and exosomal miRs in the peripheral serum of patients with TBI were isolated, and high-throughput sequencing was used to study the exosomal miRNA expression changes in patients with TBI with different degrees of injury. This study used bioinformatics analysis techniques to predict the targets of differential genes, revealing potential miRs in peripheral blood exosomes after TBI. Related research deepened people’s understanding of the molecular mechanism after TBI and provided potential intervention sites after pathological changes. The expression of exosomal miR in patients with TBI provides many TBI-related biomarkers for clinical research and improves the accuracy of the condition judgment and prognostic evaluation of patients with TBI.
The inclusion criteria for the patients were as follows: (1) the patient was between the ages of 18–80 years, (2) the patient got admitted within 24 h of the TBI event, and (3) patient with no surgical treatment. In addition, the following patients were excluded: (1) Complications with other system injuries (including the heart, liver, and lungs, etc.) and long bone fractures (upper limbs or femurs); and (2) Central nervous system disease, cardiovascular and cerebrovascular disease, and blood system disease have been diagnosed or documented with other medical histories.
A coagulation tube collected approximately 8–10 mL of upper arm venous blood from patients at 24 hospitals before surgery, mixed by inverting, allowed to stand for 2 h, and centrifuged at 1000 g at 4°C for 15 min. The supernatant was extracted, aliquoted into RNase-free 1 mL EP tubes, and stored at −80°C.
Frozen exosome serum samples extracted by the high-speed centrifugal method were thawed, melted in a 25°C water bath, and transferred to a centrifuge tube. Impurities were removed from the samples by centrifugation at 4°C, 3000 × g for 10 min and then again for 20 min at 10,000 × g. The impurities and cell debris were transferred to a new centrifuge tube; 4 mL phosphate buffer saline (PBS) and 1 mL Exo extraction purification reagent (Blood Pure Exo Solution, BPS) were added and mixed through a vortex oscillator for 1 min, at 2–8°C for 2 h; Centrifuge tube with the mixture was centrifuged at 10,000g for 60 min, the supernatant was discarded, and the precipitate was rich with exosome particles (note: as clean as possible); Then, 0.2 mL 1 × PBS was added to the sediment evenly after uniform suspension (recommended PBS liquid added volume: initial sample volume = 1:5); this was transferred to a new 1.5 mL centrifuge tube and centrifuged for 2 min at 4°C 12000 × g. The supernatant was moved to the EPF (Exosome Purification Filter) column upper chamber after centrifugation at 4°C, 3000 × g for 10 min and then purified using Exo particles. Purified Exo was stored in 0.05–0.1 mL portions in an −80 °C refrigerator for future experiments.
The morphological characteristics of exosomes were visualized using transmission electron microscopy (TEM; JEOL-1230; Tokyo, Japan). Briefly, 30 μL of exosome samples were placed on a sheet of parafilm, and a 100-mesh copper grid was transferred to drops of exosomes with forceps for 10 min. Phosphotungstic acid was then used to stain the grid for 15 s and dried at 23°C and 30°C. The size of exosomes was detected by nanoparticle tracking analysis (NTA) using ZetaView PMX110 (Particle Metrix, Meerbusch, Germany) and its accompanying software. Isolated exosome samples were appropriately diluted using 1X PBS buffer (Biological Industries, Israel) to measure particle size and concentration. The levels of multiple exosome markers (CD63 and TSG101) were measured by western blotting.
Total RNA was extracted from serum exosomes with TRIzol (Invitrogen, Carlsbad, CA, USA) reagent and was further purified.
The TruSeq Small RNA Prep Kit (RS-122-2002, Illumina, USA) was used to construct libraries according to the manufacturer’s protocol. The optimal loading amount was selected for sequencing on the Illumina platform.
After sequencing, clean data were filtered from raw reads using the following criteria: (a) 30% base quality < 20; (b) read length < 18 nucleotides (nt), >36 nt; (c) adaptor sequence; (d) align to the reference genome, remove unmatched sequences; (e) align to ncRNA (non-coding RNA) in Rfam, remove snRNA (small nuclearRNA), snoRNA (small nucleolar RNA), tRNA (Transfer RNA), etc.; and (f) remove repetitive sequences. The Rfam (Kalvari et al., 2018) genome sequence was compared with clean reads after de-redundancy to find known miRs and new miRs, siRNA, piRNA, and other sRNA molecules, and obtain their respective expression levels. Differential miRNA expression analysis was performed for each sample.
We performed statistical analysis on the expression level of exosome miR in each sample. We used | log2FC| (| log2Fold Change|) > 1.0 and P < 0.05 as the screening criteria and DESeq software to screen for differential expression. Based on the analysis results, the common and unique differences in genes between the TBI and control groups were screened.
Targets can include Mir Walk (Sticht et al., 2018), Mirta Base, and Miranda to predict the target genes of miRs with significant differential expression.
For target genes with differentially expressed miRNAs, we used the DAVID (Huang et al., 2009) website to perform GO (Gene ontology) function annotation analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis to screen enrichment analysis items and pathway information. Statistical significance was set at P < 0.05.
A PPI diagram of differentially expressed miR target genes was constructed using String (version 11.5) (Szklarczyk et al., 2021). A cystoscope was used to visualize the results.
The serum expression levels of candidate exosomal miRs in patients and control subjects were analyzed using the non-parametric Mann–Whitney test. All statistical analyses were performed using SPSS 22.5 software. P < 0.05 was considered a statistically significant difference. The study process is shown in Supplementary Table 1.
A total of 14 patients (male 12, female 2, mean age 58.4 ± 14.9) with TBI were admitted to our department from November 1, 2019, to January 31, 2020, age ranging from 30 to 80 years, and 6 matched healthy controls (1 female, 5 males, mean age 58.5 ± 16.4) were used for our study.
The patients with TBI were divided into two groups according to the Glasgow Coma Scale (GCS) on admission: mild TBI and moderate TBI (mTBI) (GCS = 9–15): 7 cases (35%); severe TBI (sTBI) group (GCS ≤ 8): 7 cases (35%); and control group, 6 cases (30%) (Table 1).
Serum was extracted from the peripheral blood of patients with TBI and the control group. Exosomes in serum were separated and extracted by high-speed centrifugation. After quantification using the BCA (bicinchoninic acid) protein analysis kit, exosome morphology was observed by TEM. As shown in Figure 1A, the particles isolated from the control group, sTBI, and mTBI microglia had a typical sphericity-shaped morphology and 50–200 nm diameters. The concentration and size distribution of the isolated particles were analyzed using NTA (Figure 1B). Furthermore, western blot analysis showed that characteristic exosome biomarkers, including CD63 and TSG101, were expressed in particles isolated from the TBI and control groups (Figure 1C). The above results were confirmed from the serum as an external body by the size of the extracellular vesicle, expression of the morphology, and protein markers.
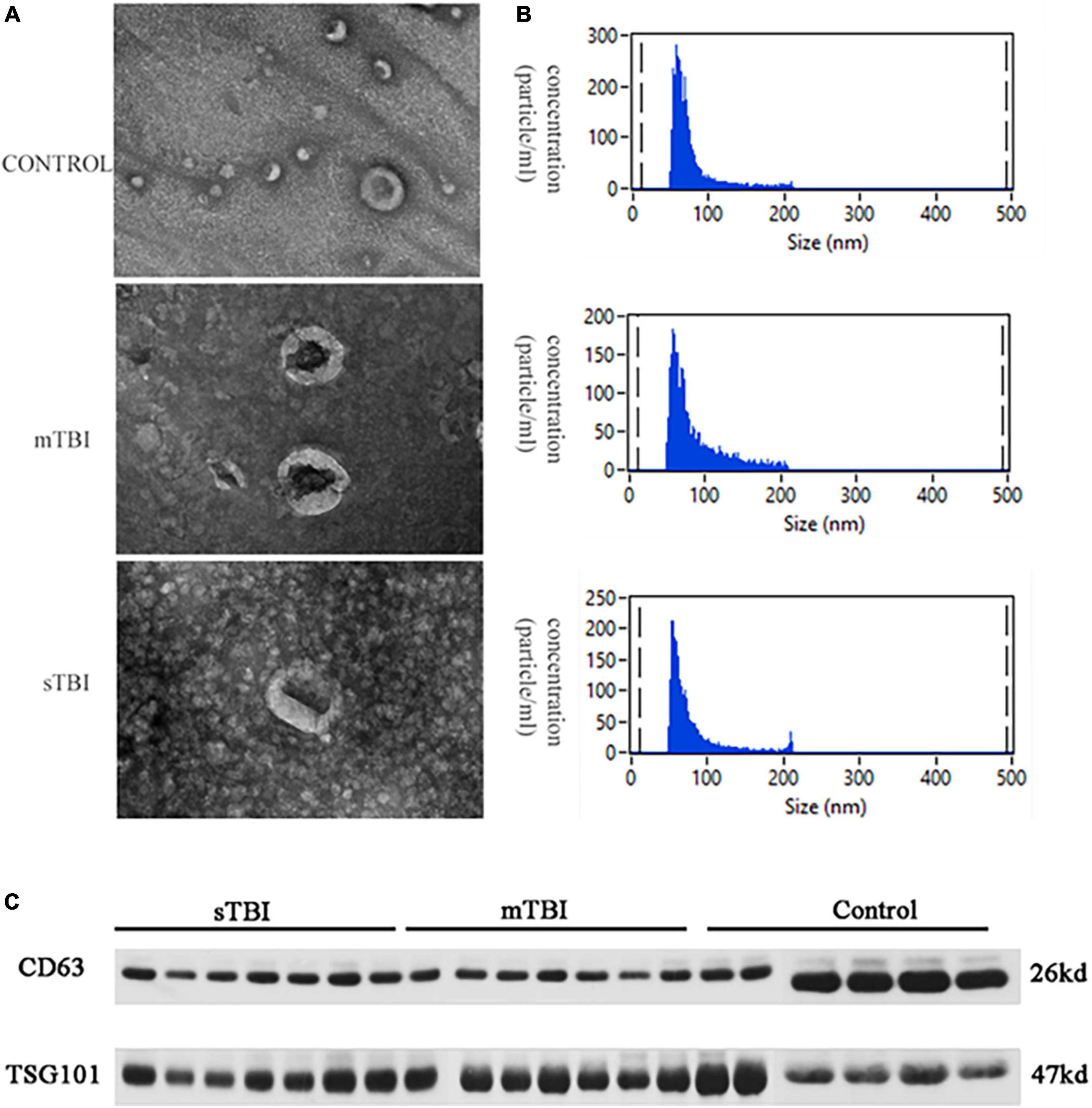
Figure 1. The characterization of the isolation exosome. (A) The TEM image shows the morphology of the exosomes. (B) NTA measured the size distribution of exosomes. (C) Western blotting of the exosomal membrane markers CD63 and TSG101.
Statistical and differential expression analyses of miRs identified in each sample were performed according to the expression data of each sample. To understand the changes in serum exosomal miRs in patients with TBI with different degrees of injury at the genetic level. Figure 2A shows that in all samples in the TBI and control groups, 522 miRs were identified in total.
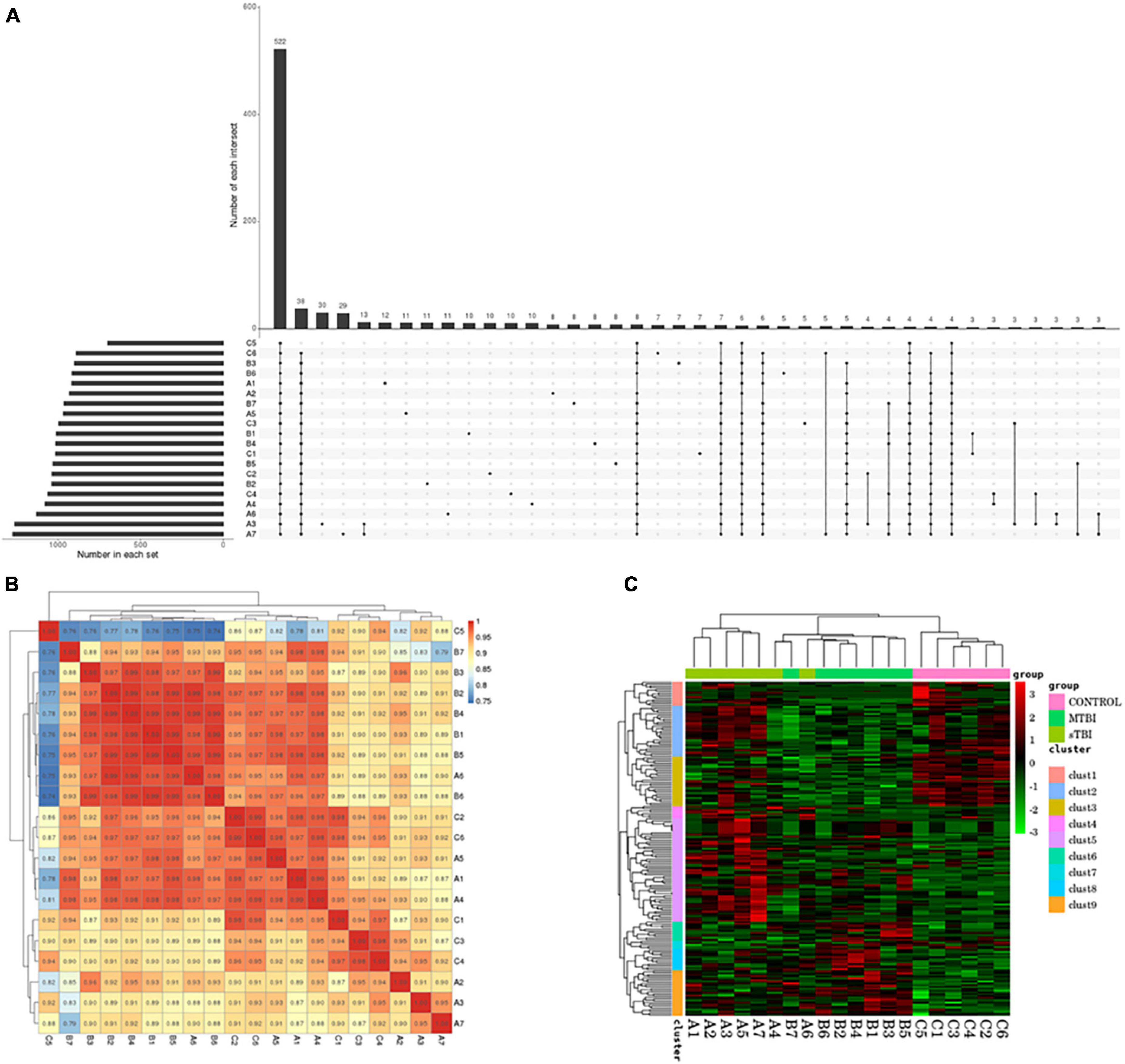
Figure 2. (A) UpSet plots obtained from each sample identification: the number in each set represents the number of all miRs identified by each sample; the number of each intersection represents the number of common miRs identified by multiple samples; and the abscissa one point represents the number of unique miRs identified by that sample item; multiple abscissa point connections represent the number of common miRs identified by multiple samples of the connection. (B) The correlation of commonly expressed genes between TBI and the control group, analyzed using Pearson’s correlation analysis: the sample is clustered on the left and upper sides, the sample names and colored squares represent the correlation between the two samples. (C) Differentially expressed miR clustering: Transsternal indicates miR; each column is one sample; red indicates high miR; green indicates low miR.
Pearson’s correlation coefficient analysis was used to analyze the correlation of miR expression levels between samples to test the experiment’s reliability and whether the sample selection was reasonable (Figure 2B).
The Euclidean method was used to calculate the distance to assess the correlation of differentially expressed miRs between samples. The hierarchical clustering-longest distance method (complete linkage) was used for clustering. Clustering was performed based on the expression level of the same miR in different samples and the expression pattern of different miRs in the same sample (Figure 2C).
DESeq software (version 1.18.0, Anders and Huber, 2010) was used to screen differentially expressed miRs and perform correlation analysis. In total, 191 differentially expressed miRs were screened. Compared with the control group, 113 miRs were differentially expressed in the mTBI group, of which 50 were down-regulated, and 63 were up-regulated (Figure 3A and Table 2). In contrast, 71 differentially expressed miRs were observed in the sTBI group, including 14 miRNAs that were down-regulated and 57 that were up-regulated (Figure 3B and Table 3). Compared with the mTBI group, 61 miRs were differentially expressed in the sTBI group, 16 miRs were down-regulated, and 45 miRs were up-regulated (Figure 3C and Table 4). Compared to the control group, the TBI group had 27 differentially expressed miRs (Figure 3D and Table 5). Three differentially expressed miRs (ExomiR-206, ExomiR-133a-3p, and ExomiR-549a-3p) co-occurred after pairwise control among the three groups (Figure 3D and Table 6).
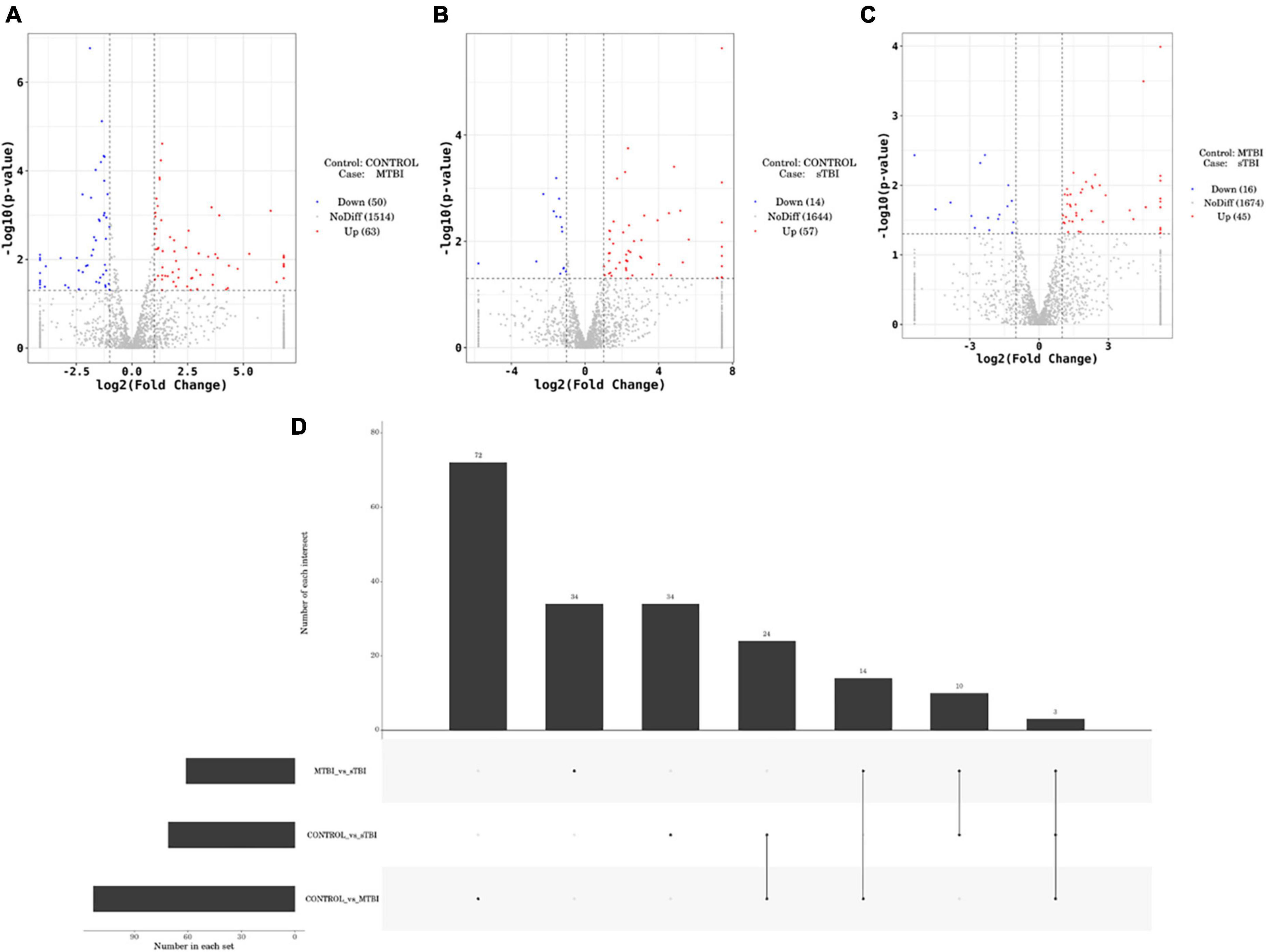
Figure 3. Volcano plots of differentially expressed genes comparing mTBI and control, sTBI and control, and sTBI and mTBI. (A) Compared to the control group, the differentially expressed miRs of mTBI. (B) Compared with the control group, the differentially expressed miRs of sTBI. (C) Compared with the mTBI group, the differential expression of miR in the sTBI group. The abscissa is |log2FoldChange|, and the ordinate is −log10 (p-value). The two vertical dashed lines in the figure are the 2-fold expression difference threshold, and the horizontal dashed line is the P = 0.05 threshold. Red dots indicate up-regulated genes in this group; blue dots indicate down-regulated genes in this group; and grey dots indicate non-significantly differentially expressed genes. (D) Different analyses of the groups revealed a common unique differential miR.
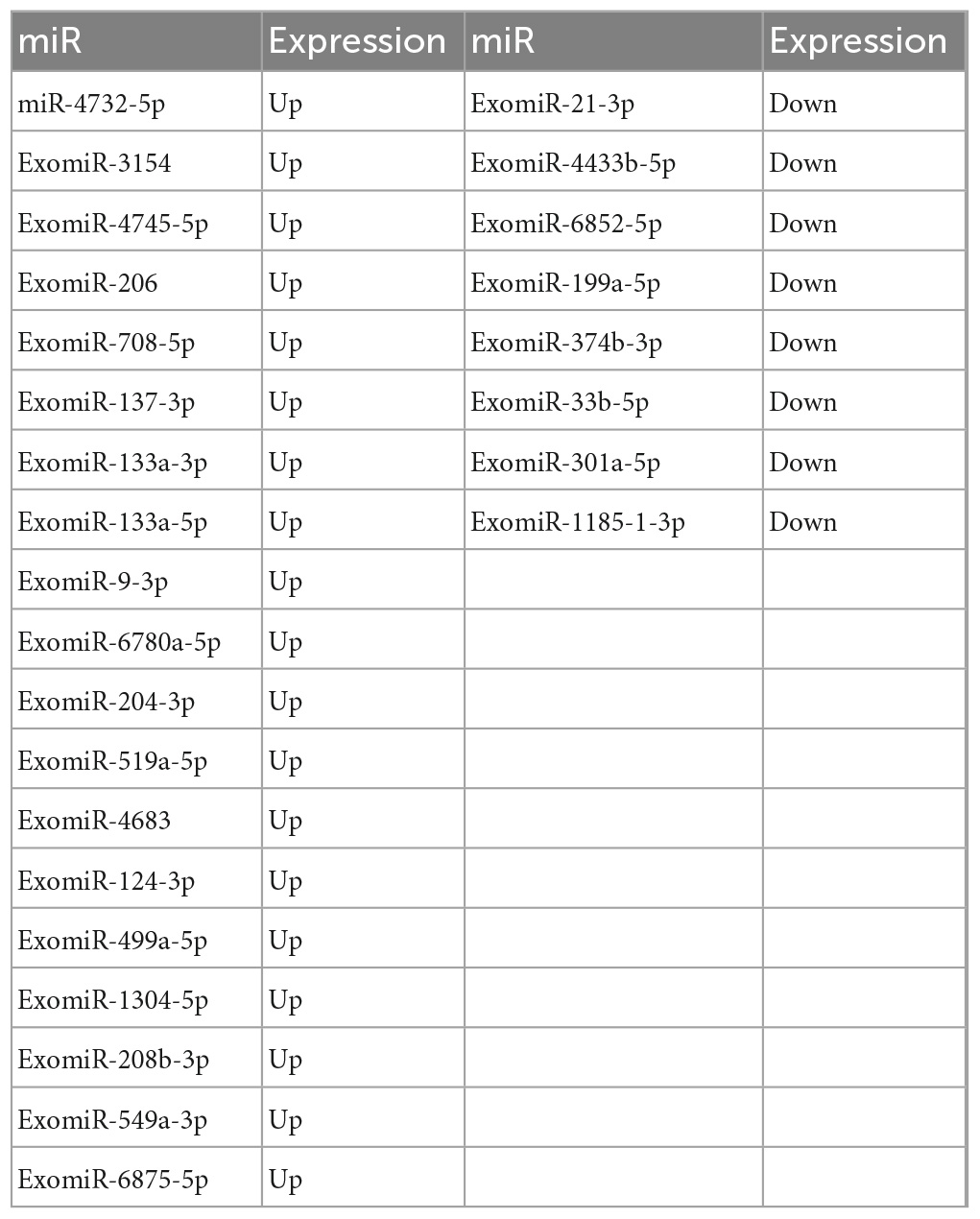
Table 5. Compared with the control group, the differential gene miR and expression shared by the mTBI and sTBI groups.
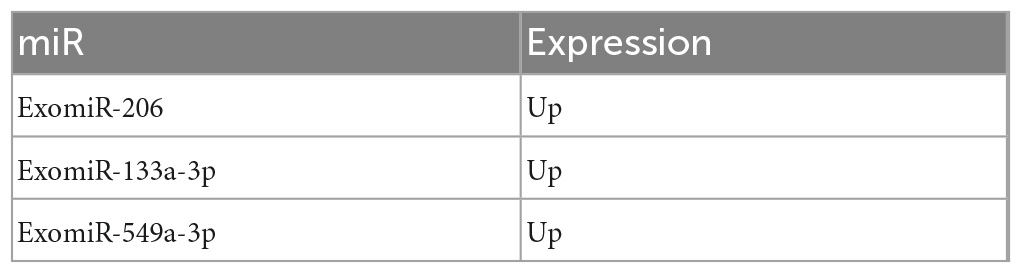
Table 6. The miRs and expression shared by the control group, mTBI, and sTBI after pairwise comparison.
Gene ontology and KEGG analyses of the host genes of differentially expressed miRs were conducted to predict miR functions and molecular interactions among genes.
Gene ontology analysis covered three domains: biological process, cellular component, and molecular function. The top 10 enriched GO terms for biological processes, cellular components, and molecular functions are shown in Figure 4. GO function annotation analyses showed that differentially expressed gene miRs are involved in the positive regulation of cellular and molecular biological processes. Compared with the control group, the mTBI group also differed in nucleotide regulation (Figure 4A). In addition, the differential gene miRs of the sTBI group are involved in the positive regulation of cellular and molecular biological processes and participate in cell adhesion and migration regulation (Figure 4B).
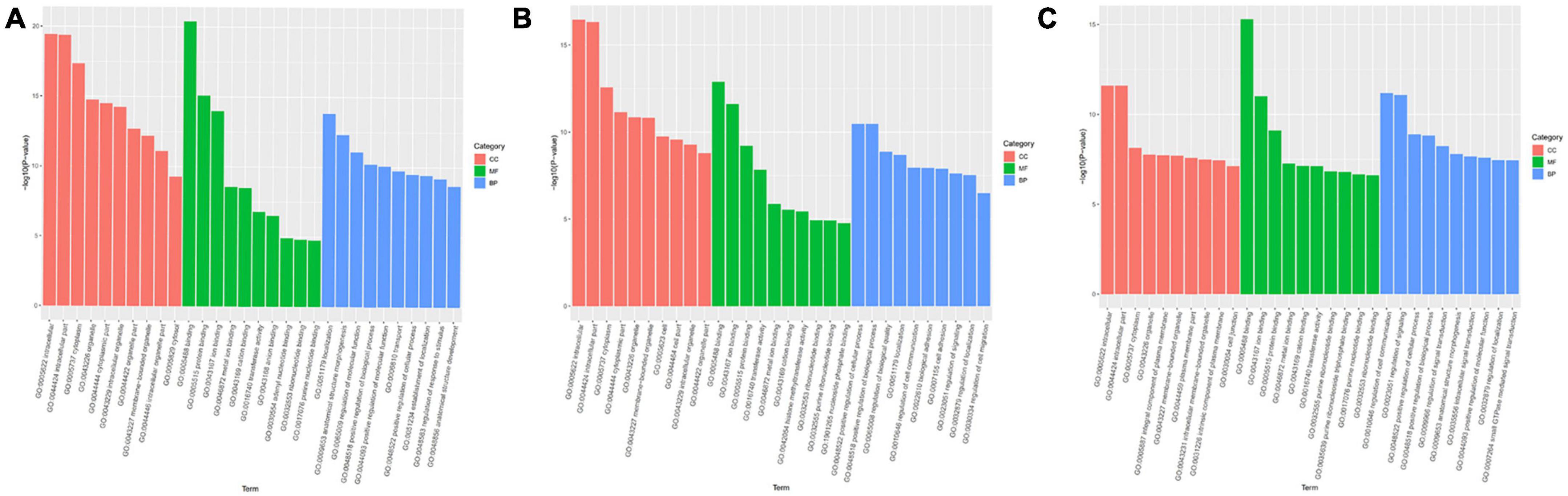
Figure 4. GO functional annotation results of differential miRs between mTBI, sTBI, and control. (A) mTBI and the control group show different gene miR GO function comment results. (B) sTBI and the control group’s differential gene miR-GO function annotation results. (C) mTBI and sTBI miR-GO function note results; CC, cellular components; MF, molecular function; BP, biological process.
Kyoto Encyclopedia of Genes and Genomes analysis showed that differentially expressed gene miRs were related to p53 signaling pathways, calcium signaling pathways, cell adhesion molecules (CAMs), Fc gamma R-mediated phagocytosis, RNA transport, the Rap1 signaling pathway, and other pathways (Figure 5). The transforming growth factor (TGF)-β signaling pathway, cAMP signaling pathway, cellular senescence, MAPK signaling pathway, and serotonergic synapse are linked to mTBI (Figure 5A). Signaling pathways related to sTBI include Staphylococcus aureus infection, cholinergic synapses, and the phospholipase D signaling pathway (Figure 5B). The three miRs (ExomiR-206, ExomiR-133a-3p, and ExomiR-549a-3p) of the three groups’ KEGG pathway enrichment analysis showed that the main enrichment pathway is the soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) in vesicle transportation, gap connection, neurotrophic factor signaling pathway, RNA transportation, sphere signal pathway, peroxisome proliferator-activated receptors, and signaling channels.
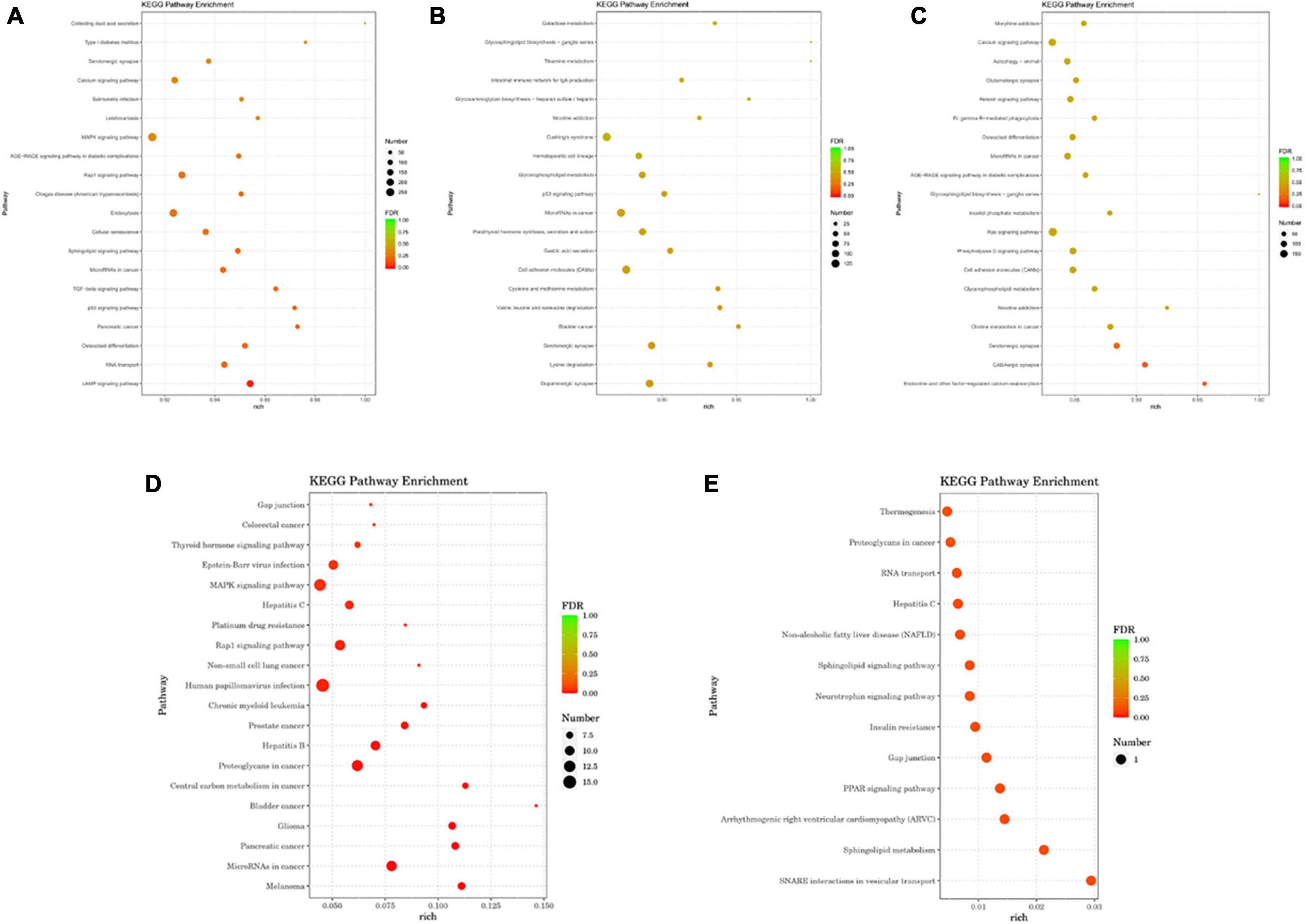
Figure 5. The analysis of the KEGG pathway comparing differential miRs between mTBI, sTBI, and control. (A) KEGG pathway analysis of the differential gene miRs between mTBI and the control group. (B) KEGG pathway analysis of the differential gene miRs between sTBI and the control group. (C) KEGG pathway analysis of the differential gene miRs between mTBI and sTBI. (D) Alignment of 27 miR KEGG functional annotation analyses of differential expression between control, mTBI, and sTBI. (E) Three miR KEGG pathway enrichment analyses showed differential expression following pairwise alignments of both controls, mTBI, and sTBI groups.
The relationship between the miR target gene was analyzed using the String database (Shrestha et al., 2019) and the PPI effect of the differential gene (Score > 0.95) was screened according to the genetic difference expression analysis results, and the PPI network was constructed. Using the Cystoscope software for visual analysis, confidence scores greater than 0.4 were obtained, and identified core modules in the network. The mTBI and control groups showed a comparative difference in expression of the miR target gene network core protein in the homology of cell division cycle 5-like (CDC5L), cell cycle divided protein 42 (CDC42), E1A binding protein (EP300), SMAD3, signal transducer and activator of transcription 3 (STAT3), and mitogen-activated protein kinase (MAPK) (Figure 6A). Compared to the control group, the sTBI group differentially expressed miRs, constructing a target gene PPI network graph core protein, mainly CDC42, MAPK (Figure 6B). In the mTBI group, the sTBI group constructing the target gene PPI network graph’s core protein was mainly CDC5L (Figure 6C).
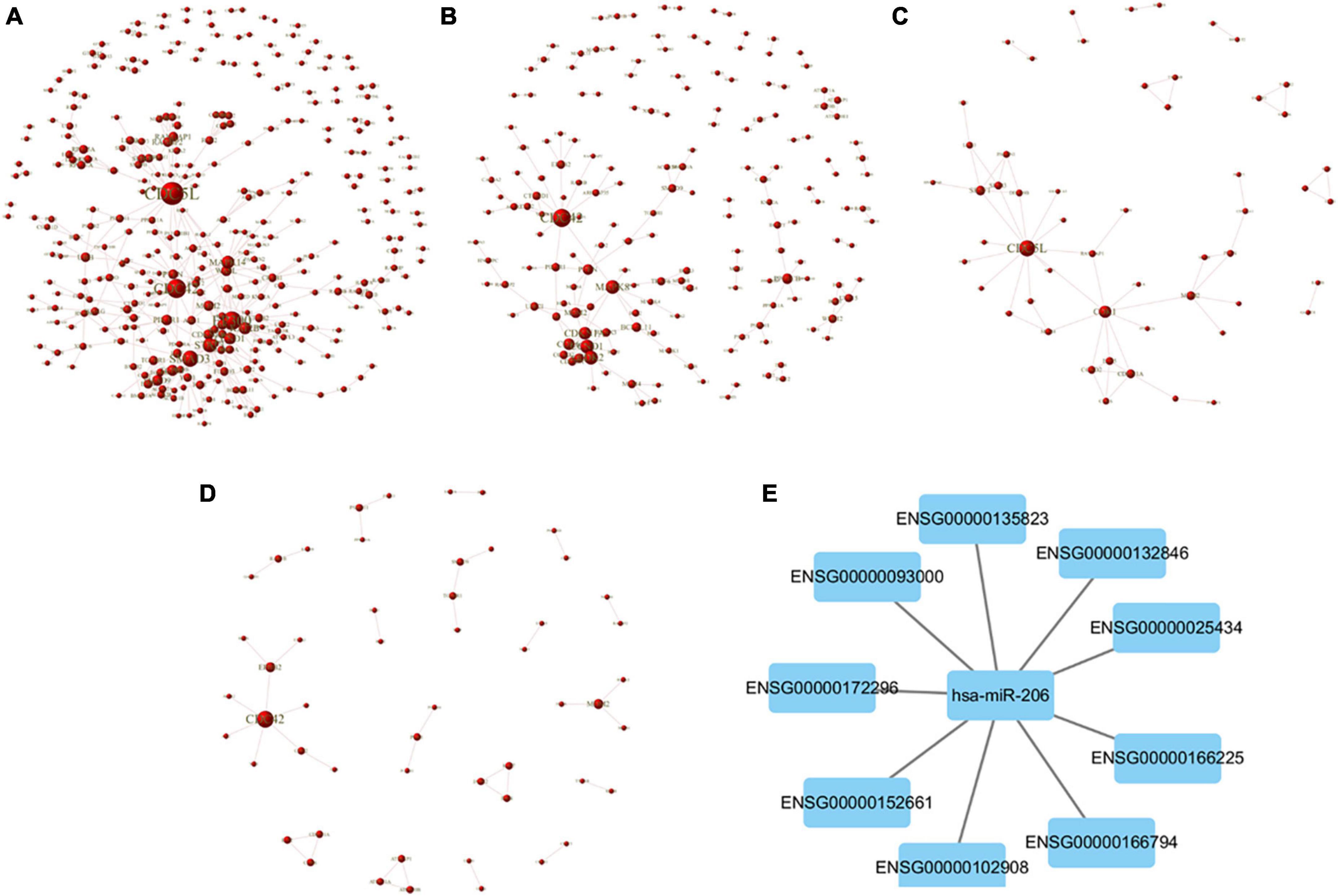
Figure 6. PPI network map of target genes. (A) Comparison of the differential expression of miRs between mTBI and control groups and a PPI network map of target genes. (B) Comparison of the sTBI group with the control group, screening out the differentially expressed miRs, and a PPI network map of target genes. (C) Comparison of the sTBI group with the mTBI group, screening out the differentially expressed miRs, and a PPI network map of target genes. (D) Comparison of the control, mTBI, and sTBI co-occurring 27 differentially expressed miRs to construct a PPI network map of target genes. (E) miR-206 and the target gene construct the PPI network diagram.
Further alignment control screening of the 27 miR constructs for differentially expressed target genes showed CDC42 as the core protein in mTBI and sTBI (Figure 6D). PPI network plots of miR-206 and its target genes in the three miRs were differentially expressed after pairwise alignments of the control, mTBI, and sTBI groups (Figure 6E and Table 7).
In this study, the analysis of serum exosomal miRNA expression profiles revealed eight up-regulated serum exosomal miRNAs (ExomiR-124-3p, ExomiR-137-3p, ExomiR-9-3p, ExomiR-133a-5p, ExomiR-204-3p, ExomiR-519a-5p, ExomiR-4732-5p, and ExomiR-206) and two down-regulated miRs (ExomiR-21-3p and ExomiR-199a-5) were regulated in concordance with TBI. miRs regulate gene expression and numerous other biological and pathological processes. These small RNAs have received increasing attention as potential biomarkers for detecting, identifying, classifying, and treating various diseases. Consistent with the results of our analysis, multiple studies have confirmed that multiple stable exosomal miRNAs can be detected in circulating body fluids and can change with disease progression (Martinez and Peplow, 2017; Zhou et al., 2018; Huang et al., 2020; Wu et al., 2020). O’Connell et al. determined that miR-124-3p, miR-137, and miR-9-3p had significantly higher serum levels in patients with TBI than in controls and that their sensitivity and specificity reached 90 and 100%, respectively (O’Connell et al., 2020). Using different sets of serum samples, we confirmed that the expression of serum exosomal miR-124-3p was higher in patients with TBI than in healthy controls. Vuokila et al. confirmed the chronic down-regulation of miR-124-3p after TBI, relevant to post-injury hippocampal pathologies in experimental models and humans (Vuokila et al., 2018). During the acute, subacute, and chronic phases after mTBI, microglial brain exosome miR-124-3p levels in damaged brain tissue were significantly altered, alleviating neurodegeneration and improving cognitive function (Ge et al., 2020). Additionally, one report reported that miR-9-5p may alleviate BBB damage and neuroinflammation after TBI (Wu et al., 2020). Ko et al. (2018) screened for miR expression profiles in patients with TBI by RNA sequencing. They revealed significant differences in miR-9-5a and mi-21 (P < 0.05) in the injured group, more accurately reflecting the heterogeneity of traumatic brain tissue injury and recovery than in the control group (Malizia and Wang, 2011; Ko et al., 2018).
They revealed significant differences in miR-9-5a and mi-21 (P < 0.05) in the injured group, more accurately reflecting the heterogeneity of traumatic brain tissue injury and recovery than in the control group (Malizia and Wang, 2011; Ko et al., 2018). In 2020, Ko et al. reported miR-206, which demonstrated positive predictive value for identifying injured patients vs. healthy controls. Recent studies have reported that increased miR-21-5p levels in the brain after TBI could improve neurological outcomes by alleviating BBB damage (Yin et al., 2020). Down-regulation of miR-21-3p levels in the injured brain could alleviate BBB damage by suppressing cellular apoptosis and the NF-κB-controlled inflammatory response, thereby improving neurological outcomes (Ge et al., 2019). Few studies have demonstrated that miR-133a (Dakhlallah et al., 2015) and miR-4732-5p (Sánchez-Sánchez et al., 2021) can act on ischemic cardiomyocytes to promote vasculature formation, reduce scarring, and prevent the deterioration of cardiac function. A literature search revealed that miR-199a-5p is highly expressed in myocardial tissues and is closely related to cardiac function (Tian et al., 2018; Hromadnikova et al., 2019). However, there is a lack of relevant studies demonstrating the association of miR-133a, ExomiR-519a-5p, ExomiR-4732-5p, and ExomiR-199a-5p with TBI. Consistent with these findings, the up-regulated expression of miRNAs in the TBI group was also monitored in this study, including ExomiR-124-3p, ExomiR-137-3p, ExomiR-9-3p, ExomiR-133a-5p, ExomiR-204-3p, ExomiR-519a-5p, ExomiR-4732-5p, and ExomiR-206, while ExomiR-21-3p, and ExomiR-199a-5 were down-regulated. Alternatively, we observed that miRs are potentially related to the pathophysiology of TBI and are a potential biomarker and novel target for drug therapy.
Gene ontology functional annotation analysis, and KEGG pathway analysis revealed that miRs were mainly associated with cellular aging, MAPK signaling, p53, TGF-β signaling, calcium signaling, cell adhesion molecules (CAMs), induction of neuronal injury, inflammatory mediator release, and damage to the BBB. Tao et al. (2018) showed that inhibition of MAPK pathways could mitigate microglial-mediated inflammatory responses and improve neural function. Yoong et al. (2006) demonstrated that the neurotrophic pathway can systematically and specifically regulate different miR expression levels, and the miR expression regulation involves the MAPK signaling pathway. Devoto et al. (2020) confirmed that dysregulated exosomal miRs correlate with inflammatory regulation pathways, neurological disease, and cell development. The expression levels of all the miRNAs studied may fluctuate (increase or decrease) depending on the regulation of particular biological processes involved in TBI. Among these miRNAs, hsa-miR-124-3p, ExomiR-137-3p, ExomiR-9-3p, ExomiR-133a-5p, ExomiR-204-3p, ExomiR-519a-5p, ExomiR-4732-5p, ExomiR-206, ExomiR-21-3p, and ExomiR-199a-5 are closely related to molecular function, biological processes, and cellular processes, respectively. Using bioinformatics technology analysis, it was observed that miR-199a-5, miR-133a-5p, miR-206, and ExomiR-21-3p can act on phosphoinositol metabolism, cellular senescence, and signaling pathways regulating stem cell pluripotency through SMAD3. miR-124-3p and miR-204-3p participate in the pathophysiology of patients with TBI, including activating MAPK signaling, cellular senescence, TNF signaling, and Rap1 signaling through MAPK14; miR-124-3p, miR-519a-5p, miR-9-3p, miR-204-3p, miR-4732-5p, miR-206, and ExomiR-199a-5 activate the Ras signaling pathway and Fc-mediated phagocytosis through CDC42. The miR-133a-5p and miR-519a-5p activate the cellular senescence, and TNF signaling pathways, and the VEGF signaling pathways through CDC5L.
High-throughput sequencing and bioinformatics analyses of the changes in serum exosomal miR expression profiles after brain tissue cell damage and the miR-acting target genes and signaling pathways related to TBI provide new research ideas for the study of TBI pathological mechanisms. However, this study has some limitations. First, the study is still in the initial stages of clinical research. Second, the differential expression of miRs may need to be quantitated further using PCR (Polymerase Chain Reaction). In addition, the therapeutic role of ExomiR-133a-5p a, ExomiR-519a-5p, ExomiR-4732 a-5p, and ExomiR vs. other miRNAs that are highly expressed in exosomes remains unclear. Finally, this study included a limited number of cases, which may have resulted in selection biases.
In this study, we extracted serum exosomal miRs from patients with TBI and identified GO functional annotation entries and KEGG pathway analysis associated with TBI. Serum exosomal miR expression profiling in patients with TBI can provide new biomarkers for the diagnosis and monitoring of TBI, which may eventually promote further research on the pathophysiological mechanism of TBI.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRINA861423.
Ethical review and approval were not required for the study of human participants per local legislation and institutional requirements. In addition, written informed consent from the patients/participants or patients/participants’ legal guardians/next of kin was not required to participate in this study per national legislation and institutional requirements.
XH and XX conceived and designed the experiments. CW, YW, YY, TY, RB, XP, FB, PL, and XX performed the experiments. XX analyzed the data. XH, XX, and XP wrote and revised the manuscript. All authors have read and approved the final manuscript.
This research was funded by the Research Project Supported by the Shanxi Scholarship Council of China (grant no: 2020-169), Fund Program for Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (grant no: 20210023), and Fundamental Research Program of Shanxi Province (grant no: 202203021211017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2023.1145307/full#supplementary-material
TBI, traumatic brain injury; miR, microRNA; NGS, next-generation sequencing; WHO, World Health Organization; CNS, central nervous system; mTBI, mild TBI and moderate TBI group; sTBI, severe TBI; PPI, protein-protein interaction network; TEM, transmission electron microscope; BP, biological process; CC, cellular component; MF, molecular function; GCS, Glasgow coma scale; NTA, nanoparticle tracking analysis; CAMs, cell adhesion molecules; TGF- β, transforming growth factor- β; CDC5L, cell division cycle 5-like; CDC42, cell cycle divided protein 42; EP300, E1A binding protein; STAT3, signal transducer and activator of transcription 3; MAPK, mitogen-activated protein kinase; BBB, blood-brain barrier; CCI, controlled cortical impact; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCR, polymerase chain reaction; BCA, bicinchoninic acid; ncRNA, non-coding RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; tRNA, Transfer RNA; EPF, Exosome Purification Filter.
Bao, N., Fang, B., Lv, H., Jiang, Y., Chen, F., Wang, Z., et al. (2018). Upregulation of miR-199a-5p protects spinal cord against ischemia/reperfusion-induced injury via downregulation of ECE1 in rat. Cell. Mol. Neurobiol. 38, 1293–1303. doi: 10.1007/s10571-018-0597-2
Cheng, P., Yin, P., Ning, P., Wang, L., Cheng, X., Liu, Y., et al. (2017). Trends in traumatic brain injury mortality in China, 2006–2013: a population-based longitudinal study. PLoS Med. 14:e1002332. doi: 10.1371/journal.pmed.1002332
Cornelius, C., Crupi, R., Calabrese, V., Graziano, A., Milone, P., Pennisi, G., et al. (2013). Traumatic brain injury: oxidative stress and neuroprotection. Antioxidants Redox Signal. 19, 836–853. doi: 10.1089/ars.2012.4981
Crupi, R., Cordaro, M., Cuzzocrea, S., and Impellizzeri, D. (2020). Management of traumatic brain injury: from present to future. Antioxidants 9:297. doi: 10.3390/antiox9040297
Dakhlallah, D., Zhang, J., Yu, L., Marsh, C. B., Angelos, M. G., and Khan, M. (2015). MicroRNA-133a engineered mesenchymal stem cells augment cardiac function and cell survival in the infarct heart. J. Cardiovasc. Pharmacol. 65, 241–251. doi: 10.1097/FJC.0000000000000183
Devoto, C., Lai, C., Qu, B. X., Guedes, V. A., Leete, J., Wilde, E., et al. (2020). Exosomal MicroRNAs in Military Personnel with Mild Traumatic Brain Injury: Preliminary Results from the Chronic Effects of Neurotrauma Consortium Biomarker Discovery Project. J. Neurotrauma 37, 2482–2492. doi: 10.1089/neu.2019.6933
Dewan, M. C., Rattani, A., Gupta, S., Baticulon, R. E., Hung, Y. C., Punchak, M., et al. (2019). Estimating the global incidence of traumatic brain injury. J. Neurosurg. 130, 1080–1097. doi: 10.3171/2017.10.JNS17352
Ge, X., Guo, M., Hu, T., Li, W., Huang, S., Yin, Z., et al. (2020). Increased microglial exosomal miR-124-3p alleviates neurodegeneration and improves cognitive outcome after rmTBI. Mol. Ther. 28, 503–522. doi: 10.1016/j.ymthe.2019.11.017
Ge, X., Li, W., Huang, S., Yin, Z., Yang, M., Han, Z., et al. (2019). Increased miR-21-3p in injured brain microvascular endothelial cells after traumatic brain injury aggravates blood-brain barrier damage by promoting cellular apoptosis and inflammation through targeting MAT2B. J. Neurotrauma 36, 1291–1305. doi: 10.1089/neu.2018.5728
Ghai, V., Fallen, S., Baxter, D., Scherler, K., Kim, T. K., Zhou, Y., et al. (2020). Alterations in plasma microRNA and protein levels in war veterans with chronic mild traumatic brain injury. J. Neurotrauma 37, 1418–1430. doi: 10.1089/neu.2019.6826
Hromadnikova, I., Kotlabova, K., Dvorakova, L., and Krofta, L. (2019). Postpartum profiling of microRNAs involved in pathogenesis of cardiovascular/cerebrovascular diseases in women exposed to pregnancy-related complications. Int. J. Cardiol. 291, 158–167. doi: 10.1016/j.ijcard.2019.05.036
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang, J. H., Xu, Y., Yin, X. M., and Lin, F. Y. (2020). Exosomes derived from miR-126-modified MSCs promote angiogenesis and neurogenesis and attenuate apoptosis after spinal cord injury in rats. Neuroscience 424, 133–145. doi: 10.1016/j.neuroscience.2019.10.043
Jiang, J. Y., Feng, H., Fu, Z., Guo-yi, G., Wei-ping, L., Wei-guo, L., et al. (2007). Violent head trauma in China: report of 2254 cases. Surg. Neurol. 68, S2–5; discussion S5. doi: 10.1016/j.surneu.2007.09.016.
Jiang, J. Y., Gao, G. Y., Feng, J. F., Mao, Q., Chen, L. G., Yang, X. F., et al. (2019). Traumatic brain injury in China. Lancet Neurol. 18, 286–295. doi: 10.1016/S1474-4422(18)30469-1
Kalvari, I., Nawrocki, E. P., Argasinska, J., Quinones-Olvera, N., Finn, R. D., Bateman, A., et al. (2018). Non-Coding RNA analysis using the RFAM database. Curr. Protoc. Bioinforma. 62:e51. doi: 10.1002/cpbi.51
Kamal, V., Agrawal, D., and Pandey, R. (2016). Epidemiology, clinical characteristics and outcomes of traumatic brain injury: evidences from integrated level 1 trauma center in India. J. Neurosci. Rural Pract. 7, 515–525. doi: 10.4103/0976-3147.188637
Ko, J., Hemphill, M., Yang, Z., Sewell, E., Na, Y. J., Sandsmark, D. K., et al. (2018). Diagnosis of traumatic brain injury using miRNA signatures in nanomagnetically isolated brain-derived extracellular vesicles. Lab Chip. 18, 3617–3630. doi: 10.1039/c8lc00672e
Li, Y., Chen, F., Zhang, J., Li, G., Yang, X., Lu, Q., et al. (2017). Epidemiological characteristics of Chinese paediatric traumatic brain injury inpatients. Brain Inj. 31, 1094–1101. doi: 10.1080/02699052.2017.1298004
Malizia, A. P., and Wang, D. Z. (2011). MicroRNAs in cardiomyocyte development. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 183–190. doi: 10.1002/wsbm.111
Martinez, B., and Peplow, P. V. (2017). MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen. Res. 12, 1749–1761. doi: 10.4103/1673-5374.219025
Nie, C., Sun, Y., Zhen, H., Guo, M., Ye, J., Liu, Z., et al. (2020). Differential expression of plasma Exo-miRNA in neurodegenerative diseases by next-generation sequencing. Front. Neurosci. 14:438. doi: 10.3389/fnins.2020.00438
O’Connell, G. C., Smothers, C. G., and Winkelman, C. (2020). Bioinformatic analysis of brain-specific miRNAs for identification of candidate traumatic brain injury blood biomarkers. Brain Inj. 34, 965–974. doi: 10.1080/02699052.2020.1764102
Rubiano, A. M., Carney, N., Chesnut, R., and Puyana, J. C. (2015). Global neurotrauma research challenges and opportunities. Nature 527, S193–S197. doi: 10.1038/nature16035
Sánchez-Sánchez, R., Gómez-Ferrer, M., Reinal, I., Buigues, M., Villanueva-Bádenas, E., Ontoria-Oviedo, I., et al. (2021). miR-4732-3p in extracellular vesicles from mesenchymal stromal cells is cardioprotective during myocardial ischemia. Front. Cell Dev. Biol. 9:734143. doi: 10.3389/fcell.2021.734143
Shrestha, S., Phay, M., Kim, H. H., Pouladvand, P., Lee, S. J., and Yoo, S. (2019). Differential regulation of brain-derived neurotrophic factor (BDNF) expression in sensory neuron axons by miRNA-206. FEBS Open Bio 9, 374–383. doi: 10.1002/2211-5463.12581
Sticht, C., De La Torre, C., Parveen, A., and Gretz, N. (2018). Mirwalk: an online resource for prediction of microrna binding sites. PLoS One 13:e0206239. doi: 10.1371/journal.pone.0206239
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi: 10.1093/nar/gkaa1074
Tao, L., Li, D., Liu, H., Jiang, F., Xu, Y., Cao, Y., et al. (2018). Neuroprotective effects of metformin on traumatic brain injury in rats associated with NF-κB and MAPK signaling pathway. Brain Res. Bull. 140, 154–161. doi: 10.1016/j.brainresbull.2018.04.008
Tian, X., Yu, C., Shi, L., Li, D., Chen, X., Xia, D., et al. (2018). Microrna-199a-5p aggravates primary hypertension by damaging vascular endothelial cells through inhibition of autophagy and promotion of apoptosis. Exp. Ther. Med. 16, 595–602. doi: 10.3892/etm.2018.6252
Vuokila, N., Lukasiuk, K., Bot, A. M., van Vliet, E. A., Aronica, E., Pitkänen, A., et al. (2018). miR-124-3p is a chronic regulator of gene expression after brain injury. Cell. Mol. Life Sci. 75, 4557–4581. doi: 10.1007/s00018-018-2911-z
Werner, C., and Engelhard, K. (2007). Pathophysiology of traumatic brain injury. Br. J. Anaesth. 99, 4–9. doi: 10.1093/bja/aem131
Wu, J., He, J., Tian, X., Luo, Y., Zhong, J., Zhang, H., et al. (2020). microRNA-9-5p alleviates blood–brain barrier damage and neuroinflammation after traumatic brain injury. J. Neurochem. 153:e14963. doi: 10.1111/jnc.14963
Xu, H., Pi, H., Ma, L., Su, X., and Wang, J. (2016). Incidence of headache after traumatic brain injury in china: a large prospective study. World Neurosurg. 88, 289–296. doi: 10.1016/j.wneu.2015.12.028
Yin, Z., Han, Z., Hu, T., Zhang, S., Ge, X., Huang, S., et al. (2020). Neuron-derived exosomes with high miR-21-5p expression promoted polarization of M1 microglia in culture. Brain. Behav. Immun. 83, 270–282. doi: 10.1016/j.bbi.2019.11.004
Yoong, L. F., Wan, G., and Too, H. P. (2006). Glial cell-line derived neurotrophic factor and neurturin regulate the expressions of distinct miRNA precursors through the activation of GFRa2. J. Neurochem. 98, 1149–1158. doi: 10.1111/j.1471-4159.2006.03959.x
Zhang, J., Li, S., Li, L., Li, M., Guo, C., Yao, J., et al. (2015). Exosome and exosomal microRNA: trafficking, sorting, and function. Genom. Proteom. Bioinforma 13, 17–24. doi: 10.1016/j.gpb.2015.02.001
Keywords: traumatic brain injury, exosome, microRNA (miR), next-generation sequencing (NGS), bioinformatics analysis
Citation: Huang X, Xu X, Wang C, Wang Y, Yang Y, Yao T, Bai R, Pei X, Bai F and Li P (2023) Using bioinformatics technology to mine the expression of serum exosomal miRNA in patients with traumatic brain injury. Front. Neurosci. 17:1145307. doi: 10.3389/fnins.2023.1145307
Received: 16 January 2023; Accepted: 13 March 2023;
Published: 18 April 2023.
Edited by:
Zilong Zhao, Department of Neurosurgery, Tianjin Medical University General Hospital, ChinaReviewed by:
Yang Shan, Michigan Medicine, University of Michigan, United StatesCopyright © 2023 Huang, Xu, Wang, Wang, Yang, Yao, Bai, Pei, Bai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xintao Huang, eGludGFvaEBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.