
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci., 22 March 2023
Sec. Neural Technology
Volume 17 - 2023 | https://doi.org/10.3389/fnins.2023.1125433
This article is part of the Research TopicInvertebrate Neurophysiology - of Currents, Cells, and CircuitsView all 15 articles
Nitric oxide (NO) is one of the most ancient and versatile signal molecules across all domains of life. NO signaling might also play an essential role in the origin of animal organization. Yet, practically nothing is known about the distribution and functions of NO-dependent signaling pathways in representatives of early branching metazoans such as Ctenophora. Here, we explore the presence and organization of NO signaling components using Mnemiopsis and kin as essential reference species. We show that NO synthase (NOS) is present in at least eight ctenophore species, including Euplokamis and Coeloplana, representing the most basal ctenophore lineages. However, NOS could be secondarily lost in many other ctenophores, including Pleurobrachia and Beroe. In Mnemiopsis leidyi, NOS is present both in adult tissues and differentially expressed in later embryonic stages suggesting the involvement of NO in developmental mechanisms. Ctenophores also possess soluble guanylyl cyclases as potential NO receptors with weak but differential expression across tissues. Combined, these data indicate that the canonical NO-cGMP signaling pathways existed in the common ancestor of animals and could be involved in the control of morphogenesis, cilia activities, feeding and different behaviors.
A free radical gas, nitric oxide (NO), is an evolutionary old and versatile signal molecule with a widespread distribution across all domains of life (Feelisch and Martin, 1995; Moroz and Kohn, 2011; Shepherd et al., 2022). NO can be synthesized by numerous non-enzymatic pathways (Moroz and Kohn, 2011) and enzymatically as parts of the nitrogen cycle and by NO synthases (NOS). NOSs were identified both in prokaryotes and eukaryotes. All NOSs catalyze the oxidation of L-arginine by molecular oxygen with several co-factors required for electron transfer from NADPH via FMN in the reductase domain to heme-/biopterin binding sites of the N-terminal oxygenase domain (Stuehr and Haque, 2019). Prokaryotic NOSs are usually truncated, often without the reductase part, and might contain additional subdomains (e.g., globin-like). Animal NOSs always have Ca2+/calmodulin-binding domains that tightly control the electron transport and NO yield in response to environmental stimuli (Stuehr and Haque, 2019).
Eukaryotic type NOS was recently discovered in Salpingoeca infusionum, the representative of Choanoflagellata (Reyes-Rivera et al., 2022) – the sister group to Metazoa. In another choanoflagellate Choanoeca flexa, with prokaryotic-type NOS, NO application induced contractions of colonies, providing a shift from feeding to swimming behaviors. Such an effect can be mediated by soluble guanylate cyclase (sGC), as in animals (Reyes-Rivera et al., 2022). These data and the presence of NOS in other eukaryotes suggest that the common ancestor of all animals might possess NOS-sGC signaling pathways. However, little is known about the distribution and functions of NO-mediated (nitrergic) signaling in early branching metazoans (Colasanti et al., 2010).
There are five major animal clades: Bilateria, Cnidaria, Placozoa, Porifera (sponges), and Ctenophora (comb jellies). The last four groups represent the earliest branching animal lineages, separated from Bilaterians more than 550 million years ago. These non-bilaterians are remarkably different in their bodyplans and tissue organizations. As a result, they are crucial to reconstruct the evolution and overall architecture of animal signaling pathways, including NO-dependent transmission. Yet only several papers deal with NO biology in Cnidaria and Porifera; and one publication discusses NO synthesis in placozoans (Moroz et al., 2020).
Ca-dependent and heat-stress-induced NO synthesis was reported in two species of desmosponges, Axinella polypoides and Petrosia ficiformis (Giovine et al., 2001). NADPH diaphorase histochemistry [a marker for NOS (Bredt et al., 1991; Moroz, 2000a)] showed specific localization of NOS activity in a particular network of dendritic cells in the sponge parenchyma (Giovine et al., 2001), but with unknown functions.
NO, and NOS control metamorphosis and symbiotic relationships in the desmosponge Amphimedon (Ueda et al., 2016; Hewitt and Degnan, 2022). There is growing evidence that NO can modulate body contractions and coordinated behaviors in freshwater sponges Ephydatia muelleri (Elliott and Leys, 2010), Spongilla lacustris (Musser et al., 2021), and the marine sponge Tethya wilhelma (Ellwanger and Nickel, 2006).
Similarly to sponges, NO signaling was implemented in cnidarian-algal symbioses (Perez and Weis, 2006; Safavi-Hemami et al., 2010; Hawkins and Davy, 2013), coral bleaching (Bouchard and Yamasaki, 2008), and apoptosis (Hawkins et al., 2013). In the hydroid polyp Hydra, non-neuronal NO/NOS is associated with regeneration (Colasanti et al., 2009) and feeding (Colasanti et al., 1995). Specific nitrergic neurons have been identified in tentacles of the hydromedusa Aglantha digitale, where the NO/cGMP pathway modulates the rhythmic swimming associated with feeding (Moroz et al., 2004). Multiples isoforms of NOSs present in the majority of sequenced cnidarian genomes, with evidence of endogenous enzymatic NO synthesis in this lineage (Morrall et al., 2000; Moroz et al., 2004; Kass-Simon and Pierobon, 2007; Cristino et al., 2008; Anctil, 2009; Colasanti et al., 2010).
The overall logic of NO signaling in early animals is still elusive. Nevertheless, both in sponges and cnidarians, we might expect tightly coupled interplays of morphogenic and behavioral functions mediated by NO, inherently linked to the ancestral feeding modes and innate immunity (Moroz, 2000b,2001). Scattered comparative data point out that the volume transmission mediated by NO in nervous systems was a relatively later innovation in evolution, (Moroz et al., 2021). Unfortunately, no data are available about NOSs and NO signaling in ctenophores or comb-jellies with well-developed neuro-muscular organization across species (Norekian and Moroz, 2016, 2019a,b, 2020, 2021).
Ctenophora is one of the earliest branching lineages of metazoans with complex tissue and organ differentiations. All extant ctenophore species have well-developed neuronal and muscular systems, which might co-evolve independently (Moroz, 2015). The recently proposed most basal position of ctenophores (as the sister group to all other animals) is still highly debated (Whelan et al., 2015, 2017; Halanych et al., 2016; Telford et al., 2016; Laumer et al., 2019; Fernandez and Gabaldon, 2020; Kapli and Telford, 2020; Li et al., 2021; Redmond and McLysaght, 2021; Giacomelli et al., 2022). Nevertheless, the presence or absence of specific signaling pathways in the ctenophore lineage reshapes our general understanding of neuronal and animal evolution. NOS was not detected in the sequenced genome of Pleurobrachia bachei (Moroz et al., 2014), but we found a putative NOS in Mnemiopsis leidyi (Moroz and Kohn, 2016) by screening its sequenced genome (Moreland et al., 2020). Here, we further explore the presence and organization of NO signaling components using Mnemiopsis as essential reference species (Moreland et al., 2020).
Figure 1 shows the phyletic relationships among basal metazoan NOSs and several eukaryotic (e.g., Amoebozoa [Physarum], Fungi [Aspergillus and kin], and Ichthyosporea [Sphaeroforma arctica]) and prokaryotic outgroups. This tree confirms a highly derived nature of NOS in ctenophores, probably reflecting their accelerated evolution and a possible bottleneck in their paleontological history around the Permian extinction (Whelan et al., 2017). The tree also highlights several independent radiation events with duplication and triplication of NOSs in lineages leading to placozoans, cnidarians, and vertebrates [see also (Moroz et al., 2020)]. However, representative species from both Porifera and Ctenophora have only one NOS gene.
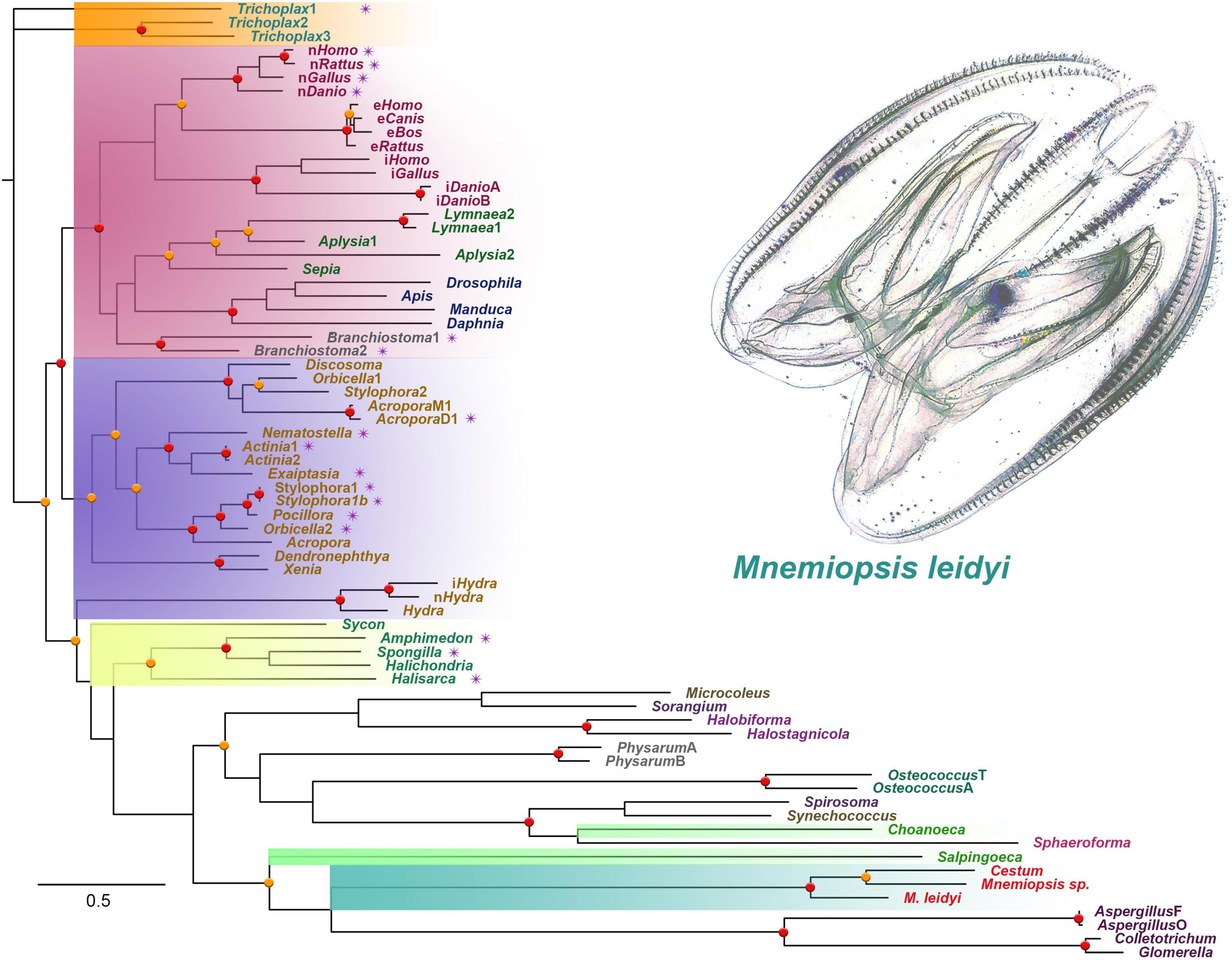
Figure 1. The phylogenetic tree of nitric oxide synthases (NOS) across different taxa, focusing on representative metazoan clades (ML analysis). Major taxons are highlighted: Ctenophora – aquamarine; Porifera – yellow; Placozoa – orange; Cnidaria – blue; Vertebrata/Bilateria – red; Choanoflagellata – bright green. The presence of PDZ domains is marked as purple stars near the species names. Bootstraps: red (90–100%) and orange (80–89%) dots.
In addition to the sequenced ctenophore genomes (Pleurobrachia bachei and Mnemiopsis leidyi), we analyzed transcriptomes from 37 ctenophore species (Whelan et al., 2017). NOSs were found only in eight ctenophores, including six representatives of Lobata: Mnemiopsis leidyi, undescribed species of Mnemiopsis sp., and their sister species Bolinopsis infundibulum [the family Bolinopsidae], the venus girdle, Cestum veneris [Cestidae], Ocyropsis crystallina [Ocyropsidae], Lobatolampea tetragona [Lobatolampeidae]. Most interesting, NOS was found in Euplokamis dunlapae [Euplokamididae] and the benthic ctenophore Coeloplana astericola [Platyctenida, Coeloplanidae] – representatives of the first and second earliest branching lineages within the phylum Ctenophora (Moroz et al., 2014). In contrast, Bolinopsis, Mnemiopsis, Cestum, Ocyropsis, Lobatolampea belong to separate branches within a highly derived (and possibly polyphyletic) clade of Lobata (Whelan et al., 2017).
NOS-type sequences were not identified in more than two dozen transcriptomes from adult tissues and different developmental stages from Pleurobrachia. Moreover, NOS was not found in the recently sequenced genome of a closely related species Hormiphora californica [Pleurobrachiidae] (Schultz et al., 2021). These data suggest that the majority of sequenced so far ctenophore lineages (including Cydippida and Beroida) lost NOS from their common ancestors. This apparently massive loss of NOS in many representatives of Ctenophora is quite unusual compared to other phyla. For example, NOS was lost in some nematodes such as C. elegans, but for most animal lineages, NOS genes are well-preserved, despite enormous ecological and morphological diversifications.
Figure 2 illustrates the domain organization of NOS across species. The Coeloplana, Bolinopsis, and Mnemiopsis NOSs revealed the canonical basal domain architecture of this class of enzymes (Figure 2). At the same time, sequences from other ctenophore species were represented mainly by the oxygenase domain and were incomplete genes, which the nature of transcriptome datasets can explain. Thus, we used M. leidyi NOS (ML074215a) as the reference sequence for these types of enzymes in ctenophores.
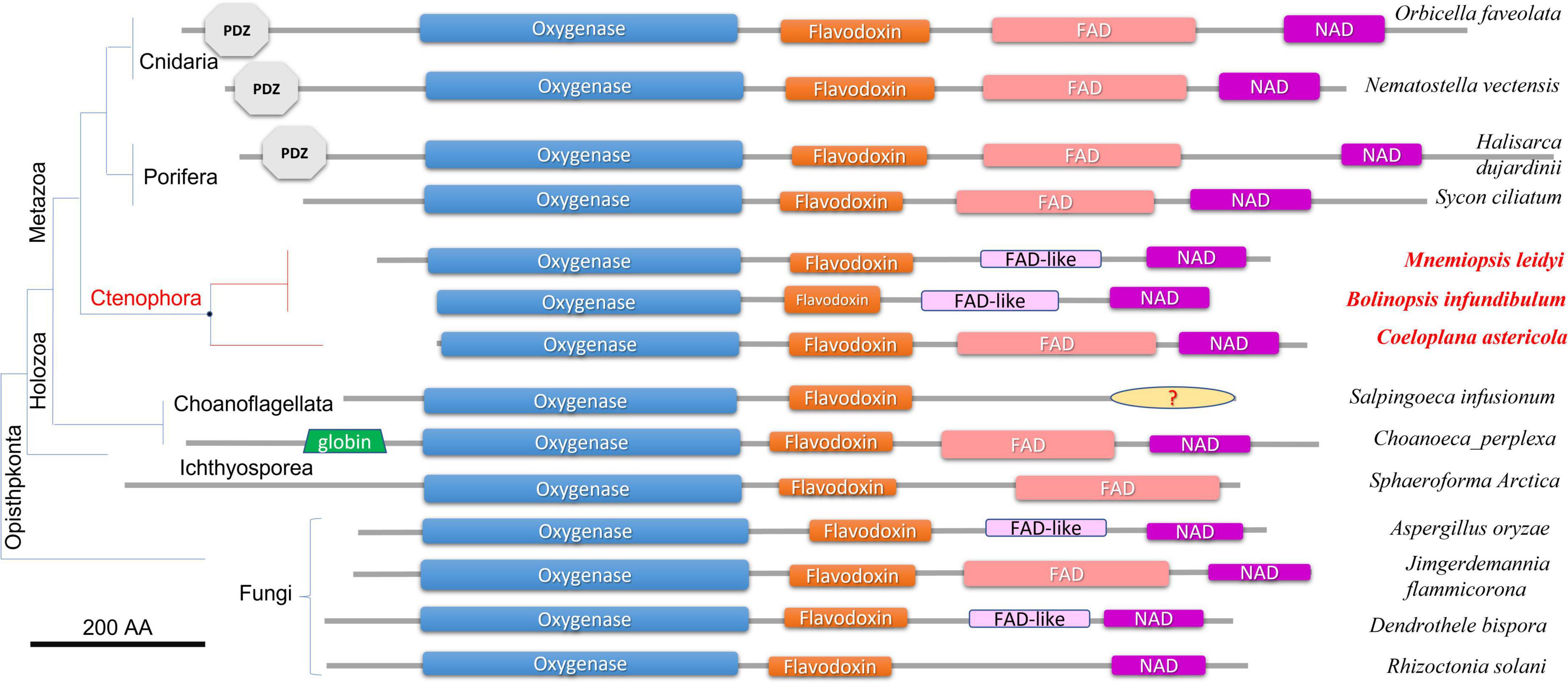
Figure 2. Domain organization of NOSs across eukaryotic phyla. All NOS are presented on the same scale, including the sizes of all domains and proteins. NOS oxygenase domain (Pfam NO_Synthase PF02898); Flavodoxin_1 (Pfam PF00258); FAD_binding_1 domain (Pfam PF00667); NAD_binding_1 (Pfam PF00175). NOSs include representatives of 3 metazoan phyla and 3 non-metazoan lineages with their respective phylogenetic relationships and species names. The references for each particular gene and/or their sequences with relevant GeneBank accession numbers are summarized in the Supplementary Table 1.
At least three features distinguish ctenophore NOSs from many other species. First, both in Bolinopsis and its sister species Mnemiopsis, a critical for NOS function FAD domain is either highly derived or might need to be better recognized. Still, their NOSs perfectly preserved NAD-binding sites (Figure 2). With this architecture NOSs from Bolinopsis and Mnemiopsis are similar to eukaryotic NOSs from the choanoflagellate Salpingoeca infusionum, and some fungi such as Aspergillus (Ascomycota) and Dendrothele (Basidiomycota). Highly modified or derived FAD-binding domains in these species might affect complex or unusual electron transport mechanisms from NADPH to heme in NOS (Stuehr and Haque, 2019). Yet, such FAD-related ctenophore modifications are likely secondarily derived. The canonical FAD-binding region is well-conserved in the benthic ctenophore Coeloplana (Figure 2), which probably represents the ancestral condition for ctenophore NOSs.
Second, Mnemiopsis leidyi NOS lacks the PDZ domain, found in many NOSs from poriferans (desmosponges Halisarca, Spongilla, and Amphimedon, but not from the calcareous sponge Sycon ciliatum), Placozoa (Trichoplax), cnidarians (corals, sea anemones), and bilaterians/chordates (Figures 1, 2). PDZ domain is best studied in mammals and is responsible for anchoring neuronal NOSs in specific membrane compartments and protein complexes, facilitating more localized and spatial control of NO release and signaling in synapses (Brenman et al., 1996; Stricker et al., 1997; Jaffrey et al., 1998; Kim and Sheng, 2004; Feng and Zhang, 2009; Sheng et al., 2018; Murciano-Calles et al., 2020). Of note, PDZ NOSs were not found in non-metazoan eukaryotes and prokaryotes and can be viewed as animal innovation coupled with their morphological and signaling complexities.
Third, Mnemiopsis leidyi NOS is shorter or truncated than known cnidarian, poriferan, and placozoans NOSs. In this case, it is also superficially similar to non-metazoan NOSs. Yet ctenophore NOSs make a highly derived branch on the phylogenetic trees with different representations of species (Figures 1, 3), suggesting an accelerated evolution within this group. A similar situation is also observed for choanoflagellates. Most choanoflagellate species apparently lose their NOSs from their common ancestor. The eukaryotic type of NOS is only preserved in one species, Salpingoeca infusionum, which is clustered with fungal NOSs (Figure 3) and lack some evolutionarily conserved regions, such as FAD- and NAD-binding (Figure 2). Other four choanoflagellate species had prokaryotic-type NOS [see (Reyes-Rivera et al., 2022) and Figures 2, 3]; these NOS might be results of a horizontal gene transfer from cyanobacteria.
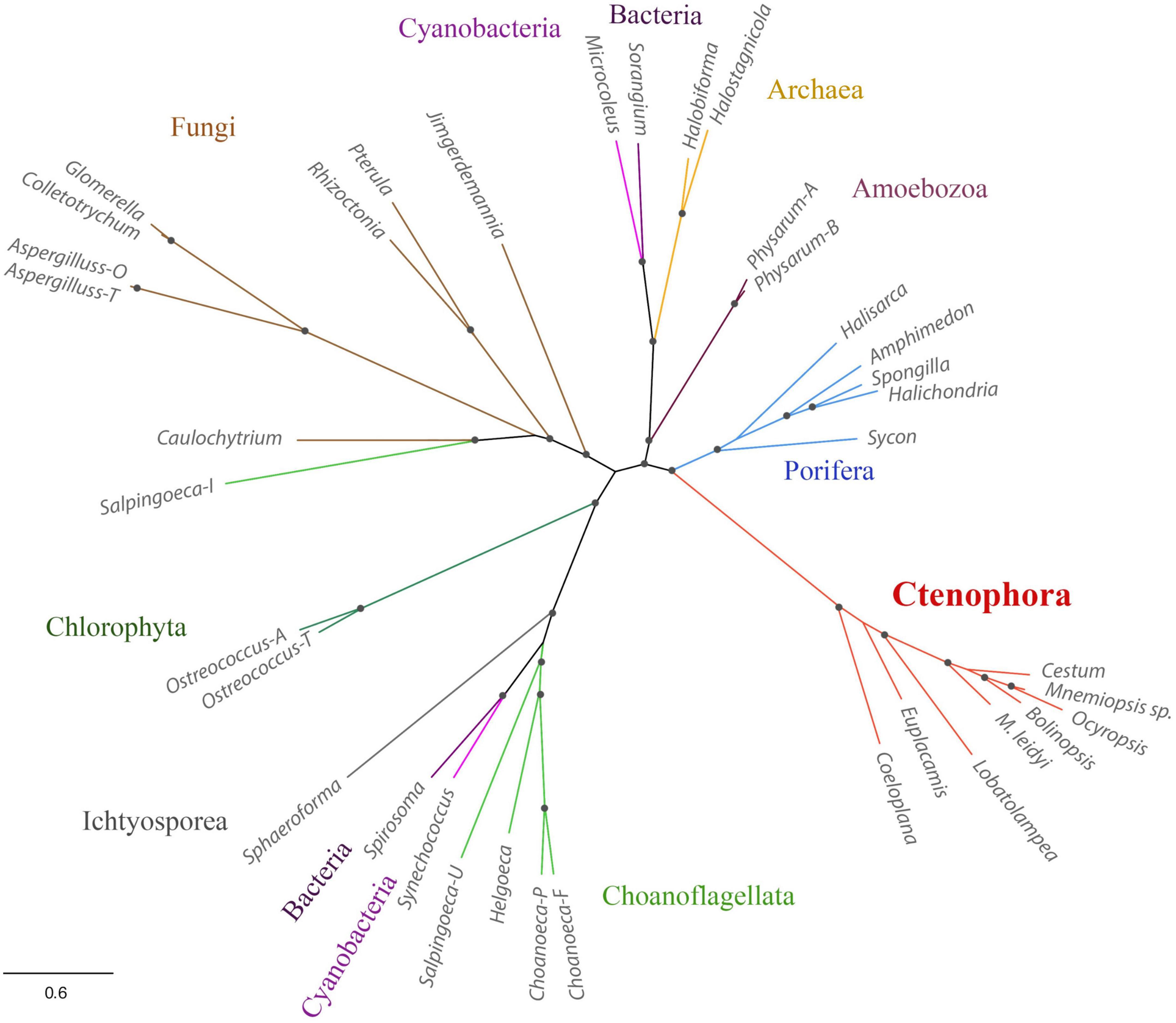
Figure 3. Phylogenetic relationships of NOSs in ctenophores, sponges, non-metazoan eukaryotes, and prokaryotes. The references for each particular gene and/or their sequences with relevant GeneBank accession numbers are summarized in the Supplementary Table 1.
Mnemiopsis leidyi NOS (ML074215a) has a recognized Ca/Calmodulin-binding region coupling oxygenase and reductase domains as in all animal NOSs (Figure 4, see Supplementary material for alignment), suggesting calcium-calmodulin dependence. In mammals, transient activation of NOS by intracellular Ca2+ is controlled by the auto-inhibitory loop/inserts (Roman and Masters, 2006; Stuehr and Haque, 2019), without sequence similarities in neuronal and endothelial isoforms. In contrast, inducible Ca-independent NOS lacks this sequence motif (Figure 4A). Thus, activation of its expression is induced by bacterial liposaccharides as a component of the innate immune defense mechanisms. Interestingly, ctenophore NOSs also lack the autoinhibitory loops (Figure 4A), and their Ca-dependence, enzymology, and expression control must be experimentally characterized. Ca-dependences of NOS in sponges and cnidarians are also unknown. However, both poriferan and cnidarian NOSs show more remarkable overall sequence similarities with their bilaterian homologs.
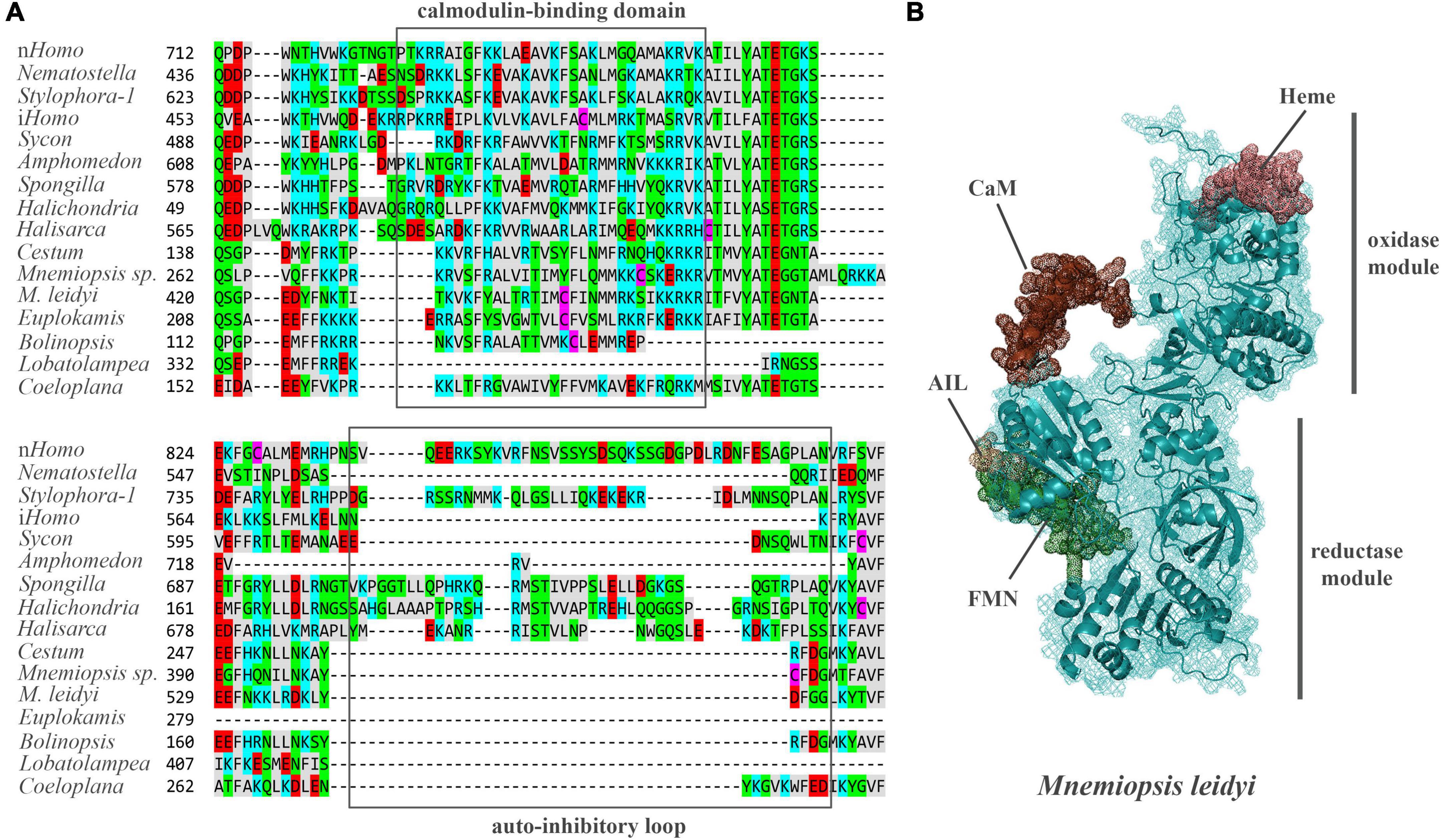
Figure 4. (A) The structure of auto-inhibitory loop and calmodulin-binding domains in NOSs. Amino acids (aa) are highlighted with different colors: red – polar negative charged aa; blue – polar positive charged aa; purple – cysteines; gray – hydrophobic aa; green – uncharged polar aa. (B) The predicted 3D structure of the NOS from Mnemiopsis leidyi with critical functional domains. Calmodulin (CaM) binding – brown, FMN – green, AIL – beige. The Supplementary Table 1 summarizes the references for each particular gene and/or their sequences with relevant GeneBank accession numbers.
Of note, we also found bacterial types of NOS in the Mnemiopsis microbiome (Mariita et al., 2021), suggesting both endogenous and exogenous sources of NO in ctenophores (e.g., from food or symbionts).
As a free radical gas, NO can interact with most biological molecules, primarily targeting SH and tyrosine groups (Holguín-Peña et al., 2007). However, in animals including cnidarians, placozoans, and sponges, NO can specifically activate soluble guanylyl cyclases [sGC, members of the adenylate cyclase superfamily (Gancedo, 2013; Bassler et al., 2018)] by binding to its heme group, which leads to conformational changes and increase of cGMP synthesis (Krumenacker et al., 2004; Martin et al., 2005; Windsor and Thompson, 2008; Gunn et al., 2012; Montfort et al., 2017; Horst and Marletta, 2018; Horst et al., 2019; Kang et al., 2019). Three orthogroups of sGCs are present in humans.
The Mnemiopsis and Pleurobrachia genomes encode four and two sGCs, respectively (Figure 5 and Supplementary Figure 1). These enzymes have the canonical heme NO binding domain and associated cyclase domain. Moreover, we also identified sGCs in several other ctenophore species with and without detectable NOS (Supplementary Figure 1). For example, there are four sGCs in the lobate Ocyropsis with NOS as in Mnemiopsis. In contrast, we have a reduced representation of sGCs in Pleurobrachia and other Cydippida and Beroida. Thus, the absence of NOS from the genomes does not exclude a possibility of NO signaling due to its non-enzymatic productions by different mechanisms (Moroz and Kohn, 2011) or from exogenous sources (environment, food, microbiomes, etc.).
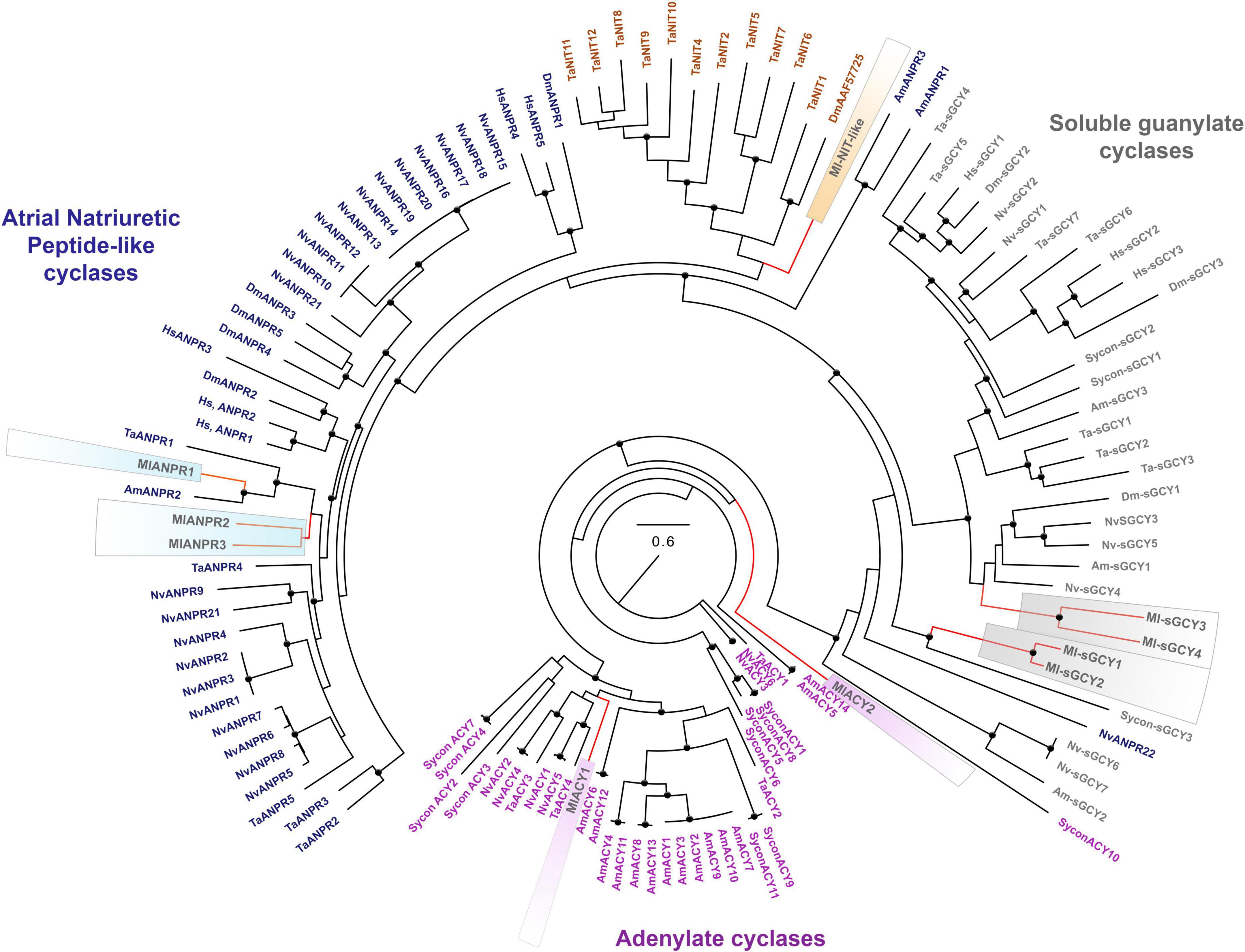
Figure 5. Phylogenomic ML analyses of cyclases across representative metazoans with the focus on Mnemiopsis leidyi and kin. Major classes of cyclases are highlighted by purple (adenylate cyclases), blue (ANPRs), brown (containing NIT-like domain cyclases), and gray (soluble guanylate cyclases). Bootstrap values are shown as black dots (90–100%). Genes of M. leidyi placed in color frames: purple (for ACYs), blue (for ANPRs), orange (for one putative NIT-like containing domain gene), gray (for sGCYs). The references for each particular gene and/or their sequences with relevant GeneBank accession numbers are summarized in the Supplementary Table 1.
Recent research also indicates that sGCs can be co-present with NOS in choanoflagellates, controlling swimming behaviors of colonies (Reyes-Rivera et al., 2022) and implying pre-metazoan origins of these signaling pathways. From this apparently simple ancestral condition, sGCs show lineage-specific radiations for many phyla, including 3 sGCs in sponges and humans, seven sGCs in placozoans and cnidarians (Figure 5).
In addition, ctenophores encode two other classes of membrane-bound cyclase candidates, such as a derived atrial natriuretic peptide receptor [ANPR] type group, probably involved in peptide sensing, and an enigmatic group of cyclases with NIT domains (Figures 5, 6). Mnemiopsis has three orthologs of ANPR-like receptors with CYC/cGMP coupling.
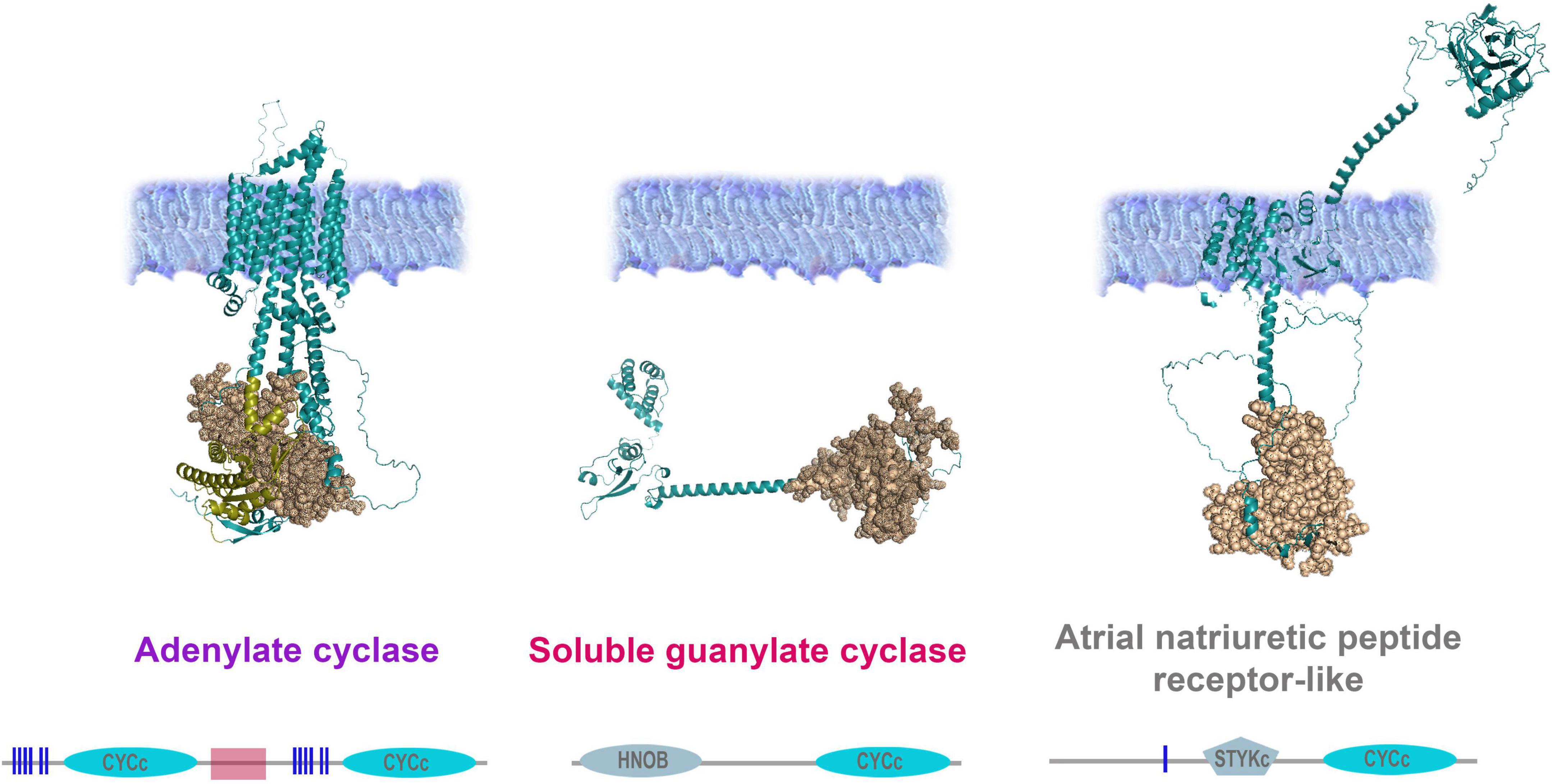
Figure 6. Organization of adenylate and guanylate cyclases in Mnemiopsis leidyi. Representative cyclases show notable structural differences: Adenylate cyclase anchor in the membrane with ten transmembrane regions and contain two intracellular catalytic CYCc domains; ANPR-like (atrial natriuretic peptide receptor-like) has the extracellular part (ANF-receptor) and two transmembrane domains, STYKc and CYCc domains are located in the intracellular space; soluble guanylate cyclase is located in the cytoplasm. All sequences were analyzed using SMART (Letunic and Bork, 2018; Letunic et al., 2021), NCBI Conserved Domain Search (Lu et al., 2020), and were built using AlphaFold2 (Jumper et al., 2021). The references for each particular gene and/or their sequences with relevant GeneBank accession numbers are summarized in the Supplementary Table 1.
Unique NIT (PF08376) domains (Oulavallickal et al., 2017), containing cyclases, were previously identified in Trichoplax (Moroz et al., 2020) as candidates for nitrite/nitrate sensors, similar to bacteria (Shu et al., 2003; Camargo et al., 2007). Mnemiopsis has one sequence (ML02033a) associated with the same cluster (Figure 5) but with a highly derived NIT-like region and unknown function.
Finally, Mnemiopsis has two adenylate cyclases (ML04963a, ML20918a) involved in the evolutionarily conserved cAMP signaling (Figures 5, 6). Surprisingly, the genome screening indicates that cnidarians and sponges possess more adenylate cyclases (5-14) than ctenophores (Figure 5).
Two groups of datasets provide initial estimates of the distribution of NOS and sGC expressions in Mnemiopsis leidyi (Figures 7–9): single cells data (scRNA-seq) from adult animals and transcriptomes (RNA-seq) of different embryonic and developmental stages (Levin et al., 2016).
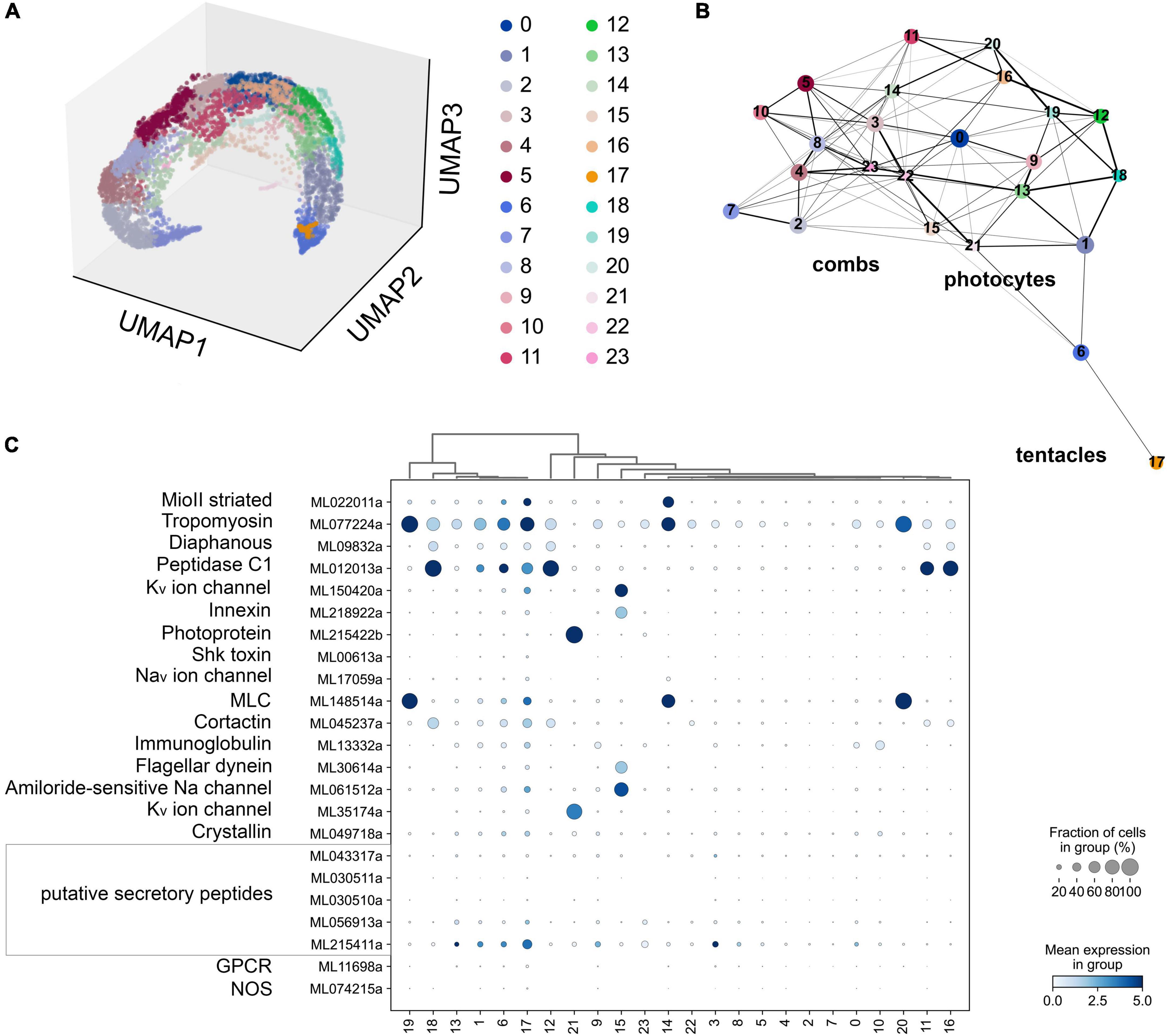
Figure 7. (A) The UMAP clustering for 6,144 cells of Mnemiopsis leidyi (Sebé-Pedrós et al., 2018) with 24 clusters. (B) The PAGA analysis placed clusters with connections based on their differential expressed genes. We found comb associated cluster (#15) using innexin, and flagellar dynein marker accordingly (Sebé-Pedrós et al., 2018). Photocyte’s cluster (#21) can be recognized using such markers as photoprotein and potassium channel. The tentacle cluster #17 is placed separately; it contains both marker genes (secretory peptides, tropomyosin, potassium and sodium channels (Sebé-Pedrós et al., 2018)) and putative NOS expressing cells. (C) Dotplots highlights cluster-specific genes (Sebé-Pedrós et al., 2018; Burkhardt and Jékely, 2021).
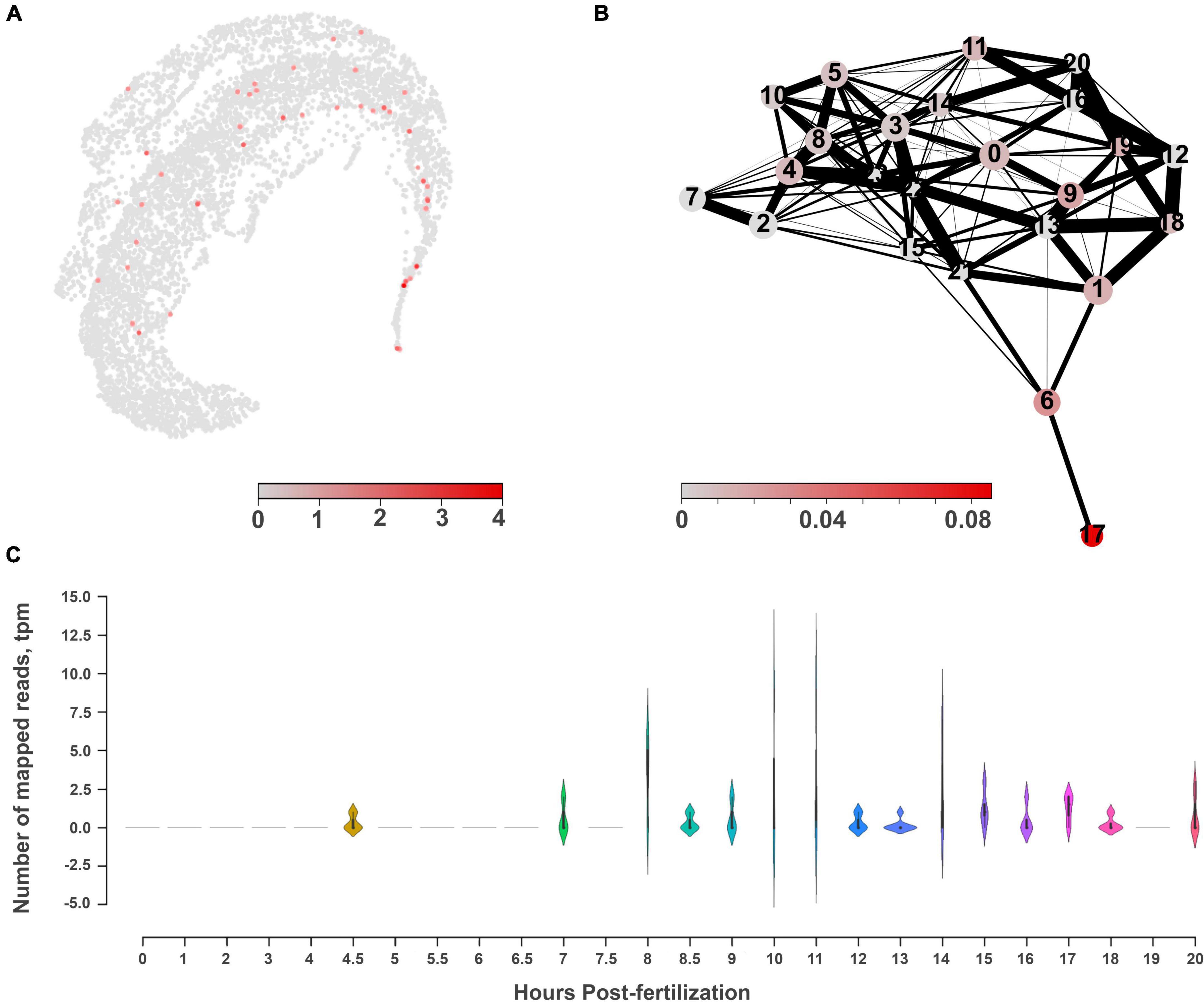
Figure 8. The molecular architecture and distribution of nitric oxide synthase (NOS) in Mnemiopsis leidyi. (A) The UMAP clustering for NOS shows a relatively diffuse distribution across several cell types with low expression levels. (B) PAGA analysis for NOS highlights its expression in cluster #17, which might belong to tentacles, according to the marker genes (Sebé-Pedrós et al., 2018). (C) RNA-seq profiling of NOS expression in development based on Levin et al. (2016), Moreland et al. (2020). NOS expression is detected after 7th hour of post-fertilization and is associated with the morphogenesis of major ctenophore organs.
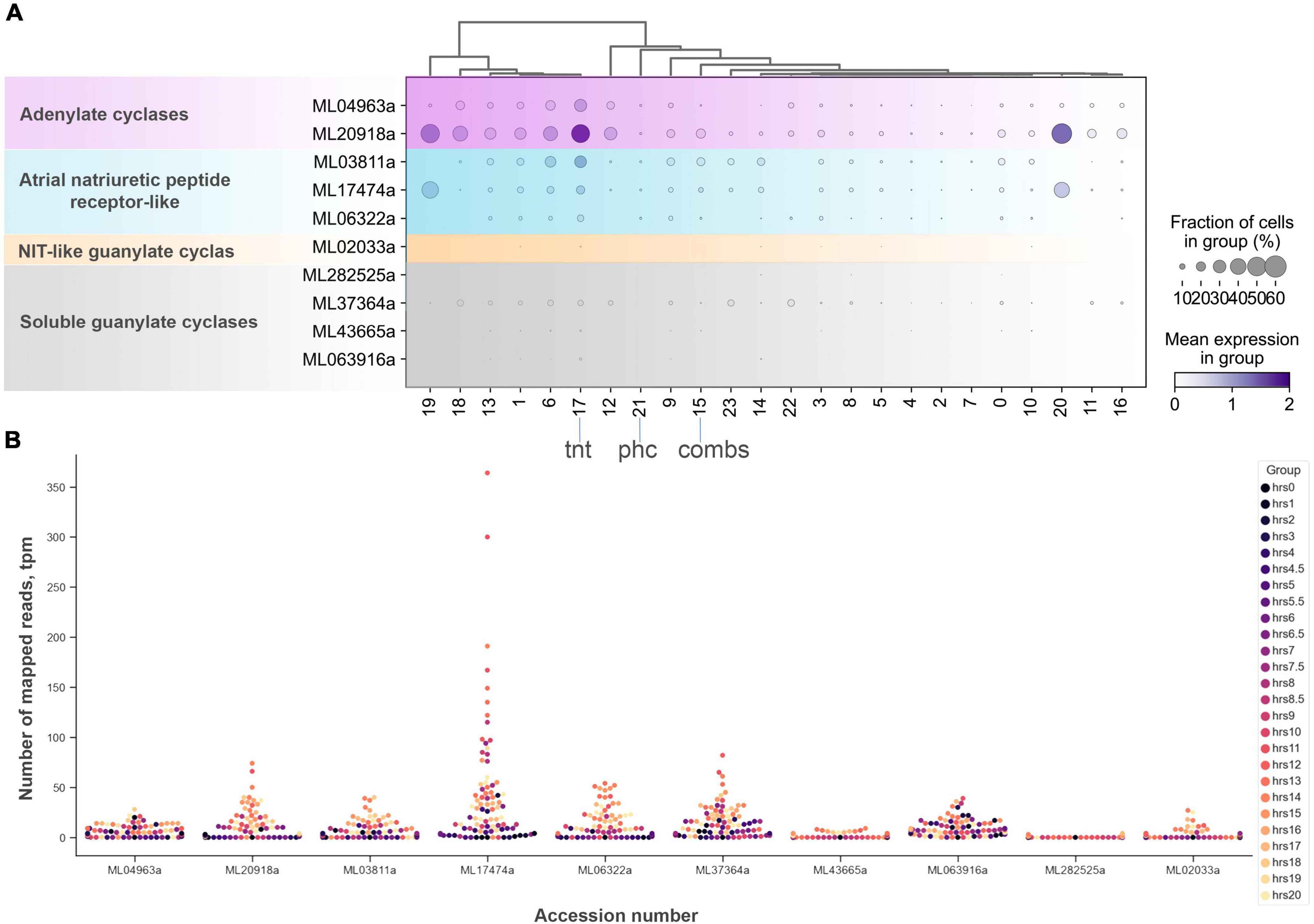
Figure 9. The diversity and expression of cyclase genes in Mnemiopsis leidyi based on scRNA-seq profiling (Sebé-Pedrós et al., 2018) (see text for details). (A) Dotplot analysis indicates that all cyclases are expressed in cluster #17 (tentacles) except one soluble guanylate cyclase (ML282525a). sGCY (ML37364a) has low expression levels in all cell clusters from an adult animal. However, the majority of sGCYs are expressed in developmental stages (B). RNA-seq profiling of expression in development is based on Levin et al. (2016), Moreland et al. (2020). Post-fertilization hours are indicated as color dots on the right.
According to the scRNA-seq datasets from 5,461 cells filtered as in Sebé-Pedrós et al. (2018), NOS was a relatively low-expressed gene, present in individual cells more or less randomly distributed in 15 clusters (out 41), including candidate clusters for some secretory cells (recognized with neuropeptide markers identified for this species). We did not detect NOS expression in combs and photocytes.
Next, considering the low abundance of NOS-containing cells and low level of expression, we performed a similar analysis without any filtration with attempts to capture rare cell types. This analysis with 6,144 cells detects a very low level of NOS expression in 15 out of 24 clusters (Figure 7), including a few cells with NOS in tentacle-associated cells/tissues, as also shown with partition-based graph abstraction (PAGA) analysis (Wolf et al., 2019).
Among all identified cyclases, adenylate cyclases and ANP-like receptors with guanylyl cyclases are the most abundantly expressed (Figure 9). Their expression is detected in most cell types, including tentacles and putative neurons. Nevertheless, sGCs are also very weakly expressed, similar to NOS. In summary, a small number of sequenced cells and an extremely low level of detected NOS/sGC expression prevent making definite conclusions about cell type specificity that might employ this signaling pathway. In future studies, this ambitious task would require multi-color in situ hybridization and immunohistochemistry experiments. A limited number of ctenophore cell-specific molecular markers also prevent more detailed annotation of these scRNA-seq data.
RNA-seq data from development contain significantly deeper sequencing than single-cell datasets. During development, NOS (ML074215a) expression became noticeable starting with the 7th hour of post-fertilization (pft), and further increased at the 10th and 11th hours, and later decreased. Here, NOS expression was associated with the specification of tissues and formation of ctenophore-specific organs such as combs, the aboral organ, and components of the digestive systems [e.g., (Simmons et al., 2012; Fischer et al., 2014; Reitzel et al., 2016)]. Expression of sGC was also weak in development and partially corresponded to the same or later stages of development compared to NOS expression (Supplementary Figure 2).
We used fixative-resistance NADPH-diaphorase (NADPH-d) histochemistry as a reporter of NOS activity (Bredt et al., 1991; Moroz, 2000a) across phyla, from cnidarians to vertebrates (Bredt et al., 1991; Kurenni et al., 1995; Moroz et al., 2000, 2004; Giovine et al., 2001; Bishop et al., 2008; Moroz and Kohn, 2011). Mnemiopsis has extremely fragile tissues, often disintegrated during conventional fixation, highly limiting in situ hybridization experiments, requiring multiple steps. However, we managed to perform simpler NADPH-d protocols using larger (2–5 cm) adult animals (n = 6). The NADPH-d reactivity (putative NOS activity) is broadly distributed across tissues in Mnemiopsis (Figure 10), including cells in the aboral organ, polar fields, base of combs, auricles, and meridional canals regions, as well as putative sensory papillae around the body surface, and in components of the digestive system.
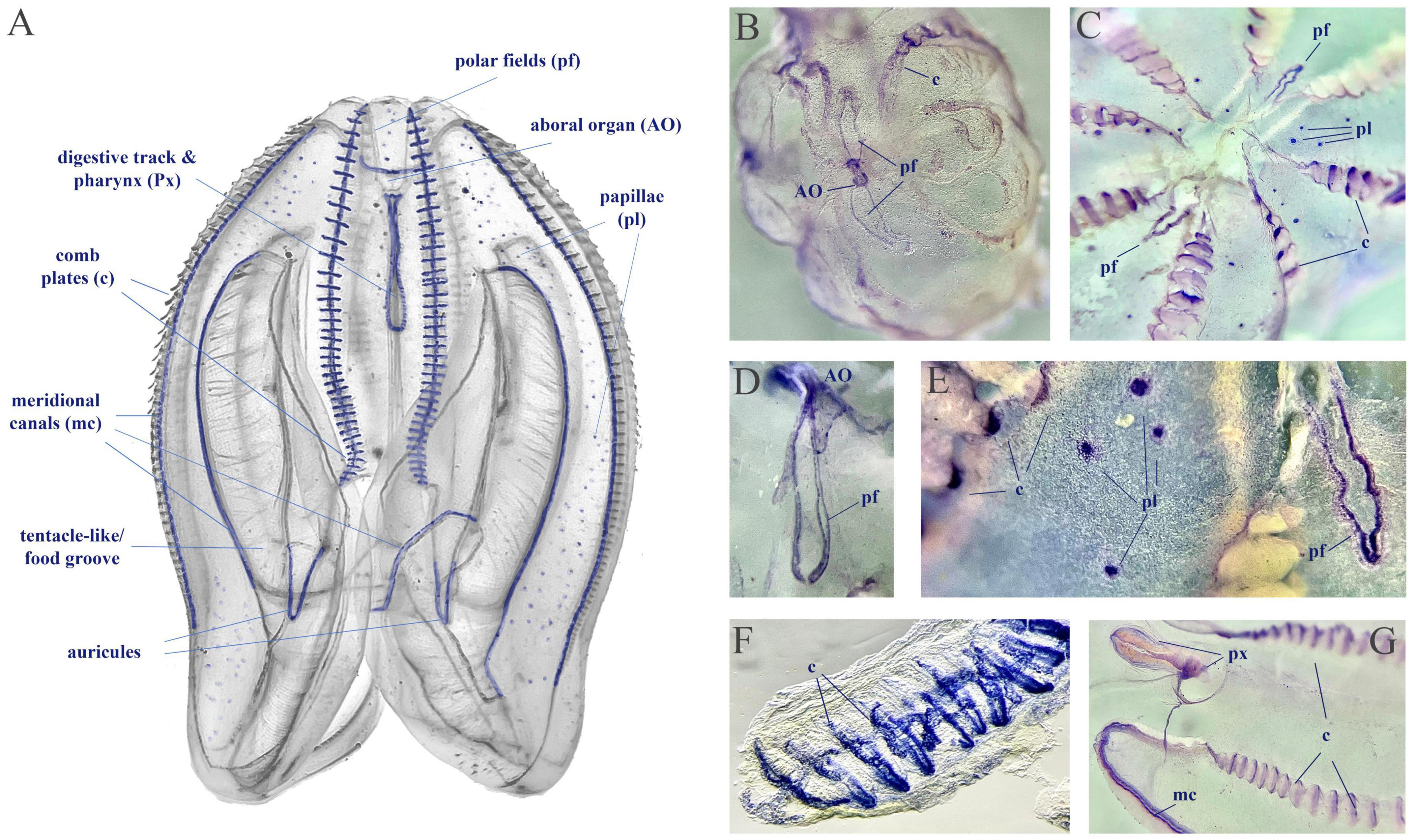
Figure 10. NADPH-diaphorase histochemistry in Mnemiopsis leidyi. (A) The distribution of putative NOS activity in Mnemiopsis. This schematics illustrate the regions with putative NOS, but not reflect quantitative estimates of tentative enzymatic activities. (B,C) Whole mount preparation with NADPH-d reactivity in the aboral organ (AO), the polar fields (pf), in comb plate areas (c), and papillae (pl). (D–G) Illustrated examples of NADPH-d reactivity across different structures, including the pharynx (px) and the meridional canal (mc) near the auricules (a feeding structure).
Furthermore, low expression levels do not imply the lack of functional role of NO-cGMP signaling because this pathway includes significant amplification cascades. Thus, we performed pilot pharmacological experiments to test the effects of NO on free-behaving animals (Figure 11).
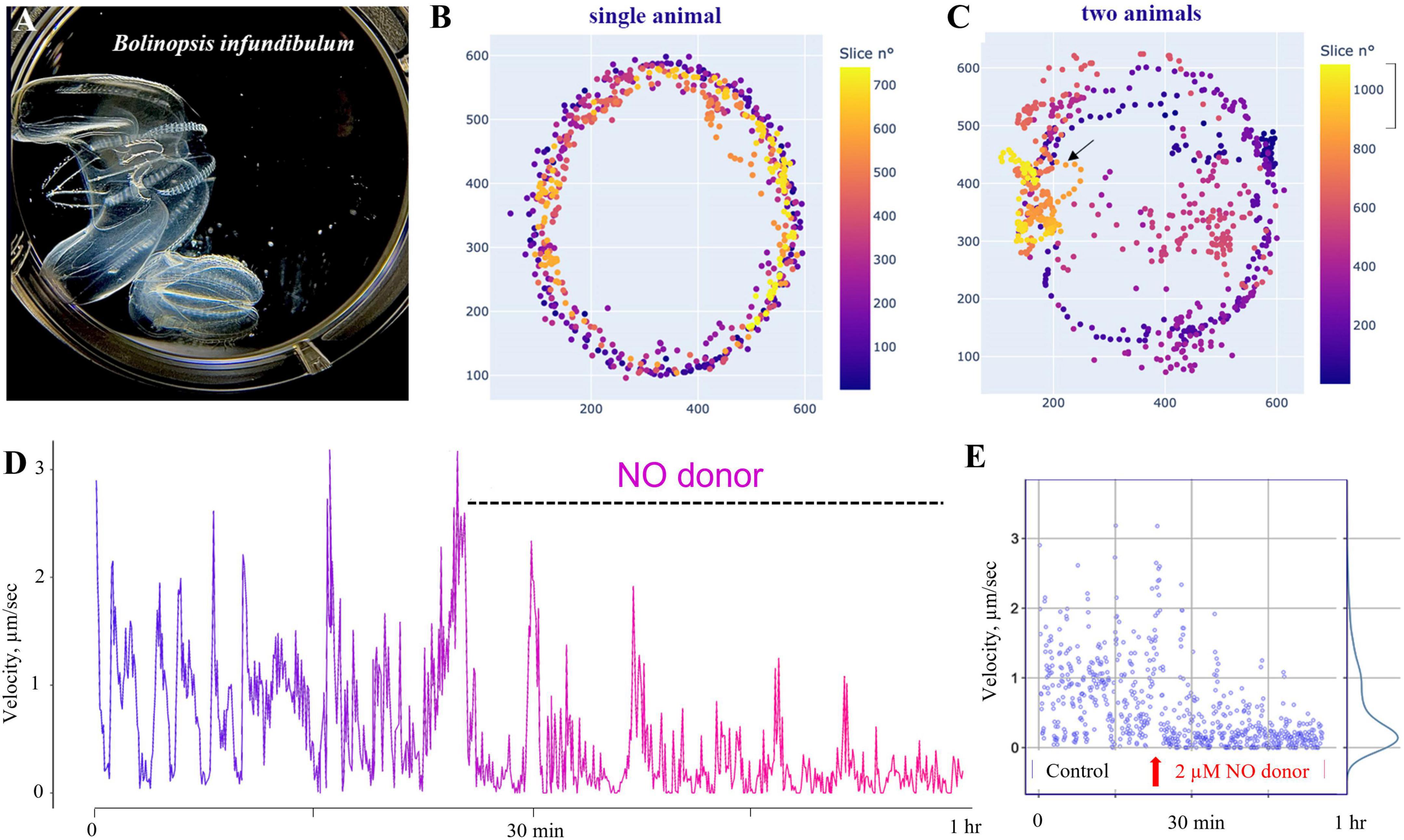
Figure 11. Nitric oxide suppresses swimming in Bolinopsis. (A) The photo of two B. influndibulum in an experimental Petri dish. (B,C) Velocity plots (color gradient at right) following the application of 2 μM of DEANO (NO donor). Illustrative tests on a single and two animals in the dish show a decrease of the speed of locomotion and travel distance following NO application. (D) Dynamic swimming shows oscillations in the velocity and gradual decay of locomotory speed in the presence of DEANO. (E) Velocity ranged from 0 to 3 in the control group, and these values gradually reduced to a nearly complete arrest of swimming in the experimental group of eight animals.
Specifically, we used Bolinopsis as a model for these initial tests. Applications of the NO donor DEA/NO 2–20 μM (n = 8 for each concentration) resulted in the suppression of locomotion within the first 10–20 min, including reduction of long-term oscillations of swimming patterns, and eventually the suppression of swimming usually within 60–90 min (n = 8). Higher concentrations of the NO donor (20–200 μM) were apparently toxic, leading to a rapid arrest of locomotion and dissociation of animals (n = 8). Of note, applying the same concentrations to Pleurobrachia and Beroe did not result in such immediate toxic effects.
This manuscript outlines the first survey of NOS distribution and functions in ctenophores. The presence of NOS and intracellular NO receptors such as sGC in ctenophores are supported by molecular evidence. However, both comparative and, especially, functional aspects of NO-mediated signaling remain to be determined, and numerous components of NO/cGMP-mediated signaling can be a particular ctenophore lineage-specific. Conclusions are summarized below.
(1) NOS is present in ctenophores with the events of secondary gene loss across several species, primarily in cydippids, for yet unknown reasons.
(2) 2–4 sGCs are also present in ctenophores, with taxonomically broader distribution than NOSs, including their presence in species without NOS. These observations suggest the occurrence of other NO synthetic pathways [enzymatic and non-enzymatic (Moroz et al., 1998; Moroz and Kohn, 2011) and environmental sources of NO (e.g., from microbiota and symbiotic organisms)]. Of note, sGC can also sense CO and potentially other molecules (Feelisch and Martin, 1995; Moroz and Kohn, 2011; Stuehr and Haque, 2019; Shepherd et al., 2022).
(3) In Mnemiopsis, NOS and sGCs are expressed at later stages during embryogenesis, and this pathway might be associated with tissue and organ specification in ctenophore development.
(4) Although NO/sGC showed a relatively low expression level in adults, NO might be involved in both localized and systemic control of locomotion with cilia as one of the potential targets of this signaling (to be experimentally validated).
We would like to stress the importance of studying the non-neuronal and systemic functions of NO in ctenophores. NADPH-d did not reveal labeling of defined neuronal populations in mesoglea or subepithelial neural nets or particular comb areas where neurons were identified in previous studies (Moroz, 2015; Norekian and Moroz, 2016, 2019a,b, 2020, 2021). Although cells at the base of combs or ciliated plates in the auricles were NADPH-d positive, they are unlikely neurons. Similarly, the labelings in the aboral organ (with the gravity sensor) and two other putative sensory structures (polar fields as well as papillae) do not convincingly occur in neurons.
These observations are consistent with our pilot findings about relatively small instant effects of NO on behaviors in two lobate ctenophores. In addition to Bolinopsis, we performed similar pilot pharmacological tests on Mnemiopsis using the same NO donor (DEA NO, 21 animals). Lower concentrations of NO donors (<1 μM, n = 7) had no noticeable effects; higher concentrations (>10–70 μM) moderately suppressed the ciliated locomotion and modulate muscle contractions.
Clearly, more systematic studies have to be performed using different NOS donors, inhibitors, and drugs affecting cGMP signaling using cilia, muscles, and secretory cells as effectors, to name a few. However, those observed and relatively “weak” effects of NO within short intervals of pharmacological testing (∼30–60 min), together with the widespread distribution of putative NOS across multiple non-neuronal cells and tissues, suggest that NO-signaling in ctenophores might have been associated with more systemic functions. For example, assuming that NADPH-d reactivity in Mnemiopsis correlates with NOS activity [as in many other preparations (Bredt et al., 1991; Kurenni et al., 1995; Moroz et al., 2000, 2004; Giovine et al., 2001; Bishop et al., 2008; Moroz and Kohn, 2011)] with such broad distribution across cells and tissues, we anticipate potential roles of ctenophore NO-mediated signaling in development, differentiation, morphogenic processes, (neuro)plasticity, immunity, etc. – similar to a plethora of diverse NO functions described in bilaterians, including the vertebrate lineage.
In summary, the studies of NO signaling in ctenophores are in their infancy. Importantly to the field, we know little about NO synthesis, regulation, compartmentalization, and evolution within the group. As critical first steps and future directions, understanding the enzymology of NOS is needed due to its unusual structure. It would be essential to determine the Ca-dependence of ctenophore NOSs, control their expression, and characterize the pharmacology of NOS inhibition, providing tools for future research. Equally important would be the characterization of NOS expression both in development and in adults using in situ hybridization and immunohistochemistry, in addition to current observations with NADPH-d histochemistry. Finally, comparative physiological analyses of NO signaling in regulating reproduction, development, feeding, immunity, and complex behavioral integration are desirable.
We performed database searches for nitric oxide synthases (NOS) and soluble guanylyl cyclases (sGC) both in animals and non-metazoan organisms. All used NOS and sGC sequences and their sources are presented in Supplementary Table 1.
Protein domains were detected using Pfam (Mistry et al., 2021), UniProt (UniProt Consortium, 2010), SMART (Letunic and Bork, 2018; Letunic et al., 2021), and NCBI Conserved Domain Search (Lu et al., 2020). The reconstruction of 3D structures was based on PDB models using Phyre2 (Kelley et al., 2015), SWISS-MODEL (Waterhouse et al., 2018), and AlphaFold2 (Jumper et al., 2021). Three-dimensional structures were analyzed in PyMol (The PyMol Molecular Graphic System, Version 1.8.6.0 Schrodinger, LLC)1,2.
NOS and sGC sequences were aligned with MAFFT v764 88, using the L-INS-i alignment algorithm with 1,000 iterations (Katoh et al., 2005). Phylogenomic ML analyses were performed by IQ-TREE ML ultrafast-bootstrap calculation (Trifinopoulos et al., 2016). The RAxML analyses of NOSs were made by a best-fit LG + F + I + G4 model, and for cyclases, we used a best-fit WAG + F + G4 model. Testing tree branches was done by SH-like aLRT with 1,000 replicates.
We screened publicly available transcriptomic (RNA-seq) datasets of 26 different development stages of Mnemiopsis (Levin et al., 2016) for NOS and sGC expression patterns. The original data were presented as TPM (log2(transcript per million (TPM)/100 + 1) and visualized using Pandas, matplotlib libraries in Python.
For analyses of cell-specific expression patters we used reference scRNA-seq data from adult Mnemiopsis (Sebé-Pedrós et al., 2018). Specifically, we visually inspected the distribution of genes, UMIs, and % mitochondrial genes across cells choosing a AnnData-file format with a final count matrix of 6,144 cells. We also fileted datasets and tested other criteria for removing cells, but we found that more stringent cutoffs yielded significantly different clustering results and eliminated low-expressed genes from the final visualization (including NOS and sGC-like genes). Standard guidelines for preprocessing, performing principal components analysis, normalization, and clustering were based on Satija et al. (2015).
We ranked differentially expressed genes in each cluster by Wilcoxon rank-sum (Mann–Whitney-U) test and t-test (Soneson and Robinson, 2018). As an alternative, we ranked genes using logistic regression (Ntranos et al., 2019). Initial annotation of clusters was based on previously suggested gene markers (Sebé-Pedrós et al., 2018). For visualization of gene expression and other variables, we used dotplots with a dendrogram implemented in Matplotlib, UMAP, and PAGA plottings.
Partition-based graph abstraction (PAGA) connectivity graphs were implemented in ScanPy, Python (Wolf et al., 2019) for mapping the expression for individual genes using Leiden algorithm as a base. Specifically, we employed PAGA to calculate connectivity between merged clusters, retaining all connections. For visualization purposes, we labeled strong edges thicker than others with the threshold 0.03. Finally, we placed the graph nodes on the median UMAP coordinates of the cells in the cluster to preserve the structure of the UMAP embedding and allow direct comparison between the PAGA graph and UMAP plots. To investigate expression changes along different paths in PAGA networks, we calculated diffusion maps and diffusion pseudotime as a proxy of the differentiation distance between all cells.
We used protocols described elsewhere in detail (Kurenni et al., 1995; Moroz, 2000a) for the visualization of NADPH-diaphorase histochemistry known as a marker for NOS activity (Bredt et al., 1991; Moroz, 2000a). Before staining, adult animals (2–5 cm) were fixed in 4% paraformaldehyde in seawater for 45–75 min at room temperature. The staining was also performed at room temperature, in the dark, for 2–5 h in the following solution: β-NADPH sodium salt (1 mM), and Nitro Blue Tetrazolium (0.5 mM) in 0.1 M Tris buffer (pH = 8.0). All reagents were from Sigma). Postfixation, embedding, and visualization were the same as described elsewhere (Moroz, 2000a; Giovine et al., 2001). Considering the highly fragile nature of ctenophore preparations, often dissociated during fixation, we processed different tissue segments separately, using care for all transfers of preparations. The data were obtained from NADPH-d labeling using six animals. The experiments were performed in the spring and summer of 2019–2021.
For behavioral assays, we used freshly collected Bolinopsis infundibulum. We used DEA NONOate (Diethylamine NONOate, Abcam, CAS Number 372965-00-9) as a nitric oxide donor. The time series for control behaviors and experimental tests were 30–60 min. We used Fiji/ImageJ and plugins (WRMTRCK, MTrack2) to track and calculate velocities, distances, and tracks. Post-analyses and visualization were done in Python environment (NumPy, Pandas, Matplotlib, SciPy).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
LM and DR designed the study. KM screened the genome datasets and 37 transcriptomes for the presence of NOS and cyclases. DR and KM was involved in the phylogenetic analyses. DR made 2D/3D protein modeling of NOS and cyclases, performed scRNA-seq and RNA-seq analyses, and involved in the behavioral analyses. LM performed the pharmacological and histochemical tests and wrote the manuscript. DR, KM, and LM confirmed the analyzed data. All authors reviewed and edited the manuscript.
This work was supported in part by the Human Frontiers Science Program (RGP0060/2017) and the National Science Foundation (1146575, 1557923, 1548121, and 1645219) grants to LM. Research reported in this publication was also supported in part by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number R01NS114491 (to LM).
We thank Drs. Arnau Sebe-Pedros and Amos Tanay for providing an original scRNA-seq dataset.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Institutes of Health and National Science Foundation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2023.1125433/full#supplementary-material
Supplementary Figure 1 | Diversification of soluble guanylyl cyclases (sGC) Ctenophora. The catalytic domain (∼190aa) of the soluble guanylate, adenylate, and membrane-type receptor (MR) guanylyl cyclase proteins was used to reconstruct the maximum-likelihood (ML) based phylogenetic tree. Ctenophore soluble cyclases are clustered within three major clades —1/2, 3/4, and NIT-like. Both Mnemiopsis leidyi (Ml) and Ocyropsis crystallina (Oc) encode five copies of GC proteins, Pleurobrachia bachei (Pb) encodes three orthologs comprising one gene product under each cluster. Pleurobrachia soluble NIT-like cyclase gene encodes two copies of the catalytic domain shown as a and b. Mnemiopsis and Pleurobrachia genes are shown in bold font in dark and light blue color. Based on the ML tree membrane receptor type (ANP-like receptor), guanylyl cyclase can be classified under three groups. Both Pleurobrachia bachei (Pb) and Mnemiopsis leidyi (Ml) encode one gene under these three groups. Similarly, both Pb and Ml encode two genes of the adenylate cyclase family. However, both these species encode two copies of the catalytic domains labeled as a and b. Proteins under each group against all other ctenophore species were retrieved from the transcriptome datasets (Whelan et al., 2015, 2017). The catalytic domain used to build the tree is given in the Supplementary Table 1. Species names are shown in the figure as well as provided in the Supplementary material.
Supplementary Figure 2 | Identifying cluster specific genes associated with NO-cGMP signaling in Mnemiopsis leidyi based on scRNA-seq data (Sebé-Pedrós et al., 2018) with filtration > 100 genes per cell. This filtration cutoff resulted produced 41 clusters (filtration n = 100, 5,461 cells). Without filtration, we produced 24 clusters with 6,144 cells (the main text and figures), and recognized digestive, epithelial, muscle, neuroid-type cells. However, here with the threshold N = 100, we lost cells with such low-expressed genes as NOS in tentacles and many cells expressing cyclase genes (see main text).
Supplementary Figure 3 | RNA-seq expression profiling of NOS and different cyclases in Mnemiopsis development the calculated values are based on Levin et al. (2016), Moreland et al. (2020). Post-fertilization hours are indicated as color dots on the right.
Supplementary Table 1 | Sequences of NOS and cyclases used for tree constructions and analyses.
Supplementary Table 2 | Sequences of different cyclases identified in comparative ctenophore RNA-seq datasets.
Supplementary Material 1 | Alignment of NOSs identified in ctenophores from RNA-seq datasets. All structural domains are marked.
Supplementary Material 2 | Alignment of NOSs identified in ctenophores from RNA-seq datasets. All structural domains are marked.
Anctil, M. (2009). Chemical transmission in the sea anemone Nematostella vectensis: A genomic perspective. Comp. Biochem. Physiol. Part D Genomics Proteomics 4, 268–289. doi: 10.1016/j.cbd.2009.07.001
Bassler, J., Schultz, J. E., and Lupas, A. N. (2018). Adenylate cyclases: Receivers, transducers, and generators of signals. Cell Signal. 46, 135–144. doi: 10.1016/j.cellsig.2018.03.002
Bishop, C., Pires, A., Norby, S., Boudko, D., Moroz, L. L., and Hadfield, M. (2008). Analysis of nitric oxide-cyclic guanosine monophosphate signaling during metamorphosis of the nudibranch Phestilla sibogae Bergh (Gastropoda: Opisthobranchia). Evol. Dev. 10, 288–299. doi: 10.1111/j.1525-142X.2008.00238.x
Bouchard, J. N., and Yamasaki, H. (2008). Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: A possible linkage between nitric oxide and the coral bleaching phenomenon. Plant Cell Physiol. 49, 641–652. doi: 10.1093/pcp/pcn037
Bredt, D., Glatt, C., Hwang, P., Fotuhi, M., Dawson, T., and Snyder, S. (1991). Nitric oxide synthase protein and mRNA are discretely localized in neuronal populations of the mammalian CNS together with NADPH diaphorase. Neuron 7, 615–624. doi: 10.1016/0896-6273(91)90374-9
Brenman, J., Chao, D., Gee, S., McGee, A., Craven, S., Santillano, D., et al. (1996). Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 84, 757–767. doi: 10.1016/s0092-8674(00)81053-3
Burkhardt, P., and Jékely, G. (2021). Evolution of synapses and neurotransmitter systems: The divide-and-conquer model for early neural cell-type evolution. Curr. Opin. Neurobiol. 71, 127–138. doi: 10.1016/j.conb.2021.11.002
Camargo, A., Llamas, A., Schnell, R., Higuera, J., González-Ballester, D., Lefebvre, P., et al. (2007). Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell. 19, 3491–3503. doi: 10.1105/tpc.106.045922
Colasanti, M., Lauro, G. M., and Venturini, G. (1995). NO in Hydra feeding response. Nature 374:505. doi: 10.1038/374505a0
Colasanti, M., Mazzone, V., Mancinelli, L., Leone, S., and Venturini, G. (2009). Involvement of nitric oxide in the head regeneration of Hydra vulgaris. Nitric Oxide 21, 164–170. doi: 10.1016/j.niox.2009.07.003
Colasanti, M., Persichini, T., and Venturini, G. (2010). Nitric oxide pathway in lower metazoans. Nitric Oxide 23, 94–100. doi: 10.1016/j.niox.2010.05.286
Cristino, L., Guglielmotti, V., Cotugno, A., Musio, C., and Santillo, S. (2008). Nitric oxide signaling pathways at neural level in invertebrates: Functional implications in cnidarians. Brain Res. 1225, 17–25. doi: 10.1016/j.brainres.2008.04.056
Elliott, G. R., and Leys, S. P. (2010). Evidence for glutamate, GABA and NO in coordinating behaviour in the sponge, Ephydatia muelleri (Demospongiae, Spongillidae). J. Exp. Biol. 213, 2310–2321. doi: 10.1242/jeb.039859
Ellwanger, K., and Nickel, M. (2006). Neuroactive substances specifically modulate rhythmic body contractions in the nerveless metazoon Tethya wilhelma (Demospongiae, Porifera). Front. Zool. 3:7. doi: 10.1186/1742-9994-3-7
Feelisch, M., and Martin, J. F. (1995). The early role of nitric oxide in evolution. Trends Ecol. Evol. 10, 496–499. doi: 10.1016/s0169-5347(00)89206-x
Feng, W., and Zhang, M. (2009). Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci. 10, 87–99. doi: 10.1038/nrn2540
Fernandez, R., and Gabaldon, T. (2020). Gene gain and loss across the metazoan tree of life. Nat. Ecol. Evol. 4, 524–533. doi: 10.1038/s41559-019-1069-x
Fischer, A. H., Pang, K., Henry, J. Q., and Martindale, M. Q. (2014). A cleavage clock regulates features of lineage-specific differentiation in the development of a basal branching metazoan, the ctenophore Mnemiopsis leidyi. Evodevo 5:4. doi: 10.1186/2041-9139-5-4
Gancedo, J. (2013). Biological roles of cAMP: Variations on a theme in the different kingdoms of life. Biol. Rev. Camb. Philos. Soc. 88, 645–668. doi: 10.1111/brv.12020
Giacomelli, M., Rossi, M. E., Lozano-Fernandez, J., Feuda, R., and Pisani, D. (2022). Resolving tricky nodes in the tree of life through amino acid recoding. iScience 25:105594. doi: 10.1016/j.isci.2022.105594
Giovine, M., Pozzolini, M., Favre, A., Bavestrello, G., Cerrano, C., Ottaviani, F., et al. (2001). Heat stress-activated, calcium-dependent nitric oxide synthase in sponges. Nitric Oxide 5, 427–431. doi: 10.1006/niox.2001.0366
Gunn, A., Derbyshire, E. R., Marletta, M. A., and Britt, R. D. (2012). Conformationally distinct five-coordinate heme-NO complexes of soluble guanylate cyclase elucidated by multifrequency electron paramagnetic resonance (EPR). Biochemistry 51, 8384–8390. doi: 10.1021/bi300831m
Halanych, K. M., Whelan, N. V., Kocot, K. M., Kohn, A. B., and Moroz, L. L. (2016). Miscues misplace sponges. Proc. Natl. Acad. Sci. U.S.A. 113, E946–E947. doi: 10.1073/pnas.1525332113
Hawkins, T. D., and Davy, S. K. (2013). Nitric oxide and coral bleaching: Is peroxynitrite generation required for symbiosis collapse? J. Exp. Biol. 216, 3185–3188. doi: 10.1242/jeb.087510
Hawkins, T. D., Bradley, B. J., and Davy, S. K. (2013). Nitric oxide mediates coral bleaching through an apoptotic-like cell death pathway: Evidence from a model sea anemone-dinoflagellate symbiosis. FASEB J. 27, 4790–4798. doi: 10.1096/fj.13-235051
Hewitt, O., and Degnan, S. (2022). Distribution and diversity of ROS-generating enzymes across the animal kingdom, with a focus on sponges (Porifera). BMC Biol. 20:212. doi: 10.1186/s12915-022-01414-z
Holguín-Peña, R., Vázquez-Juárez, R., and Martínez-Soriano, J. (2007). First report of a 16SrI-B group phytoplasma associated with a yellows-type disease affecting tomato plants in the Baja California Peninsula of Mexico. Plant Dis. 91:328. doi: 10.1094/PDIS-91-3-0328B
Horst, B. G., and Marletta, M. A. (2018). Physiological activation and deactivation of soluble guanylate cyclase. Nitric Oxide 77, 65–74. doi: 10.1016/j.niox.2018.04.011
Horst, B., Yokom, A., Rosenberg, D., Morris, K., Hammel, M., Hurley, J., et al. (2019). Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy. Elife. 8:e50634. doi: 10.7554/eLife.50634
Jaffrey, S. R., Snowman, A. M., Eliasson, M. J., Cohen, N. A., and Snyder, S. H. (1998). CAPON: A protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron 20, 115–124. doi: 10.1016/s0896-6273(00)80439-0
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kang, Y., Liu, R., Wu, J. X., and Chen, L. (2019). Structural insights into the mechanism of human soluble guanylate cyclase. Nature 574, 206–210. doi: 10.1038/s41586-019-1584-6
Kapli, P., and Telford, M. J. (2020). Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci. Adv. 6:eabc5162. doi: 10.1126/sciadv.abc5162
Kass-Simon, G., and Pierobon, P. (2007). Cnidarian chemical neurotransmission, an updated overview. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 146, 9–25. doi: 10.1016/j.cbpa.2006.09.008
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kim, E., and Sheng, M. (2004). PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5, 771–781. doi: 10.1038/nrn1517
Krumenacker, J. S., Hanafy, K. A., and Murad, F. (2004). Regulation of nitric oxide and soluble guanylyl cyclase. Brain Res. Bull. 62, 505–515. doi: 10.1016/S0361-9230(03)00102-3
Kurenni, D., Thurlow, G., Turner, R., Moroz, L., Sharkey, K., and Barnes, S. (1995). Nitric oxide synthase in tiger salamander retina. J. Comp. Neurol. 361, 525–536. doi: 10.1002/cne.903610314
Laumer, C., Fernández, R., Lemer, S., Combosch, D., Kocot, K., Riesgo, A., et al. (2019). Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. Biol. Sci. 286:20190831. doi: 10.1098/rspb.2019.0831
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Levin, M., Anavy, L., Cole, A., Winter, E., Mostov, N., Khair, S., et al. (2016). The mid-developmental transition and the evolution of animal body plans. Nature 531, 637–641. doi: 10.1038/nature16994
Li, Y., Shen, X. X., Evans, B., Dunn, C. W., and Rokas, A. (2021). Rooting the animal tree of life. Mol. Biol. Evol. 38, 4322–4333. doi: 10.1093/molbev/msab170
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M., Geer, R., Gonzales, N., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Mariita, R. M., Hossain, M. J., and Moss, A. G. (2021). Comparative genomic analysis and characterization of Staphylococcus sp. AOAB, isolated from a notoriously invasive Mnemiopsis leidyi gut revealed multiple antibiotic resistance determinants. bioRxiv [Preprint]. doi: 10.1101/2021.01.11.426108
Martin, E., Berka, V., Tsai, A. L., and Murad, F. (2005). Soluble guanylyl cyclase: The nitric oxide receptor. Methods Enzymol. 396, 478–492. doi: 10.1016/S0076-6879(05)96040-0
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G., Sonnhammer, E., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Montfort, W. R., Wales, J. A., and Weichsel, A. (2017). Structure and activation of soluble guanylyl cyclase, the nitric oxide sensor. Antioxid. Redox Signal. 26, 107–121. doi: 10.1089/ars.2016.6693
Moreland, R. T., Nguyen, A. D., Ryan, J. F., and Baxevanis, A. D. (2020). The Mnemiopsis genome project portal: Integrating new gene expression resources and improving data visualization. Database 2020:baaa029. doi: 10.1093/database/baaa029
Moroz, L. (2015). Convergent evolution of neural systems in ctenophores. J. Exp. Biol. 218(Pt 4), 598–611. doi: 10.1242/jeb.110692
Moroz, L. L. (2000a). Giant identified NO-releasing neurons and comparative histochemistry of putative nitrergic systems in gastropod molluscs. Microsc. Res. Technol. 49, 557–569. doi: 10.1002/1097-0029(20000615)49:6<557::AID-JEMT6>3.0.CO;2-S
Moroz, L. L. (2000b). “On the origin and early evolution of neuronal nitric oxide signaling: A comparative analysis,” in Nitric oxide and free radicals in peripheral neurotransmission, ed. S. B. Kalsner (Basel: Birkhauser), 1–34.
Moroz, L. L., and Kohn, A. B. (2011). Parallel evolution of nitric oxide signaling: Diversity of synthesis and memory pathways. Front. Biosci. 16:2008–2051. doi: 10.2741/3837
Moroz, L. L., and Kohn, A. B. (2016). Independent origins of neurons and synapses: Insights from ctenophores. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150041. doi: 10.1098/rstb.2015.0041
Moroz, L. L., Meech, R. W., Sweedler, J. V., and Mackie, G. O. (2004). Nitric oxide regulates swimming in the jellyfish Aglantha digitale. J. Comp. Neurol. 471, 26–36. doi: 10.1002/cne.20023
Moroz, L. L., Norekian, T. P., Pirtle, T. J., Robertson, K. J., and Satterlie, R. A. (2000). Distribution of NADPH-diaphorase reactivity and effects of nitric oxide on feeding and locomotory circuitry in the pteropod mollusc, Clione limacina. J. Comp. Neurol. 427, 274–284.
Moroz, L. L., Romanova, D. Y., and Kohn, A. B. (2021). Neural versus alternative integrative systems: Molecular insights into origins of neurotransmitters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376:20190762. doi: 10.1098/rstb.2019.0762
Moroz, L., Kocot, K., Citarella, M., Dosung, S., Norekian, T., Povolotskaya, I., et al. (2014). The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. doi: 10.1038/nature13400
Moroz, L., Norby, S., Cruz, L., Sweedler, J., Gillette, R., and Clarkson, R. (1998). Non-enzymatic production of nitric oxide (NO) from NO synthase inhibitors. Biochem. Biophys. Res. Commun. 253, 571–576. doi: 10.1006/bbrc.1998.9810
Moroz, L., Romanova, D., Nikitin, M., Sohn, D., Kohn, A., Neveu, E., et al. (2020). The diversification and lineage-specific expansion of nitric oxide signaling in Placozoa: Insights in the evolution of gaseous transmission. Sci. Rep. 10:13020. doi: 10.1038/s41598-020-69851-w
Morrall, C. E., Galloway, T. S., Trapido-Rosenthal, H. G., and Depledge, M. H. (2000). Characterisation of nitric oxide synthase activity in the tropical sea anemone Aiptasia pallida. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 125, 483–491. doi: 10.1016/s0305-0491(00)00157-7
Murciano-Calles, J., Coello, A., Cámara-Artigas, A., and Martinez, J. (2020). PDZ/PDZ interaction between PSD-95 and nNOS neuronal proteins: A thermodynamic analysis of the PSD95-PDZ2/nNOS-PDZ interaction. J. Mol. Recognit. 33:e2826. doi: 10.1002/jmr.2826
Musser, J., Schippers, K., Nickel, M., Mizzon, G., Kohn, A., Pape, C., et al. (2021). Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science 374, 717–723. doi: 10.1126/science.abj2949
Norekian, T. P., and Moroz, L. L. (2016). Development of neuromuscular organization in the ctenophore Pleurobrachia bachei. J. Comp. Neurol. 524, 136–151. doi: 10.1002/cne.23830
Norekian, T. P., and Moroz, L. L. (2019a). Neuromuscular organization of the ctenophore Pleurobrachia bachei. J. Comp. Neurol. 527, 406–436. doi: 10.1002/cne.24546
Norekian, T. P., and Moroz, L. L. (2019b). Neural system and receptor diversity in the ctenophore Beroe abyssicola. J. Comp. Neurol. 527, 1986–2008. doi: 10.1002/cne.24633
Norekian, T. P., and Moroz, L. L. (2020). Comparative neuroanatomy of ctenophores: Neural and muscular systems in Euplokamis dunlapae and related species. J. Comp. Neurol. 528, 481–501. doi: 10.1002/cne.24770
Norekian, T. P., and Moroz, L. L. (2021). Development of the nervous system in the early hatching larvae of the ctenophore Mnemiopsis leidyi. J. Morphol. 282, 1466–1477. doi: 10.1002/jmor.21398
Ntranos, V., Yi, L., Melsted, P., and Pachter, L. (2019). A discriminative learning approach to differential expression analysis for single-cell RNA-seq. Nat. Methods 16, 163–166. doi: 10.1038/s41592-018-0303-9
Oulavallickal, T., Brewster, J., McKellar, J., Fairhurst, M., Tenci, N., and Gerth, M. (2017). The Pseudomonas syringae pv. actinidiae chemoreceptor protein F (PscF) periplasmic sensor domain: Cloning, purification and X-ray crystallographic analysis. Acta Crystallogr. F Struct. Biol. Commun. 73(Pt 12), 701–705. doi: 10.1107/S2053230X17016831
Perez, S., and Weis, V. (2006). Nitric oxide and cnidarian bleaching: An eviction notice mediates breakdown of a symbiosis. J. Exp. Biol. 209(Pt 14), 2804–2810. doi: 10.1242/jeb.02309
Redmond, A. K., and McLysaght, A. (2021). Evidence for sponges as sister to all other animals from partitioned phylogenomics with mixture models and recoding. Nat. Commun. 12:1783. doi: 10.1038/s41467-021-22074-7
Reitzel, A. M., Pang, K., and Martindale, M. Q. (2016). Developmental expression of “germline”- and “sex determination”-related genes in the ctenophore Mnemiopsis leidyi. Evodevo 7:17. doi: 10.1186/s13227-016-0051-9
Reyes-Rivera, J., Wu, Y., Guthrie, B., Marletta, M., King, N., and Brunet, T. (2022). Nitric oxide signaling controls collective contractions in a colonial choanoflagellate. Curr. Biol. 32, 2539–2547.e5. doi: 10.1016/j.cub.2022.04.017
Roman, L. J., and Masters, B. S. (2006). Electron transfer by neuronal nitric-oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements. J. Biol. Chem. 281, 23111–23118. doi: 10.1074/jbc.M603671200
Safavi-Hemami, H., Young, N. D., Doyle, J., Llewellyn, L., and Klueter, A. (2010). Characterisation of nitric oxide synthase in three cnidarian-dinoflagellate symbioses. PLoS One 5:e10379. doi: 10.1371/journal.pone.0010379
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F., and Regev, A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. doi: 10.1038/nbt.3192
Schultz, D., Francis, W., McBroome, J., Christianson, L., Haddock, S., and Green, R. (2021). A chromosome-scale genome assembly and karyotype of the ctenophore Hormiphora californensis. G3 11:jkab302. doi: 10.1093/g3journal/jkab302
Sebé-Pedrós, A., Chomsky, E., Pang, K., Lara-Astiaso, D., Gaiti, F., Mukamel, Z., et al. (2018). Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2, 1176–1188. doi: 10.1038/s41559-018-0575-6
Sheng, N., Bemben, M., Díaz-Alonso, J., Tao, W., Shi, Y., and Nicoll, R. (2018). LTP requires postsynaptic PDZ-domain interactions with glutamate receptor/auxiliary protein complexes. Proc. Natl. Acad. Sci. U.S.A. 115, 3948–3953. doi: 10.1073/pnas.1800719115
Shepherd, M., Giordano, D., Verde, C., and Poole, R. K. (2022). The evolution of nitric oxide function: From reactivity in the prebiotic earth to examples of biological roles and therapeutic applications. Antioxidants 11:1222. doi: 10.3390/antiox11071222
Shu, C. J., Ulrich, L. E., and Zhulin, I. B. (2003). The NIT domain: A predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 28, 121–124. doi: 10.1016/S0968-0004(03)00032-X
Simmons, D. K., Pang, K., and Martindale, M. Q. (2012). Lim homeobox genes in the ctenophore Mnemiopsis leidyi: The evolution of neural cell type specification. Evodevo 3:2. doi: 10.1186/2041-9139-3-2
Soneson, C., and Robinson, M. D. (2018). Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 15, 255–261. doi: 10.1038/nmeth.4612
Stricker, N., Christopherson, K., Yi, B., Schatz, P., Raab, R., Dawes, G., et al. (1997). PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat. Biotechnol. 15, 336–342. doi: 10.1038/nbt0497-336
Stuehr, D. J., and Haque, M. M. (2019). Nitric oxide synthase enzymology in the 20 years after the nobel prize. Br. J. Pharmacol. 176, 177–188. doi: 10.1111/bph.14533
Telford, M. J., Moroz, L. L., and Halanych, K. M. (2016). Evolution: A sisterly dispute. Nature 529, 286–287. doi: 10.1038/529286a
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Ueda, N., Richards, G., Degnan, B., Kranz, A., Adamska, M., Croll, R., et al. (2016). An ancient role for nitric oxide in regulating the animal pelagobenthic life cycle: Evidence from a marine sponge. Sci. Rep. 6:37546. doi: 10.1038/srep37546
UniProt Consortium (2010). The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148. doi: 10.1093/nar/gkp846
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Whelan, N. V., Kocot, K. M., Moroz, L. L., and Halanych, K. M. (2015). Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5773–5778. doi: 10.1073/pnas.1503453112
Whelan, N., Kocot, K., Moroz, T., Mukherjee, K., Williams, P., Paulay, G., et al. (2017). Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746. doi: 10.1038/s41559-017-0331-3
Windsor, L., and Thompson, S. (2008). Educating homeless youth in Texas: The McKinney-Vento homeless education assistance improvements act of 2001. Educ. Real. 33, 123–130.
Keywords: Mnemiopsis, Ctenophora, nitric oxide synthase, guanylate cyclase, Pleurobrachia, nervous system evolution, Porifera, Placozoa
Citation: Moroz LL, Mukherjee K and Romanova DY (2023) Nitric oxide signaling in ctenophores. Front. Neurosci. 17:1125433. doi: 10.3389/fnins.2023.1125433
Received: 16 December 2022; Accepted: 27 February 2023;
Published: 22 March 2023.
Edited by:
James Newcomb, New England College, United StatesReviewed by:
Thibaut Brunet, Pasteur Institute, FranceCopyright © 2023 Moroz, Mukherjee and Romanova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonid L. Moroz, bW9yb3pAd2hpdG5leS51ZmwuZWR1; orcid.org/0000-0002-1333-3176
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.