
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 05 August 2022
Sec. Neurogenomics
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.967768
This article is part of the Research Topic Modulation of Neural Circuits and Plasticity Underlying Opioid Analgesia, Tolerance, and Addiction View all 9 articles
Morphine tolerance (MT) is a tricky problem, the mechanism of it is currently unknown. Circular RNAs (circRNAs) serve significant functions in the biological processes (BPs) of the central nervous system. N6-methyladenosine (m6A), as a key post-transcriptional modification of RNA, can regulate the metabolism and functions of circRNAs. Here we explore the patterns of m6A-methylation of circRNAs in the spinal cord of morphine-tolerant rats. In brief, we constructed a morphine-tolerant rat model, performed m6A epitranscriptomic microarray using RNA samples collected from the spinal cords of morphine-tolerant rats and normal saline rats, and implemented the bioinformatics analysis. In the spinal cord of morphine-tolerant rats, 120 circRNAs with different m6A modifications were identified, 54 of which were hypermethylated and 66 of which were hypomethylated. Functional analysis of these m6A circRNAs found some important pathways involved in the pathogenesis of MT, such as the calcium signaling pathway. In the m6A circRNA-miRNA networks, several critical miRNAs that participated in the occurrence and development of MT were discovered to bind to these m6A circRNAs, such as miR-873a-5p, miR-103-1-5p, miR-107-5p. M6A modification of circRNAs may be involved in the pathogenesis of MT. These findings may lead to new insights into the epigenetic etiology and pathology of MT.
Morphine, a traditional opioid analgesic, plays an important role in managing acute and chronic pain (Wiffen et al., 2016). However, chronic morphine administration often leads to the reduction of analgesic efficacy which is known as morphine tolerance (MT). MT is still a difficult issue; and it often leads to the withdrawal of treatment and many side effects, including physical dependence, constipation, vomiting, respiratory depression, and sometimes even death due to dose escalation (Paul et al., 2021). Multiple dysfunctions of biological processes (BPs) involved in the development of MT have been proposed (Chen and Sommer, 2009; Uniyal et al., 2020; Gledhill and Babey, 2021), including the opioid receptor desensitization, downregulation, internalization (Williams et al., 2013), and altered epigenetic regulation (Liang et al., 2013; Viet et al., 2017). Despite significant headway in this field, the mechanisms underlying the development of MT were not well understood.
The recent focus on the mechanism of many diseases is the role of non-coding RNAs (ncRNAs). Our previous studies reported the shreds of evidence of ncRNAs, including microRNAs (miRNAs) (Huang et al., 2019b), long non-coding RNAs (lncRNAs) (Shao et al., 2018), and circRNAs (Weng et al., 2019) in the regulation of gene expression in the spinal cord of morphine-tolerant rats. The role of miR-873a-5p (Huang et al., 2019a), and lncRNA MRAK159688 (Deng et al., 2022) have already been confirmed. CircRNAs are widely expressed in the central nervous system (Rybak-Wolf et al., 2015), they are stable and highly conserved (Jeck et al., 2013). They can regulate transcription and translation by sponge miRNAs or interacting with proteins. With these characteristics, circRNAs hold significant functions as diagnostic biomarkers and therapeutic targets for diseases, including MT (Lu et al., 2019; Mehta et al., 2020). We had previously built circRNA expression profiles in the spinal cord of morphine-tolerant rats (Weng et al., 2019). However, little is known about the upstream regulatory mechanism of circRNAs modification in MT.
M6A is one of the most common mRNA epitranscriptomic modifications (Niu et al., 2013; Meng et al., 2019), it refers to the N6 position of the adenine base is modified by a methyl group. In addition to mRNA, m6A modification also occurs in circRNAs (Ma et al., 2019; Wang et al., 2021; Wu et al., 2021). There are three factors regulating the m6A methylation: “writers,” “erasers,” and “readers.” The roles of these factors are as the name suggests (Zaccara et al., 2019). “Writers” are methyltransferases, including methyltransferase-like 3 and 14 proteins (METTL3 and METTL14) and their cofactors WT1 associated protein (WTAP), etc. “Readers” are RNA binding proteins that can specifically read m6A methylation, YTH domain-containing proteins (YTHDC1, YTHDC2, YTHDF, etc.) were identified as potential m6A-binding proteins; “Erasers” are demethylases, including fat mass and obesity-associated protein (FTO) and AlkB homologue 5 (ALKBH5). The m6A modification can affect many aspects of post-transcriptional processes of RNA, including splicing, translational efficiency, nuclear exporting, and RNA structure (Ma et al., 2019). The m6A modification in circRNAs displayed cell-type-specific methylation patterns, making m6A circRNAs more valuable in diagnosing and treating diseases (Zhou et al., 2017).
The epigenetic role of m6A circRNA in the pathogenesis of MT has not been reported. Here we hypothesized that m6A circRNAs might be associated with MT. We built an epitranscriptomic map using microarray analysis and explored the potential functions of m6A circRNAs in the spinal cord of morphine-tolerant rats, and the results might provide novel clues for epigenetic etiology and pathogenesis of MT.
Adult Sprague-Dawley (SD) male rats weighing 240–260 g were obtained from the Hunan SJA Laboratory Animal Company (Hunan, China). The animals were housed in specific pathogen-free and individually ventilated cages in a room with a 12 / 12 light-dark cycle and a temperature of 22 ± 1°C. We used the same procedure of our previous research to establish a rat model of MT (Weng et al., 2019). Briefly, the adult male SD rats were randomly divided into the normal saline group (NS group) and MT group. Rats in the MT group were administrated morphine hydrochloride injection of 10 μg / 10 μl intrathecally through the tube inserted into the sheath at 8:00 a.m. and 8:00 p.m. for 7 consecutive days, the NS group was given 10 μl of normal saline at the same time points. The tail-flick test was carried out under the condition that the water temperature allowed the basal latency of 4–6 s, and the cutoff time was set to 10 s to minimize tissue damage. The tail-flick time was converted into the percentage of maximal possible antinociceptive effect (%MPE). All animal experimental procedures were approved by the Ethics Committee for Animal Experimentation of Central South University and followed the Chinese National Institutes of Health Guide for the Care and Use of Laboratory Animals.
On the morning of the 8th day, 1 h after morphine or normal saline injection, rats were fully anesthetized with 1% pentobarbital (50mg/kg, i.p.) and sacrificed. The lumbar enlargements of spinal cords were surgically collected and were refrigerated at –80°C until used. The total RNA of lumbar enlargements was extracted following the standard protocol of TRI Reagent (Sigma, T9424), and the RNA quantity and quality were tested through NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, the United States). RNA integrity and gDNA contamination were tested by denaturing agarose gel electrophoresis.
The mRNA expression of m6A-related enzymes, including METTL3, METTL14, WTAP, FTO, ALKBH5, YTHDC2, YTHDF1, and YTHDF2 was detected separately by qRT-PCR to investigate the m6A methylation status in the spinal cord of morphine-tolerant rats. In brief, the extracted total RNAs were reverse transcribed to cDNA using Prime Script RT Reagent Kit (Takara, China) according to the manufacturer’s instructions. The primer sequences for m6A-related enzymes are shown in Supplementary Table 1. Real-time PCR reaction using PCR master mix (Arraystar, AS-MR-006-5) was performed according to the manufacturer’s manual in the ViiA7 Real-Time PCR Detection System (Applied Biosystems, the United States).
The sample preparation and microarray hybridization were performed based on Arraystar’s standard protocols. Briefly, the total RNAs were immunoprecipitated with the anti-m6A antibody (Synaptic Systems, 202003). The modified RNAs were eluted from the immunoprecipitated magnetic beads as the “IP.” The RNAs contained in the supernatant were unmodified as the “SUP.” The “IP” and “SUP” RNAs were treated with RNase R (Epicenter, RNR07250) separately to enrich circRNAs, and then labeled with Cy5 and Cy3, respectively, as cRNAs using Arraystar Super RNA Labeling Kit (Arraystar, AL-SE-005). The labeled cRNAs were combined and hybridized to Arraystar Rat CircRNA Epitranscriptomic Arrays (8x15K, Arraystar) at 65°C for 17 h in an Agilent Hybridization Oven. After washing the arrays were scanned in two-color channels by an Agilent Scanner G2505C (Agilent, the United States). The raw RNA intensities of IP (Cy5-labeled) and SUP (Cy3-labeled) were analyzed by Agilent Feature Extraction software (version 11.0.1.1) and normalized with an average of log2-scaled spike-in RNA intensities.
The normalized IP intensities represent the “m6A quantity” of the transcript comparable across samples. The m6A methylation levels were calculated as the percentage of modification (IP normalized intensities) of the input (IP and SUP normalized intensities). We also calculated the expression levels of circRNAs based on the normalized IP and SUP intensities. We used the criteria (fold change ≥ 1.5 and P < 0.05) to screen the differentially m6A-methylated or expressed circRNAs between the two groups.
To validate the microarray data, the IP and SUP RNAs from the eight spinal cords were separately reverse transcribed and performed qRT-PCR. The primers of the circRNAs are shown in Supplementary Table 1. The fraction of IP in the input was calculated using the following formula as %MERIP/Input = .
Hierarchical clustering was performed by Cluster 3.0 to show the distinguishable m6A-methylation pattern among samples, and the heat maps were generated in Java Treeview. The host genes of the differentially deregulated m6A-circRNAs were analyzed according to the pathway annotations of the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) functional enrichment using the DAVID 6.8.1 The correlation and the cumulative analysis between methylation and the expression level were performed in R v4.1.0 software. The upset graph showing the distribution patterns of circRNAs in terms of m6A quantity, m6A methylation levels, and expression was created using online tools.2 Potentially targeted miRNAs by circRNAs were predicted with Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda. The predicted circRNA/miRNA interaction networks were generated to visualize the interactions by Cytoscape (version 3.8.0).
For behavior data, two-way ANOVA followed by Bonferroni’s post-hoc test was applied to assess the differences over time. For qRT-PCR statistical analyses, a two-tailed unpaired Student’s t-test was used for two-group comparisons in GraphPad Prism 9.0 software (La Jolla, CA). P < 0.05 was considered statistically significant.
After repeated intrathecal administration of morphine for seven consecutive days, the %MPE dropped from 100% to almost 0%, and the morphine-tolerant rat model was constructed successfully (Figure 1A). In the spinal cord of morphine-tolerant rats, the mRNA expressions of m6A “eraser” (FTO and ALKBH5), m6A “readers” (YTHDC2, YTHDF1, and YTHDF2) (Figure 1B), and m6A “writers” (METTL3, METTL14, and WTAP) (Figure 1C) were tested separately, we found that the expressions of METTL3, WTAP, ALKBH5, and YTHDF1 were downregulated in the MT group compared with the NS group. These dysregulations of m6A “writers,” m6A “eraser,” and m6A “readers” may lead to a dynamic change of m6A methylation in the spinal cord of morphine-tolerant rats.
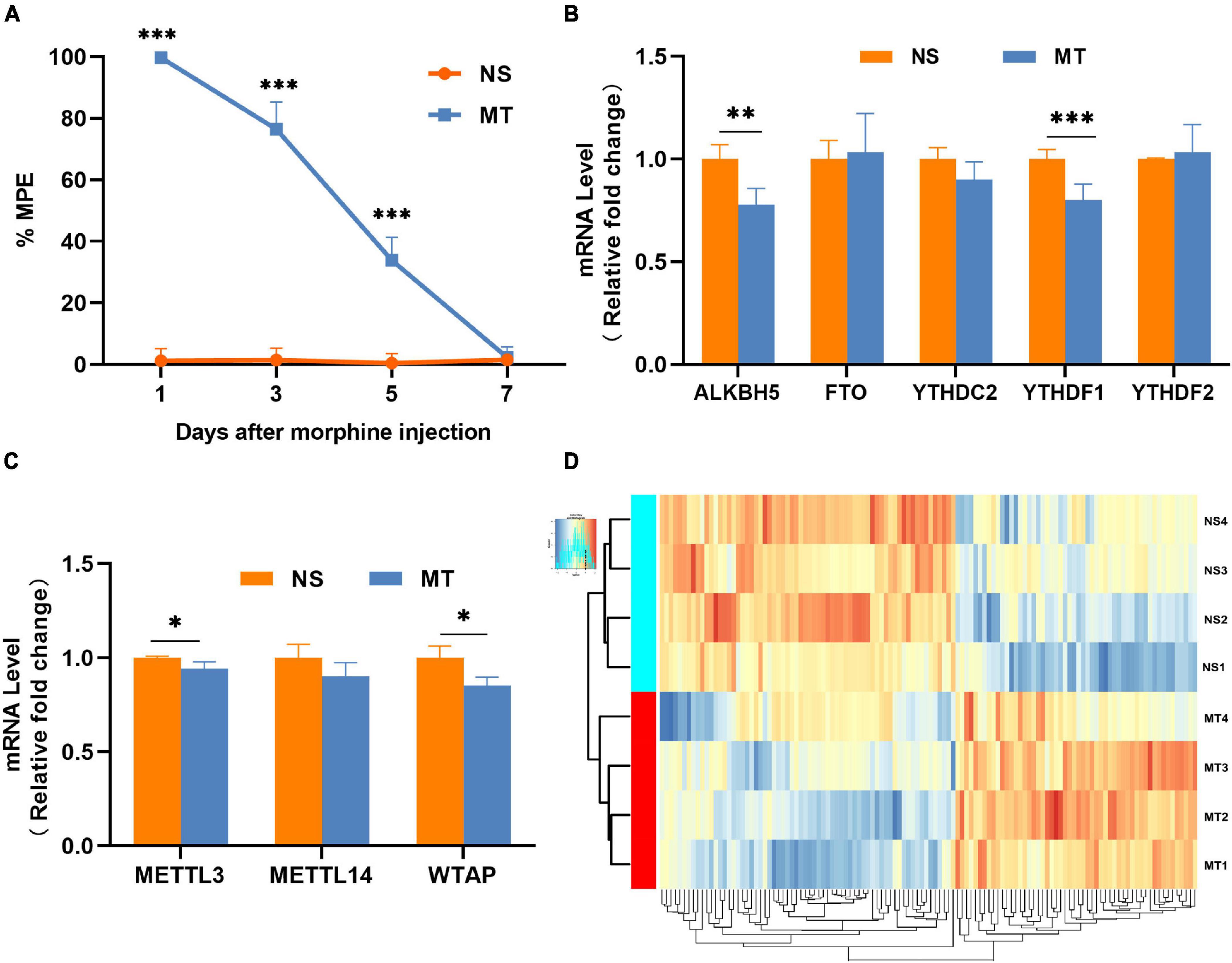
Figure 1. The animal model and the overall features of m6A-circRNA in morphine-tolerant rats. (A) Morphine-induced antinociception was assessed through the tail-flick test, and tail-flick latency was converted to %MPE (n = 4), ***P < 0.001 compared with the NS group using Two-way repeated measures of ANOVA followed by Bonferroni’s post-hoc test. (B) Expression of RNA demethylase and methyltransferase determined by qRT-PCR. (n = 4) **P < 0.01, ***P < 0.001 compared with the NS group using a two-tailed unpaired Student’s t-test. (C) Expression of RNA methylase determined by qRT-PCR (n = 4). *P < 0.05 compared with the NS group using a two-tailed unpaired Student’s t-test. (D) Hierarchical clustering shows global m6A-methylated circRNAs between the MT group and NS group (n = 4).
The m6A-circRNA microarrays were performed in the spinal cords of eight biological replicates from the morphine-tolerant and control rats (Figure 1D). All microarray results were uploaded to the GEO database (GSE199892). A total of 12,669 m6A-circRNAs were identified, compared with the control group, 120 circRNAs with different m6A modifications were identified, 54 m6A-circRNA were hypermethylated and 66 m6A-circRNAs were hypomethylated in the MT group (Figure 2A). The top 10 significantly methylated circRNAs are shown in Table 1.
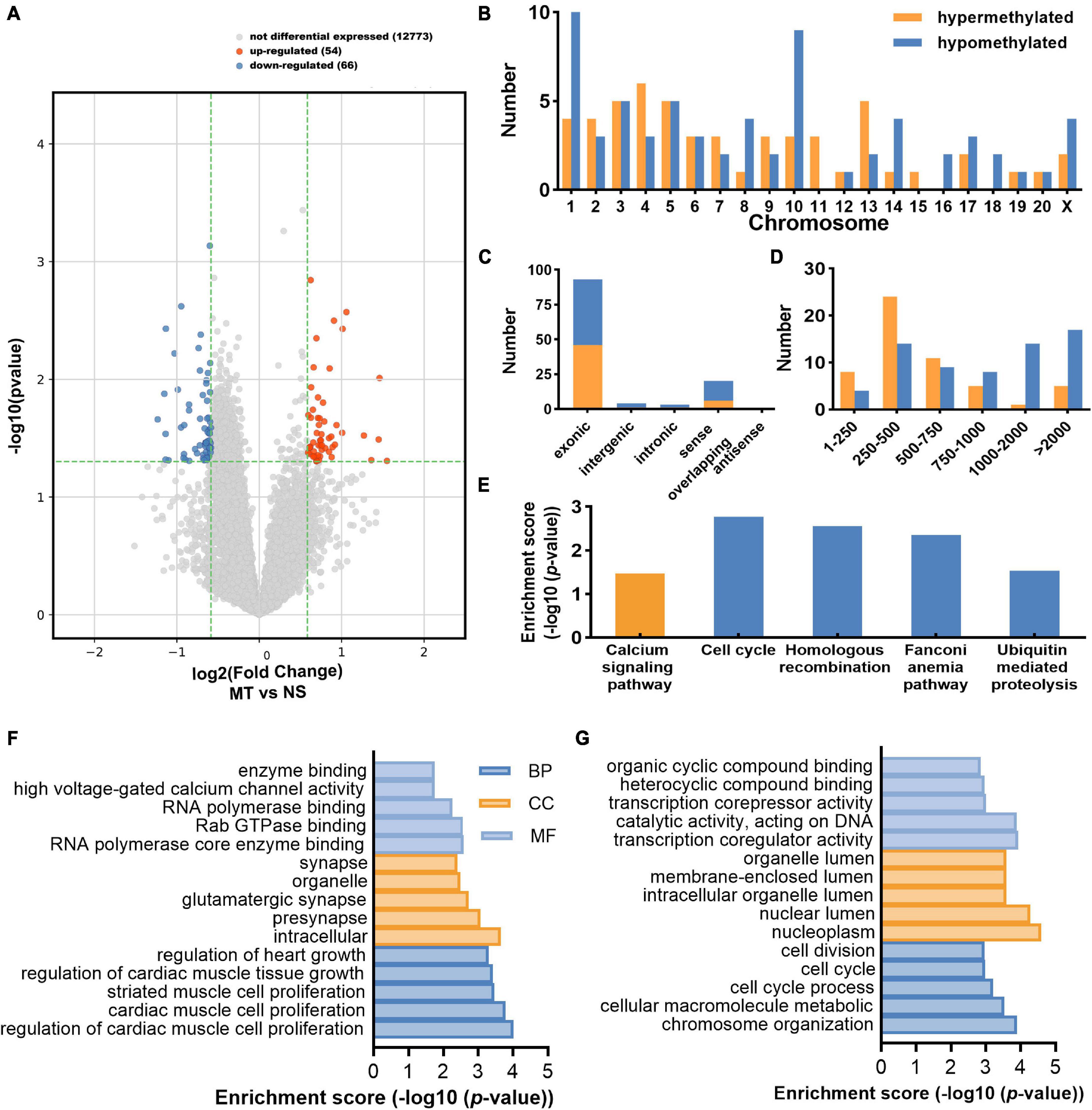
Figure 2. Distribution of differentially methylated circRNAs. (A) The volcano plot shows the distribution of differentially methylated circRNAs (Fold change ≥ 1.5 and P < 0.05) between the MT group and the NS group. (B) The chromosome’s origins for host genes of differentially m6A-methylated circRNAs. (C) The genomic origins of differentially m6A-methylated circRNAs. (D) The length distribution of differentially m6A-methylated circRNAs. (E) The enriched pathways of the host gene of the differentially m6A-methylated circRNAs through KEGG analyses. (F) The top 15 enriched GO terms of the host gene of the hypermethylated circRNAs. GO terms include biological process (BP) analysis, cellular component (CC) analysis, and molecular function (MF) analysis. (G) The top 15 enriched GO terms of the host gene of the hypomethylated circRNAs.
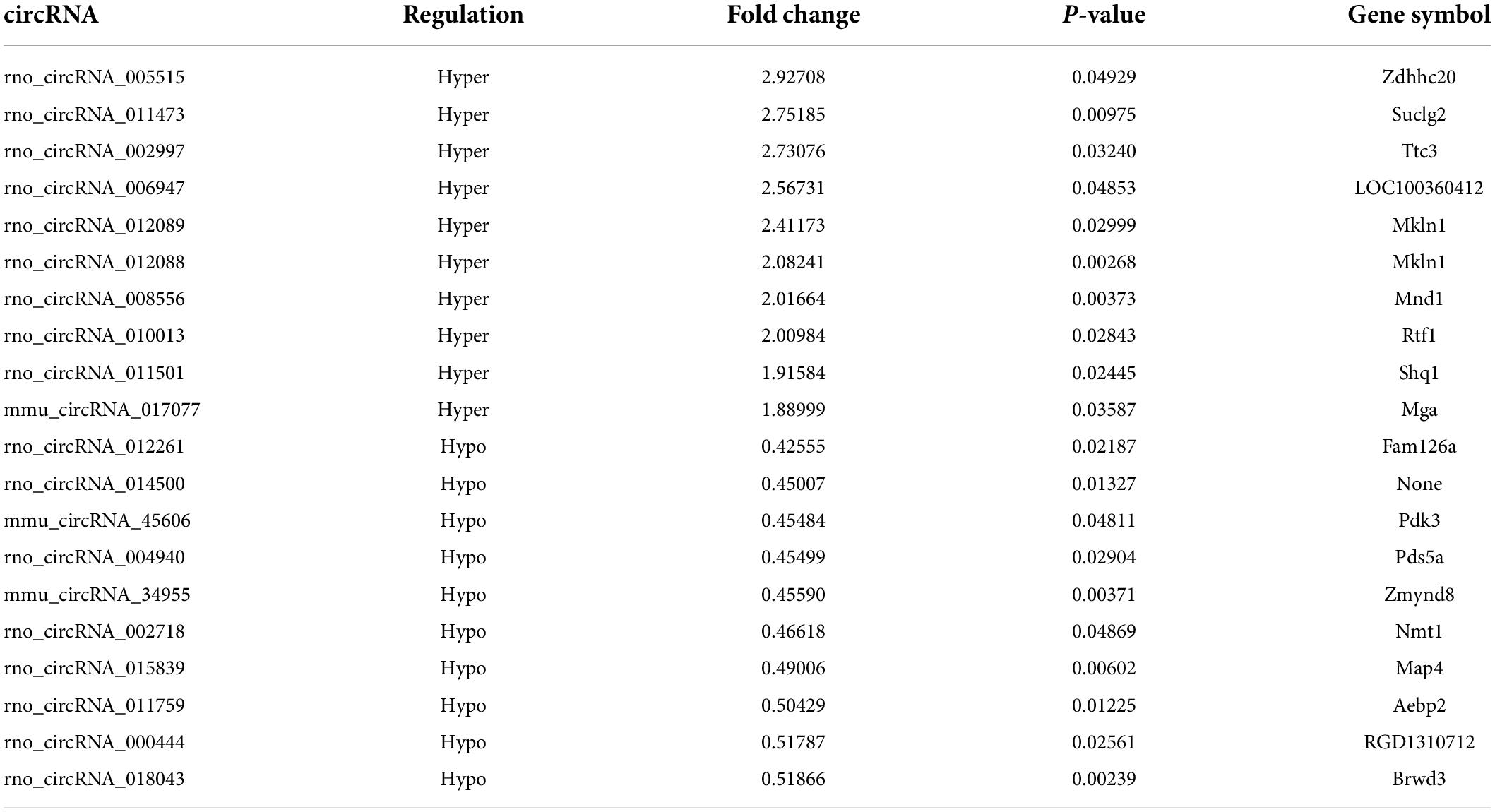
Table 1. The detailed information of significantly top 10 hypermethylated m6A-circRNA and top 10 hypomethylated m6A-circRNAs.
The different methylated (DE) m6A circRNAs were transcribed from all chromosomes, the host genes of hypermethylated circRNAs were primarily located in chromosomes 4 (6), 5 (5), and 13 (5), while the hypomethylated parts were mostly located in chromosome 1 (10), 3 (5), 5 (5), and 10 (9) (Figure 2B). Furthermore, most of the DE m6A-circRNAs were encoded by the exonic sequence of the host genes (Figure 2C). Their lengths were mostly enriched in 1–10,000 bps (Figure 2D).
To explore the physiological and pathological functions of m6A-circRNAs in MT, we performed GO and KEGG analysis for the host genes of DE m6A –circRNAs. The host genes of hypermethylated circRNAs were most enriched in the calcium signaling pathway. The host genes of hypomethylated circRNAs were most enriched in the cell cycle pathway, homologous recombination pathway, fanconi anemia pathway, ubiquitin mediated proteolysis pathway (Figure 2E). The GO terms of hypermethylated circRNAs were presented in Figure 2F. The GO terms were most enriched in regulation of cardiac muscle cell proliferation in the BP term; intracellular in the cellular component (CC) term; RNA polymerase core enzyme binding in molecular functions (MF) term. For the hypomethylated circRNAs, the GO terms were most enriched in chromosome organization in BP term, nucleoplasm in CC term, and transcription coregulator activity in MF term (Figure 2G).
Besides, the m6A modification levels of these 12,699 circRNAs were calculated as the percentage of modified RNA in all input RNAs (Supplementary Figure 1A). In total, we identified that 464 circRNAs had significantly differential modification levels in the MT group. Surprisingly, among these 464 circRNAs, only 1 circRNA had upregulated methylation level, and 463 circRNAs had downregulated methylation level (Supplementary Figure 1B).
By conjoint analysis of the results of m6A methylation and expression of circRNAs in morphine-tolerant rats, we found that 155 hypermethylated circRNAs were upregulated, 1 hypermethylated circRNA was downregulated, and 34 hypomethylated circRNAs were downregulated (Figure 3A). To further analyze how m6A methylation affects circRNA expression, 12,669 m6A-circRNAs were divided into two groups, 439 in the m6A-circRNAs group and 12,230 circRNAs in the non-m6A-circRNAs group. Then we calculated the log2 fold change of expression of them and generated cumulative curves. We found that the proportion of circRNAs not modified by m6A was higher than that of circRNAs modified by m6A in terms of the log2FC of the circRNA between 0 and 1.5 (Figure 3B).
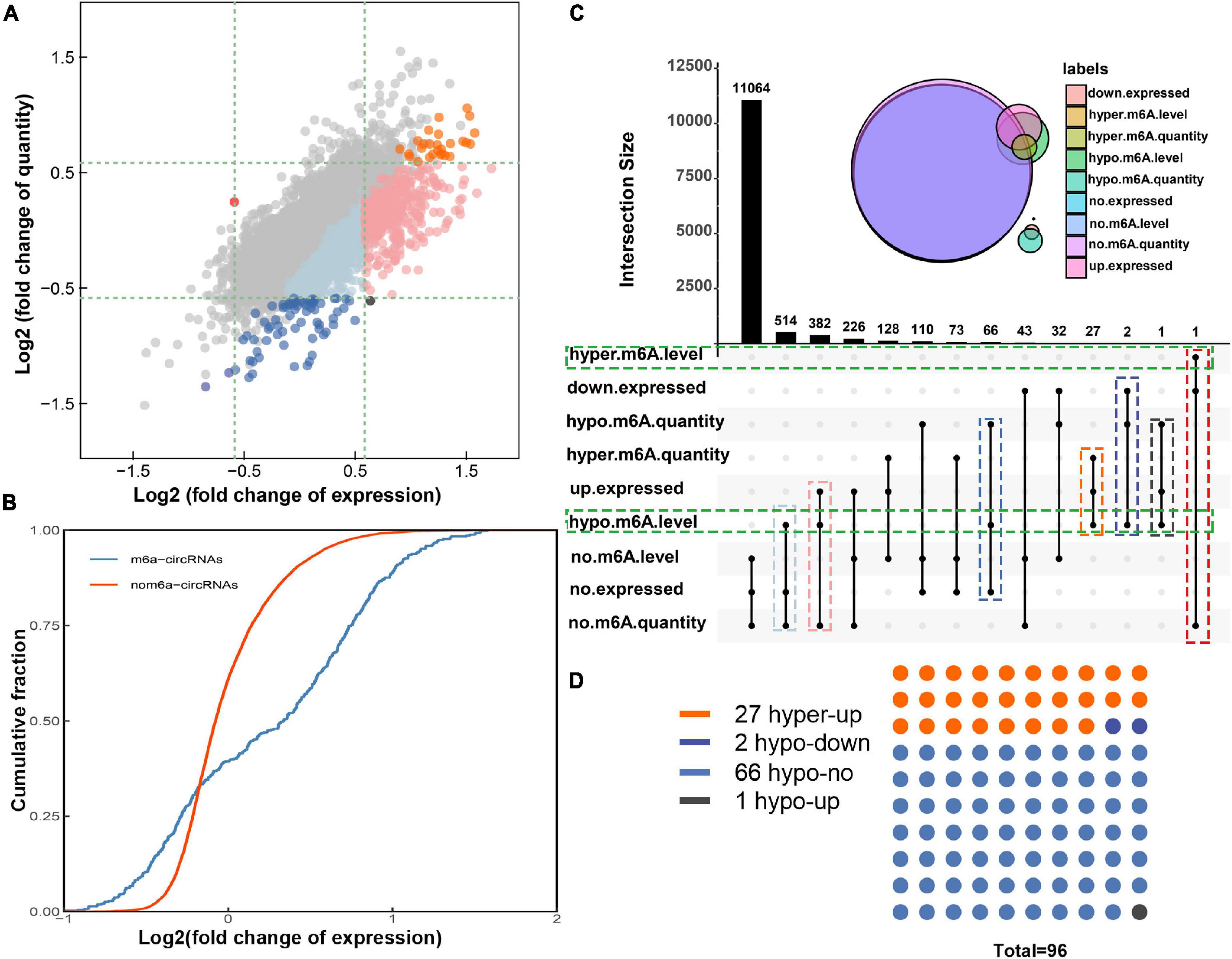
Figure 3. Joint analysis of m6A methylation and expression patterns of circRNAs in morphine tolerance. (A) The nine-quadrant graph shows the relationship between m6A quantity and the expression of circRNAs. (B) Cumulative distribution of circRNAs expression between MT group and NS group for m6A-circRNAs (blue) and non-m6A- circRNAs (red). (C) The upset diagram shows the m6A quantity, expression, and m6A level of circRNAs and presents 14 different methylation and expression patterns of circRNAs. (D) The dot plot shows the distribution of 96 circRNA with both dysregulated m6A quantity and m6A level. The orange dots (hyper-up) show 27 circRNAs were hypermethylated and up-expressed, 2 navy blue dots (hypo-down) represent circRNAs were hypomethylated and down-expressed; 1 black dot (hypo-up) represent circRNA were hypomethylated and up-expressed; the light blue dots (hypo-no) represent 66 circRNAs with decreased m6A modification quantity but unchanged expression levels.
However, studies have shown that the potential effects of RNA modifications depend not only on the m6A quantity but also on the percentage of transcripts that are modified. Therefore, the combined analysis of m6A level and expression of circRNAs was also been performed (Supplementary Figure 1C). Moreover, we studied the m6A quantity and m6A methylation level of all the 12,966 circRNAs. The upset graph showed all the circRNAs distribution in 14 different patterns considering m6A quantity, expression level and m6A methylation level (Figure 3C). Only one circRNA had increased m6A modification level with unchanged m6A quantity and decreased expression level. In addition, a total of 896 circRNAs with hypomethylated m6A level but with unchanged m6A quantity were also identified. In both cases, we need to be wary of false-positive in m6A levels simply due to altered expression levels (Figure 3C).
We focused on the remaining 96 circRNAs with hypomethylated m6A level. In detail, 27 circRNAs were hypermethylated and up-expressed, and 2 circRNA was hypomethylated and down-expressed; this may suggest that the m6A modification on these circRNAs may promote RNA stability. 1 circRNAs were hypermethylated and down-expressed; this prompted the m6A modification may inhibit circRNA stability and promote circRNA degradation. Furthermore, there were 66 circRNAs with decreased m6A modification quantity but unchanged expression levels; this suggests that m6A modifications in these circRNAs do not affect their expression and may play other roles, such as post-translational regulation (Figures 3C,D).
Given the importance of circRNA–miRNA interaction and to further explore the underlying mechanism of the above m6A-circRNAs, circRNAs with differences in m6A quantity and m6A level were selected to construct circRNA–miRNA networks. Several significant miRNAs that participated in the occurrence and development of MT were found to bind to these m6A circRNAs (Figure 4), such as miR-873a-5p, miR-103-1-5p, and miR-107-5p (Dai et al., 2018; Huang et al., 2019b). These data suggest that circRNAs with DEm6A might play a role in the pathological process of MT. Finally, to further confirm the findings of the m6A microarray data, we analyzed the m6A modification level of four circRNAs using MERIP-qPCR. The results indicate that the methylation levels (%MeRIP / Input) of these cirRNAs were downregulated and were consistent with the microarray analysis (Figure 5).
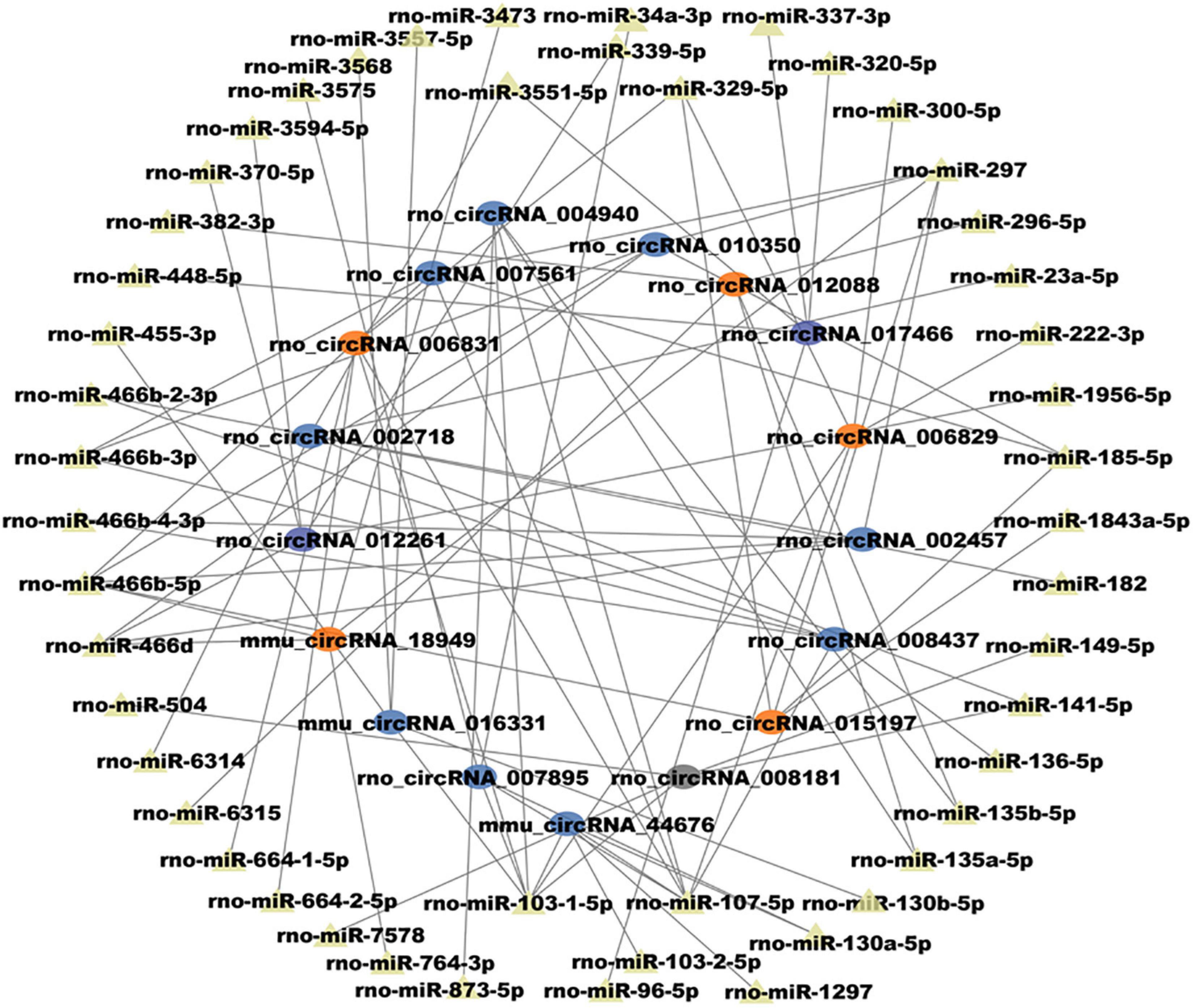
Figure 4. Construction of circRNA–miRNA networks in morphine tolerance using circRNAs with differences in both m6A quantity and m6A modification level.
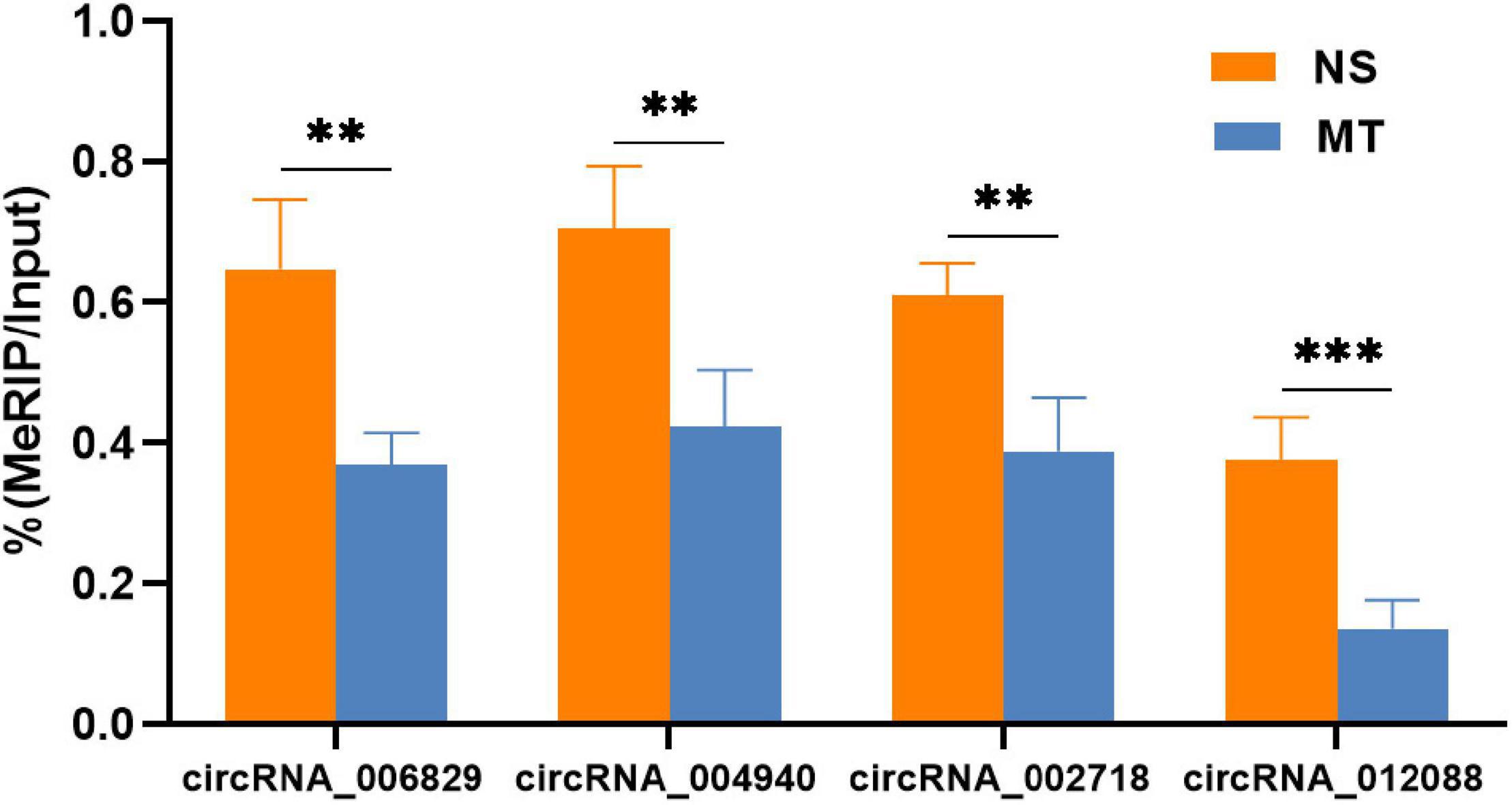
Figure 5. M6A levels of the four circRNAs involved in the circRNA–miRNA networks were verified by MeRIP-qPCR. Relative m6A methylation level was calculated as the percentage of modified transcripts in all transcripts. P-values were calculated using Student’s t-test (n = 4). **P < 0.01, ***P < 0.001 compared with the NS group using a two-tailed unpaired Student’s t-test.
In this study, we identified the epitranscriptomic map of m6A circRNAs in the spinal cord of morphine-tolerant rats. Specifically, we identified a total of 120 circRNAs with different m6A modifications including 54 hypermethylated circRNAs and 66 hypomethylated circRNAs, and found the host genes of these m6A circRNAs were involved in the key process of MT, such as the calcium signaling pathway. Meanwhile, we researched the correlation among m6A-methylation quantity, expression, and m6A level of circRNAs, and found 14 modifications and expression patterns. This study provides a deeper understanding of the functional role of circRNAs in MT and brought novel clues for epigenetic etiology and pathogenesis of MT.
The m6A modification status is probably associated with the expression of m6A “writers,” “readers,” and “erasers.” Research showed that METTL3 could regulate the global m6A level in the spinal cord of inflammatory pain (Pan et al., 2021). In our study, compared with the NS group, the expression levels of METTL3, WTAP, ALKBH5, and YTHDF1 decreased significantly in the MT group. Due to the imbalance between methyltransferase and demethylase in MT, m6A methylation microarrays resulted in both hypermethylated and hypomethylated circRNAs in the spinal cord of MT. Further research is needed to verify their specific roles and functions targeting a particular circRNA.
The chronic morphine administration could cause adaptive changes in neurons and neuronal communications (Sanna et al., 2020; Uniyal et al., 2020). The molecular adaptations include the dynamic change of opioid receptors, ion channels etc. (Williams et al., 2013). In the present study, the function analysis of DE m6A circRNAs enriched in the calcium signaling pathway. CACNA1B (Cav2.2) is one of the key genes in this pathway, it encodes an N-type voltage-gated calcium channel ubiquitously that is important in regulating neuropathic pain (Gao et al., 2021). Previous studies have demonstrated that increased intracellular levels of Ca2+ in neurons was an essential component of MT (Smith et al., 1999). Ca2+ channel antagonists enhanced morphine analgesia and prevented the development of tolerance (Michaluk et al., 1998). In addition, our previous study found that CaMKIIγ expression in the spinal cord was gradually upregulated after chronic morphine treatment, and downregulated it could alleviate tolerance (Wang et al., 2017). This evidence supports the hypothesis that m6A modification of circRNAs participated in the progression of MT.
Research has shown that m6A modification could regulate mRNA transcription, splicing, and degradation thus influencing its expression (Meyer and Jaffrey, 2014; Yue et al., 2015). Zhou et al. (2017) found low m6A level was negatively associated with circRNAs expression, while high m6A level was not linked to circRNAs expression in human embryonic stem cells and HeLa cells. In our study, the cumulative curve indicated that the proportion of circRNAs not modified by m6A was greater than that of circRNAs modified by m6A, this may suggest that methylation downregulates circRNA expression. However, the potential effects of RNA modifications not only depend on the m6A quantity but also on the percentage of transcripts that are modified (Gilbert et al., 2016). Thus, we researched modifications and expression patterns of the circRNA and found 4 patterns including 96 circRNAs with both changed m6A quantity and m6A methylation level. The relationships between the expression and m6A modification of circRNAs in these four patterns were different. We also found 897 circRNAs with hypomethylated level but with no changes in m6A quantity. The changes in their m6A levels may be additional effects caused by their expression changes and have no matter with m6A modification. Therefore, we propose that when studying the role of m6A in circRNAs, both the m6A quantity and the methylation level of this circRNA need to be considered.
As circRNAs could sponge miRNA and affect its effect on targeted genes, many miRNAs (Wang et al., 2016, 2017; Huang et al., 2019a) and genes (Dumas and Pollack, 2008) have been researched in MT. CircRNAs with differences in both m6A quantity and m6A level were selected to construct circRNA–miRNA networks. We found several important miRNAs that participated in the pathological process could bound these m6A circRNAs, including miR-873a-5p, miR-103-1-5p, and miR-107-5p. These results suggest that m6A modification of circRNAs may influence the interactions between circRNAs and miRNAs. CircRNA 007895, circRNA 006829, and circRNA 004940 were predicted to bind miR-103-1-5p and miR-107-5p. However, little research has focused on these circRNAs. It will be necessary to investigate how m6A-modification regulates them in the pathological process of MT.
Finally, some limitations of our study should be acknowledged. First, we only detected the RNA expression of m6A-related enzymes and did not confirm the results based on the protein and enzymatic activity levels. In addition, the precise mechanism of ALKBH5 and YTHDF1 during MT needs to be studied. Second, we only focused on the global situation and did not explore the specific function of m6A-circRNAs in the development of MT. Further in vivo and in vitro research will be carried out to investigate the m6A-circRNA-mediated precise regulatory mechanisms in MT.
This study is the first to reveal the m6A methylation pattern of circRNAs in the spinal cord of morphine-tolerant rats and provides a deeper understanding of the functional role of circRNAs. The results may bring novel clues for epigenetic etiology and pathogenesis of MT.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by the Central South University Ethics Committee (ethical no. 202103141).
MX and WZ conceived and designed the experiments, performed the experiments, analyzed the data, and wrote the manuscript. YS, MD, JD, and TD performed animal experiments and analyzed the data. MX and ZS completed the bioinformatics analysis. WZ revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (grant nos. 82171236 and 81974172 to WZ) and the Key Research and Development Plan of Hunan Province (2021SK2018 to WZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2022.967768/full#supplementary-material
Supplementary Figure 1. Distribution of circRNAs from the perspective of m6A-methylation level. (A) Hierarchical clustering shows global circRNAs m6A- methylation level between the MT group and NS group (n = 4). (B) The volcano plot shows the distribution of circRNAs with differentially methylation levels (Fold change ≥ 1.5 and P < 0.05) between the MT group and the NS group. (C) The nine-quadrant graph shows the relationship between the m6A methylation level and the expression of circRNAs.
m6A, N6-Methyladenosine; circRNA, circular RNA; ncRNAs, non-coding RNAs; miRNA, microRNAs; lncRNAs, long non-coding RNAs; METTL, Methyltransferase-like; WTAP, WT1 associated protein; YTHDC, YTH domain-containing; FTO, obesity-associated protein; ALKBH5, AlkB homologue 5; MT, Morphine tolerance; NS, Normal saline; %MPE, percent of the maximum possible effect; MeRIP, Methylated RNA immunoprecipitation; qRT-PCR, Quantitative real-time polymerase chain reaction; DE, different methylated; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, Biological process; CC, Cellular component; MF, Molecular functions.
Chen, Y., and Sommer, C. (2009). The role of mitogen-activated protein kinase (MAPK) in morphine tolerance and dependence. Mol. Neurobiol. 40, 101–107. doi: 10.1007/s12035-009-8074-z
Dai, Z., Chu, H., Ma, J., Yan, Y., Zhang, X., and Liang, Y. (2018). The Regulatory Mechanisms and Therapeutic Potential of MicroRNAs: From Chronic Pain to Morphine Tolerance. Front. Mol. Neurosci. 11:80. doi: 10.3389/fnmol.2018.00080
Deng, M., Zhang, Z., Xing, M., Liang, X., Li, Z., Wu, J., et al. (2022). LncRNA MRAK159688 facilitates morphine tolerance by promoting REST-mediated inhibition of mu opioid receptor in rats. Neuropharmacology 206:108938. doi: 10.1016/j.neuropharm.2021.108938
Dumas, E. O., and Pollack, G. M. (2008). Opioid tolerance development: A pharmacokinetic/pharmacodynamic perspective. AAPS J. 10, 537–551. doi: 10.1208/s12248-008-9056-1
Gao, S., Yao, X., and Yan, N. (2021). Structure of human Ca(v)2.2 channel blocked by the painkiller ziconotide. Nature 596, 143–147. doi: 10.1038/s41586-021-03699-6
Gilbert, W. V., Bell, T. A., and Schaening, C. (2016). Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412. doi: 10.1126/science.aad8711
Gledhill, L. J., and Babey, A. M. (2021). Synthesis of the Mechanisms of Opioid Tolerance: Do We Still Say NO? Cell. Mol. Neurobiol. 41, 927–948. doi: 10.1007/s10571-021-01065-8
Huang, J., Liang, X., Wang, J., Kong, Y., Zhang, Z., Ding, Z., et al. (2019a). miR-873a-5p Targets A20 to Facilitate Morphine Tolerance in Mice. Front. Neurosci. 13:347. doi: 10.3389/fnins.2019.00347
Huang, J., Wang, J., Guo, Q., and Zou, W. (2019b). Emerging roles of microRNAs in morphine tolerance. J. Pain Res. 12, 1139–1147. doi: 10.2147/jpr.S187592
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Liang, D. Y., Li, X., and Clark, J. D. (2013). Epigenetic regulation of opioid-induced hyperalgesia, dependence, and tolerance in mice. J. Pain 14, 36–47. doi: 10.1016/j.jpain.2012.10.005
Lu, S., Yang, X., Wang, C., Chen, S., Lu, S., Yan, W., et al. (2019). Current status and potential role of circular RNAs in neurological disorders. J. Neurochem. 150, 237–248. doi: 10.1111/jnc.14724
Ma, S., Chen, C., Ji, X., Liu, J., Zhou, Q., Wang, G., et al. (2019). The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 12:121. doi: 10.1186/s13045-019-0805-7
Mehta, S. L., Dempsey, R. J., and Vemuganti, R. (2020). Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 186:101746. doi: 10.1016/j.pneurobio.2020.101746
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., and Wu, M. (2019). Epigenetics in Neurodevelopment: Emerging Role of Circular RNA. Front. Cell Neurosci. 13:327. doi: 10.3389/fncel.2019.00327
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. doi: 10.1038/nrm3785
Michaluk, J., Karolewicz, B., Antkiewicz-Michaluk, L., and Vetulani, J. (1998). Effects of various Ca2+ channel antagonists on morphine analgesia, tolerance and dependence, and on blood pressure in the rat. Eur. J. Pharmacol. 352, 189–197. doi: 10.1016/s0014-2999(98)00373-2
Niu, Y., Zhao, X., Wu, Y. S., Li, M. M., Wang, X. J., and Yang, Y. G. (2013). N6-methyl-adenosine (m6A) in RNA: An old modification with a novel epigenetic function. Genom. Proteom. Bioinform. 11, 8–17. doi: 10.1016/j.gpb.2012.12.002
Pan, Z., Zhang, Q., Liu, X., Zhou, H., Jin, T., Hao, L. Y., et al. (2021). Methyltransferase-like 3 contributes to inflammatory pain by targeting TET1 in YTHDF2-dependent manner. Pain 162, 1960–1976. doi: 10.1097/j.pain.0000000000002218
Paul, A. K., Smith, C. M., Rahmatullah, M., Nissapatorn, V., Wilairatana, P., Spetea, M., et al. (2021). Opioid Analgesia and Opioid-Induced Adverse Effects: A Review. Pharmaceuticals 14:1091. doi: 10.3390/ph14111091
Rybak-Wolf, A., Stottmeister, C., Glažar, P., Jens, M., Pino, N., Giusti, S., et al. (2015). Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 58, 870–885. doi: 10.1016/j.molcel.2015.03.027
Sanna, M. D., Borgonetti, V., and Galeotti, N. (2020). μ Opioid Receptor-Triggered Notch-1 Activation Contributes to Morphine Tolerance: Role of Neuron-Glia Communication. Mol. Neurobiol. 57, 331–345. doi: 10.1007/s12035-019-01706-6
Shao, J., Wang, J., Huang, J., Liu, C., Pan, Y., Guo, Q., et al. (2018). Identification of lncRNA expression profiles and ceRNA analysis in the spinal cord of morphine-tolerant rats. Mol. Brain 11:21. doi: 10.1186/s13041-018-0365-8
Smith, F. L., Dombrowski, D. S., and Dewey, W. L. (1999). Involvement of intracellular calcium in morphine tolerance in mice. Pharmacol. Biochem. Behav. 62, 381–388. doi: 10.1016/s0091-3057(98)00168-3
Uniyal, A., Gadepalli, A., and Akhilesh Tiwari, V. (2020). Underpinning the Neurobiological Intricacies Associated with Opioid Tolerance. ACS Chem. Neurosci. 11, 830–839. doi: 10.1021/acschemneuro.0c00019
Viet, C. T., Dang, D., Aouizerat, B. E., Miaskowski, C., Ye, Y., Viet, D. T., et al. (2017). OPRM1 Methylation Contributes to Opioid Tolerance in Cancer Patients. J. Pain 18, 1046–1059. doi: 10.1016/j.jpain.2017.04.001
Wang, J., Xu, W., Shao, J., He, Z., Ding, Z., Huang, J., et al. (2017). miR-219-5p targets CaMKIIγ to attenuate morphine tolerance in rats. Oncotarget 8, 28203–28214. doi: 10.18632/oncotarget.15997
Wang, J., Xu, W., Zhong, T., Song, Z., Zou, Y., Ding, Z., et al. (2016). miR-365 targets β-arrestin 2 to reverse morphine tolerance in rats. Sci. Rep. 6:38285. doi: 10.1038/srep38285
Wang, X., Ma, R., Zhang, X., Cui, L., Ding, Y., Shi, W., et al. (2021). Crosstalk between N6-methyladenosine modification and circular RNAs: Current understanding and future directions. Mol. Cancer 20:121. doi: 10.1186/s12943-021-01415-6
Weng, Y., Wu, J., Li, L., Shao, J., Li, Z., Deng, M., et al. (2019). Circular RNA expression profile in the spinal cord of morphine tolerated rats and screen of putative key circRNAs. Mol. Brain 12:79. doi: 10.1186/s13041-019-0498-4
Wiffen, P. J., Wee, B., and Moore, R. A. (2016). Oral morphine for cancer pain. Cochrane Database Syst. Rev. 4:Cd003868. doi: 10.1002/14651858.CD003868.pub4
Williams, J. T., Ingram, S. L., Henderson, G., Chavkin, C., Von Zastrow, M., Schulz, S., et al. (2013). Regulation of μ-opioid receptors: Desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 65, 223–254. doi: 10.1124/pr.112.005942
Wu, J., Guo, X., Wen, Y., Huang, S., Yuan, X., Tang, L., et al. (2021). N6-Methyladenosine Modification Opens a New Chapter in Circular RNA Biology. Front. Cell Dev. Biol. 9:709299. doi: 10.3389/fcell.2021.709299
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. doi: 10.1101/gad.262766.115
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Keywords: morphine tolerance, circRNAs, N6-methyladenosine, microarray, bioinformatics analysis
Citation: Xing M, Deng M, Shi Y, Dai J, Ding T, Song Z and Zou W (2022) Identification and characterization of N6-methyladenosine circular RNAs in the spinal cord of morphine-tolerant rats. Front. Neurosci. 16:967768. doi: 10.3389/fnins.2022.967768
Received: 16 June 2022; Accepted: 18 July 2022;
Published: 05 August 2022.
Edited by:
Wuping Sun, Affiliated Nanshan Hospital of Shenzhen University Health Science Center, ChinaReviewed by:
Feixiang Wu, Second Military Medical University, ChinaCopyright © 2022 Xing, Deng, Shi, Dai, Ding, Song and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wangyuan Zou, d2FuZ3l1YW56b3VAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.