
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci., 03 May 2022
Sec. Neurodegeneration
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.858126
This article is part of the Research TopicThe Alzheimer's Disease Challenge, Volume IIView all 14 articles
 Rajnish Kumar1
Rajnish Kumar1 Anju Sharma2
Anju Sharma2 Athanasios Alexiou3,4
Athanasios Alexiou3,4 Anwar L. Bilgrami5,6
Anwar L. Bilgrami5,6 Mohammad Amjad Kamal7,8,9,10,11
Mohammad Amjad Kamal7,8,9,10,11 Ghulam Md Ashraf12,13*
Ghulam Md Ashraf12,13*The blood-brain barrier (BBB) is a selective and semipermeable boundary that maintains homeostasis inside the central nervous system (CNS). The BBB permeability of compounds is an important consideration during CNS-acting drug development and is difficult to formulate in a succinct manner. Clinical experiments are the most accurate method of measuring BBB permeability. However, they are time taking and labor-intensive. Therefore, numerous efforts have been made to predict the BBB permeability of compounds using computational methods. However, the accuracy of BBB permeability prediction models has always been an issue. To improve the accuracy of the BBB permeability prediction, we applied deep learning and machine learning algorithms to a dataset of 3,605 diverse compounds. Each compound was encoded with 1,917 features containing 1,444 physicochemical (1D and 2D) properties, 166 molecular access system fingerprints (MACCS), and 307 substructure fingerprints. The prediction performance metrics of the developed models were compared and analyzed. The prediction accuracy of the deep neural network (DNN), one-dimensional convolutional neural network, and convolutional neural network by transfer learning was found to be 98.07, 97.44, and 97.61%, respectively. The best performing DNN-based model was selected for the development of the “DeePred-BBB” model, which can predict the BBB permeability of compounds using their simplified molecular input line entry system (SMILES) notations. It could be useful in the screening of compounds based on their BBB permeability at the preliminary stages of drug development. The DeePred-BBB is made available at https://github.com/12rajnish/DeePred-BBB.
Neurological diseases are among the most predominant health issues, with an approximately 28% prevalence in all age groups of patients (Menken et al., 2000). Despite a decrease in communicable neurological diseases, the number of deaths due to neurological diseases has increased to 39% in the last three decades (Feigin et al., 2020). This substantial increase in the absolute number of patients indicates that available therapeutics are scarce to prevent and manage neurological diseases in the current changing global demography. Therefore, it is imperative to find novel and effective therapeutics to target the central nervous system (CNS) to meet the challenges of the ever-increasing absolute number of patients with neurological diseases. An alternative method targeting the molecular and signaling mechanisms at BBB rather than the traditional approaches has become the recent trend in drug target validation (Salman et al., 2022). Drugs must cross the blood-brain barrier (BBB) to act on the CNS. There is a higher attrition rate of drug candidates failing in clinical research due to non-permeability to the BBB compared to potency issues (Dieterich et al., 2003; Pardridge, 2005; Saunders et al., 2014; Hendricks et al., 2015). The BBB is a semipermeable and selective boundary that maintains the steady state of the CNS by protecting it from external compounds (98%) (Figure 1; Pardridge, 2002). As drugs need to enter the CNS to impart therapeutic activity, it becomes crucial to determine BBB permeability during the initial stages of CNS-acting drug design and development (Daneman, 2015; van Tellingen et al., 2015; Saxena et al., 2019).
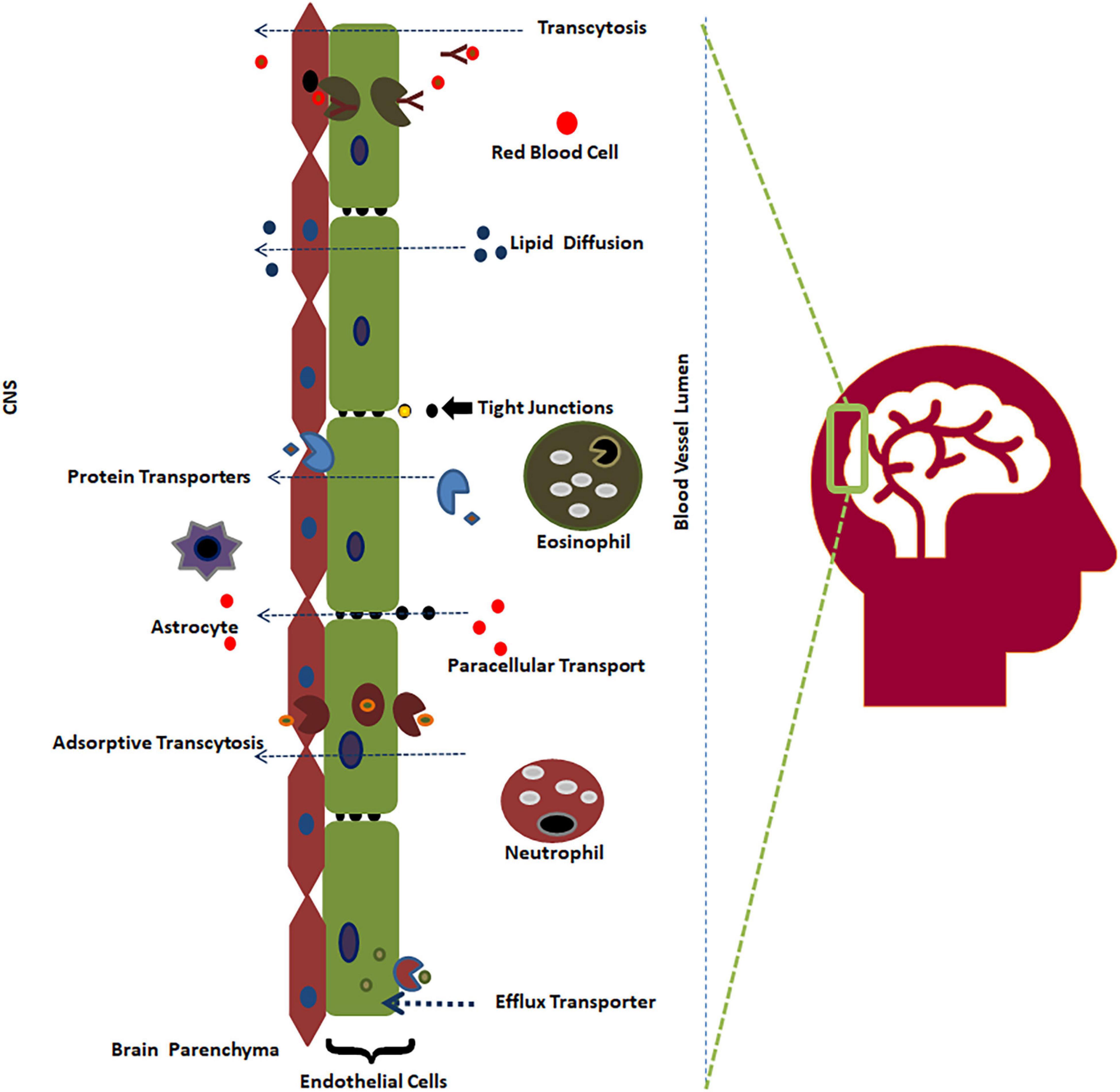
Figure 1. The BBB and its permeability mechanism (Saxena et al., 2021).
The BBB separates the CNS from the bloodstream, preventing contagions from invading the brain. Brain endothelial cells, astrocytes, neurons, and pericytes are four major components of the BBB. The largest constituent of the BBB is a layer containing brain endothelial cells, which serve as the first line of defense from the CNS surroundings. Endothelial cells are connected with tight junctions and adherence junctions, which create a strong barrier, restricting pinocytosis and decreasing vesicle-facilitated transcellular transport (Reese and Karnovsky, 1967; Tietz and Engelhardt, 2015). The BBB not only acts as a physical barrier but also serves as a metabolic barrier, transport interface, and secretory layer (Abbott et al., 2006; Rhea and Banks, 2019). Neurons reside very close to brain capillaries and play a vital role in maintaining ion balance in the local environment (Schlageter et al., 1999).
Clinical experiments to determine the BBB permeability of compounds are accurate; however, they are time-consuming and labor-intensive (Bickel, 2005; Massey et al., 2020; Schidlowski et al., 2020). Additionally, it is difficult to perform clinical experiments with diverse types of drug candidates (Main et al., 2018; Mi et al., 2020). Therefore, it is crucial to predict and forecast BBB permeability using computational algorithms or in vitro BBB mimics to elucidate the permeability of compounds across the BBB (Gupta et al., 2019). There have been numerous attempts to predict the BBB permeability of compounds since the advent of artificial intelligence (AI), primarily using machine learning (ML) algorithms such as support vector machines (SVMs), artificial neural networks (ANNs), k-nearest neighbors (kNNs), naïve Bayes (NB), and random forests (RFs) (Doniger et al., 2002; Zhang et al., 2008, 2015; Suenderhauf et al., 2012; Khan et al., 2018). In addition to above, some future directions of BBB permeability also seem promising such as application of humanized self-organized models, organoids, 3D cultures and human microvessel-on-a-chip platforms especially those which are amenable for advanced imaging such as transmission electron microscope and expansion microscopy since they enable real-time monitoring of BBB permeability (Wevers et al., 2018; Salman et al., 2020). BBB permeability prediction models developed using AI algorithms can further be assisted with the high throughput screening (Aldewachi et al., 2021), computer aided drug designing (Salman et al., 2021), and knowledge based rules, e.g., Lipinski rule of five (hydrogen bond donor ≤ 5, hydrogen bond acceptor ≤ 10, molecular weight ≤ 500, CLogP ≤ 5), Veber rule (rotatable bonds count ≤ 10, polar surface area ≤ 140), BBB rule (hydrogen bond = 8–10, molecular weight = 400–500, no acids), etc., to screen potential drug candidates with desirable end-point for prevention, mitigation and cure of neurological disorders (Veber et al., 2002; Banks, 2009; Benet et al., 2016).
In an attempt to develop the BBB permeability prediction model, Jiang et al. (2016) applied SVM with a radial basis function (RBF) kernel (Jiang et al., 2016). They used a dataset of 1,562 compounds containing 694 BBB permeable (BBB++) and 868 BBB non-permeable (BBB-) compounds. The overall accuracy, sensitivity, and specificity were reported to be more than 85%. The next year, Castillo-Garit et al. (2017) used the decision tree algorithm on 581 compounds and found that the BBB permeability prediction accuracy increased by 2.93% (Castillo-Garit et al., 2017). However, this study was performed on a much smaller dataset than Jiang et al.’s study. In another study, Yuan et al. (2018) developed SVM-based BBB prediction model using a larger dataset of 1,990 compounds with a prediction accuracy of 93.96% (Yuan et al., 2018). The sensitivity and specificity of the model were reported to be 94.3 and 91.0%, respectively. In the same year, Wang et al. (2018) applied SVM and kNN algorithms using 2,358 compounds (Wang et al., 2018). The prediction accuracy of the best-performing model was found to be 2.64% higher than that of the Yuan et al. (2018) prediction model. However, the model lagged in terms of sensitivity (0.925) and specificity (0.899). The next year, Miao et al. (2019) applied a deep learning (DL) algorithm to 462 compounds. The accuracy of the model was reported to be 97%, with decent AUC (0.98) and F1 scores (0.92). However, the dataset used for the DL study was very small compared to the earlier ML-based models for BBB prediction. Recently, Alsenan et al. (2020) proposed a recurrent neural network (RNN) algorithm-based model using 2,342 compounds for the prediction of BBB permeability (Alsenan et al., 2020). The developed model had better performance metrics with an accuracy, sensitivity, and specificity of 96.53, 94.91, and 98.09%, respectively. The Matthews correlation coefficient (MCC) (93.14) and area under the curve (AUC) (98.6) of the prediction were also found to be satisfactory. In another study, Shaker et al. (2020) applied a light gradient boosting machine algorithm to a dataset of 7,162 compounds for the prediction of BBB permeability (Shaker et al., 2020). Although the study involved a very large dataset compared to previously reported studies, the model’s accuracy was reported to be 90%, which was approximately 6.5% less than the BBB permeability prediction model proposed by Alsenan et al. (2020). In the same year, Singh et al. (2020) used random forest, multilayer perceptron, and sequential minimal optimization using 605 compounds to develop the BBB permeability prediction model (Singh et al., 2020). Upon validation of the developed model using 1,566 compounds, the prediction accuracy was found to be 86.5% only. Very recently, Saxena et al. (2021) proposed an ML-based BBB permeability prediction model using 1,978 compounds (Saxena et al., 2021). The study group found that SVM with the RBF kernel yielded an accuracy of 96.77% with AUC and F1 score values of 0.964 and 0.975, respectively, which outperformed the kNN, random forest, and naïve Bayes algorithms in the prediction of BBB permeability on the same dataset.
The major challenge while applying ML algorithms is selecting optimal features to develop predictive models based on labeled BBB permeability datasets (Hu et al., 2019; Salmanpour et al., 2020). To overcome this challenge, we applied DL algorithms and compared their performance with traditional ML algorithms.
A total of 3,971 compounds with BBB permeability classes were collected from Zhao et al. (2007); Shen et al. (2010), and Roy et al. (2019). The PubChem database1 was used to retrieve available PubChem IDs of the collected compounds. The collected datasets were checked to remove redundant compounds. After careful curation, we obtained a dataset of 3,605 non-redundant clean compounds containing 2,607 BBB permeable and 998 BBB non-permeable compounds (Table 1 and Supplementary File). The class labels for BBB non-permeable and permeable compounds were kept as “0” and “1,” respectively.
Three types of feature sets viz. physicochemical properties, molecular access system (MACCS) fingerprints and substructure fingerprints were used in this study. Physicochemical properties contain different types of physical and chemical information encoded in a compound, e.g., molecular weight, molecular volume, solubility, partition coefficient, etc. The molecular fingerprints are fixed-length vectors that indicate the presence/absence of an atom type or functional group in a compound. All features were calculated by open-source PaDel (Yap, 2011). Each compound was encoded with 1,917 features containing 1,444 physicochemical (1D and 2D) properties, 166 MACCS, and 307 substructure fingerprints. This feature set was used for the ML, DNN, and CNN-1D algorithms using Keras framework. For CNN-VGG16, the Python package RDKit was used to generate the structure images of the compounds using their Simplified molecular input line entry system (SMILES) notations (Lovrić et al., 2019; Bento et al., 2020). The Python package RDKit is a collection of ML and cheminformatic software and contains functions to modify chemical compounds. The RDKit package was used to generate 2D images of size 300 * 300 pixels (RGB) from SMILES notations of compounds. RDKit-generated images contain different colors to express the chemical information viz. carbon = black, oxygen = red, nitrogen = blue, sulfur = yellow, chlorine = green, and phosphorous = orange. Images generated by RDKit always fit the entire molecule, so there was no issue with different molecular sizes. The dataset was split into training and test sets at a ratio of 3:1. The test set was separated from the training set to avoid any bias (Table 2). To handle the data imbalance, we have already applied cost-sensitive augmentation via the class_weight argument on the fit() function when training models.
In this study, ML-based algorithms (SVM, kNN, RF, and NB) and DL-based algorithms DNN, CNN-1D were developed using keras framework with libraries; python, numpy, pandas, keras, and tensorflow on Anaconda 3–5.2. CNN (VGG16) was implemented using transfer learning through cloud-based computational resource of Google Colaboratory to develop prediction models for the BBB permeability of the compounds. Based on the performance of the generated prediction models, the DNN-based “DeePred-BBB” is proposed for BBB permeability prediction. DeePred-BBB performance was compared with ML algorithms viz. SVM, NB, kNN, RF, and DL algorithms CNN-1D and CNN (VGG16).
Support vector machine with four different kernels (RBF, polynomial, sigmoid, and linear), NB, kNN, and RF were applied to the training set of 2,704 compounds and tested with an independent set of 901 compounds. Principal component analysis (PCA) (Giuliani, 2017) was used for feature reduction. The component range (10, 20, 30, 40, 50, and 100) was used to find the best prediction accuracy for each applied ML algorithm. Tenfold cross-validation was applied to evaluate the efficacy of the model during training.
Support vector machine is among the robust ML algorithms used for classification and regression (Ghandi et al., 2014; Gaudillo et al., 2019). It searches for the optimal hyperplane with maximized margins using support vectors for classification (Ben-Hur et al., 2008). This algorithm plots the data to the N-dimensional feature space and finds a hyperplane (Θ.x + b = 0) to classify the data sets with minimized loss using the hinge loss function. The loss function is given in Eq. 1.
Support vector machine was applied using kernels to map the data to higher dimensions to linearly classify the data (Kumar et al., 2011). A penalty parameter “C” (Cost NAÏVE) adjusts the balance between training errors and forcing rigid margins. Another parameter, “γ,” regulates the kernel function amplitude (Kumar et al., 2018). Various values of C (1, 5, 10, 50, 90) and γ (0.0001, 0.0005, 0.001, 0.005, 0.01, 0.05) were tested to find the best combination. An optimized combination of C and γ was used for each SVM kernel (RBF, C = 10, γ = 0.005; polynomial, C = 1, γ = 0.005; sigmoid C = 90, γ = 0.0005; linear, C = 1, γ = 0.05). For the polynomial kernel, 2–6 values of degree (d) were applied and evaluated. The best performance of the polynomial kernel was found at d = 3.
The naïve Bayes algorithm is based on the Bayes theorem. It is a probabilistic method that works on the assumption of class conditional independence (Eq. 2) (Shen et al., 2019). Each feature present in a class is independent and individually contributes to the probability with nil dependency on other features (Wang et al., 2021a). It is fast, readily manages a large dataset, and generally produces better results than other classification techniques when features existing in a class are independent.
where P(X|Y) is the posterior probability of X (class) for a given Y (feature), P(Y|X) is the likelihood, P(X) is the prior probability of class X, and P(Y) is the marginal probability of feature Y.
k-nearest neighbor is a simple and non-parametric classifier that assumes that nearby data points are similar and tend to have similar classes. Feature similarity is used to find the class label of a new data instance. It commonly uses Euclidean distance to find the closeness of the data points, and depending upon the class matching with considered k-points, the class labels are decided (Sharma et al., 2021). Here, k is the number of neighbors. The Euclidean distance between data points x (x1, x2, x3) and y (y1, y2, y3) is calculated using Eq. 3.
To determine the optimal k, a range of k-values (1–10) was evaluated. The best-performing prediction model at k = 3 was selected for further analysis.
Random forest uses ensemble learning to create a collection of decision trees (forest) that run concurrently and classify data instances (Yao et al., 2020). Tree construction is performed using arbitrary input vectors and node division on arbitrary feature subsets. Each tree of the RF predicts a certain class, and depending upon the highest votes, the final class label is predicted (Yang et al., 2020). In the current study, the developed prediction models were tested with variable trees in a forest (4, 8, 12, 32, 64). Each decision tree’s various depths (2–5) and estimators (5, 10, 20, 30, 40) were tested to find the best performing prediction model.
Deep learning algorithms use multiple neurons and hidden layers to extract high-level functions from input data. The major advantage of DL algorithms is their inherent property of selecting the most relevant features from the training dataset. Therefore, unlike ML algorithms, separate feature selection algorithms are not required (Isensee et al., 2021). In this study, three DL algorithms, DNN, CNN-1D, and CNN-VGG16, were applied. The tenfold cross-validation method was used to assess the model’s efficiency while training. The training dataset was further divided into ten subsets, iteratively training models using all subsets except one held out to test the performance.
For DNN, 2,704 compounds, each encoded with 1,917 features (1,444 physicochemical properties, 166 MACCS, and 307 substructure fingerprints), were used to develop BBB permeability prediction models. Initial layers receive compounds encoded with feature vectors and subject them to the hidden layers. These hidden layers obtain the relevant information from the input vectors and project the freshly extracted features to the batch normalization layer. This layer increases the training process by reducing the intradata covariance. Dropout layers were applied to reduce the problem of coadaptation of neurons and overfitting (Baldi and Sadowski, 2014). These layers randomly drop the nodes as per the dropout rate. Rectified linear unit (ReLU) activation function was used, which adaptively transforms rectifier parameters. Furthermore, ReLU transforms the neuronal output by mapping it to the highest possible value or zero (if the value is negative) (Wang et al., 2021b). ReLU function is given in Eq. 4.
where xi is input for activation function f on channel “i.”
The “softmax” activation function was applied on the output layer to map the hidden layer output between 0 to 1 intervals. The Adam optimizer was used to minimize the loss value from the cross-entropy cost function.
The network performance of a DNN depends upon its depth and breadth. Therefore, it is vital to determine the optimal depth and breadth and optimize other parameters, e.g., the learning rate and dropout ratio. To achieve this, we kept other parameters fixed and evaluated the prediction accuracy by varying the hidden layers (K = 1–5) and neurons (100, 200, 300, 500, 800 neurons per layer). The DNNs were also simultaneously evaluated for five dropout ratios (0.1, 0.2, 0.3, 0.4, 0.5), and prediction accuracy was evaluated. Furthermore, various network configurations were evaluated for epochs (100, 200, 400, 500, 800) and learning rates (0.0001, 0.0002, 0.0003, 0.001, 0.002, 0.003) optimization. Table 3 summarizes the explored values of hyperparameters for the development of the DNN-based BBB permeability prediction model.
CNN is a particular type of DL that is widely used for image data classification (LeCun et al., 2015). There are three major layers in the CNN: convolutional, pooling, and fully connected layers. Cube-shaped weights and multiple filters (kernels) are applied in the convolutional layers to extract features and develop feature maps from the images (Esteva et al., 2017; Malik et al., 2021; Shan et al., 2021). The filter size may downsample the outputs; therefore, the size and number of kernels are vital (D’souza et al., 2020). To overcome the issue of downsampling, an optimized padding value is applied, which allows the filter kernels to create feature maps of the input image size.
Furthermore, other parameters of the convolutional layer also needed to be optimized, e.g., regularization type and value, activation function, and stride. Pooling layers specifically perform average or max-pooling in the filter region to lower the number of parameters and calculations by downsampling the representations. The fully connected layers flatten the output prior to the classification and are usually kept at the end. CNNs are created to process and learn from images. However, CNN-1D can be applied similarly to one-dimensional data containing physicochemical properties and fingerprints. We used three filters (15, 32, 64) to determine the local pattern in the 1,917 features, which were calculated from PaDel. After the CNN layers, dense layers (1 and 2) were tested for three dropout ratios (0.2, 0.3, 0.5). Table 4 summarizes the explored hyperparameter values for the development of the CNN-1D model.
The CNN processes the input 2D images to distinguish the image objects by allocating weights and biases. CNN captures temporal and spatial relationships using the tiny squares of input images by processing them through a series of convolution layers. Filters in each convolutional layer skid on the image to find relevant and specific features, e.g., edge detection, sharpen or blur the image and produce the feature map. The feature map’s size depends on filter numbers, filter slide-over pixels, and zero-padding (image borders are padded with zero). The 2D-array values of the feature map were subjected to the individual layer activation function (ReLU). Dimensionality reduction of each feature map is processed using pooling without any loss of information. The pooling layer’s output is sent into fully connected layers, which classify the images. The CNN with transfer learning (VGG16) was used in this study using RDKit-generated images. The images were scaled to a pixel size of 128 * 128 to develop and validate the BBB permeability prediction model.
Furthermore, image data argumentation was performed by randomly zooming (up to 10%) and flipping the images. The CNN (VGG16) hyperparameters are given in Table 5. The developed model was tested with an independent test set consisting of 901 images. Figure 2 depicts the adopted methodology to develop the DL-based prediction models.
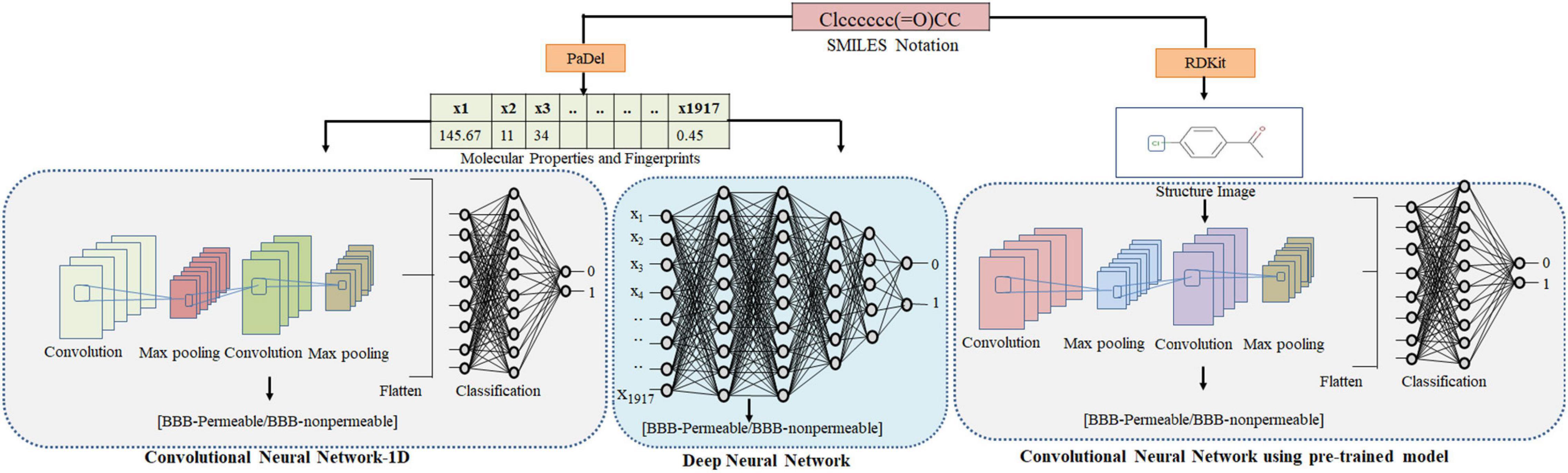
Figure 2. Methodology adopted for predicting the BBB permeability of compounds using SMILES notation. The SMILES notations were used to calculate molecular properties and fingerprints using PaDel. These features were used as input to DNN and CNN-1D to generate the BBB permeability prediction models. The 2D images of compounds were generated using RDkit and fed to the CNN (VGG16) to generate the BBB permeability prediction model.
The performance metrics of the ten developed models (ML = 7, DL = 3) for BBB permeability prediction were compared to determine the best-performing model. The performance indicators used in this study were area under the curve (AUC), area under the precision-recall curve (AUPRC), average precision (AP), F1 score and accuracy, and Hamming distance (HD) of the prediction models. Among the developed ML prediction models, the SVM (RBF kernel)-based prediction model outperformed the NB, kNN, and RF algorithms for BBB permeability prediction with test set data. The accuracy of SVM (RBF) was found to be approximately 6% higher than that of NB and RF and approximately 1% higher than that of kNN. Moreover, SVM (RBF) yielded better prediction values of other performance indicators in BBB permeability prediction on the given dataset. However, the performance metrics of SVM (polynomial) at degree 3 were found to be very comparable to the SVM (RBF).
The performance metrics of the DL algorithms were found to be very close to each other. The prediction accuracies of DNN, CNN-1D, and CNN (VGG16) were 98.07, 97.44, and 97.66, respectively. However, the DNN model was superior in AUC, AUPRC, AP, F1, and HD when compared to that of CNN-1D and CNN (VGG16) (Table 6). The comparison of receiver operating characteristic (ROC) curves between SVM (RBF), DNN, CNN-1D, and CNN (VGG16) also indicates the superiority of DNN in BBB permeability prediction with the given dataset (Figure 3). Furthermore, the accuracy and loss plots of the DNN model are given in Figure 4. The accuracy plot shows good coherence between the training (red) and test (blue) accuracy, suggesting that the model is not overfitted. Additionally, coherence in the training (red) and validation/test (blue) loss in the loss plot (binary cross-entropy loss) is indicative of an unbiased model (Figure 4).
The better performance of DL algorithms compared to ML could be due to their ability to handle the large dataset and extract the most relevant features of their own. The performance metrics of DNN, CNN-1D, and CNN (VGG16) were very comparable. To our surprise, DNN was found to be slightly better in overall performance based on accuracy and other performance indicators compared to the CNN models. DNN appears to be a better option to handle the compounds encoded with physicochemical and fingerprint features for classifications and predictions. Based on the overall performance, we selected the DNN model for the development of “DeePred-BBB.” DeePred-BBB can predict BBB permeability based on chemical SMILES notation. It uses PaDel to calculate the features from the SMILES notation and sends them as input to the DNN model. The output is either permeable or non-permeable.
Compounds can penetrate the BBB using various different mechanisms, such as transmembrane diffusion, adsorptive endocytosis, saturable transporters, and extracellular pathways. Most drugs in clinical use till date are small, lipid soluble molecules that cross the BBB by transmembrane diffusion. The prediction models to determine the mechanism of BBB permeability require a mechanism-based set of compounds with their permeability class labels for each mechanism. However, the current study deals with the prediction of the BBB permeability of compounds (irrespective of how they penetrate the BBB) using their SMILES notations. The study holds limitations in identifying the mechanism by which compounds are BBB permeable. DeePred-BBB does not take multiple SMILES notations for prediction. The user needs to input the SMILES notations of compounds one at a time for accurate prediction of BBB permeability. A comparison between DeePred-BBB and previously reported BBB permeability prediction models is given in Table 7.
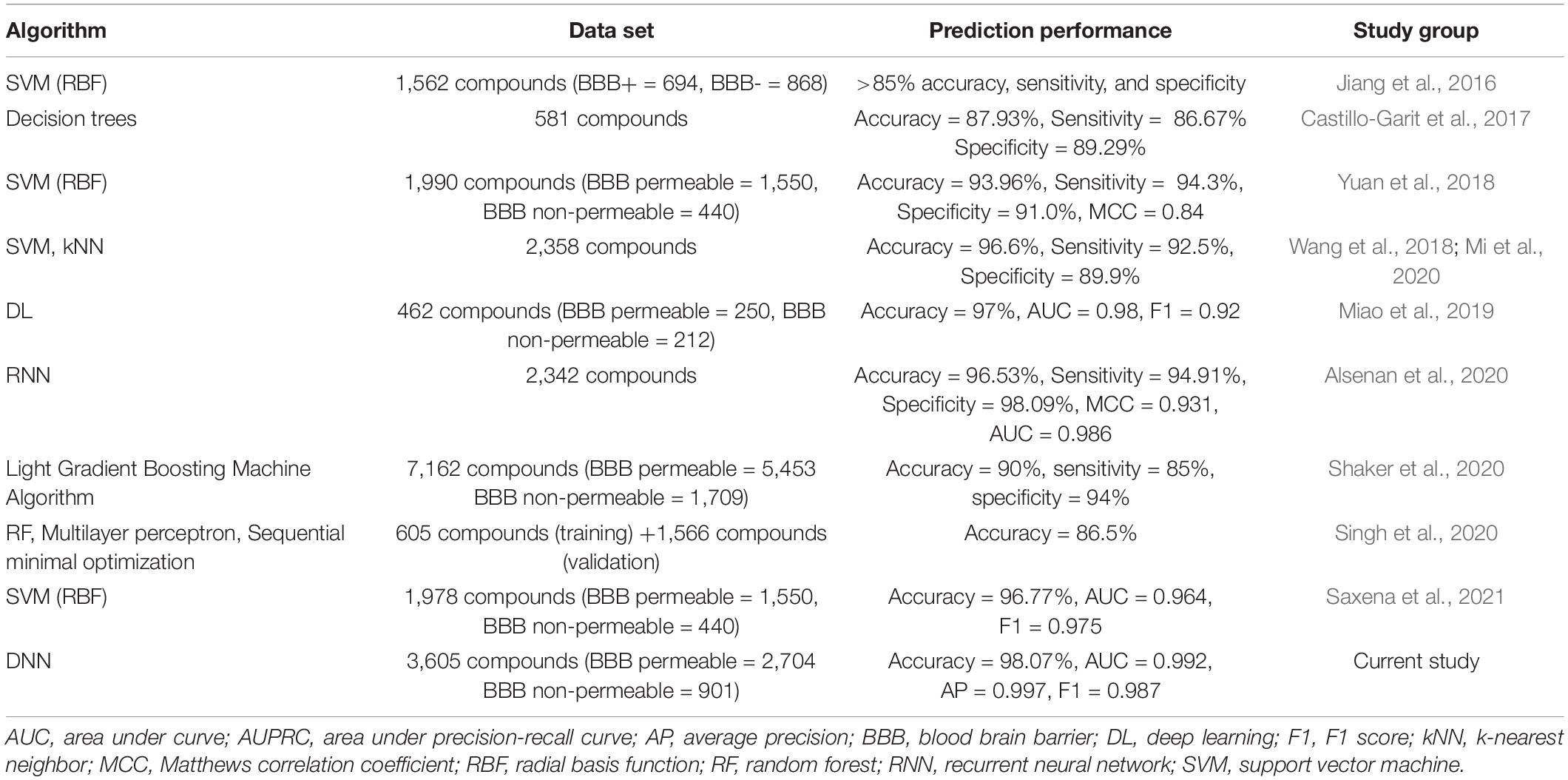
Table 7. Comparative analysis of DeePred-BBB with recently published BBB permeability prediction models.
Deep learning and machine learning algorithms were applied to a dataset of 3,605 compounds to develop a prediction model that could accurately predict the BBB permeability of compounds using their SMILES notations as input. The comparative analysis of the performance metrics of the developed models suggested that the overall performance of DNN-based BBB permeability prediction is better than that of the ML and CNN models. It was discovered that the notion of “deeper the network, better the accuracy” does not often hold true. An optimal depth of the network is required beyond which the performance of the network does not improve. A DNN model with three layers (depth) having 200, 100, and 2 nodes each was the most accurate. It was also observed that in the case of compounds, the physicochemical properties and fingerprint-based DL models yield slightly better performance than 2D-structure image-based models in BBB permeability prediction.
Based on this study, we propose the DeePred-BBB model for BBB permeability prediction of compounds using their SMILES notations as input. In DeePred-BBB, the best performing DNN model is integrated with the open-source PaDel tool to calculate features. The calculated features are automatically fed to the DNN model as input, which predicts whether the compound will be BBB permeable or non-permeable. DeePred-BBB could assist in making quality decisions regarding which compound to carry forward in subsequent drug development stages and could potentially help in reducing the attrition rate of CNS-acting drug candidates failing due to BBB non-permeability. Inevitably, such drug candidates need further in vivo validation to arrive at efficacious and safe drugs at a faster rate and lower cost. DeePred-BBB could be accessed at https://github.com/12rajnish/DeePred-BBB.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
RK and AS access to all of the data analyzed in this study, drafted the manuscript, and performed the statistical analysis. RK takes responsibility for the integrity and accuracy of the study data analysis and results. RK, AS, AA and GA involved in the study design, concept, analysis, and interpretation of data. AB, MK, AA, and GA involved in critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
The Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia, has funded this project under Grant No. KEP-36-130-42. The authors therefore acknowledge with thanks DSR’s technical and financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2022.858126/full#supplementary-material
Abbott, N. J., Ronnback, L., and Hansson, E. (2006). Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 7, 41–53. doi: 10.1038/nrn1824
Aldewachi, H., Al-Zidan, R. N., Conner, M. T., and Salman, M. M. (2021). High-throughput Screening Platforms in the discovery of novel drugs for neurodegenerative diseases. Bioengineering 8:30. doi: 10.3390/bioengineering8020030
Alsenan, S., Al-Turaiki, I., and Hafez, A. (2020). A Recurrent neural network model to predict blood-brain barrier permeability. Comput. Biol. Chem. 89:107377. doi: 10.1016/j.compbiolchem.2020.107377
Baldi, P., and Sadowski, P. (2014). The dropout learning algorithm. Artif. Intell. 210, 78–122. doi: 10.1016/j.artint.2014.02.004
Banks, W. A. (2009). Characteristics of compounds that cross the blood-brain barrier. BMC Neurol. 9:S3. doi: 10.1186/1471-2377-9-S1-S3
Benet, L. Z., Hosey, C. M., Ursu, O., and Oprea, T. I. (2016). BDDCS, the rule of 5 and drugability. Adv. Drug Deliv. Rev. 101, 89–98. doi: 10.1016/j.addr.2016.05.007
Ben-Hur, A., Ong, C. S., Sonnenburg, S., Schölkopf, B., and Rätsch, G. (2008). Support vector machines and kernels for computational biology. PLoS Comput. Biol. 4:e1000173. doi: 10.1371/journal.pcbi.1000173
Bento, A. P., Hersey, A., Félix, E., Landrum, G., Gaulton, A., Atkinson, F., et al. (2020). An open source chemical structure curation pipeline using rdkit. j. Cheminform. 12:51. doi: 10.1186/s13321-020-00456-1
Bickel, U. (2005). How to measure drug transport across the blood-brain barrier. NeuroRx 2, 15–26. doi: 10.1602/neurorx.2.1.15
Castillo-Garit, J. A., Casanola-Martin, G. M., Le-Thi-Thu, H., Pham-The, H., and Barigye, S. J. (2017). A Simple method to predict blood-brain barrier permeability of drug- like compounds using classification trees. Med. Chem. 13, 664–669. doi: 10.2174/1573406413666170209124302
Daneman, R. (2015). Prat A. the blood-brain barrier. Cold Spring Harb. Perspect. Biol. 7:a020412. doi: 10.1101/cshperspect.a020412
Dieterich, H. J., Reutershan, J., Felbinger, T. W., and Eltzschig, H. K. (2003). Penetration of intravenous hydroxyethyl starch into the cerebrospinal fluid in patients with impaired blood-brain barrier function. Anesth. Analg. 96, 1150–1154. doi: 10.1213/01.ane.0000050771.72895.66
Doniger, S., Hofmann, T., and Yeh, J. (2002). Predicting CNS permeability of drug molecules: comparison of neural network and support vector machine algorithms. J. Comput. Biol. 9, 849–864. doi: 10.1089/10665270260518317
D’souza, R. N., Huang, P. Y., and Yeh, F. C. (2020). Structural analysis and optimization of convolutional neural networks with a small sample size. Sci. Rep. 10:834. doi: 10.1038/s41598-020-57866-2
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., et al. (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118. doi: 10.1038/nature21056
Feigin, V. L., Vos, T., Nichols, E., Owolabi, M. O., Carroll, W. M., Dichgans, M., et al. (2020). The global burden of neurological disorders: translating evidence into policy. Lancet Neurol. 19, 255–265. doi: 10.1016/S1474-4422(19)30411-9
Gaudillo, J., Rodriguez, J. J. R., Nazareno, A., Baltazar, L. R., Vilela, J., Bulalacao, R., et al. (2019). Machine learning approach to single nucleotide polymorphism-based asthma prediction. PLoS One 14:e0225574. doi: 10.1371/journal.pone.0225574
Ghandi, M., Lee, D., Mohammad-Noori, M., and Beer, M. A. (2014). Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 10:e1003711. doi: 10.1371/journal.pcbi.1003711
Giuliani, A. (2017). The application of principal component analysis to drug discovery and biomedical data. Drug Discov. Today 22, 1069–1076. doi: 10.1016/j.drudis.2017.01.005
Gupta, M., Lee, H. J., Barden, C. J., and Weaver, D. F. (2019). The blood-brain barrier (BBB) Score. J. Med. Chem. 62, 9824–9836. doi: 10.1021/acs.jmedchem.9b01220
Hendricks, B. K., Cohen-Gadol, A. A., and Miller, J. C. (2015). Novel delivery methods bypassing the blood-brain and blood-tumor barriers. Neurosurg. Focus 38:E10. doi: 10.3171/2015.1.FOCUS14767
Hu, Y., Zhao, T., Zhang, N., Zhang, Y., and Cheng, L. (2019). A Review of recent advances and research on drug target identification methods. Curr. Drug Metab. 20, 209–216. doi: 10.2174/1389200219666180925091851
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J., and Maier-Hein, K. H. (2021). NNU-NET: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211. doi: 10.1038/s41592-020-01008-z
Jiang, L., Chen, J., He, Y., Zhang, Y., and Li, G. (2016). A method to predict different mechanisms for blood-brain barrier permeability of CNS activity compounds in Chinese herbs using support vector machine. J. Bioinform. Comput. Biol. 14:1650005. doi: 10.1142/S0219720016500050
Khan, A. I., Lu, Q., Du, D., Lin, Y., and Dutta, P. (2018). Quantification of kinetic rate constants for transcytosis of polymeric nanoparticle through blood-brain barrier. Biochim. Biophys Acta Gen. Subj. 1862, 2779–2787. doi: 10.1016/j.bbagen.2018.08.02016
Kumar, R., Sharma, A., Siddiqui, M. H., and Tiwari, R. K. (2018). Promises of machine learning approaches in prediction of absorption of compounds. Mini. Rev. Med. Chem. 18, 196–207. doi: 10.2174/1389557517666170315150116
Kumar, R., Sharma, A., Varadwaj, P., Ahmad, A., and Ashraf, G. M. (2011). Classification of oral bioavailability of drugs by machine learning approaches: a comparative study. J. Comp. Interdisc. Sci. 2, 179–196. doi: 10.6062/jcis.2011.02.03.0045
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Lovrić, M., Molero, J.M., Kern, R., Spark, P. Y., and Kit, R. D. (2019). Moving towards big data in cheminformatics. Mol. Inform. 38:e1800082. doi: 10.1002/minf.201800082
Main, B. S., Villapol, S., Sloley, S. S., Barton, D. J., Parsadanian, M., Agbaegbu, C., et al. (2018). Apolipoprotein E4 impairs spontaneous blood brain barrier repair following traumatic brain injury. Mol. Neurodegener 13:17. doi: 10.1186/s13024-018-0249-5
Malik, J., Kiranyaz, S., and Gabbouj, M. (2021). Self-organized operational neural networks for severe image restoration problems. Neural. Netw. 135, 201–211. doi: 10.1016/j.neunet.2020.12.014
Massey, S. C., Urcuyo, J. C., Marin, B. M., Sarkaria, J. N., and Swanson, K. R. (2020). Quantifying glioblastoma drug response dynamics incorporating treatment sensitivity and blood brain barrier penetrance from experimental data. Front. Physiol. 11:830. doi: 10.3389/fphys.2020.00830
Menken, M., Munsat, T. L., and Toole, J. F. (2000). The global burden of disease study: implications for neurology. Arch. Neurol. 57, 418–420. doi: 10.1001/archneur.57.3.418
Mi, Y., Mao, Y., Cheng, H., Ke, G., Liu, M., Fang, C., et al. (2020). Studies of blood-brain barrier permeability of gastrodigenin in vitro and in vivo. Fitoterapia 140, 104447. doi: 10.1016/j.fitote.2019.104447
Miao, R., Xia, L. Y., Chen, H. H., Huang, H. H., and Liang, Y. (2019). Improved classification of blood-brain-barrier drugs using deep learning. Sci. Rep. 9:8802. doi: 10.1038/s41598-019-44773-4
Pardridge, W. M. (2002). Why is the global CNS pharmaceutical market so under-penetrated? Drug discov. Today 7, 5–7. doi: 10.1016/s1359-6446(01)02082-7
Pardridge, W. M. (2005). The blood-brain barrier: bottleneck in brain drug development. NeuroRx 2, 3–14. doi: 10.1602/neurorx.2.1.3
Reese, T. S., and Karnovsky, M. J. (1967). Fine structural localization of a blood-brain barrier to exogenous peroxidase. J. Cell. Biol. 34, 207–217. doi: 10.1083/jcb.34.1.207
Rhea, E. M., and Banks, W. A. (2019). Role of the Blood-Brain Barrier in Central Nervous System Insulin Resistance. Front. Neurosci. 13:521. doi: 10.3389/fnins.2019.00521
Roy, D., Hinge, V. K., and Kovalenko, A. (2019). To Pass or Not To Pass: Predicting the Blood-Brain Barrier Permeability with the 3D-RISM-KH Molecular Solvation Theory. ACS Omega. 4, 16774–16780. doi: 10.1021/acsomega.9b01512
Salman, M. M., Al-Obaidi, Z., Kitchen, P., Loreto, A., Bill, R. M., and Wade-Martins, R. (2021). Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases. Int. J. Mol. Sci. 22:4688. doi: 10.3390/ijms22094688
Salman, M. M., Kitchen, P., Yool, A. J., and Bill, R. M. (2022). Recent breakthroughs and future directions in drugging aquaporins. Trends Pharmacol. Sci. 43, 30–42. doi: 10.1016/j.tips.2021.10.009
Salman, M. M., Marsh, G., Kusters, I., Delincé, M., Di Caprio, G., Upadhyayula, S., et al. (2020). Design and validation of a human brain endothelial microvessel-on-a-chip open microfluidic model enabling advanced optical imaging. Front. bioeng. biotechnol. 8:573775. doi: 10.3389/fbioe.2020.573775
Salmanpour, M. R., Shamsaei, M., Saberi, A., Klyuzhin, I. S., Tang, J., Sossi, V., et al. (2020). Machine learning methods for optimal prediction of motor outcome in parkinson’s disease.. Phys. Med. 69, 233–240. doi: 10.1016/j.ejmp.2019.12.022
Saunders, N. R., Dreifuss, J. J., Dziegielewska, K. M., Johansson, P. A., Habgood, M. D., Møllgård, K., et al. (2014). The rights and wrongs of blood-brain barrier permeability studies: a walk through 100 years of history. Front. Neurosci. 8:404. doi: 10.3389/fnins.2014.00404
Saxena, D., Sharma, A., Siddiqui, M. H., and Kumar, R. (2019). Blood brain barrier permeability prediction using machine learning techniques: an update. Curr. Pharm. Biotechnol. 20, 1163–1171. doi: 10.2174/1389201020666190821145346
Saxena, D., Sharma, A., Siddiqui, M. H., and Kumar, R. (2021). Development of machine Learning based blood-brain barrier permeability prediction models using physicochemical properties, maccs and substructure fingerprints. Curr. Bioinform. 16, 855–864. doi: 10.2174/1574893616666210203104013
Schidlowski, M., Boland, M., Rüber, T., and Stöcker, T. (2020). Blood-brain barrier permeability measurement by biexponentially modeling whole-brain arterial spin labeling data with multiple T2 -weightings. NMR Biomed. 33:e4374. doi: 10.1002/nbm.4374
Schlageter, K. E., Molnar, P., Lapin, G. D., and Groothuis, D. R. (1999). Microvessel organization and structure in experimental brain tumors: microvessel populations with distinctive structural and functional properties. Microvasc. Res. 58, 312–328. doi: 10.1006/mvre.1999.2188
Shaker, B., Yu, M. S., Song, J. S., Ahn, S., Ryu, J. Y., Oh, K. S., et al. (2020). LightBBB: Computational prediction model of blood-brain-barrier penetration based on LightGBM. Bioinformatics 37, 1135–1139. doi: 10.1093/bioinformatics/btaa918
Shan, W., Li, X., Yao, H., and Lin, K. (2021). Convolutional neural network-based virtual screening. Curr. Med. Chem. 28, 2033–2047. doi: 10.2174/0929867327666200526142958
Sharma, A., Kumar, R., Ranjta, S., and Varadwaj, P. K. (2021). SMILES to Smell: Decoding the structure-odor relationship of chemical compounds using the deep neural network approach. J. Chem. Inf. Model 61, 676–688. doi: 10.1021/acs.jcim.0c01288
Shen, J., Cheng, F., Xu, Y., Li, W., and Tang, Y. (2010). Estimation of ADME properties with substructure pattern recognition. J. Chem. Inf. Model 50, 1034–1041. doi: 10.1021/ci100104j
Shen, Y., Li, Y., Zheng, H. T., Tang, B., and Yang, M. (2019). Enhancing ontology-driven diagnostic reasoning with a symptom-dependency-aware naïve bayes classifier. BMC Bioinform 20:330. doi: 10.1186/s12859-019-2924-0
Singh, M., Divakaran, R., Konda, L. S. K., and Kristam, R. (2020). A classification model for blood brain barrier penetration. J. Mol. Graph. Model. 96:107516. doi: 10.1016/j.jmgm.2019.107516
Suenderhauf, C., Hammann, F., and Huwyler, J. (2012). Computational prediction of blood-brain barrier permeability using decision tree induction. Molecules 17, 10429–10445. doi: 10.3390/molecules170910429
Tietz, S., and Engelhardt, S. B. (2015). Brain barriers: crosstalk between complex tight junctions and adherens junctions. J. Cell. Biol. 209, 493–506. doi: 10.1083/jcb.201412147
van Tellingen, O., Yetkin-Arik, B., de Gooijer, M. C., Wesseling, P., Wurdinger, T., and de Vries, H. E. (2015). Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 19, 1–12. doi: 10.1016/j.drup.2015.02.002
Veber, D. F., Johnson, S. R., Cheng, H. Y., Smith, B. R., Ward, K. W., and Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. J.Med. Chem. 45, 2615–2623. doi: 10.1021/jm020017n
Wang, D., Zeng, J., and Lin, S. B. (2021b). Random Sketching for neural networks with relu. IEEE Trans Neural. Netw. Learn. Syst. 32, 748–762. doi: 10.1109/TNNLS.2020.2979228
Wang, M. W. H., Goodman, J. M., and Allen, T. E. H. (2021a). Machine learning in predictive toxicology: recent applications and future directions for classification models. Chem. Res. Toxicol. 34, 217–239. doi: 10.1021/acs.chemrestox.0c00316
Wang, Z., Yang, H., Wu, Z., Wang, T., Li, W., Tang, Y., et al. (2018). In silico prediction of blood-brain barrier permeability of compounds by machine learning and resampling methods. Chem. Med. Chem. 13, 2189–2201. doi: 10.1002/cmdc.201800533
Wevers, N. R., Kasi, D. G., Gray, T., Wilschut, K. J., Smith, B., van Vught, R., et al. (2018). A perfused human blood-brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids barriers CNS 15:23. doi: 10.1186/s12987-018-0108-3
Yang, L., Wu, H., Jin, X., Zheng, P., Hu, S., Xu, X., et al. (2020). Study of cardiovascular disease prediction model based on random forest in eastern China. Sci. Rep. 10:5245. doi: 10.1038/s41598-020-62133-5
Yao, D., Zhan, X., Zhan, X., Kwoh, C. K., Li, P., and Wang, J. (2020). A random forest based computational model for predicting novel lncrna-disease associations. BMC Bioinform 21:126. doi: 10.1186/s12859-020-3458-1
Yap, C. W. (2011). PADEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474. doi: 10.1002/jcc.21707
Yuan, Y., Zheng, F., and Zhan, C. G. (2018). Improved prediction of blood-brain barrier permeability through machine learning with combined use of molecular property-based descriptors and fingerprints. AAPS J. 20:54. doi: 10.1208/s12248-018-0215-8
Zhang, D., Xiao, J., Zhou, N., Zheng, M., Luo, X., Jiang, H., et al. (2015). A genetic algorithm based support vector machine model for blood-brain barrier penetration prediction. Biomed. Res. Int. 2015:292683. doi: 10.1155/2015/292683
Zhang, L., Zhu, H., Oprea, T. I., Golbraikh, A., and Tropsha, A. (2008). QSAR 21odelling of the blood-brain barrier permeability for diverse organic compounds. Pharm. Res. 25, 1902–1914. doi: 10.1007/s11095-008-9609-0
Keywords: blood-brain barrier, convolutional neural network, deep learning, machine learning, prediction, CNS-permeability
Citation: Kumar R, Sharma A, Alexiou A, Bilgrami AL, Kamal MA and Ashraf GM (2022) DeePred-BBB: A Blood Brain Barrier Permeability Prediction Model With Improved Accuracy. Front. Neurosci. 16:858126. doi: 10.3389/fnins.2022.858126
Received: 19 January 2022; Accepted: 14 March 2022;
Published: 03 May 2022.
Edited by:
Corinne Lasmezas, The Scripps Research Institute, United StatesReviewed by:
Sezen Vatansever, Icahn School of Medicine at Mount Sinai, United StatesCopyright © 2022 Kumar, Sharma, Alexiou, Bilgrami, Kamal and Ashraf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ghulam Md Ashraf, YXNocmFmLmdtQGdtYWlsLmNvbQ==, Z2FzaHJhZkBrYXUuZWR1LnNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.