- 1Department of Biology, New York University, New York, NY, United States
- 2Université de Paris, Centre National de la Recherche Scientifique, Institut Jacques Monod, Paris, France
The nervous system is one of the most sophisticated animal tissues, consisting of thousands of interconnected cell types. How the nervous system develops its diversity from a few neural stem cells remains a challenging question. Spatial and temporal patterning mechanisms provide an efficient model through which diversity can be generated. The molecular mechanism of spatiotemporal patterning has been studied extensively in Drosophila melanogaster, where distinct sets of transcription factors define the spatial domains and temporal windows that give rise to different cell types. Similarly, in vertebrates, spatial domains defined by transcription factors produce different types of neurons in the brain and neural tube. At the same time, different cortical neuronal types are generated within the same cell lineage with a specific birth order. However, we still do not understand how the orthogonal information of spatial and temporal patterning is integrated into the progenitor and post-mitotic cells to combinatorially give rise to different neurons. In this review, after introducing spatial and temporal patterning in Drosophila and mice, we discuss possible mechanisms that neural progenitors may use to integrate spatial and temporal information. We finally review the functional implications of spatial and temporal patterning and conclude envisaging how small alterations of these mechanisms can lead to the evolution of new neuronal cell types.
Spatiotemporal Patterning And Cell Fate Determination
Traditionally, neuronal types were classified by the distinct functions that they perform (Masland, 2004; Zeng and Sanes, 2017). Neurons can be categorized by their arborization and projection pattern, the neurotransmitter they use, and their electrophysiology. Neurons can also be categorized by the genes they express. The recent advent of high-throughput sequencing has demonstrated that molecular cell types and morphological/functional cell types are largely consistent (Shekhar et al., 2016; Li et al., 2017; Konstantinides et al., 2018b; Sathyamurthy et al., 2018; Delile et al., 2019; Allen et al., 2020; Özel et al., 2020; Yao et al., 2021). How different types of neurons are generated is one of the central questions in developmental neurobiology; the study of spatiotemporal patterning aims to understand the molecular developmental basis of cell type diversity. By deciphering the logic underpinning the patterning process, we can potentially understand how distinct neuronal functions are encoded genetically. Comparing different patterning mechanisms across species could also shed light on how new cell types evolve to generate a more complex nervous system (Holguera and Desplan, 2018; Konstantinides et al., 2018a).
Spatial Patterning
Early in animal development, neural progenitors are partitioned into different domains that express distinct transcription factors (Figure 1A). For example, the nervous system of flies and mammals is patterned along the anteroposterior (A–P) axis by Hox genes (Tümpel et al., 2009; Philippidou and Dasen, 2013; Estacio-Gómez and Díaz-Benjumea, 2014; Jung et al., 2014; Figure 1B). Within each segment along the A–P axis, neural progenitors are further patterned on the A-P and dorsoventral (D–V) axis, forming non-intermingling progenitor populations that will generate distinct neuronal types (Jessell, 2000; Briscoe and Ericson, 2001; Karlsson et al., 2010; Benito-Sipos et al., 2013; Birkholz et al., 2013; Figure 1C).
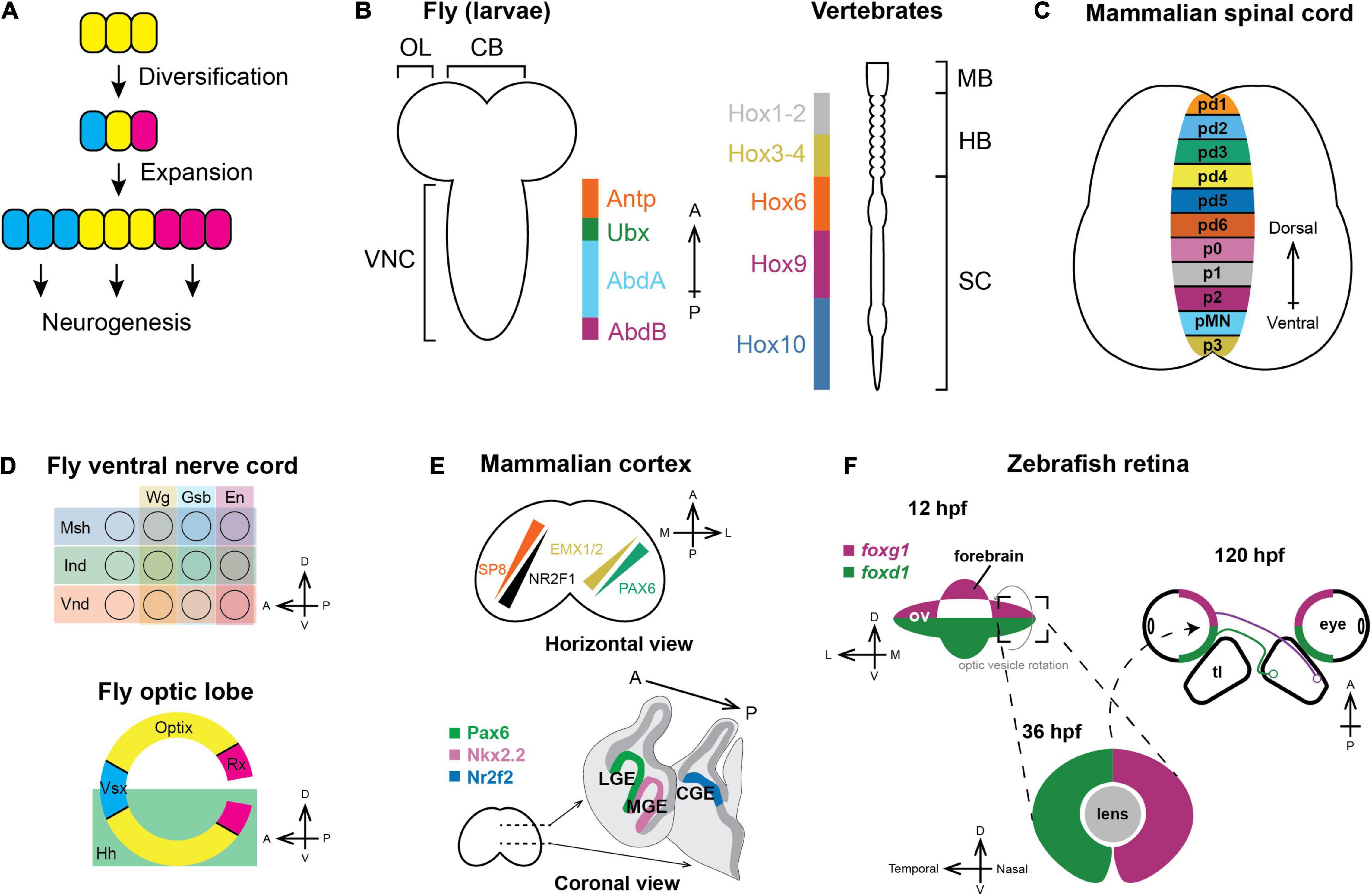
Figure 1. Spatial patterning in neurogenesis. (A) General principles of spatial patterning in neurogenesis: Progenitors expand in number and then diversify the set of neuronal types that they can generate during neurogenesis, which is represented by different colors. (B) Nervous systems of vertebrates and invertebrates are patterned along the anteroposterior axis by Hox genes. Orthologous Hox genes have the same color (McGinnis and Krumlauf, 1992). (OL: optic lobe; CB: central brain; VNC: ventral nerve cord; MB: midbrain; HB; hindbrain; SC: spinal cord). (C) Vertebral neural tubes are patterned along the dorsoventral axis and form multiple progenitor domains that will generate spinal motor neurons and various interneurons (pd1-6: Progenitors for dorsal interneurons dI1-dI6; p0-3: Progenitors for ventral interneuron V0–V3; pMN: Spinal motor neuron progenitors). (D) Each segment of the fly ventral nerve cord (top) is partitioned along both the A–P and D–V axis by the expression of spatial factors; similarly, the Outer Proliferation Center neuroepithelium generating the medulla (bottom) is compartmentalized into six distinct domains. (E) Upper panel: In mammals, the cortical protomap is defined by the graded expression of transcription factors. SP8 and NR2F1 form opposing gradients where SP8 is highest on the anteromedial side while NR2F1 is highest on the posterolateral side. PAX6 and EMX1/2 form another set of opposing gradients along the anterolateral-posteromedial axis (Reviewed in Greig et al., 2013; Cadwell et al., 2019). Lower panel: Distinct subpallial progenitor domains are marked by specific transcription factors and generate different types of cortical inhibitory interneurons (LGE: Lateral ganglionic eminence; MGE: Medial ganglionic eminence; CGE: Caudal ganglionic eminence). (F) In zebrafish, the optic vesicle is dorsoventrally patterned by the expression of foxg1 and foxd1. During developmental eye rotation, the D–V axis becomes the nasotemporal axis, and the nasal hemiretina projects to posterior tectal lobe, while the temporal hemiretina projects to the anterior tectal lobe (hpf: hours post fertilization; ov: optic vesicle; tl: tectal lobe).
In each segment of the fly ventral nerve cord, neuroepithelial cells are patterned by the expression of msh, ind, and vnd along the D–V axis (Mellerick and Nirenberg, 1995; D’Alessio and Frasch, 1996; Isshiki et al., 1997; Weiss et al., 1998) and wg, gsb, en along the A–P axis (Doe, 1992; Gutjahr et al., 1993; Skeath et al., 1995; Figure 1C). In the fly optic lobe, neuroepithelial cells of the Outer Proliferation Center form a crescent containing six domains defined by Vsx1, Optix, and Rx along the A–P axis and by Hedgehog along the D–V axis (Erclik et al., 2008, 2017; Gold and Brand, 2014; Figure 1D).
In the mammalian forebrain, morphogen gradients pattern radial glia and induce the expression of two orthogonal gradients of spatial transcription factors (Figure 1E, upper panel) that govern the potency of the neural progenitors to generate specific cell types. Different types of cortical interneurons are also made by spatially distinct progenitors. The medial ganglionic eminence expresses Nkx2.1 and is the main source of parvalbumin-and somatostatin-expressing interneurons (Xu, 2004; Flames et al., 2007; Fogarty et al., 2007; Xu et al., 2008), the caudal ganglionic eminence expresses Nr2f2 and generates serotonin-sensing interneurons (Nery et al., 2002; Tripodi et al., 2004; Kanatani et al., 2008; Lee et al., 2010), while the Pax6-expressing lateral ganglionic eminence generates interneurons in the olfactory bulb and the striatum (Wichterle et al., 2001; Kohwi et al., 2007; Figure 1E, lower panel). Another classic example of spatial patterning in vertebrates is the developing retina. In zebrafish, neuroepithelial cells in the optic vesicle are patterned along the D–V axis with foxd1 being expressed in the ventral compartment and foxg1 in the dorsal (Hatini et al., 1994; Yuasa et al., 1996; Picker and Brand, 2005). During anterior eye rotation when the optic cup forms, the D-V spatial domains become the nasal (projecting to the posterior tectum) and temporal (projecting to the anterior tectum) hemiretina, respectively (Figure 1F).
Temporal Patterning
The association between time and neural diversity has been known for a long time in mammals. Layers of morphologically distinct neurons in the mammalian cortex are born at specific embryonic days, with each layer projecting to and receiving information from different brain regions (Rakic, 1974; Raedler and Raedler, 1978; Royce, 1983). Two mechanisms could be envisaged to explain how temporal differences result in the generation of distinct neuronal types. First, temporal differences could be intrinsic to each neural progenitor, in which case each progenitor undergoes a cascade of changes to generate different types of neurons at different time points (Figure 2A). Consistent with this model, clonal lineage tracing showed that a single ventricular radial glia cell can generate neurons in all layers (Gao et al., 2014). Alternatively, the neural progenitor pool could be a mixture of different populations that differ in when they start neurogenesis. In this model, progenitors differ both in their potential of generating neurons and the time they commit to neurogenesis (Figure 2A). In support of this model, while the Olig2-expressing progenitors give rise to both early-born spinal motor neurons and late-born oligodendrocytes, there are two subtypes of progenitors: the early ones that generate motor neurons and the ones that are recruited later and only generate oligodendrocytes (Ravanelli and Appel, 2015).
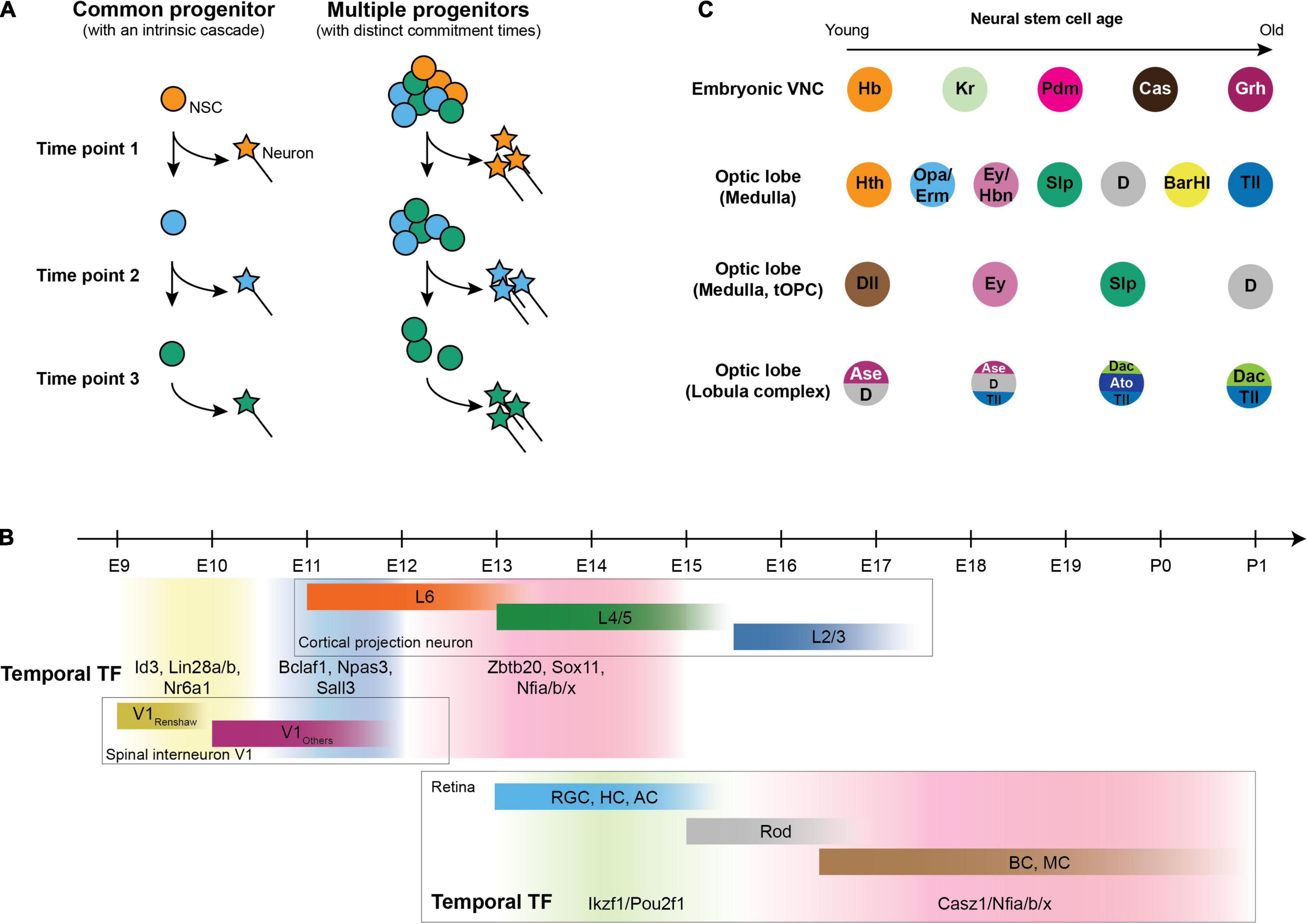
Figure 2. Temporal patterning in neurogenesis. (A) Generating distinct neurons at different stages could be either achieved by a common multipotent progenitor that undergoes a cascade (left) or by a pool of progenitors that differ both in potency and in the time to commit to neurogenesis (right). (B) Mammalian temporal patterning and neurons generated in each temporal window. Upper panel: Birth date of cortical projection neurons (Caviness, 1982) and V1 spinal interneuron (Stam et al., 2012) with temporal transcription factor expression in neural progenitors at each temporal window (Sagner et al., 2021); lower panel: Birth date of retinal cells and corresponding temporal transcription factors in retinal progenitors [RGC: retinal ganglion cell; HC: horizontal cell; AC: amacrine cell; Rod: rod cell (photoreceptor); BC: bipolar cell; MC: Müller cell] (Elliott et al., 2008; Mattar et al., 2015; Javed et al., 2020). (C) Different regions in the fly nervous system undergo different cascades of temporal factors to generate different types of neurons.
In mammals, recent studies have systematically discovered a set of birthdate markers that are shared between different cell types in the hindbrain and spinal cord of human and mouse (Delile et al., 2019; Osseward et al., 2021; Rayon et al., 2021; Figure 2B), and the expression of these markers are consistent with known subtypes within several cardinal classes of spinal neurons (Roy et al., 2012; Bikoff et al., 2016; Hayashi et al., 2018). Some of these birthdate markers are required for early-vs. late-born neuronal fate in neurons (e.g., in the mammalian cortex, Satb2 is required for later-born callosal neurons from layer 2 to 5, and Fezf2 and Ctip2 are necessary for early-born ones in layer 5) (Chen et al., 2005, 2008; Alcamo et al., 2008; Britanova et al., 2008) and in progenitors (e.g., Nfia and Nfib for the generation of late-born neurons in the retina and ventral spinal interneurons) (Xie et al., 2020; Sagner et al., 2021). Consistent with the idea that neural progenitor temporal factors specify the fate of daughter neurons, cortical neurons inherit the gene modules that are present in radial glia at the stage when the neurons were generated (Telley et al., 2019), and the late-born fate regulators Nfia/b/x directly regulate late-born fate associated genes in the mammalian retina (Clark et al., 2019; Xie et al., 2020; Lyu et al., 2021). Whether it is a general rule that these temporal window-specific gene modules specify the neuronal fates of their daughter cells warrants further investigation.
In the nervous system of flies, cascades of transcription factors expressed in the same neural stem cell underlie temporal patterning, and different regions utilize a distinct set of temporal transcription factors (Brody and Odenwald, 2000; Isshiki et al., 2001; Li et al., 2013; Suzuki et al., 2013; Bertet et al., 2014; Apitz and Salecker, 2015; Erclik et al., 2017; Mora et al., 2018; Pinto-Teixeira et al., 2018; Konstantinides et al., 2021; Zhu et al., 2021; Figure 2C). The temporal cascade in neural stem cells progresses by cross-regulation between temporal factors during neurogenesis, either by a relay of activators (Reviewed in Doe, 2017; Rossi et al., 2017) or by a more robust repressor-decay model (Averbukh et al., 2018). As a result, the expression of each temporal factor in progenitors is responsible for the generation of the neuronal types specific to each temporal window (Isshiki et al., 2001; Mettler et al., 2006; Bayraktar and Doe, 2013; Li et al., 2013; Konstantinides et al., 2021; Tang et al., 2021; Zhu et al., 2021).
Molecular Logic of Spatiotemporal Integration
The identity of a neural progenitor is defined by the spatial domain from which it originates; the neural progenitor then undergoes a temporal cascade and generates a specific subset of neuronal types within each temporal window of the cascade. How is spatiotemporal identity in progenitors translated to distinct features that define mature neuronal types? In worms, flies, and mammals, the features that define a type of neuron, including neurotransmitter choices and innervation targets, are regulated by post-mitotic regulators that are required and sufficient for the establishment and/or maintenance of the terminal features. For example, CHE-1 binds to a specific motif and defines ASE neuron fate in worms (Uchida et al., 2003; Etchberger et al., 2007), Bsh is required and sufficient for inducing Mi1 neuron fate in the fly optic lobe (Hasegawa et al., 2013), and Ngn2, Isl1, and Lhx3 specify spinal motor neurons in the mammalian spinal cord (Thaler et al., 2002; Lee and Pfaff, 2003; Son et al., 2011; Mazzoni et al., 2013).
For spatiotemporal patterning to specify neuronal types, each spatial and temporal factor must prime the expression of specific post-mitotic fate regulators. Indeed, the Drosophila Mi1 neuron is generated during the Hth-expressing temporal window, and the temporal factor Hth is required and sufficient for downstream Bsh expression (Hasegawa et al., 2011; Li et al., 2013). Similarly, Nkx6.1 and Olig2 define the motor neuron progenitor domain in the neural tube and are required and sufficient for the expression of downstream Isl1 and Lhx3 (Sander et al., 2000; Takebayashi et al., 2002).
But how do spatial and temporal factors cooperate? Several strategies could be employed:
(1) Spatial and temporal factors could directly cooperate by forming a complex and regulating downstream genes including the post-mitotic regulators (Figure 3A). This strategy is utilized to generate Ap-expressing neurons in the fly embryonic nervous system. Ap+ neurons are only present in thoracic segments while the neural stem cells, NB5-6, that generate them are present in every segment. During neurogenesis, Antp, the spatial factor defining the thoracic segments (Figure 1B) and the temporal factors, Cas and Grh (Figure 2C), bind together in a feed-forward loop to activate Col, the post-mitotic regulator for Ap+ neurons. The abdominal spatial factors Ubx, Abd-A, and Abd-B together terminate the neurogenesis before the Cas and Grh temporal window to prevent the generation of Ap+ neurons in the abdominal segment (Karlsson et al., 2010). Cis-regulatory modules targeted by spatiotemporal factors underlie the integration: The col cis-regulatory module requires binding by both Antp and Cas to activate, while Antp and Col are required to bind together for Ap and Eya to express and in turn drive the expression of Nplp1, a neuropeptide used by Ap+ neurons (Stratmann and Thor, 2017). Extrinsic signals could also introduce regulators to participate in cooperative regulation. For example, in C. elegans, AIY neurons are specified by the expression of TTX-3 and CEH-10 (Altun-Gultekin et al., 2001), which depends on iterative integration of the asymmetric Wnt/β-catenin signaling that is only active in the posterior daughter cell (Lin et al., 1998): In a REF-2-expressing progenitor that will generate four types of neurons including an AIY neuron, REF-2 activates ttx-3 in the anterior daughter cell that is not sensing Wnt (Murgan et al., 2015). When the TTX-3-expressing progenitor divides again, TTX-3 cooperates with β-catenin to activate ceh-10 and specify AIY neuron (Bertrand and Hobert, 2009).
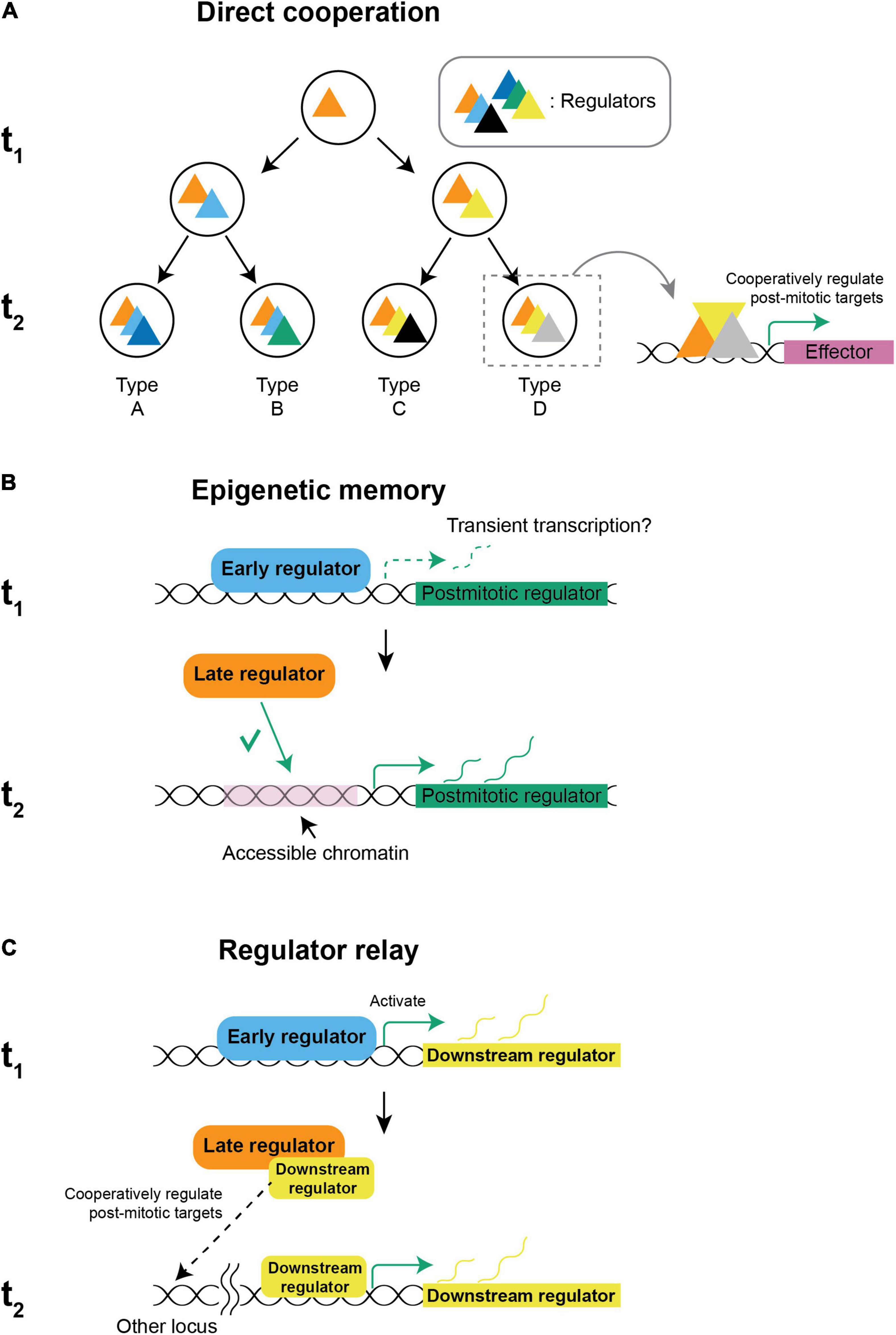
Figure 3. Possible mechanisms of spatiotemporal integration. (A) Direct cooperation: New regulators are introduced by an intrinsic cascade or by extrinsic signals and regulate target genes cooperatively with early regulators that are already present to establish distinct gene signatures for different neuronal types. (B) Epigenetic memory: Spatial and temporal factors might cooperate by early regulators establishing the chromatin landscape and defining a permissive subset of targets to be regulated by other regulators that are expressed later. The restriction of targets defines a specific set of post-mitotic regulators to be expressed after cell cycle exit. In ASE neurons, transient transcription in early development is sufficient to establish accessibility (Charest et al., 2020). (C) Regulator relay: When spatiotemporal factors are not co-expressed, the regulator that is expressed earlier might activate a downstream factor that is persistently expressed. When the later regulator is set to express, it directly cooperates with the downstream factor of the early regulator and activates a specific set of post-mitotic regulators. (t1: first time-point, e.g., neural stem cell, t2: second time-point, e.g., neuron).
(2) However, spatial and temporal factors are not always expressed at the same time, which requires regulators that are expressed at different points to integrate across time. One possibility for an earlier factor to interact with a later factor is by epigenetic memory (Figure 3B). In worms, ASER and ASEL neurons both express the same post-mitotic regulator, CHE-1, but only ASEL expresses LSY-6. The lineages giving rise to ASER/ASEL differ in their early transient expression of TBX-37/38 and become molecularly equivalent after 4 divisions. However, the seemingly equivalent lineages remember their lineage history. Early expression of TBX-37/38 in the ASEL lineage results in transient transcription of lsy-6, which is required and sufficient for keeping the locus accessible in the ASEL but not in the ASER lineage, and the difference of accessibility allows CHE-1 to activate lsy-6 in ASEL but not ASER (Charest et al., 2020). In flies, spatial factors in the ventral nerve cord and optic lobe are expressed in the neuroepithelium and disappear in the neuroblasts, while temporal factors are expressed only in neuroblasts when they commit to neurogenesis. In the ventral nerve cord neuroblasts, chromatin accessibility is regulated differently between spatial domains through the action of a spatial factor, Gsb. Reminiscent to how ASE asymmetry is regulated, the binding pattern of a temporal factor Hb, which is expressed later, is dependent on the pre-existing chromatin landscape and can only bind to the accessible regions (Sen et al., 2019). While chromatin accessibility is determined by multiple factors including histone modifications, DNA methylation (Cusack et al., 2020), pioneer factors (Voss and Hager, 2014), etc., and is used as a proxy of inheritable chromatin modifications, the molecular nature of epigenetic memory remains to be investigated.
(3) Finally, spatiotemporal factors that seem not to be co-expressed could also integrate fate information if the regulator that is expressed earlier turns on a downstream factor that continues to be expressed before eventually cooperating with a later-expressing regulator (Figure 3C).
The models for spatiotemporal integration are not mutually exclusive. On the contrary, spatiotemporal factors are likely to regulate their targets in a hybrid mode. In the fly ventral nerve cord, temporal factors not only regulate their targets directly but could also cooperate with Polycomb complexes to set the epigenetic landscape to restrict the generation of motor neurons to certain temporal windows (Touma et al., 2012). Additionally, genes that are known to be spatiotemporal factors could also play multiple roles in fate specification. For example, in Ap+ neurons in the ventral nerve cord, Antp is not only required for the activation of a post-mitotic regulator, Col, but also directly regulates Ap and Eya together with Col, effectively making Antp not only a spatial factor but also a neuronal regulator of post-mitotic features. Finally, while we only used activators as examples in this section, a regulator could serve simultaneously as an activator and a repressor for different sets of targets (Clovis et al., 2016). Therefore, repressors can have a role in all three models: a repressor can suppress the expression of some of the regulators in the complex and change the composition and target genes in the direct cooperation model; a repressive chromatin modifier could silence a locus and prevent later regulators from accessing it in the epigenetic memory model; finally, regulator relay could not only be achieved by an activator cascade but also a sequential decay of repressors (Averbukh et al., 2018).
Our current knowledge of how spatial and temporal patterning are integrated and define cell fates comes from studies of a few genes in a few cell types and is rather limited. The integration of spatial and temporal patterning is more than spatial and temporal factors functioning independently and regulating their own targets. Instead, we know that there are genes that are not activated by either a spatial or a temporal factor alone but only expressed when a specific combination of spatial and temporal factors is present. For example, in the fly optic lobe, Svp is only expressed in neurons generated from the early Hth window from the Vsx and Rx domains but neither Hth nor Vsx or Rx alone is sufficient for the expression of Svp (Erclik et al., 2017). In the ventral nerve cord, Ap neurons are only generated in the Cas window in the thoracic segment defined by Antp (Karlsson et al., 2010). Similarly in vertebrates, Calbindin is only expressed in the early-born V1 interneurons from the p1 progenitor domain but not in early-born neurons from other spatial domains of the spinal cord (Stam et al., 2012). To have a more comprehensive understanding of the molecular logic of spatiotemporal integration, models with known spatial and temporal regulators will be a great entry point, like the fly central nervous system and the mammalian retina and neural tube. To understand the origin of molecular spatiotemporal integration, we need to identify more spatiotemporal factors in various nervous tissues in different species to investigate how these mechanisms have evolved; the advent of high-throughput profiling of gene expression and chromatin landscape specific for each spatiotemporal identity holds the promise to decipher both the rules and exceptions of spatiotemporal integration.
How Does Spatiotemporal Patterning Contribute to Function?
Spatiotemporal patterning provides an elegant way to generate a wide array of neuronal types that provide the basis for nervous system function. For example, spatial patterning of the neural tube in vertebrates changed concordantly with the emergence of walking in tetrapods and new subtypes of spinal motor neurons were specified to innervate the limbs (Jung et al., 2014, 2018).
Besides providing a gene regulatory framework to enable neurons to acquire their molecular identity, temporal synchronization in neurogenesis helps limit the number of possible targets each neuron encounters and make neural wiring robust and precise during development. Alternatively, temporal patterning allows different types of daughter neurons to be generated next to each other, and this adjacency between daughter neurons could help them to preferentially connect to each other and reduce the number of possible partners by excluding neurons that are farther away. Consistent with this, neurons generated from the same neural progenitor tend to form a circuit, and when each circuit is activated, specific movements, like walking or wing waving, are generated in the fly ventral nerve cord (Harris et al., 2015).
In mammals, the projection pattern of excitatory cortical neurons has long been known to associate with their birthdate (Rakic, 1974; Raedler and Raedler, 1978; Royce, 1983), and clonally related excitatory neurons preferentially connect with each other (Yu et al., 2009, 2012; Li et al., 2012; Ohtsuki et al., 2012). Recently, the organization and projections of spinal neurons were also found to be dependent on their birthdate: Early-born spinal neurons are positioned laterally and enriched in neurons that project a long range, while late-born ones are positioned medially and enriched in neurons that project locally (Osseward et al., 2021). Spatial patterning is also shown to influence multiple neuronal features: Spatial origin is associated with electrophysiological properties of cortical interneurons (Butt et al., 2005) and defines a wide variety of interneurons and motor neurons in the spinal cord (Jessell, 2000; Sagner and Briscoe, 2019). It remains an open question if neurons born at similar stages in different regions of the mammalian nervous system, e.g., the cerebellum and spinal cord, also take advantage of the synchronicity of neurogenesis and preferentially connect and form a circuit among neurons born at similar time windows.
Little is known about the consequences of spatiotemporal patterning dysregulation. In flies, perturbation of temporal patterning by knocking down eyeless, an early temporal factor in the central complex, results in impairment of navigation (Sullivan et al., 2019). Similar disruption of navigation is observed when neurons generated during the Eyeless window are silenced (Giraldo et al., 2018; Green et al., 2018; Sullivan et al., 2019). In mice, overproduction of late-born layer 2/3 cortical neurons by prolonging the proliferation of intermediate progenitors results in compulsive behavior and deficits in social behavior reminiscent of autism spectrum disorder (Fang et al., 2013, 2014). Autism spectrum disorder (ASD), a disease involving neocortical function, has been long hypothesized to be associated with miswiring of neural circuitry, and it is tempting to speculate that patterning failure in the cortex results in autistic features if spatiotemporal patterning makes neurogenesis and wiring less error prone. Consistent with this hypothesis, mutations of mammalian cortical layer markers are often associated with ASD (Kwan, 2013). Recently, disorganized cortical lamina and changes in cortical layer thickness were also reported in ASD patients, which indicates that temporal patterning might malfunction during neurogenesis in patients with autism (Wegiel et al., 2010; Stoner et al., 2014; Trutzer et al., 2019).
Spatiotemporal Patterning and Evolution of New Cell Types
It has long been hypothesized that cell type duplication and diversification underlie nervous system evolution. In this model, the number of neurons expands and then diversifies to serve novel roles to form a more complicated nervous system. Indeed, brain volume and layer thickness differ substantially among vertebrates (Tosches and Laurent, 2019). The gain or loss of spatiotemporal factors is among the plausible mechanisms to diversify an expanded population of neurons. Consistent with this model, recent comparative studies in vertebrates have shown that evolutionary novel subtypes often share the gene expression profile with a conserved cardinal type. For example, birds have only two cerebellar nuclei, medial and interposed, while mammals have three: medial, interposed, and lateral. Neurons from each of the three nuclei in chicken, mice, and humans can be categorized into two molecular classes, while the neurons of the same class from different nuclei still differ from each other (Kebschull et al., 2020). While it remains to be determined if changes in spatiotemporal patterning are responsible for the emergence of new subtypes during evolution, it provides a feasible molecular infrastructure for duplication and diversification to occur.
The recent advent of high-throughput profiling techniques not only made possible the discovery of a shared set of temporal regulators across different vertebrate neuronal tissues (Lu et al., 2020; Sagner et al., 2021) but also opened the opportunity to understand the gene regulatory networks governed by these temporal factors (Telley et al., 2019; Lyu et al., 2021) and other birthdate-specific post-mitotic regulators (Di Bella et al., 2021). Our rapidly expanding knowledge of the molecular logic of spatiotemporal integration and fate specification in various species will set the stage for more comparative studies, such as the investigation of how the targets of conserved spatiotemporal regulators differ between species, how novel spatial domains or temporal windows emerge, and, ultimately, how new cell types are generated during development and evolution.
Author Contributions
Both authors contributed equally in the conception and writing of the manuscript.
Funding
YCC was supported by the New York University (MacCracken Fellowship and NIH R01 EY13010 to Claude Desplan). NK was supported by the Centre National de la Recherche Scientifique (CNRS) and the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program (Grant agreement No. 949500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Claude Desplan, Jennifer Malin, and Rana El-Danaf for feedback on the manuscript.
References
Alcamo, E. A., Chirivella, L., Dautzenberg, M., Dobreva, G., Fariñas, I., Grosschedl, R., et al. (2008). Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377. doi: 10.1016/j.neuron.2007.12.012
Allen, A. M., Neville, M. C., Birtles, S., Croset, V., Treiber, C. D., Waddell, S., et al. (2020). A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. eLife 9:e54074. doi: 10.7554/eLife.54074
Altun-Gultekin, Z., Andachi, Y., Tsalik, E. L., Pilgrim, D., Kohara, Y., and Hobert, O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969. doi: 10.1242/dev.128.11.1951
Apitz, H., and Salecker, I. (2015). A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nat. Neurosci. 18, 46–55. doi: 10.1038/nn.3896
Averbukh, I., Lai, S.-L., Doe, C. Q., and Barkai, N. (2018). A repressor-decay timer for robust temporal patterning in embryonic Drosophila neuroblast lineages. eLife 7:e38631. doi: 10.7554/eLife.38631
Bayraktar, O. A., and Doe, C. Q. (2013). Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498, 449–455. doi: 10.1038/nature12266
Benito-Sipos, J., Baumgardt, M., and Thor, S. (2013). “Chapter 31 - development of the Drosophila embryonic ventral nerve cord: from neuroectoderm to unique neurons and glia,” in Patterning and Cell Type Specification in the Developing CNS and PNS, eds J. L. R. Rubenstein and P. Rakic (Oxford: Academic Press), 627–644. doi: 10.1016/b978-0-12-397265-1.00073-3
Bertet, C., Li, X., Erclik, T., Cavey, M., Wells, B., and Desplan, C. (2014). Temporal patterning of neuroblasts controls notch-mediated cell survival through regulation of hid or reaper. Cell 158, 1173–1186. doi: 10.1016/j.cell.2014.07.045
Bertrand, V., and Hobert, O. (2009). Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev. Cell 16, 563–575. doi: 10.1016/j.devcel.2009.02.011
Bikoff, J. B., Gabitto, M. I., Rivard, A. F., Drobac, E., Machado, T. A., Miri, A., et al. (2016). Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell 165, 207–219. doi: 10.1016/j.cell.2016.01.027
Birkholz, O., Vef, O., Rogulja-Ortmann, A., Berger, C., and Technau, G. M. (2013). Abdominal-B and caudal inhibit the formation of specific neuroblasts in the Drosophila tail region. Development 140, 3552–3564. doi: 10.1242/dev.096099
Briscoe, J., and Ericson, J. (2001). Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11, 43–49. doi: 10.1016/S0959-4388(00)00172-0
Britanova, O., de Juan Romero, C., Cheung, A., Kwan, K. Y., Schwark, M., Gyorgy, A., et al. (2008). Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392. doi: 10.1016/j.neuron.2007.12.028
Brody, T., and Odenwald, W. F. (2000). Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev. Biol. 226, 34–44. doi: 10.1006/dbio.2000.9829
Butt, S. J. B., Fuccillo, M., Nery, S., Noctor, S., Kriegstein, A., Corbin, J. G., et al. (2005). The Temporal and Spatial Origins of Cortical Interneurons Predict Their Physiological Subtype. Neuron 48, 591–604. doi: 10.1016/j.neuron.2005.09.034
Cadwell, C. R., Bhaduri, A., Mostajo-Radji, M. A., Keefe, M. G., and Nowakowski, T. J. (2019). Development and arealization of the cerebral cortex. Neuron 103, 980–1004. doi: 10.1016/j.neuron.2019.07.009
Caviness, V. S. (1982). Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Dev. Brain Res. 4, 293–302. doi: 10.1016/0165-3806(82)90141-9
Charest, J., Daniele, T., Wang, J., Bykov, A., Mandlbauer, A., Asparuhova, M., et al. (2020). Combinatorial action of temporally segregated transcription factors. Dev. Cell 55, 483.e7–499.e7. doi: 10.1016/j.devcel.2020.09.002
Chen, B., Schaevitz, L. R., and McConnell, S. K. (2005). Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 102, 17184–17189. doi: 10.1073/pnas.0508732102
Chen, B., Wang, S. S., Hattox, A. M., Rayburn, H., Nelson, S. B., and McConnell, S. K. (2008). The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 105, 11382–11387. doi: 10.1073/pnas.0804918105
Clark, B. S., Stein-O’Brien, G. L., Shiau, F., Cannon, G. H., Davis-Marcisak, E., Sherman, T., et al. (2019). Single-cell RNA-Seq analysis of retinal development identifies nfi factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111.e5–1126.e5. doi: 10.1016/j.neuron.2019.04.010
Clovis, Y. M., Seo, S. Y., Kwon, J., Rhee, J. C., Yeo, S., Lee, J. W., et al. (2016). Chx10 consolidates V2a interneuron identity through two distinct gene repression modes. Cell Rep. 16, 1642–1652. doi: 10.1016/j.celrep.2016.06.100
Cusack, M., King, H. W., Spingardi, P., Kessler, B. M., Klose, R. J., and Kriaucionis, S. (2020). Distinct contributions of DNA methylation and histone acetylation to the genomic occupancy of transcription factors. Genome Res. 30, 1393–1406. doi: 10.1101/gr.257576.119
D’Alessio, M., and Frasch, M. (1996). msh may play a conserved role in dorsoventral patterning of the neuroectoderm and mesoderm. Mech. Dev. 58, 217–231. doi: 10.1016/S0925-4773(96)00583-7
Delile, J., Rayon, T., Melchionda, M., Edwards, A., Briscoe, J., and Sagner, A. (2019). Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 146:dev173807. doi: 10.1242/dev.173807
Di Bella, D. J., Habibi, E., Stickels, R. R., Scalia, G., Brown, J., Yadollahpour, P., et al. (2021). Molecular logic of cellular diversification in the mouse cerebral cortex. Nature 595, 554–559. doi: 10.1038/s41586-021-03670-5
Doe, C. Q. (1992). Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116, 855–863. doi: 10.1242/dev.116.4.855
Doe, C. Q. (2017). Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219–240. doi: 10.1146/annurev-cellbio-111315-125210
Elliott, J., Jolicoeur, C., Ramamurthy, V., and Cayouette, M. (2008). Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60, 26–39. doi: 10.1016/j.neuron.2008.08.008
Erclik, T., Hartenstein, V., Lipshitz, H. D., and McInnes, R. R. (2008). Conserved role of the vsx genes supports a monophyletic origin for bilaterian visual systems. Curr. Biol. 18, 1278–1287. doi: 10.1016/j.cub.2008.07.076
Erclik, T., Li, X., Courgeon, M., Bertet, C., Chen, Z., Baumert, R., et al. (2017). Integration of temporal and spatial patterning generates neural diversity. Nature 541, 365–370. doi: 10.1038/nature20794
Estacio-Gómez, A., and Díaz-Benjumea, F. J. (2014). Roles of Hox genes in the patterning of the central nervous system of Drosophila. Fly 8, 26–32. doi: 10.4161/fly.27424
Etchberger, J. F., Lorch, A., Sleumer, M. C., Zapf, R., Jones, S. J., Marra, M. A., et al. (2007). The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21, 1653–1674. doi: 10.1101/gad.1560107
Fang, W.-Q., Chen, W.-W., Fu, A. K. Y., and Ip, N. Y. (2013). Axin directs the amplification and differentiation of intermediate progenitors in the developing cerebral cortex. Neuron 79, 665–679. doi: 10.1016/j.neuron.2013.06.017
Fang, W.-Q., Chen, W.-W., Jiang, L., Liu, K., Yung, W.-H., Fu, A. K. Y., et al. (2014). Overproduction of upper-layer neurons in the neocortex leads to autism-like features in mice. Cell Rep. 9, 1635–1643. doi: 10.1016/j.celrep.2014.11.003
Flames, N., Pla, R., Gelman, D. M., Rubenstein, J. L. R., Puelles, L., and Marin, O. (2007). Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007
Fogarty, M., Grist, M., Gelman, D., Marin, O., Pachnis, V., and Kessaris, N. (2007). Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 27, 10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007
Gao, P., Postiglione, M. P., Krieger, T. G., Hernandez, L., Wang, C., Han, Z., et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788. doi: 10.1016/j.cell.2014.10.027
Giraldo, Y. M., Leitch, K. J., Ros, I. G., Warren, T. L., Weir, P. T., and Dickinson, M. H. (2018). Sun navigation requires compass neurons in Drosophila. Curr. Biol. 28, 2845.e4–2852.e4. doi: 10.1016/j.cub.2018.07.002
Gold, K. S., and Brand, A. H. (2014). Optix defines a neuroepithelial compartment in the optic lobe of the Drosophila brain. Neural Dev. 9:18. doi: 10.1186/1749-8104-9-18
Green, J., Vijayan, V., Mussells Pires, P., Adachi, A., and Maimon, G. (2018). Walking Drosophila aim to maintain a neural heading estimate at an internal goal angle. bioRxiv [Preprint]. doi: 10.1101/315796
Greig, L. C., Woodworth, M. B., Galazo, M. J., Padmanabhan, H., and Macklis, J. D. (2013). Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769. doi: 10.1038/nrn3586
Gutjahr, T., Patel, N. H., Li, X., Goodman, C. S., and Noll, M. (1993). Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and activates gooseberry neuro. Development 118, 21–31. doi: 10.1242/dev.118.1.21
Harris, R. M., Pfeiffer, B. D., Rubin, G. M., and Truman, J. W. (2015). Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. eLife 4:e04493. doi: 10.7554/eLife.04493
Hasegawa, E., Kaido, M., Takayama, R., and Sato, M. (2013). Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Dev. Biol. 377, 90–99. doi: 10.1016/j.ydbio.2013.02.012
Hasegawa, E., Kitada, Y., Kaido, M., Takayama, R., Awasaki, T., Tabata, T., et al. (2011). Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development 138, 983–993. doi: 10.1242/dev.058370
Hatini, V., Tao, W., and Lai, E. (1994). Expression of winged helix genes, BF-1 and BF-2, define adjacent domains within the developing forebrain and retina. J. Neurobiol. 25, 1293–1309. doi: 10.1002/neu.480251010
Hayashi, M., Hinckley, C. A., Driscoll, S. P., Moore, N. J., Levine, A. J., Hilde, K. L., et al. (2018). Graded arrays of spinal and supraspinal V2a interneuron subtypes underlie forelimb and hindlimb motor control. Neuron 97, 869.e5–884.e5. doi: 10.1016/j.neuron.2018.01.023
Holguera, I., and Desplan, C. (2018). Neuronal specification in space and time. Science 362, 176–180. doi: 10.1126/science.aas9435
Isshiki, T., Pearson, B., Holbrook, S., and Doe, C. Q. (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521. doi: 10.1016/S0092-8674(01)00465-2
Isshiki, T., Takeichi, M., and Nose, A. (1997). The role of the msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neuroectoderm. Development 124, 3099–3109. doi: 10.1242/dev.124.16.3099
Javed, A., Mattar, P., Lu, S., Kruczek, K., Kloc, M., Gonzalez-Cordero, A., et al. (2020). Pou2f1 and Pou2f2 cooperate to control the timing of cone photoreceptor production in the developing mouse retina. Development 147:dev188730. doi: 10.1242/dev.188730
Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. doi: 10.1038/35049541
Jung, H., Baek, M., D’Elia, K. P., Boisvert, C., Currie, P. D., Tay, B.-H., et al. (2018). The ancient origins of neural substrates for land walking. Cell 172, 667.e15–682.e15. doi: 10.1016/j.cell.2018.01.013
Jung, H., Mazzoni, E. O., Soshnikova, N., Hanley, O., Venkatesh, B., Duboule, D., et al. (2014). Evolving hox activity profiles govern diversity in locomotor systems. Dev. Cell 29, 171–187. doi: 10.1016/j.devcel.2014.03.008
Kanatani, S., Yozu, M., Tabata, H., and Nakajima, K. (2008). COUP-TFII is preferentially expressed in the caudal ganglionic eminence and is involved in the caudal migratory stream. J. Neurosci. 28, 13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008
Karlsson, D., Baumgardt, M., and Thor, S. (2010). Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biol. 8:e1000368. doi: 10.1371/journal.pbio.1000368
Kebschull, J. M., Richman, E. B., Ringach, N., Friedmann, D., Albarran, E., Kolluru, S. S., et al. (2020). Cerebellar nuclei evolved by repeatedly duplicating a conserved cell-type set. Science 370:eabd5059. doi: 10.1126/science.abd5059
Kohwi, M., Petryniak, M. A., Long, J. E., Ekker, M., Obata, K., Yanagawa, Y., et al. (2007). A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J. Neurosci. 27, 6878–6891. doi: 10.1523/JNEUROSCI.0254-07.2007
Konstantinides, N., Kapuralin, K., Fadil, C., Barboza, L., Satija, R., and Desplan, C. (2018b). Phenotypic convergence: distinct transcription factors regulate common terminal features. Cell 174, 622.e13–635.e13. doi: 10.1016/j.cell.2018.05.021
Konstantinides, N., Degabriel, S., and Desplan, C. (2018a). Neuro-evo-devo in the single cell sequencing era. Curr. Opin. Syst. Biol. 11, 32–40. doi: 10.1016/j.coisb.2018.08.001
Konstantinides, N., Rossi, A. M., Escobar, A., Dudragne, L., Chen, Y.-C., Tran, T., et al. (2021). A comprehensive series of temporal transcription factors in the fly visual system. bioRxiv [Preprint] doi: 10.1101/2021.06.13.448242
Kwan, K. Y. (2013). Transcriptional dysregulation of neocortical circuit assembly in ASD. Int. Rev. Neurobiol. 113, 167–205. doi: 10.1016/B978-0-12-418700-9.00006-X
Lee, S., Hjerling-Leffler, J., Zagha, E., Fishell, G., and Rudy, B. (2010). The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J. Neurosci. 30, 16796–16808. doi: 10.1523/JNEUROSCI.1869-10.2010
Lee, S.-K., and Pfaff, S. L. (2003). Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38, 731–745. doi: 10.1016/S0896-6273(03)00296-4
Li, H., Horns, F., Xie, Q., Xie, Q., Li, T., Luginbuhl, D. J., et al. (2017). Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell 171, 1206–1207. doi: 10.1016/j.cell.2017.10.019
Li, X., Erclik, T., Bertet, C., Chen, Z., Voutev, R., Venkatesh, S., et al. (2013). Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456–462. doi: 10.1038/nature12319
Li, Y., Lu, H., Cheng, P., Ge, S., Xu, H., Shi, S.-H., et al. (2012). Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature 486, 118–121. doi: 10.1038/nature11110
Lin, R., Hill, R. J., and Priess, J. R. (1998). POP-1 and anterior–posterior fate decisions in C. elegans embryos. Cell 92, 229–239. doi: 10.1016/S0092-8674(00)80917-4
Lu, Y., Shiau, F., Yi, W., Lu, S., Wu, Q., Pearson, J. D., et al. (2020). Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev. Cell 53, 473.e9–491.e9. doi: 10.1016/j.devcel.2020.04.009
Lyu, P., Hoang, T., Santiago, C. P., Thomas, E. D., Timms, A. E., Appel, H., et al. (2021). Gene regulatory networks controlling temporal patterning, neurogenesis, and cell-fate specification in mammalian retina. Cell Rep. 37:109994. doi: 10.1016/j.celrep.2021.109994
Masland, R. H. (2004). Neuronal cell types. Curr. Biol. 14, R497–R500. doi: 10.1016/j.cub.2004.06.035
Mattar, P., Ericson, J., Blackshaw, S., and Cayouette, M. (2015). A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 85, 497–504. doi: 10.1016/j.neuron.2014.12.052
Mazzoni, E. O., Mahony, S., Closser, M., Morrison, C. A., Nedelec, S., Williams, D. J., et al. (2013). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16, 1219–1227. doi: 10.1038/nn.3467
McGinnis, W., and Krumlauf, R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302. doi: 10.1016/0092-8674(92)90471-N
Mellerick, D. M., and Nirenberg, M. (1995). Dorsal-ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Dev. Biol. 171, 306–316. doi: 10.1006/dbio.1995.1283
Mettler, U., Vogler, G., and Urban, J. (2006). Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by seven-up and prospero. Development 133, 429–437. doi: 10.1242/dev.02229
Mora, N., Oliva, C., Fiers, M., Ejsmont, R., Soldano, A., Zhang, T. T., et al. (2018). A temporal transcriptional switch governs stem cell division, neuronal numbers, and maintenance of differentiation. Dev. Cell 45, 53.e5–66.e5. doi: 10.1016/j.devcel.2018.02.023
Murgan, S., Kari, W., Rothbächer, U., Iché-Torres, M., Mélénec, P., Hobert, O., et al. (2015). Atypical transcriptional activation by TCF via a zic transcription factor in C. elegans neuronal precursors. Dev. Cell 33, 737–745. doi: 10.1016/j.devcel.2015.04.018
Nery, S., Fishell, G., and Corbin, J. G. (2002). The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat. Neurosci. 5, 1279–1287. doi: 10.1038/nn971
Ohtsuki, G., Nishiyama, M., Yoshida, T., Murakami, T., Histed, M., Lois, C., et al. (2012). Similarity of visual selectivity among clonally related neurons in visual cortex. Neuron 75, 65–72. doi: 10.1016/j.neuron.2012.05.023
Osseward, P. J., Amin, N. D., Moore, J. D., Temple, B. A., Barriga, B. K., Bachmann, L. C., et al. (2021). Conserved genetic signatures parcellate cardinal spinal neuron classes into local and projection subsets. Science 372, 385–393. doi: 10.1126/science.abe0690
Özel, M. N., Simon, F., Jafari, S., Holguera, I., Chen, Y.-C., Benhra, N., et al. (2020). Neuronal diversity and convergence in a visual system developmental atlas. Nature 589, 88–95. doi: 10.1038/s41586-020-2879-3
Philippidou, P., and Dasen, J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80, 12–34. doi: 10.1016/j.neuron.2013.09.020
Picker, A., and Brand, M. (2005). Fgf signals from a novel signaling center determine axial patterning of the prospective neural retina. Development 132, 4951–4962. doi: 10.1242/dev.02071
Pinto-Teixeira, F., Koo, C., Rossi, A. M., Neriec, N., Bertet, C., Li, X., et al. (2018). Development of concurrent retinotopic maps in the fly motion detection circuit. Cell 173, 485.e11–498.e11. doi: 10.1016/j.cell.2018.02.053
Raedler, E., and Raedler, A. (1978). Autoradiographic study of early neurogenesis in rat neocortex. Anat. Embryol. 154, 267–284. doi: 10.1007/BF00345657
Rakic, P. (1974). Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425–427. doi: 10.1126/science.183.4123.425
Ravanelli, A. M., and Appel, B. (2015). Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 29, 2504–2515. doi: 10.1101/gad.271312.115
Rayon, T., Maizels, R. J., Barrington, C., and Briscoe, J. (2021). Single cell transcriptome profiling of the human developing spinal cord reveals a conserved genetic programme with human specific features. bioRxiv [Preprint]. doi: 10.1101/2021.04.12.439474
Rossi, A. M., Fernandes, V. M., and Desplan, C. (2017). Timing temporal transitions during brain development. Curr. Opin. Neurobiol. 42, 84–92. doi: 10.1016/j.conb.2016.11.010
Roy, A., Francius, C., Rousso, D. L., Seuntjens, E., Debruyn, J., Luxenhofer, G., et al. (2012). Onecut transcription factors act upstream of Isl1 to regulate spinal motoneuron diversification. Development 139, 3109–3119. doi: 10.1242/dev.078501
Royce, G. J. (1983). Cortical neurons with collateral projections to both the caudate nucleus and the centromedian-parafascicular thalamic complex: a fluorescent retrograde double labeling study in the cat. Exp. Brain Res. 50, 157–165. doi: 10.1007/BF00239179
Sagner, A., and Briscoe, J. (2019). Establishing neuronal diversity in the spinal cord: a time and a place. Development 146:dev182154. doi: 10.1242/dev.182154
Sagner, A., Zhang, I., Watson, T., Lazaro, J., Melchionda, M., and Briscoe, J. (2021). A shared transcriptional code orchestrates temporal patterning of the central nervous system. PLoS Biol. 19:e3001450. doi: 10.1371/journal.pbio.3001450
Sander, M., Paydar, S., Ericson, J., Briscoe, J., Berber, E., German, M., et al. (2000). Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 14, 2134–2139. doi: 10.1101/gad.820400
Sathyamurthy, A., Johnson, K. R., Matson, K. J. E., Dobrott, C. I., Li, L., Ryba, A. R., et al. (2018). Massively parallel single nucleus transcriptional profiling defines spinal cord neurons and their activity during behavior. Cell Rep. 22, 2216–2225. doi: 10.1016/j.celrep.2018.02.003
Sen, S. Q., Chanchani, S., Southall, T. D., and Doe, C. Q. (2019). Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. eLife 8:e44036. doi: 10.7554/eLife.44036
Shekhar, K., Lapan, S. W., Whitney, I. E., Tran, N. M., Macosko, E. Z., Kowalczyk, M., et al. (2016). Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308.e30–1323.e30. doi: 10.1016/j.cell.2016.07.054
Skeath, J. B., Zhang, Y., Holmgren, R., Carroll, S. B., and Doe, C. Q. (1995). Specification of neuroblast identity in the Drosophila embryonic central nervous system by gooseberry-distal. Nature 376, 427–430. doi: 10.1038/376427a0
Son, E. Y., Ichida, J. K., Wainger, B. J., Toma, J. S., Rafuse, V. F., Woolf, C. J., et al. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218. doi: 10.1016/j.stem.2011.07.014
Stam, F. J., Hendricks, T. J., Zhang, J., Geiman, E. J., Francius, C., Labosky, P. A., et al. (2012). Renshaw cell interneuron specialization is controlled by a temporally restricted transcription factor program. Development 139, 179–190. doi: 10.1242/dev.071134
Stoner, R., Chow, M. L., Boyle, M. P., Sunkin, S. M., Mouton, P. R., Roy, S., et al. (2014). Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 370, 1209–1219. doi: 10.1056/NEJMoa1307491
Stratmann, J., and Thor, S. (2017). Neuronal cell fate specification by the molecular convergence of different spatio-temporal cues on a common initiator terminal selector gene. PLoS Genet. 13:e1006729. doi: 10.1371/journal.pgen.1006729
Sullivan, L. F., Warren, T. L., and Doe, C. Q. (2019). Temporal identity establishes columnar neuron morphology, connectivity, and function in a Drosophila navigation circuit. eLife 8:e43482. doi: 10.7554/eLife.43482
Suzuki, T., Kaido, M., Takayama, R., and Sato, M. (2013). A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev. Biol. 380, 12–24. doi: 10.1016/j.ydbio.2013.05.002
Takebayashi, H., Nabeshima, Y., Yoshida, S., Chisaka, O., Ikenaka, K., and Nabeshima, Y. (2002). The basic helix-loop-helix factor Olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 12, 1157–1163. doi: 10.1016/S0960-9822(02)00926-0
Tang, J. L. Y., Hakes, A. E., Krautz, R., Suzuki, T., Contreras, E. G., Fox, P. M., et al. (2021). NanoDam identifies novel temporal transcription factors conserved between the Drosophila central brain and visual system. Dev. Biol. [Epub ahead of print]. doi: 10.1101/2021.06.07.447332
Telley, L., Agirman, G., Prados, J., Amberg, N., Fièvre, S., Oberst, P., et al. (2019). Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364:eaav2522. doi: 10.1126/science.aav2522
Thaler, J. P., Lee, S.-K., Jurata, L. W., Gill, G. N., and Pfaff, S. L. (2002). LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 110, 237–249. doi: 10.1016/S0092-8674(02)00823-1
Tosches, M. A., and Laurent, G. (2019). Evolution of neuronal identity in the cerebral cortex. Curr. Opin. Neurobiol. 56, 199–208. doi: 10.1016/j.conb.2019.04.009
Touma, J. J., Weckerle, F. F., and Cleary, M. D. (2012). Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development 139, 657–666. doi: 10.1242/dev.071589
Tripodi, M., Filosa, A., Armentano, M., and Studer, M. (2004). The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development 131, 6119–6129. doi: 10.1242/dev.01530
Trutzer, I. M., García-Cabezas, M. Á, and Zikopoulos, B. (2019). Postnatal development and maturation of layer 1 in the lateral prefrontal cortex and its disruption in autism. Acta Neuropathol. Commun. 7:40. doi: 10.1186/s40478-019-0684-8
Tümpel, S., Wiedemann, L. M., and Krumlauf, R. (2009). Chapter 8 hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 88, 103–137. doi: 10.1016/S0070-2153(09)88004-6
Uchida, O., Nakano, H., Koga, M., and Ohshima, Y. (2003). The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130, 1215–1224. doi: 10.1242/dev.00341
Voss, T. C., and Hager, G. L. (2014). Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 15, 69–81. doi: 10.1038/nrg3623
Wegiel, J., Kuchna, I., Nowicki, K., Imaki, H., Wegiel, J., Marchi, E., et al. (2010). The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 119, 755–770. doi: 10.1007/s00401-010-0655-4
Weiss, J. B., Von Ohlen, T., Mellerick, D. M., Dressler, G., Doe, C. Q., and Scott, M. P. (1998). Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 12, 3591–3602. doi: 10.1101/gad.12.22.3591
Wichterle, H., Turnbull, D. H., Nery, S., Fishell, G., and Alvarez-Buylla, A. (2001). In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development 128, 3759–3771. doi: 10.1242/dev.128.19.3759
Xie, H., Zhang, W., Zhang, M., Akhtar, T., Li, Y., Yi, W., et al. (2020). Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC-derived retinal organoids. Sci. Adv. 6:eaay5247. doi: 10.1126/sciadv.aay5247
Xu, Q. (2004). Origins of cortical interneuron subtypes. J. Neurosci. 24, 2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004
Xu, Q., Tam, M., and Anderson, S. A. (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29. doi: 10.1002/cne.21529
Yao, Z., van Velthoven, C. T. J., Nguyen, T. N., Goldy, J., Sedeno-Cortes, A. E., Baftizadeh, F., et al. (2021). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222.e26–3241.e26. doi: 10.1016/j.cell.2021.04.021
Yu, Y.-C., Bultje, R. S., Wang, X., and Shi, S.-H. (2009). Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458, 501–504. doi: 10.1038/nature07722
Yu, Y.-C., He, S., Chen, S., Fu, Y., Brown, K. N., Yao, X.-H., et al. (2012). Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature 486, 113–117. doi: 10.1038/nature10958
Yuasa, J., Hirano, S., Yamagata, M., and Noda, M. (1996). Visual projection map specified by topographic expression of transcription factors in the retina. Nature 382, 632–635. doi: 10.1038/382632a0
Zeng, H., and Sanes, J. R. (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546. doi: 10.1038/nrn.2017.85
Keywords: neuronal diversity, spatial patterning, temporal patterning, evolution of developmental mechanisms, fate specification, Drosophila, vertebrates
Citation: Chen YC and Konstantinides N (2022) Integration of Spatial and Temporal Patterning in the Invertebrate and Vertebrate Nervous System. Front. Neurosci. 16:854422. doi: 10.3389/fnins.2022.854422
Received: 13 January 2022; Accepted: 15 February 2022;
Published: 22 March 2022.
Edited by:
Idoia Quintana-Urzainqui, European Molecular Biology Laboratory Heidelberg, GermanyReviewed by:
Teresa Rayon, Babraham Institute (BBSRC), United KingdomAlberto Stolfi, Georgia Institute of Technology, United States
Copyright © 2022 Chen and Konstantinides. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yen-Chung Chen, eWNjNTIwQG55dS5lZHU=; Nikolaos Konstantinides, bmlrb3Mua29uc3RhbnRpbmlkZXNAaWptLmZy
 Yen-Chung Chen
Yen-Chung Chen Nikolaos Konstantinides
Nikolaos Konstantinides