- 1Murdoch Children’s Research Institute, The Royal Children’s Hospital Melbourne, Parkville, VIC, Australia
- 2Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia
- 3Department of Surgery, Royal Melbourne Hospital, The University of Melbourne, Parkville, VIC, Australia
- 4Department of Biochemistry and Medical Genetics, Max Rady College of Medicine, Rady Faculty of Health Sciences, University of Manitoba, Winnipeg, MB, Canada
- 5Institute of Cardiovascular Sciences, St. Boniface Hospital Albrechtsen Research Centre, Winnipeg, MB, Canada
- 6Department of Medical Genetics, University of Alberta, Edmonton, AB, Canada
- 7Department of Pediatrics, University of Alberta, Edmonton, AB, Canada
- 8Department of Oncology, Faculty of Medicine & Dentistry, University of Alberta, Edmonton, AB, Canada
Forebrain development in vertebrates is regulated by transcription factors encoded by homeobox, bHLH and forkhead gene families throughout the progressive and overlapping stages of neural induction and patterning, regional specification and generation of neurons and glia from central nervous system (CNS) progenitor cells. Moreover, cell fate decisions, differentiation and migration of these committed CNS progenitors are controlled by the gene regulatory networks that are regulated by various homeodomain-containing transcription factors, including but not limited to those of the Pax (paired), Nkx, Otx (orthodenticle), Gsx/Gsh (genetic screened), and Dlx (distal-less) homeobox gene families. This comprehensive review outlines the integral role of key homeobox transcription factors and their target genes on forebrain development, focused primarily on the telencephalon. Furthermore, links of these transcription factors to human diseases, such as neurodevelopmental disorders and brain tumors are provided.
Introduction
Overview of Forebrain Development
Early brain development is marked by the formation of different compartments through the segmentation of the neural tube that is guided and defined by specific regional expression of transcription factors. The developing brain is sectioned into three contiguous parts, the prosencephalon in the most anterior area, which then matures into the forebrain; the mesencephalon following posteriorly, which give rises to the midbrain; and further posteriorly the rhombencephalon, the early form of the hindbrain. These areas further partition, where the prosencephalon separates into primary prosencephalon (diencephalon) and secondary prosencephalon (telencephalon) (Puelles, 2013, 2018), and the rhombencephalon divides into the metencephalon and myelencephalon. In contrast to the other two regions, the mesencephalon does not divide (Stiles, 2008). Within the forebrain, the prosomeric model depicts the division of this area into 7 segments called the prosomeres (Rubenstein et al., 1994; Puelles and Rubenstein, 2003). The diencephalon develops into 3 prosomeres (p1, p2, p3), which are then recognized as the pretectum, thalamus and pre-thalamus. The secondary prosencephalon develops into two hypothalamo-telencephalic prosomeres (hp1, hp2), later giving rise to the hypothalamus and telencephalon. The mesencephalon contributes to two prosomeres (m1, m2) (Puelles, 2018).
The regions adjacent to the ventricular surface in the brain are the ventricular zone (VZ), followed by the subventricular zone (SVZ), and the mantle zone (MZ) (Figure 1A). The VZ contains radial glia, which then differentiate into intermediate neural progenitors that populate the SVZ, where both of these cell types can give rise to neurons (Miyata et al., 2001; Noctor et al., 2001, 2004; Haubensak et al., 2004). The telencephalon can be divided into the dorsal (pallium) and ventral (subpallium) telencephalon, where the neocortex and the ganglionic eminences (GE) are located, respectively. The anatomic region separating the dorsal and ventral telencephalon is often referred to as the pallio-subpallial boundary (PSB). The GE is divided into lateral, medial, and caudal GE (LGE; MGE; CGE), and ventral to the MGE is the preoptic area (PoA) (Figure 1A). The LGE can be further separated in the ventral LGE (vLGE), where striatal projection neurons originate, and the dorsal LGE (dLGE) that gives rise to intercalated cells of the amygdala and neurons in the olfactory bulb along with the lateral LGE wall (Yun et al., 2001; Stenman et al., 2003; Waclaw et al., 2010). The LGE is a local source of retinoic acid, a morphogen that regulates cortical patterning and regionalization (see Shibata et al., 2021; Ziffra et al., 2021 for more details) (Toresson et al., 1999; Molotkova et al., 2007; Shibata et al., 2021; Ziffra et al., 2021).
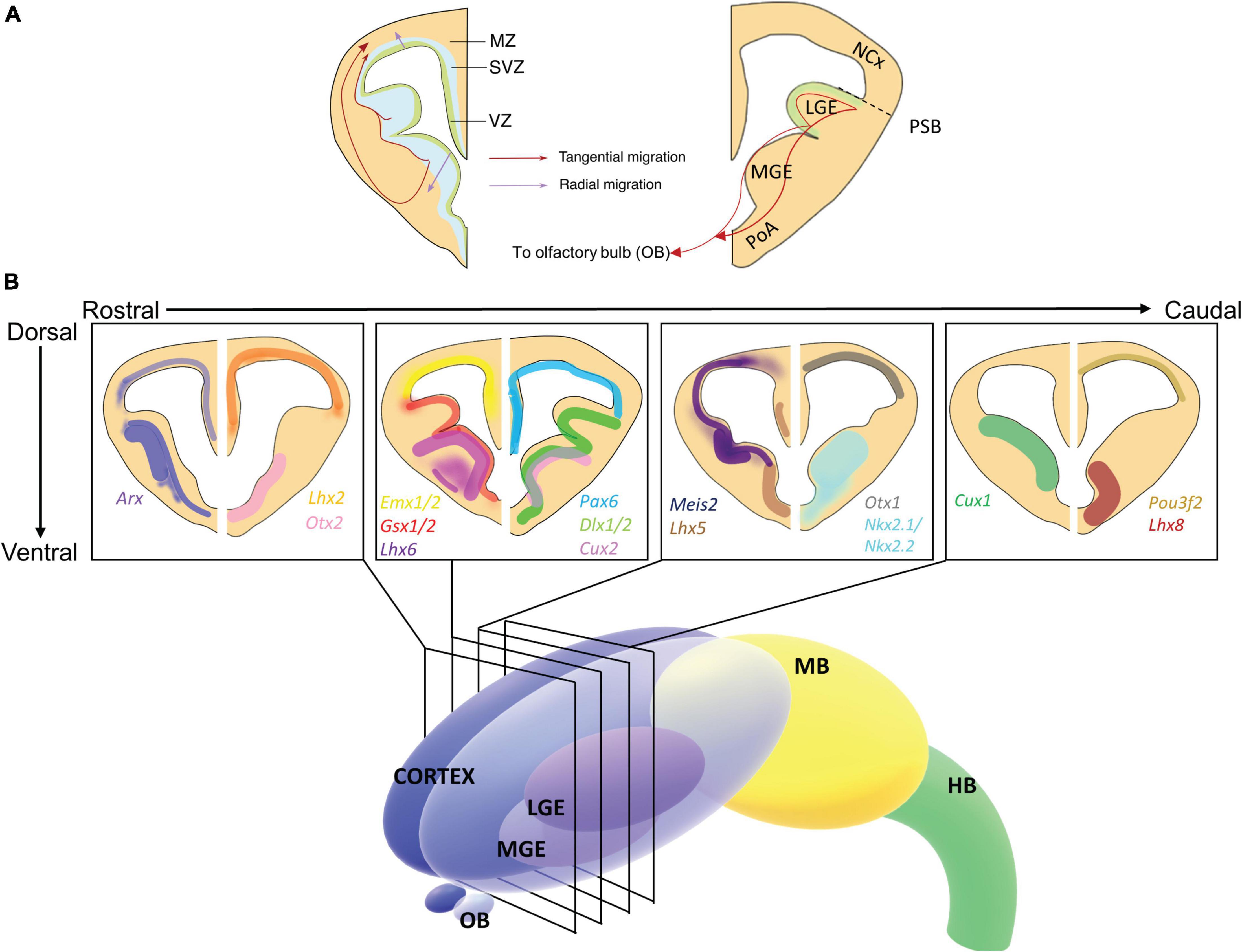
Figure 1. Expression of homeobox genes in the developing embryonic mouse forebrain. (A) Schematic illustration of coronal section of E13.5 forebrain depicting ventricular zone (VZ), subventricular zone (SVZ), and mantle zone (MZ) on the left-hand side and neocortex (NCx), lateral ganglionic eminence (LGE), and medial ganglionic eminence (MGE) on the right-hand side. The VZ and SVZ are the proliferative zones, comprised of progenitor cells. Depending on the identity of these differentiated cells, the cells migrate either tangentially (red arrows) or radially (purple arrows) into the MZ and proceed to mature (Left-hand side). Migration toward the olfactory bulb from the VZ of the LGE also occurs (Right-hand side). (B) 3-dimensional schematic of the developing forebrain. The LGE and MGE are contained within the cortex, above the olfactory bulbs (OB). The midbrain (MB) and hindbrain (HB) are also labeled. Insets show schematic representations of 4 coronal sections taken from the forebrain depicting the expression of key homeobox gene expression patterns from rostral to caudal at embryonic time point E13.5. Gene name colors correspond to the expression color shown in the section. Transcription factor expression can be overlapping or structurally distinct and is related to the function of the individual transcription factor (Allen Institute for Brain Science, 2019). Arx and Meis2, to an extent, are expressed throughout the forebrain, whereas Lhx2, Emx1/2, Pax6, Otx1, and Pou3f2 are expressed in the neocortex and pallium. Dlx1/2, Gsx1, Otx2, and Cux1 are expressed in the GE, Gsx2 is expressed specifically in the LGE, and Nkx2.1, Cux2, Lhx6, and Lhx8 in the MGE. Irx3 is not depicted here as it is expressed in the thalamus (not shown). For detailed depictions of gene expression patterns, readers are encouraged to review the cited primary references or the Allen Brain Atlas: Developing Mouse Brain (Allen Institute for Brain Science, 2019). NCx, neocortex; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; V, ventricle; VZ, ventricular zone; SVZ, subventricular zone; MZ, mantle zone.
Origin of Cortical and Striatal Neurons
Excitatory and inhibitory neuronal activities need to be balanced in order for the nervous system to maintain homeostasis and to optimally process information; these are governed by projection and inhibitory neurons in the brain, respectively. Neuronal progenitor cells (NPC) are produced in both dorsal and ventral telencephalon; NPCs from the dorsal telencephalon give rise to projection neurons (glutamatergic) and NPCs from the ventral telencephalon differentiate into inhibitory interneurons (γ-amino butyric (GABA)-ergic) (Anderson et al., 1997a, 2002b). These neuronal origin sites are conserved amongst mammals, as shown through studies in primates, rodents, and humans, in which some cortical interneurons could be generated locally in the dorsal telencephalon (Letinic et al., 2002; Hansen et al., 2013; Ma et al., 2013). Glutamatergic neurons make up ∼ 70% of the neuronal population in the mouse, with the remaining ∼ 30% being GABAergic interneurons (Hendry et al., 1987). Within the ventral telencephalon, GABAergic interneurons are produced mainly from Nkx2.1 expressing progenitor cells in the MGE and PoA (Fogarty et al., 2007; Gelman et al., 2009), and migrate tangentially to reach the neocortex (Marín and Rubenstein, 2003). These ventral telencephalic interneurons mainly consist of parvalbumin (pva+), somatostatin (sst+), and 5ht3a+ interneurons subtypes (Rudy et al., 2011). Many sst+ interneurons arise and migrate from the CGE, while other interneuron subtypes arise from progenitor cells in the LGE and CGE, including the vasoactive intestinal peptide and cholecystokinin expressing interneurons which reside in the MZ (Anderson et al., 2001; Nery et al., 2002; Miyoshi et al., 2010). The main population of striatal projection neurons comprises the GABAergic medium spiny neurons (MSNs) which arise from progenitors in the LGE, and account for ∼ 80% of the striatal neuron population in primates and rodents (Graveland and DiFiglia, 1985). Some key marker genes for MSN differentiation include Foxp1/2, Ascl1, Ebf1, and Meis2 (Garel et al., 1999; Toresson et al., 1999; Carri et al., 2013). The differentiation of MSNs is dependent on the temporal expression of a set of transcription factors, particularly the repressive function of Dlx1/2 on Ascl1 at specific timepoints, to promote differentiation and migration of striatal neurons (Anderson et al., 1997b; Yun et al., 2002). EBF1 then controls later differentiation and migration from the SVZ to the MZ (Garel et al., 1999).
Olfactory Bulb Neurogenesis
In mice, olfactory bulb neurogenesis occurs from embryonic until early postnatal stages, and is dependent on the neuronal types (Alvarez-Buylla and Lim, 2004; Tucker et al., 2006; Figueres-Oñate and López-Mascaraque, 2016). Initially, projection neurons are generated by E12.5, followed by the development of inhibitory interneurons by E14.5 (Bayer, 1983; Tucker et al., 2006). The olfactory bulb projection neurons, mitral/tufted (M/T) cells, originate from progenitor cells in the pallium and are differentiated from Pax6+ radial glia (Whitman and Greer, 2009; Imamura and Greer, 2013). M/T cells can adopt both radial and tangential migration. Earlier born neurons predominantly migrate radially and populate the deeper cortical layers, while later born projection neurons are more likely to migrate tangentially to the superficial cortical layer (Imamura et al., 2011). Migration of these projection neurons is regulated by a number of transcription factors, such as PAX6 and LHX2, which are also crucial for cortical neuron migration (Nomura et al., 2007; Saha et al., 2007). Transcription factors specific for olfactory bulb projection neuron migration include Ap2-epsilon, Arx, and FezF1, which are all important for proper orientation of M/T cells, as well as the expression of Tbr1/2 (Yoshihara et al., 2005; Feng et al., 2009; Shimizu and Hibi, 2009; Imamura and Greer, 2013).
Olfactory bulb interneurons, in contrast to cortical interneurons, are derived from the dLGE, and postnatally in the SVZ, with the exception of Emx1+ pallial progenitors (Wichterle et al., 2001; Stenman et al., 2003). Subsequently, these interneurons tangentially migrate through to the olfactory bulb, postnatally through the rostral migratory stream (Kriegstein and Alvarez-Buylla, 2009). Although born in neuroanatomic regions distinct from cortical interneurons, olfactory bulb interneuron migration is regulated by a similar set of factors. Some of these include Dlx1/2, Ascl1, and Robo-Slit (Andrews et al., 2006; Long et al., 2007). Upon reaching the olfactory bulb, the interneurons differentiate into GABAergic interneurons and subsequently, subtype specification takes place (Lois and Alvarez-Buylla, 1994; Sequerra, 2014) which is itself dependent on the developmental stage, i.e., whether born at an embryonic or postnatal stage (De Marchis et al., 2007; Batista-Brito et al., 2008). Examples of transcription factors that regulate interneuron development are Sp8/Sp9 which are essential for olfactory bulb development (Li et al., 2017). For a more in-depth discussion about olfactory bulb development refer to a recent review from Tufo et al. (2022).
Radial and Tangential Migration of Neurons
There are two modes of neuronal migration, radial and tangential, classified by the axis of migration (Figure 1B). Cells move from the VZ toward the MZ generally by radial migration, and can descend within the VZ before migrating toward the MZ. Radial migration occurs during the development of the cerebral cortex, spinal cord, striatum and thalamus (Ayala et al., 2007). Morphological changes of interneurons mark the start of radial migration, whereas restriction of such changes also impairs the migration of these interneurons (LoTurco and Bai, 2006). Two different modes of movements are adopted during radial migration (Nadarajah et al., 2001). Interneurons migrate by somal translocation, by attaching to the outer surface of the developing brain (pial surface) and as microtubules shorten, the nucleus is pulled forward (Franco et al., 2011). Locomotion, on the other hand, allows interneurons to be guided by radial glial cells toward the destination during the radial migration through complex forebrain structures (Rakic, 1972).
Tangential migration is adopted by cortical interneurons born in the GE, as these cells need to migrate from the GE to the neocortex while avoiding movement toward the striatum (DeDiego et al., 1994). Despite being derived in different areas, interneurons arising from the MGE, CGE and preoptic area have a similar transcriptome (Mayer et al., 2018), which could contribute to the similar migration pattern these interneurons adopt. Transcription factors tightly regulate the migration fate of interneurons, such as the expression or repression of Nkx2.1 determines whether interneurons migrate into the striatum or neocortex, respectively (Nóbrega-Pereira et al., 2008). There are two major paths for interneurons to migrate from the GE to the developing neocortex, through a superficial route that bypasses the MZ or a deeper route that passes through the SVZ (Figure 1A; Wichterle et al., 2001). These migration paths are guided by signaling molecules such as the chemokine CXCL12, which attract interneurons, and its receptor CXCR4. Studies have shown that disruption of CXCL12 or its receptor CXCR4 led to interneuronal mislocalization (Stumm et al., 2003; López-Bendito et al., 2008; Wang et al., 2011b). Furthermore, Tbr2+ cortical intermediate progenitor cells may actively attract interneuron migration into the cortex, which is concurrently modulated by CXCL12 signaling (Sessa et al., 2010). Another chemokine, Neuregulin 3 (Nrg3), mediated by ErbB4 attracts and regulates the final destination of GABAergic interneurons in the cortex (Rakić et al., 2015). Similarly, repulsive guidance cues Semaphorin 3A and 3F also play a role in guiding interneuron tangential migration, where their expression in the LGE prevents interneuron migration toward the basal area (Chen et al., 2008). This repulsion is achieved by the interactions between these molecules and their receptors neuropilin-1 (Nrp1) and neuropilin-2 (Nrp2), which are expressed in migrating interneurons (Marín et al., 2001). Some other extrinsic factors act as mitogens to provide motility and control the rate of migration for interneurons, such as the hepatocyte growth factor/scatter factor (Powell et al., 2001). Furthermore, GABA itself can act as a motogen and accelerate tangential migration (Inada et al., 2011). These processes that direct neuron fate determination are ultimately regulated by members of the homeobox and basic helix-loop-helix (bHLH) transcription factor families (Table 1).
Homeobox Genes
Homeobox genes are an important gene family for embryonic development, defined by a conserved homeodomain (HD) containing a helix-loop-helix-turn-helix structure (Gehring et al., 1994; Noyes et al., 2008). The 60 amino acid HD is commonly located at the carboxyl terminal end of the protein, and binds DNA primarily through the 50th residue, usually being a glutamine, allowing homeobox genes to function as transcription factors (Figure 2; Kappen, 2000). This DNA binding motif is located in the second and third helices, which recognizes and binds to the major groove of DNA at specified consensus sites (Table 2). Further, the N-terminal arm contributes to the binding strength through interactions with the DNA minor groove, typically through a basic residue such as arginine at the 5th residue in the HD (Rohs et al., 2009). Apart from the consensus binding sequence, other important factors for DNA binding specificity include cofactors and additional DNA binding domains, such as the paired domain (PRD) in PAX superfamily members. Water molecules have been shown to be crucial for the HD to bind DNA (Billeter et al., 1996). Protein-protein interactions driven by the flanking regions around HD also increase the specificity of DNA binding (Li et al., 1995; Amin et al., 2015; Merabet and Lohmann, 2015). Homeobox proteins often contain other domains apart from the HD, which provide additional DNA specificity for these proteins, and have allowed characterization of homeobox proteins into 11 different classes, such as the Antennapedia (ANTP), Paired (PRD), LIM and NK classes (Holland et al., 2007), and can be further divided into different families within these classes. Large functional and comparative genomics studies have enabled analyses of these proteins, and allowed accurate annotation, naming and classification of homeobox genes (Holland et al., 2007).
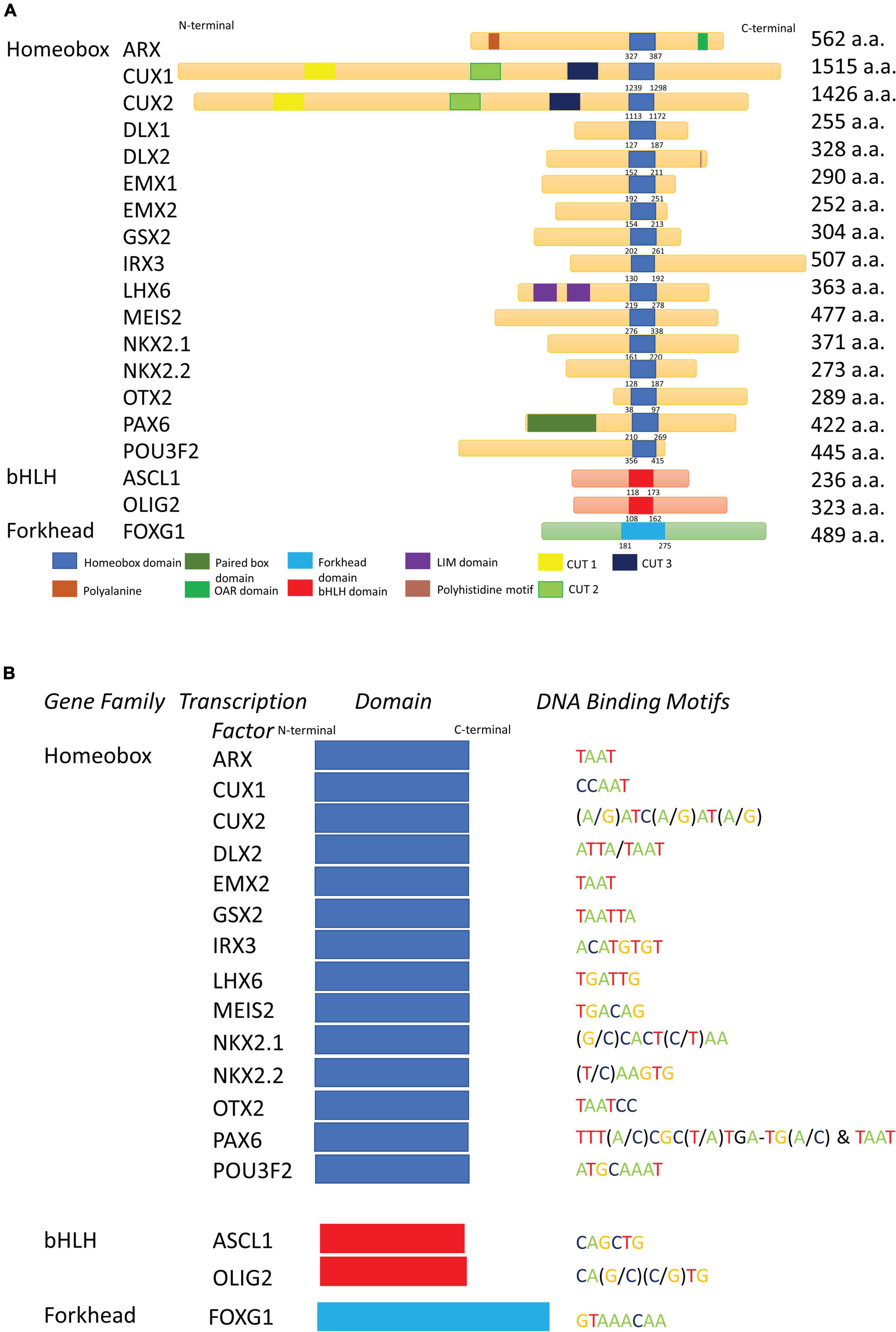
Figure 2. Homeobox transcription factors and their key functional domains. (A) Schematic depiction of the domain structure of selected homeobox, bHLH and forkhead transcription factors illustrating the highly conserved nature of the homeodomains and other DNA-binding domains of these transcription factors. Other significant functional domains are also shown. DLX2, but not DLX1, contains a polyhistidine motif in its C-terminus. PAX6 contains both a paired domain, as well as the HD. ARX, along with a HD, also contains a polyalanine repeat and an Aristaless domain. a.a., amino acid. (B) The DNA binding sites for Homeobox, bHLH and Forkhead transcription factors from (A). A consensus DNA binding motif for DLX1 and EMX1 is not available.
Basic Helix-Loop-Helix Genes
Basic helix-loop-helix (bHLH) proteins are another superfamily of transcription factors present in most eukaryotes, with critical functions during embryonic development, such as neurogenesis and myogenesis. bHLH domains contains two alpha helices, helix 1 and helix 2. These helices are connected by a short loop, and at the amino-terminal end of helix 1 is a basic region (Murre et al., 1989). This basic region binds DNA by recognizing a core CANNTGG motif, known as an E-box motif, and is specific for different transcription factors (Table 2). Upon binding, the basic region is fitted into the major grove of the DNA. The HLH domain interacts with other proteins, forming different homo- or hetero-dimeric complexes that are required for DNA binding (Ellenberger et al., 1994). The unique combinations of these bindings give rise to the diverse transcriptional regulatory functions of bHLH proteins during development. bHLH proteins can be roughly divided into those that are either cell-type specific or widely expressed where the group of transcription factors governing neuron development are often referred to as proneural proteins (Lee, 1997; Srivastava et al., 1997).
Selected Transcription Factors Encoded by Homeobox Genes
In the following major section of this comprehensive review, detailed summaries of 21 homeobox genes (in alphabetical order) that encode homeodomain containing transcription factors are provided. These genes were selected due to their essential role in forebrain development. However, we acknowledge that this selection of genes excludes several other important homeobox genes as well as key bHLH (Ascl1, Olig1, Olig2, and Olig3) and forkhead (Foxg1) genes required for neurodevelopment. For this reason, we have included Ascl1, Olig1/2/3, and Foxg1 in Figure 2 and the Tables.
A brief note about gene and protein nomenclature is useful. By consensus: mouse gene, Dlx; zebrafish gene, dlx; human gene, DLX; mouse and human protein, DLX.
Aristaless Related Homeobox Gene
The Aristaless related homeobox (Arx) paired-like HD transcription factor is the vertebrate homolog of the Drosophila aristaless (al) gene, which is essential for appendage formation (Miura et al., 1997). The gene is located on human chromosome Xp22.13 and is reported to be involved in neurological disorders such as X-linked intellectual disabilities (Table 3; Bienvenu et al., 2002; Friocourt and Parnavelas, 2010). In vertebrate embryogenesis, Arx transcriptionally regulates interneuron specification and migration (Fulp et al., 2008; Friocourt and Parnavelas, 2010; Olivetti and Noebels, 2012). ARX contains multiple structural domains and motifs, including the HD, a PRD-like domain, an N-terminal octapeptide domain, a central acidic domain and the C-terminal aristaless domain as well as three nuclear localization sequences and four polyalanine (polyA) tracts (Miura et al., 1997; Figure 2). ARX binds the transcriptional co-repressor TLE1, through the TLE1 octapeptide domain, and recognizes DNA at TAAT sites (Jennings et al., 2006; McKenzie et al., 2007; Cho et al., 2012). In vitro assays show that although ARX can be phosphorylated at multiple sites, it is unclear whether ARX functions are regulated by its phosphorylation state (Mattiske et al., 2018; Shi et al., 2020).
Aristaless Related Homeobox is expressed in various parts of the developing forebrain, such as the SVZ in developing GE and the VZ in the neocortex (Figure 1B; Miura et al., 1997; Colombo et al., 2004). Arx expression in the neocortex is limited to the proliferating neural progenitor cells, and is suppressed in cells radially migrating from the VZ (Friocourt et al., 2006), whereas in the GE Arx is continually expressed after neuronal differentiation and migration. Embryonic mice with homozygous Arx mutations have small brains with a thin neocortex and die upon birth, which may be related to defective tangential migration of cortical interneurons (Kitamura et al., 2002; Colombo et al., 2007; Friocourt et al., 2008). Targeted conditional deletion of Arx in the neocortex results in intermediate progenitor cell proliferation, with a reduced population of cortical neural progenitors. ARX also directly regulates cortical progenitor cell expansion through transcriptional regulation of CDKN1C, a cell cycle progression inhibitor in cortical VZ and SVZ (Colasante et al., 2015). The expression pattern of Arx also reveals its contribution to establishing the dorsoventral identity of the developing brain, where Arx suppresses ventralization in the dorsal forebrain by repressing Olig2 expression. Olig2 is a ventral specific gene and its expression is induced through Sonic Hedgehog (SHH) signaling. The expression of SHH downstream targets, Gli1 and Ptch3, are increased in Arx cKO mice dorsal telencephalon. Thus ARX represses these SHH downstream signals, and in turn represses Olig2 expression (Lim et al., 2019). Both inactivation, through shRNA, and overexpression of Arx impact GABAergic interneuron tangential migration to the neocortex from the MGE (Colombo et al., 2007). Furthermore, Arx is a direct regulatory target of DLX2, another homeobox transcription factor that regulates tangential migration, where overexpression of Dlx2 increases Arx levels and reduction of Dlx2 expression reduces Arx in the GE (Cobos et al., 2005a). By gain- and loss-of-function analysis, Arx was demonstrated to mediate the tangential interneuronal migration driven by DLX2, but not GABAergic neuron specification (Colasante et al., 2008). Conditional deletion of Arx in the ventral telencephalon further supports a role for Arx in tangential migration resulting in an overall reduction in the number of mature interneurons (Marsh et al., 2016). Additionally, ARX has been shown to transcriptionally regulate genes important for migration, such as Cxcr4, Cxcr7, Ebf3, and Lhx7 (Fulp et al., 2008; Colasante et al., 2009; Quille et al., 2011).
Aristaless Related Homeobox mutations can lead to severe neurological diseases, including X-linked intellectual disability, epilepsy, as well as structural brain malformations (Table 3; Friocourt and Parnavelas, 2010), and these mutations have been studied extensively using mouse models (Kitamura et al., 2002, 2009; Marsh et al., 2009; Price et al., 2009). The phenotypes related to ARX mutations can be grouped based on whether there is a corresponding malformation. Disorders in the malformation group include X-linked lissencephaly associated with abnormal genitalia (Kitamura et al., 2002) and Proud syndrome (Kato et al., 2004), whereas the non-malformation group includes epilepsy, non-syndromic X-linked intellectual disability, and X-linked Infantile Spasms Syndrome (Bienvenu et al., 2002; Kitamura et al., 2009; Price et al., 2009) and different epilepsy syndromes such as West syndrome (Strømme et al., 2002; Kato et al., 2003). Many mutations in ARX have been found in the first two polyA tracts, where the polyA tracts are expanded by insertion of either additional alanine or other residues (Kitamura et al., 2009). A common mutation consists of an in-frame 24bp duplication (Szczaluba et al., 2006), whilst longer mutations, 27bp, and 33bp have also be reported (Demos et al., 2009; Reish et al., 2009). The longest known mutation exhibits the addition of eleven alanine residues, resulting in Ohtahara syndrome (Kato et al., 2007). Other intellectual disability, seizures related disorders have also been observed (Turner et al., 2002). In summary, these ARX mutations disrupt DNA and protein binding ability, perturbing the transcriptional activity of ARX, thereby affecting cortical development (Nasrallah et al., 2012; Siehr et al., 2020).
Cut-Like Homeobox Genes
The Cut-like homeobox genes encode a transcription factor family [Cux homeobox 1/2 (Cux1/2)], previously called CCAAT-displacement protein (CDP) or Cut-like homeobox 1/2 (Cut1/2), that are the mammalian homologs of the Drosophila gene cut locus (ct) (Blochlinger et al., 1988). Ct is responsible for controlling the fate of neuronal progenitor cells in the peripheral nervous system and external sensory organs in Drosophila (Bodmer et al., 1987; Blochlinger et al., 1988) and plays a crucial role in dendritic arborization of specific sensory neurons (Grueber et al., 2003). CUX1 is located on human chromosome 7q22 and is frequently rearranged in cancers (Scherer et al., 1993), while CUX2 is on chromosome band 12q24.11-q24.12 (Craddock et al., 1993). CUX transcription factors contain up to four DNA binding regions, comprised of one HD, including a histidine residue at the 9th amino acid of the third helix (Blochlinger et al., 1988), and one, two, or three highly homologous Cut repeats of approximately 70 amino acids (CR1, CR2, CR3) (Figure 2A; Nepveu, 2001). However, individual Cut repeats are unable to bind to DNA on their own but interact with other Cut repeats or with the Cut HD to bind DNA (Moon et al., 2000). CR1/CR2 mediate transient binding to DNA (Moon et al., 2000) and the CR3 repeat and the HD have been reported to form bipartite high affinity DNA binding interactions (Harada et al., 1994, 1995). Cux1 and Cux2 splice variants encode for protein isoforms with different combinations of DNA binding domains (Weiss and Nieto, 2019). Proteolytic cleavage of the full length p200 CUX1 protein generates a p110 protein which contains CR2, CR3 and the HD (Goulet et al., 2004). While the full-length p200 protein acts as a transcriptional repressor, p110 can act as repressor or activator depending on the type of promoter it interacts with (Yoon and Chikaraishi, 1994; Truscott et al., 2004, 2007; Harada et al., 2007). CUX proteins can act as transcriptional repressors either indirectly by competing with transcriptional activators for binding to target sites, or actively suppressing transcription via a mechanism that involves recruiting histone deacetylases through the Ala, Pro-enriched carboxyl domain (Cowell and Hurst, 1994; Mailly et al., 1996; Nepveu, 2001). CUX transcriptional activity is regulated by post-translational modifications at the Cut repeats which include acetylation, proteolysis (Sansregret et al., 2010), and phosphorylation by PKC (Coqueret et al., 1996), CKII (Coqueret et al., 1998), cAMP-dependent protein kinase (Michl and Downward, 2006), and cyclin A/Cdk1 (Santaguida et al., 2001), which repress transcriptional activity.
Cux1 expression is detected widely in embryonic and adult tissues (Nieto et al., 2004), while Cux2 is more specifically expressed in the nervous system (Quaggin et al., 1996) as well as the limb buds and urogenital system (Iulianella et al., 2003). Cux1 and Cux2 are expressed early during brain development in neural progenitor cells in the ventral and dorsal telencephalon, as early as E14 for Cux1 and E10.5 for Cux2, specifically Cux1 is expressed in the VZ and SVZ of whole GE (Nieto et al., 2004; Zimmer et al., 2004; Figure 1). In contrast, Cux2 is solely expressed in the SVZ of the MGE, and is enriched in tangentially migrating cortical interneurons (Nieto et al., 2004; Zimmer et al., 2004). Indeed, Cux2 is mostly expressed in SVZ/IZ early during development while it is later expressed across most of the cortex (Zimmer et al., 2004). Furthermore, Cux2 expression distinguishes two cortical neuronal subpopulations with different origins, migration models, and phenotypic characteristics: a population of tangentially migrating GABAergic cortical interneurons and another DLX-negative neuronal population produced in the pallium, which migrates radially, divides in the SVZ and accumulates in the IZ (Zimmer et al., 2004).
In addition to controlling neural specification and differentiation in upper cortical layers, CUX proteins can act as repressors for developmental processes such as dendritic arborization (Grueber et al., 2003; Cubelos et al., 2010; Li et al., 2010). Overexpression of Cux1, but not Cux2, results in decreased dendritic arborization in cultured cortical pyramidal neurons, whereas dendritic complexity increases upon reduction of Cux1 (Li et al., 2010). A mechanism whereby Cux1 transcriptionally represses dendritic arborization is through suppression of the cyclin-dependent kinase inhibitor p27Kip7 and further plays a role in proliferating cells by repressing the p21 cyclin kinase inhibitor (Coqueret et al., 1998).
Cux2 is regulated by PAX6 and contributes to determining the upper layers (II-IV) of the cortex (Zimmer et al., 2004). Deletion of either Cux1 or Cux2 in mice does not alter overall cortical and brain organization (Cubelos et al., 2008a), whereas most Cux1 and Cux2 double homozygous mutants die prior to birth (Cubelos et al., 2008b). Although, the few pups that survive P0 do not display defects in neuronal migration or in layer specific protein expression (Cubelos et al., 2008b), Cux1/Cux2 double knockout (DKO) mice display abnormal dendrites and synapses indicating a critical role for Cux genes in dendritogenesis (Cubelos et al., 2010). The formation of cortical interneurons in Cux single and double mutants is impaired while loss of Reelin expression is only observed in upper cortical layers II-IV in double mutants (Cubelos et al., 2008b).
Cux2 deficient mice display increased brain volume, cell density and thickness of the upper cortical layers (II-IV), caused by an increase in the number of neuronal progenitor cells (Cubelos et al., 2008a). CUX1 target genes include Nfib, Fezf2, Pou6f2 and Sox5 which are all transcriptional regulators highly expressed in lower layers of the cortex (Gray et al., 2017). In addition to regulating upper cortical layer formation, Cux2 has also been shown to control cell cycle exit (Cubelos et al., 2008a). Therefore, Cux1 and Cux2 regulate neuronal proliferation of intermediate neuron precursors in SVZ, as well as the proliferation rate of neuronal precursor cells fated to form pyramidal cortical neurons in the upper layers of the cortex (Cubelos et al., 2008a,b) and in the spinal cord (Iulianella et al., 2008).
Mutations in CUX1 have been associated with global developmental delay with or without impaired intellectual development (GDI) (Platzer et al., 2018) while CUX2 is associated with intellectual disorders, seizures, autism spectrum disorder and bipolar affective disorder (Glaser et al., 2005; Barington et al., 2018). CUX1 has also been shown to undergo inactivating mutations and loss of heterozygosity (LOH) in a number of human cancers (Ramdzan and Nepveu, 2014; Wong et al., 2014). Loss of CUX1 activates the phosphoinositide-3-kinase (PI3K) signaling pathway as a result of transcriptional downregulation of the PI3K inhibitor, PIK3Ip1 (Wong et al., 2014). This mutation in CUX1 results in increased tumor growth and increased susceptibility to PI3K-Akt inhibition (Wong et al., 2014). CUX1 has also been implicated in the regulation of proteosome-mediated degradation of the Src tyrosine kinase resulting in altered tumor cell migration and invasion (Aleksic et al., 2007).
Distalless Genes
Distalless (dll) was discovered in Drosophila for its essential role in limb development (Cohen et al., 1989). Dlx genes are the vertebrate orthologs of dll; six members of this gene family can be found in humans and mice, occurring as bigenic clusters (Dlx1/2, Dlx3/4, and Dlx5/6); however, only Dlx1, Dlx2, Dlx5, and Dlx6 are expressed in the forebrain (Figure 1B). Dlx1/2 and Dlx5/6 are located on mouse chromosomes 2 and 6, and on human chromosomes 2q31.1 and 7q21.3, respectively (Stock et al., 1996). These bigenic clusters are organized from tail-to-tail, with highly conserved intergenic enhancers located between the two genes. Dlx1/2 and Dlx5/6 each contain two intergenic enhancers: i12a and i12b for Dlx1/2, and i56a and i56b for Dlx5/6 (Ghanem et al., 2003; Ruest et al., 2003). These cis-regulatory elements, although dissimilar in sequence, have overlapping activity and are essential for the expression of these genes (Fazel Darbandi et al., 2016). Dlx5/6 expression is regulated by Dlx1/2, where the absence of Dlx1/2 reduces Dlx5/6 expression through the intergenic enhancer, revealed using gene reporter systems (Zerucha et al., 2000; Zhou et al., 2004). Likewise, removing the intergenic enhancers with a targeted mutation attenuates Dlx5/6 expression in the forebrain, suggesting these intergenic enhancers are necessary for Dlx expression (Robledo et al., 2002; Ghanem et al., 2003). Dlx transcription factors are expressed in the developing GE and are essential for forebrain development (Pleasure et al., 2000). From embryonic day 9.5 (E9.5), expression is induced in the order of Dlx2, Dlx1, Dlx5, and Dlx6 (Eisenstat et al., 1999). In mice, Dlx1/2 are expressed in the VZ in the GE, and are clearly separated at the pallio-subpallial boundary (Figure 1B). Dlx5/6 are expressed in the MZ of the ventral telencephalon, and additionally, Dlx1, Dlx2, and Dlx5 are expressed in the SVZ in an overlapping manner, coinciding with regions where GABAergic interneurons are produced (Liu et al., 1997; Acampora et al., 1999; Depew et al., 1999; Robledo et al., 2002; Cobos et al., 2005b; Weinschutz Mendes et al., 2020).
Dlx1 and Dlx2 single gene homozygous knockout (KO) mice die prematurely at postnatal day 0 (P0) with minor abnormalities in GABAergic neuron formation, demonstrating DLX1 and DLX2 are somewhat functionally redundant (Qiu et al., 1997). Cortical neurons are reduced in postnatal Dlx1 KO mice which can lead to seizures (Cobos et al., 2005b). Dlx1/2 and Dlx5/6 double homozygous mutants also die at P0 with a more significant forebrain defect compared to the single KO mice. Tangential interneuron migration from the MGE to the neocortex is blocked in Dlx1/2 double homozygous mutants both in mice and zebrafish, hindering GABAergic interneuron development (Anderson et al., 1997a,b; MacDonald et al., 2013). Dlx1/2 double homozygous mutants also have reduced Dlx5/6 expression, which results in altered progenitor cell fate in the dorsal and ventral telencephalon (Pla et al., 2017). Dlx5/6 double homozygous mutant mice also exhibit tangential migration defects, with poor specification of parvalbumin GABAergic interneuron subtypes (Wang et al., 2010). Therefore, Dlx genes are essential for the differentiation of GABAergic neurons and their subsequent tangential migration (Anderson et al., 1997a,b; Marin et al., 2000).
Distalless genes transcription factors promote interneuron production by regulating transcription of various downstream targets in the ventral telencephalon, binding to the core HD DNA binding motif ATTA/TAAT (Zhou et al., 2004; Table 2). A recent report has found that DLX2 binds preferentially to transcription factors to mediate both its’ repression and activation functions (Lindtner et al., 2019). GABA is synthesized by glutamic acid decarboxylases 1 and 2 (GAD1; GAD2) which are co-expressed with Dlx1/2 in the VZ and SVZ of the GE in mouse, zebrafish, and humans (Erlander et al., 1991; Liu et al., 1997; Martin et al., 2000; MacDonald et al., 2010; Al-Jaberi et al., 2015). Gad1 and Gad2 expression is dependent on the DLX factors, where DLX1/2 bind directly to the promoters of Gad1/2 in vivo and induce Gad1/2 expression; Gad expression is reduced in Dlx1/2 double homozygous mutant mice (Stühmer et al., 2002; MacDonald et al., 2010; Li et al., 2012a; Le et al., 2017). However, Gad expression is not completely ablated in these mutants, which could be due to the compensatory function of residually expressed DLX5/6. Additionally, DLX proteins promote the differentiation of GABAergic and cholinergic interneuron subtypes through regulation of Lhx6 and Lhx8, where both Lhx genes have reduced expression in Dlx1/2 double homozygous mutants (Petryniak et al., 2007; Long et al., 2009). DLX2 also downregulates Olig2 expression to repress oligodendrocyte development in early neurogenesis, and hence may control the balance between oligodendrocyte and neuron production (Petryniak et al., 2007; Jiang et al., 2020). ASCL1, in turn, represses Dlx2 expression in later developmental stages, to allow the expression of Olig2 and promote oligodendrocyte production (Petryniak et al., 2007; Poitras et al., 2007). The repression of Olig2 by DLX2 also represses the promotion of the progenitor cell states, and likewise DLX2 downregulates several other transcription factors with similar functions, such as Gsx2, Otx2, and Pax6 (Yun et al., 2001; Hoch et al., 2015; Lindtner et al., 2019). SMAD transcription factors, which are part of the transforming growth factor-β (TGF-β) signaling pathway, interact with DLX2 in binding to the promoter regions of DLX2 target genes in the telencephalon. Although expression of TGF-β signaling components is unaffected in the Dlx1/2 double mutants, the interaction between DLX2 and SMAD factors indicate TGF-β could play a role in GABAergic interneuron differentiation (Shi and Massagué, 2003; Maira et al., 2010).
In addition to cell differentiation, DLX transcription factors also regulate interneuron tangential migration. DLX1/2 regulates this process by repressing terminal differentiation of interneurons (Cobos et al., 2007). Interneurons develop axons and dendrites post migration, promoted by proteins that regulate cytoskeleton and cell motility such as MAP2 and PAK3 (Anderson et al., 1997b; Bokoch, 2003; Dehmelt and Halpain, 2004). In Dlx1/2 DKO mice, interneurons have significantly reduced migration, increased neurite length, and upregulated expression of genes which are normally expressed post-migration. Hence, DLX1/2 represses these genes to enable tangential migration of interneurons to the cortex (Cobos et al., 2007). Nrp2, encoding for a Semaphorin-3A and 3F receptor, is also repressed by DLX1/2, as evident in the marked increase of NRP2 expression in the forebrains of Dlx1/2 DKO mice (Le et al., 2017). In Dlx5/6 double homozygous mutant mice, a receptor for tangential migration Cxcr4 is downregulated in the SVZ, which likely contributes to the impaired tangential migration observed in these mutants (Wang et al., 2010, 2011b).
Although DLX transcription factors have not been directly linked to any neurological diseases, many associations have been made between DLX mutations and neurodevelopmental defects (Table 3). Epilepsy and Rett syndrome had been linked to Dlx mutations in mouse models. Furthermore, DLX1/2 and DLX5/6 are found on chromosomes 2q and 7q, which are autism susceptibility loci (Cobos et al., 2005b; Hamilton et al., 2005; Horike et al., 2005). By site-directed mutagenesis of the Dlx1/2 intergenic enhancer regions, transgenic mice with autism-like phenotypes were generated, showing the possible role of disrupted Dlx1/2 in autism development (Poitras et al., 2007). Several neurodevelopmental disorders have been related to Dlx genes due to the importance of this gene family in regulating GABAergic interneuron production and migration (Kato and Dobyns, 2004; Verret et al., 2012). A DLX2 direct target Grin2b is linked to schizophrenia, epilepsy, intellectual disability, and autism, which provides evidence that DLX2 may contribute to neural diseases (Endele et al., 2010; Pan et al., 2019). DLX2 regulation of transcription factors such as Arx and Olig2 also support that DLX factors may potentially contribute to neurological disease (Lindtner et al., 2019).
Empty Spiracle Genes
Empty spiracles homeobox (Emx) genes are the mammalian homologues of the Drosophila gene empty spiracle (ems), which is responsible for head structure development (Younossi-Hartenstein et al., 1997). Emx1 and Emx2 are homeobox genes important for dorsal patterning in the forebrain. From mouse studies, Emx2 is shown to be expressed earlier, from E8.5, whereas Emx1 is expressed from E9.5 (Simeone et al., 1992; Gulisano et al., 1996; Medina and Abellán, 2009). The expression of Emx2 is regulated by two sets of enhancers, one at the 5′ region and the other at the 3′ region (Theil et al., 2002; Suda et al., 2010; García-Moreno and Molnár, 2015). Emx2 expression is directly promoted by DMRT5 and downregulated by the Emx2 antisense transcript Emx2OS (Spigoni et al., 2010; Saulnier et al., 2013). Both Emx genes are expressed in the dorsal telencephalon, with the highest level of expression rostrolaterally, and decreased expression in a gradient caudomedially (Mallamaci et al., 1998). Emx1 expression is nested within Emx2 expression, and only Emx2 is expressed in the caudomedial part of dorsal telencephalon (Simeone et al., 1992; Yoshida et al., 1997). While Emx2 expression is restricted to progenitor cells, Emx1 is expressed in both progenitor and differentiated cells (Gulisano et al., 1996).
Both Emx1 and Emx2 are necessary for the development of the archipallium in the dorsal telencephalon, and especially the development of the hippocampus and cortex in later stages (Simeone et al., 1993; Pellegrini et al., 1996; Yoshida et al., 1997; Hamasaki et al., 2004). Emx1 and Emx2 double homozygous mutants do not develop the dorsomedial telencephalon, whilst this phenotype is not observed in Emx1 or Emx2 single homozygous mutants (Bishop et al., 2003; Shinozaki et al., 2004). The impaired development of the neocortex could also be due to impaired tangential migration, as interneurons in Emx1/Emx2 double mutant cannot migrate out of the GE into the cortex (Shinozaki et al., 2002). Homozygous Emx1 mutants do not develop significant defects in the embryonic neocortex (Yoshida et al., 1997; Bishop et al., 2002). However, postnatal studies have shown that Emx1 could play a role in cortical patterning, as rostral areas were expanded and caudal areas were reduced in the Emx1 null mice (Stocker and O’Leary, 2016). Emx1 homozygous mutants also lack development of the corpus callosum, and heterozygous Emx1 mutants exhibit partial penetrance (Qiu et al., 1996). However, Emx2 homozygous mutants have reduced neocortex size by E11.5, with defective dorsal telencephalon development, including aberrant hippocampus formation and impaired radial migration of neurons (Pellegrini et al., 1996; Yoshida et al., 1997; Mallamaci et al., 2000). In these mutants, there is ventralization of the dorsal telencephalon with reduced dorsal gene marker expression (Ngn1, Ngn2, and Emx1) and increased ventral marker gene expression (Gsx2, Ascl1, and Dlx1/2). Emx2/Pax6 double homozygous mutants demonstrate a stronger phenotype with a lack of dorsal identity, showing these two homeobox factors function cooperatively to specify dorsal telencephalic identity (Muzio et al., 2002a,b). Furthermore, reciprocal inhibition is observed between Emx2 and Pax6, where the cKO of one factor results in the upregulation of the other (Muzio et al., 2002b).
EMX1 and EMX2 regulate a number of factors required to specify dorsal telencephalic identity (Table 2). An important aspect of EMX2 function is its’ regulation of the formation of the PSB, along with PAX6 and GSX2 (Yun et al., 2001; Muzio et al., 2002a,b). EMX2 cooperates with DMRT5 and DMRT3 to repress Gsx2 expression, with all three proteins binding directly to the ventral telencephalon specific Gsx2 enhancer, thereby contributing to the development of the PSB (Desmaris et al., 2018). A mutual repressive relationship between EMX2 and FGF8 also promotes the differentiation of neural progenitor identity, where EMX2 downregulates FGF8 to promote differentiation, whilst FGF8 represses EMX2 to promote anterior-posterior patterning (Fukuchi-Shimogori and Grove, 2001, 2003; Cholfin and Rubenstein, 2008). In addition, EMX2 represses Sox2 by inhibiting positive regulators from binding to Sox2 enhancers. Sox2 cKO mutants have a defective hippocampal phenotype, rescued when one Emx2 allele is lost (Mariani et al., 2012). This demonstrates that EMX2 regulates hippocampal development, consistent with the observed Emx2 homozygous phenotype (Pellegrini et al., 1996). Wnt signaling promotes Emx2 expression through activation of an Emx2 telencephalic enhancer, through the Wnt downstream factor GLI3 (Theil et al., 1999, 2002; Muzio et al., 2005). Additionally, EMX2 restricts Wnt-1 expression in the forebrain, which is essential for maintaining normal neuronal radial migration (Iler et al., 1995; Ligon et al., 2003). EMX1 regulates Nrp1, an axonal guidance receptor that regulates cortical connectivity (Wright et al., 2007; Lim et al., 2015). Furthermore, EMX2 regulates Teneurin-1, a transmembrane protein that also functions in axonal guidance, through binding to an alternative promoter (Table 3) and promoting the transcription of an alternative transcript (Drabikowski et al., 2005; Li et al., 2006; Beckmann et al., 2011).
While EMX2 functions in promoting cell differentiation in the developing brain, it is considered as a possible tumor suppressor in different cancers, such as sarcoma, colorectal cancer, gastric tumors, and glioblastoma (Table 3; Li et al., 2012b; Aykut et al., 2017; Jimenez-García et al., 2021a,b). In many tumors, EMX2 expression is downregulated due to methylation of the EMX2 promoter (Okamoto et al., 2010; Qiu et al., 2013). EMX2 over-expression blocks cell proliferation through inhibiting the canonical Wnt pathway, and also leads to cell cycle arrest with increased cell death of glioblastoma cells (Falcone et al., 2016; Monnier et al., 2018; Jimenez-García et al., 2021a).
Genomic Screened Homeobox Genes
Genomic screened homeobox (Gsx, formerly Gsh) genes encode a family of transcription factors important for patterning of the ventral telencephalon. Gsx genes are the mammalian orthologues of the Drosophila intermediate neuroblasts defective (ind) genes; mutation in Drosophila induces a loss of intermediate neuroblasts (Weiss et al., 1998). GSX proteins bind to DNA via the homeobox domain (Figure 2), and GSX2 activity may depend on its dimerization state, where homodimers promote gene activation, and monomers enhance gene repression (Salomone et al., 2021).
Gsx1 and Gsx2 are widely expressed in the neural progenitors found in the VZ of the LGE (Toresson et al., 2000). Gsx2 is mostly expressed in the dLGE with lower levels in the vLGE with complementary patterns for Gsx1, localizing to the vLGE and MGE (Toresson and Campbell, 2001; Yun et al., 2001). Gsx1 and Gsx2 are partially functionally redundant, due to similarities in their consensus DNA binding sites (Hsieh-Li et al., 1995; Valerius et al., 1995; Toresson and Campbell, 2001; Pei et al., 2011). Gsx2 and Gsx1/Gsx2 DKO mice have a reduced LGE size, with decreased number of olfactory bulb neurons as well as GABAergic interneurons (Yun et al., 2003). Dorsal markers including Pax6 and Ngn2 also expanded ventrally in Gsx2 homozygous mutants (Szucsik et al., 1997; Corbin et al., 2000; Toresson et al., 2000; Yun et al., 2001). The expression patterns of Pax6 and Gsx2 are complementary, separated by the PSB, and these genes function cooperatively to define the dorsoventral identity of the developing forebrain (Figure 1B; Yun et al., 2001; Carney et al., 2009). Gsx2 expression is repressed by a number of genes in the dorsal telencephalon, including Pax6, Emx2, Dmrt3 and Dmrt5 (Muzio et al., 2002a,b; Desmaris et al., 2018).
Gsx2 regulates specification of neurons, oligodendrocytes and glial cells in the LGE (Kessaris et al., 2006; Fogarty et al., 2007; Chapman et al., 2018). Neurogenesis and oligodendrogenesis take place in the dLGE and vLGE, respectively, and are tightly controlled by Gsx2 in a time-dependent manner. Conditional knockout (cKO) of Gsx2 upregulates the oligodendrocyte precursor cell (OPC) marker Pdgfrα, and promotes premature oligodendrocyte differentiation (Corbin et al., 2003; Chapman et al., 2013, 2018). Ascl1, a bHLH transcription factor crucial for neurogenesis, has reduced expression levels in Gsx2 homozygous mutant mice (Chapman et al., 2013). Studies have shown that Gsx2 upregulates Ascl1 in earlier embryo stages, promoting neuronal differentiation in early embryonic stages (Méndez-Gómez and Vicario-Abejón, 2012; Chapman et al., 2013; Wang et al., 2013). Since Ascl1 promotes the NPCs to differentiate into interneurons, GSX2 inhibits ASCL1 activity to regulate the balance between progenitor cell proliferation and differentiation (Roychoudhury et al., 2020). Whilst GSX2 upregulates Ascl1, it inhibits the homo- and heterodimer formation of ASCL1 essential for its DNA binding ability (Johnson et al., 1992; Nakada et al., 2004; Roychoudhury et al., 2020). In earlier embryonic stages (E9-11), Gsx2 promotes striatal projection neuron specification from the vLGE, and in later embryonic stages (E12.5–E15) olfactory bulb interneurons are specified in the dLGE (Waclaw et al., 2010).
Overexpression of Gsx2 from E13.5 promotes the specification of dLGE over vLGE, and subsequently favors neurogenesis over oligodendrogenesis (Waclaw et al., 2010; Pei et al., 2011; Chapman et al., 2013). GSX1 in Ascl1 expressing progenitor cells represses Gsx2 and promotes the maturation of NPCs by transitioning these cells from the VZ to the SVZ and induces differentiation (Pei et al., 2011). Gsx1/Gsx2 DKO mice have expanded OPCs comparable to Gsx2 homozygous mutants (Chapman et al., 2018). However, the reduced proliferation of OPCs in the Gsx1/2 DKO compared to Gsx2 homozygous mutants suggests GSX1 functions in promoting OPC proliferation in the ventral telencephalon (Chapman et al., 2018). Hence, Gsx2 regulates neurogenesis through repressing Gsx1, and blocks oligodendrogenesis in early embryonic stages. Furthermore, downregulation of Gsx2 in late embryonic stages is essential for oligodendrogenesis to proceed, which could be a result of negative autoregulation (Salomone et al., 2021).
Along with promoting Ascl1 expression, GSX2 regulates neural differentiation via increasing Dlx1 and Dlx2 expression in the LGE (Table 2; Corbin et al., 2000; Toresson et al., 2000; Wang et al., 2013). Dlx1/Dlx2 are part of the gene regulatory network downstream of Ascl1 and in turn negatively regulate Gsx1 and Gsx2 expression (Yun et al., 2002; Long et al., 2009; Wang et al., 2013). The activation of Gsx1 and Gsx2 regulates the patterning of LGE, and later silencing of these two genes by Dlx1/2 promotes subcortical neural differentiation (Anderson et al., 1997b; Cobos et al., 2005b). Furthermore, GSX2 also represses Dbx1, a homeobox transcription factor expressed in the hindbrain and spinal cord that regulates dorsoventral brain patterning and specification of Cajal-Retzius cells (Yun et al., 2001; Bielle et al., 2005; Winterbottom et al., 2010). DBX1 has also been suggested to repress Gsx1 in the ventral telencephalon; however, further studies are necessary to validate this relationship (Poiana et al., 2020).
Congenital brain malformations may result from mutations in the GSX2 gene (Table 3). Whole exome sequencing of patients with basal ganglia malformations reveals a homozygous missense mutation in GSX2 HD that impair its transcriptional activity (De Mori et al., 2019). These patients have similar phenotypes to homozygous mutant mice models, with malformations or defective structures derived from the LGE and MGE (putamen, globus pallidus, caudate nucleus and olfactory bulb), as well as maldevelopment of the forebrain midbrain junction (De Mori et al., 2019). These anatomical defects are also associated with a range of neurological disorders, such as Parkinson’s and Huntington’s Diseases (Zuccoli et al., 2015; Table 2).
Iroquois-Related Homeobox 3 Gene
The iroquois-related homeobox 3 (Irx3) is a TALE HD containing transcription factor (Figure 2), orthologous to the Iroquois-complex genes in Drosophila, which are responsible for the development of sensory organ, body-wall and wing identity (Gómez-Skarmeta et al., 1996; Bürglin, 1997; Diez del Corral et al., 1999). Irx genes in vertebrates are organized into two clusters, IrxA and IrxB, each containing 3 genes from the family. The IrxA cluster consists of Irx1, Irx2, and Irx4, whereas IrxB contains Irx3, Irx5, and Irx6, located on mouse chromosome 8 and human chromosome 16 (Peters et al., 2000).
Iroquois-related homeobox 3 is important for thalamic patterning in the diencephalon (Robertshaw et al., 2013). Irx3 is predominantly expressed in the midbrain, hindbrain, and spinal cord in early neurogenesis (E7.5–E9.5), and expression shifts rostrally to the diencephalon from E10.5 (Bosse et al., 1997). Notably, the expression patterns of Irx3 and Ascl1 during early neurogenesis are similar, which may suggest a regulatory relationship between the two transcription factors (Cohen et al., 2000). Similar to Dlx1/2/5 and Nkx2.1/2.2, Irx3 expression is posterior to the zona limitans intrathalamica (ZLI), a region in the diencephalon that releases SHH signaling molecules for the patterning of prethalamus and thalamus (Kitamura et al., 1997; Eisenstat et al., 1999; Robertshaw et al., 2013; Murcia-Ramón et al., 2020a). High levels of SHH signaling induces rostral thalamus, and subsequently the production of GABAergic interneurons, while a low level of SHH promotes caudal thalamus specification and glutamatergic interneurons production (Kiecker and Lumsden, 2005). Consistent with this, ectopic expression of Irx3 promotes the expression of thalamus differentiation markers Sox14 and Gbx2, both in the prethalamus and the dorsal telencephalon in response to SHH signaling (Kiecker and Lumsden, 2004; Robertshaw et al., 2013). However, such markers were not expressed upon Irx3 ectopic expression in the ventral telencephalon, which may be due to SIX3 repression of Irx3, which restricts its activity to specify thalamus identity (Kobayashi et al., 2002; Robertshaw et al., 2013). In Xenopus models, knockdown of Irx3 reduces midbrain size, and caudally shifts the forebrain-midbrain boundary, illustrating its function in ensuring the normal patterning of the diencephalon (Rodríguez-Seguel et al., 2009). A key co-regulator of thalamus patterning is PAX6, which is expressed anterior to the forebrain-midbrain boundary and specifies the caudal thalamus. The overlapping expression patterns of Irx3 and Pax6 (see Figure 2A) mark the region of thalamus patterning, while caudal and rostral thalamus identity is determined by levels of SHH signaling (Robertshaw et al., 2013).
Iroquois-related homeobox 3 is considered to be a determinant for obesity, in relation to the fat mass and obesity associated (FTO) genes, due to the role of Irx3 in neurogenesis at the paraventricular nucleus of the hypothalamus, developed from the anterior hypothalamus (Smemo et al., 2014). Single-minded 1 (Sim1), a bHLH transcription factor in the hypothalamus represses Irx3 expression, as Sim1 KO mice exhibit ectopic expression of Irx3 in the anterior hypothalamus (Caqueret et al., 2006; Son et al., 2021b). Sim1 homozygous mutant mice are perinatal lethal, whereas Sim1 heterozygous mutant mice exhibit neurodevelopmental defects and hyperphagia, as Sim1 is important for neurogenesis in the hypothalamus (Michaud et al., 1998; Holder et al., 2004). The neurogenesis defects in these mutant mice are due to the ectopic expression of Irx3 and Irx5 in the anterior hypothalamus (Son et al., 2021a,b). In Sim1/Irx3/Irx5 triple heterozygous KO mice, the neuronal population at the anterior hypothalamus is restored. Similarly, cKO of Irx3 at the paraventricular nucleus of the hypothalamus partially rescues the neuronal disruption observed in Sim1 heterozygous mutant mice, with no observable differences in body weight or hyperphagic phenotype (Son et al., 2021b).
Lhx (LIM-HD Family) Genes
The Lhx transcription factors belong to the LIM-HD family of homeobox genes that have both a LIM zinc finger domains and a HD (Figure 2; Dawid et al., 1998; Bach, 2000). The LIM zinc finger domain is named after the first three genes discovered in the family, Lin-11, Isl1 and Mec-3, and participates in protein-protein binding (Way and Chalfie, 1988; Freyd et al., 1990; Karlsson et al., 1990). Of the various members of the Lhx gene family found in both mouse and humans, Lhx1, Lhx2, Lhx5, Lhx6, and Lhx8 (i.e., L3/Lhx7) are important for differentiation and migration of interneuron in the developing telencephalon (Alifragis et al., 2004; Abellán et al., 2010; Godbole et al., 2018). Mutant mice studies had provided insights into the importance of these Lhx genes for forebrain development (Wanaka et al., 1997).
Lhx1 homozygous mutant mice have an increased number of PoA-derived interneurons and glia cells, suggesting Lhx1 regulates the survival of these cells by regulating the balance between apoptosis and proliferation. Also the PoA-derived interneurons in Lhx1 null mice migrate through the ventral telencephalon, compared to a more controlled migration in the wild-type mice through the developing neocortex (Symmank et al., 2019). Lim5 is expressed in the forebrain of zebrafish and Xenopus, and Lhx5, the Lhx1 paralog, is the murine ortholog. Lhx5 is expressed predominantly in the hindbrain at E8, and the developing forebrain starting at E9.5. After E11.5, Lhx5 is exclusively expressed in the ventral telencephalon, hypothalamus and diencephalon, which is complementary to Dlx5 expression (Figure 1B; Sheng et al., 1997). Both Lhx1/5 are expressed in the rostral area of the ZLI in the diencephalon, but only Lhx1 is expressed in the caudal ZLI (Nakagawa and Leary, 2001). Lhx5 homozygous mutant mice are defective in hippocampus development, where progenitor cells can proliferate but fail to exit the cell cycle to migrate or differentiate (Zhao et al., 1999). Cajal-Retzius neurons are responsible for the organization of the neocortex through the secretion of reelin (Soriano and Del Río, 2005). In mice, Lhx5 regulates the development and migration of Cajal-Retzius cells, which could be critical to the malformation of the hippocampus in Lhx5 null mutants (Abellán et al., 2010). Lhx1 likewise is expressed in some Cajal-Retzius cells, but limited to the septal area, and lateral olfactory to caudomedial zones (Miquelajáuregui et al., 2010). Additionally, Lhx5 can regulate forebrain development by suppressing Wnt signaling in zebrafish embryos, via promoting the expression of Wnt inhibitors Sfrp1a and Sfrp5, supported by the increase of Wnt signaling in zebrafish embryos lacking Lhx5 expression (Peng and Westerfield, 2006). There is some evidence of Lhx5 inhibiting Wnt5a in murine hypothalamus, promoting the growth of the mamillary body; however, more studies are required to confirm this regulatory effect and mechanism. Another possible target of Lhx5 is Lmo1 (LIM-only1), where Lmo1 competes with Lhx5 to bind with the Lhx binding partner LDB, thereby inhibiting Lhx function (Bach, 2000; Heide et al., 2015).
Lhx2 is the mammalian ortholog of the Drosophila apterous gene, first described in 1913, as an essential gene for Drosophila wing development (Metz, 1914; Butterworth and King, 1965). Lhx2 homozygous mutants have reduced forebrain volume, but expanded neocortex and PSB composing the entire forebrain (Porter et al., 1997; Bulchand et al., 2001; Monuki et al., 2001). Lhx2 plays a role in suppressing hippocampus (hem) and PSB (antihem) development up to E9.5 and E10.5, respectively (Roy et al., 2014; Godbole et al., 2018). Suppression of hippocampal development is regulated by interactions between Lhx2 and other transcription factors, namely Foxg1 and Pax6. Foxg1 has been shown to directly regulate Lhx2 expression, where the loss of Foxg1 also results in a loss of Lhx2 at E9.5. cKO of Lhx2 after E9.5 did not alter hippocampus development unless Foxg1 was also knocked out (Godbole et al., 2018). Pax6 is expressed in a lateral medial gradient in the neocortex, which is opposite to that of Lhx2. In Pax6/Lhx2 DKO, the hippocampus expands more so in the forebrain compared to Lhx2 null mice, suggesting Pax6 also suppresses the formation of hippocampus (Godbole et al., 2017).
Lhx6 and Lhx8 are structurally related and have synergistic functions. Lhx6 shares 75% homology with Lhx8, which is also known as L3 or Lhx7 (Matsumoto et al., 1996; Grigoriou et al., 1998). Both these genes are expressed overlappingly in the MGE but are not expressed in the LGE (Figure 1B). Lhx6 is expressed predominantly in the SVZ and the MZ, whilst Lhx8 is expressed in the MZ (Matsumoto et al., 1996). The expression of both these genes is regulated by Nkx2.1, another homeobox transcription factor that specifies ventral telencephalon development (Sandberg et al., 2016).
Lhx6 has similar functions to Lhx1. Lhx6 promotes expression of receptors that regulate cortical interneuron migration and transcription factors that control interneuron production, thereby regulating these events (Alifragis et al., 2004; Zhao et al., 2008; Neves et al., 2013). Tangential migration of GABAergic interneurons from the MGE into the neocortex are blocked in embryonic mice lacking Lhx6; normally these interneurons express Lhx6 in wildtype mice (Lavdas et al., 1999; Alifragis et al., 2004; Liodis et al., 2007). Such migration defects prevent the formation of functional connections between these neurons and their post-synaptic targets. Since Lhx6 has restricted expression in MGE progenitor cells, it does not regulate the migration of all cortical interneurons during development, especially at later stages where tangentially migrating neurons are born in the LGE (Marin et al., 2000; Nery et al., 2002). Production of GABAergic interneurons and their migration within the MGE are not affected in Lhx6 mutants, but interneuron subtype specification is dependent on the expression of Lhx6 (Neves et al., 2013). MGE-derived cortical interneurons are unable to differentiate into sst+ and pva+ subtypes, shown by a drastic reduction in the number of these neurons in Lhx6 null mutants. Lhx6 KOs had a greater effect on sst+ interneuron differentiation than pva+ interneuron differentiation, where pva+ interneuron differentiation was affected restrictively in the hippocampus (Liodis et al., 2007; Zhao et al., 2008; Yuan et al., 2018).
Lhx8, unlike Lhx6, is expressed in cholinergic neurons instead of GABAergic neurons (Lopes et al., 2012). Lhx8 is essential for the differentiation and specification of cholinergic interneurons, shown by the reduction of cholinergic neurons in Lhx8 homozygous mutant mice (Zhao et al., 2003; Fragkouli et al., 2005). Progenitor cells proliferate in Lhx8 homozygous mutant mice; however, they are unable to differentiate into cholinergic interneurons or glutamatergic neurons (Manabe et al., 2007, 2008). Cholinergic neurons are derived from progenitor cells in the MGE, where LHX8 promotes the expression of Isl1 upon cholinergic commitment, which in turn represses Lhx6 expression (Zhao et al., 2003). Lhx8 forms a hexamer with Isl1 and promotes cholinergic neuron expression by binding to specific motifs in the cholinergic enhancer sequence (Park et al., 2012). The formation of hexamers is necessary for DNA binding and subsequently cholinergic gene expression, whilst LHX8 or ISL1 alone does not bind to cholinergic enhancer sequences and are unable to promote cholinergic interneuron differentiation (Cho et al., 2014). NPCs in the striatum differentiate into GABAergic interneurons instead of cholinergic neurons in Lhx8 homozygous mutants. This is due to an upregulation of Lhx6 as a result of a lack of Isl1, suggesting the necessity of Lhx8 in cholinergic neuron specification (Manabe et al., 2005; Bachy and Rétaux, 2006). Additionally, Lhx6 acts cooperatively with Lhx8 to promote shh expression in the MGE, regulating the production of interneuron progenitors, as well as inhibiting Nkx2.1 expression in cortical neurons (Flandin et al., 2011). The Lhx6 and Lhx8/Isl1 regulatory network is therefore essential for regulating the differentiation of GABAergic and cholinergic neurons in the ventral telencephalon.
The LIM-domain transcription factor family is functionally important for the specification, differentiation and migration of neurons in the developing forebrain, and mutations in these genes can result in genetic diseases (Table 3). LHX2 mutations can result in pituitary hormone deficiency, although it is uncommon that a mutation in LHX2 alone can cause pituitary deficiency and developmental ocular abnormalities (Prez et al., 2012). The importance of Lhx6 on the differentiation of interneurons into sst+ and pva+ subtypes have a pathological link to schizophrenia (Volk et al., 2014; Donegan et al., 2020). There is reduced LHX6 expression in schizophrenic subjects who also have reduced expression of GAD1 (otherwise known as GAD67, a GABA synthesizing enzyme), sst, and pva expression. Reduction in GAD1 does not downregulate LHX6 and vice versa; hence, upstream factors likely contribute to the regulation of these genes (Volk et al., 2012). Moreover, a decrease in both GABAergic and cholinergic interneurons in the ventral telencephalon has been reported in Tourette Syndrome, suggesting LHX6 and LHX8 correlation with Tourette Syndrome due to their role in GABAergic and cholinergic interneuron specification in the striatum (Pagliaroli et al., 2020).
Myeloid Ectopic Viral Integration Site 2 Gene
The myeloid ectopic viral integration site (Meis) gene family belongs to the TALE class of homeobox proteins, a homolog of the Drosophila homothorax gene, which is essential for directing the localization of Pbx Drosophila homologue extradenticle (Rieckhof et al., 1997). There are three mammalian MEIS transcription factors (Meis1, Meis2, and Meis3), and all contain a conserved homothorax domain (Figure 2A), which promotes the interaction between MEIS and pre-B cell leukemia homeobox proteins (PBX), a transcription factor known for its regulatory role in organogenesis (Nakamura et al., 1996; Chang et al., 1997; Golonzhka et al., 2015). MEIS proteins are characterized by a three residue loop insertion between helices 1 and 2 of the HD, an important feature for protein-protein interactions (Bürglin, 1997). Out of the three Meis genes, only Meis1 and Mei2 are expressed in the developing telencephalon (Figure 1B). Meis2 in particular is an important player for striatal progenitors and neuron differentiation, as well as postnatal neuronal differentiation in the olfactory bulb (Toresson et al., 1999; Agoston et al., 2014).
Myeloid ectopic viral integration site 2 is expressed in the VZ of the entire telencephalon from E10.5, and is enriched in the LGE compared to the MGE from E12.5 to E18.5 (Figure 1B). From E14.5, MEIS2 is also expressed in the ventral thalamus and the anterior hypothalamus (Cecconi et al., 1997; Toresson et al., 1999, 2000). Additionally, the expression pattern of MEIS2 is similar in the human fetal forebrain, where MEIS2 is expressed in the proliferative zones (Larsen et al., 2010a). In the telencephalon, MEIS2 was initially considered as an LGE-specific marker due to its predominant expression in the LGE; however, MEIS2 is also widely expressed in the CGE progenitors (Toresson et al., 1999; Frazer et al., 2017). Postnatally, interneurons born and derived from the olfactory bulb express MEIS2, as it plays a crucial role, along with other transcription factors, in neuronal differentiation and specification in early postnatal stages (Allen et al., 2007; Agoston et al., 2014).
Myeloid ectopic viral integration site 2 forms complexes with various other transcription factors to cooperatively facilitate the expression of genes required for neurogenesis. As mentioned, MEIS2 interacts with PBX1 proteins and forms heteromeric complexes, which regulate the DNA binding ability of the two transcription factors (Liu et al., 2001; Longobardi et al., 2014). The MEIS2-PBX1 complex further recruits other transcription factors, such as the Kruppel-like factor 4 (Klf4) to modulate MEIS2 transcriptional activities (Bjerke et al., 2011). Other than PBX1, MEIS2 also functions synergistically with HOX and PAX homeobox factors, regulating the gene expression of other targets in the midbrain and hindbrain (Agoston et al., 2012). Mechanisms for the interactions between MEIS2 and other factors have been extensively reviewed; notably, MEIS2 recognizes and binds to a specific DNA motif TGACAG (Table 2; Chang et al., 1997; Longobardi et al., 2014; Schulte, 2014).
Myeloid ectopic viral integration site 2 controls gene expression and promotes neuronal migration and differentiation during forebrain development. There are three types of serotonin receptor 3a expressing (Htr3a+) GABAergic interneurons, which populate different regions of the brain. Type I Htr3a+ are enriched in transcription factors expressed in the LGE, including MEIS2, and these interneurons populate the deep cortical layers (von Engelhardt et al., 2011; Frazer et al., 2017). These interneurons originate from the PSB and migrate through to the cortex, contrasting with other types of Htr3a+ interneurons which are born from the CGE and populate the superficial cortical layers. Ectopic expression of Meis2 in CGE born interneurons resulted in a shift of differentiated Htr3a+ interneurons to the deep cortical layers, indicating that MEIS2 induces the migration of the LGE-derived interneurons (Frazer et al., 2017). Alternatively, MEIS2 can regulate expression of the Dlx family, by interacting with the intergenic enhancers in the Dlx bigenic clusters (Ghanem et al., 2003). MEIS2 binds to the I12b intergenic enhancer of Dlx1/2 and the I56ii intergenic enhancer of Dlx5/6. MEIS2 can activate reporter gene transcription with a I56ii promoter sequence in vitro (Yang et al., 2000; Poitras et al., 2007; Ghanem et al., 2008). Subsequently, the removal of I56ii sequence reduced Meis2 and Dlx5/6 expression, suggesting that there may be a positive feedback loop between MEIS2 and DLX5/6, further regulating interneuron migration (Fazel Darbandi et al., 2016). Furthermore, dopamine receptor expressing (D1/D2) MSNs are promoted by MEIS2 in the LGE, where deletion of Meis2 blocked differentiation of neural progenitors and reduced the medium-spiny neuron population (Su et al., 2022). MEIS2 regulates specification of these striatal projection neurons through the promotion of Zfp503 and Six3 expression, while Meis2 expression itself is regulated by DLX1/2 (Su et al., 2022). Likewise, in the prethalamus, DLX2 drives GABAergic interneuron determination through promoting Meis2 expression, and SOX14 represses Meis2 expression to maintain rostral thalamus identity (Sellers et al., 2014). In postnatal stages, interneurons continue to arise from the olfactory bulb SVZ generated neuroblasts where these differentiation events are dependent on the activity of MEIS2 and its interaction with PAX6 and DLX2 (Ming and Song, 2011; Agoston et al., 2014). Indeed, cKO of Meis2 in the olfactory bulb blocks dopaminergic neuron differentiation, as MEIS2 promotes expression of Dcx and Th, both crucial genes for dopaminergic neuron subtype specification (Agoston et al., 2014; Kim et al., 2020).
Nkx2.1/2.2 Genes
Nkx2.1 and Nkx2.2, homeobox transcription factors of the vertebrate Nkx family, are important for the regulation of embryonic telencephalon and diencephalon patterning (Price et al., 1992; Sussel et al., 1999). Nkx2.1 is the mammalian homolog of the Drosophila scarecrow (scro), and is also known as the thyroid transcription factor 1 and thyroid specific enhancer binding protein, since it also plays a role in thyroid, lung and pituitary development (Guazzi et al., 1990; Mizuno et al., 1991; Kimura et al., 1996; Maurel-Zaffran and Treisman, 2000). Nkx2.2 is homologous to the Drosophila ventral nervous system defective (vnd) gene (Kim and Nirenberg, 1989; Price et al., 1992; Jimenez et al., 1995). Nkx2.1 and Nkx2.2 encode both a HD and a NK2 box domain (Figure 2). In embryonic forebrain, Nkx2.1 is expressed in progenitor and post-mitotic cells in the MGE and PoA, and is essential for the patterning of these areas (Xu et al., 2005). Nkx2.2 is localized to the MGE, the VZ of the thalamus and MZ of the diencephalon; however, Nkx2.2 expression can vary in different mammalian species (Ericson et al., 1997; Flames et al., 2007; Vue et al., 2007; Bardet et al., 2010; Domínguez et al., 2015). The dorsoventral expression pattern of Nkx2.1 (ventral) is complementary to that of Pax6 (dorsal) (Figure 1B). Within the thalamus, Nkx2.2 expression is induced by SHH signaling in the rostral thalamus along with Ascl1, resulting in the specification of GABAergic neurons that populate the thalamus; as a result, Nkx2.2 is often co-expressed with SHH (Briscoe et al., 1999; Vue et al., 2007; Robertshaw et al., 2013).
Nkx2.1 expressing progenitor cells give rise to GABAergic and cholinergic neurons, which populate the neocortex and striatum, respectively (Anderson et al., 2001; Magno et al., 2017). Nkx2.1 expression in the GABAergic interneurons then diminishes after they tangentially migrate toward the neocortex, but is sustained in the cholinergic neurons (Marin et al., 2000). In the MGE, Nkx2.1 silencing is necessary for interneurons to tangentially migrate. Nkx2.1 silencing promotes the expression of Nrp1 and Nrp2, which then initiates neural migration (Nóbrega-Pereira et al., 2008; Kanatani et al., 2015). Nkx2.1 expressing neurons in the hypothalamus tangentially migrate into the diencephalon, and develop into GABAergic interneurons (Murcia-Ramón et al., 2020b). Additionally, NKX2.1 regulates astrocyte differentiation in the MGE and PoA from E14.5 to E16.5 in mice, and oligodendrocyte differentiation from E12.5 (Kessaris et al., 2006; Minocha et al., 2015, 2017; Orduz et al., 2019). Transcriptional activity is dependent on epigenetic states (Attanasio et al., 2014; Sandberg et al., 2016).
Nkx2.1 homozygous mutant mice die at birth with lung, thyroid, pituitary and ventral telencephalon defects (Kimura et al., 1996; Takuma et al., 1998; Sussel et al., 1999). In these mutant mice, the MGE is respecified into LGE, and exhibits reduced numbers of GABAergic and cholinergic neurons (Sussel et al., 1999; Fragkouli et al., 2009). However, ∼50% of GABAergic interneurons remain, suggesting that NKX2.1 is not the sole factor required for GABAergic interneuron specification (Sussel et al., 1999). cKO of Nkx2.1 at E10.5 and E12.5 results in altered identity of the MGE-derived interneurons subtypes. The MGE progenitor cells of these mutants were respecified into calretinin and vasointestinal peptide (VIP) expressing interneuron subtypes, resembling interneuron populations derived from the caudal GE (Xu et al., 2004; Butt et al., 2005), as opposed to pva+ or sst+ subtypes (Butt et al., 2008). GABAergic interneuron differentiation, especially pva+ and sst+ subtypes, is tightly regulated by Lhx6 and Lhx8 in the MGE, and both genes are downstream targets of NKX2.1 (Du et al., 2008; Flandin et al., 2011; Sandberg et al., 2016; Kim et al., 2021). Lhx6 and Lhx8 are activated by NKX2.1 expression in the SVZ through the recognition of epigenetic markers, and are essential for the specification of pva+ and sst+ interneuron subtypes (Du et al., 2008; Kim et al., 2021). Furthermore, NKX2.1 regulates MGE identity through repression of genes in the SHH, Wnt, and BMP signaling pathways required for cell differentiation and patterning. This repression is likely achieved by recruitment of Gro/TLE, a complex that reduces epigenetic-mediated repression, and induces activation (Patel et al., 2012; Sandberg et al., 2016). Conversely, SHH can induce the expression of Nkx2.1 in the MGE to specify ventral identity (Ericson et al., 1995). To establish ventral identity in the telencephalon, NKX2.1 also represses Pax6 expression in the GE, as the Nkx2.1 cKO showed a dorsal to ventral expansion and ectopic expression of Pax6 ventrally (Manoli and Driever, 2014). Pax6, a dorsal telencephalon specifying gene, in turn represses the expression of Nkx2.1 in the neocortex. The existence of this mechanism of mutual repression is supported by the complementary expression patterns of these two transcription factors (Sussel et al., 1999; Stoykova et al., 2000).
As NKX2.1 is essential for formation of various organs, mutations in this gene are linked to multiple phenotypes and diseases, including neurological disease, lung defects and thyroid dysfunction (Table 3; Thorwarth et al., 2014). NKX2.1 may play a role in Hirschsprung disease, a disorder of the developing enteric nervous system, through its interaction with SOX10 and PAX3. Sex-determining factor SRY is reported to displace SOX10’s interaction with NKX2.1 and PAX3, thereby promoting a Hirschsprung disease phenotype (Li et al., 2015). Furthermore, hereditary chorea, also known as brain-lung-thyroid disease, is linked to mutations in NKX2.1 with symptoms such as impaired coordination or speech development (Krude et al., 2002; Monti et al., 2015). Subsequently, NKX2.1 has also been related to the development of schizophrenia, as Nkx2.1 regulates GABAergic and cholinergic neuron specification (Sussel et al., 1999; Fragkouli et al., 2009; Malt et al., 2016). The cholinergic specification function of Nkx2.1 correlates with learning and memory, where the absence of Nkx2.1 in the septal area results in cognitive impairments (Magno et al., 2017).
Orthodenticle Homeobox Genes
Orthodenticle homeobox (Otx) is an ortholog of the Drosophila orthodenticle transcription factor, with OTX1 located in the human chromosome region 2p13 and OTX2 located in the human chromosome region 14q21-22 (Kastury et al., 1994). Crx is another member of the Otx family, but its expression is restricted to the retina, and all three Otx genes share a common OTX tail domain at the C-terminal (Figure 2A; Furukawa et al., 1997). Otx1 plays an important role in cortical neurogenesis, and along with Otx2, both genes are important for forebrain patterning and specification, as well as retinal development (Larsen et al., 2010b). Otx2 is essential during gastrulation for forebrain specification, and Otx2 expression continues in both dorsal and ventral telencephalon, diencephalon, and mesencephalon (Acampora et al., 1995; Rhinn et al., 1998; Tian et al., 2002; Kurokawa et al., 2004; Sakurai et al., 2010). The midbrain/hindbrain boundary marks the caudal limit of Otx2 expression. Otx2 expression is repressed in the hindbrain and spinal cord (Frantz et al., 1994). This pattern is regulated by fibroblast-growth-factor (Fgf)-8 and Gbx2, another homeobox gene that is required for caudal brain patterning and formation (Garda et al., 2001). GBX2 recognizes a conserved enhancer sequence in Otx2, thereby downregulating Otx2 in the hindbrain (Kurokawa et al., 2006; Inoue et al., 2012). Otx1 and Otx2 exhibit a similar expression pattern early in embryogenesis, and Otx1 expression is nested within the Otx2 expressing regions (Simeone et al., 1993). From E8.5, Otx2 expression starts to diminish in the rostral forebrain. At E11.5, Otx2 is expressed in the VZ in the GE, and promotes the ventral identity of the MGE. Otx1 expression patterns change to become complementary to Otx2; it is predominantly expressed in the VZ of the dorsal telencephalon and is expressed at lower levels in the dLGE (Hoch et al., 2015; Huang et al., 2018).
Mice lacking Otx1 survive to birth but develop spontaneous epilepsy and seizures (Acampora et al., 1996). cKO of Otx1 in the developing neocortex reduces the size of the neocortex as well as the overall cellular population (Pantò et al., 2004). Deletion of Otx1 reduced the generation of neurons by repressing neural differentiation from cortical NPCs, while NPC proliferation was promoted, subsequently increasing the population of neurons (Table 3). This suggests Otx1 promotes cell cycle exit in cortical NPCs, thereby maintaining the balance between differentiation and proliferation (Huang et al., 2018).
Deletion of both Otx1 and Otx2 in mice results in a gastrulation defect and is embryonic lethal (Acampora et al., 1995). Heterozygous double mutants exhibit a range of phenotypes, including different degrees of craniofacial malformations, ocular defects, abnormalities in central nervous system, pituitary glands dysfunction, and developmental delay (Matsuo et al., 1995; Ang et al., 1996; Ragge et al., 2005; Tajima et al., 2009; Dateki et al., 2010; Mortensen et al., 2015). cKO of Otx2 at different embryonic developmental timepoints and locations has shown a range of phenotypes indicating the essential role for Otx2 in processes such as septum formation, specification of the neocortex, neurogenesis and early oligodendrogenesis and NPC fate (Acampora et al., 1997; Puelles and Rubenstein, 2003; Puelles et al., 2006; Silbereis et al., 2014; Hoch et al., 2015). The disruption of septum formation and cortex specification following cKO of Otx2 after gastrulation suggests that Otx2 could be regulating specification through FGF signaling (Acampora et al., 1997; Puelles and Rubenstein, 2003; Hoch et al., 2015). MGE interneuron markers such as Dlx1, Arx, and Gbx were downregulated in MGE-deleted Otx2, as well as the expression of oligodendrogenesis promoting genes Olig1 and Olig2 demonstrating a requirement for Otx2 in neurogenesis and oligodendrogenesis (Silbereis et al., 2014; Hoch et al., 2015). Furthermore, Lhx6 and Lhx8 expression were reduced, suggesting Otx2 plays a role in regulating cholinergic neurons. Otx2 deletion in the thalamus resulted in a switch in NPC fate from glutamatergic neurons to GABAergic interneurons, demonstrating a requirement for Otx2 in glutamatergic neuron specification (Puelles et al., 2006).
OTX1 and OTX2 are overexpressed in medulloblastoma (Boon et al., 2005; Zakrzewska et al., 2013), which is a malignant pediatric brain tumor located in the posterior fossa that is divided into four molecular groups based on genomic and transcriptomic alterations: Wnt, SHH, Group 3, and Group 4 (Rudin et al., 2009; Northcott et al., 2011; Taylor et al., 2012). OTX2 is overexpressed in over 60% of medulloblastoma, usually in Groups 3 and 4 (Boon et al., 2005; Bunt et al., 2010). It has been postulated that the cellular context dependent nature of OTX2 expression could attribute to its overexpression in some groups of medulloblastoma (Kaur et al., 2015). As MYC, another oncogene is also overexpressed in Group 3 medulloblastoma, OTX2 may promote tumorigenesis by cooperatively binding with MYC to target genes (Bunt et al., 2011). Furthermore, OTX2 promotes the proliferation of tumors in Groups 3 and 4 (Lu et al., 2017; Zagozewski et al., 2020). The overexpression pattern observed may be a result of autoregulation. Chromatin accessibility is altered in medulloblastoma, where histone modifications may allow for increased OTX2 expression, and hence a positive feedback loop for OTX2 (Wortham et al., 2014). Recent studies have also shown OTX2 is potentially required for tumor proliferation in the SHH group, although not necessarily for tumor formation (El Nagar et al., 2018). In Group 3 medulloblastoma, OTX2 represses PAX3 and PAX6. Overexpression of PAX3 and PAX6 is associated with increased patient survival (Zagozewski et al., 2020). Additionally, OTX1 and OTX2 have been shown to act as oncogenes, promoting tumorigenesis and proliferation for cancers such as hepatocellular carcinoma, breast cancer, and Hodgkin or non-Hodgkin lymphomas (Omodei et al., 2009; Terrinoni et al., 2011; Nagel et al., 2015; Li et al., 2016; Tu et al., 2020).
Paired Box 6 Gene
The highly conserved Pax6 transcription factor was first identified as a member of the Paired box (Pax) gene family based on its homology to the Drosophila gene eyeless. There are nine PAX transcription factors identified in mammals; all contain a paired domain and can be further categorized according to the presence or absence of additional domains, usually a HD. Pax6 contains two DNA binding domains, a paired domain and a HD, as well as a proline-serine/threonine rich domain in the carboxyl-terminal (Figure 2A; Glaser et al., 1992; Duan et al., 2013). Hence, PAX6 binds to paired-HD and HD consensus DNA binding motifs (Sun et al., 2015). The Pax6 homologue eyeless was first described in Drosophila as a gene essential for segmentation and eye development (Walther et al., 1991; Gehring, 1996), and in mammals it is important for the development of the CNS, eyes, pancreas, and pituitary gland (Dohrmann et al., 2000; Jones et al., 2002). In mice, Pax6 expression begins from E8, and is then expressed in the forebrain, hindbrain, and spinal cord by E10 (Stoykova and Gruss, 1994; Inoue et al., 2000). Mice with homozygous mutation of Pax6 die upon birth with malformation in the cerebral cortex (Tyas et al., 2003), whereas heterozygous mutation results in development of a thinner cortex, and have small or reduced eyes (Hill et al., 1991; Schmahl et al., 1993; Fukuda et al., 2000; Haubst et al., 2004; Quinn et al., 2007). Mice with cortex-specific KO of Pax6 have reduced cortical size and an increased volume of the caudal cortex but without affecting thalamocortical identity (Piñon et al., 2008).
Within the telencephalon, Pax6 is expressed in the VZ of the dorsal telencephalon, as well as the PSB, with a rostral-caudal gradient (Figure 1B; Bishop et al., 2000; Hirata et al., 2002). Pax6 expression is repressed in the ventral telencephalon by OLIG2, which ensures the ventral identity of the forebrain (Lim et al., 2019). PAX6 promotes the expression of Ngn2 in the dorsal telencephalon, together specifying dorsal identity (Scardigli et al., 2003). This pattern of expression is largely complimentary to that of genes specifying ventral identity such as Ascl1, Dlx1/2, and Gsx2. The only overlapping areas in which these genes are co-expressed are the PSB and the VZ of the LGE (Puelles et al., 1999; Cocas et al., 2011). Pax6 is necessary for specification of the PSB, maintaining the physical boundary as well as a genetic boundary separating the dorsal and ventral telencephalon (Stenman et al., 2003). Pax6 homozygous mutants fail to develop such a boundary, with upregulation of ventral genes such as Gsx2, and downregulation of dorsal genes like Ngn2 in the dorsal telencephalon (Stoykova et al., 1996, 2000; Toresson et al., 2000; Yun et al., 2001; Quinn et al., 2007). As a result, the ventral telencephalon, in particular the dLGE, is expanded into the dorsal telencephalon, crossing over the PSB. During the course of development, Gsx2 expressing progenitor cells in the dorsal LGE can change fate by expressing Pax6, distinguishing either Pax6-expressing (dorsal) and Gsx2-expressing (ventral) progenitor cells at the PSB (Cocas et al., 2011). Pax6 may regulate the formation of this boundary via regulation of cell adhesion molecules (Tyas et al., 2003). Progenitor cells in dorsal and ventral telencephalon expresses R-cadherin and cadherin-6, respectively (Matsunami and Takeichi, 1995; Inoue et al., 1997). Absence of PAX6 in the dorsal telencephalon reduces the expression of R-cadherin, allowing the dorsal and ventral cells to aggregate more readily and consequently disrupts the PSB (Stoykova et al., 1997).
Thalamic patterning is also partly regulated by PAX6 within the dorsal thalamus, where Pax6 homozygous mutants display altered expression of factors that dictate dorsal identity patterning (Pratt et al., 2000). The patterning function of PAX6 is via regulation of neurogenin2 (Ngn2), a bHLH transcription factor (Wang et al., 2011a). NGN1/2 are required for maintaining the normal population of basal progenitor cells, and in Pax6 homozygous mutants, there is a reduction in Ngn2 expression, and subsequently reduced a number of basal progenitor cells (Wang et al., 2011a).
Disruption of the PSB also affects the tangential migration of GABAergic interneurons. The PSB functions to limit the number of interneurons arriving at the neocortex, with increased interneurons observed in the neocortex of Pax6 homozygous mutants along with a loss of PSB (Neyt et al., 1997; Chapouton et al., 1999). Although this outcome may be due to impaired tangential migration, it could also result from ventralization of progenitor cells (Kroll and O’Leary, 2005; Quinn et al., 2007). In these mutants, ventral GABAergic interneuron markers Dlx1/2, Ascl1, Gsx2, and Gad1 are expressed dorsally, and promote differentiation of GABAergic interneurons instead of glutamatergic neurons in cortical progenitors (Kroll and O’Leary, 2005; Long et al., 2009; Wang et al., 2013). This also suggests that Pax6 represses these dorsal specifying transcription factors, in order to promote the generation of cortical glutamatergic neurons. Unlike early corticogenesis (E12.5), in late corticogenesis (E15.5) there is an addition of differentiated neurons acquiring a GABAergic interneuron phenotype (Schuurmans et al., 2004). These results suggest that Pax6 controls the differentiation of glutamatergic neurons, whilst suppressing GABAergic interneuron production in late corticogenesis. However, in the diencephalon, Pax6 also promotes the development of GABAergic interneurons (Robertshaw et al., 2013).
Furthermore, Pax6 controls the balance between NPCs proliferation and differentiation through regulation of the cell cycle (Manuel and Price, 2005; Georgala et al., 2011). Pax6 directly regulates various genes that promote neurogenesis, and represses genes essential for non-neuronal fates depending on the histone modifications at the target promoters (Table 3; Sun et al., 2015; Thakurela et al., 2016). Pax6 homozygous mutants have shortened cell cycles at the start of corticogenesis, but as corticogenesis progresses cell cycle length increases (Estivill-Torrus et al., 2002; Mi et al., 2013). This phenomenon was observed in the cells with the longest cell cycles in the wildtype; in these cells in the Pax6 mutant mice the shortening of the cell cycle was associated with increased neuronal differentiation (Sansom et al., 2009; Mi et al., 2013; Walcher et al., 2013). Overexpressing Pax6 increased differentiation of cortical neural stem cells into basal progenitor cells (Sansom et al., 2009). As a result, neural stem cell proliferation is disrupted, and the quantity of neurons is also reduced (Heins et al., 2002; Jones et al., 2002; Hack et al., 2004; Georgala et al., 2011). Hence, an optimal level of Pax6 expression is necessary for the normal growth and development of the cortex. Further evidence indicates that the balance between proliferation and differentiation is Foxg1 dependent; Foxg1 determines whether Pax6 promotes proliferation or differentiation (Quintana-Urzainqui et al., 2018). Pax6 itself is regulated by the lncRNA PAUPAR in human embryonic stem cells, and such regulation is necessary for cortical differentiation (Xu et al., 2021).
Paired box 6 mutation in humans can result in neurological diseases, more commonly as a result of heterozygous mutations, including intellectual disability, autism, and impaired audition (Malandrini et al., 2001; Davis et al., 2008), likely to be related to reduced cerebral cortex size (Sisodiya et al., 2001; Ellison-Wright et al., 2004). Conversely, only four patients were reported to have mutations in both PAX6 alleles, of which two survived postnatally (Glaser et al., 1994; Schmidt-Sidor et al., 2009; Solomon et al., 2009). All cases exhibited cerebral cortical malformation, and in the two cases that died before birth, the cerebral cortex was only one-third the size of a normally developed cerebral cortex (Schmidt-Sidor et al., 2009; Table 3).
Pit-Oct-Unc Class 3 Homeobox 2 Gene
The POU (Pit-1, Oct-1/2, and Unc-86) gene family encodes a transcription factor family (Pou1f-Pou6f) of which Pou3f2 (Brn2) encodes a neural transcription factor that is necessary for mammalian CNS development and also for the production of corticotropin-releasing hormone (McEvilly et al., 2002; Castro et al., 2006). Pou3f2 regulates neuronal differentiation, migration, and upper cortical layer formation during mammalian embryogenesis (McEvilly et al., 2002; Sugitani et al., 2002; Castro et al., 2006; Dominguez et al., 2012; Chen et al., 2018). The protein contains a conserved POU domain composed of 150–160 amino acids, shared by the mammalian transcription factors Pituitary-specific PIT1, Octamer transcription factor proteins OCT1/2, and the nematode neural transcription factor UNC-86 (He et al., 1989; Ryan et al., 1997; Figure 2A). The DNA binding region of the POU protein is composed of two elements, a POU domain of approximately 75 amino acids present near the N-terminal and a classical HD of 60 amino acids located near the C-terminal separated by a short linker sequence (Figure 2A; Sumiyama et al., 1996; Ryan et al., 1997). Both domains are comprised of a helix-turn-helix structure (4 alpha helices in the POU domain and 3 alpha helices in the HD), which enables DNA recognition and confers DNA-binding specificity at the third helix (Klemm et al., 1994; Cook et al., 2008).
Interactions between the POU domain and its target sequence occur by recognition followed by specific binding to the canonical ATGCAAAT octameric sequence (Figure 2B). However, the linker region between the POU domain and HD is flexible (Herr and Cleary, 1995). The POU3F linker can fold as an alpha-helix which allows homo- or heterodimerization with the target DNA sequence (Blaud et al., 2004). POU3F2 has been reported to form homodimers on an octamer-like sequence of the L-amino acid decarboxylase (AADC), corticotropin (CRRH) and aldose C gene promoters in a non-cooperative fashion (Blaud et al., 2004). Pou3f2 is located on human chromosome 6q16.1 and dysregulation of this gene has been reported in disorders such as schizophrenia and bipolar disorder, as well as in melanoma (Table 3; Goodall et al., 2004a; Simmons et al., 2017; Chen et al., 2018; Ding et al., 2021).
The onset expression of Pou3f2 occurs in the VZ of the whole cortical lateral-to-medial axis during early brain development and in the paraventricular nuclei (PVN) of the hypothalamus (Figure 1B; He et al., 1989; Nakai et al., 1995; Dominguez et al., 2012). Using an antibody that detects both BRN1 and BRN2, POU3F2 expression was detected in radial migrating cells from the VZ up to the superficial cortical layers at P0 in mouse brain (Dominguez et al., 2012). Embryonic mice with homozygous Pou3f2 mutations exhibit hypothalamic and pituitary deficiencies, such as hypoplastic posterior lobe of the pituitary gland and failure to express corticotropin-releasing hormone in the PVN, and die soon after birth (Nakai et al., 1995; Schonemann et al., 1995). Pou3f2/3 (Brn1/2) DKO mice display an abnormal brain phenotype with decreased neocortical thickness and significant reduction of upper layer cells (Sugitani et al., 2002). The olfactory bulb is hypoplastic, the cerebellum is less foliated, accompanied by loosely packed Purkinje cells. Therefore, failure of radial migration results in cortical laminar inversion in the mutant mice (McEvilly et al., 2002; Sugitani et al., 2002). Therefore, Pou3f2/3 transcription factors redundantly regulate cortical neuron migration and therefore layer production, in addition to neuronal differentiation (Castro et al., 2006).
Two potential mechanisms have been suggested to explain the disruption in the cortical layering defect: via Pou3f2/3 regulation of CDK5 regulatory subunits p35 and p39 in migrating neurons (McEvilly et al., 2002) or through Pou3f2/3 regulation of Dab1 (Sugitani et al., 2002). The Pou3f2/3 double mutant display similar phenotypic abnormalities (McEvilly et al., 2002) to Cdk5-null mutants and p35/p39-null mutants (Ko et al., 2001). However, Pou3f2/3 expression is observed in both a late pool of neural precursor cells as well as in postmitotic neurons, including Tbr1+ cells in the cortical plate (Dominguez et al., 2012). Interestingly, when Pou3f2 is downregulated, there is an excessive number of Tbr1+/NeuroD1+ cells accumulating within the IZ (Dominguez et al., 2012). POUF3F2 may also regulate Dab1 as the loss of Dab1+ cells in neurons was observed at a later phase (McEvilly et al., 2002; Sugitani et al., 2002; Dominguez et al., 2012). Furthermore, Dab1 expression was markedly reduced in the Pou3f2/3 DKO mice at a late stage during which neurons fail to reach the marginal zone and remain beneath the cortical subplate (Sugitani et al., 2002).
Pit-Oct-Unc class 3 homeobox 2 interacts co-operatively with other transcription factors to regulate a number of neurodevelopmental genes, including Ascl1 in the regulation of Notch signaling, thereby controlling cell cycle exit of progenitors in addition to neuronal differentiation and radial migration in the embryonic telencephalon (Artavanis-Tsakonas et al., 1999). Disruption of POU3F2 binding was shown to prevent transcription of Notch pathway target genes, Delta1 and Hes5-1 (Castro et al., 2006). In contrast, overexpression of Pou3f2 and Ascl1 in chick neural tube resulted in excessive migration of electroporated cells in the marginal zone of the neural tube and disrupted neuronal differentiation (Castro et al., 2006).
Pit-Oct-Unc class 3 homeobox 2 dysregulation can have severe neurodevelopmental impacts, contributing to brain malformation, neurodevelopmental delays, and neuropsychiatric disorders (Table 3; Castro et al., 2006; Chen et al., 2018; Hashizume et al., 2018; Westphal et al., 2018; Ding et al., 2021). POU3f2 has been found to be associated with schizophrenia and bipolar disorder, as a hub for a gene regulatory network related to these disorders (Potkin et al., 2009; Mühleisen et al., 2014; Chen et al., 2018; Pearl et al., 2019; Ding et al., 2021). When POU3F2 is overexpressed in NSCs, several genes which are differentially expressed in the prefrontal cortex of people suffering from schizophrenia and bipolar disorder, are dysregulated. This confirms the role of POU3F2 as a key regulator of gene expression in these disorder (Pearl et al., 2019). POU3F2 and PAX6 were found to regulate the transcription of TRIM8 (Ding et al., 2021) as well as the VRK2 (Pearl et al., 2019), other genes associated with schizophrenia and bipolar disorder (Gandal et al., 2018; Li et al., 2018; Ding et al., 2021). While POU3F2 is crucial in regulating genes involved in CNS development, it is also a lineage-determining transcription factor crucial for the regulation of melanocytic lineage. It is overexpressed in many cancer types including carcinomas, neuroblastomas, and melanomas (Schreiber et al., 1990, 1992; Thomson et al., 1995; Leonard and Bell, 1997). Upregulation of POU3F2 represses Microphthalmia-associated transcription factor (MITF) expression in some melanomas by binding to its promoter region, which drives the cells to adopt a more stem-like and aggressive phenotype (Goodall et al., 2004a; Bonvin et al., 2012). This upregulation is due to the activation of BRAF, a key component of the mitogen-activated protein (MAP) kinase signaling pathway (Goodall et al., 2004b).
Non-cell Autonomous and Combinatorial Roles of Homeodomain-Containing Transcription Factors
Other than the regulatory functions discussed above, homeodomain-containing transcription factors can regulate forebrain development through non-cell autonomous roles as well as by combinatorial modes of action. As an example, PAX6 exhibits non-cell autonomous activity in the development of other organs such as the eye and spinal cord (Collinson et al., 2004; Lesaffre et al., 2007; Di Lullo et al., 2011). This activity is due to two short sequences found within the HD, which are considered essential for secretion and internalization (Prochiantz and Joliot, 2003; Joliot and Prochiantz, 2004). This suggests that some homeobox genes encode transcription factors that have the ability to act as signaling molecules, and are capable of intercellular transfer. Disruption of extracellular PAX6 has functional consequences, leading to defective eye development, with reduction in eye size (Lesaffre et al., 2007). PAX6 extracellular activities can affect cell migration in the embryonic chick spinal cord (Di Lullo et al., 2011). OPCs, a highly migratory cell population, were studied, also due to their delayed specification and dorsal shift in Pax6 mutants (Sun et al., 1998). OPCs were observed to be in close proximity to PAX6+ cells, and the ablation of extracellular PAX6 resulted in reduced migration of the OPC population (Di Lullo et al., 2011). Another transcription factor shown to have non-cell autonomous activity is OTX2 in the visual cortex, during the regulation of the timing of heightened plasticity, an important timepoint for proper visual development (Lee et al., 2017; Apulei et al., 2018). Extracellular OTX2 has been shown to regulate expression of Gadd45b, a gene that may play a role in epigenetic gene activation, as a downstream target for modulating visual cortex plasticity (Ma et al., 2009; Apulei et al., 2018). These examples demonstrate that homeobox genes, through their encoded transcription factors can also function non-cell autonomously. However, this role is yet to be fully understood for the majority of homeobox genes.
In addition, although transcription factors display highly specific expression patterns, many are co-expressed at early stages of development, and work in a combinatorial manner. This concept of a combinatorial code has been well documented and studied in the spinal cord (Sugimori et al., 2007; Sagner and Briscoe, 2019). The organization and patterning of the spinal cord is initiated during the development of the neural tube, in accordance with the activities of various morphogens that induce different transcription factor families (Briscoe et al., 2000; Jessell, 2000). A similar code is under active study for forebrain development, where cortical regionalization and patterning are tightly regulated by a transcriptional network consisting of transcription factors and their regulatory elements (Ypsilanti et al., 2021). In particular, cortical expression of transcriptional network members at E11.5 in the mouse forebrain is either in a gradient or in homogenous patterns, with coregulation by transcription factors such as Pax6, Emx2, and Nr2f1 (Muzio et al., 2002b; Ypsilanti et al., 2021). Also, transcription factors expressed in the pallium have been implicated in co-binding to regulatory elements through chromatin conformation analysis (Ypsilanti et al., 2021). Future directions employing co-immunoprecipitation, ATACseq, ChIPseq and ChIP-re-ChIP experiments will enable increased understanding of how transcription factors work cooperatively in the regulation of forebrain patterning and regionalization.
Conclusion
In this review of vertebrate forebrain development (Figure 1), selected transcription factors from the HD, paired, POU and TALE HD gene families necessary for forebrain development have been discussed and summarized (Table 1 and Figure 2). Where known, gene targets of these transcription factors have been specified (Table 2) and correlations to human diseases, including neurodevelopmental disorders and brain tumors have been briefly outlined (Table 3). Further studies are necessary to delineate protein-protein interactions and to identify and characterize post-translational modifications, such as phosphorylation, sumoylation, and ubiquitination, which regulate transcription factor function and link these modifications to signaling pathways in CNS development and disease.
Author Contributions
RL, AG, ER, MF, JW, and DE conceived and wrote the manuscript. RL, MF, and AG designed and generated the figures. All authors contributed to editing and approved the final version of the manuscript.
Funding
RL is supported by a Melbourne Research Scholarship, University of Melbourne, Parkville, VIC, Australia. DE is supported by an establishment grant from The Royal Children’s Hospital Foundation, Parkville, VIC, Australia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abellán, A., Vernier, B., Rétaux, S., and Medina, L. (2010). Similarities and differences in the forebrain expression of Lhx1 and Lhx5 between chicken and mouse: insights for understanding telencephalic development and evolution. J. Comp. Neurol. 518, 3512–3528. doi: 10.1002/cne.22410
Acampora, D., Avantaggiato, V., Tuorto, F., and Simeone, A. (1997). Genetic control of brain morphogenesis through Otx gene dosage requirement. Development 124, 3639–3650. doi: 10.1242/dev.124.18.3639
Acampora, D., Mazan, S., Lallemand, Y., Avantaggiato, V., Maury, M., Simeone, A., et al. (1995). Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121, 3279–3290. doi: 10.1242/dev.121.10.3279
Acampora, D., Mazan, S., Avantaggiato, V., Barone, P., Tuorto, F., Lallemand, Y., et al. (1996). Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat. Genet. 14, 218–222. doi: 10.1038/ng1096-218
Acampora, D., Merlo, G. R., Paleari, L., Zerega, B., Postiglione, M. P., Mantero, S., et al. (1999). Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126, 3795–3809. doi: 10.1242/dev.126.17.3795
Agoston, Z., Heine, P., Brill, M. S., Grebbin, B. M., Hau, A.-C., Kallenborn-Gerhardt, W., et al. (2014). Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141, 28–38. doi: 10.1242/dev.097295
Agoston, Z., Li, N., Haslinger, A., Wizenmann, A., and Schulte, D. (2012). Genetic and physical interaction of Meis2, Pax3 and Pax7 during dorsal midbrain development. BMC Dev. Biol. 12:12. doi: 10.1186/1471-213X-12-10
Allen Institute for Brain Science. (2019). Atlas of the Developing Human Brain. Seattle, WA, Allen Institute for Brain Science.
Aleksic, T., Bechtel, M., Krndija, D., von Wichert, G., Knobel, B., Giehl, K., et al. (2007). CUTL1 promotes tumor cell migration by decreasing proteasome-mediated Src degradation. Oncogene 26, 5939–5949. doi: 10.1038/sj.onc.1210398
Alifragis, P., Liapi, A., and Parnavelas, J. G. (2004). Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J. Neurosci. 24:5643. doi: 10.1523/JNEUROSCI.1245-04.2004
Al-Jaberi, N., Lindsay, S., Sarma, S., Bayatti, N., and Clowry, G. J. (2015). The early fetal development of human neocortical GABAergic interneurons. Cereb. Cortex 25, 631–645. doi: 10.1093/cercor/bht254
Allen, Z. J., Waclaw, R. R., Colbert, M. C., and Campbell, K. (2007). Molecular identity of olfactory bulb interneurons: transcriptional codes of periglomerular neuron subtypes. J. Mol. Histol. 38, 517–525. doi: 10.1007/s10735-007-9115-4
Alvarez-Buylla, A., and Lim, D. A. (2004). For the long run: maintaining germinal niches in the adult brain. Neuron 41, 683–686. doi: 10.1016/s0896-6273(04)00111-4
Amin, S., Donaldson, I. J., Zannino, D. A., Hensman, J., Rattray, M., Losa, M., et al. (2015). Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 32, 265–277. doi: 10.1016/j.devcel.2014.12.024
Anderson, D. J., Choi, G., and Zhou, Q. (2002a). Olig genes and the genetic logic of CNS neural cell fate determination. Clin. Neurosci. Res. 2, 17–28. doi: 10.1016/s1566-2772(02)00014-2
Anderson, S., Eisenstat, D., Shi, L., and Rubenstein, J. (1997a). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476. doi: 10.1126/science.278.5337.474
Anderson, S., Marín, O., Horn, C., Jennings, K., and Rubenstein, J. (2001). Distinct cortical migrations from the medial and lateral ganglionic eminences. Development 128, 353–363. doi: 10.1242/dev.128.3.353
Anderson, S. A., Kaznowski, C. E., Horn, C., Rubenstein, J. L., and McConnell, S. K. (2002b). Distinct origins of neocortical projection neurons and interneurons in vivo. Cereb. Cortex 12, 702–709. doi: 10.1093/cercor/12.7.702
Anderson, S. A., Qiu, M., Bulfone, A., Eisenstat, D. D., Meneses, J., Pedersen, R., et al. (1997b). Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19, 27–37. doi: 10.1016/s0896-6273(00)80345-1
Andrews, W., Liapi, A., Plachez, C., Camurri, L., Zhang, J., Mori, S., et al. (2006). Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development 133, 2243–2252. doi: 10.1242/dev.02379
Ang, S.-L., Jin, O., Rhinn, M., Daigle, N., Stevenson, L., and Rossant, J. (1996). A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 122, 243–252. doi: 10.1242/dev.122.1.243
Apulei, J., Kim, N., Testa, D., Ribot, J., Morizet, D., Bernard, C., et al. (2018). Non-cell autonomous otx2 homeoprotein regulates visual cortex plasticity through Gadd45b/g. Cereb. Cortex 29, 2384–2395. doi: 10.1093/cercor/bhy108
Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770–776. doi: 10.1126/science.284.5415.770
Attanasio, C., Nord, A. S., Zhu, Y., Blow, M. J., Biddie, S. C., Mendenhall, E. M., et al. (2014). Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 24, 920–929. doi: 10.1101/gr.168930.113
Ayala, R., Shu, T., and Tsai, L.-H. (2007). Trekking across the brain: the journey of neuronal migration. Cell 128, 29–43. doi: 10.1016/j.cell.2006.12.021
Aykut, B., Ochs, M., Radhakrishnan, P., Brill, A., Höcker, H., Schwarz, S., et al. (2017). EMX2 gene expression predicts liver metastasis and survival in colorectal cancer. BMC Cancer 17:555. doi: 10.1186/s12885-017-3556-2
Bach, I. (2000). The LIM domain: regulation by association. Mech. Dev. 91, 5–17. doi: 10.1016/s0925-4773(99)00314-7
Bachy, I., and Rétaux, S. (2006). GABAergic specification in the basal forebrain is controlled by the LIM-hd factor Lhx7. Dev. Biol. 291, 218–226. doi: 10.1016/j.ydbio.2005.10.023
Bardet, S., Ferran, J., Sanchez-Arrones, L., and Puelles, L. (2010). Ontogenetic expression of sonic hedgehog in the chicken subpallium. Front. Neuroanat. 4:28. doi: 10.3389/fnana.2010.00028
Barington, M., Risom, L., Ek, J., Uldall, P., and Ostergaard, E. (2018). A recurrent de novo CUX2 missense variant associated with intellectual disability, seizures, and autism spectrum disorder. Eur. J. Hum. Genet. 26, 1388–1391. doi: 10.1038/s41431-018-0184-5
Batista-Brito, R., Close, J., Machold, R., and Fishell, G. (2008). The distinct temporal origins of olfactory bulb interneuron subtypes. J. Neurosci. 28:3966. doi: 10.1523/JNEUROSCI.5625-07.2008
Bayer, S. A. (1983). 3H-thymidine-radiographic studies of neurogenesis in the rat olfactory bulb. Exp. Brain Res. 50, 329–340. doi: 10.1007/BF00239197
Beckmann, J., Vitobello, A., Ferralli, J., Brož, D. K., Rijli, F. M., and Chiquet-Ehrismann, R. (2011). Human teneurin-1 is a direct target of the homeobox transcription factor EMX2 at a novel alternate promoter. BMC Dev. Biol. 11:35. doi: 10.1186/1471-213X-11-35
Bertrand, N., Castro, D. S., and Guillemot, F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. doi: 10.1038/nrn874
Bienvenu, T., Poirier, K., Friocourt, G., Bahi, N., Beaumont, D., Fauchereau, F., et al. (2002). ARX. Hum. Mol. Genet. 11, 981–991.
Bielle, F., Griveau, A., Narboux-Nême, N., Vigneau, S., Sigrist, M., Arber, S., et al. (2005). Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat. Neurosci. 8, 1002–1012. doi: 10.1038/nn1511
Bilioni, A., Craig, G., Hill, C., and McNeill, H. (2005). Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc. Natl. Acad. Sci. 102, 14671–14676. doi: 10.1073/pnas.0502480102
Billeter, M., Güntert, P., Luginbühl, P., and Wüthrich, K. (1996). Hydration and DNA recognition by homeodomains. Cell 85, 1057–1065. doi: 10.1016/s0092-8674(00)81306-9
Bishop, K. M., Garel, S., Nakagawa, Y., Rubenstein, J. L., and O’Leary, D. D. (2003). Emx1 and Emx2 cooperate to regulate cortical size, lamination, neuronal differentiation, development of cortical efferents, and thalamocortical pathfinding. J. Comp. Neurol. 457, 345–360. doi: 10.1002/cne.10549
Bishop, K. M., Goudreau, G., and O’Leary, D. D. M. (2000). Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science 288, 344–349. doi: 10.1126/science.288.5464.344
Bishop, K. M., Rubenstein, J. L., and O’Leary, D. D. (2002). Distinct actions of Emx1, Emx2, andPax6 in regulating the specification of areas in the developing neocortex. J. Neurosci. 22, 7627–7638. doi: 10.1523/JNEUROSCI.22-17-07627.2002
Bjerke, G. A., Hyman-Walsh, C., and Wotton, D. (2011). Cooperative transcriptional activation by Klf4, Meis2, and Pbx1. Mol. Cell. Biol. 31, 3723–3733. doi: 10.1128/MCB.01456-10
Blaud, M., Vossen, C., Joseph, G., Alazard, R., Erard, M., and Nieto, L. (2004). Characteristic patterns of N Oct-3 binding to a set of neuronal promoters. J. Mol. Biol. 339, 1049–1058. doi: 10.1016/j.jmb.2004.04.035
Blochlinger, K., Bodmer, R., Jack, J., Jan, L. Y., and Jan, Y. N. (1988). Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature 333, 629–635. doi: 10.1038/333629a0
Bodmer, R., Barbel, S., Sheperd, S., Jack, J. W., Jan, L. Y., and Jan, Y. N. (1987). Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell 51, 293–307. doi: 10.1016/0092-8674(87)90156-5
Bokoch, G. M. (2003). Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781. doi: 10.1146/annurev.biochem.72.121801.161742
Bosse, A., Zülch, A., Becker, M. B., Torres, M., ómez-Skarmeta, J. L. G., Modolell, J., et al. (1997). Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech. Dev. 69, 169–181. doi: 10.1016/s0925-4773(97)00165-2
Bonvin, E., Falletta, P., Shaw, H., Delmas, V., and Goding, C. R. (2012). A phosphatidylinositol 3-kinase-Pax3 axis regulates Brn-2 expression in melanoma. Mol. Cell. Biol. 32, 4674–4683. doi: 10.1128/MCB.01067-12
Boon, K., Eberhart, C. G., and Riggins, G. J. (2005). Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 65, 703–707.
Briata, P., Ilengo, C., Bobola, N., and Corte, G. (1999). Binding properties of the human homeodomain protein OTX2 to a DNA target sequence. FEBS Lett. 445, 160–164. doi: 10.1016/s0014-5793(99)00113-1
Briscoe, J., Pierani, A., Jessell, T. M., and Ericson, J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445. doi: 10.1016/s0092-8674(00)80853-3
Briscoe, J., Sussel, L., Serup, P., Hartigan-O’Connor, D., Jessell, T. M., Rubenstein, J. L., et al. (1999). Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398, 622–627. doi: 10.1038/19315
Britz, O., Mattar, P., Nguyen, L., Langevin, L.-M., Zimmer, C., Alam, S., et al. (2006). A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex 16, i138–i151. doi: 10.1093/cercor/bhj168
Bulstrode, H., Johnstone, E., Marques-Torrejon, M. A., Ferguson, K. M., Bressan, R. B., Blin, C., et al. (2017). Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes Dev. 31, 757–773. doi: 10.1101/gad.293027.116
Bunt, J., de Haas, T. G., Hasselt, N. E., Zwijnenburg, D. A., Koster, J., Versteeg, R., et al. (2010). Regulation of cell cycle genes and induction of senescence by overexpression of OTX2 in medulloblastoma cell lines. Mol. Cancer Res. 8, 1344–1357. doi: 10.1158/1541-7786.MCR-09-0546
Bunt, J., Hasselt, N. E., Zwijnenburg, D. A., Koster, J., Versteeg, R., and Kool, M. (2011). Joint binding of OTX2 and MYC in promotor regions is associated with high gene expression in medulloblastoma. PLoS One 6:e26058. doi: 10.1371/journal.pone.0026058
Butterworth, F., and King, R. (1965). The developmental genetics of apterous mutants of Drosophila melanogaster. Genetics 52, 1153. doi: 10.1093/genetics/52.6.1153
Butt, S. J., Fuccillo, M., Nery, S., Noctor, S., Kriegstein, A., Corbin, J. G., et al. (2005). The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron 48, 591–604. doi: 10.1016/j.neuron.2005.09.034
Butt, S. J., Sousa, V. H., Fuccillo, M. V., Hjerling-Leffler, J., Miyoshi, G., Kimura, S., et al. (2008). The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron 59, 722–732. doi: 10.1016/j.neuron.2008.07.031
Bulchand, S., Grove, E., Porter, F., and Tole, S. (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 100, 165–175. doi: 10.1016/s0925-4773(00)00515-3
Bürglin, T. R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173–4180. doi: 10.1093/nar/25.21.4173
Caqueret, A., Boucher, F., and Michaud, J. L. (2006). Laminar organization of the early developing anterior hypothalamus. Dev. Biol. 298, 95–106. doi: 10.1016/j.ydbio.2006.06.019
Carney, R. S., Cocas, L. A., Hirata, T., Mansfield, K., and Corbin, J. G. (2009). Differential regulation of telencephalic pallial–subpallial boundary patterning by Pax6 and Gsh2. Cereb. Cortex 19, 745–759. doi: 10.1093/cercor/bhn123
Carri, A. D., Onorati, M., Lelos, M. J., Castiglioni, V., Faedo, A., Menon, R., et al. (2013). Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development 140, 301–312. doi: 10.1242/dev.084608
Castro, D. S., Skowronska-Krawczyk, D., Armant, O., Donaldson, I. J., Parras, C., Hunt, C., et al. (2006). Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell 11, 831–844. doi: 10.1016/j.devcel.2006.10.006
Cau, E., Casarosa, S., and Guillemot, F. (2002). Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development 129, 1871–1880. doi: 10.1242/dev.129.8.1871
Cecconi, F., Proetzel, G., Alvarez-Bolado, G., Jay, D., and Gruss, P. (1997). Expression of Meis2, a Knotted-related murine homeobox gene, indicates a role in the differentiation of the forebrain and the somitic mesoderm. Dev. Dyn. 210, 184–190. doi: 10.1002/(SICI)1097-0177(199710)210:2<184::AID-AJA10>3.0.CO;2-E
Chang, C. P., Jacobs, Y., Nakamura, T., Jenkins, N. A., Copeland, N. G., and Cleary, M. L. (1997). Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell Biol. 17, 5679–5687. doi: 10.1128/MCB.17.10.5679
Chapman, H., Riesenberg, A., Ehrman, L. A., Kohli, V., Nardini, D., Nakafuku, M., et al. (2018). Gsx transcription factors control neuronal versus glial specification in ventricular zone progenitors of the mouse lateral ganglionic eminence. Dev. Biol. 442, 115–126. doi: 10.1016/j.ydbio.2018.07.005
Chapman, H., Waclaw, R. R., Pei, Z., Nakafuku, M., and Campbell, K. (2013). The homeobox gene Gsx2 controls the timing of oligodendroglial fate specification in mouse lateral ganglionic eminence progenitors. Development 140, 2289–2298. doi: 10.1242/dev.091090
Chapouton, P., Gartner, A., and Gotz, M. (1999). The role of Pax6 in restricting cell migration between developing cortex and basal ganglia. Development 126, 5569–5579. doi: 10.1242/dev.126.24.5569
Chen, C., Meng, Q., Xia, Y., Ding, C., Wang, L., Dai, R., et al. (2018). The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci. Transl. Med. 10:eaat8178. doi: 10.1126/scitranslmed.aat8178
Chen, G., Sima, J., Jin, M., Wang, K.-y., Xue, X.-j., Zheng, W., et al. (2008). Semaphorin-3A guides radial migration of cortical neurons during development. Nat. Neurosci. 11, 36–44. doi: 10.1038/nn2018
Cho, G., Nasrallah, M. P., Lim, Y., and Golden, J. A. (2012). Distinct DNA binding and transcriptional repression characteristics related to different ARX mutations. Neurogenetics 13, 23–29. doi: 10.1007/s10048-011-0304-7
Cho, M. H., McDonald, M.-L. N., Zhou, X., Mattheisen, M., Castaldi, P. J., Hersh, C. P., et al. (2014). Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir. Med. 2, 214–225. doi: 10.1016/S2213-2600(14)70002-5
Cholfin, J. A., and Rubenstein, J. L. (2008). Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J. Comp. Neurol. 509, 144–155. doi: 10.1002/cne.21709
Cobos, I., Borello, U., and Rubenstein, J. L. R. (2007). Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 54, 873–888. doi: 10.1016/j.neuron.2007.05.024
Cobos, I., Broccoli, V., and Rubenstein, J. L. (2005a). The vertebrate ortholog of Aristaless is regulated by Dlx genes in the developing forebrain. J. Comp. Neurol. 483, 292–230. doi: 10.1002/cne.20405
Cobos, I., Calcagnotto, M. E., Vilaythong, A. J., Thwin, M. T., Noebels, J. L., Baraban, S. C., et al. (2005b). Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat. Neurosci. 8, 1059–1068. doi: 10.1038/nn1499
Cocas, L. A., Georgala, P. A., Mangin, J.-M., Clegg, J. M., Kessaris, N., Haydar, T. F., et al. (2011). Pax6 is required at the telencephalic pallial–subpallial boundary for the generation of neuronal diversity in the postnatal limbic system. J. Neurosci. 31, 5313–5324. doi: 10.1523/JNEUROSCI.3867-10.2011
Cohen, D. R., Cheng, C. W., Cheng, S. H., and Hui, C.-c (2000). Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech. Dev. 91, 317–321. doi: 10.1016/s0925-4773(99)00263-4
Cohen, S. M., Brönner, G., Küttner, F., Jürgens, G., and Jäckle, H. (1989). Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature 338, 432–434. doi: 10.1038/338432a0
Colasante, G., Collombat, P., Raimondi, V., Bonanomi, D., Ferrai, C., Maira, M., et al. (2008). Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J. Neurosci. 28, 10674–10678. doi: 10.1523/JNEUROSCI.1283-08.2008
Colasante, G., Sessa, A., Crispi, S., Calogero, R., Mansouri, A., Collombat, P., et al. (2009). Arx acts as a regional key selector gene in the ventral telencephalon mainly through its transcriptional repression activity. Dev. Biol. 334, 59–67. doi: 10.1016/j.ydbio.2009.07.014
Colasante, G., Simonet, J. C., Calogero, R., Crispi, S., Sessa, A., Cho, G., et al. (2015). ARX regulates cortical intermediate progenitor cell expansion and upper layer neuron formation through repression of Cdkn1c. Cereb. Cortex 25, 322–333. doi: 10.1093/cercor/bht222
Collinson, J. M., Chanas, S. A., Hill, R. E., and West, J. D. (2004). Corneal development, limbal stem cell function, and corneal epithelial cell migration in the pax6 +/- mouse. Invest. Ophthalmol. Vis. Sci. 45, 1101–1108. doi: 10.1167/iovs.03-1118
Colombo, E., Collombat, P., Colasante, G., Bianchi, M., Long, J., Mansouri, A., et al. (2007). Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J. Neurosci. 27, 4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007
Colombo, E., Galli, R., Cossu, G., Gécz, J., and Broccoli, V. (2004). Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev. Dyn. 231, 631–639. doi: 10.1002/dvdy.20164
Conforto, T. L., Zhang, Y., Sherman, J., and Waxman, D. J. (2012). Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol. Cell. Biol. 32, 4611–4627. doi: 10.1128/MCB.00886-12
Cook, A. L., Sturm, R. A., and domain, P. O. U. (2008). transcription factors: BRN2 as a regulator of melanocytic growth and tumourigenesis. Pigment Cell Melanoma Research 21, 611–626. doi: 10.1111/j.1755-148X.2008.00510.x
Coqueret, O., Bérubé, G., and Nepveu, A. (1996). DNA binding by cut homeodomain proteins is down-modulated by protein kinase C*. J. Biol. Chem. 271, 24862–24868. doi: 10.1074/jbc.271.40.24862
Coqueret, O., Martin, N., Bérubé, G., Rabbat, M., Litchfield, D. W., and Nepveu, A. (1998). DNA binding by cut homeodomain proteins is down-modulated by casein kinase II*. J. Biol. Chem. 273, 2561–2566. doi: 10.1074/jbc.273.5.2561
Corbin, J. G., Gaiano, N., Machold, R. P., Langston, A., and Fishell, G. (2000). The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development 127, 5007–5020. doi: 10.1242/dev.127.23.5007
Corbin, J. G., Rutlin, M., Gaiano, N., and Fishell, G. (2003). Combinatorial function of the homeodomain proteins Nkx2. 1 and Gsh2 in ventral telencephalic patterning. Development 130, 4895–4906. doi: 10.1242/dev.00717
Cowell, I. G., and Hurst, H. C. (1994). Transcriptional repression by the human bZIP factor E4BP4: definition of a minimal repression domain. Nucleic Acids Res. 22, 59–65. doi: 10.1093/nar/22.1.59
Craddock, N., Dawson, E., Burge, S., Parfitt, L., Mant, B., Roberts, Q., et al. (1993). The gene for Darier’s disease maps to chromosome 12q23-q24. Hum. Mol. Genet. 2, 1941–1943. doi: 10.1093/hmg/2.11.1941
Cubelos, B., Sebastián-Serrano, A., Beccari, L., Calcagnotto, M. E., Cisneros, E., Kim, S., et al. (2010). Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron 66, 523–535. doi: 10.1016/j.neuron.2010.04.038
Cubelos, B., Sebastián-Serrano, A, Kim, S., Moreno-Ortiz, C., Redondo, J. M., Walsh, C. A., et al. (2008a). Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb. Cortex 18, 1758–1770. doi: 10.1093/cercor/bhm199
Cubelos, B., Sebastián-Serrano, A., Kim, S., Redondo, J. M., Walsh, C., and Nieto, M. (2008b). Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Dev. Neurobiol. 68, 917–925. doi: 10.1002/dneu.20626
Dai, S., Li, J., Zhang, H., Chen, X., Guo, M., Chen, Z., et al. (2020). Structural basis for DNA recognition by FOXG1 and the characterization of disease-causing FOXG1 mutations. J. Mol. Biol. 432, 6146–6156. doi: 10.1016/j.jmb.2020.10.007
Dateki, S., Kosaka, K., Hasegawa, K., Tanaka, H., Azuma, N., Yokoya, S., et al. (2010). Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J. Clin. Endocrinol. Metab. 95, 756–764. doi: 10.1210/jc.2009-1334
Davis, L., Meyer, K., Rudd, D., Librant, A., Epping, E., Sheffield, V., et al. (2008). Pax6 3’ deletion results in aniridia, autism and mental retardation. Hum. Genet. 123, 371–378. doi: 10.1007/s00439-008-0484-x
Dawid, I. B., Breen, J. J., and Toyama, R. (1998). LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14, 156–162. doi: 10.1016/s0168-9525(98)01424-3
DeDiego, I., Smith-Fernández, A., and Fairén, A. (1994). Cortical cells that migrate beyond area boundaries: characterization of an early neuronal population in the lower intermediate zone of prenatal rats. Eur. J. Neurosci. 6, 983–997. doi: 10.1111/j.1460-9568.1994.tb00593.x
Dehmelt, L., and Halpain, S. (2004). The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 6:204.
Demos, M. K., Fullston, T., Partington, M. W., Gécz, J., and Gibson, W. T. (2009). Clinical study of two brothers with a novel 33 bp duplication in the ARX gene. Am. J. Med. Genet. Part A 149, 1482–1486. doi: 10.1002/ajmg.a.32851
De Marchis, S., Bovetti, S., Carletti, B., Hsieh, Y.-C., Garzotto, D., Peretto, P., et al. (2007). Generation of distinct types of periglomerular olfactory bulb interneurons during development and in adult mice: implication for intrinsic properties of the subventricular zone progenitor population. J. Neurosci. 27, 657–664. doi: 10.1523/JNEUROSCI.2870-06.2007
De Mori, R., Severino, M., Mancardi, M. M., Anello, D., Tardivo, S., Biagini, T., et al. (2019). Agenesis of the putamen and globus pallidus caused by recessive mutations in the homeobox gene GSX2. Brain 142, 2965–2978. doi: 10.1093/brain/awz247
Depew, M. J., Liu, J. K., Long, J. E., Presley, R., Meneses, J. J., Pedersen, R. A., et al. (1999). Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126, 3831–3846. doi: 10.1242/dev.126.17.3831
Desmaris, E., Keruzore, M., Saulnier, A., Ratié, L., Assimacopoulos, S., De Clercq, S., et al. (2018). DMRT5, DMRT3, and EMX2 cooperatively repress GSX2 at the pallium–subpallium boundary to maintain cortical identity in dorsal telencephalic progenitors. J. Neurosci. 38, 9105–9121. doi: 10.1523/JNEUROSCI.0375-18.2018
Diez del Corral, R., Aroca, P., mez-Skarmeta, J. L. G., Cavodeassi, F., and Modolell, J. (1999). The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes Dev. 13, 1754–1761. doi: 10.1101/gad.13.13.1754
Ding, C., Zhang, C., Kopp, R., Kuney, L., Meng, Q., Wang, L., et al. (2021). Transcription factor POU3F2 regulates TRIM8 expression contributing to cellular functions implicated in schizophrenia. Mol. Psychiatry 26, 3444–3460. doi: 10.1038/s41380-020-00877-2
Di Lullo, E., Haton, C., Le Poupon, C., Volovitch, M., Joliot, A., Thomas, J.-L., et al. (2011). Paracrine Pax6 activity regulates oligodendrocyte precursor cell migration in the chick embryonic neural tube. Development 138, 4991–5001. doi: 10.1242/dev.066282
Dohrmann, C., Gruss, P., and Lemaire, L. (2000). Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mech. Dev. 92, 47–54. doi: 10.1016/s0925-4773(99)00324-x
Dominguez, M. H., Ayoub, A. E., and Rakic, P. (2012). POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb. Cortex 23, 2632–2643. doi: 10.1093/cercor/bhs252
Domínguez, L., González, A., and Moreno, N. (2015). Patterns of hypothalamic regionalization in amphibians and reptiles: common traits revealed by a genoarchitectonic approach. Front. Neuroanat. 9:3. doi: 10.3389/fnana.2015.00003
Donegan, J. J., Boley, A. M., Glenn, J. P., Carless, M. A., and Lodge, D. J. (2020). Developmental alterations in the transcriptome of three distinct rodent models of schizophrenia. PLoS One 15:e0232200. doi: 10.1371/journal.pone.0232200
Drabikowski, K., Trzebiatowska, A., and Chiquet-Ehrismann, R. (2005). ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev. Biol. 282, 27–38. doi: 10.1016/j.ydbio.2005.02.017
Du, T., Xu, Q., Ocbina, P. J., and Anderson, S. A. (2008). NKX2. 1 specifies cortical interneuron fate by activating Lhx6. Development 135, 1559–1567. doi: 10.1242/dev.015123
Duan, D., Fu, Y., Paxinos, G., and Watson, C. (2013). Spatiotemporal expression patterns of Pax6 in the brain of embryonic, newborn, and adult mice. Brain Struct. Funct. 218, 353–372. doi: 10.1007/s00429-012-0397-2
Eisenstat, D. D., Liu, J. K., Mione, M., Zhong, W., Yu, G., Anderson, S. A., et al. (1999). DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J. Comp. Neurol. 414, 217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i
Ellenberger, T., Fass, D., Arnaud, M., and Harrison, S. C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980. doi: 10.1101/gad.8.8.970
Ellison-Wright, Z., Heyman, I., Frampton, I., Rubia, K., Chitnis, X., Ellison-Wright, I., et al. (2004). Heterozygous PAX6 mutation, adult brain structure and fronto-striato-thalamic function in a human family. Eur. J. Neurosci. 19, 1505–1512. doi: 10.1111/j.1460-9568.2004.03236.x
El Nagar, S., Chakroun, A., Le Greneur, C., Figarella-Branger, D., Di Meglio, T., Lamonerie, T., et al. (2018). Otx2 promotes granule cell precursor proliferation and shh-dependent medulloblastoma maintenance in vivo. Oncogenesis 7, 1–12. doi: 10.1038/s41389-018-0070-6
Emery, B., and Lu, Q. R. (2015). Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb. Perspect. Biol. 7:a020461. doi: 10.1101/cshperspect.a020461
Endele, S., Rosenberger, G., Geider, K., Popp, B., Tamer, C., Stefanova, I., et al. (2010). Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 42, 1021–1026. doi: 10.1038/ng.677
Ericson, J., Muhr, J., Placzek, M., Lints, T., and Jessel, T. á, and Edlund, T. (1995). Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell 81, 747–756. doi: 10.1016/0092-8674(95)90536-7
Ericson, J., Rashbass, P., Schedl, A., Brenner-Morton, S., Kawakami, A., Van Heyningen, V., et al. (1997). Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180. doi: 10.1016/s0092-8674(00)80323-2
Erlander, M. G., Tillakaratne, N. J., Feldblum, S., Patel, N., and Tobin, A. J. (1991). Two genes encode distinct glutamate decarboxylases. Neuron 7, 91–100. doi: 10.1016/0896-6273(91)90077-d
Estivill-Torrus, G., Pearson, H., van Heyningen, V., Price, D. J., and Rashbass, P. (2002). Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development 129, 455–466. doi: 10.1242/dev.129.2.455
Falcone, C., Daga, A., Leanza, G., and Mallamaci, A. (2016). Emx2 as a novel tool to suppress glioblastoma. Oncotarget 7:41005. doi: 10.18632/oncotarget.9322
Fazel Darbandi, S., Poitras, L., Monis, S., Lindtner, S., Yu, M., Hatch, G., et al. (2016). Functional consequences of I56ii Dlx enhancer deletion in the developing mouse forebrain. Dev. Biol. Online ahead of print, doi: 10.1016/j.ydbio.2016.10.015,
Figueres-Oñate, M., and López-Mascaraque, L. L. (2016). Adult olfactory bulb interneuron phenotypes identified by targeting embryonic and postnatal neural progenitors. Front. Neurosci. 10:194. doi: 10.3389/fnins.2016.00194
Filbin, M. G., Tirosh, I., Hovestadt, V., Shaw, M. L., Escalante, L. E., Mathewson, N. D., et al. (2018). Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335. doi: 10.1126/science.aao4750
Flames, N., Pla, R., Gelman, D. M., Rubenstein, J. L., Puelles, L., and Marín, O. (2007). Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007
Flandin, P., Zhao, Y., Vogt, D., Jeong, J., Long, J., Potter, G., et al. (2011). Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron 70, 939–950. doi: 10.1016/j.neuron.2011.04.020
Florian, C., Bahi-Buisson, N., and Bienvenu, T. (2011). FOXG1-related disorders: from clinical description to molecular genetics. Mol. Syndromol. 2, 153–163. doi: 10.1159/000327329
Feng, W., Simoes-de-Souza, F., Finger, T. E., Restrepo, D., and Williams, T. (2009). Disorganized olfactory bulb lamination in mice deficient for transcription factor AP-2epsilon. Mol. Cell Neurosci. 42, 161–171. doi: 10.1016/j.mcn.2009.06.010
Fode, C., Ma, Q., Casarosa, S., Ang, S.-L., Anderson, D. J., and Guillemot, F. (2000). A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14, 67–80. doi: 10.1101/gad.14.1.67
Fogarty, M., Grist, M., Gelman, D., Marín, O., Pachnis, V., and Kessaris, N. (2007). Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 27, 10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007
Fragkouli, A., Hearn, C., Errington, M., Cooke, S., Grigoriou, M., Bliss, T., et al. (2005). Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur. J. Neurosci. 21, 2923–2938. doi: 10.1111/j.1460-9568.2005.04141.x
Franco, S. J., Martinez-Garay, I., Gil-Sanz, C., Harkins-Perry, S. R., and Müller, U. (2011). Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron 69, 482–497. doi: 10.1016/j.neuron.2011.01.003
Frantz, G. D., Weimann, J. M., Levin, M. E., and McConnell, S. K. (1994). Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J. Neurosci. 14, 5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994
Frazer, S., Prados, J., Niquille, M., Cadilhac, C., Markopoulos, F., Gomez, L., et al. (2017). Transcriptomic and anatomic parcellation of 5-HT(3A)R expressing cortical interneuron subtypes revealed by single-cell RNA sequencing. Nat. Commun. 8, 14219–14219. doi: 10.1038/ncomms14219
Freyd, G., Kim, S. K., and Horvitz, H. R. (1990). Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-II. Nature 344, 876–879. doi: 10.1038/344876a0
Fragkouli, A., van Wijk, N. V., Lopes, R., Kessaris, N., and Pachnis, V. (2009). LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development 136, 3841–3851. doi: 10.1242/dev.038083
Friocourt, G., Kanatani, S., Tabata, H., Yozu, M., Takahashi, T., Antypa, M., et al. (2008). Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J. Neurosci. 28, 5794–5780. doi: 10.1523/JNEUROSCI.1067-08.2008
Friocourt, G., Poirier, K., Rakić, S., Parnavelas, J. G., and Chelly, J. (2006). The role of ARX in cortical development. Eur. J. Neurosci. 23, 869–876. doi: 10.1111/j.1460-9568.2006.04629.x
Friocourt, G., and Parnavelas, J. G. (2010). Mutations in ARX result in several defects involving GABAergic neurons. Front. Cell. Neurosci. 4:4. doi: 10.3389/fncel.2010.00004
Fukuchi-Shimogori, T., and Grove, E. A. (2003). Emx2 patterns the neocortex by regulating FGF positional signaling. Nat. Neurosci. 6, 825–831. doi: 10.1038/nn1093
Fukuchi-Shimogori, T., and Grove, E. A. (2001). Neocortex patterning by the secreted signaling molecule FGF8. Science 294, 1071–1074. doi: 10.1126/science.1064252
Fukuda, T., Kawano, H., Osumi, N., Eto, K., and Kawamura, K. (2000). Histogenesis of the cerebral cortex in rat fetuses with a mutation in the Pax-6 gene. Dev. Brain Res. 120, 65–75. doi: 10.1016/s0165-3806(99)00187-x
Furukawa, T., Morrow, E. M., and Cepko, C. L. (1997). Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiatiosn. Cell 91, 531–541. doi: 10.1016/s0092-8674(00)80439-0
Furusho, M., Ono, K., Takebayashi, H., Masahira, N., Kagawa, T., Ikeda, K., et al. (2006). Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev. Biol. 293, 348–357. doi: 10.1016/j.ydbio.2006.01.031
Fulp, C. T., Cho, G., Marsh, E. D., Nasrallah, I. M., Labosky, P. A., and Golden, J. A. (2008). Identification of Arx transcriptional targets in the developing basal forebrain. Hum. Mol. Genet. 17, 3740–3760. doi: 10.1093/hmg/ddn271
Gandal, M. J., Haney, J. R., Parikshak, N. N., Leppa, V., Ramaswami, G., Hartl, C., et al. (2018). Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697.
García-Moreno, F., and Molnár, Z. (2015). Subset of early radial glial progenitors that contribute to the development of callosal neurons is absent from avian brain. Proc. Natl. Acad. Sci. 112, E5058–E5067. doi: 10.1073/pnas.1506377112
Garda, A.-L., Echevarría, D., and Martínez, S. (2001). Neuroepithelial co-expression of Gbx2 and Otx2 precedes Fgf8 expression in the isthmic organizer. Mech. Dev. 101, 111–118. doi: 10.1016/s0925-4773(00)00567-0
Garel, S., Marín, F., Grosschedl, R., and Charnay, P. (1999). Ebf1 controls early cell differentiation in the embryonic striatum. Development 126, 5285–5294. doi: 10.1242/dev.126.23.5285
Gehring, W. J., Affolter, M., and Bürglin, T. (1994). Homeodomain proteins. Annu. Rev. Biochem. 63, 487–526.
Gelman, D. M., Martini, F. J., óbrega-Pereira, S. N., Pierani, A., Kessaris, N., and Marín, O. (2009). The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J. Neurosci. 29, 9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009
Georgala, P. A., Carr, C. B., and Price, D. J. (2011). The role of Pax6 in forebrain development. Dev. Neurobiol. 71, 690–709. doi: 10.1002/dneu.20895
Ghanem, N., Jarinova, O., Amores, A., Long, Q., Hatch, G., Park, B. K., et al. (2003). Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 13, 533–543. doi: 10.1101/gr.716103
Ghanem, N., Yu, M., Poitras, L., Rubenstein, J. L. R., and Ekker, M. (2008). Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia through the activity of the I56ii enhancer. Dev. Biol. 322, 415–424. doi: 10.1016/j.ydbio.2008.07.029
Gherzi, R., Briata, P., Boncinelli, E., Ponassi, M., Querzè, G., Viti, F., et al. (1997). The human homeodomain protein OTX2 binds to the human tenascin-C promoter and trans-represses its activity in transfected cells. DNA Cell Biol. 16, 559–567. doi: 10.1089/dna.1997.16.559
Glaser, T., Jepeal, L., Edwards, J. G., Young, S. R., Favor, J., and Maas, R. L. (1994). PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 7, 463–471. doi: 10.1038/ng0894-463
Glaser, B., Kirov, G., Green, E., Craddock, N., and Owen, M. J. (2005). Linkage disequilibrium mapping of bipolar affective disorder at 12q23-q24 provides evidence for association at CUX2 and FLJ3235. Am. J. Med. Genet. B Neuropsychiatr. Genet. 132b, 38–45. doi: 10.1002/ajmg.b.30081
Glaser, T., Walton, D. S., and Maas, R. L. (1992). Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat. Genet. 2, 232–239. doi: 10.1038/ng1192-232
Godbole, G., Roy, A., Shetty, A. S., and Tole, S. (2017). Novel functions of LHX2 and PAX6 in the developing telencephalon revealed upon combined loss of both genes. Neural Dev. 12, 1–8. doi: 10.1186/s13064-017-0097-y
Godbole, G., Shetty, A. S., Roy, A., D’Souza, L., Chen, B., Miyoshi, G., et al. (2018). Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development 145:dev154583. doi: 10.1242/dev.154583
Goodall, J., Martinozzi, S., Dexter, T. J., Champeval, D., Carreira, S., Larue, L., et al. (2004a). Brn-2 expression controls melanoma proliferation and is directly regulated by β-catenin. Mol. Cell. Biol. 24, 915–2922.
Goodall, J., Wellbrock, C., Dexter, T. J., Roberts, K., Marais, R., and Goding, C.R. (2004b). The brn-2 transcription factor links activated braf to melanoma proliferation. Mol. Cell. Biol. 24, 2923–2931. doi: 10.1128/MCB.24.7.2923-2931.2004
Golonzhka, O., Nord, A., Tang, P. L. F., Lindtner, S., Ypsilanti, A. R., Ferretti, E., et al. (2015). Pbx regulates patterning of the cerebral cortex in progenitors and postmitotic neurons. Neuron 88, 1192–1207. doi: 10.1016/j.neuron.2015.10.045
Gómez-Skarmeta, J.-L., del Corral, R. D., de la Calle-Mustienes, E., Ferrés-Marcó, D., and Modolell, J. (1996). Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85, 95–105. doi: 10.1016/s0092-8674(00)81085-5
Goulet, B., Baruch, A., Moon, N.-S., Poirier, M., Sansregret, L. L., Erickson, A., et al. (2004). A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14, 207–219. doi: 10.1016/s1097-2765(04)00209-6
Graveland, G., and DiFiglia, M. (1985). The frequency and distribution of medium-sized neurons with indented nuclei in the primate and rodent neostriatum. Brain Res. 327, 307–311. doi: 10.1016/0006-8993(85)91524-0
Gray, L. T., Yao, Z., Nguyen, T. N., Kim, T. K., Zeng, H., and Tasic, B. (2017). Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. Elife 6:e2188. doi: 10.7554/eLife.21883
Grigoriou, M., Tucker, A. S., Sharpe, P. T., and Pachnis, V. (1998). Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 125, 2063–2074. doi: 10.1242/dev.125.11.2063
Grueber, W. B., Jan, L. Y., and Jan, Y. N. (2003). Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112, 805–818. doi: 10.1016/s0092-8674(03)00160-0
Guazzi, S., Price, M., De Felice, M., Damante, G., Mattei, M.-G., and Di Lauro, R. (1990). Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 9, 3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x
Gulisano, M., Broccoli, V., Pardini, C., and Boncinelli, E. (1996). Emx1 and Emx2 show different patterns of expression during proliferation and differentiation of the developing cerebral cortex in the mouse. Eur. J. Neurosci. 8, 1037–1050. doi: 10.1111/j.1460-9568.1996.tb01590.x
Gehring, W. J. (1996). The master control gene for morphogenesis and evolution of the eye. Genes Cells 1, 11–15. doi: 10.1046/j.1365-2443.1996.11011.x
Hack, M., Sugimori, M., Lundberg, C., Nakafuku, M., and Götz, M. (2004). Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol. Cell. Neurosci. 25, 664–678. doi: 10.1016/j.mcn.2003.12.012
Hamasaki, T., Leingärtner, A., Ringstedt, T., and O’Leary, D. D. (2004). EMX2 regulates sizes and positioning of the primary sensory and motor areas in neocortex by direct specification of cortical progenitors. Neuron 43, 359–372. doi: 10.1016/j.neuron.2004.07.016
Hamilton, S. P., Woo, J. M., Carlson, E. J., Ghanem, N., Ekker, M., and Rubenstein, J. L. R. (2005). Analysis of four DLX homeobox genes in autistic probands. BMC Genet. 6:52. doi: 10.1186/1471-2156-6-52
Hanashima, C., Shen, L., Li, S. C., and Lai, E. (2002). Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J. Neurosci. 22, 6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002
Hansen, D. V., Lui, J. H., Flandin, P., Yoshikawa, K., Rubenstein, J. L., Alvarez-Buylla, A., et al. (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576–1587. doi: 10.1038/nn.3541
Harada, R., Bérubé, G., Tamplin, O. J., Denis-Larose, C., and Nepveu, A. (1995). DNA-binding specificity of the cut repeats from the human cut-like protein. Mol. Cell. Biol. 15, 129–140. doi: 10.1128/MCB.15.1.129
Harada, R., Dufort, D., Denis-Larose, C., and Nepveu, A. (1994). Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J. Biol. Chem. 269, 2062–2067. doi: 10.1016/s0021-9258(17)42135-1
Harada, R., Vadnais, C., Sansregret, L., Leduy, L., Bérubé, G., Robert, F., et al. (2007). Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. 36, 189–202. doi: 10.1093/nar/gkm970
Hashizume, K., Yamanaka, M., and Ueda, S. (2018). POU3F2 participates in cognitive function and adult hippocampal neurogenesis via mammalian-characteristic amino acid repeats. Genes Brain Behav. 17, 118–125. doi: 10.1111/gbb.12408
Haubensak, W., Attardo, A., Denk, W., and Huttner, W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 3196–3201. doi: 10.1073/pnas.0308600100
Haubst, N., Berger, J., Radjendirane, V., Graw, J., Favor, J., Saunders, G. F., et al. (2004). Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development 131, 6131–6140. doi: 10.1242/dev.01524
Haydar, T. F., and Reeves, R. H. (2012). Trisomy 21 and early brain development. Trends Neurosci. 35, 81–91. doi: 10.1016/j.tins.2011.11.001
He, X., Treacy, M. N., Simmons, D. M., Ingraham, H. A., Swanson, L. W., and Rosenfeld, M. G. (1989). Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340, 35–42. doi: 10.1038/340035a0
Heide, M., Zhang, Y., Zhou, X., Zhao, T., Miquelajáuregui, A., Varela-Echavarría, A., et al. (2015). Lhx5 controls mamillary differentiation in the developing hypothalamus of the mouse. Front. Neuroanat. 9:113. doi: 10.3389/fnana.2015.00113
Heins, N., Malatesta, P., Cecconi, F., Nakafuku, M., Tucker, K. L., Hack, M. A., et al. (2002). Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 5, 308–315. doi: 10.1038/nn828
Hendry, S. H., Schwark, H., Jones, E., and Yan, J. (1987). Numbers and proportions of GABA-immunoreactive neurons in different areas of monkey cerebral cortex. J. Neurosci. 7, 1503–1519. doi: 10.1523/JNEUROSCI.07-05-01503.1987
Herr, W., and Cleary, M. A. (1995). The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 9, 1679–1693. doi: 10.1101/gad.9.14.1679
Hill, R. E., Favor, J., Hogan, B. L., Ton, C. C., Saunders, G. F., Hanson, I. M., et al. (1991). Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354, 522–525. doi: 10.1038/354522a0
Hirata, T., Nomura, T., Takagi, Y., Sato, Y., Tomioka, N., Fujisawa, H., et al. (2002). Mosaic development of the olfactory cortex with Pax6-dependent and-independent components. Dev. Brain Res. 136, 17–26. doi: 10.1016/s0165-3806(02)00304-8
Hoch, R. V., Lindtner, S., Price, J. D., and Rubenstein, J. L. R. (2015). OTX2 transcription factor controls regional patterning within the medial ganglionic eminence and regional identity of the septum. Cell Rep. 12, 482–494. doi: 10.1016/j.celrep.2015.06.043
Holder, J. L. Jr., Zhang, L., Kublaoui, B. M., DiLeone, R. J., Oz, O. K., Bair, C. H., et al. (2004). Sim1 gene dosage modulates the homeostatic feeding response to increased dietary fat in mice. Am. J. Physiol. Endocrinol. Metab. 287, E105–E113. doi: 10.1152/ajpendo.00446.2003
Holland, P. W., Booth, H. A. F., and Bruford, E. A. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5:47. doi: 10.1186/1741-7007-5-47
Horike, S.-i, Cai, S., Miyano, M., Cheng, J.-F., and Kohwi-Shigematsu, T. (2005). Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37, 31–40. doi: 10.1038/ng1491
Hou, P.-S., Miyoshi, G., and Hanashima, C. (2019). Sensory cortex wiring requires preselection of short-and long-range projection neurons through an Egr-Foxg1-COUP-TFI network. Nat. Commun. 10, 1–18. doi: 10.1038/s41467-019-11043-w
Hsieh-Li, H. M., Witte, D. P., Szucsik, J. C., Weinstein, M., Li, H., and Potter, S. S. (1995). Gsh-2, a murine homeobox gene expressed in the developing brain. Mech. Dev. 50, 177–186. doi: 10.1016/0925-4773(94)00334-j
Huang, B., Li, X., Tu, X., Zhao, W., Zhu, D., Feng, Y., et al. (2018). OTX1 regulates cell cycle progression of neural progenitors in the developing cerebral cortex. J. Biol. Chem. 293, 2137–2148. doi: 10.1074/jbc.RA117.001249
Ide, M., Yamada, K., Toyota, T., Iwayama, Y., Ishitsuka, Y., Minabe, Y., et al. (2005). Genetic association analyses of PHOX2B and ASCL1 in neuropsychiatric disorders: evidence for association of ASCL1 with Parkinson’s disease. Hum. Genet. 117, 520–527. doi: 10.1007/s00439-005-1342-8
Iler, N., Rowitch, D. H., Echelard, Y., McMahon, A. P., and Abate-Shen, C. (1995). A single homeodomain binding site restricts spatial expression of Wnt-1 in the developing brain. Mech. Dev. 53, 87–96. doi: 10.1016/0925-4773(95)00427-0
Imamura, F., Ayoub, A. E., Rakic, P., and Greer, C. A. (2011). Timing of neurogenesis is a determinant of olfactory circuitry. Nat. Neurosci. 14, 331–337. doi: 10.1038/nn.2754
Imamura, F., and Greer, C. A. (2013). Pax6 regulates Tbr1 and Tbr2 expressions in olfactory bulb mitral cells. Mol. Cell Neurosci. 54, 58–70. doi: 10.1016/j.mcn.2013.01.002
Inada, H., Watanabe, M., Uchida, T., Ishibashi, H., Wake, H., Nemoto, T., et al. (2011). GABA regulates the multidirectional tangential migration of GABAergic interneurons in living neonatal mice. PLoS One 6:e27048. doi: 10.1371/journal.pone.0027048
Inoue, T., Chisaka, O., Matsunami, H., and Takeichi, M. (1997). Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev. Biol. 183, 183–194. doi: 10.1006/dbio.1996.8501
Inoue, T., Nakamura, S., and Osumi, N. (2000). Fate mapping of the mouse prosencephalic neural plate. Dev. Biol. 219, 373–383. doi: 10.1006/dbio.2000.9616
Inoue, F., Kurokawa, D., Takahashi, M., and Aizawa, S. (2012). Gbx2 directly restricts Otx2 expression to forebrain and midbrain, competing with class III POU factors. Mol. Cell. Biol. 32, 2618–2627. doi: 10.1128/MCB.00083-12
Iulianella, A., Sharma, M., Durnin, M., Vanden Heuvel, G. B., and Trainor, P. A. (2008). Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development 135, 729–741. doi: 10.1242/dev.013276
Iulianella, A., Vanden Heuvel, G., and Trainor, P. (2003). Dynamic expression of murine Cux2 in craniofacial, limb, urogenital and neuronal primordia. Gene Expr. Patterns 3:571–577. doi: 10.1016/s1567-133x(03)00123-6
Jennings, B. H., Pickles, L. M., Wainwright, S. M., Roe, S. M., Pearl, L. H., and Ish-Horowicz, D. (2006). Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell 22, 645–655. doi: 10.1016/j.molcel.2006.04.024
Jessell, T.M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. doi: 10.1038/35049541
Jiang, Q., Zagozewski, J., Godbout, R., and Eisenstat, D. D. (2020). Distal-less genes Dlx1/Dlx2 repress oligodendrocyte genesis through transcriptional inhibition of Olig2 expression in the developing vertebrate forebrain. bioRxiv [Preprint].
Jimenez, F., Martin-Morris, L. E., Velasco, L., Chu, H., Sierra, J., Rosen, D., et al. (1995). vnd, a gene required for early neurogenesis of Drosophila, encodes a homeodomain protein. EMBO J. 14, 3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x
Jimenez-García, M. P., Lucena-Cacace, A., Otero-Albiol, D., and Carnero, A. (2021a). Empty spiracles homeobox genes EMX1 and EMX2 regulate WNT pathway activation in sarcomagenesis. J. Exp. Clin. Cancer Res. 40, 1–18. doi: 10.1186/s13046-021-02048-9
Jimenez-García, M. P., Lucena-Cacace, A., Otero-Albiol, D., and Carnero, A. (2021b). Regulation of sarcomagenesis by the empty spiracles homeobox genes EMX1 and EMX2. Cell Death Dis. 12, 1–17. doi: 10.1038/s41419-021-03801-w
Johnson, J. E., Birren, S. J., Saito, T., and Anderson, D. J. (1992). DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc. Natl. Acad. Sci. 89:3596. doi: 10.1073/pnas.89.8.3596
Joliot, A., and Prochiantz, A. (2004). Transduction peptides: from technology to physiology. Nat. Cell Biol. 6, 189–196. doi: 10.1038/ncb0304-189
Jones, L., ópez-Bendito, G. L., Gruss, P., Stoykova, A., and Molnár, Z. (2002). Pax6 is required for the normal development of the forebrain axonal connections. Development 129, 5041–5052. doi: 10.1242/dev.129.21.5041
Kanatani, S., Honda, T., Aramaki, M., Hayashi, K., Kubo, K.-i., Ishida, M., et al. (2015). The COUP-TFII/Neuropilin-2 is a molecular switch steering diencephalon-derived GABAergic neurons in the developing mouse brain. Proc. Natl. Acad. Sci. 112, E4985–E4994. doi: 10.1073/pnas.1420701112
Karlsson, O., Thor, S., Norberg, T., Ohlsson, H., and Edlund, T. (1990). Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo-and a Cys–His domain. Nature 344, 879–882. doi: 10.1038/344879a0
Kappen, C. (2000). The homeodomain: an ancient evolutionary motif in animals and plants. Comput. Chem. 24, 95–103. doi: 10.1016/s0097-8485(99)00049-2
Kastury, K., Druck, T., Huebner, K., Barletta, C., Acampora, D., Simeone, A., et al. (1994). Chromosome locations of human EMX and OTX genes. Genomics 22, 41–45. doi: 10.1006/geno.1994.1343
Kato, M., and Dobyns, W. B. (2004). X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: proposal for a new term, “interneuronopathy”. J. Child Neurol. 19, 392–397. doi: 10.1177/08830738050200042001
Kato, M., Das, S., Petras, K., Kitamura, K., Morohashi, K. i., Abuelo, D. N., et al. (2004). Mutations of ARX are associated with striking pleiotropy and consistent genotype–phenotype correlation. Hum. Mutat. 23, 147–155. doi: 10.1002/humu.10310
Kato, M., Das, S., Petras, K., Sawaishi, Y., and Dobyns, W. B. (2003). Polyalanine expansion of ARX associated with cryptogenic West syndrome. Neurology 61, 267–277. doi: 10.1212/01.wnl.0000068012.69928.92
Kato, M., Saitoh, S., Kamei, A., Shiraishi, H., Ueda, Y., Akasaka, M., et al. (2007). A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome). Am. J. Hum. Genet. 81, 361–366. doi: 10.1086/518903
Kaur, R., Aiken, C., Morrison, L. C., Rao, R., Del Bigio, M. R., Rampalli, S., et al. (2015). OTX2 exhibits cell-context-dependent effects on cellular and molecular properties of human embryonic neural precursors and medulloblastoma cells. Dis. Models Mech. 8, 1295–1309. doi: 10.1242/dmm.020594
Kessaris, N., Fogarty, M., Iannarelli, P., Grist, M., Wegner, M., and Richardson, W. D. (2006). Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 9, 173–179. doi: 10.1038/nn1620
Kiecker, C., and Lumsden, A. (2004). Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat. Neurosci. 7, 1242–1249. doi: 10.1038/nn1338
Kiecker, C., and Lumsden, A. (2005). Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 6, 553–564. doi: 10.1038/nrn1702
Kim, Y., and Nirenberg, M. (1989). Drosophila NK-homeobox genes. Proc. Natl. Acad. Sci. 86, 7716–7720. doi: 10.1073/pnas.86.20.7716
Kim, D. W., Liu, K., Wang, Z. Q., Zhang, Y. S., Bathini, A., Brown, M. P., et al. (2021). Gene regulatory networks controlling differentiation, survival, and diversification of hypothalamic Lhx6-expressing GABAergic neurons. Commun. Biol. 4, 1–16. doi: 10.1038/s42003-020-01616-7
Kim, D. W., Washington, P. W., Wang, Z. Q., Lin, S. H., Sun, C., Ismail, B. T., et al. (2020). The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat. Commun. 11:4360.
Kimura, S., Hara, Y., Pineau, T., Fernandez-Salguero, P., Fox, C. H., Ward, J. M., et al. (1996). The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69. doi: 10.1101/gad.10.1.60
Kitamura, K., Itou, Y., Yanazawa, M., Ohsawa, M., Suzuki-Migishima, R., Umeki, Y., et al. (2009). Three human ARX mutations cause the lissencephaly-like and mental retardation with epilepsy-like pleiotropic phenotypes in mice. Hum. Mol. Genet. 18, 3708–3802. doi: 10.1093/hmg/ddp318
Kitamura, K., Miura, H., Yanazawa, M., Miyashita, T., and Kato, K. (1997). Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2. 2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech. Dev. 67, 83–96. doi: 10.1016/s0925-4773(97)00110-x
Kitamura, K., Yanazawa, M., Sugiyama, N., Miura, H., Iizuka-Kogo, A., Kusaka, M., et al. (2002). Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 32, 359–369. doi: 10.1038/ng1009
Klemm, J. D., Rould, M. A., Aurora, R., Herr, W., and Pabo, C. O. (1994). Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77, 21–32. doi: 10.1016/0092-8674(94)90231-3
Ko, J., Humbert, S., Bronson, R. T., Takahashi, S., Kulkarni, A. B., Li, E., et al. (2001). p35 and p39 are essential for cyclin-dependent kinase 5 function during neurodevelopment. J. Neurosci. 21, 6758–71. doi: 10.1523/JNEUROSCI.21-17-06758.2001
Kobayashi, D., Kobayashi, M., Matsumoto, K., Ogura, T., Nakafuku, M., and Shimamura, K. (2002). Early subdivisions in the neural plate define distinct competence for inductive signals. Development 129, 83–93. doi: 10.1242/dev.129.1.83
Kriegstein, A., and Alvarez-Buylla, A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184. doi: 10.1146/annurev.neuro.051508.135600
Kroll, T. T., and O’Leary, D. D. (2005). Ventralized dorsal telencephalic progenitors in Pax6 mutant mice generate GABA interneurons of a lateral ganglionic eminence fate. Proc. Natl. Acad. Sci. 102, 7374–7379. doi: 10.1073/pnas.0500819102
Krude, H., Schütz, B., Biebermann, H., Von Moers, A., Schnabel, D., Neitzel, H., et al. (2002). Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J. Clin. Invest. 109, 475–480. doi: 10.1172/JCI14341
Kumamoto, T., and Hanashima, C. (2017). Evolutionary conservation and conversion of Foxg1 function in brain development. Dev. Growth Differ. 59, 258–269. doi: 10.1111/dgd.12367
Kurokawa, D., Kiyonari, H., Nakayama, R., Kimura-Yoshida, C., Matsuo, I., and Aizawa, S. (2004). Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development 131, 3319–3331. doi: 10.1242/dev.01220
Kurokawa, D., Sakurai, Y., Inoue, A., Nakayama, R., Takasaki, N., Suda, Y., et al. (2006). Evolutionary constraint on Otx2 neuroectoderm enhancers-deep conservation from skate to mouse and unique divergence in teleost. Proc. Natl. Acad. Sci. 103, 19350–19355. doi: 10.1073/pnas.0604686103
Küspert, M., Hammer, A., Bösl, M. R., and Wegner, M. (2011). Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 39, 1280–1293. doi: 10.1093/nar/gkq951
Larsen, K. B., Lutterodt, M. C., Laursen, H., Graem, N., Pakkenberg, B., Møllgård, K., et al. (2010a). Spatiotemporal distribution of PAX6 and MEIS2 expression and total cell numbers in the ganglionic eminence in the early developing human forebrain. Dev. Neurosci. 32, 149–162. doi: 10.1159/000297602
Larsen, K. B., Lutterodt, M. C., Møllgård, K., and Møller, M. (2010b). Expression of the homeobox genes OTX2 and OTX1 in the early developing human brain. J. Histochem. Cytochem. 58, 669–678. doi: 10.1369/jhc.2010.955757
Lavdas, A. A., Grigoriou, M., Pachnis, V., and Parnavelas, J. G. (1999). The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. 19, 7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999
Le, T. N., Zhou, Q.-P., Cobos, I., Zhang, S., Zagozewski, J., Japoni, S., et al. (2017). GABAergic interneuron differentiation in the basal forebrain is mediated through direct regulation of glutamic acid decarboxylase isoforms by dlx homeobox transcription factors. J. Neurosci. 37, 8816–8829. doi: 10.1523/JNEUROSCI.2125-16.2017
Lee, J. E. (1997). Basic helix-loop-helix genes in neural development. Curr. Opin. Neurobiol. 7, 13–20. doi: 10.1016/s0959-4388(97)80115-8
Lee, H. H. C., Bernard, C., Ye, Z., Acampora, D., Simeone, A., Prochiantz, A., et al. (2017). Genetic Otx2 mis-localization delays critical period plasticity across brain regions. Mol. Psychiatry 22, 680–688. doi: 10.1038/mp.2017.1
Lesaffre, B., Joliot, A., Prochiantz, A., and Volovitch, M. (2007). Direct non-cell autonomous Pax6 activity regulates eye development in the zebrafish. Neural Dev. 2:2. doi: 10.1186/1749-8104-2-2
Li, H., Bishop, K. M., and O’Leary, D. D. (2006). Potential target genes of EMX2 include Odz/Ten-M and other gene families with implications for cortical patterning. Mol. Cell. Neurosci. 33, 136–149. doi: 10.1016/j.mcn.2006.06.012
Li, H., Hader, A. T., Han, Y. R., Wong, J. A., Babiarz, J., Ricupero, C. L., et al. (2012a). Isolation of a novel rat neural progenitor clone that expresses dlx family transcription factors and gives rise to functional gabaergic neurons in culture. Dev. Neurobiol. 72, 805–820. doi: 10.1002/dneu.20977
Li, H., Lu, Y., Smith, H. K., and Richardson, W. D. (2007). Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J. Neurosci. 27, 14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007
Li, H., Miao, Q., Xu, C.-w., Huang, J.-h., Zhou, Y.-f., and Wu, M.-j. (2016). OTX1 contributes to hepatocellular carcinoma progression by regulation of ERK/MAPK pathway. J. Korean Med. Sci. 31, 1215–1223. doi: 10.3346/jkms.2016.31.8.1215
Li, J., Mo, M., Chen, Z., Chen, Z., Sheng, Q., Mu, H., et al. (2012b). Adenoviral delivery of the EMX2 gene suppresses growth in human gastric cancer. PLoS One 7:e45970. doi: 10.1371/journal.pone.0045970
Li, J., Wang, C., Zhang, Z., Wen, Y., An, L., Liang, Q., et al. (2017). Transcription factors Sp8 and Sp9 coordinately regulate olfactory bulb interneuron development. Cereb. Cortex 28, 3278–3294. doi: 10.1093/cercor/bhx199
Li, M., Santpere, G., Kawasawa, Y. I., Evgrafov, O. V., Gulden, F. O., Pochareddy, S., et al. (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362:eaat7615. doi: 10.1126/science.aat7615
Li, N., Zhao, C.-T., Wang, Y., and Yuan, X.-B. (2010). The transcription factor Cux1 regulates dendritic morphology of cortical pyramidal neurons. PLoS One 5:e1059. doi: 10.1371/journal.pone.0010596
Li, T., Stark, M. R., Johnson, A. D., and Wolberger, C. (1995). Crystal structure of the MATa1/MATα2 homeodomain heterodimer bound to DNA. Science 270, 262–269. doi: 10.1126/science.270.5234.262
Li, Y., Kido, T., Garcia-Barcelo, M. M., Tam, P. K., Tabatabai, Z. L., and Lau, Y.-F. C. (2015). SRY interference of normal regulation of the RET gene suggests a potential role of the Y-chromosome gene in sexual dimorphism in Hirschsprung disease. Hum. Mol. Genet. 24, 685–697. doi: 10.1093/hmg/ddu488
Ligon, K. L., Echelard, Y., Assimacopoulos, S., Danielian, P. S., Kaing, S., Grove, E. A., et al. (2003). Loss of Emx2 function leads to ectopic expression of Wnt1 in the developing telencephalon and cortical dysplasia. Development 130, 2275–2287. doi: 10.1242/dev.00421
Lim, J. W., Donahoo, A.-L. S., Bunt, J., Edwards, T. J., Fenlon, L. R., Liu, Y., et al. (2015). EMX1 regulates NRP1-mediated wiring of the mouse anterior cingulate cortex. Development 142, 3746–3757. doi: 10.1242/dev.119909
Lim, Y., Cho, I.-T., Shi, X., Grinspan, J. B., Cho, G., and Golden, J. A. (2019). Arx expression suppresses ventralization of the developing dorsal forebrain. Sci. Rep. 9:226. doi: 10.1038/s41598-018-36194-6
Lindtner, S., Catta-Preta, R., Tian, H., Su-Feher, L., Price, J. D., Dickel, D. E., et al. (2019). Genomic resolution of DLX-orchestrated transcriptional circuits driving development of forebrain GABAergic neurons. Cell Rep. 28, 2048.e8–2063.e8. doi: 10.1016/j.celrep.2019.07.022
Liodis, P., Denaxa, M., Grigoriou, M., Akufo-Addo, C., Yanagawa, Y., and Pachnis, V. (2007). Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 27, 3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007
Liu, J. K., Ghattas, I., Liu, S., Chen, S., and Rubenstein, J. L. R. (1997). Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev. Dyn. 210, 498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3
Liu, Y., MacDonald, R. J., and Swift, G. H. (2001). DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J. Biol. Chem. 276, 17985–17993. doi: 10.1074/jbc.M100678200
Leonard, J. H., and Bell, J. R. (1997). Insights into the Merkel cell phenotype from Merkel cell carcinoma cell lines. Australas J. Dermatol. 38, (Suppl 1), S91–S98. doi: 10.1111/j.1440-0960.1997.tb01019.x
Letinic, K., Zoncu, R., and Rakic, P. (2002). Origin of GABAergic neurons in the human neocortex. Nature 417, 645–649. doi: 10.1038/nature00779
Lois, C., and Alvarez-Buylla, A. (1994). Long-distance neuronal migration in the adult mammalian brain. Science 264, 1145–1148. doi: 10.1126/science.8178174
Long, J. E., Cobos, I., Potter, G. B., and Rubenstein, J. L. R. (2009). Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb. Cortex 19(Suppl. 1), i96–i106.
Long, J. E., Garel, S., Alvarez-Dolado, M., Yoshikawa, K., Osumi, N., Alvarez-Buylla, A., et al. (2007). Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J. Neurosci. 27:3230. doi: 10.1523/JNEUROSCI.5265-06.2007
Longobardi, E., Penkov, D., Mateos, D., De Florian, G., Torres, M., and Blasi, F. (2014). Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev. Dyn. 243, 59–75. doi: 10.1002/dvdy.24016
LoTurco, J. J., and Bai, J. (2006). The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 29, 407–413. doi: 10.1016/j.tins.2006.05.006
López-Bendito, G., Sánchez-Alcaniz, J. A., Pla, R., Borrell, V., Picó, E., Valdeolmillos, M., et al. (2008). Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 28, 1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008
Lopes, R., van Wijk, N. V., Neves, G., and Pachnis, V. (2012). Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proc. Natl. Acad. Sci. 109, 3119–3124. doi: 10.1073/pnas.1109251109
Lowenstein, E.D., Rusanova, A., Stelzer, J., Hernaiz-Llorens, M., Schroer, A. E., Epifanova, E., et al. (2021). Olig3 regulates early cerebellar development. Elife 10:e64684. doi: 10.7554/eLife.64684
Lu, Q. R., Sun, T., Zhu, Z., Ma, N., Garcia, M., Stiles, C. D., et al. (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86. doi: 10.1016/s0092-8674(02)00678-5
Lu, Q. R., Yuk, D.-i., Alberta, J. A., Zhu, Z., Pawlitzky, I., Chan, J., et al. (2000). Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317–329. doi: 10.1016/s0896-6273(00)80897-1
Lu, Y., Labak, C. M., Jain, N., Purvis, I. J., Guda, M. R., Bach, S. E., et al. (2017). OTX2 expression contributes to proliferation and progression in Myc-amplified medulloblastoma. Am. J. Cancer Res. 7:647.
Ma, D. K., Jang, M.-H., Guo, J. U., Kitabatake, Y., Chang, M.-l., Pow-Anpongkul, N., et al. (2009). Neuronal activity–induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077. doi: 10.1126/science.1166859
Ma, T., Wang, C., Wang, L., Zhou, X., Tian, M., Zhang, Q., et al. (2013). Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 16, 1588–1597. doi: 10.1038/nn.3536
MacDonald, R. B., Debiais-Thibaud, M., Talbot, J. C., and Ekker, M. (2010). The relationship between dlx and gad1 expression indicates highly conserved genetic pathways in the zebrafish forebrain. Dev. Dyn. 239, 2298–2306. doi: 10.1002/dvdy.22365
MacDonald, R. B., Pollack, J. N., Debiais-Thibaud, M., Heude, E., Coffin Talbot, J., and Ekker, M. (2013). The ascl1a and dlx genes have a regulatory role in the development of GABAergic interneurons in the zebrafish diencephalon. Dev. Biol. 381, 276–285. doi: 10.1016/j.ydbio.2013.05.025
Magno, L., Barry, C., Schmidt-Hieber, C., Theodotou, P., Häusser, M., and Kessaris, N. (2017). NKX2-1 is required in the embryonic septum for cholinergic system development, learning, and memory. Cell Rep. 20, 1572–1584. doi: 10.1016/j.celrep.2017.07.053
Mailly, F., Bérubé, G., Harada, R., Mao, P. L., Phillips, S., and Nepveu, A. (1996). The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol. Cell. Biol. 16, 5346–5357. doi: 10.1128/MCB.16.10.5346
Maira, M., Long, J. E., Lee, A. Y., Rubenstein, J. L. R., and Stifani, S. (2010). Role for TGF-beta superfamily signaling in telencephalic GABAergic neuron development. J. Neurodev. Disord. 2, 48–60. doi: 10.1007/s11689-009-9035-6
Malandrini, A., Mari, F., Palmeri, S., Gambelli, S., Berti, G., Bruttini, M., et al. (2001). PAX6 mutation in a family with aniridia, congenital ptosis, and mental retardation. Clin. Genet. 60, 151–154. doi: 10.1034/j.1399-0004.2001.600210.x
Mallamaci, A., Iannone, R., Briata, P., Pintonello, L., Mercurio, S., Boncinelli, E., et al. (1998). EMX2 protein in the developing mouse brain and olfactory area. Mech. Dev. 77, 165–172. doi: 10.1016/s0925-4773(98)00141-5
Mallamaci, A., Mercurio, S., Muzio, L., Cecchi, C., Pardini, C. L., Gruss, P., et al. (2000). The lack of Emx2 causes impairment of reel in signaling and defects of neuronal migration in the developing cerebral cortex. J. Neurosci. 20, 1109–1118. doi: 10.1523/JNEUROSCI.20-03-01109.2000
Malt, E. A., Juhasz, K., Malt, U. F., and Naumann, T. (2016). A role for the transcription factor Nk2 homeobox 1 in schizophrenia: convergent evidence from animal and human studies. Front. Behav. Neurosci. 10:59. doi: 10.3389/fnbeh.2016.00059
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390. doi: 10.1016/j.cell.2015.06.034
Mariani, J., Favaro, R., Lancini, C., Vaccari, G., Ferri, A. L., Bertolini, J., et al. (2012). Emx2 is a dose-dependent negative regulator of Sox2 telencephalic enhancers. Nucleic Acids Res. 40, 6461–6476. doi: 10.1093/nar/gks295
Marín, O., and Rubenstein, J. L. (2003). Cell migration in the forebrain. Annu. Rev. Neurosci. 26, 441–483. doi: 10.1146/annurev.neuro.26.041002.131058
Marín, O., Yaron, A., Bagri, A., Tessier-Lavigne, M., and Rubenstein, J. L. (2001). Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science 293, 872–875. doi: 10.1126/science.1061891
Marin, O., Anderson, S. A., and Rubenstein, J. L. (2000). Origin and molecular specification of striatal interneurons. J. Neurosci. 20, 6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000
Martin, D. L., Liu, H., Martin, S. B., and Wu, S. J. (2000). Structural features and regulatory properties of the brain glutamate decarboxylases. Neurochem. Int. 37, 111–119. doi: 10.1016/s0197-0186(00)00014-0
Martynoga, B., Morrison, H., Price, D. J., and Mason, J. O. (2005). Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127. doi: 10.1016/j.ydbio.2005.04.005
Marsh, E., Fulp, C., Gomez, E., Nasrallah, I., Minarcik, J., Sudi, J., et al. (2009). Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain 132, 1563–1567. doi: 10.1093/brain/awp107
Marsh, E. D., Nasrallah, M. P., Walsh, C., Murray, K. A., Sunnen, C. N., McCoy, A., et al. (2016). Developmental interneuron subtype deficits after targeted loss of Arx. BMC Neurosci. 17:3. doi: 10.1186/s12868-016-0265-8
Mattiske, T., Tan, M. H., Dearsley, O., Cloosterman, D., Hii, C. S., Gécz, J., et al. (2018). Regulating transcriptional activity by phosphorylation: a new mechanism for the ARX homeodomain transcription factor. PLoS One 13:e0206914. doi: 10.1371/journal.pone.0206914
Matsumoto, K., Tanaka, T., Furuyama, T., Kashihara, Y., Mori, T., Ishii, N., et al. (1996). L3, a novel murine LIM-homeodomain transcription factor expressed in the ventral telencephalon and the mesenchyme surrounding the oral cavity. Neurosci. Lett. 204, 113–116. doi: 10.1016/0304-3940(96)12341-7
Matsunami, H., and Takeichi, M. (1995). Fetal brain subdivisions defined by R-and E-cadherin expressions: evidence for the role of cadherin activity in region-specific, cell–cell adhesion. Dev. Biol. 172, 466–478. doi: 10.1006/dbio.1995.8029
Matsuo, I., Kuratani, S., Kimura, C., Takeda, N., and Aizawa, S. (1995). Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 9, 2646–2658. doi: 10.1101/gad.9.21.2646
Manabe, T., Tatsumi, K., Inoue, M., Matsuyoshi, H., Makinodan, M., Yamauchi, T., et al. (2008). Knockdown of the L3/Lhx8 gene suppresses cholinergic differentiation of murine embryonic stem cell-derived spheres. Int. J. Dev. Neurosci. 26, 249–252. doi: 10.1016/j.ijdevneu.2007.11.004
Manabe, T., Tatsumi, K., Inoue, M., Matsuyoshi, H., Makinodan, M., Yokoyama, S., et al. (2005). L3/Lhx8 is involved in the determination of cholinergic or GABAergic cell fate. J. Neurochem. 94, 723–730. doi: 10.1111/j.1471-4159.2005.03261.x
Manabe, T., Tatsumi, K., Inoue, M., Makinodan, M., Yamauchi, T., Makinodan, E., et al. (2007). L3/Lhx8 is a pivotal factor for cholinergic differentiation of murine embryonic stem cells. Cell Death Differ. 14, 1080–1085. doi: 10.1038/sj.cdd.4402106
Manoli, M., and Driever, W. (2014). nkx2. 1 and nkx2. 4 genes function partially redundant during development of the zebrafish hypothalamus, preoptic region, and pallidum. Front. Neuroanat. 8:145. doi: 10.3389/fnana.2014.00145
Manuel, M., and Price, D. J. (2005). Role of Pax6 in forebrain regionalization. Brain Res. Bull. 66, 387–393. doi: 10.1016/j.brainresbull.2005.02.006
Maurel-Zaffran, C., and Treisman, J. E. (2000). pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development 127, 1007–1016. doi: 10.1242/dev.127.5.1007
Mayer, C., Hafemeister, C., Bandler, R. C., Machold, R., Brito, R. B., Jaglin, X., et al. (2018). Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462. doi: 10.1038/nature25999
McEvilly, R. J., de Diaz, M. O., Schonemann, M. D., Hooshmand, F., and Rosenfeld, M. G. (2002). Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295, 1528–1532. doi: 10.1126/science.1067132
McKenzie, O., Ponte, I., Mangelsdorf, M., Finnis, M., Colasante, G., Shoubridge, C., et al. (2007). Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience 146, 236–247. doi: 10.1016/j.neuroscience.2007.01.038
Medina, L., and Abellán, A. (2009). Development and evolution of the pallium. Semin. Cell Dev. Biol. 20, 698–711. doi: 10.1016/j.semcdb.2009.04.008
Méndez-Gómez, H. R. M., and Vicario-Abejón, C. (2012). The homeobox gene Gsx2 regulates the self-renewal and differentiation of neural stem cells and the cell fate of postnatal progenitors. PLoS One 7:e29799. doi: 10.1371/journal.pone.0029799
Merabet, S., and Lohmann, I. (2015). Toward a new twist in Hox and TALE DNA-binding specificity. Dev. Cell 32, 259–261. doi: 10.1016/j.devcel.2015.01.030
Metz, C. W. (1914). An apterous Drosophila and its genetic behavior. Am. Nat. 48, 675–692. doi: 10.1086/279439
Mi, D., Carr, C. B., Georgala, P. A., Huang, Y.-T., Manuel, M. N., Jeanes, E., et al. (2013). Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb. Neuron 78, 269–284. doi: 10.1016/j.neuron.2013.02.012
Michaud, J. L., Rosenquist, T., May, N. R., and Fan, C. M. (1998). Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 12, 3264–3275. doi: 10.1101/gad.12.20.3264
Michl, P., and Downward, J. (2006). CUTL1: a key mediator of TGFbeta-induced tumor invasion. Cell Cycle 5, 132–144. doi: 10.4161/cc.5.2.2311
Ming, G.-L., and Song, H. (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702. doi: 10.1016/j.neuron.2011.05.001
Minocha, S., Valloton, D., Arsenijevic, Y., Cardinaux, J.-R., Guidi, R., Hornung, J.-P., et al. (2017). Nkx2. 1 regulates the generation of telencephalic astrocytes during embryonic development. Sci. Rep. 7, 1–20. doi: 10.1038/srep43093
Minocha, S., Valloton, D., Ypsilanti, A. R., Fiumelli, H., Allen, E. A., Yanagawa, Y., et al. (2015). Nkx2. 1-derived astrocytes and neurons together with Slit2 are indispensable for anterior commissure formation. Nat. Commun. 6, 1–15. doi: 10.1038/ncomms7887
Miquelajáuregui, A., Varela-Echavarría, A., Ceci, M. L., García-Moreno, F., Ricano, I., Hoang, K., et al. (2010). LIM-homeobox gene Lhx5 is required for normal development of Cajal–Retzius cells. J. Neurosci. 30, 10551–10562. doi: 10.1523/JNEUROSCI.5563-09.2010
Miura, H., Yanazawa, M., Kato, K., and Kitamura, K. (1997). Expression of a novel aristaless related homeobox gene ‘Arx’in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 65, 99–109. doi: 10.1016/s0925-4773(97)00062-2
Miyata, T., Kawaguchi, A., Okano, H., and Ogawa, M. (2001). Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron 31, 727–741. doi: 10.1016/s0896-6273(01)00420-2
Miyoshi, G., Hjerling-Leffler, J., Karayannis, T., Sousa, V. H., Butt, S. J., Battiste, J., et al. (2010). Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J. Neurosci. 30, 1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010
Mizuno, K., Gonzalez, F. J., and Kimura, S. (1991). Thyroid-specific enhancer-binding protein (T/EBP): cDNA cloning, functional characterization, and structural identity with thyroid transcription factor TTF-1. Mol. Cell. Biol. 11, 4927–4933. doi: 10.1128/mcb.11.10.4927-4933.1991
Molotkova, N., Molotkov, A., and Duester, G. (2007). Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev. Biol. 303, 601–610. doi: 10.1016/j.ydbio.2006.11.035
Moon, N. S., Bérubé, G., and Nepveu, A. (2000). CCAAT displacement activity involves CUT repeats 1 and 2, not the cut homeodomain*. J. Biol. Chem. 275, 31325–31334. doi: 10.1074/jbc.M002912200
Monnier, A., Boniface, R., Bouvet, R., Etcheverry, A., Aubry, M., Avril, T., et al. (2018). The expression of EMX2 lead to cell cycle arrest in glioblastoma cell line. BMC Cancer 18:1213. doi: 10.1186/s12885-018-5094-y
Monti, S., Nicoletti, A., Cantasano, A., Krude, H., and Cassio, A. (2015). NKX2. 1-Related Disorders: a novel mutation with mild clinical presentation. Italian J. Pediatr. 41, 1–5. doi: 10.1186/s13052-015-0150-6
Monuki, E. S., Porter, F. D., and Walsh, C. A. (2001). Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32, 591–604. doi: 10.1016/s0896-6273(01)00504-9
Mortensen, A. H., Schade, V., Lamonerie, T., and Camper, S. A. (2015). Deletion of OTX2 in neural ectoderm delays anterior pituitary development. Hum. Mol. Genet. 24, 939–953. doi: 10.1093/hmg/ddu506
Mühleisen, T.W., Leber, M., Schulze, T.G., Strohmaier, J., Degenhardt, F., Treutlein, J., et al. (2014). Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 5:3339. doi: 10.1038/ncomms4339
Murcia-Ramón, R., Company, V., Juárez-Leal, I., Andreu-Cervera, A., Almagro-García, F., Martínez, S., et al. (2020a). Neuronal tangential migration from Nkx2.1-positive hypothalamus. Brain Struct. Funct. 225, 2857–2869.
Murcia-Ramón, R., Juárez-Leal, I., Andreu-Cervera, A., Almagro-García, F., Martínez, S., Echevarría, D., et al. (2020b). Neuronal tangential migration from Nkx2. 1-positive hypothalamus. Brain Struct. Funct. 225, 2857–2869. doi: 10.1007/s00429-020-02163-x
Murre, C., McCaw, P. S., Vaessin, H., Caudy, M., Jan, L., Jan, Y., et al. (1989). Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58, 537–544. doi: 10.1016/0092-8674(89)90434-0
Muzio, L., Di Benedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., and Mallamaci, A. (2002a). Conversion of cerebral cortex into basal ganglia in Emx2−/− Pax6 Sey/Sey double-mutant mice. Nat. Neurosci. 5, 737–745. doi: 10.1038/nn892
Muzio, L., Di Benedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., and Mallamaci, A. (2002b). Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb. Cortex 12, 129–139. doi: 10.1093/cercor/12.2.12
Muzio, L., Soria, J., Pannese, M., Piccolo, S., and Mallamaci, A. (2005). A mutually stimulating loop involving emx2 and canonical wnt signalling specifically promotes expansion of occipital cortex and hippocampus. Cereb. Cortex 15, 2021–2028. doi: 10.1093/cercor/bhi077
Nadarajah, B., Brunstrom, J. E., Grutzendler, J., Wong, R. O., and Pearlman, A. L. (2001). Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 4, 143–150. doi: 10.1038/83967
Nagel, S., Ehrentraut, S., Meyer, C., Kaufmann, M., Drexler, H. G., and MacLeod, R. A. (2015). Aberrantly expressed OTX homeobox genes deregulate B-cell differentiation in Hodgkin lymphoma. PLoS One 10:e0138416. doi: 10.1371/journal.pone.0138416
Nakada, Y., Hunsaker, T. L., Henke, R. M., and Johnson, J. E. (2004). Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development 131, 1319–1330. doi: 10.1242/dev.01008
Nakagawa, Y., and Leary, D. D. M. (2001). Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J. Neurosci. 21:2711. doi: 10.1523/JNEUROSCI.21-08-02711.2001
Nakai, S., Kawano, H., Yudate, T., Nishi, M., Kuno, J., Nagata, A., et al. (1995). The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9, 3109–3121. doi: 10.1101/gad.9.24.3109
Nakamura, T., Jenkins, N. A., and Copeland, N. G. (1996). Identification of a new family of Pbx-related homeobox genes. Oncogene 13, 2235–2242.
Nasrallah, M. P., Cho, G., Simonet, J. C., Putt, M. E., Kitamura, K., and Golden, J. A. (2012). Differential effects of a polyalanine tract expansion in Arx on neural development and gene expression. Hum. Mol. Genet. 21, 1090–1098. doi: 10.1093/hmg/ddr538
NCBI Datasets (2021). NCBI Datasets Gene, Bethesda, MD: National Center for Biotechnology Information.
Nery, S., Fishell, G., and Corbin, J. G. (2002). The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat. Neurosci. 5, 1279–1287. doi: 10.1038/nn971
Neul, J. L., Kaufmann, W. E., Glaze, D. G., Christodoulou, J., Clarke, A. J., Bahi-Buisson, N., et al. (2010). Rett syndrome: revised diagnostic criteria and nomenclature. Ann. Neurol. 68, 944–950. doi: 10.1002/ana.22124
Nepveu, A. (2001). Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270, 1–15. doi: 10.1016/s0378-1119(01)00485-1
Neves, G., Shah, M. M., Liodis, P., Achimastou, A., Denaxa, M., Roalfe, G., et al. (2013). The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb. Cortex 23, 1811–1823. doi: 10.1093/cercor/bhs159
Neyt, C., Welch, M., Langston, A., Kohtz, J., Fishell, G., and Short-Range Signal, A. (1997). Restricts cell movement between telencephalic proliferative zones. J. Neurosci. 17:9194. doi: 10.1523/JNEUROSCI.17-23-09194.1997
Nieto, M., Monuki, E. S., Tang, H., Imitola, J., Haubst, N., Khoury, S. J., et al. (2004). Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J. Comp. Neurol. 479, 168–180. doi: 10.1002/cne.20322
Nieto, M., Schuurmans, C., Britz, O., and Guillemot, F. (2001). Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29, 401–413. doi: 10.1016/s0896-6273(01)00214-8
Nóbrega-Pereira, S., Kessaris, N., Du, T., Kimura, S., Anderson, S. A., and Marín, O. (2008). Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron 59, 733–745. doi: 10.1016/j.neuron.2008.07.024
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S., and Kriegstein, A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720. doi: 10.1038/35055553
Noctor, S. C., Martínez-Cerdeño, V., Ivic, L., and Kriegstein, A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144. doi: 10.1038/nn1172
Nomura, T., Haba, H., and Osumi, N. (2007). Role of a transcription factor Pax6 in the developing vertebrate olfactory system. Dev. Growth Differ. 49, 683–690. doi: 10.1111/j.1440-169X.2007.00965.x
Northcott, P. A., Korshunov, A., Witt, H., Hielscher, T., Eberhart, C. G., Mack, S., et al. (2011). Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29:1408. doi: 10.1200/jco.2009.27.4324
Noyes, M. B., Christensen, R. G., Wakabayashi, A., Stormo, G. D., Brodsky, M. H., and Wolfe, S. A. (2008). Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277–1289. doi: 10.1016/j.cell.2008.05.023
Olivetti, P. R., and Noebels, J. L. (2012). Interneuron, interrupted: molecular pathogenesis of ARX mutations and X-linked infantile spasms. Curr. Opin. Neurobiol. 22, 859–865. doi: 10.1016/j.conb.2012.04.006
Okamoto, J., Hirata, T., Chen, Z., Zhou, H., Mikami, I., Li, H., et al. (2010). EMX2 is epigenetically silenced and suppresses growth in human lung cancer. Oncogene 29, 5969–5975. doi: 10.1038/onc.2010.330
Omodei, D., Acampora, D., Russo, F., De Filippi, R., Severino, V., Di Francia, R., et al. (2009). Expression of the brain transcription factor OTX1 occurs in a subset of normal germinal-center B cells and in aggressive Non-Hodgkin Lymphoma. Am. J. Pathol. 175, 2609–2617. doi: 10.2353/ajpath.2009.090542
Ono, K., Takebayashi, H., Ikeda, K., Furusho, M., Nishizawa, T., Watanabe, K., et al. (2008). Regional-and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev. Biol. 320, 456–468. doi: 10.1016/j.ydbio.2008.06.001
Orduz, D., Benamer, N., Ortolani, D., Coppola, E., Vigier, L., Pierani, A., et al. (2019). Developmental cell death regulates lineage-related interneuron-oligodendroglia functional clusters and oligodendrocyte homeostasis. Nat. Commun. 10, 1–13. doi: 10.1038/s41467-019-11904-4
Pagliaroli, L., Vereczkei, A., Padmanabhuni, S. S., Tarnok, Z., Farkas, L., Nagy, P., et al. (2020). Association of genetic variation in the 3’UTR of LHX6, IMMP2L, and AADAC with tourette syndrome. Front. Neurol. 11:803. doi: 10.3389/fneur.2020.00803
Pan, Y.-H., Wu, N., and Yuan, X.-B. (2019). Toward a better understanding of neuronal migration deficits in autism spectrum disorders. Front. Cell Dev. Biol. 7:205. doi: 10.3389/fcell.2019.00205
Pantò, M. R., Zappalà, A., Tuorto, F., and Cicirata, F. (2004). Role of the Otx1 gene in cell differentiation of mammalian cortex. Eur. J. Neurosci. 19, 2893–2902. doi: 10.1111/j.0953-816X.2004.03326.x
Park, M., Jeon, S., Jeong, J.-H., Park, M., Lee, D.-R., Yoon, T. K., et al. (2012). Identification and characterization of LHX8 DNA binding elements. Dev. Reprod. 16, 379–384. doi: 10.12717/DR.2012.16.4.379
Park, N. I., Guilhamon, P., Desai, K., McAdam, R.F., Langille, E., O’Connor, M., et al. (2017). ASCL1 reorganizes chromatin to direct neuronal fate and suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell 21, 209.e7–224.e7.
Patel, S. R., Bhumbra, S. S., Paknikar, R. S., and Dressler, G. R. (2012). Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol. Cell 45, 185–195. doi: 10.1016/j.molcel.2011.11.007
Pratt, T., Vitalis, T., Warren, N., Edgar, J. M., Mason, J. O., and Price, D. J. (2000). A role for Pax6 in the normal development of dorsal thalamus and its cortical connections. Development 127, 5167–5178. doi: 10.1242/dev.127.23.5167
Pearl, J. R., Colantuoni, C., Bergey, D. E., Funk, C. C., Shannon, P., and Basu, B. (2019). Genome-scale transcriptional regulatory network models of psychiatric and neurodegenerative disorders. Cell Systems 8, 122.e7–135.e7. doi: 10.1016/j.cels.2019.01.002
Pei, Z., Wang, B., Chen, G., Nagao, M., Nakafuku, M., and Campbell, K. (2011). Homeobox genes Gsx1 and Gsx2 differentially regulate telencephalic progenitor maturation. Proc. Natl. Acad. Sci. 108:1675. doi: 10.1073/pnas.1008824108
Pellegrini, M., Mansouri, A., Simeone, A., Boncinelli, E., and Gruss, P. (1996). Dentate gyrus formation requires Emx2. Development 122, 3893–3898. doi: 10.1242/dev.122.12.3893
Peng, G., and Westerfield, M. (2006). Lhx5 promotes forebrain development and activates transcription of secreted Wnt antagonists. Development 133, 191–200. doi: 10.1242/dev.02485
Peters, T., Dildrop, R., Ausmeier, K., and Rüther, U. (2000). Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 10, 1453–1462. doi: 10.1101/gr.144100
Petryniak, M. A., Potter, G. B., Rowitch, D. H., and Rubenstein, J. L. R. (2007). Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55, 417–433. doi: 10.1016/j.neuron.2007.06.036
Piñon, M. C., Tuoc, T. C., Ashery-Padan, R., Molnár, Z., and Stoykova, A. (2008). Altered molecular regionalization and normal thalamocortical connections in cortex-specific Pax6 knock-out mice. J. Neurosci. 28, 8724–8734. doi: 10.1523/JNEUROSCI.2565-08.2008
Pla, R., Stanco, A., Howard, M. A., Rubin, A. N., Vogt, D., Mortimer, N., et al. (2017). Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb. Cortex 28, 3797–3815. doi: 10.1093/cercor/bhx241
Platzer, K., Cogné, B., Hague, J., Marcelis, C. L., Mitter, D., Oberndorff, K., et al. (2018). Haploinsufficiency of CUX1 causes nonsyndromic global developmental delay with possible catch-up development. Ann. Neurol. 84, 200–207. doi: 10.1002/ana.25278
Pleasure, S. J., Anderson, S., Hevner, R., Bagri, A., Marin, O., Lowenstein, D. H., et al. (2000). Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron 28, 727–740. doi: 10.1016/s0896-6273(00)00149-5
Poiana, G., Gioia, R., Sineri, S., Cardarelli, S., Lupo, G., and Cacci, E. (2020). Transcriptional regulation of adult neural stem/progenitor cells: tales from the subventricular zone. Neural Regen. Res. 15:1773. doi: 10.4103/1673-5374.280301
Poitras, L., Ghanem, N., Hatch, G., and Ekker, M. (2007). The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development 134, 1755–1765. doi: 10.1242/dev.02845
Porter, F. D., Drago, J., Xu, Y., Cheema, S. S., Wassif, C., Huang, S.-P., et al. (1997). Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124, 2935–2944. doi: 10.1242/dev.124.15.2935
Potkin, S. G., Turner, J. A., Guffanti, G., Lakatos, A., Fallon, J. H., Nguyen, D. D., et al. (2009). A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull. 35, 96–108. doi: 10.1093/schbul/sbn155
Prez, C., Dastot-Le Moal, F., Collot, N., Legendre, M., Abadie, I., Bertrand, A.-M., et al. (2012). Screening of LHX2 in patients presenting growth retardation with posterior pituitary and ocular abnormalities. Eur. J. Endocrinol. 167:85. doi: 10.1530/EJE-12-0026
Price, M., Lazzaro, D., Pohl, T., Mattei, M., Rüther, U., Olivo, J.-C., et al. (1992). Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron 8, 241–255. doi: 10.1016/0896-6273(92)90291-k
Price, M. G., Yoo, J. W., Burgess, D. L., Deng, F., Hrachovy, R. A., Frost, J. D., et al. (2009). A triplet repeat expansion genetic mouse model of infantile spasms syndrome, Arx (GCG) 10+ 7, with interneuronopathy, spasms in infancy, persistent seizures, and adult cognitive and behavioral impairment. J. Neurosci. 29, 8752–8756. doi: 10.1523/JNEUROSCI.0915-09.2009
Prochiantz, A., and Joliot, A. (2003). Can transcription factors function as cell–cell signalling molecules? Nat. Rev. Mol. Cell Biol. 4, 814–819. doi: 10.1038/nrm1227
Powell, E. M., Mars, W. M., and Levitt, P. (2001). Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron 30, 79–89. doi: 10.1016/s0896-6273(01)00264-1
Puelles, E., Acampora, D., Gogoi, R., Tuorto, F., Papalia, A., Guillemot, F., et al. (2006). Otx2 controls identity and fate of glutamatergic progenitors of the thalamus by repressing GABAergic differentiation. J. Neurosci. 26, 5955–5964. doi: 10.1523/JNEUROSCI.1097-06.2006
Puelles, L., Kuwana, E., Puelles, E., and Rubenstein, J. (1999). Comparison of the mammalian and avian telencephalon from the perspective of gene expression data. Eur. J. Morphol. 37, 139–150. doi: 10.1076/ejom.37.2.139.4756
Puelles, L. (2013). “Plan of the developing vertebrate nervous system,” in Pattering and Cell Type Specification in the Developing CNS and PNS, eds J. Rubenstein and P. Rakic (Amsterdam: Elsevier), 187–209. doi: 10.1016/b978-0-12-397265-1.00118-0
Puelles, L. (2018). Developmental studies of avian brain organization. Int. J. Dev. Biol. 62, 207–224. doi: 10.1387/ijdb.170279LP
Puelles, L., and Rubenstein, J. L. (2003). Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 26, 469–476. doi: 10.1016/S0166-2236(03)00234-0
Qiu, H., Yan, Q., Luo, X., Zhang, H., Bao, W., and Wan, X. (2013). EMX2 is downregulated in endometrial cancer and correlated with tumor progression. Int. J. Gynecol. Pathol. 32, 193–198. doi: 10.1097/PGP.0b013e31825d8049
Qiu, M., Anderson, S., Chen, S., Meneses, J. J., Hevner, R., Kuwana, E., et al. (1996). Mutation of theEmx-1Homeobox gene disrupts the corpus callosum. Dev. Biol. 178, 174–178. doi: 10.1006/dbio.1996.0207
Qiu, M., Bulfone, A., Ghattas, I., Meneses, J. J., Christensen, L., Sharpe, P. T., et al. (1997). Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and −2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 185, 165–184. doi: 10.1006/dbio.1997.8556
Quaggin, S. E., Heuvel, G. B. V., Golden, K., Bodmer, R., and Igarashi, P. (1996). Primary structure, neural-specific expression, and chromosomal localization of cux-2, a second murine homeobox gene related to Drosophila cut*. J. Biol. Chem. 271, 22624–22634. doi: 10.1074/jbc.271.37.22624
Quille, M.-L., Carat, S., Quéméner-Redon, S., Hirchaud, E., Baron, D., Benech, C., et al. (2011). High-throughput analysis of promoter occupancy reveals new targets for Arx, a gene mutated in mental retardation and interneuronopathies. PLoS One 6:e2518. doi: 10.1371/journal.pone.0025181
Quinn, J.C., Molinek, M., Martynoga, B.S., Zaki, P.A., Faedo, A., Bulfone, A., Hevner, R.F., West, J.D., Price, D.J., et al. (2007). Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Developmental biology 302, 50–65.
Quintana-Urzainqui, I., Kozić, Z., Mitra, S., Tian, T., Manuel, M., Mason, J. O., et al. (2018). Tissue-specific actions of Pax6 on proliferation and differentiation balance in developing forebrain are Foxg1 dependent. Iscience 10, 171–191. doi: 10.1016/j.isci.2018.11.031
Ragge, N. K., Brown, A. G., Poloschek, C. M., Lorenz, B., Henderson, R. A., Clarke, M. P., et al. (2005). Heterozygous mutations of OTX2 cause severe ocular malformations. Am. J. Hum. Genet. 76, 1008–1022. doi: 10.1086/430721
Rakić, S., Kanatani, S., Hunt, D., Faux, C., Cariboni, A., Chiara, F., et al. (2015). Cdk5 phosphorylation of ErbB4 is required for tangential migration of cortical interneurons. Cereb. Cortex 25, 991–1003. doi: 10.1093/cercor/bht290
Rakic, P. (1972). Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 145, 61–83. doi: 10.1002/cne.901450105
Ramdzan, Z. M., and Nepveu, A. (2014). CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat. Rev. Cancer 14, 673–682. doi: 10.1038/nrc3805
Reish, O., Fullston, T., Regev, M., Heyman, E., and Gecz, J. (2009). A novel de novo 27 bp duplication of the ARX gene, resulting from postzygotic mosaicism and leading to three severely affected males in two generations. Am. J. Med. Genet. Part A 149, 1655–1660. doi: 10.1002/ajmg.a.32842
Rieckhof, G. E., Casares, F., Ryoo, H. D., Abu-Shaar, M., and Mann, R. S. (1997). Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91, 171–183. doi: 10.1016/s0092-8674(00)80400-6
Rhinn, M., Dierich, A., Shawlot, W., Behringer, R. R., Le Meur, M., and Ang, S.-L. (1998). Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development 125, 845–856. doi: 10.1242/dev.125.5.845
Robertshaw, E., Matsumoto, K., Lumsden, A., and Kiecker, C. (2013). Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc. Natl. Acad. Sci. 110, E3919–E3926. doi: 10.1073/pnas.1304311110
Robledo, R. F., Rajan, L., Li, X., and Lufkin, T. (2002). The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 16, 1089–1101. doi: 10.1101/gad.988402
Rodríguez-Seguel, E., Alarcón, P., and ómez-Skarmeta, J. L. G. (2009). The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev. Biol. 329, 258–268. doi: 10.1016/j.ydbio.2009.02.028
Rohs, R., West, S. M., Sosinsky, A., Liu, P., Mann, R. S., and Honig, B. (2009). The role of DNA shape in protein–DNA recognition. Nature 461, 1248–1253. doi: 10.1038/nature08473
Roy, A., Gonzalez-Gomez, M., Pierani, A., Meyer, G., and Tole, S. (2014). Lhx2 regulates the development of the forebrain hem system. Cereb. Cortex 24, 1361–1372. doi: 10.1093/cercor/bhs421
Roychoudhury, K., Salomone, J., Qin, S., Cain, B., Adam, M., Potter, S. S., et al. (2020). Physical interactions between Gsx2 and Ascl1 balance progenitor expansion versus neurogenesis in the mouse lateral ganglionic eminence. Development 147:dev185348. doi: 10.1242/dev.185348
Rubenstein, J. L., Martinez, S., Shimamura, K., and Puelles, L. (1994). The embryonic vertebrate forebrain: the prosomeric model. Science 266, 578–580. doi: 10.1126/science.7939711
Rudin, C. M., Hann, C. L., Laterra, J., Yauch, R. L., Callahan, C. A., Fu, L., et al. (2009). Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. New Engl. J. Med. 361, 1173–1178. doi: 10.1056/NEJMoa0902903
Rudy, B., Fishell, G., Lee, S., and Hjerling-Leffler, J. (2011). Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 71, 45–61. doi: 10.1002/dneu.20853
Ruest, L.-B., Hammer, R. E., Yanagisawa, M., and Clouthier, D. E. (2003). Dlx5/6-enhancer directed expression of Cre recombinase in the pharyngeal arches and brain. Genesis 37, 188–194. doi: 10.1002/gene.10247
Ryan, A. K., Rosenfeld, M. G., and domain, P. O. U. (1997). family values: flexibility, partnerships, and developmental codes. Genes Dev. 11, 1207–1225. doi: 10.1101/gad.11.10.1207
Sagner, A., and Briscoe, J. (2019). Establishing neuronal diversity in the spinal cord: a time and a place. Development 146:dev182154. doi: 10.1242/dev.182154
Saha, B., Hari, P., Huilgol, D., and Tole, S. (2007). Dual role for LIM-homeodomain gene Lhx2 in the formation of the lateral olfactory tract. J. Neurosci. 27, 2290–2297. doi: 10.1523/JNEUROSCI.5571-06.2007
Salomone, J., Qin, S., Fufa, T. D., Cain, B., Farrow, E., Guan, B., et al. (2021). Conserved Gsx2/Ind homeodomain monomer versus homodimer DNA binding defines regulatory outcomes in flies and mice. Genes Dev. 35, 157–174. doi: 10.1101/gad.343053.120
Sandberg, M., Flandin, P., Silberberg, S., Su-Feher, L., Price, J. D., Hu, J. S., et al. (2016). Transcriptional networks controlled by NKX2-1 in the development of forebrain GABAergic neurons. Neuron 91, 1260–1275. doi: 10.1016/j.neuron.2016.08.020
Sansregret, L., Gallo, D., Santaguida, M., Leduy, L., Harada, R., and Nepveu, A. (2010). Hyperphosphorylation by cyclin B/CDK1 in mitosis resets CUX1 DNA binding clock at each cell cycle. J. Biol. Chem. 285, 32834–32843. doi: 10.1074/jbc.M110.156406
Sansom, S. N., Griffiths, D. S., Faedo, A., Kleinjan, D.-J., Ruan, Y., Smith, J., et al. (2009). The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 5:e1000511. doi: 10.1371/journal.pgen.1000511
Santaguida, M., Ding, Q., Bérubé, G., Truscott, M., Whyte, P., and Nepveu, A. (2001). Phosphorylation of the ccaat displacement protein (cdp)/cux transcription factor by cyclin A-Cdk1 modulates its dna binding activity in G2*. J. Biol. Chem. 276, 45780–45789. doi: 10.1074/jbc.M107978200
Sakurai, Y., Kurokawa, D., Kiyonari, H., Kajikawa, E., Suda, Y., and Aizawa, S. (2010). Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Dev. Biol. 347, 392–403. doi: 10.1016/j.ydbio.2010.08.028
Saulnier, A., Keruzore, M., De Clercq, S., Bar, I., Moers, V., Magnani, D., et al. (2013). The doublesex homolog Dmrt5 is required for the development of the caudomedial cerebral cortex in mammals. Cereb. Cortex 23, 2552–2567. doi: 10.1093/cercor/bhs234
Scardigli, R., Bäumer, N., Gruss, P., Guillemot, F., and Le Roux, I. (2003). Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130, 3269–3281. doi: 10.1242/dev.00539
Scherer, S. W., Neufeld, E. J., Lievens, P. M. J., Orkin, S. H., Kim, J., and Tsui, L. C. (1993). Regional localization of the CCAAT displacement protein gene (CUTL1) to 7q22 by analysis of somatic cell hybrids. Genomics 15, 695–706. doi: 10.1006/geno.1993.1130
Schonemann, M. D., Ryan, A. K., McEvilly, R. J., O’Connell, S. M., Arias, C. A., Kalla, K. A., et al. (1995). Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9, 3122–3135. doi: 10.1101/gad.9.24.3122
Schmahl, W., Knoedlseder, M., Favor, J., and Davidson, D. (1993). Defects of neuronal migration and the pathogenesis of cortical malformations are associated with Small eye (Sey) in the mouse, a point mutation at the Pax-6-locus. Acta Neuropathologica 86, 126–135. doi: 10.1007/BF00334879
Schmidt-Sidor, B., Szymanska, K., Williamson, K., van Heyningen, V., Roszkowski, T., Wierzba-Bobrowicz, T., et al. (2009). Malformations of the brain in two fetuses with a compound heterozygosity for two PAX6 mutations. Folia Neuropathol. 47, 372–382.
Schreiber, E., Harshman, K., Kemler, I., Malipiero, U., Schaffner, W., and Fontana, A. (1990). Astrocytes and glioblastoma cells express novel octamer-DNA binding proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Res. 18, 5495–5503. doi: 10.1093/nar/18.18.5495
Schreiber, E., Himmelmann, A., Malipiero, U., Tobler, A., Stahel, R., and Fontana, A. (1992). Human small cell lung cancer expresses the octamer DNA-binding and nervous system-specific transcription factor N-Oct 3 (brain-2). Cancer Res. 52, 6121–6124.
Schulte, D. (2014). Meis: new friends of Pax. Neurogenesis 1:e976014. doi: 10.4161/23262133.2014.976014
Schuurmans, C., Armant, O., Nieto, M., Stenman, J. M., Britz, O., Klenin, N., et al. (2004). Sequential phases of cortical specification involve Neurogenin-dependent and-independent pathways. EMBO J. 23, 2892–2902. doi: 10.1038/sj.emboj.7600278
Sellers, K., Zyka, V., Lumsden, A. G., and Delogu, A. (2014). Transcriptional control of GABAergic neuronal subtype identity in the thalamus. Neural Dev. 9:14. doi: 10.1186/1749-8104-9-14
Sequerra, E. B. (2014). Subventricular zone progenitors in time and space: generating neuronal diversity. Front. Cell. Neurosci. 8:434. doi: 10.3389/fncel.2014.00434
Sessa, A., Mao, C.-A., Colasante, G., Nini, A., Klein, W. H., and Broccoli, V. (2010). Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev. 24, 1816–1826. doi: 10.1101/gad.575410
Sheng, H. Z., Bertuzzi, S., Chiang, C., Shawlot, W., Taira, M., Dawid, I., et al. (1997). Expression of murine Lhx5 suggests a role in specifying the forebrain. Dev. Dyn. 208, 266–277. doi: 10.1002/(SICI)1097-0177(199702)208:2<266::AID-AJA13>3.0.CO;2-1
Shi, X., Lin, W., Gao, X., Xie, W., Golden, J. A., and Tao, T. (2020). Identification and validation of the phosphorylation sites on Aristaless-related homeobox protein. Biosci. Rep. 40:BSR20194513. doi: 10.1042/BSR20194513
Shi, Y., and Massagué, J. (2003). Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700. doi: 10.1016/s0092-8674(03)00432-x
Shibata, M., Pattabiraman, K., Lorente-Galdos, B., Andrijevic, D., Kim, S.-K., Kaur, N., et al. (2021). Regulation of prefrontal patterning and connectivity by retinoic acid. Nature 598, 483–488. doi: 10.1038/s41586-021-03953-x
Shimizu, T., and Hibi, M. (2009). Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev. Growth Differ. 51, 221–231. doi: 10.1111/j.1440-169X.2009.01088.x
Shinozaki, K., Miyagi, T., Yoshida, M., Miyata, T., Ogawa, M., Aizawa, S., et al. (2002). Absence of Cajal-Retzius cells and subplate neurons associated with defects of tangential cell migration from ganglionic eminence in Emx1/2 double mutant cerebral cortex. Development 129, 3479–3492. doi: 10.1242/dev.129.14.3479
Shinozaki, K., Yoshida, M., Nakamura, M., Aizawa, S., and Suda, Y. (2004). Emx1 and Emx2 cooperate in initial phase of archipallium development. Mech. Dev. 121, 475–489. doi: 10.1016/j.mod.2004.03.013
Siehr, M. S., Massey, C. A., and Noebels, J. L. (2020). Arx expansion mutation perturbs cortical development by augmenting apoptosis without activating innate immunity in a mouse model of X-linked infantile spasms syndrome. Dis. Models Mech. 13:dmm04251. doi: 10.1242/dmm.042515
Silbereis, J. C., Nobuta, H., Tsai, H.-H., Heine, V. M., McKinsey, G. L., Meijer, D. H., et al. (2014). Olig1 function is required to repress dlx1/2 and interneuron production in Mammalian brain. Neuron 81, 574–587. doi: 10.1016/j.neuron.2013.11.024
Simeone, A., Acampora, D., Gulisano, M., Stornaiuolo, A., and Boncinelli, E. (1992). Nested expression domains of four homeobox genes in developing rostral brain. Nature 358, 687–690. doi: 10.1038/358687a0
Simeone, A., Acampora, D., Mallamaci, A., Stornaiuolo, A., D’Apice, M. R., Nigro, V., et al. (1993). A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 12, 2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x
Simmons, J. L., Pierce, C. J., Al-Ejeh, F., and Boyle, G. M. (2017). MITF and BRN2 contribute to metastatic growth after dissemination of melanoma. Sci. Rep. 7:10909. doi: 10.1038/s41598-017-11366-y
Sisodiya, S. M., Free, S. L., Williamson, K. A., Mitchell, T. N., Willis, C., Stevens, J. M., et al. (2001). PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat. Genet. 28, 214–216. doi: 10.1038/90042
Smemo, S., Tena, J. J., Kim, K.-H., Gamazon, E. R., Sakabe, N. J., Gómez-Marín, C. G., et al. (2014). Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375. doi: 10.1038/nature13138
Son, J. E., Dou, Z., Kim, K.-H., Wanggou, S., Cha, V. S. B., Mo, R., et al. (2021a). Irx3 and Irx5 in Ins2-Cre+ cells regulate hypothalamic postnatal neurogenesis and leptin response. Nat. Metab. 3, 701–713. doi: 10.1038/s42255-021-00382-y
Son, J. E., Dou, Z., Wanggou, S., Chan, J., Mo, R., Li, X., et al. (2021b). Ectopic expression of Irx3 and Irx5 in the paraventricular nucleus of the hypothalamus contributes to defects in Sim1 haploinsufficiency. Sci. Adv. 7:eabh4503. doi: 10.1126/sciadv.abh4503
Solomon, B. D., Pineda-Alvarez, D. E., Balog, J. Z., Hadley, D., Gropman, A. L., Nandagopal, R., et al. (2009). Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am. J. Med. Genet. Part A 149, 2543–2546. doi: 10.1002/ajmg.a.33081
Soriano, E., and Del Río, J. A. (2005). The cells of cajal-retzius: still a mystery one century after. Neuron 46, 389–394. doi: 10.1016/j.neuron.2005.04.019
Spigoni, G., Gedressi, C., and Mallamaci, A. (2010). Regulation of Emx2 expression by antisense transcripts in murine cortico-cerebral precursors. PLoS One 5:e8658. doi: 10.1371/journal.pone.0008658
Srivastava, D., Thomas, T., Lin, Q., Kirby, M. L., Brown, D., and Olson, E. N. (1997). Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 16, 154–160. doi: 10.1038/ng0697-154
Stenman, J., Toresson, H., and Campbell, K. (2003). Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J. Neurosci. 23, 167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003
Stiles, J. (2008). The Fundamentals of Brain Development: Integrating Nature and Nurture. Cambridge, MA: Harvard University Press.
Stock, D. W., Ellies, D. L., Zhao, Z., Ekker, M., Ruddle, F. H., and Weiss, K. M. (1996). The evolution of the vertebrate Dlx gene family. Proc. Natl. Acade. Sci. 93, 10858–10863. doi: 10.1073/pnas.93.20.10858
Stocker, A. M., and O’Leary, D. D. (2016). Emx1 is required for neocortical area patterning. PLoS One 11:e0149900. doi: 10.1371/journal.pone.0149900
Stoykova, A., and Gruss, P. (1994). Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J. Neurosci. 14, 1395–1412. doi: 10.1523/JNEUROSCI.14-03-01395.1994
Stoykova, A., Fritsch, R., Walther, C., and Gruss, P. (1996). Forebrain patterning defects in small eye mutant mice. Development 122, 3453–3465. doi: 10.1242/dev.122.11.3453
Stoykova, A., Gotz, M., Gruss, P., and Price, J. (1997). Pax6-dependent regulation of adhesive patterning, R-cadherin expression and boundary formation in developing forebrain. Development 124, 3765–3777. doi: 10.1242/dev.124.19.3765
Stoykova, A., Treichel, D., Hallonet, M., and Gruss, P. (2000). Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 20, 8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000
Striano, P., Paravidino, R., Sicca, F., Chiurazzi, P., Gimelli, S., Coppola, A., et al. (2011). West syndrome associated with 14q12 duplications harboring FOXG1. Neurology 76, 1600–1602. doi: 10.1212/WNL.0b013e3182194bbf
Strømme, P., Mangelsdorf, M. E., Scheffer, I. E., and Gécz, J. (2002). Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in Aristaless related homeobox gene, ARX. Brain Dev. 24, 266–288. doi: 10.1016/s0387-7604(02)00079-7
Stühmer, T., Anderson, S. A., Ekker, M., and Rubenstein, J. L. R. (2002). Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development 129, 245–252. doi: 10.1242/dev.129.1.245
Stumm, R. K., Zhou, C., Ara, T., Lazarini, F., Dubois-Dalcq, M., Nagasawa, T., et al. (2003). CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 23, 5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003
Su, Z., Wang, Z., Lindtner, S., Yang, L., Shang, Z., Tian, Y., et al. (2022). Dlx1/2-dependent expression of Meis2 promotes neuronal fate determination in the mammalian striatum. Development 149:dev200035. doi: 10.1242/dev.200035
Suda, Y., Kokura, K., Kimura, J., Kajikawa, E., Inoue, F., and Aizawa, S. (2010). The same enhancer regulates the earliest Emx2 expression in caudal forebrain primordium, subsequent expression in dorsal telencephalon and later expression in the cortical ventricular zone. Development 137, 2939–2949. doi: 10.1242/dev.048843
Sugimori, M., Nagao, M., Bertrand, N., Parras, C.M., Guillemot, F.o., and Nakafuku, M. (2007). Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development 134, 1617–1629. doi: 10.1242/dev.001255
Sugitani, Y., Nakai, S., Minowa, O., Nishi, M., Jishage, K.-i., Kawano, H., et al. (2002). Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16, 1760–1765. doi: 10.1101/gad.978002
Sumiyama, K., Washio-Watanabe, K., Saitou, N., Hayakawa, T., and Ueda, S. (1996). Class III POU genes: generation of homopolymeric amino acid repeats under GC pressure in mammals. J. Mol. Evol. 43, 170–178. doi: 10.1007/BF02338824
Sun, J., Rockowitz, S., Xie, Q., Ashery-Padan, R., Zheng, D., and Cvekl, A. (2015). Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 43, 6827–6846. doi: 10.1093/nar/gkv589
Sun, T., Pringle, N. P., Hardy, A. P., Richardson, W. D., and Smith, H. K. (1998). Pax6 influences the time and site of origin of glial precursors in the ventral neural tube. Mol. Cell. Neurosci. 12, 228–239. doi: 10.1006/mcne.1998.0711
Sussel, L., Marin, O., Kimura, S., and Rubenstein, J. (1999). Loss of Nkx2. 1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370. doi: 10.1242/dev.126.15.3359
Symmank, J., Gölling, V., Gerstmann, K., and Zimmer, G. (2019). The transcription factor LHX1 regulates the survival and directed migration of POA-derived cortical interneurons. Cereb. Cortex 29, 1644–1658. doi: 10.1093/cercor/bhy063
Szczaluba, K., Nawara, M., Poirier, K., Pilch, J., Gajdulewicz, M., Spodar, K., et al. (2006). Genotype-phenotype associations for ARX gene duplication in X-linked mental retardation. Neurology 67, 2073–2075. doi: 10.1212/01.wnl.0000247833.29314.5b
Szucsik, J. C., Witte, D. P., Li, H., Pixley, S. K., Small, K. M., and Potter, S. S. (1997). Altered forebrain and hindbrain development in mice mutant for the Gsh-2 homeobox gene. Dev. Biol. 191, 230–242. doi: 10.1006/dbio.1997.8733
Tajima, T., Ohtake, A., Hoshino, M., Amemiya, S., Sasaki, N., Ishizu, K., et al. (2009). OTX2 loss of function mutation causes anophthalmia and combined pituitary hormone deficiency with a small anterior and ectopic posterior pituitary. J. Clin. Endocrinol. Metab. 94, 314–319. doi: 10.1210/jc.2008-1219
Takebayashi, H., Nabeshima, Y., Yoshida, S., Chisaka, O., Ikenaka, K., and Nabeshima, Y.-i. (2002). The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 12, 1157–1163. doi: 10.1016/s0960-9822(02)00926-0
Takebayashi, H., Yoshida, S., Sugimori, M., Kosako, H., Kominami, R., Nakafuku, M., et al. (2000). Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech. Dev. 99, 143–148. doi: 10.1016/s0925-4773(00)00466-4
Takuma, N., Sheng, H. Z., Furuta, Y., Ward, J. M., Sharma, K., Hogan, B., et al. (1998). Formation of Rathke’s pouch requires dual induction from the diencephalon. Development 125, 4835–4840. doi: 10.1242/dev.125.23.4835
Tao, W., and Lai, E. (1992). Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron 8, 957–966. doi: 10.1016/0896-6273(92)90210-5
Taylor, M. D., Northcott, P. A., Korshunov, A., Remke, M., Cho, Y.-J., Clifford, S. C., et al. (2012). Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathologica 123, 465–472. doi: 10.1007/s00401-011-0922-z
Tekki-Kessaris, N., Woodruff, R., Hall, A. C., Gaffield, W., Kimura, S., Stiles, C. D., et al. (2001). Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development 128, 2545–2554. doi: 10.1242/dev.128.13.2545
Terrinoni, A., Pagani, I., Zucchi, I., Chiaravalli, A., Serra, V., Rovera, F., et al. (2011). OTX1 expression in breast cancer is regulated by p53. Oncogene 30, 3096–3103. doi: 10.1038/onc.2011.31
Thakurela, S., Tiwari, N., Schick, S., Garding, A., Ivanek, R., Berninger, B., et al. (2016). Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discovery 2, 1–22. doi: 10.1038/celldisc.2015.45
Theil, T., Alvarez-Bolado, G., Walter, A., and Ruther, U. (1999). Gli3 is required for Emx gene expression during dorsal telencephalon development. Development 126, 3561–3571. doi: 10.1242/dev.126.16.3561
Theil, T., Aydin, S.l., Koch, S., Grotewold, L., and Rüther, U. (2002). Wnt and Bmp signalling cooperatively regulate graded Emx 2 expression in the dorsal telencephalon. Development 129, 3045–3054. doi: 10.1242/dev.129.13.3045
Thomson, J. A., Murphy, K., Baker, E., Sutherland, G. R., Parsons, P. G., Sturm, R. A., et al. (1995). The brn-2 gene regulates the melanocytic phenotype and tumorigenic potential of human melanoma cells. Oncogene 11, 691–700.
Thorwarth, A., Schnittert-Hübener, S., Schrumpf, P., Müller, I., Jyrch, S., Dame, C., et al. (2014). Comprehensive genotyping and clinical characterisation reveal 27 novel NKX2-1 mutations and expand the phenotypic spectrum. J. Med. Genet. 51, 375–387. doi: 10.1136/jmedgenet-2013-102248
Tian, E., Kimura, C., Takeda, N., Aizawa, S., and Matsuo, I. (2002). Otx2 is required to respond to signals from anterior neural ridge for forebrain specification. Dev. Biol. 242, 204–223. doi: 10.1006/dbio.2001.0531
Toresson, H., Mata de Urquiza, A., Fagerstrom, C., Perlmann, T., and Campbell, K. (1999). Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. Development 126, 1317–1326. doi: 10.1242/dev.126.6.1317
Toresson, H., Parmar, M., and Campbell, K. (2000). Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech. Dev. 94, 183–187. doi: 10.1016/s0925-4773(00)00324-5
Toresson, H. K., and Campbell, K. (2001). A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development 128, 4769–4780. doi: 10.1242/dev.128.23.4769
Truscott, M., Denault, J. B., Goulet, B., Leduy, L., Salvesen, G. S., and Nepveu, A. (2007). Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J. Biol. Chem. 282, 30216–30226. doi: 10.1074/jbc.M702328200
Truscott, M., Raynal, L., Wang, Y., Bérubé, G., Leduy, L., and Nepveu, A. (2004). The n-terminal region of the CCAAT displacement protein (CDP)/cux transcription factor functions as an autoinhibitory domain that modulates DNA binding*. J. Biol. Chem. 279, 49787–40794. doi: 10.1074/jbc.M409484200
Tu, X.-P., Li, H., Chen, L.-S., Luo, X.-N., Lu, Z.-M., Zhang, S.-Y., et al. (2020). OTX1 exerts an oncogenic role and is negatively regulated by miR129-5p in laryngeal squamous cell carcinoma. BMC Cancer 20:794. doi: 10.1186/s12885-020-07279-1
Tucker, E. S., Polleux, F., and LaMantia, A. S. (2006). Position and time specify the migration of a pioneering population of olfactory bulb interneurons. Dev. Biol. 297, 387–401. doi: 10.1016/j.ydbio.2006.05.009
Tufo, C., Poopalasundaram, S., Dorrego-Rivas, A., Ford, M. C., Graham, A., and Grubb, M. S. (2022). Development of the mammalian main olfactory bulb. Development 149:dev200210. doi: 10.1242/dev.200210
Turner, G., Partington, M., Kerr, B., Mangelsdorf, M., and Gecz, J. (2002). Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am. J. Med. Genet. 112, 405–411. doi: 10.1002/ajmg.10714
Tyas, D. A., Pearson, H., Rashbass, P., and Price, D. J. (2003). Pax6 regulates cell adhesion during cortical development. Cereb. Cortex 13, 612–619. doi: 10.1093/cercor/13.6.612
Valerius, M. T., Li, H., Stock, J. L., Weinstein, M., Kaur, S., Singh, G., et al. (1995). Gsh-1: a novel murine homeobox gene expressed in the central nervous system. Dev. Dyn. 203, 337–351. doi: 10.1002/aja.1002030306
Verret, L., Mann, E. O., Hang, G. B., Barth, A. M. I., Cobos, I., Ho, K., et al. (2012). Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in alzheimer model. Cell 149, 708–721. doi: 10.1016/j.cell.2012.02.046
Volk, D. W., Edelson, J. R., and Lewis, D. A. (2014). Cortical inhibitory neuron disturbances in schizophrenia: role of the ontogenetic transcription factor Lhx6. Schizophr. Bull. 40, 1053–1061. doi: 10.1093/schbul/sbu068
Volk, D. W., Matsubara, T., Li, S., Sengupta, E. J., Georgiev, D., Minabe, Y., et al. (2012). Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am. J. Psychiatry 169, 1082–1091. doi: 10.1176/appi.ajp.2012.12030305
von Engelhardt, J., Khrulev, S., Eliava, M., Wahlster, S., and Monyer, H. (2011). 5-HT(3A) receptor-bearing white matter interstitial GABAergic interneurons are functionally integrated into cortical and subcortical networks. J. Neurosci. 31:16844. doi: 10.1523/JNEUROSCI.0310-11.2011
Vue, T. Y., Aaker, J., Taniguchi, A., Kazemzadeh, C., Skidmore, J. M., Martin, D. M., et al. (2007). Characterization of progenitor domains in the developing mouse thalamus. J. Comp. Neurol. 505, 73–91. doi: 10.1002/cne.21467
Waclaw, R. R., Ehrman, L. A., Pierani, A., and Campbell, K. (2010). Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J. Neurosci. 30:6944. doi: 10.1523/JNEUROSCI.5772-09.2010
Walcher, T., Xie, Q., Sun, J., Irmler, M., Beckers, J., Öztürk, T., et al. (2013). Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development 140, 1123–1136. doi: 10.1242/dev.082875
Walther, C., Guenet, J.-L., Simon, D., Deutsch, U., Jostes, B., Goulding, M. D., et al. (1991). Pax: a murine multigene family of paired box-containing genes. Genomics 11, 424–434. doi: 10.1016/0888-7543(91)90151-4
Wang, B., Long, J. E., Flandin, P., Pla, R., Waclaw, R. R., Campbell, K., et al. (2013). Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J. Comp. Neurol. 521, 1561–1584. doi: 10.1002/cne.23242
Wang, L., Bluske, K. K., Dickel, L. K., and Nakagawa, Y. (2011a). Basal progenitor cells in the embryonic mouse thalamus - their molecular characterization and the role of neurogenins and Pax6. Neural Dev. 6, 35–35. doi: 10.1186/1749-8104-6-35
Wang, Y., Dye, C. A., Sohal, V., Long, J. E., Estrada, R. C., Roztocil, T., et al. (2010). Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J. Neurosci. 30, 5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010
Wang, Y., Li, G., Stanco, A., Long, J. E., Crawford, D., Potter, G. B., et al. (2011b). CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron 69, 61–76. doi: 10.1016/j.neuron.2010.12.005
Wanaka, A., Matsumoto, K., Kashihara, Y., Furuyama, T., Tanaka, T., Mori, T., et al. (1997). LIM-homeodomain gene family in neural development. Dev. Neurosci. 19, 97–100. doi: 10.1159/000111191
Way, J. C., and Chalfie, M. (1988). mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54, 5–16. doi: 10.1016/0092-8674(88)90174-2
Webb, A. E., Pollina, E. A., Vierbuchen, T., Urbán, N., Ucar, D., Leeman, D. S., et al. (2013). FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 4, 477–491. doi: 10.1016/j.celrep.2013.06.035
Weinschutz Mendes, H., Taktek, M., Duret, T., and Ekker, M. (2020). Expression of dlx genes in the normal and regenerating brain of adult zebrafish. PLoS One 15:e0229549.
Weiss, L. A., and Nieto, M. (2019). The crux of Cux genes in neuronal function and plasticity. Brain Res. 1705, 32–42. doi: 10.1016/j.brainres.2018.02.044
Weiss, J. B., Von Ohlen, T., Mellerick, D. M., Dressler, G., Doe, C. Q., and Scott, M. P. (1998). Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 12, 3591–3602. doi: 10.1101/gad.12.22.3591
Westphal, D.S., Riedhammer, K.M., Kovacs-Nagy, R., Meitinger, T., Hoefele, J., and Wagner, M. (2018). A de novo missense variant in pou3f2 identified in a child with global developmental delay. Neuropediatrics 49, 401–404. doi: 10.1055/s-0038-1669926
Whitman, M. C., and Greer, C. A. (2009). Adult neurogenesis and the olfactory system. Prog. Neurobiol. 89, 162–175. doi: 10.1016/j.pneurobio.2009.07.003
Wichterle, H., Turnbull, D. H., Nery, S., Fishell, G., and Alvarez-Buylla, A. (2001). In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development 128, 3759–3771. doi: 10.1242/dev.128.19.3759
Winterbottom, E. F., Illes, J. C., Faas, L., and Isaacs, H. V. (2010). Conserved and novel roles for the Gsh2 transcription factor in primary neurogenesis. Development 137, 2623–2631. doi: 10.1242/dev.047159
Won, H., de La Torre-Ubieta, L., Stein, J. L., Parikshak, N. N., Huang, J., Opland, C. K., et al. (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 doi: 10.1038/nature19847
Wortham, M., Guo, C., Zhang, M., Song, L., Lee, B.-K., Iyer, V. R., et al. (2014). Chromatin accessibility mapping identifies mediators of basal transcription and retinoid-induced repression of OTX2 in medulloblastoma. PLoS One 9:e107156. doi: 10.1371/journal.pone.0107156
Wright, A. G., Demyanenko, G. P., Powell, A., Schachner, M., Enriquez-Barreto, L., Tran, T. S., et al. (2007). Close homolog of L1 and neuropilin 1 mediate guidance of thalamocortical axons at the ventral telencephalon. J. Neurosci. 27, 13667–13679. doi: 10.1523/JNEUROSCI.2888-07.2007
Wong, C. C., Martincorena, I., Rust, A. G., Rashid, M., Alifrangis, C., Alexandrov, L. B., et al. (2014). Inactivating CUX1 mutations promote tumorigenesis. Nat. Genet. 46, 33–38. doi: 10.1038/ng.2846
Xu, Q., Cobos, I., De La Cruz, E., Rubenstein, J. L., and Anderson, S. A. (2004). Origins of cortical interneuron subtypes. J. Neurosci. 24, 2612–2622. doi: 10.1523/jneurosci.5667-03.2004
Xu, Q., Wonders, C. P., and Anderson, S. A. (2005). Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development 132, 4987–4998. doi: 10.1242/dev.02090
Xu, Y., Xi, J., Wang, G., Guo, Z., Sun, Q., Lu, C., et al. (2021). PAUPAR and PAX6 sequentially regulate human embryonic stem cell cortical differentiation. Nucleic Acids Res. 49, 1935–1950. doi: 10.1093/nar/gkab030
Xuan, S., Baptista, C. A., Balas, G., Tao, W., Soares, V. C., and Lai, E. (1995). Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14, 1141–1152. doi: 10.1016/0896-6273(95)90262-7
Yang, Y., Hwang, C. K., D’Souza, U. M., Lee, S. H., Junn, E., and Mouradian, M. M. (2000). Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J. Biol. Chem. 275, 20734–20741. doi: 10.1074/jbc.M908382199
Yang, Y., Shen, W., Ni, Y., Su, Y., Yang, Z., and Zhao, C. (2017). Impaired interneuron development after Foxg1 disruption. Cereb. Cortex 27, 793–808. doi: 10.1093/cercor/bhv297
Yoshihara, S., Omichi, K., Yanazawa, M., Kitamura, K., and Yoshihara, Y. (2005). Arx homeobox gene is essential for development of mouse olfactory system. Development 132, 751–762. doi: 10.1242/dev.01619
Yoshida, M., Suda, Y., Matsuo, I., Miyamoto, N., Takeda, N., Kuratani, S., et al. (1997). Emx1 and Emx2 functions in development of dorsal telencephalon. Development 124, 101–111. doi: 10.1242/dev.124.1.101
Yoon, S. O., and Chikaraishi, D. M. (1994). Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J. Biol. Chem. 269, 18453–18462. doi: 10.1016/s0021-9258(17)32330-x
Younossi-Hartenstein, A., Green, P., Liaw, G.-J., Rudolph, K., Lengyel, J., and Hartenstein, V. (1997). Control of early neurogenesis of thedrosophilabrain by the head gap genestll, otd, ems, andbtd. Dev. Biol. 182, 270–283. doi: 10.1006/dbio.1996.8475
Ypsilanti, A. R., Pattabiraman, K., Catta-Preta, R., Golonzhka, O., Lindtner, S., Tang, K., et al. (2021). Transcriptional network orchestrating regional patterning of cortical progenitors. Proc. Natl. Acad. Sci. 118:e2024795118 doi: 10.1073/pnas.2024795118
Yuan, F., Chen, X., Fang, K.-H., Wang, Y., Lin, M., Xu, S.-B., et al. (2018). Induction of human somatostatin and parvalbumin neurons by expressing a single transcription factor LIM homeobox 6. Elife 7:e37382. doi: 10.7554/eLife.37382
Yun, K., Fischman, S., Johnson, J., de Angelis, M. H., Weinmaster, G., and Rubenstein, J. L. R. (2002). Modulation of the notch signaling by Mash1 and Dlx1/2regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development 129, 5029–5040.
Yun, K., Garel, S., Fischman, S., and Rubenstein, J. L. R. (2003). Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J. Comp. Neurol. 461, 151–165. doi: 10.1002/cne.10685
Yun, K., Potter, S., and Rubenstein, J. (2001). Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128, 193–205. doi: 10.1242/dev.128.2.193
Zagozewski, J., Shahriary, G. M., Morrison, L. C., Saulnier, O., Stromecki, M., Fresnoza, A., et al. (2020). An OTX2-PAX3 signaling axis regulates Group 3 medulloblastoma cell fate. Nat. Commun. 11, 1–18. doi: 10.1038/s41467-020-17357-4
Zakrzewska, M., Grešner, S. M., Zakrzewski, K., Zalewska-Szewczyk, B., and Liberski, P. P. (2013). Novel gene expression model for outcome prediction in paediatric medulloblastoma. J. Mol. Neurosci. 51, 371–379. doi: 10.1007/s12031-013-0016-6
Zerucha, T., Stühmer, T., Hatch, G., Park, B. K., Long, Q., Yu, G., et al. (2000). A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between dlx genes in the embryonic forebrain. J. Neurosci. 20:709. doi: 10.1523/JNEUROSCI.20-02-00709.2000
Zhao, Y., Guo, Y.-J., Tomac, A. C., Taylor, N. R., Grinberg, A., Lee, E. J., et al. (1999). Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc. Natl. Acad. Sci. 96, 15002–15006. doi: 10.1073/pnas.96.26.15002
Zhao, Y., Flandin, P., Long, J. E., Cuesta, M. D., Westphal, H., and Rubenstein, J. L. (2008). Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J. Comp. Neurol. 510, 79–99. doi: 10.1002/cne.21772
Zhao, Y., Marín, O., Hermesz, E., Powell, A., Flames, N., Palkovits, M., et al. (2003). The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc. Natl. Acad. Sci. U.S.A. 100, 9005–9010. doi: 10.1073/pnas.1537759100
Zhou, Q. P., Le, T. N., Qiu, X., Spencer, V., de Melo, J., Du, G., et al. (2004). Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 32, 884–892. doi: 10.1093/nar/gkh233
Ziffra, R. S., Kim, C. N., Ross, J. M., Wilfert, A., Turner, T. N., Haeussler, M., et al. (2021). Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213. doi: 10.1038/s41586-021-03209-8
Zimmer, C., Tiveron, M.-C., Bodmer, R., and Cremer, H. (2004). Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb. Cortex 14, 1408–1420. doi: 10.1093/cercor/bhh102
Keywords: forebrain, development, homeobox, bHLH factor, forkhead (Fkh) transcription factors, DNA binding domain
Citation: Leung RF, George AM, Roussel EM, Faux MC, Wigle JT and Eisenstat DD (2022) Genetic Regulation of Vertebrate Forebrain Development by Homeobox Genes. Front. Neurosci. 16:843794. doi: 10.3389/fnins.2022.843794
Received: 30 December 2021; Accepted: 14 March 2022;
Published: 25 April 2022.
Edited by:
Anthony LaMantia, Virginia Tech, United StatesReviewed by:
Tomasz Nowakowski, University of California, San Francisco, United StatesKenneth Campbell, Cincinnati Children’s Hospital Medical Center, United States
Copyright © 2022 Leung, George, Roussel, Faux, Wigle and Eisenstat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David D. Eisenstat, ZGF2aWQuZWlzZW5zdGF0QG1jcmkuZWR1LmF1
 Ryan F. Leung
Ryan F. Leung Ankita M. George
Ankita M. George Enola M. Roussel
Enola M. Roussel Maree C. Faux
Maree C. Faux Jeffrey T. Wigle
Jeffrey T. Wigle David D. Eisenstat
David D. Eisenstat