
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 10 March 2023
Sec. Neurodegeneration
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.1081124
This article is part of the Research Topic Olfactory Neuroepithelium-derived Cellular Models to Study Neurological and Psychiatric Disorders View all 5 articles
 Marina Mihaljevic1
Marina Mihaljevic1 Max Lam2,3,4,5
Max Lam2,3,4,5 Carlos Ayala-Grosso6
Carlos Ayala-Grosso6 Finn Davis-Batt7
Finn Davis-Batt7 David J. Schretlen7
David J. Schretlen7 Koko Ishizuka7
Koko Ishizuka7 Kun Yang7
Kun Yang7 Akira Sawa1,7,8,9,10,11*
Akira Sawa1,7,8,9,10,11*“Druggable genome” is a novel concept that emphasizes the importance of using the information of genome-wide genetic studies for drug discovery and development. Successful precedents of “druggable genome” have recently emerged for some disorders by combining genomic and gene expression profiles with medical and pharmacological knowledge. One of the key premises for the success is the good access to disease-relevant tissues from “living” patients in which we may observe molecular expression changes in association with symptomatic alteration. Thus, given brain biopsies are ethically and practically difficult, the application of the “druggable genome” approach is challenging for neuropsychiatric disorders. Here, to fill this gap, we propose the use of olfactory neuronal cells (ONCs) biopsied and established via nasal biopsy from living subjects. By using candidate genes that were proposed in a study in which genetic information, postmortem brain expression profiles, and pharmacological knowledge were considered for cognition in the general population, we addressed the utility of ONCs in the “druggable genome” approach by using the clinical and cell resources of an established psychosis cohort in our group. Through this pilot effort, we underscored the chloride voltage-gated channel 2 (CLCN2) gene as a possible druggable candidate for early-stage psychosis. The CLCN2 gene expression was associated with verbal memory, but not with other dimensions in cognition, nor psychiatric manifestations (positive and negative symptoms). The association between this candidate molecule and verbal memory was also confirmed at the protein level. By using ONCs from living subjects, we now provide more specific information regarding molecular expression and clinical phenotypes. The use of ONCs also provides the opportunity of validating the relationship not only at the RNA level but also protein level, leading to the potential of functional assays in the future. Taken together, we now provide evidence that supports the utility of ONCs as a tool for the “druggable genome” approach in translational psychiatry.
Cognitive deficits are one of the key domains of psychopathology in psychotic disorders, and are present throughout the illness course, affecting social and occupational functioning, and leading to poorer outcomes (Fett et al., 2011; Cowman et al., 2021). Nevertheless, currently approved antipsychotic drugs have modest or negligible effects on cognitive improvement (Meltzer and McGurk, 1999; Baldez et al., 2021). Thus, it is warranted that the discovery and development of new drugs that primarily target cognitive deficits are taken place in a mechanism-driven manner.
“Druggable genome” is a novel concept, emerged in the post-genomic era, that emphasizes the significance of using the information of genome-wide genetic studies for drug discovery and development (Hopkins and Groom, 2002). Many studies have supported that promising molecular candidates for drug discovery are underscored in genome-wide genetic studies in a wide range of diseases, including brain disorders (Lencz and Malhotra, 2015; Wain et al., 2017; Lempiainen et al., 2018; Gaziano et al., 2021; Hegvik et al., 2021; Storm et al., 2021; Valette et al., 2021; Chia et al., 2022). Successful precedents of “druggable genome” have recently emerged for some disorders by combining genomic and gene expression profiles with medical and pharmacological knowledge (Nabirotchkin et al., 2020). One of the key premises for the success is the good access to disease-relevant tissues from “living” patients in which molecular expression changes are associated with symptomatic alteration.
In addition to the studies for each specific disorder, some genome-wide association studies have targeted specific phenotypes, such as cognitive impairment. For example, a recent study used the currently largest dataset of general cognitive ability for a genome-wide association study (GWAS) in general populations (Lam et al., 2021). In this study, the final candidates were narrowed from the GWAS hits by examining postmortem gene expression databases in association with genetic variation and cognition, considering whether their encoded proteins may mediate pharmacological actions of currently available medicines by literature mining. As a result, the study finalized 16 candidate genes.
The study by Lam and colleagues (Lam et al., 2021) may be one of the first “druggable genome” efforts for brain conditions, but it also includes a potential limitation in expanding this approach in neuropsychiatric disorders. To step forward with the “druggable genome” approach, the expression of candidate molecules defined by this approach are expected to be different between patients and healthy controls (HCs) in disease-relevant tissues, and/or to be correlated with some clinical and cognitive symptoms in “living” patients. Different from other physical disorders, such as cancers in which the “druggable genome” approach has been effectively used, it is difficult for neuropsychiatric disorders to evaluate molecular expression changes in disease-relevant tissues (e.g., brain cells and neuronal cells) in association with symptomatic alteration in “living” patients. This validation is essential for further pharmacological assessment.
To overcome this dilemma, we hypothesized that olfactory neuronal cells (ONCs) biopsied and established via nasal biopsy from living subjects might be a useful tool to fill the gap. Several groups have established similar but slightly different protocols for preparing ONCs, but all these cells are cultured in vitro and can be stored until further use. These cells share molecular signatures relevant to neurons (Fan et al., 2013; Kano et al., 2013; English et al., 2015; Feron et al., 2016; Lavoie et al., 2017; Rhie et al., 2018; Sumitomo et al., 2018; Doostparast Torshizi et al., 2019; Evgrafov et al., 2020; Takayanagi et al., 2021; Tee and Mackay-Sim, 2021; Yang et al., 2021). There are advantages of using ONCs to fill the gap in the “druggable genome” approach: first, we can conduct symptomatic and cognitive assessment in parallel to nasal biopsy and molecular analysis of ONCs from the same subjects; second, at least by our own protocol, ONCs can be used for biochemical and pharmacological studies because of their homogeneity shown at the histochemical levels (Kano et al., 2013; Takayanagi et al., 2021).
In the present study, we tested our hypothesis and aimed to prove the utility of ONCs in expanding the “druggable genome” approach in neuropsychiatric disorders. To address this, we referred to the aforementioned report (Lam et al., 2021), examined the expressions of these candidate genes in ONCs, and looked for their correlations with clinical and cognitive manifestations in patients with early-stage psychosis. Through these efforts, we now highlight chloride voltage-gated channel 2 (CLCN2) as a promising target for a specific dimension of cognition in early-stage psychosis, further supporting the utility of ONCs as a tool for the “druggable genome” approach in translational psychiatry.
We studied 64 HCs and 56 patients with psychotic disorders within 5 years after onset, the majority of whom (52 subjects) within 2 years after onset. As described in recent publications that addressed different scientific questions (Kamath et al., 2018; Faria et al., 2019; Wang et al., 2019; Narita et al., 2021; Etyemez et al., 2022; Yang et al., 2022), diagnoses of psychotic disorders were confirmed using the Structural Clinical Interview for DSM-IV Patient Edition (SCID) (Williams et al., 1992). The spectrum of psychotic disorders included schizophrenia (26 patients), bipolar disorder with psychotic features (14 patients), schizoaffective disorder (seven patients), major depressive disorder with psychotic features (five patients), not otherwise specified psychotic disorder (two patients), and schizophreniform disorder (two patients). Exclusion criteria for all participants included a history of head trauma, nasal trauma, nasal surgery, neurologic disorder, cancer, viral infection, and a reported history of intellectual disability. Individuals who were pregnant or taking anti-inflammatory agents or experiencing any other psychiatric conditions were also excluded. In addition, participants with an estimated IQ below 70 on the Hopkins Adult Reading Test were excluded from this study (Schretlen et al., 2009). Patients who reported active substance abuse or produced a urine drug screen positive for illicit substance use, except cannabis, were excluded. This study was approved by the Johns Hopkins Medicine Institutional Review Boards and in accordance with The Code of Ethics of the World Medical Association (1964 Declaration of Helsinki). All study participants provided written informed consent. Parental consent and assent were obtained for all participants under the age of 18 years.
The presence and severity of positive and negative symptoms of patients were evaluated by study physicians through the Scale for the Assessment of Negative Symptoms (SANS) and the Assessment of Positive Symptoms (SAPS) (Andreasen, 1990). Each symptom category includes a global rating score. The total global score, the sum of the global ratings, was used to calculate positive and negative symptoms.
The antipsychotic dosages were converted to chlorpromazine equivalents (CPZ) using the defined daily dose method (Leucht et al., 2016). Duration of illness (DOI) (between the onset and nasal biopsy), and CPZ doses were obtained through self-reports and confirmed through the Johns Hopkins electronic medical database. Note that we did not include data of cannabis use in the analysis, as the data were based on mere self-reports and they did not have quantitative value (answers were yes/no).
We used a comprehensive neuropsychological battery which we previously developed and have used in our publications (Faria et al., 2019; Coughlin et al., 2021). We obtained cognitive scores scaled in normally distributed standardized units that covered six domains: (1) processing speed (calculated from the combined scores of the Grooved Pegboard test and the Salthouse test); (2) attention / working memory (Digit Span and Brief Attention Memory test); (3) verbal learning and memory (Hopkins Verbal Learning test); (4) visual learning and memory (Brief Visuospatial Memory test); (5) ideational fluency (Ideational Fluency assessment for Word Fluency and Acceptable Designs); and (6) executive functioning (Modified Wisconsin Card Sorting test). The median of the time gap between neurocognitive assessment and nasal biopsy for cell collection was 22 days.
The nasal biopsy was conducted at the Johns Hopkins Otolaryngology Clinic, and ONCs were prepared from nasal biopsied tissues obtained from patients and HCs in the Johns Hopkins Schizophrenia Center according to previous publications (Kano et al., 2013; Mor et al., 2013; Sumitomo et al., 2018; Takayanagi et al., 2021; Jaaro-Peled et al., 2022; Namkung et al., 2022). We collected 1-mm nasal biopsied tissues including olfactory neuroepithelium. The tissues were first incubated with 2.4 U/mL Dispase II for 45 min at 37°C, and then mechanically minced into small pieces. The tissue pieces were further treated with 0.25 mg/mL collagenase A for 10 min at 37°C. Dissociated cells were gently suspended and centrifuged to obtain pellets. Cell pellets were resuspended in D-MEM/F12 supplemented with 10% FBS and antibiotics (D-MEM/F12 medium), and tissue debris was removed by transferring only the cell suspension into a new tube. Cells were then plated on 6-well plates in fresh D-MEM/F12 medium (Day 0 plate). Cells floating were collected in fresh D-MEM/F12 medium on day 2 (Day 2 plate) and day 7 (Day 7 plate). Through these processes, connective tissue-origin cells and non-neuronal cells that are attached in Day 0 and 2 plates were removed. Cells in Day 7 plate were cultured until they reached confluency, collected, and stored in a liquid nitrogen tank until further use as ONCs. ONCs are nearly homogenous with a βIII-tubulin, a marker for immature neurons (Kano et al., 2013). Consistently, a genome-wide expression analysis of ONCs in reference to various pubic databases for gene expression also indicated that the general signature of ONCs is similar to that of developing neurons (Horiuchi et al., 2013).
We have previously published the method for the RNA-sequencing study that was also employed in the present study (Jaaro-Peled et al., 2022). In brief, total RNA was isolated from the ONCs using the RNeasy Plus Mini Kit (Qiagen). RNA libraries were prepared from 500 ng total RNA using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (E7760 and E7490) following the NEBNext Poly(A) mRNA Magnetic Isolation Module protocol. Libraries were then normalized to 4 nM and pooled in equimolar amounts. Paired-End Sequencing was performed using Illumina's NovaSeq6000 S4 200 cycle kit.
FastQC (Wingett and Andrews, 2018) was used to check the quality of reads. High-quality data were obtained from raw data by using cutadapt (Martin, 2011) to remove adapters, primers, and reads with low quality (option -q 10) or shorter than 20 nt. Hisat2 was used to map the clean reads to the human genome (Pertea et al., 2016), version GRCh38 (Genome Reference Consortium Human Build 38). Stringtie (Pertea et al., 2016) was used to assemble and merge transcripts and estimate transcript abundances. A Python script (prepDE.py) provided by Stringtie developer was used to create count tables for further analysis.
ClC2 levels were measured in whole lysates of ONCs. ONCs obtained from 20 patients were lysed in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing proteinase inhibitor and phosphatase inhibitor cocktails and sonicated. Cell lysate samples (30 μg of protein) were run on NuPAGE 10% bis-tris gels (Invitrogen, NP0302BOX), transferred to PVDF membranes (Millipore, IPVH00010), and blocked for 1 h at room temperature in 5% milk in Tris buffered saline with 0.1% Tween 20. Membranes were incubated with an antibody against CLCN2 (Alomone Labs, ACL-002) (1:200 dilution) or with an antibody against GAPDH (Santa Cruz Biotech, sc-32233) (1:4000 dilution) overnight at 4°C, washed, and incubated in a horseradish peroxidase-conjugated secondary antibody (GE Healthcare, NA934V for CLCN2; NA931V for GAPDH) (1:5000 dilution) for 1 h at room temperature. After washing, membranes were developed using the Super Signal West Dura Chemiluminescent Substrate (Thermo Scientific). Protein band intensity was measured using ImageJ software (National Institutes of Health, Bethesda, MD). The ratio CLCN2/GAPDH was calculated and is plotted as mean ± SEM of independent samples.
SPSS v24.0 was used to perform statistical analysis. Group comparisons of demographic data were calculated using independent t-tests for continuous variables, and chi-squared tests for categorical variables. ANCOVA (analysis of covariance) controlling for age, sex, and race was used to compare the expressional levels of the gene candidates between HCs and patients. ANCOVA controlling for age, sex, race, and education was used to compare neurocognitive functions between HC s and patients. Linear regression with age, sex, and race as covariates was used to evaluate the correlation between the molecular expressions and clinical factors such as SANS/SAPS, CPZ, and DOI.
We conducted a one-tailed linear regression with age, sex, race, and the years of education as covariates to test whether the expression level of each candidate genes/protein is correlated with neurocognitive function in the same direction of correlation that was indicated in the study by Lam and colleagues (Lam et al., 2021). Bonferroni procedure is used for multiple comparison correction.
The demographic, clinical, and neurocognitive information for HCs and patients are summarized in Table 1 and Supplementary Table S1. There were differences in sex and the years of education, but not in age and race, between HCs and patients. All the demographic data were controlled in downstream analyses. There were differences in all neurocognitive domains, except IQ measures, between HCs and patients.
We first tested whether 16 gene candidates underscored by the published study (Lam et al., 2021) were expressed in ONCs. By examining our RNA-sequencing dataset, we confirmed that eight genes are expressed in ONCs. These included carbonic anhydrase 13 (CA13), chloride voltage-gated channel 2 (CLCN2), dihydroorotate dehydrogenase (DHODH), dipeptidyl peptidase 4 (DPP4), 5-hydroxytryptamine receptor 1D (HTR1D), phosphodiesterase 4D (PDE4D), proteasome 20S subunit alpha 5 (PSMA5), and thyroid hormone receptor beta (THRB). However, expression levels of these eight genes themselves were not different between patients and HCs in the group comparison (Table 2). This suggested that the impact of these genes, if exists, may be more specific to distinct phenotype(s), such as symptomatic and neurocognitive manifestations.
Accordingly, we decided to study the relationship between molecular expression and clinical phenotypes for these eight genes. First, we addressed whether the molecular expressions might be associated with symptomatic manifestations, such as positive and negative symptoms. We did not observe any associations between SAPS/SANS scores and molecular expression (Supplementary Table S2), indicating that the molecular expression and symptomatic manifestations are unrelated. We also tested whether CPZ doses and DOI were associated with the molecular expression and did not observe any associations (Supplementary Table S2). These results support the idea that medication and DOI (potential clinical confounders) do not influence the expression of these candidates.
Thus, we looked for possible associations between neurocognitive domains and molecular expressions. Our analysis highlighted 2 correlations: CLCN2 expression with verbal memory in patients and CLCN2 expression with executive function in HCs (Table 3).
As described above, expression of CLCN2 at RNA levels (RNA-seq data) was positively associated with verbal memory in patients and executive function in HCs. Based on our perspectives of the “druggable genome” approach as described in the Introduction, we focused on the association between CLCN2 gene expression and verbal memory in patients (Figure 1). Accordingly, we tried to validate the observations not only at the RNA levels but also at the protein levels. We examined the ClC-2 levels from patients and tested its association with neuropsychological domains. Results showed a strong association between ClC-2 level and verbal memory (Figure 2). The whole model explained around 60% of verbal memory variance (R2 = 0.617). The association had the same positive direction as that in the association between the CLCN2 gene expression level and verbal memory. No association was found for any other neuropsychological domains.
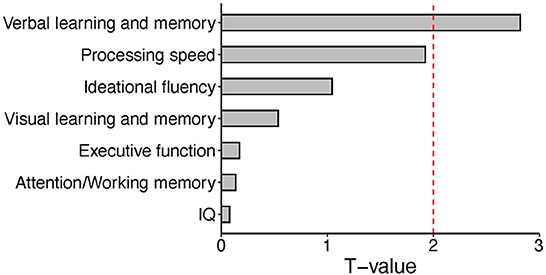
Figure 1. Bar plot of T values from the linear regression analysis between expression levels of the CLCN2 gene and each neuropsychological domain. We tested the association between CLCN2 expressional levels and all neuropsychological domains in patients. A significant correlation was observed for verbal memory. The red dashed line represents the significant cut-off (p < 0.05).

Figure 2. Bar plot of T values from the linear regression analysis between expression levels of the CLC-2 protein and each neuropsychological domain. We tested the association between CLC-2 and all neuropsychological domains in patients. A significant correlation was observed for verbal memory. The red dashed line represents the significant cut-off (p < 0.05).
The goal of the present study is to test the utility of ONCs in expanding the “druggable genome” approach in neuropsychiatric disorders from the information of genetic studies. We used the data from a report that combined the information of GWAS and postmortem expression study in association with general cognitive ability in the general population, as the initial information to explore the present study that includes molecular expression in ONCs. We examined the molecular expression with psychiatric manifestations (positive symptoms and negative symptoms) and 6 distinct cognitive domains in patients and HCs, and underscored CLCN2 as a candidate for future drug discovery.
We regard that the study from which we obtained the initial information (Lam et al., 2021) is a respectful, pioneered effort of exploring the “druggable genome” approach in brain conditions. However, without the information on molecular expression and clinical manifestations from the same set of living subjects, the study was limited by the design of connecting genomic data with postmortem transcriptomics and cognitive data from different populations. In contrast, by studying the same subjects for ONC molecular expression and clinical manifestations, we could observe a correlation of CLCN2 gene expression specific with verbal memory. We did not observe correlations with other dimensions in cognition, nor psychiatric manifestations (positive and negative symptoms). The association between this candidate molecule and verbal memory was also confirmed at the protein level. By using ONCs from living subjects, we now provide more specific information regarding molecular expression and clinical phenotypes, compared with the study we quoted for the initial resource. The use of ONCs also provides the opportunity of validating the relationship not only at the RNA level but also protein level, leading to the potential of functional assays in the future. Taken together, we now provide evidence that supports the utility of ONCs as a tool for the “druggable genome” approach in translational psychiatry.
ONCs did not express all genes that were underscored in the study we used for the initial resource (Lam et al., 2021). This indicates that, although ONCs are useful as described above, we may not be able to validate all genes for the “druggable genome” approach through this experimental system. We acknowledge that this is a limitation in the use of ONCs.
As described in the Introduction section, we defined the criteria of candidate genes in the present study to be different in their expression between patients and healthy controls (HCs), and/or to show a correlation of the expression with clinical and cognitive symptoms in patients. We have to admit that the CLCN2 gene satisfied the second criterion but not the first one. We observed a correlation between the CLCN2 gene expression and executive function in HCs, but not in patients. Transparently speaking, we have no way of reasonably interpreting the data at least at present. However, although we focus our target only on patients based for the goal of “druggable genome” in the present study, we acknowledge that this is an interesting topic in future studies.
Furthermore, sense of smell is an important feature of the olfactory system, which include many factors and elements (cells in the OE, connectivity of the OE and olfactory blub, as well as neural network of OB, olfactory primary cortex, and higher cortex). However, we used ONCs as a surrogate neuronal tissue of analyzing gene expression level related to “druggable genes” for cognition. Exploring a potential role of ONCs in smell is a separate scientific question, which is an important future topic. Finally, we acknowledge that the sample size of the present cohort is relatively small. We hope that this pioneered study focusing on CLCN2 with a modest sample size, nevertheless clearly providing the utility of this “druggable genes” approach, may encourage future studies with cohorts of a larger sample size. If this is the case, the potential occurrence and possible implication of genetic variations in the CLCN2 gene may be a future topic. Protein study with targets other than CLCN2/ClC2 will also be explored.
ClC-2 is a voltage-gated chloride channel that regulates chloride homeostasis in multiple tissues, such as the gastric tissue, testis, and retina (Goppner et al., 2021; Hanke-Gogokhia et al., 2021). Although its role in the brain remains elusive, the ClC-2 deficiency has been associated with leukoencephalopathy that accompanies psychiatric manifestation and cognitive impairment (Depienne et al., 2013; Guo et al., 2019). Some genetic studies have reported a possible association between CLCN2 variants and neurodevelopmental conditions such as autistic spectrum disorders and epilepsy (Kleefuss-Lie et al., 2009; Cukier et al., 2014). Recent GWAS have reported its association with cognitive traits (Lee et al., 2018; Davies et al., 2019; Demange et al., 2021). From the “druggable genome” perspective, we note that Lubiprostone, a prostaglandin metabolite prostone, can activate ClC-2 Cl− currents and Cl− transport with an EC50 of ~18–24 nM in a protein kinase A-independent manner (Cuppoletti et al., 2004, 2014). Oral administration of Lubiprostone is effective to the treatment of constipation and irritable bowel in clinical settings specifically targeting CIC-2. Meanwhile, the trial of this medicine for patients with neuropsychiatric disorders was made only for the management of constipation (Pedrosa Carrasco et al., 2018; Xu et al., 2021). In contrast, our study has highlighted the potential implication of CIC-2 (CLCN2-encoded protein) in the context of a specific domain of cognition. Repurposing Lubiprostone in cognitive and neuropsychiatric disorders may be a promising strategy. If this is the case, this will be a proof of concept for the “druggable genome” approach.
Datasets are available on request: The raw data supporting the conclusions of this article will be available upon requests to the authors.
The studies involving human participants were reviewed and approved by Johns Hopkins Institute Review Board. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
The current research was designed by AS and MM. The analytic pipeline was designed by KY, ML, and MM. The data was analyzed by MM, ML, and FD-B. Data analysis and interpretation regarding clinical scales were assisted by MM and DS. Data analysis and interpretation regarding molecular data were assisted by KI and CA-G. The manuscript was drafted by MM, KY, and AS. All authors contributed to the discussion of the results and have approved the final manuscript to be published.
This study was supported by NIH grants MH-092443, MH-094268, MH-105660, and MH-107730; foundation grants from Stanley and RUSK/S-R (to AS), an award from Brain and Behavior Research Foundation (to KY), and a Fulbright fellowship (to MM).
The authors appreciate Yukiko Lema for organizing the manuscript.
Under an agreement with Psychological Assessment Resources, Inc., DS is entitled to a share of royalty on sales of a test used in the study described in this article. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2022.1081124/full#supplementary-material
Andreasen, N. C. (1990). Methods for assessing positive and negative symptoms. Mod. Probl. Pharmacopsychiatry 24, 73–88. doi: 10.1159/000418013
Baldez, D. P., Biazus, T. B., Rabelo-da-Ponte, F. D., Nogaro, G. P., Martins, D. S., Kunz, M., et al. (2021). The effect of antipsychotics on the cognitive performance of individuals with psychotic disorders: Network meta-analyses of randomized controlled trials. Neurosci Biobehav. Rev. 126, 265–275. doi: 10.1016/j.neubiorev.2021.03.028
Chia, R., Saez-Atienzar, S., Murphy, N., Chio, A., Blauwendraat, C., International Myasthenia Gravis Genomics, C., et al. (2022). Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study. Proc. Natl. Acad. Sci. USA. 3, 119. doi: 10.1073/pnas.2108672119
Coughlin, J. M., Yang, K., Marsman, A., Pradhan, S., Wang, M., Ward, R. E., et al. (2021). A multimodal approach to studying the relationship between peripheral glutathione, brain glutamate, and cognition in health and in schizophrenia. Mol. Psychiatry 26, 3502–3511. doi: 10.1038/s41380-020-00901-5
Cowman, M., Holleran, L., Lonergan, E., O'Connor, K., Birchwood, M., and Donohoe, G. (2021). Cognitive predictors of social and occupational functioning in early psychosis: a systematic review and meta-analysis of cross-sectional and longitudinal data. Schizophr Bull. 47, 1243–1253. doi: 10.1093/schbul/sbab033
Cukier, H. N., Dueker, N. D., Slifer, S. H., Lee, J. M., Whitehead, P. L., Lalanne, E., et al. (2014). Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol. Autism 5, 1. doi: 10.1186/2040-2392-5-1
Cuppoletti, J., Chakrabarti, J., Tewari, K. P., and Malinowska, D. H. (2014). Differentiation between human ClC-2 and CFTR Cl- channels with pharmacological agents. Am. J. Physiol. Cell Physiol. 307, C479–492. doi: 10.1152/ajpcell.00077.2014
Cuppoletti, J., Malinowska, D. H., Tewari, K. P., Li, Q. J., Sherry, A. M., Patchen, M. L., et al. (2004). SPI-0211 activates T84 cell chloride transport and recombinant human ClC-2 chloride currents. Am. J. Physiol. Cell. Physiol. 287, C1173–1183. doi: 10.1152/ajpcell.00528.2003
Davies, G., Lam, M., Harris, S. E., Trampush, J. W., Luciano, M., Hill, W. D., et al. (2019). Author Correction: Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 10, 2068. doi: 10.1038/s41467-019-10160-w
Demange, P. A., Malanchini, M., Mallard, T. T., Biroli, P., Cox, S. R., Grotzinger, A. D., et al. (2021). Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction. Nat. Genet. 53, 35–44. doi: 10.1038/s41588-020-00754-2
Depienne, C., Bugiani, M., Dupuits, C., Galanaud, D., Touitou, V., Postma, N., et al. (2013). Brain white matter oedema due to ClC-2 chloride channel deficiency: an observational analytical study. Lancet Neurol. 12, 659–668. doi: 10.1016/S1474-4422(13)70053-X
Doostparast Torshizi, A., Armoskus, C., Zhang, H., Forrest, M. P., Zhang, S., Souaiaia, T., et al. (2019). Deconvolution of transcriptional networks identifies TCF4 as a master regulator in schizophrenia. Sci. Adv. 5, eaau4139. doi: 10.1126/sciadv.aau4139
English, J. A., Fan, Y., Focking, M., Lopez, L. M., Hryniewiecka, M., Wynne, K., et al. (2015). Reduced protein synthesis in schizophrenia patient-derived olfactory cells. Transl. Psychiatry 5, e663. doi: 10.1038/tp.2015.119
Etyemez, S., Narita, Z., Mihaljevic, M., Ishizuka, K., Kamath, V., Yang, K., et al. (2022). Olfactory dysfunction and face processing of social cognition in first-episode psychosis. Neurosci. Res. 176, 79–84. doi: 10.1016/j.neures.2021.10.003
Evgrafov, O. V., Armoskus, C., Wrobel, B. B., Spitsyna, V. N., Souaiaia, T., Herstein, J. S., et al. (2020). Gene Expression in Patient-Derived Neural Progenitors Implicates WNT5A Signaling in the Etiology of Schizophrenia. Biol. Psychiatry 88, 236–247. doi: 10.1016/j.biopsych.2020.01.005
Fan, Y., Abrahamsen, G., Mills, R., Calderon, C. C., Tee, J. Y., Leyton, L., et al. (2013). Focal adhesion dynamics are altered in schizophrenia. Biol. Psychiatry 74, 418–426. doi: 10.1016/j.biopsych.2013.01.020
Faria, A. V., Crawford, J., Ye, C., Hsu, J., Kenkare, A., Schretlen, D., et al. (2019). Relationship between neuropsychological behavior and brain white matter in first-episode psychosis. Schizophr. Res. 208, 49–54. doi: 10.1016/j.schres.2019.04.010
Feron, F., Gepner, B., Lacassagne, E., Stephan, D., Mesnage, B., Blanchard, M.P., et al. (2016). Olfactory stem cells reveal MOCOS as a new player in autism spectrum disorders. Mol. Psychiatry 21, 1215–1224. doi: 10.1038/mp.2015.106
Fett, A. K., Viechtbauer, W., Dominguez, M. D., Penn, D. L., van Os, J., and Krabbendam, L. (2011). The relationship between neurocognition and social cognition with functional outcomes in schizophrenia: a meta-analysis. Neurosci. Biobehav. Rev. 35, 573–588. doi: 10.1016/j.neubiorev.2010.07.001
Gaziano, L., Giambartolomei, C., Pereira, A. C., Gaulton, A., Posner, D. C., Swanson, S. A., et al. (2021). Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat. Med. 27, 668–676. doi: 10.1038/s41591-021-01310-z
Goppner, C., Soria, A. H., Hoegg-Beiler, M. B., and Jentsch, T. J. (2021). Cellular basis of ClC-2 Cl(-) channel-related brain and testis pathologies. J. Biol. Chem. 296, 100074. doi: 10.1074/jbc.RA120.016031
Guo, Z., Lu, T., Peng, L., Cheng, H., Peng, F., Li, J., et al. (2019). CLCN2-related leukoencephalopathy: a case report and review of the literature. BMC Neurol. 19, 156. doi: 10.1186/s12883-019-1390-7
Hanke-Gogokhia, C., Lehmann, G. L., Benedicto, I., de la Fuente-Ortega, E., Arshavsky, V. Y., Schreiner, R., et al. (2021). Apical CLC-2 in retinal pigment epithelium is crucial for survival of the outer retina. FASEB J. 35, e21689. doi: 10.1096/fj.202100349R
Hegvik, T. A., Waloen, K., Pandey, S. K., Faraone, S. V., Haavik, J., and Zayats, T. (2021). Druggable genome in attention deficit/hyperactivity disorder and its co-morbid conditions. New avenues for treatment. Mol. Psychiatry 26, 4004–4015. doi: 10.1038/s41380-019-0540-z
Hopkins, A. L., and Groom, C. R. (2002). The druggable genome. Nat. Rev. Drug. Discov. 1, 727–730. doi: 10.1038/nrd892
Horiuchi, Y., Kano, S., Ishizuka, K., Cascella, N. G., Ishii, S., Talbot, C. C., et al. (2013). Olfactory cells via nasal biopsy reflect the developing brain in gene expression profiles: utility and limitation of the surrogate tissues in research for brain disorders. Neurosci. Res. 77, 247–250. doi: 10.1016/j.neures.2013.09.010
Jaaro-Peled, H., Landek-Salgado, M. A., Cascella, N. G., Nucifora, F. C. Jr., Coughlin, J. M., Nestadt, G., et al. (2022). Sex-specific involvement of the Notch-JAG pathway in social recognition. Transl. Psychiatry 12, 99. doi: 10.1038/s41398-022-01867-4
Kamath, V., Lasutschinkow, P., Ishizuka, K., and Sawa, A. (2018). Olfactory functioning in first-episode psychosis. Schizophr Bull. 44, 672–680. doi: 10.1093/schbul/sbx107
Kano, S., Colantuoni, C., Han, F., Zhou, Z., Yuan, Q., Wilson, A., et al. (2013). Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol. Psychiatry 18, 740–742. doi: 10.1038/mp.2012.120
Kleefuss-Lie, A., Friedl, W., Cichon, S., Haug, K., Warnstedt, M., Alekov, A., et al. (2009). CLCN2 variants in idiopathic generalized epilepsy. Nat. Genet. 41, 954–955. doi: 10.1038/ng0909-954
Lam, M., Chen, C. Y., Ge, T., Xia, Y., Hill, D. W., Trampush, J. W., et al. (2021). Identifying nootropic drug targets via large-scale cognitive GWAS and transcriptomics. Neuropsychopharmacology 46, 1788–1801. doi: 10.1038/s41386-021-01023-4
Lavoie, J., Gasso Astorga, P., Segal-Gavish, H., Wu, Y. C., Chung, Y., Cascella, N. G., et al. (2017). The olfactory neural epithelium as a tool in neuroscience. Trends. Mol. Med. 23, 100–103. doi: 10.1016/j.molmed.2016.12.010
Lee, J. J., Wedow, R., Okbay, A., Kong, E., Maghzian, O., Zacher, M., et al. (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121. doi: 10.1038/s41588-018-0147-3
Lempiainen, H., Braenne, I., Michoel, T., Tragante, V., Vilne, B., Webb, T. R., et al. (2018). Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci. Rep. 8, 3434. doi: 10.1038/s41598-018-20721-6
Lencz, T., and Malhotra, A. K. (2015). Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol. Psychiatry 20, 820–826. doi: 10.1038/mp.2015.28
Leucht, S., Samara, M., Heres, S., and Davis, J. M. (2016). Dose equivalents for antipsychotic drugs: the DDD method. Schizophr. Bull. 42(Suppl 1), S90–94. doi: 10.1093/schbul/sbv167
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 3. doi: 10.14806/ej.17.1.200
Meltzer, H. Y., and McGurk, S. R. (1999). The effects of clozapine, risperidone, and olanzapine on cognitive function in schizophrenia. Schizophr. Bull. 25, 233–255. doi: 10.1093/oxfordjournals.schbul.a033376
Mor, E., Kano, S., Colantuoni, C., Sawa, A., Navon, R., and Shomron, N. (2013). MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol. Dis. 55, 1–10. doi: 10.1016/j.nbd.2013.03.011
Nabirotchkin, S., Peluffo, A. E., Rinaudo, P., Yu, J., Hajj, R., and Cohen, D. (2020). Next-generation drug repurposing using human genetics and network biology. Curr. Opin. Pharmacol. 51, 78–92. doi: 10.1016/j.coph.2019.12.004
Namkung, H., Yukitake, H., Fukudome, D., Lee, B. J., Tian, M., Ursini, G., et al. (2022). The miR-124-AMPAR pathway connects polygenic risks with behavioral changes shared between schizophrenia and bipolar disorder. Neuron 3, 31. doi: 10.1016/j.neuron.2022.10.031
Narita, Z., Yang, K., Kuga, H., Piancharoen, P., Etyemez, S., Faria, A., et al. (2021). Face processing of social cognition in patients with first episode psychosis: Its deficits and association with the right subcallosal anterior cingulate cortex. Schizophr. Res. 238, 99–107. doi: 10.1016/j.schres.2021.09.027
Pedrosa Carrasco, A. J., Timmermann, L., and Pedrosa, D. J. (2018). Management of constipation in patients with Parkinson's disease. NPJ Parkinson's Dis. 4, 6. doi: 10.1038/s41531-018-0042-8
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Rhie, S. K., Schreiner, S., Witt, H., Armoskus, C., Lay, F. D., Camarena, A., et al. (2018). Using 3D epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation. Sci. Adv. 4, eaav8550. doi: 10.1126/sciadv.aav8550
Schretlen, D. J., Winicki, J. M., Meyer, S. M., Testa, S. M., Pearlson, G. D., and Gordon, B. (2009). Development, psychometric properties, and validity of the hopkins adult reading test (HART). Clin. Neuropsychol. 23, 926–943. doi: 10.1080/13854040802603684
Storm, C. S., Kia, D. A., Almramhi, M. M., Bandres-Ciga, S., Finan, C., International Parkinson's Disease Genomics, C., et al. (2021). Finding genetically-supported drug targets for Parkinson's disease using Mendelian randomization of the druggable genome. Nat. Commun. 12, 7342. doi: 10.1038/s41467-021-26280-1
Sumitomo, A., Yukitake, H., Hirai, K., Horike, K., Ueta, K., Chung, Y., et al. (2018). Ulk2 controls cortical excitatory-inhibitory balance via autophagic regulation of p62 and GABAA receptor trafficking in pyramidal neurons. Hum. Mol. Genet. 27, 3165–3176. doi: 10.1093/hmg/ddy219
Takayanagi, Y., Ishizuka, K., Laursen, T. M., Yukitake, H., Yang, K., Cascella, N. G., et al. (2021). From population to neuron: exploring common mediators for metabolic problems and mental illnesses. Mol. Psychiatry 26, 3931–3942. doi: 10.1038/s41380-020-00939-5
Tee, J. Y., and Mackay-Sim, A. (2021). Directional persistence of cell migration in schizophrenia patient-derived olfactory cells. Int. J. Mol. Sci. 22, 1977. doi: 10.3390/ijms22179177
Valette, K., Li, Z., Bon-Baret, V., Chignon, A., Berube, J. C., Eslami, A., et al. (2021). Prioritization of candidate causal genes for asthma in susceptibility loci derived from UK Biobank. Commun. Biol. 4, 700. doi: 10.1038/s42003-021-02227-6
Wain, L. V., Shrine, N., Artigas, M. S., Erzurumluoglu, A. M., Noyvert, B., Bossini-Castillo, L., et al. (2017). Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat. Genet. 49, 416–425. doi: 10.1038/ng.3787
Wang, A. M., Pradhan, S., Coughlin, J. M., Trivedi, A., DuBois, S. L., Crawford, J. L., et al. (2019). Assessing brain metabolism with 7-T proton magnetic resonance spectroscopy in patients with first-episode psychosis. JAMA Psychiatry 76, 314–323. doi: 10.1001/jamapsychiatry.2018.3637
Williams, J. B., Gibbon, M., First, M. B., Spitzer, R. L., Davies, M., Borus, J., et al. (1992). The structured clinical interview for DSM-III-R (SCID). II. multisite test-retest reliability. Arch. Gen. Psychiatry 49, 630–636. doi: 10.1001/archpsyc.1992.01820080038006
Wingett, S. W., and Andrews, S. (2018). FastQ screen: a tool for multi-genome mapping and quality control. F1000Res 7, 1338. doi: 10.12688/f1000research.15931.2
Xu, Y., Amdanee, N., and Zhang, X. (2021). Antipsychotic-induced constipation: a review of the pathogenesis, clinical diagnosis, and treatment. CNS Drugs 35, 1265–1274. doi: 10.1007/s40263-021-00859-0
Yang, K., Hua, J., Etyemez, S., Paez, A., Prasad, N., Ishizuka, K., et al. (2021). Volumetric alteration of olfactory bulb and immune-related molecular changes in olfactory epithelium in first episode psychosis patients. Schizophr Res. 235, 9–11. doi: 10.1016/j.schres.2021.07.016
Keywords: olfactory neuronal cells, cognition, nalas biopsy, psychosis, drug discovery, CLCN2
Citation: Mihaljevic M, Lam M, Ayala-Grosso C, Davis-Batt F, Schretlen DJ, Ishizuka K, Yang K and Sawa A (2023) Olfactory neuronal cells as a promising tool to realize the “druggable genome” approach for drug discovery in neuropsychiatric disorders. Front. Neurosci. 16:1081124. doi: 10.3389/fnins.2022.1081124
Received: 26 October 2022; Accepted: 26 December 2022;
Published: 10 March 2023.
Edited by:
Greg Sutherland, The University of Sydney, AustraliaReviewed by:
Francois Feron, Aix Marseille Université, FranceCopyright © 2023 Mihaljevic, Lam, Ayala-Grosso, Davis-Batt, Schretlen, Ishizuka, Yang and Sawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akira Sawa, YXNhd2ExQGpobWkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.