- 1Department of Computer Science and Mathematics, Zhejiang Normal University, Jinhua, China
- 2School of Computer Science, Huazhong University of Science and Technology, Wuhan, China
- 3Division of Biomedical Imaging, Department of Biomedical Engineering and Health Systems, KTH Royal Institute of Technology, Stockholm, Sweden
- 4Department of Computer Science, National University of Modern Languages, Islamabad, Pakistan
- 5Department of Computer Science, University of Agriculture, Sub Campus Burewala-Vehari, Faisalabad, Pakistan
- 6Department of Radiology and Diagnostic Imaging, University of Alberta, Edmonton, AB, Canada
- 7Department of Physics, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates
- 8Department of Radiology, Emory Brain Health Center-Neurosurgery, School of Medicine, Emory University, Atlanta, GA, United States
- 9Key Laboratory of Intelligent Education Technology and Application of Zhejiang Province, Zhejiang Normal University, Jinhua, China
Alzheimer's is an acute degenerative disease affecting the elderly population all over the world. The detection of disease at an early stage in the absence of a large-scale annotated dataset is crucial to the clinical treatment for the prevention and early detection of Alzheimer's disease (AD). In this study, we propose a transfer learning base approach to classify various stages of AD. The proposed model can distinguish between normal control (NC), early mild cognitive impairment (EMCI), late mild cognitive impairment (LMCI), and AD. In this regard, we apply tissue segmentation to extract the gray matter from the MRI scans obtained from the Alzheimer's Disease National Initiative (ADNI) database. We utilize this gray matter to tune the pre-trained VGG architecture while freezing the features of the ImageNet database. It is achieved through the addition of a layer with step-wise freezing of the existing blocks in the network. It not only assists transfer learning but also contributes to learning new features efficiently. Extensive experiments are conducted and results demonstrate the superiority of the proposed approach.
1. Introduction
Alzheimer's is one of the most crucial causes of dementia all over the world. Several neurological illnesses, including dementia, afflict a sizable portion of the global population. Patients with Alzheimer show more clear symptoms after the age of 60. However, in some cases, as a result of some gene abnormalities, the symptoms may start to show up at a young age (30–50). Alzheimer's gives rise to functional and structural changes in the brain (Hampel et al., 2021). The progression of Alzheimer's disease (AD) from normal control (NC) spans over a number of years with some intermediate stages ranging from the development of early mild cognitive impairment (EMCI) to late mild cognitive impairment (LMCI). These changes can be observed through MRI images and blood plasma spectroscopy (Pan et al., 2020; Palmer et al., 2021).
Visualizing the MRI scans can somehow allow the physicians to detect the contraction of the gray matter. However, it is a complex process to detect these changes manually. Machine learning-based techniques for the classifications, such as support vector machines (SVMs), artificial neural networks (ANNs), and deep learning-based convolutional neural networks (CNNs), remained very useful in the detection of these minor tissue-level changes (Mehmood et al., 2021). It is important to note that SVM and ANN give local and global optimization-based solutions. However, deep learning-based CNNs consider the feature extraction and learning in the model itself and are considered to be more useful in medical image analysis (Pan et al., 2020; Cheng et al., 2022). But these methods are data-hungry and demand large-scale training datasets to learn the task (i.e., classification in this case) from scratch (Chen et al., 2022; Khan et al., 2022).
Recent advancements in imaging technology, including computerized axial tomography (CT), magnetic resonance images (MRI), and positron emission tomography (PET) (Masdeu et al., 2005; Han et al., 2022) images, have revolutionized the detection of Alzheimer's. Due to ionization effects and cost and computational complexity, gathering a large-scale data set for a particular task is very challenging. The 3D MRI images produced through high-dimensional diagnostic equipment contain several images in a single voxel. These voxels can help to diagnose Alzheimer's at an early stage (Zhang et al., 2019).
A deep Siamese convolution neural network (SCNN) for the multiclass classification of AD is proposed by Mehmood et al. (2020). A natural image-based network to represent neuroimaging data (NIBR-Net) is another significant approach in the target domain, based on a sparse autoencoder (Gupta et al., 2013), where the network learns from a set of bases from natural images with the help of convolution to extract features from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. This method selects the useful features in a single hierarchy while iteratively filtering the undesirable features. Considering the multiple classes of Alzheimer a deep learning-based multiclass classifier is proposed by Farooq et al. (2017). Apart from this, several computer-aided techniques have been suggested for diagnosing AD, especially in the case of severe dementia (Petot and Friedland, 2004; Acharya et al., 2021). Similarly, the issue of the imbalance of class data is handled by Murugan et al. (2021) with the help of a deep DEMNET by using the pre-processed dataset.
Training a neural network with a small-scale dataset (i.e., MRIs) with a higher prediction rate and a higher accuracy is a great challenge. In this study, we propose to utilize the pre-trained VGG16 and VGG19 models to predict NC, EMCI, LMCI, and AD. We extract the gray matter (GM) from the brain MRIs because using entire voxels or raw data directly to train neural networks also presents data management. We applied skull striping and tissue segmentation operations to segregate the full brain MRIs. Considering the data-hungry nature of the neural networks, we proposed to apply data augmentation to the extracted GM slices. It also resolves the challenges of overfitting, which is often aroused due to the unavailability of large-scale dataset. In addition, we step-wise freeze the blocks and add layers to transfer the features for accurate predictions on four classes of the input data, i.e., NC, EMCI, LMCI, and AD. Thus, we resolve the challenges of prospective fluctuations and imbalanced data size and class variation problems. The overall experimental evaluations and comparison with several state-of-the-art approaches demonstrate that the proposed transfer learning method outperformed the extant techniques, making it more suitable for future interactive Alzheimer's applications.
2. Related work
The fundamental causes of Alzheimer's are still unknown, and it is believed to be genetic (Adami et al., 2018). Alzheimer's affects a number of social cognitive capacities and results in several neurological conditions that are memory-related (Ramzan et al., 2020). According to an estimate, by the year 2050, 131.5 million individuals will be globally affected by Alzheimer's disease (AD) (Prince et al., 2015). It will become the top cause of death for older people as the number of patients grows daily. High-dimensional data from imaging modalities like MRI, fMRI, PET, amyloid-PET, diffusion tensor imaging, and neurological tests are essential parts of the existing strategies for the diagnosis (Tanveer et al., 2020; Afzal et al., 2021). However, differentiating between the patterns with radiological readings is still quite a difficult task due to the complexity of the minute patterns. As a result, it is difficult to establish an early diagnosis of Alzheimer's. Recently, several deep learning and machine learning-based techniques to enhance picture quality have been presented (Khan et al., 2019; Alenezi and Santosh, 2021). Additionally, the feature extraction- and classification-based approaches can be regularly employed to create prediction models for applications based on intelligent and expert systems (Mehmood et al., 2021; Shams et al., 2021; El-Hasnony et al., 2022).
The progression of Alzheimer's took several years, ranging from NC to MCI and AD. The development of MCI is also considered as early and late MCI (Sperling et al., 2011; Huang et al., 2018). It is imperative to diagnose the disease at an early stage, which is only possible through an accurate classification of various stages of disease (Khan et al., 2022). The recent developments in machine learning and deep learning-based methods can significantly contribute to the target domain (Tanveer et al., 2020). The extraction of the feature and identification of these features based on these techniques can ease the burden on the healthcare system (Razzak et al., 2022). The main objective of the binary class and multi-class classification is to distinguish the features of normal images from impaired images to detect the stage of the disease. Support vector machines (SVM), K-nearest neighbors (KNNs), fuzzy learning, decision trees, random forests, and dimensionality reduction algorithms, like principal component analysis, are readily used methods in traditional machine learning research (Vecchio et al., 2020). The feature extraction through CNNs and deployment of CNNs have revolutionized the whole process (Bi et al., 2020; Liang and Gu, 2021) and can yield acceptable results for the early detection of AD. CNN-based methods can adeptly learn the input images' features to identify a particular disease stage. Hao et al. (2020) suggested a multi-modal framework to extract neurological information from the MRIs in order to classify the various phases of dementia. In order to classify AD, NC, and MCI, Tong et al. developed a multimodal classification framework based on MRI, FDG, and PET scans (Tong et al., 2017). Other recent approaches for the classification of Alzheimer's (Tajbakhsh et al., 2016; Jain et al., 2019; Khan et al., 2019, 2022) can somehow resolve the challenges and some other tries to introduce some transfer learning based approaches (Afzal et al., 2021; Mehmood et al., 2021). However, it is still challenging to extract accurate features to distinguish the images and diagnose the disease at an early stage.
Considering the MRIs, a multi-modal framework is proposed to extract neurological features for the classification of various stages of dementia (Hao et al., 2020). Tong et al. proposed a multimodal classification for AD based on MRI, FDG, and PET images to classify AD, NC, and MCI. A multi-modal learning-based network (Liu et al., 2014) based on PET, and MRI images, a multi-modal stack-net (Shi et al., 2017), and a similarity matrix-based method are proposed by Zhu et al. (2014) based PET, CSF, and MRI biomarkers to distinguish various stages of AD. Some methods rely on the extraction of 2D slices, and some others utilize the whole voxel to distinguish several categories of disease (Payan and Montana, 2015; Islam and Zhang, 2018). A 3D deeply supervised adaptable convolutional neural network (CNN-3D) is proposed by Hosseini-Asl et al. (2018) to predict AD without relying on skull striping with generic feature learning through bio-markers. However, the data management challenges are still complex when handling medical images. In the case of the target problem gathering, a large-scale dataset is a great challenge as compared to ordinary computer vision and image processing tasks as we know that deep learning-based models are data-hungry and demand a large-scale dataset. Therefore, transfer learning approaches (Aderghal et al., 2018; Li et al., 2018; Basaia et al., 2019) are preferred, which utilize the weights from the pretrained models on the large scale datasets such as ImageNet (Mehmood et al., 2021).
To categorize the various stages of the disease with transfer learning, a deep Siamese convolution neural network (SCNN) for the multiclass classification of AD is proposed by Mehmood et al. (2020). A natural image-based network to represent neuroimaging data (NIBR-Net) is another significant approach in the target domain, based on sparse autoencoder (Gupta et al., 2013), where the network learns from a set of bases from natural images with the help of convolution to extract features from the ADNI dataset. A sparse multi-tasking deep learning-based method is proposed by Suk et al. (2016) with a feature adaptive weighting system. This method selects the useful features in a single hierarchy while iteratively filtering the undesirable features. Considering the multiple classes of Alzheimer, a deep learning-based multiclass classifier is proposed by Farooq et al. (2017). Apart from this, several computer-aided techniques have been suggested for diagnosing AD, especially in the case of severe dementia (Petot and Friedland, 2004). These models can assist the physicians in combination with computer-aided intelligent and expert systems (Jo et al., 2019) In this article, we proposed a multi-class classification network by using transfer learning through VGG architecture. We applied a step-wise block freezing strategy to the VGG-16 and VGG-19 models with some additional layers. The proposed method achieves higher accuracy and is capable of working with a small-scale dataset. The overall experimental evaluations demonstrate the superiority of the proposed method as compared to the state-of-the-art approaches.
3. The proposed methodology
In this study, we proposed a transfer learning-based multi-class classification for the early diagnosis of AD. The patients' data in this study was obtained from the ADNI database. We gathered 315-T1 weighted MRI images of four classes: NC, EMCI, LMCI, and AD. The processing overview of these images is shown in Figure 1. We extracted the gray matter through these 3D voxels and utilize these GM slices to train VGG architectures, as shown in Figure 2. We utilized the weights from the pre-trained network on ImageNet and adopted a layer-wise transfer learning while step-wise freezing the blocks. The proposed framework successfully classifies various stages of AD and yields significant results by using only a small-scale dataset.
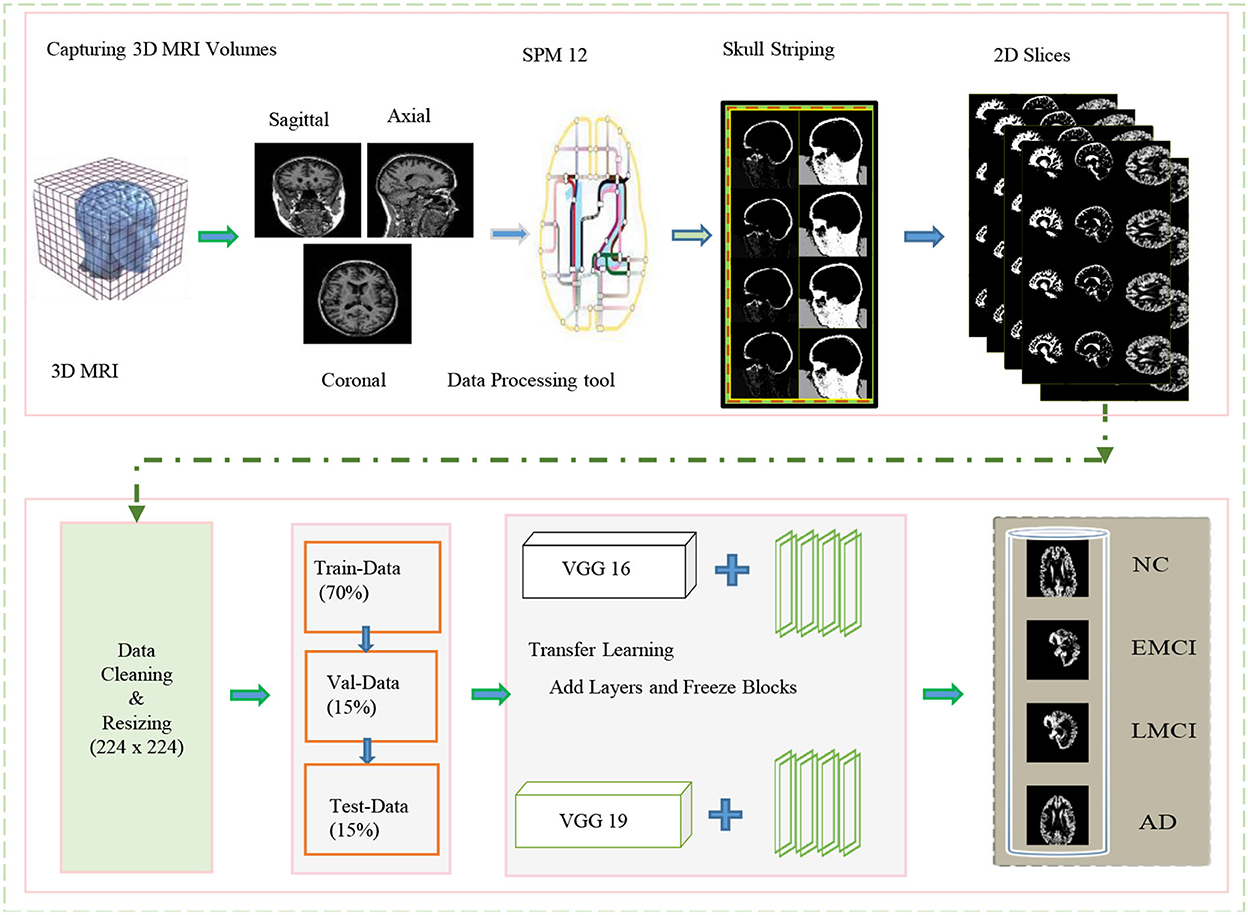
Figure 1. Overview of the proposed framework for processing data and training in the network for classification.
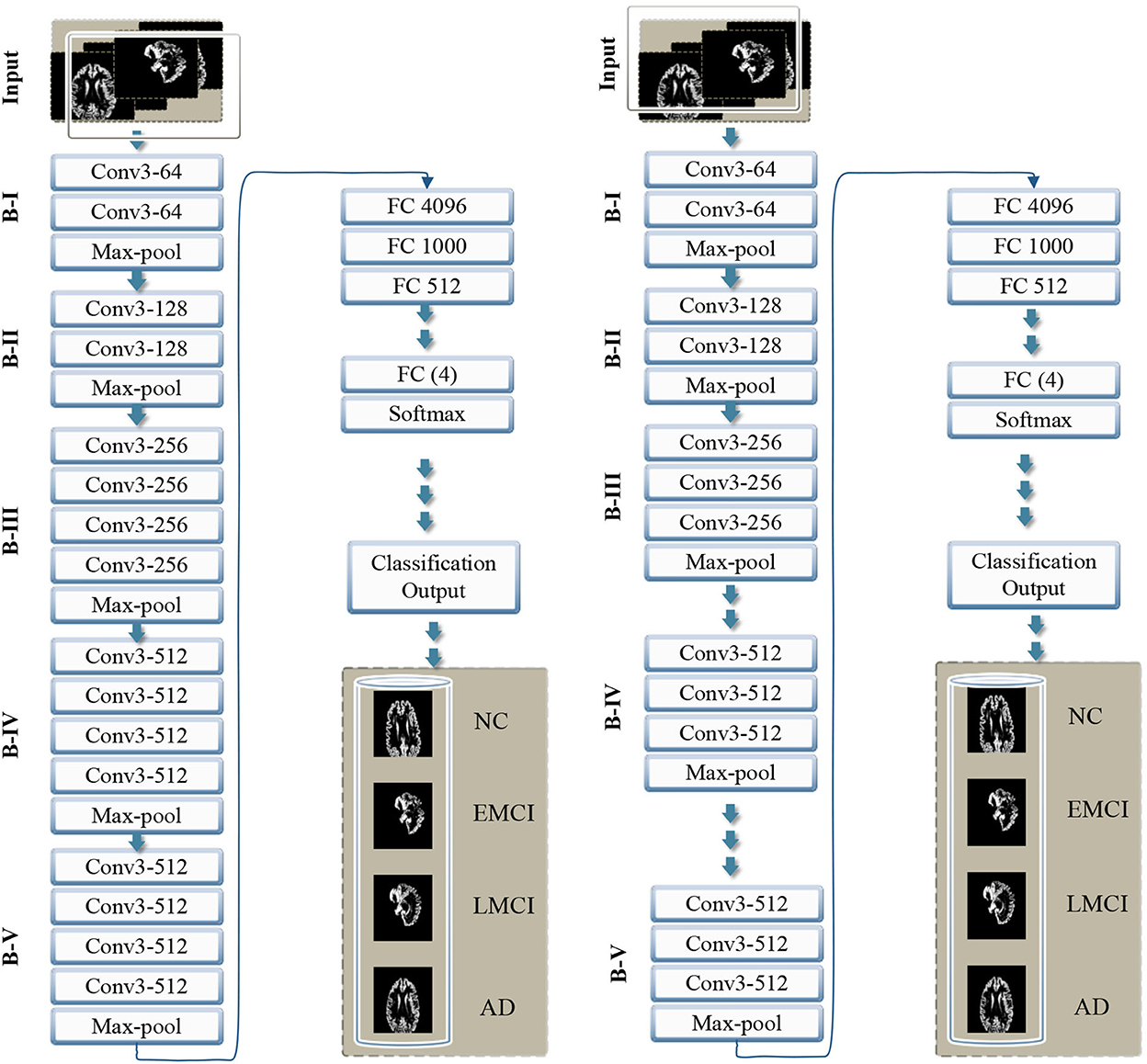
Figure 2. Overview of the proposed framework with step-wise block (B) representation as B-I to B-V. The left side shows VGG 16, and the right side shows the VGG 19 model with the proposed layer-wise transfer learning in both networks, blocks are frozen from B-I to B-V alternatively.
3.1. Datasets and pre-processing on data
The magnetic resonance imaging technology is continually being improved and developed by researchers, and it has revolutionized the way neurological disorders are discovered and diagnosed. Whereas, the inconsistent intensity scale of the image makes it difficult to visualize and evaluate the data manually (Lundervold and Lundervold, 2019). It is essential to provide the correct information during learning if learning-based technologies are to produce accurate predictions. Thus, data preprocessing is a crucial step to manage to increase the contrast and pixel intensity. We applied several pre-processing operations on the available MRI scans of several patients obtained from the ADNI data repository. The specification of the dataset utilized in this work is shown in Table 1. A different number of subjects are selected to handle the class imbalance problem. We mainly exclude the opening and closing slices of the MRI due to the limited information contained in these slices. Images (i.e., 3D MRI scans) obtained from ADNI are available in neuroimaging informatics technology initiative (NIFTI) format. We pre-process these images by utilizing the statistical parametric mapping tool (SPM12) for the tissue-wise segmentation of the input into gray matter, white matter, and cerebrospinal fluid (CSF). We perform several operations (i.e., skull striping, registration, normalization, and segmentation) to extract the 2D-MRI PNG images from the available MRI scans.

Table 1. Sample size of the dataset with specifications and number of subjects utilized for each class.
In this study, we focused only on the gray-matter slices to detect the memory loss changes in early AD detection. We consider the ICBM space template for affine regularization and a bias regularization of 1e−3 with a full bias width at half maximum of 60 mm cutoff. Once these prepossessing operations are complete, we resize all the images to a size of 224 × 224. These images are suitable for training and testing and analogous to the size of ImageNet. The subjects involved in this dataset are scanned with respect to different durations of visits of the patients. Every scan is a different subject and contains the GM, WM, and CSF slices; of which, GM slices are fed to the network to interpret the usable information extracted through MRI volumes. In this regard, we split our dataset into training (70%), testing (15%), and validation data (15%).
4. Neural networks and transfer learning framework
Convolutional neural networks perform convolutional operations to extract the features from the input data. These features are learned during the training process. Later on, the network's prediction behavior determines the network's learning quality. In artificial intelligence (AI), CNNs are distinguished from the other type of networks due to the superior performance of these networks in the multiple-image processing and visualization domains. The main type of layers in these networks are convolutional layers. These are the first layers that extract the features which are pooled with pooling layers. Finally, the fully connected layers are applied. Considering the recent advancements in AI and neural networks for feature extraction and associated task, we employed VGG 19 and VGG 16 architecture with a transfer learning approach for the target problem.
Transfer learning is a method for developing a predictive model for a separate but related problem that can then be utilized partially or entirely to speed up training and ultimately enhance the model's performance for the problem. It involves applying a previously trained model to new problems. Transfer learning-based techniques are now very popular in the field of medical image processing. Considering the advantages of automatic feature extraction and identification with the help of a pre-trained model have proved to be very useful in the target domain. Using a pre-trained model saves the time and effort of building a new model from scratch. It is also difficult to train a substantially large-scale network without amassing millions of annotated images. Therefore, using the pre-trained weights of a network with precise tuning on fresh data is a distinct and advantageous approach. Two models adopted in this study for transfer learning are pre-trained on the ImageNet database comprising millions of images.
The existing models, for example, Inception-Net with 23.62 million parameters, Xception-Net with 22.85 million, and ResNet with 23 million parameters, can also be used for transfer learning. However, considering the importance of the target problem, we selected VGG architecture with 138 million parameters. Feature transfer can be problematic in particular circumstances, such as when the datasets are small or imbalanced. Because transfer learning will not be effective if the final classification layer's characteristics are insufficient to distinguish the classes for a particular problem. Thus if the datasets are not comparable, the feature will hardly be transferred. We present transfer learning results in two categories, i.e., VGG 16 and VGG 19. Thus in each case, we propose to freeze some of the blocks in the network while freezing some fully connected layers.
5. VGG model with proposed transfer learning and experimental evaluations
The pretrained VGG-16 and VGG-19 models utilized in this study are trained on the ImageNet database. We consider freezing the weights and leveraging a pretrained convolutional base. We also include fully connected classification layers to transform the network for our multi-class classification task. The first layer in the model learn feature extraction. The initial layers learn to extract the generic features, and the final layers learn the target-oriented features. We added the new fully connected dense layers and performed several experiments with the rectified linear unit (ReLU) activation function. The main objective of the activation function is to induce non-linearity in the data. It can be expressed for an input value (v) as below.
The final layers in both of the models (i.e., VGG 16 and VGG 19) use the Softmax function. The number of neurons is reduced to 4 due to four classes (i.e., NC, EMCI, LMCI, and AD) in the target problem. The softmax function is the generalization of logistic regression, which is utilized in the classification of mutually exclusive classes. It converts the values to a normalized probability distribution of input for user display. Therefore, it is utilized at the final layer of the network and expressed as σ for input vector vi and standard exponential e followed by a normalization factor with a summation ∑ at the bottom for the K number of classes in the multiclass classifier.
The loss function utilized in this regard is categorical cross-entropy loss which exponentially penalizes error in the probability prediction. The target problem involves predictions for more than one class, i.e., NC, EMCI, LMCI, and AD, and involves 4 labels. Therefore, loss function for the network also varies accordingly, and we consider categorical cross-entropy loss for these multiple-class classification problems. The categorical cross-entropy loss with pi probabilities for ith labels with truth values ti, for the N number of classes is expressed as .
Our goal is to reduce the loss as much as possible. In each case, the cross-entropy for the i number of classes was estimated for the dementia's di, where i = 1....5 in this case. The probability of the output y for each class can be estimated for dementia, where the cross-entropy for each category NC, EMCI, LMCI, and AD is CENC, CEEMCI, CELMCI, and CEAD, respectively.
5.1. Experiments and evaluation metrics
We categorize our experiments based on VGG 16 and VGG 19 models. In each category, we perform several experiments. The overall performance is measured in terms of confusion matrices (Deng et al., 2016) where true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) give an overview of the accuracy, specificity, sensitivity, and precision. We also measure f1-score in addition to accuracy, specificity, sensitivity, and precision. Each column in the confusion matrix is a visual tool to understand the predicted score, where columns and rows show the true and predicted labels, respectively.
5.2. Evaluation metrics
In order to evaluate the performance of the proposed framework, the confusion matrix consists of TP, FP, TN, and FN. Thus, an overview of the confusion matrix gives a comprehensive overview of the F1-score, specificity (Sp), sensitivity (Se), accuracy (Ac), and precision (Pr) value because all of these metrics follow TP, TN, FP, and FN. Witten and Frank (2002). The mathematical expressions below depict this relationship explicitly.
5.2.1. Positive predictive value
The positive predictive value is called the precision and shows the portion of real positive cases.
5.2.2. Sensitivity
Sensitivity is the recall value that shows the actual positive and the correctly predicted portion of values. This metric reflects correctly anticipated cases and depicts the coverage of real positive cases, also termed the true positive rate (TPR).
5.2.3. Specificity
Specificity is associated with the likelihood of a negative test rate in the absence of the condition and is considered a true negative rate.
5.2.4. Accuracy
Classification accuracy is a statistical measurement that evaluates the performance of a classification model by dividing the number of correct predictions by the total number of predictions.
5.2.5. F1 measurement
The F-Metric is a method for combining accuracy and recall into a single measure that encompasses both and is widely utilized in classification tasks.
In addition, we also present graphical and numeric results in terms of accuracy for each class.
5.3. Experimental settings and results
Experimental results for precision, accuracy, and F1 score of VGG 16 and VGG 19 (with and without data augmentation) are presented in the Table 2. In this table, we show the results for NC, EMCI, LMCI, and AD, respectively. The overall performance of the network with and without data augmentation is also shown in this table. The average accuracy of VGG 16 with and without data augmentation for all four classes is 96.39%. Similarly, the average accuracy of the VGG 19 with and without data augmentation for all four classes is 96.81%. This comparison demonstrates that VGG 19 performs slightly better than VGG 16. We also demonstrate the fact with the help of box plots shown in Figure 3. The results are shown for VGG 16 and VGG 19 with data augmentation (WDA) and without data augmentation (WODA).
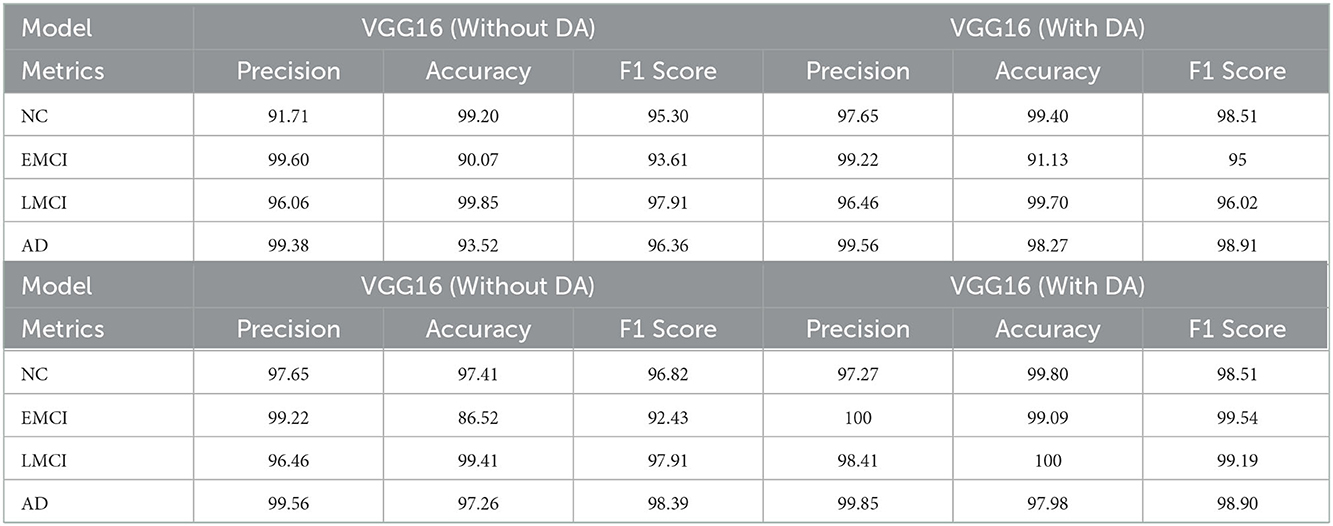
Table 2. The comparison of the evaluation metrics in terms of precision, accuracy, and F1 score for VGG16 and VGG19 with and without data augmentation.
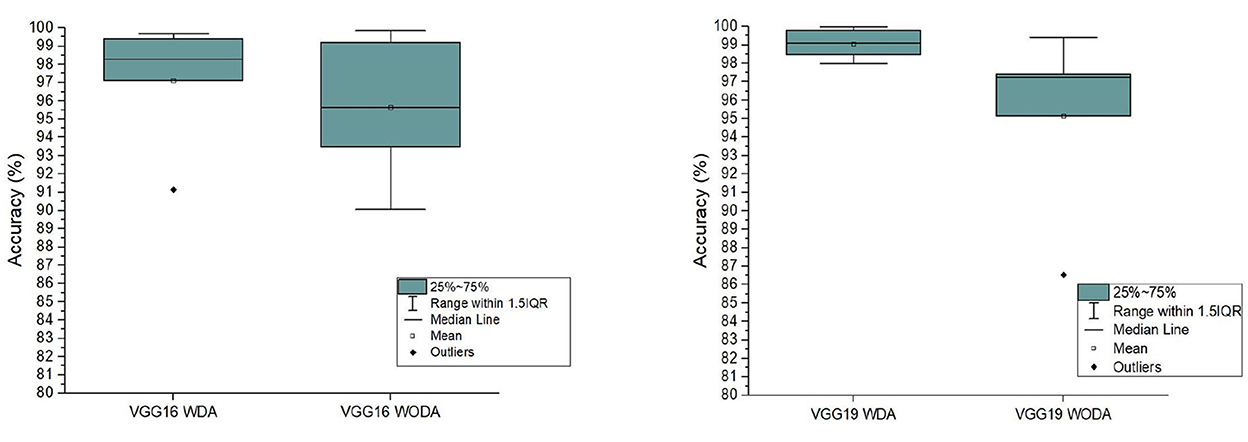
Figure 3. The comparison of the accuracy of the proposed frameworks with data augmentation (WDA) and without data augmentation (WODA).
The proposed framework's overall performance is much better compared to the existing methods. To demonstrate the fact we present the comparison of the proposed method with several state-of-the-art approaches. The results of this comparison are shown in Table 3. In addition, we also compare the performance of the proposed framework with other state-of-the-art models, including DenseNet, Inception ResNet V2, AlexNet, Inception V3, ResNet101, and ResNet50. The results for comparison of these methods with the proposed framework are shown in Table 4. This table shows that the proposed framework (i.e., VGG19+ALFB, VGG16+ALFB) outperformed the other models. To demonstrate the performance of our method with respect to the existing state-of-the-art approaches, we provide a comprehensive comparison with several methods. The competitor approaches include a deep sparse multi-task learning for feature selection in the diagnosis of AD (DSMAD-Net) (Suk et al., 2016), CNN-expedited (Wang et al., 2017) classification of AD on imaging modalities (CAIM) (Aderghal et al., 2018), a transfer learning approach for the early diagnosis of AD on MRI image (TLEDA-Net) (Mehmood et al., 2021), multi-domain transfer learning (TL), a matrix similarity-based method (Zhu et al., 2014), multimodal learning (Liu et al., 2014), multimodal stacked Net (Shi et al., 2017), shape-attributes of brain structures as biomarkers for AD (SABS-AD) (Glozman et al., 2017), detecting AD on small dataset (Li et al., 2018), multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of AD (MMSPN-AD) (Basaia et al., 2019), multimodal classification of AD diagnosis (MMC-AD) (Tong et al., 2017), predicting AD with a 3D neural network (3D-CNN-PAD) (Payan and Montana, 2015), natural image bases to represent neuroimaging data (NIBR-Net) (Gupta et al., 2013), a differential diagnosis strategy (Sørensen et al., 2017), a transfer learning bases method (Naz et al., 2022), a convolutional neural networks-based MRI image analysis for the AD prediction from MCI (Lin et al., 2018), and a novel end-to-end hybrid network for AD detection using 3D CNN and 3D CLSTM (Xia et al., 2020). We performed several experiments, while the freezing pre-trained base and step-wise tested the performance for several blocks in VGG 16 and VGG 19. We include additional layers (ADL) and perform experiments with a block-freezing strategy. We independently present the results of the effects of the proposed changes on VGG16 and VGG19.

Table 3. Comparison of the accuracy of the various state-of-the-art approaches with the proposed work.
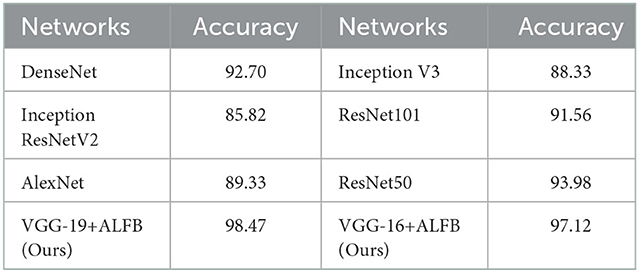
Table 4. Comparison of the proposed architectures with adding layer and freezing blocks (ALFB) in VGG16 and VGG-19, with the existing state-of-the-art architectures in terms of accuracy.
The results for the training loss and test accuracy with additional layers and frozen blocks in the VGG16 model is shown in Figure 4. The resulting confusion matrices to evaluate the network's performance in terms of true and predicted labels are shown in Figure 5. The confusion matrices are shown for (Figure 5A) without data augmentation and (Figure 5B) with data augmentation. The comparison of the accuracy is shown in Table 2 for corresponding confusion matrices, the data augmentation slightly improves the network performance.
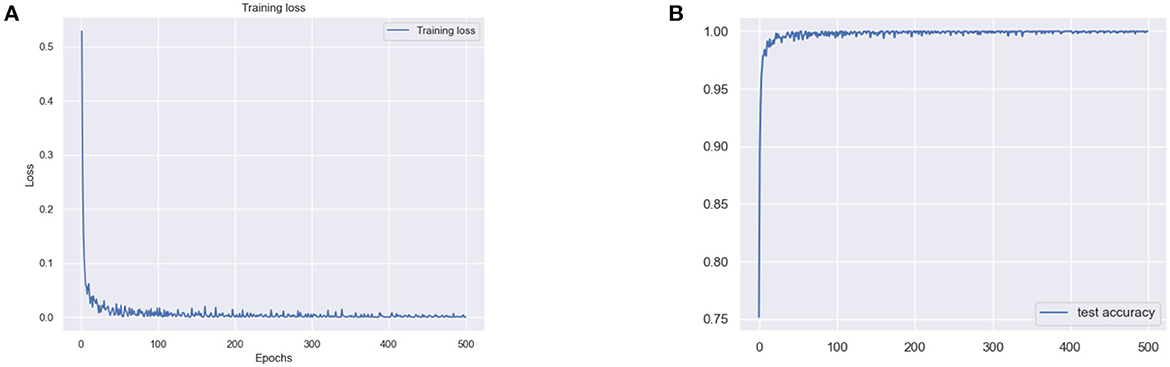
Figure 4. (A) Training loss and (B) test accuracy of the VGG 16 model with additional layer and freezing the block.
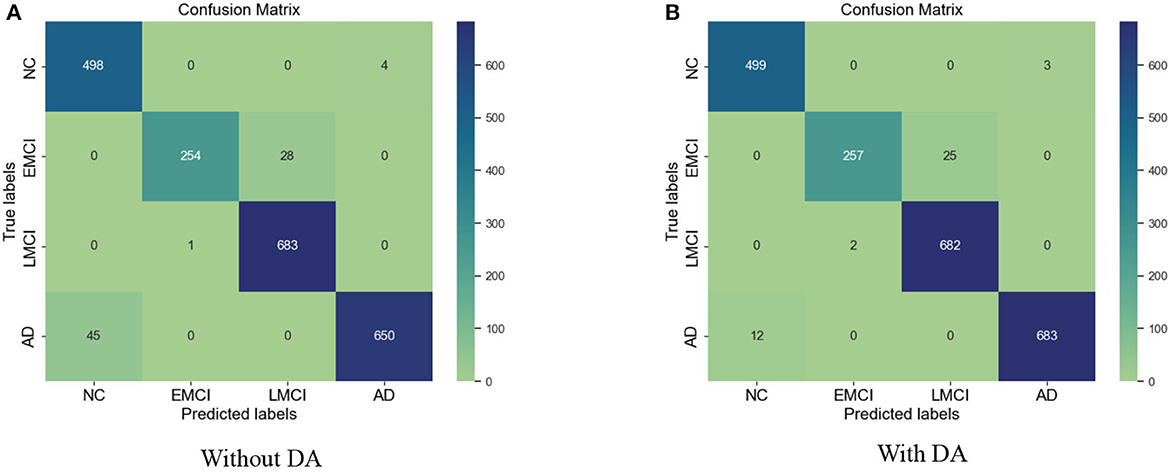
Figure 5. (A) Confusion matrix without augmentation. (B) Confusion matrix with augmentation for the VGG 16.
Similarly, for the VGG 19 model, we show the results for the training loss and testing accuracy in Figure 6, whereas the corresponding confusion matrices are shown in Figure 7. The comparison of the confusion matrices and the accuracy in Table 2 demonstrate that VGG 19 performs slightly better than VGG 16. Comparing the confusion matrices for both networks depicts that the data augmentation slightly improves the overall performance of the networks during transfer learning. In Table 2 we present the results for the specificity, sensitivity, and f1-score for VGG-16 and VGG19 with additional layers and freezing blocks. It depicts the performance and overall efficiency of the proposed method. To improve the robustness and fully-fine tune the model, we propose to increase the data size with data augmentation. It proved to be useful because gathering a large-scale dataset in the target domain is a great challenge. In comparison, the network-based methods are heavily dependent on large-scale datasets. The data augmentation improves the data's size significantly and resolves the challenges of class imbalance and over-fitting.
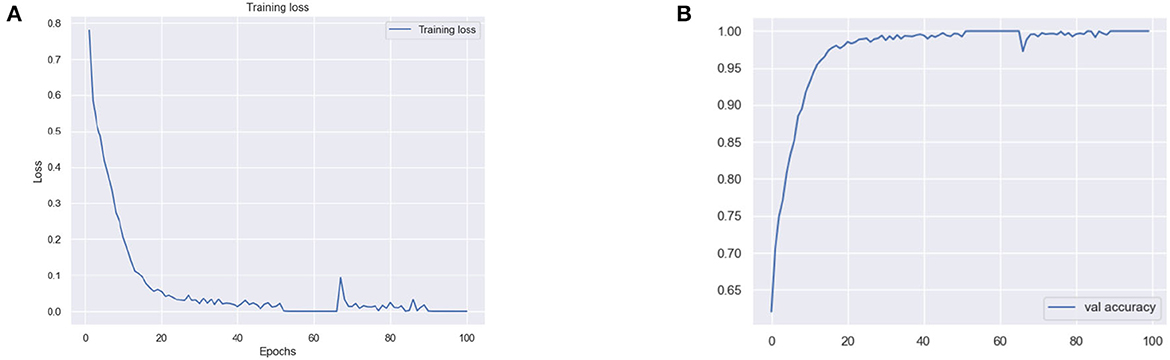
Figure 6. (A) Training loss and (B) validation (val) accuracy of the VGG 19 model with additional layer and freezing the blocks.
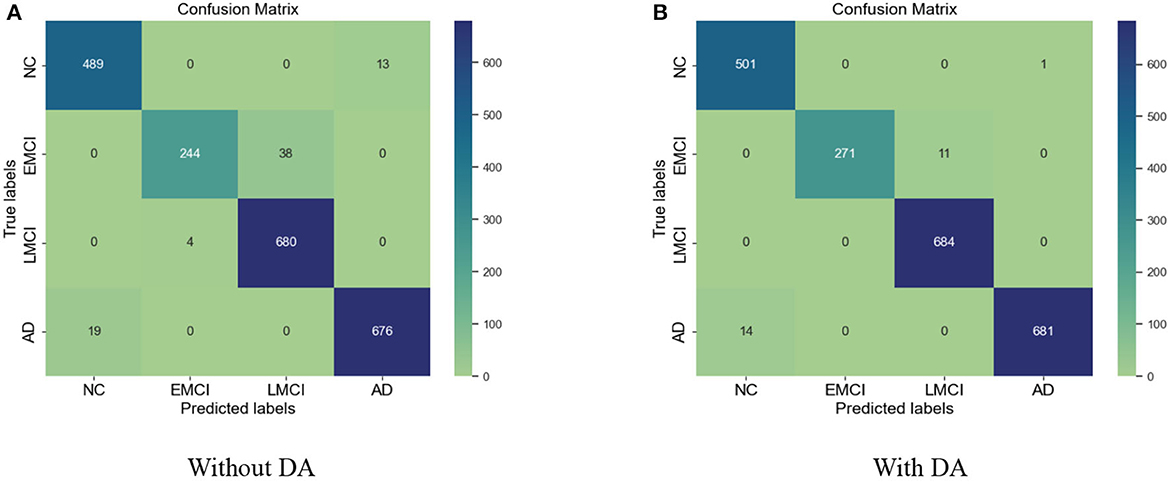
Figure 7. (A) Confusion matrix without data augmentation (DA). (B) Confusion matrix with DA for VGG 19.
6. Conclusion
This study presents a transfer learning-based approach to diagnose various stages of AD. We intend to automate the detection of various stages of Alzheimer's to ease the burden on the healthcare system. We use the pre-trained VGG architectures, i.e., VGG 16 and VGG 19, and propose a layer-wise transfer learning while adding layers and step-wise freezing the blocks in the pre-trained architecture. We leverage the pre-trained convolutional base, fine-tune the model for our 4-way classification on MRI images, and obtain state-of-the-art results in the target domain. The proposed layer-wise transfer learning significantly improves the framework's performance, where we also resolve the challenges of class imbalance and small data samples. The comparison with existing methods demonstrates that the proposed framework is superior in terms of accuracy and prediction. We achieved an accuracy of 97.89% for the proposed 4-way classification.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
RK contributes to the conceptualization, methodology, data curation, and writing of the original draft preparation with software and validation process. All authors also conceived in the process and approved the manuscript for submission.
Funding
This work was funded by the National Natural Science Foundation of China (NSFC62272419 and 11871438), the Natural Science Foundation of Zhejiang Province ZJNSFLZ22F020010, and Zhejiang Normal University Research Fund ZC304022915.
Acknowledgments
Data collection and sharing for this project was funded by the ADNI database (National Institutes of Health Grant U01 AG024904) and Department of Defense (DOD) ADNI (award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acharya, H., Mehta, R., and Singh, D. K. (2021). “Alzheimer disease classification using transfer learning,” in 2021 5th International Conference on Computing Methodologies and Communication (ICCMC) (Erode: IEEE), 1503–1508.
Adami, R., Pagano, J., Colombo, M., Platonova, N., Recchia, D., Chiaramonte, R., et al. (2018). Reduction of movement in neurological diseases: effects on neural stem cells characteristics. Front. Neurosci. 12, 336. doi: 10.3389/fnins.2018.00336
Aderghal, K., Khvostikov, A., Krylov, A., Benois-Pineau, J., Afdel, K., and Catheline, G. (2018). “Classification of Alzheimer disease on imaging modalities with deep cnns using cross-modal transfer learning,” in 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS) (Karlstad: IEEE), 345–350.
Afzal, S., Maqsood, M., Khan, U., Mehmood, I., Nawaz, H., Aadil, F., et al. (2021). Alzheimer disease detection techniques and methods: a review. Int. J. Interact. Multimedia Artif. Intell. 6, 5. doi: 10.9781/ijimai.2021.04.005
Alenezi, F. S., and Santosh, K. C. (2021). Geometric regularized hopfield neural network for medical image enhancement. Int. J. Biomed. Imaging 2021, 6664569. doi: 10.1155/2021/6664569
Basaia, S., Agosta, F., Wagner, L., Canu, E., Magnani, G., Santangelo, R., et al. (2019). Automated classification of Alzheimer's disease and mild cognitive impairment using a single mri and deep neural networks. Neuroimage Clin. 21, 101645. doi: 10.1016/j.nicl.2018.101645
Bi, X., Li, S., Xiao, B., Li, Y., Wang, G., and Ma, X. (2020). Computer aided Alzheimer's disease diagnosis by an unsupervised deep learning technology. Neurocomputing 392, 296–304. doi: 10.1016/j.neucom.2018.11.111
Chen, Z., Wang, Z., Zhao, M., Zhao, Q., Liang, X., Li, J., et al. (2022). A new classification network for diagnosing Alzheimer's disease in class-imbalance mri datasets. Front. Neurosci. 16, 807085. doi: 10.3389/fnins.2022.807085
Cheng, B., Liu, M., Shen, D., Li, Z., and Zhang, D. (2017). Multi-domain transfer learning for early diagnosis of Alzheimer's disease. Neuroinformatics 15, 115–132. doi: 10.1007/s12021-016-9318-5
Cheng, D., Chen, C., Yanyan, M., You, P., Huang, X., Gai, J., et al. (2022). Self-supervised learning for modal transfer of brain imaging. Front. Neurosci. 16, 920981. doi: 10.3389/fnins.2022.920981
Deng, X., Liu, Q., Deng, Y., and Mahadevan, S. (2016). An improved method to construct basic probability assignment based on the confusion matrix for classification problem. Inf. Sci. 340, 250–261. doi: 10.1016/j.ins.2016.01.033
El-Hasnony, I. M., Elzeki, O. M., Alshehri, A., and Salem, H. (2022). Multi-label active learning-based machine learning model for heart disease prediction. Sensors 22, 1184. doi: 10.3390/s22031184
Farooq, A., Anwar, S., Awais, M., and Rehman, S. (2017). “A deep cnn based multi-class classification of Alzheimer's disease using MRI,” in 2017 IEEE International Conference on Imaging Systems and Techniques (IST) (Beijing: IEEE), 1–6.
Glozman, T., Solomon, J., Pestilli, F., Guibas, L., Initiative, A. D. N., et al. (2017). Shape-attributes of brain structures as biomarkers for Alzheimer's disease. J. Alzheimers Dis. 56, 287–295. doi: 10.3233/JAD-160900
Gupta, A., Ayhan, M., and Maida, A. (2013). “Natural image bases to represent neuroimaging data,” in International Conference on machiNE Learning (PMLR), 987–994.
Hampel, H., Cummings, J., Blennow, K., Gao, P., Jack, C. R., and Vergallo, A. (2021). Developing the atx (n) classification for use across the Alzheimer disease continuum. Nat. Rev. Neurol. 17, 580–589. doi: 10.1038/s41582-021-00520-w
Han, H., Li, X., Gan, J. Q., Yu, H., Wang, H., Initiative, A. D. N., et al. (2022). Biomarkers derived from alterations in overlapping community structure of resting-state brain functional networks for detecting Alzheimer's disease. Neuroscience 484, 38–52. doi: 10.1016/j.neuroscience.2021.12.031
Hao, X., Bao, Y., Guo, Y., Yu, M., Zhang, D., Risacher, S. L., et al. (2020). Multi-modal neuroimaging feature selection with consistent metric constraint for diagnosis of Alzheimer's disease. Med. Image Anal. 60, 101625. doi: 10.1016/j.media.2019.101625
Hosseini-Asl, E., Ghazal, M., Mahmoud, A. H., Aslantas, A., Shalaby, A. M., Casanova, M. F., et al. (2018). Alzheimer's disease diagnostics by a 3d deeply supervised adaptable convolutional network. Front. Biosci. 23, 584–596. doi: 10.2741/4606
Huang, J., Chen, S., Hu, L., Niu, H., Sun, Q., Li, W., et al. (2018). Mitoferrin-1 is involved in the progression of Alzheimer's disease through targeting mitochondrial iron metabolism in a caenorhabditis elegans model of Alzheimer's disease. Neuroscience 385, 90–101. doi: 10.1016/j.neuroscience.2018.06.011
Islam, J., and Zhang, Y. (2018). Brain mri analysis for Alzheimer's disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Inform. 5, 1–14. doi: 10.1186/s40708-018-0080-3
Jain, R., Jain, N., Aggarwal, A., and Hemanth, D. J. (2019). Convolutional neural network based Alzheimer's disease classification from magnetic resonance brain images. Cogn. Syst. Res. 57, 147–159. doi: 10.1016/j.cogsys.2018.12.015
Jo, T., Nho, K., and Saykin, A. J. (2019). Deep learning in Alzheimer's disease: diagnostic classification and prognostic prediction using neuroimaging data. Front. Aging Neurosci. 11, 220. doi: 10.3389/fnagi.2019.00220
Khan, N. M., Abraham, N., and Hon, M. (2019). Transfer learning with intelligent training data selection for prediction of Alzheimer's disease. IEEE Access 7, 72726–72735. doi: 10.1109/ACCESS.2019.2920448
Khan, R., Qaisar, Z. H., Mehmood, A., Ali, G., Alkhalifah, T., Alturise, F., et al. (2022). A practical multiclass classification network for the diagnosis of Alzheimer's disease. Appl. Sci. 12, 6507. doi: 10.3390/app12136507
Li, W., Zhao, Y., Chen, X., Xiao, Y., and Qin, Y. (2018). Detecting Alzheimer's disease on small dataset: a knowledge transfer perspective. IEEE J. Biomed. Health Inform. 23, 1234–1242. doi: 10.1109/JBHI.2018.2839771
Liang, S., and Gu, Y. (2021). Computer-aided diagnosis of Alzheimer's disease through weak supervision deep learning framework with attention mechanism. Sensors 21, 220. doi: 10.3390/s21010220
Lin, W., Tong, T., Gao, Q., Guo, D., Du, X., Yang, Y., et al. (2018). Convolutional neural networks-based mri image analysis for the Alzheimer's disease prediction from mild cognitive impairment. Front. Neurosci. 12, 777. doi: 10.3389/fnins.2018.00777
Liu, S., Liu, S., Cai, W., Che, H., Pujol, S., Kikinis, R., et al. (2014). Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer's disease. IEEE Trans. Biomed. Eng. 62, 1132–1140. doi: 10.1109/TBME.2014.2372011
Lundervold, A. S., and Lundervold, A. (2019). An overview of deep learning in medical imaging focusing on mri. Zeitschrift für Medizinische Physik 29, 102–127. doi: 10.1016/j.zemedi.2018.11.002
Masdeu, J. C., Zubieta, J. L., and Arbizu, J. (2005). Neuroimaging as a marker of the onset and progression of Alzheimer's disease. J. Neurol Sci. 236, 55–64. doi: 10.1016/j.jns.2005.05.001
Mehmood, A., Maqsood, M., Bashir, M., and Shuyuan, Y. (2020). A deep siamese convolution neural network for multi-class classification of Alzheimer disease. Brain Sci. 10. doi: 10.3390/brainsci10020084
Mehmood, A., Yang, S., Feng, Z., Wang, M., Ahmad, A. S., Khan, R., et al. (2021). A transfer learning approach for early diagnosis of Alzheimer's disease on mri images. Neuroscience 460, 43–52. doi: 10.1016/j.neuroscience.2021.01.002
Murugan, S., Venkatesan, C., Sumithra, M., Gao, X.-Z., Elakkiya, B., Akila, M., et al. (2021). Demnet: a deep learning model for early diagnosis of Alzheimer diseases and dementia from mr images. IEEE Access 9, 90319–90329. doi: 10.1109/ACCESS.2021.3090474
Naz, S., Ashraf, A., and Zaib, A. (2022). Transfer learning using freeze features for Alzheimer neurological disorder detection using adni dataset. Multimedia Syst. 28, 85–94. doi: 10.1007/s00530-021-00797-3
Palmer, W. C., Park, S. M., and Levendovszky, S. R. (2021). Brain state transition analysis using ultra-fast fmri differentiates MCI from cognitively normal controls. Front. Neurosci. 16, 975305. doi: 10.3389/fnins.2022.975305
Pan, D., Zeng, A., Jia, L., Huang, Y., Frizzell, T., and Song, X. (2020). Early detection of Alzheimer's disease using magnetic resonance imaging: a novel approach combining convolutional neural networks and ensemble learning. Front. Neurosci. 14, 259. doi: 10.3389/fnins.2020.00259
Payan, A., and Montana, G. (2015). Predicting Alzheimer's disease - a neuroimaging study with 3d convolutional neural networks. ArXiv, abs/1502.02506. doi: 10.48550/arXiv.1502.02506
Petot, G. J., and Friedland, R. P. (2004). Lipids, diet and Alzheimer disease: an extended summary. J. Neurol Sci. 226, 31–33. doi: 10.1016/j.jns.2004.09.007
Prince, M. J., Wimo, A., Guerchet, M., Ali, G.-C., Wu, Y.-T., and Prina, M. A. (2015). World Alzheimer Report 2015 - The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends. Available online at: https://www.alzint.org/u/WorldAlzheimerReport2015.pdf
Ramzan, F., Khan, M. U. G., Rehmat, A., Iqbal, S., Saba, T., Rehman, A., et al. (2020). A deep learning approach for automated diagnosis and multi-class classification of Alzheimer's disease stages using resting-state fmri and residual neural networks. J. Med. Syst. 44, 1–16. doi: 10.1007/s10916-019-1475-2
Razzak, I., Naz, S., Ashraf, A., Khalifa, F., Bouadjenek, M. R., and Mumtaz, S. (2022). Mutliresolutional ensemble partialnet for Alzheimer detection using magnetic resonance imaging data. Int. J. Intell. Syst. 37, 6613–6630. doi: 10.1002/int.22856
Shams, M. Y., Elzeki, O. M., Abouelmagd, L. M., Hassanien, A. E., Abd Elfattah, M., and Salem, H. (2021). Hana: a healthy artificial nutrition analysis model during covid-19 pandemic. Comput. Biol. Med. 135, 104606. doi: 10.1016/j.compbiomed.2021.104606
Shi, J., Zheng, X., Li, Y., Zhang, Q., and Ying, S. (2017). Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer's disease. IEEE J. Biomed. Health Inform. 22, 173–183. doi: 10.1109/JBHI.2017.2655720
Sørensen, L., Igel, C., Pai, A., Balas, I., Anker, C., Lillholm, M., et al. (2017). Differential diagnosis of mild cognitive impairment and Alzheimer's disease using structural mri cortical thickness, hippocampal shape, hippocampal texture, and volumetry. Neuroimage Clin. 13, 470–482. doi: 10.1016/j.nicl.2016.11.025
Sperling, R. A., Aisen, P. S., Beckett, L. A., Bennett, D. A., Craft, S., Fagan, A. M., et al. (2011). Toward defining the preclinical stages of Alzheimer's disease: recommendations from the national institute on aging-Alzheimer's association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 7, 280–292. doi: 10.1016/j.jalz.2011.03.003
Suk, H.-I., Lee, S.-W., and Shen, D. (2016). Deep sparse multi-task learning for feature selection in Alzheimer's disease diagnosis. Brain Struct. Funct. 221, 2569–2587. doi: 10.1007/s00429-015-1059-y
Tajbakhsh, N., Shin, J. Y., Gurudu, S. R., Hurst, R. T., Kendall, C. B., Gotway, M. B., et al. (2016). Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging 35, 1299–1312. doi: 10.1109/TMI.2016.2535302
Tanveer, M., Richhariya, B., Khan, R. U., Rashid, A. H., Khanna, P., Prasad, M., et al. (2020). Machine learning techniques for the diagnosis of Alzheimer's disease: a review. ACM Trans. Multimedia Comput. Commun. Appl. 16, 1–35. doi: 10.1145/3344998
Tong, T., Gray, K., Gao, Q., Chen, L., Rueckert, D., Initiative, A. D. N., et al. (2017). Multi-modal classification of Alzheimer's disease using nonlinear graph fusion. Pattern Recognit. 63, 171–181. doi: 10.1016/j.patcog.2016.10.009
Vecchio, F., Miraglia, F., Alù, F., Menna, M., Judica, E., Cotelli, M. S., et al. (2020). Classification of Alzheimer's disease with respect to physiological aging with innovative eeg biomarkers in a machine learning implementation. J. Alzheimers Dis. 75, 1253–1261. doi: 10.3233/JAD-200171
Wang, S., Shen, Y., Chen, W., Xiao, T., and Hu, J. (2017). “Automatic recognition of mild cognitive impairment from MRI images using expedited convolutional neural networks,” in International Conference on Artificial Neural Networks (Springer), 373–380.
Witten, I. H., and Frank, E. (2002). Data mining: practical machine learning tools and techniques with java implementations. ACM Sigmod Rec. 31, 76–77. doi: 10.1145/507338.507355
Xia, Z., Yue, G., Xu, Y., Feng, C., Yang, M., Wang, T., et al. (2020). “A novel end-to-end hybrid network for Alzheimer's disease detection using 3D CNN and 3D CLSTM,” in 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) (Iowa City, IA: IEEE), 1–4.
Zhang, F., Tian, S., Chen, S., Ma, Y., Li, X., and Guo, X. (2019). Voxel-based morphometry: improving the diagnosis of Alzheimer's disease based on an extreme learning machine method from the adni cohort. Neuroscience 414, 273–279. doi: 10.1016/j.neuroscience.2019.05.014
Keywords: Alzheimer's disease, multiclass classification, deep learning, MRI, early diagnosis of AD
Citation: Khan R, Akbar S, Mehmood A, Shahid F, Munir K, Ilyas N, Asif M and Zheng Z (2023) A transfer learning approach for multiclass classification of Alzheimer's disease using MRI images. Front. Neurosci. 16:1050777. doi: 10.3389/fnins.2022.1050777
Received: 22 September 2022; Accepted: 05 December 2022;
Published: 09 January 2023.
Edited by:
Fahmi Khalifa, Mansoura University, EgyptReviewed by:
Nuwan Madusanka, Inje University, South KoreaSubrata Bhattacharjee, Inje University, South Korea
Copyright © 2023 Khan, Akbar, Mehmood, Shahid, Munir, Ilyas, Asif and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rizwan Khan,  aW1yaXp2YW5raGFuQGdtYWlsLmNvbQ==
aW1yaXp2YW5raGFuQGdtYWlsLmNvbQ==
 Rizwan Khan
Rizwan Khan Saeed Akbar2
Saeed Akbar2 Atif Mehmood
Atif Mehmood M. Asif
M. Asif