- 1Department of Neurobiology, University of Chicago, Chicago, IL, United States
- 2University of Chicago Neuroscience Institute, Chicago, IL, United States
- 3Committee on Development, Regeneration, and Stem Cell Biology, University of Chicago, Chicago, IL, United States
- 4Committee on Neurobiology, University of Chicago, Chicago, IL, United States
A remarkable diversity of cell types characterizes every animal nervous system. Previous studies provided important insights into how neurons commit to a particular fate, migrate to the right place and form precise axodendritic patterns. However, the mechanisms controlling later steps of neuronal development remain poorly understood. Hox proteins represent a conserved family of homeodomain transcription factors with well-established roles in anterior-posterior (A-P) patterning and the early steps of nervous system development, including progenitor cell specification, neuronal migration, cell survival, axon guidance and dendrite morphogenesis. This review highlights recent studies in Caenorhabditis elegans, Drosophila melanogaster and mice that suggest new roles for Hox proteins in processes occurring during later steps of neuronal development, such as synapse formation and acquisition of neuronal terminal identity features (e.g., expression of ion channels, neurotransmitter receptors, and neuropeptides). Moreover, we focus on exciting findings suggesting Hox proteins are required to maintain synaptic structures and neuronal terminal identity during post-embryonic life. Altogether, these studies, in three model systems, support the hypothesis that certain Hox proteins are continuously required, from early development throughout post-embryonic life, to build and maintain a functional nervous system, significantly expanding their functional repertoire beyond the control of early A-P patterning.
Introduction
Nervous system development is a multi-step process that generates a multitude of cell types. Dividing progenitor cells, or neural stem cells, will ultimately give rise to distinct types of neurons and glia. Newly born, post-mitotic neurons face a number of early challenges before participating into a functional neural circuit. They need to be molecularly specified, migrate to the right place, and acquire distinct axo-dendritic morphologies. Studies in all major model organisms suggest that these early steps of nervous system development are often controlled by Hox proteins, a conserved family of homeodomain transcription factors critical for anterior-posterior (A-P) patterning and formation of the animal body plan. The roles of Hox proteins during the early steps of nervous system development have been summarized in excellent reviews (Di Bonito et al., 2013a; Philippidou and Dasen, 2013; Parker and Krumlauf, 2020). Here, we highlight recent studies in Caenorhabditis elegans, Drosophila melanogaster, and mice that uncovered new roles for Hox in the last steps of neuronal development. We define as “last steps” the processes of synapse formation and acquisition of neuronal terminal identity features (e.g., expression of neurotransmitter [NT] receptors, neuropeptides, ion channels) because such processes represent the final events that lead to the establishment of a functional neural circuit. Perhaps more strikingly, a number of Hox proteins are continuously expressed in post-mitotic neurons of invertebrate and vertebrate nervous systems (discussed herein). Depletion of Hox gene activity at later stages of development and post-embryonic life supports the emerging hypothesis that Hox proteins are required not only to establish, but also maintain synaptic structures and terminal identity features. This review will focus on these exciting studies, offering new insights into the function of Hox proteins in the final steps of neuronal development.
The Role of Hox Genes in Late Stages of Caenorhabditis elegans Nervous System Development
The C. elegans genome contains six Hox genes. The anterior Hox (ceh-13/Lab/Hox1) together with the mid-body Hox (lin-39/Scr/Dfd/Hox3-5 and mab-5/Antp/Hox6-8) and posterior Hox (egl-5/AbdB/Hox9-13) genes were identified 30 years ago (Costa et al., 1988; Clark et al., 1993; Van Auken et al., 2000), whereas two additional posterior Hox genes (nob-1, php-3) were discovered in 2000 (Van Auken et al., 2000). C. elegans Hox genes are organized in three different sub-clusters located on Chromosome III (Figure 1A). Previous work revealed that C. elegans Hox genes control A-P patterning and the development of lateral epidermis and ventral ectoderm (Kenyon, 1986; Costa et al., 1988; Chisholm, 1991; Cowing and Kenyon, 1992; Clark et al., 1993; Wang et al., 1993; Wittmann et al., 1997; Brunschwig et al., 1999). Critical roles for Hox genes have also been described during the early steps of C. elegans nervous system development, that is, in the specification and survival of neuronal progenitors (Fixsen et al., 1985; Kenyon, 1986; Clark et al., 1993; Salser et al., 1993; Wang et al., 1993; Kalis et al., 2014), cell migration (Chisholm, 1991; Salser and Kenyon, 1992; Harris et al., 1996; Sym et al., 1999; Tihanyi et al., 2010; Wang et al., 2013), and neurite/axonal growth (Jia and Emmons, 2006; Zheng et al., 2015a). Below, we focus on recent studies uncovering functions for C. elegans Hox genes in processes occurring during later stages of neuronal development, such as synapse formation and acquisition of terminal identity.
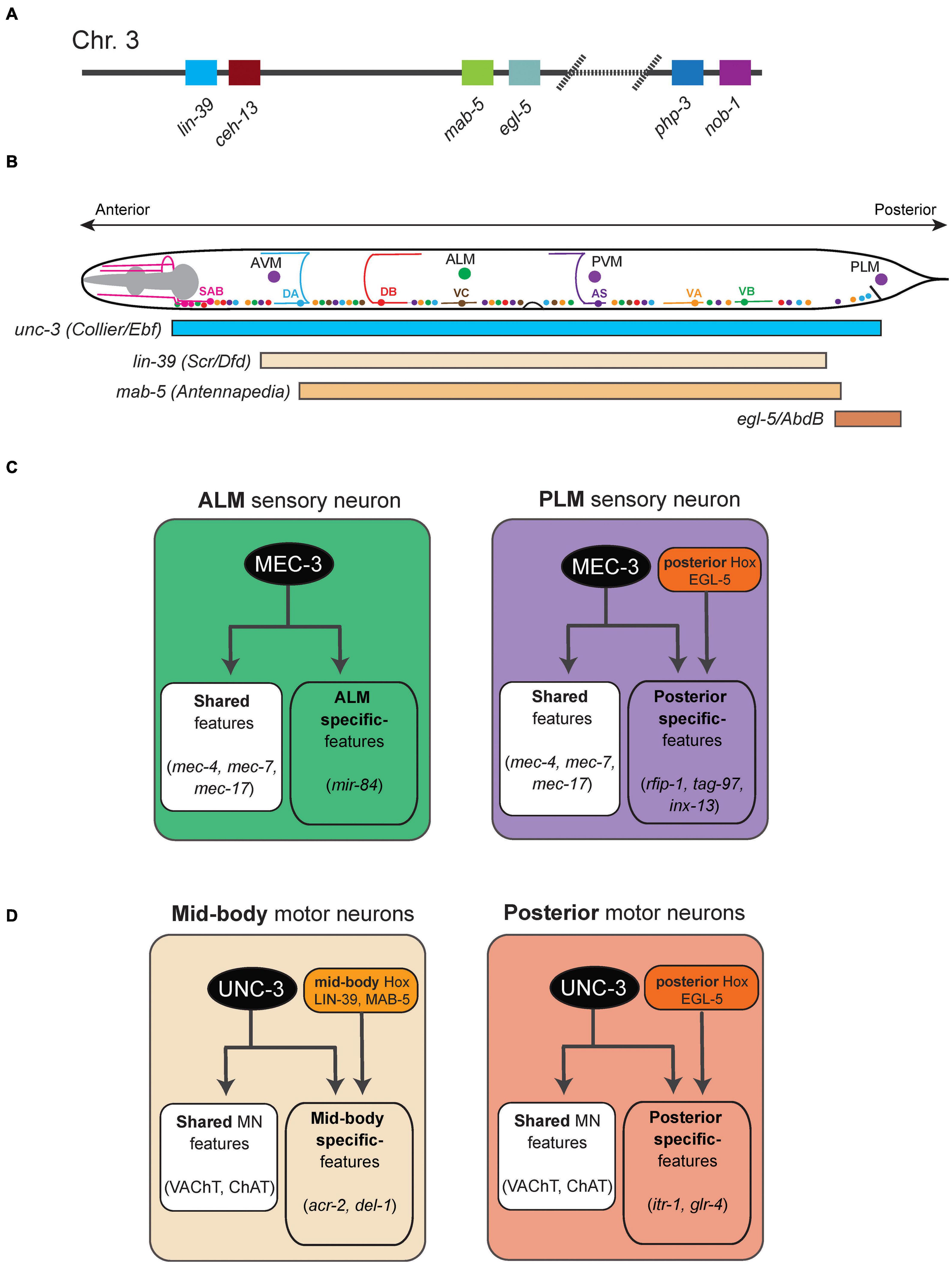
Figure 1. Hox gene functions in mechanosensory and motor neurons in C. elegans. (A) Schematic of the C. elegans Hox gene cluster. (B) Schematic showing the cell body location of mechanosensory neurons (AVM, ALM, PVM, PLM) and cholinergic MNs in the ventral nerve cord (SAB, DA, DB, VA, VB, VC, AS). The GABAergic MNs are not shown. (C) The terminal selector MEC-3 controls ALM and PLM terminal identity. The activity of the posterior Hox gene egl-5 diversifies PLM from ALM. Examples of terminal identity genes are shown in italics. (D) An intersectional strategy for the control of terminal identity of midbody (UNC-3, LIN-39, MAB-5) and posterior (UNC-3, EGL-5) MNs along the A-P axis of the C. elegans ventral nerve cord. See text for details.
Control of Synapse Formation/Maturation in Caenorhabditis elegans
The posterior Hox gene egl-5 is necessary for migration of the hermaphrodite-specific motor neuron (HSN) from the tail to the vulva, where it stimulates vulva muscle contraction resulting in egg laying (Baum et al., 1999). In posteriorly located sensory neurons, egl-5 controls neurite outgrowth (Zheng et al., 2015a). Besides its involvement in cell migration and neurite outgrowth, egl-5 also controls the wiring of the posteriorly located cholinergic motor neuron DA9 (Kratsios et al., 2017). In wild-type animals, the DA9 axon extends circumferentially to reach the dorsal body wall muscle and form en passant neuromuscular synapses. In egl-5 mutants, these DA9 synapses are generated at the “wrong” place; they are found more anteriorly when compared to wild-type animals, suggesting a synaptic specificity defect. Interestingly, split GFP reporter technology (GRASP) also revealed that the DA9 synaptic input (received by the AVG interneuron) fails to be maintained in adult egl-5 mutants, despite being properly established at earlier larval stages, indicating a critical role for egl-5 in synapse maintenance. Together, these findings suggest that the posterior Hox gene egl-5 controls both synaptic input and output of a posterior cholinergic motor neuron (DA9) in C. elegans.
Control of Neuronal Terminal Identity by Caenorhabditis elegans Hox Genes
Once post-mitotic neurons have established synapses, the function of every neuronal circuit critically relies on the ability of its constituent neurons to communicate with each other via neurotransmitters and/or neuropeptides, as well as to display neuron type-specific electrophysiological signatures. These function-defining features are determined by the expression of neurotransmitter (NT) biosynthesis proteins, ion channels, neuropeptides, NT receptors, and cell adhesion molecules. Genes coding for such proteins have been termed “terminal identity” genes (Hobert, 2008; Hobert and Kratsios, 2019), and are expressed continuously – from late developmental stages through adulthood – to determine the final (mature) identity and function of each neuron type. Recent studies on two different neuron types in C. elegans, namely the touch receptors and nerve cord motor neurons, revealed a new role for Hox genes in the control of neuronal terminal identity (Table 1). We highlight these studies below.
Hox Genes Control Terminal Identity Features of Caenorhabditis elegans Touch Receptor Neurons
In C. elegans, there are six touch receptor neurons (TRNs) mediating sensory responses to light touch. TRNs are classified into four subtypes: (a) bilaterally symmetrical pairs of ALM and PLM neurons are located at the midbody and tail region, respectively and (b) single AVM and PVM neurons are located in the midbody (Figure 1B). ALM and PLM are born embryonically, while AVM and PVM are generated post-embryonically. TRNs synapse onto and provide input to command interneurons (PVC, AVB, AVD, AVA), which stimulate downstream motor neurons, thus generating touch reflex responses. Early specification and differentiation of TRNs have been well investigated (Bounoutas and Chalfie, 2007), but how each TRN subtype acquires its unique terminal identity remains poorly understood.
At the behavioral level, animals lacking egl-5 (posterior Hox) gene activity are touch-insensitive at the tail, suggesting defects in the development of the posteriorly located PLM neuron (Chisholm, 1991). Later studies indeed demonstrated that egl-5 is necessary for PLM terminal identity (Toker et al., 2003; Zheng et al., 2015a; Figure 1C). In addition to egl-5, two other poster Hox genes (nob-1, php-3) control PLM development; nob-1 is necessary for the generation of PLM precursors, whereas php-3 together with egl-5 diversifies PLM from its more anteriorly located counterpart, the ALM neuron. Lastly, egl-5 controls PLM morphological characteristics, such as neurite length, by repressing anterior Hox genes (lin-39, mab-5) and TALE cofactors (Zheng et al., 2015a). The case of egl-5 highlights a recurring theme of Hox gene action across model systems, that is, Hox genes are required for various facets of development of a specific neuron type.
In the more anteriorly located ALM neurons, the anterior Hox gene ceh-13 regulates ALM terminal identity, as evidenced by reduced expression of a handful of terminal identity genes (mec-4, mec-7, mec-17, mec-18) in ceh-13 mutant animals. Mechanistically, these studies proposed that CEH-13 and EGL-5 function as transcriptional guarantors by controlling the levels of expression of the terminal selector gene mec-3, which in turn is required for terminal identity of both ALM and PLM neurons (Figure 1C; Zheng et al., 2015a,b). CEH-13 and EGL-5 increase the probability of mec-3 transcriptional activation by the POU-homeodomain transcription factor UNC-86 via the same Hox/Pbx binding site in ALM and PLM neurons respectively. This molecular mechanism ensures robustness of TRN terminal differentiation.
A multifaceted role of Hox genes is evident during the development of the C. elegans touch-reflex circuit: (a) Hox genes are involved in both early (e.g., generation of TRN precursor cells) and late steps of TRN development (e.g., terminal identity). (b) All six C. elegans Hox genes affect TRN development in various ways: ceh-13 regulates ALM terminal identity; lin-39 and mab-5 regulate the migration of AVM/PVM precursor; egl-5 and php-3 regulate PLM terminal identity; nob-1 is necessary for the generation of PLM precursors. (c) Although it remains mechanistically unclear how they control TRN-specific terminal identity genes (e.g., NT receptors, ion channels, neuropeptides), two Hox proteins (CEH-13, EGL-5) appear to act directly as transcriptional guarantors of mec-3, the terminal selector for all C. elegans TRNs. (d) Intriguingly, Hox genes control the development of neurons at different layers (sensory, interneuron) of the touch-reflex circuit. That is, sensory TRN terminal identity requires Hox gene function, whereas the identity of the PVC command interneurons (which receive sensory input from the PLM touch receptors) requires egl-5 gene activity (Chisholm, 1991; Zheng et al., 2015a).
Hox Genes Control Terminal Identity Features of Ventral Nerve Cord Motor Neurons
Similar to the touch receptor studies described above, the availability of terminal identity markers for ventral nerve cord motor neurons (MNs) in C. elegans has critically advanced our mechanistic understanding of Hox gene function in the nervous system. Nine distinct classes of MNs are found in the nerve cord of C. elegans hermaphrodite animals. Based on neurotransmitter usage, they can be classified into two categories: cholinergic (SAB, DA, DB, VA, VB, AS, VC) and GABAergic (DD, VD) MNs (Figure 1B). The SAB, DA, DB, and DD classes are generated embryonically, whereas the VA, VB, VC, VD, and AS neurons are generated post-embryonically (Von Stetina et al., 2006). The terminal identity of most cholinergic MN classes in the nerve cord (SAB, DA, DB, VA, VB, AS) critically depends on the terminal selector UNC-3, member of the conserved family of Collier/Olf/Ebf(COE) family of TFs (Prasad et al., 1998, 2008; Kratsios et al., 2011, 2015). Mechanistically, UNC-3 binds directly to the cis-regulatory region of terminal identity genes (e.g., acetylcholine [ACh] biosynthesis proteins, ion channels, neuropeptides) and activates their transcription. The homeodomain TF UNC-30 (PITX) acts in an analogous manner in GABAergic (DD, VD) MNs (Jin et al., 1994; Eastman et al., 1999).
In the context of both cholinergic and GABAergic MNs, recent work demonstrated that Hox genes act as cofactors of terminal selectors (Kratsios et al., 2017; Feng et al., 2020). In GABAergic MNs, the mid-body Hox genes lin-39 and mab-5 collaborate with unc-30 to control terminal identity gene expression. In cholinergic MNs, lin-39 and mab-5 collaborate with unc-3 to activate expression of several terminal identity genes (unc-129, del-1, acr-2, dbl-1, unc-77, slo-2) (Figure 1D). Like UNC-3, chromatin immunoprecipitation experiments suggest that LIN-39 and MAB-5 act directly (Kratsios et al., 2017; Feng et al., 2020). Apart from this UNC-3 co-factor role, lin-39 is also the rate-limiting factor for ensuring cholinergic MN identity. In the absence of unc-3, LIN-39 no longer binds to the cis-regulatory region of cholinergic MN genes. Instead, it relocates and switches targets, resulting in ectopic activation of alternative identity genes (Feng et al., 2020). Hence, the terminal selector UNC-3 prevents a Hox transcriptional switch to safeguard cholinergic MN identity.
Are Hox genes required during adulthood to maintain terminal identity features and thereby ensure continuous functionality of individual neuron types? Inducible, protein depletion experiments using the auxin inducible degradation (AID) system demonstrated that the midbody Hox protein LIN-39 is required in adult life to maintain MN terminal identity features (Feng et al., 2020; Li et al., 2020). This finding was somewhat unexpected because Hox genes are mostly thought to act early during animal development. Additional work on Hox is needed in C. elegans and other model systems to rigorously test whether maintenance of neuronal terminal identity is a key feature of Hox gene function in the nervous system.
The organization of cholinergic MNs into distinct subtypes along the A-P axis also offers an opportunity to dissect the molecular mechanisms underlying neuronal subtype identity. For example, the DA class of nine MNs can be subdivided into four subtypes based on cell boy position: DA1 is located at the anterior ganglion (retrovesicular ganglion [RVG]), DA2–7 are located at the VNC, and DA8–9 are found at the posterior ganglion (preanal ganglion [PAG]). In addition to their position, cholinergic MN subtypes do show distinct connectivity features and expression profiles of terminal identity genes (Kratsios et al., 2017). Hox genes control cholinergic MN subtype identity along the A-P axis of the C. elegans nervous system via an intersectional strategy that involves the terminal selector UNC-3 (Kratsios et al., 2017). For example, UNC-3 is expressed in all 9 DA neurons, but collaborates with the mid-body Hox genes lin-39 and mab-5 in mid-body DA2-7 neurons to control their terminal identity (Figure 1D). Similarly, UNC-3 and the posterior Hox gene egl-5 determine posterior MN (DA9) terminal identity (Figure 1D). Although the molecular mechanism of egl-5 activity in posterior MNs is unknown, biochemical evidence suggests that LIN-39 – like UNC-3 – acts directly by binding on the cis-regulatory region of terminal identity genes. This direct mode of regulation further extends to intermediary TFs (cfi-1/Arid3a, bnc-1/Bnc1/2) responsible for MN subtype identity (Kerk et al., 2017; Li et al., 2020).
The role of the anterior Hox gene ceh-13 during C. elegans neuronal terminal differentiation is largely elusive, partly due to the early larval lethality of ceh-13 mutants (Brunschwig et al., 1999). A recent study suggested ceh-13 controls terminal identity features of GABAergic motor neurons (DD1, DD2) located in the anterior ganglion, but the underlying mechanisms remain unknown (Aquino-Nunez et al., 2020).
Posterior Hox Gene egl-5 Controls the Identity of Serotonergic and Dopaminergic Neurons
In addition to its role on posterior MNs, the posterior Hox gene egl-5 controls the terminal identity of two other neuron types. The hermaphrodite specific neurons (HSNs) partially lose their ability to produce serotonin in egl-5 mutants (Chisholm, 1991). Moreover, egl-5 acts in tail sensory neurons of the C. elegans male. Upon egl-5 genetic removal, these neurons do not adopt dopaminergic fate and cannot be induced to express dopamine (Lints and Emmons, 1999).
The Role of Hox Genes in Late Stages of Drosophila Nervous System Development
Eight Hox genes are embedded in the genome of the fruit fly Drosophila melanogaster: labial (lab), proboscipedia (pb), Deformed (Dfd), Sex combs reduced (Scr), Antennapedia (Antp), Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B) (Figure 2A). Hox genes were first discovered in Drosophila during the 20th century; genetic experiments identified mutants with dramatic phenotypes caused by homeotic transformations (e.g., legs instead of antennae in Antp mutants, duplication of thoracic segments in Ubx mutants) (Nusslein-Volhard and Wieschaus, 1980). Subsequent studies showed that the principles of Hox gene function and their role in establishing the body plan along the A-P axis are conserved across species.
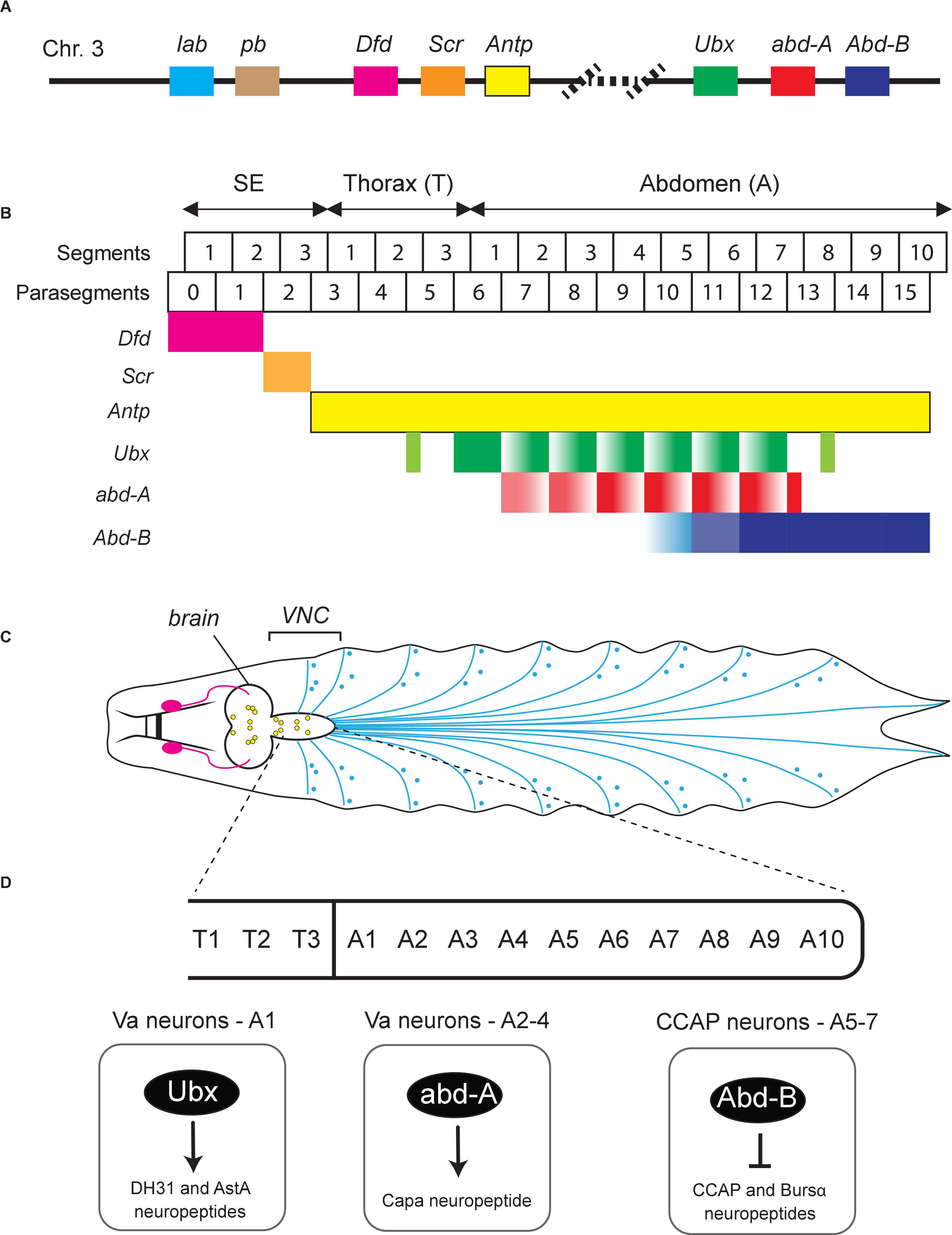
Figure 2. Hox gene expression in the Drosophila nerve cord. (A) Schematic of the Drosophila Hox gene cluster. (B) Six Hox genes (Dfd, Scr, Antp, Ubx, abd-A, Abd-B) are expressed in the Drosophila ventral nerve cord (VNC). Their expression pattern along the A-P axis is color-coded. SE, subesophagus; Th, thorax; Ab, abdomen. Adapted from Estacio-Gomez and Diaz-Benjumea (2014). (C) Schematic of the nervous system in Drosophila larvae showing the brain, VNC, and the peripheral nervous system at the late 3rd instar stage. Adapted from Sokabe et al. (2016). (D) Examples of Drosophila Hox genes that control terminal identity features of VNC neurons. See text for details.
Based on their chromosomal location, Drosophila Hox genes are organized in two gene complexes. The Antennapedia complex or Antp-C (consisting of lab, pb, Dfd, Scr, and Antp) specifies the anterior body plan from the head to the anterior thorax, while the Bithorax complex or Bx-C (consisting of Ubx, abd-A, and Abd-B) specifies the segments in posterior thorax and abdomen (Figure 2B). An important characteristic of Hox gene expression is their “temporal and spatial collinearity,” that is, the genes located at the 3′ end of a complex/cluster are expressed earlier and more rostrally than those residing at the 5′ end (Kmita and Duboule, 2003; Gaunt, 2015). This appears to be a highly conserved property of Hox genes and has been found in Drosophila and many other species (Gaunt, 2015).
During development, neurons in Drosophila arise from neuroblasts (NBs) located in three thoracic (T1–T3) and eight abdominal (A1–A8) segments of the ventral nerve cord (VNC) (Figures 2C,D). These NBs possess the potency to generate any neuron type, but they give rise to unique types of neuronal progenies depending on their location along the A-P axis. This spatial pattern of distinct neuronal types correlates with the combinatorial expression pattern of Hox genes along the A-P axis of the Drosophila body. During neurogenesis, Hox gene activity guides NBs to exit the cell cycle and promotes (or blocks) apoptosis, eventually leading to a spatial map of unique neuron types (Estacio-Gomez and Diaz-Benjumea, 2014; Gummalla et al., 2014). Moreover, Drosophila Hox genes control additional steps during early nervous system development, such as neuronal specification and axo-dendritic morphogenesis (Reichert and Bello, 2010; Baek et al., 2013; Estacio-Gomez and Diaz-Benjumea, 2014). We will discuss below recent studies suggesting Hox genes also control later steps of Drosophila nervous system development, such as synapse formation and neuronal terminal identity (Table 1).
Control of Synapse Formation/Maturation by Drosophila Hox Genes
Compelling evidence suggests that the Drosophila Hox gene Ubx controls neuromuscular synapse formation in the embryo (Hessinger et al., 2017). Interestingly, it does so by acting both in muscles and motor neurons. In abdominal segments A1–A7 of wild-type embryos, RP motor neurons innervate the ventrolateral muscles VL1-4. However, these motor neurons fail to make correct contacts with muscle VL1 in Ubx mutant embryos. Mechanistically, this study provides an intriguing link between Hox and Wnt signaling pathway – Wnt is instrumental for neuromuscular synapse formation across species (Klassen and Shen, 2007; Strochlic et al., 2012; Kerr et al., 2014). The authors proposed a model in which Ubx controls, in VL2 muscles, Wnt4 expression. Upon its secretion, Wnt4 is sensed by motor neurons (destined to innervate the VL1 muscles) via the Wnt receptor Fz-2. The Ubx-dependent Wnt4 signal from VL2 muscles triggers the repulsion of arriving growth cones belonging to motor neurons, hence these neurons innervate different muscles (VL1). Although the precise mechanism of Ubx function in these motor neurons remains obscure, rescue experiments clearly demonstrated that Ubx orchestrates the interaction between two cell types, muscles and motor neurons, to regulate the establishment of neuromuscular synapses in the fly embryo.
A second example of Hox gene involvement in Drosophila synapse formation comes from head motor neurons (MNs) that innervate the mouth hook elevator (MHE) and depressor (MHD) muscles, which coordinate the elevation and depression of the mouth hook (MH). The anterior Hox gene Dfd is expressed in a subset of MNs that specifically innervate the MHE (Friedrich et al., 2016). These Dfd-expressing MNs play a critical role in controlling the MH-dependent motor behaviors, including hatching at the end of embryogenesis and feeding in larval stages. In Dfd mutants, while the number of these MNs remains unchanged, they fail to extend axonal projections to their muscle targets, resulting in failure to hatch. Intriguingly, removing Dfd after the establishment of synaptic connections also results in impaired MH movements in larvae, suggesting Dfd is continuously required for the normal functions of these MNs (Friedrich et al., 2016). Genetically, Dfd acts upstream of a microtubule-organizing complex which is important for synapse stability even after their establishment. Dfd is continuously required to maintain the expression of Ankyrin2 extra large (Ank2-XL), which is known to be involved in determining the physical properties of synapses. Importantly, synaptic specificity is dependent on actions of Dfd both in motor neurons and muscles, reminiscent of the Ubx case discussed above (Hessinger et al., 2017). Altogether, the above studies on Drosophila Ubx and Dfd support the hypothesis that Hox genes, in addition to their well-documented roles in motor neuron specification, survival and axonal pathfinding (Baek et al., 2013; Philippidou and Dasen, 2013), also control the establishment and maintenance of neuromuscular synapses.
Control of Neuronal Terminal Identity by Drosophila Hox Genes
Much of our current understanding of Hox gene function in the Drosophila nervous system derives from studies on the abdominal leucokinergic neurons (ABLKs), which express the neuropeptide Leucokinin (Lk) and are often used as a model system to study both embryonic and post-embryonic neurogenesis (Estacio-Gomez and Diaz-Benjumea, 2014). During embryonic neurogenesis, the NB5-5 progenitor gives rise to 7 pairs of embryonic ABLKs (eABLKs), one in each of the first 7 abdominal segments (A1–7) of the VNC (Figures 2B,C). Lk is not initially expressed in the eABLKs when they are born but becomes detectable at later developmental stages (first instar larva). Later, additional post-embryonic ABLKs (pABLKs) are generated (third instar larva), and express Lk in pupal stages. The cell type-specific expression of the terminal identity gene encoding Lk is critically dependent on Bx-C (Ubx, abd-A, Abd-B) gene activity (Estacio-Gomez et al., 2013). Although Lk is a single Hox-dependent terminal identity gene, this study does suggest a later role for Hox in Drosophila neurons.
Hox genes are also expressed in the neuroectoderm at early development, but then become silenced when NBs delaminate and are reactivated at later stages in specific neurons. The posterior abdominal Hox genes, Ubx and abd-a, are expressed in post-mitotic eABLKs in the first instar larvae, where they are redundantly required for the expression of Lk. Moreover, when both Ubx and abd-A are knocked down specifically from early second instar larvae, it results in loss of Lk expression in late third instar larvae (Estacio-Gomez et al., 2013), suggesting maintenance of Lk in eABLKs relies on continuous expression of Ubx and abd-A. On the other hand, the other posterior Hox gene Abd-B represses Lk expression in non-ABLK cells during both embryonic and larval neurogenesis. Similarly, Abd-B is continuously required to maintain the repression of Lk, as removing Abd-B from first instar larvae results in de-repression of Lk and increased number of ABLKs in third star larvae and adults. Another study on Abd-B yielded similar results in the context of the crustacean cardioactive peptide (CCAP)-expressing neurons, which control ecdysis (Moris-Sanz et al., 2015). In A5–7 segments, the CCAP efferent neurons are defined by the expression of two terminal identity markers – the neuropeptides CCAP and Bursicon α (Bursα). Using a hypomorphic allele, the authors found that Abd-B represses CCAP/Bursα in early larvae. Hence, the precise onset of CCAP/Bursα expression critically relies on Abd-B-mediated repression (Figure 2D). Moreover, RNAi-induced knocked down of Abd-B in the first instar larve results in expression of the CCAP/Bursα neuropeptides, suggesting Abd-B is continuously required to maintain repression of CCAP/Bursα (Moris-Sanz et al., 2015).
The Hox genes Ubx and abd-A are also necessary to diversify the terminal identity of distinct neuropeptidergic neurons in the first four abdominal (A1–A4) segments of the fly VNC (Gabilondo et al., 2018). In A1, Ubx controls the identity of ventral abdominal (Va) neurons expressing the neuropeptides DH31 and AstA (Figure 2D). In A2–A4, abd-A controls the identity of distinct Va neurons expressing the neuropeptide Capa (Figure 2D). The diversification of these neuropeptidergic neurons is a product of regionalized, segment-specific Hox gene expression. For example, abd-A is not expressed in A1, whereas Ubx is expressed in all abdominal segments. Regionalized expression in the nervous system is a common feature between invertebrate and vertebrate Hox genes. In particular, the case of Va neuropeptidergic neurons where segment-specific Hox genes control segment-specific Va neuron terminal identity is reminiscent of a Hox-based strategy used by C. elegans MNs. In that case, mid-body (lin-39, mab-5) and posterior (egl-5) Hox genes control the terminal identity of mid-body and posterior MNs, respectively (Figure 1D; Kratsios et al., 2017).
Although the underlying mechanisms remain unknown, these studies strongly suggest that Hox genes can establish and maintain terminal identity features of post-mitotic neurons in Drosophila.
The Role of Hox Genes in Late Stages of Mouse Nervous System Development
During early vertebrate evolution, the single Hox gene cluster of vertebrate ancestors was duplicated twice, eventually giving rise to four clusters – HoxA, HoxB, HoxC, and HoxD in mammals (Soshnikova et al., 2013). In mice, these four clusters contain 39 Hox genes, which are further categorized into 13 paralog groups (PG) based on their relative position within the clusters and gene sequence (Figure 3A). The majority of these Hox genes are expressed in the mouse central nervous system (CNS) during development (Krumlauf et al., 1993; Briscoe and Wilkinson, 2004; Philippidou and Dasen, 2013).
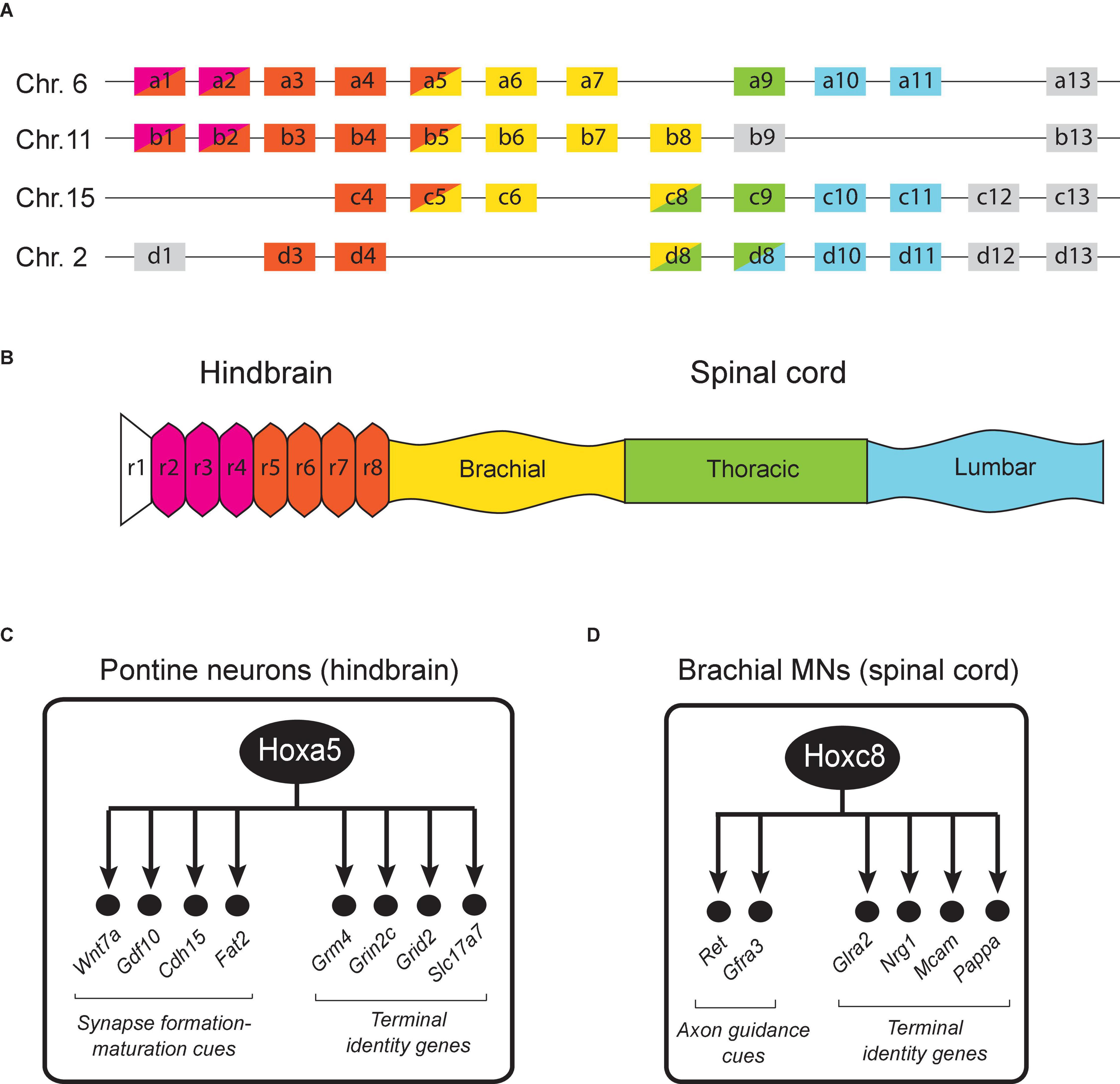
Figure 3. Hox gene expression in the mouse hindbrain and spinal cord. (A) The 39 Hox genes in mice are distributed in four clusters (a, b, c, d). (B) Region-specific Hox gene expression is shown along the rostrocaudal axis of the embryonic nervous system. Adapted from Philippidou and Dasen (2013). The dynamic nature of Hox gene expression is not illustrated for simplicity. (C) Schematic summary of putative Hoxa5 target genes in pontine neurons. See text for details. (D) Schematic summary of Hoxc8 target genes in brachial MNs. See text for details.
A large body of work in the mouse hindbrain and spinal cord has uncovered critical roles for Hox genes in defining segment identity and establishing spatial gene expression patterns necessary for neuronal differentiation during early embryogenesis (Narita and Rijli, 2009; Di Bonito et al., 2013a; Philippidou and Dasen, 2013; Parker and Krumlauf, 2020). These early Hox roles appear conserved in the zebrafish nervous system as well (Ghosh and Sagerstrom, 2018). The expression of Hox genes in the mouse hindbrain between embryonic day 7.5 (E7.5) to E9.5 appears strictly restricted within territories defined by rhombomere (transiently divided segments of the developing neural tube) boundaries (Figure 3B). Rhombomere boundaries create a series of anterior limits for Hox gene expression along the A-P axis. In the hindbrain, Hox1-2 genes have more anterior boundaries compared to Hox3-4 genes. On the other hand, Hox5-13 genes are mainly expressed in the spinal cord, which is posterior to the hindbrain. The overall map of Hox gene expression along the A-P axis therefore displays spatial collinearity (Figures 3A,B). With a number of exceptions (discussed below), most Hox studies in the mouse hindbrain and spinal cord have focused on early steps of neuronal development, and thereby uncovered crucial roles for Hox in progenitor and neuronal cell fate specification, cell migration, neuronal survival, as well as axo-dendritic growth and pathfinding (Narita and Rijli, 2009; Di Bonito et al., 2013a; Philippidou and Dasen, 2013; Parker and Krumlauf, 2020).
Although Hox gene expression is well documented in early embryonic stages, their expression in late embryonic and postnatal stages is poorly characterized. Interestingly, a number of studies in the mouse hindbrain showed that the segmental Hox gene expression pattern in postmitotic neurons is also maintained in late embryonic and early postnatal stages (Pasqualetti et al., 2007; Geisen et al., 2008; Di Bonito et al., 2013b; Karmakar et al., 2017; Lizen et al., 2017a). Two systematic expression studies on the 39 mouse Hox genes revealed that the majority of Hox genes remain expressed in the hindbrain after birth and until adulthood (Hutlet et al., 2016; Tomas-Roca et al., 2016). Hutlet et al. (2016) found that the 24 Hox genes that are normally active during early development of the hindbrain continue to be expressed during adulthood. Neuroanatomical localization analysis revealed that these Hox genes are still expressed in adult post-mitotic neurons derived from rhombomeres, with visible anterior boundaries restricting individual Hox genes along the A-P axis. This indicates that the spatial collinearity rule is also maintained in adult hindbrain. Intriguingly, transcripts of some Hox genes were also identified in more anterior regions (forebrain) where they are not expressed during embryogenesis, suggesting Hox gene neo-expression in the adult CNS. More specifically, Hoxb1, Hoxb3, Hoxb4, Hoxd3, and Hoxa5 transcripts were detected in both neocortex and the thalamus. Temporal analysis showed that their expression starts as early as the second postnatal week but becomes more robust only in the third postnatal week. In a separate study, Coughlan et al. have reported that the expression of Hox9-11 genes is maintained and remains robust in spinocerebellar neurons until P7 (Coughlan et al., 2019), that is weeks after neuronal progenitor specification occurs. Of note, analysis of HOX expression in human samples showed that 15 genes are expressed in the adult brain (Takahashi et al., 2004). As in the hindbrain, Hox gene expression in the mouse embryonic spinal cord has been detected in progenitor cells and postmitotic neurons (Dasen et al., 2003, 2005; Dasen, 2009; Sweeney et al., 2018; Baek et al., 2019). A recent study focused on the brachial domain of the spinal cord found that Hox4-8 expression is maintained in postmitotic neurons during early postnatal stages (Catela et al., 2021).
The maintained Hox expression in the mouse hindbrain and spinal cord prompts the question of what are the biological functions of mouse Hox genes in post-mitotic neurons during late developmental and postnatal stages? Below, we highlight studies on the role of mouse Hox genes in synapse formation/maturation and neuronal terminal identity; these late-occurring processes critically determine the functionality of neural circuits located in the hindbrain and spinal cord (Table 1).
Control of Synapse Formation/Maturation by Mouse Hox Genes
The expression of Hox genes in neuronal progenitors and postmitotic neurons necessitates the employment of conditional and temporally controlled gene inactivation strategies to discriminate between early and late Hox gene functions.
The first temporally controlled Hox gene inactivation study was conducted in the trigeminal system, which relays somatosensory stimuli (e.g., touch, pain) from the face to the cortex. A key structure for such relay is the principal trigeminal nucleus in the hindbrain. Oury et al. (2006) used a tamoxifen-inducible Cre/loxP strategy to inactivate Hoxa2 at different developmental stages in postmitotic neurons of the principal trigeminal nucleus. The authors found that late removal of Hoxa2 leads to topographic connectivity defects of these neurons. Consistently, Hoxa2 ectopic expression experiments suggested that maintained Hoxa2 expression is sufficient to direct topographic axon targeting and synaptic specificity defects, potentially implicating Hoxa2 in the regulation of molecules acting at the presynapse (Bechara et al., 2015; Gofflot and Lizen, 2018).
Apart from its role in the trigeminal system, Hoxa2 is involved in synaptic refinement of connectivity within the brainstem auditory circuit (Karmakar et al., 2017). Hoxa2 and Hoxb2 are expressed throughout embryonic and postnatal life (at least up to 2 months of age) in neurons of the anterior ventral cochlear nucleus (AVCN) (Narita and Rijli, 2009). In wild-type mice, glutamatergic neurons in the AVCN, called “Bushy cells,” receive a single axonal input from one spiral ganglion neuron that forms a unique and large synapse, the endbulb of Held (Gofflot and Lizen, 2018). In mice lacking Hoxa2 and Hoxb2 gene activity specifically in postmitotic AVCN Bushy cells, multiple receiving inputs were observed, suggesting an involvement for these Hox genes in synapse (endbulb of Held) elimination/maturation (Karmakar et al., 2017). Importantly, these connectivity defects resulted in behavioral defects (failure to discriminate two close pure-tone frequencies) (Karmakar et al., 2017). At the molecular level, a comparative transcriptomic analysis revealed Wnt3a and multiple cadherins (Cdh4, Cdh11, Cdh13, Cdh7) as downstream targets of HOXA2/HOXB2 in AVNC Bushy cells. Given the prominent role of WNT signaling and cadherins in synapse formation and maintenance (Dickins and Salinas, 2013; Basu et al., 2015), these downstream targets could at least partially explain the connectivity defects observed in Bushy cells of mice lacking Hoxa2 and Hoxb2.
In the mouse brainstem, Hoxa5 is continuously expressed from embryonic to adult stages (Lizen et al., 2017a), suggesting its involvement at different stages of neuronal development. To test whether Hoxa5 is functionally required in brainstem neurons, Lizen et al. (2017b) used an inducible Cre/loxP approach to inactivate Hoxa5 at postnatal days 1–4 (P1–4) and then conducted an unbiased transcriptomic (RNA-Seq) analysis. Because Hoxa5 expression is enriched in brainstem neurons that belong to the precerebellar system, called “pontine neurons,” it is likely this RNA-Seq approach primarily uncovered changes in RNA expression in these neurons. This study identified several genes with known roles in synapse formation and maturation as Hoxa5 targets, such as the secreted molecules Wnt7a and GDF10 (member of TGFβ superfamily) and the cell adhesion molecules Cdh15 and Fat2 (Gofflot and Lizen, 2018; Figure 3C). Consistent with these observations, a more recent study found that postmitotic Hoxa5 expression specifies pontine neuron connectivity (Maheshwari et al., 2020).
Similar to their roles in the C. elegans and Drosophila nervous systems, mouse Hox genes can affect neural circuit formation in various ways by acting at different stages. Besides controlling synapse formation and specificity, they can also regulate axonal pathfinding which eventually leads to a failure to establish a functional neural circuit. Supporting this possibility, several mouse studies have shown that correct expression of guidance cue receptors is often co-regulated by Hox genes (Oury et al., 2006; Geisen et al., 2008; Di Bonito et al., 2013a; Maheshwari et al., 2020). For example, in the precerebellar anterior extramural migrating stream, Hox5 genes repress the repulsive Netrin receptor Unc5b, while Hox2 genes positively regulate it (Di Meglio et al., 2013). Moreover, Hoxa2 is required for the expression of Slit receptor Robo3 in commissural neurons in the hindbrain and Robo2 in precerebellar pontine neurons (Di Bonito et al., 2013a).
In spinal cord circuits, several studies revealed connectivity defects upon Hox gene inactivation (Dasen et al., 2005, 2008; Catela et al., 2015, 2016; Baek et al., 2019). For example, Hox5 genes are required for proper connectivity of phrenic motor neurons to premotor interneurons and the diaphragm muscle (Philippidou et al., 2012; Vagnozzi et al., 2020). The phrenic motor neurons express a unique combination Hox5-dependent cell adhesion molecules of the Cadherin (Cdh) family (Vagnozzi et al., 2020), which is known to control neuronal connectivity across model systems. Importantly, early or late genetic removal of Hox5 in mice affects diaphragm innervation, suggesting a continuous Hox requirement for establishment and maintenance of neuronal wiring (Philippidou et al., 2012).
Conditional inactivation studies of Hoxc8 also revealed striking connectivity defects in spinal neurons. That is, Hoxc8 removal specifically in sensory neurons affects sensory-motor connectivity (Shin et al., 2020), whereas motor neuron-specific depletion of Hoxc8 affects forelimb muscle innervation (Catela et al., 2016). Mechanistically, Hoxc8 controls expression of axon molecules Ret and Gfrα to establish proper muscle innervation (Figure 3D). Besides their role in axon guidance, many of the aforementioned axon guidance molecules are also required for synapse formation and plasticity. This leads to the possibility that Hox genes may also maintain synaptic plasticity at post-natal stages, as suggested by the aforementioned Hoxa5 study in pontine neurons (Lizen et al., 2017b).
Control of Neuronal Terminal Identity by Mouse Hox Genes
Recent work suggests that mouse Hox genes, similar to their C. elegans and Drosophila counterparts, control terminal identity features (e.g., NT biosynthesis components, NT receptors, ion channels) of post-mitotic neurons. In the context of pontine neurons in the brainstem, Hoxa5 appears necessary for the maintained expression of genes encoding several glutamate receptor subunits (Grm4, Grin2c, Grid2), which are required for glutamatergic input by pyramidal cells (Lizen et al., 2017b). Moreover, Hoxa5 also ensures the maintained expression of Slc17a7 (VGLUT1), which is crucial for loading synaptic vesicles with glutamate – a key step for the synaptic output of pontine neurons onto granule cells of the cerebellum (Lizen et al., 2017b; Gofflot and Lizen, 2018; Figure 3C). In the mouse spinal cord, Hoxc8 is required for the induction and maintenance of several terminal identity genes (Nrg1, Mcam, Pappa) in motor neurons of the brachial region (Catela et al., 2021; Figure 3D). Interestingly, while these terminal differentiation genes require Hoxc8 for both initiation and maintenance of their expression, not all Hoxc8 target genes behave in the same way. In fact, the suite of Hoxc8 targets in brachial MNs is dynamic across different life stages. For example, the glycine receptor subunit alpha-2 (Glra2) appears significantly downregulated upon conditional knockout of Hoxc8 at postnatal day 8 (p8) but is unaffected upon Hoxc8 knockout at embryonic day 12 (e12). One possible explanation for this phenomenon is that Glra2 is redundantly regulated by additional transcription factors at early stages, whose expression fades away later on and Hoxc8-mediated regulation becomes necessary for maintenance. Although the underlying mechanisms remain elusive, these findings suggest that Hox genes are continuously required in the mouse nervous system to establish and maintain neuronal terminal identity features.
Conclusion
A large body of work has uncovered critical roles for Hox genes in the early steps of nervous system development, such as progenitor cell specification, neuronal migration, cell survival and axo-dendritic growth. This review highlights recent studies in C. elegans, Drosophila, and mice that identified later functions for Hox genes in post-mitotic neurons, such as the control of synapse formation/maturation and neuronal terminal identity. These studies strongly suggest that Hox proteins multitask over time within a neuronal lineage by acting at the level of progenitors and/or post-mitotic neurons. Precisely controlled, temporal inactivation of Hox gene activity is necessary to continue uncovering the breadth of Hox gene functions in the nervous system. The realization of this ambitious goal critically relies on inducible genetic approaches coupled with powerful transcriptomic, biochemical, and behavioral methods.
Author Contributions
WF and YL: literature search, writing — original draft, and review and editing. PK: supervision, literature search, writing — original draft, and review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Institute of Neurological Disorders and Stroke (NINDS) of the NIH (Award Numbers R01NS116365 and R21NS108505) to PK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the reviewers for their constructive comments. We apologize to colleagues for not including their work herein due to space constraints.
References
Aquino-Nunez, W., Mielko, Z. E., Dunn, T., Santorella, E. M., Hosea, C., Leitner, L., et al. (2020). cnd-1/NeuroD1 functions with the homeobox gene ceh-5/Vax2 and hox gene ceh-13/labial to specify aspects of RME and DD neuron fate in Caenorhabditis elegans. G3 10, 3071–3085. doi: 10.1534/g3.120.401515
Baek, M., Enriquez, J., and Mann, R. S. (2013). Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila. Development 140, 2027–2038. doi: 10.1242/dev.090902
Baek, M., Menon, V., Jessell, T. M., Hantman, A. W., and Dasen, J. S. (2019). Molecular logic of spinocerebellar tract neuron diversity and connectivity. Cell Rep. 27, 2620.e4–2635.e4. doi: 10.1016/j.celrep.2019.04.113
Basu, R., Taylor, M. R., and Williams, M. E. (2015). The classic cadherins in synaptic specificity. Cell Adh. Migr. 9, 193–201. doi: 10.1080/19336918.2014.1000072
Baum, P. D., Guenther, C., Frank, C. A., Pham, B. V., and Garriga, G. (1999). The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the Hsn motor neuron. Genes Dev. 13, 472–483. doi: 10.1101/gad.13.4.472
Bechara, A., Laumonnerie, C., Vilain, N., Kratochwil, C. F., Cankovic, V., Maiorano, N. A., et al. (2015). Hoxa2 selects barrelette neuron identity and connectivity in the mouse somatosensory brainstem. Cell Rep. 13, 783–797. doi: 10.1016/j.celrep.2015.09.031
Bounoutas, A., and Chalfie, M. (2007). Touch sensitivity in Caenorhabditis elegans. Pflugers. Arch. 454, 691–702. doi: 10.1007/s00424-006-0187-x
Briscoe, J., and Wilkinson, D. G. (2004). Establishing neuronal circuitry: Hox genes make the connection. Genes Dev. 18, 1643–1648. doi: 10.1101/gad.1227004
Brunschwig, K., Wittmann, C., Schnabel, R., Burglin, T. R., Tobler, H., and Muller, F. (1999). Anterior organization of the Caenorhabditis elegans embryo by the labial-like Hox gene ceh-13. Development 126, 1537–1546. doi: 10.1242/dev.126.7.1537
Catela, C., Shin, M. M., and Dasen, J. S. (2015). Assembly and function of spinal circuits for motor control. Annu. Rev. Cell Dev. Biol. 31, 669–698. doi: 10.1146/annurev-cellbio-100814-125155
Catela, C., Shin, M. M., Lee, D. H., Liu, J. P., and Dasen, J. S. (2016). Hox proteins coordinate motor neuron differentiation and connectivity programs through Ret/Gfralpha genes. Cell Rep. 14, 1901–1915. doi: 10.1016/j.celrep.2016.01.067
Catela, C., Weng, Y., Wen, K., Feng, W., and Kratsios, P. (2021). Control of spinal motor neuron terminal differentiation through sustained Hoxc8 gene activity. bioRxiv [Preprint]. doi: 10.1101/2021.05.26.445841v1
Chisholm, A. (1991). Control of cell fate in the tail region of C. elegans by the gene egl-5. Development 111, 921–932. doi: 10.1242/dev.111.4.921
Clark, S. G., Chisholm, A. D., and Horvitz, H. R. (1993). Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell 74, 43–55. doi: 10.1016/0092-8674(93)90293-y
Costa, M., Weir, M., Coulson, A., Sulston, J., and Kenyon, C. (1988). Posterior pattern formation in C. elegans involves position-specific expression of a gene containing a homeobox. Cell 55, 747–756. doi: 10.1016/0092-8674(88)90131-6
Coughlan, E., Garside, V. C., Wong, S. F. L., Liang, H., Kraus, D., Karmakar, K., et al. (2019). A hox code defines spinocerebellar neuron subtype regionalization. Cell Rep. 29, 2408.e4–2421.e4. doi: 10.1016/j.celrep.2019.10.048
Cowing, D. W., and Kenyon, C. (1992). Expression of the homeotic gene mab-5 during Caenorhabditis elegans embryogenesis. Development 116, 481–490. doi: 10.1242/dev.116.2.481
Dasen, J. S. (2009). Transcriptional networks in the early development of sensory-motor circuits. Curr. Top. Dev. Biol. 87, 119–148. doi: 10.1016/S0070-2153(09)01204-6
Dasen, J. S., De Camilli, A., Wang, B., Tucker, P. W., and Jessell, T. M. (2008). Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316. doi: 10.1016/j.cell.2008.06.019
Dasen, J. S., Liu, J. P., and Jessell, T. M. (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933. doi: 10.1038/nature02051
Dasen, J. S., Tice, B. C., Brenner-Morton, S., and Jessell, T. M. (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491. doi: 10.1016/j.cell.2005.09.009
Di Bonito, M., Glover, J. C., and Studer, M. (2013a). Hox genes and region-specific sensorimotor circuit formation in the hindbrain and spinal cord. Dev. Dyn. 242, 1348–1368. doi: 10.1002/dvdy.24055
Di Bonito, M., Narita, Y., Avallone, B., Sequino, L., Mancuso, M., Andolfi, G., et al. (2013b). Assembly of the auditory circuitry by a Hox genetic network in the mouse brainstem. PLoS Genet. 9:e1003249. doi: 10.1371/journal.pgen.1003249
Di Meglio, T., Kratochwil, C. F., Vilain, N., Loche, A., Vitobello, A., Yonehara, K., et al. (2013). Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science 339, 204–207. doi: 10.1126/science.1229326
Dickins, E. M., and Salinas, P. C. (2013). Wnts in action: from synapse formation to synaptic maintenance. Front. Cell Neurosci. 7:162. doi: 10.3389/fncel.2013.00162
Eastman, C., Horvitz, H. R., and Jin, Y. (1999). Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 19, 6225–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999
Estacio-Gomez, A., and Diaz-Benjumea, F. J. (2014). Roles of Hox genes in the patterning of the central nervous system of Drosophila. Fly 8, 26–32. doi: 10.4161/fly.27424
Estacio-Gomez, A., Moris-Sanz, M., Schafer, A. K., Perea, D., Herrero, P., and Diaz-Benjumea, F. J. (2013). Bithorax-complex genes sculpt the pattern of leucokinergic neurons in the Drosophila central nervous system. Development 140, 2139–2148. doi: 10.1242/dev.090423
Feng, W., Li, Y., Dao, P., Aburas, J., Islam, P., Elbaz, B., et al. (2020). A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life. eLife 9:e50065. doi: 10.7554/eLife.50065
Fixsen, W., Sternberg, P., Ellis, H., and Horvitz, R. (1985). Genes that affect cell fates during the development of Caenorhabditis elegans. Cold. Spring Harb. Symp. Quant. Biol. 50, 99–104. doi: 10.1101/sqb.1985.050.01.014
Friedrich, J., Sorge, S., Bujupi, F., Eichenlaub, M. P., Schulz, N. G., Wittbrodt, J., et al. (2016). Hox function is required for the development and maintenance of the drosophila feeding motor unit. Cell Rep. 14, 850–860. doi: 10.1016/j.celrep.2015.12.077
Gabilondo, H., Rubio-Ferrera, I., Losada-Perez, M., Del Saz, D., Leon, Y., Molina, I., et al. (2018). Segmentally homologous neurons acquire two different terminal neuropeptidergic fates in the Drosophila nervous system. PLoS One 13:e0194281. doi: 10.1371/journal.pone.0194281
Gaunt, S. J. (2015). The significance of Hox gene collinearity. Int. J. Dev. Biol. 59, 159–170. doi: 10.1387/ijdb.150223sg
Geisen, M. J., Di Meglio, T., Pasqualetti, M., Ducret, S., Brunet, J. F., Chedotal, A., et al. (2008). Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 6:e142. doi: 10.1371/journal.pbio.0060142
Ghosh, P., and Sagerstrom, C. G. (2018). Developing roles for Hox proteins in hindbrain gene regulatory networks. Int. J. Dev. Biol. 62, 767–774. doi: 10.1387/ijdb.180141cs
Gofflot, F., and Lizen, B. (2018). Emerging roles for HOX proteins in synaptogenesis. Int. J. Dev. Biol. 62, 807–818. doi: 10.1387/ijdb.180299fg
Gummalla, M., Galetti, S., Maeda, R. K., and Karch, F. (2014). Hox gene regulation in the central nervous system of Drosophila. Front. Cell Neurosci. 8:96. doi: 10.3389/fncel.2014.00096
Harris, J., Honigberg, L., Robinson, N., and Kenyon, C. (1996). Neuronal cell migration in C. elegans: regulation of Hox gene expression and cell position. Development 122, 3117–3131. doi: 10.1242/dev.122.10.3117
Hessinger, C., Technau, G. M., and Rogulja-Ortmann, A. (2017). The Drosophila Hox gene Ultrabithorax acts in both muscles and motoneurons to orchestrate formation of specific neuromuscular connections. Development 144, 139–150. doi: 10.1242/dev.143875
Hobert, O. (2008). Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. U.S.A. 105, 20067–20071. doi: 10.1073/pnas.0806070105
Hobert, O., and Kratsios, P. (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr. Opin. Neurobiol 56, 97–105. doi: 10.1016/j.conb.2018.12.006
Hutlet, B., Theys, N., Coste, C., Ahn, M. T., Doshishti-Agolli, K., Lizen, B., et al. (2016). Systematic expression analysis of Hox genes at adulthood reveals novel patterns in the central nervous system. Brain Struct. Funct. 221, 1223–1243. doi: 10.1007/s00429-014-0965-8
Jia, L., and Emmons, S. W. (2006). Genes that control ray sensory neuron axon development in the Caenorhabditis elegans male. Genetics 173, 1241–1258. doi: 10.1534/genetics.106.057000
Jin, Y., Hoskins, R., and Horvitz, H. R. (1994). Control of type-D GABAERGIC neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372, 780–783. doi: 10.1038/372780a0
Kalis, A. K., Kissiov, D. U., Kolenbrander, E. S., Palchick, Z., Raghavan, S., Tetreault, B. J., et al. (2014). Patterning of sexually dimorphic neurogenesis in the Caenorhabditis elegans ventral cord by Hox and Tale homeodomain transcription factors. Dev. Dyn. 243, 159–171. doi: 10.1002/dvdy.24064
Karmakar, K., Narita, Y., Fadok, J., Ducret, S., Loche, A., Kitazawa, T., et al. (2017). Hox2 genes are required for tonotopic map precision and sound discrimination in the mouse auditory brainstem. Cell Rep. 18, 185–197. doi: 10.1016/j.celrep.2016.12.021
Kenyon, C. (1986). A gene involved in the development of the posterior body region of C. elegans. Cell 46, 477–487. doi: 10.1016/0092-8674(86)90668-9
Kerk, S. Y., Kratsios, P., Hart, M., Mourao, R., and Hobert, O. (2017). Diversification of C. elegans motor neuron identity via selective effector gene repression. Neuron 93, 80–98. doi: 10.1016/j.neuron.2016.11.036
Kerr, K. S., Fuentes-Medel, Y., Brewer, C., Barria, R., Ashley, J., Abruzzi, K. C., et al. (2014). Glial wingless/Wnt regulates glutamate receptor clustering and synaptic physiology at the Drosophila neuromuscular junction. J. Neurosci. 34, 2910–2920. doi: 10.1523/JNEUROSCI.3714-13.2014
Klassen, M. P., and Shen, K. (2007). Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell 130, 704–716. doi: 10.1016/j.cell.2007.06.046
Kmita, M., and Duboule, D. (2003). Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333. doi: 10.1126/science.1085753
Kratsios, P., Kerk, S. Y., Catela, C., Liang, J., Vidal, B., Bayer, E. A., et al. (2017). An intersectional gene regulatory strategy defines subclass diversity of C. elegans motor neurons. eLife 6:e25751. doi: 10.7554/eLife.25751
Kratsios, P., Pinan-Lucarre, B., Kerk, S. Y., Weinreb, A., Bessereau, J. L., and Hobert, O. (2015). Transcriptional coordination of synaptogenesis and neurotransmitter signaling. Curr. Biol. 25, 1282–1295. doi: 10.1016/j.cub.2015.03.028
Kratsios, P., Stolfi, A., Levine, M., and Hobert, O. (2011). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15, 205–14. doi: 10.1038/nn.2989
Krumlauf, R., Marshall, H., Studer, M., Nonchev, S., Sham, M. H., and Lumsden, A. (1993). Hox homeobox genes and regionalisation of the nervous system. J. Neurobiol. 24, 1328–1340. doi: 10.1002/neu.480241006
Li, Y., Osuma, A., Correa, E., Okebalama, M. A., Dao, P., Gaylord, O., et al. (2020). Establishment and maintenance of motor neuron identity via temporal modularity in terminal selector function. eLife 9:e59464. doi: 10.7554/eLife.59464
Lints, R., and Emmons, S. W. (1999). Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development 126, 5819–5831. doi: 10.1242/dev.126.24.5819
Lizen, B., Hutlet, B., Bissen, D., Sauvegarde, D., Hermant, M., Ahn, M. T., et al. (2017a). HOXA5 localization in postnatal and adult mouse brain is suggestive of regulatory roles in postmitotic neurons. J. Comp. Neurol. 525, 1155–1175. doi: 10.1002/cne.24123
Lizen, B., Moens, C., Mouheiche, J., Sacre, T., Ahn, M. T., Jeannotte, L., et al. (2017b). Conditional loss of Hoxa5 function early after birth impacts on expression of genes with synaptic function. Front. Mol. Neurosci. 10:369. doi: 10.3389/fnmol.2017.00369
Maheshwari, U., Kraus, D., Vilain, N., Holwerda, S. J. B., Cankovic, V., Maiorano, N. A., et al. (2020). Postmitotic Hoxa5 expression specifies pontine neuron positional identity and input connectivity of cortical afferent subsets. Cell Rep. 31:107767. doi: 10.1016/j.celrep.2020.107767
Moris-Sanz, M., Estacio-Gomez, A., Sanchez-Herrero, E., and Diaz-Benjumea, F. J. (2015). The study of the Bithorax-complex genes in patterning CCAP neurons reveals a temporal control of neuronal differentiation by Abd-B. Biol. Open 4, 1132–1142. doi: 10.1242/bio.012872
Narita, Y., and Rijli, F. M. (2009). Hox genes in neural patterning and circuit formation in the mouse hindbrain. Curr. Top. Dev. Biol. 88, 139–167. doi: 10.1016/S0070-2153(09)88005-8
Nusslein-Volhard, C., and Wieschaus, E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. doi: 10.1038/287795a0
Oury, F., Murakami, Y., Renaud, J. S., Pasqualetti, M., Charnay, P., Ren, S. Y., et al. (2006). Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science 313, 1408–1413. doi: 10.1126/science.1130042
Parker, H. J., and Krumlauf, R. (2020). A Hox gene regulatory network for hindbrain segmentation. Curr. Top. Dev. Biol. 139, 169–203. doi: 10.1016/bs.ctdb.2020.03.001
Pasqualetti, M., Diaz, C., Renaud, J. S., Rijli, F. M., and Glover, J. C. (2007). Fate-mapping the mammalian hindbrain: segmental origins of vestibular projection neurons assessed using rhombomere-specific Hoxa2 enhancer elements in the mouse embryo. J. Neurosci. 27, 9670–9681. doi: 10.1523/JNEUROSCI.2189-07.2007
Philippidou, P., and Dasen, J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80, 12–34. doi: 10.1016/j.neuron.2013.09.020
Philippidou, P., Walsh, C. M., Aubin, J., Jeannotte, L., and Dasen, J. S. (2012). Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat. Neurosci. 15, 1636–1644. doi: 10.1038/nn.3242
Prasad, B., Karakuzu, O., Reed, R. R., and Cameron, S. (2008). unc-3-dependent repression of specific motor neuron fates in Caenorhabditis elegans. Dev. Biol. 323, 207–215. doi: 10.1016/j.ydbio.2008.08.029
Prasad, B. C., Ye, B., Zackhary, R., Schrader, K., Seydoux, G., and Reed, R. R. (1998). unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125, 1561–1568. doi: 10.1242/dev.125.8.1561
Reichert, H., and Bello, B. (2010). Hox genes and brain development in Drosophila. Adv. Exp. Med. Biol. 689, 145–153.
Salser, S. J., and Kenyon, C. (1992). Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature 355, 255–258. doi: 10.1038/355255a0
Salser, S. J., Loer, C. M., and Kenyon, C. (1993). Multiple HOM-C gene interactions specify cell fates in the nematode central nervous system. Genes Dev. 7, 1714–1724. doi: 10.1101/gad.7.9.1714
Shin, M. M., Catela, C., and Dasen, J. (2020). Intrinsic control of neuronal diversity and synaptic specificity in a proprioceptive circuit. eLife 9:e56374. doi: 10.7554/eLife.56374
Sokabe, T., Chen, H. C., Luo, J., and Montell, C. (2016). A switch in thermal preference in Drosophila larvae depends on multiple rhodopsins. Cell Rep. 17, 336–344. doi: 10.1016/j.celrep.2016.09.028
Soshnikova, N., Dewaele, R., Janvier, P., Krumlauf, R., and Duboule, D. (2013). Duplications of hox gene clusters and the emergence of vertebrates. Dev. Biol. 378, 194–199. doi: 10.1016/j.ydbio.2013.03.004
Strochlic, L., Falk, J., Goillot, E., Sigoillot, S., Bourgeois, F., Delers, P., et al. (2012). Wnt4 participates in the formation of vertebrate neuromuscular junction. PLoS One 7:e29976. doi: 10.1371/journal.pone.0029976
Sweeney, L. B., Bikoff, J. B., Gabitto, M. I., Brenner-Morton, S., Baek, M., Yang, J. H., et al. (2018). Origin and segmental diversity of spinal inhibitory interneurons. Neuron 97, 341.e3–355.e3. doi: 10.1016/j.neuron.2017.12.029
Sym, M., Robinson, N., and Kenyon, C. (1999). MIG-13 positions migrating cells along the anteroposterior body axis of C. elegans. Cell 98, 25–36. doi: 10.1016/S0092-8674(00)80603-0
Takahashi, Y., Hamada, J., Murakawa, K., Takada, M., Tada, M., Nogami, I., et al. (2004). Expression profiles of 39 Hox genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp. Cell Res. 293, 144–153. doi: 10.1016/j.yexcr.2003.09.024
Tihanyi, B., Vellai, T., Regos, A., Ari, E., Muller, F., and Takacs-Vellai, K. (2010). The C. elegans Hox gene ceh-13 regulates cell migration and fusion in a non-colinear way. Implications for the early evolution of Hox clusters. BMC Dev. Biol. 10:78. doi: 10.1186/1471-213X-10-78
Toker, A. S., Teng, Y., Ferreira, H. B., Emmons, S. W., and Chalfie, M. (2003). The Caenorhabditis elegans spalt-like gene sem-4 restricts touch cell fate by repressing the selector Hox gene egl-5 and the effector gene mec-3. Development 130, 3831–3840. doi: 10.1242/dev.00398
Tomas-Roca, L., Corral-San-Miguel, R., Aroca, P., Puelles, L., and Marin, F. (2016). Crypto-rhombomeres of the mouse medulla oblongata, defined by molecular and morphological features. Brain Struct. Funct. 221, 815–838. doi: 10.1007/s00429-014-0938-y
Vagnozzi, A. N., Garg, K., Dewitz, C., Moore, M. T., Cregg, J. M., Jeannotte, L., et al. (2020). Phrenic-specific transcriptional programs shape respiratory motor output. eLife 9:e52859. doi: 10.7554/eLife.52859
Van Auken, K., Weaver, D. C., Edgar, L. G., and Wood, W. B. (2000). Caenorhabditis elegans embryonic axial patterning requires two recently discovered posterior-group Hox genes. Proc. Natl. Acad. Sci. U.S.A. 97, 4499–4503. doi: 10.1073/pnas.97.9.4499
Von Stetina, S. E., Treinin, M., and Miller, D. M. III (2006). The motor circuit. Int. Rev. Neurobiol. 69, 125–167.
Wang, B. B., Muller-Immergluck, M. M., Austin, J., Robinson, N. T., Chisholm, A., and Kenyon, C. (1993). A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell 74, 29–42. doi: 10.1016/0092-8674(93)90292-x
Wang, X., Zhou, F., Lv, S., Yi, P., Zhu, Z., Yang, Y., et al. (2013). Transmembrane protein MIG-13 links the Wnt signaling and Hox genes to the cell polarity in neuronal migration. Proc. Natl. Acad. Sci. U.S.A. 110, 11175–11180. doi: 10.1073/pnas.1301849110
Wittmann, C., Bossinger, O., Goldstein, B., Fleischmann, M., Kohler, R., Brunschwig, K., et al. (1997). The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity. Development 124, 4193–4200.
Zheng, C., Diaz-Cuadros, M., and Chalfie, M. (2015a). Hox genes promote neuronal subtype diversification through posterior induction in Caenorhabditis elegans. Neuron 88, 514–527. doi: 10.1016/j.neuron.2015.09.049
Keywords: neuronal development, terminal identity, Hox genes, transcription factors, terminal selectors, synapse formation, synapse maturation
Citation: Feng W, Li Y and Kratsios P (2022) Emerging Roles for Hox Proteins in the Last Steps of Neuronal Development in Worms, Flies, and Mice. Front. Neurosci. 15:801791. doi: 10.3389/fnins.2021.801791
Received: 25 October 2021; Accepted: 31 December 2021;
Published: 04 February 2022.
Edited by:
Filipe Pinto-Teixeira, FR3743 Centre de Biologie Intégrative (CBI), FranceReviewed by:
Marie Kmita, Institut de Recherches Cliniques de Montréal (IRCM), CanadaPolyxeni Philippidou, Case Western Reserve University, United States
Françoise Gofflot, Catholic University of Louvain, Belgium
Copyright © 2022 Feng, Li and Kratsios. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paschalis Kratsios, cGtyYXRzaW9zQHVjaGljYWdvLmVkdQ==
†These authors have contributed equally to this work
 Weidong Feng
Weidong Feng Yinan Li
Yinan Li Paschalis Kratsios
Paschalis Kratsios