- 1School of Artificial Intelligence and Computer Science, Jiangnan University, Wuxi, China
- 2Jiangsu Key Laboratory of Media Design and Software Technology, Wuxi, China
- 3Department of Nuclear Medicine, Nanjing Medical University, Affiliated Wuxi People's Hospital, Wuxi, China
18F-FDG positron emission tomography (PET) imaging of brain glucose use and amyloid accumulation is a research criteria for Alzheimer's disease (AD) diagnosis. Several PET studies have shown widespread metabolic deficits in the frontal cortex for AD patients. Therefore, studying frontal cortex changes is of great importance for AD research. This paper aims to segment frontal cortex from brain PET imaging using deep neural networks. The learning framework called Frontal cortex Segmentation model of brain PET imaging (FSPET) is proposed to tackle this problem. It combines the anatomical prior to frontal cortex into the segmentation model, which is based on conditional generative adversarial network and convolutional auto-encoder. The FSPET method is evaluated on a dataset of 30 brain PET imaging with ground truth annotated by a radiologist. Results that outperform other baselines demonstrate the effectiveness of the FSPET framework.
1. Introduction
Alzheimer's disease (AD) is a progressive disease that destroys memory and other important mental functions. As of 2019, it ranked as the sixth leading cause of death in China (Vos et al., 2020). There are more than 10 million patients with AD in China, a country with the most AD patients in the world (Jia et al., 2020).
AD is usually diagnosed based on the clinical manifestation. Nowadays, medical imaging including computed tomography (CT) or magnetic resonance imaging (MRI), and with single-photon emission computed tomography (SPECT) or positron emission tomography (PET), can be used to help doctors understand the pathophysiology of AD, for example, Aβ plaques, neurofibrillary tangles, and neuroinflammation. Moreover, the pathophysiology of AD is believed that starts years ahead of the of clinical observation, and helps detect AD earlier than conventional diagnostic tools (Marcus et al., 2014).
Among the above medical imaging technique, PET/CT is a nuclear medicine technique that combines a PET scanner and a CT scanner to acquire sequential images from both devices in the same session, which are combined into a single superposed image. Figure 1 shows the brain PET/CT fusion image. The first line is the PET imaging, and the second line is the CT imaging. The fusion imaging of PET/CT is list in the third line. Each line from left to right is (a) coronal section, (b) median sagittal section, and (c) transverse section.
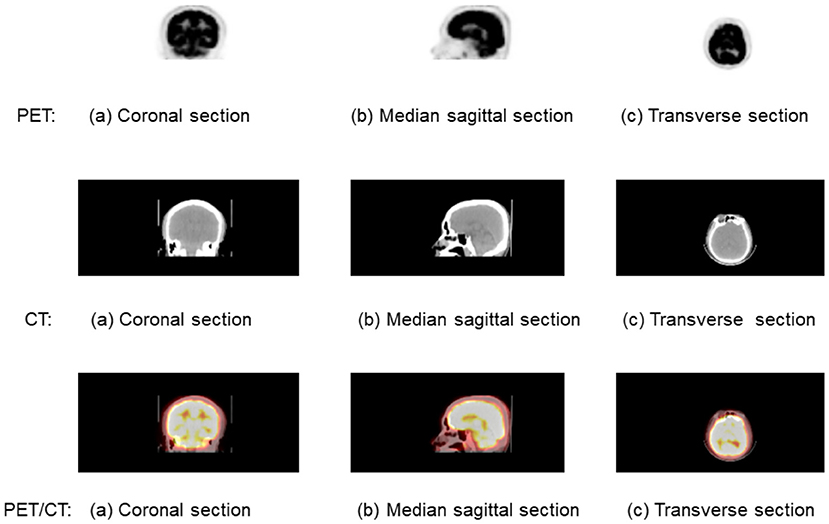
Figure 1. A brain positron emission tomography (PET)/computed tomography (CT) fusion image. The first line is the PET imaging, and the second line is the CT imaging. The fusion imaging of PET/CT is list in the third line. Each line from left to right is (a) coronal section, (b) median sagittal scan, and (c) transverse section.
18F-FDG PET imaging of brain glucose use and amyloid accumulation is a research criteria for AD diagnosis (Berti et al., 2011). Several 18F-FDG PET studies have been conducted to estimate AD-related brain changes. They have consistently shown widespread metabolic deficits in the neocortical association areas, such as frontal cortex. Further studies have demonstrated that CMRglc in frontal cortex suffers an average decline of 16−19% over a 3-year period (Smith et al., 1992; Mielke et al., 1994). Frontal cortex covers frontal lobe and contains most of the dopamine neurons. Figure 2 shows the location of frontal cortex in the brain. The yellow part of the left subfigure is its anatomical location, while the red contour in the right subfigure indicates its location in 18F-FDG PET imaging. Due to its sensitive to detect frontal cortex changes over time, 18F-FDG PET imaging can be used not only for AD diagnosis but also to monitor dementia progression and therapeutic interventions. Therefore, PET imaging are valuable in the assessment of patients with AD. Moreover, the frontal cortex segmentation of PET imaging is crucial for understanding AD progression on AD-related regions in brain.

Figure 2. The frontal cortex in the brain: the left is anatomical location, and the right is for 18F-FDG positron emission tomography (PET) imaging.
Although frontal cortex segmentation is an important problem for AD research. However as far as we know, this paper is the first work that studies the frontal cortex segmentation problem for PET imaging. Unlike organ or tumor, which is different from other tissue with gray-level, texture, gradients, edges, shape, etc., frontal cortex is a part of brain without obvious boundaries. Moreover, supervised learning frameworks need segmentation ground truth from professional doctor, and it is difficult to get large number of annotated imaging. All these makes frontal cortex segmentation a tough problem.
Since manual segmentation is time consuming, automatic semantic segmentation for medical images, which makes pathological structures changes clear in images, becomes one of the hottest research topic in image processing. Currently, more and more machine learning technologies have been used in medical applications, such as medical single processing, medical image processing, medical data analyzing, and so on (Jiang et al., 2021a,b; Yang et al., 2021). Brain and brain tumor segmentation is one of the most popular medical image segmentation tasks (Szilagyi et al., 2003; Tu and Bai, 2009; Zhang et al., 2015; Jiang et al., 2019). Many approaches have been proposed to address this problem, such as thresholding (Sujji et al., 2013), edge detection (Tang et al., 2000), Markov random fields (MRF) (Held et al., 1997), and support vector machine (SVM) (Akselrod-Ballin et al., 2006).
Due to the rapid development of deep learning, neural networks, which can extract hierarchical feature of images, become one of the most effective technique in brain imaging segmentation (Fakhry et al., 2016; Işın et al., 2016; Zhao et al., 2018). U-net (Ronneberger et al., 2015) and its 3D version V-Net (Milletari et al., 2016) are the most well-known deep learning architecture in medical image segmentation. Recently, organ and tissue's shape and position priors are combined into the segmentation algorithm to improve the accuracy. (Oktay et al., 2017) proposes a training framework ACNN, which incorporates cardiac anatomical prior into CNN. Boutillon et al. (2019) combines scapula bone anatomical prior into a conditional adversarial learning method.
The related work has made large progress in semantic segmentation in medical imaging. However, they are not designed for frontal cortex segmentation in PET imaging, and they cannot be utilized directly for this problem. Motivated by this, in this paper, we propose the supervised segmentation framework: Frontal cortex Segmentation model of brain PET imaging (FSPET). The FSPET model based on both conditional generative adversarial network (cGAN) and convolutional auto-encoder (CAE) incorporates the anatomical prior to improve the prediction accuracy.
The contribution of FSPET dedicated to frontal cortex segmentation is threefold. First, the CAE is used to find the embedding of frontal cortex shape priors in latent space. Second, the segmentation method based on U-net, as the generator of cGAN, learns the feature of frontal cortex to generate the binary mask in PET imaging. Third, the anatomical prior is fused into the discriminator model in cGAN to get more precise prediction. Extensive experiments demonstrate the effectiveness of the proposed FSPET model.
2. Methods
In this section, we will introduce the proposed FSPET framework in detail, which combines the prior of frontal cortex shape in the deep neural networks, as shown in Figure 3. FSPET contains two parts: cGAN and CAE.
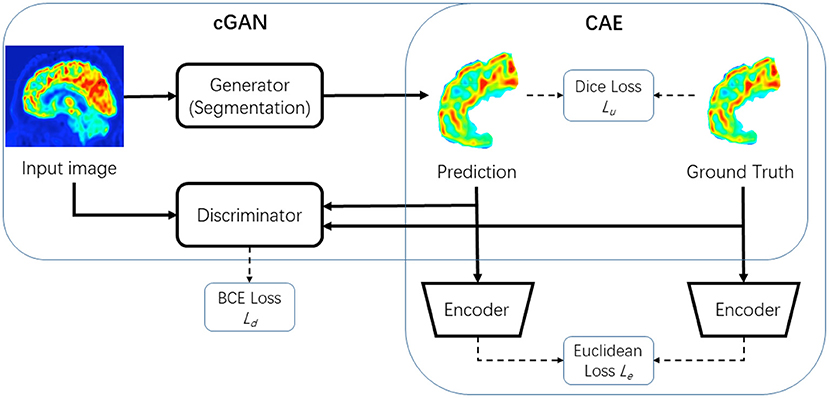
Figure 3. Framework of proposed model FSPET based on conditional generative adversarial network (cGAN) and convolutional auto-encoder (CAE).
2.1. Conditional Generative Adversarial Networks
Generative Adversarial Network (GAN) (Goodfellow et al., 2014) is widely used for data augmentation by generating new images. Since PET imaging shows low contrast, low resolution, and blurred boundaries between different tissues, GAN is becoming a popular method for medical image segmentation (Luc et al., 2016; Son et al., 2017; Souly et al., 2017).
cGAN (Mirza and Osindero, 2014) is an extension of GAN, which is used as a machine learning framework for training generative models. The proposed FSPET model adopts the framework in Conze et al. (2021) based on cGAN, which consists of two neural networks: the generator G and the discriminator D.
The generator G of cGAN in the FSPET model is the segmentation framework, which learns the feature of frontal cortex to generate the binary mask in PET imaging. Formally, let x be the source image and y be the ground truth image of class label . The generator learns the mapping between images and labels by optimizing the loss function using stochastic gradient descent. The generator of cGAN is often based on U-net framework. The network consists of a contracting path and an expansive path (shown in Figure 4B). The contracting path is a convolutional network that consists of repeated application of 3 × 3 convolutions, each followed by a rectified linear unit (ReLU) and a 2 × 2 max pooling operation. Dice loss is used in U-net to compare the prediction G(x) and ground truth y, in which the loss function is as follows:

Figure 4. FSPET architecture: (A) convolutional auto-encoder (CAE), (B) Generator (G), and (C) Discriminator (D).
The discriminator D in FSPET (shown in Figure 4C) inputs are source images and prediction to be evaluated. D distinguishes the given boundary by the generator from the realistic segmentation. The output is a binary prediction as to whether the image is real (class = 1) or fake (class = 0). In cGAN, binary cross entropy (BCE) loss is used to determine the loss function:
2.2. Convolutional Auto-Encoder
Auto-encoder is a type of neural networks used to learn a representation (encoding) for a set of data. It imposes a bottleneck in the network, which forces a compressed knowledge representation of the original input. An auto-encoder consists of two parts, the encoder and the decoder, which can be defined as f and g such that:
where is the input and h is usually referred to as code, the latent representation of the input. Motivated by Oktay et al. (2017), we utilized the CAE to find the embedding of frontal cortex shape priors in latent space (shown in Figure 4A). The BCE loss is minimized in the CAE framework with the ground truth y as input:
As shown in Figure 4A, after CAE is fixed, we use its encoder part f for segmentation training. By conducting CAE low-dimensional projection on both prediction and ground truth, we can minimize the loss function as:
2.3. Fusion
We have obtained three loss functions from different parts of the FSPET model respectively, whose information are listed in Table 1. Finally, we fuse the U-net segmentation method with frontal cortex shape priors. In the backward propagation, the loss function of generator G is:
where λ1 and λ2 are the weighting factor. Minimizing Lu tends to provide rough frontal cortex shape prediction, while maximizing log(D(x, G(x))) is designed to improve contour delineations. At the same time, the latent loss Le guarantees the global consistent and precise prediction similar to the original segmentation.
Additionally, the loss function of discriminator D is:
It maximizes log(D(x, y), which is the loss between input and ground truth. Simultaneously, it minimizes loss value for generated −log(1 − D(x, G(x))) masks. The optimization proceeds in alternative periods on G and D using stochastic gradient descent.
3. Experiments
In this section, we conduct extensive experiments to validate the effectiveness of FSPET.
3.1. Validation Setup
Dataset: We collected 30 18F-FDG PET images from different patients and their sensitive information was erased. The brain PET images were acquired using a PET/CT (Discovery STE, General Electric, Waukesha, USA) approximately 1 h after an intravenous injection of 18F-FDG (10 mCi). The original data were stored in Digital Imaging and Communications in Medicine (DICOM) format. Images were then resampled with a resolution 512 × 512 pixels. Frontal cortex in all images were annotated by a radiologist with 7 years experience to obtain the ground truth and also the shape priors.
Baselines: We compare FSPET with different baseline methods in frontal cortex segmentation for brain PET imaging. The comparison methods used in the experiments include:
• U-net (Ronneberger et al., 2015): U-net is the classical segmentation algorithm for medical images, and the generator G of FSPET is based on U-net. The architecture (shown in Figure 4B) contains contraction path and symmetric expanding path. The former path is used to capture the context in the image and the latter one is used to enable precise localization using transposed convolutions.
• ACNN (Oktay et al., 2017): It utilizes the auto-encoder and T-L network to combine anatomical prior knowledge into CNNs. These regularizers make predictions that are in agreement with the shape priors.
• cGAN-Unet (Singh et al., 2018): It is proposed for breast mass segmentation in mammography. The cGAN is used in the segmentation framework, in which the generative network learns the features of tumors and the adversarial network guarantees the contour to be similar to the ground truth.
Measurements: With the definition of true positive (TP), true negative (TN), false positive (FP), and false negative (FN), the following metrics are used to provide an overall assessment of all methods:
• Dice coefficient (dice): It is a similarity measure over prediction and ground truth. It ranges between 0 and 1.
• Jaccard index (Jaccard): It is another similarity metric with range [0, 1].
• Sensitivity: It is a measure of how well a test can identify true positives.
• Specificity: It is a measure of how well a test can identify true negatives.
• Hausdorff distance (HD): It is the greatest of all the distances from a point in one set to the closest point in the other set. HD measures how far two contours of prediction and ground truth are from each other. With A and B are the set of non-zero voxels in labels images, HD is defined as:
Training Detail: In this paper, we use Adam optimization (Kingma and Ba, 2014), which is a stochastic gradient descent method that is based on adaptive estimation of first-order and second-order moments. To train the FSPET model, the CAE with BCE loss is first optimized based on (3). With the learning rate of 0.01 for 30 epochs, the batch size is fixed at 32. Then the optimization of cGAN proceeds in alternative periods on G and D according to (5) and (6). In (5), the weight and . With the learning rate of 10−4, batch size 32 and 30 epochs enjoyed the best performance.
3.2. Results
Quantitative metric and score values are provided in Table 2 for frontal cortex segmentation. When comparing U-net and ACNN, the dice score improves from 71.03% to 74.57%, and HD score decreases from 38.73 to 35.48. This demonstrates that extending U-net with CAE allows the model taking advantage of latent representation of shape priors. Moreover, significant improvements can be noticed using cGAN-Unet comparing with U-net (38.73 to 30.32 on HD), which indicates the appropriateness of embedding U-net into a cGAN pipeline. Combining CAE and cGAN networks, the proposed FSPET model discriminates more efficiently frontal cortex from surrounding structures by achieving the best score with regard to dice, Jaccard index, sensitivity, and HD. In particular, large gains in terms of Jaccard index (55.04–71.47%) and HD (38.73–28.05) are reported between U-net and FSPET.
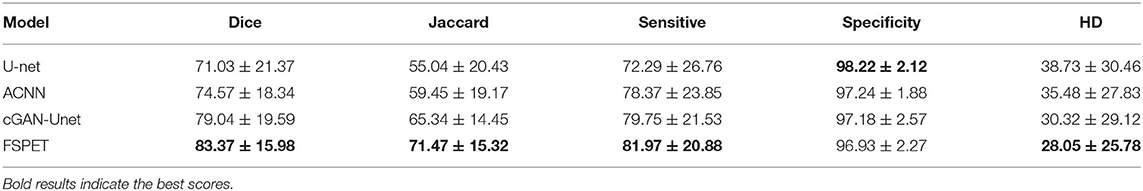
Table 2. Quantitative assessment of U-net (Ronneberger et al., 2015), ACNN (Oktay et al., 2017), cGAN-Unet (Singh et al., 2018), and the FSPET model.
Qualitative results for frontal cortex segmentation in median sagittal section of brain PET imaging are displayed in Figure 5. Compared to U-net, ACNN, and cGAN-Unet, which are prone to under- or over-segmentation, sometimes combined with unrealistic shapes, better contour adherence and shape consistency are reached by the FSPET model. We also take one example (shown in bottom right of each subfigure) for comparison, and the FSPET captures more complex shape and subtle contours compared to other frameworks. This reveals the importance of combining both adversarial networks and shape priors in the segmentation task.
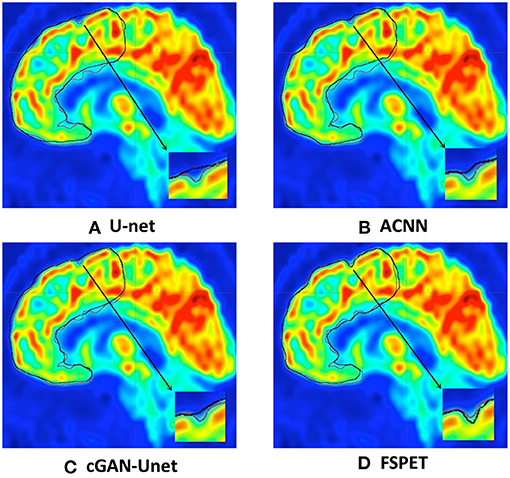
Figure 5. Frontal cortex segmentation in median sagittal section of brain positron emission tomography (PET) imaging using U-net, ACNN, cGAN-Unet, and the FSPET model. Ground truth and predicted contour are in red and black, respectively. (A) U-net. (B) ACNN. (C) cGAN-Unet. (D) FSPET.
4. Conclusion
This paper propose a deep learning framework to segment frontal cortex from brain PET imaging. The model based on both cGAN and CAE incorporates the anatomical prior to improve the prediction accuracy. Future work will utilize the proposed method to detect other parts of AD-related brain area, such as hippocampus.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by Jiangnan University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
QZ contributed to the conception of the study and wrote the manuscript. YuanyuanL performed the experiment. YuanL helped to perform the analysis with constructive discussions. WH contributed significantly to data preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akselrod-Ballin, A., Galun, M., Gomori, M. J., Basri, R., and Brandt, A. (2006). “Atlas guided identification of brain structures by combining 3D segmentation and SVM classification,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Copenhagen: Springer), 209–216. doi: 10.1007/11866763_26
Berti, V., Pupi, A., and Mosconi, L. (2011). PET/CT in diagnosis of dementia. Ann. N. Y. Acad. Sci. 1228:81. doi: 10.1111/j.1749-6632.2011.06015.x
Boutillon, A., Borotikar, B., Burdin, V., and Conze, P. H. (2019). “Combining shape priors with conditional adversarial networks for improved scapula segmentation in MR images,” in 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) (Iowa City, IA). doi: 10.1109/ISBI45749.2020.9098360
Conze, P.-H., Kavur, A. E., Cornec-Le Gall, E., Gezer, N. S., Le Meur, Y., Selver, M. A., et al. (2021). Abdominal multi-organ segmentation with cascaded convolutional and adversarial deep networks. Artif. Intell. Med. 117:102109. doi: 10.1016/j.artmed.2021.102109
Fakhry, A., Peng, H., and Ji, S. (2016). Deep models for brain EM image segmentation: novel insights and improved performance. Bioinformatics 32, 2352–2358. doi: 10.1093/bioinformatics/btw165
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). “Generative adversarial nets,” in Advances in Neural Information Processing Systems, 27.
Held, K., Kops, E. R., Krause, B. J., Wells, W. M., Kikinis, R., and Muller-Gartner, H.-W. (1997). Markov random field segmentation of brain MR images. IEEE Trans. Med. Imaging 16, 878–886. doi: 10.1109/42.650883
Işın, A., Direkoğlu, C., and Şah, M. (2016). Review of MRI-based brain tumor image segmentation using deep learning methods. Proc. Comput. Sci. 102, 317–324. doi: 10.1016/j.procs.2016.09.407
Jia, L., Du, Y., Chu, L., Zhang, Z., Li, F., Lyu, D., et al. (2020). Prevalence, risk factors, and management of dementia and mild cognitive impairment in adults aged 60 years or older in China: a cross-sectional study. Lancet Public Health 5, e661–e671. doi: 10.1016/S2468-2667(20)30185-7
Jiang, Y., Gu, X., Wu, D., Hang, W., Xue, J., Qiu, S., et al. (2021a). A novel negative-transfer-resistant fuzzy clustering model with a shared cross-domain transfer latent space and its application to brain ct image segmentation. IEEE/ACM Trans. Comput. Biol. Bioinform. 18, 40–52. doi: 10.1109/TCBB.2019.2963873
Jiang, Y., Zhang, Y., Lin, C., Wu, D., and Lin, C.-T. (2021b). EEG-based driver drowsiness estimation using an online multi-view and transfer TSK fuzzy system. IEEE Trans. Intell. Transport. Syst. 22, 1752–1764. doi: 10.1109/TITS.2020.2973673
Jiang, Y., Zhao, K., Xia, K., Xue, J., Zhou, L., Ding, Y., et al. (2019). A novel distributed multitask fuzzy clustering algorithm for automatic MR brain image segmentation. J. Med. Syst. 43, 1–9. doi: 10.1007/s10916-019-1245-1
Kingma, D. P., and Ba, J. (2014). Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980.
Luc, P., Couprie, C., Chintala, S., and Verbeek, J. (2016). Semantic segmentation using adversarial networks. arXiv preprint arXiv:1611.08408.
Marcus, C., Mena, E., and Subramaniam, R. M. (2014). Brain pet in the diagnosis of Alzheimer's disease. Clin. Nuclear Med. 39, 423–426. doi: 10.1097/RLU.0000000000000547
Mielke, R., Herholz, K., Grond, M., Kessler, J., and Heiss, W. (1994). Clinical deterioration in probable Alzheimer's disease correlates with progressive metabolic impairment of association areas. Dement. Geriatr. Cogn. Disord. 5, 36–41. doi: 10.1159/000106692
Milletari, F., Navab, N., and Ahmadi, S.-A. (2016). “V-Net: fully convolutional neural networks for volumetric medical image segmentation,” in 2016 Fourth International Conference on 3D Vision (3DV) (Stanford, CA: IEEE), 565–571. doi: 10.1109/3DV.2016.79
Mirza, M., and Osindero, S. (2014). Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784.
Oktay, O., Ferrante, E., Kamnitsas, K., Heinrich, M., Bai, W., Caballero, J., et al. (2017). Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE Trans. Med. Imaging 37, 384–395. doi: 10.1109/TMI.2017.2743464
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-Net: convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Munich: Springer), 234–241. doi: 10.1007/978-3-319-24574-4_28
Singh, V. K., Romani, S., Rashwan, H. A., Akram, F., Pandey, N., Sarker, M. M. K., et al. (2018). “Conditional generative adversarial and convolutional networks for x-ray breast mass segmentation and shape classification,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Granada: Springer), 833–840. doi: 10.1007/978-3-030-00934-2_92
Smith, G. S., de Leon, M. J., George, A. E., Kluger, A., Volkow, N. D., McRae, T., et al. (1992). Topography of cross-sectional and longitudinal glucose metabolic deficits in Alzheimer's disease: pathophysiologic implications. Arch. Neurol. 49, 1142–1150. doi: 10.1001/archneur.1992.00530350056020
Son, J., Park, S. J., and Jung, K.-H. (2017). Retinal vessel segmentation in fundoscopic images with generative adversarial networks. arXiv preprint arXiv:1706.09318.
Souly, N., Spampinato, C., and Shah, M. (2017). “Semi supervised semantic segmentation using generative adversarial network,” in Proceedings of the IEEE International Conference on Computer Vision (Venice), 5688–5696. doi: 10.1109/ICCV.2017.606
Sujji, G. E., Lakshmi, Y., and Jiji, G. W. (2013). MRI brain image segmentation based on thresholding. Int. J. Adv. Comput. Res. 3:97.
Szilagyi, L., Benyo, Z., Szilágyi, S. M., and Adam, H. (2003). “MR brain image segmentation using an enhanced fuzzy C-means algorithm,” in Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (Cancun: IEEE), 724–726. doi: 10.1109/IEMBS.2003.1279866
Tang, H., Wu, E., Ma, Q., Gallagher, D., Perera, G., and Zhuang, T. (2000). MRI brain image segmentation by multi-resolution edge detection and region selection. Comput. Med. Imaging Graph. 24, 349–357. doi: 10.1016/S0895-6111(00)00037-9
Tu, Z., and Bai, X. (2009). Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 32, 1744–1757. doi: 10.1109/TPAMI.2009.186
Vos, T., Lim, S. S., Abbafati, C., Abbas, K. M., Abbasi, M., Abbasifard, M., et al. (2020). Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the global burden of disease study 2019. Lancet 396, 1204–1222. doi: 10.1016/S0140-6736(20)30925-9
Yang, H., Lu, X., Wang, S.-H., Lu, Z., Yao, J., Jiang, Y., et al. (2021). Synthesizing multi-contrast MR images via novel 3d conditional variational auto-encoding gan. Mobile Netw. Appl. 26, 415–424. doi: 10.1007/s11036-020-01678-1
Zhang, W., Li, R., Deng, H., Wang, L., Lin, W., Ji, S., et al. (2015). Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage 108, 214–224. doi: 10.1016/j.neuroimage.2014.12.061
Keywords: brain image segmentation, convolutional auto-encoder, conditional generative adversarial network, PET, Alzheimer's disease
Citation: Zhan Q, Liu Y, Liu Y and Hu W (2021) Frontal Cortex Segmentation of Brain PET Imaging Using Deep Neural Networks. Front. Neurosci. 15:796172. doi: 10.3389/fnins.2021.796172
Received: 16 October 2021; Accepted: 01 November 2021;
Published: 08 December 2021.
Edited by:
Yuanpeng Zhang, Nantong University, ChinaReviewed by:
Yanhui Zhang, Hebei University of Chinese Medicine, ChinaQiusheng Shen, Changshu Affiliated Hospital of Nanjing University of Traditional Chinese Medicine, China
Copyright © 2021 Zhan, Liu, Liu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hu, aHV3ZWlAbmptdS5lZHUuY24=
 Qianyi Zhan
Qianyi Zhan Yuanyuan Liu
Yuanyuan Liu Yuan Liu1,2
Yuan Liu1,2