- Department of Neurology, First Affiliated Hospital of Anhui Medical University, Hefei, China
The methylenetetrahydrofolate reductase (MTHFR) gene has been associated with Alzheimer’s disease (AD) pathogenesis. Amnestic mild cognitive impairment (aMCI) represents a prodromal stage of dementia and involves a high risk of progression into AD. Although the effects of the apolipoprotein E (APOE) gene on structural alterations in aMCI have been widely investigated, the effects of MTHFR C677T and interaction effects of MTHFR × APOE genotypes on gray matter atrophy in aMCI remain largely unknown. In the present study, 60 aMCI patients and 30 healthy controls were enrolled, and voxel-based morphometry analysis was performed to inspect the effects of diagnosis, different genotypes, and their interactions on gray matter atrophy. The results showed that aMCI patients had significant gray matter atrophy involving the bilateral hippocampus, the right parahippocampal gyrus, and the left superior temporal gyrus compared with healthy controls. Besides, a substantial reduction in gray matter volume was observed in the right hippocampus region in APOE ε4 carriers from the aMCI group, compared with APOE ε4 non-carriers. A significant interaction was found between diagnosis and MTHFR C677T genotype on the right precuneus in healthy controls and aMCI patients not carrying APOE ε4 allele. Our findings may provide new evidence substantiating the genetic effects of MTHFR C677T on brain structural alternation in patients with aMCI.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder globally, accounting for 60 to 80% of dementia cases. Individuals who suffer from AD experience progressive memory decline and impairment of other cognitive functions (Scheltens et al., 2021). Amnestic mild cognitive impairment (aMCI) is a transitional state between normal age-related cognitive decline and dementia, being the prodromal stage of AD (Petersen et al., 1999), and entails a high risk of progression into AD, with an estimated rate of 10 to 15% per year, compared with a rate of 1 to 2% per year for healthy older people (Petersen et al., 1999).
Apolipoprotein E (APOE) ε4 allele is an important genetic factor that increases susceptibility to sporadic AD (Scheltens et al., 2021) and has been associated with amyloid deposition, tau hyperphosphorylation, cholesterol transport (Scheltens et al., 2021), which can reportedly accelerate the progression to AD. However, the APOE ε4 allele can only account for the heritability of AD to a certain extent (Ridge et al., 2013; Bellenguez et al., 2017). More than 40 AD-associated risk alleles have been found in genome-wide association studies (Jansen et al., 2019; Sims et al., 2020).
The methylenetetrahydrofolate reductase (MTHFR) gene is the major genetic modifier that affects the folate cycle and homocysteine metabolism. MTHFR C677T (rs1801133) is one of the most common polymorphisms of the MTHFR gene and is thought to be involved in the pathogenesis of AD (Hu et al., 2016). The C677T polymorphism, which refers to the substitution of cytosine (C) at base 677 of the MTHFR-encoding gene with thymine (T) and alanine at codon 222 replaced by valine, has been reported to result in decreased MTHFR enzyme activity (Frosst et al., 1995), leading to elevated homocysteine concentrations. Hyperhomocysteinemia is widely acknowledged to be a risk factor of AD (Smith et al., 2018), which influences DNA methylation (Coppede, 2010), DNA repair (Bednarska-Makaruk et al., 2016), oxidative stress (Deep et al., 2019), amyloid β aggregation (Chung et al., 2016; Hoffman et al., 2018), tau phosphorylation (Sontag et al., 2014), vascular endothelial dysfunction (Esse et al., 2019; Wu et al., 2019), and the secretion of inflammatory mediators, especially tumor necrosis factor α, nuclear factor κB, interleukin 6 (IL-6), and IL-8 (Di Meco et al., 2019). Moreover, epigenetic level refers to the mutual gene interaction between the MTHFR gene and other genes, which may play a pivotal role in the development of AD.
Many studies have investigated the influence of single-nucleotide polymorphism of APOE on the heterogeneity in cognitive and neuroimaging findings. However, few neuroimaging studies have focused on how MTHFR polymorphism could influence brain abnormalities in aMCI patients. Interestingly, it has been reported that MTHFR C677T variant might be associated with regional brain volume reduction in white matter (WM) brain areas in MCI patients (Rajagopalan et al., 2012). In addition, the risk T allele of the MTHFR C677T was associated with a significantly increased annual rate of progressive right periventricular parietal WM atrophy, up to 1.5% per year per “T” allele. In contrast, the effect of MTHFR C677T polymorphism on gray matter (GM) atrophy in aMCI subjects remains largely unknown.
Furthermore, numerous studies have described gene–gene and gene–environment interactions in AD in recent years. Most researchers advocate that interactions between genetic factors promote progression to AD and associated brain abnormalities; however, much heterogeneity was found in the results obtained. For instance, in a study by Peng et al. (2015), where subgroup analysis was based on APOE ε4 status, MTHFR C677T polymorphism was found to be only associated with the risk of AD in APOE ε4 carriers, but not in APOE ε4 non-carriers, indicating a synergic effect between MTHFR C677T polymorphism and the APOE ε4 allele. In contrast, Stoccoro et al. (2017) documented that the MTHFR TT genotype increased the likelihood of AD in both APOE ε4 carriers and non-carriers. Nonetheless, these studies investigated only the interactions between genes at the clinical level, whereas the effects of MTHFR C677T and MTHFR × APOE genotypes interactions on GM atrophy in aMCI were largely unexplored. Given that both APOE and MTHFR genes share similar pathogenic mechanisms in AD, we sought to explore the effect of the interaction between APOE and MTHFR genes on the brain structure of aMCI patients, in addition to the influence of MTHFR C677T itself on the GM structure.
Accordingly, the main purpose of our research was to explore the effect of MTHFR C677T and the influence of the interaction between MTHFR and APOE genotypes on GM atrophy in aMCI patients. We sought to deepen our understanding of the role of susceptible genes such as MTHFR and APOE and the outcome of their interactions on GM atrophy in aMCI disease, which could importantly help to develop new strategies for disease prevention and early therapy.
Materials and Methods
Participants
Sixty-eight aMCI patients were recruited in the present study. The clinical diagnosis of aMCI was based on the National Institute on Aging and Alzheimer’s Association criteria (Albert et al., 2011), which included (1) subjective cognitive impairment reported by participants or their caregivers; (2) objective cognitive impairment, but no dementia; (3) Clinical Dementia Rating score of 0.5; (4) a memory function score of 1.0 to 1.5 standard deviations below the mean for their age- and education-matched peers on culturally appropriate normative data. The exclusion criteria included patients with a history of stroke, severe depression, or other neurological or psychiatric illness; WM hyperintensity with Fazekas grade ≥ 2 or a modified Hachinski ischemic score > 4; previous treatment with cholinesterase inhibitors, folate, and vitamin B to improve cognition; severe visual or auditory abnormalities; and patients with magnetic resonance contraindications.
Thirty-two healthy controls (HCs) were recruited during the same period. The healthy subjects had no underlying neurological or psychiatric disorders, no subjective or objective cognitive deficits, and no psychoactive medication use. All participants in our study were Chinese Han and right-handed. Ten participants were excluded for the following reasons: failed genotyping (n = 3), severe WM hyperintensity (n = 5), and poor quality of magnetic resonance imaging (MRI; n = 2). Finally, 60 aMCI patients (67.32 ± 9.44 years old; 27 males) and 30 HCs (64.76 ± 8.01 years old; 13 males) matched by gender, age, and education were eventually enrolled in the study. Our research was approved by the Institutional Ethics Committees of the First Affiliated Hospital of Anhui Medical University, and written informed consent was signed by each participant.
Genotyping
Two milliliters of fasting blood was drawn from all participants in EDTA tubes and stored at −80°C. These blood samples were sent to Anhui Jinzhun Gene Biotechnology company for APOE gene and MTHFR C677T sequencing and typing. Genomic DNA was extracted from blood samples using Magen HiPure Blood DNA Mini Kit (D3111-03). Genotyping of rs1801133 (MTHFR), rs429358, and rs7412 (APOE) in each subject was performed using the penta-primer amplification refractory mutation system method. Primers for MTHFR and APOE genotype amplification are shown in Supplementary Table 1. After polymerase chain reaction, the plates were read by a TECAN M1000 infinite reader, and DNA sequences were analyzed using the online software snp decoder1. The χ2 test was used to assess whether the allele frequency was consistent with expectation in Hardy–Weinberg equilibrium. The statistical significance level was set at p < 0.05.
Because of the limited number of MTHFR TT and APOE ε4ε4 genotypes in our sample, we adopted the most appropriated genetic model (dominant model), as previously described (Chang et al., 2017, 2019). MTHFR rs1801133 polymorphism in the aMCI group was distributed between T allele carriers (n = 40 [66.7%], TT = 12, CT = 28), and homozygous wild-type (CC) carriers (n = 20 [33.3%], CC = 20). MTHFR rs1801133 polymorphism in the HC group consisted of T allele carriers (n = 19 [63.3%], TT = 4, CT = 15), and homozygous wild-type (CC) carriers (n = 11 [36.7%], CC = 11). Similarly, the distribution of APOE rs429358, rs7412 variants in the aMCI group was distributed between APOE ε4 carriers (n = 22 [36.7%], ε3ε4 = 16, ε2ε4 = 4, ε4ε4 = 2), and APOE ε4 non-carriers (n = 38 [63.3%], ε2ε2 = 0 ε2ε3 = 4, ε3ε3 = 34). In the HC group, there were 8 APOE ε4 carriers (n = 8 [26.7%], ε3ε4 = 8, ε2ε4 = 0, ε4ε4 = 0), and 22 APOE ε4 non-carriers (n = 22 [73.3%], ε2ε2 = 0, ε2ε3 = 3, ε3ε3 = 19). Allelic frequencies were arrived at Hardy–Weinberg equilibrium.
Clinical, Biochemical, and Neuropsychological Assessments
The general condition and medical history of each participant were recorded. Three milliliters of fasting blood was collected from all participants in collection tubes for biochemical assays. The serum samples were extracted after centrifugation, and then serum homocysteine was detected by chemical luminescent immunoassay, and serum folate and vitamin B12 were detected by electrochemiluminescence immunoassay. All subjects were interviewed with the following questionnaires: the Mini-Mental State Examination and Montreal Cognitive Assessment were used to assess general cognitive functions; the Cambridge Cognitive Examination—Chinese version was used to assess episodic memory, attention, executive function, and visuospatial skills; the Clinical Dementia Rating was used to assess the severity of the disease; the Geriatric Depression Scale was used to assess emotion; and the Activities of Daily Living scale was used to evaluate independent living skills.
Neuroimaging Acquisition
A 3.0-T MRI scanner (Signa HDxt, GE, Milwaukee, WI, United States) equipped with an 8-channel head coil was used to acquire three-dimensional (3D) T1-weighted structural images. Foam paddings were used to minimize head motion and earplugs to reduce scanner noise. Three-dimensional high spatial resolution T1-weighted images were obtained using the following parameters: repetition time (TR) = 9.5 ms; echo time (TE) = 3.9 ms; flip angle (FA) = 20°; 176 slices; slice thickness = 1 mm; matrix size = 512 × 512; and field of view (FOV) = 256 × 256 mm2. T2-weighted and T2 fluid-attenuated inversion recovery (FLAIR) sequences were performed simultaneously for each subject, the sequences were as follows: T2-weighted images (TR = 3,500 ms; TE = 85 ms; echo train length = 15; FA = 90°; axial slice thickness = 5 mm; matrix size = 512 × 512; and FOV = 230 × 184 mm2); T2-FLAIR images (TR = 11 s; TE = 120 ms; FA = 90°; axial slice thickness = 5 mm; matrix size = 512 × 512; and FOV = 230 × 230 mm2).
MRI Data Processing and Analysis
All images were visually inspected for artifacts, motion problems, or structural abnormalities. We carried out voxel-based morphometry (VBM) analysis using the VBM8 toolbox2 based on SPM8 (Statistical Parametric Mapping, Wellcome Department of Imaging Neuroscience, London, United Kingdom; available online at http://www.fil.ion.ucl.ac.uk/spm), according to the following procedure. First, the 3D T1 images of the whole brain were segmented into GM, WM, and cerebrospinal fluid (CSF). Then, the segments were iteratively registered using Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra (DARTEL) toolbox, and the GM images were normalized and modulated into the standard Montreal Neurological Institute space. Finally, the modulated spatial normalized GM images were smoothed with an 8-mm full-width at half maximum kernel. The total intracranial volume (TIV) was calculated by summing up the total GM, WM, and CSF volume. REST3 software was used to extract the GM volume of specific brain regions.
Statistical Analysis
SPSS 22.0 software (IBM SPSS Inc., Chicago, IL, United States) was used to analyze the demographic, behavioral, and genotype data. Comparison between groups was performed by χ2 test for categorical variables, and two-sample t test and Mann-Whitney U test for continuous variables. Normally distributed data were expressed as mean ± standard deviation, whereas non-parametric data were represented as median and interquartile range [M (QU – QL)]. A two-tailed p < 0.05 was statistically significant.
A full factorial analysis of covariance was applied to analyze the effect of interactions on GM volume. First, it was performed in all study participants to explore the main effects of the diagnosis (aMCI vs. HC), MTHFR C677T genotype (CC vs. CT-TT), APOE ε4 carrier status (ε4 + vs. ε4−), and interactions between these factors. Then, it was conducted, respectively, in the aMCI and HC group to study the effects of genotype (MTHFR, APOE, and MTHFR × APOE). Age, gender, years of education, and TIV were set as covariates.
To further investigate the potential interaction effects between the MTHFR C677T and APOE genotypes, we conducted stratified analyses to detect the effects of diagnosis, MTHFR genotype, and diagnosis × MTHFR genotype among subjects with and without the APOE ε4 allele separately. Furthermore, we defined the brain regions with GM volume significantly affected by interaction effects as regions of interest and extracted their corresponding GM volume for further analysis.
Multiple comparison correction was performed using the cluster-level family wise error method, with statistical significance set to p < 0.05 corrected at the cluster level and p < 0.001 at the voxel-level.
Results
Participant Characteristics
The demographics, clinical, neuropsychological, and genotype characteristics are summarized in Table 1. No significant difference in gender (χ2 = 0.023, p = 0.881), age (t = 1.318, p = 0.191), and years of education (Z = −1.257, p = 0.209) was found between the aMCI group and the HC group. With regard to the vascular risk factors, the prevalence of smoking, alcohol, hypertension, diabetes, heart disease, or hyperlipidemia in both groups was comparable (p > 0.05). No significant difference in serum homocysteine (Z = −0.320, p = 0.749), folate (t = −0.001, p = 0.999), or vitamin B12 (Z = −0.293, p = 0.770) was observed between the aMCI and HC groups.
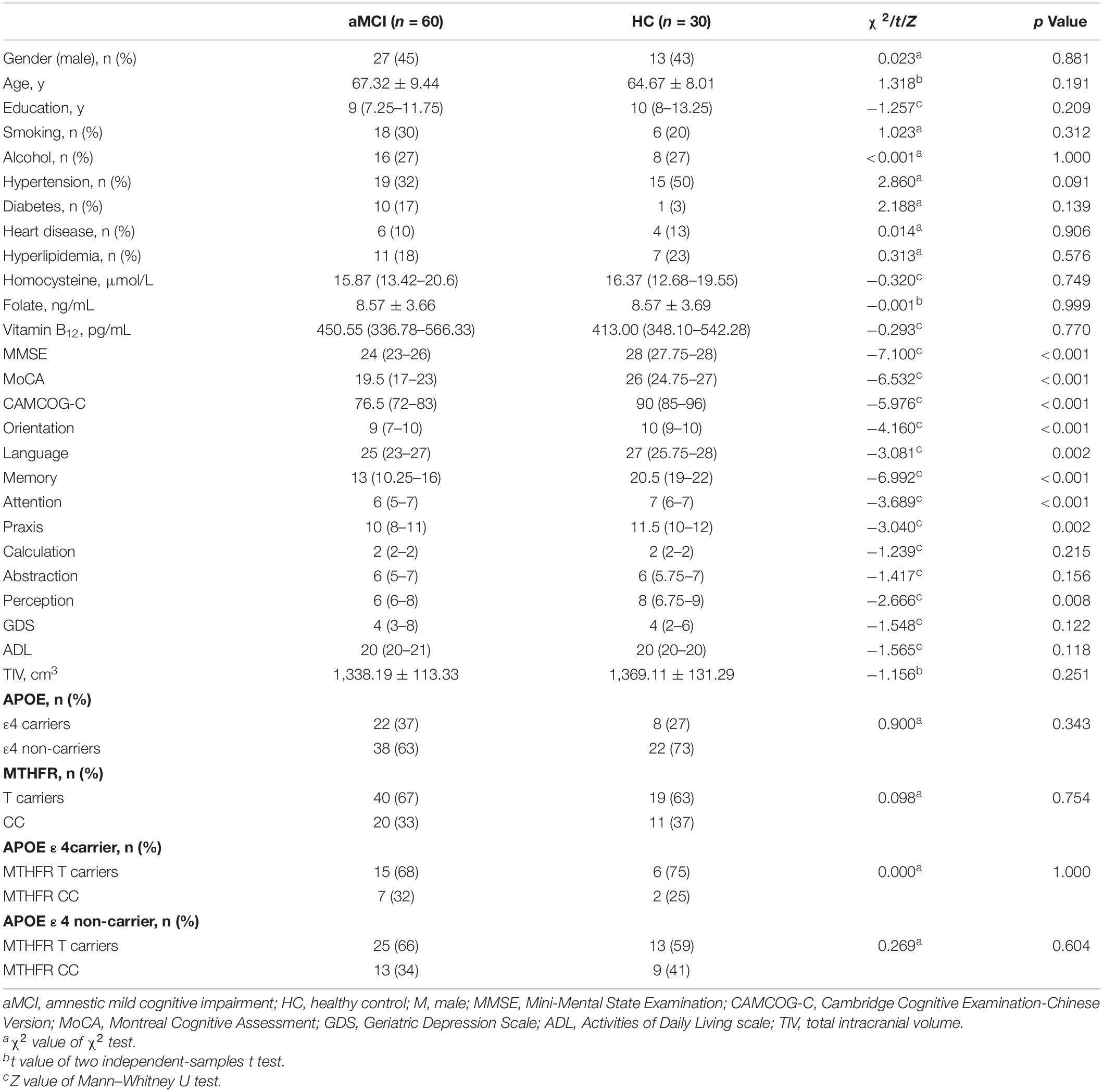
Table 1. Comparison of demographic, clinical, genotype, and neuropsychological characteristics between the amnestic mild cognitive impairment (aMCI) group and the healthy control (HC) group.
Moreover, in terms of cognitive function, the aMCI group exhibited significantly lower MMSE score (Z = −7.100, p < 0.001), MoCA score (Z = −6.532, p < 0.001), total CAMCOG-C score (Z = −5.976, p < 0.001), and its subitem orientation score (Z = −4.160, p < 0.001), language score (Z = −3.081, p = 0.002), memory score (Z = −6.992, p < 0.001), attention score (Z = −3.689, p < 0.001), praxis score (Z = −3.040, p = 0.002), and perception score (Z = −2.666, p = 0.008) compared with the HC group, and no significant difference was observed in calculation score (Z = −1.239, p = 0.215), abstraction score (Z = −1.417, p = 0.156), GDS score (Z = −1.548, p = 0.122), and Activities of Daily Living score (Z = −1.565, p = 0.118) between the two groups. TIV in the two groups was similar (t = −1.156, p = 0.251).
The distributions of the APOE and MTHFR C677T genotypes in the aMCI group and HC group were comparable (χ2 = 0.900, p = 0.343; χ2 = 0.098, p = 0.754). In addition, no significant difference in the prevalence of MTHFR C677T variant was found in the aMCI group and HC group even when the subjects were stratified by APOE ε4 status (χ2 = 0.000, p = 1.000; χ2 = 0.269, p = 0.604).
The Interactions of APOE-MTHFR on GM Volume
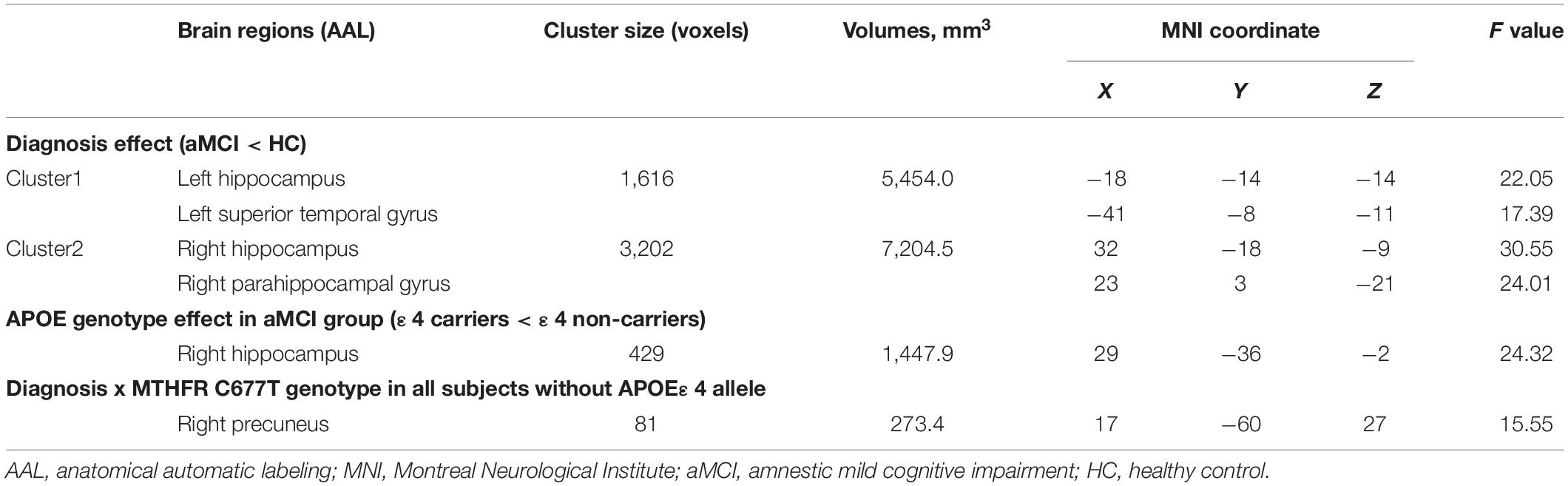
After controlling potential confounding factors, including age, gender, years of education, and TIV, the analysis involving all participants revealed a significant main effect of diagnosis (HC > aMCI) on the bilateral hippocampus, right parahippocampal gyrus, and left superior temporal gyrus (p < 0.05, corrected) (Table 2 and Figure 1). In contrast, the MTHFR C677T and APOE genotypes were not individually significantly associated with GM volume. Moreover, MTHFR × APOE and diagnosis × genotype interactions (diagnosis × MTHFR; diagnosis × APOE; diagnosis × MTHFR × APOE) had no significant effects on GM volume.
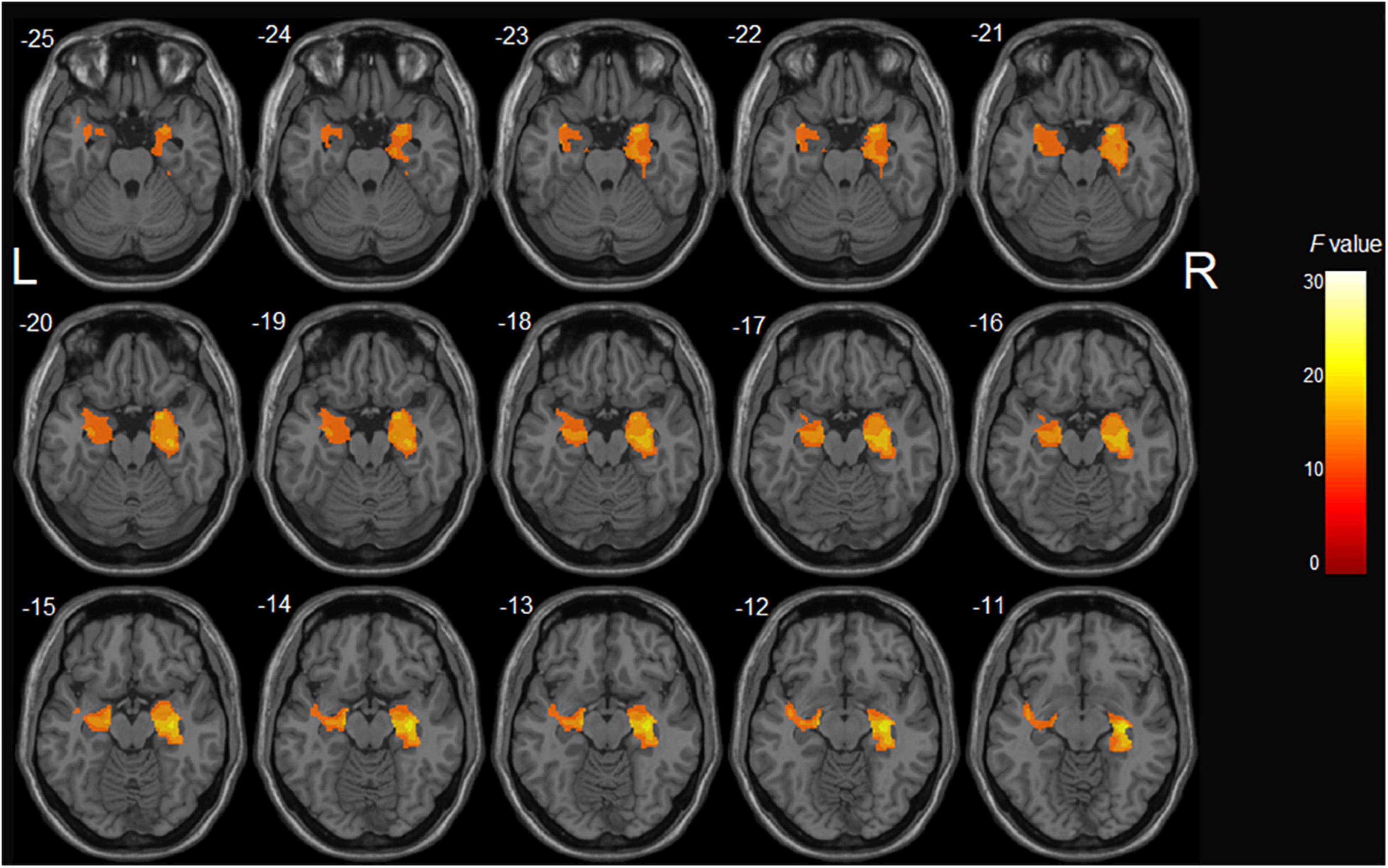
Figure 1. The main effect of diagnosis on gray matter volume in all participants: aMCI patients demonstrated significant gray matter atrophy in the bilateral hippocampus, right parahippocampal gyrus, and left superior temporal gyrus compared with healthy controls.
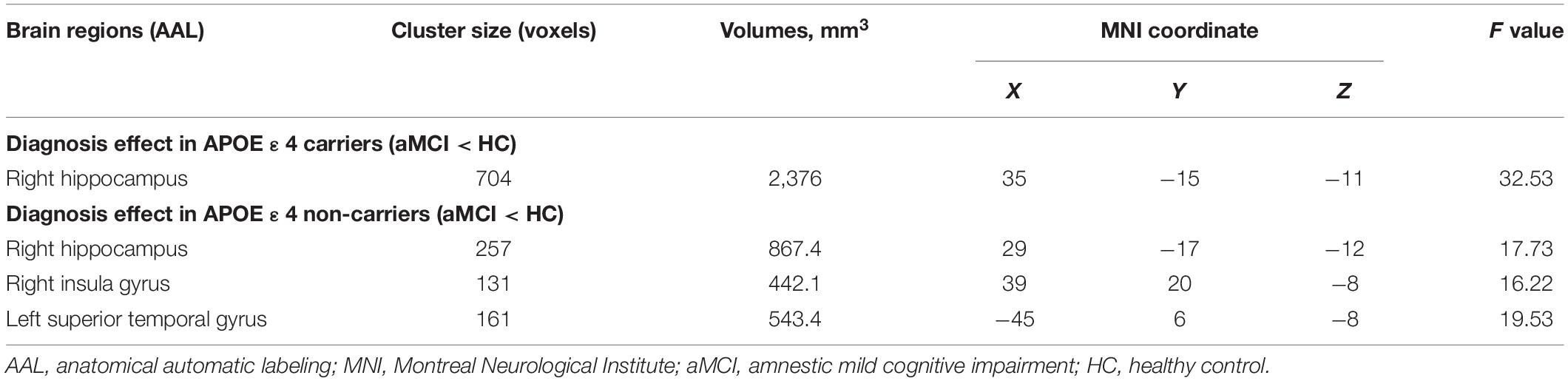
In the aMCI group, analysis showed significant GM volume reduction of the right hippocampus in APOE ε4 carriers compared with non-carriers (p < 0.001, uncorrected) (Table 2 and Figure 2). However, the main effect of MTHFR C677T genotype and epistatic interactions between MTHFR and APOE genotypes on the GM volume were not found. Furthermore, in the HC group, the MTHFR C677T and APOE genotypes and their epistatic interactions had no significant effects on GM volume.
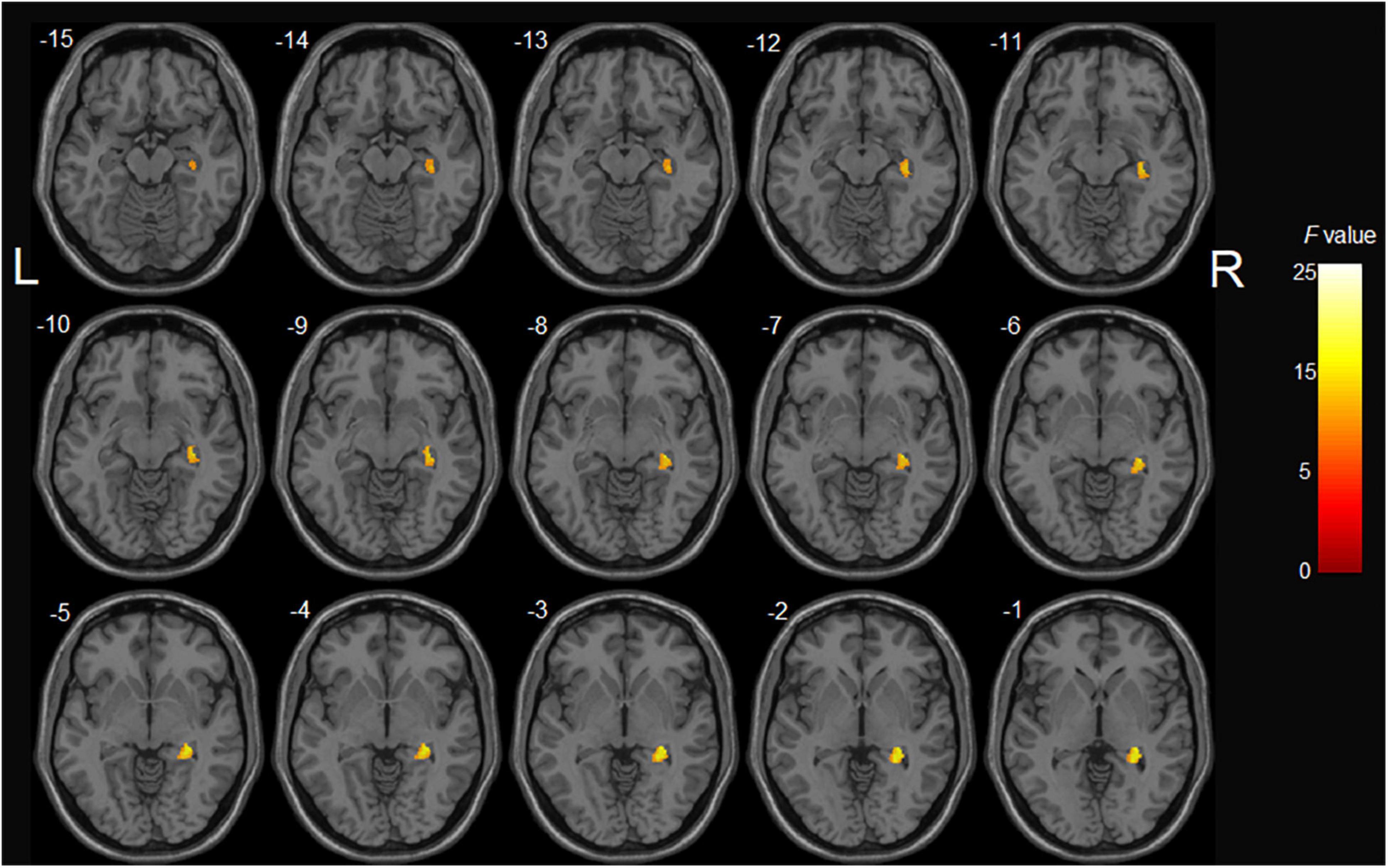
Figure 2. The main effect of APOE genotype on GMV in aMCI patients: among aMCI patients, APOE ε4 carriers demonstrated decreased gray matter volume of the right hippocampus compared with APOE ε4 non-carriers.
The effects of diagnosis, MTHFR C677T, and diagnosis × MTHFR C677T interaction in different APOE genotype subgroups were further investigated. We initially found a significant main effect of diagnosis in the subgroup consisting of APOE ε4 carriers. aMCI patients with the APOE ε4 allele presented a significantly smaller right hippocampus volume than HCs that were APOE ε4 carriers (p < 0.05, corrected) (Table 3). However, no significant effects of the MTHFR C677T genotype itself and interaction of diagnosis × MTHFR genotype on the GM volume were found. Moreover, the analysis was conducted in a subgroup of APOE ε4 non-carriers, and a significant main effect of diagnosis was found (HC > aMCI, p < 0.001, uncorrected) (Table 3); however, the effect size was weaker than that found in the subgroup of APOE ε4 carriers. In addition, when we assessed the main effect of MTHFR C677T polymorphism on GM volume, no significant difference was found in any brain regions between the T carriers and CC homozygotes. Subsequently, we estimated the interaction effect on GM volume between diagnosis and MTHFR genotype and revealed a significant interaction on the right precuneus (p < 0.001, uncorrected) (Table 2 and Figure 3A).
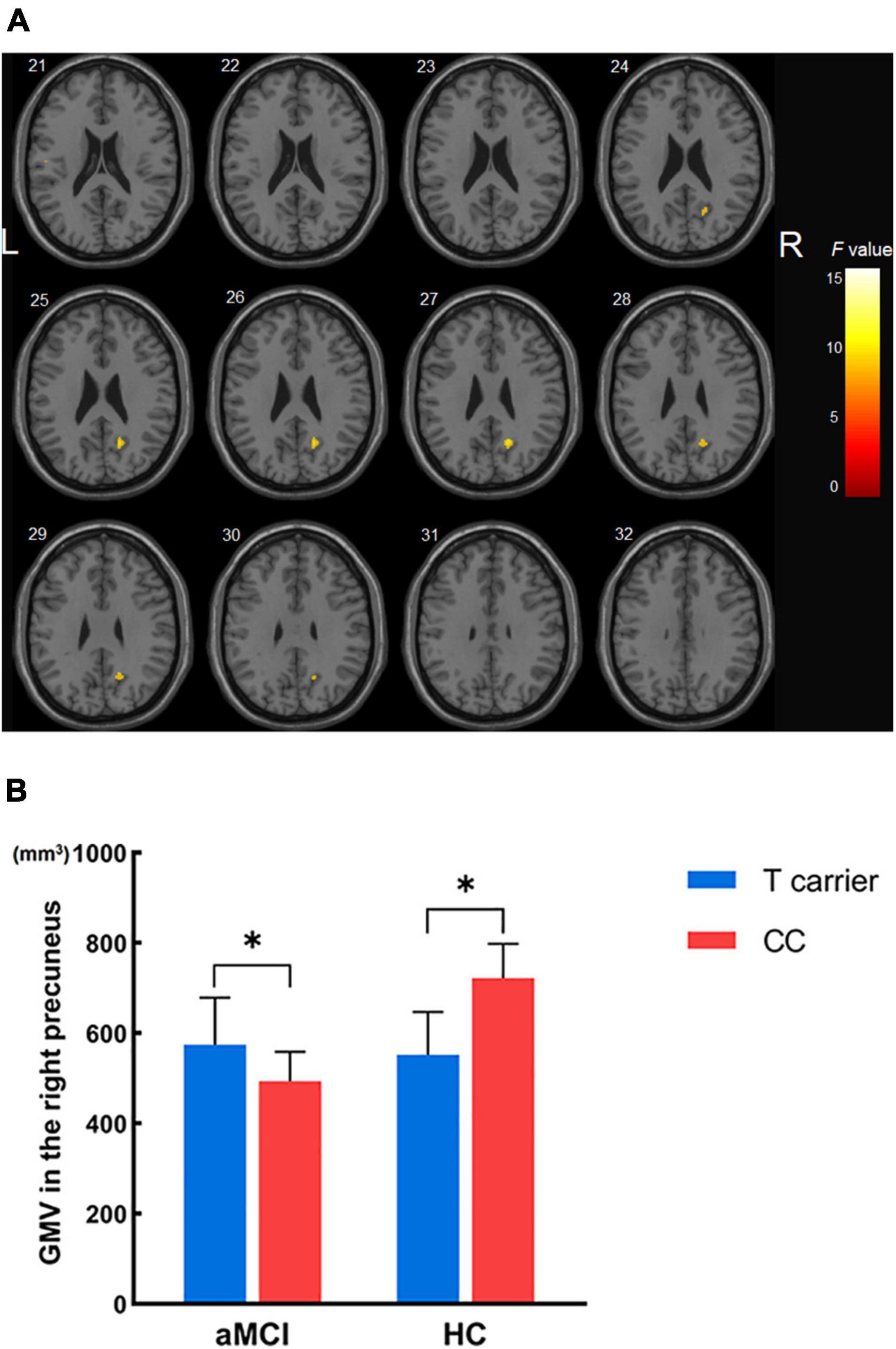
Figure 3. The interaction effects of diagnosis × MTHFR C677T genotype on gray matter volume in all subjects not carrying the APOE ε4 allele. (A) Significant interactions were found in the right precuneus. (B) Gray matter volume of the right precuneus across the four groups. *p < 0.05; GMV, gray matter volume.
Finally, we used simple effect tests to further explore the nature of the interaction effect. The right precuneus was defined as the region of interest and extracted its corresponding GM volume for further analysis as it was the brain region most significantly influenced by diagnosis × MTHFR genotype interaction. Importantly, our analysis revealed that in APOE ε4 non-carriers, a smaller right precuneus volume was found in HCs carrying the MTHFR T variant than in MTHFR CC homozygotes. In contrast with the HC group, a larger right precuneus volume was found in aMCI patients carrying the MTHFR T variant compared with MTHFR CC homozygotes (Figure 3B).
Discussion
This is the first study to document the effects of epistatic interactions between the MTHFR C677T and APOE polymorphisms on GM atrophy in aMCI patients and healthy subjects to the best of our knowledge. We observed a significant main effect of APOE genotype on GM atrophy in the right hippocampus of aMCI patients, and the right precuneus could be influenced by the interaction between diagnosis and MTHFR C677T genotype in aMCI and HC subjects not carrying the APOE ε4 allele. These results provide structural imaging evidence of the effects of genetics in aMCI. Furthermore, the findings provide new insights into the complex roles of multiple genes in aMCI.
The bulk of research showed that GM abnormalities exist in patients with aMCI. Studies have demonstrated that aMCI patients exhibit more GM shrinkage of the hippocampus, parahippocampus gyrus, and temporal lobes (Whitwell et al., 2007). In line with these studies, the main effect of diagnosis in our study showed that aMCI patients experienced significant GM volume reduction of the hippocampus bilaterally, the right parahippocampal gyrus, and the left superior temporal gyrus, compared with HCs. Indeed, the hippocampus and parahippocampal gyrus belong to the medial temporal lobe (MTL) structure and are widely perceived as the initial brain regions affected in pathological studies of AD (Braak and Braak, 1991). GMV atrophy in the MTL is regarded as the neuroimaging hallmark of aMCI and has importantly been reported to be a neurostructural biomarker predicting the progression of aMCI to AD (Ferreira et al., 2011). Furthermore, functional neuroimaging studies also found that activation in the MTL and functional connectivity in subregions in aMCI patients were decreased compared with normal controls (Chen et al., 2016). Functionally, the above structures are generally considered to be the critical rendezvous points in a widespread cerebral network including the ventrolateral temporal lobe, medial and lateral parietal lobes, medial and lateral frontal lobes, and other cortical brain regions and are involved in the episodic memory encoding and retrieval (Dickerson and Eichenbaum, 2010). Notwithstanding that GM atrophy in aMCI has been extensively explored, little is currently known on the changes in GM volume caused by genetic factors.
Among multiple genetic factors associated with sporadic AD, the APOE gene is regarded as the most critical risk factor for AD. Most importantly, among the three isoform alleles (ε2, ε3, and ε4), APOE ε4 has been reported as the primary genetic risk factor (Liu et al., 2013) and associated with amyloid-β aggregation, synaptic plasticity, cholesterol homeostasis, neuroinflammation, and neurovascular functions (Bell et al., 2012). Several neuroimaging studies on AD and aMCI have shown smaller hippocampus volume and a faster rate of hippocampus atrophy in APOE ε4 carriers than non-carriers (Hashimoto et al., 2001; Schuff et al., 2009; Spampinato et al., 2011; Manning et al., 2014). Consistently, we found significant GM atrophy in the right hippocampus of APOE ε4 carriers compared with non-carriers in aMCI patients. It has been widely recognized that MTHFR C677T polymorphism is related to AD and brain structural alterations. In this regard, the MTHFR C677T variant has been associated with smaller regional brain volume in MCI patients (Rajagopalan et al., 2012). Furthermore, this variant has been associated with local brain atrophy in brain areas involved in intellectual and emotional functions, especially the medial orbitofrontal cortex (Roussotte et al., 2017). In addition, the MTHFR C677T variant can reportedly promote brain atrophy by increasing homocysteine levels (Rajagopalan et al., 2012; Roussotte et al., 2017). Interestingly, previous imaging studies showed that older adults with elevated homocysteine levels had more pronounced regional brain atrophy (Rajagopalan et al., 2011) and thinner cortical GM (Madsen et al., 2014). Besides, hyperhomocysteinemia has been associated with hippocampal atrophy in cognitively normal older adults (Williams et al., 2002; den Heijer et al., 2003) and more severe MTL atrophy in AD (Hooshmand et al., 2013).
Given that AD is a polygenic disorder, it is a matter of debate whether the interactions between MTHFR and APOE genes are related to the pathogenesis of AD (Tysoe et al., 1997; Chapman et al., 1998; Brunelli et al., 2001; Seripa et al., 2003; Wang et al., 2005; Keikhaee et al., 2006; Styczyńska et al., 2008; Bi et al., 2009; Mansouri et al., 2013). Extensive research has identified a correlation between MTHFR C677T polymorphism and the risk of AD, which, however, varied with the APOE ε4 carrier status (Bi et al., 2009; Zhang et al., 2010; Peng et al., 2015; Rai, 2017). This correlation has been documented to be stronger in APOE ε4 carriers (Bi et al., 2009; Roussotte et al., 2016). Interestingly, some studies found that the MTHFR C677T variant could influence susceptibility to AD in APOE ε4 non-carriers (Wang et al., 2005; Kim et al., 2008). One such meta-analysis based on 40 case–control studies revealed that MTHFR C677T polymorphism might contribute to the risk of AD, particularly in APOE ε4 carriers (Peng et al., 2015), whereas another meta-analysis, which included only well-designed case–control studies and strict diagnostic criteria, held that MTHFR C677T variant might also influence susceptibility to AD in APOE ε4 non-carriers (odds ratio = 1.27; 95% confidence interval = 0.97–2.02) (Zhang et al., 2010). In light of this, it would not be surprising that the correlation between MTHFR polymorphism and GM atrophy in aMCI patients was present only in APOE ε4 non-carriers in our current study. This finding can be explained by the following reasons. First, studies have shown that APOE ε4 is not only the most important genetic risk factor but also plays a dominant role in pathogenesis of sporadic AD, whereas MTHFR C677T is a relatively weak genetic risk factor for AD (Zuin et al., 2021). Our findings suggest that in APOE ε4 carriers, the MTHFR gene does not influence GM volume. Furthermore, we found the main effect of APOE gene on GM atrophy in aMCI patients; however, the main effect of MTHFR genotype on GM volume was not found in aMCI patients, nor did we find an interaction between MTHFR and diagnosis on GMV in participants with APOE ε4 carriers. Nevertheless, the results implicated the influence of the MTHFR genotype on the right precuneus volume in both aMCI patients and HCs not carrying the APOE ε4 allele. Second, the MTHFR CC genotype exerted a protective effect against AD, whereas the MTHFR 677T variant could accelerate AD progression by impairing homocysteine metabolism and promoting oxidative stress. In this regard, previous studies suggested that folate, a major regulatory factor for MTHFR activity and homocysteine levels, could quench oxidative damage (Shea and Rogers, 2002; Shea et al., 2002). Accordingly, abnormal metabolism of homocysteine and/or inappropriate folate intake may impair the oxidative stress response. The allele-specific antioxidant potential of APOE (ε2 > ε3 > ε4) has been substantiated in biological studies (Tamaoka et al., 2000; Colton et al., 2002), suggesting that the MTHFR CC genotype in APOE ε4 non-carriers leads to synergistic beneficial effects against oxidative stress. These findings suggest that individuals without the APOE ε4 allele are more susceptible to be influenced by MTHFR C677T polymorphism.
Herein, we found significant interaction effects between MTHFR C677T polymorphism and diagnosis on the right precuneus. The precuneus is a region of the posteromedial parietal lobe (Cavanna and Trimble, 2006) that plays an essential role in visuospatial imagery, episodic memory retrieval, self-processing, and consciousness and has various reciprocal connections with frontal, temporal, and parietal cortices (Cavanna and Trimble, 2006). Interestingly, it has been shown that changes in precuneus function may have pathophysiological relevance in aMCI development, with accumulating evidence indicating the presence of cortical thinning, metabolic alterations, and early amyloid deposition in the precuneus of aMCI patients (Bailly et al., 2015a,b; Coutinho et al., 2015; Csukly et al., 2016). In all APOE ε4 non-carriers of present study, a smaller right precuneus volume was found in healthy subjects who were MTHFR T carriers than healthy subjects that were MTHFR CC homozygotes. Intriguingly, opposite results were found in the aMCI group, with a larger right precuneus volume in MTHFR T carriers than MTHFR CC homozygotes. We hypothesized that this phenomenon might be related to adaptation and compensation during the process of aMCI development. Most importantly, our results corroborated that individuals without the APOE ε4 allele are more vulnerable to the effect of MTHFR C677T polymorphism, and structural alterations to the right precuneus could be influenced by the MTHFR C677T genotype in both aMCI patients and HCs who do not carry the APOE ε4 allele. MTHFR C677T polymorphism is reportedly the major genetic modifier associated with disorders of the folate cycle and homocysteine metabolism, which can be treated by supplementations of folate and vitamin B (Fohr et al., 2002; Du et al., 2018; Huang et al., 2018). There is still an ongoing debate on whether folate and/or vitamin B supplementation can improve cognition or slow the rate of brain atrophy in AD and/or MCI by lowing homocysteine levels (Durga, 2007; Kang et al., 2008; Smith et al., 2010; Ford and Almeida, 2012; Douaud et al., 2013; Clarke et al., 2014; Ma et al., 2019). Heterogeneity in findings of these studies may be influenced by the limitations during the selection of subjects, different lengths of intervention, and inconsistent inclusion criteria (Smith and Refsum, 2016). In addition, we speculate that the absence of APOE and MTHFR polymorphisms in these intervention trials may also be an important factor of poor intervention effects. Accordingly, future studies exploring the efficiency of homocysteine therapy tailored to different genetic backgrounds are needed to confirm our assumption.
There were several limitations in this study that should be considered. First, our sample size was limited, emphasizing the need to validate our findings in clinical studies with large sample sizes. Furthermore, given the limited number of aMCI patients enrolled, we could not further classify the aMCI patients into single-domain (aMCI-SD) or multidomain (aMCI-MD) (Petersen et al., 1999, 2009), which could impact the outcome of our study. Moreover, it should be noted that other genes associated with AD, including SCIMP, SLC2A4, CLU, and PICALM, were not taken into consideration in this study. Finally, we acknowledge that the observational imaging-genetic approach used in this cross-sectional study limits the causal inference.
Conclusion
In summary, the current research provides preliminary evidence indicating that the interaction between diagnosis and MTHFR C677T polymorphism has a structural effect on the right precuneus in HCs and aMCI patients without the APOE ε4 allele. Confirmation of our findings in larger non-APOE ε4 cohorts is required. More studies should be conducted in the future, with study subjects stratified according to different genetic background, environmental exposure, or other risk factors, to clarify the possible role of MTHFR C677T polymorphism in the pathogenesis of AD and/or aMCI.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committees of the First Affiliated Hospital of Anhui Medical University (Reference no, Quick-PJ 2021-13-18). The participants provided their written informed consent to participate in this study.
Author Contributions
MY performed the analysis and wrote the manuscript. MY and XZho conceived and designed the experiments. WY, KW, and WZha helped to collect neuropsychological data. CL, ML, and WZhu helped to perform MRI and collect MRI data. XZhu and ZS designed and supervised the study. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81771154), the Key Research and Development Projects of Anhui Province (Grant No. 202104j07020031), the Natural Science Foundation of Anhui province (Grant No. 1908085QH322), and the Basic and Clinical Cooperative Research Promotion Plan of Anhui Medical University (Grant No. 2020xkjT026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the subjects for their participation in our study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2021.778123/full#supplementary-material
Footnotes
- ^ http://www.snpway.com/snpdecoder/
- ^ http://dbm.neuro.uni-jena.de/vbm8/
- ^ http://www.restfmri.net/forum/REST_V1.8
References
Albert, M. S., DeKosky, S. T., Dickson, D., Dubois, B., Feldman, H. H., Fox, N. C., et al. (2011). The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging- Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 270–279. doi: 10.1016/j.jalz.2011.03.008
Bailly, M., Destrieux, C., Hommet, C., Mondon, K., Cottier, J.-P., Beaufils, E., et al. (2015a). Precuneus and cingulate cortex atrophy and hypometabolism in patients with Alzheimer’s disease and mild cognitive impairment: MRI and 18 F-FDG PET quantitative analysis using FreeSurfer. Biomed. Res. Int. 2015:583931. doi: 10.1155/2015/583931
Bailly, M., Ribeiro, M. J. S., Vercouillie, J., Hommet, C., Gissot, V., Camus, V., et al. (2015b). 18F-FDG and 18F-Florbetapir PET in clinical practice: regional analysis in mild cognitive impairment and alzheimer disease. Clin. Nucl. Med. 40, e111–e116. doi: 10.1097/RLU.0000000000000666
Bednarska-Makaruk, M., Graban, A., Sobczyńska-Malefora, A., Harrington, D. J., Mitchell, M., Voong, K., et al. (2016). Homocysteine metabolism and the associations of global DNA methylation with selected gene polymorphisms and nutritional factors in patients with dementia. Exp. Gerontol. 81, 83–91. doi: 10.1016/j.exger.2016.05.002
Bell, R. D., Winkler, E. A., Singh, I., Sagare, A. P., Deane, R., Wu, Z., et al. (2012). Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516. doi: 10.1038/nature11087
Bellenguez, C., Charbonnier, C., Grenier-Boley, B., Quenez, O., Le Guennec, K., Nicolas, G., et al. (2017). Contribution to Alzheimer’s disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol. Aging 59, 220.e1–220.e9. doi: 10.1016/j.Neurobiolaging.2017.07.001
Bi, X.-H., Zhao, H.-L., Zhang, Z.-X., and Zhang, J.-W. (2009). Association of RFC1 A80G and MTHFR C677T polymorphisms with Alzheimer’s disease. Neurobiol. Aging 30, 1601–1607. doi: 10.1016/j.neurobiolaging.2007.12.010
Braak, H., and Braak, E. (1991). Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259. doi: 10.1007/BF00308809
Brunelli, T., Bagnoli, S., Giusti, B., Nacmias, B., Pepe, G., Sorbi, S., et al. (2001). The C677T methylenetetrahydrofolate reductase mutation is not associated with Alzheimer’s disease. Neurosci. Lett. 315, 103–105. doi: 10.1016/S0304-3940(01)02316-3
Cavanna, A. E., and Trimble, M. R. (2006). The precuneus: a review of its functional anatomy and behavioural correlates. Brain 129, 564–583. doi: 10.1093/brain/awl004
Chang, Y.-T., Hsu, S.-W., Huang, S.-H., Huang, C.-W., Chang, W.-N., Lien, C.-Y., et al. (2019). ABCA7 polymorphisms correlate with memory impairment and default mode network in patients with APOEε4-associated Alzheimer’s disease. Alzheimers Res. Ther. 11:103. doi: 10.1186/s13195-019-0563-3
Chang, Y.-T., Hsu, S.-W., Tsai, S.-J., Chang, Y.-T., Huang, C.-W., Liu, M.-E., et al. (2017). Genetic effect of MTHFR C677T polymorphism on the structural covariance network and white-matter integrity in Alzheimer’s disease: MTHFR C677T Polymorphism in Alzheimer’s Disease. Hum. Brain Mapp. 38, 3039–3051. doi: 10.1002/hbm.23572
Chapman, J., Wang, N., Treves, T. A., Korczyn, A. D., and Bornstein, N. M. (1998). ACE, MTHFR, factor V Leiden, and APOE polymorphisms in patients with vascular and Alzheimer’s dementia. Stroke 29, 1401–1404. doi: 10.1161/01.STR.29.7.1401
Chen, J., Duan, X., Shu, H., Wang, Z., Long, Z., Liu, D., et al. (2016). Differential contributions of subregions of medial temporal lobe to memory system in amnestic mild cognitive impairment: insights from fMRI study. Sci. Rep. 6:26148. doi: 10.1038/srep26148
Chung, Y. C., Kruyer, A., Yao, Y., Feierman, E., Richards, A., Strickland, S., et al. (2016). Hyperhomocysteinemia exacerbates Alzheimer’s disease pathology by way of the β-amyloid fibrinogen interaction. J. Thromb. Haemost. 14, 1442–1452. doi: 10.1111/jth.13340
Clarke, R., Bennett, D., Parish, S., Lewington, S., Skeaff, M., Eussen, S. J., et al. (2014). Effects of homocysteine lowering with B vitamins on cognitive aging: meta-analysis of 11 trials with cognitive data on 22,000 individuals. Am. J. Clin. Nutr. 100, 657–666. doi: 10.3945/ajcn.113.076349
Colton, C., Brown, C., Cook, D., Needham, L., Xu, Q., Czapiga, M., et al. (2002). APOE and the regulation of microglial nitric oxide production: a link between genetic risk and oxidative stress. Neurobiol. Aging 23, 777–785.
Coppede, F. (2010). One-Carbon metabolism and Alzheimer’s disease: focus on epigenetics. Curr. Genomics 11, 246–260. doi: 10.2174/138920210791233090
Coutinho, A. M. N., Porto, F. H. G., Duran, F. L. S., Prando, S., Ono, C. R., Feitosa, E. A. A. F., et al. (2015). Brain metabolism and cerebrospinal fluid biomarkers profile of non-amnestic mild cognitive impairment in comparison to amnestic mild cognitive impairment and normal older subjects. Alzheimers Res. Ther. 7:58. doi: 10.1186/s13195-015-0143-0
Csukly, G., Sirály, E., Fodor, Z., Horváth, A., Salacz, P., Hidasi, Z., et al. (2016). The differentiation of amnestic type MCI from the non-amnestic types by structural MRI. Front. Aging Neurosci. 8:52. doi: 10.3389/fnagi.2016.00052
Deep, S. N., Mitra, S., Rajagopal, S., Paul, S., and Poddar, R. (2019). GluN2A-NMDA receptor–mediated sustained Ca 2+ influx leads to homocysteine-induced neuronal cell death. J. Biol. Chem. 294, 11154–11165. doi: 10.1074/jbc.RA119.008820
den Heijer, T., Vermeer, S. E., Clarke, R., Oudkerk, M., Koudstaal, P. J., Hofman, A., et al. (2003). Homocysteine and brain atrophy on MRI of non-demented elderly. Brain 126, 170–175. doi: 10.1093/brain/awg006
Di Meco, A., Li, J.-G., Barrero, C., Merali, S., and Praticò, D. (2019). Elevated levels of brain homocysteine directly modulate the pathological phenotype of a mouse model of tauopathy. Mol. Psychiatry 24, 1696–1706. doi: 10.1038/s41380-018-0062-0
Dickerson, B. C., and Eichenbaum, H. (2010). The episodic memory system: neurocircuitry and disorders. Neuropsychopharmacology 35, 86–104. doi: 10.1038/npp.2009.126
Douaud, G., Refsum, H., de Jager, C. A., Jacoby, R., Nichols, T. E., Smith, S. M., et al. (2013). Preventing Alzheimer’s disease-related gray matter atrophy by B-vitamin treatment. Proc. Natl. Acad. Sci. U.S.A. 110, 9523–9528. doi: 10.1073/pnas.1301816110
Du, B., Tian, H., Tian, D., Zhang, C., Wang, W., Wang, L., et al. (2018). Genetic polymorphisms of key enzymes in folate metabolism affect the efficacy of folate therapy in patients with hyperhomocysteinaemia. Br. J. Nutr. 119, 887–895. doi: 10.1017/S0007114518000508
Durga, J. (2007). E?ect of 3-year folic acid supplementation on cognitive function in older adults in the FACIT trial: a randomised, double blind, controlled trial. Lancet 369, 208–216. doi: 10.1016/S0140-6736(07)60109-3
Esse, R., Barroso, M., Tavares de Almeida, I., and Castro, R. (2019). The contribution of homocysteine metabolism disruption to endothelial dysfunction: state-of-the-art. Int. J. Mol. Sci. 20:867. doi: 10.3390/ijms20040867
Ferreira, L. K., Diniz, B. S., Forlenza, O. V., Busatto, G. F., and Zanetti, M. V. (2011). Neurostructural predictors of Alzheimer’s disease: a meta-analysis of VBM studies. Neurobiol. Aging 32, 1733–1741. doi: 10.1016/j.neurobiolaging.2009.11.008
Fohr, I. P., Prinz-Langenohl, R., Brönstrup, A., Bohlmann, A. M., Nau, H., Berthold, H. K., et al. (2002). 5,10-Methylenetetrahydrofolate reductase genotype determines the plasma homocysteine-lowering effect of supplementation with 5-methyltetrahydrofolate or folic acid in healthy young women. Am. J. Clin. Nutr. 75, 275–282. doi: 10.1093/ajcn/75.2.275
Ford, A. H., and Almeida, O. P. (2012). Effect of homocysteine lowering treatment on cognitive function: a systematic review and meta-analysis of randomized controlled trials. Drugs Aging 29, 133–149. doi: 10.3233/JAD-2012-111739
Frosst, P., Blom, H. J., Milos, R., Goyette, P., Sheppard, C. A., Matthews, R. G., et al. (1995). A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 10, 111–113. doi: 10.1038/ng0595-111
Hashimoto, M., Yasuda, M., Tanimukai, S., Matsui, M., Hirono, N., Kazui, H., et al. (2001). Apolipoprotein E 4 and the pattern of regional brain atrophy in Alzheimer’s disease. Neurology 57, 1461–1466. doi: 10.1212/WNL.57.8.1461
Hoffman, A., Taleski, G., Qian, H., Wasek, B., Arning, E., Bottiglieri, T., et al. (2018). Methylenetetrahydrofolate reductase deficiency deregulates regional brain amyloid-β protein precursor expression and phosphorylation levels. J. Alzheimers Dis. 64, 223–237. doi: 10.3233/JAD-180032
Hooshmand, B., Polvikoski, T., Kivipelto, M., Tanskanen, M., Myllykangas, L., Erkinjuntti, T., et al. (2013). Plasma homocysteine, Alzheimer and cerebrovascular pathology: a population-based autopsy study. Brain 136, 2707–2716. doi: 10.1093/brain/awt206
Hu, Q., Teng, W., Li, J., Hao, F., and Wang, N. (2016). Homocysteine and Alzheimer’s disease: evidence for a causal link from Mendelian randomization. J. Alzheimers Dis. 52, 747–756. doi: 10.3233/JAD-150977
Huang, X., Qin, X., Yang, W., Liu, L., Jiang, C., Zhang, X., et al. (2018). MTHFR gene and serum folate interaction on serum homocysteine lowering: prospect for precision folic acid treatment. Arterioscler Thromb. Vasc. Biol. 38, 679–685. doi: 10.1161/ATVBAHA.117.310211
Jansen, I. E., Savage, J. E., Watanabe, K., Bryois, J., Williams, D. M., Steinberg, S., et al. (2019). Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413. doi: 10.1038/s41588-018-0311-9
Kang, J. H., Cook, N., Manson, J., Buring, J. E., Albert, C. M., and Grodstein, F. (2008). A trial of B vitamins and cognitive function among women at high risk of cardiovascular disease. Am. J. Clin. Nutr. 88, 1602–1610. doi: 10.3945/ajcn.2008.26404
Keikhaee, M. R., Hashemi, S. B., Najmabadi, H., and Noroozian, M. (2006). C677T methylentetrahydrofulate reductase and angiotensin converting enzyme gene polymorphisms in patients with Alzheimer’s disease in Iranian population. Neurochem. Res. 31, 1079–1083. doi: 10.1007/s11064-006-9119-6
Kim, J.-M., Stewart, R., Kim, S.-W., Yang, S.-J., Shin, I.-S., Shin, H.-Y., et al. (2008). Methylenetetrahydrofolate reductase gene and risk of Alzheimer’s disease in Koreans. Int. J. Geriat. Psychiatry 23, 454–459. doi: 10.1002/gps.1903
Liu, C.-C., Kanekiyo, T., Xu, H., and Bu, G. (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118. doi: 10.1038/nrneurol.2012.263
Ma, F., Li, Q., Zhou, X., Zhao, J., Song, A., Li, W., et al. (2019). Effects of folic acid supplementation on cognitive function and Aβ-related biomarkers in mild cognitive impairment: a randomized controlled trial. Eur. J. Nutr. 58, 345–356. doi: 10.1007/s00394-017-1598-5
Madsen, S. K., Rajagopalan, P., Joshi, S. H., Toga, A. W., and Thompson, P. M. (2014). Higher homocysteine associated with thinner cortical gray matter in 803 participants from the Alzheimer’s Disease Neuroimaging Initiative. Neurobiol. Aging 36(Suppl. 1), S203–S210. doi: 10.1016/j.neurobiolaging.2014.01.154
Manning, E. N., Barnes, J., Cash, D. M., Bartlett, J. W., Leung, K. K., Ourselin, S., et al. (2014). APOE ε4 Is associated with disproportionate progressive hippocampal atrophy in AD. PLoS One 9:e97608. doi: 10.1371/journal.pone.0097608
Mansouri, L., Fekih-Mrissa, N., Klai, S., Mansour, M., Gritli, N., and Mrissa, R. (2013). Association of methylenetetrahydrofolate reductase polymorphisms with susceptibility to Alzheimer’s disease. Clin. Neurol. Neurosurg. 115, 1693–1696. doi: 10.1016/j.clineuro.2013.03.015
Peng, Q., Lao, X., Huang, X., Qin, X., Li, S., and Zeng, Z. (2015). The MTHFR C677T polymorphism contributes to increased risk of Alzheimer’s Disease: evidence based on 40 case-control studies. Neurosci. Lett. 586, 36–42. doi: 10.1016/j.neulet.2014.11.049
Petersen, R. C., Roberts, R. O., Knopman, D. S., Boeve, B. F., Geda, Y. E., Ivnik, R. J., et al. (2009). Mild cognitive impairment: ten years later. Arch. Neurol. 66, 1447–1455. doi: 10.1001/archneurol.2009.266
Petersen, R. C., Smith, G. E., Waring, S. C., Ivnik, R. J., Tangalos, E. G., and Kokmen, E. (1999). Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 56, 303–308. doi: 10.1001/archneur.56.3.303
Rai, V. (2017). Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and Alzheimer disease risk: a meta-analysis. Mol. Neurobiol. 54, 1173–1186. doi: 10.1007/s12035-016-9722-8
Rajagopalan, P., Hua, X., Toga, A. W., Jack, C. R. Jr., Weiner, M. W., and Thompson, P. M. (2011). Homocysteine effects on brain volumes mapped in 732 elderly individuals. Neuroreport 22, 391–395. doi: 10.1097/WNR.0b013e328346bf85
Rajagopalan, P., Jahanshad, N., Stein, J. L., Hua, X., Madsen, S. K., Kohannim, O., et al. (2012). Common folate gene variant, MTHFR C677T, is associated with brain structure in two independent cohorts of people with mild cognitive impairment. Neuroimage Clin. 1, 179–187. doi: 10.1016/j.nicl.2012.09.012
Ridge, P. G., Mukherjee, S., Crane, P. K., Kauwe, J. S. K. and Alzheimer’s Disease Genetics Consortium (2013). Alzheimer’s disease: analyzing the missing heritability. PLoS One 8:e79771. doi: 10.1371/journal.pone.0079771
Roussotte, F. F., Hua, X., Narr, K. L., Small, G. W., and Thompson, P. M. (2017). The C677T variant in MTHFR modulates associations between brain integrity, mood, and cognitive functioning in old age. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2, 280–288. doi: 10.1016/j.bpsc.2016.09.005
Roussotte, F. F., Narr, K. L., Small, G. W., and Thompson, P. M. (2016). The C677T variant in MTHFR modulates associations between blood-based and cerebrospinal fluid biomarkers of neurodegeneration. NeuroReport 27, 948–951. doi: 10.1097/WNR.0000000000000636
Scheltens, P., De Strooper, B., Kivipelto, M., Holstege, H., Chételat, G., Teunissen, C. E., et al. (2021). Alzheimer’s disease. Lancet 397, 1577–1590. doi: 10.1016/S0140-6736(20)32205-4
Schuff, N., Woerner, N., Boreta, L., Kornfield, T., Shaw, L. M., Trojanowski, J. Q., et al. (2009). MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain 132, 1067–1077. doi: 10.1093/brain/awp007
Seripa, D., Forno, G. D., Matera, M. G., Gravina, C., Margaglione, M., Palermo, M. T., et al. (2003). Methylenetetrahydrofolate reductase and angiotensin converting enzyme gene polymorphisms in two genetically and diagnostically distinct cohort of Alzheimer patients. Neurobiol. Aging 24, 933–939. doi: 10.1016/S0197-4580(03)00040-X
Shea, T. B., and Rogers, E. (2002). Folate quenches oxidative damage in brains of apolipoprotein E-deficient mice: augmentation by vitamin E. Brain Res. Mol. Brain Res. 108, 1–6. doi: 10.1016/S0169-328X(02)00412-6
Shea, T. B., Rogers, E., Ashline, D., Ortiz, D., and Sheu, M.-S. (2002). Apolipoprotein E deficiency promotes increased oxidative stress and compensatory increases in antioxidants in brain tissue. Free Radic. Biol. Med. 33, 1115–1120. doi: 10.1016/S0891-5849(02)01001-8
Sims, R., Hill, M., and Williams, J. (2020). The multiplex model of the genetics of Alzheimer’s disease. Nat. Neurosci. 23, 311–322. doi: 10.1038/s41593-020-0599-5
Smith, A. D., and Refsum, H. (2016). Homocysteine, B vitamins, and cognitive impairment. Annu. Rev. Nutr. 36, 211–239. doi: 10.1146/annurev-nutr-071715-050947
Smith, A. D., Refsum, H., Bottiglieri, T., Fenech, M., Hooshmand, B., McCaddon, A., et al. (2018). Homocysteine and dementia: an international consensus statement. J. Alzheimers Dis. 62, 561–570. doi: 10.3233/JAD-171042
Smith, A. D., Smith, S. M., de Jager, C. A., Whitbread, P., Johnston, C., Agacinski, G., et al. (2010). Homocysteine-Lowering by B vitamins slows the rate of accelerated brain atrophy in mild cognitive impairment: a randomized controlled trial. PLoS One 5:e12244. doi: 10.1371/journal.pone.0012244
Sontag, E., Sontag, J. M., Wasek, B., Taleski, G., Smith, J., Arning, E., et al. (2014). Altered protein phosphatase 2A methylation and Tau phosphorylation in the young and aged brain of methylenetetrahydrofolate reductase (MTHFR) deficient mice. Front. Aging Neurosci. 6:214. doi: 10.3389/fnagi.2014.00214
Spampinato, M. V., Rumboldt, Z., Hosker, R. J., Mintzer, J. E. and Alzheimer’s Disease Neuroimaging Initiative. (2011). Apolipoprotein E and gray matter volume loss in patients with mild cognitive impairment and Alzheimer disease. Radiology 258, 843–852. doi: 10.1148/radiol.10100307
Stoccoro, A., Tannorella, P., Salluzzo, M. G., Ferri, R., Romano, C., Nacmias, B., et al. (2017). The methylenetetrahydrofolate reductase C677T polymorphism and risk for late-onset Alzheimer’s disease: further evidence in an Italian multicenter study. J. Alzheimers Dis. 56, 1451–1457. doi: 10.3233/JAD-161081
Styczyńska, M., Strosznajder, J. B., Religa, D., Chodakowska-Zebrowska, M., Pfeffer, A., Gabryelewicz, T., et al. (2008). Association between genetic and environmental factors and the risk of Alzheimer’s disease. Folia Neuropathol. 46, 249–254.
Tamaoka, A., Miyatake, F., Matsuno, S., Ishii, K., Nagase, S., Sahara, N., et al. (2000). Apolipoprotein E allele–dependent antioxidant activity in brains with Alzheimer’s disease. Neurology 54, 2319–2321. doi: 10.1212/WNL.54.12.2319
Tysoe, C., Galinsky, D., Robinson, D., Brayne, C. E., Easton, D. F., Huppert, F. A., et al. (1997). Analysis of alpha-1 antichymotrypsin, presenilin-1, angiotensin-converting enzyme, and methylenetetrahydrofolate reductase loci as candidates for dementia. Am. J. Med. Genet. 74, 207–212.
Wang, B., Jin, F., Kan, R., Ji, S., Zhang, C., Lu, Z., et al. (2005). Association of MTHFR gene polymorphism C677T with susceptibility to late-onset Alzheimer’s disease. J. Mol. Neurosci. 27, 23–27.
Whitwell, J. L., Przybelski, S. A., Weigand, S. D., Knopman, D. S., Boeve, B. F., Petersen, R. C., et al. (2007). 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain 130, 1777–1786. doi: 10.1093/brain/awm112
Williams, J. H., Pereira, E. A., Budge, M. M., and Bradley, K. M. (2002). Minimal hippocampal width relates to plasma homocysteine in community-dwelling older people. Age Ageing 31, 440–444. doi: 10.1093/ageing/31.6.440
Wu, X., Zhang, L., Miao, Y., Yang, J., Wang, X., Wang, C., et al. (2019). Homocysteine causes vascular endothelial dysfunction by disrupting endoplasmic reticulum redox homeostasis. Redox Biol. 20, 46–59. doi: 10.1016/j.redox.2018.09.021
Zhang, M.-Y., Miao, L., Li, Y.-S., and Hu, G.-Y. (2010). Meta-analysis of the methylenetetrahydrofolate reductase C677T polymorphism and susceptibility to Alzheimer’s disease. Neurosci. Res. 68, 142–150. doi: 10.1016/j.neures.2010.06.011
Keywords: methylenetetrahydrofolate reductase, Alzheimer’s disease, amnestic mild cognitive impairment, apolipoprotein E, voxel-based morphometry
Citation: You M, Zhou X, Yin W, Wan K, Zhang W, Li C, Li M, Zhu W, Zhu X and Sun Z (2021) The Influence of MTHFR Polymorphism on Gray Matter Volume in Patients With Amnestic Mild Cognitive Impairment. Front. Neurosci. 15:778123. doi: 10.3389/fnins.2021.778123
Received: 16 September 2021; Accepted: 05 November 2021;
Published: 30 November 2021.
Edited by:
Noorbakhsh Amiri Golilarz, Southern Illinois University Carbondale, United StatesReviewed by:
Nafisa M. Jadavji, Midwestern University, United StatesDana Niedowicz, University of Kentucky, United States
Copyright © 2021 You, Zhou, Yin, Wan, Zhang, Li, Li, Zhu, Zhu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongwu Sun, c3Vuemh3dUAxMjYuY29t
 Mengzhe You
Mengzhe You Xia Zhou
Xia Zhou Wenwen Yin
Wenwen Yin Ke Wan
Ke Wan Xiaoqun Zhu
Xiaoqun Zhu Zhongwu Sun
Zhongwu Sun