- 1Medical School of Chinese PLA, Beijing, China
- 2Department of Neurosurgery, The First Medical Centre, Chinese PLA General Hospital, Beijing, China
- 3Department of Information Technology, Xian Janssen Pharmaceutical Ltd., Beijing, China
- 4Department of Neurosurgery, The Seventh Medical Center of PLA General Hospital, Beijing, China
Purpose: To explore molecular alterations and their correlation with the survival of patients with glioblastoma (GBM) with corpus callosum (CC) involvement (ccGBM).
Methods: Electronic medical records were reviewed for glioma patients tested for molecular alterations and treated at our hospital between January 2016 and July 2020. ccGBM was compared to GBM without CC involvement (non-ccGBM) to identify differences in molecular alterations. Clinical outcomes and survival were compared between ccGBM and non-ccGBM patients, as well as among patients with ccGBM with different molecular alteration statuses. ccGBM was also compared to diffuse midline glioma (DMG) to clarify their correlation in molecular alterations, the progression-free survival (PFS), and overall survival (OS).
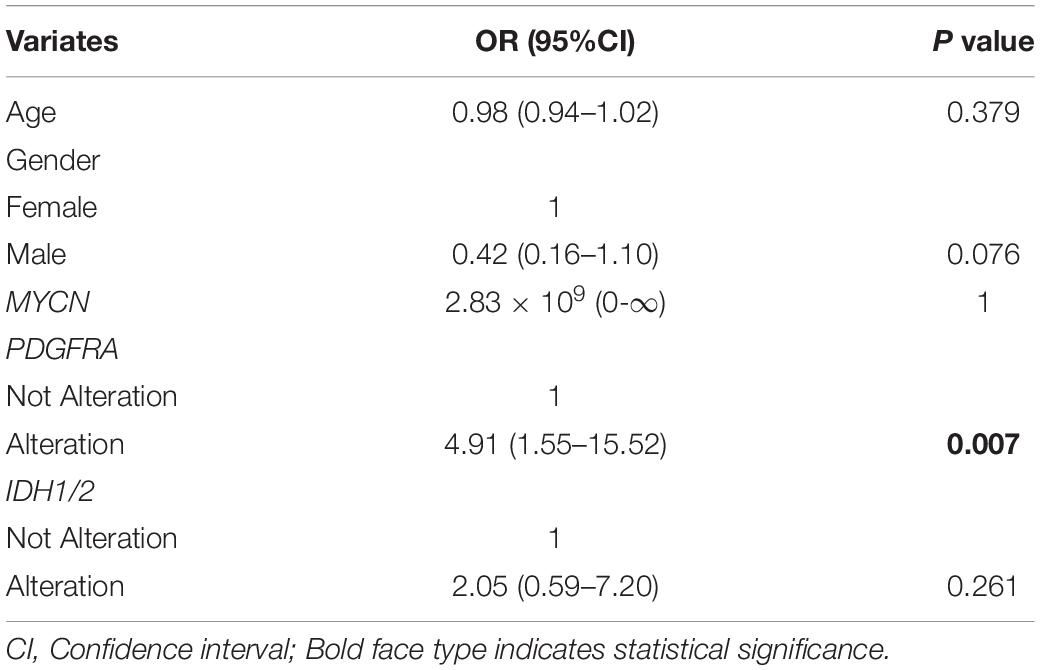
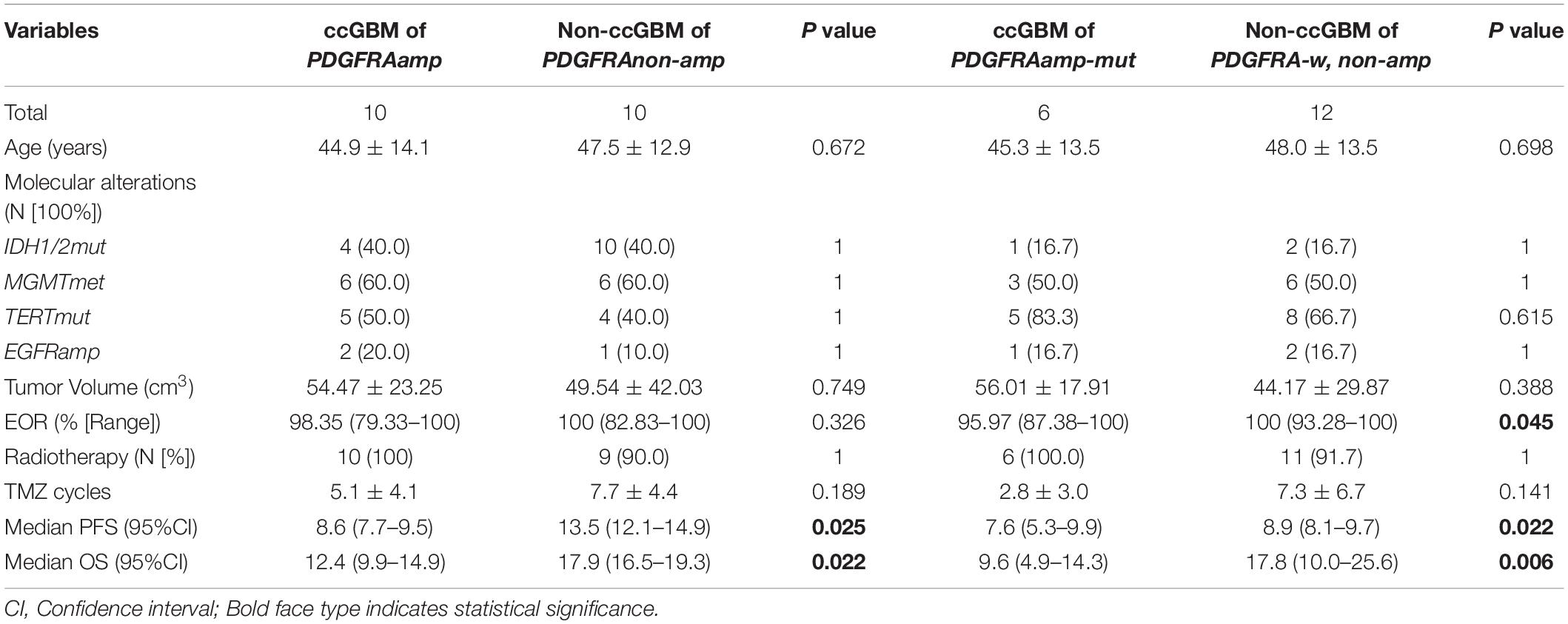
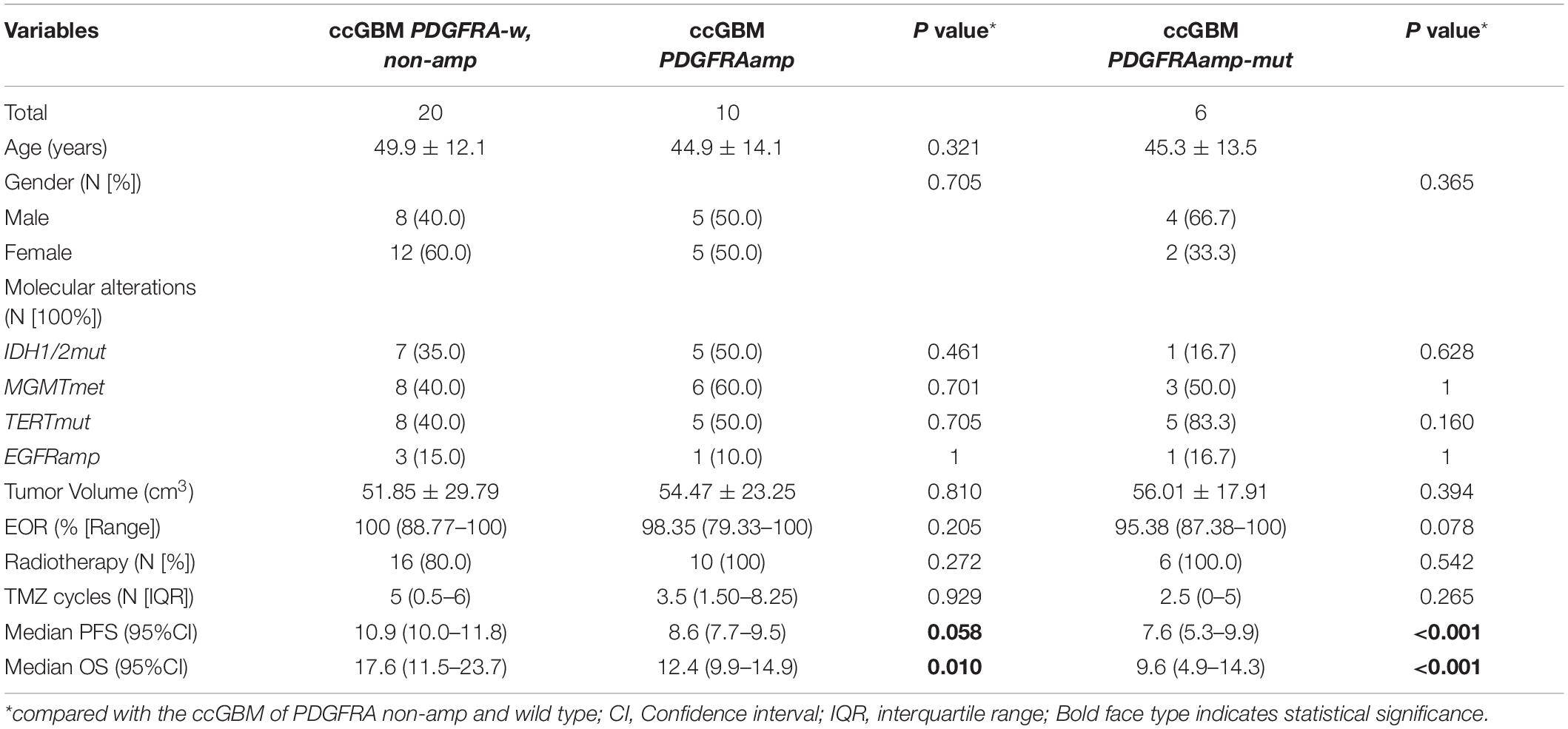
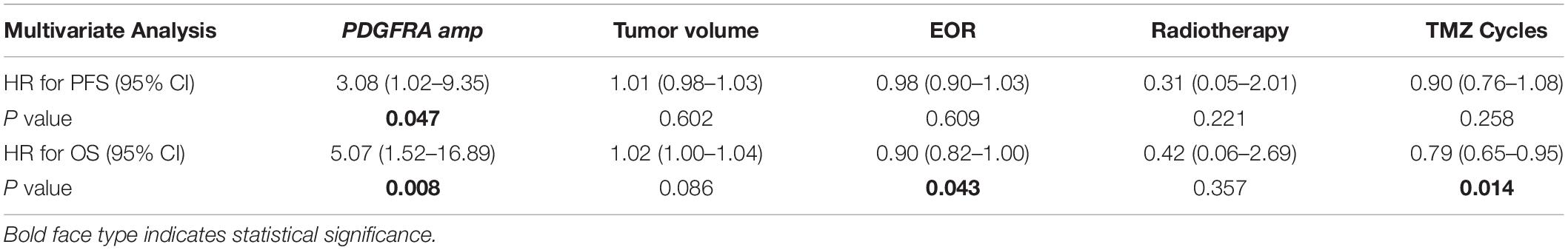
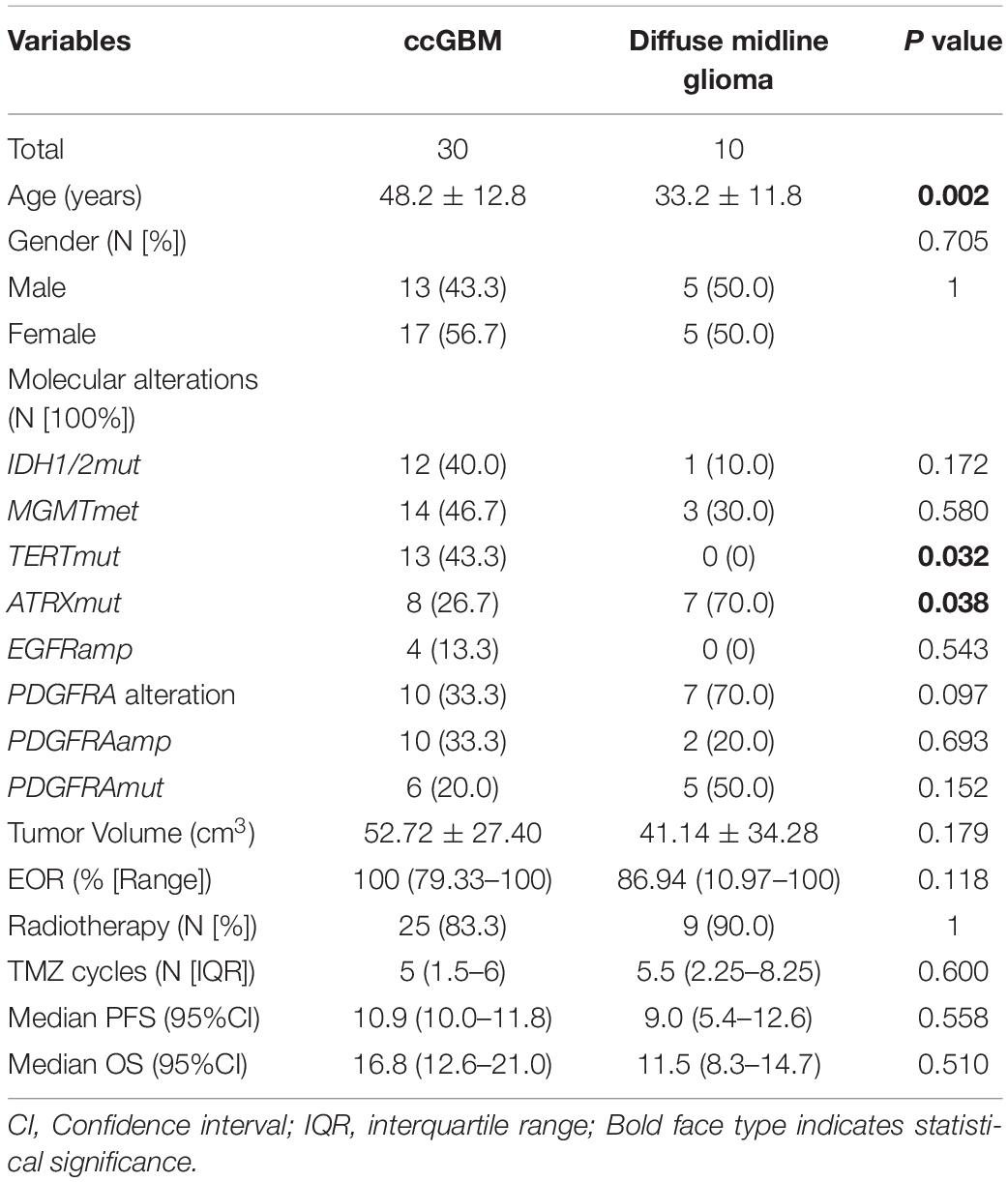
Results: Thirty ccGBM and 88 non-ccGBM patients were included. PDGFRA amplification (PDGFRAamp, 33.3 vs. 9.1%, P = 0.004) and missense mutation (PDGFRAmut, 20.0 vs. 3.4%, P = 0.011) both had higher incidences in ccGBM than in non-ccGBM. PDGFRA alteration was associated with the occurrence of ccGBM (OR = 4.91 [95% CI: 1.55–15.52], P = 0.007). ccGBM with PDGFRAamp resulted in a shorter median PFS (8.6 vs. 13.5 months, P = 0.025) and OS (12.4 vs. 17.9 months, P = 0.022) than non-ccGBM with PDGFRAnon-amp. ccGBM with PDGFRAamp combined with PDGFRAmut (PDGFRAamp-mut) had a shorter median PFS (7.6 vs. 8.9 months, P = 0.022) and OS (9.6 vs. 17.8 months, P = 0.006) than non-ccGBM with wild-type PDGFRA and no amplification (PDGFRA-w, non-amp). Compared to ccGBM with PDGFRA-w, non-amp, ccGBM with PDGFRAamp and PDGFRAamp-mut both had a shorter median PFS and OS (P < 0.05). The hazard ratios (HRs) of PDGFRAamp for PFS and OS in ccGBM were 3.08 (95% CI: 1.02–9.35, P = 0.047) and 5.07 (1.52–16.89, P = 0.008), respectively, and the HRs of PDGFRAamp-mut for PFS and OS were 13.16 (95% CI: 3.19–54.40, P < 0.001) and 16.36 (2.66–100.70, P = 0.003). ccGBM may have similar incidences of PDGFRAamp or mut (PDGFRAamp/mut) as DMG, and they also had similar median PFS (10.9 vs. 9.0 months, P = 0.558) and OS (16.8 vs. 11.5 months, P = 0.510).
Conclusion: PDGFRA alterations are significantly associated with the occurrence and poor prognosis of ccGBM. ccGBM with PDGFRAamp/mut may be classified as a single subtype of GBM that has a similar survival rate to DMG. PDGFR inhibitors may be a promising treatment method for ccGBM.
Introduction
Glioblastoma (GBM) is highly malignant tumor (World Health Organization [WHO] grade IV), with an annual incidence of 3.1 per 100,000 and a 5-year survival rate of less than 3% (Ohgaki, 2009; Wirsching et al., 2016). GBM is very invasive, typically infiltrating along white matter tracts (Giese and Westphal, 1996; Giese et al., 2003). As the largest interhemispheric fiber bundle in the human brain, the corpus callosum (CC) is frequently invaded by GBM (Nazem-Zadeh et al., 2012). GBM with CC involvement (ccGBM) can be classified into two types. In one type, the lesion invades only one side of the hemisphere and the CC (non-butterfly ccGBM). The other type is known as butterfly GBM (bGBM) and involves the corpus callosum and both cerebral hemispheres (Louis et al., 2007; Nazem-Zadeh et al., 2012; Ho et al., 2013; Komori, 2017). ccGBM has a poorer prognosis than GBM without CC involvement (non-ccGBM) and is associated with incomplete resection or residual tumor after surgery (Talos et al., 2006). Whether the overall survival (OS) rate of ccGBM can be prolonged by surgical resection has been controversial (Dziurzynski et al., 2012; Chaichana et al., 2014; Dayani et al., 2018; Opoku-Darko et al., 2018; Franco et al., 2020). The present consensus is that ccGBM has a poorer prognosis than common GBM regardless of whether resection is performed (Chen et al., 2015; Liang et al., 2016).
At present, the WHO classification of glioma is based on pathological diagnosis and analysis of molecular alterations, which provides a more precise diagnosis and better comprehension of the type of glioma. Previous studies have revealed the association between some molecular alterations and the survival rate of glioma. For example, glioma with IDH or TERT mutation has a better prognosis, and glioma with H3F3A or HIST1H3B/C mutation is classified as diffuse midline glioma and has a poorer prognosis (Onizuka et al., 2020). While the molecular characteristics of non-ccGBM are established, the molecular characteristics of ccGBM were unknown until now. In this study, we aimed to explore the molecular alterations in ccGBM and determine their association with survival.
Materials and Methods
Patient Selection
Retrospective clinical data of 1232 glioma patients were obtained from electronic medical records (EMRs) in the Department of Neurosurgery at our hospital between January 2016 and July 2020. The enrollment criteria were patients 18–80 years of age who underwent resection at our hospital, who were diagnosed with supratentorial WHO II-IV grade gliomas by pathology, received testing of the molecular alteration status of the tumor, had extent of resection (EOR) values calculated and attended postoperative follow-up regularly. The exclusion criteria were as follows: (1) patients with infratentorial glioma, (2) patients who underwent only biopsy, (3) patients for whom the pathological diagnosis could not be confirmed, (4) patients with diffuse midline glioma, and (5) patients lost to follow-up. This study was carried out according to the principles of the Helsinki Declaration and approved by our institutional ethics committee (No. S2014-096-01). Written informed consent for surgery and molecular testing was previously provided by all patients or their relatives.
Patient Groupings
The included patients were divided into the glioma invading the corpus callosum (ccGlioma) group and the non-ccGlioma control group. The incidences of molecular alterations between the two groups were compared to identify the molecular differences and the significant molecular alterations in ccGBM. Then, the patients with WHO grade IV GBM were divided into the ccGBM group and non-ccGBM group. To explore the association between significant molecular alterations and the survival rate of GBM, these two groups were matched according to age, sex, IDH mutation (IDHmut) status, TERT mutation (TERTmut) status, MGMT promoter methylation (MGMTmet) status, EOR value, radiotherapy, and cycles of temozolomide (TMZ) chemotherapy. The ccGBM group was also divided into different groups according to significant molecular alterations to identify the effect of different molecular characteristics on the survival rate. ccGBM was also compared to diffuse midline glioma (DMG) to clarify the correlation between molecular alterations and survival.
Volumetric Analysis
MRI was performed for all patients using a 1.5 Tesla (T) scanner (Siemens Espree, Erlangen, Germany). The digital imaging and communications in medicine (DICOM) data of all MRI sequences were transferred to iPlan software 2.6 (Brainlab Feldkirchen, Germany). Pre-and postoperative tumor volumes (cm3) were calculated automatically by the software based on the tumor tissue seen on postcontrast 3D T1-weighted MR images of GBM. The EOR was defined as follows: (preoperative tumor volume – postoperative residual tumor volume)/preoperative tumor volume × 100.
Next-Generation DNA Sequencing (NGS)
Tumor samples were examined by neuropathologists to confirm the histopathological diagnosis and WHO grade based on the 2016 WHO classification system (Louis et al., 2016). Genomic DNA (gDNA) was extracted from fresh tumor tissues using a QIAamp DNA Mini Kit (Qiagen 51306, Germany) and quantified by UV absorption (Nanodrop, Thermo Scientific, Wilmington, DE, United States). Sixty-eight gene mutation statuses were tested in this study (Genetron Health; Beijing, China). Libraries of gDNA were enriched for the tested regions using a custom-designed capture probe. Prepared libraries (750 ng) were hybridized with two different hybridization reagents in blocking agents using the SureSelectXT Target Enrichment System (Agilent Technologies). The enriched libraries were amplified with the P5/P7 primer. After performing a quality control analysis with a 2200 Bioanalyzer and quantification with a Qbit3 and a qPCR NGS library quantification kit (Agilent Technologies), the libraries were sequenced on a HiSeq X10 platform (Illumina, San Diego, CA, United States).
Primary processing of NGS data for tissue samples was performed using trimmomatic methods (0.36), including demultiplexing and masking of dual-index adapter sequences. Sequence reads were aligned against the human reference genome (version GRCh37/hg19) using BWA (version 0.7.10). Candidate somatic mutations consisting of point mutations, small insertions, and deletions were identified by SAMtools (version 1.3.1) and pindel (version 0.2.5b8, 20151210) across the targeted regions of interest. Integrative Genomics Viewer (IGV) was applied to filter alignment and sequencing artifacts.
Postoperative Treatment and Follow-Up
Radiotherapy plus concomitant and adjuvant TMZ chemotherapy for GBM were recommended (Stupp et al., 2005; Duffau and Taillandier, 2015). Regular MRI scans and follow-up were performed every 3 months. The end point of follow-up was March 2021. The progression-free survival and overall survival rates were calculated according to the results of the follow-up.
Statistical Analysis
Statistical analysis was performed using SPSS 21.0 software (SPSS Inc., Chicago, IL, United States). The Shapiro-Wilk test was used to test the normality of the data. Student’s t and χ2 (or Fisher’s exact test) tests were used to compare continuous parametric and categorical variables between groups, respectively. The Mann-Whitney U-test was used to compare continuous non-parametric variables, such as the EOR value, between groups. Univariate and multivariate logistic regression were used to evaluate the correlation between ccGBM and clinical factors and molecular alterations. Survival curves were estimated by the Kaplan–Meier method and were compared by the log-rank test. Cox proportional hazard models were used to identify significant prognostic factors for PFS and OS. A P value < 0.05 was considered statistically significant.
Results
Molecular Alterations of ccGlioma and Non-ccGlioma
A total of 223 patients who underwent molecular alteration testing were finally included, among whom 47 had gliomas with CC involvement. The incidences of molecular alterations are presented in Table 1, in which the ccGlioma group and the non-ccGlioma group are compared. The mutation incidences of BCOR, PPM1D, and TERT in the patients in the ccGlioma group with WHO grade III tumors were higher than those in the non-ccGlioma group. The mutation incidence of IDH 1 or 2 in the ccGBM group was higher than that in the non-ccGBM group. The alteration incidences of MYCN and PDGFRA in the ccGBM group were higher than those in the non-ccGBM. The ccGBM group had a higher incidence of PDGFRA amplification (PDGFRAamp) (33.3 vs. 9.1%, P = 0.004) and PDGFRA mutation (PDGFRAmut) (20.0 vs. 3.4%) than the non-ccGBM group. Among 10 ccGBM patients with PDGFRA alterations, 10 patients had PDGFRAamp, and 6 patients had 7 missense mutations. All ccGBM patients with PDGFRAmut had PDGFRAamp. Other molecular alterations that were significantly associated with the prognosis of glioma are also shown in Table 1, but none of them were identified to be different between the ccGlioma group and non-ccGlioma group (Figure 1). The multivariate analysis showed that only the PDGFRA alteration was associated with the occurrence of ccGBM (odds ratio [OR]: 4.91 [1.55–15.52], P = 0.007) (Table 2). The detailed results of molecular alterations in ccGBM are shown in Figure 2.
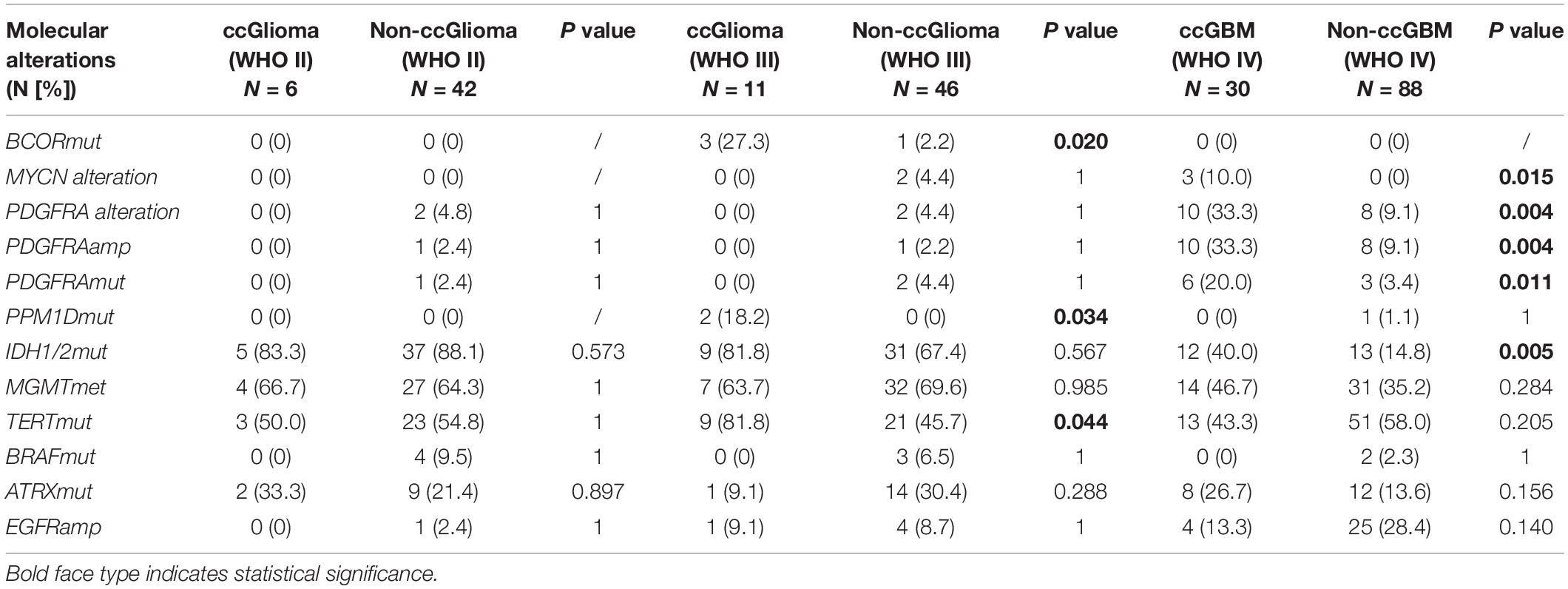
Table 1. Molecular alterations of ccGlioma and non-ccGlioma of different WHO grades by univariate logistic regression.
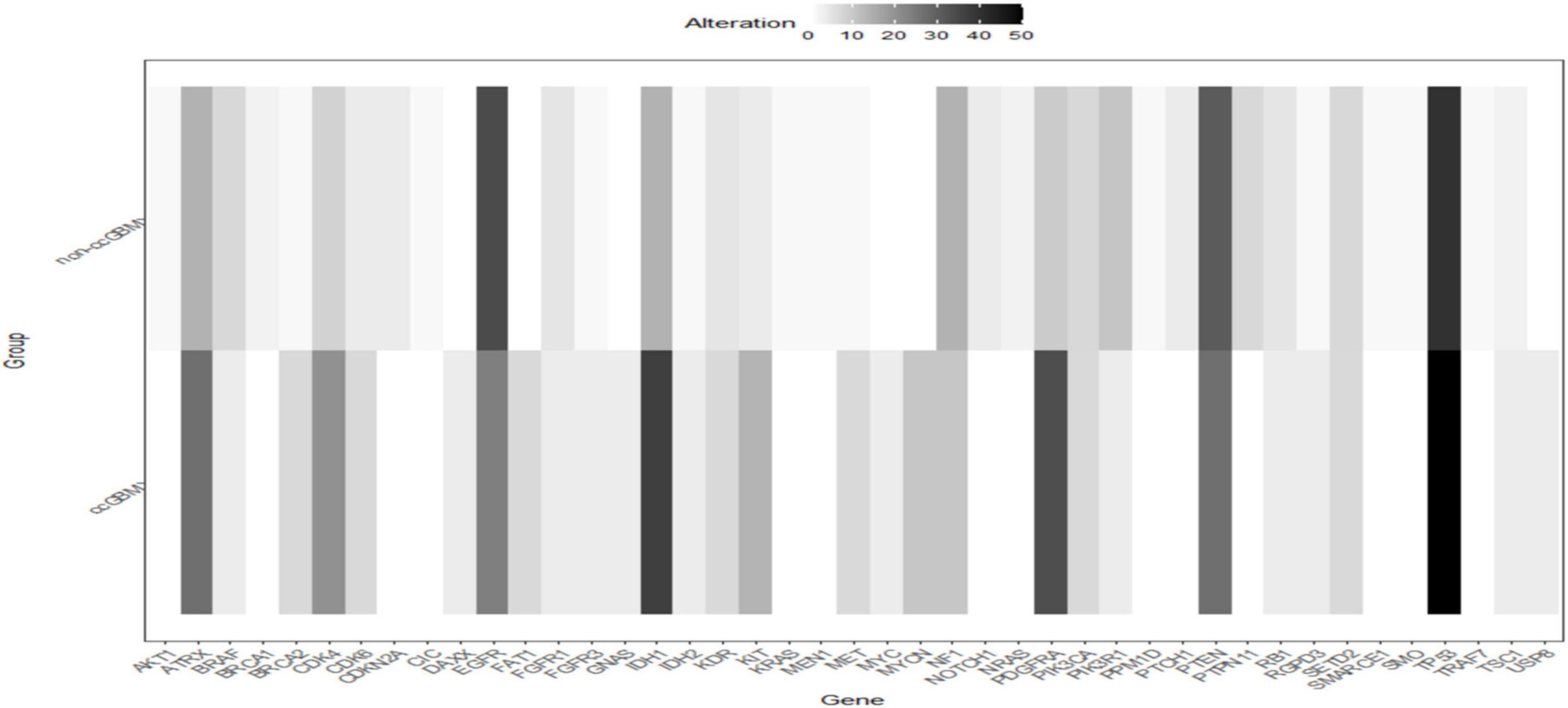
Figure 1. Heatmap of incidences of molecular alterations between ccGBM and non-ccGBM. The horizontal axis represented different genes, the longitudinal axis represented the two groups of ccGBM and non-ccGBM. Low incidences of alterations were shown in white, high incidences in black.
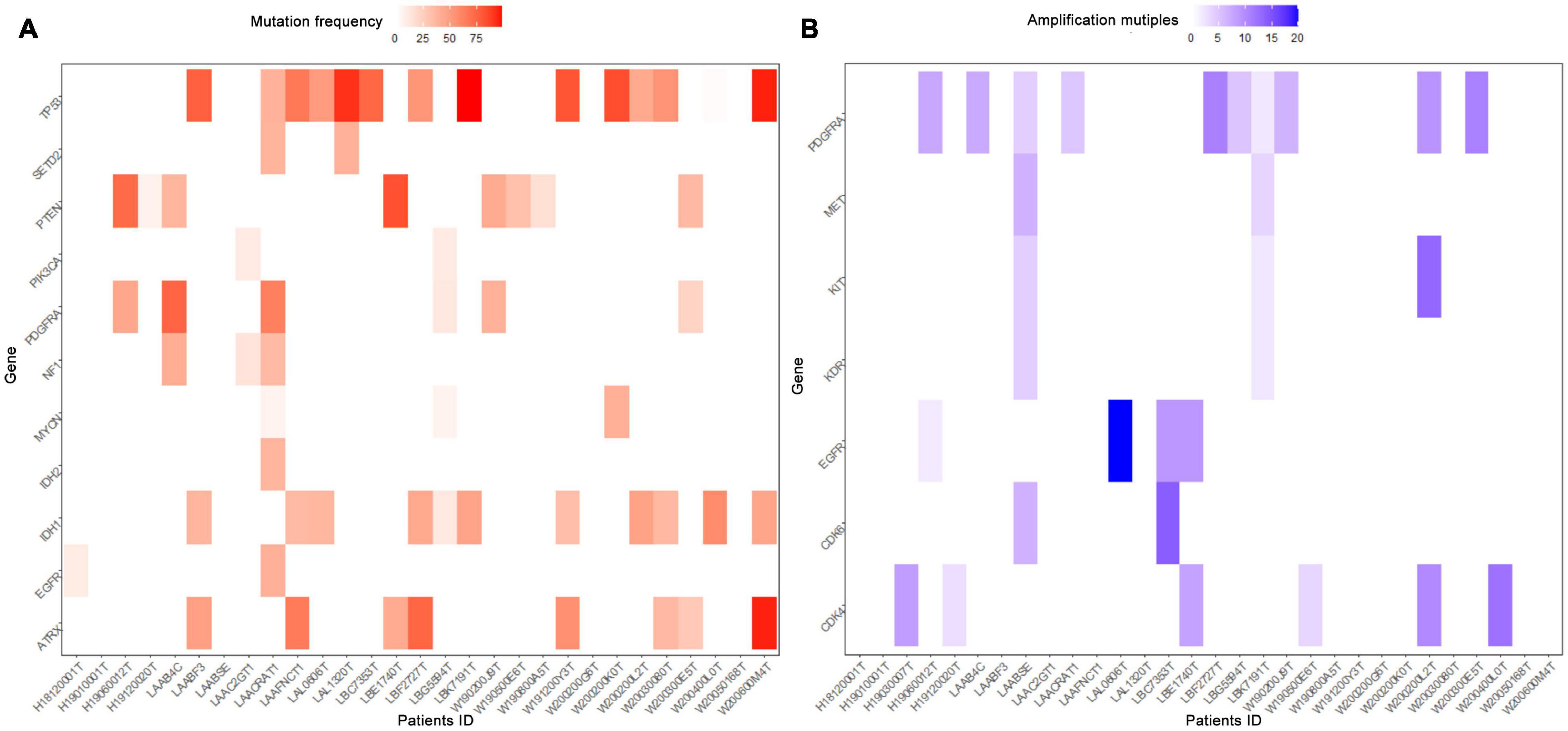
Figure 2. Heatmap of molecular mutations frequencies (A) and amplification multiples (B) in ccGBM. The horizontal axis represented the identification number of tumor samples, the longitudinal axis represented related genes. Low frequencies and multiples were shown in white, high frequencies and multiples in red and blue, respectively.
Association of PDGFRA Alterations With the Survival Rate of GBM Patients
Because of the small sample size of patients with ccGBM MYCN alterations (only three patients), we only analyzed the association between PDGFRA alteration and the survival rate of GBM patients. The GBM patients were divided into the ccGBM with PDGFRA amplified group (ccGBM with PDGFRAamp), the non-ccGBM with no PDGFRA amplification group (non-ccGBM with PDGFRAnon-amp), the ccGBM with PDGFRA mutation combined with amplification group (ccGBM of PDGFRAamp-mut), and the non-ccGBM with wild type PDGFRA with no amplification (non-ccGBM with PDGFRA-w, non-amp) group. To avoid the influence of other factors, these patients were matched according to IDH 1/2, TERT, MGMTmet, and EGFRamp status. Other clinical characteristics were also matched. The ccGBM with PDGFRAamp group had a shorter median PFS (8.6 vs. 13.5 months, P = 0.025) and OS (12.4 vs. 17.9 months, P = 0.022) rate than the non-ccGBM with PDGFRAnon-amp group. The ccGBM with PDGFRAamp-mut group had a shorter median PFS (7.6 vs. 8.9 months, P = 0.022) and OS (9.6 vs. 17.8 months, P = 0.006) rate than the ccGBM with PDGFRA-w, non-amp group. The ccGBM with PDGFRAamp-mut group also had a lower median EOR value than the non-ccGBM with PDGFRA-w, non-amp group (95.97 vs. 100%, P = 0.045) (Table 3).
Association Between PDGFRA Alterations and the Survival Rate of ccGBM Patients
To test the influence of PDGFRA alterations on the survival rate of ccGBM patients, ccGBM patients were divided into the PDGFRAamp group, the PDGFRAamp-mut group and the PDGFRA-w, and non-amp group. The clinical outcomes and survival rates were compared between the groups (Table 4). The ccGBM with PDGFRAamp group had a shorter median PFS (8.6 vs. 10.9 months, P = 0.058) and OS (12.4 vs. 17.6 months, P = 0.010) rate than the PDGFRA-w, non-amp group. The ccGBM with PDGFRAamp-mut group had a shorter median PFS (7.6 vs. 10.9 months, P < 0.001) and OS (9.6 vs. 17.6 months, P < 0.001) rate than the PDGFRA-w, non-amp group. The survival curves are presented in Figure 3.
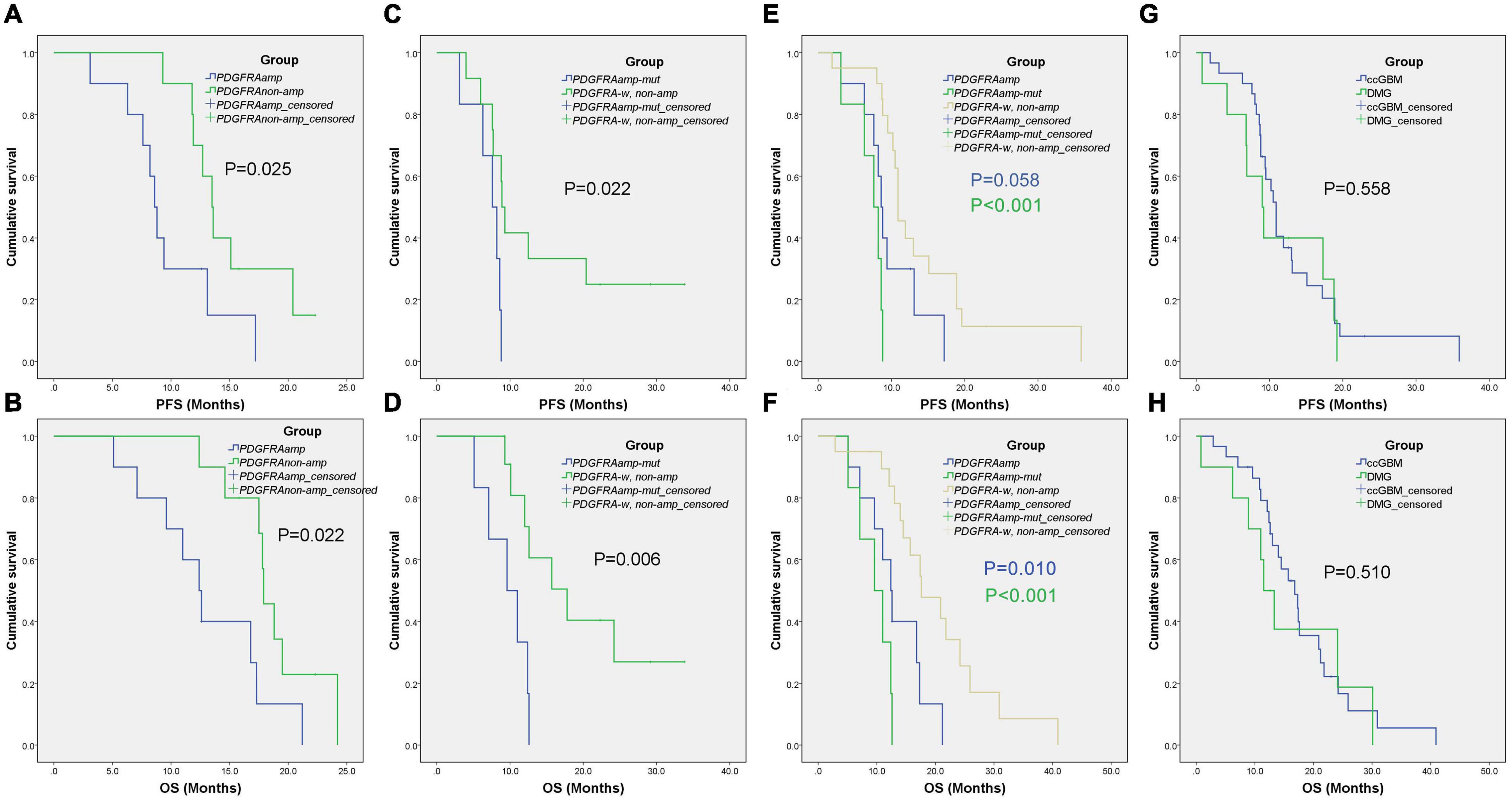
Figure 3. Kaplan-Meier survival curves of different groups. The ccGBM of PDGFRAamp vs. non-ccGBM of PDGFRAnon-amp (A,B), ccGBM of PDGFRAamp-mut vs. non-ccGBM of PDGFRA-w, non-amp (C,D), ccGBM of PDGFRA-w, non-amp compared to ccGBM of PDGFRAamp and PDGFRAamp-mut (E,F), and ccGBM vs. DMG (G,H).
Influence of PDGFRA Alterations on ccGBM Prognosis
Multivariate analysis showed that PDGFRAamp, the EOR value and number of TMZ cycles correlated with the OS rate of ccGBM patients (P < 0.05). PDGFRAamp caused a shorter OS rate (HR = 5.07 [95% CI: 1.52–16.89], P = 0.008). PDGFRAamp was also significantly associated with the PFS rate of ccGBM patients (HR = 3.08 [95% CI: 1.02–9.35], P = 0.047). PDGFRAamp-mut also had a significant negative influence on the PFS rate (HR = 13.16 [95% CI: 3.19–54.40], P < 0.001) and OS rate (HR = 16.36 [95% CI: 2.66–100.70], P = 0.003) of ccGBM patients in the multivariate Cox regression analysis (Tables 5, 6).
Comparison of ccGBM With Diffuse Midline Glioma
Molecular alterations were compared between ccGBM and diffuse midline glioma (DMG). The results demonstrated that the incidences of PDGFRA alterations were not significantly different (33.3 vs. 70.0%, P = 0.097) between the two groups. The incidences of PDGFRAamp or mut (PDGFRAamp/mut) were also similar between groups (P > 0.05). The TERT mutation incidence of ccGBM was higher than that of DMG, while the ATRX mutation incidence of ccGBM was lower than that of DMG. The median PFS and OS rates were not significantly different between the two groups (Table 7 and Figure 3).
Discussion
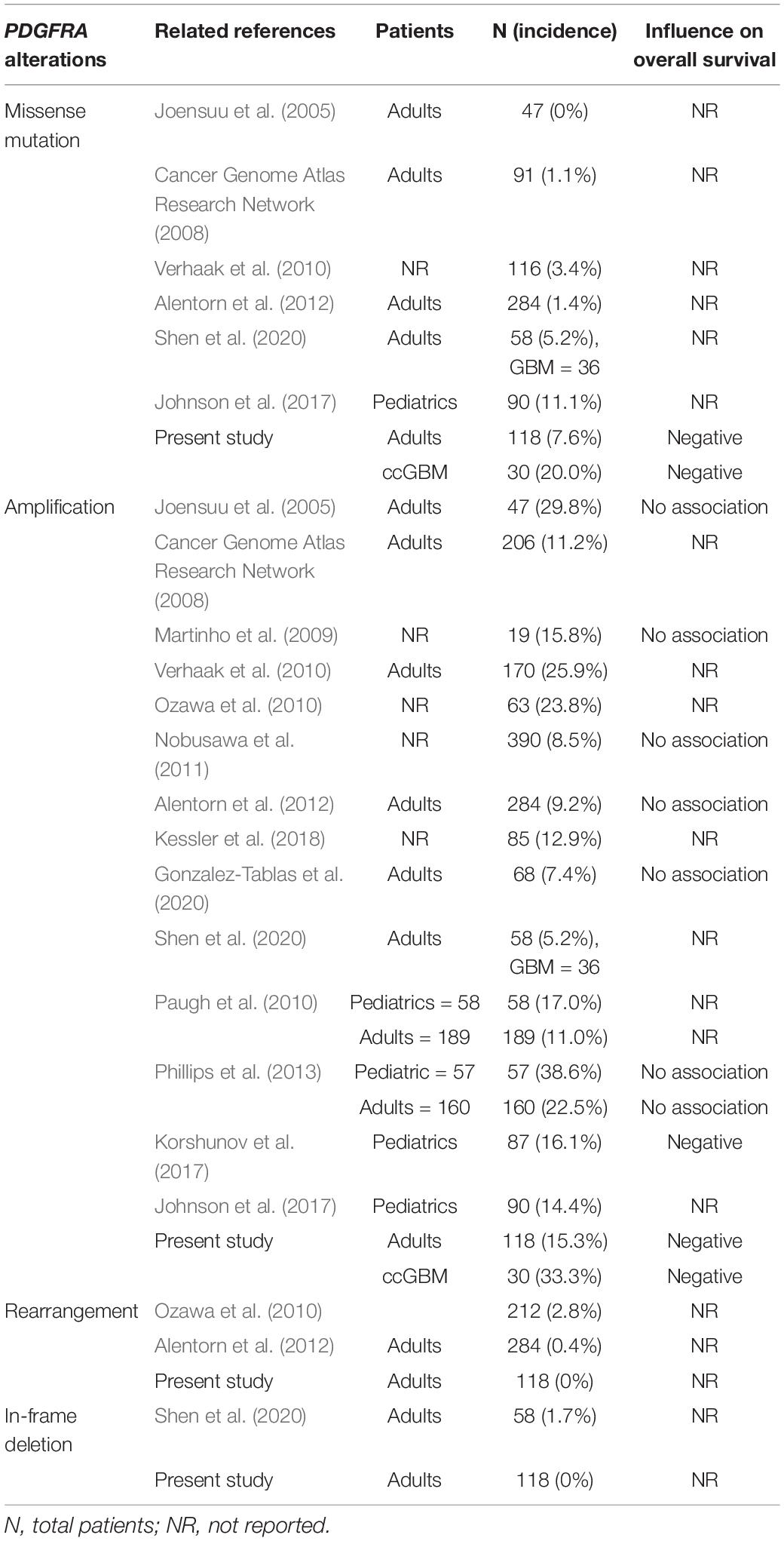
Platelet-derived growth factor receptor α (PDGFRα) is a transmembrane receptor encoded by PDGFRA. It has 5 immunoglobulin-like repeats in its extracellular domain and a tyrosine kinase (TK) in its intracellular domain. The activation of PDGFRα by its ligand can induce multiple downstream signal transduction pathways that promote oncogenesis, including the PI3K/AKT, RAS/MAP kinase, Src kinase family, JAK/STAT, and PLC/PKC pathways (Blume-Jensen and Hunter, 2001; Verhaak et al., 2010). PDGFRA is the second most frequently mutated receptor tyrosine kinase (RTK) in GBM, followed by EGFR (Verhaak et al., 2010). Several PDGFRA alterations have been detected in gliomas, including amplification, missense mutation, frameshift mutation, in-frame insertion or deletion, and rearrangement (Martinho et al., 2009; Ozawa et al., 2010). Some previous studies have demonstrated a high incidence of PDGFRAamp in GBM of adults, ranging from 7.4 to 29.8% (Joensuu et al., 2005; Cancer Genome Atlas Research Network, 2008; Martinho et al., 2009; Ozawa et al., 2010; Verhaak et al., 2010; Nobusawa et al., 2011; Alentorn et al., 2012; Kessler et al., 2018; Gonzalez-Tablas et al., 2020; Shen et al., 2020). Pediatric (<18 years old) GBM had the higher incidence of PDGFRAamp which ranged from 14.4 to 38.6% (Paugh et al., 2010; Phillips et al., 2013; Johnson et al., 2017; Korshunov et al., 2017). The incidence of PDGFRAmut in GBM of adults ranged from 0 to 5.2% (Cancer Genome Atlas Research Network, 2008; Verhaak et al., 2010; Alentorn et al., 2012; Shen et al., 2020). The pediatric GBM also had the higher incidence of PDGFRAmut in a previous study (11.1%) (Johnson et al., 2017). Neither PDGFRAamp nor PDGFRAmut were found to be associated with the survival rate of adult GBM patients in previous studies. Only Puputti et al. (2006) and Alentorn et al. (2012) reported the negative influence of PDGFRAamp on the survival rate of glioma patients with WHO grade II and III tumors (Table 8). Some previous studies demonstrated the higher incidence of PDGFRA alterations of pediatric GBM compared to adult GBM. The negative influence of PDGFRAamp on survival of pediatric GBM was also reported in a previous study (Korshunov et al., 2017). We only included adult patients with GBM in our cohort and found the incidences of PDGFRAamp/mut were similar with those of previous studies (15.3 and 7.6%, respectively). While the ccGBM in our cohort had the higher incidences of PDGFRAamp/mut (33.3 and 20.0%, respectively) which were similar with those of pediatric GBM. Because the previous studies of pediatric GBM also included many infra-tentorial tumors that invaded brain stem or cerebellum, we inferred that higher incidences of PDGFRA alterations were not only associated with the younger age of patients, but also the midline structures invasion (such as corpus callosum, thalamus and brainstem) of tumor.
Diffuse midline glioma invading midline structures (thalamus, pons, and medulla oblongata, etc.) was identified to have a much higher incidence of PDGFRAamp/mut in previous studies [30.2% in Paugh et al. (2011), 21.3% in Castel et al. (2015) and 33.3% in Paugh et al. (2011) and Dono et al. (2020)]. As the largest interhemispheric fiber bundle, the corpus callosum is another midline structure that is often invaded by glioma. ccGlioma was also considered to have a higher incidence of PDGFRA alterations than non-ccGlioma in a previous study (Shen et al., 2020). Furthermore, ccGlioma was indicated to have a poorer prognosis than non-ccGlioma, and ccGBM had an even shorter median OS, ranging from 7.0 to 15.0 months after surgery (Dziurzynski et al., 2012; Chaichana et al., 2014; Chen et al., 2015; Burks et al., 2017; Dayani et al., 2018; Opoku-Darko et al., 2018; Forster et al., 2020; Franco et al., 2020). Therefore, we hypothesized that PDGFRA alterations may play an important role in GBM and lead to a higher incidence of CC invasion and a poorer prognosis.
In our analysis of our study data, ccGBM indeed had a higher incidence of PDGFRA alterations than the non-ccGBM. Shen et al. (2020) only demonstrated the higher incidence of PDGFRA alterations in ccGlioma. We found that most PDGFRA alterations occurred in ccGBM, and no PDGFRA alterations were found in ccGlioma classified as WHO grade II and III. Thus, ccGBM, but not grade II and III ccGliomas, can be characterized by PDGFRA alterations (OR = 4.97, P = 0.007). Furthermore, among the PDGFRA alterations, ten patients had PDGFRAamp, and six patients had PDGFRA missense mutations. Frameshift mutations, in-frame insertions or deletions, and rearrangements were not identified in our study. Although these types of alterations were reported in some previous studies, their incidences were very low in GBM (Ozawa et al., 2010; Shen et al., 2020). Only PDGFRA amplification and missense mutation were thought to drive CC invasion in GBM in our study. Interestingly, PDGFRAamp accounted for 100% of PDGFRA alterations in ccGBM or non-ccGBM, which may indicate the more important initial effect of PDGFRAamp in causing CC invasion. Previous studies did not identify a correlation between PDGFRA alterations and the survival rate of GBM patients. Our analysis showed that the ccGBM with PDGFRAamp group had shorter median PFS and OS rate than the non-ccGBM with PDGFRAnon-amp group. The ccGBM with PDGFRAamp-mut group had a shorter median PFS and OS rate than the non-ccGBM of PDGFRA-w, non-amp group. The ccGBM with PDGFRAamp-mut group also had lower EOR values. These results demonstrated that PDGFRA alterations, especially PDGFRAamp combined with PDGFRAmut, can cause poorer prognosis in GBM patients by increasing the invasive ability of tumors (invading CC) and increasing the rate of incomplete resection or residual tumors. The ccGBM with PDGFRAamp or PDGFRAamp-mut group also had a significantly shorter PFS and OS rate than the ccGBM with PDGFRA-w, non-amp group, which highlighted the obvious negative influence of PDGFRA alterations on the survival rate of ccGBM patients.
Compared to the 10 patients with DMG, although the ccGBM had no statistical differences of PDGFRAamp/mut and total PDGFRA alteration incidences, because of the limited samples of DMG, we cannot conclude that they both had similar molecular characteristics of PDGFRA. But we can infer that the ccGBM group had a characteristic change in PDGFRA, which can predict the invasion of midline structures (such as the CC) and a poorer prognosis than that associated with common GBM. Castel et al. (2015) introduced a subtype of DMG with a K27M mutation in H3.3 (H3F3A) that had a prometastatic gene expression signature with PDGFRA activation and was also located in midline regions other than the pons. PDGFRAamp/mut was seen in this subtype of DMG but not in the H3.1 subtype with a HIST1H3B/C mutation. Along with PDGFRAamp/mut, H3.3 was a more common subgroup of DMG and had a poorer prognosis than H3.1 (Castel et al., 2015). In our series of DMG patients, all ten had H3.3 H3F3A mutations and had a high incidence of PDGFRAamp/mut. ccGBM had a similar incidence of PDGFRAamp/mut and survival rate to DMG. Therefore, it can be hypothesized that ccGBM with PDGFRAamp/mut is a subtype of GBM that has a poor prognosis similar to DMG with H3.3 mutations. Although PDGFRAamp and PDGFRAamp-mut both influenced the PFS and OS rate of ccGBM patients in the multivariate prognostic analysis, a higher EOR value and more TMZ cycles is indicative of an improved OS time in these patients. Radiotherapy was also found to improve the PFS (HR = 0.20 [95% CI: 0.07–0.60, P = 0.003]) and OS (HR = 0.15 [95% CI: 0.05–0.47], P = 0.001) rates of ccGBM patients in the univariate analysis but was not found to be associated with survival in the multivariate analysis. We hypothesize that this is because only five patients did not receive postoperative radiotherapy, which may have led to the underestimation of the effect of radiotherapy. Therefore, it is still suggested to perform aggressive resection and postoperative radiochemotherapy for patients with ccGBM with PDGFRAamp/mut.
PDGFRAamp/mut can cause overexpression of PDGFRα, thus promoting the proliferation and survival of tumor cells through the multiple signaling pathways mentioned above. PDGFR inhibitors are a promising drug to treat glioma. Several antitumor agents targeting PDGFR have been developed, such as imatinib, sorafenib, and nilotinib. These inhibitors have been shown to successfully inhibit GBM cells in vitro and in animal experiments (Ren et al., 2009; Ranza et al., 2010). Some clinical trials failed to prove the antitumor effects of PDGFR inhibitors in GBM, which may be due to the emergence of resistance (De Witt Hamer, 2010; Song et al., 2018). Because ccGBM was identified to have a higher incidence of PDGFRAamp/mut than non-ccGBM in this study, PDGFR inhibitors may be effective for patients with this specific type of GBM. Further clinical trials are needed to explore the effect of PDGFR inhibitors on ccGBM, especially ccGBM with PDGFRAamp/mut.
Some limitations existed in this study. First, not all patients underwent molecular tests, which may lead to selection bias. Second, the incidences of MYCN alterations were significantly different between ccGBM and non-ccGBM. However, because of the small sample size of patients with these alterations (only three patients), the association between MYCN alteration and the occurrence of ccGBM and patient survival rates could not be analyzed. Third, the limited samples of DMG made it difficult to conclude the similarity of molecular and survival between DMG and ccGBM. Last, further analysis of TCGA data for molecular alterations in ccGBM were needed to validate the findings of this study in our future work.
Conclusion
Platelet derived growth factor receptor alpha alterations are associated with the occurrence of ccGBM. The ccGBM with PDGFRAamp/mut group had a poorer prognosis than the non-ccGBM and ccGBM with PDGFRA-w, non-amp groups. ccGBM may have a similar incidence of PDGFRAamp/mut to DMG with an H3F3A mutation, and a similar survival rate. ccGBM with PDGFRAamp/mut can be classified as a single subtype of GBM that still benefits from gross total resection and radiochemotherapy. PDGFR inhibitors may be a promising treatment method for this type of GBM. Large-sample studies are needed to clarify the molecular mechanisms of ccGMB and potential treatment strategies.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://datadryad.org/stash/landing/show?id=doi%3A10.5061%2Fdryad.7d7wm37v8.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics committee of Chinese PLA General Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MC: conceptualization, methodology, software, formal analysis, resources, data curation, investigation, writing-original draft, writing-reviewing and editing, and visualization. XG: data curation, investigation, and writing-reviewing and editing. YC: software, data curation, visualization, and writing-reviewing and editing. MZ: software, validation, data curation, and investigation. HL and HC: data curation and investigation. CS: resources, supervision, and writing-reviewing and editing. XM: conceptualization, methodology, validation, resources, supervision, project administration, and writing-reviewing and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
YC was employed by the company Xian Janssen Pharmaceutical Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Genetron Health who provided the next-generation sequencing platform. We thank to the members of the Department of Neurosurgery, the First Medical Centre of Chinese PLA General Hospital (Guochen Sun, MD; Jialin Liu, MD) for their collaboration.
Abbreviations
ATRX, ATRX chromatin remodeler; BCOR, BCL6 corepressor; BRAF, B-Raf proto-oncogene, serine/threonine kinase; CC, corpus callosum; DMG, diffuse midline glioma; DNA, deoxyribonucleic acid; EGFR, epidermal growth factor receptor; EOR, extent of resection; GBM, glioblastoma multiforme; H3F3A, H3 histone, family 3A; HIST1H3B/C, histone cluster 1, H3B/C; HR, hazard ratio; IDH, isocitrate dehydrogenase; MGMT, O-6-methylguanine-DNA methyltransferase; MYCN, MYCN proto-oncogene, bHLH transcription factor; NGS, next-generation DNA sequencing; OR, odds ratio; OS, overall survival; PDGFRA, platelet derived growth factor receptor alpha; PFS, progression-free survival; PPM1D, protein phosphatase, Mg2+/Mn2+ dependent 1D; RTK, receptor tyrosine kinase; TERT, telomerase reverse transcriptase; TMZ, temozolomide; WHO, World Health Organization.
References
Alentorn, A., Marie, Y., Carpentier, C., Boisselier, B., Giry, M., Labussiere, M., et al. (2012). Prevalence, clinico-pathological value, and co-occurrence of PDGFRA abnormalities in diffuse gliomas. Neuro Oncol. 14, 1393–1403. doi: 10.1093/neuonc/nos217
Blume-Jensen, P., and Hunter, T. (2001). Oncogenic kinase signalling. Nature 411, 355–365. doi: 10.1038/35077225
Burks, J. D., Bonney, P. A., Conner, A. K., Glenn, C. A., Briggs, R. G., Battiste, J. D., et al. (2017). A method for safely resecting anterior butterfly gliomas: the surgical anatomy of the default mode network and the relevance of its preservation. J. Neurosurg. 126, 1795–1811. doi: 10.3171/2016.5.JNS153006
Cancer Genome Atlas Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. doi: 10.1038/nature07385
Castel, D., Philippe, C., Calmon, R., Le Dret, L., Truffaux, N., Boddaert, N., et al. (2015). Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 130, 815–827. doi: 10.1007/s00401-015-1478-0
Chaichana, K. L., Jusue-Torres, I., Lemos, A. M., Gokaslan, A., Cabrera-Aldana, E. E., Ashary, A., et al. (2014). The butterfly effect on glioblastoma: is volumetric extent of resection more effective than biopsy for these tumors? J. Neurooncol. 120, 625–634. doi: 10.1007/s11060-014-1597-9
Chen, K. T., Wu, T. W., Chuang, C. C., Hsu, Y. H., Hsu, P. W., Huang, Y. C., et al. (2015). Corpus callosum involvement and postoperative outcomes of patients with gliomas. J. Neurooncol. 124, 207–214. doi: 10.1007/s11060-015-1823-0
Dayani, F., Young, J. S., Bonte, A., Chang, E. F., Theodosopoulos, P., McDermott, M. W., et al. (2018). Safety and outcomes of resection of butterfly glioblastoma. Neurosurg. Focus 44:E4. doi: 10.3171/2018.3.FOCUS1857
De Witt Hamer, P. C. (2010). Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol. 12, 304–316. doi: 10.1093/neuonc/nop068
Dono, A., Takayasu, T., Ballester, L. Y., and Esquenazi, Y. (2020). Adult diffuse midline gliomas: clinical, radiological, and genetic characteristics. J. Clin. Neurosci. 82(Pt A), 1–8. doi: 10.1016/j.jocn.2020.10.005
Duffau, H., and Taillandier, L. (2015). New concepts in the management of diffuse low-grade glioma: proposal of a multistage and individualized therapeutic approach. Neuro Oncol. 17, 332–342. doi: 10.1093/neuonc/nou153
Dziurzynski, K., Blas-Boria, D., Suki, D., Cahill, D. P., Prabhu, S. S., Puduvalli, V., et al. (2012). Butterfly glioblastomas: a retrospective review and qualitative assessment of outcomes. J. Neurooncol. 109, 555–563. doi: 10.1007/s11060-012-0926-0
Forster, M. T., Behrens, M., Lortz, I., Conradi, N., Senft, C., Voss, M., et al. (2020). Benefits of glioma resection in the corpus callosum. Sci. Rep. 10:16630. doi: 10.1038/s41598-020-73928-x
Franco, P., Delev, D., Cipriani, D., Neidert, N., Kellner, E., Masalha, W., et al. (2020). Surgery for IDH1/2 wild-type glioma invading the corpus callosum. Acta Neurochir. (Wien) 163, 937–945. doi: 10.1007/s00701-020-04623-z
Giese, A., Bjerkvig, R., Berens, M. E., and Westphal, M. (2003). Cost of migration: invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 21, 1624–1636. doi: 10.1200/JCO.2003.05.063
Giese, A., and Westphal, M. (1996). Glioma invasion in the central nervous system. Neurosurgery 39, 235–250; discussion 250–232. doi: 10.1097/00006123-199608000-00001
Gonzalez-Tablas, M., Arandia, D., Jara-Acevedo, M., Otero, A., Vital, A. L., Prieto, C., et al. (2020). Heterogeneous EGFR, CDK4, MDM4, and PDGFRA gene expression profiles in primary gbm: no association with patient survival. Cancers (Basel) 12:231. doi: 10.3390/cancers12010231
Ho, M. L., Moonis, G., Ginat, D. T., and Eisenberg, R. L. (2013). Lesions of the corpus callosum. AJR Am. J. Roentgenol. 200, W1–W16. doi: 10.2214/ajr.11.8080
Joensuu, H., Puputti, M., Sihto, H., Tynninen, O., and Nupponen, N. N. (2005). Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J. Pathol. 207, 224–231. doi: 10.1002/path.1823
Johnson, A., Severson, E., Gay, L., Vergilio, J. A., Elvin, J., Suh, J., et al. (2017). Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist 22, 1478–1490. doi: 10.1634/theoncologist.2017-0242
Kessler, T., Sahm, F., Sadik, A., Stichel, D., Hertenstein, A., Reifenberger, G., et al. (2018). Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro Oncol. 20, 367–379. doi: 10.1093/neuonc/nox160
Komori, T. (2017). The 2016 WHO classification of tumours of the central nervous system: the major points of revision. Neurol. Med. Chir. (Tokyo) 57, 301–311. doi: 10.2176/nmc.ra.2017-0010
Korshunov, A., Schrimpf, D., Ryzhova, M., Sturm, D., Chavez, L., Hovestadt, V., et al. (2017). H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 134, 507–516. doi: 10.1007/s00401-017-1710-1
Liang, T. H., Kuo, S. H., Wang, C. W., Chen, W. Y., Hsu, C. Y., Lai, S. F., et al. (2016). Adverse prognosis and distinct progression patterns after concurrent chemoradiotherapy for glioblastoma with synchronous subventricular zone and corpus callosum invasion. Radiother. Oncol. 118, 16–23. doi: 10.1016/j.radonc.2015.11.017
Louis, D. N., Ohgaki, H., Wiestler, O. D., Cavenee, W. K., Burger, P. C., Jouvet, A., et al. (2007). The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109. doi: 10.1007/s00401-007-0243-4
Louis, D. N., Perry, A., Reifenberger, G., von Deimling, A., Figarella-Branger, D., Cavenee, W. K., et al. (2016). The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820. doi: 10.1007/s00401-016-1545-1
Martinho, O., Longatto-Filho, A., Lambros, M. B., Martins, A., Pinheiro, C., Silva, A., et al. (2009). Expression, mutation and copy number analysis of platelet-derived growth factor receptor a (PDGFRA) and its ligand PDGFA in gliomas. Br. J. Cancer 101, 973–982. doi: 10.1038/sj.bjc.6605225
Nazem-Zadeh, M.-R., Saksena, S., Babajani-Fermi, A., Jiang, Q., Soltanian-Zadeh, H., Rosenblum, M., et al. (2012). Segmentation of corpus callosum using diffusion tensor imaging: validation in patients with glioblastoma. BMC Med. Imaging 12:10. doi: 10.1186/1471-2342-12-10
Nobusawa, S., Stawski, R., Kim, Y. H., Nakazato, Y., and Ohgaki, H. (2011). Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: a population-based study. Neuropathology 31, 583–588. doi: 10.1111/j.1440-1789.2011.01204.x
Ohgaki, H. (2009). Epidemiology of brain tumors. Methods Mol. Biol. 472, 323–342. doi: 10.1007/978-1-60327-492-0_14
Onizuka, H., Masui, K., and Komori, T. (2020). Diffuse gliomas to date and beyond 2016 WHO classification of tumours of the central nervous system. Int. J. Clin. Oncol. 25, 997–1003. doi: 10.1007/s10147-020-01695-w
Opoku-Darko, M., Amuah, J. E., and Kelly, J. J. P. (2018). Surgical resection of anterior and posterior butterfly glioblastoma. World Neurosurg. 110, e612–e620. doi: 10.1016/j.wneu.2017.11.059
Ozawa, T., Brennan, C. W., Wang, L., Squatrito, M., Sasayama, T., Nakada, M., et al. (2010). PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 24, 2205–2218. doi: 10.1101/gad.1972310
Paugh, B. S., Broniscer, A., Qu, C., Miller, C. P., Zhang, J., Tatevossian, R. G., et al. (2011). Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J. Clin. Oncol. 29, 3999–4006. doi: 10.1200/JCO.2011.35.5677
Paugh, B. S., Qu, C., Jones, C., Liu, Z., Adamowicz-Brice, M., Zhang, J., et al. (2010). Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J. Clin. Oncol. 28, 3061–3068. doi: 10.1200/JCO.2009.26.7252
Phillips, J. J., Aranda, D., Ellison, D. W., Judkins, A. R., Croul, S. E., Brat, D. J., et al. (2013). PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 23, 565–573. doi: 10.1111/bpa.12043
Puputti, M., Tynninen, O., Sihto, H., Blom, T., Mäenpää, H., Isola, J., et al. (2006). Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol Cancer Res. 4, 927–934. doi: 10.1158/1541-7786.MCR-06-0085
Ranza, E., Mazzini, G., Facoetti, A., and Nano, R. (2010). In-vitro effects of the tyrosine kinase inhibitor imatinib on glioblastoma cell proliferation. J. Neurooncol. 96, 349–357. doi: 10.1007/s11060-009-9975-4
Ren, H., Tan, X., Dong, Y., Giese, A., Chou, T. C., Rainov, N., et al. (2009). Differential effect of imatinib and synergism of combination treatment with chemotherapeutic agents in malignant glioma cells. Basic Clin. Pharmacol. Toxicol. 104, 241–252. doi: 10.1111/j.1742-7843.2008.00371.x
Shen, S., Feng, S., Liu, H., Jiang, J., and Yu, X. (2020). Associations of histological and molecular alterations with invasion of the corpus callosum in gliomas. Acta Neurochir. (Wien) 162, 1691–1699. doi: 10.1007/s00701-020-04376-9
Song, K., Yuan, Y., Lin, Y., Wang, Y. X., Zhou, J., Gai, Q. J., et al. (2018). ERBB3, IGF1R, and TGFBR2 expression correlate with PDGFR expression in glioblastoma and participate in PDGFR inhibitor resistance of glioblastoma cells. Am. J. Cancer Res. 8, 792–809.
Stupp, R., Mason, W. P., van den Bent, M. J., Weller, M., Fisher, B., Taphoorn, M. J., et al. (2005). Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996. doi: 10.1056/NEJMoa043330
Talos, I.-F., Zou, K. H., Ohno-Machado, L., Bhagwat, J. G., Kikinis, R., Black, P. M., et al. (2006). Supratentorial low-grade glioma resectability: statistical predictive analysis based on anatomic MR features and tumor characteristics. Radiology 239, 506–513. doi: 10.1148/radiol.2392050661
Verhaak, R. G., Hoadley, K. A., Purdom, E., Wang, V., Qi, Y., Wilkerson, M. D., et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110. doi: 10.1016/j.ccr.2009.12.020
Keywords: glioblastoma, corpus callosum, PDGFRA amplification, PDGFRA mutation, survival
Citation: Cui M, Gao X, Chi Y, Zhang M, Lin H, Chen H, Sun C and Ma X (2021) Molecular Alterations and Their Correlation With the Survival of Glioblastoma Patients With Corpus Callosum Involvement. Front. Neurosci. 15:701426. doi: 10.3389/fnins.2021.701426
Received: 28 April 2021; Accepted: 08 June 2021;
Published: 30 July 2021.
Edited by:
Rossen Donev, MicroPharm Ltd., United KingdomReviewed by:
Pankaj Pathak, National Institutes of Health (NIH), United StatesGhulam Md Ashraf, King Abdulaziz University, Saudi Arabia
Copyright © 2021 Cui, Gao, Chi, Zhang, Lin, Chen, Sun and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Ma, eGlhb2RvbmdtQGhvdG1haWwuY29t; Caihong Sun, Y2FpaG9uZ2JlaWppbmdAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Meng Cui
Meng Cui Xin Gao1,2†
Xin Gao1,2† Xiaodong Ma
Xiaodong Ma