- 1Department of Biology, University of Toronto Mississauga, Mississauga, ON, Canada
- 2Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
The suprachiasmatic nucleus (SCN) of the hypothalamus is the central circadian clock of mammals. It is responsible for communicating temporal information to peripheral oscillators via humoral and endocrine signaling, ultimately controlling overt rhythms such as sleep-wake cycles, body temperature, and locomotor activity. Given the heterogeneity and complexity of the SCN, its genesis is tightly regulated by countless intrinsic and extrinsic factors. Here, we provide a brief overview of the development of the SCN, with special emphasis on the murine system.
Introduction
On this rhythmic planet, we are surrounded by countless environmental oscillations of varying frequencies. The most notable of all rhythms is the 24 h day-night cycle, which results in predictable changes in the availability of light, warmth, and sustenance. In order to thrive, organisms must be able to anticipate and prepare for daily rhythmic events in the environment, rather than simply reacting to them upon their detection. From bacteria to humans, organisms evolved circadian clocks to keep track of time, enabling them to prepare for external challenges and opportunities through temporal coordination of behavior and physiology.
In mammals, circadian rhythms are driven by a hierarchy of tissue-specific oscillators throughout the body, orchestrated by a central circadian pacemaker, the hypothalamic suprachiasmatic nuclei (SCN). Individual SCN neurons synthesize neuropeptides and neurotransmitters to coordinate endogenous oscillations and entrainment at the tissue level (Aton et al., 2005; Maywood et al., 2011; Mieda et al., 2015). The SCN is unique within the clock hierarchy, as it is the only clock to respond to light directly (Morin and Allen, 2006). The SCN (oscillator) receives and integrates time cues (input) from the environment, and communicates temporal information to peripheral oscillators via humoral and endocrine signaling. Ultimately, the SCN coordinates and controls overt rhythms (outputs) such as sleep-wake cycles, body temperature, osmoregulation, hormone secretion, and gastrointestinal, hepatic, and cardiac functions (Roenneberg and Merrow, 2016). As the biological clock coordinates nearly all physiological processes, perturbation of the circadian system constitutes a risk factor for a myriad of disorders, including obesity, diabetes, cardiovascular disease, cancer, and neurodegeneration (Kurose et al., 2011; Uth and Sleigh, 2014; Broussard and Van Cauter, 2016; Melo et al., 2016; Khaper et al., 2018; Musiek et al., 2018). Conversely, many pathological conditions (e.g., Alzheimer’s disease, cancer) contribute to circadian disruption, which further exacerbates them (Lim et al., 2014). By expanding our knowledge and mechanistic understanding of the circadian clock, we position ourselves to develop new strategies that leverage the circadian system to promote better physical and mental health.
Transcription-Translation Feedback Loop/The Molecular Clock
The molecular clock machinery relies on a series of transcription-translation feedback loops (TTFLs) that generate rhythmic expression of “clock genes” through a negative feedback mechanism. In mammals, the positive limb of the core feedback loop consists of the basic helix-loop-helix PAS transcription factors, Circadian locomotor output cycles kaput (CLOCK) and Brain and muscle ARNT-like protein 1 (BMAL1) (Figure 1). During the subjective day, CLOCK and BMAL1 dimerize and bind to the E-box elements in Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) promoters, inducing their transcription (Shearman et al., 1997; Gekakis et al., 1998; Kume et al., 1999; Bunger et al., 2000; Vitaterna et al., 2006). During the subjective night, PER and CRY protein heterodimers translocate from the cytoplasm to the nucleus, where they repress their own gene transcription through inhibition of CLOCK:BMAL1 (Kume et al., 1999; Vitaterna et al., 1999; Zheng et al., 1999, 2001; Shearman et al., 2000). PER proteins are degraded during the late subjective night, but CRY continues to accumulate in the nucleus and maintains the repressive phase of the cycle (Ye et al., 2014). In the early subjective day, the degradation of CRY results in the derepression of CLOCK:BMAL1-mediated transcription and thus a new round of E-box-dependent gene expression (Gekakis et al., 1998; Kume et al., 1999). This core clock circuitry is regulated by secondary feedback loops. For example, the transcription of Bmal1 is positively and negatively regulated by the nuclear orphan receptors, ROR (α, β, and γ) and REV-ERB (α and β), respectively, which are themselves E-box-containing genes and are therefore controlled by the primary feedback loop (Preitner et al., 2002; Sato et al., 2004; Guillaumond et al., 2005). Together, the primary and secondary TTFLs drive the ∼24 h oscillation of the molecular clock, while additional layers of regulation (described in the following sections) ensure its stability and robustness.
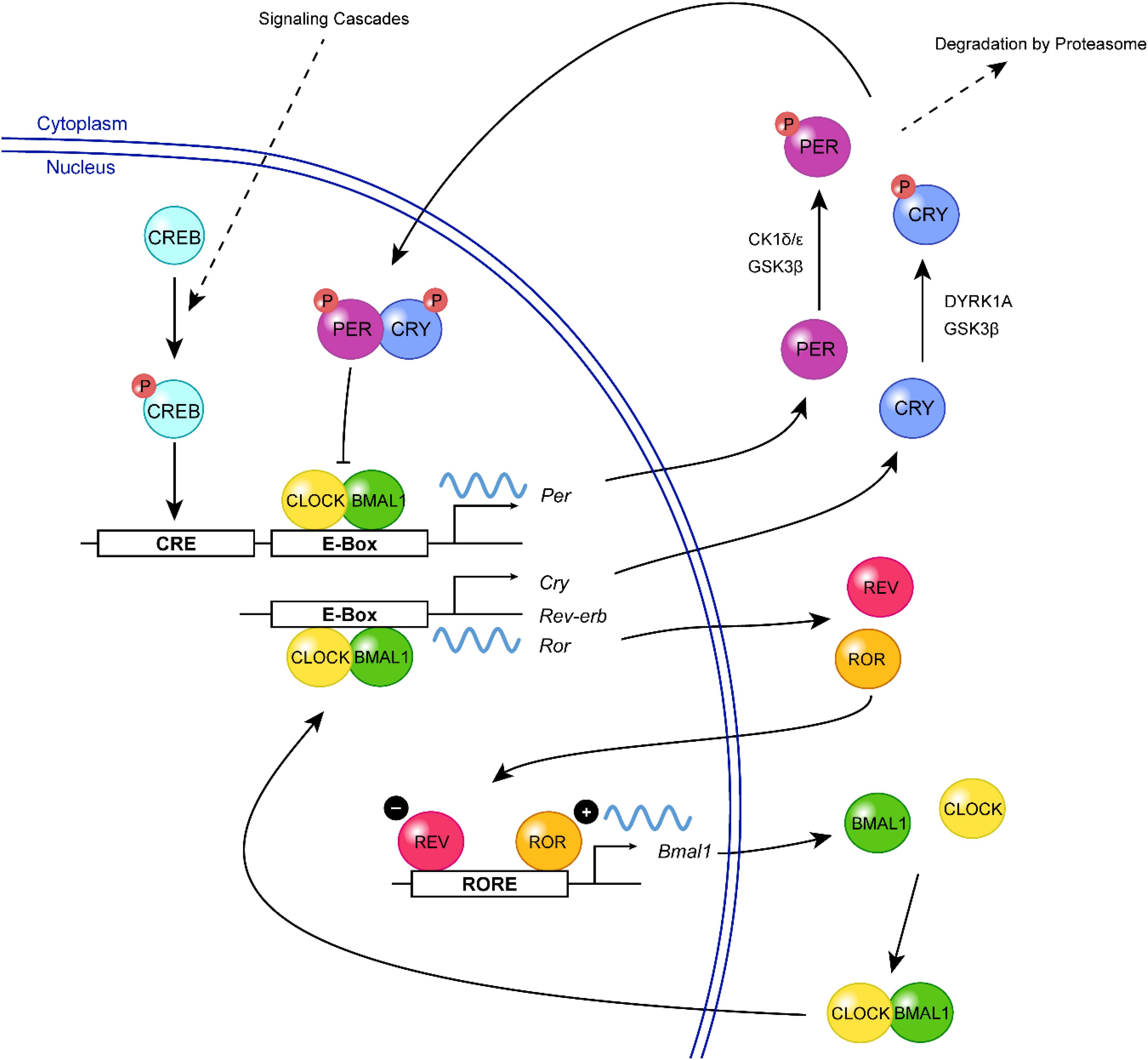
Figure 1. A simplified view of the mammalian molecular clock. In the positive limb of the primary feedback loop, CLOCK (yellow) and BMAL1 (green) form a heterodimer and bind to the E-box elements in the promoter regions of Per and Cry, triggering their transcription. Following their translation, PER (purple) and CRY (blue) proteins are phosphorylated (red, P) by various kinases including CK1δ/ε, GSK3β, and DYRK1A, which can regulate their turnover or nuclear entry. In the negative limb, PER:CRY heterodimers translocate to the nucleus, where they inhibit CLOCK:BMAL1-mediated transcription, thereby repressing their own gene expression. The transcription of Bmal1 is further regulated by a second feedback loop involving two E-box-regulated genes, Rev-Erb and Ror. REV-ERB (magenta) inhibits the transcription of Bmal1 by competing with the transcriptional activator, ROR (orange), for binding of the ROR-element within the Bmal1 promoter. Extracellular signals (e.g., neurotransmitters, neuropeptides) can activate signaling cascades resulting in the phosphorylation of CREB (turquoise), which mediates Per transcription and resetting of the clock.
SCN Structure and Connectivity
As it is currently understood, the SCN is responsible for interpreting photic and non-photic signals that it receives from afferent projections, and ultimately produces a coherent temporal output to peripheral oscillators through humoral and neuroendocrine mechanisms. Each individual SCN neuron harbors the clock machinery and is able to maintain robust molecular rhythms on a single-cell level. Through neuropeptide, neurotransmitter, and synaptic signaling, SCN neurons form an intricately connected oscillatory network with astounding precision and resilience.
The SCN is a pair of nuclei located in the anterior hypothalamus, situated directly dorsal to the optic chiasm and lateral to the third ventricle. It is comprised of approximately 20,000 heterogenous neurons that secrete dozens of neuropeptides, neurotransmitters, and cytokines, many of which can be at least partially co-expressed by certain populations of SCN neurons (Figure 2; Abrahamson and Moore, 2001; Cheng et al., 2002; Antle and Silver, 2005; Todd et al., 2020; Wen et al., 2020). The SCN is classically divided into two subregions, a light-responsive ventrolateral “core” and a rhythmic dorsomedial “shell,” based on the neurochemical nature of cells in each area and its physiological function (Aton et al., 2005). SCN core neurons are characterized by expression of vasoactive intestinal peptide (VIP), gastrin releasing peptide (GRP), calbindin, calretinin, neuromedin S (NMS), and neurotensin (Abrahamson and Moore, 2001; Lee et al., 2015). In contrast, SCN shell neurons express arginine vasopressin (AVP), calbindin, NMS, angiotensin II, and met-enkephalin (Abrahamson and Moore, 2001; Lee et al., 2015). All SCN neurons synthesize γ-aminobutyric acid (GABA) as the main neurotransmitter in addition to the neuropeptidergic signals (Moore and Speh, 1993; Abrahamson and Moore, 2001).
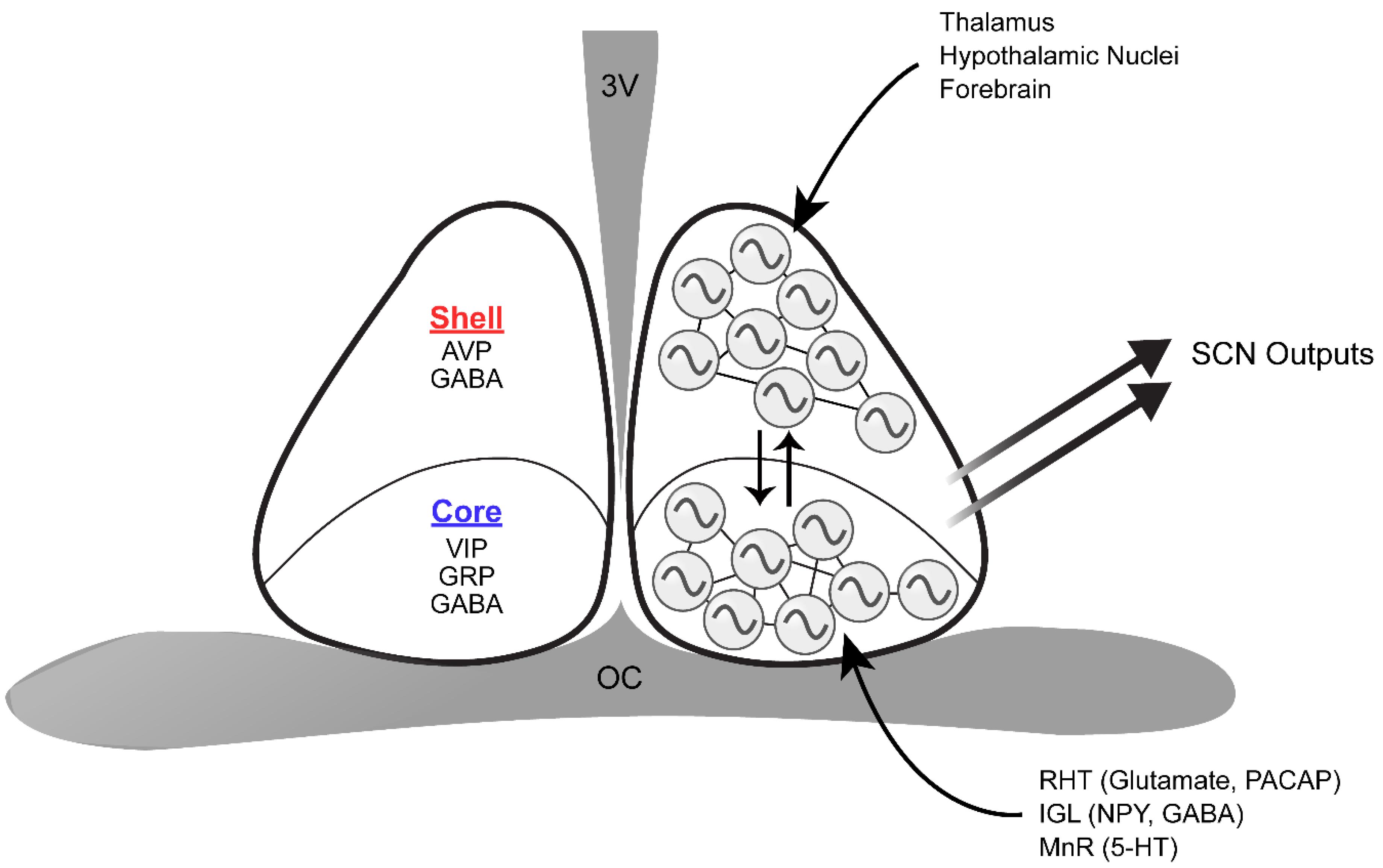
Figure 2. Schematic of the structure and organization of the SCN. The dorsomedial SCN (shell) expresses AVP and GABA, whereas the ventrolateral SCN (core) synthesizes VIP, GRP, and GABA. The retinohypothalamic tract (RHT), intergeniculate leaflet (IGL), and median raphe nucleus (MnR) directly innervate the core. On the other hand, inputs from the thalamus, various hypothalamic nuclei, and the forebrain are mainly received in the shell. Core and shell SCN neurons are synchronized through various means of intercellular communication, and are thus capable of producing coherent outputs to peripheral clocks. 3V, third ventricle; OC, optic chiasm.
In addition to neurons, astrocytes in the murine SCN also contribute to circadian timekeeping. Astrocytes have been shown to display daily rhythms in structural protein expression, morphology, metabolic function, and clock gene expression (Prolo et al., 2005; Becquet et al., 2008; Cheng et al., 2009; Burkeen et al., 2011). Astrocyte-specific ablation of Bmal1 lengthens the period of clock gene oscillations and locomotor behavior (Barca-Mayo et al., 2017; Tso et al., 2017). Furthermore, excision of the short-period CK1ε tau mutation specifically from SCN astrocytes lengthens molecular and behavioral rhythms (Brancaccio et al., 2017; Tso et al., 2017). It has been shown that SCN astrocytes control circadian period by regulating GABA uptake and glutamatergic signaling (Barca-Mayo et al., 2017; Brancaccio et al., 2017, 2019). Recently, Sominsky et al. (2021) reported that microglia are another important component for maintaining clock gene expression and behavioral rhythms. By expressing the diphtheria toxin (DT) receptor specifically in fractalkine receptor-positive cells (Cx3cr1+), 94% of SCN microglia was acutely ablated in transgenic Wistar rats (Sominsky et al., 2021). This resulted in pronounced disruption of behavioral rhythms, circadian temperature profiles, and Per1 and BMAL1 expression (Sominsky et al., 2021).
As the master circadian clock, the SCN is intricately connected with many regions of the brain to regulate the phase and period of circadian rhythms. The SCN has three major afferent connections: retinohypothalamic tract (RHT) projections from the retina, geniculohypothalamic tract (GHT) projections from the intergeniculate leaflet (IGL), and serotonergic projections from the median raphe nucleus (MnR) in the brainstem (Meyer-Bernstein and Morin, 1996; Mintz et al., 1997; Abrahamson and Moore, 2001). Photic information received by intrinsically photosensitive retinal ganglion cells (ipRGCs) is delivered to the SCN through the RHT (Do and Yau, 2010). Although the terminal fields of the RHT can be found in all parts of the murine SCN, the core region has denser retinal innervation compare to the SCN shell (Morin et al., 2006). IpRGC input is essential for SCN light entrainment, which occurs through the release of glutamate, aspartate, and the neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP) (Chen et al., 1999; Guido et al., 1999; Kawaguchi et al., 2003; Hannibal et al., 2008). In contrast, the IGL innervates the SCN with neuropeptide Y (NPY) and GABA terminals (Moore et al., 2002). As with RHT terminals, NPY terminals are concentrated in the ventro-central region of the SCN and are sparser in the dorsomedial region (Morin et al., 2006). The IGL plays a significant role in relaying photic and non-photic information to the SCN, as either IGL lesions or NPY infusion into the SCN can alter circadian rhythms (Albers and Ferris, 1984; Pickard et al., 1987). The SCN also receives serotonergic projections from the MnR, where the plexus appears to be densest along the medial and ventral SCN border, grading to sparse innervation centrally and dorsolaterally (Morin et al., 2006). Notably, all three major SCN inputs significantly overlap in the core SCN, reflecting its key role in integrating luminance information from the retina and non-photic input from the midbrain arousal center during entrainment. In addition to these three major afferents, ∼35 brain regions have been shown to project to the SCN: these include other hypothalamic nuclei, the amygdalohippocampal zone, and brainstem nuclei (Krout et al., 2002). However, relatively few SCN afferent systems have been explored in terms of their rhythm-related functions. On the other hand, SCN efferents innervate ∼15 brain regions, likely carrying circadian rhythm phase information to distal targets. Notably, the SCN projects to many hypothalamic nuclei including the preoptic area, the paraventricular nucleus, the subparaventricular zone, the retrochiasmatic area, the dorsomedial and ventromedial nuclei, and the premammillary area (Watts and Swanson, 1987; Abrahamson and Moore, 2001). These efferent projections from the SCN have been implicated in circadian regulation of body temperature, locomotor activity, sleep-wake cycles, and feeding (Lu et al., 2001; Chou et al., 2003; Abrahamson and Moore, 2006). Neuropeptides such as prokineticin 2 (PROK2) have been shown to serve as functional outputs of the SCN, communicating phase information to other brain regions (Cheng et al., 2002). Mice deficient in either PROK2 or its cognate receptor, prokineticin receptor 2 (PROKR2), have a pronounced redistribution of locomotor activity from early night to late night with significantly dampened amplitude (Li et al., 2006; Prosser et al., 2007).
Intra-SCN Communication and Signaling
In vivo or in organotypic cultures, oscillations of SCN neurons are synchronized and coherent, yet follow a consistent pattern of distinct phases and amplitudes. Bioluminescence imaging of SCN explants in vitro using Per1-LUC or PER2:LUC reporters have shown that the shell region has much more pronounced PER oscillations than the core (Yamaguchi et al., 2003). Circadian cycling of PER expression begins in the dorsomedial periventricular region of the shell, propagates ventrally and laterally to the center of the shell after 4–8 h, and ends in the ventral SCN after 12–15 h (Yamaguchi et al., 2003). However, a minority of cells have been shown to remain ∼12 h out-of-phase with the population mean, potentially allowing multiple, variously phased output signals to be generated (Herzog et al., 1997; Nakamura et al., 2001). This oscillation pattern is preserved from cycle-to-cycle, as well as after pharmacological manipulations that delay or stop the clock, suggesting that SCN coupling is mediated by specific neural circuits instead of a homogeneous coupling scheme (Liu et al., 1997). When challenged by an abrupt phase shift (jet lag), the shell and core SCN exhibit desynchronization and appear to be out-of-phase initially (Nagano et al., 2003; Albus et al., 2005; Nakamura et al., 2005). The phase of the ventral region, measured by either clock gene expression or impulse activity of SCN neurons, shifts rapidly, whereas the dorsal region requires more days to shift and to align with the ventral SCN (Nagano et al., 2003). This finding is consistent with the notion that the core SCN is the compartment that receives photic cues and directs resynchronization of the shell. When cultured at low density, individual SCN neurons can still express robust rhythms autonomously for weeks, showing that they do not require rhythmic input from other cells to oscillate (Welsh et al., 1995). However, dispersed clock cells in the same culture dish display rhythms with varying periods and progressively distinct phase relationships. Dissociating SCN cells thus removes the coupling forces that normally maintain intercellular synchrony at the tissue level.
Three modes of intercellular communication have been established to maintain network stability and coupling of the SCN: chemical synapses, electrical synapses (gap junctions), and paracrine signaling. The most common neurotransmitter in the SCN is GABA, which is present in all SCN neurons (Moore and Speh, 1993). Although GABA usually elicits spontaneous inhibitory post-synaptic potentials, it may be excitatory in some instances (De Jeu and Pennartz, 2002; Albus et al., 2005; Hee et al., 2008). GABA-mediated excitation is most common at night in the dorsal region of the SCN, a process mediated by Na+-K+-Cl– cotransporter 1 (NKCC1) and K+-Cl– cotransporters (KCCs) (De Jeu and Pennartz, 2002; Hee et al., 2008). In Syrian hamsters, blocking excitatory responses of GABA by inhibiting NKCC1 has been shown to attenuate light-induced phase delays during the early subjective night (McNeill et al., 2018). In the SCN, many cellular responses of GABA are driven by ionotropic GABAA and Gαi/o−coupled GABAB receptors (Jiang et al., 1997b; Strecker et al., 1997; Liu and Reppert, 2000; Gribkoff et al., 2003; Belenky et al., 2008). GABAA receptor-mediated signaling causes phase shifts when applied to dissociated SCN neurons, and daily pulses of GABA synchronizes them (Liu and Reppert, 2000). In addition, GABA signaling is essential for shell and core re-synchronization after a jet lag treatment, as well as for modulating phase shifting responses after a light pulse (Ralph and Menaker, 1985, 1986, 1989; Gillespie et al., 1997, 1999; Albus et al., 2005; Albers et al., 2017). On the other hand, blocking both GABAA and GABAB receptors does not affect oscillatory amplitude and synchrony of neurons in SCN slices (Aton et al., 2006). Recently, Barca-Mayo et al. (2017) showed that astrocyte-mediated GABA signaling modulates clock gene expression of cortical neurons in vitro (Barca-Mayo et al., 2017).
It is widely accepted that communication between neurons is mediated primarily by Ca2+-dependent synaptic transmission. However, when chemical synaptic transmission is blocked using Ca2+-free medium, periodic and synchronized bursts of action potentials in a large population of SCN neurons can still be detected, indicating that mechanisms other than chemical synaptic transmission may modulate SCN synchrony (Bouskila and Dudek, 1993). It was also noted that metabolic rhythms in the embryonic SCN precede chemical synaptogenesis in rats, indicating that non-synaptic mechanisms may be important in coordinating circadian rhythms (Reppert and Schwartz, 1984). Subsequently, gap junctions (electrical synapses) have been identified in the SCN and found to mediate neuronal coupling (Jiang et al., 1997a; Colwell, 2000; Long et al., 2005; Rash et al., 2007; Wang et al., 2014). Gap junction channels allow the passage of ions and other small molecules between coupled cells and function to connect cells electrically and metabolically. They are formed by two hemichannels, each composed of 6 connexin proteins (Cheung et al., 2014). The majority of neuronal gap junctions in the SCN are miniature gap junctions that are composed of less than 50 connexons comprised primarily of connexin-36 (Cx36) subunits (Rash et al., 2007). Possibly due to the small number of large gap junctions and the predominance of mini-gap junctions, there is limited electrotonic coupling and coupling-mediated spike-for-spike synchronization between SCN neurons (Rash et al., 2007). Cx36 knockout mice have deficits in circadian behavior and electrical coupling between SCN neurons; however, the results are complicated by the global nature of the Cx36 ablation (Long et al., 2005). Blocking gap junctions in SCN slices with carbonoxolone also weakens synchrony of the SCN network (Wang et al., 2014).
The SCN expresses a plethora of neuropeptides, many of which are strongly implicated in SCN coupling. The core SCN and the most prevalent neuropeptide intrinsic to this region—VIP—are vital for maintaining coupling within the SCN. In mice, VIP is released rhythmically from the core and acts through the G-protein coupled, vasoactive intestinal peptide receptor 2 (VPAC2, also known as VIPR2), which is expressed in both the core and the shell SCN (Shinohara et al., 2000; Dardente et al., 2004). Vip or Vpac2 knockout mice display weak behavioral rhythms and often become arrhythmic after a few days in constant darkness (Harmar et al., 2002; Colwell et al., 2003; Aton et al., 2005). Organotypic SCN slices from these mice show suppressed neuronal firing, low amplitude clock gene rhythms, and desynchrony among cells (Cutler et al., 2003; Aton et al., 2005; Brown et al., 2007; Hughes et al., 2008; Maywood et al., 2011). VIP evokes phase shifts in locomotor activity, AVP release, multiunit firing rate, and PER2:LUC rhythms in a phase- and dose-dependent manner (Piggins et al., 1995; Watanabe et al., 2000; Reed et al., 2001; An et al., 2011). Molecularly, VIP-mediated phase shifting requires PKA, PLC, and MAPK signaling pathways, which ultimately converge on the activation of CRE-mediated transcription of clock genes (Nielsen et al., 2002; Meyer-Spasche and Piggins, 2004). Furthermore, VIP has been shown to modulate the strength of electrical synapses, which in turn regulate intercellular coupling (Wang et al., 2014). Intriguingly, studies have demonstrated that VIP can promote network plasticity by destabilizing intercellular synchrony in addition to its role as a synchronizing factor. For instance, application of exogenous VIP at concentrations greater than 100 nM desynchronizes and broadens the phase distribution of cells within the SCN, in particular during the nadir of PER2 expression (An et al., 2013). When microinjected into the SCN during the early subjective day, a phase when VIP does not induce phase shifts, VIP accelerates entrainment of locomotor rhythms to an advanced LD cycle in a jet lag paradigm (An et al., 2013). Although Vip–/– and Vpac2–/– mice have weak behavioral rhythms in DD, their behavioral rhythmicity can be restored by long-term exposure to constant light (An et al., 2013; Hughes et al., 2015). Detailed analyses of Vpac2–/– mice revealed that exposure to LL diminishes the intercellular signaling deficit in these animals, resulting in both improved behavioral rhythms and increased cellular synchrony (Hughes et al., 2015). This is in stark contrast with the disruptive effect of LL on neuronal function and physiological rhythms in animals with a fully functional SCN, where the elevated level of VIP induced by LL might destabilize the circadian pacemaker (Ohta et al., 2005; An et al., 2011).
The core SCN also produces another type of neuropeptide that participates in maintaining cellular synchrony, behavioral rhythmicity, and entrainment—namely, GRP. In mice, GRP is expressed rhythmically under a light-dark cycle and act through the G-protein coupled receptor bombesin receptor 2 (BB2, also known as GRPR) (McArthur et al., 2000; Karatsoreos et al., 2006). Similar to VIP, application of exogenous GRP to the SCN produces phase shifts in a phase-dependent manner both in vitro and in vivo (Piggins et al., 1995; McArthur et al., 2000; Aida et al., 2002). This light-like response to GRP stimulation is accompanied by the upregulation of Per1 and c-Fos expression in the dorsal SCN (Aida et al., 2002). GRP can also act as a secondary synchronizing neuropeptide when VIP-VPAC2 signaling is defective, since addition of GRP to Vpac2–/– SCN explants can transiently restore network synchrony (Brown et al., 2005; Maywood et al., 2011). Recent single cell RNAseq studies have found that GRP-expressing neurons are a subpopulation of VIP neurons: approximately one-quarter of VIP+ cells in the SCN co-express GRP (Todd et al., 2020; Wen et al., 2020).
Neuropeptides expressed by the shell SCN have also been shown to modulate circadian timekeeping. The most prevalent neuropeptide expressed by the shell is AVP. Avp is rhythmically expressed in mice, as its promoter contains E-box motifs for CLOCK:BMAL1 transactivation (Jin et al., 1999). When the AVP receptors V1a and V1b are genetically ablated, mutant mice become resistant to jetlag, re-entraining abruptly to shifts in the light-dark cycles (Yamaguchi et al., 2013). Examination of clock gene expression coupled with bioluminescence imaging of SCN explants revealed that V1a–/–V1b–/– SCN neurons show severely permutated phase order, and loss of intercellular synchrony following phase resetting (Yamaguchi et al., 2013). These results suggest that AVP-mediated interneuronal communication provides buffering toward abrupt external perturbations, and disruption of AVP-V1a/b signaling leads to a weakened oscillator. A mouse model with Bmal1 deletion specifically in AVPergic neurons shows enhanced re-entrainment and lengthened behavioral period, possibly due to the combined attenuation of Avp, Prok2, and Rgs16 expression in the SCN shell of these conditional knockout mice (Mieda et al., 2015). Deletion of CK1δ in AVP neurons also results in lengthened behavioral period and altered spatiotemporal pattern of PER2:LUC oscillations in SCN slices, indicating that AVP neurons can regulate SCN pacemaking (Mieda et al., 2016).
The majority of VIP+ and AVP+ neurons co-express the neuropeptide neuromedin S (NMS) (Todd et al., 2020; Wen et al., 2020). Although mice lacking NMS retain normal circadian rhythms in vivo, blocking vesicular transmission from NMS+ neurons disrupted the network synchrony of the SCN (Lee et al., 2015). Manipulating the expression of core clock genes, such as Bmal1 and Per2, within NMS-expressing neurons is sufficient to disrupt molecular oscillations and behavioral rhythms (Lee et al., 2015). Conversely, overexpression of the ClockΔ19 transgene in NMS-expressing neurons can lengthen circadian period in vivo, indicating that periodicity is dictated by this subset of SCN neurons (Lee et al., 2015). Similar pace-setting effects have been reported for DRD1a+ cells in the SCN, which represent ∼60% of all SCN cells (Smyllie et al., 2016). When floxed Ck1εTau alleles are excised from DRD1a+ neurons, ∼60% of the Drd1acre/+:Ck1εTau/Tau temporally chimeric mice exhibited a free-running period that resembles wildtype animals (Ck1εWT/WT), whereas ∼30% of the chimeric mice displayed a short period similar to Ck1εTau/Tau animals (Smyllie et al., 2016). The majority of chimeric SCN slices also displayed significantly lengthened PER2:LUC period, suggesting that DRD1a+ cells play a dominant role in period determination within the SCN (Smyllie et al., 2016).
Embryonic Development of the Hypothalamus
Hypothalamic histogenesis follows the same general pattern as that observed in other neural tube-derived brain regions, with dividing progenitors residing in the ventricular zone and producing neuronal and glial precursors that migrate laterally into the parenchyma (Bedont and Blackshaw, 2015; Xie and Dorsky, 2017). It was originally thought that hypothalamic development follows an outside−in pattern, where lateral hypothalamic nuclei are generated first and displaced outward by medial nuclei that are born later (Shimada and Nakamura, 1973; Altman and Bayer, 1978). However, more recent research has found that in some hypothalamic regions, including the arcuate nucleus and dorsolateral anterior hypothalamus, cells occupying different medial-lateral locations are born during the same interval (Markakis and Swanson, 1997; Padilla et al., 2010).
After neural plate formation following gastrulation, diffusible morphogens generated by the mesodermal domains such as the notochord and prechordal plate (PCP) begin patterning the developing nervous system, including the presumptive hypothalamus. These morphogenic cues modulate important processes during hypothalamic induction and the establishment of regional identity (Bedont and Blackshaw, 2015). Sonic hedgehog (SHH) is a lipid−linked polypeptide signal that is first released from the prechordal plate, is necessary for induction of the hypothalamus, and drives Shh expression in the ventral diencephalon (Shimamura and Rubenstein, 1997). Through a Glioma associated oncogene(GLI)-mediated signaling cascade, PCP-derived SHH activates markers of hypothalamic identity along the overlying ventral diencephalic midline (Dale et al., 1997; Motoyama et al., 2003; Manning et al., 2006). Studies have shown that SOX2 and SOX3 can activate and maintain Shh transcription in the hypothalamic neuroepithelium through direct binding to the long-range Shh forebrain enhancer-2 (SBE2), whereas T-box transcription factors 2 (TBX2) and TBX3 repress Shh in the caudal hypothalamus by sequestering SOX2 away from the SBE2 (Zhao et al., 2012; Trowe et al., 2013). Sineoculis homeobox homolog 3 (SIX3) likewise targets the SBE2 to directly activate Shh transcription (Geng et al., 2008; Jeong et al., 2008). Using the Nkx2.1-cre mouse line, selective deletion of Shh from the basal plate domain of the developing hypothalamus results in ablation of markers of tuberal and anterior hypothalamic nuclei, along with thinning of the telencephalic and hypothalamic neuroepithelium (Shimogori et al., 2010). Shh deletion in the zona limitans interthalamica (ZLI) leads to a complete loss of prethalamic markers such as LIM homeobox 1 (Lhx1) and Gastrulation brain homeobox 2 (Gbx2) (Szabó et al., 2009a, b). Shh expression is also dependent on Retina and anterior neural fold homeobox (Rax), as Rax–/– mouse embryos show a downregulation of Shh expression in the dorsomedial portion of the hypothalamus along with underdevelopment of the hypothalamic neuroepithelium (Orquera et al., 2016).
In addition to SHH, modulators of Wingless/Int-1 (WNT) signaling also regulate the patterning of the diencephalon and hypothalamus (Bedont and Blackshaw, 2015). The WNTs are a diverse family of secreted, palmitoleoylated signaling glycoproteins well known for their role in regulating anteroposterior patterning (Mikels and Nusse, 2006). WNTs released by the posterior neurectoderm and somites promote hindbrain fate, whereas the PCP and anterior neurectoderm produce WNT inhibitors to antagonize WNT signaling. Wnt8b is expressed in the mouse posterior hypothalamus beginning at ∼ E8.5, consistent with a role in patterning (Braun et al., 2003; Shimogori et al., 2010; Martinez-Ferre et al., 2013). Wnt7a and Wnt7b are expressed selectively in prethalamic and hypothalamic GABAergic neuronal progenitors around E12.5, suggesting a role in interneuron development; however, their function has not been well characterized (Shimogori et al., 2010). When the transcriptional repressor of WNT targets, Transcription factor 7-like 1 (Tcf7l1), is conditionally knocked out in the mouse hypothalamus and pituitary, the developing hypothalamus is posteriorized (Gaston-Massuet et al., 2016). In contrast, loss-of-function of β-catenin (encoded by Ctnnb1) results in the anteriorization of the hypothalamus as seen in Foxd1-Cre; Ctnnb1lox/lox and Nkx2.1-Cre; Ctnnb1lox/lox mice (Newman et al., 2018b). Loss of SIX3, a direct Wnt1 repressor, results in rostral expansion of caudal diencephalic markers as well as prosencephalon truncation (Lagutin et al., 2003). Lastly, Lhx2, a potential upstream inhibitor of WNT signaling (Peukert et al., 2011), has also been shown to play a role in the patterning of the telencephalic-optic-hypothalamic field and to specify SCN neurons at the expense of neuroendocrine fates (Roy et al., 2013).
Embryonic Development of the SCN
SCN neurogenesis begins at ∼60% of gestation in rodents (E12 for mice) (Figure 3A; Shimada and Nakamura, 1973; Antle et al., 2005; Kabrita and Davis, 2008). By ∼70% of gestation (E13.5), most ventrolateral neurons have already been produced, while dorsomedial neurogenesis is still underway. The final major burst of SCN neurogenesis (including a number of ventrolateral neurons) occurs at ∼80% of gestation (E15). Neurogenesis in the SCN along its rostral/caudal axis also shows heterochronicity, with neurogenesis in the medial SCN peaking at E12, in the caudal SCN peaking at E13.5, and in the rostral SCN peaking at E14. After immature SCN neurons are generated, they will continue to develop and ultimately express signature neuropeptides such as VIP, GRP, and AVP. Consistent with the regional differences in the peak timing of neurogenesis, cell types that are concentrated in the core SCN (e.g., VIP, GRP, and calbindin expressing cells) are mostly born early. In comparison, AVP neurons of the shell SCN are generated consistently during the period of neurogenesis that extends into later embryonic ages, with those in the middle-posterior regions generated prior to those situated in the anterior pole (Antle et al., 2005). It has been suggested that SCN core and shell neurons derive from distinct progenitor pools in the neuroepithelium (Altman and Bayer, 1986), but the precise mechanisms that regulate cell−type differences in the timing of SCN neurogenesis are not well understood.
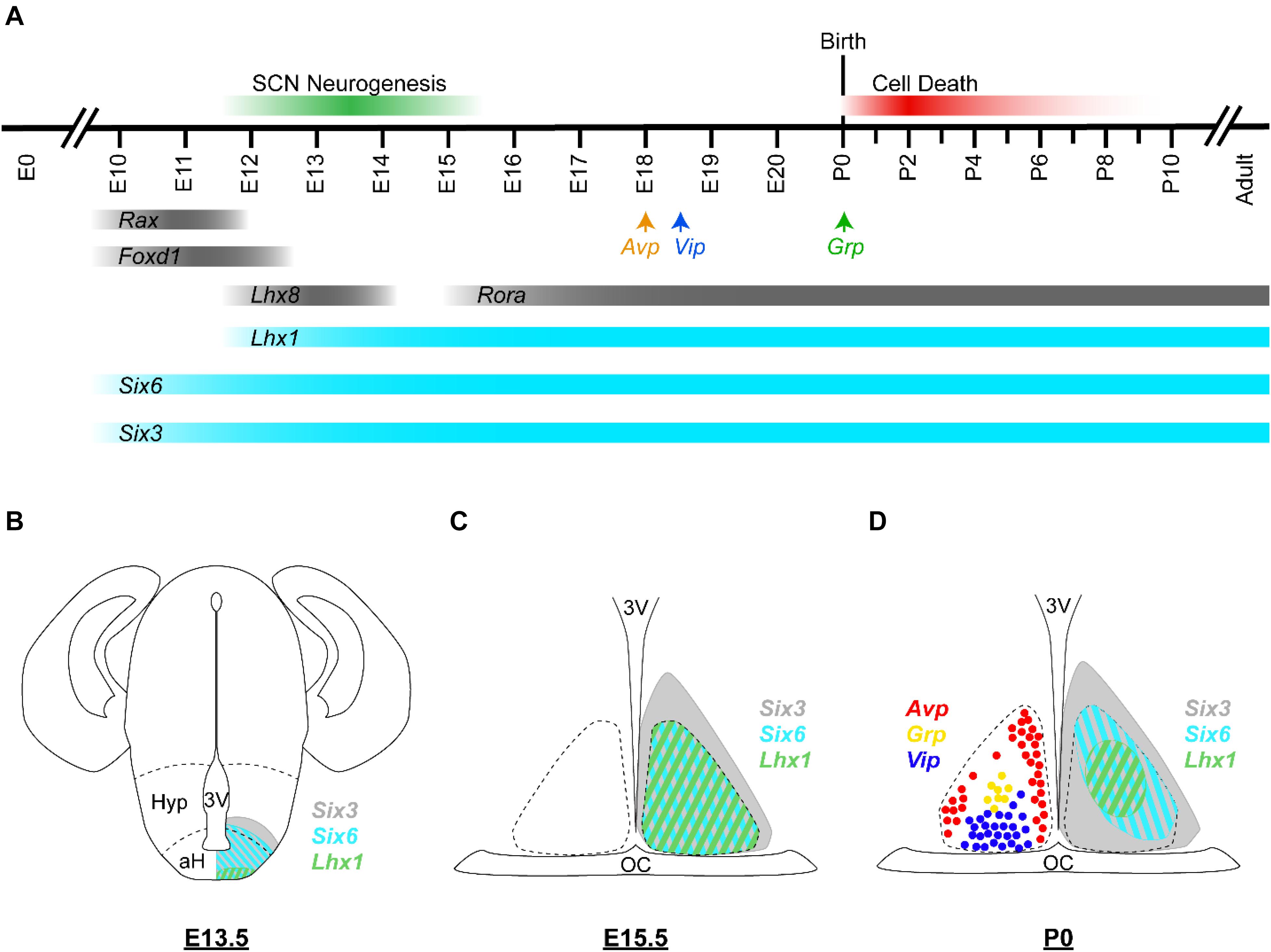
Figure 3. Embryonic development of the murine SCN. (A) Developmental timeline of the murine SCN. In mice, SCN neurogenesis begins around E12 and is considered complete by E15 (green bar). Avp, Vip, and Grp transcripts are first detected in the developing SCN at E18, E18.5, and P0, respectively (arrows). Many SCN cells are lost during P0–P8 by apoptotic cell death (red bar). Expression timeline of selected hypothalamus- and SCN-enriched transcription factors are shown; those with known developmental functions are depicted in turquoise and the others are depicted in gray. (B–D) Schematic illustrations of the murine brain/SCN showing the spatiotemporal expression patterns of Six3 (gray), Six6 (turquoise), and Lhx1 (green) at (B) E13.5, (C) E15.5, and (D) P0. Cells expressing Avp (red), Grp (yellow), and Vip (blue) at P0 are depicted as colored circles. Dashed lines in (A) indicate the boundaries of the anterior hypothalamus (aH) and the hypothalamus (Hyp). Dashed lines in (C,D) indicate the margins of the SCN. Expression data are from Shimogori et al. (2010), VanDunk et al. (2011), Bedont et al. (2014), Newman et al. (2018a); unpublished observation (AH Cheng and HYM Cheng). 3V, third ventricle; OC, optic chiasm.
SCN development is modulated by morphogen signaling. For instance, the WNT receptor frizzled 5 (Fzd5) is detected at E10.5–13.5 in mitotic cells (VanDunk et al., 2011). Fzd5 is later downregulated, coinciding with the induction of distal-less homeobox 2 (Dlx2), which is a selector gene important for GABAergic neuron development (Pla et al., 2018). In addition, members of the fibroblast growth factor (Fgf) family are known to control the development of the hypothalamus and the SCN (Tsai et al., 2011). Fgf8, one of the 22 Fgf ligands, is expressed in the developing hypothalamus by E9.5 in developing mouse embryos, with robust expression in regions surrounding the optic chiasm (Fon Tacer et al., 2010). In homozygous Fgf8 hypomorphic mice (∼54% reduction of Fgf8 mRNA) at postnatal day (P)0, SCN volume as well as the expression of AVP and VIP are severely reduced, indicating that normal SCN development relies heavily on Fgf8 signaling (Brooks et al., 2010; Tsai et al., 2011; Miller et al., 2016). Furthermore, hypomorphism in the cognate tyrosine kinase receptor of FGF8, Fgfr1, also causes reduction in VIP expression but does not affect SCN volume. The lesser dependence of SCN development on Fgfr1 suggests possible compensation by other FGF receptors, as Fgfr1, 2, and 3 are all expressed along the proliferative ventricular zone, but Fgf8 is the only ligand expressed robustly in the ventral diencephalon near the presumptive SCN (Wanaka et al., 1990; Crossley and Martin, 1995; Belluardo et al., 1997; Bansal et al., 2003). Hence, the consequence of Fgf8 expression deficit is more severe, as other compensatory factors may not be available during SCN development.
Neuronal Differentiation In The Developing SCN
Many transcription factors such as Rax, forkhead box D1 (Foxd1), Nkx2.2, Lhx2, Six3, Six6, and ventral anterior homeobox 1 (Vax1) are expressed in the ventral anterior hypothalamic neuroepithelium prior to the onset of SCN neurogenesis, although expression of many of these factors become restricted to specific anatomical regions or is lost entirely as the SCN develops (Shimogori et al., 2010; VanDunk et al., 2011).
A subset of these genetic markers such as Lhx2, Rax, Foxd1, and Nkx2.2 are expressed transiently in the ventral anterior hypothalamus and are gradually lost as the SCN develops. Foxd1 is broadly expressed in the developing hypothalamus from E11.5 to E13.5 and has been shown to be necessary for SCN development (Newman et al., 2018a). At the early stages of SCN neurogenesis, Foxd1-deficient mice display mild developmental deficits, with reduced expression of Vax1 and Six3 and a decrease in cellular proliferation (Newman et al., 2018a). By P0.5, more severe defects are observed in the SCN of Foxd1-deficient mice, including reduced expression of genetic markers and agenesis (Newman et al., 2018a). Rax is expressed in the murine ventral hypothalamus between E10.5 and E12.5 (Pak et al., 2014). Deletion of Rax prior to E8.5 disrupts Shh expression and is accompanied by a patterning defect of the mediobasal hypothalamus as described in the previous section, whereas later deletion causes misspecification of ventral medial hypothalamic nucleus (VMH) neurons (Lu et al., 2013; Orquera et al., 2016). Lhx2 is required for specification of the SCN, as Lhx2–/– mice lack expression of multiple central anterior markers at E12.5, including Lhx1 and Vax1 (Roy et al., 2013). In contrast to many early anterior hypothalamic markers that are down-regulated in the SCN later in development, Six3 and Six6 expression in the SCN persists throughout the lifespan (Shimogori et al., 2010; VanDunk et al., 2011; Clark et al., 2013). Both Six3 and Six6 are required for initial SCN specification, as both Nestin-cre; Six3fl/fl and Six6–/– mice fail to form a morphologically recognizable SCN or to express SCN-specific markers (VanDunk et al., 2011; Clark et al., 2013). Collectively, these observations suggest that early genetic markers have a long-lasting impact on SCN development by regulating the activation of other transcription factors. Moreover, they may be able to prime the activation of cis-regulatory elements that control the expression of cell type-specific genes during differentiation.
Downstream of Six3 and Six6, Lhx1 is a crucial regulator of SCN terminal differentiation (Figure 3; Bedont et al., 2014). Lhx1 is considered to be the only known transcription factor involved in SCN differentiation and the earliest selective marker expressed throughout the developing SCN (VanDunk et al., 2011; Bedont et al., 2014). Six3-cre; Lhx1fl/fl SCN retains expression and proper compartmentalization of most markers initially expressed prior to E16.5, but neuropeptides with important roles in circadian function, including VIP, GRP, and AVP, are absent in the adult SCN (Bedont et al., 2014). By P4, significantly fewer SCN neurons are present in Lhx1-deficient SCN due to increased cell death from P0 to P4 (Bedont et al., 2014). Through bioinformatic and luciferase analyses, LHX1 has been shown to directly regulate the expression of Vip and potentially Nms, Prok2, and proenkephalin (Bedont et al., 2014; Hatori et al., 2014). Given that Lhx1 is first detectable at E11.5, the relatively normal development of Lhx1-deficient SCN from E11.5 to E16.5 suggests that Lhx1 is not necessary for early cell fate decisions or regional patterning of the SCN. Instead, Lhx1 is likely regulating terminal differentiation of SCN neurons. It is also possible that the loss of Lhx1 is compensated by one or more factors during this period; one potential candidate is Lhx8, an Lhx1 homolog that is co-expressed with LHX1 in the SCN from E11.5 to E16.5 (Shimogori et al., 2010). Alternatively, Creb3l1, an Lhx1-regulated gene, is a regulator of AVP expression in the SCN, although its role in SCN development has yet to be explored (Greenwood et al., 2014; Hatori et al., 2014).
Zfhx3 is another genetic marker with known roles in certain aspects of SCN function. Zfhx3 is highly and almost exclusively expressed in the adult SCN (Lein et al., 2007). While the constitutive knockout of Zfhx3 results in preweaning sublethality in heterozygous mice and the dominant missense mutation of Zfhx3, Short circuit (Sci), is homozygous perinatal lethal, adult mice with a single allele of Sci show downregulated neuropeptide expression and shortened period of wheel-running activity (Parsons et al., 2015). Furthermore, ablating Zfhx3 in adult mice using a tamoxifen-inducible transgenic line recapitulated the circadian behavioral deficit in Zfhx3Sci/+ mice, indicating that Zfhx3 acts to regulate SCN function in adulthood by activating transcription at AT motifs (Parsons et al., 2015; Wilcox et al., 2017). Its function in the developing SCN remains unclear.
The clock gene Rora is expressed in the ventral SCN starting at E14.5, throughout the SCN at E17.5, and in a pattern more restricted to the SCN shell by P21 (VanDunk et al., 2011; Newman et al., 2018a). Staggerer mice (Rorasg/sg) containing a deletion of the fifth exon of Rora have a morphologically normal SCN with nominal VIP and AVP expression; however, more characterization is necessary to conclude the function (or the lack thereof) of Rora during SCN development, as only the expression of VIP and AVP were examined in Rorasg/sg mice (VanDunk et al., 2011).
Neuronal Loss, Glial Development, and Synaptogenesis
A large number of SCN neurons are lost during the perinatal period through the activation of caspases and pro-apoptotic proteins of the Bcl-2 family. However, the mechanisms that initiate apoptosis in SCN neurons are largely unclear. SCN cell death begins as synapse formation increases, with substantial death between P1–P7 in mice (Ahern et al., 2013; Mosley et al., 2017). SCN neuronal survival might be dependent on intercellular communication, as apoptotic neurons are isolated from the neuronal clusters (Moore and Bernstein, 1989). Although peak cell death occurs in the murine SCN by P7, an additional 20% of cells are lost by adulthood (Bedont et al., 2014).
It has been estimated that the adult rat SCN contains at least 108 synapses with the majority of these being intra-SCN connections (Güldner, 1976; van den Pol, 1980; Moore and Bernstein, 1989). Anatomical studies investigating synaptology in the rat or hamster SCN have revealed important insights on the timing of synapse formation within the SCN (Lenn et al., 1977; Moore and Bernstein, 1989; Laemle et al., 1991; Speh and Moore, 1993). Extensive studies in rats demonstrated that synaptic development is largely a postnatal event with a spike of synaptogenesis occurring between P4 and P10 (Moore and Bernstein, 1989). At E19, 2 days after the end of rat SCN neurogenesis, the neuropil surrounding SCN neurons is sparse and contains large and medium-sized dendritic profiles (Moore and Bernstein, 1989). Immature synapses with very few synaptic vesicles are also present at this time (Lenn et al., 1977; Moore and Bernstein, 1989). Synaptic number and diversity gradually increase from E21 to P2, and then expand rapidly from P2 to P10 (Moore and Bernstein, 1989). Another 30% of the synapses are formed after P10 (Moore and Bernstein, 1989). The window of SCN synaptogenesis overlaps with the arrival of prominent afferent projections. For both mice and rats, terminals from the RHT begin to sparsely innervate the ventrolateral SCN ipsilaterally at birth, followed by the first appearance of contralateral projections in the ventromedial SCN at P4 (McNeill et al., 2011). By P10, the morphology of the axon terminals and the density of the RHT projections are considered to be adult-like (McNeill et al., 2011). Other SCN afferents such as the raphe nuclei and the IGL also innervate the nuclei postnatally and slowly mature in the following weeks (Takatsuji et al., 1995; Migliarini et al., 2013).
Similar to other brain regions, astrogliogenesis follows neurogenesis in the SCN, with the astrocyte marker glial fibrillary acidic protein (GFAP) first detectable shortly before birth, at E15 in hamsters and E20 in rats (Botchkina and Morin, 1995; Munekawa et al., 2000). In both species, there is a postnatal increase in GFAP+ processes that complements the decreasing expression of vimentin (a marker of radial glia) (Botchkina and Morin, 1995; Munekawa et al., 2000). The first major increase in GFAP expression within the SCN, indicative of astrocytic maturation, occurs around P3-P4 in rats (Munekawa et al., 2000).
Expression of Clock Genes and Neuropeptides
During late gestation, circadian rhythms of clock gene expression gradually emerge in a staggered fashion. PER proteins are generally considered to be the earliest core clock components to exhibit rhythmicity. PER2:LUC rhythms are detected as early as E13.5 in SCN slice cultures, but histological data collected from in vivo studies have placed the onset of Per2 and PER2 rhythm at a later stage, around E17 (Shimomura et al., 2001; Ansari et al., 2009; Wreschnig et al., 2014; Landgraf et al., 2015; Carmona-Alcocer et al., 2018). Per1 gene and protein expression begin to show daily oscillations at around the same developmental age as Per2, or ∼E18 (Shimomura et al., 2001; Ansari et al., 2009). In comparison, Cry1 and Cry2 show robust rhythmicity after birth, with Cry1, CRY1, and CRY2 beginning to oscillate at P1, P10, and P2, respectively (Ansari et al., 2009; Huang et al., 2010). In contrast to the highly dynamic expression of PER and CRY, CLOCK and BMAL1 are constitutively expressed in the adult or developing murine SCN, respectively (Von Gall et al., 2003; Ansari et al., 2009). CLOCK is detectable at low levels at E18 and gradually rises to reach adult-level expression by P10 (Ansari et al., 2009). On the other hand, BMAL1 is robustly expressed in a large proportion of SCN cells at E18 and remains at a constantly high level during later developmental stages (Ansari et al., 2009). However, rhythmic Bmal1 expression in the SCN can be detected at ∼P3 (Huang et al., 2010). The transcriptional activator of Bmal1, Rora, is first expressed at E13.5 and is found throughout the murine SCN by E17.5; therefore, Rora might contribute to the expression of BMAL1 at E18 (Ansari et al., 2009; VanDunk et al., 2011).
In mice, Vip expression is first detected ∼3.5 days after the end of neurogenesis in the SCN core, at E18.5 (Kabrita and Davis, 2008; VanDunk et al., 2011). VPAC2 and VIP reach detectable levels shortly after birth (P0 to P2) (Carmona-Alcocer et al., 2018). Further increases in the level of VIP expression as well as VIP-containing projections have been reported as the SCN matures (Herzog et al., 2000). Similarly, Avp is first detected in the murine SCN at E18 and robust expression of AVP is evident by P0 (Hyodo et al., 1992; VanDunk et al., 2011; Bedont et al., 2014). AVP expression continues to rise as the animal matures to ∼P30, at which point an adult level of expression is achieved (Herzog et al., 2000).
Intriguingly, mature SCN neurons continue to express genes that are commonly associated with maintenance of the stem cell states, even though the SCN does not undergo adult neurogenesis. These genes include doublecortin-like (DCL), transportin 1, Six3, Lhx1, and Sox2 (Sato et al., 2011; VanDunk et al., 2011; Saaltink et al., 2012; Hoefflin and Carter, 2014; Brown et al., 2017; Beligala et al., 2018; Cheng et al., 2019). SCN neurons also conspicuously exhibit low levels of NeuN, a marker for mature neurons (Morin et al., 2011). This raises the possibility that SCN neurons are not fully committed to a differentiated state, thus retaining a degree of plasticity that presumably allows it to rearrange neuronal circuitry in order to entrain and adapt to a dynamic environment. It is also possible that SCN neurons have “re-purposed” these stem cell or developmental genes for circadian rhythm regulation once their canonical role has been fulfilled.
Author Contributions
AHC wrote the manuscript. H-YMC edited the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
H-YMC was supported by the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council (NSERC) of Canada. AHC was supported by the NSERC post-graduate scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abrahamson, E. E., and Moore, R. Y. (2001). Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191. doi: 10.1016/s0006-8993(01)02890-6
Abrahamson, E. E., and Moore, R. Y. (2006). Lesions of suprachiasmatic nucleus efferents selectively affect rest-activity rhythm. Mol. Cell. Endocrinol. 252, 46–56. doi: 10.1016/j.mce.2006.03.036
Ahern, T. H., Krug, S., Carr, A. V., Murray, E. K., Fitzpatrick, E., Bengston, L., et al. (2013). Cell death atlas of the postnatal mouse ventral forebrain and hypothalamus: Effects of age and sex. J. Comp. Neurol. 521, 2551–2569. doi: 10.1002/cne.23298
Aida, R., Moriya, T., Araki, M., Akiyama, M., Wada, K., Wada, E., et al. (2002). Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol. Pharmacol. 61, 26–34. doi: 10.1124/mol.61.1.26
Albers, H. E., and Ferris, C. F. (1984). Neuropeptide Y: role in light-dark cycle entrainment of hamster circadian rhythms. Neurosci. Lett. 50, 163–168. doi: 10.1016/0304-3940(84)90480-4
Albers, H. E., Walton, J. C., Gamble, K. L., McNeill, J. K., and Hummer, D. L. (2017). The dynamics of GABA signaling: Revelations from the circadian pacemaker in the suprachiasmatic nucleus. Front. Neuroendocrinol. 44:35–82. doi: 10.1016/j.yfrne.2016.11.003
Albus, H., Vansteensel, M. J., Michel, S., Block, G. D., and Meijer, J. H. (2005). A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr. Biol. 15, 886–893. doi: 10.1016/j.cub.2005.03.051
Altman, J., and Bayer, S. A. (1978). Development of the diencephalon in the rat. I. Autoradiographic study of the time of origin and settling patterns of neurons of the hypothalamus. J. Comp. Neurol. 182, 945–971. doi: 10.1002/cne.901820511
An, S., Harang, R., Meeker, K., Granados-Fuentes, D., Tsai, C. A., Mazuski, C., et al. (2013). A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc. Natl. Acad. Sci. U.S.A. 110, E4355–E4361.
An, S., Irwin, R. P., Allen, C. N., Tsai, C., and Herzog, E. D. (2011). Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J. Neurophysiol. 105, 2289–2296. doi: 10.1152/jn.00966.2010
Ansari, N., Agathagelidis, M., Lee, C., Korf, H.-W., and von Gall, C. (2009). Differential maturation of circadian rhythms in clock gene proteins in the suprachiasmatic nucleus and the pars tuberalis during mouse ontogeny. Eur. J. Neurosci. 29, 477–489. doi: 10.1111/j.1460-9568.2008.06605.x
Antle, M. C., LeSauter, J., and Silver, R. (2005). Neurogenesis and ontogeny of specific cell phenotypes within the hamster suprachiasmatic nucleus. Dev. Brain Res. 157, 8–18. doi: 10.1016/j.devbrainres.2005.02.017
Antle, M. C., and Silver, R. (2005). Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 28, 145–151. doi: 10.1016/j.tins.2005.01.003
Aton, S. J., Colwell, C. S., Harmar, A. J., Waschek, J., and Herzog, E. D. (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 8, 476–483. doi: 10.1038/nn1419
Aton, S. J., Huettner, J. E., Straume, M., and Herzog, E. D. (2006). GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc. Natl. Acad. Sci. U.S.A. 103, 19188–19193. doi: 10.1073/pnas.0607466103
Bansal, R., Lakhina, V., Remedios, R., and Tole, S. (2003). Expression of FGF receptors 1, 2, 3 in the embryonic and postnatal mouse brain compared with Pdgfra, Olig2 and Plp/dm20: Implications for oligodendrocyte development. Dev. Neurosci. 25, 83–95. doi: 10.1159/000072258
Barca-Mayo, O., Pons-Espinal, M., Follert, P., Armirotti, A., Berdondini, L., and De Pietri Tonelli, D. (2017). Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 8, 1–14.
Becquet, D., Girardet, C., Guillaumond, F., François-Bellan, A.-M., and Bosler, O. (2008). Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia 56, 294–305. doi: 10.1002/glia.20613
Bedont, J. L., and Blackshaw, S. (2015). Constructing the suprachiasmatic nucleus: a watchmaker’s perspective on the central clockworks. Front. Syst. Neurosci. 9:74. doi: 10.3389/fnsys.2015.00074
Bedont, J. L., LeGates, T. A., Slat, E. A., Byerly, M. S., Wang, H., Hu, J., et al. (2014). Lhx1 Controls Terminal Differentiation and Circadian Function of the Suprachiasmatic Nucleus. Cell Rep. 7, 609–622. doi: 10.1016/j.celrep.2014.03.060
Belenky, M. A., Yarom, Y., and Pickard, G. E. (2008). Heterogeneous expression of γ-aminobutyric acid and γ-aminobutyric acid-associated receptors and transporters in the rat suprachiasmatic nucleus. J. Comp. Neurol. 506, 708–732. doi: 10.1002/cne.21553
Beligala, D. H., De, A., and Geusz, M. E. (2018). A meta-analysis characterizing stem-like gene expression in the suprachiasmatic nucleus and its circadian clock. Biomed Res. Int. 2018:3610603.
Belluardo, N., Wu, G., Mudo, G., Hansson, A. C., Pettersson, R., and Fuxe, K. (1997). Comparative localization of fibroblast growth factor receptor-1, -2, and -3 mRNAs in the rat brain: In situ hybridization analysis. J. Comp. Neurol. 379, 226–246. doi: 10.1002/(sici)1096-9861(19970310)379:2<226::aid-cne5>3.0.co;2-5
Botchkina, G. I., and Morin, L. P. (1995). Ontogeny of radial glia, astrocytes and vasoactive intestinal peptide immunoreactive neurons in hamster suprachiasmatic nucleus. Dev. Brain Res. 86, 48–56. doi: 10.1016/0165-3806(95)00017-8
Bouskila, Y., and Dudek, F. E. (1993). Neuronal synchronization without calcium-dependent synaptic transmission in the hypothalamus. Proc. Natl. Acad. Sci. U.S.A. 90, 3207–3210. doi: 10.1073/pnas.90.8.3207
Brancaccio, M., Edwards, M. D., Patton, A. P., Smyllie, N. J., Chesham, J. E., Maywood, E. S., et al. (2019). Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192. doi: 10.1126/science.aat4104
Brancaccio, M., Patton, A. P., Chesham, J. E., Maywood, E. S., and Hastings, M. H. (2017). Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 93, 1420–1435.e5.
Braun, M. M., Etheridge, A., Bernard, A., Robertson, C. P., and Roelink, H. (2003). Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 130, 5579–5587. doi: 10.1242/dev.00685
Brooks, L. R., Chung, W. C. J., and Tsai, P.-S. (2010). Abnormal hypothalamic oxytocin system in fibroblast growth factor 8-deficient mice. Endocrine 38, 174–180. doi: 10.1007/s12020-010-9366-9
Broussard, J. L., and Van Cauter, E. (2016). Disturbances of sleep and circadian rhythms. Curr. Opin. Endocrinol. Diabetes Obes. 23, 353–359.
Brown, L. A., Williams, J., Taylor, L., Thomson, R. J., Nolan, P. M., Foster, R. G., et al. (2017). Meta-analysis of transcriptomic datasets identifies genes enriched in the mammalian circadian pacemaker. Nucleic Acids Res. 45, 9860–9873. doi: 10.1093/nar/gkx714
Brown, T. M., Colwell, C. S., Waschek, J. A., and Piggins, H. D. (2007). Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J. Neurophysiol. 97, 2553–2558. doi: 10.1152/jn.01206.2006
Brown, T. M., Hughes, A. T., and Piggins, H. D. (2005). Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J. Neurosci. 25, 11155–11164. doi: 10.1523/jneurosci.3821-05.2005
Bunger, M. K., Wilsbacher, L. D., Moran, S. M., Clendenin, C., Radcliffe, L. A., Hogenesch, J. B., et al. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. doi: 10.1016/s0092-8674(00)00205-1
Burkeen, J. F., Womac, A. D., Earnest, D. J., and Zoran, M. J. (2011). Mitochondrial calcium signaling mediates rhythmic extracellular ATP accumulation in suprachiasmatic nucleus astrocytes. J. Neurosci. 31, 8432–8440. doi: 10.1523/jneurosci.6576-10.2011
Carmona-Alcocer, V., Abel, J. H., Sun, T. C., Petzold, L. R., Doyle, F. J., Simms, C. L., et al. (2018). Ontogeny of circadian rhythms and synchrony in the suprachiasmatic nucleus. J. Neurosci. 38, 1326–1334. doi: 10.1523/jneurosci.2006-17.2017
Chen, D., Buchanan, G. F., Ding, J. M., Hannibal, J., and Gillette, M. U. (1999). Pituitary adenylyl cyclase-activating peptide: a pivotal modulator of glutamatergic regulation of the suprachiasmatic circadian clock. Proc. Natl. Acad. Sci. U.S.A. 96, 13468–13473. doi: 10.1073/pnas.96.23.13468
Cheng, A. H., Bouchard-Cannon, P., Hegazi, S., Lowden, C., Fung, S. W., Chiang, C. K., et al. (2019). SOX2-dependent transcription in clock neurons promotes the robustness of the central circadian pacemaker. Cell Rep. 26, 3191–3202.e8.
Cheng, H. Y. M., Alvarez-Saavedra, M., Dziema, H., Choi, Y. S., Li, A., and Obrietan, K. (2009). Segregation of expression of mPeriod gene homologs in neurons and glia: Possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum. Mol. Genet. 18, 3110–3124. doi: 10.1093/hmg/ddp252
Cheng, M. Y., Bullock, C. M., Li, C., Lee, A. G., Bermak, J. C., Belluzzi, J., et al. (2002). Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417, 405–410. doi: 10.1038/417405a
Cheung, G., Chever, O., and Rouach, N. (2014). Connexons and pannexons: Newcomers in neurophysiology. Front. Cell. Neurosci. 8:348. doi: 10.3389/fncel.2014.00348
Chou, T. C., Scammell, T. E., Gooley, J. J., Gaus, S. E., Saper, C. B., and Lu, J. (2003). Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms. J. Neurosci. 23, 10691–10702. doi: 10.1523/jneurosci.23-33-10691.2003
Clark, D. D., Gorman, M. R., Hatori, M., Meadows, J. D., Panda, S., and Mellon, P. L. (2013). Aberrant Development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein Six6. J. Biol. Rhythms 28, 15–25. doi: 10.1177/0748730412468084
Colwell, C. S. (2000). Rhythmic coupling among cells in the suprachiasmatic nucleus. J. Neurobiol. 43, 379–388. doi: 10.1002/1097-4695(20000615)43:4<379::aid-neu6>3.0.co;2-0
Colwell, C. S., Michel, S., Itri, J., Rodriguez, W., Tam, J., Lelievre, V., et al. (2003). Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285, 939–949.
Crossley, P. H., and Martin, G. R. (1995). The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439–451.
Cutler, D. J., Haraura, M., Reed, H. E., Shen, S., Sheward, W. J., Morrison, C. F., et al. (2003). The mouse VPAC 2 receptor confers suprachiasmatic nuclei cellular rhythmicity and responsiveness to vasoactive intestinal polypeptide in vitro. Eur. J. Neurosci. 17, 197–204. doi: 10.1046/j.1460-9568.2003.02425.x
Dale, J. K., Vesque, C., Lints, T. J., Sampath, T. K., Furley, A., Dodd, J., et al. (1997). Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell 90, 257–269. doi: 10.1016/s0092-8674(00)80334-7
Dardente, H., Menet, J. S., Challet, E., Tournier, B. B., Pévet, P., and Masson-Pévet, M. (2004). Daily and circadian expression of neuropeptides in the suprachiasmatic nuclei of nocturnal and diurnal rodents. Mol. Brain Res. 124, 143–151. doi: 10.1016/j.molbrainres.2004.01.010
De Jeu, M., and Pennartz, C. (2002). Circadian modulation of GABA function in the rat suprachiasmatic nucleus: excitatory effects during the night phase. J. Neurophysiol. 87, 834–844. doi: 10.1152/jn.00241.2001
Do, M. T. H., and Yau, K.-W. (2010). Intrinsically Photosensitive Retinal Ganglion Cells. Physiol. Rev. 90, 1547–1581.
Fon Tacer, K., Bookout, A. L., Ding, X., Kurosu, H., John, G. B., Wang, L., et al. (2010). Research resource: comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol. Endocrinol. 24, 2050–2064. doi: 10.1210/me.2010-0142
Gaston-Massuet, C., McCabe, M. J., Scagliotti, V., Young, R. M., Carreno, G., Gregory, L. C., et al. (2016). Transcription factor 7-like 1 is involved in hypothalamo-pituitary axis development in mice and humans. Proc. Natl. Acad. Sci. U.S.A. 113, E548–E557.
Gekakis, N., Staknis, D., Nguyen, H. B., Davis, F. C., Wilsbacher, L. D., King, D. P., et al. (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569. doi: 10.1126/science.280.5369.1564
Geng, X., Speirs, C., Lagutin, O., Inbal, A., Liu, W., Solnica-Krezel, L., et al. (2008). Haploinsufficiency of Six3 Fails to activate sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev. Cell 15, 236–247. doi: 10.1016/j.devcel.2008.07.003
Gillespie, C. F., Mintz, E. M., Marvel, C. L., Huhman, K. L., and Albers, H. E. (1997). GABA(A) and GABA(B) agonists and antagonists alter the phase-shifting effects of light when microinjected into the suprachiasmatic region. Brain Res. 759, 181–189. doi: 10.1016/s0006-8993(97)00235-7
Gillespie, C. F., Van Der Beek, E. M., Mintz, E. M., Mickley, N. C., Jasnow, A. M., Huhman, K. L., et al. (1999). GABAergic regulation of light-induced c-Fos immunoreactivity within the suprachiasmatic nucleus. J. Comp. Neurol. 411, 683–692. doi: 10.1002/(sici)1096-9861(19990906)411:4<683::aid-cne12>3.0.co;2-j
Greenwood, M., Bordieri, L., Greenwood, M. P., Rosso Melo, M., Colombari, D. S. A., Colombari, E., et al. (2014). Transcription factor CREB3L1 regulates vasopressin gene expression in the rat hypothalamus. J. Neurosci. 34, 3810–3820. doi: 10.1523/jneurosci.4343-13.2014
Gribkoff, V. K., Pieschl, R. L., and Dudek, F. E. (2003). GABA receptor-mediated inhibition of neuronal activity in rat SCN in vitro: Pharmacology and influence of circadian phase. J. Neurophysiol. 90, 1438–1448. doi: 10.1152/jn.01082.2002
Guido, M. E., De Guido, L., Goguen, D., Robertson, H. A., and Rusak, B. (1999). Differential effects of glutamatergic blockade on circadian and photic regulation of gene expression in the hamster suprachiasmatic nucleus. Mol. Brain Res. 67, 247–257. doi: 10.1016/s0169-328x(99)00074-1
Guillaumond, F., Dardente, H., Giguère, V., and Cermakian, N. (2005). Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403. doi: 10.1177/0748730405277232
Güldner, F. H. (1976). Synaptology of the rat suprachiasmatic nucleus. Cell Tissue Res. 165, 509–544.
Hannibal, J., Brabet, P., and Fahrenkrug, J. (2008). Mice lacking the PACAP type I receptor have impaired photic entrainment and negative masking. Am. J. Physiol. Integr. Comp. Physiol. 295, R2050–R2058.
Harmar, A. J., Marston, H. M., Shen, S., Spratt, C., West, K. M., Sheward, W. J., et al. (2002). The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508. doi: 10.1016/s0092-8674(02)00736-5
Hatori, M., Gill, S., Mure, L. S., Goulding, M., O’leary, D. D. M., and Panda, S. (2014). Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. Elife 3, 1–16.
Hee, J. C., Lee, C. J., Schroeder, A., Yoon, S. K., Seung, H. J., Jeong, S. K., et al. (2008). Excitatory actions of GABA in the suprachiasmatic nucleus. J. Neurosci. 28, 5450–5459. doi: 10.1523/jneurosci.5750-07.2008
Herzog, E. D., Geusz, M. E., Khalsa, S. B. S., Straume, M., and Block, G. D. (1997). Circadian rhythms in mouse suprachiasmatic nucleus explants on multimicroelectrode plates. Brain Res. 757, 285–290. doi: 10.1016/s0006-8993(97)00337-5
Herzog, E. D., Grace, M. S., Harrer, C., Williamson, J., Shinohara, K., and Block, G. D. (2000). The role of Clock in the developmental expression of neuropeptides in the suprachiasmatic nucleus. J. Comp. Neurol. 424, 86–98. doi: 10.1002/1096-9861(20000814)424:1<86::aid-cne7>3.0.co;2-w
Hoefflin, S., and Carter, D. A. (2014). Neuronal expression of SOX2 is enriched in specific hypothalamic cell groups. J. Chem. Neuroanat. 61, 153–160. doi: 10.1016/j.jchemneu.2014.09.003
Huang, J., Lu, C., Chen, S., Hua, L., and Qian, R. (2010). Postnatal ontogenesis of clock genes in mouse suprachiasmatic nucleus and heart. Lipids Health Dis. 9, 2–7. doi: 10.1007/3-540-27789-7_2
Hughes, A. T. L., Croft, C. L., Samuels, R. E., Myung, J., Takumi, T., and Piggins, H. D. (2015). Constant light enhances synchrony among circadian clock cells and promotes behavioral rhythms in VPAC2-signaling deficient mice. Sci. Rep. 5:14044.
Hughes, A. T. L., Guilding, C., Lennox, L., Samuels, R. E., McMahon, D. G., and Piggins, H. D. (2008). Live imaging of altered period1 expression in the suprachiasmatic nuclei of Vipr2-/- mice. J. Neurochem. 106, 1646–1657. doi: 10.1111/j.1471-4159.2008.05520.x
Hyodo, S., Yamada, C., Takezawa, T., and Urano, A. (1992). Expression of provasopressin gene during ontogeny in the hypothalamus of developing mice. Neuroscience 46, 241–250. doi: 10.1016/0306-4522(92)90024-v
Jeong, Y., Leskow, F. C., El-Jaick, K., Roessler, E., Muenke, M., Yocum, A., et al. (2008). Regulation of a remote Shh forebrain enhancer by the Six3 homeoprotein. Nat. Genet. 40, 1348–1353. doi: 10.1038/ng.230
Jiang, Z. G., Yang, Y. Q., and Allen, C. N. (1997a). Tracer and electrical coupling of rat suprachiasmatic nucleus neurons. Neuroscience 77, 1059–1066. doi: 10.1016/s0306-4522(96)00539-8
Jiang, Z. G., Yang, Y. Q., Liu, Z. P., and Allen, C. N. (1997b). Membrane properties and synaptic inputs of suprachiasmatic nucleus neurons in rat brain slices. J. Physiol. 499, 141–159. doi: 10.1113/jphysiol.1997.sp021917
Jin, X., Shearman, L. P., Weaver, D. R., Zylka, M. J., de Vries, G. J., and Reppert, S. M. (1999). A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57–68. doi: 10.1016/s0092-8674(00)80959-9
Kabrita, C. S., and Davis, F. C. (2008). Development of the mouse suprachiasmatic nucleus: Determination of time of cell origin and spatial arrangements within the nucleus. Brain Res. 1195, 20–27. doi: 10.1016/j.brainres.2007.12.020
Karatsoreos, I. N., Romeo, R. D., McEwen, B. S., and Silver, R. (2006). Diurnal regulation of the gastrin-releasing peptide receptor in the mouse circadian clock. Eur. J. Neurosci. 23, 1047–1053. doi: 10.1111/j.1460-9568.2006.04633.x
Kawaguchi, C., Tanaka, K., Isojima, Y., Shintani, N., Hashimoto, H., Baba, A., et al. (2003). Changes in light-induced phase shift of circadian rhythm in mice lacking PACAP. Biochem. Biophys. Res. Commun. 310, 169–175. doi: 10.1016/j.bbrc.2003.09.004
Khaper, N., Bailey, C. D. C., Ghugre, N. R., Reitz, C., Awosanmi, Z., Waines, R., et al. (2018). Implications of disturbances in circadian rhythms for cardiovascular health: A new frontier in free radical biology. Free Radic. Biol. Med. 119, 85–92. doi: 10.1016/j.freeradbiomed.2017.11.006
Krout, K., Kawano, J., Mettenleiter, T., and Loewy, A. (2002). CNS inputs to the suprachiasmatic nucleus of the rat. Neuroscience 110, 73–92. doi: 10.1016/s0306-4522(01)00551-6
Kume, K., Zylka, M. J., Sriram, S., Shearman, L. P., Weaver, D. R., Jin, X., et al. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. doi: 10.1016/s0092-8674(00)81014-4
Kurose, T., Yabe, D., and Inagaki, N. (2011). Circadian rhythms and diabetes. J. Diabetes Investig. 2, 176–177.
Laemle, L. K., Repke, K. B., Hawkes, R., and Rice, F. L. (1991). Synaptogenesis in the rat suprachiasmatic nucleus: a light microscopic immunocytochemical survey. Brain Res. 544, 108–117. doi: 10.1016/0006-8993(91)90891-x
Lagutin, O. V., Zhu, C. C., Kobayashi, D., Topczewski, J., Shimamura, K., Puelles, L., et al. (2003). Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 17, 368–379. doi: 10.1101/gad.1059403
Landgraf, D., Achten, C., Dallmann, F., and Oster, H. (2015). Embryonic development and maternal regulation of murine circadian clock function. Chronobiol. Int. 32, 416–427. doi: 10.3109/07420528.2014.986576
Lee, I. T., Chang, A. S., Manandhar, M., Shan, Y., Fan, J., Izumo, M., et al. (2015). Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85, 1086–1102. doi: 10.1016/j.neuron.2015.02.006
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Lenn, N. J., Beebe, B., and Moore, R. Y. (1977). Postnatal development of the suprachiasmatic hypothalamic nucleus of the rat. Cell Tissue Res. 178, 463–475.
Li, J.-D., Hu, W.-P., Boehmer, L., Cheng, M. Y., Lee, A. G., Jilek, A., et al. (2006). Attenuated Circadian rhythms in mice lacking the prokineticin 2 Gene. J. Neurosci. 26, 11615–11623. doi: 10.1523/jneurosci.3679-06.2006
Lim, M. M., Gerstner, J. R., and Holtzman, D. M. (2014). The sleep-wake cycle and Alzheimer’s disease: what do we know? Neurodegener. Dis. Manag. 4, 351–362. doi: 10.2217/nmt.14.33
Liu, C., and Reppert, S. M. (2000). GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron 25, 123–128. doi: 10.1016/s0896-6273(00)80876-4
Liu, C., Weaver, D. R., Strogatz, S. H., and Reppert, S. M. (1997). Cellular construction of a circadian clock: Period determination in the suprachiasmatic nuclei. Cell 91, 855–860. doi: 10.1016/s0092-8674(00)80473-0
Long, M. A., Jutras, M. J., Connors, B. W., and Burwell, R. D. (2005). Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat. Neurosci. 8, 61–66. doi: 10.1038/nn1361
Lu, F., Kar, D., Gruenig, N., Zhang, Z. W., Cousins, N., Rodgers, H. M., et al. (2013). Rax is a selector gene for mediobasal hypothalamic cell types. J. Neurosci. 33, 259–272. doi: 10.1523/jneurosci.0913-12.2013
Lu, J., Zhang, Y. H., Chou, T. C., Gaus, S. E., Elmquist, J. K., Shiromani, R., et al. (2001). Contrasting effects of ibotenate lesions of the paraventricular nucleus and subparaventricular zone on sleep-wake cycle and temperature regulation. J. Neurosci. 21, 4864–4874. doi: 10.1523/jneurosci.21-13-04864.2001
Manning, L., Ohyama, K., Saeger, B., Hatano, O., Wilson, S. A., Logan, M., et al. (2006). Regional morphogenesis in the hypothalamus: a BMP-Tbx2 pathway coordinates fate and proliferation through Shh downregulation. Dev. Cell 11, 873–885. doi: 10.1016/j.devcel.2006.09.021
Markakis, E. A., and Swanson, L. W. (1997). Spatiotemporal patterns of secretomotor neuron generation in the parvicellular neuroendocrine system. Brain Res. Rev. 24, 255–291. doi: 10.1016/s0165-0173(97)00006-4
Martinez-Ferre, A., Navarro-Garberi, M., Bueno, C., and Martinez, S. (2013). Wnt signal specifies the intrathalamic limit and its organizer properties by regulating Shh induction in the alar plate. J. Neurosci. 33, 3967–3980. doi: 10.1523/jneurosci.0726-12.2013
Maywood, E. S., Chesham, J. E., O’Brien, J. A., and Hastings, M. H. (2011). A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. U.S.A. 108, 14306–14311. doi: 10.1073/pnas.1101767108
McArthur, A. J., Coogan, A. N., Ajpru, S., Sugden, D., Biello, S. M., and Piggins, H. D. (2000). Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro. J. Neurosci. 20, 5496–5502. doi: 10.1523/jneurosci.20-14-05496.2000
McNeill, D. S., Sheely, C. J., Ecker, J. L., Badea, T. C., Morhardt, D., Guido, W., et al. (2011). Development of melanopsin-based irradiance detecting circuitry. Neural Dev. 6:8. doi: 10.1186/1749-8104-6-8
McNeill, J. K., Walton, J. C., and Albers, H. E. (2018). Functional significance of the excitatory effects of GABA in the suprachiasmatic nucleus. J. Biol. Rhythms 33, 376–387. doi: 10.1177/0748730418782820
Melo, M. C. A., Garcia, R. F., Linhares Neto, V. B., Sá, M. B., de Mesquita, L. M. F., de Araújo, C. F. C., et al. (2016). Sleep and circadian alterations in people at risk for bipolar disorder: a systematic review. J. Psychiatr. Res. 83, 211–219. doi: 10.1016/j.jpsychires.2016.09.005
Meyer-Bernstein, E. L., and Morin, L. P. (1996). Differential serotonergic innervation of the suprachiasmatic nucleus and the intergeniculate leaflet and its role in circadian rhythm modulation. J. Neurosci. 16, 2097–2111. doi: 10.1523/jneurosci.16-06-02097.1996
Meyer-Spasche, A., and Piggins, H. D. (2004). Vasoactive intestinal polypeptide phase-advances the rat suprachiasmatic nuclei circadian pacemaker in vitro via protein kinase A and mitogen-activated protein kinase. Neurosci. Lett. 358, 91–94. doi: 10.1016/j.neulet.2003.12.114
Mieda, M., Okamoto, H., and Sakurai, T. (2016). Manipulating the Cellular circadian period of arginine vasopressin neurons alters the behavioral circadian period. Curr. Biol. 26, 2535–2542. doi: 10.1016/j.cub.2016.07.022
Mieda, M., Ono, D., Hasegawa, E., Okamoto, H., Honma, K.-I., Honma, S., et al. (2015). Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85, 1103–1116. doi: 10.1016/j.neuron.2015.02.005
Migliarini, S., Pacini, G., Pelosi, B., Lunardi, G., and Pasqualetti, M. (2013). Lack of brain serotonin affects postnatal development and serotonergic neuronal circuitry formation. Mol. Psychiatry 18, 1106–1118. doi: 10.1038/mp.2012.128
Mikels, A. J., and Nusse, R. (2006). Wnts as ligands: Processing, secretion and reception. Oncogene 25, 7461–7468. doi: 10.1038/sj.onc.1210053
Miller, A. V., Kavanaugh, S. I., and Tsai, P. S. (2016). Disruption of the suprachiasmatic nucleus in fibroblast growth factor signaling-deficient mice. Front. Endocrinol. (Lausanne) 7:11. doi: 10.3389/fendo.2016.00011
Mintz, E. M., Gillespie, C. F., Marvel, C. L., Huhman, K. L., and Albers, H. E. (1997). Serotonergic regulation of circadian rhythms in Syrian hamsters. Neuroscience 79, 563–569. doi: 10.1016/s0306-4522(96)00696-3
Moore, R. Y., and Bernstein, M. E. (1989). Synaptogenesis in the rat suprachiasmatic nucleus demoinstrated by electron microscopy and synapsin I immunoreactivity. J. Neurosci. 9, 2151–2162. doi: 10.1523/jneurosci.09-06-02151.1989
Moore, R. Y., and Speh, J. C. (1993). GABA is the principal neurotransmitter of the circadian system. Neurosci. Lett. 150, 112–116. doi: 10.1016/0304-3940(93)90120-a
Moore, R. Y., Speh, J. C., and Leak, R. K. (2002). Suprachiasmatic nucleus organization. Cell Tissue Res. 309, 89–98.
Morin, L. P., and Allen, C. N. (2006). The circadian visual system, 2005. Brain Res. Rev. 51, 1–60. doi: 10.1016/j.brainresrev.2005.08.003
Morin, L. P., Hefton, S., and Studholme, K. M. (2011). Neurons identified by NeuN/Fox-3 immunoreactivity have a novel distribution in the hamster and mouse suprachiasmatic nucleus. Brain Res. 1421, 44–51.
Morin, L. P., Shivers, K. Y., Blanchard, J. H., and Muscat, L. (2006). Complex organization of mouse and rat suprachiasmatic nucleus. Neuroscience 137, 1285–1297. doi: 10.1016/j.neuroscience.2005.10.030
Mosley, M., Shah, C., Morse, K. A., Miloro, S. A., Holmes, M. M., Ahern, T. H., et al. (2017). Patterns of cell death in the perinatal mouse forebrain. J. Comp. Neurol. 525, 47–64. doi: 10.1002/cne.24041
Motoyama, J., Milenkovic, L., Iwama, M., Shikata, Y., Scott, M. P., and Hui, C. C. (2003). Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev. Biol. 259, 150–161. doi: 10.1016/s0012-1606(03)00159-3
Munekawa, K., Tamada, Y., Iijima, N., Hayashi, S., Ishihara, A., Inoue, K., et al. (2000). Development of astroglial elements in the suprachiasmatic nucleus of the rat: with special reference to the involvement of the optic nerve. Exp. Neurol. 166, 44–51. doi: 10.1006/exnr.2000.7490
Musiek, E. S., Bhimasani, M., Zangrilli, M. A., Morris, J. C., Holtzman, D. M., and Ju, Y.-E. S. (2018). Circadian Rest-activity pattern changes in aging and preclinical Alzheimer disease. JAMA Neurol. 75:582. doi: 10.1001/jamaneurol.2017.4719
Nagano, M., Adachi, A., Nakahama, K., Nakamura, T., Tamada, M., Meyer-Bernstein, E., et al. (2003). An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J. Neurosci. 23, 6141–6151. doi: 10.1523/jneurosci.23-14-06141.2003
Nakamura, W., Honma, S., Shirakawa, T., and Honma, K. (2001). Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 14, 666–674. doi: 10.1046/j.0953-816x.2001.01684.x
Nakamura, W., Yamazaki, S., Takasu, N. N., Mishima, K., and Block, G. D. (2005). Differential response of Period 1 expression within the suprachiasmatic nucleus. J. Neurosci. 25, 5481–5487. doi: 10.1523/jneurosci.0889-05.2005
Newman, E. A., Kim, D. W., Wan, J., Wang, J., Qian, J., and Blackshaw, S. (2018a). Foxd1 is required for terminal differentiation of anterior hypothalamic neuronal subtypes. Dev. Biol. 439, 102–111. doi: 10.1016/j.ydbio.2018.04.012
Newman, E. A., Wu, D., Taketo, M. M., Zhang, J., and Blackshaw, S. (2018b). Canonical Wnt signaling regulates patterning, differentiation and nucleogenesis in mouse hypothalamus and prethalamus. Dev. Biol. 442, 236–248. doi: 10.1016/j.ydbio.2018.07.021
Nielsen, H. S., Hannibal, J., and Fahrenkrug, J. (2002). Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur. J. Neurosci. 15, 570–574. doi: 10.1046/j.0953-816x.2001.01882.x
Ohta, H., Yamazaki, S., and McMahon, D. G. (2005). Constant light desynchronizes mammalian clock neurons. Nat. Neurosci. 8, 267–269. doi: 10.1038/nn1395
Orquera, D. P., Nasif, S., Low, M. J., Rubinstein, M., and de Souza, F. S. J. (2016). Essential function of the transcription factor Rax in the early patterning of the mammalian hypothalamus. Dev. Biol. 416, 212–224. doi: 10.1016/j.ydbio.2016.05.021
Padilla, S. L., Carmody, J. S., and Zeltser, L. M. (2010). Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat. Med. 16, 403–405. doi: 10.1038/nm.2126
Pak, T., Yoo, S., Miranda-Angulo, A. M., Wang, H., and Blackshaw, S. (2014). Rax-CreERT2knock-in mice: a tool for selective and conditional gene deletion in progenitor cells and radial glia of the retina and hypothalamus. PLoS One 9:e90381. doi: 10.1371/journal.pone.0090381
Parsons, M. J., Brancaccio, M., Sethi, S., Maywood, E. S., Satija, R., Edwards, J. K., et al. (2015). The regulatory factor ZFHX3 Modifies circadian function in SCN via an AT motif-driven axis. Cell 162, 607–621. doi: 10.1016/j.cell.2015.06.060
Peukert, D., Weber, S., Lumsden, A., and Scholpp, S. (2011). Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 9:e1001218. doi: 10.1371/journal.pbio.1001218
Pickard, G. E., Ralph, M. R., and Menaker, M. (1987). The Intergeniculate leaflet partially mediates effects of light on circadian rhythms. J. Biol. Rhythms 2, 35–56. doi: 10.1177/074873048700200104
Piggins, H., Antle, M., and Rusak, B. (1995). Neuropeptides phase shift the mammalian circadian pacemaker. J. Neurosci. 15, 5612–5622. doi: 10.1523/jneurosci.15-08-05612.1995
Pla, R., Stanco, A., Howard, M. A., Rubin, A. N., Vogt, D., Mortimer, N., et al. (2018). Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb. Cortex 28, 3797–3815. doi: 10.1093/cercor/bhx241
Preitner, N., Damiola, F., Luis-Lopez-Molina Zakany, J., Duboule, D., Albrecht, U., and Schibler, U. (2002). The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. doi: 10.1016/s0092-8674(02)00825-5
Prolo, L. M., Takahashi, J. S., and Herzog, E. D. (2005). Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 25, 404–408. doi: 10.1523/jneurosci.4133-04.2005
Prosser, H. M., Bradley, A., Chesham, J. E., Ebling, F. J. P., Hastings, M. H., and Maywood, E. S. (2007). Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc. Natl. Acad. Sci. U.S.A. 104, 648–653. doi: 10.1073/pnas.0606884104
Ralph, M. R., and Menaker, M. (1985). Bicuculline blocks circadian phase delays but not advances. Brain Res. 325, 362–365. doi: 10.1016/0006-8993(85)90341-5
Ralph, M. R., and Menaker, M. (1986). Effects of diazepam on circadian phase advances and delays. Brain Res. 372, 405–408. doi: 10.1016/0006-8993(86)91154-6
Ralph, M. R., and Menaker, M. (1989). GABA regulation of circadian responses to light. I. Involvement of GABA(A)-benzodiazepine and GABA(B) receptors. J. Neurosci. 9, 2858–2865. doi: 10.1523/jneurosci.09-08-02858.1989
Rash, J. E., Olson, C. O., Pouliot, W. A., Davidson, K. G. V., Yasumura, T., Furman, C. S., et al. (2007). Connexin36 vs. connexin32, “miniature” neuronal gap junctions, and limited electrotonic coupling in rodent suprachiasmatic nucleus. Neuroscience 149, 350–371. doi: 10.1016/j.neuroscience.2007.06.052
Reed, H. E., Meyer-Spasche, A., Cutler, D. J., Coen, C. W., and Piggins, H. D. (2001). Vasoactive intestinal polypeptide (VIP) phase-shifts the rat suprachiasmatic nucleus clock in vitro. Eur. J. Neurosci. 13, 839–843. doi: 10.1046/j.0953-816x.2000.01437.x
Reppert, S. M., and Schwartz, W. J. (1984). The suprachiasmatic characterization nuclei of the fetal rat: of a functional circadian. J. Neurosci. 4, 1677–1682. doi: 10.1523/jneurosci.04-07-01677.1984
Roenneberg, T., and Merrow, M. (2016). The circadian clock and human health. Curr. Biol. 26, R432–R443.
Roy, A., de Melo, J., Chaturvedi, D., Thein, T., Cabrera-Socorro, A., Houart, C., et al. (2013). LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J. Neurosci. 33, 6877–6884. doi: 10.1523/jneurosci.4216-12.2013
Saaltink, D. J., Håvik, B., Verissimo, C. S., Lucassen, P. J., and Vreugdenhil, E. (2012). Doublecortin and doublecortin-like are expressed in overlapping and non-overlapping neuronal cell population: implications for neurogenesis. J. Comp. Neurol. 520, 2805–2823. doi: 10.1002/cne.23144
Sato, M., Mizoro, Y., Atobe, Y., Fujimoto, Y., Yamaguchi, Y., Fustin, J. M., et al. (2011). Transportin 1 in the mouse brain: appearance in regions of neurogenesis, cerebrospinal fluid production/sensing, and circadian clock. J. Comp. Neurol. 519, 1770–1780. doi: 10.1002/cne.22600
Sato, T. K., Panda, S., Miraglia, L. J., Reyes, T. M., Rudic, R. D., McNamara, P., et al. (2004). A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron 43, 527–537. doi: 10.1016/j.neuron.2004.07.018
Shearman, L. P., Sriram, S., Weaver, D. R., Maywood, E. S., Chaves, I., Zheng, B., et al. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019. doi: 10.1126/science.288.5468.1013
Shearman, L. P., Zylka, M. J., Weaver, D. R., Kolakowski, L. F., and Reppert, S. M. (1997). Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269. doi: 10.1016/s0896-6273(00)80417-1
Shimada, M., and Nakamura, T. (1973). Time of neuron origin in mouse hypothalamic nuclei. Exp. Neurol. 41, 163–173. doi: 10.1016/0014-4886(73)90187-8
Shimamura, K., and Rubenstein, J. L. R. (1997). Inductive interactions direct early regionalization of the mouse forebrain. Development 124, 2709–2718.
Shimogori, T., Lee, D. A., Miranda-Angulo, A., Yang, Y., Wang, H., Jiang, L., et al. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775. doi: 10.1038/nn.2545
Shimomura, H., Moriya, T., Sudo, M., Wakamatsu, H., Akiyama, M., Miyake, Y., et al. (2001). Differential daily expression of Per1 and Per2 mRNA in the suprachiasmatic nucleus of fetal and early postnatal mice. Eur. J. Neurosci. 13, 687–693. doi: 10.1046/j.0953-816x.2000.01438.x
Shinohara, K., Funabashi, T., Mitushima, D., and Kimura, F. (2000). Effects of gap junction blocker on vasopressin and vasoactive intestinal polypeptide rhythms in the rat suprachiasmatic nucleus in vitro. Neurosci. Res. 38, 43–47. doi: 10.1016/s0168-0102(00)00141-3
Smyllie, N. J., Chesham, J. E., Hamnett, R., Maywood, E. S., and Hastings, M. H. (2016). Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 113, 3657–3662. doi: 10.1073/pnas.1511351113
Sominsky, L., Dangel, T., Malik, S., De Luca, S. N., Singewald, N., and Spencer, S. J. (2021). Microglial ablation in rats disrupts the circadian system. FASEB J. 35, 1–10.
Speh, J. C., and Moore, R. Y. (1993). Retinohypothalamic tract development in the hamster and rat. Dev. Brain Res. 76, 171–181. doi: 10.1016/0165-3806(93)90205-o
Strecker, G. J., Wuarin, J. P., and Dudek, F. E. (1997). GABAA-mediated local synaptic pathways connect neurons in the rat suprachiasmatic nucleus. J. Neurophysiol. 78, 2217–2220. doi: 10.1152/jn.1997.78.4.2217
Szabó, N. E., Zhao, T., Çankaya, M., Theil, T., Zhou, X., and Alvarez-Bolado, G. (2009a). Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J. Neurosci. 29, 6989–7002. doi: 10.1523/jneurosci.1089-09.2009
Szabó, N. E., Zhao, T., Zhou, X., and Alvarez-Bolado, G. (2009b). The Role of sonic hedgehog of neural origin in thalamic differentiation in the mouse. J. Neurosci. 29, 2453–2466. doi: 10.1523/jneurosci.4524-08.2009
Takatsuji, K., Oyamada, H., and Tohyama, M. (1995). Postnatal development of the substance P-, neuropeptide Y- and serotonin-containing fibers in the rat suprachiasmatic nucleus in relation to development of the retino-hypothalamic projection. Dev. Brain Res. 84, 261–270. doi: 10.1016/0165-3806(94)00209-i
Todd, W. D., Venner, A., Anaclet, C., Broadhurst, R. Y., De Luca, R., Bandaru, S. S., et al. (2020). Suprachiasmatic VIP neurons are required for normal circadian rhythmicity and comprised of molecularly distinct subpopulations. Nat. Commun. 11:4410.
Trowe, M.-O., Zhao, L., Weiss, A.-C., Christoffels, V., Epstein, D. J., and Kispert, A. (2013). Inhibition of Sox2-dependent activation of Shh in the ventral diencephalon by Tbx3 is required for formation of the neurohypophysis. Development 140, 2299–2309. doi: 10.1242/dev.094524
Tsai, P.-S., Brooks, L. R., Rochester, J. R., Kavanaugh, S. I., and Chung, W. C. J. (2011). Fibroblast growth factor signaling in the developing neuroendocrine hypothalamus. Front. Neuroendocrinol. 32:95–107. doi: 10.1016/j.yfrne.2010.11.002
Tso, C. F., Simon, T., Greenlaw, A. C., Puri, T., Mieda, M., and Herzog, E. D. (2017). Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr. Biol. 27, 1055–1061. doi: 10.1016/j.cub.2017.02.037
Uth, K., and Sleigh, R. (2014). Deregulation of the circadian clock constitutes a significant factor in tumorigenesis: a clockwork cancer. Part II. In vivo studies. Biotechnol. Biotechnol. Equip. 28, 379–386. doi: 10.1080/13102818.2014.925298
van den Pol, A. N. (1980). The hypothalamic suprachiasmatic nucleus of rat: Intrinsic anatomy. J. Comp. Neurol. 191, 661–702. doi: 10.1002/cne.901910410
VanDunk, C., Hunter, L. A., and Gray, P. A. (2011). Development, Maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J. Neurosci. 31, 6457–6467. doi: 10.1523/jneurosci.5385-10.2011
Vitaterna, M. H., Ko, C. H., Chang, A. M., Buhr, E. D., Fruechte, E. M., Schook, A., et al. (2006). The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc. Natl. Acad. Sci. U.S.A. 103, 9327–9332. doi: 10.1073/pnas.0603601103
Vitaterna, M. H., Selby, C. P., Todo, T., Niwa, H., Thompson, C., Fruechte, E. M., et al. (1999). Differential regulation of mammalian Period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 96, 12114–12119. doi: 10.1073/pnas.96.21.12114
Von Gall, C., Noton, E., Lee, C., and Weaver, D. R. (2003). Light does not degrade the constitutively expressed BMAL1 protein in the mouse suprachiasmatic nucleus. Eur. J. Neurosci. 18, 125–133. doi: 10.1046/j.1460-9568.2003.02735.x
Wanaka, A., Johnson, E. M., and Milbrand, J. (1990). Localization of FGF receptor mRNA in the adult rat central nervous system by in situ hybridization. Neuron 5, 267–281. doi: 10.1016/0896-6273(90)90164-b
Wang, M. H., Chen, N., and Wang, J. H. (2014). The coupling features of electrical synapses modulate neuronal synchrony in hypothalamic superachiasmatic nucleus. Brain Res. 1550, 9–17. doi: 10.1016/j.brainres.2014.01.007
Watanabe, K., Vanecek, J., and Yamaoka, S. (2000). In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res. 877, 361–366. doi: 10.1016/s0006-8993(00)02724-4
Watts, A. G., and Swanson, L. W. (1987). Efferent projections of the suprachiasmatic nucleus: II. Studies using retrograde transport of fluorescent dyes and simultaneous peptide immunohistochemistry in the rat. J. Comp. Neurol. 258, 230–252. doi: 10.1002/cne.902580205
Welsh, D. K., Logothetis, D. E., Meister, M., and Reppert, S. M. (1995). Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706. doi: 10.1016/0896-6273(95)90214-7
Wen, S., Ma, D., Zhao, M., Xie, L., Wu, Q., Gou, L., et al. (2020). Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat. Neurosci. 23, 456–467. doi: 10.1038/s41593-020-0586-x
Wilcox, A. G., Vizor, L., Parsons, M. J., Banks, G., and Nolan, P. M. (2017). Inducible knockout of mouse Zfhx3 emphasizes its key role in setting the pace and amplitude of the adult circadian clock. J. Biol. Rhythms 32, 433–443. doi: 10.1177/0748730417722631
Wreschnig, D., Dolatshad, H., and Davis, F. C. (2014). Embryonic development of circadian oscillations in the mouse hypothalamus. J. Biol. Rhythms 29, 299–310. doi: 10.1177/0748730414545086
Xie, Y., and Dorsky, R. I. (2017). Development of the hypothalamus: conservation, modification and innovation. Development 144, 1588–1599. doi: 10.1242/dev.139055
Yamaguchi, S., Isejima, H., Matsuo, T., Okura, R., Yagita, K., Kobayashi, M., et al. (2003). Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412. doi: 10.1126/science.1089287
Yamaguchi, Y., Suzuki, T., Mizoro, Y., Kori, H., Okada, K., Chen, Y., et al. (2013). Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342, 85–90. doi: 10.1126/science.1238599
Ye, R., Selby, C. P., Chiou, Y., Ozkan-Dagliyan, I., Gaddameedhi, S., and Sancar, A. (2014). Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 28, 1989–1998. doi: 10.1101/gad.249417.114
Zhao, L., Zevallos, S. E., Rizzoti, K., Jeong, Y., Lovell-Badge, R., and Epstein, D. J. (2012). Disruption of SoxB1-dependent sonic hedgehog expression in the hypothalamus causes septo-optic dysplasia. Dev. Cell 22, 585–596. doi: 10.1016/j.devcel.2011.12.023
Zheng, B., Albrecht, U., Kaasik, K., Sage, M., Lu, W., Vaishnav, S., et al. (2001). Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694. doi: 10.1016/s0092-8674(01)00380-4
Keywords: suprachiasmatic nucleus, neurogenesis, development, neuronal differentiation, transcription factors, neuropeptides
Citation: Cheng AH and Cheng H-YM (2021) Genesis of the Master Circadian Pacemaker in Mice. Front. Neurosci. 15:659974. doi: 10.3389/fnins.2021.659974
Received: 28 January 2021; Accepted: 04 March 2021;
Published: 23 March 2021.
Edited by:
Daisuke Ono, Nagoya University, JapanReviewed by:
Michihiro Mieda, Kanazawa University, JapanCharles N. Allen, Oregon Health and Science University, United States
Copyright © 2021 Cheng and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arthur H. Cheng, YWhoLmNoZW5nQG1haWwudXRvcm9udG8uY2E=; Hai-Ying Mary Cheng, aGFpeWluZy5jaGVuZ0B1dG9yb250by5jYQ==
 Arthur H. Cheng
Arthur H. Cheng Hai-Ying Mary Cheng
Hai-Ying Mary Cheng