- 1Department of Basic Medicine Sciences, and Department of Orthopaedics of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Neurosurgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Department of Basic Medicine Sciences, and Department of Obstetrics of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
The central nervous system (CNS) post-traumatic injury can cause severe nerve damage with devastating consequences. However, its pathophysiological mechanisms remain vague. There is still an urgent need for more effective treatments. Circular RNAs (circRNAs) are non-coding RNAs that can form covalently closed RNA circles. Through second-generation sequencing technology, microarray analysis, bioinformatics, and other technologies, recent studies have shown that a number of circRNAs are differentially expressed after traumatic brain injury (TBI) or spinal cord injury (SCI). These circRNAs play important roles in the proliferation, inflammation, and apoptosis in CNS post-traumatic injury. In this review, we summarize the expression and functions of circRNAs in CNS in recent studies, as well as the circRNA–miRNA–mRNA interaction networks. The potential clinical value of circRNAs as a therapeutic target is also discussed.
Introduction
Central nervous system (CNS) post-traumatic injury often leads to persistent neurological damage and devastating consequences, but there is still a lack of effective methods for its treatment (Leng et al., 2019; Tsujioka and Yamashita, 2019). It includes traumatic brain injury (TBI) and spinal cord injury (SCI) and is one of the world's major disease burdens (Warnock et al., 2020). TBI is complex and severe, resulting in neuroinflammation and neurodegeneration (Kempuraj et al., 2020; Zheng et al., 2020). It is a leading cause of death and disability (Yu et al., 2017; Radabaugh et al., 2020). Similarly, SCI is a destructive and traumatic injury that involves primary and secondary injury (Wang and Song, 2019; Ma et al., 2020). It can lead to loss of sensory, motor, and autonomic nerve function (Chen et al., 2019a; Reis et al., 2020; Wang et al., 2020c). The regeneration capacity of the CNS is very limited (Ma et al., 2019; Radabaugh et al., 2020). Recently, more and more studies have explored the expression patterns and functional characteristics of microRNA (miRNA) and lncRNA in CNS post-traumatic injury. It has been found that they are involved in the process of post-traumatic inflammation, apoptosis, and autophagy of the CNS through epigenetic regulation (Nagalakshmi et al., 2018; Yang et al., 2018a; Ge et al., 2019; Li et al., 2019; Yi et al., 2019; Wang et al., 2020b). However, the endogenous mechanisms that mediate CNS post-traumatic injury are still obscure.
Circular RNAs (circRNAs) are one of the noncoding RNAs. They have no 5′-3′ polarity, 5′ capping, and 3′ polyadenylation. Noncanonical alternative splicing connects the splicing donor to the upstream splicing recipient, forming a covalent closed ring (Li et al., 2016). In mammalian cells, circRNAs are endogenous, abundant, conserved, and stable (Wu and Yang, 2015). Although most functions remain unknown, circRNAs have been shown to function as templates for viroid and viral replication, as intermediates in RNA processing reactions, as regulators of transcription in cis, as snoRNAs, and as miRNA sponges (Lasda and Parker, 2014; Enuka et al., 2016).
In the past few years, due to the advance in second-generation sequencing (Enuka et al., 2016), microarray analysis, and bioinformatics, more and more studies have explored the expression and functions of circRNAs in CNS diseases, such as Alzheimer's disease (Zhang et al., 2019), cerebral ischemic stroke (Han et al., 2018), brain tumor (Yang et al., 2018b), and multiple sclerosis (Cardamone et al., 2017). Therefore, researchers speculate that circRNAs may also play important roles in CNS post-traumatic injury. They designed experiments to explore this. The results showed that circRNAs differentially expressed in both TBI and SCI models, and these studies further explored the functions of circRNAs.
In this review, we discussed the role of circRNAs in TBI and SCI and compared their expression profiles. These circRNAs play important roles in the proliferation, inflammation, apoptosis, and post-traumatic injury. We also discussed the clinical significance of using circRNAs as therapeutic targets for CNS post-traumatic injury.
Circular Expression Profiling in CNS Post-Traumatic Injury
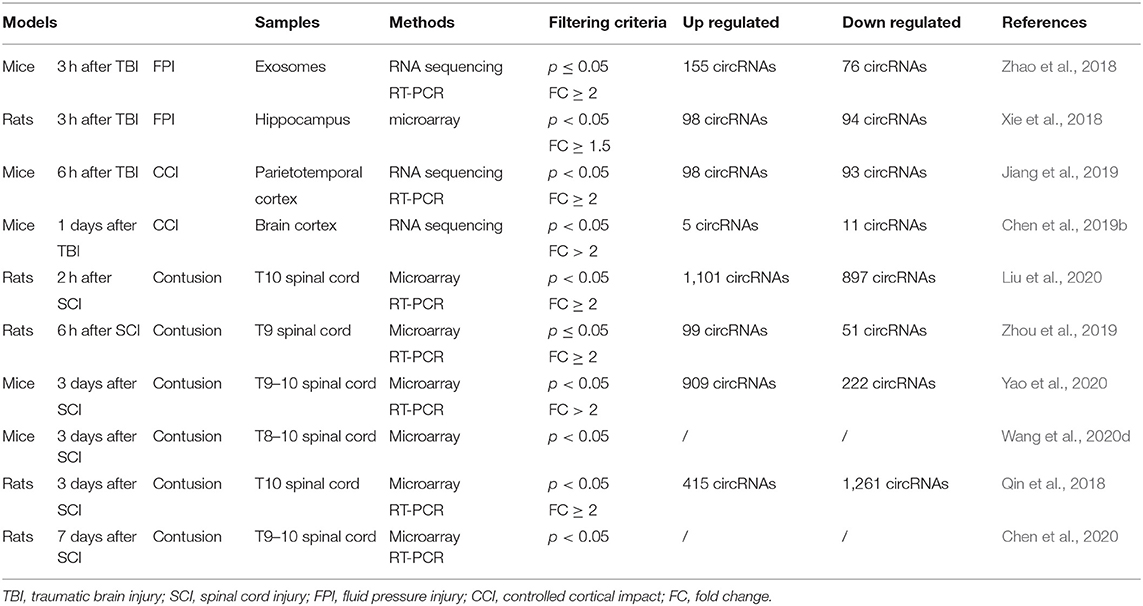
In most studies, the expression profile of circRNAs was obtained by RNA sequencing or microarray and then verified by reverse transcription-quantitative PCR (RT-qPCR). Different kinds of models are used and the results are shown in Table 1.
CircRNA Expression Profile in TBI
The fluid pressure injury (FPI) method was adopted to make the TBI model. After drilling a hole in the skull, a metal pendulum was lowered to strike the piston, causing a moderate fluid pressure injury by injecting a small amount of saline into the closed skull cavity. The two studies mentioned below took this approach (Xie et al., 2018; Zhao et al., 2018). Zhao and colleagues extracted mice exosomes from the brain extracellular space 3 h after TBI. Through high-throughput sequencing, they found 231 circRNAs with significantly differentially expressed expressions, of which 155 expressions were upregulated and 76 expressions were downregulated (Zhao et al., 2018). In another study, Xie et al. also explored what happened 3 h after TBI using rat hippocampus. Through circRNA microarray, they found 192 circRNAs with significantly differentially expressed expressions, of which 98 expressions were upregulated and 94 expressions were downregulated (Xie et al., 2018). Bioinformatics analysis including GO terms and KEGG pathway showed that circRNAs extracted from exosomes were involved in “dendrite development,” “cytoplasmic,” “protein binding,” and “glutamatergic synapse,” while in the hippocampus, they participated in the “generation of neurons,” “synapse,” “small GTPase binding,” and “axon guidance.” These results suggest that circRNAs are actively involved in regulating the repair process of neurons, dendrites, and synapses after TBI.
In addition to FPI, the controlled cortical impact (CCI) approach has also been adopted. The electromagnetic device is set in advance for speed, strike depth, and dwell time to complete the CCI. In two experiments using CCI on the cerebral cortex, circRNAs were also differentially expressed after TBI in mice. Six hours after TBI on the left parietotemporal cortex, 98 circRNAs were upregulated and 93 circRNAs were downregulated (Jiang et al., 2019). They were involved in “histone deacetylation,” “histone methyltransferase complex,” “repressing transcription factor binding,” and “natural killer cell-mediated cytotoxicity.” One day after TBI between the bregma and lambda (Rola et al., 2006), 5 circRNAs were upregulated and 11 circRNAs were downregulated (Chen et al., 2019b), participating in “innate immune response,” “extracellular region,” “protein binding,” and “natural killer cell-mediated cytotoxicity.” However, the top 10 upregulated or downregulated circRNAs in these researches about TBI were completely different (Xie et al., 2018; Zhao et al., 2018; Jiang et al., 2019; Chen et al., 2019b). The bioinformatics analysis here suggests that circRNAs might regulate the transcription process and immune system to influence the repair process after TBI.
CircRNA Expression Profile in SCI
In the SCI model, an impactor is dropped at a certain height onto the spinal cord after a laminectomy. Contusion injury can be caused by regulating different weights and falling heights. The injured spinal cord segments were concentrated in T8–T10 in various studies (Qin et al., 2018; Zhou et al., 2019; Chen et al., 2020; Liu et al., 2020; Wang et al., 2020d; Yao et al., 2020). Two studies explored the expression of circRNAs within a short period after SCI. Liu and colleagues extracted T10 spinal cord of rats 2 h after SCI. Through microarray, they found 1,998 circRNAs with significantly differentially expressed expressions, of which 1,101 were upregulated and 897 were downregulated. According to the enrichment analysis, these differentially expressed circRNAs are involved in “intracellular signal transduction,” “synapse,” “protein binding,” and “mitogen-activated protein kinase (MAPK) signaling” (Liu et al., 2020). MAPK can facilitate neuronal degeneration after SCI and promote cell proliferation (Ye et al., 2020). Zhou et al. extracted T9 spinal cord of rats 6 h after SCI. They found 99 upregulated and 51 downregulated circRNAs. According to the enrichment analysis, the upregulated circRNAs are involved in “gene silencing by RNA,” “centromeric region,” “siRNA binding,” and “peroxisome proliferator-activated receptor (PPAR) signaling pathway.” The PPARγ pathway is related with astrocyte-derived neuroinflammation in CCI rats (Zhong et al., 2019). Activation of PPARs can reduce the proinflammatory cascade (Shafi et al., 2019). Meanwhile, the downregulated circRNAs are involved in the “regulation of synaptic vesicle exocytosis,” “synapse,” “protein kinase binding,” and “extracellular matrix-receptor interaction” (Zhou et al., 2019).
Three studies examined the expression of circRNAs 3 days after SCI, which may indicate that this is a critical time point. The researchers focused on the T8–10 spinal cord segments. Yao et al. found 1,131 circRNAs with significantly differentially expressed expressions, of which 909 expressions were upregulated and 222 expressions were downregulated. These differentially expressed circRNAs are involved in “mitosis,” “dendrite,” “calcium ion binding,” and “glycosphingolipid biosynthesis and extracellular matrix-receptor” (Yao et al., 2020). In another analysis by Wang and colleagues, the differentially expressed circRNAs participated in “protein phosphorylation,” “cytoplasm,” and “protein binding” (Wang et al., 2020d). Meanwhile, Qin et al. found 415 circRNAs were upregulated and 1,261 circRNAs were downregulated. According to the enrichment analysis, the target mRNAs of five candidate circRNAs are involved in “metabolic process,” “intracellular,” “anion binding,” and “adenosine 5′ monophosphate-activated protein kinase (AMPK) signaling pathway” (Qin et al., 2018). AMPK controls the axonal regeneration of sensory neurons in the dorsal root ganglion after SCI and plays an important role in energy metabolism and cellular protein homeostasis (Zhao et al., 2017; Kong et al., 2020). There was also a study of 7 days after SCI. Results showed that differentially expressed circRNAs related with “metabolic process,” “intracellular organelles,” “nucleic acid binding,” and “MAPK signaling pathway” (Chen et al., 2020). These results suggest that circRNAs participate in the transcription, metabolism, and other crucial biological processes in cells to regulate the process of repair after SCI.
The expression of some circRNAs decreased significantly at different time points. CircRNA_009608 decreased significantly at both 2 h and 3 days after SCI (Qin et al., 2018; Liu et al., 2020). The fold change was 11.9 at 2 h and 9.3 at 3 days. The gene symbol of circRNA_009608 is Gnaz, which is capable of coupling neurotransmitter receptors to ion channels in sympathetic neurons (Jeong and Ikeda, 1998). The expression of circRNA_015152 decreased 2 h after SCI and the fold change was 10.2 (Zhou et al., 2019), while the expression of circRNA_015151 also decreased 3 days after SCI and the fold change was 9.5 (Qin et al., 2018). CircRNA_015152 and circRNA_015151 have the same gene symbol Anks1b, which is a major component of the postsynaptic density in excitatory neurons (Dosemeci et al., 2016). The decreased expression of these circRNAs may be a manifestation of continuous inhibition of neurological function, but we need more data to observe their expression trend at different time points.
Comparison of Similarities and Differences of CircRNAs Between TBI and SCI
SCI and TBI are closely related in the pathological mechanism (Baek et al., 2017). Indeed, studies have shown similarities and differences in the research for circRNAs in these two different kinds of injuries. First, various time points are selected in the experiments. In the modeling of these studies, although they are all processes simulating acute injuries, the time points of sample analysis after TBI injuries are mostly earlier than SCI. Second, the sampling location varies. The samples adopted by TBI include the exosomes, hippocampus, brain cortex, etc., while the SCI samples are mostly concentrated in the spinal segments of T8–10. There are also some common points in the findings of bioinformatics analysis. Some results of TBI and SCI are similar in the existing reports on enrichment analysis of differentially expressed circRNAs. For example, circRNAs in both TBI and SCI participate in “organelle organization” (Zhao et al., 2018; Chen et al., 2020), “synapse” (Xie et al., 2018; Liu et al., 2020), and “protein binding” (Zhao et al., 2018; Liu et al., 2020) in the GO term analysis. They also participated in the “axon guidance” (Xie et al., 2018; Chen et al., 2019b, Chen et al., 2020) in the KEGG pathways.
We collected circRNAs that differentially expressed top 10 or top 5. A total of 25 are upregulated and 30 are downregulated in TBI. Thirty-five are upregulated and 35 are downregulated in SCI. The distribution of gene symbols of these circRNAs on chromosomes is shown in Figure 1A. They are distributed across almost every chromosome. In TBI, chromosomes 7, 10, and 11 are mainly concentrated, while in SCI, chromosomes 6, 2, and 3 are mainly concentrated. The gene symbols of upregulated circRNAs are shown in Figure 1B and the gene symbols of downregulated circRNAs are shown in Figure 1C. CircRNA_00180 was significantly decreased 6 h after TBI and its fold change was 5.4 (Jiang et al., 2019). Circ_0008614 was also significantly decreased 3 days after SCI and its fold change was 2.68 (Yao et al., 2020). They share the same gene symbol Col19a1, which is related with encoding nonfibrillar collagen XIX (Su et al., 2010).
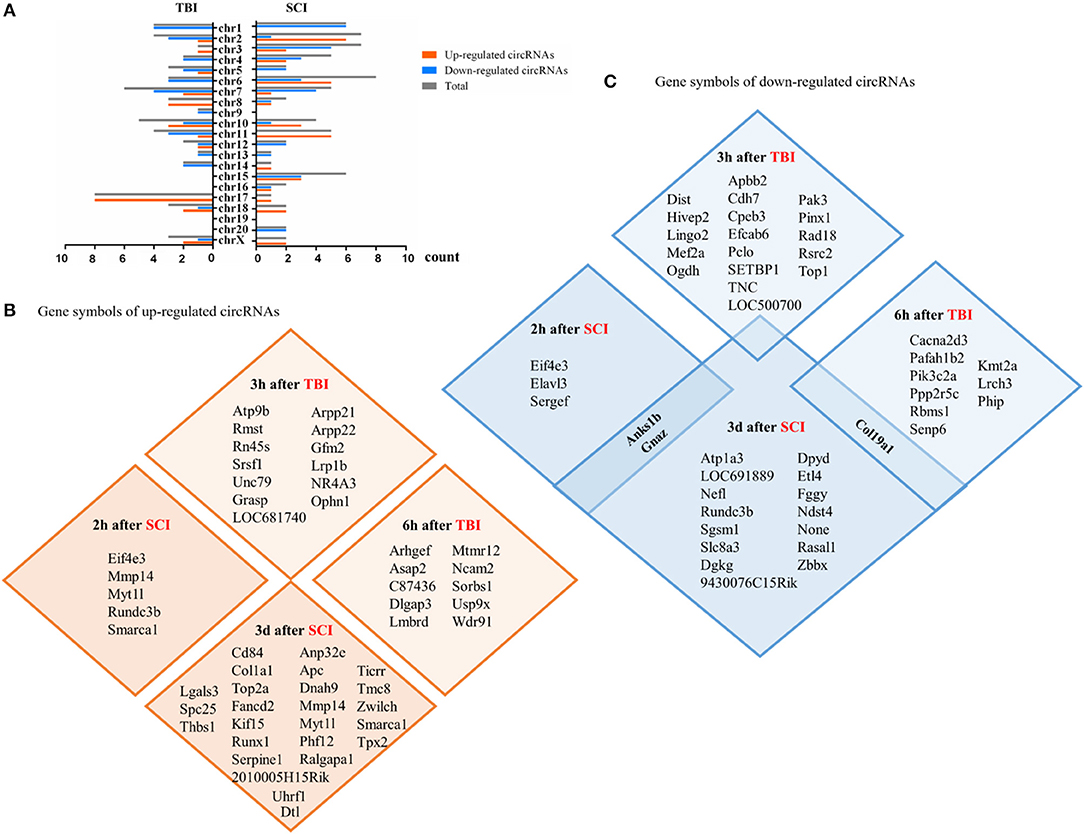
Figure 1. Differentially expressed circular RNAs (circRNAs) (within the top 5–10 upregulated or downregulated in different studies) in CNS post-traumatic injury. (A) The distribution of differentially expressed circRNAs on chromosomes. (B) The gene symbols of upregulated circRNAs at different time points in the injury site. (C) The gene symbols of downregulated circRNAs at different time points in the injury site.
Functional Roles of Specific Circrnas in CNS Post-Traumatic Injury
Upregulated CircRNAs
The expression of circRNAchr8_87,859,283–87,904,548 increased significantly in the mouse model 6 h after TBI. Its expression rose about fourfold in the damaged areas around the damaged cerebral cortex. The study proved that circRNAchr8_87,859,283–87,904,548 can play a role in increasing the CXCR2 protein by sponging mmu-let-7a-5p. CXCR2 can cause neuropathic pain and inflammatory pain through the interaction between astrocytes and neurons (Cao et al., 2016). Thus, the increase of the circRNAchr8_87,859,283–87,904,548 promotes neuroinflammatory responses and further inhibits the recovery of nervous system function. This suggests that inhibiting the expression of circRNAchr8_87,859,283–87,904,548 may contribute to the recovery after TBI through anti-inflammatory effects (Chen et al., 2019b).
Three days after SCI, the expression of cicRNA_7079 increased significantly in mice. Ying Yao et al. found that cicRNA_7079 could act on the cicRNA_7079–mmu-Mir-6953-5p–Lgals3 axis (Yao et al., 2020). Recent studies have found that Gal-3 can modulate the severity of neuroinflammation after SCI by enhancing the activation of the ROS/TXNIP/NLRP3 signaling pathway (Ren et al., 2019). Meanwhile, in NSC-34 motor neurons, deletion of cicRNA_7079 increases neuronal apoptosis. The results further proves that cicRNA_7079 could play an anti-apoptotic role. This study helps elucidate the molecular mechanisms associated with apoptosis in SCI (Yao et al., 2020).
The study of Jun Chen et al. showed that 7 days after SCI, circRNA_2960 was the top 1 upregulated circRNA. CircRNA_2960 downregulated miRNA_124 and functioned through the interaction network of circRNA_2960–miRNA_124. It was proved that circRNA_2960 could aggravate the inflammatory response and induce apoptosis at the site of injury. The study further demonstrated that blocking the expression of circRNA_2960 could be beneficial to the recovery of spinal cord injured tissues (Chen et al., 2020). However, the functional relationship between miRNA-124 and mRNA remains to be explored by subsequent researches.
In summary, both circRNAchr8_87,859,283–87,904,548 in the TBI model and circRNA_2960 in the SCI model play roles in promoting inflammation. The three circRNAs mentioned above are upregulated in traumatic CNS injuries (Table 2 and Figure 2). They act on different circRNA–miRNA–mRNA axes to function and are related to the process of inflammation or apoptosis. Other unexplored upregulated circRNAs might play important roles in similar or different biological processes in CNS post-traumatic injury. They may also act in ways other than miRNA sponges, which need to be confirmed by further investigation.
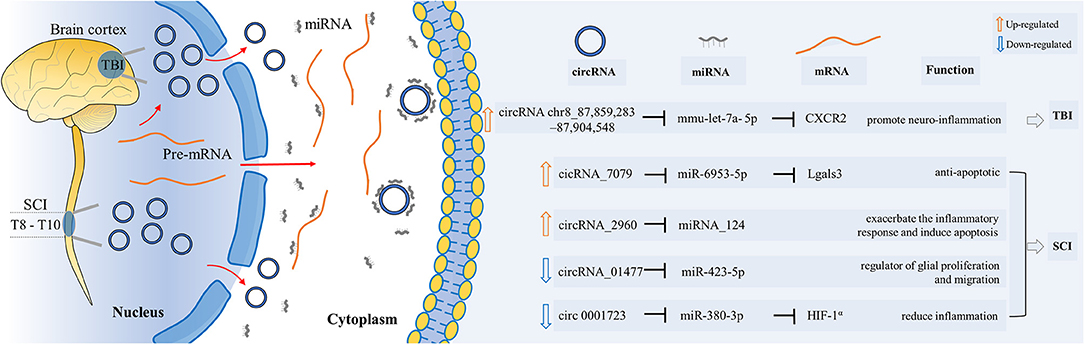
Figure 2. Functional roles of specific circRNAs in CNS post-traumatic injury. CircRNAs play their roles as miRNA sponges through the circRNA–miRNA–mRNA interaction networks. They influenced the biological processes of proliferation, inflammation, apoptosis, etc.
Downregulated CircRNAs
CircRNA_16895 was significantly decreased in mice 6 h after TBI, and it was predicted to act on the circRNA_16895–miRNA–Myo10 axis. It might be able to regulate the fragment crystallizable gamma receptor-mediated phagocytosis pathway (Jiang et al., 2019). However, this pathway is still at the predicted stage and has not been confirmed by experiments yet. Future studies are supposed to figure out the function mechanism of circRNA_16895.
The expression of circRNA_01477 decreased significantly in rats after SCI, which serves as a regulator of glial proliferation and migration by acting on the axis of circRNA_01477–miR-423-5p. In this study, Wu et al. (2019) prevented the expression of circRNA_01477 through small interfering RNA. It stunted the proliferation and migration of astrocytes. Subsequently, microarray assay was used to explore the circRNA_01477–miRNA–mRNA network. They found that among the seven validated mRNAs with decreased expression, four were regulated by miRNA-423-5p. Meanwhile, miRNA-423-5p was significantly increased after circRNA_01477 interference. This further indicates that the regulation of the circRNA_01477/miR-423-5p axis may be very important for the restoration of environmental regulation after SCI.
In the study of Li and colleagues, the expression of Circ 0001723 decreased significantly in rats 1 day after SCI, while the expression of miR-380-3p increased significantly. In vitro experiments showed that the decrease of Circ 0001723 promoted the expression of proinflammatory factors such as TNF-α and IL-1β, while the expression of miR-380-3p increased. This process inhibits the expression of HIF-1 and leads to the expression of NLRP3 and caspase-1 proteins (Li et al., 2020). Therefore, the Circ 0001723–miR-380-3p–HIF-1α axis plays a role in the regulation of inflammatory response. Circ 0001723 might be an important target related to the regulation of inflammation in SCI recovery.
The expression of these three circRNAs decreased in traumatic CNS injury (Table 2 and Figure 2). According to current research results, they all function as miRNA sponges. These three studies used different animal models. CircRNAs mentioned above are involved in different circRNA–miRNA–mRNA networks. They act on various biological processes such as inflammation, proliferation, and migration. Future research needs to confirm and explore their functions to see if they can be used clinically as therapeutic targets for the recovery from CNS post-traumatic injuries.
Conclusion and Future Perspective
CNS post-traumatic injury is a serious public health problem. At present, there is still great value in the study of circRNAs in CNS post-traumatic injury. More and more circRNAs are being explored. Previous studies have shown that they participated in the processes of proliferation, inflammation, and apoptosis in TBI or SCI. As summarized by this review, emerging lines of evidence have described their different expressions (e.g., circRNA_16895, circRNAchr8_87,859,283–87,904,548, circRNA_01477, etc.). They act as miRNA sponges to influence the corresponding downstream mRNAs. Despite its strong clinical potential, the best way to intervene with circRNAs has not yet been defined.
Subsequent studies can be devoted to finding and confirming more functions of various circRNAs. With further research, their clinical research potential could be fully explored in the future. First, studies on the cellular level are supposed to be designed. Most of the current researches are limited to the homogenization of the whole tissue. Therefore, single-cell sequencing and other cutting-edge technologies can be applied to explore the functions of circRNAs in specific types of nerve cells or immune cells. Second, circRNAs in neuron stem cells are worth studying to find possible clinical therapeutic targets. Researches have investigated the effect of circRNA regulation in neural stem cells on functional recovery after ischemic stroke (Wang et al., 2020a). The methods could also be used in CNS post-traumatic injury. Third, the different roles of circRNAs in different recovery stages are worth exploring and comparing. Thus, we are able to find better opportunities and methods for regulation. Fourth, there is a lack of knowledge on chronic and regeneration stage of trauma. Most of the present studies have investigated the expression differences of circRNAs in the acute phase, but the chronic stage is still unknown. Fifth, circRNAs may also be used as biomarkers to monitor prognosis due to their great stability and abundance. They have been used as potential diagnostic biomarkers in other diseases such as prediabetes and tumors (Xie et al., 2017). Sixth, studies of human tissue samples have not been reported. Researches will not be confined to experimental animals in the future. The development of clinical studies is helpful to fully explore the clinical intervention value of circRNAs. Overall, future researches could maximize the benefits of circRNAs for patients suffering from CNS post-traumatic injury.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
LX, XY, and L-lW wrote the draft manuscript, designed the figures, and tables, all co-authors (LX, XY, JZ, Y-yC, and L-lW) contributed substantially with revision and improvement of the manuscript. The authors read and approved the final manuscript.
Funding
This study was supported in part by grants from the National Natural Science Foundation of China (Nos. 81572229 and 81972138).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Baek, A., Cho, S. R., and Kim, S. H. (2017). Elucidation of gene expression patterns in the brain after spinal cord injury. Cell Transplant. 26, 1286–1300. doi: 10.1177/0963689717715822
Cao, D. L., Qian, B., Zhang, Z. J., Gao, Y. J., and Wu, X. B. (2016). Chemokine receptor CXCR2 in dorsal root ganglion contributes to the maintenance of inflammatory pain. Brain Res. Bull. 127, 219–225. doi: 10.1016/j.brainresbull.2016.09.016
Cardamone, G., Paraboschi, E. M., Rimoldi, V., Duga, S., Solda, G., and Asselta, R. (2017). The characterization of GSDMB splicing and backsplicing profiles identifies novel isoforms and a circular RNA that are dysregulated in multiple sclerosis. Int. J. Mol. Sci. 18. doi: 10.3390/ijms18030576
Chen, H., Tan, Q., Xie, C., Li, C., Chen, Y., Deng, Y., et al. (2019a). Application of olfactory ensheathing cells in clinical treatment of spinal cord injury: meta-analysis and prospect. J. Neurorestoratol. 7, 70–81. doi: 10.26599/JNR.2019.9040008
Chen, J., Fu, B., Bao, J., Su, R., Zhao, H., and Liu, Z. (2020). Novel circular RNA 2960 contributes to secondary damage of spinal cord injury by sponging miRNA-124. J. Comp. Neurol. 2020, 1–9. doi: 10.1002/cne.25030
Chen, Z., Wang, H., Zhong, J., Yang, J., Darwazeh, R., Tian, X., et al., (2019b). Significant changes in circular RNA in the mouse cerebral cortex around an injury site after traumatic brain injury. Exp. Neurol. 313, 37–48. doi: 10.1016/j.expneurol.2018.12.003
Dosemeci, A., Toy, D., Burch, A., Bayer, K. U., and Tao-Cheng, J. H. (2016). CaMKII-mediated displacement of AIDA-1 out of the postsynaptic density core. FEBS Lett. 590, 2934–2939. doi: 10.1002/1873-3468.12334
Enuka, Y., Lauriola, M., Feldman, M. E., Sas-Chen, A., Ulitsky, I., and Yarden, Y. (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383. doi: 10.1093/nar/gkv1367
Ge, X. T., Li, W. Z., Huang, S., Yin, Z. Y., Yang, M. C., Han, Z. Y., et al. (2019). Increased miR-21-3p in injured brain microvascular endothelial cells after traumatic brain injury aggravates blood-brain barrier damage by promoting cellular apoptosis and inflammation through targeting MAT2B. J. Neurotrauma. 36, 1291–1305. doi: 10.1089/neu.2018.5728
Han, B., Zhang, Y., Zhang, Y. H., Bai, Y., Chen, X. F., Huang, R. R., et al. (2018). Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy 14, 1164–1184. doi: 10.1080/15548627.2018.1458173
Jeong, S. W., and Ikeda, S. R. (1998). G protein alpha subunit G alpha z couples neurotransmitter receptors to ion channels in sympathetic neurons. Neuron 21, 1201–1212. doi: 10.1016/S0896-6273(00)80636-4
Jiang, Y. J., Cao, S. Q., Gao, L. B., Wang, Y. Y., Zhou, B., Hu, X., et al. (2019). Circular ribonucleic acid expression profile in mouse cortex after traumatic brain injury. J. Neurotrauma. 36, 1018–1028. doi: 10.1089/neu.2018.5647
Kempuraj, D., Ahmed, M. E., Selvakumar, G. P., Thangavel, R., Raikwar, S. P., Zaheer, S. A., et al. (2020). Acute traumatic brain injury-induced neuroinflammatory response and neurovascular disorders in the brain. Neurotox Res. 39, 359–368. doi: 10.1007/s12640-020-00288-9
Kong, G., Zhou, L., Serger, E., Palmisano, I., De Virgiliis, F., Hutson, T. H., et al. (2020). AMPK controls the axonal regenerative ability of dorsal root ganglia sensory neurons after spinal cord injury. Nat. Metab. 2, 918–933. doi: 10.1038/s42255-020-0252-3
Lasda, E., and Parker, R. (2014). Circular RNAs: diversity of form and function. RNA. 20, 1829–1842. doi: 10.1261/rna.047126.114
Leng, Z., Kethidi, N., Chang, A. J., Sun, L., Zhai, J., Yang, Y., et al. (2019). Muse cells and neurorestoratology. J. Neurorestoratol. 7, 18–25. doi: 10.26599/JNR.2019.9040005
Li, D., Huang, S., Yin, Z. Y., Zhu, J. L., Ge, X. T., Han, Z. L., et al. (2019). Increases in miR-124-3p in microglial exosomes confer neuroprotective effects by targeting FIP200-mediated neuronal autophagy following traumatic brain Injury. Neurochem. Res. 44, 1903–1923. doi: 10.1007/s11064-019-02825-1
Li, L. J., Huang, Q., Pan, H. F., and Ye, D. Q. (2016). Circular RNAs and systemic lupus erythematosus. Exp. Cell Res. 346, 248–254. doi: 10.1016/j.yexcr.2016.07.021
Li, X., Lou, X., Xu, S., Du, J., and Wu, J. (2020). Hypoxia inducible factor-1 (HIF-1alpha) reduced inflammation in spinal cord injury via miR-380-3p/ NLRP3 by Circ 0001723. Biol. Res. 53:35. doi: 10.1186/s40659-020-00302-6
Liu, Y. D., Liu, J. F., and Liu, B. (2020). Identification of circular RNA expression profiles and their implication in spinal cord injury rats at the immediate phase. J. Mol. Neurosci. 70, 1894–1905. doi: 10.1007/s12031-020-01586-9
Ma, C., Zhang, P., and Shen, Y. (2019). Progress in research into spinal cord injury repair: Tissue engineering scaffolds and cell transdifferentiation. J. Neurorestoratol. 7, 196–206. doi: 10.26599/JNR.2019.9040024
Ma, Z., Lu, Y., Yang, F., Li, S., He, X., Gao, Y., et al. (2020). Rosmarinic acid exerts a neuroprotective effect on spinal cord injury by suppressing oxidative stress and inflammation via modulating the Nrf2/HO-1 and TLR4/NF-kappaB pathways. Toxicol Appl. Pharmacol. 397:115014. doi: 10.1016/j.taap.2020.115014
Nagalakshmi, B., Sagarkar, S., and Sakharkar, A. J. (2018). Epigenetic mechanisms of traumatic brain injuries. Prog. Mol. Biol. Transl. Sci. 157, 263–298. doi: 10.1016/bs.pmbts.2017.12.013
Qin, C., Liu, C. B., Yang, D. G., Gao, F., Zhang, X., Zhang, C., et al. (2018). Circular RNA Expression alteration and bioinformatics analysis in rats after traumatic spinal cord Injury. Front. Mol. Neurosci. 11:497. doi: 10.3389/fnmol.2018.00497
Radabaugh, H. L., Bonnell, J., Dietrich, W. D., Bramlett, H. M., Schwartz, O., and Sarkar, D. (2020). “Development and evaluation of machine learning models for recovery prediction after treatment for traumatic brain injury,” in 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (Montreal, QC), 2416–2420. doi: 10.1109/EMBC44109.2020.9175658
Reis, K. P., Sperling, L. E., Teixeira, C., Sommer, L., Colombo, M., Koester, L. S., et al. (2020). VPA/PLGA microfibers produced by coaxial electrospinning for the treatment of central nervous system injury. Braz. J. Med. Biol. Res. 53:e8993. doi: 10.1590/1414-431x20208993
Ren, Z. L., Liang, W. D., Sheng, J., Xun, C. H., Xu, T., Cao, R., et al. (2019). Gal-3 is a potential biomarker for spinal cord injury and Gal-3 deficiency attenuates neuroinflammation through ROS/TXNIP/NLRP3 signaling pathway. Biosci. Rep. 39:BSR20192368. doi: 10.1042/BSR20192368
Rola, R., Mizumatsu, S., Otsuka, S., Morhardt, D. R., Noble-Haeusslein, L. J., Fishman, K., et al. (2006). Alterations in hippocampal neurogenesis following traumatic brain injury in mice. Exp. Neurol. 202, 189–199. doi: 10.1016/j.expneurol.2006.05.034
Shafi, S., Gupta, P., Khatik, G. L., and Gupta, J. (2019). PPARgamma: potential therapeutic target for ailments beyond diabetes and its natural agonism. Curr. Drug Targets 20, 1281–1294. doi: 10.2174/1389450120666190527115538
Su, J. M., Gorse, K., Ramirez, F., and Fox, M. A. (2010). Collagen XIX is expressed by interneurons and contributes to the formation of hippocampal synapses. J. Compar. Neurol. 518, 229–253. doi: 10.1002/cne.22228
Tsujioka, H., and Yamashita, T. (2019). Comparison of gene expression profile of the spinal cord of sprouting-capable neonatal and sprouting-incapable adult mice. BMC Genomics 20:619. doi: 10.1186/s12864-019-5974-9
Wang, G., Han, B., Shen, L., Wu, S., Yang, L., Liao, J., et al. (2020a). Silencing of circular RNA HIPK2 in neural stem cells enhances functional recovery following ischaemic stroke. EBioMed. 52:102660. doi: 10.1016/j.ebiom.2020.102660
Wang, N., He, L., Yang, Y., Li, S., Chen, Y., Tian, Z., et al. (2020b). Integrated analysis of competing endogenous RNA (ceRNA) networks in subacute stage of spinal cord injury. Gene 726:144171. doi: 10.1016/j.gene.2019.144171
Wang, W., Li, J., Zhang, Z., Ma, H., Li, Q., Yang, H., et al. (2020c). Genome-wide analysis of acute traumatic spinal cord injury-related RNA expression profiles and uncovering of a regulatory axis in spinal fibrotic scars. Cell Prolif. 54:e12951. doi: 10.1111/cpr.12951
Wang, W., Wang, S., Zhang, Z., Li, J., Xie, W., Su, Y., et al. (2020d). [Identification of potential traumatic spinal cord injury related circular RNA-microRNA networks by sequence analysis]. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 34, 213–219. doi: 10.7507/1002-1892.201905079
Wang, Z. Z. F., and Song, M. (2019). The cell repair research of spinal cord injury: a review of cell transplantation to treat spinal cord injury. J. Neurorestoratol. 7, 55–62. doi: 10.26599/JNR.2019.9040011
Warnock, A., Toomey, L. M., Wright, A. J., Fisher, K., Won, Y., Anyaegbu, C., et al. (2020). Damage mechanisms to oligodendrocytes and white matter in central nervous system injury: The Australian Context. J. Neurotrauma. 37, 739–769. doi: 10.1089/neu.2019.6890
Wu, N., and Yang, B. B. (2015). The biological functions of non-coding RNAs: from a line to a circle. Discoveries (Craiova). 3:e48. doi: 10.15190/d.2015.40
Wu, R., Mao, S., Wang, Y., Zhou, S., Liu, Y., Liu, M., et al. (2019). Differential crcular RNA expression profiles following spinal cord injury in rats: a temporal and experimental analysis. Front. Neurosci. 13:1303. doi: 10.3389/fnins.2019.01303
Xie, B. S., Wang, Y. Q., Lin, Y., Zhao, C. C., Mao, Q., Feng, J. F., et al., (2018). Circular RNA expression profiles alter significantly after traumatic brain injury in rats. J. Neurotrauma. 35, 1659–1666. doi: 10.1089/neu.2017.5468
Xie, L., Mao, M., Xiong, K., and Jiang, B. (2017). Circular RNAs: a novel player in development and disease of the central nervous system. Front. Cell. Neurosci. 11:354. doi: 10.3389/fncel.2017.00354
Yang, L., Ge, D., Chen, X., Jiang, C., and Zheng, S. (2018a). miRNA-544a regulates the inflammation of spinal cord injury by inhibiting the expression of NEUROD4. Cell Physiol. Biochem. 51, 1921–1931. doi: 10.1159/000495717
Yang, Y., Gao, X., Zhang, M., Yan, S., Sun, C., Xiao, F., et al. (2018b). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110, 304–315. doi: 10.1093/jnci/djx166
Yao, Y., Wang, J., He, T., Li, H., Hu, J., Zheng, M., et al., (2020). Microarray assay of circular RNAs reveals cicRNA.7079 as a new anti-apoptotic molecule in spinal cord injury in mice. Brain Res. Bull. 164, 157–171. doi: 10.1016/j.brainresbull.2020.08.004
Ye, J., Xue, R., Ji, Z. Y., Zou, C. J., Chen, Y. Q., Wang, J. J., et al. (2020). Effect of NT-3 on repair of spinal cord injury through the MAPK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 2165–2172. doi: 10.26355/eurrev_202003_20481
Yi, M., Dai, X., Li, Q., Xu, X., Chen, Y., and Wang, D. (2019). Downregulated lncRNA CRNDE contributes to the enhancement of nerve repair after traumatic brain injury in rats. Cell Cycle 18, 2332–2343. doi: 10.1080/15384101.2019.1647024
Yu, P., Li, S. J., Zhang, Z. F., Wen, X. L., Quan, W., Tian, Q. L., et al. (2017). Progesterone-mediated angiogenic activity of endothelial progenitor cell and angiogenesis in traumatic brain injury rats were antagonized by progesterone receptor antagonist. Cell Prolif. 50:12362. doi: 10.1111/cpr.12362
Zhang, Y., Yu, F., Bao, S., and Sun, J. (2019). Systematic characterization of circular RNA-associated CeRNA network identified novel circRNA biomarkers in Alzheimer's disease. Front. Bioeng Biotechnol. 7:222. doi: 10.3389/fbioe.2019.00222
Zhao, H., Chen, S., Gao, K., Zhou, Z., Wang, C., Shen, Z., et al., (2017). Resveratrol protects against spinal cord injury by activating autophagy and inhibiting apoptosis mediated by the SIRT1/AMPK signaling pathway. Neuroscience 348, 241–251. doi: 10.1016/j.neuroscience.2017.02.027
Zhao, R. T., Zhou, J., Dong, X. L., Bi, C. W., Jiang, R. C., Dong, J. F., et al. (2018). Circular ribonucleic acid expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. J. Neurotrauma. 35, 2056–2066. doi: 10.1089/neu.2017.5502
Zheng, F., Zhou, Y. T., Li, P. F., Hu, E., Li, T., Tang, T., et al. (2020). Metabolomics analysis of hippocampus and cortex in a rat model of traumatic brain injury in the subacute Phase. Front. Neurosci. 14:876. doi: 10.3389/fnins.2020.00876
Zhong, Y., Chen, J. L., Chen, J., Chen, Y. H., Li, L., and Xie, Y. B. (2019). Crosstalk between Cdk5/p35 and ERK1/2 signalling mediates spinal astrocyte activity via the PPARy pathway in a rat model of chronic constriction injury. J. Neurochem. 151, 166–184. doi: 10.1111/jnc.14827
Keywords: central nervous system, traumatic brain injury, spinal cord injury, circular RNAs, inflammation, apoptosis
Citation: Xu L, Ye X, Zhong J, Chen Y-y and Wang L-l (2021) New Insight of Circular RNAs' Roles in Central Nervous System Post-Traumatic Injury. Front. Neurosci. 15:644239. doi: 10.3389/fnins.2021.644239
Received: 20 December 2020; Accepted: 04 February 2021;
Published: 23 March 2021.
Edited by:
Domenico De Berardis, Azienda Usl Teramo, ItalyReviewed by:
Felice Iasevoli, University of Naples Federico II, ItalyAmaresh Chandra Panda, Institute of Life Sciences (ILS), India
Copyright © 2021 Xu, Ye, Zhong, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin-lin Wang, d2FuZ2xpbmxpbkB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Lvwan Xu1†
Lvwan Xu1† Lin-lin Wang
Lin-lin Wang