- Department of Anesthesiology, Shengjing Hospital of China Medical University, Shenyang, China
Axons in the central nervous system often fail to regenerate after injury due to the limited intrinsic regeneration ability of the central nervous system (CNS) and complex extracellular inhibitory factors. Therefore, it is of vital importance to have a better understanding of potential methods to promote the regeneration capability of injured nerves. Evidence has shown that non-coding RNAs play an essential role in nerve regeneration, especially long non-coding RNA (lncRNA), microRNA (miRNA), and circular RNA (circRNA). In this review, we profile their separate roles in axon regeneration after CNS injuries, such as spinal cord injury (SCI) and optic nerve injury. In addition, we also reveal the interactive networks among non-coding RNAs.
Introduction
Central nerve injuries are commonly seen in clinical practice. They can severely affect quality of life and may be fatal. Unfortunately, there is still no known cure for these kinds of injuries because of their complex pathophysiology. Damaged axons in the central nervous system (CNS) rarely regenerate, mainly due to the inhibitory environment around the damaged nerves and a lack of intrinsic neuronal growth ability (Goncalves et al., 2015). In the search for a cure for CNS injuries, researchers have made a leap in progress in terms of the mechanisms of neuronal regeneration. Evidence shows that multiple transcriptional regulatory pathways can promote the capacity of neurite regrowth (Sun et al., 2011; Apara et al., 2017; Wang Z. et al., 2018). Non-coding RNAs, which used to be regarded as transcription noise, are now proven to be important transcriptional and post-transcriptional regulators in the development of the nervous system and neurological diseases (Wu et al., 2013). In recent years, the functions of non-coding RNAs in axon regeneration in the peripheral nervous system (PNS) have continuously been revealed, which has provided clues about CNS axon regeneration. Researchers have found that multiple non-coding RNAs are differentially expressed after CNS injuries, such as spinal cord injury (SCI) and optic nerve injury, which implies that non-coding RNAs may have the potential to become brand new biomarkers and targeted therapies for axon regeneration in the CNS. Therefore, in this review, we first profile the extrinsic and intrinsic mechanisms for nerve regeneration; then, we give a brief introduction on long non-coding RNA (lncRNA), microRNA (miRNA), and circular RNA (circRNA), which are widely associated with axon regeneration. Finally, we focus on some typical non-coding RNAs to demonstrate the separate and interactive functions of non-coding RNAs in nerve regeneration. Last but not least, we also suggest potential directions for future research.
Extrinsic and Intrinsic Mechanisms for Nerve Regeneration
After nerve damage in nervous systems, successful compensation may include axonal sprouting from intact neurons or regeneration from injured axons, therefore, bringing about function recovery. Unlike the robust regrowth in the peripheral nervous system (PNS) following injury, the central nervous system (CNS) appears to have defects related to nerve regeneration (Oh et al., 2018). In mammals, this process relies both on the improvement of a permissive extracellular environment (Tom et al., 2004) and intrinsic growth ability (Goncalves et al., 2015). In the last century, Aguayo et al. revealed that axons in an injured spinal cord and brainstem could successfully regrow over long distances when the CNS glial environment was replaced by peripheral nerves (David and Aguayo, 1981), showing that the inhibitory microenvironment attributes to the failure of axon regeneration in the CNS. Over time, varieties of extrinsic factors were discovered to have a relationship with the regeneration process after CNS injury. Glial scars, myelin debris, and axonal growth inhibitors form significant impediments to axonal regeneration (Li et al., 2020). Glial scars, which serve a primary role in preventing inflammatory processes from propagating to healthy tissue (Cregg et al., 2014), also create a physical barrier in which axonal tips get trapped. Confronted with the scar tissue, the ends of regenerating axons cease to extend and turn into abnormal growth cones that can remain for years without robust regeneration (Tom et al., 2004). Moreover, demyelination occurs after nerve damage, which severely impairs axon survival as the myelinating cells of the CNS, oligodendrocytes, are lost through injury. Under permissive conditions, they can be replaced by oligodendrocyte progenitor cells and differentiate into mature oligodendrocytes. However, myelin debris inhibits the differentiation process, so it is of vital importance to clear the myelin debris following demyelination (Church et al., 2017). Apart from that, a variety of neurite growth inhibitory factors were found to strongly suppress the sprouting and regeneration of injured neurites after CNS injury. In addition to myelin-associated glycoprotein (MAG), Nogo-A, and oligodendrocyte-myelin glycoprotein (OMgp), which are expressed by the myelin sheath in injured axons (Gribble and Scott, 2002), chondroitin sulfate proteoglycans (CSPGs), released by hypertrophic astrocytes at the lesioned site, present a potent barrier to axon regeneration (Yiu and He, 2006). Although we can remove molecules that restrict regrowth in the extracellular environment, the effect of regeneration is insufficient (Holmes, 2017). As a result, more attention has been focused on how to promote the inherent growing ability of the lesioned axons. As is known, gene transcription and protein translation play a key role in this course. Researchers have found that a series of genes take part in intrinsic regulation following CNS injury, including Pten, Klf4/9, Socs3, B-RAF, c-Myc, GSK3, and Lin28 (Moore et al., 2009; Luo and Park, 2012; Saijilafu et al., 2013; O’Donovan et al., 2014; Wang X.W. et al., 2018; Zhang et al., 2018; Ma et al., 2019). Moreover, many regeneration-associated signaling pathways, such as cAMP/PKA,DLK/JNK, are continually being uncovered (Neumann et al., 2002; Larhammar et al., 2017). However, with the development of transcriptomics, mounting studies suggest that non-coding RNAs, especially lncRNA, miRNA, and circRNA, exert their function as regulators in multiple biological programs. More importantly, they are widely found to be differentially expressed in the central nervous system after an injury (Yao et al., 2015), which indicates that they have the potential to become clinical strategies for axon regeneration after nerve injury.
Classification and Biogenesis of Non-coding RNAs
There are two types of non-coding RNAs. The first type, constitutive non-coding RNAs, include transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and small cytoplasmic RNA (scRNA). Their abundance is constant, and they play an important role in maintaining the normal physiological function of species. The second type, regulatory non-coding RNAs, include lncRNA, miRNA, circRNA, siRNA, and piRNA. Their abundance changes with the external environment and cell characteristics, which play an important role in regulating gene expression. In this review, we focus on three members of the non-coding RNA family–lncRNA, microRNA, and circRNA–and describe their functions in nerve regeneration as well as their combined effects.
lncRNAs are a cluster of non-coding RNAs of more than 200 nucleotides in length (Paraskevopoulou and Hatzigeorgiou, 2016). These non-coding RNAs are poorly conserved and play a significant role at different levels, including chromatin remodeling, transcriptional control, and post-transcriptional processing. Such variable regulatory patterns suggest that lncRNA might have both complex biological and pathological functions. microRNAs are 20∼25 nucleotides in length, and they often regulate gene expression at the post-transcription level. Mature miRNA functions by binding to the complementary sequences in the 3′ untranslated regions of the target messenger RNA (mRNA). As a result, the miRNA silences the gene through mRNA degradation or transcriptional repression (Butz et al., 2012). circRNAs are a novel class of non-coding RNAs discovered in 2012. Unlike traditional linear non-coding RNAs with 5′ cap and 3′ tail structures, circRNAs are characterized by a closed-loop structure, which is more stable (Wu et al., 2019). These RNA molecules are rich in binding sites for miRNAs and serve as miRNA sponges in cells (Hansen et al., 2013). By binding to different miRNAs, they remove miRNA inhibition to the target mRNA, and expression of the mRNA increases as a result. This type of RNA is called competing endogenous RNA (ceRNA). By working with miRNAs, circRNAs play a crucial role in biological and pathological processes (Figure 1).
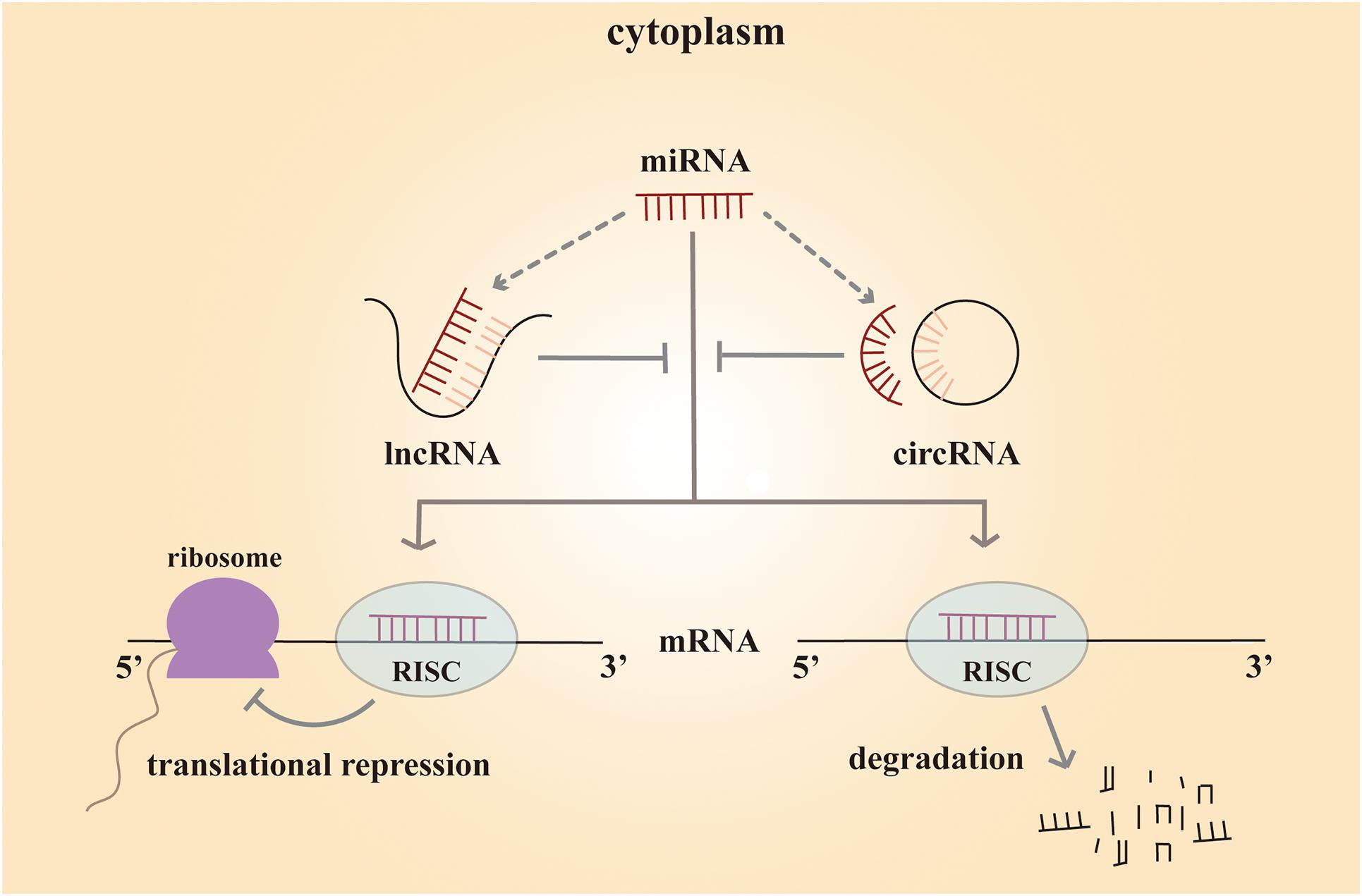
Figure 1. Mechanism scheme of ceRNA network. The mature miRNA regulates the gene expression by forming a RISC (RNA-induced silencing complex) and binding to the complementary sequences in their 3′ untranslated regions of the target mRNA. As a result, the miRNA silences the gene through mRNA degradation or transcriptional repression. Both lncRNAs and circRNAs are rich in binding sites for miRNAs and serve as miRNA sponges in cells. They compete with mRNA for opportunities to bind with miRNAs. In this way, lncRNAs and circRNAs remove the miRNA’s inhibition to the target mRNA, and the expression of the mRNA increases as a consequence. This mechanism is called competing endogenous RNA (ceRNA).
Non-coding RNAs in Central Nerve Regeneration
Spinal Cord Injury
SCI is a devastating traumatic event that damages sensory, motor, or autonomic functions and, therefore, affects quality of life (Fehlings et al., 2017). Every year, up to a million people experience spinal cord injury. More than half of them suffer from complete injury, losing function completely in the area controlled by the lesioned nerves. The others only recover part of their body function (Holmes, 2017). The management of acute SCI requires long-term, high-level care resources, and it brings substantial financial burden at both individual and societal levels (Fehlings et al., 2017). As a result, quite a lot of attention has been paid to the mechanisms of SCI to seek better repairing strategies.
As far as we are concerned, the pathophysiology of SCI temporally changes, consisting of two injury phases. The primary mechanical injury phase damages the axons, blood vessels, and cell membranes. Within hours or days after the injury, it enters the secondary injury phase with ischemia, edema, inflammation, and vascular changes (Rowland et al., 2008; Rouanet et al., 2017). The pathophysiological changes of SCI are so complicated that a curable strategy remains a big challenge. Despite the fact that a few repair studies are making some progress in animal models, to date, clinical translation to patients has not been convincing (Dietz, 2016). Mounting evidence has shown that multiple non-coding RNAs are differentially expressed after injury. Changes in lncRNA, miRNA, and circRNA are so evident that much attention has been focused on them (Chen Y. et al., 2016; Zhou et al., 2018; Wu et al., 2019). The non-coding RNAs are strongly connected to the various pathophysiological processes of SCI, such as inflammation, apoptosis, axon regeneration, oxidative stress, and astrocyte proliferation and activation (Li et al., 2016; Gu et al., 2017; Xia et al., 2018; Yao and Yu, 2019). Moreover, they have been found to play a potential role in SCI complications, including neuropathic pain (Wang F. et al., 2019). Here, we review recent studies on non-coding RNAs and their mechanisms of axon regeneration after spinal cord injury.
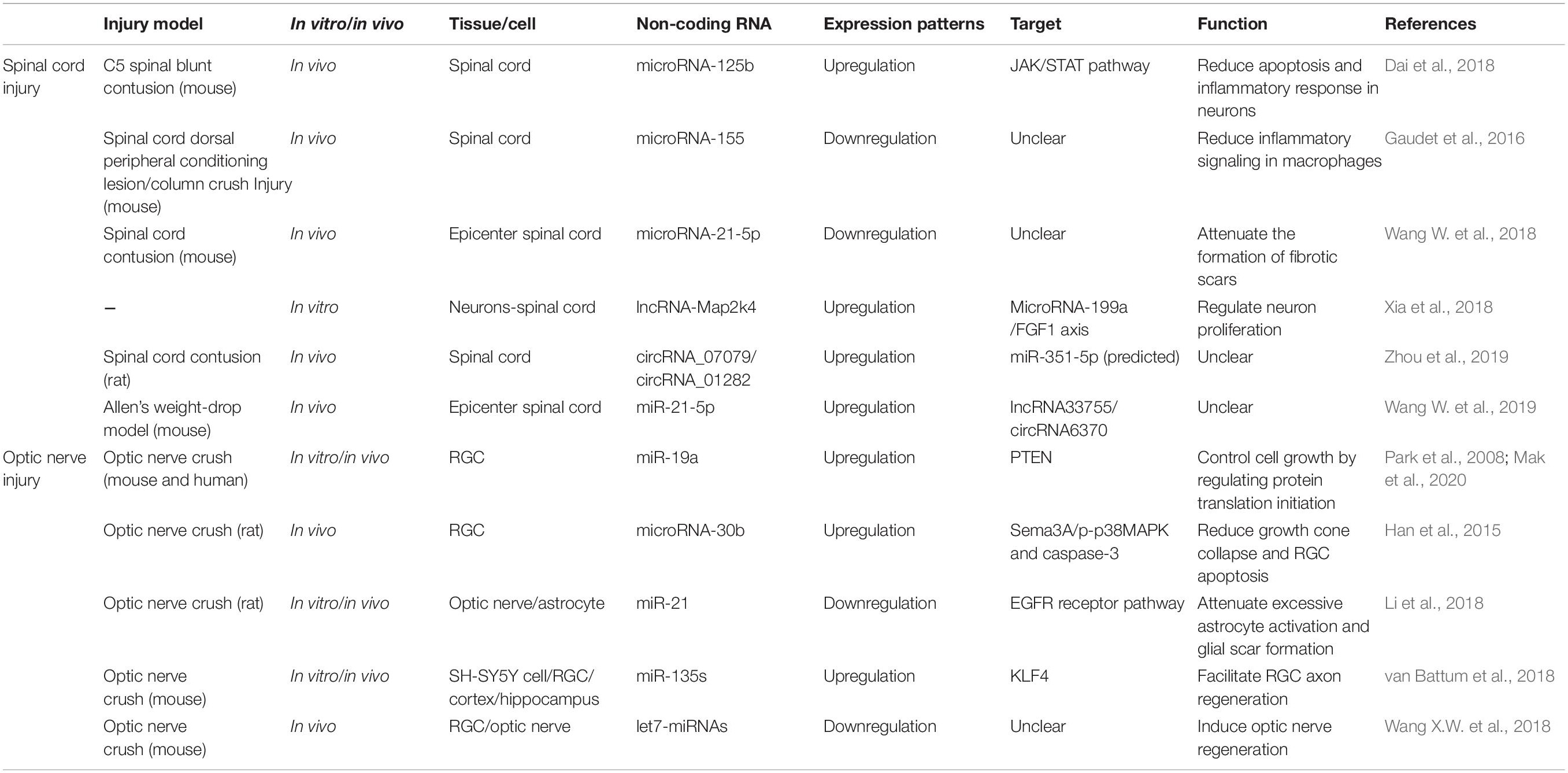
Accumulated evidence suggests that miRNAs are strongly related to the regulation process after SCI. Bioinformatics analysis demonstrated expression changes of a cluster of miRNAs in vivo, concluding that miRNAs may take part in promoting self-repair in SCI (Liu et al., 2016; Wang W. et al., 2019). In previous studies, micro-125b showed a significant decrease in expression level after SCI, and overexpression of microRNA-125b promotes axon regeneration following spinal cord injury by regulating the JAK/STAT pathway. Furthermore, micro-125b displays a positive neuronal protective effect by reducing apoptosis and inflammatory response in neurons (Dai et al., 2018). microRNA-155 deletion has been shown to reduce inflammatory signaling in macrophages, which enhances axon regeneration dynamics. In addition, microRNA-155 knockout neurons in adults also show independent enhanced axon growth (Gaudet et al., 2016). Moreover, some researchers point out that, regulated by TGF-β1, microRNA-21-5p mediates the expression of fibrosis-related genes. microRNA-21-5p knockdown attenuates the formation of fibrotic scars, which are thought to be the main physical obstacle for axonal regeneration following SCI (Wang W. et al., 2018). These results demonstrate that microRNAs might be a novel target for axon regeneration in SCI patients.
lncRNAs have been shown to have significant effects on the development of the brain (Goff et al., 2015), nervous systems (Zhang et al., 2019), and a variety of neurological diseases, such as brain tumors (Voce et al., 2019), neurodegenerative diseases (Chanda et al., 2018; Cao et al., 2019), and ischemic strokes (Hu et al., 2020). Moreover, lncRNAs also serve as a crucial regulator in SCI, affecting its initiation and progression. Several studies have demonstrated that they mostly perform functions via cell autophagy and apoptosis (Salah et al., 2020). However, some evidence tells us that lncRNAs probably also play a role in regulating axonal regrowth following SCI. Some previous studies have suggested that lncRNAs are of vital importance to neurite outgrowth, which is an early step in neuronal regeneration. It has been shown that lncRNA Malat1 plays a crucial role in neurite outgrowth in N2a cells by activating the MAPK/ERK signaling pathway. The MAPK family is thought to take part in a variety of biological processes such as development and apoptosis. Furthermore, the data show that elevated expression of lncRNA Malat1 promotes the average neurite length and thus enhances neuronal differentiation (Chen L. et al., 2016). Moreover, lncRNA-Map2k4 was found to have a high expression level and decline in SCI pathology. Data shows that lncRNA-Map2k4 regulates neuron proliferation through the microRNA-199a/FGF1 axis. Knockdown of lncRNA-Map2k4 inhibited FGF1 expression by upregulating microRNA-199a (Lv, 2017). As is known, FGF1 is a neurotrophic factor with a potent neuroregenerative ability in the nervous system (Ding et al., 2016), so this finding may provide some new ideas for the treatment of SCI.
In recent decades, more investigations into circRNAs have taken place, and circRNAs have been found to have a variety of functions in various disorders, such as cancers, cardiovascular diseases, diabetes, and ischemic strokes (Siede et al., 2017; Bai et al., 2018; Li et al., 2019; Abbaszadeh-Goudarzi et al., 2020). A number of studies using bioinformatics analysis have revealed that circRNAs are differentially expressed in central nerve injuries. One study explored the implication of circRNA expression profiles in spinal cord injury in rats at the immediate phase. Using microarray analysis, researchers found that 1,101 circRNAs were upregulated and 897 circRNAs were downregulated in SCI rats at the immediate phase compared with sham control rats. Additionally, enrichment analysis displayed that dysregulated circRNAs were enriched in many biological processes, such as spinal cord development and synapse assembly. They were also enriched in signaling pathways related to neuronal signal transduction and inflammation, such as the MAPK signaling pathway and axon guidance (Liu et al., 2020). In another study, 150 circRNAs were differentially expressed in spinal cords from an SCI group compared with the sham-injured control group. Among these, 99 circRNAs were upregulated, and 51 were downregulated. Further analysis disclosed that circRNA_07079 and circRNA_01282, two of the dysregulated circRNAs, were associated with SCI and might play a role in the pathophysiology of SCI (Zhou et al., 2019).
Optic Nerve Regeneration
Like the spinal cord, the optic nerve is also a part of the CNS, which has little ability to regenerate after it has been injured. As a result, optic nerve injuries and some degenerative diseases, such as traffic trauma, ischemic injury, and glaucoma, have become leading causes of irreversible blindness worldwide. Up to now, there are still no satisfactory treatments for this type of visual impairment, which is primarily due to the complex mechanisms underlying the pathological process (Li et al., 2017). Retina ganglion cells (RGCs) make up the innermost layer of neurons in the retina, of which the axons extend out to form the optic nerve. It has been reported that the irreversible loss of vision is attributed to RGCs losing the ability to regenerate axons (Mak et al., 2020). Therefore, axon regeneration becomes critical to recovering function after optic nerve impairment. While many transcription factors and proteins have had their functions related to axon regeneration after optic nerve injury uncovered, some mechanisms still remain elusive (Park et al., 2008; Duan et al., 2015; Li et al., 2015). Subsequently, researchers found that non-coding RNAs, especially microRNAs (miRs), are differentially expressed during RGC development and affect axon regeneration.
One previous study found that the developmental decline of axon regenerative ability in RGCs is in accordance with the expression levels of the miR-17-92 family members. Their data also showed that the developmental downregulation of miR-19a, one of the family members, is related to the upregulated expression of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) in RGCs. It is believed that miR-19a promotes axon regeneration in both mature rodent RGCs and human adult RGCs by suppressing PTEN, a negative regulator of the mammalian target of the rapamycin (mTOR) pathway, which controls cell growth by regulating cap-dependent protein translation initiation (Park et al., 2008). In addition, with the development of RGCs, miR-19a expression is drastically reduced, which contributes to the decline of axon regenerative capacity (Mak et al., 2020). In another study, microRNA-30b (miR-30b) was found to inhibit Semaphorin3A (Sema3A) expression in RGCs by bonding to its 3′ UTR. Sema3A is a very potent repulsive molecule and an important inhibitory factor involved in CNS repair following damage. miR-30b can promote axon regeneration by reducing the binding ability of Sema3A to NRP1/PlexA1, both of which are receptors, in order to transmit a signal of growth cone collapse. Moreover, miR-30b also reduces RGC apoptosis by inhibiting the expression of p-p38MAPK and active caspase 3 (Han et al., 2015). Some findings have demonstrated that inhibition of miR-21 attenuates excessive astrocyte activation and glial scar formation by regulating the epidermal growth factor receptor (EGFR) pathway, thereby promoting axon regeneration in RGCs and function recovery in flash visual evoked potential (F-VEP) in the rat model of optic nerve injury (Li et al., 2018). Apart from these results, miR-135s has also been proven to facilitate RGC axon regeneration after optic nerve injury in adult mice, in part by repressing Krüppel-like factor 4 (KLF4), which is a well-known intrinsic inhibitor of axon regeneration (van Battum et al., 2018). In addition, another study suggests that let7-miRNAs act downstream of Lin28a/b and serve as negative regulators of axon regeneration. Overexpression of Lin28a can induce robust and sustainable optic nerve regeneration without affecting the survival rate of RGCs (Wang W. et al., 2018). All the above highlight the potential of microRNAs to serve as a new tool in the treatment of optic nerve impairments.
Many circRNAs are expressed in different stages in the retina. One study pointed out that silencing circTulp4 would lead to a thin outer nuclear layer and defective retinal function. In addition, they found that circRNAs were dysregulated at a much earlier time point than disease onset in a retinal degeneration model (Chen et al., 2020). These results provide some clues for investigating the underlying mechanisms and potential repairing targets of CNS injuries. However, the functions of circRNAs in axon regeneration are still elusive and require further investigation.
Networks of Non-coding RNAs
With the development of high-throughput sequencing technology, we can obtain a lot of complete transcriptome information at one time. Through bioinformatics analysis, interestingly, researchers found that, apart from the separate functions of the non-coding RNAs in central nerve regeneration, they can also interact with each other to produce a marked effect. Recently, miRNA has been verified to be affected by the presence of miRNA sponge transcripts, the so-called competing endogenous RNAs (Hansen et al., 2013). Either lncRNA or circRNA can be miRNA sponges and thus affect the expression of target mRNAs, which is true of interactive networks among the two non-coding RNAs.
Using microarray analysis, Liu et al. (2020) constructed a circRNA-miRNA network of 10 candidate circRNAs in a spinal cord injury rat model using miRanda and found that most of the dysregulated circRNAs have a number of target miRNAs. In another study, results showed that retinal circRNAs could act as miRNA sponges. They also discovered that circTulp4 functioned as a ceRNA by acting as a sponge for miR-204-5p and miR-26a-5p to regulate retina development (Chen et al., 2020). Wang et al. found that lncRNA33755 and circRNA6370 were targets of miR-21-5p after SCI and displayed an lncRNA/circRNA-miRNA-mRNA axis (Wang W. et al., 2019), which implies the possibility of an analogous axis in central nerve regeneration. One research team suggested that lncRNA-Map2k4 was the target gene of miR-199a. Finally, they regulated FGF1 expression, which is essential to axon regeneration, as we discussed above (Lv, 2017).
Discussion
In this review, we summarized the extrinsic and intrinsic mechanisms for central nerve regeneration, especially non-coding RNAs. After central nerve injury, a quantity of non-coding RNAs perform differential expression, which implies their potential functions in repairing the nervous system. Table 1 lists recent studies with an overall profile of their roles in axon regeneration after CNS injuries, such as SCI and optic nerve injury. We not only elaborate on the separate functions of lncRNA, microRNA, and circRNA, but also reveal interactive networks among the non-coding RNAs (see Figure 2).
From this review, we can see that most of the research in this field begins with bioinformatics analysis, using microarray and RNA-seq. In this way, we can discover potential regulated non-coding RNAs and predict interactive networks as well as signaling pathways. Quantitative real-time PCR (qRT-PCR) is performed to verify the expression, and lncRNAs/circRNAs-miRNA-gene regulated networks are constructed to identify target genes of certain lncRNAs/circRNAs (Yao and Yu, 2019).
Although there has been mounting evidence to show that dysregulated non-coding RNAs have a strong relationship with axon regeneration in the CNS, there is still a long way to go before we understand the precise mechanisms and methods of translational medicine.
There are some issues to be addressed in a future investigation: ① apart from SCI and optic nerve injury, there is still a lack of research into axonal regeneration-associated non-coding RNAs in other types of CNS injuries, such as traumatic brain injury (TBI). A few studies have reported that non-coding RNAs could promote neurite regeneration after TBI by attenuating inflammatory responses (Pan et al., 2017; Huang et al., 2018), but the specific mechanisms of axon regeneration are still elusive. ② Some previous studies only proved that circRNAs were differentially expressed after CNS injuries and their specific mechanism of axon regeneration remains to be further studied in the future. ③ We still know little about the upstream factors of the non-coding RNAs involved in regeneration. ④ How can we bring non-coding RNAs into clinical application? Some scientists have pointed out that fully differentiated cells can be dedifferentiated by some reprogramming transcription factors (SOX2, Lin28, KLF4, Nanog) and revert into pluripotent stem cells (iPSC). Moreover, differentiated non-neural cells can also be reprogrammed directly into neurons without going through the stem cell phase (Qian and Zhou, 2020). One previous study showed that increased expression of the miR-200 family promotes neuronal differentiation, while decreased expression of the miR-200 family promotes neuronal proliferation by targeting SOX2 and KLF4 (Pandey et al., 2015). Perhaps more investigations should be done to clarify if non-coding RNAs can be a class of bridges to connect the reprogramming process with CNS nerve regeneration. Thus, patient-derived soma cells could be reprogrammed into neurons for future drug discovery or a source of neural grafts (Faravelli and Corti, 2018). In a series of studies, extracellular vesicle localized miRNAs have been shown to improve neurological function and promote neural health, which offers a potential therapeutic strategy for our topic (Blandford et al., 2018). Furthermore, non-invasive therapeutic approaches for delivering non-coding RNAs, such as miRNA agomir/mimics or miRNA hairpin inhibitor/antagomir by oral or intravenous injection, are more available and safer. In addition, exosomes are probably also ideal vesicles as they provide a new method for the transportation of non-coding RNAs (Schorey and Bhatnagar, 2008). We still need to verify the consistency of results in animal models and human patients. In addition, much attention should be paid to safety and ethical issues when applied to patients.
In a word, non-coding RNAs provide us with further clues to understanding the underlying mechanisms of CNS nerve regeneration, and more efforts are needed to promote the application of non-coding RNAs to become therapeutic targets after CNS injuries.
Author Contributions
PL drafted the manuscript. YJ and WT discussed the manuscript. PL, QC, and ML made the figures and tables. JJ designed the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) disclose receipt of the following financial support for the research, authorship, and/or publication of this article: the work was supported by the Natural Science Foundation of Liaoning Province (2019-MS-394) and 345 Talent Project of Shengjing Hospital.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the study participants for taking part in this study.
References
Abbaszadeh-Goudarzi, K., Radbakhsh, S., Pourhanifeh, M. H., Khanbabaei, H., Davoodvandi, A., Fathizadeh, H., et al. (2020). Circular RNA and diabetes: epigenetic regulator with diagnostic role. Curr. Mol. Med. 20, 516–526. doi: 10.2174/1566524020666200129142106
Apara, A., Galvao, J., Wang, Y., Blackmore, M., Trillo, A., Iwao, K., et al. (2017). KLF9 and JNK3 interact to suppress axon regeneration in the adult CNS. J Neurosci 37, 9632–9644. doi: 10.1523/jneurosci.0643-16.2017
Bai, Y., Zhang, Y., Han, B., Yang, L., Chen, X., Huang, R., et al. (2018). Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J. Neurosci. 38, 32–50. doi: 10.1523/jneurosci.1348-17.2017
Blandford, S. N., Galloway, D. A., and Moore, C. S. (2018). The roles of extracellular vesicle microRNAs in the central nervous system. Glia 66, 2267–2278. doi: 10.1002/glia.23445
Butz, H., Racz, K., Hunyady, L., and Patocs, A. (2012). Crosstalk between TGF-beta signaling and the microRNA machinery. Trends Pharmacol. Sci. 33, 382–393. doi: 10.1016/j.tips.2012.04.003
Cao, M., Li, H., Zhao, J., Cui, J., and Hu, G. (2019). Identification of age- and gender-associated long non-coding RNAs in the human brain with Alzheimer’s disease. Neurobiol. Aging 81, 116–126. doi: 10.1016/j.neurobiolaging.2019.05.023
Chanda, K., Das, S., Chakraborty, J., Bucha, S., Maitra, A., Chatterjee, R., et al. (2018). Altered levels of long NcRNAs Meg3 and Neat1 in cell and animal models of huntington’s disease. RNA Biol. 15, 1348–1363. doi: 10.1080/15476286.2018.1534524
Chen, L., Feng, P., Zhu, X., He, S., Duan, J., and Zhou, D. (2016). Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J. Cell Mol. Med. 20, 2102–2110. doi: 10.1111/jcmm.12904
Chen, X. J., Zhang, Z. C., Wang, X. Y., Zhao, H. Q., Li, M. L., Ma, Y., et al. (2020). The circular RNome of developmental retina in mice. Mol. Ther. Nucleic Acids 19, 339–349. doi: 10.1016/j.omtn.2019.11.016
Chen, Y., Cao, S., Xu, P., Han, W., Shan, T., Pan, J., et al. (2016). Changes in the expression of miR-34a and its target genes following spinal cord injury in rats. Med. Sci. Monit. 22, 3981–3993. doi: 10.12659/msm.900893
Church, J. S., Milich, L. M., Lerch, J. K., Popovich, P. G., and McTigue, D. M. (2017). E6020, a synthetic TLR4 agonist, accelerates myelin debris clearance, Schwann cell infiltration, and remyelination in the rat spinal cord. Glia 65, 883–899. doi: 10.1002/glia.23132
Cregg, J. M., DePaul, M. A., Filous, A. R., Lang, B. T., Tran, A., and Silver, J. (2014). Functional regeneration beyond the glial scar. Exp. Neurol. 253, 197–207. doi: 10.1016/j.expneurol.2013.12.024
Dai, J., Xu, L. J., Han, G. D., Sun, H. L., Zhu, G. T., Jiang, H. T., et al. (2018). MicroRNA-125b promotes the regeneration and repair of spinal cord injury through regulation of JAK/STAT pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 582–589. doi: 10.26355/eurrev_201802_14271
David, S., and Aguayo, A. J. (1981). Axonal elongation into peripheral nervous system “bridges” after central nervous system injury in adult rats. Science (New York N. Y.) 214, 931–933. doi: 10.1126/science.6171034
Dietz, V. (2016). Improving outcome of sensorimotor functions after traumatic spinal cord injury. F1000Res 5:8129. doi: 10.12688/f1000research.8129.1
Ding, Y., Song, Z., and Liu, J. (2016). Aberrant LncRNA expression profile in a contusion spinal cord injury mouse model. Biomed. Res. Int. 2016:9249401. doi: 10.1155/2016/9249401
Duan, X., Qiao, M., Bei, F., Kim, I. J., He, Z., and Sanes, J. R. (2015). Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron 85, 1244–1256. doi: 10.1016/j.neuron.2015.02.017
Faravelli, I., and Corti, S. (2018). MicroRNA-directed neuronal reprogramming as a therapeutic strategy for neurological diseases. Mol. Neurobiol. 55, 4428–4436. doi: 10.1007/s12035-017-0671-7
Fehlings, M. G., Tetreault, L. A., Wilson, J. R., Kwon, B. K., Burns, A. S., Martin, A. R., et al. (2017). A clinical practice guideline for the management of acute spinal cord injury: introduction, rationale, and scope. Global Spine J. 7(Suppl. 3), 84S–94S. doi: 10.1177/2192568217703387
Gaudet, A. D., Mandrekar-Colucci, S., Hall, J. C., Sweet, D. R., Schmitt, P. J., Xu, X., et al. (2016). miR-155 deletion in mice overcomes neuron-intrinsic and neuron-extrinsic barriers to spinal cord repair. J. Neurosci. 36, 8516–8532. doi: 10.1523/JNEUROSCI.0735-16.2016
Goff, L. A., Groff, A. F., Sauvageau, M., Trayes-Gibson, Z., Sanchez-Gomez, D. B., Morse, M., et al. (2015). Spatiotemporal expression and transcriptional perturbations by long non-coding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 112, 6855–6862. doi: 10.1073/pnas.1411263112
Goncalves, M. B., Malmqvist, T., Clarke, E., Hubens, C. J., Grist, J., Hobbs, C., et al. (2015). Neuronal RARbeta signaling modulates PTEN activity directly in neurons and via exosome transfer in astrocytes to prevent glial scar formation and induce spinal cord regeneration. J. Neurosci. 35, 15731–15745. doi: 10.1523/JNEUROSCI.1339-15.2015
Gribble, P. L., and Scott, S. H. (2002). Overlap of internal models in motor cortex for mechanical loads during reaching. Nature 417, 938–941. doi: 10.1038/nature00834
Gu, S., Xie, R., Liu, X., Shou, J., Gu, W., and Che, X. (2017). Long coding RNA XIST contributes to neuronal apoptosis through the downregulation of AKT phosphorylation and is negatively regulated by miR-494 in rat spinal cord injury. Int. J. Mol. Sci. 18:732. doi: 10.3390/ijms18040732
Han, F., Huo, Y., Huang, C. J., Chen, C. L., and Ye, J. (2015). MicroRNA-30b promotes axon outgrowth of retinal ganglion cells by inhibiting Semaphorin3A expression. Brain Res. 1611, 65–73. doi: 10.1016/j.brainres.2015.03.014
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Holmes, D. (2017). Spinal-cord injury:spurring regrowth. Nature 552:7550. doi: 10.1038/d41586-017-07550-9
Hu, S., Zheng, J., Du, Z., and Wu, G. (2020). Knock down of lncRNA H19 promotes axon sprouting and functional recovery after cerebral ischemic stroke. Brain Res. 1732:146681. doi: 10.1016/j.brainres.2020.146681
Huang, S., Ge, X., Yu, J., Han, Z., Yin, Z., Li, Y., et al. (2018). Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 32, 512–528. doi: 10.1096/fj.201700673R
Larhammar, M., Huntwork-Rodriguez, S., Rudhard, Y., Sengupta-Ghosh, A., and Lewcock, J. W. (2017). The Ste20 family kinases MAP4K4, MINK1, and TNIK converge to regulate stress-induced JNK signaling in neurons. J. Neurosci. 37, 11074–11084. doi: 10.1523/jneurosci.0905-17.2017
Li, F., Sami, A., Noristani, H. N., Slattery, K., Qiu, J., Groves, T., et al. (2020). Glial metabolic rewiring promotes axon regeneration and functional recovery in the central nervous system. Cell Metab. 32, 767–85.e7. doi: 10.1016/j.cmet.2020.08.015
Li, H. J., Pan, Y. B., Sun, Z. L., Sun, Y. Y., Yang, X. T., and Feng, D. F. (2018). Inhibition of miR-21 ameliorates excessive astrocyte activation and promotes axon regeneration following optic nerve crush. Neuropharmacology 137, 33–49. doi: 10.1016/j.neuropharm.2018.04.028
Li, H. J., Sun, Z. L., Yang, X. T., Zhu, L., and Feng, D. F. (2017). Exploring optic nerve axon regeneration. Curr. Neuropharmacol. 15, 861–873. doi: 10.2174/1570159X14666161227150250
Li, P., Teng, Z. Q., and Liu, C. M. (2016). Extrinsic and intrinsic regulation of axon regeneration by MicroRNAs after spinal cord injury. Neural. Plast 2016:1279051. doi: 10.1155/2016/1279051
Li, S., He, Q., Wang, H., Tang, X., Ho, K. W., Gao, X., et al. (2015). Injured adult retinal axons with Pten and Socs3 co-deletion reform active synapses with suprachiasmatic neurons. Neurobiol. Dis. 73, 366–376. doi: 10.1016/j.nbd.2014.09.019
Li, Y., Zhao, J., Yu, S., Wang, Z., He, X., Su, Y., et al. (2019). Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem. 65, 798–808. doi: 10.1373/clinchem.2018.301291
Liu, Y., Han, N., Li, Q., and Li, Z. (2016). Bioinformatics analysis of microRNA time-course expression in brown rat (Rattus norvegicus): spinal cord injury self-repair. Spine 41, 97–103. doi: 10.1097/brs.0000000000001323
Liu, Y., Liu, J., and Liu, B. (2020). Identification of circular RNA expression profiles and their implication in spinal cord injury rats at the immediate phase. J. Mol. Neurosci. 70, 1894–1905. doi: 10.1007/s12031-020-01586-9
Luo, X., and Park, K. K. (2012). Neuron-intrinsic inhibitors of axon regeneration: PTEN and SOCS3. Int. Rev. Neurobiol. 105, 141–173. doi: 10.1016/b978-0-12-398309-1.00008-1
Lv, H. R. (2017). lncRNA-Map2k4 sequesters miR-199a to promote FGF1 expression and spinal cord neuron growth. Biochem. Biophys. Res. Commun. 490, 948–954. doi: 10.1016/j.bbrc.2017.06.145
Ma, J. J., Ju, X., Xu, R. J., Wang, W. H., Luo, Z. P., Liu, C. M., et al. (2019). Telomerase reverse transcriptase and p53 regulate mammalian peripheral nervous system and CNS axon regeneration downstream of c-Myc. J. Neurosci. 39, 9107–9118. doi: 10.1523/jneurosci.0419-19.2019
Mak, H. K., Yung, J. S. Y., Weinreb, R. N., Ng, S. H., Cao, X., Ho, T. Y. C., et al. (2020). MicroRNA-19a-PTEN axis is involved in the developmental decline of axon regenerative capacity in retinal ganglion cells. Mol. Ther. Nucleic Acids 21, 251–263. doi: 10.1016/j.omtn.2020.05.031
Moore, D. L., Blackmore, M. G., Hu, Y., Kaestner, K. H., Bixby, J. L., Lemmon, V. P., et al. (2009). KLF family members regulate intrinsic axon regeneration ability. Science (New York N. Y.) 326, 298–301. doi: 10.1126/science.1175737
Neumann, S., Bradke, F., Tessier-Lavigne, M., and Basbaum, A. I. (2002). Regeneration of sensory axons within the injured spinal cord induced by intraganglionic cAMP elevation. Neuron 34, 885–893. doi: 10.1016/s0896-6273(02)00702-x
O’Donovan, K. J., Ma, K., Guo, H., Wang, C., Sun, F., Han, S. B., et al. (2014). B-RAF kinase drives developmental axon growth and promotes axon regeneration in the injured mature CNS. J. Exp. Med. 211, 801–814. doi: 10.1084/jem.20131780
Oh, Y. M., Mahar, M., Ewan, E. E., Leahy, K. M., Zhao, G., and Cavalli, V. (2018). Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration. Proc. Natl. Acad. Sci. U.S.A. 115, E12417–E12426. doi: 10.1073/pnas.1812518115
Pan, Y. B., Sun, Z. L., and Feng, D. F. (2017). The role of MicroRNA in traumatic brain injury. Neuroscience 367, 189–199. doi: 10.1016/j.neuroscience.2017.10.046
Pandey, A., Singh, P., Jauhari, A., Singh, T., Khan, F., Pant, A. B., et al. (2015). Critical role of the miR-200 family in regulating differentiation and proliferation of neurons. J. Neurochem. 133, 640–652. doi: 10.1111/jnc.13089
Paraskevopoulou, M. D., and Hatzigeorgiou, A. G. (2016). Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. (Clifton N. J.) 1402, 271–286. doi: 10.1007/978-1-4939-3378-5_21
Park, K. K., Liu, K., Hu, Y., Smith, P. D., Wang, C., Cai, B., et al. (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science (New York N. Y.) 322, 963–966. doi: 10.1126/science.1161566
Qian, C., and Zhou, F. Q. (2020). Updates and challenges of axon regeneration in the mammalian central nervous system. J. Mol. Cell Biol. doi: 10.1093/jmcb/mjaa026 [Epub ahead of print].
Rouanet, C., Reges, D., Rocha, E., Gagliardi, V., and Silva, G. S. (2017). Traumatic spinal cord injury: current concepts and treatment update. Arq. Neuropsiquiatr. 75, 387–393. doi: 10.1590/0004-282X20170048
Rowland, J. W., Hawryluk, G. W., Kwon, B., and Fehlings, M. G. (2008). Current status of acute spinal cord injury pathophysiology and emerging therapies: promise on the horizon. Neurosurgical focus 25:E2. doi: 10.3171/foc.2008.25.11.E2
Saijilafu, Hur, E. M., Liu, C. M., Jiao, Z., Xu, W. L., and Zhou, F. Q. (2013). PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat. Commun. 4:2690. doi: 10.1038/ncomms3690
Salah, S. M. M., Matboli, M., Nasser, H. E., Abdelnaiem, I. A., Shafei, A. E., and El-Asmer, M. F. (2020). Dysregulation in the expression of (lncRNA-TSIX, TP53INP2 mRNA, miRNA-1283) in spinal cord injury. Genomics 112, 3315–3321. doi: 10.1016/j.ygeno.2020.06.018
Schorey, J. S., and Bhatnagar, S. (2008). Exosome function: from tumor immunology to pathogen biology. Traffic (Copenh. Den.) 9, 871–881. doi: 10.1111/j.1600-0854.2008.00734.x
Siede, D., Rapti, K., Gorska, A. A., Katus, H. A., Altmüller, J., Boeckel, J. N., et al. (2017). Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J. Mol. Cell. Cardiol. 109, 48–56. doi: 10.1016/j.yjmcc.2017.06.015
Sun, F., Park, K. K., Belin, S., Wang, D., Lu, T., Chen, G., et al. (2011). Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 480, 372–375. doi: 10.1038/nature10594
Tom, V. J., Steinmetz, M. P., Miller, J. H., Doller, C. M., and Silver, J. (2004). Studies on the development and behavior of the dystrophic growth cone, the hallmark of regeneration failure, in an in vitro model of the glial scar and after spinal cord injury. J. Neurosci. 24, 6531–6539. doi: 10.1523/JNEUROSCI.0994-04.2004
van Battum, E. Y., Verhagen, M. G., Vangoor, V. R., Fujita, Y., Derijck, A., O’Duibhir, E., et al. (2018). An image-based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting kruppel-like factor 4. J. Neurosci. 38, 613–630. doi: 10.1523/JNEUROSCI.0662-17.2017
Voce, D. J., Bernal, G. M., Wu, L., Crawley, C. D., Zhang, W., Mansour, N. M., et al. (2019). Temozolomide treatment induces lncRNA MALAT1 in an NF-κB and p53 codependent manner in glioblastoma. Cancer Res. 79, 2536–2548. doi: 10.1158/0008-5472.Can-18-2170
Wang, F., Liu, J., Wang, X., Chen, J., Kong, Q., Ye, B., et al. (2019). The emerging role of lncRNAs in spinal cord injury. Biomed. Res. Int. 201:3467121. doi: 10.1155/2019/3467121
Wang, W., Liu, R., Su, Y., Li, H., Xie, W., and Ning, B. (2018). MicroRNA-21-5p mediates TGF-beta-regulated fibrogenic activation of spinal fibroblasts and the formation of fibrotic scars after spinal cord injury. Int. J. Biol. Sci. 14, 178–188. doi: 10.7150/ijbs.24074
Wang, W., Su, Y., Tang, S., Li, H., Xie, W., Chen, J., et al. (2019). Identification of non-coding RNA expression profiles and regulatory interaction networks following traumatic spinal cord injury by sequence analysis. Aging 11, 2352–2368. doi: 10.18632/aging.101919
Wang, X. W., Li, Q., Liu, C. M., Hall, P. A., Jiang, J. J., Katchis, C. D., et al. (2018). Lin28 signaling supports mammalian PNS and CNS axon regeneration. Cell Rep. 24, 2540–2552.e6. doi: 10.1016/j.celrep.2018.07.105
Wang, Z., Mehra, V., Simpson, M. T., Maunze, B., Chakraborty, A., Holan, L., et al. (2018). KLF6 and STAT3 co-occupy regulatory DNA and functionally synergize to promote axon growth in CNS neurons. Sci. Rep. 8:12565. doi: 10.1038/s41598-018-31101-5
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of long non-coding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull 97, 69–80. doi: 10.1016/j.brainresbull.2013.06.001
Wu, R., Mao, S., Wang, Y., Zhou, S., Liu, Y., Liu, M., et al. (2019). Differential circular RNA expression profiles following spinal cord injury in rats: a temporal and experimental analysis. Front. Neurosci. 13:1303. doi: 10.3389/fnins.2019.01303
Xia, P., Gao, X., Duan, L., Zhang, W., and Sun, Y. F. (2018). Mulberrin (Mul) reduces spinal cord injury (SCI)-induced apoptosis, inflammation and oxidative stress in rats via miroRNA-337 by targeting Nrf-2. Biomed. Pharmacother. 107, 1480–1487. doi: 10.1016/j.biopha.2018.07.082
Yao, C., Wang, J., Zhang, H., Zhou, S., Qian, T., Ding, F., et al. (2015). Long non-coding RNA uc.217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. Eur. J. Neurosci. 42, 1718–1725. doi: 10.1111/ejn.12966
Yao, C., and Yu, B. (2019). Role of long noncoding RNAs and circular RNAs in nerve regeneration. Front. Mol. Neurosci. 12:165. doi: 10.3389/fnmol.2019.00165
Yiu, G., and He, Z. (2006). Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 7, 617–627. doi: 10.1038/nrn1956
Zhang, J., Yang, D., Huang, H., Sun, Y., and Hu, Y. (2018). Coordination of necessary and permissive signals by PTEN inhibition for CNS axon regeneration. Front. Neurosci. 12:558. doi: 10.3389/fnins.2018.00558
Zhang, L., Xue, Z., Yan, J., Wang, J., Liu, Q., and Jiang, H. (2019). LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 52:e12573. doi: 10.1111/cpr.12573
Zhou, H., Shi, Z., Kang, Y., Wang, Y., Lu, L., Pan, B., et al. (2018). Investigation of candidate long non-coding RNAs and messenger RNAs in the immediate phase of spinal cord injury based on gene expression profiles. Gene 661, 119–125. doi: 10.1016/j.gene.2018.03.074
Keywords: non-coding RNA, nerve regeneration, central nervous system, optic nerve injury, spinal cord injury
Citation: Li P, Jia Y, Tang W, Cui Q, Liu M and Jiang J (2021) Roles of Non-coding RNAs in Central Nervous System Axon Regeneration. Front. Neurosci. 15:630633. doi: 10.3389/fnins.2021.630633
Received: 18 November 2020; Accepted: 05 January 2021;
Published: 01 February 2021.
Edited by:
Jan Kaslin, Australian Regenerative Medicine Institute (ARMI), AustraliaCopyright © 2021 Li, Jia, Tang, Cui, Liu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Jiang, ampqX3NqQDE2My5jb20=
 Pei Li
Pei Li Yuanyuan Jia
Yuanyuan Jia Wenbo Tang
Wenbo Tang Qingjun Cui
Qingjun Cui Ming Liu
Ming Liu Jingjing Jiang
Jingjing Jiang